Abstract
The BCL6 gene, which has been identified from the chromosomal translocation breakpoint in B-cell lymphomas, functions as a sequence-specific transcriptional repressor. We cloned a novel Bcl6-homologous gene, BAZF (encoding Bcl6-associated zinc finger protein). The predicted amino acid sequence of BAZF indicated that the BTB/POZ domain and the five repeats of the Krüppel-like zinc finger motif are located in the NH2-terminal region and the COOH-terminal region, respectively. BAZF associated with Bcl6 at the BTB/POZ domain and localized in the nucleus. Since zinc finger motifs of BAZF were 94% identical to those of Bcl6 at the amino acid level, BAZF bound specifically to the DNA-binding sequence of Bcl6 and functioned as a transcriptional repressor. The repressor activity was associated with both the BTB/POZ domain and the middle portion of BAZF. The 17-amino-acid sequence in the middle portion was completely conserved between BAZF and Bcl6, and the conserved region was critical for the repressor activity. Expression of BAZF mRNA, like that of Bcl6 mRNA, was induced in activated lymphocytes as an immediate-early gene. Therefore, the biochemical character of BAZF is similar to that of Bcl6 although the tissue expression pattern of BAZF differs from that of Bcl6. This is apparently the first report of a gene family whose members encode zinc finger proteins with the BTB/POZ domain.
Chromosomal translocations involving 3q27 have been identified in some non-Hodgkin’s lymphomas, particularly in diffuse large B-cell lymphomas (5, 35). The human proto-oncogene BCL6 has been identified from chromosomal breakpoints (25, 30, 43). Translocations lead to deregulation of the BCL6 gene as a result of the juxtaposition of the BCL6-coding region to heterologous promoters (44) and may be related to lymphomagenesis. The BCL6 gene encodes a 92- to 98-kDa nuclear phosphoprotein (6, 36) that contains the BTB/POZ domain in the NH2-terminal region and six Krüppel-type zinc finger motifs in the COOH-terminal region (17, 25, 30, 43). The BTB/POZ domain is important for protein-protein interactions (3, 47). The zinc finger motifs of BCL6 bind to specific DNA sequences in vitro (4, 24). Since the NH2-terminal half of the protein contains a repressor domain(s) in vitro (2, 7, 11, 42), BCL6 can function as a sequence-specific transcriptional repressor.
The BCL6 gene is well conserved between humans and mice, with a 100% identity of zinc finger motifs at the amino acid level (17). This gene is ubiquitously expressed in various tissues, including those of lymphatic organs, and is predominant in the germinal-center B cells (6, 18, 36). Expression of this gene is induced in activated lymphocytes as an immediate-early gene (17), and it is progressively upregulated in differentiating keratinocytes, which suggests a role in developing tissues at the terminal differentiation stage (46). To observe the physiological functions of Bcl6, this gene was disrupted in the mouse germ line (10, 18, 45); the Bcl6-deficient mice showed growth retardation, and most died at a young age. The mature lymphocytes did develop in these Bcl6-deficient mice but germinal-center formation was impaired. In addition, Bcl6-deficient mice had inflammatory responses in multiple organs, especially the heart and lung, characterized by infiltration of eosinophils, and there was an increased production of Th2-type cytokines (10). Since the BCL6-binding DNA sequence resembled the sequence motif bound by the signal transducer and activator of transcription (STAT) factors, and since interleukin-4 (IL-4) induces the differentiation of Th0 cells to Th2 cells (26), Bcl6 may repress IL-4-induced transcription by competitive binding to sites recognized by the IL-4-responsive STAT factor, STAT6 (10).
Zinc finger-type transcription factors compose a family like the GATA family (14). We attempted to identify the Bcl6-homologous gene(s) with a probe carrying Bcl6 zinc finger motifs. We found a novel homolog (BAZF), the amino acid sequence of which was 94% identical to Bcl6, at the zinc finger motifs. Thus, BAZF can associate with Bcl6 at the BTB/POZ domain, localize in the nucleus, bind to the BCL6-binding DNA sequence, and play the role of transcriptional repressor. The physiological roles of BAZF in gene regulation are discussed below.
MATERIALS AND METHODS
Animals.
(C57BL/6 × DBA/2)F1 female mice were purchased from Japan SLC Co. Ltd. (Hamamatsu, Japan).
Cloning of the BAZF cDNA.
A 213-bp fragment of the Bcl6 cDNA carrying the zinc finger motifs was obtained by PCR with the following primers: 5′-CTGGAGAAAAGCCCTACAAATGTG-3′ and 5′-CGGTATTGCACCTTGGTGTTG-3′. This fragment was used as a probe to screen a genomic library from mouse kidney, and a homologous clone (BAZF) was isolated. A KpnI fragment in the homologous clone was used as a probe to screen mouse cDNA libraries from lung (Stratagene, La Jolla, Calif.) and entire 11-day-gestated embryos (Stratagene). The 5′-most side of the BAZF cDNA, including the ATG start codon, was cloned with the poly(A)+ mRNAs from the mouse heart with the 5′ RACE system for rapid amplification of cDNA ends (GIBCO BRL, Rockville, Md.). The products of the amplification were cloned into the pGEM-T vector (Promega, Madison, Wis.). Both strands were sequenced with an automatic DNA sequencer (ALF Express; Pharmacia, Piscataway, N.J.) with vector-specific primers. Those cDNA fragments were reconstructed into pGEM-4Z (Promega) to obtain a full length of the open reading frame (pBAZF).
Construction of luciferase reporter plasmids.
To construct pBSX4-SV40-LUC, double-stranded oligonucleotides containing two copies of the BCL6-binding DNA sequence (BS) (5′-CGACATTCCTAGAAAGCATA-3′) with BamHI sites on both flanks were synthesized. The BamHI fragment was ligated into the BamHI site of pGEM-7Z, and pBSX4, which carried four copies of the BS, was isolated. The KpnI-SacI fragment of pBSX4 was inserted into the KpnI-SacI-digested pGL2 plasmid, which carries the simian virus 40 promoter and enhancer and the luciferase reporter gene (Promega), to construct pBSX4-SV40-LUC. Another luciferase reporter plasmid (p5GBS luc) (32) carrying the Gal4-binding sequence was kindly provided by K. Nakajima (Osaka University, Osaka, Japan).
Construction of fusion genes.
