Abstract
The yeast protein Spa2p localizes to growth sites and is important for polarized morphogenesis during budding, mating, and pseudohyphal growth. To better understand the role of Spa2p in polarized growth, we analyzed regions of the protein important for its function and proteins that interact with Spa2p. Spa2p interacts with Pea2p and Bud6p (Aip3p) as determined by the two-hybrid system; all of these proteins exhibit similar localization patterns, and spa2Δ, pea2Δ, and bud6Δ mutants display similar phenotypes, suggesting that these three proteins are involved in the same biological processes. Coimmunoprecipitation experiments demonstrate that Spa2p and Pea2p are tightly associated with each other in vivo. Velocity sedimentation experiments suggest that a significant portion of Spa2p, Pea2p, and Bud6p cosediment, raising the possibility that these proteins form a large, 12S multiprotein complex. Bud6p has been shown previously to interact with actin, suggesting that the 12S complex functions to regulate the actin cytoskeleton. Deletion analysis revealed that multiple regions of Spa2p are involved in its localization to growth sites. One of the regions involved in Spa2p stability and localization interacts with Pea2p; this region contains a conserved domain, SHD-II. Although a portion of Spa2p is sufficient for localization of itself and Pea2p to growth sites, only the full-length protein is capable of complementing spa2 mutant defects, suggesting that other regions are required for Spa2p function. By using the two-hybrid system, Spa2p and Bud6p were also found to interact with components of two mitogen-activated protein kinase (MAPK) pathways important for polarized cell growth. Spa2p interacts with Ste11p (MAPK kinase [MEK] kinase) and Ste7p (MEK) of the mating signaling pathway as well as with the MEKs Mkk1p and Mkk2p of the Slt2p (Mpk1p) MAPK pathway; for both Mkk1p and Ste7p, the Spa2p-interacting region was mapped to the N-terminal putative regulatory domain. Bud6p interacts with Ste11p. The MEK-interacting region of Spa2p corresponds to the highly conserved SHD-I domain, which is shown to be important for mating and MAPK signaling. spa2 mutants exhibit reduced levels of pheromone signaling and an elevated level of Slt2p kinase activity. We thus propose that Spa2p, Pea2p, and Bud6p function together, perhaps as a complex, to promote polarized morphogenesis through regulation of the actin cytoskeleton and signaling pathways.
In both unicellular and multicellular organisms, polarized cell growth is crucial for the formation of precise cell morphologies that allow cells to carry out their specialized functions (17, 19, 67, 72). For example, the development of neurites enables nerve cells to carry out sensory transduction (20), formation of microvilli enables epithelial cells to absorb nutrients (56), and growth of pollen tubes in the styles of flowers facilitates plant fertilization (6). Although the cytological events involved in polarized cell growth have been well studied, the molecular mechanisms involved in this process are not well understood.
The budding yeast Saccharomyces cerevisiae undergoes polarized cell growth in several stages of its life cycle (17, 19, 67, 72). Polarized growth is prominent during budding in vegetative and pseudohyphal growth and during projection formation in the mating response. Polarized growth in a vegetative cell begins in late G1, when a bud emerges from a specific site dictated by the mating-type locus and the pedigree of the cell (12, 28, 36, 70, 80). Cell growth occurs initially at the tip of the bud (apical growth) and then continues isotropically as the bud enlarges (47). Finally, just prior to cytokinesis, new cell wall and membrane deposition occurs at the mother-bud neck (47). When limited for nitrogen sources, yeast cells also undergo budding but adopt an elongated morphology and form chains of connected cells called pseudohyphae, which allow cells to spread across a surface to gain access to nutrients (31, 75). During mating, haploid cells respond to pheromone from cells of the opposite mating type and form projections toward their mating partners (82); these projections are important for cell fusion (30, 87).
Polarized cell growth in yeast is a complex process that involves the polarized organization of the actin cytoskeleton (19), the coordinated function of many polarity proteins (67, 72), and the regulation of signal transduction cascades (35, 45). The actin cytoskeleton appears as distinct structures during polarized cell growth (1, 41). Cortical actin patches are concentrated at sites of polarized growth, and actin cables run parallel to the polarity axis (the mother-bud axis during budding and longitudinal to the projection during mating). The actin cytoskeleton is thought to direct secretory vesicles containing growth components (e.g., cell wall and plasma membrane) to growth sites (4, 58, 60).
Many components that influence cell polarity localize to sites of polarized growth. The yeast protein Spa2p localizes to growth sites and is important for polarized morphogenesis (14, 30, 57, 71, 80, 81, 92). Spa2p can be found at the incipient bud sites of unbudded cells, the bud tips of small budded cells, the necks of cells undergoing cytokinesis, and the projection tips of mating cells. spa2Δ cells are rounder than wild-type cells and are defective in bud site selection in diploid cells, cytokinesis, projection formation during mating, and pseudohyphal growth. Spa2p is a large protein of 1,466 amino acids with several domains (30, 71), including a predicted coiled-coil region, a domain with 25 9-amino-acid repeats, and five regions that are conserved with those of a related yeast protein, Sph1p (3, 71). These five domains are named SHD-I to -V (SHD stands for Spa2p homology domain). The N-terminal SHD-I is also present in proteins from a wide variety of eukaryotes, and approximately half of SHD-II is predicted to be coiled coil (71). How these domains contribute to the function of Spa2p is not clear.
Pea2p and Bud6p (Aip3p) exhibit many similarities to Spa2p (2, 86). These proteins localize to growth sites in a fashion similar to that of Spa2p, and diploid pea2Δ and bud6Δ mutants are defective in bud site selection (86, 93). Like spa2Δ cells, pea2Δ mutants are defective in projection formation and pseudohyphal growth (14, 57); bud6 mutants form round cells and are defective in cytokinesis (2, 93). Pea2p and Bud6p are smaller than Spa2p (420 and 788 amino acids, respectively), and each has a predicted coiled-coil domain (2, 86). Spa2p fails to localize in pea2Δ mutants, and Pea2p is not stably produced in spa2Δ mutants, raising the possibility that these proteins might interact (86). Bud6p interacts with actin (2), which also participates in diploid bud site selection and cellular morphogenesis (18, 60, 88, 91). Thus, Spa2p, Pea2p, and Bud6p represent an important group of proteins that participate in many common cellular processes; perhaps these proteins help regulate the actin cytoskeleton during polarized cell growth. The similar localization patterns of Spa2p, Pea2p, and Bud6p together with the common phenotypes of cells that lack these proteins suggest that these components may function very closely, perhaps as a complex, in the same processes. Direct evidence for such a complex has not been described.
Like Spa2p, components of several mitogen-activated protein kinase (MAPK) pathways also participate in the process of polarized cell growth (35, 46). MAPK pathways are composed of a cascade of protein kinases which act sequentially to transmit signals upon perception of external stimuli or internal cues; these signals ultimately result in various cellular responses, such as cell growth or differentiation (52, 85). The MAPK cascade includes a MAPK; a MAPK kinase (MEK), which phosphorylates and activates MAPK; and a MEK kinase (MEKK), which phosphorylates and activates MEK. In the budding yeast, at least two MAPK pathways participate in cell growth and differentiation. The mating response requires the Fus3p-Kss1p pathway (59, 95), which functions downstream of the Ste20p kinase (43). This pathway is composed of two MAPKs, Fus3p and Kss1p (21, 22, 29); the MEK Ste7p (25); and the MEKK Ste11p (83). Ste11p, Ste7p, and Kss1p also function in pseudohypha formation (48, 51, 69). The other MAPK pathway, the Slt2p (Mpk1p) MAPK pathway, functions downstream of the yeast protein kinase C homolog, Pkc1p, to maintain cellular integrity during polarized growth (46). Components of this pathway include the MAPK Slt2p (Mpk1p) (53, 84); two homologous and redundant MEKs, Mkk1p and Mkk2p (37); and the MEKK Bck1p (Slk1p) (15, 44). Defects in this pathway lead to cell lysis at elevated temperatures and failure to form proper mating projections (15, 24, 45). This pathway is also activated in response to mating pheromone and at the G1-S transition when bud emergence is initiated, consistent with its role in cell polarity (24, 94). The concomitant involvement of both Spa2p and components of these MAPK pathways in mating, pseudohypha formation, and bud growth raises the possibility that Spa2p and perhaps other polarity proteins might interact with signaling components. Genetic interactions between SPA2 and BCK1 (SLK1) and between SPA2 and SLT2 (MPK1) have been demonstrated (15, 16). Other interactions have not been uncovered.
In this study, we analyzed different SPA2 deletions and investigated the interactions among a number of the different polarity proteins and signaling components. We provide evidence that Spa2p is a complex protein with many important domains. Spa2p physically interacts with Pea2p and Bud6p, and these proteins cosediment at approximate 12S, suggesting that they form a multiprotein complex. Spa2p interacts with Pea2p via the conserved SHD-II, which is important for both the stability and localization of Spa2p and Pea2p. In addition, Spa2p and Bud6p interact with components of MAPK pathways. The N-terminal 150-amino-acid MEK-interacting region of Spa2p contains a conserved domain, SHD-I, that is important for mating and other Spa2p functions. The signaling activities of two MAPK pathways are altered in spa2 mutants. Taken together, these results suggest that Spa2p, Pea2p, and Bud6p are part of a large multiprotein complex that may exert its function through regulation of the actin cytoskeleton; this complex may link polarity components and signaling pathways during polarized cell growth.
MATERIALS AND METHODS
Strains, media, and microbiological techniques.
The yeast strains used in this study are listed in Table 1. Growth media and genetic manipulation of yeast strains were as described by Sherman et al. (77). Yeast transformations were performed by the lithium acetate procedure (38) or the one-step method (13).
TABLE 1.
Yeast strains
| Straina | Genotype | Source and/or reference |
|---|---|---|
| Y1209* | MATa/MATα leu2::HISG/leu2::HISG ura3-52/ura3-52 spa2Δ::URA3/spa2Δ::URA3 | 71 |
| Y2001 | MATa/MATα leu2Δ98/leu2Δ98 ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-Δ1/trp1-Δ1 HIS3/his3-Δ200 spa2Δ3::URA3/spa2Δ3::URA3 | This study |
| Y754 | MATa leu2Δ98 ura3-52 lys2-801 ade2-101 trp1-Δ1 spa2Δ-1::TRP1 slk1-1 + pSPA2/TRP1/SUP4/CEN | 15 |
| Y831 | MATa leu2Δ98 ura3-52 lys2-801 ade2-101 trp1-Δ1 spa2Δ-1::TRP1 cdc10-10 + pSPA2/TRP1/SUP4/CEN | 27a |
| Y875 | MATa leu2Δ98 ura3-52 lys2-801 ade2-101 trp1-Δ1 spa2Δ-1::TRP1 bem2-101 + pSPA2/TRP1/SUP4/CEN | This lab |
| Y877 | MATa leu2Δ98 ura3-52 lys2-801 ade2-101 trp1-Δ1 spa2Δ-1::TRP1 swi4-100 + pSPA2/TRP1/SUP4/CEN | This lab |
| Y864* | MATa leu2-3,112 ura3 ade2 trp1-901 his3-Δ200 gal4Δ gal80Δ pLEXA-lacZ::URA3 | CTY10-5D |
| Y1213* | MATa leu2-3,112 ura3 ade2 trp1-901 his3-Δ200 gal4Δ gal80Δ pLEXA-lacZ::URA3 ste5Δ::TRP1 | This study; a derivative of Y864 |
| Y602 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 spa2Δ3::URA3 | This lab |
| Y604 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 | This lab |
| Y609 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 spa2Δ2::TRP1 | This lab |
| Y762 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 | This lab |
| Y2002 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 pea2Δ::HIS3 | This study |
| Y2003 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 PEA2::HA | This study |
| Y2004 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 PEA2::myc | This study |
| Y2005 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 spa2Δ2::TRP1 PEA2::HA | This study |
| Y2006 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 spa2Δ2::TRP1 PEA2::myc | This study |
| Y2007 | MATa leu2Δ98 ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 spa2Δ3::URA3 PEA2::myc | This study |
| Y2008 | MATa leu2Δ98 ura3-52 lys2-801 ade2-101 trp1-Δ1 spa2Δ3::URA3 | This lab |
| Y2009 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 PEA2::myc BUD6::HA | This study |
| Y2011 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 spa2(1-2, 116-1466)::HA | This study |
| Y2012 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 spa2(1-410, 531-1466)::HA | This study |
| Y2013 | MATα leu2Δ98 ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 spa2Δ3::URA3 | This study |
| Y2014 | MATa leu2Δ98 ura3-52 lys2-801 ade2-101 his3-Δ200 fus2::lacZ::LEU2 | Segregant of 105A-1 (23) |
| Y2015 | MATa leu2Δ98 ura3-52 lys2-801 ade2-101 his3-Δ200 fus2::lacZ::LEU2 spa2Δ::URA3 | This study |
| Y2016 | MATa leu2Δ98 ura3-52 lys2-801 ade2-101 his3-Δ200 fus2::lacZ::LEU2 spa2(1-2, 116-1466)::HA | This study |
All strains except those marked with an asterisk are in the same genetic background (S288c).
Construction of yeast strains.
Y2003 (PEA2::HA) and Y2004 (PEA2::myc) were constructed by epitope tagging the PEA2 gene in a haploid wild-type yeast strain, Y762, by the strategy described by Schneider et al. (76). Primers 5′-GAG GCA AAC ACC TCG CTG GCG CTT AAT AGA GAC GAT CCA CCA GAT ATG CTA AGG GAA CAA AAG CTG GAG-3′ and 5′-ATT CTT CTT ATT CTA TAT TTA TAT ATC AAT GTT TTA TAA TAA GAT GTT TAT TCA CTA TAG GGC GAA TTG-3′ were used to amplify a region of pMPY-3XHA (for the hemagglutinin [HA] tag) or pMPY-3XMYC (for the c-myc tag). The resulting PCR product of approximately 1.5 kb contains a URA3 gene flanked by direct repeats encoding three copies of the HA or c-myc epitope (76). Both ends of the PCR fragment contain about 50 bp of PEA2 sequences surrounding the site of fragment insertion. Each DNA fragment was used to transform Y762. Transformants that had integrated the fragment correctly at the chromosomal locus were identified by PCR analysis. The integrants were allowed to grow overnight in YPAD (77) and plated onto 5-fluoro-orotic acid plates. 5-Fluoro-orotic acid-resistant colonies were checked by PCR for loss of the URA3 gene through homologous recombination between the two repeated epitope-coding sequences; this leaves a single in-frame epitope-coding sequence at the site of integration. Proper formation of the epitope-tagged proteins was further confirmed by immunoblot analysis. The tagged genes contain approximately 40 additional codons for the epitope plus linker sequences, and the resulting protein is approximately 5 kDa larger than the wild-type proteins.
