Abstract
Approximately 20% of colorectal cancer (CRC) patients present with metastasis at diagnosis. Among Stage I-III CRC patients who undergo surgical resection, 18% typically suffer from distal metastasis within the first three years following initial treatment. The median survival duration after the diagnosis of metastatic CRC (mCRC) is only 9 mo. mCRC is traditionally considered to be an advanced stage malignancy or is thought to be caused by incomplete resection of tumor tissue, allowing cancer cells to spread from primary to distant organs; however, increasing evidence suggests that the mCRC process can begin early in tumor development. CRC patients present with high heterogeneity and diverse cancer phenotypes that are classified on the basis of molecular and morphological alterations. Different genomic and nongenomic events can induce subclone diversity, which leads to cancer and metastasis. Throughout the course of mCRC, metastatic cascades are associated with invasive cancer cell migration through the circulatory system, extravasation, distal seeding, dormancy, and reactivation, with each step requiring specific molecular functions. However, cancer cells presenting neoantigens can be recognized and eliminated by the immune system. In this review, we explain the biological factors that drive CRC metastasis, namely, genomic instability, epigenetic instability, the metastatic cascade, the cancer-immunity cycle, and external lifestyle factors. Despite remarkable progress in CRC research, the role of molecular classification in therapeutic intervention remains unclear. This review shows the driving factors of mCRC which may help in identifying potential candidate biomarkers that can improve the diagnosis and early detection of mCRC cases.
Keywords: Colorectal cancer, Metastasis cascade, Cancer immunity, Genomic variation, Epigenetic instability, Lifestyle factor
Core Tip: Metastatic colorectal cancer (CRC) is traditionally considered to be an advanced stage malignancy or is thought to be caused by incomplete resection of tumor tissue, allowing cancer cells to spread from primary to distant organs; however, increasing evidence suggests that this process can begin early during tumor development. CRC patients exhibit high heterogeneity and diverse cancer phenotypes that are classified based on molecular and morphological alterations. Different genomic and non-genomic events can induce sub-clone diversity, which leads to cancer and metastasis.
INTRODUCTION
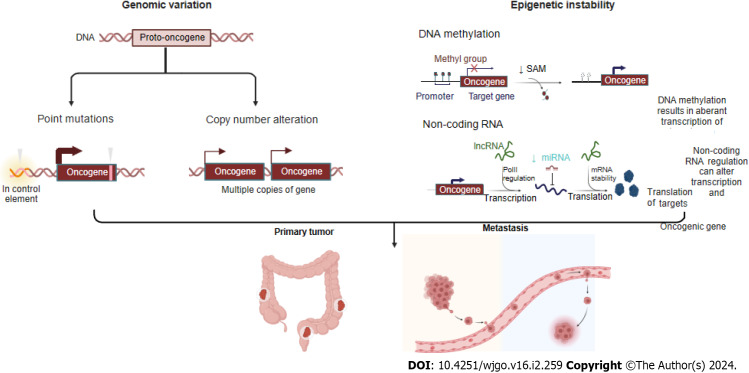
Colorectal cancer (CRC) is the second leading cause of cancer-related mortality and the third most common malignancy worldwide[1]. Aging and an unhealthy diet and lifestyle are risk factors for CRC[2]. In all metastatic CRC (mCRC) patients, the most common metastasis is found in the liver, followed by the lung and peritoneum, while metastases are found less frequently in the brain[3]. The median survival duration after diagnosis of mCRC patients is only 9 mo. Although mCRC is traditionally considered to be caused by incomplete resection of tumor tissue or late-stage malignancy-related events, increasing evidence has demonstrated that the mCRC process can begin early during tumor development and that the genomic characteristics of these metastases typically impact cancer treatment. CRC patients exhibit high heterogeneity that manifests as different cancer phenotypes, which are classified in terms of morphological and molecular variations[4]. Continuous accumulation of mutations leads to genotype diversity and promotes cancer evolution. Genomic variation, including driving point mutations, putative copy number alterations (CNVs), structural gene variants, and epigenetic instability, affects transcription factor expression, disrupts DNA methylation, and alters chromatin accessibility, which can lead to subclone divergence and thus supply energy for cancer evolution and metastasis (Figure 1). Through metastatic cascades, circulating cancer cells can move from an original tumor site to distant organs in the body and develop metastases[5]. Therefore, identifying the driving biological factors of mCRC and measuring them as biomarkers or potential treatment targets for early intervention to prevent metastasis and even for late-stage treatment of advanced mCRC are extremely important.
Figure 1.
Different genomic events, including point mutations, copy number alterations, and epigenetic instability (such as disruption to DNA methylation and differential splicing based on noncoding RNA), can engender sub clonal diversity, thus providing fuel for primary tumor and its metastasis. Created with BioRender.com.
GENOMIC VARIATION OF MCRC
Abundant molecular data on genomic variations have been obtained from both nonmCRC and mCRC samples, and some studies have focused on characterizing the molecular abnormalities in mCRC.
Driving mutations in mCRC
The mutational patterns and overall mutational burdens of primary and metastatic cancers are largely consistent, as shown by comparisons of CRC cohorts[6]. Analysis of the mutation frequencies of certain genes in early stage tumors, mCRC tumors, and metastatic mCRC tumors from distal organs revealed that most of these mutations did not lead to significant differences, but a few were associated with tumor stage[4]. Indeed, the accumulated alterations in these oncogenes play a crucial role in mCRC tumorigenesis and progression. However, whether mutations in these key oncogenes affect CRC metastasis remains unclear.
To identify oncogenes associated with the progression of metastases, Yaeger et al[7] compared the frequency of oncogenic variations in primary CRC tumors that did or did not develop metastasis in the TCGA and Memorial Sloan Kettering cohorts and identified 42 genes that were recurrently mutated to a significant degree in these cohorts. Alterations in TP53 were the only genomic mutations significantly enriched in mCRC samples, while alterations in FBXW7 were enriched in the early stage compared with mCRC samples, suggesting a potential protective role for FBXW7. A similar result was found by Vakiani et al[8] Among mCRC patients, TP53 mutations were notably more frequent, while BRAF mutations were less frequent. The frequencies of KRAS and PIK3CA mutations were not significantly different between primary CRC and mCRC samples. Li et al[9] conducted a study comparing a Chinese CRC (CCRC) patient dataset with a TCGA CRC dataset. Among CCRC patients, the proportion of mCRC patients was much greater than that in the TCGA CRC dataset (47.9% vs 14.4%), and uniquely, SMAD4, a pivotal factor in the TGF-beta signaling pathway, tended to promote distant metastasis of CRC tumors[10] and was significantly more common in mCRC patients in the CCRC dataset than in mCRC patients in the TCGA dataset (20.0% vs 6.6%; P = 0.015)[9]. A study by Huang et al[10] revealed no significant associations between mutations in APC or PIK3CA in mCRC patients. Subgroup analyses stratified by ethnicity showed that in Asian populations, KRAS, BRAF, and TP53 mutations, including lymph node and distant metastasis, were associated with mCRC, whereas only TP53 mutations promoted mCRC in Caucasians. Jo et al[11] investigated the difference between mCRC primary tumors and mCRC metastatic tissues and identified a significant concordance of KRAS mutation status in 81.18% (9/11) of patients (P = 0.03271). Only two patients showed intertumor heterogeneity. This intertumoral heterogeneity may be attributable to KRAS mutations early in disease progression from colorectal adenoma to malignant disease, leading to a continuous tumor growth advantage, whereas TP53 is mutated relatively late during CRC progression[12].
CNVs and mCRC
Increased CNVs, indicative of chromosomal instability (CIN), are strongly correlated with metastatic burden, and some chromosomes are significantly more unstable in some tumor metastases[13]. However, in CRC, Nguye et al[13] showed that CNV was not correlated with metastatic burden in colorectal adenocarcinoma. Similarly, another study showed that the most frequent CNV did not differ between mCRC and nonmCRC samples[9]. However, several studies have reported exceptions; for example, a study by Casimiro et al[14] showed a greater frequency of MTDH amplification (copy number > 1.8) in mCRC patients with lung metastasis than in nonmCRC patients (17.4% vs 100.0%, P < 0.001). Another prospective study showed that MYC was strongly amplified in mCRC patients with microsatellite stable (MSS) disease[13]. A study by Liu et al[15] did not directly show CNV differences but classified chromosomal stable (CS) and CIN. The ratio of CIN to CS was found to be significantly increased in Stage III and Stage IV samples, which may explain why high-level CNV causes CIN and thus leads to CRC metastasis.
EPIGENETIC INSTABILITY IN MCRC
Together, epigenetic instability and genetic alterations function to drive the progression of normal cells into cancer cells. Epigenetic instability is caused by several mechanisms, such as DNA methylation of cytosine bases in CG-rich sequences, which results in aberrant transcription of target genes; regulation of noncoding RNA, which can alter the transcription of oncogenic gene targets; and posttranslational modifications of histones, which regulate structural changes in packed DNA known as chromatin[16].
