Abstract
Osf2/Cbfa1, hereafter called Osf2, is a member of the Runt-related family of transcription factors that plays a critical role during osteoblast differentiation. Like all Runt-related proteins, it contains a runt domain, which is the DNA-binding domain, and a C-terminal proline-serine-threonine-rich (PST) domain thought to be the transcription activation domain. Additionally, Osf2 has two amino-terminal domains distinct from any other Runt-related protein. To understand the mechanisms of osteoblast gene regulation by Osf2, we performed an extensive structure-function analysis. After defining a short Myc-related nuclear localization signal, a deletion analysis revealed the existence of three transcription activation domains and one repression domain. AD1 (for activation domain 1) comprises the first 19 amino acids of the molecule, which form the first domain unique to Osf2, AD2 is formed by the glutamine-alanine (QA) domain, the second domain unique to Osf2, and AD3 is located in the N-terminal half of the PST domain and also contains sequences unique to Osf2. The transcription repression domain comprises the C-terminal 154 amino acids of Osf2. DNA-binding, domain-swapping, and protein interaction experiments demonstrated that full-length Osf2 does not interact with Cbfβ, a known partner of Runt-related proteins, whereas a deletion mutant of Osf2 containing only the runt and PST domains does. The QA domain appears to be responsible for preventing this heterodimerization. Thus, our results uncover the unique functional organization of Osf2 by identifying functional domains not shared with other Runt-related proteins that largely control its transactivation and heterodimerization abilities.
The Runt/Cbfa family of proteins comprises a group of transcription factors that have recently emerged as major regulators of organogenesis in invertebrates and vertebrates. This family includes Runt and Lozenge, two Drosophila proteins (7, 20), and Cbfa1, Cbfa2, and Cbfa3 in mice and humans (3, 5, 33). In addition, Runt homologs have been identified in Caenorhabditis elegans and sea urchins (6, 16). This evolutionary conservation further underscores the biological importance of these proteins. Genetic and biochemical analyses with Drosophila melanogaster, mice, and humans have shown that runt and lozenge in Drosophila and Cbfa2 and Osf2/Cbfa1, hereafter called Osf2, in mice and humans play crucial roles in organogenesis processes such as neurogenesis, eye development, hematopoiesis, and skeletogenesis, respectively (7, 10, 20, 21, 24, 29, 35, 36).
The mechanism by which each of these transcription factors controls different cell differentiation programs and organogenesis processes remains largely unknown. All of the Runt-related proteins have a common 128-amino-acid motif called the runt domain, which is their DNA-binding domain (18, 19, 25). They bind to the consensus site 5′TGT/cGGT3′ (28), found in the control regions of numerous genes involved in various developmental processes. These proteins are capable of binding to DNA as monomers, but it has been shown that both Runt and Cbfa2 (formerly known as AML1), can heterodimerize with a ubiquitously expressed partner protein called Cbfβ (13, 34). Cbfβ does not directly bind to DNA but increases the affinity of Runt and Cbfa2 for DNA (4, 13).
Our long-standing interest in understanding the molecular mechanisms controlling osteoblast differentiation led us to the recent characterization of Osf2 as an osteoblast-specific Cbfa protein (10). During embryogenesis, Osf2 transcripts appear initially in cells of the mesenchymal condensations that prefigure the skeleton and are subsequently expressed exclusively in cells of the osteoblastic lineage (10). Inactivation of the Cbfa1 gene in mice demonstrates that Osf2 is an essential factor in osteoblast differentiation (21, 36). Indeed, in its absence, mesenchymal cells are correctly positioned and proliferate but fail to differentiate into osteoblasts, resulting in a total absence of ossification. Moreover, mice heterozygous for Cbfa1 inactivation have a delay in ossification, recapitulating the phenotype of a classical mouse mutant termed cleidocranial dysplasia (ccd) (42, 43). In humans also, there is a skeletal disorder called CCD, and the phenotype of patients is similar to that observed in mouse ccd (17). CCD patients have been shown to have either deletion, insertion, or missense mutations in the CBFA1 gene that abolish binding of Osf2 to DNA (24, 29). Taken together, these results demonstrated that Osf2 is a key regulator of skeletogenesis whose function is nonredundant with the function of other genes and whose level of expression must be kept within tight limits.
In contrast to the wealth of knowledge available for other members of this family, such as runt and Cbfa2, nothing is known about the molecular mechanisms by which Osf2 controls osteoblast gene expression and differentiation. A comparison of Osf2 and the other Runt-related proteins reveals that Osf2 contains three domains that are not present in other Runt-related and Cbfa proteins (Fig. 1). The first is a stretch of 19 amino acid residues at the amino terminus absent from the original cDNA of Cbfa1 (10, 33). The second is a unique glutamine-alanine domain (QA domain) located N terminal to the runt domain. This domain contains 29 glutamine residues in a row followed by a stretch of 18 alanine residues. Mutational analysis in CCD patients suggests that the length of the alanine stretch within the QA domain influences the transcriptional activity of the protein, although the phenotype of the patients is different from the classical CCD phenotype (29). Finally, there is a stretch of 27 amino acids in the proline-serine-threonine-rich (PST) domain that has no homology with sequences present in the PST domains of other Runt-related proteins. These findings suggest that these unique structural domains may contribute to the osteoblast-specific functions of Osf2.
FIG. 1.
Alignment of the amino acid sequences of Osf2 (10) and Cbfa2 (3). Dashes denote gaps inserted to maximize sequence alignment. Identical residues are indicated by double dots, and similar residues are indicated by single dots. The N-terminal 19 amino acids (AD1) and the QA domain (AD2), which are unique to Osf2, are double-underlined. The runt domain sequence is underlined, and the C-terminal 27 amino acids of AD3 (amino acids 348 to 374) are shown in boldface type and underlined.
To understand the mechanisms by which Osf2 controls osteoblast differentiation, we embarked on a search for domains responsible for nuclear localization and transcriptional functions, as well as for domains involved in regulating heterodimerization. Our analysis shows that Osf2 has a unique functional organization among Runt-related proteins. Indeed, the first 19 amino acids and the QA domain largely control the transactivation function, and the QA domain additionally prevents heterodimerization of Osf2. This finding might now be used to decipher the mechanisms by which Osf2 controls osteoblast differentiation.
MATERIALS AND METHODS
Plasmids.
The Osf2 cDNA cloned in pBluescript KS(−) and pCMV5 (10) was used for generating deletion constructs in the pCMV5 expression vector. Osf2 lacking the 9-amino-acid nuclear localization signal (NLS) sequence (Osf2ΔNLS) was generated by a two-step PCR strategy (2) using the oligonucleotides 5′GGACGGTCCCCGGGAAGACTCTAAACCTAGTTTG3′ (NLS-F) and 5′AGGTTTAGAGTCTTCCCGGGGACCGTCCACTG3′ (NLS-R). Osf2Δ1-108 was generated by inserting a 1,275-bp NcoI fragment of the Osf2 coding sequence in pCMV5, with the ATG codon within the NcoI site serving as a translational initiator. Δ1-38 was generated by inserting an 1,856-bp PstI fragment in PstI-digested pCMV5, with the ATG codon immediately downstream of the PstI site intended to serve as a translational initiator. Δ1-19 was generated by PCR amplification of the 5′ region of the Osf2 coding sequence with the primers 5′TCAATCGATGACTATGGATCCGAGCACCAGC3′ (DEL5′F) and 5′CGGGGACCGTCCACTG3′ (R3); the PCR product was digested with BstEII, and the resulting 5′-end/BstEII fragment was ligated to Δ1-38, which was digested with MluI (end filled) and BstEII. The ATG codon (underlined) in the DEL5′F primer served as the initiator of translation. The QA domain was deleted (Δ49-96) by removing the FspI-NotI and NotI-NotI fragments from pCMV5-Osf2 followed by end filling and religation, and Δ82-96 was generated by removing the NotI fragment from pCMV5-Osf2 followed by religation. A PCR-amplified molecule that has the Osf2 coding sequence with a 24-bp internal deletion (caused by an error during amplification by Taq polymerase) was cloned in pCMV5 to get Δ89-96. Osf2Ala27 was generated by replacing the NotI fragment from the Osf2 coding region with a synthetic double-stranded oligonucleotide that would code for 27 alanine residues. Δ(1-38, 82-96) was made by removing the NotI fragment from Δ1-38 followed by religation. Δ258-528 was made by removal of the BsmI-BsmI and BsmI-XbaI fragments from pCMV5-Osf2 followed by religation.
