Abstract
The past 10 years have brought paradigm-shifting changes to clinical microbiology. This paper explores the top 10 transformative innovations across the diagnostic spectrum, including not only state of the art technologies but also preanalytic and post-analytic advances. Clinical decision support tools have reshaped testing practices, curbing unnecessary tests. Innovations like broad-range polymerase chain reaction and metagenomic sequencing, whole genome sequencing, multiplex molecular panels, rapid phenotypic susceptibility testing, and matrix-assisted laser desorption ionization time-of-flight mass spectrometry have all expanded our diagnostic armamentarium. Rapid home-based testing has made diagnostic testing more accessible than ever. Enhancements to clinician-laboratory interfaces allow for automated stewardship interventions and education. Laboratory restructuring and consolidation efforts are reshaping the field of microbiology, presenting both opportunities and challenges for the future of clinical microbiology laboratories. Here, we review key innovations of the last decade.
Introduction
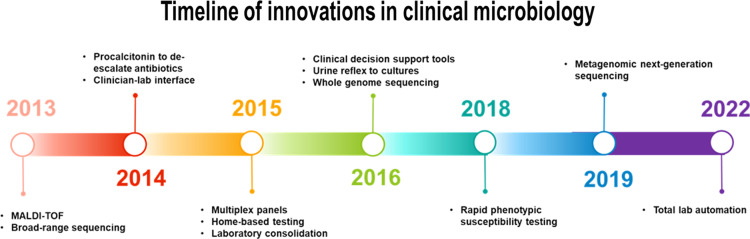
The past 10 years have brought paradigm-shifting changes to clinical microbiology. This paper explores the top 10 transformative innovations across the diagnostic spectrum, including not only state of the art technologies but also preanalytic and post-analytic advances (Table 1). Clinical decision support tools (CDST) have reshaped testing practices, curbing unnecessary tests. Innovations like broad-range polymerase chain reaction (PCR) and metagenomic sequencing, whole genome sequencing (WGS), multiplex molecular panels, rapid phenotypic susceptibility testing, and matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) have all expanded our diagnostic armamentarium. Rapid home-based testing has made diagnostic testing more accessible than ever. Enhancements to clinician–laboratory interfaces allow for automated stewardship interventions and education. Laboratory restructuring and consolidation efforts are reshaping the field of microbiology, presenting both opportunities and challenges for the future of clinical microbiology laboratories. Herein, we categorize these laboratory advances as preanalytic, analytic, post-analytic, and other to reflect how these would be implemented in clinical care. A timeline is provided to demonstrate when in the past 10 years these technologies or innovations emerged (Fig. 1).
Table 1.
Overview of the top ten innovations in clinical microbiology over the past decade, highlighting their applications, key benefits, and associated challenges
| Innovation area | Applications | Key benefits | Challenges/Limitations | |
|---|---|---|---|---|
| Preanalytic | Clinical decision support tools | Best-practice alerts Guidelines and algorithms Indication selection using guided test ordering Change in order sets |
Drive appropriate test selection Prevent overutilization of tests in low-impact situations |
Decreased testing when actually indicated Alert fatigue leading to clinicians overriding alerts Provider and IT pushback |
| Host Pathogen Response | Inflammatory markers (Procalcitonin) Urinalysis Reflex to Culture |
Antibiotic discontinuation Diagnostic stewardship |
Specificity and Reproducibility Utilization management Integration with microbiology |
|
| Analytic | Sequencing | Broad range PCR, targeted NGS Metagenomic sequencing Whole genome sequencing |
Identification of organisms directly from clinical specimens, even when culture negative High species level resolution for organism identification Determine strain relatedness for epidemiological purposes |
Sensitivity and specificity dependent on preanalytic factors Results can be difficult to interpret when commensal organisms or contaminants are identified Unknown how to report and act upon WGS data in real-time Does not provide phenotypic susceptibility data |
| Multiplex panels | Syndromic-based testing | Antibiotic stewardship Avoid decision fatigue |
Positive results not always clinically relevant Expense |
|
| Rapid susceptibility testing | Novel methods of rapid susceptibility testing | Guides early selection and use of optimal antibiotics | Requires adjudication of discrepancies between rapid AST and finalized traditional AST results | |
| MALDI TOF MS | Bacterial, fungal identification from isolates | Improved accuracy Shorter turnaround time |
Capital costs Over-reporting |
|
| Home testing | Rapid home-based antigen tests | Convenience Privacy Access |
Test performance and result interpretation Potential cost per test Quality control of the testing components Linkage to care and inclusion in the EMR Tracking of any results that are of concern for public health |
|
| Post analytic | Clinician-lab interface | Framing Cascade reporting Selective reporting Result review and feedback |
Guides appropriate decision-making following test results Automates stewardship interventions and education |
Limiting clinician’s input leading to missed diagnosis |
| Other | Laboratory consolidation | Acquisition by commercial laboratories Centralized/localized testing within a health system Total laboratory automation |
Increased cost savings and efficiency Concentration of resources/expertise/ technology within a network to provide access to highest quality across the system Uniform adherence to stewardship best practices and guidelines |
Increased turnaround time for results to remote sites Logistical challenges, such as specimen stability Risk for financial considerations to drive decisions at the expense of patient safety or quality |
Figure 1.
Timeline demonstrating the top innovations over the last 10 years. Though some technologies were developed prior to 2013, these dates reflect their emergence in mainstream clinical microbiology.
Preanalytic
Clinical decision support tools
Clinical microbiologists have known for a long time that preanalytical issues are often the most important, yet overlooked, factors in producing high-quality results. For instance, emphasizing good specimen collection using appropriate techniques and having quality criteria for working up cultures from non-sterile sites. The focus of preanalytic innovations over the past decade has shifted to behavioral economics using automated system-based CDST to nudge clinicians earlier in clinical workup for better utilization of diagnostic tests. 1,2 Antimicrobial stewardship programs combined with information technology staff have played a crucial role in the design and implementation of CDST. Some examples of these novel interventions include:
Best-practice alerts (BPAs) to stop unnecessary diagnostic testing in patients without symptoms of infection. For example, firing BPAs when ordering urine testing in asymptomatic patients led to reduced urine culture ordering and reduced antibiotic orders 3 Utilizing Electronic Medical Record (EMR) hard and soft stops if a patient is not meeting criteria for Clostridium difficile testing: <1 year old, laxatives within 48 hours, or < 3 loose stools in 24 hours led to reduced testing and positive clinician perception. 4,5 .
Guidelines and algorithms to promote appropriate testing practices. Examples include algorithms to reduce unnecessary blood culture collection 6,7 or urine culture reflex based on predefined urinalysis criteria to reduce unnecessary workup for urine cultures and administration of antibiotics. 8
Indication selection using restrictive or guided test ordering via built-in EMR algorithms to drive appropriate test selection. Examples include requiring a clinician to input an approved indication on an order set before ordering a urine culture, 9,10 endotracheal aspirate culture, 11 rapid multiplex respiratory pathogen panel, 12 or rapid multiplex meningitis panel. 13
Change in order sets—for instance, removing urine cultures from standard admission order sets to prevent overutilization in low-yield situations led to decreased urine cultures ordered 14,15
Challenges: These interventions may inadvertently reduce testing in situations where it is indicated, for instance, not obtaining urine cultures in asymptomatic pregnant women. Most importantly, implementation challenges such as “alert fatigue” due to overuse of electronic reminders disrupting usual workflow or “provider pushback” due to conflicts between CDST recommendations and provider expertise or beliefs lead to clinicians ignoring or overriding these alerts. 16,17 Resources such as information technology staff are additionally utilized during integration of these tools into clinical practice.
