Abstract
The correlations among individual exposures in the exposome, which refers to all exposures an individual encounters throughout life, are important for understanding the landscape of how exposures co-occur, and how this impacts health and disease. Exposome-wide association studies (ExWAS), which are analogous to genome-wide association studies (GWAS), are increasingly being used to elucidate links between the exposome and disease. Despite increased interest in the exposome, tools and publications that characterize exposure correlations and their relationships with human disease are limited, and there is a lack of data and results sharing in resources like the GWAS catalog. To address these gaps, we developed the PEGS Explorer web application to explore exposure correlations in data from the diverse North Carolina-based Personalized Environment and Genes Study (PEGS) that were rigorously calculated to account for differing data types and previously published results from ExWAS. Through globe visualizations, PEGS Explorer allows users to explore correlations between exposures found to be associated with complex diseases. The exposome data used for analysis includes not only standard environmental exposures such as point source pollution and ozone levels but also exposures from diet, medication, lifestyle factors, stress, and occupation. The web application addresses the lack of accessible data and results sharing, a major challenge in the field, and enables users to put results in context, generate hypotheses, and, importantly, replicate findings in other cohorts. PEGS Explorer will be updated with additional results as they become available, ensuring it is an up-to-date resource in exposome science.
Keywords: exposome-wide association study (ExWAS), Personalized Environment and Genes Study (PEGS), occupational exposures, web application, data sharing, exposome data
Introduction
The exposome, which represents all exposures over the course of an individual’s life, has substantial effects on human health and disease.1,2 As technology improves and the importance and complexity of the exposome are recognized, exposome-wide association studies (ExWAS) are increasingly being conducted to understand the associations between exposures and disease. ExWAS are analogous to genome-wide association studies (GWAS) and generate results that are similarly high dimensional. Accordingly, data-sharing efforts can magnify the impact of individual studies and maximize the return on investments in ExWAS.
ExWAS have been conducted for several important clinical phenotypes in data from the diverse, North Carolina-based Personalized Environment and Genes Study (PEGS).3-6 PEGS captures extensive environmental data and broad information on multiple diseases and both internal and external exposures through multiple questionnaires, so data from the cohort is well-suited for ExWAS. Lee et al. recently conducted ExWAS for cardiovascular-related phenotypes in PEGS data, including cardiac arrhythmia, congestive heart failure, coronary artery disease, heart attack, and stroke. Additionally, Akhtari and Lloyd et al. conducted ExWAS for type 2 diabetes, and Lloyd et al. (in this issue) conducted ExWAS for 11 high-prevalence diseases in PEGS: allergic rhinitis, asthma, bone loss, fibroids, high cholesterol, hypertension, iron-deficient anemia, lower GI polyps, migraines, ovarian cysts, and type 2 diabetes. While the study results and top findings for each disease are promising, comprehensive results, results stratified by biological variables such as sex and race/ethnicity, and results of sensitivity analyses conducted with epidemiological covariates have not been reported.
To address these limitations and support data transparency and scientific discovery, we built a public-facing web interface to disseminate results and expand the resources available for sharing existing exposome results.7,8 The PEGS Explorer web interface allows users to examine, visualize, and download ExWAS results and exposure correlations. Users can interrogate associations between individual exposures and between exposures and disease traits and examine exposure correlations in a trait-specific or trait-agnostic manner. Users can also visualize correlations through globes9 that reflect the complex mixtures that comprise the exposome. PEGS Explorer allows users to view ExWAS results described in prior work as well as stratified results to understand how exposures and traits are associated with diseases and how these associations vary by strata such as age.
While seemingly straightforward, care must be taken when calculating correlation values across data types, especially heterogeneous data such as that collected by PEGS through questionnaires that generate multiple data types (eg, binary, ordinal). When analyzing big data, correlation methods are often chosen out of convenience, without considering the data types. Typically, a conservative non-parametric approach such as Spearman’s rank correlation coefficient is used, but this can lead to incorrect summarization and subsequent interpretation. For example, many of the survey questions have “yes” or “no” answers that, without careful consideration, would typically be treated as binary. While there are two answer categories, the answers represent summaries of continuous distributions. For example, answering “yes” or “no” to “Have you ever smoked?” represents a range of practical exposures. Answering “yes” does not represent necessarily equal exposures but rather an unmeasured continuous range of exposures. Accordingly, this type of variable should be treated differently than variables that are truly binary. To estimate the correlation between two continuous latent variables (such as self-reported exposure) from two observed ordinal variables, polychoric or tetrachoric correlation methods can be used.10-12 In the current study, we rigorously calculated correlations using appropriate methods to account for the data types evaluated.
