Abstract
Hibiscus syriacus L. is a renowned ornamental plant. We constructed 95 chloroplast genomes of H. syriacus L. cultivars using a short-read sequencing platform (Illumina) and a long-read sequencing platform (Oxford Nanopore Technology). The following genome assembly, we delineate quadripartite structures encompassing large single-copy, small single-copy, and inverted repeat (IRa and IRb) regions, from 160,231 bp to 161,041 bp. Our comprehensive analyses confirmed the presence of 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes in the pan-chloroplast genome, consistent with prior research on the H. syriacus chloroplast genome. Subsequent pangenome analysis unveiled widespread genome sequence conservation alongside unique cultivar-specific variant patterns consisting of 193 single-nucleotide polymorphisms and 61 insertions or deletions. The region containing intra-species variant patterns, as identified in this study, has the potential to develop accession-specific molecular markers, enhancing precision in cultivar classification. These findings are anticipated to drive advancements in breeding strategies, augment biodiversity, and unlock the agricultural potential inherent in H. syriacus.
Subject terms: Genetics, Plant sciences
Background & Summary
H. syriacus, commonly known as rose of Sharon, is a fast-growing deciduous shrub belonging to the Malvaceae family and is renowned for its diverse applications, including culinary, ornamental, and medicinal uses1. Its wide range of flower colors makes it an attractive choice for decorative landscaping2,3. In North American countries, it has gained immense popularity as a garden tree due to its versatile properties4. However, breeding H. syriacus presents significant challenges due to its self-incompatibility, resulting in most landraces being natural hybrids5. Consequently, there have been limited reports of breeding trials aimed at developing polyploidy plants4,6. In Korea, breeding advancements have been achieved through methods such as the propagation of naturally occurring mutants, inter-generic crossings, and the induction of mutations using gamma-ray irradiation6–9. The complexities of breeding H. syriacus highlights the importance of elucidating the phylogenetic relationships among its cultivars to establish a breeding system capable of generating F1 hybrids.
Given the challenges in H. syriacus breeding, the utilization of chloroplast genomes represents a strategic approach due to their unique features. These organelles are typically maternally inherited, except in some gymnosperms where inheritance is paternally directed. Chloroplast genomes contain non-recombinant sequences and are usually inherited in a uniparental manner, allowing for lineage tracing through the maternal line and minimizing uncertainties associated with biparentally inherited nuclear genomes10–13. Furthermore, the high conservation of the chloroplast genome, including gene repertories and structures, enables comparative analyses that offer clear insights into the evolutionary trajectories and phylogenetic relationships among cultivars14–16. Previous studies on Atractylodes species and Panax ginseng demonstrated that even with low divergence, unique polymorphic chloroplast-derived markers could be developed to distinguish inter- and intra-species differences, respectively11,17–22. This highlights the potential applications of chloroplast genomes in the development of highly species-specific molecular markers, even at the intra-species level, thereby overcoming challenges posed by minimal genetic divergence. Nevertheless, the majority of studies on Hibiscus chloroplast genomes have predominantly focused on the taxonomic level of genus, leaving in-depth intra-species studies relatively unexplored10,23–25. Given the breeding challenges of H. syriacus outlined earlier, comparative studies at the intra-species level are not only crucial but indispensable. Developing more molecular markers at the intra-species level is essential to gain unparalleled insights into the evolutionary trajectory and contribute to the precise taxonomic classification of H. syriacus26–28.
In this study, we generated 94 H. syriacus chloroplast genomes using a short-read sequencing platform (Illumina) and 1 genome using a long-read sequencing platform (Oxford Nanopore Technology). Subsequent pangenome analysis of these 95 H. syriacus chloroplast genomes revealed a high degree of conservation in the majority of genome sequences, while also identifies unique cultivar-specific variant patterns. A total of 193 single-nucleotide polymorphisms (SNPs) and 61 insertions or deletions (Indels) were identified, highlighting their potential applications as intra-species molecular markers29. The development of molecular markers utilizing these regions will play a pivotal role in achieving precise classification among H. syriacus cultivars and establishing refined breeding strategies. Moreover, these results will offer essential insights for species conservation, biodiversity enhancement, and the exploration of the agricultural and ornamental potentials of H. syriacus.
Methods
Plant materials and sequencing
H. syriacus cv. Gangneung was used for long-read-based chloroplast genome assembly30. A core collection of H. syriacus from the National Institute of Forest Science was utilized for short-read-based chloroplast genome assembly. Genomic DNA was extracted from fresh leaf tissues of H. syriacus plants using the standard cetyltrimethylammonium bromide method31.
The quantity and quality of genomic DNA were assessed using a Nanodrop spectrophotometer with a quality cut-off at an OD260/280 ratio of 1.8–2.0 and a Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Massachusetts, USA). Following quality assessment, the DNA was used to generate libraries with an average insert size of 550 bp. Paired-end sequencing was performed to obtain 150-bp sequences at both ends using an Illumina NovaSeq. 6000 platform (Illumina Inc., San Diego, CA, USA).
Genome assembly and annotation
For long-read assembly, the generated reads30 were aligned to a reference chloroplast sequence obtained from prior research32, using minimap2 (v2.22) with default parameters33. Reads with a mapping coverage exceeding 80 were extracted using Seqtk (https://github.com/lh3/seqtk) v1.3. These extracted reads were then assembled into a pseudo-molecule using Flye (v.2.9)34 and subsequently polished using NextPolish (v1.4)35 to correct base errors arising from noisy long reads.
For short-read assembly, Trimmomatic (v0.39)36 was used to trim adapters and eliminate low-quality sequences from the raw reads to enhance read quality. The trimmed reads were then aligned to the reference chloroplast genomes obtained from prior studies37–44, using the Burrows–Wheeler alignment (v0.7.17) tool45 (Table 1). The mapped reads were assembled using NOVOPlasty (v4.3.1)46, which employed a 39 k-mer and default RUBP sequences as seeds for chloroplast assembly47,48. The contigs generated by NOVOPlasty were ordered and merged into a single pseudo-molecule according to the reference chloroplast genome sequence.
Table 1.
Reference chloroplast genomes for mapping.
| Species | Accession ID |
|---|---|
| Gossypium arboretum | NC_016712.1 |
| Gossypium hirsutum | NC_007944.1 |
| Gossypium raimondii | NC_116668.1 |
| Hibiscus cannabinus | NC_045873.1 |
| Hibiscus rosa-sinensis | NC_042239.1 |
| Hibiscus syriacus | NC_026909.1 |
| Hibiscus taiwanensis | NC_054167.1 |
| Hibiscus trionum | NC_060636.1 |
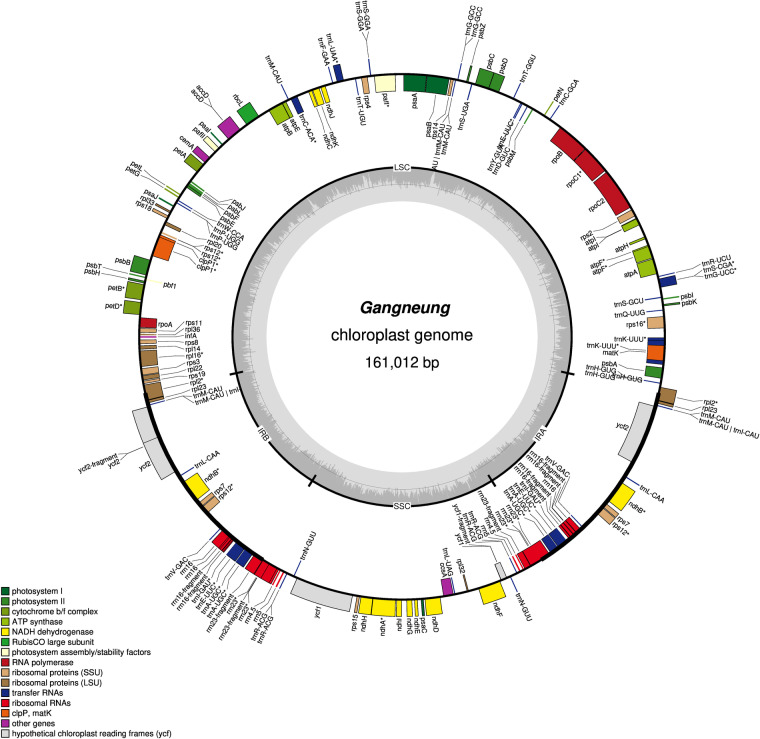
Genome annotation was performed using the GeSeq platform, which provides rapid and accurate annotation of organellar genomes49. We employed BLAT50, Chloë (v0.1.0), and HMMER51 to annotate coding sequences and rRNA, and ARAGORN (v1.2.38)52 and tRNAscan-SE (v2.0.7)53 to annotate tRNA. Annotation accuracy was validated against H. syriacus var. Baekdansim30, and any discrepancies were manually curated. The circular map representation of the chloroplast genome was generated using OGDRAW (v1.3.1)54 (Fig. 1).