Gal4 fusion genes were constructed as follows. Various fragments of Bcl6 and BAZF cDNA were obtained by PCR with pairs of primers specific for the Bcl6 or BAZF sequence linked to an EcoRI site. The PCR products were subcloned into the pGEM-T vector. An EcoRI fragment of the PCR products was subcloned into the EcoRI site of pSG424 carrying the Gal4(1–147)-coding sequence (38). Primer sequences with a new EcoRI site (underlined) used for various fragments were as follows: for Gal4-Bcl6(1–122), 5′-TTTGAATTCATGGCCTCCCCGGCTGACA-3′ and 5′-TTTGAATTCGCATGTGTCGACAACATGCT-3′; forGal4-Bcl6(1–520), 5′-TTTGAATTCGAAGAAGGTCCCATTCTCA-3′; forGal4-BAZF(1–128), 5′-TTTGAATTCATGGGTTCCACAGCGGCCCCA-3′and 5′-TTTGAATTCACACGCCTGGACTACGTGTT-3′; for Gal4-BAZF(1–319), 5′-TTTGAATTCCTCGTCCCCTGGAGCTTAGA-3′; and for Gal4-BAZF(148–319), 5′-TTTGAATTCCCCCCAAGACCCCCAACAGT-3′. TheGal4-BAZF fusion gene with a 97-bp deletion was constructed as follows. Since BAZF(1–319) contains a PstI site, the sequence (5′-TTTCTGCAGCCTAGTTGGGGAGAGTTCA-3′) with a new PstI site on the 5′ side (underlined) 97 bp downstream of the PstI site was synthesized as a primer. PCR was done between the primer and the primer for the 3′ side of Gal4-BAZF(1–319). The PstI-EcoRI fragment of Gal4-BAZF(1–319) or Gal4-BAZF(148–319) was replaced by a PstI-EcoRI fragment of the PCR product. Translation termination was provided by three-frame termination codons in the pSG424 plasmid. All PCR-derived products as well as subcloning junctions were verified by sequence analysis.
For construction of the glutathione S-transferase (GST) fusion genes, an EcoRI fragment of Gal4-Bcl6(1–122) or Gal4-BAZF(1–128) was subcloned into the EcoRI site of the pGEX1λT vector (Pharmacia) to obtain GST-Bcl6(1–122) or GST-BAZF(1–128). To prepare GST-BAZF(314–498), two primers 5′-TTTGGATCCCTCTTAGCTCCAGGGGACGAGGAC-3′ and 5′-TTTGAATTCTGCCCCTAGGGAGCGAAGTTTGGC-3′) with a BamHI or an EcoRI site (underlined) were prepared and the PCR product was obtained. The BamHI-EcoRI fragment of this product was subcloned into the BamHI-EcoRI site of the pGEX2T vector (Pharmacia).
To construct the BAZF and the Bcl6 expression plasmids, an EcoRI-XhoI fragment of BAZF or a BamHI fragment of Bcl6 was inserted into the pcDNA3 expression vector (Invitrogen, San Diego, Calif.) (pcDNA3-BAZF or pcDNA3-Bcl6). pHemagglutinin (pHA)-tagged BAZF was constructed by subcloning an EcoRI-XhoI fragment of pBAZF into the EcoRI-XhoI site of pHAKIT (33). To construct pHA-HOX11, an EcoRI-XhoI fragment of HOX11 (20) was inserted into pHAKIT.
Assays for transcriptional repressor activity of BAZF with luciferase reporter genes.
L cells maintained in Dulbecco’s modified Eagle’s medium (Life Technologies, Grand Island, N.Y.) with 5% fetal calf serum (Bioserum; CSL Ltd., Victoria, Australia) were plated at 3.3 × 105 cells per 60-mm-diameter dish 1 day before transfection. WEHI231 cells were maintained in RPMI 1640 medium (Life Technologies) with 10% fetal calf serum. The luciferase reporter plasmid (10 μg), various expression plasmids, and pRL-tk (0.5 μg) (TOYO INK, Tokyo, Japan) or pRL-SV40 (0.5 μg) (TOYO INK) as a transfection efficiency control were cotransfected into L cells by the calcium phosphate coprecipitation technique (39) or into WEHI231 cells by electroporation with a Gene Pulser (Bio-Rad, Richmond, Calif.) equipped with a capacitance extender (0.40 kV and 960 μF). The transfected L cells were incubated for 8 h, washed three times in Dulbecco’s modified Eagle’s medium, and cultured for an additional 48 h. The WEHI231 cells were cultured for 48 h. The transfected cells were harvested and lysed, and luciferase activity in the lysates was determined by standard methods with Pikkagene Dual (TOYO INK).
EMSA.
Specific DNA-binding activity of BAZF was determined by electrophoretic mobility shift assay (EMSA) (46). Briefly, the synthesized double-stranded BS was labeled with digoxigenin (DIG) with DIG oligonucleotide 3′-end labeling kits (Boehringer Mannheim, Mannheim, Germany). Binding reactions were performed in a mixture containing 1 μg of GST-BAZF(314–498), 1 μg of poly(dI-dC) (Pharmacia), and a 15 fM concentration of the DIG-labeled probe in 21 μl of reaction buffer (10 mM HEPES [pH 7.8], 50 mM KCl, 1 mM dithiothreitol, 50 μg of bovine serum albumin/ml). This mixture was then incubated for 30 min at room temperature, separated by electrophoresis on a 6% nondenaturing polyacrylamide gel, transferred to a nylon membrane (Boehringer Mannheim), and fixed by cross-linking with UV irradiation with a Spectrolinker (Schleicher & Schuell) and baking at 120°C for 30 min. The DIG-labeled probe was detected with sheep anti-DIG antibody conjugated with alkaline phosphatase. The antibody detection reaction was performed with an enhanced chemiluminescence detection system (Boehringer Mannheim). A competitive EMSA was done by adding to the mixture a 10- or 50-fold molar excess of unlabeled double-stranded oligonucleotide. Sequences of the mutant oligonucleotide (one base mismatch, underlined) were as follows: 5′-CGACATTCCTATAAAGCATA-3′ and 5′-CGACATTCCTAGCAAGCATA-3′.
Assays for interaction between BAZF and Bcl6.
The GST pull-down assay was done as described previously (9). Briefly, the GST-BAZF(1–128) and GST-Bcl6(1–122) proteins were purified as described previously (24). The BAZF full-length cDNA was subcloned into the pCITE4a vector (Novagen, Madison, Wis.). In vitro T7-driven transcription and translation of pCITE4a-BAZF were done with TNT T7 Quick Coupled Reticulocyte Lysates (Promega) with [35S]methionine (Amersham, Little Chalfont, Buckinghamshire, England). The 35S-labeled translation products (10 μl) were incubated with 10 μg of each GST fusion protein in 500 μl of Triton X-100 lysis buffer (0.5% Triton X-100, 20 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA) containing 1 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride, 1 μg of aprotinin/ml, 10 μg of leupeptin/ml, and 0.1 mM sodium orthovanadate for 2 h, washed three times in Triton X-100 lysis buffer, and separated by electrophoresis on a 12.5% nondenaturing polyacrylamide gel. The labeled band was analyzed with an image analyzer (BAS2000; Fuji Photo Film Corp., Tokyo, Japan).
The interaction was also identified by Western blotting. pHA-BAZF and pcDNA3-Bcl6 were cotransfected into L cells by the calcium phosphate coprecipitation technique (39). The transfected L cells were lysed with Nonidet P-40 (NP-40) lysis buffer (1.0% NP-40, 50 mM Tris-HCl [pH 8.0], 150 mM NaCl) with proteinase inhibitors on ice for 15 min. Total proteins were incubated with mouse monoclonal anti-HA antibody (CA125; Boehringer Mannheim) at 4°C for 1 h and further incubated with protein A-Sepharose (Pharmacia) at 4°C for 2 h with continuous mixing. The mixture was centrifuged at 7,500 × g at 4°C for 30 s, washed with 1 ml of ice-cold NP-40 lysis buffer, and centrifuged at 7,500 × g at 4°C for 30 s. This washing procedure was repeated three times. The HA-BAZF protein was fractionated on a 7.5% polyacrylamide gel and transferred to a membrane (Immobilon-P; Millipore, Bedford, Mass.) with an electroblot (Bio-Rad). The filter was incubated with rabbit anti-BCL6 antibody (18) followed by horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulin (Ig) antibody (Amersham) for 1 h for each step at room temperature. The antibody detection reaction was performed with the enhanced chemiluminescence detection system (Amersham).
