Abstract
Background
Inflammatory Bowel Disease (IBD)-associated arthritis is a frequent and potentially debilitating complication of IBD, that can affect those with or without active intestinal disease, and is often difficult to treat. The microbiome is known to play a role in IBD development and has been shown to be associated with inflammatory arthritis without concomitant IBD, but its role in IBD-associated arthritis is still unexplored. Further, disease localization is associated with development of IBD-associated arthritis, and stool compositional profiles are predictive of disease localization, yet mucosal location-specific microbiomes have not been well characterized. To address this gap in understanding, we designed a study (LOCATION-IBD) to characterize the mucosa-associated intestinal microbiome and metabolome in IBD-associated arthritis.
Methods
Adults with an established diagnosis of IBD undergoing clinical colonoscopy between May of 2021 and February of 2023 were invited to participate in this study; those interested in participation who met inclusion criteria were enrolled. Prior to enrollment, participants were stratified into those with or without IBD-associated arthritis. All participants were interviewed and had clinical and demographic data collected, and 97.8% completed clinical colonoscopy with biopsy collection.
Results and conclusion
A total of 182 participants, 53 with confirmed IBD-associated arthritis, were enrolled in this study, resulting in 1151 biopsies obtained for microbiome and metabolome analysis (median 6, mean 6.3 per participant). Clinical and demographic data obtained from the study population will be analyzed with microbiome and metabolome data obtained from biopsies, with the goal of better understanding the mechanisms underpinning the host-microbiome relationship associated the development of IBD-associated arthritis.
Keywords: Inflammatory bowel disease, IBD-Associated arthritis, Spondylarthritis, Microbiome, Metabolome
1. Introduction
Inflammatory Bowel Disease (IBD) is an umbrella term for an increasingly prevalent set of diseases causing inflammation of the gastrointestinal tract for upwards of 3 million people in the United States [1,2], and millions of others around the world. IBD-associated arthritis, the most common extra-intestinal manifestation (EIM) of IBD, is estimated to affect 25–40% of individuals with IBD [3,4]. While treatment of intestinal symptoms is often sufficient to prevent EIM occurrence or recurrence, many individuals still experience EIMs and particularly IBD-associated arthritis, even when gut disease is well-controlled [5]. Little is known about the underlying etiology of IBD-associated arthritis, though it is known to be associated with certain phenotypes of IBD, including colon involvement in Crohn's disease (CD) and extensive/pancolitis in ulcerative colitis (UC) [4,6,7].
Differences in stool bacterial composition are reported in IBD compared to the general population, and are also associated with various phenotypes of both CD and UC, specifically colonic involvement in CD and extensive or pancolitis in UC [[8], [9], [10]]. The intestinal bacterial community composition is also altered in those with inflammatory arthritis [11], including spondylarthritis in those without concomitant IBD [12]. Stool compositional changes are observable early in disease [13], and in a few case studies and murine studies, were detected prior to disease onset [9,14,15]. While many of these compositional changes between IBD and inflammatory arthritis overlap [16], there are differences that suggest potentially distinct microbial etiologies.
In spite of the spatial heterogeneity of the microbiome in the intestinal tract [[17], [18], [19], [20], [21]], and the association between disease localization and certain microbial profiles [22], little has been done to characterize the location-specific mucosal microbiome in IBD. Across the intestinal tract, both microbial density and composition vary [17]. This is important as intestinal location involvement is associated with IBD-associated arthritis and variation in one location of the GI tract may not be predictive of variation in other locations. Stool sampling provides a composite estimate of bacteria in the gut; however, beta diversity of bacterial composition in mucosal versus stool and even luminal samples from the same patient is generally high [18], and this discrepancy is increased in patients with IBD [[19], [20], [21]].
