Abstract
In Neurospora crassa, the major nitrogen regulatory protein, NIT2, a member of the GATA family of transcription factors, controls positively the expression of numerous genes which specify nitrogen catabolic enzymes. Expression of the highly regulated structural gene nit-3, which encodes nitrate reductase, is dependent upon a synergistic interaction of NIT2 with a pathway-specific control protein, NIT4, a member of the GAL4 family of fungal regulatory factors. The NIT2 and NIT4 proteins both bind at specific recognition elements in the nit-3 promoter, but, in addition, we show that a direct protein-protein interaction between NIT2 and NIT4 is essential for optimal expression of the nit-3 structural gene. Neurospora possesses at least five different GATA factors which control different areas of cellular function, but which have a similar DNA binding specificity. Significantly, only NIT2, of the several Neurospora GATA factors examined, interacts with NIT4. We propose that protein-protein interactions of the individual GATA factors with additional pathway-specific regulatory factors determine each of their specific regulatory functions.
A complex global regulatory circuit governs the entire realm of nitrogen metabolism in Neurospora crassa, Aspergillus nidulans, Saccharomyces cerevisiae, and other lower eukaryotic organisms (20, 21). In N. crassa, a major nitrogen control gene, nit-2, encodes a positive-acting regulatory protein that is a member of the GATA family of transcription factors. GATA factors are widely represented in plant and animal species, including yeasts, filamentous fungi, Caenorhabditis elegans, Dictyostelium sp., Drosophila melanogaster, tobacco, and vertebrates. GATA factors contain one or two Cys2/Cys2-type zinc fingers with a central loop of 17 (or 18) amino acids and are so named because they show sequence-specific DNA binding to elements that contain a core GATA sequence. The GATA factors of higher organisms possess two zinc finger motifs, the carboxy-terminal finger being primarily responsible for sequence-specific DNA binding (22, 26). Six distinct mouse and human GATA factors have been identified, and each appears to positively control a particular spectrum of tissue- and cell-specific gene expression (1, 26). It now appears that the DNA-binding domains of certain transcription factors are also involved in promoting protein-protein interactions with other regulatory proteins. The amino-terminal zinc finger of the mammalian GATA-1 protein mediates interactions with Sp1 and EKLF proteins (23), and this finger of GATA-2 specifically binds to the bZip factors JUN and FOS (15).
Most fungal GATA factors possess only a single zinc finger motif which is responsible for specific DNA binding. A wealth of genetic and biochemical studies have demonstrated that the A. nidulans AREA protein and the N. crassa NIT2 protein control the expression of many different structural genes that encode nitrogen metabolic enzymes (21). In Neurospora, nitrogen repression, exerted by favored nitrogen sources, e.g., ammonium ion or glutamine, prevents expression of the nitrogen-related structural genes. Expression of the nitrogen catabolic genes requires both nitrogen derepression and a functional NIT2 regulatory protein. A second major regulatory factor, NMR (nitrogen metabolic regulation), acts in a negative fashion, precluding nitrogen catabolic gene transcription during conditions of nitrogen repression (33). NMR itself is not a DNA-binding protein, but acts by binding directly to NIT2 and inhibiting the latter’s activation function (31).
Neurospora readily senses a variety of environmental cues, such as nutritional, light, and temperature signals, which elicit specific cellular responses. The regulation of nitrate assimilation represents a dual-signal regulatory system that ensures the pathway enzymes are turned on only when both nitrogen derepression and nitrate induction conditions are satisfied (10, 21). The well-characterized structural gene nit-3, which encodes NADPH-dependent nitrate reductase, is highly regulated as a member of the nitrogen control circuit. Expression of nit-3 not only requires N derepression and the globally acting regulatory protein NIT2, but also is completely dependent upon induction by nitrate, mediated by the pathway-specific positive-acting regulatory protein NIT4 (34). NIT4 is a member of the GAL4 family of fungal transcription factors which possess an amino-terminal Cys6/Zn2 domain that provides sequence-specific DNA binding (11). The nit-3 promoter region possesses several elements that serve as NIT2 binding sites and two elements for NIT4 binding, all of which are required for full expression. It is particularly significant that neither NIT2 nor NIT4 alone allows any detectable transcription of the nit-3 gene, but when both NIT2 and NIT4 are present, nit-3 is turned from “off” to “on,” resulting in a high level of expression (5, 12). The strong synergy between NIT2 and NIT4 suggested the possibility that these proteins functionally interact with one another to activate expression of the nitrate assimilatory structural genes.
Recently, it has become clear that Neurospora possesses at least five distinct GATA factors, each of which is presumed to regulate a specific set of genes within an important area of cellular function. In addition to NIT2, several other global regulators, including the white collar factors, WC1 and WC2, also possess GATA-type DNA-binding domains (2, 19). WC1 and WC2 mediate blue light regulation, which controls carotenoid biosynthesis, sexual development, and photoinduced resetting of the circadian clock (7). Recently, two additional GATA factors have been identified for N. crassa. One (NGF1), whose function is still not clear, encodes a GATA factor whose zinc finger closely resembles that of the GAT1/NIl2 and DAL80 factors of S. cerevisiae and thus may play a role in nitrogen control (10a). Another recently discovered Neurospora GATA factor, designated SRE, contains two closely related zinc fingers and functions as a negative regulator controlling iron homeostasis (35). Thus, at least five distinct Neurospora GATA factors with overlapping DNA binding specificities coexist in the same cells, which immediately raises questions as to how each of these global factors exerts functional specificity in regulating distinct sets of structural genes. Previous studies have demonstrated that fungal and vertebrate GATA factors recognize DNA elements with the identical GATA core sequence, with little preference to flanking sequences (4, 6, 17, 22). GATA sequences, which allow strong NIT2 DNA binding in vitro, appear in coding regions and promoters of genes which are not at all subject to NIT2 control. Thus, it appears that DNA binding specificity alone cannot adequately account for the stringent functional specificity of these factors.
One intriguing possibility is that the specificity displayed by each GATA factor in controlling its own set of target genes is achieved by specific interactions with other distinct regulatory proteins. Here we show that several Neurospora GATA factors, NIT2, WC1, WC2, and NGF1, overlap significantly in DNA-binding activity, such that DNA binding alone cannot explain their specificity in controlling unique gene sets. Results are presented which demonstrate that a specific protein-protein interaction occurs between NIT2 and NIT4 and is essential for activation of nit-3 gene expression in vivo.