Host–pathogen response
Integration of the host immune response to pathogens into infectious disease diagnostics has seen substantial expansion in the past decade. Two of the major areas of growth include:
Monitoring of inflammatory markers—the most notable change is the use of procalcitonin for guidance in discontinuation of antibiotic treatment in certain patient populations (critically ill, lower respiratory tract infections, and in certain pediatric populations). 18
Incorporation of cell counts and differentials in guiding specimen adequacy and likelihood of infection—this has been most notably used in urinalysis reflex to urine culture, in which demonstration of pyuria (commonly defined in urine as >10 WBC/hpf) is required before performing urine culture. This has led to a significant decrease in unnecessary workup of urine isolates. 19,20 Similar approaches to using cell counts or inflammatory markers have been utilized in the workup of meningitis/encephalitis as well as periprosthetic joint infection. Although the methodology for measurement has not changed dramatically in the past decade, their utilization in stewardship and influence on the workup of microbiology specimens has considerably increased in the past 10 years and is now the standard of care.
Challenges: Specificity remains a significant limitation to using host–pathogen response markers to guide treatment. Treatment markers such as C-reactive protein (CRP) and procalcitonin can be elevated in a variety of infectious and inflammatory conditions, and despite their long standing use, are often improperly used. 21,22 Inclusion of additional markers such as tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and other proteins have helped refine this approach but is still early in its use. 23 Another challenge is a lack of integration between the microbiology laboratory and the laboratories that monitor the host immune response (eg, CRP, procalcitonin in chemistry, cell counts in hematology). Closer interactions between clinical laboratory sections will be crucial in the successful utilization of these approaches.
Analytic
Broad-range sequencing
Various sequencing methods have become integral for identifying organisms and defining taxonomy, whether directly from specimens or from cultured isolates. 24 In microbiology labs, initial efforts are made to identify isolates using conventional techniques. However, if these conventional methods prove unsuccessful, labs have the option to either perform sequencing in house (if they have the capability) or send isolates to reference labs for a definitive identification through sequencing. Sequencing results are often used as the “gold standard” for species-level identification.
Despite being colloquially referred to as “universal PCR,” broad-range PCR is restricted to selecting ribosomal subunits specific to either fungi or bacteria. Additional broad-range targets, such as rpoB, may be used to further differentiate among groups such as acid-fast bacteria. 25 Some laboratories will require clinicians to choose the relevant targets for testing. Fresh frozen tissue is generally the preferred specimen. It is also possible to perform sequencing on formalin-fixed paraffin-embedded tissue (FFPE). The sensitivity of sequencing from FFPE specimens may be compromised or reduced after the fixation process. In general, samples with concurrent pathology that do not demonstrate histopathological evidence of infection are unlikely to yield positive results by sequencing and efforts should be made to discourage sequencing in these scenarios. 26,27
Challenges: Sequencing is not available in most clinical microbiology laboratories. Commercially available options, such as University of Washington or Mayo Clinical Laboratories, are often expensive and the overall turnaround time can range from 1 to 4 weeks. Because most laboratories only send sequencing in instances where traditional microbiological methods are negative or inconclusive, clinical laboratories must have some way of retaining, tracking, and freezing specimens where sequencing is requested, but initial results are still pending (to avoid accidentally discarding specimens). There is variability in sensitivity as compared to culture-based methods, depending on the specimen source, fixation process, and correlative histopathology. Sequencing results must be correlated clinically, usually by a clinical microbiology or infectious disease clinician, as sequencing of contaminants may occur and result in provider confusion.
Metagenomic Next-Generation Sequencing (mNGS) and Whole Genome Sequencing (WGS)
In recent years, the use of mNGS (a massively paralleled, rapid, high throughput method of sequencing all the genetic material in a clinical sample) and WGS (sequencing of entire organism genomes) has entered into mainstream clinical use. 28,29 These technologies have generally required significant bioinformatic/computational analysis and, outside of large academic medical centers, are typically confined to reference laboratories. This is starting to change with commercially available software solutions.
mNGS assays can be used directly on primary specimens. For example, NGS metagenomics can be performed on CSF and may be used for detection of RNA viruses. 30 In recent years, there has been increased interest in organism identification directly from blood samples, bypassing culture incubation steps. NGS directly from plasma is available commercially, using cell-free DNA sequencing (sequencing small fragments of DNA released into the bloodstream) to identify pathogens both within the bloodstream and at distant sites of infection. This novel concept of a “liquid biopsy” is gaining popularity, especially in identifying organisms when traditional methods are negative or where invasive biopsy is contraindicated. 31
Pathogen WGS (sequencing from pure or highly enriched isolate preparations) is also entering mainstream clinical microbiology laboratories. WGS is most routinely used to characterize bacterial specimens for purposes such as high-resolution identification of unusual isolates, investigations into novel antimicrobial resistance genes, and assessments of relatedness for infection control and epidemiological purposes. 32 Although fungal WGS is still a developing method, viral WGS became a critical tool to understand epidemiology and spread during the COVID-19 pandemic. 33
Challenges: Sequencing of clinical specimens presents a number of challenges that limit widespread use of these technologies. Besides high costs and variable turnaround times, 34 the lack of understanding of what pathogens are being tested and the optimal timing of testing may cause clinicians to not order appropriate testing (eg, when suspecting certain viruses or parasites that are not included in the reference NGS database) or miss the window of opportunity for optimal testing (eg, most arboviral infections do not exhibit detectable levels of RNA in the CSF beyond the first 1-2 weeks after the infection’s onset, limiting the sensitivity of this test in the later diagnostic stages 35,36 ). Clinicians may erroneously want to use mNGS sequencing methods to “rule out infection” 37 despite lack of data on use for this indication. Most importantly, test interpretation and reporting remain a critical problem. mNGS detection of multiple organisms within a clinical sample may often include detection of commensal or nonpathogenic organisms that may be misleading to clinicians and lead to excessive treatment 38 or additional diagnostics that would not have otherwise been ordered. 37,39,40 Adjudication by clinical microbiologists or infectious disease physicians should be performed in all cases.
WGS from cultured isolates also produces an incredibly rich data set; however, the real-time clinical utility of WGS data is still an area in need of development. Challenges include predicting the phenotype of genomic antibiotic resistance results and understanding the threshold for relatedness when comparing genomes from potentially related strains as part of outbreak investigations. There are lack of clear guidelines as to which genetic information should be reported to the clinician and little guidance on how this information should be acted on, if at all.
Multiplex panels
Multiplex PCR panels are commercially available for multiple specimen sources including upper respiratory tract, lower respiratory tract, blood, stool, prosthetic joint, abdominal, and genitourinary tract (Table 2). These panels include multiple organism targets common to a particular infectious syndrome. This can reduce cognitive error when providers are required to order multiple tests separately and their rapid turnaround time can theoretically reduce broad-spectrum antimicrobial use, though this has not consistently been found to be the case in studies unless testing is coupled with antimicrobial stewardship feedback. 41 The maximal benefit of these tests is realized if they are able to be run on all shifts, with relatively short turnaround times, which can cause significant logistical issues for laboratories, particularly given current staffing shortages. 42,43
Table 2.
