Abstract
The function(s) and RNA binding properties of vigilin, a ubiquitous protein with 14 KH domains, remain largely obscure. We recently showed that vigilin is the estrogen-inducible protein in polysome extracts which binds specifically to a segment of the 3′ untranslated region (UTR) of estrogen-stabilized vitellogenin mRNA. In order to identify consensus mRNA sequences and structures important in binding of vigilin to RNA, before vigilin was purified, we developed a modified in vitro genetic selection protocol. We subsequently validated our selection procedure, which employed crude polysome extracts, by testing natural and in vitro-selected RNAs with purified recombinant vigilin. Most of the selected up-binding mutants exhibited hypermutation of G residues leading to a largely unstructured, single-stranded region containing multiple conserved (A)nCU and UC(A)n motifs. All eight of the selected down-binding mutants contained a mutation in the sequence (A)nCU. Deletion analysis indicated that approximately 75 nucleotides are required for maximal binding. Using this information, we predicted and subsequently identified a strong vigilin binding site near the 3′ end of human dystrophin mRNA. RNA sequences from the 3′ UTRs of transferrin receptor and estrogen receptor, which lack strong homology to the selected sequences, did not bind vigilin. These studies describe an aproach to identifying long RNA binding sites and describe sequence and structural requirements for interaction of vigilin with RNAs.
The steps between gene transcription and mRNA translation, which include nuclear RNA processing, mRNA trafficking, and cytoplasmic mRNA degradation, are increasingly seen as important regulatory sites in diverse cellular processes (5, 6, 52). Many of these steps in mRNA metabolism appear to be regulated by RNA binding proteins containing K-homology (KH) domains (14). While a detailed picture of KH-domain–RNA interaction is not yet available, in several cases proteins containing KH domains have been shown to bind RNA (13, 14, 21, 57). Some KH-domain proteins are clinically significant, including FMR1 protein (57), which is involved in fragile X syndrome, the major cause of heritable human mental retardation, and Nova-1, which is important in the motor control disorder paraneoplastic opsoclonus-ataxia (12, 13). KH-domain proteins which affect nuclear RNA splicing include Mer1, SF1, and PSI (1, 46, 55), while the α-poly(C) binding protein plays a role in the cytoplasmic stability of globin mRNA (68). Prokaryotic KH-domain proteins are highly diverse and include NusA, polyribonucleotide:orthophosphate nucleotidyltransferase, CsrA, and ribosomal protein S3 (27, 40, 58). Since the RNA binding sites and mechanisms of action of many KH-domain proteins remain obscure, the question of whether KH-domain proteins can preferentially recognize and bind to specific RNA binding sites remained unresolved. In an important recent paper, Buckanovich and Darnell used in vitro genetic selection with purified recombinant Nova-1 to identify RNAs which bind Nova-1 with high affinity. Nova-1 and inhibitory glycine receptor α2 RNAs were then shown to contain consensus binding sites which bind to Nova-1 through a KH domain (12).
One important but little-understood KH-domain protein is vigilin. Vigilin initially was cloned from chicken chondrocytes and then was shown to be up-regulated in rapidly dividing cells and to be present in all vertebrate cell lines tested, in nematodes, and perhaps in yeast (35, 47, 49, 50, 54, 69). With its 14 KH domains, vigilin has been used as a model protein for the KH domains in FMR1 and other proteins in a seminal study solving the structure of a vigilin KH domain uncomplexed to RNA (45). However, the intracellular RNA targets of this ubiquitous, model protein and its functions remained elusive.
We had shown that estrogen specifically stabilizes the hepatic mRNA coding for the egg yolk precursor protein vitellogenin, increasing its half life from 16 to 500 h (10). The estrogen-mediated stabilization of vitellogenin mRNA requires estrogen receptor (48) and association of the mRNA with polysomes (9) and involves the 3′ untranslated region (UTR) of the mRNA (48). We identified an estrogen-inducible protein which binds specifically to a segment of the vitellogenin mRNA 3′ UTR (25). We recently unambiguously identified this protein as Xenopus vigilin (26). Our observations that the protein we now refer to as vigilin is found in several human cell lines and in several Xenopus cell lines and tissues and that testosterone induces vigilin in Xenopus muscle suggested a wider role for vigilin in eukaryotic mRNA metabolism (24). Despite the wide distribution of vigilin, binding sites in mRNAs other than vitellogenin mRNA had not been identified.
To allow prediction of vigilin binding sites in human mRNAs, we first defined the vigilin binding site by using iterative in vitro genetic selection. In vitro genetic selection or systematic evolution of ligands by exponential enrichment has been used to identify RNA (and DNA) ligands which interact with proteins or small molecules (28). Although an early study (8) and several subsequent studies used DNA gel mobility shifts to separate free and protein-bound DNA, in vitro genetic selection with an RNA gel mobility shift has not been described. RNA binding sites have been identified for several proteins by using in vitro genetic analysis (11, 12, 15, 31, 39, 56, 62, 63, 66). Those studies used purified RNA binding proteins. For example, in early studies, in vitro selection of RNA binding sites was used to identify the structure of the human immunodeficiency virus (HIV) Rev binding site (3) and the binding preferences of splicing factors (31, 56, 62), hnRNPs (15, 29), and the autoimmune RNA binding protein Hel-N1 (39). Using in vitro genetic selection of gel-shifted RNA-protein complexes, we have extended this technique to proteins present in relatively crude extracts. Since this work was carried out in parallel with studies aimed at the identification of the vitellogenin mRNA 3′ UTR binding protein, we had to employ a relatively crude polysome extract. We first selected RNA binding sites with an increased affinity for vigilin. We then used an RNA selected for increased binding to vigilin to carry out a novel in vitro selection for random point mutations that markedly reduced binding of vigilin.
Together with deletion analysis and RNA footprinting, the in vitro genetic analysis enabled us to identify structural and sequence elements important for the interaction of vigilin with RNA. Using this information, we scanned the human sequence database and predicted and subsequently identified a strong vigilin binding site near the 3′ end of human dystrophin mRNA. We then analyzed binding of purified recombinant human vigilin to the selected up- and down-binding RNAs and demonstrated that vigilin exhibits strong preferences for binding to specific RNAs.
MATERIALS AND METHODS
Construction of DNA template pools.
A 116-base oligonucleotide that contained 106 bases of the 3′ UTR of the Xenopus vitellogenin B1 mRNA was synthesized. Phosphoramidite mixtures were used to introduce point mutations at a frequency of 36% per position in the 70-nucleotide region shown in Fig. 3 (wild-type sequence). The synthetic oligonucleotides were purified on a 12% polyacrylamide gel and amplified by five cycles of PCR with 5′-PR1 (5′-CCGAATTCTAATACGACTCACTATAGGGAGTCTCTATATCTCTATCAA-3′) and 3′-PR2 (5′-CGCGGGATCCAAATTGAATTGTTTACAA-3′) primers to obtain an initial pool, pool 0. The templates for in vitro transcription for pool 0 consisted of >1013 unique, double-stranded DNA fragments which contained a T7 RNA polymerase initiation sequence (boldface in the 5′-PR1 primer sequence) and restriction enzyme sites on both the 5′ and 3′ ends (EcoRI and BamHI sites in the 5′-PR1 and 3′-PR2 primers, respectively) (shown in italics), as well as 18 flanking nucleotides on the 5′ and 3′ sides of the 70-nucleotide central region (underlined nucleotides in the 5′-PR1 and 3′-PR2 primer sequences, respectively).
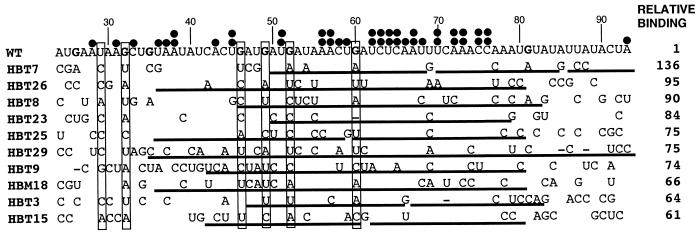
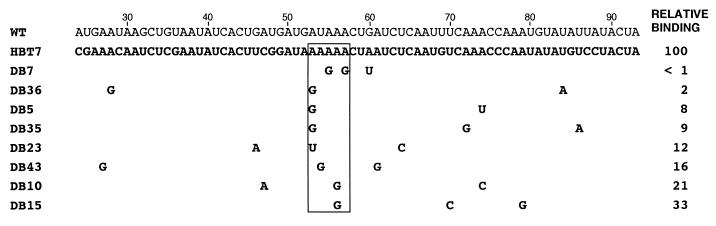
FIG. 3.
Mutation of specific Gs is common in up-binding mutants. The mutations in 70 nucleotides of the 10 strongest up-binders are shown. HBM18 is from pool 3, and the other nine mutants are from pool 10. Highly mutated nucleotides (>80%) are boxed. Gs in the wild-type (WT) sequence are shown in boldface. Two filled circles are over each strongly conserved nucleotide (more than 90% conservation), and one filled circle is above each conserved nucleotide (more than 80% conservation). G-free tracts are underlined. Wild-type RNA relative binding was set equal to 1. Relative binding is the ratio of binding of the mutant to that of the wild-type RNA. Numbers indicate the nucleotide position (the transcription start site is set to 1).
For selection of RNAs exhibiting reduced binding to vigilin (down-binders), the highest binder from the selection for up-binders, HBT7, was used as the starting material, and the primers described above were used to create the starting pool by employing two rounds of mutagenic PCR (30 cycles each) as described previously (16). The expected mutation frequency was 1.2% (an average of 0.8 nucleotide in 70 bases) without a strong bias with respect to the type of base substitution.
In vitro selections by RNA gel mobility shifts.
Extracts containing vigilin were a salt wash of polysomes from estrogen-treated Xenopus liver prepared as previously described (25).
RNAs were transcribed in vitro by using T7 RNA polymerase and templates either from PCR products or from plasmids linearized with BamHI and were labeled to a specific activity of 109 cpm/μg with [α-32P]UTP (ICN; 3,000 Ci/mmol). Incubations for the RNA gel mobility shift assays were carried out for 1 h on ice in a 10-μl total volume and the mixtures typically contained 8 U of RNasin, 10 μg of yeast tRNA, 10 μg of heparin, 0.1 ng of [32P]RNA, 1 to 2 μg of polysome extract, 10 mM Tris-HCl (pH 7.6), 1 mM Mg acetate (MgOAc), 1 mM EDTA, and 70 mM KCl (25). To select strong up-binding mutants in selection cycles 4 to 10, the following RNAs were used as nonspecific competitors: cycles 4 and 5, 50 μg of tRNA; cycles 6 to 9, 1 μg of Escherichia coli rRNA plus 10 μg of tRNA; and cycle 10, 5 μg of rRNA plus 10 μg of tRNA. The samples were then loaded on a 4% polyacrylamide gel (80:1 acrylamide-bisacrylamide in a low-ionic-strength buffer [6.7 mM Tris-HCl {pH 7.9}, 3.3 mM NaOAc, 1 mM EDTA]) and run at 300 V at 4°C. Either gels were visualized by autoradiography with X-ray film or gels were analyzed and the bands were quantitated with a Molecular Dynamics PhosphorImager. Relative binding was determined as the ratio of the intensity of the gel-shifted band to that of the gel-shifted wild-type band except where noted.