MATERIALS AND METHODS
DNA and plasmids.
DNA manipulations were carried out according to standard procedures (27). Site-directed mutagenesis (Bio-Rad Mutagene kit) to alter the NIT2 finger was done with plasmid Bluescript containing the XmnI-EcoRI nit-2 DNA fragment. Three oligonucleotides with limited nucleotide randomization (B is C, G, or T; D is A, G, or T; K is G or T; S is C or G; Y is C or T) at appropriate positions were used to introduce amino acid changes: 5′ACAACT TGCACCAACTGCSDGACGCAAACGACCCCATT3′, 5′ACCCCATTGTG GCGCCGTDGCBCAATGGGACAACCCCTCTGCAAC3′, and 5′CGCCG TAACCCAGATGGAAGCGKAGYCTGCAACGCTTGTGGCTTG3′. After determination of the exact nucleotide changes by sequencing, each nit-2 fragment with mutations was subcloned into the targeting vector pDE-nit2 by using HindIII and EcoRI (9). At least two independent mutated clones of each mutation were used for further analysis. The expression vector for His6-tagged NIT4 (residues 48 to 179) was constructed by introducing BamHI and EcoRI sites at the ends of the nit4 DNA fragment with PCR and then subcloning it into the pRSET vector (Invitrogen), resulting in pRSETNIT4(48–179). The glutathione S-transferase (GST)-NIT2 fusions were constructed by first subcloning different regions of NIT2 (see figures) into pRSET; BamHI and EcoRI sites were then used to clone each NIT2 region into the GST fusion expression vector pGEX-2T or pGEX-3X (Pharmacia) to achieve the correct reading frame. The GST-NIT4(48–179) fusion was constructed by subcloning the BamHI-EcoRI nit-4 fragment from PRSETNIT4(48–179) to the pGEX expression vector. pGEX-WC1 and pGEX-WC2 constructs (2, 19) for expression of GST-WC1 and GST-WC2 fusion proteins, respectively, were kindly provided by G. Macino and P. Ballario, University of Rome.
Antisera for NIT4 and NIT2.
A histidine-tagged NIT4 protein (residues 109 to 176) was expressed in the BL(21) Lys− Escherichia coli strain and purified to apparent homogeneity by nickel-agarose affinity chromatography. After collection of preimmune serum, about 200 μg of the purified protein was used to inject each of two rabbits. Booster shots of the same dosage were given after 28 and 60 days. Polyclonal anti-NIT4 serum was collected 3 weeks after the second boost. The development of a polyclonal anti-NIT2 antibody (to NIT2 residues 732 to 822) was described previously (31).
Protein expression.
The histidine-tagged NIT4 and NIT2 proteins were expressed in the BL(21) Lys− E. coli strain. IPTG (isopropyl-β-d-thiogalactopyranoside [1 mM]) was added when cultures reached an A600 of 0.3. The cells were incubated for an additional 4 h and harvested. Three rounds of freeze-thaw with liquid N2 in the presence of DNase I and RNase A (10 μg/ml) were used to disrupt the cells. All histidine-tagged proteins were purified on an Ni-nitrilotriacetic acid-agarose column (Qiagen) under native conditions. The proteins were eluted with 0.2 M imidazole in washing buffer. For expression of GST fusion proteins, an overnight culture of the BL(21) Lys− E. coli strain bearing the expression vector was diluted 10-fold in fresh 2× YT (yeast-tryptone) medium containing 100 μg of ampicillin per ml. After 1 h of incubation, IPTG was added to a final concentration of 1 mM, and the culture was incubated for an additional 5 h at 37°C. The cells were collected and lysed by three freeze-thaw cycles. GST fusion proteins were purified according to the method in reference 28 with 0.3 ml of resin for a 250-ml E. coli culture.
In vitro protein-binding assays.
For protein-protein interaction assays, 70 μl of glutathione-agarose resin (Sigma), which usually contained 3 to 5 μg of bound GST-NIT2, GST-NIT4, or other GST fusion proteins, was incubated with 200 μl of phosphate-buffered saline buffer (20 mM sodium phosphate [pH 7.0], 150 mM sodium chloride) containing 5 μg of histidine-tagged NIT2 or NIT4 proteins for 30 min at 0°C with occasional gentle mixing. In the various experiments, identical amounts of each protein being examined were used; the input of each was verified via gel electrophoresis. After being washed four times with 1 ml of the binding buffer at room temperature, proteins were eluted in 30 μl of 10 mM glutathione in 50 mM Tris buffer (pH 8.0). Occasionally, ethidium bromide (20 μg/ml) was added to the washing buffer to detect any possible interactions mediated by DNA. This modification and others, such as an increase in NaCl concentration or the inclusion of Triton X-100 in the binding buffer, did not affect the magnitude of observed protein-protein interactions. Approximately 1 μl of each eluate was electrophoresed in SDS–12% polyacrylamide gels, and the relevant proteins were detected via Western blotting with anti-NIT2 or anti-NIT4 antiserum; 2 to 10 μl of serum in 10 ml of TBST (50 mM Tris-Cl [pH 7.6], 150 mM sodium chloride, 0.5% Tween 20) was used for Western blotting. The horseradish peroxidase-coupled secondary antibody and ECL (enhanced chemiluminescence) light emission substrates (Amersham) were used to develop the Western blots.
Mobility shift assays.
DNA fragments used in mobility shift experiments were end labelled with 32P by the Klenow filling-in reaction (27). A 34-bp DNA probe containing a single GATA element present in the nit-3 promoter region was synthesized as a double-stranded oligonucleotide. A 60-bp al-3 promoter region containing two GATA sites was kindly provided by G. Macino and P. Ballario (19). Other probes (see figures) were from restriction digestion of plasmid pN3P containing the nit-3 promoter and were isolated from an agarose gel. Labelled DNA probes were incubated with DNA-binding proteins in binding buffer [20 mM HEPES (pH 8.0), 50 mM KCl, 2 mM MgCl2, 2 mM dithiothreitol, 20 μM ZnCl2, 1 mM EDTA, 200 μg of bovine serum albumin per ml, 0.5 to 1 μg of poly(dI-dC) per 25 ml, 10% glycerol] at room temperature for 30 min. DNA-protein complexes and free probe were resolved with 4 to 6% polyacrylamide gels in 0.25× Tris-borate-EDTA buffer by electrophoresis at 20 V/cm. The gels were dried and exposed to X-ray film.
Transformation procedures and nitrate reductase assays.