Summary of FDA approved commercially available multiplex assays including targets, source, methodology, turnaround time, and clinical caveats
| Blood | ||||||||
|---|---|---|---|---|---|---|---|---|
| Name | Manufacturer | Organism targets | Gene targets resistance genes (RGs) | Specimen source | Technique | Turnaround time | Clinical caveat | Citation |
| Biofire Blood culture identification (BCID) 2 panel | Biomerieux | 7 yeast, 26 bacterial | 10 RGs | Positive blood culture bottle | mPCR | 1 hour | Detects multiple targets in polymicrobial samples | 95 |
| ePlex BCID Gram-positive panel | GenMarkDx | 20 organisms, pan-Gram-negative and pan- Candida | 4 RGs | Positive blood culture bottle | mPCR | 1.5 hours | Detects multiple targets in polymicrobial samples | 96 |
| ePlex BCID Gram-negative panel | GenMarkDx | 21 organisms, pan-Gram- positive and pan-Candida | 7 RGs | Positive blood culture bottle | mPCR | 1.5 hours | Detects multiple targets in polymicrobial samples | 96 |
| ePlex BCID Fungal panel | GenMarkDx | 15 organisms | None | Positive blood culture bottle | mPCR | 1.5 hours | Detects multiple targets in polymicrobial samples | 96 |
| Verigene Gram-positive panel | Diasorin | 13 organisms | 3 RGs | Positive blood culture bottle | Nanoparticle probe | 2.5 hours | Unreliable for polymicrobial samples | 97 |
| Verigene Gram-negative | Diasorin | 9 organisms | 6 RGs | Positive blood culture bottle | Nanoparticle probe | 2.5 hours | Unreliable for polymicrobial samples | 97 |
| T2Bacteria panel | T2Biosystems | 5 organisms | None | Direct from blood | NMR | 3-5 hours | Lower limit of detection | 98 |
| T2Candida panel | T2Biosystems | 5 organisms | None | Direct from blood | NMR | 3-5 hours | Lower limit of detection | 99 |
| Accelerate Pheno and blood culture panel | Accelerate diagnostics | 14 bacteria, 2 yeast | Susceptibility to 17 agents | Positive blood culture bottle | PNA-FISH, morphokinetic cellular analysis | 2-7 hours | Unreliable if multiple organism morphologies or polymicrobial samples | 100 |
| Upper respiratory | ||||||||
| Name | Manu-facturer | Organism Targets | Gene targets | Specimen source | Turnaround time | Cost | ||
| Biofire FilmArray respiratory pathogen panel | Biomerieux | 17 viruses, 3 bacteria | None | Nasopharyngeal (NP) swab | mPCR | 1 hour | Requires pairing with ASP* to reduce antimicrobial utilization | 101 |
| Biofire Respiratory 2.1-EZ panel | Biomerieux | 15 viruses, 4 bacteria | None | NP swab | mPCR | 45 minutes | Requires pairing with ASP to reduce antimicrobial utilization | 102 |
| Verigene Respiratory pathogens flex | Luminex | 13 viruses and 3 bacteria | None | NP swab | mPCR | 2 hours | Requires pairing with ASP to reduce antimicrobial utilization | 103 |
| eSensor Respiratory virus panel | GenMarkDx | 14 viruses | None | NP swab | mPCR | 6 hours | 104 | |
| ePlex Respiratory virus panel 2 | GenMarkDx | 16 viruses, 2 bacteria | None | NP swab | mPCR | 3 hours | Requires pairing with ASP to reduce antimicrobial utilization | 104 |
| Lower respiratory | ||||||||
| Biofire FilmArray pneumonia panel | Biomerieux | 8 viruses, 18 bacteria | RGs | Sputum or BAL | mPCR | 1 hour | Requires pairing with ASP to reduce antimicrobial utilization | 105 |
| Gastrointestinal | ||||||||
| Biofire FilmArray GI panel | Biomerieux | 5 viruses, 11 bacteria, 4 parasites | None | Stool | mPCR | 1 hour | Cannot differentiate between live and dead organisms, high rates of colonization with unclear clinical significance | 106 |
| xTAG GI Pathogen panel | Luminex | 3 viruses, 8 bacteria, 1 bacterial toxin, 3 parasites | None | Stool | mPCR | 5 hours | Cannot differentiate between live and dead organisms, high rates of colonization with unclear clinical significance | 107 |
| BDMax Enteric bacterial panel | BD | 4 bacteria | None | Stool | mPCR | 3.5 hours | Cannot differentiate between live and dead organisms | 108 |
| BDMax Extended enteric bacterial panel | BD | 8 bacteria | None | Stool | mPCR | 3.5 hours | Cannot differentiate between live and dead organisms | 109 |
| BDMax Enteric viral panel | BD | 5 viruses | None | Stool | mPCR | 3 hours | Cannot differentiate between live and dead organisms | 110 |
| BDMax Enteric parasite panel | BD | 3 parasites | None | Stool | mPCR | 4.5 hours | Lower sensitivity than conventional methods | 111 |
| Verigene Enteric pathogens panel | Diasorin | 2 viruses, 5 bacteria, 2 bacterial toxins | None | Stool | mPCR | 2 hours | Cannot differentiate between live and dead organisms, high rates of colonization with unclear clinical significance | 112 |
| Joint | ||||||||
| Biofire joint infection panel | Biomerieux | 15 Gram-positive, 14 Gram-negative, 2 yeast | 20 RGs | Synovial fluid | mPCR | 1 hour | Missing many common causes of prosthetic joint infections | 113 |
| CNS | ||||||||
| Biofire meningitis/Encephalitis panel | Biomerieux | 7 viruses, 6 bacteria, 1 yeast | None | Cerebrospinal fluid | mPCR | 1 hour | Cannot fully replace traditional diagnostics | 114 |
Note.
ASP: antibiotic stewardship program.
Challenges: There are downsides to the use of multiplex panels. One of these is significant testing costs, which are inconsistently covered by insurance, leading to either a high bill to the patient or a high cost to the laboratory. Additionally, all components of these panels are not necessarily understood by all clinicians, which can create confusion and inappropriate additional testing or treatments without clear interpretation guidelines. Panels are often not customizable, though some companies offer panels with extension options. 44 Excessive targets may result in multiple positive targets that do not always fit the clinical picture, such as with stool multiplex panels 45 and the detection of colonizing forms of Clostridium difficile 46 or species of Escherichia coli that lack clinical guidelines regarding treatment or significance (Enteroaggregative E. coli, Enteropathogenic E. coli). Positive results may lead to confusion from patients and providers or lead to unnecessary treatment. These testing panels have significant promise to decrease testing turnaround time and improve diagnostic accuracy, but their successful implementation requires stewardship integration for test appropriateness and interpretation.