The doubly tagged strain Y2009 was constructed by tagging the 3′ coding sequence of BUD6 with HA in strain Y2004 by the procedure described above. The tag is inserted before the last 10 codons of Bud6p. The primer pair used was 5′-GAA AAT TTT GTG GGT AAT TCA AAC CTG AAA AAA TCG GGA GGC TTG AAG AAG AGG GAA CAA AAG CTG GAG-3′ and 5′-CAA AAT ATG CTC TCA AAT TTG CTT CAT CCT TCT GCT TTC TTA TCC TTT CTA TCA CTA TAG GGC GAA TTG-3′. All of the tagged strains used in this study (Y2003, Y2004, and Y2009) appear normal in growth and form mating projections similar to those of wild-type cells after pheromone treatment.
Y1213 was generated by deletion of STE5 from Y864 by a method previously described (5). Primers 5′-CTA AAA AAG GAA GAT ACA GGA TAC AGC GGA AAC AAC TTT TAA ATG ATG GAA CTG AGA GTG CAC CAT AAA-3′ and 5′-C GGG ATG CTT TCT TTT TAT TAT TGC ATA AAA TTT AGT GTA TAC TCT ATA TTG AGC TGA TTT AAC AAA AAT-3′ were used to amplify TRP1 from plasmid pRS314 (78). The resulting PCR product contained a TRP1 gene flanked by 50 bp of sequences upstream and downstream of STE5 coding region. This fragment was transformed into Y864. Correct substitution at the STE5 locus was verified by PCR. The resulting strain (Y1213) does not form mating projections after α-factor treatment.
Y2002 was constructed by deletion of PEA2 from yeast strain Y762 as described above with primers 5′-C GAA GTC CTG TGT TCG AGA GGG AGA ATA ACG GTG GAT ATC ACG TTT CAT AAG CGC GCC TCG TTC AGA ATG-3′ and 5′-C TTC TTA TTC TAT ATT TAT ATA TCA ATG TTT TAT AAT AAG ATG TTT ATT CAC TCT TGG CCT CCT CTA GTA-3′ to amplify HIS3 from pRS313 (78). Correct substitution of PEA2 was verified by PCR analysis. This strain (Y2002) forms peanut-shaped mating projections upon exposure to mating pheromone as described for typical pea2 mutants (86).
The spa2(1-2, 116-1466) alleles in Y2011 and Y2016 were generated from Y762 and Y2014 (a segregant of 105A-1 [23]), respectively, as described by Schneider et al. (76). The primers used were 5′-CG AGC CAC CGA AAC AGA ATA AAC AAA AGA AAA GAA AGA GTA AAC ATG GGT AGG GAA CAA AAG CTG GAG CTC-3′ and 5′-GG GGG CCG TGG AGC ATC CAA ATC CTT GTC GAA CCC TCT TCT CTT GAT CTC TAG GGC GAA TTG GGT ACC GGG-3′. The resulting spa2 allele has three in-frame copies of HA-coding sequence replacing codons 3 to 115 of SPA2.
HA::SPA2 deletion constructs and phenotypic assays.
A fragment containing SPA2 with a NotI site after the first codon (created by K. Madden) was cloned into pRS315, and a NotI fragment encoding three copies of an HA epitope was introduced into the NotI site, creating plasmid pBU4. The ApaI site in pBU4 was destroyed by filling in with T4 DNA polymerase, and a linker with the ApaI site and stop codons in all three frames was inserted into the HindIII-XhoI sites of this plasmid, resulting in pBU19. A series of 3′ deletions were made from pBU19 by using the Double-Stranded Nested Deletion Kit from Pharmacia, Piscataway, N.J. pHA::spa2(1-481, 1208-1466) was created by excising the internal SphI fragment of SPA2. pHA::spa2(1-13, 265-1466) and pHA::spa2(1-13, 265-552) were created by digesting pHA::SPA2(1-1466) and pHA::spa2(1-552), respectively, with EcoRV and ligating the in-frame sites. The last codon of each deletion was determined by DNA sequence analysis (W. M. Keck Foundation Biotechnology Resource Laboratory, Yale University). (The plasmid constructs generated are listed in Table 3 and diagrammed in Fig. 3.)
TABLE 3.
Summary of deletion analysesa
| HA::SPA2 deletion construct | HA::Spa2p level | Localization
|
Bud site selection | Mating projection formation | Pseudohypha formation | Complementation of spa2Δ colethality with:
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Spa2p | Pea2p | bem2 | cdc10 | slk1 | swi4 | |||||
| (1-1466) | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| (1-1410b) | ++ | ++ | ++ | − | − | − | − | − | − | − |
| (1-1380) | ++ | ++ | ++ | − | − | − | − | − | − | − |
| (1-1049) | ++ | +++ | +++ | − | − | − | − | − | − | − |
| (1-852) | ++ | +++ | +++ | NT | NT | NT | NT | NT | NT | NT |
| (1-736) | +++ | +++ | +++ | NT | − | − | NT | NT | − | NT |
| (1-552) | ++++ | + | + | − | − | − | NT | NT | NT | NT |
| (1-530) | ++++ | + | + | − | − | − | − | − | − | − |
| (1-430) | − | − | − | − | − | − | − | − | − | − |
| (1-193) | − | − | NT | NT | NT | NT | NT | NT | NT | NT |
| (1-481, 1208-1466) | ++++ | − | − | − | − | − | − | − | − | − |
| (1-13, 265-552) | ++++ | − | − | − | − | − | − | − | − | − |
| (1-99, 537-552) | − | − | − | NT | NT | NT | NT | NT | NT | NT |
| (1-13, 265-1466) | ++++ | +++ | +++ | − | − | − | − | − | − | − |
| (1-99, 537-1466) | − | − | NT | NT | NT | NT | NT | NT | NT | NT |
++++, indistinguishable from wild type; +++, slight defect; ++, modest defect; +, strong defect; −, no function or complementation; NT, not tested.
The C-terminal end of this construct is estimated.
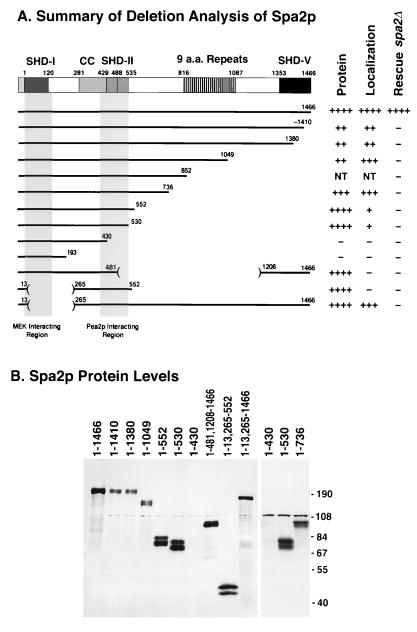
FIG. 3.
HA::SPA2 deletion constructs and their relative cellular protein levels. (A) Diagram of HA::Spa2p deletions. Each horizontal bar represents a segment of HA::Spa2p. The end residues for each segment are labeled according to the position in the wild-type protein. A summary of deletion analysis results is presented to the right of the constructs. For more detailed phenotypic analysis, see Table 3. The structure of Spa2p is presented above the deletions as described by Gehrung and Snyder (30) and Roemer et al. (71). The shaded regions below SHD-I and SHD-II correspond to the MEK-interacting region and Pea2p-interacting region, respectively (see also Fig. 2 and 6). (B) Immunoblot analysis of proteins from HA::Spa2p deletion strains. Cell lysates were prepared from a spa2Δ strain (Y2007) containing the indicated HA::SPA2 deletion constructs. Equal amounts of protein were loaded in each well and fractionated by SDS–8% PAGE. The immunoblot was prepared and probed with anti-HA MAb 16B12. Two separate samples from two different experiments are shown for the 1-430 and 1-530 constructs. The 110-kDa band is a protein that cross-reacts with 16B12 ascites; this band is also present in Fig. 1A. NT, not tested.
Plasmids were transformed into spa2Δ strains for phenotypic analyses. Protein levels in lysates prepared from the resulting strains were examined by immunoblot analysis with anti-HA monoclonal antibodies (MAb) (see below). Immunolocalization of the proteins is described below.
Complementation of the different defects was determined as follows. Complementation of defects in projection formation was carried out with yeast strains Y2007 and Y2008. Cells carrying the HA::Spa2p deletion construct were treated with α-factor mating pheromone (Sigma Chemical Co., St. Louis, Mo.) for 1.5 h as described previously (50), fixed, and observed under a light microscope. Complementation of the bud site selection defect was assessed with strain Y2001. Budding patterns were analyzed by Calcofluor staining (50). Complementation of the pseudohyphal defect was assayed in Y1209 (Σ background) as described by Roemer et al. (71). Complementation of spa2Δ colethality with cdc10-10, swi4-100, bem2-101, or slk1-1 was examined with yeast strains Y831, Y877, Y875, and Y754, respectively, by using a colony sectoring assay (15).
Two-hybrid interactions.
The plasmids used in the two-hybrid assay are listed in Table 2. Unless noted otherwise, the plasmids were constructed by amplifying the protein-coding sequences by PCR and inserting each fragment into the BamHI sites of vectors pSH2-1 (27, 32) and pACTII (22a). Typically the BamHI sites were created on both ends of the PCR fragment; for fragments containing endogenous BamHI sites, BglII sites were created and used instead. The orientations of the insert and fusion junctions were checked by restriction mapping and DNA sequence analysis (W. M. Keck Foundation Biotechnology Resource Laboratory, Yale University).
TABLE 2.
Plasmids used in the two-hybrid analyses
| Fusion and plasmida | Description | Source or reference |
|---|---|---|
| LexA fusions | ||
| pB342 (pSH2-1) | Vector; pLexA(1-87)/2μm/HIS3 | 27, 32 |
| pB551* | pLexA::SPA2(1-1466)/2μm/HIS3 | K. Madden |
| pB552 | pLexA::SPA2(1-698)/2μm/HIS3 | This study |
| pB553 | pLexA::SPA2(512-698)/2μm/HIS3 | This study |
| pB554 | pLexA::SPA2(512-1118)/2μm/HIS3 | This study |
| pB555 | pLexA::SPA2(743-1466)/2μm/HIS3 | This study |
| pB556 | pLexA::SPA2(743-1118)2μm/HIS3 | This study |
| pB557 | pLexA::SPA2(1-530)/2μm/HIS3 | This study |
| pB558 | pLexA::SPA2(1-488)/2μm/HIS3 | This study |
| pB559 | pLexA::SPA2(112-698)/2μm/HIS3 | This study |
| pB560 | pLexA::SPA2(112-530)/2μm/HIS3 | This study |
| pB561 | pLexA::SPA2(281-698)/2μm/HIS3 | This study |
| pB562 | pLexA::SPA2(281-530)/2μm/HIS3 | This study |
| pB563* | pLexA::SPA2(375-698)/2μm/HIS3 | This study |
| pB564 | pLexA::SPA2(1-150)/2μm/HIS3 | This study |
| pB565 | pLexA::BUD6(1-788)/2μm/HIS3 | This study |
| pB566 | pLexA::BUD6(272-788)/2μm/HIS3 | This study |
| pB567 | pLexA::BUD6(358-768)/2μm/HIS3 | This study |
| pB568 | pLexA::STE11/2μm/HIS3 | 14a |
| pB46* | pLexA::CIK1(20-466)/2μm/HIS3 | 62 |
| GAL4 AD fusions | ||
| pB282 (pACTII) | Vector; pAD/2μm/LEU2 | 22a |
| pB581 | pAD::PEA2(1-420)/2μm/LEU2 | This study |
| pB582 | pAD::SPA2(1-150)/2μm/LEU2 | This study |
| pB583* (p831) | pAD::BUD6(275-788)/2μm/LEU2 | 26 |
| pB584* (p688) | pAD::BUD6(478-788)/2μm/LEU2 | 26 |
| pB585 | pAD::BUD6(358-768)/2μm/LEU2 | This study |
| pB586* | pAD::ACT1(1-375)/2μm/LEU2 | This study |
| pB587* | pAD::BEM1(1-551)/2μm/LEU2 | This study |
| pB588 | pAD::MKK1(1-508)/2μm/LEU2 | This study |
| pB589 | pAD::MKK1(214-508)/2μm/LEU2 | This study |
| pB590 | pAD::MKK1(1-214)/2μm/LEU2 | This study |
| pB591 | pAD::MKK1(1-179)/2μm/LEU2 | This study |
| pB592* | pAD::MKK2(1-508)/2μm/LEU2 | G. Paravicini |
| pB331 | pAD::SLT2(1-484)/2μm/LEU2 | 16 |
| pB593 | pAD::PBS2(1-668)/2μm/LEU2 | This study |
| pB594 (pSL2091) | pAD::STE11/2μm/LEU2 | 67a |
| pB595 (pSL2165) | pAD::STE7(1-515)/2μm/LEU2 | 67a |
| pB596 (pSL2170) | pAD::STE7(172-515)/2μm/LEU2 | 67a |
| pB597 | pAD::STE7(1-172)/2μm/LEU2 | This study |
| pB598* | pAD::STE7(1-136)/2μm/LEU2 | This study |
| pB599 (pSL2175) | pAD::FUS3/2μm/LEU2 | 67a |
| pB45 | pAD::KAR3(12-515)/2μm/LEU2 | 62 |
The two-hybrid constructs created in this study were made by cloning the PCR products into the BamHI or BglII sites of vectors, except for those marked with an asterisk.
