Abstract
Human transportin1 (hTRN1) is the nuclear import receptor for a group of pre-mRNA/mRNA-binding proteins (heterogeneous nuclear ribonucleoproteins [hnRNP]) represented by hnRNP A1, which shuttle continuously between the nucleus and the cytoplasm. hTRN1 interacts with the M9 region of hnRNP A1, a 38-amino-acid domain rich in Gly, Ser, and Asn, and mediates the nuclear import of M9-bearing proteins in vitro. Saccharomyces cerevisiae transportin (yTRN; also known as YBR017c or Kap104p) has been identified and cloned. To understanding the nuclear import mediated by yTRN, we searched with a yeast two-hybrid system for proteins that interact with it. In an exhaustive screen of the S. cerevisiae genome, the most frequently selected open reading frame was the nuclear mRNA-binding protein, Nab2p. We delineated a ca.-50-amino-acid region in Nab2p, termed NAB35, which specifically binds yTRN and is similar to the M9 motif. NAB35 also interacts with hTRN1 and functions as a nuclear localization signal in mammalian cells. Interestingly, yTRN can also mediate the import of NAB35-bearing proteins into mammalian nuclei in vitro. We also report on additional substrates for TRN as well as sequences of Drosophila melanogaster, Xenopus laevis, and Schizosaccharomyces pombe TRNs. Together, these findings demonstrate that both the M9 signal and the nuclear import machinery utilized by the transportin pathway are conserved in evolution.
The heterogeneous nuclear ribonucleoprotein (hnRNP) complexes comprise a group of more than 20 abundant proteins, designated A through U, that associate with nascent RNA polymerase II transcripts and remain associated as hnRNP complexes during subsequent nuclear RNA processing reactions (10). Among them, hnRNP A1 is one of the best characterized. A1 binds with high affinity to RNA sequences that resemble pre-mRNA 3′ and 5′ splice sites (6, 37) and strongly influences pre-mRNA alternative splicing (7, 13, 23, 45). One of the most intriguing properties of A1 is that it shuttles rapidly between the nucleus and the cytoplasm in a transcription-dependent manner (31, 32). While in the cytoplasm, A1 is also bound to poly(A)+ RNA (32), and immunoelectron microscopy has shown that hnRNP A1/A2-type proteins are bound to mRNAs while the mRNAs translocate from the nucleus to the cytoplasm through the nuclear pore complexes (NPCs) (40). A1 is therefore likely to play an important role in the export of mRNAs from the nucleus (10, 40). Consistent with this, nuclear microinjections of hnRNP A1 in Xenopus laevis oocytes inhibit mRNA export (17).
The signal within hnRNP A1 that mediates its nuclear import is a 38-amino-acid domain, termed M9, near the carboxyl terminus of the protein (34, 43). M9 is a nuclear transport signal distinct from the classical nuclear localization signals (NLSs) (9), since it does not contain any stretches of basic residues characteristic of classical NLSs and functions as a nuclear export signal (NES) of A1 (24). Interestingly, M9 itself is a transcription-dependent nuclear transport signal, since pyruvate kinase (PK) fused to M9 accumulates in the cytoplasm after treatment with an RNA polymerase II inhibitor, actinomycin D, as does hnRNP A1 (35).
Nuclear import of classical NLS-bearing proteins is mediated by importin α/β heterodimers (14, 29). The nuclear import of hnRNP A1 does not utilize the importin-mediated pathway (33). Rather, human transportin1 (hTRN1), a 90-kDa protein distantly related (24% amino acid identity) to human importin β, is the nuclear import receptor for M9-bearing proteins (5, 11, 33). hTRN1 interacts directly with M9 without the necessity for an adapter equivalent to importin α, as is the case for the import pathway of classical NLS-bearing proteins (26). hTRN1 binds RanGTP through its N-terminal domain with a similar affinity to that with which importin β binds RanGTP (4, 4a), although hTRN1 does not have an obvious consensus Ran-binding motif that is present in the other importin β-related receptors (8, 15). RanGTP binding to hTRN1 causes the dissociation of A1 from hTRN1 in vitro (18, 35), a process that most likely occurs in the nucleus after translocation through NPCs, since the nucleus apparently contains high levels of RanGTP, while the cytoplasm does not. Once A1 is released from hTRN1 upon nuclear entry, it is incorporated into hnRNP complexes, where A1 serves in pre-mRNA metabolism (35). Very recently, the bidirectional translocation of hTRN1 itself through NPCs was investigated. Both nuclear import and export of hTRN1 were found to be temperature-dependent processes that do not require GTP hydrolysis, and the C-terminal region of hTRN1 was found to be an independent nuclear import module (28).
The transportin homolog in Saccharomyces cerevisiae, yeast transportin (yTRN) (33), also known as Kap104p (2), is the most closely related protein to hTRN1 (35% identity) in S. cerevisiae (26). To understand the nuclear import mediated by yTRN, we performed an exhaustive genomic two-hybrid screen, using yTRN as a bait. Among several selected targets, the S. cerevisiae nuclear RNA-binding protein Nab2p was the most highly represented. Nab2p is one of the major nuclear proteins associated with nuclear poly(A)+ RNA in this organism and contains at least two distinct types of RNA-binding motifs, an RGG box and CCCH motif repeats (3). Nab2p has been previously shown to be a substrate for yTRN (Kap104p) (2). However, the specific sequence which mediates the nuclear import of Nab2p has not been determined. In this study, we delineated a region in Nab2p, termed NAB35, which specifically interacts with yTRN in vitro. Interestingly, hTRN1 can also interact with NAB35 and can mediate its nuclear import in a mammalian import assay system. Moreover, yTRN mediates the nuclear import of NAB35-bearing proteins in the same system, indicating that the nuclear import machinery utilized by TRN is conserved in evolution. We also further define the transportin receptor family by alignment of known TRN amino acid sequences (human and S. cerevisiae) with two additional amino acid sequences we have now determined from X. laevis and Drosophila melanogaster and with a Schizosaccharomyces pombe protein sequence present in databases.