Methylation and mCRC
Many studies have attempted to identify genomic metastasis-promoting mutations. However, few metastasis-associated mutations have been found, and those that have been reported to date exhibit largely concordant mutation patterns and mutation burdens in both primary and metastatic cancers. Therefore, genomic mutations are not mediators of cascade-related metastatic progression[6]. However, several studies have shown unique features of a significantly greater frequency of CpG island hypermethylation in gastrointestinal (GI) adenocarcinomas than in non-GI adenocarcinomas[15], and marked differences in methylation were found in CRC patients. Therefore, Toyota et al[16] named one CRC type, CpG island methylated subphenotype (CIMP), after observing its association with a unique molecular pathogenesis. It is widely accepted that aberrant gene hypermethylation is associated with tumor suppressor gene silencing, which leads to cancer formation via transcriptional repression of these genes[17]. Studying mCRC, Ju et al[18] showed that the number of methylated genes was markedly greater in Stage I-III CRC than in the other two stage groups, namely, the stage IV primary mCRC and liver metastasis groups. However, when several studies compared primary CRC tumors and CRC liver metastases, the methylated phenotype was similar to that of primary cancer, which led researchers to conclude that most of the key DNA hypermethylation events associated with colorectal carcinogenesis likely occur before cancer cells spread to metastatic organs[19-21].
Furthermore, several studies have shown that differentially methylated genes, such as hypermethylated MGMT, which repairs alkylated DNA[19]; hypermethylated TIMP3, which increases the EGFR signaling pathway by inhibiting MMPs and increasing the TNF signaling pathway by inhibiting ADAM[21]; and hypomethylated nuclear element-1 (LINE-1) enables the inadvertent activation of methylation-silenced proto-oncogenes in mCRC[20,22]. In summary, aberrantly methylated genes play important roles in the initiation and progression of mCRC and can even interfere with drug responses[15].
Noncoding RNAs and mCRC
Previously, 98% of noncoding sequences were considered useless, but increasing evidence has shown that these RNA transcripts strongly impact different physiological and pathological processes[23]. To date, more than 1400 miRNAs have been identified, accounting for 2%-5% of the entire human genome and regulating 30% of human gene expression[24]. Recently, several studies have been performed to determine the regulatory roles of noncoding RNAs (ncRNAs) in different steps of the colorectal metastasis cascade and how ncRNAs coordinate a series of pathological events.
A study by Mokutani et al[25] showed that downregulation of miR-132 by targeting downstream ANO1, which is a key oncogenic factor, contributed to the CRC metastatic cascade. A study by Hu et al[26] showed that low expression of miR-744, which is a target of Notch1, was positively correlated with TNM stage; in contrast, overexpression of miR-744 significantly inhibited the proliferation and invasion of CRC cells. A study by Yan et al[27] indicated that miR-520d-5p was significantly downregulated by targeting CTHRC1, which is involved in the progression and metastasis of CRC. Additionally, it is regulated by SP1 and affects the epithelial-mesenchymal transition (EMT) by inactivating the phosphorylation of Erk1/2. A study indicated that significantly upregulated miR-425 and miR-576, which target PTEN, were key factors in CRC liver metastasis[26,28].
Liang et al[29] reported that the long non-coding RNA (lncRNA) RPPH1 was markedly upregulated in CRC tissues and RPPH1 overexpression induced EMT in CRC cells by interacting with TUB3 to prevent its ubiquitination, an outcome that has been associated with advanced TNM stage and poor prognosis. Two studies identified functional roles for CYTOR (a lncRNA also known as LINC00152) in CRC progression. The first of these two studies, conducted by Wang et al[30], showed that CYTOR forms a heterotrimeric complex with the RNA-binding proteins NCL and Sam68 through EXON1, which activates the NF-κB signaling pathway, thus promoting EMT and CRC metastasis. The second study, conducted by Lv et al[31], showed that CYTOR mediates the binding of ENO2 to large tumor suppressor 1 (LATS1) and competitively inhibits the phosphorylation of Yes-associated protein 1 (YAP1), which ultimately triggers the EMT and CRC metastasis.
MSI and mCRC
A series of studies led to the understanding that deficient DNA mismatch repair (dMMR) causes microsatellite instability (MSI); importantly, most sporadic MSI-type CRCs are derived from CIMP-type CRC[15]. A study by Tieng et al[32] showed that only 4% of mCRC patients exhibited the MSI genotype, a significantly lower frequency than that reported for nonmCRC patients, which can be explained by the MSI subtype showing a much lower tendency to metastasize. More detailed data from the study by Liu et al[15] showed that in patients with Stage IV CRC, the percentage of patients with MSI was 28%, and that of patients with MSS was 72%, with the same ratio of MSI-to-MSS in patients with Stage III CRC. These ratios were significantly different from those identified in patients with Stage I or II CRC; 42% presented with the MSI type, and 58% presented with the MSS type. Accordingly, MSI-type patients, including mCRC and nonmCRC patients, exhibited a longer OS[15]. Moreover, in the lower GI tract, CIMP-H- and MSI-type tumors were largely absent in the descending colon compared with their incidence in the upper GI tract[33].
Recent studies showing the efficacy of immune checkpoint inhibitor therapy in dMMR-MSI-high CRC patients have indicated promising effects, but its use in patients with the MSS subtype has either been unsuccessful or not widely explored[34]. The likely explanation for the discriminative efficacy of this approach in patients with the MSI genotype is because it is associated with the next-highest IFN-γ expression signature[35], and diverse immune signatures, such as high expression levels of the checkpoint protein CD276[15], can lead to the accumulation of mutations and subsequent immunogenic neoantigens[36], inducing high cytotoxic T cells (CTLs) engagement and better clinical outcomes[37]. Mechanistically, the group of patients with MSI-type CRC harbored lower WNT expression signatures than did the other CRC groups, a finding that may have been attributable to reduced metastasis[15]. Our understanding is that among mCRC patients, a much greater incidence of the MSS subtype was identified, and patients with this subtype exhibited a lower immune response (Figure 2).
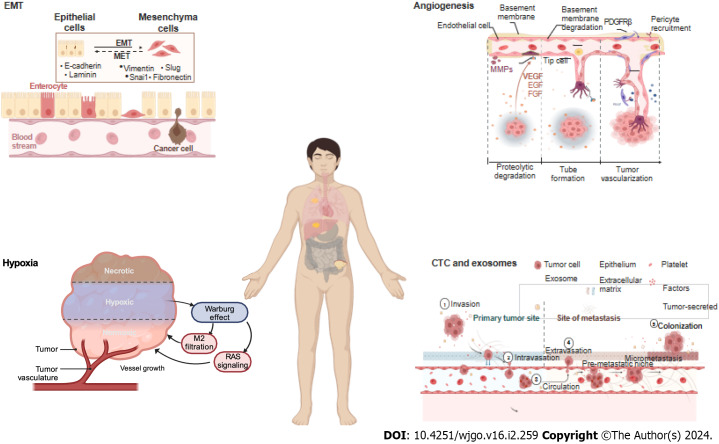
Figure 2.
Key factors of metastasis: Epithelial-mesenchymal transition, angiogenesis, hypoxia, circulating tumor cells and exosomes. EMT: Epithelial-mesenchymal transition; CTC: Circulating tumor cells. Created with BioRender.com.
MCRC AND METASTATIC CASCADES
To metastasize, a series of metastatic cascades, such as invasive migration via the circulatory system, extravasation, distal cell seeding, dormancy and reactivation, characterize cancer cells, with each step triggered by specific functions. In these cascades, EMT, angiogenesis, hypoxia, and circulating tumor cells (CTC) and exosomes are the key factors (Figure 2).
mCRC and EMT
The EMT is a critical process during cancer metastasis in which epithelial cells acquire the phenotype of mesenchymal stem cells. Cells undergoing the EMT are at the invasive front of a cancerous tumor, and the EMT drives cancer cell migration, invasion, and metastatic spread[38]. Multiple signaling pathways, such as the TGF-β, BMP, RTK, Wnt/β-catenin, Notch, Hedgehog, and STAT3 pathways as well as ECM-mediated and hypoxia-regulated pathways, are involved in the EMT[39]. Members of the Snail family of transcriptional regulators, namely, Snail1 and Snail2, and the zinc finger transcription factors Zeb1 and Zeb2 also make critical regulatory contributions to the EMT[40].
Several studies on mCRC have shown the impact of genetic and relevant signaling pathways on EMT and the role of EMT in cell migration from the original site to metastatic sites (Table 1)[41-49].
Table 1.
Epithelial-mesenchymal transition signaling pathways
|
Target gene
|
Signaling pathways
|
Transcription factor
|
Intervention
|
| Intracellular AGR2[41] | KDELR-Gs-PKA | Upregulates the expression of Snail and Slug | AGR2 upregulation mediated by the PGE2-EP4-PI3k-AKT |
| Depletion of DRD2[42] | β-catenin | Zeb1 | A DRD2 antagonist, pimozide |
| DKK1[43] | Wnt/β-catenin | S1004 (transcriptional cross-regulation) | Expression of S1004 downregulates DKK1 |
| BEX2[44] | Hedgehog | Zic2 | Negatively modulated by Zic2 retention |
| PLK4[45] | Wnt/β-catenin | N-cadherin and snail | Knockdown decreases the levels of EMT-associated factors |
| FRA1[46] | RAS-ERK and TGFβ | ||
| c-MYC[47] | |||
| SIRT1[48] | Snail | ||
| STAT3[49] | Snail and slug | / |
EMT: Epithelial-mesenchymal transition.
mCRC and angiogenesis
Angiogenesis is a hallmark of cancer and is closely related to tumor growth, cancer cell metastasis and invasion, prognosis, and recurrence. Angiogenesis, which involves the formation of new vessels from preexisting vessels, is critical for the progression of both primary and metastatic cancer[50]. At metastatic sites, angiogenesis enables malignant cells to repeat the entire sequence of events required for further metastasis[51].
