Abstract
To investigate the effects of in vivo genomic DNA double-strand breaks on the efficiency and mechanisms of gene targeting in mouse embryonic stem cells, we have used a series of insertion and replacement vectors carrying two, one, or no genomic sites for the rare-cutting endonuclease I-SceI. These vectors were introduced into the hypoxanthine phosphoribosyltransferase (hprt) gene to produce substrates for gene-targeting (plasmid-to-chromosome) or intrachromosomal (direct repeat) homologous recombination. Recombination at the hprt locus is markedly increased following transfection with an I-SceI expression plasmid and a homologous donor plasmid (if needed). The frequency of gene targeting in clones with an I-SceI site attains a value of 1%, 5,000-fold higher than that in clones with no I-SceI site. The use of silent restriction site polymorphisms indicates that the frequencies with which donor plasmid sequences replace the target chromosomal sequences decrease with distance from the genomic break site. The frequency of intrachromosomal recombination reaches a value of 3.1%, 120-fold higher than background spontaneous recombination. Because palindromic insertions were used as polymorphic markers, a significant number of recombinants exhibit distinct genotypic sectoring among daughter cells from a single clone, suggesting the existence of heteroduplex DNA in the original recombination product.
The ability to generate transgenic animals with predictable genomic alterations has opened the way for analyzing the physiological consequences of these changes in the whole animal. A significant obstacle to obtaining transgenic animals with targeted modifications of their genomes is the fact that homologous recombination in mouse embryonic stem (ES) cells is relatively infrequent (10−5 to 10−8 events per transfected cell) (8, 15, 24). Two complementary strategies have been used to circumvent this limitation: one relies on the use of positive selectable markers in the targeting DNA to identify transformants (18, 37, 38); the second relies on positive-negative selection strategies, i.e., selection strategies that eliminate cells that have undergone nontargeted events (24). Such positive-negative selection procedures have been adopted to eliminate the nonhomologous events, which are far more frequent (perhaps 10- to 105-fold) than the desired homologous recombination (23, 37, 38, 42). In each instance, however, putative targeted transformants must be screened by restriction analysis to confirm that the desired change has been made.
Rather than focusing on improved means for selection of the desired events, we have sought to overcome the difficulties stemming from the low yield of accurately targeted events by developing a way to increase the frequency of homologous targeted events. Double-strand breaks (DSBs) made by rare-cutting endonucleases have been shown to stimulate mitotic homologous recombination in yeast (reviewed in reference 13), plant (29), and mammalian cells (4, 6, 17, 33, 35). Recombinational repair of DSBs relies on the ability of an intact double-stranded DNA which is homologous to and spans the DSB site to contribute its sequence information for repairing the break (39). Relying on this fact, we have expanded an approach to facilitate the introduction of any desired modifications at a chosen chromosomal site (6, 34, 36). Our approach relies first on introducing an endonuclease cleavage site into a desired genomic locus to create a target for generating a DSB; subsequently, transient expression of the corresponding endonuclease produces a DSB which is repaired by homologous recombination.
To explore the feasibility of such an approach, the hypoxanthine phosphoribosyltransferase (hprt) gene was chosen as a model genomic target because it is X linked and therefore exists as a single copy gene in XY ES cells. Additionally, convenient selections are available for loss of hprt function (resistance to the toxic base analog 6-thioguanine) and its functional restoration (resistance to aminopterin in the presence of hypoxanthine and thymidine). Our experiments involved creating two classes of ES cell clones, each of which contained a modification of the hprt gene. In one class, the wild-type hprt genomic segment containing exons 2 and 3 and the introns between and flanking them was replaced by a homologous segment containing an expressible neo gene flanked by one or more endonuclease cleavage sites. The other class was modified by an insertion which created a duplication of the same genomic segment with the expressible neo gene and flanking endonuclease cleavage sites between the duplicate regions. In each case, the endonuclease cleavage site was the 18-bp nonpalindromic sequence that is cleaved by the I-SceI endonuclease from Saccharomyces cerevisiae mitochondria (7). The targeted genomic locus was modified in such a way that plasmid-mediated repair of the DSB, as well as DSB-promoted repair of the tandem duplication, could occur upon the introduction of a plasmid that expresses the I-SceI endonuclease (33). In the first instance, the chromosomal sequences carried by a repair template plasmid contained subtle changes in the sequence that would enable us to determine if these modifications were transferred into the repaired chromosomal site. Where the genomic substrate was a tandem duplication flanking the I-SceI site(s), each member of the duplication contained different sequence alterations to be introduced into the repaired locus. In this way, the sequences surrounding the DSB can be mutagenized according to the sequence in the recombination partner.
Our results demonstrate that the frequency of gene targeting and intrachromosomal recombination is increased to 1 and 3%, respectively, by the endonuclease cleavages directed at the modified chromosomal locus. Moreover, recombinational repair of the chromosomal DSB is accompanied by sequence changes neighboring the cleavage site; these changes mirror the sequence in the repair template. These alterations occur with decreasing frequency as the distance from the break site increases.
An interesting feature of our experiments was the finding of clones with mixed genotypes for each of the sequences in which the original genomic target and the repair template differed. These arise from the transient formation of heteroduplex DNA which fails to be resolved by the mismatch repair system and relies on genetic segregation of the two genotypes following DNA replication.
MATERIALS AND METHODS
Plasmid construction.
The targeting DNA used in these experiments is a 5.5-kb DNA fragment of the hprt gene on the mouse X chromosome. The segment is bounded by the XmnI restriction site upstream of exon 2 and the EcoRI site downstream of exon 3 and was derived from plasmid pMP8 (30). This fragment had previously been modified to replace the 3′ EcoRI site with a SalI site and was placed in pBluescript in such a way as to eliminate the XmnI site, replacing it with BamHI and NotI sites in plasmid GPD351.
The repair template plasmid GPD384 was constructed by introducing the 5.5-kb hprt segment into a modified pBluescript vector from which the entire polylinker region had been removed, leaving only BamHI and SalI sites. Self-annealing linker oligonucleotides were ligated into five sites distributed along the hprt DNA segment, destroying the original sites and replacing them with polymorphic marker sites. Site 1 is a BglII site changed to an EcoRI site with the oligonucleotide GATCCGAATTCG. Site 2 is an EcoRV site changed to a PstI site with the oligonucleotide GCATCCTGCAGGATGC. Site 3 is an XbaI site changed to a BamHI site with the oligonucleotide CTAGCGGATCCG. Site 4 is an EcoRI site changed to an XmnI site with the oligonucleotide AATTGGTTCGAACC. Site 5 is an XbaI site changed to a PvuII site with the oligonucleotide CTAGGCAGCTGC.