RNAs in shifted (up-binders) or unshifted (down-binders) bands were recovered from polyacrylamide gel slices and incubated in the elution buffer (0.5 N NH4OAc, 10 mM MgCl2, 0.5% sodium dodecyl sulfate [SDS], 1 mM EDTA) for 2 h at room temperature. The supernatants were phenol-chloroform extracted and ethanol precipitated.
The RNAs recovered after the selections were reverse transcribed by using the PR2 primer and PCR amplified by adding PR1 primer. The PCR products were gel purified for subsequent selection or digested with EcoRI and BamHI to be subcloned into the pUC19 plasmid.
Determination of the minimum length of RNA required for efficient binding by vigilin.
HBT7 RNA was end labeled at the 5′ end with [γ-32P]ATP by using T4 polynucleotide kinase, gel purified, and partially digested under mild alkaline conditions. To obtain an RNA ladder, the end-labeled RNA was incubated in 25 mM sodium bicarbonate for 20 min at 90°C. The digestion products were incubated for 1 h on ice with or without vigilin as described above for the RNA gel mobility shift. The reaction mixtures contained 50 μg of yeast tRNA and 50 μg of heparin in 50-μl volumes. The reaction mixture was filtered onto nitrocellulose, and RNAs retained with the protein on the filter were recovered as described previously (65). Briefly, the RNAs were incubated at 65°C in 0.2 ml of 7 M urea–3 mM EDTA–100 mM sodium citrate (pH 5.0) for 5 min, and RNAs were recovered by phenol-chloroform extraction and ethanol precipitation. The RNAs were then dissolved in gel loading buffer (8 M urea, 20 mM Tris [pH 7.9], 1 mM EDTA), boiled for 90 s, chilled on ice, and loaded on an 8% polyacrylamide gel containing 7 M urea next to RNA sequencing ladders made by chemical modification (diethyl pyrocarbonate and hydrazine-chloride).
HBT7/83 RNA (HBT7 RNA lacking nucleotides 84 to 116) was synthesized by in vitro transcription from a template DNA PCR amplified from HBT7 template by using the PR1 primer and HBT7/83 primer (5′-TATATTGGGTTTGACATTGA-3′). The HBT7/83 RNA was 3′ end labeled with 5′-32P-labeled cytidine 3′,5′-bisphosphate by using RNA ligase and subjected to the same analysis as the 5′-end-labeled HBT7 RNA.
RNA footprinting.
End-labeled RNAs were incubated for 1 h on ice either with extract containing vigilin (3 μg) or with buffer as described above, except a reaction volume of 30 μl and 30 μg of tRNA plus 30 μg of heparin were used. The incubation mixtures were then digested with α-sarcin. α-Sarcin was expressed and purified from E. coli and was a kind gift from A. Martínez del Pozo, Universidad Complutense de Madrid, Madrid, Spain (37). α-Sarcin was added to a final concentration of 5 μM, and the mixture was incubated for 15 min at 30°C, phenol-chloroform extracted, and ethanol precipitated. The α-sarcin digestion products were dissolved in the gel loading buffer described above and resolved by electrophoresis on 8% polyacrylamide gels containing 7 M urea.
In vitro transcription and translation and purification of human vigilin.
PCRs were performed to amplify a region of the human vigilin cDNA spanning nucleotides 73 to 3947 (GenBank accession no. M64098). The 5′ primer (5′-AAGCTTTAATACGACTCACTATAGGGAGGCGGCCTCAGGACGG-3′) was designed to incorporate a T7 promoter (boldface) upstream of the start codon. The 3′ primer (5′-GGGAAGCTTATTTATCGTCATCGTCTTTGTAGTCCCAAG GGAGGGTCTTGG-3′) was designed to incorporate an in-frame FLAG peptide sequence and a stop codon (underlined) on the 3′ end of the coding sequence of human vigilin at amino acid residue 1264. The vigilin-FLAG cDNA was amplified by using the LA PCR kit (Takara) from the plasmid pM306N containing a full-length human vigilin clone (43).
In vitro transcription-translation was performed according to the supplier’s instructions for a 1.4-ml reaction mixture by using a T7 coupled reticulocyte lysate system (TnT; Promega). The PCR-amplified vigilin-FLAG cDNA was used as the template. The in vitro-translated FLAG-tagged product was dialyzed against BC100 (20 mM Tris-HCl [pH 7.9], 20% glycerol, 0.2 mM EDTA, 100 mM KCl, 1 mM dithiothreitol, and 0.5 mM phenylmethylsulfonyl fluoride), purified by binding to anti-FLAG M2 agarose (Eastman Kodak Co.), and eluted by using the FLAG peptide (final volume, 200 μl) as described previously (20).
RNA gel mobility shift assays were carried out under the same conditions described above for assays with crude vigilin-containing extracts. One to two microliters of the elutant from the FLAG affinity purification was added to each reaction mixture. In the antibody supershift experiments, 1 μg of anti-FLAG M2 monoclonal antibody (Eastman Kodak Co.) or 1 μg of anti-human estrogen receptor H222 antibody (41) was added to the mixture in the absence of RNA probe, and the reaction mixture was incubated for 30 min on ice. The RNA probe was then added, and the reaction mixture was incubated for an additional 45 min before gel analysis.
Analysis of human mRNA 3′ UTR binding to vigilin.
The human dystrophin cDNA clone p9-13, which contains the dystrophin 3′ UTR, was a kind gift from L. M. Kunkel. A portion of the 3′ UTR was amplified by PCR with 5′-DYS1 (5′-GAATTCATTTAGGTGACACTATAGAACATTTACGAATTATTT-3′) and 3′-DYS2 (5′-AGTAAAGCAGTACTATAA-3′) primers to obtain a dystrophin 3′ UTR template (DYS). A 106-mer oligonucleotide (5′-AACATTTACA AATTATTTTTGTAAACTTCAGTTTTACTGCATTTTCGCAACATATCAT ACTTCACCAAGTATATGCCTTACTATTATATTATAGTACTGCTTTAC T-3′) was synthesized and amplified by PCR with 5′-mDYS (5′-GAATTAATTTAGGTGACACTATAGAACATTTACAAATTATTT-3′) and 3′-DYS2 primers to obtain a template for mutated dystrophin 3′ UTR (mDYS) (mutated nucleotides from dystrophin cDNA are shown in boldface). DYS and mDYS templates were in vitro transcribed with SP6 RNA polymerase (the SP6 promoter is underlined) and used in RNA gel shift assays. The 107-nucleotide transcripts contain 106 nucleotides of human dystrophin mRNA 3′ UTR (GenBank accession no. M18533; nucleotides 13820 to 13925) plus one extra G residue at the 5′ end.
Similarly, the human estrogen receptor cDNA 3′ UTR (30) and human transferrin receptor cDNA 3′ UTR (36) were PCR amplified to obtain templates for in vitro transcription with 5′-ER primer (5′-GAATTCATTTAGGTGACACTATAGCAGCTTTGCTTTGTTTA-3′) and 3′-ER primer (5′-AATAGGTTGAGAAAATTG-3′) for human estrogen receptor and 5′-TfR primer (5′-GAATTCATTTAGGTGACACTATAGTCACAATGGTAACACATTA-3′) and 3′-TfR primer (5′-CATATGGAGATCACTGTCTC-3′) for human transferrin receptor. PCR templates were in vitro transcribed with SP6 polymerase (the SP6 promoter is underlined) to generate the 106-nucleotide human estrogen receptor 3′ UTR RNA (GenBank accession no. X03635, nucleotides 6320 to 6425) and the 108-nucleotide human transferrin receptor 3′ UTR RNA (GenBank accession no. X01060, nucleotides 3869 to 3976), respectively. The labeled human estrogen receptor and human transferrin receptor RNAs were used as probes in RNA gel mobility shift assays carried out with purified recombinant vigilin as described above.
Computer analysis.
Computer programs used for the nucleotide sequence database search and RNA secondary structure analysis were from the University of Wisconsin Genetics Computer Group package. The GenBank (release 91.0) and EMBL (release 42.0) nucleotide sequence databases were scanned with the program FASTA for nucleotide sequences homologous to HBT7 RNA. RNA secondary structures at 37°C were calculated by using the MFOLD program (70) and drawn by means of the loopDloop program of D. G. Gilbert (a Macintosh program for visualizing RNA secondary structure, published electronically on the Internet and available via anonymous ftp to ftp.bio.indiana.edu). The free energy of each RNA mutant at 4°C was calculated by using the MFOLD program.
RESULTS
Selection and identification of mutant RNAs exhibiting enhanced binding to vigilin.
Mutagenesis and in vitro genetic selections were carried out on a 116-nucleotide mRNA segment which contains the vigilin binding site (25). This RNA sequence ends 2 nucleotides upstream of the vitellogenin mRNA polyadenylation signal (Fig. 1, top). A pool of variants was generated by using doped oligonucleotides to mutate the central section at a frequency of approximately 36%. This level of mutation allows for the generation of a large mutant pool, in which the mutants retain some resemblance to the wild-type sequence (3). Sequencing of 10 of the resulting pool 0 mutants showed that they exhibited an overall mutation rate of 35%, which is very close to the predicted rate of 36%. The overall base composition of the 10 pool 0 mutants was very similar to that predicted for this mutation frequency (Table 1).
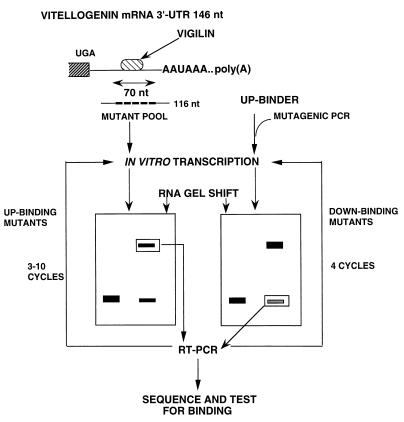
FIG. 1.