Neurospora transformation was done as described previously (30). Wild-type and nit-2 mutant genes in pBluescript were introduced into the N. crassa nit-2 rip23 mutant strain (this laboratory). Transformants were selected for growth on Vogel’s medium (8) containing 20 mM sodium nitrate as the sole nitrogen source. Duplicates for each construct were prepared, and each was transformed into the host strain at least three times. The size of the colonies obtained by transformation was scored after incubation for 5 days at 30°C. In some cases, it was desirable to obtain nit-2 transformants without selection for nit-2+ function (growth on nitrate). The pDE vector (9) containing a truncated his-3 gene and the wild-type or mutant nit-2 genes was used for targeted transformation to the his-3 locus via homologous recombination, with the double mutant nit-2 rip23 his-3′ as the recipient strain. This procedure results in the integration of a single copy of the transforming DNA. Transformants (about 10/μg of DNA) were selected on Vogel’s minimal medium (without histidine).
At least three independent transformants for each DNA sample were used to assay for nitrate reductase. After growth in 30 ml of Vogel’s −N medium with 20 mM glutamine for 12 to 16 h, the mycelia were harvested, washed with water, and divided into two flasks, each containing 20 ml of Vogel’s −N medium with either 20 mM sodium nitrate or 20 mM sodium nitrate and 20 mM glutamine as the sole nitrogen source(s). After 3 h of further incubation, mycelia (about 100 mg [wet weight]) were harvested by filtration and washed with water. Cell extracts were made by disrupting the mycelia with a Mini Beadbeater (Biospec) in extraction buffer (20 mM sodium phosphate [pH 6.0], 150 mM sodium chloride, 2 mM β-mercaptoethanol, 0.1 mM EDTA). Protein concentrations were determined with the Bio-Rad reagent according to their manual. Nitrate reductase assays were done as described previously (13). Each value was normalized with the protein concentration. For enzyme assays, two nit-2 rip host strains transformed with the wild-type gene were included as internal standards.
RESULTS
A physical interaction occurs between NIT2 and NIT4.
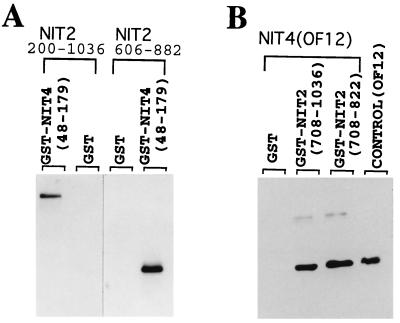
Activation of nit-3, which encodes nitrate reductase, has an absolute requirement for both the globally acting NIT2 protein and the pathway-specific NIT4 factor, suggesting the possibility that these two proteins functionally interact to turn on nit-3. To look for a possible direct interaction between NIT2 and NIT4, residues 48 to 179 of NIT4, which encompasses its entire Cys6/Zn2 DNA-binding domain (34), were expressed in E. coli as a GST fusion protein. Two different NIT2 segments were expressed as histidine-tagged proteins, purified, and examined for specific binding to the GST-NIT4 protein immobilized on glutathione-agarose (Fig. 1A). Neither of the his-tagged NIT2 proteins (residues 200 to 1036 and 606 to 822) showed any binding to GST, even when large amounts of the proteins were tested. In contrast, both NIT2 proteins exhibited a strong interaction with the GST-NIT4 fusion protein, as demonstrated via Western blot analysis (Fig. 1). These data suggest that a specific interaction occurs between NIT2 and NIT4. A reciprocal experiment also showed a specific NIT2-NIT4 interaction. Two NIT2 segments, residues 708 to 822 and 708 to 1036, were expressed as GST-NIT2 fusion proteins and immobilized on a glutathione agarose affinity column. A purified histidine-tagged NIT4 protein (OF12 [NIT4 residues 48 to 179]) was added to test for the physical association with NIT2. OF12 strongly associated with GST-NIT2 in this in vitro binding assay, but not with the GST protein control (Fig. 1B). The combined results indicate that a specific physical interaction occurs between the NIT2 and NIT4 proteins.
FIG. 1.
Specific interaction between NIT2 and NIT4 proteins. (A) Two segments of the NIT2 protein (residues 200 to 1036 and 606 to 882) were expressed in E. coli and incubated with GST (negative control) or with a GST-NIT4 fusion protein immobilized on glutathione-agarose resin, as described in Materials and Methods. Specifically bound NIT2 proteins were detected by Western blot analysis. Both NIT2 proteins (the larger one 92 kDa, the smaller one 24 kDa) were retained by the GST-NIT4 protein but not by GST. (B) In a reciprocal experiment, NIT2 segments were expressed as GST fusion proteins and immobilized on glutathione-agarose. NIT4 protein was expressed, purified, and incubated with the GST-NIT2 or GST-bound resin. The NIT4 protein (15 kDa) retained by specific binding to GST-NIT2 (but not GST alone) was detected by Western blotting with authentic NIT4 protein (OF12) serving as a positive control.
The NIT2-NIT4 interaction is mediated by their DNA-binding domains.
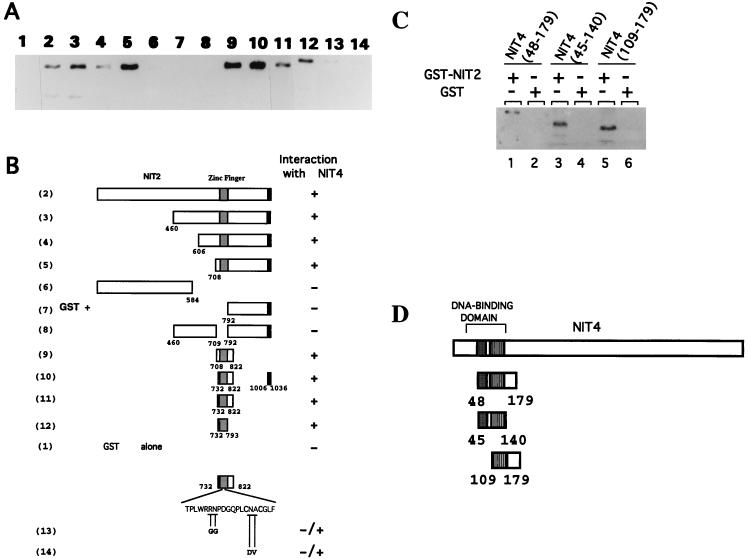
To further investigate the protein-protein binding between NIT2 and NIT4, a systematic analysis was done to map the regions of each protein that were required for the interaction. The full-length NIT2 protein, as well as different regions of NIT2, was fused to GST, and each was tested for the ability to interact with NIT4 protein OF12 (Fig. 2A and B). NIT2 regions corresponding to residues 1 to 584 and 792 to 1036, which encompass most of the protein at its N and C termini, respectively, did not display any obvious interaction with OF12 (lanes 6 and 7). However, the full-length NIT2 protein, as well as each NIT2 segment containing the DNA-binding domain, displayed strong interaction with NIT4 OF12 (lanes 2 to 5). NIT2 proteins lacking the DNA-binding domain were incapable of binding NIT4 (compare lanes 3 and 8). The minimal region of NIT2 required for interaction with NIT4 was further mapped to a region which essentially contains only the DNA-binding zinc finger motif (lanes 9 to 12). Furthermore, amino acid substitutions at conserved positions within the NIT2 zinc finger (Fig. 2, lanes 13 and 14) largely eliminated its ability to interact with NIT4. Together, the results suggest that the NIT2 DNA-binding domain is essential and sufficient to mediate its interaction with NIT4.
