Abstract
The Mdm2 oncoprotein is a well-known inhibitor of the p53 tumor suppressor, but it may also possess p53-independent activities. In search of such p53-independent activities, the yeast two-hybrid screen was employed to identify Mdm2-binding proteins. We report that in vitro and in transfected cells, Mdm2 can associate with Numb, a protein involved in the determination of cell fate. This association causes translocation of overexpressed Numb into the nucleus and leads to a reduction in overall cellular Numb levels. Through its interaction with Numb, Mdm2 may influence processes such as differentiation and survival. This could potentially contribute to the altered properties of tumor cells which overexpress Mdm2.
The mdm2 gene was first described as part of a DNA region amplified in a spontaneously transformed mouse cell line (7) and was eventually shown to be responsible for the oncogenic properties of these cells (16). The idea that mdm2 is a potential oncogene was further supported through the discovery that it is overexpressed in a variety of human tumors (11, 12, 35, 44, 52); for reviews, see references 41 and 51). In many of these tumors, the mdm2 locus is amplified, but there are also numerous examples in which overexpression of the Mdm2 protein without any gene amplification has been described. Moreover, overexpression of Mdm2 has been shown experimentally to have oncogenic consequences, ranging from immortalization of primary cells to enhanced tumorigenicity in animals (13, 16, 18, 57).
Perhaps the best-known facet of mdm2 is its intricate interaction with the p53 tumor suppressor. The Mdm2 protein forms a very tight specific association with p53 (42). This association occurs through the N-terminal transactivation domain of p53 (9, 22, 45, 50). Consequently, Mdm2 blocks the interaction of p53 with the transcriptional machinery, resulting in abrogation of the ability of p53 to transactivate its specific target genes (42, 45). Furthermore, the complex between p53 and Mdm2 can actually function directly as a potent transcriptional repressor (59). Recently, it has been found that Mdm2 can also cause a general decrease in steady-state cellular p53 levels through targeting p53 for rapid degradation via the ubiquitin-proteasome pathway (26, 33). All of the above effects of Mdm2 probably account for its ability to extinguish efficiently the biological activities of p53, such as induction of G1 arrest and apoptosis (8, 10, 25).
While Mdm2 represses the accumulation and activity of p53, p53 actually enhances the expression of Mdm2. The mdm2 gene contains within its first intron a cryptic promoter, which carries two adjacent p53 binding motifs (30, 62, 64). When p53 is induced within a cell, this cryptic promoter becomes functional and drives the accumulation of mdm2 mRNA (3, 4, 30, 62).
These interactions between p53 and Mdm2 are believed to drive an autoregulatory feedback loop, which serves to restrain the extent and duration of p53 activity under physiological conditions (4, 47–49, 62). The importance of this autoregulatory loop is highlighted by the observations that mice deficient in Mdm2 die early in development but that mice deficient for both p53 and Mdm2 develop rather normally and are viable (29, 43). Hence, at least during early development, keeping p53 in check is a critical function of Mdm2.
Nevertheless, there are also numerous indications that Mdm2 may possess additional p53-independent activities. The mdm2 gene can give rise to a complex set of Mdm2 polypeptides (46, 57), which are generated through alternative splicing, utilization of multiple translation initiation codons, and possibly also posttranslational modifications. Of note, only a subset of these polypeptides can associate with p53 (46). Moreover, the Mdm2 protein contains several distinct structural motifs (41, 51) which are dispensable for p53 binding (9, 45). Hence, at least some forms of Mdm2 are likely to fulfill roles which do not involve binding to p53.
In support of this conjecture, several p53-independent biochemical properties of Mdm2 have been described. Mdm2 binds the ribosomal L5 protein (38) and can also bind specific RNA sequences through its C-terminal RING domain (15). Perhaps related to its oncogenic potential, Mdm2 can interact with the product of the Rb tumor suppressor gene (pRB [63]) and with the E2F1/DP1 transcription factor (39); both interactions result in induction of E2F transcriptional activity, which is crucial for progression into the S phase of the cell cycle. It is thus conceivable that Mdm2 not only releases a proliferative block by silencing p53 but also positively augments proliferation by stimulating S phase entry through E2F activation.
The existence of a p53-independent role(s) for Mdm2 also gains support from biological studies. Mdm2 can transform p53-null cells and can overcome a G1 arrest induced in such cells by the pRB-related protein p107 (13). Furthermore, targeted overexpression of Mdm2 in the murine mammary gland uncouples S phase from mitosis and inhibits mammary gland development in the absence of p53 (37).
In an attempt to gain further insight into the biological activities of Mdm2 and into molecular events which regulate its function, a search for Mdm2-interacting proteins was undertaken. We report here that Mdm2 can associate in vitro and in vivo with the mammalian Numb protein. The association occurs through the N-terminal domain of Mdm2, which is a region also involved in p53 binding. Mutations in the N-terminal domain of Mdm2 strongly attenuate its interaction with Numb. Elevated Mdm2 expression can alter the subcellular localization of Numb as well as cause its accelerated degradation. The Numb protein is involved in the regulation of cell fate and in a variety of developmental processes, most notably in the nervous system. Through its interaction with Numb, Mdm2 may influence processes such as differentiation and survival. This could contribute to the altered properties of tumor cells which overexpress Mdm2.
MATERIALS AND METHODS
Cells and transfections.
293, 293T, and Saos-2 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS) in a 5% CO2 atmosphere. H1299 cells were maintained routinely in RPMI medium supplemented with 10% FCS.
Cells were replated on the day before the transfection. Saos-2 and H1299 transfections were performed by the calcium phosphate procedure in DMEM plus 10% FCS as described before (30). With 293 and 293T cells, transfections were done in the presence of 25 μM chloroquine. Eight hours posttransfection, the transfection medium was replaced by DMEM plus 10% FCS. Cells were harvested 48 to 70 h posttransfection.
Yeast two-hybrid screen.
Y190 yeast (24) were transformed with the N terminus of mouse Mdm2 (first 134 amino acids) fused to the GAL4 DNA binding domain (DBD) in plasmid pAS1-CYH2 as bait. Expression of the fusion protein was confirmed by Western blot analysis with anti-Haemophilus influenzae hemagglutinin tag antibodies (data not shown). Y190 yeast cells expressing N-terminal Mdm2 were transformed with a human peripheral lymphocyte cDNA expression library fused to the GAL4 activation domain (14). Transformants were assayed for β-galactosidase activity with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) as a substrate. A total of 28 of the 6 × 105 transformants screened were positive for β-galactosidase staining. The CYH2 gene on the bait plasmid confers cycloheximide sensitivity on the otherwise resistant Y190 strain. Therefore, positive yeast transformants were selected for loss of the bait plasmid (Mdm2-GAL4 DBD) by growth on selective medium containing cycloheximide (24). Recovered prey plasmids were used to cotransform Y190 yeast with either the N terminus of mouse Mdm2, full-length mouse Mdm2, a deleted form of p53, cyclin D, lamin, SNF1, or human immunodeficiency virus type 1 (HIV-1) Tat, all fused to the GAL4 DBD. Eleven plasmids were found to encode proteins that interacted with both the N-terminal domain of Mdm2 and full-length Mdm2, but not with unrelated control proteins. The human cDNA insert of each of these plasmids was subjected to DNA sequence analysis. Clone 24 was selected for further analysis because it contained the largest open reading frame (ORF), whereas most of the other clones contained much shorter ORFs.
Isolation of a human Numb (hNumb) cDNA clone.
A human ALL-λZAPII cDNA library (cDNA library of an acute lymphoblastic leukemia cell line; generous gift of E. Canaani) was screened with a PCR-generated 300-bp cDNA probe derived from Clone 24. Rescued positive clones were analyzed by restriction enzyme analysis and sequencing.