MATERIALS AND METHODS
Yeast two-hybrid screening.
The bait plasmids pLexA/yTRN and pGAL4/yTRN were constructed by cloning a BamHI/PstI yTRN fragment from pGBT9 (Clontech) into pBTM116 (41) and pAS2DD (12), respectively. The reading frame of the fusion protein was verified by sequencing.
Two-hybrid screens were performed by a mating strategy with either pGAL4/yTRN-transformed CG1945 cells or pLexA/yTRN-transformed L40 cells and the Y187 cells transformed with the FRYL library (12). The FRYL library contains over 3.5 million genomic inserts. His+ colonies were selected, restreaked twice on plates, and stored. Each library plasmid insert was amplified by PCR on yeast cells (42). The lengths of inserts were estimated on agarose gels, and the genomic location of fragments was precisely determined by sequencing. Identification in the yeast genome was performed by the BLAST program, the Saccharomyces Genome Database (http://genome-www.stanford.edu), and the Yeast Protein Database (http://www.proteome.com).
Plasmid construction.
The plasmids employed for in vitro transcription-translation of myc-PK, myc-PK-M9, hTRN1, and importin β were described previously (33–35). The plasmid myc-A1 was constructed by performing PCR to introduce EcoRI and XhoI sites at the 5′ and 3′ ends of hnRNP A1 full-length cDNA, digesting the PCR product with EcoRI and XhoI, and inserting the resultant fragment into myc-PK plasmid (34). With the EcoRI site for the construction, PK cDNA was removed from the plasmid. In order to construct plasmids myc-PK-NAB34 and myc-PK-NAB35, PCR was performed to introduce KpnI and XhoI sites at the 5′ and 3′ ends of the NAB34 and NAB35 regions, respectively. After digestion, the KpnI-XhoI fragments were inserted into the myc-PK plasmid (34). Since we used an internal KpnI site in the PK cDNA to construct the fusions, the fusion proteins were smaller than the PK itself (see Fig. 1A). In order to obtain yTRN cDNA, PCR was performed according to the DNA sequence available in the database (YBR017c). Composite full-length cDNA of yTRN was inserted into pET28b vector (Novagen) to construct the expression plasmid HIS-yTRN. The plasmid pGST-D0-aux was constructed by performing PCR to introduce BamHI and XhoI sites at the 5′ and 3′ ends of the cDNA corresponding to the auxiliary domain of hnRNP D0 (accession no. D55674 [19]), digesting the PCR product with BamHI and XhoI, and inserting the resultant fragment into pGEX-5X-1 (Pharmacia).
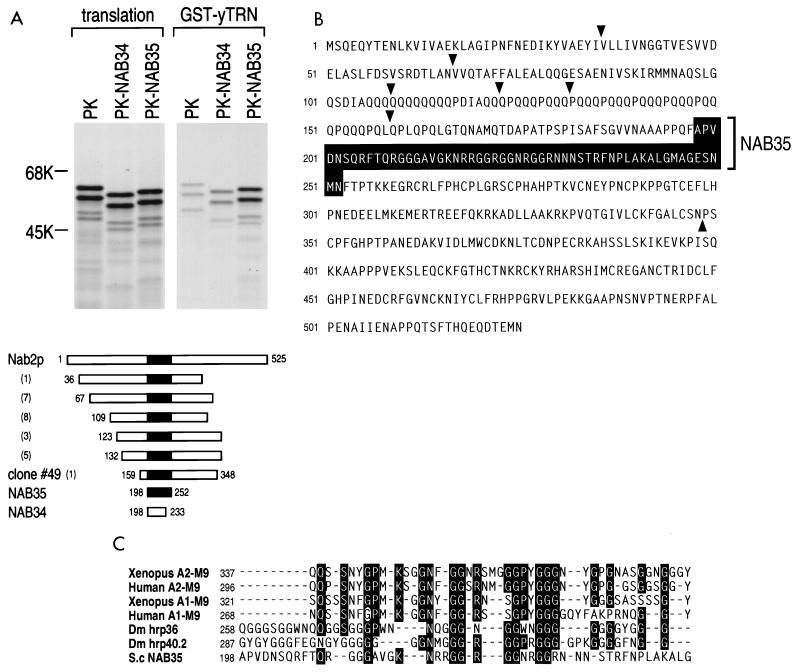
FIG. 1.
NAB35 is the target sequence of S. cerevisiae mRNA-binding protein, Nab2p, for yTRN interaction. (A) GST-yTRN on glutathione-Sepharose was incubated with myc-PK, myc-PK-NAB34 (PK-NAB34), or myc-PK-NAB35 (PK-NAB35) translated in vitro in the presence of [35S]methionine. The bound fraction was analyzed by SDS-PAGE and visualized by fluorography (GST-yTRN). Products of the translation reaction are shown as translation. Numbers at the left are molecular size standards (in kilodaltons). Schematic drawings of full-length Nab2p protein (3), Nab2p clones selected in an exhaustive genomic two-hybrid screen, NAB35 (shown by a black box), and NAB34 are shown below. The numbers in parentheses are the numbers of identical clones obtained by the screen. The numbers at the beginning and the end of the fragments indicate the first and the last amino acid numbers, respectively, for each fragment according to reference 3. (B) The NAB35 region in the Nab2p amino acid sequence (3) is shown by a black box. The starting points of clones selected by the screen and the ending point of clone 49 are indicated by arrowheads. (C) Alignment of M9 and M9-like sequences from a variety of organisms. Identical residues found in five or more sequences are boxed in black. The point mutation of Gly-274 to Ala in human A1-M9, indicated by a white circle, abolishes both NLS and NES activities of A1 (24).
GST-fusion protein binding assays.
