Abstract
14-3-3 proteins complex with many signaling molecules, including the Raf-1 kinase. However, the role of 14-3-3 in regulating Raf-1 activity is unclear. We show here that 14-3-3 is bound to Raf-1 in the cytosol but is totally displaced when Raf-1 is recruited to the plasma membrane by oncogenic mutant Ras, in vitro and in vivo. 14-3-3 is also displaced when Raf-1 is targeted to the plasma membrane. When serum-starved cells are stimulated with epidermal growth factor, some recruitment of 14-3-3 to the plasma membrane is evident, but 14-3-3 recruitment correlates with Raf-1 dissociation and inactivation, not with Raf-1 recruitment. In vivo, overexpression of 14-3-3 potentiates the specific activity of membrane-recruited Raf-1 without stably associating with the plasma membrane. In vitro, Raf-1 must be complexed with 14-3-3 for efficient recruitment and activation by oncogenic Ras. Recombinant 14-3-3 facilitates Raf-1 activation by membranes containing oncogenic Ras but reduces the amount of Raf-1 that associates with the membranes. These data demonstrate that the interaction of 14-3-3 with Raf-1 is permissive for recruitment and activation by Ras, that 14-3-3 is displaced upon membrane recruitment, and that 14-3-3 may recycle Raf-1 to the cytosol. A model that rationalizes many of the apparently discrepant observations on the role of 14-3-3 in Raf-1 activation is proposed.
Diverse genetic and biochemical studies have demonstrated that the Raf-1 kinase is an important downstream effector of Ras (32, 37). Raf-1 activates the dual-specificity kinase MEK1 by phosphorylating two regulatory serine residues. MEK1 in turn activates the mitogen-activated protein (MAP) kinases extracellular signal-regulated kinase 1 (ERK1) and ERK2 by phosphorylating regulatory tyrosine and threonine residues (33). The structural basis of ERK2 activation has recently been solved (3); in marked contrast, the mechanism of activation of Raf-1 is complex and incompletely understood.
The initial event in activation is the recruitment of Raf-1 from the cytosol to the plasma membrane (25, 44, 48, 52). The localization of Raf-1 to the plasma membrane initiates a series of events that ultimately leads to full activation. These events include tyrosine, serine, and threonine phosphorylation (4, 8, 30, 49, 54) plus interactions with Ras (7, 35, 43), phospholipids (17, 18), and 14-3-3 proteins and their associated proteins (11, 14, 15, 22, 26, 47), and possibly dimerization (12, 28). Unraveling the relative contributions of these processes and the sequence in which they come into play at the plasma membrane is presently of great interest.
There is good evidence that Raf-1 kinase activity is regulated by tyrosine phosphorylation. Raf-1 is phosphorylated in vivo on tyrosine residues 340 and 341, and replacement of these tyrosines with negatively charged aspartate (RafDD) significantly upregulates Raf-1 basal kinase activity (8, 31), although further activation occurs when RafDD is localized to the plasma membrane (30, 43). Conversely, replacement of the regulatory tyrosines with phenylalanine (RafFF) renders Raf-1 resistant to activation by Ras and membrane targeting (8, 43). In mammalian cells, but not insect cells, phosphorylation of tyrosines 340 and 341 occurs only when Raf-1 is recruited to the plasma membrane (10, 30). Thus, one consequence of membrane recruitment is the colocalization of Raf-1 with activated tyrosine kinases and the facilitation of tyrosine phosphorylation.
The minimal Ras binding domain (RBD), comprising Raf-1 residues 55 to 131, binds to the switch 1 region of activated Ras-GTP (50, 51, 56). A mutation in the Raf-RBD (R89L) that abrogates Ras binding prevents the recruitment of Raf-1 to the plasma membrane by Ras and blocks Raf-1 activation in mammalian cells (10, 30). The Ras-RBD interaction appears to serve no role in Raf-1 activation other than membrane recruitment, because the loss-of-function R89L mutation is silent in membrane-targeted RafCAAX (30, 43). The interaction between Ras and Raf-1 is, however, more complex than was initially thought. It has been shown recently that the Ras switch 2 region interacts with a second Raf-1 domain, the Raf cysteine-rich domain (Raf-CRD), comprising Raf-1 residues 130 to 184 (2, 7, 16, 20). Posttranslationally processed Ras binds more avidly to the Raf-CRD than unmodified Ras (20, 27), although it has yet to be determined whether this reflects direct binding of the Ras prenyl group to the Raf-CRD or some influence of the prenyl group on the Ras N-terminal structure. The interaction of Ras with the Raf-CRD is important because mutations in the Ras switch 2 region that impair binding to the Raf-CRD compromise Ras biological activity (7).
The Raf-CRD is not required for membrane recruitment of Raf-1 by Ras (43), but mutations that disrupt the Raf-CRD significantly reduce Raf-1 basal kinase activity and impair Raf-1 activation at the plasma membrane (27, 43). Our recent work also shows that full activation of membrane-recruited Raf-1 requires negative charges on residues 340 and 341, an intact Raf-CRD, and coexpression of Ras-GTP (43). Thus, continuing interactions between Ras and membrane-localized Raf-1 via the CRD play a critical role in Raf-1 activation.
Raf-1 contains two phosphorylation sites at S259 and S621 which match a recently identified 14-3-3 consensus binding sequence (39). 14-3-3 is also a ligand for the Raf-CRD (6, 34). Although 14-3-3 interacts with both the N-terminal regulatory and C-terminal kinase domains of Raf-1, the exact role of 14-3-3 in Raf-1 activation remains uncertain. Certain biochemical studies support a negative role for 14-3-3 in Raf-1 activation (6, 41), while genetic (5, 24) and other studies (11, 14, 22, 26) suggest that 14-3-3 may positively regulate Raf-1. For example, mutations in Raf-1 that block 14-3-3 binding to S259 facilitate Raf-1 activation (42), and mutations in the Raf-CRD that selectively interfere with 14-3-3 binding enhance Raf-1 transforming activity (6). Moreover, activated Ras competes with 14-3-3 for binding to the Raf-1 N terminus, consistent with activation requiring displacement of 14-3-3 from the regulatory domain (41). Conversely, phosphopeptides that displace 14-3-3 from Raf-1 block Raf-1-mediated Xenopus oocyte maturation (39, 40). These studies suggest that 14-3-3 plays multiple and probably competing roles in Raf-1 regulation.
In this study we have used a combination of in vivo and in vitro Raf-1 activation assays to identify positive and negative roles for 14-3-3 in Raf-1 activation. We show that 14-3-3 must be complexed with Raf-1 for efficient membrane recruitment and activation by Ras but that 14-3-3 is completely displaced from Raf-1 at the plasma membrane. We also demonstrate a role for 14-3-3 in recycling Raf-1 from the plasma membrane to the cytosol.
MATERIALS AND METHODS
COS cell transfections and fractionation.
COS cells were electroporated as described previously (21). After 54 h, cells were incubated in serum-free medium for 18 h before being harvested. Cells were washed and scraped on ice into 0.5 ml of buffer A (10 mM Tris Cl [pH 7.5], 25 mM NaF, 5 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol [DTT], 100 μM NaVO4). After 10 min on ice, cells were homogenized with 50 strokes in a tight-fitting Dounce homogenizer, and the nuclei were removed by low-speed centrifugation. The postnuclear supernatants were spun at 100,000 × g. The supernatant (S100) was removed, and the sedimented fraction (P100) was rinsed and resuspended by sonication in 100 μl of ice-cold buffer A. The P100 membrane fraction contains plasma membranes and various intracellular membranes, including the endoplasmic reticulum, Golgi apparatus, and endosomes. Protein content was measured by the Bradford reaction, and the S100 fraction and resuspended P100 fraction were snap frozen and stored in aliquots at −70°C. COS cells to be treated with epidermal growth factor (EGF) were plated at 50% confluence and 18 h later were switched to serum-free medium. After a further 18 h in serum-free medium, cells were incubated in prewarmed Dulbecco modified Eagle medium (DMEM) with 1% bovine serum albumin (BSA) plus 50 nM EGF for the specified times. The cells were then rinsed with ice-cold phosphate-buffered saline (PBS) and placed on ice for harvesting. Cell fractionation was carried out as described above.
Western blotting.