DNA sequencing.
DNA sequencing was performed by the Weizmann Institute DNA sequencing unit. Sequence analysis was carried out with the DNA sequence assembly software AutoAssembler (ABI).
Production of anti-hNumb polyclonal antibodies.
A C-terminal fragment of h-Numb, encompassing residues 385 to 549, was fused in frame to glutathione S-transferase (GST). Abundant expression of the fusion protein in Escherichia coli was detected upon a 1.5-h induction with 0.1 mM isopropyl-β-d-thiogalactopyranoside. The recombinant protein was purified by chromatography over glutathione agarose, followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and was employed to immunize New Zealand White rabbits according to standard procedures (23).
In vitro binding assays.
GST, GST-Mdm2 (encoding full-length mouse Mdm2 fused in frame to GST), GST-p53 (residues 1 to 42), or GST–c-Jun (residues 1 to 79) were overexpressed in bacteria and purified according to standard procedures. Full-length hNumb, deletion mutants of hNumb, or control expression plasmids, all of which were constructed as in-frame fusions downstream to the FLAG epitope, were transcribed and translated in vitro in the presence of [35S]methionine with the T7-coupled reticulocyte lysate system (TNT; Promega). In vitro-translated proteins were incubated with GST fusion proteins immobilized on glutathione agarose beads (Sigma). Incubation was for 2 h at 4°C in 250 μl of binding buffer (25 mM Tris-Cl [pH 7.2], 50 mM NaCl, 0.2% Nonidet P-40 [NP-40]) with continuous shaking. Following incubation, the beads were pelleted, washed three times with a large excess of washing buffer (100 mM Tris-Cl [pH 8.0], 100 mM NaCl, 1% NP-40), and boiled in protein sample buffer. Proteins released into the supernatant were resolved by SDS-PAGE and visualized by exposure to X-ray film after fluorography.
Coprecipitation and Western blot analysis.
293 cells (1.5 × 106 to 2.0 × 106 per 10-cm dish) were transfected with 5 μg of hNumb expression plasmid together with 5 μg of Mdm2 expression plasmid or vector control plasmid. At 8 h posttransfection, the transfection medium was removed, and the cells were replenished with fresh culture medium. Cells were harvested at 48 to 70 h posttransfection. Cell extracts were subjected to immunoprecipitation with the appropriate antibodies, as described before (4). When indicated, extracts were first precleared by incubation with preimmune serum. Immunoprecipitates as well as 1% of each extract were resolved by SDS-7.5% PAGE. Western blot analysis was performed with the enhanced chemiluminescence (ECL) system (Amersham).
Northern blot analysis.
Total RNA from transfected 293 cells was prepared by the RNAzol B method (Biotecx). A 10-μg amount of total RNA was resolved on a denaturing (formaldehyde) agarose gel, transferred to a Nylon membrane (Hybond N; Amersham), and hybridized with random-primed probes specific for hNumb or human GAPDH. Hybridization was at 65°C in 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM Na2HPO4, and 1 mM EDTA [pH 7.7])–2.5× Denhardt’s solution–0.25% SDS–150 μg of herring sperm DNA per ml. The blots were washed at increasing stringencies, with the final wash in 0.1× SSPE–0.1% SDS at 65°C.
Pulse-chase analysis.
Forty-eight hours posttransfection, transfected 293 cells were starved for 30 min in methionine-free medium and then metabolically labelled for 15 min with 110 mCi of [35S]methionine per 10-cm plate. Culture dishes were washed twice in phosphate-buffered saline (PBS) and replenished with nonradioactive medium. At the indicated time points (chase), cells were harvested and extracts containing equal amounts of acid-insoluble radioactivity were immunoprecipitated with anti-hNumb polyclonal serum. Immunoprecipitates were resolved by SDS-7.5% PAGE and visualized by exposure to X-ray film after fluorography.
Immunostaining.
Saos-2 cells were plated 24 h prior to transfection on glass coverslips at a density of 0.5 × 106 cells per 6-cm dish. Cells were transfected with FLAG-hNumb together with either control vector DNA or Mdm2 expression plasmid. At 24 to 48 h posttransfection, the cells were washed with ice-cold PBS and fixed in cold methanol (−20°C) for at least 20 min. The coverslips were rehydrated in ice-cold PBS for 5 min, blocked in 1% bovine serum albumin in PBS for 5 min, and incubated with anti-FLAG monoclonal antibodies M2 (1:300; Kodak) for 1 h at room temperature. The coverslips were then washed three times in PBS, incubated in the dark with Cy3-conjugated goat anti-mouse immunoglobulin G (Jackson ImmunoResearch Laboratories) and with 4′,6-diamidino-2-phenylindole (DAPI; 0.5 μg/ml; Sigma) for 30 min at room temperature, washed three times in PBS, mounted on glass slides, and viewed under a fluorescence microscope (Zeiss Axioskop).
RESULTS
Isolation of hNumb as a candidate Mdm2-binding protein.
The yeast two-hybrid screen was employed to identify proteins capable of interacting with Mdm2. The N terminus of mouse Mdm2 (first 134 amino acid residues), fused to the GAL4 DBD, was used as a bait to screen a human peripheral lymphocyte cDNA expression library fused to the GAL4 activation domain (24). One candidate clone, denoted clone 24, was found to encode a polypeptide interacting in the two-hybrid assay with the N terminus of mouse Mdm2 and with full-length mouse and human Mdm2, but not with unrelated proteins (cyclin D, lamin, SNF1, HIV-1 Tat, and a deleted form of p53; data not shown). Therefore, this clone was chosen for further analysis.
Sequence analysis of the clone 24 cDNA insert revealed that it is homologous to the Drosophila Numb gene, which is required for sensory organ formation in the developing nervous system (32, 53, 60). Numb participates in cell fate specification. It localizes asymmetrically during cell division and segregates into one daughter cell, generating two daughter cells with distinct fates.
Using the clone 24 insert as a probe, we screened a human cDNA library prepared from an acute lymphoblastic leukemia cell line (ALL-1) and isolated a clone containing the full ORF of hNumb. The predicted protein sequence is shown in Fig. 1. Indicated are a phosphotyrosine-binding (PTB) domain, which is implicated in the preferential binding of tyrosine phosphorylated proteins (6, 31, 36, 58); eight Pro-X-X-Pro motifs which might serve as putative SH3-binding elements (1); and a zinc finger-like domain. The entire ORF comprises 592 amino acid residues. It is thus shorter than the predicted product of the hNumb clone described recently by Salcini et al. (54), which contains 603 residues (hNumb-S; Fig. 1). These 11 extra residues are present in hNumb-S but not in our cDNA or in the published mouse Numb cDNA (65). It is formally possible that the human genome contains two closely related hNumb genes, encoding a shorter protein and longer protein, respectively. However, the rest of the sequence is perfectly conserved between the two human cDNA clones; hence, it is most likely that both transcripts are derived from a single gene, presumably by alternative splicing. This can be resolved through future analysis of genomic hNumb DNA. It is of note that the extra amino acids reside within the PTB domain (Fig. 1), which is important for Numb function (20). It is thus conceivable that the two clones may represent functionally distinct forms of hNumb.
FIG. 1.
Protein sequence of hNumb. Sequence of the ORF encoded by the hNumb cDNA clone isolated from the ALL-1 library (see text). hNumb-S denotes the extra 11 residues present in the hNumb clone reported by Salcini et al. (54). The putative PTB domain is underlined. Eight putative SH3-binding motifs, Pro-X-X-Pro (X being any amino acid), are underlined by empty boxes. The most C-terminal box fits the extended consensus SH3-binding motif with Pro residues at positions 2, 7, and 10. A zinc finger-like domain is underlined by dashes.
