Abstract
Growth suppression by the retinoblastoma protein (RB) is dependent on its ability to form complexes with transcription regulators. At least three distinct protein-binding activities have been identified in RB: the large A/B pocket binds E2F, the A/B pocket binds the LXCXE peptide motif, and the C pocket binds the nuclear c-Abl tyrosine kinase. Substitution of Trp for Arg 661 in the B region of RB (mutant 661) inactivates both E2F and LXCXE binding. The tumor suppression function of mutant 661 is not abolished, because this allele predisposes its carriers to retinoblastoma development with a low penetrance. In cell-based assays, 661 is shown to inhibit G1/S progression. This low-penetrance mutant also induces terminal growth arrest with reduced but detectable activity. We have constructed mutations that disrupt C pocket activity. When overproduced, the RB C-terminal fragment did not induce terminal growth arrest but could inhibit G1/S progression, and this activity was abolished by the C-pocket mutations. In full-length RB, the C-pocket mutations reduced but did not abolish RB function. Interestingly, combination of the C-pocket and 661 mutations completely abolished RB’s ability to cause an increase in the percentage of cells in G1 and to induce terminal growth arrest. These results suggest that the A/B or C region can induce a prolongation of G1 through mechanisms that are independent of each other. In contrast, long-term growth arrest requires combined activities from both regions of RB. In addition, E2F and LXCXE binding are not the only mechanisms through which RB inhibits cell growth. The C pocket also contributes to RB-mediated growth suppression.
The retinoblastoma susceptibility gene, Rb1, encodes a ubiquitously expressed nuclear retinoblastoma protein (RB) which is a negative regulator of cell proliferation (28). The growth-inhibitory activity of RB is neutralized by phosphorylation at G1/S transition in a cell division cycle (27, 28). In addition to inhibiting cell proliferation, RB has also been implicated in the regulation of terminal differentiation and apoptosis (24, 26, 34). The inactivation of RB, either through mutations of the Rb1 gene itself or through mutations that enhance RB phosphorylation, has been widely observed in tumor cells (22).
A major mechanism by which RB inhibits G1/S progression is repression of E2F-regulated genes (20, 28). This transcription repression mechanism is dependent on two distinct protein-binding sites in RB: the large A/B pocket, which binds E2F, and the A/B pocket, which binds the LXCXE peptide motif in proteins. Three recent reports have demonstrated that RB can interact with a histone deacetylase through the LXCXE-binding site (3, 14, 15). The simultaneous binding of E2F and histone deacetylase by RB can therefore lead to the assembly of a transcription repression complex at promoters containing E2F-binding sequences (3, 14, 15). The three-dimensional structure of the A/B domain has been solved (11). The LXCXE-binding site (i.e., the A/B pocket) is entirely within the B region, although its formation is dependent on multiple interactions between the A and B regions (11). Within the A/B domain crystal (which lacks the RB insert and the C-terminal region), the binding site for an E2F peptide and the binding site for the LXCXE peptide are shown to be distinct from each other by the concurrent binding of both peptides to the A/B domain (11). Thus, the three-dimensional structure of the A/B domain is consistent with the model that RB can simultaneously bind to more than one target.
Germ line mutations in Rb1 result in the development of bilateral retinoblastoma in 90% of human carriers. These high-penetrance mutant alleles produce either no RB protein or an unstable product. The importance of the E2F- and LXCXE-binding sites to the tumor suppression function of RB is underscored by the occurrence of at least four germ line point mutations in the A/B domain (11). One of these, the C706F mutation, which produces a highly unstable mutant protein, occurs in the hydrophobic core of the B region. While a majority of the germ line mutations result in 90% predisposition to bilateral retinoblastoma, a few mutant alleles have been found to cause retinoblastoma with a lower penetrance (13). These low-penetrance alleles are likely to encode RB proteins with decreased activities but activities that are sufficient to maintain threshold levels of tumor suppression. One of the low-penetrance RB mutants contains a substitution of Trp for Arg 661 in the B region of RB (R661W) (10, 13). The crystal structure shows that the side chain of Arg 661 (which is in the B region) participates in the formation a hydrogen bond network between helices of the A and B regions (11). Mutation of Arg 661 is therefore likely to disrupt the packing of these alpha helices. The R661W mutation has been shown to disrupt both the LXCXE- and E2F-binding activities of RB (10, 19). However, R661W can suppress the growth of RB-negative tumor cells (10, 19). This suppression is consistent with the ability of R661W to confer partial protection from retinoblastoma development. The partial function of R661W suggests that RB can exert some growth suppression through mechanisms that are independent of binding to E2F and LXCXE proteins (10, 19).
While the presence of the A/B domain is sufficient for RB to bind E2F subunits such as E2F-1, additional interactions provided by the C-terminal region of RB are required for binding to the functional E2F heterodimer (E2F-DP) (5, 17). The C-terminal region of RB is also required for growth suppression, as the A/B domain alone is not functional in cell-based growth suppression assays (17, 21, 32). The minimal functional domain, called the large A/B pocket, which contains RB amino acids 395 to 876, has been shown to correspond to the binding site for E2F heterodimers (5, 17). Welch and Wang have previously shown that RB can bind to the nuclear c-Abl tyrosine kinase and found that the c-Abl binding site is outside of the A/B domain and entirely within the C-terminal region of RB (31). The c-Abl binding site of RB is referred to as the C pocket. Because RB can simultaneously bind E2F and c-Abl (32), the C-terminal E2F-binding site and the C pocket are distinct from each other. Two lines of evidence have indicated that C pocket activity contributes to the growth suppression function of RB (30, 32). First, coexpression of RB with its C-terminal fragment disrupts the ability of RB to suppress growth, as was determined by the inhibition of colony formation in RB-negative Saos-2 cells (32). Second, overproduction of a c-Abl mutant (c-Abl–AS2), which does not bind RB and whose kinase activity is therefore not inhibited by RB, can override the growth suppression function of RB (30).
To directly assess the role of the C pocket in the growth suppression function of RB, we sought to isolate RB C-terminal mutations that specifically disrupt RB–c-Abl interaction without affecting RB-E2F interaction. In this report, we describe the construction and characterization of such mutations. Although the C-terminal fragment by itself cannot induce terminal growth arrest in Saos-2 cells (32), we found that the overproduction of the C-terminal fragment alone can delay G1/S transition. This G1/S-transition-inhibitory activity was disrupted by the C-pocket mutations. In the context of full-length RB, mutations of the C pocket did not affect the ability of RB to inhibit G1/S transition but reduced the growth suppression function of RB by half. In the Saos-2 cell-based assays, the low-penetrance A/B domain mutation R661W also inhibited G1/S transition but produced about one-third the level of wild-type activity in inducing terminal growth arrest. Interestingly, however, combination of C-pocket mutations with R661W led to the complete abrogation of both the G1/S-inhibitory and the growth suppression functions of RB. Taken together, these results suggest that activities from either the A/B domain or the C region can inhibit G1/S progression. However, the induction of terminal growth arrest appears to require contributions from both the A/B domain and the C region. Terminal growth arrest induced by the RB A/B/C region can occur without RB binding to the E2F and LXCXE proteins. This E2F-independent growth suppression requires the activity of the RB C pocket.
MATERIALS AND METHODS
Cell culture and transfections.
The human cervical carcinoma cells C33A and the osteosarcoma cells Saos-2 were obtained from the American Type Culture Collection. These cells were cultured in Dulbecco modified Eagle medium-high glucose supplemented with 15% heat-inactivated fetal bovine serum at 37°C in 5% CO2.
Transfections were performed by the calcium phosphate method, and flat-cell assays and neomycin-resistant colony formation assays were performed with Saos-2 cells as previously described (32). To achieve the same level of protein expression, the amount of plasmid DNA used was titrated for each construct. For the competition experiment described in Table 2, we used a 3:1 ratio of plasmid DNA expressing the C-terminal SE fragment (see Fig. 1A) to plasmid DNA expressing the wild-type RB C-terminal SE fragment.
TABLE 2.
Effects of RB C-terminal fragments on terminal growth arrest
| Transfected RB plasmid(s) | Flat cellsa | Neor coloniesb |
|---|---|---|
| pCMV | − | ++ |
| pCMV-wtRB | ++ | − |
| pCMV-SE | − | ++ |
| pCMV-SEΔ | − | ++ |
| pCMV-SE-13S | − | ++ |
| pCMV-wtRB + pCMV-SE | +/− | ND |
| pCMV-wtRB + pCMV-SEΔ | +/− | ND |
| pCMV-wtRB + pCMV-SE-13S | ++ | ND |
Numbers of flat cells formed relative to that formed by wild-type RB (++) are schematically shown. +/− indicate a number less than 10% of that formed by wild-type RB, and − indicates a number less than 1% of that formed by wild-type RB.
Numbers of neomycin-resistant colonies relative to the number of pCMV-vector-transfected colonies (++; 100%) are schematically shown. − indicates a colony number less than 10% of that of the vector control. ND, not determined.