Rapid phenotypic susceptibility
In addition to rapid detection of molecular targets, another advance has been the introduction of rapid phenotypic susceptibility testing. Traditional antimicrobial susceptibility testing (AST) can take 24–48 hours to result after isolation of cultured organisms. To expedite this process, rapid AST can be performed manually by direct use of positive blood culture broth as the inoculum for conventional methods using disk diffusion techniques with results in up to 6 hours. 47 However, this is a laborious practice and not widely used. Novel, less labor-intensive phenotypic susceptibility methods that mimic results produced by standard AST have become commercially available and represent an area of rapid growth. Novel methods include detection of changes in cell morphology induced by antimicrobials using single cell microscopy, assessing the rate of cell division, examining gene expression patterns, or detection of volatile organic compounds. 48 One commercially available system in the US performs AST approximately 7 hours from positive blood cultures using time-lapse imaging under dark-field microscopy, monitoring morphological and kinetic changes in the bacteria to determine MICs. 49 Several other rapid AST platforms are in the development pipeline. Rapid AST should be paired with rapid identification, as MIC results and their correlative breakpoints are only interpretable when paired with the organism ID. Institutions may choose platforms that pair rapid ID with rapid AST or use separate instruments to achieve this.
Challenges: Implementation of rapid AST methods requires monitoring and adjudication of discrepancies between rapid AST and finalized traditional AST results if both methods are performed. 50 Institutions should consider how they will alert providers or stewardship teams of rapid results, particularly if rapid AST is run on all shifts in institutions that are used to receiving updated susceptibility reports only during day shift hours.
MALDI-TOF MS
Perhaps the most impactful technological innovation in the clinical microbiology laboratory over the past decade has been the introduction of matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF) for the identification of routine clinical bacterial and fungal isolates. This technique involves the untargeted proteomic spectral analysis directly from bacterial or fungal colonies and rapid species-level identification through a growing highly diverse database. The first FDA-approved MALDI-TOF MS instruments were introduced in 2013, and since that time, these platforms have largely replaced many of the time and labor-intensive biochemical methods that had been the primary method of identification in laboratories for over a half century. In addition to more accurate species identification, MALDI-TOF MS provides significant reduction (12–48 hours) in turnaround times for identification. 51,52 More recent and ongoing advances involve the application of the methodology for acid-fast bacteria, Nocardia, 53 and mold identifications 54 as well as applications in epidemiological investigations and antibiotic resistance. 55
Challenges: Despite this significant impact and low reagent costs, the relatively high capital costs of MALDI-TOF MS instrumentation have slowed its adoption, particularly in smaller laboratories. Also, the discontinuation of conventional biochemical assays results in a lack of robustness during planned or unplanned instrument “downtimes.” Finally, its ease of use coupled with its rapid and accurate identification to species level has resulted in reporting of organisms, which may not have been easily reported in the past, such as certain coagulase-negative staphylococcal or alpha-hemolytic streptococcal species. This “overreporting” can lead to confusion amongst clinicians not familiar with these organisms and may lead to an increase in their unnecessary treatment. Therefore, the inclusion of laboratory and antimicrobial stewardship is recommended during implementation.
Home-based testing
The COVID-19 pandemic accelerated what was already a widespread interest in patient-centered infectious disease testing, including samples which are patient collected but analyzed by a clinical laboratory as well as fully home-based testing.
Patient-collected samples have become a popular method for diagnosing sexually transmitted infections, including Neisseria gonorrhoeae, Chlamydia trachomatis, and Trichomonas vaginalis. 56–58 With the COVID-19 pandemic, self-collection of anterior nares (AN) specimens for respiratory viral testing 59 was also utilized in many situations, though concerns for the sensitivity of AN swabs for viruses such as RSV 60 and adenovirus 61 may limit respiratory self-testing for other infections. Convenience, privacy, and access are major advantages of self-collected specimens, and analyses of self-collection tend to support both the quality of the result as well as a positive effect on test uptake/utilization. 62 Linking self-collected specimens with telemedicine (rather than traditional clinic visits with a provider) enhances access as well as speed and flexibility. 63,64 In addition to established healthcare routes that incorporate self-sampling with and without telemedicine, there are also several companies that provide self-collection kits for sexually transmitted diseases, respiratory viruses, and urinary tract infections, distributed in the context of remote providers. 65
Challenges: The discussion around home-based testing without associated telemedicine visits is far more complex. Fully home-based FDA-cleared infectious disease testing had been confined to assays for HIV before at-home COVID and COVID/Influenza tests became available under Emergency Use Authorization (EUA). To allow for kit stability, ease of use, and clear interpretation, home-based tests are usually antigen-detection lateral-flow assays that wick a patient’s sample across test and control zones and deliver a result through the appearance of a colored spot or line. Generally speaking, these tests do not have the same sensitivity (and often specificity) as molecular assays 66 and a great deal of debate has occurred over what constitutes sufficient sensitivity. 67 During the COVID-19 pandemic, at-home molecular assays were also developed and are a promising tool for future at-home testing. 68 Concerns about at-home testing include test performance and result interpretation, potential cost per test (often out of pocket), 69 quality control of the testing components/process, linkage to care and inclusion in the EMR, and tracking of any results that are of concern for public health. 70,71
The public focus on at-home testing and telemedicine during the COVID-19 pandemic has brought the issues surrounding both self-collected laboratory testing and at-home testing into the spotlight. The challenges of self-collected testing largely involve sample/transport device stability/performance and quality of sample collection. 65 The advantages of privacy, agency, convenience, and increased test uptake are so substantial that, despite concerns about quality and care linkage, it is expected that these patient-centered testing approaches will expand in the future. 72
Post-analytic
Clinician/laboratory interface
Another significant advancement in the clinical microbiology laboratory has been the enhancement of the clinician–laboratory interface with the help of nudging strategies to guide appropriate decision making while maintaining prescriber autonomy. 73 Some examples of these novel interventions include:
Framing: combines results with free text or educational materials to provide context for the results, changing their relative attractiveness. Examples include adding interpretative guidance on respiratory cultures growing normal commensal flora with a comment “no Methicillin-resistant Staphylococcus aureus or Pseudomonas aeruginosa isolated 74 ,” adding a nudge on a positive C. difficile nucleic acid amplification test with negative toxin enzyme immunoassay test to “consider colonization or early infection,” 75 adding interpretation guidance for coagulase-negative staphylococci growing in one of four blood culture bottles (one of two sets) as “possible contaminant,” and adding an interpretive comment on respiratory cultures for β-lactamase-negative Haemophilus influenzae or Moraxella catarrhalis stating, “this organism is predictably susceptible to ampicillin or amoxicillin.” 76
Cascade reporting: reports narrow-spectrum agents initially with subsequent susceptibilities reported only on resistant organisms, for example, reporting only ceftriaxone on ceftriaxone-susceptible Escherichia coli and Klebsiella species 77 and only cefazolin on cefazolin-susceptible gram-negative organisms. 78 The goal of these interventions is often to reduce the use of broad-spectrum agents like meropenem or antibiotics with high risk for C. difficile infection or other antibiotic-associated adverse events, like fluoroquinolones. 79
Selective reporting: restricts reporting of susceptibility results of certain antimicrobials based on predefined criteria (ie, broad-spectrum antimicrobials and high adverse drug events). Examples include suppression of ciprofloxacin susceptibility for Enterobacterales, for all sites of infection, when there was susceptibility to other agents on the gram-negative susceptibility testing panel, 80 or in its most extreme form, not reporting urine culture results from noncatheterized inpatients, instead requiring clinicians to call the clinical microbiology lab for results if concerns for true infection persist. 81,82
Result Review and Feedback: results of blood culture rapid diagnostics are reported with real-time decision support using antimicrobial stewardship personnel to assist in interpretation at the time of medical decision making. Studies have been published using this with staphylococcal blood-stream infections (BSI), 83 gram-negative BSI, 84,85 and all BSI. 86 These interventions are known to improve patient outcomes 87 and are cost effective. 88
Challenges: although the aim of these nudging strategies is to improve diagnostic processes to prompt timely action, there is limited evidence to show that they decrease antimicrobial use. More prescriptive interventions could limit clinician input and lead to missed diagnoses.