Fig. 1.
Circular map of the chloroplast genome in H. syriacus var. Gangneung. The center of the plot displays the cultivar name and genome length. The inner grey circle represents the GC content proportion in each region, with the line representing 50%. Genes located outside the outer circle are transcribed counterclockwise, and those inside the circle are transcribed clockwise. Genes with different functional annotations are differentiated by color.
Chloroplast genome alignment and pan-chloroplast genome-graph construction
To validate the genome assembly, we employed the chloroplast genome of H. syriacus var. Gangneung, constructed using long-read sequencing, as a reference for multiple sequence alignment. Sequence alignment was performed using MAFFT55 with default parameters. Subsequently, pairwise alignments of the chloroplast genomes were generated using MUMmer456.
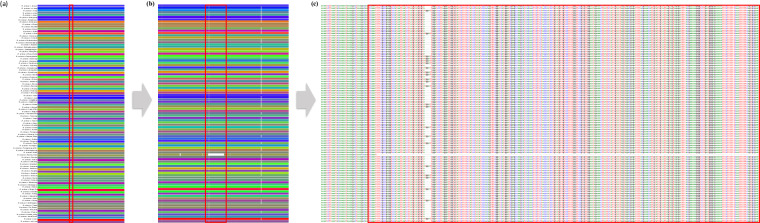
To construct a pan-chloroplast genome-graph encompassing 95 H. syriacus genomes, we utilized the Minigraph-Cactus Pangenome Pipeline (v2.6.8)57. The integration process involved the iterative addition of the remaining 94 genomes with the reference chloroplast genome. Precise base-level alignments were achieved with the Cactus-pangenome tool using the parameters “--giraffe --fa --bz --viz.” From this comprehensive graph, we employed the Cactus-graphmap (v2.6.8) tool to map the graph utilizing the default parameters. We identified a total of 193 SNPs and 61 Indels across the entire genomes, observations that offer significant potential for the future development of intra-species molecular markers29. Overall, H. syriacus cultivars exhibit similarity across all genomic regions. However, for H. syriacus var. Russian Violet, a notable divergence in similarity was observed in the regions spanning 59,000 bp to 62,000 bp (Fig. 2).
Fig. 2.
Pan-chloroplast genome-graph for 95 H. syriacus cultivars. (a) The pan-chloroplast genome-graph represents all 95 H. syriacus cultivars with the total chloroplast genome scale. (b) An enlarged view of the pan-chloroplast genome graph highlighting a region of the largest variation identified in H. syriacus var. Russian violet, indicated by red bars. (c) Multiple sequence alignment for the largest variation site among the 95 H. syriacus varieties.
Comparative genomic analysis in 95 H. syriacus chloroplast genomes
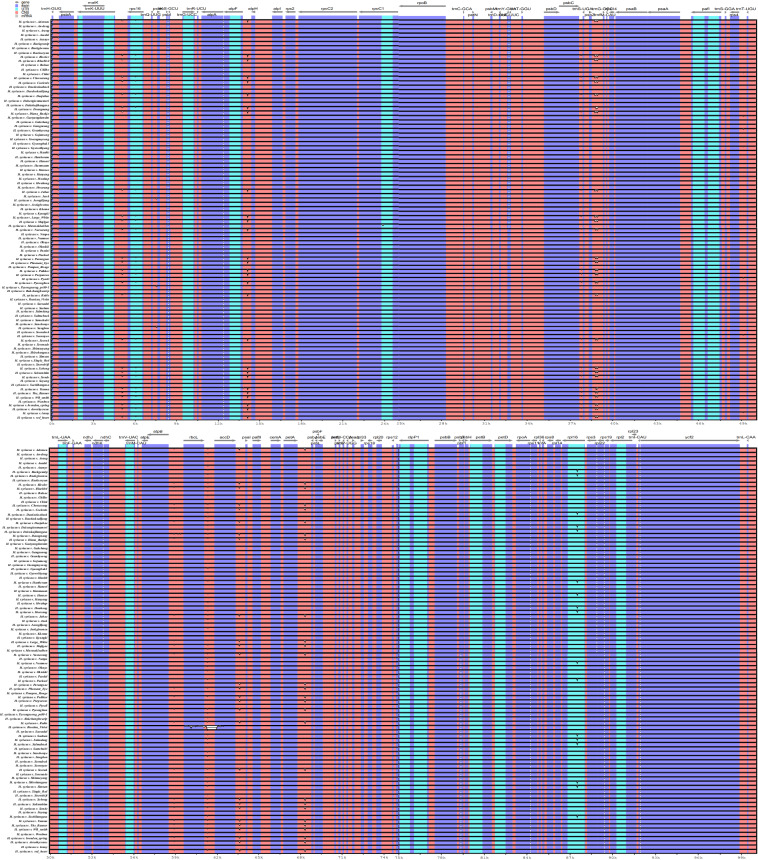
Structural similarity and gene distribution among the 95 chloroplast genomes were analyzed using mVISTA software in LAGAN mode with the default settings, with H. syriacus var. Baekdansim used as the reference58–61 (Fig. 3). This observation was consistent with the results from the pan-chloroplast genome analysis, where H. syriacus var. Russian Violet exhibits a significant deletion in specific regions.
Fig. 3.
The 95 H. syriacus accessions mVISTA map, with the Gangneung chloroplast genome as the reference. The vertical scale represents the percentage of identity, ranging from 50% to 100%. The horizontal axis corresponds to the base sequence region. Red indicates non-coding sequences(CNS), blue indicates the exons of protein-coding genes and light green indicates untranslated regions(UTR) including tRNA or rRNA.