Scheme for the in vitro genetic selection of RNAs with altered affinity for vigilin. The 6-kb Xenopus laevis vitellogenin B1 mRNA consists of a very short 5′ UTR (12 nucleotides [nt]), an approximately 6-kb coding region, and a 146-nucleotide 3′ UTR which contains a vigilin binding site (top). For the selection of up-binding mutants (left), we mutated the central 70 nucleotides of a 116-nucleotide 3′ UTR region (see Fig. 3 for the RNA sequence). After selection by RNA gel mobility shift assay for 3 and 10 cycles, individual clones were isolated, sequenced, and assayed for binding. For the selection of mutants exhibiting reduced binding to vigilin (right), mutagenic PCR was used to introduce point mutations into DNA encoding the highest-affinity up-binding mutant, HBT7. After four cycles of selection, individual clones were sequenced and analyzed. RT, reverse transcription.
TABLE 1.
Nucleotide compositions of the up-binding RNAs
| RNA | No. of nucleotidesa
|
|||
|---|---|---|---|---|
| G | A | U | C | |
| Wild type | 8 | 29 | 23 | 10 |
| Mutants | ||||
| Predicted | 13 | 23 | 20 | 14 |
| Actual | ||||
| Pool 0 (n = 10) | 12 | 24 | 21 | 13 |
| Pool 3 (n = 38) | 9 | 24 | 21 | 16 |
| Pool 10 (n = 20) | 5 | 23 | 20 | 20 |
The average number of times that each nucleotide occurs in the 70-nucleotide sequence is indicated. To generate the starting pool (pool 0), at each nucleotide there was a 12% probability of mutation to each of the other three nucleotides (a total mutation rate of 36% was predicted, and a 35% rate was obtained in pool 0). The nucleotide composition of pool 0 therefore differs from the wild type.
The scheme for in vitro genetic selection of RNAs binding to vigilin is shown schematically in Fig. 1. The pool of RNAs used in the initial selection (pool 0) contained approximately 109 independent mutant RNAs. RNAs in the original pool which bound to vigilin were recovered from a narrow gel-shifted band with the same mobility as the original vitellogenin mRNA binding site-vigilin complex (Fig. 2A). The gel-shifted band was reverse transcribed, amplified by PCR, and in vitro transcribed to generate a new pool of RNAs, pool 1. To select RNA sequences binding to vigilin and not to other RNA binding proteins in the relatively crude polysomal salt wash, we carried out the RNA gel shift assays under stringent conditions favoring vigilin-RNA complexes. To suppress binding to the mutant RNAs by other proteins, all of the selections contained at least a 100,000-fold excess of tRNA. Selection cycles 4 and 5 contained a 500,000-fold excess of tRNA, while cycles 6 to 10 contained a 100,000-fold excess of tRNA plus an increasing amount of rRNA.
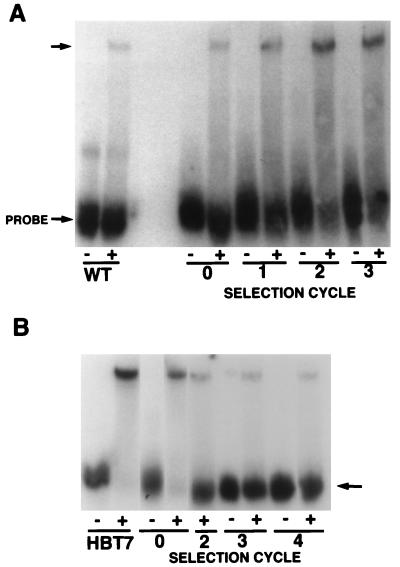
FIG. 2.
In vitro genetic selection of RNAs exhibiting increased (A) and reduced (B) binding to vigilin. RNA gel mobility shift assays were performed with different pools of RNAs. (A) Selection cycles 0, 1, 2, and 3 for up-binding mutants or wild-type (WT) RNA with (+) or without (−) vigilin-containing polysome extracts. The upper arrow indicates the RNA-vigilin complex. (B) The DNA encoding the most efficient up-binder (HBT7) was subjected to two mutational PCR cycles to generate the starting pool (cycle 0) for selection of down-binding mutants. Unshifted RNAs from selection cycles 0, 2, 3, and 4 as well as HBT7 RNA after incubation with (+) or without (−) extracts are indicated by an arrow.
Figure 2A demonstrates the progressive enrichment of up-binding RNAs in the pools as the result of the selection. After three selection cycles, the gel-shifted band showed a significant increase in intensity relative to wild-type RNA. The use of even more stringent selection conditions in cycles 4 to 10 (see above) resulted in an additional and progressive increase in the intensity of the gel-shifted band through 10 selection cycles (data not shown).
Fifty-eight clones were isolated from pools 3 and 10, subcloned, sequenced, and analyzed for RNA binding under stringent binding conditions (see Materials and Methods). The RNA sequences of the 10 mutants with the highest relative affinities for vigilin are presented in Fig. 3. It is striking that one of the mutants (HBT7) shows a 136-fold increase in binding compared to the wild-type RNA. The subset of mutants with the highest affinities for vigilin is a representative sample, since essentially the same conclusions are reached by analyzing the data from these mutants and from the entire data set of 58 mutant RNAs.
Sequence and structural features of the RNA binding site.
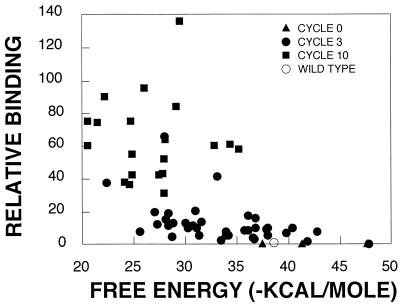
Analysis of the nucleotide compositions of the up-binding mutants indicated hypermutation of Gs and mutation of other nucleotides to C, with no change in the proportion of A and U (Table 1). There was a general correlation between the loss of G residues, increased free energy of the RNA secondary structures, and increased vigilin binding (Fig. 4). This suggests that efficient binding of vigilin to RNA might require a relatively flexible stretch of single-stranded RNA.
FIG. 4.
The free energy of RNA structures is increased for RNAs with enhanced binding to vigilin. Relative binding versus free energy was plotted for mutant RNAs which were assayed in each selection cycle (cycle 0, 3, or 10), as well as for wild-type RNA. Binding was plotted relative to that of wild-type RNA, which was set equal to 1. The free energy of each RNA was calculated by using the MFOLD program.
An important sequence feature evident in the up-binding mutants is the mutation of specific Gs to create a largely G-free stretch of RNA (Fig. 3). In the 10 mutants exhibiting the highest-affinity binding, the four Gs (G46, G49, G52, and G60) in the center of the wild-type vitellogenin mRNA binding site as well as G32 are either mutated to other nucleotides or deleted 94% of the time (47 of 50 possible cases). In contrast, the mutation frequency for the other three Gs at the periphery of the binding site (G26, G35, and G81) was only 47% (Fig. 3). As a result of hypermutation of Gs to other nucleotides, the mutant RNAs contain G-free regions up to 61 nucleotides in length (Fig. 3, HBT29).
While alignment of the sequences of the up-binders does not reveal a clear consensus sequence, both individual nucleotides and short nucleotide sequences are undermutated in the up-binding mutants (Fig. 3). The undermutated sequences cluster between positions 56 and 76 in the G-free region (Fig. 3). Several copies of the sequences (A)nCU and UC(A)n are found in this region (at positions 55 to 59, 64 to 67, and 70 to 74) and are therefore candidate sequences for interaction with vigilin.
Identification of nucleotides critical for vigilin binding by using down-binding mutants.
To more directly identify nucleotides important in creating a strong binding site, we carried out a selection for mutants with reduced ability to bind vigilin (Fig. 1, right). In this selection, we recovered unshifted bands from the RNA gel mobility shift assay. Since mutants exhibiting reduced binding and other RNAs which had simply failed to bind vigilin are both represented in this band, it was important to use conditions in which essentially all of the labeled RNA probe interacted with the binding protein. To achieve maximal interaction of the RNA with the binding protein, we used HBT7 RNA, the mutant RNA with the highest affinity for the binding protein (Fig. 2B). We used mutagenic PCR to generate an RNA pool with mutations in the central 70 nucleotides of the HBT7 template. With increasing cycles of selection, there was a progressive increase in the intensity of the unshifted band (Fig. 2B). After four selection cycles, RNA in the unshifted band was recovered, reverse transcribed, PCR amplified, subcloned, sequenced, transcribed into RNA, and analyzed for vigilin binding.
Mutants obtained after four cycles of selection showed reduced binding compared to the starting HBT7 up-binding RNA (Fig. 5). The mutagenesis was carried out under conditions in which the predicted mutation frequency was only 0.8 mutation per 70-nucleotide RNA, and only a small fraction of the RNAs contained multiple mutations. Nevertheless, all of the mutants selected contained at least two mutations. This suggests that no single point mutation was sufficient to abolish binding by vigilin, a relatively large polypeptide with a mass of approximately 155 kDa. Most, and perhaps all, of the mutations identified in the mutants shown in Fig. 5 contribute to the down-binding phenotype. All of the down-binders which had three or fewer mutations are shown in Fig. 5. Although the conditions for the mutagenesis do not show a strong bias with respect to the type of base substitution (16), approximately half (13 of 23) of the mutations in the down-binders were to G. This is consistent with our finding that mutation of Gs to other nucleotides is characteristic of up-binding mutants.
FIG. 5.
Sequences of RNAs selected for reduced binding to vigilin. The mutated nucleotides in all of the selected RNAs containing three or fewer point mutations are shown. The boxed region delineates an A tract which was mutated (usually to G) in each of the mutant RNAs. Relative binding by HBT7 RNA was set equal to 100. The wild-type vitellogenin (WT) and HBT7 RNA sequences are shown for comparison.
Both RNA sequence and structure are important in establishing a binding site.
