ABSTRACT
Since 2016, in Colombia, ongoing transmission of Candida auris has been reported in multiple cities. Here, we provide an updated description of C. auris genomic epidemiology and the dynamics of antifungal resistance in Colombia. We sequenced 99 isolates from C. auris cases with collection dates ranging from June 2016 to January 2021; the resulting sequences coupled with 103 previously generated sequences from C. auris cases were described in a phylogenetic analysis. All C. auris cases were clade IV. Of the 182 isolates with antifungal susceptibility data, 67 (37%) were resistant to fluconazole, and 39 (21%) were resistant to amphotericin B. Isolates predominately clustered by country except for 16 isolates from Bogotá, Colombia, which grouped with isolates from Venezuela. The largest cluster (N = 166 isolates) contained two subgroups. The first subgroup contained 26 isolates, mainly from César; of these, 85% (N = 22) were resistant to fluconazole. The second subgroup consisted of 47 isolates from the north coast; of these, 81% (N = 38) were resistant to amphotericin B. Mutations in the ERG11 and TAC1B genes were identified in fluconazole-resistant isolates. This work describes molecular mechanisms associated with C. auris antifungal resistance in Colombia. Overall, C. auris cases from different geographic locations in Colombia exhibited high genetic relatedness, suggesting continued transmission between cities since 2016. These findings also suggest a lack of or minimal introductions of different clades of C. auris into Colombia.
IMPORTANCE
Candida auris is an emerging fungus that presents a serious global health threat and has caused multiple outbreaks in Colombia. This work discusses the likelihood of introductions and local transmission of C. auris and provides an updated description of the molecular mechanisms associated with antifungal resistance in Colombia. Efforts like this provide information about the evolving C. auris burden that could help guide public health strategies to control C. auris spread.
KEYWORDS: Candida auris, antifungal, resistance, genomics, WGS, epidemiology
INTRODUCTION
Candida auris is a multidrug-resistant yeast capable of causing outbreaks of infections associated with high mortality rates (1). Genomic analysis of C. auris infections from global regions showed six major clades (I–VI) (2). In Colombia, genomic sequencing revealed that all C. auris cases were clade IV and suggested ongoing transmission of C. auris in multiple cities (3).
Regarding antifungal susceptibility, about 35% of C. auris isolates were resistant to fluconazole and 33% were resistant to amphotericin B (4), mainly from the north-coast region of Colombia (3). Molecular mechanisms associated with azole resistance often involve mutations in the ERG11 and TAC1B genes. For polyenes, molecular mechanisms conferring resistance are largely unknown; a previous report identified two non-synonymous substitutions related with high MIC values of amphotericin B (3).
In this study, we investigated genomic epidemiology of C. auris cases from 2016 to 2021 in Colombia to obtain an updated description of C. auris transmission dynamics and molecular characterization of antifungal resistance.
MATERIALS AND METHODS
In 2016, the Colombian Instituto Nacional de Salud (INS) published the National Alert (5), which requested that public health laboratories to send all suspected or confirmed C. auris isolates to the INS.
Additional Materials and Methods are described in the Supplemental Material. Briefly, the U.S. Centers for Disease Control and Prevention received 99 C. auris isolates and performed species identification, antifungal susceptibility testing (AFST), and whole-genome sequencing (WGS; BioProject PRJNA1003896; Tables S1 and S2). WGS data were used to generate a phylogenetic tree and pairwise distance comparisons and to identify mutations related to antifungal drug resistance.
RESULTS
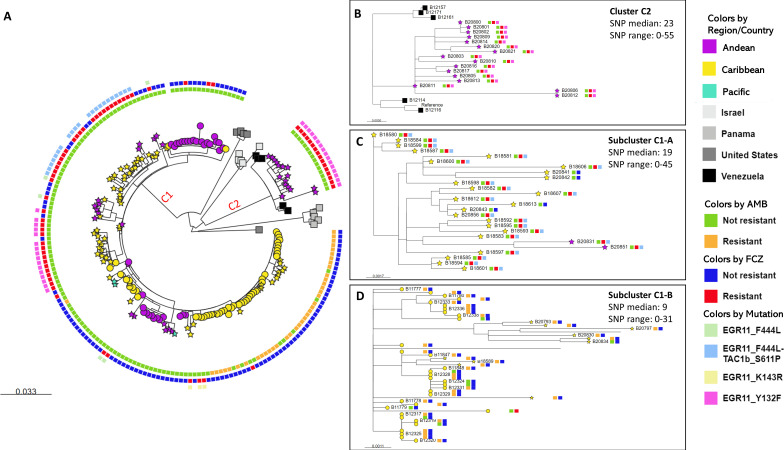
Phylogenetic analysis of 99 newly and 83 previously generated sequences revealed that all C. auris in Colombia were clade IV (maximum single nucleotide polomorphims [SNPs] difference to the reference sequence: 205 SNPs). Isolates predominately clustered by country. Colombian isolates were dispersed in two clusters (C1 and C2). Most Colombian isolates (n = 166, 91%) clustered with the previously described Colombian isolates in cluster C1 with collection dates from 2015 to 2016 (Fig. 1A).