GAL4 fusion constructs of full-length Osf2, N1-19, N49-96, and N1-108, were obtained by ligating the appropriate coding sequence downstream and in frame with a sequence coding for the GAL4 DNA-binding domain (GAL4DBD [amino acids 1 to 147]) in the vector pSG424 (38). For analysis of the functional domains in the PST region, the full-length PST region or fragments of it were also cloned in the vector pSG424 (38). Fragments of the PST coding sequence were obtained by using suitable restriction sites or were PCR amplified with appropriate primers and cloned. Construct Osf2ΔGASEL was generated by the two-step PCR strategy (2) using the oligonucleotides 5′AAGAAGAGCCAGGCAGGCCCTTTTTCAGACCCCAG3′ (ΔGASEL-1) and 5′CTGAAAAAGGGCCTGCCTGGCTCTTCTTACTGAG3′ (ΔGASEL-2).
The constructs GAL4-6× VWRPY-VP16 and GAL4-6× GASEL-VP16 were made by inserting synthetic double-stranded oligonucleotides (that would code for six copies of either the VWRPY or the GASEL sequence) at the Asp718 site, between sequences coding for GAL4DBD and the VP16 activation domain. The TLE2 expression construct was obtained by digesting a TLE2 cDNA with EcoRV and XbaI followed by ligation to pcDNA3 cut with the same enzymes. Osf2ΔC12 was generated by inserting an EcoRI fragment (obtained from pCMV5-Osf2) into pCMV5 cut with the same enzyme. The reporter plasmid p6OSE2luc has been previously described (9), and the pGAL4SVluc reporter plasmid (which has a luciferase reporter gene driven by five copies of the GAL4 upstream activation sequence [UASG] and the simian virus 40 minimal promoter) was obtained from Jennifer Philhower, Science Park Research Division, M. D. Anderson Cancer Center, Smithville, Tex.
For bacterial expression of recombinant proteins, the coding sequences for Cbfa2 and Cbfβ (amino acids 7 to 182) were cloned downstream and in frame with a sequence coding for six histidine residues in pTrcHis vectors (Invitrogen). The His-Osf2 expression construct has been previously described (10). The GST-Osf2 expression construct was generated by inserting the Osf2 coding sequence downstream of the glutathione S-transferase (GST) coding sequence in the expression vector pGEX-2T. For the domain-swapping experiment, the chimeric construct 1.2.2 was generated by PCR-amplifying fragments of Osf2 and Cbfa2 coding sequences and ligating them in frame to a sequence coding for six histidine residues in the vector pV2a (46). ΔN19.1.1 was made by removing a BamHI fragment at the 5′ end of the Osf2 coding sequence followed by religation of the His-Osf2 expression construct. Δ.runt.PST was made by inserting a 1,275-bp NcoI fragment in the pTrcHis vector. For in vitro binding assays, the Cbfβ coding sequence (amino acids 7 to 182) was cloned in frame with the GST coding sequence in pGEX-2T. The integrity of all constructs was verified by DNA sequencing.
For in vitro transcription and translation, Osf2Met69 was generated by deletion of the 189-bp 5′-end/DraI fragment of the original Osf2 cDNA. The Osf2Met1 [the original Osf2 cDNA cloned in pBluescript II KS(−)] and Osf2Met69 constructs were transcribed and translated in vitro with the TnT kit (Promega), according to the manufacturer’s instructions, and the labeled proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Cell culture and DNA transfection.
The kidney cell line COS7 was grown in Dulbecco modified Eagle medium (DMEM)–10% fetal bovine serum, and NIH 3T3 cells were grown in DMEM–10% calf serum (Gibco BRL). Cells (3 × 105/dish) were transfected by the calcium phosphate coprecipitation method (2) with 5 μg of reporter plasmid (p6OSE2luc or pGAL4SVluc), 5 μg of expression construct, and 2 μg of pRSVβgal. Following transfection, the cells were washed twice with phosphate-buffered saline (PBS) and incubated with the appropriate medium for 24 h. Cells were harvested in 0.3 ml of 0.25 M Tris-HCl, lysed by three cycles of freezing and thawing, and subjected to a colorimetric β-galactosidase activity assay, with resorufin-β-d-galactopyranoside (Sigma) as the substrate. Twenty microliters of cell extract was used for measuring luciferase activity with a Monolight 2010 luminometer (Analytical Luminescence Laboratory) using d-luciferin substrate in luciferase reaction buffer (100 mM Tris-HCl [pH 7.8], 5 mM ATP, 15 mM MgSO4, 1 mM dithiothreitol [DTT]). Luciferase activity values were adjusted to β-galactosidase values to normalize for transfection efficiency.
Generation of recombinant fusion proteins, nuclear extract preparation, DNA-binding assays, and in vitro binding assays.
For protein production, bacterial cells were induced with 2 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and the oligohistidine fusion proteins were enriched with Ni-nitrilotriacetic acid agarose resin (Qiagen) per the manufacturer’s guidelines. Nuclear extracts were prepared from COS7 cells according to the method of Schreiber et al. (41). DNA-binding assays were performed with 5 fmol of 32P-labeled double-stranded OSE2 oligonucleotides (9) in a buffer containing 20 mM Tris-HCl (pH 8.0), 10 mM NaCl, 3 mM EGTA, 0.05% Nonidet P-40 (NP-40), 5 mM DTT, and 2 μg of poly(dI-dC) · poly(dI-dC), with equivalent amounts of wild-type or mutant proteins. DNA-binding reactions with nuclear extracts were performed in a buffer containing 5% glycerol, 100 mM NaCl, 50 mM Tris-HCl (pH 7.5), 0.1% NP-40, 2 mM EDTA, 1 mM DTT, 2.5 μg of Leupeptin per ml, and 2.5 μg of Pepstatin per ml. The reaction mixtures were incubated for 10 min at room temperature and then electrophoresed on a 5% polyacrylamide gel (10). For in vitro binding assays, the GST and GST-Cbfβ proteins were eluted from glutathione-Sepharose beads with reduced glutathione or used as such bound to the beads. The proteins were checked for purity and quantified before use. 35S-labeled Osf2 and Cbfa2 were synthesized in rabbit reticulocyte lysate by coupled in vitro transcription and translation (TnT kit; Promega). Typically, 100 ng of GST or GST-Cbfβ protein bound to glutathione-agarose beads was used for each assay, while the amount of labeled protein in the assay was determined by fluorography. The in vitro binding assay was carried out as described previously (2).
Cellular fractionation, immunoblot, and immunofluorescence analyses.
COS7 cells (106) were transfected with either wild-type Osf2 or Osf2ΔNLS by the calcium phosphate coprecipitation method (2). Cytoplasmic and nuclear fractions were prepared from transfected cells, separated by SDS-PAGE, and subjected to immunoblot analysis with rabbit polyclonal anti-Osf2 antibody (generated against the peptide sequence SFFWDPSTSRRFSPPS, present at the N terminus of Osf2) and horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (IgG) followed by ECL detection (Amersham). For immunofluorescence, 2 days after transfection the cells were plated on slides, washed with PBS buffer, and fixed in 3.7% formaldehyde at room temperature for 10 min followed by permeabilization with 0.1% Triton X-100. Blocking was done for 30 min in 5% goat serum–3% bovine serum albumin. The cells were incubated with anti-Osf2 antibody at a dilution of 1:150 in blocking buffer for 1 h at room temperature, followed by a wash with blocking buffer and then with PBS. Rhodamine-conjugated goat anti-rabbit IgG was then used at a dilution of 1:10,000. Slides were mounted with 50% glycerol, and the staining pattern of Osf2 was visualized by confocal microscopy.
Expression of GAL4 fusion proteins was confirmed by immunoblot analysis of extracts from transfected cells with a 1:2,000 dilution of rabbit polyclonal anti-GAL4DBD antibody (Upstate Biotechnology, Lake Placid, N.Y.) and horseradish peroxidase-conjugated anti-rabbit IgG (1:3,000 dilution) followed by ECL detection (Amersham). For analysis of the expression of TLE2 protein in osteoblastic cells, 60 μg of ROS17/2.8 cell lysate was fractionated on an SDS–8% polyacrylamide gel, transferred to nitrocellulose, and subjected to immunoblot analysis with affinity-purified anti-TLE2 antibody (1:100 dilution) (31).
RESULTS
Identification of a Myc-related NLS in Osf2.