FIG. 2.
Mapping the regions of NIT2 and NIT4 which mediate the NIT2-NIT4 protein-protein interaction. (A) Various NIT2 regions were expressed as GST fusion proteins and immobilized on glutathione-agarose and then incubated with the purified NIT4 segment OF12 (residues 48 to 179). A Western blot was used to detect the interaction of NIT4 with any of the NIT2 segments. The numbers at the top correspond to the GST fusions described below for panel B. (B) GST-NIT2 fusion proteins. The amino acid changes in two loss-of-function nit-2 mutants are shown. Full-length NIT2 protein contains 1,036 amino acids. (C) Different NIT4 regions expressed as GST fusion proteins were immobilized to the glutathione-agarose resin and incubated with NIT2. The NIT2 protein bound by the different GST-NIT4 fusions was eluted, electrophoresed in an SDS-polyacrylamide gel, and detected by Western blotting. The amount of NIT4(48–179) loaded in lane 1 was 1/10 that used in the other lanes. (D) NIT4 regions in the GST-NIT4 fusions shown in panel C. Full-length NIT4 protein contains 1,090 amino acids. Solid boxes, zinc cluster DNA-binding domain; stippled boxes, dimerization region.
The DNA-binding domain of NIT4 constitutes a major portion of the OF12 protein and contains only about 40 additional residues at the C-terminal coiled-coil region. We speculated that the NIT4 DNA-binding domain is responsible for mediating its interaction with NIT2. Two regions of NIT4 which overlap the dimerization domain, residues 48 to 140 and 109 to 179, were tested for interaction with NIT2. Both of these NIT4 regions showed significant interaction with NIT2, as detected by an anti-NIT4 antibody (Fig. 2C and D). These results indicated that the NIT4 domain consisting of its DNA-binding and dimerization motifs is sufficient to interact with NIT2.
A NIT2-NIT4 interaction is essential for their synergistic action.
The promoter region of the nitrate reductase gene (nit-3) contains two NIT4 binding sites and seven NIT2 binding sites clustered in two regions. All of these binding sites contribute to the nit-3 expression level and display strong binding by the cognate NIT2 or NIT4 proteins in mobility shift assays in vitro. However, neither NIT2 nor NIT4 alone is able to turn on expression of the nit-3 gene. The stringent requirement for both NIT2 and NIT4 to turn nit-3 from “off” to “on” represents a strong synergy between these two proteins. We speculated that the physical interaction between NIT2 and NIT4 is essential to establish their functional synergy. Site-directed mutagenesis was employed to introduce amino acid changes in the NIT2 zinc finger region in an attempt to disrupt the NIT2-NIT4 protein-protein interaction while preserving its full DNA-binding activity. Residues within the NIT2 zinc finger motif which are not conserved among various GATA factors and which are surface located (25)—namely, Phe at position 5; Asn, Pro, and Asp at positions 15 to 17; and Gln, Pro, and Leu at positions 19 to 21—were chosen for substitution by mutagenesis. It was important that the mutated NIT2 proteins retained strong DNA-binding activity; thus, residues on the DNA recognition surface were not altered. Figure 3 shows the spectrum of substitutions.
FIG. 3.
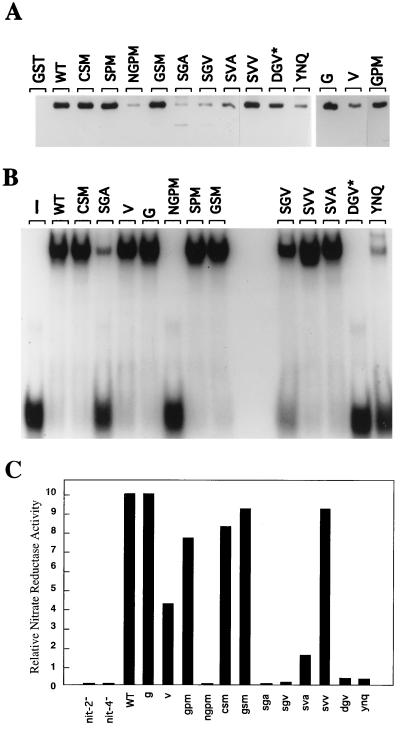
NIT2 zinc finger mutants created by site-directed mutagenesis. The sequence of the NIT2 GATA-type zinc finger (ZF) is shown with the four cysteine residues marked in boldface. The amino acid substitutions of mutants are shown below the sequence. The nit-2 genes bearing different mutations were transformed into the Neurospora nit-2 rip23 strain. The growth rate of transformants was evaluated by the size of colonies on solid medium containing nitrate as the sole N source. —, loss-of-function mutants; ∗, stop codon that resulted in a truncation of NIT2 in the DNA-binding domain.