Vascular endothelial growth factor A is a major proangiogenic factor that is associated with metastasis formation and poor prognosis in CRC patients[52]. Many studies have shown that the VEGF signaling pathway has high therapeutic value because it regulates angiogenesis in CRC patients. Nogués et al[53] reported that VEGF serum levels in mCRC patients before surgery were significantly greater than those in nonmCRC patients. Therefore, antiangiogenic therapeutic strategies are important and effective tools for improving outcomes in patients diagnosed with mCRC in specific settings[54]. Many drugs targeting VEGF, platelet-derived growth factor, fibroblast growth factor, and their receptors are marketed or are under development[55].
mCRC and hypoxia
The metastatic process exerts strong selective pressure on cancer cells, and metastatic cancer cells develop high oxidative stress. However, the Warburg effect helps cancer cells minimize oxidative stress by inhibiting mitochondrial oxidative metabolism, thereby promoting metastatic spread[56]. As a hallmark of cancer, the Warburg effect facilitates CRC metastasis by promoting angiogenesis, promoting cancer-associated fibroblast formation, and suppressing the immune system, and it can also lead to drug resistance[57,58].
Qi et al[59] analyzed a total of 1730 CRC samples and were able to classify most of them into two subgroups: a hypoxia subgroup and a normoxia subgroup. They found that hypoxia was associated with poor prognosis in CRC and was closely associated with activation of the RAS signaling pathway independent of KRAS mutation. Furthermore, hypoxia promoted M2 macrophage infiltration and was associated with poor outcomes[59]. In addition, other researchers have shown that hypoxic cell-derived miR-135a-5p exosomes promote CRC liver metastasis by suppressing the kinase 2- YAP1-matrix metalloproteinase 7 axis[60], and hypoxic cell-derived circ-133 exosomes promote cancer metastasis by acting on the miR-133a/GEF-H1/RhoA axis[61].
CTCs and exosomes
Clearly, only a small portion of CTCs can undergo metastasis, while multiple mechanisms in heterogeneous CTCs facilitate their metastatic potential by driving CTC interactions with immune and stromal cells[62]. First, Chiu et al[63] showed that, compared with carcinoembryonic antigen (CEA) alone, CTC detection increases the power of the area under the receiver operating characteristic curve for predicting mCRC (0.7800 vs 0.8378). Surprisingly, Le et al[64] showed that CTCs were not associated with the size or number of metastases, as determined after previously administered drug therapy, or disease-free survival (DFS). Only tumor marker-positive thoracic lymph nodes were associated with the presence of CTCs in pulmonary venous blood, and CTCs were present in all patients (Fisher’s exact test, P = 0.02). Moreover, researchers have shown that the results of overall and DFS analyses are not different regardless of whether CTC marker expression or CTC number is considered[64]. However, some studies have shown that heterogeneous CTCs promote metastasis. Gkountela et al[65] demonstrated that CTC clustering altered DNA methylation, specifically hypomethylation of the binding sites of OCT4, NANOG, SOX2, and SIN3A, which is similar to what has been observed in embryonic stem cells, and promoted stemness and metastasis. Hamid et al[66] showed that the potential target gene AKT1 was expressed at a significantly (P = 0.0129) greater level in single CTCs from Stage III or IV samples than in early stage samples.
Exosomes, as primary communication mediators, are extracellular vesicles with multiple biological functions[67]. Exosomes play important roles in the development of mCRC, including enhancing tumorigenicity; promoting angiogenesis, cancer cell proliferation, and endothelial cell migration; and establishing an immunosuppressive environment[68]. Several studies have demonstrated that tumor-derived exosomes and their functions in CRC metastasis. For example, miR-27b-3p-enriched exosomes increase the permeability of blood vessels and facilitate CTC generation[69], and miR-25-3p-, miR-130b-3p-, miR-425-5p-, miR-934-, and RPPH1-enriched exosomes induce M2 polarization of macrophages and promote cancer metastasis[29,70,71]. In addition, miR-200b-enriched exosomes are transferred to a targeted cell to increase CRC cell proliferation by directly targeting the 3′-UTRs of p27 and RND3[72], and ANGPTLI-enriched exosomes reprogram Kupffer cells and decrease MMP9 expression to hinder vascular leakiness in liver premetastatic niches[73]. HSPC11-enriched exosomes reprogram lipid metabolism in cancer-associated fibroblasts (CAFs) to facilitate premetastatic niche formation and liver metastasis[74]. circLONP2 exosomes modulate miR-17-5p intracellular maturation and intercellular transfer and are subsequently internalized by adjacent cells to increase their metastatic ability[75], and circPACRGL exosomes play an oncogenic role in CRC proliferation and metastasis[76].
In addition to tumor cell-derived exosomes, nontumor cell-derived exosomes also play roles in CRC metastasis. Hu et al[77] showed that CAF-derived miR-92a-3p exosomes promoted cell stemness, the EMT, metastasis and chemoresistance in CRC cells by activating the Wnt/beta-catenin signaling pathway and inhibiting mitochondrial apoptosis through FBXW7 and MOAP1 inhibition. Ren et al[78] also showed the same functions in CAF-derived H19 exosomes by activating the Wnt/beta-catenin signaling pathway and acting as competing endogenous RNA sponges for miR-141, while miR-141 inhibited the stemness of CRC cells. In addition, several researchers have initiated exosome therapy studies with CRC samples. MiR-155-enriched exosomes, also called dendritic cell (DC) immunotherapy, induce antitumor immune responses and prolong survival in a CRC mouse model by increasing the expression of principal cytokines[79].
MCRC AND IMMUNITY
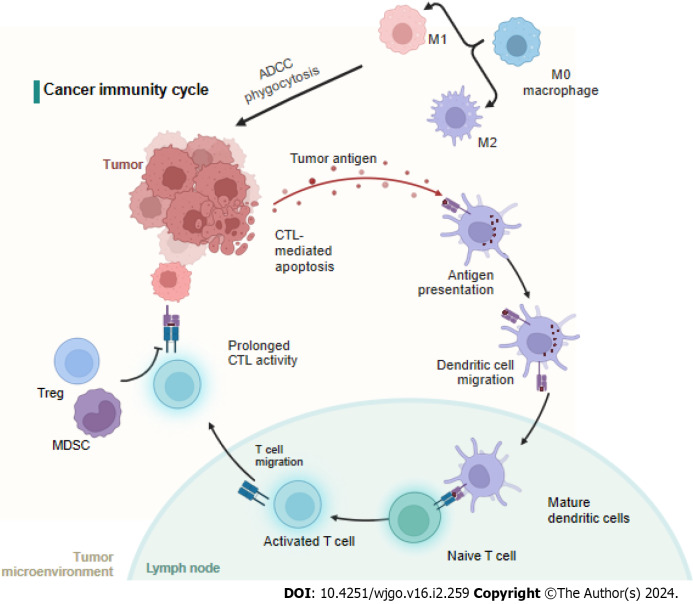
The cancer-immunity cycle includes neoantigen generation during tumorigenesis, DC processing and antigen presentation, which activate effector T cells that respond to cancer antigens and kill targeted cancer cells (Figure 3). However, in cancer patients, the cancer-immunity cycle is not optimal due to low neoantigen levels and subsequent failure to activate T cells or suppress immunity[80]. It is widely accepted that immune cells in the tumor microenvironment (TME) significantly affect the progression of CRC at the primary tumor site and metastatic sites[81]. For CRC patients, the immunoscore, which is based on quantified immune cell infiltration, has been shown to be superior to the current staging system[82,83], and immunoscore diagnostic kits have been approved for CRC patients[84] (Figure 3).
Figure 3.
Neoantigens created by oncogenesis are released and captured by dendritic cell for processing. Dendritic cells present the captured antigens on MHCI and MHCII molecules to T cells, resulting in the priming and activation of effector T-cell responses against the cancer-specific antigens. Finally, the activated effector T cells move toward and infiltrate to the tumor bed. MDSC: Myeloid-derived suppressor cells; ADCC: Antibody dependent cell mediated cytotoxicity; CTC: Circulating tumor cells; CTL: Cytotoxic T cell. Created with BioRender.com.
mCRC and innate immunity
M0 macrophages are polarized to the M1 subtype through the action of LPS and IFN-γ or to the M2 subtype after induction by IL-4, IL-10, and IL-13[85-87]. Studies have shown that the M1 subtype can increase immunity by recruiting CTLs and inducing cancer cell apoptosis through phagocytosis, antibody dependent cell mediated cytotoxicity, and the release of TNF and nitric oxide. In contrast, the M2 subtype leads to angiogenesis, the EMT, and immunosuppression to promote CRC metastasis[88]. Interestingly, the number of circulating or tumor-infiltrating NK cells is inversely associated with metastasis in CRC patients[89]. However, another study showed that NK cells rarely infiltrated tumors, but a high number of tumor-associated NK cells correlated with good clinical outcomes in CRC patients[90]. Moreover, the number of tumor-infiltrating NKT cells was positively correlated with good clinical outcomes in CRC patients[91].
mCRC and adaptive immunity
By presenting tumor antigens, DCs induce specific polarization of T lymphocytes into different subsets of cells. However, the mechanisms by which these DCs enhance invasion are unclear. However, as observed in CRC, tumor-infiltrating DCs exhibit distinct patterns of tumor infiltration according to their maturation status, which can partially explain their highly variable prognostic value[92]. One study showed DC-related outcomes in which an increased density of CD208-positive DCs was associated with worsened disease outcomes in CRC patients[83].
