Abstract
Background
Sleep restriction (SR) has been shown to upregulate neuronal reward networks in response to food stimuli, but prior studies were short-term and employed severe SR paradigms.
Objective
Our goal was to determine whether mild SR, achieved by delaying bedtimes by 1.5 h, influences neuronal networks responsive to food stimuli compared with maintained adequate sleep (AS) >7 h/night.
Methods
A randomized controlled crossover study with 2 6-wk phases, AS (≥7 h sleep/night) and SR (−1.5 h/night relative to screening), was conducted. Adults with AS duration, measured using wrist actigraphy over a 2-wk screening period, and self-reported good sleep quality were enrolled. Resting-state and food-stimulated functional neuroimaging (fMRI) was performed at the endpoint of each phase. Resting-state fMRI data analyses included a priori region-of-interest seed-based functional connectivity, whole-brain voxel-wise analyses, and network analyses. Food task-fMRI analyses compared brain activity patterns in response to food cues between conditions. Paired-sample t tests tested differences between conditions.
Results
Twenty-six participants (16 males; age 29.6 ± 5.3 y, body mass index 26.9 ± 4.0 kg/m2) contributed complete data. Total sleep time was 7 h 30 ± 28 min/night during AS compared with 6 h 12 ± 26 min/night during SR. We employed different statistical approaches to replicate prior studies in the field and to apply more robust approaches that are currently advocated in the field. Using uncorrected P value of <0.01, cluster ≥10-voxel thresholds, we replicated prior findings of increased activation in response to foods in reward networks after SR compared with AS (right insula, right inferior frontal gyrus, and right supramarginal gyrus). These findings did not survive more rigorous analytical approaches (Gaussian Random Field theory correction at 2-tailed voxel P < 0.001, cluster P < 0.05).
Conclusions
The results suggest that mild SR leads to increased reward responsivity to foods but with low confidence given the failure to meet significance from rigorous statistical analyses. Further research is necessary to inform the mechanisms underlying the role of sleep on food intake regulation.
This trial was registered at clinicaltrials.gov as NCT02960776.
Keywords: sleep duration, food, obesity, fMRI, functional connectivity
Introduction
Insufficient sleep, defined as sleeping <7 h/night, is pervasive in today’s society [1] and has been associated with increased odds of developing obesity [2]. Clinical intervention studies show that sleep restriction (SR) leads to weight gain and that this is due to increased energy intakes [3]. Indeed, several meta-analyses have now reported increases in energy intakes of ∼150 to 385 kcal/d [[4], [5], [6]] under conditions of SR relative to adequate sleep (AS).
Different mechanisms have been proposed to explain the increase in energy intakes resulting from SR, including alterations in appetite-regulating hormones, longer time available to consume foods due to longer wake periods, and increased responsivity to the appeal of food. Based on the prior available evidence, Chaput and St-Onge [7] suggested that enhanced hedonic appeal of foods was most likely responsible for the difference in energy intakes between conditions of SR and habitual AS. Indeed, short periods of severe SR [8,9] or total sleep deprivation [10,11] lead to alterations in neuronal responses to food stimuli compared to a period of sufficient sleep, indicating upregulation of reward processing networks in the sleep-deprived state. These studies support the hedonic mechanistic pathway toward increased energy intakes resulting from insufficient sleep. However, participants were subjected to no [10,11] or very little sleep (4 h time in bed) in these studies. Only one study used a milder SR protocol of 6 h time in bed in adults with usual AS to test the impact on neuronal responses to food stimuli [12]. That study also found increased activation by food stimuli of regions implicated in reward processing in the context of SR compared with AS. However, contrary to prior research [8], the study by Demos et al. [12] also showed greater food cue responsivity in the inferior frontal gyrus and ventral medial prefrontal cortex, inhibitory control decision-making regions of the brain, in the short sleep condition.
Beyond food-driven brain responses, a handful of fMRI studies without an explicit task (i.e., resting-state) also support the notion that connectivity patterns emerging using resting-state fMRI measures can be affected by circadian misalignment [13], other circadian disturbances [14], and total sleep deprivation [15].
Although available studies are concordant and supportive of a role of sleep duration in modulating reward networks involved in processing information from food stimuli that would indicate increased consumption, the SR paradigms are, for the most part, not representative of habitual short sleep duration observed in real life owing to their short duration and/or severe SR [[8], [9], [10], [11], [12],15]. It therefore remains unknown whether milder forms of SR, representing higher ecologic validity, sustained for a prolonged period of time, would elicit similar results. In addition, all studies performed functional neuroimaging assessments in the morning in the fasted state. Studies suggest that increased food intake due to SR is mostly due to increased intake at night [16]. We hypothesized that prior studies may have underevaluated the impact of SR on neuronal networks due to a potential overpowering impact of homeostatic drivers of food intake that are in effect in the fasted state. To address this question, we performed a randomized controlled crossover study of mild SR compared with AS sustained for 6 wk and conducted our assessments in the evening, at a time corresponding to our participants’ usual predinner time.
Methods
Participants
Young to middle-aged males and females (20–40 y) with BMI 25–29.9 kg/m2 or those with a BMI of 19–24.9 kg/m2 who had one parent with obesity were eligible. Recruitment occurred through flyers and online advertisements. The primary inclusion criteria included having sleep duration ≥7 h/night, assessed using wrist actigraphy (Actigraphy GT3X+, ActiLife LLC) over 2 wk, and good sleep quality, assessed by questionnaire (Pittsburgh Sleep Quality Index ≤5, Epworth Sleepiness Score <10, and low risk of sleep apnea from the Berlin Questionnaire). Exclusion criteria included smoking (unless exsmoker for ≥3 y), drug or alcohol abuse, and excessive caffeine intake (>300 mg/d), working night or rotating shifts, or having type 2 diabetes, cardiovascular disease, or depression. Individuals who experienced recent weight change or were actively participating in a weight loss program were excluded. To be enrolled, participants were required to agree not to operate a motor vehicle during the SR condition and to remain in their current time zone during each phase.