Other
Laboratory consolidation
Health system consolidation is a growing trend that has impacted clinical laboratories in recent years encompassing all phases of testing. Laboratory consolidation came into prominence with the development of large commercial laboratories. 89 As hospital laboratories have shifted from revenue to cost centers, these commercial laboratories have increasingly sought to purchase hospital laboratories. Some laboratory medicine departments have alternatively navigated a solution to consolidate various laboratory tests and functions within their growing health systems. 90 This solution often entails one central flagship laboratory taking on a larger volume of testing, allowing the health system to concentrate resources toward recent technologies and advancements mentioned earlier in this review in one location, which may allow a cost-efficient mechanism for improvements in quality to reach smaller hospitals in the network.
Thus, consolidation both depends on and facilitates related advancements, a prime example of which is total laboratory automation (TLA). TLA has been particularly impactful for microbiology laboratories, which traditionally have maintained highly complex, manual, and time-intensive test menus. Thus, TLA may allow health systems to continue offering this testing to increasing volumes of patients around the clock as the workforce of qualified technologists continues to shrink, although robust comparisons of outcomes in automated laboratories remain lacking in the literature. Consolidation provides the opportunity for coordination and standardization of such activities, as well as optimal adherence to best practices, reporting, and turnaround time across a network 91 ; TLA likewise may facilitate access to results via technologies such as remote visualization of culture plates. 89
Challenges: A major drawback of laboratory consolidation is the loss of access of providers to the laboratory. This results in disengagement between the two groups and impedes education, consultation, and the close engagement required for policy development in antimicrobial stewardship and infection prevention. Off-site centralization may lead to delays or quality of care. For example, blood culture incubation and workup are a highly complex and resource-intensive process that is increasingly targeted for centralization. Though guidelines suggest that specimens must be placed into incubation systems within two hours of collection to prevent false-negative results, 92 significant delays are common when samples are transported to centralized locations. Although technology is increasingly available to mitigate the impact of delayed incubation, 93 turnaround time remains of paramount importance for blood cultures. Throughout the process of laboratory consolidation, medical directors and administration leadership must work together to maintain an appropriate balance between financial considerations and patient safety.
Conclusion
We have summarized top advances made in the field of clinical microbiology in the past decade (Table 1) that every antimicrobial steward and infection prevention practitioner should know. To justify the costs of incorporating these novel microbiology advances into clinical practice, it is essential for clinicians to utilize these techniques appropriately. For example, developing institution-specific guidelines would be one way to support these key diagnostic and antimicrobial stewardship initiatives. 94 This would require a highly collaborative and interdisciplinary approach by working synergistically with key stakeholders including clinical microbiologists, infectious disease specialists, antimicrobial stewards, infection preventionists, hospitalists, primary care physicians, and healthcare information technology teams.
Acknowledgements
None.
Financial support
None reported.
Competing interests
All authors report no conflicts of interest relevant to this article.
References
- 1. Ku TSN, Al Mohajer M, Newton JA, et al. Improving antimicrobial use through better diagnosis: The relationship between diagnostic stewardship and antimicrobial stewardship. Infect Control Hosp Epidemiol 2023;44:1901–1908. [DOI] [PubMed] [Google Scholar]
- 2. Claeys KC, Johnson MD. Leveraging diagnostic stewardship within antimicrobial stewardship programmes. Drugs Context 2023;12. doi: 10.7573/dic.2022-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Keller SC, Feldman L, Smith J, Pahwa A, Cosgrove SE, Chida N. The use of clinical decision support in reducing diagnosis of and treatment of asymptomatic bacteriuria. J Hosp Med 2018;13:392–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mizusawa M, Small BA, Hsu Y-J, et al. Prescriber behavior in Clostridioides difficile Testing: A 3-hospital diagnostic stewardship intervention. Clin Infect Dis 2019;69:2019–2021. [DOI] [PubMed] [Google Scholar]
- 5. Rock C, Abosi O, Bleasdale S, et al. Clinical decision support systems to reduce unnecessary clostridioides difficile testing across multiple hospitals. Clin Infect Dis 2022;75:1187–1193. [DOI] [PubMed] [Google Scholar]
- 6. Fabre V, Klein E, Salinas AB, et al. A diagnostic stewardship intervention to improve blood culture use among adult nonneutropenic inpatients: the DISTRIBUTE Study. J Clin Microbiol 2020;58. doi: 10.1128/JCM.01053-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Woods-Hill CZ, Colantuoni EA, Koontz DW, et al. Association of diagnostic stewardship for blood cultures in critically ill children with culture rates, antibiotic use, and patient outcomes: Results of the Bright STAR Collaborative. JAMA Pediatr 2022;176:690–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Claeys KC, Trautner BW, Leekha S, et al. Optimal urine culture diagnostic stewardship practice-results from an expert modified-delphi procedure. Clin Infect Dis 2022;75:382–389. [DOI] [PubMed] [Google Scholar]
- 9. Yarrington ME, Reynolds SS, Dunkerson T, et al. Using clinical decision support to improve urine testing and antibiotic utilization. Infect Control Hosp Epidemiol 2023;44:1582–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Watson KJ, Trautner B, Russo H, et al. Using clinical decision support to improve urine culture diagnostic stewardship, antimicrobial stewardship, and financial cost: A multicenter experience. Infect Control Hosp Epidemiol 2020;41:564–570. [DOI] [PubMed] [Google Scholar]
- 11. Sick-Samuels AC, Linz M, Bergmann J, et al. Diagnostic stewardship of endotracheal aspirate cultures in a PICU. Pediatrics 2021;147. doi: 10.1542/peds.2020-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Escovedo C, Bell D, Cheng E, et al. Noninterruptive clinical decision support decreases ordering of respiratory viral panels during influenza season. Appl Clin Inform 2020;11:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Messacar K, Palmer C, Gregoire L, et al. Clinical and financial impact of a diagnostic stewardship program for children with suspected central nervous system infection. J Pediatr 2022;244:161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Munigala S, Jackups RR Jr, Poirier RF, et al. Impact of order set design on urine culturing practices at an academic medical centre emergency department. BMJ Qual Saf 2018;27:587–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Munigala S, Rojek R, Wood H, et al. Effect of changing urine testing orderables and clinician order sets on inpatient urine culture testing: Analysis from a large academic medical center. Infect Control Hosp Epidemiol 2019;40:281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khalifa M, Zabani I. Improving utilization of clinical decision support systems by reducing alert fatigue: Strategies and recommendations. Stud Health Technol Inform 2016;226:51–54. [PubMed] [Google Scholar]
