Abstract
Background:
Genetic changes that drive the transition from lepidic to invasive cancer development within a radiographic ground glass or semi-solid lung lesion (SSL) are not well understood. Biomarkers to predict the transition to solid, invasive cancer within SSL are needed.
Methods:
Patients with surgically resected SSL were identified retrospectively from a surgical database. Clinical characteristics and survival were compared between stage I SSL (n=65) and solid adenocarcinomas (n=120) resected during the same time period. Areas of normal lung, in situ lepidic, and invasive solid tumor were microdissected from within the same SSL specimens and next generation sequencing (NGS) and Affymetrix microarray of gene expression were performed.
Results:
There were more never smokers, Asian patients, and sub-lobar resections among SSL but no difference in 5-year survival between SSL and solid adenocarcinoma. Driver mutations found in both lepidic and solid invasive portion were EGFR (43%), KRAS (21%), and DNMT3A (5%). CEACAM5 was the most upregulated gene found in solid, invasive portions of SSL. Lepidic and invasive solid areas had many similarities in gene expression, however there were some significant differences with the gene SPP1 being a unique biomarker for the invasive component of a SSL.
Conclusions:
Common lung cancer driver mutations are present in in situ lepidic as well as invasive solid portions of a SSL, suggesting early development of driver mutations. CEACAM5 and SPP1 emerged as promising biomarkers of invasive potential in semi-solid lesions. Other studies have shown both genes to correlate with poor prognosis in lung cancer and their role in evolution of semi-solid lung lesions warrants further study.
Keywords: Lung adenocarcinoma, ground glass opacity, semi-solid lung nodules, adenocarcinoma in situ, next generation sequencing, microarray
BACKGROUND
The increasing use of cross-sectional imaging and CT scans for lung cancer screening has led to a rise in the diagnosis of incidental ground glass opacities (GGO) and semi-solid lesions (SSL) in the lung of unclear significance.1 GGO without a solid component, or SSL which are part-solid, can represent a variety of conditions from benign infection and inflammation to malignant precursors of invasive lung adenocarcinoma. Lesions on the lung adenocarcinoma spectrum persist on imaging over time and may slowly increase in either total size of the ground glass component or in the size of the solid component.2, 3 Once identified, patients with GGOs will undergo repeat CT scan surveillance4, 5 to monitor for increases in size or the development of a solid component. However only 10–20% of pure GGOs will grow on long-term surveillance while the other 80–90% remain stable.6–8 Among lesions that already have a solid component, 40% will have an increase of their solid component over that time and it is this sub-group of lesions that are likely to be early malignancy.7 Studies have shown that an invasive cancer component begins to develop when a SSL is greater than 15 mm or has a solid component on mediastinal windows of CT scan.7
There are many factors which influence the decision to treat a GGO or SSL, but many physicians use a ground glass size greater than 2 cm or a solid component size greater than 5 mm as a threshold for intervention. Studies have shown that the majority of these lesions are stage IA lung adenocarcinoma,6 long-term survival following resection of these lesions is excellent,9 and very few of these patients will ever develop a tumor recurrence, though many will develop second primary GGOs or SSLs that require subsequent followup.10 Despite a paucity of data or guidelines to support surgical resection in all patients; many patients will undergo a surgical resection to treat these lesions, which may pose no risk of systemic dissemination.
These lung lesions represent a critical area of research interest for several reasons. First, they are an increasingly common diagnosis that requires repeat CT scans over many years3 and ultimately surgery for many patients. Second, the molecular pathways that lead to the evolution of these tumors from premalignant lesion to early-stage lung cancer are not well understood. Attempts to predict a radiographic ground glass nodules degree of invasion on subsequent pathology based on radiographic density have been unsuccessful.11
Genomic characterization of the very early forms of lung adenocarcinoma and the genetic evolution that drives the development of early-stage invasive cancer remain poorly understood. Better molecular-based methods are needed to risk-stratify such lesions beyond using radiographic appearance alone as criteria for undergoing an invasive surgical resection. Such biomarkers might be used in the future for companion diagnostic assay to accompany CT imaging or have a role in potential treatment. Preliminary work has been published on the genetic landscape of early lung adenocarcinoma in studies of Asian patients12–15 where there have been shown to be high rates of EGFR mutations, particularly in SSL.15
Here we present a single institution study of clinical characteristics, long-term survival outcomes, driver mutation analysis and gene expression profiles to study differences between neighboring normal lung tissue, in situ lepidic portions, and invasive solid component in surgically resected SSL.
METHODS
Patient Selection
Patients with semi-solid lesions (SSL) diagnosed as lung adenocarcinomas on surgical pathology were retrospectively identified from a prospectively maintained surgical database at the University of California San Francisco (UCSF) Thoracic Oncology Tissue bank. Preoperative cross-sectional images from CT scans were reviewed for all pathologically proven lung adenocarcinoma. Radiographic appearance of a SSL was confirmed with all lesions possessing both a radiographic measurable “ground glass” area on lung windows and a “solid” component on mediastinal windows. These radiographic criteria with examples are shown in Figure 1A. Patients without good quality preoperative radiographic images for comparison were excluded. Sixty-five patients met the inclusion criteria underwent complete surgical resection between 2011–2018 for a SSL lesion (Figure 1B). Pathologic diagnosis of adenocarcinoma was confirmed for all patients by an expert thoracic pathologist and staging was performed using 7th edition staging criteria. Pathology reports were later reviewed and patients were restaged using the 8th edition staging system which allowed for improved separation between 1 and 2 cm lesions.16 A group of 120 patients with >75% solid stage IA lung adenocarcinomas resected during the same time period were retrospectively identified from a prospectively maintained surgical database for purposes of recurrence and survival comparison. Patients treated with neoadjuvant therapy were excluded. Clinical characteristics and disease-free survival (DFS) and overall survival (OS) were compared between patients with SSL and solid stage I adenocarcinomas. Statistical analysis was performed on SPSS Statistics Standard (V28.0 for Macintosh). Unpaired t-tests were performed for continuous variables. Chi-squared tests were performed for categorical variables with two-sided α <0.05 was defined as statistically significant. Stratified Kaplan-Meier analysis with a censored dataset and the log-rank test were used to evaluate the endpoints of DFS and OS.
Figure 1: Study design.