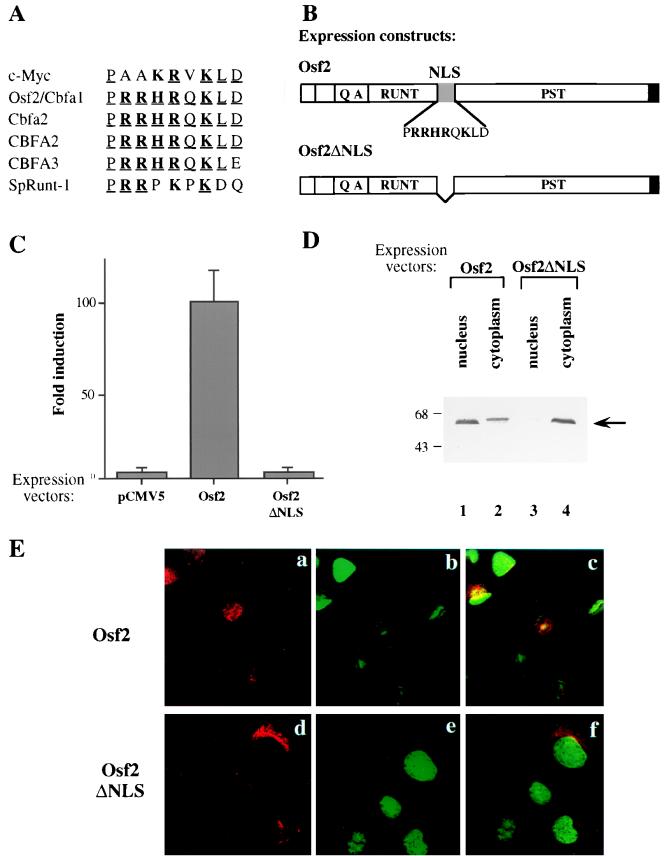
To identify a transcription activation domain(s) in Osf2 through a deletion mutagenesis approach, we first delineated the shortest possible NLS in Osf2. The NLS was originally assigned to a broad region of the protein containing the runt domain and the entire PST domain of Cbfa1 (26). To define a shorter NLS, we compared the sequence of Osf2 to known NLS sequences. Stretches of basic amino acid residues have been shown to be responsible for targeting proteins to the nucleus (8, 32). We found, overlapping the runt and PST domains of Osf2, a stretch of 9 amino acids (PRRHRQKLD), including five basic residues (in boldface type), that is highly homologous to the known NLS of c-Myc (Fig. 2A). This sequence contains a short motif, RRHR, that has been shown to be responsible for nuclear localization of various proteins (32). Moreover, this 9-amino-acid sequence is present at the same location in several other Runt-related proteins (CBFA2, Cbfa2, CBFA3, and SpRunt-1) (Fig. 2A), suggesting that this stretch of amino acids may act as a common NLS in these proteins.
FIG. 2.
Identification of a Myc-related NLS sequence in Osf2. (A) Comparison of the NLS sequences of Runt-related proteins with that of c-Myc. Basic residues are indicated in boldface type, and residues that are identical in at least three of the proteins shown are underlined. (B) Schematic representation of the wild-type Osf2 and Osf2ΔNLS expression constructs. (C) Transcriptional activity of wild-type Osf2 and Osf2ΔNLS lacking the basic 9-amino-acid stretch (NLS) in transient transfection assays done with the p6OSE2luc reporter in COS7 cells. The fold induction of luciferase activity, normalized to β-galactosidase activity, is shown. Values are means of nine independent transfection experiments. Error bars represent standard deviations of the means. (D) Immunoblot analysis of fractions from COS7 cells transfected with wild-type Osf2 or Osf2ΔNLS expression constructs. Cytoplasmic and nuclear fractions were prepared from transfected cells and subjected to immunoblot analysis with rabbit polyclonal anti-Osf2 antibody. Molecular size markers (in kilodaltons) are shown on the left. (E) Indirect immunofluorescence analysis of COS7 cells transfected with wild-type Osf2 (panels a, b, and c) or Osf2ΔNLS (panels d, e, and f) expression constructs. Subcellular localization of Osf2 was detected with rabbit polyclonal anti-Osf2 antibody and a rhodamine-conjugated secondary antibody (red staining) followed by confocal microscopy. Nuclei were stained with the DNA dye YO-PRO-1 (green staining). Panels a and d, red staining indicates proteins; panels b and e, green staining of nuclei; panels c and f, overlap of green and red staining. Note on panel c the overlap of green and red staining indicating nuclear localization of Osf2. Such an overlap is absent with Osf2ΔNLS (panel f).
To test whether this 9-amino-acid stretch acts as an NLS in Osf2, we constructed an in-frame deletion of this motif in the full-length coding sequence. This mutant Osf2 (Osf2ΔNLS) was cloned in the pCMV5 expression vector (Fig. 2B) and checked for its ability to activate transcription from p6OSE2luc, a construct containing six copies of a canonical Osf2 binding site (OSE2) (9) in COS7 cells that do not express the Cbfa genes (23). Osf2ΔNLS failed to drive expression of the luc reporter, while wild-type Osf2 did activate transcription under the same conditions (Fig. 2C). To determine if this lack of transactivation by Osf2ΔNLS was due to the inability of the mutant protein to be translocated to the nucleus, we performed cellular fractionation and immunolocalization analyses. Extracts from transfected cells were separated into nuclear and cytosolic fractions and subjected to immunoblot analysis with a polyclonal antibody directed against Osf2. The wild-type protein was found predominantly in the nuclear fraction, whereas Osf2ΔNLS was found only in the cytosolic fraction (Fig. 2D). Lastly, indirect immunofluorescence analysis of transfected cells revealed the presence of the wild-type protein in the nucleus, while Osf2ΔNLS was localized in the cytosol (Fig. 2E). Thus, these experiments identify the 9-amino-acid stretch (PRRHRQKLD) as a sequence necessary for nuclear localization of Osf2.
The first 19 amino acids comprise one activation domain.
Osf2 has a longer 5′ end than the originally described Cbfa1 (10, 33). Analysis of the genomic structure of the gene has shown that the 5′ end of Osf2 is located in an additional exon, whereas the originally proposed 5′ end of Cbfa1 is located at the 3′ end of a large intron that contains a cryptic splice acceptor site (11).
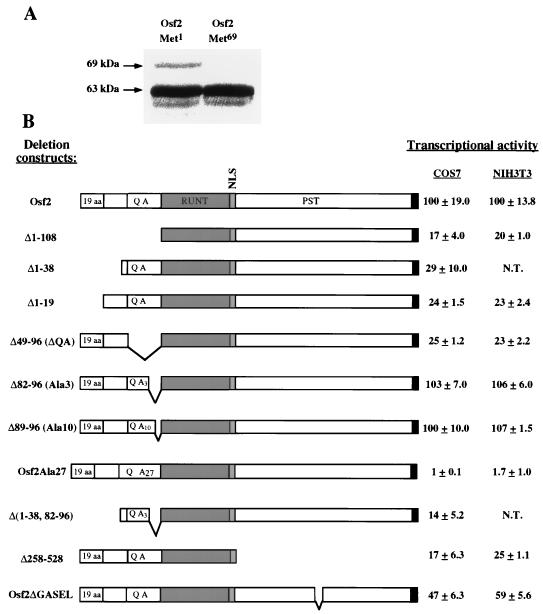
The 5′ end of Osf2 has two ATG codons in frame with the predicted coding sequence. The one at position 1 (Met1) is in a poor context for translational initiation, whereas the one at position 69 (Met69) is in an appropriate context for translational initiation (22). To test the respective efficiencies of these two potential translational initiators, we generated two constructs, one containing both ATG codons (Osf2Met1) and the other containing only the second ATG codon (Osf2Met69), and tested them in an in vitro transcription-translation assay. As shown in Fig. 3A, Met69 is by far the best translational initiator.
FIG. 3.
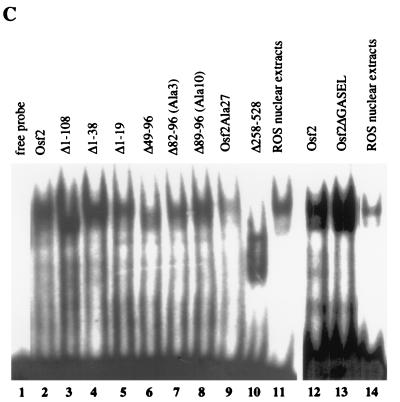
Identification of transactivation domains in Osf2. (A) In vitro transcription and translation of Osf2 cDNAs. Osf2Met1 and Osf2Met69 were constructed as described in Materials and Methods. The plasmid constructs were transcribed and translated in vitro in the presence of [35S]methionine, and the labeled proteins were subjected to SDS-PAGE followed by autoradiography. (B) Transactivation ability of Osf2 deletion mutants. Deletions of Osf2 were cloned in pCMV5 expression vector, and transfections were carried out with p6OSE2luc as a reporter in COS7 and NIH 3T3 cells. Numbers on the left indicate the amino acids deleted. Values on the right are means ± standard deviations of nine independent transfection experiments. N.T., not tested. (C) DNA-binding ability of Osf2 deletion mutants. Eight micrograms of nuclear extracts from COS7 cells transfected with the expression plasmids were incubated with radiolabeled double-stranded OSE2 oligonucleotides, and the protein-DNA complexes were analyzed by EMSA. The name of each deletion mutant is indicated at the top of the figure. ROS17/2.8 nuclear extracts were used as a positive control.
