Abstract
Schizosaccharomyces pombe cdc5p is a Myb-related protein that is essential for G2/M progression. To explore the structural and functional conservation of Cdc5 throughout evolution, we isolated Cdc5-related genes and cDNAs from Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, and Homo sapiens. Supporting the notion that these Cdc5 gene family members are functionally homologous to S. pombe cdc5+, human and fly Cdc5 cDNAs are capable of complementing the temperature-sensitive lethality of the S. pombe cdc5-120 mutant. Furthermore, S. cerevisiae CEF1 (S. cerevisiae homolog of cdc5+), like S. pombe cdc5+, is essential during G2/M. The location of the cdc5-120 mutation, as well as mutational analyses of Cef1p, indicate that the Myb repeats of cdc5p and Cef1p are important for their function in vivo. However, we found that unlike in c-Myb, single residue substitutions of glycines for hydrophobic residues within the Myb repeats of Cef1p, which are essential for maintaining structure of the Myb domain, did not impair Cef1p function in vivo. Rather, multiple W-to-G substitutions were required to inactivate Cef1p, and many of the substitution mutants were found to confer temperature sensitivity. Although it is possible that Cef1p acts as a transcriptional activator, we have demonstrated that Cef1p is not involved in transcriptional activation of a class of G2/M-regulated genes typified by SWI5. Collectively, these results suggest that Cdc5 family members participate in a novel pathway to regulate G2/M progression.
The entrance of eukaryotic cells into mitosis from G2 is a highly regulated event which involves a complex series of interactions among an evolutionarily conserved set of kinases and phosphatases (reviewed in references 41 and 57). Genetic studies with the fission yeast Schizosaccharomyces pombe have identified molecules that participate in the regulation of the G2/M transition through phosphorylation (34, 44–46), including cdc2+, cdc13+, cdc25+, wee1+, and mik1+. cdc2+ encodes the single essential cyclin-dependent kinase (CDK) required to drive fission yeast cells through the cell cycle. During G2, cdc2p is associated with the mitotic B-type cyclin encoded by cdc13+. The activity of cdc2p/cdc13p complexes is inhibited by phosphorylation of cdc2p at tyrosine 15, an event which is catalyzed by the wee1p and mik1p protein tyrosine kinases. As fission yeast cells achieve a critical size required for entry into mitosis, the cdc25p protein tyrosine phosphatase activates cdc2p/cdc13p complexes by catalyzing the dephosphorylation of tyrosine 15.
In addition to genes which regulate cdc2p function directly, two other S. pombe genes, cdc5+ and cdc28+, are known to be required during G2 for entry into mitosis. cdc28+ encodes a DEAH-box protein homologous to the Saccharomyces cerevisiae splicing factors Prp2p, Prp16p, and Prp22p, suggesting that RNA processing is in some way required to promote the G2/M transition (33). The cdc5+ gene product contains a putative DNA-binding domain (DBD) that is most similar to those contained within Myb-related proteins and is the only putative transcription factor cloned in S. pombe that performs an essential role during G2/M progression (49).
Recently identified Arabidopsis thaliana, Homo sapiens, and Xenopus laevis cDNAs encode proteins closely related to S. pombe cdc5p (4, 23, 58). Of these, the A. thaliana cDNA, AtCDC5, was found to be a functional homolog of cdc5+ since it was capable of complementing the temperature-sensitive growth defect of the fission yeast cdc5-120 mutant (23). A partial X. laevis Cdc5 cDNA clone was isolated in a screen designed to identify mitotic phosphoproteins that can be phosphorylated in vitro by cyclin B-Cdc2 (58). In addition, Bernstein and Coughlin (4) have reported that the human Cdc5 protein, PCDC5RP (pombe Cdc5-related protein), translocates from the cytoplasm to the nucleus of cultured mammalian cells stimulated with serum, implicating the human protein as a potential effector in a mitogen-activated signaling pathway.
To explore further the structural and functional conservation of cdc5p, we have isolated cDNAs and genes from S. cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, and H. sapiens that encode homologs of S. pombe cdc5p and report here an analysis of these genes. Supporting a role for this subfamily of Myb-related proteins in G2/M cell cycle progression, budding yeast cells lacking CEF1 (S. cerevisiae homolog of cdc5+) arrest growth during G2/M. Furthermore, we demonstrate that human and fly Cdc5 proteins are functionally homologous to S. pombe cdc5p because they are able to complement the temperature-sensitive lethality of the S. pombe cdc5-120 mutant.
Aside from the sequence similarity of Cdc5-related proteins to the transcriptional activator c-Myb, two lines of evidence have implicated Cdc5 in DNA binding. First, the Myb repeats of S. pombe cdc5p bind DNA-cellulose (49). Second, the Myb repeats of AtCDC5 were found to be capable of preferentially binding to the DNA sequence CTCAGCG (23). In this report, we demonstrate by mutational analysis of the Myb repeats of Cef1p and localization of the cdc5-120 mutation to the Myb repeat R1 of cdc5p that the Myb repeats of these proteins are essential for their function in vivo. In the budding yeast S. cerevisiae, the Mcm1p transcription factor, in concert with an uncloned gene encoding a protein named Sff (SWI5 transcription factor), is required for transcription of at least six genes that normally exhibit a G2/M-specific expression pattern, namely, CLB1, CLB2, CDC5, SWI5, ACE2, and ASE1 (1, 26, 35). On the basis of phenotypic analysis of S. cerevisiae cells genetically depleted of CEF1 activity and the ability of these cells to transcribe gene targets of Mcm1p/Sff, we conclude that CEF1 does not encode Sff. Our work demonstrates that fission yeast cdc5p is a member of a highly conserved subfamily of Myb-related proteins that plays a key role in cell cycle progression at the G2/M boundary.
MATERIALS AND METHODS
Strains, growth media, and genetic methods.
The S. cerevisiae and S. pombe strains used in this study are listed in Table 1. Media used to grow S. pombe and general genetic manipulations of S. pombe were described previously (39). S. pombe transformations were performed by electroporation (50). Repression of transcription from the nmt1+ promoter (37) was achieved by addition of thiamine to a concentration of 2 μM.
TABLE 1.
Yeast strains used in this study
| Strain(s) | Genotype | Reference or source |
|---|---|---|
| S. cerevisiae | ||
| YPH274 | MATa ura3-53 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 | A. Weil (Vanderbilt University, Nashville, Tenn.) |
| MATα ura3-53 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 | ||
| KGY856 and KGY857 | MATa ura3-53 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 cef1-Δ1::HIS3 | This study |
| MATα ura3-53 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 CEF1 | ||
| KGY1120 | MATα ura3-53 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 cef1-Δ1::HIS3/pRS425GAL1::CEF1 | This study |
| KGY1140 | MATα ura3-53 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 cef1-Δ1::HIS3/pRS425GAL1::CEF1 | This study |
| KGY1338 | MATα ura3-53 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 cef1-Δ1::HIS3/pRS416CEF1 | This study |
| S. pombe | ||
| KGY69 | 975 h+ | This laboratory |
| KGY155 | cdc25-22 h+ | This laboratory |
| KGY162 | cdc5-120 h+ | This laboratory |
| KGY360 | cdc5-120 leu1-32 his3-D1 h− | This laboratory |
| KGY450 | cdc5::ura4+/cdc5+ ura4-D18/ura4-D18 leu1-32/leu1-32 ade6-M210/ade6-M216 h+/h− | This laboratory |
S. cerevisiae strains were grown in either synthetic minimal medium with the appropriate nutritional supplements or complex yeast extract-peptone (YP) medium supplemented with either 2% glucose or 2% galactose–2% raffinose as the carbon source(s). Genetic methods were as described by Guthrie and Fink (21). Transformation of S. cerevisiae was performed by the lithium acetate method (25).
Isolation and molecular cloning of CEF1 and the Cdc5Ce, Cdc5Dm, and Cdc5Hs.
A 2,555-bp fragment encompassing the CEF1 gene was amplified from genomic DNA by using primers cef1.P5 (5′-CCCGGATCCGGCTTCTTATCTGTGGTC-3′) and cef1.P6 (5′-CCCCTGCAGCCGGATGGTACTTCTTAG-3′) (introduced restriction enzyme sites are italicized). cef1.P5 introduces a BamHI site onto the 5′ end of the CEF1 gene, and cef1.P6 introduces a PstI site onto the 3′ end of the CEF1 gene. The PCR product was cleaved with BamHI and PstI and ligated into pRS415 (9), which had been cut with BamHI and PstI to produce the plasmid pCEF1. The CEF1 open reading frame (ORF) was sequenced to ensure the absence of PCR-induced mutations. A URA3-CEN plasmid (pRS416) containing the CEF1 gene, which was used in all plasmid shuffle assays, was constructed by inserting the BamHI/HindIII CEF1 fragment from pCEF1 into pRS416, which had been cleaved with these enzymes. The 1,770-bp CEF1 ORF was amplified from S. cerevisiae genomic DNA by using primers SC5531 (5′-GCCGATATCTACTAGCATTTCAAGATGCCC-3′) SC5351 (5′-GAAGATATCCTATACTAATGCTATATGGAA-3′). Both primers were designed to add EcoRV restriction sites to the ends of the coding region during amplification. The amplified fragment was cut with EcoRV and cloned into the EcoRV site of pSKREG351 to produce pSKREG351CEF1. pSKREG351 is pBS(SK+) (Stratagene, LaJolla, Calif.) containing 585 bp of nucleotide sequence immediately upstream of the S. pombe cdc5+ ATG. The cdc5+ promoter sequence was amplified by using primers 5REG351 (5′-GAAGATATCAACCCTGAC-3′) and T7 and cloned into pBS(SK+) which had been linearized with EcoRV and treated with dTTP and Taq polymerase. The EcoRV fragment from pSKREG351CEF1 was subcloned into the SmaI site of the 2 μm LEU2-based shuttle vector pRS425GAL1 (42) to produce pGAL1-CEF1. pCEF1 and pGAL1-CEF1 both produce functional Cef1p since they are capable of restoring growth to a strain lacking the CEF1 gene.