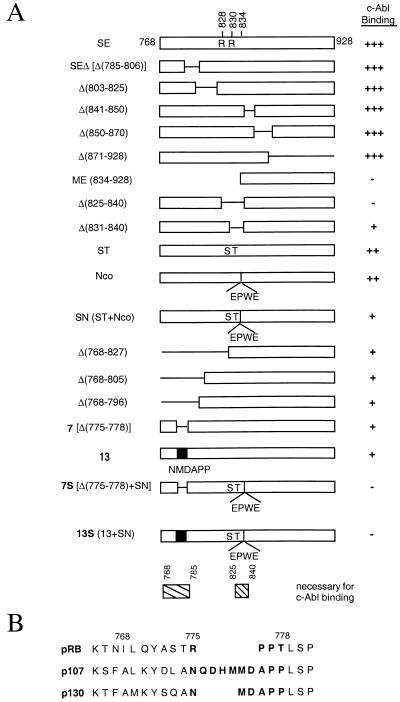
FIG. 1.
Summary of RB C-terminal mutant constructs. (A) c-Abl binding activity of C-terminal mutants of RB. The SE fragment is the wild-type RB C terminus from the SspI site to the end and contains amino acids 768 to 928. This fragment contains the C pocket, as was determined by the presence of c-Abl binding (31). Δ signifies a deletion of the indicated amino acids. The ST mutant is the result of two amino acid substitutions: R828 to S and R830 to T. The Nco mutant contains an insertion at the MunI site (amino acid 834 in exon 24) of a 4-amino-acid sequence, EPWE, and the corresponding base pair sequence introducing an NcoI restriction enzyme site. SN is a combination of the ST and Nco mutations. Mutant 13 was constructed by swapping the sequence of RB which encodes amino acids 775 to 778 (RPPT) with the corresponding p130 sequence (NMDAPP), based on amino acid alignment (see panel B). Mutant 7S is a combination of the deletion of amino acids 775 to 778 and the SN mutation. Mutant 13S is a combination of the mutant 13 and SN mutations. GST-RB mutant binding to in vitro-transcribed and -translated c-Abl is measured relative to GST-SE binding. The wild-type level of activity is indicated with +++. Mutants with 50 to 60% of the wild-type level of activity are indicated with ++. Mutants with 20 to 30% of the wild-type level of activity are indicated with +. Mutants with undetectable activity are indicated with −. (B) Partial amino acid sequence alignment of pRB, p107, and p130 C termini. pRB amino acids 775 to 778 and corresponding nonhomologous p107 and p130 sequence are indicated in boldface print.
The repression of a Gal4-E2F1 fusion transcription factor was measured as described previously, using a reporter containing five tandem repeats of Gal4-DNA binding sequences linked to chloramphenicol acetyltransferase (5×Gal4-CAT) (29). In general, 0.8 μg of 5×Gal4-CAT, 0.8 μg of Gal4-E2F, and between 8 and 16 μg of RB constructs were transfected into Saos-2 cells. The dehydrofolate reductase (DHFR)-Luc reporter constructs contained the wild-type DHFR promoter region (−270 to +20) or the same region with the E2F-binding site mutated (mutDHFR). One microgram of DHFR-Luc reporter or mutDHFR-Luc reporter and 5 to 10 μg of RB constructs were used. In all transfections, 0.5 μg of a β-galactosidase-expressing construct was included to correct for the transfection efficiencies. The total amount of transfected DNA was made up to 18 μg by adding the pCMV-neo-Bam vector.
The RB mutant constructs were made by PCR-based mutagenesis and subcloned into pCMV-neo-Bam for expression. Glutathione S-transferase (GST)-SE and GST-ME (see Fig. 1A) have been previously described (31). The additional GST-RB C-terminal mutants were subcloned in frame into pGEX-KG (Pharmacia).
Binding of RB to GST–E2F-1 and GST-E7.
GST–E2F-1 or GST-E7 was expressed in bacteria and adsorbed onto glutathione-agarose. Each was then incubated at 4°C for 2 h with lysates from C33A cells transfected with pCMV-RB expression plasmids. Preparation of lysates with NETN lysis buffer (0.5% Nonidet P-40, 1 mM EDTA, 50 mM Tris [pH 8.0], 120 mM NaCl) was previously described (9, 31). The agarose beads were washed four times with NETN, and the bound proteins were eluted with sodium dodecyl sulfate (SDS), resolved by SDS–7.5% polyacrylamide gel electrophoresis, and then transferred to Immobilon membranes. Immunoblotting was performed with monoclonal anti-RB 245 antibody (Pharmingen, San Diego, Calif.).
Abl binding and kinase inhibition.
35S-labeled in vitro-translated c-Abl was incubated for 2 h at 4°C with GST-immobilized RB C-terminal fragments expressed as GST fusion proteins (31). The agarose beads were washed four times with NETN, the bound proteins were eluted with SDS and resolved on an SDS–7% polyacrylamide gel, and the amount of GST-RB in each sample was determined by staining with Coomassie blue. The amount of Abl was determined by autoradiography after fluoro-enhancement.
For the c-Abl kinase inhibition assay, in vitro-translated hemagglutinin (HA)-tagged c-Abl was immunoprecipitated with anti-HA antibody (Babco, Berkeley, Calif.) and then incubated with occasional agitation for 30 min at 4°C with an ∼50-fold molar excess of RB C-terminal fragments. The wild-type and mutant RB C-terminal fragments were expressed and purified from bacteria as GST fusion proteins. The anti-HA immunoprecipitates were then washed four times with NETN and twice with 1× kinase buffer (20 mM Tris [pH 7.4], 10 mM MgCl2, 1 mM dithiothreitol). Kinase reactions were initiated by the addition of 100 ng of GST–C-terminal repeated domain (CTD) of RNA polymerase II (2), 5 μM cold ATP, and 20 μCi of [γ-32P]ATP (7,000 Ci/mmol; ICN Pharmaceuticals) in a final volume of 50 μl of 1× kinase buffer. Reaction was carried out at room temperature for 30 min and terminated by adding an equal volume of 3× SDS sample buffer. The products were separated by SDS–7% polyacrylamide gel electrophoresis and transferred onto Immobilon-P, and the amount of 32P incorporated into GST-CTD was determined by autoradiography. The upper portion of the Immobilon membrane corresponding to Abl’s molecular-weight region was probed with anti-Abl (8E9) antibody to determine the amount of Abl in each of the reaction mixtures. The lower portion of the Immobilon membrane corresponding to the GST-RB molecular-weight region was stained with amido black to determine the amount of purified GST-RB added to each reaction mixture.
Immunoprecipitation and immunoblotting of RB.
Immunoprecipitation and immunoblotting of RB were performed as previously described (31). Anti-RB C36 (Pharmingen) was used to immunoprecipitate RB complexes for the deoxycholate (DOC) release-gel shift assay; anti-RB 245 (Pharmingen) was used for immunoblotting RB proteins.
E2F electrophoretic mobility shift assay.
DOC release-gel shift assays were performed as previously described, with transfected C33A cell lysate as the source of RB proteins and endogenous E2F proteins (32).
Luciferase assays.
Luciferase assays were performed with the Promega Luciferase Assay System according to the manufacturer’s protocol. Luciferase activity was corrected for β-galactosidase activity to normalize the transfection efficiency.
Flow cytometry.
RB constructs (5 μg of RB, 7S, and 13S; 10.0 μg of 661, 7S-661, and 13S-661; or 15 μg of SE, SE with amino acids 785 to 806 deleted [SEΔ], and SE-13S) along with CD20 plasmid (0.5 μg) and an amount of the backbone vector sufficient to bring the amount of total DNA to 20 μg were transfected into Saos-2 or C33A cells by the calcium phosphate method. Where indicated in Table 1, cells were treated with 0.1 μg of nocadozole per ml. Cells were harvested and stained with anti-CD20 antibodies and propidium iodide according to published protocols (35). The cell cycle profile of CD20-positive cells was determined by fluorescence-activated cell sorter (FACS) analysis with either Cell Fit or ModFit software (Becton Dickinson).
TABLE 1.
Effect of RB C-terminal fragments on cell cycle progression
| Transfected RB plasmid | Nocadozolea | Cell cycle profileb
|
||
|---|---|---|---|---|
| %G1 | %S | %G2/M | ||
| pCMV | − | 41.0 | 33.9 | 25.1 |
| pCMV-wtRB | − | 69.7 | 12.1 | 18.1 |
| + | 68.0 | 12.8 | 19.1 | |
| pCMV-SE | − | 63.9 | 27.3 | 8.8 |
| + | 61.6 | 20.7 | 17.6 | |
| pCMV-SEΔ | − | 59.5 | 21.2 | 19.4 |
| + | 60.6 | 12.4 | 27.0 | |
| pCMV-SE-13S | − | 44.6 | 29.0 | 26.4 |
| + | 24.5 | 29.2 | 46.3 | |
+ indicates treatment with 0.1 μg of nocadozole per ml for 24 h as described in Materials and Methods.