Additionally, sample-size differences can dramatically affect the magnitude and interpretation of correlations. Small sample sizes can lead to unreliable and spurious correlations while large sample sizes can increase statistical power but do not guarantee the presence of significant correlations. While confidence intervals are often used to capture the uncertainty between a sample and the overall population, the correlations reported are highly significant, with very small confidence intervals. We addressed the variability in estimated correlations by developing suitable shrinkage factors that substantially reduced coefficients for correlations based on fewer observations. This approach improved the reliability of correlations and increased comparability across variable pairs.13
Methods
Completed ExWAS
Cardiovascular diseases
Results from Lee et al. from an ExWAS conducted in PEGS data for five cardiovascular outcomes, namely cardiac arrhythmia, congestive heart failure, coronary artery disease, heart attack, and stroke, are included in PEGS Explorer. Participants were assigned to the case group if they answered ‘YES’ when asked if they had been diagnosed with a particular cardiovascular outcome and the control group if they answered ‘NO’.
Highly prevalent diseases
Lloyd et al. (in this issue) examined 11 common, complex human diseases, namely allergic rhinitis, asthma, bone loss, fibroids, high cholesterol, hypertension, iron-deficient anemia, lower GI polyps, migraines, ovarian cysts, and type 2 diabetes.14 For each phenotype, we used the Health and Exposure Survey question that asked whether a participant has ever been diagnosed by a doctor or physician with the condition to define cases (response = “yes”) and controls (response = “no”). We determined additional inclusion/exclusion criteria for each phenotype from the results of a literature review.
Statistical analysis
Exposome-wide association studies (ExWAS)
The previously reported ExWAS examined the association of exposures and their combinations with each phenotype in two stages. We repeated this analysis for all phenotypes using the methods described below, adding additional exposures and strata to Lee et al.’s original for cardiovascular diseases. In the first stage, after study-specific quality control measures (as detailed in Lee et al.), we used logistic regression models to test the association of individual exposures with each phenotype, correcting for study-specific epidemiological covariates. We applied a Benjamini-Hochberg false discovery rate (FDR) of 0.10 to determine the statistical significance of exposure associations for each phenotype. In the second stage, we used a deletion/substitution/addition (DSA) algorithm to build multi-exposure models for each disease. For each phenotype, we used all significant exposures (FDR ≤ 0.10) from the first stage as input variables for the DSA analysis.15 Additional details of the analyses are found in the respective papers.
We conducted additional, updated analyses on the diseases analyzed in Lee et al. using the same covariates and stratification as the other diseases included in PEGS Explorer to maintain consistency within the results currently available.
Exposome correlations
We calculated correlations using data from PEGS survey responses comprising continuous, categorical, ordinal, and binary data types. We chose appropriate correlation methods based on the data types in a variable pair (see Table 1) and fit a best linear unbiased prediction (BLUP) model to shrink the correlation coefficients based on the sample size of the variable pair. We calculated correlations for all 1,077 exposures in our analysis, including correlations between exposures in different surveys. Leveraging the diversity of the PEGS cohort, we repeated the analyses using the same methods for age, income, race, and sex strata.
Table 1.
Correlation methods used for variable pairs by data type
| Correlation Type | Binary | Factor | Numeric | Ordered factor |
|---|---|---|---|---|
| Binary | Tetrachoric | |||
| Factor | Polychoric | Cramer's V | ||
| Numeric | Pearson | Polychoric | Spearman | |
| Ordered Factor | Polychoric | Polychoric | Polychoric | Polychoric |
To address the potential misinterpretation of correlations, we took two steps to ensure the accuracy of correlation values and avoid misinterpretation due to sample-size differences. First, we calculated correlation values using methods that are appropriate for the specific data types (eg, binomial, ordinal) comprising each variable pair. Second, we used linear models to develop suitable shrinkage factors to adjust correlations for sample size. These steps ensure the reliability of correlations and guarantee comparability across variable pairs.16 The supplement provides details of the methods used to calculate correlations.