A C. elegans λ cDNA clone, YK82fl, whose partial translation product had high sequence identity to a portion of the Myb repeats of cdc5p, was obtained from Yuji Kohara of the Gene Library Lab at the National Institute of Genetics (Mishima, Japan). The insert of this clone was subcloned and sequenced in its entirety on both strands.
The fly homolog of the cdc5+ gene was isolated by sequential nested PCR with D. melanogaster genomic DNA by using the following degenerate primers: Aout, 5′-A(G/A)A T(T/C/A)(T/C) TNA A(G/A)G CNG CNG T-3′; Ain, 5′-AA(G/A) TA(T/C) GGN AA(G/A) AA(T/C) CA(A/G) TGG-3′; Bout, 5′-NGC (T/C)TT NCC (T/C)TG NGT (A/G)TT-3′; Bin, 5′-(T/C)TC (A/G)TC (T/C)TC (A/G)TC CAT (G/A)TC (A/T/G)AT-3′. The corresponding amino acid sequences of these primers are as follows: Aout, EILKAAV; Ain, KYGNQW; Bout, NTQGKKA; Bin, IDMDEDE. Following the second round of PCR, a band of 431 bp was obtained and subcloned into the TA vector (InVitrogen, Carlsbad, Calif.). Five clones were sequenced and found to be identical. This 431-bp fragment was then used to screen an adult Drosophila Canton S strain genomic λ library (Clontech, Palo Alto, Calif.; a gift from M. Fuller). A 4.5-kb XhoI fragment encompassing the entire Cdc5Dm gene locus was subcloned from a positive λ clone and sequenced in its entirety. The amino acid sequence was inferred from the genomic sequence based on homology to other Cdc5 family members.
To isolate Cdc5Hs cDNAs, an I.M.A.G.E. clone (Consortium Clone ID 74654) that contains a 185-bp human expressed sequence tag (EST) whose translation product has high sequence identity to a portion of the Myb repeats of cdc5p was obtained from the American Type Culture Collection (Rockville, Md.) and used to screen a cDNA library constructed from a human microvascular endothelial cell line (a gift from T. O. Daniel). Eight positive clones were obtained after screening approximately 5 × 105 phage clones. The longest clone, pSKCdc5Hs.10, was cleaved with convenient restriction enzymes to generate a bank of truncated clones to facilitate sequencing. The sequence of the Cdc5Hs gene was determined on both strands by using either custom synthesized oligonucleotides (Operon, Alameda, Calif.), M13 reverse primer, or T7 primers.
Chromosomal mapping of Cdc5Hs.
To determine the chromosomal location of the Cdc5Hs gene, three PAC clones (11107, 11108, and 11109; Genome Systems, St. Louis, Mo.) were isolated by using full-length Cdc5Hs cDNA as a probe. PAC clone 11108 was labeled with digoxigenin-11-dUTP (Boehringer Mannheim, Indianapolis, Ind.) by nick translation. The labeled probe was combined with sheared human DNA and hybridized to normal metaphase chromosomes derived from phytohemagglutinin-stimulated peripheral blood lymphocytes in a solution containing 50% formamide, 10% dextran sulfate, and 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Specific hybridization signals were detected by incubating the hybridized slides in fluorescein-conjugated sheep antibodies to digoxigenin (Boehringer Mannheim). The chromosomes were then counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and analyzed. Definitive chromosomal assignment to chromosome 6 was confirmed by cohybridization of clone 11108 with a biotinylated chromosome 6 centromere-specific probe (D6Z1; Oncor Inc., Gaithersburg, Md.). Specific probe signals were detected by incubating the hybridized slides in fluorescein-conjugated sheep antibodies to digoxigenin and Texas red avidin (Vector Laboratories, Burlington, Calif.) followed by counterstaining with DAPI. Band assignment was made by conducting fractional length measurements of 10 specifically hybridized chromosomes 6.
Disruption of CEF1 and construction of KGY1120.
Deletion of the CEF1 coding sequence was performed by replacing nearly the entire ORF with a DNA fragment containing the HIS3 gene. The cef1-Δ1::HIS3 null allele was generated by PCR as described by Baudin et al. (3; see also reference 53), using the primers 5′-GCATTTCAAGATGCCCCCCGTACCAATATACGTGAGCAGATTGTACTGAG-3′ and 5′-CATGGCTTGAAGACGCTTTCTTCTACGACCCTCAAGCGCTCCTTACGCATCTG-3′ and pRS313 (55) as a template to amplify the HIS3 gene. The underlined portions of each primer correspond to the HIS3 flanking sequence. The resulting PCR product contains the complete HIS3 gene flanked by 35 bp of CEF1 5′ sequence corresponding to nucleotides −10 to +25 and 35 bp of CEF1 3′ sequence corresponding to nucleotides +1714 to +1748. When substituted at the CEF1 locus, this DNA fragment deletes the entire CEF1 gene except for the initial 8 codons and final 18 codons. The diploid strain YPH274 was transformed with this DNA fragment, and two independent transformants (KGY856 and KGY857) that contained a cef1-Δ1::HIS3 allele substituted correctly at one of the two CEF1 loci were identified by Southern analysis. These strains were sporulated, and tetrads were dissected and analyzed.
To construct KGY1120, KGY856 was transformed with pGAL-CEF1. This strain was sporulated and tetrads were dissected on synthetic medium containing 2% raffinose–2% galactose. KGY1120 is one of the His+ Leu+ colonies which arose from the tetrad analysis. KGY1140 is derived from a colony from the same tetrad that is His− Leu+.
Cytological methods and flow cytometry.
To visualize nuclei, cells were fixed with 70% ethanol overnight at 4°C, washed twice with phosphate-buffered saline, and resuspended in 1 μg of DAPI per ml. For immunostaining of tubulin, cells were processed as outlined by Pringle et al. (51) and stained with TAT-1 monoclonal antibody (65) followed by a Texas red-conjugated goat-anti mouse immunoglobulin G secondary antibody. Cells were viewed with a Zeiss Axioskop20 with the appropriate set of filters, and images were captured with a Zeiss ZVS-47DEC image-capturing system. Yeast cells were processed for flow cytometric analysis as described previously (49).
Mutagenesis of Cef1p.
To generate single amino acid substitutions in Cef1p, pCEF1 was mutagenized by using the Muta-gene in vitro mutagenesis kit (Bio-Rad Laboratories, Hercules, Calif.) or the Chameleon double-stranded mutagenesis kit (Stratagene). All mutations were confirmed by the presence of introduced restriction enzyme sites and sequencing. Double, triple, and quadruple mutants were constructed by using a unique BglII site that cuts between the codon for the third tryptophan in Myb repeat R1 and that for the second tryptophan in Myb repeat R2.
pGAL1-CEF12R contains the sequence for the first two Myb repeats of Cef1p inserted downstream of the GAL1 promoter in pRS425GAL1 (42). The coding sequence for R1R2 was amplified by using pCEF1 as the template and the primers Cef12R.5′ (5′-AAAGGATCCATGCCCCCCGTACCAATA-3′) and Cef12R.3′ (5′-TTTCTGCAGTTTCTAACTAGTTTGAGTTTCAGCGTTAGG-3′), which insert BamHI and PstI sites onto the 5′ and 3′ ends of the PCR product, respectively. pGAL1-CEF1CT contains the sequence for the C-terminal portion of Cef1p including the Myb-like repeat 3 (MLR3) sequence in pRS425GAL1. The coding sequence for this region of Cef1p was also amplified by using pCEF1 as the template and the primers Cef1CT.5′ (5′-AAAGGATCCATGGCTAGACCAGATAATGG-3′) and Cef1CT.3′ (5′-TTTCTGCAGTTTCTAACTAGTTATGGAAGAAGAAGAATTTAGCATGGCTTG-3′), which insert BamHI and PstI sites onto the 5′ and 3′ ends of the PCR product, respectively. Both of the amplification products were cloned into the BamHI and PstI sites of pRS425GAL1 and sequenced to ensure the absence of PCR-induced mutations.
The pGAL1-CEF1dR1 and pGAL1-CEF1dR2 constructs were generated as follows. The coding sequence for Cef1p lacking R1 was generated by PCR using the primers CEF1dR1 (5′-AAAGGATCCATGTTGAATTTTACAGAGTTC-3′) and Cef1CT.3′. The CEF1dR1 primer inserts a BamHI site at the 5′ end of the PCR product. The amplification product was cloned into the BamHI and PstI sites of pRS425GAL1 and sequenced to ensure the absence of PCR-induced mutations. pGAL1-CEF1dR2 was constructed by inserting HindIII sites at the end of the coding sequence for R1 and at the beginning of the coding sequence for MLR3 by using pGAL1-CEF1 as the template. The internal HindIII fragment coding for R2 was then released from the vector, and the vector was religated to itself. The mutagenesis results in sequence coding for the insertion of a single amino acid, leucine, between R1 and MLR3. The oligonucleotides used for this mutagenesis are as follows: EndR1.HIII, 5′-TGGAATGAATATTTAAATCCAAAGCTTAATTTTACAGAGTTCTCGAAGG-3′; BeginMLR3.HIII, 5′-GATATTAATCCTAACGCTGAAAAGCTTATGGCTAGACCAGATAATGGTG-3′; SWPstBam, 5′-CAAGCCATGCTAAATTCTTCTTCTTAAATAACTAGTTAGTATAGGATGGGGGATCCGAATTCGATATCAAGCTTATCG-3′.
Preparation of total RNA and Northern blot analysis.