Examples of the problems this can pose can be seen in contradictory results sometimes observed in mucosal and stool studies, particularly in some bacteria of interest. Dialister spp have been consistently associated with both isolated IBD and non-IBD associated spondyloarthritis in mucosal samples [23,24], while stool samples have provided more contradictory results, with stool from spondyloarthritis samples alone showing increases [25,26], and those from patients with IBD alone displaying decreases in abundance associated with disease and disease activity [27,28]. Lachnospiraceae genera decreased with prolonged spondyloarthritis disease duration in stool [29], and increased in stool from patients with IBD following successful treatment with adalimumab [30], while gut-adherent Lachnospiraceae have been observed to be increased in PSC-IBD versus IBD alone [31], again suggesting that for bacteria of interest, stool versus mucosal levels may result in discordant conclusions. Given that mucosally adherent bacteria that are physically closer to the intestinal barrier are likelier to impart impact both via metabolic output and triggering of the immune system, and that shedding occurs at different rates in different parts of the colon, mucosal assessment becomes more important when attempting to parse out specific mechanisms by which bacteria are directly contributing to pathogenesis of disease. Examples of metabolites that may be playing an overlapping role include short-chain fatty acids, which have been consistently demonstrated to be decreased in IBD [32], and for which supplementation led to amelioration of both IBD and spondyloarthritis symptoms in animal models [33,34]. Decreased tryptophan production has been observed in both IBD and spondyloarthritis [[35], [36], [37]].
These observations support the hypothesis that the intestinal bacteria play a role in spondyloarthritis, and specifically IBD-associated arthritis, and that the role of the microbiome in IBD-associated arthritis is distinct from its role in IBD alone; to date, however, no studies have been performed to assess the association between the microbiome and IBD-associated arthritis. Further, no research has studied these relationships in a location specific manner at the mucosal level. The goal of our study is to identify microbes that are associated with and predictive of IBD-associated arthritis by analyzing the biopsy-associated intestinal microbiome of IBD patients both with and without IBD-associated arthritis (LOCATION-IBD). Further, we aim to perform multi-omic characterization of the mucosal-associated metabolome in the same patients, with the goal of identifying metabolites that may play a role in IBD-associated arthritis. We hypothesize that a subset of microbes that differs between our groups with vs without IBD-associated arthritis are responsible for the production of identifiable metabolites that lead to this EIM. The purpose of this manuscript is to describe the protocol for enrollment, biopsy collection, and the participants characteristics that will be used to answer these questions.
2. Methods
2.1. Patient identification, recruitment, and enrollment
Adult (18 years or older) patients with a previously established diagnosis of IBD at the University of Maryland School of Medicine or Baltimore Veterans Affairs Medical Center IBD clinics who were scheduled for clinical colonoscopy were considered for recruitment. Patients who met inclusion criteria (Table 1) based on chart review were sent a recruitment letter prior to their scheduled procedure. At least 3 days after the letter was mailed, and within one week of their colonoscopy, participants were contacted via telephone to discuss their interest in participating. Those interested in participating were screened using the eligibility checklist (Table 1), and if eligible, were verbally consented and enrolled over the phone which included a discussion of study procedures, risk/benefits of participation, and confirmation of participant understanding. On the day of the colonoscopy, participants reconfirmed interest in participation, were invited to ask any additional questions regarding the study, and written consent was obtained. Patients who could not be reached prior to the procedure were approached at the endoscopy center on the day of the procedure and underwent a similar consent process which included discussion of the study procedures, risk/benefits of participation, and confirmation of understanding prior to attaining written consent. It was emphasized to all participants that participation was voluntary and choosing not to participate would have no impact on the care they received.
Table 1.
Inclusion and exclusion criteria used to screen participants.