Polyserial, polychoric, tetrachoric, and Cramer’s V correlation methods are appropriate when one or both observed variables are non-continuous. These methods assume that the observed binary, ordinal, and categorical survey responses result from polychotomous underlying real-world exposures, which are normally distributed, continuous, latent variables. For ordered-factor, binary, and numeric variables, we calculated polychoric correlations using polychor in R. For two binary variables, we calculated tetrachoric correlation using the psych package. For two ordered-factor variables, we calculated Cramer’s V using the biserial rcompanion package. For two numeric variables, we used Spearman’s rank correlation coefficient, and for a binary and a numeric variable, we used Pearson’s correlation coefficient. We repeated the analyses using the same methods for age, income, race, and sex strata with the following categories: age–under 20 years, 20-40 years, 41-60 years, and over 60 years; income–lower (under $20k/year), lower-middle ($20-$49k/year), upper-middle ($50-$79k/year), and upper (> $80k/year); race–Asian, Black, Hispanic, White, and other; and sex–male and female. We calculated correlations for 31 combinations of the five strata for each variable pair.
We subsequently calculated correlations for all variable pairs, separately adjusting for age, income, race, and sex. We calculated Spearman’s correlation coefficients between the residuals of the linear models of each variable pair for both unadjusted and adjusted models. We calculated correlations for all 1,077 exposures in our analysis, including correlations between exposures in different surveys, where there were drastic differences in sample sizes for individual exposures. To account for varied sample sizes, we fit a BLUP model to shrink the correlation coefficients. For the chosen correlation type based on an observed (non-missing) sample size , the estimated sample variance is , where r and v are the vectors of these values across all variable pairs. The quantity is the estimated underlying variance of true correlations , and is the estimated true average . For each pairwise correlation, the quantity is the best linear predictor for the true correlation . We shrunk correlation coefficients based on the smallest sample size to enable the accurate comparison of correlations.
PEGS explorer web application
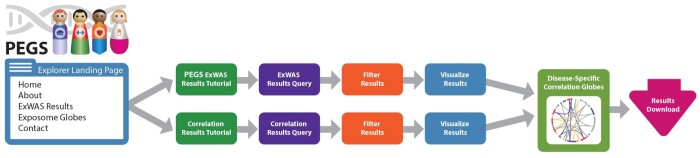
The ExWAS and correlation results are substantial and, in totality, too large for dissemination through publications and ad hoc data sharing. The PEGS Explorer web application facilitates sharing by hosting the complete results from the ExWAS and correlation analyses. The web application is located on the PEGS website hosted by the National Institute of Environmental Health Sciences. PEGS Explorer allows users to explore and download the ExWAS and correlation analysis results. The PEGS explorer website can be accessed at https://www.niehs.nih.gov/research/clinical/studies/pegs/index.cfm under the About PEGS tab. A tutorial for users is provided on the homepage, and Figure 1 demonstrates how PEGS Explorer can be used.
Figure 1.
PEGS Explorer functionality walkthrough. PEGS Explorer displays downloadable results from both ExWAS and correlation analyses. The tool includes built-in tutorials and query-generating functionality to help users quickly and easily retrieve detailed results.
Briefly, we used Java, Spring Framework (https://spring.io/), Vaadin, MariaDB, Circos.js (https://github.com/nicgirault/circosJS), JavaScript, and lit-element (https://lit.dev/) to create a production-grade solution to provide access to the analysis results. The Spring Framework provides key software infrastructure to facilitate the development of various aspects of enterprise software in a Java environment. Vaadin is a modern web application platform for Java used in conjunction with Spring Framework for user-interface development and client-server communication. The PEGS data are stored in MariaDB, an open-source database consumed by the application through the Spring Data component of the Spring Framework. We used lit-element, a web component technology, to create custom JavaScript components for exploring and visualizing the data. We built a custom exposome-globe viewer by wrapping the Circos.js library in a lit-element web component and a robust forest plot viewer with JavaScript and lit-element in conjunction with the Vaadin framework.
Results
PEGS Explorer overview
By enabling users to explore the ExWAS and correlation analysis results, PEGS Explorer addresses the lack of data and results sharing that remains a challenge in exposomics. Users can simultaneously view ExWAS results for multiple phenotypes, enabling the comparison of statistically significant exposures across phenotypes in forest plots. Results can be viewed for each survey and filtered by topical survey section to group related exposures, exposure names, and significance level.