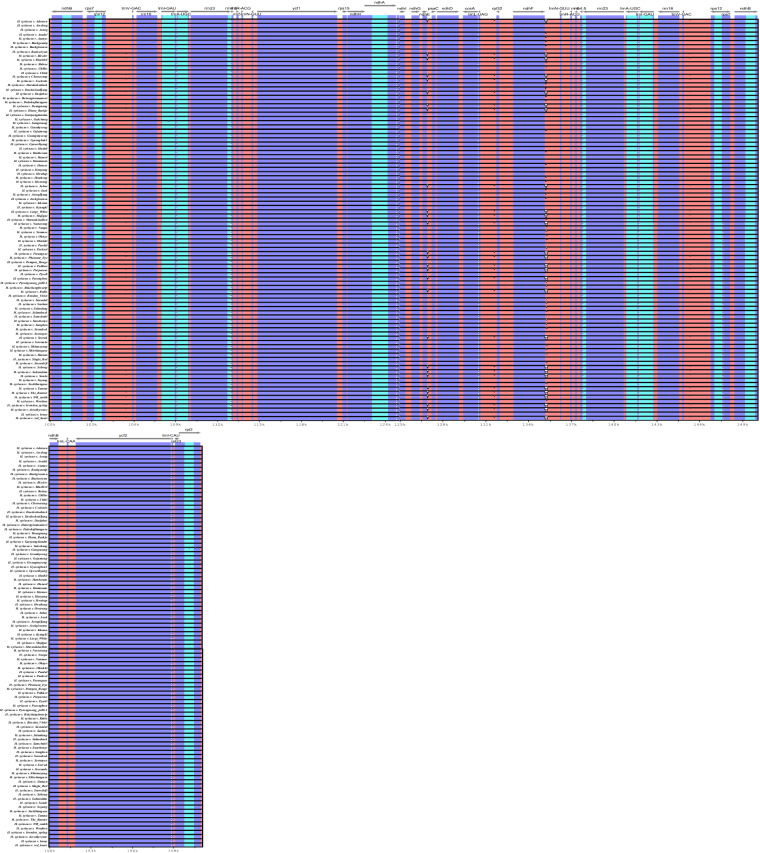
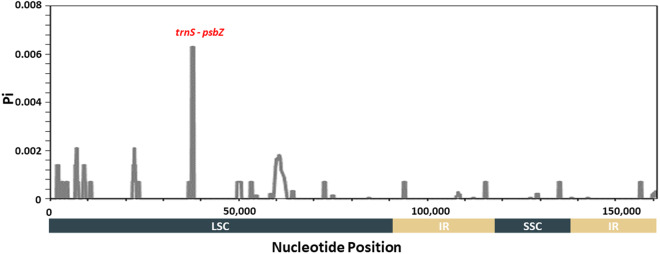
Hypervariable regions within the chloroplast genome of H. syriacus were identified using DnaSP version 6 software62. A total of 95 H. syriacus chloroplast genomes were aligned using MAFFT55 with default parameters. Nucleotide diversity was calculated through sliding window analysis, with the window size set at 600 bp with a step size of 100 bp22 (Fig. 4). The inverted repeat regions tend to be more conservative than the single copy regions. The highest nucleotide diversity was identified in the trnS-psbZ region. This region has the potential for use as a DNA barcode to facilitate distinction among the H. syriacus cultivars.
Fig. 4.
Nucleotide diversity in 95 H. syriacus chloroplast genomes. Sliding window analysis was performed with a window length of 600 bp and a step size of 100 bp. The x-axis represents nucleotide position, while the y-axis represents nucleotide diversity (Pi). Genes within the most hypervariable regions are highlighted in red.
Data Records
A total of 94 raw reads obtained through Illumina sequencing have been deposited in the NCBI Sequence Read Archive under the accession number SRP46454163. The assembled chloroplast genome sequences, accompanied by their corresponding gene annotations for the 94 cultivars have been submitted to NCBI GenBank64–157 and are detailed in Table S1. Additionally, H. syriacus var. Gangneung has been deposited in the NCBI GenBank with the accession number OR619829158.
Technical Validation
Evaluation of chloroplast genome assembly
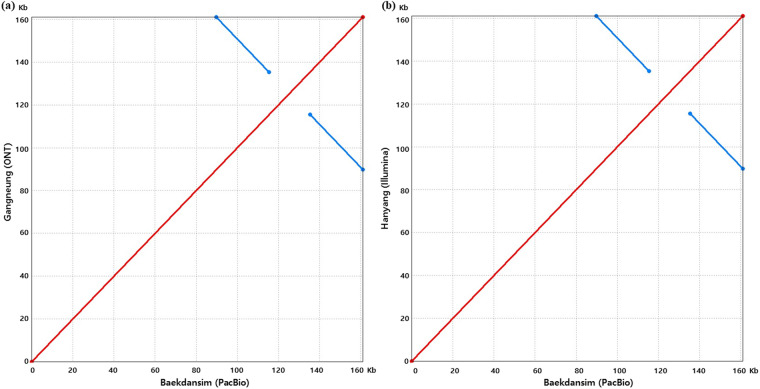
To evaluate the completeness of the chloroplast genome assembly, chloroplast reads were aligned to the chloroplast genome as described in the “Genome assembly and annotation” section. The lengths of the 95 assembled pseudo-molecules ranged from 160,231 bp to 161,041 bp, which is consistent with the observed chloroplast genome length in other members of the Malvaceae family23–27. Synteny analyses were conducted using MUMmer159 with the previously reported chloroplast genome of H. syriacus var. Baekdansim as the reference30. The dot plot revealed that the assembled genomes align cohesively with no major rearrangements observed (Fig. 5). Instead, the plot displayed inversions, represented by a blue line, corresponding to the chloroplast-specific inverted region.
Fig. 5.
Pairwise comparative analysis of chloroplast genomes in various H. syriacus cultivars with H. syriacus var. Baekdansim using MUMmer plots. (a) Comparison of chloroplast genomes constructed using ONT and PacBio long-read sequencing platforms. (b) Comparison of chloroplast genomes constructed using Illumina short-read and PacBio long-read sequencing platforms. The red lines represent collinear sequences and the blue lines represent inverted sequences.
Evaluation of gene annotation
The accuracy of the gene annotations was meticulously evaluated by comparing them to the H. syriacus var. Baekdansim61 chloroplast genome. Any discrepancies identified were refined through manual curation. In total, 113 distinct genes were identified, including 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes (Table 2).
Table 2.
Genes annotated in the chloroplast genome.
| Gene category | Gene function | Gene names |
|---|---|---|
| Photosynthesis-related genes | Photosystem I | psaA; psaB; psaC; psaI; psaJ |
| Photosystem II | psbA; psbB; psbC; psbD; psbE; psbF; psbH; psbI; psbJ; psbK; psbL; psbM; psbT; psbZ | |
| Cytochrome b/f complex | petA; petB; petD; petG; petL; petN | |
| ATP synthase | atpA; atpB; atpE; atpF; atpH; atpI | |
| NADH dehydrogenase | ndhA; ndhB*; ndhC; ndhD; ndhE; ndhF; ndhG; ndhH; ndhI; ndhJ; ndhK | |
| RubisCO large subunit | rbcL | |
| Photosystem assembly/stability Factors | pafI; pafII; pbf1 | |
| Self-replication-related genes | RNA polymerase | rpoA; rpoB; rpoC1*; rpoC2 |
| Ribosomal proteins (small subunit) | rps2; rps3; rps4; rps7*; rps8; rps11; rps12*; rps14; rps15; rps16; rps18; rps19 | |
| Ribosomal proteins (large subunit) | rpl2*; rpl14; rpl16; rpl20; rpl22; rpl23*; rpl32; rpl33; rpl36 | |
| tRNAs | trnA-UGC*; trnC-GCA; trnD-GUC; trnE-UUC; trnF-GAA; trnfM-CAU; trnG-GCC; trnG-UCC*; trnH-GUG; trnI-CAU*; trnI-GAU*; trnK-UUU; trnL-CAA*; trnL-UAA; trnL-UAG; trnM-CAU; trnN-GUU*; trnP-UGG; trnQ-UUG; trnR-ACG*; trnR-UCU; trnS-GCU; trnS-GGA; trnS-UGA; trnT-GGU; trnT-UGU; trnV-GAC*; trnV-UAC; trnW-CCA; trnY-GUA | |
| rRNAs | rrn5*; rrn4.5*; rrn16*; rrn23* | |
| Other genes | Maturase | matK |
| Envelope membrane protein | cemA | |
| Subunit of acetyl-CoA | accD | |
| C-type cytochrome synthesis gene | ccsA | |
| Protease | clpP1 | |
| Translational initiation factor | infA | |
| Unknown | Conserved hypothetical chloroplast reading frames | ycf1, ycf2* |
*Indicates duplicated genes.
The gene repertoires were consistent across all 95 cultivars, with the only observed differences being related to specific gene loci details. Our results indicate that the gene repertoire was congruent with annotations commonly observed within the Malvaceae family23,160,161, with minor variations detected in the pafI (ycf3), pafII (ycf4), and pbf1 (psbN) genes162,163.