| Inclusion Criteria | Exclusion Criteria |
|---|---|
| A previously established diagnosis of IBD | Active hepatobiliary disease (including primary sclerosing cholangitis) |
| Clinically scheduled colonoscopy | Intravenous antibiotic use or oral consumption of antibiotics within the prior 4 weeks |
| 18 years of age or older | Chronic musculoskeletal pain from osteoarthritis or fibromyalgia |
| A history of diverticulitis | |
| A history of non IBD-associated arthritis EIM without a concurrent history of IBD-associated arthritis | |
| Current therapeutic anticoagulation, history of thrombocytopenia, or coagulopathy (platelets <50,000, INR >2, or PTT>50); or previous biopsy complications (e.g., severe bleeding) |
2.2. Joint pain classification
Participants were assigned to the IBD-associated arthritis or non-IBD-associated arthritis groups through adjudication by a board-certified rheumatologist. IBD-associated arthritis was defined as axial or peripheral joint pain affecting at least one joint with swelling, or without joint swelling associated with significant morning stiffness that improves with exercise or responds to underlying IBD treatment. Because Assessment of Spondylo-Arthritis international Society (ASAS) criteria for peripheral arthritis are fulfilled via a concurrent diagnosis of IBD with arthritis, enthesitis, and/or dactylitis, all patients with peripheral arthritis met criteria for peripheral spondyloarthropathy [38]. Axial involvement required radiographic (X-ray) or non-radiographic (MRI) imaging evidence of either spondylitis and/or isolated sacroiliitis, which together with diagnosis of IBD met ASAS criteria for axial spondylarthritis [39,40]. Information regarding enthesitis and dactylitis could not be collected based on the available data; however, data on concomitant psoriasis was collected. After participant interview and prior to group stratification, participants with any history of IBD-associated arthritis, including those with peripheral mono and oligo-arthritis, sacroiliitis, and ankylosing spondylitis were screened by a rheumatologist to ensure eligibility criteria were met. Information regarding history of which and how many joints were affected, and current disease activity (whether active or inactive), were collected.
2.3. Baseline demographics and clinical data collection
Information regarding: IBD phenotype and location (inflammatory, stricturing, or fistulizing behavior, perianal involvement, upper-tract involvement, and ileal, colonic, or ileocolonic location for CD; extent of disease (i.e., proctitis, left-sided disease, or pancolitis), for UC); duration of disease; age at diagnosis; and IBD related surgical history was collected via both chart review and in the form of a HIPAA-compliant REDCap questionnaire. Clinical data such as demographics, medical history, physical exam findings, social history including alcohol consumption, smoking status, and diet, laboratory panel results, and concomitant medication usage was additionally collected both from participant interview and the electronic medical record. Regarding use of advanced therapies, the majority of patients in our cohort were on a biologic, with only three classes of biologics used, allowing us to control for biologic use in analyses. Nearly all patients with IBD-associated arthritis are on a biologic, and thus excluding this population would not be feasible; however, given that the majority of the non-IBD-associated arthritis participants were also on a biologic, they act as a reasonable control, and prevent this from being a confounding variable. Demographic information along with height, weight, blood pressure, and temperature were collected on the day of each procedure.
2.4. Sample collection and storage
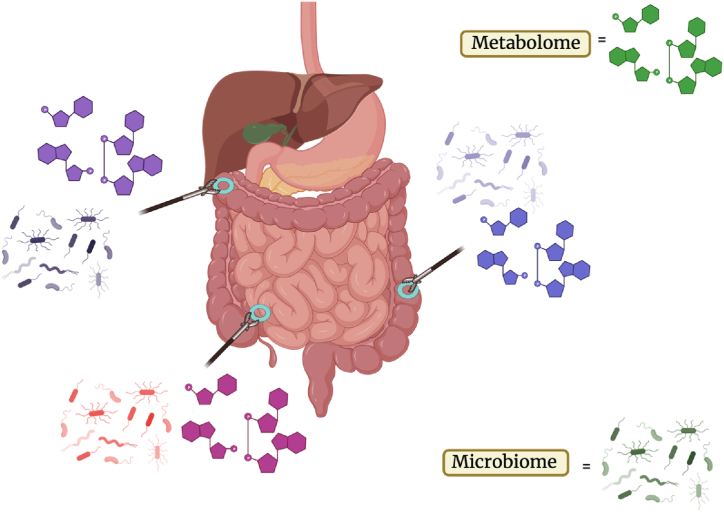
During clinically indicated colonoscopy, 6 to 12 intestinal biopsies were taken - two from each of the following sites to localize biological signals: terminal ileum, ascending/transverse colon (hepatic flexure), and sigmoid colon/rectum (Fig. 1). When applicable, biopsies were taken from inflamed tissue and adjacent, healthy-appearing mucosa within 5–10 cm of inflamed tissue; to minimize risk, biopsies were taken only if deemed safe by an experienced endoscopist. The safety of obtaining multiple endoscopic biopsies for research was previously reported in an NIH series; a mean of 38.2 biopsies were obtained per endoscopy without complication [41]. Each biopsy was taken in one pass, and samples were placed either into an empty tube for metabolomic studies or a tube with a nucleic acid protective agent (DNA/RNA Shield) for metagenomic sequencing. Within 5 h of being taken, biopsy specimens, which had been kept on ice, were transferred to storage in a −80 °C freezer until processing.
