Abstract
During ribosome biogenesis a plethora of assembly factors and essential enzymes drive the unidirectional maturation of nascent pre-ribosomal subunits. The DEAD-box RNA helicase Dbp10 is suggested to restructure pre-ribosomal rRNA of the evolving peptidyl-transferase center (PTC) on nucleolar ribosomal 60S assembly intermediates. Here, we show that point mutations within conserved catalytic helicase-core motifs of Dbp10 yield a dominant-lethal growth phenotype. Such dbp10 mutants, which stably associate with pre-60S intermediates, impair pre-60S biogenesis at a nucleolar stage prior to the release of assembly factor Rrp14 and stable integration of late nucleolar factors such as Noc3. Furthermore, the binding of the GTPase Nug1 to particles isolated directly via mutant Dbp10 bait proteins is specifically inhibited. The N-terminal domain of Nug1 interacts with Dbp10 and the methyltransferase Spb1, whose pre-60S incorporation is also reduced in absence of functional Dbp10 resulting in decreased methylation of 25S rRNA nucleotide G2922. Our data suggest that Dbp10’s helicase activity generates the necessary framework for assembly factor docking thereby permitting PTC rRNA methylation and the progression of pre-60S maturation.
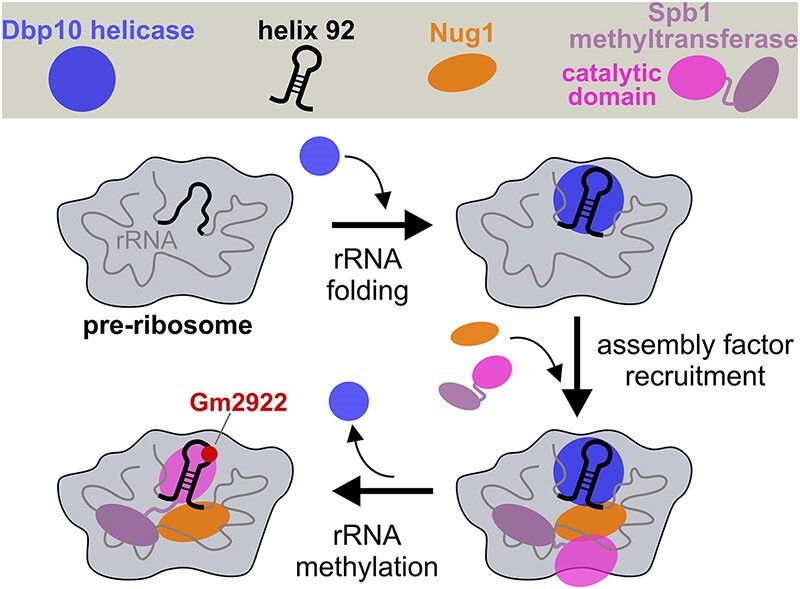
Graphical Abstract
Graphical Abstract.
Introduction
The synthesis of eukaryotic ribosomes is a highly dynamic, complex, and compartmentalized process taking place in the nucleolus, nucleoplasm, and finally in the cytoplasm. The biogenesis pathway requires >200 assembly factors and ∼80 small nucleolar RNAs (snoRNAs) that promote the efficient and failsafe production of translating ribosomes. This involves the spatially and temporally coordinated assembly of 79 ribosomal proteins (r-proteins) with four ribosomal RNAs (rRNAs) going along with rRNA processing steps and the stepwise integration and folding of the distinct rRNA domains into the pre-ribosomal protein core. The current knowledge on ribosome assembly is reviewed in (1–4).
Ribosome biogenesis starts in the nucleolus by the synthesis of a common precursor RNA transcript (the 35S pre-rRNA), which contains the sequences for the mature rRNAs of the small 40S (18S rRNA) and large 60S ribosomal subunits (5.8S and 25S rRNA) (5,6). The third rRNA of the 60S subunit, the 5S rRNA, is transcribed independently by RNA-polymerase III and incorporated into nucleolar 60S precursor particles in complex with r-proteins Rpl5 and Rpl11 (7–11). Upon endonucleolytic pre-rRNA cleavage within the internal-transcribed-spacer element 1 (ITS1) of the 35S pre-rRNA, the 40S and 60S subunits are separated and undergo independent rearrangement and maturation steps facilitated by the functions of successively associating and dissociating ribosomal assembly factors.
In the nucleolus, the first independent precursors of the 60S ribosomal subunit are shaped. These pre-60S particles contain the 27SA2/A3 pre-rRNA, which is initially highly flexible and presumably mainly unfolded (12–15) in contrast to the already more compacted conformation of the subsequent 27SB pre-rRNA species (12,16–18). For further maturation of the 27SB pre-rRNA, the pre-60S association of so-called ‘B-factors’ is required (19–21), which allows endonucleolytic cleavage within internal transcribed spacer element 2 (ITS2) generating the 7S and 26S pre-rRNAs (22–25). During these processing steps, the 60S precursors enter the nucleoplasm, where further remodelling goes along with the association of export factors and nuclear export to the cytoplasm (26–35). In the cytoplasm, final maturation steps and the release of assembly factors are required to produce the mature 60S ribosome (35–44).
The essential DEAD-box RNA helicase Dbp10 is one of the ‘B-factors’ that must associate with 60S precursors to permit 27SB pre-rRNA processing (19,21,45). Accordingly, Dbp10 binds to pre-60S particles at a nucleolar maturation stage and is associated with assembly intermediates isolated via Nsa3, Nug1, or Nsa1/Ytm1 as baits and leaves again before late nucleolar stages (16,46,47). The helicase is genetically linked to the circularly permuted GTPase Nug1 and was shown to directly interact with Nug1 in vitro (46,48). Nug1 depletion results in a failure of Dbp10 assembly into pre-60S particles and in vivo crosslinking revealed that Dbp10 and Nug1 bind at proximal and partially overlapping binding sites in the 27S pre-rRNA (48). Both proteins bind to rRNA regions around the evolving peptidyl-transferase center (PTC) (helices H89-H92 of 25S rRNA domain V), which is the assumed restructuring target of Dbp10. In agreement with this, biochemical and structural analyses of Dbp10’s bacterial homologue DbpA, revealed how the interaction of DbpA with the H89-H92 rRNA stimulates helicase activity to restructure the PTC (49–53). PTC folding by Dbp10 may also be important to allow RNA modifications by the methyltransferases Nop2 and Spb1, which methylate C2870 (in H89) and G2922 (in H92), respectively (54,55). While the G2922 methylation by Spb1, which appears important to facilitate downstream 60S maturation steps (56,57), has occurred on late nucleolar pre-60S particles (58,59), it remains open how the crucial rRNA modifications are coordinated with PTC restructuring by Dbp10 and potentially by the earlier-acting Dbp7 RNA helicase, whose rRNA binding sites overlap with those of Dbp10 (60–62).
Here, we report deeper insights into the function of Dbp10 for nucleolar pre-60S assembly. Yeast strains with mutations of the Dbp10 catalytic RecA-like domains display a dominant-lethal growth-phenotype and effectively inhibit 60S maturation upon their binding. Assembly of the late-joining nucleolar assembly factor Noc3 is reduced on particles isolated from such mutants, whereas the release of upstream factor Rrp14 is impaired. Most significantly, the pre-60S association of the GTPase Nug1 is abolished in catalytic dbp10 mutants and upon Dbp10 depletion, suggesting that the activity of the helicase specifically facilitates Nug1 assembly. Moreover, Dbp10 promotes the final incorporation of the methyltransferase Spb1 permitting methylation of G2922 within PTC helix 92.
Materials and methods
Yeast strains and plasmids
Saccharomyces cerevisiae strains used in this study are listed in Supplementary Table S1 and are derived from the W303 background (63). Strains were constructed using established gene disruption and genomic tagging methods (64,65). For yeast two-hybrid analyses, the reporter strain PJ69-4A was used (66). Plasmids used in this study are listed in Supplementary Table S2 and were constructed using standard DNA cloning techniques and verified by sequencing.
Growth analyses of mutant dbp10 yeast strains
To investigate growth phenotypes of plasmid-derived dbp10 mutant alleles, LEU2 plasmids carrying the respective constructs were transformed into a DBP10 shuffle (dbp10Δ YCplac33-PDBP10-DBP10) strain. Growth phenotypes of the mutant alleles were analysed on plates containing 1 g/l 5-fluoroorotic acid (5-FOA) (Apollo Scientific, Cat# PC4054) to select for cells that have lost the wild-type DBP10-containing URA3 plasmid. The growth phenotypes of viable dbp10 mutants at different temperatures was further analysed on YPD plates.
Yeast two-hybrid analyses
Plasmids expressing the Dbp10 or Spb1 bait protein, N-terminally fused to the GAL4 DNA-binding domain (GAL4-BD), and distinct Nug1 prey protein constructs, C-terminally fused to the GAL4 activation domain (GAL4-AD), were co-transformed into the reporter strain PJ69-4A (66). Yeast two-hybrid interactions were documented by spotting representative transformants in 10-fold serial dilution steps on SDC-Leu-Trp and SDC-Leu-Trp-His (HIS3 reporter gene) plates and incubation at 30°C for 2 and 4 days, respectively.