Expression and subcellular localization of Raf-1, 14-3-3, and Ras proteins were determined by immunoblotting. Sample loading was normalized for S100 protein content, and equal proportions of the S100 (cytosol) and P100 (membrane) fractions of each lysate were then used for blotting. Samples were resolved on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels with 10% (Raf), 12% (14-3-3), or 15% (Ras) polyacrylamide and were transferred to polyvinylidene difluoride membranes by using semidry transfer. Western blots were probed with an anti-FLAG monoclonal antibody (M2; Kodak) or with an anti-Raf-1 (C20; Santa Cruz), anti-14-3-3 (β/pan 14-3-3; Santa Cruz), or anti-Ras (Y13-259) antibody. Immunoblots were developed by enhanced chemiluminescence (SuperSignal; Pierce) and then were quantitated by phosphorimaging with a CH-screen (Bio-Rad).
When equimolar amounts of recombinant Raf-1 and 14-3-3, over a range of 0.5 to 5 pmol, were immunoblotted on the same membrane by using a 1:1,000 dilution of primary antibody and a 1:5,000 dilution of anti-rabbit horseradish peroxidase conjugate (Pierce) and were developed as described above, the signal obtained from 14-3-3 was approximately 45% of that obtained from Raf-1.
Immunoprecipitations.
S100 fractions were normalized for Raf-1 content by quantitative Western blotting or for protein content by the Bradford reaction, adjusted to 1% Nonidet P-40 (NP-40), and diluted to 400 μl with buffer B (50 mM Tris Cl [pH 7.5], 75 mM NaCl, 5 mM MgCl2, 25 mM NaF, 5 mM EGTA, 100 μM NaVO4, 1% NP-40, 1 mM DTT). P100 fractions normalized for Raf-1 content by quantitative Western blotting or for protein content by the Bradford reaction were adjusted to 1% NP-40, sonicated for 90 s at 4°C, incubated on ice for 10 min, and microcentrifuged for 5 min, and the soluble membrane extract was diluted to 400 μl with buffer B. The samples were rotated with 10 μl of anti-FLAG Sepharose beads for 2 h at 4°C and then were washed six times in buffer B. Immunoprecipitates were taken up in 1× SDS sample buffer, resolved by SDS-PAGE, and analyzed by Western blotting as described above.
Immunofluorescence analysis in BHK cells.
BHK cells were cultured at 37°C (5% CO2) in DMEM supplemented with 10% (vol/vol) bovine calf serum and 100 U (each) of penicillin and streptomycin/ml. Cells were plated onto glass coverslips at 60% confluence, and 4 h later they were transfected by using Lipofectamine (Life Technologies) according to the manufacturer’s instructions, with 1.6 μg of EXV expression plasmid for FLAG Raf, myc14-3-3, and RasG12V. Duplicate coverslips were prepared for each transfection, which included FLAG Raf plus myc14-3-3 and FLAG Raf plus myc14-3-3 plus RasG12V, as well as single transfections of FLAG Raf, myc14-3-3, and RasG12V alone. After an overnight incubation, cells were fixed with 4% paraformaldehyde, permeabilized in 0.2% Triton X-100, and blocked with 3% BSA in PBS. The cells were then incubated in undiluted 9E10 hybridoma supernatant (anti-myc), undiluted Y13-238 hybridoma supernatant (anti-Ras), or 20 μg of M2 anti-FLAG antibody/ml as required. After extensive washing in PBS, fluorescein isothiocyanate (FITC)-conjugated anti-mouse (for FLAG and myc) or anti-rat (for Y13-238) secondary antibodies (Pierce) were added at 30 μg/ml. Coverslips were washed and mounted in Moviol. Fluorescence images were taken in a Bio-Rad MRC-600 Zeiss microscope with a 63× magnification lens, a BHS filter, and blue light exciting at 488 ± 5 nm, with correction for emissions 515 nm and longer. Kalman averaging of 30 scans was used to produce the final images.
Raf-1 kinase assays.
The Raf-1 kinase assay is discussed in detail elsewhere (43). In brief, P100 aliquots, normalized for Raf-1 content by quantitative Western blotting (typically 10 to 30 μg of total protein), were adjusted to 20 μl with buffer A. Two and two-tenths microliters of 10% NP-40 was added, and the membranes were sonicated in a sonicating water bath for 2 min at 4°C. A 10-μl aliquot of sonicated P100 fraction was incubated with 6 μl of buffer A containing 0.25 μg of MEK, 1 μg of ERK, and 4 μl of 0.5 mM ATP–40 mM MgCl2 and was vortexed at 30°C. A second 10-μl aliquot of sonicated P100 fraction was incubated with 6 μl of buffer A containing 4 μl of 0.5 mM ATP–40 mM MgCl2 and 1 μg of ERK but no MEK, and the mixture was vortexed at 30°C (control tube). After 20 min, the samples were placed on ice, and 10 μl was diluted into 40 μl of ice-cold buffer C (50 mM Tris Cl [pH 7.5], 75 mM NaCl, 5 mM MgCl2, 25 mM NaF, 5 mM EGTA, 100 μM NaVO4, 1 mM DTT). Ten microliters of these diluted samples was taken into a second incubation with 5 μl of myelin basic protein (MBP; 16 μg) and 10 μl of an ATP mixture containing 0.5 mM ATP, 50 mM MgCl2, and [γ-32P]ATP (2,400 cpm/pmol). The MBP kinase reaction was performed in duplicate. After 10 min the reaction was stopped by the addition of 6 μl of 5 × SDS-PAGE sample buffer and the reaction products were resolved on SDS–15% PAGE gels. The radioactivity incorporated into MBP was measured by phosphorimaging after the gels were spotted with a known amount of radioactive [γ-32P]ATP. Background counts due to any P100-associated MEK and ERK were estimated from the control tubes and subtracted from the assay counts (<5% of total activity). To verify Raf-1 normalization, 10 μl of the initial reaction mixture incubated with MEK and ERK was reanalyzed by quantitative Western blotting.
To measure the activity of cytosolic Raf, S100 fractions were normalized for Raf-1 content by quantitative Western blotting and immunoprecipitated as described above. The immunoprecipitates were split into two aliquots and incubated with MEK and ERK, or with ERK alone, and the kinase assay was completed as described above. The beads were then collected, washed, and immunoblotted to verify the amount of Raf-1 present in the assay.
Preparation of recombinant 14-3-3.
Cultures (250 ml) of Escherichia coli containing a pGEX plasmid encoding glutathione S-transferase–14-3-3 were induced for 5 h with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were harvested, and recombinant protein was purified, as described previously (19) but with two modifications. Before sonication, cells were snap frozen and then thawed in 10 ml of lysis buffer (50 mM Tris Cl [pH 7.5], 150 mM NaCl, 1% Triton X-100, 5 mM MgCl2, 1 mM EGTA, 1 mM DTT, 10 μg of leupeptin/ml, 10 μg of aprotinin/ml, 3 mM phenylmethylsulfonyl fluoride), and after thrombin cleavage, 14-3-3 was purified to homogeneity by MonoQ chromatography.
In vitro activation assay.
Only freshly harvested cells were used for this assay, and S100 and P100 fractions, once prepared, were used immediately. COS cells were transfected, harvested, and fractionated as described above, except that each 10-cm plate was harvested in 200 μl of buffer A and the P100 fractions were resuspended by sonication in 80 μl of buffer A. The protein content was measured by the Bradford reaction, and concentrations were adjusted with buffer A. Optimal protein concentrations were 5 mg/ml (P100) and 2 mg/ml (S100). Protein concentrations of the control membranes (EXV vector transfected) and Ras-expressing membranes were always adjusted to equivalence. For the assay, 25 μl of the P100 fraction was mixed on ice with 50 μl of the S100 fraction and then incubated at 25°C on a shaking heating block for 10 min. Control incubations included EXV P100 incubated with Raf-1 S100 and Ras P100 incubated with buffer A. Samples were then spun immediately at 100,000 × g at 4°C for 15 min. The supernatant was removed, and the pellet was rinsed in buffer A, then resuspended by sonication in 45 μl of buffer A; 20 μl of the resuspended membrane pellet was assayed for Raf-1 activity as described above. Another 20 μl was adjusted to 1% NP-40 by using 10% NP-40, sonicated for 90 s, incubated on ice for 10 min, then microcentrifuged for 20 min. The soluble and insoluble fractions were both analyzed by quantitative Western blotting for Ras and Raf-1 as described above. Approximately 70% of Raf-1 and 100% of Ras in the membrane pellet were solubilized under these conditions.
RESULTS
Raf-1 recruited to the plasma membrane by Ras is not complexed with 14-3-3.