Exposome globes display stratified results with the top 1,000 correlations shown for a given stratum. The results can be filtered to show correlations for particular strata or correlations after adjusting for specific covariates. The exposome globes can also be filtered by exposure name, survey section, and correlation coefficient value. All the results can be downloaded for further analysis by the user.
Disease correlation globes show ExWAS-significant results (FDR-adjusted P < 0.10) after adjusting for age, income, race, and sex as well as correlations between ExWAS-significant exposures for each phenotype. The globes can be enlarged to show results retained in the DSA model for each survey. Users can download the results, including correlations between exposures not among the top 1,000 correlations. Multiple phenotype globes can be concurrently displayed to visualize differences in exposure correlations across phenotypes.
ExWAS results exploration
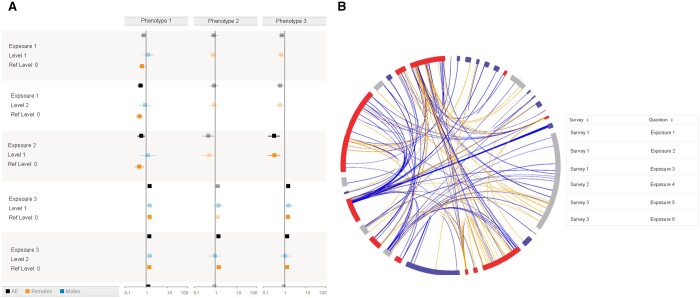
Results for traits with differing impacts on various populations can be assessed with the ExWAS results framework in PEGS Explorer. Figure 2A is an example of the PEGS Explorer interface for ExWAS results for generic exposures and phenotypes. Figure 2B is an example PEGS Explorer interface for disease-specific correlation and DSA results.
Figure 2.
(A) Example PEGS Explorer interface for ExWAS results for generic exposures and phenotypes. The results are presented in a forest plot, where black represents both sexes, orange represents females only, and blue represents males only. Solid boxes are statistically significant at a false discovery rate (FDR) of 10%. Users can choose the number of phenotypes that are displayed. (B) Example PEGS Explorer interface for disease-specific correlation and DSA results. The exposome globe displays only exposures that are statistically significant at an FDR of 10%. The table shows the DSA analysis results for each survey.
Deletion/substitution/addition (DSA) results
The web application provides DSA results for all phenotypes as tables that indicate the exposures retained in the particular DSA model. Figure 2B includes a table displaying sample DSA results.
Exposome correlations and globes
The exposome globes in PEGS Explorer, originally proposed by Patel and Manrai,9 enable users to explore the correlation structure of exposures within and across surveys. The exposome globes display the shrinkage-corrected correlations and allow users to examine how exposures are associated with demographic characteristics, diseases, and other exposures. Results are presented for the entire cohort regardless of phenotype, with options to view results stratified by sex, race, income, and age. Users can also view correlation globes for individual phenotypes that show only exposures that are significantly associated with the disease at an FDR < 0.01. The variable names shown have been simplified so they are easily understood by users, with the full text of the questions on which the display names are based available online (https://www.niehs.nih.gov/research/atniehs/labs/crb/studies/pegs/about/data/index.cfm). In addition, hovering the mouse over an exposure pair of interest brings up additional information in a pop-up text box.