To test for interactions, plasmid pairs (one LexA fusion and one activation domain [AD] fusion) were cotransformed into the reporter yeast strain Y864. Transformants or patches grown from the transformants were replica plated onto a sterile circle of Whatman filter paper on fresh SC-His-Leu solid medium to select for the pSH2-1- and pACTII-derived plasmids. Cells were allowed to grow overnight at room temperature, and after lysis with chloroform vapor for 15 min, the filters were transferred onto X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates (89). The X-Gal plates with the filters were incubated at 30°C for up to 24 h. Blue color appeared within 2 h for the very strong interactions. The strength of the interactions was determined visually by the intensity of coloration.
For quantitative β-galactosidase (β-Gal) assays most experiments were performed with strains grown in liquid medium as described by Erdman et al. (23). However, for some of the two-hybrid strains, the β-Gal activity was lost after continuous propagation in liquid medium, presumably because the constructs are deleterious for cell growth. To circumvent this problem, yeast transformants were patched on plates containing selective medium. After incubation at room temperature for 1 day, cells were scraped from the plate and transferred to Z buffer for analysis (23). β-Gal activity was normalized to protein concentration in all cases. Three samples were usually analyzed for each interaction.
Antibodies.
MAb against HA (clone 16B12) and c-myc (clone 9E10) were purchased from BAbCO, Richmond, Calif. A MAb against actin (clone C4) was purchased from ICN Biochemicals, Inc., Aurora, Ohio. Anti-Swi6p antiserum was obtained from Brenda Andrews’ laboratory and affinity purified in a study by Madden et al. (49). Affinity-purified anti-Spa2p antiserum was as described by Snyder (80). Alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G (IgG) and anti-rabbit IgG and CY3-conjugated goat antimouse antibodies were purchased from Jackson ImmunoResearch Laboratory, Inc., West Grove, Pa. Antibodies used in indirect immunofluorescence were preabsorbed with whole cells and spheroplasts of untagged wild-type yeast cells to remove nonspecific antibodies (10).
IP and immunoblot analyses.
Overnight cultures grown in YPAD or selective media were diluted to an optical density at 600 nm (OD600) of 0.1 to 0.15 in fresh YPAD and grown to early log phase (OD600 = 0.3 to 0.5). Cells were then collected, washed, and lysed with glass beads in lysis buffer as described by Kamada et al. (40). Lysates were centrifuged for 10 min at 14,000 × g to remove cell debris. The clear lysates were subjected to immunoprecipitation (IP), or 1/3 volume of 4× Laemmli sample buffer (73) was added for immunoblot analysis.
IPs were performed by using the buffer system described by Kamada et al. (40). For each IP, 100 μg of total yeast protein in 450 μl of IP buffer was incubated with antibodies for 1.5 h at 4°C. For IP with MAb 16B12 or 9E10, a 1:150 dilution of crude ascites fluid was used. For IP with affinity-purified anti-Spa2p or preimmune serum, a 1:100 dilution of the affinity-purified antibody was used. To collect the immunocomplex, 50 μl of a 1:2 IP buffer suspension of protein A-conjugated Sepharose beads (Pierce, Rockford, Ill.) was added, and the mixture was incubated for an additional 1.5 h at 4°C with constant mixing. The beads were pelleted and washed four times with IP wash buffer and then eluted with 50 μl of lysis buffer plus 25 μl of 4× Laemmli sample buffer.
For immunoblot analysis, cell lysates or immunoprecipitates in Laemmli sample buffer were fractionated by sodium dodecyl sulfate–8% polyacrylamide gel electrophoresis (SDS–8% PAGE), blotted onto Immobilon (Millipore, Bedford, Mass.), and probed with primary antibody. The reactive bands were visualized with alkaline phosphatase-conjugated antibodies and CDP-Star (Boehringer Mannheim Corp., Indianapolis, Ind.) detection reagent or the NBT–BCIP (nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate) color detection system as described by Harlow and Lane (34).
In vitro kinase assays.
Slt2p kinase activity was assayed as described by Kamada et al. (40). Cells containing pFL44-SLT2HA (94) were grown to early log phase (OD600 = 0.2 to 0.3), collected, and lysed with glass beads. For each kinase reaction, 250 μg of total yeast proteins was preincubated with protein A-conjugated Sepharose beads for 1 h, and the supernatant was immunoprecipitated with MAb 16B12. IP mixtures were then incubated with 0.25 mg of myelin basic proteins (MBP) per ml and 15 μCi of [γ-32P]ATP at 30°C for 25 min. The reaction were stopped by addition of sample buffer. The phosphorylation of MBP was analyzed by SDS–15% PAGE followed by autoradiography. Kinase activities were quantified by using Bio-Rad Multi Analyst, version 1.0.2. As a control, background phosphorylation of MBP was determined in a parallel assay with strains lacking HA-tagged Slt2p.
Indirect immunofluorescence microscopy.
Indirect immunofluorescence of yeast cells containing HA::Spa2p deletions and Pea2p::myc was performed as described previously (30). Y2007 yeast strains with plasmids were grown overnight in selective media, diluted to an OD600 of 0.1 in fresh YPAD, and grown to early log phase (OD600 = 0.3). α-Factor was added to a final concentration of 5 μg/ml as described by Madden and Snyder (50). Cells were fixed in 3.7% formaldehyde for 60 min and spheroplasted. Spheroplasts were allowed to settle on poly-l-lysine-coated slides and incubated with preabsorbed primary antibodies (MAb 16B12 for the HA epitope tag or MAb 9E10 for the c-myc epitope tag) overnight at 4°C. For detection of primary antibodies, preabsorbed CY3-conjugated goat anti-mouse IgG was used.
Sucrose gradient velocity sedimentation.
Cell lysates were prepared from strain Y2009 and subjected to velocity centrifugation. Cells were grown to early log phase (OD600 = 0.3 to 0.5) in YPAD and lysed by vortexing in modified phosphate-buffered saline buffer (0.17 M NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.2) containing glass beads and protease inhibitors. The crude extract was centrifuged at 16,000 × g at 4°C for 10 min. Prior to loading onto the gradient, 160 μl of crude extract was centrifuged at 110,000 × g at 4°C for 5 min in an A-100/18 Airfuge rotor (Beckman, Instruments., Fullerton, Calif.). Molecular size standards (40 μl of a mixture of thyroglobulin [19.4S], catalase [11.3S], aldolase [7.4S], and bovine serum albumin [4.4S] at 10 mg/ml in the same buffer) were mixed with the supernatant and then loaded onto a 5-ml linear sucrose gradient (5 to 20%, wt/vol) in modified phosphate-buffered saline buffer containing protease inhibitors. Gradients were centrifuged at 40,000 × g at 4°C for 20 h in an SW55Ti rotor (Beckman). Approximately 175-μl aliquots were withdrawn from the bottom of each tube by using a peristaltic pump and analyzed by immunoblot analysis as described above. Proteins on the immunoblots were quantified by using the National Institutes of Health Image version 1.59. Molecular size standards were resolved on 8% polyacrylamide gels stained with Coomassie blue. To test whether the migration of the 12S complex is affected by detergent, 0.1% Triton X-100 was added to the protein lysate prior to centrifugation. Analysis of the 12S complex in pheromone-treated cells was carried out by incubating yeast cells grown to an OD600 of 0.3 with 5 μg of mating pheromone per ml for 2 h. Cell lysates were prepared and analyzed as described above.
RESULTS
Spa2p and Pea2p form a stable complex in vivo.
spa2Δ and pea2Δ cells share many phenotypes, and Spa2p and Pea2p localize to the same regions of the cell (30, 80, 86). To determine whether Spa2p and Pea2p interact physically, co-IP experiments and two-hybrid analysis were performed. For the co-IP experiments, we tagged the chromosomal PEA2 gene immediately upstream of the translational stop codon with a DNA fragment encoding three copies of either an HA epitope or a c-myc epitope, using the method described by Schneider et al. (76). Yeast cells expressing the epitope-tagged PEA2 gene (PEA2::HA or PEA2::myc) form mating projections that are indistinguishable from those of isogenic wild-type cells, and the Pea2p::HA and Pea2p::myc proteins localize to polarized growth sites in a manner identical to that described for wild-type Pea2p (86) (see Fig. 4, bottom panel a, which shows the localization of Pea2p::myc). Thus, the epitope-tagged proteins are functional. Immunoblot analysis indicates that the epitope-tagged Pea2 proteins migrate as a doublet at approximately 60 kDa (see Fig. 1A for Pea2p::HA and Fig. 5 for Pea2p::myc), consistent with the predicted molecular mass. This protein is not observed in control strains (see Fig. 1A, lanes 3, 4, and 6).
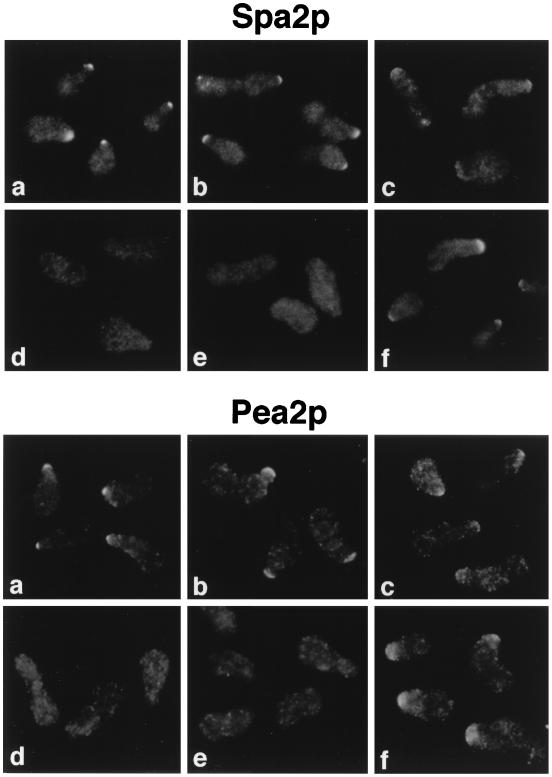
FIG. 4.
Immunolocalization of HA::Spa2p and Pea2p::myc in strains containing Spa2p deletion constructs. spa2Δ PEA2::myc strains containing the different HA::SPA2 deletion constructs were treated with α-factor and stained with anti-HA antibodies (top) or anti-c-myc antibodies (bottom). Examples of the full-length protein (a) and five different constructs, i.e., pHA::spa2(1-736) (b), pHA::spa2(1-530) (c), pHA::spa2(1-430) (d), pHA::spa2(1-13, 265-552) (e), and pHA::spa2(1-13, 265-1466) (f), are shown. Although each of the different mutants exhibits polarization defects, fields that contain polarized cells are shown.
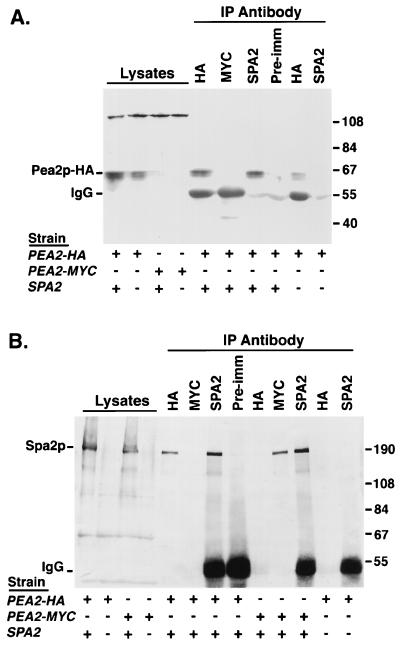
FIG. 1.
Spa2p and Pea2p coimmunoprecipitate. Proteins were prepared from SPA2 PEA2::HA, SPA2 PEA2::myc, spa2Δ PEA2::HA, and spa2Δ PEA2::myc strains (Y2003, Y2004, Y2005, and Y2006, respectively) and immunoprecipitated with the indicated antibodies. The total yeast lysates and immunoprecipitates were analyzed on immunoblots. (A) Immunoblot probed with anti-HA MAb 16B12. The doublet at approximately 60 kDa corresponds to Pea2p::HA; it is observed in lysates from the SPA2 PEA2::HA strain but not the SPA2 PEA2::myc strain. The doublet is also observed in IP with anti-HA antibody or anti-Spa2p antiserum from the SPA2 PEA2::HA lysate (fifth and seventh lanes from the left). Anti-Spa2p antiserum failed to precipitate Pea2p::HA from the spa2Δ PEA2::HA lysate (last lane). +, presence of an allele (for PEA2::HA and PEA2::myc) or wild-type copy of the gene (for SPA2). (B) Immunoblot probed with affinity-purified anti-Spa2p antiserum. Spa2p migrates as a 190-kDa band that is not present in the spa2Δ lysates. The band was detected in an IP with anti-HA antibody or anti-Spa2p antiserum from the SPA2 PEA2::HA lysate or in IP with anti-c-myc antibody or anti-Spa2p antiserum from the SPA2 PEA2::myc lysate. IgG marks the position of immunoglobulin heavy chain. Pre-imm, preimmune serum.
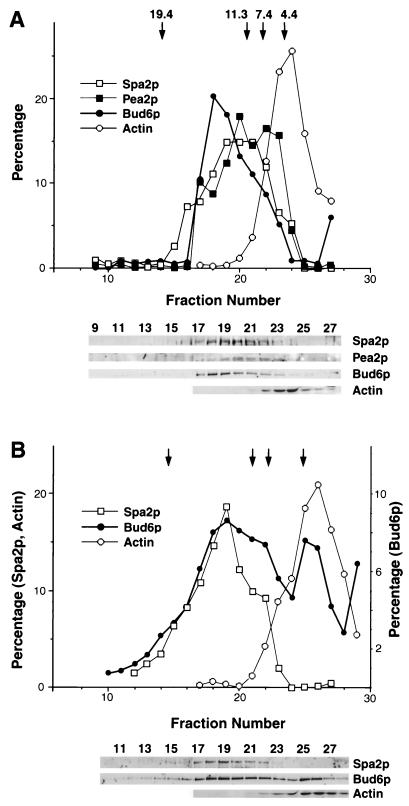
FIG. 5.