S. pombe wild-type (strain 972) and temperature-sensitive strains were grown to mid-log phase at 25°C in yeast extract medium. Aliquots of the cultures were collected while the remaining portions were shifted to 37°C. Cells at 37°C were collected following a 5-h incubation. S. cerevisiae KGY1120 cells were grown to mid-log phase in synthetic medium containing the appropriate supplements and 2% raffinose–2% galactose (SRG medium) as the carbon sources. A portion of the culture was collected for processing, while the remainder of the culture was harvested and resuspended in synthetic medium containing glucose (SD medium) and the appropriate supplements. Cells were collected for processing at 8, 10, 12, 14, and 16 h following the SRG-to-SD medium shift. Total RNA was prepared from S. cerevisiae and S. pombe cells by glass bead disruption as described by Moreno et al. (39) or by RNeasy columns (Qiagen, Chatsworth, Calif.). Twenty micrograms of total RNA was electrophoresed on 1% formaldehyde agarose gels, blotted onto Gene Screen Plus membranes (DuPont-NEN, Boston, Mass.), and fixed by incubation at 80°C under vacuum for 2 h. Hybridization was performed as described by Church and Gilbert (10). Templates for probes were prepared from genomic DNA by PCR or by releasing appropriate fragments from plasmids by restriction enzyme digestion. Probes for hybridization were prepared by using the Rediprime random primer labeling system (Amersham, Arlington Heights, Ill.).
RESULTS
S. pombe cdc5p is conserved throughout evolution: isolation of and structural analysis of Cdc5 gene homologs.
We previously reported that the S. pombe cdc5+ gene encodes a protein that has significant sequence similarity to the c-Myb family of proteins (49). More recently, genome sequencing efforts have revealed that gene products much more closely related to cdc5p than c-Myb family members exist in diverse eukaryotic organisms, including S. cerevisiae, C. elegans, and H. sapiens. Additionally, Hirayama and Shinozaki (23), Bernstein and Coughlin (4), and Stukenberg et al. (58) have reported the identification of cDNAs from A. thaliana, H. sapiens, and X. laevis, respectively, whose predicted proteins are closely related to cdc5p. To determine if the cell cycle function of S. pombe cdc5+ also has been conserved throughout evolution, we isolated presumptive homologs of cdc5+ from S. cerevisiae, C. elegans, D. melanogaster, and H. sapiens.
Comparison of the complete amino acid sequence of cdc5p with those of all the translation products of the ORFs present within the S. cerevisiae genome identified a single translation product which displays significant similarity to cdc5p. For this reason, we have designated the corresponding previously uncharacterized ORF (YMR213w) CEF1 for S. cerevisiae homolog of cdc5+. The 1,770-bp CEF1 ORF is located on chromosome XIII and encodes a protein of 590 amino acids (aa) with a predicted molecular size of 68 kDa (Fig. 1A).
FIG. 1.
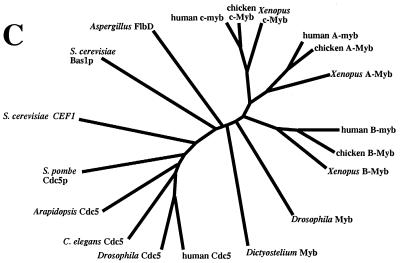
Isolation of homologs of S. pombe cdc5+. (A) Alignment of S. pombe cdc5p homologs by using ClustalW1.6 (14, 60). Residues found to be identical or similar to those of Cdc5Hs are highlighted in black or grey, respectively, to emphasize the evolutionary conservation of Cdc5 homologs. Roman numerals signify the beginning of a Myb repeat. Asterisks indicate the positions of the amino acid residues which are classically occupied by tryptophan residues in Myb repeats. Percent identities among the various Cdc5 family members are indicated at the bottom of this panel. (B) Alignment of MLR3 of Cdc5 family members with R3 of human c-, A-, and B-Myb and Drosophila Myb. Alignment was performed by using ClustalW1.6. Identical and similar residues found within the various proteins are highlighted in black and grey, respectively. Residues found to be identical between all proteins are indicated by arrows. Sp, S. pombe; Sc, S. cerevisiae; Hs, H. sapiens; Dm, D. melanogaster. (C) Phylogenetic analysis of Cdc5 and c-Myb family members.
To identify possible C. elegans homologs of cdc5+, the complete amino acid sequence of cdc5p was used to search a six-way conceptual translation of the database of expressed sequence tags (dbEST) (6). This analysis identified a 360-bp nematode EST (λ clone YK82fl) whose translation product has high sequence identity to a portion of the Myb repeats of cdc5p. The complete primary structure of the presumptive C. elegans homolog was revealed by sequence analysis of the λ clone and by sequencing of cosmid D1081 by the C. elegans genome sequencing project. The Cdc5Ce gene encodes a protein of 755 aa with a predicted molecular size of 86 kDa (Fig. 1A).
A fragment of the D. melanogaster cdc5+ homolog, the Cdc5Dm gene, was identified by sequential nested PCR using primers designed to anneal to highly conserved portions of Cdc5 family members (see Materials and Methods). This fragment was then used as a probe to isolate a genomic clone containing the entire Cdc5Dm gene coding sequence. cDNA clones were then isolated by reverse transcription-PCR and sequenced. The Cdc5Dm protein is 814 aa in length and has a predicted molecular size of 93 kDa (Fig. 1A).
By using a 185-bp human EST (Consortium CloneID 74654), a full-length cDNA clone encoding the putative human homolog of cdc5p, Cdc5Hs, was found to contain a 2,406-bp ORF capable of coding for an 802-aa protein with a predicted molecular size of 93 kDa (Fig. 1A). The predicted amino acid sequence of the Cdc5Hs gene is identical to that of PCDC5RP reported by Bernstein and Coughlin (4). In agreement with the results obtained by Bernstein and Coughlin (4), we have detected a Cdc5Hs gene transcript in a variety of human tissues and cell lines of ∼3.4 kb, which is consistent with the size of the isolated full-length cDNA clones (data not shown). To determine the chromosomal location of the Cdc5Hs gene, a human genomic PAC clone spanning the Cdc5Hs gene locus was isolated and used for fluorescence in situ hybridization (see Materials and Methods). This analysis demonstrated that the Cdc5Hs gene is localized to chromosome 6p21 (data not shown).
Comparison of the amino acid sequences of Cdc5 family members has revealed that they are structurally similar to each other throughout their entire lengths (see bottom of Fig. 1A for percent identities among the various Cdc5 family members). The first ∼100 aa of Cdc5 relatives, which encompass two complete Myb repeats, resemble classical Myb repeats in that each repeat contains three tryptophan residues that are separated by an interval of 18 or 19 aa (reviewed in reference 32). The single exception to this is that the third position in the second repeat (R2) of Cdc5 family members is occupied by a tyrosine residue (Fig. 1A). The presence of hydrophobic residues at these positions is essential for the DNA binding and transcriptional activation properties of c-Myb (27, 54). Mutational and modeling studies of c-Myb predicted that the Myb repeat forms a structure related to a helix-turn-helix motif in which each tryptophan residue forms the hydrophobic backbone of a helix (15, 17). This prediction has been confirmed through nuclear magnetic resonance solution spectroscopy, and these studies have further demonstrated that the minimal Myb DBD (R2R3) contacts DNA through the third helix of each imperfect repeat (47, 48). Mutational studies and analysis of the oncogenic avian myeloblastosis virus product, v-Myb, have demonstrated that alteration of a number of other residues within the DBD of c-Myb results in loss of DNA binding and transcriptional activation capabilities (7, 15, 17, 19, 22, 24, 43). A number of these residues are conserved between c-Myb and Cdc5 family members (49) (Fig. 1A).
Cdc5 family members contain a ∼60-aa Myb-like repeat located immediately adjacent to the second repeat (Fig. 1A and B). At the amino acid level, the third Cdc5 Myb-like repeat (labeled MLR3 in Fig. 1A) is significantly divergent from that found in c-Myb, although certain residues are conserved among all c-Myb and Cdc5 family members analyzed (Fig. 1B). Interestingly, MLR3 lacks the hydrophobic residues which form the helical backbones in a Myb repeat. As observed by Hirayama and Shinozaki (23) and Bernstein and Coughlin (4) regarding AtCDC5, human Cdc5, and S. pombe Cdc5p, the central third of Cdc5 family members harbors a significant number of potential phosphorylation sites for proline-directed kinases (S/TPXK/R or S/TPK/R), which suggests that Cdc5 activity may be regulated by phosphorylation in vivo. Consistent with this notion is the finding that X. laevis Cdc5 is phosphorylated by mitotic egg extracts and cyclin B-Cdc2 (58). Sequence similarity among Cdc5 family members decreases in the C-terminal third of the sequence, where significant identity is observed primarily among the C. elegans, D. melanogaster, A. thaliana, and H. sapiens proteins.
The temperature-sensitive growth defect of the S. pombe cdc5-120 mutant is complemented by both human and fly Cdc5 cDNAs.
To determine if CEF1 and the Cdc5Ce, Cdc5Dm, and Cdc5Hs cDNAs were capable of rescuing the temperature-sensitive lethality of the S. pombe cdc5-120 mutant (46, 49), we introduced these cDNAs into cdc5-120 cells under control of the thiamine-repressible nmt1+ promoter (37). Full-length cDNAs for CEF1 and the Cdc5Ce and Cdc5Hs cDNAs were incapable of rescuing growth of cdc5-120 cells at 36°C in both the presence and absence of thiamine (Fig. 2). In contrast, the full-length Cdc5Dm cDNA was capable of rescuing growth of cdc5-120 cells when induced (in the absence of thiamine) but not when repressed (in the presence of thiamine) (Fig. 2). Microscopic examination of cdc5-120 cells overexpressing CEF1 and the Cdc5Hs cDNA revealed that many of them were elongated at 25°C (data not shown). Overexpression of full-length CEF1 and the Cdc5Hs cDNA had the same phenotypic consequence in wild-type cells (data not shown).
