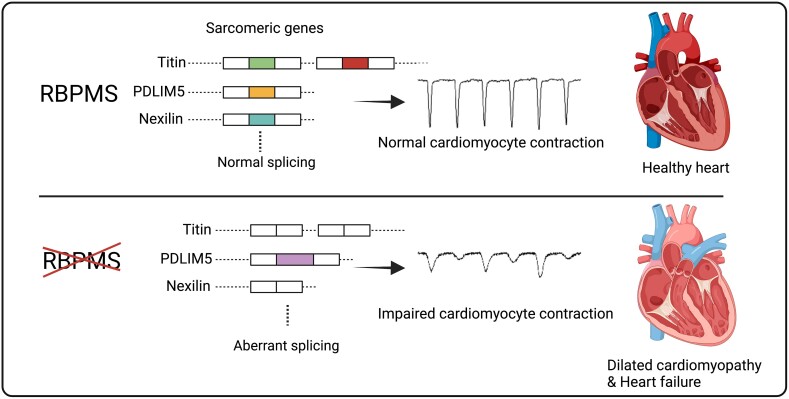
Abstract
Aims
RNA binding proteins play essential roles in mediating RNA splicing and are key post-transcriptional regulators in the heart. Our recent study demonstrated that RBPMS (RNA binding protein with multiple splicing) is crucial for cardiac development through modulating mRNA splicing, but little is known about its functions in the adult heart. In this study, we aim to characterize the post-natal cardiac function of Rbpms and its mechanism of action.
Methods and results
We generated a cardiac-specific knockout mouse line and found that cardiac-specific loss of Rbpms caused severe cardiomyocyte contractile defects, leading to dilated cardiomyopathy and early lethality in adult mice. We showed by proximity-dependent biotin identification assay and mass spectrometry that RBPMS associates with spliceosome factors and other RNA binding proteins, such as RBM20, that are important in cardiac function. We performed paired-end RNA sequencing and RT–PCR and found that RBPMS regulates mRNA alternative splicing of genes associated with sarcomere structure and function, such as Ttn, Pdlim5, and Nexn, generating new protein isoforms. Using a minigene splicing reporter assay, we determined that RBPMS regulates target gene splicing through recognizing tandem intronic CAC motifs. We also showed that RBPMS knockdown in human induced pluripotent stem cell-derived cardiomyocytes impaired cardiomyocyte contraction.
Conclusion
This study identifies RBPMS as an important regulator of cardiomyocyte contraction and cardiac function by modulating sarcomeric gene alternative splicing.
Keywords: RNA binding protein, Cardiomyocyte contraction, Dilated cardiomyopathy, Titin, Pdlim5
Graphical Abstract
Graphical Abstract.
Time of primary review: 24 days
1. Introduction
Cardiomyocyte maturation is a hallmark of heart development, involving enhanced contractility, metabolic switching, and structural maturation.1 The increase in contractility of individual cardiomyocytes supports the forceful pumping of the post-natal heart. Abnormal cardiomyocyte contractility can cause various cardiac diseases, including hypertrophic and dilated cardiomyopathy (DCM).2 Cardiomyocyte contraction is tightly regulated by the intracellular calcium concentration, a process known as excitation–contraction (E-C) coupling. The cardiomyocyte sarcomere, composed of hundreds of different sarcomeric proteins, serves as the contractile unit that senses calcium fluctuation.3 Functional maturation of the sarcomere during heart development relies on transcriptional changes and mRNA alternative splicing of sarcomeric genes,1 and much remains to be learned.
RNA post-transcriptional modifications act as an important layer of gene regulation, in addition to transcriptional regulation and epigenetic modifications. RNA splicing is a key process of post-transcriptional gene regulation, which amplifies the transcriptional diversity of the genome. RNA splicing is orchestrated by the spliceosome complex and various RNA binding proteins (RBPs), including the serine/arginine-rich (SR) protein family and the heterogeneous nuclear ribonucleoprotein (hnRNP) protein family.4 The cooperation of different spliceosome factors and RBPs finely tunes splicing in tissue-specific manners and can generate different protein products from a single gene. In recent years, mutations in RBPs, such as RBM20, have been shown to cause DCM through aberrant mRNA splicing of sarcomere genes,5 pointing towards a new aetiological mechanism of cardiovascular diseases. Rbpms is an evolutionarily conserved RBP that is highly expressed in heart and smooth muscle-enriched organs.6 Global knockout of Rbpms causes neonatal lethality in mice, which also manifests as myocardium non-compaction and patent ductus arteriosus.7 Rbpms deficiency influences RNA alternative splicing in foetal mouse heart, disrupting cardiomyocyte cytokinesis and causing premature cardiomyocyte binucleation.7 However, the cardiomyocyte-specific function of Rbpms in the post-natal heart has not been explored, due to the neonatal lethality in global knockout mice.
To address these questions, we generated a cardiac-specific Rbpms knockout mouse. We found that cardiac deletion of Rbpms caused DCM and heart failure. Unlike its role in the foetal heart, Rbpms mainly regulates cardiomyocyte contractility in the adult heart. Knockdown of RBPMS in human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) results in impaired contractility, highlighting its conserved role in mouse and human cardiomyocyte contractility. We also found that RBPMS is broadly associated with spliceosome factors and directly mediates target gene splicing. Overall, our work reveals the post-natal cardiac functions of Rbpms and provides insights into how RBP regulates cardiac function in developing and adult hearts.
2. Methods
2.1. Study approval
Animal work described was approved and conducted under the oversight of the University of Texas Southwestern Institutional Animal Care and Use Committee.
2.2. Generation of Rbpms cKO mice
A mixture of Cas9 mRNA, a pair of Rbpms sgRNAs targeting sequences 5′ and 3′ of Rbpms exon 1, and corresponding single-stranded oligodeoxynucleotide DNA (ssDNA, IDT Ultramer DNA Oligos) donors containing loxP sequence were injected into the pronucleus and cytoplasm of B6C3F1 mouse zygotes. Chimeric F0 mice were crossed to C57BL/6 mice to establish germline transmission of the conditional allele. An equal number of male and female mice were used in every animal experiment.
2.3. Mouse echocardiography
Cardiac function was evaluated by two-dimensional transthoracic echocardiography on conscious mice at 1, 2, or 5 months of age using a VisualSonics Vevo2100 imaging system as described previously.7 All measurements were performed by an experienced operator blinded to the study.
2.4. Mouse cardiomyocyte contractility and calcium transient
Isolation of cardiomyocytes, contractility, and calcium transient analysis were performed as previously described.8 Cardiomyocytes were isolated from mice at 2 months of age by Langendorff perfusion and collagenase digestion. The contractility and calcium fluctuation of individual cardiomyocytes were measured by the IonOptix Myocyte Calcium and Contractility System.
2.5. Paired-end RNA sequencing (RNA-seq) and alternative splicing analysis
RNA from 2-month-old heart ventricle tissues was extracted for paired-end RNA-seq. The analysis was performed as previously described.7
2.6. RT–PCR analysis
Total RNA was extracted from mouse heart ventricular tissues or hiPSC-CMs using Trizol and reverse transcribed using iScript Reverse Transcription Supermix (Bio-Rad) with random primers.
2.7. Minigene reporter splicing assay
Ttn exon 11, Ttn exon 47, Pdlim5 exons 5 and 6, and flanking intronic sequences were cloned into Globin minigene reporter plasmid. HEK293 cells were co-transfected with minigenes and Rbpms expression plasmid. Forty-eight hours after transfection, RNA was extracted and reverse transcribed into cDNAs. RT–PCR analysis was performed to check minigene splicing patterns.
2.8. HiPSC-CMs culture and electrophysiological analysis
All iPSC work was performed in compliance with the UT Southwestern Stem Cell Research Oversight Committee. Human iPSCs were differentiated using CHIR-99021 and WNT-C59 small molecules as previously described.7 Mature (4 months) hiPSC-CMs were plated on 35 mm glass bottom dishes for contractility and calcium fluctuation measurements.
2.9. Statistical analysis
Results are expressed as mean ± SEM. Unpaired two-tailed Student’s t-test with Welch correction was performed to determine statistical significance. P-values of <0.05 were considered significant.
More details can be found in the Supplementary Methods.
