Dear Editor,
Human coenzyme A synthase (COASY) encodes a bifunctional enzyme containing 4’PP adenyltransferase (PPAT) and dephospho-CoA kinase (DPCK) domains that catalyzes the last two steps of de novo CoA biosynthesis (Supplementary Fig. S1)1. Biallelic COASY variants have been associated with severe neurodegenerative diseases2,3. However, no muscular disorders associated with COASY have been reported until now. Here, we found that biallelic COASY variants can be a novel genetic cause of riboflavin-responsive lipid storage myopathy (RR-LSM). RR-LSM is a subtype of lipid metabolic disorders with onset age ranging from infancy to adulthood. It is characterized by muscle weakness or exercise intolerance sometimes triggered by precipitating factors, occasional extramuscular multi-system symptoms, excessive accumulation of lipid droplets (LDs) in myofibers, and dramatic responsiveness to riboflavin4. Although genetic causes of the majority of the disorders have been identified, those for late-onset RR-LSM remain unresolved.
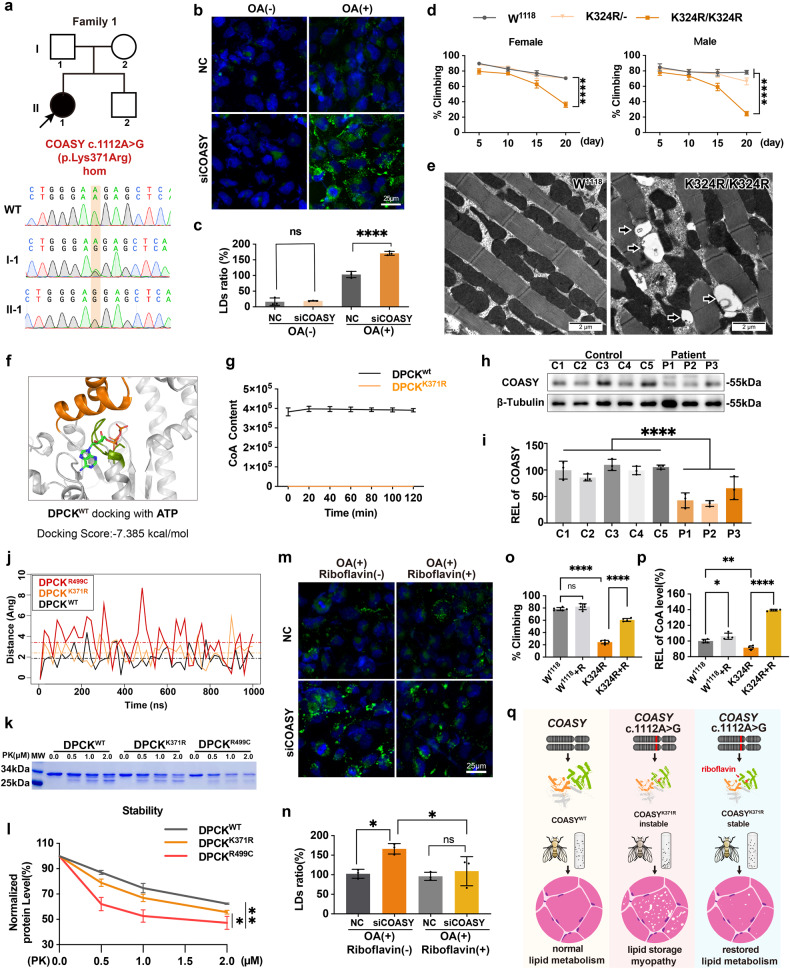
To search for the genetic cause of these unsolved RR-LSM patients, we conducted whole-exome sequencing (WES) on 28 RR-LSM patients with unidentified causative genes from 294 patients diagnosed with RR-LSM based on clinical and pathological criteria at five neuromuscular disease centers. Among these patients, 266 people carried variants in the gene encoding electron transfer flavoprotein dehydrogenase. WES data analysis revealed that 16 out of 28 patients were identified to carry biallelic COASY variants (Fig. 1a). The clinical and myopathological features of these 16 patients were illustrated in Supplementary Figs. S2–S5 and Supplementary Tables S1, S2. No iron deposits were observed in the brains of 4 patients who underwent sensitivity-weighted imaging, with the oldest patient being 43 years old. Additionally, among the 7 patients who underwent metabolic screening, 4 patients showed a metabolic profile of multiple acyl-coenzyme A dehydrogenase deficiency (MADD)5. Out of the 16 patients, 13 patients were homozygous for c.1112A > G (p.Lys371Arg), and the other 3 were compound heterozygous, with c.1112A > G in one allele and c.805_806insC(p.Leu269Profs*11), c.1018_1019insA(p.Met340Asnfs*11), or c.383C > T(p.Pro128Leu) in the other allele, respectively. The examined individuals displayed family co-segregation (Supplementary Figs. S6, S7). Four variants were novel and predicted as deleterious by in silico analysis with highly evolutional conservation (Supplementary Fig. S8). The p.Lys371Arg, located in the DPCK domain, was a hotspot variant with an allele frequency of 90.6% (29/32). Homozygosity mapping in 12 patients with c.1112A > G homozygote showed a 0.64 Mb common region containing the variant, and further SNP analysis revealed a shared haplotype in the 0.64 Mb region, suggesting a founder effect in these patients (Supplementary Fig. S9). These results suggested that the COASY gene was implicated as a new causative gene for RR-LSM.