DNA content of transfected Saos-2 cells was determined as described in Materials and Methods. Data presented are representative of results from three independent experiments. %G1, %S, and %G2/M, percentages of cells in G1, S, and G2/M, respectively.
RESULTS
Construction of RB C-pocket mutants defective in c-Abl binding.
The C pocket has been defined as the binding site for the c-Abl tyrosine kinase (31). The C pocket was previously shown to be within the C-terminal region of RB, from amino acids 768 to 928 (Fig. 1) (31). The E2F binding site, i.e., the large A/B pocket, resides within amino acids 379 to 876 of RB (5, 17). The C pocket and the large A/B pocket are functionally distinct, as it is possible for RB to simultaneously bind both E2F and c-Abl (32). An internal deletion of RB amino acids 785 to 806 was previously shown to disrupt E2F binding (5) but not c-Abl binding (32). These observations indicated that it might be possible to isolate RB mutants which have lost C pocket function but that have retained E2F binding activity.
Binding to c-Abl was determined by the ability of GST fusion proteins containing the C-terminal region of RB to interact with in vitro-translated [35S]Met-labeled c-Abl (Materials and Methods). The binding of each RB mutant was compared to that of GST–RB-SE (from the SspI site to the end) which contains wild-type RB amino acids 768 to 928 (Fig. 1A). The GST-ME fusion, containing RB amino acids 834 to 928, served as a negative control, as it has previously been shown to be defective in binding c-Abl (31, 32). As summarized in Fig. 1A, deletion of amino acids 870 to 928 does not affect either c-Abl binding or E2F binding. Internal deletions of amino acids 785 to 806 (SEΔ), 803 to 825, 841 to 850, and 850 to 870 have previously been shown to disrupt E2F binding (5), but these deletions did not have any detectable effect on c-Abl binding (Fig. 1A). Thus, RB amino acids 785 to 825 and 841 to 870 are dispensable for the formation of the C pocket.
An internal deletion of amino acids 825 to 840 decreased c-Abl binding by 60 to 70%, as did a smaller deletion eliminating the 10 amino acids encoded by RB exon 24 (amino acids 831 to 840) (Fig. 1A). Alternative manipulations of this region, such as substitutions of Arg 828 and Arg 830 for Ser and Thr, respectively (mutant ST), or an insertion of 4 amino acids at position 834 (mutant Nco), also reduced c-Abl-binding activity (Fig. 1A). Further reduction of c-Abl binding was achieved when the ST and Nco mutations were combined to create the mutant designated SN (Fig. 1A). Though RB-SN had reduced c-Abl-binding activity, this combination of mutations did not completely abolish c-Abl binding (Fig. 1A). Internal deletions of a noncontiguous region, between amino acids 768 and 805 or 768 and 796, were found to also reduce c-Abl-binding activity by 60 to 70% (Fig. 1A). Taken together, the results of the deletion analysis indicate that two separate regions, from amino acids 768 to 785 and amino acids 825 to 840, are necessary for c-Abl binding.
The RB-related p107 protein does not bind c-Abl (32a). Thus, nonhomologous amino acids between RB and p107 may be required for c-Abl binding by RB. RB and p107 have extensive homology in the A and B domains. However, the RB sequence from amino acids 775 to 778, immediately C terminal to the B domain, is not conserved in the corresponding p107 or p130 sequence (Fig. 1B). Deletion of these 4 amino acids (775 to 778; mutant 7) decreased c-Abl binding to the same level as the deletion of amino acids 768 to 796 (Fig. 1A). We also constructed a substitution mutation in which the RB sequence from amino acids 775 to 778 (RPPT) was replaced with p130 sequence (NMDAPP), which is homologous to the p107 sequence. The p130 sequence was used instead of the p107 sequence because the p130 sequence introduced fewer amino acids (Fig. 1B). The substitution mutant (designated mutant 13) had the same impact on c-Abl binding as the mutant in which amino acids 775 to 778 were deleted (Fig. 1A).
Each of the individual mutations in either the region from amino acids 775 to 778 or the region from amino acids 825 to 840 compromised but did not eliminate c-Abl binding. However, combining two of the most disruptive mutations into a single mutant—either 7S (combining mutant 7 with SN) or 13S (combining mutant 13 with SN)—was sufficient to abrogate c-Abl binding (Fig. 1A). Taken together, these data indicate that the C pocket is composed of two noncontiguous regions within RB amino acids 768 to 870. While disruption of either of these regions decreases c-Abl binding, disruption of both regions is required to completely eliminate c-Abl binding.
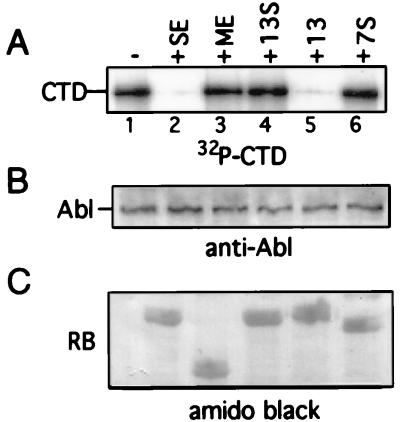
The consequence of RB binding to c-Abl is the inhibition of c-Abl tyrosine kinase activity (30, 31). This kinase inhibition can be recapitulated in vitro with purified GST–RB-C-terminus fusion proteins. GST-SE, which contains a functional C pocket, efficiently inhibits c-Abl kinase activity (Fig. 2, lane 2). A truncated RB C-terminal fragment, GST-ME, which does not bind c-Abl, does not inhibit c-Abl kinase activity (lane 3). Neither mutant 13S nor mutant 7S could inhibit c-Abl tyrosine kinase (lanes 4 and 6, respectively), providing a functional confirmation for the C-pocket binding defect in mutants 7S and 13S.
FIG. 2.
RB C-pocket mutants do not inhibit c-Abl kinase activity. The indicated GST-RB C-terminal proteins purified from bacteria were incubated with in vitro-translated c-Abl. The c-Abl proteins from these incubations were then subjected to an immune-complex kinase assay, with a GST-CTD fusion protein as the substrate. Phosphorylated CTD was detected by autoradiography (A). The immune complex was also subjected to anti-Abl immunoblot analysis (B) to measure the relative amount of in vitro-translated c-Abl in each reaction. The amount of GST-RB protein present in each reaction mixture was visualized by amido black staining of the nitrocellulose membrane after transfer (C). See Fig. 1 for explanations of the RB fragments.
E2F binding and other activities of the C-pocket mutants.
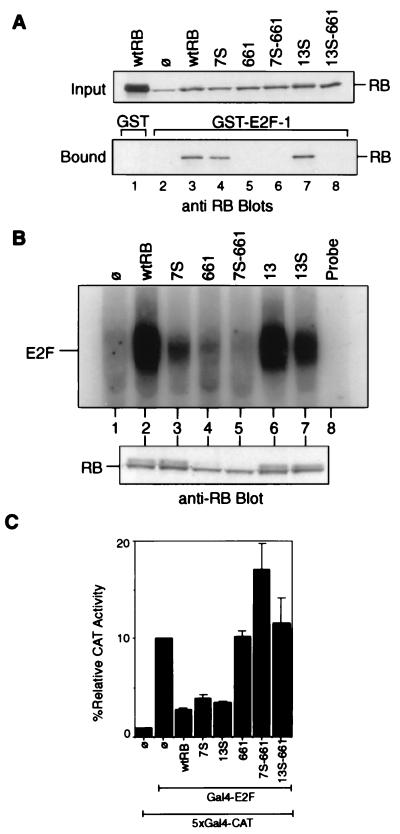
The C-terminal region of RB is required for RB’s binding to E2F (5); we therefore examined whether the 7S and 13S mutations affected E2F binding activity. First, we assayed for binding of RB proteins, transiently produced in C33A cells, to GST–E2F-1 immobilized on glutathione agarose. Total cell lysates were added to immobilized GST–E2F-1, and the input and the bound RB proteins were then detected by anti-RB immunoblotting (Fig. 3A). The binding of RB to E2F-1 is mediated by the A/B domain interaction with a C-terminal 18-amino-acid peptide in E2F-1 (11). Therefore, mutations in the C-terminal regions were not expected to interfere with the RB–E2F-1 interaction. The wild-type RB did not bind nonspecifically to GST alone (Fig. 3A, lane 1). C33A cells express a mutant RB which is unstable and does not bind to E2F-1 (lane 2) (18). The R661W mutation, which has been shown to disrupt RB–E2F interaction (19), served as an additional control (lane 5). The C-pocket mutants, 7S (lane 4) and 13S (lane 7), were able to bind GST–E2F-1 with efficiencies comparable to that of RB (lane 3). Thus, the mutations in 7S and 13S did not affect A/B domain-mediated binding to E2F-1.
FIG. 3.