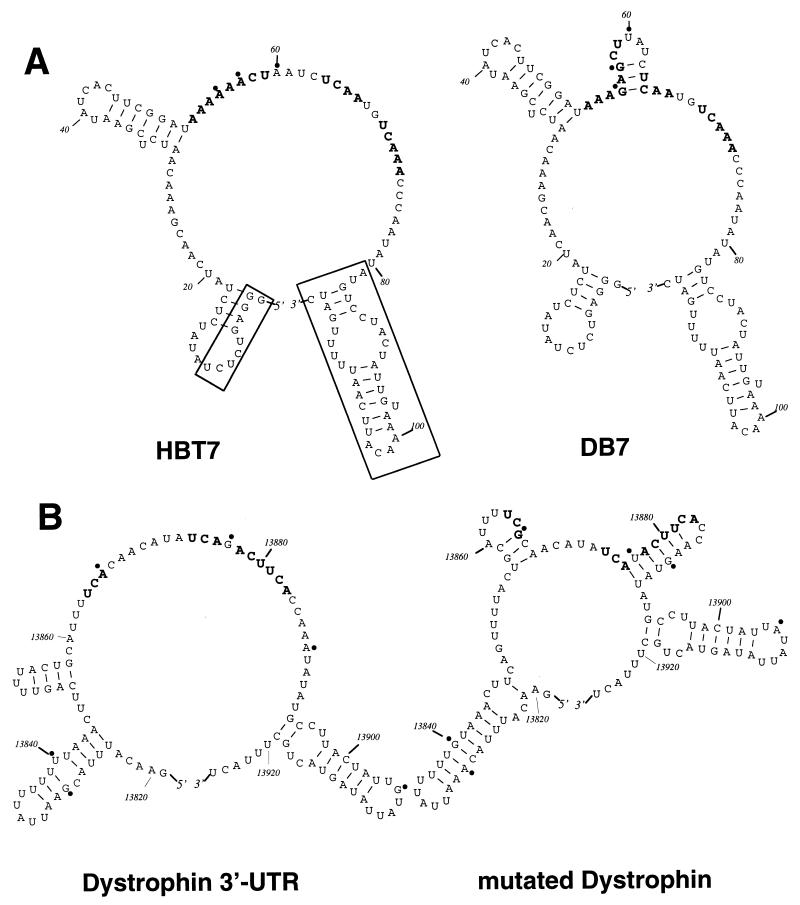
Three clustered point mutations (G55, G57, and U60) in the DB7 mutant are sufficient to virtually abolish vigilin binding (Fig. 5). A model of the secondary structure of DB7 RNA suggests that the mutations create a small stem-loop structure in the long 33-nucleotide single-stranded region near the center of the HBT7 up-binding mutant (Fig. 6A). Although the mutations in the other down-binding mutants were more dispersed than those in DB7, the importance of the central region of the RNA mutated in DB7 was shown by the fact that every other down-binding mutant contains a mutation in the A tract of the (A)nCU sequence (nucleotides 52 to 59) (Fig. 5).
FIG. 6.
Predicted secondary structures of RNAs exhibiting very strong or very weak binding to vigilin. (A) One of the predicted secondary structures for the highest-affinity RNA, HBT7 RNA (left), and for one of the down-binding mutants, DB7 RNA (right). Mutations at A55, A57, and A60 are in the single-stranded region of HBT7 RNA and are indicated by filled circles near each mutated nucleotide in HBT7 and DB7. The boxed regions delineate nucleotides in the HBT7 sequence that were determined to be nonessential for binding. (B) One of the predicted secondary structures of the segment of the 3′ UTR of human dystrophin mRNA (left) and of mutated dystrophin mRNA (right). Mutations were introduced in the six nucleotides G11, U22, A48, G59, A70, and G87 (denoted by filled circles) to obtain the mutated dystrophin RNA whose secondary structure is shown on the right. The (A)nCU and UC(A)n motifs in the central single-stranded areas of HBT7 and dystrophin RNAs and the corresponding nucleotides in the mutants are shown in boldface. Numbers are the positions of the nucleotides (the transcription start site is set to 1 for HBT7 and DB7).
The down-binding mutants provide additional support for a role in vigilin binding for the (A)nCU and UC(A)n sequences conserved in the up-binding mutants. The mutations in DB7 disrupt the (A)nCU sequence, and the small stem-loop structure created in DB7 moves a UC(A)n sequence from a single-stranded region of the RNA to a double-stranded region (Fig. 6, boldface). In addition, four of the eight down-binding mutants showed mutations in a nearby UC(A)n sequence (nucleotides 70 to 74).
The mutation data were compatible with the view that the oligo(A) tract plays a largely structural role, facilitating formation of a single-stranded region of the RNA. However, these data did not exclude the possibility that vigilin binds to the oligo(A) tract. We therefore used competition gel mobility shift assays to test the ability of poly(A) to compete for binding to vigilin. A 10,000-fold excess of poly(A) was unable to compete for binding to vigilin. These data indicate that vigilin is not a poly(A) binding protein.
Determination of the 3′ and 5′ boundaries of the RNA binding site.
The in vitro genetic analysis suggested that the RNA segment containing the (A)nCU and UC(A)n motifs in a single-stranded region of the RNA played an important role in vigilin binding. However, vigilin is a large, approximately 155-kDa protein (26) with 14 KH domains. We therefore determined the minimal length of the sequence required for efficient vigilin binding.
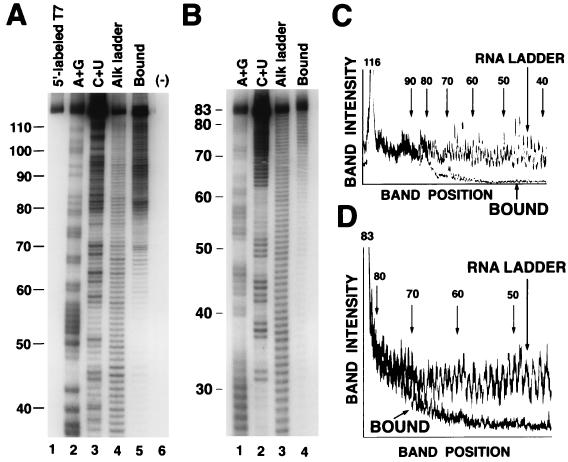
An RNA ladder was prepared by mild alkaline hydrolysis of a 116-nucleotide 5′-end-labeled HBT7 RNA (Fig. 7A, lane 4). The RNAs were incubated with vigilin, and protein-RNA complexes were isolated on a nitrocellulose filter and fractionated on a denaturing polyacrylamide gel (Fig. 7A, lane 5). Bands for RNAs smaller than 55 nucleotides (Fig. 7A, lane 5) or for RNAs incubated in the absence of the binding protein (Fig. 7A, lane 6) were undetectable. Quantitation of band intensities of both the ladder and recovered RNA revealed that binding was unimpaired with RNAs as short as 80 nucleotides (Fig. 7C). Binding then declined to undetectable levels for RNAs shorter than 55 nucleotides (Fig. 7C). Since binding was unimpaired for an 80-nucleotide RNA, in which the 3′-terminal 36 nucleotides were deleted, nucleotides 81 to 116 are not essential for binding (Fig. 6A, boxed area at the 3′ end). Our conclusion that this RNA segment is not important for vigilin binding is supported by our observations that it has the potential to form a secondary structure and is not well conserved among up-binding variants (Fig. 3). The highly conserved region lies immediately upstream of the nonessential RNA segment (Fig. 3). Deletions in this region (nucleotides 79 to 55) exhibited a progressive decline in binding to vigilin (Fig. 7C).
FIG. 7.
Determination of the minimum RNA length for efficient vigilin binding. (A) The 5′-end-labeled HBT7 RNA (lane 1 [T7]) was partially hydrolyzed with mild alkaline treatment to obtain an RNA ladder (lane 4). The RNA ladder was incubated either with (lane 5) or without (lane 6) vigilin-containing polysome extract. The RNAs bound to the protein were recovered and gel electrophoresed. To identify the sizes of the RNA bands, RNA sequence ladders were made by chemical modification (diethyl pyrocarbonate [A+G] in lane 2 and hydrazine-NaCl [C+U] in lane 3). Numbers indicate the lengths of the RNAs. (C) Band intensities were quantitated and plotted. RNA LADDER and BOUND are scans from lanes 4 and 5 of the gel in panel A, respectively. To compare the relative intensities of each band, the band intensities were corrected at nucleotide 116. (B and D) HBT7/83 RNA with the 3′ end deleted was synthesized, and a similar analysis was carried out with a 3′-end-labeled RNA.
To determine the 5′ boundary of the RNA sequence required for vigilin binding, we deleted the nonessential portion of the 3′ end of the RNA, labeled the 3′ end of the RNA, and analyzed the 5′ boundary of the RNA binding site (Fig. 7B). Binding began to decline after deletion of 10 nucleotides from the 5′ end of the RNA (Fig. 7B, lane 4). The first 10 nucleotides (Fig. 6A, boxed area at the 5′ end) were nonessential for binding, and 73 nucleotides were required for full binding. Both the 5′-end deletions and the 3′-end deletions exhibited unimpaired binding to RNAs 73 to 80 nucleotides in length, followed by a progressive decline in binding as the RNA length decreased, to no binding when the RNA length was <55 nucleotides (Fig. 7C and D). Since binding declined before deletions from the 5′ end reached the conserved sequence, we conclude that there is a sequence-independent length requirement for efficient binding to vigilin.
Analysis of RNA-protein contacts by RNA footprinting.
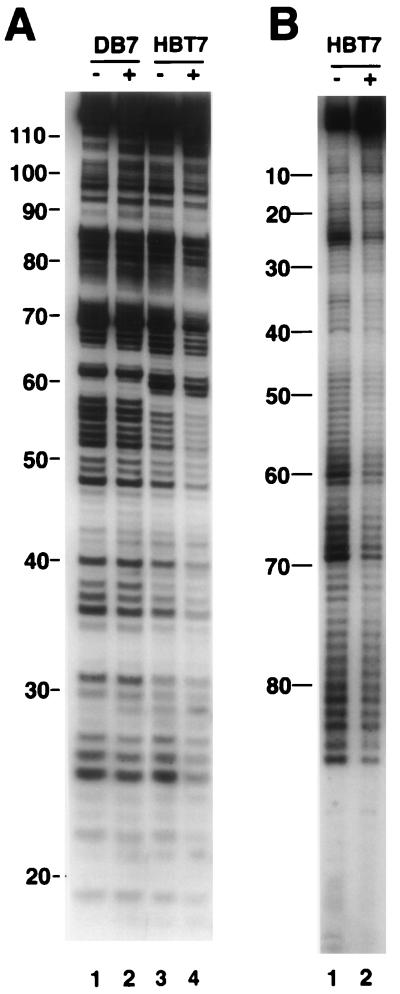
Our observation that maximal binding of vigilin required RNAs at least 73 to 80 nucleotides long suggested either that the large, approximately 155-kDa vigilin protein exhibits multiple contacts with the RNA over a stretch of 70 to 80 nucleotides or that a long unstructured RNA is required to make a short RNA binding sequence available for binding. To distinguish between these possibilities, we performed an RNA footprinting analysis with the up-binding RNA HBT7 and the down-binding mutant RNA DB7. The end-labeled RNA was incubated with (Fig. 8A, lanes 2 and 4, and B, lane 2) or without (Fig. 8A, lanes 1 and 3, and B, lane 1) the vigilin-containing polysome extract and digested with α-sarcin. α-Sarcin digests RNAs at the 3′ side of purines, independent of RNA secondary structure. Since the experiments were carried out in the presence of a large excess of tRNA, protection is due to specific interaction of vigilin with the RNA. We digested HBT7 RNA labeled either at the 5′ or 3′ end, and 5′-end-labeled nonbinding DB7 RNA in parallel, using α-sarcin (Fig. 8). Since the digestion patterns of DB7 in the presence and absence of vigilin are identical (Fig. 8A, lanes 1 and 2), the vigilin-containing extract neither protects DB7 RNA against degradation nor inhibits the activity of α-sarcin. In contrast, HBT7 RNA was partially protected from digestion from positions 18 to 87 (Fig. 8A, lanes 3 and 4, and B). The A tract of the (A)nCU sequence (nucleotides 52 to 57) shown to be important for binding by in vitro selection of down-binding mutants was also in a highly protected region of the RNA (Fig. 8A, lane 4).