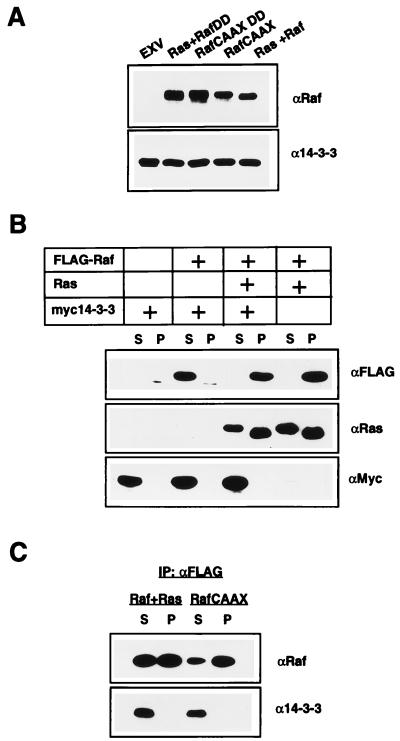
Several studies have shown that Raf-1 immunoprecipitated from whole-cell lysates is complexed with 14-3-3 proteins, but these studies have not rigorously addressed whether Raf-1 that has been recruited to the plasma membrane and activated by Ras remains bound to 14-3-3. If 14-3-3 does remain complexed with Raf, then the amount of 14-3-3 associated with the plasma membrane should increase in proportion to the amount of Raf-1 at the membrane. To test this hypothesis, the subcellular distribution of endogenous 14-3-3 in COS cells coexpressing oncogenic mutant RasG12V and epitope-tagged FLAG Raf was compared to that in COS cells transfected with empty vector. The immunoblots in Fig. 1A show that although the membrane (P100) fractions of COS cells coexpressing FLAG Raf and RasG12V contained 30-fold more Raf-1 than the mock-transfected cells, there was no corresponding increase in the amount of 14-3-3 associated with the membranes. Similarly, P100 fractions of COS cells expressing high levels of plasma membrane-targeted RafCAAX contained no more 14-3-3 than mock-transfected cells (Fig. 1A). Identical results were also obtained when a constitutively active form of Raf (RafDD) (Fig. 1A) or an inactive form of Raf (RafFF) (data not shown) was recruited to the plasma membrane.
FIG. 1.
Plasma membrane-recruited Raf-1 is not complexed with 14-3-3. (A) P100 fractions from COS cells transfected with empty vector (EXV) or an EXV expression plasmid for FLAG RafCAAX or FLAG RafCAAXDD, or cotransfected with EXV expression plasmids for FLAG Raf plus RasG12V or FLAG RafDD plus RasG12V, were normalized for protein content, resolved by SDS-PAGE, and Western blotted for Raf-1 and 14-3-3. (B) S100 and P100 fractions from COS cells transfected with combinations of EXV expression plasmids for FLAG Raf, RasG12V, and myc14-3-3β were normalized for protein content, resolved by SDS-PAGE, and Western blotted for FLAG, Ras, and myc. The Western blots in panels A and B were developed by enhanced chemiluminescence and quantitated by phosphorimaging (Bio-Rad). We conclude that less than 5% of total endogenous 14-3-3, or 2% of myc14-3-3, fractionates with cell membranes, irrespective of the amount of Raf-1 bound to the membranes. (C) S100 and solubilized P100 fractions from COS cells transfected with an EXV expression plasmid for FLAG RafCAAX or cotransfected with EXV expression plasmids for FLAG Raf plus RasG12V were normalized for protein content and immunoprecipitated by using anti-FLAG Sepharose. The anti-FLAG immunoprecipitates (IP) were resolved by SDS-PAGE and Western blotted for Raf and 14-3-3. The amount of EXV RasG12V used in this experiment was 25% of that used in the experiment for which results are shown in panel B, so that only half of the coexpressed FLAG Raf was recruited to the membrane. Only a small amount of RafCAAX can be immunoprecipitated from the cytosol, since most is localized to the plasma membrane.
Although 14-3-3 proteins are very abundant, it was possible that no 14-3-3 recruitment was seen because insufficient endogenous 14-3-3 was available to complex with the overexpressed FLAG Raf. To exclude this possibility, epitope-tagged myc14-3-3β was coexpressed with FLAG Raf and the fractionation experiments were repeated. The immunoblots in Fig. 1B show that myc14-3-3 is predominantly cytosolic both in the presence and in the absence of plasma membrane-localized FLAG Raf and that the amount of Raf-1 recruited to the plasma membrane by Ras is not altered when 14-3-3 is overexpressed. From quantitative Western blotting and phosphorimager analysis, we estimate that approximately 5% of endogenous 14-3-3 or 2% of myc14-3-3 is P100 associated, irrespective of how much Raf-1 is in the membrane. We conclude that cells expressing RasG12V and Raf-1 or RafCAAX do not have increased amounts of 14-3-3 stably associated with the plasma membrane, as would be expected if 14-3-3 remained complexed with Raf-1 after membrane recruitment.
Given these results, COS cells were then cotransfected with FLAG Raf and small amounts of RasG12V plasmid, so that the FLAG Raf expressed was approximately equally distributed between the cytosol and the membrane. FLAG Raf was then immunoprecipitated from the S100 and solubilized P100 fractions of these cells and immunoblotted for Raf-1 and 14-3-3. Figure 1C shows that immunoprecipitates of cytosolic Raf-1 contain 14-3-3 but that immunoprecipitates of membrane-recruited Raf-1 do not. Similar results were obtained with cells expressing RafCAAX: the small amount of RafCAAX expressed that remains cytosolic is complexed with 14-3-3, but RafCAAX immunoprecipitated from membranes is not (Fig. 1C).
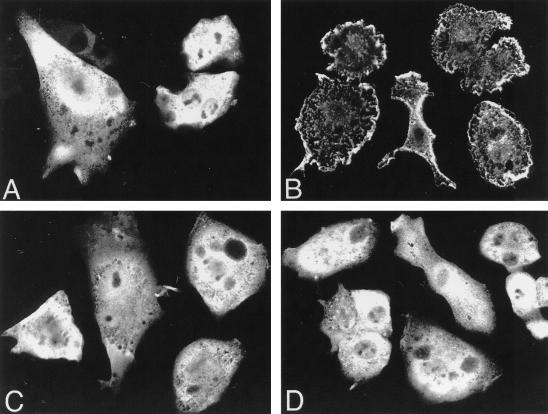
Finally, we used confocal microscopy to investigate whether any 14-3-3 could be visualized in the plasma membranes of BHK cells expressing RasG12V and Raf-1. Figure 2 shows that in the absence of RasG12V, FLAG Raf and myc14-3-3 colocalize to the cytosol (Fig. 2A and C). However, when RasG12V is also expressed, FLAG Raf localizes predominantly to the plasma membrane (Fig. 2B) (25, 44, 48) but myc14-3-3 remains cytosolic (Fig. 2D). Together these experiments demonstrate that 14-3-3 is completely displaced from Raf-1 when Raf-1 is recruited to the plasma membrane by RasG12V or when Raf-1 is targeted to the plasma membrane by using Ras localization motifs.
FIG. 2.
Activated Ras recruits Raf-1 but not 14-3-3 to the plasma membrane. BHK cells were seeded onto coverslips and cotransfected by using Lipofectamine with EXV expression plasmids for myc14-3-3 plus FLAG Raf (A and C) or for myc14-3-3 plus FLAG Raf plus RasG12V (B and D). Duplicate coverslips from each transfection were fixed after 24 h and incubated with anti-FLAG or anti-myc antisera followed by FITC-conjugated anti-mouse immunoglobulin G. The expressed proteins were visualized by indirect immunofluorescence with a confocal microscope. Transfection efficiency was 90%. FLAG Raf and myc14-3-3 both localize to the cytosol in the absence of RasG12V (A and C), but whereas coexpression of RasG12V results in recruitment of FLAG Raf to the plasma membrane (B), 14-3-3 remains in the cytosol (D). Additional control experiments (not shown, but described in Materials and Methods) were performed to visualize transfected Ras and to confirm that the staining patterns of myc14-3-3 and FLAG Raf were no different when they were coexpressed from those obtained when each protein was expressed alone.
EGF stimulation of COS cells is accompanied by transient Raf-1 and 14-3-3 recruitment but with different time courses.