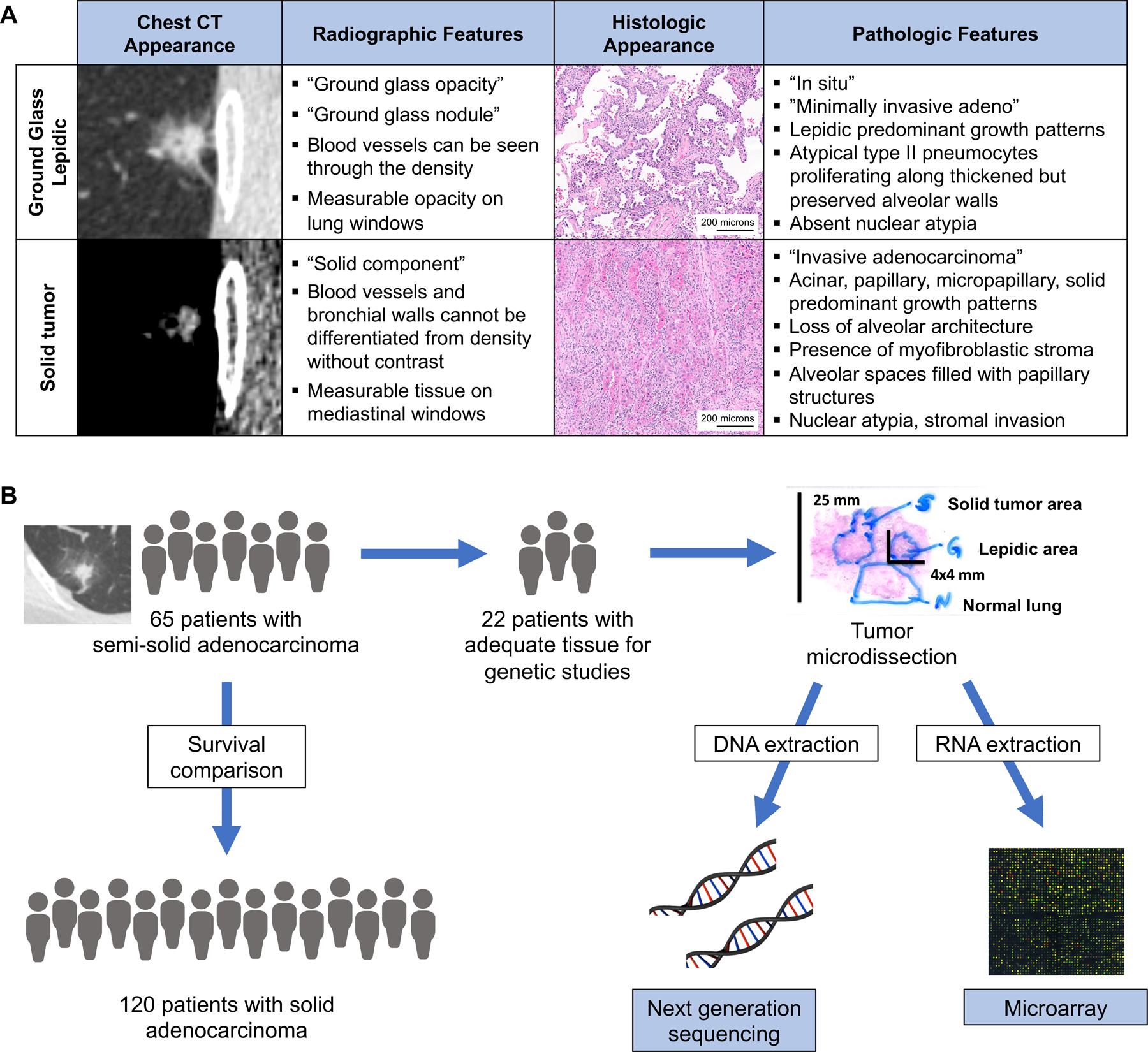
A) Representative images and criteria used to define a semi-solid lung nodule’s ground glass and solid components based on chest CT findings, and histologic criteria used to differentiate in situ lepidic verses invasive lung adenocarcinoma. B) Study design of clinical and genomic comparisons.
There were twenty-two patients with adequate tissue available for detailed genetic analysis consented for biomarker research study participation (Figure 1B). This study was approved by the UCSF institutional review board CC# 00654.
Tumor Microdissection and Sample Preparation
Following surgical resection, tumors were formalin-fixed and paraffin-embedded for pathologic diagnosis and staging. An expert thoracic pathologist then identified separate areas of in situ, lepidic predominant growth patterns, and areas of invasive, solid tumor adenocarcinoma with loss of alveolar architecture, nuclear atypia and stromal invasion from within the same tumor specimen using a standard hematoxylin and eosin stain. Special attention was taken to differentiate cases of lepidic pattern with mechanical or physiologic collapse from invasive patterns. Representative histologic images are shown in Figure 1A. Neighboring normal lung tissue from each patient was also collected to be used for baseline levels of gene expression.
The corresponding areas of normal lung, lepidic, and solid tumor were then microdissected from additional ten micron thick unstained tissue slides with care taken to avoid any-cross contamination between areas. In instances where the area of solid tumor was only a few millimeters in size, up to fifteen serial slides were combined to obtain an adequate genetic sample that did not risk cross-contamination between dissection areas. DNA and RNA were extracted from the microdissected cells using the Qiagen AllPrep FFPE kit (QIAGEN, Germantown, Maryland) according to the manufacturer’s protocol. Extracted DNA was resuspended in AE buffer and DNA concentration and purity was assessed spectrophotometrically (Nanodrop 8000; Thermo Fisher Scientific). RNA samples were suspended in nuclease free water and quantified using spectrophotometer (Nanodrop 8000; Thermo Fisher Scientific) and stored at −80° C per standard protocols.
Next Generation Sequencing
A panel of twenty-five common lung cancer driver mutations was tested on extracted DNA from matched lepidic and solid tumor samples from twenty-two patients using NGS. The genes on the panel included ABL1, AKT1, ALK, BRAF, CTNNB1, DDR2, DNMT3A, EGFR, ERBB2, ESR1, FLT3, GNA11, GNAQ, HRAS, IDH1, IDH2, KRAS, MAP2K1, NRAS, PIK3CA, PTEN, RET, SMAD4, SMO, and TSC1. DNA target library preparation and enrichment were performed using Ion AmpliSeq Library Kit 2.0. Sequencing was performed on the Ion Torrent Proton platform with Ion PGM Sequencing 200 kit v2 according to the manufacturer’s instructions.
For driver mutation comparison to a large population of stage I lung adenocarcinomas, The Cancer Genome Atlas (TCGA) data was used on all n=274 stage I (stage Ia or Ib) adenocarcinoma in TCGA for the 25 driver mutations in our NGS panel.17 TCGA data was used as there was incomplete NGS data available for the population of solid lung adenocarcinomas from our own institution.
Expression Analysis
Extracted RNA was processed with Affymetrix GeneChip WT Pico kit and analyzed using Affymetrix microarray GeneChip Human Transcriptome Array (HTA) 2.0 covering over 48,000 coding and noncoding gene transcripts. Using thresholds of fold changes of ≥ 2.0 and ANOVA p-value <0.05 for significance normal lung was compared to lepidic, lepidic to solid tumor and normal lung to solid tumor to evaluate for individual gene transcripts with significant differences in fold change between parts of SSL. Lists of genes with upregulation and downregulation were evaluated with specific attention to differences seen between normal lung and the solid component to look for biomarkers of invasive cancer and between the lepidic and the invasive solid tumor component to look for biomarkers that are unique to the differences between the preinvasive area and the area of invasive tumor.
A comparison to gene expression levels in all stage I lung adenocarcinomas (stage Ia or Ib) with gene expression data in TCGA (n=268) was performed for all top biomarkers identified as having increased or decreased expression in SSL.17
Role of the funding source
The funding source had no role in the study design, data collection, data analysis, data interpretation, or writing of this report.