Subcellular localization of the BAZF protein was examined in L cells cotransfected with pHA-BAZF and pcDNA3-Bcl6 by immunohistochemistry. Those transfected L cells were stained with mouse monoclonal anti-HA antibody followed by biotinylated rabbit anti-mouse IgG (Vector, Burlingame, Calif.) and streptavidin-Texas red (Vector) and then by rabbit anti-Bcl6 antibody (18) to sequentially facilitate fluorescein isothiocyanate-conjugated anti-rabbit IgG (Cappel, Durham, N.C.). Fluorescence in the L cells was visualized on CLSM (Zeiss) with 63× oil objectives.
Northern blot analysis.
Splenocyte cultures were prepared as described previously (17). Splenocytes from 6-week-old mice were stimulated with phorbol myristate acetate (PMA) (5 ng/ml; Sigma, St. Louis, Mo.) plus ionomycin (250 ng/ml; Sigma) in the presence or the absence of cycloheximide (10 μg/ml; Sigma). Total RNAs were extracted from cultured splenocytes or adult female mouse tissues with the TRIzol reagent (Life Technologies). Northern blot analysis was performed as described previously (17). A 0.7-kb KpnI fragment (bp 1763 to 2463) of the 3′ untranslated region of BAZF was used as a probe. The fragment was subcloned into the KpnI site of pGEM-4Z and labeled with DIG (Boehringer Mannheim) by PCR with primers T7 and SP6.
Nucleotide sequence accession number.
The BAZF sequence has been deposited with DDBJ, EMBL, and GenBank under accession no. AB011665.
RESULTS
Cloning of BAZF cDNA.
To search for a Bcl6-related gene(s), Southern blot analysis of genomic DNA was done with three probes carrying different regions of the Bcl6 cDNA: the BTB/POZ domain, the middle portion, and zinc finger motifs. When mouse genomic DNA was digested with BamHI, an extra 3.5-kb band was detected for Bcl6 zinc finger motifs at low stringency (data not shown). However, the other two probes detected no extra band at low stringency. Thus, we considered the possibility of a novel Bcl6-homologous gene carrying similar zinc finger motifs.
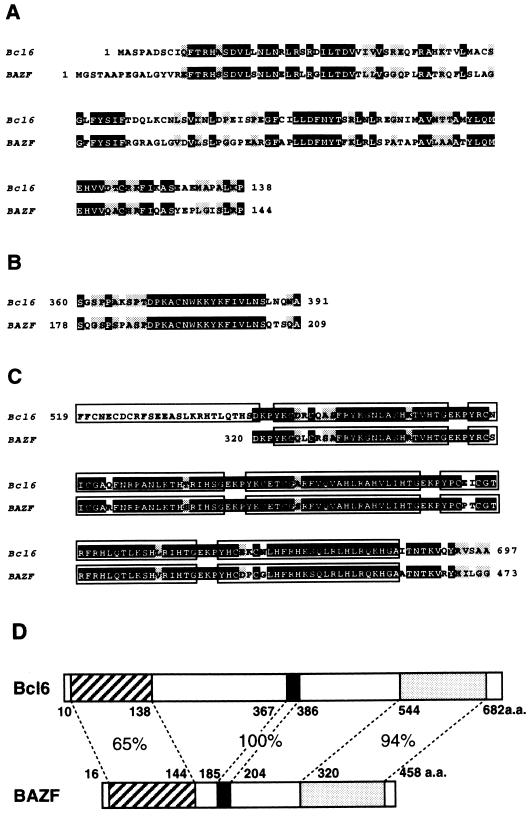
A mouse genomic library was then screened with the zinc finger probe to identify the homologous gene. A clone (BAZF) with a sequence similar to that of Bcl6 at the zinc finger motifs was selected. Putative zinc finger motifs of BAZF were further used as a probe to screen cDNA libraries. Several overlapping cDNA clones were obtained (Fig. 1A). The 5′ region of the cDNA, including the ATG start codon, was cloned with mRNAs from a normal mouse heart by the anchored reverse transcription-PCR method and the sequence was confirmed on the genomic DNA sequence. As shown in Fig. 1B, an open reading frame of 1,425 bp encoded the predicted 474 amino acids (aa). The putative amino acid sequence contained the five zinc finger motifs located at the region from aa 320 to 470 and the BTB/POZ domain at the region from aa 1 to 128. When the amino acid sequence of BAZF was compared with that of Bcl6 (Fig. 2), BAZF was found to be 94% identical to Bcl6 at zinc finger motifs, 65% identical at the BTB/POZ domain, and, surprisingly, 100% identical at the region from aa 188 to 204. The region from aa 137 to 187 was proline rich (34%), and the similar region of Bcl6 (from aa 319 to 371) was also proline rich (21%) (17).
FIG. 1.
Cloning strategy and sequence of murine BAZF cDNA. (A) Schematic diagram of the overlapping BAZF cDNA clones. The closed and hatched boxes indicate the BTB/POZ domain and the zinc finger motifs, respectively. (B) Nucleotide and predicted amino acid sequences of BAZF. The ATTTA sequences are underlined.
FIG. 2.
Comparison of the predicted amino acid sequences between Bcl6 and BAZF at three highly homologous regions. (A) The BTB/POZ domain. (B) The middle portion, including the identical 17-aa sequence. (C) The zinc finger domain. Numbers refer to the amino acids located in these proteins. Closed and shaded boxes indicate identical and conserved residues, respectively. Open boxes in panel C show the zinc finger motifs. (D) Schematic representation of the Bcl6 and BAZF proteins. Hatched, closed, and stippled boxes represent the BTB/POZ domain, the middle portion, and the zinc finger motifs, respectively. Numbers refer to the amino acids located in these proteins. Approximate percentages of sequence similarity are given.
The zinc finger domain of BAZF can bind to the BS.
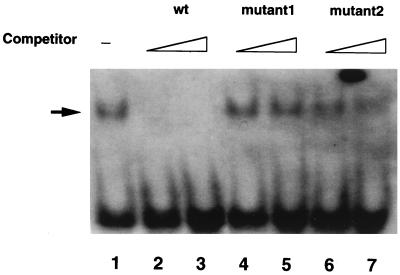
Since the α-helical region in the COOH-terminal half of Krüppel-like zinc finger motifs is important for determining sequence specificity of DNA binding (27) and the amino acid sequence of this region was identical in BAZF and Bcl6 (Fig. 2), the capacity of the zinc finger domain of BAZF to bind to the BS was examined by EMSA with the BS as a probe. The protein used for DNA binding lacked the BTB/POZ domain since inclusion of the BTB/POZ domain in a protein can affect its DNA binding (3). As shown in Fig. 3, the zinc finger domain of BAZF could form complexes with the BS. This binding was specific for the BS, since the 50-fold excess amount of cold competitors with one base mismatch did not block the binding.