FIG. 8.
RNA footprinting with α-sarcin. (A) HBT7 RNA (lanes 3 and 4) or DB7 RNA (lanes 1 and 2) was 5′ end labeled, incubated with (+) or without (−) vigilin-containing polysome extract, and digested with 5 μM α-sarcin. (B) 3′-end-labeled HBT7 RNA was incubated with (+) or without (−) extract and digested with 5 μM α-sarcin. Digested RNAs were analyzed by gel electrophoresis. Numbers are the positions of the nucleotides, as indicated in Fig. 3.
A strong vigilin binding site is located near the 3′ end of human dystrophin mRNA.
If the in vitro genetic selections truly created an artificial phylogeny of vigilin binding sites, it should be possible to use the RNA sequence and structure information obtained by in vitro genetic analysis to predict the occurrence of mRNA binding sites. The HBT7 sequence was used to screen the EMBL and GenBank databases of human DNA sequences by using the program FASTA from the University of Wisconsin Genetics Computer Group package (23). Since the natural vitellogenin binding site was in the 3′ UTR and this region has often been implicated in control of mRNA degradation, we selected sequences only from mRNA 3′ UTRs to examine. The 3′ UTR with the highest homology to HBT7 was located at the 3′ end of the human dystrophin gene. Homology between HBT7 and this region of the dystrophin 3′ UTR extended over a 69-nucleotide region (HBT7 nucleotides 30 to 98), with a 71% sequence identity over nucleotides 56 to 76, which the in vitro selections identified as the conserved region, important for vigilin binding. Subsequent sequence analysis and secondary structure modeling of the dystrophin mRNA 3′ UTR region revealed a long, largely single-stranded, G-free region (Fig. 6B) containing some of the ACU and UCA motifs (Fig. 6B, boldface) found in HBT7 and other strong up-binding mutants. Dystrophin mRNA was therefore tested for binding to vigilin, and it represents the only mRNA from the database search that we analyzed for vigilin binding.
We synthesized a 107-nucleotide RNA from a cDNA template derived from the 2.7-kb 3′ UTR of dystrophin mRNA. This RNA terminates 10 nucleotides upstream of the AAUAAA polyadenylation signal. In RNA gel shift assays, vigilin in Xenopus polysome extracts bound effectively to the human dystrophin RNA sequence (Fig. 9A, lanes 3 and 4). In addition to a gel-shifted band with the same mobility as the vitellogenin RNA-vigilin complex (Fig. 9A, lane 2), two faster-migrating bands were observed. Since only the band with the same electrophoretic mobility as the vitellogenin RNA-vigilin complex is competed by unlabeled HBT7 RNA (data not shown) and the additional bands are not seen in binding studies carried out with purified recombinant vigilin (see below), the more rapidly migrating bands represent interactions with other proteins in the extract.
FIG. 9.
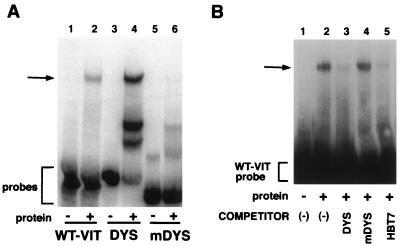
The 3′ UTR of dystrophin mRNA contains a strong vigilin binding site. (A) RNA gel shift assays of the labeled vitellogenin mRNA 3′ UTR (WT-VIT) (lanes 1 and 2), 107-nucleotides human dystrophin mRNA 3′ UTR (DYS) (lanes 3 and 4), and mutated dystrophin mRNA (mDYS) (lanes 5 and 6) with (+) and without (−) vigilin-containing polysome extract. (B) Competition gel shift assays with unlabeled dystrophin (lane 3), mutated dystrophin (lane 4), and HBT7 (lane 5) RNAs. The vigilin binding region in the vitellogenin mRNA 3′ UTR was 32P labeled, and 0.3 ng of probe was incubated with vigilin in the absence of competitor (−) or in the presence of the unlabeled competitors (DYS [100 ng], HBT7 [100 ng], or mDYS [2.5 μg]). The arrows indicate the RNA-vigilin complex. RNA probes are also indicated.
If the same type of sequence and structural information is required for interaction of vigilin with dystrophin RNA, vitellogenin RNA, and the up-binding mutants, mutations similar to those selected in the down-binding mutants should abolish binding. To test this, we replaced two As with Gs and one G with U in the single-stranded region of the dystrophin RNA predicted to be critical for binding (Fig. 6B). This not only introduced an additional stem-loop structure into the single-stranded region but also moved all the ACU and UCA motifs (Fig. 6, boldface) in the single-stranded area into RNA segments exhibiting substantial secondary structure (Fig. 6B). This mutated dystrophin RNA did not show the characteristic up-shifted band in a gel shift assay, indicating that it had lost the ability to specifically bind to vigilin (Fig. 9A, lanes 5 and 6).
To directly demonstrate that the same protein was binding to human dystrophin RNA and Xenopus vitellogenin mRNA, we did a competition gel mobility shift assay (Fig. 9B). The up-binding HBT7 mutant (lane 5) and the dystrophin RNA (lane 3) effectively blocked binding to the vitellogenin RNA. This suggests that the in vitro-selected HBT7 RNA and the dystrophin RNA use the same general RNA binding mechanism to interact with vigilin. The mutated dystrophin sequence (lane 4) was unable to compete even at a much higher concentration (HBT7 and dystrophin, 333-fold, compared to mutated dystrophin, 8,300-fold molar excess relative to the labeled vitellogenin probe). These data demonstrate that an RNA segment near the 3′ end of dystrophin mRNA contains a strong vigilin binding site.
Recombinant human vigilin binds to the HBT7 and dystrophin RNAs.
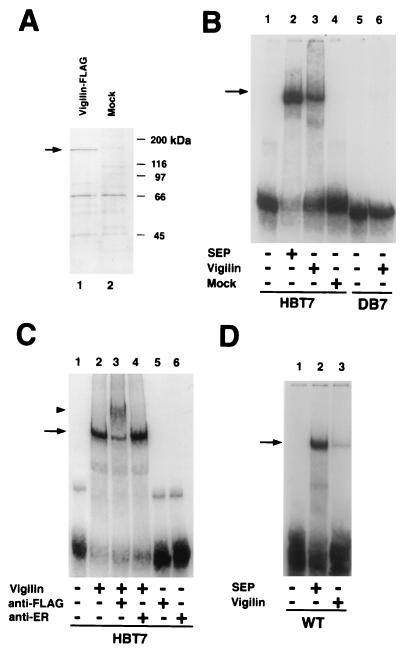
We had previously demonstrated that Xenopus vigilin was the protein binding to the vitellogenin mRNA 3′ UTR sequence (26). While the HBT7, dystrophin, and vitellogenin RNA-protein complexes exhibit the same mobility in gel shift assays and the HBT7 and dystrophin RNAs effectively competed out binding to the vitellogenin RNA (Fig. 9B), it was still important to confirm that vigilin, and not other proteins in the polysome extracts, was binding to the RNAs. We also wished to determine whether human vigilin bound the human dystrophin RNA sequence. We therefore tested the ability of purified, recombinant human vigilin to bind to in vitro-selected RNAs and the vitellogenin and dystrophin RNAs. We introduced the FLAG epitope at the C terminus of a full-length human vigilin cDNA (43), expressed the vigilin by in vitro transcription-translation in the rabbit reticulocyte lysate cell-free protein synthesis system, and isolated the epitope-tagged vigilin by anti-FLAG immunoaffinity chromatography. SDS-polyacrylamide gel analysis demonstrated that a protein of the expected 155-kDa size was purified from the lysate containing the in vitro-translated vigilin but not from the mock-purified control reticulocyte lysate (Fig. 10A). In gel mobility shift assays, the mock-purified product from the rabbit reticulocyte lysate did not bind HBT7 RNA, while the purified protein did bind the HBT7 RNA (Fig. 10B, lanes 3 and 4), confirming that vigilin is the protein purified from the lysate which is responsible for formation of the RNA-protein complex. Confirmation that the RNA-protein complexes represent binding of recombinant vigilin to the RNAs comes from an antibody supershift experiment. Antibody to the FLAG epitope, present in the recombinant vigilin, supershifted the RNA-protein complex (Fig. 10C, lanes 2 and 3). The same amount (1 μg) of anti-estrogen receptor monoclonal antibody had no effect on migration of the RNA-protein complex (Fig. 10C, lane 4). Addition of either the FLAG antibody or the estrogen receptor antibody did not alter migration of the probe (Fig. 10C, lanes 5 and 6).
FIG. 10.
HBT7 up-binding RNA binds to purified recombinant human vigilin. (A) FLAG-tagged human vigilin was produced by in vitro transcription-translation, purified by antibody affinity chromatography, and analyzed by SDS-polyacrylamide gel electrophoresis. The in vitro transcription-translation reaction was carried out in the presence (lane 1) (Vigilin-FLAG) or in the absence (lane 2) (Mock) of PCR-generated FLAG-tagged human vigilin DNA template. In vitro-translated FLAG-tagged products were affinity purified by using the FLAG epitope. Samples were separated on an SDS–7.2% polyacrylamide gel and silver stained. The sizes of molecular mass markers are indicated at the right. (B) RNA gel shift assays of the 32P-labeled HBT7 up-binding RNA (lanes 1 to 4) and DB7 down-binding RNA (lanes 5 and 6) with Xenopus vigilin-containing polysome extract (SEP) (lane 2), with purified human vigilin (lanes 3 and 6), or with mock-purified extract (lane 4). (C) Supershift of the vigilin RNA complex by anti-FLAG antibody. The RNA gel shift assay was performed with 32P-labeled HBT7 up-binding RNA and FLAG-tagged vigilin in the presence of anti-FLAG M2 antibody (1 μg) (lane 3) or anti-human estrogen receptor (anti-ER) monoclonal antibody H222 (1 μg) (lane 4) or without antibody (lane 2). HBT7 RNA was also incubated with 1 μg of the antibodies alone (anti-FLAG [lane 5] or anti-ER [lane 6]). The RNA-vigilin complex and supershifted RNA-vigilin-antibody complex are indicated by an arrow and an arrowhead, respectively. (D) RNA gel shift assays of the 32P-labeled vitellogenin mRNA 3′ UTR (WT) with Xenopus vigilin-containing polysome extract (SEP) (lane 2) or with purified recombinant human vigilin (lane 3).