3. Results
3.1. Cardiac-specific deletion of Rbpms causes early post-natal lethality and DCM in mice
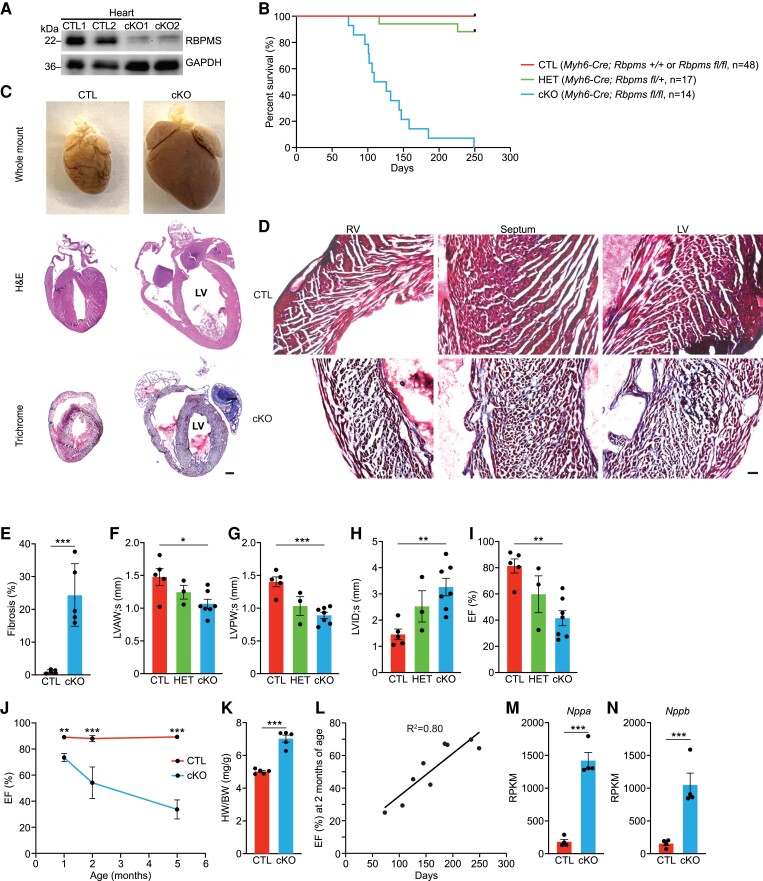
Global Rbpms knockout (KO) mice show neonatal lethality, precluding the analysis of its cardiac function in adulthood.7 To circumvent this early lethality, we generated mice carrying a conditional allele of Rbpms by inserting loxP sequences flanking Rbpms exon 1, using CRISPR-Cas9-mediated homology-directed repair (HDR) (see Supplementary material online, Figure S1A). Insertion of loxP sites was confirmed by PCR (see Supplementary material online, Figure S1A). We intercrossed Rbpmsfl/+ mice with Myh6-Cre mice to generate Myh6-Cre; Rbpmsfl/fl mice, referred to as cardiac KO (cKO) mice. Myh6-Cre; Rbpms+/+ mice and Rbpmsfl/fl mice were used as controls (CTLs). Heterozygous (HET) mice were Myh6-Cre; Rbpmsfl/+. Loss of RBPMS protein expression in cKO hearts was confirmed by western blotting in 2-month-old mice, while its expression was not changed in kidney, intestine, or liver (Figure 1A; Supplementary material online, Figure S1B).
Figure 1.
Cardiac-specific deletion of Rbpms causes early post-natal lethality and DCM in mice. (A) Western blot analysis showing RBPMS protein expression in the hearts of Rbpms CTL and cKO mice at 2 months of age (n = 2 for CTL and cKO). GAPDH is a loading control. (B) Survival curve of Rbpms CTL, HET, and cKO mice. (C) Whole mount (top panel), H&E (middle panel), and Masson’s trichrome-stained (bottom panel) coronal sections of CTL and cKO hearts at 4 months of age. Scale bar: 2 mm. (D) Masson’s trichrome staining showing regions of the left ventricle (LV), septum, and right ventricle (RV) of CTL and cKO hearts at 4 months of age. Scale bar: 50 μm. (E) Quantification of the percentage of fibrotic area over the whole myocardium in hearts at 4 months of age (n = 5 for CTL and cKO). (F–I) Measurements of LV anterior wall thickness at end cardiac systole (LVAW; s), LV posterior wall thickness at end cardiac systole (LVPW; s), LV internal dimension at end cardiac systole (LVID; s), and ejection fraction (EF%) of CTL (n = 5), HET (n = 3), and cKO (n = 7) hearts at 2 months of age by echocardiography. (J) Comparison of EF% between CTL and cKO hearts at 1, 2, and 5 months of age (n = 4–10 for CTL and cKO at each stage). (K) Quantification of heart weight to body weight ratios of CTL and cKO mice at 4 months of age. CTL (n = 5) and cKO (n = 5). (L) Scatter diagram showing the correlation between the EF% at 2 months of age and the life span of cKO mice (n = 9). (M, N) Expression levels of Nppa and Nppb mRNA in CTL and cKO hearts at 2 months of age, measured by RNA-seq (n = 4 for CTL and cKO). All data are presented as mean ± SEM. *P < 0.05, **P < 0.01; ***P < 0.001, by Student’s t-test two-tailed for E, J, K, M, N, and by one-way ANOVA and post hoc analysis with Bonferroni correction for F, G, H, I.
Rbpms cKO mice showed early lethality starting from 73 days of age (Figure 1B). All cKO mice died by 9 months of age, and their average lifespan was 129 days (Figure 1B). The ratio of female to male cKO mice at weaning was 1:1, suggesting that there was no gender-specific consequence of Rbpms ablation (see Supplementary material online, Figure S1C, D). At 4 months of age, cKO hearts were larger than those of CTL mice (Figure 1C top panel). Histological analysis revealed severely enlarged ventricular chamber volume and thinner myocardium in cKO hearts, hallmarks of DCM (Figure 1C). Masson’s trichrome staining revealed pervasive fibrosis in the left ventricle (LV), right ventricle, and septum of the cKO hearts (Figure 1C, D), accounting for ∼20% of ventricular myocardium (Figure 1E).
To evaluate cardiac function of Rbpms cKO mice, we performed echocardiographic analysis at 1 and 2 months of age (see Supplementary material online, Table S1, S2). At 1 month of age, cKO hearts showed mild reductions of left ventricular anterior and posterior wall thickness (LVAW; s and LVPW; s) and an increase of left ventricular internal diameter at end systole (LVID; s) (see Supplementary material online, Figure S1E–G), indicative of LV dilation. We also observed a mild reduction of ejection fraction (EF%) in cKO mice compared to littermate CTL mice (see Supplementary material online, Figure S1H). Consistent with these results, 1-month-old cKO hearts showed modest left ventricular chamber dilation by H&E staining (see Supplementary material online, Figure S1I). At 2 months of age, cKO hearts showed significantly reduced left ventricular wall thickness, increased internal dimension at end systole, and a significant reduction in ejection fraction (Figure 1F–I; Supplementary material online, Figure S1J). We observed a progressive worsening of cardiac contractility in cKO hearts with age, and by 5 months of age, the EF% of the cKO hearts was less than 40% (Figure 1J). At 4 months of age, cKO mice had increased heart weight to body weight ratios compared to CTLs (Figure 1K). We did not observe any obvious conduction defects in cKO hearts, except for mild sinus arrhythmia (see Supplementary material online, Figure S1K). We found a strong correlation between the EF% at 2 months of age and the lifespan of cKO mice (R2 = 0.80). Mice with higher EF% at 2 months of age lived longer than mice with lower EF% (Figure 1L). In addition, cKO hearts expressed significantly higher levels of the cardiac disease markers Nppa and Nppb at 2 months of age (Figure 1M, N). Intriguingly, we observed a modest reduction of Rbpms2 expression in cKO hearts, indicating the potential functional interaction between the two Rbpms homologues (see Supplementary material online, Figure S1L). We did not observe DCM phenotypes or reduced cardiac function in HET mice (Figure 1F–I). Taken together, Rbpms cKO mice developed DCM, reduced cardiac contractility, and died of heart failure.
3.2. Rbpms cKO hearts exhibit myocardial non-compaction and progressive ventricular dilation
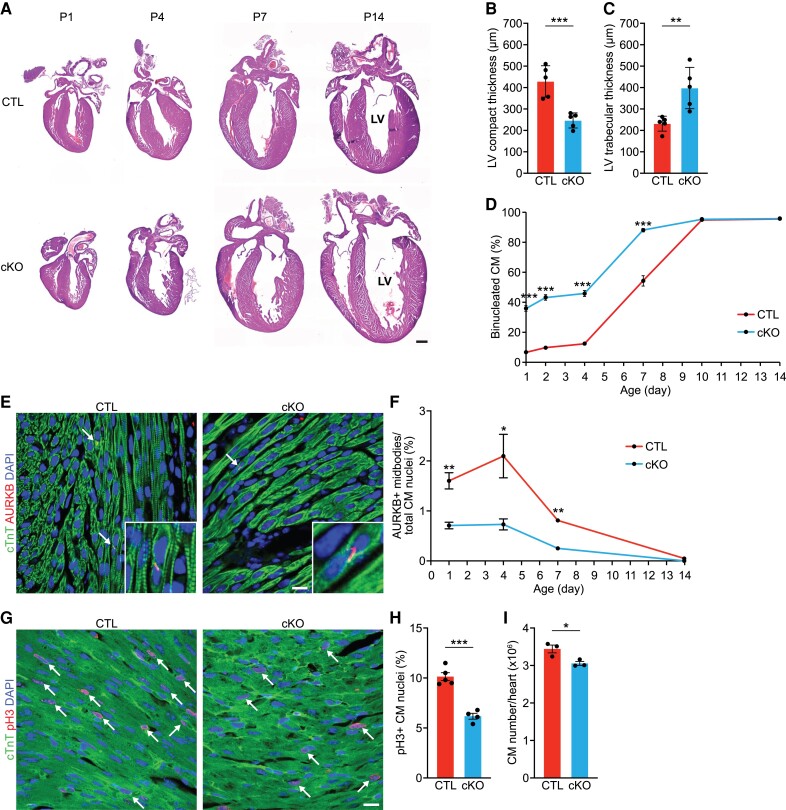
We previously demonstrated that Rbpms KO hearts develop non-compaction cardiomyopathy at birth, caused by defects in cardiomyocyte cytokinesis and premature cardiomyocyte binucleation.7 To investigate whether neonatal cKO hearts recapitulate the non-compaction cardiomyopathy phenotype, we performed histological analysis of cKO hearts at P1, P4, P7, and P14. At P1, like KO hearts, cKO hearts showed myocardium non-compaction, as evidenced by the reduced compact zone thickness and increased trabecular zone thickness in the LV (Figure 2A–C). There was no significant difference in the LV compact or trabecular zone thickness between cKO and KO hearts at P1 (see Supplementary material online, Figure S2A, B), suggesting that cKO hearts recapitulated the non-compaction cardiomyopathy of KO hearts.
Figure 2.