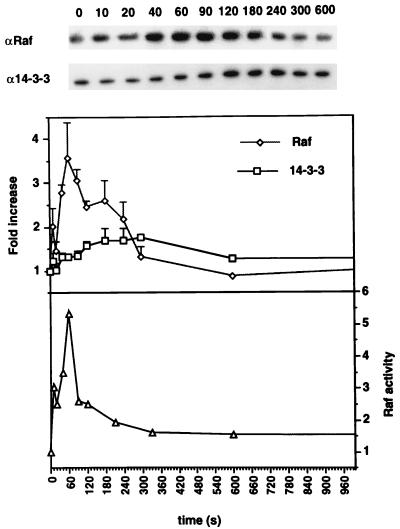
In the experiments described in the preceding section, constitutively activated Ras was used to recruit coexpressed FLAG Raf to the plasma membrane. We next examined to what extent transient activation of endogenous Ras results in recruitment of endogenous Raf-1 and whether under these conditions 14-3-3 can be observed associating with the plasma membrane. To this end, serum-starved COS cells were stimulated with EGF and the cells were harvested at 10- to 20-s, intervals during the first 5 min of EGF treatment, and then at 10 and 30 min. Cells were harvested on ice and fractionated into cytosol and membranes. Membrane fractions, normalized for protein content, were assayed for Raf-1 kinase activity and immunoblotted for Raf-1 and 14-3-3. These immunoblots were quantitated by phosphorimaging. Figure 3 shows immunoblots from a representative experiment and data pooled from multiple assays on membranes from three independent experiments.
FIG. 3.
EGF stimulates transient membrane recruitment of Raf-1 and 14-3-3. COS cells grown for 18 h in serum-free medium were treated for various times (in seconds) with 50 nM EGF. The cells were fractionated, and 10 μg of each P100 fraction was resolved by SDS-PAGE and immunoblotted for Raf-1 and 14-3-3. Immunoblots from a typical representative experiment are shown. There are some Raf-1 and 14-3-3 bound to the P100 fraction at 0 s. To allow pooling of data from independent experiments, the immunoblots were quantitated by phosphorimaging and the fold increases in P100-associated Raf-1 and 14-3-3 were calculated for each time point by using the value at 0 s as 1. Data on seven immunoblots from three independent experiments are presented in the upper graph. Raf-1 activity was assayed on the same membrane fractions in a coupled MEK-ERK assay and is shown in the lower graph.
Figure 3 shows that Raf-1 recruitment to the P100 fraction was evident at 10 s, the earliest time point assayed after EGF stimulation, and increased to a peak at 1 min. Thereafter, the amount of Raf-1 associated with the P100 fraction decreased, and it returned close to baseline by 5 min. Raf-1 activity associated with the plasma membrane followed a time course very similar to that of Raf-1 recruitment (Fig. 3). In contrast, recruitment of 14-3-3 to the P100 fraction followed a time course different from that of Raf-1; it increased slowly for 3 min, maintained a plateau for a further 2 min, and then fell to baseline approximately 10 min after the onset of EGF stimulation (Fig. 3). It is interesting that the maximum recruitment of 14-3-3 to the membrane fraction correlated most closely with Raf-1 dissociation.
Overexpression of 14-3-3 potentiates the activity of membrane-recruited Raf-1.
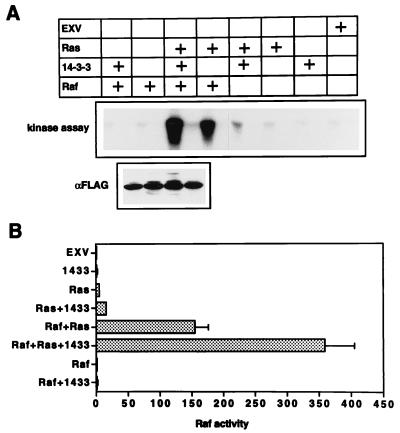
We next determined whether overexpression of 14-3-3 affects the activation of Raf-1 by Ras in vivo. COS cells coexpressing combinations of FLAG Raf, RasG12V, and myc14-3-3β were fractionated and immunoblotted as shown in Fig. 1B. The FLAG immunoblots were quantitated by phosphorimaging, and P100 fractions, normalized for FLAG Raf content, were assayed for Raf-1 activity by using a coupled MEK-ERK assay. Figure 4 shows that the specific activity of FLAG Raf recruited to the plasma membrane by Ras increased almost threefold when 14-3-3 was coexpressed. A similar effect on the activation of endogenous Raf-1 was also seen: endogenous Raf-1 activity in the control membranes expressing oncogenic Ras alone, although low, increased threefold when 14-3-3 was coexpressed (Fig. 4). In contrast, coexpression of 14-3-3 had no effect on the kinase activity of FLAG Raf immunoprecipitated from the cytosol of COS cells coexpressing RasG12V and FLAG Raf (data not shown).
FIG. 4.
Overexpression of 14-3-3 potentiates the activity of membrane-recruited Raf. (A) Membrane (P100) fractions from COS cells expressing combinations of FLAG Raf, RasG12V, and myc14-3-3 were normalized for FLAG Raf content by quantitative Western blotting (as illustrated in Fig. 1). No FLAG Raf was present in four of the control assays, so a mass of membranes equivalent to that used in the FLAG Raf plus Ras plus 14-3-3 assay was assayed. The Raf-1 kinase activity was measured in a coupled MEK-ERK assay. A representative kinase assay is shown. Each Raf-1 assay was performed both with MEK plus ERK and with ERK alone (to estimate background counts; for details, see Materials and Methods): aliquots of the MBP kinase assay from the MEK-ERK and the control incubation, respectively, are shown alongside each other in the kinase panel. The FLAG immunoblot shown is that of the kinase reaction mixtures containing FLAG Raf; it verifies the Raf-1 assay normalization. (B) Although only a single kinase assay is shown in panel A, each P100 fraction was assayed in triplicate, and each respective MBP kinase assay was performed in duplicate (see Materials and Methods). The mean (n = 6) Raf-1 activities shown have had background counts subtracted. The units are picomoles of phosphate transferred to MBP per 10 min. Results are representative of those obtained in three independent experiments.
These results indicate that the concentration of 14-3-3 in cytosol, although high, is limiting for Ras-dependent Raf-1 activation when Raf-1 or Ras is overexpressed. However, as is evident from Fig. 1, this potentiation of Raf-1 activation is not accompanied by any increase in the amount of 14-3-3 associated with the membrane.
Efficient in vitro activation of Raf-1 by Ras membranes requires 14-3-3.
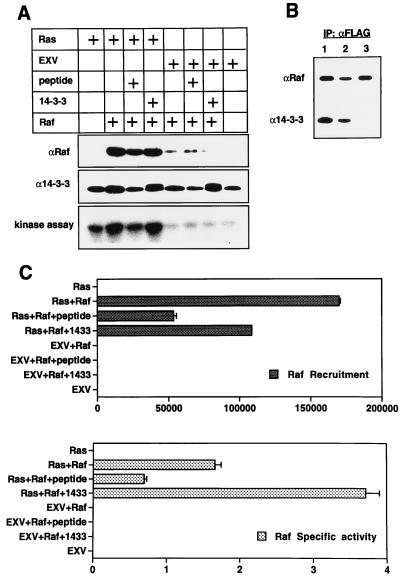
We have recently developed an assay that reproduces in vitro the Ras-dependent membrane recruitment and activation of Raf-1 that are evident in vivo. Membranes prepared from COS cells expressing RasG12V are incubated with cytosol from COS cells expressing FLAG Raf, and the membranes are reisolated by centrifugation. The amount of FLAG Raf captured is measured by quantitative Western blotting, and Raf-1 activity associated with the membranes is measured in a coupled MEK-ERK assay. A notable difference between this assay and a similar one reported recently (45) is that magnesium is present in all of our incubation buffers; we find that these assay conditions yield better Raf-1 membrane recruitment and Raf-1 activation than the magnesium-free assay conditions described elsewhere (45). Figures 5A and C show that, under the conditions of this assay, 20-fold more Raf-1 binds to membranes containing RasG12V than to control membranes containing no Ras. Moreover, the Raf-1 bound to Ras membranes undergoes activation, whereas Raf-1 bound to control membranes does not (Fig. 5A and C).
FIG. 5.
