Abstract
The nuclear matrix is thought to play an important role in the DNA replication of eukaryotic cells, although direct evidence for such a role is still lacking. A nuclear matrix-associated transcription factor, polyomavirus (Py) enhancer binding protein 2αB1 (PEBP2αB1) (AML1/Cbfa2), was found to stimulate Py replication through its cognate binding site. The minimal replication activation domain (RAD) was identified between amino acid (aa) 302 and aa 371 by using a fusion protein containing the GAL4 DNA binding domain (GAL4-RAD). In addition, the region showed affinity for the nuclear matrix and, on the basis of competition studies, binding activity for one or more proteins involved in the initiation of Py DNA replication. A leukemogenic chimeric protein, AML1/ETO(MTG8), which does not contain this region of PEBP2αB1/AML1, was also localized in the nuclear matrix fraction and competed for nuclear matrix association with PEBP2αB1 and GAL4-RAD. Moreover, AML1/ETO inhibited Py DNA replication stimulated by PEBP2αB1 and GAL4-RAD. The inhibition was specific for replication mediated by PEBP2αB1 and GAL4-RAD, and proportional to the degree of loss of these activators from the nuclear matrix, suggesting a requirement for nuclear matrix targeting in the stimulation of Py DNA replication by RAD. These results are the first to suggest a molecular link between the initiation of DNA replication and the nuclear matrix compartment.
Accumulating evidence suggests an involvement of transcription factors in the regulation of DNA replication in eukaryotic cells. The polyomavirus (Py) DNA replication system is ideal for elucidating the roles of transcription factors in DNA replication, as Py DNA replicates in the nuclei of rodent cells and relies entirely on host factors, except for a single viral protein, large T antigen (TAg). The Py origin of replication contains a transcription enhancer in addition to a core sequence of the origin (ori-core). The ori-core contains binding sites for TAg and determines where replication begins, while the enhancer stimulates Py DNA replication 200- to 1,000-fold and determines the tissue specificity of DNA replication (12). The enhancer sequence can be replaced with multiple copies of a binding site for a single transcription factor (18, 42, 51). In this case, the initiation of Py DNA replication becomes dependent on a single transcription factor, thus allowing an analysis of the properties of each transcription factor with respect to its capacity to stimulate Py DNA replication. By using this system, we and others showed that transcription factors such as AP1 (18, 42, 51), VP16, GAL4 (5, 6, 18), c-Rel/NF-κB (24), bovine papilloma virus E2 (44), and p53 (30) are able to stimulate Py DNA replication when the Py enhancer is replaced by the binding sites for these factors.
So far, all transcription factors with replication-enhancing activity appear to require an activation domain, in addition to the DNA binding domain. The activation domain for replication overlaps that for transcription in many cases. For example, a yeast transcription factor, GAL4, requires the transcriptional activation domain to stimulate Py DNA replication, and the domain can be functionally replaced with transcriptional activation domains from other transcription factors such as VP16 and c-Jun (5, 6, 18). However, both domains are not necessarily identical. We showed that one of the two replication activation domains of p53, which mapped to the C-terminal region, did not stimulate transcription, while the other domain overlapped the transcription activation domain in the N-terminal region (30). Similarly, the Rel homology region in the N-terminal region of c-Rel stimulated Py DNA replication but not transcription, in contrast to the C-terminal transactivation domain, which enhanced both transcription and DNA replication (24). By analogy with transcription activation domains, one may imagine that the replication activation domain stimulates Py DNA replication through interaction with factors participating in the initiation of Py DNA replication. Indeed, the activation domains of VP16, GAL4, E2, and p53 (both domains) were shown to interact with a single-stranded-DNA binding protein, replication protein A (RP-A), which is essential for the initiation of Py DNA replication (13, 21, 35). More recently, we showed that c-Jun interacts with TAg to stimulate the formation of the core origin-TAg initiation complex (26).
Polyomavirus enhancer binding protein 2 (PEBP2) binds to DNA within the core elements of the Py enhancer which are important for replication (29, 41). PEBP2 is a heterodimer of two subunits, α and β (45, 46). There are three genes which encode the α subunits of PEBP2: PEBP2αA (also called CBFA1 and AML3) (45), PEBP2αB (also called CBFA2 and AML1) (3), and PEBP2αC (also called CBFA3 and AML2) (4). The α subunit is a homolog of the products of the Drosophila developmental regulator genes runt and lozenge. Two Drosophila genes which are homologous to the β subunit genes, brother and big brother, were identified (17). The α subunit has DNA binding activity, and the β subunit stimulates DNA binding activity of the α subunit. There are two functional regions within the α subunit of PEBP2. The evolutionarily conserved 128-amino-acid region termed the Runt domain is responsible for binding to DNA and dimerizing with the β subunit (28, 38, 46). The region downstream of the Runt domain is required for transcription activation (3, 31). Results from homozygous disruption of PEBP2αB in the mouse indicated that the factor is essential for definitive hematopoiesis (47, 50). Indeed, many genes important in regulating growth and differentiation of hematopoietic cells have been found to be regulated by PEBP2αB. The most characteristic feature of PEBP2 is that it is involved in context-dependent transcription activation: it intimately interacts with several other transcription factors and cooperates with them for either DNA binding or transcription activation (31).
PEBP2αB corresponds to the human gene AML1 (15, 39), which is rearranged in the 8-to-21 chromosome translocation, t(8;21), the most frequent chromosomal translocation found in acute myelogenous leukemia. The t(8;21) translocation produces the AML1/ETO(MTG8) chimeric protein, in which the region between the N terminus and Runt domain of AML1 is fused to ETO (15, 39). In this paper, murine AML1 is referred to as PEBP2αB and the full-length product of 451 amino acids is referred to as PEBP2αB1.
Recent experiments suggest that many nuclear events such as transcription and DNA replication are linked to the organization of nuclear structure, and especially to the filamentous ribonucleoprotein complex. This structure is alternatively referred to as the nuclear matrix, scaffold, or skeleton, depending on the isolation procedure. For simplification, we use the term “nuclear matrix” herein. The nuclear matrix is thought to contribute to replication and transcription by localizing or concentrating the factors implicated in these processes. For example, in mammalian cells, DNA replication appears to take place in specialized nuclear substructures which can be visualized as replication foci by immunolabeling of the incorporated analog, bromodeoxyuridine (43). The foci are attached to the nuclear matrix and contain proteins involved in DNA replication, such as DNA polymerase α, proliferating cell nuclear antigen, and RP-A. Such foci have been referred to as replication “factories” (23). Although no precise role has been ascribed to such structures, one possibility is that efficient initiation of DNA replication requires the attachment of origins to the nuclear matrix. Indeed, most of the autonomously replicating sequences in Saccharomyces cerevisiae and Schizosaccharomyces pombe, which in most cases function as origins of replication in their chromosomal DNA contexts, are bound to the nuclear matrix (1, 2).
Recently, Merriman et al. reported that the PEBP2-related transcription factor NMP-2, which binds to the PEBP2 binding sequence in the osteocalcin promoter, is exclusively localized in the nuclear matrix of osseous cells (37). More recently, Zeng et al. showed that AML1 is localized in the nuclear matrix and identified a nuclear matrix targeting sequence within AML1 near the transactivation domain (54). However, the importance of the nuclear matrix localization in the function of PEBP2 is not clear.
In this study, we showed that PEBP2 can stimulate Py DNA replication by itself. This enabled us to examine the role of the nuclear matrix compartment in the stimulation of Py DNA replication. We mapped a replication activation domain (RAD) of 70 amino acids within the region C terminal to the Runt domain. Interestingly, the RAD appears to provide two properties: nuclear matrix targeting and affinity for a DNA replication protein. By using the chimeric protein AML1/ETO, which inhibited both activities, we demonstrate that nuclear matrix targeting of RAD is important for the stimulation of DNA replication by RAD.
MATERIALS AND METHODS
Cell culture and transfection.
P19 cells, a murine embryonal carcinoma cell line, were maintained in a 1:1 mixture of Dulbecco’s modified Eagle’s medium and Ham F-12 medium supplemented with 10% (vol/vol) fetal bovine serum at 37°C. P19 cells were plated at 5 × 105 cells per 100-mm-diameter dish 10 h before transfection. The indicated amount of plasmid DNA was transfected by a modified Chen-Okayama calcium phosphate procedure (7). For each dish, the total amount of DNA used for transfection was adjusted to 15 to 20 μg by the addition of the backbone vector to the effector plasmids. Sixteen hours after transfection, the precipitates were washed and the cells were incubated for a further 24 h. Then the cells were harvested for further analysis. All transfection experiments were repeated independently at least three times.
Plasmids.
The reporter plasmid pPy(AE)4OICAT was a derivative of pPyOICAT. The double-stranded oligonucleotides representing four repeats of the PEBP2 binding site derived from the A core of the polyomavirus enhancer sequence (41) were inserted between the HindIII and BglII sites of pPyOICAT. The reporter plasmid pPyG5OICAT contains five copies of the yeast GAL4 DNA-binding motif (24). Plasmids for expression of Py TAg and various deletion proteins of PEBP2αB1 were derived from pEF-BOS (40) and described previously (30, 31). A series of deletions of PEBP2αB1 (amino acid [aa] 262 to aa 371) were generated by PCR amplification by using various sets of primers containing BamHI sites at both ends. To make fusion proteins with the GAL4 DNA binding domain (1 to 147), the BamHI-BglII VP16 fragment of pSGGAL4-VP16 (16) was replaced by the PCR products digested with BamHI. The introduction of point mutations was performed with an in vitro PCR-based site-directed mutagenesis kit (Stratagene) by using pSG-GAL4/PEBP2αB1 (aa 302 to aa 371) as a template and the synthetic oligonucleotides as mutagenic primers. The DNA sequence was confirmed by dideoxy sequencing with Sequenase version 2.0 (Amersham).