Rbpms cKO hearts exhibit myocardial non-compaction and progressive ventricular dilation. (A) Representative H&E images of CTL and cKO hearts at post-natal (P) days P1, P4, P7, P14. Scale bar: 500 μm. (B, C) Measurements of left ventricle (LV) compact myocardium, and trabecular zone thicknesses in coronal sections of P1 hearts at the level of papillary muscle roots (n = 5 for CTL and cKO). (D) Percentage of binucleated cardiomyocytes in CTL and cKO hearts at the indicated post-natal days (n = 3–6 for CTL and cKO mice at each stage, average 100–200 cardiomyocytes per mouse). (E) Representative immunofluorescence images of CTL and cKO hearts at P1 stained for AURKB and cTnT. Scale bar: 25 μm. White arrows indicate AURKB-positive midbodies between nuclei. (F) Percentage of AURKB-positive midbodies in CTL and cKO hearts at the indicated post-natal days (n = 5–7 for CTL and cKO at each stage). CM, cardiomyocyte. (G, H) Immunofluorescence staining for phosphorylated histone H3 (pH3), cTnI, and DAPI of CTL and cKO myocardium at P1 (G) and percentage of pH3+ CM nuclei (H). White arrows indicate pH3-positive CM nuclei. CTL (n = 5) and cKO (n = 4). P-value represents result of t-test. Scale bar: 25 μm. (I) Total cardiomyocyte (CM) number in CTL and cKO hearts at P14 (n = 3 for CTL and cKO). All data are presented as mean ± SEM. *P < 0.05, **P < 0.01; ***P < 0.001, by Student’s t-test two-tailed for B–D, F, H, I.
Ploidy analysis revealed that cKO hearts had a higher percentage of binucleated cardiomyocytes than CTL hearts until P7, while hearts of both genotypes reached a plateau of ∼95% binucleated cardiomyocytes at P10 (Figure 2D). Intriguingly, the percentage of binucleated cardiomyocytes was similar in cKO and KO hearts at P1, indicating that premature binucleation is a result of cardiomyocyte-specific loss of Rbpms (see Supplementary material online, Figure S2C). At 2 months of age, there was no difference in the percentage of mononucleated cardiomyocytes between CTL and cKO mice, but there was a modest decrease in the percentage of binucleated cardiomyocytes and an increase in multi-nucleated cardiomyocytes in cKO mice (see Supplementary material online, Figure S2D). Similar to the global KO mice, cKO hearts also showed reduced cardiomyocyte cytokinesis until P7, as evidenced by Aurora kinase B staining (Figure 2E, F). By phospho histone H3 (pH3) staining, we also observed a significant reduction of cardiomyocyte proliferation in P1 cKO hearts compared to CTLs, suggesting a proliferation defect in cKO hearts (Figure 2G, H). Additionally, there were fewer cardiomyocytes in cKO hearts than CTLs at P14 (Figure 2I). Taken together, these results suggest that the cardiomyocyte-specific loss of Rbpms causes cardiomyocyte cytokinesis defects, premature binucleation, and non-compaction cardiomyopathy during development.
3.3. Cardiac-specific loss of Rbpms causes cardiomyocyte contractile defects
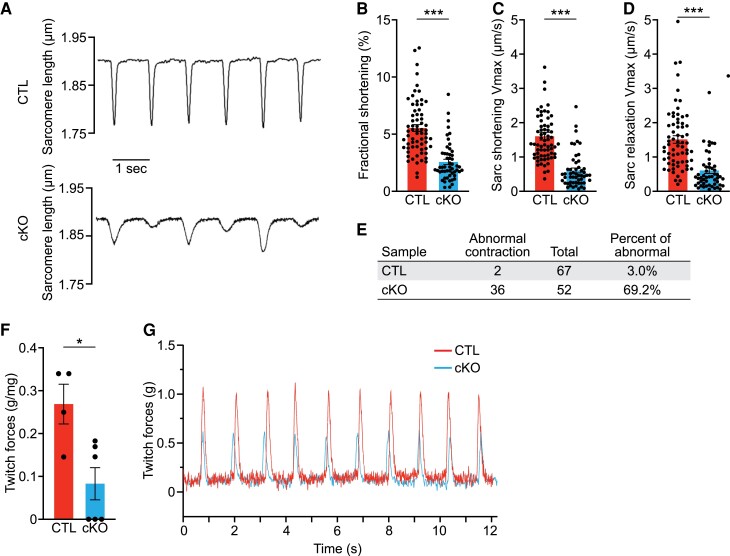
To evaluate cardiomyocyte contractility, we isolated single cardiomyocytes from cKO and CTL hearts at 2 months of age and measured their contractility using the IonOptix Calcium and Contractility System. We found a significant reduction in the fractional shortening (FS%) of cKO cardiomyocytes compared to CTLs (Figure 3A, B). cKO cardiomyocytes also showed significantly decreased maximum velocities of sarcomere shortening and relaxation, suggesting a defect in E-C coupling (Figure 3C, D; Supplementary material online, Table S3). Additionally, the irregular contraction, as exemplified in Figure 3A, appeared more frequently in cKO cardiomyocytes than CTLs (Figure 3E). The reduced contractility of cKO cardiomyocytes corroborates the reduced cardiac contractility in cKO mice at the organ level (see Supplementary material online, Table S1).
Figure 3.
Rbpms cKO hearts display cardiomyocyte contractile defects. (A) Representative sarcomere shortening images of single isolated CTL and cKO cardiomyocytes at 2 months of age. (B–D) Measurements of sarcomere fractional shortening at peak contraction, shortening velocity, and relaxation velocity of individual cardiomyocytes during the contraction cycle (n = 6 for CTL and n = 5 cKO mice at each stage, average 10–20 cardiomyocytes per mouse). (E) Quantification of the number and frequency of abnormal contraction in single isolated CTL and cKO cardiomyocytes at 2 months of age. (F) Measurements of twitch force of isolated cardiac papillary muscles from CTL and cKO mice at 2 months of age (n = 4 for CTL and n = 6 cKO mice). (G) Representative recordings of stimulated twitches of cardiac papillary muscle from CTL and cKO mice with a beating rate of 1 Hz. All data are presented as mean ± SEM. *P < 0.05; ***P < 0.001, by Student’s t-test two-tailed for B, C, D, F.
Ultrastructural analysis of cKO and CTL hearts by electron microscopy did not reveal any obvious difference in sarcomere structures at 2 months of age (see Supplementary material online, Figure S3A–D), suggesting that the impaired contraction of cKO cardiomyocytes was not caused by sarcomere disorganization. There was a mild decrease of sarcomere width in cKO cardiomyocytes, while basal sarcomere length was unchanged between cKO and CTL hearts (see Supplementary material online, Figure S3E, F). We did not observe obvious changes in mitochondrial morphology or density between cKO and CTL cardiomyocytes (see Supplementary material online, Figure S3G, H). Morphological analysis of binucleated cardiomyocytes, which represent 80–90% of adult cardiomyocytes, revealed that cKO cardiomyocytes were longer, with no difference in width compared to CTL cardiomyocytes (see Supplementary material online, Figure S3I–L). cKO cardiomyocyte area was larger than that of CTL (see Supplementary material online, Figure S3M). We reasoned that the morphological changes of cKO cardiomyocytes are an adaptive outcome due to DCM and chronic overload of the cKO hearts. We also analysed the calcium transients of cKO and CTL cardiomyocytes but did not observe any difference in calcium amplitude measured by calcium fluorescent ratio, release, or reuptake maximum velocities (see Supplementary material online, Figure S3N–Q; Supplementary material online, Table S4). Taken together, these results suggest that the absence of Rbpms does not alter calcium fluctuation in adult cardiomyocytes. To determine how the cardiomyocyte contractile defect influences the myocardial contractile force, we isolated single cardiac papillary muscles and found that cKO myocardium generated a significantly lower twitch force than CTL (Figure 3F, G). We conclude that the contractile defect of cKO cardiomyocytes causes reduced myocardial contractility, contributing to DCM.
3.4. RBPMS associates with spliceosome factors
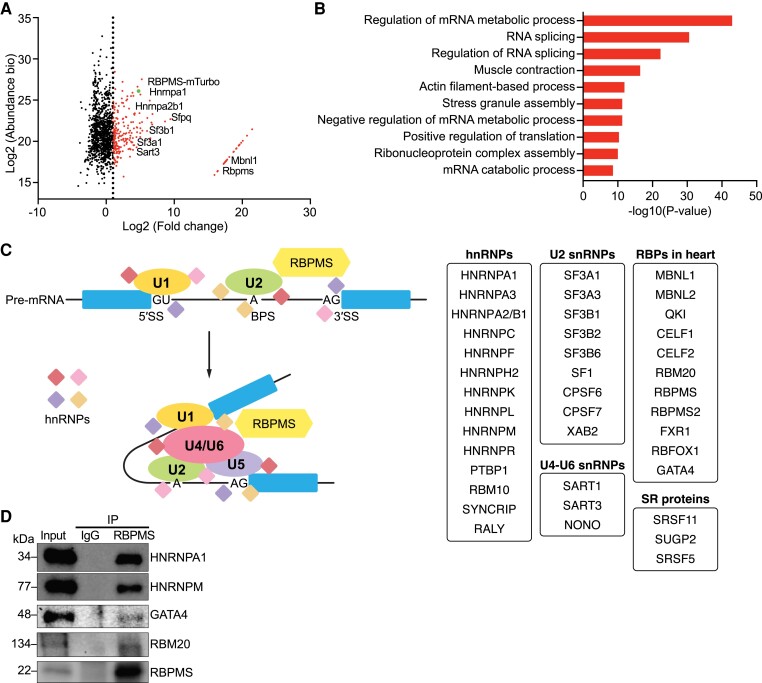
To further understand the functions of RBPMS, we performed a proximity-dependent biotin identification (BioID) assay in neonatal rat ventricular cardiomyocytes (NRVMs) to identify RBPMS interaction partners. NRVMs were infected with adenovirus expressing Rbpms fused to biotin ligase (miniTurbo), followed by biotin or vehicle treatment and mass spectrometry (see Supplementary material online, Figure S4A, B). In total, we identified 233 proteins labelled by RBPMS-miniTurbo that were at least two-fold enriched compared with controls, which were not treated with biotin (Figure 4A; Supplementary material online, Table S5).