- 17. Sutton RT, Pincock D, Baumgart DC, Sadowski DC, Fedorak RN, Kroeker KI. An overview of clinical decision support systems: benefits, risks, and strategies for success. NPJ Digit Med 2020;3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kotula JJ, Moore WS, Chopra A, Cies JJ. Association of Procalcitonin Value and Bacterial Coinfections in Pediatric Patients with Viral Lower Respiratory Tract Infections Admitted to the Pediatric Intensive Care Unit. J Pediatr Pharmacol Ther 2018;23:466–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ourani M, Honda NS, MacDonald W, Roberts J. Evaluation of evidence-based urinalysis reflex to culture criteria: Impact on reducing antimicrobial usage. Int J Infect Dis 2021;102:40–44. [DOI] [PubMed] [Google Scholar]
- 20. Claeys KC, Zhan M, Pineles L, et al. Conditional reflex to urine culture: Evaluation of a diagnostic stewardship intervention within the Veterans’ Affairs and Centers for Disease Control and Prevention Practice-Based Research Network. Infect Control Hosp Epidemiol 2021;42:176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aloisio E, Dolci A, Panteghini M. Procalcitonin: Between evidence and critical issues. Clin Chim Acta 2019;496:7–12. [DOI] [PubMed] [Google Scholar]
- 22. Paudel R, Dogra P, Montgomery-Yates AA, Coz Yataco A. Procalcitonin: A promising tool or just another overhyped test? Int J Med Sci 2020;17:332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van Houten CB, de Groot JAH, Klein A, et al. A host-protein based assay to differentiate between bacterial and viral infections in preschool children (OPPORTUNITY): a double-blind, multicentre, validation study. Lancet Infect Dis 2017;17:431–440. [DOI] [PubMed] [Google Scholar]
- 24. Heather JM, Chain B. The sequence of sequencers: The history of sequencing DNA. Genomics 2016;107:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim BJ, Lee SH, Lyu MA, et al. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB). J Clin Microbiol 1999;37:1714–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lysen C, Silva-Flannery L, Zaki SR, Gary JM, Lockhart SR. Performance evaluation of fungal DNA PCR amplification from formalin-fixed paraffin-embedded tissue for diagnosis: Experience of a tertiary reference laboratory. Mycoses 2021;64:603–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kubiak J, Morgan A, Kirmaier A, Arnaout R, Riedel S. Universal PCR for bacteria, mycobacteria, and fungi: A 10-year retrospective review of clinical indications and patient outcomes. bioRxiv 2023; https://www.medrxiv.org/content/10.1101/2023.08.02.23293145v1.full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Muhamad Rizal NS, Neoh H-M, Ramli R, et al. Advantages and limitations of 16S rRNA next - generation sequencing for pathogen identification in the diagnostic microbiology laboratory: perspectives from a middle - income country. Diagnostics (Basel) 2020;10. doi: 10.3390/diagnostics10100816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mitchell SL, Simner PJ. Next-generation sequencing in clinical microbiology: Are we there yet? Clin Lab Med 2019;39:405–418. [DOI] [PubMed] [Google Scholar]
- 30. Miller S, Naccache SN, Samayoa E, et al. Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid. Genome Res 2019;29:831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hill JA, Dalai SC, Hong DK, et al. Liquid biopsy for invasive mold infections in hematopoietic cell transplant recipients with pneumonia through next-generation sequencing of microbial cell-free DNA in plasma. Clin Infect Dis 2021;73:e3876–e3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ballard SA, Sherry NL, Howden BP. Public health implementation of pathogen genomics: the role for accreditation and application of ISO standards. Microb Genom 2023;9:mgen001097. doi: 10.1099/mgen.0.001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen Z, Azman AS, Chen X, et al. Global landscape of SARS-CoV-2 genomic surveillance and data sharing. Nat Genet 2022;54:499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gwinn M, MacCannell D, Armstrong GL. Next-generation sequencing of infectious pathogens. JAMA 2019;321:893–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Piantadosi A, Kanjilal S. Diagnostic approach for arboviral infections in the United States. J Clin Microbiol 2020;58:e01926–19. doi: 10.1128/JCM.01926-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Debiasi RL, Tyler KL. West Nile virus meningoencephalitis. Nat Clin Pract Neurol 2006;2:264–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Weiss ZF, Pyden AD, Jhaveri TA, Kanjilal S. The diagnostic and clinical utility of microbial cell-free DNA sequencing in a real-world setting. Diagn Microbiol Infect Dis 2023;107:116004. [DOI] [PubMed] [Google Scholar]
- 38. Musher DM. Problems with etiologic diagnosis of community-acquired pneumonia using plasma microbial cell-free DNA sequencing. Antimicrob Steward Healthc Epidemiol 2023;3:e206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen Y, Fan L-C, Chai Y-H, Xu J-F. Advantages and challenges of metagenomic sequencing for the diagnosis of pulmonary infectious diseases. Clin Respir J 2022;16:646–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Blauwkamp TA, Thair S, Rosen MJ, et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol 2019;4:663–674. [DOI] [PubMed] [Google Scholar]
- 41. Donner LM, Campbell WS, Lyden E, Van Schooneveld TC. Assessment of rapid-blood-culture-identification result interpretation and antibiotic prescribing practices. J Clin Microbiol 2017;55:1496–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ramanan P, Bryson AL, Binnicker MJ, Pritt BS, Patel R. Syndromic panel-based testing in clinical microbiology. Clin Microbiol Rev 2018;31:e00024–17. doi: 10.1128/CMR.00024-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weiss ZF, Cunha CB, Chambers AB, et al. Opportunities revealed for antimicrobial stewardship and clinical practice with implementation of a rapid respiratory multiplex assay. J Clin Microbiol 2019;57:e00861–19. doi: 10.1128/JCM.00861-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Simner PJ, Oethinger M, Stellrecht KA, et al. Multisite evaluation of the BD Max extended enteric bacterial panel for detection of Yersinia enterocolitica, Enterotoxigenic Escherichia coli, Vibrio, and Plesiomonas shigelloides from stool specimens. J Clin Microbiol 2017;55:3258–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Spina A, Kerr KG, Cormican M, et al. Spectrum of enteropathogens detected by the FilmArray GI Panel in a multicentre study of community-acquired gastroenteritis. Clin Microbiol Infect 2015;21:719–728. [DOI] [PubMed] [Google Scholar]
- 46. Zacharioudakis IM, Zervou FN, Phillips MS, Aguero-Rosenfeld ME. Rate and consequences of missed Clostridioides (Clostridium) difficile infection diagnosis from nonreporting of Clostridioides difficile results of the multiplex GI PCR panel: experience from two-hospitals. Diagn Microbiol Infect Dis 2021;100:115346. [DOI] [PubMed] [Google Scholar]