Fig 1.
(A) Phylogenetic tree using maximum likelihood method; the majority of isolates clustered by country. Cluster C1 consists of Colombian isolates, and cluster C2 consists of Venezuelan isolates and fluconazole-resistant Colombian isolates. Taxa color represents country or Colombian region: Andean (Antioquia, Bogotá-Cundinamarca, Norte de Santander, Huila), Caribbean (Atlántico, Bolívar, Cesar, Magdalena), and Pacific (Nariño, Valle); isolates from countries other than Colombia are shown as squares. Colombian isolates sequenced in previous studies are shown as circles, and Colombian isolates sequenced in this study are shown as stars. External circle colors represent resistant and no resistant phenotypes for fluconazole and amphotericin B; presence of mutation in EGR11 and TACb1 genes. (B) Zoom-in view of cluster C2. (C) Zoom-in view of subcluster C1-A, which contains mainly fluconazole-resistant isolates. (D) Zoom-in view of subcluster C1-B, which contains mainly amphotericin B-resistant isolates. For panels B, C, and D, the squares to the right of the label represent resistant and non-resistant phenotypes and mutation in EGR11 and TACb1 genes.
Cluster C2 was separated by 152 SNPs from cluster C1. Cluster C2 comprised 16 isolates from Bogotá collected between May and November 2020 and five isolates from Venezuela collected between 2012 and 2015 (Fig. 1A). All Colombian isolates in this cluster and two Venezuelan isolates were fluconazole-resistant (Fig. 1B; Table S1). Within cluster C1, we highlighted two subclusters (C1-A and C1-B) of isolates with bootstrap values >98% and with high (>80%) proportion of antifungal-resistant isolates (Fig.1C through D; Fig. S2).
Among all 182 Colombian isolates, 67 (37%) were resistant to fluconazole, 39 (21%) were resistant to amphotericin B, and only one was resistant to anidulafungin (Fig. S1). We found that 62 (93%) of the 67 fluconazole-resistant isolates had known mutations in ERG11 and TAC1B genes. The Y132F mutation in the ERG11 gene was the most prevalent mutation, observed in 32 (48%) azole-resistant isolates (Table S3). All fluconazole-resistant isolates in subcluster C1-A (n = 22), collected in 2016, carried the concomitant mutations F444L in the ERG11 gene and S611P in the TAC1B gene. Significant differences were observed between MIC values, supporting the cumulative effect of the concomitant mutation in ERG11 and TAC1B genes (Fig. 2B). All (n = 16) fluconazole-resistant isolates in cluster C2 carried the mutation Y132F. Of the 39 amphotericin B-resistant isolates, we found that 38 (97%) carried the substitutions S108N in FLO8 (PSK76257) gene and I139T in PSK74852 gene. Both substitutions were also found in eight susceptible isolates with amphotericin B MIC values ≥ 0.75 and in only one isolate with amphotericin B MIC value of 0.25 mg/µL.
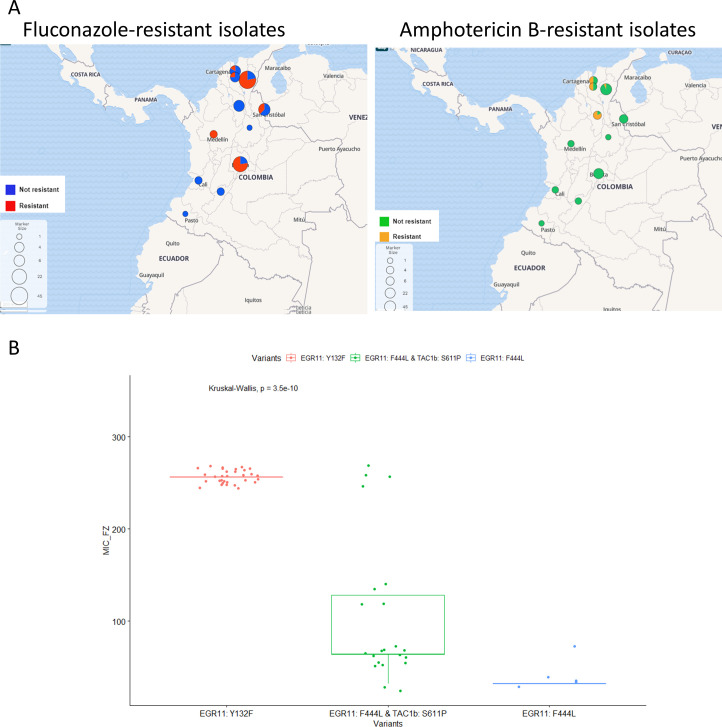
Fig 2.