Replication assay.
Reporter plasmids (0.2 μg), effector plasmids (0.5 μg for plasmids based on pEF-BOS and 2 μg for plasmids which express GAL4 fusion proteins), the Py TAg expression plasmid pEF-BOS LT (4 μg), and the DpnI-resistant pHSG398 control plasmid (0.8 μg), which was prepared from dam mutant Escherichia coli GM33, were cotransfected into P19 cells as described above. Low-molecular-weight DNAs were isolated by the Hirt procedure (22). Purified DNAs were digested with HindIII and DpnI to convert replicated molecules and the control plasmid into their linear forms and to eliminate unreplicated DNA. Digested DNA was fractionated by 0.8% agarose gel electrophoresis, blotted onto Hybond N+ (Amersham), and then detected by hybridization with the BamHI-EcoRI fragment of pPyOICAT containing a part of the chloramphenicol acetyltransferase (CAT) gene as a hybridization probe. The radioactivity in the bands representing replicated and control DNAs was quantified with a BAS2000 analyzer (Fuji) and normalized with respect to the bands of the control DpnI-resistant plasmid.
Preparation of cellular fractions and nuclear skeleton.
The isolation of the nuclear matrix/scaffold fraction and other cellular fractions was performed according to the method described by Merriman et al. (37). Briefly, transfected cells were harvested and sequentially extracted with cytoskeleton (CSK) buffer (100 mM NaCl, 300 mM sucrose, 10 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] [pH 6.8], 3 mM MgCl2, 1 mM EGTA, 0.5% Triton X-100 and 1.2 mM phenylmethylsulfonyl fluoride [PMSF]) and reticulocyte standard buffer (RSB)-Majik buffer (100 mM NaCl, 10 mM Tris [pH 7.4], 3 mM MgCl2, 1.0% Tween 40, 0.5% deoxycholate [Na salt], and 1.2 mM PMSF). The extract with CSK buffer was used as the soluble fraction. Then the pellet was incubated with digestion buffer (50 mM NaCl, 300 mM sucrose, 10 mM PIPES [pH 6.8], 3 mM MgCl2, 1 mM EGTA, 0.5% Triton X-100, and 1.2 mM PMSF) containing DNase I (100 μg/ml) and RNase A (50 μg/ml) for 20 min at room temperature. After digestion, ammonium sulfate was added to a final concentration of 250 mM, and the nuclear matrix fraction was recovered by short centrifugation (880 × g, 10 min). The remaining supernatant was used as the chromatin fraction. Different fractions of the cells and the nuclear matrix were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Nuclear skeleton was isolated as described by Hozak et al. (23). Briefly, transfected P19 cells were harvested and encapsulated in 0.5% low-melting-point agarose (FMC). Encapsulated cells were treated with streptolysin O (1,000 U/ml per 106 cells) (Sigma) in phosphate-buffered saline (PBS) and permeabilized with physiological buffer (pH 7.4). Physiological buffer contains 130 mM KCl, 10 mM Na2HPO4, 1 mM MgCl2, 1 mM Na2ATP, 1 mM dithiothreitol, and 0.1 mM PMSF, and the pH was adjusted to 7.4 by adding 100 mM KH2PO4. Alternatively, encapsulated cells were permeabilized by incubation with 0.5% Triton X-100 in physiological buffer. Encapsulated and permeabilized cells were digested with DNase I (100 μg/ml) and RNase A (50 μg/ml) in a physiological buffer and then subjected to electrophoresis in 0.8% agarose gel at 4 V/cm for 4 h. After electrophoresis, agarose beads were harvested, treated with β-agarase (1 U/200 μl) (FMC) at 45°C for 60 min, and then centrifuged (880 × g, 10 min). The pellets were considered nuclear skeleton and were subjected to SDS-PAGE for Western blot analysis.
Western blot analysis.
A total of 10% of the different fractions of the harvested cells isolated as described above were separated by SDS–12% PAGE. The proteins in the gel were transferred electrophoretically (at 40 V for 12 h) onto a reinforced cellulose nitrate membrane (Schleicher & Schuell). The blocking reaction was performed by shaking the membrane for 1 h in PBS (80 mM Na2HPO4, 20 mM NaH2PO4, and 100 mM NaCl) containing 0.1% Tween 20 and 10% nonfat dry milk. The membrane was incubated for 1 h with the 2,000-fold diluted polyclonal antibody directed against the full-length PEBP2αB1 or the polyclonal anti-yeast GAL4 antibody (UBI) in PBS containing 0.1% Tween 20 and 5% nonfat dry milk. After washing in PBS containing 0.1% Tween 20, the membrane was incubated for 1 h with 5,000-fold diluted goat anti-rabbit immunoglobulin G conjugated with peroxidase (Zymed Laboratories, Inc.). Proteins were visualized by using the ECL Western blotting analysis system (Amersham Life Science) and quantified by densitometric scanning of the films.
Electrophoretic mobility shift assay (EMSA).
AML1/ETO, CH15, and 1-185 were in vitro translated by using the TNT reticulocyte-lysate system (Promega, Madison, Wis.). Equal amounts of the translated products were used in the reaction with the 32P-labeled probe M4A (52). E. coli-produced β2 (46) was added when indicated, and the DNA binding reaction and electrophoresis were carried out as described previously (3).
RESULTS
PEBP2αB1 stimulates Py DNA replication in a binding site-dependent manner.
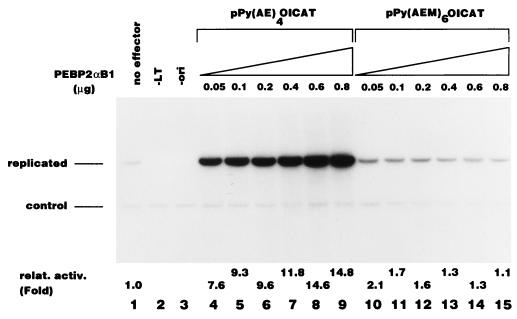
We cotransfected the reporter plasmid and an effector plasmid expressing PEBP2αB1 into P19 cells, which expresses the β subunit but not the α subunits of PEBP2 (3, 46). Figure 1 shows that PEBP2αB1 stimulated replication of Py(AE)4OICAT in a dose-dependent manner (lanes 4 to 9) but not Py(AEM)6OICAT, which contains mutations in the PEBP2 DNA binding sites (lanes 10 to 15). The replication activity of Py(AE)4OICAT shown in Fig. 1 represents authentic Py DNA replication, because this activity was completely dependent on both Py TAg (lane 2) and the core origin of replication (lane 3).
FIG. 1.
Stimulation of Py DNA replication by PEBP2αB1. Replication assays were performed as described in Materials and Methods by using pPy(AE)4OICAT (lanes 1, 2, and 4 to 9) and pPy(AEM)6OICAT (lanes 10 to 15), which contain the wild-type and mutated PEBP2 binding site, respectively, as reporter plasmids. In lane 2, the Py TAg-expressing plasmid was omitted, and in lane 3, pPy(AE)4CAT, which has a deletion in the Py core origin, was used. Indicated amounts of PEBP2αB1 expression plasmids were included in lanes 4 to 15. The bands corresponding to the replicated reporter plasmid and the control plasmid are indicated.
The C-terminal region of the PEBP2αB1 is responsible for the activation of Py DNA replication.
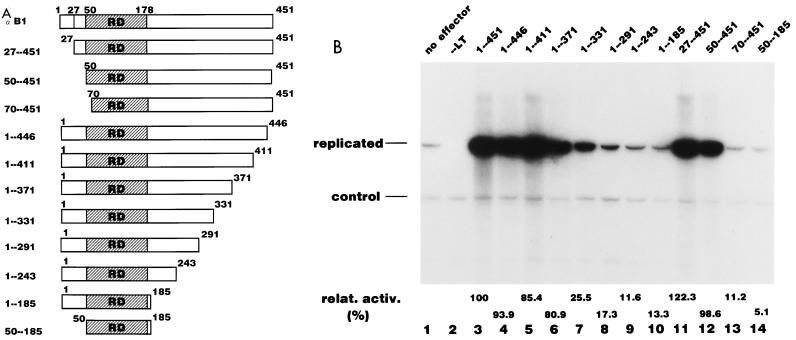
In order to identify regions of PEBP2αB1 required for the activation of Py DNA replication, a series of deletion mutants of PEBP2αB1 was constructed (Fig. 2A). We tested the DNA binding properties of all the deletion mutants by EMSA by using whole-cell extracts prepared from P19 cells transfected with each of these expression plasmids. All the mutants showed DNA binding activity except 70-451 (data not shown). 70-451 lacks the 20-aa region of the Runt domain that is essential for DNA binding activity (28). In addition, we confirmed that PEBP2αB1 and its deletion derivatives were capable of forming heterodimers with the β subunit because anti-β subunit antibody caused a supershift of each of the shifted bands in EMSA (data not shown).
FIG. 2.
The C-terminal region of the PEBP2αB1 contains the replication activation domain. (A) Schematic representation of the N-terminal and C-terminal deletion mutants of PEBP2αB1 used in the experiments shown in panel B. RD, Runt domain. (B) Replication assays were performed by using pPy(AE)4OICAT as a reporter plasmid as described in Materials and Methods. A total of 0.5 μg of plasmid of the indicated deletion derivatives of PEBP2αB1 was cotransfected. The bands corresponding to the replicated reporter plasmid and the control plasmid are indicated. Assays without effector (lane 1) or without TAg (lane 2) were also included as controls. Replication activity relative to that of the wide type is indicated below each lane.