Interactions of RB mutants with E2F. (A) In vitro binding of RB to GST–E2F-1. Lysates from C33A cells transfected with the indicated construct were collected 48 h posttransfection. These lysates were incubated with GST (lane 1) or GST–E2F-1 (lanes 2 to 8) immobilized on glutathione agarose beads. Relative amounts of the indicated RB proteins in each reaction mixture (input) were determined by anti-RB immunoblot analysis (upper blot). RB proteins bound to the glutathione beads are visualized in the lower blot. The RB band in lane 2 of the input blot is the endogenous mutant RB of C33A cells. wtRB, wild-type RB; ø, vector-transfected cells. (B) Binding of full-length RB mutants to E2F in vivo. Extracts were prepared from C33A cells transfected with the vector (lane 1) or the indicated RB expression constructs (lanes 2 to 7). Immunoprecipitation of E2F DNA-binding activity with anti-RB antibodies has been previously described (32). The immunoprecipitate was treated with DOC, and the released material was assayed for binding to a radiolabeled E2F oligonucleotide. Relative amounts of RB-bound E2F are indicated (upper gel). Relative amounts of the indicated RB proteins in each reaction mixture are determined by anti-RB immunoblot analysis (lower blot). (C) Inhibition of Gal4–E2F-1 activity by RB. Saos-2 cells were cotransfected with β-galactosidase, the 5×Gal4-CAT reporter, the Gal4–E2F-1 effector, and either the RB, 7S, 13S, 661, 7S-661, or 13S-661 CMV construct as described in Materials and Methods. CAT activity was normalized to β-galactosidase activity. The CAT activity of Gal4–E2F-1 alone without RB was considered 100%. Results shown are the means and standard deviations of results of three independent experiments.
The association of RB with the functional E2F was then examined by a coimmunoprecipitation-release assay. Mudryj et al. have shown that E2F DNA-binding activity (which is mediated by the E2F-DP heterodimers) can be immunoprecipitated with anti-RB antibody and then dissociated from RB by treatment with DOC (16). By this assay, E2F DNA-binding activity was found to coimmunoprecipitate with the wild-type RB (Fig. 3B, lane 2). The mutated RB of C33A cells did not coimmunoprecipitate with E2F (lane 1), nor did mutant 661 (lane 4). The 7S mutant, though expressed at the same level as RB (Fig. 3B [Western blot]), was compromised in its ability to coimmunoprecipitate functional E2F (lane 3). Mutant 13 coimmunoprecipitated E2F activity as efficiently as RB (lane 6). Mutant 13S also bound functional E2F (lane 7). This result suggested that deletion of amino acids 775 to 778 in 7S did affect the association of RB with functional E2F. However, a substitution of p130 sequence for RB amino acids 775 to 778 in 13S was less disruptive of RB-E2F interaction.
We next examined the C-pocket mutants for their ability to inhibit the transactivating function of a Gal4–E2F-1 fusion protein that contains the transactivating (and RB-binding) domain of E2F-1 (Fig. 3C). The 7S and the 13S mutants inhibited Gal4–E2F-1 with efficiencies similar to that of wild-type RB (Fig. 3C). The 661 mutant, unable to bind E2F, did not inhibit Gal4–E2F-1 activity (Fig. 3C). These observations are in keeping with the E2F-1-binding results (Fig. 3A) and further confirmed that the C-pocket mutations in 13S and 7S do not disrupt the ability of RB to inhibit the transactivating function of E2F-1. We have tested the 7S and 13S mutants with four other promoter constructs: an artificial luciferase reporter with three tandem E2F-binding sites (33), the adenovirus E2 promoter, the human cyclin A promoter, and the human cdc2 promoter. We found that 7S and 13S inhibited all four promoters to levels comparable to that of wild-type RB in transient-cotransfection assays conducted with either Saos-2 cells or C33A cells (not shown). These results suggested that the two C-pocket mutants, 7S and 13S, could interact with E2F at levels sufficient to block its transactivating function.
In other functional tests, we examined the LXCXE-binding activities of the 7S and 13S mutants, using a GST-E7 fusion protein as the ligand (9). Consistent with the crystal structure showing that the LXCXE-binding site resides in the B region only (11), we found that the 7S and 13S mutants bound the human papillomavirus type 16 E7 protein as well as wild-type RB did (not shown). The 7S and 13S mutations did not affect RB phosphorylation (see the anti-RB blot in Fig. 3B). The R661W mutation did interfere with RB phosphorylation in C33A cells, possibly because the LXCXE-binding site is required to recruit cyclin D, which is responsible for RB phosphorylation (22). The 7S and 13S mutants localized to the nuclei of Saos-2 and C33A cells (not shown). In addition, the 7S and 13S mutations did not affect the binding of RB to Mdm2 (33) (not shown). Taken together, the results of these analyses suggested that the 13S mutation affected only c-Abl binding but that the 7S mutations affected both biding to c-Abl and binding to E2F. We could not rule out the possibility that the 13S mutation also affected other unidentified functions of RB.
The 13S mutations abolished the G1/S-inhibitory activity associated with the RB C-terminal fragment.
Welch and Wang have previously shown that the C-terminal fragment of RB, when expressed alone, cannot suppress the growth of Saos-2 cells as determined by the induction of growth-arrested flat cells and the inhibition of colony formation (32). Expression of RB in Saos-2 cells also induces an increase in the G1 population, which can be detected within 48 h of transfection (6). To assay for an increase in the G1 population, Saos-2 cells were cotransfected with plasmids expressing the RB mutants of interest and the cell surface marker CD20. Forty-eight hours posttransfection, the DNA content of CD20-positive cells was determined by FACS analysis. When cotransfected with RB, ∼70% of the CD20-positive cells were in G1 whereas only ∼40% of the vector-cotransfected CD20-positive cells had G1 DNA content (Table 1). Expressing the C-terminal SE fragment (containing RB amino acids 768 to 928) (Fig. 1) at a level similar to that of RB did not induce an increase in the G1 population (not shown). However, when SE was overproduced to a level that was threefold higher, it caused an increase in the percentage of G1 cells (Table 1). A C-terminal fragment (SEΔ) which lacks a region required for E2F binding (5, 32) (Fig. 1) also caused an increase in the G1 population (Table 1), suggesting that E2F binding is not essential to the G1-inhibitory activity of SE. An SE fragment with the C-pocket mutation 13S (SE-13S), when expressed at levels equivalent to that of SE, was unable to cause an increase in the G1 population of Saos-2 cells (Table 1).
To test if the increase in G1 cells caused by SE and SEΔ was the result of a G1 arrest or a lengthening of the G1 phase, transfected Saos-2 cells were treated with nocadozole, which blocks cells at G2/M (Table 1). If cells are arrested in G1, nocadozole will not cause an increase in G2/M. However, if cells are slowly progressing through G1, nocadozole treatment will lead to an increase in G2/M (12). Nocadozole treatment did not affect the percentage of G1, S, or G2 cells in the RB-transfected population (Table 1), consistent with previous results demonstrating G1 arrest (12). While nocadozole treatment did not alter the percentage of G1 cells in the SE- or SEΔ-transfected population, the percentage of cells in S phase was decreased and the percentage of cells in G2/M phase was increased (Table 1), indicating a prolonged G1 phase but a slow progression through the cell cycle. The cells transfected with the SE-13S mutant showed a reduction in G1 population and a concomitant increase in G2/M population after nocadozole treatment, consistent with the inability of SE-13S to slow cells in G1 (Table 1).
Welch and Wang have previously constructed a c-Abl mutant (AS2) which does not bind RB and is therefore resistant to the inhibitory effect of RB. When overproduced, AS2 can antagonize RB in flat-cell formation and colony inhibition assays (30). Because SE, but not SE-13S, can induce an increase in the G1 population, we tested whether the binding and/or the inhibition of c-Abl tyrosine kinase was required for this activity. Towards this end, we coexpressed SE and c-Abl–AS2 in Saos-2 cells. If the G1-inhibitory activity of SE were due to the inhibition of c-Abl tyrosine kinase, then c-Abl–AS2 would be expected to override that activity. However, this was not the case. The expression of c-Abl–AS2 did not alter the cell cycle distribution of transfected, CD20-positive Saos-2 cells (not shown). Moreover, coexpression of c-Abl–AS2 did not have any detectable effect on an SE-induced increase in the percentage of cells in G1 (not shown). Taken together, these results show that the SE-mediated increase in the G1 population is disrupted by the 13S mutation; however, this effect cannot be reversed by a constitutive Abl-AS2 kinase that does not interact with RB.
The 7S, 13S, or 661 mutations do not affect the G1/S-inhibitory function of full-length RB.