Study design
This randomized, crossover, outpatient study comprised of 2 phases of 6 wk each: AS (habitual sleep ≥7 h/night) and SR (−1.5 h/night relative to habitual sleep). A total of 43 participants completed the first study phase and 36 completed the second phase (see CONSORT diagram). From those participants, fMRI data were obtained from 38 (21 males) in phase 1 and 26 (16 males) in phase 2.
During AS, participants were required to go to bed and wake up at their usual times to achieve an average nightly sleep duration of ≥7 h. During SR, participants were asked to delay their bedtimes by 1.5 h relative to screening data to achieve an average reduction in nightly sleep duration of 1.5 h. Sleep was monitored daily throughout each sleep phase with wrist actigraphy and verified with the research assistant at weekly visits. Adjustments to the sleep schedule were made to ensure that sleep duration goals for each study condition were met. Only one participant was disqualified due to nonadherence. Upon completion of phase 1, participants returned to their usual sleep patterns for a washout period (≥2 wk) prior to returning for phase 2, with the alternate sleep condition. Sleep was verified for ≥2 wk with wrist actigraphy to ensure return to prestudy sleep duration prior to phase 2. Daytime naps were not permitted during the study.
fMRI scanning occurred in the last week of each study phase. Participants provided information on their habitual dinner time and, based on this information, fMRI scanning was scheduled to start ∼1 h before habitual dinner.
The Institutional Review Boards of Columbia University Irving Medical Center and New York University Grossman School of Medicine granted ethical approval, and all participants provided written informed consent. This trial is registered on clinicaltrials.gov (NCT02960776).
Task design and experimental procedure
Each scanning session included 2 resting-state runs followed by 4 task runs. During the resting-state runs (each lasting 5 min), participants were instructed to keep their eyes open, look at the fixation cross on screen, and relax.
Task runs involved passive viewing of visual stimuli [8,17]. Each task analyzed consisted of 8 blocks of 4 s, with 4 active blocks alternating with 4 fixation blocks. Each active block contained the same type of stimuli, either food or nonfood (NF) items. Food blocks could further contain either healthy (HF) or unhealthy food (UHF) stimuli exclusively. The list of HF, UHF, and NF items from which stimuli were selected for presentation based on each individual’s top item ratings prior to the fMRI task is available in Supplementary Table S1. Identical procedures (task runs and stimulus set presentation and instructions) were used for each study phase (AS and SR) for each participant.
Self-reported sleepiness was measured using the Stanford Sleepiness Scale before and after each scanning session for n = 25 (15 males) of the participants who provided neuroimaging data for both phases.
Imaging data acquisition
Neuroimaging data were acquired using a 3T Siemens Skyra scanner (Siemens Medical Solutions; 32-channel head coil) at the New York University Langone Health Center for Biomedical Imaging. High-resolution anatomical T1-weighted images were first obtained (3D-MPRAGE: repetition time [TR] = 1900 ms, echo time [TE] = 2.52 ms, inversion time [TI] = 900 ms, flip angle = 9°, field of view [FOV] = 256 mm, isotropic 1 mm3 voxels). Functional echo planar images were then collected with the same acquisition parameters for the resting-state and the task data (TR = 2500 ms, TE = 30 ms, flip angle = 80°, FOV = 216 mm, voxel size = 3 × 3 × 3 mm, 38 slices, anterior commissure-posterior commissure aligned).
Imaging quality control
To control the quality of neuroimaging data, we considered image quality, head motion index, normalization quality, and brain coverage. Participants’ fMRI data were deemed ineligible if their structural or functional images were of poor quality, mean frame-wise displacement (FD) exceeded 0.2 mm [18], normalization was low-quality, or brain coverage was inadequate. Thus, data were analyzed and reported for a total of 26 participants (16 males) who passed all quality control measures.
Resting-state fMRI data analyses
Resting-state fMRI data were preprocessed and analyzed using DPABI V6.1 (Data Processing & Analysis for [resting-state] Brain Imaging; downloaded from http://rfmri.org/dpabi) [19]. The initial 10 functional images were discarded. We then performed slice timing correction and realignment to functional images. Linear trends, Friston 24 head motion parameters [20], the white matter signal, and the cerebrospinal fluid signal were regressed out as nuisance covariates, without global signal regression [21]. Then, we performed segmentation-based normalization using the Diffeomorphic Anatomical Registration Through Exponentiated Lie algebra tool [22]. Finally, we spatially smoothed the normalized functional images with a Gaussian kernel with full width at half maximum of 4 mm. We then applied bandpass temporal filtering (0.01–0.1 Hz) and smoothing to the normalized functional images for data preprocessing.
Next, regions of interest (ROIs) relevant to food valuation and interoception were chosen based on previous studies [8,17]; these included amygdala (right [R] and left [L] hemispheres), anterior mid-cingulate (R), brainstem (R), central opercular cortex (R and L), hypothalamus (R and L), inferior frontal gyrus (L), insula (L), medial prefrontal cortex (R), mid-cingulate (R), pons (L), posterior cingulate (L), superior temporal gyrus (R), and inferior ventral striatum (R and L). Multiple metrics were analyzed, including seed-based functional connectivity (FC), amplitude of low frequency fluctuations, fractional amplitude of low frequency fluctuations, regional homogeneity, and voxel-mirrored homotopic connectivity. Of note, regional homogeneity was calculated before smoothing [23]. Paired-sample t tests were performed to compare the 2 sleep conditions for each resting-state fMRI metric with Gaussian Random Field theory (GRF) correction (2-tailed voxel P < 0.001, cluster P < 0.05). We also considered whether SR might have differential effects by sex and repeated the same analyses and correction parameters separately for males (n = 16) and females (n = 10).