In vitro activation of Raf-1 by Ras requires 14-3-3. (A) Raf-1 cytosol prepared from COS cells expressing FLAG Raf was mixed on ice with Ras membranes prepared from COS cells expressing RasG12V or with control membranes from COS cells transfected with empty EXV plasmid. To control for endogenous Raf-1 already associated with the EXV and Ras membranes, incubations were also set up with buffer A in place of the Raf-1 cytosol. Either 6 μg of recombinant 14-3-3 (1 mg/ml in buffer A), 6 μl of peptide pS-Raf 621 (500 μM in buffer A), or 6 μl of buffer A was added, the samples were agitated for 10 min at 25°C, and membranes were reisolated by centrifugation. The immunoblots show the amounts of Raf-1 (detected by anti-FLAG) and 14-3-3 bound to the P100 fractions after the 10-min incubation. The Raf-1 activity associated with the membranes was measured in a coupled MEK-ERK assay as described in Materials and Methods. (B) FLAG Raf was immunoprecipitated with anti-FLAG Sepharose from Raf-1 cytosol containing no peptide (lane 1), 40 μM Raf-621 (nonphosphorylated Raf peptide) (lane 2), or 40 μM pS-Raf-621 (phosphorylated Raf peptide) (lane 3) and was immunoblotted for Raf-1 and 14-3-3. The 40 μM concentration was selected after a set of preliminary titration experiments using peptide concentrations in the range of 1 to 100 μM (33a). IP, immunoprecipitate. (C) The Raf-1 blots and the coupled MEK-ERK assay for membrane-associated Raf-1 activity in panel A were quantitated by phosphorimaging. Ras-dependent Raf-1 binding was calculated from the phosphorimager data by subtracting the value obtained for Raf-1 bound to control EXV membranes (treated as background). For the calculation of Raf-1 specific activities, the Raf-1 activity associated with EXV or Ras membranes after incubation in buffer A was subtracted from the Raf-1 activity associated with the respective membranes after incubation with Raf-1 cytosol to obtain the net membrane-dependent Raf-1 activation. The Raf-1 specific activity was then calculated by dividing the net Raf-1 activity by the amount of Raf-1 recruited. The total amount of 14-3-3 associated with the P100 fractions was also estimated by phosphorimaging. The amount of 14-3-3 bound to membranes was 50% greater for the Raf-plus-Ras incubation, and 30% lower for the incubation with peptide, than the amount present in Ras membranes incubated with buffer A. There was no overall increase in the amount of membrane-bound 14-3-3 when recombinant 14-3-3 was included in the incubation, although endogenous 14-3-3 was partially replaced by recombinant 14-3-3 (which has a slightly slower mobility). Similar changes were also seen in the amount of 14-3-3 bound to the control membranes.
We used this assay to determine whether 14-3-3 potentiates Ras-dependent Raf-1 activation in vitro. Raf-1 cytosol was supplemented with recombinant 14-3-3 and incubated with Ras membranes. Interestingly, the recombinant 14-3-3 had two effects: it reduced the amount of Raf-1 that bound to the Ras membranes but increased the specific activity of membrane-bound Raf-1 (Fig. 5A and C). Additional 14-3-3 had no effect on the binding of Raf-1 to control membranes and did not result in any activation of Raf-1 by these membranes (Fig. 5). Recombinant 14-3-3 also had no measurable effect on cytosolic Raf-1 activity either in the presence or in the absence of Ras membranes (data not shown).
We next investigated whether Raf-1 needs to be complexed with 14-3-3 in order to be activated by Ras. To remove 14-3-3 from Raf-1, peptides that compete with 14-3-3 for binding to Raf-1 were used. The 15-amino-acid Raf-1 phosphopeptide pS-Raf-621 was synthesized together with the control nonphosphorylated Raf-1 peptide Raf-621 (39). To confirm that the phosphorylated peptide would displace 14-3-3 from Raf-1 in whole cytosol, Raf-1 was immunoprecipitated from cytosol containing either 40 μM pS-Raf-621 or 40 μM Raf-621 (the nonphosphorylated control peptide) and was immunoblotted for 14-3-3. Figure 5B shows that pS-Raf-621 totally removed 14-3-3 from Raf, whereas the nonphosphorylated control peptide Raf-621 had no detectable effect on the interaction of 14-3-3 with Raf. To determine whether Raf-1 devoid of 14-3-3 can still be activated by Ras, Raf-1 cytosol was incubated with Ras membranes in the presence of peptide pS-Raf-621. Two distinct effects were seen: first, the recruitment of Raf-1 by Ras membranes was inhibited by 70%, and secondly, the specific activity of the membrane-recruited Raf-1 was decreased by 60% (Fig. 5A and C). The combination of these two effects significantly reduces Ras-dependent Raf-1 activation (Fig. 5A). No further inhibition of Raf-1 activation was seen when the concentration of pS-Raf-621 was increased to 100 μM (data not shown). When Raf-1 cytosol was incubated with Ras membranes in the presence of the nonphosphorylated control peptide, Raf-621, no inhibitory effect on the recruitment or activation of Raf-1 was seen (data not shown). These results demonstrate the probable mechanism for the inhibition of insulin-induced Xenopus oocyte maturation by the pS-Raf-621 peptide (39).
The Raf-1 Western blot was stripped and reprobed for 14-3-3 (Fig. 5A). The recombinant 14-3-3 used in this experiment has a short N-terminal linker sequence that results in a 1-kDa increase in molecular size; this is sufficient to allow discrimination from endogenous 14-3-3. Figure 5A shows that the amount of 14-3-3 associated with Ras membranes was only minimally increased when these membranes were incubated with Raf-1 cytosol, in contrast with the 20-fold increase in the amount of Raf-1 bound. Conversely, the presence of peptide pS-Raf-621 slightly decreased the amount of 14-3-3 associated with the Ras membranes. Interestingly, when membranes were incubated with Raf-1 cytosol supplemented with recombinant 14-3-3, the total amount of 14-3-3 bound did not change, but approximately 60% of the endogenous 14-3-3 present initially was replaced with recombinant 14-3-3 after the 10-min incubation (Fig. 5A).
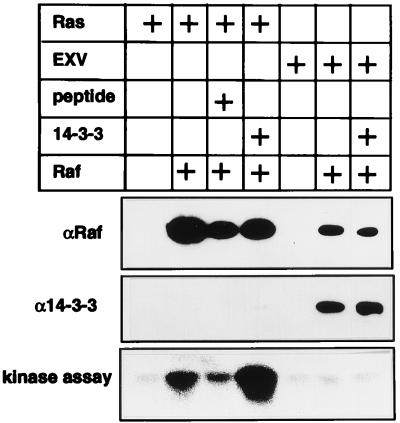
Finally, Raf-1 cytosol was incubated in vitro with Ras or control membranes, but the membranes recovered by centrifugation were solubilized in NP-40 and incubated with anti-FLAG Sepharose. Since so little FLAG Raf normally binds to the control membranes (see Fig. 5), fivefold more control (EXV) membranes and Raf-1 cytosol were used in the control incubations. The anti-FLAG immunoprecipitates were assayed for Raf-1 kinase activity and then immunoblotted for Raf-1 and 14-3-3. Figure 6 shows that Raf-1 captured by the Ras membranes is activated but not complexed with 14-3-3, whereas Raf-1 that bound nonspecifically to the control membranes is not activated and remains complexed with 14-3-3. Also, Figure 6 again shows that peptide pS-Raf-621 inhibits Raf-1 recruitment and activation by Ras membranes, whereas recombinant 14-3-3 potentiates Raf-1 activation but reduces the amount of Raf-1 associated with the Ras membranes.
FIG. 6.
Raf-1 activated by Ras in vitro is not complexed with 14-3-3. Raf-1 cytosol prepared from COS cells expressing FLAG Raf was mixed on ice with Ras membranes prepared from COS cells expressing RasG12V or with control membranes from COS cells transfected with empty EXV plasmid. EXV and Ras membranes were also incubated with buffer A in place of the Raf-1 cytosol. Given that only small amounts of Raf-1 associate with the EXV membranes (see Fig. 5), fivefold more control membranes and Raf-1 cytosol were used in the EXV incubations than in the Ras membrane incubations. Either 6 μg of recombinant 14-3-3 (1 mg/ml in buffer A), 6 μl of peptide pS-Raf 621 (500 μM in buffer A), or 6 μl of buffer A was added, the samples were agitated for 10 min at 25°C, and membranes were reisolated by centrifugation and solubilized by sonication in 1% NP-40. The FLAG Raf bound to the membranes was immunopurified with anti-FLAG Sepharose and assayed for kinase activity in a coupled MEK-ERK assay. After the kinase assay, the FLAG immunoprecipitates were immunoblotted for Raf-1 (by using anti-Raf-1 antisera) and 14-3-3. Note that Raf-1 recruited to the membranes by Ras is activated and is devoid of 14-3-3, whereas Raf-1 that has bound nonspecifically to the EXV membranes is not activated and remains complexed with 14-3-3. The Raf-1 activity associated with Ras membranes in this assay is much lower than that seen in Fig. 5 because endogenous Raf-1 is not captured by the anti-FLAG Sepharose. Quantification of the Raf-1 immunoblots and the Raf-1 kinase assay by phosphorimaging showed that the amount of FLAG Raf-1 associated with the Ras membranes decreased by 60% in the presence of peptide pS-621 and by 40% in the presence of recombinant 14-3-3. There was a fivefold stimulation of Raf-1 specific activity by recombinant 14-3-3 and a 60% inhibition of Raf-1 specific activity by peptide pS-Raf621. These figures are comparable with those derived in Fig. 5C.