Fig. 1.
Localization of biopsy sample collection. One biopsy was taken for microbiome and metabolome sampling at each site (for a total of two biopsies/site). Created with Biorender.com.
2.5. Sample size calculation
We anticipated collection of approximately 600 uniquely localized biopsy samples (2 per site for a total of 1200 biopsies) in 200 participants, with the goal of having at least 480 and 120 unique samples for metabolome and microbiome analyses total per group for IBD patients without vs with arthritis. We calculated this for a minimum of 30% of the participants in each IBD group having either UC or CD (i.e., at least 30% and no more than 70% in each group will have one disease or the other), assuming only minimum biopsy sample requirements were met. For a cross sectional study design such as this with an approximately 4:1:4:1 of UC:UC/EIM:CD:CD/EIM (80:20:80:20; a minimum of 40 with EIMs, 20 in each group), and maintaining an alpha value of no more than 5% with a statistical power of at least 80%, and based on previously published results [42], we estimated that in each of the four groups (where each IBD group can be split into UC vs CD, with and without EIMs) we would require between 100 and 150 unique samples per group to have a sufficient sample for describing both alpha and beta diversity between the groups at 80% power. The higher than anticipated rate of IBD-associated arthritis in our study population allowed us to meet our recruitment goal early.
2.6. Post colonoscopy research questionnaire
A HIPAA compliant REDCap questionnaire (Supplement 1) was administered and included questions about: disease type, phenotype, and severity; number of surgeries; arthropathy, when applicable; diet; stress levels; Manitoba index and IBD symptoms; and medications.
2.7. Proposed analytical plan
Microbiome composition data will be generated using DNA extracted from frozen tissue biopsy samples suspended in the nucleic acid protective solution using the NEB Monarch Genomic DNA Purification Kit, and bacterial DNA will be enriched using the NEB Next Microbiome DNA Enrichment Kit. Samples will undergo 16S rRNA V3V4 amplicon sequencing on the NextSeq 1000. The frozen biopsy tissues will be shipped on dry ice, and metabolite characterization will be performed utilizing coupled liquid chromatography (LC) and gas chromatography (GC) to mass spectrometry (MS) for increased resolution and confidence in metabolite identification. These analyses will be performed using a validated commercial metabolomics technology platform that adheres to industry standards. A variety of metrics will be applied to ascertain relative differences in bacterial composition across samples both among and between participants, such as Shannon index, Jensen-Shannon Divergence (JSD), Yue-Clayton (YC) coefficient of similarity, among others; these metrics will be used to ensure that different aspects of compositional differences are captured. Sparse canonical correlation analysis will be used to integrate various types of data by identifying axes of covariance between data types. The results of this analysis will offer insight into which sets of microbes and metabolites have associations, which can be incorporated into developing both uni- and multivariate models to characterize the relationship between the microbiome and the metabolome, clinical data, and presence of EIMs across IBD phenotypes (as a categorical variable), while controlling for factors like gender and age.
2.8. Ethics statement
This study was approved by the University of Maryland, Baltimore Institutional Review Board (HP-00094082) on March 18th, 2021.