Replacement plasmids were constructed by removing a 1-kb BglII-to-XhoI fragment (eliminating the splice acceptor site and the first 15 coding bases of hprt exon 3) and replacing it with the PGKneo expression cassette of the plasmid pPGKneopolyA, which was derived from pMC1neopolyA (41) by replacing the hybrid PyF441/tk promoter with the mouse pgk-1 promoter (1). The PGKneo expression cassette was flanked by 18-bp cut sites for endonuclease I-SceI (TAGGGATAACAGGGTAAT) in both forward and reverse directions to produce the configurations found in plasmids GPD371, GPD372, GPD373, GPD374, and GPD376.
Insertion plasmids were constructed by flanking the 5.5-kb hprt segment with the I-SceI site in either the forward or the reverse direction and placing the entire DNA segment (with polymorphic markers at sites 1, 2, 4, and 5) into the polylinker of plasmid PGKneopolyA to produce the configurations found in plasmids GPD377 through GPD381. Plasmid GPD383 is a derivative of GPD377 in which the EcoRI site immediately upstream of the PGKneo expression cassette has been destroyed and replaced with the I-SceI cut site.
The I-SceI endonuclease expression plasmid pPGK3XnlsI-SceI is a derivative of pCMV-I-SceI (33) and PGKneopolyA in which the coding and downstream regions of I-SceI replace the neo coding sequences under the control of the mouse pgk-1 promoter. In addition, pPGK3XnlsI-SceI contains an altered nuclear localization signal such that the N-terminal amino acids of the hybrid I-SceI protein were changed to MGSRSPKKKRKVPKKHAAPPKKKRKV, which contains three repeats (two complete and one partial) of the simian virus 40 large T-antigen nuclear localization signal (PKKKRKV).
ES cell culture.
All ES cell clones were derived from the CCE cell line (31). Cells were grown on gelatin-treated cell culture plates without feeder cells by using Dulbecco’s modified Eagle’s medium (DMEM) containing 20% heat-inactivated fetal calf serum, 0.1 mM 2-mercaptoethanol, 1× MEM nonessential amino acids and 1,000 U of recombinant murine leukemia inhibitory factor/ml (all from GIBCO/BRL).
Knockout ES clones were generated by transfecting cells with replacement plasmids that had been cut with BamHI and SalI or insertion plasmids that had been linearized with XbaI. ES cells (2 × 107) were suspended with 20 μg of cut plasmid DNA in 0.8 ml of HEPES-buffered solution (20 mM HEPES, 137 mM NaCl, 5 mM KCl, 0.7 mM Na2HPO4, 6 mM dextrose [pH 7.05]) and electroporated with a BTX electroporation unit set at 250 V, 1,100 μF, and 720 Ω. Selection was imposed by the addition of G418 medium (250 μg of active substance/ml) 48 h after transfection. On the 5th day after electroporation, 6-thioguanine (1.5 × 10−5 M) was added to the medium as well.
For recombination investigations, 5.0 × 104 ES cells containing the gene-targeting recombination substrates (plus 15 μg of pPGK3XnlsI-SceI and 15 μg of GPD384) or 2.5 × 104 ES cells containing the intrachromosomal-recombination substrates (plus 30 μg of pPGK3XnlsI-SceI) were transfected by using a calcium phosphate procedure (5) with the following modifications: the phosphate buffer was equilibrated to pH 7.15, and cells were cultured at 37°C and 5% CO2 after the calcium phosphate-DNA precipitate was added to the cells. An alternate transfection protocol was the electroporation of 3.0 × 107 cells with 30 μg of pPGK3XnlsI-SceI (plus 30 μg of circular or NotI-linearized GPD384 for cells being tested for gene targeting) in 0.8 ml of HEPES-buffered solution with a BTX electroporation unit set at 270 V, 1,100 μF, and 720 Ω. After electroporation, a small aliquot of cells was removed, diluted, and plated without selection until colonies formed in order to estimate the numbers of cells surviving the procedure. No such estimate was made in the case of calcium phosphate precipitation because cell survival was relatively unaffected by the transfection procedure. In both cases, selection was imposed by the addition of HAT medium (5 × 10−5 M hypoxanthine, 2 × 10−7 M aminopterin, and 8 × 10−6 M thymidine) 48 h after transfection. HATR colonies were picked (or stained and counted) after 14 days.
Southern blotting.
Total DNA extraction and subsequent Southern blotting were performed by standard methods. A DNA fragment to be used as a probe was prepared by performing PCR on the mouse hprt cDNA plasmid pHPT5 (ATCC 37424) (20) with the oligonucleotides GTGATTAGCGATGATGAACCAG and CCCCGTTGACTGATCATTACAGTA. This yielded a 312-bp PCR product containing the entire coding regions of hprt exons 2 and 3. This DNA fragment was then 32P labeled with the Stratagene PrimeIt II kit, with the exception that the original PCR primers were substituted for the normal random nonamers. Hybridization was done in RapidHyb (Amersham), and after washing, results were detected by using the BioMax film/screen system (Kodak).
Restriction fragment length polymorphisms in HATR clones were determined on Southern blots with the following restriction enzymes: EcoRI for analysis of sites 1 and 4, PstI for analysis of site 2, BamHI for analysis of site 3, and XbaI for analysis of sites 3 and 5. An EcoRI-PstI double digest was also performed to determine whether sites 2 and 4 were modified in a reciprocal fashion in daughter cells of clones suspected to have undergone single-strand annealing.
RESULTS
Production of gene-targeting and intrachromosomal-recombination substrates.
Using a portion of the wild-type hprt locus (Fig. 1A) to create several restriction site polymorphic markers in intronic regions around exons 2 and 3 of hprt (Fig. 1B), we have generated two distinct series of hprt knockout ES cell clones in which the I-SceI endonuclease site is integrated into the genome. The first is a series of replacement (Ω-type) clones (12, 15) in which a portion of the hprt gene is replaced by a segment containing a selectable neo marker, thereby eliminating hprt function (Fig. 2A). The second is a series of insertion (O-type) clones (12, 15) in which the integration of a neo selection plasmid bearing a portion of the hprt gene causes a duplication of that region, also knocking out hprt function (Fig. 3A).