The function of each nit-2 mutant was initially assessed by transformation to determine whether it had the ability to complement a nit-2 null mutant in supporting growth on nitrate. As expected, many of the mutations that resulted in amino acid substitutions in the zinc finger region appeared to fully complement the nit-2 mutant. Some mutants, however, failed to transform the nit-2 mutant, whereas others only partially complemented it, and the transformed colonies exhibited markedly reduced size. To gain insight into the molecular defects of these NIT2 finger mutants, we examined the ability of each mutant protein to bind DNA and to interact with the NIT4 protein (Fig. 4A and B). The expressed mutant NIT2 proteins for those mutations which fully complemented the nit-2 null mutation, namely g, gpm, csm, gsm, and svv, displayed normal DNA-binding activity and also interacted with NIT4 in a wild-type fashion. The expressed NIT2 proteins for the new finger mutants which completely failed to complement the nit-2 null strain (ngpm and sga) showed a large decrease in both DNA-binding ability and in interaction with NIT4. Of greatest interest were the third type of mutants, namely sgv and sva, which only partially complemented the nit-2 null mutant and gave rise only to tiny colonies. Significantly, sgv and sva still possess normal or close to normal DNA-binding ability, but showed detectable but markedly reduced interaction with NIT4. Another new nit-2 mutant protein, v, displayed normal DNA binding but only about one-third of the wild type’s ability to interact with NIT4; this mutant (v) exhibited moderately reduced NIT2 function in vivo. These results strongly suggest that the NIT2-NIT4 protein-protein interaction is critical for their synergistic activation of the nitrate assimilation genes.
FIG. 4.
DNA binding, NIT4 interaction, and activation of nit-3 expression by different NIT2 finger mutant proteins. (A) Protein-protein interaction between NIT4 and NIT2 zinc finger mutants. Residues 708 to 822 of NIT2 zinc finger mutants were expressed as GST fusion proteins, and identical amounts of each were immobilized to glutathione-agarose beads. GST alone served as negative control. The NIT4 protein His6-NIT4(48–179) (OF12) was tested for interaction with NIT2 proteins as described in Materials and Methods and was detected by Western blotting. GST and the GST fusion proteins used are indicated at the top of each lane. WT, wild type (B) A representative mobility shift assay with the NIT2 mutant proteins. The identical GST-NIT2 proteins used in the NIT4 protein interaction assay (described above) were also tested for DNA-binding activity. Sixty-two nanograms of each protein in 25 μl of the binding reaction mixture was incubated with the 34-bp double-stranded DNA fragment corresponding to the nit-3 promoter region containing the distal single GATA (bp −1084 to −1118 from the start codon). The NIT2 proteins used are identified on the top of the panel. The band at the bottom of the figure represents the free probe. Additional mobility shift assays with different protein concentrations confirmed the results shown. (C) Nitrate reductase assays of Neurospora strains transformed with the wild-type or mutant nit-2 gene. The nitrate reductase activities of the various mutants are compared with that of the transformant containing the wild-type nit-2+ gene, whose relative activity was arbitrarily set equal to 10. (Its specific activity was 4 U/mg of protein.) These results represent triplicate assays of at least three independent strains transformed with each gene construct. nit-2− and nit-4− represent rip mutant strains.
Analysis of a non-DNA-binding NIT2 mutant protein.
The NIT2 mutant protein dgv is truncated at the basic region of its zinc finger, and thus it completely lacks DNA-binding activity. Surprisingly, the dgv mutant still transforms the nit-2 null strain to allow weak expression of nitrate reductase (see below) and slow growth with nitrate as the sole nitrogen source. The dgv mutant NIT2 protein retains the ability to strongly interact with NIT4 (Fig. 4A and B). This finding suggests that, via its interaction with NIT4, which is bound at its DNA elements, the dgv mutant NIT2 protein can be recruited to the promoter without binding DNA.
Ability of nit-2 mutants to express nitrate reductase in vivo.
To further examine the effects of NIT2 finger mutations on gene activation, a single copy of each of the various mutant nit-2 genes was integrated at the his3 locus in a his-3 nit-2 rip N. crassa host strain by homologous recombination. The activation of the nitrate assimilation genes by each mutant NIT2 protein was determined by the expression level of nitrate reductase, in comparison with that obtained with the same host strain transformed with the wild-type nit-2+ gene.
Transformants with nit-2 mutant sva, which possesses wild-type DNA-binding activity but is greatly impaired in its interaction with NIT4, showed significantly reduced expression of nitrate reductase—only 16% of that found with nit-2+ (Fig. 4C). More severe reduction in nit-3 expression was seen with mutant sgv, whose protein showed an even weaker NIT4 interaction than that found with sva, and also has a modest loss of DNA binding. The nit-2 mutant v, with an intermediate reduction in NIT4 interaction and normal DNA binding, expressed 50% wild-type activity.
The interesting nit-2 dgv mutant, which carries a gene that encodes a truncated protein with no DNA-binding activity but a normal interaction with NIT4, expressed about 4% of the wild-type level of nitrate reductase (Fig. 4C). As expected, the nit-2 mutants ngpm and sga, which failed to show complementation and exhibited both impaired DNA binding and NIT4 binding, failed to activate nit-3 expression. These combined results demonstrate that the interaction between NIT2 and NIT4 is required for the cooperative activation of nit-3 expression.
Promoter recognition by different Neurospora GATA factors.
In addition to NIT2, several other global regulators, e.g., the white collar factors WC1 and WC2, as well as SRE and NGF1, also possess GATA-type DNA-binding domains (2, 19). WC1 and WC2 govern blue light regulation of carotenoid biosynthesis, and SRE negatively controls the biosynthesis of siderophores which function in iron transport; the function of NGF1 is still uncertain, although it may act in the nitrogen circuit. Thus, at least five distinct Neurospora GATA factors with overlapping DNA binding specificities coexist in the same cells. This feature raises questions as to how each of these GATA factors exerts functional specificity in regulating a distinct set of structural genes. Previous studies have demonstrated that fungal and vertebrate GATA factors recognize DNA elements with the identical GATA core sequence, with little preference to flanking sequences (4, 22). Thus, DNA binding specificity alone apparently cannot provide the functional specificity exerted by each of these factors. We hypothesized that the specificity inherent in regulation of a specific set of target genes by each GATA factor is dependent upon its specific interactions with other regulatory proteins.