Purified glutathione S-transferase (GST)-fusion proteins (GST-yTRN for Fig. 1A and 4, GST itself, GST-NAB35, and GST-M9 for Fig. 3A and B) were incubated with 30 μl of glutathione-Sepharose (Pharmacia) in 500 μl of binding buffer [50 mM Tris-HCl, 400 mM NaCl, 5 mM Mg(OAc)2, 2 μg of leupeptin per ml, 2 μg of pepstatin per ml, and 0.5% aprotinin (pH 7.5)]. After an incubation period of at least 1 h at 4°C, the resin was washed with binding buffer and protein products (myc-PK and myc-PK-NAB35 for Fig. 1A and 4, myc-PK-NAB34 for Fig. 1A, myc-PK-M9 and myc-A1 for Fig. 4, and hTRN1 and importin β for Fig. 3A) produced by in vitro transcription-translation of plasmids (see above) with a TnT kit (Promega) in rabbit reticulocyte lysate in the presence of [35S]methionine (Amersham) or the cytoplasmic fraction of HeLa cells (Fig. 3B) were added. After incubation for 3 h at 4°C, the resin was washed with binding buffer, and the bound fraction was eluted by boiling in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, analyzed by SDS-PAGE, and visualized by either immunoblotting with D45, an anti-hTRN1 monoclonal antibody (Fig. 3B), or fluorography (otherwise). Immunoblotting was performed as described previously (35).
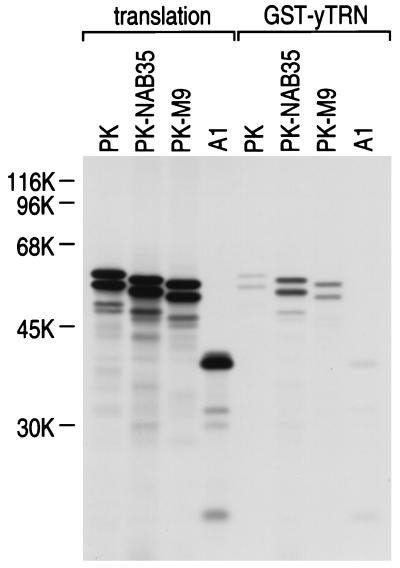
FIG. 4.
hnRNP A1 does not bind yTRN. GST-yTRN on glutathione-Sepharose was incubated with either myc-PK (PK), myc-PK-NAB35 (PK-NAB35), myc-PK-M9 (PK-M9), or myc-A1 (A1) translated in vitro in the presence of [35S]methionine. The bound fraction was analyzed by SDS-PAGE and visualized by fluorography (GST-yTRN). Products of in vitro translation are shown as translation. The positions of molecular mass markers are indicated (in kilodaltons) on the left.
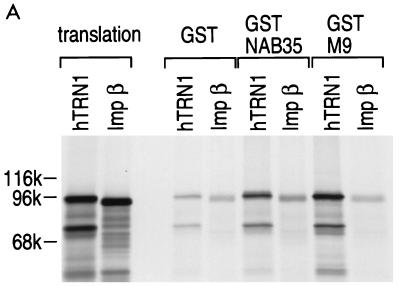
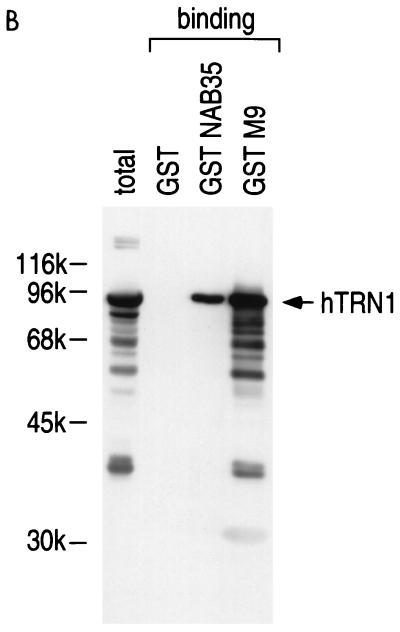
FIG. 3.
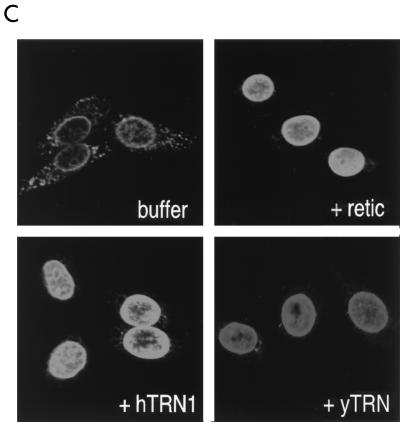
hTRN1 mediates the nuclear import of NAB35-bearing proteins. (A) GST-NAB35 binds hTRN1 in vitro. GST, GST-NAB35, or GST-M9 (24) on glutathione-Sepharose was incubated with either hTRN1 or importin β (Imp β) translated in vitro in the presence of [35S]methionine. The bound fraction was analyzed by SDS-PAGE and visualized by fluorography. Products of in vitro translation are shown as translation. The positions of molecular mass markers are indicated on the left (in kilodaltons). (B) GST, GST-NAB35, or GST-M9 on glutathione-Sepharose was incubated with the cytoplasmic fraction of HeLa cells in the presence of 400 mM NaCl. After extensive washing, the bound fraction was analyzed by SDS-PAGE and visualized by immunoblotting with D45 (binding) (35). The cytoplasmic fraction used in this experiment is indicated in the lane labeled total. The position of hTRN1 is indicated with an arrow on the right, and the positions of molecular mass markers are indicated (in kilodaltons) on the left. (C) hTRN1 and yTRN mediate the nuclear import of GST-NAB35. Digitonin-permeabilized HeLa cells were incubated with GST-NAB35 (100 μg/ml) in buffer alone (buffer) or in the presence of either rabbit reticulocyte lysate (retic), hTRN1 (50 μg/ml), or yTRN (50 μg/ml). Import was detected with an anti-GST monoclonal antibody, followed by indirect immunofluorescence with fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G.
