Abstract
This report by the European Food Safety Authority and the European Centre for Disease prevention and Control, provides an overview of the main findings of the 2021–2022 harmonised Antimicrobial Resistance (AMR) monitoring in Salmonella spp., Campylobacter jejuni and C. coli from humans and food‐producing animals (broilers, laying hens and fattening turkeys, fattening pigs and cattle under one year of age) and relevant meat thereof. For animals and meat thereof, AMR data on indicator commensal Escherichia coli, presumptive extended‐spectrum beta‐lactamases (ESBL)‐/AmpC beta‐lactamases (AmpC)−/carbapenemase (CP)‐producing E. coli, and the occurrence of methicillin‐resistant Staphylococcus aureus (MRSA) are also analysed. Generally, resistance levels differed greatly between reporting countries and antimicrobials. Resistance to commonly used antimicrobials was frequently found in Salmonella and Campylobacter isolates from humans and animals. In humans, increasing trends in resistance to one of two critically antimicrobials (CIA) for treatment was observed in poultry‐associated Salmonella serovars and Campylobacter, in at least half of the reporting countries. Combined resistance to CIA was however observed at low levels except in some Salmonella serovars and in C. coli from humans and animals in some countries. While CP‐producing Salmonella isolates were not detected in animals in 2021–2022, nor in 2021 for human cases, in 2022 five human cases of CP‐producing Salmonella were reported (four harbouring bla OXA‐48 or bla OXA‐48‐like genes). The reporting of a number of CP‐producing E. coli isolates (harbouring bla OXA‐48, bla OXA‐181, bla NDM‐5 and bla VIM‐1 genes) in fattening pigs, cattle under 1 year of age, poultry and meat thereof by a limited number of MSs (5) in 2021 and 2022, requires a thorough follow‐up. The temporal trend analyses in both key outcome indicators (rate of complete susceptibility and prevalence of ESBL‐/AmpC‐producers in E. coli) showed an encouraging progress in reducing AMR in food‐producing animals in several EU MSs over the last 7 years.
Keywords: antimicrobial resistance, ESBL, indicator bacteria, MRSA, zoonotic bacteria
SUMMARY
In 2021–2022, data on antimicrobial resistance in zoonotic and indicator bacteria submitted by 27 EU Member States (MSs), the United Kingdom (Northern Ireland) and four non‐MSs were jointly analysed by EFSA and ECDC and EFSA's contractor. Resistance in zoonotic Salmonella and Campylobacter from humans, food‐producing animals (fattening pigs, cattle under 1 year of age, broilers and fattening turkeys, and also laying hens for Salmonella) and derived meat, as well as resistance in indicator commensal Escherichia coli and methicillin‐resistant Staphylococcus aureus (MRSA) from animals and derived meat were addressed. In 2022, it was mandatory to report AMR data from poultry and derived meat, while in 2021, it was mandatory to report AMR data from fattening pigs and cattle under 1 year of age and derived meat. ‘Microbiological’ resistance in the isolate populations was assessed using epidemiological cut‐off (ECOFF) values. For the countries reporting qualitative data on human isolates, the categories of ‘clinically resistant’ (R) and ‘susceptible with increased exposure’ (I) were combined, thereby achieving close correspondence with the proportion of isolates with the ECOFF‐defined ‘microbiological’ resistance.
New legislation related to the harmonised monitoring and reporting of AMR in food‐producing animals and derived meat was introduced in 2021, requiring MSs to sample imported fresh meat at border control posts and analyse for indicator commensal E. coli and extended‐spectrum beta‐lactamases (ESBL)‐/AmpC beta‐lactamases (AmpC)−/carbapenemase (CP)‐producing E. coli. In addition, imported fresh meat from poultry should also be analysed for Salmonella. New substances were also added in the harmonised antimicrobial panels, including amikacin for Salmonella and E. coli, and chloramphenicol and ertapenem for Campylobacter spp. Additionally, from 2021, whole genome sequencing (WGS) was authorised as an alternative method to supplementary (panel 2) phenotypic testing of Salmonella and indicator E. coli isolates with resistance to extended‐spectrum cephalosporins and/or carbapenems and for presumptive ESBL‐/AmpC‐/CP‐producing isolates from the specific monitoring.
In Salmonella spp. from human cases in 2022, resistance to ampicillin, sulfonamides and tetracyclines was observed at overall high levels, while resistance to third‐generation cephalosporins was noted at overall very low to low level of 1.4% and 1.2% for cefotaxime and ceftazidime, respectively. A statistically significant decline in resistance to ampicillin and tetracycline in isolates from humans was observed in 15 and 12 countries, respectively, over the period 2013–2022. This was particularly evident in S. Typhimurium, a serovar commonly associated with pigs and calves. For cefotaxime, seven MSs reported significant declining trends compared to four MSs reporting increasing trends. A moderate occurrence of resistance to ciprofloxacin (18.7%) was observed in human cases from 2022; however, an extremely high proportion of resistant isolates was noted in S. Kentucky (72.7%) and increasing trends in ciprofloxacin resistance were observed for S. Enteritidis in 12 countries over the period 2013–2022, with this serovar predominantly being associated with poultry.
For Salmonella spp. and indicator commensal E. coli isolates recovered from food‐producing animals and poultry carcases in 2021–2022, resistance to ampicillin, tetracyclines and sulfonamides ranged from moderate to very high in most MSs. Resistance to third‐generation cephalosporins (cefotaxime and ceftazidime) was reported at low levels in Salmonella spp. isolates from cattle, broiler and turkey flocks, and at very low levels in laying hen flocks and fattening pigs. These findings mirror those observed in Salmonella isolates reported from human cases. However, very high levels of resistance to third‐generation cephalosporins were reported in imported fresh broiler and turkey meat sampled at border control posts. Resistance to (fluoro)quinolones (ciprofloxacin and nalidixic acid) was high to very high among Salmonella spp. and indicator commensal E. coli isolates recovered from broilers, fattening turkeys and poultry carcases/meat in 2022, and low or moderate levels in isolates from pigs and calves in 2021.
Resistance to amikacin, the new substance included in the harmonised panel since 2021, was very low among E. coli from all four animal populations and in Salmonella spp. isolates from all animal populations, with the exception of the isolates from cattle under 1 year of age, for which no resistance was detected. Resistance to colistin was uncommon among Salmonella spp. and E. coli isolates recovered from food‐producing animals and poultry carcases, although moderate resistance was observed in certain Salmonella serovars (S. Enteritidis) and in Salmonella isolates from cattle under 1 year of age.
Combined resistance to ciprofloxacin and cefotaxime, categorised as highest priority critically important antimicrobials, was very low in Salmonella isolates from humans and rare or very low in Salmonella isolates in almost all animal and derived meat categories, with the exception of broilers and cattle under 1 year of age where low levels were detected. However, some Salmonella serovars from poultry sources, such as S. Kentucky from broilers and S. Infantis from turkeys, had comparatively elevated levels of combined resistance to ciprofloxacin and cefotaxime. The same was observed in these serovars isolated from humans. For E. coli isolates, low levels were reported in all animal populations and cattle meat. No resistant E. coli S., C. col etc., isolates were detected in pig meat, but in poultry meat, the level of resistance was low to moderate.
In 2021 and 2022, MSs had to submit AMR data on E. coli isolates recovered from imported meat sampled at border control posts for the first time. Over the 2 years, 11 MSs contributed data from imported fresh meat. Overall, resistance was more common among isolates from poultry meat than from pig or cattle meat.
Overall, the data obtained in 2021–2022 from C. jejuni and C. coli from human and animal origins showed high to extremely high levels of resistance to fluoroquinolones. Due to these levels of resistance, fluoroquinolones can no longer be recommended for the treatment of Campylobacter infections in humans. High to extremely high resistance levels to ciprofloxacin were observed in human C. jejuni and C. coli isolates in the EU in 2022, ranging from 33.1% (and even lower in the EEA countries Iceland and Norway) to 100%. Very high levels of resistance were observed for ciprofloxacin in isolates from food‐producing animals (ranging from 41.7% to 84.1%). Overall, the levels of resistance to ciprofloxacin in isolates obtained from food‐producing animals were higher for C. coli than for C. jejuni, although the levels of resistance to ciprofloxacin obtained from C. jejuni isolates from poultry in 2022 were also high (70.9% in broilers and 78.1% in fattening turkeys). The lowest levels of resistance to ciprofloxacin in both C. jejuni and C. coli were observed in isolates from fattening pigs in 2021 (41.7% and 51.7%, respectively). Resistance to erythromycin (representing the macrolide class, a critically important antimicrobial ([CIA] for the treatment of Campylobacter infections in humans) was detected at very low levels in C. jejuni from humans (0.9%) and at low levels in C. jejuni from animals (ranging from 1.0% to 1.7%). However, higher levels of resistance were observed in C. coli isolates from humans (7.8%) and animals (range from 8.8% to 35.7%). The level of overall resistance to tetracycline ranged from high to extremely high (43.3%–90.5%) in Campylobacter from food‐producing animals, with overall higher levels in C. coli (ranging from 67.5% to 90.5%) than in C. jejuni (ranging from 43.3% to 68.8%), and was high in C. jejuni and extremely high (71.2%) in C. coli from humans.
Over the period 2013–2022, ciprofloxacin resistance in C. jejuni from humans increased in 13 countries, while erythromycin resistance decreased in seven countries. Similar trends were observed in C. jejuni from broilers over 2014–2022 for six countries where resistance to ciprofloxacin increased, and six countries where resistance to erythromycin decreased. Over the same period, resistance to ciprofloxacin in C. jejuni from turkeys increased in one country, while resistance to erythromycin decreased in three countries. Over 2013–2022 for human isolates and 2014–2022 for isolates from fattening pigs, a decrease in resistance to erythromycin was observed in C. coli from humans in six countries, and in C. coli from fattening pigs in two countries. Despite these declining trends, several of the erythromycin‐resistant isolates displayed high MIC values (128 mg/L < MIC ≤ 512 mg/L or MIC > 512 mg/L), which could indicate a presence of the transferrable macrolide resistance gene erm(B). The whole genome sequencing results reported for a total of 110 erythromycin‐resistant Campylobacter isolates recovered among fattening pigs and cattle under 1 year of age in 2021 and broilers and fattening turkeys in 2022, showed the presence of erm(B) in a single isolate of C. coli from fattening pigs in 2021. Among the remaining isolates with an observed erythromycin resistance genotype, a mutation in the 23SrRNA ribosomal gene was detected, mostly the A2075G mutation, while the A2074C mutation was detected in a single isolate of C. coli from fattening turkeys.
The occurrence of combined resistance to ciprofloxacin and erythromycin in Campylobacter spp. is considered of high public health relevance. Overall combined resistance to these antimicrobials was lower in C. jejuni isolates than in C. coli isolates from humans and food‐producing animals. Combined resistance to ciprofloxacin and erythromycin reported from C. jejuni isolates was detected in 0.7% of isolates from humans, 1.0% isolates from broilers, 1.6% of isolates from fattening turkeys, 0.8% of isolates from cattle under 1 year of age and 1.7% of fattening pig isolates. Combined resistance to ciprofloxacin and erythromycin reported from C. coli isolates was detected in 7.1% of isolates from humans in 2022, 8.2% of broiler isolates and 17.4% of isolates from fattening turkeys in 2022 and 32.7% of samples from cattle under 1 year of age and 9.1% of fattening pig isolates in 2021. The levels of combined resistance to ciprofloxacin and erythromycin increased from 4.1% in 2020 to 8.2% in 2022 in C. coli isolates from broilers, which might infer a public health concern. However, an increase in the number of countries reporting results for C. coli from broilers in 2022 (24 MSs, the United Kingdom [Northern Ireland] and three non‐MSs) compared to 2020 (seven MSs and one non‐MS) could be a reason for this finding. Multidrug resistance (MDR) levels were generally very low for C. jejuni isolated from humans (0.7%) and ranged from very low to low in the animal species considered. Compared to C. jejuni, MDR was markedly higher in C. coli, specifically occurring in 9.0% of the isolates from humans, 39.3% of isolates from cattle under 1 year of age, 16.9% of isolates from fattening turkeys, 9.5% of isolates from fattening pigs and 8.3% of isolates from broilers. These results agree with the higher levels of resistance to selected antimicrobials seen in C. coli isolates.
The prevalence of resistance to selected antimicrobials in C. jejuni and C. coli from broilers and fattening turkeys in 2022 has been estimated at country level as the product of the proportion of isolates showing microbiological resistance to each antimicrobial and the percentage of all caecal samples cultured for C. jejuni or C. coli. Between‐country variability from rare to very high or extremely high levels was observed in the prevalence of ciprofloxacin‐resistant and tetracycline‐resistant C. jejuni and C. coli from broilers. A more limited between‐country variation in the levels of prevalence of resistance to erythromycin were found among the same isolates. In Campylobacter isolates from fattening turkeys, the prevalence of resistance to ciprofloxacin and to tetracycline presented a lower between‐country variability than that observed among broiler isolates, ranging from low to high or very high. Noteworthy, two MSs presented comparably higher prevalence of resistance to erythromycin in C. coli from fattening turkeys in 2022 (41.7% and 43%). This finding is of particular public health concern since macrolides, such as erythromycin and azithromycin, have become the first‐line treatment of human campylobacteriosis.
The monitoring also included assessment of the levels of presumptive extended‐spectrum beta‐lactamase (ESBL)‐/AmpC‐/CP‐producers among Salmonella spp. from human cases, food‐producing animals and imported fresh meat; as well as among indicator commensal E. coli isolates from food‐producing animals and derived meat. At the reporting MS group level, the proportion of presumptive ESBL‐ or AmpC‐producers ranged from very low to low among Salmonella spp. isolates recovered from animals/carcases (broilers, laying hens, fattening turkeys, fattening pigs) and very low in isolates from human cases, although higher in some Salmonella serovars.
Within both the routine and specific monitoring (non‐selective and selective media, respectively), varying occurrence/prevalence rates of presumptive ESBL‐ and/or AmpC‐producing E. coli were observed in different reporting countries. Statistically significant decreasing trends are evident in the prevalence of ESBL‐producing E. coli in broilers, broiler meat and pig meat at the EU level. A larger proportion of isolates were identified as presumptive ESBL‐producers compared with AmpC‐producers based on phenotypic methods in 2021 and 2022. This was supported by WGS that revealed 2239 E. coli isolates carrying ESBL genes, 267 isolates carrying plasmid‐mediated AmpC genes and 189 isolates presenting a point mutation in the AmpC promotor.
WGS results also revealed one CP‐producing E. coli reported by Italy in the routine monitoring for indicator E. coli in 2022. The isolate originated from fattening turkeys and carried the bla OXA‐181 gene. Furthermore, in 2021, within the specific monitoring of ESBL‐/AmpC‐/CP‐producing E. coli, two CP‐producing isolates from cattle meat and one isolate from fattening pigs were detected by Hungary. Those isolates were confirmed as CP‐producers harbouring the bla NDM‐5 gene, responsible for the CP‐phenotype, by the EURL‐AR during the confirmatory testing exercise. In 2022, one CP‐producing E. coli isolate from broilers reported by Italy and two CP‐producing E. coli from broilers in Austria carried the bla VIM‐1 gene. Moreover, in 2021, within the specific monitoring of CP‐producing microorganisms (using selective media for CP‐producers), two isolates from fattening pigs detected in Spain, carried the bla OXA‐48 gene. WGS revealed additional 26 E. coli isolates from Italy (21 from fattening pigs and five from cattle under 1 year of age) and three isolates from Czechia (all from fattening pigs) carrying CP‐encoding genes under the specific CP‐monitoring. These included bla OXA‐181 (four isolates from calves and 20 from fattening pigs), bla OXA‐48 (one isolate from a fattening pig) and bla NDM‐5 (one isolate from a calf) detected in the Italian isolates, and bla NDM‐5 (three isolates from fattening pigs) reported by Czechia. In 2022, a single CP‐producing E. coli isolate was reported from fattening turkeys in Italy, carrying the bla OXA‐181 gene. CP‐producing Salmonella isolates were not detected in animals in 2021–2022, nor in 2021 for human cases but in 2022 five human cases of CP‐producing Salmonella were reported, four with bla OXA genes (bla OXA‐48 or bla OXA‐48‐like) and the fifth, reported as susceptible with increased exposure to meropenem, was not available for further testing and confirmation.
The voluntary monitoring of MRSA from food and healthy animals in 2021–2022 revealed that most of the MRSA isolates, where typing data were available, were associated with spa‐types assigned to livestock‐associated (LA‐)MRSA in both reporting years. However, spa‐types associated with community‐associated (CA‐) and hospital‐associated (HA‐)MRSA were also reported, as well as mecC‐MRSA. The occasional detection of lineages of CA‐ and HA‐MRSA primarily associated with humans is not surprising, since the sporadic interchange of strains between humans and animals may be expected. An important observation from the 2021–2022 monitoring includes the detection of rifampicin resistance in isolates from cattle, pig meat and poultry meat. Vancomycin and rifampicin are important compounds in human medicine for the treatment of MRSA. None of the isolates subjected to susceptibility testing in 2021 and 2022 displayed resistance to vancomycin nor linezolid. Belgium reported three isolates with the cfr gene, encoding linezolid resistance, but susceptibility data were not available for these isolates.
The key outcome indicators for AMR in food‐producing animals – complete susceptibility (KOICS) to the harmonised panel of antimicrobials in E. coli and the prevalence of ESBL‐/AmpC‐producing E. coli – have also been analysed over the period 2014–2022. There are marked variations in both key outcome indicators among reporting countries. Statistically significant decreasing trends in the key outcome indicator of ESBL‐ and/or AmpC‐producing E. coli (KOIESC) were observed in 22 MSs and one non‐MS. A statistically significant increasing trend was identified in three MSs, and in the remaining countries, no statistically significant trend was seen. Statistically significant increasing trends in the key outcome indicators of complete susceptibility (KOICS) were registered in 16 MSs and two non‐MS and decreasing trends in three MSs. The increasing trends in changes to CS and KOICS in indicator commensal E. coli isolates reveal a progress towards lower levels of resistance in several countries and in the MS‐group. The improvement seen in changes to CS was most pronounced in poultry. Both key outcome indicators show that encouraging progress has been registered in reducing AMR in food‐producing animals in several EU MSs over the last years.
1. INTRODUCTION
The European Union system for the monitoring and collection of information on zoonoses is based on Directive 2003/99/EC, which obliges the Member States of the European Union (EU) to collect data on the occurrence of zoonoses, zoonotic agents, antimicrobial resistance, animal populations and food‐borne outbreaks. The structure of the monitoring is further elaborated in Commission Implementing Decision 2020/1729 (EU).
EFSA is assigned the tasks of examining these data and publishing annual European Union Summary Reports in cooperation with the European Centre for Disease Prevention and Control (ECDC). ECDC provides and analyses the data on zoonotic infections in humans. These reports illustrate the evolving situation in the EU and identify the pathogens that cause the most common zoonotic infections in humans.
EU Summary annual Reports regarding zoonotic agents and AMR in zoonotic and indicator bacteria from humans, animals and food as well as dashboards and story maps regarding these matters are presented at a common site at the EFSA web, available online here.
Legal basis
Monitoring of AMR in bacteria from food‐producing animals and derived meat
Regulation (EC) 178/2002 Article 33 establishes that EFSA is responsible for examining data on antimicrobial resistance (AMR) collected from the Member States (MSs) in accordance with Directive 2003/99/EC and for preparing the EU Summary Report from the results.
Directive 2003/99/EC on monitoring zoonoses and zoonotic agents lays down the provisions for monitoring AMR in zoonotic and indicator bacteria in food‐producing animals and derived meat. The Directive obliges EU MSs to collect relevant and, where applicable, comparable data on zoonoses, zoonotic agents, AMR and food‐borne outbreaks.
Commission Implementing Decision (EU) 2020/1729 on the monitoring and reporting antimicrobial resistance in zoonotic and commensal bacteria repeals Commission Implementing Decision (EU) 2013/6524. Commission Implementing Decision 2020/1729 (EU) applies from 2021 to 2027 and sets up priorities for the monitoring of AMR from a public health perspective, prescribes a list of combinations of bacterial species, food‐producing animal populations and foodstuffs and lays down harmonised rules for the period 2021–2027, for the monitoring and reporting of AMR in food‐producing animals and food.
Terms of Reference
In accordance with the Zoonoses Directive 2003/99/EC, the EU MSs are required to assess trends and sources of zoonoses, zoonotic agents and AMR, as well as outbreaks in their territory, submitting an annual report each year by the end of May to the European Commission covering the data collected.
In accordance with Article 9 of Directive 2003/99/EC, the EFSA shall examine the submitted national reports of the MSs and publish a summary report on the trends and sources of zoonoses, zoonotic agents and AMR in the EU.
The ECDC has provided data on zoonotic infections in humans and their analyses for the EU Summary Reports since 2005. Since 2007, data on human cases have been reported through The European Surveillance System (TESSy), maintained by the ECDC.
Monitoring of AMR in bacteria from humans
Commission Implementing Decision (EU) 2018/945 on the communicable diseases and related special health issues to be covered by epidemiological surveillance as well as relevant case definitions stipulates mandatory testing and reporting of a representative subset of isolates using methods and criteria specified in the EU protocol for harmonised monitoring of antimicrobial resistance in human Salmonella and Campylobacter isolates (ECDC, 2016).
The data collection on human diseases from MSs was in 2022 conducted in accordance with Decision 1082/2013/EU on serious cross‐border threats to health.
The antimicrobial agents used in food‐producing animals and human medicine in Europe are frequently the same or belong to the same classes. The route of administration and the administered quantities of antimicrobials differ between humans and food‐producing animals.
Moreover, there are important variations between and within food‐producing animal populations and countries. Nevertheless, frequently exposing the bacterial biota in both humans and animals to antimicrobial agents might result in the development of AMR by favouring the selection of resistant bacterial clones, regardless of whether these are pathogenic, commensal or environmental bacteria. This could, over time, change the population structure of microbial communities with serious consequences for human and animal health.
Antimicrobial resistance
AMR is defined as the inability or reduced ability of an antimicrobial agent to inhibit the growth of a bacterium, which, in the case of a pathogenic organism, can lead to therapy failure. A bacterial strain can acquire resistance by mutation, by the uptake of exogenous genes by horizontal transfer from other bacterial strains or by the activation/triggering of a genetic cascade, thereby inducing the expression of resistance mechanisms (EMA and EFSA, 2017). AMR is also an acronym for the health problems arising, in humans and animals, when antimicrobial‐resistant microorganisms spread within a population or society. The development and spread of resistance can be triggered by different factors such as use of antimicrobials in human and veterinary medicine, poor hygiene conditions and practices in healthcare settings or the food chain that facilitate the transmission of resistant microorganisms. Over time, this makes antimicrobials less effective.
Antimicrobial‐resistant bacteria derived from food‐producing animals can spread to humans by ingestion of, or from handling, food contaminated with zoonotic bacteria such as Campylobacter, Salmonella or Escherichia coli (E. coli), from direct contact with animals, or rarely, by environmental contamination. Infections with antimicrobial resistant bacteria may result in treatment failures or the need for second‐line antimicrobials for therapy. The commensal bacterial flora can also form a reservoir of resistance genes, which may be transferred between bacterial species, including organisms capable of causing disease in humans and animals (EFSA, 2008).
The European Commission adopted an Action Plan to tackle AMR on 29 June 2017. 1 The Action Plan is underpinned by a One Health approach that addresses resistance in bacteria from both humans and animals. EU actions have focused on the areas with the highest added value for MSs, such as promoting the prudent use of antimicrobials via antimicrobial stewardship (AMS), enhancing cross‐sectorial work, improving infection prevention and control (IPC) and consolidating surveillance of AMR and antimicrobial consumption (AMC). AMR monitoring in zoonotic and commensal bacteria in food‐producing animals and foodstuffs entails specific and continuous data collection, analysis and reporting. It enables the understanding of the development and diffusion of resistance, the following of temporal trends in the occurrence and distribution of AMR, the identification of emerging or specific resistance patterns, it provides relevant risk assessment data and helps to evaluate targeted interventions.
This EU Summary Report (EUSR) on AMR includes data related to the occurrence of AMR in isolates from humans, animals and food. The EUSR on AMR is a collaboration between EFSA and ECDC with the assistance of EFSA's contractors. EU MSs, European Free Trade Association (EFTA), the European Commission and the relevant EU Reference Laboratory for antimicrobial resistance (EURL‐AR) are consulted while preparing the report. The efforts made by the MSs, and the other reporting countries are gratefully acknowledged.
Data on AMR collected by the EU MSs and compiled in the EUSR on AMR are also used to perform wider analyses, such as the Joint Report on Consumption of Antimicrobial Agents and AMR in animals, food and humans – Joint Interagency Antimicrobial Consumption and Resistance Analysis (JIACRA), produced by ECDC, EFSA and EMA, under a One Health approach on a regular basis (2015, 2017, 2021) (ECDC, EFSA and EMA, 2021). The JIACRA report provides evidence‐based analysis of the possible association between AMC and AMR in humans and food‐producing animals by focusing on combinations of antimicrobials and bacterial species considered important for public health.
1.1. Monitoring and reporting of antimicrobial resistance in the EU
1.1.1. Humans: Monitoring of antimicrobial resistance
The EU protocol for harmonised monitoring of AMR in human Salmonella and Campylobacter isolates was developed by ECDC in collaboration with its Food‐ and Waterborne Diseases and Zoonoses (FWD) network (ECDC, 2016). The document is targeted to the National Public Health Reference Laboratories (NPHRL) to guide the susceptibility testing required for EU surveillance and reporting to ECDC and to facilitate comparison of data across countries and with the food‐ and animal AMR monitoring. Based on the defined EU level surveillance objectives, the protocol describes the panel of antimicrobials to be tested, the methods to use (dilution or disc diffusion according to EUCAST recommended methods), how to perform screening and confirmation of ESBL, AmpC and carbapenemase‐producing Salmonella, the interpretive criteria that should be applied and the reporting format when submitting data to ECDC. It has been agreed that for the joint report with EFSA, human data should be interpreted with EUCAST epidemiological cut‐off values (ECOFFs). Countries are therefore since 2014 (2013 data collection) requested to report quantitative antimicrobial susceptibility test results per isolate. After Decision 2018/945/EU came into force in July 2018, countries are legally required to report their AMR test results to ECDC following to the methods and criteria specified in the EU protocol. The countries that still do not perform AST at the NPHRL, however, continue to report data collected from clinical laboratories, interpreted with clinical breakpoints.
As whole genome sequencing has started to replace phenotypical typing methods in the NPHRLs, ECDC has enabled the reporting of resistance predicted from whole genome sequencing since 2020 and from 2023 ECDC is encouraging countries to report the raw sequences to ECDC to allow a harmonised AST interpretation.
ECDC has provided external quality assessment (EQA) schemes via a contracting laboratory to support laboratories in implementing the recommended test methods and antimicrobials and obtaining high‐quality AST results. Further capacity building activities, including training, EQA schemes on WGS and networking activities are provided via the HaDEA funded FWD AMR RefLabCap project in 2021–2024. 2
1.1.2. Animals and food: Monitoring of antimicrobial resistance
According to Commission Implementing Decision (EU) 2020/1729, which applies from 1 January 2021 to December 2027, monitoring of AMR is mandatory in Salmonella spp., Campylobacter coli (C. coli), Campylobacter jejuni (C. jejuni) and indicator commensal E. coli, in the major domestically produced animal populations and their derived meat. Further characterisation is required for E. coli and Salmonella isolates showing resistance to extended‐spectrum cephalosporins and carbapenems. Moreover, specific monitoring of extended‐spectrum beta‐lactamases (ESBL)‐, AmpC beta‐lactamases (AmpC)‐ and carbapenemase (CP)‐producing E. coli is also required. Monitoring is performed on a rotating basis, targeting fattening pigs and cattle under 1 year of age and meat derived thereof in odd years and poultry populations (broilers, laying hens, fattening turkeys) and derived meat in even years, as specified by the legislation.
Monitoring AMR in food‐producing animals is performed in domestically produced animal populations, corresponding to different production types to collect data that could be combined with data on exposure to antimicrobials. From 2021, monitoring of imported fresh meat at border control posts shall also be undertaken to complement AMR monitoring in food‐producing animals. MSs may also voluntarily perform complementary monitoring for MRSA. Representative random sampling of food‐producing animals and derived meat is based on a generic proportionate stratified sampling and performed according to the legislation and the technical specifications issued by EFSA.
Microdilution methods for testing should be used and results interpreted using EUCAST ECOFFs to understand ‘microbiological’ resistance. The harmonised panels of antimicrobials used for Salmonella, Campylobacter and indicator commensal E. coli include substances important for human health, such as critically important antimicrobials (CIAs), and can provide clearer insight into the resistance mechanisms involved. The concentration ranges to be used encompass both the ECOFF and the clinical breakpoints (CBPs), as defined by EUCAST, allowing for comparison with data coming from humans. For Salmonella and E. coli, a supplementary panel of antimicrobial substances for testing isolates showing resistance to third‐generation cephalosporins or carbapenems in the first panel is also used. From 2021, whole genome sequencing (WGS) is authorised as an alternate method to conventional phenotypic testing for isolates obtained for the specific monitoring of ESBL‐/AmpC‐/CP‐producing E. coli and for indicator commensal E. coli or Salmonella spp. isolates showing resistance to extended‐spectrum cephalosporins and carbapenems from routine monitoring. WGS is also recommended for Campylobacter isolates expressing high levels of phenotypic resistance to erythromycin. WGS is authorised on a voluntary basis only; however, technical conditions on the WGS technique have been imposed to ensure data comparability (EFSA, 2020).
External quality assurance is provided by the EURL‐AR, which distributes panels of well‐characterised organisms to all MSs for susceptibility testing, arranges proficiency tests (PTs) trials for the National Reference Laboratories for Antimicrobial Resistance (NRLs‐AR) of the MSs every year, and, together with EFSA and the MSs, performs a reference testing exercise that includes re‐testing the antimicrobial susceptibility and WGS analysis of selected isolates (Appendix F – Materials and methods). The EURL‐AR also provides a source of reference for MSs when there are issues or problems with the susceptibility test methodology.
Data reporting is performed at the isolate level to enable analyses on the occurrence of resistance and patterns of multidrug resistance (MDR). The reporting of isolate‐based data also allows in‐depth phenotypic characterisation of certain resistance mechanisms, e.g. third‐generation cephalosporin and carbapenem resistance. The voluntary reporting of WGS data from 2021 onwards on ESBL‐/AmpC‐/CP‐producing E. coli and Salmonella isolates will facilitate an understanding of the potential contribution of food‐producing animals and derived food to the burden of AMR in humans (EFSA, 2019).
1.2. Further harmonised monitoring of antimicrobial resistance
To facilitate data comparability, the AMR surveillance methodology should be harmonised across countries as much as possible. The main issues when comparing AMR data originating from different countries are the use of various laboratory methods and different interpretive criteria for resistance. These issues have been addressed by the development of ECDC's protocol for harmonised monitoring and reporting of resistance in humans and by the legislation on harmonised monitoring in food producing animals and derived meat. To respond effectively to the constantly evolving threat of AMR, ongoing enhancement and specific adaptations will be regularly required. Under the 2017 Action Plan on AMR, the European Commission is committed to reviewing this legislation to consider new scientific developments and data collection needs. In 2019, EFSA received a mandate from the European Commission to provide recommendations on harmonised randomisation procedures for AMR monitoring. The new technical specifications were published in November 2020 (EFSA, 2020), with the new legislation on the monitoring and reporting of AMR in animals and food enacted on 17 November 2020 (see text box below).
New legislation on the monitoring and reporting of AMR in animals and food came into effect on 1 January 2021
Monitoring of AMR is essential to have comprehensive and reliable information on the development and spread of resistant bacteria and resistant determinants. AMR data provide insights to inform decision‐making and facilitate the development of appropriate strategies and actions to manage AMR at the EU level. In its Communication of 29 June 2017 to the Council and the European Parliament – A European One Health Action Plan against AMR, the Commission committed to review EU implementing legislation, namely Commission Implementing Decision (EU) 2013/652, on monitoring AMR in zoonotic and commensal bacteria in food‐producing animals and food to take into account new scientific developments and data collection needs. After this, EFSA issued the new technical specifications, for implementing updated guidelines for the harmonised monitoring of AMR in food‐producing animals and derived meat and to ensure continuity in assessing occurrence and temporal trends in resistance (EFSA, 2019).
On 17 November 2020, the European Commission laid down the new technical specifications in Commission Implementing Decision (EU) 2020/1729 and repealed Commission Implementing Decision (EU) 2013/652. The new legislation came into effect on 1 January 2021, and updates technical specifications for harmonised AMR monitoring and reporting to include the monitoring of AMR in derived meat sampled at border control posts, the testing of new substances and authorises WGS as an alternate method to phenotypic testing, for the specific monitoring of ESBL/AmpC/CP‐producing E. coli and indicator commensal E. coli and Salmonella isolates with resistance to extended cephalosporins or carbapenems. The new rules apply to monitoring performed in 2021 onwards.
1.3. The 2021–2022 EU summary report on AMR
This EUSR presents AMR data on zoonotic and indicator bacteria from humans, animals and food collected in 2021 and 2022, jointly analysed by EFSA and ECDC. This report includes an introduction section, followed by five main chapters on AMR in Salmonella, Campylobacter, indicator commensal E. coli, ESBL‐/AmpC‐/CP‐producing Salmonella and E. coli, and MRSA, with sections detailing resistance in isolates from humans, food‐producing animals and derived meat. A section on key findings is included at the beginning of each chapter. Appendices containing complementary information are located at the end of the report. The list of annexes is available at the end of the report (Appendix G), and available on the EFSA Knowledge Junction community on Zenodo at: https://doi.org/10.5281/zenodo.10528846.
With the present report, EFSA has also published interactive communication tools:
The EFSA story maps on monitoring of AMR in Campylobacter, monitoring of AMR in MRSA and monitoring of AMR in E. coli.
The EFSA dashboards on Antimicrobial Resistance and Indicators of Antimicrobial Resistance.
All story maps and dashboards can be accessed on the EFSA website. Data used in this report and in the related communication tools were extracted from the EFSA AMR database on 30 November 2023.
For food‐producing animals and derived meat, most data reported by the MSs in 2021 and 2022 comprised data collected in accordance with Commission Implementing Decision (EU) 2020/1729. Quantitative antimicrobial susceptibility data for Campylobacter, Salmonella and indicator commensal E. coli isolates from animals and food were interpreted using ECOFFs. The occurrence of resistance, complete susceptibility (CS) and MDR is reported at the country and EU level, along with the results from the phenotypic monitoring of resistance to third‐generation cephalosporins and/or carbapenems caused by presumptive ESBL‐/AmpC‐/CP‐producing Salmonella and E. coli. Results from voluntary monitoring of MRSA in food and animals are also reported for countries that contribute data.
For human data in 2022, MSs reported results from antimicrobial susceptibility testing of Salmonella spp. and Campylobacter spp. isolates from clinical cases of salmonellosis and campylobacteriosis. Phenotypic test results were reported by MSs to TESSy either as quantitative or categorical/qualitative data at the isolate level according to the EU protocol for harmonised monitoring of AMR in human Salmonella and Campylobacter (ECDC, 2016). Quantitative phenotypic data were interpreted using EUCAST ECOFFs, where available, to understand microbiological resistance. Qualitative phenotypic data were interpreted using clinical breakpoints (CBPs). CBPs enable clinicians to choose the appropriate treatment based on information relevant to the individual patient while ECOFFs help epidemiologists identify small changes in bacterial susceptibility, which may indicate emerging resistance and allow for appropriate control measures to be considered. The breakpoints for ‘clinical’ resistance are often less sensitive than the ECOFF for a specific bacterium–drug combination resulting in higher levels of ‘microbiological’ resistance than ‘clinical’ resistance. By combining the categories of ‘clinically resistant’ (R) and ‘susceptible with increased exposure’ (I) into one category, however, close correspondence with the ECOFF can be achieved. A few countries reported genotypic data, either as resistance predicted from WGS or as sequences which were analysed at ECDC with ResFinder and PointFinder. Such genetic results are considered to correspond to the ECOFF with a separation between wild‐type and non‐wild‐type isolates. For assessing MDR in Salmonella and Campylobacter, ECDC and EFSA have agreed on a harmonised panel of nine and four antimicrobial classes, respectively, for better comparison between the two sectors.
Information on the materials and methods used in this EUSR on AMR can be found in Appendix F – Materials and methods at the end of this document. Additional information on the human data reported in 2021 can also be found in the European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2020–2021 (EFSA and ECDC, 2023).
Effect of the UK withdrawal on the analysis of AMR data at the EU
As a consequence of the UK withdrawal from the EU on 31 January 2020, the entry into force of the Withdrawal Agreement was ratified by the UK and the EU. The AMR data retrieved until 2019 covered all 28 EU Member States (MSs), which during that period included the UK. From 1 February 2020, the UK withdrew and became a ‘third‐country’ in relation to the EU, with final withdrawal effective after a transition period ending on 31 December 2020. The UK's withdrawal from the EU impacts the AMR data reported at the EU level in 2020 and onwards. UK data from 2020 are shown with non‐MSs in this report. For 2021 and 2022, data at the EU level are reported in accordance with the membership of the EU (i.e. EU without the UK). In 2021 and 2022, the only United Kingdom data that were reported to EFSA were from Northern Ireland. “In accordance with the Agreement on the withdrawal of the United Kingdom of Great Britain and Northern Ireland from the European Union and the European Atomic Energy Community, and in particular Article 5(4) of the Windsor Framework (see Joint Declaration No 1/2023 of the Union and the United Kingdom in the Joint Committee established by the Agreement on the withdrawal of the United Kingdom of Great Britain and Northern Ireland from the European Union and the European Atomic Energy Community of 24 March 2023, OJ L 102, 17.4.2023, p.87) in conjunction with section 24 of Annex 2 to that Framework, for the purposes of this Regulation, references to Member States should include the United Kingdom in respect of Northern Ireland.” Hence, the European Union requirements on data sampling were also applicable to Northern Ireland and data transmitted by the United Kingdom (Northern Ireland) have been assigned to the MSs group.
2. ANTIMICROBIAL RESISTANCE IN SALMONELLA SPP.
Monitoring of non‐typhoidal Salmonellas
This section focuses on non‐typhoidal Salmonellas (NTS) and summarises the occurrence and AMR patterns of isolates recovered from several food‐producing animal populations and fresh meat of broilers and turkeys taken at the border control posts (BCPs). Typhoidal salmonellas are human host‐adapted organisms causing typhoid and paratyphoid fever. Non‐typhoidal strains can either infect or colonise a multitude of animal hosts or be host‐specific for particular animal species (Crump et al., 2015). Typhoidal salmonellas belong to Salmonella enterica subsp. enterica serovars Typhi, Paratyphi A, Paratyphi B (d‐tartrate negative) and Paratyphi C, while NTS include all other serovars within the subspecies enterica (including the d‐tartrate positive Paratyphi B variant Java).
According to the World Health Organisation (WHO), the transmission of disease‐causing bacterial infections from non‐human sources to humans is more common in specific bacteria such as non‐typhoidal Salmonella, Campylobacter spp., and E. coli (WHO, 2019). Thus, the WHO urges for the recognition of this transmission potential. In 2022, salmonellosis was the second most commonly reported food‐borne zoonosis in the European Union, with 65,208 confirmed human cases and the most frequent cause of food‐borne outbreaks accounting for 17.6% of all food‐borne outbreaks reported in 2022 (EFSA and ECDC, 2023).
2.1. Key findings
The number of reported Salmonella spp. isolates from human cases varied considerably among the 29 reporting EU/EEA countries, often reflecting differences in population size: six countries reported < 100 human isolates, while six countries reported more than 1000 isolates.
Overall resistance to ampicillin, sulfonamides and tetracyclines was observed at high levels in Salmonella spp. isolates from humans in 2022 and ranged from moderate to very high in isolates from food‐producing animals and imported poultry meat, except in laying hens where low levels of resistance were reported.
Over the period 2013–2022, declining statistically significant trends in resistance to ampicillin and tetracyclines in isolates from humans were observed in 15 and 12 countries, respectively, primarily driven by declining resistance in Salmonella Typhimurium, a serovar commonly associated with pigs and cattle under 1 year of age.
Overall resistance to fluoroquinolones (ciprofloxacin) was observed at very high levels among isolates from broiler (55.5%) and fattening turkey flocks (57.9%), and at a high level in laying hens (24.7%) in 2022, and at moderate levels in Salmonella isolates from fattening pigs (10.1%) and cattle under 1 year of age (12.7%) from data reported in 2021. In Salmonella isolates from humans reported in 2022, the overall resistance to ciprofloxacin was 18.7%, with the lowest levels observed in monophasic S. Typhimurium (9.6%) and high to extremely high levels in S. Infantis (40.1%) and S. Kentucky (72.7%).
Extremely high resistance to ciprofloxacin was also reported in S. Kentucky isolates from broilers (84.2%), laying hens (82.1%) and fattening turkeys (100%). In S. Enteritidis, the most common Salmonella serovar detected in humans, resistance to ciprofloxacin was 22.8%. Resistance trends calculated for 2013–2022 for human data showed statistically significant increasing trends in resistance to ciprofloxacin in nine countries and decreasing trends in three, with the increase most noticeable in S. Enteritidis (12 countries with increasing trends) but also in S. Typhimurium and its monophasic variant and in S. Infantis.
Resistance to amikacin, the new substance included in the harmonised panel since 2021, was very low levels in all the animal populations except for Salmonella spp. isolates from cattle under 1 year of age, where no resistance was detected.
Overall resistance to third‐generation cephalosporins was noted at very low levels in isolates from humans in 2022 (1.4% resistance to ceftazidime and 1.2% to cefotaxime on average), at very low level in laying hens (0.2% resistance to cefotaxime and ceftazidime) and pigs (0.9% resistance to cefotaxime and ceftazidime) and at low levels in broiler flocks (1.4% resistance to cefotaxime and 1.3% to ceftazidime), turkey flocks (2.2% resistance to cefotaxime and ceftazidime) and cattle under 1 year of age (2.6% resistance to cefotaxime and 1.3% to ceftazidime). Consequently, the overall proportion of presumptive ESBL‐/AmpC‐producing Salmonella spp. at MS level was generally very low or low in 2021 and 2022 among all food‐producing animal populations and very low in isolates from human cases, although higher resistance was observed in specific Salmonella serovars.
In 2021 and 2022, no Salmonella spp. isolates recovered from animal/meat origins were microbiologically resistant to meropenem. However, unlike in 2021, when no meropenem resistance was reported in Salmonella spp. isolates from humans, in 2022, the occurrence of meropenem resistance was rare (< 0.1%), with three countries reporting five (of which four were confirmed) resistant isolates.
Overall, combined resistance to fluoroquinolones and cephalosporins was very low (≤ 1.0%) in isolates from both humans and food‐producing animals but higher in certain Salmonella serovars, reaching high levels in S. Kentucky isolates from broilers (21.1%) and moderate levels in S. Infantis isolates from turkeys (16.9%). Corresponding levels in humans were moderate in S. Kentucky (12.2%) and low in S. Infantis (5.9%).
Multidrug resistance (MDR) was overall high (22.1%) among Salmonella spp. reported in human cases in the EU, ranging from low levels among S. Enteritidis (2.4%) to very high among S. Kentucky (63.7%) and monophasic S. Typhimurium 1,4,[5],12:i:‐ (68.2%). Similarly, MDR was observed at high levels in Salmonella spp. recovered from broilers and turkeys in 2022 (43.6% and 39.4%, respectively), and from fattening pigs (39.1%) and cattle under 1 year of age (30.4%) in 2021. Salmonella spp. isolates from laying hens showed a markedly lower MDR level (7.5%). At the serovar level, the occurrence of MDR was similar across human and animal populations, with the exception of S. Kentucky, which on average exhibited a higher MDR occurrence in humans and in turkeys than when recovered from other animals.
Overall, in 2022, complete susceptibility (CS) in Salmonella spp. isolates from humans was observed in 57.7% of tested isolates. For animal data, CS was high for broilers (35.4%) and turkeys (29.4%) and was found at a very high level in laying hens (69.1%). For 2021 data, CS was high in pigs (40.5%) and very high in cattle under 1 year of age (55.7%). At the serovar level, S. Enteritidis had the highest levels of CS in both humans and all food‐producing animals.
2.2. Data on AMR in Salmonella spp. addressed
Commission Implementing Decision (EU) 2020/1729 lays down detailed protocols and rules for harmonising AMR monitoring and reporting in zoonotic and commensal bacteria. In 2021, the AMR monitoring in Salmonella isolates recovered from caecal contents of fattening pigs and bovine animals under 1 year of age, taken at slaughter, was mandatory. While for 2022, it was mandatory to monitor AMR in Salmonella isolates recovered from faecal samples and/or environmental samples (boot swabs or dust) of broiler, laying hen and fattening turkey flocks collected as part of National Control Programmes (NCPs) for Salmonella in poultry, and to monitor AMR in Salmonella isolates recovered from fresh meat from broilers and turkeys sampled at the border control posts (BCPs).
This chapter describes 2022 AMR data from faecal samples and/or environmental samples (boot swabs or dust) collected from flocks of broilers, laying hens and fattening turkeys, and 2021 AMR data on Salmonella isolates from bovine animals under 1 year of age (referred to as ‘cattle under 1 year of age’) and fattening pigs (referred to as ‘pigs’). Data for Salmonella spp. isolated from human cases are reported for both 2021 and 2022. However, Section 2.3 only presents data for 2022 since 2021 data from humans were published in the EU Summary report for 2021–2022 (EFSA and ECDC, 2023). Antimicrobial susceptibility testing (AST) results in Salmonella isolates from human cases include those serovars that are more prevalent in animal species.
Results from data on Salmonella spp. isolates include all serovars reported from the different animal origins. According to Commission Implementing Decision (EU) 2020/1729, only one isolate per Salmonella serovar from the same epidemiological unit is tested for AMR each year. Since AMR can vary markedly among serovars, the relative contribution of different serovars can influence the overall resistance levels reported for Salmonella spp. in the different animal/meat origins. Therefore, results are also presented for selected serovars if they exhibit a high prevalence (i.e. a high recovery rate from samples) or if they are deemed relevant to public health.
In cases where fewer than 10 isolates were retrieved from a particular animal origin in a given country, their resistance profiles were also considered in the analysis. This approach ensures that serovars with low prevalence are not excluded, that emerging serovars are accounted for and that all relevant data are included in the analysis. Note that some figures in this chapter only display individual MS data where 10 or more Salmonella spp. isolates were reported, although the occurrence of resistance at the MS‐group level includes all reported isolates.
Variations in Salmonella prevalence from food‐producing animals and their derived carcasses
In 2021 and 2022, countries reported data on Salmonella spp. from different origins according to their national situation. Noteworthy, some MSs did not obtain any Salmonella isolates from animal or meat origins; therefore, data are not presented for those countries. In 2022, the number of countries reporting results for broilers and laying hens was considerably higher than for fattening turkeys. This difference can be attributed to the small size of the turkey sector in certain MSs, with production levels falling below the threshold at which the monitoring is mandatory. Similarly, in 2021, the number of MSs reporting data from pigs was considerably higher than MSs reporting data from cattle under 1 year of age. Additionally, the number of isolates reported by countries varied due to different Salmonella prevalence. These factors may be a source of variation in the results when considering all reporting countries.
In this chapter, the occurrence of resistance refers to microbiological resistance (i.e. determined by the ECOFF value). The level of resistance is described as either ‘rare’: < 0.1%, ‘very low’: 0.1%–1.0%, ‘low’: > 1.0%–10.0%, ‘moderate’: > 10.0%–20.0%, ‘high’: > 20.0%–50.0%, ‘very high’: > 50.0%–70.0%, ‘extremely high’: > 70.0%. The significance of a specific level of resistance depends on the antimicrobial substance and its relative importance in both human and veterinary medicine. Furthermore, when interpreting the results, special attention should be given to the small sample size of some countries.
2.3. Humans: Occurrence of antimicrobial resistance in Salmonella
2.3.1. Data reported
For 2022, 27 MSs and two non‐MSs reported data on AMR in Salmonella isolates from human cases of non‐typhoidal salmonellosis. Twenty‐two countries provided data as measured values (quantitative data), five as data interpreted with clinical breakpoints and two reported whole genome sequences that were analysed by ECDC and interpreted as predicted wild type or predicted non‐wild type. Not all countries reported results for all antimicrobials in the harmonised panel (ECDC, 2016, 2021). The reported data represented 26.2% of the confirmed human cases with non‐typhoidal Salmonella reported in the EU/EEA in 2022.
2.3.2. Occurrence of resistance to commonly used antimicrobials in human and/or veterinary medicine
In 2022, high proportions of human Salmonella isolates were resistant to ampicillin (25.2%), sulfonamides (25.6%) and tetracyclines (25.1%) (Figure 1, Table 1 and Annex A). By serovar, resistance to these compounds ranged from low (3.1%–5.1%) in S. Enteritidis to extremely high in monophasic S. Typhimurium 1,4,[5],12:i:‐ (77.3%–86.7%). The variation in the proportion of resistance was large when considering countries reporting 10 or more isolates. Overall, for all Salmonella spp., outliers in terms of high proportion of resistance were observed in Bulgaria and Italy for ampicillin (54.5% and 44.3%, respectively), Portugal and Belgium for sulfonamides (43.2% and 32.3%, respectively) and Italy for tetracycline (42.2%) (Annex A, Table 1). Outliers in terms of a low proportion of resistance in Salmonella spp. was observed in Slovenia and Sweden for sulfonamides (8.9% and 9.3%, respectively). For S. Enteritidis, outliers with a higher proportion of resistance were observed in Bulgaria, Hungary and the Netherlands (31.6%, 17.2% and 16.3%, respectively) for ampicillin; in Portugal and Greece (18.2% and 15.7%, respectively) for sulfonamides; and in Italy for tetracycline (17.4%) (Annex A, Table 2). For monophasic S. Typhimurium 1,4,[5],12:i:‐, Austria reported a lower proportion of ampicillin resistance (67.2%) compared to other countries and Austria and Spain a lower proportion of sulfonamide resistance (64.1% and 68.7%, respectively) (Annex A, Table 4). For S. Infantis, Slovakia reported a much higher proportion of resistance to ampicillin than other MS (69.2% vs. 14.9%) (Annex A, Table 5). For S. Kentucky, Belgium was an outlier in reporting lower levels of ampicillin resistance (39.1%) (Annex A, Table 6).
FIGURE 1.
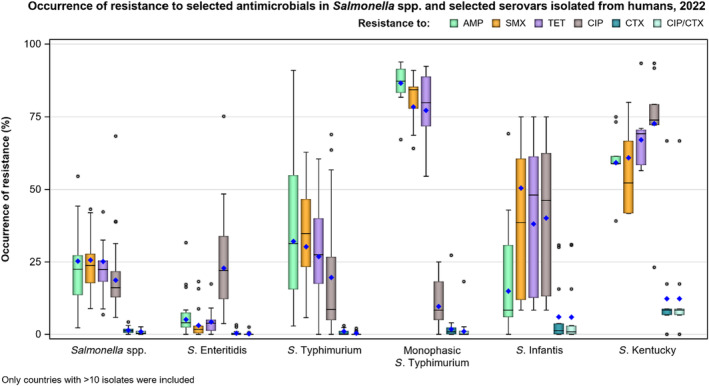
Occurrence of resistance to selected and critically important antimicrobials in Salmonella spp. and selected serovars isolated from humans, 2022.
Note: AMP, ampicillin; SMX, sulfamethoxazole; TET, tetracycline; CIP, ciprofloxacin; CTX, cefotaxime; CIP/CTX, combined ‘microbiological’ resistance to ciprofloxacin and cefotaxime; Blue diamond, resistance at the reporting MS group level; Horizontal lines represent median; Lower and upper box boundaries, 25th and 75th percentiles, respectively. Only MSs reporting data for 10 or more isolates are shown in the graph; however, all isolates are included in the calculation of resistance at the reporting MS group level.
TABLE 1.
Occurrence of resistance to selected and critically important antimicrobials in Salmonella spp. and selected serovars from humans, 2022.
| EU total | AMP | SMX | TET | CIP | CTX | Combined CIP/CTX | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | % res | N | % res | N | % res | N | % res | N | % res | N | % res | |
| Salmonella spp. (27 MSs) | 16,059 | 25.2 | 8596 | 25.6 | 14,305 | 25.1 | 15,824 | 18.7 | 15,323 | 1.4 | 15,264 | 0.9 |
| S. Enteritidis (26 MSs) | 4629 | 5.1 | 2637 | 3.1 | 3867 | 4.3 | 4431 | 22.8 | 4226 | 0.4 | 4191 | 0.2 |
| S. Typhimurium (27 MSs) | 1971 | 32.1 | 941 | 30.2 | 1633 | 26.8 | 1963 | 19.6 | 1839 | 1.0 | 1833 | 0.4 |
| Monophasic S. Typhimurium (19 MSs) | 2587 | 86.7 | 1673 | 78.5 | 2528 | 77.3 | 2588 | 9.6 | 2588 | 1.8 | 2587 | 1.0 |
| S. Infantis (26 MSs) | 705 | 14.9 | 305 | 50.5 | 656 | 38.1 | 701 | 40.1 | 698 | 6.0 | 698 | 5.9 |
| S. Kentucky (18 MSs) | 228 | 59.2 | 169 | 60.9 | 213 | 67.1 | 227 | 72.7 | 227 | 12.3 | 227 | 12.3 |
Abbreviation: %res, percentage of resistance; AMP, ampicillin; CIP, ciprofloxacin/pefloxacin; CTX, cefotaxime; N, number of Salmonella isolates tested; SMX, sulfamethoxazole; TET, tetracycline.
Overall, resistance to gentamicin was low (2.9%) and across all reported serovars (Annex A, Tables 1–6) except in S. Kentucky where gentamicin resistance was high (34.7%) at the EU level (Annex A, Table 6). Similarly, levels of trimethoprim resistance were overall low among Salmonella spp. (6.1%) (Annex A, Table 1), but moderate in monophasic S. Typhimurium 1,4,[5],12:i:‐ and S. Infantis (12.1% and 14.2%, respectively) and high in S. Kentucky (22.2%) (Annex A, Tables 4–6).
2.3.3. Occurrence of resistance to highest priority ‘critically important antimicrobials’ (CIAs) and last resort antimicrobials
The proportion of Salmonella isolates resistant to the highest priority critically important antimicrobial (hpCIA) ciprofloxacin was overall 18.7% (Figure 1; Table 1). A high proportion of resistance to ciprofloxacin was observed in isolates of S. Enteritidis (22.8%) and S. Infantis (40.1%), while an extremely high proportion was observed in S. Kentucky isolates (72.7%) (Figure 1, Annex A: Tables 2, 5, 6). At the country level, a very high proportion of ciprofloxacin resistance in Salmonella spp. was observed in Poland (68.4%), with extremely high resistance in Polish S. Enteritidis isolates (75.2%) (Annex A: Tables 1, 2). Croatia and Malta reported very high resistance to ciprofloxacin in S. Infantis isolates (68.9% and 63.6%, respectively) (Annex A: Table 5). Caution should be taken when interpreting results for some countries as they report data on a small number of isolates.
For cefotaxime and ceftazidime, representing third‐generation cephalosporins, another class of hpCIAs for Salmonella, resistance levels were generally low among Salmonella spp. (1.4% and 1.2%, respectively) (Annex A: Table 1), with low levels of resistance ranging from 0.3% to 12.3% across the serovars of interest (Annex A). Resistance was more pronounced in S. Infantis and S. Kentucky isolates (range: 5.9%–12.3%) (Table 1; Annex A: Tables 5, 6). Outliers in terms of high resistance to third‐generation cephalosporins were observed in the Netherlands and Italy for S. Infantis (30.8% and 30.0%, respectively) and very high resistance in Sweden for S. Kentucky (66.7%) (Annex A: Tables 5, 6).
Twelve countries tested resistance to last line antimicrobials azithromycin and tigecycline. Resistance was overall low among Salmonella spp. (0.6% and 4.2%, respectively), although Belgium observed a moderate resistance to tigecycline (19.2%, Annex A: Table 1). Among the individual serovars, the highest level of resistance to azithromycin was observed in S. Kentucky (6.3%). The highest proportion of isolates resistant to tigecycline was observed in S. Infantis and S. Kentucky (15.4% and 15.5%, respectively, see Annex A: Tables 5, 6). While tigecycline resistance was highest in these serovars also in 2021, the resistance increased noticeably in 2022 compared to the levels in 2021 (2.0% and 3.9%, for S. Infantis and S. Kentucky, respectively). Resistance to colistin was detected in 8.9% of Salmonella isolates, with resistance being most pronounced in S. Enteritidis isolates (29.7%), a serovar belonging to group D Salmonella which tend to show a higher natural tolerance to colistin (Agersø et al., 2012; Ricci et al., 2020).
Combined resistance to both ciprofloxacin and cefotaxime was overall very low overall in Salmonella spp. in human cases (0.9%) (Figure 2A, and Annex A: Table 7) and in the serovar S. Enteritidis, S. Typhimurium and monophasic S. Typhimurium (0.2%, 0.4% and 1.0%, respectively, Annex A: Tables 8–10). Higher levels of combined resistance were observed in S. Infantis (5.9%) and S. Kentucky (12.3%) (Figure 2B,C, and Annex A: Tables 11, 12). The Netherlands and Italy reported the highest proportion of combined resistance in S. Infantis (31.8% and 30.0%, respectively) and Sweden in S. Kentucky (66.7%). The high proportion of combined resistance in S. Kentucky in Sweden was due to an accidental transmission of a S. Kentucky with bla CTX‐M‐14b between patients in a hospital when using a contaminated instrument for gastroscopy (University hospital of Skåne, 2022).
FIGURE 2.
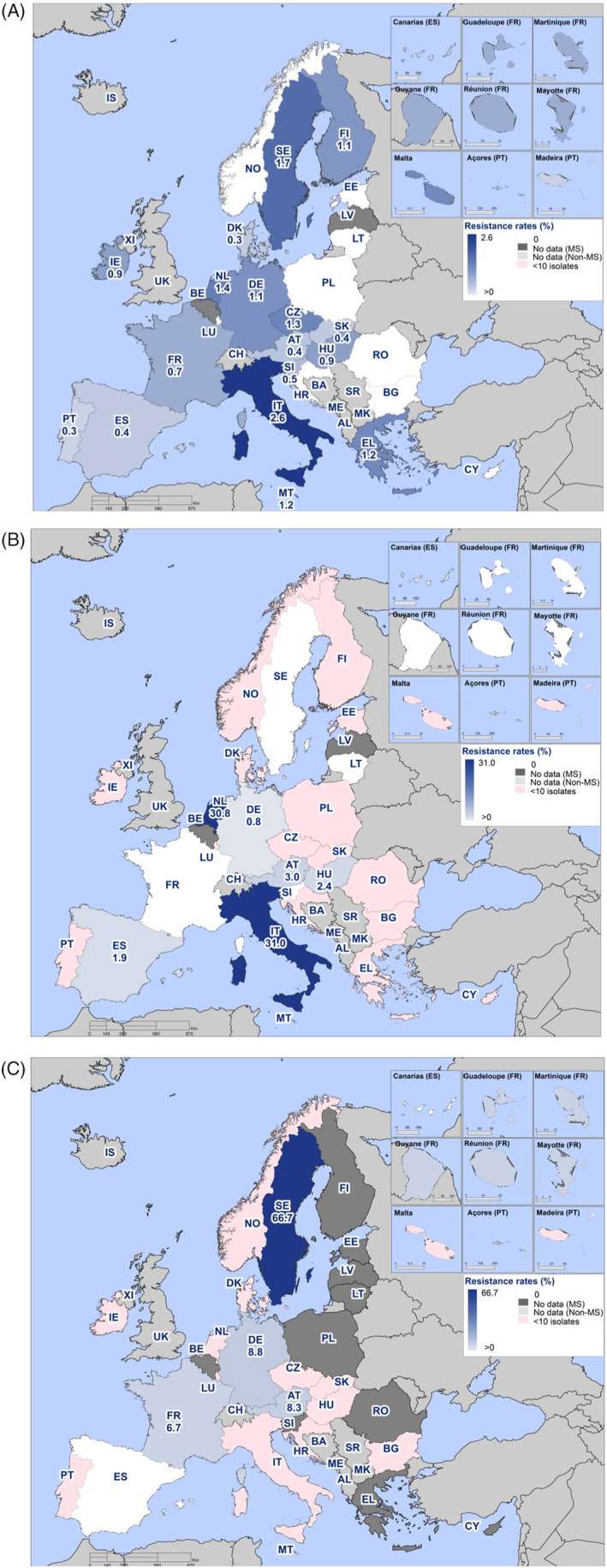
Spatial distribution of combined ‘microbiological’ resistance to ciprofloxacin and cefotaxime among (A) Salmonella spp., (B) S. Infantis and (C) S. Kentucky isolated from human cases, 2022 (pink indicates fewer than ten isolates tested).
2.3.4. ESBL‐, AmpC‐ and carbapenemase‐producing Salmonella
Among the 26 MSs and one non‐MS reporting data on third‐generation cephalosporins in 2022, resistance was either not detected (4 MSs) or found at very low to low levels. Three countries reporting cephalosporin‐resistant isolates did not provide further details on phenotypic/genotypic characterisation of ESBL/AmpC, and these isolates were excluded from analysis in Table 2 and Annex A: Table 13. Four countries had not tested all presumptive ESBL/AmpC isolates, most likely due to clinical breakpoints being used in routine AST and not ECOFFs. In Italy, there is a special focus on ESBL/AmpC monitoring in Salmonella where primary laboratories are requested to send any isolates resistant to cefotaxime and/or ceftazidime to the national public health reference laboratory for confirmation. This may have resulted in an overrepresentation of such isolates in the Salmonella AMR data set from Italy.
TABLE 2.
ESBL, AmpC and carbapenemase phenotypes and genotypes in Salmonella spp. isolates from humans by serovar in reporting EU/EEA countries, 2022.
| Serovar | Tested for CTX and/or CAZ | Res to CTX and/or CAZ | Resistance phenotype | Negative for ESBL, AmpC, CP | Genotype | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ESBL | AmpC | AmpC + ESBL | Carbapenemase | ||||||||||
| N | N | N | % | N | % | N | % | N | % | N | % | ||
| Bareilly | 48 | 2 | 1 | 2.1 | |||||||||
| Durham | 49 | 1 | 1 | 2.0 | CTX‐M‐1 (1) | ||||||||
| Enteritidis | 3927 | 10 | 4 | 0.1 | 2 | 0.1 | 4 | 0.1 | CTX‐M‐15 (2), CTX‐M (1), CMY‐2 (1), DHA‐18 (1) | ||||
| Goldcoast | 89 | 4 | 2 | 2.2 | 2 | 2.2 | CTX‐M‐1 (1), CTX‐M‐32 (1), CMY‐2 (2) | ||||||
| Haifa | 25 | 3 | 3 | 12.0 | OXA‐48‐like (3) | ||||||||
| Heidelberg | 19 | 1 | 1 | 5.3 | CMY‐2 (1) | ||||||||
| Infantis | 614 | 41 | 40 | 6.5 | 2 | 0.3 | 1 | 0.2 | CTX‐M‐1 (24), CTX‐M‐65 (8),CTX‐M‐3 (2), CTX‐M (1), SHV‐12 (1), CMY‐2 (1) | ||||
| Kentucky | 223 | 31 | 22 | 9.9 | 1 | 0.4 | 1 | 0.4 | 1 | 0.4 | CTX‐M‐243/14b (14), CTX‐M (4), CTX‐M‐9 (1), OXA‐48 (1) | ||
| London | 42 | 1 | 1 | 2.4 | CTX‐M‐15 (1) | ||||||||
| Mikawasima | 97 | 1 | 1 | 1.0 | CMY‐2 (1) | ||||||||
| Minnesota | 7 | 2 | 1 | NA | CTX‐M‐8 (1) | ||||||||
| Mishmarhaemek | 27 | 1 | 1 | 3.7 | CMY‐2 (1) | ||||||||
| Monophasic Typhimurium 1,4,[5],12:i:‐ | 2449 | 45 | 26 | 1.1 | 11 | 0.4 | 3 | 0.1 | CTX‐M‐1 (17), CTX‐M‐14 (2), SHV‐12 (4), CMY‐2 (11) | ||||
| Muenster | 33 | 2 | 1 | 2.9 | CTX‐M‐55 (1) | ||||||||
| Napoli | 131 | 1 | 1 | 1.0 | CTX‐M‐15 (1) | ||||||||
| Ohio | 22 | 1 | 1 | 4.5 | CTX‐M‐15 (1) | ||||||||
| Schwartzengrund | 31 | 3 | 2 | 6.5 | CTX‐M‐55 (2) | ||||||||
| Stanley | 110 | 1 | 1 | 0.9 | 1 | 0.9 | 1 | 0.9 | DHA‐1 and OXA‐1 (1) | ||||
| Typhimurium | 1796 | 17 | 8 | 0.4 | 3 | 0.2 | 3 | 0.2 | CTX‐M‐1 (5), CTX‐M‐9 (1), CTX‐M‐15 (1), CTX‐M‐65 (1), CMY‐2 (3) | ||||
| Virchow | 130 | 7 | 6 | 4.6 | SHV‐12 (6) | ||||||||
Abbreviations: CTX, cefotaxime; CAZ, ceftazidime; ESBL, extended spectrum beta‐lactamase. Bulgaria, Hungary and Slovakia did not perform confirmatory testing of resistant isolates and their results could therefore not be included in this table.
ESBL‐producing Salmonella were identified in 0.9% of the tested isolates, ranging by MS from 0% in Lithuania, Luxembourg and Romania to 3.1% in Italy (Annex A: Table 13). AmpC was less frequent, identified in 0.2% of tested isolates, with the highest occurrence in Italy (1.2%). Four isolates (0.03%) were reported to be both AmpC‐ and ESBL‐producing and four isolates (0.03%) carried a carbapenemase (Annex A: Table 13). ESBL was reported in 20 serovars in 2022, with the highest proportions observed in isolates of S. Kentucky (9.9%), S. Schwarzengrund (9.7%), S. Infantis (6.5%), S. Virchow (4.6%) and S. Ohio (4.5%) (Table 2). AmpC‐type beta‐lactamases were overall reported in 10 serovars, with the highest proportion observed in S. Heidelberg (5.3%) and S. Mishmarhaemek (3.7%). Four of the five Salmonella isolates reported as resistant to meropenem in 2022 could be confirmed. Three of the isolates were S. Haifa and one S. Kentucky, all carrying bla OXA‐48, (Table 2 and Annex A: Table 13). It should, however, be noted that in six of 26 reporting MSs, meropenem results were interpreted using the EUCAST clinical breakpoint (CBP), where the MIC is substantially higher (+4 dilutions) than the ECOFF.
2.3.5. Complete susceptibility (CS) and multidrug resistance (MDR)
In this report, complete susceptibility (CS) is defined as susceptibility to each of the nine antimicrobial classes tested in the harmonised panel described by the ECDC (ECDC, 2016). Multidrug resistance (MDR) is defined as resistance to three or more antimicrobial classes among Salmonella isolates from human cases.
The level of CS in 2022 was 57.7% in Salmonella spp. from humans with the highest proportion in S. Enteritidis (71.7%), S. Typhimurium (56.3%) and S. Infantis (43.4%). The lowest levels of CS were observed in S. Kentucky (20.8%) and monophasic S. Typhimurium (6.4%) (Figure 3 and Annex A: Tables 13–18).
FIGURE 3.
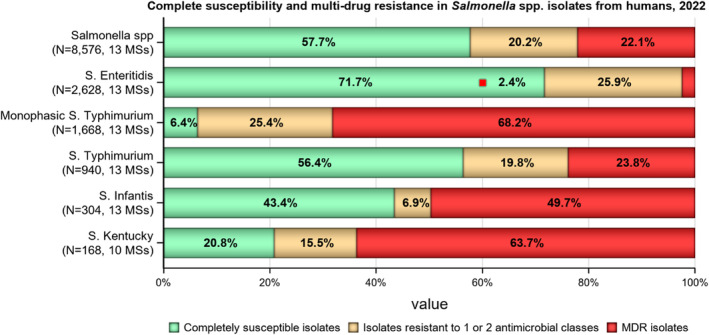
Proportion of Salmonella isolates from humans being completely susceptible, resistant to one and/or two antimicrobial classes or multidrug resistant (MDR) in 2022.
Note: MDR and complete susceptibility are expressed as percentages; N, total number of isolates reported by MSs.
MDR was high overall (22.1%, n = 8576) among Salmonella spp. (Figure 3 and Annex A: Table 13). For the investigated serovars, MDR was most frequently reported among monophasic S. Typhimurium 1,4,[5],12:i:‐ (68.2%) and S. Kentucky (63.7%), followed by S. Infantis (49.7%), S. Typhimurium (23.8%) and lastly S. Enteritidis (2.4) (Figure 3 and Annex A: Tables 14–18). Ten isolates (eight S. Infantis and one each of S. Kentucky and monophasic S. Typhimurium) were resistant to eight of the nine tested substances, only susceptible to meropenem.
2.3.6. Temporal trends
Trends in resistance over the 10‐year period 2013–2022 were assessed with logistic regression. Trends varied by country for the different serovars and antimicrobials (Table 3, Figure 4, Annex A: Figures 1–6). For Salmonella spp. overall, 15 and 12 countries out of 26 observed a statistically significant decrease in resistance to ampicillin and tetracycline, respectively, whereas one and three countries reported an increase. For cefotaxime and ciprofloxacin, seven and three countries, respectively, observed a statistically significant decrease in resistance, while four and nine countries reported an increase.
TABLE 3.
Number of countries with statistically significant (p < 0.05) increasing or decreasing trends in resistance to selected antimicrobials for Salmonella spp. and selected serovars in humans in 2013–2022.
| Serovar | Ampicillin | Cefotaxime | Ciprofloxacin/Quinolones | Tetracycline | ||||
|---|---|---|---|---|---|---|---|---|
| Incr. | Decr. | Incr. | Decr. | Incr. | Decr. | Incr. | Decr. | |
| Salmonella spp. (24 MSs + 2 non‐MS) | 1 (FI) | 15 (CY, DE, DK, EL, ES, FR, IE, IT, LT, LU, NO, PT, RO, SE, SI) | 4 (HU, IT, SE, SI) | 7 (BE, EE, ES, FR, MT, PL, SK) | 9 (AT, DE, HU, LT, NL, NO, PL, RO, SK) | 3 (ES, FR, MT) | 3 (EE, SI, SK) | 12 (DK, EL, ES, FR, HU, IE, IT, LU, NL, NO, PT, SE) |
| S. Enteritidis (23 MSs + 1 non‐MS) | 4 (AT, BE, NL, SK) | 6 (DE, ES, LT, MT, PL, RO) | – | 4 (EE, IE, NO, PL) | 12 (AT, DE, EE, HU, LT, LU, NL, NO, PL, RO, SI, SK) | 6 (BE, ES, FR, MT, PT, SE) | 7 (AT, BE, DE, IT, NL, SI, SK) | 5 (ES, FR, LT, PL, RO) |
| S. Typhimurium (23 MSs + 2 non‐MS) | – | 16 (AT, CY, DE, DK, EE, ES, FI, FR, HU, IE, IS, LU, NO, PT, RO, SI) | 2 (DE, HU) | 1 (IE) | 6 (DE, HU, LT, NO, SI, SK) | 2 (IS, MT) | – | 15 (AT, DE, EE, EL, ES, FI, FR, HU, IE, LU, NL, NO, PT, SE, SI) |
| Monophasic S. Typhimurium (15 MSs + 1 non‐MSs) | 4 (EE, IT, MT, NL) | 4 (AT, ES, HU, LU) | 2 (IT, SI) | 3 (BE, ES, LU) | 5 (AT, HU, NL, PT, SI) | 1 (NO) | 3 (DK, SE) | 7 (AT, ES, FR, HU, IE, PT, SI) |
| S. Infantis (12 MSs) | 5 (AT, BE, HU, NL, SK) | 4 (DE, ES, FR, LT) | 1 (NL) | – | 5 (BE, DE, ES, NL, SK) | 2 (HU, MT) | 3 (BE, ES, NL) | 1 (DE) |
| S. Kentucky (7 MSs) | – | 1 (BE) | 1 (BE) | 1 (MT) | – | 2 (ES, FR) | – | 2 (AT, BE) |
Abbreviations: AT, Austria; BE, Belgium, BG, Bulgaria; CY, Cyprus; DE, Germany; DK, Denmark; EE, Estonia; EL, Greece; ES, Spain; FI, Finland; HU, Hungary; FR, France; IS, Iceland; IT, Italy; LU, Luxembourg; LT, Lithuania; LV, Latvia; MT, Malta; NL, Netherlands, NO, Norway, PL, Poland; PT, Portugal; RO, Romania; SE, Sweden; SI, Slovenia, SK, Slovakia.
FIGURE 4.
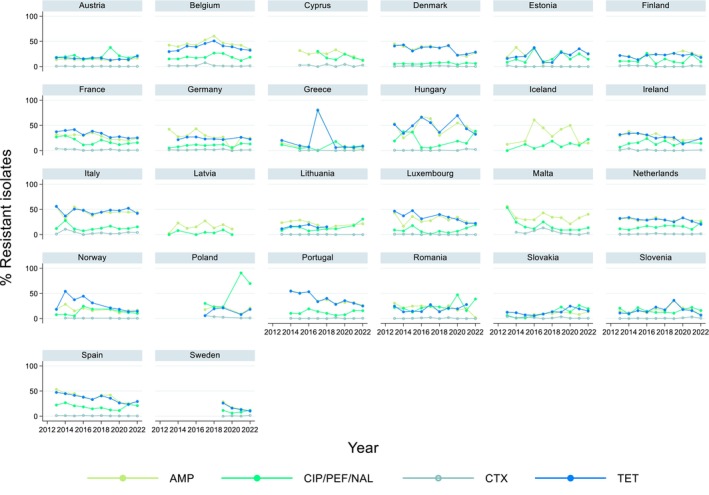
Trends in resistance to ampicillin, ciprofloxacin/pefloxacin/nalidixic acid, cefotaxime and tetracycline in Salmonella spp. from humans in 26 reporting countries, 2013–2022.
By serovar, statistically significant increasing trends in resistance to ciprofloxacin/quinolones were more commonly observed than decreasing trends in all investigated serovars except for S. Kentucky, and with the most notable increase observed in S. Enteritidis (12 countries). Respectively, 16 and 15 countries reported decreasing trends in resistance to ampicillin and tetracycline in S. Typhimurium.
2.3.7. High‐level ciprofloxacin resistance
In 2022, 1.5% (N = 7954) of Salmonella spp. from humans expressed high‐level resistance to ciprofloxacin (MIC ≥ 4 mg/L, Table 4). Such isolates were reported from seven of the 12 countries reporting MIC values for ciprofloxacin. Among the nine serovars reported with MICs of ≥ 4 mg/L, high‐level ciprofloxacin resistance was most frequently observed in S. Kentucky (in 65.4% of tested S. Kentucky) and this serovar accounted for 106 out of 121 isolates reported with high‐level MIC.
TABLE 4.
Occurrence of high‐level resistance to ciprofloxacin (MIC ≥ 4 mg/L) in Salmonella serovars from human cases in 2022.
| Serovar | N | High‐level resistance to ciprofloxacin (MIC ≥ 4 mg/L) | |
|---|---|---|---|
| n | % | ||
| S. Agona | 127 | 2 | 1.6 |
| S. Corvallis | 21 | 1 | 4.8 |
| S. Enteritidis | 1976 | 1 | 0.1 |
| S. Hadar | 36 | 2 | 5.6 |
| S. Infantis | 385 | 1 | 0.3 |
| S. Kentucky | 162 | 106 | 65.4 |
| S. Saintpaul | 35 | 2 | 5.7 |
| S. Schwarzengrund | 15 | 1 | 6.7 |
| Unspecified serovar | 184 | 5 | 2.7 |
| Other | 5013 | – | 0 |
| Total (13 MSs) | 7954 | 121 | 1.5 |
Abbreviations: N, Number of isolates tested for ciprofloxcin with dilution methods; n, number of isolates with high‐level resistance to ciprofloxacin.
2.4. Food‐producing animals and meat thereof: Occurrence of antimicrobial resistance in Salmonella
2.4.1. Data reported
In 2022, 24 MSs, the United Kingdom (Northern Ireland) and two non‐MSs reported AMR data on Salmonella isolates recovered from broiler flocks, 23 MSs, the United Kingdom (Northern Ireland) and one non‐MS reported AMR data on Salmonella isolates recovered from laying hen flocks, and 19 MSs reported AMR data on Salmonella isolates recovered from fattening turkey flocks. Additionally, five and one MSs reported data on Salmonella isolates recovered from fresh meat of broilers and turkeys sampled at the border control posts, respectively.
In 2021, 25 MSs, the United Kingdom (Northern Ireland) and one non‐MS reported AMR data on Salmonella isolates recovered from the caecal contents of pigs at slaughter, and 10 MSs reported AMR data on Salmonella isolates recovered from the caecal contents of cattle under 1 year of age at slaughter.
The reporting of isolate‐based data allows for the analysis of MDR patterns, the detection of high‐level ciprofloxacin resistance and combined resistance to ciprofloxacin and cefotaxime, which are first‐line agents critically important for treating human salmonellosis. In accordance with Commission Implementing Decision (EU) 2020/1729, MSs also included information on serovars and production type. This enabled a detailed analysis of the occurrence of resistance and MDR by serovar for the different animal/meat origins (see Appendix B).
Summary data on the occurrence of resistance to commonly used antimicrobials in veterinary medicine (ampicillin, sulfamethoxazole and tetracycline) as well as hpCIAs (ciprofloxacin, cefotaxime and combined resistance to these two antimicrobials) are displayed in Figure 5 for Salmonella isolates from broiler, laying hen and fattening turkey flocks in 2022, and cattle under 1 year of age and pigs in 2021. Annex A presents the occurrence of AMR (%), CS, MDR and combined resistance to ciprofloxacin and cefotaxime, in Salmonella spp. from broilers, laying hens, turkeys and fresh meat from broilers and turkeys sampled at the BCPs (corresponding to 2022 data), and pigs and cattle under 1 year of age (corresponding to 2021 data), at both the MS and MS‐group level (Annex A is available on the EFSA knowledge junction community on Zenodo at: https://doi.org/10.5281/zenodo.10528846).
Changes in the harmonised panel of antimicrobial substances for the monitoring of AMR in animals and food for Salmonella spp. according to the new legislation
The Commission Implementing Decision (EU) 2020/1729 lays down specific technical requirements for AMR testing and reporting in representative isolates derived from randomised sampling in food‐producing animals performed at farm level and/or at slaughter and in imported fresh meat at border control posts. The new rules apply to the monitoring performed from 2021 onwards.
In 2021, amikacin was added to the harmonised panel of antimicrobials for the monitoring and reporting of AMR in Salmonella and was included in the analyses of the occurrence of resistance, CS and MDR. While amikacin is not used in food‐producing animals, it is commonly used in hospitals to treat urinary tract infections, bacteraemia and intra‐abdominal infections caused by Gram‐negative bacteria. Amikacin was added to the harmonised panel because it is thought to improve the detection of 16S RNA methyltransferases (RMTases) (EFSA, 2019). RMTases are increasingly associated with carbapenemases, AmpC‐ or ESBL‐ enzymes and fluoroquinolone resistance in Enterobacteriaceae, particularly outside of Europe, and have recently also been detected in human cases within Europe (Arca‐Suárez et al., 2022; Fournier et al., 2022).
Additionally, under the new legislation, the epidemiological cut‐off values (ECOFFs) and clinical breakpoints (CBPs) used to determine the microbiological resistance of Salmonella isolates to some antimicrobial substances have also changed. This is the case for tigecycline, for which the ECOFF changed from > 1 to > 0.5 mg/L; for nalidixic acid, the ECOFF changed from MIC > 16 mg/L to MIC > 8 mg/; and for ciprofloxacin, the CBP changed from MIC > 1 mg/L to MIC > 0.064 mg/L.
In this report, the occurrence of resistance to tigecycline and nalidixic acid is determined using the new ECOFFs (MIC > 0.5 mg/L and MIC > 8 mg/L, respectively). Equally, for ciprofloxacin, clinical resistance was determined by the new CBP (MIC > 0.064 mg/L).
FIGURE 5.
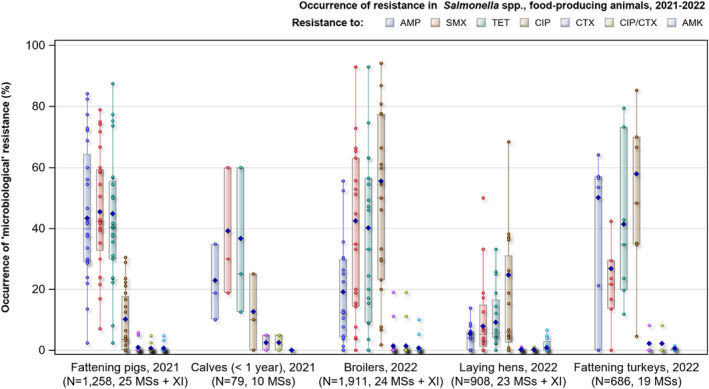
Occurrence of resistance to selected and critical important antimicrobials in Salmonella spp. recovered from broilers, laying hens, fattening turkeys in 2022, and fattening pigs and cattle under 1 year of age (calves) in 2021.
Note: AMP, ampicillin; SMX, sulfamethoxazol; TET, tetracycline; CIP, ciprofloxacin; CTX, cefotaxime; CIP/CTX, combined 'microbiological' resistance to ciprofloxacin and cefotaxime; AMK, amikacin; N, total number of Salmonella spp. isolates reported by MSs; blue diamond shows resistance at the reporting MS group level. Horizontal lines represent media; Lower and upper box boundaries, 25th and 75th percentiles, respectively. Only MSs reporting data for 10 or more isolates are shown in the graph; however, all isolates are included in the calculation of resistance at the reporting‐MS group level.
2.4.2. Occurrence of resistance to commonly used antimicrobials in veterinary medicine
Since 2014, the antimicrobial substances included in the harmonised panel designed for monitoring and reporting AMR in Salmonella from food‐producing animals and derived meat have ensured the continuity of monitoring data and epidemiological tracing of isolates (particularly serovars) exhibiting resistance patterns of interest to public health. The selection of these antimicrobial substances was done due to either their public health importance or their common use in veterinary medicine.
Antimicrobial substances such as ampicillin, sulfamethoxazole and tetracycline are widely used in veterinary medicine to treat infections in production animals. The WHO categorises ampicillin as a ‘critically important antimicrobial’ (CIA) in human medicine, while sulfamethoxazole and tetracycline are categorised as ‘highly important antimicrobials’ (HIA) in human medicine (WHO, 2019).
Food‐producing animals
In 2022, Salmonella spp. isolated from broiler flocks exhibited on average high resistance levels for tetracycline (40.2%; median = 24.4%) and sulfonamides (42.5%; median = 33.3%). Similarly, turkey flocks showed a high level of resistance to tetracycline (41.4%; median = 25.0%) and sulfonamides (26.7%; median = 16.7%). While laying hen flocks exhibited low resistance levels for tetracycline (9.1%; median = 3.9%) and sulfonamides (7.9%; median = 2.7%) (Annex A, Tables 20–22). The average resistance levels reported for ampicillin were lower than the levels of the other antimicrobials commonly used in veterinary medicine in broiler and laying hen flocks, showing moderate (19.1%, median = 12.2%) and low (5.4%; median = 0%) resistance levels, respectively. However, Salmonella isolates recovered from turkey flocks showed on average a very high resistance level to ampicillin (50.2%; median = 0%) (Figure 5; Annex A, Table 22). Resistance levels in Salmonella isolates recovered from poultry varied from low (laying hens, 3.6%; median = 0.0%) to moderate for trimethoprim (broilers, 16.2%, median = 1.8%; turkeys, 14.6%, median = 0.0%). While resistance levels for gentamicin were low in isolates from broilers and turkeys (1.2%, median = 0.0%; 3.4%, median = 0.0%, respectively) and very low in isolates recovered from laying hens (1.0%, median = 0.0%). Resistance levels to chloramphenicol in Salmonella isolates recovered from poultry were low ranging from 1.3% (laying hens) to 3.1% (turkeys) (Annex A, Tables 21, 22).In 2021, Salmonella spp. recovered from pigs and cattle under 1 year of age showed on average high resistance to tetracycline and sulfonamides. Overall, tetracycline resistance was reported at high levels for pigs and cattle under 1 year of age (44.9%; median = 40.4%, and 36.7%; median = 37.5%, respectively). Sulfamethoxazole resistance was also reported at a high level, with an average of 45.5% (median = 42.5%) and 39.2% (median = 40.0%), respectively (Annex A, Tables 23, 24). The occurrence of ampicillin resistance was similar in pigs (43.4%; median = 39.9%) and slightly lower in cattle under 1 year of age (22.8%; median = 13.3%) when compared to tetracycline and sulfamethoxazole. Resistance levels in Salmonella isolates recovered from both species varied from rare to moderate for other antimicrobial substances included in the harmonised panel (Annex A, Tables 23, 24).
2.4.3. Occurrence of resistance to highest priority critically important antimicrobials (hpCIAs) and last resort antimicrobials
Food‐producing animals
In 2022, overall resistance to ciprofloxacin was reported at a very high level for broilers (55.5%, median = 46.2%) and fattening turkeys (57.9%, median = 35.3%) and at a high level for laying hens (24.7%, median = 4.2%). Resistance to nalidixic acid was reported at similar levels, with 55.3% and 49.1% for broilers and turkeys, respectively, and 23.7% for laying hens (Figure 5; Annex A, Tables 20–22).
Of the Salmonella isolates from broilers resistant to ciprofloxacin and susceptible to nalidixic acid (n = 13), three were S. Paratyphi B (Belgium), two each were S. Anatum (Portugal), S. Virchow (Belgium) and S. enterica subsp. Enterica (Czechia), and one each were S. Derby (Belgium), S. Bovismorbificans (Spain) and S. Kedougou (Spain). Of the Salmonella isolates from laying hens resistant to ciprofloxacin and susceptible to nalidixic acid (n = 10), two each were S. Infantis (Belgium and Romania) and S. Kentucky (Italy), and one each were S. Bovismorbificans (Spain), S. Braenderup (Hungary), S. Enteritidis (Spain), S. Meleagridis (Spain), S. Molade (France) and S. Uganda (Spain). Although MSs reported a high number (N = 63) of Salmonella isolates from turkeys resistant to ciprofloxacin and susceptible to nalidixic acid, 74.6% were S. Anatum (Hungary [n = 2], Italy [n = 37], Portugal [n = 4] and Slovenia [n = 4]). Other serovars were S. Muenchen (Spain, n = 6), S. Agona (France [n = 1], Italy [n = 3]), S. Bovismorficans (Spain, n = 2), S. Cerro (Portugal, n = 1), S. Derby (Spain = 1), S. London (Spain, n = 1) and S. Uganda (Spain, n = 1).
In 2021, overall resistance to ciprofloxacin was reported at a moderate level for both pigs (10.1%; median = 4.1%) and cattle under 1 year of age (12.7%; median = 0%). Resistance to nalidixic acid was reported at the same level 9.8% (median = 4.1%) and 10.1% (median = 0%), respectively (Figure 5; Annex A, Tables 23, 24). Of the Salmonella isolates from pigs resistant to ciprofloxacin and susceptible to nalidixic acid (n = 10), four were monophasic S. Typhimurium (Spain [n = 2], Croatia [n = 1] and Poland [n = 1]); two were S. Derby (one each from Belgium and Romania); and one each was S. Typhimurium (Spain), S. Bovismorbificans (Romania), S. Uganda (Spain) and S. Rissen (Bulgaria). Only two Salmonella isolates from cattle under 1 year of age displayed ciprofloxacin resistance and nalidixic acid susceptibility (S. Agama and S. Typhimurium), both from Italy.
In 2022, no resistance to third‐generation cephalosporins (i.e. cefotaxime and ceftazidime) in Salmonella isolates from broilers, laying hens and fattening turkeys was reported by most countries (Annex A, Tables 20–22). Only Italy (cefotaxime = 11.1% and ceftazidime = 9.5%, N = 190) and Malta (19.0% for both antimicrobials, N = 21) reported low and moderate resistance to both antimicrobials, respectively, in Salmonella isolates from broilers, while Poland and The Netherlands reporting only one isolate each resistant to both antimicrobials, resulting in an overall low resistance to cefotaxime and ceftazidime in broilers (1.4%; median = 0%, 1.3%, median = 0%, respectively). In laying hens, only Italy reported two isolates (S. Infantis) resistant to both substances (N = 174). Similarly, Italy was the only country reporting 15 Salmonella isolates from turkeys (S. Infantis) resistant to both substances (N = 184) (Annex A, Tables 21, 22).