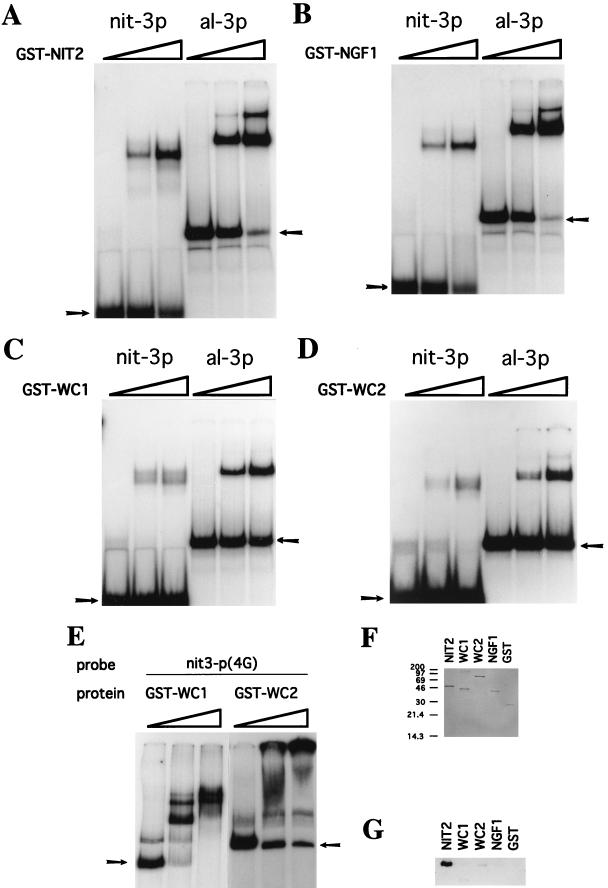
To investigate this proposed mechanism, we first compared the DNA-binding properties of four distinct Neurospora GATA factors. The GST fusion protein containing the NIT2 finger (residues 708 to 822) was tested for in vitro binding to several DNA fragments. The albino-3 (al-3) gene, whose promoter contains GATA elements, is tightly controlled by blue light regulation, mediated by WC1 and WC2. NIT2 plays no role in al-3 expression, which is completely normal in nit-2 mutant strains. Nevertheless, the GST-NIT2 protein recognizes and strongly binds to both the 34-bp nit-3 promoter element (with a single GATA) and the 60-bp al-3 gene promoter element (with two GATAs) (Fig. 5A). Similarly, to examine WC1 and WC2 DNA recognition, GST-WC1(870–980) and GST-WC2(178–530) fusion proteins were expressed in E. coli. WC1 and WC2 clearly bind to both nit-3 and al-3 promoter DNA fragments containing GATA elements (Fig. 5C and D). The fact that WC1 and WC2 contain PAS domains, which are believed to mediate homodimer or heterodimer formation (19), suggests that WC1 and WC2 might preferentially bind to double or multiple GATA sequences. In fact, both GST-WC1 and GST-WC2 bound most strongly to the distal nit-3 promoter fragment containing multiple GATA elements (Fig. 5E), and GST-WC2 was able to form larger DNA-protein complexes when bound to these multiple GATA sites. Furthermore, the GST-NGF1(1–104) fusion protein bound strongly to both nit-3 and al-3 promoters with essentially the same affinities found with NIT2 (Fig. 5A). These results demonstrate that the NIT2, WC1, WC2, and NGF1 proteins all recognize both nit-3 and al-3 promoters with little or no discrimination; i.e., although the DNA binding affinities of these proteins may differ, each protein binds to both the nit-3 and al-3 promoters. WC1 and WC2 bound the nit-3 promoter fragment (Fig. 5E) more strongly than the al-3 promoter (Fig. 5C and D), although these factors regulate al-3 but not nit-3.
FIG. 5.
Recognition of nit-3 and al-3 promoter elements and interaction with NIT4 by different Neurospora GATA factors. (A to E) Mobility shift assays of NIT2 (A), NGF1 (B), WC1 (C and E), and WC2 (D and E). nit-3p and al-3p represent the 34-bp DNA fragment with the single GATA element from the nit-3 promoter and the 60-bp fragment with two GATAs from the albino-3 promoter, respectively. nit-3p(4G) identifies a 350-bp NarI-XbaI DNA fragment from the distal region of the nit-3 promoter which contains four GATA sequences. Approximately 0.07 pmol (about 10,000 cpm) of each 32P-labeled DNA probe was used in each binding assay. The amounts of protein used for the experiments shown are as follows (by panel): A, 0, 30, and 90 ng; B, 0, 0.46, and 1.5 μg; C, 0, 30, and 90 ng; and D, 0, 0.7, and 2.1 μg. The 1:6-diluted proteins were used for panel E; namely, the amounts of protein used were 0, 73, ng and 219 ng for WC1 and 0, 120, and 360 ng for WC2. Arrows in each panel identify the free DNA probe. (F and G) NIT2, but not other GATA factors, specifically interacts with NIT4. The same GST fusion proteins of the different GATA factors used in the mobility shift experiments were used to test for binding to NIT4. A total of 4.5 μg of each GST fusion protein was immobilized on 150 μl of the affinity resin; 3 μg of NIT4 (OF12) was added, and following extensive washing, bound proteins were eluted with glutathione. (F) SDS-polyacrylamide gel stained with Coomassie blue of eluents from protein binding assays to ensure equal loading. GST alone was the negative control. (G) Western blot of the gel shown in panel F to detect NIT4. Anti-NIT4 serum was used as the primary antibody.
The Neurospora NIT2 GATA factor alone recognizes NIT4.
If DNA promoter recognition by GATA factors is not sufficient to determine their functional specificities, their specific regulatory functions might be dictated by protein-protein interactions (i.e., cooperative activation with other transactivators). This hypothesis implies that the nitrate assimilation pathway-specific protein NIT4 should specifically interact only with the NIT2 zinc finger motif, and not with the fingers in WC1, WC2, or NGF1. To directly test this concept, GST-WC1, GST-WC2, GST-NGF1, and GST-NIT2 fusion proteins were each immobilized to glutathione-agarose and incubated with the NIT4 OF12 protein. After extensive washing and elution, any retained NIT4 protein was detected by Western blotting (Fig. 5F and G). NIT2-GST consistently exhibited strong binding to NIT4 (OF12). In contrast, the WC1, WC2, and NGF1 fusion proteins showed either a barely detectable interaction or no interaction with NIT4, indicating that the interaction between NIT2 and NIT4 is very specific. It should be noted that these experiments were conducted with truncated proteins, because of poor expression of the full-length proteins. Thus, it is formally possible that other regions of the proteins might contribute to DNA binding specificity or interaction with NIT4, although we consider this quite unlikely.