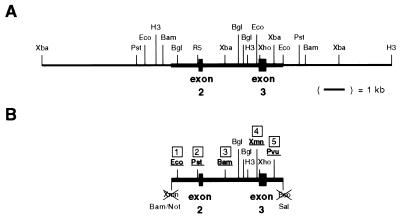
FIG. 1.
Genomic and plasmid restriction maps of hprt region. (A) The genomic region of hprt containing exons 2 and 3. (B) The 5.5-kb fragment of hprt used in these experiments (indicated by the heavy solid line in the genomic map) was modified at five sites to produce plasmid GPD384. Sites 1, 2, 3, 4, and 5 are approximately 300 bp, 1.3 kbp, 2.6 kbp, 4.2 kbp, and 5.0 kbp downstream of the XmnI site at the 5′ end of the 5.5-kb fragment, respectively. Restriction sites indicated are BamHI (Bam), BglII (Bgl), EcoRI (Eco), EcoRV (R5), HindIII (H3), NotI (Not), PstI (Pst), PvuII (Pvu), SalI (Sal), XbaI (Xba), XhoI (Xho), and XmnI (Xmn).
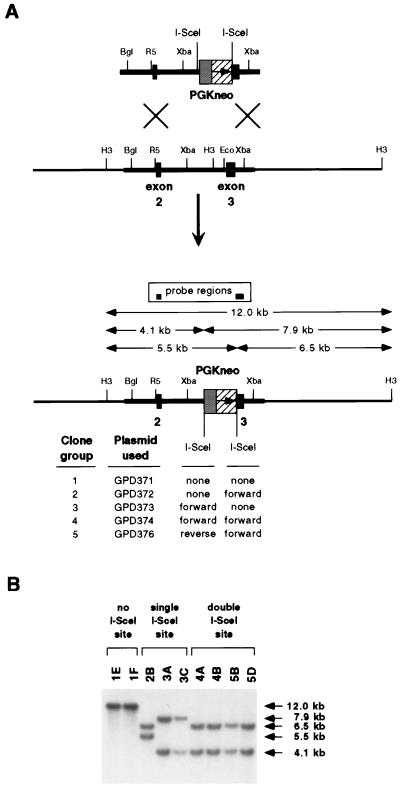
FIG. 2.
Production of replacement series of hprt knockout clones. (A) Diagram of homologous integration into the genome by a BamHI/SalI-linearized plasmid(s) carrying the PGKneo G418R marker to produce a set of hprt knockout clones varying only in the location, number, and orientation of the I-SceI-cut site(s). All possible HindIII/I-SceI digestion fragments, as detected by Southern blotting, are indicated by arrows. Restriction enzyme abbreviations are as explained for Fig. 1. (B) Southern blots of genomic DNA digested with HindIII and I-SceI from clones identified as having correct genomic integration structures were probed with hprt exons 2 and 3. Fragments containing only PGKneo or plasmid backbone-derived sequences are not detected.
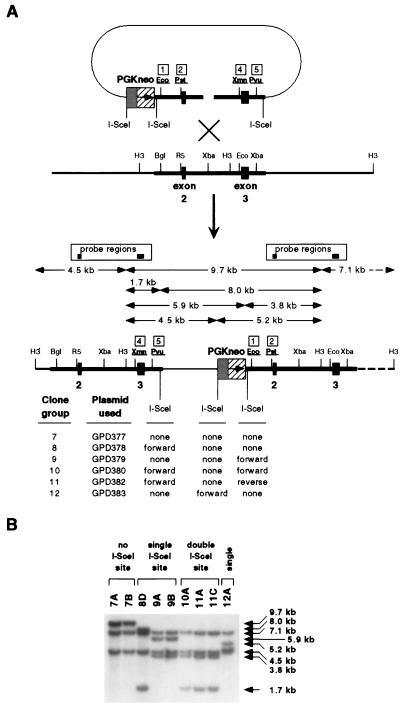
FIG. 3.
Production of insertion series of hprt knockout clones. (A) Diagram of homologous integration into the genome by an XbaI-linearized plasmid(s) carrying the PGKneo G418R marker to produce a set of hprt knockout clones varying only in the location, number, and orientation of the I-SceI cut site(s). All possible HindIII/I-SceI digestion fragments, as detected by Southern blotting, are indicated by arrows. Restriction enzyme abbreviations are as explained for Fig. 1. (B) Southern blots of genomic DNA digested with HindIII and I-SceI from clones identified as having correct genomic integration structures were probed with hprt exons 2 and 3. Fragments containing only PGKneo or plasmid backbone-derived sequences are not detected.
Colonies which were doubly resistant to G418 and 6-thioguanine were isolated as being putative hprt knockout clones. Genomic DNA from doubly resistant clones was examined by Southern blotting to confirm that the input DNA had integrated correctly into the genomic hprt locus and that the integrated I-SceI sites were intact. The restriction fragment patterns of DNA from representative clones show that clones with the expected genomic structures were isolated in all groups (Fig. 2B and 3B).
Cells from each replacement cell line were transfected with equal amounts of the I-SceI expression plasmid pPGK3XnlsI-SceI and GPD384, a repair template plasmid containing five polymorphic sites in the DNA that spans the deletion created in the replacement clones (Fig. 1B). The repair template was introduced either in circular or in linearized form. Cells from each insertion cell line were transfected with the I-SceI expression plasmid alone. Either of two methods of transfection was used: calcium phosphate precipitation or electroporation. Following transfection, recombinant HATR colonies were counted, and recombination frequencies were calculated for each clone analyzed.
Recombination frequency.