Fluorescence microscopy
Living W303 NOP58-mCherry yeast cells expressing plasmid-borne N-terminally GFP-tagged wild-type DBP10 or catalytic dbp10 mutant proteins were grown in SDC-Leu medium (supplemented with 5-fold excess of adenine) to the logarithmic growth phase and imaged by fluorescence microscopy using a Zeiss Axioskop microscope.
Affinity purification of pre-ribosomal 60S particles
For two-step affinity purifications from yeast, respective C-terminally Flag-TEV-proteinA (FTpA) or N-terminally TAP-Flag tagged bait proteins were expressed under control of their native promoter. Yeast strains were grown in synthetic dextrose complete (SDC) medium lacking leucine to select for plasmid-derived bait proteins (Figure 3C) or maintenance of plasmid-derived mutants (Figures 2 and 3A, Supplementary Figure S2) at 30°C. For depletion of auxin-inducible degron (AID) tagged chromosomal DBP10, cultures were incubated in the presence of 0.5 mM auxin (3-indoleacetic acid, Sigma-Aldrich, Cat# I2886) for 120 min prior to harvesting the cells. Cells were harvested in the logarithmic growth phase, flash frozen in liquid nitrogen and stored at -20 °C. Cell pellets were resuspended in lysis buffer containing 50 mM Tris–HCl (pH 7.5), 100 mM NaCl, 5 mM MgCl2, 0.05% NP-40, 1 mM DTT, supplemented with 1× SIGMAFAST protease inhibitor (Sigma-Aldrich), and cells were ruptured by shaking in a bead beater (Fritsch) in the presence of glass beads. Lysates were cleared by two subsequent centrifugation steps at 4 °C for 10 and 30 min at 5000 and 15 000 rpm, respectively. Supernatants were incubated with immunoglobulin G (IgG) Sepharose 6 Fast Flow beads (GE Healthcare) on a rotating wheel at 4°C for 90 min. Beads were transferred into Mobicol columns (MoBiTec) and, after washing with 15 ml of lysis buffer (per 2l culture volume), cleavage with tobacco etch virus (TEV) protease was performed at 16°C for 120 min. In a second purification step, TEV eluates were incubated with Flag agarose beads (ANTI-FlagM2 Affinity Gel, Sigma-Aldrich, Cat# A2220) for 75 min at 4°C. After washing with 5 ml of lysis buffer, bound proteins were eluted with lysis buffer containing 300 μg/ml Flag peptide (Sigma-Aldrich, Cat# F3290) at 4°C for 45 min. Flag eluates were analysed by SDS-PAGE on 4–12% polyacrylamide gels (NuPAGE, Invitrogen, Cat# NP0322BOX) with colloidal Coomassie staining (Roti-blue, Carl Roth, Cat# A152.1) or by western blotting with antibodies, as indicated in the respective figures.
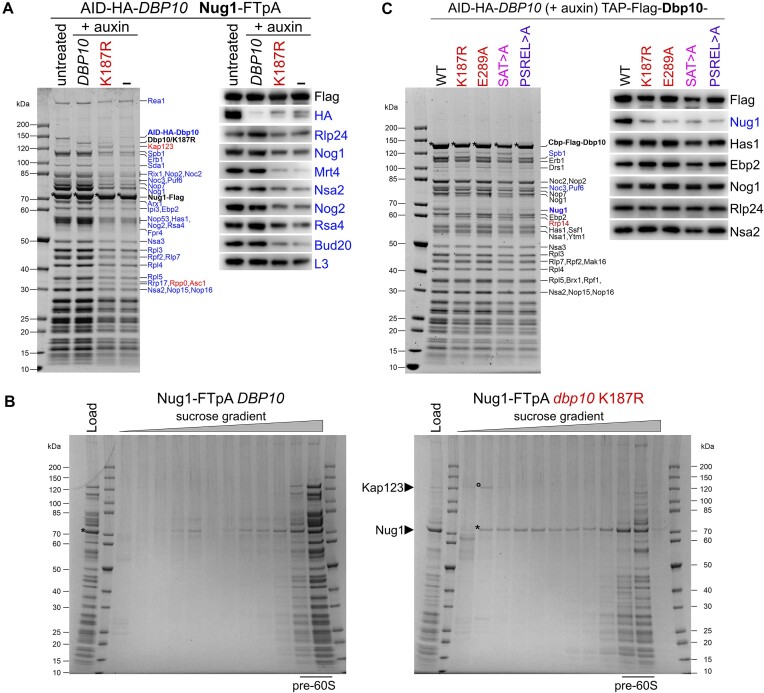
Figure 3.
Catalytic activity of Dbp10 facilitates Nug1 assembly. (A) An AID-HA-DBP10 NUG1-FTpA strain was transformed with plasmids harbouring wild-type DBP10, the dbp10 K187R mutant, or empty vector (–). Subsequently, Nug1 particles were affinity purified from cells in presence of AID-HA-Dbp10 (untreated) or upon its auxin-induced proteasomal degradation (+auxin). Final eluates were analysed by SDS-PAGE and Coomassie staining (left panel) or Western blotting using indicated antibodies (right panel). Depicted Coomassie stained protein bands were identified by mass spectrometry. Bait proteins are marked with an asterisk. Protein bands with increased or decreased intensity in the K187R mutant sample are indicated in red and blue, respectively. (B) Sucrose gradient centrifugation of Nug1 particles affinity purified from cells expressing plasmid-borne wild-type DBP10 or the dbp10 K187R mutant upon auxin induced depletion of AID-HA-Dbp10. After centrifugation, the sucrose gradient was fractionated, and the fractions were TCA-precipitated and analysed together with the input by SDS-PAGE and Coomassie staining. In the dbp10 K187R mutant sample, the importin Kap123 (circle) is present in a ribosome-free fraction together with the Nug1 bait protein (asterisk) (C) An AID-HA-DBP10 strain was transformed with plasmids harbouring TAP-Flag tagged wild-type DBP10 or indicated mutant dbp10 variants under control of their native promoter. Affinity purifications of the plasmid-borne Dbp10 variants were performed upon auxin-induced degradation of the AID-tagged wild-type protein and final eluates were analysed by SDS-PAGE and Coomassie staining (left panel) or western blotting (right panel). The Dbp10 bait proteins are marked with an asterisk and depicted Coomassie stained protein bands were identified by mass spectrometry. Dbp10 mutant variants are colour-coded as in Figure 1.
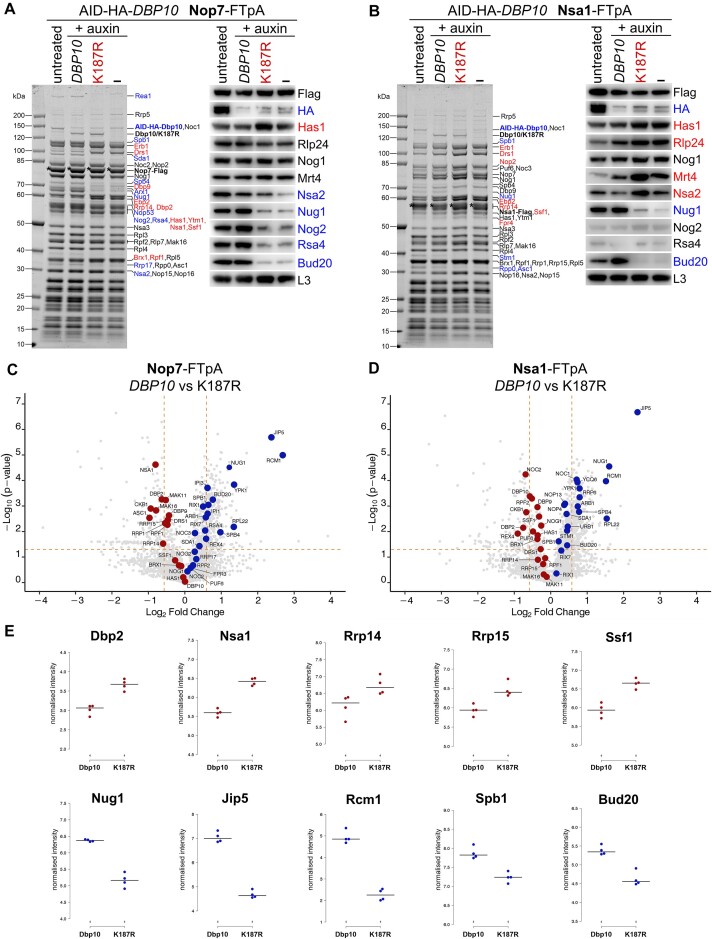
Figure 2.