FIG. 3.
Binding of the zinc finger domain of BAZF to the BS. The BS was labeled with DIG, and binding was examined by EMSA. The arrow shows the DNA-protein complex. The prominent shifted band (lane 1) disappeared after addition of excess amounts of unlabeled BS (wt; 10-fold [lane 2] and 50-fold [lane 3]) but not by after addition of excess amounts of the unlabeled BS with one base mismatch (mutant 1 [5′-CGACATTCCTATAAAGCATA-3′], 10-fold [lane 4] and 50-fold [lane 5]; mutant 2 [5′-CGACATTCCTAGCAAGCATA-3′], 10-fold [lane 6] and 50-fold [lane 7]).
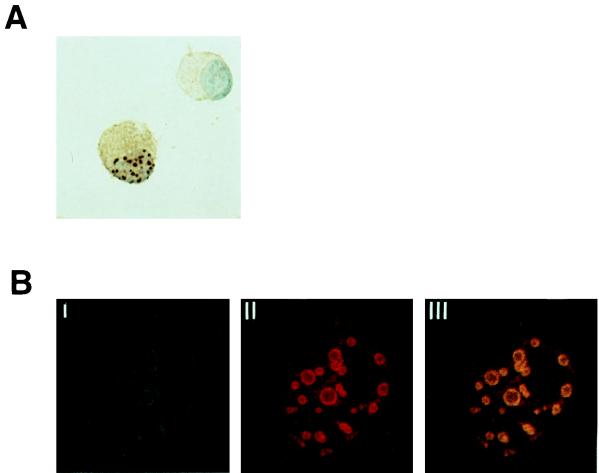
BAZF and Bcl6 colocalize in a nucleus.
Since BAZF can bind to the specific DNA sequence in vitro, BAZF may play a role in the nucleus as a transcription factor. We histologically examined localization of BAZF in a cell by transfection of pHA-BAZF into L cells (Fig. 4A). HA-BAZF was detected in a nucleus as a dot pattern. This pattern in a nucleus is supported by evidence that the BTB/POZ domain of Bcl6 is important for punctate nuclear localization (12). Distribution of Bcl6 and BAZF in a nucleus was further examined by cotransfection of pHA-BAZF and pcDNA3-Bcl6 into L cells. The intracellular localization of the two proteins was identified immunohistochemically by confocal microscopy. Both HA-BAZF and Bcl6 were detected in the nucleus as a dot pattern (Fig. 4B), and the proteins located at the same area in the nucleus (Fig. 4B, panel III). Since the BTB/POZ domain is important for protein-protein interactions (3, 47) and the BTB/POZ domain of BCL6 interacts with itself (12), this colocalization suggested interaction between Bcl6 and BAZF in the nucleus.
FIG. 4.
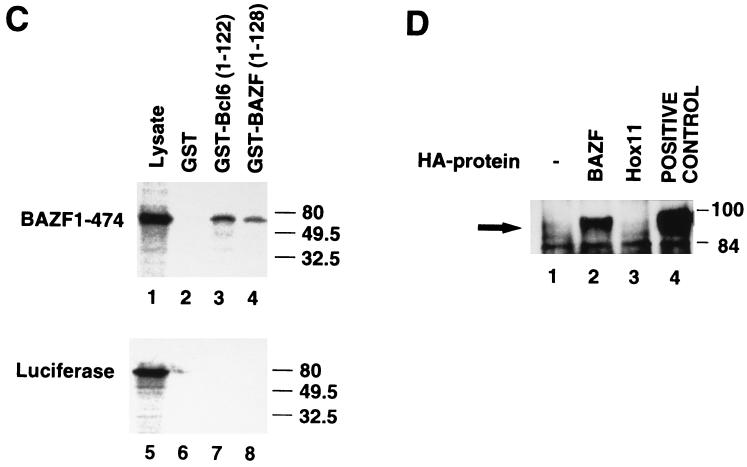
Interaction between BAZF and Bcl6. (A) Localization of BAZF in the nucleus. L cells were transfected with pHA-BAZF. The cells were stained with anti-HA antibody. BAZF was identified in the nucleus of the left cell as a nuclear dot pattern, but not in the cell on the right. Cells were counterstained with hematoxylin. (B) Colocalization of BAZF and Bcl6 in the nucleus. L cells were transfected with pHA-BAZF and pcDNA3-Bcl6. Bcl6 and HA-BAZF were detected in the nucleus with anti-Bcl6 antibody and fluorescein isothiocyanate-conjugated secondary antibody (B, panel I) and with anti-HA antibody and Texas-red conjugated secondary antibody (B, panel II), respectively. When superimposed, the green and red signals generated a yellow signal confirming colocalization of these proteins (B, panel III). (C) Interaction between BAZF and Bcl6 in vitro. Interaction between the BTB/POZ domain of GST-BAZF or GST-Bcl6 and in vitro-translated BAZF was examined by a pull-down assay. [35S]methionine-labeled BAZF was left untreated (lysate) or incubated with either immobilized GST alone, GST-Bcl6(1–122), or GST-BAZF(1–128). For the controls, [35S]methionine-labeled luciferase was left untreated (lysate) or incubated with either immobilized GST alone, GST-Bcl6(1–122), or GST-BAZF(1–128). Molecular masses (in kilodaltons) are indicated on the right. (D) Interaction between BAZF and Bcl6 in vivo. pHA-BAZF and pcDNA3-Bcl6 were cotransfected into L cells. Lysates of L cells transfected with pcDNA3-Bcl6 (−), pcDNA3-Bcl6 and pHA-BAZF (BAZF), or pcDNA3-Bcl6 and pHA-HOX11 (Hox11) were immunoprecipitated with anti-HA antibody. Interaction of Bcl6 with HA-BAZF was detected in the immunoprecipitates by Western blotting with anti-BCL6 antibody. The lysate from L cells transfected with pcDNA3-Bcl6 was immunoprecipitated with anti-Bcl6 antibody, and Bcl6 in the immunoprecipitate was immunoblotted with anti-BCL6 antibody as a positive control. The arrow indicates the Bcl6 protein. Molecular masses (in kilodaltons) are indicated on the right.
The possibility of this interaction was investigated in vitro with a pull-down assay. Immobilized GST-Bcl6(1–122) or GST-BAZF(1–128) carrying the BTB/POZ domain was incubated with the in vitro translation products of pCITE4a-BAZF (encoding the full length of BAZF), and immobilized GST-Bcl6(1–122) or GST-BAZF(1–128) was precipitated. The presence of the in vitro-translated BAZF labeled with [35S]methionine in the precipitant was analyzed autoradiographically (Fig. 4C). GST-Bcl6(1–122) and GST-BAZF(1–128) bound to the products of pCITE4a-BAZF but not to the products of pCITE4a-LUC (encoding luciferase). Since GST alone did not bind to BAZF, the BTB/POZ domain of Bcl6 and BAZF directly binds to BAZF in vitro.