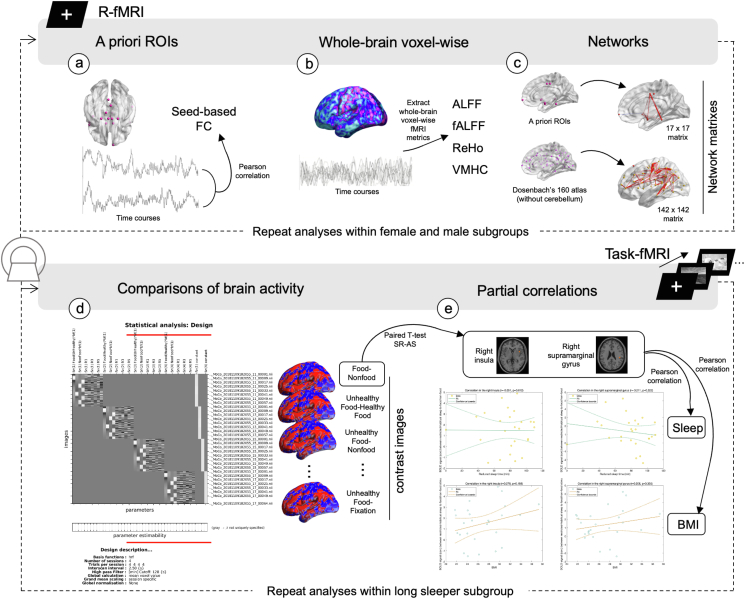
Then, we performed network analyses for the resting-state fMRI data. The network neuroscientific approaches could efficiently model the elements and interactions of brain areas as a graph [24]. Functional brain networks were constructed and analyzed by nodes and edges generated from resting-state fMRI data using DPABINet V1.1, which evolved from DPABI/DPABISurf/DPARSF (downloaded from http://rfmri.org/dpabi) [19]. We determined brain nodes using the 17 a priori ROIs listed above. Brain edges were defined by FC between nodes. Mean FC of the 2 resting-state fMRI scans per sleep condition and data from each scan in each sleep condition were separately used to construct 6 networks per participant. Thus, differences between the 2 sleep conditions were assessed 3 times by paired-sample t test at the group level using the corresponding networks for each sleep condition. Three different correction strategies, including false discovery rate (FDR) correction at a level of q < 0.05, permutation test with FDR correction at q < 0.05, and permutation test with the network-based statistic approach [25] at edge P < 0.001 and component P < 0.05, were applied separately to all inferences to control the family-wise error rate. In addition, brain nodes were also defined using Dosenbach’s 160 ROIs [26], excluding the cerebellum as an exploratory whole-brain analysis. Brain edges were the mean FC of the 2 resting-state fMRI scans corresponding to the same sleep condition. Statistical analyses and correction strategies for multiple comparisons were the same as those used to assess the 17-node networks. A schematic of the analyses is provided in Figure 1.
FIGURE 1.
Schematic diagram of the fMRI data analyses. The first row showcases the resting-state fMRI data analyses that included 3 categories of approaches. (A) A set of 17 a priori regions of interest (ROIs) were used as selected seeds to calculate seed-based functional connectivity (FC). Differences in FC for these seed regions after SR compared with AS were examined. (B) Exploratory whole-brain voxel-wise paired t test procedures were conducted for comparing a set of fMRI metrics after comparing sleep restriction (SR) with adequate sleep (AS) conditions, which included amplitude of low frequency fluctuations (ALFF), fractional ALFF (fALFF), regional homogeneity (ReHo), and voxel-mirrored homotopic connectivity (VMHC). (C) Differences in SR compared with AS were detected through network analyses. A priori ROIs were used to define AS- and SR-brain networks of each participant, and Dosenbach’s 160 atlas without the cerebellum was exploratorily used. The second row showcases the task-fMRI data analyses that included 2 categories. (D) Comparisons of brain activity patterns in response to food cues in SR and AS conditions. Brain activities in response to food minus nonfood and unhealthy food minus healthy food contrasts were the most relevant ones for our hypotheses. Brain contrast images for every other pairwise stimulus comparison were also generated and analyzed as supplementary data. Finally, (E) correlation analyses were conducted to examine the linkage between brain activities and extent of SR and between brain activities and BMI. Brain activities were extracted from 2 clusters that emerged in the threshold of uncorrected P < 0.01 paired t test for SR compared with AS of the food–nonfood contrast.
Task-based fMRI data analyses
Task-based fMRI data and structural MRI data were first preprocessed using the DPABI V6.1 toolbox [19]. The initial 4 functional images acquired for scanner stabilization were discarded. Next, slice acquisition time and head motion were corrected in functional images. Diffeomorphic Anatomical Registration through Exponentiated Lie algebra and normalization were performed using the same preprocessing procedures as for the resting-state fMRI data.
The contrast images of neuronal responses to UHF > HF stimuli and food (food: UHF and HF) > NF stimuli in each sleep condition (AS and SR) were generated for each participant by using SPM12 (Statistical Parametric Mapping; downloaded from https://www.fil.ion.ucl.ac.uk/spm/software/spm12/). Statistical analyses performed across all participants compared the SR and AS sleep conditions using the DPABI toolbox with paired-sample t tests across the whole brain. We also performed 1-sample t tests for contrasts of food > NF and UHF > HF in sleep conditions of SR and AS. GRF correction with a 2-tailed P < 0.001 threshold at the voxel level and P < 0.05 at the cluster level was used to correct for multiple comparisons. Moreover, contrasts of every other pair of stimuli types, including food > fixation (FIX), UHF > FIX, HF > FIX, NF > FIX, UHF > NF, and HF > NF cues were generated and analyzed in the same way (see Supplementary Tables, and Supplementary Figure S1 and S2).
Partial Pearson correlations were employed to evaluate the relationship between the extent of sleep reduction between SR and AS and changes in brain responses to food stimuli, while controlling for sex and age as covariates. The changes in brain responses to food were quantified by the relative blood-oxygen-level-dependent (BOLD) signal in SR compared with AS for food > NF. The quantification of SR was represented by the difference in total sleep time between AS and SR conditions. Similarly, we used the same approach to examine the relationship between BMI and changes in brain responses to food stimuli in specific brain regions.