Purified pre-60S assembly intermediates upon Dbp10 depletion and from dbp10 K187R mutant cells. AID-HA-DBP10 NOP7-FTpA and NSA1-FTpA strains were transformed with plasmids harbouring wild-type DBP10, the dbp10 K187R mutant, or empty vector (–). Subsequently, Nop7 (A) and Nsa1 (B) particles were affinity purified from cells in presence of AID-HA-Dbp10 (untreated) or upon its auxin-induced proteasomal degradation (+auxin). Final eluates were analysed by SDS-PAGE and Coomassie staining (left panels) or Western blotting using indicated antibodies (right panels). Depicted Coomassie stained protein bands were identified by mass spectrometry. Bait proteins are marked with an asterisk. Protein bands with increased or decreased intensity in the K187R mutant samples are indicated in red and blue, respectively. (C–E) Final Nop7 and Nsa1 eluates purified from DBP10 wild-type or K187R mutant cells were analysed by timsTOF mass spectrometry in four biological replicates. Median-normalised label-free quantification (LFQ) values of the co-enriched proteins were log2-transformed and visualized as volcano plots (C and D) or box plots focusing on selected assembly factors from the Nop7 affinity purifications (E). Proteins that were increased or decreased in the K187R mutant vs DBP10 wild-type samples are indicated in red and blue, respectively.
Sucrose gradient centrifugation of Nug1-FTpA eluates
FTpA-tagged Nug1 was affinity purified as described above from cells expressing plasmid-borne wild-type DBP10 or the catalytic dbp10 K187R mutant. The final eluates were loaded onto 7–30% sucrose gradients (w/v) containing 50 mM Tris–HCl (pH 7.5), 100 mM NaCl, 5 mM MgCl2, 0.05% NP-40, 0.5 mM DTT and centrifuged at 27 000 rpm for 16 h at 4°C. The gradients were fractionated into 13 fractions and subsequently precipitated by 15% (final concentration) trichloroacetic acid (TCA). TCA-precipitated proteins were resuspended in SDS sample buffer and analysed by SDS-PAGE on 4–12% polyacrylamide gels with colloidal Coomassie staining.
Split-tag affinity purification of proteinA-Dbp10 and Spb1-flag
A SPB1 shuffle (spb1Δ pRS416-PSPB1-SPB1) proteinA-DBP10 strain was transformed with a plasmid harbouring SPB1 with an internal Flag tag. After plasmid shuffling and selection for cells that have lost the pRS416-SPB1 URA3 plasmid, cells were grown in YPD medium at 30°C and harvested in the logarithmic growth phase. Cell lysis and affinity purification via IgG Sepharose 6 Fast Flow beads (proteinA-Dbp10 pull-down) followed by purification via Flag agarose beads (Spb1-Flag pull-down) were performed as described above. TEV eluates and Flag eluates were analysed by SDS-PAGE on 4–12% polyacrylamide gels with colloidal Coomassie staining. As negative control, the SPB1 shuffle proteinA-DBP10 strain (without Spb1-Flag) was used for the affinity purification.
Mass spectrometry proteomics
Sample processing
Nop7- and Nsa1-FTpA Flag-eluates were processed using S-Trap™ micro columns (Protifi, Cat# C02-micro-80) following the high recovery protocol recommended by the manufacturer and using trypsin (Pierce, Cat# 90059) as the carrier protein. After elution from the columns, the samples were lyophilized and resuspended in 7 μl 0.1% formic acid. 1 μl digest was used per injection.
Mass spectrometry
Samples were analysed on timsTOF ion mobility mass spectrometer (Bruker) in-line with UltiMate 3000 UHPLC system (Thermo). Peptides were separated on a reversed-phase C18 Aurora column (25 cm × 75 μm) with an integrated CaptiveSpray Emitter (IonOpticks, Cat# AUR3-25075C18-CSI). Mobile phases A and B were 0.1 vol% formic acid in water and 0.1 vol% formic acid in ACN, respectively, and the flow rate was 300 nl/min. The fraction of B was linearly increased from 2% to 25% in 90 min, followed by an increase to 40% in 10 min and a further increase to 80% in 10 min and re-equilibration. The spectra were recorded in DIA mode as previously described (67) as duplicates.
Data processing
The DIA data were processed with DIA-NN (DIA-NN 1.8.2 beta 22) (68), using a synthetic library created from UniProt S. cerevisiae protein database (downloaded on 03.07.2023) supplemented with the transgene protein sequences. Precursor charge range was 2–6, while the precursor and fragment m/z ranges were set to 200–2000 and 100–1800, respectively. In addition, trypsin digestion with 1 missed cleavage allowed was specified and cysteine carbamidomethylation (mass delta of 57.021464 Da) was enabled as fixed modification. Methionine oxidation (mass delta of 15.9949 Da) and N-terminal acetylation (mass delta of delta 42.0106) were included as variable modifications, with a maximum number of variable modifications set to 1. Scan window radius was 10 and mass accuracies were fixed to 20 ppm, both for MS1 and MS2. Filetering was performed at 1% FDR.
Data analysis
The Label-Free Quantification (LFQ) values were log2-transformed, median-normalised and the values for each biological replicate was averaged across the two MS runs. After averaging step, the missing values in the IP samples were treated as zeros. The two-sided t-tests for volcano plots were calculated in Perseus (version 2.0.10.0) (69) and visualised using ggVolcanoR (70).
Quantification of 25S rRNA G2922 2′-O-ribose methylation levels
Pre-ribosomal particles were first purified using Nsa1-FTpA as bait from cells lacking snR52 snoRNA and expressing either wild type DBP10 or the dbp10K187R mutant. Briefly, cell pellets corresponding to 2 l cultures at OD600 ∼0.6 were resuspended with approximately one volume of ice-cold A200-KCl buffer (20 mM Tris–HCl, pH 8.0, 5 mM magnesium acetate, 200 mM KCl, 0.2% Triton X-100) supplemented with 1 mM DTT, 1× Complete EDTA-free protease inhibitor cocktail (Roche), 0.1 U/μl RNasin (Promega). Cell suspensions were transferred into 15 ml Precellys lysing tubes prefilled with glass beads (Bertin Technologies, P000961-LYSK0-A) and cells were broken using a Cryolys Evolution cell disruptor (Bertin Instruments), 18 cycles of 30-s shaking at 6000 rpm separated by 30-s rest at 2°C. Extracts were clarified at 20 400 × g for 30 min at 4°C using a Ti50.2 rotor (Beckman) in an Optima XE-100 ultracentrifuge (Beckman Coulter). Soluble extracts were incubated for 2 h at 4°C on a rocking table with 150 μl of IgG Sepharose 6 Fast Flow slurry (Cytiva) previously equilibrated in A200-KCl buffer supplemented with 1 mM DTT. The sepharose beads were washed seven times with 10 ml of ice-cold A200-KCl buffer supplemented with 1 mM DTT. RNAs were extracted from bead pellets as follows: 340 μl of 4 M guanidium isothiocyanate solution, 4 μl of 20 mg/ml glycogen (Roche), 170 μl of [10 mM Tris–HCl, pH 8.0, 1 mM EDTA, pH 8.0, 100 mM sodium acetate], 255 μl of phenol and 255 μl of chloroform were added directly to the bead pellets. Tubes were shaken vigorously for 1 min, incubated 5 min at 65°C, and centrifuged 5 min at 16 000 × g and 4 °C. Aqueous phases (500 μl) were extracted one more time with 250 μl of phenol and 250 μl of chloroform and the resulting samples were precipitated overnight at - 20°C with 1.5 ml 100% ethanol. Final RNA pellets were resuspended with 10 μl ultrapure H2O. For the two-step purification procedure, pre-ribosomal particles bound to IgG-sepharose beads were eluted for 2 h at 16°C using 200 units of AcTEV protease (Invitrogen, 12575–023). The resulting eluates were loaded on anti-FLAG M2 affinity gel (SIGMA, A2220-5ML) for 1 h at 4°C on a rocking table. The beads were washed five times with 10 ml of ice-cold A200-KCl buffer supplemented with 1 mM DTT. RNAs were extracted from bead pellets as described above. RNAs extracted from the Nsa1-FTpA-containing pre-ribosomal particles were hybridized to primer OHA612 (see below) radiolabelled at its 5′ end using T4 polynucleotide kinase (Promega, M4103) and [γ-32P] ATP (Perkin Elmer, NEG502Z). Primer extensions were performed for 1 h at 42°C using AMV reverse transcriptase (Promega, M5101) in the presence of 1 mM (high) or 0.01 mM (low) dNTP concentrations and ribonuclease inhibitor (Promega, N2511). The resulting cDNAs were ethanol precipitated overnight in the presence of 0.1 volume of 3M NaAc and 100 μg of glycogen, and the pellets were dissolved with 20 μl of formamide loading buffer. Samples were denatured for 2 min at 95°C, loaded on 6% acrylamide:bisacrylamide gels containing 8M urea in 1x TBE and resolved by electrophoresis for 2 h in 1× TBE running buffer.
Primer OHA612 (5′-TTGCGGTAACATTCATCAGTAGGG-3′) hybridizes about 60 nucleotides downstream of G2922.