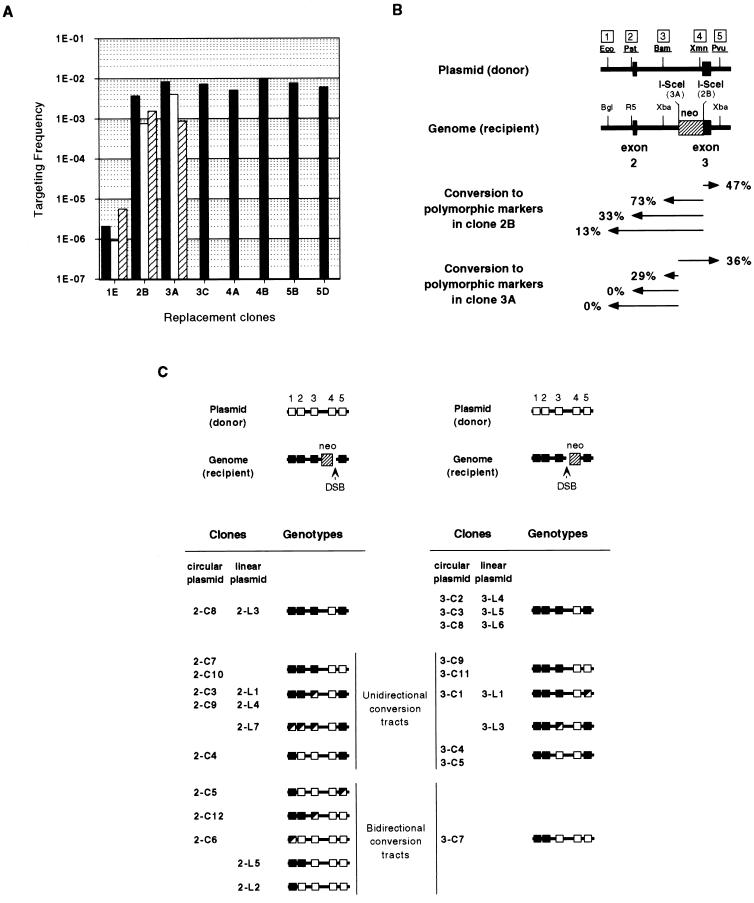
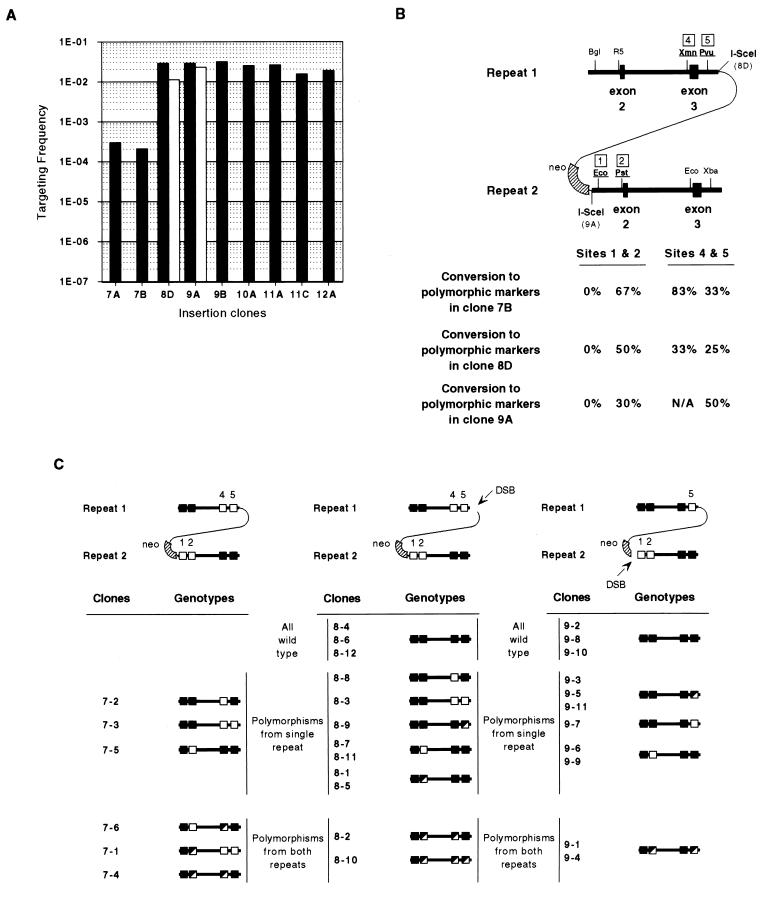
As expected, genomic DSBs stimulated recombination in all cases tested. Considering only calcium phosphate transfection with circular plasmids (where evaluation of all clones is possible) (Fig. 4A), gene targeting was increased from a base value of 2.0 × 10−6 events per transfected cell (clone 1E) to frequencies as high as 1.0 × 10−2 (clone 4B). Similarly, intrachromosomal recombination (Fig. 5A) was increased from an average spontaneous “pop-out” frequency of 2.5 × 10−4 events per transfected cell (clones 7A and 7B) to frequencies as high as 3.1 × 10−2 (clone 9B).
FIG. 4.
Targeting frequencies and genotypes of HATR recombinants from the replacement clones. (A) Targeting frequencies for replacement clones by calcium phosphate transfection with pPGK3XnlsI-SceI and circular GPD384 (solid bars), electroporation with pPGK3XnlsI-SceI and circular GPD384 (open bars), or electroporation with pPGK3XnlsI-SceI and linearized GPD384 (striped bars). (B) Overall gene conversion frequencies in HATR recombinant clones isolated after electroporation of clones 2B and 3A. Arrows indicate gene conversion from the DSBs to a particular site. (C) Summaries of individual genotype profiles for recombinants isolated from clones 2B and 3A. HATR clones are labeled to distinguish whether circular (C) or NotI-linearized (L) GPD384 was introduced as a donor substrate. Wild-type restriction sites, polymorphic restriction sites, and mixes of wild-type and polymorphic sites are represented by filled, open, and half-filled, half-open squares, respectively. Restriction enzyme abbreviations are as explained for Fig. 1.
FIG. 5.
Targeting frequencies and genotypes of HATR recombinants from the insertion clones. (A) Targeting frequencies for insertion clones by calcium phosphate transfection (solid bars) or electroporation (open bars) with pPGK3XnlsI-SceI. (B) Overall gene conversion frequencies in HATR recombinant clones isolated after spontaneous recombination in clone 7B or electroporation of clones 8D and 9A. The percent clones showing gene conversion at a particular site is indicated. (C) Summaries of individual genotype profiles for recombinants isolated from clones 7B, 8D, and 9A. Because clone 9A had not incorporated a polymorphic marker at site 4, no conclusions regarding gene conversion at that location could be drawn. Wild-type restriction sites, polymorphic restriction sites, and mixes of wild-type and polymorphic sites are represented by filled, open, and half-filled, half-open squares, respectively. Restriction enzyme abbreviations are as explained for Fig. 1.