Fig. 1. Biallelic COASY variant as a genetic cause of RR-LSM.
a The index patient from the first family carried a homozygous missense variant c.1112A > G (p.Lys371Arg) in COASY. b, c BODIPYTM staining showed no LD accumulation in normal control (NC) and COASY-knockdown HEK293T cells in normal culture medium, while numerous LDs accumulated in COASY-knockdown cells under OA overload (n = 3, ****P < 0.0001). d Female (left) and male (right) flies with homozygous p.Lys342Arg showed progressive locomotor deficits, as compared to heterozygous p.Lys342Arg or WT W1118 flies (n = 115–137, ****P < 0.0001). e Electron microscopy revealed obvious LDs accumulation in the muscles of homozygous p.Lys342Arg files at day 20 compared to WT W1118 flies. f MD analysis indicated that the P-loop (Green) containing p.Lys371 in the DPCK domain can bind with ATP. g Purified DPCKK371R protein (orange) lost the ability to produce CoA in vitro compared to WT DPCKWT (black). h, i Immunoblotting showed that the COASY levels in muscle biopsies of RR-LSM patients (P1–3) were significantly decreased by approximately half of those in age-matched controls (C1–5) (Control 100 ± 11.93 vs patient 48.43 ± 18.57, n = 3, ****P < 0.0001). j The 3 trajectories (DPCKWT, DPCKK371R and DPCKR499C) indicated the distance of the αC atom at Arg503 compared to the one of initial position during the process of 1000 ns MD simulation. k, l DPCKR499C was more susceptible to be incrementally degraded by proteinase K at different concentrations compared to DPCKWT and DPCKK371R in vitro (n = 3, *P < 0.05). m, n Riboflavin supplement significantly decreased the accumulation of LDs in COASY-knockdown cells under OA overload (*P < 0.05). o Riboflavin treatment significantly improved the locomotor of homozygous p.Lys342Arg files at day 20 (n = 139–170, ****P < 0.0001). p The CoA level in homozygous p.Lys342Arg flies was significantly lower than that of the WT W1118, but could be restored by riboflavin supplementation (P < 0.0001). q Schematic model of pathogenic mechanism in COASY-related RR-LSM.
To demonstrate whether loss-of-function COASY variants played a causative role in lipid metabolism disorder, we established cell and Drosophila melanogaster models. Our studies revealed that, although there was no significant accumulation of LDs in the COASY-knockdown cells in the normal culture medium, the LDs significantly increased after oleic acid (OA) overload (Fig. 1b, c; Supplementary Fig. S10). The dysfunction of lipid metabolism resulting from lipid overload stress was highly consistent with the clinical phenotype of COASY-related RR-LSM patients, particularly case 2, who developed muscle weakness after a high-fat diet (Supplementary Table S1). Compared to wild-type (WT) Drosophila, Drosophila homozygous for the p.Lys342Arg in Ppat-Dpck (equivalent to p.Lys371Arg in human COASY) generated by CRISPR/Cas9, exhibited substantial LD accumulation in muscle fibers and showed a significant decrease in locomotor ability (Fig. 1d, e; Supplementary Fig. S11). Neurodegeneration is usually characterized by vacuole formation in the Drosophila brain, and thus we carefully examined the brain sections. Brains from p.Lys342Arg flies displayed no detectable neurodegenerative vacuoles compared to WT flies, suggesting that the decrease in locomotor activity was associated with defects in the muscular system (Supplementary Fig. S12). Collectively, the LSM phenotype recapitulated in the cellular and Drosophila models suggested that COASY variants identified in patients were disease drivers.