When considering 2021 data, no resistance to third‐generation cephalosporins (i.e. cefotaxime and ceftazidime) in Salmonella isolates from pigs and cattle under 1 year of age was reported by most countries (Annex A, Tables 23, 24). Overall resistance to cefotaxime and ceftazidime in pigs was very low (0.9%; median = 0%, for each antimicrobial). The few countries reporting resistance to both antimicrobials in Salmonella isolates from pigs were Romania (5.8%, N = 104), Hungary (5.0%, N = 80) and Italy (1.1%, N = 91) (Annex A, Table 23). Italy was the only country reporting a single Salmonella isolate from cattle under 1 year of age (S. Ngor) resistant to both substances (N = 20), while Spain reported a single isolate (S. Derby) from cattle under 1 year of age, resistant to cefotaxime and susceptible to ceftazidime (N = 20) (Annex A, Table 24).
Overall combined resistance to ciprofloxacin and cefotaxime in Salmonella isolates from broilers, laying hens and turkey flocks in 2022 was at very low or low levels (broilers = 1.4%, median = 0%; laying hens = 0.2%, median = 0%; and turkeys = 2.2%, median = 0%), with exceptions noted only in two countries (Annex A). Specifically, when considering broiler flocks, Italy (11.1%, N = 190) and Malta (19.1%, N = 21) reported a moderate level of combined microbiological resistance to ciprofloxacin and cefotaxime. While for laying hens and fattening turkeys, Italy was the only MS reporting a low level of combined resistance (1.2%, N = 174, and 8.2%, N = 184, respectively), with all other countries reporting no resistance (Annex A, Tables 21, 22).
In 2021, overall combined resistance to ciprofloxacin and cefotaxime in Salmonella isolates from pigs and cattle under 1 year of age was also found at very low or low levels (pigs = 0.6%, median = 0%, cattle under 1 year of age = 2.5%, median = 0%). Although most countries reported no combined resistance to ciprofloxacin and cefotaxime, exceptions were noted in four countries (Annex A, Tables 23, 24). Specifically, when considering pig data, two isolates from Hungary (2.5%, N = 80) and five isolates from Romania (4.8%, N = 104) had combined microbiological resistance to ciprofloxacin and cefotaxime. For cattle under 1 year of age, a single isolate each from Italy (N = 20) and Spain (N = 20) was detected with combined resistance to both substances (Annex A, Tables 24).
In 2022, resistance to azithromycin in Salmonella isolates from broilers (0.5%, median = 0%) and turkeys (0.4%, median = 0%) was reported at a very low level, whereas for laying hen flocks, no resistance was detected (Annex A, Tables 20–22). Germany reported a high level of resistance in isolates from broilers (25.0%) but from a very small number of isolates tested (N = 8). Similarly, in 2021, overall resistance to azithromycin in Salmonella isolates from pigs was low (1.9%, median = 0%), while for cattle under 1 year of age, no resistance was detected (Annex A, Tables 23, 24). Only two countries, Portugal (21.1%, n = 4) and Slovenia (22.2%, n = 4), reported a high level of resistance in isolates from pigs but from a very small number of isolates tested (Portugal [N = 19] and Slovenia [N = 18]).
For amikacin, the new substance included in 2021, in the harmonised panel of antimicrobials tested, resistance was very low for all animal populations included in both monitoring years, except for cattle under 1 year of age for which no resistance was reported in 2021 (Annex A, Tables 20–24). Low levels of resistance to amikacin were reported in broilers (0.7%, median = 0%), only by four MSs, with Portugal, Austria, the Netherlands and Hungary reporting two (10.0%, N = 20); nine (5.3%, N = 170), one (0.8%; N = 123) and one (0.6%; N = 170) resistant isolates, respectively. In laying hens, several MSs reported low levels of resistance to amikacin (Portugal, 6.7%, N = 15; Cyprus, 4.8%, N = 21; Austria, 4.4%, N = 45; Malta, 4.0%, N = 25; and Germany, 1.7%, N = 58 and). Resistance to amikacin was reported in turkeys (0.6%, median = 0%) in only four MSs (Annex A, Table 22). While in 2021, resistance to amikacin in pigs (0.6%, median = 0%) was reported for one single isolate each from Austria (14.3%, N = 7), Germany (3.3%, N = 30), Romania (1.0%, N = 104) and Spain (0.6%, N = 170) and three isolates from the Netherlands (4.8%, N = 63) (Annex A, Table 24).
Regarding tigecycline, the new ECOFF (MIC > 0.5 mg/L), laid down in the current legislation, was used for the analysis of the AMR data. Considering all MSs, overall resistance to tigecycline in 2022 was reported at a high level in broilers (25.0%, median = 8.8%), at a moderate level in turkeys (18.4%, median = 0%) and at a low level in laying hens (2.9%, median = 0%). While in 2021, resistance to tigecycline was reported at a low level for pigs (7.0%, median = 4.4%) and at a moderate level for cattle under 1 year of age (10.1%, median = 0%). Lowering the tigecycline ECOFF from MIC > 1 mg/L to MIC > 0.5 mg/L may have contributed in part to a higher than expected level of resistance in Salmonella isolates from broilers, turkeys, pigs and cattle under 1 year of age. Indeed, many of these isolates were reported with MICs within one dilution range of the new ECOFF. For instance, 409 out of 478 (85.6%), 84 out of 126 (66.7%), 19 out of 26 (73.1%) and 73 out of 88 (83.0%) tigecycline‐resistant isolates from broilers, turkeys, laying hens and pigs, respectively, had a MIC of 1 mg/L. If considering the previous legislation, these isolates would be categorised as susceptible to tigecycline. Furthermore, the instability of tigecycline in the Mueller‐Hinton broth medium used in MIC‐testing can lead to inconsistencies in MIC values (Bradford et al., 2005).
In 2022, overall resistance to colistin in Salmonella isolates from broilers (3.5%, median = 0%) and laying hens (5.4%, median = 0%) was low, and very low in isolates from fattening turkeys (0.3%, median = 0%; Annex A, Tables 20–22). While in 2021, overall resistance to colistin in Salmonella isolates from pigs was low (1.0%, median = 0%) but moderate for cattle under 1 year of age (11.4%, median = 7.5%) (Annex A, Tables 23, 24). The small overall sample size of Salmonella isolates from cattle under 1 year of age (N = 79) compared to the other animal groups (i.e. broilers, N = 1911; laying hens N = 908; turkeys, N = 686; and pigs, N = 1258) should be taken into consideration when interpreting these results. Across all animal populations, most individual countries reported no resistance or very low levels of resistance to colistin. However, some exceptions occurred where MSs reporting > 10 isolates reported, showed moderate to high levels of resistance to colistin. These countries included Bulgaria (26.7%, N = 15) and Poland (19.2%, N = 167) for broiler flocks, and Cyprus (23.8%, N = 21), Czechia (16.7%, N = 12), the Netherlands (26.9%, N = 26) and Poland (17.3%, N = 81) for laying hen flocks (Annex A, Tables 20, 21).
2.4.4. Tigecycline and colistin resistance in Salmonella serovars
Tigecycline resistance in Salmonella spp.
Mechanisms of tigecycline resistance
Tigecycline is categorised as a CIA by the WHO (WHO, 2019) and is considered a last resort antimicrobial for the treatment of serious infections in adults caused by MDR bacteria. Tigecycline is structurally related to the tetracycline class of antibiotics and is active against Gram‐positive and Gram‐negative bacteria, as well as tetracycline‐resistant bacteria and some anaerobes (Yaghoubi et al., 2022).
Resistance mechanisms to tigecycline include non‐mobile tet(X) and mobile‐plasmid‐mediated transmissible tet(X) and resistance–nodulation–division (RND) efflux pump mediated tmexCD‐toprJ genes (Anyanwu et al., 2022). The transferable plasmid‐mediated spread of tigecycline resistance genes, is of the highest concern, such as tet(X3) and tet(X4) Which confer high levels of resistance to all tetracyclines, including tigecycline (MICs of ≥ 32 mg/L). Two recent studies investigating the global distribution, evolution pattern and spread of tet(X) genes, identified isolates carrying tet(X) genes from over 20 countries across five continents (Pan et al., 2020; Wang et al., 2021).
The first report of transferable high‐level tigecycline (HLT) resistance by tet(X3) and tet(X4) genes in Enterobacteriaceae from food animals, meat and the environment came from China in 2019 (He et al., 2019). Another study from China in the same year identified tet(X4) MDR E. coli isolates from retail pork samples (Bai et al., 2019). The tet(X4) gene in these isolates was located on several conjugative plasmids of different replicon types, indicating that the gene may be captured by a range of mobile genetic elements circulating among bacterial strains. Since then, additional plasmid‐mediated tet(X) genes, including tet(X5) and tet(X6), have been identified in over 10 different Gram‐negative species, although rarely in Salmonella spp. (Wang et al., 2021).
While tigecycline is not used in food‐producing animals, it is postulated that the excessive use of tetracyclines in food‐producing animals may contribute to the emergence of plasmid‐mediated tet(X) genes, with the potential to spread to human bacterial species (Anyanwu et al., 2022; Pan et al., 2020). The potential for other bacteria within the Enterobacteriaceae family (such as Salmonella) to acquire such transferable tigecycline resistance genes is highlighted, and the importance of monitoring tigecycline resistance through the determination of MICs or by molecular investigation such as WGS is further underlined.
The number and percentage of tigecycline‐resistant Salmonella isolates detected from poultry flocks, pigs and cattle under 1 year of age and the predominant serovars accounting for this resistance, by MSs, are shown in Figure 6. Particular serovars displayed microbiological resistance to tigecycline, suggesting a clonal expansion of microbiologically resistant strains of these serovars.
FIGURE 6.
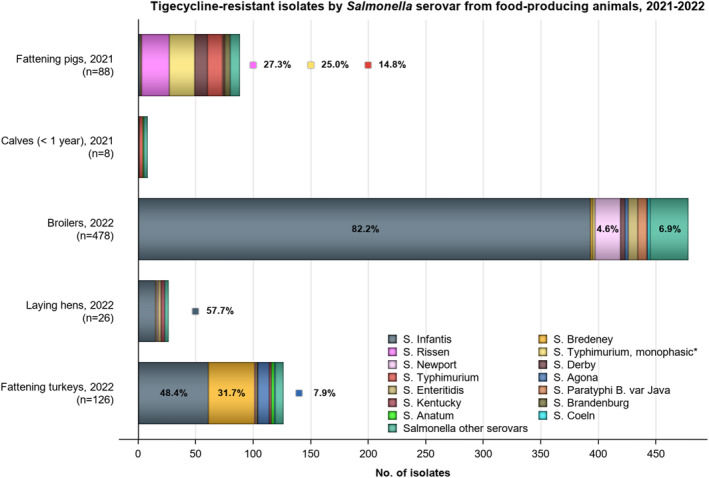
Breakdown of the number of tigecycline‐resistant Salmonella isolates by serovar from broilers, fattening turkeys, laying hens in 2022 and fattening pigs and cattle under year old (calves) in 2021, using harmonised ECOFFs.
Note: n, Total number of tigecycline‐resistant isolates reported by MSs; predominant serovars are also expressed as a percentage; *Monophasic S. Typhimurium includes all antigenic formulas; salmonellas in the legend are listed according to their predominance within all the animal/origins. The ECOFF used to determine tigecycline resistance was MIC > 0.5 mg/L.
In 2022, serovar Infantis accounted for most of the tigecycline‐resistant isolates recovered from broilers (82.2%, n = 393), laying hens (57.7%, n = 15) and turkeys (48.4%, n = 61). In laying hens, the second most predominat serovar among the tigecycline‐resistant Salmonella isolates were S. Enteritidis and S. Kentucky, each 11.5% (n = 3), and in Turkeys was S. Bredeney 31.7% (n = 40) followed by S. Agona, 7.9% (n = 10) (Figure 6).
In 2021, S. Rissen (27.3%, n = 24), monophasic S. Typhimurium (25.0%, n = 22), S. Typhimurium (14.8%, n = 13) and S. Derby (12.5%, n = 11) accounted for most of the resistant isolates recovered from pigs. For cattle under 1 year of age, of the eight Salmonella isolates resistant to tigecycline, three were serotyped as S. Typhimurium and the remaining were S. Infantis, S. Dublin, S. Anatum, S. Agama and S. Muenster, one each (Figure 6).
Considering individual countries, the highest levels of tigecycline resistance in Salmonella isolates from broilers were reported by Hungary (54.1%, n = 92), Slovenia (50.0%, n = 44), Poland (46.7%, n = 78) and Romania (40.6%, n = 69), while the highest levels of tigecycline in Salmonella isolates recovered from turkeys were reported by Hungary (42.9%, n = 73) and Italy (21.2%, n = 39). The reporting countries responsible for most of the tigecycline‐resistant isolates in pigs (n = 88) were Ireland (14.6%), Spain (10.0%) and Romania (13.5%). While in cattle under 1 year of age, Italy reported four of the eight tigecycline‐resistant isolates (Annex A, Table 24).
Among certain serovars within the different food‐producing animal populations, MDR was often a feature where tigecycline resistance was reported. For instance, among broilers, of all tigecycline‐resistant S. Infantis isolates 97.5% were multidrug‐resistant, with resistance to ciprofloxacin, nalidixic acid, sulfamethoxazole and tetracycline (CIP‐NAL‐SUL‐TET) being a common feature of all these isolates. This is a resistance pattern typical of some MDR broiler clones of S. Infantis (Alba et al., 2020; Alvarez et al., 2023; Nógrády et al., 2012). For laying hens, 20 tigecycline‐resistant isolates (76.9%, n = 26) were MDR, with a similar resistance pattern to that described above for broilers.
Among turkeys, of all the tigecycline‐resistant isolates, 98.4% (124 out of 126) were multidrug‐resistant, with ampicillin, nalidixic acid, ciprofloxacin and tetracycline resistance being a feature. For pigs, MDR was present in 16 of the 24 tigecycline‐resistant S. Rissen isolates, with most having ampicillin, nalidixic acid, sulfamethoxazole and tetracycline resistance. For cattle under 1 year of age, all tigecycline‐resistant S. Typhimurium isolates (n = 3) were MDR, showing resistance to sulfamethoxazole and tetracycline.
Colistin resistance in Salmonella spp.
Mechanisms of colistin resistance
Colistin belongs to the polymyxin antimicrobial class and is considered a highest priority CIA (hpCIA) and a last resort antimicrobial for treating serious human infections caused by several Gram‐negative bacteria (WHO, 2019). Although not frequently used in human medicine due to its nephrotoxic effects, colistin has been widely used in veterinary medicine for prophylactic/metaphylactic treatment (Kieffer et al., 2017). Several mechanisms of polymyxin resistance in Gram‐negative bacteria have been described (lipopolysaccharide modifications, efflux pumps, capsule formation and overexpression of membrane protein, Hamel et al., 2021). Furthermore, transferable mobile colistin resistance (mcr) genes have also been detected in Salmonella isolates (Portes et al., 2022).
Group D Salmonella serovars
Among Salmonella isolates recovered from poultry in 2022, resistance to colistin (i.e. MIC > 2 mg/L) was generally observed in S. Enteritidis isolates. This serovar accounted for 86.6% and 87.8% of the colistin‐resistant isolates recovered from broilers and laying hens, respectively (Figure 7). Only two colistin‐resistant isolates were reported from turkeys one of them S. Napoli. In 2021, of the 12 Salmonella isolates from pigs with resistance to colistin, 41.7% were serotyped as S. Enteritidis (n = 5), three isolates were S. Panama (25%) and one isolate was S. Dublin. All nine colistin‐resistant isolates from cattle under 1 year of age were serotyped as S. Dublin.
FIGURE 7.
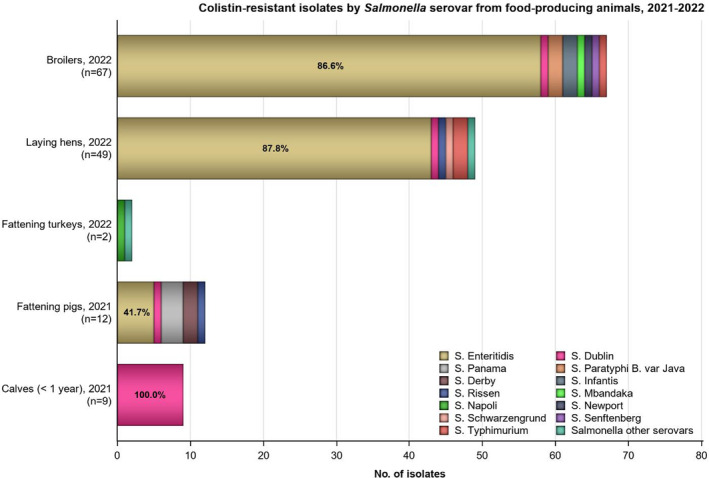
Breakdown of the number of colistin‐resistant Salmonella isolates by serovar, where detected among the animal origins by reporting MSs in 2021/2022.
Note: n, Total number of colistin‐resistant isolates reported by the MSs; predominant serovars are expressed as a percentage. Salmonellas in the legend are listed according to their predominance within all the animal origins.
Notably, S. Enteritidis, S. Dublin and S. Panama are group D salmonellas (serogroup O9). Salmonella belonging to group D tend to show a decreased susceptibility to colistin without having any known acquired or mutational colistin resistance mechanisms (Agersø et al., 2012; Ricci et al., 2020). This is exemplified by the proportion of colistin‐resistant isolates belonging to S. Enteritidis and S. Dublin in 2022 and 2021, respectively. Figure 7 presents the number and percentage of colistin‐resistant isolates detected from the different food‐producing animal species reported by MSs and the predominant serovars accounting for this resistance. Together with S. Enteritis and S. Dublin, the serovars Napoli and Panama are group D salmonella serovars. The remaining serovars listed do not belong to group D (serogroup O9).
Other Salmonella serovars
Among the colistin‐resistant isolates from broilers, S. Paratyphi B and S. Infantis (3.0%, n = 2, each) were the most reported serovars that did not belong to group D Salmonella. The other serovars reported, one each, were S. Mbandaka, S. Newport, S. Senftenberg and S. Typhimurium. While for laying hens, four isolates (8.2%) were not group D salmonellas. Namely, two S. Typhimurium isolates (S. 1,4,5,12:‐:1,2), S. Rissen and S. Schwarzengrund. In 2021, two of the Salmonella isolates recovered from pigs were serotyped as S. Derby (16.7%) and another as S. Rissen (8.3%). Across all poultry data, several MSs reported some colistin‐resistant Salmonella isolates as ‘Other serovars’.
A very small number of isolates reported markedly elevated colistin MICs (i.e. MIC ≥ 16 mg/L), including one S. Enteritidis isolate from laying hens in 2022, and two isolates from pigs in 2021: one S. Rissen and one S. Derby.
2.4.5. Complete susceptibility (CS) and multidrug resistance (MDR)
For the analysis of the AMR data from Salmonella spp. isolates, the new antimicrobial substance amikacin included in the harmonised panel for the monitoring of AMR according to the Commission Implementing Decision (EU) 2020/1729 was included in the assessment of CS and MDR (see Appendix F). This list of antimicrobials included in the CS and MDR analysis are the following: amikacin/gentamicin (aminoglycosides), ampicillin, cefotaxime/ceftazidime (third‐generation cephalosporins), chloramphenicol, ciprofloxacin/nalidixic acid ((fluoro)quinolones), meropenem, sulfamethoxazole, tetracycline/tigecycline (glycylcyclines) and trimethoprim. CS is defined as complete susceptibility to the selected antimicrobial substances listed above. MDR is defined as resistance to three or more antimicrobial classes listed above. Data from countries submitting less than 10 Salmonella isolates from the different animal origins are excluded from some of the analyses described in this section.
Food‐producing animals
The levels of CS and MDR among Salmonella isolates recovered from food‐producing animals are shown in Figure 8 In this figure, only MSs with 10 or more isolates tested are included in the analysis. Annex A includes Tables with overall and individual country MDR and CS. Overall MDR at MS level was observed at high levels in isolates from broilers (43.6%, median = 33.3%), turkeys (39.4%, median = 21.2%), pigs (39.1%, median = 35.1%) and cattle under 1 year of age (30.4%, median = 17.7%), and at a low level in isolates from laying hens (7.5%, median = 2.7%). Few countries reported zero MDR in any of the food‐producing animal populations. Across all MSs submitting data from broilers in 2022 extremely high levels of MDR were reported by Cyprus (92.9%, N = 14) and Austria (72.9%, N = 170). MDR in isolates from turkeys was reported at very high levels by Hungary (69.4%, N = 170), Poland (66.7%, N = 6) and Romania (57.1%, N = 7). When considering laying hens, there were 12 countries reporting zero MDR isolates. In 2021, MDR in isolates from pigs was reported at extremely high levels by Portugal (78.9%, N = 19), Croatia (74.2%, N = 31), Slovenia (72.2%, N = 18) and Austria (71.4%, N = 7) (Annex A, Tables 23, 24).
FIGURE 8.
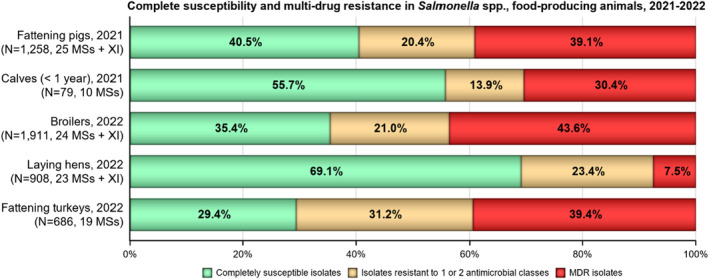
MDR and CS Salmonella spp. isolates recovered from broilers, laying hens, fattening turkeys, fattening pigs and cattle under 1 year of age (calves) for all reporting MSs, 2021–2022, using harmonised EUCAST ECOFFs.
Note: The MDR analysis of animal isolates included the following antimicrobials: amikacin/gentamicin, ampicillin, azithromycin, cefotaxime/ceftazidime, chloramphenicol, ciprofloxacin/nalidixic acid, meropenem, sulfamethoxazole, tetracycline/tigecycline and trimethoprim. MDR and complete susceptibility are expressed as percentages; N, total number of Salmonella spp. reported by MSs. Only MSs with 10 or more isolates are included in the MDR analysis. The ECOFFs used to determine microbiological resistance are from the current legislation (2020/1729/EU).
Overall CS at the MS level was observed at high levels for broilers (35.4%, median = 47.6%), turkeys (29.4%. median = 41.2%) and pigs (40.5%, median = 42.7%) and very high levels for laying hens (69.1%, median = 82.6%) and cattle under 1 year of age (55.7%, median = 50.0%) (Annex A, Tables 20–24). However, the levels of CS varied widely among reporting countries, particularly in broiler, turkey and pig populations. For 2022 broilers' data across all MSs, both Finland and Sweden reported the highest level of CS (100%, N = 2 and N = 7, respectively), followed closely by France (93.5%, N = 168), while Cyprus (0%, N = 14), Hungary (5.3%, N = 170) and Slovakia (10.5%, N = 38) reported the lowest percentage of CS in Salmonella isolates (Annex A, Table 20). For turkeys, five countries reported 100% CS, but from a very low number of isolates (Belgium, N = 1; Denmark, N = 2; Estonia, N = 1; Germany, N = 3; and Slovakia, N = 1). France also reported an extremely high level of CS (77.3%, N = 66). While Cyprus (0.0%, N = 1), Czechia (0.0%, N = 2) and Slovenia (0.0%, N = 7) reported the lowest percentage of CS in Salmonella isolates. For laying hens, the median CS was extremely high at 82.6%, with almost every country reporting CS above 60% except for Italy (28.7%, N = 174), Belgium (37.5%, N = 16), Luxembourg (50%, N = 2) and Poland (56.8%, N = 81). While for data from 2021, for pigs, the median CS was 42.7% with Latvia (92.9%, N = 42), Luxembourg (75.7%, N = 37) and Greece (70.0%, N = 10), reporting the highest levels of CS. For cattle under 1 year of age, the median CS was 50.0%; however, only three countries submitted AMR data on more than 10 isolates.
The spatial distribution of CS across all reporting countries can be seen in Figure 9. The proportions of isolates that were completely susceptible and multidrug resistant among particular Salmonella serovars within the different food‐producing animal species are presented in Appendix B – Occurrence of antimicrobial resistance at the Salmonella serovar level.
FIGURE 9.
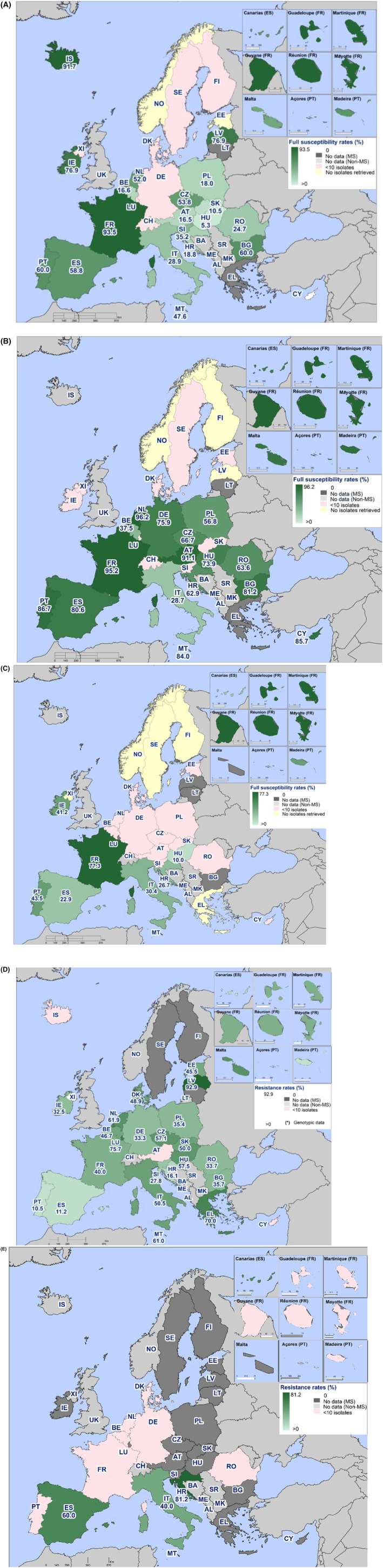
Spatial distributions of complete susceptibility to the selected antimicrobials tested among Salmonella spp. from (A) broilers, (B) laying hens and (C) fattening turkeys, (D) fattening pigs and (E) cattle under 1 year of age using harmonised ECOFFs, 2021–2022.
2.4.6. High‐level resistance to ciprofloxacin (CIP) in Salmonella spp.
The distribution of ciprofloxacin‐resistant isolates displaying levels of microbiological resistance or high‐level resistance to ciprofloxacin within each of the food‐producing animal species is illustrated in Figure 10. Notably, the distribution of MICs provided is only for ciprofloxacin‐resistant isolates and the total number of Salmonella isolates ciprofloxacin resistant is also provided in the figure (n). High‐level resistance to ciprofloxacin is considered in isolates with an MIC ≥ 4 mg/L. In 2021, according to the new implementing decision 2020/1729 the ciprofloxacin CBP was changed from MIC > 1 mg/L to MIC > 0.06 mg/L, which aligns with the EUCAST ECOFF (MIC > 0.06 mg/L), and therefore unlike in the previous reports, it is no longer necessary to distinguish between clinical and microbiological ciprofloxacin resistance.
FIGURE 10.
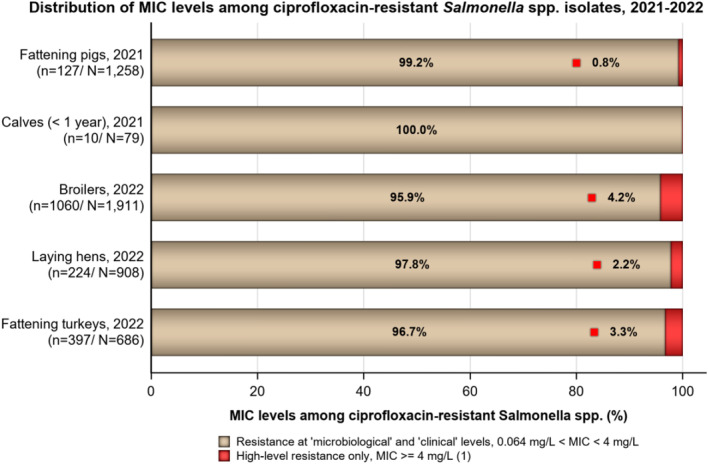
Distribution of MIC levels among ciprofloxacin‐resistant Salmonella spp. from broilers, laying hens, fattening turkeys, fattening pigs and cattle under 1 year of age (calves), for all reporting EU MSs, 2021–2022.
Note: N, Total number of Salmonella isolates tested; n, number of ciprofloxacin resistant Salmonella isolates.
Among the Salmonella isolates displaying ciprofloxacin resistance, 4.2% of the isolates from broilers (n = 44, of which 15 S. Kentucky), 2.2% of the isolates from laying hens (n = 5, one S. Kentucky) and 3.3% of the isolates from turkeys (n = 13, all S. Kentucky), exhibited MICs of ≥ 4 mg/L (Figure 10). In 2021, 127 Salmonella isolates from pigs, (10.1%, N = 1258) recorded resistance to ciprofloxacin, with just a single isolate (S. Kentucky) from Malta displaying high‐level resistance to ciprofloxacin (and nalidixic acid). For cattle under 1 year of age, 10 Salmonella isolates (12.7%; N = 79) were resistant to ciprofloxacin, with none displaying high‐level resistance (Figure 10).
The serovars displaying high‐level resistance to fluoroquinolones are of interest from both the epidemiological and the public/animal health perspectives. S. Kentucky was the most reported serovar exhibiting high‐level ciprofloxacin resistance in poultry populations. A detailed analysis of the high‐level resistance to ciprofloxacin in S. Kentucky and other Salmonella serovars is presented in Appendix A – High‐level resistance to ciprofloxacin among certain Salmonella serovars.
2.4.7. Phenotypic characterisation of third‐generation cephalosporin and carbapenem resistance in Salmonella spp.
According to Decision 2020/1729/EU, any Salmonella isolate from food‐producing animals or imported fresh meat showing resistance to cefotaxime or ceftazidime or meropenem (i.e. presumptive ESBL‐/AmpC‐/CP‐producing Salmonella isolates) should be further tested with a second panel of harmonised antimicrobial substances to confirm the phenotypic resistance to third‐generation cephalosporins or carbapenems.
The proportion of presumptive ESC‐resistant Salmonella spp. at the MS level collected within the routine monitoring based on phenotypic results was generally very low or low in 2021 and 2022 (ranging between 0% and 2.5%, Annex A, Tables 20–24). The occurrence of ESC‐resistant Salmonella in a specific animal population may be greatly influenced by the prevalence of different Salmonella serovars in some countries. At the reporting MS‐group level, the occurrence of presumptive ESBL‐/AmpC‐producing Salmonella was 1.4% in broilers, 2.2% in fattening turkeys, 0.2% in laying hens, 0.9% in fattening pigs and 2.5% in cattle under 1 year of age. Detailed data per country and matrix are presented in Annexes D2 and D3. An overview of presumptive ESBL‐/AmpC‐/CP‐producing Salmonella spp. reported in 2021 and 2022 is given in Tables 5, 6.
TABLE 5.
Summary of presumptive ESBL‐/AmpC‐/CP‐producing Salmonella spp. from food‐producing animals collected within the routine monitoring by serovar, 2021–2022.
| Year | Matrix | Serotype | n | ESBL a | AmpC b | ESBL + AmpC c |
|---|---|---|---|---|---|---|
| 2021 | Cattle under 1 year of age (n = 2) | S. Derby | 1 | – | – | 1 |
| S. Ngor | 1 | 1 | – | – | ||
| Fattening pigs (n = 11) | S. Derby | 2 | 1 | – | 1 | |
| S. Infantis | 2 | 2 | – | – | ||
| S. London | 1 | 1 | – | – | ||
| S. Kedougou | 1 | 1 | – | – | ||
| S. Rissen | 4 | – | – | 4 | ||
| S. Typhimurium, monophasic | 1 | 1 | – | – | ||
| 2022 | Broilers (n = 26) | S. Infantis | 18 | 18 | – | – |
| S. Mbandaka | 2 | 2 | – | – | ||
| S. Kentucky | 4 | 4 | – | – | ||
| S. Blockley | 1 | 1 | – | – | ||
| S. Senftenberg | 1 | 1 | – | – | ||
| Fattening turkeys (n = 15) | S. Infantis | 15 | 15 | – | – | |
| Laying hens (n = 2) | S. Infantis | 2 | 2 | – | – |
Abbreviations: n, number of presumptive ESBL‐ and /or AmpC‐ producing isolates, ESBL, extended spectrum b‐lactamase, AmpC, AmpC beta lactamase.
All isolates showing clavulanate synergy with CTX or CAZ or both, suggesting ESBL phenotype, or reported presence of ESBL‐encoding gene.
Isolates with cefoxitin resistance, suggesting AmpC phenotype, or reported presence of AmpC‐encoding gene.
Isolates showing synergy with CTX or CAZ and cefoxitin resistance, suggesting ESBL‐ and AmpC‐enzymes in the same isolates, or both ESBL‐ and AmpC‐encoding genes reported.
TABLE 6.
Summary of the presumptive ESBL‐, AmpC‐ or CP‐producing Salmonella spp. from humans and food‐producing animals, subjected to supplementary testing (panel 2) or whole genome sequencing, EU MSs, 2021–2022.
| Matrix | ESBL and/or AmpC a | ESBL b | AmpC c | ESBL + AmpC d | CP e |
|---|---|---|---|---|---|
| n (% R) | n (% R) | n (% R) | n (% R) | n (% R) | |
| Humans 2021 (N = 9787, 14 MSs) | 88 (0.9) | 76 (0.8) | 12 (0.1) | 0 (0) | 0 (0) |
| Humans 2022 (N = 14,058, 26 MSs) | 150 (1.1) | 122 (0.9) | 24 (0.2) | 4 (< 0.1) | 4 (< 0.1) |
| Fattening pigs, 2021 (N = 1258, 25 MSs + XI) | 11 (0.9) | 9 (0.7) | 0 (0) | 2 (0.2) | 0 (0) |
| Calves, 2021 (N = 79, 10 MSs) | 2 (2.5) | 1 (1.3) | 0 (0) | 1 (1.3) | 0 (0) |
| Broilers, 2022 (N = 1911, 24 MSs + XI) | 26 (1.4) | 26 (1.4) | 0 (0) | 0 (0) | 0 (0) |
| Fattening turkeys, 2022 (N = 686, 19 MSs) | 15 (2.2) | 15 (2.2) | 0 (0) | 0 (0) | 0 (0) |
| Laying hens, 2022 (N = 908, 23 MSs + XI) | 2 (0.2) | 2 (0.2) | 0 (0) | 0 (0) | 0 (0) |
Abbreviations: AmpC, AmpC beta‐lactamase; CP, carbapenemase; ESBL, extended‐spectrum beta‐lactamase; N, total of isolates reported; n, number of isolates with the phenotype; %R, percentage of isolates resistant.
According to EUCAST guidelines (EUCAST, 2017), only isolates showing MIC > 1 mg/L for CTX and/or CAZ or reported presence of ESBL‐/AmpC‐encoding gene were considered (see Appendix F ).
All isolates showing clavulanate synergy with CTX or CAZ or both, suggesting ESBL phenotype, or reported presence of ESBL‐encoding gene.
Isolates with cefoxitin resistance, suggesting AmpC phenotype, or reported presence of AmpC‐encoding gene.
Isolates showing synergy with CTX or CAZ and cefoxitin resistance, suggesting ESBL‐ and AmpC‐ enzymes in the same isolates, or both ESBL‐ and AmpC‐encoding genes reported.
Isolates with meropenem resistance or CP‐encoding gene reported.
WGS data for Salmonella spp. were reported by three MSs (Germany, Italy and the Netherlands) in 2022. Italy reported bla CTX‐M‐1 in 18 S. Infantis, one S. Blockley, one S. Mbandaka and one S. Senftenberg isolated from broiler flocks, 15 S. Infantis isolates from fattening turkeys and two S. Infantis from laying hens. Germany and the Netherlands reported WGS data for imported meat sampled at BCPs (see specific Textbox regarding Monitoring AMR in imported fresh meat sampled at BCP).
In 2021, one MS (Italy) reported WGS for Salmonella spp. Italy reported one monophasic S. Typhimurium isolated from fattening pigs that carried bla SHV‐12 , and one S. Ngor from cattle under 1 year of age harbouring bla CTX‐M‐3 .
2.4.8. Carbapenem resistance in Salmonella spp. from food‐producing animals
Carbapenems are recognised as hpCIAs (WHO, 2019). This antimicrobial class includes meropenem, an antimicrobial agent specified in the antimicrobial panel for monitoring and reporting AMR in Salmonella spp. as stipulated by the Commission Implementing Decision (EU) 2020/1729. This class of antimicrobials is not therapeutically used in food‐producing animals but is reserved for use in humans.
In 2022 and 2021, none of the Salmonella isolates recovered from any of the animal populations or imported meat exhibited microbiological resistance to meropenem. This is consistent with data from animal or carcase origins in 2020 and 2021.
2.4.9. Resistance exhibited by dominant Salmonella serovars
The detailed reporting of results at the serovar level highlights the contribution of a few serovars to the overall occurrence of resistance when considering aggregated data for Salmonella spp. The resistance patterns associated with these different serovars have a marked influence on the overall resistance levels in Salmonella spp., as the proportion of completely susceptible and multidrug‐resistant isolates may vary significantly among serovars recovered from each of the studied food‐producing animal populations and derived meat thereof. The analysis of antimicrobial resistance at the serovar level is presented in Appendix B .
2.5. Comparison of resistance data in Salmonella from human and food‐producing animals
The prevalence of particular Salmonella serovars within countries and animal populations, and their associated patterns of resistance, may explain some of the observed differences in the occurrence of AMR and MDR. Indeed, the spread of resistant clones and the presence of resistance genes within these clones can be exacerbated by selective pressure from using antimicrobials in human and animal populations. However, it should be noted that relating the occurrence of AMR in human Salmonella isolates to that in isolates from food‐producing animals and extrapolating directionality or causality should be done with caution because other sources of Salmonella occur, and such evaluations should be performed and interpreted considering the complex epidemiology of salmonellosis.
Levels of CS and MDR
The occurrence of MDR, CS and resistance to one or two antimicrobial classes across humans in 2022 and food‐producing animals for the years 2021–2022 in Salmonella spp. isolates and particular serovars: S. Typhimurium and its monophasic variant, S. Infantis, S. Enteritidis and S. Kentucky is summarised in Figure 11.
FIGURE 11.
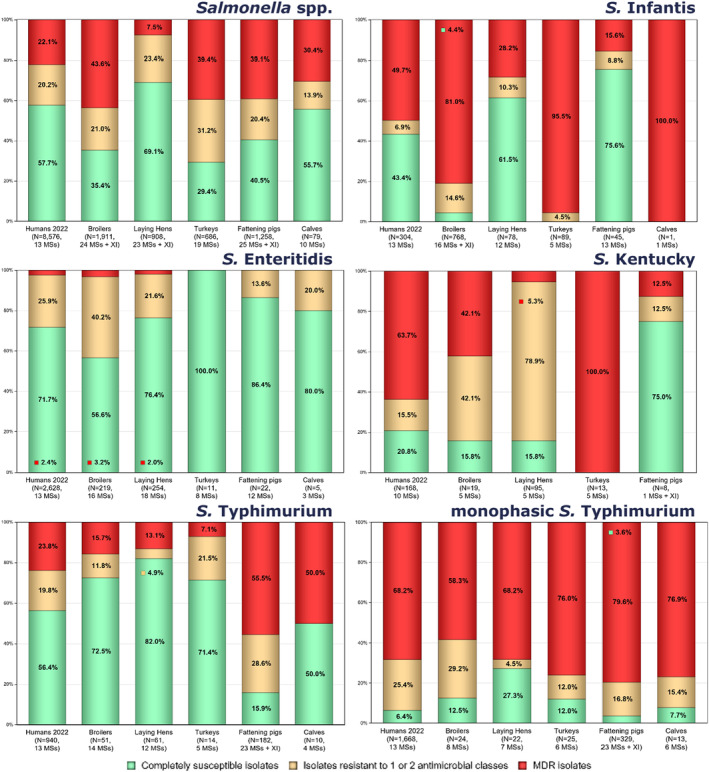
MDR and CS Salmonella spp. from selected serovars recovered from humans in 2022, broiler, turkey and laying hen flocks in 2022, and fattening pigs and cattle under 1 year of age (calves) in 2021.
Note: The MDR analysis of animal isolates included the following antimicrobials: amikacin/gentamicin, ampicillin, azithromycin, cefotaxime/ceftazidime, chloramphenicol, ciprofloxacin/nalidixic acid, meropenem, sulfamethoxazole, tetracycline/tigecycline and trimethoprim. MDR and complete susceptibility are expressed as percentages; N: total number of Salmonella isolates reported by MSs. Only MSs with 10 or more isolates are included in the MDR analysis. The ECOFFs used to determine microbiological resistance are from the current legislation (2020/1729/EU).
In 2022, MDR was overall high (22.1%) among Salmonella spp. reported in human cases in the EU, ranging from low levels among S. Enteritidis (2.4%, N = 2628) to very high among S. Kentucky (63.7%, N = 168) and monophasic S. Typhimurium 1,4,[5],12:i:‐ (68.2%, N = 1668). S. Typhimurium and S. Infantis exhibited high MDR levels (23.8%, N = 940 and 49.7%, N = 304, respectively). Similarly, MDR was observed at high levels in Salmonella spp. recovered from broilers and turkeys (43.6% and 39.4%, respectively), pigs (39.1%) and cattle under 1 year of age (30.4%), while Salmonella spp. isolates from laying hens showed a markedly lower MDR level (7.5%).
In general, when compared to human data, Salmonella Enteritidis from poultry populations showed a similar occurrence of MDR. Namely, S. Enteritidis isolates from broilers (3.2%, N = 219) and laying hens (2.0%, N = 254) exhibited low MDR levels, while in turkeys (N = 11), as well as in pigs (N = 22) and cattle under 1 year of age (N = 5), no MDR was reported.
MDR in monophasic S. Typhimurium and S. Infantis isolates from broilers (58.3%, N = 24 and 81.0%, N = 768, respectively) and turkeys (76.0%, N = 25 and 95.5%, N = 89, respectively) was reported at very high to extremely high levels, showing generally higher MDR values than those serovars recovered from humans. Monophasic S. Typhimurium isolates recovered from pigs (79.6%, N = 329) and cattle under 1 year of age (76.9%, N = 13) also exhibited extremely high MDR levels. In contrast, MDR levels in S. Infantis from laying hens (28.2%, N = 78) and pigs (15.6%, N = 45) were lower than those in humans.
MDR levels in S. Typhimurium were reported in 23.8% (N = 940), 55.5% (N = 182) and 50.0% (N = 10) of the isolates from humans, pigs and cattle under 1 year of age, respectively. While in poultry populations MDR levels in S. Typhimurium were lower (broilers, 15.7%, N = 51; laying hens, 13.1%, N = 61; turkeys, 7.1%, N = 14).
Differences in the occurrence of MDR levels in S. Kentucky are evident. All S. Kentucky isolates from turkeys (N = 13) were MDR followed by very high levels in humans (63.7%, N = 168), while broilers (42.1%, N = 19) exhibited high MDR levels, and laying hens (5.3%, N = 95) showed low MDR levels. Only one out of the eight S. Kentucky isolates from pigs, was found MDR while no S. Kentucky isolates were found in cattle under 1 year of age.
Overall, in 2022, CS in Salmonella spp. isolates from humans was 57.7%. For animal data CS was high for broilers (35.4%), turkeys (29.4%) and pigs (40.5%) and at very high level in laying hens (69.1%) and in cattle under 1 year of age (55.7%).
Resistance to selected antimicrobials
Overall resistance to ampicillin, sulfonamides and tetracyclines was observed at high levels in Salmonella spp. isolates from humans in 2022 and ranged from moderate to very high in isolates from food‐producing animals, except in laying hens where low levels of resistance were reported (Figure 12). Ciprofloxacin resistance was observed at moderate levels in humans (18.7%), very high levels among isolates from broiler (55.5%) and fattening turkey flocks (57.9%) and at a high level in laying hens (24.7%) in 2022, while moderate levels were reported in Salmonella spp. isolates from fattening pigs (10.1%) and cattle under 1 year of age (12.7%) in 2021 (Figure 12). Overall resistance to cefotaxime was noted at very low levels in isolates from humans in 2022 (1.2%) and was seldomly detected in food‐producing animals in 2021–2022, except in broiler flocks (1.4%),turkey flocks (2.2%) and cattle under 1 year of age (2.6%, Figure 12).
FIGURE 12.
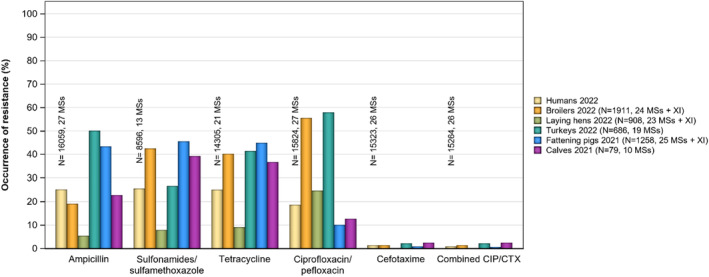
Occurrence of resistance to selected antimicrobials in Salmonella spp. from humans (2022) and animal populations (2021–2022), all reporting MSs.
The comparison of the occurrence of resistance to selected antimicrobials in different Salmonella serovars between humans and food‐producing animals in 2022 is presented in Appendix C .
Presumptive ESBL‐/AmpC‐CP‐producing Salmonella spp.
The overall proportion of presumptive ESBL‐/AmpC‐producing Salmonella spp. at MS level was generally very low or low in 2021 and 2022 among all food‐producing animal populations and very low in isolates from human cases (Table 6). In 2021 and 2022, no Salmonella spp. isolates recovered from animal origins were microbiologically resistant to meropenem. However, unlike in 2021 when no meropenem resistance was reported in Salmonella spp. isolates from humans, in 2022 the occurrence of meropenem resistance was rare (< 0.1%, Table 6), with three countries reporting resistant isolates.
2.6. Discussion
The continuous monitoring of AMR in Salmonella is essential to identify new and emerging resistance mechanisms in both human and animal populations. The harmonised monitoring of Salmonella from food‐producing animals and imported fresh meat using phenotypic antimicrobial susceptibility testing, aims to identify emerging risks. For instance, this monitoring helped identify MDR S. Kentucky with high resistance to fluoroquinolones and S. Infantis isolates exhibiting combined resistance to extended‐spectrum cephalosporins, fluoroquinolones and colistin (EFSA, 2019).
In 2021 and 2022, the detection of resistant Salmonella isolates varied markedly based on their animal origins, serovars and reporting countries. These factors may introduce a source of variability in the results when considering data from all reporting countries. Thus, results should be interpreted cautiously. Moreover, due to the changes in legislation that led to an alteration of the reporting requirements, the comparison of AMR data reported for Salmonella isolates from poultry, and pigs and cattle under 1 year of age in 2022/2020 and 2021/2019, respectively, should be made with caution. Additionally, the changes in the AMR monitoring and the limited number of Salmonella isolates reported by some countries at serovar level is insufficient for conducting a comprehensive statistical trend analysis of the occurrence of resistance at the Salmonella serovar level.
Regarding data from humans, resistance predicted from WGS has been an accepted test method since 2019. WGS is increasingly recognised as a powerful tool for epidemiological surveillance of AMR in the ‘One Health’ context (WHO, 2020). It complements phenotypic methods by providing information on molecular determinants and mechanisms, genetic factors that facilitate transmission and geographical distributions of resistance genes. Several studies on Enterobacterales, including Salmonella, have reported a high concordance rate between WGS data and phenotypic data (Hendriksen et al., 2019; McDermott et al., 2016).
Occurrence of resistance to commonly used antimicrobials in veterinary medicine
In 2022, both broiler and fattening turkey flocks exhibited high levels of resistance to tetracycline and sulfonamides. On the other hand, laying hen flocks displayed comparatively lower resistance levels to both antimicrobials. Resistance to ampicillin was reported at lower levels in broiler and laying hen flocks. In contrast, turkey flocks showed very high resistance to ampicillin. In 2021, a similar pattern of resistance was observed in Salmonella isolates recovered from pigs and cattle under 1 year of age, with high resistance to tetracyclines, sulfonamides and ampicillin.
Occurrence of resistance to highest priority critically important antimicrobials (hpCIAs) and last resort antimicrobials
Third‐generation cephalosporins and fluoroquinolones are categorised as hpCIAs because they are commonly used in treatment of gastrointestinal infections, including Salmonella infections, in humans (WHO, 2019). This sets the rationale for monitoring combined resistance to these antimicrobial classes within food‐producing animals.
From the monitoring of food‐producing animals in 2022 and 2021, the overall resistance levels to azithromycin ranged from zero, with no resistance observed in Salmonella isolates from laying hen flocks and cattle under 1 year of age, to very low in turkeys and broilers and low in pigs. From the monitoring of all poultry populations, the highest levels of resistance were generally noted to ciprofloxacin/nalidixic acid, while moderate levels of resistance to ciprofloxacin/nalidixic acid were observed in isolates from pigs and cattle under 1 year of age. Among Salmonella isolates recovered from broiler and fattening turkey flocks, S. Infantis exhibited the highest levels of resistance to ciprofloxacin and nalidixic acid, whereas in isolates from laying hens, this was seen in S. Kentucky. This likely reflects the spread of resistant clones belonging to these serovars. From human data reported in 2022, S. Infantis and S. Kentucky also showed the highest resistance to these substances. Resistance to ciprofloxacin/nalidixic acid, sulfamethoxazole and tetracycline are typical of a clone of S. Infantis prevalent in Europe in broilers (Alba et al., 2020). Among isolates from broilers and turkeys, ciprofloxacin resistance was observed at equally high levels and was generally very similar to nalidixic acid resistance. However, Salmonella isolates exhibiting ciprofloxacin resistance and nalidixic acid susceptibility, indicating the occurrence of plasmid‐mediated quinolone resistance (PMQR) mechanisms, were mostly evident in turkeys. From the monitoring of pigs, S. Rissen, S. Typhimurium and monophasic S. Typhimurium, showed the highest levels of resistance to ciprofloxacin and nalidixic acid.
Resistance to third‐generation cephalosporins, cefotaxime and ceftazidime in Salmonella isolates recovered from food‐producing animals was either rare or detected at very low/low levels in most of the reporting MSs in 2021 and 2022 (ranging between 0% and 2.5%). In Salmonella spp. isolated from human cases the levels of resistance to cefotaxime and ceftazidime were also low, at 1.4% and 1.2%, respectively.
In 2022, the overall combined resistance to ciprofloxacin and cefotaxime in Salmonella isolates from broilers, laying hens and turkey flocks was very low or low, with most countries reporting no resistant isolates. From human cases in 2022, combined resistance to cefotaxime and ciprofloxacin was very low overall (0.9%) but higher in S. Infantis (5.9%) and S. Kentucky (12.3%). Sweden observed a very high proportion of combined resistance in S. Kentucky in 2022 due to nosocomial transmission of a S. Kentucky with bla CTX‐M‐14b which was accidentally transmitted between patients in a hospital when using a contaminated instrument for gastroscopy (University hospital of Skåne, 2022). Hawkey et al. (2019) recently documented that MDR S. Kentucky multilocus sequence type (ST) 198 is a globally disseminated clone, capable of rapid spread and accumulation of resistance determinants to last‐line antimicrobials. Acquisition of Salmonella genomic island 1 (SGI1) and plasmids, as well as mutations in the Quinolone Resistance‐determining regions (QRDR), were the only genetic features found during this study to explain the global epidemiological success of the MDR S. Kentucky ST198 lineage which is highly resistant to ciprofloxacin. In the WGS data submitted to ECDC from human isolates for 2022, no less than four mutations in the QRDR were identified in all S. Kentucky ST198 (see Appendix A – High‐level resistance to ciprofloxacin among certain Salmonella serovars). Coipan et al. (2020) describe how a clone of this lineage, with an ESBL‐gene (bla CTX‐M‐14b) incorporated on the chromosome, has emerged in Europe. In contrast to plasmid‐mediated resistance, chromosomal‐mediated resistance is most likely to be maintained without antibiotic pressure, especially if it incurs no fitness cost for the bacteria.
Regarding S. Infantis isolates from humans, particularly high proportions of combined resistance were observed among S. Infantis isolates from Italy (a similar observation for Italy was made in 2020 and 2021). Similarly, Italy reported a moderate level of combined microbiological resistance to ciprofloxacin and cefotaxime (17.5%) in broiler flocks and it was the only MS reporting a higher level of combined resistance (34.9%) in fattening turkeys for S. Infantis. Several scientific publications in Europe highlight the involvement of plasmids, which appear to be responsible for resistance in many European MDR S. Infantis isolates (Alba et al., 2020; Alvarez et al., 2023; Franco et al., 2015; Nógrády et al., 2012).
In 2021, the overall combined resistance to ciprofloxacin and cefotaxime in Salmonella isolates from pigs and cattle under 1 year of age was also predominantly very low or low.
In 2021, the ECOFF for tigecycline was changed under the new legislation from > 1 mg/L to > 0.5 mg/L. Where tigecycline resistance was reported among food‐producing animal populations, most isolates displayed MICs that were one dilution above the ECOFF, meaning that these would have been considered susceptible under the previous legislation. Indeed, when compared to poultry data reported in 2020, there was a substantial increase in the occurrence of tigecycline resistance in broilers (1.0%–25.0%), laying hens (0.2%–2.9%) and in turkeys (5.5%–18.4%). In Salmonella isolated from humans, a similar increase occurred. Serovar Infantis accounted for a very high proportion of these resistant isolates in broilers (82.2%) and laying hens (57.7%). Additionally, a considerable proportion of tigecycline‐resistant isolates in turkeys was attributed to S. Infantis (48.4%) and S. Bredeney (31.7%). Tigecycline resistance in pigs was mostly found in S. Rissen (27.3%) and monophasic S. Typhimurium (25.0%). In 2022, S. Infantis and S. Kentucky were the serovars exhibiting the highest tigecycline resistance in humans. Multidrug resistance was a common feature among tigecycline‐resistant Salmonella serovars in humans and animal populations. Microbiological resistance to tigecycline may suggest clonal expansion of microbiologically resistant strains belonging to these serovars. Determining the susceptibility to tigecycline is not straightforward as this compound can be inactivated by oxidation and exposure to light, which may lead to falsely elevated MIC values. In addition, upregulation of normal cell pathways or processes may also contribute to elevated tigecycline MIC values at levels above the ECOFF in Enterobacteriaceae (He et al., 2016). Two transferable plasmid‐mediated tigecycline resistance genes, tet(X3) and tet(X4), conferring higher levels of tigecycline resistance (MICs of ≥ 16 mg/L), have been reported in numerous Enterobacteriaceae from animals and meat (chicken and pork) in China (Bai et al., 2019; He et al., 2019). These findings suggest the possibility of Salmonella acquiring transferable tigecycline resistance genes, emphasising the importance of monitoring tigecycline resistance through molecular investigations such as WGS. To note, no such genes have been reported in Salmonella spp. in the European AMR monitoring in animals or the surveillance in humans so far.
Resistance to colistin (MIC > 2 mg/L) was predominantly observed in S. Enteritidis. This serovar accounted for a substantial portion of the colistin‐resistant isolates recovered from humans (84.2%), broilers (86.6%) and laying hens (87.8%). Group D salmonellas (serogroup O9) tend to exhibit intrinsic decreased susceptibility to colistin without having known acquired or mutational colistin resistance mechanisms. This trend is exemplified by the high proportion of colistin‐resistant isolates belonging to S. Enteritidis in humans and poultry populations in 2022 and pigs in 2021 and S. Dublin in cattle under 1 year of age in 2021. Phenotypical testing for colistin is complicated, and EUCAST recommends performing testing using microbroth dilution or specific PCR. For that reason, colistin results from humans are only available from a third of the reporting MSs. Additionally, due to the possibility of different intrinsic resistance to colistin at serovar level, EUCAST has temporarily removed the Salmonella‐specific ECOFF until more comprehensive data are available. A tentative ECOFF has, however, been suggested for S. Dublin where an MIC of ≤ 16 mg/L would be considered wild type (EUCAST, 2023). Because S. Enteritidis belongs to the same antigen O group as S. Dublin (O:9) the occurrence of colistin resistance would decrease, making it easier to correctly identify isolates with acquired resistance. Further molecular characterisation of colistin‐resistant isolates obtained from the EU AMR monitoring to determine the underlying genetic mechanisms would assist in identifying the emergence and dissemination of colistin‐resistant Salmonella clones in human and animal populations.
For the first time in 2021, amikacin was included in the harmonised panel for testing food‐producing animals and derived meat. Amikacin is an aminoglycoside and is considered a high priority CIA for human medicine, it also improves the detection of 16S rRNA methyltransferase enzymes (RMTases). In both monitoring years, resistance to amikacin was very low for all animal populations, except for cattle under 1 year of age for which no resistance was reported in 2021.
Multidrug resistance (MDR)
In 2022, MDR in Salmonella spp. isolates, varied between reporting countries and animal populations, with overall MDR ranging from a low level of 7.5% in laying hens to the highest level of 43.6% in broilers. In 2021, MDR was higher among Salmonella spp. from pigs (39.1%) compared to cattle under 1 year of age (30.4%). Resistance levels varied among serovars which may exhibit particular MDR patterns, so the relative contribution of individual serovars within the different animal origins and between countries should be considered when comparing the situation between reporting countries. For example, the overall lower level of MDR among isolates from laying hens in comparison to those reported from broilers and turkeys most likely reflects in part the predominance of S. Enteritidis, which accounted for 28.0% of Salmonella isolates from laying hens reported by MSs, and where 76.4% of S. Enteritidis isolates exhibited complete susceptibility. Additionally, only a limited number of antimicrobials are authorised for the treatment of laying hens in many EU countries, and this factor may also be reflected in overall AMR levels in Salmonella isolates from this sector.
In Salmonella spp. strains from human cases, MDR was detected in 22.1% of the isolates, ranging by serovar from the lowest in S. Enteritidis (2.4%) to the highest in S. Kentucky (63.7%) and monophasic S. Typhimurium (68.2%). Apparent differences in MDR levels in S. Kentucky between human and animal populations were observed. S. Kentucky isolates were most prevalent in laying hens where MDR was reported at low levels (5.3%, N = 95). However, all S. Kentucky isolates from turkeys (N = 13) were MDR, which suggests a higher risk can be attributed to this source. Although European studies where genotypic AMR among human and animal populations are lacking, a recent US study concluded that MDR and fluoroquinolone‐resistant ST198 infections in humans may be linked to the consumption of food products that are imported or consumed while travelling (Tate et al., 2022). Similar studies at EU level are needed to elucidate these differences.
High‐level resistance to ciprofloxacin
In 2021, high‐level resistance to ciprofloxacin (MIC ≥ 4 mg/L) was detected only in one isolate (S. Kentucky) from a pig sample from Malta, and not detected in cattle under 1 year of age. In 2022 on the contrary, high‐level resistance to ciprofloxacin was observed among several isolates from poultry (see – High‐level resistance to ciprofloxacin among certain Salmonella serovars). While many serovars (including Bredeney, Derby and Infantis) exhibited high‐level resistance among poultry, S. Kentucky accounted for most of the high‐level resistant Salmonella isolates recovered from the poultry origins. The same finding was also noted among isolates from human cases in 2022, where high‐level ciprofloxacin resistance was most commonly found in S. Kentucky (in 87.6% of S. Kentucky isolates) among the 13 countries reporting MIC data.
S. Kentucky isolates exhibiting high‐level ciprofloxacin resistance are likely to belong to the above‐mentioned ST198 clone. This was demonstrated by that all eight S. Kentucky ST198 isolates reported with WGS data from humans in 2022 carried four separate mutations in the QRDR region (see Appendix A – High‐level resistance to ciprofloxacin among certain Salmonella serovars). Notably, in 2022, the occurrence of this serovar exhibiting high‐level resistance in poultry was observed by eight MSs; however, sequence typing is not performed on these isolates; thus, further studies are needed to understand if these isolates also belong to the above‐mentioned ST198 clone. Furthermore, a very high proportion of the poultry S. Kentucky isolates displaying ciprofloxacin MICs of ≥ 4 mg/L were also multidrug‐resistant, primarily showing resistance to ampicillin, gentamicin, nalidixic acid, sulfamethoxazole and tetracycline. The same observation was also noted among S. Kentucky isolates from human cases. The sole S. Kentucky isolate showing high resistance to ciprofloxacin from pigs was also resistant to nalidixic acid but no other antimicrobial substances.
Phenotypic characterisation of third‐generation cephalosporin and carbapenemase resistance
For data from animal origin in both reporting years, very small numbers of isolates were determined to be presumptive ESBL, AmpC‐ or ESBL + AmpC‐producers. The highest number observed in poultry populations was in turkey flocks (2.2%, n = 15), followed by broiler (1.4%) and laying hen flocks (0.2%). Since 2021, further testing of third‐generation cephalosporin‐resistant isolates can be undertaken using WGS as an alternative to broth‐microdilution, with Italy submitting WGS data in 2021 and Germany, Italy and the Netherlands in 2022.
Among poultry populations in 2022, only ESBL phenotypes were reported. These were associated primarily with S. Infantis, suggesting the possible clonal expansion of this particular serovar. Except for one MS, presumptive ESBL‐ producers were identified at rare or low levels. Italy, however, reported the ESBL phenotype in 21 Salmonella isolates from broilers (80.7%) and 15 Salmonella isolates from turkeys (100%). Of these, S. Infantis accounted for all turkey isolates and 18 of the broiler isolates, all harbouring bla CTX‐M‐1 . Similarly, most ESBL‐producing S. Infantis isolates from humans were also reported by Italy which accounted for 22/24 reported S. Infantis isolates with bla CTX‐M‐1. In a recent Italian study, S. Infantis strains harbouring pESI‐like plasmids, carrying bla CTX‐M‐1 genes were reported and core genome multilocus sequence typing (cgMLST) and single‐nucleotide polymorphisms (SNP)‐based analysis revealed the presence of one main cluster composed of strains isolated from the environment, animals, food and humans (Russo et al., 2024), suggesting clonal spread among animal populations and humans. The emergence of this S. Infantis clone remains hard to explain, because the use of third‐generation cephalosporins is not permitted in the EU within the poultry sector (Franco et al., 2015). Still, a recent systematic review of the global emergence of MDR S. Infantis suggests that broiler and chicken meat may be the primary sources of MDR S. Infantis (Alvarez et al., 2023). A study by Zhao et al. (2021) found MDR S. Infantis from dead‐in‐shell chicken embryos, indicating that the initial source of contagion could be the breeding grandparents or hatchery surfaces.
In humans in 2022, ESBL was reported in 20 Salmonella serovars, with the highest proportions observed in isolates of S. Kentucky (9.9%), S. Schwarzengrund (9.7%) and S. Infantis (6.5%). Considering the monitoring performed in 2021, the ESBL or ESBL + AmpC phenotype was detected in 10 isolates representing five serovars among pig isolates: namely, S. Derby, S. Infantis, S. London, S. Rissen and S. Kedougou. These isolates from pigs were reported by just two countries, Romania (n = 6) and Hungary (n = 4). Presumptive ESBL‐producing Salmonella (n = 8) was more frequently reported than presumptive ESBL + AmpC‐producing Salmonella isolates (n = 2). In cattle under 1 year of age, only a single S. Derby isolate from Spain displayed an ESBL + AmpC phenotype.
Moreover, both in 2021 and 2022, no Salmonella spp. isolates recovered from animal/meat origins were microbiologically resistant to meropenem. Unlike in 2021, Salmonella spp. isolates from humans recovered in several countries (Bulgaria, Germany and Malta) were resistant to meropenem, although at MS level, the occurrence of meropenem resistance was rare (< 0.1%).
In conclusion, it is important to note that zoonotic Salmonella infections in humans tend to be self‐limiting diseases, rarely leading to clinical conditions where patients should receive antibiotic treatment. However, considering the relatively high number of people that each year fall ill with salmonellosis, the level of resistance to commonly used antimicrobials or antimicrobials used for critically ill patients such as the CIAs needs to be continuously watched as these still may be of importance. In addition, the monitoring of the resistance levels in Salmonella from food‐producing animals and meat derived thereof allows the assessment and the evaluation of the occurrence of AMR in the EU and help to identify the achievements in the efforts to tackle AMR.
3. ANTIMICROBIAL RESISTANCE IN CAMPYLOBACTER SPP.
3.1. Key findings
For 2022, 22 MSs and two non‐MS (Iceland and Norway) reported data on AMR in Campylobacter jejuni and Campylobacter coli from humans. In the same year, data on AMR in C. jejuni from broilers and from fattening turkeys were reported by 26 MSs and the United Kingdom (Northern Ireland) and by 10 MSs, respectively, whereas data on AMR in C. coli from broilers and from fattening turkeys were reported by 24 MSs and the United Kingdom (Northern Ireland) and by 11 MSs, respectively. In 2021, data related to the antimicrobial susceptibility of C. jejuni from cattle under 1 year of age and from fattening pigs were reported by 10 and 12 MSs, respectively, whereas data on AMR in C. coli from cattle under 1 year of age and from fattening pigs were reported by 10 MSs and 26 MSs and the United Kingdom (Northern Ireland), respectively.
Resistance rates differed greatly between reporting countries, between antimicrobials and between the two Campylobacter species.
Levels of resistance to ciprofloxacin ranged from high and very high to extremely high, respectively, in C. jejuni and C. coli isolates, recovered from humans and food‐producing animals in the EU. In 2022, levels of resistance to ciprofloxacin in human C. jejuni isolates ranged from 20.6% to 96.6% among the EU countries, while for C. coli isolates, 12 out of 17 reporting countries found levels of ciprofloxacin resistance higher than 70%. In food‐producing animals, the highest levels of resistance to ciprofloxacin were observed in C. coli isolates, ranging from 51.7% in fattening pigs to 84.1% in fattening turkeys. The levels of resistance to ciprofloxacin obtained from C. jejuni isolates from poultry in 2022 were also extremely high (78.2% in fattening turkeys and 70.9% in broilers).
Resistance to erythromycin was either not detected or very low to low in C. jejuni from humans and food‐producing animals but was higher in C. coli, ranging from 7.8% in humans to 35.7% in cattle under 1 year of age.
The whole genome sequencing results reported for erythromycin‐resistant C. jejuni and C. coli isolates recovered from food‐producing animals in 2021–2022, mostly those highly resistant (MIC ≥ 512 mg/L), showed detection of the mutation A2075G in the 23SrRNA gene and no detection of the transferable erm(B) gene in most isolates. A single isolate of C. coli from fattening pigs was reported positive to the presence of erm(B). In 15.5% of the sequenced isolates, neither presence of erm(B) nor mutation in the 23SrRNA gene were reported.
Combined resistance to both ciprofloxacin and erythromycin, which are considered critically important antimicrobials for the treatment of campylobacteriosis, was generally rare to low in C. jejuni from humans and food‐producing animals. The combined resistance was low in C. coli from humans (7.1%), broilers (8.2%) and fattening pigs (9.1%), moderate in C. coli from fattening turkeys (17.4%) and high in C. coli from cattle under 1 year of age (32.7%). This finding may be a cause for public health concern.
The observed levels of resistance to gentamicin and ertapenem in C. coli isolated from cattle under 1 year of age in 2021 (12.4% and 29.1%, respectively), and the moderate to very high levels of resistance to ertapenem in C. jejuni and C. coli isolated from poultry in 2022 might be a cause for public health concern as those are recommended antimicrobials for treatment in severe invasive Campylobacter infections in humans. Gentamicin resistance in C. coli from humans was observed at low levels except in one Member State, while ertapenem is not (yet) included in the priority panel for Campylobacter monitoring of human isolates at EU level. However, findings on ertapenem resistance should be interpreted with caution as the epidemiological cut‐off for ertapenem used by EFSA is still under discussion and there is not yet a validated threshold for resistance to ertapenem established by EUCAST.
The prevalence of resistance to selected antimicrobials in C. jejuni and C. coli from broilers and fattening turkeys in 2022 has been estimated at country‐level. Between‐country variability, from rare, low or moderate to extremely high levels, was observed in the prevalence of ciprofloxacin‐resistant and tetracycline‐resistant C. jejuni and C. coli isolates. Interestingly, a more limited between‐country variability and notably lower levels of prevalence of resistance were found for erythromycin‐resistant Campylobacter.
Overall, complete susceptibility (CS), defined in this report as susceptibility to ciprofloxacin, erythromycin, tetracycline and gentamicin, was higher in C. jejuni than in C. coli isolates. The overall CS observed from C. jejuni isolates was 26.4% in humans in 2022, and among C. jejuni from food‐producing animals, it was lowest in fattening turkeys (16.5%) and highest in fattening pigs (48.3%). Regarding overall CS among C. coli isolates, it was moderate in humans (13.7%), broilers (13.1%) and fattening pigs (19.9%), and low in fattening turkeys (4.4%) and cattle under 1 year of age (6.3%).
Multidrug resistance (MDR), defined in this report as resistance to at least three antimicrobials among ciprofloxacin, erythromycin, tetracycline and gentamicin, was generally very low for C. jejuni isolated from humans (0.7%) and ranged from very low to low in the animal species considered. Compared to C. jejuni, MDR was markedly higher in C. coli, specifically occurring in 9.0% of the isolates from humans, 39.3% of isolates from cattle under one year of age, 16.9% of isolates from fattening turkeys, 9.5% of isolates from fattening pigs, and 8.3% of isolates from broilers. These results agree with the higher levels of resistance to selected antimicrobials seen in C. coli isolates.
Over the period 2013–2022, resistance to ciprofloxacin in C. jejuni from humans increased in 13 MSs and decreased in three reporting countries (two MSs and one non‐MS). Resistance to ciprofloxacin in C. coli from humans increased in two MSs and decreased in one MS in the same period. In C. jejuni from broilers in the period 2014–2022, resistance to ciprofloxacin increased in six MSs and decreased in three MSs. During the same period, an increasing trend in resistance to ciprofloxacin was also observed in one MS each for C. coli from broilers, C. jejuni from fattening turkeys and C. coli from fattening pigs.
In the same two periods, erythromycin resistance decreased in C. jejuni from humans in seven countries (six MSs and one non‐MSs), from broilers in six MSs and from fattening turkeys in three MSs. Erythromycin resistance also decreased in C. coli from humans in six MSs, and from fattening pigs in two countries (one MSs and one non‐MS).
3.2. Data on AMR in Campylobacter spp. addressed
The two main Campylobacter species responsible for human infections are C. jejuni, which is a predominant species in poultry, followed by C. coli (Jehanne et al., 2020), frequently found in pigs and in poultry, sometimes at higher rates than C. jejuni (Pergola et al., 2017). C. coli is often more resistant than C. jejuni to several important antimicrobials and may contain and transfer resistance genes to C. jejuni.
In 2024, EFSA has published for the first time (together with the present report) a story map on AMR in Campylobacter, where further information on the topic can be found (see text box below).
EFSA story map on Monitoring AMR in Campylobacter
The EFSA story map on antimicrobial resistance in Campylobacter is a new interactive communication tool published by EFSA in 2024, tailored to the general public, available online (here). This story map provides general information on the pathogen, why it is important to monitor resistance in Campylobacter, and the main mechanisms of resistance and modes of resistance spread in C. jejuni and C. coli. In addition, this story map also illustrates the activities implemented in the EU for the monitoring of antimicrobial resistance in C. jejuni and C. coli in humans and animals, and the role of EFSA with respect to these activities. Furthermore, the story map shows the key findings of the 2021–2022 EU monitoring of occurrence of antimicrobial resistance in C. jejuni and C. coli and informs on how to prevent antimicrobial resistance. Users can easily display and explore the content of the different sections in the story map, browsing the dynamic infographic, text and plots.
Campylobacter AMR data from human infections either derive from monitoring programmes set up by national public health reference laboratories/services or are collected from primary or regional laboratories and integrated with the case information in the national surveillance of human Campylobacter infections. This report covers AMR data for C. jejuni and C. coli from human cases from 2022. Data from 2021 are presented in the 2020–2021 report (EFSA and ECDC, 2023).
In the framework of the Commission Implementing Decision (EU) 2020/1729, the monitoring of AMR in Campylobacter spp. from food‐producing animals is focused on the species C. jejuni and C. coli. Since 2021, the AMR monitoring has been mandatory, at biannual basis, in both Campylobacter species from caecal samples from broilers and fattening pigs, and in countries where the national production of cattle meat or turkey meat is more than 10,000 tonnes per year, from cattle under 1 year of age and fattening turkeys, respectively.
This chapter includes data on C. jejuni and C. coli in broilers and fattening turkeys from 2022, and in cattle under 1 year of age and fattening pigs in 2021 resulting from mandatory monitoring. In addition, the voluntary monitoring of AMR in Campylobacter isolates recovered from meat samples (of broilers, fattening turkeys, cattle, fattening pigs and other animal species) at retail was performed in 2021 and 2022 and related data are presented in the Annex B.
In 2024, EFSA has published for the first time a dedicated dashboard on AMR in Campylobacter, an online data visualisation tool where information on the occurrence of resistance in C. jejuni and C. coli from animals can be visualised interactively (see text box below).
EFSA dashboard on AMR in Campylobacter
The EFSA dashboard on Antimicrobial Resistance (available online here). is a graphical user interface for searching and querying the data on the monitoring of AMR in Campylobacter from animals reported to EFSA by EU MSs and other reporting countries according with Commission Implementing Decision (EU) 2020/1729. In the dashboard, monitoring data and related statistics on the occurrence of resistance to selected antimicrobials in C. jejuni and C. coli from broilers, fattening turkeys, fattening pigs and cattle under 1 year of age can be visualised interactively through maps and graphs. Temporal trends for the period 2014–2022 are also shown using dynamic graphs. In this tool, the main statistics can also be viewed and downloaded in tabular format. Links to the dashboard are available in the relevant sections of this chapter.
Detailed information on antimicrobial resistance data reporting including requirements, sample descriptions and codes for mandatory and voluntary reporting are presented in EFSA's manual for reporting AMR data within the framework of Directive 2003/99/EC and Commission Implementing Decision (EU) 2020/1729 (EFSA, 2023).
Further consideration on the data used and the methodology applied in the analysis can be found in Appendix F – Materials and methods.
3.3. Humans: Occurrence of antimicrobial resistance in Campylobacter
3.3.1. Data reported
For 2022, 22 MSs and two non‐MS (Iceland and Norway) reported data on AMR from Campylobacter jejuni and C. coli isolated from human cases of campylobacteriosis. Greece reported Campylobacter AMR data for the first time for 2022. The five Member States that did not report have no surveillance system in place for Campylobacter AMR, i.e. they do not collect AMR data from clinical laboratories or routinely perform Campylobacter AST at the national public health reference laboratory.
Seventeen countries (two more than in 2021) reported measured values, five reported results interpreted as susceptible standard dosing regimen, susceptible increased exposure, or resistant (SIR) according to the clinical breakpoints (CBPs) applied, and two countries reported results that were categorised as predicted wild type or predicted non‐wild type based on analysis of bacterial genomes (Ireland and the Netherlands, both providing interpreted data) (Annex B, Tables 1, 2). Not all countries reported results for all antimicrobials in the harmonised panel (ECDC, 2016).
The reported data represented 22.1% and 26.2% of the confirmed human cases with Campylobacter jejuni and Campylobacter coli, respectively, reported in the EU/EEA in 2022.
3.3.2. Occurrence of resistance
In 2022, reported resistance data indicated high to extremely high resistance levels to ciprofloxacin in human Campylobacter jejuni isolates ranging from around 20.6% and 25.6% in, respectively, Norway and Iceland to levels approaching nearly 100% in Portugal (96.6%), Lithuania (96.2%), Cyprus (94%) and Poland (93.8%) (Annex B, Table 1). The rest of the reporting countries obtained results that indicated high, very high, to extremely high resistance levels to ciprofloxacin resulting in an overall resistance level of 69.1% in the EU (Figure 13; Table 7). The overall ciprofloxacin resistance level in C. coli in the EU was similar (70.6%) to the overall level found in C. jejuni; however, the median level was higher due to all countries – except for one – reporting very high to extremely high ciprofloxacin resistance levels (Figure 13; Table 7; Annex B, Table 2). Both overall levels were higher than the levels reported in the past 2 years. It must be noted that only 17 EU countries reported at least 10 C. coli isolates and were included in the analysis. Similar to 2021, the highest resistance levels in C. coli isolates were reported by Estonia (100%) and Portugal (98.7%). In addition, 12 out of 17 countries reported extremely high resistance levels to ciprofloxacin (> 70%) (Annex B, Table 2). The lowest resistance level in C. coli was reported by the Netherlands (0%) among 55 isolates. This is considered a large decrease compared with 2021 when the Netherlands reported 71.7% resistance among 92 isolates. (Annex B, Table 2). However, it must be noted that the Netherlands previously reported SIR data according to clinical breakpoints but are depending on the results of WGS for 2022.
FIGURE 13.
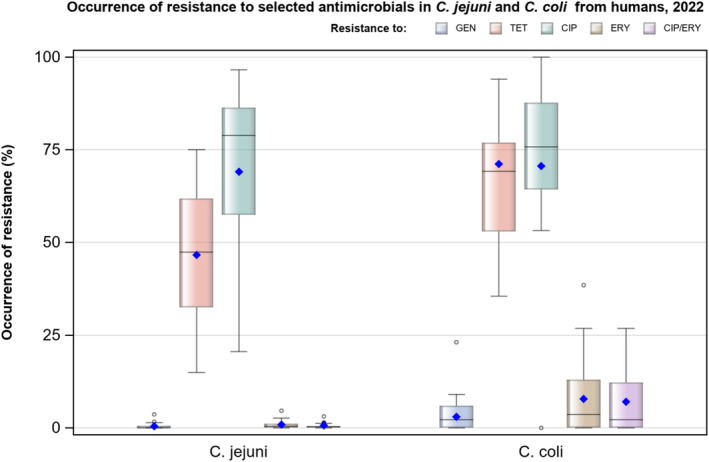
Boxplot of the occurrence of resistance to a selection of antimicrobials in Campylobacter jejuni and C. coli isolates from humans, 2022.
Note: GEN, gentamicin; TET, tetracycline; CIP, ciprofloxacin; ERY, erythromycin; CIP/ERY, combined resistance to ciprofloxacin and erythromycin. Horizontal line represents the median; blue diamond: overall resistance in the EU. Only countries reporting ≥ 10 isolates per species are included in the graph.
TABLE 7.
Overall resistance levels in the European Union in Campylobacter jejuni and C. coli (22 MSs).
| Campylobacter species | Ciprofloxacin | Erythromycin | Tetracyclines | Gentamicin | Combined CIP/ERY | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | % res | N | % res | N | % res | N | % res | N | % res | |
| C. jejuni (22 MSs) | 15,499 | 69.1 | 15,721 | 0.9 | 14,339 | 46.6 | 10,240 | 0.5 | 15,467 | 0.7 |
| C. coli (22 MSs) | 2291 | 70.6 | 2313 | 7.8 | 2160 | 71.2 | 1615 | 3.0 | 2289 | 7.1 |
The overall level of resistance to erythromycin in human C. jejuni isolates in the EU was very low or rare, except for Spain (4.6%), Greece (2.6%) and Finland (2.1%) where low levels of resistance were observed (Annex B, Table 1). Only one country (Romania) did not report at least 10 isolates to be included in the analysis. The overall level of erythromycin resistance in human C. coli isolates in the EU was low (7.8%, Figure 13; Table 7), but higher compared with resistance in human C. jejuni (0.9%, Table 7). In addition, the percentages resistance varied quite considerably between countries ranging from 0% in Austria, Estonia, Ireland and Malta to 38.5% in Greece. Twelve out of 17 countries providing at least 10 C. coli isolates reported rare to low levels of resistance (Annex B, Table 2).
In 2022, overall, EU countries reported high tetracycline resistance in human C. jejuni (46.6%, Table 7) with levels ranging from moderate (15%, Norway) to extremely high (75%, Poland). Twelve out of 22 countries reported high levels of resistance (Annex B, Table 1). The overall level of tetracycline resistance in human C. coli and in the EU (71.2%) was higher than in C. jejuni and isolate resistance levels varied from high (35.5%, Slovenia) to extremely high (approx. 94%, Estonia and Portugal). Seven out of 15 countries reporting at least 10 isolates obtained extremely high tetracycline resistance levels (Annex B, Table 2).
Thirteen and 10 countries reported data from more than 10 isolates on gentamicin resistance in, respectively, C. jejuni and C. coli (Annex B, Tables 1, 2). For C. jejuni, resistance levels in the EU were very low in general (0.5%, Table 7) with seven countries reporting 0% resistance and only three countries reporting low levels of resistance (Spain, Italy and Greece). Overall gentamicin resistance levels in C. coli are somewhat higher in the EU, but are still considered low (3.0%, Table 7 ). All countries reporting resistance in C. coli in at least 10 isolates reported rare to low levels of resistance, except for one country (Greece, 23.1%).
Few countries reported at least 10 isolates tested in relation to co‐amoxiclav resistance (n = 5: C. jejuni and C. coli). Both for C. jejuni and C. coli the overall EU resistance level was found to be low (Annex B, Tables 1, 2). However, out of five countries reporting resistance in more than 10 C. jejuni and 10 C. coli isolates, respectively, three and four countries reported high levels of co‐amoxiclav resistance (Annex B, Tables 1, 2).
3.3.3. Combined resistance to ciprofloxacin and erythromycin
Combined resistance to both ciprofloxacin and erythromycin, which are considered critically important antimicrobials for the treatment of campylobacteriosis, was overall very low at the EU level in C. jejuni (0.7%) and low (7.1%) in C. coli (Annex B, Tables 3, 4). These percentages were very similar to the numbers reported in 2021. The levels of combined resistance to both ciprofloxacin and erythromycin in human C. jejuni isolates ranged from 0% to 3.1% (including Iceland and Norway), with the highest level being reported by Spain. The levels of combined resistance to both ciprofloxacin and erythromycin in human C. coli isolates ranged from 0% to 26.9% (including Iceland and Norway); the highest level being reported by Portugal and Greece (Figure 14; Annex B, Tables 3, 4).
FIGURE 14.
Spatial distribution of combined resistance to ciprofloxacin and erythromycin in (A) Campylobacter jejuni and (B) C. coli isolates from humans, 2022.
3.3.4. Complete susceptibility and multidrug resistance
Analyses of complete susceptibility (CS) and multidrug resistance (MDR) focus on critically important antimicrobials in humans, and the target substances were agreed by EFSA and ECDC to include ciprofloxacin (class: fluoroquinolones), erythromycin (class: macrolides), gentamicin (class: aminoglycosides) and tetracycline. It must be noted that the MDR analysis is based on data from fewer reporting countries as not all countries test gentamicin susceptibility. As the aim of such analyses is to compare CS and MDR in animals and humans, the related findings are only presented in Section 3.5 on the comparison of human and animal data.
Detailed results at country level on the occurrence of MDR and CS in C. jejuni and C. coli isolates from humans are presented in Annex B, Tables 5, 6.
3.3.5. Temporal trends
Temporal trends were analysed for countries reporting data for at least 3 years over the period 2013–2022 using logistic regression (Appendix F – Materials and methods). Trends in antimicrobial resistance to C. jejuni and C. coli varied by country depending on the respective antimicrobial (Table 8; Figure 15, 16). Statistically significant (p < 0.05) increasing trends of ciprofloxacin resistance were observed in C. jejuni isolates from 13 EU MSs and in C. coli isolates from Slovenia and Slovakia. Statistically significant decreasing trends in C. jejuni were found for Spain, Finland and Norway, while for France, a decreasing trend was found in C. coli. Only one country showed a significant increase in erythromycin resistance in C. jejuni isolates. Statistically significant decreases in C. jejuni and C. coli were, respectively, noted in seven (including non‐MS Norway) and six countries. Finally, tetracycline resistance in C. jejuni significantly increased for six MSs and decreased for three MSs. For C. coli, increasing trends in time were seen for three countries (France, the Netherlands and Slovakia) and decreasing trends in one country (Finland).
TABLE 8.
Number of countries with significantly a increasing or decreasing trends in resistance to selected antimicrobials for Campylobacter jejuni and C. coli in humans, 2013–2022.
| Campylobacter species | Ciprofloxacin | Erythromycin | Tetracycline | |||
|---|---|---|---|---|---|---|
| Incr. | Decr. | Incr. | Decr. | Incr. | Decr. | |
| C. jejuni (19 MS + 2 non‐MS) | 13 (AT, BG, CY, DK, EE, FR, IT, LT, MT, NL, PL, SI, SK) | 3 (ES, FI, NO) | 1 (ES) | 7 (DK, FI, IT, MT, NO, PT, SI) | 6 (AT, DK, EE, NL, SI, SK) | 6 (ES, FI, FR, LT, NO, PT) |
| C. coli (13) | 2 (SI, SK) | 1 (FR) | – | 6 (EE, ES, FR MT, PT, SK) | 3 (FR, NL, SK) | 1 (FI) |
p < 0.05, logistic regression.
FIGURE 15.
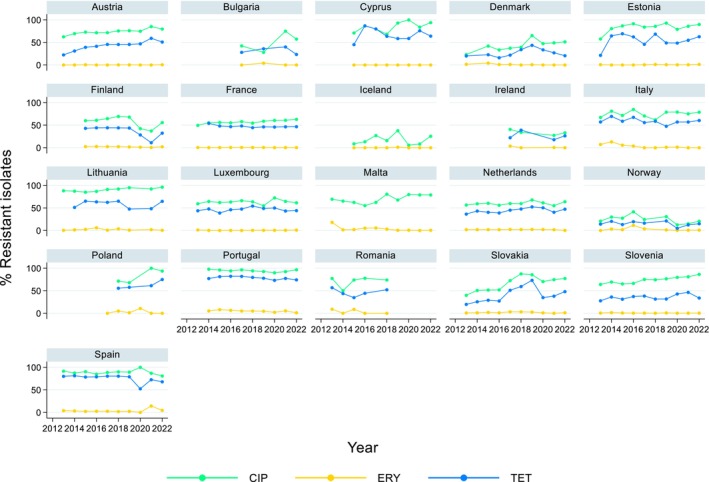
Trends in ciprofloxacin (CIP), erythromycin (ERY) and tetracycline (TET) resistance in Campylobacter jejuni from humans in 21 reporting countries, 2013–2022.
FIGURE 16.
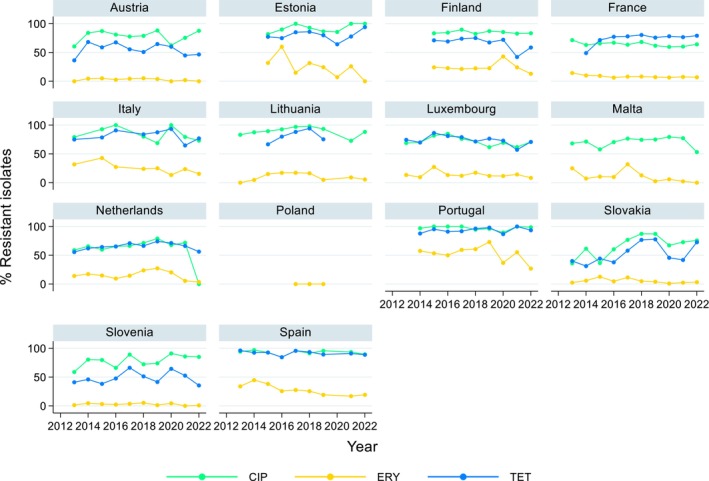
Trends in ciprofloxacin (CIP), erythromycin (ERY) and tetracycline (TET) resistance in Campylobacter coli from humans in 14 reporting countries, 2013–2022.