The interaction was further investigated in vivo by cotransfection of pHA-BAZF and pcDNA3-Bcl6 into L cells. HA-BAZF in the transfected L cells was immunoprecipitated with anti-HA antibody, and the presence of Bcl6 in the immunoprecipitate was examined by Western blotting with an anti-BCL6 antibody (Fig. 4D). Bcl6 was detected in the immunoprecipitate as 92- to 98-kDa molecules. These molecules were not detected in the immunoprecipitate from nontransfected L cells or in pHA-HOX11-transfected L cells, thereby showing the specificity of the interaction.
BAZF functions as a transcriptional repressor.
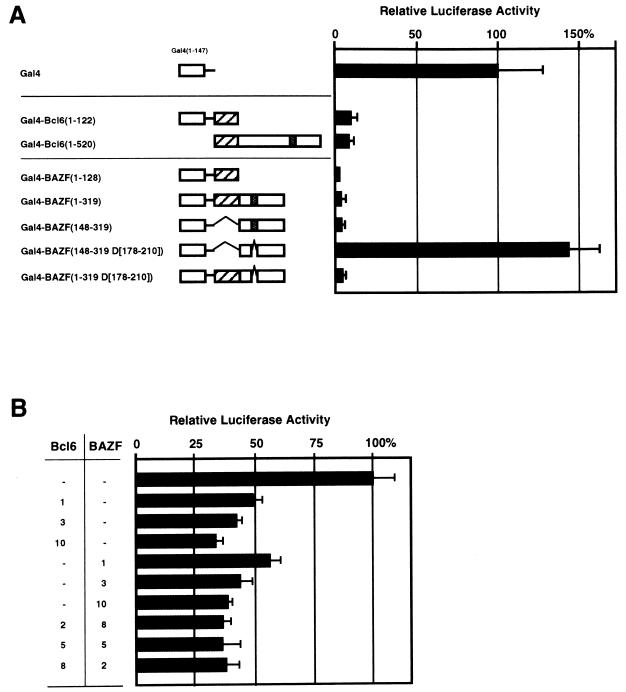
BCL6 was found to be a transcriptional repressor since two noncontiguous regions of BCL6 (aa 10 to 138 in the BTB/POZ domain and aa 267 to 395 in the middle portion) contain transrepressor activity (7). These two regions were also homologous to the region from aa 16 to 144 in the BTB/POZ domain and to aa 178 to 209 in the middle portion of BAZF (Fig. 2). The transcriptional repressor activity of BAZF was then examined by cotransfection of the Gal4-BAZF gene and the p5GBS luc reporter gene into L cells. The p5GBS luc gene carries the Gal4-binding sequence and the luciferase gene controlled by the E1b promoter (32). Figure 5A shows that repressor activity of Gal4-BAZF associated with the BTB/POZ domain (from aa 1 to 122) or the middle portion (from aa 148 to 319) of BAZF. However, the middle portion of BAZF, lacking the 100% conserved region (from aa 188 to 204), lost repressor activity but slightly enhanced expression of the luciferase reporter gene.
FIG. 5.
Transcriptional repressor activity of BAZF. (A) Schematic representation of Gal4-BAZF fusion proteins and their repression of transcription of the luciferase gene from the p5GBS luc reporter plasmid in L cells. Gal4-BAZF (10 μg) was cotransfected with the p5GBS luc reporter plasmid (10 μg) into L cells. pRL-tk was also cotransfected in each experiment to serve as an internal control for transfection efficiency. The luciferase activity in the lysates was measured with a luminometer 48 h after transfection and compensated for by luciferase activity from pRL-tk. The activity is expressed as a percentage of the luciferase activity in L cells transfected with Gal4 and the p5GBS luc plasmid. The data presented are representative of three independent experiments, and error bars are shown. In the schematic representation, hatched and stippled boxes indicate the BTB/POZ domain and the conserved 17 aa in the middle portion, respectively. (B) Repression by BAZF and Bcl6 of transcription of the luciferase gene from the pBSX4-SV40-LUC reporter plasmid in WEHI231 cells. Various amounts of pcDNA3-BAZF and pcDNA3-Bcl6 were cotransfected with the pBSX4-SV40-LUC reporter plasmid (10 μg) into WEHI231 cells. Numbers refer to the amount (in micrograms) of plasmid transfected. The total amount of transfected DNA was kept constant (20 μg) in each experiment by adding pcDNA3. pRL-SV40 was also cotransfected in each experiment to serve as an internal control for transfection efficiency. Luciferase activities in lysates were measured with a luminometer 48 h after transfection and compensated for by luciferase activity from pRL-SV40. The activity was expressed as a percentage of the luciferase activity in WEHI231 cells transfected with the pBSX4-SV40-LUC reporter plasmid. The data presented are representative of three independent experiments, and error bars are shown.
The repressor activity of BAZF was further confirmed with a different assay system. Since BAZF can bind to the BS, as determined by EMSA (Fig. 3), we cotransfected the pBSX4-SV40-LUC reporter plasmid and pcDNA3-BAZF (encoding the full-length BAZF protein) into WEHI231 cells (Fig. 5B). The pBSX4-SV40-LUC gene carries four repeats of the BS and the luciferase gene controlled by the simian virus 40 promoter and enhancer. BAZF as well as Bcl6 repressed expression of the luciferase gene in a dose-dependent manner. When the reporter plasmid without the BS (pSV40-LUC) was cotransfected with pcDNA3-BAZF, transcriptional repression was not detected (data not shown), indicating that this repression is specific for the BS. Since BAZF and Bcl6 may form a complex in a nucleus (Fig. 4B), the functional collaboration of both proteins was further examined by cotransfection of the pBSX4-SV40-LUC gene and the BAZF and Bcl6 genes at different ratios into WEHI231 cells. The presence of both BAZF and Bcl6 displayed additive but not synergistic effects on the repressor activity (Fig. 5B), indicating that the two proteins are functionally similar.
Expression of BAZF mRNA is strong in mouse heart and lung, and BAZF is induced in lymphocytes as an immediate-early gene.
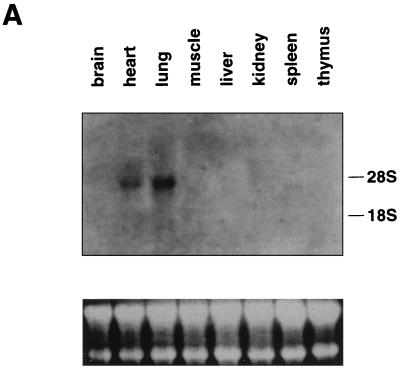
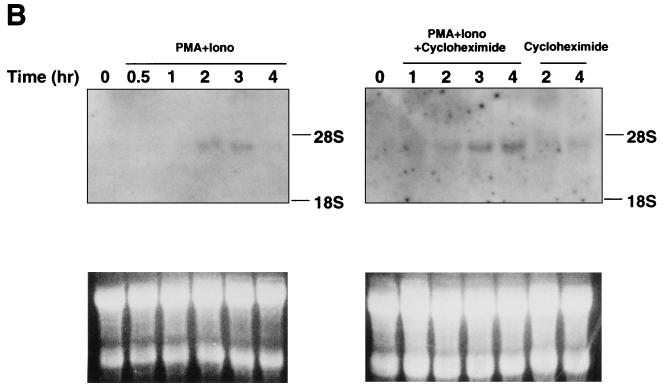
Expression of BAZF mRNA was examined in normal tissues from an adult mouse by Northern blotting with the 3′ untranslated region of BAZF as a probe (Fig. 6A). A 3.8-kb band was detected in heart and lung, and expression was ubiquitous in every tissue examined by reverse transcription-PCR (data not shown). Since the Bcl6 gene is induced in lymphocytes activated with PMA plus ionomycin as an immediate-early gene (17), the inducibility of BAZF mRNA in splenocytes stimulated with PMA and ionomycin was analyzed by Northern blotting. As shown in Fig. 6B, BAZF mRNA was induced in splenocytes from 1 h, increased for up to 2 h, and downregulated from 4 h after stimulation. This induction was also examined in splenocytes stimulated with PMA plus ionomycin in the presence of cycloheximide. The cycloheximide treatment did not block the induction but did increase the expression. Therefore, BAZF also plays a role in activated splenocytes as an immediate-early gene.