The location of the DSB did not have a great effect on the recombination frequency in cells containing either replacement or insertion modifications. DSBs at either end of the selectable marker in the replacement clones 2B, 3A, and 3C stimulated recombination to similar extents. DSBs at the edge of the homology-nonhomology boundary in clones 8D, 9A, and 9B promoted recombination to frequencies comparable to those seen in clone 12A, where the DSB was located well within the nonhomology of the selection plasmid backbone. Although a double-site clone (4B) did exhibit the highest recombination frequency of any replacement clone, the range of frequencies in the other double-site clones (4A, 5B, 5D, 10A, 11A, and 11C) indicates that no particular advantage exists with two such DSB sites in either replacement or insertion clones. Once cell survival after electroporation was used to calculate corrected frequencies, calcium phosphate transfection of cells seemed to show no more than a two- to fivefold advantage over electroporation (Fig. 4A and 5A). Because cell survival seemed to be 50% or greater with the calcium phosphate procedure, no adjustments were made to those frequencies.
We, as well as others (18, 38), have observed that linearization of input DNA substrates normally increases the frequency of plasmid-to-chromosome gene targeting. This is supported by the electroporation data of clone 1E, where linearization of GPD384 consistently increased homologous recombination over that seen with circular GPD384. However, no clear trend could be observed with linear DNA substrates in cells where a DSB was introduced into the genome. The electroporation data reported for clones 2B and 3A indicate that linearization could depress or increase recombination frequencies in the presence of genomic DSBs, but no more than fivefold.
Chromosomal arrangement at the hprt locus in recombinants.
Although we presumed that the HATR colonies arose by homologous recombination and contained the normal arrangement of the hprt region, there remained the possibility that some of the “corrected” clones arose by anomalous events. To examine that possibility, genomic DNA from independent HATR colonies selected after electroporation (cell lines 2B, 3A, 8D, and 9A) or spontaneous recombination (cell line 7B) were isolated. Southern blots were prepared from DNA digested with a variety of diagnostic enzymes and probed with a labeled fragment comprising hprt exons 2 and 3. In fact, 32 of 35 clones isolated from the repair of replacement cell lines 2B and 3A had correct genomic structures. The three remaining clones were excluded from analysis because they contained structures which were not open to a simple interpretation. An additional clone had clearly undergone two separate recombination events and was also excluded. All clones isolated from the repair of insertion cell lines 7B, 8D, and 9A had correct genomic hprt structures.
The extent of conversion tracts.
The extent of gene conversion accompanying recombinational repair of the DSB was determined by restricting the recombinant clone’s DNA with enzymes that could distinguish between the sites in the wild-type genomic DNA and those on the repair plasmid or on the introduced arms of the tandem duplication. Both 2B and 3A recombinants showed only contiguous conversion tracts which extended unidirectionally or bidirectionally away from the break repaired during recombination (Fig. 4B). The major difference between the two groups was in the extent of the conversion tracts in the direction leading away from the DSB toward exon 2 of hprt. The gene conversion events in clone 2B recombinants extended as far as site 1, but clone 3A recombinants never converted beyond site 3. Recombinants isolated from clones 2B and 3A were the result of transfection with either circular or NotI-linearized DNA, but this made no substantial difference in the types of conversion tracts recovered.
Recombinants isolated from insertion clones 7B, 8D, and 9A displayed a variety of conversion genotypes, with no obvious directionality (Fig. 5B). Site 1 was never converted in any of the clones analyzed. The observed clones included simultaneous conversions at sites unique to each of the duplicated regions of hprt.
Recombination often generates heteroduplex products that yield clones containing both wild-type and converted genotypes.
Mixtures of both wild-type and polymorphic genotypes were observed in 11 of 34 gene-targeting recombinants and 13 of 29 intrachromosomal recombinants (Fig. 4C and 5C). In the gene-targeting recombinants, mixed genotypes were observed at the ends of conversion tracts but were never seen simultaneously at both termini in any clone examined. In the intrachromosomal recombinants, mixed conversions were observed at sites unique to each of the duplicated regions of hprt. Interestingly, there were no discernible differences in the spectrum of conversion patterns or in the frequency of colonies containing mixed genotypes between recombinants resulting from recombination events that occurred spontaneously and those resulting from homologous recombination induced by a DSB (Fig. 5C). The origin of colonies with mixed genotypes and the explanation of how they arise are considered in the discussion.
DISCUSSION
Our results confirm and extend observations that have been reported before (6, 21, 34, 35), namely, that expression of the I-SceI endonuclease in cells greatly stimulates homologous recombination between recombination substrates having one or more I-SceI sites. Recombinational repair of the DSBs in the replacement clones by an intact segment that overlaps the DSB approached 1% of the transfected cells, an increase of 5,000-fold over the spontaneous frequency. Moreover, with the insertion clones, where the recombination partners are tandem repeats separated by I-SceI sites, the fraction of homologous recombinants was about 3%, or 120-fold more than the spontaneous pop-out frequency. However, the efficiency of the recombination events may be even greater because they are expressed on the basis of the number of transfected cells, not all of which may have received and expressed the I-SceI endonuclease. Another issue which confounds these estimates is that we do not know if DSBs are invariably created in those cells that take up and express the I-SceI endonuclease. Quite possibly, however, competing nonhomologous processes may limit the efficiency of the homologous recombination events (17, 21, 35).
Cleavage of the I-SceI site leaves complementary single-strand ends, and thereby the likelihood of a very efficient rejoining process is increased. In the event that there are short repeated sequences in the neighborhood of the DSB, nuclease activity could also create complementary single strands which would favor intramolecular rejoining rather than intermolecular recombinational repair (34). Even in the absence of such sequence repeats, nonconservative end-to-end joining processes probably compete with the recombinational repair of DSBs. Recent studies have demonstrated an increased frequency of recombination following the creation of DSBs at the adenine phosphoribosyltransferase gene by I-SceI endonuclease and revealed that nonhomologous recombination at that site was also stimulated (35). Although we had no way to detect such nonhomologous end joining, the efficiency of homologous recombination would dictate that the ratio of nonhomologous to homologous events probably does not exceed 100. We expect that the relative frequencies of these events will vary in different cell types, at different genomic locations, and perhaps with the efficiency of DNA uptake.