To identify regions of Osf2 responsible for transcriptional activation, we generated a series of deletion mutants that all contained the NLS and assayed their abilities to transactivate the OSE2-dependent luciferase reporter construct (p6OSE2luc) in DNA cotransfection experiments with COS7 cells that do not express the Cbfa genes (23) and in NIH 3T3 cells that express the Cbfa genes (26). A construct containing both ATG codons was nearly as active as a construct containing only Met69 (data not shown). Hence, in the rest of the study, we considered the protein initiating from Met69 as the full-length protein. Surprisingly, a deletion of the entire N-terminal end of Osf2 that left only the runt and PST domains intact (Δ1-108) resulted in a fivefold decrease in the transactivation ability of the protein in both cell lines (Fig. 3B). Given the fact that haploinsufficiency of Cbfa1 causes ccd in mice and humans, this decrease in the transactivation function of Osf2 is biologically significant. This result suggested that, unlike what has been proposed for Cbfa2 (4), the transactivation function of Osf2 is located not only in the PST domain but also in the N-terminal part of the molecule, prompting us to generate additional deletion mutants of this region of Osf2.
A deletion of the first 38 amino acid residues (Δ1-38), which left only the QA, runt, and PST domains intact, led to a 3.4-fold decrease in transactivation. This region is made up of two parts: the first 19 amino acids, which are unique to Osf2 and are not present in the partial Cbfa1 cDNA initially identified (33), and the next 21 amino acids, which show a high degree of similarity (85%) with the corresponding amino acids of Cbfa2 (Fig. 1). Interestingly, deletion of the first 19 amino acids (Δ1-19) resulted in a fourfold decrease in the transactivation ability of Osf2 in both cell lines, indicating that they constitute a transactivation domain unique to Osf2, called AD1 (for activation domain 1). Electrophoretic mobility shift assays (EMSA) performed with equal amounts of nuclear extracts from COS7 cells transfected with the various deletion constructs showed that all of the mutant proteins were expressed to comparable levels and were capable of binding to the DNA (Fig. 3C).
The QA domain is a second activation domain.
Next, we analyzed the transactivation function of another domain unique to Osf2, the QA domain. Deletion of the QA domain alone (Δ49-96) resulted in a fourfold decrease in the transactivation ability of the protein, indicating that the QA domain has a transactivation function. For that reason, we called this region AD2. To assess the relative importance of the glutamine and alanine residues in the QA domain, we generated additional mutants of Osf2 (Fig. 3B). A deletion of 8 of the 18 alanines (Δ89-96) did not affect the transactivation function of Osf2. This is in agreement with genetic analysis demonstrating that a similar polymorphism does not cause phenotypic abnormalities in humans (29). A deletion of 15 of the 18 alanine residues (Δ82-96) also had no effect on the transactivation function of Osf2. In contrast, an expansion of the alanine stretch (27 alanine residues), identical to that observed in a CCD patient (29), led to a complete abrogation of Osf2 transactivation function in both cell lines, indicating that the mechanism by which this mutation causes a CCD phenotype is an alteration of the transactivation function of Osf2.
Next, we deleted the entire N-terminal part of Osf2 (AD1) and 15 of the 18 alanine residues, leaving in place only the glutamine residues [Δ(1-38, 82-96)]. This deletion mutant had the same weak transactivation ability as Δ1-38, which contains the QA, runt, and PST domains, demonstrating that within the wild-type QA domain it is the glutamine stretch that bears most, if not all, of the transactivation function. Again, all of the mutant proteins tested above were found to be expressed in similar amounts and were capable of binding to the OSE2 element (Fig. 3C).
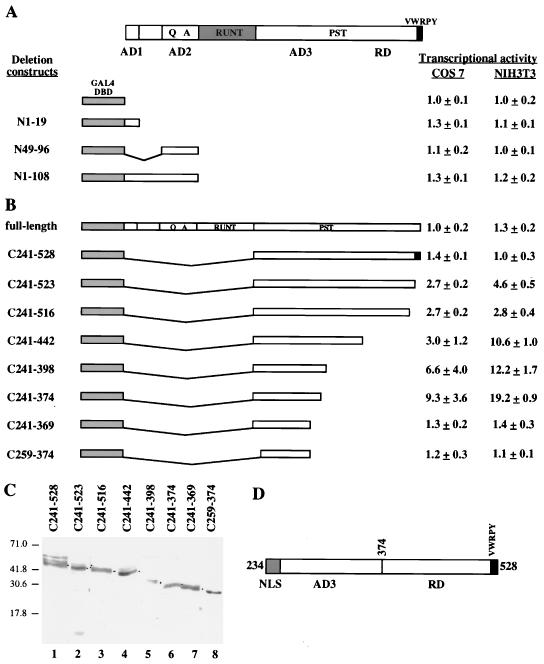
To determine whether AD1 and AD2 could act as autonomous activation domains, we fused these two domains separately or together to the heterologous DNA-binding domain of the yeast transcription factor GAL4 (GAL4DBD [amino acids 1 to 147]) (Fig. 4A) and tested the ability of the constructs to transactivate a luciferase reporter gene driven by five copies of the GAL4 upstream activation sequence (UASG) and the simian virus 40 minimal promoter in pGAL4SVluc. In this assay, AD1 and AD2 alone or together had no transcriptional activity (Fig. 4A), indicating that they need other domains of Osf2 to transactivate. Activation domains that are functional only in the context of the native protein are known to be present in other transcription factors, such as USF2 (27).
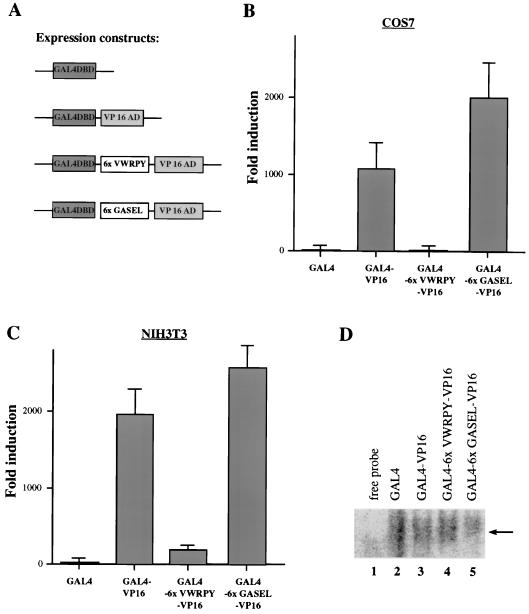
FIG. 4.
Identification of activation and repression domains in Osf2 with a heterologous DNA-binding domain. (A) AD1 and AD2 cannot activate transcription autonomously. GAL4 fusion constructs of AD1 and/or AD2 were tested for the ability to transactivate the GAL4-based luciferase reporter construct (pGAL4SVluc) in DNA cotransfection experiments in COS7 and NIH 3T3 cells. Luciferase activity values shown are averages of three independent transfection experiments done in triplicate. (B) Transcriptional activities of different GAL4-Osf2 fusion proteins. DNA cotransfection experiments were performed in COS7 and NIH 3T3 cells with pGAL4SVluc as the reporter construct. Luciferase activity in cell extracts was assayed as described in Materials and Methods. Values shown are averages of four independent transfection experiments done in triplicate. (C) Expression of GAL4 fusion proteins in transfected cells. Extracts from transfected cells were subjected to immunoblot analysis with anti-GAL4DBD antibody, and the fusion proteins were visualized by ECL detection. Molecular size markers (in kilodaltons) are shown on the left. (D) Schematic representation of the various functional domains in the PST region of Osf2.
Identification of activation and repression domains in the PST region of Osf2.