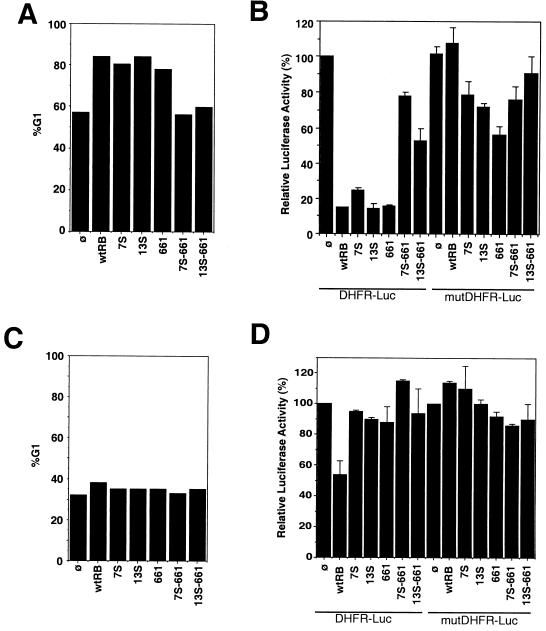
The 13S mutation had no effect on the G1/S-inhibitory activity of the full-length RB. The percentages of G1 cells of one representative experiment are shown in Fig. 4A. Overall, CD20-positive cells cotransfected with RB were between 70 and 85% G1 whereas vector-cotransfected cells were between 40 and 55% G1. Both the 7S and 13S mutants caused consistent increases in percentages of G1 cells to levels comparable to that of wild-type RB (Fig. 4A). The 661 mutant also induced an increase in G1 cells comparable to that of RB in Saos-2 cells (Fig. 4A). In these experiments, different amounts of expression plasmids were used to achieve the same levels of RB accumulation at 48 h posttransfection (for example, see Fig. 5C). Thus, neither the A/B domain mutation nor the C-region mutations affected the ability of full-length RB to induce an increase in the percentage of cells in G1.
FIG. 4.
Abilities of RB mutants to induce G1 arrest. (A) Induction of an increase in the G1 population of Saos-2 cells. Saos-2 cells were cotransfected with CMV–β-galactosidase, CD20, the DHFR-Luc reporter, or the mutDHFR-Luc reporter and either the vector, wild-type RB (wtRB), 7S, 13S, 661, 7S-661, or 13S-661 (as described in Materials and Methods). One-half of each transfection mixture was used for cell cycle analysis, and the other half was used for a luciferase assay (B). Transfected cells were stained for CD20 and DNA (see Materials and Methods). The gated CD20-positive cells were analyzed by FACS analysis for DNA content. Results of a representative experiment are shown. Similar results were obtained from three additional independent experiments. %G1, percent increase in number of cells in G1. (B) Repression of the DHFR promoter in Saos-2 cells. One-half of the transfected Saos-2 cells in panel A were analyzed for luciferase activity, which was normalized to cotransfected β-galactosidase activity to correct for transfection efficiency. The normalized luciferase activities are represented as percentages of the luciferase activity of vector-cotransfected cells (ø), which is considered 100%. Results shown are the means and standard deviations of results from four independent experiments. (C) Inability of RB to induce an increase in the G1 population of C33A cells. C33A cells were cotransfected with CMV–β-galactosidase, CD20, the DHFR-Luc reporter, or the mutDHFR-Luc reporter and either the vector, RB, 7S, 13S, 661, 7S-661, or 13S-661 as described for panel A. Transfected cells were stained for CD20 and DNA, and the percentages of cells in G1 were determined as described in Materials and Methods. (D) Repression of the DHFR promoter in C33A cells. C33A cells were analyzed as described for panel B. Normalized luciferase activities are represented as percentages of the luciferase activity of vector-cotransfected cells (ø), which is considered 100%. Results shown are the means and standard deviations of results from four independent experiments.
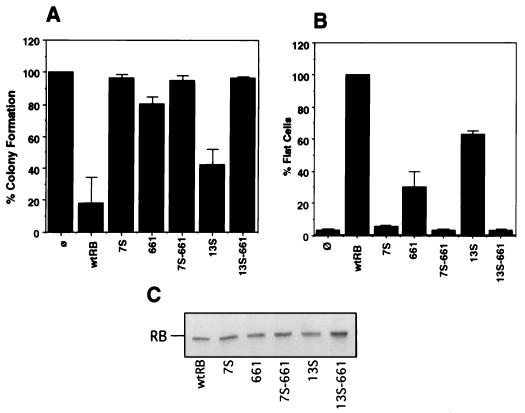
FIG. 5.
Effects of C-pocket mutations on terminal growth arrest. (A) Inhibition of neomycin-resistant colony formation. The vector used to express RB and the various mutants contained a neomycin resistance gene. The expression plasmid DNA was transfected into Saos-2 cells. Transfectants were subjected to G418 selection for 2 to 3 weeks. The number of neomycin-resistant colonies arising from each transfectant was counted and expressed as a percentage relative to the number of such colonies in the vector-alone transfectant, which was considered 100%. Results shown are the means and standard deviations of results of four independent experiments. wtRB, wild-type RB; ø, vector-cotransfected cells. (B) Induction of flat-cell formation. Saos-2 cells were cotransfected with pCMVneo-RB, -7S, -13S, -661, –7S-661, –13S-661, or -vector and a puromycin resistance plasmid. Transfected cells were selected with puromycin for 7 days. Resulting flat cells were counted in each dish, and the number of flat cells formed for each transfectant was expressed a percentage of the number of flat cells formed by RB, which was considered 100%. Results shown are the means and standard deviations of results from four independent experiments. (C) Protein expression levels. The amount of plasmid transfected was titrated to yield equal levels of RB protein expression. Saos-2 cells transfected with RB, 7S, 661, 7S-661, 13S, or 13S-661 were divided into three equal parts: one-third of the cells was used for the colony formation assay (A), one-third was used for the flat-cell assay (B), and one-third was collected at 48 h posttransfection for protein analysis (C). Equal amounts of total protein from lysates collected from the transfected cells were separated on an SDS–7.5% polyacrylamide gel. RB proteins were detected with anti-RB 245 antibody (Pharmingen). The transfected pCMV-RB expression construct for each lysate is indicated.
The ability to cause an increase in the G1 population correlated with the repression of the DHFR promoter by the RB mutants. The DHFR promoter contains an E2F-binding sequence which is responsible for the repression of this promoter in G0/G1 cells (7, 20, 23). The effect of RB mutants on DHFR promoter activity was examined with a reporter that contains the region from −270 to +20 of the DHFR gene. A mutant promoter lacking the E2F-binding site was also examined for comparison (Fig. 4B). Mutation of the E2F-binding site did not abolish promoter activities in Saos-2 cells or C33A cells (Fig. 4B and D, compare the ø samples of DHFR-Luc and mutDHFR-Luc). These results showed that transcription from this promoter fragment in Saos-2 and C33A cells is stimulated by factors other than E2F (20, 23). Cotransfection with RB repressed DHFR promoter activity in Saos-2 cells (Fig. 4B, lane with wild-type RB and DHFR-Luc), and this repression was abolished by the mutation of the E2F site (Fig. 4B, lane with wild-type RB and mutDHFR-Luc). In Saos-2 cells, 7S and 13S also repressed the DHFR promoter and this was again dependent on the E2F site (Fig. 4B). The 661 mutant, although unable to bind E2F (19), also repressed the DHFR promoter in Saos-2 cells (Fig. 4B). Mutation of the E2F site reduced but did not abolish the repressive activity of 661 on the DHFR promoter (Fig. 4B, compare lanes with 661 to lanes with wild-type RB), suggesting that 661 might repress the DHFR promoter through a mechanism that does not require a stable RB-E2F interaction.
The repression of the DHFR promoter may be mediated by one of two mechanisms: (i) a direct RB-E2F interaction at the promoter and (ii) an increase in the G1 population that leads to repression, possibly through endogenous p130–p107-E2F complexes (7, 23). To distinguish between these two possibilities, we repeated the DHFR promoter assays with the RB-negative C33A cells, which do not become arrested in G1 with the introduction of exogenous RB (35). As shown in Fig. 4C, expression of RB or the various mutants did not increase the G1 population. In C33A cells, RB was able to repress the DHFR promoter, albeit at only a twofold reduction, compared to the eightfold reduction observed with Saos-2 cells (compare lanes with wild-type RB and DHFR-Luc in Fig. 4B and D). Interestingly, 7S, 13S, and 661 were unable to repress the DHFR promoter in these C33A cells (Fig. 4D). These results suggest that the repression of the DHFR promoter by the RB mutants is the result of an increase in the G1 population in Saos-2 cells. Since the E2F-binding sites are involved, it is possible that this repression is mediated by endogenous p107-p130. Because the 7S, 13S, and 661 mutants did not repress the DHFR promoter in C33A cells, these results also suggest that both the A/B domain and the C pocket are required to achieve the twofold repression of the DHFR promoter by wild-type RB in C33A cells.
The 7S, 13S, and 661 mutations affect the growth suppression function to various degrees.
