Abstract
In the yeast Saccharomyces cerevisiae, the SWI-SNF complex has been proposed to antagonize the repressive effects of chromatin by disrupting nucleosomes. The SIN genes were identified as suppressors of defects in the SWI-SNF complex, and the SIN1 gene encodes an HMG1-like protein that has been proposed to be a component of chromatin. Specific mutations (sin mutations) in both histone H3 and H4 genes produce the same phenotypic effects as do mutations in the SIN1 gene. In this study, we demonstrate that Sin1 and the H3 and H4 histones interact genetically and that the C terminus of Sin1 physically associates with components of the SWI-SNF complex. In addition, we demonstrate that this interaction is blocked in the full-length Sin1 protein by the N-terminal half of the protein. Based on these and additional results, we propose that Sin1 acts as a regulatable bridge between the SWI-SNF complex and the nucleosome.
Genetic studies have shown that chromatin structure in the yeast Saccharomyces cerevisiae affects gene expression (11, 47). The study of mutations that suppress transcriptional defects caused by Ty or δ insertion mutations at HIS4 or LYS2 (named SPT for suppressor of Ty [46]) identified a group of genes whose products are involved in chromatin structure and its regulation. These include histones H2A and H2B (SPT11 and SPT12) (8), the SPT2 gene, which encodes an HMG1-like protein (14, 31), and genes whose activity has been proposed to affect nucleosome assembly (SPT4, SPT5, and SPT6) (7, 20, 43). The ability of this group of genes to affect transcription suggested an important role for chromatin in the control of gene expression.
A second group of genetic screens, which identified SWI-SNF components, were obtained from an analysis of the HO gene (required for mating type switching; SWI stands for switching [39]) and the SUC2 gene (encoding an invertase required for growth on sucrose and raffinose; SNF stands for sucrose nonfermenting [24]). Genetic and biochemical studies (reviewed in reference 29) have shown that the SWI-SNF products form a complex composed of at least 11 polypeptides, including SWI1-ADR6, SWI2-SNF2, SW13, SNF5, SNF6, SNF11, TFG3, and SWP73 (5, 6, 16, 17, 27, 44). The link between the SWI-SNF complex and chromatin was identified by the study of suppressors of defects in components of this complex. Deletion of one of the two loci that encode histones H2A and H2B suppresses transcriptional defects caused by loss of the SWI-SNF complex (12). The SIN (for switch independent) genes were identified as suppressors of the swi phenotype (23, 40). Two of them, sin1 and sin2, partially suppress mutants of the SWI1, SWI2, and SWI3 genes (14, 15, 40). The sin2-1 mutation was found to lie in the HHT1 gene, which encodes histone H3. Five additional point mutations, two in histone H3 and three in histone H4, also displayed a Sin− phenotype in that they partially suppress the requirements for SWI genes in transcriptional activation (15, 20). These mutations change residues believed to contact DNA or to be involved in histone-histone interactions within the histone octamer and thus might affect nucleosome stability (45). SIN1 was found to be allelic to SPT2 and encodes an HMG1-like protein (14). Furthermore, other spt mutants are able to suppress defects in the SWI-SNF complex (47), lending additional support to the idea that the SWI-SNF complex is involved in chromatin remodeling.
In this study, we address the role of SIN1-SPT2. As outlined above, this gene was obtained by two different screens and encodes a protein with sequence similarities to mammalian HMG1 proteins. The localization of the protein to the nucleus, its ability to bind DNA nonspecifically, and its relatively high abundance (14) suggest that Sin1 also encodes a protein similar to the mammalian HMG1 proteins. Though the precise role of the mammalian HMG1 proteins is not known, they have been implicated in transcriptional processes and chromatin assembly (3, 35). In yeast, Sin1 has been defined genetically as a negative regulator of transcription, but its precise role and specific targets in the cell are not known. Here we provide evidence that the Sin1 protein interacts in a regulated way with both histones and with components of the SWI-SNF complex, and we suggest that Sin1 mediates the effects of the SWI-SNF complex on chromatin.
MATERIALS AND METHODS
Strains and genetic methods.
The S. cerevisiae strains used in this study, described in Table 1, are derivatives of JJY10 (26), MATa ura3-52 leu2Δ1 trp1 his4-912δ lys2-128δ HO-lacZ. Standard yeast genetic methods were used (32). The sin1Δ::TRP1 allele was constructed by one-step gene replacement with the plasmid pUC-SIN1Δ-TRP1 (14). The HO-lacZ fusion allele is described in reference 33. The histone mutations were introduced into the chromosome by a two-step replacement procedure (34) with integrating plasmids marked with the URA3 gene (obtained from R. K. Tabtiang and I. Herskowitz). A strain carrying a swi5::LEU2 null allele was generated as described in reference 41.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| JJY10 | Mata ura3-52 leu2Δ1 trp1 his4-912δ lys2-128δ HO-lacZ |
| JJY14 | same as JJY10 plus swi5::hisG |
| JJY15 | same as JJY14 plus sin1Δ::TRP1 |
| JJY19 | same as JJY14 plus hhf2-7 |
| JJY20 | same as JJY14 plus hhf2-8 |
| JJY21 | same as JJY14 plus hhf2-13 |
| JJY22 | same as JJY14 plus sin2-1 |
| JJY23 | same as JJY10 plus sin1Δ::TRP1 |
| JJY24 | same as JJY10 plus hhf2-7 |
| JJY25 | same as JJY10 plus hhf2-8 |
| JJY26 | same as JJY10 plus hhf2-13 |
| JJY27 | same as JJY10 plus sin2-1 |
| JJY29 | same as JJY15 plus hhf2-7 |
| JJY31 | same as JJY15 plus hhf2-8 |
| JJY33 | same as JJY15 plus hhf2-13 |
| JJY35 | same as JJY15 plus sin2-1 |
Expression vectors.
pLL10 is a 2μm vector (YEp13) carrying the LEU2 marker and the wild-type SIN1 locus (18). pBD1 is a 2μm vector (YEp24) carrying the URA3 marker and the wild-type SWI1 locus. pBD12 is an ARS vector (YCp50) carrying the URA3 marker and the wild-type SWI1 locus (38).
