Abstract
The human β-globin locus control region (LCR) consists of five erythroid-lineage-specific DNase I-hypersensitive sites (HSs) and is required for activation of the β-globin locus chromatin domain and globin gene expression. Each DNase I-HS of the LCR consists of a highly conserved core element and flanking sequences. To analyze the functional role of the core elements of the HSs, we deleted a 234-bp fragment encompassing the core of HS3 (HS3c) from a β-globin locus residing on a 248-kb β-locus yeast artificial chromosome and analyzed its function in F2 progeny of transgenic mice. Human ɛ-globin gene expression was absent at day 10 and severely reduced in the day 12 embryonic erythropoiesis of mice lacking HS3c. In contrast, γ-globin gene expression was normal in embryonic erythropoiesis but it was absent in definitive erythropoiesis in the fetal liver. These results indicate that the core element of HS3 is necessary for ɛ-globin gene transcription in embryonic cells and for γ-globin gene transcription in definitive cells. Normal γ-globin gene expression in embryonic cells and the absence of γ-globin gene expression in definitive cells show that different HSs interact with γ-globin gene promoters in these two stages of development. Such results provide direct evidence for developmental stage specificity of the interactions between the core elements of HSs and the promoters of the globin genes.
The human β-globin locus contains five actively transcribed genes that are arranged in their developmental order of expression. High-level expression of the β-globin gene cluster is dependent on the presence of the locus control region (LCR) (18), an element characterized by a series of five DNase I-hypersensitive sites (HSs) located 6 to 22 kb upstream of the ɛ-globin gene (9, 10, 18, 44). Naturally occurring deletions of this element result in changes in chromatin structure that extend at least 200 kb 3′ of the deletion, transcriptional silencing of the β-globin locus, and a phenotype of β thalassemia (4, 5, 12, 22). Functional properties of the LCR include activation of the β-globin locus (10, 18), restriction of globin gene expression to cells of the erythroid cell lineage (18, 45), enhancement of globin gene expression (11, 18, 39), and protection from position effects of globin genes transferred in transgenic mice (13, 18, 25, 41).
Transgenic mice have been extensively used to study the developmental control of the β-globin genes, the function of the LCR, and the role of individual HSs in β-globin gene regulation. Linkage of individual HSs to individual globin genes have shown that HS2, HS3, and HS4 are capable of conferring position-independent expression of globin genes, with stronger activation of expression at a specific stage of development (14, 25). Several observations have led to a model suggesting that the HSs form a complex that directly interacts with globin gene promoters by looping of the intervening DNA (7, 28, 46). HS2, HS3, and HS4 have 200- to 400-bp core regions that are able to provide position-independent expression in transgenic mice (27, 34, 35, 37, 42). These HS core regions may be indispensable components of the LCR complex; deletions of the HS3 or HS4 core elements result in disruption of HS function and reduction of globin gene expression (3).
Discernment of the function of individual HSs and analysis of how the LCR interacts with individual genes during development require studies in the context of intact, native β-globin loci. Entire β-globin loci have been used to generate transgenic mice, by ligating two cosmids to produce a 70-kb fragment (40) or by using 248-kb (30) or 150-kb (15, 36) yeast artificial chromosomes harboring the β-globin locus (β-YACs). Mice carrying β-YACs show correct regulation of the human globin genes, presumably because all the human cis-regulatory elements are present in the transferred sequences of the β-globin locus and are properly recognized by the murine trans-acting environment. In β-YAC transgenic mice, the ɛ-globin gene is expressed during the embryonic stage of development and is confined to primitive erythropoiesis in the yolk sac. The γ-globin genes are also expressed in the embryonic yolk sac, but unlike their murine homologous gene, βh1, γ-globin gene expression continues in the fetal liver stage of erythropoiesis. Human β-globin gene expression occurs only in the cells of definitive erythropoiesis.
To delineate the role of HS3 in LCR function and globin gene expression during development, we produced β-YAC transgenic mice carrying either large deletions of LCR sequences containing the individual HSs or the core elements of these sites. We have previously reported results obtained from extensive deletions of HS3 and HS2 (32). In the study summarized in this paper, we deleted the core element of HS3 of the LCR from a β-globin locus residing on a 248-kb β-YAC and used this β-YAC to produce transgenic mice. In the embryonic yolk sac of these mice, ɛ-globin gene expression was absent but γ-globin gene expression was normal. However, γ-globin gene expression was totally silent in erythroid cells originating in fetal liver. The levels of β-globin gene expression were decreased and varied among the transgenic lines, indicating that β-globin gene expression was influenced by the position of integration of the β-YAC transgene into the murine genome. Based on these results, we propose that the core of the HS3 directly interacts with the globin gene promoters during embryonic and fetal development, resulting in activation of ɛ-globin gene expression in the yolk sac and of γ-globin gene expression in the fetal liver. Indirectly, our results also suggest that the LCR changes conformations during the course of development.
MATERIALS AND METHODS
Construction of a β-YAC lacking HS3c (ΔHS3c).
Plasmid pIII(0.7) contains a 784-bp PstI fragment encompassing the 225-bp 5′ HS3c element and 559 bp of flanking DNA sequence (GenBank coordinates 4348 to 5132). Plasmid pIII(0.7) DNA was digested with the restriction enzyme PstI, and the 784-bp insert was subcloned into PstI-digested plasmid pALTER-1 (Promega, Madison, Wis.) to generate pALHS3(0.7). Two primers, each with two nucleotide substitutions, were synthesized and used to create EcoRI sites flanking the 5′ HS3c element (GenBank coordinates 4541 to 4776) by site-directed mutagenesis with the Altered Sites II in vitro mutagenesis system (Promega) according to the manufacturer’s protocol. The primer DNA sequences were 5′ CCCTCACGGTGAATTCGCGAGCTGG 3′ (proximal) and 5′ GTAGTAGAATGAAGAATCTGCTATGC 3′ (distal); the nucleotide substitutions are in boldface type and the EcoRI sites are underlined. Plasmid pIII(4.4), containing a 225-bp HpaI-KpnI fragment encompassing 5′ HS3 (GenBank coordinates 3379 to 7764), was digested with restriction enzyme HindIII, and a 1.8-kb HindIII fragment containing 5′ HS3 sequence was isolated and subcloned into HindIII-digested pUC19 in which the EcoRI and PstI sites had been ablated. The resultant plasmid, pUCHS3(1.8), which contained 5′ HS3 (GenBank coordinates 3379 to 5172), was digested with restriction enzyme PstI to remove the 784-bp PstI fragment containing 5′ HS3 sequence, and the mutagenized 784-bp fragment from plasmid pALHS3(0.7) was subcloned in its place to generate plasmid pUCHS3m. Plasmid pUCHS3m was digested with restriction enzyme EcoRI to remove the 234-bp EcoRI fragment containing the 5′ HS3c element and circularized by the addition of T4 DNA ligase (Boehringer Mannheim, Indianapolis, Ind.) to generate plasmid pUCΔHS3c(1.6). Digestion of pUCΔHS3c(1.6) with restriction enzyme HindIII generated a 1.6-kb HindIII fragment containing the 5′ HS3 core deletion that was subcloned into the yeast-integrating-plasmid (YIP) vector pRS406 (Stratagene, La Jolla, Calif.), from which the SpeI restriction site had been deleted to produce plasmid pRSΔHS3c(1.6). One microgram of pRSΔHS3c(1.6) was linearized with SpeI at a unique site 3′ of the 5′ HS3 deletion and transformed into yeast spheroplasts (16). Transformants were selected for uracil prototrophy on complete medium lacking uracil, and proper intergration of the YIP in isolates containing YACs was determined by Southern blot hybridization analysis. Spontaneous excision of the YIP via homologous recombination was permitted by overnight growth in nonselective rich medium (yeast-peptone-dextrose) (47). Aliquots of the culture were plated on 5-fluoroorotic acid plates to select for loss of the URA3 gene due to excision of the YIP vector, which resulted in 5-fluoroorotic acid resistance. Deletion of the 5′ HS3c element was determined by Southern blot hybridization analysis. (The approach used is summarized in Fig. 1.)