Western blotting
Western blot analysis was performed using the following antibodies: anti-Nog1 antibody (1:5000), anti-Nog2 antibody (1:5000), anti-Nsa2 antibody (1:5000), anti-Rlp24 antibody (1:2000), provided by Micheline Fromont-Racine, anti-Nug1 antibody (1:10 000), anti-Bud20 antibody (1:5000), provided by Vikram Panse, anti-Has1 antibody (1:10 000) provided by Patrick Linder, anti-Ebp2 antibody (1:10 000), provided by Keiko Mizuta, anti-Rpl3 antibody (1:5000), provided by Jonathan Warner, anti-Rsa4 antibody (1:10 000), provided by Miguel Remacha, anti-Mrt4 antibody (1:1000) provided by Juan Pedro Ballesta, horseradish-peroxidase-conjugated anti-Flag antibody (1:10 000; Sigma-Aldrich, Cat# A8592, RRID:AB_439702), horseradish-peroxidase-conjugated anti-HA antibody (1:5000; Roche, Cat# 12013819001; RRID:AB_390917), secondary horseradish-peroxidase-conjugated goat anti-rabbit antibody (1:2000; Bio-Rad, Cat# 166-2408EDU, RRID:AB_11125345), secondary horseradish-peroxidase-conjugated goat anti-mouse antibody (1:2000; Bio-Rad, Cat# STAR105P, RRID:AB_323002).
Results
Lethal and dominant dbp10 mutations of the RecA-like helicase core
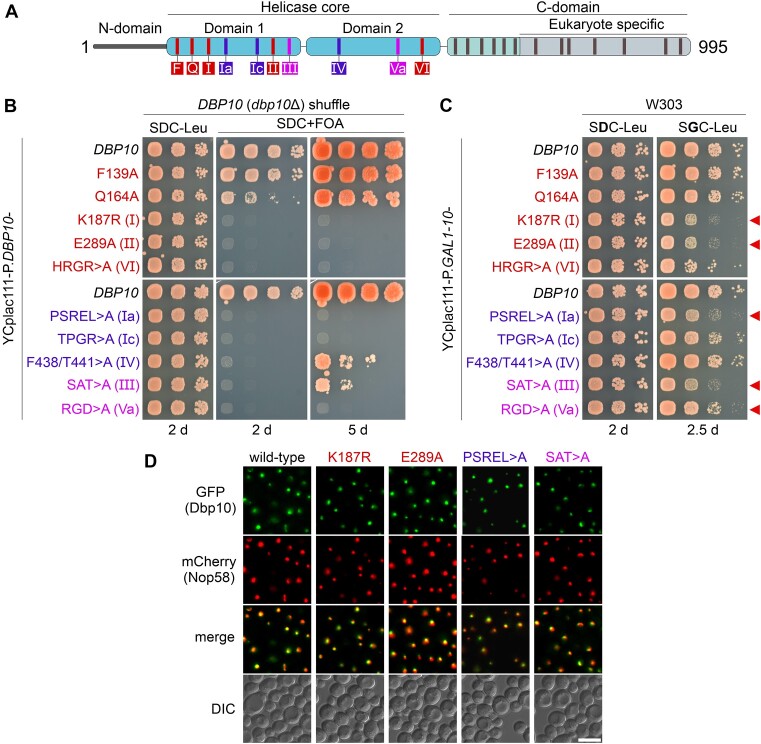
Genetic depletion of the RNA helicase Dbp10 in previous studies revealed that the protein is required for the efficient processing of the 27SB to the 25.5S and 7S pre-rRNAs (21,45). To assess the importance of Dbp10’s catalytic activity for this process, we introduced several point mutations within conserved helicase-core signature motifs suggested to play a role in ATP-binding or hydrolysis, substrate RNA-binding, and communication between the ATP- and RNA-binding sites (71,72) (Figure 1A, Supplementary Figure S1A). Most of the introduced mutations yielded a non-viable or slow-growth phenotype asserting that an intact catalytic core is essential for the protein function (Figure 1B, Supplementary Figure S1B). Moreover, several mutants, including single-point-mutations in the Walker A ATP-binding motif (K187R – motif I) and the Walker B ATP-hydrolysis motif (E289A – motif II) were dominant-negative when over-expressed from a galactose-inducible promoter in a DBP10 wild-type strain (Figure 1C). This is a noteworthy difference to motif I and II mutants of the related DEAD-box helicase Spb4 (59), which were not dominant upon over-expression (Supplementary Figure S1C) (58). As a dominant-negative phenotype indicates functional binding to pre-60S particles, these findings suggest that in contrast to Spb4, ATP-binding and hydrolysis may not be required for an efficient integration of Dbp10 into 60S subunit precursor particles. All tested catalytic Dbp10 mutant variants, similar to the wild-type protein, displayed a predominantly nucleolar localization when N-terminally fused to GFP (Figure 1D), agreeing with a functioning pre-60S incorporation at a nucleolar maturation stage.
Figure 1.
Genetic analyses of catalytic dbp10 mutants. (A) Domain organization of the DEAD-box helicase Dbp10. Conserved motifs involved in ATP-binding/hydrolysis, RNA-binding, and communication between RNA-binding and ATP-binding motifs that were mutated in this study are indicated in red, blue, and pink, respectively. (B) A DBP10 (dbp10Δ) shuffle strain was transformed with plasmids harbouring wild-type DBP10 or indicated dbp10 mutant alleles under transcriptional control of the DBP10 promoter. Transformants were spotted in 10-fold serial dilutions on SDC-Leu or 5-FOA containing plates (SDC + FOA) and growth was monitored after incubation at 30 °C for the indicated days. (C) Over-expression of DBP10 and indicated dbp10 catalytic mutants under the control of the galactose-inducible GAL1-10 promoter. Transformants were spotted in 10-fold serial dilution on plates containing glucose (SDC-Leu; repressed condition) or galactose (SGC-Leu; induced condition) and growth was monitored after incubation at 30°C for the indicated days. Red arrows indicate dominant-negative dbp10 mutants. (D) The subcellular distribution of GFP-tagged wild-type Dbp10 and indicated catalytic Dpp10 mutant variants in yeast cells was monitored by fluorescence microscopy. Co-localization with the nucleolar marker Nop58-mCherrry revealed a predominant nucleolar protein localization of all tested Dbp10 variants. DIC: differential interference contrast. Scale bar is 5 μm.
The helicase activity of Dbp10 promotes nucleolar pre-60S maturation
The dominant-negative growth phenotype of catalytic dbp10 mutants indicated that the mutant proteins could be capable of binding to pre-60S particles and that for a subsequent disassembly the catalytic activity of the helicase is required. To directly assess whether catalytic mutants are associated with pre-60S, we aimed to biochemically analyse nucleolar 60S assembly intermediates derived from cells expressing the dominant-lethal dbp10 K187R allele. Therefore, we constructed yeast strains harbouring Flag-TEV-proteinA (FTpA) tagged nucleolar assembly factors NOP7 or NSA1 in combination with an auxin-inducible degron (AID) tag fused to endogenous DBP10. Upon transformation with plasmids expressing wild-type DBP10, the dbp10 K187R mutant (both under control of the native DBP10 promoter), or an empty plasmid, we affinity-purified Nop7-FTpA and Nsa1-FTpA particles from cells in presence of AID-Dbp10 (untreated) or after its auxin-induced depletion (+auxin) (Figure 2). While the AID-tagged Dbp10 protein was efficiently degraded in presence of auxin (Figure 2A and B), both plasmid-borne wild-type and mutant Dbp10 variants assembled to Nop7 and Nsa1 particles and became co-enriched with similar efficiency, as obvious by a similar intensity of the Coomassie stained Dbp10 bands (Figure 2A and B). In contrast, the pattern of various other assembly factors significantly changed in presence of the K187R mutant or in absence of any Dbp10 protein. To obtain more quantitative and comprehensive insights into the protein composition of the isolated pre-60S particles, we subjected the entire eluates upon Nop7 and Nsa1 affinity-purification to mass spectrometry analyses (in four replicates for each bait protein) (Figure 2C–E). The accumulation of early pre-60S factors such as Ebp2, Rrp14, Dbp9 and Drs1 on Nop7-derived particles suggests that maturation was inhibited before these factors dissociate, whereas the association of late nucleolar Spb4 and Rrp17, as well as downstream nucleoplasmic factors (e.g. Rsa4, Bud20, Sda1) was reduced. Interestingly, also the co-enrichment of the nucleolar methyltransferase Spb1 and the GTPase Nug1 with Nop7 and Nsa1 particles derived from dbp10 K187R mutant cells was reduced (Figure 2A–E). Furthermore, the association of the poorly characterized WD40 repeat protein Jip5 was significantly impaired (Figure 2C–E) indicating that this factor could play a role on nucleolar pre-60S intermediates after Dbp10-dependent PTC structuring. Indeed, the human Jip5 homologue WDR55 was recently visualized bound to a late DDX54 (Dbp10)-containing nucleolar human 60S intermediate chaperoning H57 of 25S rRNA domain III and preventing an inter-domain rRNA interaction with domain VI at this stage (73). The mass spectrometry analyses further revealed a strong decrease of the methyltransferase Rcm1 on Dbp10 K187R containing pre-ribosomes (Figure 2C–E), indicating that Rcm1 is joining downstream of Dbp10’s activity to introduce a m5C modification at C2278 (55).