DISCUSSION
The experiments reported here show directly that the specific activity of Raf-1 recruited to the plasma membrane by activated Ras in mammalian cells is significantly potentiated when 14-3-3 is coexpressed. This is consistent with similar results with Xenopus oocytes and Saccharomyces cerevisiae (11, 22). Our results also confirm earlier studies, which did not measure Raf-1 kinase activity directly but showed that Raf-1 mediated activation of AP-1 and NF-κB in NIH3T3 cells, or PC12 cell differentiation driven by an isolated Raf-1 kinase domain, is potentiated by coexpression of 14-3-3 (26). A critical issue is how this potentiation of Raf-1 activity by 14-3-3 is effected: are cellular 14-3-3 levels limiting for the Ras–Raf-1 interactions, or are they limiting for some other signaling pathway feeding into Raf-1 activation?
We and others (26, 29, 46) have shown that 14-3-3 does not directly activate Raf-1, because Raf-1 immunoprecipitated from the cytosol of cells overexpressing 14-3-3 is not activated. Moreover, we have shown here that 14-3-3 is completely displaced from Raf-1 when Raf-1 is recruited to the plasma membrane, because (i) there is no corresponding increase in membrane bound 14-3-3 when Raf-1 membrane levels increase 20- to 40-fold, (ii) 14-3-3 coimmunoprecipitates with cytosolic but not with membrane-recruited Raf-1, and (iii) no 14-3-3 can be visualized by confocal microscopy in the plasma membranes of cells coexpressing Raf-1 and activated Ras. This displacement of 14-3-3 from membrane-bound Raf-1 occurs in COS and BHK cells expressing activated Ras and Raf-1, in cells transiently activated by EGF, and also in vitro, when Raf-1 is recruited and activated by Ras membranes. Our data are consistent with previous results for NIH3T3 cells which showed that all active Raf-1 is localized to the plasma membrane (23) and that 14-3-3 is complexed only with inactive Raf-1 (26), therefore implying that plasma membrane-localized Raf-1 is not complexed with 14-3-3.
Various data strongly support a role for 14-3-3 as a positive regulator of Ras and Raf-1 signaling. For example, dominant-negative mutations in 14-3-3ɛ suppress the Drosophila rough-eye phenotype caused by activated Ras or membrane-targeted D-Raf, and a 50% reduction in 14-3-3ɛ levels enhances the lethality of a weak loss-of-function allele of D-Raf (5). Together these data imply that 14-3-3ɛ facilitates some aspect of Ras and Raf signaling. Kockel et al. (1997) describe a lethal 14-3-3ζ mutation that is rescued by activated membrane-localized Raf-1 but not by activated Ras, demonstrating a critical requirement for 14-3-3 in Raf-1 activation (24).
A body of data is also accumulating which demonstrates that 14-3-3 negatively regulates Raf-1 and that displacement of 14-3-3 from the Raf-1 N terminus is accompanied by activation. For example, S259A and S259D mutations in the N-terminal 14-3-3 binding domain abrogate the interaction of 14-3-3 with the isolated Raf-1 N terminus but do not affect 14-3-3 interactions with the Raf-1 C terminus (42). These mutations increase Raf-1 basal kinase activity five- to sixfold (34, 42), although mutant Raf S259A remains sensitive to further activation by Ras (42). In vivo, Raf S259A has an activated phenotype in Xenopus oocytes (34), and the equivalent mutation in D-Raf is activating for R7 cell formation in Drosophila eye development (1, 42). It has been postulated that the S259A mutation mimics one effect of Ras, in that binding of activated Ras also displaces 14-3-3 from the isolated Raf-1 N terminus in vitro (41). The isolated Raf-CRD also interacts with 14-3-3 (6), although the interaction may be cryptic in larger Raf-1 fragments because the S259A mutation is sufficient to completely abrogate 14-3-3 binding to the whole Raf-1 N terminus (comprising residues 1 to 330). Nevertheless, mutations that decrease 14-3-3 binding to the Raf-CRD weakly activate Raf-1 transforming activity and compensate for loss-of-function mutations in the Ras switch 2 domain that interacts with the Raf-CRD. These results support a role for Ras in displacing 14-3-3 from the CRD (6).
However, 14-3-3 displacement from Raf-1 cannot be wholly explained by an interaction with Ras because this only affects the interaction with the N terminus and Raf-1 recruited to the plasma membrane is no longer complexed with 14-3-3. Moreover, membrane-targeted RafCAAX, which is activated independently of Ras, is also devoid of 14-3-3. Thus, the interaction of Raf-1 with the plasma membrane must also be important in displacing 14-3-3. This brings into focus a possible role for phospholipid interactions with the CRD in displacing 14-3-3 from the Raf-1 N terminus, which in turn may explain how RafCAAX is activated. How 14-3-3 is displaced from the Raf-1 C terminus remains unclear: it occurs at the cell membrane and cannot simply be a consequence of activation, because Raf S259A and RafDD, which are both constitutively active in the cytosol, remain complexed with 14-3-3 (33b, 42).
The results from our in vitro experiments throw some additional light on the positive and negative roles for 14-3-3 in Raf-1 activation. Recombinant 14-3-3 potentiates Raf-1 activation by Ras membranes, as in vivo, and Raf-1 devoid of 14-3-3 is poorly recruited by Ras membranes and fails to undergo significant activation at the membrane. An attractive interpretation of these results is that 14-3-3 plays a critical permissive role in maintaining Raf-1 in a conformation that is optimal for plasma membrane recruitment and subsequent activation by Ras.
It is also interesting that maximal recruitment of 14-3-3 to the membrane fraction following EGF stimulation coincides with Raf-1 dissociation. While these events may be completely independent, they raise the possibility that 14-3-3 may also have a role in removing Raf-1 from the plasma membrane, perhaps analogous to the removal of phosphorylated BAD from the mitochondrial membrane by 14-3-3 (55). In this context, it is worth noting that phosphorylation of S621 and S624 is responsible for the mobility shift which occurs after Raf-1 activation (13) and that mobility-shifted Raf-1 binds poorly to membranes (53). Phosphorylation of S621 also downregulates Raf-1 kinase activity (36), and S621 has been identified as a 14-3-3 binding site (39). Taking all these results together with our data, it is tempting to speculate that phosphorylation of S621 simultaneously downregulates Raf-1 kinase activity and creates a 14-3-3 binding site at the C terminus, which allows 14-3-3 to rebind and extract Raf-1 from the membrane. This interpretation can also rationalize the opposing effects of recombinant 14-3-3 seen in vitro, namely, an increase in the specific activity of membrane-recruited Raf-1 but a decrease in the amount of Raf-1 associated with the Ras membranes. The latter effect may reflect increased removal of Raf-1 from the membrane rather than an inhibition of Raf-1 recruitment, with the net effect of reducing the total amount of Raf-1 at the membrane. Increased turnover of Raf-1 at the membrane may also be the mechanism by which 14-3-3 potentiates Raf-1 activity: inactive Raf-1 is removed, allowing Ras to recruit and reactivate Raf-1 at a greater rate. Consistent with this interpretation, turnover of the 14-3-3 membrane pool was clearly evident in vitro. Implicit in this model of a dual role for 14-3-3 as permissive for recruitment and activation, yet involved in the membrane extraction of Raf, is the concept that dephosphorylation of S621 on initial membrane recruitment may contribute both to Raf-1 activation and to loss of 14-3-3 from the Raf-1 C terminus.
Previous studies have shown that mutation of S621 generates a Raf-1 protein that cannot be activated and that functions as a dominant-negative molecule (9, 38). Since mutation of S621 results in a kinase-defective Raf-1 (38), one explanation of this phenotype is that the kinase-inactive mutant competitively inhibits access to Ras by wild-type Raf-1. However, there are other interesting possibilities. To explain our results, we propose recruitment of Raf-1 and 14-3-3 by Ras, displacement of 14-3-3 at the plasma membrane, activation of Raf-1, rebinding of 14-3-3, and recycling of Raf-1 back to the cytosol. Since this process is dynamic, mutants that are arrested at any stage of the cycle could potentially tie up the activation and recycling machinery and block activation of wild-type Raf-1. For example, Raf S621A may not be folded properly around 14-3-3, so that displacement of 14-3-3 does not occur upon membrane recruitment. This may leave Raf S621A in a dead-end complex with Ras at the membrane. Alternatively, the release machinery may get backed up if 14-3-3 cannot bind to the C terminus of membrane-recruited Raf S621A. Future studies with S621A (and S259A) Raf-1 mutants will address these speculations.