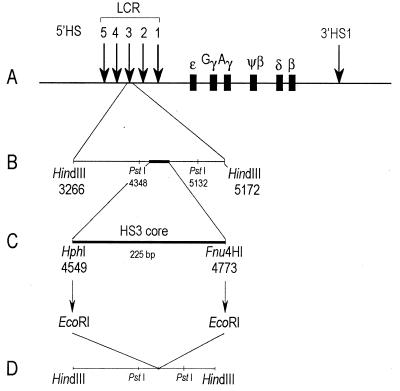
FIG. 1.
β-Globin locus and the location of the HS3c deletion. (A) Five DNase I-HSs are located 6 to 22 kb 5′ of the ɛ-globin gene, and a single HS (3′ HS1) is located approximately 20 kb 3′ of the β-globin gene. (B) A 784-bp PstI fragment containing the HS3c was subcloned into pALTER-1 for site-directed mutagenesis. (C) The HphI and Fnu4HI restriction sites, flanking the core element of HS3, were mutagenized to create EcoRI sites. Digestion with EcoRI simultaneously eliminated the core element of HS3 and left a diagnostic EcoRI restriction site. (D) The 1.6-kb HindIII fragment was subcloned into the YIP used to introduce the deletion into the β-YAC via homologous recombination.
YAC purification and production of transgenic mice.
The ΔHS3c β-YAC yeast strain was grown and agarose plugs were prepared as previously described (20). Preparative plugs were loaded on a 0.5% MP agarose gel (Boehringer Mannheim), and the DNA was fractionated by pulsed-field gel electrophoresis (PFGE) (CHEF DRII apparatus; Bio-Rad, Hercules, Calif.) in 0.5× TBE (44.5 mM Tris, 44.5 mM boric acid, 1 M EDTA) at 200 V with a 60-s switch for 20 h at 12°C. A portion of the gel was stained with ethidium bromide to determine the migration distance of the ΔHS3c β-YAC, and the YAC DNA was cut from the gel. The gel slice containing the YAC DNA and a second slice containing a yeast chromosome were rotated 90° relative to their original directions of mobility and electrophoresed in a 4% low-melting-point agarose (LMPA) (NuSieve GTG; FMC, Rockland, Maine) in 0.5× TBE at 47 V for 15 h to concentrate the YAC. The yeast lane was stained with ethidium bromide to determine the migration distance of the β-YAC DNA into the 4% LMPA. A slice of approximately 8 mm was weighed and equilibrated in a 100× volume of 10 mM Tris-HCl (pH 7.5)–250 μM EDTA–100 mM NaCl for 1 h at room temperature without agitation. The gel slice was placed in a microcentrifuge tube, and the agarose was melted at 68°C for 10 min and then immediately placed at 42.5°C for 5 min. Two units of β-agarase (New England Biolabs, Beverly, Mass.) per 100 mg of agarose was added and digested overnight at 42.5°C. Integrity of the YAC DNA was determined by PFGE as described above prior to its injection into fertilized mouse eggs. DNA concentration was determined by fluorometry (Pharmacia, Piscataway, N.J.), and the YAC DNA was diluted to a final concentration of 2.0 ng/μl with a solution containing 10 mM Tris-HCl (pH 7.5), 250 μM EDTA, and 100 mM NaCl and filtered through a 0.22-μm-pore-size Acrodisk (Gelman, Ann Arbor, Mich.) just prior to injection.
Structural analysis of ΔHS3c β-YAC transgenic mice.
Transgenic founder (F0) animals were identified by hybridization of tail DNA slot blots with a γ-globin gene probe. Founders were bred to produce F1 progeny for structure-function analysis. Fresh liver cell suspensions were prepared as follows. Liver was cut into small pieces and then mechanically sheared by successive passage through a 16-gauge syringe. The cells were washed twice with Dulbecco’s phosphate-buffered saline and resuspended at a concentration of 3 × 107 cells/ml in phosphate-buffered saline. An equal volume of 2% LMPA (Seaplaque GTG agarose) was added to the liver suspension, and plugs were cast. The plugs were incubated in LDS solution (1% lithium dodecyl sulfate, 100 mM EDTA [pH 8.0], 10 mM Tris-HCl [pH 8.0]) at 37°C for 1 h, followed by a second incubation overnight. The plugs were then washed twice for 30 min in 0.2× NDS (0.2% lauryl sarcosinate, 100 mM EDTA, 2 mM Tris base [pH 9.5]), followed by three 30-min washes in TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0]). The plugs were stored at 4°C in TE.
Twelve agarose plugs were digested overnight at 50°C with 20 U of SfiI (Boehringer Mannheim) in a total volume of 200 μl after preequilibration in 200 μl of 1× SfiI buffer. The DNAs were fractionated by PFGE with a 1% agarose gel (SeaKem Gold GTG; FMC) at 200 V with a 14-s switch for 22 h at 14°C in 0.5× TBE. The gels were capillary blotted overnight onto nylon membranes (Hybond N+; Amersham, Arlington Heights, Ill.) in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The nylon membrane was cut into strips representing individual lanes, and each strip was hybridized with a different probe spanning the β-globin locus from 5′ HS3 to the hereditary persistence of fetal hemoglobin type 6 (HPFH6) breakpoint. After overnight hybridization and washing, the strips were reassembled and subjected to autoradiography. A 140-kb SfiI fragment is the expected size for an intact ΔHS3c β-YAC, and fragments of different sizes indicate that ΔHS3c β-YAC copies have deletions. The probes used were as follows: 0.7-kb PstI 5′ HS3, 1.9-kb HindIII 5′ HS2, the 3.7-kb EcoRI ɛ-globin gene, the 2.4-kb EcoRI 3′ Aγ-globin gene, 1.0-kb EcoRV ψβ, the 2.1-kb PstI 5′ δ-globin gene, the 0.9-kb EcoRI-BamHI β-globin gene, 1.4-kb XbaI DF10 (3′ HS1), 1.9-kb BglII HPFH3, 0.5-kb HindIII H500, and 1.5-kb EcoRI-BglII HPFH6. All fragments were radiolabeled with a Decaprime II random labeling kit (Ambion, Austin, Tex.). The 5′ δ-globin gene, HPFH3, and HPFH6 probe templates were gifts of N. P. Anagnou (University of Crete), DF10 was a gift from D. Fleenor (Duke University), and H500 was a gift from D. Mager (University of British Columbia).
Copy number determination.