Deletion of the entire PST domain of Osf2 (Δ258-528) also resulted in a four- to fivefold decrease in the transactivation ability of the protein (Fig. 3B), indicating the presence of an additional transactivation domain(s) within the PST region. This is in agreement with the genetic analysis performed in humans (29). To localize this third activation domain more precisely and to determine if it could act independently of AD1 and AD2, we fused the Osf2 PST domain to GAL4DBD (Fig. 4B) and tested the ability of this construct to transactivate pGAL4SVluc. These experiments were performed in two different cell lines, COS7 and NIH 3T3. In this assay, the PST domain (C241-528) had no transcriptional activity. This was also the case for the GAL4 full-length Osf2 fusion protein. Since the PST domain is important for the function of the native protein in vivo (29) and in transient transfection assays (Fig. 3B [Δ258-528]), we hypothesized that the absence of transactivation by the PST domain in this assay might reflect the existence of multiple activation and repression subdomains and generated more deletion mutants.
Deletions were made initially from the C-terminal end of the PST domain. Deletion of the last 5 amino acid residues (VWRPY) (C241-523), which are identical in all known Runt-related proteins (1), led to a significant and reproducible increase in activation (Fig. 4B), suggesting that this short motif has a repression function in itself. Further C-terminal deletions extending to amino acid 374 (C241-374) resulted in a progressive increase in the level of expression of the reporter gene (Fig. 4B), indicating that the repression domain (RD) is 154 amino acids long and located between amino acids 374 and 528. Removal of amino acids 370 to 374 (GASEL) resulted in a total loss of transcriptional activity in both cell lines (Fig. 4B), demonstrating that these 5 amino acids are a significant part of the transactivation domain. Interestingly, this GASEL motif is part of a short region of the Osf2 PST domain (amino acids 348 to 374) that is the most divergent with the corresponding region of Cbfa2 (Fig. 1). To determine the importance of the GASEL motif, we made an in-frame deletion of the sequence encoding these 5 amino acids in Osf2 and tested the effect of this internal deletion on transactivation by the native protein. As shown in Fig. 3B, deletion of the GASEL motif alone led to a reproducible decrease in the transactivation ability of Osf2 in both COS7 and NIH 3T3 cells, indicating that this short motif is involved in the transactivation function of AD3.
A deletion of 18 amino acid residues from the N-terminal end of the PST domain (C259-374) led to a complete loss of transactivation, suggesting that the entire N-terminal half of the PST domain, up to the GASEL motif (135 amino acids long), is required for transactivation. We termed this region AD3. Unlike the case for AD1 and AD2, AD3 can function in a heterologous system.
To determine if the GAL4 fusion proteins were being made, extracts from transfected COS7 cells were used for immunoblot analysis with a polyclonal antibody directed against GAL4DBD. Figure 4C shows that cells transfected with a plasmid coding for each of the chimeric proteins tested above expressed the recombinant protein. A schematic representation of the various functional domains identified within the PST region of Osf2 is shown in Fig. 4D.
The VWRPY motif can act as a transcriptional repression domain.
The analysis presented above suggested that the last 5 amino acids of Osf2 (VWRPY) might repress transcription (Fig. 4B). This motif is conserved in all known Runt-related proteins (16). To demonstrate the repression function of these 5 amino acids, we cloned in frame, between GAL4DBD and the VP16 activation domain, six copies of the VWRPY coding sequence (Fig. 5A) and tested their functions in a DNA cotransfection assay, also performed with the same two cell lines. This multimer of VWRPY led to a 280-fold decrease in the transactivation ability of VP16 in COS7 cells (Fig. 5B) and a 10-fold decrease in NIH 3T3 cells (Fig. 5C). In a control experiment, we cloned, at the same location, an oligonucleotide that would code for six copies of the 5-amino-acid motif (GASEL) located at the C-terminal end of AD3 (Fig. 5A). This construct resulted in a nearly twofold increase in the transactivation ability of VP16 in COS7 cells (Fig. 5B). This is consistent with the effect of the internal deletion of the GASEL motif presented in Fig. 3B. These results demonstrate that the VWRPY motif can act as a repressor of transcription. EMSA was performed with equal amounts of nuclear extracts from transfected COS7 cells to show that the fusion proteins were being produced (Fig. 5D).
FIG. 5.
Analysis of the repression function of the VWRPY motif. (A) Schematic representation of the GAL4-VP16 constructs used to determine the function of the VWRPY motif. (B) Fold induction of luciferase activity in extracts from COS7 cells transfected with the GAL4-VP16 constructs shown in panel A. (C) Luciferase activity in extracts from NIH 3T3 cells transfected with the GAL4-VP16 constructs. Values are means of nine independent transfection experiments. (D) Expression of GAL4 fusion proteins in transfected cells. EMSA was done with nuclear extracts from transfected COS7 cells by using radiolabeled UASG (GAL4-binding element). Only the protein-DNA complexes are shown.
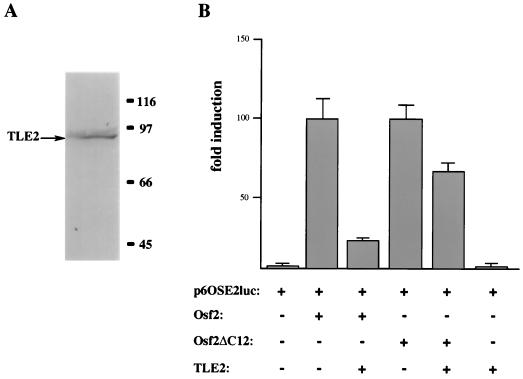
It has been proposed that in Drosophila Runt, the VWRPY motif also acts as a repressor of transcription partly by interacting with Groucho (1). Thus, we asked whether TLE2 (45), a mammalian homolog of Groucho that is expressed in osteoblastic cells (Fig. 6A), could affect the transactivation ability of Osf2. Cotransfection of TLE2 with Osf2 led to an eightfold decrease in Osf2 transactivation function, whereas cotransfection of TLE2 with Osf2ΔC12 (which lacks the last 12 amino acids, including VWRPY) resulted in a weak but reproducible decrease in the transactivation function of this Osf2 deletion mutant (Fig. 6B). This observation supports the hypothesis that one mechanism by which TLE2 may inhibit the transactivation function of Osf2 is through an interaction with the VWRPY motif.
FIG. 6.
Effects of TLE2 on the transactivation abilities of Osf2 and Osf2ΔC12. (A) TLE2 protein is expressed in osteoblastic cells. ROS17/2.8 cell lysate was subjected to SDS-PAGE followed by immunoblot analysis with affinity-purified anti-TLE2 antibody (31). (B) DNA cotransfection experiments were performed in COS7 cells with Osf2 or Osf2ΔC12 and p6OSE2luc reporter in the presence or absence of the TLE2 expression construct pcDNA3-TLE2. Values indicate fold induction of luciferase activity and are means of nine independent transfection experiments.
The QA domain prevents heterodimerization of full-length Osf2 with Cbfβ.
Cbfa2 and the Drosophila Runt protein can heterodimerize with the widely expressed Cbfβ protein in vertebrates (44) and two homologs of Cbfβ, Brother and Big Brother, in Drosophila (13). Moreover, deletion of Cbfβ or Cbfa2 in mice results in an identical phenotype, underscoring the importance of the Cbfa2-Cbfβ interaction in vivo (35, 39, 47, 48). Since Cbfβ is also expressed in osteoblasts, we tested whether Cbfβ is also a partner for Osf2 by using in vitro protein association assays with a purified recombinant GST-Cbfβ fusion protein. 35S-labeled Cbfa2 was bound by immobilized GST-Cbfβ but not by GST alone (Fig. 7A, lanes 4 to 6). In contrast, 35S-labeled Osf2 was not bound by immobilized GST-Cbfβ (Fig. 7A, lanes 1 to 3). To further establish that full-length Osf2 could not interact directly with Cbfβ, we performed EMSA. His-Osf2 alone formed a specific doublet with OSE2, and the addition of Cbfβ resulted in intensification of the Osf2-DNA complex but not in the appearance of a slower-migrating protein-DNA complex (Fig. 7B [compare lanes 1 and 2]). In contrast, when using His-Cbfa2 as a positive control, we always detected heterodimerization with Cbfβ, resulting in a protein-DNA complex of lower mobility (Fig. 7B [compare lanes 3 and 4]). These two results strongly suggest that under the conditions of this assay, Osf2 and Cbfβ are not able to heterodimerize whereas Cbfβ heterodimerizes with the positive control, Cbfa2.