Figure 4.
RBPMS is broadly associated with spliceosome factors. (A) Scatter plot showing the relative abundance and fold change over control of proteins identified in the BioID assay labelled by RBPMS. The red dots indicate proteins with log2(Fold Change) > 2 (n = 233), and the green dot indicates RBPMS-miniTurbo protein as a positive control. (B) Top GO terms for all identified RBPMS-interacting proteins with log2(Fold Change) > 2. (C) The identified RBPMS-interacting proteins are grouped according to snRNP association and RBPs in heart. (D) Western blot analysis showing the co-IP assay to validate identified RBPMS-interacting partners in hiPSC-derived cardiomyocyte lysate with Rbpms overexpression. RBPMS was immunoprecipitated by an RBPMS antibody, and blots were probed by different target antibodies.
Gene Ontology (GO) analysis revealed that RBPMS-interacting proteins were significantly enriched in regulation of mRNA metabolic process, RNA splicing, and regulation of RNA splicing (Figure 4B). We clustered the identified binding partners according to their molecular functions and found that RBPMS was broadly associated with hnRNPs that participate in pre-mRNA splicing, such as HNRNPA1 and HNRNPA2/B1. RBPMS also interacted with multiple U2 small nuclear ribonucleoprotein (snRNP) factors, such as SF3A1 and SF3B1, and some U4-U6 snRNPs, such as SART3 (Figure 4C). Intriguingly, RBPMS was in close proximity to many RBPs that are known to regulate heart development and cardiac function, such as MBNL1,9 QKI,10 CELF1/2,11 FXR1,12 RBM205, RBFOX1,13 and GATA4, a cardiogenic transcription factor that has been shown to mediate RNA splicing in cardiac progenitor cells14 (Figure 4C). To confirm the interaction of RBPMS with the identified binding partners, we overexpressed RBPMS via adenovirus infection in hiPSC-CMs and performed co-immunoprecipitation (co-IP) for RBPMS. We validated that HNRNPA1, HNRNPM, GATA4, and RBM20 were bound to RBPMS (Figure 4D). These results demonstrated that RBPMS is broadly associated with spliceosome factors in both murine and human cardiomyocytes, supporting its roles in mediating RNA splicing. Additionally, we found that RBPMS is widely associated with other cardiac RBPs, suggesting that RBPMS may function synergistically with these RBPs.
3.5. RBPMS regulates alternative RNA splicing in adult hearts
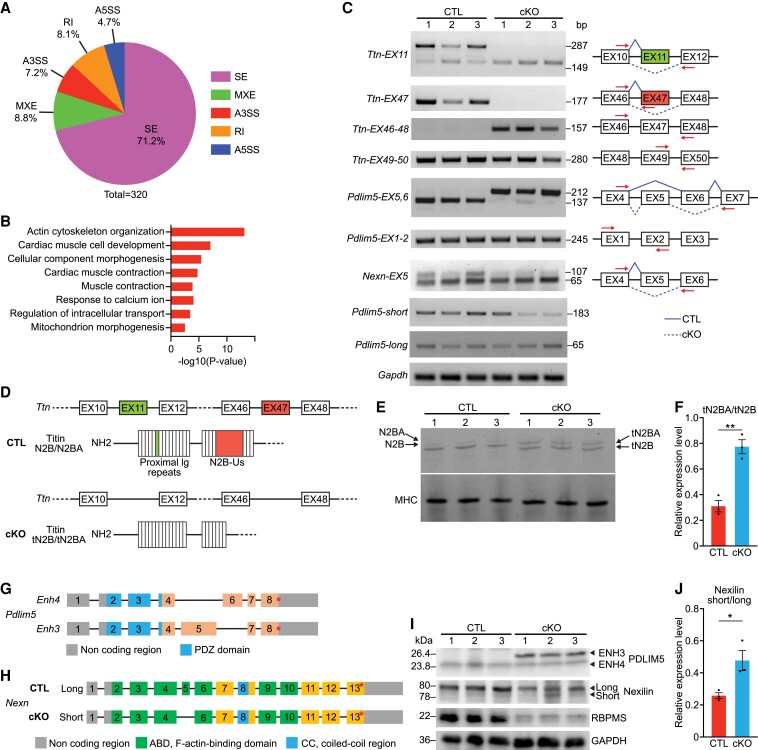
RBPMS mediates alternative RNA splicing in the neonatal heart and smooth muscle.7,15 To determine whether RBPMS also regulates RNA splicing in adult hearts, we performed paired-end RNA sequencing with CTL and cKO hearts (n = 4 per group) at 2 months of age. A total of 207 genes were down-regulated, and 436 genes were up-regulated in cKO hearts compared to CTLs (see Supplementary material online, Figure S5A). GO analysis revealed that extracellular matrix organization pathways were the most up-regulated in cKO hearts, while pathways related to heart rhythm and cardiac conduction were down-regulated (see Supplementary material online, Figure S5B). Replicate multivariate analysis of transcript splicing (rMATS) identified a total of 320 differential alternative splicing events (ASEs) from 245 genes between CTL and cKO hearts (see Supplementary material online, Figure S5C). The most common ASEs were skipped exons, representing 71.2% of the total ASEs, followed by mutually exclusive exons (MXEs, 8.8%), retained introns (RI, 8.1%), alternative 3′ splice sites (A3SS, 7.2%), and alternative 5′ splice sites (A5SS, 4.7%) (Figure 5A). GO analysis of the differential ASE gene targets revealed that actin cytoskeleton organization, muscle contraction, and cardiac muscle contraction were the most enriched biological pathways (Figure 5B). The top-ranked ASE targets included Ttn, Pdlim5, Nexn, Cacna2d1, and Camk2d, which are central to cardiomyocyte contractility (see Supplementary material online, Figure S5C). We validated these ASEs in CTL and cKO hearts by RT–PCR analysis with primer pairs that distinguish different splicing variants (Figure 5C; Supplementary material online, Figure S5D).
Figure 5.
RBPMS mediates alternative RNA splicing in adult hearts. (A) Pie chart showing the percentage of different ASEs between CTL and cKO mice (FDR < 0.01). SE, skipped exon; MXE, mutually exclusive exon; RI, retained intron; A3SS, alternative 3′ splice site; A5SS, alternative 5′ splice site. (B) Top GO terms for differential ASE genes identified by rMATS between CTL and cKO. (C) Representative RT–PCR confirmation of splicing events at 2 months of age, and schematic diagram of SE events based on rMATS analysis. Solid blue lines indicate normal splicing events in CTL hearts, and dashed blue lines indicate abnormal events in cKO hearts. Red arrows indicate the locations of primers used for the RT–PCR analysis (n = 3 for CTL and cKO). (D) Schematic diagram showing Ttn exon skipping events based on rMATS analysis and protein variants encoded in CTL and cKO hearts, respectively. (E) SDS-polyacrylamide gel electrophoresis blot showing high molecular-weight proteins in CTL and cKO hearts at 2 months of age (n = 3 for CTL and cKO). Black arrows in the top panel indicate Titin N2BA/N2B (tN2BA/tN2B) isoforms, and the bottom panel shows the MHC expression as a loading control. (F) Quantification of the relative expression ratio of Titin N2BA to N2B isoforms in CTL and cKO hearts from (E) (n = 3 for CTL and cKO). (G) Schematic diagram showing Pdlim5 exon mutually exclusive event based on rMATS analysis, and the protein domain encoded by each exon. (H) Schematic diagram showing Nexn exon skipping event based on rMATS analysis and the protein domain encoded by each exon. (I) Western blot analysis showing PDLIM5, Nexilin, RBPMS protein expression in the hearts of CTL and cKO mice at 2 months of age (n = 3 for CTL and cKO). GAPDH is a loading control. (J) Quantification of the relative expression ratio of Nexilin long to short isoforms in CTL and cKO hearts from I (n = 3 for CTL and cKO). All data are presented as mean ± SEM. *P < 0.05, **P < 0.01, by Student’s t-test two-tailed for F, J.
To investigate how ASEs in adult cKO hearts influence protein expression, we first looked into Ttn. In cKO hearts, both exons 11 and 47 of Ttn were skipped, generating a shorter splice isoform of Ttn missing the two exons (Figure 5D). rMATS analysis revealed that the per cent of Ttn exon 11 inclusion in cKO hearts was 20.1% compared to 94.6% in CTLs (see Supplementary material online, Figure S5E). Similarly, the per cent of Ttn exon 47 inclusion was 16.3% in cKO hearts, compared to 99.7% in CTLs (see Supplementary material online, Figure S5F). Ttn exons 11 and 47 skipping events in cKO hearts were confirmed by sequencing of the RT–PCR products (see Supplementary material online, Figure S5G, H). Exon 11 encodes part of the proximal Ig domains that anchor Titin to the Z-line, while exon 47 encodes the N2B-unique sequence (N2B-Us) that modulates elasticity of the Titin filament of the sarcomere (Figure 5D). Next, we performed SDS-agarose gel electrophoresis to evaluate expression of Titin protein. We found that cKO hearts expressed truncated Titin N2BA and N2B isoforms (tN2BA and tN2B), indicating that the skipping of exons 11 and 47 in cKO hearts leads to expression of shorter Titin proteins (Figure 5E). Intriguingly, we found that the ratio of the foetal isoform tN2BA to the adult isoform tN2B was significantly increased in cKO hearts (Figure 5F). Overall, we found that the absence of Rbpms in adult hearts caused skipping of Ttn exons 11 and 47, generating truncated forms of Titin tN2BA and tN2B. The absence of Rbpms also caused increased expression of the foetal isoform tN2BA, which could potentially impair cardiomyocyte contraction.