To overexpress SIN1, a 1-kb EcoRI-SalI fragment carrying the SIN1 open reading frame was amplified by PCR and subcloned into the plasmid pRD53 (YCp vector, GAL1 promoter, URA3 marked; R. Deshaies, California Institute of Technology) under the control of the GAL1 promoter to create plasmid pRD-SIN1. Sequences encoding SIN1Δ189–333 (N-terminal [Nt] half) and SIN1Δ1–188 (C-terminal [Ct] half) were produced by PCR amplification of a 0.57- and 0.43-kb EcoRI-SalI fragment, respectively, with the SIN1 gene and appropriate oligonucleotides. The PCR products were cloned into pRD53 or pJL602 (YCp vector, GAL1 promoter, LEU2 marked, J. Li; University of California, San Francisco), to give pRD-SIN1Nt and pRD-SIN1Ct or pJL-SIN1Nt and pJL-SIN1Ct, respectively. All PCR products were verified by sequencing.
Glutathione S-transferase (GST) fusion proteins were expressed in yeast with the plasmid pRD56 (YCp vector, URA3 marked; R. Deshaies), which contains the GAL1 promoter followed by the GST coding region. The various GST-SIN1 gene fusions were produced by subcloning the EcoRI-SalI fragments from pRD-SIN1, pRD-SIN1Nt, and pRD-SIN1Ct into pRD56. In yeast, these fusions produced the same phenotypes as did their non-GST-fused counterparts.
GST purifications and Western blotting.
GST purifications were carried out as described previously (21). Briefly, overexpressing strains were constructed by transforming JJY23 (sin1 Δ::TRP1) with the respective GST plasmids. Cultures (100 ml each) of yeast cells expressing GST, GST-SIN1, GST-SIN1 Nt and GST-SIN1 Ct fusions were grown in selective media containing 2% galactose to an A660 of 1. Cells were harvested and lysed with glass beads, and a protein lysate was prepared in buffer A (50 mM HEPES [pH 7.6], 10% glycerol, 10 mM EDTA, 0.2 M NaCl, 1% Triton X-100, 5 mM dithiothreitol, 2 mg [each] of leupeptin, bestatin, and pepstatin per ml, 5 mM benzamidine-HCl, and 1 mM phenylmethylsulfonyl fluoride). After a high-speed spin, the supernatant was saved, and 400 μl of the lysates was incubated with 200 μl of a 50% slurry of glutathione agarose (Sigma) beads. Reaction mixtures were incubated at 4°C on an end-over-end mixer for 1 h and centrifuged at 2,000 rpm for 2 min. The beads were washed twice with buffer A and once more with buffer A lacking Triton X-100, resuspended in 100 μl of 2× Laemmli sample buffer, and boiled for 5 min. Ten microliters of each reaction mixture was applied to sodium dodecyl sulfate (SDS) polyacrylamide gels, followed by electrophoresis and transfer to a polyvinylidene difluoride membrane (Millipore). Blots were incubated with GST antibody or SIN1 antibody (a gift of R. K. Tabtiang) or with SWI1-ADR6 antibody (a gift of E. T. Young) followed by antirabbit antibody coupled with horseradish peroxidase. Western detection was performed by the Amersham enhanced chemiluminescence system.
RNA analysis.
Strains were grown to mid-log phase in yeast extract-peptone-dextrose medium. Total yeast RNA was isolated and fractionated on formaldehyde gels, transferred to nylon membranes (Genescreen; DuPont), and hybridized with random-primed 32P-labeled fragments. The DNA probes used were obtained as PCR fragments by amplification of the desired open reading frame with specific primers.
Other methods.
Yeast cells were transformed by the lithium acetate method (10). β-Galactosidase assays were performed as described previously (32). For growth in toxic conditions (i.e., overexpression of the Sin1 Ct domain), both assay and control cells were grown on plates for 3 to 4 days, and a similar number of cells were scratched from the plate, washed twice with Z buffer, resuspended to a similar optical density at 600 nm in Z buffer, and subjected to β-galactosidase assay (32).
RESULTS
Sin1 interacts genetically with histones H3 and H4.
The sin2-1 mutation (which lies in one of the two genes encoding histone H3) was recovered in the same screen as the original sin1 mutation. Both mutations were identified by their ability to suppress swi defects (40). Five additional point mutations (histone sin mutations), two in the histone H3 and three in the histone H4 genes, also displayed a Sin− phenotype in that they partially suppress the requirements for SWI genes in transcriptional activation (15, 30). Both sin1 and histone sin mutations allow growth in medium lacking lysine or histidine of a strain carrying the mutant alleles lys2-128δ and his4-912δ (Spt− phenotype [15, 31]). These mutations also permit the expression of the HO gene (quantified as β-galactosidase activity produced by a HO-lacZ gene fusion) in a strain carrying a disruption of the gene SWI5 (one of the regulators of this promoter [22]) (Sin− phenotype; see reference 15). Furthermore, sin1 and sin2-1 mutations both suppress gcn5 defects (26) as well as transcriptional defects caused by partial deletions of the Ct domain of the largest subunit of RNA polymerase II (Srb− phenotype [28]). These results suggest that Sin1 and the histones H3 and H4 may be involved in the same process. To test this idea, we measured ability to suppress the Sin phenotype by the combination of a deletion in the SIN1 gene and several sin histone alleles (sin2-1, hhf2-7, hhf2-8, and hhf2-13; all these mutations are partially dominant [15]). We found (Table 2) that the double mutants with sin1Δsin histone mutations have the same degree of effect (quantitated as β-galactosidase activity) as do the single sin histone mutants, suggesting that SIN1 and the histone genes work together in the same genetic pathway.
TABLE 2.