CTLs are important components of antitumor immunity, and the number of CTLs among infiltrating cells correlates with low recurrence in CRC patients[93,94]. Similarly, Lazarus et al[95] showed that an increased number of CTLs is more frequently associated with epithelial cells in the tumor microenvironment of MMR-deficient mCRC and prolongs overall survival. Bindea et al[96] reported that increased T-cell infiltration was associated with reduced metastasis, which likely reflects ongoing adaptive immune pressure on tumor development and spread. In general, CD4+ helper T cells modulate the positive effects of the cancer immune response by secreting cytokines[97]. T-helper 1 (TH1) cells enhance CTL effectors, and T-helper 17 (TH17) cells exhibit immunosuppressive effects, leading to poor clinical outcomes[98]. Amicarella et al[99] demonstrated that IL-17 derived from TH17 cells promoted the production of protumor genetic factors, while IL-8 derived from TH17 cells induced cytotoxic CCR5+CCR6+CD8+ T-cell infiltration into the CRC tumor microenvironment and recruited neutrophils, CC-chemokine ligand 5 (CCL5), and CCL20. Moreover, two studies have shown that, in mCRC samples, the T-cell density is lower and the B-cell density is greater than those in nonmCRC tumor microenvironments[100,101].
Regulatory T cells (Tregs) characterized by CD25 and FoxP3 expression are considered potent mediators of immunosuppression. However, in CRC, Tregs have complex effects on the TME, and evidence suggests that their protumorigenic and antitumorigenic functions are context specific[81]. The number of myeloid-derived suppressor cells in peripheral blood was positively associated with cancer stage in CRC patients[102,103]. Furthermore, fully differentiated neutrophils with increased granule density and increased CD66b+ neutrophils were associated with better outcomes in CRC patients[104,105].
MCRC AND LIFESTYLE FACTORS
In addition to internal biological factors, external lifestyle factors, such as overweight/obesity, physical inactivity, cigarette smoking, alcohol consumption and inappropriate dietary patterns, also influence metabolism, cell survival, tumor progression and metastasis in CRC patients[106].
mCRC and body mass index
A high body mass index (BMI) is a convincing risk factor for the development of CRC, and the overall CRC risk is estimated to increase by 3% for every five kilograms of weight gain[107]. Among the mCRC patient cohort, 63% and 27% were overweight with a BMI > 25 kg/m2 and BMI > 30 kg/m2, respectively[108]. Mechanistically, this could be explained by the fact that the adipose tissue of patients releases more unfavorable factors, such as TNF-α, IL-1, IL-6, IL-7, and IL-8, which inhibit apoptosis, promote oxidative stress, suppress the immune response, and reduce the activity of the IGF-1 axis; these factors are also associated with cancer development and progression[109]. Another study showed that overweight and obese mCRC patients who were receiving therapies targeting VEGF had poorer outcomes with bevacizumab[108]. However, a low BMI is associated with an increased risk of progression and death among the patients enrolled in mCRC trials, while there is no increased risk for an elevated BMI, in contrast to the adjuvant setting in mCRC patients[110].
mCRC and dietary intake
Studies have shown that regular consumption of red and processed meat is an important risk factor that may increase the risk of mCRC by approximately 17% for every 100 g portion of red meat and approximately 18% for every 50 g of processed meat eaten daily; moreover, it was shown that high consumption of dietary fiber could reduce the risk of CRC development by up to 50%[109]. However, more than 80% of Stage III and metastatic CRC patients fail to meet the US FDA recommended daily intake of vegetables, fruits, and milk products[107]. Mechanistically, harmful substances, such as heterocyclic amines and polycyclic aromatic hydrocarbons, generated from grilled and roasted meat have the potential to cause point mutations (deletions, insertions, and substitutions). Similarly, nitrosamines and nitrosamides are potent carcinogenic agents that can react with DNA. In contrast, the potential mechanism of the protective effect of fiber consumption on CRC development occurs through reducing contact between carcinogenic substances and the colonic epithelium as well as stimulating the growth of beneficial gut microbiota.
mCRC and lifestyle habits
Tobacco smoke is an established risk factor for the development of many types of cancer, including CRC. Smoking cigarettes increases the risk of developing CRC by 2- to 3-fold compared with nonsmokers[111]. The mCRC cohort represented 9% of the smokers, while the Stage III cohort represented 10% of the smokers; these two cohorts were not significantly different[107]. Tobacco contains a mixture of thousands of chemicals, more than 60 of which are well-established carcinogens that are known to damage DNA and lead to mutations.
Alcohol intake is another contributor to CRC development. However, the data show that CRC risk has no significant correlation with light to moderate alcohol consumption[112]. Although approximately half of the patients in the two cohorts (47% of Stage III patients and 43% of metastatic patients) reported no alcohol intake during the 3 mo prior to questionnaire completion[108], the risk of heavy drinking is remarkable, and people who drink more than 4 times have a 52% risk of developing CRC[113]. Furthermore, a Canadian study reported that among subjects who consumed an alcohol beverage at least once a week for 6 mo or longer, those with a BMI > 30 had an overall CRC OR of 2.2[114]. Mechanically, alcohol oxidation and nonoxidative metabolism and the formation of byproducts, such as ROS and metabolites, can lead to a constellation of genetic, epigenetic, cell signaling, and immune processes[115]. Similarly, decreased miR-135 expression in response to ethanol exposure plays a role in colon carcinogenesis and enhances metastasis via APC suppression[116,117]. It has also been shown in an HCT116 cell model that ethanol inactivates GSK3β, leading to increased nuclear translocation of β-catenin and induction of cancer stem cell metastasis via the production of MCP-1/CCR-2[118].
CONCLUSION
Despite remarkable progress in CRC research, the role of molecular classification in therapeutic intervention has not been fully elucidated. This review highlights the driving factors of mCRC and may help in identifying potential candidate biomarkers that can improve the diagnosis and early detection of mCRC, thereby prolonging the overall survival and clinical outcomes of CRC patients. Using molecular alterations to predict CRC risk is a promising approach, but further research is needed to determine whether aberrant mutations, methylation patterns, CNVs, epigenetic marks, immune cell infiltration, and lifestyle factors can be used as reliable and accurate indicators of mCRC risk.
Footnotes
Conflict-of-interest statement: The authors declare that they have no conflicts of interest.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Peer-review started: October 1, 2023
First decision: December 11, 2023
Article in press: January 8, 2024
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ogino S, United States; Perez-Holanda S, Spain S-Editor: Chen YL L-Editor: A P-Editor: Zhao YQ
Contributor Information
Shuai-Xing An, Department of Pharmacology, School of Pharmacy, China Medical University, Shenyang 110122, Liaoning Province, China; Liaoning Key Laboratory of Molecular Targeted Antitumor Drug Development and Evaluation, Liaoning Cancer Immune Peptide Drug Engineering Technology Research Center, Key Laboratory of Precision Diagnosis and Treatment of Gastrointestinal Tumors (China Medical University), Ministry of Education, Shenyang 110122, Liaoning Province, China; BD Department, Greenpine Pharma Group Co., Ltd, Tianjin 300020, China.
Zhao-Jin Yu, Department of Pharmacology, School of Pharmacy, China Medical University, Shenyang 110122, Liaoning Province, China; Liaoning Key Laboratory of Molecular Targeted Antitumor Drug Development and Evaluation, Liaoning Cancer Immune Peptide Drug Engineering Technology Research Center, Key Laboratory of Precision Diagnosis and Treatment of Gastrointestinal Tumors (China Medical University), Ministry of Education, Shenyang 110122, Liaoning Province, China.
Chen Fu, Department of Pharmacology, School of Pharmacy, China Medical University, Shenyang 110122, Liaoning Province, China; Liaoning Key Laboratory of Molecular Targeted Antitumor Drug Development and Evaluation, Liaoning Cancer Immune Peptide Drug Engineering Technology Research Center, Key Laboratory of Precision Diagnosis and Treatment of Gastrointestinal Tumors (China Medical University), Ministry of Education, Shenyang 110122, Liaoning Province, China.
Min-Jie Wei, Department of Pharmacology, School of Pharmacy, China Medical University, Shenyang 110122, Liaoning Province, China; Liaoning Key Laboratory of Molecular Targeted Antitumor Drug Development and Evaluation, Liaoning Cancer Immune Peptide Drug Engineering Technology Research Center, Key Laboratory of Precision Diagnosis and Treatment of Gastrointestinal Tumors (China Medical University), Ministry of Education, Shenyang 110122, Liaoning Province, China.
Long-Hai Shen, Center of Oncology, Genertec Liaoyou Gem Flower Hospital, PanJin 124010, Liaoning Province, China. 154571964@qq.com.