Figure 2B shows the results of the replication assay of the deletion constructs. A 50-aa deletion from the N terminus did not affect stimulatory activity (lanes 11 and 12). However, activity was abolished when the deletion extended into the Runt domain (lane 13). The C-terminal truncation up to aa 371 did not affect the activity significantly: 1-371 still retained more than 80% of the activity of the full length PEBP2αB1 (Fig. 2A and B, lane 6). When the C-terminal deletion extended as far as aa 331, the activity was reduced to 25% of the full-length protein (Fig. 2B, lane 7). Further deletion up to aa 291 completely abolished the activity. These results indicated that the region between aa 291 and 371 was primarily responsible for the activity. Both 1-185 and 50-185 could bind to the β subunit, and the resultant complexes bound to DNA but could not stimulate Py replication (Fig. 2B, lanes 10 and 14), indicating that the β subunit does not independently contribute to the stimulation of DNA replication (data not shown).
The replication activation domain can function when it is fused to the GAL4 DNA binding domain.
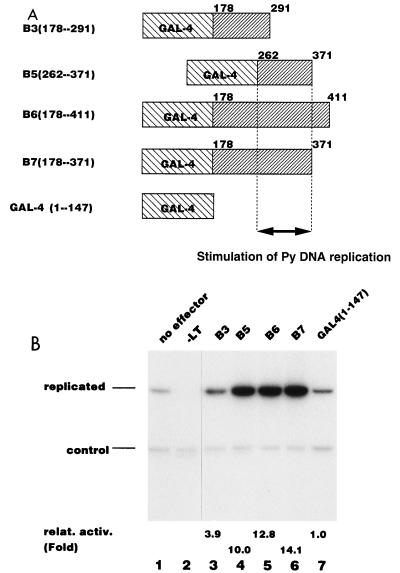
In order to see whether the replication activation activity can function when the activation domain is fused to a heterologous DNA binding domain, as was observed with c-Rel (24), we dissected the C-terminal region of PEBP2αB1 into several subregions and fused them to the GAL4 DNA binding domain (Fig. 3A). We confirmed that each fusion protein was expressed at a comparable level in P19 cells by Western blotting (data not shown).
FIG. 3.
Stimulation of Py DNA replication by GAL4-PEBP2αB1 fusion proteins. (A) Schematic representation of the GAL4-PEBP2αB1 fusion constructs shown in panel B. (B) Replication assays for GAL4-PEBP2αB1 fusion constructs shown in panel A were performed by using pPyG5OICAT as a reporter plasmid. The bands corresponding to the replicated reporter and the control plasmid are indicated. Replication activity relative to that of the GAL4 DNA binding domain (1-147) is indicated below each lane.
To test the replication stimulation activity of these GAL4 fusion proteins, we used the reporter plasmid pPyG5OICAT in which PEBP2 sites are replaced by 5 copies of the yeast GAL4 DNA binding motif. The GAL4 DNA binding domain itself did not stimulate Py DNA replication (Fig. 3B, lane 7), but the C-terminal region of PEBP2αB1 fused to the GAL4 DNA binding domain (B6 [aa 178 to 411]) activated Py DNA replication (lane 5). We further delineated the activation domain by using GAL4 fusion constructs. Consistent with the results shown above, the deletion from the C terminus to aa 371 did not affect the activity (B7 [Fig. 3B, lane 6]), but the deletion extending up to aa 291 (B3 [Fig. 3B, lane 3]) eliminated the activity. The smallest fusion protein, B5 (aa 262 to 371), strongly activated DNA replication (compare lane 4 with lane 7): B5 showed 10-fold-higher activity than the control, which is comparable to the level of the full-length PEBP2αB1 (Fig. 1). From the results of these experiments, we concluded that the region spanning aa 262 to 371 harbors the main part of the domain responsible for stimulation of Py DNA replication.
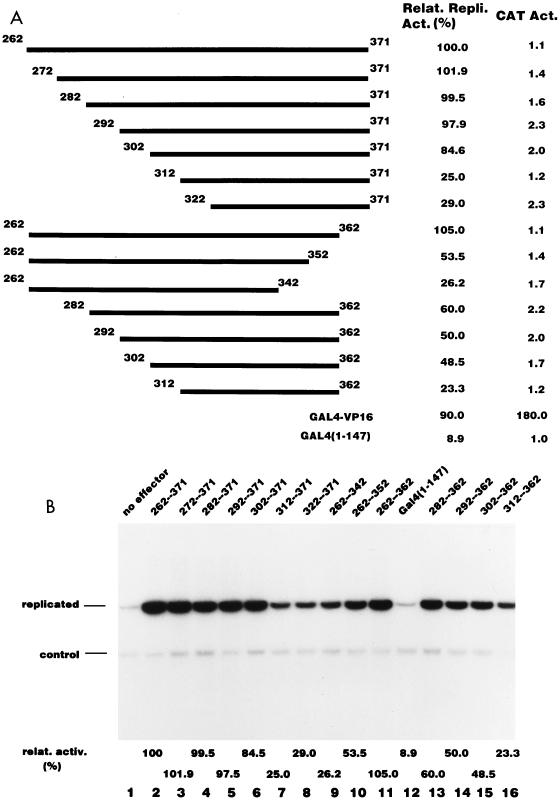
To narrow down the replication activation region further, we constructed a series of deletion mutants of B5. Figure 4A illustrates the structure of the 14 deletion constructs used. We confirmed that all the fusion proteins showed DNA binding activities by EMSA (data not shown). We tested the stimulatory activity of each fusion protein for Py DNA replication as well as for transcription, and the results are shown in Fig. 4A and B. The N-terminal deletion up to aa 302 (Fig. 4B, lane 6) did not affect replication activity significantly. The truncated fragment, aa 302 to 371, still retained about 80% of the activity compared to the entire fragment. A further N-terminal deletion up to aa 312 (Fig. 4B, lane 7) sharply reduced the activity to 25% of that of B5. From the C terminus, a deletion up to aa 362 (Fig. 4B, lane 11) had little effect on the activity. Further deletions resulted in a gradual reduction in activity (Fig. 4B, lanes 9 and 10). However, the 60-aa fragment between aa 302 and 362 retained only about half of the activity of the original fragment (Fig. 4B, lane 15). The most likely interpretation is that functional redundancy exists between aa 262 to 302 and aa 362 to 371. One of these regions was required for maximum stimulation of DNA replication (compare the activities of the regions from aa 262 to 362, 302 to 371, and 302 to 362). From these results, we chose the 70 aa from 302 to 371 as a minimal RAD.
FIG. 4.
Determination of the replication activation domain. (A) Schematic representation of the deletion mutants used in the experiments shown in panel B. Each mutant was fused to the GAL4 DNA binding domain. The relative replication activity of each deletion mutant measured in panel B and the level of transcription from the Py early promoter on pPyG5OICAT are indicated. The activities of GAL4-VP16 construct are also indicated for comparison. (B) The replication activity of each fusion protein shown in panel A was assayed as described in Materials and Methods by using pPyG5OICAT as a reporter plasmid. The bands corresponding to the replicated reporter and the control plasmid are indicated. Control experiments without effector (lane 1) or with an effector expressing the GAL4 DNA binding domain (1-147) are also indicated (lane 12).
In sharp contrast, none of the fusion proteins stimulated transcription from the Py early promoter (Fig. 4A). On the other hand, GAL4-VP16, which stimulated Py DNA replication to the same extent as the B5 fragment, strongly stimulated transcription (Fig. 4A). Unlike many other transcription factors, VP16 is highly cooperative, and the level of activation achieved by GAL4-VP16 increases dramatically when an increasing number of GAL4 sites are used in the reporter. In order to eliminate the possibility that the observed difference between GAL4-VP16 and GAL4-RAD in stimulation of transcription from the Py early promoter is due to the difference in cooperativity, we compared the ability to stimulate transcription of GAL4-VP16 and GAL4-RAD by using the reporter containing a single GAL4 site linked to the thymidine kinase promoter-driven luciferase gene. Even under such conditions, VP16 stimulated luciferase activity strongly, whereas RAD did not do so at a detectable level (data not shown). These results indicate that the region from aa 302 to 371 preferentially stimulates Py DNA replication but not transcription in P19 cells.
RAD is attached to the nuclear matrix, and AML1/ETO interferes with its attachment.
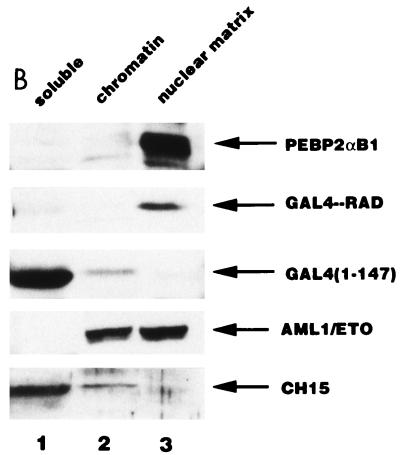
Zeng et al. reported that AML1 is exclusively localized in the nuclear matrix and that the nuclear matrix targeting signal maps to the region between aa 351 and 381, which corresponds to aa 324 and 353, respectively, in our numbering system (3). This targeting signal is comprised within RAD. We first examined the subnuclear localization of PEBP2αB1 and found that it was mainly present in the nuclear matrix fraction, with only a small portion appearing in the chromatin fraction; more than 80% of the protein was in the nuclear matrix fraction (Fig. 5B). Consistent with the observation that RAD contains the nuclear matrix targeting signal, more than 70% of the GAL4-RAD fusion protein was found to be present in the nuclear matrix fraction, whereas less than 25% of the protein containing only the GAL4 DNA binding domain was in the same fraction (Fig. 5B), as observed by Zeng et al. (54).