β-Galactosidase activity of sin1 and histone sin mutants
| Strain | Relevant genotype | HO-lacZ activity (Miller units) |
|---|---|---|
| JJY10 | SWI5 | 110 |
| JJY14 | swi5 | 0.1 |
| JJY15 | swi5 sin1Δ | 33.8 |
| JJY19 | swi5 hhf2-7 | 40.9 |
| JJY29 | swi5 sin1Δ hhf2-7 | 32.8 |
| JJY20 | swi5 hhf2-8 | 45.3 |
| JJY31 | swi5 sin1Δ hhf2-8 | 35 |
| JJY21 | swi5 hhf2-13 | 41.9 |
| JJY33 | swi5 sin1Δ hhf2-13 | 34.9 |
| JJY22 | swi5 sin2-1 | 45 |
| JJY35 | swi5 sin1Δ sin2-1 | 38 |
In support of this last idea, we found that a high-copy-number plasmid carrying the SIN1 gene can suppress the Spt− and Sin− phenotypes produced by the sin histone mutations (Fig. 1). This suppression is specific in that the same plasmid was unable to suppress other mutations that have the same range of phenotypes, including spt4, spt5, and spt6, and high and low doses of H2A-H2B or H3-H4 gene pairs (data not shown).
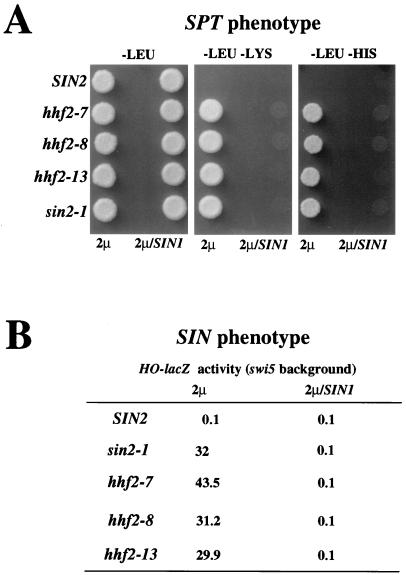
FIG. 1.
SIN1 is a high-copy suppressor of the sin mutations in histone H3 and H4 genes. Wild-type (SIN2; JJY14) or H4 histone mutant (hhf2-7, JJY19; hhf2-8, JJY20; hhf2-13, JJY21) and H3 histone mutant (sin2-1; JJY22) cells were transformed with YEp13 (2μm) or YEp13-SIN1 (2μm/SIN1). (A) Spt phenotype. Two microliters of a cell suspension (approximately 5 × 106 cells/ml) from each culture was spotted onto minimal-medium plates (lacking leucine, lacking leucine and lysine, or lacking leucine and histidine) and incubated at 30°C for 3 days. Cells with a histone sin mutation show an Spt− phenotype (i.e., the ability to suppress a δ element insertion in the HIS4 and LYS2 promoters) and are able to grow without lysine or histidine. A high dose of the SIN1 gene suppresses this phenotype. (B) Sin phenotype. Exponentially growing cultures were assayed for β-galactosidase activity, expressed as Miller units. Cells with a histone sin mutation show a Sin− phenotype; that is, they are able to express the HO gene in the absence of the Swi5 protein, one of the activators of this promoter (note that all strains used in the experiment shown in this figure carry the swi5::hisG mutation). This ability is suppressed by a high-copy vector carrying the SIN1 gene.
Histone mutations reduce the level of Sin1 protein.
During the course of this analysis, we noticed that the strains carrying the histone sin mutations have lower levels of Sin1 protein (Fig. 2A) than do strains that carry normal histone genes. The histone mutant strains have a wild-type copy of the SIN1 gene and produce normal levels of SIN1 mRNA (Fig. 2C), suggesting that Sin1 is made but rapidly degraded. These results help explain why the sin histone alleles produce many of the same phenotypes as does a deletion of the SIN1 gene and why the overexpression of Sin1 (Fig. 2B) suppresses the effects produced by the histone sin mutations.
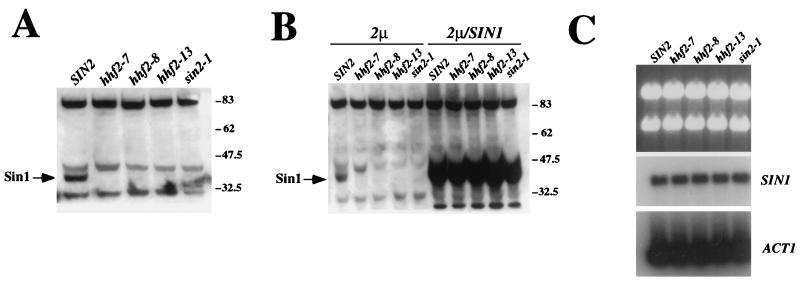
FIG. 2.
Sin1 levels are lower in histone sin mutants. (A) Western blot with an anti-Sin1 polyclonal antibody from wild-type (SIN2; JJY14), H4 histone mutant (hhf2-7, JJY19; hhf2-8, JJY20; hhf2-13, JJY21), and H3 histone mutant (sin2-1; JJY22) cells. The arrow indicates the Sin1 protein. (B) Overexpression of Sin1 protein in histone sin mutant cells. The strains used are the same as those shown in panel A but were transformed with a control high-copy-number plasmid (2 μm) or the same plasmid carrying the wild-type SIN1 locus (2μm/SIN1). (C) Northern analysis of strains used in panel A. The upper panel shows the agarose gel stained with ethidium bromide, while the middle and bottom panels show blots of the same gel after hybridization with a SIN1 (middle) or an ACT1 (bottom) probe. Numbers to the right of panels A and B indicate molecular weight standards (in thousands).
Overexpression of the Ct end of Sin1 is toxic to cells.