Agarose plugs containing transgenic mouse liver DNA were digested overnight with the restriction enzyme AccI, and the DNAs were fractionated by agarose gel electrophoresis and blotted to a nylon membrane as described above. Copy number was determined by comparing human Aγ-globin gene and murine Thy1.1 (gift from R. Perlmutter) hybridization signals by Southern blot hybridization. Thy1.1 serves as an internal diploid control. To ensure equal levels of labeling of both the Aγ-globin gene and Thy1.1 fragments, the following construct was synthesized. A 753-bp HindIII fragment containing sequences 3′ of the Aγ-globin gene (GenBank coordinates 41382 to 42135) was cloned into pW126, a pBluescript (Stratagene) plasmid containing a 544-bp BamHI Thy1.1 cDNA, to produce pThy1.1/3′ Aγ(753). Digestion with XbaI and XhoI released a 1.3-kb fragment that was labeled with a Decaprime II random labeling kit (Ambion). As an internal control during Southern blot hybridization, we digested pThy1.1/3′ Aγ(753) with PstI and ScaI to release a 2.6-kb Aγ fragment and a 1.6-kb Thy1.1 fragment. Approximately 10 pg of this control was electrophoresed alongside the digested mouse genomic DNAs. Hybridization signals were quantitated with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). The ratio of Aγ to Thy1.1 was calculated from the plasmid control, and this ratio was used to correct for differences in specific activities between the two probes. The corrected Thy1.1 values were divided by 2 to obtain a single copy value for each lane containing genomic DNA. ΔHS3c β-YAC copy number was determined by dividing the Aγ signal by the corrected Thy1.1 single-copy value.
Measurement of globin mRNA synthesis.
Total RNA was isolated from F2 transgenic tissues with the RNAgents Isolation system (Promega). Human and murine globin RNA were quantitated by RNase protection analysis with an RPA II kit (Ambion). RNA probes were synthesized with a MAXIscript transcription kit (Ambion). Template DNAs used to measure human ɛ-, γ-, and β-globin mRNAs were pT7H ɛ(188), pT7Aγm(170), and pT7βm, respectively (26). Template DNAs used to measure murine α and ζ globin mRNAs were pT7Mα and pT7Mζ, respectively (1). RNAs were isolated from day 10 yolk sac (1,000 ng), day 12 liver (500 ng), day 12 blood (80 ng), day 14 liver (500 ng), and adult blood (80 ng). Signals were quantitated with a PhosphorImager (Molecular Dynamics).
Immunofluorescent detection of human globin chains.
Globin chains were visualized by staining fixed cells with ɛ-, γ-, or β-globin-specific monoclonal antibodies. Cytocentrifuge smears were fixed in methanol and incubated with appropriate antibodies. A second antibody [goat F(ab′)2 anti-mouse fluorescein isothiocyanate-conjugated immunoglobulin G; Dupont, Wilmington, Del.] that is reactive to the mouse monoclonal antibodies was added to allow color detection of the globin proteins.
RESULTS
Structural analysis of transgenic mice.
Previous studies have shown that YAC transgenes frequently have deletions of the 5′ and 3′ sequences, thus requiring detailed structural analysis of the YAC DNA integrated in transgenic mice (30–33). Identification of mice bearing intact β-globin loci is an essential prerequisite to functional studies. The continuity of the β-globin locus within individual YAC copies was determined as described in Materials and Methods and shown in Fig. 2. Four lines had at least one intact 140-kb SfiI fragment and were used in this study. The presence of HS4, which resides upstream of the 5′ SfiI site used in our structural analysis, was confirmed in all lines (Fig. 3). Line A has an intact 140-kb fragment and an additional 120-kb fragment containing a β-globin locus with a deletion 3′ to the δ-globin gene. Thus, line A has two copies of ɛ-, Gγ-, Aγ-, and δ-globin genes but a single β-globin gene. Line B has a single 140-kb fragment containing the entire β-globin locus. Line C has two intact β-globin loci of 150 and 160 kb. Line D contains an intact β-globin locus on a 135-kb fragment which is missing sequences downstream of the β-globin gene, including 3′ HS1; the deletion in the 135-kb fragment, however, spares the enhancer element (2, 23, 43), which is located 0.4 kb 3′ of the β-globin locus (Fig. 4). Structural rearrangements of some YAC copies (deletions of the 5′ or 3′ end or both ends) are common in β-YAC transgenics (31–33). However, if the β-globin locus itself is intact (i.e., from 5′ HS4 through the β-globin gene enhancer), developmental expression of the globin genes is normal both temporally and spatially and there is position-independent, copy-number-dependent expression of the globin genes (33).
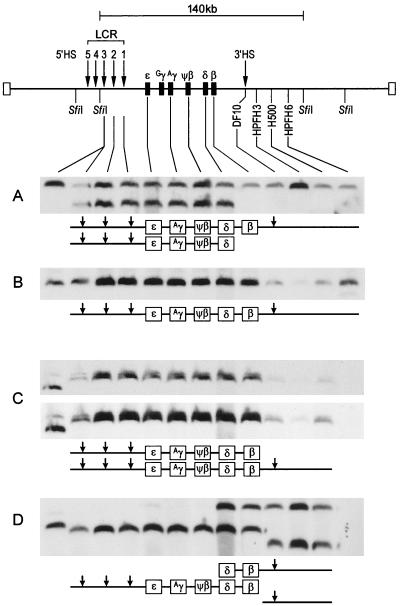
FIG. 2.
Structures of the β-YAC globin loci of ΔHS3c β-YAC transgenic mouse lines. The upper part of the figure shows the 140-kb SfiI fragment encompassing most of the β-globin locus from 5′ HS3 to the breakpoint of HPFH6 approximately 53 kb downstream of the β-globin gene. Arrows indicate HSs. Agarose plugs containing high-molecular-weight DNA from liver tissue were isolated from four ΔHS3c β-YAC transgenic lines. The plugs were digested with SfiI, fractionated by PFGE, and subjected to Southern hybridization analyses. The location of each of the probes used in the structural analysis is identified on the map. Schematic representations of the structures of the SfiI fragments are drawn below each autoradiogram. (A) Line A has an intact 140-kb fragment and an additional 120-kb fragment which is deleted from sequences 3′ of the δ-globin gene. (B) Line B has a single 140-kb fragment identified by each of the probes. (C) Line C has a 150-kb fragment containing the intact locus from 5′ HS3 to position H500 (placed between the breakpoints of HPFH3 and HPFH6). A second 160-kb fragment extends from 5′ HS3 to the β-globin gene. This fragment can be seen in the doublets apparent in the lighter exposure of the same autoradiogram in the upper portion of panel C. (D) Line D has a 140-kb fragment containing an intact β-globin locus and two additional fragments of 120 and 170 kb, from both of which most of the β-globin locus is deleted. The first lane in each of the autoradiograms contains control DNA from a mouse erythroleukemia cell line containing a single intact β-YAC, as determined by structural analysis and fluorescent in situ hybridization.
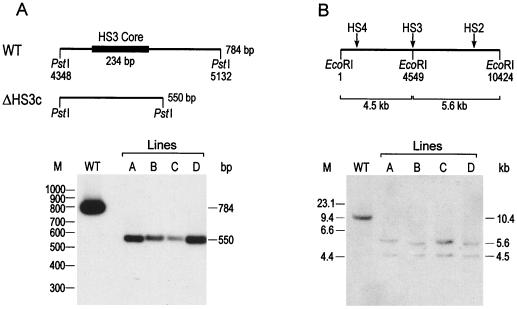
FIG. 3.