Supplementary information
Acknowledgements
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2021R1I1A2044678) to Y.-M.K.
Author contributions
Y.-M.K. designed and organized the project. S.G., H.K., M.J., S.H. and G.Y. conducted the data analysis. S.G., H.K. and Y.-M.K. wrote the draft of the manuscript. All authors reviewed, revised, and approved the final version of the manuscript.
Code availability
All software used for data processing was implemented following the manual provided by the bioinformatic software cited in the method section. When specific parameters for the software were not detailed, the default settings were utilized.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sangjin Go, Hyunjin Koo.
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-024-03077-7.
References
- 1.Lee SJ, et al. An antioxidant lignan and other constituents from the root bark of Hibiscus syriacus. Planta Med. 1999;65:658–660. doi: 10.1055/s-2006-960841. [DOI] [PubMed] [Google Scholar]
- 2.Akpan GA. Cytogenetic characteristics and the breeding system in six Hibiscus species. Theor. Appl. Genet. 2000;100:315–318. doi: 10.1007/s001220050041. [DOI] [Google Scholar]
- 3.Zhang P, et al. Identification and quantitative analysis of anthocyanins composition and their stability from different strains of Hibiscus syriacus L. flowers. Ind. Crops Prod. 2022;177:114457. doi: 10.1016/j.indcrop.2021.114457. [DOI] [Google Scholar]
- 4.Contreras RN, Friddle M, Lattier JD. Relative fertility and ploidy levels of selected rose of sharon cultivars. SNA Research Conference. 2013;58:232–236. [Google Scholar]
- 5.Song, H.-S., Kim, J.-K., Lee, K.-U. & Lim, Y.-T. Studies on mutation breeding of Hibiscus syriacus. Report No. KAERI/RR-1669/96 (Korea Atomic Energy Research Institute, 1997).
- 6.Van Laere K, Van Huylenbroeck J, Van Bockstaele E. Breeding strategies to increase genetic variability within Hibiscus syriacus. Acta Hortic. 2006;714:75–82. doi: 10.17660/ActaHortic.2006.714.9. [DOI] [Google Scholar]
- 7.Park H-S, Cho Y-J, Chung H-G, Jang Y-S, Chung D-J. Variation in flower color among hybrids of jeoktanshim Hibiscus syriacus L. Korean Journal of Plant Resources. 2005;8:250–257. [Google Scholar]
- 8.Park YM, Lee S-W, Kwon S-H, Kwon HY. ‘Haeoreum’, A new Hibiscus syriacus cultivar for street trees. HortScience. 2020;55:1159–1161. doi: 10.21273/HORTSCI15035-20. [DOI] [Google Scholar]
- 9.Kim, S. H., Sim, S. J., Nam, J. I. & Kwon, H. Y. Korean Hibiscus Variety Catalog, (The National Institute of Forest Science (NIFoS) Press, 2014).
- 10.Abdullah, et al. Chloroplast genome of Hibiscus rosa-sinensis (Malvaceae): comparative analyses and identification of mutational hotspots. Genomics. 2020;112:581–591. doi: 10.1016/j.ygeno.2019.04.010. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, et al. Chloroplast genome variation and phylogenetic relationships of Atractylodes species. BMC Genomics. 2021;22:103. doi: 10.1186/s12864-021-07394-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong W, Xu C, Cheng T, Lin K, Zhou S. Sequencing angiosperm plastid genomes made easy: a complete set of universal primers and a case study on the phylogeny of saxifragales. Genome Biol. Evol. 2013;5:989–997. doi: 10.1093/gbe/evt063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park HS, et al. Inheritance of chloroplast and mitochondrial genomes in cucumber revealed by four reciprocal F(1) hybrid combinations. Sci. Rep. 2021;11:2506. doi: 10.1038/s41598-021-81988-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shinozaki K, et al. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. The EMBO Journal. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang F, Li W, Gao CW, Zhang D, Gao LZ. Deciphering tea tree chloroplast and mitochondrial genomes of Camellia sinensis var. assamica. Sci. Data. 2019;6:209. doi: 10.1038/s41597-019-0201-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y, et al. Comprehensive analysis of complete chloroplast genome and phylogenetic aspects of ten Ficus species. BMC Plant Biol. 2022;22:253. doi: 10.1186/s12870-022-03643-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hultquist SJ, et al. and nuclear DNA content variations among cultivars of switchgrass. Crop Sci. 1996;36:1049–1052. doi: 10.2135/cropsci1996.0011183X003600040039x. [DOI] [Google Scholar]
- 18.Li R, et al. Genetic diversity in Chinese sorghum landraces revealed by chloroplast simple sequence repeats. Genet. Resour. and Crop Evol. 2009;57:1–15. doi: 10.1007/s10722-009-9446-y. [DOI] [Google Scholar]
- 19.Cho KS, et al. Determination of cytoplasmic male sterile factors in onion plants (Allium cepa L.) using PCR-RFLP and SNP markers. Mol. Cells. 2006;21:411–417. doi: 10.1016/S1016-8478(23)12871-8. [DOI] [PubMed] [Google Scholar]
- 20.Nikiforova SV, Cavalieri D, Velasco R, Goremykin V. Phylogenetic analysis of 47 chloroplast genomes clarifies the contribution of wild species to the domesticated apple maternal line. Mol. Biol. Evol. 2013;30:1751–1760. doi: 10.1093/molbev/mst092. [DOI] [PubMed] [Google Scholar]
- 21.Kim K, et al. Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLoS One. 2015;10:e0117159. doi: 10.1371/journal.pone.0117159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joh HJ, et al. Authentication of Golden-Berry P. ginseng cultivar ‘Gumpoong’ from a landrace ‘Hwangsook’ based on pooling method using chloroplast-derived markers. Plant Breed. and Biotechnol. 2017;5:16–24. doi: 10.9787/PBB.2017.5.1.16. [DOI] [Google Scholar]
- 23.Cheng Y, Zhang L, Qi J, Zhang L. Complete chloroplast genome sequence of Hibiscus cannabinus and comparative analysis of the Malvaceae family. Front. Genet. 2020;11:227. doi: 10.3389/fgene.2020.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu XR, Zhou SD, Shi XQ. The complete chloroplast genome of Hibiscus Taiwanensis (Malvaceae) Mitochondrial DNA B: Resour. 2019;4:2533–2534. doi: 10.1080/23802359.2019.1640084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon S-H, Park Y, Jang YL, Kwon H-Y. The complete chloroplast genome sequence of Hibiscus sabdariffa (Malvaceae) Korean Journal of Plant Taxonomy. 2022;52:123–126. doi: 10.11110/kjpt.2022.52.2.123. [DOI] [Google Scholar]
- 26.Koo H, Shin AY, Hong S, Kim YM. The complete chloroplast genome of Hibiscus syriacus using long-read sequencing: comparative analysis to examine the evolution of the tribe Hibisceae. Front. Plant Sci. 2023;14:1111968. doi: 10.3389/fpls.2023.1111968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim YS, et al. The complete chloroplast genome sequence of Hibiscus syriacus L. ‘Mamonde’ (Malvaceae) Mitochondrial DNA Part B. 2019;4:558–559. doi: 10.1080/23802359.2018.1553526. [DOI] [PubMed] [Google Scholar]