FIG. 6.
Expression of BAZF mRNA in various mouse tissues. (A) Northern blot analysis of BAZF expression in mouse tissues. The expression was analyzed in total RNA (25 μg) from the indicated tissues of adult mice. rRNA positions are indicated on the right. The lower part of the panel shows 28S and 18S rRNA from the ethidium bromide-stained gel as the loading control. (B) Inducibility of the BAZF gene in splenocytes after stimulation. Splenocytes were stimulated with PMA plus ionomycin (Iono) in the presence or absence of cycloheximide. Total RNA (25 μg) was isolated from the splenocytes at the indicated times after stimulation. Expression of the BAZF gene in these RNAs was analyzed by Northern blotting. rRNA positions are indicated on the right. The lower part of the panel shows 28S and 18S rRNA from the ethidium bromide-stained gel as the loading control.
DISCUSSION
We identified BAZF, a homolog of Bcl6. Three sections of the predicted amino acid sequence are highly conserved (Fig. 2). The zinc finger motifs of BAZF are 94% identical to those of Bcl6. Zinc finger motifs of transcription factors determine the specificity of binding to DNA (15). Although BAZF contains five repeats of the zinc finger motif instead of the six repeats in Bcl6, BAZF can bind to the BS (Fig. 3). Among transcription factors with zinc finger motifs, each zinc finger will make contact with a base or a phosphate of DNA or make no contact at all with DNA. For example, GLI (37) has five repeats of a zinc finger motif, and one in the NH2-terminal-most side does not make contact with DNA. PLZF (28) has nine repeats of a zinc finger motif and binds to DNA through seven repeats located on the carboxyl-terminal-most side. Therefore, five repeats of the zinc finger motif in the carboxyl-terminal-most side of Bcl6 may be important to bind to the DNA sequence. These observations also suggest the presence of a large number of genes regulated by BAZF and Bcl6.
The 17-aa sequence in the region from aa 188 to 204 of BAZF is 100% identical to that in the region from aa 370 to 386 of Bcl6. The repressor activity of BAZF associates with the middle portion (aa 148 to 319), which includes this conserved region (Fig. 5A). The middle section of Bcl6 contains a proline-rich region (21% proline content) in the region from aa 319 to 371. BAZF also has a proline-rich region (34% proline content) in the region from aa 137 to 189, indicating that this proline-rich region followed by the conserved 17 aa is shared between them. Thus, the shared sequence in the middle portion of BAZF may be important for transcriptional regulation. Indeed, as BAZF lost repressor activity by deletion of the region from aa 178 to 210 (Fig. 5A), the conserved 17-aa sequence is likely to be critical for the activity. This sequence has no homology to the known activity site of transcriptional factors and may be a novel activity domain of transcriptional repressors.
The BTB/POZ domain is also conserved (65% identity). This domain has homology to the NH2-terminal regions of some zinc finger-type DNA-binding proteins, such as the Drosophila tramtrack (Ttk) (19) and the human KUP proteins (8). This shared domain is important for specific protein-protein interactions. For example, the BTB/POZ domain of Ttk can interact efficiently with that of Ttk itself and that of GAGA, but not with that of the distinct zinc finger protein ZID (3). We found that the BTB/POZ domain of BAZF can bind to BAZF itself and to Bcl6 (Fig. 4C). Using the yeast two-hybrid system, we confirmed that the BTB/POZ domain of BAZF can interact with the BTB/POZ domain of BAZF itself and that of Bcl6 (unpublished data).
The repressor activity of BAZF also associates with the BTB/POZ domain (from aa 1 to 122). Other zinc finger-type DNA-binding proteins with the BTB/POZ domain, including ZF5 (34) and PLZF (28), have transrepressor activity. This activity may depend on the cellular environment because the BTB/POZ domain of ZF5 is required to activate the human immunodeficiency virus type 1 long terminal repeat and to repress the β-actin promoter (23). Several DNA-binding proteins with the BTB/POZ domain form higher-order macromolecular complexes involved in chromatin folding (1). Since the corepressor SMRT binds to the BTB/POZ domain of Bcl6 (13) and PLZF (21), the repressor activity of Bcl6 and PLZF may be explained by remodeling of the chromatin structure. SMRT can form a ternary complex with mSin3A and HDAC-1 (31). The BTB/POZ domain of Bcl6 or PLZF may also associate with SMRT-mSin3A-HDAC-1 and form a multimeric repressor complex involving histone deacetylation activity. BAZF may also be involved in the formation of a complex, suggesting the mechanism of repressor activity in the BTB/POZ domain of BAZF.
A Drosophila zinc finger-type transcriptional factor, Krüppel, can both activate and repress gene expression in a concentration-dependent manner (40). A homodimer or a monomer of Krüppel is dominantly formed at a higher or lower concentration, respectively. Although the homodimer binds to the same DNA sequence as does the monomer, transcriptional activity of the homodimer is opposite to that of the monomer (41). Since BAZF can form a complex with BAZF and Bcl6 (Fig. 4), we examined the transcriptional activity of BAZF in various complexes in cotransfection experiments (Fig. 5B). The repressor activity was not modulated but instead increased in a concentration-dependent manner, thereby suggesting a functional similarity to Bcl6.
The inducibility of mRNA expression in lymphocytes is also similar for BAZF and Bcl6. Expression of BAZF is transiently upregulated in splenocytes stimulated with PMA plus ionomycin in the presence of cycloheximide (Fig. 6B), suggesting that BAZF, like Bcl6, plays a role in activated lymphocytes as an immediate-early gene. Since the amount of BAZF mRNA in splenocytes increased after cycloheximide treatment, the BAZF gene may be transcribed in those cells. These results also suggest that BAZF mRNA undergoes relatively rapid degradation compared with mRNAs from the constitutively expressed genes. The AUUUA sequence in lymphokine mRNAs in the 3′ untranslated region has been demonstrated to confer instability on mature mRNAs (29). The two AUUUA sequences (318 and 1,622 bases from the stop codon) in the 3′ untranslated region of BAZF (Fig. 1B) may contribute to the instability of this mRNA. This biochemical character of BAZF mRNA is also similar to that of Bcl6 mRNA (17).