Velocity sedimentation analysis of Spa2p, Pea2p, and Bud6p. Cell lysates of a PEA2::myc BUD6::HA strain were prepared in the absence (A) or presence (B) of detergent and subjected to centrifugation in a 5 to 20% sucrose gradients. Fractions were collected and probed with anti-Spa2p antibodies, anti-c-myc antibodies (to detect Pea2p::myc), anti-HA antibodies (to detect Bud6p::HA), or an anti-actin MAb (C4). The S values of markers included in the same gradients are indicated at the top; these are thyroglobulin (19.4S), catalase (11.3S), aldolase (7.4S), and bovine serum albumin (4.4S). The amount of each immunoreactive protein was quantified for each fraction and is shown above the immunoblots. Only the top two-thirds of the gradient is shown, as no immunoreactive material is detected in the bottom third of the gradient. Note that in panel A the Pea2p peak contains material with a lower S value than Spa2p and Bud6p; this is likely to reflect its association with a degradation product of Spa2p that is not shown.
To test whether Spa2p and Pea2p associate in vivo, protein extracts prepared from PEA2::HA and PEA2::myc strains were immunoprecipitated with either anti-HA MAb, anti-c-myc MAb, affinity-purified anti-Spa2p antiserum, or preimmune serum. The resulting immunoprecipitates were then analyzed on immunoblots probed with anti-HA, anti-c-myc, and anti-Spa2p antibodies. As expected, Pea2p::HA was immunoprecipitated by an antibody that recognizes the HA tag but not by an anti-c-myc antibody (Fig. 1A); similarly, Pea2p::myc can be immunoprecipitated by an anti-c-myc MAb but not an anti-HA MAb (data not shown). When affinity-purified anti-Spa2p antiserum was used for immunoprecipitation of the PEA2::HA lysates, Pea2p::HA was also detected in the precipitate (Fig. 1A, seventh lane from the left). The preimmune serum was unable to immunoprecipitate Pea2p::HA from the same cell lysate (Fig. 1A, eighth lane from the left). Similarly, affinity-purified anti-Spa2p antiserum, but not the preimmune serum, can precipitate Pea2p::myc from the PEA2::myc extract (data not shown; the results were identical to those shown in Fig. 1A). Anti-Spa2p antiserum failed to precipitate Pea2p::HA with protein extracts prepared from a PEA2::HA spa2Δ strain (Fig. 1A, rightmost lane). Reciprocally, the anti-HA or anti-c-myc MAb can specifically precipitate Spa2p from PEA2::HA and PEA2::myc lysates, respectively, but not from extracts of untagged control strains (Fig. 1B). Thus, Spa2p and Pea2p are tightly associated with each other in a complex in vivo.
We also tested whether Spa2p and Pea2p interact by two-hybrid analysis. The coding sequence of SPA2 was fused to the sequence encoding the LexA DNA binding domain (LexA), and the sequence of PEA2 was fused to the sequence of the Gal4p transcriptional AD. Coexpression of LexA::Spa2p and AD::Pea2p results in an 18-fold increase in β-Gal activity in a LexA-responsive lacZ reporter yeast strain (see Table 4). The interactions between Spa2p and Pea2p fusion proteins are specific, since these fusions do not activate transcription either on their own, when coexpressed with other fusions, or in the presence of vectors alone (Fig. 2A and data not shown). We could not test whether LexA::Pea2p interacts with AD::Spa2p because the LexA::Pea2p construct activates transcription in the absence of AD plasmids. Nevertheless, the co-IP and two-hybrid results both demonstrate that Spa2p interacts with Pea2p.
TABLE 4.
Two-hybrid interactions between Spa2p and other polarity protins
| LexA DNA binding domain fusion | Gal4p AD fusion | Relative β-Gal activity (mean ± SD)a |
|---|---|---|
| Spa2p(1-1466) | Vector | 1.00 ± 0.09, 1.00 ± 0.10b |
| Pea2p(1-420) | 17.85 ± 4.58b | |
| Bud6p(275-788) | 8.30 ± 3.85 | |
| Bud6p(358-768) | 5.37 ± 1.28 | |
| Bud6p(478-788) | 2.35 ± 1.89 | |
| Bem1p(1-551) | 1.37 ± 0.67 | |
| Act1p(1-375) | 1.66 ± 0.12 | |
| Bud6p(1-788) | Vector | 0.97 ± 0.18 |
| Bud6p(272-788) | Vector | 2.66 ± 0.18 |
| Bud6p(1-788) | Pea2p(1-420) | 1.98 ± 0.32 |
| Bud6p(272-788) | Pea2p(1-420) | 3.05 ± 0.51 |
| Vector | Pea2p(1-420) | 1.10 ± 0.21 |
| Bud6p(275-788) | 1.06 ± 0.07 | |
| Bud6p(358-768) | 1.06 ± 0.14 |
The relative β-Gal activities are normalized to that for Spa2p-vector.
Value obtained from analysis of cells grown on plates (see Materials and Methods).
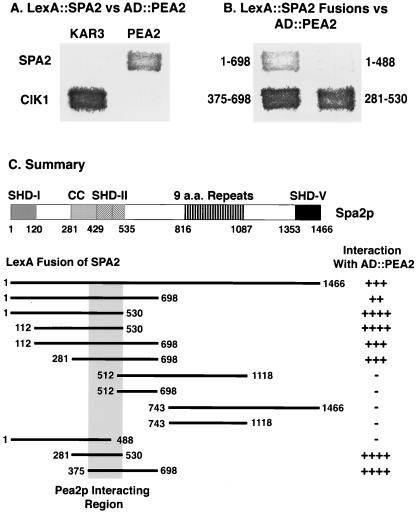
FIG. 2.
Spa2p interacts with Pea2p through a region containing SHD-II. (A and B) Filters showing the two-hybrid interactions. The dark patches indicate protein-protein interactions that result in expression of β-Gal. (A) LexA::Spa2p and AD::Pea2p interact with each other but not AD::Kar3p or LexA::Cik1p, two coiled-coil proteins. (B) Examples of interactions between AD::Pea2p and LexA fusions containing different regions of Spa2p. (C) Summary of mapping of the Pea2p-interacting region of Spa2p. Each horizontal bar represent a segment of Spa2p-coding sequence fused to the LexA plasmid (pSH2-1); the end residues for each Spa2p segment are labeled. These fusions were tested for interaction with the AD::PEA2 construct, and the strength of interaction were assigned as ++++ for the strongest interaction and − for interaction not detectable above background as judged from replicas of whole transformation plates. The summarized Pea2p-interacting region is shaded. The structure of Spa2p is presented above the fusions as described by Roemer et al. (71). CC, coiled coil A from residue 281 to 428; SHD-II, residue 429 to 535; a.a., amino acid. The first subregion of SHD-II is predicted to be coiled coil as well (coiled coil B).
Spa2p interacts with Pea2p through SHD-II, which contains coiled-coil sequence.
To delineate the region of Spa2p that interacts with Pea2p, we constructed LexA fusions containing various fragments of SPA2 coding sequence and used the two-hybrid system to test for interaction with AD::Pea2p (Fig. 2B and C). LexA::Spa2p(1-698), when coexpressed with AD::Pea2p, resulted in an increase of β-Gal activity, indicating a Pea2p-interacting region in the first 698 residues of Spa2p. This interaction was not observed with LexA::Spa2p(512-1118) or LexA::Spa2p(743-1466).
There is a predicted coiled-coil region in Spa2p from amino acid 281 to 488 (30). Part of this region is unique to Spa2p; however, residues 429 to 488 also belong to a domain (i.e., SHD-II [residues 429 to 535]) that is shared with a Spa2p-related protein, Sph1p (71). To find out whether the coiled-coil region of Spa2p is involved in Pea2p interaction, we further delineated the Pea2p-interacting region within the first 698 amino acids. Coexpression of AD::Pea2p with either LexA::Spa2p(1-530) or LexA::Spa2p(281-530), but not LexA::Spa2p(1-488), resulted in a strong increase in β-Gal (Fig. 2B and C). Thus, the C-terminal boundary for Pea2p interaction resides between residues 488 and 530. Because LexA::Spa2p(375-698) interacts with AD::Pea2p, the N-terminal boundary likely lies after residue 375 (Fig. 2C). The region from residue 375 to 530 could not be directly tested for interaction in this assay because a LexA fusion containing only this region activates transcription in the absence of an AD construct. These results indicate that the region necessary for Pea2p interaction lies between residues 375 and 530. This region is predicted to have mostly a coiled-coil structure and includes SHD-II.
It is possible that the interaction between Spa2p and Pea2p is through nonspecific interactions between coiled-coil sequences. This does not appear be the case, because neither Spa2p nor Pea2p interacts with the coiled-coil proteins Cik1p and Kar3p in parallel two-hybrid tests (54, 63) (Fig. 2A and data not shown); furthermore, Spa2p failed to interact with two other coiled-coil proteins, Nuf1p and NuMA (42, 55, 81a, 90), and Spa2p failed to interact with itself in similar assays (data not shown). Thus, Spa2p specifically interacts with Pea2p through a region predicted to contain coiled-coil sequence.
Deletion analysis reveals multiple regions involved in proper localization of Spa2p.
Spa2p is a large protein, and spa2Δ phenotypes suggest that it is involved in many aspects of polarized cell growth (30, 80). To determine which region of Spa2p is important for its localization and whether there are separable functional domains in the protein, we generated and analyzed a series of SPA2 deletion mutants. To facilitate detection of Spa2p, a DNA segment encoding three copies of HA was fused immediately before the SPA2 coding sequence corresponding to the N terminus. Full-length HA::Spa2p(1-1466) localizes in a pattern indistinguishable from that of wild-type Spa2p and is fully functional because it complements all spa2Δ defects, including projection formation, bud site selection, pseudohyphal growth, and spa2Δ colethality, with cdc10-10, swi4-100, bem2-101, and slk1-1 (Table 3). A series of deletion constructs were prepared from the pHA::SPA2(1-1466) construct (Fig. 3A and Table 3). The levels of HA::Spa2p produced from these constructs were examined by immunoblot analysis (Fig. 3B). Constructs lacking either part or all of the coiled-coil domain and the Pea2p-interacting SHD-II failed to produce detectable amounts of HA::Spa2p [e.g., pHA::spa2(1-430), -(1-193), -(1-99, 537-552), and -(1-99, 537-1466)]. In contrast, pHA::spa2(1-13, 266-552), which contains almost exclusively the entire coiled-coil domain and SHD-II, produces protein at a level comparable to that for the full length construct (Fig. 3B). These observations suggest that an intact coiled-coil domain and SHD-II are important for the stability of Spa2p.
We also determined the regions of Spa2p that are important for its localization. HA::Spa2p produced from the different pHA::spa2 deletion constructs was analyzed by indirect immunofluorescence microscopy in both vegetative and mating pheromone-treated cells. Cells carrying pHA::spa2 constructs that lacked portions or all of the coiled-coil domain and SHD-II did not stain above the background level, since these cells lacked detectable Spa2 protein by immunoblot analysis. As shown in Fig. 4 for pheromone-treated cells, HA::Spa2p(1-13, 266-552), which contains primarily the coiled-coil region and the Pea2p-interacting SHD-II, failed to localize to the projection tips (Fig. 4, upper panel, e), indicating that these regions are not sufficient for localization to growth sites. There are two other regions flanking the Spa2p coiled-coil domain that are also important for localization. All constructs containing residues 1 to 530 localize to projection tips (Fig. 4, upper panels a to c, and data not shown), although the shortest construct, HA::Spa2p(1-530), exhibits weaker staining (Fig. 4, upper panel c). These results indicate that a region from residue 14 to 265, which is N terminal to the coiled-coil domain, contributes to Spa2p targeting to growth sites. However, this region is not absolutely required for localization in the presence of another portion of Spa2p; HA::Spa2p(1-13, 265-1466), which lacks the region from residue 14 to 265 but has all the sequences C terminal to the coiled-coil domain, is able to localize to projection tips. Despite their ability to localize at growth sites, the Spa2p deletion constructs differ from the full-length protein in that they are more diffuse along the projection tip region rather than localized as a tight patch at the very tip (Fig. 4, upper panels, compare panel a to panels b, c, and f). Identical results for Spa2p localization at the incipient bud site and bud tip were obtained with vegetative cells (data not shown). These data indicate that in addition to the coiled-coil domain and Pea2p-interacting region, at least two other regions of Spa2p are important and redundant for localizing Spa2p to polarized growth sites.
Proper localization of Spa2p is required for the localization of Pea2p to growth sites.
It was reported previously that in spa2Δ cells, Pea2p was not detectable by immunoblot analysis with anti-Pea2p antiserum (86). Thus, it was not possible to determine whether Spa2p only affected the stability of Pea2p or whether Spa2p was directly involved in Pea2p localization. However, Pea2p::HA and Pea2p::myc are readily detected by immunoblot analysis when SPA2 is deleted, albeit at a lower level (Fig. 1A, second lane) (data not shown for Pea2p::myc). This allows analysis of Pea2p localization in the absence of Spa2p or in the presence of Spa2p deletion constructs.
Pea2p::myc localization in spa2Δ strains carrying the pHA::spa2 deletion constructs described above was analyzed (Fig. 4, bottom panels). All strains that localize HA::Spa2p properly also localize Pea2p::myc. For example, a deletion construct with most of the carboxy terminus of Spa2p removed [Spa2p(1-736)] still localizes Pea2p::myc to the growth sites (Fig. 4, bottom panel b). These results indicate that full-length Spa2p is not required for Pea2p::myc localization. Moreover, constructs that exhibit weak Spa2p localization [e.g., Spa2p(1-530)] at growth sites also exhibit very weak Pea2p::myc staining, suggesting a correlation between the efficiency of Spa2p localization and that of Pea2p::myc. Finally, all deletion constructs tested with either part of the Pea2p-interacting region removed [e.g., HA::Spa2p(1-481, 1208-1466)] or the localization of Spa2p disrupted [e.g., HA::Spa2p(1-13, 265-552)] also fail to localize Pea2p::myc. These results indicate Pea2p localization at growth sites is dependent upon both its interaction with Spa2p and the Spa2p localization sequences.
Full-length Spa2p is required for all aspects of Spa2p function.