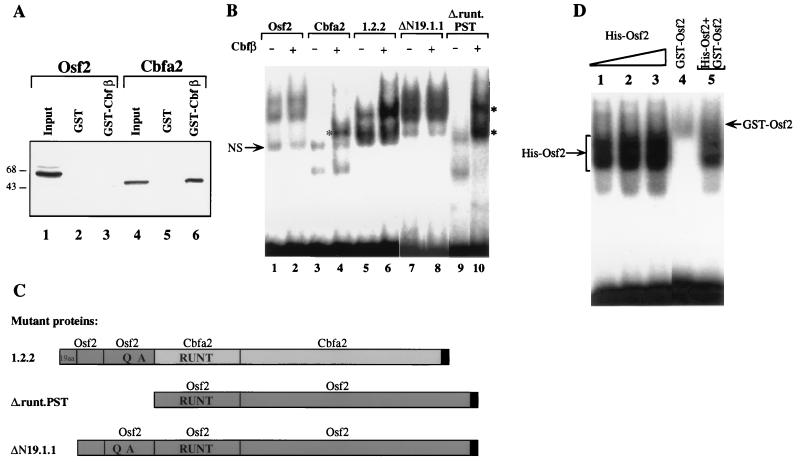
FIG. 7.
The QA domain prevents heterodimerization of Osf2 with Cbfβ. (A) In vitro binding assay. GST protein and GST-Cbfβ fusion protein immobilized on glutathione-agarose beads were incubated with in vitro-translated 35S-labeled Osf2 (lanes 2 and 3) or Cbfa2 (lanes 5 and 6). The bound proteins were then analyzed on an SDS-polyacrylamide gel followed by autoradiography. The input amounts of 35S-labeled Osf2 (lane 1) and Cbfa2 (lane 4) are shown. (B) EMSA done with labeled double-stranded OSE2 oligonucleotide and equivalent amounts of histidine-tagged wild-type and mutant proteins in the presence or absence of Cbfβ. The nonspecific band (NS) is indicated by an arrow. Asterisks indicate the supershifted complexes seen in lanes 4 and 10. (C) Schematic representation of the Osf2 and Cbfa2 chimeric constructs. (D) Osf2 protein does not homodimerize. EMSA was done with increasing amounts of His-Osf2 alone (lanes 1 to 3), GST-Osf2 alone (lane 4), or a combination of His-Osf2 and GST-Osf2 (lane 5). The specific protein-DNA complexes are indicated.
We reasoned that this inhibition of heterodimerization could be due to one or both of the two major Osf2-specific domains, AD1 and AD2 (the QA domain). To test this hypothesis, we swapped the entire amino-terminal region of Cbfa2 with the amino-terminal region of Osf2 (Fig. 7C). In EMSA, this chimeric protein (1.2.2) could not heterodimerize with Cbfβ (Fig. 7B [compare lanes 5 and 6]). ΔN19.1.1, another deletion mutant of Osf2 that lacks AD1 (the first 19 amino acids) alone, also could not heterodimerize with Cbfβ (Fig. 7B, lane 8), while a deletion mutant containing only the runt and PST domains of Osf2 (Δ.runt.PST) could heterodimerize with Cbfβ (Fig. 7B, lane 10). This indicated that it is the QA domain (AD2) that prevents heterodimerization of the native protein with Cbfβ.
The absence of heterodimerization of Osf2 with Cbfβ prompted us to analyze whether Osf2 could homodimerize. We first used His-tagged Osf2 and performed EMSA. The protein-DNA complex migrated as a doublet. This doublet was observed whether we used low or high concentrations of Osf2, but no additional complexes suggestive of homodimer formation were observed with high concentrations of Osf2 (Fig. 7D, lanes 1 to 3). Secondly, we did an EMSA with His-Osf2 and GST-Osf2, which forms a lower-mobility complex than His-Osf2. Coincubation of these two proteins with the probe did not generate a third complex of lower mobility (not detectable even when we used more GST-Osf2 protein), suggesting that under the conditions of this assay, Osf2 is unable to homodimerize (Fig. 7C, lanes 4 and 5).
DISCUSSION
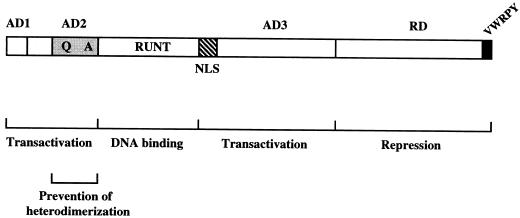
Osf2 is one mammalian member of the Runt-related family of transcription factors. Its critical function during skeletogenesis (10, 21, 24, 29, 36) and the presence of stretches of amino acids in this molecule that are distinct from those in most other Runt-related proteins (Fig. 1) suggest that it has functional domains which might specify its unique role in osteoblast differentiation. An extensive structure-function analysis of Osf2 revealed a novel functional organization for this family of proteins, demonstrating that the N-terminal end and the QA domain control to a large extent its transactivation and dimerization abilities. These findings are summarized in Fig. 8.
FIG. 8.
Schematic representation of the various functional domains of Osf2. The locations of the different activation domains and the repression domain, as well as the N-terminal region comprising the QA domain, which is thought to prevent heterodimerization of Osf2 with Cbfβ, are shown.
Definition of a short NLS in Osf2.
Previous analyses of Cbfa1 had indicated that the NLS spans a broad region covering the runt and PST domains. This was based on a study of the subcellular localization of a series of deletion mutants of Cbfa1 (26). Here, we show that in the context of the wild-type protein, the NLS of Osf2 is much shorter. A 9-amino-acid sequence located at the junction of the runt and PST domains is necessary for nuclear localization of the protein. This sequence is rich in basic residues known to be important for nuclear localization of some proteins (32) and is present in other Runt-related proteins as well, implying that it might perform the same function in these proteins. Recently a short NLS, present at the same location as in Osf2, was identified in Cbfa2 (21a). More generally, it is conceivable that in other Runt-related proteins, this region, rather than the entire PST domain, may act as a sequence necessary for nuclear localization.
Existence of an efficient transactivation domain in the N-terminal end of Osf2.
We generated N-terminal deletion mutants of Osf2, and, to our surprise, a deletion that left the runt and PST domains intact gave a very low level of activation in our transactivation assay. To our knowledge, this is the first demonstration of a transactivation function in the N-terminal end of any Runt-related protein. The sequence of the N-terminal end of Osf2 is substantially different from that of the homologous regions in Cbfa2 and Runt. It contains two subdomains that are unique to Osf2. One includes the first 19 amino acid residues, and the other comprises the QA domain; both of these domains have a transactivation function. The fact that AD1 and AD2 alone or together could not activate transcription in a GAL4 based assay suggests the existence of a critical interaction of these two domains with other regions of Osf2. Dependence of transactivation on the context of the native protein has also been shown for one activation domain in USF2 (27).
What are the implications of these findings for the functional organization of other Runt-related proteins? The functional organizations of the N-terminal ends of other Runt-related proteins have not been fully analyzed. However, AD1 and AD2 sequences are unique to Osf2, suggesting that the presence of transactivation domains at the N-terminal end may be specific to Osf2.
Analysis of the transactivation ability of the QA domain.
Osf2 is the only Runt-related protein to have consecutive glutamine and alanine stretches. Our analyses show that the QA domain has an important transactivation function and that within the QA domain the stretch of 29 glutamine residues is responsible for most, if not all, of the transactivation function. This is in agreement with studies showing that glutamine stretches have a transactivation function in several other transcription factors (12). Our studies also indicate that although the alanine stretch has no detectable transactivation function per se (since it can be removed without affecting the transactivation ability of the protein), an expansion to 27 alanines results in a loss of function of Osf2. This study provides a molecular explanation for the phenotype of one CCD patient (29). In the case of another transcription factor, HOXD13, it has also been postulated that an expansion of the alanine stretch causes a loss of transactivation function resulting in synpolydactyly (30). This is in agreement with studies showing that long alanine-rich regions have a repression function in several transcription factors (14, 15).
To our knowledge there is no other vertebrate transcription factor with such a compact QA domain. In Drosophila, there are several transcription factors that have glutamine- and alanine-rich regions, and they may serve as examples to predict the function of the QA domain in Osf2. Bicoid is a Drosophila factor that has been intensively studied and shown to activate transcription through an interaction between its glutamine-alanine-rich region and TATA box-binding protein-associated factors (TAFs) TAF110 and TAF60 (40). Thus, it is possible that the QA domain of Osf2 may interact with the TAFs and/or other proteins of the general transcription machinery. Alternatively, the QA domain may also interact with cell-specific coactivators.
A third activation domain is present in the PST domain.