Interestingly, the C-terminal region of hNumb (residues 355 to 592) is identical to clone S171, a partial cDNA clone assigned previously to human chromosomal region 14q24.3, which is associated with familial Alzheimer’s disease (56). Most likely, S171 is indeed a partial hNumb clone, implying that the hNumb gene is part of this Alzheimer’s disease-associated chromosomal region. Taken together with the documented involvement of Numb in neuronal development in Drosophila (53, 60) and its proposed role in mammalian neurogenesis (61, 65), this raises the provocative possibility that defective hNumb function may contribute to human neurodegenerative disorders.
The hNumb protein associates with Mdm2 in vitro and in vivo.
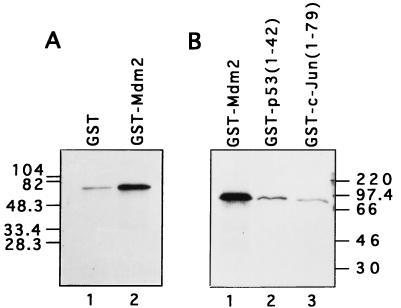
Full-length hNumb protein was produced by translation in a reticulocyte lysate, giving rise to a protein of about 65 kDa, as expected from its amino acid sequence (Fig. 2). This hNumb protein was tested for binding in vitro to GST-Mdm2 and to a series of control proteins, i.e., GST, GST-p53 (amino acids 1 to 42) and GST–c-Jun (amino acids 1 to 79). In agreement with the data obtained in the yeast two-hybrid screen, hNumb was found to bind preferentially to GST-Mdm2 (Fig. 2). Under the same conditions, a number of other in vitro-translated proteins failed to exhibit preferential binding to GST-Mdm2 (data not shown); these included c-abl, the TFIIH-associated protein p62, and the p53-inducible protein PAG608 (28). This supports the existence of a specific direct interaction between Mdm2 and hNumb.
FIG. 2.
hNumb binds specifically to Mdm2 in vitro. Full-length hNumb was produced by in vitro translation in a rabbit reticulocyte lysate in the presence of [35S]methionine. Equal amounts of radiolabeled hNumb were incubated with the indicated GST fusion proteins or with GST alone. Bound proteins were eluted in protein sample buffer, resolved by SDS-PAGE, and visualized by exposure to X-ray film after fluorography. Positions and molecular masses of protein molecular size markers are indicated. (A and B) Results are from separate experiments.
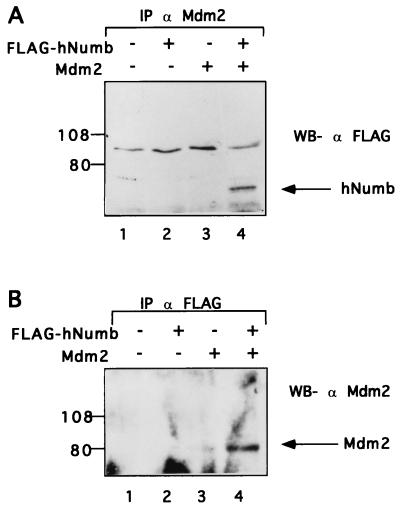
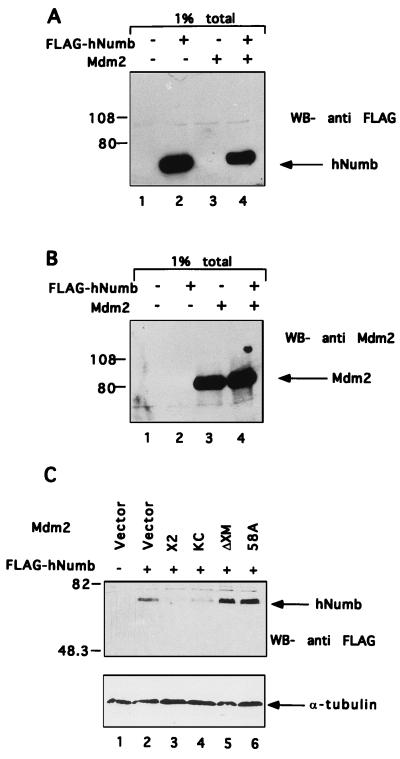
To determine whether Mdm2 and hNumb can associate in vivo, hNumb was cloned in frame downstream to a FLAG epitope and overexpressed transiently in human 293 cells with or without cotransfected Mdm2. Cell extracts were precleared with preimmune serum, followed by immunoprecipitation with anti-Mdm2 polyclonal serum. Immunoprecipitates were subjected to Western blot analysis with an anti-FLAG monoclonal antibody. As seen in Fig. 3A, hNumb was brought down from extracts of cells cotransfected with an Mdm2 expression plasmid (lane 4), but not with empty vector DNA (lane 2). Hence, the two proteins can associate specifically within living cells as well.
FIG. 3.
Mdm2 associates with hNumb in vivo. Mdm2 and FLAG-hNumb were overexpressed in 293 cells by transient transfection. (A) Cell extracts were precleared with preimmune serum, followed by immunoprecipitation with anti-Mdm2 polyclonal serum. Immunoprecipitates were subjected to Western blot (WB) analysis with anti-FLAG monoclonal antibodies. (B) Cell extracts were precleared with preimmune serum, followed by immunoprecipitation with anti-FLAG monoclonal antibodies. Immunoprecipitates (IP) were subjected to Western blot analysis with anti-Mdm2 polyclonal serum. Positions of protein molecular size markers are indicated on the left.
A similar picture was revealed when identical extracts were first immunoprecipitated with anti-FLAG monoclonal antibodies and then subjected to Western blot analysis with anti-Mdm2 polyclonal serum (Fig. 3B). Again, coprecipitation of hNumb and Mdm2 was clearly seen when both proteins were present (lane 4). In that case, a minute amount of Mdm2 was also coprecipitated from extracts of cells transfected with Mdm2 alone (lane 3). This implies the existence of some nonspecific background; however, the specific signal was reproducibly much stronger (Fig. 3B and data not shown). These data extend the initial observations in the yeast two-hybrid screen and imply that hNumb can interact with Mdm2 in vitro and in vivo.
The hNumb protein contains at least two Mdm2-binding regions.
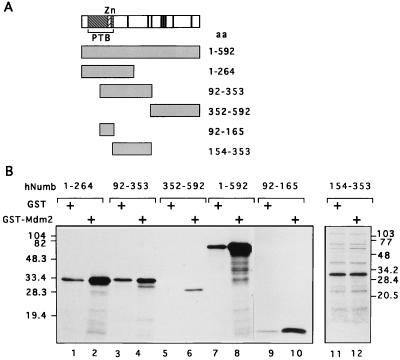
To identify the region(s) of hNumb involved in interaction with Mdm2, a series of hNumb deletion mutants were generated (Fig. 4A; indicated at the top are the PTB domain, the zinc finger-like motif, and the eight putative SH3-binding motifs). Each deletion mutant was produced by translation in a reticulocyte lysate in the presence of [35S]methionine and tested for binding in vitro to either GST or GST-Mdm2. The results (Fig. 4B) indicate that the hNumb protein contains at least two Mdm2-binding domains, one confined to the C-terminal half of the PTB domain (residues 92 to 165) and the other residing in the C terminus of the protein (residues 352 to 592), while the central part of the hNumb polypeptide (residues 154 to 353) does not exhibit preferential binding to Mdm2. Interestingly, an important activity of the Drosophila Numb (dNumb) protein, i.e., inhibition of Notch signaling, was also reported to involve both the PTB and the C-terminal domains (20). While the PTB domain of dNumb was shown to bind directly to Notch (21), such interaction was not demonstrated for the C-terminal domain.