We also analysed the splicing of other sarcomeric genes related to cardiomyocyte contractility, including Pdlim5 and Nexn. Two Pdlim5 short isoforms are expressed in the adult heart, as a result of MXE of exon 5 and exon 6: Enh4, which contains exon 6, and Enh3 containing exon 516 (Figure 5G). Enh4 is shorter than Enh3, because exon 6 is smaller than exon 5, and Enh4 is the more abundant isoform in adult heart. We detected a prominent Pdlim5 exon 5 and exon 6 MXE between CTL and cKO hearts. In CTL hearts, exon 6 was included in 98.1% of Pdlim5 transcripts, while only 1.9% of exon 5 was included, consistent with the Enh4 being abundant in the adult hearts. In contrast, only 6.3% of exon 6 inclusion was observed in cKO hearts, while the exon 5 inclusion per cent was 93.7% (see Supplementary material online, Figure S5I). These results were confirmed by RT–PCR (Pdlim5-EX5,6) and sequencing (Figure 5C; Supplementary material online, Figure S5J). We performed western blotting to evaluate the expression of PDLIM5 in adult cKO and CTL hearts. As expected, we detected the larger ENH3 isoform in cKO hearts, but not in CTLs (Figure 5I). However, functional differences between ENH3 and ENH4 are not understood.
In cKO hearts, Nexn exon 5 inclusion was 16.7%, which was significantly lower than that of CTLs (37.1%) (see Supplementary material online, Figure S5K, L). Nexn exon 5 encodes a 14-amino acid region located in the first F-actin–binding domain of Nexilin, influencing its cross-linking activity with F-actin17 (Figure 5H). Nexn with exon 5 encodes a longer isoform (long), and Nexn without exon 5 encodes a shorter isoform (short), both of which are identifiable by western blotting. As expected, cKO hearts expressed higher levels of the short isoform than CTLs (Figure 5I, J). Taken together, the presence of new protein variants of Titin, PDLIM5, and Nexilin protein variants confirmed the alternative RNA splicing events identified from paired-end RNA sequencing and rMATS analysis.
In our previous study, we found that RBPMS mainly regulates the splicing of cytoskeleton-associated genes and cardiomyocyte cytokinesis.7 We performed RT–PCR analysis to determine whether the ASEs identified from the adult cKO hearts also exist in P1 cKO hearts. Pdlim5 was identified as an alternatively spliced gene in both adult and P1 hearts. Consistent with our previous findings,7 the Pdlim5 short isoform was expressed at significantly higher levels in P1 cKO hearts due to the inclusion of exon 8 compared to CTL, while no difference was observed in the long isoform (see Supplementary material online, Figure S5M). However, in the adult heart, we did not detect any difference in the Pdlim5 long or short isoforms in adult cKO and CTL hearts, suggesting that RBPMS does not mediate Pdlim5 exon 8 splicing in adult hearts (Figure 5C). For Pdlim5 exon 5 and 6 MXEs, we detected a unique 137 bp band by RT–PCR in cKO hearts due to exon 6 inclusion (see Supplementary material online, Figure S5M). This pattern is different from adult hearts, in which exon 6 was predominantly preserved in CTL, while exon 5 was predominantly preserved in the cKO (Figure 5C; Supplementary material online, Figure S5I). Therefore, the splicing of Pdlim5 is distinct between P1 and adult cKO hearts. Ttn is another gene manifesting different ASEs between P1 and adult cKO hearts. We found that Ttn contained exon 47 in both cKO and CTL transcripts at P1 (see Supplementary material online, Figure S5M). These results demonstrate that RBPMS exerts a stage-specific influence on target gene splicing between neonatal and adult hearts.
3.6. RBPMS directly modulates exon splicing of target genes
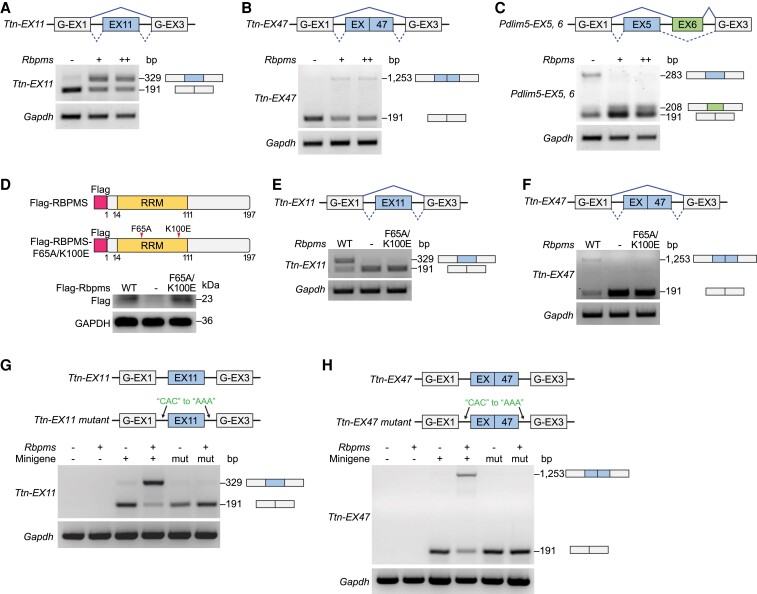
To address whether RBPMS directly regulates exon splicing of target genes, we developed a minigene reporter assay. Briefly, Ttn exon 11, exon 47, Pdlim5 exons 5 and 6, and their surrounding intronic sequences were cloned into minigene reporter plasmids, containing splicing reporter exons (G-EX1, G-EX3) flanking testing exons (Figure 6A–C). We co-transfected HEK293 cells with low (+) or high (++) amounts of the RBPMS expression plasmids and minigene reporter plasmids of target exons. Consistent with findings in the heart (Figure 5C), RBPMS promoted the inclusion of Ttn exon 11 and exon 47 in the minigene splicing assay (Figure 6A, B; Supplementary material online, Figure S6A). We also observed that Pdlim5 exon 5 was skipped and exon 6 was preserved by the expression of RBPMS (Figure 6C), which recapitulated the observations in the adult mouse heart (Figure 5C). These results suggest that RBPMS directly modulates Ttn and Pdlim5 exon splicing in vitro. To ensure the specificity of RBPMS in the minigene assay, we co-transfected HEK293 cells with the complete minigene reporter containing three reporter exons (G-EX1, G-EX2, G-EX3) and the RBPMS expression plasmid. RBPMS did not influence the splicing of the reporter, suggesting that the exon splicing by RBPMS is target-specific (see Supplementary material online, Figure S6B).
Figure 6.
RBPMS directly modulates exon splicing of target genes. (A–C) Schematic of the Ttn exon 11 (EX11), Ttn exon 47 (EX47), and Pdlim5 exon 5 and 6 (EX5, EX6) minigene reporter constructs. Minigene reporter constructs were transfected alone or with low (+), high (++)-dose Rbpms-expressing vector in HEK293 cells. Exon inclusion events were evaluated by RT–PCR analysis. (D) Schematic of RBPMS and RBPMS-F65A/K100E mutant protein structures. Western blot analysis showing RBPMS and mutant protein expression in HEK293 cells. (E, F) RT–PCR analysis showing minigene reporter splicing results of Ttn-EX11 and Ttn-EX47 by wildtype (WT) or F65A/K100E mutant RBPMS. (G, H) Schematic of the minigene reporter constructs containing wildtype and intronic CAC-motif mutated Ttn exon 11 (EX11) and Ttn exon 47 (EX47). Minigene reporter constructs were transfected alone or with Rbpms-expression vector in HEK293 cells. Exon inclusion event was evaluated by RT–PCR analysis.
RBPMS contains an RNA recognition motif (RRM), which facilitates its recognition of RNAs. Crystallographic structural analysis of the RBPMS RRM domain revealed the direct binding of the RRM to RNA.18 RBPMS RRM mutagenesis demonstrated that Phe65 (F65) and Lys100 (K100) are essential for RNA binding.18 To determine whether the target splicing activity of RBPMS requires its RNA binding capacity, we created a mutant RBPMS expression construct carrying two RRM-mutations (F65A/K100E) that abolish its RNA binding capacity (Figure 6D). It has been shown that these two mutations do not change the subcellular localization of RBPMS.18 RBPMS-F65A/K100E was stably expressed in HEK293 cells (Figure 6D). We found that RBPMS-F65A/K100E was unable to preserve Ttn exon 11 or exon 47 in the minigene reporter assay (Figure 6E, F), suggesting that RBPMS modulates alternative RNA splicing through the RRM-RNA binding domain. RBPMS recognizes and binds to RNAs through the tandem CAC clusters in the flanking intronic regions of target exons.15,18,19 To gain more mechanistic insights into how RBPMS modulates target exon splicing, we mutated all CAC motifs to AAA in the flanking intronic regions of Ttn exon 11 and exon 47 minigenes. In both cases, mutations of CAC motifs completely ablated the inclusion of target exons (Figure 6G, H). Together, these results show that RBPMS directly regulates exon splicing of its target genes through recognizing the intronic CAC motifs.