In agreement with a reduced binding of Nug1 to pre-60S particles associated with Dbp10 K187R, purification of Nug1-FTpA from the dbp10 K187R mutant or from cells in absence of any Dbp10 protein resulted in a clear reduction in the co-enrichment of several pre-60S factors with the Nug1 bait protein (Figure 3A). In addition, the Dbp10 K187R protein band was strongly reduced in the Nug1 eluates and Nug1 appeared to be associated with the importin Kap123 (Figure 3A). Similar to Nug1, the nucleoplasmic/cytoplasmic factor Arx1 did not efficiently co-purify pre-ribosomes, when isolated from dbp10 K187R cells (Supplementary Figure S2). Prompted by the association of the Kap123 importin with Nug1, we analysed Nug1-derived particles by sucrose gradient centrifugation, which allows the separation of pre-ribosomal particles from smaller protein complexes. This further corroborated that in dbp10 K187R mutant cells, Nug1 is less efficiently incorporated into 60S precursors and instead partly associates with Kap123 (Figure 3B). Hence, Kap123, which is also the major importin for ribosomal proteins (74), may promote the nuclear import of Nug1, likely by recognition of a previously described nuclear localization signal (NLS) within the Nug1 N-terminal domain (46).
Taken together, the very strong decrease of Nug1 on pre-60S particles in absence of functional Dbp10 suggests that nucleolar 60S maturation could be stalled shortly before assembly of the GTPase into the particles, a step which might be facilitated by Dbp10.
Nug1 fails to associate with catalytic Dbp10-mutant-derived assembly intermediates
To obtain more insight into the interaction network of Dbp10, we directly affinity-purified N-terminally TAP-Flag-tagged wild-type Dbp10 or catalytic Dbp10 mutant variants upon depletion of the endogenously encoded AID-Dbp10 protein. All plasmid-derived wild-type and mutant Dbp10 bait proteins efficiently co-purified a set of ribosomal assembly factors typical for nucleolar pre-60S assembly intermediates (e.g. Erb1, Ytm1, Nsa1, Has1) (Figure 3C). While the pattern of assembly factors associated with wild-type and mutant particles was in general similar, the amount of late nucleolar Noc3, Puf6 and Spb1 was reduced, whereas Rrp14 was enriched on particles isolated via the mutant Dbp10 variants. Thus, Rrp14, which exhibits an important stabilizing role on nucleolar 60S precursors bridging 25S rRNA domains II and VI (17), may be disassembled dependent on Dbp10’s restructuring activity. This assumption is further supported by recent cryo-EM structures of human pre-60S particles (73) (see Discussion). The most striking difference in comparison of the wild-type and mutant Dbp10 affinity-purifications was the almost complete absence of Nug1 from mutant-derived particles (Figure 3C). This strongly suggests that Dbp10’s catalytic activity is required for the incorporation of Nug1 into 60S precursors.
Dbp10 interacts with the N-terminal domain of Nug1
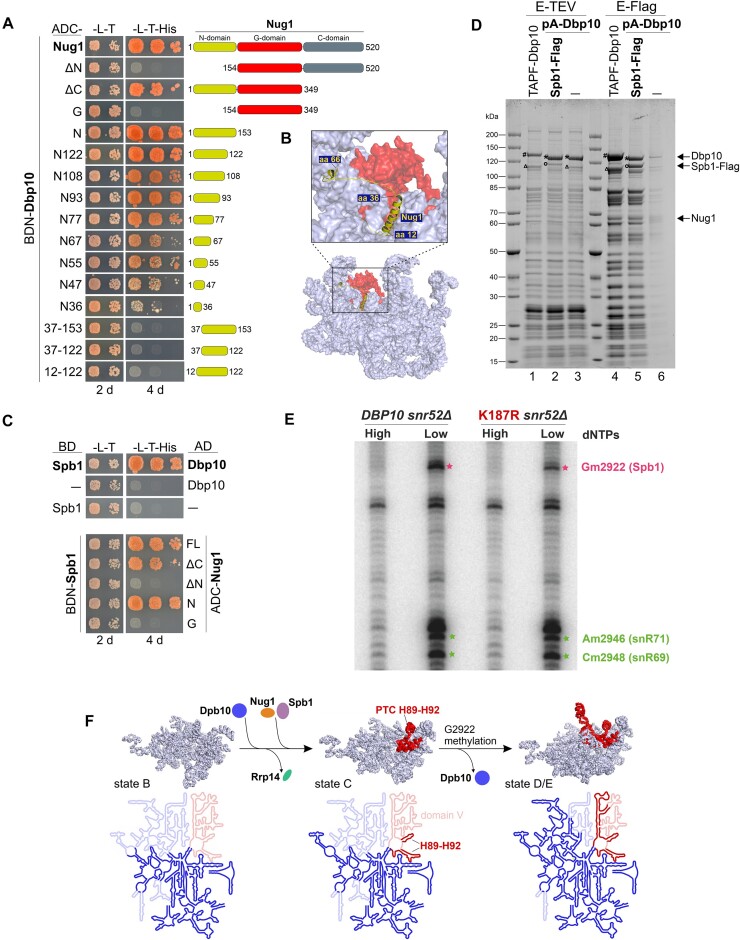
Given the observed importance of Dbp10 for Nug1 assembly, we wanted to find out more about the interplay between these two assembly factors. Since it was previously shown that the two proteins can interact in vitro (48), we performed yeast two-hybrid (Y2H) assays to reveal the Nug1 protein domain responsible for binding to Dbp10. The Y2H analyses confirmed a direct Nug1–Dbp10 interaction and furthermore suggested that the Nug1 N-terminal domain (NTD) (amino acids 1–153) is necessary and sufficient to establish the interaction with the helicase (Figure 4A). It was possible to C-terminally truncate the Nug1 NTD until amino acid 67 (construct N67) before the interaction was reduced, and almost abolished by truncating further (construct N36). Moreover, removal of only the N-terminal eleven Nug1 amino acids completely abolished the interaction, further underscoring the importance of the very N-terminal Nug1 amino acids for Dbp10-binding. The very N-terminal Nug1 region is also the only visible part of the protein on available yeast pre-60S cryo-EM structures bound in proximity to the H89-H92 PTC rRNA (Figure 4B) (16,31,34). According to our Y2H data, both the α-helix (amino acids 12–36) and the following unstructured region (amino acids 37–66) contribute to establish the Nug1–Dbp10 interaction.
Figure 4.
Pre-ribosomal Dbp10–Nug1–Spb1 interaction and G2922 methylation by the Spb1 methyltransferase. (A) Indicated Nug1 constructs fused to the Gal4 activation domain (AD) were tested for a yeast two-hybrid interaction with Dbp10 fused to the Gal4 DNA-binding domain (BD). Cells were spotted in 10-fold serial dilutions on SDC-Leu-Trp (-L-T) and SDC-Leu-Trp-His (-L-T-His) plates and growth was monitored upon incubation at 30°C for two and four days, respectively (B) The interacting N-terminal protein part of Nug1 (yellow) resolved on a nucleoplasmic pre-60S cryo-EM structure (PDB 6YLH). The 25S rRNA is shown as surface representation with PTC rRNA helices H89-H92 coloured in red. (C) Spb1 fused to the Gal4 DNA-binding domain (BD) was tested for a Y2H interaction with Dbp10 (upper panel) and indicated Nug1 constructs (lower panel) fused to the Gal4 activation domain (AD). Cells were spotted in 10-fold serial dilutions on SDC-Leu-Trp (-L-T) and SDC-Leu-Trp-His (-L-T-His) plates and growth was monitored upon incubation at 30°C for the indicated days. (D) Split-tag affinity purification via endogenous proteinA-TEV-Dbp10 (1st purification step) and plasmid-borne Spb1-Flag (2nd purification step). The TEV (proteinA) eluate (lane 2) and Flag eluate (lane 5) were analysed by SDS page and Coomassie staining. As comparison a TAP-Flag-Dbp10 construct (lanes 1 and 4) and proteinA-TEV-Dbp10 alone (lanes 3 and 6; negative control) were used for affinity purifications. Relevant proteins are indicated: #Cbp-Flag-Dbp10, *Dbp10 (after removal of tag by TEV protease), °Spb1-Flag, ΔSpb1 (untagged). (E) Primer extension analyses using RNAs co-purified with Nsa1-FTpA from cells lacking the snR52 snoRNA and expressing DBP10 WT or the dbp10 K187R mutant. Primer extensions were performed using AMV reverse transcriptase at high (1 mM) or low (0.01 mM) dNTP concentration to detect and quantify the methylation levels of G2922 and two neighbouring nucleotides, A2946 and C2948, the ribose of which are methylated by the box C/D snoRNPs snR71 and snR69, respectively. (F) Dbp10-dependent Nug1/Spb1 assembly and PTC H89-H92 rRNA folding. Upper panel: the pre-rRNA (including 25S, 5.8S, and ITS2) of state B (PDB 6EM4), state C (PDB 6EM1), and state E (PDB 6ELZ) pre-60S cryo-EM structures is shown in light blue with 25S domain V rRNA coloured in red. The proposed assembly stage of Dbp10/Nug1/Spb1 to these nucleolar pre-60S intermediates and Rrp14 release, as well as the stage of G2922 methylation are indicated. Lower panel: the 25S rRNA folding state within the depicted particles is illustrated schematically. Already folded/visible rRNA regions are coloured in bright blue/red, unfolded/invisible rRNA regions in faint blue/red. The 25S domain V rRNA including the H89-H92 Dbp10 target is coloured in red.