Because the increase in the G1 population at 48 h posttransfection may have been the result of a lengthening of the G1 phase but not a permanent G1 arrest, we tested the mutants by two other assays: a flat cell induction assay and a colony formation inhibition assay (Fig. 5). When transfected into Saos-2 cells, RB can inhibit the formation of drug-resistant colonies, measured at 14 days posttransfection (17, 32) (Fig. 5A). In our assay, cotransfection with RB led to a fivefold reduction in colony numbers relative to the number for the vector control (Fig. 5A, bar wtRB). The 13S mutant was compromised in its ability to suppress G418-resistant colonies, producing ∼50% of the level of activity produced by the wild type in repeated experiments (Fig. 5A, bar 13S). The 7S mutant, though capable of inducing an increase in the G1 population (Fig. 4A), was unable to suppress colony formation (Fig. 5A, bar 7S) at an expression level equivalent to that of RB (Fig. 5C, lane 7S). The 661 mutant exhibited consistent but weak activity in colony suppression, producing only 25 to 30% of the level of activity produced by the wild type (Fig. 5A).
Expression of RB in Saos-2 cells also induced the formation of large flat cells whose state appeared to be senescent and/or differentiated (19, 25). Previous work showed that both the A/B and C regions of RB are required for flat-cell formation (5, 17, 30). In our flat-cell assay, 13S exhibited 60% of the level of activity exhibited by the wild type whereas 7S was completely defective (Fig. 5B). The 661 mutant was capable of inducing flat cells with approximately 30% efficiency relative to that of RB (Fig. 5B). Taken together, these results showed that the induction of flat cells and the inhibition of colony formation by RB are differentially affected by the 7S, 13S, and 661 mutations (Fig. 5). This finding is in direct contrast to results from induction of an increase in the G1 population, which is not affected by any of the three mutations alone (Fig. 4).
Combining the 7S or 13S mutations with R661W abrogates the G1/S and the growth suppression activities of RB.
The tumor suppression function of 661 is compromised; however, this mutant retains sufficient activity to protect against retinoblastoma in some carriers. As the C-terminal region of 661 is intact, the residual biological function of 661 may depend on the function of the C pocket. To test this idea, we inactivated the C pocket of 661 through the construction of 7S-661 and 13S-661.
The 7S-661 and 13S-661 mutants could be stably expressed in Saos-2 cells (Fig. 5C) and C33A cells (Fig. 3B). At expression levels comparable to that of RB, 7S-661 and 13S-661 did not increase the G1 population at 48 h after transfection (Fig. 4A) or repress transcription from the DHFR promoter (Fig. 4B). These mutants did not suppress colony formation (Fig. 5A), and they did not induce flat-cell formation (Fig. 5B). Abrogation of the G1/S-inhibitory activity of 661 by 7S or 13S suggests that an intact C-terminal region is required for this activity. This suggestion is consistent with the observations that SE alone can cause an increase in the G1 population and that this increase is inactivated by the 13S mutations. Conversely, the G1/S-inhibitory activity of 13S must require an intact A/B domain, which is disrupted by 661. Regarding the growth suppression functions as measured by flat-cell formation or colony inhibition, the 7S-661 mutant was uninformative because 7S itself has no detectable growth suppression function. Because the C-terminal fragment alone did not have any growth suppression function, the partial activity of 661 could not be attributed to the C region alone. However, an intact C region is required for the biological activity of this low-penetrance retinoblastoma mutation because 661-13S is defective in growth suppression assays.
Fusion with the E2F-1 DNA-binding domain does not rescue the 13S-661 mutant.
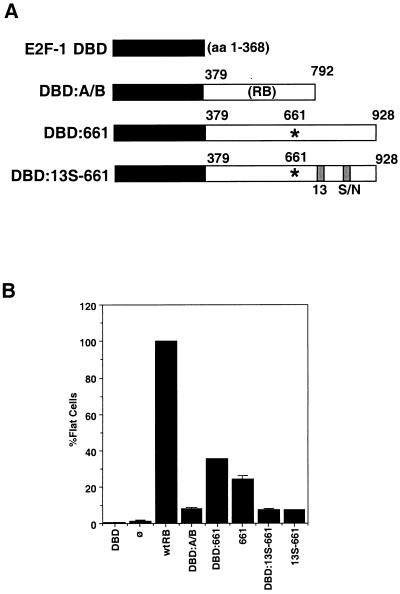
Although 661 did not detectably bind to E2F (Fig. 3), the residual activity of 661 in the growth suppression assays may have been due to weak binding to E2F mediated by the mutant’s C-terminal region. It might be argued that the defect of 13S-661 was due to the disruption of this weak E2F interaction by the 13S mutation. If this were the case, then the fusion of the E2F-1 DNA-binding domain with 13S-661 would be expected to rescue the 13S-661 mutant. Sellers et al. previously demonstrated that fusion of the A/B domain of RB with the DNA-binding and dimerization domain of E2F-1 generates a transcription repressor that can induce an increase in the G1 population of Saos-2 cells (21). Following their strategy, we constructed two fusion proteins in which the E2F-1 DNA-binding and dimerization domain (E2F-1 amino acids 1 to 368 [DBD]) was linked in frame with the A/B/C region (amino acids 379 to 928) of either 661 or 13S-661 (DBD-661 and DBD–13S-661, respectively) (Fig. 6A). As a control, we used the fusion construct previously described by Sellers et al. DBD-A/B (21), in which the DNA-binding domain of E2F-1 is linked in frame with RB amino acids 379 to 793 (Fig. 6A). The DBD-A/B fusion, therefore, lacks a functional C pocket.
FIG. 6.
Loss of function of the RB 13S-661 mutant cannot be rescued by fusion with the E2F DNA-binding domain. (A) E2F-RB mutant fusion constructs. An E2F-1 fragment from amino acids (aa) 1 to 368 containing the E2F DNA binding domain was fused in frame to RB amino acids 379 to 928 containing either the 661 mutation (DBD-661) or the 661 mutation plus the 13S C-pocket mutation (DBD–13S-661). The E2F-1 DBD and DBD-A/B have been previously described (21). (B) Fusion of an E2F DNA binding domain does not enhance the formation of flat cells. DBD, RB, DBD-A/B, DBD-661, 661, DBD–13S-661, 13S-661, or pCMVneo was cotransfected with a puromycin resistance plasmid into Saos-2 cells. Cells were selected for 7 days in medium containing puromycin, and flat cells were counted. The number of flat cells formed for each transfectant was expressed as a percentage of the number of flat cells formed by RB, which was considered 100%. Results shown are the means and standard deviations of results from three independent experiments. ø, vector-transfected cells.
These fusion constructs were tested for their ability to induce flat Saos-2 cells (Fig. 6B). Both DBD-661 and 661 induced flat-cell formation at ∼30% of the wild-type level of efficiency (Fig. 6B). Thus, the addition of the E2F DNA-binding domain did not increase the growth suppression function of 661. The E2F DBD–13S-661 fusion protein, like 13S-661, did not induce flat-cell formation (Fig. 6B). Thus, the addition of the E2F-1 DNA-binding domain did not rescue the defect of 13S-661. Sellers et al. have previously reported that the A/B fragment alone (amino acids 379 to 792) does not have any biological activities but that the fusion protein DBD-A/B can inhibit E2F-dependent transcription and induce an increase in the G1 population (21). In our hands, DBD-A/B also caused an increase in the G1 population (not shown). However, this DBD-A/B fusion did not induce flat-cell formation (Fig. 6B). Taken together, these results showed that the tethering of 13S-661 to E2F sites did not restore the growth suppression function. In addition, the lack of C-pocket activity was correlated with the inability of DBD-A/B and 13S-661 to induce flat Saos-2 cells.
Coexpression of SE, but not SE-13S, disrupts the growth suppression activity of wild-type RB.
The partial growth suppression activity of 661 could not be attributed entirely to its intact C region. This is because the C-terminal region alone cannot induce flat cells and it cannot inhibit colony formation (32). Consistently with previous results (32), expression of the SE, SEΔ, and SE-13S C-terminal fragments did not induce flat cells or inhibit neomycin-resistant-colony formation (Table 2). Welch and Wang have reported that coexpression with SE or SEΔ can abrogate the growth suppression activity of full-length RB and showed that SE and SEΔ could compete with RB for binding to c-Abl (32). If binding to c-Abl is required for SE and SEΔ to disrupt the function of RB, then SE-13S should not interfere with RB. This was indeed the case (Table 2). Using the flat-cell formation assay, we found that the coexpression of SE or SEΔ with RB caused a 10- to 20-fold reduction in the number of flat cells. However, coexpression with SE-13S did not reduce the number of flat cells induced by RB (Table 2). These observations show that the ability to bind c-Abl is necessary for the C-terminal RB fragment to exert a dominant negative effect on the growth suppression function of wild-type RB.
While SE and SEΔ can interfere with RB in flat-cell and colony formation assays, they did not interfere with the induction of an increase in the G1 population by RB (not shown). This result is consistent with the observations that SE and SEΔ alone can induce an increase in the number of cells in G1 and that the DBD-A/B fusion of Sellers et al. can also induce such an increase (21). Thus, the A/B and C regions of RB appear to be able to cause an increase in the G1 population independently of each other.
DISCUSSION
Definition of the RB C pocket.