FIG. 5.
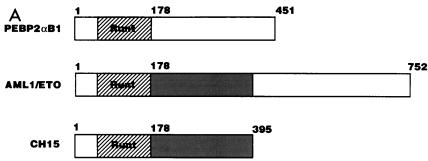
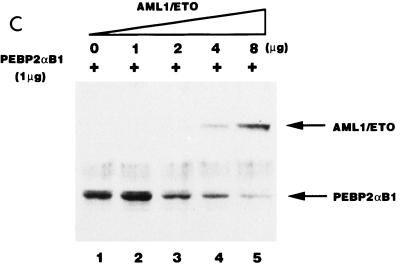
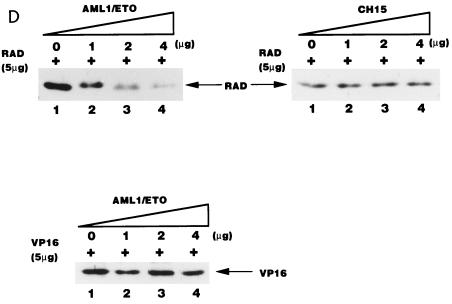
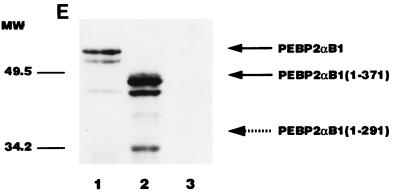
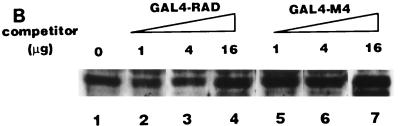
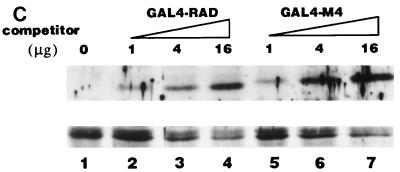
AML1/ETO inhibited the nuclear matrix localization activity of PEBP2αB1 and GAL4-RAD but not that of VP16. (A) Schematic structure of PEBP2αB1 and two forms of the AML1(PEBP2αB1)/ETO fusion protein. (B) The plasmids expressing the indicated proteins were transfected into P19 cells. Twenty-four hours after transfection, soluble, chromatin, and nuclear matrix fractions were isolated as described in Materials and Methods. Proteins in each fraction were analyzed by Western blotting. (C) Increasing amounts of AML1/ETO expression plasmids were cotransfected with a constant amount of PEBP2αB1 expression plasmid. The nuclear matrix fraction was isolated and analyzed by Western blotting. (D) Increasing amounts of plasmids expressing AML1/ETO or CH15 were cotransfected with a constant amount of plasmid expressing GAL4-RAD (upper panel), and increasing amounts of plasmids expressing AML1/ETO were cotransfected with a constant amount of plasmid expressing GAL4-VP16 (lower panel). The nuclear matrix fraction was isolated and analyzed by Western blotting. (E) Nuclear skeleton. Five micrograms of plasmids expressing full-length PEBP2αB1 (lane 1), 1-371 (lane 2) and 1-291 (lane 3) was transfected into P19 cells. The nuclear skeleton fractions of the transfected cells were isolated as described in Materials and Methods and analyzed by Western blotting. The positions of three proteins are indicated, and the expected position of PEBP2αB1(1-291) is indicated by a dotted arrow.
In order to determine the significance of RAD attachment to the nuclear matrix, we examined the possibility that the fusion protein AML1/ETO interferes with AML1 affinity for the nuclear matrix. To do so, we first examined the subnuclear localization of AML1/ETO. The results showed that AML1/ETO was distributed nearly equally between the chromatin fraction and the nuclear matrix fraction (Fig. 5B). Since the AML1 portion of the chimeric protein does not localize to the nuclear matrix (31, 54), ETO must have nuclear matrix localization activity. CH15 is a short variant of AML1/ETO which lacks the C-terminal 358 aa of the long form (Fig. 5A) (14, 34). About 75% of the CH15 protein was recovered in the soluble fraction, with a small amount appearing in the chromatin fraction. CH15 was hardly detected in the nuclear matrix fraction (Fig. 5B). We transfected increasing amounts of the plasmid expressing the full-length chimeric protein, AML1/ETO, in the presence of a constant amount of the PEBP2αB1 or GAL4-RAD expressing plasmid and analyzed their relative amounts in the nuclear matrix fraction. The results showed that increasing amounts of AML1/ETO progressively decreased the amount of PEBP2αB1 in the nuclear matrix fraction (Fig. 5C). The amount of PEBP2αB1 did not vary appreciably throughout the experiment (data not shown). Coexpression of AML1/ETO (Fig. 5D) but not CH15 (Fig. 5D) interfered with the nuclear matrix localization of GAL4-RAD. In contrast, the nuclear matrix localization of GAL4-VP16 was not affected by coexpression of AML1/ETO, showing that the competition by the chimeric protein was specific for PEBP2αB1 and RAD (Fig. 5D). The significance of this observation will be discussed below.
One of the reasons why physiological roles of the nuclear matrix have not been established is that the nuclear matrix fraction is prepared under harsh conditions, and therefore, it may not represent biologically active material (10). On the other hand, the “nuclear skeleton” fraction is prepared under physiological conditions (27). We therefore examined whether PEBP2αB1 can be recovered in the nuclear skeleton fraction. As shown in Fig. 5E, full-length PEBP2αB1 was retained in the nuclear skeleton fraction (lane 1). The C-terminal truncation up to aa 371 did not affect the property (lane 2). However, the C-terminal deletion up to aa 291 completely abolished the ability of PEBP2αB1 to associate with the nuclear skeleton (lane 3). The result was entirely compatible with those obtained by using the nuclear matrix fraction (31) and suggests that PEBP2αB1 is tightly attached to the nuclear insoluble material under physiological conditions.
AML1/ETO inhibited PEBP2αB1-dependent Py DNA replication.
Since AML1/ETO was able to compete with PEBP2αB1 for nuclear matrix attachment, we tested whether the chimeric protein could inhibit PEBP2αB1 DNA replication activity (Fig. 6). In this study, we compared the properties of AML1/ETO, CH15, and 1-185, all of which have been shown to be nuclear proteins (reference 31 and data not shown).
FIG. 6.
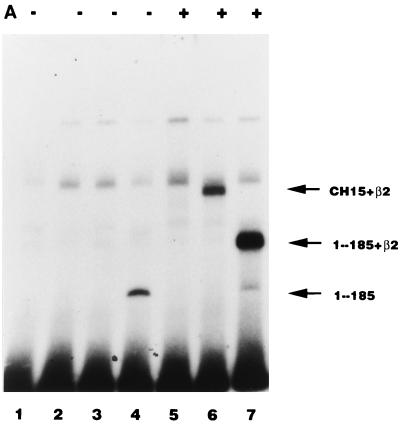
Effect of the AML1/ETO chimeric proteins on the replication stimulation activity of PEBP2αB1 and GAL4-RAD. (A) Binding of the in vitro-translated AML1/ETO (lanes 2 and 5), CH15 (lanes 3 and 6), and PEBP2αB1(1-185) (lanes 4 and 7) proteins to the PEBP2 site in the absence (lanes 2 to 4) or presence (lanes 5 to 7) of β2 (10 ng). (B) Replication assays for AML1/ETO chimeric proteins were performed with pPy(AE)4OICAT as a reporter plasmid. In lane 2, the TAg-expressing plasmid was omitted. The indicated amounts of plasmids expressing the long form (lanes 3 to 6) or the short form (lanes 7 to 10) of AML1/ETO, PEBP2αB1 lacking the region C-terminal to the runt domain (lanes 11 to 14), or wild-type PEBP2αB1 (lane 15) were cotransfected. (C) A constant amount (0.5 μg) of the plasmid expressing full-length PEBP2αB1 and the reporter plasmid, pPy(AE)4OICAT (0.2 μg), was cotransfected with increasing amounts of the plasmids expressing the long form of AML1/ETO (lanes 2 to 5), the short form of AML1/ETO (lanes 6 to 9), or truncated PEBP2αB1 that lacked the C-terminal region (1-185) (lanes 10 to 13). Replication of the reporter plasmid was measured as described in Materials and Methods. The bands corresponding to replicated reporter and the control plasmid are indicated. Activities relative to the wild-type domain are indicated below each lane. (D and E) Effect of the chimeric proteins and truncated protein on the replication stimulation activity of GAL4-RAD (D) or GAL4-VP16 (E) was measured as described for panel C. Activity relative to that of the wild-type domain is indicated below each lane.