RESULTS
Semi-solid lesion demographics and outcomes
There were demographic differences in the 65 patients with SSL compared to the 120 with solid stage I adenocarcinoma resected during the same time period. Patients with SSL were significantly older at the time of surgical resection with a mean ages of 70.6 years verses 66.6 years among the stage I solid adenocarcinoma patients (p=0.0045). Patients with SSL were also more likely to identify as Asian race 29% verses 16% (p=0.0312) though the majority (71%) of patients in our SSL population were non-Asian. SSL patients were more likely to be never smokers than those with solid stage I adenocarcinoma (52% vs 32%, p-value 0.0119). Regarding surgical management, wedge resection was more commonly performed in patients with SSL than those with solid adenocarcinoma (72% vs 50%, p-value=0.0034). More patients with SSL compared to solid adenocarcinoma had tumors with very early-stage pathology of either pT1mi or pT1a (31% of all SSL), while patients with solid adenocarcinoma were more likely to have pT1b or pT1c stage tumors (88% of all solid tumors, p=0.0007) (Table 1).
Table 1:
Clinical characteristics of SSL vs. solid stage I adenocarcinoma
| SSL (n=65) | Solid adeno (n=120) | P-value* | |
|---|---|---|---|
| Demographics | |||
| Mean Age (years ± SE) | 70.6 (±0.9) | 66.6 (±0.9) | 0.0045 |
| Men | 37% | 37% | 0.9203 |
| Women | 63% | 63% | |
| Race | |||
| Asian | 29% | 16% | 0.0312 |
| Non-Asian | 71% | 84% | |
| Smoking history | |||
| Never smoker | 52% | 32% | 0.0119 |
| Current or former smoker | 48% | 67% | |
| Surgical Resection | |||
| Sub-lobar Resection | 72% | 50% | 0.0034 |
| Lobectomy | 28% | 50% | |
| Pathologic Stage, 8th Edition** | |||
| pT1mi | 11% | 0% | 0.0007 |
| pT1a | 20% | 11% | |
| pT1b | 46% | 52% | |
| pT1c | 23% | 38% | |
| Follow up | |||
| Mean follow up time (months) | 56.1 (±2.7) | 54.7 (±2.8) | 0.7368 |
| Recurrence | 7.6% | 14.2% | 0.5120 |
| Cancer-related mortality | 0% | 3% | 0.3126 |
| All-cause mortality | 7.6% | 10% | 0.6033 |
| 5-year Survival | |||
| 5-year Disease Free Survival (± SE) | 95.1% | 86.2% | 0.1281 |
| 5-year Overall Survival (± SE) | 93.4% | 94.2% | 0.7097 |
Unpaired t-tests of equal variance performed for continuous variables. Pearson Chi-squared tests performed for categorical variables. Kaplan-Meier log-rank p-value was calculated for 5-year survival.
Tumors were retrospectively restaged based on 8th edition criteria. Pearson Chi-squared test comparison of (pTmi, pT1a) vs (pT1b, pT1c)
These differences translated into slight, statistically insignificant differences in risk of recurrence, risk of cancer-related mortality, and risk of all-cause mortality (Table 1). Minor differences were seen in the risk of recurrence of 7.6% among SSL and 14.2% among solid adenocarcinomas (p=0.5120), and in the 5-year disease-free survival (DFS) of 95.1% vs 86.2% (log-rank p=0.1281) but these were not statistically significant and did not translate into differences in survival. Five-year OS was 93% among SSL and 94.2% among solid adenocarcinoma (log-rank p=0.7097), demonstrating excellent outcomes for both subsets of stage I adenocarcinoma (Figure 2).
Figure 2: Survival outcomes.
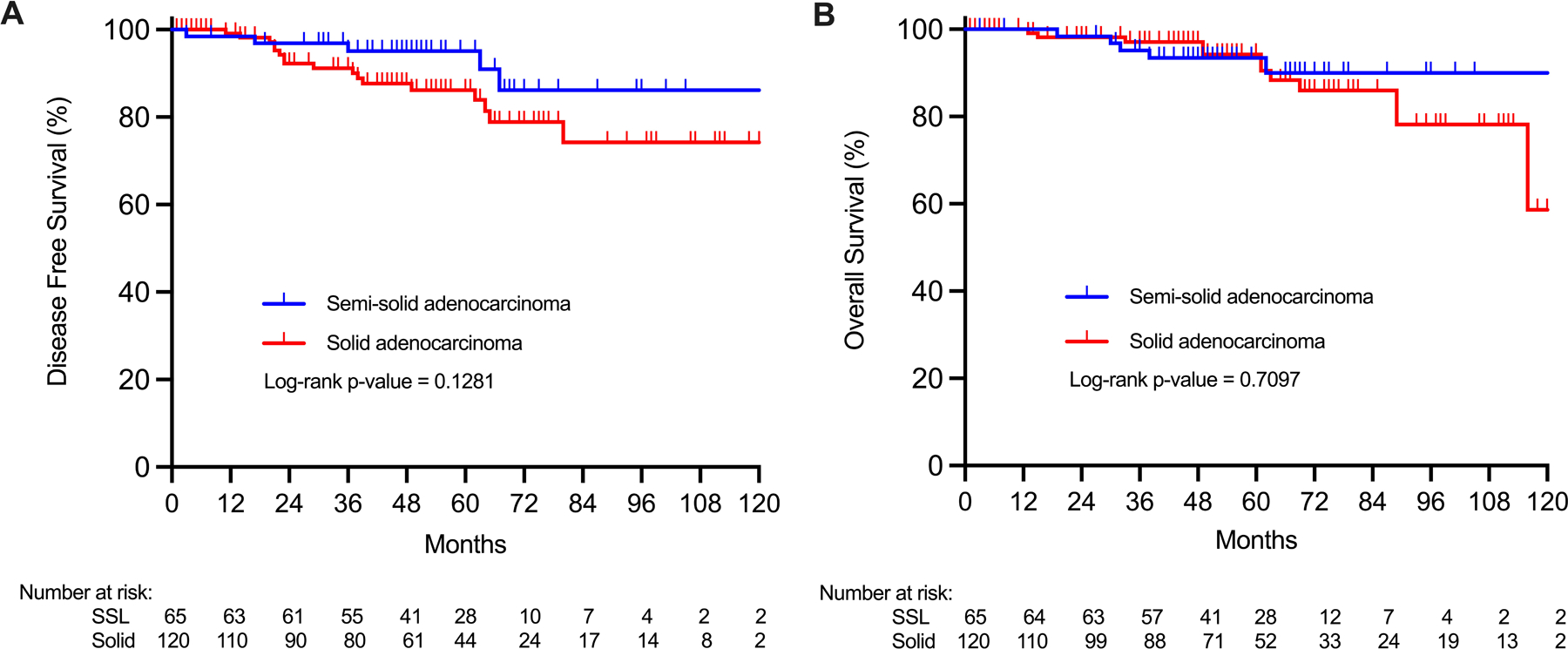
A) Disease free survival and B) overall survival of surgically resected lung adenocarcinoma showed no statistically significant survival differences between semi-solid and pure solid adenocarcinomas.
A comparison of demographics and survival outcomes was also performed between all patients with SSL (n=65) and those with adequate SSL tissue available for genetic study (n=22). There were no significant differences in age, sex, race, smoking history, pathologic stage, recurrence rates, DFS or OS in the patients with tissue available for study. There were significantly more surgical lobectomy performed in the group with tissue available (55% vs 28% lobectomy rate, p=0.0219) and significantly shorter mean follow up (47 vs 56 months, p=0.0018) in patients with adequate tissue samples available for additional study (Supplemental Table 1).