References
- 1.Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. 2019;16:713–732. doi: 10.1038/s41575-019-0189-8. [DOI] [PubMed] [Google Scholar]
- 2.Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467–1480. doi: 10.1016/S0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- 3.Riihimäki M, Hemminki A, Sundquist J, Hemminki K. Patterns of metastasis in colon and rectal cancer. Sci Rep. 2016;6:29765. doi: 10.1038/srep29765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Testa U, Castelli G, Pelosi E. Genetic Alterations of Metastatic Colorectal Cancer. Biomedicines. 2020;8 doi: 10.3390/biomedicines8100414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huebner A, Dietzen M, McGranahan N. SnapShot: Tumor evolution. Cell. 2021;184:1650–1650.e1. doi: 10.1016/j.cell.2021.02.024. [DOI] [PubMed] [Google Scholar]
- 6.Ganesh K, Massagué J. Targeting metastatic cancer. Nat Med. 2021;27:34–44. doi: 10.1038/s41591-020-01195-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yaeger R, Chatila WK, Lipsyc MD, Hechtman JF, Cercek A, Sanchez-Vega F, Jayakumaran G, Middha S, Zehir A, Donoghue MTA, You D, Viale A, Kemeny N, Segal NH, Stadler ZK, Varghese AM, Kundra R, Gao J, Syed A, Hyman DM, Vakiani E, Rosen N, Taylor BS, Ladanyi M, Berger MF, Solit DB, Shia J, Saltz L, Schultz N. Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell. 2018;33:125–136.e3. doi: 10.1016/j.ccell.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vakiani E, Janakiraman M, Shen R, Sinha R, Zeng Z, Shia J, Cercek A, Kemeny N, D'Angelica M, Viale A, Heguy A, Paty P, Chan TA, Saltz LB, Weiser M, Solit DB. Comparative genomic analysis of primary versus metastatic colorectal carcinomas. J Clin Oncol. 2012;30:2956–2962. doi: 10.1200/JCO.2011.38.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li C, Sun YD, Yu GY, Cui JR, Lou Z, Zhang H, Huang Y, Bai CG, Deng LL, Liu P, Zheng K, Wang YH, Wang QQ, Li QR, Wu QQ, Liu Q, Shyr Y, Li YX, Chen LN, Wu JR, Zhang W, Zeng R. Integrated Omics of Metastatic Colorectal Cancer. Cancer Cell. 2020;38:734–747.e9. doi: 10.1016/j.ccell.2020.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Huang D, Sun W, Zhou Y, Li P, Chen F, Chen H, Xia D, Xu E, Lai M, Wu Y, Zhang H. Mutations of key driver genes in colorectal cancer progression and metastasis. Cancer Metastasis Rev. 2018;37:173–187. doi: 10.1007/s10555-017-9726-5. [DOI] [PubMed] [Google Scholar]
- 11.Jo P, Bernhardt M, Nietert M, König A, Azizian A, Schirmer MA, Grade M, Kitz J, Reuter-Jessen K, Ghadimi M, Ströbel P, Schildhaus HU, Gaedcke J. KRAS mutation status concordance between the primary tumor and the corresponding metastasis in patients with rectal cancer. PLoS One. 2020;15:e0239806. doi: 10.1371/journal.pone.0239806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palomba G, Colombino M, Contu A, Massidda B, Baldino G, Pazzola A, Ionta M, Capelli F, Trova V, Sedda T, Sanna G, Tanda F, Budroni M Sardinian Translational Oncology Group (STOG), Palmieri G, Cossu A, Contu M, Cuccu A, Farris A, Macciò A, Mameli G, Olmeo N, Ortu S, Petretto E, Pusceddu V, Virdis L. Prevalence of KRAS, BRAF, and PIK3CA somatic mutations in patients with colorectal carcinoma may vary in the same population: clues from Sardinia. J Transl Med. 2012;10:178. doi: 10.1186/1479-5876-10-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen B, Fong C, Luthra A, Smith SA, DiNatale RG, Nandakumar S, Walch H, Chatila WK, Madupuri R, Kundra R, Bielski CM, Mastrogiacomo B, Donoghue MTA, Boire A, Chandarlapaty S, Ganesh K, Harding JJ, Iacobuzio-Donahue CA, Razavi P, Reznik E, Rudin CM, Zamarin D, Abida W, Abou-Alfa GK, Aghajanian C, Cercek A, Chi P, Feldman D, Ho AL, Iyer G, Janjigian YY, Morris M, Motzer RJ, O'Reilly EM, Postow MA, Raj NP, Riely GJ, Robson ME, Rosenberg JE, Safonov A, Shoushtari AN, Tap W, Teo MY, Varghese AM, Voss M, Yaeger R, Zauderer MG, Abu-Rustum N, Garcia-Aguilar J, Bochner B, Hakimi A, Jarnagin WR, Jones DR, Molena D, Morris L, Rios-Doria E, Russo P, Singer S, Strong VE, Chakravarty D, Ellenson LH, Gopalan A, Reis-Filho JS, Weigelt B, Ladanyi M, Gonen M, Shah SP, Massague J, Gao J, Zehir A, Berger MF, Solit DB, Bakhoum SF, Sanchez-Vega F, Schultz N. Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell. 2022;185:563–575.e11. doi: 10.1016/j.cell.2022.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casimiro S, Fernandes A, Oliveira AG, Franco M, Pires R, Peres M, Matias M, Tato-Costa J, Guerra N, Ramos M, Cruz J, Costa L. Metadherin expression and lung relapse in patients with colorectal carcinoma. Clin Exp Metastasis. 2014;31:689–696. doi: 10.1007/s10585-014-9659-0. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Sethi NS, Hinoue T, Schneider BG, Cherniack AD, Sanchez-Vega F, Seoane JA, Farshidfar F, Bowlby R, Islam M, Kim J, Chatila W, Akbani R, Kanchi RS, Rabkin CS, Willis JE, Wang KK, McCall SJ, Mishra L, Ojesina AI, Bullman S, Pedamallu CS, Lazar AJ, Sakai R Cancer Genome Atlas Research Network, Thorsson V, Bass AJ, Laird PW. Comparative Molecular Analysis of Gastrointestinal Adenocarcinomas. Cancer Cell. 2018;33:721–735.e8. doi: 10.1016/j.ccell.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toyota M, Issa JP. CpG island methylator phenotypes in aging and cancer. Semin Cancer Biol. 1999;9:349–357. doi: 10.1006/scbi.1999.0135. [DOI] [PubMed] [Google Scholar]
- 17.Toh TB, Lim JJ, Chow EK. Epigenetics in cancer stem cells. Mol Cancer. 2017;16:29. doi: 10.1186/s12943-017-0596-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ju HX, An B, Okamoto Y, Shinjo K, Kanemitsu Y, Komori K, Hirai T, Shimizu Y, Sano T, Sawaki A, Tajika M, Yamao K, Fujii M, Murakami H, Osada H, Ito H, Takeuchi I, Sekido Y, Kondo Y. Distinct profiles of epigenetic evolution between colorectal cancers with and without metastasis. Am J Pathol. 2011;178:1835–1846. doi: 10.1016/j.ajpath.2010.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011;8:686–700. doi: 10.1038/nrgastro.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orjuela S, Menigatti M, Schraml P, Kambakamba P, Robinson MD, Marra G. The DNA hypermethylation phenotype of colorectal cancer liver metastases resembles that of the primary colorectal cancers. BMC Cancer. 2020;20:290. doi: 10.1186/s12885-020-06777-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konishi K, Watanabe Y, Shen L, Guo Y, Castoro RJ, Kondo K, Chung W, Ahmed S, Jelinek J, Boumber YA, Estecio MR, Maegawa S, Kondo Y, Itoh F, Imawari M, Hamilton SR, Issa JP. DNA methylation profiles of primary colorectal carcinoma and matched liver metastasis. PLoS One. 2011;6:e27889. doi: 10.1371/journal.pone.0027889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hur K, Cejas P, Feliu J, Moreno-Rubio J, Burgos E, Boland CR, Goel A. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut. 2014;63:635–646. doi: 10.1136/gutjnl-2012-304219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5–18. doi: 10.1038/nrc.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mokutani Y, Uemura M, Munakata K, Okuzaki D, Haraguchi N, Takahashi H, Nishimura J, Hata T, Murata K, Takemasa I, Mizushima T, Doki Y, Mori M, Yamamoto H. Down-Regulation of microRNA-132 is Associated with Poor Prognosis of Colorectal Cancer. Ann Surg Oncol. 2016;23:599–608. doi: 10.1245/s10434-016-5133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu X, Chen Q, Guo H, Li K, Fu B, Chen Y, Zhao H, Wei M, Li Y, Wu H. Identification of Target PTEN-Based miR-425 and miR-576 as Potential Diagnostic and Immunotherapeutic Biomarkers of Colorectal Cancer With Liver Metastasis. Front Oncol. 2021;11:657984. doi: 10.3389/fonc.2021.657984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan L, Yu J, Tan F, Ye GT, Shen ZY, Liu H, Zhang Y, Wang JF, Zhu XJ, Li GX. SP1-mediated microRNA-520d-5p suppresses tumor growth and metastasis in colorectal cancer by targeting CTHRC1. Am J Cancer Res. 2015;5:1447–1459. [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H, Tian Y, Li J, Zhang G, Liu Q, Yang M, Yue L, Cao Q, Cheng Y, Kong N, Fang L, Li S, Sun Q. Identification and functional analysis of lncRNAs and mRNAs between tumorigenesis and metastasis in CRC. Aging (Albany NY) 2021;13:25859–25885. doi: 10.18632/aging.203775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang ZX, Liu HS, Wang FW, Xiong L, Zhou C, Hu T, He XW, Wu XJ, Xie D, Wu XR, Lan P. LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes-mediated macrophage M2 polarization. Cell Death Dis. 2019;10:829. doi: 10.1038/s41419-019-2077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Yu H, Sun W, Kong J, Zhang L, Tang J, Wang J, Xu E, Lai M, Zhang H. The long non-coding RNA CYTOR drives colorectal cancer progression by interacting with NCL and Sam68. Mol Cancer. 