3. Results and Conclusion
3.1. Results
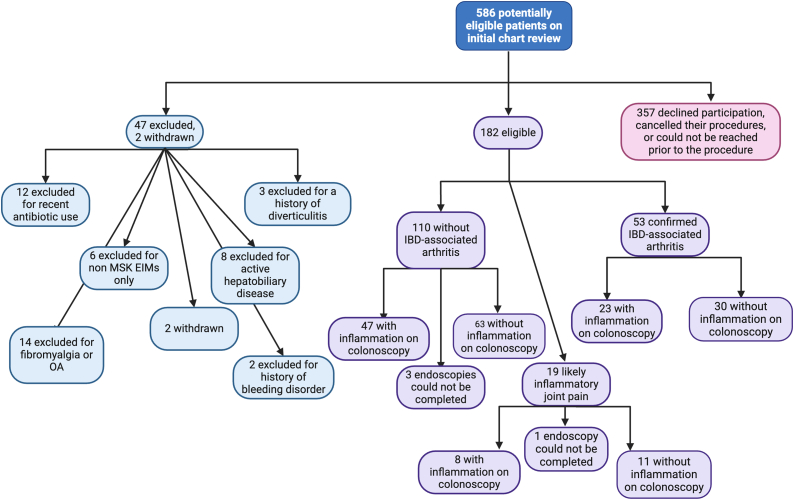
Through chart review, 586 patients were identified as potentially eligible and mailed a recruitment letter. Upon patient interview, some individuals were deemed ineligible due to a history of diverticulitis (n = 3), recent systemic antibiotic use (n = 12), a history of non-musculoskeletal EIM without concurrent IBD-associated arthritis (n = 6), active hepatobiliary disease (n = 8), active joint pain due to fibromyalgia or osteoarthritis (n = 14), or a history of bleeding disorder (n = 2); 357 patients declined participation, rescheduled or cancelled their colonoscopy, or could not be reached prior to the procedure. Of the remaining 184 participants, two were withdrawn after enrollment due to screen failure (recent antibiotic use), leaving a final study population of 182 participants, 52 with confirmed IBD-associated arthritis, and 19 with potential IBD-associated arthritis (Fig. 2). A total of 1151 biopsies were collected (one metabolome terminal ileum sample was not collected), 134 from inflamed tissue and 1017 from healthy tissue, with a proportional distribution between the IBD-associated arthritis and non-IBD-associated arthritis groups as shown in Fig. 2.
Fig. 2.
Participant screening, recruitment, and enrollment. Created with Biorender.com.
Table 2 contains a description of participant demographic information. The mean age of participants was 41.8 years (IQR: 31,52), with an age range of 19–77 years. Participants were predominantly White (75.3%) or Black (19.2%), with fewer participants identifying as Asian (4.4%) or another race (1.1%). Nearly half of participants (48.9%) identified as female. Biopsies were obtained from most participants (97.8%) but could not be obtained from 4 participants due to inadequate bowel preparation; inflammation was observed in 42.9% of colonoscopies.
Table 2.
Demographic characteristics of the study population.