FIG. 4.
In vitro interaction of Mdm2 with hNumb deletion mutants. (A) Schematic representation of the hNumb deletion mutants; all mutants were constructed in pcDNA3FLAG. Indicated are the positions of the PTB domain, the zinc finger-like domain (Zn), and the eight putative SH3-binding motifs (vertical bars). (B) hNumb polypeptides described in panel A were translated in vitro in the presence of [35S]methionine and analyzed for binding to either GST or GST-Mdm2 as described in the legend to Fig. 2. aa, amino acids.
Binding to hNumb requires an intact N-terminal domain of Mdm2.
The bait employed for identifying hNumb in the yeast two-hybrid screen contained only the first 134 residues of Mdm2. This implies that the N terminus of Mdm2 is sufficient for hNumb binding. To further explore the importance of the N-terminal domain for this interaction, a series of Mdm2 mutants were tested for binding to hNumb in vivo.
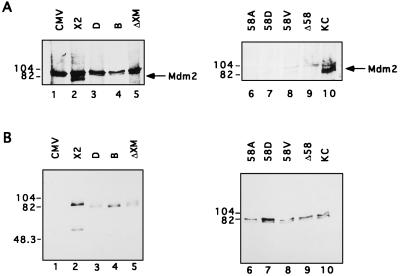
The first set of mutants encodes proteins with short N-terminal truncations. The first four in-frame methionines are located at amino acid positions +1, +6, +50, and +62 of the mouse Mdm2 protein (3, 16). Plasmid Mdm2-X2 expresses full-length Mdm2 protein. Plasmid Mdm2-B, Mdm2-D, and Mdm2-ΔXM all encode polypeptides initiating at positions +50 and/or +62 (3); all of these N-terminally truncated polypeptides fail to bind p53 (3, 46). Each of the mutants, as well as intact Mdm2 (X2) or an empty cytomegalovirus (CMV) promoter vector control, was cotransfected together with a hNumb expression plasmid into 293 cells. Cell extracts were immunoprecipitated with anti-hNumb polyclonal serum. Immunoprecipitates (Fig. 5A) as well as 1% of each total extract (Fig. 5B) were subjected to Western blot analysis with an anti-Mdm2 polyclonal serum. Coprecipitation of Mdm2 with hNumb was readily observed in cells overexpressing full-length Mdm2 (Fig. 5A, lane 2; the band immediately above Mdm2 represents a nonspecific contaminant, which is brought down with preimmune serum as well). However, comparable coprecipitation was not obtained with any of the N-terminally deleted Mdm2 polypeptides (Fig. 5A, lanes 3 to 5), even if one corrects for variations in total levels of transfected Mdm2 (Fig. 5B). The crystal structure of the N-terminal domain of Mdm2 complexed to a 15-residue p53 peptide reveals that Mdm2 possesses a deep hydrophobic cleft, to which p53 binds as an amphipathic α helix (34). This structure further predicts that residue Gly58 of Mdm2, which is located within this cleft, is critical for proper interaction with p53. We generated four mouse Mdm2 variants in which Gly58 was either deleted completely (Δ58) or mutated to Ala, Asp, or Val (58A, 58D, and 58V, respectively). All mutants were constructed on the basis of plasmid Mdm2-KC, which was optimized for efficient expression of full-length Mdm2 protein (3). As expected and in agreement with earlier observations (19), none of these mutants retains significant p53 binding (56a).
FIG. 5.
In vivo interaction of hNumb with Mdm2 mutants. Expression plasmids encoding different forms of Mdm2 were transfected into 293 cells together with an hNumb expression plasmid. Cell extracts were immunoprecipitated with anti-hNumb polyclonal serum. Immunoprecipitates (A) as well as 1% of each direct cell extract (B) were subjected to Western blot analysis with anti-Mdm2 polyclonal serum. X2 and KC encode full-length Mdm2 protein; D, B, and ΔXM encode Mdm2 polypeptides lacking the N-terminal portion of the protein; and 58A, 58D, 58V, and Δ58 encode Mdm2 proteins with alterations at amino acid position 58 (see text). The positions of protein molecular size markers are indicated on the left. The band present slightly above Mdm2 in panel A, particularly in lanes 1 to 5, corresponds to a nonspecific contaminant observable also when immunoprecipitation is performed with preimmune serum (data not shown). Lanes 1 to 5 and 6 to 10, respectively, are from separate experiments differing in nonspecific background.
Following transfection into 293 cells, the wild-type protein encoded by Mdm2-KC exhibited easily detectable coprecipitation with cotransfected hNumb (Fig. 5A, lane 10). All mutations at position 58 of Mdm2 resulted in a marked attenuation of this association (lanes 6 to 9), despite the comparable levels of all Mdm2 forms in the transfected cells (Fig. 5B, lanes 6 to 10). Hence, efficient binding to hNumb depends on the integrity of the N-terminal domain of Mdm2.
Mdm2 reduces the steady-state level of hNumb.
As a routine control for efficient expression of transfected plasmids, aliquots of transfected cell extracts were subjected directly to gel electrophoresis, followed by Western blot analysis. A typical result is depicted in Fig. 6A and B; samples were derived from the experiment described in Fig. 3, with each lane corresponding to 1% of the total amount of extract employed for immunoprecipitation in Fig. 3. It is evident that coexpression of Mdm2 caused a significant decrease in the steady-state level of the transfected hNumb protein (Fig. 6A; compare lanes 2 and 4). This effect was observed in three separate experiments with 293 cells, as well as with 293T cells (derived from 293 by stable transfection with the simian virus 40 large T antigen). In contrast, steady-state levels of Mdm2 were actually augmented by cotransfection with hNumb (Fig. 6B).
FIG. 6.
Mdm2 reduces the steady-state level of hNumb. (A and B) 1% of each cell extract employed for immunoprecipitation in Fig. 3 was subjected directly to Western blot analysis with either anti-FLAG monoclonal antibodies (A) or anti-Mdm2 polyclonal serum (B). hNumb and Mdm2 are indicated by arrows. Positions of protein molecular size markers are shown on the left. ECL exposure time is identical to that used in Fig. 3. (C) Expression plasmids encoding different forms of Mdm2 (see Fig. 5 and text) were transfected into Saos-2 cells, together with a FLAG-hNumb expression plasmid. Total cell extracts were subjected to Western blot (WB) analysis with anti-FLAG monoclonal antibodies. hNumb is indicated by an arrow. The positions of protein molecular size markers are shown on the left. Equal loading was confirmed by reprobing the same blot with an anti-α-tubulin antibody (bottom panel).
A similar ability of full-length Mdm2 to decrease the steady-state levels of hNumb was also observed with Saos-2 human osteosarcoma cells (Fig. 6C; compare lane 3 and 4 to lane 2). In order to find out whether this activity depends on the binding of Mdm2 to hNumb, a similar analysis was also performed with N-terminally mutated forms of Mdm2 (ΔXM and 58A) defective in hNumb binding (Fig. 5). As seen in Fig. 6C, lanes 5 and 6, both mutants failed to decrease the cellular levels of hNumb; rather, they actually even caused a mild increase. Hence, the ability of Mdm2 to downregulate steady-state hNumb levels most probably requires a direct interaction between the two proteins.