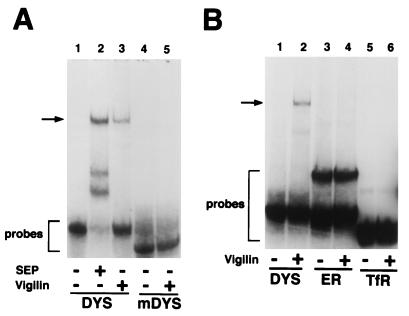
The RNA-protein complexes formed with purified recombinant vigilin or with the protein in the polysome extracts exhibited the same electrophoretic mobility with the vitellogenin (Fig. 10D, lanes 2 and 3), HBT7 (Fig. 10B, lanes 2 and 3), and dystrophin (Fig. 11A, lanes 2 and 3) RNAs. The failure of the mutated dystrophin and DB7 down-binding RNAs, which do not bind the protein in the polysome extract (Fig. 5 and 9), to bind to purified recombinant vigilin (Fig. 10B, lane 6, and 11A, lane 5) provides additional evidence that vigilin is the protein in the polysome extracts binding both to the selected RNAs and to the dystrophin and vitellogenin RNAs. Taken together, these data demonstrate that the RNAs generated by the in vitro selection did indeed select for higher binding to vigilin.
FIG. 11.
Purified human vigilin binds to the human dystrophin mRNA 3′ UTR but not to the human estrogen receptor or transferrin receptor mRNA 3′ UTR. (A) RNA gel shift assays of the labeled dystrophin mRNA 3′ UTR (DYS) (lanes 1 to 3) and mutated dystrophin mRNA (mDYS) (lanes 4 and 5) with (+) and without (−) affinity-purified FLAG-tagged human vigilin or with (+) and without (−) Xenopus vigilin-containing polysome extract (SEP). (B) RNA gel mobility shift assays with 32P-labeled 107-nucleotide human dystrophin mRNA 3′ UTR (lanes 1 and 2), 106-nucleotide human estrogen receptor mRNA 3′ UTR (ER) (lanes 3 and 4), and 108-nucleotide human transferrin receptor mRNA 3′ UTR (TfR) (lanes 5 and 6) with (lanes 2, 4, and 6) and without (lanes 1, 3, and 5) human vigilin. RNA-vigilin complexes are indicated by arrows. Uncomplexed RNA probes are also indicated.
While purified recombinant vigilin and vigilin in the polysome extracts form complexes with the dystrophin mRNA 3′ UTR exhibiting identical electrophoretic mobility, the two faster-migrating complexes are not seen with purified vigilin (Fig. 11A, lanes 2 and 3), suggesting that these bands are due to binding of other proteins in the polysome extract.
The sequence criteria for vigilin binding identified through the in vitro genetic selections were used successfully to predict a binding site in the 3′ UTR of dystrophin RNA, and this site can be destroyed by inserting only three point mutations. To further examine the specificity of vigilin binding, we investigated the ability of vigilin to bind to segments of the 3′ UTRs of two additional RNAs, which were chosen without use of a database search. We chose a segment of human transferrin receptor mRNA because it contains iron response elements which bind a cellular protein important in regulating the stability of human transferrin receptor mRNA (17, 44). We also tested a segment close to the 3′ end of human estrogen receptor mRNA because this is the same relative position as the vitellogenin and dystrophin mRNA 3′-UTR binding sites. While several studies have reported that the stability of human estrogen receptor mRNA is regulated (51, 53), RNA sequences important in human estrogen receptor mRNA degradation have not been identified. The RNAs were designed so that transcription would yield labeled probes whose lengths (106 to 108 nucleotides) were similar to those of the selected RNAs and the natural RNAs. Neither the human estrogen receptor RNA nor the human transferrin receptor RNA exhibited detectable binding to purified recombinant vigilin (Fig. 11B).
DISCUSSION
In vitro genetic analysis for up-binding and down-binding mutants.
The structural and functional diversity of RNA molecules has been elegantly exploited by the development of in vitro genetic methods for screening large populations of RNAs (15, 28, 29, 62). We have successfully adapted this technology to identify the RNA binding site of the multi-KH-domain protein vigilin before the protein was purified. To carry out this selection, we separated the RNA-vigilin complexes from other RNA-protein complexes by using an RNA gel mobility shift assay carried out under unusually stringent conditions and selecting a very narrow upshifted band. To confirm that this strategy was successful in that vigilin was the protein in the extracts binding to the RNAs, we showed that the RNA-protein complexes formed by purified recombinant vigilin and by the protein in the polysome extract exhibit the same electrophoretic mobility. Additionally, we confirmed that purified recombinant vigilin and the binding protein in the polysome extracts display similar binding preferences with five in vitro-selected and natural RNAs. Most compelling was our finding that the RNA binding protein we identified as vigilin in the polysome extract and purified recombinant vigilin both bind effectively to the selected HBT7 RNA (Fig. 10B) and do not bind to the very similar selected down-binding mutant DB7; both bind to the RNA segment at the 3′ end of dystrophin RNA and do not exhibit binding to the very similar mutated dystrophin RNA (Fig. 9A and 11A). This methodology may be useful for characterizing the binding sites of nucleic acid binding proteins which are difficult to purify and express or which require protein cofactors or multiprotein complexes to bind DNA or RNA.
One unusual feature of our approach is that we were able to obtain additional important information about the RNA binding site by first isolating high-affinity binding sites and then mutating a high-affinity site. This enabled us to carry out a novel genetic selection for down-binding RNA mutants. This type of selection had not been previously reported, presumably because of the high background produced by RNAs which failed to bind the protein of interest. By using our most effective up-binding mutant (HBT7), we were able to develop conditions under which all of the HBT7 RNA probe was bound to vigilin and shifted up the gel. After only four selection cycles, we did not observe a background of nonmutated HBT7 RNA sequences which had simply failed to bind the protein. One problem which did emerge was that while the mutation rate in the original pool of RNAs was less than one mutation per RNA molecule, the powerful selective pressure for isolation of nonbinding RNAs led to the selection of rare RNAs with numerous mutations. Nevertheless, the information provided by the down-binding mutants was important to the identification of binding motifs.
Vigilin exhibits strong preferences for binding to specific RNAs.
The in vitro selections reported here demonstrate that vigilin shows strong sequence preferences for binding to specific RNAs and does not simply bind RNA nonspecifically. This conclusion is supported by several lines of evidence. (i) After 10 selection cycles in the presence of high concentrations of nonspecific RNAs, 2 of the 20 selected up-binding mutants were identical and 2 others differed by only 2 nucleotides. From these data we calculate that there are no more than a few hundred RNAs which exhibit strong binding to vigilin in the original pool of 109 mutants. Since <1 in 106 RNAs exhibits high-affinity binding, the length, sequence, and structural requirements for high-affinity binding to vigilin are quite stringent. (ii) All of our RNA gel mobility shift assays (and the footprinting studies) were carried out in the presence of a 100,000-fold excess of tRNA. This is a significantly higher level of tRNA than was used in several other studies demonstrating sequence-specific binding to RNAs (13, 18, 22, 33). While we have not carried out detailed kinetic measurements, we used purified vigilin and the above-described RNA binding conditions to carry out a preliminary assessment of the binding of several concentrations of vigilin to the HBT7 RNA. The recombinant vigilin exhibited an approximate KD for HBT7 of ∼1 nM (data not shown), which is in the same of order of magnitude as the KDs of many sequence-specific RNA and DNA binding proteins. (iii) When we searched the human sequence database for sequences in the 3′ UTRs of mRNAs homologous to HBT7, the sequence with the highest homology, which was located at the 3′ end of human dystrophin RNA, exhibited strong binding to vigilin (Fig. 9A and 11A). We did not explore the possibility that other 3′ UTR RNAs with lower but still significant homology to HBT7 would still bind to vigilin. However, two additional 3′ UTR RNAs, the human transferrin receptor and human estrogen receptor RNAs, chosen without reference to a database search, did not bind vigilin (Fig. 11B). (iv) Probably the most direct demonstration that vigilin exhibits sequence-specific binding to RNAs is the demonstration that the selected DB7 RNA, which differs by only 3 nucleotides (of 116) from the strong binder HBT7, exhibits no binding to either crude or recombinant vigilin (Fig. 5 and 10B). Similarly, using the information obtained in the in vitro selections for up- and down-binding RNAs, we found that mutating only three widely spaced nucleotides (with three compensatory changes outside the core binding region to maintain the identical RNA base composition) abolished vigilin binding to the dystrophin RNA (Fig. 9A and 11A, mDYS). Taken together, these data provide clear evidence that vigilin exhibits strong sequence preferences for binding to specific RNAs.
The in vitro-selected RNAs and the natural RNAs appear to bind to vigilin in a similar fashion.
By using large pools of random RNA sequences, it is possible to select RNA receptors (aptamers) that can bind to proteins which do not contain known RNA binding motifs (7, 28, 32). However, in this work, we created a mutant pool which retained some resemblance to the natural vitellogenin RNA binding site, which may bias the selection toward RNA sequences that bind to vigilin in a fashion similar to that of the vitellogenin 3′-UTR RNA. Perhaps the most direct evidence that the selected RNAs and the naturally occurring RNAs bind vigilin through the same general mechanism comes from the competition gel mobility shift assays. The in vitro-selected HBT7 RNA and the natural dystrophin RNA sequence, but not the mutated dystrophin RNA, effectively compete with the labeled vitellogenin RNA for binding to vigilin (Fig. 9B). These data support the view that the vitellogenin RNA 3′ UTR sequence and the selected HBT7 RNA use similar mechanisms and either share the same binding site on vigilin or use substantially overlapping binding sites. Additional support for the view that the selected and natural RNAs interact with vigilin through similar mechanisms comes from our data indicating that the same types of mutations which abolish binding in the selected DB7 RNA also abolish binding in the naturally occurring dystrophin RNA.