Deletion analysis showed that the PST domain contains a third transactivation domain, which we termed AD3. This is in agreement with what is already known of other Runt-related proteins, such as Cbfa2 (4). Our results identifying AD3 within the PST domain provide a molecular explanation for another CCD mutation, as a nonsense mutation in the PST domain that causes CCD in humans is located in AD3 (29). AD3 is highly homologous to the corresponding region of Cbfa2 (3) except for its C-terminal 27 amino acids (amino acids 348 to 374). Experiments conducted with human OSF2 have shown that this small region of the PST domain is also required for optimum transactivation (11). It will be important to determine in vivo the role of this small region in controlling osteoblast gene expression. The fact that deletion of each of the activation domains in Osf2 results in a similar decrease in the transactivation function of the protein indicates that these domains are functionally dependent on each other and that they may interact together with common coactivators.
Existence of a large repression subdomain in the PST domain.
Another surprising result of this analysis of the Osf2 protein is that the PST domain as a whole had no transactivation function in a GAL4-based cotransfection assay. This is due, at least in part, to the presence of a relatively large repression domain (RD) that comprises the last 154 amino acids. Further studies will be required to understand the importance of the repression domain in Osf2. Given the sequence homology between Osf2 and Cbfa2 in this repression domain, it would be important to determine if the corresponding region in Cbfa2 also has a repression function. This repression domain includes the VWRPY motif, the last 5 amino acids of the molecule.
We have shown that six copies of the VWRPY motif were able to inhibit the transactivation function of VP16. In Drosophila, this motif of Runt interacts with Groucho and leads to transcriptional repression (1). TLE2, a mammalian homolog of Groucho (45), also inhibits transactivation by Osf2, and this inhibition requires the last 5 amino acids. Interestingly, TLE2 is expressed in osteoblasts and therefore may have the same function in vivo. TLE2 could cause a slight inhibition of transcription even in the absence of the VWRPY motif. This is consistent with the fact that the repression domain extends further towards the amino terminus of the PST domain and suggests that once recruited by Osf2, TLE2 might also modify chromatin structure and thereby modulate Osf2 function (37).
Lack of heterodimerization of full-length Osf2 with Cbfβ.
Cbfa2 heterodimerizes with Cbfβ (4), and the Drosophila Runt protein interacts with Cbfβ homologs called Brother and Big Brother (13). Moreover, deletion of the Cbfβ gene in mice leads to a phenotype identical to that caused by inactivation of Cbfa2 (35, 39, 47, 48), indicating that the interaction between Cbfβ and Cbfa2 is functionally important in vivo. Therefore, we were surprised when we failed to detect any interaction between Osf2 and Cbfβ in DNA-binding assays, while in a control experiment Cbfβ heterodimerized with Cbfa2.
Several lines of evidence suggest that this absence of interaction is specific and occurs in vivo and that the QA domain may be responsible for preventing this heterodimerization. First, in our control experiments, we always observed heterodimerization of Cbfa2 and Cbfβ. Second, Cbfβ colocalizes to the nucleus only with a deletion mutant of the Cbfa1 protein lacking its N-terminal end (26). This finding is in agreement with the absence of skeletal abnormalities in mice heterozygous for the Cbfβ deletion (39, 48) and with the observation that the N-terminal part of Cbfa1 prevents its heterodimerization with Cbfβ (26). Lastly, deletion and domain-swapping experiments strongly suggest that the QA domain is responsible for preventing heterodimerization with Cbfβ. Our deletion removed AD1 (ΔN19.1.1), leaving in place the amino-terminal part of Osf2, which is highly homologous to the corresponding region of Cbfa2, and the QA domain (AD2). Full-length Cbfa2 heterodimerizes readily with Cbfβ; therefore, it is likely that it is the QA domain that prevents heterodimerization of Osf2 with Cbfβ. This is possibly due to conformational changes imposed on the molecule by the QA domain. Alternatively, Osf2 might heterodimerize with as yet unknown and possibly cell-specific proteins.
In summary, the results of our structure-function analysis of Osf2 reveal the existence of domains and functions that are unique to this Runt-related protein. Indeed, they indicate that the first 19 amino acids (AD1) and the QA domain (AD2) have critical functions as activators of transcription and that the QA domain functions additionally as an inhibitor of heterodimerization. The latter function may be best understood when the three-dimensional structure of the protein becomes available. This unique functional organization among Runt-related proteins might reflect the particular function of Osf2 during skeletogenesis.
ACKNOWLEDGMENTS
K.T. thanks Patricia Ducy for pointing out the NLS, for providing various reagents, and for suggestions throughout the course of the study. We thank Yoshiaki Ito (Kyoto University, Kyoto, Japan) for providing the Cbfβ cDNA clone, Marilyn Szentirmay for the pSG424 and GAL4VP16 expression vectors, and Jennifer Philhower for the pGAL4SVluc reporter construct. Thanks are due to Brendan Lee and Thorsten Schinke for critical reading of the manuscript and to members of the Karsenty lab for helpful suggestions.
S.S. is a scholar of the Fonds de la Recherche en Sante du Quebec and a Killam Scholar of the Montreal Neurological Institute. This work was supported by a grant from the Medical Research Council of Canada, PG11473, to S.S. and by grants from the National Institutes of Health, DE11290 and HD97006, and a Basic Science Award of the March of Dimes Foundation to G.K.
REFERENCES
- 1.Aronson B D, Fisher A L, Blechman K, Caudy M, Gergen J P. Groucho-dependent and -independent repression activities of Runt domain proteins. Mol Cell Biol. 1997;17:5581–5587. doi: 10.1128/mcb.17.9.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 3.Bae S C, Yamaguchi-Iwai Y, Ogawa E, Maruyama M, Inuzuka M, Kagoshima H, Shigesada K, Satake M, Ito Y. Isolation of PEBP2αB cDNA representing the mouse homolog of human acute myeloid leukemia gene, AML1. Oncogene. 1993;8:809–814. [PubMed] [Google Scholar]
- 4.Bae S-C, Ogawa E, Maruyama M, Oka H, Satake M, Shigesada K, Jenkins N A, Gilbert D J, Copeland N G, Ito Y. PEBP2αB/Mouse AML1 consists of multiple isoforms that possess differential transactivation potentials. Mol Cell Biol. 1994;14:3242–3252. doi: 10.1128/mcb.14.5.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bae S C, Takahasi E, Zhang Y W, Ogawa E, Shigesada K, Namba Y, Satake M, Ito Y. Cloning, mapping and expression of PEBP2αC, a third gene encoding the mammalian Runt domain. Gene. 1995;159:245–248. doi: 10.1016/0378-1119(95)00060-j. [DOI] [PubMed] [Google Scholar]
- 6.Coffman J A, Kirchhamer C V, Harrington M G, Davidson E H. SpRunt-1, a new member of the runt domain family of transcription factors, is a positive regulator of the aboral ectoderm-specific CyIIIA gene in sea urchin embryos. Dev Biol. 1996;174:43–54. doi: 10.1006/dbio.1996.0050. [DOI] [PubMed] [Google Scholar]
- 7.Daga A, Karlovich C A, Dumstrei K, Banerjee U. Patterning of cells in the Drosophila eye by Lozenge, which shares homologous domains with AML1. Genes Dev. 1996;10:1194–1205. doi: 10.1101/gad.10.10.1194. [DOI] [PubMed] [Google Scholar]
- 8.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus. Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 9.Ducy P, Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of the mouse osteocalcin gene. Mol Cell Biol. 1995;15:1858–1869. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ducy P, Zhang R, Geoffroy V, Ridall A L, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 11.Geoffroy V, Corral D A, Zhou L, Lee B, Karsenty G. Genomic organization, expression of the human CBFA1 gene, and evidence for an alternative splicing event affecting protein function. Mamm Genome. 1998;9:54–57. doi: 10.1007/s003359900679. [DOI] [PubMed] [Google Scholar]
- 12.Gerber H-P, Seipel K, Georgiev O, Hofferer M, Hug M, Rusconi S, Schaffner W. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science. 1994;263:808–811. doi: 10.1126/science.8303297. [DOI] [PubMed] [Google Scholar]