Results
A total of 26 participants contributed complete data for these analyses (mean age 29.6 ± 5.3 y and BMI 26.9 ± 4.0 kg/m2; 11 had a BMI <25 kg/m2). Average daily time in bed was 8 h 16 min ± 26 min during AS compared with 6 h 42 min ± 25 min during SR. Total sleep time was 7 h 30 min ± 28 min/night for AS compared with 6 h 12 min ± 26 min/night for SR. Sleep efficiency was 90.5% for AS and 91.8% for SR.
Self-reported sleepiness ratings were assessed for pre- compared with postscanning session changes after SR compared with AS conditions using a linear mixed model with factors condition (SR compared with AS), time (pre- compared with postscanning), and their interaction and subject as a random effect. As expected, participants reported greater sleepiness in the SR condition (mean ± SD: 3.22 ± 1.52) than in the AS condition (2.36 ± 1.24), and greater sleepiness after each scan relative to before the scan (3.34 ± 1.52 compared with 2.24 ± 1.13). Importantly, the interaction between condition and time was not significant (F1,72 = 1.01; P = 0.32), suggesting that between-condition brain activity differences were unlikely to be merely driven by a differential increase in sleepiness in the SR compared with AS condition.
Resting-state fMRI results
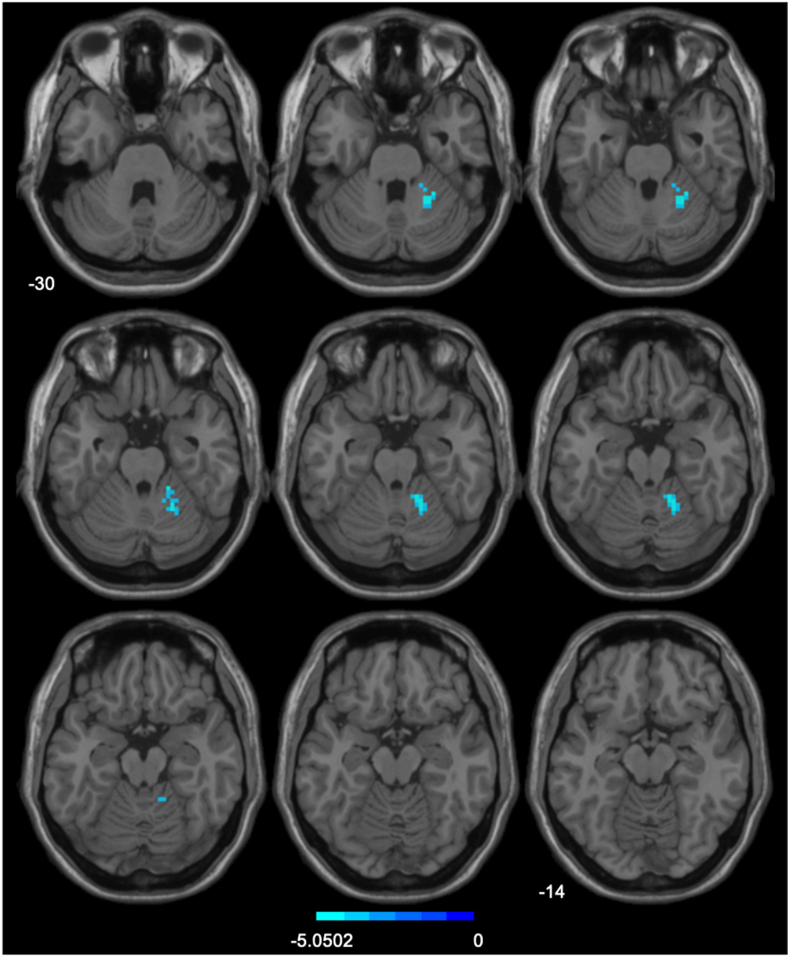
After thresholding each statistical map of the 17 seed-based FC significance tests for the 2 sleep states using a 2-tailed GRF correction procedure of P < 0.001 at the voxel level and P < 0.05 at the cluster level, only FC between the right hypothalamus seed and right cerebellar hemispheric lobules IV, V, and VI were observed to decline with an effect size of Cohen’s d = −0.62 (Figure 2, Supplementary Table S2). Given that the family-wise error rate across the series of significance tests was over 10 times greater than our current threshold for a single test, we consider this a likely false-positive result.
FIGURE 2.
Comparison of seed-based functional connectivity in sleep restriction > adequate sleep. The region of interest is the right hypothalamus. Gaussian Random Field theory correction with 2-tailed voxel-wise P < 0.001, cluster-wise P < 0.05.
When we applied the same correction strategy parameters for other whole-brain voxel-wise resting-state fMRI metrics to compare SR and AS, no significant differences were observed. As for network analyses, no significant edges from either the 17 × 17 a priori ROIs network matrix or the 142 × 142 Dosenbach atlas network matrix survived when comparing the 2 sleep conditions. Moreover, when we explored if SR > AS differed by sex, no significant results emerged from within-sex analyses.
Task-based fMRI results
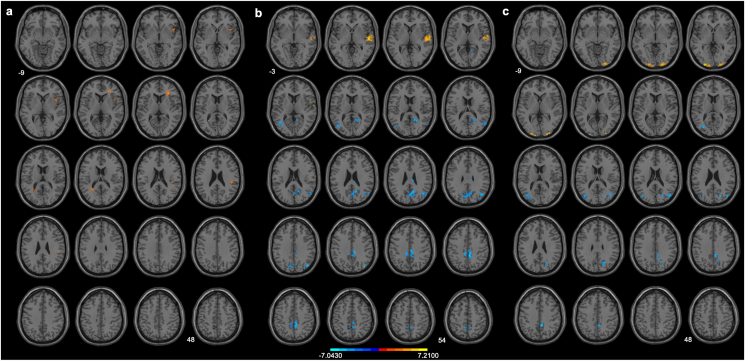
When brain responses to all food > NF contrasts were compared after 6 wk of SR with AS, 2 clusters in the right insula, right inferior frontal gyrus, and right supramarginal gyrus exhibited greater activation with uncorrected P < 0.01, cluster ≥10 voxels, and an effect size of Cohen’s d = 0.16 (Figure 3A and Supplementary Table S3), from which intensities were extracted for follow-up correlation analyses. However, these clusters did not survive GRF correction. Further, when brain responses to all the stimuli contrasts were compared after 6 wk of SR with AS, no results survived statistical correction.