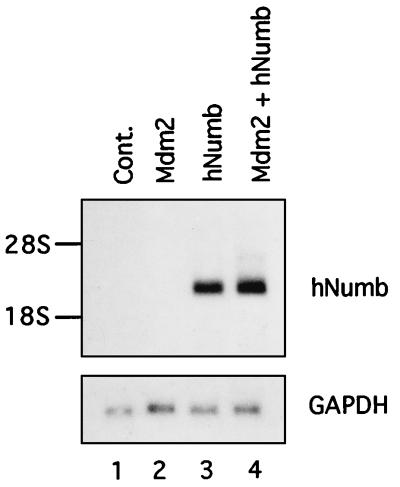
Quantitative analysis of hNumb mRNA in transfected 293 cells revealed that the amounts of these transcripts were not reduced by Mdm2 coexpression (Fig. 7). Hence, the decrease in the amount of hNumb protein was not due to an effect of Mdm2 on transcription or RNA stability.
FIG. 7.
Mdm2 does not alter hNumb mRNA levels. 293 cells were transfected with the indicated plasmids. Total cellular RNA was prepared 70 h posttransfection, and 10 μg of each sample was subjected to Northern blot analysis. The blots were hybridized sequentially with probes corresponding to hNumb and to human GAPDH. The positions of 28S and 18S rRNA are indicated. Cont., control.
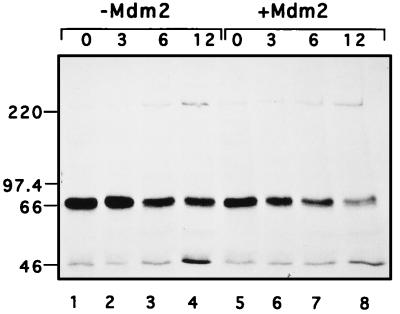
Mdm2 was recently found to promote the degradation of p53 (26, 33), with which Mdm2 also associates through its N-terminal domain. This raised the interesting possibility that Mdm2 might enhance hNumb proteolysis too. To further explore this idea, a pulse-chase experiment was performed. 293 cells were transiently transfected with hNumb, together with either Mdm2 or empty vector (Fig. 8 [+Mdm2 and −Mdm2, respectively]). The cells were metabolically labelled with [35S]methionine for 15 min and then chased in nonradioactive medium for the numbers of hours indicated above each lane. Extracts containing equal amounts of acid-insoluble radioactivity were immunoprecipitated with anti-hNumb polyclonal serum and were resolved by SDS-PAGE. As seen in Fig. 8, coexpression of Mdm2 accelerated the disappearance of hNumb. Densitometric scanning indicated that the half-life of hNumb was reduced from 11 h in the absence of extra Mdm2 to 6.5 h in its presence (data not shown). Thus, Mdm2 indeed appears capable of targeting hNumb for faster degradation, although the effect is not as dramatic as that for p53 (26, 33), probably because Mdm2 associates with p53 more efficiently than with hNumb. It is of note that Mdm2 overexpression caused a slight reduction in the amount of label incorporated into hNumb during the pulse (compare lanes 1 and 5); in this particular experiment, the reduction was by 29%. Thus, although the main effect of Mdm2 on hNumb is likely to be exerted on protein stability, a change in translation efficiency cannot presently be ruled out. In this regard, it is noteworthy that Mdm2 binds to the L5 ribosomal protein and its associated 5S rRNA (38), as well as to specific RNA sequences or structures (15), suggesting a possible involvement of Mdm2 in translational regulation.
FIG. 8.
Pulse-chase analysis of transfected hNumb in the absence or presence of excess Mdm2. 293 cells were transiently transfected with an hNumb expression plasmid, together with either Mdm2 expression plasmid (+Mdm2) or control vector (−Mdm2). Cells were metabolically labelled by incubation with [35S]methionine for 15 min and then chased in nonradioactive medium for the numbers of hours indicated at the top. Portions of each extract, containing equal amounts of acid-insoluble radioactivity, were immunoprecipitated with anti-hNumb polyclonal serum. The immunoprecipitates were resolved by SDS-7.5% PAGE and visualized by exposure to X-ray film after fluorography. The positions of protein molecular size markers are indicated.
Mdm2 can modulate the subcellular localization of hNumb.
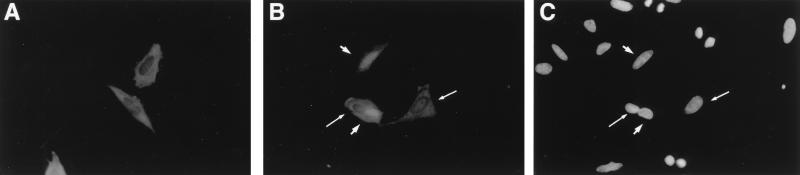
Mdm2 is predominantly nuclear (4, 41, 46). On the other hand, in both Drosophila and mouse cells, Numb was found associated with the plasma membrane (32, 65). Rat Numb was reported to be present primarily in the soluble cytoplasmic fraction as well as in the particulate membrane fraction (61). The fact that Mdm2 and Numb appear to reside in totally different subcellular compartments would seem inconsistent with the in vivo interactions described above. Therefore, we asked whether any of these two proteins could influence the other’s localization. To that end, Saos-2 cells were transiently transfected with expression plasmids encoding Mdm2 and FLAG-hNumb, either each alone or both together.
The subcellular localization of Mdm2 was not visibly affected by hNumb expression; Mdm2 remained primarily in the nucleus (data not shown). However, we did observe a reproducible effect of Mdm2 on hNumb localization. In cells transfected with FLAG-hNumb alone, staining was exclusively cytoplasmic (Fig. 9A). On the other hand, when Mdm2 was included in the transfection, approximately 20% of all of the cells staining positive for transfected hNumb revealed conspicuous accumulation of FLAG-hNumb in the nucleus. Figure 9B depicts a field of doubly transfected cells, which were stained for FLAG-hNumb; DAPI staining of the same field, which displays the nuclei of all of the cells in that field, is shown in Fig. 9C. Whereas two of the four positive transfectants in Fig. 9B retain characteristic cytoplasmic staining (long arrows), the other two display copious amounts of FLAG-hNumb in the nucleus (arrowheads). While potentially of great interest, the reason why Mdm2 affects the localization of hNumb only in some cells but not in others is presently unknown. A similar effect of Mdm2 on the subcellular localization of FLAG-hNumb was observed in transfected H1299 cells (data not shown). Thus, Mdm2 is capable of directing hNumb to the nucleus, at least when both are overexpressed in transfected cells. The ability of Mdm2 to modulate the subcellular localization of hNumb argues in favor of the idea that interactions between the two proteins can occur in vivo and might actually affect hNumb function.
FIG. 9.
Mdm2 affects the subcellular localization of hNumb. Saos-2 cells were transfected transiently with FLAG-hNumb expression plasmid DNA together with either vector control (A) or Mdm2 expression plasmid (B and C). Cells were fixed with methanol, stained with anti-FLAG monoclonal antibody (A and B), and visualized by indirect immunofluorescence. Arrowheads and long arrows identify cells with predominantly nuclear and predominantly cytoplasmic hNumb staining, respectively. (C) DAPI staining of the field shown in panel B. The photographs were taken at a magnification of ×400.
Enhanced coprecipitation of Mdm2 with hNumb in the presence of a tyrosine phosphatase inhibitor.
Screening of a peptide library has revealed that the PTB domain of dNumb binds preferentially to phosphopeptides containing the motif YIGPYF, where F represents any hydrophobic residue (36). Interestingly, Mdm2 contains a related stretch of amino acids (YIGQYI in mouse Mdm2 [16]). This stretch corresponds to residues 56 to 61 of mouse Mdm2. It is located at the heart of the p53-binding cleft of Mdm2 (34), within the N-terminal region which also mediates hNumb binding. Moreover, the G in this motif corresponds to Gly58 of Mdm2, whose importance for hNumb binding is documented in Fig. 5. This segment of Mdm2 is, therefore, an attractive candidate for playing a key role in the recognition of hNumb through its PTB domain.