Dbp10–Nug1–Spb1 interaction on pre-60S intermediates
The methyltransferase Spb1 was shown to methylate the A-site loop nucleotide G2922 of 25S rRNA H92 within the assumed restructuring target region of Dbp10 at the evolving PTC (54); however, the precise timing and pre-requisites to introduce this RNA modification remained unclear. Recent cryo-EM structures showed how Spb1’s methyltransferase domain (MTD) embraces H92 and revealed that G2922 is already methylated on late nucleolar assembly intermediates purified via RNA helicase Spb4 (58,59). Due to the physical proximity of the sites of action of Spb1, Dbp10 and Nug1, we hypothesized that Dbp10 and/or Nug1 might engage in a direct interaction with Spb1. Indeed, Y2H analyses revealed that both Nug1 and Dbp10 interact with Spb1 (Figure 4C) and that the Nug1 N-terminus promotes both, the observed interaction with Dbp10 (Figure 4A), as well as the interaction with Spb1 (Figure 4C, lower panel). Spb1 and Nug1 were also co-enriched with pre-ribosomal intermediates purified directly via Dbp10 as bait protein (Figures 3C and 4D, lane 4) and both proteins were less efficiently assembled in catalytic dbp10 mutants (Figures 2 and 3). The observation of the mutual interactions of Dbp10, Nug1 and Spb1 suggested that all three proteins are bound together to the same pre-60S particles (rather than Dbp10 and Spb1 competing for the same binding region of Nug1) (13). To collect further evidence for this model, we performed a split-tag affinity purification in which in the first step pre-ribosomal particles were purified via Dbp10 and, subsequently, via Spb1 in a second purification step (Figure 4D). Analyses of the final Spb1-Flag eluate clearly revealed a stoichiometric association of Dbp10 with Spb1-containing pre-60S particles (Figure 4D, lane 5). Thus, while the MTD of Spb1 only becomes structurally visible on late nucleolar assembly intermediates where it embraces H92 (58,59), the protein assembles already earlier and at a similar stage as Dbp10 and Nug1. Moreover, Nug1 was co-enriched in the Dbp10/Spb1 split-tag purification, confirming that it is jointly bound to pre-60S particles with Dbp10 and Spb1 (Figure 4D, lane 5).
Dbp10 facilitates the rRNA methylation of G2922 by Spb1
The methyltransferase Spb1 catalyzes 2′-O-ribose-methylation of nucleotide G2922 within H92 of the PTC (54). Since our data suggest that Dbbp10’s activity is required for the efficient assembly of Spb1, we wanted to determine the methylation status of G2922 on pre-ribosomes isolated from the dbp10 K187R helicase mutant. In addition to G2922, Spb1 can methylate the adjacent U2921, which is however in wild-type conditions methylated by the snR52 C/D box snoRNP during an earlier maturation step (54,75). Therefore, we affinity-purified on IgG-sepharose Nsa1-FTpA particles associated with wild-type Dbp10 or the K187R mutant from a snr52Δ strain to unambiguously discriminate for Spb1 activity at the H92 rRNA region by primer extension analyses. After extraction of the co-purified 27S pre-rRNA, primer extension experiments were performed using an oligonucleotide hybridizing about 60 nucleotides downstream of G2922, at high and low dNTP concentrations to reveal the position and modification levels of 2′-O-ribose methylations. These experiments revealed a substantial decrease of G2922 methylation on Nsa1 particles isolated from the dbp10 K187R mutant compared to the wild-type condition (Figure 4E). Signal quantifications (Supplementary Figure S3A and S3B) revealed a close to 50% decrease in the signal corresponding to the 2′-O-methylation of G2922, whereas other non-specific reverse transcription signals in the neighbourhood, used as loading controls, remained little affected. Interestingly, the methylation of two neighbouring nucleotides, A2946 and A2948, also seemed slightly affected in the mutant particles, suggesting that the dbp10 K187R mutant might perturb the methylation of additional sites. We further confirmed these results using pre-ribosomes isolated from wild-type or K187R mutant cells following a two-step purification procedure (IgG-sepharose followed by anti-FLAG affinity gel) instead of the one-step purification shown in Figure 4E. Despite a higher background of non-specific reverse transcription stops, this approach confirmed the decrease in Gm2922 2′-O-methylation levels in cells expressing the dbp10 K187R mutant (Supplementary Figure S3C and S3D).
Taken together, our data reveal that the catalytic activity of Dbp10 is required to allow G2922 methylation by Spb1, which likely occurs upon the release of the helicase and liberation of the accurately folded H92.
Discussion
Based on the data from this study and available pre-60S cryo-EM structures we propose the following timeline for crucial Dbp10-dependent maturation events around the evolving PTC: The H89-H92 rRNA restructuring target of Dbp10 becomes folded and visible at structural state C of nucleolar pre-60S particles (16), hence, the helicase may associate during the transition from state B to state C (Figure 4F). Its helicase activity is essential to allow pre-60S incorporation of Nug1 and the efficient association of the methyltransferase Spb1 around this stage, which in turn methylates nucleotide G2922 within the PTC. Dbp10 and Nug1 have partially overlapping binding sites around the H89-H92 area (48) and, as suggested by the strong pre-60S association of catalytic Dbp10 mutant protein variants, the catalytic RecA domains of Dbp10 may tightly interact with its rRNA substrate until its pre-60S release. Thus, the release of Dbp10 after PTC restructuring would subsequently allow the Spb1 MTD to snap in at the now liberated and folded H92 to methylate G2922 as observed on Spb4/state E structural intermediates (58,59).
In the affinity-purifications utilizing catalytic Dbp10 mutant proteins directly as bait, Nug1 was almost absent, whereas Spb1 and late nucleolar Noc3 were decreased compared to wild-type Dbp10 purifications. In contrast, Rrp14 accumulated on particles isolated via catalytic Dbp10 mutants, implying that the helicase promotes the release of this assembly factor. Our biochemical data are in line with a most recent publication showing cryo-EM structures of a series of nuclear human pre-60S intermediates (73). The structures reveal how the human homologue of Dbp10, DDX54, interacts with PTC H92 on pre-60S particles and acts as a recruitment platform for the FTSJ3 methyltransferase (Spb1) and the Noc2–Noc3 complex. Furthermore, the structures unveil that, besides PTC restructuring, DDX54 in a second function remodels root helices of 25S rRNA domains III and IV. The rearrangement of domain IV H61 thereby disrupts the binding site of the SURF6 (Rrp14)–RRP15–SSF1 module illuminating the observed Rrp14 accumulation on pre-60S particles lacking functional Dbp10. Moreover, on human pre-60S structures, the long C-terminal Rrp14 α-helix occupies the binding site of a C-terminal Spb1 segment, further explaining the inefficient pre-60S association of the methyltransferase in dbp10 mutants and block of G2922 methylation.
While our study reveals that the activity of Dbp10 on pre-60S intermediates is a pre-requisite for GTPase Nug1 assembly, it was previously shown that the presence of Nug1 appears vice versa required for Dbp10 and also Spb1 assembly (48). Thus, while further investigations are needed to illuminate these reciprocal interdependencies, this demonstrates the complex interplay of assembly factors during highly dynamic ribosomal maturation events in order to shape crucial rRNA regions such as the PTC.
Supplementary Material
Acknowledgements
We thank Petra Ihrig and Jürgen Reichert from the BZH Heidelberg MS facility for performing mass spectrometry analyses and Helmut Bergler and Brigitte Pertschy for critical reading of the manuscript.
Author contributions: Valentin Mitterer: Funding acquisition; Conceptualization; Methodology; Validation; Formal analysis; Investigation; Resources; Data curation; Visualization; Writing – original draft preparation; Writing – review & editing, Hussein Hamze: Methodology; Validation; Formal analysis; Data curation; Visualization; Writing – review & editing, Natalia Kunowska: Methodology; Validation; Formal analysis; Data curation; Visualization; Writing – review & editing, Ulrich Stelzl: Methodology; Validation; Writing – review & editing, Anthony Henras: Funding acquisition; Methodology; Validation; Formal analysis; Data curation; Writing – review & editing, Ed Hurt: Funding acquisition; Writing – review & editing.
Contributor Information
Valentin Mitterer, Biochemistry Center, University of Heidelberg, 69120 Heidelberg, Germany; Institute of Molecular Biosciences, University of Graz, 8010 Graz, Austria.
Hussein Hamze, Molecular, Cellular and Developmental Biology Unit (MCD), Center for Integrative Biology (CBI), CNRS, University of Toulouse, 31062 Toulouse, France.
Natalia Kunowska, Institute of Pharmaceutical Sciences, Pharmaceutical Chemistry, University of Graz, 8010 Graz, Austria.
Ulrich Stelzl, Institute of Pharmaceutical Sciences, Pharmaceutical Chemistry, University of Graz, 8010 Graz, Austria; BioTechMed-Graz, 8010 Graz, Austria; Field of Excellence BioHealth, University of Graz, 8010 Graz, Austria.