Southern analysis of the HS3c deletion showing the presence of HS4 in the ΔHS3c β-YAC transgenic lines. (A) A map of the wild-type 784-bp PstI fragment (GeneBank coordinates indicated) and the 550-bp PstI fragment deleted from the HS3c elements is shown above the autoradiograph. All DNA samples were digested with PstI. Lane WT contains DNA from a transgenic mouse line carrying a wild-type β-YAC. Lanes A to D correspond to the four lines of Fig. 2. Each line has the predicted 550-bp fragment which is diagnostic of the HS3c deletion. The probe was the 784-bp PstI HS3 fragment. M, molecular weight. (B) HS2 to -4 reside on a 10.4-kb EcoRI fragment (GeneBank coordinates indicated) in the wild-type β-globin locus. As shown in the diagram, the creation of an EcoRI site by site-directed mutagenesis and subsequent deletion of the HS3c element results in a 4.5-kb EcoRI fragment containing HS4 and a 5.6-kb fragment containing HS2. The autoradiogram shows the results of EcoRI digestion of samples from the control (lane WT) and lines A to D. Notice that all lines contain a 5.6- and a 4.5-kb fragment, indicating that HS4 is linked to the HS3c deletion. The probe was the 1.4-kb SpeI-HindIII fragment spanning the HS3c element. M, molecular size (in kilobases).
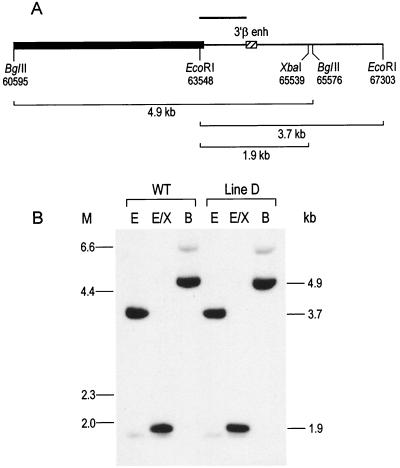
FIG. 4.
Detection of the 3′ β enhancer (3′β enh) in line D by Southern analysis. (A) Map of the location of the 260-bp PstI fragment containing the 3′ β enhancer (indicated by the hatched box) relative to the β-globin gene (indicated by the black box). Restriction enzyme sites and GenBank coordinates are shown below the map. The fragment sizes in wild-type DNA indicated below the map are 4.9 kb for BglII, 3.7 kb for EcoRI, and 1.9 kb for EcoRI-XbaI. (B) Southern analysis of DNAs from the wild-type (WT) β-YAC line and line D digested with the restriction enzymes indicated above each lane as follows: B, BglII; E, EcoRI; and E/X, EcoRI-XbaI. The probe was a 771-bp EcoRI-PstI fragment indicated in panel A as a solid bar. Migration positions of λHindIII size markers are indicated to the right of the autoradiogram, and positions of molecular size markers (M) (in kilobases) are indicated to the left.
Deletion of the HS3 core element abolishes ɛ-globin gene expression during embryonic erythropoiesis.
Transgenic mice carrying a wild-type β-YAC express human ɛ-globin mRNA in the yolk sac stage of erythropoiesis. ɛ-Globin mRNA ranges from 10 to 20% of murine α- plus ζ-globin mRNA (per copy) in the yolk sac, and it continues to be synthesized in circulating embryonic erythroblasts. Synthesis peaks in the embryonic erythroblasts at about day 12 of development.
Developmental studies were performed with yolk sac and blood samples from day 10 F2 embryos and with blood samples from day 12 and 14 F2 fetuses. Staining of fixed yolk sac or blood preparations with anti-human ɛ-globin fluorescent monoclonal antibody failed to detect any ɛ-globin in the primitive erythroblasts of the ΔHS3c transgenic embryos (not shown). Total RNA from yolk sac and blood samples from multiple embryos of the same litter were subjected to RNase protection analysis with human ɛ- and γ-globin and mouse α- and ζ-globin antisense RNA probes. In contrast to the wild-type β-YAC controls, ɛ-globin mRNA was undetectable in day 10 yolk sac from all four ΔHS3c β-YAC lines (Fig. 5). In the day 12 blood, where peak values of ɛ-globin mRNA are normally found, ɛ-globin mRNA was not detected in one of the lines and was 1.5% or less in the other three lines (Fig. 6). These data suggest that the HS3 core element is necessary for ɛ-globin gene transcription during the embryonic stage of erythropoiesis.
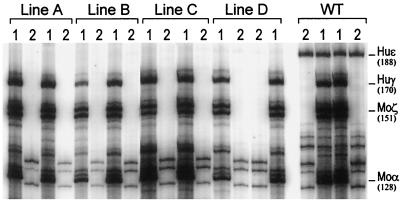
FIG. 5.
Expression of the human ɛ-globin gene in day 10 embryonic yolk sacs from F2 progeny of β-YAC transgenic lines containing the deletion of the HS3c element. Two littermates were examined from each transgenic line. Lanes 1, RNase protections with antisense probes for human ɛ- and γ-globin and murine α- and ζ-globin mRNAs; lanes 2, RNase protections with only a human ɛ-globin mRNA antisense probe in order to unambiguously test for the presence of ɛ-globin mRNA. Notice the presence of ɛ-globin mRNA in the control (wild type [WT]) and its complete absence in the lanes of lines A to D. The migrations of the protected fragments are shown to the right of the autoradiogram; the size of each fragment (in bases) is shown in parentheses. Huɛ, human ɛ-globin; Huγ, human γ-globin; Moα, mouse α-globin; moζ, mouse ζ-globin.
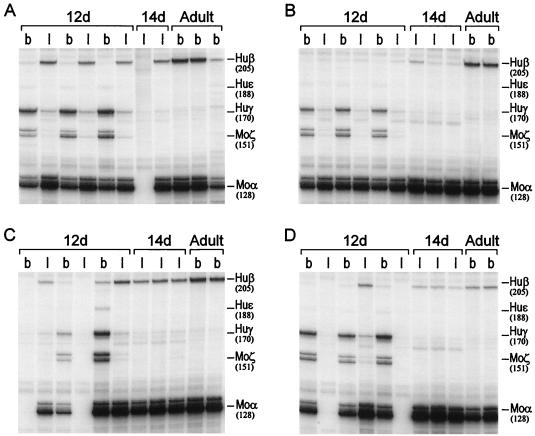
FIG. 6.
mRNA analysis of F2 progeny of β-YAC transgenic mice carrying a deletion of the HS3c element. Total RNA was isolated from day 12 liver and blood, day 14 liver, and adult blood from two or three F2 littermates from each line and subjected to RNase protection analysis. The migrations of the protected fragments are shown to the right of each autoradiogram; the size of each fragment (in bases) is shown in parentheses. Panels A to D correspond to lines A to D. Lanes b contain blood mRNA; lanes l contain liver RNA. Notice in the day 12 (12d) samples of all lines the normal levels of γ-globin mRNA in the blood (see also Table 1) and the striking decreases in levels of γ-globin mRNA in the day 12 and 14 liver mRNA preparations. Also notice the variation in β-globin mRNA levels in the day 12 or 14 liver samples as well as in the adult blood. See the legend to Fig. 5 for clarification of the abbreviations.
Normal γ-globin gene expression in the embryonic cells of ΔHS3c transgenic mice.
In contrast to the severe reduction of ɛ-globin gene expression, γ-globin gene expression was normal in embryonic erythrocytes. Multiple embryos from the same litter were used for RNase protection assays to minimize experimental error and determine sample variation. As shown in Table 1, all lines displayed, in the day 10 yolk sac, levels of γ-globin mRNA that were similar to those observed in control wild-type β-YAC mice. Similar results were obtained with day 12 fetal blood (Fig. 6), which consisted mostly of nucleated erythrocytes of yolk sac origin. Mean levels of γ-globin mRNA were 71 and 72% of those of the controls on days 10 and 12, respectively, but the difference from levels in the wild-type β-YAC control mice was not statistically significant. Most importantly, there was only a small degree of variation in the per copy levels of γ-globin mRNA between lines; levels of γ-globin mRNA in ΔHS3c embryos varied by 1.5-fold, indicating the absence of position effects. A statistical measure of variability is the coefficient of variation (ς/μ) in the levels of per copy expression between lines. Coefficients of variation smaller than 0.5 in levels of globin gene expression between lines denote a small, statistically insignificant degree of variation (25, 35). As calculated from the data of Table 1, the coefficients of variation in the levels of per copy γ-globin gene expression were 0.16 and 0.26 for day 10 and 12 embryonic erythropoiesis in the ΔHS3c lines and 0.26 and 0.27 for day 10 and 12 embryonic erythropoiesis in the wild-type β-YAC controls. These results show that the level of γ-globin gene expression in the embryonic cells of the ΔHS3c mice was nearly normal and that it was not influenced by the position of integration of the transgene.