The indirect immunofluorescence analysis showed that only a portion of Spa2p is required for localization of itself and Pea2p. We also analyzed the abilities of the truncated Spa2 proteins to complement various defects of the spa2Δ mutant. The results of these studies are summarized in Table 3 and Fig. 3. Only the full-length Spa2 protein is capable of rescuing the different defects of spa2 mutants, including the bud site selection defect, projection formation defect, pseudohypha formation defect, and colethality with bem2-101, cdc10-10, slk1-1, and swi4-100. These results suggest that full-length Spa2p is required for multiple aspects of its biological functions. Alternatively, because the larger HA::Spa2p derivatives are present at lower levels (Fig. 3B), the level of Spa2p may be important for its function.
Spa2p interacts with Bud6p (Aip3p).
We also tested whether Spa2p interacts with other polarity components by using the two-hybrid system. LexA::Spa2p(1-1466) was tested for interaction with AD fusions of Bud6p (Aip3p), Bem1p, Cdc24p, Cdc42p, Chs5p, or Act1p (actin) (see references 2, 7, 39, 61, 65, 74, and 79 for publications describing these proteins). No reproducible interaction was observed with Bem1p, Cdc24p, Cdc42p, Chs5p, or Act1p. However, interaction was observed with the AD::Bud6p(275-788) and AD::Bud6p(358-768) constructs. A low level of interaction was observed with a shorter Bud6p construct, AD::Bud6p(478-788) (Table 4). These results suggest that the region of Bud6p from residue 275 to 478 contributes to its interaction with Spa2p. Attempts to coimmunoprecipitate Spa2p and Bud6p epitope tagged with HA or c-myc at either its N terminus or C terminus were not successful. However, as demonstrated below, a significant portion of Bud6p cosediments in a similar-size complex with Spa2p, raising the possibility that these two proteins associate in vivo.
Sedimentation of Spa2p, Pea2p, and Bud6p.
The interactions described above suggest that Spa2p, Pea2p, and Bud6p may be part of a large multiprotein complex. To test this possibility, velocity sedimentation experiments were performed. For detection of all three proteins, we constructed an SPA2 BUD6::HA PEA2::myc strain. This strain has a normal growth rate and forms pointed mating projections similar to those of wild-type cells after pheromone treatment. Cell lysates were prepared from this strain, and cell debris was removed. The resulting lysates containing the soluble material were separated in a 5 to 20% sucrose gradient. Fractions from the gradient were collected and probed with anti-Spa2p antiserum, an anti-HA MAb (to detect Bud6p::HA), an anti-c-myc MAb (to detect Pea2p::myc), and an antiactin MAb. As shown in Fig. 5, most of the Spa2p, Pea2p::myc, and Bud6p::HA cosediment and peak at approximately 12S. A small amount of Pea2p migrates slightly slower than Spa2p; this is likely due to cosedimentation with Spa2p degradation products. Most of Bud6p cosediments with Spa2p, although a small portion sediments at a higher S value (e.g., see fraction 18). Actin present in these lysates remains at the top of the gradient. Similar results for Spa2p and HA::Bud6p were observed when cell lysates from an HA::BUD6 strain (containing Bud6p tagged at the N terminus) were analyzed. Control experiments analyzing another complex, the Cik1p-Kar3p microtubule motor complex (62), have shown that this motor complex sediments much more slowly than Spa2p, indicating that the 12S complex is relatively specific for these proteins.
The sedimentation of Spa2p, Pea2p, and Bud6p was also analyzed in protein samples treated with the nonionic detergent Triton X-100. The sedimentation of Spa2p (Fig. 5B) and Pea2p (not shown) is not affected by the presence of detergent. However, a portion of Bud6p reproducibly migrates to low-S fractions (approximately 4.4S) of the gradients, while the majority cosediments with Spa2p (Fig. 5B) and Pea2p (not shown). These data suggest that the majority of Spa2p, Bud6p, and Pea2p cosediment as part of a multiprotein complex that is not associated with vesicles and that a fraction of Bud6p is associated with vesicles. The size of the Spa2p-associated complex was similar in cells treated with mating pheromone. These results are consistent with the hypothesis that Spa2p, Pea2p, and Bud6p constitute part of a large multiprotein complex.
Spa2p interacts with components of two MAPK pathways involved in polarized morphogenesis.
Two MAPK signaling pathways, the Fus3p Kss1p mating pathway and the Slt2p cell integrity pathway, have been implicated in yeast polarized cell growth (35, 46). We tested whether Spa2p interacts with components of these two pathways by using the two-hybrid system. As shown in Table 5 and Fig. 6A, LexA::Spa2p(1-1466) interacts with AD fusions of Mkk1p and Mkk2p, the MEKs of the Slt2p pathway, and Ste7p and Ste11p, the respective MEK and MEKK needed for the mating response and pseudohypha formation. The scaffolding protein Ste5p (14a, 67a) is not required for the interaction between Spa2p and Ste11p or Ste7p, since these interactions still occur in a ste5Δ strain (Table 5). Spa2p does not interact in this assay with other signaling components, including Pkc1p, Ste20p, Slk1p, Slt2p, and Pbs2p (Pbs2p is the MEK of the Hog1p pathway, which mediates responses to high external osmolarity [8]) (Table 5, Fig. 6A, and data not shown). Attempts to coimmunoprecipitate Spa2p and its interacting kinases produced either at wild-type levels (Ste7p, Mkk1p, and Mkk2p), at high copy (Mkk1p), in either the presence or absence of mating pheromone, or from a GAL1 promoter (Ste11p) were not successful. This suggests that these interactions may be either weak or transient in vivo or that they require different physiological conditions.
TABLE 5.
Spa2p interacts with components of MAPK pathways
| Reporter strain | LexA DNA binding domain fusion | Gal4p AD fusion | Relative β-Gal activity (mean ± SD)a |
|---|---|---|---|
| Wild type | Spa2p(1-1466) | Vector | 1.00 ± 0.09, 1.00 ± 0.10b |
| Ste11p(22-738) | 15.21 ± 1.88b | ||
| Ste7p(1-515) | 679.34 ± 30.36b | ||
| Fus3p(1-353) | 1.95 ± 0.36b | ||
| Mkk1p(1-508) | 2,883.54 ± 1,204.52b | ||
| Mkk2p(1-506) | 35.96 ± 8.03 | ||
| Slt2p(1-484) | 0.67 ± 0.04 | ||
| Pbs2p(1-668) | 0.70 ± 0.02 | ||
| ste5Δ mutant | Spa2p(1-1466) | Vector | 1.00 ± 0.14, 1.00 ± 0.10b |
| Ste11p(22-738) | 8.17 ± 1.21b | ||
| Ste7p(1-515) | 117.51 ± 7.11b | ||
| Fus3p(1-353) | 1.91 ± 0.25b | ||
| Mkk1p(1-508) | 963.06 ± 157.88b | ||
| Mkk2p(1-506) | 115.19 ± 12.16 |
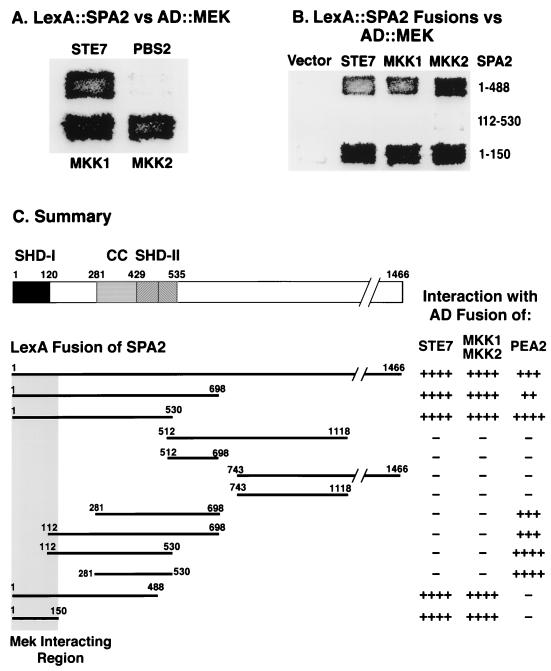
FIG. 6.
Spa2p interacts with MEKs through its N-terminal region containing the conserved domain SHD-I. (A and B) Filters of two-hybrid assays. (A) Interaction of LexA::Spa2p with AD constructs of Ste7p, Pbs2p, Mkk1p, and Mkk2p. (B) Different Spa2p fusions were tested for interaction with each of the AD::MEK constructs in panel A and with AD vector. (C) Summary of the interaction analysis for the different fusions. For each construct, the interactions were tested on colonies of hundreds of transformants and on patches from at least six transformants. Not shown is the interaction of AD::Spa2p(1-150) with LexA::Ste7p; these fusions interact strongly.
The MEK-interacting domain of Spa2p resides in the conserved N-terminal region.
To determine the MEK-interacting region of Spa2p, we tested for interactions between LexA fusions of different Spa2p domains and the AD fusions of Mkk1p, Mkk2p, and Ste7p. As shown in Figure 6C, LexA::Spa2p(1-698), which contains the Pea2p-interacting region, also interacted with all three MEKs. However, the fusion LexA::Spa2p(1-488), which did not interact with AD::Pea2p, still interacts with AD fusions of all three MEKs; another fusion, LexA::Spa2p(112-530), which is positive for Pea2p interaction, failed to interact with any of these three MEKs. A shorter fusion, LexA::Spa2p(1-150), containing only the first 150 residues of Spa2p interacts strongly with each of the AD::MEKs, although this construct activates transcription slightly on its own (Fig. 6B). There was no such increase with LexA::Spa2p(1-150) when the vector control plasmid was tested (Fig. 6B). In addition, AD::Spa2p(1-150), which does not activate transcription alone, also interacts with LexA::Ste7p (data not shown). Thus, the MEK-interacting region of Spa2p lies within the first 150 residues (Fig. 6B). This region contains a highly conserved sequence (SHD-I) with two repeats that are shared with Sph1p, a Spa2p homolog, and several unknown proteins in other organisms (71).
To determine if the MEK-interacting region is important for Spa2p function, we substituted codons 3 to 115 with a sequence containing three copies of the HA-coding region by using the PCR method (76). The resulting strain produces a protein, Spa2p(1-2, 116-1466)::HA, that is expressed at levels indistinguishable from those produced by wild-type cells (data not shown). In mating assays, strains containing this protein are defective in mating, similar to spa2Δ cells (Table 6). These cells also display defects in bud site selection and mating projection formation similar to those of spa2Δ cells (data not shown). Thus, the conserved region of the MEK-interacting domain of Spa2p is important for its function.
TABLE 6.
The MEK-interacting region of Spa2p is important for matinga
| Strain(s) | Relevant genotype | % Mating efficiency (mean ± SD) |
|---|---|---|
| Y604 and Y762 | Wild type | 1.22 ± 0.35 |
| Y609 | spa2Δ | 0.15 ± 0.02 |
| Y2011 | spa2(1-2, 116-1466)::HA | 0.19 ± 0.06 |
| Y2012 | spa2(1-410, 531-1466)::HA | 0.21 ± 0.08 |
The quantitative mating assays were performed as described previously (30). A spa2Δ strain (Y2013) was used as the mating tester.
The N-terminal nonkinase domains of Ste7p and Mkk1p interact with Spa2p.
Ste7p and Mkk1p each contain a kinase domain at their C termini and putative regulatory domain at their N termini (37, 82). To determine the Spa2p-interacting region of each of these kinases, AD fusions containing N-terminal and C-terminal regions of both Mkk1p and Ste7p were constructed and tested for interactions with LexA::Spa2p. AD::Ste7p(172-515), which contains the kinase domain, did not interact with LexA::Spa2p, whereas AD::Ste7p(1-172) and AD::Ste7p(1-136), which contain the nonkinase regions of Ste7p, both interacted strongly with Spa2p (Fig. 7B). Thus, Spa2p interacts with the amino-terminal nonkinase region of Ste7p.
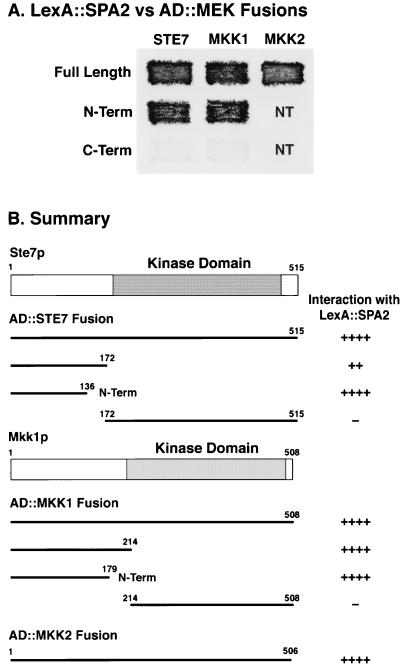
FIG. 7.
The N-terminal nonkinase domains of MEKs interact with Spa2p. (A) LexA::Spa2p fusions were tested for interactions with full-length, N-terminal (N-Term), or C-terminal (C-Term) AD constructs of Ste7p, Mkk1p, and Mkk2p. (B) Specific constructs from panel A and all fusions tested. The kinase domains of Ste7p and Mkk1p are indicated. NT, not tested.
AD fusions of different Mkk1p regions were also tested for interactions with LexA-Spa2p. As found for Ste7p, LexA-Spa2p interactions occurred with fusions containing the nonkinase domain, AD::Mkk1p(1-214) and AD::Mkk1p(1-179), but not with AD::Mkk1p(214-508), which contained the kinase domain (Fig. 7B). Therefore, the Spa2p-interacting region of Mkk1p also lies in the N-terminal noncatalytic region of the kinase.
Effects of Spa2p on the MAPK signaling pathways.