The suppression of sin histone alleles by a high dose of SIN1 requires the presence of a functional carboxy-terminal end in the protein; that is, point mutations or small deletions of the Ct end prevented this suppression. This observation suggests that this region of the protein may be involved in the interaction with histones. To determine the consequences of overexpression of this portion of Sin1, we placed full-length Sin1, the Nt domain of Sin1, and the Ct domain of Sin1 under the control of the GAL1 promoter (Fig. 3A). We found (Fig. 3B) that overexpression of the full-length Sin1 produced no apparent effects in the cell, whereas overexpression of the Nt half of Sin1 produced a dominant negative sin1 mutant phenotype (data not shown) in accordance with published reports (18). In contrast, overexpression of the Ct half of Sin1 produced a spectrum of unanticipated phenotypes, including slow growth and low expression of the HO gene (Fig. 3B).
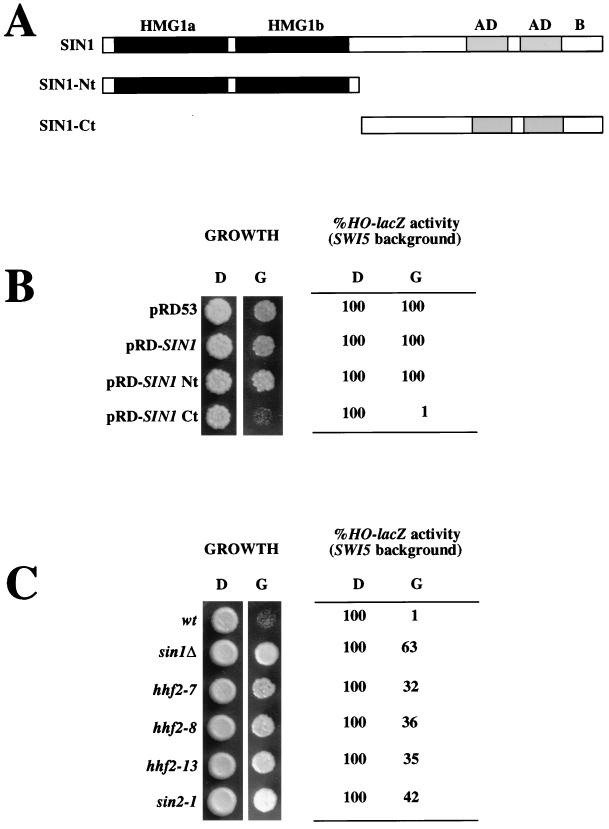
FIG. 3.
SIN1 Ct half overexpression causes cell growth defects and low HO expression. (A) Schematic representation of the Sin1 protein with its most relevant characteristics. The Nt half includes two regions with similarities to mammalian HMG1 (HMG1a and HMG1b) protein. The Ct includes two regions rich in acidic domains (similar to those found in several other HMG-like proteins [AD]) and a region rich in positively and negatively charged residues (B). The panel also shows the Nt and Ct derivatives. (B) Overexpression of the Sin1 Ct causes slow growth and low HO expression. To score growth, 2 μl of a suspension (approximately 5 × 106 cells/ml) of JJY10 cells transformed with the indicated plasmids was spotted onto minimal medium lacking uracil and containing either dextrose (D) or galactose (G). Plates were incubated at 30°C for 3 days. HO-lacZ activity was determined by assaying for β-galactosidase activity in exponentially growing liquid cultures in either dextrose or galactose. The expression of the HO-lacZ reporter was normalized to that of the control (JJY10 transformed with pRD53 in dextrose), which ranged between 110 and 90 Miller units. β-Galactosidase activity of cells grown in galactose was similar to that of cells grown in dextrose. (C) The defects produced by Sin1 Ct overexpression are alleviated by the same kinds of mutations that suppress swi defects. The plasmid pRD-SIN1Ct was introduced in JJY10 (wild type [wt]), JJY23 (sin1Δ::TRP1), JJY24 (hhf2-7), JJY25 (hhf2-8), JJY26 (hhf2-13), and JJY27 (sin2-1) cells, and cultures of these strains were scored for both growth and HO-lacZ expression as described above. The Sin1 Ct protein was expressed at similar levels in all strains, as assessed by Western blotting (data not shown).
The effects of Sin1 Ct overexpression are alleviated by any of the sin histone alleles described in this study and by a deletion of the chromosomal copy of the SIN1 gene. The levels of the Sin1 Ct half are the same whether or not the endogenous full-length gene is present (data not shown) and in strains carrying sin histone mutations (Fig. 3C). We also found that overexpression of the Sin1 Ct half is nontoxic in spt4, spt5, and spt6 mutant strains (data not shown).
Defects caused by Sin1 Ct overexpression can be suppressed by a high dose of the SWI1 gene.
The effects of overexpression of the Ct domain of Sin1 resembled those produced by the loss of function mutations in components of the SNF-SWI complex. It therefore seemed plausible that Sin1 Ct inhibited the activity of the SWI-SNF complex. This idea is consistent with the observation that deletion of the chromosomal copy of SIN1 suppressed the defects produced by the Sin1 Ct overexpression, because SIN1 deletions suppress defects produced by SWI-SNF mutations. Consistent with this idea, we found that a high dose of the SWI1 gene specifically suppresses the defects produced by the Sin1 Ct overexpression (Fig. 4). Neither the SWI1 gene present on a low-copy plasmid (ARS) nor an SWI2 or SWI3 gene present on high-copy-number plasmids efficiently suppressed the observed defects (data not shown).
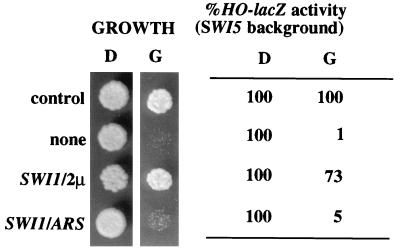
FIG. 4.
High dose of SWI1 suppresses the Sin1 Ct-associated defects. JJY10 cells were transformed with pJL602 and YEp24 (control), pJL-SIN1Ct and YEp24 (none), pJL-SIN1Ct and pBD1 (SWI1/2μm), and pJL-SIN1Ct and pBD12 (SWI1/ARS). Cell growth and HO-lacZ activity were scored as in Fig. 3.
Physical association between the Sin1 Ct half and SWI-SNF components.