TABLE 1.
Human globin mRNA levels per copy of transgene and copy of endogenous murine α-globin gene in ΔHS3c mice and wild-type β-YAC control mice
| Line or mouse type | Copy no. | % of murine α- plus ζ-globin mRNA (mean ± SD)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ɛ-Globin mRNA during embryonic erythropoiesis
|
γ-Globin mRNA during:
|
β-Globin mRNA during definitive erythropoiesis
|
||||||||
| Embryonic erythropoiesis
|
Definitive erythropoiesis
|
|||||||||
| Day 10 yolk sac | Day 12 blood | Day 10 yolk sac | Day 12 blood | Day 12 liver | Day 14 liver | Day 12 liver | Day 14 liver | Adult blood | ||
| Aa | 2 | 0 | 1.1 ± 0.2 | 17.4 ± 2.4 | 23.9 ± 1.0 | 2.2 ± 0.5 | 0 | 20.6 ± 1.6 | 13.7 ± 6.4 | 59.4 ± 5.8 |
| B | 3 | 0 | 0 | 12.3 ± 2.9 | 25.4 ± 1.8 | 1.9 ± 0.4 | 0 | 3.1 ± 0.8 | 0.8 ± 0.2 | 3.0 ± 0.5 |
| C | 2 | 0 | 1.3 ± 0.1 | 15.7 ± 5.0 | 11.5 ± 1.0 | 1.3 ± 0.4 | 0 | 11.7 ± 1.2 | 8.6 ± 0.6 | 17.1 ± 0.2 |
| D | 1 | 0 | 1.5 ± 0.3 | 19.2 ± 5.4 | 23.9 ± 1.0 | 2.2 ± 0.5 | 0 | 2.0 ± 0.2 | 2.7 ± 1.3 | 27.8 ± 0.2 |
| Mean of ΔHS3c line | 16.1 ± 2.5 | 21.2 ± 5.6 | 1.9 ± 0.4 | 9.3 ± 7.5 | 6.4 ± 5.1 | 26.8 ± 20.8 | ||||
| Wild-type β-YAC control mice | 10.2 ± 1.4 | 22.4 ± 6.5 | 22.7 ± 5.8 | 29.5 ± 7.9 | 16.1 ± 6.7 | 6.4 ± 3.4 | 38.9 ± 12 | 58.0 ± 26 | 117.0 ± 41 | |
Expression levels are based on two copies of the ɛ-, Gγ-, and Aγ-globin genes but a single copy of the β-globin gene (see Fig. 2).
Deletion of the HS3 core abolishes γ-globin gene expression in definitive erythroid cells.
The definitive stage of erythropoiesis begins in the murine fetal liver on day 10.5, and it is characterized by the exclusive transcription of the two adult globins, β-major and β-minor. In transgenic mice carrying a wild-type human β-YAC, the γ-globin genes were active in the fetal liver and the mean γ-globin mRNA level in the livers of the day 12 wild-type β-YAC fetuses was 16.1% ± 6.7% of the murine α- and ζ-globin mRNA levels (Table 1). In contrast, levels of γ-globin mRNA in the day 12 livers of ΔHS3c transgenic fetuses were strikingly reduced and ranged from 1.3 to 2.2% of levels of murine α- and ζ-globin (Fig. 6 and Table 1). In wild-type β-YAC transgenics there is a rapid switch from γ- to β-globin, so that by day 14 the mean level of γ-globin mRNA in fetal liver is 6.4% ± 3.4% of the levels of murine α- and ζ-globin mRNA (Table 1). γ-Globin mRNA was not detectable by the RNase protection assay in the livers of the 14-day-old fetuses of the four ΔHS3c lines (Table 1).
Embryonic erythroblasts contaminate the fetal livers of transgenic mice and contribute to the species of globin mRNAs detected in the fetal liver preparations from early fetuses (reference 38 and unpublished data). It was therefore possible that the low levels of γ-globin mRNA present in the liver RNA samples of the day 12 ΔHS3c fetuses derived from contaminating embryonic erythroblasts. To test this possibility, day 12 fetal liver preparations were stained with anti-γ-globin-chain monoclonal antibodies. As shown in Fig. 7A and B, only embryonic erythroblasts of yolk sac origin, characterized by their large size, their large cytoplasm/nucleus ratio, and their pycnotic nucleus, stained with the anti-γ-globin-chain fluorescent antibody. There was no fluorescent labeling of the definitive erythroblasts other than background staining. These results suggest that the γ-globin mRNA measured in the livers of the day 12 ΔHS3c fetuses derived from embryonic erythroblasts and that there was no detectable γ-globin gene expression in the definitive erythroid cells of fetal liver origin.
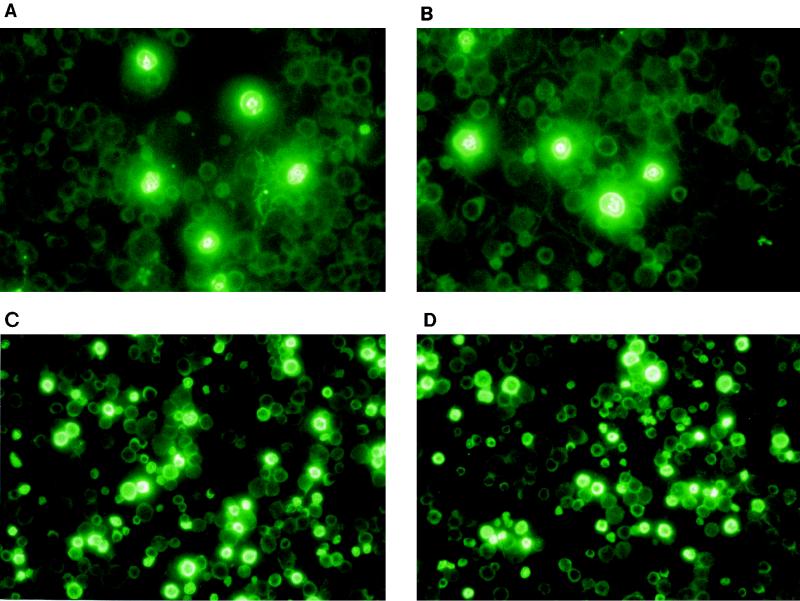
FIG. 7.
Staining with fluoroscein isothiocyanate conjugated anti-human globin-chain antibodies of day 12 fetal liver preparations of F2 transgenic mice carrying the HS3c deletion. (A and B) Staining with anti-γ-globin-chain antibodies. Notice that only the embryonic erythroblasts, recognized by their large size and high cytoplasm/nucleus ratio, are stained. (C and D) Staining with the anti-β-globin-chain antibodies. Notice the heterogeneity of β-globin-chain synthesis among the erythroblasts of definitive erythropoiesis; several nonstained embryonic erythrocytes can be seen in the background of panels C and D.
β-Globin gene expression in ΔHS3c β-YAC transgenic mice is decreased and position dependent.