Tumor mutations
Of the twenty-two SSL with adequate surgical specimens for DNA and RNA extraction, nineteen produced DNA samples with adequate reads and coverage for NGS analysis. The in situ lepidic and the invasive solid paired portions of SSL were analyzed as separate specimens, however no differences in mutation patterns between lepidic and solid component were identified. When a driver mutation was identified it was found to be present in both the preinvasive lepidic (Figure 3A) and the solid portions (Figure 3B) in all nineteen SSL specimens analyzed. The targeted NGS panel of lung cancer driver mutations identified common lung cancer driver mutations in thirteen (68%) of the nineteen SSL. The most commonly mutated gene was EGFR, mutated in 42%, followed by KRAS which was mutated in 21%. Mutations were identified in the lepidic portions of SSL as follows EGFR L858R (5 patients, 26%), EGFR exon 19 deletion (3 patients 16%), KRAS Codon 12 (4 patients, 21%), DNMT3A – P904L (1 patient, 5%) no identified mutation (6 patients, 32%). (Figure 3). This distribution of driver mutations was present in the lepidic portions of SSL and the solid portions of SSL.
Figure 3: Semi-solid adenocarcinoma mutations.
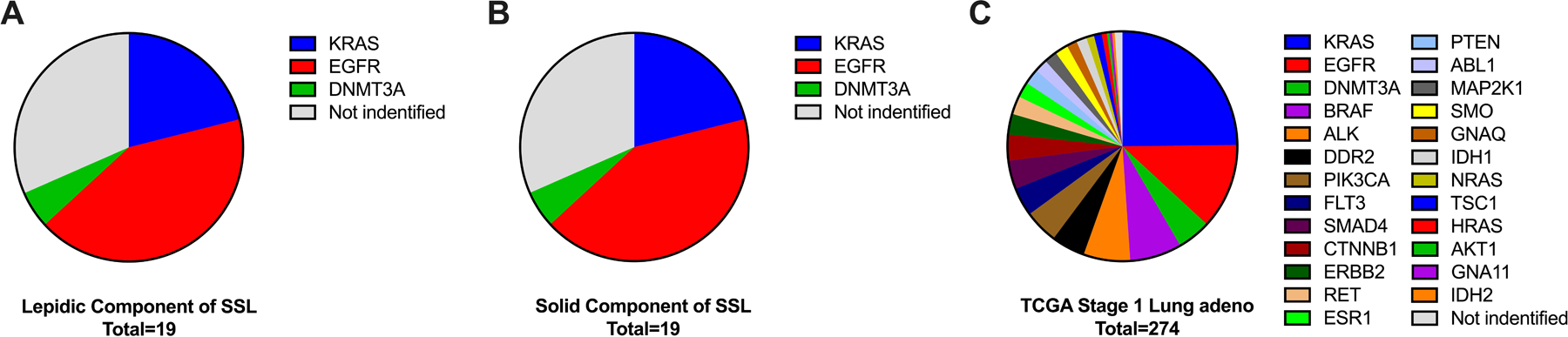
Driver mutations detected by NGS panel in the lepidic component of SSL (A) were all detected in the matched solid components (B) from the same tumors. C) Comparison to TCGA mutation frequently from stage I lung adenocarcinoma showed similar prevalence of KRAS mutations but more frequent EGFR mutations among SSL than all types stage I lung adenocarcinomas (42% vs 12%, χ2 p=0.001).
There was incomplete NGS sequencing data available as a comparison from our larger population of solid lung adenocarcinomas to compare to the mutations found in our SSL. To avoid bias, The Cancer Genome Atlas (TCGA) data was used as a comparison for driver mutations in stage I (stage Ia or Ib) lung adenocarcinoma. In TCGA there were n=274 stage I (stage Ia or Ib) lung adenocarcinoma patients with NGS data available.17 Comparison to TCGA mutation frequency from stage I lung adenocarcinoma showed similar prevalence of KRAS mutations (21% vs 25%, p=NS) but more frequent EGFR mutations among SSL than all types stage I lung adenocarcinomas (42% vs 12%, p=0.001) from the TCGA. DNMT3A mutations were seen with similar frequency (5% vs 5%, p=NS) between our set of SSL and TCGA stage I adenocarcinomas (Figure 3C).
Our sample size was too small to properly evaluate if driver mutations of SSL in our study had any correlation with demographic characteristics, recurrence, or survival outcomes.
Expression profiles
In situ lepidic and invasive solid portions of SSL overall had more downregulation of genes than upregulation of genes compared with normal lung. There were 105 upregulated genes between normal lung and lepidic and 282 downregulated genes (Figure 4A). Hierarchical clustering showed clear differences between normal lung and abnormal portion of the SSL, though the lepidic and solid portions of SSL were very similar with only 32 differentially expressed genes between lepidic and solid portions of SSL (Figure 4B). Volcano plot analyses were performed for comparisons of normal lung to lepidic, lepidic to invasive solid, and normal lung to invasive solid (Figure 5A). The top ten upregulated genes between normal lung and lepidic were MUC21, CEACAM5, CD24, XAGE1B, ABCC3, IGKV1, BCAT1, NPC2, CTSE, and GDF15 (Figure 5B) and the top ten downregulated genes were SLC6A4, RTKN2, CASS4, CALCRL, S100A12, GBP4, MME, SLCO2A1, WIF1, and SVEP1 (Figure 5C). Similarly, there was also more downregulation of genes between the normal lung tissue and the invasive solid portion of the tumors with 128 genes upregulated and 319 downregulated between normal lung and the solid portion of the tumor (Figure 4A). The top ten upregulated genes between normal lung and solid tumor were CEACAM5, XAGE1B, SPP1, SPINK1, MUC21, CRABP2, FDCSP, IGKV1–5, COL10A1, TOX3 (Figure 5B) and the top ten downregulated genes were SLC6A4, RTKN2, WIF1, GKN2, CA4, FOSB, S100A12, FENDRR, MME, CDH5 (Figure 4C). In general, genes with the greatest changes in expression levels were significantly and similarly altered relative to normal lung in both the lepidic and the invasive solid components of tumors as shown in Figure 5B and 5C where we have highlighted any gene which fell into the top ten expression level changes for lepidic or solid components of SSL tumors.
Figure 4: Differential gene expression.