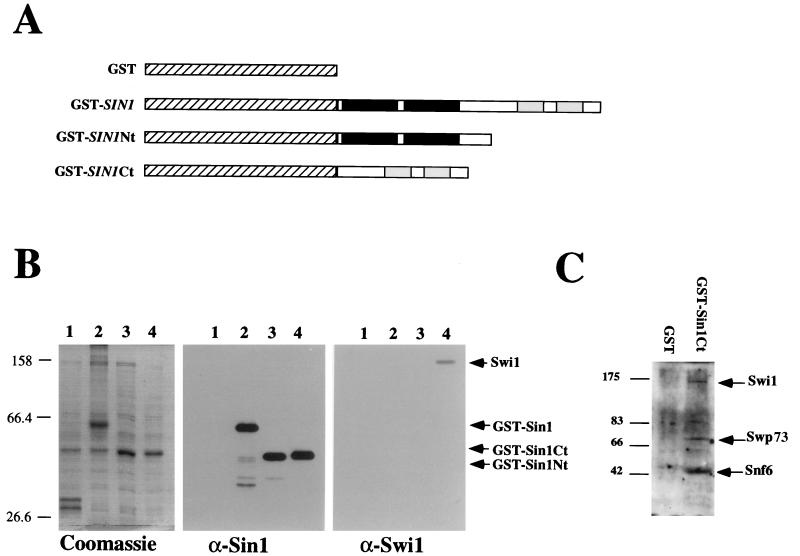
The genetic interactions described above suggest that the Ct end of Sin1 interacts with the SWI-SNF complex and blocks its activity. Therefore, we investigated whether the Ct half of Sin1 interacts physically with the SWI-SNF complex. We overexpressed several GST-Sin1 protein fusions in sin1Δ hosts (Fig. 5A), purified these proteins by affinity chromatography, and determined whether the Swi1 protein was associated with them. We found that Swi1 protein associates with a GST-Sin1 Ct protein fusion but not with a GST, a GST-Sin1 Nt fusion, or a GST-Sin1 full-length fusion (Fig. 5B). Using antibodies against Snf6 and Swp73 (two additional components of the SWI-SNF complex), we found that these proteins also associated with the GST-Sin1 Ct fusion (Fig. 5C), indicating that the SWI-SNF complex (and not just Swi1) associates with the Sin1 Ct.
FIG. 5.
Interactions between the Ct half of Sin1 and the Swi1 protein. (A) Schematic representation of fusion proteins used. GST portions are indicated as hatched boxes. The proposed HMG1 boxes in Sin1 are highlighted in black, and the two acidic tracts are shaded. (B) The Swi1 protein copurifies with the Ct half of Sin1. Proteins obtained by the GST-affinity purification procedure (see Materials and Methods) were separated on SDS–10% polyacrylamide gels and either stained with Coomassie (left panel) or transferred to Immobilon membrane (Millipore), probed with the antiserum indicated, and detected with a secondary antibody and the Amersham enhanced chemiluminescence detection kit (center and right panels). The samples were obtained from cells overexpressing GST alone (lane 1), GST-SIN1 fusion (lane 2), GST-Sin1 Nt fusion (lane 3), and GST-Sin1 Ct fusion (lane 4). In spite of its lower estimated molecular weight, the GST-Sin1 Ct fusion migrates more slowly than does the GST-Sin1 Nt fusion, perhaps due to its high content of charged residues. (C) Western blot of GST-affinity eluates from cells overexpressing GST or the GST-Sin1 Ct fusion probed with anti-Swi1, anti-Swp73, and anti-Snf6. The numbers to the left of the blots are molecular weight standards (in thousands).
The Nt half of Sin1 masks the ability of the Sin1 Ct to interact with SWI-SNF components.
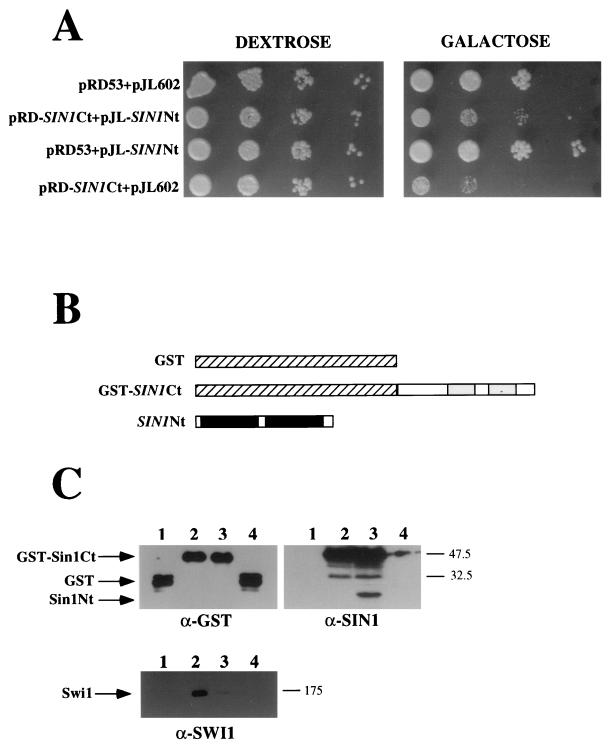
The fact that Swi1 associates with the GST-Sin1 Ct protein fusion but not with the GST-Sin1 full-length fusion is consistent with the fact that overexpression of full-length Sin1 did not produce any detectable phenotype, whereas overexpression of the Ct domain caused a range of swi/snf-like phenotypes. One model to explain these results is that in the full-length Sin1 protein, the Nt half masks the Ct half and thereby prevents its interaction with the Swi1 protein. To test this model, we overexpressed both halves of Sin1 as independent polypeptides in the cell at the same time. We found that overexpression of the Sin1 Nt half alleviates the defects associated with overexpression of the Sin1 Ct half alone (Fig. 6A). In addition, we found by affinity chromatography that the Nt half of Sin1 specifically associates with the Ct half (Fig. 6B and C). Moreover, the presence of the Nt half of Sin1 in the cell impairs the binding of Swi1 protein to the Sin1 Ct half (Fig. 6C), further supporting the idea that intramolecular masking prevents the full-length Sin1 protein from interacting with the Swi1 protein.