2018;17:110. doi: 10.1186/s12943-018-0860-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lv C, Yu H, Wang K, Chen C, Tang J, Han F, Mai M, Ye K, Lai M, Zhang H. ENO2 Promotes Colorectal Cancer Metastasis by Interacting with the LncRNA CYTOR and Activating YAP1-Induced EMT. Cells. 2022;11 doi: 10.3390/cells11152363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tieng FYF, Baharudin R, Abu N, Mohd Yunos RI, Lee LH, Ab Mutalib NS. Single Cell Transcriptome in Colorectal Cancer-Current Updates on Its Application in Metastasis, Chemoresistance and the Roles of Circulating Tumor Cells. Front Pharmacol. 2020;11:135. doi: 10.3389/fphar.2020.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malki A, ElRuz RA, Gupta I, Allouch A, Vranic S, Al Moustafa AE. Molecular Mechanisms of Colon Cancer Progression and Metastasis: Recent Insights and Advancements. Int J Mol Sci. 2020;22 doi: 10.3390/ijms22010130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lichtenstern CR, Ngu RK, Shalapour S, Karin M. Immunotherapy, Inflammation and Colorectal Cancer. Cells. 2020;9 doi: 10.3390/cells9030618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, Bot BM, Morris JS, Simon IM, Gerster S, Fessler E, De Sousa E Melo F, Missiaglia E, Ramay H, Barras D, Homicsko K, Maru D, Manyam GC, Broom B, Boige V, Perez-Villamil B, Laderas T, Salazar R, Gray JW, Hanahan D, Tabernero J, Bernards R, Friend SH, Laurent-Puig P, Medema JP, Sadanandam A, Wessels L, Delorenzi M, Kopetz S, Vermeulen L, Tejpar S. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vilar E, Gruber SB. Microsatellite instability in colorectal cancer-the stable evidence. Nat Rev Clin Oncol. 2010;7:153–162. doi: 10.1038/nrclinonc.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maby P, Galon J, Latouche JB. Frameshift mutations, neoantigens and tumor-specific CD8(+) T cells in microsatellite unstable colorectal cancers. Oncoimmunology. 2016;5:e1115943. doi: 10.1080/2162402X.2015.1115943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Babaei G, Aziz SG, Jaghi NZZ. EMT, cancer stem cells and autophagy; The three main axes of metastasis. Biomed Pharmacother. 2021;133:110909. doi: 10.1016/j.biopha.2020.110909. [DOI] [PubMed] [Google Scholar]
- 39.Beerling E, Seinstra D, de Wit E, Kester L, van der Velden D, Maynard C, Schäfer R, van Diest P, Voest E, van Oudenaarden A, Vrisekoop N, van Rheenen J. Plasticity between Epithelial and Mesenchymal States Unlinks EMT from Metastasis-Enhancing Stem Cell Capacity. Cell Rep. 2016;14:2281–2288. doi: 10.1016/j.celrep.2016.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, Zhang Y, Li Q, Wang Y. Transgelins: Cytoskeletal Associated Proteins Implicated in the Metastasis of Colorectal Cancer. Front Cell Dev Biol. 2020;8:573859. doi: 10.3389/fcell.2020.573859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang H, Chi J, Hu J, Ji T, Luo Z, Zhou C, Huang L, Dai Z, Li J, Wang G, Wang L, Wang Z. Intracellular AGR2 transduces PGE2 stimuli to promote epithelial-mesenchymal transition and metastasis of colorectal cancer. Cancer Lett. 2021;518:180–195. doi: 10.1016/j.canlet.2021.06.025. [DOI] [PubMed] [Google Scholar]
- 42.Lee H, Shim S, Kong JS, Kim MJ, Park S, Lee SS, Kim A. Overexpression of dopamine receptor D2 promotes colorectal cancer progression by activating the β-catenin/ZEB1 axis. Cancer Sci. 2021;112:3732–3743. doi: 10.1111/cas.15026. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Dahlmann M, Monks A, Harris ED, Kobelt D, Osterland M, Khaireddine F, Herrmann P, Kemmner W, Burock S, Walther W, Shoemaker RH, Stein U. Combination of Wnt/β-Catenin Targets S100A4 and DKK1 Improves Prognosis of Human Colorectal Cancer. Cancers (Basel) 2021;14 doi: 10.3390/cancers14010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan Y, Hu Y, Xiao Q, Tang Y, Chen H, He J, Chen L, Jiang K, Wang Z, Yuan Y, Ding K. Silencing of brain-expressed X-linked 2 (BEX2) promotes colorectal cancer metastasis through the Hedgehog signaling pathway. Int J Biol Sci. 2020;16:228–238. doi: 10.7150/ijbs.38431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao Z, Zhang H, Fan P, Huang Q, Dong K, Qi Y, Song J, Chen L, Liang H, Chen X, Zhang Z, Zhang B. [Corrigendum] High PLK4 expression promotes tumor progression and induces epithelialmesenchymal transition by regulating the Wnt/βcatenin signaling pathway in colorectal cancer. Int J Oncol. 2022;60 doi: 10.3892/ijo.2021.5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diesch J, Sanij E, Gilan O, Love C, Tran H, Fleming NI, Ellul J, Amalia M, Haviv I, Pearson RB, Tulchinsky E, Mariadason JM, Sieber OM, Hannan RD, Dhillon AS. Widespread FRA1-dependent control of mesenchymal transdifferentiation programs in colorectal cancer cells. PLoS One. 2014;9:e88950. doi: 10.1371/journal.pone.0088950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jackstadt R, Röh S, Neumann J, Jung P, Hoffmann R, Horst D, Berens C, Bornkamm GW, Kirchner T, Menssen A, Hermeking H. AP4 is a mediator of epithelial-mesenchymal transition and metastasis in colorectal cancer. J Exp Med. 2013;210:1331–1350. doi: 10.1084/jem.20120812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee YH, Kim SJ, Fang X, Song NY, Kim DH, Suh J, Na HK, Kim KO, Baek JH, Surh YJ. JNK-mediated Ser27 phosphorylation and stabilization of SIRT1 promote growth and progression of colon cancer through deacetylation-dependent activation of Snail. Mol Oncol. 2022;16:1555–1571. doi: 10.1002/1878-0261.13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rokavec M, Kaller M, Horst D, Hermeking H. Pan-cancer EMT-signature identifies RBM47 down-regulation during colorectal cancer progression. Sci Rep. 2017;7:4687. doi: 10.1038/s41598-017-04234-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saman H, Raza SS, Uddin S, Rasul K. Inducing Angiogenesis, a Key Step in Cancer Vascularization, and Treatment Approaches. Cancers (Basel) 2020;12 doi: 10.3390/cancers12051172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fidler IJ. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 52.Canavese M, Ngo DT, Maddern GJ, Hardingham JE, Price TJ, Hauben E. Biology and therapeutic implications of VEGF-A splice isoforms and single-nucleotide polymorphisms in colorectal cancer. Int J Cancer. 2017;140:2183–2191. doi: 10.1002/ijc.30567. [DOI] [PubMed] [Google Scholar]
- 53.Nogués A, Gallardo-Vara E, Zafra MP, Mate P, Marijuan JL, Alonso A, Botella LM, Prieto MI. Endoglin (CD105) and VEGF as potential angiogenic and dissemination markers for colorectal cancer. World J Surg Oncol. 2020;18:99. doi: 10.1186/s12957-020-01871-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lopez A, Harada K, Vasilakopoulou M, Shanbhag N, Ajani JA. Targeting Angiogenesis in Colorectal Carcinoma. Drugs. 2019;79:63–74. doi: 10.1007/s40265-018-1037-9. [DOI] [PubMed] [Google Scholar]
- 55.Capdevila J, Carrato A, Tabernero J, Grande E. What could Nintedanib (BIBF 1120), a triple inhibitor of VEGFR, PDGFR, and FGFR, add to the current treatment options for patients with metastatic colorectal cancer? Crit Rev Oncol Hematol. 2014;92:83–106. doi: 10.1016/j.critrevonc.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 56.Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 57.Lu J. The Warburg metabolism fuels tumor metastasis. Cancer Metastasis Rev. 2019;38:157–164. doi: 10.1007/s10555-019-09794-5. [DOI] [PubMed] [Google Scholar]
- 58.Zhong X, He X, Wang Y, Hu Z, Huang H, Zhao S, Wei P, Li D. Warburg effect in colorectal cancer: the emerging roles in tumor microenvironment and therapeutic implications. J Hematol Oncol. 2022;15:160. doi: 10.1186/s13045-022-01358-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qi L, Chen J, Yang Y, Hu W. Hypoxia Correlates With Poor Survival and M2 Macrophage Infiltration in Colorectal Cancer. Front Oncol. 2020;10:566430. doi: 10.3389/fonc.2020.566430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun H, Meng Q, Shi C, Yang H, Li X, Wu S, Familiari G, Relucenti M, Aschner M, Wang X, Chen R. Hypoxia-Inducible Exosomes Facilitate Liver-Tropic Premetastatic Niche in Colorectal Cancer. Hepatology. 2021;74:2633–2651. doi: 10.1002/hep.32009. [DOI] [PubMed] [Google Scholar]