3.3.6. High‐level resistance to erythromycin
High‐level resistance to erythromycin (MIC > 128 mg/L) was assessed as a potential indication for transferrable erythromycin resistance due to the potential presence of the erm(B) gene (Qin et al., 2014). The results indicated that 0.4% of the C. jejuni isolates (N = 3664, seven MSs and one non‐MS) showed MIC values higher than 128 mg/L while for C. coli the percentage was higher, 2.6% (N = 493, eight MSs and one non‐MS) (Figure 17). These results were lower than compared to 2021 (EFSA and ECDC, 2023). It should, however, be noted that one MS only tested a range up to 64 mg/L and higher MIC results could therefore not properly be assigned and were categorised as MIC 128 mg/L. This may have resulted in an underestimation of the proportion of isolates with MIC > 128 mg/L. Similarly, in 1.0% (N = 3664, eight MSs and one non‐MS) of C. jejuni and 10.5% (N = 493, eight MSs and one non‐MS) of C. coli tested with disc diffusion no inhibition zone could be observed (6 mm zone equals the disc size), which corresponds to an MIC of ≥ 128 mg/L for C. jejuni and 64–≥ 128 mg/L for C. coli (EUCAST, 2023).
FIGURE 17.
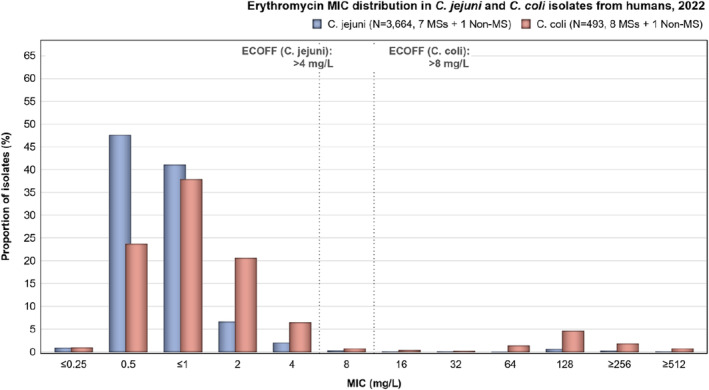
Erythromycin minimum inhibitory concentration (MIC) distribution in Campylobacter jejuni and C. coli isolates from humans, 2022.
Note: ≤ 1 mg/L could potentially include values smaller than or equal to 0.5 and 0.25 mg/L. ≥ 256 mg/L could potentially include values greater than or equal to 512 mg/L.
3.4. Food‐producing animals: Occurrence and prevalence of antimicrobial resistance in Campylobacter
3.4.1. Data reported
In the present report, the 2021 antimicrobial resistance data on Campylobacter isolates from fattening pigs and cattle under 1 year of age are considered for comparison with 2022 resistance data on Campylobacter from broilers and fattening turkeys.
In 2021, 12 MSs (Bulgaria, Cyprus, Denmark, Germany, Ireland, Italy, Latvia, Lithuania, Luxemburg, Malta, the Netherlands and Portugal) and one non‐MS (Norway) reported mandatory data on C. jejuni isolates recovered from fattening pigs (N = 60 and N = 17, respectively). Ten MSs (Belgium, Croatia, Denmark, France, Germany, Italy, the Netherlands, Portugal, Romania and Spain) and two non‐MSs (Norway and Switzerland) reported data on C. jejuni isolates recovered from cattle under 1 year of age (N = 1198 and N = 270, respectively). Twenty‐six MSs (all MSs, except Czechia) and the United Kingdom (Northern Ireland), as well as three non‐MSs (Iceland, Norway and Switzerland) reported mandatory data on C. coli isolates recovered from fattening pigs (N = 3546 and N = 624, respectively). Ten MSs (Belgium, Croatia, Denmark, France, Germany, Italy, the Netherlands, Portugal, Romania and Spain) reported data on C. coli isolates recovered from cattle under 1 year of age (N = 443) in 2021. The complete overview of the data reported in 2021, is presented in Annex B (Table 7).
In the year 2022, mandatory data on C. jejuni isolates recovered from caecal samples of broilers were reported by 26 MSs (all MSs, except Greece) and the United Kingdom (Northern Ireland), as well as by three non‐MSs (Iceland, Norway and Switzerland) (N = 2927 and N = 325, respectively). Data on C. jejuni isolates recovered from caecal samples of fattening turkeys were reported by 10 MSs (Austria, Croatia, France, Germany, Ireland, Italy, Poland, Portugal, Romania and Spain) in 2022 (N = 929) (Annex B, Table 7, 9). Twenty‐four MSs (all MSs, except Finland, Greece and Lithuania) and the United Kingdom (Northern Ireland), as well as three non‐MS (Norway, Republic of North Macedonia and Switzerland) reported mandatory data on C. coli isolates recovered from caecal samples of broilers (N = 1565 and N = 64, respectively). Furthermore, 11 MSs (Austria, Croatia, France, Germany, Hungary, Ireland, Italy, Poland, Portugal, Romania and Spain) reported data on C. coli isolates recovered from caecal samples of fattening turkeys (N = 1381) (Annex B, Table 7, 8).
Resistance data concerning food‐producing animals, reported in 2021 and 2022, are presented in the following sections. Only data collected in the framework of the Commission Implementing Decision (EU) 2020/1729 are presented. The complete overview of all reported data (mandatory and voluntary) from food‐producing animals and meat derived thereof, reported in 2021 and 2022, is presented in Annex B (Tables 7–11), which is available as supporting documentation in Zenodo (https://doi.org/10.5281/zenodo.10528846).
The new EU rules for the AMR monitoring in Campylobacter in place from 2021 (Commission Implementing Decision (EU) 2020/1729) implemented changes to the harmonised panel of antimicrobials, consisting in the removal of streptomycin and nalidixic acid and in the inclusion of two new substances: ertapenem and chloramphenicol. Therefore, the mandatory antimicrobials to be reported for C. jejuni and C. coli from 2021 onward are: chloramphenicol, ciprofloxacin, ertapenem, erythromycin, gentamicin and tetracycline. Furthermore, in accordance with Commission Implementing Decision (EU) 2020/1729, the range of concentrations (mg/L) has been reduced for gentamicin and increased for erythromycin and ciprofloxacin, compared to indications provided in the old Commission Implementing Decision (EU) 2013/652. As this report presents both 2021 and 2022 data, all data are reported in accordance with the new AMR monitoring rules laid down in Commission Implementing Decision (EU) 2020/1729.
3.4.2. Occurrence of resistance in poultry, fattening pigs and cattle under 1 year of age
Comparison of resistance data between bacterial and animal species should be done with caution due to the dispersion of resistance rates between countries and because numbers of isolates and reporting countries vary, especially for voluntary monitoring and reporting. Data reported in 2022 for C. jejuni and C. coli isolates from legislative and non‐legislative categories are presented in Annex B, Tables 8–11.
In this Report, data on the occurrence of resistance in Campylobacter species in caecal samples from fattening pigs (2021), cattle under 1 year of age (2021), broilers (2022) and fattening turkeys (2022) are presented in Table 9 and Figure 18. The detailed country‐level information on the occurrence of resistance is presented in Annex B (Tables 13–20).
TABLE 9.
Occurrence of resistance (%) to selected antimicrobials in Campylobacter coli and C. jejuni in caecal samples from broilers, fattening turkeys, fattening pigs and cattle under 1 year of age using harmonised ECOFFs, 27 EU MSs and United Kingdom.
| Campylobacter species | Categories | Year | No. of isolates | Reporting countries (N) | GEN | CHL | ETP | CIP | ERY | TET | CIP/ERY |
|---|---|---|---|---|---|---|---|---|---|---|---|
| C. jejuni | Broilers | 2022 | 2927 | AT, BE, BG, CY, CZ, DE, DK, EE, ES, FI, FR, HR, HU, IE, IT, LT, LU, LV, MT, NL, PL, PT, RO, SE, SI, SK, XI (27) | 0.1 | 0.0 | 10.1 | 70.9 | 1.5 | 50.7 | 1.0 |
| Fattening turkeys | 2022 | 929 | AT, DE, ES, FR, HR, IE, IT, PL, PT, RO (10) | 0 | 0.1 | 15.1 | 78.1 | 1.7 | 59 | 1.6 | |
| Cattle under 1 year of age | 2021 | 1198 | BE, DE, DK, ES, FR, HR, IT, NL, PT, RO (10) | 0.5 | 0.1 | 1.0 | 54.7 | 1.0 | 68.8 | 0.8 | |
| Fattening Pigs | 2021 | 60 | BG, CY, DE, DK, IE, IT, LT, LU, LV, MT, NL, PT (12) | 1.7 | 0.0 | 0.0 | 41.7 | 1.7 | 43.3 | 1.7 | |
| C. coli | Broilers | 2022 | 1565 | AT, BE, BG, CY, CZ, DE, DK, EE, ES, FR, HR, HU, IE, IT, LU, LV, MT, NL, PL, PT, RO, SE, SI, SK, XI (25) | 2.0 | 0.7 | 43.1 | 73.0 | 8.8 | 67.5 | 8.2 |
| Fattening turkeys | 2022 | 1381 | AT, DE, ES, FR, HR, HU, IE, IT, PL, PT, RO (11) | 0.6 | 0.0 | 58.1 | 84.1 | 18.2 | 79.8 | 17.5 | |
| Cattle under 1 year of age | 2021 | 443 | BE, DE, DK, ES, FR, HR, IT, NL, PT, RO (10) | 12.4 | 3.4 | 29.1 | 79.7 | 35.7 | 90.5 | 32.7 | |
| Fattening Pigs | 2021 | 3546 | AT, BE, BG, CY, DE, DK, EE, ES, FI, FR, GR, HR, HU, IE, IT, LT, LU, LV, MT, NL, PL, PT, RO, SE, SI, SK, XI (27) | 2.6 | 0.5 | 1.3 | 51.7 | 12.2 | 69.3 | 9.1 |
Abbreviations: CIP, ciprofloxacin; CIP/ERY, combined ‘microbiological’ resistance to ciprofloxacin and erythromycin; CS, complete susceptibility to the four antimicrobial classes (ciprofloxacin, erythromycin, tetracycline and gentamicin); CHL, chloramphenicol; ETP, ertapenem; ERY, erythromycin; GEN, gentamicin; TET, tetracycline; N, Total number of reporting Member States (MSs); XI, United Kingdom (Northern Ireland).
FIGURE 18.
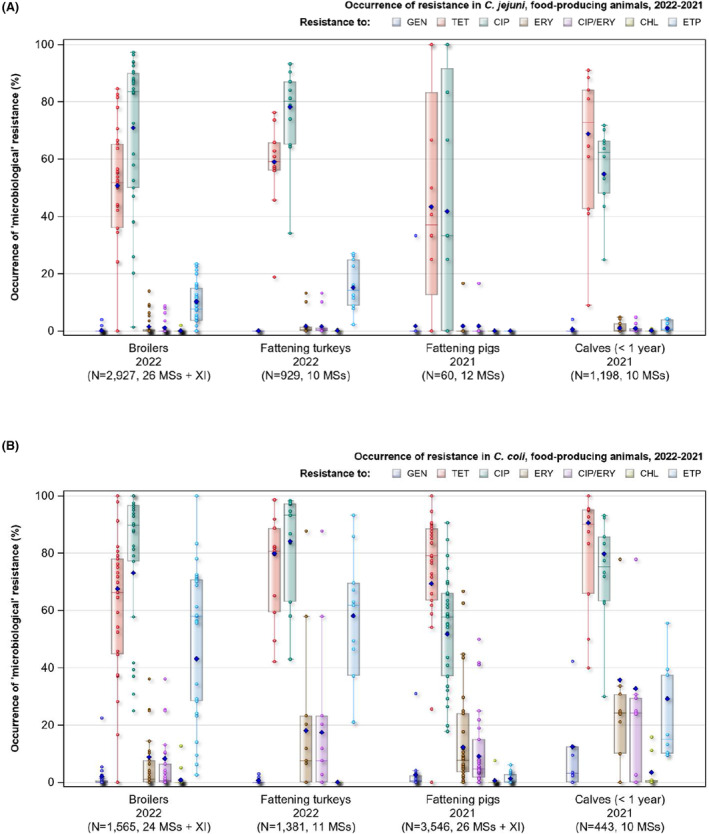
Occurrence of resistance to antimicrobials in (A) Campylobacter jejuni and (B) C. coli from food‐producing animals, 2021/2022.
Note: GEN, gentamicin; TET, tetracycline; CIP, ciprofloxacin (CIP); ERY, erythromycin; CIP/ERY, combined resistance to ciprofloxacin and erythromycin; CHL, chloramphenicol; ETP, ertapenem. Horizontal line represents the median; blue diamond: overall resistance in the EU; dots represent resistance in the different countries.
Based on the reported animal data, the level of resistance to tetracycline ranged from high to very high (43.3%–68.8%) in C. jejuni and from very high to extremely high (67.5%–90.5%) in C. coli isolates among all the animal categories considered. The highest levels of resistance to tetracycline were observed in C. coli isolated from cattle under 1 year of age (90.5%) in 2021 (data from 10 MSs) and from fattening turkeys (79.8%) in 2022 (data from 11 MSs). Overall, the levels of resistance to tetracycline were higher than those observed for the other selected antimicrobials in C. coli and C. jejuni isolates from cattle under 1 year of age and from fattening pigs in 2021.
High to extremely high levels of resistance were also observed to ciprofloxacin in both C. jejuni (range from 41.7% to 78.2%) and C. coli (range from 51.7% to 84.1%) isolates among all the animal categories considered. The highest average level of resistance to ciprofloxacin was observed in C. coli isolated from fattening turkeys in 2022 (84.1%; data from 11 MSs). Likewise, an extremely high average level of resistance to ciprofloxacin (79.7%) was obtained from C. coli isolated from cattle under 1 year of age in 2021. Overall, the levels of resistance to ciprofloxacin were higher for C. coli than for C. jejuni within an animal species, although the average level of resistance to ciprofloxacin obtained from C. jejuni isolates from broilers (70.9%) and fattening turkeys (78.2%) in 2022 were also extremely high (data from 26 MSs and the United Kingdom (Northern Ireland) and from 10 MSs, respectively). The lowest levels of resistance to ciprofloxacin were in both C. jejuni and C. coli obtained from fattening pigs in 2021 (41.7% in C. jejuni and 51.7% in C. coli; data from 12 MSs and from 26 MSs and the United Kingdom (Northern Ireland), respectively).
Resistance to gentamicin ranged from absent to very low among C. jejuni isolates from broilers in 2022 and from cattle under 1 year of age in 2021, as well as in both C. jejuni and C. coli from fattening turkeys in 2022 (average 0.0%–0.6%). In 2022, the overall average levels of resistance to this antimicrobial were 0.1% for C. jejuni in broilers and 0% in fattening turkeys. Higher average occurrence of resistance to gentamicin was observed in C. coli isolates from broilers (2%) and from fattening turkeys (0.6%). The highest average level of resistance to gentamicin was observed in C. coli from cattle under 1 year of age in 2021 (12.4%; data from 10 MSs).
Average resistance to erythromycin was detected at low levels in C. jejuni isolates for the different animal categories (range from 1% to 1.7%). Notably higher levels of resistance were reported in C. coli isolates (range from 8.8% to 35.7%). The highest average level of resistance to erythromycin was observed in C. coli isolates recovered from cattle under 1 year of age (35.7%) in 2021, followed by fattening turkeys (18.0%) in 2022, fattening pigs (12.2%) in 2021 and broilers (8.8%) in 2022.
Resistance to chloramphenicol in isolates from all animal species considered in 2021 and 2022 varied from absent to very low (average range from 0% to 0.7%), except for low average resistance in C. coli isolates from cattle under 1 year of age in 2021 (3.4%).
Resistance to ertapenem in C. jejuni isolates varied from absent to moderate (average range from 0% to 15.1%). Notably, C. jejuni isolated from fattening turkeys (15.1%) and from broilers (10.2%) in 2022 presented higher level of resistance compared to C. jejuni isolated from cattle under 1 year of age (1%) and from fattening pigs in 2021 (0%). It is noteworthy that a higher level of average resistance to ertapenem was reported in C. coli, varying from low to very high (range 1.3% to 58.2%). Average ertapenem resistance in C. coli was highest in fattening turkeys (58.2%), followed by broilers (43.1%) and by cattle under 1 year of age (29.1%) and lowest in fattening pigs (1.3%) (see specific textbox on ‘further considerations on the detected levels of ertapenem resistance in C. jejuni and C. coli from food‐producing animals’).
Further considerations on the detected levels of ertapenem resistance in C. jejuni and C. coli from food‐producing animals
Due to the absence of a validated threshold for resistance to ertapenem in C. jejuni and C. coli recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST), an epidemiological cut‐off (ECOFF) of 0.5 mg/L was used by EFSA in 2021 and 2022 in agreement with the EU Reference Laboratory for AMR (EFSA, 2023). The choice of the ECOFF has a direct impact on the interpretation of an isolate as susceptible or resistant to a selected antimicrobial.
MIC distribution (%) in ertapenem resistant and susceptible C. coli and C. jejuni isolates from broilers and fattening turkeys in 2022 and from fattening pigs and cattle under 1 year of age in 2021, reported by MSs and non‐MSs.
| Animals/year | MIC (mg/L) | % resistant isolates | ||||||
| ≤ 0.125 | 0.25 | 0.5 * | 1 | 2 | 4 | > 4 | ||
| Campylobacter coli | ||||||||
| Broilers, 2022 (N = 1629) | 24.1% | 12.6% | 21.2% | 18.8% | 14.7% | 6.4% | 2.0% | 42.1% |
| Fattening turkeys, 2022 (N = 1381) | 13.0% | 11.1% | 17.9% | 17.9% | 22.8% | 12.2% | 5.1% | 58.1% |
| Fattening pigs, 2021 (N = 4170) | 76.2% | 17.6% | 5.1% | 0.8% | 0.3% | 0.0% | 0.0% | 1.1% |
| Cattle under 1 year of age, 2021 (N = 443) | 30.0% | 17.6% | 23.3% | 23.3% | 5.2% | 0.7% | 0.0% | 29.1% |
| Campylobacter jejuni | ||||||||
| Broilers, 2022 (N = 3252) | 63.0% | 15.2% | 12.6% | 4.0% | 2.5% | 1.9% | 0.8% | 9.2% |
| Fattening turkeys, 2022 (N = 929) | 52.0% | 17.8% | 15.2% | 5.4% | 5.1% | 3.7% | 1.0% | 15.1% |
| Fattening pigs, 2021 (N = 77) | 93.5% | 3.9% | 2.6% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Cattle under 1 year of age, 2021 (N = 1468) | 86.8% | 9.2% | 2.9% | 0.8% | 0.1% | 0.1% | 0.1% | 1.2% |
Epidemiological cut‐off.
The unexpected level of ertapenem resistance reported in Campylobacter from food‐producing animals in 2021 and 2022, as well as the ECOFF chosen for interpretation of ertapenem resistance in C. jejuni and C. coli will be further investigated in 2024 in the project CarbaCamp, a collaboration between EFSA, ECDC, EUCAST and the European Reference Laboratory for Antimicrobial Resistance (EURL‐AR). The project will address several points regarding ertapenem resistance in Campylobacter, including assessing (1) if the present ECOFF for interpretation is set correctly; (2) if ertapenem is the best carbapenem to be included in the antimicrobial test panel; (3) the effect of using different recommended test media; (4) if there are differences in the wild‐type resistance distribution between Campylobacter species and animal populations; (5) if there are emerging clones with ertapenem resistance; (6) if a resistance mechanism can be identified being responsible for the observed results.
3.4.3. Combined resistance to ciprofloxacin and erythromycin
Since resistance to fluoroquinolones is common in C. jejuni and C. coli, macrolides are recognised as critically important antimicrobials (CIAs) for the treatment of Campylobacter infections in humans. Therefore, the occurrence of combined resistance to ciprofloxacin and erythromycin in Campylobacter spp. from food‐producing animals is of great importance to public health, since it might hamper the treatment of human campylobacteriosis (Friedrich, 2019; WHO, 2019).
Overall combined resistance to these antimicrobials was lower in C. jejuni isolates than in C. coli isolates for all animal species tested (Table 9; Figure 18). Very low to low average levels of combined resistance to ciprofloxacin and erythromycin were reported in C. jejuni isolates from cattle under 1 year of age (0.8%) and fattening pigs (1.7%) in 2021 (Table 9; Figure 18; see also Annex B, Tables 33, 35), and from broilers (1.1%) and fattening turkeys (1.6%) in 2022 (Table 9; Figure 18; see also Annex B, Tables 29, 31). The highest average levels of combined resistance were reported in C. coli isolates from cattle under 1 year of age (32.7%) in 2021 and from fattening turkeys (17.5%) in 2022, followed by fattening pigs (9.1%) in 2021 and broilers (8.2%) in 2022 (Table 9; Figure 18; see also Annex B, Tables 30, 32, 34, 36). The average occurrence of combined resistance to ciprofloxacin and erythromycin observed in C. coli isolates from broilers notably increased from 4.1% in 2020 to 8.2% in 2022, whereas a decrease was observed in C. coli isolates from fattening turkeys, from 21.2% in 2020 to 17.4% in 2022. This comparison shall be interpreted with caution, since the number of reporting countries differed considerably from 2020 to 2022 (data from seven and from 24 MSs and the United Kingdom (Northern Ireland) for broilers, and from three and 11 MSs for fattening turkeys, in 2020 and 2022, respectively).
Combined resistance to ciprofloxacin and erythromycin in C. jejuni isolated from broilers was detected in five MSs (Belgium, Bulgaria, Czechia, Portugal and Romania) out of 26 MSs and the United Kingdom (Northern Ireland) reporting data in 2022, ranging from 0.5% to 8.8%, with the highest level of combined resistance reported by Romania (Figure 19A; Annex B, Table 29). Co‐resistance to ciprofloxacin and erythromycin detected in C. coli isolates from broilers was reported from 12 MSs out of 25 countries reporting data in 2022 (Belgium, Bulgaria, Cyprus, France, Germany, Ireland, Italy, Malta, Netherlands, Portugal, Romania, Spain and United Kingdom (Northern Ireland)). The levels of combined resistance ranged from 1.2% reported by Cyprus to 36.1% reported by Portugal (Figure 20A; Annex B, Table 30).
FIGURE 19.
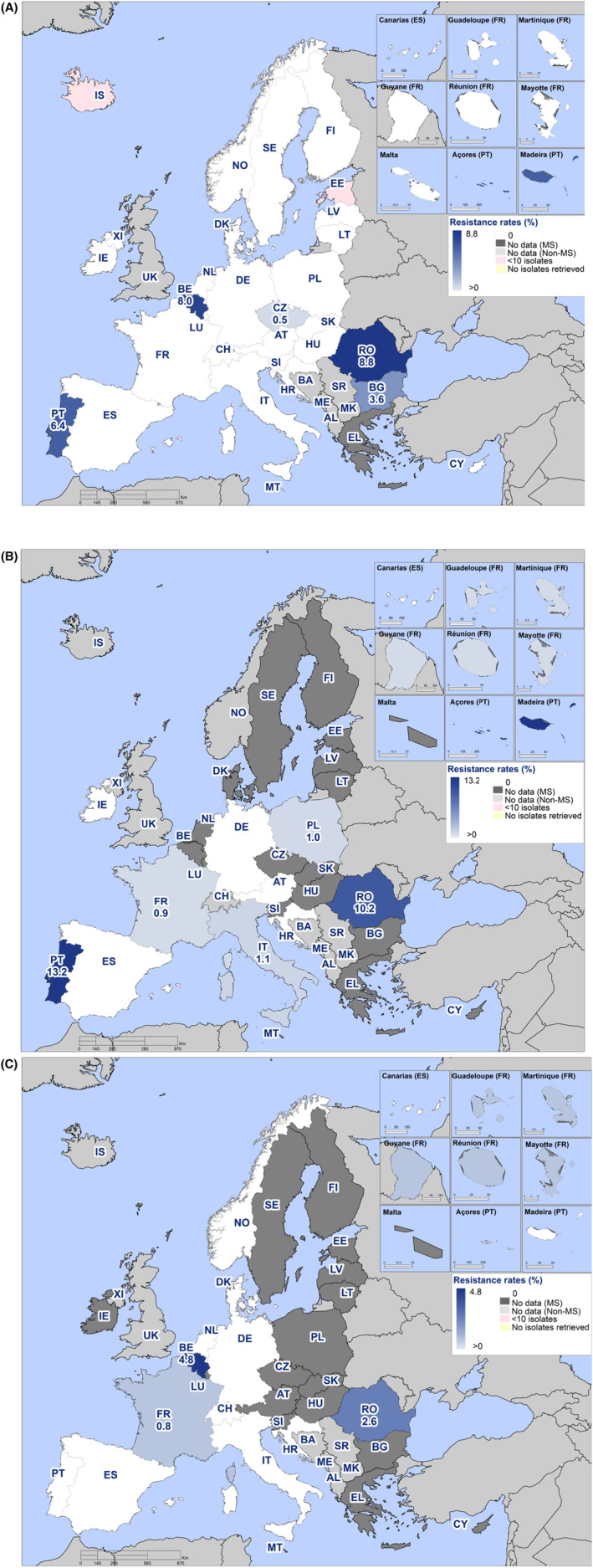
Spatial distribution of combined resistance to ciprofloxacin and erythromycin in Campylobacter jejuni isolates from (A) broilers (26 MSs, the United Kingdom (Northern Ireland) and three non‐MSs, 2022); (B) fattening turkeys (10 EU MSs, 2022); (C) cattle under 1 year of age (10 EU MSs and two non‐MSs, 2021).
Note: Maps are presented only when at least four Member States reported data. Countries that reported less than 10 isolates are displayed in pink (and their % of resistance is not shown).
FIGURE 20.
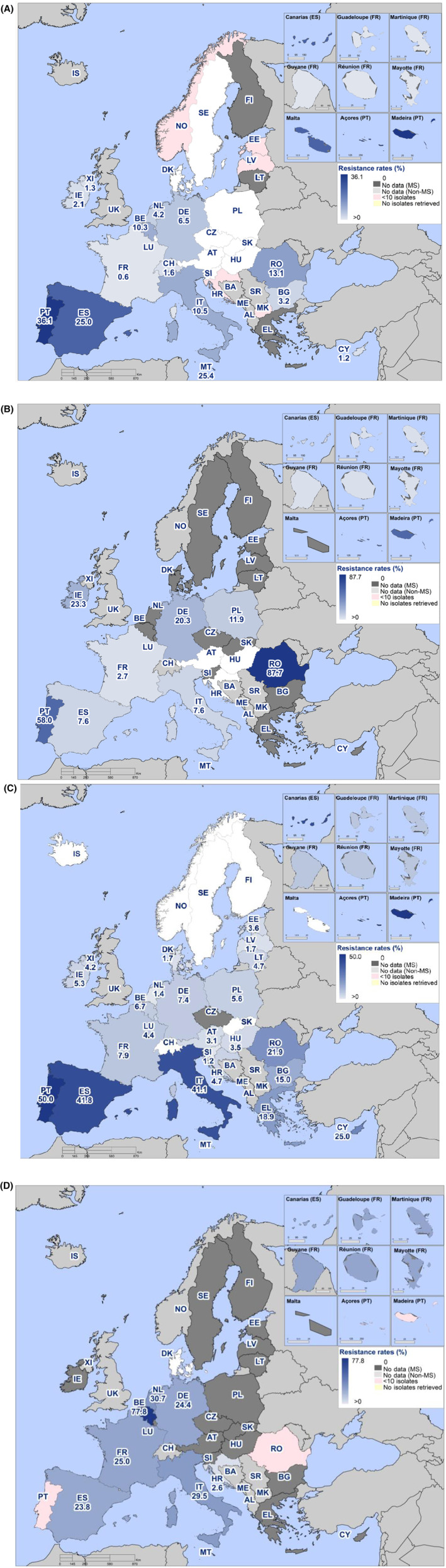
Spatial distribution of combined resistance to ciprofloxacin and erythromycin in Campylobacter coli isolates from (A) broilers (24 MSs, the United Kingdom (Northern Ireland) and three non‐MS, 2022); (B) fattening turkeys (11 EU MSs, 2022); (C) fattening pigs (26 EU MSs, the United Kingdom (Northern Ireland), and three non‐MSs, 2021); and (D) cattle under 1 year of age (10 EU MSs, 2021).
Note: Maps are presented only when at least four Member States reported data. Countries that reported less than 10 isolates are displayed in pink (and their % of resistance is not shown).
Combined resistance to ciprofloxacin and erythromycin in C. jejuni from fattening turkeys was detected in five MSs (France, Italy, Poland, Portugal and Romania), out of 10 reporting MSs in 2022 (Figure 19B; Annex B, Table 31), ranging from 1.0% to 13.2%, with the highest level of combined resistance reported by Portugal. Combined resistance to ciprofloxacin and erythromycin in C. coli isolates from fattening turkeys in 2022 was detected by eight MSs (France, Germany, Ireland, Italy, Poland, Portugal, Romania and Spain) out of 11 MSs reporting 2022 data, with levels of co‐resistance ranging from 3.6% in France, up to 58.0% and 87.7% in Portugal and Romania, respectively (Figure 20B, Annex B, Table 32).
Data on C. jejuni isolated from fattening pigs were reported by 12 MSs and one non‐MS in 2021 (Annex B, Table 33), however only very few isolates were reported by each country (ranging from one to six isolates per country, except for Malta who reported 27 isolates). Only one country (Lithuania) reported co‐resistance to ciprofloxacin and erythromycin in C. jejuni in 16.7% of pig isolates (N = 6) in 2021. Data on combined resistance to ciprofloxacin and erythromycin in C. coli isolated from fattening pigs was reported by 26 MSs (all except Czechia), the United Kingdom (Northern Ireland) and three non‐MSs (Iceland, Norway and Switzerland) (Figure 20C; Annex B, Table 34). All MSs detected combined resistance to these antimicrobials, except Finland, Malta, Slovakia and Sweden. The combined resistance levels ranged from 1.2% reported by Slovenia to 50% reported by Portugal. No combined resistance in C. coli isolated from fattening pigs was reported by the three non‐MSs who submitted data.
Data on combined resistance to both ciprofloxacin and erythromycin in C. jejuni isolated from cattle under 1 year of age were reported by 10 MSs and two non‐MSs in 2021 (Figure 19C; Annex B, Table 35). Three out of the 12 reporting countries found combined resistance to these antimicrobials, with levels ranging from 0.8% reported by France to 2.6% reported by Romania and 4.8% reported by Belgium. Data on combined resistance to both ciprofloxacin and erythromycin in C. coli isolated from cattle under 1 year of age were reported by 10 MSs in 2021 (Figure 20D; Annex B, Table 36). Seven MSs detected combined resistance to both antimicrobials in C. coli isolates, with combined resistance levels ranging from 2.6% reported by Croatia to 77.8% reported by Belgium.
The combined resistance to ciprofloxacin and erythromycin in C. jejuni obtained from broilers and fattening turkeys tested in 2022 increased from very low to low in comparison with the levels reported from these animal species in 2020. The average levels of combined resistance increased from 0.7% to 1.1% in C. jejuni from broilers (data from 27 MSs in 2020 and 26 MSs and the United Kingdom (Northern Ireland) in 2022) and from 0.8% to 1.6% in C. jejuni from fattening turkeys (data from nine and 10 MSs in 2020 and 2022, respectively). The combined resistance to ciprofloxacin and erythromycin in C. coli isolated from broilers and fattening turkeys also changed from 2020 to 2022, especially in C. coli isolates from broilers, which could be partly explained by the higher number of reporting countries and isolates in 2022 (due to sampling frequency and changes in legislative requirement based on Commission Implementing Decision (EU) 2020/1729). The average levels of combined resistance to ciprofloxacin and erythromycin in C. coli isolates from broilers increased from 4.1% in 2020 to 8.2% in 2022 (data from seven MSs and from 24 MSs and the United Kingdom (Northern Ireland) in 2020 and 2022, respectively). The average levels of combined resistance to ciprofloxacin and erythromycin in C. coli isolates from fattening turkeys decreased from 21.2% in 2020 to 17.5% in 2022.
The spatial distribution of combined resistance to both ciprofloxacin and erythromycin in C. jejuni and C. coli isolates from different animal categories is presented in Figures 19, 20 when more than four countries have reported data. Detailed data on combined resistance to ciprofloxacin and erythromycin in C. jejuni and C. coli isolates from food‐producing animals are presented in Annex B (Tables 29–36).
3.4.4. Prevalence of resistance to selected antimicrobials in C. jejuni and C. coli from poultry
The prevalence of resistance to selected antimicrobials in C. jejuni and C. coli from broilers and fattening turkeys has been estimated at country level as the product of the prevalence of C. jejuni or C. coli in caecal samples from the given animal species (Annex B, Tables 21–24) and the percent occurrence of resistance in the corresponding isolates (Annex B, Tables 17–20). Monitoring the prevalence of resistant Campylobacter enables to address together both evolving temporal trends in the prevalence of C. jejuni and C. coli and the occurrence of resistance in both species recovered from the different food‐producing animals, through a unique indicator. This indicator is primarily intended to follow up trends over time at the country level. The country level estimates of the prevalence of resistance to selected antimicrobials in C. jejuni and C. coli from caecal samples of broilers and fattening turkeys are presented in Tables 25–28 of Annex B.
The isolation method for Campylobacter has been harmonised across the EU in accordance with the protocol of the EURL for Campylobacter . Therefore, the prevalence of C. coli can be considered comparable between reporting countries. Similarly, the harmonised implementation of the antimicrobial susceptibility testing in C. jejuni and C. coli isolates allows comparability of the estimates of prevalence of resistance in those species of Campylobacter from all monitored food‐producing animals across the EU. In 2022 a number of MSs (seven for broilers and two for fattening turkeys) did not report prevalence of Campylobacter at species level, i.e. separately for C. jejuni and for C. coli, thus prevalence of resistance could not be estimated for those countries, which have consequently been excluded from the analysis.
Although the sampling design for the monitoring of antimicrobial resistance in C. jejuni and C. coli from food‐producing animals is harmonised according to the Commission Implementing Decision (EU) 2020/1729, still differences in the intensity of sampling effort exist across the EU MSs, as shown in the numbers of tested samples included in Tables 21–24 of Annex B. To account for between‐country variation in the intensity of sampling effort, 95% confidence intervals have been calculated together with the estimated prevalence of resistance, as presented in Tables 25–28 of Annex B, as well as in Figures 21–22, 23, 24.
FIGURE 21.
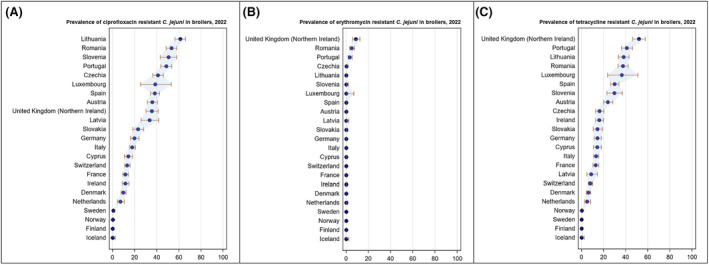
Prevalence of resistance to ciprofloxacin (A), erythromycin (B), tetracycline (C) and related 95% confidence intervals in Campylobacter jejuni from broilers, per reporting country, 2022.
Note: Only countries reporting prevalence of Campylobacter at species level, i.e. prevalence of C. jejuni in broilers were included in the analysis.
FIGURE 22.
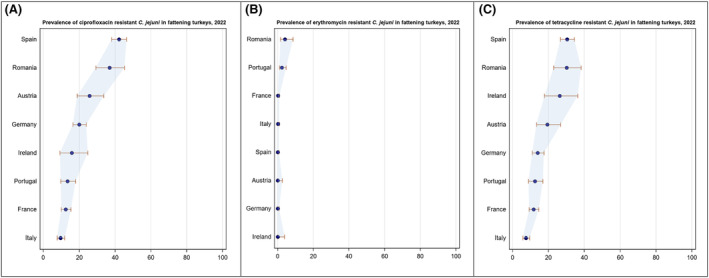
Prevalence of resistance to ciprofloxacin (A), erythromycin (B), tetracycline (C) and related 95% confidence intervals in Campylobacter jejuni from fattening turkeys, per reporting country, 2022.
Note: Only countries reporting prevalence of Campylobacter at species level, i.e. prevalence of C. jejuni in fattening turkeys were included in the analysis.
FIGURE 23.
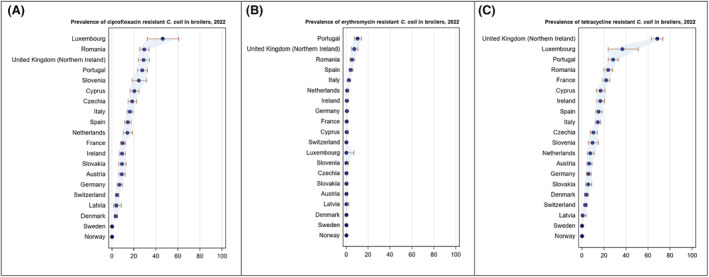
Prevalence of resistance to ciprofloxacin (A), erythromycin (B), tetracycline (C) and related 95% confidence intervals in Campylobacter coli from broilers, per reporting country, 2022.
Note: Only countries reporting prevalence of Campylobacter at species level, i.e. prevalence of C. coli in broilers were included in the analysis. Republic of North Macedonia (not included in this graph) reported data only on five samples.
FIGURE 24.
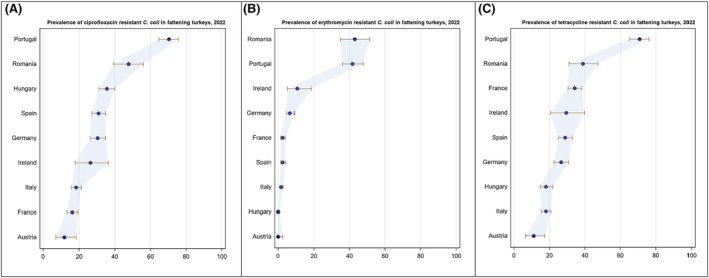
Prevalence of resistance to ciprofloxacin (A), erythromycin (B), tetracycline (C) and related 95% confidence intervals in Campylobacter coli from fattening turkeys, per reporting country, 2022.
Note: Only countries reporting prevalence of Campylobacter at species level, i.e. prevalence of C. coli in fattening turkeys were included in the analysis.
The country level prevalence of resistance to ciprofloxacin, erythromycin and tetracycline in C. jejuni isolates is presented in Figure 21 for broilers and Figure 22 for fattening turkeys, together with the 95% confidence intervals that provide an indication of the uncertainty around the point prevalence estimates.
Between‐country variability in the levels of prevalence of resistance in C. jejuni from broilers was from rare to extremely high for ciprofloxacin resistance (ranging from 0% to 61.2%; Figure 21A) and from rare to very high for tetracycline resistance (ranging from 0.0% to 51.9%; Figure 21C). Erythromycin‐resistant C. jejuni from broilers were detected in seven out of 23 reporting countries (19 MSs, the United Kingdom [Northern Ireland] and three non‐MSs), ranging from 0.3% to 8.7% (Figure 21B). Noteworthy, in one reporting country (United Kingdom [Northern Ireland]) C. jejuni from broilers also presented resistance to gentamicin (prevalence of 1.7%) (Annex B, Table 25).
Between‐country variability from low to very high was observed in the prevalence of resistance to ciprofloxacin (ranging from 9.5% to 42.1%; Figure 22A) and tetracycline (ranging from 7.4% to 30.5%; Figure 22C) in C. jejuni from fattening turkeys. Prevalence of erythromycin resistance in C. jejuni from fattening turkeys was detected in four out of eight reporting MSs, with values ranging from very low to low (0.1%–4.0%; Figure 22B). A wider between‐country variability was found for the prevalence of resistance to ertapenem in C. jejuni from fattening turkeys, ranging from 1.1% to 10.7% among the eight reporting MSs (Annex B, Table 26).
The country‐level prevalence of resistance to ciprofloxacin, erythromycin and tetracycline in C. coli isolates is presented in Figure 23 for broilers and Figure 24 for fattening turkeys, also including 95% confidence intervals. The prevalence of resistance in C. coli from broilers presented high between‐country variability among the 21 reporting countries (17 MSs, United Kingdom (Northern Ireland) and three non‐MSs) for ciprofloxacin resistance, tetracycline resistance and ertapenem resistance, ranging from rare to very high or extremely high (0%–46.2% for ciprofloxacin, 0%–68.3% for tetracycline and 0%–26.9% for ertapenem) (Figure 23A, C and Annex B, Table 27). The lowest between‐country variability for prevalence of resistance in C. coli from broilers was observed for gentamicin resistance, with detection of gentamicin resistant C. coli in broilers in only six reporting countries (varying between 0.2% prevalence from Czechia and 1.2% prevalence from United Kingdom [Northern Ireland]). A similar level of between‐country variability was observed for prevalence of erythromycin resistant C. coli (range from 0% to 10.3%) from broilers (Figure 23B and Annex B, Table 27).
Similar between‐country variability from moderate to extremely high levels was observed in the prevalence of resistance to ciprofloxacin (ranging from 11.8% to 70.3%; Figure 24A) and to tetracycline (ranging from 11.1% to 70.8%; Figure 24C) in C. coli from fattening turkeys. A more limited between‐country variability and notably lower levels of prevalence of resistance to erythromycin were found in C. coli from fattening turkeys, which have been estimated to be below 10.5% for seven out of the nine MSs who reported data, while being estimated at 41.7% and 43% for two MSs (Portugal and Romania, respectively) (Figure 24B and Annex B, Table 28).
Overall, the prevalence of resistance in Campylobacter isolates from broilers and fattening turkeys in 2022 presented a high between‐country variability for tetracycline and ciprofloxacin resistance, and a low to moderate between‐country variability for resistance to erythromycin, gentamicin, chloramphenicol and ertapenem, with a few exceptions. Notably, the prevalence of resistance to ertapenem varied moderately and greatly between countries in C. jejuni isolates from fattening turkeys and C. coli isolates from broilers, respectively. Furthermore, in C. coli from broilers, there was a moderate between‐country variability in the prevalence of resistance to erythromycin and gentamicin.
3.4.5. Complete susceptibility and multidrug resistance
Analyses of complete susceptibility (CS) and multidrug resistance (MDR) focus on critically important antimicrobials in humans, and the target substances were agreed by EFSA and ECDC to include ciprofloxacin (class: fluoroquinolones), erythromycin (class: macrolides), gentamicin (class: aminoglycosides) and tetracycline. As the aim of such analyses is to compare CS and MDR in animals and humans, the related findings are only presented in Section 3.5 on the comparison of human and animal data.
Detailed data on complete susceptibility and multidrug resistance in C. jejuni and C. coli isolates from different animal categories and in the different reporting countries are presented in Annex B (Tables 29–36).
3.4.6. Temporal trends in resistance
Evaluation of temporal trends in resistance to selected antimicrobials in Campylobacter isolates recovered from food‐producing animals to ciprofloxacin, erythromycin and tetracycline was performed for countries reporting data in the context of Commission Implementing Decision (EU) 2020/1729 for at least 3 years (three data points) separated by not more than a one‐year gap between them, within the period 2014–2022, and with a minimum of 10 isolates per data point The updated criteria of only allowing the inclusion of data reported in accordance with Commission Implementing Decision (EU) 2020/1729, and from 2014 onwards, differs from the corresponding criteria applied in previous reports. Thus, some of the trends here reported may differ from the ones presented previously, due to this change in criteria for data inclusion. When interpreting the results, it is important to note that trend analyses may be driven by particularly high or low levels of resistance reported in one or few data points leading to unexpected findings (e.g. detection of significant increasing or decreasing trends where the observed data do not seem to show any clear trend over the entire period). It is relevant to note that between‐year oscillations in the occurrence of resistance may not be captured in the evaluation of the trend for the entire period (2014–2020) and that very recent decreasing or increasing trends may therefore be masked by the overall trend. Moreover, trend results based on very few data points shall be interpreted with caution, and more data and further analyses will be needed in the future for a more robust evaluation.
Temporal trends in resistance in C. jejuni and C. coli isolates from broilers
The results from the analysis of temporal trends in resistance in C. jejuni isolated from broilers over the period 2014–2022 were obtained using data from 23 reporting MSs and three non‐MSs (Figure 25; Table 10; see also Annex B, Table 37).
FIGURE 25.
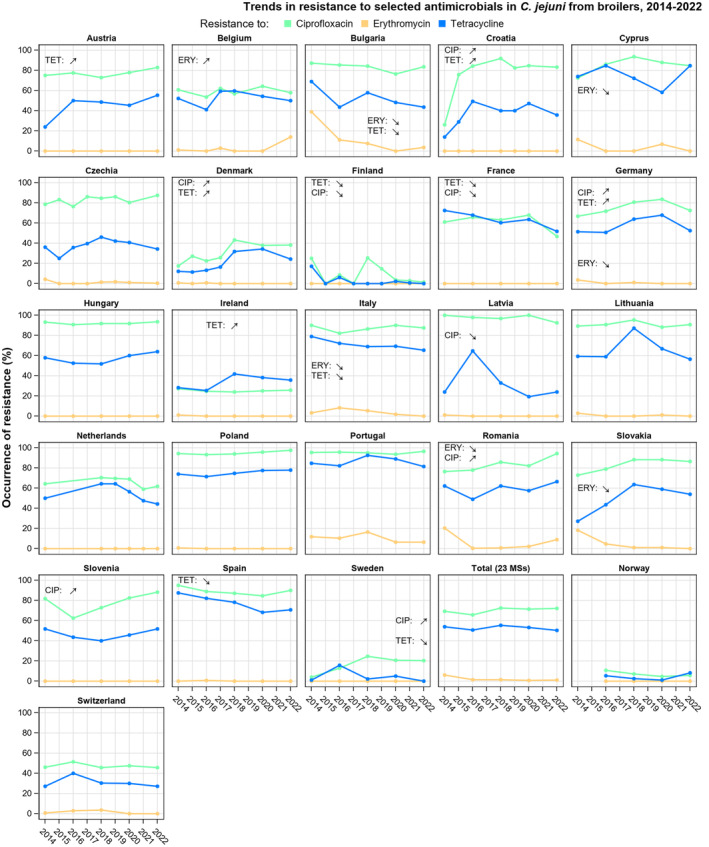
Trends in ciprofloxacin (CIP), erythromycin (ERY) and tetracycline (TET) resistance in Campylobacter jejuni from broilers, 2014–2022.
Note: Only countries that reported data fulfilling all inclusion criteria explained in the text are shown. Overall temporal trend (shown in box 'Total (23 MSs)') is presented only for Member States and for even years, when the monitoring of antimicrobial resistance in poultry population in EU is mandatory according to Decision (EU) 2020/1729.
TABLE 10.
Number of countries with significantly increasing or decreasing trends in resistance to selected antimicrobials for Campylobacter jejuni and C. coli from humans, 2013–2022, and broilers, fattening turkeys and fattening pigs, 2014–2022.
| Campylobacter species | Origin | Ciprofloxacin | Erythromycin | Tetracycline | |||
|---|---|---|---|---|---|---|---|
| Increase | Decrease | Increase | Decrease | Increase | Decrease | ||
| C. jejuni | Humans (19 MSs +2 non‐MSs) | 13 (AT, BG, CY, DK, EE, FR, IT, LT, MT, NL, PL, SI, SK) | 3 (ES, FI, NO) | 1 (ES) | 7 (DK, FI, IT, MT, NO, PT, SI) | 6 (AT, DK, EE, NL, SI, SK) | 6 (ES, FI, FR, LT, NO, PT) |
| Broilers (23 MSs + 3 non‐MSs) | 6 (DK, DE, HR, RO, SI, SE) | 3 (FI, FR, LV) | – | 6 (BG, CY, DE, IT, RO, SK) | 5 (AT, DK, DE, HR, IE) | 7 (BG, FI, FR, EL, IT, ES, SE) | |
| Fattening Turkeys (8 MSs) | 1 (PL) | 1 (PT) | – | 3 (DE, IT, ES) | – | 3 (FR, DE, ES) | |
| C. coli | Humans (13 MSs) | 2 (SI, SK) | 1 (FR) | – | 6 (EE, ES, FR MT, PT, SK) | 3 (FR, NL, SK) | 1 (FI) |
| Broilers (3 MSs) | 1 (NL) | – | – | – | 1 (CZ) | 1 (SI) | |
| Fattening Pigs (5 MSs + 2 non‐MS) | 1 (DE) | – | – | 2 (ES, CH) | 1 (EE) | 2 (ES, SE) | |
Abbreviations: AT, Austria; BG, Bulgaria; CH, Switzerland; CY, Cyprus; DE, Germany; DK, Denmark; EE, Estonia; EL, Greece; ES, Spain; FI, Finland; FR, France; HR, Croatia; IE, Ireland; IT, Italy; LT, Lithuania; LV, Latvia; MT, Malta; MSs, Member States; NL, Netherlands; NO, Norway; PL, Poland; PT, Portugal; RO, Romania; SE, Sweden; SI, Slovenia; SK, Slovakia.
The analysis of temporal trends in resistance to ciprofloxacin in C. jejuni from broilers indicated that, over the period 2014–2022, a significant increase was reported in six MSs (Croatia, Denmark, Germany, Romania, Slovenia and Sweden). On the other hand, a significant decrease in resistance to ciprofloxacin in C. jejuni from broilers was detected in Finland, France and Latvia. A significant decrease in resistance to erythromycin was detected in six MSs (Bulgaria, Cyprus, Germany, Italy, Romania and Slovakia), while a significant decrease in resistance to tetracycline was seen in seven MSs (Bulgaria, Finland, France, Greece, Italy, Spain and Sweden). Interestingly, a significant increase in resistance was detected for tetracycline in five MSs (Austria, Croatia, Denmark, Germany and Ireland).
The results from the analysis of temporal trends in resistance in C. coli from broilers over the period 2014–2022 were obtained using data from only three MSs (Czechia, Netherlands and Slovenia) who reported data fulfilling the criteria to be included in the analysis (Table 10; see also Annex B, Figure 1A and Table 37). Three significant temporal trends were identified in C. coli isolated from broilers, including the decrease in tetracycline resistance in Slovenia, the increase in tetracycline resistance in Czechia and the increase in ciprofloxacin resistance in the Netherlands.
Temporal trends in resistance in C. jejuni isolates from fattening turkeys
Temporal trends of resistance to selected antimicrobials in C. jejuni isolates from fattening turkeys during the period 2014–2022 are displayed in Figure 26 and in Table 10 (see also Annex B, Table 37). The results from the analysis of temporal trends in resistance of C. jejuni isolated from fattening turkeys were obtained using data from eight reporting MSs (Austria, France, Germany, Hungary, Italy, Poland, Portugal and Spain) (Table 10; Annex B, Table 37). Significant temporal trends of resistance in C. jejuni isolated from fattening turkeys were detected for ciprofloxacin in two MSs (increasing trend in Poland and decreasing trend in Portugal), as well as for tetracycline in three MSs (decreasing trend in France, Germany and Spain), and for erythromycin in three MSs (decreasing trend in Germany, Italy and Spain). No significant increase of resistance to tetracycline or erythromycin was detected.
FIGURE 26.
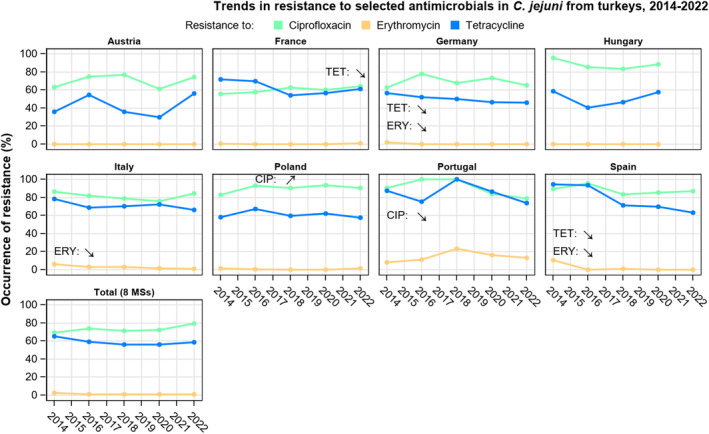
Trends in ciprofloxacin (CIP), erythromycin (ERY) and tetracycline (TET) resistance in Campylobacter jejuni from fattening turkeys, 2014–2022.
Note: Only countries that reported data fulfilling all inclusion criteria explained in the text are shown. Overall temporal trend (shown in box 'Total (8 MSs)') is presented only for Member States and for even years when the monitoring of antimicrobial resistance in EU in poultry population is mandatory according to Decision (EU) 2020/1729.
Due to the scarcity of historical data on C. coli from fattening turkeys, the temporal trends of resistance to selected antimicrobials were not analysed for C. coli in this animal population. Comparable data will be available in the coming years thanks to the implementation of the monitoring requirements laid down in Commission Implementing Decision (EU) 2020/1729.
Temporal trends in resistance in C. coli from fattening pigs
Temporal trends showing resistance to selected antimicrobials in C. coli from fattening pigs, for the period 2014–2022, are shown in Table 10 (see also in Annex B, Table 37 and Figure 1B). A significant increasing trend of resistance to ciprofloxacin was observed in a single country (Germany). No significantly increasing trend of resistance to erythromycin was observed among the reporting countries, while a significantly decreasing trend of resistance to erythromycin was found in Spain and Switzerland. A significantly increasing trend of resistance to tetracycline was observed in Estonia, while the results indicated significantly decreasing tetracycline resistance in Spain and Sweden. Due to the change in criteria for the inclusion of data in the temporal trend analysis in the present report, these results partially differ from the temporal trend results for resistance in C. coli from fattening pigs presented in the previous report.
Temporal trends in resistance in Campylobacter isolates from cattle under 1 year of age
Due to the scarcity of comparable historical data on C. jejuni and C. coli from cattle under 1 year of age, the temporal trends of resistance to selected antimicrobials were not analysed for this animal population. Comparable data will be available in the coming years thanks to the implementation of the monitoring requirements laid down in Commission Implementing Decision (EU) 2020/1729.
3.4.7. High‐level resistance to erythromycin
The distribution of MIC values related to erythromycin resistance in Campylobacter spp. recovered from caecal samples of food‐producing animals following legislative requirements in 2021 and 2022 are shown in Figure 27. It is interesting to note that even though MIC values were reported at low and moderate levels (ECOFF < MIC ≤ 128 mg/L), several isolates displayed high MIC (> 128 mg/L), particularly C. coli isolates from cattle under 1 year of age (2021) and fattening turkeys (2022).
FIGURE 27.
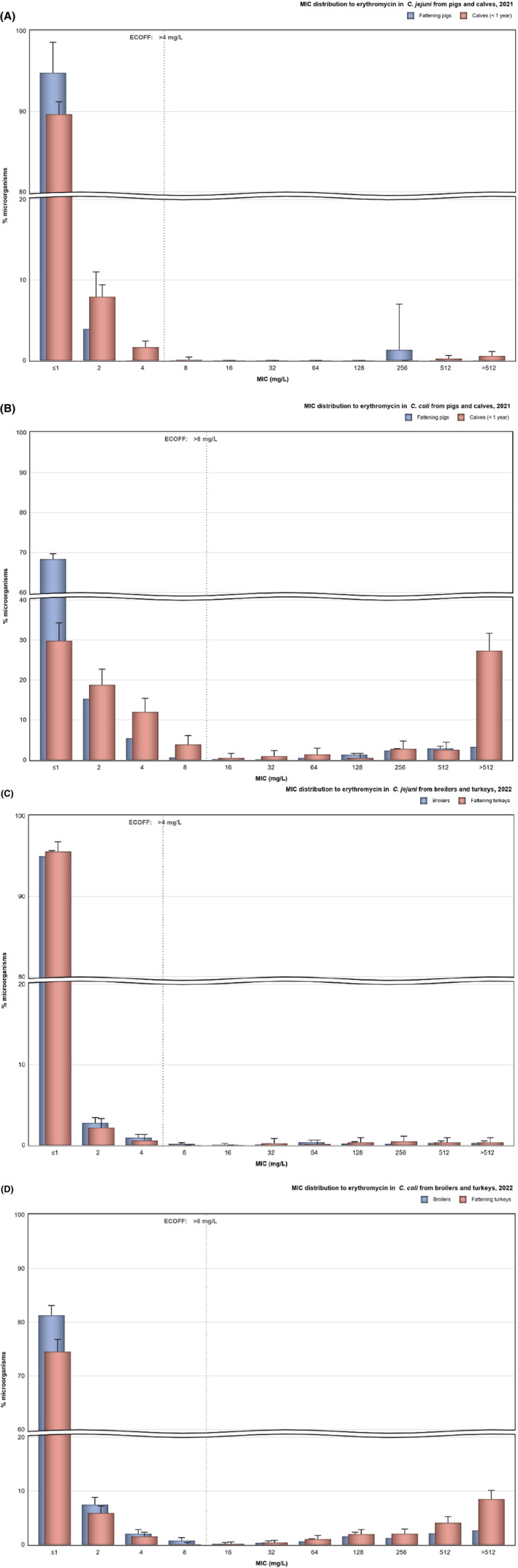
Distribution of minimum inhibitory concentration (MIC) values related to erythromycin resistance in (A) Campylobacter jejuni from fattening pigs and cattle under 1 year of age, (B) C. coli from fattening pigs and cattle under 1 year of age, (C) C. jejuni from broilers and fattening turkeys and (D) C. coli from broilers and fattening turkeys, in reporting EU MSs, the United Kingdom (Northern Ireland), and non‐EU MSs, 2021 and 2022.
Figure 28 (see also Annex B, Table 38) shows the number and proportion of erythromycin‐resistant isolates (based on ECOFF values for C. jejuni: MIC > 4 mg/L and for C. coli: MIC > 8 mg/L) reported by MSs and non‐MSs displaying resistance below or equal to 128 mg/L in comparison to those displaying high‐level erythromycin resistance (128 mg/L < MIC ≤ 512 mg/L) and the highest level of erythromycin resistance (MIC > 512 mg/L) within each of the animal categories. As shown in Figure 28, a notable proportion of erythromycin‐resistant isolates displayed high MIC values in both Campylobacter species, particularly in C. coli isolated from fattening turkeys (34.3% of isolates with MIC between 128 and 512 mg/L; 46.6% of isolates with MIC > 512 mg/L), from fattening pigs (49.4% of isolates with MIC between 128 and 512 mg/L; 31.2% of isolates with MIC > 512 mg/L) and from cattle under 1 year of age (14.6% of isolates with MIC between 128 and 512 mg/L; 76.6% of isolates with MIC > 512 mg/L). A very high level of erythromycin resistance was also observed in C. jejuni erythromycin‐resistant isolates from cattle under 1 year of age, although it is to be noted that only few C. jejuni isolates (n = 12) from cattle under 1 year of age were resistant to erythromycin.
FIGURE 28.
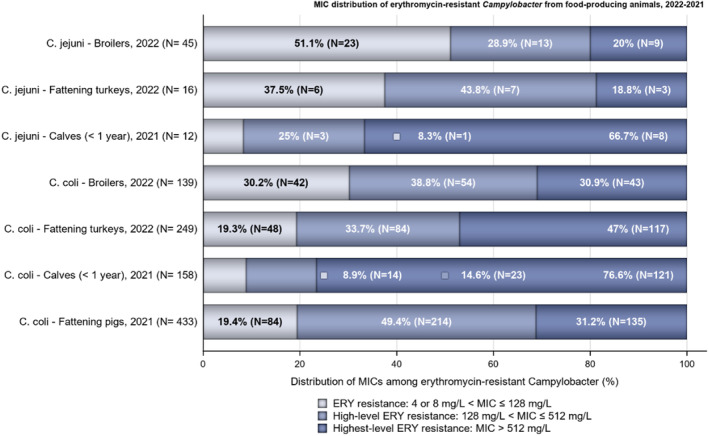
Number of isolates (and proportion) exhibiting different levels of erythromycin resistance in broilers, fattening turkeys, fattening pigs and cattle under 1 year of age in reporting EU MSs, the United Kingdom (Northern Ireland) and non‐EU MSs, 2021–2022.
C. jejuni isolates from broilers with the highest level of erythromycin resistance (MIC > 512 mg/L) were obtained in 2022 from samples from Romania (nine isolates) and Belgium (two isolates). Two of the three erythromycin highest‐level resistant C. jejuni isolates from fattening turkeys were also detected in Romania, and the other was detected in Poland. In 2022, highest‐level erythromycin‐resistant C. coli isolates from broilers were reported by 10 out of 28 reporting countries. Twenty‐four out of 52 C. coli isolates from broilers with the highest‐level of erythromycin resistance were reported together by Portugal (14 isolates) and Romania (10 isolates). Additionally, Malta and Spain reported eight and seven isolates, respectively, while six other countries reported less than five isolates. Notably, in 2022, the highest level of erythromycin resistance was detected in a high proportion of the erythromycin‐resistant C. coli isolates from fattening turkeys (117 out of a total of 249 erythromycin‐resistant isolates). The majority of the highest‐level resistant isolates were reported among four MSs, specifically Germany (20 isolates), Poland (12 isolates), Portugal (34 isolates) and Romania (46 isolates). The remaining three MSs who detected highest‐level erythromycin resistance in C. coli isolates from fattening turkeys reported between one and three isolates with MIC > 512 mg/L.
In 2021, a very high level of erythromycin resistance (MIC > 512 mg/L) was obtained in several C. coli isolates from calves reported by Belgium (54 isolates) and the Netherlands (42 isolates), followed by Italy and Germany (9 isolates each) and France (6 isolates). Several countries reported high‐level resistant isolates among the erythromycin‐resistant C. coli from fattening pigs obtained in 2021. The largest number of very high‐level resistant isolates (MIC > 512 mg/L) was reported by Germany (21 isolates), Spain (20 isolates) and Italy (18 isolates), followed by Romania (13 isolates), Ireland and Portugal (9 isolates each).
In 2021 and 2022, whole genome sequencing (WGS) information was reported at voluntary basis by certain countries to further investigate the present of genes (e.g. erm(B) gene) or gene mutations (e.g. point mutations in the 23SrRNA ribosomal gene) that may confer high resistance against erythromycin. In 2021, 37 isolates with high‐level erythromycin resistance (30 C. coli from fattening pigs from 5 MSs and 7 C. coli from cattle under 1 year of age from 2 MSs) were subject to whole genome sequencing to determine their erythromycin resistance genotype. Among C. coli isolates from fattening pigs, of the 29 isolates with MIC > 512 mg/L, 23 presented a A2075G mutation in the 23SrRNA ribosomal gene, while the ermB gene was detected in a single isolate reported by Spain. Additionally, Austria reported a single C. coli isolate from fattening pigs with MIC 512 mg/L with a A2075G mutation. Also in 2021, among the 7 C. coli isolates from cattle under 1 year of age, only the four isolates reported by Germany contained an erythromycin‐resistance genotype – the A2075G mutation.
In 2022, a total of 73 erythromycin‐resistant isolates (5 C. jejuni from broilers from a single MS, 5 C. jejuni from fattening turkeys from 2 MSs, 29 C. coli from broilers from 4 MSs and 34 C. coli from fattening turkeys from 4 MSs) were also sequenced. Among the five C. jejuni isolates from broilers and the four C. jejuni isolates from fattening turkeys reported by Portugal, the erm(B) gene was not detected (3 isolates with MIC 256 mg/L and 6 isolates with MIC 512 mg/L). Interestingly, the sequenced single C. jejuni isolate from fattening turkeys reported by Italy contained a A2075G mutation, even though it presented an MIC value of 64 mg/L. The A2075G mutation was equally detected in all 29 sequenced C. coli isolates from broilers, including three isolates with MIC 512 mg/L and eight isolates with MIC > 512 mg/L. Notably, the 18 C. coli isolates from broilers reported by Italy with the A2075G mutation presented MIC < 512 mg/L (15 isolates with MIC 128 mg/L and 3 isolates with MIC 64 mg/L). Among the 34 sequenced C. coli isolates from fattening turkeys, all but one presented the A2075G mutation, while a single isolate from Spain, with MIC > 512 mg/L presented the mutation A2074C in the same gene. Sixteen of the remaining C. coli isolates from turkeys with the A2075G mutation presented MIC < 512 mg/L (4 isolates with MIC 512 mg/L and 12 with MIC > 512 mg/L), while 17 presented lower MIC values (12 isolates from Italy with MIC 128 mg/L, and one and four isolates, from Italy and Ireland, respectively, with MIC 256 mg/L).
3.5. Comparison of human and animal data on Campylobacter spp.
In 2021–2022, quantitative human data were interpreted using EUCAST epidemiological cut‐off (ECOFF) values, where available, in the same way as for the animal data. Figure 29 illustrates the clinical breakpoints (CBPs) and ECOFFs used to interpret the MIC data reported for Campylobacter spp. from humans and animals. In the absence of CBPs from EUCAST (i.e. gentamicin), CBPs from the French Society for Microbiology (SFM) were applied (CASFM/EUCAST, 2022). For qualitative data interpreted with clinical breakpoints (S = susceptible, I = susceptible with increased exposure and R = resistant), I + R results were combined into one category. It can be observed that there is agreement across interpretive categories, with the exception of the EUCAST CBP for tetracycline in C. jejuni which is one dilution above the EUCAST ECOFF.
FIGURE 29.
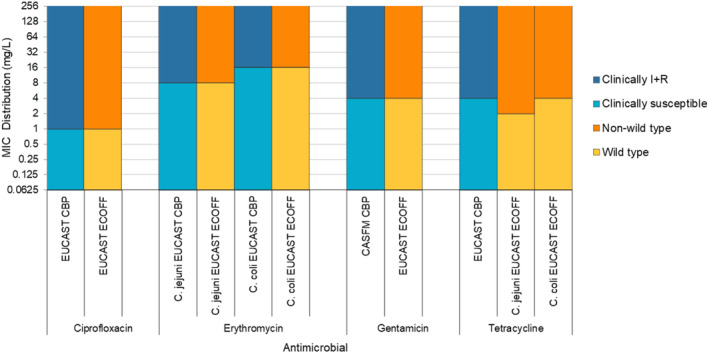
Comparison of clinical breakpoints (CBPs) and epidemiological cut‐off values (ECOFFs) used to interpret MIC data reported for Campylobacter spp. from humans and food‐producing animals.
The comparison of occurrence of resistance to selected antimicrobials and combined resistance to erythromycin and ciprofloxacin in C. jejuni and C. coli isolates between humans (2022) and food‐producing animals (2021 and 2022) is presented in Figures 30 and 31, respectively.
FIGURE 30.
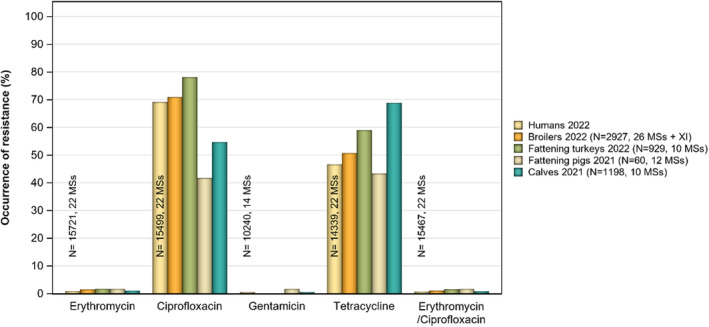
Comparison of Campylobacter jejuni occurrence of resistance between humans and food‐producing animals, EU MSs, 2021/2022.
FIGURE 31.
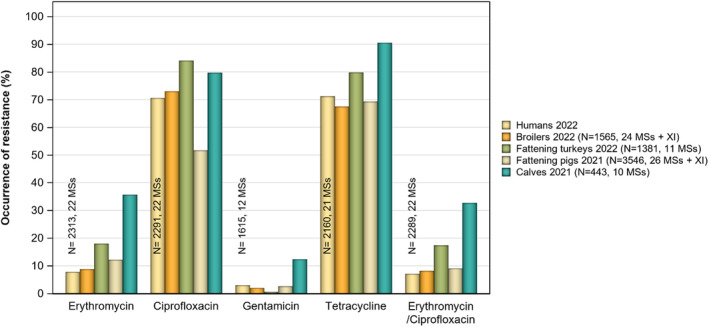
Comparison of Campylobacter coli occurrence of resistance between humans and food‐producing animals, EU MSs, 2021/2022.
High to extremely high levels of resistance to ciprofloxacin were observed both in humans (69.1% in C. jejuni by 22 MSs; 70.6% in C. coli by 17 MS) and food‐producing animals (ranging from 41.7% to 84.1%). In both humans and animals, the level of resistance were higher for C. coli than for C. jejuni. The highest levels of resistance to ciprofloxacin were observed in C. coli from fattening turkeys in 2022 (84.1%), followed by C. coli from cattle under 1 year of age in 2021 (79.7%). However, C. jejuni from broilers (70.9%) and fattening turkeys (78.2%) presented high occurrence as well. The lowest levels of resistance to ciprofloxacin were in both C. jejuni and C. coli from fattening pigs in 2021 (41.7% and 51.7%, respectively).
Overall resistance to erythromycin was reported at low levels for C. jejuni from both humans (0.9%) and food‐producing animals (range from 1% to 1.7%). Higher levels of resistance were observed in C. coli isolates from both humans (7.8%) and food‐producing animals (range from 8.8% to 35.7%). Overall, erythromycin resistance among animals was observed at the highest levels in C. coli isolates recovered from cattle under 1 year of age (35.7%) in 2021, followed by fattening turkeys (18.0%) in 2022, fattening pigs (12.2%) in 2021 and broilers (8.8%) in 2022.
Overall resistance to gentamicin was either absent or detected at very low levels among Campylobacter isolated from food‐producing animals (range from 0.0% to 0.6%), except for moderate occurrence in C. coli from cattle under 1 year of age in 2021 (12.4%). Similarly, overall resistance to gentamicin in human C. jejuni isolates was very low (0.5%), and higher levels of resistance to gentamicin were reported in C. coli isolates from humans.
The overall levels of resistance to tetracycline were high to very high in C. jejuni isolates from both humans (46.6%) and food‐producing animals (ranging from 43.3% to 68.8%). Very high to extremely high levels of resistance to tetracycline were detected in C. coli isolates from humans (71.2%) and food‐producing animals (ranging from 67.5% to 90.5%). The highest levels of resistance to tetracycline were observed in C. coli isolated from cattle under 1 year of age (90.5%) in 2021 and from fattening turkeys (79.8%) in 2022.
Overall levels of combined resistance to ciprofloxacin and erythromycin in C. jejuni were very low to low in both humans and food‐producing animals. Conversely, notable higher levels of co‐resistance were observed in C. coli from both humans and animals, with the highest levels reported in isolates from cattle under 1 year of age (32.7%) in 2021, followed by fattening turkeys (17.4%) in 2022.
The analyses of complete susceptibility (CS) and multidrug resistance (MDR) in both humans and food‐producing animals focus mainly on critically important antimicrobials in humans. For this reason, the target substances were agreed by EFSA and ECDC and include ciprofloxacin (class: fluoroquinolones), erythromycin (class: macrolides), gentamicin (class: aminoglycosides) and tetracycline. Levels of CS and MDR in Campylobacter isolates recovered from humans in 2022 and from food‐producing animals in 2021–2022 by EU MSs are displayed in Figure 32. Detailed results on occurrence of complete susceptibility and multidrug resistance in C. jejuni and C. coli isolates from humans and different animal categories and in the different reporting countries are presented in Annex B (Tables 5, 6, 29–36).
FIGURE 32.
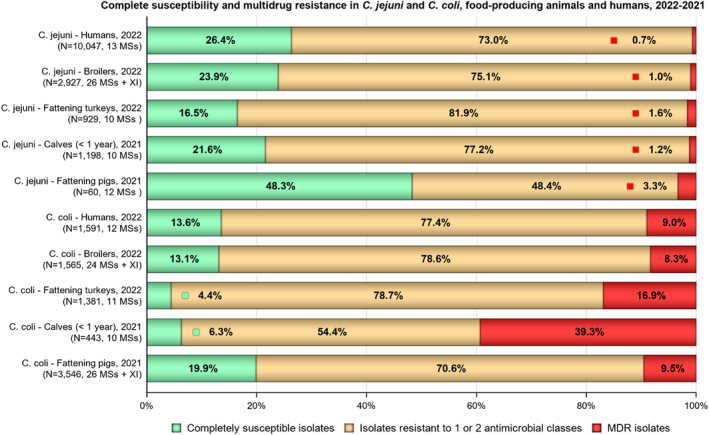
Proportion of isolates completely susceptible, resistant to one or two antimicrobial classes and multidrug resistant (MDR) among Campylobacter jejuni and C. coli from humans, broilers, fattening turkeys, fattening pigs and cattle under 1 year of age, in reporting EU MSs, 2021–2022.
Complete susceptibility to the four target antimicrobials was reported at levels of 26.4% in C. jejuni isolates from humans and in C. coli isolates at levels of 13.6% (Figure 32). Similarly, in food‐producing animals, the observed overall complete susceptibility was higher in C. jejuni than in C. coli isolates. In C. jejuni isolates, the highest levels of complete susceptibility were observed in fattening pigs (48.3%) in 2021, followed by broilers (23.9%) in 2022, cattle under 1 year of age (21.6%) in 2021 and fattening turkeys (16.5%) in 2022. Regarding C. coli, the overall complete susceptibility observed was low for isolates from fattening turkeys (4.4%) in 2022 and cattle under 1 year of age (6.3%) in 2021 while it was higher for broilers (13.1%) in 2022 and fattening pigs (19.9%) in 2021. The average complete susceptibility observed in C. jejuni and C. coli from broilers and fattening turkeys were at levels like those observed in 2020.
It is also relevant to consider that all countries used EUCAST ECOFFs (MIC > 1 mg/L) to determine resistance to tetracycline in C. jejuni isolates from animals, whereas five of 22 MSs reported SIR results using clinical breakpoints (MIC > 2 mg/L) regarding resistance to tetracycline in C. jejuni isolates from humans (Figure 29).
Multidrug resistance (MDR), defined as resistance to at least three antimicrobials among the four target substances, was reported at levels of 0.7% in C. jejuni isolates and 9.0% in C. coli isolates from humans in 2022. Similarly, in food‐producing animals, the observed overall MDR was higher in C. coli than in C. jejuni isolates (Figure 32). Multidrug resistance was observed at low levels in C. jejuni isolates from broilers (1%) and fattening turkeys (1.6%) in 2022, and from cattle under 1 year of age (1.2%) and fattening pigs (3.3%) in 2021. The highest levels of multidrug resistance were reported in C. coli isolates from cattle under 1 year of age (39.3%), followed by fattening turkeys (16.9%), fattening pigs (9.5%) and broilers (8.3%). It is relevant to note that the multidrug resistance levels observed in C. coli from broilers in 2022 were higher than the corresponding levels reported in 2020, as well as the multidrug resistance levels observed in C. jejuni from fattening turkeys, although to a lesser extent.
Interestingly, the proportions of completely susceptible‐ and MDR C. jejuni and C. coli isolates recovered from humans showed the most similar results to isolates from broilers for both Campylobacter species, followed by isolates from cattle under 1 year of age for C. jejuni and by isolates from fattening pigs for C. coli.
The most common resistance pattern among MDR isolates in C. jejuni and C. coli isolates from humans was resistance to ciprofloxacin, erythromycin and tetracycline (in 52.9% and 69.2%, respectively). In C. jejuni, the second most common pattern was resistance to gentamicin, ciprofloxacin and tetracycline (30.9%), while in C. coli it was resistance against all four classes (18.9%). In C. jejuni, resistance against all four classes was observed in 13.2% of the MDR isolates (n = 9). Among isolates recovered from food‐producing animals, the most common MDR pattern across both Campylobacter species and all monitored animal categories was resistance to ciprofloxacin, erythromycin and tetracycline (ranging between 50.0% in fattening pigs and 100% in fattening turkeys for C. jejuni and 69.5% in cattle under 1 year of age and 97.4% in fattening turkeys for C. coli). The second most common MDR pattern across both Campylobacter species and all but one monitored animal categories was resistance to gentamicin, ciprofloxacin and tetracycline. Notably, the second most common pattern in C. jejuni and C. coli isolates from broilers was however combined resistance to all four target substances (13.4% in C. coli reported by four MSs and 7.1% in C. jejuni reported by one MS). In C. coli isolates recovered from the other three animal categories, resistance to the four substances appeared as the third most common MDR pattern (0.4% in fattening turkeys, 13.8% in cattle under 1 year of age and 13.9% in fattening pigs). The least frequent MDR pattern was combined resistance to gentamicin, erythromycin and tetracycline (1.5% in C. coli from broilers and 0.6% in C. coli from cattle under 1 year of age). Overall, a higher variation in the spectra of MDR patterns was observed among C. coli than among C. jejuni recovered from food‐producing animals. Detailed results on the MDR patterns reported by individual countries, by Campylobacter species and animal category can be found in dedicated tables available in Zenodo (https://doi.org/10.5281/zenodo.10528846).
Table 10 presents countries with significantly increasing or decreasing trends in occurrence of resistance to selected antimicrobials (ciprofloxacin, erythromycin and tetracycline) from human isolates (2013–2022) and isolates from food‐producing animals (2014–2022). Factors such as time periods considered, data collected and antibiotic usage may explain the variability observed between countries.
The most frequently detected country‐level trends among C. jejuni isolates from both humans and food‐producing animals were the increase in ciprofloxacin resistance (13 MSs for humans, six MSs for broilers and one MS for fattening turkeys), and the decrease in erythromycin resistance (seven MSs for humans, six MSs for broilers and three MSs for fattening turkeys). Significant decrease of ciprofloxacin resistance or increase of erythromycin resistance were rarely detected among the reporting countries included in the temporal trend analysis. No clear pattern in tetracycline resistance trends was observed among the reporting countries. (Table 4).
3.6. Discussion
Campylobacter bacteria can be found widespread in the environment and animals, including food‐producing animals, which represents a risk for transmission to humans. Indeed, Campylobacter is an important food‐borne zoonotic agent. Moreover, Campylobacter strains resistant to antibiotics may interfere with the treatment of human campylobacteriosis (Garcia & Heredia, 2013; Moore et al., 2006) and represent an important public health concern. The main species responsible for human infections is C. jejuni, which is usually predominant in poultry, followed by C. coli (Jehanne et al., 2020). Because of this, the monitoring of AMR in Campylobacter spp. from food‐producing animals and food derived thereof focuses on these two species, as laid down in the Commission Implementing Decision (EU) 2020/1729.
In the framework of Commission Implementing Decision (EU) 2020/1729, the monitoring of AMR in C. jejuni and C. coli in food‐producing animals provides comparable data on the occurrence and enables to assess the trends of AMR in these Campylobacter species in animal production. However, it must be highlighted that the number of MSs that report data on AMR in C. jejuni and C. coli from food‐producing animals and the number of reported isolates vary considerably, also due to differences in the national production of meat thereof.
Campylobacter monitoring programmes, surveillance programmes and epidemiological studies aim to improve the knowledge in relation to the prevalence of C. jejuni and C. coli in animal production and food of animal origin. Increasing the knowledge on the occurrence of resistance in these bacterial species will contribute to a better understanding of the epidemiology of resistant Campylobacter infections in humans.
Resistance in bacteria isolated from humans has been associated with resistance in bacteria from food‐producing animals, and with antimicrobial consumption in both humans and animals. In the third joint inter‐agency JIACRA report of the three EU agencies ECDC, EFSA and EMA, providing data from the respective networks on antimicrobial consumption and resistance in isolates from humans and animals, a statistically significant association was observed between resistance to fluoroquinolones, macrolides and tetracycline in Campylobacter spp. isolates from animals and isolates from humans in the EU (ECDC, EFSA, and EMA, 2021). This finding is biologically plausible as Campylobacter in meat products is a major source of food‐borne infections in humans.
Overall, the data obtained in 2021–2022 from C. jejuni and C. coli from human and animal origins showed high to extremely high levels of resistance to fluoroquinolones (in this report represented by ciprofloxacin). Moreover, increasing trends of resistance of C. jejuni to ciprofloxacin were detected from human isolates in 13 reporting countries (between 2013 and 2022) and from isolates from broilers in six reporting countries (between 2014 and 2022). An increase in fluoroquinolone resistance in Campylobacter from human infections has indeed been observed at the global level, with evidence of zoonotic transmission of resistant strains (Inglis et al., 2021; Yang et al., 2019). High resistance levels in animal isolates point to a continued high use of fluoroquinolones in food‐producing animals and particularly in poultry production, where large numbers of animals could be affected by flock antimicrobial treatment. This might not only affect resistance levels in Campylobacter but also select for other resistant food‐borne microorganisms as well (e.g. Escherichia coli and Salmonella). The high level of ciprofloxacin resistance in Campylobacter is worrisome, since fluoroquinolones are among the most widely used antibiotics for the treatment of diarrhoea in humans (Espinoza et al., 2020). In general, antibiotic treatment of human campylobacteriosis is discouraged, since these infections tend to cause self‐limiting disease, however treatment may be necessary when patients are immunocompromised or have other co‐morbidities (Yang et al., 2019). Due to the high level of fluoroquinolone resistance in Campylobacter, the use of ciprofloxacin in the treatment of Campylobacter infections is not advised due to the risk of treatment failure, unless prior testing can confirm susceptibility. As the use of ciprofloxacin is no longer advised for the treatment of human campylobacteriosis, even low levels of resistance to other critically important antimicrobials are a cause for concern in public health.
Resistance to quinolones and fluoroquinolones has been found to be mainly due to the C257T mutation on the gyrA gene (Elhadidy et al., 2020; Espinoza et al., 2020). Modifications in the expression of the Campylobacter multidrug efflux (Cme) pump CmeABC may result in higher MIC values of various structurally unrelated antimicrobials, including fluoroquinolones, while facilitating the survival of Campylobacter in bile salts and its successful inhabitation of the host intestine (Lekshmi et al., 2023). Highly resistant isolates bearing a transferrable ‘super’ efflux pump variant of CmeABC (RE‐CmeABC) were described in China (Yao et al., 2016), and their MIC values of ciprofloxacin, along with florfenicol, chloramphenicol, erythromycin and tetracycline, were found to be increased.
Considering that fluoroquinolones are no longer an obvious option for treatment of severe human campylobacteriosis in Europe, macrolides are now the main class of antibiotics used as first‐line treatment of these infections. Resistance to erythromycin, belonging to macrolides, was either not detected or detected at very low levels in C. jejuni from humans, poultry and cattle under 1 year of age, but was higher in C. coli isolates from humans (overall, 7.8%), cattle under 1 year of age (overall, 35.7%), fattening turkeys (overall, 18.2%), fattening pigs (overall, 12.2%) and broilers (overall, 8.8%). High variability can be observed between countries, with some countries reporting high to extremely high levels of resistance to erythromycin in isolates from humans and food‐producing animals, e.g. 26.9% and 38.5% erythromycin resistance in C. coli isolates from humans reported from Portugal and Greece; between 36.1% and 66.7% erythromycin resistance in C. coli isolates from food‐producing animals from Portugal; and 77.8% resistance in C. coli isolates from cattle under 1 year of age from Belgium.
Interestingly, erythromycin resistance significantly decreased in C. jejuni from humans in seven reporting countries (over the period 2013–2022), from broilers in six MSs (over 2014–2022) and from fattening turkeys in three MSs (period 2014–2022). Regarding C. coli, decreasing trends of resistance to erythromycin were observed in six MSs for humans, and in two reporting countries for fattening pigs and three MSs for fattening turkeys (period 2014–2022). An increasing trend of resistance of C. jejuni to erythromycin has only been detected in humans in one MS (Spain), with no detection in animals. For C. coli, increasing trends of erythromycin resistance were also not detected in the periods under analysis. However, it is to be noted that a high proportion of erythromycin‐resistant isolates displayed very high MIC values (MIC > 512 mg/L), particularly in C. coli isolated from food‐producing animals, with the highest level or resistance reported in cattle under 1 year of age (76.6%). Common mechanisms of resistance to macrolides are mutations in one or several copies of the ribosomal gene 23S rRNA, such as A2074G, A2074C and A2075G (Lekshmi et al., 2023; Luangtongkum et al., 2009). In addition, the transferable erm(B) gene, encoding an rRNA methylase, usually present on multidrug resistance islands (MDRI) or plasmids in Campylobacter, may confer a high level of resistance to macrolides, lincosamides, and/or streptogramin B antibiotics (Wang et al., 2014). These MDRI also harbour the ‘super’ efflux pump RE‐CmeABC (Liu et al., 2019). Initially described in Asia, this emerging resistance mechanism has also been detected in patients and in animal isolates in Europe (Elhadidy et al., 2019; Florez‐Cuadrado et al., 2017; Jehanne et al., 2021). An increase of the tested concentrations of erythromycin in susceptibility testing (up to 512 mg/L instead of 128 mg/L) has been implemented since 2021 according to Comiision Implementing Decision (EU) 2020/1729, allowing for a better screening of isolates which may carry erm(B). The increase in the panel's concentrations enables differentiation of isolates with an MIC < 128 mg/L, which are likely to be caused by modifications of the L4 and L22 ribosomal proteins or the expression of the CmeABC efflux pump alone, from those with a higher MIC (≥ 512 mg/L) that have an erythromycin resistance phenotype more likely associated with possession of transferable erm(B), even though mutational resistance cannot be ruled out (Wang et al., 2014). Point mutations in the 23S rRNA gene may confer levels of high resistance against erythromycin similar to those observed in ermB‐mediated resistance in Campylobacter, especially in combination with the activity of the efflux pump CmeABC (Bejaoui et al., 2022; Wei & Kang, 2018; Wieczorek & Osek, 2013). The whole genome sequencing results reported by MSs for C. jejuni and C. coli isolates, mostly those highly resistant to erythromycin (MIC ≥ 512 mg/L), recovered from food‐producing animals in 2021/2022, showed detection of a mutation in the 23SrRNA gene and no detection of the erm(B) gene in 92 out of 110 isolates (91 isolates with the A2075G mutation and one isolate with the A2074C mutation). A single isolate of C. coli from fattening pigs from Spain in 2021 was reported positive to the presence of erm(B). Furthermore, for 17 out of the 110 isolates with whole genome sequencing results neither presence of erm(B) nor mutation in the 23SrRNA gene were reported.
The resistance to tetracycline ranged from moderate to extremely high levels in humans and food‐producing animals based on data reported in 2021 and 2022. Particularly high levels of resistance were reported in C. jejuni from cattle under 1 year of age in 2021 (68.8%) and for C. coli isolated from all food‐producing animals (ranging between 67.5% in broilers in 2022 and 90.5% in cattle under 1 year of age in 2021).
The prevalence of resistance to selected antimicrobials in Campylobacter isolates from food‐producing animals in 2021 and 2022 has been estimated at country level as the product between the proportion of C. jejuni or C. coli isolates showing microbiological resistance and the percentage of all caecal samples positive for the corresponding Campylobacter species. Monitoring the prevalence of resistance in Campylobacter enables to address together both evolving temporal trends in the prevalence of C. jejuni or C. coli and in the occurrence of resistance in each species in the different animal populations, through a unique indicator. This indicator is primarily intended to follow up trends over time at the country level. However, it should be noticed that various factors, such as rearing conditions, feed, climate, etc., may also affect the true prevalence of resistance. Between‐country variability from rare to very or extremely high levels was observed in the prevalence of ciprofloxacin‐resistant and tetracycline‐resistant C. jejuni and of ciprofloxacin‐resistant, tetracycline‐resistant and ertapenem‐resistant C. coli from broilers. The variability for prevalence of ciprofloxacin‐resistant and tetracycline‐resistant C. jejuni and C. coli from fattening turkeys was from low to very high and from moderate to extremely high, respectively. Interestingly, a more limited between‐country variability and notably lower levels of prevalence of resistance to erythromycin were found in C. jejuni and C. coli from broilers and fattening turkeys. However, prevalence of erythromycin resistance in C. coli still ranged between absent and moderate levels in broilers and between low in most reporting countries and high in two MSs (Portugal and Romania) in fattening turkeys. Noteworthy, one of the MSs (Portugal) with the highest prevalence of resistance to erythromycin in C. coli from fattening pigs in 2021 and fattening turkeys in 2022 also had extremely high levels of erythromycin resistance in C. coli from humans, suggesting that both pigs and turkeys could be a reservoir of erythromycin resistance for humans. The potential for zoonotic transmission of erythromycin‐resistant Campylobacter is of particular public health concern since, as commented above, macrolides, such as erythromycin and azithromycin, have become the first‐line treatment of human campylobacteriosis.