In wild-type β-YAC mice, β-globin gene expression is similar to that of the endogenous murine genes and there is relatively small variation in the levels of per copy β-globin gene expression among lines. β-Globin gene expression is copy number dependent, indicating that the genes of the β-YAC are protected from position effects (31–33). The small degree of variation in levels of β-globin gene expression in the wild-type β-YAC mice is reflected in the small coefficient of variation (0.35) of the control lines, shown in Table 1. β-Globin gene expression in the ΔHS3c mice was significantly lower than that in control mice, and per copy levels of β-globin gene expression displayed striking variation. The per copy levels of β-globin mRNA varied, among the four lines, 10- and 17-fold in the day 12 and 14 fetal liver definitive erythroblasts and 19.8-fold in adult erythrocytes (Table 1), indicating that β-globin gene expression is strongly influenced by the position of integration of the transgene. Coefficients of variation for the levels of per copy β-globin gene expression for the day 12 and 14 fetuses and the adult mice were 0.8, 0.79, and 0.77, respectively. The presence of strong position effects was also reflected in the striking heterogeneity in the staining of the definitive erythroblasts of the fetal liver preparations with the anti-β-globin-chain fluorescent antibody (Fig. 7C and 7D).
DISCUSSION
Developmental specificity of the interaction between the core of HS3 and globin genes.
Our results show that deletion of the core sequence of DNase I-hypersensitive site 3 of the LCR results in the absence of ɛ-globin gene expression in day 10 embryonic cells and in the absence of γ-globin gene expression in the cells of definitive erythropoiesis in the fetal liver. These results provide direct evidence that there is developmental specificity in the interactions between the LCR and the globin genes: in the presence of an otherwise intact LCR, the core of HS3 is necessary for the activation of the ɛ-globin gene in embryonic cells and for activation of the γ-globin gene in definitive cells.
That the HSs of the LCR may display developmental specificities was first shown by Fraser et al. in experiments with transgenic mice (14). Those authors produced a series of recombinant constructs in which LCR sequences containing DNase I-HS1, -2, -3, and -4 were linked to an ∼36-kb cosmid containing the region of the β-globin locus from the Gγ-globin gene to the β-globin gene (14). Transgenic mice carrying the HS2-Gγ to -β cosmid expressed the γ-globin gene in the yolk sac and the β-globin gene in the adult cells, suggesting that HS2 interacts equally well with the γ- and β-globin genes. In contrast, the mice carrying the HS3-Gγ to -β cosmid were characterized by high γ-globin expression in embryos and fetuses and low β-globin expression in adults, indicating a preferential interaction of HS3 with the γ-globin gene. The opposite phenotype was observed in HS4-Gγ to -β transgenics (14). Developmental specificity of HS3 was also demonstrated in a study of transgenic mice carrying HS3-γβ and HS2-γβ constructs that showed a qualitative difference between HS2 and HS3 with respect to γ-globin gene expression (25). Our results support these previous conclusions and, further, provide evidence that specific sequences of the LCR interact with specific globin genes at specific stages of development.
Bungert et al. (3) have produced transgenic mice carrying either a deletion of HS3c or a replacement of HS3c with HS4c in the context of a 150-kb β-YAC. One ΔHS3c line showed nearly normal γ-globin gene expression and the near absence of ɛ-globin gene expression in the yolk sac cells. Two lines with substitutions of HS4c for HS3c had a significant reduction of ɛ-globin gene expression, as would be expected if HS3c is necessary for activation of the ɛ-globin gene in embryonic cells.
The LCR may change conformations during development.
Studies of γ- and β-globin primary transcripts in fetal erythroid cells of transgenic mice carrying a normal β-globin locus have shown that the LCR interacts with only one gene at any given time and that it switches back and forth between the two genes in a flip-flop type of mechanism (46). Similar results have been obtained by Fraser et al. (12a) with embryonic cells in which both the ɛ- and the γ-globin gene are transcribed from a single locus. Such oscillations of the interactions of the LCR with the ɛ- and the γ-globin genes should occur in the embryonic cells of transgenic mice carrying the ΔHS3c β-YAC. If the γ-globin genes of the embryonic cells interacted with the mutant HS3 whose core is deleted, these γ-globin genes should have been transcriptionally inactive. Since γ-globin gene expression was normal in the embryonic cells of the ΔHS3c mice, an HS of the LCR other than HS3 should have engaged the γ-globin gene and activated its promoter. These results suggest that different HSs of the LCR interact with the γ- or the ɛ-globin gene in an embryonic cell. On the basis of the transcriptional behavior of the γ-globin genes in embryonic cells (normal expression) and in fetal cells (no expression), we can also conclude that different HSs of the LCR interact with the γ-globin genes of the embryonic cells or with the γ-globin genes of the fetal cells.
The prevailing hypothesis of the structure-function relationship of the LCR is that LCR sequences interact with various constitutive and erythroid-lineage-specific transcriptional factors in a way that leads to formation of a complex (holocomplex [46]). Perhaps this complex attains a specific conformation which is optimal for interaction of the LCR with a globin gene. The strikingly different phenotypes of γ-globin gene expression in embryonic and fetal cells raise the possibility that the LCR attains different conformations in order to interact with the γ-globin genes of embryonic cells or with the γ-globin genes of definitive cells. The conformation of the LCR may change as the transcriptional milieu of the erythroid cells changes during the course of development.
Role of the core of HS3 in the opening of the β-globin locus chromatin domain in embryonic and in definitive erythroid cells.
Although β-globin mRNA was present in the ΔHS3c adult mice, β-globin mRNA levels varied by 19.8-fold among lines, indicating that β-globin gene expression is strongly influenced by the position of integration of the transgene. These results support previous suggestions that the core of HS3 is required for opening of the globin chromatin domain in cells of definitive (liver-stage) erythropoiesis (6). In contrast to the position-sensitive β-globin gene expression in definitive erythroid cells, there was minimal variation in levels of γ-globin mRNA among the ΔHS3c lines, indicating that γ-globin gene expression in the embryonic cells of the ΔHS3c transgenic mice is not influenced by the position of the integration of the transgene. These results suggest that, in contrast to the adult cells, the core of HS3 is not an important contributor to the function of the LCR, which opens the globin locus domain in embryonic cells. Apparently, in the ΔHS3c mice the domain-opening function of the LCR is conducted by other DNase I HSs.
The phenotypes of deletions that remove the core of HS3 as well as the sequences flanking the core.
Peterson et al. (32) have deleted 2.3 kb of HS3 (including the HS3 core) in the context of a 248-kb β-YAC. Transgenic mice carrying these ΔHS3 β-YACs displayed, in embryonic cells, about a threefold reduction in ɛ-globin gene expression compared to that of controls but no reduction of γ-globin gene expression in the fetal cells. Hug et al. (19) deleted 2.3 kb of murine HS3 through homologous recombination in embryonic stem cells and analyzed murine globin gene expression in chimeric mice; there was only a small (about 20%) reduction in expression of the murine globin genes in embryonic or in definitive erythropoiesis. It thus appears that the specific effects on ɛ- and γ-globin gene expression produced by the HS3 core deletions are not observed when the sequences flanking the core are also deleted. One way of reconciling these results is to assume that the core of HS3 and the HS3 flanking sequences possess different, but complementary, functions. The sequences flanking the core of HS3 may function by engaging a globin gene and positioning it in a way that allows optimal interaction of the gene with the HS3 core element; the HS3 core element, on the other hand, may interact with the transcriptional complex of the gene, thus activating globin gene expression. When the core element is deleted, as in the ΔHS3c β-YAC, the HS3 flanking region still interacts with the globin gene but activation of transcription does not occur. When the whole HS3 is deleted, another HS interacts with the γ-globin gene and there is minimal effect on gene expression. Redundancy of the functions of HSs is supported by several observations (8, 19, 32), and there is evidence from various experiments suggesting that the flanking sequences of HSs have a role in HS function (21, 24, 29).