Cell culture, transfection, and immunofluorescence.
COS7 cells were cultured at 37°C to subconfluent densities in Dulbecco modified Eagle medium (DME) supplemented with penicillin and streptomycin and 10% calf serum. COS7 cells grown on glass coverslips in 30-mm-diameter dishes were transfected with myc-PK-NAB35 and myc-PK-NAB34 (5 μg each) as described previously (35). Forty-eight hours after transfection, immunofluorescence was performed as described previously (35) with an anti-myc antibody.
Nuclear import assays.
Nuclear import reactions were performed as described previously (1) except that GTP was added to 0.1 mM. The transport substrates were added at a concentration of 100 μg/ml and for the TRN-mediated import assays, 50 μg of HIS-hTRN1 per ml (33) or 50 μg of HIS-yTRN per ml was added to the import substrates in transport buffer plus ATP-regenerating system. After the import reacted for 20 min at room temperature, the nuclei were fixed and processed for immunostaining as described previously (35). Import of the GST substrate (GST-NAB35) was detected with an anti-GST monoclonal antibody (Santa Cruz Biotechnology). Laser confocal fluorescence microscopy was performed with a confocal microscope (TCS NT; Leica).
Far Western blotting.
The pGST-D0-aux plasmid was transformed and expressed in BL21(DE3) cells, and the GST-D0-aux fusion protein was partially purified according to the manufacturer’s instructions. Purified GST, GST-M9, and partially purified GST-D0 aux were analyzed by SDS-PAGE and transferred to a nitrocellulose membrane, which was then blocked with 5% nonfat milk in phosphate-buffered saline, probed with hTRN1 produced by in vitro transcription-translation of plasmid, and exposed to X-ray film.
cDNA isolation of dTRN and xTRN.
A GenBank database search with the amino acid sequence of hTRN1 as a probe led to the identification of six expressed sequence tag (EST) clones (accession no. AA202234, AA202375, AA391031, AA391593, AA246323, and AA201572) corresponding to a D. melanogaster gene significantly similar to hTRN1. Based on the sequence information of these tags, two different partial coding sequences were amplified by PCR on oligo(dT)-primed cDNA of D. melanogaster. One of the resultant PCR fragments was about 1.5 kb, containing the N-terminal half of D. melanogaster TRN (dTRN). The other fragment, containing the central part of dTRN (0.3 kb), was employed as a probe to screen a Lambda ZAPII D. melanogaster embryo cDNA library (Stratagene) to obtain the C-terminal half of dTRN. One of two positive clones obtained from the screening contained the C-terminal half of dTRN plus ca. 100 bp of 3′ untranslated region. In order to obtain X. laevis TRN (xTRN) cDNA, a Lambda ZAPII X. laevis oocyte cDNA library (Stratagene) was screened with hTRN1 cDNA as a probe. Four positive clones were obtained and sequenced. The longest clones contained an almost-full-length xTRN coding region, missing six amino acids at the N terminus compared to the hTRN1 amino acid sequence (33).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences reported in this paper are AF059613 (dTRN) and AF059614 (xTRN).
RESULTS
An M9-like nuclear localization sequence in S. cerevisiae, NAB35.
We used the yeast two-hybrid system to identify proteins that interact with yTRN (YBR017c [also known as Kap104p]) (2, 26, 33). The pGAL4-yTRN fusion exhibited a strong toxicity when expressed in the CG1945 cells. A first attempt to carry out an exhaustive genomic two-hybrid screen with this bait led to very few diploids (8 × 106) compared to expected numbers, and only a single His+ clone was recovered. This clone was also found to be positive for the LacZ reporter. It was identified by sequencing as a Nab2p genomic fragment (3). Due to this high toxicity, we cloned and expressed yTRN as a fusion protein with the LexA DNA binding domain. Thirty million diploids were screened, and 54 His+ colonies were selected. The interactions found with these candidates were assayed with the Gal4-yTRN bait fusion and found to be positive for both the HIS3 and the LacZ reporter genes. Clones corresponding to four different open reading frames were selected at least twice, and clones corresponding to seven additional open reading frames were selected once. Twenty-five of the 54 positive clones of proteins interacting with the yTRN bait were fragments encoding overlapping portions of the Nab2 protein (3). This family of interactors contained six different inserts, which are schematically shown in Fig. 1A. Most of these inserts were selected several times, showing a good redundancy of the screen (Fig. 1A). Comparison of the 5′ junctions and the sizes of the fragments allowed the definition of the minimal domain necessary for the interaction with yTRN protein. Clone 49 corresponds to the insert showing the minimal region.
This Nab2p binding to yTRN is in agreement with data which have been published in the meantime (2). The interaction of yTRN with Nab2p observed in the two-hybrid system was confirmed by performing in vitro protein binding assays. Full-length yTRN was expressed as a fusion protein with GST. Portions of clone 49 were produced and labeled with [35S]methionine by in vitro transcription-translation in rabbit reticulocyte lysate as a fusion protein with myc-tagged PK (34). The purified yTRN immobilized on glutathione-Sepharose beads was incubated with these 35S-labeled myc-PK peptides or with myc-PK alone as a control. After the beads were washed, bound proteins were analyzed by SDS-PAGE. Figure 1A shows that one region of Nab2p, termed NAB35 (Fig. 1A and B), consisting of 55 amino acids (amino acids 198 to 252, according to reference 3), bound specifically to the immobilized yTRN but that a smaller, C-terminally deleted region, NAB34 (amino acids 198 to 233), exhibited a much-reduced binding activity. An alignment of NAB35 with M9 and M9-like sequences found in various organisms is shown in Fig. 1C. We note that Gly-274 is conserved among all the sequences presented in Fig. 1C and that this Gly residue is essential for A1-M9 activity, as its replacement by Ala abolishes both its NLS and NES activities (24). The figure also includes the putative M9 sequences of two D. melanogaster proteins, hrp36 and hrp40.2 (see Discussion).
hTRN1 interacts with NAB35 and mediates its nuclear import.