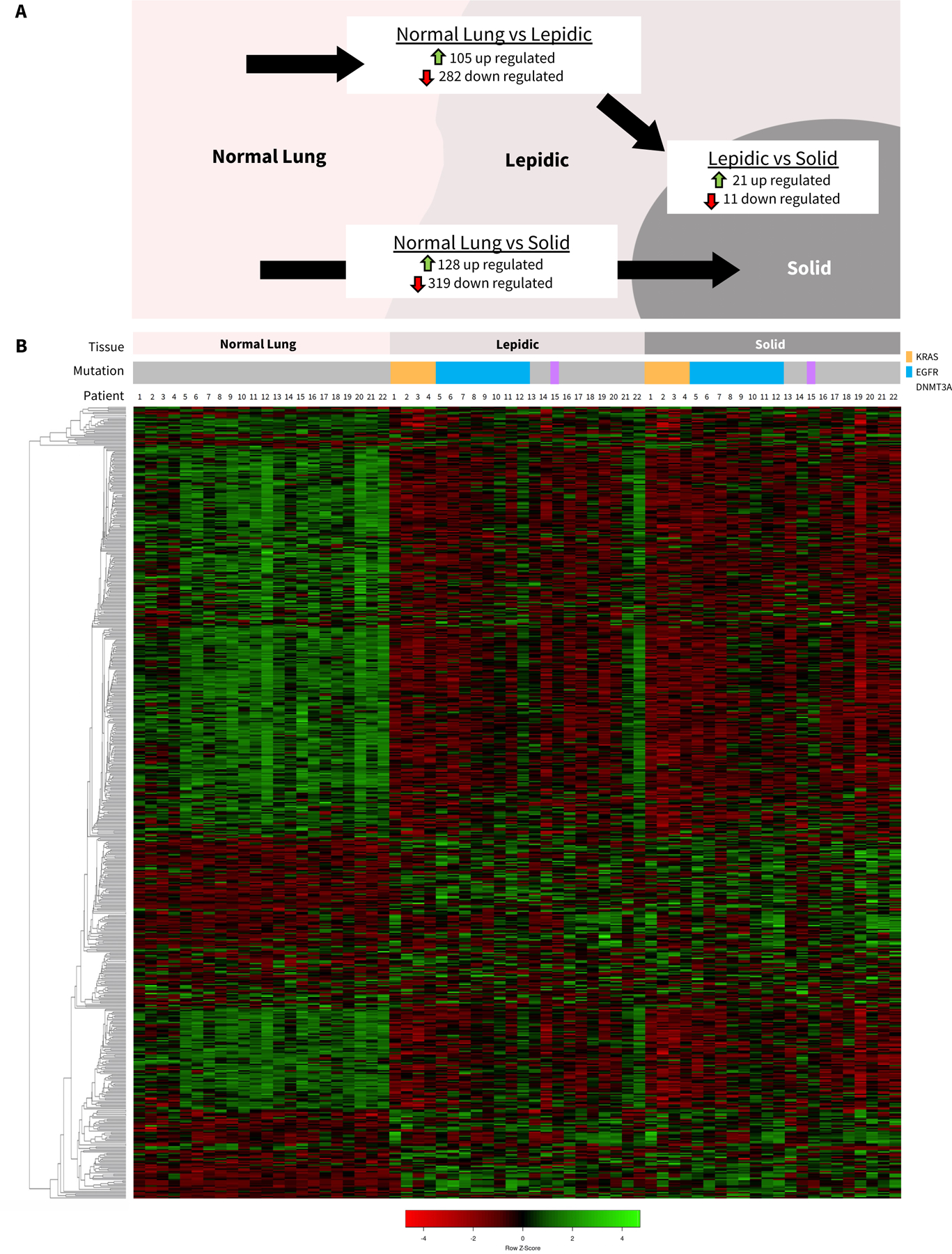
A) Number of significantly up-regulated and down-regulated genes with fold changes >2.0 and between paired normal lung, lepidic, and solid components of SSL. B) Hierarchical clustering heatmap of genes with significant fold changes between normal, lepidic, and solid sorted by tissue type and patient.
Figure 5: Potential biomarkers: Selected differentially expressed genes within semi-solid lung lesions.
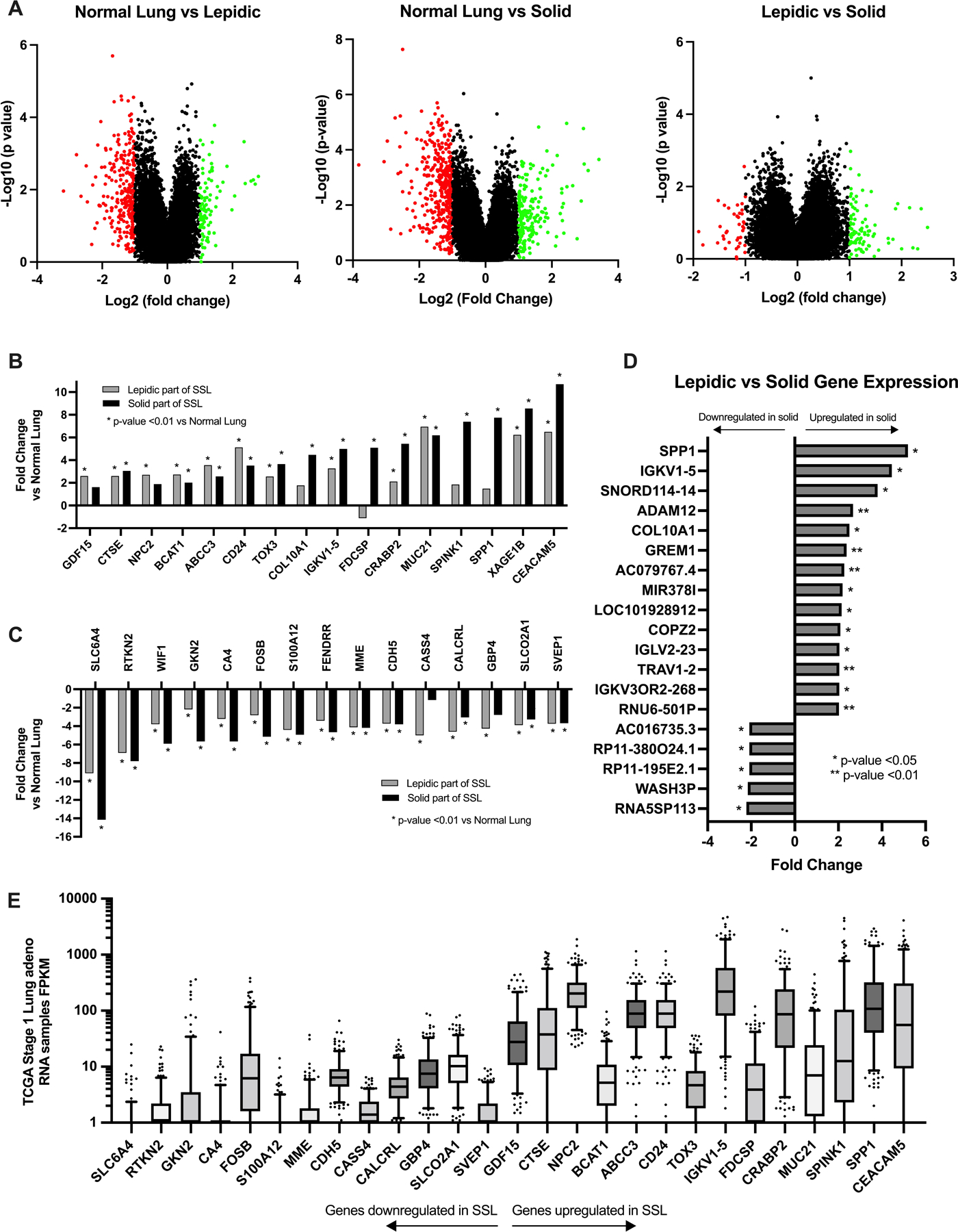
A) Volcano plots of all gene transcripts examined showing fold change comparisons between lepidic and normal lung, solid tumor and normal lung, and lepidic and solid tumor. B) Top genes with the most upregulation of gene expression in either lepidic or solid component compared with normal lung. *=p-value<0.01 compared to normal lung. C) Top genes with the most downregulation of gene expression in either lepidic or solid component compared with normal lung. *=p-value<0.01 compared to normal lung. D) All coding genes with significant differences in gene expression between lepidic and solid tumor. E) TCGA RNA sample data, fragments per kilobase of transcript per million mapped reads (FPKM) for all stage I lung adenocarcinomas (n=268) of the top downregulated and upregulated genes identified in the SSL data.