The major difference between BAZF and Bcl6 is the tissue expression pattern. Expression of Bcl6 mRNA is ubiquitous in various mouse tissues (17), while BAZF mRNA was detected in lung and heart by Northern blotting (Fig. 6A). Furthermore, expression of BAZF mRNA was induced in a macrophage line after activation although that of Bcl6 remained at a constant level after activation (data not shown). Bcl-2 and Bcl-xL are equivalent in their potentials to inhibit cell death, but the patterns and levels of expression are not identical (22), suggesting that the difference between Bcl-2 and Bcl-xL reflects the need to tune in the control of apoptosis. BAZF and Bcl6 may also affect the control of gene expressions in a cell-type- and differentiation stage-specific manner. Since the nuclear expression pattern (diffuse and microgranular) of Bcl6 under physiological conditions (6) differed from that (punctate nuclear localization) under gene transfection conditions (16) (Fig. 4), the activity of BAZF and Bcl6 seen in gene transfection experiments may not reflect all the physiological functions of BAZF and Bcl6.
In summary, this is apparently the first report of a gene family whose members encode the zinc finger-type DNA-binding proteins carrying the BTB/POZ domain. We cloned a novel Bcl6-homologous gene (BAZF) that can specifically bind to the BS with zinc finger motifs and that functions as a transcriptional repressor. As the repressor activity associates with both the BTB/POZ domain and the middle portion of BAZF, the biochemical character of BAZF is similar to that of Bcl6.
ACKNOWLEDGMENTS
We thank M. Ptashne, K. Nakajima, and H. Nakano for gifts of plasmids. We also thank M. Maekawa for help with the micrographs, H. Satake for skillful technical assistance, N. Fujita for secretarial services, and M. Ohara for comments on the manuscript.
This work was supported in part by Grants-in-Aid for Cancer Research from the Ministry of Education, Science, Sports and Culture of Japan, special coordination funds for the promotion of science and technology from the Science and Technology Agency of Japan, grants from the Ministry of Health and Welfare of Japan, and a grant from the Japanese Cardiovascular Research Foundation.
REFERENCES
- 1.Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D. The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 1995;6:1193–1198. [PubMed] [Google Scholar]
- 2.Albagli O, Dhordain P, Bernardin F, Quief S, Kerckaert J P, Leprince D. Multiple domains participate in distance-independent LAZ3/BCL6-mediated transcriptional repression. Biochem Biophys Res Commun. 1996;220:911–915. doi: 10.1006/bbrc.1996.0505. [DOI] [PubMed] [Google Scholar]
- 3.Bardwell V J, Treisman R. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 4.Baron B W, Stanger R R, Hume E, Sadhu A, Mick R, Kerckaert J P, Deweindt C, Bastard C, Nucifora G, Zeleznik-Le N, McKeithan W. BCL6 encodes a sequence-specific DNA-binding protein. Genes Chromosomes Cancer. 1995;13:221–224. doi: 10.1002/gcc.2870130314. [DOI] [PubMed] [Google Scholar]
- 5.Bastard C, Tilly H, Lenormand B, Bigorgne C, Boulet D, Kunlin A, Monconduit M, Piguet H. Translocations involving band 3q27 and Ig gene regions in non-Hodgkin’s lymphoma. Blood. 1992;79:2527–2531. [PubMed] [Google Scholar]
- 6.Cattoretti G, Chang C C, Cechova K, Zhang J, Ye B H, Falini B, Louie D C, Offit K, Chaganti R S, Dalla-Favera R. BCL-6 protein is expressed in germinal-center B cells. Blood. 1995;86:45–53. [PubMed] [Google Scholar]
- 7.Chang C C, Ye B H, Chaganti R S, Dalla-Favera R. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc Natl Acad Sci USA. 1996;93:6947–6952. doi: 10.1073/pnas.93.14.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chardin P, Courtois G, Mattei M G, Gisselbrecht S. The KUP gene, located on human chromosome 14, encodes a protein with two distant zinc fingers. Nucleic Acids Res. 1991;19:1431–1436. doi: 10.1093/nar/19.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chin H, Nakamura N, Kamiyama R, Miyasaka N, Ihle J N, Miura O. Physical and functional interactions between Stat5 and the tyrosine-phosphorylated receptors erythropoietin and interleukin-3. Blood. 1996;88:4415–4425. [PubMed] [Google Scholar]
- 10.Dent A L, Shaffer A L, Yu X, Allman D, Staudt L M. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 11.Deweindt C, Albagli O, Bernardin F, Dhordain P, Quief S, Lantoine D, Kerckaert J P, Leprince D. The LAZ3/BCL6 oncogene encodes a sequence-specific transcriptional inhibitor: a novel function for the BTB/POZ domain as an autonomous repressing domain. Cell Growth Differ. 1995;6:1495–1503. [PubMed] [Google Scholar]
- 12.Dhordain P, Albagli O, Ansieau S, Koken M H, Deweindt C, Quief S, Lantoine D, Leutz A, Kerckaert J P, Leprince D. The BTB/POZ domain targets the LAZ3/BCL6 oncoprotein to nuclear dots and mediates homomerisation in vivo. Oncogene. 1995;11:2689–2697. [PubMed] [Google Scholar]
- 13.Dhordain P, Albagli O, Lin R J, Ansieau S, Quief S, Leutz A, Kerckaert J P, Evans R M, Leprince D. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc Natl Acad Sci USA. 1997;94:10762–10767. doi: 10.1073/pnas.94.20.10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorfman D M, Wilson D B, Bruns G A, Orkin S H. Human transcription factor GATA-2. Evidence for regulation of preproendothelin-1 gene expression in endothelial cells. J Biol Chem. 1992;267:1279–1285. [PubMed] [Google Scholar]
- 15.Evans R M, Hollenberg S M. Zinc fingers: gilt by association. Cell. 1988;52:1–3. doi: 10.1016/0092-8674(88)90522-3. [DOI] [PubMed] [Google Scholar]
- 16.Flenghi L, Ye B H, Fizzotti M, Bigerna B, Cattoretti G, Venturi S, Pacini R, Pileri S, Lo Coco F, Pescarmona E, Pelicci P G, Dalla-Favera R, Falini B. A specific monoclonal antibody (PG-B6) detects expression of the BCL-6 protein in germinal center B cells. Am J Pathol. 1995;147:405–411. [PMC free article] [PubMed] [Google Scholar]
- 17.Fukuda T, Miki T, Yoshida T, Hatano M, Ohashi K, Hirosawa S, Tokuhisa T. The murine BCL6 gene is induced in activated lymphocytes as an immediate early gene. Oncogene. 1995;11:1657–1663. [PubMed] [Google Scholar]