FIG. 6.
The Sin1 Nt half interacts with the Sin1 Ct half. (A) Overexpression of the Sin1 Nt half alleviates the defects associated with the overexpression of Sin1 Ct. Cells (JJY10) carrying the following plasmids—pRD53 and pJL602 (controls), pRD-SIN1Ct and pJL-SIN1Nt (Sin1 Ct and Sin1 Nt as independent polypeptides), pRD53 and pJL-SIN1Nt (Sin1 Nt alone), pRD-SIN1Ct and pJL602 (Sin1 Ct alone)—were spotted in 10-fold serial dilutions into media lacking uracil and leucine with either dextrose or galactose and incubated for 3 days at 30°C. (B) Scheme showing the protein fusions used in panel C. Note that Sin1 Nt half was not fused to GST protein in the following experiments. (C) Sin1 Nt interacts with GST-Sin1 Ct. Proteins obtained by the GST-affinity purification procedure were loaded into an SDS–12% polyacrylamide gel (GST and Sin1 blots) or an SDS–8.5% polyacrylamide gel (Swi1 blot) and treated as described in the legend for Fig. 4B. The samples were obtained from cells expressing GST (lane 1), GST-Sin1 Ct (lane 2), GST-Sin1 Ct and SIN1 Nt (lane 3), or GST and Sin1 Nt (lane 4). The numbers to the right of the blots are molecular weight standards (in thousands).
DISCUSSION
In this study, we investigated the functional relations between Sin1, histones H3 and H4, and the SWI-SNF complex. Our results, taken together with those of previous studies (14, 15, 40), indicate that the Sin1 protein interacts with both the nucleosome and the SWI-SNF complex.
Sin1 is highly charged and shows two regions of similarity to the mammalian HMG1 protein. The HMG proteins were originally described as nonhistone components of chromatin, and it is well established that the mammalian proteins are able to bind to assembled nucleosomes (1, 35). The sequence characteristics of the Sin1 protein, its nuclear localization, its abundance, and its ability to bind DNA in a nonspecific way (14) suggested that this protein may be a chromatin component, and the results presented in this study, summarized below, support this idea. In addition, we found, using hydroxyapatite fractionation of a yeast nuclear extract, that Sin1 elutes at the same salt conditions as do histones H3 and H4 (data not shown).
The specific suppression of the sin histone mutations by a high dose of SIN1, as well as the sin1 and sin histone allele double-mutant analysis, indicates that the two genes function together, a conclusion that is consistent with the similarity of phenotypes between sin1 and histone sin mutations (14, 15, 28). Furthermore, the observation that the Sin1 protein is present at significantly reduced levels in a strain carrying sin histone alleles further supports a physical association between Sin1 and histones H3 and H4. This result also suggests that the ability of sin histone mutations to suppress swi defects could be mediated by the effects of the levels of Sin1. Since SIN1 mRNA levels are unchanged in the histone mutants, the reduction in Sin1 levels must occur posttranscriptionally. A likely possibility is that Sin1 that is not complexed in chromatin is degraded. It has been reported that the Nt domain of Sin1 interacts with Cdc23 (37), a component of the APC ubiquitin ligase and a protein with homologies to the AAA family of proteasome components (19). It is possible that these factors affect the stability of Sin1.
Our experiments also demonstrate genetic and physical interactions between the Sin1 protein and the SWI-SNF complex. The defects observed when the Ct half of Sin1 is overexpressed, as well as the scope of the mutations which suppress such defects, indicate that the overexpression of the Sin1 Ct half interferes with the SWI-SNF complex. Furthermore, the ability of high levels of SWI1 to correct these defects supports this view. Finally, results of copurification experiments indicate that the Ct half of Sin1 is physically associated with at least three components of the SWI-SNF complex.
Our results do not address the question of whether the Sin1-SWI-SNF interaction is direct or whether it occurs through one or more intermediates. We think it unlikely that DNA could serve as an intermediate, because the ability of Sin1 to bind DNA is located in the Nt domain of Sin1 (14, 48) and the interaction with SWI-SNF was seen in the absence of this domain. In addition, the finding that full-length Sin1 and the Nt half of Sin1 (both of which contain the DNA binding domain) do not interact with the SWI-SNF complex supports the view that the interaction is not mediated just through DNA. We suggest that the simplest explanation for these observations is a direct interaction between Sin1 and the SWI-SNF complex.
An unexpected feature of the Sin1-SWI-SNF interaction is that it is observed only with the Ct half and not with the full-length Sin1 protein. The simplest interpretation of this result is the existence of a masking domain in the Sin1 protein. Intramolecular masking domains, which are released in response to stimuli or interactions with the appropriate partner, have many precedents (9, 13, 25). We propose, therefore, that Sin1 is able to interact with the SWI-SNF complex only when its Ct domain is released from the interaction with the Nt half of the protein. The suppression of the growth defect associated with Sin1 Ct overexpression by the Nt half of Sin1 (Fig. 6A) as well as the association of the Nt and Ct domains expressed as separated polypeptides (Fig. 6C) supports this interpretation. Furthermore, an interaction between the basic Nt half and the acidic Ct half in mammalian HMG1 proteins has been demonstrated (36, 42), reinforcing the similarities between Sin1 and the mammalian HMG1 proteins. In the case of HMG1 proteins, the Nt-Ct interaction is released by the binding to DNA (36, 42). We do not know what signal might release the interaction between the two domains in Sin1 protein, but an appealing possibility is that this release occurs as a consequence of interaction of Sin1 with the nucleosome.
The genetic and biochemical results described in this study all point to Sin1 as a target of the SWI-SNF complex. One plausible scenario is that nucleosome disruption by the SWI-SNF complex involves not only the removal of H2A-H2B dimers, as has been proposed previously (29), but also the specific removal of other chromatin-associated proteins, such as Sin1.