Another important aspect to highlight in this report is linked to the multidrug resistance (MDR) levels in isolates from humans and food‐producing animals. MDR levels were generally lower for C. jejuni than for C. coli isolated both from humans and from the animal species considered. MDR in human isolates tested for four antimicrobial classes (fluoroquinolones, macrolides, tetracyclines and aminoglycosides) was overall very low in C. jejuni (0.7%) and low in C. coli (9.0%). Among animal C. jejuni isolates tested for the same antimicrobial classes, MDR levels were very low for broilers and low for the remaining animal species considered, while C. coli isolates presented MDR levels varying from low in broilers and fattening pigs to moderate in fattening turkeys and high in cattle under 1 year of age. Interestingly, the comparison of the proportions of completely susceptible‐ and MDR C. jejuni and C. coli isolates recovered from humans and from the different animal species considered showed the most similar results between humans and broilers for both Campylobacter species, suggesting that broilers might be also an important reservoir for the transmission of resistant C. coli to humans. Indeed, chicken has been recently identified as the main source of C. jejuni and C. coli human campylobacteriosis in France (Jehanne et al., 2021). The implementation of harmonised EU monitoring of AMR in C. jejuni and C. coli from food‐producing animals according with Commission Implementing Decision (EU) 2020/1729, will improve our knowledge of the prevalence and occurrence of antimicrobial resistance in C. jejuni and C. coli isolated from animals in the reporting countries. Moreover, the modifications of the panel of antimicrobials and concentrations tested will enable a better detection of the emerging and threatening resistance mechanisms already mentioned (RE‐CmeABC, erm(B)) and other ones such as the cfr(C) gene, borne on a conjugative plasmid and conferring resistance to phenicols, lincosamides, pleuromutilins and oxazolidinones (Tang, Fang, et al., 2017). These mechanisms (efflux pumps) and/or their genetic support (plasmids, MDRI) confer resistance to one or several families of antimicrobials of major importance for human treatment strategies (macrolides, fluoroquinolones or aminoglycosides) and could favour co‐selection of resistant clones or plasmids.
According to Commission Implementing Decision (EU) 2020/1729, two new substances, chloramphenicol and ertapenem, were added to the panel of antimicrobials and related findings were reported for the first time mandatorily for poultry isolates in 2022, and for isolates from fattening pigs and cattle under 1 year of age in 2021. In 2021, resistance to chloramphenicol and ertapenem in Campylobacter from fattening pigs and cattle under 1 year of age were either absent or very low, except for C. coli from cattle under 1 year of age, which presented low resistance to chloramphenicol (3.4%) and high resistance to ertapenem (29.1%). In 2022, chloramphenicol resistance varied between absent and very low among both C. jejuni and C. coli from broilers and fattening turkeys. However, the occurrence of ertapenem resistance was moderate in C. jejuni from broilers and fattening turkeys, high in C. coli from broilers and very high in C. coli from fattening turkeys. These findings are of public health concern as carbapenems, together with aminoglycosides, are the antimicrobial classes recommended for treatment of invasive Campylobacter infections in humans (Dai et al., 2020; EFSA, 2019). However, since the epidemiological cut‐off for ertapenem used by EFSA is still under discussion as there is not yet a validated threshold for resistance to ertapenem established by EUCAST, the results presented in this report should be interpreted with caution. At present, only few comparable data on the resistance to ertapenem in C. coli from food‐producing animals are available, and therefore, further investigations are needed in the coming years. Still, there are already recent reports of occurrence of ertapenem resistance in C. coli from broilers outside of this report (Metreveli et al., 2022; Rafei et al., 2019).
Additional data will be available in the coming years because of the implementation of the harmonised AMR monitoring in accordance with Commission Implementing Decision (EU) 2020/1729 and in the context of the EU project CarbaCamp, a collaboration between EFSA, ECDC, EUCAST and EURL‐AR, which will focus on the detailed evaluation and validation of the appropriate threshold for resistance to ertapenem in both C. coli and C. jejuni from humans and from food‐producing animals.
Furthermore, WGS of isolates, particularly those with multidrug resistance, high‐level resistance to erythromycin or ciprofloxacin, or resistance to gentamicin or ertapenem is strongly encouraged to provide further evidence of involved antimicrobial resistance genes, their genetic origin (chromosome, plasmid, transposon, integrative and conjugative element, genomic islands) and their potential of horizontal transmission (Mourkas et al., 2019). WGS of isolates can also contribute to the detection of prevalent resistant lineages or subtypes (Mouftah et al., 2021; Webb et al., 2018) in the various sources and for comparison of animal and human isolates.
4. ANTIMICROBIAL RESISTANCE IN INDICATOR COMMENSAL E. COLI
The monitoring of antimicrobial resistance (AMR) in indicator commensal E. coli collected from the intestinal flora of healthy food‐producing animals and food provides information on the reservoirs of resistant bacteria that could potentially be transferred between animal populations and humans. It also provides indirect information on the reservoirs of resistance genes that could be transferred to bacteria that are pathogenic to animals and/or humans. Such monitoring has relevance to both public health and animal health. The occurrence of AMR in indicator E. coli likely depends on several factors, including the selective pressure exerted by the use of antimicrobials in food‐producing animal populations, clonal spread of resistant organisms, dissemination of genetic elements, such as plasmids, and the effects of co‐selection in bacteria exhibiting multidrug resistance (MDR).
4.1. Key findings
Resistance to ampicillin, sulfamethoxazole, trimethoprim or tetracycline was common and the median 3 level of resistance was high or very high in all animal categories in 2021/2022. Resistance to quinolones was common in broilers and fattening turkeys with very high and high median level of resistance, respectively. Resistance to other antimicrobials was less common.
Resistance to meropenem was detected in one isolate from broilers in 2021/2022. The isolate was reported by Italy and was confirmed to carry the bla OXA‐181 gene.
Large differences in the levels of resistance were observed between countries. Generally, lower levels were reported in Northern Europe, although countries in other parts of Europe also reported low levels of resistance, for example in isolates from cattle under 1 year of age.
Complete susceptibility (CS) was more common in isolates from fattening pigs and cattle under 1 year of age than in those from broilers and fattening turkeys. Conversely, multidrug resistance (MDR) was more common in isolates from broilers and turkeys than in those from pigs and cattle under 1 year of age.
Marked differences in the levels of CS and MDR were observed between countries. The antimicrobials most often represented in the patterns of MDR isolates were tetracycline, ampicillin, sulfamethoxazole, trimethoprim and additionally, quinolones in broilers and turkeys.
The key outcome indicator of complete susceptibility (KOI CS ), accounting for differences in the relative size of food‐producing animal populations in a country, varied widely between countries ranging from < 20% to > 80%. Lower KOICS were generally observed in countries in Eastern and Southern Europe and the highest in countries in the Northern part.
Resistance to highest priority critically important antimicrobials (hpCIA) in human medicine was uncommon for colistin, azithromycin and third‐generation cephalosporins (cefotaxime or ceftazidime), and median levels ranged between rare and low in all animal categories. Median levels of resistance to ciprofloxacin were low among pigs and cattle under 1 year of age but very high among broilers and high among fattening turkeys. Combined resistance to third‐generation cephalosporins and fluoroquinolones was generally uncommon in all animal categories but more common in imported fresh meat from broilers (moderate median level) and turkeys (low median level).
Statistically significant decreasing temporal trends in resistance to ampicillin, ciprofloxacin, cefotaxime, tetracycline and colistin, as well as increasing trends in CS and KOICS reveal progress towards lower levels of resistance in several countries. Improvement in the situation has been the most pronounced in broilers and fattening turkeys.
In 2022, the new requirements for the harmonised monitoring of AMR, valid since 2021, were applied for the first time on the monitoring of resistance in broilers and turkeys, as well as on imported fresh meat from these animal categories.
4.2. Data on AMR in indicator commensal E. coli addressed
Since 2014, Commission implementing decision (EU) 2013/652 and subsequent Commission implementing decision (EU) 2020/1729 have provided detailed requirements for the harmonised monitoring and reporting of AMR in zoonotic and commensal bacteria. The antimicrobial substances included in the harmonised panel for the monitoring of AMR in E. coli from food‐producing animals have provided continuity of monitoring data and epidemiological tracing of isolates with resistance patterns of interest to public health. The antimicrobials included in the panel have been selected because they are either of public health importance, of epidemiological relevance, or are commonly used in veterinary medicine.
In 2022, monitoring AMR in indicator commensal E. coli isolates recovered from caecal contents of domestically produced broilers was mandatory. Furthermore, for the MSs with consistent production of fattening turkeys over a certain tonnage per annum, monitoring of indicator E. coli was also mandatory. Additionally, for the first time in 2022, the monitoring of fresh meat from broilers and turkeys imported from third countries and sampled at border control posts (BCPs) was performed. In 2021, equivalent data from pigs, cattle under 1 year of age, as well as pig and cattle meat were similarly reported. The occurrence of resistance to third‐generation cephalosporins (cefotaxime and/or ceftazidime) among E. coli from all sample types are reported in this chapter. More in‐depth data, including prevalence and occurrence of the phenotypes suggestive of presumptive ESBL‐, AmpC‐ or CP‐producing E. coli are reported in chapter 5.
With the Commission implementing decision (EU) 2020/1729, amikacin 4 was added to the harmonised panel for the monitoring of AMR in indicator E. coli and was included in the analyses of the occurrence of resistance, complete susceptibility (CS) and multidrug resistance (MDR). The harmonised susceptibility testing of E. coli isolates derived from fresh meat imported from third countries improves our understanding of the dissemination of AMR through food and provides information that may help reduce the impact of AMR. In 2022, imported broiler and turkey meat was sampled. A limited number of countries reported data on isolates from meat samples collected at border control posts (BCPs), as only a small proportion of meat was imported into their country.
From 2021, whole genome sequencing (WGS) has been authorised as an alternate method to phenotypic testing for E. coli isolates obtained from routine monitoring that shows resistance to extended‐spectrum cephalosporins and/or carbapenems. Information on data obtained with WGS of these isolates can be found in chapter 5 Extended‐spectrum β‐lactamase (ESBL)‐, AmpC‐ and /or carbapenemase (CP)‐producing Salmonella and Escherichia coli.
4.3. AMR in poultry, porcine and cattle populations
4.3.1. Data reported
In 2022, 27 MSs, the United Kingdom (Northern Ireland) and four non‐MSs reported data on isolates from broilers, and 13 MSs and one non‐MSs reported data on isolates from fattening turkeys. For E. coli isolates recovered from fresh meat samples taken at BCPs, eight MSs provided data from broiler meat and three MSs provided data from turkey meat. Results from imported meat sampled at BCPs in 2022 are presented in a specific Textbox regarding Monitoring AMR in imported fresh meat sampled at BCP.
In 2021, 27 MSs and 4 non‐MSs reported data on indicator E. coli from pigs and 11 MSs and two non‐MSs submitted data on isolates from cattle under 1 year of age. For E. coli isolates recovered from fresh meat samples taken at BCPs, four MSs provided data from pig meat and six MSs provided data from cattle meat. Of those MSs, most submitted AMR data on fewer than 10 isolates. Results from imported meat sampled at BCPs in 2021 and 2022 are presented in a specific Textbox regarding Monitoring AMR in imported fresh meat sampled at BCP. In 2023, EFSA has publhised for the first time a story map on the monitoring of AMR in indicator commensal E. coli, where further information on the topic can be found (see text box below).
EFSA Story map on monitoring AMR in indicator commensal E. coli
The EFSA story map on antimicrobial resistance in indicator commensal E. coli is a new interactive communication tool published by EFSA in 2023, tailored to the general public, available online (here). This story map provides general information on the bacteria, why it is important to monitor resistance in indicator commensal E. coli. In addition, this story map also illustrates the activities implemented in the EU for the monitoring of antimicrobial resistance in indicator commensal E. coli in food‐producing animals, and the role of EFSA with respect to these activities. Furthermore, the story map shows the key findings of the 2021–2022 EU monitoring of occurrence of antimicrobial resistance in indicator commensal E. coli and informs on how to prevent antimicrobial resistance. Users can easily display and explore the content of the different sections in the story map, browsing the dynamic infographic, text, and plots.
EFSA dashboard on AMR in indicator commensal E. coli
The EFSA dashboard on Antimicrobial Resistance (available online here) is a graphical user interface for searching and querying the large amount of data collected each year by EFSA from EU MSs and other reporting countries based on Decision (EU) 2020/1729. In 2023, data on the occurrence of resistance and temporal trends in resistance to selected antimicrobials in E. coli from broilers, fattening turkeys, fattening pigs and cattle under 1 year of age became available to the users. Data and trends in AMR in E. coli can be displayed interactively using charts, graphs and maps in the online EFSA dashboard, which allows the user to select for reporting year, reporting country, animal population and antimicrobial substance. In this tool, the main statistics can also be viewed and downloaded in tabular format. Detailed information on the use and features of the Antimicrobial Resistance dashboard can be found in the user guide (a video embedded in the dashboard).
Summary data on the occurrence of resistance to commonly used antimicrobials in veterinary medicine 5 as well as hpCIAs 6 are described in this chapter. Additionally, results from the analysis of MDR patterns and combined resistance to ciprofloxacin and cefotaxime are reported here. Annex C presents detailed information on the occurrence of AMR, MDR and combined resistance in E. coli from broilers and turkeys (2022 data), as well as pigs and cattle (2021 data), at the MS and MS‐group level.
4.3.2. The occurrence of AMR
Under the new legislation (Commission implementing decision (EU) 2020/1729), changes were made to the ECOFFs and clinical breakpoints for several antimicrobial substances included in the harmonised panel for testing of E. coli isolates. The substances include tigecycline, where the ECOFF changed from MIC > 1 to > 0.5 mg/L and the clinical breakpoint changed from MIC > 2 to > 0.5 mg/L. Also, the ECOFF for nalidixic acid changed from > 16 to > 8 mg/L. For ciprofloxacin, the clinical breakpoint changed from > 1 to > 0.5 mg/L. Also, in 2021, a new substance, amikacin, was added to the harmonised panel with the ECOFF > 8 mg/L and the clinical breakpoint > 16 mg/L. See Appendix F – Materials and methods for more details on the changes to the harmonised panel of substances tested for E. coli isolates from the routine monitoring programme.
Resistance to ampicillin, sulfamethoxazole, trimethoprim and tetracycline was common among all four investigated animal categories, with high to very high median levels of resistance considering reporting MSs (Figure 33; Annex C). Large differences in resistance levels to ampicillin, sulfamethoxazole, trimethoprim and tetracycline between countries were, however, observed in all animal categories, ranging 0.0%–88.2%, 0.0%–74.0%, 0.0%–62.0% and 5.0%–78.8%, respectively (Figure 1; Annex C, Tables 1–4).
FIGURE 33.
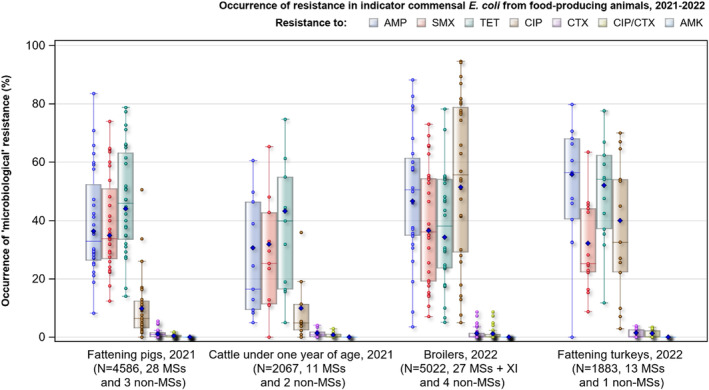
Distribution of the occurrence of resistance to selected antimicrobials in indicator commensal E. coli isolates recovered from fattening pigs and cattle under 1 year of age in 2021, and from broilers and fattening turkeys in 2022, from EU MSs, XI and non‐MSs, 2022.
Note: AMP, ampicillin; SMX, sulfamethoxazol; TET, tetracycline; CIP, ciprofloxacin; CTX, cefotaxime; CIP/CTX, combined 'microbiological' resistance to ciprofloxacin and cefotaxime; AMK, amikacin; N, total number of indicator commensal E. coli isolates reported by MSs; blue diamond shows resistance at the reporting MS group level. Horizontal lines represent media; Lower and upper box boundaries, 25th and 75th percentiles, respectively.
In poultry, resistance to ciprofloxacin and nalidixic acid was common, with very high median levels in broilers and high median levels in turkeys (Figure 33; Annex C, Tables 1, 2). In contrast, in pigs and cattle under 1 year of age, median resistance levels to both these antimicrobials were low (Figure 33; Annex C, Tables 3,4). For both ciprofloxacin and nalidixic acid resistance, there were large differences between reporting countries, ranging from 0.0% to 94.5% and 0.0% to 90.0%, respectively (Figure 33; Annex C). Furthermore, most countries reported nalidixic acid resistance at lower levels than ciprofloxacin resistance. This was most notable for pigs and cattle under 1 year of age, and at the MS‐group level, nalidixic acid resistance was approximately half that for ciprofloxacin for both these animal categories.
For chloramphenicol, the overall median resistance level for all reporting MSs was low to moderate for all animal categories (range 9.7%–15.1%). Some countries, however, reported high to very high resistance (Annex C, Tables 1–4). The overall median level of resistance to gentamicin was very low or low in all four animal categories (range 1.1%–3.7%). Some countries reported, however, moderate level of resistance and one country reported high level of resistance among isolates from broilers (Annex C, Tables 1–4). Resistance to cefotaxime, ceftazidime, colistin and azithromycin were rare, very low or low in all four animal categories (Annex C, Tables 1–4). Meropenem resistance was only detected in one isolate of indicator E. coli from broilers reported by Italy (Annex C, Tables 1–4). That isolate was confirmed to carry the bla OXA‐181 gene. Level of resistance to tigecycline were very low or low in all four animal categories (Annex C, Tables 1–4). For amikacin, the new substance included in the monitoring from 2021, the levels of resistance were very low (0.1%–0.3%), in all four animal categories (Annex C, Tables 1–4).
Resistance to highest priority critically important antimicrobials (hpCIA)
Among the antimicrobials tested in the mandatory monitoring of indicator commensal E. coli, ciprofloxacin (fluoroquinolones), cefotaxime and ceftazidime (third‐generation cephalosporins), colistin (polymyxins) and azithromycin (macrolides) are categorised by the WHO as hpCIA (WHO, 2019).
Considering all reporting MSs, overall median colistin resistance was rare in all four animal categories (Annex C, Tables 1–4), with the only exception of the data from turkeys reported by Poland and Portugal whose level of resistance was moderate. Trends were not assessed at the EU MS‐group level due to changes in the handling of data from UK. Resistance to azithromycin was generally uncommon with overall median levels of resistance in broilers (0.6%) and turkeys (0.2%) at very low level, and at a low level in pigs (1.6%) and cattle under 1 year of age (1.2%). Resistance to both colistin and azithromycin was not observed in most countries (Annex C, Tables 1–4). For both substances, countries with median levels of resistance in specific animal categories were, however, registered.
High to very high levels of resistance to fluoroquinolones/quinolones were recorded in isolates from broilers (median: ciprofloxacin 51.4%; nalidixic acid 48.0%), and high levels in isolates from turkeys (median: ciprofloxacin 40.0%; nalidixic acid 30.2%) (Figure 33; Annex C, Tables 1, 2). In contrast, overall median resistance to ciprofloxacin and nalidixic acid were reported at low levels in isolates from pigs (median 6.4% and 4.5%, respectively) and cattle under 1 year of age (median 9.9% and 5.6%, respectively) (Figure 1; Annex C, Tables 3, 4). Large variations in resistance levels were observed among reporting countries for each of the animal categories monitored. In broilers, the registered levels ranged from low to extremely high, in turkeys, from low to very high, in pigs, from rare to very high, and in cattle under 1 year of age, from rare to high. (Figure 33; Annex C, Tables 1–4). In all animal categories monitored, overall median resistance to third generation cephalosporins (cefotaxime and/or ceftazidime) was rare or very low (range 0.0–0.9%) (Figure 33; Annex C, Tables 1–4).
Combined resistance to ciprofloxacin and cefotaxime
In most reporting countries, microbiological combined resistance (i.e. defining resistance by epidemiological cut‐off values 7 ) to ciprofloxacin and cefotaxime was either not detected or detected at low to very low levels in all four animal categories monitored (Figure 34; Annex C, Tables 1–4). The overall median of microbiological combined resistance to ciprofloxacin and cefotaxime was rare in all four animal categories. Clinical combined resistance (i.e. defining resistance by clinical breakpoint) was not detected in isolates from any of the four animal categories in most countries, and where it was, the level of resistance was low to very low (Annex C, Tables 1–4). Isolates exhibiting combined resistance were more common in broilers and fattening turkeys than in pigs and cattle of less than 1 year.
FIGURE 34.
Spatial distribution of microbiological combined resistance to cefotaxime and ciprofloxacin in indicator commensal E. coli from (A) fattening pigs, 2021; (B) cattle under 1 year of age, 2021; (C) broilers, 2022; and (D) fattening turkeys, 2022, EU MSs and non‐MSs.
4.3.3. Temporal trends in resistance
Temporal trends in resistance to ampicillin, ciprofloxacin, cefotaxime and tetracyclines in indicator E. coli from pigs, cattle under 1 year of age, broilers and turkeys were assessed for countries having provided data for three or more years over the period 2010–2022. Furthermore, trends in resistance to colistin in indicator E. coli from pigs, cattle under 1 year of age, broilers and turkeys were also assessed and are presented in Appendix D – Trends in colistin resistance in indicator commensal E. coli from broilers, turkeys, fattening pigs and cattle under 1 year of age.
Ampicillin and tetracycline are the most used antimicrobials in food‐producing animals in Europe (EMA, 2023) and monitoring temporal trend in resistance is of particular relevance. Decreasing trends in resistance to those substances are believed to primarily reflect changes in usage (ECDC, EFSA, EMA, 2021). Resistance trends in the hpCIAs, ciprofloxacin and cefotaxime were also addressed, as the presence of such resistance in E. coli from food‐producing animals may impact human health. The statistical significance (p < 0.05) of trends was tested by logistic regression (see Appendix F – Materials and methods for details on methodological approach).
Sufficient data for assessing temporal trends were available from 31 countries for pigs, 29 for broilers, 12 for cattle under 1 year of age and 11 for turkeys. Thus, 332 different animal/substance combinations were available and analysed for statistical significance of trends in resistance to ampicillin, ciprofloxacin, cefotaxime and tetracyclines. For broiler and turkey data in 2022, 160 analyses were performed, and in total, 75 combinations had statistically significant (p < 0.05) decreasing trends, while 12 combinations had statistically significant increasing trends (Table 12). For pig and calf data in 2021, 172 analyses were performed, and in total, 21 combinations had statistically significant (p < 0.05) decreasing trends, while nine combinations had statistically significant increasing trends (Table 2). It is, however, important to note that in several countries, levels of resistance were stable over time at low levels, and major changes cannot be expected.
TABLE 12.
Summary of temporal trends in resistance to ampicillin (AMP), cefotaxime (CTX), ciprofloxacin (CIP) and tetracyclines (TET) in indicator commensal E. coli from broilers and fattening turkeys for the period 2009–2022, and fattening pigs and cattle under 1 year of age for the period 2009–2021, EU MSs and non‐MSs.
| Animal population | AMP | CTX | CIP | TET | Total | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ↓ | ↑ | ↔ | ↓ | ↑ | ↔ | ↓ | ↑ | ↔ | ↓ | ↑ | ↔ | ↓ | ↑ | ↔ | |
| Broilers, 2022 26 MSs, 3 non‐MSs | 13 | 4 | 13 | 17 | 1 | 11 | 13 | 5 | 11 | 11 | 2 | 16 | 54 | 12 | 51 |
| Turkeys, 2022 10 MSs, 1 non‐MSs | 6 | 0 | 5 | 3 | 0 | 8 | 6 | 0 | 5 | 6 | 0 | 5 | 21 | 0 | 23 |
| Total, 2022 | 18 | 4 | 18 | 20 | 1 | 19 | 19 | 5 | 16 | 17 | 2 | 21 | 75 | 12 | 74 |
| Pigs, 2021 27 MSs, 4 non‐MSs | 2 | 2 | 27 | 3 | 2 | 26 | 2 | 2 | 27 | 14 | 2 | 15 | 21 | 8 | 95 |
| Cattle <1 year, 2021 10 MSs, 2 non‐MSs | 1 | 0 | 11 | 0 | 0 | 12 | 2 | 0 | 10 | 5 | 1 | 6 | 8 | 1 | 45 |
| Total, 2021 | 3 | 2 | 38 | 3 | 2 | 38 | 4 | 2 | 37 | 19 | 3 | 21 | 29 | 9 | 140 |
Abbreviations: ↓, statistically significant decreasing trends; ↑, statistically significant increasing trends; ↔, statistically non‐significant trends; AMP, ampicillin; CTX, cefotaxime; CIP, ciprofloxacin; TET, Tetracycline.
Fattening pigs
Of the 31 countries reporting AMR data on indicator E. coli isolates from pigs between 2011 and 2021, a total of 21 decreasing and eight increasing trends were registered (Table 12, Figure 35). In two countries, Cyprus and the Republic of North Macedonia, decreasing trends for three (ampicillin, ciprofloxacin, tetracycline) of the four antimicrobials were seen. There was no country in which increasing trend in resistance was seen to more than one substance. Notably, tetracycline resistance decreased in 14 countries and increased in just two. In nine countries, no statistically significant changes in trends were observed. It is, however, important to note that in some countries, a favourable situation has been seen over time, and major improvements cannot be expected.
FIGURE 35.
Trends in resistance to ampicillin (AMP), cefotaxime (CTX), ciprofloxacin (CIP) and tetracyclines (TET) in indicator commensal E. coli from fattening pigs (pigs), EU MSs and non‐MSs, 2011–2021.
Cattle under 1 year of age
In the 12 countries reporting data on indicator E. coli from cattle under 1 year of age between 2011 and 2021, a total of eight decreasing and one increasing trend was observed (Table 12; Figure 36). In two countries, France and Switzerland, decreasing trends for two and three, respectively, of the four antimicrobials were registered (ciprofloxacin and tetracycline in both countries and in addition ampicillin in Switzerland). An increasing trend was detected in only one country (tetracycline in Denmark). In six countries, no statistically significant changes in trends were observed. It is, however, important to note that in some countries, a favourable situation has been seen over time, and major improvements cannot be expected.
FIGURE 36.
Trends in resistance to ampicillin (AMP), cefotaxime (CTX), ciprofloxacin (CIP) and tetracyclines (TET) in indicator commensal E. coli from cattle under 1 year of age, EU MSs and non‐MSs 2013–2021.
Broilers
In the 29 countries reporting data on isolates from broilers, 54 decreasing and 12 increasing trends were registered (Table 12; Figure 37). In seven countries (24%), decreasing trends for all four antimicrobials were detected and in four countries (14%), decreasing trends for three of the four antimicrobials were observed. In only two countries (7%), Cyprus and Norway, increasing trends for more than one substance was seen. In five countries, no statistically significant changes in trends were observed. It is, however, important to note that in some countries, a favourable situation has been seen over time, and major improvements cannot be expected.
FIGURE 37.
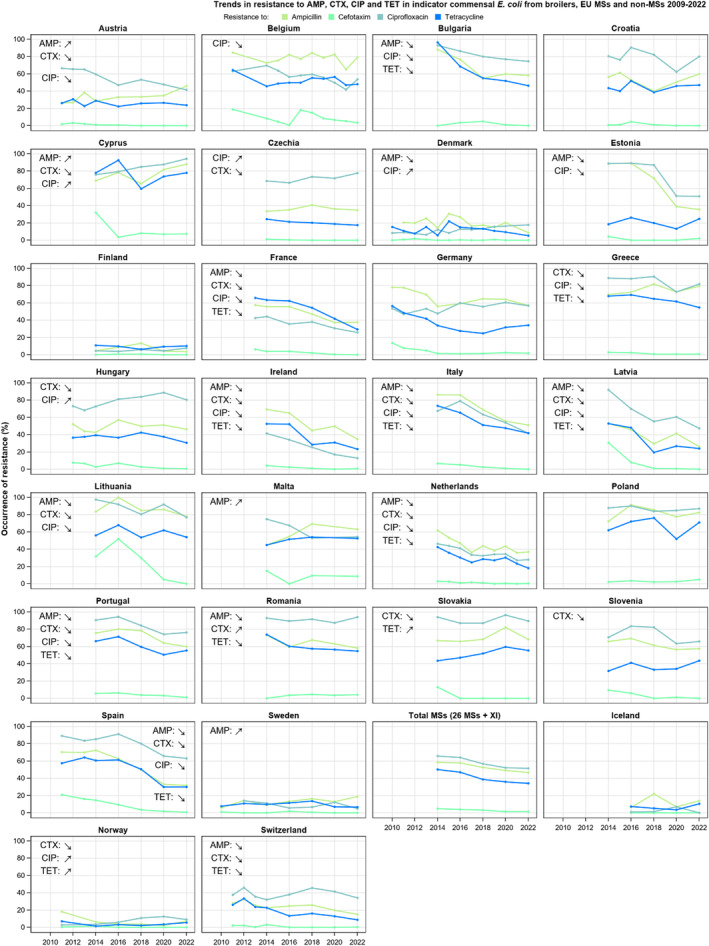
Trends in resistance to ampicillin (AMP), cefotaxime (CTX), ciprofloxacin (CIP) and tetracyclines (TET) in indicator commensal E. coli from broilers, EU MSs and non‐MSs, 2010–2022.
Fattening turkeys
In the 11 countries reporting data on isolates from fattening turkeys, there were 21 decreasing, and no increasing trends observed (Table 12; Figure 38). In one country (Spain) decreasing trends for all four antimicrobials were reported, and in three countries (France, Italy and Portugal) decreasing trends for three of the four antimicrobials (ampicillin, ciprofloxacin, tetracycline) were registered. In two countries, Poland and Norway, no statistically significant changes in trends were observed. It is, however, important to note that in some countries, a favourable situation has been seen over time, and major improvements cannot be expected.
FIGURE 38.
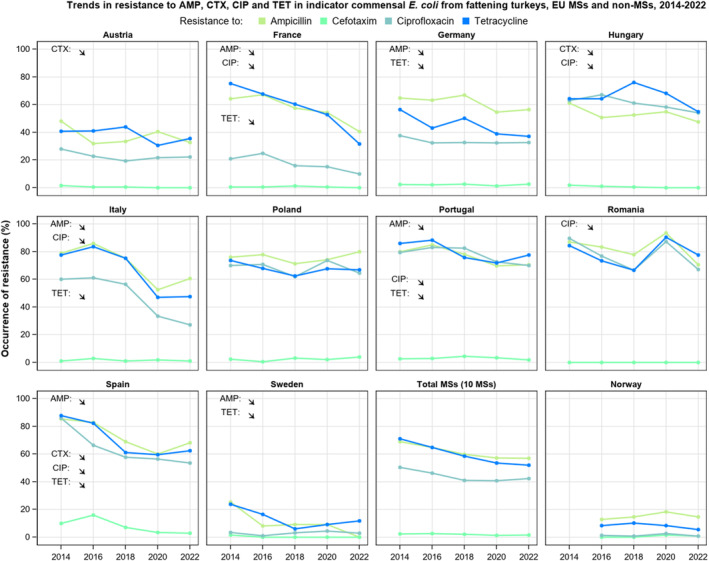
Trends in resistance to ampicillin (AMP), cefotaxime (CTX), ciprofloxacin (CIP) and tetracycline (TET) in indicator commensal E. coli from fattening turkeys (turkeys), EU MSs and non‐MSs, 2014–2022.
4.3.4. Phenotypic characterisation of third‐generation cephalosporin and carbapenem resistance
A low number of indicator E. coli isolates from caecal samples from broilers and turkeys in 2022 and from pigs and cattle under 1 year of age in 2021 were phenotypically resistant to third‐generation cephalosporins (cefotaxime and/or ceftazidime) (Table 13). In countries reporting resistant isolates, the levels of resistance were very low or low. No isolates of indicator E. coli from caecal samples from pigs, cattle under 1 year of age or turkeys reported by MSs and non‐MSs in 2021/2022 showed microbiological resistance to carbapenems (meropenem). One isolate of indicator E. coli from broilers with phenotypic resistance to meropenem was, however, reported by Italy (Annex C, Tables 1–4). That isolate was confirmed to carry the bla OXA‐181 gene.
TABLE 13.
Occurrence of resistance to third‐generation cephalosporins in indicator commensal E. coli isolates from fattening pigs (pigs), cattle under 1 year of age, broilers and fattening turkeys (turkeys). EU MSs and non‐MSs, 2021/2022.
| Animal category | No. of MSs/No. non‐MSs | N | Cefotaxime | Ceftazidime | ||
|---|---|---|---|---|---|---|
| n | % R | n | % R | |||
| Broilers, 2022 | 27 + XI/4 | 5022 | 57 | 1.1 | 56 | 1.1 |
| Turkeys, 2022 | 13/1 | 1883 | 27 | 1.4 | 20 | 1.1 |
| Pigs, 2021 | 28/3 | 4586 | 46 | 1.0 | 43 | 0.9 |
| Cattle < 1 year of age, 2021 | 11/2 | 2067 | 31 | 1.5 | 26 | 1.3 |
Abbreviations: N, total number of indicator commensal E.coli isolates reported; n, total number of indicator commensal E.coli isolates resistant isolates; % R, percentage of resistance.
Further phenotypic characterisation of all isolates resistant to third‐generation cephalosporins and/or carbapenems for identification of presumptive production of ESBL‐, AmpC‐ and/or CP‐enzymes (on a second panel) was performed. The results of these investigations are reported in chapter 5 (ESBL‐, AmpC‐ and/or CP‐producing Salmonella and Escherichia coli).
4.3.5. Multidrug resistance
Multidrug resistant isolates
Multidrug resistance (MDR) is defined as microbiological resistance to three or more antimicrobial classes of the harmonised panel tested.
In 2022, the median MDR among MSs was 38.8% among indicator E. coli isolates from broilers and 43.5% among isolates from turkeys, while in 2021, the median MDR observed among E. coli isolates from pigs was 31.2%, and 18.8% among isolates from cattle under 1 year of age. Large variations in MDR between reporting countries were observed, ranging from 3.5%–81.5% in broilers and 0.0%–75.3% in turkeys, 8.2%–78.8% in pigs and 0.0%–62.9% in cattle under 1 year of age (Annex C).
MDR patterns
A wide variety of resistance patterns were observed in MDR isolates recovered from all four animal categories. The antimicrobials most often represented in the MDR patterns of isolates from broilers were ampicillin, ciprofloxacin and nalidixic acid, often also in combination with tetracycline. In addition to this combination, many isolates were also resistant to sulfamethoxazole and trimethoprim. The addition of resistance to chloramphenicol was also common. Among isolates from turkeys, resistance to ampicillin, tetracycline and ciprofloxacin, sometimes also together with resistance against nalidixic acid, were the most common traits.
Among isolates from pigs, the most common MDR patterns were resistance to ampicillin, sulfamethoxazole and trimethoprim. This pattern was in most cases also extended with resistance to tetracycline, and sometimes also with resistance against chloramphenicol. Among isolates from cattle under 1 year of age, by far the most common MDR pattern was resistance to ampicillin, sulfamethoxazole, trimethoprim and tetracycline, sometimes also together with resistance against chloramphenicol.
None of the MDR‐resistant patterns included resistance to azithromycin or tigecycline and in only nine isolates (4 from pigs, 3 from broilers, 1 from turkey and 1 from cattle under 1 year of age) did the MDR pattern include resistance to amikacin. Resistance to colistin was a part of the MDR‐resistant pattern in a few isolates from broilers (n = 37), pigs (n = 6) and cattle under 1 year of age (n = 2), but in a higher proportion (7.3%, n = 56) of MDR isolates from turkeys.
4.3.6. Complete susceptibility
Completely susceptible isolates
Resistance can also be addressed by considering the proportion of indicator E. coli isolates exhibiting susceptibility to all the 15 antimicrobials tested in the harmonised panel, using ECOFFs for interpretation. Such isolates are here called completely susceptible isolates (CS). Considering all reporting MSs, the median CS among E. coli isolates was 18.9% in broilers, 29.4% in turkeys, 38.3% in pigs and 54.1% in cattle under 1 year of age (Annex C, Tables 1–4). For all animal categories, CS varied widely between reporting countries and ranged from 2.4%–80.0% in broilers, 8.8%–85.3% in turkeys, 6.5%–77.68% in pigs and 20.6%–95.0% in cattle under 1 year of age (Figure 39; Annex C, Tables 1–4).
FIGURE 39.
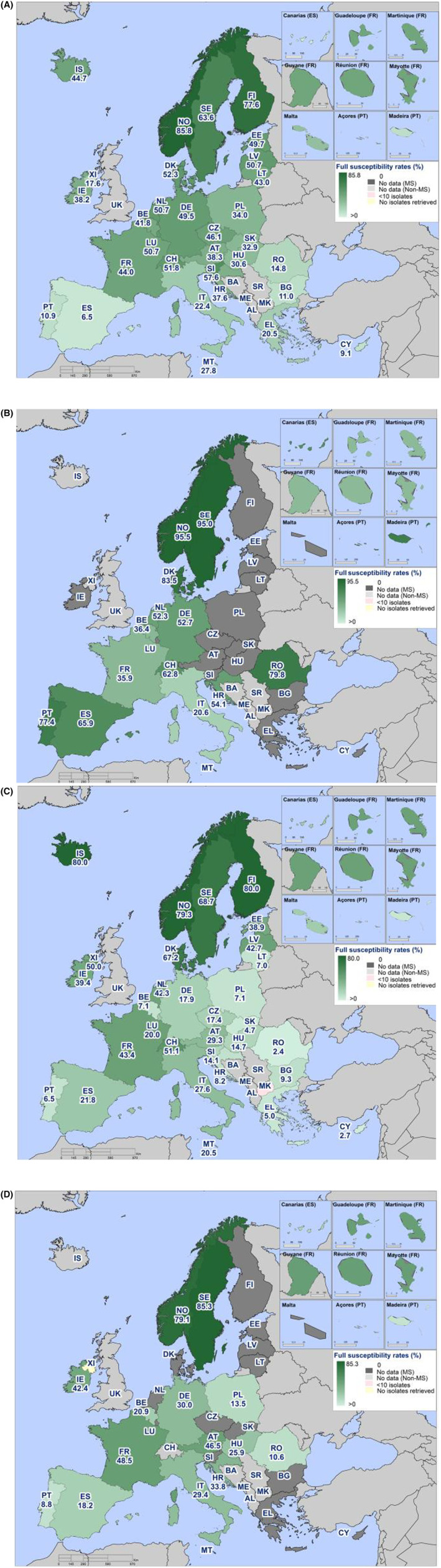
Spatial distribution of complete susceptibility to the antimicrobials tested in indicator commensal E. coli. (A) fattening pigs (pigs) 2021; (B) cattle under 1 year of age (calves) 2021; (C) broilers 2022; (D) fattening turkeys (turkeys) 2022, EU MSs and non‐EU MSs.
Typically, the highest levels of CS in all four animal categories were observed in isolates from the Nordic countries, with levels generally decreasing in a north‐to‐south gradient and, to a lesser extent, in a west‐to‐east gradient.
Trends in complete susceptibility
Trends in complete susceptibility in indicator E. coli from broilers, turkeys, pigs and cattle under 1 year of age were assessed for countries providing data for three or more years over the period 2014–2022.
For the four animal categories, a total of 26 statistically significant increasing and five decreasing trends were seen among all the reporting countries (Figures 40, 41). Among broilers, and especially turkeys, increasing trends were more common than decreasing ones. Notably, for turkeys and cattle under 1 year of age no countries reported decreasing trends.
FIGURE 40.
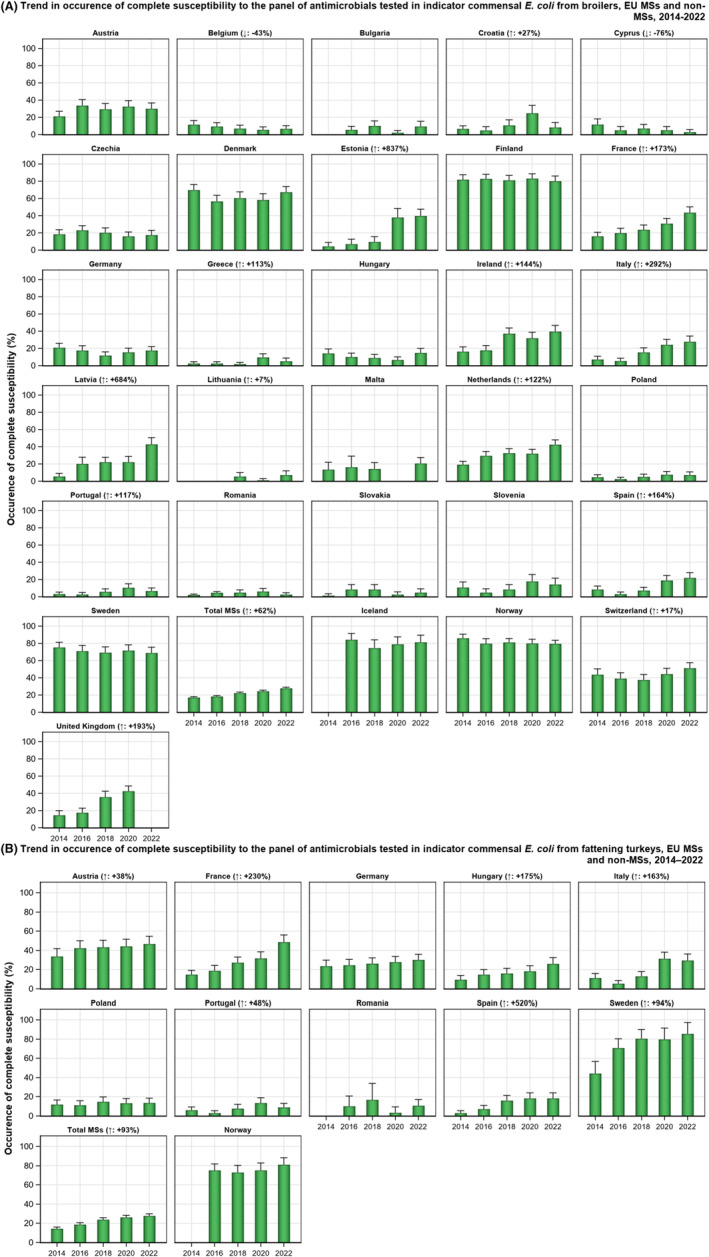
Trends in the occurrence of complete susceptibility to the panel of antimicrobials tested in indicator commensal E. coli from (A) broilers and (B) fattening turkeys (turkeys), EU MSs and non‐MSs, 2014–2022.
FIGURE 41.
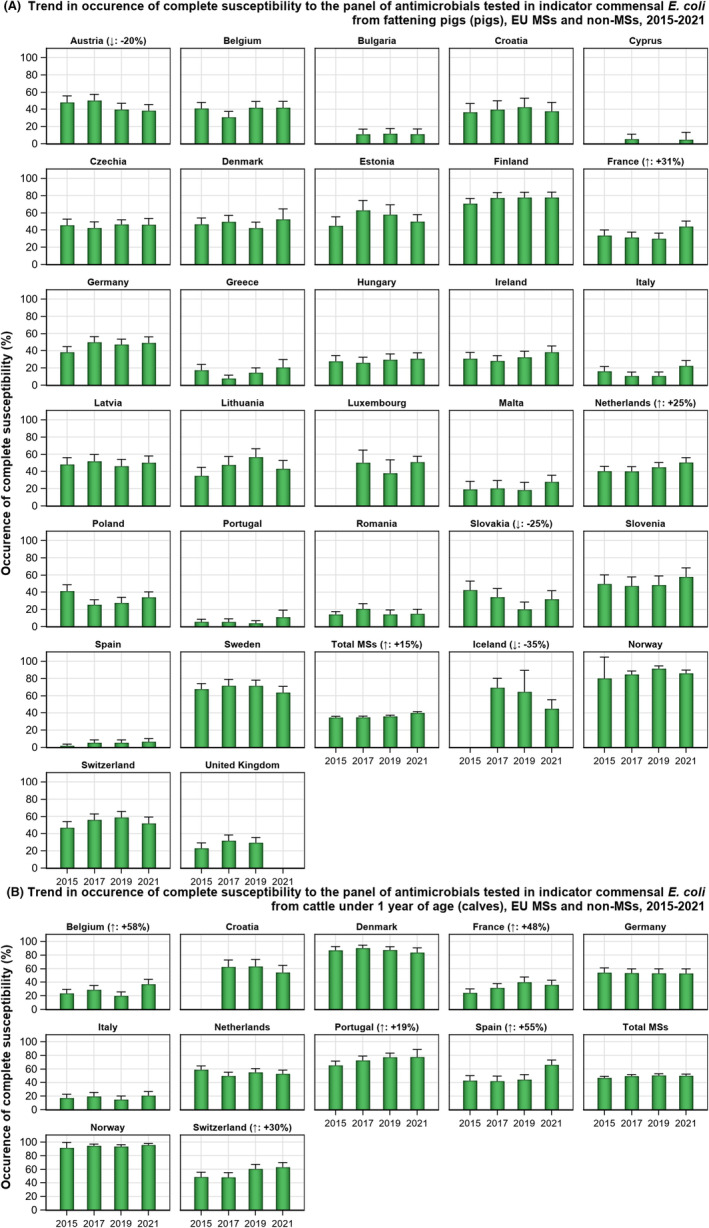
Trends in the occurrence of complete susceptibility to the panel of antimicrobials tested in indicator commensal E. coli from (A) fattening pigs and (B) cattle under 1 year of age, EU MSs and non‐MSs, 2015–2021.
However, it is of note that some countries have reported levels of complete susceptibility at high levels during many years and major changes, especially not increasing trends, cannot be expected.
4.3.7. Key outcome indicator of complete susceptibility (KOICS )
The occurrence of completely susceptible indicator E. coli isolates from the most important food‐producing animals (broilers, fattening turkeys, fattening pigs, cattle under 1 year of age) in a country is used as a key outcome indicator (KOICS) for the overall AMR situation in food‐producing animals. To account for differences in the relative size of food‐producing animal populations in a country, the KOICS was calculated as the weighted mean of the proportions of completely susceptible indicator E. coli isolates in each of the four animal categories monitored. For the calculation of the KOICS, the occurrence of CS in each animal population was weighted in relation to the relative size of the populations within a country using the ‘population correction unit’ (PCU).
To calculate the KOICS, data reported in two consecutive years was used. The KOIcs values were calculated from CS data on broilers and fattening turkeys reported in even‐numbered years and data on fattening pigs and cattle under 1 year of age reported in the immediately preceding/following odd‐numbered years. Data for broilers and pigs were included in the calculation for each country, while data for turkeys and cattle under 1 year of age were included in the calculation only in those countries reporting such data.
Marked variations in KOICS were observed among the 30 countries reporting consistently over the study period (Figure 42). In 2021–2022, levels of KOICS were < 20% in 14 countries, 20%–40% in six countries, 40%–60% in five countries, 60%–80% in four countries (Denmark, Finland, Norway and Sweden) and > 80% in one country (Iceland). The lowest KOICS were generally observed in countries in Eastern and Southern Europe and the highest in countries in Northern Europe.
The KOICS has been stable at a high level over the period in some countries and in others at a low level. Statistically significant increasing trends in KOICS were registered in 18 countries (16 MSs and 2 non‐MSs) and decreasing trends in three countries (all MSs).
FIGURE 42.
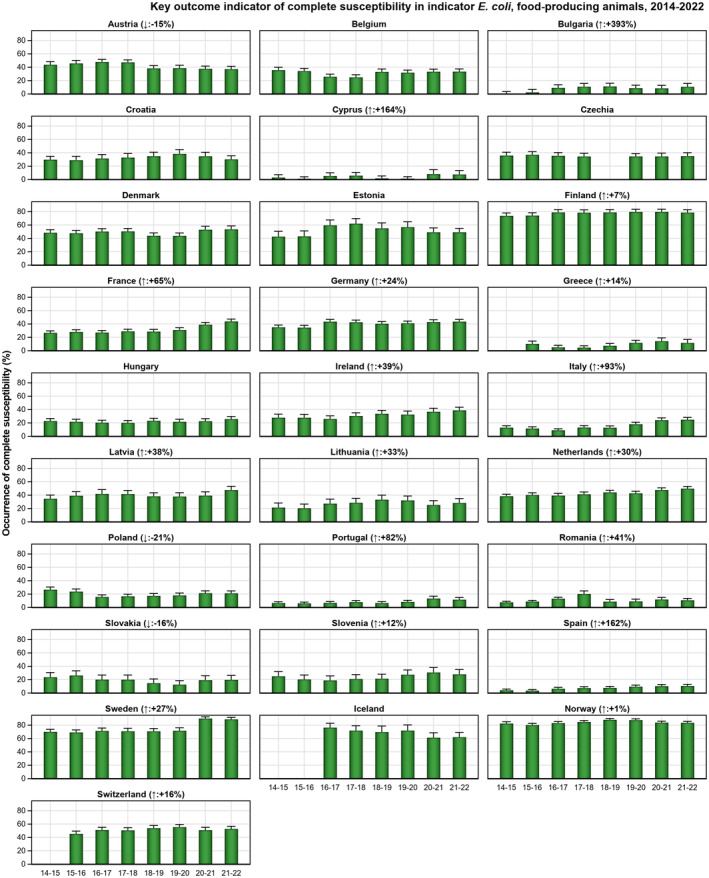
Trends in the key outcome indicator of complete susceptibility (KOICS) in indicator commensal E. coli from food‐producing animals (broilers, fattening turkeys, fattening pigs and cattle under 1 year of age), 27 EU MSs and 3 non‐MSs, 2014–2022.
4.4. Discussion
Studying AMR in indicator commensal E. coli sampled from the caecal contents of healthy food‐producing animals domestically produced provides information on the reservoirs of resistant bacteria that could potentially be transferred between animals and humans. Such monitoring is therefore relevant for both animal and public health. Resistance among indicator E. coli is likely to depend on several factors, such as selective pressure from the use of antimicrobials in food‐producing animals, co‐selection of bacteria harbouring multiple resistance, clonal spread of resistant bacteria and dissemination of genetic elements, such as plasmids, between bacteria.
Representative monitoring
The data on AMR in indicator E coli used in the present report were collected over the years 2014–2022 in accordance with Commission implementing decision (EU) 2013/652 (up until December 2020) and the replacement legislation, Commission implementing decision (EU) 2020/1729, which came into force on 1 January 2021. The new legislation enlarged the scope of the monitoring, with the inclusion of imported fresh meat from third countries, the addition of a new substance (amikacin) to the harmonised testing panel and the option for MSs to perform WGS as an alternative to extended phenotypic testing of isolates resistant to third generation cephalosporins and/or carbapenems. The data are harmonised with respect to representative sampling design, laboratory methodologies, reporting and interpretation of resistance. In addition, as data on indicator E. coli from pigs (2021) and broilers (2022) were reported by all MSs and as data from turkeys (13 MSs in 2022) and cattle under 1 year of age (11 MSs in 2021) included those MSs which are the main producers of meat derived thereof in the EU, AMR data can be considered representative.
General observations
At the MS‐group level, resistance to ampicillin, sulfamethoxazole, trimethoprim and tetracyclines was generally common in indicator E. coli and the median levels of resistance among reporting countries were high or very high for all animal categories. In poultry, resistance to ciprofloxacin and nalidixic acid was also common, and the median levels of resistance were very high in broilers and high in fattening turkeys. In contrast, in pigs and cattle under 1 year of age, both median levels of resistance to these antimicrobials were low.
The frequent occurrence of resistance to these substances in indicator commensal E. coli isolates from animal origins likely reflects the widespread past and present use of these antimicrobials in food‐producing animals in several MSs. The observed differences between animal species likely reflect the diversity of the quantities of antimicrobials used among categories of animals, but possibly also the mode of administration. In poultry, flock treatment is almost exclusively practised, whereas pigs and cattle under 1 year of age are typically treated individually in some countries.
Marked spatial differences were notably observed in the occurrence of resistance to most antimicrobials as well as in the occurrence of MDR, complete susceptibility and for the KOICS. Regarding pigs and broilers, the situation was generally more favourable in Northern Europe than in Southern and Eastern Europe. For turkeys, data indicate a similar spatial distribution as that for pigs and broilers was observed, albeit less marked. Contrastingly, for cattle under 1 year of age, the picture was more complex, and although the most favourable situation was registered in the Nordic countries (Norway, Denmark, Sweden), countries in other parts of Europe (e.g. Portugal, Romania) also reported favourable situations in comparison with neighbouring countries in these regions. This is also of interest, as it indicates that the situation in one animal production system in a specific country is not necessarily representative of the situation in other systems in the same country.
Meropenem resistance was reported in one isolate of indicator E. coli from broilers in Italy. This indicates that carbapenem resistance is still uncommon among indicator E. coli from food‐producing animals in Europe. This finding, however, relates to commensal E. coli isolated using non‐selective culture methods. Within the mandatory monitoring, samples of caecal contents and fresh meat are also cultured on selective media to specifically detect the presence of E. coli resistant to carbapenems. The results of these analyses are presented in Section 5 Extended‐spectrum β‐lactamase (ESBL)‐, AmpC‐ and/or carbapenemase (CP)‐producing Salmonella and Escherichia coli.
Resistance to highest‐priority critically important antimicrobials (hpCIA)
Of the antimicrobials tested within the mandatory monitoring of indicator E. coli from caecal contents and imported fresh meat, ciprofloxacin (fluoroquinolones), cefotaxime and ceftazidime (third‐generation cephalosporins), colistin (polymyxins) and azithromycin (macrolides) have been categorised by the WHO as hpCIA (WHO, 2019). Isolates from food‐producing animals and derived meat exhibiting resistance to these antimicrobials are therefore of particular interest considering the risk of possible spread to humans along the food chain. At the EU level, no significant differences in the occurrence of resistance to cefotaxime, ceftazidime, azithromycin and colistin were observed between the four animal categories monitored.
The findings regarding third‐generation cephalosporins (cefotaxime and ceftazidime) relate to E. coli which are isolated using non‐selective culture methods. Within the mandatory monitoring, samples of caecal contents and fresh meat are also cultured on selective media to specifically detect the presence of E. coli resistant to third‐generation cephalosporins. The results of these analyses are presented in Section 5 Extended‐spectrum β‐lactamase (ESBL)‐, AmpC‐ and /or carbapenemase (CP)‐producing Salmonella and Escherichia coli.
The median levels of resistance to ciprofloxacin and nalidixic acid among E. coli isolates from pigs and cattle under 1 year of age were low at the EU level. In contrast, the median levels of ciprofloxacin and nalidixic acid resistance were high to very high in broilers and high in turkeys. Notably, a substantial proportion of isolates from all animal categories, especially turkeys and cattle under 1 year of age, exhibited resistance to ciprofloxacin but not to nalidixic acid; a resistance pattern which generally indicates the presence of transmissible genes mediating quinolone resistance (Jacoby et al., 2014).
Although the median levels of azithromycin resistance were low to very low in all four animal categories, certain countries (i.e. broilers from Germany, Malta and Romania; turkeys from Romania; pigs from Cyprus, Greece and Portugal; cattle under 1 year of age from France) reported higher levels of resistance. Azithromycin, which is an azalide, a subclass of macrolide antimicrobials, is not used in animals. The selective pressure exerted by the use of other related macrolides in food‐producing animals may have favoured the emergence of azithromycin resistance.
Considering all reporting MSs, the median colistin resistance was rare in all four animal categories. However, certain countries reported higher levels of resistance. Statistically significant decreasing trends in the level of colistin resistance were reported among isolates from specific animal categories by some countries. This is in line with the fact that sales of polymyxins (e.g. colistin) for use in animals has decreased between 2017 and 2022 by over 40% in Europe (EMA, 2023).
Complete susceptibility and multidrug resistance in all reporting countries
Considering all reporting countries, the median percentage of indicator commensal E. coli isolates exhibiting complete susceptibility to all antimicrobial classes tested were lower in broilers (18.9%) and turkeys (29.4%) than in pigs (38.3%) and cattle under 1 year of age (54.1%). Conversely, MDR isolates were more common in broilers (median 38.8%) and turkeys (median 43.5%) than in pigs (median 31.2%) and cattle under 1 year of age (median 18.8%). In all four animal categories, there were marked differences in the levels of CS, as well as in MDR between countries. Generally, completely susceptible isolates from pigs, broilers, and to some extent, turkeys were more common in Northern than in Southern and Eastern Europe, whereas the converse situation was observed for MDR. For cattle under 1 year of age, the picture was more complex, and a favourable situation was reported in the Nordic countries (Norway, Denmark, Sweden) as well as in countries in other parts of Europe (Portugal, Romania).
Tetracycline, ampicillin, sulfamethoxazole and trimethoprim were antimicrobials often represented in the patterns of MDR isolates from all animal categories. Among isolates from broilers and turkeys, resistance to fluoroquinolones/quinolones (ciprofloxacin/nalidixic acid) was also a common trait in the patterns of MDR isolates. The frequent occurrence of these substances as a core component of MDR patterns in E. coli from broilers and turkeys likely reflects an extensive usage in several countries over many years and that the genes conferring resistance to these substances often are linked on mobile genetic elements, resulting in co‐selection.
Key outcome indicator of complete susceptibility
An abundant and ubiquitous commensal bacterial species, E. coli has been selected as a reporting organism because it is considered more relevant in representing the overall resistance situation, including transmissible genes, than less abundant zoonotic bacterial species. The assumption underlying the choice of key outcome indicator of complete susceptibility (KOICS), as indicator, is that E. coli that is rarely, if ever, exposed to antimicrobials will tend to be fully susceptible (Martinez, 2014). As KOICS accounts for differences in the relative size of food animal populations in a country, it is relevant in the overall assessment of risks related to AMR in food‐producing animals. Marked variations in KOICS were registered from < 20% in 14 MSs to > 80% in one country (Iceland). To fully appreciate the situation within a country, the evaluation of KOICS should be complemented with data on resistance and CS available at the level of each animal population monitored so that any positive or negative trends occurring in one animal population of small relative size may not go unnoticed. The KOICS can be used to assess the development of AMR in relation to the total use of antimicrobials in food‐producing animals (ECDC, EFSA, and EMA, 2017; Queenan et al., 2016). Therefore, it is to be expected that a reduction in the use of antimicrobials in food‐producing animals would eventually result in an improvement in this indicator.
Temporal trends in resistance
Overall, in several countries, statistically significant trends towards reduction of resistance in indicator E. coli, notably in broilers and turkeys, were revealed. Considering data on pigs and cattle under 1 year of age in 2021, there were 29 decreasing and nine increasing trends in resistance to ampicillin, cefotaxime, ciprofloxacin and tetracycline in the period 2011–2021. Considering data on broilers and fattening turkeys in 2022, there were 76 decreasing and 12 increasing trends in resistance to ampicillin, cefotaxime, ciprofloxacin and tetracycline in the period 2009–2022. For several antimicrobials, statistically significant associations were demonstrated between use of antimicrobials in food‐producing animals and the occurrence of resistance in indicator E. coli from these animals (ECDC, EFSA, and EMA, 2021). The positive decreasing trends in several countries are, therefore, believed to be due to the overall decline in sales of antimicrobials for use in animals since 2011, as documented in the ESVAC report (EMA, 2023).
As for the individually assessed antimicrobials above, improving trends were seen for CS in many individual countries. Furthermore, improving trends regarding the KOICS can be seen in some countries.
Statistically significant decreasing trends in resistance to individual substances reveal progress towards reduced resistance in several countries. This is further highlighted by statistically significant trends towards higher levels of complete susceptibility and KOICS in a few countries. It should, however, be noted that, in some countries, the situation regarding antimicrobial resistance has been already favourable for a number of years and major changes, especially not improving trends, cannot be expected. Efforts to maintain, and even further improve the situation should however be made also in those countries.
Monitoring AMR in imported fresh meat sampled at BCP
Salmonella spp.
In 2022 , Salmonella spp. isolates from imported fresh meat from broilers sampled at the BCPs were reported by five MSs: Germany (N = 5), Ireland (N = 8), the Netherlands (N = 32), Poland (N = 19) and Spain (N = 12). Overall, Salmonella isolates exhibited very high to extremely high levels of resistance to tetracycline (63.2%), sulfonamides (71.1%) and ampicillin (60.5%). While resistance to gentamicin (4.0%), chloramphenicol (5.3%) and trimethoprim (7.8%) was low. Resistance to ciprofloxacin and nalidixic acid was reported at an extremely high level (83%, N = 76, each antimicrobial). Furthermore, among the Salmonella isolates displaying ciprofloxacin resistance, one isolate (S. Heidelberg), exhibited high‐level ciprofloxacin resistance (MIC ≥ 4 mg/L). Regarding resistance to third‐generation cephalosporins, Salmonella isolates from imported broiler meat exhibited very high resistance (at 55.3%, each antimicrobial). Resistance to tigecycline in Salmonella isolates from imported fresh meat from broilers was reported at a high level (38.2%, N = 76). Low levels of colistin resistance were reported from broiler meat (4.0%).
Only the Netherlands reported data on fresh meat from turkeys sampled at BCPs (N = 3). All isolates were resistant to tetracycline, while two out of three isolates were resistant to sulfonamides and ampicillin, and only one to chloramphenicol. All three isolates were susceptible to gentamicin and trimethoprim. For fresh meat from turkeys, all isolates (N = 3) were resistant to both ciprofloxacin and nalidixic acid. Regarding resistance to third‐generation cephalosporins, Salmonella isolates from turkey meat exhibited very high resistance (at 66.7%, each antimicrobial). No tigecycline‐resistant nor colistin‐resistant Salmonella isolates from fresh meat from turkeys were reported (N = 3).
Lastly, no resistance to azithromycin nor amikacin was reported in Salmonella isolates from meat sampled at BCPs.
Indicator E. coli
Occurrence of resistance
In total, 11 MSs reported data on 576 indicator E. coli isolates from imported fresh meat of the four kinds monitored, the majority coming from broiler meat (N = 399) and cattle meat (N = 110) (Table 14; Annex C, Tables 5–8). Among isolates from imported fresh broiler meat, resistance was high to very high for ampicillin, ciprofloxacin and nalidixic acid, sulfamethoxazole, trimethoprim and tetracycline. Furthermore, resistance to cefotaxime and ceftazidime was moderate. Somewhat similar, among isolates from imported fresh turkey meat (N = 54), resistance to ampicillin, ciprofloxacin, nalidixic acid, sulfamethoxazole and tetracycline was high to very high. For cefotaxime and trimethoprim, resistance was moderate.
TABLE 14.
Combined resistance to ciprofloxacin and cefotaxime in indicator E. coli from imported fresh meat from broilers, turkeys, pigs and cattle, applying ECOFFs and clinical breakpoints, as issued by EUCAST, EU MSs and non‐MSs, 2021–2022.
| Imported fresh meat | Microbiological combined resistance to CIP & CTX (using ECOFFs) | ‘Clinical’ combined resistance to CIP & CTX (using clinical breakpoints) | ||||
|---|---|---|---|---|---|---|
| N | % R | 95% CI | N | % R | 95% CI | |
| Pig meat, 2021 (N = 13, 4 MSs) | 0 | 0 | 0.0, 22.8 | 0 | 0.0 | 0.0, 22.8 |
| Bovine meat, 2021 (N = 110, 6 MSs) | 1 | 0.9 | 0, 5.0 | 1 | 0.9 | 0, 5.0 |
| Broiler meat, 2022 (N = 399, 8 MSs) | 47 | 11.8 | 9.0, 15.3 | 27 | 6.8 | 4.7, 9.7 |
| Turkey meat, 2022 (N = 54, 3 MSs) | 5 | 9.3 | 4.0, 19.9 | 2 | 3.7 | 1.0, 12.5 |
Abbreviations: CIP, ciprofloxacin (fluoroquinolones); CTX, cefotaxime (third‐generation cephalosporins); N, number of isolates; % R, percentage of resistance; 95% CI, 95% confidence interval.
Among isolates from imported cattle meat, resistance was uncommon, with the exception of resistance to tetracycline, which was the most common trait registered at the moderate level of 10.9%. For the remaining substances, resistance was rare, very low or low. Among the few isolates obtained from imported pig meat (N = 13), resistance to ampicillin and tetracycline were the most common traits, being reported at a high level.
Resistance to amikacin or meropenem was not detected in any isolates from imported fresh meat.
Resistance to colistin and azithromycin was not detected in indicator E. coli isolates from imported pig or turkey meat (Annex C, Tables 6, 7). In imported bovine meat, one isolate (0.9%) resistant to azithromycin but no isolates resistant to colistin were detected (Annex C, Table 8). From imported broiler meat, five isolates (1.3%) resistant to azithromycin and 10 isolates (2.5%) resistant to colistin were detected (Annex C, Table 5).
For nalidixic acid, the overall level of resistance was rare for isolates from imported cattle meat (1.8%) and not detected in imported bovine meat, while resistant to ciprofloxacin was rare in imported bovine meat (1.8%) but low in imported pig meat (7.7%) (Annex C, Tables 7, 8). In contrast, the overall levels of resistance for both substances were very high among isolates from fresh broiler meat and high among isolates from fresh turkey meat, 54.6% and 31.5% for nalidixic acid and 58.7% and 38.9% for ciprofloxacin, respectively (Annex C, Tables 5, 6). A similar situation was seen for cefotaxime and ceftazidime where the overall levels of resistance in imported cattle meat were very low for both substances, and no resistance was reported for imported pig meat, whereas the overall levels of resistance in fresh broiler and turkey meat were moderate and low, respectively (Annex C, Tables 5–8).
Combined resistance to ciprofloxacin and cefotaxime
Microbiological combined resistance to ciprofloxacin and cefotaxime was moderate in fresh broiler meat (11.8%) and low in fresh turkey meat (9.3%). Conversely, microbiological combined resistance was not detected in any E. coli isolates recovered from imported fresh pig meat, and only one isolate from fresh bovine meat exhibited microbiological combined resistance (Table 14; Annex C, Tables 5–8).
No E. coli isolates from imported fresh pig meat (n = 13) displayed phenotypic resistance to third‐generation cephalosporins, while a single isolate from imported fresh cattle meat (n = 110) was resistant to both cefotaxime and ceftazidime. Conversely, among isolates from imported fresh broiler and turkey meat, resistance was moderate and low, respectively (Table 15).
TABLE 15.
Occurrence of resistance to third‐generation cephalosporins in indicator E. coli isolates from imported fresh meat from broilers, turkeys, pigs and cattle, EU MSs, 2020–2021.
| Imported fresh meat | No. of MSs | N | Cefotaxime | Ceftazidime | ||
|---|---|---|---|---|---|---|
| n | % R | n | % R | |||
| Broiler meat, 2022 | 8 | 399 | 60 | 15.0 | 60 | 15.0 |
| Turkey meat, 2022 | 3 | 54 | 5 | 9.3 | 3 | 5.6 |
| Pig meat, 2021 | 4 | 13 | 0 | 0 | 0 | 0 |
| Cattle meat, 2021 | 6 | 110 | 1 | 0.9 | 1 | 0.9 |
Abbreviations: N, number of E.coli isolates tested; n, number of indicator E. coli resistant isolates; % R, percentage of resistance.
Multidrug resistance
The level of MDR among indicator E. coli isolates from imported broiler and turkey meat was high; 48.4% and 48.1%, respectively. Contrastingly, the level of MDR among isolates from imported cattle meat was low (3.6%), while the level of MDR among isolates from fresh pig meat was moderate (15.4%).
The antimicrobials most often represented in the MDR patterns of isolates from fresh broiler meat were ampicillin, sulfamethoxazole and trimethoprim, often also in combination with tetracycline. Resistance to ciprofloxacin and nalidixic acid was also a common trait among MDR‐resistant isolates, sometimes without the inclusion of sulfamethoxazole and trimethoprim. Among isolates from imported turkey meat, the most common MDR patterns were resistance to ampicillin and tetracycline in combination with sulfamethoxazole or ciprofloxacin and nalidixic acid.
The resistance patterns of the two MDR E. coli isolates from imported fresh pig meat featured sulfamethoxazole and tetracycline, along with other substances. One of the two isolates also featured resistance to ciprofloxacin. The resistance patterns for the four MDR E. coli isolates from fresh cattle meat also featured one or more of the substances: ampicillin, sulfamethoxazole, trimethoprim or tetracycline. One of the isolates was resistant to six antimicrobial classes, including third‐generation cephalosporins and fluoroquinolones. No resistance to tigecycline or amikacin was recorded for either imported meat origin among MDR isolates.
None of the MDR‐resistant patterns included resistance to amikacin, azithromycin or tigecycline. Resistance to colistin was a part of the MDR‐resistant pattern in nine isolates from fresh broiler meat but not in any isolates from fresh pig, cattle or turkey meat.
Considering all reporting MSs, the median CS among E. coli isolates from imported fresh meat was 14.7% from broiler meat, 9.1% from turkey meat, 45.0% from pig meat and 95.2% from cattle meat (Annex C, Tables 5–8).
Routine monitoring: ESBL‐/AmpC/CP‐producing Salmonella spp .
ESBL‐/AmpC‐/CP‐producing Salmonella spp. isolates were detected in 42 samples of imported broiler meat and two samples of imported turkey meat collected at BCPs in 2022 (Tables 16, 17). Germany reported one S. Heidelberg and one S. Minnesota from imported broiler meat carrying bla CMY‐2 . The Netherlands detected bla CMY‐2 in two S. Heidelberg from imported fresh meat from turkeys, 13 S. Minnesota and 12 S. Heidelberg from imported fresh meat from broilers sampled at BCPs, and bla CTX‐M‐2 in two S. Minnesota from imported meat from broilers.
TABLE 16.
Summary of presumptive ESBL‐/AmpC‐/CP‐producing Salmonella spp. from imported fresh meat collected at BCP – routine monitoring, 2022.
| Year | Imported fresh meat | Serotype | n | ESBL a | AmpC b | ESBL+AmpC c |
|---|---|---|---|---|---|---|
| 2022 | Meat from broilers, at BCP (N = 42) | S. Minnesota | 17 | 2 | 15 | – |
| S. Heidelberg | 14 | 14 | – | |||
| S. Infantis | 1 | 1 | – | |||
| S. 21:‐:‐ | 3 | – | 3 | – | ||
| S. 1,4,12:‐:‐ | 1 | – | 1 | – | ||
| Unspecified | 6 | 1 | – | 5 | ||
| Meat from turkeys, at BCP (N = 2) | S. Heidelberg | 2 | – | 2 | – |
Abbreviations: AmpC, AmpC beta‐lactamase; ESBL, extended‐spectrum beta‐lactamase; n, number of isolates with the phenotype; N, total number of isolates.
According to EUCAST guidelines (EUCAST, 2017), only isolates showing MIC >1 mg/L for CTX and/or CAZ or reported presence of ESBL‐/AmpC‐encoding gene were considered (see Appendix G).
All isolates showing clavulanate synergy with CTX or CAZ or both, suggesting ESBL phenotype, or reported presence of ESBL‐encoding gene.
Isolates with cefoxitin resistance, suggesting AmpC phenotype, or reported presence of AmpC‐encoding gene.
Isolates showing synergy with CTX or CAZ and cefoxitin resistance, suggesting ESBL‐ and AmpC‐enzymes in the same isolates, or both ESBL‐ and AmpC‐encoding genes reported.
Isolates with meropenem resistance or CP‐encoding gene reported.
TABLE 17.
Presumptive ESBL‐, AmpC‐ or CP‐producing Salmonella spp. and indicator E. coli from imported fresh meat at BCPs, routine monitoring, EU MSs, 2021–2022.
| Matrix | ESBL and/or AmpC a | ESBL b | AmpC c | ESBL + AmpC d | CP e |
|---|---|---|---|---|---|
| n (% R) | n (% R) | n (% R) | n (% R) | n (% R) | |
| Salmonella spp. | |||||
| Imported meat from broilers at BCP, 2022 (N = 76, 5 MSs) | 42 (55.3) | 4 (5.3) | 38 (50) | 0 (0) | 0 (0) |
| Imported meat from turkeys at BCP, 2022 (N = 44, 1 MS) | 2 (4.5) | 0 (0) | 2 (4.5) | 0 (0) | 0 (0) |
| Indicator Escherichia coli | |||||
| Imported meat from pigs at BCP, 2021 (N = 13, 4 MSs) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Imported meat from cattle at BCP, 2021 (N = 110, 6 MS) | 1 (0.9) | 1 (0.9) | 0 (0) | 0 (0) | 0 (0) |
| Imported meat from broilers at BCP, 2022 (N = 399, 8 MSs) | 60 (15) | 49 (12.3) | 7 (1.8) | 4 (1.0) | 0 (0) |
| Imported meat from turkeys at BCP, 2022 (N = 54, 3 MSs) | 5 (9.3) | 3 (5.6) | 1 (1.9) | 1 (1.9) | 0 (0) |
Abbreviations: AmpC, AmpC beta‐lactamase; CP, carbapenemase; ESBL, extended‐spectrum beta‐lactamase; n, number of isolates with the phenotype; N, total number of samples investigated.
According to EUCAST guidelines (EUCAST, 2017), only isolates showing MIC > 1 mg/L for CTX and/or CAZ or reported presence of ESBL‐/AmpC‐encoding gene were considered (see Appendix G).
All isolates showing clavulanate synergy with CTX or CAZ or both, suggesting ESBL phenotype, or reported presence of ESBL‐encoding gene.
Isolates with cefoxitin resistance, suggesting AmpC phenotype, or reported presence of AmpC‐encoding gene.
Isolates showing synergy with CTX or CAZ and cefoxitin resistance, suggesting ESBL‐ and AmpC‐enzymes in the same isolates, or both ESBL‐ and AmpC‐encoding genes reported.
Isolates with meropenem resistance or CP‐encoding gene reported.
Routine monitoring: ESBL‐/AmpC‐/CP‐producing indicator E. coli
ESC‐resistant indicator E. coli were detected in imported broiler meat, turkey meat and cattle meat sampled at BCPs (Table 17). The occurrence of ESBL‐/AmpC‐producing isolates was lower in imported meat from pigs and cattle compared with imported meat from broilers and turkeys. This is in correspondence with findings of the specific monitoring of ESBL‐/AmpC‐/CP‐producing E. coli in meat sampled at retail. Only a limited number of samples for pig meat, cattle meat and turkey meat were collected at BCPs.
In 2021, the Netherlands reported WGS data on imported cattle meat. One isolate carried the bla CTX‐M‐15 gene. In 2022, Germany, Ireland and the Netherlands reported WGS data on imported broiler meat. In total, 21 isolates carried bla CTX‐M‐55 , 12 isolates carried bla CTX‐M‐2 , five isolates carried bla CTX‐M‐8 , while bla CTX‐M‐14 , and bla CTX‐M‐65 were detected in one isolate each. Also, bla CMY‐2 was reported in three isolates. Of note, one of the above mentioned isolates harboured both genes bla CTX‐M‐55 and bla CTX‐M‐8 , at the same time. The Netherlands also reported WGS data for imported turkey meat. bla CTX‐M‐8 , bla CTX‐M‐15 and bla SHV‐12 were detected in one isolate each as well as bla CMY‐2 in another isolate from imported turkey meat.
Specific monitoring of ESBL‐/AmpC/CP‐producing E. coli
Ten MSs (Belgium, Denmark, Finland, Germany, Ireland, Italy, Poland, Portugal, Spain and Sweden) and nine MSs (Estonia, France, Germany, Hungary, Ireland, the Netherlands, Portugal, Romania and Spain) collected samples of imported fresh meat at BCPs in 2021 and 2022, respectively. No ESBL‐/AmpC‐/CP‐producing E. coli was detected in imported pig meat sampled at BCPs, whereas only one single isolate was isolated from imported cattle meat sampled at BCPs in 2021. The occurrence of ESBL‐/AmpC‐/CP‐producing E. coli in broiler meat/turkey meat in 2022 was higher/comparable to that observed in meat samples collected at retail. Overall, 204 out of 332 (61.4%) samples from broiler meat and 32 out of 45 (71.1%) samples from turkey meat tested positive. It is of note that not all positive/presumptive samples necessarily contained an ESBL‐/AmpC‐/CP‐producing E. coli isolate.
WGS data for E. coli isolated from imported meat sampled at BCPs were reported by Germany and the Netherlands. Germany detected bla CMY‐2 (n = 3), bla CTX‐M‐2 (n = 12), bla CTX‐M‐8 (n = 8), bla CTX‐M‐15 (n = 1), bla CTX‐M‐55 (n = 20) and bla CTX‐M‐55 and bla CMY‐2 (n = 1) in isolates from imported broiler meat. Similarly, the Netherlands reported bla CMY‐2 (n = 3), bla CTX‐M‐1 (n = 2), bla CTX‐M‐2 (n = 9), bla CTX‐M‐8 (n = 16), bla CTX‐M‐14 (n = 1), bla CTX‐M‐15 (n = 4), bla CTX‐M‐55 (n = 24) and bla SHV‐12 (n = 2) in isolates from imported broiler meat, and bla CTX‐M‐8 (n = 11), bla CTX‐M‐55 (n = 9), bla SHV‐12 (n = 2), bla CTX‐M‐15 (n = 2), bla CTX‐M‐2 (n = 1) as well as bla CMY‐2 (n = 2) in isolates from imported turkey meat.
The ESBL‐encoding genes dominating among isolates from imported broiler meat and turkey meat were different from those reported in meat produced in Europe. For imported broiler meat, bla CTX‐M‐55 was the most frequently reported, followed by bla CTX‐M‐8. For imported turkey meat, bla CTX‐M‐8 dominated, followed by bla CTX‐M‐55. The majority of imported meat originated from Brazil, where these genes are commonly occurring (Egervärn et al., 2014; Casella et al., 2018; Adur et al., 2022; Soncini et al., 2022).
Specific monitoring of CP‐producing E. coli
Within the framework of the specific monitoring of CP‐producing E. coli, a total of 20 samples from imported pig meat and 231 samples from imported cattle meat were collected in 2021, and 192 samples from imported broiler meat and six samples from imported turkey meat collected in 2022. No presumptive CP‐producing E. coli were detected in the investigated samples.
5. EXTENDED‐SPECTRUM β‐LACTAMASE (ESBL)‐, AmpC‐ AND/OR CARBAPENEMASE (CP)‐PRODUCING SALMONELLA AND ESCHERICHIA COLI
All ESBL‐, AmpC‐ and/or CP‐producers prevalence and occurrence tables from the 2021 and 2022 monitoring, as well as tables on occurrence of resistance (panel 1 and panel 2) mentioned in this chapter can be found in Annex D and the Salmonella spp., E. coli or ESBL documents available on Zenodo (https://doi.org/10.5281/zenodo.10528846). Materials and methods are presented in Appendix F – Materials and methods.
5.1. Key findings
The occurrence of presumptive ESBL‐/AmpC‐/CP‐producing Salmonella spp. and indicator E. coli was generally very low or low in 2021 and 2022.
In 2021, whole genome sequencing was introduced as an alternative method to the phenotypic testing of E. coli and Salmonella isolates displaying phenotypic resistance to extended‐spectrum cephalosporins and/or carbapenems in Panel 1, or in the specific monitoring of ESBL‐/AmpC‐/CP‐producing E. coli. In 2021, four countries reported WGS data, namely Czechia, Finland, Germany and Italy. In 2022, WGS data were reported by seven countries; Czechia, Finland, Germany, Italy, the Netherlands, Norway and Sweden.
A variety of ESBL‐ and AmpC‐encoding genes were reported in both 2021 and 2022. The most commonly reported ESBL‐encoding gene overall in E. coli was bla CTX‐M‐1 followed by bla CTX‐M‐15 . Although, the bla SHV‐12 gene was also frequently reported in isolates from poultry and their derived meat. The most commonly reported AmpC‐encoding gene was bla CMY‐2 . The mutation C‐42T in the chromosomal ampC gene causing an AmpC phenotype was also frequently reported, and was the most common cause of the AmpC phenotype in isolates from fattening pigs, fattening turkeys and turkey meat in 2021 and 2022.
For isolates where both phenotypic and genotypic data were reported, there was a high correspondence between phenotype determined by testing at the laboratory and phenotype predicted from genotype.
The prevalence of ESBL‐/AmpC‐producing E. coli was still high in some countries. However, statistically significant decreasing trends were observed in several countries and animal categories.
There were major differences in the spatial distributions of ESBL‐/AmpC‐producing E. coli for all animal and food categories tested.
The ESBL phenotype was overall more common than the AmpC phenotype in all animals and food categories tested. The same was observed for countries reporting genotypic data.
Statistically significant decreasing trends in the key outcome indicator of ESBL‐ and/or AmpC‐producing E. coli (KOIESC) were observed in 19 MSs and three non‐MSs. In three, a statistically significant increasing trend was observed. No statistically significant trend was observed in the remaining countries.
In 2022, one E. coli isolate originating from fattening turkeys, detected in the routine monitoring, displayed carbapenem resistance and was reported to harbour the CP‐encoding gene bla OXA‐181 .
Two E. coli isolates from bovine meat and one from pig meat collected in the specific monitoring of ESBL‐/AmpC‐/CP‐producing E. coli in 2021 were reported to carry the CP‐encoding gene bla NDM‐5 . In 2022, three CP‐producing isolates were reported in the specific monitoring of ESBL‐/AmpC‐/CP‐producing E. coli. They originated from broilers, and all carried the bla VIM‐1 gene.
In the specific monitoring of CP‐producing E. coli in 2021, two isolates from fattening pigs carrying bla OXA‐48 were reported based on MIC. Furthermore, 24 isolates from fattening pigs and five from cattle under 1 year of age were reported as CP‐producers using WGS. These isolates carried bla OXA‐181 (four isolates from cattle and 20 from pigs), bla OXA‐48 (one fattening pig) or bla NDM‐5 (one isolate from cattle and tree from pigs). In 2022, a single CP‐producing E. coli was reported based on WGS data in the specific monitoring of CP‐producing E. coli. The isolate was from fattening turkeys and carried the bla OXA‐181 gene.
5.2. Data on ESBL‐, AmpC‐ and/or CP‐producing Salmonella and Escherichia coli addressed
The presence of ESBL‐, AmpC‐ and/or CP‐producing bacteria in the intestinal flora of animals is undesired, due to the potential risk of transmission of resistant bacteria from animals and food to humans. Bacteria from animals harbouring such resistance are a reservoir of resistance genes, which may be transferred to other bacteria, including food‐borne zoonoses such as Salmonella spp. This further adds to the public health concern. Although no statistically significant association between resistance to extended‐spectrum cephalosporins in indicator E. coli from animals and invasive E. coli from humans was observed in the latest JIACRA report (ECDC, EFSA, EMA, 2021), the complexity of ESBL‐, AmpC‐ and/or CP‐producing E. coli epidemiology should not be underestimated. Thus, it is important to continue to improve the monitoring system to study their prevalence in healthy food‐producing animals and food derived thereof in order to increase our knowledge and understanding of their epidemiology.
Routine monitoring of indicator E. coli and Salmonella spp. and the specific monitoring of ESBL‐/AmpC‐/CP‐producing E. coli in caecal samples from animals and their derived meat products are mandatory. In 2022, monitoring samples collected from the caeca of broilers and fattening turkeys at slaughter, as well as broiler meat and turkey meat collected at retail was mandatory. While in 2021, the mandatory monitoring included caecal samples collected from fattening pigs and cattle under 1 year of age at slaughter, and of pig meat and cattle meat collected at retail. In addition, since 2021, it was mandatory to report AMR data from fresh meat samples from pigs and cattle (2021), and broiler and turkey (2022) imported from third countries collected at border control posts (BCPs). In 2022, the specific monitoring was performed by 26 MSs, the United Kingdom (Northern Ireland) and three non‐MSs for broilers, by 12 MSs and one non‐MS for fattening turkeys, by 26 MSs and two non‐MSs for broiler meat at retail and by 22 MSs and two non‐MSs for turkey meat at retail. In 2021, the specific monitoring was performed by 11 MSs and two non‐MSs for cattle under 1 year of age, by 26 MSs, the United Kingdom (Northern Ireland) and three non‐MSs for fattening pigs, 27 MSs and two non‐MS for meat from pigs, and by 27 MSs and one non‐MS for meat from cattle under 1 year of age, both sampled at retail.
Furthermore, in 2022, nine MSs reported results from imported broiler meat and three MSs for imported turkey meat collected at BCPs, while in 2021, five MSs reported results from imported pig meat and nine MSs for imported cattle meat collected at BCPs. Results from imported meat sampled at BCPs in 2021 and 2022 are presented in a specific Textbox.
In 2021, WGS was introduced as an alternative method to the phenotypic testing of E. coli and Salmonella isolates displaying phenotypic resistance to extended‐spectrum cephalosporins and/or carbapenems in Panel 1, and in the specific monitoring of ESBL‐/AmpC‐/CP‐producing E. coli. The harmonised protocols developed by the EURL‐AR have been followed when using the WGS technique, to ensure data comparability between countries reporting WGS data. In 2021, four MSs; Czechia, Germany, Italy and Finland, reported WGS results for the specific monitoring of ESBL‐/AmpC‐/CP‐producing E. coli in samples from cattle meat at retail (Czechia, Germany and Italy), cattle under 1 year of age (Germany and Italy), meat from pigs at retail (Czechia, Germany and Italy) and fattening pigs (Czechia, Germany, Finland and Italy). In 2022, six MSs and one non‐MSs (Czechia, Germany, Finland, Italy, the Netherlands, Sweden and Norway) reported WGS results for the specific monitoring of ESBL‐/AmpC‐/CP‐producing E. coli in samples from broilers (Czechia, Germany, Finland, Italy, the Netherlands, Sweden and Norway), turkeys (Germany, Italy and Norway), broiler meat at retail (Czechia, Germany, Finland, Italy, the Netherlands and Sweden) and turkey meat at retail (Czechia, Germany and Italy). In addition, the Netherlands and Germany reported WGS data for imported broiler meat and the Netherlands for imported turkey meat collected at BCPs. Reporting WGS data will facilitate and enhance understanding of the potential role and contribution of food‐producing animals and food derived thereof to the burden of AMR in humans (EFSA, 2019).
EFSA dashboard on ESBL‐, AmpC‐ and/or CP‐producing E. coli
The EFSA dashboard on Antimicrobial Resistance (available online here) is a graphical user interface for searching and querying the large amount of data collected each year by EFSA from EU MSs and other reporting countries based on Decision (EU) 2020/1729. In 2022, data on the occurrence of resistance and temporal trends of prevalence of ESBL‐, AmpC‐ and/or CP‐producing E. coli in broilers, fattening turkeys, fattening pigs and cattle under 1 year of age became available to the users. The data and trends of occurrence of ESBL‐, AmpC‐ and/or CP‐producing E. coli can be displayed interactively using charts, graphs and maps in the online EFSA dashboard, which allows the user to select for reporting year, reporting country and animal population. In this tool, the main statistics can also be viewed and downloaded in tabular format. Detailed information on the use and features of the Antimicrobial Resistance dashboard can be found in the user guide (a video embedded in the dashboard) and can also be downloaded from the online tool.
5.3. Food‐producing animals and derived meat: Routine antimicrobial resistance monitoring. Presumptive ESBL‐, AmpC‐ and/or CP‐producers and related WGS data
In 2021 and 2022, extended‐spectrum cephalosporin (ESC)‐resistance was investigated in Salmonella spp. from broilers, laying hens, fattening pigs, cattle under 1 year of age and from imported meat from broilers and turkeys, as well as in indicator E. coli from broilers, fattening turkeys, fattening pigs, cattle under 1 year of age, and in imported meat from broilers, turkeys and cattle, within the framework of the routine monitoring. 8 Isolates collected in the routine monitoring of E. coli and Salmonella spp. displaying phenotypic resistance to third‐generation cephalosporins in the first panel of antimicrobials tested, were subjected to susceptibility testing using the second panel of antimicrobials or WGS according to the updated legislation.
ESC resistance and ESBL/AmpC/CP phenotypes and genotypes in Salmonella spp.
The proportion of presumptive ESC‐resistant Salmonella spp. at the MS level collected within the routine monitoring based on phenotypic results, was generally very low or low in 2021 and 2022 (ranging between 0 and 2.5%, see Annex A2, Appendix G). The occurrence of ESC‐resistant Salmonella in a specific animal population may be greatly influenced by the prevalence of different Salmonella serovars in some countries. At the reporting MS‐group level, the occurrence of presumptive ESBL‐/AmpC‐producing Salmonella was 0.9% in fattening pigs, 2.5% in cattle under 1 year of age, 1.4% in broilers, 2.2% in fattening turkeys and 0.2% in laying hens. Detailed data per country and matrix is presented in Annex D2 and D3, Appendix G. An overview of presumptive ESBL‐/AmpC‐/CP‐producing Salmonella spp. reported in 2021 and 2022 is given in Table 18. Further information and WGS results regarding presumptive ESBL‐/AmpC‐/CP‐producing Salmonella spp. from food‐producing animals collected within the routine monitoring are presented in Chapter 2 Antimicrobial resistance in Salmonella spp.
TABLE 18.