- 18.Fukuda T, Yoshida T, Okada S, Hatano M, Miki T, Ishibashi K, Okabe S, Koseki H, Hirosawa S, Taniguchi M, Miyasaka N, Tokuhisa T. Disruption of the Bcl6 gene results in an impaired germinal center formation. J Exp Med. 1997;186:439–448. doi: 10.1084/jem.186.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrison S D, Travers A A. The tramtrack gene encodes a Drosophila finger protein that interacts with the ftz transcriptional regulatory region and shows a novel embryonic expression pattern. EMBO J. 1990;9:207–216. doi: 10.1002/j.1460-2075.1990.tb08097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatano M, Roberts C W, Minden M, Crist W M, Korsmeyer S J. Deregulation of a homeobox gene, HOX11, by the t(10;14) in T cell leukemia. Science. 1991;253:79–82. doi: 10.1126/science.1676542. [DOI] [PubMed] [Google Scholar]
- 21.Hong S H, David G, Wong C W, Dejean A, Privalsky M L. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor alpha (RARalpha) and PLZF-RARalpha oncoproteins associated with acute promyelocytic leukemia. Proc Natl Acad Sci USA. 1997;94:9028–9033. doi: 10.1073/pnas.94.17.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang D C, Cory S, Strasser A. Bcl-2, Bcl-XL and adenovirus protein E1B19kD are functionally equivalent in their ability to inhibit cell death. Oncogene. 1997;14:405–414. doi: 10.1038/sj.onc.1200848. [DOI] [PubMed] [Google Scholar]
- 23.Kaplan J, Calame K. The ZiN/POZ domain of ZF5 is required for both transcriptional activation and repression. Nucleic Acids Res. 1997;25:1108–1116. doi: 10.1093/nar/25.6.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawamata N, Miki T, Ohashi K, Suzuki K, Fukuda T, Hirosawa S, Aoki N. Recognition DNA sequence of a novel putative transcription factor, BCL6. Biochem Biophys Res Commun. 1994;204:366–374. doi: 10.1006/bbrc.1994.2468. [DOI] [PubMed] [Google Scholar]
- 25.Kerckaert J P, Deweindt C, Tilly H, Quief S, Lecocq G, Bastard C. LAZ3, a novel zinc-finger encoding gene, is disrupted by recurring chromosome 3q27 translocations in human lymphomas. Nat Genet. 1993;5:66–70. doi: 10.1038/ng0993-66. [DOI] [PubMed] [Google Scholar]
- 26.Kopf M, Gros G L, Bachmann M, Lamers M C, Bluethmann H, Kohler G. Disruption of the murine IL-4 gene blocks Th2 cytokine responses. Nature. 1993;362:245–248. doi: 10.1038/362245a0. [DOI] [PubMed] [Google Scholar]
- 27.Lee M S, Gippert G P, Soman K V, Case D A, Wright P E. Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science. 1989;245:635–637. doi: 10.1126/science.2503871. [DOI] [PubMed] [Google Scholar]
- 28.Li J Y, English M A, Ball H J, Yeyati P L, Waxman S, Licht J D. Sequence-specific DNA binding and transcriptional regulation by the promyelocytic leukemia zinc finger protein. J Biol Chem. 1997;272:22447–22455. doi: 10.1074/jbc.272.36.22447. [DOI] [PubMed] [Google Scholar]
- 29.Lindsten T, June C H, Ledbetter J A, Stella G, Thompson C B. Regulation of lymphokine messenger RNA stability by a surface-mediated T cell activation pathway. Science. 1989;244:339–342. doi: 10.1126/science.2540528. [DOI] [PubMed] [Google Scholar]
- 30.Miki T, Kawamata N, Hirosawa S, Aoki N. Gene involved in the 3q27 translocation associated with B-cell lymphoma, BCL5, encodes a Krüppel-like zinc-finger protein. Blood. 1994;83:26–32. [PubMed] [Google Scholar]
- 31.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 32.Nakae K, Nakajima K, Inazawa J, Kitaoka T, Hirano T. ERM, a PEA3 subfamily of Ets transcription factors, can cooperate with c-Jun. J Biol Chem. 1995;270:23795–23800. doi: 10.1074/jbc.270.40.23795. [DOI] [PubMed] [Google Scholar]
- 33.Nakano H, Oshima H, Chung W, Williams-Abbott L, Ware C F, Yagita H, Okumura K. TRAF5, an activator of NF-κB and putative signal transducer for the lymphotoxin-β receptor. J Biol Chem. 1996;271:14661–14664. doi: 10.1074/jbc.271.25.14661. [DOI] [PubMed] [Google Scholar]
- 34.Numoto M, Niwa O, Kaplan J, Wong K K, Merrell K, Kamiya K, Yanagihara K, Calame K. Transcriptional repressor ZF5 identifies a new conserved domain in zinc finger proteins. Nucleic Acids Res. 1993;21:3767–3775. doi: 10.1093/nar/21.16.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Offit K, Jhanwar S, Ebrahim S, Filippa D, Clarkson B, Chaganti R. t(3;22)(q27;q11): a novel translocation associated with diffuse non-Hodgkin’s lymphoma. Blood. 1989;74:1876–1879. [PubMed] [Google Scholar]
- 36.Onizuka T, Moriyama M, Yamochi T, Kuroda T, Kazama A, Kanazawa N, Sato K, Kato T, Mori S. BCL-6 gene product, a 92- to 98-kD nuclear phosphoprotein, is highly expressed in germinal center B cells and their neoplastic counterparts. Blood. 1995;86:28–37. [PubMed] [Google Scholar]
- 37.Pavletich N P, Pabo C O. Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science. 1993;261:1701–1707. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]
- 38.Sadowski I, Ptashne M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Sauer F, Jackle H. Concentration-dependent transcriptional activation or repression by Krüppel from a single binding site. Nature. 1991;353:563–566. doi: 10.1038/353563a0. [DOI] [PubMed] [Google Scholar]
- 41.Sauer F, Jackle H. Dimerization and the control of transcription by Krüppel. Nature. 1993;364:454–457. doi: 10.1038/364454a0. [DOI] [PubMed] [Google Scholar]
- 42.Seyfert V L, Allman D, He Y, Staudt L M. Transcriptional repression by the proto-oncogene BCL-6. Oncogene. 1996;12:2331–2342. [PubMed] [Google Scholar]
- 43.Ye B H, Lista F, Lo Coco F, Knoeles R, Offit K, Chaganti R S K, Dalla-Favera R. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science. 1993;262:747–750. doi: 10.1126/science.8235596. [DOI] [PubMed] [Google Scholar]
- 44.Ye B H, Chaganti S, Chang C C, Niu H, Corradini P, Chaganti R S, Dalla-Favera R. Chromosomal translocations cause deregulated BCL6 expression by promoter substitution in B cell lymphoma. EMBO J. 1995;14:6209–6217. doi: 10.1002/j.1460-2075.1995.tb00311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye B H, Cattoretti G, Shen Q, Zhang J, Hawe N, de Waard R, Leung C, Nouri-Shirazi M, Orazi A, Chaganti R S, Rothman P, Stall A M, Pandolfi P P, Dalla-Favera R. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet. 1997;16:161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- 46.Yoshida T, Fukuda T, Okabe S, Hatano M, Miki T, Hirosawa S, Miyasaka N, Isono K, Tokuhisa T. The BCL6 gene is predominantly expressed in keratinocytes at their terminal differentiation stage. Biochem Biophys Res Commun. 1996;228:216–220. doi: 10.1006/bbrc.1996.1642. [DOI] [PubMed] [Google Scholar]
- 47.Zollman S, Godt D, Prive G G, Couderc J L, Laski F A. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc Natl Acad Sci USA. 1994;91:10717–10721. doi: 10.1073/pnas.91.22.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]