The experiments described in this study, the previous work of others (14, 18), and the comparison of Sin1 with the better-studied mammalian HMG1 proteins (4) all suggest a provisional model for Sin1 function. We propose that in solution, Sin1 is folded back on itself as the result of interactions between its Nt and Ct halves. This interaction would prevent Sin1 from interacting with the SWI-SNF complex in solution. The binding of Sin1 to the nucleosome (either to DNA or to histones) would, according to this model, release the inhibition.
Since Sin1 is formally a repressor of transcription, while the SWI-SNF components are formally activators, our proposal that the two function together may seem paradoxical. However, it is possible that Sin1 functions both to stabilize chromatin, perhaps by interacting with the nucleosome core, and to destabilize it by recruiting the SWI-SNF complex. According to this view, Sin1 would function to maintain the balance between chromatin assembly and disassembly.
ACKNOWLEDGMENTS
We thank R. K. Tabtiang for providing indispensable strains, plasmids, antibodies, and advice throughout the course of this work; E. T. Young for providing anti-SWI1 antibodies; B. Cairns for providing anti-SNF6 and anti-SWP73 antibodies; D. Moazed for expert advice on affinity purification procedures; R. Smith for proposing the experiment shown in Fig. 6A; and B. Braun, I. Herskowitz, D. Moazed, R. K. Tabtiang, and F. Winston for comments on the manuscript.
This study was supported by an NIH grant to A.D.J. and an EMBO long-term postdoctoral fellowship to J.P.-M.
REFERENCES
- 1.Bernues J, Querol E, Martinez P, Barrios A, Espel E, Llobevas J. Detection by chemical cross-linking of the interaction between high mobility group protein 1 and histone oligomers in free solution. J Biol Chem. 1983;258:11020–11024. [PubMed] [Google Scholar]
- 2.Bianchi M E, Falciola L, Ferrari S, Lilley D M J. The DNA binding site of HMG1 protein is composed of two similar segments (HMG boxes), both of which have counterparts in other eukaryotic regulatory proteins. EMBO J. 1992;3:1055–1063. doi: 10.1002/j.1460-2075.1992.tb05144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonne-Andrea C, Harper F, Sobczac J, de Recondo A M. Rat liver HMG1: a physiological nucleosome assembly factor. EMBO J. 1984;3:1193–1199. doi: 10.1002/j.1460-2075.1984.tb01950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bustin M, Lehn D A, Landsman D. Structural features of the HMG chromosomal proteins and their genes. Biochim Biophys Acta. 1990;1049:231–243. doi: 10.1016/0167-4781(90)90092-g. [DOI] [PubMed] [Google Scholar]
- 5.Cairns B R, Henry N L, Kornberg R D. TFG3/TF30/ANC1, a component of the yeast SWI/SNF complex that is similar to the leukemogenic proteins ENL and AF-9. Mol Cell Biol. 1996;16:3308–3316. doi: 10.1128/mcb.16.7.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cairns B R, Levinson R S, Yamamoto K R, Kornberg R D. Essential role of Swp73p in the function of yeast Swi/Snf complex. Genes Dev. 1996;10:2131–2144. doi: 10.1101/gad.10.17.2131. [DOI] [PubMed] [Google Scholar]
- 7.Clark-Adams C D, Winston F. The SPT6 gene is essential for growth and is required for δ-mediated transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:679–686. doi: 10.1128/mcb.7.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark-Adams C D, Norris D, Osley M A, Fassler J S, Winston F. Changes in histone gene dosage alter transcription in yeast. Genes Dev. 1988;2:150–159. doi: 10.1101/gad.2.2.150. [DOI] [PubMed] [Google Scholar]
- 9.Dombroski A J, Walter W A, Gross C A. Amino-terminal amino acids modulate sigma-factor DNA-binding activity. Genes Dev. 1993;7:2446–2455. doi: 10.1101/gad.7.12a.2446. [DOI] [PubMed] [Google Scholar]
- 10.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grunstein M. Histone function in transcription. Annu Rev Cell Biol. 1990;6:643–678. doi: 10.1146/annurev.cb.06.110190.003235. [DOI] [PubMed] [Google Scholar]
- 12.Hirschhorn J N, Brown S A, Clark C D, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 13.Jonsen M D, Petersen J M, Xu Q P, Graves B J. Characterization of the cooperative function of inhibitory sequences in Ets-1. Mol Cell Biol. 1996;16:2065–2073. doi: 10.1128/mcb.16.5.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kruger W, Herskowitz I. A negative regulator of HO transcription, SIN1 (SPT2), is a nonspecific DNA-binding protein related to HMG1. Mol Cell Biol. 1991;11:4135–4146. doi: 10.1128/mcb.11.8.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kruger W, Peterson C L, Sil A, Coburn C, Arents G, Moudrianakis E N, Herskowitz I. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 1995;9:2770–2779. doi: 10.1101/gad.9.22.2770. [DOI] [PubMed] [Google Scholar]
- 16.Laurent B C, Carlson M. Yeast SNF2/SWI2, SNF5, and SNF6 proteins function coordinately with the gene-specific transcriptional activators GAL4 and Bicoid. Genes Dev. 1992;6:1707–1715. doi: 10.1101/gad.6.9.1707. [DOI] [PubMed] [Google Scholar]
- 17.Laurent B C, Treitel M A, Carlson M. The SNF5 protein of Saccharomyces cerevisiae is a glutamine- and proline-rich transcriptional activator that affects expression of a broad spectrum of genes. Mol Cell Biol. 1990;10:5616–5625. doi: 10.1128/mcb.10.11.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lefevbre L, Smith M. Mutational and functional analysis of dominant SPT2 (SIN1) suppressor alleles in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:5393–5407. doi: 10.1128/mcb.13.9.5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liberzon A, Shpungin S, Bangio H, Yona E, Katcoff D J. Association of yeast SAP1, a novel member of the AAA ATPase family of proteins with the chromatin protein SIN1. FEBS Lett. 1996;388:5–10. doi: 10.1016/0014-5793(96)00500-5. [DOI] [PubMed] [Google Scholar]
- 20.Malone E A, Fassler J S, Winston F. Molecular and genetic characterization of SPT4, a gene important for transcription initiation in Saccharomyces cerevisiae. Mol Gen Genet. 1993;237:449–459. doi: 10.1007/BF00279450. [DOI] [PubMed] [Google Scholar]