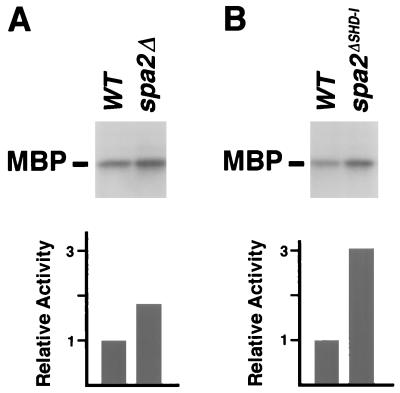
The interaction of Spa2p with Mkk1p and with Mkk2p raises the possibility that Spa2p might affect the signaling activity of the Slt2p MAPK. To test this possibility, we determined the relative Slt2p kinase activities in wild-type and spa2 mutant strains. An HA epitope-tagged Slt2p was immunoprecipitated from SPA2, spa2Δ, and spa2(1-2, 116-1466) cells (the latter lack the MEK-interacting domain) and incubated with [γ-32P]ATP and MBP, a MAPK substrate. As shown in Fig. 8, Slt2p proteins immunoprecipitated from the spa2 mutant strains exhibit a two- to threefold increase in in vitro phosphorylation of MBP relative to that for wild-type control strains. These results demonstrate that the activity of the Slt2p MAPK pathway is elevated in the spa2 mutants.
FIG. 8.
Slt2p kinase activities in wild-type cells (WT) and spa2 mutants. (A) In vitro phosphorylation of MBP by Slt2p immunoprecipitated from spa2Δ and congenic wild-type cells. (B) MBP phosphorylation by Slt2p immunoprecipitated from spa2(1-2, 116-1466) strain (shown as spa2ΔSHD-I) and an isogenic wild-type strain. The relative kinase activities were quantified and are shown below each autoradiograph.
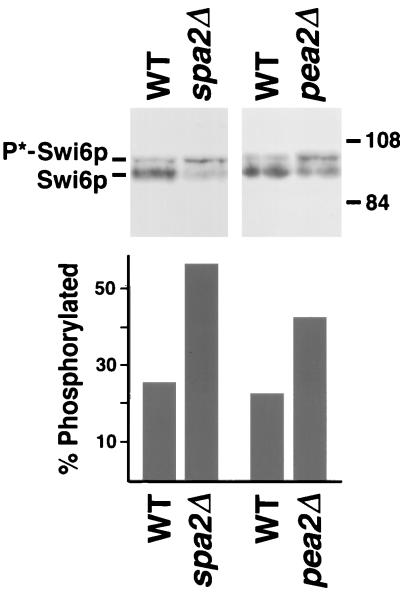
To further examine the activity of the Slt2p pathway, the phosphorylation state of Swi6p, a downstream target of the Slt2p MAPK (49), was analyzed in wild-type and spa2Δ cells. As shown in Fig. 9, the slow-mobility isoform of Swi6p, which is the phosphorylated form of the protein (49), is more intense in the protein extract prepared from a spa2Δ strain than in that from wild-type cells. Quantification indicates that the level of Swi6p hyperphosphorylation is about 25% in wild-type cells and 50% in spa2Δ cells (Fig. 9). The same phenomenon was observed when the phosphorylation patterns of Swi6p in diploid wild-type and spa2Δ/spa2Δ mutant cells were compared (data not shown). These results suggest that deletion of SPA2 results in a higher activity of the Slt2p pathway which causes hyperphosphorylation of Swi6p. Hyperphosphorylation of Swi6p was also observed in pea2Δ cells (Fig. 9). This is consistent with the observation that Spa2p and Pea2p function as a complex in the same process.
FIG. 9.
Levels of hyperphosphorylated Swi6p in wild-type (WT), spa2Δ, and pea2Δ strains. Equal amounts of proteins from cell lysates of wild-type (Y604 and Y762), spa2Δ (Y602), and pea2Δ (Y2002) strains grown to early log phase (OD600 = 0.3) were fractionated by SDS–8% PAGE. The immunoblot was prepared and probed with affinity-purified anti-Swi6p antiserum. The percentages of hyperphosphorylated Swi6p are shown below the blots. The relative amounts of proteins in the upper and lower bands were quantified by transmittence-reflectance scanning densitometry.
The interaction of Spa2p with Ste7p and Ste11p also raises the possibility that Spa2p might affect the Ste11p-Ste7p signaling pathways. Therefore, we analyzed the expression of two mating signaling reporter constructs, fus2::lacZ and fig1::lacZ (23), in spa2Δ and wild-type strains after treatment with different concentrations of mating pheromone. spa2Δ fus2::lacZ strains either treated with low levels of mating pheromone (0.6 nM) or not treated with pheromone reproducibly exhibit a reduced level of expression (approximately two- to threefold) relative to that of wild-type cells (Table 7). Treatment of cells with high levels of mating pheromone or analysis of fig1::lacZ strains revealed little or no reproducible difference between spa2Δ and wild-type cells (Table 7 and data not shown). spa2(1-2, 116-1466) cells, which lack SHD-I, also exhibit reduced fus2::lacZ expression in the presence of a low concentration of or no mating pheromone, similar to that of a spa2Δ mutant (Table 7). These experiments indicate that SHD-I is important for optimal MAPK signaling.
TABLE 7.
Expression of fus2::lacZ in wild-type and spa2Δ cells treated with different concentrations of mating pheromone
| Concn of α-factor (nM) | Relative β-Gal activity (mean ± SD)
|
||
|---|---|---|---|
| SPA2 cells | spa2Δ cells | spa2(1-2,116-1466) cells | |
| 0 | 1.00 ± 0.08 | 0.32 ± 0.03 | 0.34 ± 0.02 |
| 0.6 | 1.06 ± 0.06 | 0.48 ± 0.06 | 0.52 ± 0.02 |
| 6 | 6.25 ± 0.21 | 4.02 ± 0.44 | 4.54 ± 0.17 |
| 60 | 37.56 ± 0.44 | 23.40 ± 2.15 | 21.19 ± 1.69 |
| 6,000 | 831.25 ± 14.58 | 633.33 ± 29.17 | 627.08 ± 14.58 |
SPA2, spa2Δ, and spa2(1-2,116-1466) cells containing a fus2::lacZ reporter were grown to early log phase (OD600 = 0.3) and treated with the indicated concentrations of α-factor mating pheromone for 1.5 h. Cells were collected, and β-Gal activity was measured as nanomoles of o-nitrophenyl-β-d-galatopyranoside hydrolyzed per minute per milligram of total yeast proteins. The activities presented here are fold differences relative to the activity of vegetative SPA2 cells without pheromone treatment. The spa2 mutants (Y2015 and Y2016) used in this experiment are direct derivatives of a SPA2 fus2::lacZ strain (Y2014 [Table 1]). Two independent isolates each of spa2Δ and spa2(1-2,116-1466) cells were analyzed here.
Bud6p interacts with Ste11p.
We also used the two-hybrid system to test for interactions between Pea2p, Bud6p, and components of the signaling pathways described above. AD::Pea2p did not interact with LexA::Ste11p, LexA::Ste7p, or LexA::Fus3p (not shown). We could not test for Pea2p interaction with Mkk1p, since LexA::Mkk1p, like LexA::Pea2p, activates reporter transcription by itself. However, AD::Bud6p(358-768) and AD::Bud6p(275-788) showed strong transcription activation when coexpressed with LexA::Ste11p (Table 8), suggesting an interaction between Bud6p and Ste11p. The level of interaction between AD::Bud6p(478-788) and LexA::Ste11p was similar to the background level. These interactions suggest that the region from residue 358 to 478 contributes to the Ste11p interaction. The Bud6p fusions did not interact, as determined by two-hybrid analysis, with other signaling components tested, including Ste7p, Fus3p, Mkk1p, Mkk2p, Slt2p, and Pbs2p. Co-IP between Myc::Bud6p and HA::Ste11p was not successful, suggesting that the interaction may be transient or weak in vivo.
TABLE 8.
Two-hybrid interaction between Bud6p and Ste11p
| LexA DNA binding domain fusion | Gal4p AD fusion | Relative β-Gal activity (mean ± SD)a |
|---|---|---|
| Bud6p(1-788) | Vector | 0.84 ± 0.08, 0.88 ± 0.05b |
| Ste11p(22-738) | 217.06 ± 45.94b | |
| Ste7p(1-515) | 0.84 ± 0.07 | |
| Fus3p(1-353) | 0.75 ± 0.05 | |
| Mkk1p(1-508) | 0.74 ± 0.07 | |
| Mkk2p(1-506) | 0.98 ± 0.01 | |
| Slt2p(1-484) | 0.72 ± 0.08 | |
| Pbs2p(1-668) | 0.61 ± 0.04 | |
| Ste11p | Vector | 1.05 ± 0.17 |
| Bud6p(358-768) | 661.53 ± 300.73 | |
| Bud6p(478-788) | 5.84 ± 0.47 |
DISCUSSION
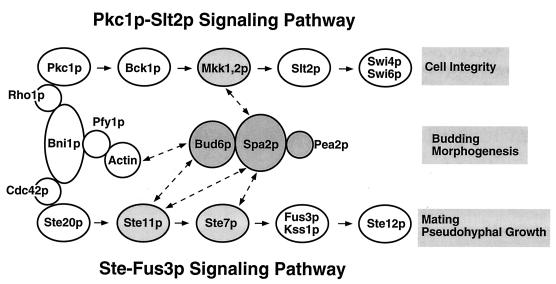
In this study, we demonstrated that the 1,466-amino-acid protein Spa2p contains multiple domains that interact with the polarity proteins Pea2p and Bud6p. Together, these polarity proteins may constitute a large multiprotein complex that functions to promote polarized cell growth through regulation of the actin cytoskeleton (Fig. 10). We also showed that components of this putative multiprotein complex interact with constituents of two MAPK signaling pathways involved in budding, mating, and pseudohypha formation (Fig. 10). These different interactions may provide links by which polarity components and signaling pathways can coordinate the complex processes of polarized growth.
FIG. 10.
Summary of the different interactions between constituents of the 12S complex and signaling components. Proteins that contact one another have been shown to be physically associated by co-IP or sedimentation analysis. Proteins that interact as determined by the two-hybrid system are indicated by lines with arrowheads at each end. Components that were analyzed in this study are shaded. For a further description of the different components, see reference 17.
Spa2p contains multiple protein-interacting domains that are important for its function.
Sequence analysis of Spa2p has revealed a variety of domains within this protein (30, 71). There are three large regions of over 100 amino acids (SHD-I, -II, and -V) that are homologous to Sph1p, a related protein in yeast. The second of these, SHD-II (amino acids 429 to 535), contains a large putative coiled-coil segment (71). In addition, Spa2p has two regions that are unique: a coiled-coil domain from residue 281 to 428 as well as a stretch of 9-amino-acid repeats between residues 816 and 1087 (30).
We have found that Spa2p interacts with Pea2p and that the Pea2p-interacting domain maps to a region between residues 375 and 530. This region is composed mostly of SHD-II, which is primarily coiled coil, and a small portion of the coiled-coil region that is unique to Spa2p. Pea2p also possesses a putative coiled-coil domain from residue 236 to 327 (86). The interaction of the Spa2p coiled-coil segments with Pea2p raises the possibility that the two polarity proteins might associate with each other via a coiled-coil interaction. Although much of the Pea2p-interacting region of Spa2p is homologous to Sph1p, Pea2p and Sph1p do not interact by using the two-hybrid system (71). Thus, it is likely either that the region outside the SHD-II region contributes to interaction with Spa2p or that the SHD-II regions of Spa2p and Sph1p interact with different proteins.
The Pea2p-interacting region of Spa2p is important for stability, localization, and function of the protein. Deletions that remove most of this region result in loss of detectable Spa2p (Fig. 3 and Table 3), and immunolocalization analyses indicate that this region in conjunction with one of two other regions of Spa2p is required for localization to growth sites within the cell (Fig. 4). Arkowitz and Lowe (3) have also mapped a localization domain of Spa2p to this same region (see below), and Valtz and Herskowitz have found that loss of Pea2p results in mislocalization of Spa2p (86). Thus, interaction of this region with Pea2p is critical for Spa2p localization and function.
We have also found a region within the N-terminal 150 amino acids of Spa2p that interacts with the yeast MEKs Ste7p, Mkk1p, and Mkk2p (Fig. 6) and an as-yet-unidentified region that interacts with Ste11p. All of these kinases belong to signaling pathways that have been implicated in polarized morphogenesis and mating (25, 37, 48, 69, 83, 94). The MEK-interacting region of Spa2p overlaps with SHD-I, which is homologous to domains in Sph1p and proteins in other organisms, such as humans and nematodes (71). In addition, Roemer et al. have found that Sph1p also interacts with these same MEKs (71). SHD-I contains two conserved repeated sequences called SDRs (for Spa2 direct repeat). We speculate that the conserved SDRs interact with MEKs in a wide variety of organisms.
Several other domains exist in the Spa2p protein. SHD-V and at least two other smaller SHDs are predicted from the Spa2p sequence, in addition to the Spa2p unique coiled-coil domain segment and 9-amino-acid repeats. We have found that Bud6p and Ste11p interact with Spa2p as determined by the two-hybrid system, and Roemer et al. found that Sph1p also interacts with these two proteins (71). Thus, it is likely that other SHDs (III, IV, and/or V) bind to Bud6p and Ste11p. Presumably other proteins also bind Spa2p domains. One additional candidate is Bni1p; bni1Δ mutants have phenotypes similar to those of spa2Δ cells, and Bni1p localizes to growth sites similarly to Spa2p (26).
Additional localization and functional domains of Spa2p.
In addition to the Pea2p-interacting domain, our analysis of HA::Spa2p deletions also revealed two other, less defined regions that are important for proper Spa2p localization at growth sites. One contains the sequence located between residues 14 and 264, and the other resides between residues 552 and 1466. Either region, when combined with the Pea2p-interacting SHD-II, is sufficient for targeting Spa2p to growth sites (Table 3; Fig. 3 and 4). It is likely that these two regions either contain additional sequences that facilitate Spa2p targeting or help Spa2p adopt a conformation that is competent for localization to growth sites. The different sequences that help localize Spa2p also are critical for localization of Pea2p (Table 3 and Fig. 4).
As this paper was being prepared, Arkowitz and Lowe reported that a region of Spa2p from amino acids 397 to 549 is sufficient for targeting a Spa2-green fluorescent protein (GFP) fusion to growth sites (3). Although this region overlaps well with the Pea2p-interacting region/localization domain that we identified, they did not identify the other regions of the proteins that are important for Spa2p localization. The discrepancy between our results and theirs may be due to the difference in sensitivity of detection between GFP and indirect immunofluorescence microscopy or to differences in the stability of Spa2p-GFP and HA::Spa2p proteins. Regardless, our data indicate that other regions of Spa2p are important for its proper localization.