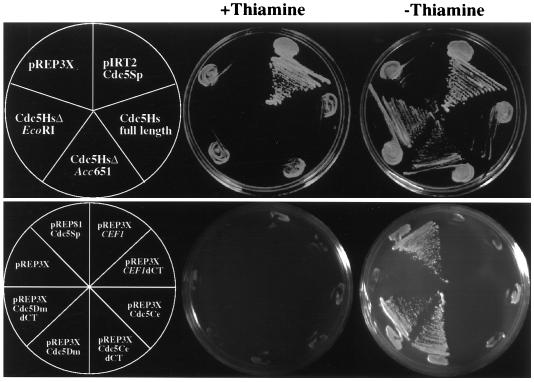
FIG. 2.
The temperature-sensitive growth defect of cdc5-120 is complemented by human and fly Cdc5 proteins. Full-length cDNAs for CEF1 and the Cdc5Ce, Cdc5Dm, and Cdc5Hs cDNAs were placed under control of the nmt1+ promoter in pREP3X. In addition, sequences for products of these cDNAs were truncated at their C termini by 129 aa (CEF1dCT), 337 aa (Cdc5Ce dCT), 293 aa (Cdc5Dm dCT), 441 (ΔEcoRI), and 196 (ΔAcc651) aa (Cdc5Hs) and placed under control of the nmt1+ promoter in pREP3X. S. pombe cdc5+ is pIRT2 (39) harboring full-length cdc5+ cDNA under control of 585 bp of the cdc5+ promoter sequence. pREP81cdc5Sp is pREP81 (2) harboring full-length cdc5+ cDNA. KGY360 transformants carrying Cdc5Hs constructs were struck to minimal medium with or without thiamine and incubated for 18 h at 25°C to induce or repress expression of the inserts, respectively, and then shifted to 36°C for 3 days. KGY360 transformants carrying CEF1, Cdc5Ce cDNA, and Cdc5Dm cDNA constructs were struck to minimal medium with or without thiamine at 36°C and grown for 3.5 days.
We have previously shown that the Myb repeats of S. pombe cdc5p are sufficient to rescue the temperature-sensitive lethality of cdc5-120 (49). Since overproduction of full-length Cef1p and Cdc5Hs proteins appeared to be toxic in S. pombe, 3′-truncated versions of both CEF1 and Cdc5Hs cDNA ORFs were constructed and placed under control of the nmt1+ promoter. 3′-Truncated versions of both the Cdc5Dm and Cdc5Ce cDNAs placed under control of nmt1+ were also constructed. The ability of these deletion mutants to rescue growth of cdc5-120 cells at 36°C was then tested. As shown in Fig. 2, C-terminal truncations of 441 aa (ΔEcoRI) or 196 aa (ΔAcc651) in Cdc5Hs were able to complement the temperature-sensitive growth defect of cdc5-120 cells in the absence of thiamine but not in the presence of thiamine. Cdc5Dm truncated by 293 aa at the C terminus was also capable of complementing the temperature-sensitive growth defect of cdc5-120 in the absence of thiamine but not in the presence of thiamine. A CEF1 ORF coding for a protein truncated by 150 aa at the C terminus, which did not cause elongation of S. pombe cells in the absence of thiamine (data not shown), was unable to restore growth of cdc5-120 cells at 36°C (Fig. 2). Cdc5Ce truncated by 337 aa was also unable to rescue growth of cdc5-120 cells at 36°C (Fig. 2). The inability of Cef1p and Cdc5Ce to restore growth of cdc5-120 cells at 36°C could be due to inappropriate levels of protein expression and/or insufficient structural similarity to S. pombe cdc5p. The ability of the Cdc5Dm cDNA and truncated versions of the Cdc5Hs cDNA to rescue growth of cdc5-120 cells at 36°C, however, clearly establishes these genes as functional homologs of S. pombe cdc5+.
CEF1 is essential during G2/M.
To help determine the role of CEF1 in budding yeast, a complete deletion of one genomic copy of the gene was created in a diploid strain (see Materials and Methods). Precise replacement of one of the copies of CEF1 by a CEF1::HIS3 disruption fragment was confirmed by Southern blot analysis and PCR (data not shown). Two independent cef1-Δ1::HIS3/CEF1 heterozygotes were sporulated, and tetrads were dissected. In each case, only two His− colonies grew, demonstrating that CEF1 is essential for viability in S. cerevisiae. His+ haploid colonies could be recovered following sporulation of the cef1-Δ1::HIS3/CEF1 diploid heterozygote if a plasmid carrying the CEF1 gene was introduced into the strain prior to sporulation. Neither the human Cdc5 cDNA nor S. pombe cdc5+ was found to be capable of complementing the lethality of S. cerevisiae cef1-Δ1::HIS3 cells. Microscopic observation of cef1-Δ1::HIS3 cells in dissected tetrads revealed that they underwent one or two rounds of cell division and then experienced growth arrest as large budded cells (data not shown).
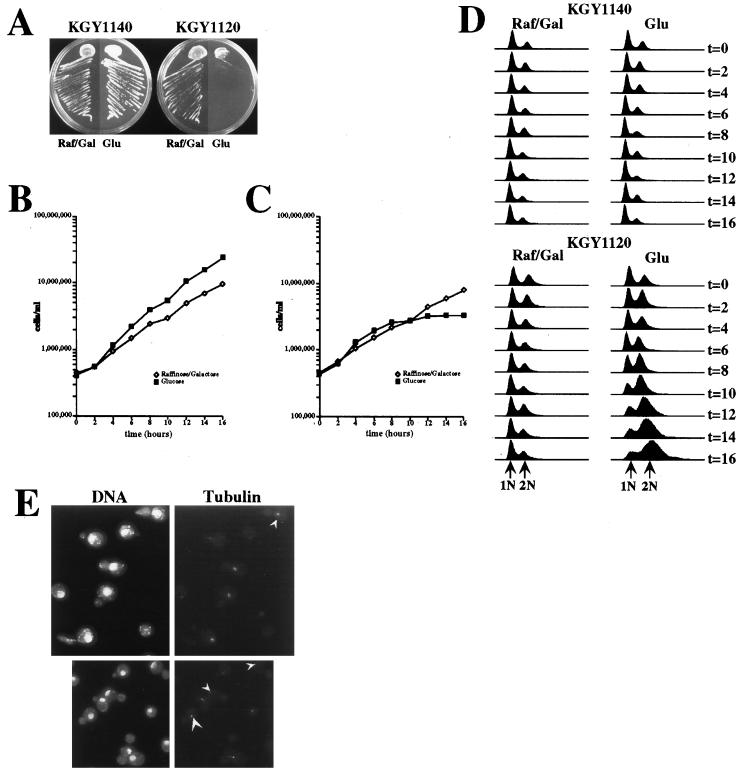
To analyze more carefully the phenotype of cells lacking CEF1 function, a strain in which CEF1 expression could be conditionally inactivated was constructed (see Materials and Methods). In this strain, KGY1120, CEF1 expression was driven by the galactose-inducible GAL1 promoter. Analysis of the KGY1120 strain and an otherwise isogenic wild-type CEF1 strain (KGY1140) in liquid SRG medium revealed no differences in growth rate. Both strains grew with a doubling time of approximately 3.5 h at 32°C (Fig. 3B and C). However, as expected, KGY1120 cells failed to grow on solid medium containing glucose (Fig. 3A) and, upon transfer to liquid SD medium, underwent growth arrest 10 h following the SRG-to-SD medium shift (Fig. 3C).
FIG. 3.
Deletion of CEF1 blocks cell growth at G2/M. (A) KGY1120 fails to grow on medium containing glucose as a carbon source. KGY1120 and the isogenic wild-type strain, KGY1140, were struck onto solid medium containing either 2% glucose or 2% raffinose–2% galactose as a carbon source(s) and incubated at 30°C for 4 days. (B and C) Growth of KGY1140 (B) or KGY1120 (C) cells in liquid SD medium or SRG medium is shown. KGY1140 or KGY1120 cells were grown to mid-log phase in SRG medium at 32°C and then shifted to SD medium (▪), or maintained in SRG (◊). The medium shift was performed at time (t) 0 h. (D) Cells from experiments shown in panels B and C were collected every 2 h, stained with propidium iodide, and subjected to flow cytometric analysis as described in Materials and Methods. Linear fluorescence histograms show relative DNA content in arbitrary units on the horizontal axis and cell number on the vertical axis. Arrows indicate the positions of 1N and 2N DNA peaks. (E) Growth-arrested KGY1120 cells contain short intranuclear spindles. Microtubules were detected by indirect immunofluorescence using the α-tubulin monoclonal antibody TAT-1 (65) followed by staining with Texas red-conjugated secondary antibodies. DNA was visualized by using DAPI. Arrowheads indicate cells that contain a single tubulin plaque instead of a short intranuclear spindle. Raf/Gal, medium containing 2% raffinose–2% galactose; Glu, medium containing 2% glucose.
To determine if growth arrest of KGY1120 cells in SD medium occurred during a specific stage of the cell cycle, we used flow cytometric analysis to determine the DNA content of KGY1120 cells at 2-h intervals following the SRG-to-SD medium shift. By 6 h following the shift, KGY1120 cells began to exhibit a noticeably higher percentage of cells which contained 2N DNA (49%) than did isogenic wild-type KGY1140 cells (34%) (Fig. 3D). By 10 h following the SRG-to-SD medium shift, greater than 80% of KGY1120 cells exhibited a 2N content of DNA whereas KGY1140 cells displayed no change in DNA content profile. The percentage of KGY1120 cells with a 2N content of DNA did not change appreciably between 10 and 16 h following the SRG-to-SD medium shift (Fig. 3D).