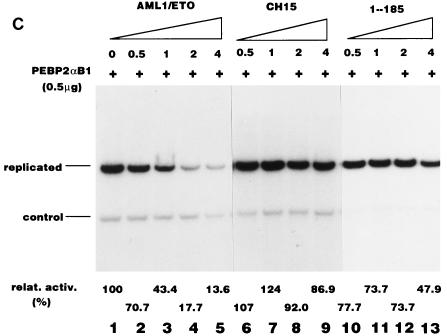
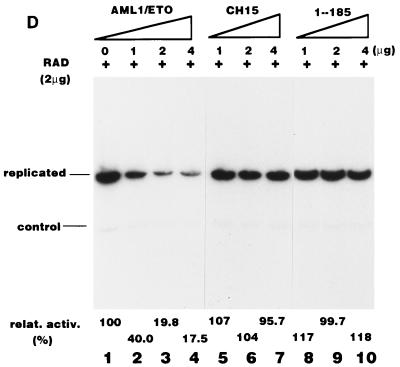
Before considering the influence of the nuclear matrix, we examined whether the structures of these three proteins, all of which contain the intact Runt domain, might alter their DNA binding activities. We examined by EMSA the DNA binding abilities of AML1/ETO, CH15, and 1-185 in the absence or the presence of the β subunit, because the β subunit dimerizes with the Runt domain and enhances the DNA binding activity of the α subunit. The results are shown in Fig. 6A. 1-185 by itself was able to bind strongly to DNA. The β subunit supershifted the band and increased its intensity. Despite the fact that AML1/ETO contains the intact Runt domain, it hardly interacted with DNA, even in the presence of the β subunit (Fig. 6A). CH15, on the other hand, bound poorly to DNA, but its binding was enhanced by the β subunit (Fig. 6A). None of these proteins were able to stimulate Py DNA replication by themselves (Fig. 6B), which is consistent with the notion that they lack RAD. Cotransfection of increasing amounts of plasmids expressing AML1/ETO progressively inhibited PEBP2αB1-dependent Py DNA replication (Fig. 6C, lanes 1 to 5). However, inhibition by CH15 and 1-185 was far less effective than by AML1/ETO (Fig. 6C, lanes 6 to 9 and 10 to 13, respectively). When the ratio of the plasmids expressing chimeric or truncated proteins to PEBP2αB1 was 4 to 1, AML1/ETO, CH15 and 1-185 reduced replication activity to 18, 92, and 74%, respectively (Fig. 6C, compare lanes 4, 8, and 12). From these results we concluded that a mechanism other than competition for a common DNA binding site was involved in the inhibition by the chimeric protein. The results also indicated that the C-terminal 357-aa region of ETO was required for the inhibitory effect.
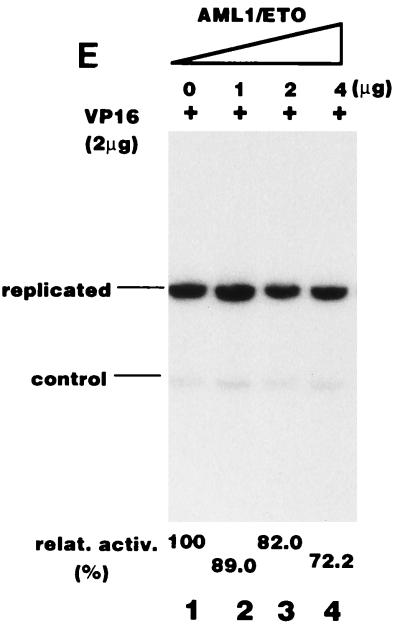
Consistent with this conclusion, AML1/ETO effectively inhibited the replication of pPyG5OICAT stimulated by GAL4-RAD, whereas neither CH15 nor 1-185 had any effect on replication (Fig. 6D). Since pPyG5OICAT does not have the PEBP2 binding site, the results clearly showed that AML1/ETO inhibited RAD-dependent replication without binding to the reporter plasmid. However, the possibility remained that the effect of the chimeric protein was indirect and nonspecific. For example, expression of AML1/ETO might inhibit the expression of TAg or might be toxic for the cells. To test this possibility, we examined the influence of the chimeric protein on replication activated by GAL4-VP16. AML1/ETO hardly inhibited the replication of the reporter plasmid (Fig. 6E), indicating that AML1/ETO specifically inhibited RAD-dependent replication. Moreover, the fact that AML1/ETO did not inhibit basal replication observed in the absence of effectors confirmed this conclusion (Fig. 6B, lanes 3 to 6). The results shown in Fig. 5 and 6 suggest that nuclear matrix localization is necessary for the stimulation of Py DNA replication by PEBP2αB1 and that inhibition of replication activity by AML1/ETO is, at least partly, due to the elimination of PEBP2αB1 from the nuclear matrix compartment by the chimeric protein.
Mutational analysis of RAD.
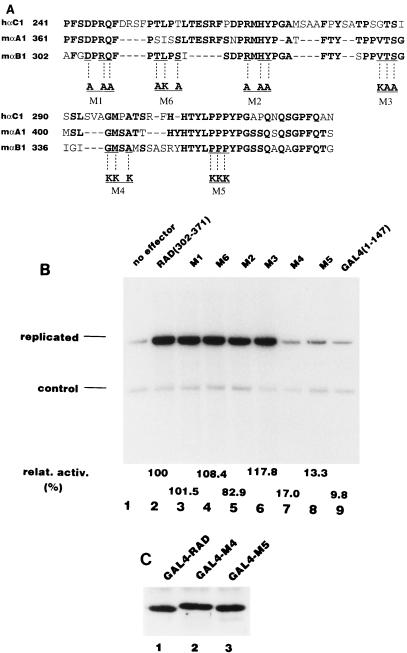
In order to determine which amino acids in the activation domain are important for replication activation, we introduced a series of mutations into RAD. We chose highly conserved amino acids common to PEBP2αB1, PEBP2αA1, and PEBP2αC1 as targets for mutation, since PEBP2αA1 and PEBP2αC1 also strongly stimulate Py DNA replication (data not shown). Figure 7A shows a sequence comparison of RAD with the corresponding regions of PEBP2αA1 and PEBP2αC1 and the sites of each of the mutations. We made six mutants, termed M1 through M6. Each mutant had three amino acid substitutions. The mutated 70-aa fragments were fused to the GAL4 DNA binding domain and were tested for replication stimulation activity. As shown in Fig. 7B, M1, M2, M3, and M6 showed almost the same activity as the wild type, whereas two mutants, M4 and M5, displayed only 10 to 20% of the activity of the wild type. Since all six proteins bound to the GAL4 binding site as efficiently as the wild-type fusion protein as revealed by EMSA (data not shown) and both M4 and M5 still localized in the nuclear matrix (Fig. 7C), the decrease in the activities of M4 and M5 would appear to be due to the loss of affinity for some replication related protein(s) required for Py DNA replication (see below).
FIG. 7.
Effects of mutations in the replication activation domain on DNA replication. (A) Sequence comparison of PEBP2αB1, αA1, and αC1 in RAD. The amino acids conserved in this family of proteins are shown in bold, and the modified amino acids in each mutant (M1 to M6) are indicated below. (B) Replication assay for the mutants. Each mutant was fused to the GAL4 DNA binding domain and assayed for stimulation of Py DNA replication as described in Materials and Methods. The bands corresponding to the replicated reporter and the control plasmid are indicated. Activity relative to that of the wild-type RAD is indicated below each lane. (C) Binding of the M4 and M5 mutants to the nuclear matrix. Five micrograms each of the plasmids expressing GAL4-RAD (lane 1), M4 (lane 2), and M5 (lane 3) was transfected into P19 cells, and the nuclear matrix fraction was analyzed by Western blotting.
RAD competed with the full-length PEBP2αB1 for activation of Py DNA replication.
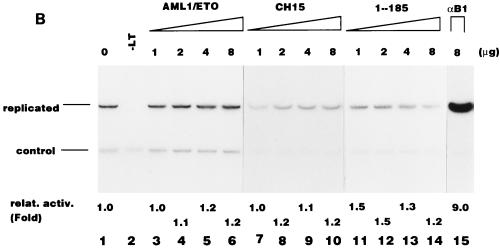
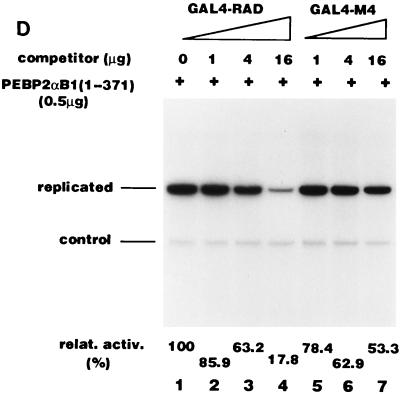
Based on our earlier observations (26) and the results of mutational analysis shown above, we assumed that RAD interacts with proteins involved in DNA replication. If this assumption is correct, overexpression of the non-DNA binding activation domain should compete with PEBP2αB1 for the putative target replication protein(s), resulting in the inhibition of the PEBP2αB1-dependent Py DNA replication. We cotransfected a constant amount of full-length PEBP2αB1 expression plasmids and Py(AE)4OICAT, together with increasing amounts of plasmids expressing GAL4-RAD, into the cells (Fig. 8A). The stimulation activity of PEBP2αB1 was observed to gradually decrease with the increase in the amount of GAL4-RAD (Fig. 8A, lanes 1 to 6). This was in contrast to the absence of any effect when the M4 (Fig. 8A, lanes 8 to 13) and M5 mutant (data not shown) proteins were used to replace GAL4-RAD under the same conditions. Since the reporter plasmid, pPy(AE)4OICAT, did not contain the GAL4 binding site, the overexpressed GAL4 fusion protein did not stimulate the replication activity of the reporter plasmid by itself (Fig. 8A, lanes 7 and 14). The results suggest that PEBP2αB1 interacts with replication protein(s) for its activity to stimulate replication, if the nuclear matrix localization of PEBP2αB1 is not affected by cotransfected GAL4-RAD or M4. In fact, overexpression of GAL4-RAD, M4, or M5 mutant protein did not compete with PEBP2αB1 for the nuclear matrix localization (Fig. 8B and data not shown for M5). The reason for the lack of competition for the nuclear matrix localization is clarified below.