The HS3 core and embryonic expression of the γ-globin gene.
Several species have genes which are orthologous to the γ-globin genes of primates, but they are expressed only in embryonic cells. The βh1 gene of the mouse is such an example. The expression of the γ-globin genes of primates was initially limited to the embryonic stage of erythropoiesis until the so-called fetal recruitment of the γ-globin genes; i.e., the expression of γ-globin genes in the definitive cells of the fetal liver stage of erythropoiesis occurred about 30 to 50 million years ago (17). As we show here, when the core of HS3 is deleted, the γ-globin gene becomes an embryonic gene, i.e., it is expressed exclusively in embryonic cells. This reversion of the γ-globin gene to its ancestral developmental pattern raises the possibility that the sequences of the core of HS3 may have contributed to the recruitment of its expression in fetal erythroid cells. Mutations that accumulated, during the evolution of primates, either in the γ-globin gene promoter, in the HS3 core sequence, or in both may have created the protein binding site(s) which allows the interaction between the core of HS3 and the γ-globin gene to occur in the cells of fetal liver erythropoiesis.
ACKNOWLEDGMENTS
We thank Richard Swank for comments on the manuscript.
This work was supported by National Institutes of Health grants DK45365, HL53750, and HL20899.
REFERENCES
- 1.Baron M H, Maniatis T. Rapid reprogramming of globin gene expression in transient heterokaryons. Cell. 1986;46:591–602. doi: 10.1016/0092-8674(86)90885-8. [DOI] [PubMed] [Google Scholar]
- 2.Behringer R R, Hammer R E, Brinster R L, Palmiber R D, Townes T M. Two 3′ sequences direct adult erythroid-specific expression of human β-globin gene in transgenic mice. Proc Natl Acad Sci USA. 1987;84:7056–7060. doi: 10.1073/pnas.84.20.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bungert J, Dave U, Lim K-C, Lieuw K H, Shavit J A, Liu Q, Engel J D. Synergistic regulation of human β-globin gene switching by locus control region elements HS3 and HS4. Genes Dev. 1995;9:3083–3096. doi: 10.1101/gad.9.24.3083. [DOI] [PubMed] [Google Scholar]
- 4.Curtin P, Pirastu M, Kan Y W, Gobert-Jones J A, Stephens A D, Lehmann H. A distant gene deletion affects β-globin gene function in an atypical γδβ-thalassemia. J Clin Invest. 1985;76:1554–1558. doi: 10.1172/JCI112136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Driscoll M C, Dobkin C S, Alter B P. γδβ-Thalassemia due to a de novo mutation deleting the 5′ β-globin gene activation-region hypersensitive sites. Proc Natl Acad Sci USA. 1989;86:7470–7474. doi: 10.1073/pnas.86.19.7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellis J, Tan-Un K C, Harper A, Michalovich D, Yannoutsos N, Philipsen S, Grosveld F. A dominant chromatin-opening activity in 5′ hypersensitive site 3 of the human β-globin locus control region. EMBO J. 1996;15:562–568. [PMC free article] [PubMed] [Google Scholar]
- 7.Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature (London) 1992;355:219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- 8.Fiering S, Epner E, Robinson K, Zhuang Y, Telling A, Hu M, Martin D I, Enver T, Ley T J, Groudine M. Targeted deletion of 5′HS2 of the murine β-globin LCR reveals that it is not essential for proper regulation of the β-globin locus. Genes Dev. 1995;9:2203–2213. doi: 10.1101/gad.9.18.2203. [DOI] [PubMed] [Google Scholar]
- 9.Forrester W C, Thompson C, Elder J T, Groudine M. A developmentally stable chromatin structure in the human β-globin gene cluster. Proc Natl Acad Sci USA. 1986;83:1359–1363. doi: 10.1073/pnas.83.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forrester W C, Takegawa S, Papayannopoulou T, Stamatoyannopoulos G, Groudine M. Evidence for a locus activation region: the formation of developmentally stable hypersensitive sites in globin-expressing hybrids. Nucleic Acids Res. 1987;15:10159–10177. doi: 10.1093/nar/15.24.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forrester W C, Novak U, Gelinas R, Groudine M. Molecular analysis of the human β-globin locus activation region. Proc Natl Acad Sci USA. 1989;86:5439–5443. doi: 10.1073/pnas.86.14.5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forrester W C, Epner E, Driscoll M C, Enver T, Brice M, Papayannopoulou T, Groudine M. A deletion of the human β-globin locus activation region causes a major alteration in chromatin structure and replication across the entire β-globin locus. Genes Dev. 1990;4:1637–1649. doi: 10.1101/gad.4.10.1637. [DOI] [PubMed] [Google Scholar]
- 12a.Fraser, P., et al. Personal communication.
- 13.Fraser P, Hurst J, Collis P, Grosveld F. DNaseI hypersensitive sites 1, 2 and 3 of the human β-globin dominant control region direct position-independent expression. Nucleic Acids Res. 1990;18:3503–3508. doi: 10.1093/nar/18.12.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fraser P, Pruzina S, Antoniou M, Grosveld F. Each hypersensitive site of the human β-globin locus control region confers a different developmental pattern of expression on the globin genes. Genes Dev. 1993;7:106–113. doi: 10.1101/gad.7.1.106. [DOI] [PubMed] [Google Scholar]
- 15.Gaensler K M L, Kitamura M, Kan Y W. Germ-line transmission and developmental regulation of a 150-kb yeast artificial chromosome containing the human β-globin locus in transgenic mice. Proc Natl Acad Sci USA. 1993;90:11381–11385. doi: 10.1073/pnas.90.23.11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gnirke A, Huxley C, Peterson K, Olson M V. Microinjection of intact 200- to 500-kb fragments of YAC DNA into mammalian cells. Genomics. 1993;15:659–667. doi: 10.1006/geno.1993.1121. [DOI] [PubMed] [Google Scholar]
- 17.Goodman M, Czelusniak J, Koop B F, Tagle D A, Slighton J L. Globin: a case study in molecular phylogeny. Cold Spring Harbor Symp Quant Biol. 1987;52:875–900. doi: 10.1101/sqb.1987.052.01.096. [DOI] [PubMed] [Google Scholar]
- 18.Grosveld F, van Assendelft G B, Greaves D R, Kollias G. Position-independent, high-level expression of the human β-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 19.Hug B A, Wesselschmidt R L, Fiering S, Bender M A, Epner E, Groudine M, Ley T J. Analysis of mice containing a targeted deletion of β-globin locus control region 5′ hypersensitive site 3. Mol Cell Biol. 1996;16:2906–2912. doi: 10.1128/mcb.16.6.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huxley C, Gnirke A. Transfer of yeast artificial chromosomes from yeast to mammalian cells. Bioessays. 1991;13:545–550. doi: 10.1002/bies.950131009. [DOI] [PubMed] [Google Scholar]
- 21.Jackson J D, Petrykowska H, Philipsen S, Miller W, Hardison R. Role of DNA sequences outside of the cores of DNase hypersensitive sites (HSs) in functions of the β-globin locus control region. Domain opening and synergism between HS2 and HS3. J Biol Chem. 1996;271:11871–11878. doi: 10.1074/jbc.271.20.11871. [DOI] [PubMed] [Google Scholar]