Meanwhile, to investigate the impact of COASY deficiency on CoA biosynthesis, we selected the hotspot variant, p.Lys371Arg, for functional experiments. Molecular docking (MD) analysis indicated that p.Lys371 is located in the P-loop motif (365–372: GISGSGKS) of enzymatic active pocket in the DPCK domain, which forms the binding site for ATP, suggesting that the p.Lys371Arg variant could directly affect ATP hydrolysis in DPCK (Fig. 1f; Supplementary Fig. S13). HPLC analysis revealed that purified human DPCKK371R was unable to convert dephospho-CoA to CoA in vitro, suggesting that CoA biosynthesis dependent on COASY pathway might be disrupted by the p.Lys371Arg variant (Fig. 1g; Supplementary Fig. S14). Consistently, the level of CoA in the p.Lys342Arg knock-in flies was significantly lower than that in the WT flies (Fig. 1p). These data suggested that COASY variants related to RR-LSM caused insufficient DPCK enzyme activity, leading to an impairment in CoA biosynthesis.
CoA, primarily synthesized intracellularly through the COASY protein, plays an important role as an acyl-group carrier and carbonyl activator in fatty acid metabolism6. Previous studies have shown that a complete loss of COASY was linked to lethal perinatal-onset pontocerebellar hypoplasia type 123, while 5% of COASY preservation was associated with early-onset COASY protein-associated neurodegeneration (CoPAN)2 (Supplementary Table S3). Immunoblotting on biopsied muscles from COASY-related RR-LSM patients with homozygous p.Lys371Arg variant showed a significant reduction in COASY protein levels by approximately half of those in age-matched controls (Fig. 1h, i). The results suggested that the level of residual mutant COASY proteins might be associated with the heterogeneous phenotype of COASY-related disorders.
Subsequently, to investigate the mechanisms underlying the different levels of residual COASY proteins caused by different variants, we conducted protein stability studies on the p.Lys371Arg variant and the CoPAN-associated variant p.Arg499Cys as a positive control7. Under the ff14SB force field simulated by molecular dynamics, the trajectories of the αC atom at residue Arg503 in the catalytic lid domain (LID) of the three systems (DPCKWT, DPCKK371R and DPCKR499C) indicated that variants could influence the flexibility of the LID region and expand the catalytic pocket, ultimately leading to increased instability of the mutant protein (Fig. 1j). Consistently, in vitro experiments of limited digestion by proteinase K demonstrated that the protein level of p.Lys371Arg DPCK exhibited only a slight decrease compared to the WT, while DPCK with p.Arg499Cys was much more susceptible to proteinase K digestion compared to the WT or p.Lys371Arg variant (Fig. 1k, l). These findings suggested that different COASY variants exerted varying impacts on protein stability, consequently contributing to divergent clinical manifestations. Notably, COASY protein also exhibited several other biological functions for mitotic fidelity and DNA damage repair in tumor cells8,9. Furthermore, it was reasonably proposed that in CoPAN patients, the severity of central nervous system symptoms might be profound, potentially overshadowing muscle-related symptoms and associated pathological changes2,3. Collectively, the phenotypes of COASY-related diseases might not solely hinge on residual enzyme activity but also be contingent upon the level of residual COASY protein.