The mutational analysis described here has further characterized the RB C pocket, defined as the c-Abl binding site. First, the C pocket is within the minimal functional domain of RB (RB amino acids 395 to 876). Second, two nonconsecutive amino acid sequences in the C-terminal region, from 768 to 785 and from 825 to 840, are important for the c-Abl-binding function (Fig. 1). Third, the c-Abl-binding function requires RB amino acids that are not conserved in p107-p130. The mutational analysis supports the previous conclusion that c-Abl and E2F can simultaneously bind to RB (32), as mutations that disrupt E2F binding do not affect c-Abl binding; conversely, mutations that disrupt c-Abl binding do not disrupt E2F binding.
The C-terminal region of RB also binds to Mdm2 (33). Janicke et al. have recently shown that Mdm2 binding requires the extreme C-terminal sequences of RB (8). RB is cleaved at a caspase consensus site at amino acid 884 during apoptosis (reviewed in reference 24). Caspase-cleaved RB, which is shortened by only 44 amino acids, does not bind Mdm2 (8, 24). Cleavage by caspase at amino acid 884 does not affect c-Abl binding (23a), consistent with the mapping results shown in Fig. 1. Because Mdm2 binding requires amino acids C terminal to the minimal functional domain of RB (amino acids 395 to 876), the RB-Mdm2 interaction may not contribute to growth suppression but may be important in the regulation of apoptosis (reviewed in references 24 and 26).
Induction of an increase in the percentage of cells in G1 can be distinguished from the growth suppression function of RB.
Previous studies of RB mutations have generated a simple picture in which the loss of E2F binding is correlated with the complete inactivation of RB (28). Results obtained from the analyses of the 7S, 13S, and 661 mutations (see Table 3 for a summary), however, have allowed the separation of two distinct functions of RB in Saos-2 cell-based assays: (i) inhibition of G1/S progression, as measured by an increase in the percentage of cells in G1, and (ii) long-term growth arrest, as measured by the inhibition of colony formation and the induction of flat cells.
TABLE 3.
Summary of growth-inhibitory activities of RB mutants
(i) The A/B and C regions of RB can each induce an increase in the percentage of cells in G1.
Three different types of RB mutants—7S, the DBD-A/B fusion (21), and a C-terminal fragment (SE or SEΔ)—can cause an increase in the G1 population but are unable to induce terminal growth arrest (Table 3). The 7S mutant does not bind c-Abl and is compromised for binding to E2F (Fig. 2 and 3). The DBD-A/B fusion does not bind to c-Abl or E2F; however, it can be tethered to E2F sites through fusion with E2F-1 DBD (21). The SE and SEΔ fragments do not contain the A/B domain. The fact that all three types of mutants can cause an increase in the percentage of cells in G1 suggests (i) that the A/B domain can increase the G1 population when it is tethered to E2F sites without the C-terminal region and (ii) that the C-terminal region can increase the G1 population without the A/B domain. The conclusion that the A/B and C regions can each inhibit G1/S progression is further supported by the following observations. Disruption of the A/B domain by 661 did not affect G1/S-inhibitory activity. Disruption of the C pocket by 13S also had no detectable effect on this activity of RB. However, combination of these two mutations completely abolished G1/S-inhibitory activity (Table 3).
The negative effect of RB on G1/S progression has been attributed to the repression of E2F-regulated genes (28). A recent study conducted with Rb-deficient mouse embryo fibroblasts demonstrated that the expression of many E2F-regulated genes was not significantly altered; however, the G1 phase was shortened in the absence of RB function (7). It thus appears that critical genes involved in G1/S progression and controlled by RB are yet to be identified. The recent demonstration of RB binding to a histone deacetylase can account for the RB-mediated repression of promoters containing E2F sites (3, 14, 15). The 7S and the DBD-A/B mutants contain a functional LXCXE-binding site, and they may inhibit G1/S progression by recruiting histone deacetylase to E2F-regulated promoters. Sellers et al. have shown that introduction of either the C706F or the R661W mutation into the DBD-A/B fusion can inactivate its function (21). Thus, an intact LXCXE-binding site is important for the A/B domain to cause an increase in the percentage of cells in G1 when the domain is tethered to E2F sites.
The observation that 661 and SE, both of which do not bind E2F or LXCXE proteins, can also cause an increase the G1 population suggests that RB may do so through mechanisms other than the binding to E2F and histone deacetylase. Introduction of the 13S mutation can eliminate the G1/S-inhibitory functions of both 661 and SE (Table 3). Introduction of the mutation deleting amino acids 785 to 806, which disrupts E2F binding but not c-Abl binding, does not affect this activity of SE (Table 3). Thus, the G1/S-inhibitory activity associated with the C-terminal region of RB can be correlated with binding to c-Abl. Since constitutively active c-Abl–AS2 (30) cannot overcome SE, the inhibition of c-Abl kinase activity is not important for the lengthening of G1 phase. This, however, does not rule out the possibility that binding to c-Abl is required for SE to induce an increase in the G1 population. In addition to binding RB, the c-Abl protein contains two distinct binding sites for CTD of RNA polymerase II and has been shown to associate with RNA polymerase II in vivo (1, 2, 4). Binding to c-Abl may be required to bring the SE fragment to the transcription machinery. At present, we cannot rule out that the 13S mutations may have also inactivated other functions of RB. Welch and Wang have previously argued that c-Abl binding is not the only biologically important function of the C region of RB (30, 32). The G1-inhibitory activity of the RB SE fragment may well be mediated through as yet unidentified mechanisms unrelated to RB–c-Abl interaction.
(ii) The A/B and C regions cooperate to induce growth arrest.
The minimal functional domain of RB includes the A/B and C regions of RB, between amino acids 395 and 876 (5, 17). This functional domain can bind to E2F, histone deacetylase, c-Abl, and other targets (reviewed in references 20, 26, and 27). Because the tethering of a functional A/B domain to E2F sites (achieved with the DBD-A/B fusion) cannot induce flat-cell formation, the C region is required for growth suppression (Table 3). Because the C-terminal fragment of RB is also unable to induce flat cells, the A/B domain is required as well (Table 3). Moreover, coexpression of the SE fragment with RB disrupts its growth suppression function (32). These results suggest that activities associated with the A/B and C regions may have to cooperate to induce growth arrest. The A/B and C regions clearly cooperate to bind E2F (5, 17), and E2F binding has been proposed to be the only reason why the C region is required for growth suppression. This model cannot explain how 661, which does not bind to E2F, can induce growth arrest (see also the report of Sellers et al. [19]). Sellers et al. have proposed that 661 suppresses Saos-2 growth by inducing a terminal differentiation program (19). We show here that the 661 function can be inactivated by the 13S mutations (Table 3). The precise mechanism by which 661 suppresses growth is presently unknown. Analyses performed in this study suggest that the A/B and C regions of 661 also cooperate to provide the reduced but significant growth suppression function.
The role of C pocket in growth suppression.
Three lines of evidence have argued for the importance of an RB–c-Abl interaction in the induction of growth arrest. First, overproduction of c-Abl–AS2 can override the growth suppression function of RB (30). Second, coexpression with SE, but not SE-13S, can disrupt the growth suppression function of RB. Third, the tethering of the A/B domain to E2F sites (DBD-A/B) can induce an increase in the G1 population but not increase the number of flat cells. However, those results did not prove the C pocket to be essential. The contribution of C-pocket activity can be assessed only through analyses of the C-pocket mutations.
Effects of the C-pocket mutations on the biological activities of RB are complex. In the context of the C region alone, which by itself can delay G1/S progression, disruption of the C pocket inactivated this biological function. In the context of the full-length RB, however, the disruption of RB–c-Abl interaction did not have a significant effect on growth suppression. The 7S mutations abolished growth suppression (Table 3), but the severity of the 7S mutations may be due to a combined defect in c-Abl and E2F binding (Fig. 2 and 3). It should be noted that 7S binds E2F better than 661 (Fig. 3), yet 7S is defective and 661 has partial function. The combined mutations of 13S and 661 also completely abolished biological activities. Taken together, these results show that C-pocket activity becomes essential to growth suppression in the context of other RB mutations that weaken the RB-E2F interaction.
A possible interpretation of the various phenotypes of these RB mutants is to consider that the A/B and C regions of RB can also bind targets other than E2F, histone deacetylase, and c-Abl. Each of these multiple targets of RB may contribute to the growth suppression activities measured in Saos-2 cell-based assays. The crippled A/B domain of 661 may retain some activity in binding to proteins other than E2F and histone deacetylase. In the context of 661, an intact C region becomes critical for either the formation or the functioning of these alternative protein complexes that cause growth inhibition. In the context of wild-type RB, the C pocket is less important as the assembly of E2F and LXCXE protein complexes may be sufficient to suppress growth. In any event, the complete loss of function associated with the 13S-661 double mutant supports a role for the RB C pocket in the induction of long-term growth arrest.