FIG. 8.
Demonstration of two types of competition. (A) Overexpression of GAL4-RAD inhibited the replication stimulation activity of PEBP2αB1. A total of 0.5 μg of the PEBP2αB1 expression plasmid and 0.2 μg of the reporter plasmid, pPy(AE)4OICAT, were cotransfected into P19 cells together with increasing amounts of the plasmids expressing GAL4-RAD (lanes 1 to 7) or M4 (lanes 8 to 14). In lanes 7 and 14, 4 μg of GAL4-RAD and M4 expression plasmids was transfected, respectively, without PEBP2αB1 expression plasmid. The replicated reporter and the control plasmid are indicated. Relative replication activity is indicated below each lane. (B) Overexpression of GAL4-RAD and M4 did not inhibit the nuclear matrix localization activity of PEBP2αB1. Increasing amounts of the GAL4-RAD (lanes 2 to 4) and M4 (lanes 5 to 7) expression plasmids were cotransfected with a constant amount of PEBP2αB1 expression plasmid into P19 cells. The nuclear matrix fraction was analyzed by Western blotting. Only the band representing PEBP2αB1 is shown. (C) Overexpression of GAL4-RAD and M4 inhibited the nuclear matrix binding activity of PEBP2αB1(1-371). Increasing amounts of GAL4-RAD (lanes 2 to 4) or M4 (lanes 5 to 7) expression plasmids were cotransfected with a constant amount of PEBP2αB1(1-371) expression plasmid into P19 cells. The nuclear matrix fraction was analyzed by Western blotting. The upper panel shows the blot probed with anti-GAL4 polyclonal antibody, while the lower panel shows the same blot probed with anti-PEBP2αB1 polyclonal antibody. (D) Effect of overexpression of GAL4-RAD and M4 on the replication stimulation activity of PEBP2αB1(1-371). A total of 0.5 μg of PEBP2αB1(1-371) expression plasmid and 0.2 μg of reporter plasmid, pPy(AE)4OICAT, were cotransfected into P19 cells together with increasing amounts of the plasmids expressing GAL4-RAD (lanes 2 to 4) or M4 (lanes 5 to 7). The replicated reporter and the control plasmid are indicated. Relative replication activity is indicated below each lane. (E) GAL4-VP16 did not compete with PEBP2αB1 for the replication stimulation activity. A total of 0.5 μg of the PEBP2αB1 expression plasmid and 0.2 μg of the reporter plasmid, pPy(AE)4OICAT, were cotransfected into P19 cells together with increasing amounts of the plasmids expressing GAL4-VP16 (lanes 2 to 4) or GAL4-RAD (lanes 5 to 7). The replicated reporter and the control plasmid are indicated. Relative replication activity is indicated below each lane.
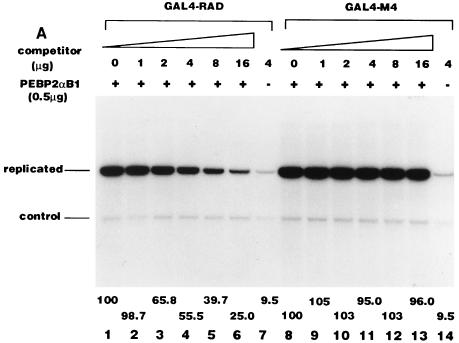
We previously observed that RAD is not the only region of PEBP2αB1 which has the nuclear matrix localization activity. There is the second nuclear matrix localization activity between aa 371 and 451 (31). This explains why overexpression of GAL4-RAD or its mutant did not inhibit the nuclear matrix localization of the full-length PEBP2αB1 as shown in Fig. 8B: RAD may not have competed with the second nuclear matrix localization signal. In order to test whether this explanation is genuine, we examined whether overexpression of GAL4-RAD or its mutant would inhibit the nuclear matrix localization of the truncated PEBP2αB1 lacking the second nuclear matrix localization signal (Fig. 8C). As expected, GAL4-RAD (Fig. 8C, lanes 2 to 4), M4 (lanes 5 to 7), and M5 (data not shown) all inhibited the nuclear matrix localization of PEBP2αB1(1-371) in a dose-dependent manner. The extent of inhibition was 54% (Fig. 8C, lane 4) and 49% (lane 7) at the highest amount of the competitor (16 μg) for GAL4-RAD and M4, respectively, suggesting that the M4 mutation does not affect the nuclear matrix targeting of RAD. Then, we examined the replication stimulation activity of PEBP2αB1(1-371) (Fig. 8D). We observed that the replication activity was about 18% at the highest amount of the competitor (16 μg) in the presence of GAL4-RAD (Fig. 8D, lane 4), whereas it was about 53% in the presence of M4 (lane 7).
The results shown in Fig. 8D appear to reflect two types of competition. The first is the competition at the level of nuclear matrix binding: the reduction of replication activity observed with M4 to 53% (Fig. 8D, lane 7) was well correlated with the reduction of the amount of M4 in the nuclear matrix fraction to 49% under the same conditions (Fig. 8C, lane 7). The amount of GAL4-RAD was also reduced to about half under the same conditions (Fig. 8C, lane 4), yet the replication activity was down to 18% (Fig. 8D, lane 4). The additional reduction of the replication activity of GAL4-RAD must be due to the competition of the replication protein(s) interacting with RAD. This set of the results shown in Fig. 8C and D once again reinforced the conclusion described above that the initiation of replication depends on the presence of RAD in the nuclear matrix.
The results shown in Fig. 8 altogether strongly suggested that the PEBP2αB1 must interact with one or more proteins necessary for Py DNA replication through the region of RAD affected by the M4 and M5 mutations.
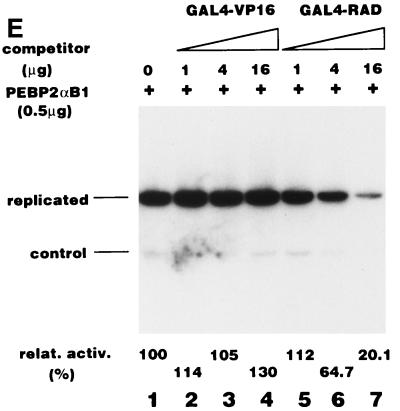
GAL4-VP16 stimulated Py DNA replication as efficiently as GAL4-RAD (Fig. 4A). VP16 was shown to interact with RP-A, and the interaction is thought to be important for the stimulation of Py DNA replication (35). Overexpression of GAL4-VP16, however, did not affect the replication stimulated by PEBP2αB1 (Fig. 8E, lanes 2 to 4), whereas GAL4-RAD severely inhibited this activity (lanes 5 to 7). This result suggested that the target protein of PEBP2αB1 is different from RP-A or other VP16 binding proteins.
In addition to RP-A, TAg is a possible target of transcription factors that stimulate Py DNA replication (26). We tested whether RAD and TAg interact directly by using surface plasmon resonance measurements in a BIAcore instrument. However, no direct interaction between RAD and TAg could be detected under conditions that allow detection of the interaction between c-Jun and TAg. In addition, under conditions that allowed detection of the interaction between VP16 and RP-A, we were unable to observe any interaction of RAD with RP-A (data not shown).
DISCUSSION
A growing body of evidence suggests the importance of the nuclear matrix in DNA replication. However, there exists relatively little evidence to show that the nuclear matrix domains are important for the control of DNA replication. This report shows that the replication activity of a transcription factor, PEBP2αB1, depends on its location in the nuclear matrix. PEBP2αB1 stimulated the initiation of Py DNA replication, and the minimal RAD was found to correspond to a region of 70 aa between aa 302 and 371. In addition the RAD was found to harbor a region responsible for the association of PEBP2αB1 with the nuclear matrix, which was inhibited by the chimeric protein AML1/ETO. Furthermore, the nuclear matrix association of RAD appears to be essential for the ability of GAL4-RAD to stimulate Py DNA replication. Thus, this report is the first indication of a strong correlation between the initiation of DNA replication and the targeting of a protein with replication stimulation activity to the nuclear matrix compartment. This simple system should be useful to further investigate the role of the nuclear matrix in DNA replication.
It is not known at present whether Py DNA replication takes place in a subnuclear structure such as the replication foci observed in cellular DNA replication. However, it is conceivable that Py DNA replication also takes place in such a structure on the nuclear matrix, since the DNA replication of herpes simplex virus and adenovirus takes place in the subnuclear structure (33, 53). If so, targeting of the replication origin to the replication factory would be an important step for the initiation of Py DNA replication. Therefore, we suggest that the nuclear matrix localization activity of RAD enhances Py DNA replication by recruiting the origin to the replication factory on the nuclear matrix. Alternatively, it is possible that the targeting to the nuclear matrix is required for RAD to interact properly with a protein present in the replication factory. The finding that VP16 is also localized in the nuclear matrix may suggest that targeting to the nuclear matrix is a common feature of transcription factors that stimulate Py DNA replication.
At this stage, it is not clear how PEBP2αB1 and GAL4-RAD are targeted to the nuclear matrix. Direct or indirect interaction between the signal sequence and the nuclear matrix protein(s) may be involved in the localization. It is interesting that both VP16 and RAD interact with the nuclear matrix, yet only the association of the latter is inhibited by AML1/ETO. We assume that more than one nuclear matrix-targeting signal exists and that each of them interacts with a different set of targets. Further analysis of the nuclear matrix targeting signals will be required to clarify the mechanism. Of particular importance will be the necessity to identify the protein(s) that associates with the nuclear matrix-targeting sequence.