DISCUSSION
NIT2, a member of the GATA family of transcription factors, is a globally acting nitrogen regulatory protein that positively activates the expression of many nitrogen catabolic genes. We have presented results which demonstrate that a specific protein-protein interaction of NIT2 with the nitrate assimilatory pathway-specific factor NIT4 is required for full expression of nit-3, the structural gene that encodes nitrate reductase. Perhaps surprisingly, the regions of NIT2 and NIT4 which contain their respective DNA-binding motifs also mediate the specific binding interaction between these two proteins. A critical observation was the finding that several NIT2 mutant proteins with amino acid substitutions in the zinc finger region that possessed wild-type DNA-binding activity but a clearly reduced interaction with NIT4 in vitro also showed significant loss of function in vivo in activation of the nitrate reductase gene. Thus, it now appears clear that sequence-specific DNA binding of NIT2 and of NIT4 to their respective recognition elements in the nit-3 promoter and a specific interaction between these two transcription factors are both required for their synergistic activation of the nitrate reductase structural gene. A reduction or loss of either DNA binding for either factor or of the NIT2-NIT4 protein-protein interaction leads to a significant decline in nit-3 expression in vivo. The ability to recognize cognate DNA sites is critically important for specific gene activation by NIT2. Although it appears that NIT4 can recruit a truncated NIT2 protein devoid of DNA-binding activity to the nit-3 promoter, gene activation in this case is very weak.
Neurospora possesses at least five different GATA factors which serve to regulate distinct major areas of cell function, and yet these factors, as demonstrated here, overlap in their DNA-binding activities. The finding that only NIT2, of the several GATA factors we examined, is capable of specifically binding to NIT4 and that the NIT2-NIT4 interaction is essential for activation of the highly regulated nit-3 gene in vivo is very instructive. These features imply that specific interactions that occur between DNA-bound proteins in the context of a promoter represent a molecular mechanism that allows each GATA factor to act only at its own set of target genes; moreover, these specific protein-protein interactions may be critical to provide synergy between the factors for gene activation.
An intriguing question is why does the strong synergy between NIT2 and NIT4 require their physical interaction. It is possible that the NIT2-NIT4 interaction is required for their efficient DNA binding in vivo, even though in vitro both NIT2 and NIT4 individually bind strongly to their cognate DNA sites. In vivo, the promoters are packed as nucleosomes in highly ordered structures, along with accessory chromatin proteins, and thus may not be readily accessible to DNA-binding proteins. A physical interaction between transactivators may be required to overcome a general repression mechanism to form a stable transcription initiation complex. A requirement that NIT2 and NIT4 must interact for productive DNA binding in vivo would allow the cellular content of NIT2 to be used efficiently, since nonproductive binding to GATA elements throughout the genome would not occur. An alternative possibility is that the NIT2-NIT4 interaction is not required for the binding of these factors to their cognate elements but rather is required for their effective communication with each other and with the general transcriptional machinery, possibly to overcome repression by ubiquitous chromatin proteins that otherwise maintain the genes in an inactive state (16, 32).
The concept that highly specific interactions occur between regulatory proteins can help explain how each of the multiple GATA factors in higher organisms mediates tissue- and cell-specific gene activation (3, 14, 23, 24). At least six GATA factors occur in mammalian species, including human beings (1, 17). A new multitype zinc finger protein, FOG (friend of GATA-1), has been shown to synergize with GATA-1 to activate transcription of hematopoietic-specific genes (29). GATA-1 appears to interact with at least three other proteins, FOG, Sp1, and EKLF (23, 29), and GATA-2 interacts with the bZip factors JUN and FOS (15). Such interactions can provide considerable specificity; e.g., C/EBPβ, but not the closely related C/EBPα factor, can synergize with the Sp1 protein (18). The ability of transcription factors to participate in functional protein-protein interactions appears to be important for gene regulation in a wide range of organisms, including fungi, other lower eukaryotes, and higher plants and animals.
It is of particular significance that NIT2 of Neurospora and the homologous GATA factor, AREA, of Aspergillus are responsible for activation of entire sets of genes of different pathways that encode nitrogen catabolic enzymes. The structural genes of the distinct pathways for nitrate assimilation, purine metabolism, amino acid metabolism, protein catabolism, and acetamide utilization all require a functional NIT2 (or AREA) protein for expression. Expression of the genes of each metabolic pathway also requires activation by a pathway-specific factor that mediates induction of just the particular pathway. Thus, the Aspergillus AREA protein and NIRA, the Aspergillus equivalent of NIT4, are both needed to turn on the nitrate assimilatory genes, whereas AREA and a distinct factor, UAY, are both required to activate purine catabolic genes; similarly, AREA and PRNA are needed for proline catabolic gene expression. Nearly all of the pathway-specific factors of the nitrogen regulatory circuit of Aspergillus and Neurospora are members of the GAL4 family of fungal regulatory proteins. It is tempting to speculate that a functional interaction of NIT2 (or AREA) with each of the pathway-specific factors is essential for the cooperative activation of the distinct sets of structural genes. If this concept proves to be correct, the global regulatory proteins NIT2 and AREA are remarkable in their ability to interact with an entire series of pathway-specific factors to integrate the expression of distinct sets of genes which lie within the realm of nitrogen metabolism.
ACKNOWLEDGMENTS
We thank Giuseppe Macino and Paola Ballario, University of Rome, who generously provided expression vectors for GST-WC1 and GST-WC2 and al-3 promoter DNA.
This work was supported by grant GM23367 from the National Institutes of Health.