While these data support the view that vigilin binding to the selected and natural RNAs occurs by similar mechanisms, the precise role of the 14 vigilin KH domains in RNA binding remains to be established. Using the boundaries for the KH domains described by Musco et al. (45), who solved the crystal structure of a vigilin KH domain, the 14 KH domains of vigilin represent approximately 80% of its 155-kDa mass, with the remainder largely in linker regions between the KH domains. The very large number of KH domains and the potential complexity of their combinatorial interactions with the long RNA binding site we describe will make it difficult to demonstrate an explicit role for vigilin’s KH domains in RNA binding. We therefore cannot exclude the formal possibility that vigilin binds RNAs not through its 14 KH domains but by still-unidentified RNA binding sequences or RNA binding domains.
The vigilin binding site.
The RNA binding properties of KH-domain proteins have often been assessed primarily by binding to simple RNAs (1, 38, 42, 58, 59, 67, 68). While it had been suggested that these proteins bind RNA nonspecifically (27), several KH-domain proteins have been shown to bind to specific RNAs (2, 13, 34, 46, 55, 60), but the precise nature of these RNA binding sites, except in the case of the Nova-1 site (12), has generally not been established.
Our data indicate that the length of the RNA, the absence of secondary structure, and the presence of specific RNA sequence motifs all contribute to high-affinity vigilin binding. While a specific binding motif has emerged for the α-globin stability complex, binding involves several proteins which do not all contain KH domains (68). However, the identical electrophoretic mobility of the RNA-protein complexes formed by crude vigilin and the purified recombinant vigilin indicates that the vigilin-RNA complexes do not contain other tightly bound proteins.
Perhaps because it contains 14 KH domains, the sequence and structural requirements for vigilin binding to RNA are different from those of many other RNA binding proteins which bind short sequence motifs (15, 19, 29, 61, 64). In contrast, our observations from the minimum-size determination experiment (Fig. 7) indicate that an unusually long, 75-nucleotide RNA is required for efficient vigilin binding.
Our finding that efficient binding requires a long single-stranded region of RNA suggests that interaction of the RNA with vigilin may allow the protein to deform the RNA and create specific contacts. A similar conclusion was reached from interaction of the HIV Rev protein with its binding site (4). Although both the HIV Rev protein and vigilin may deform RNA on binding, the HIV Rev protein and the heavily studied iron response element binding protein, which regulates transferrin receptor mRNA stability (17, 44), differ from vigilin in that they both use RNA secondary structure in their recognition motifs.
The selection of strong up-binding mutants led to the identification of RNA sequence motifs which were candidate sites for the interaction of vigilin with RNA. We identified an RNA sequence containing several undermutated nucleotides, whose deletion we previously showed to abolish binding (25). It seemed probable that this region (nucleotides 53 to 76) played an important role in generating a high-affinity binding site. The undermutated sequences which were candidate vigilin binding sites included (A)nCU and UC(A)n, which are clustered in this area. The selection of mutant RNAs which had lost the ability to bind effectively to vigilin strengthened this conclusion. All of the eight down-binding mutants which had three or fewer mutations contained mutations in an A tract in the (A)nCU sequence (Fig. 5). In the down-binding mutant DB7, the three mutations altered the sequence of one (A)nCU element to (A)nGCU and moved a UC(A)n sequence into a double-stranded stem. Analysis of the other down-binding mutants confirmed that insertion of Gs into the central single-stranded area can both destroy the (A)nCU and UC(A)n sequence motifs and eliminate the single-stranded structure of this region, both of which appear to be important for vigilin binding. Interestingly, the Nova-1 protein, which contains three KH domains, binds to multiple separated copies of the motif UCAU in a single-stranded region (12). However, the UCAU motifs recognized by Nova-1 are part of the loop of a stem-loop structure, while the vigilin binding sites contain extended, largely unstructured regions.
We conclude that in HBT7 RNA a 75-nucleotide RNA containing multiple (A)nCU and UC(A)n motifs in a largely G-free region of single-stranded RNA provides a strong binding site. However, other vigilin binding motifs may be possible. Indeed, the ability of vigilin to exhibit a range of affinities for different RNAs may be functionally important. This spectrum of RNA binding sites would allow for coordinate regulation of diverse RNAs and for both regulated and constitutive binding to mRNAs for which vigilin has different affinities.
The identification of a binding site in dystrophin mRNA by homology to the HBT7 RNA site confirmed our prediction that vigilin could bind other mRNAs. While the Becker form of muscular dystrophy is characterized by a reduced level of dystrophin, there is as yet no direct evidence for an in vivo role for vigilin in the metabolism of dystrophin mRNA. The tight in vitro binding of vigilin to the dystrophin mRNA binding site raises the possibility that this is a biologically significant interaction, as does the presence of vigilin in muscle and the ability of testosterone to induce vigilin binding activity in Xenopus muscle (24). To directly assess the potential role of vigilin in dystrophin mRNA metabolism will require technically difficult studies with normal and dystrophic muscle.
In this work we describe an elaboration of in vitro genetic analysis in which a relatively crude nucleic acid binding protein was used to identify sequence and structural determinants important in the interaction of the multi-KH-domain protein vigilin with mRNA. The validity and utility of this analysis were demonstrated by our ability to use information gained from this study to successfully predict the presence of a strong binding site near the 3′ end of human dystrophin mRNA, while two RNAs which were chosen by using other criteria did not bind vigilin. These data provide an analysis of the interaction of this large and complex KH-domain protein with RNA and describe a strategy for the identification of the still-unknown binding sites of other RNA binding proteins.
ACKNOWLEDGMENTS
We are grateful to L. Kunkel for the dystrophin cDNA clone, to A. Martínez del Pozo for the gift of α-sarcin, and to R. Gumport for helpful comments on the manuscript.
This research was supported by NIH grants DK-50080 and HD-16720.
REFERENCES
- 1.Arning S, Grüter P, Bilbe G, Krämer A. Mammalian splicing factor SF1 is encoded by variant cDNAs and binds to RNA. RNA. 1996;2:794–810. [PMC free article] [PubMed] [Google Scholar]
- 2.Ashley C T, Jr, Wilkinson K D, Reines D, Warren S T. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- 3.Bartel D P, Zapp M L, Green M R, Szostak J W. HIV-1 Rev regulation involves recognition of non-Watson-Crick base pairs in viral RNA. Cell. 1991;67:529–536. doi: 10.1016/0092-8674(91)90527-6. [DOI] [PubMed] [Google Scholar]
- 4.Battiste J L, Tan R, Frankel A D, Williamson J R. Binding of an HIV Rev peptide to Rev responsive element RNA induces formation of purine-purine base pairs. Biochemistry. 1994;33:2741–2747. doi: 10.1021/bi00176a001. [DOI] [PubMed] [Google Scholar]
- 5.Beelman C A, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 6.Belasco J, Brawerman G. Control of messenger RNA stability. San Diego, Calif: Academic Press Inc.; 1993. [Google Scholar]
- 7.Binkley J, Allen P, Brown D M, Green L, Tuerk C, Gold L. RNA ligands to human nerve growth factor. Nucleic Acids Res. 1995;23:3198–3205. doi: 10.1093/nar/23.16.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blackwell T K, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- 9.Blume J E, Shapiro D J. Ribosome loading, but not protein synthesis, is required for estrogen stabilization of Xenopus laevis vitellogenin mRNA. Nucleic Acids Res. 1989;17:9003–9014. doi: 10.1093/nar/17.22.9003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brock M L, Shapiro D J. Estrogen stabilizes vitellogenin mRNA against cytoplasmic degradation. Cell. 1983;34:207–214. doi: 10.1016/0092-8674(83)90151-4. [DOI] [PubMed] [Google Scholar]
- 11.Brown D, Brown J, Kang C, Gold L, Allen P. Single-stranded RNA recognition by the bacteriophage T4 translational repressor, regA. J Biol Chem. 1997;272:14969–14974. doi: 10.1074/jbc.272.23.14969. [DOI] [PubMed] [Google Scholar]
- 12.Buckanovich R J, Darnell R B. The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol Cell Biol. 1997;17:3194–3201. doi: 10.1128/mcb.17.6.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buckanovich R J, Yang Y Y, Darnell R B. The onconeural antigen Nova-1 is a neuron-specific RNA-binding protein, the activity of which is inhibited by paraneoplastic antibodies. J Neurosci. 1996;16:1114–1122. doi: 10.1523/JNEUROSCI.16-03-01114.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 15.Burd C G, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cadwell R C, Joyce G F. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 17.Casey J L, Koeller D M, Ramin V C, Klausner R D, Harford J B. Iron regulation of transferrin receptor mRNA levels requires iron-responsive elements and a rapid turnover determinant in the 3′ untranslated region of the mRNA. EMBO J. 1989;8:3693–3699. doi: 10.1002/j.1460-2075.1989.tb08544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chagnovich D, Fayos B E, Cohn S L. Differential activity of ELAV-like RNA-binding proteins in human neuroblastoma. J Biol Chem. 1996;271:33587–33591. doi: 10.1074/jbc.271.52.33587. [DOI] [PubMed] [Google Scholar]
- 19.Chen C Y, Shyu A B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 20.Chiang C M, Roeder R G. Expression and purification of general transcription factors by FLAG epitope-tagging and peptide elution. Peptide Res. 1993;6:62–64. [PubMed] [Google Scholar]
- 21.Dejgaard K, Leffers H. Characterisation of the nucleic-acid-binding activity of KH domains. Different properties of different domains. Eur J Biochem. 1996;241:425–431. doi: 10.1111/j.1432-1033.1996.00425.x. [DOI] [PubMed] [Google Scholar]
- 22.DeMaria C T, Brewer G. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J Biol Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- 23.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dodson R E, Acena M R, Shapiro D J. Tissue distribution, hormone regulation and evidence for a human homologue of the estrogen-inducible Xenopus laevis vitellogenin mRNA binding protein. J Steroid Biochem Mol Biol. 1995;52:505–515. doi: 10.1016/0960-0760(95)00018-u. [DOI] [PubMed] [Google Scholar]