Anthony K Henras, Molecular, Cellular and Developmental Biology Unit (MCD), Center for Integrative Biology (CBI), CNRS, University of Toulouse, 31062 Toulouse, France.
Ed Hurt, Biochemistry Center, University of Heidelberg, 69120 Heidelberg, Germany.
Data availability
The data underlying this article are available in the article and in its online supplementary material. Uncropped gels and blots are provided in Supplementary File 1. Yeast strains and plasmids are available from the corresponding author upon request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (76) partner repository with the dataset identifier PXD046068.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Austrian Science Fund [P37114 to V.M.]; German Research Council [HU363/15-2]; European Research Council [ADG 741781 GLOWSOME to E.H.]; ANR PRC grant [REA1COM to A.K.H.]; MS-proteomics was supported by the Field of Excellence BioHealth – University of Graz. Funding for open access charge: Austrian Science Fund (FWF).
Conflict of interest statement. None declared.
References
- 1. Baßler J., Hurt E. Eukaryotic ribosome assembly. Annu. Rev. Biochem. 2019; 88:281–306. [DOI] [PubMed] [Google Scholar]
- 2. de la Cruz J., Karbstein K., Woolford J.L. Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu. Rev. Biochem. 2015; 84:93–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Klinge S., Woolford J.L. Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 2019; 20:116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kressler D., Hurt E., Baßler J. A puzzle of life: crafting ribosomal subunits. Trends Biochem. Sci. 2017; 42:640–654. [DOI] [PubMed] [Google Scholar]
- 5. Fernández-Pevida A., Kressler D., de la Cruz J. Processing of preribosomal RNA in Saccharomyces cerevisiae. Wiley Interdiscip Rev. RNA. 2015; 6:191–209. [DOI] [PubMed] [Google Scholar]
- 6. Turowski T.W., Tollervey D. Cotranscriptional events in eukaryotic ribosome synthesis. Wiley Interdiscip Rev. RNA. 2015; 6:129–139. [DOI] [PubMed] [Google Scholar]
- 7. Calviño F.R., Kharde S., Ori A., Hendricks A., Wild K., Kressler D., Bange G., Hurt E., Beck M., Sinning I. Symportin 1 chaperones 5S RNP assembly during ribosome biogenesis by occupying an essential rRNA-binding site. Nat. Commun. 2015; 6:6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Castillo Duque de Estrada N.M., Thoms M., Flemming D., Hammaren H.M., Buschauer R., Ameismeier M., Baßler J., Beck M., Beckmann R., Hurt E. Structure of nascent 5S RNPs at the crossroad between ribosome assembly and MDM2-p53 pathways. Nat. Struct. Mol. Biol. 2023; 30:1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kharde S., Calviño F.R., Gumiero A., Wild K., Sinning I. The structure of Rpf2-Rrs1 explains its role in ribosome biogenesis. Nucleic Acids Res. 2015; 43:7083–7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kressler D., Bange G., Ogawa Y., Stjepanovic G., Bradatsch B., Pratte D., Amlacher S., Strauß D., Yoneda Y., Katahira J. et al. Synchronizing nuclear import of ribosomal proteins with ribosome assembly. Science. 2012; 338:666–671. [DOI] [PubMed] [Google Scholar]
- 11. Madru C., Lebaron S., Blaud M., Delbos L., Pipoli J., Pasmant E., Réty S., Leulliot N. Chaperoning 5S RNA assembly. Genes Dev. 2015; 29:1432–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Burlacu E., Lackmann F., Aguilar L.-C., Belikov S., Nues R.v., Trahan C., Hector R.D., Dominelli-Whiteley N., Cockroft S.L., Wieslander L. et al. High-throughput RNA structure probing reveals critical folding events during early 60S ribosome assembly in yeast. Nat. Commun. 2017; 8:714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ismail S., Flemming D., Thoms M., Gomes-Filho J.V., Randau L., Beckmann R., Hurt E. Emergence of the primordial pre-60S from the 90S pre-ribosome. Cell Rep. 2022; 39:110640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Joret C., Capeyrou R., Belhabich-Baumas K., Plisson-Chastang C., Ghandour R., Humbert O., Fribourg S., Leulliot N., Lebaron S., Henras A.K. et al. The Npa1p complex chaperones the assembly of the earliest eukaryotic large ribosomal subunit precursor. PLoS Genet. 2018; 14:e1007597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sanghai Z.A., Piwowarczyk R., Broeck A.V., Klinge S. A co-transcriptional ribosome assembly checkpoint controls nascent large ribosomal subunit maturation. Nat. Struct. Mol. Biol. 2023; 30:594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kater L., Thoms M., Barrio-Garcia C., Cheng J., Ismail S., Ahmed Y.L., Bange G., Kressler D., Berninghausen O., Sinning I. et al. Visualizing the assembly pathway of nucleolar pre-60S ribosomes. Cell. 2017; 171:1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sanghai Z.A., Miller L., Molloy K.R., Barandun J., Hunziker M., Chaker-Margot M., Wang J., Chait B.T., Klinge S. Modular assembly of the nucleolar pre-60S ribosomal subunit. Nature. 2018; 556:126–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou D., Zhu X., Zheng S., Tan D., Dong M.-Q., Ye K. Cryo-EM structure of an early precursor of large ribosomal subunit reveals a half-assembled intermediate. Protein Cell. 2018; 10:120–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gamalinda M., Jakovljevic J., Babiano R., Talkish J., de la Cruz J., Woolford J.L. Yeast polypeptide exit tunnel ribosomal proteins L17, L35 and L37 are necessary to recruit late-assembling factors required for 27SB pre-rRNA processing. Nucleic Acids Res. 2013; 41:1965–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pöll G., Braun T., Jakovljevic J., Neueder A., Jakob S., Woolford J.L., Tschochner H., Milkereit P. rRNA maturation in yeast cells depleted of large ribosomal subunit proteins. PLoS One. 2009; 4:e8249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Talkish J., Zhang J., Jakovljevic J., Horsey E.W., Woolford J.L. Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res. 2012; 40:8646–8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fang F., Phillips S., Butler J.S. Rat1p and Rai1p function with the nuclear exosome in the processing and degradation of rRNA precursors. RNA. 2005; 11:1571–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fromm L., Falk S., Flemming D., Schuller J.M., Thoms M., Conti E., Hurt E. Reconstitution of the complete pathway of ITS2 processing at the pre-ribosome. Nat. Commun. 2017; 8:1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gasse L., Flemming D., Hurt E. Coordinated ribosomal ITS2 RNA processing by the Las1 complex integrating endonuclease, polynucleotide kinase, and exonuclease activities. Mol. Cell. 2015; 60:808–815. [DOI] [PubMed] [Google Scholar]
- 25. Schillewaert S., Wacheul L., Lhomme F., Lafontaine D.L.J. The evolutionarily conserved protein Las1 is required for pre-rRNA processing at both ends of ITS2. Mol. Cell. Biol. 2012; 32:430–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Altvater M., Chang Y., Melnik A., Occhipinti L., Schütz S., Rothenbusch U., Picotti P., Panse V.G. Targeted proteomics reveals compositional dynamics of 60S pre-ribosomes after nuclear export. Mol. Syst. Biol. 2012; 8:628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bassler J., Klein I., Schmidt C., Kallas M., Thomson E., Wagner M.A., Bradatsch B., Rechberger G., Strohmaier H., Hurt E. et al. The conserved Bud20 zinc finger protein is a new component of the ribosomal 60S subunit export machinery. Mol. Cell. Biol. 2012; 32:4898–4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gadal O., Strauss D., Kessl J., Trumpower B., Tollervey D., Hurt E. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 2001; 21:3405–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ho J.H., Kallstrom G., Johnson A.W. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 2000; 151:1057–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hung N.-J., Lo K.-Y., Patel S.S., Helmke K., Johnson A.W. Arx1 is a nuclear export receptor for the 60S ribosomal subunit in yeast. Mol. Biol. Cell. 2008; 19:735–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kater L., Mitterer V., Thoms M., Cheng J., Berninghausen O., Beckmann R., Hurt E. Construction of the central protuberance and L1 stalk during 60S subunit biogenesis. Mol. Cell. 2020; 79:615–628. [DOI] [PubMed] [Google Scholar]
- 32. Liang X., Zuo M.-Q., Zhang Y., Li N., Ma C., Dong M.-Q., Gao N. Structural snapshots of human pre-60S ribosomal particles before and after nuclear export. Nat. Commun. 2020; 11:3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Matsuo Y., Granneman S., Thoms M., Manikas R.-G., Tollervey D., Hurt E. Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export. Nature. 2014; 505:112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu S., Tutuncuoglu B., Yan K., Brown H., Zhang Y., Tan D., Gamalinda M., Yuan Y., Li Z., Jakovljevic J. et al. Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature. 2016; 534:133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhou Y., Musalgaonkar S., Johnson A.W., Taylor D.W. Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome. Nat. Commun. 2019; 10:958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Greber B.J., Gerhardy S., Leitner A., Leibundgut M., Salem M., Boehringer D., Leulliot N., Aebersold R., Panse V.G., Ban N. Insertion of the biogenesis factor Rei1 probes the ribosomal tunnel during 60S maturation. Cell. 2016; 164:91–102. [DOI] [PubMed] [Google Scholar]