| Total group (n = 182) | CD (n = 123) | UC (n = 49) | IC (n = 10) | |
|---|---|---|---|---|
| Age (mean, IQR) | 41.9 (31,52) | 40.3 (30.0,49.5) | 44.2 (34.0,54.0) | 50.1 (37.3,68.5) |
| (range) | (19,77) | (19,71) | (21,77) | (23,73) |
| Race | ||||
|
136 (74.7%) | 91 (74.0%) | 36 (73.5%) | 9 (90.0%) |
|
34 (18.7%) | 24 (19.5%) | 9 (18.4%) | 1 (10.0%) |
|
8 (4.4%) | 5 (4.1%) | 3 (6.1%) | 0 (0%) |
|
4 (1.1%) | 3 (2.4%) | 1 (2.0%) | 0 (0%) |
| Sex (female) | 88 (48.4%) | 62 (50.4%) | 21 (42.9%) | 5 (50.0%) |
| Gender (female) | 89 (48.9%) | 62 (50.4%) | 22 (44.9%) | 5 (50.0%) |
| Completed colonoscopy | 178 (97.8%) | 120 (97.6%) | 48 (98%) | 10 (100%) |
| Inflammation on colonoscopy | 78 (42.9%) | 55 (44.8%) | 21 (42.9%) | 1 (10%) |
| IBD-associated arthritis | ||||
|
53 (29.1%) | 41 (33.3%) | 7 (14.3%) | 5 (50.0%) |
|
110 (60.4%) | 68 (55.3%) | 38 (77.6%) | 4 (40.0%) |
|
19 (10.4%) | 14 (11.4%) | 4 (8.2%) | 1 (10.0%) |
3.2. Conclusion
Here, we describe the LOCATION-IBD study population at the center of our study aimed at exploring the role of the mucosal intestinal microbiome in IBD-associated arthritis using an integrative multi-omics approach. Analysis of the data collected in this study will characterize associations between microbes and metabolites, and IBD-associated arthritis, allowing us to better elucidate the mechanistic underpinnings of host-microbiome relationships using a localized approach. The combining of large clinical datasets with biopsy-based omics data is critical to answering these questions, particularly given the importance of disease localization in severity of disease, likelihood of complications, and development of EIMs. This work should lead to the identification of actionable targets that could be tested in the laboratory setting. Further, the use of mucosal samples in characterization of these relationships allows us to address spatial heterogeneity in composition throughout the colon, which is likely important given the association between localization of disease and development of EIMs. IBD-associated arthritis affects a large proportion of patients with IBD, and can drastically negatively impact quality of life, yet there remains a limited number of therapeutic options that adequately address both intestinal symptoms and EIMs [43]. Samples and data collected using this protocol are being and will continue to be analyzed with the eventual goal of identifying diagnostic and therapeutic targets, using both clinical and multi-omic data.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. MA was supported by the National Institutes of Diabetes and Digestive and Kidney Diseases, of the National Institutes of Health, under award number T32DK067872; the sponsor had no role in the design, execution, or interpretation of this study, nor in the writing of this report or decision to submit the article for publication. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
CRediT authorship contribution statement
Madeline Alizadeh: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Visualization, Writing – original draft, Writing – review & editing. Uni Wong: Conceptualization, Investigation, Methodology, Project administration, Writing – review & editing. Bernadette C. Siaton: Investigation, Writing – review & editing. Seema A. Patil: Writing – review & editing. Lauren George: Writing – review & editing. Jean-Pierre Raufman: Writing – review & editing, Investigation. William H. Scott: Writing – review & editing. Erik C. von Rosenvinge: Conceptualization, Investigation, Project administration, Writing – review & editing. Jacques Ravel: Conceptualization, Funding acquisition, Investigation, Methodology, Resources, Writing – original draft, Writing – review & editing. Raymond K. Cross: Investigation, Methodology, Project administration, Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.The authors have no financial interests/personal relationships which may be considered as potential competing interests.
Acknowledgments
We would like to thank the participants for their time and enthusiastic participation in this study. We would also like to thank Danielle Brown, Abigail Noyes, Samia Siddiqui, and Ethel Wiggleton for their help in ensuring a smooth recruitment process.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e26571.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Dahlhamer J.M., et al. MMWR Morb Mortal Wkly Rep; 2016. Prevalence of Inflammatory Bowel Disease Among Adults Aged ≥18 Years — United States, 2015. [DOI] [PubMed] [Google Scholar]