Localization of Spa2p to the growth sites is not sufficient for its function. Only the full-length HA::Spa2p construct is capable of rescuing spa2 mutant defects (Table 3 and Fig. 3). Even the deletions removing fewer than 90 residues at the C terminus [i.e., HA::Spa2p(1-∼1410) and HA::Spa2p(1-1380)] fail to complement the spa2 defects. Since this region overlaps with SHD-V, it is likely that SHD-V interacts with another component that functions in the same processes as does Spa2p. Alternatively, it is also possible that SHD-V contributes to the stability of the protein so that proteins lacking this region are present at lower levels [which is the case for HA::Spa2p(1-∼1410) and HA::Spa2p(1-1380)] and are thus unable to provide proper Spa2p function. It is also important to note that although many Spa2p deletion constructs are capable of localizing to growth sites, their localization patterns are more diffuse over a broad region instead of forming a tight patch at growth sites as does the full-length protein. This is especially evident in pheromone-treated cells, in which the projection is wider in these mutants (Fig. 4); the wide projection might explain in part the broad distribution of the protein. Alternatively, as discussed further below, Spa2p is part of a multiprotein complex that functions early in the process of polarized cell growth. Thus, multiple regions of Spa2p are essential for the proper localization and function of Spa2p.
Spa2p interacts with other polarity proteins to form a multiprotein complex.
The results presented in this study indicate that Spa2p interacts with at least two other polarity proteins, Pea2p and Bud6p, and that a large portion of these proteins cosediment in sucrose gradients as a 12S complex (Fig. 5), which we call the 12S polarisome. These data are consistent with the similar localization patterns of these proteins and the similar phenotypes of spa2Δ, pea2Δ, and bud6Δ cells (2, 14, 30, 81, 86, 93).
A 12S sedimentation coefficient corresponds to a size of approximately 300 kDa, assuming a globular conformation (11). However, if the complex adopts an asymmetrical conformation, the mass might be greater. Given the predicted sizes of Spa2p (160 kDa), Pea2p::myc (53 kDa), and Bud6p::HA (90 kDa), it is likely that many of the components of the 12S complex are already accounted for. There may be additional components in this complex, such as Bni1p, which has been found to interact with Bud6p (26). The steady-state levels of Spa2p, Pea2p, Bud6p, and Bni1p in cell extracts appear to be similar (data not shown) suggesting that these proteins function at similar stochiometric ratios.
All of the proteins of the 12S complex may not associate with one another with similar affinity. Pea2p and Spa2p coimmunoprecipitate readily, whereas we have been unable to immunoprecipitate a complex containing both Bud6p and Spa2p. Perhaps the latter two proteins are more weakly associated with one another. Consistent with this interpretation, some of the Bud6p material does not appear to cosediment with Spa2p and Pea2p. A small amount of Bud6p migrates faster than Spa2p in cell lysates prepared in the absence of detergent (Fig. 5A), whereas a similar amount of material migrates much more slowly than Spa2p in detergent-treated extracts (Fig. 5B). Perhaps the association of Bud6p with Spa2p is spatially and/or temporally regulated.
The 12S polarisome may function as a regulator of the actin cytoskeleton. In support of this hypothesis, Bud6p was identified as an actin-interacting protein (2). In addition, the round shape of the spa2Δ and bud6Δ mutant cells and the tips of their mating projections are consistent with that expected for a mutant with an altered cytoskeletal structure (reference 80 and data not shown). However, the 12S complex is probably not always associated with actin, because actin does not comigrate with it in a sucrose gradient and actin has a localization pattern distinct from that of the 12S complex components. Furthermore, the localization of Spa2p and Bud6p to growth sites does not require an intact actin cytoskeleton (4, 81). Spa2p, Pea2p, Bud6p, and actin each affect bud site selection in diploids and other polarized growth processes, and all of these proteins localize to the incipient bud site, suggesting that they act very early in establishing and maintaining polarity and may work together in some processes. The 12S complex may function to regulate the actin cytoskeleton at an early step in morphogenesis, perhaps directing the actin cytoskeleton to specified growth sites in diploid cells and/or maintaining it in a more concentrated region once polarized growth has been initiated. All components of the proposed 12S complex, as well as the actin cytoskeleton, are also important for projection formation during mating (14, 30, 68, 86), which is a dynamic process that requires tracking and responding to pheromone gradients. Thus, we hypothesize that the 12S complex is particularly important for targeting, maintaining, and/or remodeling the actin cytoskeleton during dynamic growth periods.
Although our results indicate that Spa2p and Bud6p may function together in the same complex to promote polarized cell growth, they are likely to have different roles in this process, as suggested by three lines of evidence. First, a subset of Bud6p does not cofractionate with Spa2p and is likely affiliated with vesicles, indicating that these two proteins are not always associated. Second, we found that GFP-Spa2p is able to localize to growth sites in the bud6Δ cells (data not shown). This result indicates that, unlike Pea2p, Bud6p is not required for Spa2p localization (86). Third, although both spa2Δ and bud6Δ cells are round and each form less-polarized mating projections than wild-type cells, bud6Δ cells have less-severe polarity defects than those of spa2Δ cells; the mating projections of bud6Δ mutant cells are more pointed (and polarized) than those of spa2Δ cells (data not shown). Moreover, the phenotype of the spa2Δ bud6Δ double mutant is more severe than that of single mutants. Cells with both genes deleted appear to be larger and grow slower than cells with either gene deleted(data not shown), and a higher proportion of spa2Δ bud6Δ cells than single mutant cells do not form mating projections upon α-factor treatment (data not shown). The additive morphological defects of the double mutant indicate that Spa2p and Bud6p act in distinct steps in the process of polarized morphogenesis.
Interactions between Spa2p and signaling components.
Spa2p interacts with Ste7p, Ste11p, Mkk1p, and Mkk2p and plays a role in cell signaling. Another putative 12S complex component, Bud6p, interacts with Ste11p. Ste7p and Mkk1p interact with Spa2p through their N-terminal putative regulatory domains. Given the similar amino acid sequences and functions of Mkk1p and Mkk2p (37), it is likely that Mkk2p interacts through this region as well. The Spa2p-interacting region of Ste7p lies within the N-terminal 136-amino-acid region, and that of Mkk1p is within its first 179 residues. However, Ste7p and Mkk1p lack obvious similarity in the primary and secondary features in these regions. It is possible that the similarity between the two MEKs occurs in a higher-order structure or through very short or degenerate sequences.
Several lines of evidence suggest that the interactions between Spa2p and MEKs are functionally important. First, deletion of SHD-I, which lies in the MEK-interacting region of Spa2p, decreases mating efficiency and results in defects in other SPA2 functions (bud site selection or proper mating projection formation) (Table 6 and data not shown). Second, expression of the portions of Mkk1p or Ste7p that interact with Spa2p from two-hybrid constructs in wild-type cells results in a higher proportion of cells unable to produce mating projections or cells producing mating projections of aberrant morphology after treatment with mating pheromone (data not shown). Third, spa2Δ and spa2(1-2, 116-1466) strains have modest defects in pheromone-induced expression of a fus2::lacZ reporter, particularly at low pheromone concentrations (Table 7), and these cells appear to be hyperactive for activation of the Slt2p pathway as indicated by increased Slt2p kinase activity and hyperphosphorylation of Swi6p, a substrate for Slt2p (Fig. 8 and 9). Thus, Spa2p may help activate the Fus3p-Kss1p pathway and repress the Slt2p pathway. Alternatively, for the latter case, loss of Spa2p may damage cell integrity and may thereby activate the Slt2p pathway for compensation.
Because the defects in signaling activity are rather modest relative to the loss in mating efficiency, it is likely that Spa2p and its associated complex has another, as-yet-unidentified effect on the MAPK signaling pathways. One possibility is that 12S complex components help localize these signaling proteins to sites of polarized cell growth. Perhaps Ste11p, Ste7p, Mkk1p, and Mkk2p are concentrated and/or activated at growth sites, as is the case for Ste20p, which acts upstream of Ste11p (64). Mkk1p and Mkk2p have been shown to be important for polarized cell growth in yeast, and Ste7p and Ste11p are important for mating and pseudohyphal growth, both of which involve morphological changes. Localization of these different components at growth sites may be important for their function in these processes. A second possibility, not mutually exclusive with the first, is that the 12S complex serves as a scaffold for facilitating the interactions among these different components. Spa2p interaction with Ste7p and Ste11p is independent of Ste5p (Table 5). Thus, Spa2p may help mediate interactions of the different kinases and/or prevent the kinases from interacting with components of other MAPK pathways. Finally, Spa2p (and Bud6p) may mediate crosstalk between two signaling pathways. For example, it is possible that the initial response of the Slt2p pathway to the mating pheromone is through activation of Mkk1p or Mkk2p by components of the mating pathway (Ste11p and/or Ste7p) and that Spa2p interacts with both mating pathway signaling components and Mkk1p and thereby facilitates this activation. Consistent with this possibility, Zarzov et al. (94) have found that the activating tyrosine phosphorylation of Slt2p in response to mating pheromone is delayed in a ste11 mutant, and a similar delay can also been found in spa2Δ cells (9). Moreover, deletion of BCK1 (SLK1) still results in pheromone-induced activation of Slt2p kinase, suggesting the existence of another upstream activator (9). Crosstalk between different MAPK pathways has recently been reported in many cases. Ste11p can function in the Hog1p pathway by activating Pbs2p (66), and the Hog1p pathway can repress the activity of the mating pathway (33). Perhaps Spa2p facilitates or inhibits crosstalk between components of different signaling pathways.
ACKNOWLEDGMENTS
We thank G. Sprague, E. Elion, C. Boone, and G. Paravicini for plasmids. T. Roemer, N. Burns, K. Madden, B. Manning, and Y. Barral provided critical comments on the manuscript, and D. Diener and D. Cole provided advice on the sedimentation studies.
This research was supported by grant GM36494 to M. Snyder. B. Santos was supported by a postdoctoral fellowship from the Ministerio de Educación y Ciencia, Madrid, Spain.
REFERENCES
- 1.Adams A, Pringle J. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amberg D C, Zahner J E, Mulholland J W, Pringle J R, Botstein D. Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol Biol Cell. 1997;8:729–753. doi: 10.1091/mbc.8.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arkowitz R A, Lowe N. A small conserved domain in the yeast Spa2p is necessary and sufficient for its polarized localization. J Cell Biol. 1997;138:17–36. doi: 10.1083/jcb.138.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayscough K R, Stryker J, Pokala N, Sanders M, Crews P, Drubin D G. High rate of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bedinger P A, Hardeman K J, Loukides C A. Traveling in style: the cell biology of pollen. Trends Cell Biol. 1994;4:132–138. doi: 10.1016/0962-8924(94)90068-x. [DOI] [PubMed] [Google Scholar]
- 7.Bender A, Pringle J R. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:1295–1305. doi: 10.1128/mcb.11.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brewster J L, Valoir T D, Dwyer N D, Winter E, Gustin M C. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- 9.Buehrer B M, Errede B. Coordination of the mating and cell integrity mitogen-activated protein kinase pathways in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6517–6525. doi: 10.1128/mcb.17.11.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burns N, Grimwade B, Ross-Macdonald P B, Choi E-Y, Finberg K, Roeder G S, Snyder M. Large-scale characterization of gene expression, protein localization and gene disruption in Saccharomyces cerevisiae. Genes Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- 11.Canter C R, Schimmel P R. Biophysical chemistry, part II. Techniques for the study of biological structure and function. San Francisco, Calif: W. H. Freeman and Company; 1980. pp. 591–641. [Google Scholar]
- 12.Chant J, Pringle J R. Patterns of bud site selection in the yeast Saccharomyces cerevisiae. J Cell Biol. 1995;129:751–765. doi: 10.1083/jcb.129.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen D-C, Yang B-C, Kuo T-T. One step-transformation of yeast in stationary phase. Curr Genet. 1992;21:83–84. doi: 10.1007/BF00318659. [DOI] [PubMed] [Google Scholar]
- 14.Chenevert J, Valtz N, Herskowitz I. Identification of genes required for normal pheromone-induced cell polarization in Saccharomyces cerevisiae. Genetics. 1994;136:1287–1297. doi: 10.1093/genetics/136.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Choi K-Y, Satterberg B, Lyons D, Elion E A. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- 15.Costigan C, Gehrung S, Snyder M. A synthetic lethal screen identifies SLK1, a novel protein kinase homolog implicated in yeast cell morphogenesis and cell growth. Mol Cell Biol. 1992;12:1162–1178. doi: 10.1128/mcb.12.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costigan C, Kolodrubetz D, Snyder M. NHP6A and NHP6B, which encode HMG1-like proteins, function downstream in the yeast SLT2 MAPK pathway. Mol Cell Biol. 1994;14:2391–2403. doi: 10.1128/mcb.14.4.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costigan, C., and M. Snyder. Cell polarity in the budding yeast, Saccharomyces cerevisiae. Adv. Mol. Cell Biol., in press.
- 18.Drubin D G, Jones H D, Wertman K F. Actin structure and function: roles in mitochondrial organization and morphogenesis in budding yeast and identification of the phalloidin-binding site. Mol Biol Cell. 1993;4:1277–1294. doi: 10.1091/mbc.4.12.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drubin D G, Nelson W J. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- 20.Eisen J S. Development of motoneuronal phenotype. Annu Rev Neurol. 1994;17:1–30. doi: 10.1146/annurev.ne.17.030194.000245. [DOI] [PubMed] [Google Scholar]
- 21.Elion E A, Grisafi P L, Fink G R. FUS3 encodes a cdc2+/CDC28-related kinase required for the transition from mitosis into conjugation. Cell. 1990;60:649–664. doi: 10.1016/0092-8674(90)90668-5. [DOI] [PubMed] [Google Scholar]
- 22.Elion E A, Satterberg B, Kranz J E. FUS3 phosphorylates multiple components of the mating signaling transduction cascade: evidence for STE12 and FAR1. Mol Biol Cell. 1993;4:495–510. doi: 10.1091/mbc.4.5.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Elledge, S. J. Personal communication.