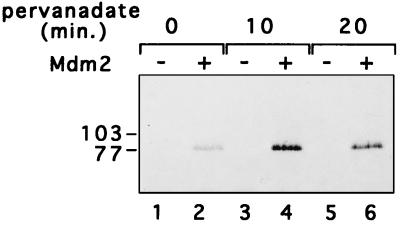
The fact that hNumb-Mdm2 interactions can be easily detected with recombinant GST-Mdm2 (Fig. 2) argues that phosphorylation of Mdm2 is not required for binding to hNumb. Similarly, despite its name (phosphotyrosine binding), the PTB domain of dNumb specifically recognizes the YIGPYF motif in the absence of tyrosine phosphorylation (36). However, when peptides carrying this motif are phosphorylated on the tyrosine at position 5, this significantly enhances their binding to the dNumb PTB domain (36). Therefore, we monitored Mdm2-hNumb interactions under conditions which increase the extent of tyrosine phosphorylation on cellular proteins. To that end, 293 cells were cotransfected with expression plasmids for hNumb and Mdm2 and either left untreated or treated with H2O2-sodium orthovanadate (pervanadate) for 10 or 20 min prior to harvesting. Pervanadate treatment strongly inhibits phosphotyrosine phosphatase activity, augmenting the fraction of protein molecules phosphorylated on tyrosine (27). Cells were then harvested, extracted, and analyzed for Mdm2-hNumb interaction as in Fig. 3. As seen in Fig. 10, pervanadate treatment increased the amount of Mdm2 that could be precipitated with hNumb (compare lane 2 with lanes 4 and 6). While rather modest, this increase was seen reproducibly (four of four experiments; data not shown). This suggests that the interaction between Mdm2 and hNumb may be regulated positively, directly or indirectly, by tyrosine phosphorylation.
FIG. 10.
Effect of pervanadate on the in vivo association of Mdm2 and hNumb. 293 cells were transiently transfected with a hNumb expression plasmid, together with either Mdm2 expression plasmid (+) or control vector (−). The cells were pretreated with pervanadate (1 mM Na3VO4, 3 mM H2O2) for 0, 10, or 20 min prior to harvesting. Cell extracts were precleared with preimmune serum, followed by immunoprecipitation with anti-hNumb polyclonal serum. Immunoprecipitates were subjected to Western blot analysis with anti-Mdm2 polyclonal serum. The positions of protein molecular size markers are indicated on the left.
DISCUSSION
Although there exists extensive information on the relationship between Mdm2 and p53, far less is known about possible interactions of Mdm2 with other proteins. The present study describes a novel specific association between Mdm2 and hNumb, which is a protein involved in cell fate determination. This association can be detected in vitro as well as with transfected cells. In transfected cells, Mdm2 overexpression causes a decrease in overall hNumb levels, due at least in part to an accelerated rate of hNumb protein degradation. Moreover, excess Mdm2 causes nuclear accumulation of hNumb in a subset of cells.
While we have not determined the precise boundaries of the hNumb binding site within the Mdm2 protein, the data argue strongly that this site may at least partially overlap the p53 binding domain; point mutations within the p53-binding cleft of Mdm2, which eliminate p53 binding, also interfere strongly with hNumb binding. It is conceivable that this cleft has evolved primarily to accommodate p53, which binds Mdm2 very tightly and is subject to stringent regulation by Mdm2. All of the residues which contribute to this cleft may thus be required for ensuring a tight fit with the p53 transactivation domain. It is nevertheless of note that this cleft includes a sequence (YIGQYI and YLGQYI in mouse and human Mdm2, respectively) which significantly resembles the YIGPYF motif, which was recently shown to confer selective binding to the dNumb PTB domain (36). Of the six residues comprising this motif, only the Gly at position 58 of Mdm2 and the Ile at position 61 are directly engaged in interactions with p53 (34). It is tempting to speculate that the stringent conservation of the entire stretch may also serve to maintain the association with other Mdm2 partners, including Numb.
Also of potential interest is the observation that the amount of Mdm2 found in association with hNumb increases upon treatment of transfected cells with an inhibitor of protein tyrosine phosphatases. One attractive possibility is that Mdm2 is directly phosphorylated on tyrosine under these conditions, thereby increasing its avidity for the hNumb PTB domain. However, there is so far no description of tyrosine phosphorylation on Mdm2. Therefore, one must also consider indirect mechanisms through which an overall increase in cellular tyrosine phosphorylation affects the formation of Mdm2-hNumb complexes. Interestingly, exposure of cells to basic fibroblast growth factor elicits a p53-independent increase in Mdm2 protein levels (55). The basic fibroblast growth factor receptor possesses tyrosine kinase activity, and its stimulation induces specific protein tyrosine phosphorylation. Thus, in addition to being transcriptionally regulated by p53, Mdm2 may also be responsive to signaling pathways which involve tyrosine phosphorylation.
The functional significance of the interaction between Mdm2 and Numb is presently unclear. At first glance, the relevance of this interaction is challenged by the fact that each of these proteins seems to operate in a totally different subcellular compartment, i.e., Mdm2 in the nucleus and Numb in the plasma membrane. However, the ability of Mdm2 to direct hNumb to the nucleus, at least in cells overexpressing both proteins, suggests that there may exist situations in which the two proteins not only see each other but even affect each other’s function. There is now a growing list of proteins which were initially considered nonnuclear and are now known to reach the nucleus under defined circumstances and contribute to specific biochemical and biological processes; one such example is β-catenin, a cytoplasmic protein involved in the coupling of cell adhesion molecules to the cytoskeleton, which has recently been shown to participate directly in transcriptional regulation through interaction with transcription factors (5, 40). By directing Numb into the nucleus, Mdm2 may conceivably either interfere with the normal activity of the former or, alternatively, allow it to exert a distinct nuclear activity. Of note, Mdm2 itself shuttles very efficiently between the cytoplasm and nucleus, and this shuttling is required for the efficient promotion of p53 degradation (53a). It is thus conceivable that some of the effects of Mdm2 on other proteins are attained through translocation of these proteins into or out of the nucleus, as part of a complex with Mdm2. Our data are consistent with a model in which the subcellular localization of Numb is also subject to a similar type of modulation by Mdm2.
Mdm2 has recently been proposed to possess an E3 ubiquitin protein ligase activity, through which it can directly elicit p53 ubiquitination (27a) and thereby target p53 for proteasomal degradation. It is tempting to speculate that, presumably via its N-terminal pocket, Mdm2 associates with a variety of other proteins which are consequently ubiquitinated through that E3 activity and then degraded. Our data suggest that Numb is another target of this activity. The approach employed here, which uses the N-terminal domain of Mdm2 as a bait in the yeast two-hybrid screen, is likely to reveal additional proteins which are subject to a similar type of regulation by Mdm2.
A complementary approach should involve the identification of other proteins, which through binding to Mdm2 may either facilitate or inhibit its protein-destabilizing activity. Regulatory proteins of this type may interact with other domains of Mdm2, including those required for its putative E3 enzymatic activity. In this regard, it is of note that the p19Arf tumor suppressor protein associates specifically with Mdm2 and prevents it from promoting the rapid degradation of p53 (51a, 64a). Moreover, this interaction, which requires structural elements within the C-terminal part of Mdm2 and is not abrogated by mutations within its N-terminal p53-binding domain, also accelerates the degradation of Mdm2 itself. Hence, Mdm2 appears to reside at the heart of a complex network of protein-protein interactions which is greatly modulated through changes in protein turnover.