FIGURE 3.
Comparison of regional brain activation in response to food > nonfood of (A) sleep restriction (SR) > adequate sleep (AS), (B) SR, and (C) AS. The statistical maps of SR and AS were thresholded using 2-tailed Gaussian Random Field theory (GRF) correction with voxel size P < 0.001 and cluster size P < 0.05. The comparison of the sleep conditions was thresholded using uncorrected P < 0.01 with a cluster size of ≥10 voxels. When stringent GRF correction parameters were applied, no clusters remained significant.
To further explain the nonsignificant results and to see if a common brain activity pattern to food cues across sleep conditions exists, we quantified brain activity patterns for food > NF and UHF > HF contrasts in each sleep condition through a series of 1-sample t tests. Brain activity patterns for food > NF exhibited differently in sleep conditions of SR and AS. After 6 wk of SR, increased activation in the right superior temporal gyrus, along with decreased activation in the right precuneus, right cuneus, bilateral middle cingulate gyri, right middle occipital gyrus, and left middle temporal gyrus, was observed (corrected P < 0.001 at voxel level and P < 0.05 at cluster level; effect size of Cohen’s d = 0.04; Figure 3B and Supplementary Table S4). After 6 wk of AS, increased activation in response to food > NF was observed in the left middle occipital gyrus and right inferior occipital gyrus, along with decreased activation in the right precuneus, right cuneus, right middle cingulate gyrus, and right middle occipital gyrus (effect size of Cohen’s d = −0.11; Figure 3C and Supplementary Table S5). As for UHF > HF stimuli, although the uncorrected 1-sample t test procedures seemed to reveal different brain activity patterns in the condition of SR and of AS especially in the left frontal gyrus (respective effect sizes of Cohen’s d = −0.17 and = −0.14; Supplementary Figure S1, Supplementary Tables S6 and S7), these clusters did not survive stringent statistical correction.
For the correlation analyses, we detected nominal brain activation differences in response to food > NF stimuli after SR > AS in 2 clusters: right insula and right supramarginal gyrus. There were no correlations between the extent of sleep reduction between SR and AS and changes in brain responses to food stimuli in these 2 clusters. Similarly, we did not detect correlations between participants’ BMI and their changes in brain responses to food between conditions (Supplementary Figure S3).
Discussion
To our knowledge, this study is the longest SR intervention to date to test the impact of ecologically-valid SR in adults with adequate habitual sleep on neuronal responses to food stimuli. Using current rigorous procedures to correct for multiple comparisons, none of our contrasts of interest yielded significant activations or deactivations in this sample of 26 participants, all of whom were scanned twice, with both task-based and resting-state fMRI, at the conclusion of two 6-wk long sleep conditions. No differences were noted in sex-specific analyses.
In this study, resting-state FC analyses based on a priori ROIs revealed increased FC between the right hypothalamus and cerebellum under the SR condition compared with the AS condition. However, we note that we did not correct for the number of a priori ROIs (i.e., a priori hypotheses), and neither a priori ROI-based nor whole-brain node-based network analyses yielded significant results. Therefore, we conservatively conclude that a mild reduction in sleep over 6 wk does not cause meaningful changes in FC between the hypothalamus and cerebellum in the resting state.
Using stringent analytical thresholds, we observed that foods specifically elicited different brain activity patterns after SR and AS. Although increased activation in the occipital gyrus and decreased activation in the right middle cingulate gyrus were observed after AS, we noted increased activation in the right superior temporal gyrus and right insula and decreased activation in the bilateral middle cingulate gyri elicited by foods relative to NF after SR. However, no differences between SR and AS were detected in neuronal responses to foods after pairwise between-group analyses with stringent correction.
Based on our prior studies, we expected increased activation in the insula, nucleus accumbens, and superior frontal gyrus in response to foods compared with NF after SR relative to AS [8]. However, those results were obtained using more liberal thresholds that were standard practice at the time. To verify whether our earlier results would be replicated in our current, mild SR paradigm, we repeated the analyses using those thresholds. When this was done, we observed similar activation in the right insula, supramarginal gyrus, and inferior frontal gyrus in response to foods but not NF after SR relative to AS. Furthermore, we replicated our findings of activation by UHF relative to HF after SR in the inferior, middle, and superior frontal gyrus and inferior parietal gyrus [17]. That these findings failed to emerge in our present, more stringent, analyses suggests that the prior results either included type I errors, as suggested by Eklund et al. [27], or that the milder, albeit longer, duration of SR was insufficient to produce reliable changes in brain BOLD responses.
Of note is a body of recent research questioning published MRI brain imaging studies by highlighting issues, such as high false-positive rates, low reproducibility, and insufficient statistical power [27,28]. Thus, we employed adequate confound regression strategies and multiple comparison corrections in this study, including decreasing head motion effects through a 24-parameter regression model, GRF correction strategies, and network-based statistic and FDR-based correction for network analyses, according to recommendations for best practices from studies in the field of MRI methodology [[28], [29], [30], [31], [32], [33], [34], [35]]. Thus, our findings reflect up-to-date, state-of-the-art methodologic approaches for sound fMRI data analyses.
Strengths
To our knowledge, this study is the longest study to date to evaluate the influence of SR on neuronal appetitive networks. Moreover, contrary to prior research, we employed a mild degree of SR, which is reflective of average short sleepers in the general population. Sleep was verified on a weekly basis using wrist actigraphy in conjunction with sleep diaries, and compliance was high.
We conducted exploratory whole-brain network analyses and whole-brain voxel-wise analyses, analyzing various metrics of resting-state fMRI. Whole-brain analyses are comprehensive and not affected by researcher bias and can complement and confirm the results of ROI-based analysis. In addition, because the functional systems of the human brain have features of an intricate network with multiple temporal and spatial levels, network and graph theory analyses can serve as an effective new approach to help us characterize and understand the integration, cooperation, and information exchange among different regions of the brain [24,36,37]. This study made exploratory attempts in this regard, and we believe that these attempts can provide new ideas and possibilities for future research in this field.