The cell and nuclear morphologies of growth-arrested KGY1120 cells were analyzed by phase-contrast microscopy and staining with the DNA-binding dye DAPI. After 8 h in glucose-containing medium, KGY1120 cells began to accumulate as round budded cells (Table 2). This accumulation was more apparent after a 12-h incubation in SD medium when greater than 80% of KGY1120 cells were round budded, whereas only 35% of KGY1140 cells were budded in SD medium (Table 2). An unusual phenotype was present in nearly 25% of budded, growth-arrested KGY1120 cells. These cells contained a single nucleus present at either the end of the mother cell most distal to the mother-bud neck or immediately adjacent to the cell wall offset from the mother-bud neck (Fig. 3E and Table 2). In many cases, these nuclei were compressed against the cell wall and thus appeared to be crescent shaped. These abnormal nuclear morphologies were never observed in the isogenic KGY1140 strain.
TABLE 2.
Quantitation of cell and nuclear morphologies of cells lacking CEF1 activitya
| Strain | Medium | Time (h) | % of total cells scored with indicated morphologyb
|
nc |
|---|---|---|---|---|
| KGY1140 | Raf/Gal | 6814 611 | 500 | |
| Glucose | 65101015 | 500 | ||
| KGY1120 | Glucose | 8 | 3911181022 | 521 |
| Glucose | 10 | 311524 91911 | 513 | |
| Glucose | 12 | 182121 92524 | 648 | |
| Glucose | 14 | 192918102013 | 525 | |
| Glucose | 16 | 152818102315 | 523 |
KGY1120 or the isogenic wild-type strain KGY1140 (Table 2) was grown to the logarithmic phase at 32°C in synthetic complete medium containing 2% raffinose–2% galactose (SRG medium) as the carbon source and then shifted into synthetic complete medium containing 2% glucose (SD medium) (Glucose) or maintained in SRG medium (Raf/Gal). The nuclear and cellular morphologies of the two strains were scored at the indicated time points.
The criteria used for each morphologic class scored are shown schematically.
n = total cells scored.
Arrested KGY1120 cells were stained with the antitubulin monoclonal antibody TAT-1 (65) to view microtubule structures. As shown in Fig. 3, KGY1120 cells cultivated in SD medium contained short intranuclear spindles. A portion of arrested KGY1120 cells appeared to contain a single spindle pole body “plaque” (Fig. 3E), which most likely represents duplicated spindle pole bodies that had not separated to a degree detectable by fluorescence microscopy. Collectively, the cytological and fluorescence-activated cell sorter analyses of KGY1120 cells indicated that CEF1 activity is required predominantly during G2/M in S. cerevisiae.
Mutational analysis of Cef1p.
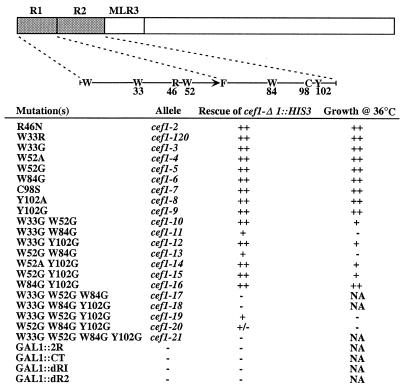
The importance of the two Myb repeats and MLR3 of Cdc5 family members to their function is predicted by the high degree of sequence identity among them (Fig. 1A). As expected, Myb repeats R1 and R2 of Cef1p are essential for its function in vivo; when expressed under control of the GAL1 promoter, a truncated version of Cef1p lacking the R1 and R2 Myb repeats (pGAL1-CEF1CT) is incapable of rescuing the growth of the cef1-Δ1::HIS3 mutant (Fig. 4). Furthermore, the mutation responsible for causing the temperature sensitivity of the S. pombe cdc5-120 mutant was sequenced and found to be present in the first Myb repeat (R1). In cdc5-120p, the second tryptophan of R1 (W29) is converted to an arginine residue as a result of a T:A to C:G mutation at the corresponding codon (TGG to CGG). Although these results underscore the importance of the Cef1p and cdc5p Myb repeats, a construct containing only R1 and R2 of Cef1p under control of the GAL1 promoter (pGAL1-CEF12R) is incapable of rescuing the growth of the cef1-Δ1::HIS3 mutant, indicating that other regions of Cef1p are essential for function as well (Fig. 4).
FIG. 4.
Mutational analysis of Cef1p. Amino acids predicted to be critical for maintaining the structure of the Myb repeats of Cef1p were replaced with the indicated amino acid(s) as described in Materials and Methods. Substitution mutations were constructed in pCEF1, which is pRS415 (9) harboring a 2.5-kb genomic DNA fragment encompassing the CEF1 coding region. In addition to performing site-directed mutagenesis of the Cef1p gene, various domains of Cef1p were removed. pGAL1-CEF12R, Cef1p R1R2; pGAL1-CEF1CT, Cef1p CT including MLR3; pGAL1-CEF1dR1, Cef1p lacking R1; pGAL1-CEF1dR2, Cef1p lacking R2. These mutants were assayed for their abilities to rescue growth of the cef1-Δ1::HIS3 mutant by tetrad analysis and/or plasmid shuffle. Plasmid shuffle with the pRS425GAL1-based plasmids was performed on 5-fluoroorotic acid medium containing raffinose and galactose as the carbon sources. ++, rescue or growth comparable to that of wild-type CEF1; +, reduced rescue of growth and colony formation; +/−, poor rescue of growth; −, failure to rescue growth of the cef1-Δ1::HIS3 mutant or no growth; NA, not applicable.
As discussed above, Myb domains are comprised of three regularly spaced hydrophobic residues, each of which forms the backbone of a helix. Mutation of any one of the six tryptophan residues in c-Myb repeats R2 and R3, which constitute the minimal DBD of c-Myb, to alanine or glycine results in loss of DNA binding and transcriptional activation (27, 54). Furthermore, in R1 of Cdc5 family members and R2 of c-Myb, a basic residue is present at position −6 relative to the third tryptophan. Substitution of this lysine with an asparagine in c-Myb results in loss of DNA binding (15), and mutational analysis of this residue in Petunia MYB.Ph3 indicates that this residue plays a role in determining sequence-specific binding of MYB.Ph3 to DNA (56). Finally, Cdc5 family members contain a cysteine residue at position −4 relative to the third tryptophan in both R1 and R2 (Fig. 1A). Cef1p is exceptional in this case because it contains a serine in place of a cysteine in R1. In c-Myb, replacement of a cysteine residue in a homologous position in R2 with a serine results in a dramatic reduction of DNA binding in vitro (20, 43), raising the possibility that modulation of the oxidation state of this cysteine may be a way of regulating the DNA-binding activity of c-Myb.
To analyze the in vivo consequences of altering these key residues within the Cef1p Myb repeats which are conserved among Cdc5 family members and c-Myb, we generated mutations within the Myb repeats of Cef1p by site-directed mutagenesis (see Materials and Methods) and tested the ability of these mutants to restore growth of cells which lack CEF1. All of the single mutations that were constructed result in nonconservative amino acid substitutions and are located in positions that are important for maintaining structure of the second (W33R, W33G, W84G) and third (R46N, W52A, W52G, C98S, Y102A, Y102G) helices in the corresponding Myb repeats (Fig. 4) (15, 17, 27, 43). Various double, triple, and quadruple mutations which are predicted to disrupt conformation of the second and third helices in both of the canonical Myb repeats of Cef1p were also constructed.
As shown in Fig. 4, all of the single mutants were found to be fully capable of rescuing the growth of the cef1-Δ1::HIS3 mutant, demonstrating that the substitutions in these mutants do not abolish Cef1p function in vivo. Interestingly, reconstruction of the cdc5-120 mutation in CEF1 (cef1-120) does not produce a temperature-sensitive phenotype in S. cerevisiae. All of the double-mutant combinations were also able to rescue growth of cells lacking CEF1. Notably, however, the W33G W84G and W52G W84G mutants were not able to rescue the growth of cef1-Δ1::HIS3 cells as well as the wild-type CEF1 protein at 30°C. Furthermore, cef1-Δ1::HIS3 cells harboring these double mutants were temperature sensitive (Fig. 4) and underwent growth arrest following two to four cell divisions at 36°C as large-budded cells (data not shown). Of the four triple mutants that were analyzed, only two, W33G W52G Y102G and W52G W84G Y102G, rescued the growth of the cef1-Δ1::HIS3 mutant, and the latter conferred only poor growth to cells lacking the CEF1 gene. Similar to the W33G W84G and W52G W84G mutants, these triple mutants were incapable of restoring growth to cells lacking CEF1 at 36°C. The W33G W52G W84G Y102G quadruple mutant was unable to rescue growth of the cef1-Δ1::HIS3 mutant.
The ability of the W33G W52G and W84G Y102G double mutants, which are predicted to disrupt the second and third helices in R1 and R2, respectively, to rescue growth of cells lacking CEF1 raised the issue of whether R1 and R2 are functionally redundant in vivo. We tested this directly by individually removing R1 (pGAL1-CEF1dR1) and R2 (pGAL1-CEF1dR2) from the pGAL1-CEF1 construct. Neither of the deletion mutants was capable of rescuing the growth of the cef1-Δ1::HIS3 mutant, demonstrating that the Cef1p Myb repeats are not functionally redundant. Collectively, these results indicate that the Myb repeats of Cef1p perform an essential function and that structural alteration of the Myb repeats is capable of eliciting a temperature-sensitive phenotype or abolishing function of Cef1p in vivo.
CEF1 and S. pombe cdc5+ are not required for expression of genes which are known to be transcriptionally up-regulated during G2/M.