Multiple I-SceI-cut sites at or near the recombination target did not influence the yield of recombinants over that obtained with a single site in either the gene-targeting or the intrachromosomal-recombination substrates, but we do not know if DSBs are created in every cell that takes up and expresses the I-SceI endonuclease. At the very least, two cut sites might increase the probability of a DSB in a region. When the two nonpalindromic cut sites are in opposite orientations relative to one another, simultaneous cuts at both sites leave ends with noncomplementary overhangs which cannot easily be joined together; this configuration might favor the initiation of recombinational repair. In the gene-targeting clones especially, simultaneous cutting at both sites would remove the entire selectable marker and remove large regions of nonhomology from either exposed end, favoring rejoining by the annealing of complementary single-strand ends. Although the modifications made to the nuclear localization signal of I-SceI increased the amount of nuclearly localized endonuclease activity (9), we have no way of verifying whether simultaneous cuts occurred in cells that had two I-SceI sites, so the potential advantage of such an arrangement has not yet been adequately explored.
The placement of a cut site made a considerable difference in the products produced by the plasmid-chromosome gene conversion events. The directional bias in terms of the lengths of the conversion tracts extending from the DSB toward exon 2 is obvious in comparisons between clones 2B and 3A (Fig. 4B). The large block of nonhomology represented by the PGKneo expression cassette may hinder the ability of an end to pair with its recombination partner or to be extended by DNA synthesis by using the homologous partner as a template if pairing has occurred. In this manner, cleavage of clone 2B’s I-SceI site would produce an end that could easily pair and be extended in the direction of exon 2, while this would not be possible with clone 3A. Such recombination-directed replication has been proposed to operate in a number of systems (for a review, see reference 19).
Although the frequency of recombination between two direct tandem repeats was strikingly increased over that of spontaneous events, the structures of the recombinants in each instance were remarkably similar, and the distribution of gene conversion events showed no particular trends, regardless of presence or placement of the cut site (Fig. 5B). Additionally, simultaneous conversion at polymorphic sites present in both repeats was found both in, spontaneous recombinants (e.g., 7B [3 of 6 examined]) and in clones arising by induction of DSBs (e.g., clones 8D and 9A [4 of 23 examined]). Because of the arrangement of the two groups of polymorphic sites, sites 1 or 2 and sites 4 or 5 in the parental clones, a simple crossover between the two repeats would not produce recombinants with polymorphisms from both groups.
One noteworthy observation in the recombination leading to removal of the duplication is that site 1 was never gene converted in any of the clones analyzed (Fig. 5C). This might be explained by the fact that site 1 is within 300 bp of the end of the homology shared between the two duplications in the insertion clones. If the formation of an extensive heteroduplex was responsible for the recombination events that were detected, then irrespective of the way in which the recombination is initiated, site 1 is always immediately adjacent either to a long unpaired tail of nonhomology (consisting of the intervening plasmid backbone) or to an exposed single-stranded end. The proximity of mismatched sequences to a point of resolution might prevent the succeeding events leading to gene conversion at that site.
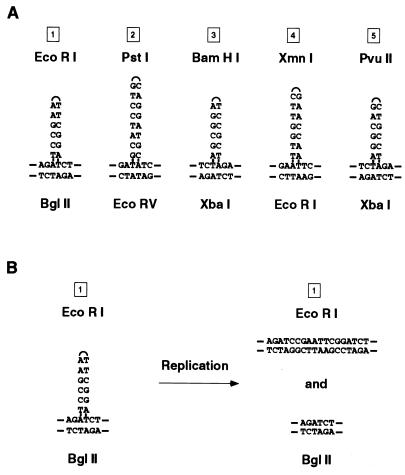
Our finding of mixed genotypes in many of the recombinant clones was initially puzzling, but because they were observed both for the infrequently derived spontaneous recombinants and for the frequently derived DSB-induced recombinants, it is unlikely that they were generated from two independent recombination events. A single induced “clone” did contain more than two genotypes (a total of four) and likely arose from two independent events (data not shown). How, then, can recombination give rise to colonies containing cells with mixed genotypes? In discussing the probable origin of such genotypically mixed clones, it should be noted that the restriction sites used to create the polymorphic markers used in these experiments are palindromic in nature and could form hairpin loops when base paired in heteroduplexes with their wild-type counterparts (Fig. 6A). Such palindromic insertions have been reported to avoid the surveillance of mismatch repair systems (28). Where such heteroduplexes have been studied in mammalian cells (3), they produce two distinct daughter genotypes after replication (Fig. 6B). Any colony derived from a cell in which a heteroduplex was formed as a product of the recombination would then appear to have a mixed genotype by maintaining two populations of progeny cells, one with the wild-type site and one with the polymorphism, as has been seen in genotypically sectored yeast colonies after postmeiotic segregation (11) or mitotic recombination (25, 32).
FIG. 6.
Preservation of heteroduplex at palindromic restriction site insertion polymorphisms. (A) Heteroduplex formed between palindromic insertion polymorphism and wild-type restriction markers at sites 1 through 5 with no mismatched base pairs. (B) Demonstration of mixed genotypes of daughter strands after replication through a heteroduplex at site 1.
The long conversion tracts (up to 3 kb) and mixed genotypes seen here with palindromic insertions differ from the findings of recent studies examining the incorporation of silent single-base mutations during repair of genomic DSBs (10, 40), in which mixed genotypes are rarely observed. Such variation in recombinational repair may be due to differences in length of overall homology between recombination substrates, the dissimilar repair of the two different types of mismatches when they are present in heteroduplex DNA, or the total number of mismatches that might be produced within a relatively short region of heteroduplex DNA. Indeed, increased divergence due to single-base mutations between recombination substrates has been shown to correlate with decreased numbers of recombination events (10).