- 13.Golling G, Li L-H, Pepling M, Stebbins M, Gergen J P. Drosophila homologs of the proto-oncogene product PEBP2/CBFβ regulate the DNA-binding properties of Runt. Mol Cell Biol. 1996;16:932–942. doi: 10.1128/mcb.16.3.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han K, Manley J L. Transcriptional repression by the Drosophila even-skipped protein: definition of a minimal repression domain. Genes Dev. 1993;7:491–503. doi: 10.1101/gad.7.3.491. [DOI] [PubMed] [Google Scholar]
- 15.Han K, Manley J L. Functional domains of the Drosophila Engrailed protein. EMBO J. 1993;12:2723–2733. doi: 10.1002/j.1460-2075.1993.tb05934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito Y, Bae S C. The runt domain transcription factor, PEBP2/CBF, and its involvement in human leukemia. In: Yaniv M, Ghysdael J, editors. Cell cycle regulators and chromosomal translocation. Basel, Switzerland: Birkhauser Verlag; 1997. pp. 108–132. [Google Scholar]
- 17.Jones K L. Smith’s recognizable patterns of human malformation. W. B. Philadelphia, Pa: Saunders Company; 1997. [Google Scholar]
- 18.Kagoshima H, Satake M, Miyoshi H, Ohki M, Pepling M, Gergen J P, Shigesada K, Ito Y. The runt domain identifies a new family of heteromeric transcriptional regulators. Trends Genet. 1993;9:338–341. doi: 10.1016/0168-9525(93)90026-e. [DOI] [PubMed] [Google Scholar]
- 19.Kagoshima H, Akamatsu Y, Ito Y, Shigesada K. Functional dissection of the α and β subunits of transcription factor PEBP2α and the redox susceptibility of its DNA-binding activity. J Biol Chem. 1996;271:33074–33082. doi: 10.1074/jbc.271.51.33074. [DOI] [PubMed] [Google Scholar]
- 20.Kania M A, Bonner A S, Duffy J B, Gergen J P. The Drosophila segmentation gene runt encodes a novel regulatory protein that is also expressed in the developing nervous system. Genes Dev. 1990;4:1701–1713. doi: 10.1101/gad.4.10.1701. [DOI] [PubMed] [Google Scholar]
- 21.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson R T, Gao Y-H, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 21a.Kanno T, Kanno Y, Chen L-F, Ogawa E, Kim W-Y, Ito Y. Intrinsic transcriptional activation-inhibition domains of the polymavirus enhancer binding protein 2/core binding factor α subunit revealed in the presence of the β subunit. Mol Cell Biol. 1998;18:2444–2454. doi: 10.1128/mcb.18.5.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozak M. An analysis of 5′-noncoding sequences of 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurokawa M, Tanaka T, Tanaka K, Hirano N, Ogawa S, Mitani K, Yazaki Y, Hirai H. A conserved cysteine residue in the runt homology domain of AML1 is required for the DNA-binding ability and the transforming activity on fibroblasts. J Biol Chem. 1996;271:16870–16876. doi: 10.1074/jbc.271.28.16870. [DOI] [PubMed] [Google Scholar]
- 24.Lee B, Thirunavukkarasu K, Zhou L, Pastore L, Baldini A, Hecht J, Geoffroy V, Ducy P, Karsenty G. Missense mutations abolishing DNA-binding of the osteoblast-specific transcription factor OSF2/CBFA1 in cleidocranial dysplasia. Nat Genet. 1997;16:307–310. doi: 10.1038/ng0797-307. [DOI] [PubMed] [Google Scholar]
- 25.Lenny N, Meyers S, Hiebert S W. Functional domains of the t(8;21) fusion protein, AML-1/ETO. Oncogene. 1995;11:1761–1769. [PubMed] [Google Scholar]
- 26.Lu J, Maruyama M, Satake M, Bae S-C, Ogawa E, Kagoshima H, Shigesada K, Ito Y. Subcellular localization of the α and β subunits of the acute myeloid leukemia-linked transcription factor PEBP2/CBF. Mol Cell Biol. 1995;15:1651–1661. doi: 10.1128/mcb.15.3.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo X, Sawadogo M. Functional domains of the transcription factor USF2: atypical nuclear localization signals and context-dependent transcriptional activation domains. Mol Cell Biol. 1996;16:1367–1375. doi: 10.1128/mcb.16.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyers S, Downing J R, Hiebert S W. Identification of AML-1 and the (8;21) translocation protein (AML-1/ETO) as sequence-specific DNA-binding proteins: the runt homology domain is required for DNA binding and protein-protein interactions. Mol Cell Biol. 1993;13:6336–6345. doi: 10.1128/mcb.13.10.6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mundlos S, Otto F, Mundlos C, Mulliken J B, Aylsworth A S, Albright S, Lindhout D, Cole W G, Henn W, Knoll J H M, Owen M J, Mertelsmann R, Zabel B U, Olsen B R. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- 30.Muragaki Y, Mundlos S, Upton J, Olsen B R. Altered growth and branching patterns in synpolydactyly caused by mutations in HOXD13. Science. 1996;272:548–551. doi: 10.1126/science.272.5261.548. [DOI] [PubMed] [Google Scholar]
- 31.Nakayama H, Liu Y, Stifani S, Cross J C. Developmental restriction of Mash-2 expression in trophoblasts correlates with potential activation of the Notch-2 pathway. Dev Genet. 1997;21:21–30. doi: 10.1002/(SICI)1520-6408(1997)21:1<21::AID-DVG3>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 32.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 33.Ogawa E, Maruyama M, Kagoshima H, Inuzuka M, Lu J, Satake M, Shigesada K, Ito Y. PEBP2/PEA2 represents a new family of transcription factors homologous to the products of the Drosophila runt and the human AML1 gene. Proc Natl Acad Sci USA. 1993;90:6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogawa E, Inuzuka M, Maruyama M, Satake M, Naito-Fujimoto M, Ito Y, Shigesada K. Molecular cloning and characterization of PEBP2β, the heterodimeric partner of a novel Drosophila Runt-related DNA-binding protein, PEBP2α. Virology. 1993;194:314–331. doi: 10.1006/viro.1993.1262. [DOI] [PubMed] [Google Scholar]
- 35.Okuda T, van Deursen J, Hiebert S W, Grosveld G, Downing J R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 36.Otto F, Thornell A P, Crompton T, Denzel A, Gilmour K C, Rosewell I R, Stamp W H, Beddington R S P, Mundlos S, Olsen B R, Selby P B, Owen M J. Cbfa1, a candidate gene for the cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 37.Palaparti A, Baratz A, Stifani S. The Groucho/transducin-like Enhancer of Split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J Biol Chem. 1997;272:26604–26610. doi: 10.1074/jbc.272.42.26604. [DOI] [PubMed] [Google Scholar]
- 38.Sadowski I, Ptashne M. A vector for expressing GAL4 (1-147) fusion in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sasaki K, Yagi H, Bronson R T, Tominaga K, Matsunashi T, Deguchi K, Tani Y, Kishimoto T, Komori T. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor β. Proc Natl Acad Sci USA. 1996;93:12359–12363. doi: 10.1073/pnas.93.22.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sauer F, Hansen S K, Tjian R. Multiple TAFIIs directing synergistic activation of transcription. Science. 1995;270:1783–1788. doi: 10.1126/science.270.5243.1783. [DOI] [PubMed] [Google Scholar]
- 41.Schreiber E, Matthias P, Muller M, Schaffner W. Rapid detection of octamer binding proteins with “mini-extracts” prepared from small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Selby P B, Selby P R. Gamma-ray-induced dominant mutations that cause skeletal abnormalities in mice. II. Description of proved mutations. Mutat Res. 1978;51:199–236. doi: 10.1016/s0027-5107(78)80019-0. [DOI] [PubMed] [Google Scholar]
- 43.Sillence D O, Ritchie H E, Selby P B. Animal model: skeletal anomalies in mice with cleidocranial dysplasia. Am J Med Genet. 1987;27:75–85. doi: 10.1002/ajmg.1320270109. [DOI] [PubMed] [Google Scholar]
- 44.Speck N A, Stacy T. A new transcription factor family associated with human leukemias. Crit Rev Eukaryot Gene Expr. 1995;5:337–364. doi: 10.1615/critreveukargeneexpr.v5.i3-4.60. [DOI] [PubMed] [Google Scholar]
- 45.Stifani S, Blaumueller C M, Redhead N J, Hill R E, Artavanis-Tsakonas S. Human homologs of a Drosophila Enhancer of Split gene product define a novel family of nuclear proteins. Nat Genet. 1992;2:119–126. doi: 10.1038/ng1092-119. [DOI] [PubMed] [Google Scholar]
- 46.Van Dyke M W, Sirito M, Sawadogo M. Single-step purification of bacterially expressed polypeptides containing an oligo-histidine domain. Gene. 1992;111:99–104. doi: 10.1016/0378-1119(92)90608-r. [DOI] [PubMed] [Google Scholar]
- 47.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe A H, Speck N A. Disruption of the cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci USA. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Q, Stacy T, Miller J D, Lewis A F, Gu T-L, Huang X, Bushweller J H, Bories J-C, Alt F W, Ryan G, Liu P P, Wynshaw-Boris A, Binder M, Marin-Padilla M, Sharpe A H, Speck N A. The CBFβ subunit is essential for CBFα2 (AML1) function in vivo. Cell. 1996;87:697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]