- 23.Erdman S, Lin L, Malczynski M, Snyder M. Pheromone-regulated genes required for yeast mating differentiation. J Cell Biol. 1998;140:461–483. doi: 10.1083/jcb.140.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Errede B, Cade R M, Yasar B M, Kamada Y, Levin D E, Irie K, Matsumoto K. Dynamics and organization of MAP kinase signal pathways. Mol Reprod Dev. 1995;42:477–485. doi: 10.1002/mrd.1080420416. [DOI] [PubMed] [Google Scholar]
- 25.Errede B, Gartner A, Zhou Z, Nasmyth K, Ammerer G. MAP kinase-related FUS3 from S. cerevisiae is activated by STE7 in vitro. Nature. 1993;362:261–264. doi: 10.1038/362261a0. [DOI] [PubMed] [Google Scholar]
- 26.Evangelista M, Blundell K, Longtine M S, Chow C J, Adames N, Pringle J R, Peter M, Boone C. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- 27.Fields S, Song O-K. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 27a.Flescher E G, Madden K, Snyder M. Components required for cytokinesis are important for bud site selection in yeast. J Cell Biol. 1993;122:373–386. doi: 10.1083/jcb.122.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freifelder D. Bud position in Saccharomyces cerevisiae. J Bacteriol. 1960;124:511–523. doi: 10.1128/jb.80.4.567-568.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gartner A, Nasmyth K, Ammerer G. Signal transduction in Saccharomyces cerevisiae requires tyrosine and threonine phosphorylation of FUS3 and KSS1. Genes Dev. 1992;6:1280–1292. doi: 10.1101/gad.6.7.1280. [DOI] [PubMed] [Google Scholar]
- 30.Gehrung S, Snyder M. The SPA2 gene of Saccharomyces cerevisiae is important for pheromone-induced morphogenesis and efficient mating. J Cell Biol. 1990;111:1451–1464. doi: 10.1083/jcb.111.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 32.Golemis E A, Brent R. Fused protein domains inhibit DNA binding by LexA. Mol Cell Biol. 1992;12:3006–3014. doi: 10.1128/mcb.12.7.3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall J P, Cherkasova V, Elion E, Guitin M C, Winter E. The osmoregulatory pathway represses mating pathway activity in Saccharomyces cerevisiae: isolation of a FUS3 mutant that is insensitive to the repression mechanism. Mol Cell Biol. 1996;16:6715–6723. doi: 10.1128/mcb.16.12.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 35.Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 36.Hicks J B, Strathern J N, Herskowitz I. Interconversion of yeast mating types. III. Action of the homothallism (HO) gene in cells homozygous for the mating type locus. Genetics. 1977;85:395–405. doi: 10.1093/genetics/85.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Irie K, Takase M, Lee K, Levin D, Araki H, Matsumoto K, Oshima Y. MKK1 and MKK2, which encode Saccharomyces cerevisiae mitogen-activated protein kinase-kinase homologs, function in the pathway mediated by protein kinase C. Mol Cell Biol. 1993;13:3076–3083. doi: 10.1128/mcb.13.5.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson D I, Pringle J R. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J Cell Biol. 1990;111:143–152. doi: 10.1083/jcb.111.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamada Y, Jung U S, Piotrowski J, Levin D E. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 1995;9:1559–1571. doi: 10.1101/gad.9.13.1559. [DOI] [PubMed] [Google Scholar]
- 41.Kilmartin J V, Adams A E M. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kilmartin J V, Dyos S L, Kershaw D, Finch J T. A spacer protein in the Saccharomyces cerevisiae spindle pole body whose transcript is cell cycle regulated. J Cell Biol. 1993;123:1175–1184. doi: 10.1083/jcb.123.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leberer E, Dignard D, Harcus D, Thomas D Y, Whiteway M. The protein kinase homologue STE20p is required to link the yeast pheromone response G-protein βγ subunits to downstream signalling components. EMBO J. 1992;11:4815–4824. doi: 10.1002/j.1460-2075.1992.tb05587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee K, Levin D. Dominant mutations in a gene encoding a putative protein kinase (BCK1) bypass the requirement for a Saccharomyces cerevisiae protein kinase C homolog. Mol Cell Biol. 1992;12:172–182. doi: 10.1128/mcb.12.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levin D, Errede B. A multitude of MAP kinase activation pathways. J NIH Res. 1993;5:49–52. [Google Scholar]
- 46.Levin D E, Errede B. The proliferation of MAP kinase signaling pathways in yeast. Curr Opin Cell Biol. 1995;7:197–202. doi: 10.1016/0955-0674(95)80028-x. [DOI] [PubMed] [Google Scholar]
- 47.Lew D J, Reed S I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu H, Styles C A, Fink G R. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 49.Madden K, Sheu Y-J, Baetz K, Andrews B, Snyder M. SBF cell cycle regulator as a target of the yeast SLT2 MAP kinase pathway. Science. 1997;275:1781–1784. doi: 10.1126/science.275.5307.1781. [DOI] [PubMed] [Google Scholar]
- 50.Madden K, Snyder M. Specification of sites of polarized growth in Saccharomyces cerevisiae and the influence of external factors on site selection. Mol Biol Cell. 1992;3:1025–1035. doi: 10.1091/mbc.3.9.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Madhani H D, Styles C A, Fink G R. MAP kinases with distinct inhibitory function impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 52.Marshall C J. MAP kinase kinase kinase, MAP kinase kinase, and MAP kinase. Curr Opin Genet Dev. 1994;4:82–89. doi: 10.1016/0959-437x(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 53.Mazzoni C, Zarzov P, Rambourg A, Mann C. The SLT2 (MPK1) MAP kinase homolog is involved in polarized cell growth in Saccharomyces cerevisiae. J Cell Biol. 1993;123:1821–1833. doi: 10.1083/jcb.123.6.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meluh P B, Rose M D. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell. 1990;60:1029–1941. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- 55.Mirzayan C, Copeland C, Snyder M. The NUF1 gene encodes a coiled-coil related protein that is a potential component of the yeast nucleoskeleton. J Cell Biol. 1992;116:1319–1332. doi: 10.1083/jcb.116.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mooseker M S. Organization, chemistry, and assembly of the cytoskeletal apparatus of the intestinal brush border. Annu Rev Cell Biol. 1985;1:209–241. doi: 10.1146/annurev.cb.01.110185.001233. [DOI] [PubMed] [Google Scholar]
- 57.Mosch H-U, Fink G R. Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics. 1997;145:671–684. doi: 10.1093/genetics/145.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mulholland J, Preuss D, Moon A, Wong A, Drubin D, Botstein D. Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J Cell Biol. 1994;125:381–391. doi: 10.1083/jcb.125.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neiman A M, Herskowitz I. Reconstitution of a yeast protein kinase cascade in vitro: activation of the yeast MEK homologue STE7 by STE11. Proc Natl Acad Sci USA. 1994;91:3398–3402. doi: 10.1073/pnas.91.8.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Novick P, Botstein D. Phenotypic analysis of temperature-sensitive yeast actin mutants. Cell. 1985;40:405–416. doi: 10.1016/0092-8674(85)90154-0. [DOI] [PubMed] [Google Scholar]
- 61.Ohya Y, Miyamoto S, Ohsumi Y, Anraku Y. Calcium-sensitive cls4 mutant of Saccharomyces cerevisiae with a defect in bud formation. J Bacteriol. 1986;165:28–33. doi: 10.1128/jb.165.1.28-33.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Page B D, Satterwhite L L, Rose M D, Snyder M. Localization of the KAR3 kinesin heavy chain-like protein requires the CIK1 interacting protein. J Cell Biol. 1994;124:507–519. doi: 10.1083/jcb.124.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Page B D, Snyder M. CIK1: a developmentally regulated spindle pole body-associated protein important for microtubule functions in Saccharomyces cerevisiae. Genes Dev. 1992;6:1414–1429. doi: 10.1101/gad.6.8.1414. [DOI] [PubMed] [Google Scholar]
- 64.Peter M, Neiman A M, Park H-O, van Lohuizen M, Herskowitz I. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO. 1996;15:7046–7059. [PMC free article] [PubMed] [Google Scholar]
- 65.Peterson J, Zheng Y, Bender L, Myers A, Cerione R, Bender A. Interactions between the bud emergence proteins Bem1p and Bem2p and Rho-type GTPases in yeast. J Cell Biol. 1994;127:1395–1406. doi: 10.1083/jcb.127.5.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 67.Pringle J, Bi E, Harkins H, Zahner J, Devirgilio C, Chant J, Corado K, Fares H. Establishment of cell polarity in yeast. Cold Spring Harbor Symp Quant Biol. 1995;60:729–744. doi: 10.1101/sqb.1995.060.01.079. [DOI] [PubMed] [Google Scholar]
- 67a.Printen J A, Sprague G F., Jr Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics. 1994;138:609–619. doi: 10.1093/genetics/138.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Read E B, Okamura H H, Drubin D G. Actin- and tubulin-dependent functions during Saccharomyces cerevisiae mating projection formation. Mol Biol Cell. 1992;3:429–444. doi: 10.1091/mbc.3.4.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roberts R, Fink G R. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 70.Roemer T, Madden K, Chang J, Snyder M. Selection of axial growth sites in yeast requires Axl2p, a novel plasma membrane glycoprotein. Genes Dev. 1996;10:777–793. doi: 10.1101/gad.10.7.777. [DOI] [PubMed] [Google Scholar]
- 71.Roemer T, Vallier L, Sheu Y-J, Snyder M. The Spa2-related protein, Sph1, is important for polarized growth in yeast. J Cell Sci. 1998;111:479–494. doi: 10.1242/jcs.111.4.479. [DOI] [PubMed] [Google Scholar]
- 72.Roemer T, Vallier L, Snyder M. Selection of polarized growth sites in yeast. Trends Cell Biol. 1996;6:434–441. doi: 10.1016/s0962-8924(96)10039-8. [DOI] [PubMed] [Google Scholar]
- 73.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 74.Santos B, Snyder M. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J Cell Biol. 1997;136:95–110. doi: 10.1083/jcb.136.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scherr G H, Weaver R H. The dimorphism phenomenon in yeasts. Bacteriol Rev. 1953;17:51–92. doi: 10.1128/br.17.1.51-92.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schneider B L, Seufert W, Steiner B, Yang Q H, Futcher A B. Use of PCR epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- 77.Sherman F, Fink G, Hicks J. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 78.Sikorski R, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sloat B, Pringle J. A mutant of yeast defective in cellular morphogenesis. Science. 1978;200:1171–1173. doi: 10.1126/science.349694. [DOI] [PubMed] [Google Scholar]
- 80.Snyder M. The SPA2 protein of yeast localizes to sites of cell growth. J Cell Biol. 1989;108:1419–1429. doi: 10.1083/jcb.108.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Snyder M, Gehrung S, Page B D. Studies concerning the temporal and genetic control of cell polarity in Saccharomyces cerevisiae. J Cell Biol. 1991;114:515–532. doi: 10.1083/jcb.114.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81a.Sobel, S., S. McPherson, and M. Snyder. Unpublished data.
- 82.Sprague G F, Thorner J. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. pp. 657–744. [Google Scholar]
- 83.Stevenson B J, Rhodes N, Errede B, Sprague G F. Constitutive mutants of the protein kinase STE11 activate the yeast pheromone response pathway in the absence of G protein. Genes Dev. 1992;6:1293–1304. doi: 10.1101/gad.6.7.1293. [DOI] [PubMed] [Google Scholar]
- 84.Torres L, Martin H, Garcia-Saez M I, Arroyo J, Molina M, Sanchez M, Nombela C. A protein kinase gene complements the lytic phenotype of Saccharomyces cerevisiae lyt2 mutants. Mol Microbiol. 1991;5:2845–2854. doi: 10.1111/j.1365-2958.1991.tb01993.x. [DOI] [PubMed] [Google Scholar]
- 85.Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- 86.Valtz N, Herskowitz I. Pea2 protein of yeast is localized to sites of polarized growth and is required for efficient mating and bipolar budding. J Cell Biol. 1996;135:725–739. doi: 10.1083/jcb.135.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Valtz N, Peter M, Herskowitz I. FAR1 is required for oriented polarization of yeast cells in response to mating pheromones. J Cell Biol. 1995;131:863–873. doi: 10.1083/jcb.131.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wertman K F, Drubin D G, Botstein D. Systematic mutational analysis of the yeast ACT1 gene. Genetics. 1992;132:337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xie K, Lambie E, Snyder M. Nuclear dot antigens may specify transcriptional domains in the nucleus. Mol Cell Biol. 1993;13:6170–6179. doi: 10.1128/mcb.13.10.6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang C H, Lambie E J, Snyder M. NuMA: an unusually large coiled-coil protein in the mammalian nucleus. J Cell Biol. 1992;116:1303–1317. doi: 10.1083/jcb.116.6.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang S, Ayscough K R, Drubin D G. A role for the actin cytoskeleton of Saccharomyces cerevisiae in bipolar bud-site selection. J Cell Biol. 1997;136:111–123. doi: 10.1083/jcb.136.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yorihuzi T, Ohsumi Y. Saccharomyces cerevisiae MATα mutant cells defective in pointed projection formation in response to α-factor at high concentrations. Yeast. 1994;10:579–594. doi: 10.1002/yea.320100503. [DOI] [PubMed] [Google Scholar]
- 93.Zahner J E, Harkins H A, Pringle J R. Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1857–1870. doi: 10.1128/mcb.16.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zarzov P, Mazzoni C, Mann C. The SLT2 (MPK1) MAP kinase is activated during periods of polarized cell growth in yeast. EMBO. 1996;15:83–91. [PMC free article] [PubMed] [Google Scholar]
- 95.Zhou Z, Gartner A, Cade R, Ammerer G, Errede B. Pheromone-induced signal transduction in Saccharomyces cerevisiae requires the sequential function of three protein kinases. Mol Cell Biol. 1993;13:2069–2080. doi: 10.1128/mcb.13.4.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]