The gene expression profiles of lepidic and solid components were similar with relatively few genes with significant differences in expression. Of the genes with significant differences between lepidic and solid portions of SSL, upregulation was more common than down-regulation with 21 upregulated and 11 significantly downregulated genes between the lepidic and solid tumor. (Figure 4A). The genes with significant upregulation between the lepidic and solid portion of tumors were SPP1, IGKV1–5, SNORD114–14, ADAM12, COL10A1, GREM1, AC079767.4, MIR378I, LOC101928912, COPZ2, IGLV2–23, TRAV1–2, IGKV3OR2–268, and RNU6–501P and downregulation of genes RNA5SP113, WASH3P, RP11, and AC016735.3 in the solid tumor verses the lepidic portions of the tumors (Figure 5D).
For comparison, genes that were found to have significant upregulation and downregulation in SSL were then compared to TCGA gene expression data seen in stage I adenocarcinoma (n=268). Genes with increased expression in SSL had in general higher expression in stage I lung adenocarcinoma and genes that were downregulated in SSL had generally lower expression in stage I lung adenocarcinomas from TCGA (Figure 5E).
DISCUSSION
This is a comprehensive study of semi-solid lung adenocarcinomas detailing the demographics, survival, driver mutation profiles, and gene expression changes that occur in the lepidic and invasive solid components of these lesions. There is a growing need to better understand these common, incidental, and slowly invasive lung lesions and to better predict which patients require intervention.18–21 Future management of semi-solid lung lesions could be individualized based on needle biopsy or serum biomarkers which correlate with more invasive or biologically aggressive tumor profiles.
The demographic profile of the patients with SSL show that these patients are slightly older, more likely to be Asian and never smokers than those with solid stage I adenocarcinoma at our institution, which corroborates what has been shown in other studies.14, 22 We did not observe any differences in the frequency between men and women here, perhaps due to small sample size, though other studies have shown a female predominance in GGO and SSL.9 In this patient population surgical sublobar resection was the most common operation performed for SSL, comprising 72% of all surgical resections. Though there were five recurrences (7.6% of the SSL patients) in this group, there were no cancer related mortalities with a mean follow-up time of almost 5 years. This supports recent data from large trials such as CALGB140503 and JCOG0802 which have shown that sub-lobar and wedge resection are adequate oncologic operations for stage I NSCLC.23, 24 The proportion of patients who experienced cancer-related mortality was low among both SSL (0%) and solid stage I adenocarcinomas (3%), and 5-year overall survival rates were similar for both groups (93% vs 94%). Large studies of survival of ground glass and partially solid, SSL, lung nodules have shown that part-solid nodules have better survival probability that solid stage I lung cancers,9 and our study is likely under-powered to show any differences. However, it may also be that cancer-related morality from both solid stage I lung cancer and SSL are low, and mortality from other causes may mask these differences. These 5-year survival rates from solid tumors in our study are higher than stage I 5-year survival rates seen in the 8th edition staging project which found 5-year OS rates of 90% (stage IA1), 85% (stage IA2), and 80% (stage IA3) for pathologic stage I patients25 which may be explained by difference in patient populations, and a relatively healthy patient population at our institution.
Among our samples that underwent NGS, we observed at 42% rate of EGFR mutations and a 21% rate of KRAS mutations. EGFR mutations are known to be associated with never smoking status, female gender, and Asian race26 and this rate of mutations is consistent with the demographics of the SSL patients in our study. Two large studies from China performed NGS on surgically resected SSL and found 50–77% of SSL had and EGFR mutation and 5–8% had a KRAS mutation, reflecting the well-established EGFR mutation predominance among Asian patients, and additionally SSL lesions which came from a predominately (79%) never smoking population.15, 27
We also observed that if present, the driver mutation was present in both the lepidic and the solid component of the SSL. Our mutation findings corroborate findings from other published studies which have demonstrated mutation development early in the emergence of GGO, also termed atypical adenomatous hyperplasia (AAH) and minimally invasive adenocarcinoma (MIA) lesions and heterogeneity of driver mutation development with EGFR, KRAS, and BRAF being the most common mutations.12, 28 A large study of the genomics of lung cancer in never smokers found alterations in EGFR in 30% and KRAS in 7% also observed the occurrence of driver mutations in tumors with slow growth suggesting early driver mutation occurrence.29 These data and ours show that the in situ, preinvasive lepidic portion of a semi-solid lesion has the same mutation as the invasive solid component, suggesting that the mutation occurs early, however there are no data in the literature to prove this with serial tissue sampling of the same lesion over time.
A Korean study of ten samples which included both pure GGO without a solid component and mixed GGO with a solid component found a 20% EGFR mutation rate and zero KRAS mutation in their samples.30 These slight differences from mutation frequencies seen in our study of may be relate to small sample size, demographic differences in the patient populations, or reflect a different mutation frequencies in pure GGO, non-solid lung lesions.
We propose several candidate biomarkers including CEACAM5, XAGE1B, SPP1, SPINK1, and MUC21 which have upregulated gene expression in solid and lepidic portions of semi-solid lung lesions but have the greatest expression in the solid invasive portion, and therefore may be the most impactful as a potential biomarker to influence treatment decisions. Among our SSL samples, CEACAM5 expression levels were significantly increased in both the lepidic component and the solid component, with the greatest fold change over normal lung in the solid component of SSL of any of 48,000 transcripts we studied. CEACAM5 is most frequently associated with gastrointestinal, colorectal and pancreatic cancers where it is upregulated in 90% of tumors.31 In NSCLC, CEACAM5 has been shown to promote cell proliferation and migration,32 can be detected via immunohistochemistry, and targeted with a monoclonal antibody.33 CEACAM5 is also promising as a biomarker in NSCLC given its potential to be used as a serum marker the way CEA is used as a serum biomarker in colorectal cancer.34 In large studies of resected GGO and SSL from Japan, serum CEA levels have been shown to be significantly more elevated in solid-dominant lung lesions compared with non-solid GGO,9 which supports our finding. A maytansinoid-loaded monoclonal antibody to CEACAM5 (SAR408701, Sanofi)33 is currently in multiple clinical trials in metastatic non-squamous NSCLC (NCT04154956, NCT04394624, NCT04524689)35 and may potentially demonstrate a benefit of inhibition of CEACAM5 in lung adenocarcinoma.