- 47. Jhaveri TA, Taqi A, Pearson JC, Kanjilal S. Impact of direct disk-diffusion testing on time to optimal antibiotic therapy. Antimicrob Steward Healthc Epidemiol 2023;3:e59. [Google Scholar]
- 48. Kuil SD, Hidad S, Schneeberger C, et al. Susceptibility testing by volatile organic compound detection direct from positive blood cultures: A proof-of-principle laboratory study. Antibiotics (Basel) 2022;11:705. doi: 10.3390/antibiotics11060705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Banerjee R, Humphries R. Rapid antimicrobial susceptibility testing methods for blood cultures and their clinical impact. Front Med 2021;8:635831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Charnot-Katsikas A, Tesic V, Love N, et al. Use of the accelerate pheno system for identification and antimicrobial susceptibility testing of pathogens in positive blood cultures and impact on time to results and workflow. J Clin Microbiol 2018;56:e01166–17. doi: 10.1128/JCM.01166-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lo CK-F, Mertz D, Yamamura D, Loeb M. Assessing impact of MALDI mass spectroscopy on reducing directed antibiotic coverage time for Gram-negative organisms. PLoS One 2020;15:e0228935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rychert J. Benefits and limitations of MALDI-TOF mass spectrometry for the identification of microorganisms. J Infectiology 2019;2:1–5. [Google Scholar]
- 53. Marín M, Ruiz A, Iglesias C, et al. Identification of Nocardia species from clinical isolates using MALDI-TOF mass spectrometry. Clin Microbiol Infect 2018;24:1342.e5–1342.e8. [DOI] [PubMed] [Google Scholar]
- 54. Normand A-C, Blaize M, Imbert S, et al. Identification of molds with matrix-assisted laser desorption ionization-time of flight mass spectrometry: Performance of the newly developed MSI-2 application in comparison with the bruker filamentous fungi database and MSI-1. J Clin Microbiol 2021;59:e0129921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Florio W, Baldeschi L, Rizzato C, Tavanti A, Ghelardi E, Lupetti A. Detection of antibiotic-resistance by MALDI-TOF mass spectrometry: An expanding area. Front Cell Infect Microbiol 2020;10:572909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Barbee LA, Golden MR, Thibault CS, McNeil CJ, Soge OO. Performance of patient-collected specimens for Neisseria gonorrhoeae culture. Clin Infect Dis 2021;73:e3196–e3200. [DOI] [PubMed] [Google Scholar]
- 57. Barnes P, Vieira R, Harwood J, Chauhan M. Self-taken vaginal swabs versus clinician-taken for detection of candida and bacterial vaginosis: a case-control study in primary care. Br J Gen Pract 2017;67:e824–e829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lunny C, Taylor D, Hoang L, et al. Self-collected versus clinician-collected sampling for chlamydia and gonorrhea screening: A systemic review and meta-analysis. PLoS One 2015;10:e0132776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gadenstaetter AJ, Mayer CD, Landegger LD. Nasopharyngeal versus nasal swabs for detection of SARS-CoV-2: a systematic review. Rhinology 2021;59:410–421. [DOI] [PubMed] [Google Scholar]
- 60. Stensballe LG, Trautner S, Kofoed P-E, et al. Comparison of nasopharyngeal aspirate and nasal swab specimens for detection of respiratory syncytial virus in different settings in a developing country. Trop Med Int Health 2002; 7:317–321. [DOI] [PubMed] [Google Scholar]
- 61. Blaschke AJ, Allison MA, Meyers L, et al. Non-invasive sample collection for respiratory virus testing by multiplex PCR. J Clin Virol 2011;52:210–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kpokiri EE, Marley G, Tang W, et al. Diagnostic infectious diseases testing outside clinics: a global systematic review and meta-analysis. Open Forum Infect Dis 2020;7:ofaa360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gilbert M, Thomson K, Salway T, et al. Differences in experiences of barriers to STI testing between clients of the internet-based diagnostic testing service GetCheckedOnline.com and an STI clinic in Vancouver, Canada. Sex Transm Infect 2019;95:151–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Greenland KE, Op de Coul ELM, van Bergen JEAM, et al. Acceptability of the internet-based Chlamydia screening implementation in the Netherlands and insights into nonresponse. Sex Transm Dis 2011;38:467–474. [DOI] [PubMed] [Google Scholar]
- 65. Kersh EN, Shukla M, Raphael BH, Habel M, Park I. At-home specimen self-collection and self-testing for sexually transmitted infection screening demand accelerated by the COVID-19 pandemic: A review of laboratory implementation issues. J Clin Microbiol 2021;59:e0264620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Soni A, Herbert C, Lin H, et al. Performance of Rapid Antigen Tests to Detect Symptomatic and Asymptomatic SARS-CoV-2 Infection: A Prospective Cohort Study. Ann Intern Med 2023;176:975–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Prestedge J, Williamson DA. The performance of rapid antigen tests against SARS-CoV-2 variants. Lancet Infect Dis 2023;23:883–884. [DOI] [PubMed] [Google Scholar]
- 68. Zahavi M, Rohana H, Azrad M, Shinberg B, Peretz A. Rapid SARS-CoV-2 detection using the Lucira™ Check It COVID-19 test kit. Diagnostics (Basel) 2022;12:1877. doi: 10.3390/diagnostics12081877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kirby RP, Maimaran M, Palamountain KM. Is there a ‘price that’s right’ for at-home COVID tests? PLoS One 2023;18:e0282043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rader B, Gertz A, Iuliano AD, et al. Use of at-home COVID-19 tests - United States, August 23, 2021-March 12, 2022. MMWR Morb Mortal Wkly Rep 2022;71:489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Benda A, Zerajic L, Ankita A, Cleary E, Park Y, Pandey S. COVID-19 testing and diagnostics: a review of commercialized technologies for cost, convenience and quality of tests. Sensors 2021;21:6581. doi: 10.3390/s21196581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Adalja AA. At-home infectious disease testing: An idea whose time has come. Antimicrob Steward Healthc Epidemiol 2022;2:e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Langford BJ, Leung E, Haj R, et al. Nudging In MicroBiology Laboratory Evaluation (NIMBLE): A scoping review. Infect Control Hosp Epidemiol 2019;40:1400–1406. [DOI] [PubMed] [Google Scholar]
- 74. Musgrove MA, Kenney RM, Kendall RE, et al. Microbiology Comment Nudge Improves Pneumonia Prescribing. Open Forum Infect Dis 2018;5:ofy162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Herman DJ, Sarabia A, Chan H, Graham C. Changing results to change results: nudging antimicrobial prescribing for Clostridium difficile. Open Forum Infect Dis 2021;8:ofaa605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Arena CJ, Kenney RM, Kendall RE, Tibbetts RJ, Veve MP. Respiratory culture nudge improves antibiotic prescribing for Moraxella catarrhalis and Haemophilus influenzae lower respiratory tract infections. Antimicrob Steward Healthc Epidemiol 2023;3:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Liao S, Rhodes J, Jandarov R, DeVore Z, Sopirala MM. Out of sight-out of mind: impact of cascade reporting on antimicrobial usage. Open Forum Infect Dis 2020;7:ofaa002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Johnson LS, Patel D, King EA, Maslow JN. Impact of microbiology cascade reporting on antibiotic de-escalation in cefazolin-susceptible Gram-negative bacteremia. Eur J Clin Microbiol Infect Dis 2016;35:1151–1157. [DOI] [PubMed] [Google Scholar]