- 61.Yang H, Zhang H, Yang Y, Wang X, Deng T, Liu R, Ning T, Bai M, Li H, Zhu K, Li J, Fan Q, Ying G, Ba Y. Hypoxia induced exosomal circRNA promotes metastasis of Colorectal Cancer via targeting GEF-H1/RhoA axis. Theranostics. 2020;10:8211–8226. doi: 10.7150/thno.44419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Castro-Giner F, Aceto N. Tracking cancer progression: from circulating tumor cells to metastasis. Genome Med. 2020;12:31. doi: 10.1186/s13073-020-00728-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chiu SY, Hsieh CH, You JF, Chu PY, Hung HY, Chu PH, Wu MH. Enhancing Prediction Performance by Add-On Combining Circulating Tumor Cell Count, CD45(neg) EpCAM(neg) Cell Count on Colorectal Cancer, Advance, and Metastasis. Cancers (Basel) 2021;13 doi: 10.3390/cancers13112521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Le UT, Bronsert P, Picardo F, Riethdorf S, Haager B, Rylski B, Czerny M, Beyersdorf F, Wiesemann S, Pantel K, Passlick B, Kaifi JT, Schmid S. Intraoperative detection of circulating tumor cells in pulmonary venous blood during metastasectomy for colorectal lung metastases. Sci Rep. 2018;8:8751. doi: 10.1038/s41598-018-26410-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gkountela S, Castro-Giner F, Szczerba BM, Vetter M, Landin J, Scherrer R, Krol I, Scheidmann MC, Beisel C, Stirnimann CU, Kurzeder C, Heinzelmann-Schwarz V, Rochlitz C, Weber WP, Aceto N. Circulating Tumor Cell Clustering Shapes DNA Methylation to Enable Metastasis Seeding. Cell. 2019;176:98–112.e14. doi: 10.1016/j.cell.2018.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hamid FB, Gopalan V, Matos M, Lu CT, Lam AK. Genetic Heterogeneity of Single Circulating Tumour Cells in Colorectal Carcinoma. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21207766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nabariya DK, Pallu R, Yenuganti VR. Exosomes: The protagonists in the tale of colorectal cancer? Biochim Biophys Acta Rev Cancer. 2020;1874:188426. doi: 10.1016/j.bbcan.2020.188426. [DOI] [PubMed] [Google Scholar]
- 68.Guo Y, Ji X, Liu J, Fan D, Zhou Q, Chen C, Wang W, Wang G, Wang H, Yuan W, Ji Z, Sun Z. Effects of exosomes on pre-metastatic niche formation in tumors. Mol Cancer. 2019;18:39. doi: 10.1186/s12943-019-0995-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dou R, Liu K, Yang C, Zheng J, Shi D, Lin X, Wei C, Zhang C, Fang Y, Huang S, Song J, Wang S, Xiong B. EMT-cancer cells-derived exosomal miR-27b-3p promotes circulating tumour cells-mediated metastasis by modulating vascular permeability in colorectal cancer. Clin Transl Med. 2021;11:e595. doi: 10.1002/ctm2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang D, Wang X, Si M, Yang J, Sun S, Wu H, Cui S, Qu X, Yu X. Exosome-encapsulated miRNAs contribute to CXCL12/CXCR4-induced liver metastasis of colorectal cancer by enhancing M2 polarization of macrophages. Cancer Lett. 2020;474:36–52. doi: 10.1016/j.canlet.2020.01.005. [DOI] [PubMed] [Google Scholar]
- 71.Zhao S, Mi Y, Guan B, Zheng B, Wei P, Gu Y, Zhang Z, Cai S, Xu Y, Li X, He X, Zhong X, Li G, Chen Z, Li D. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J Hematol Oncol. 2020;13:156. doi: 10.1186/s13045-020-00991-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Z, Xing T, Chen Y, Xiao J. Exosome-mediated miR-200b promotes colorectal cancer proliferation upon TGF-β1 exposure. Biomed Pharmacother. 2018;106:1135–1143. doi: 10.1016/j.biopha.2018.07.042. [DOI] [PubMed] [Google Scholar]
- 73.Jiang K, Chen H, Fang Y, Chen L, Zhong C, Bu T, Dai S, Pan X, Fu D, Qian Y, Wei J, Ding K. Exosomal ANGPTL1 attenuates colorectal cancer liver metastasis by regulating Kupffer cell secretion pattern and impeding MMP9 induced vascular leakiness. J Exp Clin Cancer Res. 2021;40:21. doi: 10.1186/s13046-020-01816-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang C, Wang XY, Zhang P, He TC, Han JH, Zhang R, Lin J, Fan J, Lu L, Zhu WW, Jia HL, Zhang JB, Chen JH. Cancer-derived exosomal HSPC111 promotes colorectal cancer liver metastasis by reprogramming lipid metabolism in cancer-associated fibroblasts. Cell Death Dis. 2022;13:57. doi: 10.1038/s41419-022-04506-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Han K, Wang FW, Cao CH, Ling H, Chen JW, Chen RX, Feng ZH, Luo J, Jin XH, Duan JL, Li SM, Ma NF, Yun JP, Guan XY, Pan ZZ, Lan P, Xu RH, Xie D. CircLONP2 enhances colorectal carcinoma invasion and metastasis through modulating the maturation and exosomal dissemination of microRNA-17. Mol Cancer. 2020;19:60. doi: 10.1186/s12943-020-01184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, Chen C, Chang W, Ping Y, Ji P, Wu J, Quan W, Yao Y, Zhou Y, Sun Z, Li D. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-β1 axis. Mol Cancer. 2020;19:117. doi: 10.1186/s12943-020-01235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hu JL, Wang W, Lan XL, Zeng ZC, Liang YS, Yan YR, Song FY, Wang FF, Zhu XH, Liao WJ, Liao WT, Ding YQ, Liang L. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol Cancer. 2019;18:91. doi: 10.1186/s12943-019-1019-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen S, Wang Y, Wang T, Hou Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8:3932–3948. doi: 10.7150/thno.25541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Asadirad A, Baghaei K, Hashemi SM, Dehnavi S, Ghanbarian H, Mortaz E, Anissian A, Asadzadeh Aghdaei H, Amani D. Dendritic cell immunotherapy with miR-155 enriched tumor-derived exosome suppressed cancer growth and induced antitumor immune responses in murine model of colorectal cancer induced by CT26 cell line. Int Immunopharmacol. 2022;104:108493. doi: 10.1016/j.intimp.2021.108493. [DOI] [PubMed] [Google Scholar]
- 80.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39:1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 81.Chandra R, Karalis JD, Liu C, Murimwa GZ, Voth Park J, Heid CA, Reznik SI, Huang E, Minna JD, Brekken RA. The Colorectal Cancer Tumor Microenvironment and Its Impact on Liver and Lung Metastasis. Cancers (Basel) 2021;13 doi: 10.3390/cancers13246206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pagès F, Mlecnik B, Marliot F, Bindea G, Ou FS, Bifulco C, Lugli A, Zlobec I, Rau TT, Berger MD, Nagtegaal ID, Vink-Börger E, Hartmann A, Geppert C, Kolwelter J, Merkel S, Grützmann R, Van den Eynde M, Jouret-Mourin A, Kartheuser A, Léonard D, Remue C, Wang JY, Bavi P, Roehrl MHA, Ohashi PS, Nguyen LT, Han S, MacGregor HL, Hafezi-Bakhtiari S, Wouters BG, Masucci GV, Andersson EK, Zavadova E, Vocka M, Spacek J, Petruzelka L, Konopasek B, Dundr P, Skalova H, Nemejcova K, Botti G, Tatangelo F, Delrio P, Ciliberto G, Maio M, Laghi L, Grizzi F, Fredriksen T, Buttard B, Angelova M, Vasaturo A, Maby P, Church SE, Angell HK, Lafontaine L, Bruni D, El Sissy C, Haicheur N, Kirilovsky A, Berger A, Lagorce C, Meyers JP, Paustian C, Feng Z, Ballesteros-Merino C, Dijkstra J, van de Water C, van Lent-van Vliet S, Knijn N, Mușină AM, Scripcariu DV, Popivanova B, Xu M, Fujita T, Hazama S, Suzuki N, Nagano H, Okuno K, Torigoe T, Sato N, Furuhata T, Takemasa I, Itoh K, Patel PS, Vora HH, Shah B, Patel JB, Rajvik KN, Pandya SJ, Shukla SN, Wang Y, Zhang G, Kawakami Y, Marincola FM, Ascierto PA, Sargent DJ, Fox BA, Galon J. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. 2018;391:2128–2139. doi: 10.1016/S0140-6736(18)30789-X. [DOI] [PubMed] [Google Scholar]
- 83.Bruni D, Angell HK, Galon J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat Rev Cancer. 2020;20:662–680. doi: 10.1038/s41568-020-0285-7. [DOI] [PubMed] [Google Scholar]
- 84.Angell HK, Bruni D, Barrett JC, Herbst R, Galon J. The Immunoscore: Colon Cancer and Beyond. Clin Cancer Res. 2020;26:332–339. doi: 10.1158/1078-0432.CCR-18-1851. [DOI] [PubMed] [Google Scholar]
- 85.Shi Y, Luo P, Wang W, Horst K, Bläsius F, Relja B, Xu D, Hildebrand F, Greven J. M1 But Not M0 Extracellular Vesicles Induce Polarization of RAW264.7 Macrophages Via the TLR4-NFκB Pathway In Vitro. Inflammation. 2020;43:1611–1619. doi: 10.1007/s10753-020-01236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Oishi S, Takano R, Tamura S, Tani S, Iwaizumi M, Hamaya Y, Takagaki K, Nagata T, Seto S, Horii T, Osawa S, Furuta T, Miyajima H, Sugimoto K. M2 polarization of murine peritoneal macrophages induces regulatory cytokine production and suppresses T-cell proliferation. Immunology. 2016;149:320–328. doi: 10.1111/imm.12647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Su B, Han H, Gong Y, Li X, Ji C, Yao J, Yang J, Hu W, Zhao W, Li J, Zhang G, Zhou L. Let-7d inhibits intratumoral macrophage M2 polarization and subsequent tumor angiogenesis by targeting IL-13 and IL-10. Cancer Immunol Immunother. 2021;70:1619–1634. doi: 10.1007/s00262-020-02791-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Y, Zhao Y, Li Q, Wang Y. Macrophages, as a Promising Strategy to Targeted Treatment for Colorectal Cancer Metastasis in Tumor Immune Microenvironment. Front Immunol. 2021;12:685978. doi: 10.3389/fimmu.2021.685978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.López-Soto A, Gonzalez S, Smyth MJ, Galluzzi L. Control of Metastasis by NK Cells. Cancer Cell. 2017;32:135–154. doi: 10.1016/j.ccell.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 90.Melero I, Rouzaut A, Motz GT, Coukos G. T-cell and NK-cell infiltration into solid tumors: a key limiting factor for efficacious cancer immunotherapy. Cancer Discov. 2014;4:522–526. doi: 10.1158/2159-8290.CD-13-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tachibana T, Onodera H, Tsuruyama T, Mori A, Nagayama S, Hiai H, Imamura M. Increased intratumor Valpha24-positive natural killer T cells: a prognostic factor for primary colorectal carcinomas. Clin Cancer Res. 2005;11:7322–7327. doi: 10.1158/1078-0432.CCR-05-0877. [DOI] [PubMed] [Google Scholar]