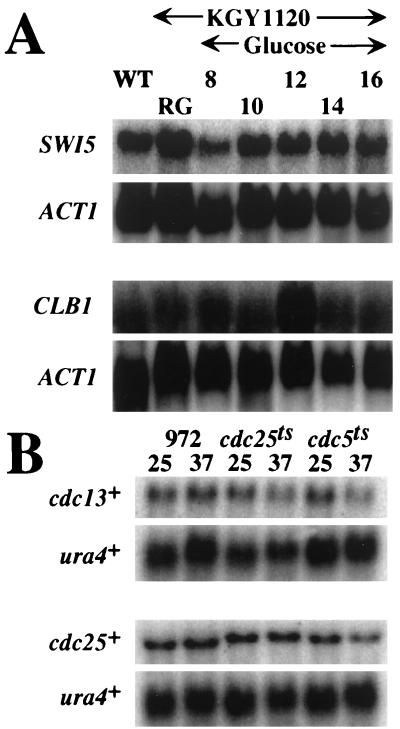
The G2/M and G2 growth arrest phenotypes of S. cerevisiae cells lacking CEF1 activity and S. pombe cdc5-120, respectively, and the similarity of Cdc5 family members to c-Myb prompted us to examine a potential involvement of these proteins in the transcriptional activation of genes known to be up-regulated during G2/M. In S. cerevisiae, at least six genes show a G2/M-specific pattern of transcription: SWI5, CLB1, CLB2, ACE2, CDC5, and ASE1 (1, 26, 35). The first five of these genes, and very likely the sixth as well, require the activity of the MADS-box transcription factor encoded by MCM1 and an uncloned protein termed Sff (1, 26, 35). To test whether Cef1p was required for transcriptional activation of these genes, total RNA was prepared from cells depleted of CEF1 activity and subjected to Northern blot analysis using portions of CLB1 and SWI5 ORFs as probes. As shown in Fig. 5A, CLB1 and SWI5 transcripts persist in cells depleted of CEF1 activity, suggesting that CEF1 is not required for expression of these genes.
FIG. 5.
CEF1 and cdc5+ are not required for transcriptional activation of genes known to be transcriptionally up-regulated during G2/M. (A) CEF1 activity is not required for transcription of SWI5 and CLB1. KGY1120 cells were grown under permissive (SRG medium) or restrictive (SD medium) nutritional conditions as described in the legend to Fig. 3 and Materials and Methods. Total RNA was isolated from cells at 8, 10, 12, 14, and 16 h and subjected to Northern blot analysis using portions of ACT1, SWI5 and CLB1 ORFs as probes. ACT1 transcript levels served as a loading control. WT, wild type; RG, synthetic medium containing 2% galactose and 2% raffinose as carbon sources. (B) cdc5-120 is not defective for transcription of cdc13+ and cdc25+. Logarithmically growing wild-type (strain 972) or temperature-sensitive (ts) strains cdc25-22 or cdc5-120 were collected during growth at 25°C or after incubation at 37°C for 5 h. Total RNA was prepared from these cells and subjected to Northern blot analysis using portions of ura4+, cdc13+, and cdc25+ ORFs as probes. ura4+ transcript levels served as a loading control.
In S. pombe, cdc13p and cdc25p are required during G2 for entry into mitosis. The levels of these proteins oscillate throughout the cell cycle, with maximal amounts present during G2 and mitosis. The amount of cdc25+ transcript in the cell mirrors cdc25p levels, indicating that transcriptional activation of cdc25+ is an important aspect of its regulation (40). cdc13p protein levels are primarily determined at the posttranslational level (12, 66). To determine if a functional cdc5+ product was required for transcription of cdc13+ or cdc25+, total RNA was isolated from wild-type, cdc5-120, and cdc25-22 cells grown at either 25 or 37°C for 5 h and subjected to Northern analysis using portions of cdc13+ and cdc25+ as probes. The cdc25-22 strain, which, like cdc5-120, blocks during G2, was used as a control to identify any nonspecific changes in transcript levels induced by a G2 arrest. As shown in Fig. 5B, cdc25+ and cdc13+ transcript levels are not significantly altered in the cdc5-120 mutant grown at 37°C.
DISCUSSION
In this study, we have reported the identification and characterization of cDNAs and genes from S. cerevisiae, C. elegans, D. melanogaster, and H. sapiens that encode proteins closely related to S. pombe cdc5p. Bernstein and Coughlin (4) have independently reported the isolation of human Cdc5 (PCDC5RP). In addition, Hirayama and Shinozaki (23) have reported the isolation and characterization of AtCDC5, a gene which encodes the A. thaliana homolog of fission yeast cdc5p. Stukenberg et al. (58) have identified an X. laevis relative, and genome sequencing efforts have revealed the existence of a Cdc5-related protein in mice (for an example, see the EST under GenBank accession no. AA269568). When subjected to phylogenetic analysis, all of the analyzed Cdc5 family members display significantly greater similarity to S. pombe cdc5p than to any other Myb-related protein (Fig. 1C). cdc5p is thus a member of a family of Myb-related proteins that has been highly conserved throughout the eukaryotic lineage. In S. cerevisiae, only one gene, CEF1, encodes a protein that bears any significant similarity to fission yeast cdc5p. This also appears to be the case in complex metazoan systems, since all of about 20 Cdc5Hs-related human sequences in dbEST appear to represent overlapping segments of a single gene which we have mapped to chromosome 6p21. Since mammalian Cdc5 activity is likely encoded by a single genetic locus, it is possible that inappropriate modulation of Cdc5 activity may be associated with the onset or progression of tumorigenesis. Although no obvious previously described cancer or disease genes have been mapped to this locus, knowledge of its chromosomal location provides an opportunity to investigate such a possibility directly.
Several lines of evidence suggest that the cell cycle function of S. pombe cdc5+ has also been conserved throughout the eukaryotic lineage. Like S. pombe cdc5+, S. cerevisiae CEF1 is essential for viability. Haploid cells lacking a functional CEF1 gene undergo growth arrest with a 2N DNA content and short intranuclear spindles, indicating that they are arrested during G2/M. Interestingly, a noticeable population of cells lacking CEF1 activity display abnormal nuclear morphologies, indicating that Cef1p may be important for maintenance of nuclear integrity prior to or during the process of nuclear division. Similarly, in S. pombe, approximately 30% of fission yeast cells lacking the cdc5+ gene also display aberrant nuclear morphologies (49). Despite the sequence identity possessed by Cef1p and cdc5p, CEF1 is unable to complement the temperature-sensitive growth defect of S. pombe cdc5-120 at its restrictive temperature of 36°C. Similar to AtCDC5 (23), H. sapiens and S. cerevisiae Cdc5 cDNAs are unable to rescue growth of S. pombe cells harboring a deletion of cdc5+. Human and fission yeast Cdc5 proteins were also observed to be incapable of rescuing the lethality of S. cerevisiae cef1Δ cells. The inability of various Cdc5 family members to rescue growth of cef1Δ or cdc5Δ cells is not surprising given that the C termini of Cdc5 family members are less well conserved than the putative DBDs and that the C termini of S. pombe cdc5p and S. cerevisiae Cef1p have indispensable functions (49, 49a) (Fig. 4). Full-length Cdc5Dm and partially truncated human Cdc5, however, were capable of rescuing growth of cdc5-120 cells at 36°C, thus demonstrating that Cdc5Hs and Cdc5Dm are functional homologs of fission yeast cdc5p. The inability of the Cdc5Ce cDNA and S. cerevisiae CEF1 to rescue growth of cdc5-120 is most simply explained by a limited degree of sequence identity possessed by the Myb repeats of S. pombe cdc5p, Cdc5Dm, and Cdc5Hs (Fig. 1A). Recently, Bernstein and Coughlin (4) have reported that an epitope-tagged version of human Cdc5 translocates from the cytoplasm to the nuclei of serum-deprived cultured mammalian cells upon stimulation with serum. Based on these results, the authors speculated that Cdc5 may function in a mitogen-activated signaling pathway during the G0/G1 transition. Although a definitive role(s) for Cdc5 in the mammalian cell cycle requires further analysis, our data indicate that an essential, conserved function of Cdc5 in the eukaryotic cell cycle is at the G2/M transition.
In S. cerevisiae, the Mcm1p transcription factor, in combination with the uncloned SWI5 factor (Sff), is required during G2/M to activate transcription of at least six genes which are necessary for entry into and completion of mitosis: CLB1, CLB2, ACE2, SWI5, CDC5, and ASE1 (1, 26, 35). Sff is absolutely required for transcription of SWI5 because mutation of the Sff binding site results in the loss of SWI5 upstream activating sequence activity (35). S. cerevisiae cells lacking MCM1 activity undergo growth arrest with replicated DNA and elongated buds (1), a phenotype commonly associated with S. cerevisiae mutants which possess low Clb-associated Cdc28p (Cdk1) kinase activity (31, 52, 59). Cells lacking CEF1 undergo arrest with a round-budded phenotype, which indicates the presence of Clb-associated Cdk1 activity (31). The phenotypic difference between cells lacking MCM1 and CEF1 activities indicates that CEF1 is unlikely to encode Sff. We have confirmed this hypothesis by demonstrating that cells depleted of CEF1 activity are capable of accumulating transcripts of Mcm1p and Sff target genes.
In S. pombe, cdc13p and cdc25p levels oscillate in a cell cycle-dependent manner, with maximal levels observed near the G2/M transition (12, 40). cdc25+ transcript is most abundant near the G2/M transition, indicating that accumulation of cdc25p during G2 is achieved, at least in part, through its transcriptional activation (40). The cdc5-120 mutant is not defective for transcription of these mitotic regulators. We conclude from these results and those obtained from Cef1p-depleted cells that Cdc5 does not regulate the G2/M transition in eukaryotic cells at the level of transcription of these known mitotic regulators.