In all analyses, our index of head motion was low (mean FD_Jekinson < 0.2 mm). Additionally, the Friston 24-parameter model was used to regress out head motion confounds, and mean FD was used as a covariate to address the residual effects of motion in group analyses [18,20]. These procedures to control head motion effect are important because previous studies have indicated that even minor head movements can cause changes in the FC maps of fMRI data [[38], [39], [40]].
Finally, our analyses utilized stringent statistical and multiple comparison correction strategies in replicating and comparing to previously published studies. As a result, we were able to replicate prior findings using less stringent analytical practices, which are no longer gold-standard. However, when we applied more stringent approaches, findings became null. Given the milder SR protocol employed here, we are faced with the following 2 potential interpretations: 1) that prior analyses would also fail to produce significant results using more stringent analytical methods; or 2) that milder SR does not produce a robust impact on neuronal responses to food stimuli and FC networks. Additional work in this field is needed to provide definitive answers.
Limitations
Sample size standards have changed, and the quantity of BOLD data produced in this study, especially for resting-state fMRI, was less than optimal for detecting brain–behavior relationships [[41], [42], [43], [44]]. An inadequate sample size has been proposed to dampen the reproducibility of MRI results and brain-wide associations [45]. Though a robust within-participant study can afford a smaller sample size, the reproducibility of its condition effects at a classical sample size (the median sample size of neuroimaging studies is <30) across fMRI metrics and multiple comparison correction strategies is low to moderate [29,46].
Moreover, in such a long-term study spanning several months, data loss is almost inevitable, and we lost as many as a dozen participants. Unfortunately, as a high-dimensional, block-missing dataset, we currently lack established methods for imputing missing data in fMRI datasets [[47], [48], [49], [50]]. Instead, we described the characteristics of the lost participant group using other available information (Supplementary Table S8). We assume that the missing data can be treated as missing at random and excluded, considering that their distribution does not show substantial differences from the overall included participant data.
When considering the statistical power, it indicates that our sample size may not have been sufficient to detect small effects, especially in the cases that most treatment effects in this study have effect sizes of Cohen’s d < 0.2. Therefore, the nonsignificant results may indeed be attributed to both the small effect size and the study's statistical power. However, although power is influenced by sample size, Cohen’s d allows us to gauge the magnitude of the effect [51,52], which is a crucial consideration. The small Cohen's d values suggest that even if our study were highly powered by a larger sample size, the observed effects would likely remain relatively modest [51].
Conclusions
Although we observed expected brain responses to “low-level” contrasts, these effects did not survive current standards for multiple comparison correction in an examination of the effects of ∼1.5 h of SR sustained over 6 wk in a sample of 26 healthy participants. These null results could indicate that a milder form of SR does not lead to meaningful alterations in food-based neuronal activation patterns. Alternatively, they may indicate that prior reported findings may need to be reconsidered, as at least some of them may reflect type I errors based on then-prevailing statistical correction methods that are now considered insufficiently rigorous.
Acknowledgments
We thank the research assistants (Ismael Salazar and Mehreen Bhatti) who contributed to this work.
Author contributions
The authors’ responsibilities were as follows – MPSO, FXC, YY: designed research; MPSO, YY: conducted research; XYL, CGY: analyzed data; XYL, MPSO, YY, FXC: wrote the paper; MPSO: had primary responsibility for final content; and all authors: read and approved the final manuscript.
Conflict of interest
MPSO reports financial support was provided by National Heart Lung and Blood Institute and reports a relationship with Nestle Research & Development that includes: consulting or advisory. All other authors report no conflicts of interest.
Funding
This work was supported by NIH grant R01HL128226 and NIH R35HL155670 (MPSO) and by an NIH Clinical and Translational Science Award (CTSA) through its Center for Advancing Translational Sciences, grant no. UL1TR001873, the Sci-Tech Innovation 2030 - Major Project of Brain Science and Brain-inspired Intelligence Technology (grant number: 2021ZD0200600), National Key R&D Program of China (grant number: 2017YFC1309902), the National Natural Science Foundation of China (grant numbers: 82122035, 81671774, 81630031), the 13th Five-year Informatization Plan of Chinese Academy of Sciences (grant number: XXH13505), the Key Research Program of the Chinese Academy of Sciences (grant NO. ZDBS-SSW-JSC006), Beijing Nova Program of Science and Technology (grant number: Z191100001119104), and the Scientific Foundation of Institute of Psychology, Chinese Academy of Sciences (grant number: E2CX4425YZ). The supporting sources had no involvement or restriction regarding publication.
Data availability
Data described in the manuscript, code book, and analytic code will be made available upon request.