- 28.Wang R, Park SY, Park SW, Puja AM, Kim Y-J. Development of a molecular marker based on chloroplast gene for specific identification of Korean Hibiscus (Hibiscus syriacus ‘Simbaek’) Appl. Biol. Chem. 2021;64:96. doi: 10.1186/s13765-021-00669-4. [DOI] [Google Scholar]
- 29.Kim Y-M. 2023. SNPs and Indels in pan-chloroplast genomes of 95 Hibiscus syriacus cultivars. figshare. [DOI]
- 30.Koo, H. et al. Two long read-based genome assembly and annotation of polyploidy woody plants, Hibiscus syriacus L. using PacBio and Nanopore platforms. Sci. Data10, 713 (2023). [DOI] [PMC free article] [PubMed]
- 31.Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 2006;1:2320–2325. doi: 10.1038/nprot.2006.384. [DOI] [PubMed] [Google Scholar]
- 32.Kim J-H, Lee H, Kwon H-Y, Kim S-H. 2015. Hibiscus syriacus chloroplast, complete genome. GenBank. KP688069.1
- 33.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolmogorov M, Yuan J, Lin Y, Pevzner PA. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019;37:540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- 35.Hu J, Fan J, Sun Z, Liu S. NextPolish: a fast and efficient genome polishing tool for long-read assembly. Bioinformatics. 2020;36:2253–2255. doi: 10.1093/bioinformatics/btz891. [DOI] [PubMed] [Google Scholar]
- 36.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Q, 2023. Gossypium arboreum chloroplast, complete genome. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/NC_016712.1/
- 38.Lee SB, 2023. Gossypium hirsutum chloroplast, complete genome. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/NC_007944.1/
- 39.Xu Q, 2023. Gossypium raimondii chloroplast, complete genome. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/NC_016668.1/
- 40.Cheng Y, Zhang L, Qi J, Zhang L. 2023. Hibiscus cannabinus chloroplast, complete genome. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/NC_045873.1
- 41.Abdullah, 2023. Gossypium raimondii chloroplast, complete genome. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/NC_042239.1
- 42.Kim J-H, Lee H, Kwon H-Y, Kim S-H. 2023. Hibiscus syriacus chloroplast, complete genome. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/NC_026909.1
- 43.Wang J-H, Moore MJ, Wang H, Zhu Z-X, Wang H-F. 2023. Hibiscus taiwanensis chloroplast, complete genome. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/NC_054167.1
- 44.Kwon S-H, Park YM, Jang YL, Kwon H-Y. 2023. Hibiscus trionum chloroplast, complete genome. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/NC_060636.1
- 45.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dierckxsens N, Mardulyn P, Smits G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017;45:e18. doi: 10.1093/nar/gkw955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin J-J, et al. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020;21:241. doi: 10.1186/s13059-020-02154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yun S, Kim H. Insight into the phylogenetic relationships and evolutionary history of pepper cultivars (Capsicum annuum L.) through comparative analyses of plastomes. Horticulturae. 2023;9:1092. doi: 10.3390/horticulturae9101092. [DOI] [Google Scholar]
- 49.Tillich M, et al. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45:W6–W11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–6. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chan PP, Lin BY, Mak AJ, Lowe TM. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021;49:9077–9096. doi: 10.1093/nar/gkab688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greiner S, Lehwark P, Bock R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019;47:W59–W64. doi: 10.1093/nar/gkz238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Bio. and Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marçais G, et al. MUMmer4: a fast and versatile genome alignment system. PLOS Comput. Biol. 2018;14:e1005944. doi: 10.1371/journal.pcbi.1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hickey, G. et al. Pangenome graph construction from genome alignments with Minigraph-Cactus. Nat. Biotechnol. (2023). [DOI] [PMC free article] [PubMed]
- 58.Brudno M, et al. Glocal alignment: finding rearrangements during alignment. Bioinformatics. 2023;19:i54–i62. doi: 10.1093/bioinformatics/btg1005. [DOI] [PubMed] [Google Scholar]
- 59.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song W, et al. Comparative analysis the complete chloroplast genomes of nine Musa species: genomic features, comparative analysis, and phylogenetic implications. Plant Sci. 2022;13:832884. doi: 10.3389/fpls.2022.832884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koo H, 2022. Hibiscus syriacus cultivar Baekdansim chloroplast, complete genome. GenBank. OP874596.1
- 62.Rozas J, et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 63.2023. NCBI Seqeunce Read Archive. SRP464541
- 64.Koo H, 2023. Hibiscus syriacus cultivar Adenseu plastid, complete genome. GenBank. OR350231.1
- 65.Koo H, 2023. Hibiscus syriacus cultivar An-dong plastid, complete genome. GenBank. OR164399.1
- 66.Koo H, 2023. Hibiscus syriacus cultivar Arang plastid, complete genome. GenBank. OR397843.1
- 67.Koo H, 2023. Hibiscus syriacus cultivar Asadal plastid, complete genome. GenBank. OR397844.1
- 68.Koo H, 2023. Hibiscus syriacus cultivar Asanyo plastid, complete genome. GenBank. OR397845.1
- 69.Koo H, 2023. Hibiscus syriacus cultivar Bicolor plastid, complete genome. GenBank. OR397846.1
- 70.Koo H, 2023. Hibiscus syriacus cultivar Bluebird plastid, complete genome. GenBank. OR397847.1
- 71.Koo H, 2023. Hibiscus syriacus cultivar Bulsae plastid, complete genome. GenBank. OR397848.1
- 72.Koo H, 2023. Hibiscus syriacus cultivar Chilbo plastid, complete genome. GenBank. OR397849.1
- 73.Koo H, 2023. Hibiscus syriacus cultivar Chini plastid, complete genome. GenBank. OR397850.1
- 74.Koo H, 2023. Hibiscus syriacus cultivar Chossarang plastid, complete genome. GenBank. OR397851.1
- 75.Koo H, 2023. Hibiscus syriacus cultivar Coelestis plastid, complete genome. GenBank. OR397852.1
- 76.Koo H, 2023. Hibiscus syriacus cultivar Daisengionmamori plastid, complete genome. GenBank. OR397853.1
- 77.Koo H, 2023. Hibiscus syriacus cultivar Daitokujihanagasa plastid, complete genome. GenBank. OR397854.1
- 78.Koo H, 2023. Hibiscus syriacus cultivar Gaeryangdansim plastid, complete genome. GenBank. OR397855.1
- 79.Koo H, 2023. Hibiscus syriacus cultivar Geunhyeong plastid, complete genome. GenBank. OR397856.1
- 80.Koo H, 2023. Hibiscus syriacus cultivar Gojumong plastid, complete genome. GenBank. OR397857.1
- 81.Koo H, 2023. Hibiscus syriacus cultivar Gwangmyeong plastid, complete genome. GenBank. OR397858.1