Summary of presumptive ESBL‐/AmpC‐/CP‐producing Salmonella spp. and indicator commensal E. coli subjected to supplementary testing (panel 2) or whole genome sequencing, EU MSs, 2021‐2022.
| Matrix | ESBL and/or AmpC a | ESBL b | AmpC c | ESBL + AmpC d | CP e |
|---|---|---|---|---|---|
| n (% R) | n (% R) | n (% R) | n (% R) | n (% R) | |
| Salmonella | |||||
| Fattening pigs, 2021 (N = 1258, 25 MSs + XI) | 11 (0.9) | 9 (0.7) | 0 (0) | 2 (0.2) | 0 (0) |
| Calves, 2021 (N = 79, 10 MSs) | 2 (2.5) | 1 (1.3) | 0 (0) | 1 (1.3) | 0 (0) |
| Broilers, 2022 (N = 1911, 24 MSs + XI) | 26 (1.4) | 26 (1.4) | 0 (0) | 0 (0) | 0 (0) |
| Fattening turkeys, 2022 (N = 686, 19 MSs) | 15 (2.2) | 15 (2.2) | 0 (0) | 0 (0) | 0 (0) |
| Laying hens, 2022 (N = 908, 23 MSs + XI) | 2 (0.2) | 2 (0.2) | 0 (0) | 0 (0) | 0 (0) |
| Indicator commensal E. coli | |||||
| Pigs, 2021 (N = 4013; 27 MSs + XI) | 42 (1.0) | 31 (0.8) | 11 (0.3) | 0 (0) | 0 (0) |
| Calves, 2021 (N = 1599; 11 MSs) | 20 (1.3) | 19 (1.2) | 1 (0.1) | 0 (0) | 0 (0) |
| Broilers, 2022 (N = 4341; 27 MSs + XI) | 48 (1.1) | 39 (0.9) | 6 (0.1) | 3 (0.1) | 0 (0) |
| Fattening turkeys, 2022 (N = 1773; 13 MSs) | 18 (1.0) | 17 (1.0) | 1 (0.1) | 0 (0) | 1 (0.1) |
Abbreviations: N: total number of isolates tested; n: number of presumptive ESBL‐/AmpC‐/CP‐producing isolates; %R: percentage of resistant isolates; ESBL, extended‐spectrum beta‐lactamase; AmpC, AmpC beta‐lactamase; CP, carbapenemase.
According to EUCAST guidelines (EUCAST, 2017), only isolates showing MIC > 1 mg/L for CTX and/or CAZ or reported presence of ESBL‐/AmpC‐encoding gene were considered (see Appendix F – Materials and methods).
All isolates showing clavulanate synergy with CTX or CAZ or both, suggesting ESBL phenotype, or reported presence of ESBL‐encoding gene.
Isolates with cefoxitin resistance, suggesting AmpC phenotype, or reported presence of AmpC‐encoding gene.
Isolates showing synergy with CTX or CAZ and cefoxitin resistance, suggesting ESBL‐ and AmpC‐enzymes in the same isolates, or both ESBL‐ and AmpC‐encoding genes reported.
Isolates with meropenem resistance or CP‐encoding gene reported.
ESC resistance and ESBL/AmpC/CP phenotypes and genotypes in indicator commensal E. coli
The proportion of ESC‐resistant indicator E. coli (tested with panel 2) collected within the routine monitoring was generally low in 2021 and 2022. Among the reporting MSs, the occurrence of ESC resistance ranged from 0% to 8.7% in broilers, from 0% to 4.7% in fattening turkeys, from 0% to 5.5% in fattening pigs and from 0% to 4.1% in cattle under 1 year of age. At the MS‐group level, the occurrence of presumptive ESBL‐ and/or AmpC‐producers in indicator E. coli was 1.1% in broilers, 1.0% in fattening turkeys, 1.0% in fattening pigs and 1.3% in cattle under 1 year of age. The ESBL phenotype was more common than the AmpC phenotype in all matrices investigated (Table 18). Detailed data per country is available in Annexes D3 and D4.
Italy and Germany reported WGS data for indicator E. coli displaying ESC resistance in 2021. In Germany, bla CTX‐M‐1 (n = 3), bla CTX‐M‐32 (n = 1) and bla SHV‐12 (n = 1) were reported in E. coli from cattle under 1 year of age. Italy reported bla CTX‐M in eight isolates from cattle under 1 year of age, and bla SHV‐12 in one E. coli from fattening pigs.
Germany, Italy and the Netherlands reported WGS data for ESC‐resistant indicator E. coli in 2022. Germany reported bla SHV‐12 in three isolates from broilers and bla CTX‐M‐27 in one isolate from fattening turkeys. The Netherlands reported one isolate from broilers harbouring the bla TEM‐52B . In Italy, two isolates from fattening turkeys carried bla SHV‐12 and bla OXA‐181 , respectively. The detection of the CP‐encoding gene bla OXA‐181 is especially concerning, as carbapenems are last line antimicrobials used to treat severe human infections. The fact that it was detected without the use of selective isolation method is also worrying, as it might indicate a high occurrence of the CP‐resistant E. coli in the investigated sample.
5.4. Food‐producing animals and derived meat: Specific monitoring of presumptive ESBL‐/AmpC‐producing Escherichia coli and related WGS data
5.4.1. Prevalence and occurrence of presumptive ESBL‐/AmpC‐/CP‐producing Escherichia coli
The specific monitoring of ESBL‐/AmpC‐/CP‐producers includes culturing of samples on selective media containing 1 mg/L cefotaxime as recommended by EUCAST, enabling the detection of very low numbers of resistant isolates present within a sample. 9 The prevalence of presumptive ESBL‐ or AmpC‐producing E. coli for all matrices tested in 2021 and 2022 is shown in Figure 43. Detailed data on prevalence and occurrence per country and matrix is presented in Annex D.2–D.4, and the overall prevalence at MS level is presented in Table 19.
FIGURE 43.
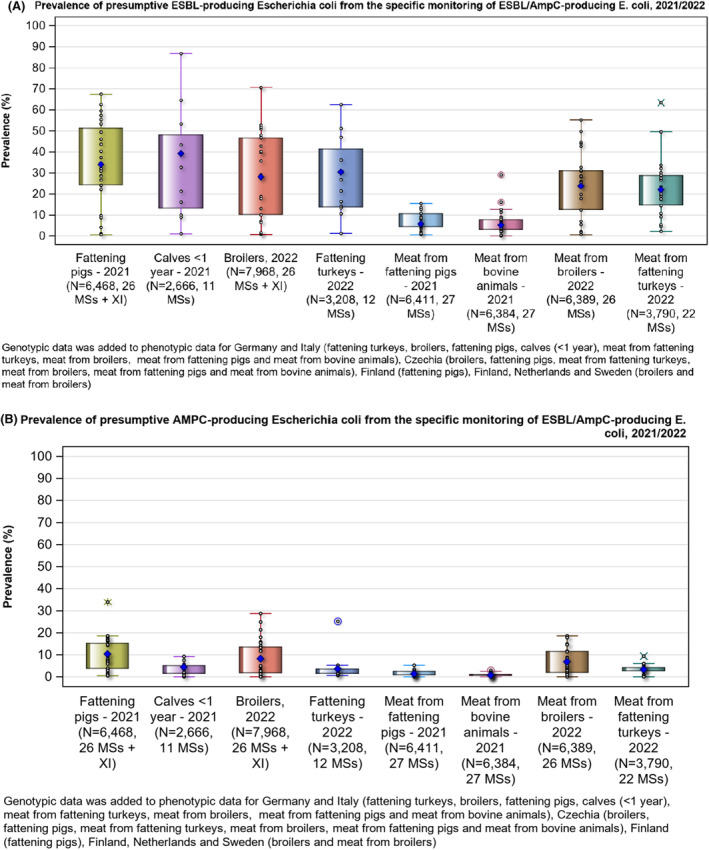
Prevalence of presumptive (A) ESBL‐producing and (B) AmpC‐producing Escherichia coli from the specific monitoring of ESBL‐/AmpC‐producing Escherichia coli, 2021/2022.
Note: N, number of samples tested; diamonds with white outline are the data (one data point per country); blue diamond is Total EU. Outliers (> 1.5 IQR from 75 percentile) are spotted using a different symbol for each matrix (i.e. square for fattening turkeys).
TABLE 19.
Summary of the presumptive ESBL‐and/or AmpC ‐producing Escherichia coli isolates from food‐producing animals and derived meat, specific monitoring, EU MSs, 2021/2022.
| Matrix | ESBL and/or AmpC producers a | ESBL producers b | AmpC producers c | ESBL + AmpC producers d | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Occ % | Prev% | n | Occ % | Prev% | n | Occ % | Prev% | n | Occ % | Prev% | |
| Pig meat at retail 2021 (Ns = 6411; N = 437; 27 MSs) | 435 | 99.5 | 6.9 | 353 | 80.8 | 5.6 | 75 | 17.2 | 1.2 | 7 | 1.6 | 0.1 |
| Bovine meat at retail 2021 (Ns = 6384; N = 361; 25 MSs) | 357 | 98.9 | 5.8 | 312 | 86.4 | 5.0 | 40 | 11.1 | 0.6 | 5 | 1.4 | 0.1 |
| Broiler meat at retail 2022 (Ns = 6389; N = 1915; 26 MSs) | 1882 | 98.3 | 29.6 | 1460 | 76.2 | 22.9 | 382 | 20 | 6.0 | 60 | 3.1 | 0.9 |
| Turkey meat at retail 2022 (Ns = 3790; N = 923; 22 MSs) | 915 | 99.1 | 24.2 | 800 | 86.7 | 21.2 | 92 | 10.0 | 2.4 | 35 | 3.8 | 0.9 |
| Pigs 2021 (Ns = 6468; N = 2714; 26 MSs + XI) | 2690 | 99.1 | 43.3 | 2050 | 75.5 | 33.0 | 578 | 21.3 | 9.3 | 64 | 2.4 | 1.0 |
| Calves, 2021 (Ns = 2666; N = 1125; 11 MSs) | 1118 | 99.4 | 42.5 | 1002 | 89.1 | 38.1 | 88 | 7.8 | 3.3 | 28 | 2.5 | 1.1 |
| Broilers, 2022 (Ns = 7968; N = 2804; 26 MSs + XI) | 2768 | 98.7 | 35.0 | 2123 | 75.7 | 26.8 | 556 | 19.8 | 7.0 | 95 | 3.4 | 1.2 |
| Fattening turkeys, 2022 (Ns = 3208; N = 1028; 12 MSs) | 1021 | 99.3 | 32.1 | 908 | 88.3 | 28.6 | 56 | 5.4 | 1.8 | 57 | 5.5 | 1.8 |
Abbreviations: Ns, total number of samples tested, N, number of isolates tested; n, number of presumptive ESBL‐/AmpC‐/CP‐producing isolates; Occ %, percentage of cephalosporin‐resistant isolates presenting a presumptive phenotype; Prev %, percentage of samples harbouring a presumptive ESBL‐/AmpC‐producing E. coli; ESBL, extended‐spectrum beta lactamase; AmpC, AmpC beta lactamase; CP, carbapenemase.
Note: Prevalence was calculated using the formula for genotypic prevalence presented in Appendix F – Materials and methods.
According to EUCAST guidelines (EUCAST, 2017), only isolates showing MIC > 1 mg/L for CTX and/or CAZ or reported presence of ESBL‐/AmpC‐encoding gene were considered (see Appendix F – Materials and methods).
All isolates showing clavulanate synergy with CTX or CAZ or both, suggesting ESBL phenotype, or reported presence of ESBL‐encoding gene.
Isolates with cefoxitin resistance, suggesting AmpC phenotype, or reported presence of AmpC‐encoding gene.
Isolates showing synergy with CTX or CAZ and cefoxitin resistance, suggesting ESBL‐ and AmpC‐enzymes in the same isolates, or both ESBL‐ and AmpC‐encoding genes reported.
Regarding food‐producing animals, the prevalence of presumptive ESBL‐ and/or AmpC‐producing 10 E. coli varied markedly between MSs during 2021–2022. When considering reported MICs, it ranged from 1.3% (Denmark) to 86% (Slovakia) in broilers, from 0% (Sweden) to 63.1% (Spain) in fattening turkeys, from 8.3% (Sweden) to 77.1% (Spain) in fattening pigs and from 5.8% (Denmark) to 59.1% (Belgium) in cattle under 1 year of age (Annexes D.2–D.4). Differences between MSs were also clear when the prevalence of ESBL‐ or AmpC‐producing E. coli was assessed separately (Annexes D.2–D.4). The prevalence of ESC‐resistant E. coli also varied among MSs reporting WGS data. In 2021, the prevalence ranged from 6.5% (Finland) to 80.7% (Italy) in fattening pigs and from 65.2% (Germany) to 90.3% (Italy) in cattle under 1 year of age. In 2022, the prevalence ranged from 1.3% (Finland) to 46.8% (Italy) in broilers and from 29.2% (Italy) to 39% (Germany) in fattening turkeys. The prevalence of presumptive ESBL‐ and/or AmpC‐producing E. coli in meat at retail also varied markedly between MSs. When considering MICs, it ranged from 3.4% (Denmark) to 61.3% (Hungary) in broiler meat, from 0% (Finland and Sweden) to 64% (Spain) in turkey meat, from 0% (Cyprus, Finland and Sweden) to 18.8% (Slovakia) in pig meat and from 0% (Cyprus and Finland) to 30.7% (Hungary) for cattle meat at retail (Annexes D.2–D.4). Differences between MSs were also clear when the prevalence of ESBL‐ or AmpC‐producing E. coli was assessed separately (Annexes D.2–D.4). For MSs reporting WGS data, the prevalence of ESC‐resistant E. coli ranged from 5.1% (Germany) to 8.2% (Czechia) in pig meat, and from 2.2% (Germany) to 7.4% (Czechia) in cattle meat in 2021. In 2022, the prevalence ranged from 1.7% (Sweden and Finland) to 40.1% (Italy) in broiler meat, and from 17.1% (Czechia) to 35.1% (Germany) in turkey meat Figures 45, 46. Additionally, an overview of meat samples collected at retail and BCPs is presented in Annex D.6, Table D.25.
FIGURE 45.
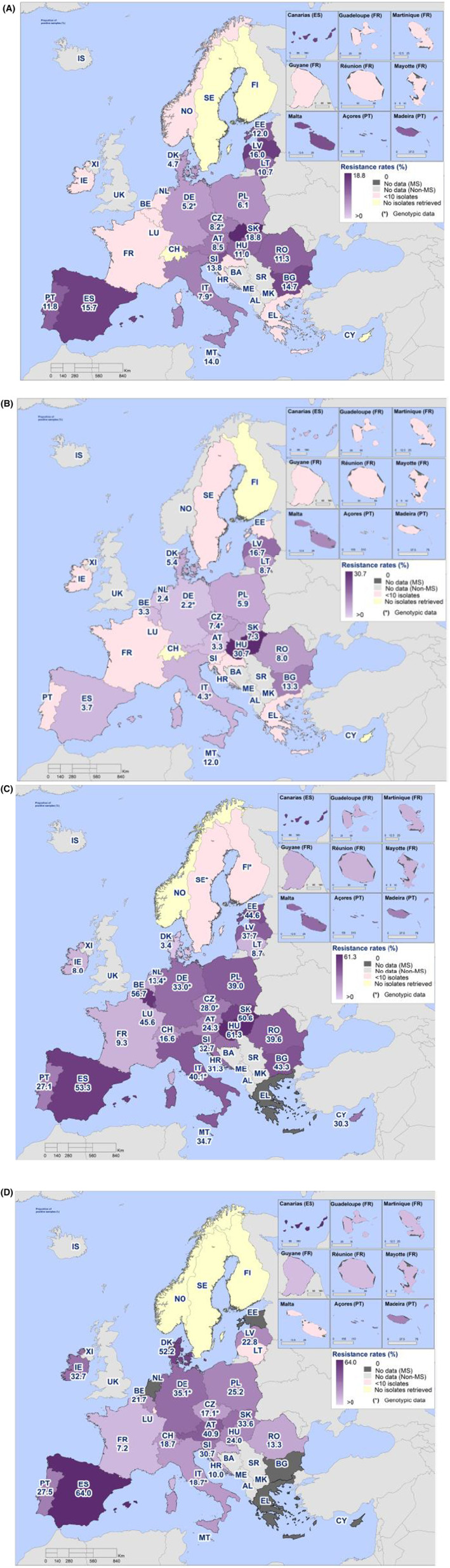
Spatial distribution of the prevalence of presumptive ESBL‐ and/or AmpC‐producing Escherichia coli from (A) pig meat in 2021, (B) cattle meat in 2021, (C) broiler meat in 2022 and (D) turkey meat in 2022, EU MSs and non‐MSs, 2021/2022.
FIGURE 46.
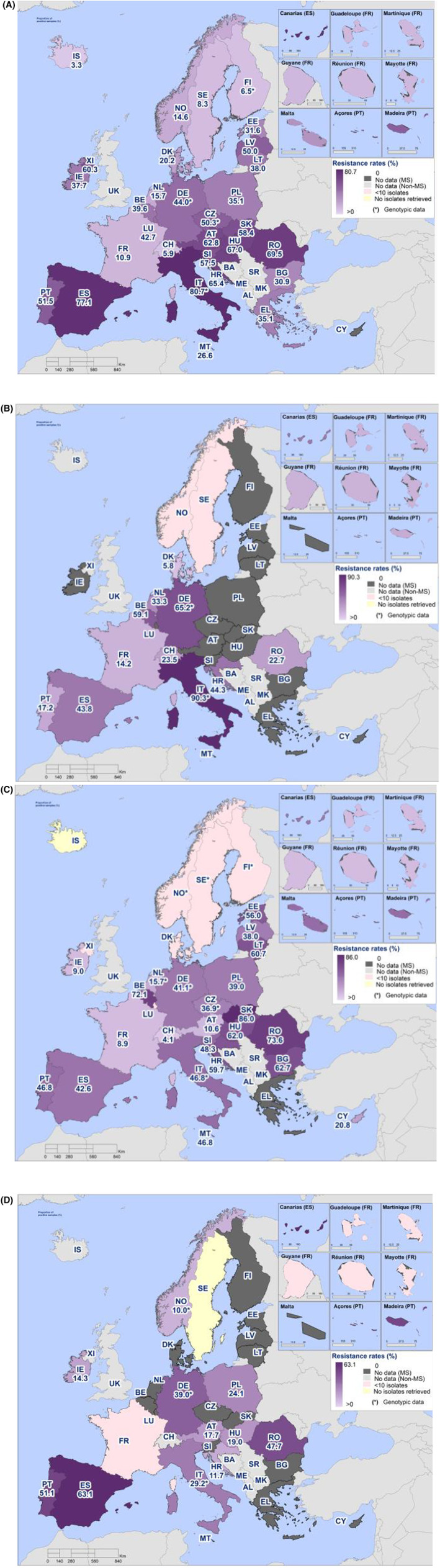
SpatialY distribution of the prevalence of presumptive ESBL‐ and/or AmpC‐producing Escherichia coli from (A) fattening pigs in 2021, (B) cattle under 1 year of age in 2021, (C) broilers in 2022 and (D) fattening turkeys in 2022, EU MSs and non‐MSs, 2021/2022.
ESBL‐, AmpC‐ and/or CP‐results based on WGS
Within the specific monitoring of ESBL‐/AmpC‐/CP‐producing E. coli, ESBL‐ and AmpC‐ encoding genes were reported by four MSs in 2021 (Czechia, Finland, Germany and Italy) and seven reporting countries in 2022 (Czechia, Germany, Finland, Italy, the Netherlands, Norway and Sweden). In 2021, WGS results were provided for isolates from fattening pigs (n = 579), cattle under 1 year (n = 473), cattle meat (n = 45) and pig meat (n = 70), while in 2022, WGS results were provided for isolates from broilers (n = 531), fattening turkeys (n = 260), broiler meat (n = 387) and turkey meat (n = 232). The majority of the isolates were reported to harbour ESBL‐encoding genes, followed by AmpC‐encoding genes. This is in concordance with the phenotypic findings, as presumptive ESBL‐producers were more commonly reported than AmpC‐producers in all animal and food matrices. A summary of CP‐producers identified in 2021 and 2022 is presented in Section 5.5 Monitoring of carbapenemase‐producing Escherichia coli.
Countries reporting WGS data reported several different genes responsible for the ESC‐resistance phenotypes (Appendix E and Figure 44). bla CTX‐M‐1 followed by bla CTX‐M‐15 were the most frequently reported ESBL gene in isolates from cattle under 1 year, cattle meat at retail, fattening pigs and pig meat at retail. In broilers, bla CTX‐M‐1 was also the most frequently reported ESBL‐encoding gene, but in this case followed by bla SHV‐12 , and vice versa for broiler meat at retail. bla CTX‐M‐15 and bla SHV‐12 , followed by bla CTX‐M‐1 were most frequently reported in fattening turkeys and bla CTX‐M‐15 followed by bla CTX‐M‐1 and bla SHV‐12 in turkey meat at retail.
For the isolates with AmpC‐phenotypes, bla CMY‐2 was the most commonly reported plasmid‐mediated gene in all animal populations and derived meat at retail, while the chromosomal point mutation C‐42T causing up‐regulation of the ampC gene, was the most frequently reported resistance mechanism responsible for the AmpC‐phenotype in fattening pigs, fattening turkeys and meat thereof.
FIGURE 44.
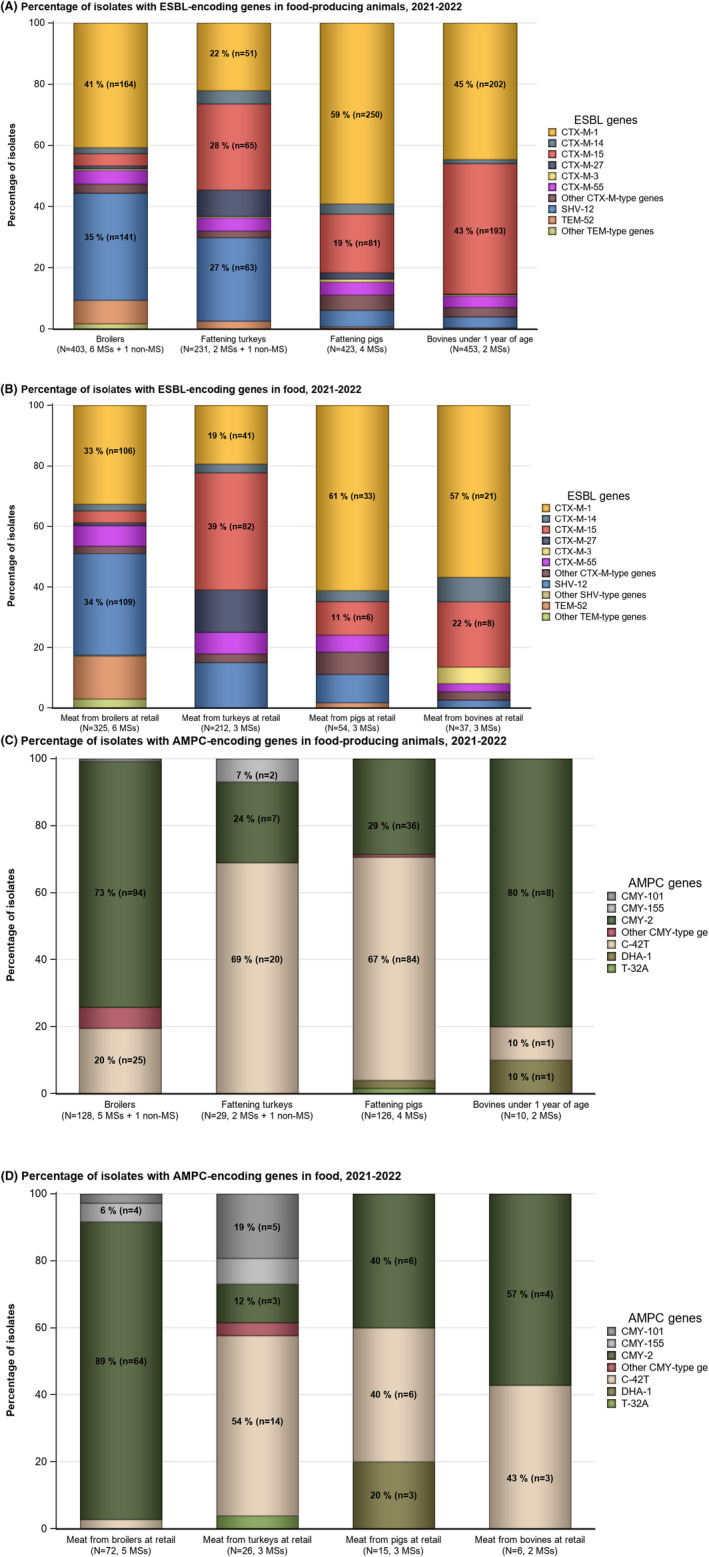
Escherichia coli isolates harbouring (A) ESBL‐encoding genes in animals, (B) ESBL‐encoding genes in retail meat, (C) AmpC‐encoding genes and AmpC‐chromosomal point mutations in animals and (D) AmpC‐encoding genes and AmpC‐chromosomal point mutations in retail meat.
Note: The figures include data from countries reporting WGS data to be used for analysis instead of MIC values. This excludes countries that provided both MIC results and WGS results voluntarily. ESBL, extended‐spectrum beta‐lactamase; AmpC, AmpC beta‐lactamase; N, number of isolates harbouring an ESBL or AmpC‐ encoding gene; n, number of isolates harbouring a specific gene.
In 2022, Austria, Finland, Italy, Ireland and Sweden reported both phenotypic and genotypic data for ESBL‐/AmpC‐/CP‐producing E. coli. In general, there was good correspondence between the presence of an ESBL‐/AmpC‐encoding gene and the predicted phenotype based on MIC data. A table showing reported genes and corresponding MIC values for cephalosporins and carbapenems is available on Zenodo (https://doi.org/10.5281/zenodo.10528846).
5.4.2. Relative abundance (occurrence) of presumptive ESBL‐/AmpC‐/CP‐producing Escherichia coli
If both ESBL‐ and AmpC‐producing E. coli isolates are present in a sample, the probability of detecting E. coli with either phenotype is influenced by the relative abundance of the phenotypes present in the investigated sample, as only a single isolate is further investigated and characterised. In the monitored animal populations and food matrices, the occurrence of presumptive ESBL‐producing E. coli exceeds that of presumptive AmpC‐producing E. coli in most countries (Figure 47–48, 49, 50 and Annex D.2). However, the occurrence of the different phenotypes varied considerably among the reporting countries for certain matrices. When excluding countries with less than 10 presumptive ESBL‐/AmpC‐producing isolates tested, the occurrence of isolates with an ESBL phenotype ranged from 24.6% (Latvia) to 97.7% (Germany) in broilers, 10% (Finland) to 98.7% (Latvia) in fattening pigs and 16.7% (Denmark) to 98.5% (Germany) in cattle under 1 year of age (Annex D.3 and D.4). For the remaining matrices, the differences were less pronounced and ranged between 47.1% (Romania) to 97.5% (Spain) in fattening turkeys, 50.8% (Latvia) to 96.2% (Luxembourg) in broiler meat, 62.5% (France) to 94.3% (Latvia) in turkey meat, 61.9% (Slovenia) to 92.6% (Austria) in pig meat and 53.3% (Denmark) to 100% (Belgium) in cattle meat. Cattle under 1 year of age and fattening turkeys are only investigated by some countries, at is it not mandatory for those with limited production. The overall prevalence at the MS level of presumptive ESBL‐/AmpC‐producers in meat from pigs (6.9%) and cattle (5.8%), was low, while it was high in broiler meat and turkey meat with 29.2% and 24.2%, respectively. A high overall prevalence at the MS level was reported for all animal populations investigated, with 32.1% in fattening turkeys, 34.9% in broilers, 42.5% in cattle under 1 year of age and 43.3% in fattening pigs.
FIGURE 47.
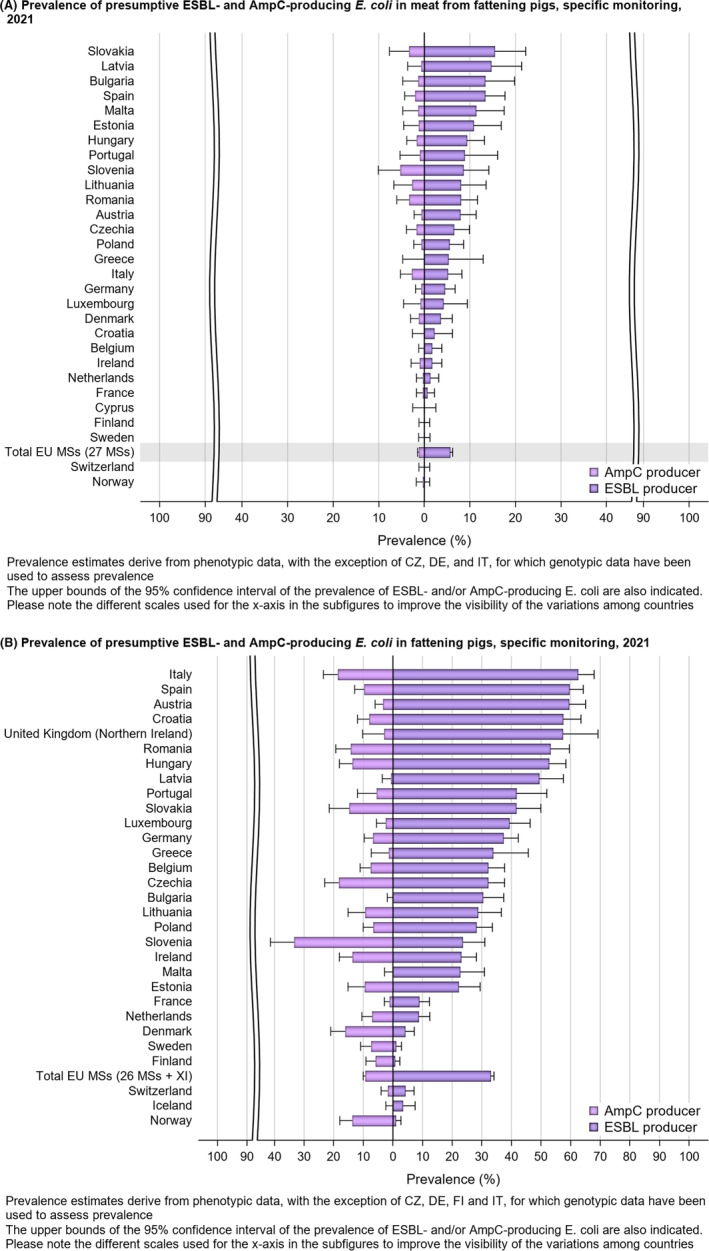
Prevalence of presumptive ESBL‐producing versus AmpC‐producing Escherichia coli from (A) pig meat and (B) fattening pigs, EU MSs and non‐EU MSs, 2021.
FIGURE 48.
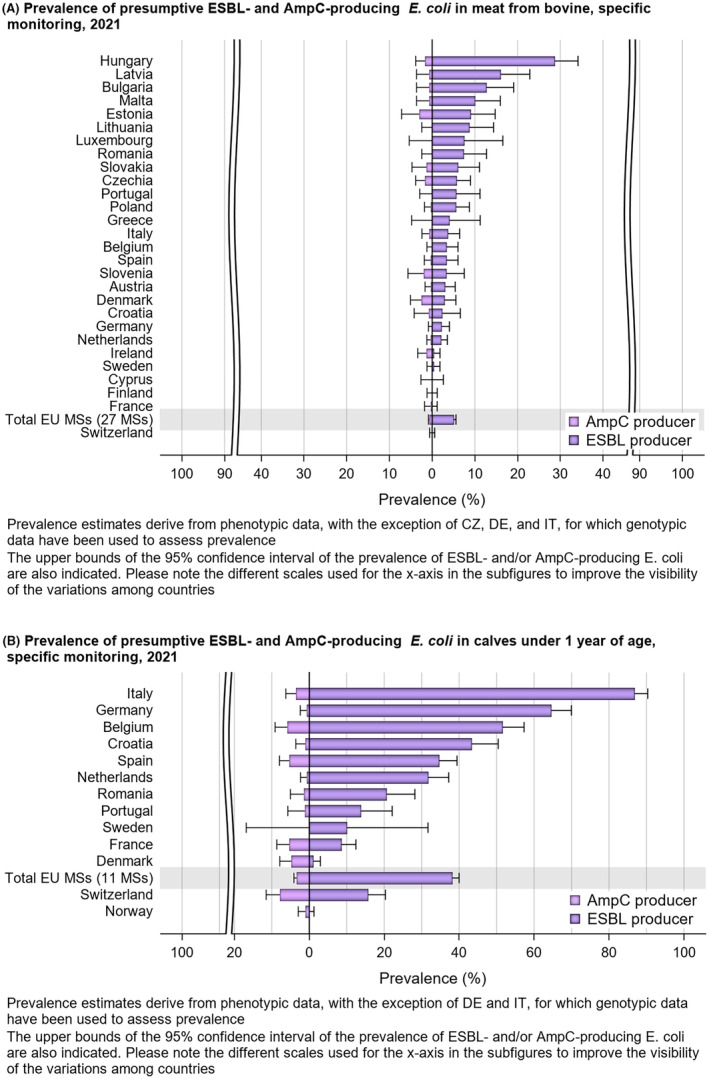
Prevalence of presumptive ESBL‐producing versus AmpC‐producing Escherichia coli from (A) cattle meat and (B) cattle under 1 year of age, EU MSs and non‐EU MSs, 2021.
FIGURE 49.
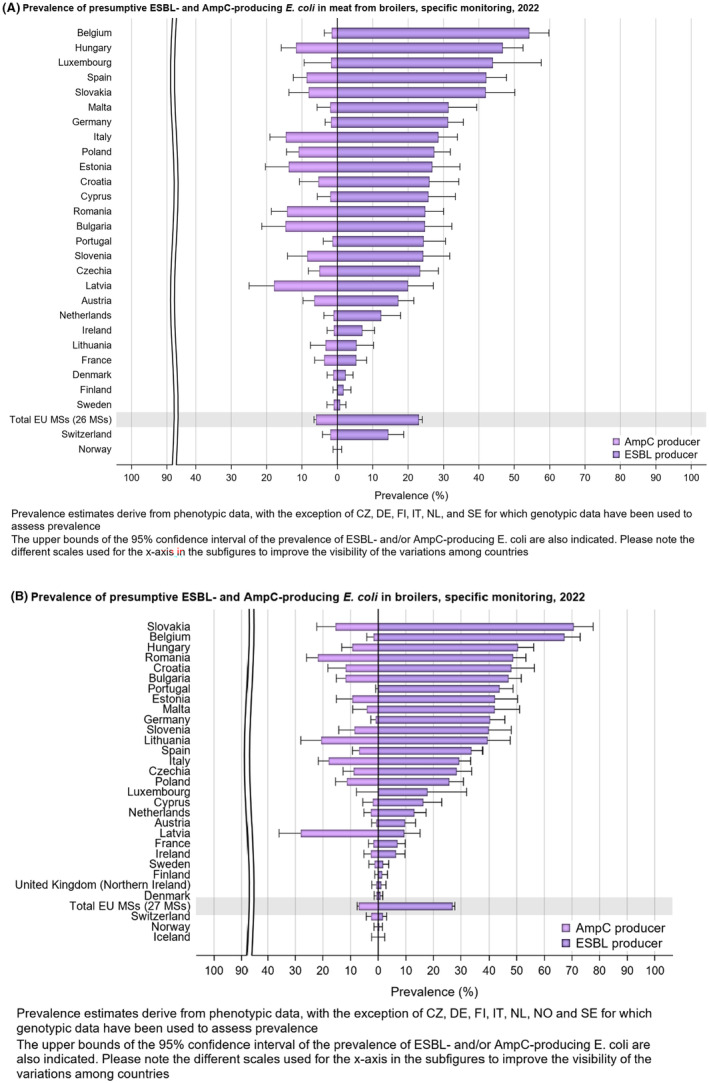
Prevalence of presumptive ESBL‐producing versus AmpC‐producing Escherichia coli from (A) broiler meat and (B) broilers, EU MSs and non‐EU MSs, 2022.
FIGURE 50.
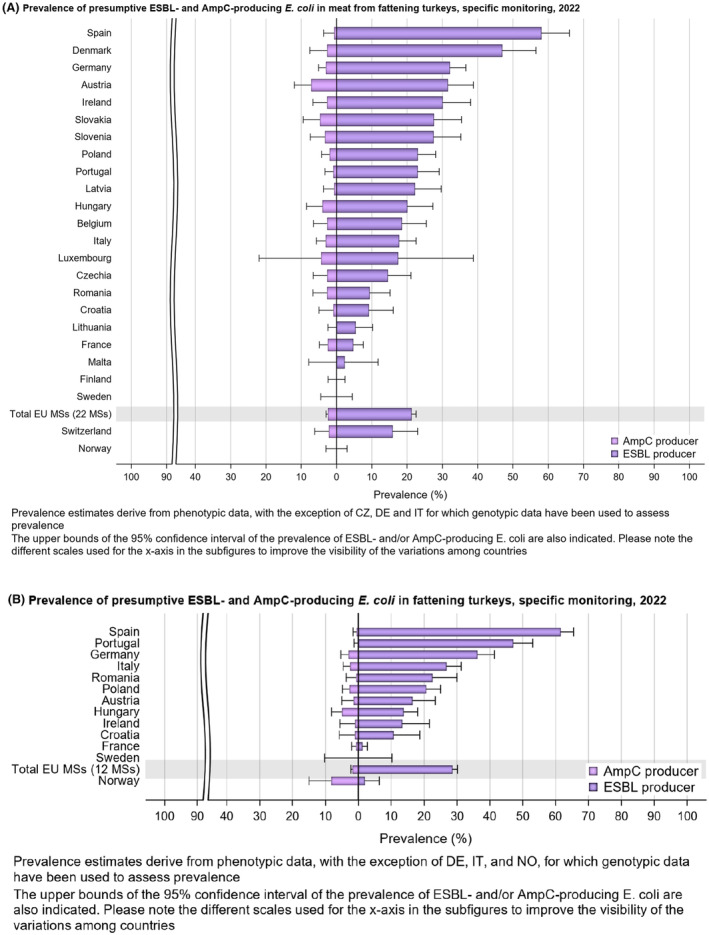
Prevalence of presumptive ESBL‐producing versus AmpC‐producing Escherichia coli from (A) turkey meat and (B) fattening turkeys, EU MSs and non‐ MSs, 2022.
Countries displayed in yellow colour include countries collecting samples without reporting any positive findings (0% prevalence) and countries displayed in grey colour include countries not reporting any data.
5.4.3. Temporal trends in prevalence of presumptive ESBL‐/AmpC‐/CP‐producers
Temporal trends in the prevalence of presumptive ESBL‐/AmpC‐producing E. coli in each separate animal population and meat category since the start of the mandatory harmonised monitoring is presented at both country and MS‐group level in Figures 51, 52.
FIGURE 51.
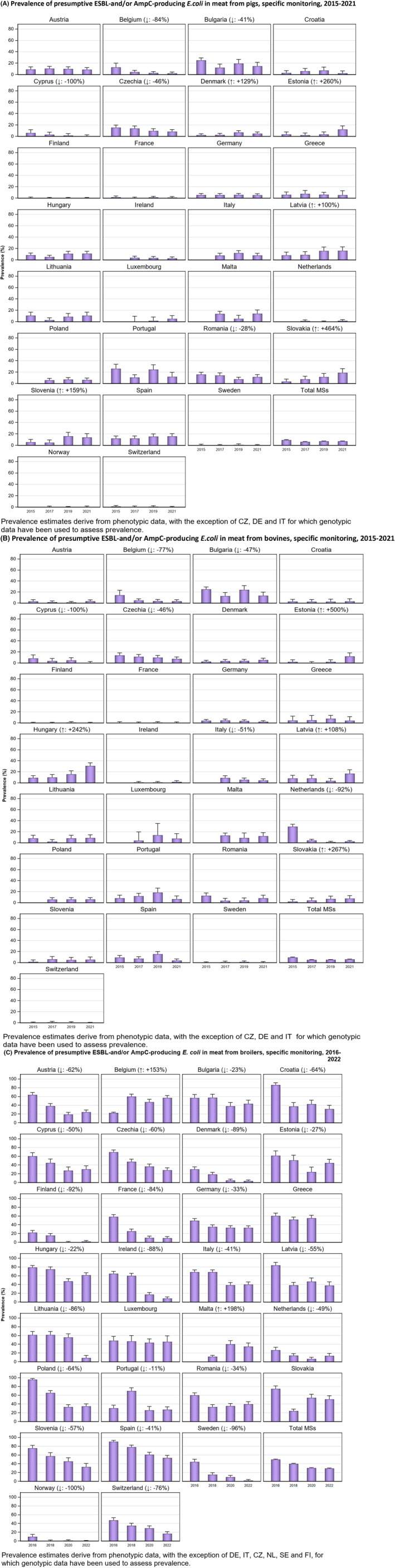
Trends on the prevalence of presumptive ESBL‐ and/or AmpC‐producing Escherichia coli in (A) pig meat, (B) cattle meat and (C) broiler meat, EU MSs and non‐MSs, 2016–2022. Arrows (↓/↑): indicate statistically significant decreasing/increasing trends over the period.
FIGURE 52.
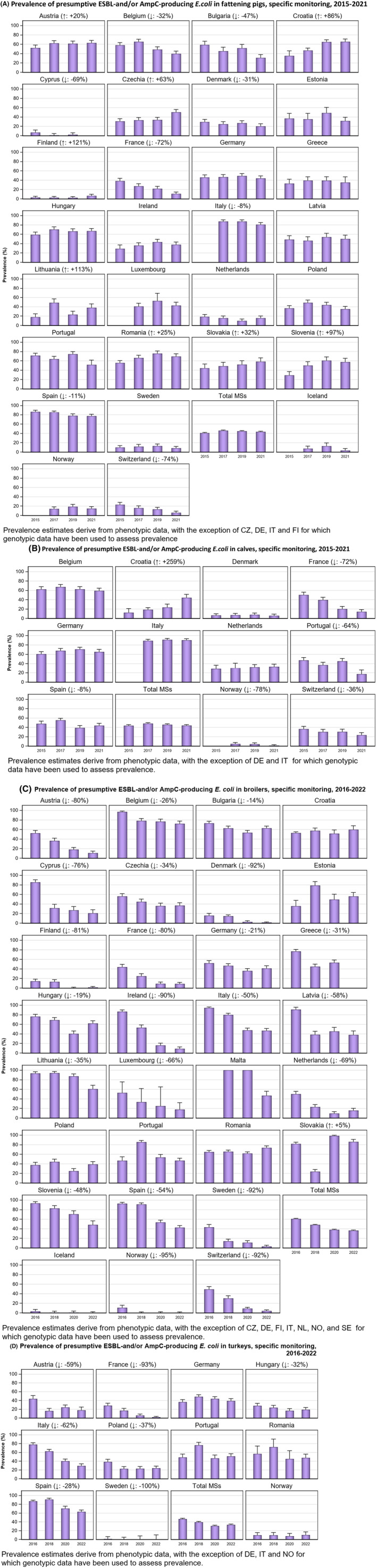
Trends on the prevalence of presumptive ESBL and/or AmpC‐producing Escherichia coli in (a) fattening pigs, (b) cattle under 1 year of age and (c) broilers, (d) fattening turkeys, EU MSs and non‐MSs, 2016–2022. Arrows (↓/↑): indicate statistically significant decreasing/increasing trends over the period.
Overall, the prevalence of presumptive ESBL‐/AmpC‐producing E. coli in broilers and broiler meat has slightly decreased at the MS‐group level. In 2022, the prevalence was at 34.9% and 29.2% at the MS‐group level for broilers and broiler meat, respectively, compared to 38.0% and 30.6% in 2020 (EFSA and ECDC, 2023). Austria, Belgium, Bulgaria, Cyprus, Czechia, Denmark, Finland, France, Germany, Greece, Hungary, Ireland, Latvia, Lithuania, Luxembourg, the Netherlands, Portugal, Slovenia, Spain, Sweden, Norway and Switzerland reported statistically significant decreasing trends in broilers. Further, Austria, Bulgaria, Croatia, Cyprus, Czechia, Denmark, Estonia, Finland, France, Hungary, Ireland, Latvia, Lithuania, the Netherlands, Poland, Portugal, Romania, Slovenia, Spain, Sweden, Norway and Switzerland reported statistically significant decreasing trends in broiler meat. Also, the overall prevalence in fattening turkeys at the MS level has decreased from 47% in 2016 to 32.1% in 2022. Statistically decreasing trends were seen in six countries (Austria, France, Hungary, Poland, Spain and Sweden).
In pig meat (6.9%) and cattle meat (5.8%), the prevalence of presumptive ESBL‐/AmpC‐producing E. coli at the MS‐group level in 2021 is comparable with that assessed in previous years. However, a significant increase has been reported in pig meat in Denmark, Estonia, Latvia, Slovakia and Slovenia, and in cattle meat in Estonia, Hungary, Latvia and Slovakia from 2015 to 2021.
In fattening pigs and cattle under 1 year of age, the prevalence of presumptive ESBL‐/AmpC‐producing E. coli assessed at the MS‐level in 2021 was also comparable to that in previous years. Eight countries (Belgium, Bulgaria, Cyprus, Denmark, France, Italy, Spain and Switzerland) reported statistically decreasing trends for fattening pigs, while five countries (France, Portugal, Spain, Norway and Switzerland) reported significant decreasing trends for cattle under 1 year of age from 2015 to 2021.
Decreasing trends in prevalence of ESBL‐/AmpC‐producing E. coli have been detected in several countries for all animal populations and meat matrices investigated, and some have reported considerable improvement during the period. However, some MSs have reported increasing or consistently high prevalence. Detailed data on the prevalence per country, animal population and meat category for 2021 and 2022 are presented in Figures 51, 52, and in Annex D. Details on historical data can be found in previously published reports.
5.4.4. Key outcome indicator of prevalence of ESBL‐/AmpC‐producing Escherichia coli (KOIESC )
The proportion of samples from broilers, fattening turkeys, fattening pigs and cattle under 1 year of age, weighted by PCU, that are positive for presumptive ESBL‐ and/or AmpC‐producing E. coli according to Commission Implementing Decision 2020/1729/EU has been retained as a summary indicator.
To account for differences in the relative size of food animal populations in a country, a weighted key outcome indicator of the prevalence of ESBL‐ and/or AmpC‐producing E. coli (KOIESC) was calculated. The indicator is the weighted mean of the prevalence of ESBL‐ and/or AmpC‐producing E. coli in each of the four animal populations monitored. For the calculation of the mean, the value for each population was weighted in relation to the relative size of the populations within a country using the ‘population correction unit’ (PCU). PCU is a technical unit of measurement used as an indicator of animal population size and was developed by the EMA, primarily to estimate sales of antimicrobials corrected by the animal population in individual countries. The data sources and the methodology for the calculation of PCU are comprehensively described in EMA's report ‘Sales of veterinary antimicrobial agents in 31 European countries in 2019 and 2020’ (EMA, 2023). For each country, KOIESC was calculated using data reported for two consecutive years. Thus, values for 2014–2015 were calculated from data for broilers and fattening turkeys in 2014 and on data for fattening pigs and cattle under 1 year in 2015. Likewise, values for 2015–2016 were calculated from data reported for fattening pigs and cattle under 1 year in 2015 and data reported for broilers and fattening turkeys in 2016, and so on. The changes in KOIESC are shown in Figure 11.
A positive development as shown by a decreasing trend in the KOIESC is seen in 22 countries. However, some of these countries started from very high or extremely high levels. For four countries, the trends in KOIESC has remained stable, with no statistically significant trend discerned in any direction, over the study period. Still, in three countries (Croatia, Romania), a statistically significant increasing trend in KOIESC was seen.
5.4.5. Discussion
Plasmid‐mediated ESBL genes are regarded as significant from a public health perspective (EFSA BIOHAZ Panel, 2011). AmpC beta‐lactamases can be produced due to both up‐regulation and overexpression of the chromosomal ampC gene and as a result of the presence of plasmid‐mediated AmpC genes in E. coli (Jacoby, 2009; Pfeifer et al., 2010). AmpC‐producers may also carry chromosomal mutations resulting in hyperexpression of the chromosomal ampC gene. Thus, not all classified isolates necessarily carry transferable genes. There are many different enzymes capable of conferring ESC resistance, with a corresponding number of genes encoding them (Chong et al., 2011; Pimenta et al., 2014). Plasmids may be transferred between E. coli strains (Händel et al., 2015), and it has been suggested that ESC‐resistant E. coli from agricultural settings may be of importance for human health (Manges, 2016; Törneke et al., 2015). Most countries classify isolates as presumptive ESBL‐, AmpC‐ or ESBL + AmpC‐producers based on phenotypes. Therefore, molecular characterisation is required to determine whether transferable genes encoding ESC resistance are present. Since 2021, WGS is recognised as an alternative method to phenotypic testing of isolates. In 2021, four MSs reported WGS data, while six MSs and one non‐MS reported WGS data in 2022.
In the routine monitoring of indicator commensal E. coli and Salmonella spp., the prevalence of ESBL‐/AmpC‐/CP‐producers was still low in 2021 and 2022.
In the specific monitoring of ESBL‐ and/or AmpC‐producing E. coli , at the reporting MS‐group level, the ESBL phenotype was more common than the AmpC phenotype in all monitored animal populations and meat categories. The same was the case in the majority of the reporting countries. Furthermore, the vast majority of reported genes were ESBL‐encoding, followed by AmpC‐encoding. However, the prevalence of the different phenotypes varied considerably between countries. In some countries, the AmpC phenotype dominated, such as in broilers in Latvia, cattle under 1 year of age in Denmark and fattening pigs in Norway. Only one presumptive ESBL‐/AmpC‐producing isolate per sample is further investigated. Thus, the relative abundance of E. coli with an ESBL and/or AmpC phenotype in the sample will influence the probability of detecting either phenotype.
Interestingly, the prevalence of presumptive ESBL‐/AmpC‐producing E. coli in pig meat and cattle meat was lower than that in fattening pigs and cattle under 1 year of age. In poultry, similar prevalence of presumptive ESBL‐/AmpC‐producing E. coli were seen in broilers and fattening turkeys and their derived meats. This observation may indicate that the ESBL‐/AmpC‐producing E. coli present in the animal intestine do not contaminate the carcases during the slaughter process. This may be influenced by the differences in the slaughter process for broilers compared to those for pigs and cattle. Another alternative is that the bacteria contaminating the carcases are somehow removed at a later stage in the process, before reaching the consumer.
Based on the WGS findings, bla CTX‐M‐1 and bla CTX‐M‐15 were the most commonly reported ESBL‐encoding genes in isolates from fattening pigs, pig meat, cattle under 1 year of age and cattle meat. In broilers, bla CTX‐M‐1 was the most frequently reported ESBL‐encoding gene, followed by bla SHV‐12 , while bla SHV‐12 followed by bla CTX‐M‐1 were the most frequently reported genes in broiler meat. bla CTX‐M‐15 and bla SHV‐12 were most frequently reported in fattening turkeys, and bla CTX‐M‐15 followed by bla CTX‐M‐1 in turkey meat. The frequent occurrence of bla CTX‐M‐1 and bla CTX‐M‐15 is supported by previous reports from food‐producing animals and derived meat in Europe (Day et al., 2016; Ewers et al., 2012; Ewers et al., 2021; Käsbohrer et al., 2019). Further, bla SHV‐12 has previously been reported as a common cause of ESC resistance in E. coli from poultry (Ewers et al., 2012; Ewers et al., 2021). Although this gene was reported in only a limited number of isolates from fattening pigs (n = 23), cattle under 1 year of age (n = 16), pig meat (n = 5) and cattle meat (n = 1) in 2021, it was frequently reported in broilers, fattening turkeys and their derived meat. This might indicate that bla SHV‐12 is more predominant in poultry than in pigs and cattle. In 2022, Germany reported 16 different ESBL‐encoding genes in poultry and derived meat. bla SHV‐12 was the most commonly occurring ESBL‐encoding gene in isolates from broilers (66/333) and broiler meat (68/467) in Germany, and it was also commonly occurring in isolates from fattening turkeys (20/351) and turkey meat (11/439). The bla SHV‐12 gene was also the second most common gene reported in broilers, broiler meat and turkey meat in Italy. Furthermore, the bla CTX‐M‐27 gene was frequently reported in fattening turkeys (20/351) and turkey meat (21/439) in Germany and turkey meat from Czechia (8/152), while only a single isolate from turkey meat in Italy carried this gene. On the other hand, Germany and the Netherlands were the only countries reporting the bla TEM‐52 gene. This was present in 31/333 isolates from broilers, 46/467 isolates from broiler meat and 6/351 isolates from fattening turkeys in Germany, and in 7/187 isolates from broiler meat and 4/300 isolates from broilers in the Netherlands. The observed differences in the distribution of ESBL‐encoding genes are likely a result of differences in the variety of genes present in different animal populations in different countries. This may be a result of variations in animal husbandry, animal breeds, antimicrobial use etc. However, further investigations are needed in order to elucidate the cause of the observed differences.
In all matrices investigated, bla CMY‐2 was the most common plasmid‐mediated AmpC‐encoding gene reported. The mutation C‐42T in the chromosomal AmpC gene was also frequently reported from fattening pigs, broilers and fattening turkeys. Both bla CTX‐M‐1 and bla CMY‐2 have persisted in the pig and broiler production in Europe for several years, and studies have suggested that this may be related to dissemination of epidemic plasmids carrying bla CTX‐M‐1 or bla CMY‐2 , respectively (Bevan et al., 2017; Mo et al., 2016).
The specific monitoring highlighted the presence of presumptive ESBL‐ and/or AmpC‐producing E. coli in caecal samples from all animal populations monitored. However, the prevalence of ESBL‐/AmpC‐producing E. coli at the MS‐level has decreased significantly in both broiler and fattening turkeys from 2014 to 2022. This is probably linked to the discontinuation of the off‐label use of extended‐spectrum cephalosporins in poultry (EMA/CVMP, 2018).
5.5. Monitoring of carbapenemase‐producing Escherichia coli
5.5.1. Routine monitoring of indicator Escherichia coli
In 2022, one CP‐producing indicator E. coli was reported from fattening turkeys in Italy. Following susceptibility testing using the first panel of antimicrobials, the isolate displayed meropenem resistance. Subsequently, it was subjected to WGS, and the carbapenemase‐encoding gene bla OXA‐181 was detected.
5.5.2. Specific monitoring of ESBL‐, AmpC‐ and/or CP‐producing Escherichia coli
The specific monitoring of ESBL‐/AmpC‐/CP‐producing E. coli on selective media (supplemented with 1 mg/L cefotaxime) also enables detection of isolates with some mechanisms of carbapenem resistance.
In 2021, Hungary reported the CP phenotype in one E. coli isolated from pig meat and two E. coli isolated from cattle meat collected at retail. The confirmatory testing exercise performed by the EURL‐AR confirmed that the three isolates harboured the bla NDM‐5 gene responsible for the CP phenotype (EFSA and ECDC, 2023).
In 2022, three CP‐producing isolates were reported under the specific monitoring of ESBL‐/AmpC‐/CP‐producing E. coli. One isolate originated from broilers in Italy, and two from broilers in Austria. All three isolates harboured the bla VIM‐1 gene.
5.5.3. Specific monitoring of carbapenemase‐producing Escherichia coli
Specific monitoring of CP‐producing bacteria using selective media for CP‐producers, in accordance with the protocol developed by the EURL‐AR 11 , 12 was made mandatory from 2021 (Appendix F – Materials and methods). In 2021 and 2022, 28 countries (25 MSs and 3 non‐MSs) investigated 8077 samples from broilers, 3031 samples from fattening turkeys, 5889 samples from fattening pigs, 2803 samples from cattle under 1 year of age, 6279 samples from broiler meat at retail, 3567 samples from turkey meat at retail, 5221 samples from pig meat at retail and 5126 samples from cattle meat at retail (Annex D.5).
In 2021, presumptive CP‐producers were detected in two samples from fattening pigs (Spain), (Annex D.5) based on phenotypic testing. The Spanish isolates from fattening pigs carried the bla OXA‐48 gene. Furthermore, two countries (Czechia and Italy) reported CP‐producing E. coli in 28 samples from fattening pigs and cattle under 1 year of age based on WGS data (Appendix E, Table E.3). The CP‐encoding gene bla OXA‐181 was identified in 20 E. coli isolates from fattening pigs and four E. coli isolates from cattle under 1 year of age (Italy), bla OXA‐48 in one E. coli from fattening pigs from Italy, and bla NDM‐5 in three E. coli from fattening pigs (Czechia) and one E. coli from cattle under 1 year of age (Italy). In 2022, a single CP‐producing E. coli was reported based on WGS data in the specific monitoring of CP‐producing E. coli. The isolate was from fattening turkeys sampled in Italy and carried the bla OXA‐181 gene (Appendix E, Table E.6). The CP‐encoding genes reported in 2021 and 2022 are summarised in Table 20.
TABLE 20.
Summary table on carbapenemase‐encoding genes reported in Escherichia coli sampled in the routine monitoring, the specific monitoring of ESBL‐/AmpC‐/CP‐producers and the specific monitoring of CP‐producers in 2021 and 2022.
| Year | Matrix | Gene | Number of isolates | Number of countries detecting the isolates |
|---|---|---|---|---|
| Routine monitoring of indicator E. coli | ||||
| 2022 | Fattening turkeys | bla OXA‐181 | 1 | 1 (IT) |
| Specific monitoring of ESBL‐/AmpC‐/CP‐producing E. coli | ||||
| 2021 | Pig meat at retail | bla NDM‐5 | 1 | 1 (HU) |
| Cattle meat at retail | bla NDM‐5 | 2 | 1 (HU) | |
| 2022 | Broilers | bla VIM‐1 | 3 | 2 (AT, IT) |
| Specific monitoring of CP‐producing E. coli | ||||
| 2021 | Fattening pigs | bla OXA‐48 | 3 | 2 (ES, IT) |
| bla OXA‐181 | 20 | 1 (IT) | ||
| bla NDM‐5 | 3 | 1 (CZ) | ||
| Cattle under 1 year of age | bla NDM‐5 | 1 | 1 (IT) | |
| bla OXA‐181 | 4 | 1 (IT) | ||
| 2022 | Fattening turkeys | bla OXA‐181 | 1 | 1 (IT) |
5.5.4. Discussion
Among the 47,874 samples included in the specific monitoring for ESBL‐/AmpC‐/CP‐producing E. coli and 39,993 samples included in the specific monitoring for carbapenemase‐producing E. coli in 2021 and 2022, 39 CP‐producing E. coli were detected. Six of these (one from pig meat, two from cattle meat and three from broilers) were reported in the specific monitoring of ESBL‐/AmpC‐/CP‐producing E. coli, and 32 (26 from fattening pigs, five from cattle under 1 year of age and one from fattening turkeys) in the specific monitoring of CP‐producing E. coli. In addition, a single CP‐producing isolate was detected in the routine monitoring of indicator E. coli in 2022, originating from fattening turkeys carrying the bla OXA‐181 gene. This is worrying, as carbapenems are considered as last line of therapy and are used for treatment of severe human infections. The detection of CP‐producing bacteria using non‐selective methods is worrying, as it may indicate that a high proportion of E. coli of isolates with this genotype in the particular sample. E. coli carrying bla OXA‐181 were also reported in fattening pigs and cattle under 1 year of age in 2021, but they were isolated using selective detection methods (Carfora et al., 2022; EFSA and ECDC, 2023).
The reported numbers of CP‐producing E. coli are still low. However, an increasing number of isolates has been observed compared with previous years (EFSA and ECDC, 2022). In a total of 48,184 samples investigated using selective methods for detection of CP‐producing microorganisms in 2015–2017, a total of three CP‐producing E. coli were reported. In 2019, three isolates reported by Germany were confirmed to carry bla VIM‐1 (pig meat), as well as bla OXA‐48 (fattening pigs) and bla GES‐5 (fattening pigs), while one isolate from broilers in Austria was confirmed to carry bla VIM‐1 in 2020. This isolate was collected as part of the specific monitoring of ESBL‐/AmpC‐/CP‐producing E. coli (EFSA and ECDC, 2022). The specific monitoring of CP‐producing microorganisms has only been mandatory and harmonised since 2021. When the harmonised monitoring will be carried out for some years, it will be possible to evaluate the trends in occurrence, as is done for ESBL‐/AmpC‐/CP‐producing E. coli. The occurrence of CP‐producing E. coli is higher in pigs and cattle and their meat products compared to poultry and their meat products.
In both, the specific monitoring of ESBL‐/AmpC‐/CP‐producers and the specific monitoring of CP‐producers, bla OXA‐181 (n = 24) was the most frequently reported CP‐encoding gene, followed by bla NDM‐5 (n = 7) and bla OXA‐48 (n = 3) in 2021 (Carfora et al., 2022; EFSA and ECDC, 2023). In 2022, bla VIM‐1 (n = 3) was the most commonly reported gene, followed by bla OXA‐181 (n = 2). The bla VIM‐1 gene was present in E. coli in broilers in Austria, as was also the case in 2020 (EFSA and ECDC, 2023). The class D carbapenemase bla OXA‐181 has mainly been associated with humans, and its first isolation from porcine E. coli was reported from Italy (Pulss et al., 2017). Further, the New Delhi metallo‐beta‐lactamase bla NDM‐5 has previously been isolated from dairy cows in Algeria, as well as dogs and humans, from pigs and poultry in China, and cattle in India (Ramírez‐Castillo et al., 2023). A structured survey of carbapenemase‐ and/or colistin‐resistant Enterobacterales conducted in 36 European countries in 2019 showed that bla NDM‐5 was the most frequently reported carbapenemase‐encoding gene in E. coli in humans in Europe (ECDC, 2018, 2023a, 2023b). The increasing occurrence of E. coli with bla NDM‐5 in Europe suggests that there is an ongoing global expansion of certain E. coli STs carrying this gene, and that this is now a significant concern in EU/EEA countries (ECDC, 2023a). Based on nucleotide similarity between plasmids carrying CP‐encoding genes in human and food‐producing animal sources, it has been suggested that specific plasmids have been circulating among E. coli in different species, possibly increasing the risk of zoonotic transmission of carbapenem resistance (Ramírez‐Castillo et al., 2023). However, further studies including epidemiological and genetic analyses are required to elucidate the transmission dynamics.
CP‐producing Enterobacterales have not only been reported among isolates collected in the harmonised EU monitoring (Bortolaia et al., 2021; Carfora et al., 2022; Diaconu et al., 2020; EFSA and ECDC, 2023; Garcia‐Graells et al., 2020), but also from companion animals (Ramírez‐Castillo et al., 2023; Rincon‐Real & Suárez‐Alfonso, 2022), food‐producing animals and derived meat, seafood and vegetables (Brouwer et al., 2018; Irrgang et al., 2019; Irrgang et al., 2020; Köck et al., 2018; Liu et al., 2018; Ramírez‐Castillo et al., 2023; Slettemeås et al., 2017; Touati et al., 2017; Zurfluh et al., 2015). Carbapenems are not authorised for use in animals in the EU, and spill‐over from CP‐encoding genes and/or bacteria from humans has been suggested as a potential source of CP‐producers in the food production (Irrgang et al., 2020; Madec et al., 2017). Dissemination of similar or closely related CP‐encoding genes in isolates from different farms in certain regions could also indicate horizontal/vertical spread within the food chain (Carfora et al., 2022). As CP‐producing Enterobacterales are probably still emerging in the matrices investigated in the harmonised EU monitoring, actions to preserve the situation can hopefully still be effective, preventing food‐producing animals and food from becoming an important source of CP‐producing bacteria for humans.
Carbapenem resistance in Enterobacterales from companion animals
Carbapenems are last‐resort antibiotics used to treat complicated infections in humans. The use of carbapenems in animals has been prohibited in the EU since 2022 (European Commission, 2022). No products have been approved for animals, but off‐licence use for companion animals was previously possible according to the ‘cascade’ principle. Internationally, two studies from the USA and Canada reported carbapenem use in around 0.5% of their prescriptions (Cole et al., 2022; Cousto et al., 2020). However, in one case, this use might have led to an outbreak of NDM‐5‐producing E. coli, prompting veterinarians not to use this class of antibiotics.
In Europe, despite the rarity of use, carbapenem‐resistant Enterobacterales are sporadically but regularly reported in companion animals (Figure 1). The type of enzyme reported often mirrors the human epidemiology in the same country, as for example VIM‐1 reported in a dog in Spain, where this enzyme is frequently found in hospitals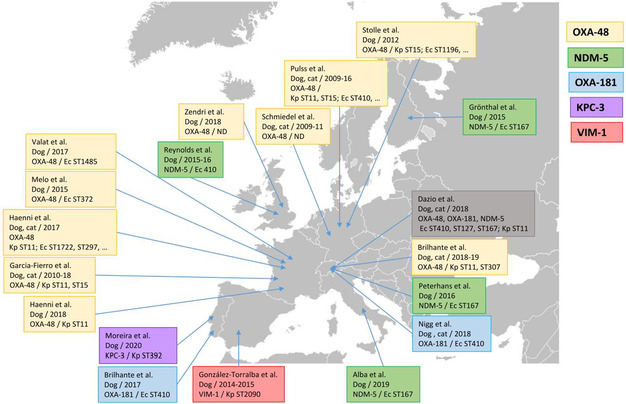 .
.
In France, the first carbapenem‐resistant OXA‐48‐producing E. coli was isolated from a dog in 2007 (Melo et al., 2017). Two additional studies identified the OXA‐48 enzyme in E. coli, Klebsiella pneumoniae or Enterobacter hormaechei in a veterinary clinic, in the environment and in hospitalised dogs, and the community (stools collected in parks) (Haenni, Boulouis, et al., 2022; Haenni, Métayer, et al., 2022). In parallel, OXA‐48‐producing Enterobacterales, mostly K. pneumoniae, have been regularly collected since 2014 through the Resapath network (García‐Fierro et al., 2022). This network, counting 108 member laboratories throughout France, gathers disc‐diffusion antibiogram data from both livestock and companion animals (https://resapath.anses.fr/Site_RESAPATH). Through this system, bacterial isolates can be collected based on their phenotype, thus offers the possibility of a continuous monitoring.
In Europe, other specific monitoring systems regarding AMR in bacteria from companion animals are in place, as in Sweden. There, carbapenemase‐producing Enterobacterales is notifiable according to legislation from the Board of agriculture. To increase the probability of detecting such bacteria, the Board of agriculture finances the confirmation of clinical isolates with resistance to extended‐spectrum cephalosporins and/or carbapenems. This confirmation is performed by the Swedish Veterinary Agency in Sweden. Every year, around 100 isolates, mainly from dogs and horses are received for confirmation and about 40–50 of those are confirmed as ESBL‐ or AmpC‐producers. So far, no carbapenemase‐producing Enterobacterales has been confirmed in domestic animals, whether food‐producing or companion animals, in Sweden.
Ignoring the problem of carbapenem resistance in companion animals on the grounds that these antibiotics are not used in the veterinary field would ignore a potential reservoir. Indeed, even if humans might be the origin on the contamination, carbapenem resistance genes are often carried by small plasmids (IncL, IncX3) that spread with a very high efficiency so that secondary reservoirs can arise in animals (Guo et al., 2022; Pitout et al., 2019). Resistant bacteria can further be transmitted to other animals or humans in contact, and disseminated to the close environment of the carrier. Moreover, scientific data show that these resistances are disseminating worldwide, also in livestock. Consequently, monitoring and characterising carbapenem‐resistant isolates in all animal sectors is important to help avoiding any diffusion. Furthermore, carbapenemase‐producing Enterobacterales are often also resistant to other antibiotic substances. Hence, a (further) general reduction in the use of antibiotics in all veterinary medicine is needed to minimise the risk of co‐selection of carbapenemase‐producing Enterobacterales.
The presence of CP‐producing E. coli in food‐producing animals and their meat products is concerning. The public health importance of CP‐producing E. coli and Salmonella spp. as both pathogens and vectors for resistance mechanisms, underlines the need to closely monitor and follow up further possible developments in this area. The occurrence of CP‐producing E. coli in food‐producing animals underlines the critical importance of having monitoring programmes specifically designed to detect these isolates, even when present in only small numbers. Amplification of CP‐producers in high‐intensity animal production systems may result in food‐producing animals becoming an additional source for human acquisition of such bacteria, which is highly unwanted (Carfora et al., 2022). It seems like the introduction of WGS has facilitated the detection of more CP‐producing E. coli than the use of phenotypic methods. Thus, the authorisation of WGS as an alternative method to phenotypic testing will hopefully facilitate the detection of CP‐producing isolates present in the investigated samples.
6. ANTIMICROBIAL RESISTANCE IN METHICILLIN‐RESISTANT STAPHYLOCOCCUS AUREUS (MRSA)
6.1. Key findings
Although not harmonised, the monitoring of MRSA in 2021 and 2022 provided useful information regarding the occurrence of MRSA in food‐producing animals and food. The results show a need for continued monitoring and molecular characterisation of MRSA in order to follow up on trends and evaluate the potential human health effects of MRSA in food‐producing animals and food.
Most MRSA isolates subjected to molecular typing were assigned to the livestock‐associated (LA) clonal complex (CC) 398. This was by far the dominating CC reported in MRSA from both animals and food in 2021 and 2022. However, spa‐types associated with community‐associated (CA) and hospital‐associated (HA) MRSA were also detected.
MRSA assigned to other CCs than CC398 were more common in food than in food‐producing animals.
The majority of MRSA isolates subjected to antimicrobial susceptibility testing in 2021 and 2022 were multidrug resistant (i.e. resistant to three or more antimicrobial classes), and extremely high levels of resistance to tetracycline were reported.
Resistance to the critically important antimicrobials linezolid and vancomycin were not reported in any of the isolates tested. However, Belgium reported the presence of the cfr‐gene, encoding linezolid resistance, in three isolates from pigs. Antimicrobial susceptibility data were not available for these isolates.
Resistance to the critical important antimicrobial rifampicin was detected in isolates from cattle, pig meat and poultry meat in 2021 and 2022.
6.2. Data on MRSA addressed
Methicillin‐resistant Staphylococcus aureus (MRSA) can be found on the skin and mucosa of humans and animals and may cause severe infections. MRSA can be divided into three broad categories: community‐associated (CA‐) MRSA, healthcare‐associated (HA‐) MRSA and livestock‐associated (LA‐) MRSA. These categories differ in their epidemiology, but the separation between them is not strict. HA‐MRSA and CA‐MRSA include strains predominantly affecting humans, and these strains are less frequently reported from food‐producing animals. LA‐MRSA have been detected in most farm‐animal species, including those covered by the AMR monitoring according to Decision 2020/1729/EU. Further information on MRSA can be found in specific sections of the dedicated EFSA story map on MRSA published together with the present report (see text box below).
EFSA story map on monitoring MRSA
The EFSA story map on monitoring MRSA is an interactive communication tool with the general public as the target audience. The story map is available online (here). This story map includes information on Staphylococcus aureus and MRSA, its epidemiology, transmission routes, occurrence of MRSA and mechanisms for AMR in MRSA and preventive measures. Also, the story map describes monitoring activities implemented for MRSA, and the role EFSA has in these activities. The story map allows users to easily explore the content of the different sections including both text and dynamic images.
Periodic monitoring of MRSA in food‐producing animals and food facilitates identification of trends in the dissemination and evolution of potentially zoonotic MRSA. In conjunction with systematic surveillance of MRSA in humans, potential transmission of MRSA from food‐producing animals to the human population can be analysed (EFSA, 2009a, 2009b, 2012). Thus, in addition to determining the prevalence of MRSA in different food chains, representative isolates should be characterised to determine molecular subtype and AMR. As the monitoring of MRSA and related AMR in Europe is voluntary, only a limited number of countries reported MRSA occurrence data to EFSA in 2021 and 2022. Some countries also reported data on spa‐ and/or sequence types, clonal complex and/or antimicrobial susceptibility. In 2024, a dedicated dashboard on MRSA was published for the first time. This is an online data visualisation tool where information on the occurrence of MRSA in animals and food can be visualised interactively (see text box below).
EFSA dashboard on MRSA
The EFSA dashboard on MRSA (available online here) is a graphical user interface searching and querying the data collected each year by EFSA from EU MSs and other reporting countries. The dashboard shows summary statistics on occurrence of MRSA. Further, data on spa‐types can be visualised. In the online EFSA dashboard, the data on the occurrence of MRSA can be displayed interactively using graphs, and the main statistics can also be viewed and downloaded in tabular format. Due to the scarcity of the information reported, data on temporal trends and on the occurrence of resistance in MRSA are not included in the online dashboard.
Following the EFSA's technical specifications for a baseline study on the prevalence of MRSA in fattening pigs in EU (EFSA, 2022), the Commission Implementing Decision (EU) 2023/1017 13 published on 23 May 2023 amended the EU/2020/1729 14 as regards the EU monitoring of MRSA in fattening pigs that shall be carried out in 2025. The purpose of this monitoring is to estimate the MRSA prevalence in the European population of fattening pigs. The target population is represented by healthy fattening pigs sampled at slaughter level. Details on the sampling design and testing requirements can be found in Decision (EU) 2023/1017 (available online).
As MRSA monitoring is not mandatory and harmonised, the availability of comparable data over time is limited. This is due to a scarce number of countries reporting data on MRSA from different animal populations and food matrices. Furthermore, the countries reporting MRSA data differ between years, as does the matrices tested. In addition, there are differences in both sampling strategies and types of samples collected between countries. Samples may also be analysed using different isolation methods with differences in sensitivity and specificity. Due to these data limitations, it is not possible to assess the temporal trends in MRSA occurrence and therefore, no visualisation of the temporal trends of MRSA is presented in the current report or in the EFSA's MRSA dashboard.
Findings on MRSA from humans are not specifically addressed in this chapter, but these are presented in the ‘Antimicrobial resistance in the EU/EEA (EARS‐Net) – Annual epidemiological report for 2022’ published by ECDC (ECDC, 2023a, 2023b). Antimicrobial susceptibility in invasive S. aureus from humans is, as of data collected in 2023, reported by all EU Member States and EEA countries to the European Antimicrobial Resistance Surveillance Network (EARS‐Net) hosted by ECDC. MRSA typing data are not reported, and possible links to the animal reservoir of LA‐MRSA are therefore not easily detected at the European level. The EU/EEA (excluding the United Kingdom) population‐weighted mean percentage of MRSA among invasive S. aureus isolates reported to EARS‐Net decreased significantly from 17.8% in 2018 to 15.2% in 2022. The estimated EU incidence of bloodstream infections with MRSA also decreased significantly from 5.80 per 100,000 population in 2018 to 4.94 per 100,000 population in 2022. Still, levels of MRSA were high in several European countries, and combined resistance to other antimicrobial groups was quite common. Thus, MRSA remains an important human pathogen in the EU/EEA (ECDC, 2023b).
6.3. Food and animals: MRSA
This section summarises the occurrence of MRSA and its susceptibility to antimicrobials in various food categories, in food‐producing animals excluding clinical investigations, and in clinical investigations in food‐producing and companion animals in 2021 and 2022. Additional tables on MRSA in food and food‐producing animals are included in Annex E that is available as supporting documentation in Zenodo (https://doi.org/10.5281/zenodo.10528846). Data on the occurrence of MRSA in foods and animals can also be visualised interactively in the online EFSA dashboard on MRSA (here).
In 2022, six MSs (Belgium, Germany, Italy, the Netherlands, Slovakia and Spain) and one non‐MS (Norway) reported data on the occurrence of MRSA in food and animals. Isolate‐based data on antimicrobial susceptibility was reported by Germany and Spain. In 2021, occurrence data on MRSA was reported by five MSs (Austria, Belgium, Finland, Germany and the Netherlands) and two non‐MSs (Norway and Switzerland). Furthermore, Austria, Belgium, Germany and Switzerland reported isolate‐based data on antimicrobial susceptibility of MRSA. It is important to keep in mind that findings and related comparison between countries should be interpreted with caution, as the monitoring of MRSA is not harmonised. Differences in the sensitivity of methods used for MRSA isolation as well as the sampling strategies, types of samples collected and sampling stage can influence the MRSA prevalence rates and should be taken into account when interpreting the results discussed below.
6.3.1. Food: Monitoring of MRSA
In 2021 and 2022, a very low number of countries reported data on the occurrence of MRSA in food (4 and 3, respectively). In 2021, Austria, Finland, Germany and the Netherlands provided data. The occurrence of MRSA was investigated in meat from cattle (Austria, Germany and the Netherlands) and meat from pigs (Austria, Finland and Germany). The Netherlands also provided data on MRSA detected in meat from broilers, turkeys, sheep, ducks and deer (Annex E, Table 1). In 2022, only Germany, the Netherlands and Spain provided data on the monitoring of MRSA in food. The occurrence of MRSA was investigated in meat from broilers (Germany, the Netherlands and Spain) and meat from turkey (Germany and the Netherlands). In addition, Germany reported data on MRSA in meat from ducks, and the Netherlands reported data on MRSA in meat from cattle, pigs, deer, farmed game, wild game (birds) and from fruits (Annex E, Table 2). Monitoring results on the occurrence of MRSA in food reported in 2021 and 2022 are presented in Figure 53, 54. It is of note that only food origins where positive isolates were obtained are included in the figure.
FIGURE 53.
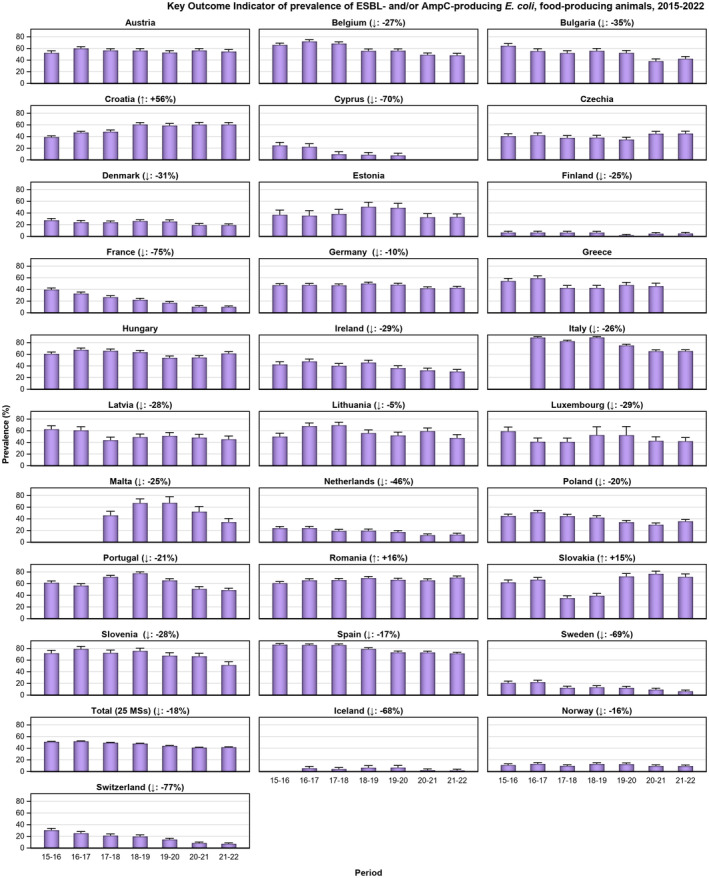
Changes in key outcome indicator of ESBL‐ and/or AmpC‐producing Escherichia coli (KOIESC), 27 MSs and 2 non‐MSs, 2015‐2022.
FIGURE 54.
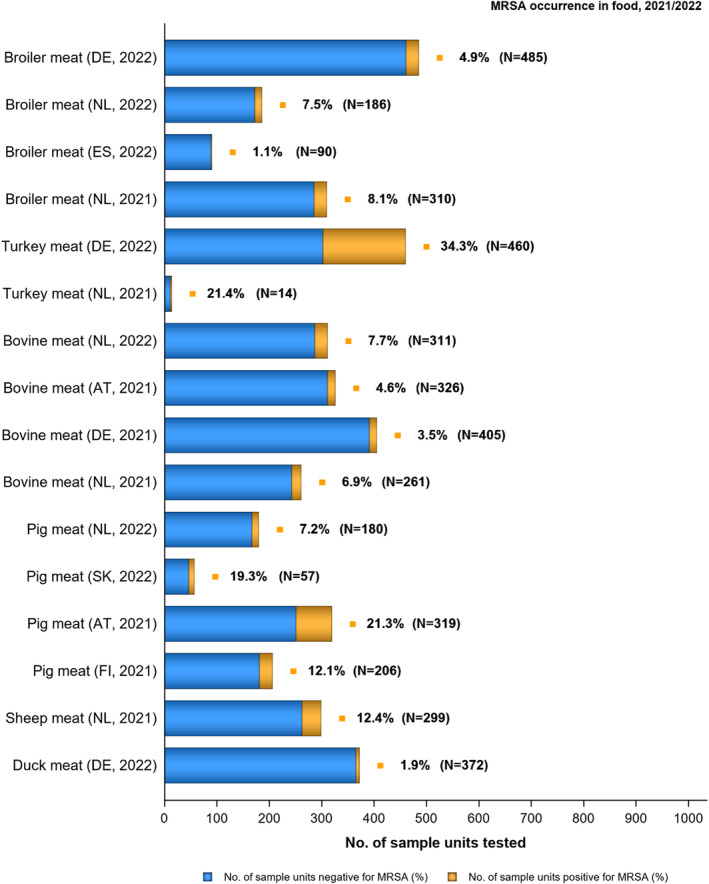
Methicillin‐resistant Staphylococcus aureus in food‐producing animals, 2021/2022.
Note: Only food‐producing animal categories where positive isolates were obtained and countries investigating > 10 samples are presented in this graph. The isolation method used for detection of MRSA is not considered in this analysis. N, Total number of sample units tested; BE, Belgium; CH, Switzerland; NL, the Netherlands; SK, Slovakia. Blue: number of units negative for MRSA (%). Orange: number of units positive for MRSA (%).
- Broiler meat (DE, 2022): t011 (2), t034 (17), t571 (2), t2011 (1), t 2330 (1), t10485 (1), all CC398
- Broiler meat (ES, 2022): CC398 spa‐type t6228 (1).
- Turkey meat (DE, 2022): t008 CC8 (1), t127 CC5 (1), CC398 spa‐types t011 (16), t034 (85), t899 (26), t1255 (1), t1422 (3), t1430 (3), t1580 (1), t2011 (2), t5452 (2), t10204 (2).
- Bovine meat (AT, 2021): CC45 spa‐type t095 (7), CC398 spa‐types t011 (4), t034 (2), CC121 spa‐type t898 (1), CC1 spa‐type t588 (1).
- Bovine meat (DE, 2021): CC1 spa‐types t174 (1), t559 (1), CC9 spa‐type t1430 (1), CC97 spa‐types t359 (1), CC130 spa‐type t843 mecC positive (1), CC9/CC398 spa‐type t899 (1), CC398 spa‐types t011 (4), t034 (4), t1451 (1).
- Pig meat (AT, 2021): CC9/CC398 spa‐type t899 (5), CC9 spa‐type t1430 (2), CC45 spa‐type t095 (11), CC398 spa‐types t011 (24), t034 (12), t1793 (1), t2576 (1), t9013 (1), t588 (1), t571 (1), no spa‐type reported (1).
- Pig meat (FI, 2021): CC45 spa‐type t728 (1), CC398 spa‐types t034 (14), t899 (1), t2741 (9), t4677 (1).
Note: sheep meat reported by NL in 2021 were from meat preparation (n = 44) and fresh meat (n = 255); bovine meat reported by NL in 2022 were from meat preparation (n = 147) and fresh meat (n = 164). In 2021, MRSA was also detected in meat from duck (n = 1). In 2022, MRSA was also detected in meat from wild gamebirds (n = 1), meat from farmed game‐land mammals (n = 2) and meat from other animal species or not specified (n = 1): as only a limited number of samples were collected and these categories are not included in the figure.
In both years, the highest occurrence of MRSA was reported from turkey meat, 34.3% reported in 2022 by Germany, 50% and 21.4% reported by the Netherlands in 2022 and 2021, respectively. However, it should be noted that only a limited number of samples (n = 8 and n = 14, respectively) were tested in 2022 and 2021 by the Netherlands. The MRSA occurrence in pig meat ranged from 7.2% in the Netherlands (2022) to 14.3% in Germany (2021, though only in seven samples) and 21.3% in Austria (2021). In general, the occurrence of MRSA was lower in meat from broilers and cattle. In broiler meat, the occurrence varied from 1.1% (Spain, 2022) to 8.1% (the Netherlands, 2021). In the Netherlands, a small decrease in the occurrence was seen from 2021 (8.1%) to 2022 (7.5%). In bovine meat, the MRSA occurrence varied from 3.7% in Germany (2021) to 4.6% in Austria (2021) and 9.1% in the Netherlands (2022). MRSA was detected in 12.4% of samples form sheep meat in the Netherlands (2021).
6.3.2. Animals: Monitoring of MRSA
6.3.2.1. Monitoring of MRSA in healthy animals
MRSA occurrence data from food‐producing animals (excluding clinical investigations) were reported by two MSs (Belgium and the Netherlands) and two non‐MSs (Norway and Switzerland) in 2021 and by three MSs (Belgium, the Netherlands and Slovakia) and one non‐MS (Norway) in 2022 (Annex E, Table 3). The data originated from different frameworks, including voluntary monitoring, surveillance, control programmes and specific surveys. The specific survey included veal calves in the Netherlands (2022). In 2021/2022, MRSA monitoring was performed on fattening pigs, breeding pigs, cattle under 1 year of age, dairy cows and meat production cattle.
In 2021 and 2022, MRSA was most frequently detected in pigs (Figure 55). In fattening pigs, the occurrence of MRSA ranged from 12.5% in Slovakia (2022), to 53.6% in Switzerland (2021) and 80% in Belgium (2022). In 2022, Belgium also reported MRSA in 45.3% of the breeding pig herds. In contrast, MRSA was not detected in any pig herds in Norway in 2021 and 2022 (see Annex E, Table 4). These findings are in line with results from previous years. However, as some countries changed the isolation method, direct comparison should be done with caution.
FIGURE 55.
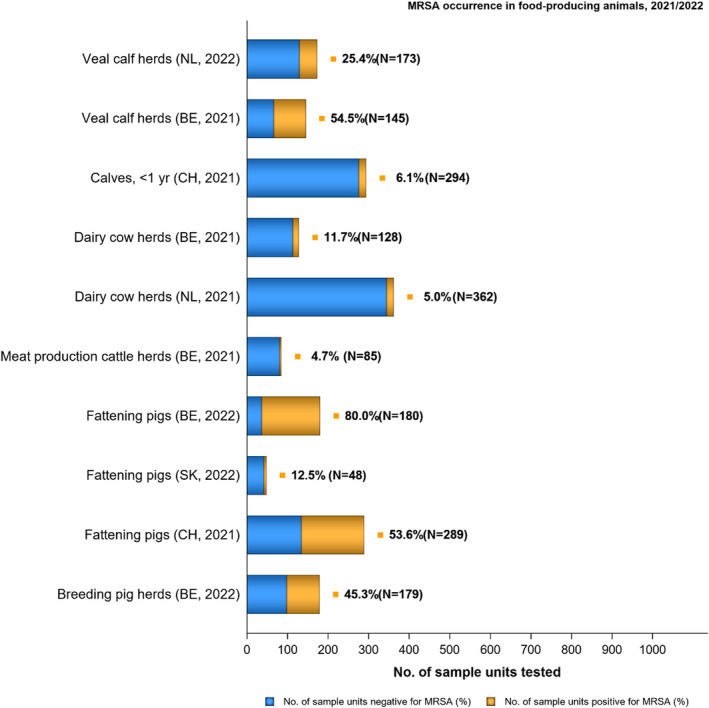
Methicillin‐resistant Staphylococcus aureus occurrence in food, 2021/2022.
Note: Only food origins where positive isolates were obtained and countries investigating > 10 samples are presented in this graph. The isolation method used for detection of MRSA is not considered in this analysis. N, Total number of sample units tested; AT, Austria; DE, Germany; FI, Finland; NL, the Netherlands; ES, Spain. Blue: number of units negative for MRSA (%). Orange: number of units positive for MRSA (%).
- Veal calves (BE, 2021): CC1 spa‐type t386 (1), CC398 spa‐types t011 (65), t034 (6), t1451 (1), t1456 (1), t2346 (1), t2370 (1), t3423 (1), t5210 (1), t6228 (1).
- Dairy cows (BE, 2021): CC8 spa‐type t037 (3), CC398 spa‐types t011 (10), t034 (2).
- Production cattle (BE, 2021): CC8 spa‐type t037 (2), CC398 spa‐type t011 (2).