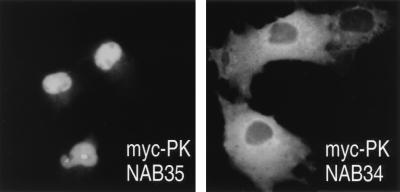
In order to assess the NLS activity of NAB35, we expressed NAB35 fused to myc-tagged PK in monkey COS7 cells and examined its localization by immunofluorescence microscopy. As shown in Fig. 2, the NAB35-bearing protein localizes to the nucleus. In contrast, a deletion mutant, NAB34, which does not interact significantly with yTRN in vitro (Fig. 1A), was excluded from the nucleus and accumulated in the cytoplasm.
FIG. 2.
NAB35-bearing protein is localized in the nucleus of mammalian cells. Transfection of COS7 cells was carried out with either myc-PK-NAB35 or myc-PK-NAB34, and immunofluorescence microscopy was carried out with an anti-myc antibody. Note that myc-PK-NAB35 is clearly localized in the nucleus in mammalian cells in contrast to myc-PK-NAB34.
The capacity of NAB35 to interact with hTRN1 was investigated by testing whether GST-NAB35 immobilized on glutathione-Sepharose could interact with 35S-labeled hTRN1 in the presence of 400 mM NaCl. As a control, we tested NAB35 binding to 35S-labeled importin β, since hTRN1 and importin β are of similar size and distantly related to each other (26, 33). As expected, although the interaction appears to be weaker than that of M9, the GST-NAB35 fusion protein specifically interacted with hTRN1 (Fig. 3A). To further confirm the interaction of NAB35 with hTRN1, we prepared a cytoplasmic extract from HeLa cells and carried out protein binding assays in the presence of 400 mM NaCl, using the same GST-NAB35 and GST-M9 fusion proteins on glutathione-Sepharose beads, followed by immunoblotting with D45, an anti-hTRN1 monoclonal antibody (35). When GST alone did not show any detectable binding to hTRN1, a strong interaction between hTRN1 and GST-NAB35 was observed (Fig. 3B).
To determine directly if hTRN1 is the nuclear import mediator of NAB35 in mammalian cells, we performed in vitro nuclear import assays, using a digitonin-permeabilized HeLa cell system (1) in which hTRN1 mediates the nuclear import of M9-bearing proteins (26, 33). GST-NAB35 is not imported into the nucleus in the absence of exogenous factors. However, the addition of either rabbit reticulocyte lysate or recombinant hTRN1 facilitates the nuclear import of GST-NAB35 (Fig. 3C), demonstrating that hTRN1 mediates NAB35-bearing protein nuclear import in mammalian cells. Interestingly, yTRN also mediates the nuclear import of GST-NAB35 in mammalian cells, although the signal intensity by immunofluorescence is slightly lower than that of hTRN1 (Fig. 3C), indicating that the nuclear import machinery and the components of NPCs utilized by TRN are highly conserved in evolution. The lower intensity with yTRN is likely a result of a slightly lower affinity of yTRN for human NPC components.
yTRN is not capable of interacting with hnRNP A1.
hnRNP A1 expressed in either S. cerevisiae or S. pombe does not localize to the nucleus, whereas hnRNP C1, which contains a classical bipartite basic NLS (27, 36), does accumulate in these nuclei (25). The capacity of yTRN to interact with hnRNP A1 was assessed by protein binding assays with a GST-yTRN fusion protein with 35S-labeled proteins produced by in vitro transcription-translation (PK, myc-PK-NAB35, myc-PK-M9, and myc-hnRNP A1). The interaction of myc-PK-M9 with yTRN is much weaker than that of myc-PK-NAB35 with yTRN, and no myc-hnRNP A1 interaction with yTRN was detected in this assay (Fig. 4). The failure of yTRN to interact with hnRNP A1 likely explains why hnRNP A1 cannot be imported into the nucleus in yeast.
hnRNP D0 is an additional candidate for hTRN1 nuclear import substrate.
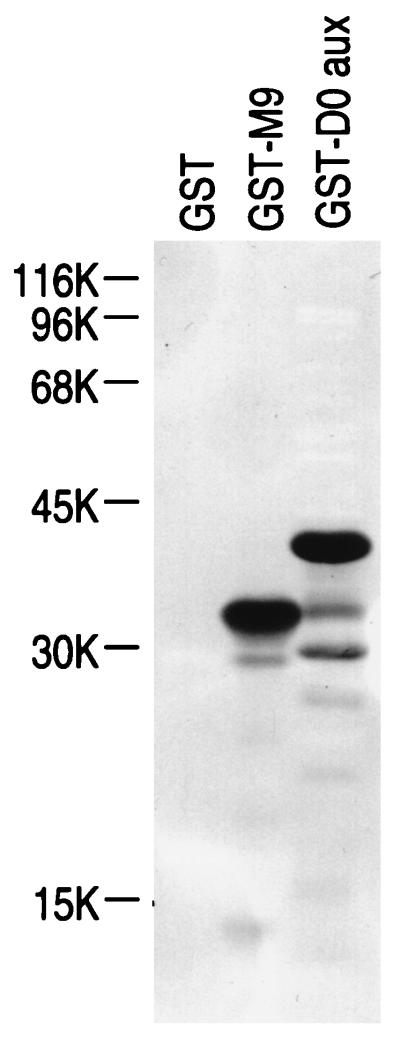
Previously, we have shown by far-Western blotting that hTRN1 can interact with several proteins (likely mostly hnRNP proteins [30, 38]) that can be enriched by single-stranded DNA binding from the nuclei of HeLa cells (35). The candidate proteins in the profile included not only hnRNP A1 and F, which were demonstrated to be nuclear import substrates of hTRN1 (26, 33, 35), but also hnRNP D proteins (19) according to their mobility on SDS-polyacrylamide gels (10, 30). The possibility that hnRNP D could interact with hTRN1 in vitro was investigated by testing whether purified hnRNP D0 Gly-rich auxiliary domain fused to GST (GST-D0 aux) could interact with 35S-labeled hTRN1. As shown in Fig. 5, under conditions in which GST-M9 is capable of interacting with hTRN1, there is a strong interaction between GST-D0 aux and hTRN1. The amino acid sequence of the D0 auxiliary domain (residue 328 to the end of the protein; 112 amino acids) is rich in Gly, Asn, Tyr, and Arg, but no other obvious similarity between M9/NAB35 and hnRNP D0 was found.