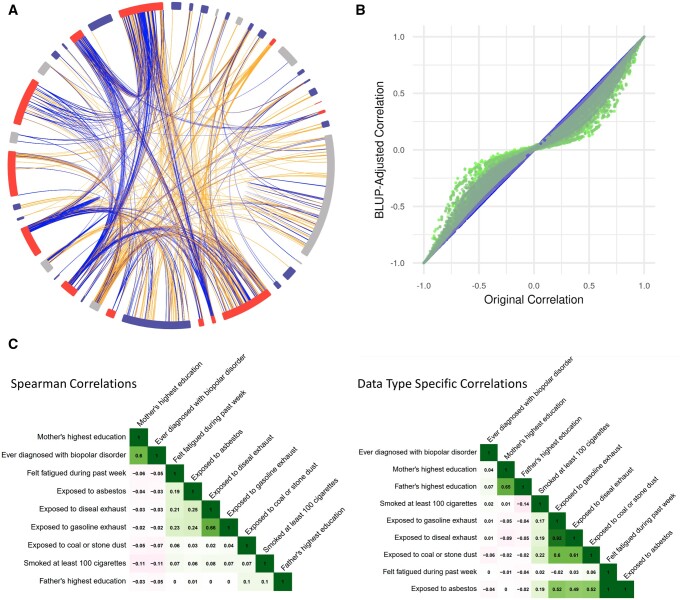
Figure 3A is an overall exposome globe for all surveys. Figure 3B shows how shrinkage affects the correlation coefficients for all pair-wise correlations across all three surveys Many coefficients drastically changed after fitting a BLUP model to account for sample size. Figure 3C shows the change in data-specific correlation coefficients and the structure of hierarchal clustering applied to the correlations. For example, using Spearman’s correlation coefficient, paternal and maternal education are not highly correlated, which is contrary to the expected results. When polychoric correlation is applied, the factors are highly correlated as expected. Additionally, for exposures already found to be correlated, the magnitude of the correlation coefficient changed when we applied an appropriate correlation method. When we applied hierarchical clustering to the correlations, the clustering structure changed dramatically with the use of data-specific methods.
Figure 3.
(A) Overall exposome globe for all surveys. The colors around the edge of the globe indicate the survey from which the sections come. The lines represent pairwise positive (blue) and negative (orange) correlations. Interactive globes can be viewed on the PEGS Explorer website. (B) We calculated correlations using appropriate methods for the data types in each variable pair and then shrunk the correlations to control for sample size (small = green; large = blue) by fitting a best linear unbiased prediction (BLUP) model. Original correlations (x-axis) and shrunken correlations (y-axis) are plotted for roughly 3.6M pairwise correlations. (C) Differences correlations calculated using Spearman’s rank correlation coefficient (left) and appropriate methods for the specific variable types (right) are shown for exemplar exposures in the External Exposome Survey.
Discussion
PEGS Explorer is an easy-to-use interactive website that allows users to explore ExWAS and correlation results from multiple studies. The straightforward globe visualizations in PEGS Explorer can assist both scientists and the general public in understanding correlations among exposures and allow for the examination of which exposures are associated with multiple diseases. Stratifying correlation results by race, sex, and other demographic categories highlights how some groups experience and are affected differently by exposures. Stratified results also allow users to explore biological hypotheses based on biological factors such as sex, enabling deeper investigations into questions surrounding issues such as sex differences in environmental risk factors for a particular disease. Providing results and the tools to interpret them will expand understanding of the exposome and its effects on human health and demonstrate what data are available and how they can be analyzed.
Disseminating the ExWAS results through PEGS Explorer will advance the field of exposome science in several meaningful ways. First, sharing ExWAS results through the publicly accessible web application will allow domain experts with knowledge of diseases and exposures to access and interpret the results and place them in the context of other work in their fields. Second, the findings can be used for hypothesis generation and further analyses, functional validation, and replication and could lead to the discovery of novel exposures involved in disease pathways. Third, transparently sharing ExWAS results and exposome correlations in a single online resource will facilitate the coordination and planning of future analyses and collaborations. Fourth, sharing the code used to build the PEGS Explorer visualizations and the underlying methods will enable researchers to recreate the results and incorporate them into their work. This commitment to data sharing and transparency is essential to advance the field of exposomics and is crucial to the field reaping the full benefits of substantial investments in ExWAS studies.
It is possible to apply principles used in genomics to characterize correlations in exposomics as well, such as HapMap17 and similar methods. HapMap characterizes human genetic variation and the different frequencies and correlations across race/ethnicity and ancestry similarity groups.18,19 Here, we apply these principles to present correlation results for exposures.20 Sharing references from multiple studies may be especially helpful when considering additional aspects of the exposome such as biological and chemical measurements of pollutants, data from wearable technology, and individual biological samples. While PEGS Explorer currently displays results from analyses of survey data, the underlying methods for the ExWAS, correlations, and exposome globe visualizations could be applied to these additional types of exposure data. For example, ongoing efforts include conducting ExWAS in PEGS using recently linked geospatial estimates of exposure, and PEGS Explorer can be expanded to disseminate those results.