3.7. RBPMS knockdown in hiPSC-CMs causes contractile defects
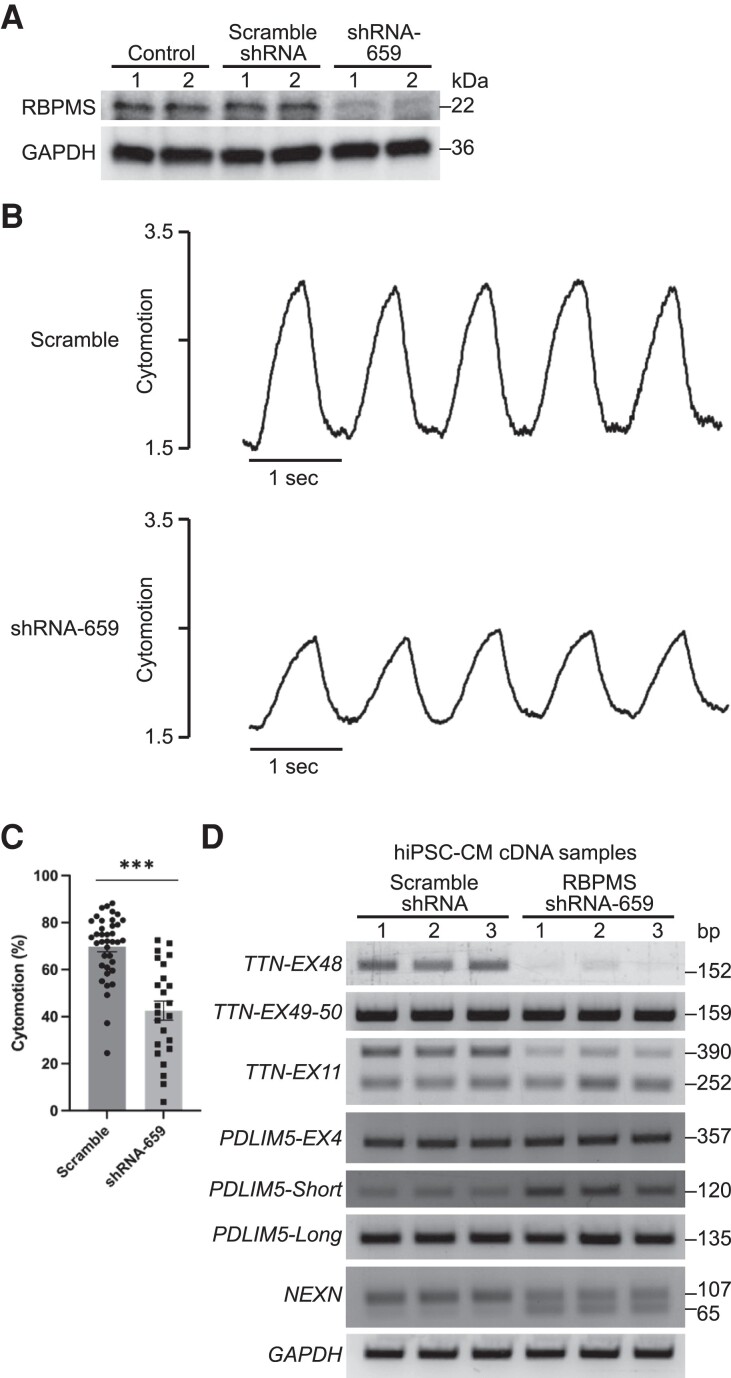
To further understand how RBPMS influences cardiomyocyte contractility, we performed acute RBPMS knockdown by small hairpin RNA (shRNA) in hiPSC-CMs. Among the five shRNAs tested, we identified shRNA-659 to be the most efficient in reducing RBPMS mRNA to around 10% of control levels (see Supplementary material online, Figure S7A). We infected WT hiPSC-CMs with retrovirus expressing shRNA-659 and confirmed the knockdown of RBPMS protein by western blot analysis (Figure 7A). Individual hiPSC-CM contractility was measured by the cytomotion module of the IonOptix Myocyte Calcium and Contractility System. We observed significantly reduced cardiomyocyte contractility upon RBPMS knockdown (Figure 7B, C), suggesting that the acute loss of RBPMS in mature cardiomyocytes impairs contractility. Consistent with observations from adult mouse cardiomyocytes (see Supplementary material online, Figure S3N–Q), we did not see obvious defects in calcium handling in RBPMS knockdown hiPSC-CMs (see Supplementary material online, Figure S7B, C), confirming that RBPMS deficiency does not disrupt cardiomyocyte calcium handling. To investigate how RBPMS knockdown influences alternative RNA splicing in hiPSC-CMs, we performed RT–PCR analysis for ASEs identified from adult cKO hearts. We found that the knockdown of RBPMS in hiPSC-CMs recapitulated some ASEs in cKO hearts, such as TTN exon 48 (equivalent to mouse Ttn exon 47), TTN exon 11, and NEXN exon 5 (Figure 7D, 5C). RBPMS knockdown in hiPSC-CMs did not recapitulate the PDLIM5 exon 5 and exon 6 MXE in mouse heart. In WT and RBPMS knockdown hiPSC-CMs, we observed equal amounts of PDLIM5 exon 4 (equivalent to mouse Pdlim5 exon 5) inclusion (Figure 7D). We conclude that these shared ASEs in mouse and human cardiomyocytes contribute to the impaired cardiomyocyte contractility in RBPMS deficiency.
Figure 7.
RBPMS knockdown in hiPSC-CMs causes contractile defects. (A) Western blot analysis showing RBPMS protein expression in control hiPSC-CMs or hiPSC-CMs infected with retrovirus expressing scramble shRNA or shRNA-659. (B) Representative recording traces of individual hiPSC-CM treated with scramble shRNA and shRNA-659 (48 h) following field stimulation of 1 Hz. (C) Quantification of hiPSC-CM contractility by IonOptix cytomotion software (n = 38 for scramble group and n = 24 for shRNA-659 group). (D) Representative RT–PCR results of splicing events in hiPSC-CMs. All data are presented as mean ± SEM. ***P < 0.001, by Student’s t-test two-tailed for C.
4. Discussion
A major conclusion of this work is that Rbpms is essential for cardiomyocyte contractility and post-natal cardiac function. Mechanistically, RBPMS is a key regulator of sarcomeric gene splicing that modulates cardiomyocyte contraction.
4.1. RBPMS plays essential roles in DCM and cardiomyocyte contractility
Mutations in sarcomere genes, such as TTN (14.6%), MYH7 (5.3%), TNNT2 (2.9%), and TPM1 (1.9%), are common causes of DCM.2,20–23 Instead of influencing a single sarcomere gene, RBPs can exert profound effects on the splicing of a variety of genes. Indeed, Rbpms deletion causes abnormal splicing of multiple genes related to sarcomere structure and cardiomyocyte contractility such as Ttn, Pdlim5, and Nexn, generating novel protein isoforms. Titin serves as a molecular spring spanning the Z disk and the M-band of the cardiomyocyte sarcomere and is a major determinant of cardiomyocyte passive tension during contraction.24 Truncation mutations of TTN are common genetic causes of DCM in humans, accounting for ∼25% of familial cases of idiopathic DCM and 18% of sporadic cases.25,26 In the heart, the two major isoforms of Titin: N2BA (foetal isoform) and N2B (adult isoform) both contain the N2B-Us encoded by exon 48, a unique region of cardiac Titin.27–31 Besides acting as a molecular spring, N2B-Us is also a key signalling hub regulated by various kinases that affect its mechanical properties.32 Mutations in the N2B-Us region cause DCM.25,33 We demonstrated that RBPMS is required to maintain the inclusion of Ttn exon 47 (equivalent to human TTN exon 48) specifically in adult hearts, while the absence of Rbpms causes shorter Titin proteins missing the N2B-Us. It has been shown that the deletion of N2B-Us in the mouse heart altered cardiac contractility.30 Thus, it is likely that the truncated Titin proteins contribute to the contractile defects in adult cKO cardiomyocytes. Besides Ttn, RBPMS also regulates splicing of other sarcomeric and cytoskeletal genes. Therefore, cardiomyocyte contractile defects in cKO hearts are likely a combinatorial result of various abnormal ASEs, contributing to DCM and heart failure. We did not observe changes of calcium handling or sarcomere ultrastructure in cKO hearts. One recent study reported the loss-of-function effects of RBPMS2, a homologue of RBPMS, on cardiomyocyte myofibril structure and calcium handling.34 Therefore, it is likely that RBPMS and RBPMS2 work synergistically and complementarily in regulating cardiomyocyte physiology, highlighting the essential roles of the RBPMS gene family in the heart.
4.2. RBPMS associates with spliceosomes to mediate ASEs
Among the 233 identified binding partners of RBPMS, we highlighted two major families of splicing factors: the hnRNPs and U2 snRNPs. hnRNPs represent a large family of RBPs regulating RNA alternative splicing, stabilization, and translation.35 RBPMS was found to interact with nearly every member of splicing-related hnRNPs, such as HNRNPA1, HNRNPA2/B1, and HNRNPM. hnRNPs often bind to exonic splicing silencers (ESSs) that inhibit inclusion of the exon they reside in.36 The U2 snRNPs are essential components of the spliceosome, which binds to the intron’s branch point during splicing,37 and RBPMS is associated with the core U2 snRNP components SF3A1 and SF3B1. These results indicate that RBPMS is directly associated with splicing factors. We therefore speculate that RBPMS mediates RNA alternative splicing by interacting with hnRNPs and U2 snRNPs to determine splicing sites, and thus the exclusion or inclusion of exons. Further investigation is needed to elucidate the detailed mechanism whereby RBPMS interacts with those factors in regulating splicing. Intriguingly, we found that RBPMS also interacts with GATA4, a canonical cardiac transcription factor and a non-canonical splicing regulator.14,38 GATA4 dysfunction causes various human congenital heart defects, including ventricular septal defects and non-compaction cardiomyopathy,39,40 which were also seen in Rbpms KO mice.7 It will be interesting to investigate the functional interactions between RBPMS and GATA4 as they might relate to congenital heart defects. According to the RBPMS interactome, RBPMS may not only regulate RNA splicing but also involve in other aspects of RNA processing, including synthesis, folding/unfolding, modification, processing, and degradation (Figure 4B). Actually, a previous study demonstrated a potential role of RBPMS in regulating mRNA subcellular localization in stress granules.19 Another study proposed a role of RBPMS in sequestering RNAs in germinal granules and repression of translation.41 We also observed ‘stress granule assembly’ among the top GO terms from RBPMS-interacting partners. Further study is needed to explore the additional roles of RBPMS in RNA processing in the heart.