In summary, the data presented here, together with the studies outlined in the introduction, lead to the following model for Raf-1 activation at the plasma membrane. The Ras-RBD interaction brings the Raf–14-3-3 complex to the membrane and sets in train subsequent activation events: CRD-Ras interactions then act in concert with CRD-phosphatidylserine interactions and lead to (i) partial uncovering and activation of the kinase domain, (ii) displacement of 14-3-3 from the N terminus, and (iii) more favorable presentation of Y340 and Y341 for phosphorylation. At some early point in the activation process, 14-3-3 is also displaced from the Raf-1 C terminus; displacement of 14-3-3 allows for dephosphorylation of S259 and S621. Successful completion of all these events is required for full Raf-1 activation. A continuing interaction between activated Raf-1 and 14-3-3 is not required to maintain the activity of Raf-1 at the plasma membrane, because such an interaction cannot be demonstrated in vivo. Following rephosphorylation of S621 and/or S259, 14-3-3 rebinds to inactive Raf-1 and sequesters it to the cytosol. This model explains why 14-3-3 functions as a negative regulator in some assays (because it must be displaced from Raf-1 for activation and may be involved in removing Raf-1 from the plasma membrane) but appears to be essential for Ras-to-Raf-1 signaling in genetic assays (because it is permissive for Ras-dependent membrane recruitment and activation).
ACKNOWLEDGMENTS
We thank Ellen Freed and Frank McCormick of Onyx Pharmaceuticals for the GST-14-3-3β and Colin McQueen of the Department of Vision, Touch and Hearing, University of Queensland, for help with the confocal imaging.
This work was supported by a grant to J.F.H. from the National Health and Medical Research Council of Australia. J.F.H. is also supported by the Royal Children’s Hospital Foundation (Queensland).
REFERENCES
- 1.Baek K H, Fabian J R, Sprenger F, Morrison D K, Ambrosio L. The activity of D-raf in torso signal transduction is altered by serine substitution, N-terminal deletion, and membrane targeting. Dev Biol. 1996;175:191–204. doi: 10.1006/dbio.1996.0107. [DOI] [PubMed] [Google Scholar]
- 2.Brtva T R, Drugan J K, Ghosh S, Terrell R S, Campbell Burk S, Bell R M, Der C J. Two distinct Raf domains mediate interaction with Ras. J Biol Chem. 1995;270:9809–9812. doi: 10.1074/jbc.270.17.9809. [DOI] [PubMed] [Google Scholar]
- 3.Canagarajah B J, Khokhlatchev A, Cobb M H, Goldsmith E J. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell. 1997;90:859–869. doi: 10.1016/s0092-8674(00)80351-7. [DOI] [PubMed] [Google Scholar]
- 4.Carroll M P, May W S. Protein kinase C-mediated serine phosphorylation directly activates Raf-1 in murine hematopoietic cells. J Biol Chem. 1994;269:1249–1256. [PubMed] [Google Scholar]
- 5.Chang H C, Rubin G M. 14-3-3 epsilon positively regulates Ras-mediated signaling in Drosophila. Genes Dev. 1997;11:1132–1139. doi: 10.1101/gad.11.9.1132. [DOI] [PubMed] [Google Scholar]
- 6.Clark G J, Drugan J K, Rossman K L, Carpenter J W, Rogers-Graham K, Fu H, Der C J, Campbell S L. 14-3-3ζ negatively regulates Raf-1 activity by interactions with the Raf-1 cysteine-rich domain. J Biol Chem. 1997;272:20990–20993. doi: 10.1074/jbc.272.34.20990. [DOI] [PubMed] [Google Scholar]
- 7.Drugan J K, Khosravi Far R, White M A, Der C J, Sung Y J, Hwang Y W, Campbell S L. Ras interaction with two distinct binding domains in Raf-1 may be required for Ras transformation. J Biol Chem. 1996;271:233–237. doi: 10.1074/jbc.271.1.233. [DOI] [PubMed] [Google Scholar]
- 8.Fabian J R, Daar I O, Morrison D K. Critical tyrosine residues regulate the enzymatic and biological activity of Raf-1 kinase. Mol Cell Biol. 1993;13:7170–7179. doi: 10.1128/mcb.13.11.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabian J R, Morrison D K, Daar I O. Requirement for Raf and MAP kinase function during the meiotic maturation of Xenopus oocytes. J Cell Biol. 1993;122:645–652. doi: 10.1083/jcb.122.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fabian J R, Vojtek A B, Cooper J A, Morrison D K. A single amino acid change in Raf-1 inhibits Ras binding and alters Raf-1 function. Proc Natl Acad Sci USA. 1994;91:5982–5986. doi: 10.1073/pnas.91.13.5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fantl W J, Muslin A J, Kikuchi A, Martin J A, MacNicol A M, Gross R W, Williams L T. Activation of Raf-1 by 14-3-3 proteins. Nature. 1994;371:612–614. doi: 10.1038/371612a0. [DOI] [PubMed] [Google Scholar]
- 12.Farrar M A, Alberol I, Perlmutter R M. Activation of the Raf-1 kinase cascade by coumermycin-induced dimerization. Nature. 1996;383:178–181. doi: 10.1038/383178a0. [DOI] [PubMed] [Google Scholar]
- 13.Ferrier A F, Lee M, Anderson W B, Benvenuto G, Morrison D K, Lowy D R, DeClue J E. Sequential modification of serines 621 and 624 in the Raf-1 carboxyl terminus produces alterations in its electrophoretic mobility. J Biol Chem. 1997;272:2136–2142. doi: 10.1074/jbc.272.4.2136. [DOI] [PubMed] [Google Scholar]
- 14.Freed E, Symons M, Macdonald S G, McCormick F, Ruggieri R. Binding of 14-3-3 proteins to the protein kinase Raf and effects on its activation. Science. 1994;265:1713–1716. doi: 10.1126/science.8085158. [DOI] [PubMed] [Google Scholar]
- 15.Fu H, Xia K, Pallas D C, Cui C, Conroy K, Narsimhan R P, Mamon H, Collier R J, Roberts T M. Interaction of the protein kinase Raf-1 with 14-3-3 proteins. Science. 1994;266:126–129. doi: 10.1126/science.7939632. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh S, Bell R M. Identification of discrete segments of human Raf-1 kinase critical for high affinity binding to Ha-Ras. J Biol Chem. 1994;269:30785–30788. [PubMed] [Google Scholar]
- 17.Ghosh S, Strum J C, Sciorra V A, Daniel L, Bell R M. Raf-1 kinase possesses distinct binding domains for phosphatidylserine and phosphatidic acid. Phosphatidic acid regulates the translocation of Raf-1 in 12-O-tetradecanoylphorbol-13-acetate-stimulated Madin-Darby canine kidney cells. J Biol Chem. 1996;271:8472–8480. doi: 10.1074/jbc.271.14.8472. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh S, Xie W Q, Quest A F, Mabrouk G M, Strum J C, Bell R M. The cysteine-rich region of Raf-1 kinase contains zinc, translocates to liposomes, and is adjacent to a segment that binds GTP-ras. J Biol Chem. 1994;269:10000–10007. [PubMed] [Google Scholar]
- 19.Hancock J F, Hall A. A novel role for RhoGDI as inhibitor of GAP proteins. EMBO J. 1993;12:1915–1921. doi: 10.1002/j.1460-2075.1993.tb05840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu C D, Kariya K, Tamada M, Akasaka K, Shirouzu M, Yokoyama S, Kataoka T. Cysteine-rich region of Raf-1 interacts with activator domain of post-translationally modified Ha-Ras. J Biol Chem. 1995;270:30274–30277. doi: 10.1074/jbc.270.51.30274. [DOI] [PubMed] [Google Scholar]
- 21.Huang D C S, Marshall C J, Hancock J F. Plasma membrane-targeted ras GTPase-activating protein is a potent suppressor of p21ras function. Mol Cell Biol. 1993;13:2420–2431. doi: 10.1128/mcb.13.4.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irie K, Gotoh Y, Yashar B M, Errede B, Nishida E, Matsumoto K. Stimulatory effects of yeast and mammalian 14-3-3 proteins on the Raf protein kinase. Science. 1994;265:1716–1719. doi: 10.1126/science.8085159. [DOI] [PubMed] [Google Scholar]