By providing tools to analyze exposome data from the diverse PEGS cohort and disseminating the ExWAS results through an accessible platform, PEGS Explorer supports a collaborative approach to in-depth analysis of the exposome and examination of the associations of exposures with multiple phenotypes. It allows researchers to evaluate associations and correlation statistics without the upfront investment needed to gain access to, download, and clean data and run analyses themselves.
Supplementary Material
Acknowledgments
We wish to thank Hannah Collins Cakar for assistance with manuscript submission. We would like to thank Dr Yihui Zhao for sharing the R function to shrink correlations.
Contributor Information
Dillon Lloyd, Biostatistics and Computational Biology Branch, National Institute of Environmental Health Sciences, Durham, NC, USA.
John S House, Biostatistics and Computational Biology Branch, National Institute of Environmental Health Sciences, Durham, NC, USA.
Farida S Akhtari, Biostatistics and Computational Biology Branch, National Institute of Environmental Health Sciences, Durham, NC, USA.
Charles P Schmitt, Division of Translational Toxicology, National Institute of Environmental Health Sciences, Durham, NC, USA.
David C Fargo, Office of the Director, National Institute of Environmental Health Sciences, Durham, NC, USA.
Elizabeth H Scholl, Sciome LLC, Durham, NC, USA.
Jason Phillips, Sciome LLC, Durham, NC, USA.
Shail Choksi, Sciome LLC, Durham, NC, USA.
Ruchir Shah, Sciome LLC, Durham, NC, USA.
Janet E Hall, Clinical Research Branch, National Institute of Environmental Health Sciences, Durham, NC, USA.
Alison A Motsinger-Reif, Biostatistics and Computational Biology Branch, National Institute of Environmental Health Sciences, Durham, NC, USA.
Supplementary data
Supplementary material is available at Exposome online.
Funding
Financial support was received from intramural funds from the National Institutes of Health, National Institute of Environmental Health Sciences.
Author contributions
Dillon Lloyd (Data curation [equal], Formal analysis [equal], Writing—original draft [lead]), John S House (Data curation [equal], Project administration [equal], Writing—review & editing [equal]), Farida S Akhtari (Data curation [equal], Writing—review & editing [equal]), Charles Schmitt (Data curation [equal], Writing—review & editing [equal]), David C. Fargo (Data curation [equal], Writing—review & editing [equal]), Elizabeth H. Scholl (Data curation [equal], Formal analysis [equal], Visualization [equal], Writing—review & editing [equal]), Jason Phillips (Data curation [equal], Software [equal], Visualization [equal], Writing—review & editing [equal]), Shail Choksi (Software [equal], Visualization [equal], Writing—review & editing [equal]), Ruchir Shah (Project administration [equal], Writing—review & editing [equal]), Janet E. Hall (Funding acquisition [equal], Writing—review & editing [equal]), and Alison A. Motsinger-Reif (Conceptualization [equal], Funding acquisition [equal], Project administration [equal], Writing—review & editing [equal])
Data availability
The data underlying this article are available to researchers engaging in collaborative projects with PEGS. Information about submitting proposals for collaborative research is available at: https://www.niehs.nih.gov/research/clinical/studies/pegs/collaboration/proposal/index.cfm. Results from the ExWAS conducted as part of this work can be explored with the PEGS Explorer web application that can be accessed at: https://www.niehs.nih.gov/research/clinical/studies/pegs/index.cfm under the About PEGS tab.
Conflict of interest statement
None declared.