- Fattening pigs (BE, 2022): CC398 spa‐types t011 (63), t034 (26), t779 (1), t1255 (1), t1451 (1), t1580 (2), t2011 (2), t3423 (1), no spa‐type reported (1). Breeding pigs (BE, 2022): CC398 spa‐types t011 (46), t034 (16), t588 (1), t1451 (1), t1457 (1), t2011 (3), t3423 (1), t5104 (1), t6575 (1), t15528 (1).
Note: Norway tested a high number of pig herds in 2021 (n = 763) and 2022 (n = 591), but no MRSA were detected.
MRSA was detected in 5% of sampled dairy herds in the Netherlands (2021) and 11.7% of dairy herds in Belgium (2021). The MRSA occurrence in calves ranged from 6.1% in Switzerland (2021) to 25.4% in the Netherlands (2022) and 54.5% in Belgium (2021). In contrast, only 4.7% of meat production cattle in Belgium were MRSA positive (2021).
6.3.2.2. Monitoring of MRSA in animals following clinical investigations
Clinical investigations in food‐producing animals typically differ from monitoring studies as selective culturing methods are normally not used, and the sampling is biased. These data can therefore not be used to infer occurrence or be extrapolated to the population level. However, it is still considered relevant to report animal species/populations affected, and MRSA lineages detected. In 2022, data on the occurrence of MRSA from clinical investigations were reported by the Netherlands and Italy, while in 2021 such data were reported only by the Netherlands (Annex E, Tables 5–7).
The Netherlands reported data following clinical investigations for MRSA in cats, dogs (sampled on veterinary clinics) and horses (sampled in farms) in both 2021 and 2022. The highest rates of MRSA were reported in horses (1.9% in 2021; 4.6% in 2022), followed by cats (0.8% in 2021; 2% in 2022) and dogs (0.1% in 2021; 0.2% in 2022) (Annex E.4, Tables 6, 7). In 2022, Italy reported clinical findings from two cattle, where MRSA was isolated from milk sampled on farm. No molecular data were provided.
6.3.3. Animals and food: Results of molecular typing of isolates
In 2021, molecular typing data were reported for 133 isolates from food originating from 3 MSs (Austria, Germany and Finland) and 270 isolates from food‐producing animals in one MS and one non‐MS (Belgium and Switzerland) (Annex E, Table 8). The MRSA dashboard (here) can be used to explore the spa‐type results interactively.
In 2022, results of molecular typing were reported for 447 (out of 544) MRSA isolates originating from food (Annex E, Table 9), submitted by Germany (n = 446) and Spain (n = 1). Also, molecular data were reported for 170 out of 275 MRSA isolates from food‐producing animals, all originating from Belgium (Annex E, Table 9). MRSA CC398 was the most commonly reported clonal complex in both food and food‐producing animals in both 2021 and 2022. The molecular typing results are presented in Figure 56, and Tables 21, 22.
FIGURE 56.
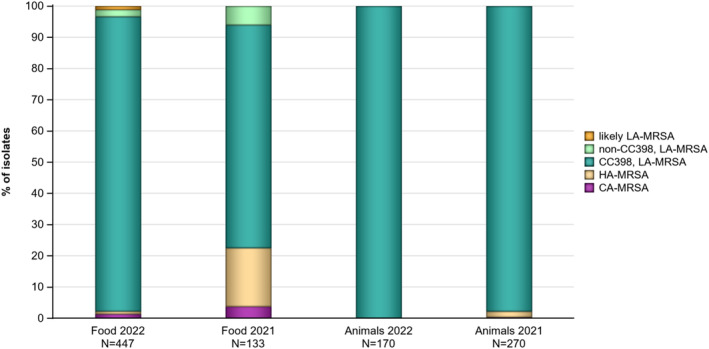
Methicillin‐resistant Staphylococcus aureus types reported from food and animals in 2021 and 2022, inferred from molecular typing data.
Note: N, number of reported isolates with typing data. HA‐MRSA, hospital‐associated MRSA; CA‐MRSA, community‐associated MRSA; LA‐MRSA, livestock‐associated MRSA; CC: clonal complex. The category ‘likely LA‐MRSA’ includes CC692 associated with birds (Monecke et al., 2016). The category ‘non‐CC398, LA‐MRSA’ includes CC9, CC97, CC121 and CC130 (Bal et al., 2016). One CC130 isolate categorised as ‘non‐CC398, LA‐MRSA’, carried the mecC gene. The category ‘HA‐MRSA’ includes CC5, CC8 ST247, CC8 t008, CC8 t009 and CC45 (Bal et al., 2016; Boost et al., 2012; Cuny et al., 2016). The category ‘CA‐MRSA’ includes CC1 and CC8 ST72 (Bal et al., 2016; Earls et al., 2021). Molecular typing data included spa‐type information, except for one isolate from pigs in Belgium, two isolates from turkey meat in Germany, nine isolates from pig meat in Austria, 17 isolates from cattle under 1 year of age from Switzerland and 155 isolates from pigs in Switzerland where only information on CC and/or ST was provided. If data on CC was not provided, the CC was inferred based on findings in the literature. Detailed information on mec‐genes, spa‐types, STs and CCs reported and CCs inferred are shown in Tables 8, 9 in Annex E.
TABLE 21.
spa‐types of CC398 and their detection in animals and food, 2021 and 2022.
| Spa‐type | Year | Animals (number of isolates) | Tot | Food (number of isolates) | Tot |
|---|---|---|---|---|---|
| t011 | 2022 | Pigs (109) | 109 | Broiler meat (34), Turkey meat (39) | 73 |
| 2021 | Calves (65), Dairy cattle (10), Cattle (2) | 77 | Pig meat (24), Bovine meat (8) | 32 | |
| t034 | 2022 | Pigs (42) | 42 | Broiler meat (39), Turkey meat (227) | 266 |
| 2021 | Calves (6), Dairy cattle (2) | 8 | Pig meat (26), Bovine meat (6) | 32 | |
| t538 | 2022 | 0 | Turkey meat (1) | 1 | |
| t571 | 2022 | 0 | Broiler meat (3) | 3 | |
| 2021 | 0 | Pig meat (1) | 1 | ||
| t588 | 2022 | Pigs (1) | 1 | Turkey meat (2) | 2 |
| 2021 | Pig meat (1) | 1 | |||
| t779 | 2022 | Pigs (1) | 1 | 0 | |
| t899 a | 2022 | 0 | Broiler meat (1), Turkey meat (46) | 47 | |
| 2021 | 0 | Pig meat (6), Bovine meat (1) | 7 | ||
| t1255 | 2022 | Pigs (1) | 1 | Turkey meat (4) | 4 |
| t1451 | 2022 | Pigs (2) | 2 | Broiler meat (1), Turkey meat (1) | 2 |
| 2021 | Calves (1) | 1 | Bovine meat (1) | 1 | |
| t1456 | 2021 | Calves (1) | 1 | 0 | |
| t1457 | 2022 | Pigs (1) | 1 | 0 | |
| t1580 | 2022 | Pigs (2) | 2 | Turkey meat (1) | 1 |
| t1793 | 2022 | 0 | Turkey meat (1) | 1 | |
| 2021 | 0 | Pig meat (1) | 1 | ||
| t2011 | 2022 | Pigs (5) | 5 | Broiler meat (2), Turkey meat (7) | 9 |
| t2330 | 2022 | 0 | Broiler meat (1) | 1 | |
| t2346 | 2022 | 0 | Turkey meat (1) | 1 | |
| 2021 | Calves (1) | 1 | 0 | ||
| t2370 | 2021 | Calves (1) | 1 | 0 | |
| t2576 | 2022 | 0 | Turkey meat (1) | 1 | |
| 2021 | 0 | Pig meat (1) | 1 | ||
| t2741 | 2021 | 0 | Pig meat (9) | 9 | |
| t2922 | 2022 | 0 | Turkey meat (1) | 1 | |
| t3423 | 2022 | Pigs (2) | 2 | 0 | |
| 2021 | Calves (1) | 1 | 0 | ||
| t4677 | 2021 | 0 | Pig meat (1) | 1 | |
| t5104 | 2022 | Pigs (1) | 1 | 0 | |
| t5210 | 2021 | Calves (1) | 1 | 0 | |
| t5452 | 2022 | 0 | Turkey meat (2) | 2 | |
| t6228 | 2022 | 0 | Broiler meat (1) | 1 | |
| 2021 | Calves (1) | 1 | 0 | ||
| t6575 | 2022 | Pigs (1) | 1 | Broiler meat (1) | 1 |
| t9013 | 2021 | 0 | Pig meat (1) | 1 | |
| t10485 | 2022 | 0 | Broiler meat (1) | 1 | |
| t14089 | 2022 | 0 | Turkey meat (1) | 1 | |
| t15528 | 2022 | Pigs (1) | 1 | 0 | |
| t21217 | 2022 | 0 | Turkey meat (1) | 1 |
Note: Calves are cattle under 1 year of age. Cattle are meat production cattle.
t899 may also be attributed to CC9.
TABLE 22.
spa‐types of other clonal complexes and their occurrence in animals and food, 2021 and 2022.
| Spa‐type | Clonal complex (sequence type) | Year | Animals (number of isolates) | Tot | Food (number of isolates) | Tot |
|---|---|---|---|---|---|---|
| t002 | 5 a | 2021 | Bovine meat (2) | 2 | ||
| t008 | 8 a | 2022 | Turkey meat (1) | 1 | ||
| 2021 | Bovine meat (2) | 2 | ||||
| t009 | 8 a | 2022 | Turkey meat (1) | 1 | ||
| t037 | 8 | 2021 | Dairy cows (3), Cattle (2) | 5 | ||
| t095 | 45 | 2021 | Bovine meat (7), Pig meat (11) | 18 | ||
| t127 | 5 (1) a | 2022 | Turkey meat (5) | 5 | ||
| t174 | 1 | 2021 | Bovine meat (1) | 1 | ||
| t235 | 1 a | 2022 | Turkey meat (1) | 1 | ||
| t242 | 5 (5) a | 2022 | Turkey meat (2) | 2 | ||
| t311 | 5 a | 2021 | Bovine meat (2) | 2 | ||
| t359 | 97 a | 2022 | Bovine meat (1) | 1 | ||
| t386 | 1 | 2021 | Calves (1) | 1 | ||
| t559 | 1 a | 2021 | Bovine meat (1) | 1 | ||
| t588 | 1 | 2021 | Bovine meat (1) | 1 | ||
| t728 | 45 | 2021 | Pig meat (1) | 1 | ||
| t843 | 130 a | 2021 | Bovine meat (1) | 1 | ||
| t898 | 121 | 2021 | Bovine meat (1) | 1 | ||
| t1346 | 8 | 2021 | Bovine meat (1) | 1 | ||
| t1422 | 692 a | 2022 | Turkey meat (5) | 5 | ||
| t1430 | 9 a | 2022 | Broiler meat (1), Turkey meat (6) | 7 | ||
| 2021 | Bovine meat (1), Pig meat (3) | 4 | ||||
| t2112 | 97 a | 2021 | Bovine meat (1) | 1 | ||
| t10204 | 9 a | 2022 | Turkey meat (3) | 3 |
The CC associated with the spa‐type is based on information from the literature, and not data reported by the countries. Calves are cattle under 1 year of age. Cattle are meat production cattle.
The most commonly reported CC was CC398, frequently associated with spa‐types t011 and t034. Spa‐types t011 and t034 were reported in 151 of 169 (89.3%) CC398 with spa‐types reported from food‐producing animals and in 339 of 420 (80.7%) CC398 with spa‐types reported from food in 2022. In 2021, t011 and t034 were reported in 85 of 92 (92.4%) CC398 isolates with spa‐types reported from food‐producing animals, and in 64 of 87 (73.6%) CC398 isolates with spa‐type information from food. This is in concordance with reports from previous years (EFSA and ECDC, 2022, 2023). In addition to t011 and t034, 30 other spa‐types were reported among CC398 isolates (Table 21). The most common was spa‐type t899, which was reported from pig meat and cattle meat in 2021 and from broiler meat and turkey meat in 2022. This spa‐type can be associated with both CC398 and CC9 as it can be a mosaic strain. The mosaic strain has the backbone of CC398 and has also acquired the CC9 region with the spa gene (Guardabassi et al., 2009; Larsen et al., 2016). Several spa‐types associated with CC398 originating from food and food‐producing animals were observed in the EU baseline in breeding pigs in 2008 (EFSA, 2009a). This underlines the possibility that these strains persist in the animal production. A complete overview of spa‐types associated with MRSA CC398 can be found in Table 21.
Isolates that did not belong to CC398 included a range of spa‐types associated with a number of different CCs (Annex E.5, Tables 8, 9). The highest number of MRSA not belonging to CC398 were reported from turkey meat (n = 24), bovine meat (n = 20) and pig meat (n = 15). The most frequently reported spa‐type among non‐CC398 isolates was t095 (n = 18) belonging to CC45, followed by t1430 (n = 7) belonging to CC9. Isolates belonging to t095 were reported from bovine meat and pig meat in 2021, while t1430 were reported in isolates from bovine meat and pig meat in 2021, and from broiler meat and turkey meat in 2022. A complete overview of spa‐types not associated with CC398 reported in 2021 and 2022 can be found in Table 22.
For isolates where only spa‐type or ST was reported, the CC was inferred based on reports in the literature. The following classifications were made:
spa‐types t011, t034, t571, t588, t1255, t1451, t1580, t1793, t2011, t2330, t2346, t2546, t5452, t6228, t6575 and t10485 were classified as CC398 (EFSA, 2009a; Kinross et al., 2017; Köck et al., 2013; Pauly et al., 2019).
spa‐type t899 were classified as CC9/398 (EFSA, 2009a; Guardabassi et al., 2009; Larsen et al., 2016).
spa‐types t127 and t559 were classified as CC1 (EFSA, 2009a; Kinross et al., 2017; Shukla et al., 2010).
spa‐types t002, t242 and t311 were classified as CC5 (Asanin et al., 2019; Köck et al., 2013; Silva et al., 2023).
spa‐types t008 and t009 were classified as CC8 (Boost et al., 2012; Cuny et al., 2016).
spa‐types t1430 and t10204 were classified as CC9 (EFSA, 2009a; Köck et al., 2013).
spa‐types t359 and t2112 were classified as CC97 (Boost et al., 2012; EFSA, 2009a).
spa‐type t843 was classified as CC130 (Bengtsson et al., 2017).
spa‐type t1422 was classified as CC692 (Silva et al., 2020).
STs 400, 402 and 8325 were classified as CC1 by PubMed. 15
ST 399 was classified as CC121 by PubMed.15
6.4. Summary data on the occurrence and susceptibility of MRSA
Determination of the susceptibility of MRSA isolates to antimicrobials, including those of medical importance, such as mupirocin, linezolid and vancomycin, provides valuable information on the MRSA situation in animals and food. It is important to monitor AMR patterns in different MRSA lineages as there is potential for multiple resistance genes harboured by less virulent strains to spread to other S. aureus strains (Sahibzada et al., 2017).
Data on antimicrobial susceptibility of MRSA were reported by Germany and Spain in 2022, and by Austria, Belgium, Germany and Switzerland in 2021 (Annex E, Tables 10, 11). All countries used a broth microdilution method and applied EUCAST epidemiological cut‐offs (ECOFFs) to determine the susceptibility of isolates. As expected, all MRSA isolates were resistant to penicillin and cefoxitin.
6.4.1. Susceptibility data of MRSA isolates from meat and food‐producing animals (excluding clinical investigations)
Figure 57 presents the overall resistance to selected antimicrobials of MRSA from meat in 2021 and 2022. Susceptibility data for MRSA from food‐producing animals were only provided in 2021, and the overall resistance to selected antimicrobials for these isolates can be found in Annex E, Figure 1. Also, detailed data on the occurrence of AMR in MRSA from food‐producing animals and meat reported in 2021 and 2022 are presented in Annex E: Tables 10, 11.
FIGURE 57.
Antimicrobial resistance in methicillin‐resistant Staphylococcus aureus in food, 2021 and 2022.
Note: AT: Austria; DE: Germany; ES: Spain. All isolates were resistant to penicillin and cefoxitin.
None of the MRSA isolates subjected to susceptibility testing in 2021 and 2022 were resistant to the critically important antimicrobials linezolid, vancomycin and mupirocin. Also, resistance to fusidic acid was not reported in any of the tested isolates. However, resistance to rifampicin, which is critically important for treatment of human patients (WHO, 2019) and may improve clinical outcomes when combined with other antimicrobials in treatment of MRSA (Nandhini et al., 2022), was reported in a limited number of isolates from dairy cows, meat production cattle and meat from pigs, broilers and turkeys in 2021 and 2022.
Extremely high levels of tetracycline resistance were reported in MRSA isolated from most animal populations and food matrices tested (Figure 57 and Annex E: Figure 1, Tables 10, 11). This is linked to most of the MRSA isolates belonging to CC398, in which tetracycline resistance is common (Crombé et al., 2012). In MRSA from food‐producing animals, 268 of 271 (98.9%) isolates tested were tetracycline resistant. In isolates from food, 70 of 107 (65.4%) isolates tested in 2021 and 419 of 447 (93.7%) isolates tested in 2022 displayed tetracycline resistance. For the other antimicrobial substances, the occurrence of resistance varied markedly between animal populations and food matrices.
In meat from broilers (fresh and carcase) from Germany (n = 84) and Spain (n = 1), extremely high resistance to erythromycin (81.2%), clindamycin (95.3%), quinupristin/dalfopristin (74.1%), tiamulin (77.6%) and trimethoprim (90.6%) were reported in 2022. A similar trend was seen in turkey meat (fresh and carcase) from Germany (n = 362) in 2022. Resistance to ciprofloxacin (44.5%), erythromycin (80.3%), clindamycin (81.2%), quinupristin/dalfopristin (69.3%), tiamulin (78.7%) and trimethoprim (79.6%) were reported at high or extremely high levels.
In 2021, Austria (n = 15) and Germany (n = 23) reported susceptibility data for MRSA from bovine meat. It is important to keep in mind that only a limited number of samples were tested when interpreting the data. High levels of resistance were reported to streptomycin (33.3%), ciprofloxacin (26.7%), erythromycin (26.7%) and clindamycin (26.7%) in isolates form Austria. Germany reported high levels of resistance to ciprofloxacin (34.8%), erythromycin (30.4%) and trimethoprim (21.7%).
Susceptibility data were reported for cattle under 1 year of age by Belgium (n = 79) and Switzerland (n = 18) in 2021. Very high to extremely high levels of resistance to gentamicin (67.1%), kanamycin (70.9%), clindamycin (93.7%), erythromycin (93.7%) and trimethoprim (96.2%) were reported from Belgium. Switzerland reported high to very high levels of resistance to streptomycin (50%), ciprofloxacin (55.6%), erythromycin (55.6%) and clindamycin (61.1%). Belgium also reported susceptibility data for MRSA from dairy cows (n = 15) and meat production cattle (n = 4) in 2021. Only a small number of isolates were tested. For dairy cows, high to very high levels of resistance were reported for kanamycin (60%), streptomycin (26.7%), erythromycin (60%), clindamycin (46.7%), quinupristin/dalfopristin (33.3%), tiamulin (26.7%) and trimethoprim (66.7%). In meat production cattle, high to extremely high levels of resistance were reported for gentamicin (50%), kanamycin (100%), streptomycin (50%), chloramphenicol (50%), rifampicin (50%), erythromycin (50%), sulfamethoxazole (50%) and trimethoprim (50%). Overall, the occurrence of resistance to gentamicin, kanamycin, streptomycin, erythromycin, clindamycin, quinupristin/dalfopristin, trimethoprim and tetracycline was higher in cattle than in cattle meat.
MRSA isolates originating from pig meat were susceptibility tested by Austria (n = 68) and Germany (n = 1) in 2021. Overall, high levels of resistance to ciprofloxacin (24.6%), erythromycin (37.7%), clindamycin (46.4%), quinupristin/dalfopristin (24.6%), tiamulin (24.6%) and trimethoprim (33.3%) were reported. MRSA from fattening pigs in Switzerland (n = 155) were susceptibility tested in 2021. High levels of resistance were reported for streptomycin (25.2%), ciprofloxacin (31.6%), erythromycin (29.7%), clindamycin (43.2%), quinupristin/dalfopristin (42.6%), tiamulin (43.2%) and trimethoprim (47.7%). The overall occurrence of resistance to gentamicin, kanamycin, streptomycin, chloramphenicol, ciprofloxacin, quinupristin/dalfopristin, tiamulin, trimethoprim and tetracycline was higher in fattening pigs than in pig meat.
6.4.2. Multidrug resistance (MDR) in MRSA isolates from meat and food‐producing animals (excluding clinical investigations)
All MRSA isolates were resistant to the beta‐lactams penicillin and cefoxitin. Moreover, most isolates also showed resistance to three or more antimicrobial groups and were considered multidrug resistant (MDR). In this analysis, penicillins and cephalosporins are considered as two separate antimicrobial groups.
Substantial variability was observed in the resistance patterns of MDR MRSA isolates (Annex E, Tables 12, 13). In 2021, 93.7% (354/378) of MRSA reported from cattle (calves, dairy cows and meat production animals), pigs and meat thereof from Austria, Belgium, Germany and Switzerland were MDR.
All MRSA isolated from fresh broiler meat and 59 of 60 (98.3%) of MRSA isolated from broiler carcases in 2022 were MDR. In addition, all isolates from fresh turkey meat and turkey carcases sampled in 2022 were MDR. The most common MDR pattern for isolates from both broiler meat and turkey meat (including both fresh meat and carcases) was penicillin, cefoxitin, erythromycin, clindamycin, quinupristin/dalfopristin, tiamulin, trimethoprim and tetracycline. This MDR pattern was reported in 13 of 25 (52%) MDR isolates from broiler meat, in 31 of 59 (52.5%) MDR isolates from broiler carcases, in 44 of 143 (30.8%) isolates from fresh turkey meat and in 78 of 219 (35.6%) of isolates from turkey carcases. The MDR pattern penicillin, cefoxitin, ciprofloxacin, erythromycin, clindamycin, quinupristin/dalfopristin, tiamulin, trimethoprim and tetracycline was also commonly reported in isolates from fresh turkey meat (28.0%) and turkey carcases (21.5%).
MRSA from pig meat tested in 2021 were also frequently reported to be MDR, with 58 of 69 (84.1%) displaying resistance to three or more antimicrobial classes. The most frequently reported MDR pattern was penicillin, cefoxitin and tetracycline, reported in 11 of the 58 (19.0%) MDR isolates, followed by penicillin, cefoxitin, erythromycin, clindamycin and tetracycline, reported in six of the 58 (10.3%) MDR isolates.
Of the 38 MRSA isolates from bovine meat, 25 (65.8%) were MDR. A variety of MDR patterns were observed, but the most common were penicillin, cefoxitin and ciprofloxacin (n = 4, 16.0%), penicillin, cefoxitin and tetracycline (n = 3, 7.9%), penicillin, cefoxitin, gentamicin and kanamycin (n = 2, 8.0%) and penicillin, cefoxitin, kanamycin, streptomycin, erythromycin and tetracycline (n = 2, 8.0%).
In MRSA from cattle, all isolates subjected to susceptibility testing were MDR. The MDR pattern penicillin, cefoxitin, gentamicin, kanamycin, erythromycin, clindamycin, trimethoprim and tetracycline was detected in 28 (28.9%) isolates from cattle under 1 year of age. In dairy cows, the MDR pattern penicillin, cefoxitin, kanamycin, streptomycin, chloramphenicol, rifampicin, erythromycin, sulfamethoxazole and tetracycline was reported in three isolates (20.0%), and the patterns penicillin, cefoxitin, gentamicin, kanamycin, erythromycin, clindamycin, trimethoprim and tetracycline and penicillin, cefoxitin, ciprofloxacin, trimethoprim and tetracycline were reported for two (13.3%) isolates each. Two isolates (50.0%) from meat production animals were resistant to penicillin, cefoxitin, gentamicin, kanamycin, trimethoprim and tetracycline, and two (50.0%) were resistant to penicillin, cefoxitin, kanamycin, streptomycin, chloramphenicol, rifampicin, erythromycin, sulfamethoxazole and tetracycline.
6.5. Discussion
As monitoring of MRSA in animals and food is voluntary, only a limited number of countries report data on the occurrence of MRSA to EFSA. Also, monitoring of MRSA is not harmonised, and comparison of MRSA occurrence in animal populations is therefore complicated. Countries may have different sampling strategies, different sampling locations, and also collect different sample types and number of samples. In addition, isolation methods may vary between countries. All these aspects may affect the estimation of MRSA prevalence. This should be kept in mind when evaluating the results from different countries (Mesa Varona et al., 2020).
Several types of food were investigated for the presence of MRSA in 2021 and 2022, including meat from cattle, broilers, pig, turkeys, deer (venison), duck, sheep, farmed‐ and wild game and fruits. MRSA were detected in all matrices at varying levels, except fruit and meat from deer. Meat is not considered as a major source for human MRSA acquisition (Tang, Larsen, et al., 2017; Wendlandt et al., 2013), and detection methods often involves the use of selective culture techniques, which may detect very low levels of contamination (Pauly et al., 2019). Nevertheless, the presence of MRSA in food products is considered a public health concern (Lozano et al., 2020).
Data on MRSA from animals were collected through monitoring programmes, surveys and after clinical investigations in 2021 and 2022. In 2021 and 2022, cattle under 1 year of age, dairy cows, meat production cattle, breeding pigs and fattening pigs were investigated for the presence of MRSA as part of monitoring or surveillance programmes. MRSA were detected in all animal populations investigated, but at varying levels.
Molecular typing data on spa‐types, STs and/or CCs were reported by some countries. The most frequently reported CC overall was CC398, which is in concordance with findings from previous years (EFSA and ECDC, 2023). Among isolates from animals with molecular typing data reported, only six of the 270 (2.2%) isolates collected in 2021 belonged to other CCs than CC398. All isolates reported with typing data from food‐producing animals collected in 2022 were CC398. The six isolates from 2021 that were non‐CC398 were all from cattle reported by Belgium. Five of them were assigned to CC8, while one was reported to be CC1. CC1 is CA‐MRSA, while CC8 can be both CA‐ and HA‐MRSA (Bal et al., 2016). Both CCs have also been reported in cattle previously (Alba et al., 2015; Resch et al., 2013; Sakwinska et al., 2011). It has been suggested that both CC1 and CC8 have made a host shift from humans to cattle (Resch et al., 2013; Sakwinska et al., 2011). CC398 was the only CC reported from pigs in both 2021 and 2022. Since the first description of LA‐MRSA CC398 in 2005 (Armand‐Lefevre et al., 2005; Voss et al., 2005), this CC has shown a remarkable ability to persist in the European pig production. A significant association between the presence of a single‐nucleotide polymorphism (SNP) on chromosome 12 of pigs and nasal carriage of MRSA has been reported (Skallerup et al., 2015), which may partly explain the ability of MRSA to persist in the pig production. In food, CC398 was also the most frequently reported CC, reported in 72% of isolates in 2021 and 94% of isolates in 2022. It is of note that the variation of CCs reported in food matrices is higher compared to animal populations. The reason for this is unknown but may be related to contamination of meat post slaughter (Li et al., 2019). Several different CCs were reported from meat in 2021 and 2022, including CC1, CC8, CC9, CC97, CC130, CC5, CC45, CC121 and CC692. MRSA CC1 is often associated with the community, but has also been reported in livestock (Alba et al., 2015; Elstrøm et al., 2019; Silva et al., 2022) and wild animals in Europe (Silva et al., 2020). Further, the CA/HA‐MRSA CC8 and HA‐MRSA CC5 and CC45 have been described in several animal species, including cattle and pigs, but also companion animals (Silva et al., 2022). CC9, CC97, CC121, CC130 and CC692 can be regarded as LA‐MRSA (Bal et al., 2016). Among humans, LA‐MRSA are predominantly carried by people with occupational contact with colonised livestock (Kinross et al., 2017; Chen & Wu, 2020). The proportion of LA‐MRSA carried by humans in Europe is estimated to be small (3.9%), although higher proportions have been found in Belgium, Denmark, Germany, the Netherlands, Slovenia and Spain (Ceballos et al., 2019; Cuny et al., 2015; Kinross et al., 2017; Sieber et al., 2018). Studies have revealed distinct LA‐MRSA strains disseminating into the community, also in the absence of livestock contact (Cuny et al., 2015; Kinross et al., 2017; Sieber et al., 2019). In addition, loss of AMR genes and acquisition of virulence genes specific to human strains suggest that they are adapting to the human hosts (Sieber et al., 2019; Walther et al., 2018). Providing further typing data for MRSA strains, including the presence of virulence genes, could contribute to assessment of the origin of the isolates and public health significance of the isolates. However, such data are currently not provided routinely, and therefore, it is difficult to draw further conclusions.
The mecC gene was reported in a single isolate originating from bovine meat sampled at retail in 2021. Some mecC‐MRSA from animals have been reported to carry genes belonging to the immune evasion cluster (IEC) (Abdullahi et al., 2021; Lozano et al., 2020), which is considered an adaption enabling colonisation and infection of humans (Cuny et al., 2015). This further underlines the importance of molecular typing of isolates to evaluate public health significance.
Resistance to the critically important antimicrobials mupirocin, linezolid and vancomycin were not reported in any of the MRSA isolates subjected to susceptibility testing. Furthermore, none of the isolates displayed resistance to fusidic acid. The majority of isolates tested were MDR. Resistance to tetracycline was commonly reported in all animals and food matrices tested, which is probably linked to the fact that CC398 is commonly associated with tetracycline resistance (Crombé et al., 2012). Overall, the occurrence of resistance to antimicrobials used for treatment and control of MRSA infections in humans were rare or very low, although rifampicin resistance was reported in some isolates from both food‐producing animals and meat. Belgium reported the presence of the cfr‐gene in three MRSA isolates, one from sows and two from fattening pigs. This gene is known to confer resistance to the critically important antimicrobial linezolid (Kehrenberg et al., 2005). However, antimicrobial susceptibility data were not reported for these isolates, and it is therefore not known whether phenotypic linezolid resistance was expressed by the isolates.
Antimicrobial use for food‐producing animals in Europe has decreased in recent years (EMA, 2023). As there is a strong correlation between antimicrobial use and occurrence of resistant bacteria (Chantziaras et al., 2014), it would be tempting to assume that the occurrence of resistant bacteria, including MRSA, would also decrease. However, this is not the case for all countries. For example, the occurrence of MRSA in fattening pigs increased from 36.3% in 2012 to 53.6% in 2021 in Switzerland. Studies have suggested that other reservoirs of MRSA, such as colonised farm workers or veterinarians, could contribute to the persistence, and in some cases increase of MRSA in animal populations (Crespo‐Piazuelo & Lawlor, 2021). Human to animal transmission was also identified as the source of MRSA introduction and MRSA outbreaks in the Norwegian pig population (Elstrøm et al., 2019; Grøntvedt et al., 2016). Furthermore, transmission of MRSA between animals may occur during transportation to slaughterhouses and during lairage (Bangerter et al., 2016; Grøntvedt et al., 2016). This underlines the critical importance of maintaining a high level of biosecurity and hygiene at all levels of animal production to minimise the likelihood of introduction and/or transmission of MRSA (Grøntvedt et al., 2016; Komodromos et al., 2022).
ABBREVIATIONS
- %
Percentage
- % Occ
Percentage of cephalosporin‐resistant isolates presenting a presumptive phenotype
- % Prev
Percentage of samples harbouring a presumptive ESBL/AmpC‐producing E. coli; ESBL; extended‐spectrum β‐lactamase
- % f
Percentage frequency of isolates tested
- % Res
Percentage of resistant isolates
- AMC
Antimicrobial consumption
- AMR
Antimicrobial resistance
- AMS
Antimicrobial stewardship
- AST
Antimicrobial susceptibility test
- CA‐MRSA
Community associated MRSA
- BCP
Border control posts
- CASFM
Comité de l'Antibiogramme de la Société Française de Microbiologie
- CBP
Clinical breakpoints
- CC
Clonal complex
- CI
Confidence interval
- CIA
Critically important antimicrobial
- CLSI
Clinical and Laboratory Standards Institute
- CS
Complete susceptibility
- CP
Carbapenemase
- DD
Disc diffusion method
- DL
Dilution/dilution method
- DLG
Dilution with gradient step
- EARS‐Net
European Antimicrobial Resistance Surveillance Network
- ECDC
European Centre for Disease Prevention and Control
- ECOFF
Epidemiological cut‐off value
- EEA
European Economic Area
- EFTA
European Free Trade Association
- EMA
European Medicines Agency
- ESBL
Extended spectrum beta‐lactamase
- ESC
Extended spectrum cephalosporins
- EUCAST
European Committee on Antimicrobial Susceptibility Testing
- EURL‐AR
EU Reference Laboratory for Antimicrobial Resistance
- EUSR
European Union Summary Report
- FWD
Food‐ and Waterborne Diseases and Zoonoses
- HaDEA
European Health and Digital Executive Agency
- HA‐MRSA
Hospital associated MRSA
- hpCIA
Highest‐priority critically important antimicrobials
- I
Intermediate
- IEC
Immune evasion cluster
- IPC
Infection, prevention, and control
- KOICS
Key outcome indicator of completely susceptibility (susceptible to all tested substances) E. coli
- KOIESC
Key outcome indicator of ESBL‐ and/or AmpC‐producing E. coli
- LA‐MRSA
Livestock associated MRSA
- MDR
Multi‐drug resistant
- MDRI
Multi‐drug resistant islands
- MIC
Minimum inhibitory concentration
- MLST
Multi‐locus sequence typing
- MRSA
Methicillin‐resistant Staphylococcus aureus
- MS
Member State
- NA
Not applicable/not available
- NCP
National Control Programme
- NRL
National Reference Laboratory
- NTS
Non‐typhoidal Salmonellas
- OECD
Organisation for Economic Cooperation and Development
- PCR
Polymerase chain reaction
- PMQR
Plasmid‐mediated quinolone resistance
- PCU
Population correction unit
- PVL
Panton valentine leukocidin
- Q
Quantitative
- QRDR
Quinolone resistance‐determining regions
- R
Resistant
- RCs
Reporting countries
- res1–res9
Resistance to one antimicrobial substance/resistance to nine antimicrobial substances of the common set for Salmonella
- S
Susceptible
- SIR
Susceptible, intermediate, resistant
- ST
Sequence type
- SYN
Synergy
- TESSy
The European Surveillance System
- WGS
Whole genome sequencing
- WHO
World Health Organisation
ANTIMICROBIAL SUBSTANCES
- AMC
Amoxicillin/clavulanate
- AMK
Amikacin
- AMP
Ampicillin
- AZM
Azithromycin
- CLA
Clavulanate
- CTZ/CAZ
Ceftazidime
- CHL
Chloramphenicol
- CIP
Ciprofloxacin
- CLI
Clindamycin
- COL
Colistin
- CTX
Cefotaxime
- ERT/ETP
Ertapenem
- ERY
Erythromycin
- CFT/FOX
Cefoxitin
- FUS
Fusidic acid
- FEP
Cefepime
- GEN
Gentamicin
- IMI
Imipenem
- KAN
Kanamycin
- LZD
Linezolid
- MEM
Meropenem
- MUP
Mupirocin
- NAL
Nalidixic acid
- PEN/PNC
Penicillin
- PEF
Pefloxacin
- QD
Quinupristin/Dalfopristin
- RIF
Rifampicin
- SMX
Sulfonamides
- STR
Streptomycin
- SUL
Sulfonamides
- SXT
Sulfamethoxazole
- TEM
Temocillin
- TET/TCY
Tetracycline
- TGC
Tigecycline
- TIA
Tiamulin
- TMP
Trimethoprim
MSs OF THE EU AND OTHER REPORTING COUNTRIES
- AT
Austria
- BE
Belgium
- BG
Bulgaria
- HR
Croatia
- CY
Cyprus
- CZ
Czechia
- DK
Denmark
- EE
Estonia
- FI
Finland
- FR
France
- DE
Germany
- EL
Greece
- HU
Hungary
- IE
Ireland
- IT
Italy
- LV
Latvia
- LT
Lithuania
- LU
Luxembourg
- MT
Malta
- NL
Netherlands
- PL
Poland
- PT
Portugal
- RO
Romania
- SK
Slovakia
- SI
Slovenia
- ES
Spain
- SE
Sweden
- XI
United Kingdom (Northern Ireland)
NON‐MSs REPORTING COUNTRIES
- AL
Albania
- IS
Iceland
- MK
Republic of North Macedonia
- NO
Norway
- CH
Switzerland
- UK
United Kingdom
DEFINITIONS
| ‘Antimicrobial‐resistant isolate’ |
In the case of quantitative data, an isolate was defined as ‘resistant’ to a selected antimicrobial when its minimum inhibitory concentration (MIC) value (in mg/L) was above the cut‐off value or the disc diffusion diameter (in mm) was below the cut‐off value. The cut‐off values, used to interpret MIC distributions (mg/L) for bacteria from animals and food, are shown in Appendix F ‘Material and methods’, Tables F.5, F.6, F.7 In the case of qualitative data, an isolate was regarded as resistant when the country reported it as resistant using its own cut‐off value or break point |
| ‘Level of antimicrobial resistance’ | The percentage of resistant isolates among the tested isolates |
| ‘Reporting MS group’ | Member States (MSs) that provided data and were included in the relevant table for antimicrobial resistance data for the bacteria–food/animal category–antimicrobial combination |
| Terms used to describe the levels of antimicrobial resistance |
Rare: < 0.1% Very low: 0.1%–1.0% Low: > 1.0%–10.0% Moderate: > 10.0%–20.0% High: > 20.0%–50.0% Very high: > 50.0%–70.0% Extremely high: > 70.0% |
CONFLICT OF INTEREST
If you wish to access the declaration of interests of any expert contributing to an EFSA scientific assessment, please contact interestmanagement@efsa.europa.eu.
REQUESTOR
European Commission
QUESTION NUMBER
EFSA‐Q‐2021‐00769
COPYRIGHT FOR NON‐EFSA CONTENT
EFSA may include images or other content for which it does not hold copyright. In such cases, EFSA indicates the copyright holder and users should seek permission to reproduce the content from the original source.
MAP DISCLAIMER
The designations employed and the presentation of material on any maps included in this scientific output do not imply the expression of any opinion whatsoever on the part of the European Food Safety Authority concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries.
ACKNOWLEDGEMENTS
EFSA and ECDC wish to thank the members of the Scientific Network for Zoonoses Monitoring Data (EFSA) and the Food‐ and Waterborne Diseases and Zoonoses Network (ECDC) who provided the data and reviewed the report and the members of the Scientific Network for Zoonoses Monitoring Data, for their endorsement of this scientific output. Also, the contribution of EFSA staff members: Raquel García‐Fierro, Pierre‐Alexandre Beloeil, Giusi Amore, Valentina Rizzi, Anca‐Violeta Stoicescu, Luca Pasinato, Maria Sanz Zapata, Beatriz Guerra Roman, Ernesto Liebana, and the contributions of ECDC staff members: Therese Westrell, Liese Van Gompel and Hanna Merck, and the contributions of EFSA's contractor: Norwegian Veterinary Institute (Solveig Sølverød Mo, Jannice Schau Slettemeås, Madelaine Norström, Anne Margrete Urdahl and Marianne Sunde); Swedish Veterinary Agency (Oskar Nilsson and Anna Werinder); Technical University of Denmark (Ana Sofia Ribeiro Duarte, Joana Pessoa and Marianne Sandberg); Soladis (Florian Kroell, Frédéric Chavanel, Thomas Brière, Olivier Omraam Agbohouto, Agnès Iltis, Diane Plouchart, Gwenneg Kerdivel and Catherine Pahon) and CVO‐EFOR (Patrick Etiévant) for the support provided to this scientific output. Also, the contribution of external expert from the French Agency for Food, Environmental and Occupational Health & Safety (Marisa Haenni) for contributing to the text box on Carbapenem resistance in Enterobacterales from companion animals. As well as the support from the EURL‐AR, specifically, Birthe S. Rosenqvist Lund, Elif Seyda Tosun, Sarah Marvig Johansson, Natasia Rebekka Thornval, Mirena Ivanova, Jette Sejer Kjeldgaard and Rene S. Hendriksen for the confirmatory testing are gratefully acknowledged.
APPENDIX A. High‐level resistance to ciprofloxacin among certain Salmonella serovars
High‐level resistance to ciprofloxacin in S. Kentucky
Considering individual serovars, in 2022, Salmonella Kentucky accounted for most of the Salmonella isolates recovered from the poultry origins which exhibited high level of resistance to ciprofloxacin, with MIC ≥ 4 mg/L. This serovar accounted for 34.1%, 20.0% and 100% of the total number of isolates displaying MICs of ≥ 4 mg/L from broiler, laying hen and turkey flocks, respectively. Additionally, in 2021, a single isolate recovered from fattening pigs displayed high‐level ciprofloxacin resistance and was serotyped as S. Kentucky. No Salmonella spp. isolates from cattle under 1 year of age displayed high‐level resistance to ciprofloxacin. The single isolate from fattening pigs showing high resistance to ciprofloxacin was also resistant to nalidixic acid (with an MIC > 64 mg/L for nalidixic acid).
Figure A.1 presents the overall AMR levels among MDR Salmonella Kentucky isolates from broilers, laying hens, turkeys and pigs that exhibited high‐level ciprofloxacin resistance.
FIGURE A.1.
Resistance levels among MDR Salmonella Kentucky isolates exhibiting high‐level ciprofloxacin resistance from food‐producing animals, reported by MSs in 2021–2022.
FIGURE A.2.
Number of Salmonella isolates displaying high‐level ciprofloxacin resistance by serovar, reported from the different food‐producing animal populations by MSs in 2021–2022.
From the monitoring of human cases in 2022, high to extremely high levels of resistance were noted to gentamicin (34.7%), ampicillin (59.2%), sulfonamides (60.9%), tetracyclines (67.1%), ciprofloxacin (72.7%) and nalidixic acid (72.4%); consistent with the multidrug resistance patterns observed in isolates from the monitoring of poultry in 2022, and the possible dissemination of the S. Kentucky ST198 strain within Europe. Of 7954 Salmonella isolates from humans where ciprofloxacin MIC data were available, 121 of these (1.5%) exhibited MICs of ≥ 4 mg/L, of which S. Kentucky accounted for 106 (87.6%). Twelve S. Kentucky isolates were reported by the two countries submitting sequences as their official Salmonella AMR data set to ECDC for 2022. Eight of these were of ST198 and they all displayed two mutations each in parC and gyrA, which explains the high‐level resistance to ciprofloxacin in ST198. The other four isolates, three ST314 and one ST696, only had the parC T57S mutation which on its own normally does not result in phenotypic resistance to fluoroquinolones (Chang et al., 2021).
High‐level resistance to ciprofloxacin among other Salmonella serovars
In 2022, while S. Kentucky generally accounted for an important number of the Salmonella isolates exhibiting high‐level resistance to ciprofloxacin, there was also a significant contribution from Salmonella Infantis and Salmonella Newport in broilers (27.3% and 31.8%, respectively) and laying hens (60.0% and 20.0%, respectively). Figure A.2 shows the number of isolates exhibiting high‐level resistance to ciprofloxacin by serovar within each of the poultry origins.
data from humans and food
APPENDIX B. Occurrence of antimicrobial resistance at the Salmonella serovar level
Breakdown of the most prevalent serovars in food‐producing animal populations
Considering all reporting countries (including non‐MSs), the relative contribution of some of the most dominant serovars recovered from each of the food‐producing animal populations is illustrated in Figure B.1. In broilers, two serovars (Infantis and Enteritidis) accounted for 51.4% of Salmonella isolates, while in laying hens, five serovars (Enteritidis, Kentucky, Infantis, Typhimurium and Mbandaka) accounted for 57.5% of isolates; and in turkeys, serovars Agona, Infantis, Derby and Bredeney accounted for 52.6% of Salmonella isolates. Additionally, in pigs, four serovars (Derby, monophasic Typhimurium, Typhimurium and Rissen) accounted for 78.0% of Salmonella spp., while in cattle under 1 year of age, serovars monophasic Typhimurium, Typhimurium, Dublin, Bredeney, Stanleyville and Enteritidis accounted for 62.0% of the total Salmonella spp. recovered from this origin.
FIGURE B.1.
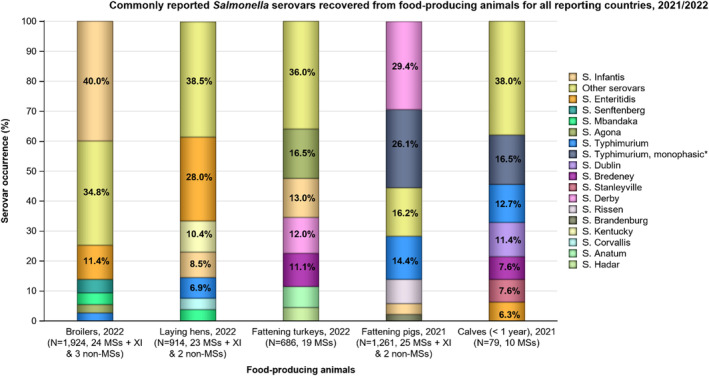
Commonly reported Salmonella serovars recovered from broilers, laying hens, fattening turkeys, fattening pigs and cattle under 1 year of age (calves), for all reporting countries, 2021–2022.
Note: *Monophasic S. Typhimurium includes all antigenic formulas; serovars in the legend are listed according to their predominance within all the animal origins.
FIGURE B.2.
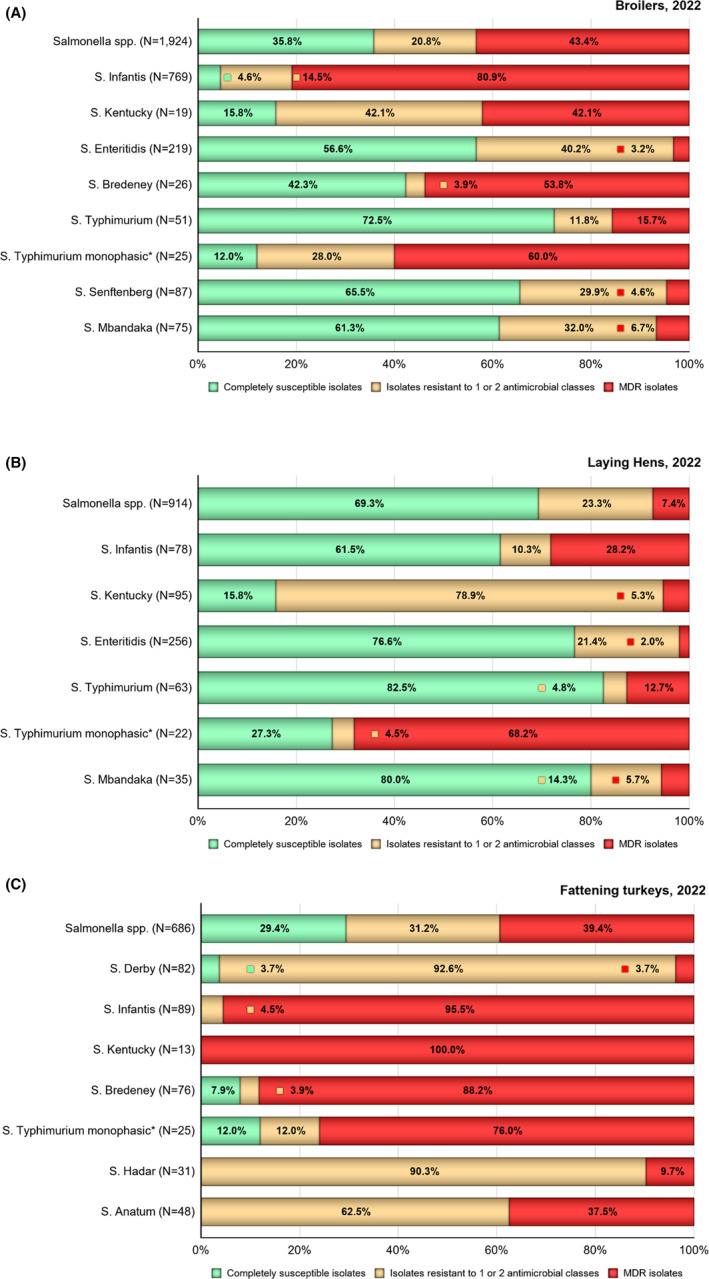
Proportions of isolates completely susceptible (green), resistant to one or two antimicrobial classes (gold) and MDR (red) in Salmonella spp. and certain serovars recovered from (A) broilers, (B) laying hens and (C) fattening turkeys for all reporting countries, 2022.
Note: N, total number of Salmonella spp. isolates or total number of particular serovars tested; *monophasic S. Typhimurium includes antigenic formulas. The MDR analysis of animal isolates included the following antimicrobials: ampicillin, cefotaxime/ceftazidime, chloramphenicol, ciprofloxacin/nalidixic acid, gentamicin/amikacin, meropenem, sulfamethoxazole, tetracycline/ tigecycline and trimethoprim.
FIGURE B.3.
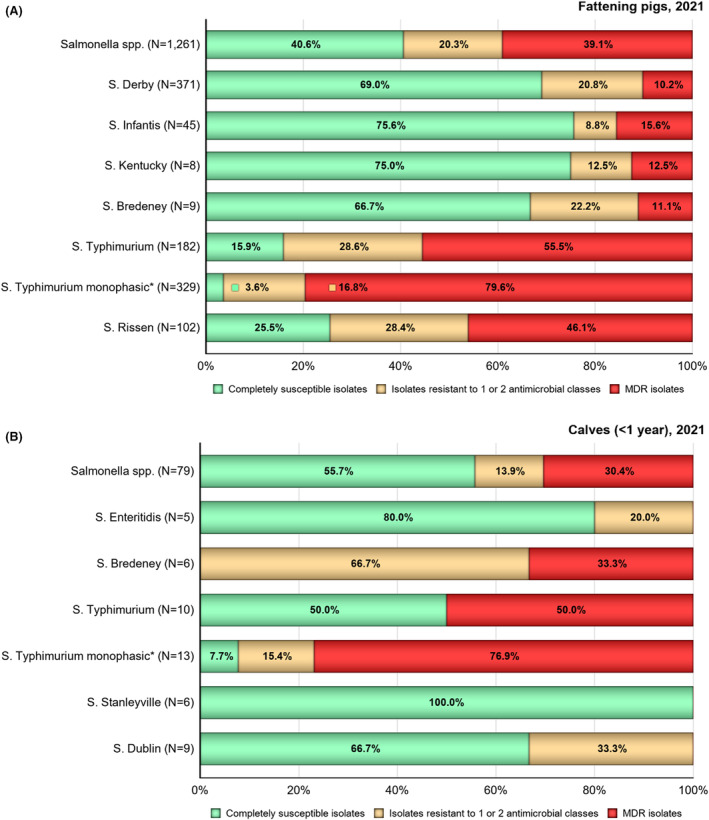
Proportions of isolates completely susceptible (green), resistant to one or two antimicrobial classes (gold) and MDR (red) in Salmonella spp. and certain serovars recovered from (A) fattening pigs, and (B) cattle under 1 year of age (calves), for all reporting countries, 2021.
Note: N, total number of Salmonella spp. isolates or total number of particular serovars tested; *monophasic S. Typhimurium includes antigenic formulas. The MDR analysis of animal isolates included the following antimicrobials: ampicillin, cefotaxime/ceftazidime, chloramphenicol, ciprofloxacin/nalidixic acid, gentamicin/amikacin, meropenem, sulfamethoxazole, tetracycline/tigecycline and trimethoprim.
FIGURE B.4.
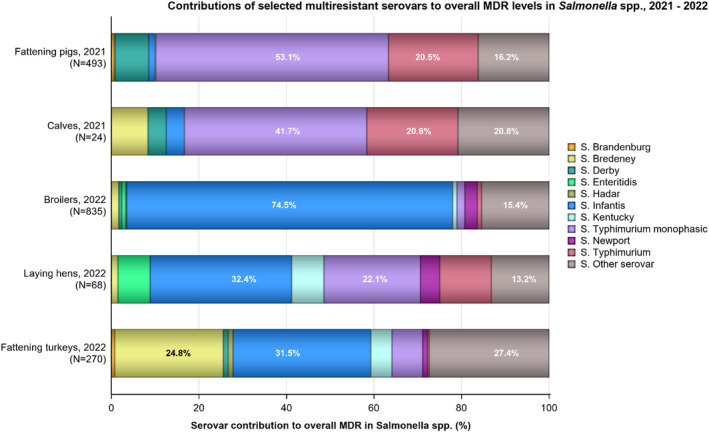
Proportions of certain serovars exhibiting multidrug resistance to overall MDR levels in Salmonella spp. recovered from each of the food‐producing animal populations, for all reporting countries in 2021–2022.
Note: N: Total number of Salmonella isolates exhibiting MDR; serovars contributing the highest levels of MDR to overall MDR levels in Salmonella spp. are illustrated with a percentage; *monophasic S. Typhimurium includes antigenic formulas; serovars in the legend are listed according to their predominance within all the animal origins.
Complete susceptibility and multidrug resistance
The patterns of resistance associated with these different serovars influenced the overall resistance levels in Salmonella isolates, and Figures B.2, B.3 summarise the proportion of CS and MDR isolates among particular serovars recovered from each of these food‐producing animal populations considering all reporting countries (including non‐MSs).
High proportions of MDR isolates were reported in the different food‐producing animal populations. For instance, 55.8% of the S. Typhimurium isolated in pigs were MDR. Similarly, the Salmonella Infantis isolates from broilers were largely MDR (81.0) and were the leading contributors to overall MDR (74.5%; Figures B.2, B.4). Like in broilers, S. Infantis was the lead serovar contributing with MDR in laying hens (32.4%, MDR = 28.2%); while in turkeys, MDR isolates contributing most to overall MDR were mainly S. Infantis (31.5%; MDR = 95.5%) and Bredeney (24.8%; MDR = 88.2%).
Resistance profiles exhibited by particular serovars
Salmonella Infantis was the most frequently recovered serovar from broilers (40.0%) in 2022 (Figures B.1, B.2). Additionally, this serovar was the fourth most reported in humans (1193 cases), and the second and third most reported in turkeys (13.0%) and in laying hens (8.5%), respectively.
Although a wide range of MDR patterns were reported among S. Infantis isolates from poultry, the most frequent MDR pattern was TGC‐CIP‐NAL‐SMX‐TET. The percentage of S. Infantis isolates with this specific resistance profile (resistance to tigecycline, ciprofloxacin, nalidixic acid, sulfamethoxazole and tetracycline) out of the MDR isolates at the MS level was 29.8% in isolates recovered from broilers, 12.5% from laying hens and 28.0% from turkeys. Resistance to five antimicrobial classes was noted among isolates from all poultry origins. A single S. Infantis isolate recovered from broilers with an MDR pattern AMK‐AMP‐TGC‐NAL‐CIP‐SMX‐TMP was reported, while 12, 19 and 44 isolates exhibited resistance to AMP‐CTX‐CAZ‐NAL‐CIP‐SMX‐TMP‐TET, AMP‐CTX‐CAZ‐TGC‐NAL‐CIP‐SMX‐TMP‐TET and AMP‐TGC‐NAL‐CIP‐SMX‐TMP‐TET, respectively. Four S. Infantis recovered from laying hens also exhibited an MDR pattern AMP‐TGC‐NAL‐CIP‐SMX‐TMP‐TET. While 14, 16 and 8 S. Infantis isolates recovered from turkeys exhibited the following MDR patterns: AMP‐CTX‐CAZ‐NAL‐CIP‐SMX‐TMP‐TET, AMP‐CTX‐CAZ‐TGC‐NAL‐CIP‐SMX‐TMP‐TET and AMP‐TGC‐NAL‐CIP‐SMX‐TMP‐TET.
In 2021, S. Infantis represented only 3.6% of Salmonella isolated from pigs. Seven isolates from pigs showed seven different MDR patterns. Three out of the seven isolates from pigs showed resistance to quinolones, and only one MDR isolate was resistant to tigecycline. Resistance to third‐generation cephalosporins was detected in one out of seven MDR isolates from pigs. A single S. Infantis isolate from a calf was MDR with the pattern CIP‐NAL‐SMX‐TET‐TGC (i.e. showing quinolone and tigecycline resistance).
In human isolates in 2022, 49.7% of the serovar S. Infantis were MDR (Annex A1: Table 18). The human isolates showed resistance to nalidixic acid (33.2%), ciprofloxacin (40.1%), sulfamethoxazole (50.5%) and tetracycline (38.1%) (Annex A1: Table 5). Tigecycline resistance was observed in 15.4% of the isolates. Combined resistance to ciprofloxacin and cefotaxime was reported in 5.9% (Annex A1: Table 11).
TABLE 11.
Combined resistance to ciprofloxacin and cefotaxime in indicator E. coli from broilers, fattening turkeys (turkeys), fattening pigs (pigs) and cattle under 1 year of age applying ECOFFs and clinical breakpoints, as issued by EUCAST, EU MSs and non‐MSs, 2021–2022.
| Food‐producing animal population | ‘Microbiological’ combined resistance to CIP & CTX (using ECOFFs) | ‘Clinical’ combined resistance to CIP & CTX (using clinical breakpoints) | ||||
|---|---|---|---|---|---|---|
| N | % R | 95% CI | N | % R | 95% CI | |
| Pigs, 2021 1 | 17 | 0.4 | 0.2, 0.6 | 6 | 0.1 | 0, 0.3 |
| Cattle < 1 year, 2021 2 | 11 | 0.5 | 0, 1.2 | 2 | 0.1 | 0, 0.5 |
| Broilers, 2022 3 | 52 | 1.0 | 0.8, 1.4 | 21 | 0.4 | 0.3, 0.6 |
| Turkeys, 2022 4 | 23 | 1.2 | 0.8, 1.8 | 9 | 0.5 | 0.3, 0.9 |
Abbreviations: CIP, ciprofloxacin (fluoroquinolones); CTX, cefotaxime (third‐generation cephalosporins); N, number of isolates; % R, percentage of resistance; 95% CI, 95% confidence interval.
N = 4586, 28 MSs, 3 non‐MSs.
N = 2067, 11 MSs, 2 non‐MSs.
N = 5022, 28 MSs, 4 non‐MSs.
N = 1883, 13 MSs, 1 non‐MSs.
In contrast, Salmonella Enteritidis isolates exhibited much lower multidrug resistance. This serovar was the most frequently reported in laying hens (28.0%) and the second most reported in broilers (11.4%; Figures B.1, B.2). Also in humans, it is the most common Salmonella serovar with 26,811 cases reported in the EU/EEA in 2022. While complete susceptibility was observed at 56.6% in S. Enteritidis isolates from broilers, in isolates recovered from laying hens and turkeys, the vast majority of isolates exhibited complete susceptibility (76.6% and 100%, respectively). S. Enteritidis belongs to group D Salmonella (serogroup O9) which tend to show elevated colistin MICs, a phenomenon considered to reflect slightly decreased intrinsic susceptibility of wild‐type isolates belonging to Group D (Agersø et al., 2012). This is exemplified by the proportion of colistin‐resistant isolates attributed to S. Enteritidis from laying hens (87.8%) and broilers (86.6%) in comparison to other serovars belonging to different serogroups. In 2021, S. Enteritidis were recovered in 1.7% and 6.3% of Salmonella isolates originating from pigs and cattle under 1 year of age, respectively (Figure 11).
In human isolates, only 2.4% of the serovar S. Enteritidis were MDR (Annex A1: Table 15). However, the highest proportions of resistance in these isolates were reported to nalidixic acid (26.0%), ciprofloxacin (22.8%) and colistin (27.9%) (Annex A1: Table 2).
Salmonella Kentucky was the second most reported serovar in laying hens, the 13th in turkeys and the 18th in broilers, accounting for 10.4%, 1.9% and 1.0% of Salmonella spp. recovered from these poultry origins, respectively (Figure B.1). In Salmonella infections in humans 2022, S. Kentucky was the 11th most common serovar in 2022, with 344 cases reported in EU/EEA countries. All S. Kentucky isolates from turkeys were MDR, while the proportion of MDR isolates recovered from broilers was high (42.1%) but low for laying hens (5.3%). A wide range of MDR patterns were reported among S. Kentucky isolates from broilers, laying hens and turkeys. The most frequent core pattern of resistance was GEN‐AMP‐NAL‐CIP‐SMX‐TET (n = 8) in isolates recovered from turkeys, while the most common MDR pattern in broiler isolates was AMP‐CTX‐CAZ‐NAL‐CIP (n = 3). In 2021, eight isolates of S. Kentucky were recovered from pigs (0.6%). Of these, a single isolate exhibited the resistance pattern TGC‐NAL‐CIP‐SMX‐TMP‐TET demonstrating resistance to tigecycline and quinolones. No S. Kentucky was reported in cattle under 1 year of age in 2021.
In human isolates, 63.7% of the serovar S. Kentucky were MDR (Annex A1: Table 19). The human isolates showed most resistance to gentamicin (34.7%), ampicillin (59.2%), nalidixic acid (72.4%), ciprofloxacin (72.7%), sulfamethoxazole (60.9%) and tetracycline (67.1%) (Annex A1: Table 6). Resistance to the last resort antimicrobials tigecycline and azithromycin was 15.5% and 6/5%, respectively. Combined resistance to both clinically important antimicrobials ciprofloxacin and cefotaxime was reported in 12.3% of the isolates (Annex A1: Table 12).
Salmonella Bredeney was the fourth most reported serovar in turkeys (11.1%, n = 76) and accounted for 1.4% (n = 26) and 0.6% (n = 5) of the Salmonella isolates from broilers and laying hens, respectively. In 2021, this serovar was the fourth most reported (7.6%) in cattle under 1 year of age (Figure B.1). In humans, 114 cases of S. Bredeney were reported in the EU/EEA in 2022, ranking it on place 37th among all serovars. Multidrug resistance was extremely high in this serovar among isolates recovered from turkeys (88.2%, n = 67) and very high in isolates from broilers (53.9%, n = 14), while only one S. Bredeney isolate from laying hens was MDR (Figure B.2). In 2021, one isolate from a calf reported by Italy showed quinolone resistance (NAL‐CIP‐SMX‐TET), while tigecycline resistance was reported by Poland in the only MDR isolate from pigs (AMP‐TGC‐SMX‐TMP‐TET). Notably, among the isolates recovered from turkeys, the level of MDR was greatly influenced by one MS, with Hungary reporting 64 multidrug‐resistant isolates. Among MDR S. Bredeney isolates recovered from turkeys, the most frequent pattern of resistance was to AMP‐NAL‐CIP‐TET (n = 23, 34.3%), closely followed by the same pattern but with the addition of tigecycline and trimethoprim (AMP‐TGC‐NAL‐CIP‐TMP‐TET; n = 22, 32.8%), and AMP‐TGC‐NAL‐CIP‐TET (n = 18, 26.9%), all reported by Hungary.
Salmonella Derby was the third most common serovar recovered from turkeys (12.0%, N = 82), and it accounted for 1.2% and 0.3% in broilers (N = 23) and laying hens (N = 3), respectively (Figure B.2). In 2021, S. Derby was the most reported serovar from pigs (29.4%, N = 371), while only two isolates of this serovar were recovered from cattle under 1 year of age (Figure B.3). In humans, it was the sixth most common serovar in 2022 with 551 cases reported in the EU/EEA. Multidrug resistance was reported at low levels in S. Derby isolates from turkeys (3.7%), at moderate levels in pigs (10.2%) and at high levels in broilers (26.1%), while no MDR was reported in laying hens and one out of two isolates from cattle under 1 year of age was MDR (Figures B.2, B.3). Among S. Derby isolates recovered from these animal populations, different core resistance patterns predominated. The three MDR S. Derby isolates recovered from turkeys exhibited three different patterns: AMP‐NAL‐CIP‐SMX‐TMP‐TET, AMP‐CIP‐SMX‐TMP‐TET (both reported by Spain) and CHL‐AMP‐TGC‐NAL‐CIP‐SMX‐TET (Poland). Whereas the MDR isolates from broilers were CHL‐AMP‐TGC‐NAL‐CIP‐SMX‐TET (n = 3, Poland), CHL‐AMP‐TGC‐SMX‐TET (n = 1, Poland), AMP‐SMX‐TMP (n = 1, Ireland) and CHL‐SMX‐TMP (n = 1, Netherlands). In pigs, the most common resistance pattern in MDR S. Derby was AMP‐SMX‐TET, and was seen in isolates reported by Denmark, Estonia, Germany, Poland and Slovenia. Five of the MDR isolates from pigs were resistant to up to five classes of antimicrobials. The MDR S. Derby isolated from cattle under 1 year of age was reported by Spain with the pattern GEN‐CHL‐AMP‐CTX‐NAL‐CIP‐SMX‐TMP‐TET (including a third‐generation cephalosporin).
MDR with quinolone resistance (ciprofloxacin/nalidixic acid) was reported in isolates from all animal populations from which MDR S. Derby was recovered, while tigecycline resistance was observed in one isolate from turkeys, four MDR isolates from broilers and six isolates from pigs. No S. Derby isolates exhibited combined resistance to quinolones and third‐generation cephalosporins, except for the single MDR isolate from cattle under 1 year of age. Recent studies on S. Derby originating from the pork and poultry sectors in France identified four major host‐specific lineages, corresponding to multilocus sequence typing (MLST) profiles ST39, ST40, ST71 and ST682 (Sevellec et al., 2018, 2020). The lineages ST39, ST40 and ST682 were associated with pork, and ST71 with poultry. While ST71 and ST682 isolates were generally devoid of resistance genes, ST40 isolates commonly harboured several resistance genes, with Clade 2 of this lineage characterised by SGI1 and the pattern of resistance to streptomycin, sulfonamides and tetracycline. Additionally, a resistance gene for fosfomycin was detected in all genomes from ST39 (Sevellec et al., 2020).
Monophasic S. Typhimurium was recovered in 1.3%, 2.4% and 3.6% of the Salmonella isolates from broilers, laying hens and turkeys, respectively (Figure B.2), while in 2021, it was the main serovar recovered in cattle under 1 year of age (16.5%) and the second most frequently reported in pigs (26.1%; Figure B.3). In humans, it was the third most common serovar with 5585 cases in 2022. The proportion of all Salmonella isolates showing MDR in all food‐producing animal populations was influenced by the occurrence of multidrug resistance monophasic S. Typhimurium, particularly in laying hens (22.2%), in cattle under 1 year of age (41.7%) and in pigs (53.1%; Figure B.4). MDR in Monophasic S. Typhimurium isolates was reported at very high levels in broilers (58.3%) and in laying hens (68.2%), and at extremely high levels in turkeys (76.0%), cattle under 1 year of age (76.9%) and pigs (79.3%). Resistance to ampicillin, sulfamethoxazole and tetracycline (AMP‐SMX‐TET) was the most frequent pattern in all food‐producing animal populations. The second leading pattern in turkeys, laying hens and pigs was an addition of ciprofloxacin and nalidixic acid to the core resistance pattern (AMP‐NAL‐CIP‐SMX‐TET).
Monophasic S. Typhimurium has spread widely among European pig populations. There are particular MDR patterns associated with monophasic S. Typhimurium and because this serovar is prevalent in many countries, these patterns greatly influenced the overall resistance figures. This is exemplified by the resistance pattern AMP‐SMX‐TET which occurred as the main MDR pattern in broilers (13/15; 86.7%), laying hens (11/15; 73.3%), turkeys (15/19; 79.0%) and pigs 159/493 (32.2%). This resistance pattern (together with resistance to streptomycin) is typical of the European clone of monophasic S. Typhimurium (Hopkins et al., 2010). The genes conferring resistance to these antimicrobials are commonly found in association together with IS26 mobile genetic elements, responsible for their integration at different chromosomal locations, as described in European strains of monophasic S. Typhimurium (Sun et al., 2020). It is noteworthy that multidrug resistance in the European clone of monophasic S. Typhimurium appears to have originated from the integration of MDR plasmids into the chromosome, facilitated by the presence of these IS26 mobile genetic elements (Sun et al., 2020).
Resistance to five or more classes of antimicrobials was observed among two monophasic S. Typhimurium isolates from broilers (CHL‐AMP‐TGC‐NAL‐CIP‐SMX‐TMP‐TET and CHL‐AMP‐TGC‐NAL‐SMX‐TMP‐TET, both reported by Germany) and in 31 isolates from pigs, with nine of them exhibiting resistance to six antimicrobial classes.
In human isolates, 68.2% of monophasic S. Typhimurium were MDR (Annex A1: Table 17). This is confirmed in Table 4 where human isolates showed mainly resistance to ampicillin (86.7%), sulfamethoxazole (78.5%) and tetracycline (77.3%). Combined resistance to ciprofloxacin and cefotaxime was observed in 1.0% of isolates (Annex A1: Table 10). Interestingly, in isolates from food‐producing animals, resistance to quinolones (ciprofloxacin/nalidixic acid) was only reported in broiler and pig isolates, accounting for 13.3% (2 out of 15) and 13.7% (36 out of 262) of all MDR monophasic S. Typhimurium isolated from these origins, respectively. Similarly, tigecycline resistance was only reported in isolates from broilers and pigs. Combined resistance to quinolones and third‐generation cephalosporins was not reported in any food‐producing animal population.
Salmonella Typhimurium was recovered in 2.7%, 6.7% and 2.0% of the Salmonella isolates from broilers, laying hens and turkeys, respectively (Figure B.2), while in 2021, it was the second and third most frequently reported serovar from cattle under 1 year of age (12.7%) and from pigs (14.4%), respectively (Figure B.3). In humans, S. Typhimurium was the second most common serovar with 5809 cases reported in 2022. Among S. Typhimurium isolates, MDR was reported at moderate levels in broilers (15.7%, n = 8), laying hens (12.7%, n = 8), at a low level in turkeys (7.1%, n = 1) and at a high level and a very high level in cattle under 1 year of age (50.0%) and pigs (55.5%), respectively (Figure B.2, B.3 and Annex A, Table 22). Notably, the proportion of all Salmonella isolates showing MDR in laying hens, cattle under 1 year of age and pigs was influenced by the occurrence of multidrug‐resistant S. Typhimurium (Figure B.4). Four MDR S. Typhimurium isolates from broilers, the single MDR isolate from turkeys and most of the pig isolates exhibited resistance to AMP‐SMX‐TET with addition of CHL to this pattern as the second most encountered pattern in pigs. Moreover, of the eight S. Typhimurium isolates from laying hens, four different MDR patterns were reported, namely GEN‐SMX‐TET (n = 4), CHL‐AMP‐SMX‐TET (n = 2), AMP‐SMX‐TET (n = 1) and CHL‐AMP‐NAL‐CIP‐SMX‐TET (n = 1). Moreover, two S. Typhimurium isolates from broilers also displayed the MDR pattern including CHL‐AMP‐NAL‐CIP‐SMX‐TET, and one each exhibited resistance to GEN‐CHL‐AMP‐SMX‐TMP and GEN‐CHL‐AMP‐SMX‐TMP‐TET. Of the five MDR S. Typhimurium isolates recovered from cattle under 1 year of age, four different resistance patterns were noted, with the pattern TGC‐SMX‐TET observed in two isolates. Mobile genetic elements that could account for this resistance pattern in S. Typhimurium isolates have previously been described. Salmonella genomic island 1 (SGI1), known to contain a multidrug resistance region located on a complex class 1 integron designated In104, confers pentavalent resistance (i.e. ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, tetracycline resistance phenotype – AMP‐CHL‐STR‐SMX‐TET) and has been widely documented in a range of Salmonella serovars.
Resistance to (fluoro)quinolones (ciprofloxacin/nalidixic acid) was reported in a single isolate from laying hens and cattle under 1 year of age, two isolates from broilers, while in pig isolates 20.8% of the 101 MDR S. Typhimurium isolates exhibited resistance to this antimicrobial class. Tigecycline resistance among multidrug‐resistant S. Typhimurium isolates from poultry was not reported; however, three isolates from cattle under 1 year of age and 11.9% of the 101 MDR pig isolates were resistant to tigecycline. In 2022, no isolate was resistant to five or more AM classes. While in 2021, a single isolate from a calf was resistant to at least five classes of antimicrobials and 20.8% (21/101) MDR S. Typhimurium from pigs showed resistance to five or more antimicrobial classes. No MDR S. Typhimurium isolates recovered from any of the food‐producing animal populations displayed resistance to third‐generation cephalosporins.
In human isolates, 23.8% of the serovar S. Typhimurium were MDR (Annex A: Table 16). The human isolates showed mainly resistance to ampicillin (32.1%), sulfamethoxazole (30.2%), tetracycline (26.8%) but also ciprofloxacin (19.6%) (Annex A: Table 3). A few of the isolates (0.4%) showed combined resistance to ciprofloxacin and cefotaxime (Annex A: Table 9).
Multidrug resistant serovars
Considering all reporting countries (including non‐MSs), the contributions of particular multidrug resistant serovars to overall MDR levels in Salmonella spp. from each of the animal categories are illustrated in Figure B.4. Overall, the figure shows that monophasic S. Typhimurium and S. Infantis were the lead serovars contributing to multidrug resistance among all Salmonella spp. isolated from food‐producing animals in 2021–2022.
APPENDIX C. Comparison of Salmonella data from humans and food‐producing animals at the serovar level
C.1.
In 2021–2022, the quantitative human data were interpreted using EUCAST ECOFF values (categorised into wild type and non‐wild type), when available, in the same way as for the animal and food data. For 2021–2022 data, this resulted in one dilution difference (e.g. defining a higher number of samples to be of the non‐wild type) compared to Commission Implementing Decision (EU) 2020/1729 regarding ampicillin (Figure C.1). Also, from 2021 onwards, the clinical breakpoint for ciprofloxacin for animal data was changed from > 1 to > 0.064 mg/L with the new Implementing Decision and now aligns with the EUCAST ECOFF (> 0.064 mg/L). Where ECOFFs do not exist, EUCAST or Clinical and Laboratory Standards Institute (CLSI) CBPs were applied. Notably, for sulfamethoxazole/sulfonamides, there is no EUCAST interpretative criterion for Salmonella, and therefore, a threshold of > 256 mg/L was applied to both the human and animal data. For qualitative data interpreted with clinical breakpoints (S = susceptible, I = susceptible with increased exposure and R = resistant), I + R results were combined into one category, except for tetracycline where only R was used. When aligning susceptible isolates with wild‐type isolates and I + R isolates with non‐wild‐type isolates, there is generally close concordance across categories (Figure C.1). An exception is meropenem where the EUCAST CBP is substantially higher (+4 dilutions) than the ECOFF.
FIGURE C.1.
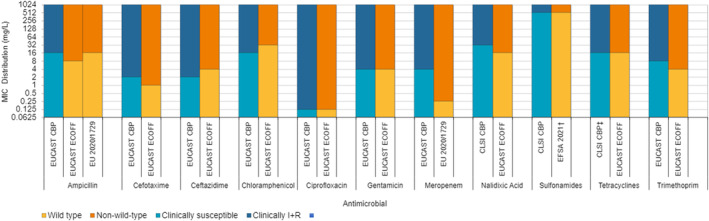
Comparison of CBPs and ECOFFs used to interpret MIC data reported for Salmonella spp. from humans, animals or food.
Note: †EFSA Manual for reporting 2022 antimicrobial resistance data within the framework of Directive 2003/99/EC and Decision 2020/1729/EU. ‡Only R category included.
FIGURE C.2.
Occurrence of resistance to selected antimicrobials in S.Infantis from humans (2022) and animal populations reporting ≥ 10 isolates (2021–2022), all reporting MSs.
FIGURE C.3.
Occurrence of resistance to selected antimicrobials in S. Enteritidis from humans (2022) and animal populations reporting ≥ 10 isolates (2021–2022), all reporting MSs.
FIGURE C.4.
Occurrence of resistance to selected antimicrobials in Salmonella Kentucky from humans (2022) and animal populations reporting ≥ 10 isolates (2021–2022), all reporting MSs.
FIGURE C.5.
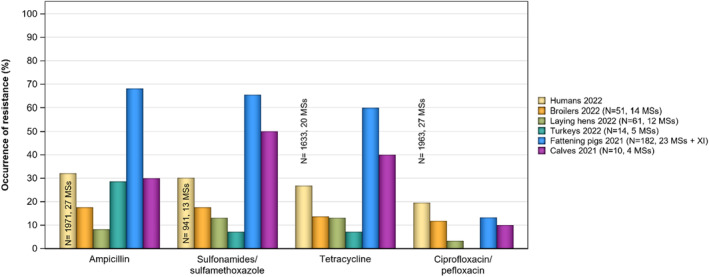
Occurrence of resistance to selected antimicrobials in Salmonella Typhimurium from humans (2022) and animal populations reporting ≥ 10 isolates (2021–2022), all reporting MSs.
FIGURE C.6.
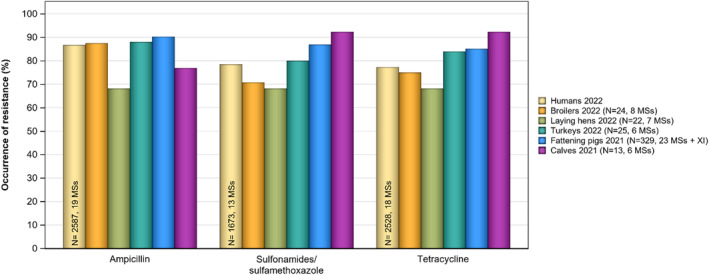
Occurrence of resistance to selected antimicrobials in monophasic Salmonella Typhimurium from humans (2022) and animal populations reporting ≥ 10 isolates (2021–2022), all reporting MSs.
It is of note that the countries reporting data on particular Salmonella serovars from human cases are not always the same as those reporting corresponding serovar data within the animal categories. Additionally, the number of isolates reported from human cases and from the animal origins varied, both at the MS and MS‐group level. Furthermore, Salmonella isolates have been derived from different scenarios, for human data are from clinical cases while animal data come from healthy animals. All of these factors mentioned above may introduce a source of variation to results when comparing overall percentage resistance to particular antimicrobials and MDR levels among human and animal isolates. The panel of nine antimicrobial classes comprising the MDR analysis of human isolates includes ampicillin, cefotaxime/ceftazidime, chloramphenicol, ciprofloxacin/pefloxacin/nalidixic acid, gentamicin, meropenem, sulfonamides/sulfamethoxazole, tetracyclines and trimethoprim/trimethoprim‐sulfamethoxazole (co‐trimoxazole) and does not include azithromycin and tigecycline. Not all MSs use the full panel when resistance testing clinical Salmonella isolates; when the data originate from testing done in clinical laboratories, the antimicrobials tested reflect local or national prescribing habits and/or treatment guidelines. For animal isolates, the MDR analysis included the same nine antimicrobial classes, as well as tigecycline (glycylcycline class). Tigecycline was addressed together with tetracycline from the MDR analysis of animal isolates to align with the panel analysed from humans. As tigecycline resistance is less common than tetracycline resistance, this procedure has very limited effect on the MDR outputs and negligible effect on human and animal comparisons.
Within a given MS, attempts to relate the occurrence of AMR in human Salmonella isolates to that in isolates from food/food‐producing animals are complicated, as much of the food consumed in a MS may have originated from other MSs or non‐member countries. Salmonella infections can also be associated with foreign travel, other types of animal contact (such as pet reptiles) or the environment. Additionally, some human infections may result from human‐to‐human transmission and, although known travel‐associated isolates from human cases were excluded from the analysis, a large proportion of cases lacked information on travel status. Such circumstances may influence the human AMR data at the reporting MS level. Furthermore, the local medical and diagnostic practices and policies for referral to clinical laboratories may vary between countries, which may result in the reporting of various clinical or regional subsets of isolates from humans.
Resistance to selected antimicrobials by serovar
Resistance to ciprofloxacin/pefloxacin and nalidixic acid in S. Infantis from humans at the MS level in 2022 was on average 40.1% and 33.2%, respectively. When looking at the occurrence of resistance in poultry populations, only laying hens exhibited similar resistance levels (33.3% and 30.8%, respectively). Resistance levels to ciprofloxacin and nalidixic acid in S. Infantis isolated from both broiler and turkey flocks were extremely high. Generally, resistance levels to sulfonamides and tetracycline were lowest in laying hens (29.5% and 28.2%, respectively), reaching high to very high levels in humans (50.5% and 38.1%, respectively) and extremely high levels in broilers (78.8% and 79.2%, respectively) and turkeys (94.4% for both antimicrobials). These high resistance levels to fluoroquinolones, sulfonamides and tetracycline in broilers and turkeys align with the circulations of a clone of S. Infantis which is prevalent in Europe. Figure C.2 presents the resistance levels to these four antimicrobials considering all reporting MSs.
Salmonella Enteritidis was the most common Salmonella serovar identified in human cases in 2022, with 26,811 cases reported in the EU/EEA. While MDR was uncommon among S. Enteritidis isolates (from both humans and poultry populations), the highest levels of resistance in S. Enteritidis from humans were noted to ciprofloxacin/pefloxacin (22.8%), nalidixic acid (26.0%) and colistin (29.7%). Similar resistance levels to these AMs were reported in S. Enteritidis isolated from laying hens (20.9%, 20.5% and 16.9%, respectively), while in broilers ciprofloxacin and nalidixic acid, resistance levels were reported at a higher level (42.0% for both AMs). Colistin resistance among S. Enteritidis is not uncommon, since this serovar belongs to group D salmonellas (serogroup O9) which tend to show decreased intrinsic susceptibility to colistin (Agersø et al., 2012; Ricci et al., 2020). Figure C.3 presents the resistance levels to these antimicrobials considering all reporting MSs.
Considering S. Kentucky, the 11th most reported serovar from human cases in 2022 with 344 cases reported in the EU/EEA, very high to extremely high levels of resistance were noted to ampicillin (59.2%), ciprofloxacin/pefloxacin (72.7%), nalidixic acid (72.4%), gentamicin (34.7%), sulfonamides (60.9%) and tetracycline (67.1%). Figure C.4 presents the resistance levels to these antimicrobials in human and animal isolates considering all reporting MSs. The number of S. Kentucky isolates recovered from broilers and turkeys was relatively low (N = 19, N = 13, respectively). While for laying hens, a higher number of isolates were recovered (N = 95). Resistance levels in S. Kentucky in laying hens were generally lower when compared to those reported in human isolates but reached higher resistance levels to ciprofloxacin and nalidixic acid (82.1% and 81.1%, respectively). Additionally, S. Kentucky accounted for most of the Salmonella isolates recovered from poultry populations exhibiting high‐level resistance to ciprofloxacin, with MIC of ≥ 4 mg/L. Similarly, S. Kentucky isolated from humans in 2022 accounted for 87.6% of the Salmonella isolates exhibiting high ciprofloxacin resistance. S. Kentucky isolates exhibiting high‐level ciprofloxacin resistance are likely to belong to the multilocus sequence type (ST) 198 clone, which has shown epidemic spread, first in Africa, then the Middle East, Asia and finally Europe (Hawkey et al., 2019; Le Hello et al., 2011, 2013). All the S. Kentucky isolates exhibiting high‐level ciprofloxacin resistance from poultry in 2022 displayed MICs of ≥ 8 mg/L.
Salmonella Typhimurium was the second most common Salmonella serovar identified in human cases in 2022, with 5809 cases reported in the EU/EEA. Considering all reporting MSs, the highest levels of resistance in S. Typhimurium from humans were observed for ampicillin (32.1%), sulfonamides (30.2%) and tetracycline (26.8%). S. Typhimurium isolates from pigs (N = 182) showed considerably higher resistance levels to these AMs. On the other hand, S. Typhimurium isolates from laying hens (N = 61) exhibited lower resistance levels to ampicillin (8.2%), sulfonamides and tetracycline (13.1%, each). Resistance levels to ciprofloxacin were moderate in humans (19.6), broilers (11.8%) and pigs (13.2%), and lower in laying hens (3.3%) and cattle under 1 year of age (10.0%). Notably, all turkey isolates were susceptible to ciprofloxacin (N = 14). Figure C.5 presents the resistance levels to these compounds considering all reporting MSs.
Monophasic S. Typhimurium was the third most common serovar reported from human cases in 2022, with 5585 registered cases in the EU/EEA. Considering all reporting MSs, the highest levels of resistance in monophasic S. Typhimurium from humans were observed for ampicillin (86.7%), sulfonamides (78.5%) and tetracycline (77.3%); as were also for isolates from poultry populations and pigs and cattle under 1 year of age. Notably, this resistance pattern (together with resistance to streptomycin) is typical of monophasic S. Typhimurium (Hopkins et al., 2010). Figure C.6 presents resistance levels to these compounds in human and animal populations considering all reporting MSs.
APPENDIX D. Trends in colistin resistance in indicator commensal Escherichia coli from broilers, turkeys, fattening pigs and cattle under 1 year of age
D.1.
Colistin (polymyxin E) is an antimicrobial of the polymyxin group that has been used extensively in farm animals all over the world, including in Europe. In human medicine, the use of colistin has historically been limited, but an increased usage has been recorded to account for the need for last resort antimicrobials to treat infections caused by multidrug‐resistant Gram‐negative bacteria. Consequently, polymyxins are now among the five antimicrobials listed by the WHO as critically important and of highest priority for human medicine. The discovery of transferable genetic elements (e.g. mcr‐genes) conferring resistance to colistin further underlines the importance of monitoring such resistance in food‐producing animals. The mandatory monitoring, in accordance with Commission Implementing Decision (EU) 2020/1729, is based on phenotypic susceptibility testing whereas molecular investigations would be required for clarifying the underlying mechanisms of resistance. i.e. if it is due to transferable genetic elements or not.
During the period 2021–2022, the level of resistance to colistin among indicator Escherichia coli from broilers, fattening pigs and cattle under 1 year of age was very low, and from turkeys, low (Annex C, Tables 1–4). For all animal populations, the majority of countries (62%–82%) did not detect any resistant isolates. Only among isolates from turkeys reported by Poland (19.1%) and Portugal (11.2%), the level of resistance was moderate (Annex C, Tables 1–4).
Colistin resistance was not detected among E coli isolates from imported fresh pig, cattle or turkey meat. However, among isolates from imported fresh broiler meat, the overall level of resistance was low (2.5%) and four out of eight countries reporting data had detected such isolates (Annex C, Tables 5–8).
The statistical significance (p < 0.05) of trends in colistin resistance in indicator commensal E. coli from broilers, fattening turkeys, fattening pigs and cattle under 1 year of age was analysed by logistic regression for countries reporting data for 3 years or more in the period 2014–2022 for broilers and turkeys and 2014–2021 for pigs and cattle under 1 year of age (see Appendix F – Materials and methods for details on methodology). The temporal trend analyses are presented in Figures D.1–D.4 below.
FIGURE D.1.
Temporal trends in resistance to colistin in indicator commensal E. coli from caecal samples from broilers, 26 EU MSs and 3 non‐MSs, 2009–2022.
FIGURE D.2.
Temporal trends in resistance to colistin in indicator commensal E. coli from caecal samples from fattening turkeys (turkeys), eight EU MSs and one non‐MS, 2014–2022.
FIGURE D.3.
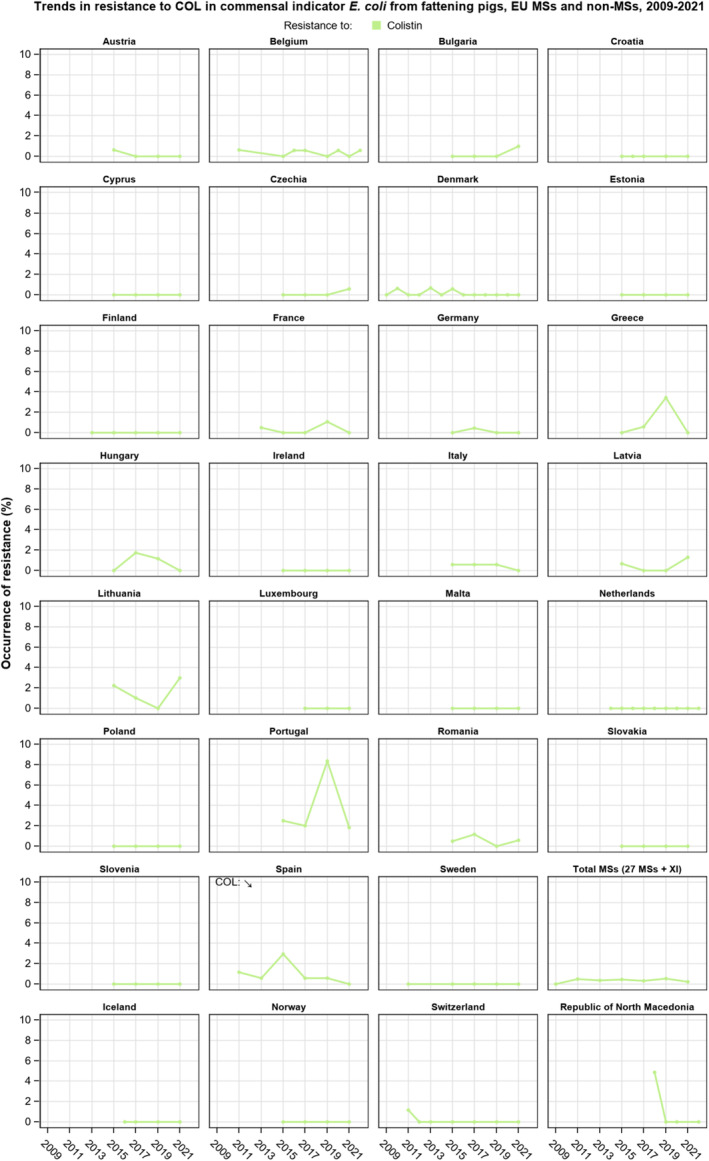
Temporal trends in resistance to colistin in indicator commensal E. coli from caecal samples from fattening pigs, 27 EU MSs and 4 non‐MSs, 2009–2021.