- 82.Koo H, 2023. Hibiscus syriacus cultivar Gyeongbuk1 plastid, complete genome. GenBank. OR397859.1
- 83.Koo H, 2023. Hibiscus syriacus cultivar Gyewolhyang plastid, complete genome. GenBank. OR397860.1
- 84.Koo H, 2023. Hibiscus syriacus cultivar Hanbit plastid, complete genome. GenBank. OR397861.1
- 85.Koo H, 2023. Hibiscus syriacus cultivar Hanboram plastid, complete genome. GenBank. OR397862.1
- 86.Koo H, 2023. Hibiscus syriacus cultivar Haneol plastid, complete genome. GenBank. OR397863.1
- 87.Koo H, 2023. Hibiscus syriacus cultivar Hanmaum plastid, complete genome. GenBank. OR397864.1
- 88.Koo H, 2023. Hibiscus syriacus cultivar Hanseo plastid, complete genome. GenBank. OR397865.1
- 89.Koo H, 2023. Hibiscus syriacus cultivar Hanyang plastid, complete genome. GenBank. OR397866.1
- 90.Koo H, 2023. Hibiscus syriacus cultivar Hwahap plastid, complete genome. GenBank. OR397867.1
- 91.Koo H, 2023. Hibiscus syriacus cultivar Hwahong plastid, complete genome. GenBank. OR397868.1
- 92.Koo H, 2023. Hibiscus syriacus cultivar Jaok plastid, complete genome. GenBank. OR397869.1
- 93.Koo H, 2023. Hibiscus syriacus cultivar Kkoma plastid, complete genome. GenBank. OR397870.1
- 94.Koo H, 2023. Hibiscus syriacus cultivar Kyungki plastid, complete genome. GenBank. OR397871.1
- 95.Koo H, 2023. Hibiscus syriacus cultivar Large_White plastid, complete genome. GenBank. OR397872.1
- 96.Koo H, 2023. Hibiscus syriacus cultivar Mujigae plastid, complete genome. GenBank. OR397873.1
- 97.Koo H, 2023. Hibiscus syriacus cultivar Murasakisaiben plastid, complete genome. GenBank. OR397874.1
- 98.Koo H, 2023. Hibiscus syriacus cultivar Naesarang plastid, complete genome. GenBank. OR397875.1
- 99.Koo H, 2023. Hibiscus syriacus cultivar Nanpa plastid, complete genome. GenBank. OR397876.1
- 100.Koo H, 2023. Hibiscus syriacus cultivar Nunmoe plastid, complete genome. GenBank. OR397877.1
- 101.Koo H, 2023. Hibiscus syriacus cultivar Oknyo plastid, complete genome. GenBank. OR397878.1
- 102.Koo H, 2023. Hibiscus syriacus cultivar Oktokki plastid, complete genome. GenBank. OR397879.1
- 103.Koo H, 2023. Hibiscus syriacus cultivar Paedal plastid, complete genome. GenBank. OR397880.1
- 104.Koo H, 2023. Hibiscus syriacus cultivar Paeksol plastid, complete genome. GenBank. OR397881.1
- 105.Koo H, 2023. Hibiscus syriacus cultivar Parangsae plastid, complete genome. GenBank. OR397882.1
- 106.Koo H, 2023. Hibiscus syriacus cultivar Pheasant_Eye plastid, complete genome. GenBank. OR397883.1
- 107.Koo H, 2023. Hibiscus syriacus cultivar Pompon_Rouge plastid, complete genome. GenBank. OR397884.1
- 108.Koo H, 2023. Hibiscus syriacus cultivar Pulkkot plastid, complete genome. GenBank. OR397885.1
- 109.Koo H, 2023. Hibiscus syriacus cultivar Purpureus plastid, complete genome. GenBank. OR397886.1
- 110.Koo H, 2023. Hibiscus syriacus cultivar Pyeli plastid, complete genome. GenBank. OR397887.1
- 111.Koo H, 2023. Hibiscus syriacus cultivar Pyeonghwa plastid, complete genome. GenBank. OR397888.1
- 112.Koo H, 2023. Hibiscus syriacus cultivar Rubis plastid, complete genome. GenBank. OR397889.1
- 113.Koo H, 2023. Hibiscus syriacus cultivar Russian_Violet plastid, complete genome. GenBank. OR397890.1
- 114.Koo H, 2023. Hibiscus syriacus cultivar Saeasadal plastid, complete genome. GenBank. OR397891.1
- 115.Koo H, 2023. Hibiscus syriacus cultivar Saehan plastid, complete genome. GenBank. OR397892.1
- 116.Koo H, 2023. Hibiscus syriacus cultivar Salmabaek plastid, complete genome. GenBank. OR397893.1
- 117.Koo H, 2023. Hibiscus syriacus cultivar Samchulri plastid, complete genome. GenBank. OR397894.1
- 118.Koo H, 2023. Hibiscus syriacus cultivar Sanchonye plastid, complete genome. GenBank. OR397895.1
- 119.Koo H, 2023. Hibiscus syriacus cultivar Seondeok plastid, complete genome. GenBank. OR397896.1
- 120.Koo H, 2023. Hibiscus syriacus cultivar Seonnyeo plastid, complete genome. GenBank. OR397897.1
- 121.Koo H, 2023. Hibiscus syriacus cultivar Seorak plastid, complete genome. GenBank. OR397898.1
- 122.Koo H, 2023. Hibiscus syriacus cultivar Serenade plastid, complete genome. GenBank. OR397899.1
- 123.Koo H, 2023. Hibiscus syriacus cultivar Shintaeyang plastid, complete genome. GenBank. OR397900.1
- 124.Koo H, 2023. Hibiscus syriacus cultivar Shirohanagasa plastid, complete genome. GenBank. OR397901.1
- 125.Koo H, 2023. Hibiscus syriacus cultivar Single_Red plastid, complete genome. GenBank. OR397902.1
- 126.Koo H, 2023. Hibiscus syriacus cultivar Snowdrift plastid, complete genome. GenBank. OR397903.1
- 127.Koo H, 2023. Hibiscus syriacus cultivar Sobong plastid, complete genome. GenBank. OR397904.1
- 128.Koo H, 2023. Hibiscus syriacus cultivar Soltanshim plastid, complete genome. GenBank. OR397905.1
- 129.Koo H, 2023. Hibiscus syriacus cultivar Sonde plastid, complete genome. GenBank. OR397906.1
- 130.Koo H, 2023. Hibiscus syriacus cultivar Soyang plastid, complete genome. GenBank. OR397907.1
- 131.Koo H, 2023. Hibiscus syriacus cultivar Suchihanagasa plastid, complete genome. GenBank. OR397908.1
- 132.Koo H, 2023. Hibiscus syriacus cultivar Tamna plastid, complete genome. GenBank. OR397909.1
- 133.Koo H, 2023. Hibiscus syriacus cultivar The_Banner plastid, complete genome. GenBank. OR397910.1
- 134.Koo H, 2023. Hibiscus syriacus cultivar Wonhwa plastid, complete genome. GenBank. OR397911.1
- 135.Koo H, 2023. Hibiscus syriacus cultivar Baekgeunip plastid, complete genome. GenBank. OR625140.1
- 136.Koo H, 2023. Hibiscus syriacus cultivar Baekgiwonsu plastid, complete genome. GenBank. OR625141.1
- 137.Koo H, 2023. Hibiscus syriacus cultivar Baeksoryun plastid, complete genome. GenBank. OR625142.1
- 138.Koo H, 2023. Hibiscus syriacus cultivar bredon_spring plastid, complete genome. GenBank. OR625143.1
- 139.Koo H, 2023. Hibiscus syriacus cultivar Daedeoksaback plastid, complete genome. GenBank. OR625144.1
- 140.Koo H, 2023. Hibiscus syriacus cultivar Daedeoksailjung plastid, complete genome. GenBank. OR625145.1
- 141.Koo H, 2023. Hibiscus syriacus cultivar Diana_Baekjo plastid, complete genome. GenBank. OR625148.1
- 142.Koo H, 2023. Hibiscus syriacus cultivar dorothycranc plastid, complete genome. GenBank. OR625149.1
- 143.Koo H, 2023. Hibiscus syriacus cultivar Gakchang plastid, complete genome. GenBank. OR625150.1
- 144.Koo H, 2023. Hibiscus syriacus cultivar Jeogiljung plastid, complete genome. GenBank. OR625153.1
- 145.Koo H, 2023. Hibiscus syriacus cultivar Jeokgiwonsu plastid, complete genome. GenBank. OR625154.1
- 146.Koo H, 2023. Hibiscus syriacus cultivar lenny plastid, complete genome. GenBank. OR625155.1