REFERENCES
- 1.Arceci R J, King A A J, Simon M C, Orkin S H, Wilson D B. Mouse GATA-4: a retinoic acid-inducible GATA-binding transcription factor expressed in endodermally derived tissues and heart. Mol Cell Biol. 1993;13:2235–2246. doi: 10.1128/mcb.13.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballario P, Vittorioso P, Magrelli A, Talora C, Cabibbo A, Macino G. White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J. 1996;15:1650–1657. [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng X, Reginato M J, Andrews N C, Lazar M A. The transcriptional integrator CREB-binding protein mediates positive cross talk between nuclear hormone receptors and the hematopoietic bZip protein p45/NF-E2. Mol Cell Biol. 1997;17:1407–1416. doi: 10.1128/mcb.17.3.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiang T Y, Marzluf G A. DNA recognition by the NIT2 nitrogen regulatory protein: importance of the number, spacing, and orientation of GATA core elements and their flanking sequences upon NIT2 binding. Biochemistry. 1994;33:576–582. doi: 10.1021/bi00168a024. [DOI] [PubMed] [Google Scholar]
- 5.Chiang T-Y, Marzluf G A. Binding affinity and functional significance of NIT2 and NIT4 binding sites in the promoter of the highly regulated nit-3 gene, which encodes nitrate reductase in Neurospora crassa. J Bacteriol. 1995;177:6093–6099. doi: 10.1128/jb.177.21.6093-6099.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiang T Y, Rai R, Cooper T G, Marzluf G A. DNA binding site specificity of the Neurospora global nitrogen regulatory protein NIT2: analysis with mutated binding sites. Mol Gen Genet. 1994;245:512–516. doi: 10.1007/BF00302264. [DOI] [PubMed] [Google Scholar]
- 7.Crosthwaite S K, Dunlap J C, Loros J J. Neurospora wc-1 and wc-2: transcription, photoresponses, and the origins of circadian rhythmicity. Science. 1997;276:763–769. doi: 10.1126/science.276.5313.763. [DOI] [PubMed] [Google Scholar]
- 8.Davis R H, deSerres F J. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 1970;17A:79–143. [Google Scholar]
- 9.Ebbole D. Vectors for construction of translational fusions to β-galactosidase. Neurospora Newsl. 1990;37:15–16. [Google Scholar]
- 10.Exley G E, Colandene J D, Garrett R H. Molecular cloning, characterization, and nucleotide sequence of nit-6, the structural gene for nitrite reductase in Neurospora crassa. J Bacteriol. 1993;175:2379–2392. doi: 10.1128/jb.175.8.2379-2392.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Feng, B., H. Haas, and G. A. Marzluf. Unpublished observations.
- 11.Fu Y H, Feng B, Evans S, Marzluf G A. Sequence-specific DNA binding by NIT4, the pathway-specific regulatory protein which mediates nitrate induction in Neurospora. Mol Microbiol. 1995;15:935–942. doi: 10.1111/j.1365-2958.1995.tb02362.x. [DOI] [PubMed] [Google Scholar]
- 12.Fu Y H, Marzluf G A. Molecular cloning and analysis of the regulation of nit-3, the structural gene for nitrate reductase in Neurospora crassa. Proc Natl Acad Sci USA. 1987;84:8243–8247. doi: 10.1073/pnas.84.23.8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrett R H, Cove E J. Formation of NADPH-nitrate reductase activity in vitro from Aspergillus nidulans niaD and cnx mutants. Mol Gen Genet. 1967;149:179–186. doi: 10.1007/BF00332887. [DOI] [PubMed] [Google Scholar]
- 14.Kaushal S, Schneider J W, Nadal-Ginard B, Mahdavi V. Activation of the myogenic lineage by MEF2A, a factor that induces and cooperates with myoD. Science. 1994;266:1236–1240. doi: 10.1126/science.7973707. [DOI] [PubMed] [Google Scholar]
- 15.Kawana M, Lee M-E, Quertermous E E, Quertermous T. Cooperative interaction of GATA-2 and AP1 regulates transcription of the endothelin-1 gene. Mol Cell Biol. 1995;15:4225–4231. doi: 10.1128/mcb.15.8.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keleher C A, Redd M, Schultz J, Carlson M, Johnson A D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- 17.Ko L J, Engel J D. DNA-binding specificities of the GATA transcription factor family. Mol Cell Biol. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee Y-H, Williams S C, Baer M, Sterneck E, Gonzalez F J, Johnson P F. The ability of C/EBPβ but not C/EBPα to synergize with an Sp1 protein is specified by the leucine zipper and activation domain. Mol Cell Biol. 1997;17:2038–2047. doi: 10.1128/mcb.17.4.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linden H, Macino G. White collar 2, a partner in blue light signal transduction, controlling expression of light-regulated genes in Neurospora crassa. EMBO J. 1997;16:98–107. doi: 10.1093/emboj/16.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marzluf G A. Regulation of sulfur and nitrogen metabolism in filamentous fungi. Annu Rev Microbiol. 1993;47:31–55. doi: 10.1146/annurev.mi.47.100193.000335. [DOI] [PubMed] [Google Scholar]
- 21.Marzluf G A. Genetic regulation of nitrogen metabolism in the fungi. Microbiol Mol Biol Rev. 1997;61:17–32. doi: 10.1128/mmbr.61.1.17-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merika M, Orkin S H. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merika M, Orkin S H. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Krüppel family proteins SP1 and EKLF. Mol Cell Biol. 1995;15:2437–2447. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molkentin J D, Black B L, Martin J F, Olson E N. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 25.Omichinski J G, Clore G M, Schaad O, Felsenfeld G, Trainor C, Appella E, Stahl S J, Gronenborn A M. NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science. 1993;261:438–446. doi: 10.1126/science.8332909. [DOI] [PubMed] [Google Scholar]
- 26.Orkin S H. GATA-binding transcription factors in hematopoietic cells. Blood. 1992;80:575–581. [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 28.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in E. coli as fusion with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 29.Tsang A P, Visvader J E, Turner C A, Fujiwara Y, Yu C, Weiss M J, Crossley M, Orkin S H. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- 30.Vollmer S J, Yanofsky C. Efficient cloning of genes of Neurospora crassa. Proc Natl Acad Sci USA. 1986;83:4869–4873. doi: 10.1073/pnas.83.13.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao X, Fu Y H, Marzluf G A. The negative-acting NMR regulatory protein of Neurospora crassa binds to and inhibits the DNA-binding activity of the positive-acting nitrogen regulatory protein NIT2. Biochemistry. 1995;34:8861–8868. doi: 10.1021/bi00027a038. [DOI] [PubMed] [Google Scholar]
- 32.Yamashiro C T, Ebbole D J, Lee B-U, Brown R E, Bourland C, Madi L, Yanofsky C. Characterization of rco-1 of Neurospora crassa, a pleiotropic gene affecting growth and development that encodes a homolog of Tup1 of Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:6218–6228. doi: 10.1128/mcb.16.11.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young J L, Jarai G, Fu Y H, Marzluf G A. Nucleotide sequence and analysis of nmr, a negative-acting regulatory gene in the nitrogen circuit of Neurospora crassa. Mol Gen Genet. 1990;222:120–128. doi: 10.1007/BF00283032. [DOI] [PubMed] [Google Scholar]
- 34.Yuan G-F, Fu Y-H, Marzluf G A. nit-4, a pathway-specific regulatory gene of Neurospora crassa, encodes a protein with a putative binuclear zinc DNA-binding domain. Mol Cell Biol. 1991;11:5735–5745. doi: 10.1128/mcb.11.11.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou, L. W., H. Haas, and G. A. Marzluf. Submitted for publication.