References
- 1. Wild CP. The exposome: from concept to utility. Int J Epidemiol. 2012;41(1):24-32. https://orcid.org/10.1093/ije/dyr236. [DOI] [PubMed] [Google Scholar]
- 2. Wild CP. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev. 2005;14(8):1847-1850. https://orcid.org/10.1158/5–9965.Epi-5–0456. [DOI] [PubMed] [Google Scholar]
- 3. Akhtari FS, Lloyd D, Burkholder A, et al. Questionnaire-based polyexposure assessment outperforms polygenic scores for classification of type 2 diabetes in a multiancestry cohort. Diabetes Care. 2022;46(5):929-937. https://orcid.org/10.2337/dc22-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee EY, Choi W, Burkholder AB, et al. Race/ethnicity-stratified fine-mapping of the MHC locus reveals genetic variants associated with late-onset asthma. Front Genet. 2023;14:1173676.https://orcid.org/10.3389/fgene.2023.1173676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lowe ME, Akhtari FS, Potter TA, et al. The skin is no barrier to mixtures: Air pollutant mixtures and reported psoriasis or eczema in the Personalized Environment and Genes Study (PEGS). J Expo Sci Environ Epidemiol. 2023;33(3):474-481. https://orcid.org/10.1038/s41370-2–00502-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ayala-Ramirez M, MacNell N, McNamee LE, et al. Association of distance to swine concentrated animal feeding operations with immune-mediated diseases: an exploratory gene-environment study. Environ Int. 2023;171:107687. https://orcid.org/10.1016/j.envint.2022.107687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Neveu V, Moussy A, Rouaix H, et al. Exposome-explorer: a manually-curated database on biomarkers of exposure to dietary and environmental factors. Nucleic Acids Res. 2016;45(D1):D979-D984. https://orcid.org/10.1093/nar/gkw980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vineis P, Chadeau-Hyam M, Gmuender H, et al. The exposome in practice: design of the EXPOsOMICS project. Int J Hyg Environ Health. 2017;220(2 Pt A):142-151. https://orcid.org/10.1016/j.ijheh.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Patel CJ, Manrai AK.. Development of exposome correlation globes to map out environment-wide associations. In: Proceedings of the Pacific Symposium on Biocomputing. World Scientific Publishing; 2015;20:231-242. [PMC free article] [PubMed] [Google Scholar]
- 10. Timofeeva A, Khailenko E. Generalizations of the polychoric correlation approach for analyzing survey data. In: International Forum on Strategic Technology. Institute of Electrical Engineers (IEEE) 2016:254–258.
- 11. Drasgow F. Polychoric and polyserial correlations. In: Kotz S, Read, BC, Balakrishnan N, et al., eds. Wiley StatsRef: Statistics Reference Online. 2014. https://orcid.org/10.1002/0471667196.ess2014.pub2. [Google Scholar]
- 12. Bonett DG, Price RM.. Inferential methods for the tetrachoric correlation coefficient. J Educ Behav Stat. 2005;30(2):213-225. [Google Scholar]
- 13. Mandal M, Levy J, Ives C, et al. Correlation analysis of variables from the Atherosclerosis Risk in Communities Study. Front Pharmacol. 2022;13:883433. https://orcid.org/10.3389/fphar.2022.883433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee EY, Akhtari F, House JS, et al. Questionnaire-based exposome-wide association studies (ExWAS) reveal expected and novel risk factors associated with cardiovascular outcomes in the Personalized Environment and Genes Study. Environ Res. 2022;212(Pt D):113463. https://orcid.org/10.1016/j.envres.2022.113463. [DOI] [PubMed] [Google Scholar]
- 15. Sinisi SE, van der Laan MJ.. Deletion/substitution/addition algorithm in learning with applications in genomics. Stat Appl Genet Mol Biol. 2004;3:Article 18. https://orcid.org/10.2202/4–6115.1069. [DOI] [PubMed] [Google Scholar]
- 16. McCulloch CE, Searle SR.. Generalized, Linear, and Mixed Models. John Wiley & Sons; 2004. [Google Scholar]
- 17. International HapMap Consortium. The International Hap Map Project. Nature. 2003;426:789–796. https://orcid.org/10.1038/mature02168. [DOI] [PubMed] [Google Scholar]
- 18. Manolio TA, Brooks LD, Collins FS.. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118(5):1590-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Manolio TA, Collins FS.. The HapMap and genome-wide association studies in diagnosis and therapy. Annu Rev Med. 2009;60:443-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang P, Carlsten C, Chaleckis R, et al. Defining the scope of exposome studies and research needs from a multidisciplinary perspective. Environ Sci Technol Lett. 2021;8(10):839-852. https://orcid.org/10.1021/acs.estlett.1c00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available to researchers engaging in collaborative projects with PEGS. Information about submitting proposals for collaborative research is available at: https://www.niehs.nih.gov/research/clinical/studies/pegs/collaboration/proposal/index.cfm. Results from the ExWAS conducted as part of this work can be explored with the PEGS Explorer web application that can be accessed at: https://www.niehs.nih.gov/research/clinical/studies/pegs/index.cfm under the About PEGS tab.