- 147.Koo H, 2023. Hibiscus syriacus cultivar Rakchanghwarip plastid, complete genome. GenBank. OR625157.1
- 148.Koo H, 2023. Hibiscus syriacus cultivar Sangbon plastid, complete genome. GenBank. OR625160.1
- 149.Koo H, 2023. Hibiscus syriacus cultivar WR_smith plastid, complete genome. GenBank. OR625161.1
- 150.Koo H, 2023. Hibiscus syriacus cultivar Hwarang plastid, complete genome. GenBank. OR625151.1
- 151.Koo H, 2023. Hibiscus syriacus cultivar Saimdang plastid, complete genome. GenBank. OR625159.1
- 152.Koo H, 2023. Hibiscus syriacus cultivar Daejabae plastid, complete genome. GenBank. OR625146.1
- 153.Koo H, 2023. Hibiscus syriacus cultivar Deungnang plastid, complete genome. GenBank. OR625147.1
- 154.Koo H, 2023. Hibiscus syriacus cultivar Jabae plastid, complete genome. GenBank. OR625152.1
- 155.Koo H, 2023. Hibiscus syriacus cultivar Pyeongseong_ps80-1 plastid, complete genome. GenBank. OR625156.1
- 156.Koo H, 2023. Hibiscus syriacus cultivar red_heart plastid, complete genome. GenBank. OR625158.1
- 157.Koo H, 2023. Hibiscus syriacus cultivar Simsan plastid, complete genome. GenBank. OR619828.1
- 158.Koo H, 2023. Hibiscus syriacus cultivar Gangneung plastid, complete genome. GenBank. OR619829.1
- 159.Delcher AL, Salzberg SL, Phillippy AM. Using MUMmer to identify similar regions in large sequence sets. Curr. Protoc. in Bioinform. 2003;00:10.3.1–10.3.18. doi: 10.1002/0471250953.bi1003s00. [DOI] [PubMed] [Google Scholar]
- 160.Abdullah, et al. Correlations among oligonucleotide repeats, nucleotide substitutions, and insertion–deletion mutations in chloroplast genomes of plant family Malvaceae. J. of Syst. and Evol. 2020;59:388–402. doi: 10.1111/jse.12585. [DOI] [Google Scholar]
- 161.Yu J, Jin W, Wang Y, Yuan Q. The complete chloroplast genome of a medicinal plant, Abutilon theophrasti Medik. (Malvaceae) Mitochondrial DNA B: Resour. 2020;5:3777–3778. doi: 10.1080/23802359.2020.1835582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 2011;76:273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zupok A, et al. A photosynthesis operon in the chloroplast genome drives speciation in evening primroses. The Plant Cell. 2021;33:2583–2601. doi: 10.1093/plcell/koab155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Kim Y-M. 2023. SNPs and Indels in pan-chloroplast genomes of 95 Hibiscus syriacus cultivars. figshare. [DOI]
- Kim J-H, Lee H, Kwon H-Y, Kim S-H. 2015. Hibiscus syriacus chloroplast, complete genome. GenBank. KP688069.1
- Xu Q, 2023. Gossypium arboreum chloroplast, complete genome. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/NC_016712.1/
- Lee SB, 2023. Gossypium hirsutum chloroplast, complete genome. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/NC_007944.1/
- Xu Q, 2023. Gossypium raimondii chloroplast, complete genome. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/NC_016668.1/
- Cheng Y, Zhang L, Qi J, Zhang L. 2023. Hibiscus cannabinus chloroplast, complete genome. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/NC_045873.1
- Abdullah, 2023. Gossypium raimondii chloroplast, complete genome. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/NC_042239.1
- Kim J-H, Lee H, Kwon H-Y, Kim S-H. 2023. Hibiscus syriacus chloroplast, complete genome. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/NC_026909.1
- Wang J-H, Moore MJ, Wang H, Zhu Z-X, Wang H-F. 2023. Hibiscus taiwanensis chloroplast, complete genome. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/NC_054167.1
- Kwon S-H, Park YM, Jang YL, Kwon H-Y. 2023. Hibiscus trionum chloroplast, complete genome. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/NC_060636.1
- Koo H, 2022. Hibiscus syriacus cultivar Baekdansim chloroplast, complete genome. GenBank. OP874596.1
- 2023. NCBI Seqeunce Read Archive. SRP464541
- Koo H, 2023. Hibiscus syriacus cultivar Adenseu plastid, complete genome. GenBank. OR350231.1
- Koo H, 2023. Hibiscus syriacus cultivar An-dong plastid, complete genome. GenBank. OR164399.1
- Koo H, 2023. Hibiscus syriacus cultivar Arang plastid, complete genome. GenBank. OR397843.1
- Koo H, 2023. Hibiscus syriacus cultivar Asadal plastid, complete genome. GenBank. OR397844.1
- Koo H, 2023. Hibiscus syriacus cultivar Asanyo plastid, complete genome. GenBank. OR397845.1
- Koo H, 2023. Hibiscus syriacus cultivar Bicolor plastid, complete genome. GenBank. OR397846.1
- Koo H, 2023. Hibiscus syriacus cultivar Bluebird plastid, complete genome. GenBank. OR397847.1
- Koo H, 2023. Hibiscus syriacus cultivar Bulsae plastid, complete genome. GenBank. OR397848.1
- Koo H, 2023. Hibiscus syriacus cultivar Chilbo plastid, complete genome. GenBank. OR397849.1
- Koo H, 2023. Hibiscus syriacus cultivar Chini plastid, complete genome. GenBank. OR397850.1
- Koo H, 2023. Hibiscus syriacus cultivar Chossarang plastid, complete genome. GenBank. OR397851.1
- Koo H, 2023. Hibiscus syriacus cultivar Coelestis plastid, complete genome. GenBank. OR397852.1
- Koo H, 2023. Hibiscus syriacus cultivar Daisengionmamori plastid, complete genome. GenBank. OR397853.1
- Koo H, 2023. Hibiscus syriacus cultivar Daitokujihanagasa plastid, complete genome. GenBank. OR397854.1
- Koo H, 2023. Hibiscus syriacus cultivar Gaeryangdansim plastid, complete genome. GenBank. OR397855.1
- Koo H, 2023. Hibiscus syriacus cultivar Geunhyeong plastid, complete genome. GenBank. OR397856.1
- Koo H, 2023. Hibiscus syriacus cultivar Gojumong plastid, complete genome. GenBank. OR397857.1
- Koo H, 2023. Hibiscus syriacus cultivar Gwangmyeong plastid, complete genome. GenBank. OR397858.1
- Koo H, 2023. Hibiscus syriacus cultivar Gyeongbuk1 plastid, complete genome. GenBank. OR397859.1
- Koo H, 2023. Hibiscus syriacus cultivar Gyewolhyang plastid, complete genome. GenBank. OR397860.1
- Koo H, 2023. Hibiscus syriacus cultivar Hanbit plastid, complete genome. GenBank. OR397861.1
- Koo H, 2023. Hibiscus syriacus cultivar Hanboram plastid, complete genome. GenBank. OR397862.1
- Koo H, 2023. Hibiscus syriacus cultivar Haneol plastid, complete genome. GenBank. OR397863.1
- Koo H, 2023. Hibiscus syriacus cultivar Hanmaum plastid, complete genome. GenBank. OR397864.1
- Koo H, 2023. Hibiscus syriacus cultivar Hanseo plastid, complete genome. GenBank. OR397865.1
- Koo H, 2023. Hibiscus syriacus cultivar Hanyang plastid, complete genome. GenBank. OR397866.1
- Koo H, 2023. Hibiscus syriacus cultivar Hwahap plastid, complete genome. GenBank. OR397867.1
- Koo H, 2023. Hibiscus syriacus cultivar Hwahong plastid, complete genome. GenBank. OR397868.1
- Koo H, 2023. Hibiscus syriacus cultivar Jaok plastid, complete genome. GenBank. OR397869.1
- Koo H, 2023. Hibiscus syriacus cultivar Kkoma plastid, complete genome. GenBank. OR397870.1