Although cdc5p and Cef1p do not appear to function through activating transcription of genes known to be up-regulated during G2/M, it is still tempting to assign these proteins a role as DNA-binding proteins for the following reasons: (i) all Cdc5 family members analyzed thus far contain two highly conserved Myb repeats, (ii) the S. pombe cdc5-120 mutation converts the second tryptophan in R1 of cdc5p to an arginine residue, and (iii) the Myb repeats of AtCDC5 have been shown to preferentially bind a specific nucleotide sequence (23). For these reasons, we have tried to identify a preferential DNA-binding sequence for S. pombe cdc5p and S. cerevisiae Cef1p. Specifically, we employed the cyclic amplification and selection of target sequence (CASTing) approach with the binding (23, 62) and washing (16) conditions described previously. The N-terminal 201 amino acids of Cef1p, which contain R1, R2, and MLR3, were fused downstream of glutathione S-transferase (GST), and this fusion protein was used for CASTing. An oligonucleotide library containing a core of 20 random nucleotides was used as a probe. A similar approach was used to identify a target sequence of AtCDC5 (23). While it is clear that the Myb repeats of Cef1p and cdc5p have the ability to bind DNA, we have not been able to select a high-affinity binding sequence for these proteins although under the same conditions a fusion protein between GST and the DBD of AML was able to select the sequence TGT/cGGT, the known high-affinity target sequence of AML (38, 49, 49b). In a separate experiment, a hexahistidine-tagged version of the DBD of Cdc5Dm was used in an electrophoretic mobility shift assay with the AtCDC5 binding sequence as a probe. We reasoned that since the DBDs of these proteins have 80% identity, they may be capable of binding the same sequence. While a modest binding of Cdc5Dm to this probe was observed, the binding was abrogated in the presence of increasing amounts of nonspecific competitor, indicating that this interaction occurs with low affinity (37a). In this regard, it is noteworthy that AtCDC5 binding to the sequence CTCAGCG was also reduced with nonspecific competitor DNA (23). While these data may argue that Cdc5 does not bind a particular DNA sequence with high affinity, we cannot exclude the possibility that additional regions of these proteins are required for sequence-specific DNA binding or that they require additional factors to bind DNA with high affinity.
In this study, we have analyzed the in vivo consequences of altering residues within the Cef1p Myb repeats which are predicted to disturb the conformation of these domains. Of particular relevance to this work, mutational studies of c-Myb have demonstrated that hydrophobic residues forming the backbones of the DBD helices are essential for the DNA-binding activity of c-Myb in vitro and its trans-activation activity in reporter assays (27, 54). Surprisingly, we found that the Cef1p Myb repeats are quite resiliant to similar changes; single, double, or even certain triple mutant combinations in residues which form the hydrophobic backbones of the second or third helices in both R1 and R2 do not abolish Cef1p function in vivo, although many produce a temperature-sensitive growth defect. Although these results are unexpected, it is important to bear in mind that our assay determined the functional effects of altering helical backbone residues in vivo and is therefore not directly comparable to the DNA binding, transcriptional activation, and differentiation assays that have traditionally been used in mutational analyses of c-Myb. To our knowledge, systematic mutational analyses of Myb domains in other proteins, such as those presented here, have yet to be reported. Thus, it is possible that interaction of Cef1p with other proteins in vivo may stabilize the conformation of mutant Myb repeats, thus allowing Cef1p to bind DNA.
An intriguing, equally plausible interpretation is that in vivo, Cef1p may cooperate with a second DNA-binding protein whose activity compensates for a diminution of Cef1p DNA-binding activity. Evidence does exist for Myb-related proteins functioning in heterodimeric complexes. c-Myb itself cooperates with heat shock factor 3 (HSF3) to activate transcription from the hsp70 promoter (28, 29). In S. cerevisiae and Zea mays, the Myb-related proteins Bas1p and C1, respectively, are thought to act in concert with a second DNA-binding protein to activate transcription of target genes; Bas1p cooperates with the homeobox protein Bas2p, and C1 is thought to dimerize with basic helix-loop-helix proteins such as B, R, SN, and LC (13, 18, 36, 61). In the cases of C1 and c-Myb, heterodimerization has been shown to require the Myb domains (18, 28). The in vivo activity of Cef1p clearly requires both R1 and R2. Perhaps, as in the case of C1, R1R2 of Cef1p functions as both a protein-protein interaction domain and a DBD. Evidence also exists for heterodimeric transcription factor complexes that require only one active DNA-binding subunit. For example, mutational studies have demonstrated that elimination of the sequence-specific DNA-binding activity of the yeast homeodomain protein α-2 does not destroy its ability to bind DNA in concert with a1 (64). Additionally, c-Myb, in concert with HSF3, stimulates transcription from the hsp70 promoter through the heat shock element without apparently binding to this sequence (28, 29).
Although the precise function of Cdc5 family members is not understood presently, it is clear that the product of the cdc5+ gene in S. pombe and its homolog in S. cerevisiae are required during G2/M. At present, all available evidence indicates a role for this protein family in DNA binding. However, it is quite possible that Cdc5 family members do not function as transcriptional activators. Indeed, there is precedence for Myb-related proteins which play roles in biological processes other than activation of transcription. For example, human TRF1 and TRF2 and fission yeast taz1p are Myb-related proteins that negatively regulate telomere length (5, 8, 11, 63) and the Myb-related budding yeast protein Reb1p functions in transcriptional termination as well as transcriptional activation of rRNA genes (30). An approach utilizing a combination of genetic, biochemical, and cytological analyses of the isolated Cdc5 family members should lead to an understanding of how Cdc5 facilitates the G2/M transition in eukaryotic cells.
ACKNOWLEDGMENTS
We thank members of the laboratories of A. P. Weil and J. Flick for yeast strains and expression plasmids and T. O. Daniel and E. Stein for the human microvascular endothelial cell line library and invaluable technical advice. S. Hiebert kindly provided the plasmid to produce GST-AML. We are grateful to J. Price for flow cytometric analyses. All members of the Gould laboratory, including M. K. Balasubramanian and D. McCollum, are appreciated for valuable discussions and technical advice.
This work was supported by NIH grant GM47728 to K.L.G. and USPHS grants RO1 CA43592 and PO1 CA70404 to J.S.L. and CA71907 to A.T.L. S.M. was supported by USPHS grant NRSA 5T32 CA09302. K.L.G. is an assistant investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Althoefer H, Schleiffer A, Wassmann K, Nordheim A, Ammerer G. Mcm1 is required to coordinate G2-specific transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:5917–5928. doi: 10.1128/mcb.15.11.5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- 3.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein H S, Coughlin S R. Pombe Cdc5-related protein. A putative human transcription factor implicated in mitogen-activated signaling. J Biol Chem. 1997;272:5833–5837. doi: 10.1074/jbc.272.9.5833. [DOI] [PubMed] [Google Scholar]
- 5.Bilaud T, Brun C, Ancelin K, Koering C E, Laroche T, Gilson E. Telomeric localization of TRF2, a novel human telobox protein. Nat Genet. 1997;17:236–239. doi: 10.1038/ng1097-236. [DOI] [PubMed] [Google Scholar]
- 6.Boguski M S, Lowe T M, Tolstoshev C M. dbEST—database for “expressed sequence tags”. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 7.Brendeford E M, Myrset A H, Hegvold A B, Lundin M, Gabrielsen O S. Oncogenic point mutations induce altered conformation, redox sensitivity, and DNA binding in the minimal DNA binding domain of avian myeloblastosis virus v-Myb. J Biol Chem. 1997;272:4436–4443. doi: 10.1074/jbc.272.7.4436. [DOI] [PubMed] [Google Scholar]
- 8.Broccoli D, Smogorzewska A, Chong L, de Lange T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet. 1997;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- 9.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 10.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper J P, Nimmo E R, Allshire R C, Cech T R. Regulation of telomere length and function by a Myb-domain protein in fission yeast [see comments] Nature. 1997;385:744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- 12.Creanor J, Mitchison J M. The kinetics of the B cyclin p56cdc13 and the phosphatase p80cdc25 during the cell cycle of the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1996;109:1647–1653. doi: 10.1242/jcs.109.6.1647. [DOI] [PubMed] [Google Scholar]
- 13.Daignan-Fornier B, Fink G R. Coregulation of purine and histidine biosynthesis by the transcriptional activators BAS1 and BAS2. Proc Natl Acad Sci USA. 1992;89:6746–6750. doi: 10.1073/pnas.89.15.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng D F, Doolittle R F. Progressive alignment and phylogenetic tree construction of protein sequences. Methods Enzymol. 1990;183:375–387. doi: 10.1016/0076-6879(90)83025-5. [DOI] [PubMed] [Google Scholar]