FIG. 5.
hTRN1 interaction with hnRNP D0 protein. GST, GST-M9, and GST-D0 aux (2 μg in each lane) bound to nitrocellulose blots were probed with 35S-labeled hTRN1. Note that GST-D0 aux can interact with hTRN1 under conditions in which GST-M9 is capable of binding hTRN1. The positions of molecular mass markers are indicated (in kilodaltons) on the left.
TRNs are highly conserved in divergent organisms.
To better characterize the M9 nuclear import pathway and facilitate further experiments in additional systems, we identified and cloned the TRNs of X. laevis and D. melanogaster. Both clones are highly similar to hTRN1 (xTRN and dTRN show 95 and 73% identity, respectively). The amino acid sequence alignment of xTRN and dTRN with hTRN1 (5, 11, 33), yTRN (2, 33), and an S. pombe protein that shows high similarity to all these TRNs (accession no. g2656009) is shown in Fig. 6. It is apparent that the entire primary structures of all the TRN proteins are highly similar.
FIG. 6.
Sequence alignment of TRNs from divergent organisms. hTRN1 (Human) (5, 11, 33) was aligned with TRNs from X. laevis (Xenopus), D. melanogaster (Drosophila), Schizosaccharomyces pombe (pombe; accession no. g2656009), and Saccharomyces cerevisiae (cerevisiae) (2, 33). Identical amino acids found in four or more sequences are indicated with gray boxes. The numbers to the left of the amino acid sequences indicate amino acid positions.
DISCUSSION
In this study, we show that Nab2p, a yeast protein selected in an exhaustive genomic two-hybrid screen performed with yTRN (also known as YBR017c or Kap104p), specifically interacts with this protein, and we have identified an S. cerevisiae M9-like nuclear import sequence, NAB35. NAB35 has weak but significant similarity to M9 of human and Xenopus A1/A2 proteins. The expanded consensus that this sequence comparison afforded suggests that two D. melanogaster hnRNP proteins, hrp36 and hrp40.2 (21), also contain M9 signals (Fig. 1C). Both hrp36 and hrp40.2 are nuclear pre-mRNA/mRNA-binding proteins. Interestingly, another alternative splicing form of hrp40, hrp40.1, excludes the M9 sequence found in hrp40.2 (21) and is cytoplasmic (20a). The most prominent feature of M9 is its richness in Gly and Asn, but its primary sequence is otherwise not highly conserved. We have previously shown that hTRN1 is capable of interacting not only with hnRNP A1 but also with other hnRNP proteins, including hnRNP F, and it mediates the nuclear import of hnRNP F in vitro (35). In this study, we show that the auxiliary domain of hnRNP D0 protein also interacts avidly with hTRN1 (Fig. 5) and is likely to be an hTRN1 nuclear import substrate. The amino acid sequence of the D0 auxiliary domain, especially the region containing amino acids 276 to 324 (19), seems to show homology to M9, since both are rich in Gly, Asn, Tyr, and Arg, but as is the case for hnRNP F (22), no obvious alignment of M9/NAB35 with hnRNP D0 was found. The observed interaction of hTRN1 with the D0 auxiliary domain strengthens the idea that TRN recognizes its import substrates not only by primary sequence but also by secondary and/or tertiary structural features (35).
Nab2p is an hnRNP protein in S. cerevisiae, since it strongly and specifically associates with nuclear poly(A)+ RNA in vivo (3). The nuclear import of Nab2p is mediated by yTRN (this study and reference 2). However, of the known hnRNP proteins, Nab2p is not the closest structural homolog of hnRNP A1 in S. cerevisiae, since the overall structural features of Nab2p are barely similar to those of A1. Moreover, other than its NLS, the structural and functional properties of NAB35 are different from those of M9; A1 consists of two RNA-binding domains and an auxiliary Gly-rich domain which contains an RGG box, another type of RNA-binding motif (20), and the bidirectional nuclear transport signal, M9. The RGG box and M9 are located in tandem in A1 and are not overlapping. Moreover, M9 does not have any detectable RNA-binding activity (24). Nab2p, in contrast, has an RGG box in the middle of the protein. The RGG box and the M9-related sequences are intermingled in the NAB35 sequence, and this sequence likely has RNA-binding activity (3). It will be of interest to determine whether NAB35 is also an NES and/or a transcription-dependent nuclear import signal, as is M9 (24, 35).
Neither human M9 nor hnRNP A1 interacts well with yTRN in vitro (Fig. 4). We have previously shown that hnRNP A1 does not accumulate in the nucleus when expressed in yeast, whereas hnRNP C1, containing a classical bipartite basic NLS which binds Rch1 (a homolog of importin α [44]) in the yeast two-hybrid system (28a), localizes in the nucleus in yeast as well as in mammalian cells (25). The lack of nuclear accumulation of hnRNP A1 in yeast is likely due to its inability to bind yTRN. It seems that yTRN is more constrained than hTRN1 in its capacity to interact with various members of the M9-containing import substrates, although yTRN does apparently bind Nab4p (YOL123w or Hrp1p) (2), another nuclear mRNA-binding protein in S. cerevisiae. This protein also contains an area rich in Gly, Arg, and Asn near the C terminus, especially in the regions containing amino acids 328 to 370 and 483 to the end of the protein (16), but no obvious alignment with M9/NAB35 was found in those regions.