- 25.Dodson R E, Shapiro D J. An estrogen-inducible protein binds specifically to a sequence in the 3′ untranslated region of estrogen-stabilized vitellogenin mRNA. Mol Cell Biol. 1994;14:3130–3138. doi: 10.1128/mcb.14.5.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dodson R E, Shapiro D J. Vigilin, a ubiquitous protein with 14 K homology domains, is the estrogen-inducible vitellogenin mRNA 3′-untranslated region-binding protein. J Biol Chem. 1997;272:12249–12252. doi: 10.1074/jbc.272.19.12249. [DOI] [PubMed] [Google Scholar]
- 27.Gibson T J, Thompson J D, Heringa J. The KH domain occurs in a diverse set of RNA-binding proteins that include the antiterminator NusA and is probably involved in binding to nucleic acid. FEBS Lett. 1993;324:361–366. doi: 10.1016/0014-5793(93)80152-k. [DOI] [PubMed] [Google Scholar]
- 28.Gold L, Polisky B, Uhlenbeck O, Yarus M. Diversity of oligonucleotide functions. Annu Rev Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 29.Görlach M, Burd C G, Dreyfuss G. The determinants of RNA-binding specificity of the heterogeneous nuclear ribonucleoprotein C proteins. J Biol Chem. 1994;269:23074–23078. [PubMed] [Google Scholar]
- 30.Green S, Walter P, Kumar V, Krust A, Bornert J M, Argos P, Chambon P. Human oestrogen receptor cDNA: sequence, expression and homology to v-erb-A. Nature. 1986;320:134–139. doi: 10.1038/320134a0. [DOI] [PubMed] [Google Scholar]
- 31.Heinrichs V, Baker B S. The Drosophila SR protein RBP1 contributes to the regulation of doublesex alternative splicing by recognizing RBP1 RNA target sequences. EMBO J. 1995;14:3987–4000. doi: 10.1002/j.1460-2075.1995.tb00070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jellinek D, Lynott C K, Rifkin D B, Janjic N. High-affinity RNA ligands to basic fibroblast growth factor inhibit receptor binding. Proc Natl Acad Sci USA. 1993;90:11227–11231. doi: 10.1073/pnas.90.23.11227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kjems J, Calnan B J, Frankel A D, Sharp P A. Specific binding of a basic peptide from HIV-1 Rev. EMBO J. 1992;11:1119–1129. doi: 10.1002/j.1460-2075.1992.tb05152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kruse C, Grünweller A, Notbohm H, Kügler S, Purschke W G, Müller P K. Evidence for a novel cytoplasmic tRNA-protein complex containing the KH-multidomain protein vigilin. Biochem J. 1996;320:247–252. doi: 10.1042/bj3200247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kügler S, Grünweller A, Probst C, Klinger M, Müller P K, Kruse C. Vigilin contains a functional nuclear localisation sequence and is present in both the cytoplasm and the nucleus. FEBS Lett. 1996;382:330–334. doi: 10.1016/0014-5793(96)00204-9. [DOI] [PubMed] [Google Scholar]
- 36.Kühn L C, McClelland A, Ruddle F H. Gene transfer, expression, and molecular cloning of the human transferrin receptor gene. Cell. 1984;37:95–103. doi: 10.1016/0092-8674(84)90304-0. [DOI] [PubMed] [Google Scholar]
- 37.Lacadena J, Martínez del Pozo A, Barbero J L, Mancheño J M, Gasset M, Oñaderra M, López-Otín C, Ortega S, García J, Gavilanes J G. Overproduction and purification of biologically active native fungal alpha-sarcin in Escherichia coli. Gene. 1994;142:147–151. doi: 10.1016/0378-1119(94)90370-0. [DOI] [PubMed] [Google Scholar]
- 38.Leffers H, Dejgaard K, Celis J E. Characterisation of two major cellular poly(rC)-binding human proteins, each containing three K-homologous (KH) domains. Eur J Biochem. 1995;230:447–453. [PubMed] [Google Scholar]
- 39.Levine T D, Gao F, King P H, Andrews L G, Keene J D. Hel-N1: an autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol Cell Biol. 1993;13:3494–3504. doi: 10.1128/mcb.13.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu M Y, Yang H, Romeo T. The product of the pleiotropic Escherichia coli gene csrA modulates glycogen biosynthesis via effects on mRNA stability. J Bacteriol. 1995;177:2663–2672. doi: 10.1128/jb.177.10.2663-2672.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lorincz M A, Holt J A, Greene G L. Monoclonal antibody recognition of multiple forms of estrogen receptor tagged with [125I]methoxy-iodovinyl estradiol in ovarian carcinomas. J Clin Endocrinol Metab. 1985;61:412–417. doi: 10.1210/jcem-61-3-412. [DOI] [PubMed] [Google Scholar]
- 42.Matunis M J, Michael W M, Dreyfuss G. Characterization and primary structure of the poly(C)-binding heterogeneous nuclear ribonucleoprotein complex K protein. Mol Cell Biol. 1992;12:164–171. doi: 10.1128/mcb.12.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKnight G L, Reasoner J, Gilbert T, Sundquist K O, Hokland B, McKernan P A, Champagne J, Johnson C J, Bailey M C, Holly R, O’Hara P J, Oram J F. Cloning and expression of a cellular high density lipoprotein-binding protein that is up-regulated by cholesterol loading of cells. J Biol Chem. 1992;267:12131–12141. [PubMed] [Google Scholar]
- 44.Müllner E W, Neupert B, Kühn L C. A specific mRNA binding factor regulates the iron-dependent stability of cytoplasmic transferrin receptor mRNA. Cell. 1989;58:373–382. doi: 10.1016/0092-8674(89)90851-9. [DOI] [PubMed] [Google Scholar]
- 45.Musco G, Stier G, Joseph C, Castiglione Morelli M A, Nilges M, Gibson T J, Pastore A. Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome. Cell. 1996;85:237–245. doi: 10.1016/s0092-8674(00)81100-9. [DOI] [PubMed] [Google Scholar]
- 46.Nandabalan K, Roeder G S. Binding of a cell-type-specific RNA splicing factor to its target regulatory sequence. Mol Cell Biol. 1995;15:1953–1960. doi: 10.1128/mcb.15.4.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neu-Yilik G, Zorbas H, Gloe T R, Raabe H M, Hopp-Christensen T A, Müller P K. Vigilin is a cytoplasmic protein. A study on its expression in primary cells and in established cell lines of different species. Eur J Biochem. 1993;213:727–736. doi: 10.1111/j.1432-1033.1993.tb17813.x. [DOI] [PubMed] [Google Scholar]
- 48.Nielsen D A, Shapiro D J. Estradiol and estrogen receptor-dependent stabilization of a minivitellogenin mRNA lacking 5,100 nucleotides of coding sequence. Mol Cell Biol. 1990;10:371–376. doi: 10.1128/mcb.10.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Plenz G, Gan Y, Raabe H M, Müller P K. Expression of vigilin in chicken cartilage and bone. Cell Tissue Res. 1993;273:381–389. doi: 10.1007/BF00312841. [DOI] [PubMed] [Google Scholar]
- 50.Plenz G, Kügler S, Schnittger S, Rieder H, Fonatsch C, Müller P K. The human vigilin gene: identification, chromosomal localization and expression pattern. Hum Genet. 1994;93:575–582. doi: 10.1007/BF00202827. [DOI] [PubMed] [Google Scholar]
- 51.Ree A H, Knutsen H K, Landmark B F, Eskild W, Hansson V. Down-regulation of messenger ribonucleic acid (mRNA) for the estrogen receptor (ER) by phorbol ester requires ongoing RNA synthesis but not protein synthesis. Is hormonal control of ER mRNA degradation mediated by an RNA molecule? Endocrinology. 1992;131:1810–1814. doi: 10.1210/endo.131.4.1382961. [DOI] [PubMed] [Google Scholar]
- 52.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saceda M, Knabbe C, Dickson R B, Lippman M E, Bronzert D, Lindsey R K, Gottardis M M, Martin M B. Post-transcriptional destabilization of estrogen receptor mRNA in MCF-7 cells by 12-O-tetradecanoylphorbol-13-acetate. J Biol Chem. 1991;266:17809–17814. [PubMed] [Google Scholar]
- 54.Schmidt C, Henkel B, Poschl E, Zorbas H, Purschke W G, Gloe T R, Müller P K. Complete cDNA sequence of chicken vigilin, a novel protein with amplified and evolutionary conserved domains. Eur J Biochem. 1992;206:625–634. doi: 10.1111/j.1432-1033.1992.tb16967.x. [DOI] [PubMed] [Google Scholar]
- 55.Siebel C W, Admon A, Rio D C. Soma-specific expression and cloning of PSI, a negative regulator of P element pre-mRNA splicing. Genes Dev. 1995;9:269–283. doi: 10.1101/gad.9.3.269. [DOI] [PubMed] [Google Scholar]
- 56.Singh R, Valcarcel J, Green M R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 57.Siomi H, Choi M, Siomi M C, Nussbaum R L, Dreyfuss G. Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell. 1994;77:33–39. doi: 10.1016/0092-8674(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 58.Siomi H, Matunis M J, Michael W M, Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siomi M C, Siomi H, Sauer W H, Srinivasan S, Nussbaum R L, Dreyfuss G. FXR1, an autosomal homolog of the fragile X mental retardation gene. EMBO J. 1995;14:2401–2408. doi: 10.1002/j.1460-2075.1995.tb07237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Siomi M C, Zhang Y, Siomi H, Dreyfuss G. Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them. Mol Cell Biol. 1996;16:3825–3832. doi: 10.1128/mcb.16.7.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Swanson M S, Dreyfuss G. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol Cell Biol. 1988;8:2237–2241. doi: 10.1128/mcb.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tacke R, Manley J L. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takagaki Y, Manley J L. RNA recognition by the human polyadenylation factor CstF. Mol Cell Biol. 1997;17:3907–3914. doi: 10.1128/mcb.17.7.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsai D E, Harper D S, Keene J D. U1-snRNP-A protein selects a ten nucleotide consensus sequence from a degenerate RNA pool presented in various structural contexts. Nucleic Acids Res. 1991;19:4931–4936. doi: 10.1093/nar/19.18.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tuerk C, Eddy S, Parma D, Gold L. Autogenous translational operator recognized by bacteriophage T4 DNA polymerase. J Mol Biol. 1990;213:749–761. doi: 10.1016/S0022-2836(05)80261-X. [DOI] [PubMed] [Google Scholar]
- 66.Wang J, Dong Z, Bell L R. Sex-lethal interactions with protein and RNA. Roles of glycine-rich and RNA binding domains. J Biol Chem. 1997;272:22227–22235. doi: 10.1074/jbc.272.35.22227. [DOI] [PubMed] [Google Scholar]
- 67.Wang L L, Richard S, Shaw A S. P62 association with RNA is regulated by tyrosine phosphorylation. J Biol Chem. 1995;270:2010–2013. doi: 10.1074/jbc.270.5.2010. [DOI] [PubMed] [Google Scholar]
- 68.Wang X, Liebhaber S A. Complementary change in cis determinants and trans factors in the evolution of an mRNP stability complex. EMBO J. 1996;15:5040–5051. [PMC free article] [PubMed] [Google Scholar]
- 69.Wintersberger U, Kühne C, Karwan A. Scp160p, a new yeast protein associated with the nuclear membrane and the endoplasmic reticulum, is necessary for maintenance of exact ploidy. Yeast. 1995;11:929–944. doi: 10.1002/yea.320111004. [DOI] [PubMed] [Google Scholar]
- 70.Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]