- Koo H, 2023. Hibiscus syriacus cultivar Kyungki plastid, complete genome. GenBank. OR397871.1
- Koo H, 2023. Hibiscus syriacus cultivar Large_White plastid, complete genome. GenBank. OR397872.1
- Koo H, 2023. Hibiscus syriacus cultivar Mujigae plastid, complete genome. GenBank. OR397873.1
- Koo H, 2023. Hibiscus syriacus cultivar Murasakisaiben plastid, complete genome. GenBank. OR397874.1
- Koo H, 2023. Hibiscus syriacus cultivar Naesarang plastid, complete genome. GenBank. OR397875.1
- Koo H, 2023. Hibiscus syriacus cultivar Nanpa plastid, complete genome. GenBank. OR397876.1
- Koo H, 2023. Hibiscus syriacus cultivar Nunmoe plastid, complete genome. GenBank. OR397877.1
- Koo H, 2023. Hibiscus syriacus cultivar Oknyo plastid, complete genome. GenBank. OR397878.1
- Koo H, 2023. Hibiscus syriacus cultivar Oktokki plastid, complete genome. GenBank. OR397879.1
- Koo H, 2023. Hibiscus syriacus cultivar Paedal plastid, complete genome. GenBank. OR397880.1
- Koo H, 2023. Hibiscus syriacus cultivar Paeksol plastid, complete genome. GenBank. OR397881.1
- Koo H, 2023. Hibiscus syriacus cultivar Parangsae plastid, complete genome. GenBank. OR397882.1
- Koo H, 2023. Hibiscus syriacus cultivar Pheasant_Eye plastid, complete genome. GenBank. OR397883.1
- Koo H, 2023. Hibiscus syriacus cultivar Pompon_Rouge plastid, complete genome. GenBank. OR397884.1
- Koo H, 2023. Hibiscus syriacus cultivar Pulkkot plastid, complete genome. GenBank. OR397885.1
- Koo H, 2023. Hibiscus syriacus cultivar Purpureus plastid, complete genome. GenBank. OR397886.1
- Koo H, 2023. Hibiscus syriacus cultivar Pyeli plastid, complete genome. GenBank. OR397887.1
- Koo H, 2023. Hibiscus syriacus cultivar Pyeonghwa plastid, complete genome. GenBank. OR397888.1
- Koo H, 2023. Hibiscus syriacus cultivar Rubis plastid, complete genome. GenBank. OR397889.1
- Koo H, 2023. Hibiscus syriacus cultivar Russian_Violet plastid, complete genome. GenBank. OR397890.1
- Koo H, 2023. Hibiscus syriacus cultivar Saeasadal plastid, complete genome. GenBank. OR397891.1
- Koo H, 2023. Hibiscus syriacus cultivar Saehan plastid, complete genome. GenBank. OR397892.1
- Koo H, 2023. Hibiscus syriacus cultivar Salmabaek plastid, complete genome. GenBank. OR397893.1
- Koo H, 2023. Hibiscus syriacus cultivar Samchulri plastid, complete genome. GenBank. OR397894.1
- Koo H, 2023. Hibiscus syriacus cultivar Sanchonye plastid, complete genome. GenBank. OR397895.1
- Koo H, 2023. Hibiscus syriacus cultivar Seondeok plastid, complete genome. GenBank. OR397896.1
- Koo H, 2023. Hibiscus syriacus cultivar Seonnyeo plastid, complete genome. GenBank. OR397897.1
- Koo H, 2023. Hibiscus syriacus cultivar Seorak plastid, complete genome. GenBank. OR397898.1
- Koo H, 2023. Hibiscus syriacus cultivar Serenade plastid, complete genome. GenBank. OR397899.1
- Koo H, 2023. Hibiscus syriacus cultivar Shintaeyang plastid, complete genome. GenBank. OR397900.1
- Koo H, 2023. Hibiscus syriacus cultivar Shirohanagasa plastid, complete genome. GenBank. OR397901.1
- Koo H, 2023. Hibiscus syriacus cultivar Single_Red plastid, complete genome. GenBank. OR397902.1
- Koo H, 2023. Hibiscus syriacus cultivar Snowdrift plastid, complete genome. GenBank. OR397903.1
- Koo H, 2023. Hibiscus syriacus cultivar Sobong plastid, complete genome. GenBank. OR397904.1
- Koo H, 2023. Hibiscus syriacus cultivar Soltanshim plastid, complete genome. GenBank. OR397905.1
- Koo H, 2023. Hibiscus syriacus cultivar Sonde plastid, complete genome. GenBank. OR397906.1
- Koo H, 2023. Hibiscus syriacus cultivar Soyang plastid, complete genome. GenBank. OR397907.1
- Koo H, 2023. Hibiscus syriacus cultivar Suchihanagasa plastid, complete genome. GenBank. OR397908.1
- Koo H, 2023. Hibiscus syriacus cultivar Tamna plastid, complete genome. GenBank. OR397909.1
- Koo H, 2023. Hibiscus syriacus cultivar The_Banner plastid, complete genome. GenBank. OR397910.1
- Koo H, 2023. Hibiscus syriacus cultivar Wonhwa plastid, complete genome. GenBank. OR397911.1
- Koo H, 2023. Hibiscus syriacus cultivar Baekgeunip plastid, complete genome. GenBank. OR625140.1
- Koo H, 2023. Hibiscus syriacus cultivar Baekgiwonsu plastid, complete genome. GenBank. OR625141.1
- Koo H, 2023. Hibiscus syriacus cultivar Baeksoryun plastid, complete genome. GenBank. OR625142.1
- Koo H, 2023. Hibiscus syriacus cultivar bredon_spring plastid, complete genome. GenBank. OR625143.1
- Koo H, 2023. Hibiscus syriacus cultivar Daedeoksaback plastid, complete genome. GenBank. OR625144.1
- Koo H, 2023. Hibiscus syriacus cultivar Daedeoksailjung plastid, complete genome. GenBank. OR625145.1
- Koo H, 2023. Hibiscus syriacus cultivar Diana_Baekjo plastid, complete genome. GenBank. OR625148.1
- Koo H, 2023. Hibiscus syriacus cultivar dorothycranc plastid, complete genome. GenBank. OR625149.1
- Koo H, 2023. Hibiscus syriacus cultivar Gakchang plastid, complete genome. GenBank. OR625150.1
- Koo H, 2023. Hibiscus syriacus cultivar Jeogiljung plastid, complete genome. GenBank. OR625153.1
- Koo H, 2023. Hibiscus syriacus cultivar Jeokgiwonsu plastid, complete genome. GenBank. OR625154.1
- Koo H, 2023. Hibiscus syriacus cultivar lenny plastid, complete genome. GenBank. OR625155.1
- Koo H, 2023. Hibiscus syriacus cultivar Rakchanghwarip plastid, complete genome. GenBank. OR625157.1
- Koo H, 2023. Hibiscus syriacus cultivar Sangbon plastid, complete genome. GenBank. OR625160.1
- Koo H, 2023. Hibiscus syriacus cultivar WR_smith plastid, complete genome. GenBank. OR625161.1
- Koo H, 2023. Hibiscus syriacus cultivar Hwarang plastid, complete genome. GenBank. OR625151.1
- Koo H, 2023. Hibiscus syriacus cultivar Saimdang plastid, complete genome. GenBank. OR625159.1
- Koo H, 2023. Hibiscus syriacus cultivar Daejabae plastid, complete genome. GenBank. OR625146.1
- Koo H, 2023. Hibiscus syriacus cultivar Deungnang plastid, complete genome. GenBank. OR625147.1
- Koo H, 2023. Hibiscus syriacus cultivar Jabae plastid, complete genome. GenBank. OR625152.1
- Koo H, 2023. Hibiscus syriacus cultivar Pyeongseong_ps80-1 plastid, complete genome. GenBank. OR625156.1
- Koo H, 2023. Hibiscus syriacus cultivar red_heart plastid, complete genome. GenBank. OR625158.1
- Koo H, 2023. Hibiscus syriacus cultivar Simsan plastid, complete genome. GenBank. OR619828.1
- Koo H, 2023. Hibiscus syriacus cultivar Gangneung plastid, complete genome. GenBank. OR619829.1
Supplementary Materials
Data Availability Statement
All software used for data processing was implemented following the manual provided by the bioinformatic software cited in the method section. When specific parameters for the software were not detailed, the default settings were utilized.