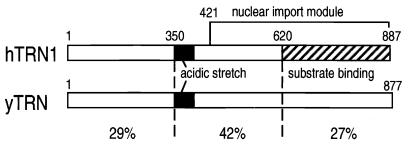
Using an in vitro nuclear import assay, we have been able to test whether hTRN1 and yTRN are indeed functional analogs (orthologs). Although the efficiency is lower than that of hTRN1, yTRN can clearly mediate the nuclear import of NAB35-bearing proteins in the mammalian cell system (Fig. 3C). This demonstrates the high degree of evolutionary conservation of function of the transportin pathway. To our knowledge, this may be the first demonstration that a yeast transport receptor can function in mammalian cells. Very recently, we showed that the C-terminal half of hTRN1, comprising amino acids 421 to 887, is a nuclear transport module that can function independently and does not require GTP hydrolysis for its translocation (28). The region of highest homology between hTRN1 and yTRN is located in the central portion of the proteins (42% identity for about 300 amino acids [Fig. 7]), which mostly overlaps with the nuclear transport module in hTRN1, indicating that this region may be important for translocation through NPCs. It has been reported that haploid cells carrying a disrupted copy of yTRN are severely impaired in their growth at 23°C and are inviable when transferred to 30°C (2). In addition, the temperature shift in one of the temperature-sensitive mutants leads to depletion of yTRN and is coincident with redistribution of Nab2p from the nucleus to the cytoplasm (2). It will be of interest to see whether expression of hTRN1 in yeast cells can complement the loss of endogenous yTRN function.
FIG. 7.
Schematic representation of hTRN1 and yTRN. The structures of hTRN1 and yTRN are schematically shown, the solid boxes representing the acidic stretch in both proteins, and the cross-hatched box representing the substrate (M9) binding domain (11, 33) in hTRN1. The nuclear import module (28) in hTRN1 is also indicated. The percent identities of three regions of hTRN1 and yTRN are shown.
After this manuscript was submitted for publication, Truant et al. (39) reported the identification and characterization of a novel NLS in Nab2p. These authors demonstrate by a two-hybrid assay that the core yTRN binding domain of Nab2p maps to residues 191 to 251 (residues 198 to 252 in our NAB35 sequence) and that a region including these residues functions as an NLS in vivo. Using an in vitro import assay, they also show that yTRN can mediate the nuclear import of Nab2p NLS-containing substrates. One discrepancy between our data and those of Truant et al., however, is that Truant et al. (39) found that human importin β, but not hTRN1, interacts with the Nab2p NLS and mediates its nuclear import, whereas we found that human importin β does not interact with NAB35 and that hTRN1 is the NAB35 nuclear import mediator by an in vitro import assay. The reasons for these disparities are not yet clear but may be differences in the specific conditions of the assays.
ACKNOWLEDGMENTS
We thank Haruhiko Siomi for transfecting COS7 cells, Amy Hreha for performing far Western blotting, Jennifer L. Bachorik for screening the D. melanogaster cDNA library, and all the members of our laboratory, especially Haruhiko Siomi, Paul Eder, and Sara Nakielny, for critically reading the manuscript. We also thank Susan Kelchner for her secretarial assistance.
This work was supported by a grant from the National Institutes of Health and by the Howard Hughes Medical Institute (G.D.), by grant Biotech 950009 from the European Union (P.L.), and by a long-term fellowship from the Japan Science and Technology Corporation (M.C.S.).
REFERENCES
- 1.Adam S A, Marr R S, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aitchison J D, Blobel G, Rout M P. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- 3.Anderson J T, Wilson S M, Datar K V, Swanson M S. NAB2: a yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol Cell Biol. 1993;13:2730–2741. doi: 10.1128/mcb.13.5.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bischoff F R, Görlich D. RanBP1 is crucial for the release of RanGTP from importin β-related nuclear transport factors. FEBS Lett. 1997;419:249–254. doi: 10.1016/s0014-5793(97)01467-1. [DOI] [PubMed] [Google Scholar]
- 4a.Bischoff, F. R., S. Nakielny, and G. Dreyfuss. Unpublished results.
- 5.Bonifaci N, Moroianu J, Radu A, Blobel G. Karyopherin β2 mediates nuclear import of a mRNA binding protein. Proc Natl Acad Sci USA. 1997;94:5055–5060. doi: 10.1073/pnas.94.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burd C G, Dreyfuss G. RNA binding specificity of hnRNP A1: hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caceres J F, Stamm S, Helfman D M, Krainer A R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 8.Chi N C, Adam S A. Functional domains in nuclear import factor p97 for binding the nuclear localization sequence receptor and the nuclear pore. Mol Biol Cell. 1997;8:945–956. doi: 10.1091/mbc.8.6.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 10.Dreyfuss G, Matunis M J, Piñol-Roma S, Burd C G. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 11.Fridell R A, Truant R, Trorne L, Benson R E, Cullen B R. Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-β. J Cell Sci. 1997;110:1325–1331. doi: 10.1242/jcs.110.11.1325. [DOI] [PubMed] [Google Scholar]
- 12.Fromont-Racine M, Rain J-C, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 13.Fu X-D, Mayeda A, Maniatis T, Krainer A R. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5′ and 3′ splicing site selection. Proc Natl Acad Sci USA. 1992;89:11224–11228. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Görlich D, Mattaj I W. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 15.Görlich D, Dabrowski M, Bischoff F R, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henry M, Borland C Z, Bossie M, Silver P. Potential RNA-binding proteins in Saccharomyces cerevisiae identified as suppressors of temperature-sensitive mutations in NPL3. Genetics. 1996;142:103–115. doi: 10.1093/genetics/142.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Izaurralde E, Jarmolowski A, Beisel C, Mattaj I W, Dreyfuss G, Fischer U. A role of the M9 transport signal of hnRNP A1 in mRNA nuclear export. J Cell Biol. 1997;137:27–35. doi: 10.1083/jcb.137.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Izaurralde E, Kutay U, Cayetano K, Mattaj I W, Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kajita Y, Nakayama J, Aizawa M, Ishikawa F. The UUUA-specific RNA-binding protein, heterogeneous nuclear ribonucleoprotein D0; common modular structure and binding properties of the 2×RBD-Gly family. J Biol Chem. 1995;270:22167–22175. doi: 10.1074/jbc.270.38.22167. [DOI] [PubMed] [Google Scholar]
- 20.Kiledjian M, Dreyfuss G. Primary structure of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992;11:2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Matunis, E. L., and G. Dreyfuss. Unpublished data.