- 22.Kioussis D, Vanin E, deLange T, Flavell R A, Grosveld F G. β-Globin gene inactivation by DNA translocation in γβ-thalassaemia. Nature (London) 1983;306:662–666. doi: 10.1038/306662a0. [DOI] [PubMed] [Google Scholar]
- 23.Kollias G, Hurst J, deBoer E, Grosveld F. The human β-globin gene contains a downstream developmental specific enhancer. Nucleic Acids Res. 1987;15:5739–5747. doi: 10.1093/nar/15.14.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q, Zhou B, Powers P, Enver T, Stamatoyannopoulos G. β-Globin locus activation regions: conservation of organization, structure, and function. Proc Natl Acad Sci USA. 1990;87:8207–8211. doi: 10.1073/pnas.87.21.8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Q, Stamatoyannopoulos J A. Position independence and proper developmental control of γ-globin gene expression require both a 5′ locus control region and a downstream sequence element. Mol Cell Biol. 1994;14:6087–6096. doi: 10.1128/mcb.14.9.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q, Clegg C, Peterson K, Shaw S, Raich N, Stamatoyannopoulos G. Binary transgenic mouse model for studying the trans control of globin gene switching: evidence that GAGA-1 is an in vivo repressor of human ɛ gene expression. Proc Natl Acad Sci USA. 1997;94:2444–2448. doi: 10.1073/pnas.94.6.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu D, Chang J C, Moi P, Liu W, Kan Y W, Curtin P T. Dissection of the enhancer activity of β-globin 5′ DNase I-hypersensitive site 2 in transgenic mice. Proc Natl Acad Sci USA. 1992;89:3899–3903. doi: 10.1073/pnas.89.9.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milot E, Strouboulis J, Trimborn T, Wijgerde M, de Boer E, Langeveld A, Tan-Un K, Vergeer W, Yannoutsos N, Grosveld F, Fraser P. Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell. 1996;87:105–114. doi: 10.1016/s0092-8674(00)81327-6. [DOI] [PubMed] [Google Scholar]
- 29.Moon A M, Ley T J. Conservation of the primary structure, organization, and function of the human and mouse β-globin locus-activating regions. Proc Natl Acad Sci USA. 1990;87:7693–7697. doi: 10.1073/pnas.87.19.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peterson K R, Clegg C H, Huxley C, Josephson B M, Haugen H S, Furukawa T, Stamatoyannopoulos G. Transgenic mice containing a 248-kb yeast artificial chromosome carrying the human β-globin locus display proper developmental control of human globin genes. Proc Natl Acad Sci USA. 1993;90:7593–7597. doi: 10.1073/pnas.90.16.7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peterson K R, Li Q, Clegg C H, Furukawa T, Navas P A, Norton E J, Kimbrough T G, Stamatoyannopoulos G. Use of yeast artificial chromosomes (YACs) in studies of mammalian development: production of β-globin locus YAC mice carrying human globin developmental mutants. Proc Natl Acad Sci USA. 1995;92:5655–5659. doi: 10.1073/pnas.92.12.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peterson K R, Clegg C H, Navas P A, Norton E J, Kimbrough T G, Stamatoyannopoulos G. Effect of deletion of 5′HS3 or 5′HS2 of the human β-globin locus control region on the developmental regulation of globin gene expression in β-globin locus yeast artificial chromosome transgenic mice. Proc Natl Acad Sci USA. 1996;93:6605–6609. doi: 10.1073/pnas.93.13.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterson, K. R., P. A. Navas, Q. Li, C. H. Clegg, and G. Stamatoyannopoulos. Human β-globin locus YAC transgenic mice: relationship of transgene structural integrity to globin gene function using 248 kb and 155 kb β-globin YACs. Submitted for publication.
- 34.Philipsen S, Talbot D, Fraser P, Grosveld F. The β-globin dominant control region: hypersensitive site 2. EMBO J. 1990;9:2159–2167. doi: 10.1002/j.1460-2075.1990.tb07385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Philipsen S, Pruzina S, Grosveld F. The minimal requirements for activity in transgenic mice of hypersensitive site 3 of the β-globin locus control region. EMBO J. 1993;12:1077–1085. doi: 10.1002/j.1460-2075.1993.tb05749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porcu S, Kitamura M, Witkowska E, Zhang Z, Mutero A, Lin C, Chang J, Gaensler K M L. The human β globin locus introduced by YAC transfer exhibits a specific and reproducible pattern of developmental regulation in transgenic mice. Blood. 1997;90:4602–4609. [PubMed] [Google Scholar]
- 37.Pruzina S, Hanscombe O, Whyatt D, Grosveld F, Philipsen S. Hypersensitive site 4 of the human β-globin locus control region. Nucleic Acids Res. 1991;19:1413–1419. doi: 10.1093/nar/19.7.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raich N, Enver T, Nakamoto B, Josephson B, Papayannopoulou T, Stamatoyannopoulos G. Autonomous developmental control of human embryonic globin gene switching in transgenic mice. Science. 1990;250:1147–1149. doi: 10.1126/science.2251502. [DOI] [PubMed] [Google Scholar]
- 39.Ryan T M, Behringer R R, Martin N C, Townes T M, Palmiter R D, Brinster R L. A single erythroid-specific DNase I super-hypersensitive site activates high levels of human β-globin gene expression in transgenic mice. Genes Dev. 1989;3:314–323. doi: 10.1101/gad.3.3.314. [DOI] [PubMed] [Google Scholar]
- 40.Strouboulis J, Dillon N, Grosveld F. Developmental regulation of a complete 70-kb human β-globin locus in transgenic mice. Genes Dev. 1992;6:1857–1864. doi: 10.1101/gad.6.10.1857. [DOI] [PubMed] [Google Scholar]
- 41.Talbot D, Collis P, Antoniou M, Vidal M, Grosveld F, Greaves D R. A dominant control region from the human β-globin locus conferring integration site-independent gene expression. Nature (London) 1989;338:352–355. doi: 10.1038/338352a0. [DOI] [PubMed] [Google Scholar]
- 42.Talbot D, Philipsen S, Fraser P, Grosveld F. Detailed analysis of the site 3 region of the human β-globin dominant control region. EMBO J. 1990;9:2169–2177. doi: 10.1002/j.1460-2075.1990.tb07386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trudel M, Costantini F. A 3′ enhancer contributes to the stage-specific expression of the human β-globin gene. Genes Dev. 1987;1:954–961. doi: 10.1101/gad.1.9.954. [DOI] [PubMed] [Google Scholar]
- 44.Tuan D, Solomon W, Li Q, London I M. The “β-like-globin” gene domain in human erythroid cells. Proc Natl Acad Sci USA. 1985;82:6384–6388. doi: 10.1073/pnas.82.19.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Assendelft G B, Hanscombe O, Grosveld F, Greaves D R. The β-globin dominant control region activates homologous and heterologous promoters in a tissue-specific manner. Cell. 1989;56:969–977. doi: 10.1016/0092-8674(89)90630-2. [DOI] [PubMed] [Google Scholar]
- 46.Wijgerde M, Grosveld F, Fraser P. Transcription complex stability and chromatin dynamics in vivo. Nature (London) 1995;377:209–213. doi: 10.1038/377209a0. [DOI] [PubMed] [Google Scholar]
- 47.Winston F, Chumley F, Fink G R. Eviction and transplacement of mutant genes in yeast. Methods Enzymol. 1983;101:211–228. doi: 10.1016/0076-6879(83)01016-2. [DOI] [PubMed] [Google Scholar]