- 92.Sandel MH, Dadabayev AR, Menon AG, Morreau H, Melief CJ, Offringa R, van der Burg SH, Janssen-van Rhijn CM, Ensink NG, Tollenaar RA, van de Velde CJ, Kuppen PJ. Prognostic value of tumor-infiltrating dendritic cells in colorectal cancer: role of maturation status and intratumoral localization. Clin Cancer Res. 2005;11:2576–2582. doi: 10.1158/1078-0432.CCR-04-1448. [DOI] [PubMed] [Google Scholar]
- 93.Giraldo NA, Becht E, Remark R, Damotte D, Sautès-Fridman C, Fridman WH. The immune contexture of primary and metastatic human tumours. Curr Opin Immunol. 2014;27:8–15. doi: 10.1016/j.coi.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 94.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, Tosolini M, Camus M, Berger A, Wind P, Zinzindohoué F, Bruneval P, Cugnenc PH, Trajanoski Z, Fridman WH, Pagès F. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 95.Lazarus J, Maj T, Smith JJ, Perusina Lanfranca M, Rao A, D'Angelica MI, Delrosario L, Girgis A, Schukow C, Shia J, Kryczek I, Shi J, Wasserman I, Crawford H, Nathan H, Pasca Di Magliano M, Zou W, Frankel TL. Spatial and phenotypic immune profiling of metastatic colon cancer. JCI Insight. 2018;3 doi: 10.1172/jci.insight.121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, Bruneval P, Fridman WH, Becker C, Pagès F, Speicher MR, Trajanoski Z, Galon J. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 97.Ahrends T, Spanjaard A, Pilzecker B, Bąbała N, Bovens A, Xiao Y, Jacobs H, Borst J. CD4(+) T Cell Help Confers a Cytotoxic T Cell Effector Program Including Coinhibitory Receptor Downregulation and Increased Tissue Invasiveness. Immunity. 2017;47:848–861.e5. doi: 10.1016/j.immuni.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 98.Tosolini M, Kirilovsky A, Mlecnik B, Fredriksen T, Mauger S, Bindea G, Berger A, Bruneval P, Fridman WH, Pagès F, Galon J. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011;71:1263–1271. doi: 10.1158/0008-5472.CAN-10-2907. [DOI] [PubMed] [Google Scholar]
- 99.Amicarella F, Muraro MG, Hirt C, Cremonesi E, Padovan E, Mele V, Governa V, Han J, Huber X, Droeser RA, Zuber M, Adamina M, Bolli M, Rosso R, Lugli A, Zlobec I, Terracciano L, Tornillo L, Zajac P, Eppenberger-Castori S, Trapani F, Oertli D, Iezzi G. Dual role of tumour-infiltrating T helper 17 cells in human colorectal cancer. Gut. 2017;66:692–704. doi: 10.1136/gutjnl-2015-310016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Van den Eynde M, Mlecnik B, Bindea G, Galon J. Multiverse of immune microenvironment in metastatic colorectal cancer. Oncoimmunology. 2020;9:1824316. doi: 10.1080/2162402X.2020.1824316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Van den Eynde M, Mlecnik B, Bindea G, Fredriksen T, Church SE, Lafontaine L, Haicheur N, Marliot F, Angelova M, Vasaturo A, Bruni D, Jouret-Mourin A, Baldin P, Huyghe N, Haustermans K, Debucquoy A, Van Cutsem E, Gigot JF, Hubert C, Kartheuser A, Remue C, Léonard D, Valge-Archer V, Pagès F, Machiels JP, Galon J. The Link between the Multiverse of Immune Microenvironments in Metastases and the Survival of Colorectal Cancer Patients. Cancer Cell. 2018;34:1012–1026.e3. doi: 10.1016/j.ccell.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 102.Sun HL, Zhou X, Xue YF, Wang K, Shen YF, Mao JJ, Guo HF, Miao ZN. Increased frequency and clinical significance of myeloid-derived suppressor cells in human colorectal carcinoma. World J Gastroenterol. 2012;18:3303–3309. doi: 10.3748/wjg.v18.i25.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang B, Wang Z, Wu L, Zhang M, Li W, Ding J, Zhu J, Wei H, Zhao K. Circulating and tumor-infiltrating myeloid-derived suppressor cells in patients with colorectal carcinoma. PLoS One. 2013;8:e57114. doi: 10.1371/journal.pone.0057114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Deng D, Luo X, Zhang S, Xu Z. Immune cell infiltration-associated signature in colon cancer and its prognostic implications. Aging (Albany NY) 2021;13:19696–19709. doi: 10.18632/aging.203380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Galdiero MR, Bianchi P, Grizzi F, Di Caro G, Basso G, Ponzetta A, Bonavita E, Barbagallo M, Tartari S, Polentarutti N, Malesci A, Marone G, Roncalli M, Laghi L, Garlanda C, Mantovani A, Jaillon S. Occurrence and significance of tumor-associated neutrophils in patients with colorectal cancer. Int J Cancer. 2016;139:446–456. doi: 10.1002/ijc.30076. [DOI] [PubMed] [Google Scholar]
- 106.Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol. 2019;14:89–103. doi: 10.5114/pg.2018.81072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Van Loon K, Wigler D, Niedzwiecki D, Venook AP, Fuchs C, Blanke C, Saltz L, Goldberg RM, Meyerhardt JA. Comparison of dietary and lifestyle habits among stage III and metastatic colorectal cancer patients: findings from CALGB 89803 and CALGB 80405. Clin Colorectal Cancer. 2013;12:95–102. doi: 10.1016/j.clcc.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Patel GS, Ullah S, Beeke C, Hakendorf P, Padbury R, Price TJ, Karapetis CS. Association of BMI with overall survival in patients with mCRC who received chemotherapy versus EGFR and VEGF-targeted therapies. Cancer Med. 2015;4:1461–1471. doi: 10.1002/cam4.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sawicki T, Ruszkowska M, Danielewicz A, Niedźwiedzka E, Arłukowicz T, Przybyłowicz KE. A Review of Colorectal Cancer in Terms of Epidemiology, Risk Factors, Development, Symptoms and Diagnosis. Cancers (Basel) 2021;13 doi: 10.3390/cancers13092025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Renfro LA, Loupakis F, Adams RA, Seymour MT, Heinemann V, Schmoll HJ, Douillard JY, Hurwitz H, Fuchs CS, Diaz-Rubio E, Porschen R, Tournigand C, Chibaudel B, Falcone A, Tebbutt NC, Punt CJ, Hecht JR, Bokemeyer C, Van Cutsem E, Goldberg RM, Saltz LB, de Gramont A, Sargent DJ, Lenz HJ. Body Mass Index Is Prognostic in Metastatic Colorectal Cancer: Pooled Analysis of Patients From First-Line Clinical Trials in the ARCAD Database. J Clin Oncol. 2016;34:144–150. doi: 10.1200/JCO.2015.61.6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Giovannucci E. An updated review of the epidemiological evidence that cigarette smoking increases risk of colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2001;10:725–731. [PubMed] [Google Scholar]
- 112.Cai S, Li Y, Ding Y, Chen K, Jin M. Alcohol drinking and the risk of colorectal cancer death: a meta-analysis. Eur J Cancer Prev. 2014;23:532–539. doi: 10.1097/CEJ.0000000000000076. [DOI] [PubMed] [Google Scholar]
- 113.Marley AR, Nan H. Epidemiology of colorectal cancer. Int J Mol Epidemiol Genet. 2016;7:105–114. [PMC free article] [PubMed] [Google Scholar]
- 114.Zhao J, Zhu Y, Wang PP, West R, Buehler S, Sun Z, Squires J, Roebothan B, McLaughlin JR, Campbell PT, Parfrey PS. Interaction between alcohol drinking and obesity in relation to colorectal cancer risk: a case-control study in Newfoundland and Labrador, Canada. BMC Public Health. 2012;12:94. doi: 10.1186/1471-2458-12-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rossi M, Jahanzaib Anwar M, Usman A, Keshavarzian A, Bishehsari F. Colorectal Cancer and Alcohol Consumption-Populations to Molecules. Cancers (Basel) 2018;10 doi: 10.3390/cancers10020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yeligar S, Tsukamoto H, Kalra VK. Ethanol-induced expression of ET-1 and ET-BR in liver sinusoidal endothelial cells and human endothelial cells involves hypoxia-inducible factor-1alpha and microrNA-199. J Immunol. 2009;183:5232–5243. doi: 10.4049/jimmunol.0901084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink JA, Bolijn A, Meijer GA, Agami R. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68:5795–5802. doi: 10.1158/0008-5472.CAN-08-0951. [DOI] [PubMed] [Google Scholar]
- 118.Xu M, Wang S, Qi Y, Chen L, Frank JA, Yang XH, Zhang Z, Shi X, Luo J. Role of MCP-1 in alcohol-induced aggressiveness of colorectal cancer cells. Mol Carcinog. 2016;55:1002–1011. doi: 10.1002/mc.22343. [DOI] [PMC free article] [PubMed] [Google Scholar]