Due to the therapeutic effectiveness of riboflavin for COASY-related LSM, we further examined the mechanism of flavin adenine dinucleotide (FAD) on mutant COASY protein. COASY protein comprises a Rossmann-like fold domain, in which an αβα fold structure can bind to the FAD molecule10. MD scores indicated that FAD could effectively bind to the catalysis pocket of p.Lys371Arg viriant, while the p.Arg499Cys located in the LID region disrupted the conformation of the substrate-binding pocket and blocked the binding of FAD (Supplementary Fig. S15). Consistently, the FAD precursor riboflavin significantly increased the level of COASY proteins in p.Lys371Arg expressing cells, but no detectable effects in p.Arg499Cys expressing cells (Supplementary Fig. S16). Furthermore, riboflavin treatment resulted in a significant reduction in the accumulation of LDs in COASY-knockdown cells under OA overload (Fig. 1m, n). In Ppat-DpckK324R knock-in flies, supplementation with 50 μM riboflavin resulted in a substantial reduction in the accumulation of LDs in muscle, a significant increase in CoA levels, and a marked improvement in motility performance (Fig. 1o, p; Supplementary Fig. S17). These findings suggested that FAD attenuated the clinical phenotype of COASY-related RR-LSM by stabilizing the p.Lys371Arg COASY protein (Fig. 1q). Additionally, considered that multiple flavoproteins play fundamental roles in cellular energy metabolism, riboflavin supplementation has the potential to enhance the conformation of variable flavoproteins and improve their stability. Therefore, longer follow-up and more functional assays are required for the efficacy of riboflavin in these patients.
In summary, we identified biallelic variants in COASY as a new genetic cause in 16 out of 28 genetically unresolved RR-LSM patients. By modeling the loss-of-function variant in cells and Drosophila, we have demonstrated that COASY defects lead to LSM. Importantly, our findings showed that riboflavin could stabilize the mutant COASY protein, offering therapeutic benefits for COASY-related RR-LSM, as well as a potential intervention for COASY-related neurodegenerative diseases.
Supplementary information
COASY variant as a new genetic cause of riboflavin-responsive lipid storage myopathy
Acknowledgements
This work was supported by the National Natural Science Foundation of China (82071409, 82171846, 82271439, U20A20356), the Natural Science Foundation of Jiangxi province (20202BAB206029), Beijing Nova Program (20220484017), the High Quality Clinical Research Project of PKUFH (2023HQ03), and the Double thousand talents program of Jiangxi province (jxsq2019101021).
Author contributions
X.L., J.D., Z.W., and D.H. conceptualized and supervised the study. Y.L.Z. and T.LF. performed the experiments. C.Z., Y.Q., W.Q.Z., Y.L., J.P., and Y.H.Z. analyzed the data. B.W., L.L., W.Z., W.C., C.H.Z., Y.Y., G.W., C.Y., X.L., Z.W., and D.H. performed clinical assessments. Y.L.Z., T.LF., B.W., C.Z., and L.L. wrote the manuscript.
Conflict of interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yilei Zheng, Tongling Liufu, Bing Wen, Chao Zhou, Lingchun Liu
Contributor Information
Xin Lu, Email: luxin_n@163.com.
Jianwen Deng, Email: jianwendeng@pkufh.com.
Zhaoxia Wang, Email: drwangzx@163.com.
Daojun Hong, Email: hongdaojun@hotmail.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41421-023-00641-0.
References
- 1.Aghajanian S, Worrall DM. Biochem. J. 2002;365:13–18. doi: 10.1042/bj20020569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dusi S, et al. Am. J. Hum. Genet. 2014;94:11–22. doi: 10.1016/j.ajhg.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Dijk T, et al. Eur. J. Hum. Genet. 2018;26:1752–1758. doi: 10.1038/s41431-018-0233-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wen B, et al. J. Neurol. Neurosurg. Psychiatry. 2010;81:231–236. doi: 10.1136/jnnp.2009.176404. [DOI] [PubMed] [Google Scholar]
- 5.Olsen RK, et al. Brain. 2007;130:2045–2054. doi: 10.1093/brain/awm135. [DOI] [PubMed] [Google Scholar]
- 6.Theodoulou FL, Sibon OC, Jackowski S, Gout I. Biochem. Soc. Trans. 2014;42:1025–1032. doi: 10.1042/BST20140176. [DOI] [PubMed] [Google Scholar]
- 7.Evers C, et al. Am. J. Med. Genet. A. 2017;173:1878–1886. doi: 10.1002/ajmg.a.38252. [DOI] [PubMed] [Google Scholar]
- 8.Lin CC, et al. Nat. Commun. 2018;9:1039. doi: 10.1038/s41467-018-03422-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrandon S, et al. Cancer Res. 2020;80:334–346. doi: 10.1158/0008-5472.CAN-19-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Longo LM, Hirai H, McGlynn SE. Protein Sci. 2022;31:e4463. doi: 10.1002/pro.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
COASY variant as a new genetic cause of riboflavin-responsive lipid storage myopathy