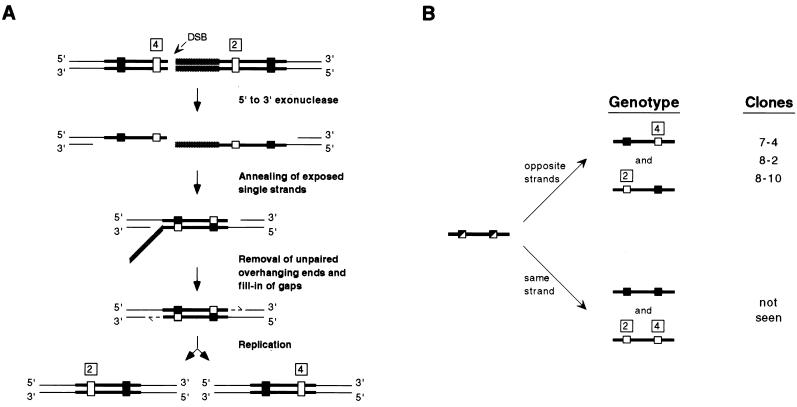
In the case of intrachromosomal-recombination events, formation of a Holliday junction (16) followed by extensive branch migration and resolution could produce such mixed genotypes, but one might expect that the initiation of such a one-sided strand invasion event by a free DNA end would result in an obvious recombination pattern difference between clones, depending upon the location of the endonuclease cut site. Because no conspicuous differential bias was observed, it is possible that most of the recombinants do not result from a crossover but from heteroduplex DNA formed by a single-strand annealing mechanism of recombination. The single-strand annealing model (22) predicts that when heteroduplex DNA is formed, polymorphisms from each direct repeat reside on strands opposite one another. This would generate a colony containing two different genotypes following replication and cell division (Fig. 7A). Such mixed colonies were obtained in clones 7-4, 8-2, and 8-10, in which polymorphic markers proved to reside on opposite strands (rather than on the same strand) through the analysis of subclones as well as original recombinants (Fig. 7B). Indeed, the placement of the I-SceI sites between the direct repeats (even at the homology-nonhomology junction between the insertion plasmid backbone and a direct repeat) should have favored recombination via single-strand annealing. However, the high frequency of putative single-strand annealing events, even when recombination events occurred spontaneously, is interesting as well.
FIG. 7.
Generation of trans heteroduplex in HATR clones containing mixed genotypes at sites 2 and 4. (A) The single-strand annealing model is diagrammed to explain the generation of simultaneous mixed genotypes at both sites 2 and 4 during intrachromosomal recombination. (B) The two possible outcomes of replicational resolution of unknown mixed genotypes at sites 2 and 4 are diagrammed. Resulting clones are identified on the basis of same-strand or opposite-strand orientation of polymorphisms. Wild-type restriction sites, polymorphic restriction sites, and mixes of wild-type and polymorphic sites are represented by filled, open, and half-filled, half-open rectangles, respectively.
In the case of gene-targeting events, the production of mixed genotypes is not as easily unraveled. The location of the DSB site does seem to influence the structure of the final gene-targeting recombination product, indicating that free DNA ends might initiate a strand invasion of homologous DNA present in the targeting plasmid. Whether these recombinants were the result of “one-sided” invasion events initiated by a single free DNA end (2, 34) or “two-sided” invasion events involving both free DNA ends (39) is not open to interpretation from these data, although both unidirectional and bidirectional conversion tracts were observed. Migration of a Holliday structure (16) formed during strand invasion and/or replicative extension (26, 39) followed by resolution of the junction would serve to trap exchanged DNA strands on opposite homologous-recombination partners in a heteroduplex. This trapped heteroduplex between wild-type and palindromic restriction site markers, like the single-strand annealing events during intrachromosomal recombination, would result in genotypically mixed cells arising from a single original cell that had undergone gene targeting.
The procedures we describe here, both gene-targeting and intrachromosomal recombination, are efficient in yielding homologous recombinants with sequence changes within several kilobases of a DSB. Both procedures require that the I-SceI site first be integrated by spontaneous homologous recombination into the genome, which entails isolation of that event before endonuclease-enhanced homologous recombination can be performed. However, this is what must be done in any procedure relying on spontaneous “hit and run” recombination (14, 43). To use this DSB-induced approach with a nonselectable autosomal locus, the DNA bearing the I-SceI site would carry markers for positive and negative selection (17). Although the homologous chromosome may be used as a repair template (27), targeted clones could be differentiated from such events by the use of marked gene-targeting vectors. Such screening could also be used to distinguish targeted clones from those clones arising from nonhomologous loss of a negative-selection marker.
The intrachromosomal-recombination procedure requires only an expression plasmid and has consistently generated a higher frequency of homologous recombination than the gene-targeting reaction. However, once an insertion vector has integrated to produce the cell line with tandem repeats surrounding the DSB site, the spectrum of modifications that can be generated by that line is limited by the number and type of modifications that were present in the transfected DNA. With the gene-targeting procedure, a single cell line can be used in conjunction with a variety of recombination plasmids (spanning a defined deletion of any size) to generate a large collection of modified cell lines. We foresee the value of such a capability where a single cell line carrying a specifically located endonuclease cleavage site can be mutagenized locally in virtually unlimited ways depending on the structure of the DNA used to repair the DSB.
ACKNOWLEDGMENTS
We thank Laura Hink Reid for her generous gift of pMP8. G.D. thanks Marianne Dieckmann, Eugeni Namsaraev, J. You, Sharon Hays, and other members of the Berg laboratory for many helpful discussions and comments.
This work was supported by a grant from the National Institutes of Health (GM13235) to P.B. and a grant from the National Science Foundation (MCB-9419507) to M.J.