The data presented above raise the intriguing possibility that Mdm2-Numb interactions may couple the p53-Mdm2 pathway with processes regulated by Numb. There are numerous indications that p53 may contribute to proper differentiation (2). Abnormally high activity of Mdm2, which occurs in various tumors, might therefore disrupt normal tissue differentiation through neutralizing the resident p53. Another positive regulator of differentiation which is recognized and presumably inactivated by Mdm2 is pRB (63). Furthermore, Mdm2 overexpression was experimentally demonstrated to interfere with the differentiation of myogenic cells in culture (17). If the binding of Mdm2 to Numb results in inactivation of the latter, this could represent yet another mechanism through which excess Mdm2 might inhibit differentiation. Such a situation is likely to be particularly relevant in cells of neuroectodermal origin, in which Numb appears to play a key role in cell fate determination. This putative ability to maintain cells in a nondifferentiated state might contribute to the oncogenic properties of Mdm2. It is noteworthy that Mdm2 gene amplification occurs in human neuroblastomas, albeit at low frequency (12). It will be interesting to find out whether Numb function is defective in such tumors and whether they are relatively less differentiated than similar tumors retaining normal Mdm2 expression.
ACKNOWLEDGMENTS
We thank P. Wieder-Salamon and S. Wilder for excellent technical assistance. We thank S. Elledge for plasmids and cDNA library, E. Canaani and members of the Canaani lab for cDNA library and helpful suggestions, and A. Bershadsky and Y. Shay for helpful advice.
This research was supported in part by PHS grant RO1 CA 40099 from the National Cancer Institute and by the Leo and Julia Forchheimer Center for Molecular Genetics.
REFERENCES
- 1.Alexandropoulos K, Cheng G, Baltimore D. Proline-rich sequences that bind to Src homology 3 domains with individual specificities. Proc Natl Acad Sci USA. 1995;92:3110–3114. doi: 10.1073/pnas.92.8.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almog N, Rotter V. Involvement of p53 in cell differentiation and development. Biochim Biophys Acta. 1997;1333:F1–F27. doi: 10.1016/s0304-419x(97)00012-7. [DOI] [PubMed] [Google Scholar]
- 3.Barak Y, Gottlieb E, Juven-Gershon T, Oren M. Regulation of mdm2 expression by p53: alternative promoters produce transcripts with nonidentical translation potential. Genes Dev. 1994;8:1739–1749. doi: 10.1101/gad.8.15.1739. [DOI] [PubMed] [Google Scholar]
- 4.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behrens J, von Kris J P, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 6.Blaikie P, Immanuel D, Wu J, Li N, Yajnik V, Margolis B. A region in Shc distinct from the SH2 domain can bind tyrosine-phosphorylated growth factor receptors. J Biol Chem. 1994;269:32031–32034. [PubMed] [Google Scholar]
- 7.Cahilly-Snyder L, Yang F T, Francke U, George D L. Molecular analysis and chromosomal mapping of amplified genes isolated from a transformed mouse 3T3 cell line. Somat Cell Mol Genet. 1987;13:235–244. doi: 10.1007/BF01535205. [DOI] [PubMed] [Google Scholar]
- 8.Chen C Y, Oliner J D, Zhan Q, Fornace A J, Vogelstein B, Kastan M B. Interactions between p53 and MDM2 in a mammalian cell cycle checkpoint pathway. Proc Natl Acad Sci USA. 1994;91:2684–2688. doi: 10.1073/pnas.91.7.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, Marechal V, Levine A J. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, Wu X, Lin J, Levine A J. mdm-2 inhibits the G1 arrest and apoptosis functions of the p53 tumor suppressor protein. Mol Cell Biol. 1996;16:2445–2452. doi: 10.1128/mcb.16.5.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cordon-Cardo C, Latres E, Drobnjak M, Oliva M R, Pollack D, Woodruff J M, Marechal V, Chen J, Brennan M F, Levine A J. Molecular abnormalities of mdm2 and p53 genes in adult soft tissue sarcomas. Cancer Res. 1994;54:794–799. [PubMed] [Google Scholar]
- 12.Corvi R, Savelyeva L, Breit S, Wenzel A, Handgretinger R, Barak J, Oren M, Amler L, Schwab M. Non-syntenic amplification of MDM2 and MYCN in human neuroblastoma. Oncogene. 1995;10:1081–1086. [PubMed] [Google Scholar]
- 13.Dubs-Poterszman M, Tocque B, Wasylyk B. MDM2 transformation in the absence of p53 and abrogation of the p107 G1 cell-cycle arrest. Oncogene. 1995;11:2445–2449. [PubMed] [Google Scholar]
- 14.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A E, Lee W H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 15.Elenbaas B, Dobbelstein M, Roth J, Shenk T, Levine A J. The MDM2 oncoprotein binds specifically to RNA through its RING finger domain. Mol Med. 1996;2:439–451. [PMC free article] [PubMed] [Google Scholar]
- 16.Fakharzadeh S S, Trusko S P, George D L. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J. 1991;10:1565–1569. doi: 10.1002/j.1460-2075.1991.tb07676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiddler T A, Smith L, Tapscott S J, Thayer M J. Amplification of MDM2 inhibits MyoD-mediated myogenesis. Mol Cell Biol. 1996;16:5048–5057. doi: 10.1128/mcb.16.9.5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finlay C A. The mdm-2 oncogene can overcome wild-type p53 suppression of transformed cell growth. Mol Cell Biol. 1993;13:301–306. doi: 10.1128/mcb.13.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freedman D A, Epstein C B, Roth J C, Levine A J. A genetic approach to mapping the p53 binding site in the MDM2 protein. Mol Med. 1997;3:248–259. [PMC free article] [PubMed] [Google Scholar]
- 20.Frise E, Knoblich J A, Younger S S, Jan L Y, Jan Y N. The Drosophila Numb protein inhibits signaling of the Notch receptor during cell-cell interaction in sensory organ lineage. Proc Natl Acad Sci USA. 1996;93:11925–11932. doi: 10.1073/pnas.93.21.11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo M, Jan L Y, Jan Y N. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron. 1996;17:27–41. doi: 10.1016/s0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 22.Haines D S, Landers J E, Engle L J, George D L. Physical and functional interaction between wild-type p53 and mdm2 proteins. Mol Cell Biol. 1994;14:1171–1178. doi: 10.1128/mcb.14.2.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harlow E, Lane P. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 24.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 25.Haupt Y, Barak Y, Oren M. Cell type-specific inhibition of p53-mediated apoptosis by mdm2. EMBO J. 1996;15:1596–1606. [PMC free article] [PubMed] [Google Scholar]
- 26.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 27.Heffetz D, Bushkin I, Dror R, Zick Y. The insulinomimetic agents H2O2 and vanadate stimulate protein tyrosine phosphorylation in intact cells. J Biol Chem. 1990;265:2896–2902. [PubMed] [Google Scholar]
- 27a.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 28.Israeli D, Tessler E, Haupt Y, Elkeles A, Wilder S, Amson R, Telerman A, Oren M. A novel p53-inducible gene, PAG608, encodes a nuclear zinc finger protein whose overexpression promotes apoptosis. EMBO J. 1997;16:4384–4392. doi: 10.1093/emboj/16.14.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones S N, Roe A E, Donehower L A, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 30.Juven T, Barak Y, Zauberman A, George D L, Oren M. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene. 1993;8:3411–3416. [PubMed] [Google Scholar]