In targeting differences between the indolent lepidic and the more invasive solid component, the greatest upregulated gene was in SPP1, also known as osteoponin. SPP1 upregulation has been found in gastric cancer, osteosarcoma, ovarian cancer, and oral squamous cell carcinoma as well as NSCLC.36, 37 In NSCLC SPP1 expression correlates with increased tumor grade, worse prognosis, and cisplatin resistance. It has been proposed that DNA methylation is responsible for regulating the expression of SPP1 in lung cancer.36 SPP1 has also been shown to mediate macrophage polarization and lung cancer immune evasion.37 Interestingly, a major regulator of SPP1 are the DNA methyltransferases and DNMT3A was the only other gene mutation besides EGFR and KRAS detected in our cohort. The occurrence of a DNMT3A mutation in a single tumor in our study is not sufficient evidence to draw any conclusions about its role in semi-solid lung adenocarcinoma. As background, DNMT1, DNMT2, DNMT3A, and DNMT3B are the major enzymes of DNA methylation and it is believed that increased expression of DNMT3A increases DNA methylation level which can inhibit gene expression.38 DNMT3A mutations are primarily associated with acute myloid leukemia (AML) where it is mutated in 22% of patients,39 and it is found in only 5% of lung adenocarcinoma.17 Increased DNA methylation via DNMT3A is associated with activation of the Wnt/β-catenin signaling pathway and silencing of DNMT3A with siRNA has been shown to increase SPP1.40 Additional studies are needed to more clearly link and define the role of DNMT3A and SPP1 in semi-solid lung adenocarcinoma.
Limitations of this study include a small sample size which was underpowered to detect differences in survival. The smaller sample size may have impacted our ability to detect less common driver mutations by NGS. We therefore cannot say if only detecting a DNMT3A mutation among mutations other than KRAS or EGFR is significant or just related to small sample size. We also used an NGS panel of only twenty-five driver mutation genes and perhaps additional mutations might have been discovered by a larger panel. Our findings therefore should be validated in subsequent experiments with larger sample sizes and a more comprehensive genome sequencing.
In summary, in this single institution study, SSL patients were more common in Asian never smokers and had excellent 5-year overall survival outcomes following sublobar resection. EGFR (42%) followed by KRAS (21%) were the most common gene mutations which is different from large studies of Asian SSL tumors which have EGFR (50%−77%) and KRAS (5–8%). Our expression data yields targets for further exploration in GGOs and SSL development, specifically CEACAM5 and SPP1. These results should be validated in subsequent larger studies as we work towards identifying biomarkers and molecular-based diagnostics to risk-stratify indolent SSL to observation and more aggressive SSL for surgical resection or perhaps targeted therapies.
Supplementary Material
Novelty and Impact Statement:
This study is the largest gene expression and driver mutation sequencing study of microdissected areas of lepidic and solid tumor from the same semi-solid lung lesions, particularly in a predominately non-Asian population. Driver mutation rates in this study population were 42% EGFR mutations and 21% KRAS mutations, with lower rates of EGFR mutations and higher rates of KRAS mutations than have been demonstrated in Asian patients with semi-solid lung lesions. All driver mutations, if present, were found in both the lepidic and solid portions of the same lesion suggesting early development of driver mutations. Invasive sold and preinvasive, lepidic portions of the same lesions have similar patterns of gene expression, however several genes with increased expression were identified which may prove to be potential biomarkers for future investigation.
Funding:
This research was primarily funded by a private grant from the Braude Foundation. Dr. Woodard’s research is funded in part by the Yale Cancer Center, #IRG 17-172-57 from the American Cancer Society, a Career Enhancement Program Grant from the Yale SPORE in Lung Cancer (P50 CA196530), and by the International Lung Cancer Foundation.
Footnotes
CRediT Authorship Contribution Statement:
GAW - Conceptualization, Data curation, Data analysis, Writing Original Draft, Final approval
VD – Conceptualization, Data curation, Data analysis, Writing reviewing and editing, Final approval; CC – Data analysis, Writing reviewing and editing, Final approval; NRB – Data curation, Writing reviewing and editing, Final approval; JRK - Data analysis, Writing reviewing and editing, Final approval; KDJ - Data curation, Writing reviewing and editing, Final approval;
DMJ – Conceptualization, Supervision, Data analysis, Writing reviewing and editing, Final approval.
Conflicts of Interest:
The authors declare no relevant conflicts of interest. GAW reports participation in advisory boards for Roche Genentech and AstraZeneca. JRK reports consultant work for Oncocyte.
Data Availability:
Dataset associated with this study, with all identifying patient information removed, are available from the authors upon request.
REFERENCES
- 1.Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. The New England journal of medicine. 2011;365:395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kakinuma R, Noguchi M, Ashizawa K, Kuriyama K, Maeshima AM, Koizumi N, et al. Natural History of Pulmonary Subsolid Nodules: A Prospective Multicenter Study. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2016;11:1012–1028. [DOI] [PubMed] [Google Scholar]
- 3.Tang EK, Chen CS, Wu CC, Wu MT, Yang TL, Liang HL, et al. Natural History of Persistent Pulmonary Subsolid Nodules: Long-Term Observation of Different Interval Growth. Heart Lung Circ. 2018. [DOI] [PubMed] [Google Scholar]
- 4.Lee HY, Lee KS. Ground-glass opacity nodules: histopathology, imaging evaluation, and clinical implications. Journal of thoracic imaging. 2011;26:106–118. [DOI] [PubMed] [Google Scholar]
- 5.Lee HY, Choi YL, Lee KS, Han J, Zo JI, Shim YM, et al. Pure ground-glass opacity neoplastic lung nodules: histopathology, imaging, and management. AJR. American journal of roentgenology 2014;202:W224–233. [DOI] [PubMed] [Google Scholar]
- 6.Chang B, Hwang JH, Choi YH, Chung MP, Kim H, Kwon OJ, et al. Natural history of pure ground-glass opacity lung nodules detected by low-dose CT scan. Chest. 2013;143:172–178. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi Y, Mitsudomi T. Management of ground-glass opacities: should all pulmonary lesions with ground-glass opacity be surgically resected? Translational lung cancer research. 2013;2:354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JH, Park CM, Lee SM, Kim H, McAdams HP, Goo JM. Persistent pulmonary subsolid nodules with solid portions of 5 mm or smaller: Their natural course and predictors of interval growth. European radiology. 2016;26:1529–1537. [DOI] [PubMed] [Google Scholar]
- 9.Nakao M, Oikado K, Sato Y, Hashimoto K, Ichinose J, Matsuura Y, et al. Prognostic Stratification According to Size and Dominance of Radiologic Solid Component in Clinical Stage IA Lung Adenocarcinoma. JTO Clin Res Rep. 2022;3:100279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao RW, Berry MF, Kunder CA, Khuong AA, Wakelee H, Neal JW, et al. Survival and risk factors for progression after resection of the dominant tumor in multifocal, lepidic-type pulmonary adenocarcinoma. The Journal of thoracic and cardiovascular surgery. 2017;154:2092–2099.e2092. [DOI] [PubMed] [Google Scholar]
- 11.Fu F, Zhang Y, Wang S, Li Y, Wang Z, Hu H, et al. Computed tomography density is not associated with pathological tumor invasion for pure ground-glass nodules. The Journal of thoracic and cardiovascular surgery. 2021;162:451–459.e453. [DOI] [PubMed] [Google Scholar]
- 12.Sivakumar S, Lucas FAS, McDowell TL, Lang W, Xu L, Fujimoto J, et al. Genomic Landscape of Atypical Adenomatous Hyperplasia Reveals Divergent Modes to Lung Adenocarcinoma. Cancer research. 2017;77:6119–6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakamoto H, Shimizu J, Horio Y, Ueda R, Takahashi T, Mitsudomi T, et al. Disproportionate representation of KRAS gene mutation in atypical adenomatous hyperplasia, but even distribution of EGFR gene mutation from preinvasive to invasive adenocarcinomas. The Journal of pathology. 2007;212:287–294. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi Y, Ambrogio C, Mitsudomi T. Ground-glass nodules of the lung in never-smokers and smokers: clinical and genetic insights. Translational lung cancer research. 2018;7:487–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Li X, Li H, Zhao Y, Liu Z, Sun K, et al. Genomic characterisation of pulmonary subsolid nodules: mutational landscape and radiological features. The European respiratory journal. 2020;55: 1901409. [DOI] [PubMed] [Google Scholar]
- 16.Travis WD, Asamura H, Bankier AA, Beasley MB, Detterbeck F, Flieder DB, et al. The IASLC Lung Cancer Staging Project: Proposals for Coding T Categories for Subsolid Nodules and Assessment of Tumor Size in Part-Solid Tumors in the Forthcoming Eighth Edition of the TNM Classification of Lung Cancer. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2016;11:1204–1223. [DOI] [PubMed] [Google Scholar]
- 17.National Cancer Institute. The Cancer Genoma Atlas (TCGA). https://www.cancer.gov/tcga. Accessed August 8, 2022.2022.