Our data clearly indicate that nuclear matrix targeting alone is not enough for the stimulation of Py DNA replication, since mutant RADs (M4 and M5) that had lost DNA replication activity were still localized in the nuclear matrix. From the competition experiments, we concluded that the activation domain of PEBP2αB1 interacts with protein(s) involved in the initiation of Py DNA replication and that M4 and M5 mutations disturb these interactions. What is the target protein of RAD for the stimulation of replication?
Initiation of Py DNA replication consists of multiple steps. First, chromatin structure around the origin must be opened for the binding of TAg. TAg forms a double hexamer in an ATP-dependent manner which induces a structural change in the DNA at the ori-core . Then, unwinding of double-stranded DNA starts in the presence of RP-A. Finally, DNA polymerase α-primase starts DNA synthesis on the unwound DNA covered with RP-A. Some of the transcription factors that stimulate Py DNA replication were shown to interact with the proteins involved in the initiation steps. VP16, p53, E2, and GAL4 interact with RP-A (13, 21, 35). Recently, we showed that c-Jun interacts with TAg and stimulates the formation of the TAg-origin complex (26). In this study, we were unable to detect any interaction of RAD with RP-A or TAg, suggesting that RAD interacts with proteins involved in another step(s) such as the one before the formation of the TAg-origin complex or after unwinding. One of the candidates could be DNA polymerase α-primase. However, we were unable to detect a direct interaction between RAD and DNA polymerase α-primase in the assay using the BIAcore (data not shown).
A factor involved in chromatin remodeling could be a target protein. Experiments using the in vitro simian virus 40 system indicated that the assembly of the origin DNA into chromatin structure inhibits the binding of simian virus 40 TAg to the origin, with a consequent negative effect on the initiation of replication (8, 25). Moreover, Cheng and Kelly showed that prebinding of the transcription factor (NFI and GAL4-VP16) relieved the inhibitory effect of chromatin in an activation domain-dependent manner (8, 9). In the case of the initiation of transcription, chromatin formation is also inhibitory, and thus, remodeling of chromatin is important for gene activation. For example, chromatin-remodeling factors, such as SWI/SNF and NURF, have been identified as transcriptional regulators (48). More recently, the coactivator-adapter complexes for transcription were shown to contain histone acetyltransferase activity which can alter chromatin structure by acetylating histone tails (49). Therefore, it is attractive to speculate that RAD interacts with proteins that alter chromatin structure to assist the binding of TAg or other replication proteins to the origin.
We showed that the GAL4-RAD fusion protein did not stimulate transcription in P19 cells from the Py early promoter containing the GAL4 site. However, it should be noted that the transcription activation domain (TAD) of PEBP2αB1 was mapped to a region between aa 291 and 371 by using a GAL4 fusion protein in Jurkat or U937 cells and a reporter plasmid containing a GAL4 binding site linked to the herpesvirus thymidine kinase gene (31). In other words, TAD (aa 291 to 371) largely overlaps RAD (aa 302 to 371). In addition, using the same reporter, we detected weak but reproducible transcription activity of the same GAL4 fusion protein containing aa 291 to 371 in P19 cells while GAL4-RAD containing aa 302 to 371 barely showed the activity (data not shown). We speculate that TAD interacts with transcription-related protein(s) to exert its function while RAD interacts with replication-related proteins. However, the fact that TAD and RAD regions are coincident suggests that the activities have a common molecular basis. This might simply be nuclear matrix targeting. If so, this 80-aa region may represent a major nuclear matrix targeting signal. However, we found that a region closer to the C terminus is equally effective in nuclear matrix targeting. Alternatively, interaction with a chromatin remodeling factor(s) might provide a common mechanism. In any case, further analysis is required to clarify the nature of the underlying mechanism.
We used Py DNA replication as a model system for cellular DNA replication. The critical question is whether transcription factors actually participate in the initiation of cellular DNA replication. Participation of transcription factors in cellular DNA replication was first indicated for S. cerevisiae ARS1 function. In ARS1, the activation domains of transcription factors such as ABF1, RAP1, and GAL4 were found to stimulate replication (36). Interestingly, RAP1 was shown to be localized in the nuclear scaffold (32). In addition, autonomously replicating sequences are associated with the nuclear matrix in yeast. Therefore, it is conceivable that cellular DNA replication in yeast takes place in the nuclear matrix and that transcription factors play an important role in initiation. In mammalian cells, many putative replication origins have been identified. In almost all cases, the origin maps to a promoter-enhancer region or includes multiple transcription factor binding sites (11). It was also shown at the beginning of S phase that a significant number of replication foci and transcription foci coincide (19). This result is consistent with the long-standing observation that transcriptionally active genes tend to replicate early in S phase (20). In general, these results are in good agreement with the involvement of transcription factors in the regulation of DNA replication in mammalian cells.
It is still not clear how the leukemogenic chimeric protein AML1/ETO induces leukemia. From this point of view, it will be worth examining whether the ability of AML1/ETO to interfere with RAD is related to its leukemogenic potential. It is possible that through the inhibition of nuclear matrix localization, the chimeric protein disturbs the control of DNA replication and transcription mediated by PEBP2αB1, eventually resulting in the leukemogenic state of the cell. A more detailed analysis of the relationship between nuclear matrix association and stimulation of replication and transcription, in addition to the identification of the target protein of RAD, should provide us with important clues about the mechanism of leukemogenesis induced by these chimeric proteins.
ACKNOWLEDGMENTS
We thank T. Era (Kumamoto University) for generously providing the plasmids for the expression of the AML1/ETO and CH15.
This study was supported in part by a grant-in-aid for Priority Area on Cancer Research from the Minister of Education, Science and Culture, Japan, to Y.I. (contract no. 09253220).
REFERENCES
- 1.Amati B B, Gasser S M. Chromosomal ARS and CEN elements bind specifically to the yeast nuclear scaffold. Cell. 1988;54:967–978. doi: 10.1016/0092-8674(88)90111-0. [DOI] [PubMed] [Google Scholar]
- 2.Amati B B, Gasser S M. Drosophila scaffold-attached regions bind nuclear scaffolds and can function as ARS elements in both budding and fission yeasts. Mol Cell Biol. 1990;10:5442–5454. doi: 10.1128/mcb.10.10.5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae S C, Ogawa E, Maruyama M, Oka H, Sataka M, Shigesada K, Jenkins N A, Gilbert D J, Copeland N G, Ito Y. PEBP2αB/mouse AML1 consists of multiple isoforms that possess differential transactivation potentials. Mol Cell Biol. 1994;14:3242–3252. doi: 10.1128/mcb.14.5.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bae S C, Takahashi E, Zhang Y W, Ogawa E, Shigesada K, Namba Y, Sataka M, Ito Y. Cloning, mapping and expression of PEBP2αC, a third gene encoding the mammalian Runt domain. Gene. 1995;159:245–248. doi: 10.1016/0378-1119(95)00060-j. [DOI] [PubMed] [Google Scholar]
- 5.Baru M, Shlissel M, Manor H. The yeast GAL4 protein transactivates the polyomavirus origin of DNA replication in mouse cells. J Virol. 1991;65:3494–3503. doi: 10.1128/jvi.65.7.3496-3503.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett-Cook E R, Hassell J A. Activation of polyomavirus DNA replication by yeast GAL4 is dependent on its transcriptional activation domains. EMBO J. 1991;10:959–969. doi: 10.1002/j.1460-2075.1991.tb08030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng L Z, Kelly T J. Transcriptional activator nuclear factor 1 stimulates the replication of SV40 minichromosomes in vivo and in vitro. Cell. 1989;59:541–551. doi: 10.1016/0092-8674(89)90037-8. [DOI] [PubMed] [Google Scholar]
- 9.Cheng L Z, Workman J L, Kingston R E, Kelly T J. Regulation of DNA replication in vitro by the transcription activation domain of GAL4-VP16. Proc Natl Acad Sci USA. 1992;89:589–593. doi: 10.1073/pnas.89.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook P R. The nuclear skeleton: artefact, passive framework or active site? J Cell Sci. 1988;90:1–6. doi: 10.1242/jcs.90.1.1. [DOI] [PubMed] [Google Scholar]
- 11.DePamphilis M L. Eukaryotic DNA replication: anatomy of an origin. Annu Rev Biochem. 1993;62:29–63. doi: 10.1146/annurev.bi.62.070193.000333. [DOI] [PubMed] [Google Scholar]
- 12.de Villiers J, Schaffner W, Tyndall C, Lupton S, Kamen R. Polyoma virus DNA replication requires an enhancer. Nature. 1984;312:242–246. doi: 10.1038/312242a0. [DOI] [PubMed] [Google Scholar]