- 37. Kappel L., Loibl M., Zisser G., Klein I., Fruhmann G., Gruber C., Unterweger S., Rechberger G., Pertschy B., Bergler H. Rlp24 activates the AAA-ATPase Drg1 to initiate cytoplasmic pre-60S maturation. J. Cell Biol. 2012; 199:771–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kargas V., Castro-Hartmann P., Escudero-Urquijo N., Dent K., Hilcenko C., Sailer C., Zisser G., Marques-Carvalho M.J., Pellegrino S., Wawiórka L. et al. Mechanism of completion of peptidyltransferase centre assembly in eukaryotes. eLife. 2019; 8:e44904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Klingauf-Nerurkar P., Gillet L.C., Portugal-Calisto D., Oborská-Oplová M., Jäger M., Schubert O.T., Pisano A., Peña C., Rao S., Altvater M. et al. The GTPase Nog1 co-ordinates the assembly, maturation and quality control of distant ribosomal functional centers. eLife. 2020; 9:e52474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lo K.-Y., Li Z., Bussiere C., Bresson S., Marcotte E.M., Johnson A.W. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol. Cell. 2010; 39:196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ma C., Wu S., Li N., Chen Y., Yan K., Li Z., Zheng L., Lei J., Woolford J.L., Gao N. Structural snapshot of cytoplasmic pre-60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1. Nat. Struct. Mol. Biol. 2017; 24:214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Malyutin A.G., Musalgaonkar S., Patchett S., Frank J., Johnson A.W. Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis. EMBO J. 2017; 36:854–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Prattes M., Grishkovskaya I., Hodirnau V.-V., Hetzmannseder C., Zisser G., Sailer C., Kargas V., Loibl M., Gerhalter M., Kofler L. et al. Visualizing maturation factor extraction from the nascent ribosome by the AAA-ATPase Drg1. Nat. Struct. Mol. Biol. 2022; 29:942–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Weis F., Giudice E., Churcher M., Jin L., Hilcenko C., Wong C.C., Traynor D., Kay R.R., Warren A.J. Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat. Struct. Mol. Biol. 2015; 22:914–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Burger F., Daugeron M.C., Linder P. Dbp10p, a putative RNA helicase from Saccharomyces cerevisiae, is required for ribosome biogenesis. Nucleic Acids Res. 2000; 28:2315–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bassler J., Kallas M., Hurt E. The NUG1 GTPase reveals and N-terminal RNA-binding domain that is essential for association with 60 S pre-ribosomal particles. J. Biol. Chem. 2006; 281:24737–24744. [DOI] [PubMed] [Google Scholar]
- 47. Nissan T.A., Bassler J., Petfalski E., Tollervey D., Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002; 21:5539–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Manikas R.-G., Thomson E., Thoms M., Hurt E. The K+-dependent GTPase Nug1 is implicated in the association of the helicase Dbp10 to the immature peptidyl transferase centre during ribosome maturation. Nucleic Acids Res. 2016; 44:1800–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hardin J.W., Hu Y.X., McKay D.B. Structure of the RNA binding domain of a DEAD-box helicase bound to its ribosomal RNA target reveals a novel mode of recognition by an RNA recognition motif. J. Mol. Biol. 2010; 402:412–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Karginov F.V., Uhlenbeck O.C. Interaction of Escherichia coli DbpA with 23S rRNA in different functional states of the enzyme. Nucleic Acids Res. 2004; 32:3028–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nicol S.M., Fuller-Pace F.V. The ‘DEAD box’ protein DbpA interacts specifically with the peptidyltransferase center in 23S rRNA. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:11681–11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tsu C.A., Kossen K., Uhlenbeck O.C. The Escherichia coli DEAD protein DbpA recognizes a small RNA hairpin in 23S rRNA. 2001; 7:702–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wurm J.P., Glowacz K.-A., Sprangers R. Structural basis for the activation of the DEAD-box RNA helicase DbpA by the nascent ribosome. Proc. Natl. Acad. Sci. USA. 2021; 118:e2105961118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lapeyre B., Purushothaman S.K. Spb1p-directed formation of Gm2922 in the ribosome catalytic center occurs at a late processing stage. Mol. Cell. 2004; 16:663–669. [DOI] [PubMed] [Google Scholar]
- 55. Sharma S., Yang J., Watzinger P., Kötter P., Entian K.-D. Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res. 2013; 41:9062–9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sekulski K., Cruz V.E., Weirich C.S., Erzberger J.P. rRNA methylation by Spb1 regulates the GTPase activity of Nog2 during 60S ribosomal subunit assembly. Nat. Commun. 2023; 14:1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yelland J.N., Bravo J.P.K., Black J.J., Taylor D.W., Johnson A.W. A single 2’-O-methylation of ribosomal RNA gates assembly of a functional ribosome. Nat. Struct. Mol. Biol. 2023; 30:91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cruz V.E., Sekulski K., Peddada N., Sailer C., Balasubramanian S., Weirich C.S., Stengel F., Erzberger J.P. Sequence-specific remodeling of a topologically complex RNP substrate by Spb4. Nat. Struct. Mol. Biol. 2022; 29:1228–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mitterer V., Thoms M., Buschauer R., Berninghausen O., Hurt E., Beckmann R. Concurrent remodeling of nucleolar 60S subunit precursors by the Rea1 ATPase and Spb4 RNA helicase. eLife. 2023; 12:e84877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Aquino G.R.R., Hackert P., Krogh N., Pan K.-T., Jaafar M., Henras A.K., Nielsen H., Urlaub H., Bohnsack K.E., Bohnsack M.T. The RNA helicase Dbp7 promotes domain V/VI compaction and stabilization of inter-domain interactions during early 60S assembly. Nat. Commun. 2021; 12:6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jaafar M., Contreras J., Dominique C., Martín-Villanueva S., Capeyrou R., Vitali P., Rodríguez-Galán O., Velasco C., Humbert O., Watkins N.J. et al. Association of snR190 snoRNA chaperone with early pre-60S particles is regulated by the RNA helicase Dbp7 in yeast. Nat. Commun. 2021; 12:6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mitterer V., Pertschy B. RNA folding and functions of RNA helicases in ribosome biogenesis. RNA Biol. 2022; 19:781–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Thomas B.J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989; 56:619–630. [DOI] [PubMed] [Google Scholar]
- 64. Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E. et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004; 21:947–962. [DOI] [PubMed] [Google Scholar]
- 65. Longtine M.S., McKenzie A., Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998; 14:953–961. [DOI] [PubMed] [Google Scholar]
- 66. James P., Halladay J., Craig E.A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996; 144:1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Meier F., Brunner A.D., Frank M., Ha A., Bludau I., Voytik E., Kaspar-Schoenefeld S., Lubeck M., Raether O., Bache N. et al. diaPASEF: parallel accumulation–serial fragmentation combined with data-independent acquisition. Nat. Methods. 2020; 17:1229–1236. [DOI] [PubMed] [Google Scholar]
- 68. Demichev V., Messner C.B., Vernardis S.I., Lilley K.S., Ralser M. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods. 2020; 17:41–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016; 13:731–740. [DOI] [PubMed] [Google Scholar]
- 70. Mullan K.A., Bramberger L.M., Munday P.R., Goncalves G., Revote J., Mifsud N.A., Illing P.T., Anderson A., Kwan P., Purcell A.W. et al. ggVolcanoR: a Shiny app for customizable visualization of differential expression datasets. Comput. Struct. Biotechnol. J. 2021; 19:5735–5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Granneman S., Bernstein K.A., Bleichert F., Baserga S.J. Comprehensive mutational analysis of yeast DEXD/H box RNA helicases required for small ribosomal subunit synthesis. Mol. Cell. Biol. 2006; 26:1183–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Linder P., Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011; 12:505–516. [DOI] [PubMed] [Google Scholar]
- 73. Vanden Broeck A., Klinge S. Principles of human pre-60S biogenesis. Science. 2023; 381:eadh3892. [DOI] [PubMed] [Google Scholar]
- 74. Rout M.P., Blobel G., Aitchison J.D. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997; 89:715–725. [DOI] [PubMed] [Google Scholar]
- 75. Bonnerot C., Pintard L., Lutfalla G. Functional redundancy of Spb1p and a snR52-dependent mechanism for the 2’-O-ribose methylation of a conserved rRNA position in yeast. Mol. Cell. 2003; 12:1309–1315. [DOI] [PubMed] [Google Scholar]
- 76. Perez-Riverol Y., Bai J., Bandla C., García-Seisdedos D., Hewapathirana S., Kamatchinathan S., Kundu D.J., Prakash A., Frericks-Zipper A., Eisenacher M. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022; 50:D543–D552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material. Uncropped gels and blots are provided in Supplementary File 1. Yeast strains and plasmids are available from the corresponding author upon request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (76) partner repository with the dataset identifier PXD046068.