Footnotes
Marie-Pierre St-Onge is an Editor for The Journal of Nutrition and played no role in the Journal’s evaluation of the manuscript.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tjnut.2023.12.016.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Tsao C.W., Aday A.W., Almarzooq Z.I., Anderson C.A.M., Arora P., Avery C.L., et al. Heart disease and stroke statistics-2023 update: a report from the American Heart Association. Circulation. 2023;147(8) doi: 10.1161/CIR.0000000000001123. e93–e621, [DOI] [PubMed] [Google Scholar]
- 2.Guimarães K.C., Silva C.M., Latorraca C.O.C., Oliveira R.Á., Crispim C.A. Is self-reported short sleep duration associated with obesity? A systematic review and meta-analysis of cohort studies. Nutr. Rev. 2022;80(5):983–1000. doi: 10.1093/nutrit/nuab064. [DOI] [PubMed] [Google Scholar]
- 3.Covassin N., Singh P., McCrady-Spitzer S.K., St Louis E.K., Calvin A.D., Levine J.A., et al. Effects of experimental sleep restriction on energy intake, energy expenditure, and visceral obesity. J. Am. Coll. Cardiol. 2022;79(13):1254–1265. doi: 10.1016/j.jacc.2022.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al Khatib H.K., Harding S.V., Darzi J., Pot G.K. The effects of partial sleep deprivation on energy balance: a systematic review and meta-analysis. Eur. J. Clin. Nutr. 2017;71(5):614–624. doi: 10.1038/ejcn.2016.201. [DOI] [PubMed] [Google Scholar]
- 5.Fenton S., Burrows T.L., Skinner J.A., Duncan M.J. The influence of sleep health on dietary intake: a systematic review and meta-analysis of intervention studies. J. Hum. Nutr. Diet. 2021;34(2):273–285. doi: 10.1111/jhn.12813. [DOI] [PubMed] [Google Scholar]
- 6.González-Ortiz A., López-Bautista F., Valencia-Flores M., Espinosa Cuevas Á. Partial sleep deprivation on dietary energy intake in healthy population: a systematic review and meta-analysis. Nutr. Hosp. 2020;37(5):1052–1060. doi: 10.20960/nh.03108. [DOI] [PubMed] [Google Scholar]
- 7.Chaput J.P., St-Onge M.P. Increased food intake by insufficient sleep in humans: are we jumping the gun on the hormonal explanation? Front. Endocrinol. 2014;5:116. doi: 10.3389/fendo.2014.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.St-Onge M.P., McReynolds A., Trivedi Z.B., Roberts A.L., Sy M., Hirsch J. Sleep restriction leads to increased activation of brain regions sensitive to food stimuli. Am. J. Clin. Nutr. 2012;95(4):818–824. doi: 10.3945/ajcn.111.027383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiFrancesco M.W., Alsameen M., St-Onge M.P., Duraccio K.M., Beebe D.W. Altered neuronal response to visual food stimuli in adolescents undergoing chronic sleep restriction. Sleep. 2023 doi: 10.1093/sleep/zsad036. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benedict C., Brooks S.J., O’Daly O.G., Almèn M.S., Morell A., Åberg K., et al. Acute sleep deprivation enhances the brain's response to hedonic food stimuli: an fMRI study. J. Clin. Endocrinol. Metab. 2012;97(3):E443–E447. doi: 10.1210/jc.2011-2759. [DOI] [PubMed] [Google Scholar]
- 11.Rihm J.S., Menz M.M., Schultz H., Bruder L., Schilbach L., Schmid S.M., et al. Sleep deprivation selectively upregulates an amygdala-hypothalamic circuit involved in food reward. J. Neurosci. 2019;39(5):888–899. doi: 10.1523/JNEUROSCI.0250-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demos K.E., Heatherton T.F., Kelley W.M. Individual differences in nucleus accumbens activity to food and sexual images predict weight gain and sexual behavior. J. Neurosci. 2012;32(16):5549–5552. doi: 10.1523/JNEUROSCI.5958-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoncheva Y.N., Castellanos F.X., Pizinger T., Kovtun K., St-Onge M.P. Sleep and meal-time misalignment alters functional connectivity: a pilot resting-state study. Int. J. Obes. (Lond.) 2016;40(11):1813–1816. doi: 10.1038/ijo.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kyeong S., Choi S.H., Eun Shin J., Lee W.S., Yang K.H., Chung T.S., et al. Functional connectivity of the circadian clock and neural substrates of sleep-wake disturbance in delirium. Psychiatry Res. Neuroimaging. 2017;264:10–12. doi: 10.1016/j.pscychresns.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 15.Fang Z., Spaeth A.M., Ma N., Zhu S., Hu S., Goel N., et al. Altered salience network connectivity predicts macronutrient intake after sleep deprivation. Sci. Rep. 2015;5:8215. doi: 10.1038/srep08215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spaeth A.M., Dinges D.F., Goel N. Effects of experimental sleep restriction on weight gain, caloric intake, and meal timing in healthy adults. Sleep. 2013;36(7):981–990. doi: 10.5665/sleep.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.St-Onge M.P., Wolfe S., Sy M., Shechter A., Hirsch J. Sleep restriction increases the neuronal response to unhealthy food in normal-weight individuals. Int. J. Obes. (Lond.) 2014;38(3):411–416. doi: 10.1038/ijo.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenkinson M., Bannister P., Brady M., Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17(2):825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 19.Yan C.G., Wang X.D., Zuo X.N., Zang Y.F. DPABI: data processing & analysis for (resting-state) brain imaging. Neuroinformatics. 2016;14(3):339–351. doi: 10.1007/s12021-016-9299-4. [DOI] [PubMed] [Google Scholar]
- 20.Friston K.J., Williams S., Howard R., Frackowiak R.S., Turner R. Movement-related effects in fMRI time-series. Magn. Reson. Med. 1996;35(3):346–355. doi: 10.1002/mrm.1910350312. [DOI] [PubMed] [Google Scholar]
- 21.Murphy K., Fox M.D. Towards a consensus regarding global signal regression for resting state functional connectivity MRI. Neuroimage. 2017;154:169–173. doi: 10.1016/j.neuroimage.2016.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38(1):95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Zang Y., Jiang T., Lu Y., He Y., Tian L. Regional homogeneity approach to fMRI data analysis. Neuroimage. 2004;22(1):394–400. doi: 10.1016/j.neuroimage.2003.12.030. [DOI] [PubMed] [Google Scholar]
- 24.Bassett D.S., Sporns O. Network neuroscience. Nat. Neurosci. 2017;20(3):353–364. doi: 10.1038/nn.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zalesky A., Fornito A., Bullmore E.T. Network-based statistic: identifying differences in brain networks. Neuroimage. 2010;53(4):1197–1207. doi: 10.1016/j.neuroimage.2010.06.041. [DOI] [PubMed] [Google Scholar]