- 13.Dutta A, Ruppert J M, Aster J C, Winchester E. Inhibition of DNA replication factor RPA by p53. Nature. 1993;365:79–82. doi: 10.1038/365079a0. [DOI] [PubMed] [Google Scholar]
- 14.Era T, Asou N, Kunisada T, Yamasaki H, Asou H, Kamada N, Nishikawa S, Yamaguchi K, Takatsuki K. Identification of two transcripts of AML1/ETO-fused gene in t(8;21) leukemic cells and expression of wild-type ETO gene in hematopoietic cells. Genes Chromosomes Cancer. 1995;13:25–33. doi: 10.1002/gcc.2870130105. [DOI] [PubMed] [Google Scholar]
- 15.Erickson P, Gao J, Chang K-S, Look T, Whisenant E, Raimondi S, Lasher R, Trujillo J, Rowley J, Drabkin H. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992;80:1825–1831. [PubMed] [Google Scholar]
- 16.Fujii M, Tsuchiya H, Seiki M. HTLV-1 Tax has distinct but overlapping domains for transcriptional activation and enhancer specificity. Oncogene. 1991;6:2349–2352. [PubMed] [Google Scholar]
- 17.Golling G, Li L-H, Pepling M, Stabbins M, Gergen J P. Drosophila homologs of the proto-oncogene product PEBP/CBFβ regulate the DNA-binding properties of Runt. Mol Cell Biol. 1996;16:932–942. doi: 10.1128/mcb.16.3.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo Z-S, DePamphilis M L. Specific transcription factors stimulate simian virus 40 and polyomavirus origins of DNA replication. Mol Cell Biol. 1992;12:2514–2524. doi: 10.1128/mcb.12.6.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hassan A B, Errington R J, White N S, Jackson D A, Cook P R. Replication and transcription sites are colocalized in human cells. J Cell Sci. 1994;107:425–434. doi: 10.1242/jcs.107.2.425. [DOI] [PubMed] [Google Scholar]
- 20.Hatton K S, Dhar V, Brown E H, Iqbal M A, Stuart S, Didamo V T, Schildkraut C L. Replication program of active and inactive multigene families in mammalian cells. Mol Cell Biol. 1988;8:2149–2158. doi: 10.1128/mcb.8.5.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Z, Brinton B T, Greenblatt J, Hassell J A, Ingles C J. The transactivation proteins VP16 and GAL4 bind replication factor A. Cell. 1993;73:1223–1232. doi: 10.1016/0092-8674(93)90650-f. [DOI] [PubMed] [Google Scholar]
- 22.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:265–269. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 23.Hozak P, Hassan A B, Jackson D A, Cook P R. Visualization of replication factories attached to a nucleoskeleton. Cell. 1993;73:361–373. doi: 10.1016/0092-8674(93)90235-i. [DOI] [PubMed] [Google Scholar]
- 24.Ishikawa H, Asano M, Kanda T, Kumar S, Gelinas C, Ito Y. Two novel functions associated with the Rel oncoproteins: DNA replication and cell-specific transcriptional activation. Oncogene. 1993;8:2889–2896. [PubMed] [Google Scholar]
- 25.Ishimi Y. Preincubation of LT antigen with DNA overcomes repression of SV40 DNA replication by nucleosome assembly. J Biol Chem. 1992;267:10910–10913. [PubMed] [Google Scholar]
- 26.Ito K, Asano M, Hugues P, Kohzaki H, Masutani C, Hanaoka F, Kerppola T, Curran T, Murakami Y, Ito Y. c-Jun stimulates origin-dependent DNA unwinding by polyomavirus large T antigen. EMBO J. 1996;15:5636–5646. [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson D A, Yuan J, Cook P R. A gentle method for preparing of cyto- and nucleo-skeletons and associated chromatin. J Cell Sci. 1988;90:365–378. doi: 10.1242/jcs.90.3.365. [DOI] [PubMed] [Google Scholar]
- 28.Kagoshima H, Satake M, Miyoshi H, Ohki M, Pepling M, Gergen P, Shigesada K, Ito Y. The Runt domain identifies a new family of heteromeric transcription regulators. Trends Genet. 1993;9:338–341. doi: 10.1016/0168-9525(93)90026-e. [DOI] [PubMed] [Google Scholar]
- 29.Kamachi Y, Ogawa E, Asano M, Ishida S, Murakami Y, Satake M, Ito Y, Shigesada K. Purification of a mouse nuclear factor that binds to both the A and B cores of the polyomavirus enhancer. J Virol. 1990;64:4808–4819. doi: 10.1128/jvi.64.10.4808-4819.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanda T, Segawa K, Ohuchi N, Mori S, Ito Y. Stimulation of polyomavirus DNA replication by wild-type p53 through the DNA-binding site. Mol Cell Biol. 1994;14:2651–2663. doi: 10.1128/mcb.14.4.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanno T, Kanno Y, Chen L-F, Ogawa E, Kim W-Y, Ito Y. Intrinsic transcription activation-inhibition domains of the polyomavirus enhancer binding protein 2/Core binding factor α subunit revealed in the presence of the β subunit. Mol Cell Biol. 1998;18:2444–2454. doi: 10.1128/mcb.18.5.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klein F, Laroche T, Cardenas M E, Hofman J F, Schweizer D, Gasser S M. Localization of RAP1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J Cell Biol. 1992;117:935–948. doi: 10.1083/jcb.117.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kops A B, Knipe D M. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell. 1988;55:857–868. doi: 10.1016/0092-8674(88)90141-9. [DOI] [PubMed] [Google Scholar]
- 34.Kozu T, Miyoshi H, Shimizu K, Maseki N, Kaneko Y, Asou H, Kamada N, Ohki M. Junctions of the AML1/MTG8(ETO) fusion are constant in t(8;21) acute myeloid leukemia detected by reverse transcription polymerase chain reaction. Blood. 1993;82:1270–1276. [PubMed] [Google Scholar]
- 35.Li R, Botchan M R. The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell. 1993;73:1207–1221. doi: 10.1016/0092-8674(93)90649-b. [DOI] [PubMed] [Google Scholar]
- 36.Marahrens Y, Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- 37.Merriman H L, van Wijnen A J, Hiebert S, Bidwell J P, Fey E, Lian J, Stein J, Stein G S. The tissue-specific nuclear matrix protein, NMP-2, is a member of the AML/CBF/PEBP2/runt domain transcription factor family: interactions with the osteocalcin gene promoter. Biochemistry. 1995;34:13125–13132. doi: 10.1021/bi00040a025. [DOI] [PubMed] [Google Scholar]
- 38.Meyers S, Downing J R, Hiebert S W. Identification of AML-1 and the (8;21) translocation protein (AML-1/ETO) as sequence-specific DNA-binding proteins: the runt homology domain is required for DNA binding and protein-protein interactions. Mol Cell Biol. 1993;13:6336–6345. doi: 10.1128/mcb.13.10.6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyoshi H, Kozu T, Shimizu K, Enomoto K, Maseki N, Kaneko Y, Kamada N, Ohki M. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J. 1993;12:2715–2721. doi: 10.1002/j.1460-2075.1993.tb05933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murakami Y, Asano M, Satake M, Ito Y. A tumor promoting phorbol ester, TPA, enhances polyomavirus DNA replication by activating the function of viral enhancer. Oncogene. 1990;5:5–13. [PubMed] [Google Scholar]
- 42.Murakami Y, Satake M, Yamaguchi-Iwa Y, Sakai M, Muramatsu M, Ito Y. The nuclear protooncogenes c-jun and c-fos as regulators of DNA replication. Proc Natl Acad Sci USA. 1991;88:3947–3951. doi: 10.1073/pnas.88.9.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakamura H, Morita T, Sato C. Structural organization of replicon domains during DNA synthetic phase in mammalian nucleus. Exp Cell Res. 1986;165:291–297. doi: 10.1016/0014-4827(86)90583-5. [DOI] [PubMed] [Google Scholar]
- 44.Nilsson M, Forsberg M, You Z, Westin G, Magnusson G. Enhancer effect of bovine papillomavirus E2 protein in replication of polyomavirus DNA. Nucleic Acids Res. 1991;19:7061–7065. doi: 10.1093/nar/19.25.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogawa E, Maruyama M, Kagoshima H, Inuzuka M, Lu J, Satake M, Shigesada K, Ito Y. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci USA. 1993;90:6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogawa E, Inuzuka M, Maruyama M, Satake M, Naito-Fujimoto M, Ito Y, Shigesada K. Molecular cloning and characterization of PEBP2β, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2α. Virology. 1993;194:314–331. doi: 10.1006/viro.1993.1262. [DOI] [PubMed] [Google Scholar]
- 47.Okuda T, van-Deursen J, Hiebert S W, Grosveld G, Downing J R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 48.Tsukiyama T, Daniel C, Tamkun J, Wu C. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell. 1995;83:1021–1026. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- 49.Wade P A, Wolffe A P. Histone acetyltransferases in control. Curr Opin Biol. 1997;7:82–84. doi: 10.1016/s0960-9822(06)00042-x. [DOI] [PubMed] [Google Scholar]
- 50.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe A H, Speck N A. Disruption of the cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci USA. 1996;92:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wasylyk C, Schneikert J, Wasylyk B. Oncogene v-jun modulates DNA replication. Oncogene. 1990;5:1055–1058. [PubMed] [Google Scholar]
- 52.Yamaguchi Y, Satake M, Ito Y. Two overlapping sequence motifs within the polyomavirus enhancer are independently the targets of stimulation by both the tumor promoter 12-O-tetradecanoylphorbol-13-acetate and the Ha-ras oncogene. J Virol. 1989;63:1040–1048. doi: 10.1128/jvi.63.3.1040-1048.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Younghusband H B, Maundrell K. Adenovirus DNA is associated with nuclear matrix of infected cells. J Virol. 1982;43:705–713. doi: 10.1128/jvi.43.2.705-713.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeng C, van Wijnen A J, Stein J L, Meyers S, Sun W, Shopland L, Lawrence J B, Penman S, Lian J B, Stein G S, Hiebert S W. Identification of a nuclear matrix targeting signal in the leukemia and bone-related AML/CBF-α transcription factors. Proc Natl Acad Sci USA. 1997;94:6746–6751. doi: 10.1073/pnas.94.13.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]