ACKNOWLEDGMENTS
We thank Paolo Vigneri, Lauren Wood, and Pam Woodring for critically reading the manuscript. We also thank the following researchers for the generous provision of reagents: B. Vogelstein (Johns Hopkins University); E. Harlow (Massachusetts General and Harvard); H. Land (Imperial Cancer Research Fund, London, United Kingdom); W. Krek, D. Livingston, W. Sellers, and W. Kaelin, Jr. (Dana-Farber Cancer Institute); S. Hiebert (St. Jude Children’s Hospital); C. Murre (University of California, San Diego); and A. Means and P. Farnham (McArdle Laboratory for Cancer Research, University of Wisconsin Medical School).
H.S. was supported by National Institutes of Health grant CA66314. R.B. was supported by Council for Tobacco Research grant CTR3762. This research was supported by National Institutes of Health grant CA58320 to J.Y.J. Wang.
Footnotes
In memory of Laura L. Whitaker, 25 February 1998.
REFERENCES
- 1.Baskaran R, Chiang G, Mysliwiec T, Krul G, Wang J Y J. Tyrosine phosphorylation of RNA polymerase II carboxyl-terminal domain by Abl-related gene (Arg) encoded tyrosine kinase. J Biol Chem. 1997;272:18905–18909. doi: 10.1074/jbc.272.30.18905. [DOI] [PubMed] [Google Scholar]
- 2.Baskaran R, Chiang G G, Wang J Y J. Identification of a binding site in c-Abl tyrosine kinase for the C-terminal repeated domain of RNA polymerase II. Mol Cell Biol. 1996;16:3361–3369. doi: 10.1128/mcb.16.7.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brehm A, Miska E, McCane D, Reid J, Bannister A, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 4.Duyster J, Baskaran R, Wang J Y J. Src homology 2 domain as a specificity determinant in the c-Abl mediated tyrosine phosphorylation of the RNA polymerase II carboxyl-terminal repeated domain. Proc Natl Acad Sci USA. 1995;92:1555–1559. doi: 10.1073/pnas.92.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hiebert S W. Regions of the retinoblastoma gene product required for its interaction with the E2F transcription factor are necessary for E2 promoter repression and pRb-mediated growth suppression. Mol Cell Biol. 1993;13:3384–3391. doi: 10.1128/mcb.13.6.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinds P W, Mittnacht S, Dulic V, Arnold A, Reed S I, Weinberg R A. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 7.Hurford R K, Cobrinik D, Lee M-H, Dyson N. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 1997;11:1447–1463. [Google Scholar]
- 8.Janicke R U, Walker P A, Lin X Y, Porter A G. Specific cleavage of the retinoblastoma protein by an ICE-like protease in apoptosis. EMBO J. 1996;15:6969–6978. [PMC free article] [PubMed] [Google Scholar]
- 9.Knudsen E S, Wang J Y J. Differential regulation of retinoblastoma protein function by specific cdk phosphorylation sites. J Biol Chem. 1996;271:8313–8320. doi: 10.1074/jbc.271.14.8313. [DOI] [PubMed] [Google Scholar]
- 10.Kratzke R A, Otterson G A, Hogg A, Coxon A B, Geradts J, Cowell J K, Kaye F J. Partial inactivation of the RB product in a family with incomplete penetrance of familial retinoblastoma and benign retinal tumors. Oncogene. 1994;9:1321–1326. [PubMed] [Google Scholar]
- 11.Lee J-O, Russo A, Pavletich N. Structure of the retinoblastoma tumor suppressor pocket domain bound to a peptide from HPV E7. Nature. 1998;391:859–865. doi: 10.1038/36038. [DOI] [PubMed] [Google Scholar]
- 12.Lees E, Faha B, Dulic V, Reed S I, Harlow E. Cyclin E/cdk2 and cyclin A/cdk2 kinases associate with p107 and E2F in a temporally distinct manner. Genes Dev. 1992;6:1874–1885. doi: 10.1101/gad.6.10.1874. [DOI] [PubMed] [Google Scholar]
- 13.Lohmann D, Brandt B, Hooping W, Passarge E, Horsthemke B. Distinct Rb1 gene mutations with low penetrance in hereditary retinoblastoma. Hum Genet. 1994;94:349–354. doi: 10.1007/BF00201591. [DOI] [PubMed] [Google Scholar]
- 14.Luo R, Postigo A, Dean D. RB interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 15.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Villain J L, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 16.Mudryj M, Devoto S H, Hiebert S W, Hunter T, Pines J, Nevins J R. Cell cycle regulation of the E2F transcription factor involves an interaction with cyclin A. Cell. 1991;65:1243–1253. doi: 10.1016/0092-8674(91)90019-u. [DOI] [PubMed] [Google Scholar]
- 17.Qin X-Q, Chittenden T, Livingston D M, Kaelin W G., Jr Identification of a growth suppression domain within the retinoblastoma gene product. Genes Dev. 1992;6:953–964. doi: 10.1101/gad.6.6.953. [DOI] [PubMed] [Google Scholar]
- 18.Scheffner M, Munger K, Byrne J, Howley P. The state of the p53 and retinoblastoma genes in human cervical carcinoma cell lines. Proc Natl Acad Sci USA. 1991;88:5523–5527. doi: 10.1073/pnas.88.13.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sellers W, Novitch B, Miyake S, Heith A, Otterson G, Kaye F, Lassar A, Kaelin W G., Jr Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation and suppress tumor cell growth. Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sellers W R, Kaelin W G., Jr RB as a modulator of transcription. Biochim Biophys Acta. 1996;1288:1–5. doi: 10.1016/0304-419x(96)00014-5. [DOI] [PubMed] [Google Scholar]
- 21.Sellers W R, Rodgers J W, Kaelin W G., Jr A potent transrepression domain in the retinoblastoma protein induces a cell cycle arrest when bound to E2F sites. Proc Natl Acad Sci USA. 1995;92:11544–11548. doi: 10.1073/pnas.92.25.11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherr C. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 23.Slanksy J E, Farnham P J. Introduction to the E2F family: protein structure and gene regulation. Curr Top Microbiol Immunol. 1996;208:1–30. doi: 10.1007/978-3-642-79910-5_1. [DOI] [PubMed] [Google Scholar]
- 23a.Tan, X.-Q., H. Su, and J. Y. J. Wang. Unpublished data.
- 24.Tan X-Q, Wang J Y J. The caspase-RB connection in cell death. Trends Cell Biol. 1998;8:116–120. doi: 10.1016/s0962-8924(97)01208-7. [DOI] [PubMed] [Google Scholar]
- 25.Templeton D J, Park S H, Lanier L, Weinberg R A. Nonfunctional mutants of the retinoblastoma protein are characterized by defects in phosphorylation, viral oncoprotein association, and nuclear tethering. Proc Natl Acad Sci USA. 1991;88:3033–3037. doi: 10.1073/pnas.88.8.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J Y J. Retinoblastoma protein in growth suppression and death protection. Curr Opin Genet Dev. 1997;7:39–45. doi: 10.1016/s0959-437x(97)80107-4. [DOI] [PubMed] [Google Scholar]
- 27.Wang J Y J, Knudsen E S, Welch P J. The retinoblastoma tumor suppressor protein. Adv Cancer Res. 1994;64:25–85. doi: 10.1016/s0065-230x(08)60834-9. [DOI] [PubMed] [Google Scholar]
- 28.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 29.Weintraub S J, Chow K N B, Luo R X, Zhang S H, He S, Dean D C. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 30.Welch P J, Wang J Y J. Abrogation of retinoblastoma protein function by c-Abl through tyrosine kinase-dependent and -independent mechanisms. Mol Cell Biol. 1995;15:5542–5551. doi: 10.1128/mcb.15.10.5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welch P J, Wang J Y J. A C-terminal protein-binding domain in the retinoblastoma protein regulates nuclear c-Abl tyrosine kinase in the cell cycle. Cell. 1993;75:779–790. doi: 10.1016/0092-8674(93)90497-e. [DOI] [PubMed] [Google Scholar]
- 32.Welch P J, Wang J Y J. Disruption of retinoblastoma protein function by coexpression of its C pocket fragment. Genes Dev. 1995;9:31–46. doi: 10.1101/gad.9.1.31. [DOI] [PubMed] [Google Scholar]
- 32a.Welch, P. J., and J. Y. J. Wang. Unpublished data.
- 33.Xiao Z-X, Chen J, Levine A, Modjtahedi N, Xing J, Sellers W R, Livingston D M. Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature. 1995;375:694–698. doi: 10.1038/375694a0. [DOI] [PubMed] [Google Scholar]
- 34.Zacksenhaus E, Jiang Z, Chung D, Marth J D, Phillips R A, Gallie B L. pRb controls proliferation, differentiation, and death of skeletal muscle cells and other lineages during embryogenesis. Genes Dev. 1996;10:3051–3064. doi: 10.1101/gad.10.23.3051. [DOI] [PubMed] [Google Scholar]
- 35.Zhu L, van den Heuvel S, Helin K, Fattaey A, Ewen M, Livingston D, Dyson N, Harlow E. Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein. Genes Dev. 1993;7:1111–1125. doi: 10.1101/gad.7.7a.1111. [DOI] [PubMed] [Google Scholar]