- 26.Dosenbach N.U., Nardos B., Cohen A.L., Fair D.A., Power J.D., Church J.A., et al. Prediction of individual brain maturity using fMRI. Science. 2010;329(5997):1358–1361. doi: 10.1126/science.1194144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eklund A., Nichols T.E., Knutsson H. Cluster failure: why fMRI inferences for spatial extent have inflated false-positive rates. Proc. Natl. Acad. Sci. U. S. A. 2016;113(28):7900–7905. doi: 10.1073/pnas.1602413113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bennett C., Miller M., Wolford G. Neural correlates of interspecies perspective taking in the post-mortem Atlantic Salmon: an argument for multiple comparisons correction. Neuroimage. 2009;47(suppl 1):S125. doi: 10.1016/S1053-8119(09)71202-9. [DOI] [Google Scholar]
- 29.Chen X., Lu B., Yan C.G. Reproducibility of R-fMRI metrics on the impact of different strategies for multiple comparison correction and sample sizes. Hum. Brain Mapp. 2018;39(1):300–318. doi: 10.1002/hbm.23843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nichols T., Hayasaka S. Controlling the familywise error rate in functional neuroimaging: a comparative review. Stat. Methods Med. Res. 2003;12(5):419–446. doi: 10.1191/0962280203sm341ra. [DOI] [PubMed] [Google Scholar]
- 31.Genovese C.R., Lazar N.A., Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage. 2002;15(4):870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- 32.Ciric R., Wolf D.H., Power J.D., Roalf D.R., Baum G.L., Ruparel K., et al. Benchmarking of participant-level confound regression strategies for the control of motion artifact in studies of functional connectivity. Neuroimage. 2017;154:174–187. doi: 10.1016/j.neuroimage.2017.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan C.G., Cheung B., Kelly C., Colcombe S., Craddock R.C., Di Martino A., et al. A comprehensive assessment of regional variation in the impact of head micromovements on functional connectomics. Neuroimage. 2013;76:183–201. doi: 10.1016/j.neuroimage.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ciric R., Rosen A.F.G., Erus G., Cieslak M., Adebimpe A., Cook P.A., et al. Mitigating head motion artifact in functional connectivity MRI. Nat. Protoc. 2018;13(12):2801–2826. doi: 10.1038/s41596-018-0065-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Power J.D., Mitra A., Laumann T.O., Snyder A.Z., Schlaggar B.L., Petersen S.E. Methods to detect, characterize, and remove motion artifact in resting state fMRI. Neuroimage. 2014;84:320–341. doi: 10.1016/j.neuroimage.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bullmore E., Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 2009;10(3):186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- 37.Dale A.M., Fischl B., Sereno M.I. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9(2):179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 38.Power J.D., Barnes K.A., Snyder A.Z., Schlaggar B.L., Petersen S.E. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59(3):2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Satterthwaite T.D., Wolf D.H., Loughead J., Ruparel K., Elliott M.A., Hakonarson H., et al. Impact of in-scanner head motion on multiple measures of functional connectivity: relevance for studies of neurodevelopment in youth. Neuroimage. 2012;60(1):623–632. doi: 10.1016/j.neuroimage.2011.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Dijk K.R., Sabuncu M.R., Buckner R.L. The influence of head motion on intrinsic functional connectivity MRI. Neuroimage. 2012;59(1):431–438. doi: 10.1016/j.neuroimage.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bijsterbosch J., Harrison S.J., Jbabdi S., Woolrich M., Beckmann C., Smith S., et al. Challenges and future directions for representations of functional brain organization. Nat. Neurosci. 2020;23(12):1484–1495. doi: 10.1038/s41593-020-00726-z. [DOI] [PubMed] [Google Scholar]
- 42.Ioannidis J.P. Why most published research findings are false. PLoS Med. 2005;2(8):e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Button K.S., Ioannidis J.P., Mokrysz C., Nosek B.A., Flint J., Robinson E.S., et al. Power failure: why small sample size undermines the reliability of neuroscience. Nat. Rev. Neurosci. 2013;14(5):365–376. doi: 10.1038/nrn3475. [DOI] [PubMed] [Google Scholar]
- 44.Zuo X.N., Xu T., Milham M.P. Harnessing reliability for neuroscience research. Nat. Hum. Behav. 2019;3(8):768–771. doi: 10.1038/s41562-019-0655-x. [DOI] [PubMed] [Google Scholar]
- 45.Marek S., Tervo-Clemmens B., Calabro F.J., Montez D.F., Kay B.P., Hatoum A.S., et al. Reproducible brain-wide association studies require thousands of individuals. Nature. 2022;603(7902):654–660. doi: 10.1038/s41586-022-04492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szucs D., Ioannidis J.P. Sample size evolution in neuroimaging research: an evaluation of highly-cited studies (1990-2012) and of latest practices (2017-2018) in high-impact journals. Neuroimage. 2020;221 doi: 10.1016/j.neuroimage.2020.117164. [DOI] [PubMed] [Google Scholar]
- 47.Eekhout I., Enders C.K., Twisk J.W., de Boer M.R., de Vet H.C., Heymans M.W. Including auxiliary item information in longitudinal data analyses improved handling missing questionnaire outcome data. J. Clin. Epidemiol. 2015;68(6):637–645. doi: 10.1016/j.jclinepi.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 48.Enders C.K. Multiple imputation as a flexible tool for missing data handling in clinical research. Behav. Res. Ther. 2017;98:4–18. doi: 10.1016/j.brat.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 49.Ibrahim J.G., Molenberghs G. Missing data methods in longitudinal studies: a review. Test (Madr.) 2009;18(1):1–43. doi: 10.1007/s11749-009-0138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoyez H., Schockaert C., Rambach J., Mirbach B., Stricker D. Unsupervised image-to-image translation: a review. Sensors (Basel) 2022;22(21):8540. doi: 10.3390/s22218540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peterson S.J., Foley S. Clinician’s guide to understanding effect size, alpha level, power, and sample size. Nutr. Clin. Pract. 2021;36(3):598–605. doi: 10.1002/ncp.10674. [DOI] [PubMed] [Google Scholar]
- 52.Faul F., Erdfelder E., Lang A.G., Buchner A. G∗Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods. 2007;39(2):175–191. doi: 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data described in the manuscript, code book, and analytic code will be made available upon request.