4.3. Stage-specific RNA splicing by RBPMS
Our study demonstrated that Rbpms has a significant impact on alternative RNA splicing in both neonatal and adult hearts, but the target profiles are highly different between the two stages. Phenotypic analysis revealed that Rbpms regulates cardiomyocyte cytokinesis and proliferation exclusively in neonatal hearts, and influences cardiomyocyte contractility in adult hearts. These results highlight the stage-specific roles of RBPMS in the heart.
In our previous study, we validated Pdlim5 as a major splicing target of RBPMS in foetal cardiomyocytes and found that RBPMS mediates the exclusion of Pdlim5 exon 8, which contains a stop codon and determines expression of the long and short isoforms.7 The current study revealed that the Pdlim5 exon 8 ASE does not occur in adult cKO hearts. Splicing of Pdlim5 is not only mediated by RBPMS but also by other RBPs. For instance, QKI mediates the splicing of Pdlim5 exons 9, 10, and 11,10 MBNL1 mediates the splicing of Pdlim5 exons 7 and 8,42 and RBM20 and RBM24 cooperatively mediate the inclusion of Pdlim5 exon 8.43 Ttn is another top-ranked splicing target of RBPMS, and we demonstrated that RBPMS mediates Ttn exon 47 inclusion exclusively in adult hearts. Ttn is also a common splicing target of different cardiac RBPs. For instance, Rbm20 loss-of-function causes retention of Ttn exon 8 of the PEVK region, generating a pathological Titin isoform;5 QKI and RBM24 have also been shown to mediate Ttn splicing in the heart.10,44 Interestingly, we found that RBPMS is in close proximity with all these RBPs in cardiomyocytes, indicating that they may work synergistically in mediating splicing of different exons of a gene. Given that the expression of different RBPs is not synchronized during development, the specificity of the spliceosome may be altered due to different dosage contributions of RBPs at different stages, which potentially leads to the stage-specific alternative splicing by each RBP. It would be interesting to investigate how different RBPs cooperate in driving pre-mRNA splicing and maturation in the heart, which will broaden our understanding of the regulatory network underlying heart development and disease.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors’ contributions
P.G., N.L., R.B.-D., and E.N.O. designed the experiments and overall study. P.G., N.L., and E.N.O. wrote the manuscript. P.G. and J. M. designed and created the Rbpms cKO mouse line. P.G. and Z.W. performed the RNA sequencing data analysis and rMATS analysis. P.G., S.B., W.T., and H.L. performed the experiments. All authors discussed the results and participated in the manuscript preparation and editing. Correspondence and requests for materials should be addressed to E.N.O. (eric.olson@utsouthwestern.edu) and N.L. (Ning.Liu@utsouthwestern.edu).
Supplementary Material
Acknowledgements
We thank the Molecular Histopathology Core managed by John Shelton for help with histology. We thank Jose Cabrera for graphics. We thank Dr Jian Huang and Dr Pradeep P.A. Mammen for their support on cardiac papillary muscle twitch force measurements. We thank Joseph J. Wingate for his technical support on the BioID assay. We thank Drs Jian Xu and Yoon Jung Kim from the Next Generation Sequencing Core Facility at Children’s Research Institute for performing the Illumina sequencing, and Stephen Johnson for help with high-performance computing. We thank Dr Andrew Lemoff from the Proteomics Core for help with the proteomics analysis. We thank members of the Olson laboratory for helpful discussions.
Contributor Information
Peiheng Gan, Department of Molecular Biology, Hamon Center for Regenerative Science and Medicine, University of Texas Southwestern Medical Center, 6000 Harry Hines Blvd., Dallas, TX 75390, USA.
Zhaoning Wang, Department of Molecular Biology, Hamon Center for Regenerative Science and Medicine, University of Texas Southwestern Medical Center, 6000 Harry Hines Blvd., Dallas, TX 75390, USA; Department of Cellular and Molecular Medicine, University of California, San Diego School of Medicine, La Jolla, CA 92093, USA.
Svetlana Bezprozvannaya, Department of Molecular Biology, Hamon Center for Regenerative Science and Medicine, University of Texas Southwestern Medical Center, 6000 Harry Hines Blvd., Dallas, TX 75390, USA.
John R McAnally, Department of Molecular Biology, Hamon Center for Regenerative Science and Medicine, University of Texas Southwestern Medical Center, 6000 Harry Hines Blvd., Dallas, TX 75390, USA.
Wei Tan, Department of Molecular Biology, Hamon Center for Regenerative Science and Medicine, University of Texas Southwestern Medical Center, 6000 Harry Hines Blvd., Dallas, TX 75390, USA.
Hui Li, Department of Molecular Biology, Hamon Center for Regenerative Science and Medicine, University of Texas Southwestern Medical Center, 6000 Harry Hines Blvd., Dallas, TX 75390, USA.
Rhonda Bassel-Duby, Department of Molecular Biology, Hamon Center for Regenerative Science and Medicine, University of Texas Southwestern Medical Center, 6000 Harry Hines Blvd., Dallas, TX 75390, USA.
Ning Liu, Department of Molecular Biology, Hamon Center for Regenerative Science and Medicine, University of Texas Southwestern Medical Center, 6000 Harry Hines Blvd., Dallas, TX 75390, USA.
Eric N Olson, Department of Molecular Biology, Hamon Center for Regenerative Science and Medicine, University of Texas Southwestern Medical Center, 6000 Harry Hines Blvd., Dallas, TX 75390, USA.
Funding
This work was supported by grants from the National Institutes of Health (NIH) (HL-130253, AR-071980, and HD-087351), the Fondation Leducq Transatlantic Networks of Excellence in Cardiovascular Research, and the Robert A. Welch Foundation (grant 1-0025 to E.N.O.). P.G. was supported by a post-doctoral fellowship from the American Heart Association (825635). Z.W. is a DDBrown Awardee of the Life Sciences Research Foundation.
Data availability
All data presented in this study are available in the main text or the Supplementary materials. Raw and analysed RNA sequencing data generated during this study are available in the Gene Expression Omnibus (GEO) repository 6 (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession number GSE218672. Additional methods information is available in the Supplementary methods.
Translational perspective.
RNA binding proteins are associated with cardiovascular diseases through modulating RNA post-transcriptional modifications. Our previous study revealed that Rbpms deficiency caused non-compaction cardiomyopathy and patent ductus arteriosus in mice. Our current study demonstrates that Rbpms mainly regulates cardiomyocyte contraction in the adult heart, as cardiac-specific Rbpms knockout leads to dilated cardiomyopathy and heart failure. RBPMS mediates the alternative splicing of key sarcomeric components in the adult heart, including Ttn, Pdlim5, and Nexn. These findings indicate that RBPMS is an essential regulator of cardiac function. A better understanding of its function will provide new therapeutic opportunities for cardiovascular diseases.