- 2.Xu F., et al. Trends and factors associated with Hospitalization costs for inflammatory bowel disease in the United States. Appl. Health Econ. Health Pol. 2019;17(1):77–91. doi: 10.1007/s40258-018-0432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine J.S., Burakoff R. Extraintestinal manifestations of inflammatory bowel disease. Gastroenterol. Hepatol. 2011;7(4):235–241. [PMC free article] [PubMed] [Google Scholar]
- 4.Vavricka S.R., et al. Extraintestinal manifestations of inflammatory bowel disease. Inflamm. Bowel Dis. 2015;21(8):1982–1992. doi: 10.1097/MIB.0000000000000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greuter T., et al. Emerging treatment options for extraintestinal manifestations in IBD. Gut. 2021;70(4):796–802. doi: 10.1136/gutjnl-2020-322129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vavricka S.R., et al. Frequency and risk factors for extraintestinal manifestations in the Swiss inflammatory bowel disease cohort. Am. J. Gastroenterol. 2011;106(1):110–119. doi: 10.1038/ajg.2010.343. [DOI] [PubMed] [Google Scholar]
- 7.Schorr-Lesnick B., Brandt L.J. Selected rheumatologic and dermatologic manifestations of inflammatory bowel disease. Am. J. Gastroenterol. 1988;83(3):216–223. [PubMed] [Google Scholar]
- 8.Ni J., et al. Gut microbiota and IBD: causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 2017;14(10):573–584. doi: 10.1038/nrgastro.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gevers D., et al. The treatment-Naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15(3):382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morgan X.C., et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13(9):R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaahtovuo J., et al. Fecal microbiota in early rheumatoid arthritis. J. Rheumatol. 2008;35(8):1500–1505. [PubMed] [Google Scholar]
- 12.Jethwa H., Abraham S. The evidence for microbiome manipulation in inflammatory arthritis. Rheumatology. 2017;56(9):1452–1460. doi: 10.1093/rheumatology/kew374. [DOI] [PubMed] [Google Scholar]
- 13.El Mouzan M.I., et al. Microbiota profile in new-onset pediatric Crohn's disease: data from a non-Western population. Gut Pathog. 2018;10(1):49. doi: 10.1186/s13099-018-0276-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomez A., et al. Loss of sex and age driven differences in the gut microbiome characterize arthritis-susceptible 0401 mice but not arthritis-resistant 0402 mice. PLoS One. 2012;7(4) doi: 10.1371/journal.pone.0036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobranowski P.A., et al. Compositional changes to the ileal microbiome precede the onset of spontaneous ileitis in SHIP deficient mice. Gut Microb. 2019;10(5):578–598. doi: 10.1080/19490976.2018.1560767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salem F., et al. Gut microbiome in chronic rheumatic and inflammatory bowel diseases: similarities and differences. United European Gastroenterol J. 2019;7(8):1008–1032. doi: 10.1177/2050640619867555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z., et al. Spatial heterogeneity and co-occurrence patterns of human mucosal-associated intestinal microbiota. Isme j. 2014;8(4):881–893. doi: 10.1038/ismej.2013.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bashir M., et al. Effects of high doses of vitamin D3 on mucosa-associated gut microbiome vary between regions of the human gastrointestinal tract. Eur. J. Nutr. 2016;55(4):1479–1489. doi: 10.1007/s00394-015-0966-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lo Presti A., et al. Fecal and mucosal microbiota profiling in irritable bowel syndrome and inflammatory bowel disease. Front. Microbiol. 2019;10(1655) doi: 10.3389/fmicb.2019.01655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prast-Nielsen S., et al. Investigation of the skin microbiome: swabs vs. biopsies. Br. J. Dermatol. 2019;181(3):572–579. doi: 10.1111/bjd.17691. [DOI] [PubMed] [Google Scholar]
- 21.Shobar R.M., et al. The effects of bowel preparation on microbiota-related metrics differ in Health and in inflammatory bowel disease and for the mucosal and luminal microbiota compartments. Clin. Transl. Gastroenterol. 2016;7(2):e143. doi: 10.1038/ctg.2015.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willing B.P., et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139(6):1844–1854.e1. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 23.Tito R.Y., et al. Brief report: dialister as a microbial marker of disease activity in spondyloarthritis. Arthritis Rheumatol. 2017;69(1):114–121. doi: 10.1002/art.39802. [DOI] [PubMed] [Google Scholar]