- 15.Frampton J, Gibson T J, Ness S A, Doderlein G, Graf T. Proposed structure for the DNA-binding domain of the Myb oncoprotein based on model building and mutational analysis. Protein Eng. 1991;4:891–901. doi: 10.1093/protein/4.8.891. [DOI] [PubMed] [Google Scholar]
- 16.Funk W D, Pak D T, Karas R H, Wright W E, Shay J W. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol Cell Biol. 1992;12:2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabrielsen O S, Sentenac A, Fromageot P. Specific DNA binding by c-Myb: evidence for a double helix-turn-helix-related motif. Science. 1991;253:1140–1143. doi: 10.1126/science.1887237. [DOI] [PubMed] [Google Scholar]
- 18.Goff S A, Cone K C, Chandler V L. Functional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev. 1992;6:864–875. doi: 10.1101/gad.6.5.864. [DOI] [PubMed] [Google Scholar]
- 19.Grasser F A, LaMontagne K, Whittaker L, Stohr S, Lipsick J S. A highly conserved cysteine in the v-Myb DNA-binding domain is essential for transformation and transcriptional trans-activation. Oncogene. 1992;7:1005–1009. [PubMed] [Google Scholar]
- 20.Guehmann S, Vorbrueggen G, Kalkbrenner F, Moelling K. Reduction of a conserved Cys is essential for Myb DNA-binding. Nucleic Acids Res. 1992;20:2279–2286. doi: 10.1093/nar/20.9.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guthrie C, Fink G R, editors. Guide to yeast genetics and molecular biology. Vol. 194. San Diego, Calif: Academic Press, Inc.; 1991. [Google Scholar]
- 22.Hegvold A B, Gabrielsen O S. The importance of the linker connecting the repeats of the c-Myb oncoprotein may be due to a positioning function. Nucleic Acids Res. 1996;24:3990–3995. doi: 10.1093/nar/24.20.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirayama T, Shinozaki K. A cdc5+ homolog of a higher plant, Arabidopsis thaliana. Proc Natl Acad Sci USA. 1996;93:13371–13376. doi: 10.1073/pnas.93.23.13371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Introna M, Golay J, Frampton J, Nakano T, Ness S A, Graf T. Mutations in v-myb alter the differentiation of myelomonocytic cells transformed by the oncogene. Cell. 1990;63:1289–1297. doi: 10.1016/0092-8674(90)90424-d. [DOI] [PubMed] [Google Scholar]
- 25.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Juang Y L, Huang J, Peters J M, McLaughlin M E, Tai C Y, Pellman D. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 1997;275:1311–1314. doi: 10.1126/science.275.5304.1311. [DOI] [PubMed] [Google Scholar]
- 27.Kanei-Ishii C, Sarai A, Sawazaki T, Nakagoshi H, He D N, Ogata K, Nishimura Y, Ishii S. The tryptophan cluster: a hypothetical structure of the DNA-binding domain of the myb protooncogene product. J Biol Chem. 1990;265:19990–19995. [PubMed] [Google Scholar]
- 28.Kanei-Ishii C, Tanikawa J, Nakai A, Morimoto R I, Ishii S. Activation of heat shock transcription factor 3 by c-Myb in the absence of cellular stress. Science. 1997;277:246–248. doi: 10.1126/science.277.5323.246. [DOI] [PubMed] [Google Scholar]
- 29.Kanei-Ishii C, Yasukawa T, Morimoto R I, Ishii S. c-Myb-induced trans-activation mediated by heat shock elements without sequence-specific DNA binding of c-Myb. J Biol Chem. 1994;269:15768–15775. [PubMed] [Google Scholar]
- 30.Lang W H, Morrow B E, Ju Q, Warner J R, Reeder R H. A model for transcription termination by RNA polymerase I. Cell. 1994;79:527–534. doi: 10.1016/0092-8674(94)90261-5. [DOI] [PubMed] [Google Scholar]
- 31.Lew D J, Reed S I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lipsick J S. One billion years of Myb. Oncogene. 1996;13:223–235. [PubMed] [Google Scholar]
- 33.Lundgren K, Allan S, Urushiyama S, Tani T, Ohshima Y, Frendewey D, Beach D. A connection between pre-mRNA splicing and the cell cycle in fission yeast: cdc28+ is allelic with prp8+ and encodes an RNA-dependent ATPase/helicase. Mol Biol Cell. 1996;7:1083–1094. doi: 10.1091/mbc.7.7.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lundgren K, Walworth N, Booher R, Dembski M, Kirschner M, Beach D. mik1 and wee1 cooperate in the inhibitory tyrosine phosphorylation of cdc2. Cell. 1991;64:1111–1122. doi: 10.1016/0092-8674(91)90266-2. [DOI] [PubMed] [Google Scholar]
- 35.Lydall D, Ammerer G, Nasmyth K. A new role for MCM1 in yeast: cell cycle regulation of SW15 transcription. Genes Dev. 1991;5:2405–2419. doi: 10.1101/gad.5.12b.2405. [DOI] [PubMed] [Google Scholar]
- 36.Martin C, Paz-Ares J. MYB transcription factors in plants. Trends Genet. 1997;13:67–73. doi: 10.1016/s0168-9525(96)10049-4. [DOI] [PubMed] [Google Scholar]
- 37.Maundrell K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- 37a.McCann, S., and J. S. Lipsick. Unpublished observations.
- 38.Meyers S, Downing J R, Hiebert S W. Identification of AML-1 and the (8;21) translocation protein (AML-1/ETO) as sequence-specific DNA-binding proteins: the runt homology domain is required for DNA binding and protein-protein interactions. Mol Cell Biol. 1993;13:6336–6345. doi: 10.1128/mcb.13.10.6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 40.Moreno S, Nurse P, Russell P. Regulation of mitosis by cyclic accumulation of p80cdc25 mitotic inducer in fission yeast. Nature. 1990;344:549–552. doi: 10.1038/344549a0. [DOI] [PubMed] [Google Scholar]
- 41.Morgan D O. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 42.Mumberg D, Muller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myrset A H, Bostad A, Jamin N, Lirsac P N, Toma F, Gabrielsen O S. DNA and redox state induced conformational changes in the DNA-binding domain of the Myb oncoprotein. EMBO J. 1993;12:4625–4633. doi: 10.1002/j.1460-2075.1993.tb06151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nasmyth K, Nurse P. Cell division cycle mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1981;182:119–124. doi: 10.1007/BF00422777. [DOI] [PubMed] [Google Scholar]
- 45.Nurse P, Thuriaux P. Regulatory genes controlling mitosis in the fission yeast Schizosaccharomyces pombe. Genetics. 1980;96:627–637. doi: 10.1093/genetics/96.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- 47.Ogata K, Hojo H, Aimoto S, Nakai T, Nakamura H, Sarai A, Ishii S, Nishimura Y. Solution structure of a DNA-binding unit of Myb: a helix-turn-helix-related motif with conserved tryptophans forming a hydrophobic core. Proc Natl Acad Sci USA. 1992;89:6428–6432. doi: 10.1073/pnas.89.14.6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell. 1994;79:639–648. doi: 10.1016/0092-8674(94)90549-5. [DOI] [PubMed] [Google Scholar]
- 49.Ohi R, McCollum D, Hirani B, Den Haese G J, Zhang X, Burke J D, Turner K, Gould K L. The Schizosaccharomyces pombe cdc5+ gene encodes an essential protein with homology to c-Myb. EMBO J. 1994;13:471–483. doi: 10.1002/j.1460-2075.1994.tb06282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49a.Ohi, R., and K. L. Gould. Unpublished observations.
- 49b.Ohi, R., C. G. Burns, and K. L. Gould. Unpublished observations.
- 50.Prentice H L. High efficiency transformation of Schizosaccharomyces pombe by electroporation. Nucleic Acids Res. 1992;20:621. doi: 10.1093/nar/20.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pringle J, Adams A E M, Drubin D G, Haarer B K. Immunofluorescence methods for yeast. In: Guthrie C, Fink G R, editors. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press, Inc.; 1991. pp. 565–601. [Google Scholar]
- 52.Richardson H, Lew D J, Henze M, Sugimoto K, Reed S I. Cyclin-B homologs in Saccharomyces cerevisiae function in S phase and in G2. Genes Dev. 1992;6:2021–2034. doi: 10.1101/gad.6.11.2021. [DOI] [PubMed] [Google Scholar]
- 53.Rothstein R J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 54.Saikumar P, Murali R, Reddy E P. Role of tryptophan repeats and flanking amino acids in Myb-DNA interactions. Proc Natl Acad Sci USA. 1990;87:8452–8456. doi: 10.1073/pnas.87.21.8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Solano R, Fuertes A, Sanchez-Pulido L, Valencia A, Paz-Ares J. A single residue substitution causes a switch from the dual DNA binding specificity of plant transcription factor MYB.Ph3 to the animal c-MYB specificity. J Biol Chem. 1997;272:2889–2895. doi: 10.1074/jbc.272.5.2889. [DOI] [PubMed] [Google Scholar]
- 57.Solomon M J. Activation of the various cyclin/cdc2 protein kinases. Curr Opin Cell Biol. 1993;5:180–186. doi: 10.1016/0955-0674(93)90100-5. [DOI] [PubMed] [Google Scholar]
- 58.Stukenberg P T, Lustig K D, McGarry T J, King R W, Kuang J, Kirschner M W. Systematic identification of mitotic phosphoproteins. Curr Biol. 1997;7:338–348. doi: 10.1016/s0960-9822(06)00157-6. [DOI] [PubMed] [Google Scholar]
- 59.Surana U, Robitsch H, Price C, Schuster T, Fitch I, Futcher A B, Nasmyth K. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell. 1991;65:145–161. doi: 10.1016/0092-8674(91)90416-v. [DOI] [PubMed] [Google Scholar]
- 60.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tice-Baldwin K, Fink G R, Arndt K T. BAS1 has a Myb motif and activates HIS4 transcription only in combination with BAS2. Science. 1989;246:931–935. doi: 10.1126/science.2683089. [DOI] [PubMed] [Google Scholar]
- 62.Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell. 1993;5:1529–1539. doi: 10.1105/tpc.5.11.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1 [see comments] Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 64.Vershon A K, Jin Y, Johnson A D. A homeo domain protein lacking specific side chains of helix 3 can still bind DNA and direct transcriptional repression. Genes Dev. 1995;9:182–192. doi: 10.1101/gad.9.2.182. [DOI] [PubMed] [Google Scholar]
- 65.Woods A, Sherwin T, Sasse R, MacRae T H, Baines A J, Gull K. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J Cell Sci. 1989;93:491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
- 66.Yamano H, Gannon J, Hunt T. The role of proteolysis in cell cycle progression in Schizosaccharomyces pombe. EMBO J. 1996;15:5268–5279. [PMC free article] [PubMed] [Google Scholar]