References
- 1. Guo Y, Pu WT. Cardiomyocyte maturation: new phase in development. Circ Res 2020;126:1086–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McNally EM, Mestroni L. Dilated cardiomyopathy: genetic determinants and mechanisms. Circ Res 2017;121:731–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eisner DA, Caldwell JL, Kistamas K, Trafford AW. Calcium and excitation–contraction coupling in the heart. Circ Res 2017;121:181–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van den Hoogenhof MM, Pinto YM, Creemers EE. RNA splicing: regulation and dysregulation in the heart. Circ Res 2016;118:454–468. [DOI] [PubMed] [Google Scholar]
- 5. Guo W, Schafer S, Greaser ML, Radke MH, Liss M, Govindarajan T, Maatz H, Schulz H, Li S, Parrish AM, Dauksaite V, Vakeel P, Klaassen S, Gerull B, Thierfelder L, Regitz-Zagrosek V, Hacker TA, Saupe KW, Dec GW, Ellinor PT, MacRae CA, Spallek B, Fischer R, Perrot A, Ozcelik C, Saar K, Hubner N, Gotthardt M. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med 2012;18:766–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Akerberg AA, Burns CE, Burns CG. Exploring the activities of RBPMS proteins in myocardial biology. Pediatr Cardiol 2019;40:1410–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gan P, Wang Z, Morales MG, Zhang Y, Bassel-Duby R, Liu N, Olson EN. RBPMS is an RNA-binding protein that mediates cardiomyocyte binucleation and cardiovascular development. Dev Cell 2022;57:959–973.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gan P, Baicu C, Watanabe H, Wang K, Tao G, Judge DP, Zile MR, Makita T, Mukherjee R, Sucov HM. The prevalent I686T human variant and loss-of-function mutations in the cardiomyocyte-specific kinase gene TNNI3K cause adverse contractility and concentric remodeling in mice. Hum Mol Genet 2021;29:3504–3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. LeMasters KE, Blech-Hermoni Y, Stillwagon SJ, Vajda NA, Ladd AN. Loss of muscleblind-like 1 promotes invasive mesenchyme formation in endocardial cushions by stimulating autocrine TGFbeta3. BMC Dev Biol 2012;12:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen X, Liu Y, Xu C, Ba L, Liu Z, Li X, Huang J, Simpson E, Gao H, Cao D, Sheng W, Qi H, Ji H, Sanderson M, Cai CL, Li X, Yang L, Na J, Yamamura K, Liu Y, Huang G, Shou W, Sun N. QKI is a critical pre-mRNA alternative splicing regulator of cardiac myofibrillogenesis and contractile function. Nat Commun 2021;12:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci U S A 2008;105:20333–20338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mientjes EJ, Willemsen R, Kirkpatrick LL, Nieuwenhuizen IM, Hoogeveen-Westerveld M, Verweij M, Reis S, Bardoni B, Hoogeveen AT, Oostra BA, Nelson DL. Fxr1 knockout mice show a striated muscle phenotype: implications for Fxr1p function in vivo. Hum Mol Genet 2004;13:1291–1302. [DOI] [PubMed] [Google Scholar]
- 13. Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K, Yasuda K, Inoue K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J 2003;22:905–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhu L, Choudhary K, Gonzalez-Teran B, Ang YS, Thomas R, Stone NR, Liu L, Zhou P, Zhu C, Ruan H, Huang Y, Jin S, Pelonero A, Koback F, Padmanabhan A, Sadagopan N, Hsu A, Costa MW, Gifford CA, van Bemmel JG, Huttenhain R, Vedantham V, Conklin BR, Black BL, Bruneau BG, Steinmetz L, Krogan NJ, Pollard KS, Srivastava D. Transcription factor GATA4 regulates cell type-specific splicing through direct interaction with RNA in human induced pluripotent stem cell-derived cardiac progenitors. Circulation 2022;146:770–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nakagaki-Silva EE, Gooding C, Llorian M, Jacob AG, Richards F, Buckroyd A, Sinha S, Smith CW. Identification of RBPMS as a mammalian smooth muscle master splicing regulator via proximity of its gene with super-enhancers. Elife 2019;8:e46327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yamazaki T, Walchli S, Fujita T, Ryser S, Hoshijima M, Schlegel W, Kuroda S, Maturana AD. Splice variants of enigma homolog, differentially expressed during heart development, promote or prevent hypertrophy. Cardiovasc Res 2010;86:374–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ohtsuka T, Nakanishi H, Ikeda W, Satoh A, Momose Y, Nishioka H, Takai Y. Nexilin: a novel actin filament-binding protein localized at cell-matrix adherens junction. J Cell Biol 1998;143:1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Teplova M, Farazi TA, Tuschl T, Patel DJ. Structural basis underlying CAC RNA recognition by the RRM domain of dimeric RNA-binding protein RBPMS. Q Rev Biophys 2016;49:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Farazi TA, Leonhardt CS, Mukherjee N, Mihailovic A, Li S, Max KE, Meyer C, Yamaji M, Cekan P, Jacobs NC, Gerstberger S, Bognanni C, Larsson E, Ohler U, Tuschl T. Identification of the RNA recognition element of the RBPMS family of RNA-binding proteins and their transcriptome-wide mRNA targets. RNA 2014;20:1090–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Walsh R, Thomson KL, Ware JS, Funke BH, Woodley J, McGuire KJ, Mazzarotto F, Blair E, Seller A, Taylor JC, Minikel EV, Exome Aggregation C, MacArthur DG, Farrall M, Cook SA, Watkins H. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med 2017;19:192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hassel D, Dahme T, Erdmann J, Meder B, Huge A, Stoll M, Just S, Hess A, Ehlermann P, Weichenhan D, Grimmler M, Liptau H, Hetzer R, Regitz-Zagrosek V, Fischer C, Nurnberg P, Schunkert H, Katus HA, Rottbauer W. Nexilin mutations destabilize cardiac Z-disks and lead to dilated cardiomyopathy. Nat Med 2009;15:1281–1288. [DOI] [PubMed] [Google Scholar]
- 22. Liu C, Spinozzi S, Chen JY, Fang X, Feng W, Perkins G, Cattaneo P, Guimaraes-Camboa N, Dalton ND, Peterson KL, Wu T, Ouyang K, Fu XD, Evans SM, Chen J. Nexilin is a new component of junctional membrane complexes required for cardiac T-tubule formation. Circulation 2019;140:55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cheng H, Kimura K, Peter AK, Cui L, Ouyang K, Shen T, Liu Y, Gu Y, Dalton ND, Evans SM, Knowlton KU, Peterson KL, Chen J. Loss of enigma homolog protein results in dilated cardiomyopathy. Circ Res 2010;107:348–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. LeWinter MM, Granzier H. Cardiac titin: a multifunctional giant. Circulation 2010;121:2137–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Herman DS, Lam L, Taylor MR, Wang L, Teekakirikul P, Christodoulou D, Conner L, DePalma SR, McDonough B, Sparks E, Teodorescu DL, Cirino AL, Banner NR, Pennell DJ, Graw S, Merlo M, Di Lenarda A, Sinagra G, Bos JM, Ackerman MJ, Mitchell RN, Murry CE, Lakdawala NK, Ho CY, Barton PJ, Cook SA, Mestroni L, Seidman JG, Seidman CE. Truncations of titin causing dilated cardiomyopathy. N Engl J Med 2012;366:619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roberts AM, Ware JS, Herman DS, Schafer S, Baksi J, Bick AG, Buchan RJ, Walsh R, John S, Wilkinson S, Mazzarotto F, Felkin LE, Gong S, MacArthur JA, Cunningham F, Flannick J, Gabriel SB, Altshuler DM, Macdonald PS, Heinig M, Keogh AM, Hayward CS, Banner NR, Pennell DJ, O’Regan DP, San TR, de Marvao A, Dawes TJ, Gulati A, Birks EJ, Yacoub MH, Radke M, Gotthardt M, Wilson JG, O’Donnell CJ, Prasad SK, Barton PJ, Fatkin D, Hubner N, Seidman JG, Seidman CE, Cook SA. Integrated allelic, transcriptional, and phenomic dissection of the cardiac effects of titin truncations in health and disease. Sci Transl Med 2015;7:270ra276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lahmers S, Wu Y, Call DR, Labeit S, Granzier H. Developmental control of titin isoform expression and passive stiffness in fetal and neonatal myocardium. Circ Res 2004;94:505–513. [DOI] [PubMed] [Google Scholar]
- 28. Opitz CA, Leake MC, Makarenko I, Benes V, Linke WA. Developmentally regulated switching of titin size alters myofibrillar stiffness in the perinatal heart. Circ Res 2004;94:967–975. [DOI] [PubMed] [Google Scholar]
- 29. Warren CM, Krzesinski PR, Campbell KS, Moss RL, Greaser ML. Titin isoform changes in rat myocardium during development. Mech Dev 2004;121:1301–1312. [DOI] [PubMed] [Google Scholar]
- 30. Radke MH, Peng J, Wu Y, McNabb M, Nelson OL, Granzier H, Gotthardt M. Targeted deletion of titin N2B region leads to diastolic dysfunction and cardiac atrophy. Proc Natl Acad Sci U S A 2007;104:3444–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Labeit S, Kolmerer B. Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science 1995;270:293–296. [DOI] [PubMed] [Google Scholar]
- 32. LeWinter MM, Granzier HL. Titin is a major human disease gene. Circulation 2013;127:938–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fomin A, Gartner A, Cyganek L, Tiburcy M, Tuleta I, Wellers L, Folsche L, Hobbach AJ, von Frieling-Salewsky M, Unger A, Hucke A, Koser F, Kassner A, Sielemann K, Streckfuss-Bomeke K, Hasenfuss G, Goedel A, Laugwitz KL, Moretti A, Gummert JF, Dos Remedios CG, Reinecke H, Knoll R, van Heesch S, Hubner N, Zimmermann WH, Milting H, Linke WA. Truncated titin proteins and titin haploinsufficiency are targets for functional recovery in human cardiomyopathy due to TTN mutations. Sci Transl Med 2021;13:eabd3079. [DOI] [PubMed] [Google Scholar]
- 34. Akerberg AA, Trembley M, Butty V, Schwertner A, Zhao L, Beerens M, Liu X, Mahamdeh M, Yuan S, Boyer L, MacRae C, Nguyen C, Pu WT, Burns CE, Burns CG. RBPMS2 is a myocardial-enriched splicing regulator required for cardiac function. Circ Res 2022;131:980–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Geuens T, Bouhy D, Timmerman V. The hnRNP family: insights into their role in health and disease. Hum Genet 2016;135:851–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 2008;14:802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van der Feltz C, Hoskins AA. Structural and functional modularity of the U2 snRNP in pre-mRNA splicing. Crit Rev Biochem Mol Biol 2019;54:443–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xin M, Olson EN, Bassel-Duby R. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat Rev Mol Cell Biol 2013;14:529–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, Matsuoka R, Cohen JC, Srivastava D. GATA4 Mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 2003;424:443–447. [DOI] [PubMed] [Google Scholar]
- 40. Blinder JJ, Martinez HR, Craigen WJ, Belmont J, Pignatelli RH, Jefferies JL. Noncompaction of the left ventricular myocardium in a boy with a novel chromosome 8p23.1 deletion. Am J Med Genet A 2011;155A:2215–2220. [DOI] [PubMed] [Google Scholar]
- 41. Aguero T, Zhou Y, Kloc M, Chang P, Houliston E, King ML. Hermes (Rbpms) is a critical component of RNP complexes that sequester germline RNAs during oogenesis. J Dev Biol 2016;4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dixon DM, Choi J, El-Ghazali A, Park SY, Roos KP, Jordan MC, Fishbein MC, Comai L, Reddy S. Loss of muscleblind-like 1 results in cardiac pathology and persistence of embryonic splice isoforms. Sci Rep 2015;5:9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ito J, Iijima M, Yoshimoto N, Niimi T, Kuroda S, Maturana AD. RBM20 and RBM24 cooperatively promote the expression of short enh splice variants. FEBS Lett 2016;590:2262–2274. [DOI] [PubMed] [Google Scholar]
- 44. Lu SH, Lee KZ, Hsu PW, Su LY, Yeh YC, Pan CY, Tsai SY. Alternative splicing mediated by RNA-binding protein RBM24 facilitates cardiac myofibrillogenesis in a differentiation stage-specific manner. Circ Res 2022;130:112–129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data presented in this study are available in the main text or the Supplementary materials. Raw and analysed RNA sequencing data generated during this study are available in the Gene Expression Omnibus (GEO) repository 6 (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession number GSE218672. Additional methods information is available in the Supplementary methods.