- 31.Kavanaugh W M, Williams L T. An alternative to SH2 domains for binding tyrosine-phosphorylated proteins. Science. 1994;266:1862–1865. doi: 10.1126/science.7527937. [DOI] [PubMed] [Google Scholar]
- 32.Knoblich J A, Jan L Y, Jan Y N. Asymmetric segregation of Numb and Prospero during cell division. Nature. 1995;377:624–627. doi: 10.1038/377624a0. [DOI] [PubMed] [Google Scholar]
- 33.Kubbutat M H, Jones S N, Vousden K H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 34.Kussie P H, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine A J, Pavletich N P. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 35.Leach F S, Tokino T, Meltzer P, Burrell M, Oliner J D, Smith S, Hill D E, Sidransky D, Kinzler K W, Vogelstein B. p53 Mutation and MDM2 amplification in human soft tissue sarcomas. Cancer Res. 1993;53:2231–2234. [PubMed] [Google Scholar]
- 36.Li S C, Songyang Z, Vincent S J, Zwahlen C, Wiley S, Cantley L, Kay L E, Forman K J, Pawson T. High-affinity binding of the Drosophila Numb phosphotyrosine-binding domain to peptides containing a Gly-Pro-(p)Tyr motif. Proc Natl Acad Sci USA. 1997;94:7204–7209. doi: 10.1073/pnas.94.14.7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lundgren K, Montes de Oca Luna R, McNeill Y B, Emerick E P, Spencer B, Barfield C R, Lozano G, Rosenberg M P, Finlay C A. Targeted expression of MDM2 uncouples S phase from mitosis and inhibits mammary gland development independent of p53. Genes Dev. 1997;11:714–725. doi: 10.1101/gad.11.6.714. [DOI] [PubMed] [Google Scholar]
- 38.Marechal V, Elenbaas B, Piette J, Nicolas J C, Levine A J. The ribosomal L5 protein is associated with mdm-2 and mdm-2–p53 complexes. Mol Cell Biol. 1994;14:7414–7420. doi: 10.1128/mcb.14.11.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin K, Trouche D, Hagemeier C, Sorensen T S, La Thangue N B, Kouzarides T. Stimulation of E2F/DP1 transcriptional activity by MDM2 oncoprotein. Nature. 1995;375:691–694. doi: 10.1038/375691a0. [DOI] [PubMed] [Google Scholar]
- 40.Molenaar M, van de Wetering M, Oosterwegel M, Peterson M J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 41.Momand J, Zambetti G P. Mdm-2: “big brother” of p53. J Cell Biochem. 1997;64:343–352. [PubMed] [Google Scholar]
- 42.Momand J, Zambetti G P, Olson D C, George D, Levine A J. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 43.Montes de Oca Luna R, Wagner D S, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 44.Oliner J D, Kinzler K W, Meltzer P S, George D L, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 45.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 46.Olson D C, Marechal V, Momand J, Chen J, Romocki C, Levine A J. Identification and characterization of multiple mdm-2 proteins and mdm-2–p53 protein complexes. Oncogene. 1993;8:2353–2360. [PubMed] [Google Scholar]
- 47.Otto A, Deppert W. Upregulation of mdm-2 expression in Meth A tumor cells tolerating wild-type p53. Oncogene. 1993;8:2591–2603. [PubMed] [Google Scholar]
- 48.Perry M E, Piette J, Zawadzki J A, Harvey D, Levine A J. The mdm-2 gene is induced in response to UV light in a p53-dependent manner. Proc Natl Acad Sci USA. 1993;90:11623–11627. doi: 10.1073/pnas.90.24.11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Picksley S M, Lane D P. The p53-mdm2 autoregulatory feedback loop: a paradigm for the regulation of growth control by p53? Bioessays. 1993;15:689–690. doi: 10.1002/bies.950151008. [DOI] [PubMed] [Google Scholar]
- 50.Picksley S M, Vojtesek B, Sparks A, Lane D P. Immunochemical analysis of the interaction of p53 with MDM2;--fine mapping of the MDM2 binding site on p53 using synthetic peptides. Oncogene. 1994;9:2523–2529. [PubMed] [Google Scholar]
- 51.Piette J, Neel H, Marechal V. Mdm2: keeping p53 under control. Oncogene. 1997;15:1001–1010. doi: 10.1038/sj.onc.1201432. [DOI] [PubMed] [Google Scholar]
- 51a.Pomerantz J, Schreiber-Agus N, Liegeois N, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H-W, Cordon-Cardo C, DePinho R A. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 52.Reifenberger G, Liu L, Ichimura K, Schmidt E E, Collins V P. Amplification and overexpression of the MDM2 gene in a subset of human malignant gliomas without p53 mutations. Cancer Res. 1993;53:2736–2739. [PubMed] [Google Scholar]
- 53.Rhyu M S, Jan L Y, Jan Y N. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 53a.Roth J, Dobbelstein M, Freedman D A, Levine A J. Nucleocytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salcini A E, Confalonieri S, Doria M, Santolini E, Tassi E, Minenkova O, Cesareni G, Pelicci P G, Di F P. Binding specificity and in vivo targets of the EH domain, a novel protein-protein interaction module. Genes Dev. 1997;11:2239–2249. doi: 10.1101/gad.11.17.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shaulian E, Reznitzky D, Shifman O, Blandino G, Amsterdam A, Yayon A, Oren M. Induction of Mdm2 and enhancement of cell survival by bFGF. Oncogene. 1997;15:2717–2725. doi: 10.1038/sj.onc.1201453. [DOI] [PubMed] [Google Scholar]
- 56.Sherrington R, Rogaev E I, Liang Y, Rogaeva E A, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- 56a.Shifman, O., T. Juven-Gershon, and M. Oren. Unpublished results.
- 57.Sigalas I, Calvert A H, Anderson J J, Neal D E, Lunec J. Alternatively spliced mdm2 transcripts with loss of p53 binding domain sequences: transforming ability and frequent detection in human cancer. Nat Med. 1996;2:912–917. doi: 10.1038/nm0896-912. [DOI] [PubMed] [Google Scholar]
- 58.Songyang Z, Margolis B, Chaudhuri M, Shoelson S E, Cantley L C. The phosphotyrosine interaction domain of SHC recognizes tyrosine phosphorylated NPXY motif. J Biol Chem. 1995;270:14863–14866. doi: 10.1074/jbc.270.25.14863. [DOI] [PubMed] [Google Scholar]
- 59.Thut C J, Goodrich J A, Tjian R. Repression of p53-mediated transcription by MDM2: a dual mechanism. Genes Dev. 1997;11:1974–1986. doi: 10.1101/gad.11.15.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uemura T, Shepherd S, Ackerman L, Jan L Y, Jan Y N. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell. 1989;58:349–360. doi: 10.1016/0092-8674(89)90849-0. [DOI] [PubMed] [Google Scholar]
- 61.Verdi J M, Schmandt R, Bashirullah A, Jacob S, Salvino R, Craig C G, Program A E, Lipshitz H D, McGlade C J. Mammalian NUMB is an evolutionarily conserved signaling adapter protein that specifies cell fate. Curr Biol. 1996;6:1134–1145. doi: 10.1016/s0960-9822(02)70680-5. [DOI] [PubMed] [Google Scholar]
- 62.Wu X, Bayle J H, Olson D, Levine A J. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 63.Xiao Z, Chen J, Levine A J, Modjtahedi N, Xing J, Sellers W R, Livingston D M. Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature. 1995;375:694–698. doi: 10.1038/375694a0. [DOI] [PubMed] [Google Scholar]
- 64.Zauberman A, Flusberg D, Haupt Y, Barak Y, Oren M. A functional p53-responsive intronic promoter is contained within the human mdm2 gene. Nucleic Acids Res. 1995;23:2584–2592. doi: 10.1093/nar/23.14.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64a.Zhang Y, Xiong Y, Yarbrough W G. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 65.Zhong W, Feder J N, Jiang M, Jan L Y, Jan Y N. Assymetric localization of a mammalian Numb homolog during mouse cortical neurogenesis. Neuron. 1996;17:43–53. doi: 10.1016/s0896-6273(00)80279-2. [DOI] [PubMed] [Google Scholar]