- 21.Moazed D, Kistler A, Axelrod A, Rine J, Johnson A D. Silent information regulatory protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc Natl Acad Sci USA. 1997;94:2186–2191. doi: 10.1073/pnas.94.6.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nasmyth K. Regulating the HO endonuclease in yeast. Curr Opin Genet Dev. 1993;3:286–294. doi: 10.1016/0959-437x(93)90036-o. [DOI] [PubMed] [Google Scholar]
- 23.Nasmyth K, Stillman D J, Kipling D. Both positive and negative regulators of HO transcription are required for mother-cell-specific mating-type switching in yeast. Cell. 1987;48:579–587. doi: 10.1016/0092-8674(87)90236-4. [DOI] [PubMed] [Google Scholar]
- 24.Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984;108:845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pérez-Martín J, de Lorenzo V. The amino-terminal domain of the prokaryotic enhancer-binding protein XylR is a specific intramolecular repressor. Proc Natl Acad Sci USA. 1995;92:9392–9396. doi: 10.1073/pnas.92.20.9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pérez-Martín J, Johnson A D. Mutations in chromatin components suppress a defect of Gcn5 protein in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:1049–1054. doi: 10.1128/mcb.18.2.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peterson C L, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 28.Peterson C L, Kruger W, Herskowitz I. A functional interaction between the C-terminal domain of RNA polymerase II and the negative regulator SIN1. Cell. 1991;64:1135–1143. doi: 10.1016/0092-8674(91)90268-4. [DOI] [PubMed] [Google Scholar]
- 29.Peterson C L, Tamkun J W. The SWI-SNF complex: a chromatin remodeling machine? Trends Biochem Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- 30.Prelich G, Winston F. Mutations that suppress the deletion of an upstream activating sequence in yeast: involvement of a protein kinase and histone H3 in repressing transcription in vivo. Genetics. 1993;135:665–676. doi: 10.1093/genetics/135.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roeder G S, Beard C, Smith M, Keranen S. Isolation and characterization of the SPT2 gene, a negative regulator of Ty-controlled yeast gene expression. Mol Cell Biol. 1985;5:1543–1553. doi: 10.1128/mcb.5.7.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rose M D, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 33.Russell D W, Jensen R, Zoller M J, Burke J, Errede B, Smith B M, Herskowitz I. Structure of the Saccharomyces cerevisiae HO gene and analysis of its upstream regulatory sequences. Mol Cell Biol. 1986;6:4281–4294. doi: 10.1128/mcb.6.12.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scherer S, Davis R W. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci USA. 1979;76:4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schroter H, Bode J. The binding sites for large and small high mobility group (HMG) proteins: studies on HMG-nucleosome interactions in vitro. Eur J Biochem. 1982;127:429–436. doi: 10.1111/j.1432-1033.1982.tb06890.x. [DOI] [PubMed] [Google Scholar]
- 36.Sheflin L G, Fucile N W, Spaulding S W. The specific interactions of HMG1 and 2 with negatively supercoiled DNA are modulated by their acidic C-terminal domains and involve cysteine residues in their HMG 1/2 boxes. Biochemistry. 1993;32:3238–3248. doi: 10.1021/bi00064a005. [DOI] [PubMed] [Google Scholar]
- 37.Shpungin S, Liberzon A, Bangio H, Yona E, Katcoff D J. Association of yeast SIN1 with the tetratricopeptide repeats of CDC23. Proc Natl Acad Sci USA. 1996;93:8274–8277. doi: 10.1073/pnas.93.16.8274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stern M. Genes controlling the expression of the HO gene in yeast. Ph.D. thesis. University of California, San Francisco; 1985. [Google Scholar]
- 39.Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- 40.Sternberg P W, Stern J M, Clark I, Herskowitz I. Activation of the yeast HO gene by release from multiple negative controls. Cell. 1987;48:567–577. doi: 10.1016/0092-8674(87)90235-2. [DOI] [PubMed] [Google Scholar]
- 41.Stillman D J, Bankier A T, Seddon A, Groenhout E G, Nasmyth K. Characterization of a transcription factor involved in mother cell specific transcription of the yeast HO gene. EMBO J. 1988;7:485–494. doi: 10.1002/j.1460-2075.1988.tb02836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stros M, Stokrova J, Thomas J O. DNA looping by the HMG-box domains of HMG1 and modulation of DNA binding by the acidic C-terminal domain. Nucleic Acids Res. 1994;22:1044–1051. doi: 10.1093/nar/22.6.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swanson M S, Malone E A, Winston F. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol Cell Biol. 1991;11:3009–3019. doi: 10.1128/mcb.11.6.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Treich I, Cairns B R, Santos T, Brewster E, Carlson M. SNF11, a new component of the yeast SNF-SWI complex that interacts with a conserved region of SNF2. Mol Cell Biol. 1995;15:4240–4248. doi: 10.1128/mcb.15.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wechser M A, Kladde M P, Alfieri J A, Peterson C L. Effects of Sin− versions of histone H4 on yeast chromatin structure and function. EMBO J. 1997;16:2086–2095. doi: 10.1093/emboj/16.8.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winston F, Chaleff D T, Valent B, Fink G R. Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics. 1984;107:179–197. doi: 10.1093/genetics/107.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winston F, Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 48.Yona E, Bangio H, Friedman Y, Shpungin S, Katcoff D J. Characterization of a short unique sequence in the yeast HO gene promoter that regulates HO transcription in a SIN1 dependent manner. FEBS Lett. 1996;382:97–100. doi: 10.1016/0014-5793(96)00159-7. [DOI] [PubMed] [Google Scholar]