FIGURE D.4.
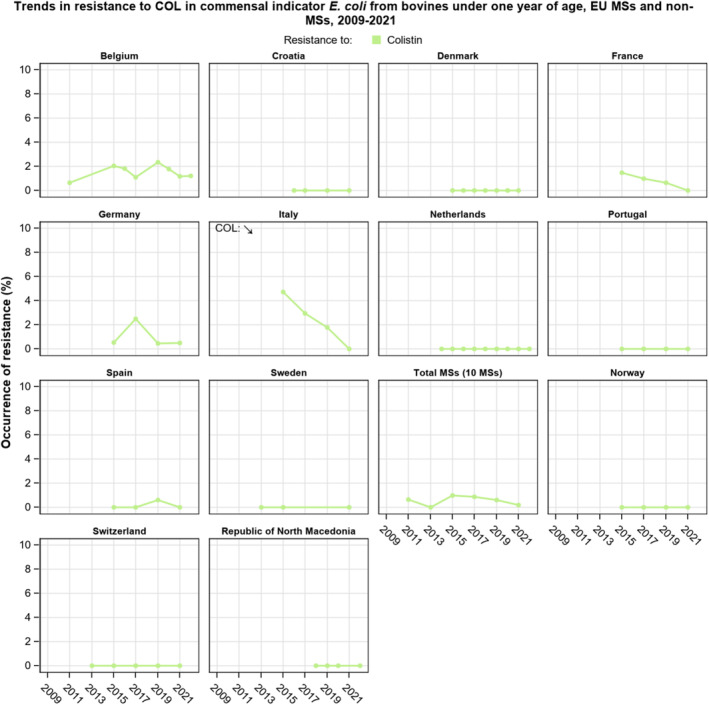
Temporal trends in resistance to colistin in indicator commensal E. coli from caecal samples from cattle under 1 year of age, 10 EU MSs and 3 non‐MSs, 2009–2021.
For broilers and turkeys, five and four MSs, respectively, reported statistically significant decreasing trends whereas one MS (Poland) reported increasing trends for both these animal populations (Figures D.1, D.2). For each of the two animal populations pigs and cattle under 1 year of age, one country (Spain for pigs and Italy for cattle under 1 year of age) reported decreasing trends (Figures D.3, D.4). No countries reported increasing trends among pigs or cattle under 1 year of age.
APPENDIX E. ESBL‐, AmpC‐ and CP‐encoding genes identified during the specific monitoring of ESBL‐/AmpC‐/CP‐producing E. coli isolates using whole genome sequencing (WGS)
E.1.
In 2021, four MSs (Czechia, Finland, Germany and Italy) reported WGS data for isolates detected in the specific monitoring of ESBL‐/AmpC‐/CP‐producing E. coli, whereas six MSs (Czechia, Germany, Finland, Italy, the Netherlands and Sweden) and one non‐MS (Norway) reported WGS data in 2022.
A variety of ESBL genes were identified in all countries reporting WGS data (Tables E.1, E.4). The most frequently reported ESBL‐encoding gene overall was bla CTX‐M‐1. This gene was detected in fattening pigs (n = 250), cattle under 1 year of age (n = 202) and in pig meat (n = 33) and cattle meat (n = 21) in 2021. In 2022, bla CTX‐M‐1 was reported in broilers (n = 164), fattening turkeys (n = 51), broiler meat (n = 106) and turkey meat (n = 41). In fattening pigs and cattle under 1 year of age and their derived meats, bla CTX‐M‐15 was the second most commonly detected ESBL‐encoding gene. It was reported from 81 fattening pigs and 193 cattle under 1 year of age. Notably, the occurrence in meat was lower, with only six and eight isolates from pig meat and cattle meat, respectively, reported to carry bla CTX‐M‐15. In poultry, the second most common ESBL‐encoding gene reported was bla SHV‐12. It was reported in broilers (n = 141), fattening turkeys (n = 63), broiler meat (n = 109) and turkey meat (n = 32).
TABLE E.1.
ESBL‐encoding genes detected in Escherichia coli isolates from the specific monitoring of ESBL‐/AmpC‐/CP‐producers in food‐producing animals and derived meat, by country in 2021.
| ESBL‐encoding genes | Cattle under 1 year of age | Cattle meat | Fattening pigs | Pig meat | ||||
|---|---|---|---|---|---|---|---|---|
| Escherichia coli | N | n | N | n | N | n | N | n |
| Czechia | ||||||||
| bla CTX‐M‐1 | – | – | 298 | 12 | 302 | 64 | 294 | 12 |
| bla CTX‐M‐3 | – | – | 298 | 1 | 302 | 1 | 294 | – |
| bla CTX‐M‐8 | – | – | 298 | – | 302 | 3 | 294 | – |
| bla CTX‐M‐14 | – | – | 298 | 1 | 302 | 1 | 294 | 1 |
| bla CTX‐M‐15 | – | – | 298 | 1 | 302 | 18 | 294 | 2 |
| bla CTX‐M‐27 | – | – | 298 | – | 302 | 5 | 294 | – |
| bla CTX‐M‐32 | – | – | 298 | 1 | 302 | – | 294 | 1 |
| bla CTX‐M‐55 | – | – | 298 | 1 | 302 | 3 | 294 | 1 |
| bla CTX‐M‐125 | – | – | 298 | – | 302 | 1 | 294 | – |
| bla SHV‐12 | – | – | 298 | – | 302 | 1 | 294 | 1 |
| bla TEM‐52 | – | – | 298 | – | 302 | – | 294 | 1 |
| bla TEM‐126 | – | – | 298 | – | 302 | 1 | 294 | – |
| Finland | ||||||||
| bla CTX‐M‐15 | – | – | – | – | 307 | 2 | – | – |
| Germany | ||||||||
| bla CTX‐M‐1 | 299 | 103 | 418 | 4 | 382 | 94 | 466 | 16 |
| bla CTX‐M‐2 | 299 | 1 | 418 | – | 382 | – | 466 | – |
| bla CTX‐M‐3 | 299 | 2 | 418 | 1 | 382 | 2 | 466 | – |
| bla CTX‐M‐8 | 299 | – | 418 | – | 382 | 3 | 466 | – |
| bla CTX‐M‐9 | 299 | – | 418 | – | 382 | 1 | 466 | – |
| bla CTX‐M‐14 | 299 | 2 | 418 | 2 | 382 | 4 | 466 | 1 |
| bla CTX‐M‐15 | 299 | 60 | 418 | 2 | 382 | 25 | 466 | 2 |
| bla CTX‐M‐27 | 299 | – | 418 | – | 382 | 1 | 466 | – |
| bla CTX‐M‐32 | 299 | 6 | 418 | – | 382 | 2 | 466 | – |
| bla CTX‐M‐55 | 299 | 6 | 418 | – | 382 | 2 | 466 | 1 |
| bla CTX‐M‐65 | 299 | 4 | 418 | – | 382 | 1 | 466 | – |
| bla SHV‐12 | 299 | 5 | 418 | – | 382 | 2 | 466 | – |
| bla TEM‐52 | 299 | 1 | 418 | – | 382 | 2 | 466 | – |
| bla TEM‐207 | 299 | 1 | 418 | – | 302 | – | 466 | – |
| Italy * | ||||||||
| bla CTX‐M‐1 | 310 | 99 | 301 | 5 | 301 | 92 | 305 | 5 |
| bla CTX‐M‐2 | 310 | 1 | 301 | – | – | – | 305 | – |
| bla CTX‐M‐3 | 310 | – | 301 | – | 301 | 1 | 305 | – |
| bla CTX‐M‐8 | 310 | – | 301 | – | 301 | 4 | 305 | 1 |
| bla CTX‐M‐14 | 310 | 4 | 301 | – | 301 | 9 | 305 | – |
| bla CTX‐M‐15 | 310 | 133 | 301 | 5 | 301 | 36 | 305 | 2 |
| bla CTX‐M‐27 | 310 | 1 | 301 | – | 301 | 3 | 305 | – |
| bla CTX‐M‐32 | 310 | 1 | 301 | – | 301 | 3 | 305 | 1 |
| bla CTX‐M‐55 | 310 | 11 | 301 | – | 301 | 13 | 305 | 1 |
| bla CTX‐M‐65 | 310 | 1 | 301 | – | 301 | 3 | 305 | 1 |
| bla SHV‐12 | 310 | 11 | 301 | 1 | 301 | 20 | 305 | 4 |
Abbreviations: N, Total number of samples tested per country; n, number of isolates harbouring the gene; –, No data reported for this category.
Italy also reported the presence of CTX‐M in seven isolates form cattle under 1 year of age and four isolates from fattening pigs. The isolates are not included in the table, as only the CTX‐M‐ family was reported, and not the specific genes.
TABLE E.2.
AmpC‐encoding genes detected in Escherichia coli isolates from the specific monitoring of ESBL‐/AmpC‐/CP‐producers in food‐producing animals and derived meat, by country in 2021.
| AmpC‐encoding genes | Cattle under 1 year of age | Cattle meat | Fattening pigs | Pig meat | ||||
|---|---|---|---|---|---|---|---|---|
| Escherichia coli | N | n | N | n | N | n | N | n |
| Czechia | ||||||||
| bla CMY‐2 | – | – | 298 | 3 | 302 | 9 | 294 | 2 |
| C‐42T mutation | – | – | 298 | 2 | 302 | 46 | 294 | 3 |
| Finland | ||||||||
| bla CMY‐2 | – | – | – | – | 307 | 1 | – | – |
| C‐42T mutation | – | – | – | – | 307 | 16 | – | – |
| T‐32A mutation | – | – | – | – | 307 | 1 | – | – |
| Germany | ||||||||
| bla CMY‐2 | 299 | 1 | – | – | 382 | 2 | 466 | – |
| C‐42T mutation | 299 | 1 | – | – | 382 | 22 | 466 | 3 |
| T‐32A mutation | 299 | – | – | – | 382 | 1 | 466 | – |
| Italy * | ||||||||
| bla CMY‐2 | 310 | 7 | 301 | 1 | 301 | 24 | 305 | 4 |
| bla CMY‐113 | 310 | – | 301 | – | 301 | 1 | 305 | – |
| bla DHA‐1 | 310 | 1 | 301 | – | 301 | 3 | 305 | 3 |
| C‐42T mutation | 310 | – | 301 | 1 | 301 | – | 305 | – |
Abbreviations: N, Total number of samples tested per country; n, number of isolates harbouring the gene; –, no data reported for this category.
Italy also reported the presence of AmpC phenotype/genotype for 32 isolates, three from cattle under 1 year of age, 28 from fattening pigs and one from retail pig meat. These isolates are not included in the table.
TABLE E.3.
CP‐encoding genes detected in Escherichia coli isolates from the specific monitoring of CP‐producers in food‐producing animals and derived meat, by country in 2021.
| CP‐encoding genes | Cattle under 1 year of age | Fattening pigs | ||
|---|---|---|---|---|
| Escherichia coli | N | n | N | n |
| Italy | ||||
| bla NDM‐5 | 310 | 1 | 301 | – |
| bla OXA‐181 | 310 | 4 | 301 | 20 |
| bla OXA‐48 | 310 | – | 301 | 1 |
| Czechia | ||||
| bla NDM‐5 | – | – | 302 | 3 |
Abbreviations: N, Total number of samples tested per country; n, number of isolates harbouring the gene; –, no data reported for this category.
TABLE E.4.
ESBL‐encoding genes detected in Escherichia coli isolates from the specific monitoring of ESBL‐/AmpC‐/CP‐producers in food‐producing animals and derived meat, by country in 2022.
| ESBL‐encoding genes | Broilers | Broiler meat | Fattening turkeys | Turkey meat | ||||
|---|---|---|---|---|---|---|---|---|
| Escherichia coli | N | n | N | n | N | n | N | n |
| Czechia | ||||||||
| bla CTX‐M‐1 | 293 | 70 | 296 | 48 | – | – | 152 | 4 |
| bla CTX‐M‐14 | 293 | 1 | 296 | 5 | – | – | 152 | 1 |
| bla CTX‐M‐15 | 293 | – | 296 | 3 | – | – | 152 | 3 |
| bla CTX‐M‐27 | 293 | – | 296 | 2 | – | – | 152 | 8 |
| bla CTX‐M‐32 | 293 | 1 | 296 | 1 | – | – | 152 | – |
| bla CTX‐M‐55 | 293 | 1 | 296 | 6 | – | – | 152 | 1 |
| bla CTX‐M‐65 | 293 | – | 296 | – | – | – | 152 | 1 |
| bla SHV‐12 | 293 | 10 | 296 | 4 | – | – | 152 | 4 |
| Finland | ||||||||
| bla CTX‐M‐1 | 301 | 4 | 300 | 5 | – | – | – | – |
| Germany | ||||||||
| bla CTX‐M‐1 | 333 | 17 | 467 | 11 | 351 | 17 | 439 | 17 |
| bla CTX‐M‐2 | 333 | – | 467 | 1 | 351 | – | 439 | – |
| bla CTX‐M‐3 | 333 | 1 | 467 | – | 351 | 1 | 439 | – |
| bla CTX‐M‐8 | 333 | – | 467 | – | 351 | – | 439 | 1 |
| bla CTX‐M‐9 | 333 | 1 | 467 | – | 351 | – | 439 | – |
| bla CTX‐M‐14 | 333 | – | 467 | – | 351 | 7 | 439 | 4 |
| bla CTX‐M‐15 | 333 | 3 | 467 | 4 | 351 | 40 | 439 | 69 |
| bla CTX‐M‐27 | 333 | 4 | 467 | – | 351 | 20 | 439 | 21 |
| bla CTX‐M‐32 | 333 | – | 467 | 1 | 351 | 2 | 439 | 2 |
| bla CTX‐M‐55 | 333 | 5 | 467 | 7 | 351 | 9 | 439 | 12 |
| bla CTX‐M‐65 | 333 | – | 467 | – | 351 | 1 | 439 | 1 |
| bla SHV‐12 | 333 | 66 | 467 | 68 | 351 | 20 | 439 | 11 |
| bla TEM‐15 | 333 | – | 467 | 1 | 351 | – | 439 | – |
| bla TEM‐52 | 333 | 31 | 467 | 46 | 351 | 6 | 439 | – |
| bla TEM‐106 | 333 | 3 | 467 | 1 | 351 | – | 439 | – |
| bla TEM‐207 | 333 | 1 | 467 | – | 351 | – | 439 | – |
| Italy | ||||||||
| bla CTX‐M‐1 | 479 | 56 | 302 | 39 | 397 | 34 | 294 | 20 |
| bla CTX‐M‐8 | 479 | 4 | 302 | – | 397 | 1 | 294 | – |
| bla CTX‐M‐14 | 479 | 7 | 302 | 2 | 397 | 3 | 294 | 1 |
| bla CTX‐M‐15 | 479 | 7 | 302 | 5 | 397 | 23 | 294 | 10 |
| bla CTX‐M‐27 | 479 | – | 302 | 1 | 397 | – | 294 | 1 |
| bla CTX‐M‐32 | 479 | – | 302 | – | 397 | 1 | 294 | |
| bla CTX‐M‐55 | 479 | 6 | 302 | 6 | 397 | 1 | 294 | 2 |
| bla CTX‐M‐65 | 479 | 3 | 302 | 2 | 397 | – | 294 | 1 |
| bla CTX‐M‐138 | 479 | 1 | 302 | – | 397 | – | 294 | – |
| bla SHV‐12 | 479 | 56 | 302 | 30 | 397 | 43 | 294 | 17 |
| bla SHV129 | 479 | – | 302 | 1 | 397 | – | 294 | – |
| Netherlands | ||||||||
| bla CTX‐M‐1 | 300 | 12 | 187 | 3 | – | – | – | – |
| bla CTX‐M‐3 | 300 | 1 | 187 | – | – | – | – | – |
| bla CTX‐M‐9 | 300 | 1 | 187 | – | – | – | – | – |
| bla CTX‐M‐15 | 300 | 6 | 187 | 1 | – | – | – | – |
| bla CTX‐M‐32 | 300 | 1 | 187 | 1 | – | – | – | – |
| bla CTX‐M‐55 | 300 | 5 | 187 | 2 | – | – | – | – |
| bla CTX‐M‐65 | 300 | – | 187 | 1 | – | – | – | – |
| bla SHV‐12 | 300 | 9 | 187 | 7 | – | – | – | – |
| bla TEM‐20 | 300 | – | 187 | 1 | – | – | – | – |
| bla TEM‐52B | 300 | 1 | 187 | 6 | – | – | – | – |
| bla TEM‐52C | 300 | 3 | 187 | 1 | – | – | – | – |
| Norway | ||||||||
| bla CTX‐M‐15 | 363 | – | – | – | 110 | 2 | – | – |
| bla CTX‐M‐55 | 363 | 1 | – | – | 110 | – | – | – |
| Sweden | ||||||||
| bla CTX‐M‐1 | 302 | 5 | 296 | – | – | – | – | – |
| bla CTX‐M‐32 | 302 | – | 296 | 1 | – | – | – | – |
| bla CTX‐M‐55 | 302 | – | 296 | 1 | – | – | – | – |
Abbreviations: N, Total number of samples tested per country; n, number of isolates harbouring the gene; –, No data reported for this category.
TABLE E.5.
AmpC‐encoding genes detected in Escherichia coli isolates from the specific monitoring of ESBL‐/AmpC‐/CP‐producers in food‐producing animals and derived meat, by country in 2022.
| AmpC‐encoding genes | Broilers | Broiler meat | Fattening turkeys | Turkey meat | ||||
|---|---|---|---|---|---|---|---|---|
| Escherichia coli | N | n | N | n | N | n | N | n |
| Czechia | ||||||||
| bla CMY‐2 | 293 | 12 | 296 | 15 | – | – | 152 | 1 |
| C‐42T mutation | 293 | 15 | 296 | – | – | – | 152 | 3 |
| Germany | ||||||||
| bla CMY‐2 | 333 | 2 | 467 | 7 | – | – | 439 | 1 |
| C‐42T mutation | 333 | 1 | 467 | 1 | 351 | 10 | 439 | 11 |
| T‐32A mutation | – | – | – | – | – | – | 439 | 1 |
| Italy | ||||||||
| bla CMY‐2 | 479 | 80 | 302 | 38 | 397 | 7 | 294 | 1 |
| bla CMY‐101 | 479 | 1 | 302 | 2 | 397 | – | 294 | 5 |
| bla CMY‐113 | 479 | 2 | 302 | – | 397 | – | 294 | 1 |
| bla CMY‐155 | 479 | – | 302 | 4 | 397 | 2 | 294 | 2 |
| C‐42T mutation | 479 | 3 | 302 | – | 397 | 1 | 294 | – |
| Netherlands | ||||||||
| bla CMY‐2 | 300 | – | 187 | 2 | – | – | – | – |
| bla CMY‐102 | 300 | 6 | 187 | – | – | – | – | – |
| C‐42T mutation | 300 | 2 | 187 | – | – | – | – | – |
| Norway | ||||||||
| C‐42T mutation | 363 | 1 | – | – | 110 | 9 | – | – |
| Sweden | ||||||||
| bla CMY‐2 | 302 | – | 296 | 2 | – | – | – | – |
| C‐42T mutation | 302 | 4 | 296 | 1 | – | – | – | – |
Abbreviations: N, Total number of samples tested per country; n, number of isolates harbouring the gene; –, No data reported for this category.
TABLE E.6.
CP‐encoding genes detected in Escherichia coli isolates from the specific monitoring of CP‐producers in food‐producing animals and derived meat, by country in 2022.
| CP‐encoding genes | Fattening turkey | |
|---|---|---|
| Escherichia coli | N | n |
| Italy | ||
| bla OXA‐181 | 397 | 1 |
Abbreviations: N, Total number of samples tested per country; n, number of isolates harbouring the gene.
Six different plasmid‐mediated AmpC‐encoding genes were reported during 2021 and 2022 (Tables E.2, E.5). These included bla CMY‐2 (all tested matrices in 2021 and 2022), bla CMY‐101 (broilers, broiler meat and turkey meat), bla CMY‐102 (broilers), bla CMY‐113 (fattening pigs, broilers, turkey meat), bla CMY‐155 (fattening turkeys, broiler meat and turkey meat), bla DHA‐1 (cattle under 1 year of age, fattening pigs and pig meat). The bla CMY‐2 gene was the most commonly reported plasmid‐mediated AmpC‐encoding gene overall. In addition, chromosomal mutations causing an AmpC phenotype were frequently reported. In fattening pigs, the C‐42T mutation was the most common cause of the AmpC phenotype in 2021, while bla CMY‐2 dominated in poultry.
In the CP‐specific monitoring, four different CP‐encoding genes were reported in 2021, and one reported in 2022 (Tables E.3, E.6). In 2021, the bla OXA‐181 gene was the most frequently reported, detected in fattening pigs (n = 20) and cattle under 1 year of age (n = 4) from Italy. bla NDM‐5 was detected in cattle under 1 year of age (n = 1) in Italy and in fattening pigs (n = 3) in Czechia. In addition, bla OXA‐48 was detected in fattening pigs (n = 1) in Italy. In 2022, a single CP‐producing E. coli was reported in the CP‐specific monitoring. This isolate originated from fattening turkey in Italy and was reported to harbour the bla OXA‐181 gene. Furthermore, a single CP‐producing E. coli carrying bla OXA‐181 from fattening turkey was detected in the routine monitoring of indicator E. coli. Also, three CP‐producing isolates from broilers carrying bla VIM‐1 collected in the specific monitoring of ESBL‐/AmpC‐/CP‐producing E. coli were reported. Further details are presented in Section 5.5 Monitoring of carbapenemase‐producing Escherichia coli. In 2022, Austrian, Finland, Italy, Ireland, Norway and Sweden reported both phenotypic and genotypic data for ESBL‐/AmpC‐/CP‐producing E. coli.
Some MSs reported WGS data in addition to phenotypic data in both routine monitoring and the specific monitoring of ESBL‐/AmpC‐/CP‐producers. These data are presented in Annex F.
APPENDIX F. Materials and methods
Antimicrobial susceptibility data from humans available in 2022
Data reported to The European Surveillance System (TESSy)
EU MSs report results from antimicrobial susceptibility testing of Salmonella spp. and Campylobacter spp. isolated from clinical cases to ECDC on an annual basis. Data can be submitted to ECDC and The European Surveillance System (TESSy) in different formats. Phenotypic test results can be reported either as measured values (inhibition zone diameters or minimum inhibitory concentrations (MIC)) in the isolate‐based reporting or as results interpreted with clinical breakpoints via the case‐based reporting of Salmonella and Campylobacter infections. Genomic‐based test results can be submitted either as the laboratory's prediction of the phenotype from sequencing of the bacterial genome, or from year 2023, submission of sequences directly to TESSy for analysis of resistance determinants and predicted phenotype in ECDC using ResFinder and PointFinder. The reporting of phenotypic quantitative data via the isolate‐based reporting is so far the preferred format, as stipulated in the EU protocol for harmonised monitoring of AMR in human Salmonella and Campylobacter isolates (ECDC, 2016).
| Salmonella spp.: For 2022, 27 MSs, plus Iceland and Norway provided data on antimicrobial resistance (AMR) in human Salmonella isolates. Twenty‐two countries reported measured values and five reported results interpreted as susceptible standard dosing regimen, susceptible increased exposure or resistant (SIR) according to the clinical breakpoints (CBPs) applied. Two countries reported whole genome sequences that were analysed by ECDC and interpreted as predicted wild type or predicted non‐wild type (Table F.1) | Campylobacter spp.: For 2022, 22 MSs, plus Iceland and Norway provided AMR data from human isolates. Seventeen countries reported measured values, five reported results interpreted as susceptible standard dosing regimen, susceptible increased exposure or resistant (SIR) according to the clinical breakpoints (CBPs) applied and two countries reported results that were categorised as predicted wild type or predicted non‐wild type based on analysis of bacterial genomes (one country reporting interpreted data and one country submitting whole genome sequences which were analysed at ECDC) (Table F.2) |
Harmonised testing
Most laboratories follow the ‘EU protocol for harmonised monitoring of antimicrobial resistance in human Salmonella and Campylobacter isolates’ (ECDC, 2016) on the antimicrobial panel to be tested. The antimicrobials tested, the method used (dilution, disk diffusion, gradient strip, WGS), the type of data provided and the interpretive criteria applied are presented in Table F.1 for Salmonella and in Table F.2 for Campylobacter. For Salmonella, eight MSs, plus Iceland and Norway used only disk diffusion methods (DDs) for their AST, seven MSs used dilution methods (DLs) and another 10 MSs used various combinations of DD and DL or dilution with gradient strip (DLG) methods. Two countries used sequencing (Table F.1). For Campylobacter, 10 MSs and Iceland used only DDs for their AST, five MSs used DL, two MSs and Norway used DLG and one MS used a combination of DD and DLG. Two countries used sequencing and bioinformatics tools to predict phenotypic resistance from the genome. Two MSs did not provide the methodology (Table F.2). All data on measured MIC or zone mm values were results of AST performed at the national public health reference laboratories, with the exception of Italy for Salmonella where two regional laboratories also contributed, and Finland for Campylobacter where the quantitative data had been collected from regional laboratories. Data interpreted with clinical breakpoints were normally from local or regional laboratories and reported together with the information on the clinical case. In these cases, AST had primarily been performed with the purpose of treatment of the case rather than AMR monitoring. For this reason, the number of tests per antimicrobial varied.
Salmonella test panel
The national public health laboratories within the Food‐ and Waterborne Diseases and Zoonoses (FWD) network has agreed on a panel of priority antimicrobials and optional antimicrobials to test for and report to ECDC (ECDC, 2016). Compared with earlier recommendations, a second beta‐lactam (ceftazidime) and a carbapenem (meropenem) were added. For 2022, all but one MS reported results on meropenem and all but two for ceftazidime. Three last‐line antimicrobials – azithromycin, colistin and tigecycline – are also included in the priority list. For colistin, however, the methodology is complicated due to chemical properties of the substance and a joint EUCAST and Clinical and Laboratory Standards Institute (CLSI) subcommittee confirmed that broth microdilution is so far the only valid method for colistin susceptibility testing (CLSI and EUCAST, 2016). Disk diffusion does not work because of poor diffusion of the large colistin molecule in the agar and tested gradient strips also underestimate colistin MIC values, again most likely due to poor diffusion in the agar (Matuschek et al., 2018). Therefore, only countries performing broth microdilution (or those predicting resistance from WGS) should report on colistin resistance. Eleven MSs reported AST results on colistin, 12 on azithromycin and 13 on tigecycline for 2022.
Due to the problems in detecting low‐level fluoroquinolone resistance in Salmonella spp. using disk diffusion, nalidixic acid was, for a long time, used as a marker for fluoroquinolone resistance. After the discovery that plasmid‐mediated fluoroquinolone resistance is often not detected using nalidixic acid, EUCAST studied alternative disks and concluded that pefloxacin was an excellent surrogate marker (except for isolates having the aac(6′)‐Ib‐cr gene as the only resistance determinant) (Skov & Monnet, 2016). Since 2014, EUCAST has recommended this agent for screening of low‐level fluoroquinolone resistance in Salmonella with disk diffusion (EUCAST, 2014) and, since June 2016, this is also reflected in the EU protocol. In 2022, all countries reporting measured values for disk diffusion tested with pefloxacin instead of ciprofloxacin. Fifteen countries reported the combination drug co‐trimoxazole (trimethoprim–sulfamethoxazole) in addition to, or instead of, testing the substances separately, partly because this combination is used for clinical treatment and partly because no EUCAST interpretive criterion exists for sulfamethoxazole for Salmonella.
Campylobacter test panel
The antimicrobials included in the 2022 report followed the panel of antimicrobials from the EU protocol for harmonised monitoring of AMR in human Salmonella and Campylobacter isolates (ECDC, 2016). The priority panel for Campylobacter includes ciprofloxacin, erythromycin, tetracyclines and gentamicin. Gentamicin was added in 2016 and is recommended for screening of invasive isolates. Co‐amoxiclav (combination drug with amoxicillin and clavulanic acid) was included from the list of optional antimicrobials. In 2022, all reporting countries tested the isolates against the three main antimicrobials ciprofloxacin, erythromycin and tetracycline (although one country tested less than 10 isolates). In relation to C. jejuni isolates, 15 reporting countries also tested for gentamicin and seven tested for co‐amoxiclav. With regard to C. coli isolates, 13 reporting countries also tested these isolates for gentamicin and five tested for co‐amoxiclav (Annex B, Section B.1: Tables 1 and 2).
Analyses of antimicrobial resistance testing
Harmonised interpretation of data with animal and food data
Data reported as measured values were interpreted by ECDC based on the EUCAST ECOFF values, when available. For MIC data, the same criteria as used by EFSA were applied (Tables F.5, F.6) except for when EUCAST had changed the ECOFF after the regulation for animal and food AMR monitoring had been implemented, e.g. as for ampicillin in 2021 for Salmonella enterica (MIC lowered with one dilution). Where EUCAST had removed the ECOFF (colistin, meropenem, tigecycline and trimethoprim–sulfamethoxazole for Salmonella), the same criteria were applied as recommended by EFSA (EFSA, 2023). For zone diameter data, corresponding EUCAST disk diffusion ECOFF values were applied with a few exceptions (Tables F.1, F.2). Regarding data reported as SIR values, the categories of ‘susceptible, increased exposure’ (I) and ‘clinically’ resistant (R) were combined into one group, except for tetracycline and Salmonella. Alignment of the susceptible category with the ‘wild type’ category based on epidemiological cut‐off values (ECOFFs) and of the I + R category with the ECOFF‐based ‘non‐wild type’ category provides better comparability and more straightforward interpretation of the data for most antimicrobial agents included (Figure 19 in Section 3.5 and Figure C.1 in Appendix C). For Salmonella, this procedure results in good concordance (± 1 dilution) across categories with the exception of meropenem where the MIC for non‐susceptible category is substantially higher (+ 4 dilutions) than the ECOFF. For Campylobacter, there was full concordance across interpretive categories with this procedure, except for the EUCAST CBP for C. jejuni for tetracyclines, which is one dilution step higher than the EUCAST ECOFF.
Separation by species or serovar
As resistance levels differ substantially between Salmonella serovars, results are presented separately for selected serovars of importance in humans. The serovars presented in the report are S. Enteritidis, S. Typhimurium, monophasic S. Typhimurium, S. Infantis and S. Kentucky. AMR data on the 10 most common serovars in human cases in the last years are also available in the ECDC Surveillance Atlas for Infectious Diseases (https://atlas.ecdc.europa.eu/public/index.aspx). For Campylobacter, resistance levels differ quite substantially between the two most important Campylobacter species, C. jejuni and C. coli, and data are therefore presented by species. The proportion of resistant isolates is only shown when at least 10 isolates were reported from an MS.
Exclusion of travel‐associated cases
To better assess the impact from food consumed within each reporting country on the AMR levels found in human isolates, cases known to have travelled outside of the country during the incubation period were excluded from the analysis. However, as several countries had not provided any information on travel status of their cases, cases with unknown travel status were also included in addition to domestically acquired cases. The exception to this is Denmark and Finland, where it has been agreed that cases of unknown travel status should be excluded from analyses for Salmonella spp. and for Denmark also for Campylobacter spp. The proportions of travel‐associated, domestic and cases with unknown travel status among the tested isolates are presented in Tables F.3, F.4.
Temporal trends in resistance
Temporal trends in the proportion of resistant human isolates to selected antimicrobials over the period 2013–2022 were analysed by country. The statistical significance was assessed with logistic regression in Stata 17.0 and a p‐value of < 0.05 was considered to be significant. Only countries testing at least 10 isolates per year and for at least 3 years in the 10‐year period were included. For Salmonella, the antimicrobials analysed were ciprofloxacin/pefloxacin/nalidixic acid, cefotaxime, ampicillin and tetracycline. For Campylobacter, the corresponding antimicrobials were ciprofloxacin, erythromycin and tetracycline (Annex B, Section B.1: Figures 1, 2).
Maps for critically important antimicrobials resistance
For Salmonella, the proportions of human isolates resistant to both of the critically important antimicrobials for treatment of severe Salmonella infections (WHO, 2019), fluoroquinolones (ciprofloxacin/pefloxacin) and third‐generation cephalosporins (cefotaxime), were presented in maps to provide an overview of the geographical distribution of resistance in the EU/EEA. Combined ‘microbiological resistance’ was presented for Salmonella spp. and selected serovars (tables with combined resistance are also available in Annex A). For Campylobacter, the proportions of human isolates resistant to both critically important antimicrobials for treatment of severe Campylobacter infections (WHO, 2019), fluoroquinolones (ciprofloxacin) and macrolides (erythromycin), were presented in maps to provide an overview of the geographical distribution of this combined resistance in the EU/EEA. Combined ‘microbiological’ resistance (using EUCAST ECOFFs and EUCAST CBPs) was presented for C. jejuni and C. coli.
Analysis of multidrug resistance
Multidrug resistance (MDR) of human Salmonella spp. to nine antimicrobial classes was analysed. Multidrug resistance of an isolate was defined as resistance or non‐susceptibility to at least three different antimicrobial classes (Magiorakos et al., 2012). The antimicrobials included were ampicillin, cefotaxime/ceftazidime, chloramphenicol, ciprofloxacin/pefloxacin/nalidixic acid, gentamicin, meropenem, sulfonamides/sulfamethoxazole, tetracyclines and trimethoprim/trimethoprim–sulfamethoxazole (co‐trimoxazole). Resistance to nalidixic acid, ciprofloxacin and pefloxacin were addressed together, as they belong to the same class of antimicrobials: quinolones. Isolates that were non‐wild type or I + R to any of these antimicrobials were classified as microbiologically resistant to the class of quinolones. The same method was applied to the two third‐generation cephalosporins cefotaxime and ceftazidime. Trimethoprim and co‐trimoxazole were also addressed together, as a few countries had only tested for susceptibility to the combination. This approach was considered appropriate because among the countries that provided data on both trimethoprim alone and the combination co‐trimoxazole, the proportion of resistant or non‐susceptibles corresponded closely between the two. Multidrug resistance of a C. jejuni or C. coli isolate was defined as resistance or non‐susceptibility to at least three different antimicrobial classes (Magiorakos et al., 2012). The antimicrobials in the MDR analysis were harmonised between EFSA and ECDC and included ciprofloxacin, erythromycin, gentamicin and tetracycline.
Analysis of ESBL, AmpC and carbapenemase‐production in Salmonella
All countries except one reported results from AST of third‐generation cephalosporins in 2023. Those which reported findings of ESBL and/or AmpC or non‐wild type results to third‐generation cephalosporins and ampicillin were contacted by mail to provide further details on phenotypic and/or genotypic results. Of the 20 MSs and one non‐MS reporting such isolates, all except three could provide further information.
TABLE F.1.
Antimicrobials reported, method used, type of data reported and interpretive criteria applied by MSs for Salmonella isolates from humans in 2022.
| Country | Gentamicin | Chloramphenicol | Ampicillin | Cefotaxime | Ceftazidime | Meropenem | Tigecycline | Nalidixic acid | Ciprofloxacin/pefloxacin | Azithromycin | Colistin | Sulfonamides | Trimethoprim | Trimethoprim–sulfamethoxazole | Tetracyclines | Method used | Quantitative (Q) or categorical (SIR or PWT/PNWT) | Interpretive criteria |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Austria | ● | ● | ● | ● | ● | ● | ● | ● | ● a | ● | ● | ● | ● | DD | Q | Interpreted by ECDC. EUCAST ECOFFs for all except CLSI CBP for SUL | ||
| Belgium | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | DL | Q | Interpreted by ECDC, as for Austria | |||
| Bulgaria | ● | ● | ● | ● | ● | ● | ● | ● | DL | SIR | EUCAST CBP | |||||||
| Croatia | ● | ● | ● | ● | ● | ● | ● a | ● | DD | Q | Interpreted by ECDC, as for Austria | |||||||
| Cyprus | ● | ● | ● | ● | ● | ● | ● | ● | DL/DLG | Q | Interpreted by ECDC, as for Austria, except for CTX, MEM and SXT where EUCAST CBP were used | |||||||
| Czechia | ● | ● | ● | ● | ● | ● | ● | ● a | ● | ● | ● | ● | ● | DL/DD | Q | Interpreted by ECDC, as for Austria | ||
| Denmark | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | DL | Q | Interpreted by ECDC, as for Austria | |
| Estonia | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | DL | Q | Interpreted by ECDC, as for Austria | ||
| Finland | ● | ● | ● | ● | ● | ● | ● a | ● | ● | DD | Q | Interpreted by ECDC, as for Austria | ||||||
| France | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | DL | Q | Interpreted by ECDC, as for Austria | |
| Germany | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | DL | Q | Interpreted by ECDC, as for Austria | ||||
| Greece | ● | ● | ● | ● | ● | ● | ● | ● a | ● | ● | ● | ● | ● | DD/DL | Q | Interpreted by ECDC, as for Austria | ||
| Hungary | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | DD | SIR | EUCAST CBP except CLSI CBP for TET | |||||
| Iceland | ● | ● | ● | ● | ● | ● a | ● | ● | ● | ● | DD | Q | Interpreted by ECDC, as for Austria | |||||
| Ireland | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | WGS | PWT/PNWT | Interpreted by ECDC using ResFinder and PointFinder | |
| Italy | ● | ● | ● | ● | ● | ● | ● | ● | ● a | ● | ● | ● | ● | ● | DD/DL | Q | Interpreted by ECDC, as for Austria | |
| Latvia | ● | ● | ● | ● | DD | SIR | No recent information on guidelines used | |||||||||||
| Lithuania | ● | ● | ● | ● | ● | ● | ● | ● | ● | DL/DD | SIR | EUCAST CBP | ||||||
| Luxembourg | ● | ● | ● | ● | ● | ● | ● a | ● | ● | DD/DLG | Q | Interpreted by ECDC, as for Austria | ||||||
| Malta | ● | ● | ● | ● | ● | ● | ● a | ● | DL/DD/DLG | Q | Interpreted by ECDC, as for Austria, except for CTX and MEM where EUCAST CBP were used | |||||||
| Netherlands | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | DL | Q | Interpreted by ECDC, as for Austria | |
| Norway | ● | ● | ● | ● | ● | ● a | ● | DD | Q | Interpreted by ECDC, as for Austria | ||||||||
| Poland | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | DD/DLG/DL | Q | Interpreted by ECDC, as for Austria | |
| Portugal | ● | ● | ● | ● | ● | ● | ● | ● | ● a | ● | ● | ● | ● | DD | Q | Interpreted by ECDC, as for Austria | ||
| Romania | ● | ● | ● | ● | ● | ● | ● | ● a | ● | ● | ● | DD | Q | Interpreted by ECDC, as for Austria | ||||
| Slovakia | ● | ● | ● | ● | ● | ● | ● | ● | DD/DL | SIR | EUCAST CBP except CLSI CBP for TET | |||||||
| Slovenia | ● | ● | ● | ● | ● | ● | ● a | ● | ● | ● | ● | DD/DLG | Q | Interpreted by ECDC, as for Austria | ||||
| Spain | ● | ● | ● | ● | ● | ● | ● | ● a | ● | ● | ● | DD | Q | Interpreted by ECDC, as for Austria | ||||
| Sweden | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | WGS | PWT/PNWT | Interpreted by ECDC using ResFinder and PointFinder |
Abbreviations: CBP, clinical breakpoint; CLSI, Clinical and Laboratory Standards Institute; CTX, cefotaxime; DD, disc diffusion; DL, dilution; DLG, dilution with gradient strip; ECDC, European Centre for Disease Prevention and Control; ECOFF, epidemiological cut‐off; EUCAST, European Committee on Antimicrobial Susceptibility Testing; MEM, meropenem; NAL, nalidixic acid; PWT/PNWT, predicted wild type/predicted non‐wild type (categorical); Q, quantitative data; SIR, susceptible standard dosing regimen, susceptible increased exposure, resistant (categorical data); SUL, sulfonamides; TET, tetracycline; WGS, whole genome sequencing.
Pefloxacin used in disc diffusion.
TABLE F.2.
Antimicrobials reported, method used, type of data reported and interpretive criteria applied by MSs for Campylobacter isolates from humans in 2022.
| Country | Gentamicin | Co‐amoxiclav | Ciprofloxacin | Erythromycin | Tetracyclines | Method used | Quantitative (Q) or categorical (SIR) | Interpretive criteria |
|---|---|---|---|---|---|---|---|---|
| Austria | ● | ● | ● | ● | DL | Q | Interpreted by ECDC. EUCAST ECOFF (CIP, ERY, GEN, TET) | |
| Bulgaria | ● | ● | ● | DD | SIR | No information available | ||
| Cyprus | ● | ● | ● | DD | Q | Interpreted by ECDC, as for Austria | ||
| Denmark | ● | ● | ● | ● | DL | Q | Interpreted by ECDC, as for Austria | |
| Estonia | ● | ● | ● | ● | DL | Q | Interpreted by ECDC, as for Austria | |
| Finland | ● | ● | ● | DD/DLG | Q | Interpreted by ECDC, as for Austria. | ||
| France | ● | ● | ● | ● | ● | DD | SIR | EUCAST CBP (CIP, ERY, TET), CA‐SFM CBP (AMC, GEN) |
| Germany | ● | ● | ● | ● | ● | DL | Q | Interpreted by ECDC, as for Austria, plus CA‐SFM CBP 2022 (AMC) |
| Greece | ● | ● | ● | ● | ● | DD | Q | Interpreted by ECDC, as for Germany |
| Hungary | ● | ● | ● | No information | SIR | No information available | ||
| Ireland | ● | ● | ● | WGS | PWT/PNWT | Interpreted by IE using ResFinder and BioNumerics | ||
| Iceland | ● | ● | ● | DD | Q | Interpreted by ECDC, as for Austria | ||
| Italy | ● | ● | ● | ● | DD | Q | Interpreted by ECDC, as for Austria | |
| Lithuania | ● | ● | ● | DD | SIR | EUCAST CBP | ||
| Luxembourg | ● | ● | ● | ● | ● | DD | Q | Interpreted by ECDC, as for Germany |
| Malta | ● | ● | ● | ● | ● | DLG | Q | Interpreted by ECDC, as for Germany |
| Netherlands | ● | ● | ● | ● | WGS | PWT/PNWT | Interpreted by NL using in‐house pipeline based on PointFinder and Resfinder | |
| Norway | ● | ● | ● | ● | DLG | Q | Interpreted by ECDC, as for Austria | |
| Poland | ● | ● | ● | ● | DLG | Q | Interpreted by ECDC, as for Austria | |
| Portugal | ● | ● | ● | ● | DD | Q | Interpreted by ECDC, as for Austria | |
| Romania | ● | ● | ● | DD | Q | Interpreted by ECDC, as for Austria | ||
| Slovakia | ● | ● | ● | ● | ● | No information | SIR | No information available |
| Slovenia | ● | ● | ● | DD | Q | Interpreted by ECDC, as for Austria | ||
| Spain | ● | ● | ● | ● | DL | Q | Interpreted by ECDC, as for Austria |
Abbreviations: AMC, amoxicillin/clavulanate; CA‐SFM, French Society for Microbiology; CBP, clinical breakpoint; CIP, ciprofloxacin; DD, disc diffusion; DL, dilution; DLG, dilution with gradient strip; ECDC, European Centre for Disease Prevention and Control; ECOFF, epidemiological cut‐off; ERY, erythromycin; EUCAST, European Committee on Antimicrobial Susceptibility Testing; GEN, gentamicin; MSs, Member States; PWT/PNWT, predicted wild type/predicted non‐wild type (categorical); Q, quantitative data; SIR, susceptible standard dosing regimen, susceptible increased exposure, resistant (categorical data); TET, tetracycline; WGS, whole genome sequencing.
TABLE F.3.
Proportion of tested Salmonella spp. isolates from human cases associated with travel, domestic cases and cases with unknown travel information by country, 2022.
| Country | Total Salmonella tested | Travel‐associated | Domestic | Unknown |
|---|---|---|---|---|
| N | % | % | % | |
| Austria | 1157 | 0.0 | 0.0 | 100.0 |
| Belgium | 1044 | 7.8 | 16.4 | 75.9 |
| Bulgaria | 77 | 0.0 | 0.0 | 100.0 |
| Croatia | 654 | 0.0 | 0.0 | 100.0 |
| Cyprus | 94 | 0.0 | 0.0 | 100.0 |
| Czechia | 242 | 5.4 | 90.5 | 4.1 |
| Denmark | 416 | 29.8 | 70.2 | 0.0 |
| Estonia | 148 | 12.2 | 59.5 | 28.4 |
| Finland | 102 | 7.8 | 92.2 | 0.0 |
| France | 1242 | 11.5 | 13.0 | 75.4 |
| Germany | 3683 | 1.8 | 98.2 | 0.0 |
| Greece | 160 | 0.0 | 0.0 | 100.0 |
| Hungary | 1087 | 0.1 | 99.9 | 0.0 |
| Ireland | 325 | 0.0 | 0.0 | 100.0 |
| Italy | 1055 | 0.9 | 4.1 | 95.0 |
| Latvia | 1 | 0.0 | 100.0 | 0.0 |
| Lithuania | 226 | 5.8 | 92.9 | 1.3 |
| Luxembourg | 125 | 0.0 | 0.0 | 100.0 |
| Malta | 161 | 0.0 | 0.0 | 100.0 |
| Netherlands | 711 | 19.5 | 0.0 | 80.5 |
| Poland | 152 | 0.0 | 0.0 | 100.0 |
| Portugal | 377 | 0.5 | 6.6 | 92.8 |
| Romania | 90 | 0.0 | 0.0 | 100.0 |
| Slovakia | 857 | 1.3 | 98.7 | 0.0 |
| Slovenia | 384 | 3.6 | 32.8 | 63.5 |
| Spain | 1600 | 0.5 | 80.1 | 19.4 |
| Sweden | 651 | 9.2 | 87.9 | 2.9 |
| Total (27 MSs) | 16,821 | 4.2 | 52.5 | 43.3 |
| Iceland | 54 | 50.0 | 38.9 | 11.1 |
| Norway | 380 | 32.4 | 49.2 | 18.4 |
Abbreviations: MSs, Member States; N, number of isolates tested.
TABLE F.4.
Proportion of tested Campylobacter jejuni and Campylobacter coli isolates from human cases associated with travel, domestic cases and cases with unknown travel information by country in 2022.
| Country | Total C. jejuni & C. coli tested | Travel‐associated | Domestic | Unknown |
|---|---|---|---|---|
| N | % | % | % | |
| Austria | 498 | 8.0 | 90.2 | 1.8 |
| Bulgaria | 51 | 0.0 | 0.0 | 100.0 |
| Cyprus | 59 | 0.0 | 0.0 | 100.0 |
| Denmark | 288 | 18.1 | 81.9 | 0.0 |
| Estonia | 243 | 2.5 | 46.1 | 51.4 |
| Finland | 1624 | 0.0 | 0.0 | 100.0 |
| France | 7846 | 0.0 | 0.0 | 100.0 |
| Germany | 1971 | 0.2 | 99.8 | 0.0 |
| Greece | 89 | 0.0 | 0.0 | 100.0 |
| Hungary | 556 | 0.0 | 100.0 | 0.0 |
| Ireland | 212 | 0.0 | 2.4 | 97.6 |
| Italy | 148 | 2.0 | 18.2 | 79.7 |
| Lithuania | 433 | 1.2 | 98.4 | 0.5 |
| Luxembourg | 237 | 0.0 | 0.0 | 100.0 |
| Malta | 237 | 0.0 | 0.0 | 100.0 |
| Netherlands | 460 | 1.3 | 0.0 | 98.7 |
| Poland | 35 | 0.0 | 0.0 | 100.0 |
| Portugal | 431 | 0.0 | 100.0 | 0.0 |
| Romania | 7 | 0.0 | 100.0 | 0.0 |
| Slovakia | 1097 | 0.0 | 100.0 | 0.0 |
| Slovenia | 938 | 2.2 | 28.0 | 69.7 |
| Spain | 750 | 0.0 | 62.3 | 37.7 |
| Total | 18,210 | 0.8 | 33.2 | 66.1 |
| Iceland | 100 | 56.0 | 28.0 | 16.0 |
| Norway | 350 | 31.7 | 47.7 | 20.6 |
Abbreviations: MSs, Member States; N, number of isolates tested.
TABLE F.5.
Panel of antimicrobial substances included in AMR monitoring, thresholds for interpreting resistance and concentration ranges tested in Salmonella spp. and indicator commensal E. coli (first panel) based on Commission Implementing Decision (EU) 2020/1729 and EFSA Technical Report 2021.
| Antimicrobial | Salmonella EU surveillance 2021/2022 EUCAST ECOFF* (mg/L) | E. coli EU surveillance 2021/2022 EUCAST ECOFF* (mg/L) | Concentration range, mg/L (no. of wells) |
|---|---|---|---|
| Amikacin | > 4 a | > 8 | 4–128 (6) |
| Ampicillin | > 8 | > 8 | 1–32 (6) |
| Azithromycin | NA b | NA b | 2–64 (6) |
| Cefotaxime | > 0.5 | > 0.25 | 0.25–4 (5 |
| Ceftazidime | > 2 | > 0.5 | 0.25–8 (6) |
| Chloramphenicol | > 16 | > 16 | 8–64 (4) |
| Ciprofloxacin | > 0.06 | > 0.06 | 0.015–8 (10) |
| Colistin | NA c | > 2 | 1–16 (5) |
| Gentamicin | > 2 | > 2 | 0.5–16 (6) |
| Meropenem | > 0.125 | > 0.125 | 0.03–16 (10) |
| Nalidixic acid | > 8 | > 8 | 4–64 (5) |
| Sulfamethozaxole | NA d | > 64 | 8–512 (7) |
| Tetracycline | > 8 | > 8 | 2–32 (5) |
| Tigecycline | NA e | NA e | 0.25–8 (6) |
| Trimethoprim | > 2 | > 2 | 0.25–16 (7) |
Abbreviations: AMR, antimicrobial resistance; ECOFFs, epidemiological cut‐off values; EUCAST, European Committee on Antimicrobial Susceptibility Testing; NA, not available.
EUCAST epidemiological cut‐off values. ‘>’ than the ECOFF, criteria used to determine microbiological resistance.
EUCAST epidemiological cut‐off (ECOFF) value for Salmonella is tentative.
EUCAST epidemiological cut‐off (ECOFF) not available, > 16 mg/L was used.
EUCAST epidemiological cut‐off (ECOFF) value for Salmonella spp. not available, > 2 mg/L was used.
EUCAST epidemiological cut‐off (ECOFF) not available, > 256 mg/L was used.
EUCAST epidemiological cut‐off (ECOFF) not available, > 0.5 mg/L was used.
TABLE F.6.
Panel of antimicrobial substances included in AMR monitoring, thresholds for interpreting resistance and concentration ranges tested in C. jejuni and C. coli based on Commission Implementing Decision (EU) 2020/1729 and EFSA Technical Report 2021.
| Antimicrobial | C. jejuni EU surveillance 2021/2022 EUCAST ECOFF* (mg/L) | C. coli EU surveillance 2021/2022 EUCAST ECOFF* (mg/L) | Concentration range, mg/L (no. of wells) |
|---|---|---|---|
| Chloramphenicol | > 16 | > 16 | 2–64 (6) |
| Ciprofloxacin | > 0.5 | > 0.5 | 0.12–32 (9) |
| Ertapenem | > 0.5** | > 0.5** | 0.125–4 (6) |
| Erythromycin | > 4 | > 8 | 1–512 (10) |
| Gentamicin a | > 2 | > 2 | 0.12–16 (7) |
| Tetracycline | > 1 | > 2 | 0.5–64 (8) |
Abbreviations: AMR, antimicrobial resistance; ECOFFs, epidemiological cut‐off values; EUCAST, European Committee on Antimicrobial Susceptibility Testing.
The updated EUCAST epidemiological cut‐off (ECOFF) value for both species is at 1 mg/L.
EUCAST epidemiological cut‐off values. ‘>’ than the ECOFF, criteria used to determine microbiological resistance.
Due to the absence of a validated threshold for resistance to ertapenem in C. jejuni and C. coli recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST), an ECOFF of 0.5 mg/L was used by EFSA in 2021 and 2022 in agreement with the EU Reference Laboratory for AMR (EFSA, 2023).
TABLE F.7.
Panel of antimicrobial substances, EUCAST ECOFFs and concentration ranges used for testing Salmonella spp. and indicator commensal E. coli isolates resistant to cefotaxime, ceftazidime or meropenem (second panel).
| Antimicrobial | Salmonella EU surveillance 2021/2022 EUCAST ECOFF* (mg/L) | E. coli EU surveillance 2021/2022 EUCAST ECOFF* (mg/L) | Concentration range, mg/L (no. of wells) |
|---|---|---|---|
| Cefepime a | > 0.125 | > 0.125 | 0.06–32 (10) |
| Cefotaxime | > 0.5 | > 0.25 | 0.25–64 (9) |
| Cefotaxime + clavulanic acid | > 0.5 | > 0.25 | 0.06–64 (11) |
| Cefoxitin | > 8 | > 8 | 0.5–64 (8) |
| Ceftazidime | > 2 | > 0.5 | 0.25–128 (9) |
| Ceftazidime + clavulanic acid | > 2 | > 0.5 | 0.125–128 (11) |
| Ertapenem b | > 0.06 | > 0.06 | 0.015–2 (8) |
| Imipenem | > 1 | > 0.5 | 0.12–16 (8) |
| Meropenem | > 0.125 | > 0.125 | 0.03–16 (10) |
| Temocillin | > 16 | > 16 | 0.5–128 (9) |
Abbreviations: AMR, antimicrobial resistance; ECOFFs, epidemiological cut‐off values; EUCAST, European Committee on Antimicrobial Susceptibility Testing.
EUCAST epidemiological cut‐off values. ‘>’ than the ECOFF, criteria used to determine microbiological resistance.
EUCAST epidemiological cut‐off (ECOFF) value for E. coli is 0.25 mg/L.
EUCAST epidemiological cut‐off (ECOFF) value for E. coli is tentative 0.03 mg/L.
Antimicrobial susceptibility data from animals and food in 2021–2022
Data reported under Directive 2003/99/EC, Commission Implementing Decision (EU) 2020/1729
EU MSs reported mandatory data collected following AMR monitoring programmes during 2021 and 2022. ‘Directive 2003/99/EC requires Member States to ensure that monitoring provides comparable data on the occurrence of antimicrobial resistance (‘AMR’) in zoonotic agents and, in so far as they present a threat to public health, other agents’. ‘Directive 2003/99/EC also requires Member States to assess the trends and sources of AMR in their territory and to transmit a report every year covering data collected in accordance with that Directive to the Commission.’ Furthermore, some non‐EU countries reported AMR data and both some EU and non‐EU reporting countries (RCs) also reported voluntary data from samples that were not included in the mandatory programmes per reporting year.
The Commission Implementing Decision (EU) 2020/1729 of 17 November 2020 lays down the rules for antimicrobial resistance monitoring performed from 2021 onwards. This decision specifies harmonised rules for the period 2021–2027 for the monitoring and reporting of AMR to be carried out by Member States in accordance with EU Regulations. It also determines specific technical requirements for AMR testing and reporting in relation to sampling in food‐producing animals and derived meat (at retail and at border control posts).
The Commission Implementing Decision (EU) 2020/1729 indicates that the monitoring and reporting of AMR shall cover the following bacteria: (a) Salmonella spp.; (b) Campylobacter coli (C. coli); (c) Campylobacter jejuni (C. jejuni); (d) Indicator commensal Escherichia coli (E. coli); (e) Salmonella spp. and E. coli producing the following enzymes: (i) Extended Spectrum β‐Lactamases (ESBL); (ii) AmpC β‐Lactamases (AmpC); (iii) Carbapenemases (CPs). Therefore, during 2021 and 2022, AMR data were collected from the bacteria listed above.
Countries can also report AMR data from other agents of public health importance such as methicillin‐resistant Staphylococcus aureus (MRSA). According to Commission Implementing Decision (EU) 2020/1729, the monitoring and reporting of AMR may also cover indicator commensal Enterococcus faecalis (E. faecalis) and Enterococcus faecium (E. faecium).
A scientific report published by EFSA in 2019 included technical specifications on the harmonised monitoring and reporting of antimicrobial resistance in methicillin‐resistant Staphylococcus aureus (MRSA) in food‐producing animals and food (EFSA, 2019). Detailed rules were specified for harmonised monitoring and reporting on the prevalence of resistant microorganisms in food‐producing animals and food, in particular as regards the microorganisms to be included, the origin of the isolates, the number of isolates to be tested, the antimicrobial susceptibility tests to be used, the specific monitoring of MRSA and ESBL‐/AmpC‐/CP‐producing bacteria and the collection and reporting of the data. Comparison between human data and data from food‐producing animals and the food sector was ensured by the involvement of ECDC.
The Commission Implementing Decision (EU) 2020/1729 specifies that the monitoring and reporting of AMR shall cover the following food‐producing animal populations and food: (a) broilers; (b) laying hens; (c) fattening turkeys; (d) bovine animals under 1 year of age; (e) fattening pigs; (f) fresh meat from broilers; (g) fresh meat from turkeys; (h) fresh meat from pigs; (i) fresh meat from bovine animals. Fresh meat includes meat sampled at retail and imported meat from third countries sampled at border control posts (BCPs). This European Commission Decision indicates the sampling frequency for MSs to carry out the AMR monitoring and reporting in accordance with the following rotational system: (a) In the years 2021, 2023, 2025 and 2027: in fattening pigs, bovine animals under 1 year of age, pig meat and bovine meat. (b) In the years 2022, 2024 and 2026: in laying hens, broilers, fattening turkeys and fresh meat derived from broilers and turkeys.
Therefore, following relevant EU legislation, AMR data presented in this report were collected from poultry populations and derived meat thereof in 2022 and from fattening pigs and cattle under 1 year of age in 2021.
The Commission Implementing Decision (EU) 2020/1729 lays down detailed rules for sampling design and sample size as well as for antimicrobial susceptibility testing for the different bacteria. This European Commission Decision indicates the analytical methods for detection and antimicrobial susceptibility testing that shall be performed by the laboratories referred to in Article 3(2). AMR testing shall be performed by using the broth microdilution method according to the reference method ISO 20776‐1:2019.
For AMR testing, isolates were obtained through harmonised national programmes. The broth microdilution testing method was widely used for susceptibility testing following EU legislation.
On 17 November 2020, the European Commission laid down the new technical specifications in Commission Implementing Decision (EU) 2020/1729. The new legislation came into effect on 1 January 2021, and updates technical specifications for harmonised AMR monitoring and reporting to include the monitoring of AMR in derived meat sampled at border control posts, and the testing of new substances. The new legislation also authorises WGS as an alternative method to extended phenotypic testing of isolates with resistance to third‐generation cephalosporins and/or carbapenems. The new rules apply until December 2027. 16
The resulting quantitative isolate‐based data were reported to EFSA and considered for this report. Resistance was interpreted using EUCAST ECOFF values (see text box below for further information). The antimicrobials incorporated in this report were selected based on their public health relevance and as representatives of different antimicrobial classes. Data on methicillin resistant Staphylococcus aureus (MRSA) and other microorganisms apart from those required by legislation were reported on a voluntary basis.
Harmonised representative sampling and monitoring
Representative sampling and AMR monitoring should be performed following the current legislation and the technical specifications published by EFSA (EFSA, 2019). Regulation (EC) No 2073/2005 Article 4 indicates that: ‘Food business operators shall perform testing as appropriate against the microbiological criteria set out in Annex I, when they are validating or verifying the correct functioning of their procedures based on HACCP principles and good hygiene practice.’ 17
The Commission Implementing Decision (EU) 2020/1729 lays down rules for antimicrobial resistance monitoring performed from 2021 onwards. The Commission Implementing Decision (EU) 2020/ 1729 determines specific technical requirements for AMR testing and reporting in relation to sampling in food‐producing animals and derived meat (at retail and at border control posts).
Salmonella spp.
In 2022, MSs collected representative Salmonella spp. isolates for AMR monitoring from the populations of broilers, laying hens and fattening turkeys sampled following the Salmonella National Control Programmes (NCPs) set up in accordance with Article 5(1) of the Regulation (EC) No. 2169/2003. For the purposes of sampling design and representativeness, no more than one isolate per Salmonella serovar from the same epidemiological unit (herd/holding/flock of birds) per year should be included in the AMR monitoring programme. Moreover, samples of imported fresh meat from broilers and fattening turkeys were collected at the border control posts. In most MSs, the isolates tested for AMR formed a representative subsample of the total Salmonella isolates available at the National Reference Laboratory (NRL) and/or other laboratories involved. The sampling was performed in such a way as to ensure geographical representativeness and even distribution over the year. However, when sampling from low prevalence areas, all the Salmonella isolates available should be tested for susceptibility.
In 2021, MSs collected representative Salmonella spp. isolates for AMR monitoring from samples of caecal content taken at slaughter from cattle under 1 year of age where the production of meat of those bovines in the MSs was more than 10,000 tonnes per year sampled for testing and verification of compliance, in accordance with point 2.1.3 of chapter 2 of Annex I of Regulation (EC) No 2073/2005. Also in 2021, representative Salmonella isolates for AMR monitoring were obtained by MSs from samples of caecal content taken at slaughter from fattening pigs sampled for testing and verification of compliance, in accordance with point 2.1.4 of chapter 2 of Annex I of Regulation (EC) No 2073/2005. In compliance with EU legislation and EFSA guidelines, MSs sampled the caecal contents of fattening pigs and of cattle under 1 year of age at the slaughterhouse. MSs employed a two‐stage stratified sampling design (with slaughterhouses as primary sampling units and carcases as secondary units) based on proportional allocation of the number of samples to the annual throughput of the slaughterhouse.
Campylobacter spp.
C. coli and C. jejuni isolates were collected by MSs as part of their national monitoring programme of AMR according to the provisions of the Commission Implementing Decision (EU) 2020/1729.
In 2022, MSs collected at least 170 isolates of the nationally most prevalent species of Campylobacter (among C. coli and C. jejuni) from samples obtained from broilers and fattening turkeys following regulations and technical requirements for AMR testing, and up to 170 isolates of the least prevalent Campylobacter species (among C. coli and C. jejuni). By way of derogation, where Member States have a national annual production of less than 100,000 tonnes of broiler meat, they may decide to test a minimum of 85 isolates instead of 170 isolates of the nationally most prevalent species of Campylobacter (among C. coli and C. jejuni) and up to 85 isolates of the least prevalent species (among C. coli and C. jejuni). The sample collection was approximately evenly distributed over the year 2022. One representative caecal sample (single or pooled) per epidemiological unit (i.e. batch of birds sent to the slaughterhouse) was gathered to account for clustering. Isolates were recovered from caecal samples (single or pooled) in accordance with EFSA's recommendations (EFSA, 2019, 2020).
In 2021, MSs collected at least 170 isolates of the nationally most prevalent species of Campylobacter (among C. coli and C. jejuni) from samples obtained from cattle under 1 year of age and up to 170 isolates of the nationally least prevalent Campylobacter species following regulations and technical requirements for AMR testing. Moreover, MSs collected at least 170 isolates of C. coli from samples obtained from fattening pigs and up to 170 isolates from C. jejuni following regulations and technical requirements for AMR testing. By way of derogation, where Member States have a national annual production of less than 100,000 tonnes of pig meat, they may decide to test a minimum of 85 isolates instead of 170 isolates. The sample collection was approximately evenly distributed over the year 2021. One representative caecal sample (single or pooled) per epidemiological unit (i.e. batch of animals sent to the slaughterhouse) was gathered to account for clustering. Isolates were recovered from caecal samples (single or pooled) in accordance with EFSA's recommendations (EFSA, 2019, 2020).
Indicator commensal E. coli
Routine monitoring of indicator commensal E. coli
Indicator commensal E. coli isolates were collected by MSs as part of their national monitoring programme of AMR according to the provisions of the Commission Implementing Decision (EU) 2020/1729. In 2021, MSs collected indicator E. coli isolates based on random sampling of caecal samples gathered at slaughter from fattening pigs and calves under 1 year of age where the production of cattle meat in the MSs is more than 10,000 tonnes slaughtered per year as specified in Annex Part A paragraph 1(c) (iv). In 2022, MSs collected indicator E. coli isolates based on random sampling of caecal samples gathered at slaughter from broilers and fattening turkeys where the production of turkey meat in the MSs is more than 10,000 tonnes slaughtered per year as specified in Annex Part A paragraph 1(c) (ii). One representative caecal sample (single or pooled) per epidemiological unit (herds) was gathered to account for clustering. Isolates were recovered from caecal samples (single or pooled), in accordance with regulations and EFSA's recommendations (EFSA, 2019, 2020).
As per Regulations, from 2021 MSs shall test at least 170 isolates obtained from samples referred to in points 1(c)(i‐iv). By way of derogation, where MSs have a national annual production of less than 100,000 tonnes of pig meat, less than 100,000 tonnes of broiler meat or less than 100,000 tonnes of turkey meat, they may test a minimum of 85 isolates instead of 170 isolates for each specific animal population (depending on the mandatory testing every year).
A two‐stage stratified sampling design was applied in the reporting countries, with slaughterhouses as primary sampling units and carcasses as secondary units, accounting for proportional allocation of the number of samples to the annual throughput of the slaughterhouse. Only one representative caecal sample (single or pooled) per epidemiological unit (batch of carcasses deriving from the same flock) was gathered to account for clustering. Isolates were recovered from caecal contents samples (single or pooled), in accordance with EFSA's recommendations (EFSA, 2019). The sample collection was approximately evenly distributed over the respective years.
Specific monitoring of ESBL/AmpC/CP‐producing Escherichia coli
In 2022, MSs obtained caecal samples from broilers and from fattening turkeys at slaughter, in those MSs where the production of turkey meat was more than 10,000 tonnes slaughtered per year. Moreover, samples of fresh meat from broilers and fattening turkeys were collected at retail and at the border control posts. In 2021, MSs collected caecal samples from fattening pigs and cattle under 1 year of age at slaughter, where the production of cattle meat was more than 10,000 tonnes slaughtered per year and samples of fresh pig meat and cattle meat gathered at retail and at border control posts. Only one representative caecal sample (single or pooled) per epidemiological unit (batch of carcases deriving from the same herd/flock) was collected to account for clustering. Isolates were recovered from caecal contents samples (single or pooled), in accordance with relevant Regulations and EFSA's recommendations (EFSA, 2014, 2019, 2020). The sample collection as described above was approximately evenly distributed over the years 2021 and 2022. The same sampling design was used to collect indicator E. coli isolates, whether dedicated to the routine monitoring of AMR or the specific monitoring of ESBL‐/AmpC‐/CP‐producing E. coli. EUSR on AMR in zoonotic and indicator bacteria from humans, animals and food in 2021–2022.
Epidemiological cut‐off values (ECOFFs) and clinical breakpoints (CBPs)
A microorganism is defined as ‘clinically’ resistant when the degree of resistance shown is associated with a high likelihood of therapeutic failure. The microorganism is categorised as resistant by applying the appropriate CBP in a defined phenotypic test system, and this breakpoint may alter with legitimate changes in circumstances (e.g. alterations in dosing regimen, drug formulation, patient factors). A microorganism is defined as wild type for a bacterial species when no acquired or mutational resistance mechanisms are present to the antimicrobial in question. A microorganism is categorised as wild type for a given bacterial species when presenting a lower MIC to the antimicrobial in question than the appropriate ECOFF in a defined phenotypic test system. This cut‐off value will not be altered by changing circumstances (such as alterations in frequency of antimicrobial administration). Wild‐type microorganisms may or may not respond clinically to antimicrobial treatment. A microorganism is defined as non‐wild type for a given bacterial species by the presence of an acquired or mutational resistance mechanism to the antimicrobial in question. A microorganism is categorised as non‐wild type for a given bacterial species by applying the appropriate ECOFF value in a defined phenotypic test system; non‐wild type organisms are considered to show ‘microbiological’ resistance (as opposed to ‘clinical’ resistance). CBPs and ECOFFs may be the same, although it is often the case that the ECOFF is lower than the CBP. EUCAST has defined CBPs and ECOFFs.
Clinical breakpoints (clinical resistance)
The clinician, or veterinarian, choosing an antimicrobial agent to treat humans or animals with a bacterial infection requires information that the antimicrobial selected is effective against the bacterial pathogen. Such information will be used, together with clinical details such as the site of infection, ability of the antimicrobial to reach the site of infection, formulations available and dosage regimes, when determining an appropriate therapeutic course of action. In 2019, EUCAST updated their definitions of susceptibility testing categories susceptible (S), intermediate (I) and resistant (R) (EUCAST, 2019). The updated definitions are as follows:
S – Susceptible, standard dosage regimen: When there is a likelihood of therapeutic success using a standard dosing regimen of the agent.
I – Susceptible, increased exposure: When there is a high likelihood of therapeutic success because exposure to the agent is increased adjusting the dosing regimen or by its concentration at the site of infection.
R – Resistant: When there is a high likelihood of therapeutic failure even when there is increased exposure.
The in vitro susceptibility of the bacterial pathogen can be determined and CBPs used to ascertain whether the organism is likely to respond to treatment. CBPs will take into account the distribution of the drug in the tissues of the body following administration and assume that a clinical response will be obtained if the drug is given as recommended and there are no other adverse factors which affect the outcome. Different dosing regimens can lead to the development of different CBPs, as occurs in some countries for certain antimicrobials where different therapeutic regimes are in place. Although the rationale for the selection of different CBPs may be clear, their use makes the interpretation of results from different countries problematic, as the results are not directly comparable between those different countries.
Epidemiological cut‐off values (microbiological resistance)
For a given bacterial species, the pattern of the MIC distribution (i.e. the frequency of occurrence of each given MIC plotted against the MIC value) can enable the separation of the wild‐type population of microorganisms from those populations that show a degree of acquired resistance. The wild‐type susceptible population is assumed to have no acquired or mutational resistance and commonly shows a normal distribution. When bacteria acquire resistance by a clearly defined and efficient mechanism, such as a plasmid bearing a gene which produces an enzyme destroying the antimicrobial, the MIC distribution commonly shows two major subpopulations. One is a fully susceptible normal distribution of isolates and the other a resistant population which has acquired the resistance mechanism. Resistance may be achieved by a series of small steps, such as changes in the permeability of the bacterial cell wall to the antimicrobial or other mechanisms which confer a degree of resistance. In this case, there may be populations of organisms which occur lying between the fully susceptible population and more resistant populations. The ECOFF value indicates the MIC or zone diameter above which the pathogen has some detectable reduction in susceptibility. ECOFFs are derived by testing an adequate number of isolates to ensure that the wild‐type population can be confidently identified for a given antimicrobial. The clinical breakpoint, which is set to determine the therapeutic effectiveness of the antimicrobial, may fail to detect emergent resistance. Conversely, the ECOFF detects any deviation in susceptibility from the wild‐type population, although it may not be appropriate for determining the likelihood of success or failure for clinical treatment.
MRSA
MRSA isolates may have been collected by reporting countries using different monitoring approaches, either by active surveillance and monitoring of animals and foods or, in some cases, by passive monitoring (e.g. based on diagnostic submission of samples from clinical cases of disease in animals, or from foods sampled as part of investigatory work). Furthermore, countries may apply different sampling strategies and collect different types of samples from different animal populations and food matrices. Isolation methods also differ between countries.
Harmonised antimicrobial susceptibility testing
Routine monitoring antimicrobial susceptibility
MSs followed Commission Implementing Decision (EU) 2020/1729 and recommendations from EFSA regarding the use of epidemiologic cut‐off values for AMR monitoring. MSs tested antimicrobials and interpreted the results using the ECOFFs and concentration ranges shown in Tables F.5, F.6 to determine the susceptibility of the following microorganisms: Salmonella spp., C. coli, C. jejuni and indicator commensal E. coli. Under the new legislation (Commission Implementing (EU) 2020/1729), changes were made to the ECOFFs and clinical breakpoints for several antimicrobial substances included in the harmonised panel for testing of Salmonella spp. and E. coli. The substances with changes to ECOFFs and/or clinical breakpoints included tigecycline, nalidixic acid and ciprofloxacin (Table F.5). For 2021 and 2022 data from pigs, calves, poultry and meat from BCPs, the occurrence of resistance to tigecycline, nalidixic acid and ciprofloxacin is determined using the new ECOFFs and clinical breakpoints. Also, in 2021, a new substance, amikacin, was added to the harmonised panel for both Salmonella spp. and E. coli. For Campylobacter spp., no changes were made to ECOFFs and clinical breakpoints for the substances included in the harmonised panel. However, two new substances were added (chloramphenicol and ertapenem) and two substances were removed (nalidixic acid and streptomycin).
Presumptive ESBL‐/AmpC‐/CP‐producing E. coli identified through the specific monitoring, as well as presumptive ESBL‐/AmpC‐/CP‐producing Salmonella spp. and E. coli from the routine monitoring should be further tested with a second panel of antimicrobial substances (Table F.7) or investigated using WGS. The second panel includes cefoxitin, cefepime and clavulanic acid in combination with cefotaxime and ceftazidime for the detection of presumptive ESBL‐ and AmpC‐producing isolates. Moreover, the second panel contains imipenem, meropenem and ertapenem to phenotypically verify presumptive CP‐producers.
Specific monitoring of ESBL‐/AmpC‐/CP‐producing E. coli
To isolate presumptive ESBL‐/AmpC‐/CP‐producing E. coli in the specific monitoring, samples were first subjected to a non‐selective pre‐enrichment step followed by inoculation on selective MacConkey agar. The selective agar contains 1 mg/L cefotaxime, in accordance with the detailed protocol for standardisation of the EU Reference Laboratory for Antimicrobial Resistance (EURL‐AR). 18 Following this protocol, presumptive CP‐producing isolates can also be recovered. If available, one presumptive ESBL‐/AmpC‐/CP‐producing E. coli isolate obtained from each positive caecal sample and meat sample was tested for its antimicrobial susceptibility to the first panel of antimicrobials (Table F.5). This step was performed to confirm the microbiological resistance to cefotaxime (expected as the antimicrobial is present in the isolation medium at a concentration higher than the ECOFF), as well as to identify possible resistance to cefotaxime and/or ceftazidime and/or meropenem. In a second step, the isolate should be tested using the second panel of antimicrobials (Table F.6) to infer the presumptive ESBL‐/AmpC‐/CP‐producing phenotypes according to the beta‐lactams resistance phenotypes obtained (Figure F.1). WGS can be used as a replacement for the phenotypic testing of presumptive ESBL‐/AmpC‐/CP‐producing E. coli from the specific monitoring.
Specific monitoring of CP‐producing microorganisms
From 2021, the specific monitoring of CP‐producing microorganisms was made mandatory. For the specific monitoring of CP‐producing microorganisms, isolation required the use of non‐selective pre‐enrichment and subsequent selective plating on carbapenem‐containing media, in accordance with the most recent version of the detailed protocol of the EURL‐AR. The microbial species was identified using appropriate methods. If available, one presumptive CP‐producing isolate (primarily E. coli, but also Salmonella) obtained from each positive caecal sample and meat sample should be tested for its antimicrobial susceptibility to the first panel of antimicrobials (Table F.5: Panel of antimicrobial substances included in AMR monitoring, thresholds for interpreting resistance and concentration ranges tested in Salmonella spp. and indicator commensal E. coli (first panel) based on Commission Implementing Decision (EU) 2020/1729 and EFSA Technical Report 2021 Table F.5) to confirm the microbiological resistance to meropenem and to identify possible resistance to other antimicrobials such as cefotaxime and/or ceftazidime. In a second step, the isolate should be tested against the second panel of antimicrobials (Table F.7) to infer the presumptive CP‐producer phenotype according to the beta‐lactam resistance phenotypes obtained (Figure F.1). The EUCAST epidemiological cut‐off values applied for the antimicrobial susceptibility testing (Tables F.5, F.6, F.7) are based on Commission Implementing Decision (EU) 2020/1729.
Data validation
Validation against business rules
The reported data were first checked for usability against a series of ‘business rules’, which were automatically applied in the EFSA data collection system once a file was sent. This automatic data validation process refers to the first validation of incoming data. Quality checks are related to a specific business only. The positive result of the automatic validation process places the file in a valid state and makes it available for further steps of validation performed by EFSA.
Scientific data validation
The scientific validation of the data collected by the MSs/non‐MSs and submitted to EFSA consisted of the data reviewing and making comparisons between data reported for the same antimicrobials when tested by different panels. Special attention was given to carbapenems, colistin, azithromycin, tigecycline and to possible discrepancies between results for antimicrobials present in both panels (i.e. cefotaxime, ceftazidime, meropenem). MSs were contacted by EFSA asking for clarifications. If necessary, MSs were asked to confirm the MIC results and the species identification of the reported isolates.
Reference testing
To ensure the quality of the data submitted, a reference testing exercise was run by the EURL‐AR in close collaboration with the MSs. The exercise consisted in re‐doing the AST of the isolates received using both Panel 1 and Panel 2 of antimicrobials, as well as WGS analyses of the isolates. Based on the data submitted to EFSA, a selection of 362 isolates was made. The selection of these isolates was based on different criteria:
Isolates not showing resistance to any of the tested substances.
Escherichia coli isolates showing resistance to colistin (MIC >2 mg/L).
Salmonella spp. and E. coli isolates showing high resistance to amikacin (MIC >128 mg/L).
Salmonella spp. and E. coli isolates showing resistance to both gentamicin (MIC > 16 mg/L) and amikacin (MIC > 128 mg/L).
Salmonella spp. and E. coli isolates showing resistance to tigecycline (MIC > 0.5) but susceptibility to tetracycline (MIC < 8).
Campylobacter coli and C. jejuni isolates showing the highest level resistance to erythromycin (MIC > 512 mg/L).
Campylobacter coli and C. jejuni isolates showing resistance to both gentamicin (MIC > 16 mg/L) and erythromycin (MIC > 512 mg/L).
Isolates showing multidrug resistance to the highest number of substances (≥ 6 substances for Salmonella spp. and E. coli, ≥ 6 substances for Campylobacter coli and C. jejuni).
Isolates representing the categorisations presumptive ESBL‐, AmpC‐ and ESBL + AmpC producers or with such genes reported.
Isolates representing the categorisations presumptive CP‐producers or with such genes reported.
Isolates where phenotypic and genotypic data are not in accordance with each other.
MSs sent the selected isolates to the EURL‐AR, where they were retested. EFSA, EURL‐AR and MSs liaised together to address possible discrepancies found.
Analyses of antimicrobial resistance data
Data are reported in separate sections dedicated to each microorganism. Clinical investigation data were not accounted for in this report.
Overview tables of the resistance data reported
Data generated from the antimicrobial susceptibility testing and reported as quantitative at the isolate level by MSs have been described in the overview tables included in the Annexes A–E published on the EFSA Knowledge Junction community on Zenodo (https://doi.org/10.5281/zenodo.10528846). The tables also display complete susceptibility, multidrug resistance and co‐resistance. These analyses are described in Section 2.4.
Epidemiological cut‐off values and the occurrence of resistance
ECOFFs, as listed in Commission Implementing Decision (EU) 2020/1729, have been used in this report to interpret the isolate based reported MIC data and determine non‐wild‐type organisms also termed ‘microbiologically’ resistant organisms (i.e. displaying a decreased susceptibility), and to ensure that results from different MSs are comparable. In this report, ‘microbiologically’ resistant organisms are referred to as ‘resistant’ for brevity. This report also incorporates re‐evaluation of the historical data accounting for the revised EU legislation, which included the revised ECOFFs. Under the new legislation, the ECOFF used to determine microbiological resistance of Salmonella isolates to tigecycline changed from > 1 to > 0.5 mg/L. To note for tigecycline testing is the instability of the substance in aged (> 12 h old) Mueller‐Hinton broth medium used in MIC‐testing, which may result in elevated MIC for some isolates (Bradford et al., 2005). The reported instability of tigecycline during testing, coupled with the lower ECOFF, may result in elevated reporting of resistance to tigecycline in Salmonella and E. coli isolates with MICs within one dilution range of the ECOFF.
Starting in 2021, new legislative requirements listed in Commission Implementing Decision (EU) 2020/1729, require MSs to test target bacterial isolates for new substances. For Salmonella spp., and E. coli, the new substance to be tested is amikacin. For Campylobacter spp., the new substances are chloramphenicol and ertapenem. Nalidixic acid and streptomycin were removed from the harmonised panel for Campylobacter spp.
The occurrence of resistance to a number of antimicrobials was determined for Salmonella, Campylobacter and indicator commensal E. coli isolates and are tabulated at the production‐type level in this report. The occurrence of resistance (i.e. resistance levels) in reporting MS groups was calculated as totals (the total number of resistant isolates out of the total number of tested isolates across reporting MSs) and in the E. coli chapter, also as weighted means to account for the animal population sizes.
Data description
Throughout the report, level or occurrence of AMR means the percentage of resistant isolates as a proportion of the isolates tested of that microorganism. MSs reporting group means the MSs that provided data and were included in the relevant table of antimicrobial resistance for that bacterium–food or animal category–antimicrobial combination. Terms used to describe the levels or occurrence of antimicrobial resistance are ‘rare’: < 0.1%, ‘very low’: 0.1%–1.0%, ‘low’: > 1%–10.0%, ‘moderate’: > 10.0%–20.0%, ‘high’: > 20.0%–50.0%, ‘very high’: > 50.0%–70.0%, ‘extremely high’: > 70.0%. Although these terms are applied to all antimicrobials, the significance of a given level of resistance depends on the particular antimicrobial and its importance in human and veterinary medicine.
Temporal trends in resistance
Temporal trends show the resistance to different antimicrobials over time, by plotting the level of resistance for each year of sampling and for diverse species such as humans and different food‐producing animals. Trend graphs were generated for data meeting the minimum criteria for inclusion, e.g. 10 or more isolates reported and with three or more time points since 2014. Additional criteria, where applicable, are indicated in the specific chapters. When historical data were available, the trends could also cover these; otherwise, the time period could be different. To assess the statistical significance of temporal trends, the proportions of resistance were modelled using logistic regression. Logistic regression models used resistance as the outcome variable (with resistant (1)/non‐resistant (0)) and year as a covariate. This analysis was carried out using the PROC LOGISTIC of SAS 9.2. for each country reporting at least 10 total tested isolates, where there were 3 years or more of available data to use in the model. The PROC LOGISTIC function uses a logit transformation to model the proportions against year and provides estimates for both intercepts and slope. Models where the likelihood ratio test suggested it to be meaningful and resulting in a p‐value associated with a slope of < 0.05 were significant (linear model fit). It is important to note that between‐year fluctuations in the occurrence resistance (%) may not be captured in the evaluation of the linear trend over the entire time period (2009–2021) and that very recent decreasing or increasing trends may therefore be masked by the overall trend. Also, when interpreting the results, it is important to note that trend analyses may be driven by particularly high or low levels of resistance reported in one or few data points leading to unexpected findings (e.g. detection of significant increasing or decreasing trends where the observed data do not show any clear trend over the entire period). The withdrawal of the UK from the EU has an impact on the AMR data reported at the EU level in 2020. In this report, data at the EU level are reported in accordance with the membership of the EU, whether before 2020 (EU including the UK) or after 2020 (EU without the UK). However, Northern Ireland is counted as an EU MS in this report. As monitoring of MRSA is not mandatory and harmonised, the availability of comparable data over time is limited. Thus, temporal trends were not assessed for MRSA.
Spatial analysis of resistance through maps
MS‐specific AMR levels for selected bacterium–food category/animal population combinations were plotted in blue, purple or green shaded maps for 2021 and 2022, using ArcGIS 9.3. In the maps, resistance levels are presented with colours reflecting the continuous scale of resistance to the antimicrobial of interest among reporting MSs; so there might be some apparent discrepancies between the colours and resistance levels between maps.
Resistance in Salmonella serovars of public health importance
In this report, AMR in tested Salmonella isolates were aggregated to give a value for Salmonella spp. for each country and animal/meat category. In addition, the most prevalent Salmonella serovars were also reported separately for each animal category. Additional tables were included in this report to describe the occurrence of AMR among selected Salmonella serovars of public health importance or of high prevalence in animals. To present a complete overview of the animal populations categories in which specific Salmonella serovars of public health importance have been recovered, all the data reported (derived even from fewer than four reporting countries and less than 10 isolates tested) have been included.
Prevance of resistance to selected antimicrobials in C. coli and C. jejuni
The prevalence of resistance to selected antimicrobials in C. jejuni and C. coli from broilers and fattening turkeys has been estimated at country level as the product of the prevalence of C. jejuni or C. coli in caecal samples from the given animal species and the percent occurrence of resistance in the corresponding isolates. The prevalence of resistance is an indicator that addresses together both the prevalence of C. jejuni and C. coli and the occurrence of resistance in both species recovered from the different food‐producing animals. This indicator is primarily intended to follow up trends over time at the country level.
Analysis of multidrug resistance, complete susceptibility and co‐resistance data
The analysis of MDR and co‐resistance data is important considering the emergence of multiresistant bacteria. The intention was to focus mainly on multi/co‐resistance patterns involving critically important antimicrobials (WHO, 2019), such as cephalosporins, fluoroquinolones and macrolides, and to summarise important information in the EU Summary Report. The occurrence of the isolates of a serotype/resistance pattern of interest was studied both at the MS level and at the EU level (by grouping data for all MSs and where also relevant for MSs and other reporting countries), as the overall picture for all MSs might show a more definite pattern of emergence and spread.
Analysis of MDR and complete susceptibility
For the analysis of MDR and complete susceptibility, a multiresistant isolate is one defined as resistant to at least three of the antimicrobial substances. In contrast, a completely susceptible isolate is one defined as non‐resistant (MIC < ECOFF) to these antimicrobial substances. For indicator commensal E. coli and Salmonella spp., the following substances from the harmonised test panel laid out in Commission Implementing Decision (EU) 2020/1729 were included in the assessment of MDR, as done in the previous EUSR on AMR including 2020–2021 data:
For E. coli, the substances included were amikacin/gentamicin (assessed together as aminoglycoside antimicrobial class), ampicillin, cefotaxime/ceftazidime (assessed together as third‐generation cephalosporin), chloramphenicol, ciprofloxacin/nalidixic acid (assessed together as quinolone antimicrobial class), colistin, meropenem, sulfamethoxazole, tetracycline/tigecycline (assessed together as glycylcycline antimicrobial class) and trimethoprim.
For Salmonella spp., the substances included were amikacin/gentamicin (assessed together as aminoglycoside antimicrobial class), ampicillin, cefotaxime/ceftazidime (assessed together as third‐generation cephalosporin), chloramphenicol, ciprofloxacin/nalidixic acid (assessed together as quinolone antimicrobial class), meropenem, sulfamethoxazole, tetracycline/tigecycline (assessed together as glycylcycline antimicrobial class) and trimethoprim.
For C. coli and C. jejuni, the substances included were ciprofloxacin, erythromycin (macrolide antimicrobial class), gentamicin and tetracycline.
Key Outcome Indicators
To support EU countries in their progress to reduce use of antimicrobials and AMR a list of key outcome indicators has been jointly published by ECDC, EFSA and EMA (ECDC, EFSA, and EMA, 2017). Two of these key outcome indicators (KOI) are included in the report: (1) The key outcome indicator of complete susceptibility (KOICS) in indicator commensal E. coli; and (2) the key outcome indicator of the prevalence of ESBL‐ and/or AmpC‐producing E. coli (KOIESC). KOICS is the proportion of fully susceptible indicator E. coli isolates, weighted by the size of the populations of the most important production animals (broilers, fattening turkeys, fattening pigs, cattle under 1 year of age) and is used as an indicator (KOICS) for the overall AMR situation in food‐producing animals. KOIESC is the weighted mean of the prevalence of ESBL‐ and/or AmpC‐producing E. coli in each of the four animal populations monitored. The KOICS and KOIESC account for differences in the relative size of food animal populations in a country and are therefore relevant in evaluation of risks related to resistance in food animals. These KOIs are displayed in bar charts showing changes in KOI over the years. The statistical significance of the trends was analysed using a simple linear regression over time. The F‐test was used to assess the overall significance of the models (p‐value < 0.05).