- 23.Jelinek T, Dent P, Sturgill T W, Weber M J. Ras-induced activation of Raf-1 is dependent on tyrosine phosphorylation. Mol Cell Biol. 1996;16:1027–1034. doi: 10.1128/mcb.16.3.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kockel L, Vorbruggen G, Jackle H, Mlodzik M, Bohmann D. Requirement for Drosophila 14-3-3 zeta in Raf-dependent photoreceptor development. Genes Dev. 1997;11:1140–1147. doi: 10.1101/gad.11.9.1140. [DOI] [PubMed] [Google Scholar]
- 25.Leevers S J, Paterson H F, Marshall C J. Requirement for Ras in Raf activation is overcome by targeting Raf to the plasma membrane. Nature. 1994;369:411–414. doi: 10.1038/369411a0. [DOI] [PubMed] [Google Scholar]
- 26.Li S, Janosch P, Tanji M, Rosenfeld G C, Waymire J C, Mischak H, Kolch W, Sedivy J M. Regulation of Raf-1 kinase activity by the 14-3-3 family of proteins. EMBO J. 1995;14:685–696. doi: 10.1002/j.1460-2075.1995.tb07047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo Z, Diaz B, Marshall M, Avruch J. An intact zinc finger is required for optimal binding to processed Ras and for Ras-dependent Raf activation in situ. Mol Cell Biol. 1997;17:46–53. doi: 10.1128/mcb.17.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo Z, Tzivion G, Belshaw P J, Vavvas D, Marshall M, Avruch J. Oligomerization activates c-Raf-1 through a Ras-dependent mechanism. Nature. 1996;383:181–185. doi: 10.1038/383181a0. [DOI] [PubMed] [Google Scholar]
- 29.Luo Z J, Zhang X F, Rapp U, Avruch J. Identification of the 14.3.3 zeta domains important for self-association and Raf binding. J Biol Chem. 1995;270:23681–23687. doi: 10.1074/jbc.270.40.23681. [DOI] [PubMed] [Google Scholar]
- 30.Marais R, Light Y, Paterson H F, Marshall C J. Ras recruits Raf-1 to the plasma membrane for activation by tyrosine phosphorylation. EMBO J. 1995;14:3136–3145. doi: 10.1002/j.1460-2075.1995.tb07316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marais R, Light Y, Paterson H F, Mason C S, Marshall C J. Differential regulation of Raf-1, A-Raf, and B-Raf by oncogenic ras and tyrosine kinases. J Biol Chem. 1997;272:4378–4383. doi: 10.1074/jbc.272.7.4378. [DOI] [PubMed] [Google Scholar]
- 32.Marais R, Marshall C J. Control of the ERK MAP kinase cascade by Ras and Raf. Cancer Surv. 1996;27:101–125. [PubMed] [Google Scholar]
- 33.Marshall C J. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 33a.McPherson, R. A. Unpublished data.
- 33b.McPherson, R. A., and J. F. Hancock. Unpublished data.
- 34.Michaud N R, Fabian J R, Mathes K D, Morrison D K. 14-3-3 is not essential for Raf-1 function: identification of Raf-1 proteins that are biologically activated in a 14-3-3- and Ras-independent manner. Mol Cell Biol. 1995;15:3390–3397. doi: 10.1128/mcb.15.6.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mineo C, Anderson R G, White M A. Physical association with ras enhances activation of membrane-bound raf (RafCAAX) J Biol Chem. 1997;272:10345–10348. doi: 10.1074/jbc.272.16.10345. [DOI] [PubMed] [Google Scholar]
- 36.Mischak H, Seitz T, Janosch P, Eulitz M, Steen H, Schellerer M, Philipp A, Kolch W. Negative regulation of Raf-1 by phosphorylation of serine 621. Mol Cell Biol. 1996;16:5409–5418. doi: 10.1128/mcb.16.10.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morrison D K, Cutler R E. The complexity of Raf-1 regulation. Curr Opin Cell Biol. 1997;9:174–179. doi: 10.1016/s0955-0674(97)80060-9. [DOI] [PubMed] [Google Scholar]
- 38.Morrison D K, Heidecker G, Rapp U R, Copeland T D. Identification of the major phosphorylation sites of the Raf-1 kinase. J Biol Chem. 1993;268:17309–17316. [PubMed] [Google Scholar]
- 39.Muslin A J, Tanner J W, Allen P M, Shaw A S. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 40.Radziwill G, Steinhusen U, Aitken A, Moelling K. Inhibition of Raf/MAPK signaling in Xenopus oocyte extracts by Raf-1-specific peptides. Biochem Biophys Res Commun. 1996;227:20–26. doi: 10.1006/bbrc.1996.1461. [DOI] [PubMed] [Google Scholar]
- 41.Rommel C, Radziwill G, Lovric J, Noeldeke J, Heinicke T, Jones D, Aitken A, Moelling K. Activated Ras displaces 14-3-3 protein from the amino terminus of c-Raf-1. Oncogene. 1996;12:609–619. [PubMed] [Google Scholar]
- 42.Rommel C, Radziwill G, Moelling K, Hafen E. Negative regulation of Raf activity by binding of 14-3-3 to the amino terminus of Raf in vivo. Mech Dev. 1997;64:95–104. doi: 10.1016/s0925-4773(97)00052-x. [DOI] [PubMed] [Google Scholar]
- 43.Roy S, Lane A, Yan J, McPherson R, Hancock J F. Activity of plasma membrane recruited Raf-1 is regulated by Ras via the Raf zinc finger. J Biol Chem. 1997;272:20139–20145. doi: 10.1074/jbc.272.32.20139. [DOI] [PubMed] [Google Scholar]
- 44.Stokoe D, Macdonald S G, Cadwallader K, Symons M, Hancock J F. Activation of Raf as a result of recruitment to the plasma membrane. Science. 1994;264:1463–1467. doi: 10.1126/science.7811320. [DOI] [PubMed] [Google Scholar]
- 45.Stokoe D, McCormick F. Activation of c-Raf-1 by Ras and Src through different mechanisms: activation in vivo and in vitro. EMBO J. 1997;16:2384–2396. doi: 10.1093/emboj/16.9.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suen K L, Bustelo X R, Barbacid M. Lack of evidence for the activation of the Ras/Raf mitogenic pathway by 14-3-3 proteins in mammalian cells. Oncogene. 1995;11:825–831. [PubMed] [Google Scholar]
- 47.Therrien M, Michaud N R, Rubin G M, Morrison D K. KSR modulates signal propagation within the MAPK cascade. Genes Dev. 1996;10:2684–2695. doi: 10.1101/gad.10.21.2684. [DOI] [PubMed] [Google Scholar]
- 48.Traverse S, Cohen P, Paterson H, Marshall C, Rapp U, Grand R J. Specific association of activated MAP kinase kinase kinase (Raf) with the plasma membranes of ras-transformed retinal cells. Oncogene. 1993;8:3175–3181. [PubMed] [Google Scholar]
- 49.Ueda Y, Hirai S, Osada S, Suzuki A, Mizuno K, Ohno S. Protein kinase C activates the MEK-ERK pathway in a manner independent of Ras and dependent on Raf. J Biol Chem. 1996;271:23512–23519. doi: 10.1074/jbc.271.38.23512. [DOI] [PubMed] [Google Scholar]
- 50.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 51.Warne P H, Viciana P R, Downward J. Direct interaction of Ras and the amino-terminal region of Raf-1 in vitro. Nature. 1993;364:352–355. doi: 10.1038/364352a0. [DOI] [PubMed] [Google Scholar]
- 52.Wartmann M, Davis R J. The native structure of the activated Raf protein kinase is a membrane-bound multi-subunit complex. J Biol Chem. 1994;269:6695–6701. [PubMed] [Google Scholar]
- 53.Wartmann M, Hofer P, Turowski P, Saltiel A R, Hynes N E. Negative modulation of membrane localization of the Raf-1 protein kinase by hyperphosphorylation. J Biol Chem. 1997;272:3915–3923. doi: 10.1074/jbc.272.7.3915. [DOI] [PubMed] [Google Scholar]
- 54.Yao B, Zhang Y, Delikat S, Mathias S, Basu S, Kolesnick R. Phosphorylation of Raf by ceramide-activated protein kinase. Nature. 1995;378:307–310. doi: 10.1038/378307a0. [DOI] [PubMed] [Google Scholar]
- 55.Zha J, Harada H, Yang E, Jockel J, Korsmeyer S J. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L) Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 56.Zhang X, Settleman J, Kyriakis J M, Takeuchi-Suzuki E, Elledge S J, Marshall M S, Bruder J T, Rapp U R, Avruch J. Normal and oncogenic p21ras proteins bind to the amino-terminal regulatory domain of c-Raf-1. Nature. 1993;364:308–365. doi: 10.1038/364308a0. [DOI] [PubMed] [Google Scholar]