- 21.Matunis E L, Matunis M J, Dreyfuss G. Characterization of the major hnRNP proteins from Drosophila melanogaster. J Cell Biol. 1992;116:257–269. doi: 10.1083/jcb.116.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matunis M J, Xing J, Dreyfuss G. The hnRNP F protein: unique primary structure, nucleic acid-binding properties, and subcellular localization. Nucleic Acids Res. 1994;22:1059–1067. doi: 10.1093/nar/22.6.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayeda A, Krainer A R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- 24.Michael W M, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 25.Michael W M, Siomi H, Choi M, Piñol-Roma S, Nakielny S, Liu Q, Dreyfuss G. Signal sequences that target nuclear import and nuclear export of pre-mRNA-binding proteins. Cold Spring Harbor Symp Quant Biol. 1995;60:663–668. doi: 10.1101/sqb.1995.060.01.071. [DOI] [PubMed] [Google Scholar]
- 26.Nakielny S, Siomi M C, Siomi H, Michael W M, Pollard V, Dreyfuss G. Transportin: nuclear transport receptor of a novel nuclear protein import pathway. Exp Cell Res. 1996;229:261–266. doi: 10.1006/excr.1996.0369. [DOI] [PubMed] [Google Scholar]
- 27.Nakielny S, Dreyfuss G. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J Cell Biol. 1996;134:1365–1373. doi: 10.1083/jcb.134.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakielny S, Dreyfuss G. Import and export of the nuclear protein import receptor, transportin, by a mechanism independent of GTP hydrolysis. Curr Biol. 1998;8:89–95. doi: 10.1016/s0960-9822(98)70039-9. [DOI] [PubMed] [Google Scholar]
- 28a.Nakielny, S., and G. Dreyfuss. Unpublished data.
- 29.Pante N, Aebi U. Toward the molecular dissection of protein import into nuclei. Curr Opin Cell Biol. 1996;8:397–406. doi: 10.1016/s0955-0674(96)80016-0. [DOI] [PubMed] [Google Scholar]
- 30.Piñol-Roma S, Choi Y D, Matunis M J, Dreyfuss G. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 1988;2:215–227. doi: 10.1101/gad.2.2.215. [DOI] [PubMed] [Google Scholar]
- 31.Piñol-Roma S, Dreyfuss G. Transcription-dependent and transcription-independent nuclear transport of hnRNP proteins. Science. 1991;253:312–314. doi: 10.1126/science.1857966. [DOI] [PubMed] [Google Scholar]
- 32.Piñol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 33.Pollard V, Michael W M, Nakielny S, Siomi M C, Wang F, Dreyfuss G. A novel receptor-mediated nuclear import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 34.Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129:551–559. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siomi M C, Eder P S, Kataoka N, Wan L, Liu Q, Dreyfuss G. Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J Cell Biol. 1997;138:1181–1192. doi: 10.1083/jcb.138.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swanson M S, Nakagawa T Y, LeVan K, Dreyfuss G. Primary structure of human nuclear ribonucleoprotein particle C proteins: conservation of sequence and domain structures in heterogeneous nuclear RNA, mRNA, and pre-mRNA-binding proteins. Mol Cell Biol. 1987;7:1731–1739. doi: 10.1128/mcb.7.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swanson M S, Dreyfuss G. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by DNA binding proteins. Mol Cell Biol. 1988;3:2237–2241. doi: 10.1128/mcb.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swanson M S, Dreyfuss G. RNA binding specificity of hnRNP proteins: a subset bind to the 3′ end of introns. EMBO J. 1988;7:3519–3529. doi: 10.1002/j.1460-2075.1988.tb03228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Truant R, Fridell R A, Benson R E, Bogerd H, Cullen B R. Identification and characterization of a novel nuclear localization signal present in the yeast Nab2 poly(A)+ RNA binding protein. Mol Cell Biol. 1998;18:1449–1458. doi: 10.1128/mcb.18.3.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Visa N, Alzhanova-Ericsson A T, Sun X, Kiseleva E, Bjorkroth B, Wurtz T, Daneholt B. A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell. 1996;84:253–264. doi: 10.1016/s0092-8674(00)80980-0. [DOI] [PubMed] [Google Scholar]
- 41.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian Ras directly interacts with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 42.Wang H, Kohalmi S E, Cutler A J. An improved method for polymerase chain reaction using whole yeast cells. Anal Biochem. 1996;237:145–146. doi: 10.1006/abio.1996.0213. [DOI] [PubMed] [Google Scholar]
- 43.Weighardt F, Biamonti G, Riva S. Nucleo-cytoplasmic distribution of human hnRNP proteins: a search for the targeting domains in hnRNP A1. J Cell Sci. 1995;108:545–555. doi: 10.1242/jcs.108.2.545. [DOI] [PubMed] [Google Scholar]
- 44.Weis K, Mattaj I W, Lamond A I. Identification of hSRP1α as a functional receptor for nuclear localization sequences. Science. 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- 45.Yang W, Bani M R, Ju S J, Rowan S, Ben-David Y, Chabot B. The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc Natl Acad Sci USA. 1994;81:6924–6928. doi: 10.1073/pnas.91.15.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]