Combined resistance patterns of interest
The term combined resistance is used in this report to indicate phenotypic resistance to two or more different classes of antimicrobials, exhibited by the same bacterial isolate. In Salmonella and E. coli isolates, combined resistance to cefotaxime (CTX) and ciprofloxacin (CIP) was estimated, as these two antimicrobials are of particular interest in human medicine. In 2021, co‐resistance was addressed using both ECOFFs (CTX > 0.25 mg/L and CIP > 0.064 mg/L) and CBPs (CTX > 2 mg/L and CIP > 0.064 mg/L) for E. coli. In C. jejuni and C. coli isolates, co‐resistance to ciprofloxacin and erythromycin (ERY) was estimated, as these two antimicrobials are of particular interest in human medicine in the treatment of severe campylobacteriosis. The interpretive ECOFFs used to address co‐resistance to ciprofloxacin and erythromycin were, for C. jejuni, CIP > 0.5 mg/L and ERY > 4 mg/L and, for C. coli, CIP > 0.5 mg/L and ERY > 8 mg/L. These values may be considered as very similar to CBPs.
Identification of presumptive ESBL‐, AmpC‐ and/or CP‐producers
Definition of ESBL, AmpC, ESBL + AmpC, CP phenotypes
The categorisation of isolates resistant to third‐generation cephalosporins and/or carbapenems in presumptive ESBL‐, AmpC‐ or CP‐producers was carried out based on the EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance (EUCAST, 2017). In these expert guidelines and, based on other EUCAST and CLSI guidelines to detect ESBL/AmpC producers, a screening breakpoint of > 1 mg/L is recommended for cefotaxime and ceftazidime (EUCAST, 2017). This screening breakpoint is higher than the ECOFFs applied for antimicrobial susceptibility of both antimicrobials for E. coli, and to cefotaxime for Salmonella. For this report, a first condition for classifying isolates as presumptive ESBL/AmpC producers related to their MIC for either cefotaxime or ceftazidime was to apply this screening breakpoint of MICs > 1 mg/L. Only isolates which presented MIC values accomplishing with this requisite (as expected for most of the ESBL/AmpC producers) were further considered. In total, for the third‐generation cephalosporin‐ and/or carbapenem‐resistant isolates, five main categorisations are made: (1) ESBL phenotype; (2) AmpC phenotype; (3) ESBL + AmpC phenotype; (4) CP‐phenotype; and (5) other phenotypes (Figure F.1).
To detect the production of ESBLs, a synergy test for cefotaxime and ceftazidime, in combination with clavulanic acid, was performed. An eightfold reduction in the MIC for the cephalosporin combined with clavulanic acid compared with that obtained for the cephalosporin alone was interpreted as a positive synergy test. In all other cases, the synergy test was considered negative. For the present report, isolates with MICs > 1 mg/L for cefotaxime and/or ceftazidime and a synergy test positive for any of these antimicrobials, together with susceptibility to cefoxitin (≤ 8 mg/L) and meropenem (MEM ≤ 0.125 mg/L, see CP phenotype) were classified as ESBL phenotype (Figure F.1).
For the AmpC phenotype, the combination MIC > 8 mg/L (ECOFF) for cefoxitin together with MICs > 1 mg/L for cefotaxime and/or ceftazidime was used as phenotypic criteria to investigate the presence of AmpC production in E. coli. It should be also underlined that there are a few AmpC enzymes that do not confer resistance to cefoxitin (i.e. ACC‐1), and that there are other mechanisms (porin loss, presence of carbapenemases, a few ESBLs like cefotaxime (CTX‐M‐5)) that could generate similar MIC values for the different antimicrobials (EFSA, 2012; EUCAST, 2017). Phenotypic AmpC confirmation tests (i.e. cloxacillin synergy) were not required for the present monitoring. For the present report, isolates with MICs > 1 mg/L for cefotaxime and/or ceftazidime and cefoxitin MIC > 8 mg/L, together with negative synergy test for both cefotaxime and ceftazidime/clavulanic acid, together with susceptibility to meropenem (MEM ≤ 0.125 mg/L) were classified in the AmpC phenotype category. No distinction between acquired AmpC and chromosomal AmpC was made (Figure F.1).
For the present report, isolates with MICs > 1 mg/L for cefotaxime and/or ceftazidime, positive synergy tests for any of these antimicrobials with clavulanic acid and cefoxitin MIC > 8 mg/L, together with susceptibility to meropenem (MEM ≤ 0.125 mg/L) were classified under the ESBL + AmpC phenotype category (Figure F.1). In some isolates, several mechanisms can be present at the same time, making it very difficult to differentiate the phenotypes. Also the high‐level expression of AmpC beta‐lactamases can mask the presence of ESBLs. AmpC can also be present in isolates with positive ESBL tests (clavulanic acid synergy). In this case, the cefepime/clavulanic acid synergy test should be used to overturn/confirm the presence of ESBLs in these isolates (EUCAST, 2017) but, unfortunately, the combination of cefepime/clavulanic acid was not included among the substances tested for monitoring. The inclusion of resistance to cefepime with an MIC value ≥ 4 mg/L as an additional criterion proposed elsewhere (EFSA, 2014), could be useful to as certain the presence of an ESBL‐producer.
For the classification of isolates into the putative carbapenem producers (CPs), a meropenem screening cut‐off of > 0.125 mg/L (which coincides with the harmonised ECOFF) was chosen (Figure F.1). It is known that other mechanisms (i.e. hyperproduction or combination of ESBLs and/or AmpC and porin loss) can also affect to the MIC values generated for the different carbapenems, especially for ertapenem. The confirmation of the carbapenemase production recommended by the EUCAST guidelines cannot be inferred from the carbapenem susceptibility testing data reported but needs further phenotypic or molecular testing. MSs that reported data suggesting the presence of putative CPs were recommended to validate the results by performing further confirmatory testing, and the EURL‐AR offered to apply WGS of the isolates. For the present report, isolates with MIC > 0.125 mg/L for meropenem would be considered as presumptive CP producers and were classified under the CP phenotype. The presence of other resistance mechanisms (ESBLs, AmpC, etc.) within the isolates placed in this group cannot be ruled out.
In this group, phenotypes not included in the categorisations defined above were included: isolates with an MIC > 0.125 for ertapenem and/or MIC > 1 mg/L for imipenem (EUCAST screening cut‐offs, one dilution step higher than the currently defined ECOFFs) but no resistance to meropenem (MIC < 0.125 mg/L) were classified under the category ‘other phenotype’. Finally, isolates with MICs ≤ 1 mg/L for cefotaxime and ceftazidime would be considered as not ESBL and/or AmpC producers. This implied that some isolates considered as microbiologically resistant (MICs over the ECOFFs) would not be further classified, as probably other mechanisms or technical issues in the MIC testing (i.e. MIC value close to the ECOFF) would be responsible for the MIC values obtained. For the present report, cefotaxime‐ and ceftazidime‐resistant isolates with MICs ≤ 1 mg/L for both antimicrobials were considered as putative on‐ESBL/AmpC producers and were classified under the category ‘other phenotype’ (Figure F.1).
We are aware that without a further molecular characterisation of the isolates, it will not be possible to know exactly which resistance mechanisms are present. For epidemiological purposes and based on the EUCAST guidelines, the classification of ‘presumptive’ producers for the different mechanisms conferring resistance to third‐generation cephalosporins and/or carbapenems was considered. Molecular characterisation of these mechanisms is recommended.
FIGURE F.1.
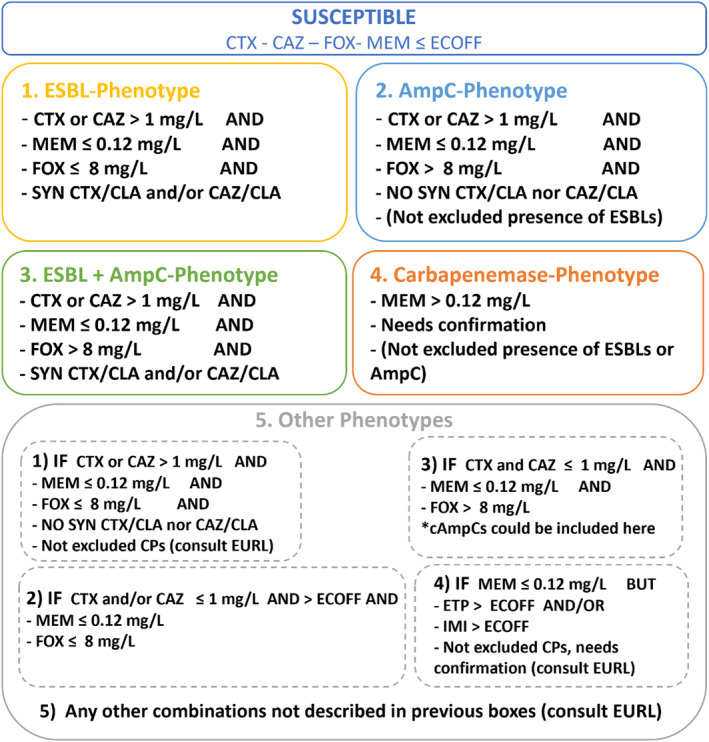
Phenotypes inferred based on the resistance to the beta‐lactams included in Panel 2.
For the occurrence and prevalence tables, as well as the violet shaded maps and graphics shown in Section ‘Extended‐spectrum β‐lactamase (ESBL)‐, AmpC‐ and/or CP‐producing Salmonella and Escherichia coli’, presumptive ESBL producers were considered as those exhibiting an ESBL and/or ESBL + AmpC phenotype and presumptive AmpC‐producers, those with an AmpC and/or ESBL + AmpC phenotype (see below).
For the present report, the terms:
‘Presumptive ESBL‐/AmpC‐producers’ refers to those isolates who present an ESBL and/or and AmpC and/or an ESBL + AmpC phenotype (presumptive ESBL producers and/or presumptive AmpC‐producers).
‘Presumptive ESBL‐producers’ refers to those isolates with MICs > 1 mg/L for cefotaxime and/or ceftazidime and a synergy test positive for any of these antimicrobials and susceptibility to meropenem (MEM ≤ 0.125 mg/L, see CP phenotype). These isolates may also harbour other resistance mechanisms (e.g. AmpC‐encoding genes).
‘Presumptive ESBL‐cefotaximase‐producers’ refers to those presumptive ESBL‐producers with MICs > 1 mg/L for cefotaxime and a synergy test positive for cefotaxime only. These isolates may also harbour other resistance mechanisms.
‘Presumptive ESBL‐ceftazidimase‐producers’ refers to those presumptive ESBL‐producers with MICs > 1 mg/L for ceftazidime and synergy test positive for ceftazidime only. These isolates may also harbour other resistance mechanisms.
‘Presumptive AmpC‐producers’ refers to isolates with MICs > 1 mg/L for cefotaxime and/or ceftazidime and cefoxitin MIC > 8 mg/L together with susceptibility to meropenem (MEM ≤ 0.125 mg/L, see CP phenotype). No distinction between plasmid‐mediated AmpC and chromosomal AmpC was made. These isolates may also harbour other resistance mechanisms (e.g. ESBL‐encoding genes).
‘Presumptive ESBL + AmpC‐producers’ refers to isolates with the ESBL + AmpC phenotype described above.
‘Presumptive CP‐producers’ refers to those isolates with the CP phenotype described above.
Data on ESBL/AmpC/CP‐genes
From 2021, MSs and non‐MSs can choose to report WGS data for characterisation of presumptive ESBL‐/AmpC‐/CP‐producing E. coli and Salmonella spp. isolates from the routine monitoring (if resistance to cefotaxime, ceftazidime and/or meropenem was detected in the first panel) or from the specific ESBL‐/AmpC‐/CP‐producing E. coli monitoring. Definitions for genotypic interpretation of AMR data for 2021 and 2022 are listed below:
Positive isolate is an isolate where at least one ESBL‐ or AmpC‐ or CP‐gene was detected using WGS.
Negative isolate is an isolate where no ESBL‐ or AmpC‐ or CP‐genes are detected using WGS.
It is important to highlight that genotypic complete susceptibility is not the same as phenotypic complete susceptibility because not all genes that are detected are phenotypically expressed.
For the analysis with genotypic data, the following definitions are applied:
genotypic prevalence will be defined using the following formula:
genotypic occurrence will be defined as the proportion (%) of ESBL‐/AmpC‐/CP‐producing E. coli or Salmonella positive isolates (associated with at least one ESBL‐/AmpC‐/CP‐gene) divided by the total number of ESBL‐ or AmpC‐ or CP‐ isolates tested.
Bar charts were also used to present the WGS data.
The list of ESBL‐/AmpC‐/CP‐encoding genes used for the analysis of the ESBL‐/AmpC‐/CP‐producing isolates can be consulted in the catalogue browser https://github.com/openefsa/catalogue‐browser/wiki.
Data on methicillin‐resistant Staphylococcus aureus (MRSA)
The occurrence of MRSA and its susceptibility to antimicrobials in various food categories (including meat samples from various species) and food‐producing animals was reported by few MSs. MRSA occurrence data reported from clinical investigations of food‐producing and companion animals in 2021–2022 were also reported. Details of the antimicrobials selected are provided in the section on MRSA. For further information on reported MIC distributions and the number of resistant isolates, refer to the submitted and validated MS data published on the EFSA website. The methods for collecting and testing samples for MRSA are not harmonised between MSs. The different methods employed for MRSA monitoring are explained in detail within the section on MRSA to enable readers to better follow the procedures carried out by individual countries.
APPENDIX G. Additional information and supporting data
List of Annexes
The annexes are available on the EFSA Knowledge Junction community on Zenodo at: https://doi.org/10.5281/zenodo.10528846
The annexes contain the following information:
Annex A. Data reported on antimicrobial resistance in Salmonella spp.
The annex contains tables on antimicrobial resistance data:
Antimicrobial resistance in Salmonella spp. from humans, 2021–2022;
Occurrence of resistance to selected antimicrobials in Salmonella spp. from animals, 2021–2022;
Occurrence of resistance (%) to selected antimicrobials in specific Salmonella serovars.
Annex B. Data reported on antimicrobial resistance in Campylobacter spp.
The annex contains tables and figures showing antimicrobial resistance data:
Antimicrobial resistance in Campylobacter spp. from humans, 2022 and trends for 2016–2022 period;
Data reported on antimicrobial resistance and occurrence of resistance to selected antimicrobials in C. jejuni and C. coli from food‐producing animals and derived meat, for 2021–2022 and trends of resistance in broilers and in fattening turkeys (2014–2022).
Annex C. Data reported on AMR in indicator commensal Escherichia coli from food‐producing animals and derived meat
The annex contains tables on data reported on AMR in indicator commensal Escherichia coli from food‐producing animals and derived meat (data from poultry from 2022 and data from pigs and cattle under 1 year from 2021).
Annex D. Data on presumptive ESBL‐, AmpC‐ and/or carbapenemase‐producing microorganisms (routine and specific monitoring)
The annex contains tables with the data reported on presumptive ESBL‐, AmpC‐ and/or carbapenemase‐producing microorganisms for poultry (2022), pigs and cattle (2021) and meat thereof, and their resistance occurrence (routine and specific monitoring):
ESBL‐, AmpC‐producers prevalence and occurrence tables – pigs and cattle and meat thereof, 2021;
ESBL‐, AmpC‐, carbapenemase‐producers prevalence and occurrence tables – poultry 2022;
Specific carbapenemase‐producing E. coli monitoring 2021–2022;
ESBL‐, AmpC‐producers prevalence maps – pigs and cattle and meat thereof, 2021; ESBL‐, AmpC‐ producers, carbapenemase‐producers prevalence maps – poultry 2022.
Annex E. Data reported on antimicrobial resistance in MRSA from food‐producing animals and derived meat
The annex contains tables and data reported on the prevalence, genetic diversity and antimicrobial resistance of MRSA from food‐producing animals and derived meat collected in 2021 and 2022.
Annex F. WGS data reported complementary to MIC values
The annex contains tables with WGS data reported complementary to MIC values for presumptive ESBL‐/AmpC‐/CP‐producing E. coli collected in the routine monitoring (when isolates resistant to cefotaxime, ceftazidime and/or meropenem were detected in the first panel), as well as for the specific monitoring of ESBL‐/AmpC‐/CP‐producing E. coli.
Supporting data
All tables produced for the European Union Summary Report on Antimicrobial Resistance in Zoonotic and Indicator Bacteria from Humans, Animals and Food in 2021/2022 are available on the EFSA Knowledge Junction community on Zenodo at: https://doi.org/10.5281/zenodo.10528846.
The aggregated data set submitted on the negative results for extended‐spectrum β‐lactamases (ESBL) and carbapenemase‐producers is also available on the Knowledge Junction at: https://doi.org/10.5281/zenodo.105288462022 prevalence of MRSA aggregated dataset is also available on the Knowledge Junction at: https://doi.org/10.5281/zenodo.10528846.
A table showing ESBL‐/AmpC‐/CP‐encoding genes and corresponding MIC values for cephalosporins and carbapenems reported in 2022 is also available on the knowledge junction at: https://doi.org/10.5281/zenodo.10528846.
Country data sets
All country data sets containing the tables on the occurrence of antimicrobial resistance per each country are available on the EFSA Knowledge Junction community on Zenodo – please see below the list and corresponding link to the data sets.
The countries that submitted data sets on the 2021–2022 monitoring years are: the 27 EU Member States and United Kingdom (Northern Ireland), the three non‐EU Member States and Republic of North Macedonia as pre‐accession countries.
EFSA (European Food Safety Authority) & ECDC (European Centre for Disease Prevention and Control) . (2024). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2021–2022. EFSA Journal, 22, e8583. 10.2903/j.efsa.2024.8583
Approved: 19 January 2024
Note: A plain language summary of this scientific report is available at https://efsa.onlinelibrary.wiley.com/doi/10.2903/j.efsa.2024.p220202
Notes
A European One Health Action Plan against Antimicrobial Resistance (AMR), https://health.ec.europa.eu/system/files/2020‐01/amr_2017_action‐plan_0.pdf
The median level of all reporting EU‐MSs for each animal category.
While amikacin is not used in food‐producing animals, it is commonly used in people to treat urinary tract infections, bacteremia and intra‐abdominal infections caused by Gram‐negative bacteria. The addition of amikacin to the harmonised panel is thought to improve the detection of 16S RNA methyltransferases (RMTases) (EFSA, 2019).
Ampicillin, sulfamethoxazole and tetracyclines.
Ciprofloxacin, cefotaxime and combined resistance to these two antimicrobials.
See Appendix F – Materials and methodsfor definition of epidemiological cut‐off value (ECOFF).
All Salmonella and indicator E. coli isolates tested with the harmonised panel of antimicrobial substances (Panel 1) and exhibiting microbiological resistance to cefotaxime, ceftazidime or meropenem were subsequently subjected to further testing using a supplementary panel of substances (Panel 2) to obtain more detailed phenotypic characterisation (see Appendix F, materials and methods).
The method is described in Appendix F and protocols are available at: https://www.eurl‐ar.eu/protocols.aspx
E. coli isolates exhibiting an ESBL‐, AmpC‐ or ESBL + AmpC‐producing phenotype.
Commission Implementing Decision (EU) 2023/1017 of 23 May 2023 amending Implementing decision (EU) 2020/1729 as regards the monitoring of methicillin‐resistant Staphylococcus aureus (MRSA) in fattening pigs, https://eur‐lex.europa.eu/eli/dec_impl/2023/1017/oj
Commission Implementing Decision (EU) 2020/1729 of 17 November 2020 on monitoring and reporting of antimicrobial resistance in zoonotic and commensal bacteria and repealing Implementing Decision 2013/652/EU, https://eur‐lex.europa.eu/legal‐content/EN/TXT/PDF/?uri=CELEX:32020D1729&rid=4
‘Quantitative data’derived from dilution methods consisted of the number of isolates having a specific MIC value (measured in mg/L) relative to the total number of isolates tested, for each antimicrobial agent and specific food/animal category
Commission Regulation (EC) No 2073/2005 of 15 November 2005 on microbiological criteria for foodstuffs. OJ L 338,22.12.2005, p. 1–26
Available online: www.eurl‐ar.eu
Contributor Information
European Food Safety Authority (EFSA), Email: zoonoses@efsa.europa.eu.
European Centre for Disease Prevention and Control (ECDC), Email: fwd@ecdc.europa.eu.
REFERENCES
- Abdullahi, I. N. , Fernández‐Fernández, R. , Juárez‐Fernández, G. , Martínez‐Álvarez, S. , Eguizábal, P. , Zarazaga, M. , Lozano, C. , & Torres, C. (2021). Wild animals are reservoirs and sentinels of Staphylococcus aureus and MRSA clones: A problem with “one health” concern. Antibiotics, 10(12), 1556. 10.3390/antibiotics10121556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adur, M. A. , Châtre, P. , Métayer, V. , Drapeau, A. , Pillonetto, M. , Penkal, M. L. , Lopes, J. K. , Beirão, B. C. B. , Madec, J.‐Y. , Freitas de Macedo, R. E. , & Haenni, M. (2022). Escherichia coli ST224 and IncF/blaCTX‐M‐55 plasmids drive resistance to extended‐spectrum cephalosporins in poultry flocks in Parana, Brazil. International Journal of Food Microbiology, 380(109885). 10.1016/j.ijfoodmicro.2022.109885 [DOI] [PubMed] [Google Scholar]
- Agersø, Y. , Torpdahl, M. , Zachariasen, C. , Seyfarth, A. , Hammerum, A. M. , & Nielsen, E. M. (2012). Tentative colistin epidemiological cut‐off value for salmonella spp. Foodborne Pathogens and Disease, 9(4), 367–369. 10.1089/fpd.2011.1015 [DOI] [PubMed] [Google Scholar]
- Alba, P. , Feltrin, F. , Cordaro, G. , Porrero, M. C. , Kraushaar, B. , Argudín, M. A. , Nykäsenoja, S. , Monaco, M. , Stegger, M. , Aarestrup, F. M. , Butaye, P. , Franco, A. , & Battisti, A. (2015). Livestock‐associated methicillin resistant and methicillin susceptible Staphylococcus aureus sequence type (CC)1 in European farmed animals: High genetic relatedness of isolates from Italian cattle herds and humans. PLoS One, 10(8), e0137143. 10.1371/journal.pone.0137143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba, P. , Leekitcharoenphon, P. , Carfora, V. , Amoruso, R. , Cordaro, G. , Di Matteo, P. , Ianzano, A. , Iurescia, M. , Diaconu, E. L. , Engage‐Eurl‐Ar Network Study Group , Pedersen, S. K. , Guerra, B. , Hendriksen, R. S. , Franco, A. , & Battisti, A. (2020). Molecular epidemiology of salmonella Infantis in Europe: Insights into the success of the bacterial host and its parasitic pESI‐like megaplasmid. Microbial. Genomics, 6(5), e000365. 10.1099/mgen.0.000365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez, D. M. , Barrón‐Montenegro, R. , Conejeros, J. , Rivera, D. , Undurraga, E. A. , & Moreno‐Switt, A. I. (2023). A review of the global emergence of multidrug‐resistant salmonella enterica subsp. enterica Serovar Infantis. International Journal of Food Microbiology, 110297. 10.1016/j.ijfoodmicro.2023.110297 [DOI] [PubMed] [Google Scholar]
- Anyanwu, M. U. , Nwobi, O. C. , Okpala, C. O. R. , & Ezeonu, I. M. (2022). Mobile tigecycline resistance: An emerging health catastrophe requiring urgent one health global intervention. Frontiers Microbiology, 13, 808744. 10.3389/fmicb.2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armand‐Lefevre, L. , Ruimy, R. , & Andremont, A. (2005). Clonal comparison of Staphylococcus aureus isolates fromhealthy pig farmers, human controls, and pigs. Emerging Infectious Diseases, 11(5), 711–714. 10.3201/eid1105.040866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arca‐Suarez, J. , Rodiño‐Janeiro, B. K. , Pérez, A. , Guijarro‐Sánchez, A. , Vázquez‐Ucha, J. C. , Cruz, F. , Gómez‐Garrido, J. , Alioto, T. S. , Álvarez‐Tejado, M. , Gut, M. , Gut, I. , Oviaño, M. , Beceiro, A. , Bou, G. , & GEMARA‐SEIMC/REIPI Enterobacterales Study Group . (2022). Emergence of 16S rRNA methyltransferases among carbapenemase‐producing Enterobacterales in Spain studied by whole‐genome sequencing. International Journal of Antimicrobial Agents, 59(1), 106456. 10.1016/j.ijantimicag.2021.106456 [DOI] [PubMed] [Google Scholar]
- Asanin, J. , Misic, D. , Aksentijevic, K. , Tambur, Z. , Rakonjac, B. , Kovacevic, I. , Spergser, J. , & Loncaric, I. (2019). Genetic profiling and comparison of human and animal methicillin‐resistant Staphylococcus aureus (MRSA) isolates from Serbia. Antibiotics, 8(1), 26. 10.3390/antibiotics8010026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, L. , du, P. , du, Y. , Sun, H. , Zhang, P. , Wan, Y. , Lin, Q. , Fanning, S. , Cui, S. , & Wu, Y. (2019). Detection of plasmid‐mediated tigecycline‐resistant gene tet(X4) in Escherichia coli from pork, Sichuan and Shandong provinces, China, February 2019. Eurosurveillance, 24(25), 1900340. 10.2807/1560-7917.ES.2019.24.25.1900340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bal, A. M. , Coombs, G. W. , Holden, M. T. G. , Lindsay, J. A. , Nimmo, G. R. , Tattevin, P. , & Skov, R. L. (2016). Genomic insights into the emergence and spread of international clones of healthcare‐, community‐ and livestock‐associated methicillin‐resistant Staphylococcus aureus: Blurring of the traditional definitions. Journal of Global Antimicrobial Resistance, 6, 95–101. 10.1016/j.jgar.2016.04.004 [DOI] [PubMed] [Google Scholar]
- Bangerter, P. D. , Sidler, X. , Perreten, V. , & Overesch, G. (2016). Longitudinal study on the colonisation and transmission of methicillin‐resistant Staphylococcus aureus in pig farms. Veterinary Microbiology, 183, 125–134. 10.1016/j.vetmic.2015.12.007 [DOI] [PubMed] [Google Scholar]
- Bejaoui, A. , Gharbi, M. , Bitri, S. , Nasraoui, D. , Ben Aziza, W. , Ghedira, K. , Rfaik, M. , Marzougui, L. , Ghram, A. , & Maaroufi, A. (2022). Virulence profiling, multidrug resistance and molecular mechanisms of campylobacter strains from chicken carcasses in Tunisia. Antibiotics, 11, 830. 10.3390/antibiotics11070830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson, B. , Persson, L. , Ekström, K. , Unnerstad, H. E. , Uhlhorn, H. , & Börjesson, S. (2017). High occurrence of mecC‐MRSA in wild hedgehogs (Erinaceus europaeus) in Sweden. Veterinary Microbiology, 207, 103–107. 10.1016/j.vetmic.2017.06.004 [DOI] [PubMed] [Google Scholar]
- Bevan, E. R. , Jones, A. M. , & Hawkey, P. M. (2017). Global epidemiology of CTX‐M β‐lactamases: Temporal and geographical shifts in genotype. Journal of Antimicrobial Chemotherapy, 72(8), 2145–2155. 10.1093/jac/dkx146 [DOI] [PubMed] [Google Scholar]
- Boost, M. , Ho, J. , Guardabassi, L. , & O'Donoghue, M. (2012). Colonization of butchers with livestock‐associated methicillin‐resistant Staphylococcus aureus . Zoonoses and Public Health, 60(8), 572–576. 10.1111/zph.12034 [DOI] [PubMed] [Google Scholar]
- Bortolaia, V. , Ronco, T. , Romascu, L. , Nicorescu, I. , Milita, N. M. , Vaduva, A. M. , Leekitcharoenphon, P. , Kjeldgaard, J. S. , Hansen, I. M. , Svendsen, C. A. , Mordhorst, H. , Guerra, B. , Beloeil, P. A. , Hoffmann, M. , & Hendriksen, R. S. (2021). Co‐localization of carbapenem (bla OXA‐162) and colistin (mcr‐1) resistance genes on a transferable IncHI2 plasmid in Escherichia coli of chicken origin. Journal of Antimicrobial Chemotheraphy, 76, 3063–3065. 10.1093/jac/dkab285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, P. A. , Petersen, P. J. , Young, M. , Jones, C. H. , Tischler, M. , & O'Connell, J. (2005). Tigecycline MIC testing by broth dilution requires use of fresh medium or addition of the biocatalytic oxygen‐reducing reagent oxyrase to standardize the test method. Antimicrobial Agents and Chemotherapy, 49(9), 3903–3909. 10.1128/AAC.49.9.3903-3909.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer, M. S. M. , Rapallini, M. , Geurts, Y. , Harders, F. , Bossers, A. , Mevius, D. J. , Wit, B. , & Veldman, K. T. (2018). Enterobacter cloacae complex isolated from shrimps from Vietnam carrying bla IMI‐1 resistant to carbapenems but not cephalosporins. Antimicrobial Agents and Chemotherapy, 62(7), e00398‐18. 10.1128/AAC.00398-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carfora, V. , Diaconu, E. L. , Ianzano, A. , Di Matteo, P. , Amoruso, R. , Dell'Aira, E. , Sorbara, L. , Bottoni, F. , Guarneri, F. , Campana, L. , Franco, A. , Alba, P. , & Battisti, A. (2022). The hazard of carbapenemase (OXA‐181)‐producing Escherichia coli spreading in pig and veal calf holdings in Italy in the genomics era: Risk of spill over and spill back between humans and animals. Frontiers in Microbiology, 13, 1016895. 10.3389/fmicb.2022.1016895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casella, T. , Haenni, M. , Madela, N. K. , Kellen de Andrade, L. , Pradela, L. K. , Neves de Andrade, L. , da Costa Darini, A. L. , Madec, J.‐Y. , & Nogueira, M. C. L. (2018). Extended‐spectrum cephalosporin‐resistant Escherichia coli isolated from chickens and chicken meat in Brazil is associated with rare and complex resistance plasmids and pandemic ST lineages. Journal of Antimicrobial Chemotherapy, 73(12), 3293–3297. 10.1093/jac/dky335 [DOI] [PubMed] [Google Scholar]
- CASFM/EUCAST . (2022). CASFM/EUCAST: French Society for Microbiology v1.0. French Society for Microbiology. https://www.sfm‐microbiologie.org/wp‐content/uploads/2022/05/CASFM2022_V1.0.pdf
- Ceballos, S. , Aspiroz, C. , Ruiz‐Ripa, L. , Reynaga, E. , Azcona‐Gutiérrez, J. M. , Rezusta, A. , Seral, C. , Antoñanzas, F. , Torres, L. , López, C. , López‐Cerero, L. , Cercenado, E. , Zarazaga, M. , Torres, C. , & Study Group of clinical LA‐MRSA . (2019). Epidemiology of MRSA CC398 in hospitals located in Spanish regions with different pig‐farming densities: A multicentre study. Journal of Antimicrobial Chemotherapy, 74(8), 2157–2161. 10.1093/jac/dkz180 [DOI] [PubMed] [Google Scholar]
- Chang, M.‐X. , Zhang, J.‐F. , Sun, Y.‐H. , Li, R.‐S. , Lin, X.‐L. , Yang, L. , Webber, M. A. , & Jiang, H.‐X. (2021). Contribution of different mechanisms to ciprofloxacin resistance in salmonella spp. Frontiers in Microbiology, 12, 663731. 10.3389/fmicb.2021.663731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantziaras, I. , Boyen, F. , Callens, F. , & Dewulf, J. (2014). Correlation between veterinary antimicrobial use and antimicrobial resistance in food‐producing animals: A report on seven countries. Journal of Antimicrobial Chemotherapy, 69, 827–834. 10.1093/jac/dkt443 [DOI] [PubMed] [Google Scholar]
- Chen, C. , & Wu, F. (2020). Livestock‐associated methicillin‐resistant Staphylococcus aureus(LA‐MRSA) colonization and infection among livestock workers and veterinarians: A systematic review and meta‐analysis. Occupational and Environmental Medicine. 10.1136/oemed-2020-106418 [DOI] [PubMed] [Google Scholar]
- Chong, Y. , Ito, Y. , & Kamimura, T. (2011). Genetic evolution and clinical impact in extended‐spectrum β‐lactamaseproducing Escherichia coli and Klebsiella pneumoniae . Infection, Genetics and Evolution, 11(7), 1499–1504. 10.1016/j.meegid.2011.06.001 [DOI] [PubMed] [Google Scholar]
- CLSI and EUCAST . (2016). Recommendations for MIC determination of colistin (polymyxin E) as recommended by the joint CLSI‐EUCAST Polymyxin Breakpoints Working Group. 22 March 2016. Clinical and Laboratory Standards Institute and European Committee on Antimicrobial Susceptibility Testing. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/General_documents/Recommendations_for_MIC_determination_of_colistin_March_2016.pdf
- Coipan, C. E. , Westrell, T. , van Hoek, A. , Alm, E. , Kotila, S. , Berbers, B. , de Keersmaecker, S. C. J. , Ceyssens, P. J. , Borg, M. L. , Chattaway, M. , McCormick, J. , Dallman, T. J. , & Franz, E. (2020). Genomic epidemiology of emerging ESBL‐producing salmonella Kentucky blaCTX‐M‐14b in Europe. Emerging Microbes & Infections, 9(1), 2124–2135. 10.1080/22221751.2020.1821582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, S. D. , Perez‐Bonilla, D. , Hallowell, A. , & Redding, L. E. (2022). Carbapenem prescribing at a veterinary teaching hospital before an outbreak of carbapenem‐resistant Escherichia coli . The Journal of Small Animal Practice, 63(6), 442–446. [DOI] [PubMed] [Google Scholar]
- Cousto, L. M. , Weese, J. S. , & Bateman, S. W. (2020). Prescribing patterns and comparison of culture versus empiric‐based selection of meropenem in cats and dogs in a veterinary teaching hospital (2011–2018). The Canadian Veterinary Journal = La Revue Veterinaire Canadienne, 61(3), 274–280. [PMC free article] [PubMed] [Google Scholar]
- Crespo‐Piazuelo, D. , & Lawlor, P. G. (2021). Livestock‐associated methicillin‐resistant Staphylococcus aureus (LA‐MRSA) prevalence in humans in close contact with animals and measures to reduce on‐farm colonisation. Irish Veterinary Journal, 74(1), 21. 10.1186/s13620-021-00200-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crombé, F. , Willems, G. , Dispas, M. , Hallin, M. , Denis, O. , Suetens, C. , Gordts, B. , Struelens, M. , & Butaye, P. (2012). Prevalence and antimicrobial susceptibility of methicillin‐resistant Staphylococcus aureus among pigs in Belgium. Microbial Drug Resistance, 18(2), 125–131. 10.1089/mdr.2011.0138 [DOI] [PubMed] [Google Scholar]
- Crump, J. A. , Sjolund‐Karlsson, M. , Gordon, M. A. , & Parry, C. M. (2015). Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive salmonella infections. Clinical Microbiology Reviews, 28(4), 901–937. 10.1128/CMR.00002-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuny, C. , Abdelbary, M. M. H. , Köck, R. , Layer, F. , Scheidmann, W. , Werner, G. , & Witte, W. (2016). Methicillin‐resistant Staphylococcus aureus from infections in horses in Germany are frequent colonizers of veterinarians but rare among MRSA from infections in humans. One Health, 2, 11–17. 10.1016/j.onehlt.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuny, C. , Wieler, L. H. , & Witte, W. (2015). Livestock‐associated MRSA: The impact on humans. Antibiotics, 4(4), 521–543. 10.3390/antibiotics4040521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, L. , Sahin, O. , Grover, M. , & Zhang, Q. (2020). New and alternative strategies for the prevention, control, and treatment of antibiotic‐resistant campylobacter. Translational Research, 223, 76–88. 10.1016/j.trsl.2020.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day, M. J. , Rodríguez, I. , van Essen‐Zandbergen, A. , Dierikx, C. , Kadlec, K. , Schink, A.‐K. , Wu, G. , Chattaway, M. A. , DoNascimento, V. , Wain, J. , Helmuth, R. , Guerra, B. , Schwarz, S. , Threlfall, J. , Woodward, M. J. , Coldham, N. , Mevius, D. , & Woodford, N. (2016). Diversity of STs, plasmids and ESBL genes among Escherichia coli from humans, animals and food in Germany, The Netherlands and the UK. Journal of Antimicrobial Chemotherapy, 71(5), 1178–1182. 10.1093/jac/dkv485 [DOI] [PubMed] [Google Scholar]
- Diaconu, E. L. , Carfora, V. , Alba, P. , Di Matteo, P. , Stravino, F. , Buccella, C. , Dell'Aira, E. , Onorati, R. , Sorbara, L. , Battisti, A. , & Franco, A. (2020). Novel IncFII plasmid harbouring bla NDM‐4 in a carbapenem‐resistant Escherichia coli of pig origin, Italy. Journal of Antimicrobial Chemotheraphy, 75, 3475–3479. 10.1093/jac/dkaa374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earls, M. R. , Steining, E. J. , Monecke, S. , Castruita, J. A. S. , Simbeck, A. , Schneider‐Brachert, W. , Vremera, T. , Dorneanu, O. S. , Loncaric, I. , Bes, M. , Lacoma, A. , Aymerich, C. P. , Wernery, U. , Armengol‐Porta, M. , Blomfeldt, A. , Duchene, S. , Bartels, M. D. , Ehricht, R. , & Coleman, D. C. (2021). Exploring the evolution and epidemiology of European CC1‐MRSA‐IV: Tracking a multidrug‐resistant community‐associated methicillin‐resistant Staphylococcus aureus clone. Microbial Genomics, 7(7). 10.1099/mgen.0.000601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ECDC (European Centre for Disease Prevention and Control) . (2016). EU protocol for harmonised monitoring of antimicrobial resistance in human salmonella and campylobacter isolates – June 2016. European Centre for Disease Prevention and Control. https://www.ecdc.europa.eu/en/publications‐data/eu‐protocol‐harmonised‐monitoring‐antimicrobial‐resistance‐human‐salmonella‐and‐0 [Google Scholar]
- ECDC (European Centre for Disease Prevention and Control) . (2018). ECDC study protocol for genomic‐based surveillance of carbapenem‐resistant and/or colistin‐resistant Enterobacteriaceae at the EU level. Version 2.0. https://www.ecdc.europa.eu/sites/default/files/documents/Protocol‐genomic‐surveillance‐resistant‐Enterobacteriaceae‐v2_0.pdf
- ECDC (European Centre for Disease Prevention and Control) , 2023a. Increase in Escherichia coli isolates carrying bla NDM‐5 in the European Union/European economic area, 2012–2022. https://www.ecdc.europa.eu/sites/default/files/documents/Increase‐E‐coli‐isolates‐blaNDM‐5‐EU‐EEA‐may2023.pdf [Google Scholar]
- ECDC (European Centre for Disease Prevention and Control) , 2023b. Antimicrobial resistance in the EU/EEA (EARS‐net) ‐ annual epidemiological report 2022. https://www.ecdc.europa.eu/en/publications‐data/surveillance‐antimicrobial‐resistance‐europe‐2022 [Google Scholar]
- ECDC, EFSA and EMA (European Centre for Disease Prevention and Control, European Food Safety Authority and European Medicines Agency) . (2017). ECDC/EFSA/EMA second joint report on the integrated analysis of the consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from humans and food‐producing animals. EFSA Journal, 15(7), e04872. 10.2903/j.efsa.2017.4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ECDC, EFSA and EMA (European Centre for Disease Prevention and Control, European Food Safety Authority and European Medicines Agency) . (2021). Third joint inter‐agency report on integrated analysis of consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from humans and food‐producing animals in the EU/EEA, JIACRA III. In 2016–2018. Parma; European Centre for Disease Prevention and Control European Food Safety Authority and European medicines agency (Vol. 19). 10.2903/j.efsa.2021.6712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA (European Food Safety Authority) . (2008). Report from the task force on Zoonoses data collection including guidance for harmonized monitoring and reporting of antimicrobial resistance in commensal Escherichia coli and enterococcus spp. from food animals. EFSA Journal, 6(4), 141. 10.2903/j.efsa.2008.141r [DOI] [Google Scholar]
- EFSA (European Food Safety Authority) . (2009a). Analysis of the baseline survey on the prevalence of methicillin‐resistant Staphylococcus aureus (MRSA) in holdings with breeding pigs, in the EU, 2008. EFSA Journal, 7(11), 1376. 10.2903/j.efsa.2009.1376 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority) . (2009b). Assessment of the public health significance of meticillin resistant Staphylococcus aureus (MRSA) in animals and foods. EFSA Journal, 7(3), 993. 10.2903/j.efsa.2009.993 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority) . (2012). Technical specifications on the harmonised monitoring and reporting of antimicrobial resistance in methicillin‐resistant Staphylococcus aureus in food‐producing animals and food. EFSA Journal, 10(10), 2987. 10.2903/j.efsa.2012.2897 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority) . (2014). Technical specifications on randomised sampling for harmonisedmonitoring of antimicrobial resistance in zoonotic and commensal bacteria. EFSA Journal, 12(5), 3686. 10.2903/j.efsa.2014.3686 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority) . (2019). Technical specifications on harmonised monitoring of antimicrobial resistance in zoonotic and indicator bacteria from food‐producing animals and food. EFSA journal, 17(6), 5709. 10.2903/j.efsa.2019.5709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA (European Food Safety Authority) . (2020). Technical specifications on a randomisation of sampling for the purpose of antimicrobial resistance monitoring from food‐producing animals and food as from 2021. EFSA Journal, 18(12), 6364. 10.2903/j.efsa.2020.6364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA (European Food Safety Authority) . (2022). Technical specifications for a baseline survey on the prevalence of methicillin‐resistant Staphylococcus aureus (MRSA) in pigs. EFSA Journal, 20(10), 7620. 10.2903/j.efsa.2022.7620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA (European Food Safety Authority) . (2023). Manual for reporting 2022 antimicrobial resistance data within the framework of directive 2003/99/EC and decision 2020/1729/EU. EFSA supporting publication, EN‐7826. 10.2903/sp.efsa.2023.EN-7826 [DOI] [Google Scholar]
- EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control) . (2022). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2019–2020. EFSA Journal, 20(3), 7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control) . (2023). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2020–2021. EFSA Journal, 21(3), 7867. 10.2903/j.efsa.2023.7867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA BIOHAZ Panel (Panel on Biological Hazards) . (2011). Scientific opinion on the public health risks of bacterial strains producing extended‐spectrum β‐lactamases and/or AmpC β‐lactamases in food and food‐producing animals. EFSA Journal, 9(8), 2322. 10.2903/j.efsa.2011.2322 [DOI] [Google Scholar]
- Egervärn, M. , Börjesson, S. , Byfors, S. , Finn, M. , Kaipe, C. , Englund, S. , & Lindblad, M. (2014). Escherichia coli with extended‐spectrum beta‐lactamases or transferable AmpC beta‐lactamases and Salmonella on meat imported into Sweden. International Journal of Food Microbiology, 171, 8–14. 10.1016/j.ijfoodmicro.2013.11.005 [DOI] [PubMed] [Google Scholar]
- Elhadidy, M. , Ali, M. M. , El‐Shibiny, A. , Miller, W. G. , Elkhatib, W. F. , Botteldoorn, N. , & Dierick, K. (2020). Antimicrobial resistance patterns and molecular resistance markers of campylobacter jejuni isolates from human diarrheal cases. PLoS One, 15(1), e0227833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhadidy, M. , Miller, W. G. , Arguello, H. , Alvarez‐Ordo'nez, A. , Dierick, K. , & Botteldoorn, N. (2019). Molecular epidemiology ˜ and antimicrobial resistance mechanisms of campylobacter coli from diarrhoeal patients and broiler carcasses in Belgium. Transboundary and Emerging Diseases, 66(1), 463–475. 10.1111/tbed.13046 [DOI] [PubMed] [Google Scholar]
- Elstrøm, P. , Grøntvedt, C. A. , Gabrielsen, C. , Stegger, M. , Angen, Ø. , Åmdal, S. , Enger, H. , Urdahl, A. M. , Jore, S. , Steinbakk, M. , & Sunde, M. (2019). Livestock‐associated MRSA CC1 in Norway; introduction to pig farms, zoonotic transmission, and eradication. Frontiers in Microbiology, 10. 10.3389/fmicb.2019.00139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EMA (European Medicines Agency) . (2023). Surveillance of Veterinary Antimicrobial Consumption, 2022. Sales of veterinary antimicrobial agents in 31 European countries in 2022. EMA/299538/2023. 13th ESVAC Report. European Medicines Agency. https://www.ema.europa.eu/en/documents/report/sales‐veterinary‐antimicrobial‐agents‐31‐european‐countries‐2022‐trends‐2010‐2022‐thirteenth‐esvac_en.pdf
- EMA and EFSA (European Medicines Agency and European Food Safety Authority) . (2017). EMA and EFSA joint scientific opinion on measures to reduce the need to use antimicrobial agents in animal husbandry in the European Union, and the resulting impacts on food safety (RONAFA). EFSA Journal, 15(1), 4666. 10.2903/j.efsa.2017.4666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EMA/CVMP (European Medicines Agency's/Committee for Veterinary Medicinal Products) . (2018). Opinion Following an Article 35 Referral for All Veterinary Medicinal Products Containing Systemically Administered (Parenteral and Oral) 3rd and 4th Generation Cephalosporins Intended for Use in Food Producing Species. European Medicines Agency. https://www.ema.europa.eu/en/medicines/veterinary/referrals/cephalosporins [Google Scholar]
- Espinoza, N. , Rojas, J. , Pollett, S. , Meza, R. , Patiño, L. , Leiva, M. , Camiña, M. , Bernal, M. , Reynolds, N. D. , Maves, R. , Tilley, D. H. , Kasper, M. , & Simons, M. P. (2020). Validation of the T86I mutation in the gyrA gene as a highly reliable real time PCR target to detect fluoroquinolone‐resistant campylobacter jejuni. BMC Infectious Disease, 20, 518. 10.1186/s12879-020-05202-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EUCAST (European Committee for Antimicrobial Susceptibility Testing), 2014. Screening for Fluoroquinolone Resistance in Salmonella spp. with Pefloxacin 5 ug. Tentative Quality Control Criteria for Users and Disk Manufacturers. European Committee for Antimicrobial Susceptibility Testing Available online : (2014). The EUCAST Guidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance. Version 2. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/QC/Tentative_QC_criteria_for_pefloxacin_5_g.pdf
- EUCAST (European Committee for Antimicrobial Susceptibility Testing) . (2017). The EUCAST Guidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance. Version 2. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_170711.pdf
- EUCAST (European Committee for Antimicrobial Susceptibility Testing) . (2019). New definitions of S, I and R from 2019. https://www.eucast.org/newsiandr/
- EUCAST (European Committee for Antimicrobial Susceptibility Testing) . (2023). EUCAST: Clinical breakpoints and dosing of antibiotics. https://www.Eucast.Org/clinical_breakpoints/
- European Commission , 2022. AMR one health network meeting of 25–26 January 2022 MINUTES. European Commission: Directorate General for Health and Food Safety. [Google Scholar]
- Ewers, C. , Bethe, A. , Semmler, T. , Guenther, S. , & Wieler, L. H. (2012). Extended‐spectrum β‐lactamase‐producing and AmpC‐producing Escherichia coli from livestock and companion animals, and their putative impact on public health: A global perspective. Clinical Microbiology and Infection, 18(7), 646–655. 10.1111/j.1469-0691.2012.03850.x [DOI] [PubMed] [Google Scholar]
- Ewers, C. , de Jong, A. , Prenger‐Berninghoff, E. , El Garch, F. , Leidner, U. , Tiwari, S. K. , & Semmler, T. (2021). Genomic diversity and virulence potential of ESBL and AmpC‐β‐lactamase‐producing Escherichia coli strains from healthy food animals across Europe. Frontiers in Microbiology, 12, 626774. 10.3389/fmicb.2021.626774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florez‐Cuadrado, D. , Ugarte‐Ruiz, M. , Meric, G. , Quesada, A. , Porrero, M. C. , Pascoe, B. , Saez‐Llorente, J. L. , Orozco, G. L. , Dom'ınguez, L. , & Sheppard, S. K. (2017). Genome comparison of erythromycin resistant campylobacter from turkeys identifies hosts and pathways for horizontal spread of erm(B) genes. Frontiers in Microbiology, 8, 2240. 10.3389/fmicb.2017.02240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier, C. , Poirel, L. , Despont, S. , Kessler, J. , & Nordmann, P. (2022). Increasing trends of association of 16S rRNA methylases and carbapenemases in enterobacterales clinical isolates from Switzerland, 2017–2020. Microorganisms, 10(3), 615. 10.3390/microorganisms10030615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco, A. , Leekitcharoenphon, P. , Feltrin, F. , Alba, P. , Cordaro, G. , Iurescia, M. , Tolli, R. , D'Incau, M. , Staffolani, M. , di Giannatale, E. , Hendriksen, R. S. , & Battisti, A. (2015). Emergence of a clonal lineage of multidrug‐resistant ESBL producing salmonella Infantis transmitted from broilers and broiler meat to humans in Italy between 2011 and 2014. PLoS One, 10(12), e0144802. 10.1371/journal.pone.0144802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich, M. J. (2019). Antimicrobial resistance on the rise in zoonotic bacteria in Europe. Journal of the American Medical Association, 321(15), 1448. [DOI] [PubMed] [Google Scholar]
- Garcia, S. , & Heredia, N. L. (2013). Campylobacter. In Labbe R. G. & Garcia S. (Eds.), Guide to foodborne pathogens (pp. 188–196). John Wiley & Sons. [Google Scholar]
- García‐Fierro, R. , Drapeau, A. , Dazas, M. , Saras, E. , Rodrigues, C. , Brisse, S. , Madec, J. Y. , & Haenni, M. (2022). Comparative phylogenomics of ESBL‐, AmpC‐ and carbapenemase‐producing Klebsiella pneumoniae originating from companion animals and humans. The Journal of Antimicrobial Chemotherapy, 77(5), 1263–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Graells, C. , Berbers, B. , Verhaegen, B. , Vanneste, K. , Marchal, K. , Roosens, N. H. C. , Botteldoorn, N. , & De Keersmaecker, S. C. J. (2020). First detection of a plasmid located carbapenem resistant bla VIM‐1 gene in E. Coli isolated from meat products at retail in Belgium in 2015. International Journal of Food and Microbiology, 324, 108624. 10.1016/j.ijfoodmicro.2020.108624 [DOI] [PubMed] [Google Scholar]
- Grøntvedt, C. A. , Elstrøm, P. , Stegger, M. , Skov, R. L. , Andersen, P. S. , Larssen, K. W. , Urdahl, A. M. , Angen, Ø. , Larsen, J. , Åmdal, S. , Løtvedt, S. M. , Sunde, M. , & Bjørnholt, J. V. (2016). Methicillin‐resistant Staphylococcus aureus CC398 in humans and pigs in Norway: A “one health” perspective on introduction and transmission. Clinical Infectious Diseases, 63(11), 1431–1438. 10.1093/cid/ciw552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardabassi, L. , O'Donoghue, M. , Moodley, A. , Ho, J. , & Boost, M. (2009). Novel lineage of methicillin‐resistant Staphylococcus aureus . Hong Kong. Emerging Infectious Diseases, 15(12), 1998–2000. 10.3201/eid1512.090378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, X. , Chen, R. , Wang, Q. , Li, C. , Ge, H. , Qiao, J. , & Li, Y. (2022). Global prevalence, characteristics, and future prospects of IncX3 plasmids: A review. Frontiers in Microbiology, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenni, M. , Boulouis, H. J. , Lagrée, A. C. , Drapeau, A. , Va, F. , Billet, M. , Chatre, P. , & Madec, J. Y. (2022). Enterobacterales high‐risk clones and plasmids spreading blaESBL/AmpC and blaOXA‐48 genes within and between hospitalized dogs and their environment. The Journal of Antimicrobial Chemotherapy, 77(10), 2754–2762. [DOI] [PubMed] [Google Scholar]
- Haenni, M. , Métayer, V. , Lupo, A. , Drapeau, A. , & Madec, J. Y. (2022). Spread of the bla(OXA‐48)/IncL plasmid within and between dogs in City parks, France. Microbiolial Spectrum, 10(3), e0040322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel, M. , Rolain, J. M. , & Baron, S. A. (2021). The history of Colistin resistance mechanisms in bacteria: Progress and challenges. Microorganisms, 9(2), 442. 10.3390/microorganisms9020442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Händel, N. , Otte, S. , Jonker, M. , Brul, S. , & ter Kuile, B. (2015). Factors that affect transfer of the IncI1 ¨ β‐lactam resistance plasmid pESBL‐283 between E. Coli strains. PLoS One, 10(4), e0123039. 10.1371/journal.pone.0123039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkey, J. , le Hello, S. , Doublet, B. , Granier, S. A. , Hendriksen, R. S. , Fricke, W. F. , Ceyssens, P. J. , Gomart, C. , Billman‐Jacobe, H. , Holt, K. E. , & Weill, F. X. (2019). Global phylogenomics of multidrug‐resistant salmonella enterica serotype Kentucky ST198. Microbial. Genomics, 5(7). 10.1099/mgen.0.000269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, F. , Xu, J. , Wang, J. , Chen, Q. , Hua, X. , Fu, Y. , & Yu, Y. (2016). Decreased susceptibility to tigecycline mediated by a mutation in mlaA in Escherichia coli strains. Antimicrobial Agents and Chemotherapy, 60(12), 7530–7531. 10.1128/AAC.01603-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, T. , Wang, R. , Liu, D. , Walsh, T. R. , Zhang, R. , Lv, Y. , Ke, Y. , Ji, Q. , Wei, R. , Liu, Z. , Shen, Y. , Wang, G. , Sun, L. , Lei, L. , Lv, Z. , Li, Y. , Pang, M. , Wang, L. , Sun, Q. , … Wang, Y. (2019). Emergence of plasmid‐mediated high‐level tigecycline resistance genes in animals and humans. Nature Microbiology, 4(9), 1450–1456. 10.1038/s41564-019-0445-2 [DOI] [PubMed] [Google Scholar]
- Hendriksen, R. S. , Bortolaia, V. , Tate, H. , Tyson, G. H. , Aarestrup, F. M. , & McDermott, P. F. (2019). Using genomics to track global antimicrobial resistance. Frontiers in public health, 7. https://www.Frontiersin.Org/articles/10.3389/fpubh.2019.00242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins, K. L. , Kirchner, M. , Guerra, B. , Granier, S. A. , Lucarelli, C. , Porrero, M. C. , Jakubczak, A. , Threlfall, E. J. , & Mevius, D. J. (2010). Multiresistant salmonella enterica serovar 4,[5],12:i:‐ in Europe: A new pandemic strain? Euro surveillance: Bulletin Europeen Sur les maladies Transmissibles = European communicable disease bulletin, 15(22), 19580. [PubMed] [Google Scholar]
- Inglis, D. G. , Taboada, E. N. , & Boras, V. F. (2021). Rates of fluoroquinolone resistance in domestically acquired campylobacter jejuni are increasing in people living within a model study location in Canada. Canadian Journal of Microbiology, 67(1), 37–52. [DOI] [PubMed] [Google Scholar]
- Irrgang, A. , Pauly, N. , Tenhagen, B. A. , Grobbel, M. , Kaesbohrer, A. , & Hammerl, A. J. A. (2020). Spill‐over from public health? First detection of an OXA‐48‐producing Escherichia coli in a German pig farm. Microorganisms, 8(6), 855. 10.3390/microorganisms8060855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irrgang, A. , Tenhagen, B. A. , Pauly, N. , Schmoger, S. , Kaesbohrer, A. , & Hammerl, J. A. (2019). Characterization of VIM‐1‐ producing E. Coli isolated from a German fattening pig farm by an improved isolation procedure. Frontiers in Microbiology, 10, 2256. 10.3389/fmicb.2019.02256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby, G. A. (2009). AmpC β‐lactamases. Clinical Microbiology Reviews, 22(1), 161–182. 10.1128/cmr.00036-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby, G. A. , Strahilevitz, J. , & Hooper, D. C. (2014). Plasmid‐mediated quinolone resistance. Microbiology. Spectrum, 2(5). 10.1128/microbiolspec.PLAS-0006-2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehanne, Q. , Benéejat, L. , Ducournau, A. , Domingues‐Martins, C. , Cousinou, T. , Bessede, T. , & Lehours, P. (2021). Emergence of erythromycin resistance methyltransferases in campylobacter coli strains in France. Antimicrobial Agents and Chemotherapy, 65(11), e01124‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehanne, Q. , Pascoe, B. , Benejat, L. , Ducournau, A. , Buissonniere, A. , Mourkas, E. , Megraud, F. , Bessede, E. , Sheppard, S. K. , & Lehours, P. (2020). Genome‐wide identification of host‐segregating single‐nucleotide polymorphisms for source attribution of clinical campylobacter coli isolates. Applied and Environmental Microbiology, 86(24), e01787‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käsbohrer, A. M. , Bakran‐Lebl, K. , Irrgang, A. , Fischer, J. , Kämpf, P. , Schiffmann, A. , Werckenthin, C. , Busch, M. , Kreienbrock, L. , & Hille, K. (2019). Diversity in prevalence and characteristics of ESBL/pAmpC producing E. Coli in food in Germany. Veterinary Microbiology, 233, 52–60. 10.1016/j.vetmic.2019.03.025 [DOI] [PubMed] [Google Scholar]
- Kehrenberg, C. , Schwartz, S. , Jacobsen, L. , Hansen, L. H. , & Vester, B. (2005). A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: Methylation of 23S ribosomal RNA at A2503. Molecular Microbiology, 57(4), 1064–1073. 10.1111/j.1365-2958.2005.04754.x [DOI] [PubMed] [Google Scholar]
- Kieffer, N. , Aires‐de‐Sousa, M. , Nordmann, P. , & Poirel, L. (2017). High rate of MCR‐1–producing Escherichia coli and Klebsiella pneumoniae among pigs, Portugal. Emerging Infectious Diseases, 23(12), 2023–2029. 10.3201/eid2312.170883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinross, P. , Petersen, A. , Skov, R. , van Hauwermeiren, E. , Pantosti, A. , Laurent, F. , Voss, A. , Kluytmans, J. , Struelens, M. J. , Heuer, O. , Monnet, D. L. , & European Human LA‐MRSA Study Group . (2017). Livestock‐associated methicillin resistant Staphylococcus aureus (MRSA) among human MRSA isolates, European Union/European economic area countries, 2013. Eurosurveillance, 22(44). 10.2807/1560-7917.ES.2017.22.44.16-00696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köck, R. , Daniels‐Haardt, I. , Becker, K. , Mellmann, A. , Friedrich, A. W. , Mevius, D. , Schwarz, S. , & Jurke, A. (2018). Carbapenem‐resistant Enterobacteriaceae in wildlife, food‐producing, and companion animals: A systematic review. Clinical Microbiology and Infection, 24(12), 1241–1250. 10.1016/j.cmi.2018.04.004 [DOI] [PubMed] [Google Scholar]
- Köck, R. , Schaumburg, F. , Mellmann, A. , Köksal, M. , Jurke, A. , Becker, K. , & Friedrich, A. W. (2013). Livestock‐associated methicillin‐resistant Staphylococcus aureus (MRSA) as causes of huiman infection and colonization in Germany. PLoS One, 8(2), e55040. 10.1371/journal.pone.0055040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komodromos, D. , Kotzamanidis, C. , Giantzi, V. , Pappa, S. , Papa, A. , Zdragas, A. , Angelidis, A. , & Sergelidis, D. (2022). Prevalence, infectious characteristics and genetic diversity of Staphylococcus aureus and methicillin‐resistant Staphylococcus aureus (MRSA) in two raw‐meat processing establishments in northern Greece. Pathogens, 11, 1370. 10.3390/pathogens11111370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen, J. , Stegger, M. , Andersen, P. S. , Petersen, A. , Larsen, A. R. , Westh, H. , Agersø, Y. , Fetsch, A. , Kraushaar, B. , Käsbohrer, A. , Feβler, A. T. , Schwarz, S. , Cuny, C. , Witte, W. , Butaye, P. , Denis, O. , Haenni, M. , Madec, J. Y. , Jouy, E. , … Skov, R. L. (2016). Evidence for human adaptation and foodborne transmission of livestock‐associated methicillin‐resistant Staphylococcus aureus . Clinical Infectious Diseases, 63(10), 1349–1352. 10.1093/cid/ciw532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hello, S. , Bekhit, A. , Granier, S. A. , Barua, H. , Beutlich, J. , Zajac, M. , Munch, S. , Sintchenko, V. , Bouchrif, B. , Fashae, K. , Pinsard, J. L. , Sontag, L. , Fabre, L. , Garnier, M. , Guibert, V. , Howard, P. , Hendriksen, R. S. , Christensen, J. P. , Biswas, P. K. , … Weill, F. X. (2013). The global establishment of a highly fluoroquinolone resistant salmonella enterica serotype Kentucky ST198 strain. Frontiers in Microbiology, 4. 10.3389/fmicb.2013.00395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hello, S. , Hendriksen, R. S. , Doublet, B. , Fisher, I. , Nielsen, E. M. , Whichard, J. M. , Bouchrif, B. , Fashae, K. , Granier, S. A. , Jourdan‐da Silva, N. , Cloeckaert, A. , Threlfall, E. J. , Angulo, F. J. , Aarestrup, F. M. , Wain, J. , & Weill, F. X. (2011). International spread of an epidemic population of salmonella enterica serotype kentucky ST198 resistant to ciprofloxacin. The Journal of Infectious Diseases, 204(5), 675–684. 10.1093/infdis/jir409 [DOI] [PubMed] [Google Scholar]
- Lekshmi, M. , Kumar, S. H. , Nayak, B. B. , & Varela, M. F. (2023). Antimicrobial resistance in food‐borne campylobacter spp. In Mothadaka M. P., Vaiyapuri M., Rao Badireddy M., Nagarajrao Ravishankar C., Bhatia R., & Jena J. (Eds.), Handbook on antimicrobial resistance. Springer. 10.1007/978-981-16-9723-4_16-1 [DOI] [Google Scholar]
- Li, H. , Andersen, P. S. , Stegger, M. , Sieber, R. N. , Ingmer, H. , Staubrand, N. , Dalsgaard, A. , & Leisner, J. J. (2019). Antimicrobial resistance and virulence gene profiles of methicillin‐resistant and ‐susceptible Staphylococcus aureus from food products in Denmark. Frontiers in Microbiology, 10, 2681. 10.3389/fmicb.2019.02681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B.‐T. , Zhang, X. Y. , Wan, S. W. , Hao, J. J. , Jiang, R. D. , & Song, F. J. (2018). Characteristics of carbapenem‐resistant Enterobacteriaceae in ready‐to‐eat vegetables in China. Frontiers in Microbiology, 9, 1147. 10.3389/fmicb.2018.01147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D. , Liu, W. , Lv, Z. , Xia, J. , Li, X. , Hao, Y. , Zhou, Y. , Yao, H. , Liu, Z. , Wang, Y. , Shen, J. , Ke, Y. , & Shen, Z. (2019). Emerging erm (B)‐mediated macrolide resistance associated with novel multidrug resistance genomic islands in campylobacter. Antimicrobial Agents and Chemotherapy, 63(7), e00153‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano, C. , Fernández‐Fernández, R. , Ruiz‐Ripa, L. , Gómez, P. , Zarazaga, M. , & Torres, C. (2020). Human mecC‐carrying MRSA: Clinical implications and risk factors. Microorganisms, 8(10), 1615. 10.3390/microorganisms8101615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luangtongkum, T. , Jeon, B. , Han, J. , Plummer, P. , Logue, C. M. , & Zhang, Q. (2009). Antibiotic resistance in campylobacter: Emergence, transmission and persistence. Future Microbiology, 4(2), 189–200. 10.2217/17460913.4.2.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madec, J.‐Y. , Haenni, M. , Nordmann, P. , & Poirel, L. (2017). Extended‐spectrum β‐lactamase/AmpC‐ and carbapenemase‐producing Enterobacteriaceae in animals: A threat for humans? Clinical Microbiology and Infection, 23(11), 826–833. 10.1016/j.cmi.2017.01.013 [DOI] [PubMed] [Google Scholar]
- Magiorakos, A. P. , Srinivasan, A. , Carey, R. B. , Carmeli, Y. , Falagas, M. E. , Giske, C. G. , Harbarth, S. , Hindler, J. F. , Kahlmeter, G. , Olsson‐Liljequist, B. , Paterson, D. L. , Rice, L. B. , Stelling, J. , Struelens, M. J. , Vatopoulos, A. , Weber, J. T. , & Monnet, D. L. (2012). Multidrug‐resistant, extensively drug‐resistant and pandrug‐resistant bacteria: An international expertproposal for interim standard definitions for acquired resistance. Clinical Microbiology and Infection, 18, 268–281. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- Manges, A. R. (2016). Escherichia coli and urinary tract infections: The role of poultry meat. Clinical Microbiology and Infection, 22(2), 122–129. 10.1016/j.cmi.2015.11.010 [DOI] [PubMed] [Google Scholar]
- Martinez, J. L. (2014). General principles of antibiotic resistance in bacteria. Drug Discovery Today: Technologies, 11, 33–39. 10.1016/j.ddtec.2014.02.001 [DOI] [PubMed] [Google Scholar]
- Matuschek, E. , Åhman, J. , Webster, C. , Kahlmeter, G. (2018). Antimicrobial susceptibility testing of colistin ‐ evaluation of seven commercial MIC products against standard broth microdilution for Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter spp. Clin Microbiol Infect , 24(8), 865–870. 10.1016/j.cmi.2017.11.020 [DOI] [PubMed] [Google Scholar]
- McDermott, P. F. , Tyson, G. H. , Kabera, C. , Chen, Y. , Li, C. , Folster, J. P. , Ayers, S. L. , Lam, C. , Tate, H. P. , & Zhao, S. (2016). Whole genome sequencing for detecting antimicrobial resistance in nontyphoidal salmonella. Antimicrobial Agents Chemotheraphy, 60, 5515–5520. 10.1128/AAC.01030-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo, L. C. , Boisson, M. N. , Saras, E. , Medaille, C. , Boulouis, H. J. , Madec, J. Y. , & Haenni, M. (2017). OXA‐48‐producing ST372 Escherichia coli in a French dog. The Journal of Antimicrobial Chemotherapy, 72(4), 1256–1258. [DOI] [PubMed] [Google Scholar]
- Mesa Varona, O. , Chaintarli, K. , Muller‐Pebody, B. , Anjum, M. F. , Eckmanns, T. , Norström, M. , Boone, I. , & Tenhagen, B. A. (2020). Monitoring antimicrobial resistance and drug usage in the human and livestock sector and foodborne antimicrobial resistance in six European countries. Infection and Drug Resistance, 13, 957–993. 10.2147/IDR.S237038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metreveli, M. , Bulia, S. , Tevzadze, L. , Tsanava, S. , Zarske, M. , Goenaga, J. C. , Preuß, S. , Lomidze, G. , Koulouris, S. , Imnadze, P. , & Stingl, K. (2022). Comparison of antimicrobial susceptibility profiles of Thermotolerant campylobacter spp. Isolated from Human and Poultry Samples in Georgia (Caucasus). Antibiotics, 11, 1419. 10.3390/antibiotics11101419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo, S. S. , Slettemeås, J. S. , Berg, E. S. , Norström, M. , & Sunde, M. (2016). Plasmid and host strain characteristics of Escherichia coli resistant to extended‐Spectrum Cephalosporins in the Norwegian broiler production. PLoS One, 11(4), e0154019. 10.1371/journal.pone.0154019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monecke, S. , Gavier‐Widén, D. , Hotzel, H. , Peters, M. , Guenther, S. , Lazaris, A. , Loncaric, I. , Müller, E. , Reissig, A. , Ruppelt‐Lorz, A. , Shore, A. C. , Walter, B. , Coleman, D. C. , & Ehricht, R. (2016). Diversity of Staphylococcus aureus isolates in European wildlife. PLoS ONE, 11(12), e0168433. 10.1371/journal.pone.0168433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, J. E. , Barton, M. D. , Blair, I. S. , Corcoran, D. , Dooley, J. S. , Fanning, S. , Kempf, I. , Lastovica, A. J. , Lowery, C. J. , Matsuda, M. , McDowell, D. , McMahon, A. , Millar, B. C. , Rao, J. R. , Rooney, P. J. , Seal, B. S. , Snelling, W. J. , & Tolba, O. (2006). The epidemiology of antibiotic resistance in Campylobacter. Microbes and Infection, 8(7), 1955–1966. [DOI] [PubMed] [Google Scholar]
- Mouftah, S. F. , Cobo‐D'ıaz, J. F. , Alvarez‐Ord'o'nez, A. , Elserafy, M. , Saif, N. A. , Sadat, A. , el‐Shibiny, A. , & Elhadidy, M. (2021). High‐throughput sequencing reveals genetic determinants associated with antibiotic resistance in campylobacter spp. from farm‐to‐fork. PLoS One, 16(6), e0253797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourkas, E. , Florez‐Cuadrado, D. , Pascoe, B. , Calland, J. K. , Bayliss, S. C. , Mageiros, L. , Meric, G. , Hitchings, M. D. , Quesada, A. , Porrero, C. , Ugarte‐Ruiz, M. , Gutierrez‐Fernandez, J. , Domınguez, L. , & Sheppard, S. K. (2019). Gene pool transmission of multidrug resistance among campylobacter from livestock, sewage and human disease. Environmental Microbiology, 21(12), 4597–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandhini, P. , Kumar, P. , Mickymaray, S. , Alothaim, A. S. , Somasundaram, J. , & Rajan, M. (2022). Recent developments in methicillin‐resistant Staphylococcus aureus (MRSA) treatment: A review. Antibiotics (Basel), 11(5), 606. 10.3390/antibiotics11050606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nógrády, N. , Király, M. , Davies, R. , & Nagy, B. (2012). Multidrug resistant clones of ´ salmonella infantis of broiler origin in Europe. International Journal of Food Microbiology, 157(1), 108–112. 10.1016/j.ijfoodmicro.2012.04.007 [DOI] [PubMed] [Google Scholar]
- Pan, Y. , Awan, F. , Zhenbao, M. , Zhang, X. , Zeng, J. , Zeng, Z. , & Xiong, W. (2020). Preliminary view of the global distribution and spread of the tet(X) family of tigecycline resistance genes. Journal of Antimicrobial Chemotherapy, 75(10), 2797–2803. 10.1093/jac/dkaa284 [DOI] [PubMed] [Google Scholar]
- Pauly, N. , Wichmann‐Schauer, H. , Ballhausen, B. , Torres Reyes, N. , Fetsch, A. , & Tenhagen, B. A. (2019). Detection and quantification of methicillin‐resistant Staphylococcus aureus in fresh broiler meat at retail in Germany. International Journal of Food Microbiology, 292, 8–12. 10.1016/j.ijfoodmicro.2018.11.025 [DOI] [PubMed] [Google Scholar]
- Pergola, S. , Franciosini, M. P. , Comitini, F. , Ciani, M. , de Luca, S. , Bellucci, S. , Menchetti, L. , & Casagrande Proietti, P. (2017). Genetic diversity and antimicrobial resistance profiles of campylobacter coli and campylobacter jejuni isolated from broiler chicken in farms and at time of slaughter in central Italy. Journal of Applied Microbiology, 122(5), 1348–1356. [DOI] [PubMed] [Google Scholar]
- Pfeifer, Y. , Cullik, A. , & Witte, W. (2010). Resistance to cephalosporins and carbapenems in gram‐negative bacterial pathogens. International Journal of Medical Microbiology, 300(6), 371–379. 10.1016/j.ijmm.2010.04.005 [DOI] [PubMed] [Google Scholar]
- Pimenta, A. C. , Fernandes, R. , & Moreira, I. S. (2014). Evolution of drug resistance: Insight on TEM β‐lactamases structure and activity and β‐lactam antibiotics. Mini Reviews in Medicinal Chemistry, 14(2), 111–122. [DOI] [PubMed] [Google Scholar]
- Pitout, J. D. D. , Peirano, G. , Kock, M. M. , Strydom, K.‐A. , & Matsumura, Y. (2019). The global ascendency of OXA‐48‐type carbapenemases. Clinical Microbiology Reviews, 33(1), e00102‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portes, A. B. , Rodrigues, G. , Leitão, M. P. , Ferrari, R. , Conte Junior, C. A. , & Panzenhagen, P. (2022). Global distribution of plasmid‐mediated colistin resistance mcr gene in salmonella: A systematic review. Journal of Applied Microbiology, 132(2), 872–889. 10.1111/jam.15282 [DOI] [PubMed] [Google Scholar]
- Pulss, S. , Semmler, T. , Prenger‐Berninghoff, E. , Bauerfeind, R. , & Ewers, C. (2017). First report of an Escherichia coli strain from swine carrying an OXA‐181 carbapenemase and the colistin resistance determinant MCR‐1. International Journal of Antimicrobial Agents, 50(2), 232–236. 10.1016/j.ijantimicag.2017.03.014 [DOI] [PubMed] [Google Scholar]
- Qin, S. , Wang, Y. , Zhang, Q. , Zhang, M. , Deng, F. , Shen, Z. , Wu, C. , Wang, S. , Zhang, J. , & Shen, J. (2014). Report of ribosomal RNA methylase gene erm (B) in multidrug‐resistant campylobacter coli. Journal of Antimicrobial Chemotherapy, 69(4), 964–968. [DOI] [PubMed] [Google Scholar]
- Queenan, K. , Hasler, B. , & Rushton, J. (2016). A one health approach to antimicrobial resistance surveillance: Is there a business case for it? International Journal of Antimicrobial Agents, 48(4), 422–427. 10.1016/j.ijantimicag.2016.06.014 [DOI] [PubMed] [Google Scholar]
- Rafei, R. , Al Kassaa, I. , Osman, M. , Dabboussi, F. , & Hamze, M. (2019). Molecular epidemiology of campylobacter isolates from broiler slaughterhouses in Tripoli. North of Lebanon, British Poultry Science, 60(6), 675–682. 10.1080/00071668.2019.1645945 [DOI] [PubMed] [Google Scholar]
- Ramírez‐Castillo, F. Y. , Guerro‐Barrera, A. L. , & Avelar‐González, F. J. (2023). An overview of carbapenem‐resistant organisms from food‐producing animals, seafood, aquaculture, companion animals and wildlife. Frontiers in Veterinary Science, 10, 1158588. 10.3389/fvets.2023.1158588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch, G. , François, P. , Morisset, D. , Stojanov, M. , Bonetti, E. J. , Schrenzel, J. , Sakwinska, O. , & Moreillon, P. (2013). Humanto‐bovine jump of Staphylococcus aureus CC8 is associated with the loss of a β‐hemolysin converting prophage and the acquisition of a new staphylococcal cassette chromosome. PLoS One, 8(3), e58187. 10.1371/journal.pone.0058187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci, V. , Zhang, D. , Teale, C. , & Piddock, L. J. V. (2020). The O‐antigen epitope governs susceptibility to colistin in salmonella enterica. mBio, 11(1), e02831‐19. 10.1128/mBio.02831-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincon‐Real, A. A. , & Suárez‐Alfonso, M. C. (2022). Carbapenem resistance in critically important human pathogens isolated from companion animals: A systematic literature review. Osong Public Health and Research Perspectives, 13, 407–423. 10.24171/j.phrp.2022.0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo, I. , Fischer, J. , Uelze, L. , Napoleoni, M. , Schiavano, G. F. , Andreoni, F. , & Amagliani, G. (2024). From farm to fork: Spread of a multidrug resistant salmonella Infantis clone encoding blaCTX‐M‐1 on pESI‐like plasmids in Central Italy. International Journal of Food Microbiology, 110490. 10.1016/j.ijfoodmicro.2023.110490 [DOI] [PubMed] [Google Scholar]
- Sahibzada, S. , Abraham, S. , Coombs, G. W. , Pang, S. , Hernandez‐Jover, M. , Jordan, D. , & Heller, J. (2017). Transmission of highly virulent community‐associated MRSA ST93 and livestock‐associated MRSA ST398 between humans and pigs in Australia. Scientific Reports, 7(1), 5273. 10.1038/s41598-017-04789-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakwinska, O. , Giddey, M. , Moreillon, M. , Morisset, D. , Waldvogel, A. , & Moreillon, P. (2011). Staphylococcus aureus host range and human‐bovine host shift. Applied and Environmental Microbiology, 77(17), 5908–5915. 10.1128/AEM.00238-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevellec, Y. , Granier, S. A. , Le Hello, S. , Weill, F. X. , Guillier, L. , Mistou, M. Y. , & Cadel‐Six, S. (2020). Source attribution study of sporadic Salmonella derby cases in France. Frontiers in Microbiology, 11. 10.3389/fmicb.2020.00889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevellec, Y. , Vignaud, M. L. , Granier, S. A. , Lailler, R. , Feurer, C. , le Hello, S. , Mistou, M. Y. , & Cadel‐Six, S. (2018). Polyphyletic nature of salmonella enterica serotype derby and lineage‐specific host‐association revealed by genome‐wide analysis. Frontiers in Microbiology, 9, 891. 10.3389/fmicb.2018.00891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla, S. K. , Karow, M. E. , Brady, J. M. , Stemper, M. E. , Kislow, J. , Moore, N. , Wroblewski, K. , Chyou, P.‐H. , Warshauer, D. M. , Reed, K. D. , Lynfield, R. , & Schwan, W. R. (2010). Virulence genes and genotypic associations in nasal carriage, community‐associated methicillin‐susceptible and methicillin‐resistant USA400 Staphylococcus aureus isolates. Journal of Clinical Microbiology, 48(10), 3582–3592. 10.1128/JCM.00657-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber, R. N. , Larsen, A. R. , Urth, T. R. , Iversen, S. , Møller, C. H. , Skov, R. L. , Larsen, J. , & Stegger, M. (2019). Genome investigations show host adaptation and transmission of LA‐MRSA CC398 from pigs into Danish healthcare institutions. Scientific Reports, 9, 18655. 10.1038/s41598-019-55086-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber, R. N. , Skov, R. L. , Nielsen, J. , Schulz, J. , Price, L. B. , Aarestrup, F. M. , Larsen, A. R. , Stegger, M. , & Larsen, J. (2018). Drivers and dynamics of methicillin‐resistant livestock‐associated Staphylococcus aureus CC398 in pigs and humans in Denmark. mBio, 9(6), e02142‐18. 10.1128/mBio.02142-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, V. , Araújo, S. , Monteiro, A. , Eira, J. , Pereira, J. E. , Maltez, L. , Igrejas, G. , Lemsaddek, T. S. , & Poeta, P. (2023). Staphylococcus aureus and MRSA in livestock: Antimicrobial resistance and genetic lineages. Microorganisms, 11(1), 124. 10.3390/microorganisms11010124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, V. , Capelo, J. L. , Igrejas, G. , & Poeta, P. (2020). Molecular epidemiology of Staphylococcus aureus lineages in wild animals in Europe: A review. Antibiotics, 9(3). 10.3390/antibiotics9030122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, V. , Monteiro, A. , Pereira, J. E. , Maltez, L. , Igrejas, G. , & Poeta, P. (2022). MRSA in humans, pets and livestock in Portugal: Where we came from and where we are going. Pathogens, 11, 1110. 10.3390/pathogens11101110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skallerup, P. , Espinosa‐Gongora, C. , Jørgensen, C. B. , Guardabassi, L. , & Fredholm, M. (2015). Genome‐wide association study reveals a locus for nasal carriage of Staphylococcus aureus in Danish crossbred pigs. BMC Veterinary Research, 11, 290. 10.1186/s12917-015-0599-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skov, R. L. , & Monnet, D. L. (2016). Plasmid‐mediated colistin resistance (mcr‐1 gene): Three months later, the story unfolds. Eurosurveillance, 21(9). 10.2807/1560-7917.ES.2016.21.9.30155 [DOI] [PubMed] [Google Scholar]
- Slettemeås, J. S. , Urdahl, A. M. , Mo, S. S. , Johannessen, G. S. , Grave, K. , Norström, M. , Steinbakk, M. , & Sunde, M. (2017). Imported food and feed as contributors to the introduction of plasmid‐mediated colistin‐resistant Enterobacteriaceae to a “low prevalence” country. Journal of Antimicrobial Chemotherapy, 72(9), 2675–2677. 10.1093/jac/dkx161 [DOI] [PubMed] [Google Scholar]
- Soncini, J. G. M. , Cerdeira, L. , Sano, E. , Koga, V. L. , Tizura, A. T. , Tano, Z. N. , Nakazato, G. , Kobayashi, R. K. T. , Aires, C. A. M. , Lincopan, N. , & Vespero, E. C. (2022). Genomic insights of high‐risk clones of ESBL‐producing Escherichia coli isolated from community infections and commercial meat in southern Brazil. Scientific Reports, 12(9354). 10.1038/s41598-022-13197-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H. , Wan, Y. , Du, P. , & Bai, L. (2020). The epidemiology of monophasic salmonella Typhimurium. Foodborne Pathogens and Disease, 17(2), 87–97. 10.1089/fpd.2019.2676 [DOI] [PubMed] [Google Scholar]
- Tang, Y. , Fang, L. , Xu, C. , & Zhang, Q. (2017). Antibiotic resistance trends and mechanisms in the foodborne pathogen, Campylobacter. Animal Health Research Reviews, 18(2), 87–98. [DOI] [PubMed] [Google Scholar]
- Tang, Y. , Larsen, J. , Kjeldgaard, J. , Andersen, P. S. , Skov, R. , & Ingmer, H. (2017). Methicillin‐resistant and –susceptible Staphylococcus aureus from retail meat in Denmark. International Journal of Food Microbiology, 249, 72–76. 10.1016/j.ijfoodmicro.2017.03.001 [DOI] [PubMed] [Google Scholar]
- Tate, H. , Hsu, C. H. , Chen, J. C. , Han, J. , Foley, S. L. , Folster, J. P. , & Zhao, S. (2022). Genomic diversity, antimicrobial resistance, and virulence gene profiles of salmonella Serovar Kentucky isolated from humans, food, and animal ceca content sources in the United States. Foodborne Pathogens and Disease, 19(8), 509–521. [DOI] [PubMed] [Google Scholar]
- Törneke, K. , Torren‐Edo, J. , Grave, K. , & Mackay, D. K. (2015). The management of risk arising from the use of antimicrobial agents in veterinary medicine in EU/EEA countries – A review. Journal of Veterinary Pharmacology and Therapeutics, 38(6), 519–528. 10.1111/jvp.12226 [DOI] [PubMed] [Google Scholar]
- Touati, A. , Mairi, A. , Baloul, Y. , Lalaoui, R. , Bakour, S. , Thighilt, L. , Gharout, A. , & Rolain, J. M. (2017). First detection of Klebsiella pneumoniae producing OXA‐48 in fresh vegetables from Bejaïa city, Algeria. Journal of Global Antimicrobial Resistance, 9, 17–18. 10.1016/j.jgar.2017.02.006 [DOI] [PubMed] [Google Scholar]
- University hospital of Skåne . (2022). Skånes universitetssjukhus utreder smitta av salmonella [Internet]. Press release (in Swedish). Malmö: 8 Apr 2022 (cited 4 Dec 2023). https://www.mynewsdesk.com/se/sus/pressreleases/skaanes‐universitetssjukhus‐utreder‐smitta‐av‐salmonella‐3174767
- Voss, A. , Loeffen, F. , Bakker, J. , Klaassen, C. , & Wulf, M. (2005). Methicillin‐resistant Staphylococcus aureus in pig farming. Emerging Infectious Diseases, 11(12), 1965–1966. 10.3201/eid1112.050428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, B. , Klein, K.‐S. , Barton, A.‐K. , Semmler, T. , Huber, C. , Merle, R. , Tedin, K. , Mitrach, F. , Lübke‐Becker, A. , & Gehlen, H. (2018). Equine methicillin‐resistant sequence type 398 Staphylococcus aureus (MRSA) harbor mobile genetic elements promoting host adaptation. Frontiers in Microbiology, 9, 2516. 10.3389/fmicb.2018.02516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Liu, F. , Xu, X. , Huang, H. , Lyu, N. , Ma, S. , Chen, L. , Mao, M. , Hu, Y. , Song, X. , Li, J. , Pan, Y. , Wang, A. , Zhang, G. , Zhu, B. , & Gao, G. F. (2021). Detection of plasmid‐mediated tigecycline resistance gene tet(X4) in a salmonella enterica serovar llandoff isolate. Infectious Microbes & Diseases, 3(4), 198–204. 10.1097/IM9.0000000000000077 [DOI] [Google Scholar]
- Wang, Y. , Zhang, M. , Deng, F. , Shen, Z. , Wu, C. , Zhang, J. , Zhang, Q. , & Shen, J. (2014). Emergence of multidrug‐resistant campylobacter species isolates with a horizontally acquired rRNA methylase. Antimicrobial Agents and Chemotherapy, 58(9), 5405–5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, A. L. , Selinger, L. B. , Taboada, E. N. , & Inglis, G. D. (2018). Subtype‐specific selection for resistance to fluoroquinolones but not to tetracyclines is evident in campylobacter jejuni isolates from beef cattle in confined feeding operations in southern Alberta, Canada. Applied and Environmental Microbiology, 84(7), e02713‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, B. , & Kang, M. (2018). Molecular basis of macrolide resistance in campylobacter strains isolated from poultry in South Korea. BioMed Research International, 2018, 4526576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendlandt, S. , Schwartz, S. , & Silley, P. (2013). Methicillin‐resistant Staphylococcus aureus: A food‐borne pathogen? Annual Review of Food Science and Technology, 4, 117–139. 10.1146/annurev-food-030212-182653 [DOI] [PubMed] [Google Scholar]
- WHO (World Health Organization) . (2019a). Critically important antimicrobials for human medicine. 6th revision. World Health Organisation. https://apps.who.int/iris/bitstream/handle/10665/312266/9789241515528‐eng.pdf?ua=1 [Google Scholar]
- WHO (World Health Organization) . (2020). GLASS whole‐genome sequencing for surveillance of antimicrobial resistance. World Health Organisation. https://www.who.int/publications‐detail‐redirect/9789240011007 [Google Scholar]
- Wieczorek, K. , & Osek, J. (2013). Antimicrobial resistance mechanisms among campylobacter. Biomed Research International, 2013, 340605. 10.1155/2013/340605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaghoubi, S. , Zekiy, A. O. , Krutova, M. , Gholami, M. , Kouhsari, E. , Sholeh, M. , Ghafouri, Z. , & Maleki, F. (2022). Tigecycline antibacterial activity, clinical effectiveness, and mechanisms and epidemiology of resistance: Narrative review. European Journal of Clinical Microbiology & Infectious Diseases, 41(7), 1003–1022. 10.1007/s10096-020-04121-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Feye, K. M. , Shi, Z. , Pavlidis, H. O. , Kogut, M. , Ashworth, J. A. , & Ricke, S. C. (2019). A historical review on antibiotic resistance of foodborne campylobacter. Frontiers in Microbiology, 10, 1509. 10.3389/fmicb.2019.01509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, H. , Shen, Z. , Wang, Y. , Deng, F. , Liu, D. , Naren, G. , Dai, L. , Su, C. C. , Wang, B. , Wang, S. , Wu, C. , Yu, E. W. , Zhang, Q. , & Shen, J. (2016). Emergence of a potent multidrug efflux pump variant that enhances campylobacter resistance to multiple antibiotics. MBio, 7(5), e01543‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X. , Ju, Z. , Wang, G. , Yang, J. , Wang, F. , Tang, H. , Zhao, X. , & Sun, S. (2021). Prevalence and antimicrobial resistance of salmonella isolated from dead‐in‐Shell chicken embryos in Shandong, China. Frontiers of Veterinary Science, 16(8), 581946. 10.3389/fvets.2021.581946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurfluh, K. , Poirel, L. , Nordmann, P. , Klumpp, J. , & Stephan, R. (2015). First detection of Klebsiella variicola producing OXA‐181 carbapenemase in fresh vegetable imported from Asia to Switzerland. Antimicrobial Resistance and Infection Control, 4(1), 38. 10.1186/s13756-015-0080-5 [DOI] [PMC free article] [PubMed] [Google Scholar]