- 79. Vissichelli NC, Orndahl CM, Cecil JA, et al. Impact of cascade reporting of antimicrobial susceptibility on fluoroquinolone and meropenem consumption at a Veterans’ Affairs medical center. Infect Control Hosp Epidemiol 2022;43:199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Langford BJ, Seah J, Chan A, Downing M, Johnstone J, Matukas LM. Antimicrobial Stewardship in the Microbiology Laboratory: Impact of Selective Susceptibility Reporting on Ciprofloxacin Utilization and Susceptibility of Gram-Negative Isolates to Ciprofloxacin in a Hospital Setting. J Clin Microbiol 2016;54:2343–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Daley P, Garcia D, Inayatullah R, Penney C, Boyd S. Modified reporting of positive urine cultures to reduce inappropriate treatment of asymptomatic bacteriuria among nonpregnant, noncatheterized inpatients: A randomized controlled trial. Infect Control Hosp Epidemiol 2018;39:814–819. [DOI] [PubMed] [Google Scholar]
- 82. Leis JA, Rebick GW, Daneman N, et al. Reducing antimicrobial therapy for asymptomatic bacteriuria among noncatheterized inpatients: A proof-of-concept study. Clin Infect Dis 2014;58:980–983. [DOI] [PubMed] [Google Scholar]
- 83. Wenzler E, Wang F, Goff DA, et al. An Automated, pharmacist-driven initiative improves quality of care for Staphylococcus aureus bacteremia. Clin Infect Dis 2017;65:194–200. [DOI] [PubMed] [Google Scholar]
- 84. Claeys KC, Heil EL, Hitchcock S, Johnson JK, Leekha S. Management of gram-negative bloodstream infections in the era of rapid diagnostic testing: impact with and without antibiotic stewardship. Open Forum Infect Dis 2020;7:ofaa427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Rivard KR, Athans V, Lam SW, et al. Impact of antimicrobial stewardship and rapid microarray testing on patients with Gram-negative bacteremia. Eur J Clin Microbiol Infect Dis 2017;36:1879–1887. [DOI] [PubMed] [Google Scholar]
- 86. Banerjee R, Teng CB, Cunningham SA, et al. Randomized trial of rapid multiplex polymerase chain reaction-based blood culture identification and susceptibility testing. Clin Infect Dis 2015;61:1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Timbrook TT, Morton JB, McConeghy KW, Caffrey AR, Mylonakis E, LaPlante KL. The effect of molecular rapid diagnostic testing on clinical outcomes in bloodstream infections: A systematic review and meta-analysis. Clin Infect Dis 2017;64:15–23. [DOI] [PubMed] [Google Scholar]
- 88. Mponponsuo K, Leal J, Spackman E, Somayaji R, Gregson D, Rennert-May E. Mathematical model of the cost-effectiveness of the BioFire FilmArray Blood Culture Identification (BCID) Panel molecular rapid diagnostic test compared with conventional methods for identification of Escherichia coli bloodstream infections. J Antimicrob Chemother 2022;77:507–516. [DOI] [PubMed] [Google Scholar]
- 89. Vandenberg O, Durand G, Hallin M, et al. Consolidation of clinical microbiology laboratories and introduction of transformative technologies. Clin Microbiol Rev 2020;33:e00057–19. doi: 10.1128/CMR.00057-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Jensen KJ, Stallone R, Eller M, et al. Northwell Health Laboratories: the 10-year outcomes after deciding to keep the lab. Arch Pathol Lab Med 2019;143:1517–1530. [DOI] [PubMed] [Google Scholar]
- 91. Sautter RL, Thomson RB Consolidated clinical microbiology laboratories. J Clin Microbiol 2015;53:1467–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Guideline M47 CC. Principles and Procedures for Blood Cultures; 2022.
- 93. Adamik M, Hutchins A, Mangilit J, Katzin B, Totty H, Deol P. Effect of delayed entry on performance of the BACT/ALERT FAN PLUS bottles in the BACT/ALERT VIRTUO blood culture system. Eur J Clin Microbiol Infect Dis 2021;40:699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Pierce VM, Bhowmick T, Simner PJ. Guiding antimicrobial stewardship through thoughtful antimicrobial susceptibility testing and reporting strategies: an updated approach in 2023. J Clin Microbiol 2023:e0007422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.BioFire Blood Culture Identification (BCID) panels, 2018. https://www.biofiredx.com/products/the-filmarray-panels/filmarraybcid/. Accessed 28 September 2023.
- 96.Blood Culture Identification (BCID) panels. https://www.genmarkdx.com/panels/eplex-panels/bcid-panels/. Accessed 28 September 2023.
- 97.Homepage. https://int.diasorin.com/en. Accessed 25 October 2023.
- 98.T2Bacteria panel, 2017. https://www.t2biosystems.com/products-technology/t2bacteria-panel/. Accessed 25 October 2023.
- 99.T2Candida panel. 2017. https://www.t2biosystems.com/products-technology/t2candida-panel/. Accessed 25 October 2023.
- 100.Accelerate diagnostics. https://acceleratediagnostics.com/products/accelerate-phenotest-bc/. Accessed 25 October 2023.
- 101.BioFire® FilmArray® respiratory 2.1 Panel, 2021. https://www.biofiredx.com/products/the-filmarray-panels/filmarrayrp/. Accessed 25 October 2023.
- 102.POC respiratory panel test, 2020. https://www.biofiredx.com/products/the-filmarray-panels/filmarray-respiratory-panel-ez/. Accessed 27 September 2023.
- 103.VERIGENE® Respiratory Pathogen Flex Test. Luminex Corporation - A DiaSorin Company, 2016. https://www.luminexcorp.com/respiratory-pathogens-flex-test/. Accessed 27 September 2023.
- 104.Respiratory pathogen panels. https://www.genmarkdx.com/panels/eplex-panels/respiratory-pathogen-panel/. Accessed 26 September 2023.
- 105.The BioFire® FilmArray® Pneumonia panel, 2018. https://www.biofiredx.com/products/the-filmarray-panels/filmarray-pneumonia/. Accessed 27 September 2023.
- 106.The BioFire® FilmArray® gastrointestinal (GI) Panel, 2018. https://www.biofiredx.com/products/the-filmarray-panels/filmarraygi/. Accessed 27 September 2023.
- 107.XTAG® gastrointestinal pathogen panel. Luminex Corporation - A DiaSorin Company, 2014. https://www.luminexcorp.com/gastrointestinal-pathogen-panel/. Accessed 27 September 2023.
- 108.BD MAX™ enteric bacterial panel, 2017. https://moleculardiagnostics.bd.com/syndromic-solutions/enteric-solutions/enteric-bacterial-panel/. Accessed 25 October 2023.
- 109.BD MAX™ extended enteric bacterial panel, 2017. https://moleculardiagnostics.bd.com/syndromic-solutions/enteric-solutions/extended-enteric-bacterial-panel/. Accessed 25 October 2023.
- 110.BD MAX™ enteric viral panel, 2019. https://moleculardiagnostics.bd.com/syndromic-solutions/enteric-solutions/bd-max-evp/. Accessed 25 October 2023.
- 111.BD MAX™ enteric parasite panel, 2017. https://moleculardiagnostics.bd.com/syndromic-solutions/enteric-solutions/enteric-parasite-panel/. Accessed 25 October 2023.
- 112.VERIGENE® enteric pathogens test. https://us.diasorin.com/en/molecular-diagnostics/kits-reagents/verigene-enteric-pathogens-test. Accessed 25 October 2023.
- 113.BioFire® joint infection (JI) panel, 2020. https://www.biofiredx.com/products/the-filmarray-panels/ji/. Accessed 27 September 2023.
- 114.The BioFire® FilmArray® meningitis/encephalitis (ME) Panel, 2018. https://www.biofiredx.com/products/the-filmarray-panels/filmarrayme/. Accessed 27 September 2023.