- 24.Čipčić Paljetak H., et al. Gut microbiota in mucosa and feces of newly diagnosed, treatment-naïve adult inflammatory bowel disease and irritable bowel syndrome patients. Gut Microb. 2022;14(1) doi: 10.1080/19490976.2022.2083419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manasson J., et al. Gut microbiota perturbations in reactive arthritis and postinfectious spondyloarthritis. Arthritis Rheumatol. 2018;70(2):242–254. doi: 10.1002/art.40359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L., et al. Gut microbiota changes in patients with spondyloarthritis: a systematic review. Semin. Arthritis Rheum. 2022;52 doi: 10.1016/j.semarthrit.2021.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Taylor H., et al. Multiomic features associated with mucosal healing and inflammation in paediatric Crohn's disease. Aliment. Pharmacol. Ther. 2020;52(9):1491–1502. doi: 10.1111/apt.16086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marie J., et al. Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut. 2011;60(5):631. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 29.Marie V., et al. Characterisation of gut microbiota composition in patients with axial spondyloarthritis and its modulation by TNF inhibitor treatment. RMD Open. 2023;9(1) doi: 10.1136/rmdopen-2022-002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ribaldone D.G., et al. Adalimumab therapy improves intestinal dysbiosis in Crohn's disease. J. Clin. Med. 2019;8(10):1646. doi: 10.3390/jcm8101646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quraishi M.N., et al. The gut-adherent microbiota of PSC-IBD is distinct to that of IBD. Gut. 2017;66(2):386–388. doi: 10.1136/gutjnl-2016-311915. [DOI] [PubMed] [Google Scholar]
- 32.Lloyd-Price J., et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569(7758):655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asquith M., et al. Intestinal metabolites are profoundly altered in the context of HLA–B27 expression and functionally modulate disease in a rat model of spondyloarthritis. Arthritis Rheumatol. 2017;69(10):1984–1995. doi: 10.1002/art.40183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russo E., et al. Immunomodulating activity and therapeutic effects of short chain fatty acids and tryptophan post-biotics in inflammatory bowel disease. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.02754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoll M.L., et al. Fecal metabolomics in pediatric spondyloarthritis implicate decreased metabolic diversity and altered tryptophan metabolism as pathogenic factors. Gene Immun. 2016;17(7):400–405. doi: 10.1038/gene.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berlinberg A.J., et al. Multi ‘omics analysis of intestinal tissue in ankylosing spondylitis identifies alterations in the tryptophan metabolism pathway. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.587119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S., et al. The involvement of intestinal tryptophan metabolism in inflammatory bowel disease identified by a meta-analysis of the transcriptome and a systematic review of the metabolome. Nutrients. 2023;15(13):2886. doi: 10.3390/nu15132886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rudwaleit M., et al. The Assessment of SpondyloArthritis international Society classification criteria for peripheral spondyloarthritis and for spondyloarthritis in general. Ann. Rheum. Dis. 2011;70(1):25. doi: 10.1136/ard.2010.133645. [DOI] [PubMed] [Google Scholar]
- 39.Rudwaleit M., et al. The development of Assessment of SpondyloArthritis international Society classification criteria for axial spondyloarthritis (part II): validation and final selection. Ann. Rheum. Dis. 2009;68(6):777. doi: 10.1136/ard.2009.108233. [DOI] [PubMed] [Google Scholar]
- 40.Rudwaleit M., et al. The development of Assessment of SpondyloArthritis international Society classification criteria for axial spondyloarthritis (part I): classification of paper patients by expert opinion including uncertainty appraisal. Ann. Rheum. Dis. 2009;68(6):770. doi: 10.1136/ard.2009.108217. [DOI] [PubMed] [Google Scholar]
- 41.Yao M.D., et al. Multiple endoscopic biopsies in research subjects: safety results from a National Institutes of Health series. Gastrointest. Endosc. 2009;69(4):906–910. doi: 10.1016/j.gie.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casals-Pascual C., et al. Microbial diversity in clinical microbiome studies: sample size and statistical power considerations. Gastroenterology. 2020;158(6):1524–1528. doi: 10.1053/j.gastro.2019.11.305. [DOI] [PubMed] [Google Scholar]
- 43.Fragoulis G.E., et al. Inflammatory bowel diseases and spondyloarthropathies: from pathogenesis to treatment. World J. Gastroenterol. 2019;25(18):2162–2176. doi: 10.3748/wjg.v25.i18.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.