- 18.Zhao ZR, Lau RWH, Long H, Mok TSK, Chen GG, Underwood MJ, et al. Novel method for rapid identification of micropapillary or solid components in early-stage lung adenocarcinoma. The Journal of thoracic and cardiovascular surgery. 2018. [DOI] [PubMed] [Google Scholar]
- 19.Garzelli L, Goo JM, Ahn SY, Chae KJ, Park CM, Jung J, et al. Improving the prediction of lung adenocarcinoma invasive component on CT: Value of a vessel removal algorithm during software segmentation of subsolid nodules. European journal of radiology. 2018;100:58–65. [DOI] [PubMed] [Google Scholar]
- 20.Lee KH, Goo JM, Park SJ, Wi JY, Chung DH, Go H, et al. Correlation between the size of the solid component on thin-section CT and the invasive component on pathology in small lung adenocarcinomas manifesting as ground-glass nodules. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2014;9:74–82. [DOI] [PubMed] [Google Scholar]
- 21.Ahn H, Lee KH, Kim J, Kim J, Kim J, Lee KW. Diameter of the Solid Component in Subsolid Nodules on Low-Dose Unenhanced Chest Computed Tomography: Measurement Accuracy for the Prediction of Invasive Component in Lung Adenocarcinoma. Korean journal of radiology. 2018;19:508–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seow WJ, Matsuo K, Hsiung CA, Shiraishi K, Song M, Kim HN, et al. Association between GWAS-identified lung adenocarcinoma susceptibility loci and EGFR mutations in never-smoking Asian women, and comparison with findings from Western populations. Human molecular genetics. 2017;26:454–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altorki N PL03.06 Lobar or Sub-lobar Resection for Peripheral Clinical Stage IA = 2cm Non-small Cell Lung Cancer (NSCLC): Results From an International Randomized Phase III Trial (CALGB 140503 [Alliance]). World Conference on Lung Cancer. Vienna, Austria: 2022. [Google Scholar]
- 24.Saji H, Okada M, Tsuboi M, Nakajima R, Suzuki K, Aokage K, et al. Segmentectomy versus lobectomy in small-sized peripheral non-small-cell lung cancer (JCOG0802/WJOG4607L): a multicentre, open-label, phase 3, randomised, controlled, non-inferiority trial. Lancet (London, England). 2022;399:1607–1617. [DOI] [PubMed] [Google Scholar]
- 25.Detterbeck FC, Boffa DJ, Kim AW, Tanoue LT. The Eighth Edition Lung Cancer Stage Classification. Chest. 2017;151:193–203. [DOI] [PubMed] [Google Scholar]
- 26.Sholl LM, Aisner DL, Varella-Garcia M, Berry LD, Dias-Santagata D, Wistuba II, et al. Multi-institutional Oncogenic Driver Mutation Analysis in Lung Adenocarcinoma: The Lung Cancer Mutation Consortium Experience. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2015;10:768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Sun Z, Li Y, Qi Q, Huang H, Wang X, et al. Disparate genomic characteristics of patients with early-stage lung adenocarcinoma manifesting as radiological subsolid or solid lesions. Lung Cancer. 2022;166:178–188. [DOI] [PubMed] [Google Scholar]
- 28.Izumchenko E, Chang X, Brait M, Fertig E, Kagohara LT, Bedi A, et al. Targeted sequencing reveals clonal genetic changes in the progression of early lung neoplasms and paired circulating DNA. Nature communications. 2015;6:8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang T, Joubert P, Ansari-Pour N, Zhao W, Hoang PH, Lokanga R, et al. Genomic and evolutionary classification of lung cancer in never smokers. Nat Genet. 2021;53:1348–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim D, Lee JY, Yoo JY, Cho JY. Genetic Features of Lung Adenocarcinoma with Ground- Glass Opacity: What Causes the Invasiveness of Lung Adenocarcinoma? Korean J Thorac Cardiovasc Surg. 2020;53:250–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson JA, Grunert F, Zimmermann W. Carcinoembryonic antigen gene family: molecular biology and clinical perspectives. J Clin Lab Anal. 1991;5:344–366. [DOI] [PubMed] [Google Scholar]
- 32.Zhang X, Han X, Zuo P, Zhang X, Xu H. CEACAM5 stimulates the progression of non-small-cell lung cancer by promoting cell proliferation and migration. J Int Med Res. 2020;48:300060520959478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Decary S, Berne PF, Nicolazzi C, Lefebvre AM, Dabdoubi T, Cameron B, et al. Preclinical Activity of SAR408701: A Novel Anti-CEACAM5-maytansinoid Antibody-drug Conjugate for the Treatment of CEACAM5-positive Epithelial Tumors. Clin Cancer Res. 2020;26:6589–6599. [DOI] [PubMed] [Google Scholar]
- 34.Primrose JN, Perera R, Gray A, Rose P, Fuller A, Corkhill A, et al. Effect of 3 to 5 years of scheduled CEA and CT follow-up to detect recurrence of colorectal cancer: the FACS randomized clinical trial. Jama. 2014;311:263–270. [DOI] [PubMed] [Google Scholar]
- 35.National Institute of Health. www.clinicaltrials.gov. Accessed September 20, 2022.
- 36.Tang H, Chen J, Han X, Feng Y, Wang F. Upregulation of SPP1 Is a Marker for Poor Lung Cancer Prognosis and Contributes to Cancer Progression and Cisplatin Resistance. Front Cell Dev Biol. 2021;9:646390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Du W, Chen Z, Xiang C. Upregulation of PD-L1 by SPP1 mediates macrophage polarization and facilitates immune escape in lung adenocarcinoma. Exp Cell Res. 2017;359:449–457. [DOI] [PubMed] [Google Scholar]
- 38.Anteneh H, Fang J, Song J. Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation. Nature communications. 2020;11:2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. The New England journal of medicine. 2010;363:2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang M, Gao Y, Li Q, Cao H, Yang J, Cai X, et al. Downregulation of DNA methyltransferase-3a ameliorates the osteogenic differentiation ability of adipose-derived stem cells in diabetic osteoporosis via Wnt/β-catenin signaling pathway. Stem Cell Res Ther. 2022;13:397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Dataset associated with this study, with all identifying patient information removed, are available from the authors upon request.