REFERENCES
- 1.Adra C N, Boer P H, McBurney M W. Cloning and expression of the mouse pgk-1 gene and the nucleotide sequence of its promoter. Gene. 1987;60:65–74. doi: 10.1016/0378-1119(87)90214-9. [DOI] [PubMed] [Google Scholar]
- 2.Belmaaza A, Chartrand P. One-sided invasion events in homologous recombination at double-strand breaks. Mutat Res. 1994;314:199–208. doi: 10.1016/0921-8777(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 3.Bollag R J, Elwood D R, Tobin E D, Godwin A R, Liskay R M. Formation of heteroduplex DNA during mammalian intrachromosomal gene conversion. Mol Cell Biol. 1992;12:1546–1552. doi: 10.1128/mcb.12.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenneman M, Gimble F S, Wilson J H. Stimulation of intrachromosomal homologous recombination in human cells by electroporation with site-specific endonucleases. Proc Natl Acad Sci USA. 1996;93:3608–3612. doi: 10.1073/pnas.93.8.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choulika A, Perrin A, Dujon B, Nicolas J F. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colleaux L, D’Auriol L, Galibert F, Dujon B. Recognition and cleavage site of the intron-encoded omega transposase. Proc Natl Acad Sci USA. 1988;85:6022–6026. doi: 10.1073/pnas.85.16.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doetschman T, Gregg R G, Maeda N, Hooper M L, Melton D W, Thompson S, Smithies O. Targeted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature. 1987;330:576–578. doi: 10.1038/330576a0. [DOI] [PubMed] [Google Scholar]
- 9.Donoho G P. Ph.D. thesis. Stanford, Calif: Stanford University; 1996. [Google Scholar]
- 10.Elliott B, Richardson C, Winderbaum J, Nickoloff J A, Jasin M. Gene conversion tracts from double-strand break repair in mammalian cells. Mol Cell Biol. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esposito M S. Postmeiotic segregation in Saccharomyces. Mol Gen Genet. 1971;111:297–299. doi: 10.1007/BF00433113. [DOI] [PubMed] [Google Scholar]
- 12.Gregg R G, Smithies O. Targeted modification of human chromosomal genes. Cold Spring Harbor Symp Quant Biol. 1986;51:1093–1099. doi: 10.1101/sqb.1986.051.01.127. [DOI] [PubMed] [Google Scholar]
- 13.Haber J E. In vivo biochemistry: physical monitoring of recombination induced by site-specific endonucleases. Bioessays. 1995;17:609–620. doi: 10.1002/bies.950170707. [DOI] [PubMed] [Google Scholar]
- 14.Hasty P, Ramirez-Solis R, Krumlauf R, Bradley A. Introduction of a subtle mutation into the Hox-2.6 locus in embryonic stem cells. Nature. 1991;350:243–246. doi: 10.1038/350243a0. [DOI] [PubMed] [Google Scholar]
- 15.Hasty P, Rivera-Perez J, Chang C, Bradley A. Target frequency and integration pattern for insertion and replacement vectors in embryonic stem cells. Mol Cell Biol. 1991;11:4509–4517. doi: 10.1128/mcb.11.9.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holliday R. A mechanism for gene conversion in fungi. Genet Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- 17.Jasin M. Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet. 1996;12:224–228. doi: 10.1016/0168-9525(96)10019-6. [DOI] [PubMed] [Google Scholar]
- 18.Jasin M, Berg P. Homologous integration in mammalian cells without target gene selection. Genes Dev. 1988;2:1353–1363. doi: 10.1101/gad.2.11.1353. [DOI] [PubMed] [Google Scholar]
- 19.Kogoma T. Recombination by replication. Cell. 1996;85:625–627. doi: 10.1016/s0092-8674(00)81229-5. [DOI] [PubMed] [Google Scholar]
- 20.Konecki D S, Brennand J, Fuscoe J C, Caskey C T, Chinault A C. Hypoxanthine-guanine phosphoribosyltransferase genes of mouse and Chinese hamster: construction and sequence analysis of cDNA recombinants. Nucleic Acids Res. 1982;10:6763–6775. doi: 10.1093/nar/10.21.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang F, Romanienko P J, Weaver D T, Jeggo P A, Jasin M. Chromosomal double-strand break repair in Ku80-deficient cells. Proc Natl Acad Sci USA. 1996;93:8929–8933. doi: 10.1073/pnas.93.17.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin F L, Sperle K, Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984;4:1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin F L, Sperle K, Sternberg N. Recombination in mouse L cells between DNA introduced into cells and homologous chromosomal sequences. Proc Natl Acad Sci USA. 1985;82:1391–1395. doi: 10.1073/pnas.82.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mansour S L, Thomas K R, Capecchi M R. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- 25.McDonald J P, Rothstein R. Unrepaired heteroduplex DNA in Saccharomyces cerevisiae is decreased in RAD1 RAD52-independent recombination. Genetics. 1994;137:393–405. doi: 10.1093/genetics/137.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meselson M S, Radding C M. A general model for genetic recombination. Proc Natl Acad Sci USA. 1975;72:358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moynahan M E, Jasin M. Loss of heterozygosity induced by a chromosomal double-strand break. Proc Natl Acad Sci USA. 1997;94:8988–8993. doi: 10.1073/pnas.94.17.8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nag D K, White M A, Petes T D. Palindromic sequences in heteroduplex DNA inhibit mismatch repair in yeast. Nature. 1989;340:318–320. doi: 10.1038/340318a0. [DOI] [PubMed] [Google Scholar]
- 29.Puchta H, Dujon B, Hohn B. Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc Natl Acad Sci USA. 1996;93:5055–5060. doi: 10.1073/pnas.93.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reid L H, Shesely E G, Kim H S, Smithies O. Cotransformation and gene targeting in mouse embryonic stem cells. Mol Cell Biol. 1991;11:2769–2777. doi: 10.1128/mcb.11.5.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robertson E, Bradley A, Kuehn M, Evans M. Germ-line transmission of genes introduced into cultured pluripotential cells by retroviral vector. Nature. 1986;323:445–448. doi: 10.1038/323445a0. [DOI] [PubMed] [Google Scholar]
- 32.Ronne H, Rothstein R. Mitotic sectored colonies: evidence of heteroduplex DNA formation during direct repeat recombination. Proc Natl Acad Sci USA. 1988;85:2696–2700. doi: 10.1073/pnas.85.8.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci USA. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sargent R G, Brenneman M A, Wilson J H. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol Cell Biol. 1997;17:267–277. doi: 10.1128/mcb.17.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smih F, Rouet P, Romanienko P J, Jasin M. Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res. 1995;23:5012–5019. doi: 10.1093/nar/23.24.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith A J, Berg P. Homologous recombination between defective neo genes in mouse 3T6 cells. Cold Spring Harbor Symp Quant Biol. 1984;49:171–181. doi: 10.1101/sqb.1984.049.01.020. [DOI] [PubMed] [Google Scholar]
- 38.Smithies O, Koralewski M A, Song K Y, Kucherlapati R S. Homologous recombination with DNA introduced into mammalian cells. Cold Spring Harbor Symp Quant Biol. 1984;49:161–170. doi: 10.1101/sqb.1984.049.01.019. [DOI] [PubMed] [Google Scholar]
- 39.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 40.Taghian D G, Nickoloff J A. Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol Cell Biol. 1997;17:6386–6393. doi: 10.1128/mcb.17.11.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas K R, Capecchi M R. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 42.Thomas K R, Folger K R, Capecchi M R. High frequency targeting of genes to specific sites in the mammalian genome. Cell. 1986;44:419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- 43.Valancius V, Smithies O. Testing an “in-out” targeting procedure for making subtle genomic modifications in mouse embryonic stem cells. Mol Cell Biol. 1991;11:1402–1408. doi: 10.1128/mcb.11.3.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]