(A) Geographic distribution of antifungal-resistant isolates. Maps were generated from metadata containing AFST data for each sample using Microreact (http://microreact.org). (B) R boxplot on common mutation in ERG11 and TAC1B genes and MIC values (mg/µL) of fluconazole-resistant isolates. Isolates with mutation Y132F in the ERG11 gene had significantly higher fluconazole MIC values (MIC value median = 256) than isolates with concomitant mutations F444L in the ERG11 gene and S611P in the TAC1B gene (MIC value median = 64). The isolates with concomitant mutations also had significantly higher fluconazole MIC values than isolates with the single mutation F444L in the ERG11 gene (MIC value median = 32). Significant differences between MIC values support the cumulative effect of the concomitant mutation in ERG11 and TAC1B genes. Isolates B11790 and B11087 with mutations ERG11 K143R were excluded from the graphic. jitter R function was used to add minimal and random noise to the MIC values to visualize the number of isolates at each value.
DISCUSSION
Here, we provided an update on the genomic epidemiology of C. auris in Colombia. All C. auris cases in Colombia continue to be of clade IV. This contrasts with several countries experiencing C. auris outbreaks that have identified multiple C. auris clades (6). The uniformity of clade IV isolates could be due to (1) lack of introductions of other clades or (2) lack of subsequent transmission after introductions of other clades. Alternatively, isolates from other C. auris clades could have been missed by our sampling.
Previously, a global description of C. auris reported genetic clustering by country within clade IV (6). Here, we observed a weaker phylogeographic structure as isolates from Colombia were dispersed into two clusters. Cluster C2 included fluconazole-resistant isolates collected in 2020 from cases in Bogotá and Venezuela. The remaining Colombian isolates clustered with cases from Colombia (cluster C1). Interestingly, all clinical isolates from Bogotá collected before 2020 were susceptible; this could suggest a later introduction of fluconazole-resistant cases from Venezuela. We hypothesize that an introduction of clade IV cases occurred in Colombia, possibly from Venezuela where C. auris is circulating since 2012 (4). Such a recent introduction could have given rise to the C2 cluster. Additional phylogeographic and molecular clock analyses are needed to estimate when this described introduction could have occurred.
Low median SNP differences were observed among isolates in both subclusters C1-A and C1-B (Fig. 1) spanning a 6-year period, suggesting that most cases in Colombia (cluster C1) are the result of ongoing transmission since 2016.
We concluded that amphotericin B-resistant isolates clustered (subcluster C1-B) and were predominately collected from the northern region of the country, which was previously reported by Escandon et al. (3). Additionally, the fluconazole-resistant isolates from Colombia were predominately collected from the northeast region and Bogotá, in the central region. We observed that the most common mutation, ERG11 Y132F, was associated with the highest MICs for fluconazole compared to other mutations (Fig. 2B). Y132F was present in all isolates in cluster C2 from 2020. The second most common genotype associated with fluconazole resistance had both mutations in ERG11 (F444L) and TAC1B (S611P; Fig. 2B). Interestingly, the genotypes ERG11 F444L and TAC1B S611P have been reported only in Colombian clade IV isolates to date (7) and were restricted to C1-A, which were collected in 2016 or later. The emergence of a new mechanism of fluconazole resistance in 2016 and the introduction of resistant C. auris from another country in 2020 could explain the increase in MIC values of fluconazole-resistant isolates observed by Escandon et al. (4).
In contrast with the observed increase in MIC values of fluconazole-resistant isolates, resistance to amphotericin B has not changed significantly over time (Table S4). In amphotericin B-resistant isolates, we observed the substitution S108N in FLO8 (PSK76257) and the substitution I139T in PSK74852, which were also previously reported in Colombian isolates (3). If and how the described substitutions lead to a reduction in amphotericin B susceptibility are unknown.
One limitation is that the patient history of antifungal treatment was unknown. Some cases with resistant infections could have acquired resistance in response to therapy rather than transmission of drug-resistant C. auris. However, given the low genetic differences between some cases and the common phylogeny, transmission seems more likely. A second limitation is that specimens were collected by convenience sampling and therefore are not representative of C. auris cases in Colombia.
In conclusion, the findings provided evidence of ongoing spread and a better understanding of molecular mechanisms associated with C. auris antifungal resistance in Colombia. This work contributed to the understanding of the C. auris epidemiology and factors associated with transmission in Colombia and could be useful to guide prevention and control strategies.
ACKNOWLEDGMENTS
We give special thanks to Anastasia P. Litvintseva for the insightful discussions and Centers for Disease Control and Prevention’s Office of Advanced Molecular Detection. We acknowledge Oak Ridge Institute for Science and Education for financial support for E.M.
The findings and the conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention. Use of trade names is for identification only and does not imply endorsement.
Contributor Information
Nancy A. Chow, Email: yln3@cdc.gov.
Rebecca S. Shapiro, University of Guelph, Canada
DATA AVAILABILITY
The data underlying this study are available in the GenBank Nucleotide Database at https://www.ncbi.nlm.nih.gov/bioproject/ , and can be accessed with the accession number PRJNA1003896.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/msphere.00577-23.
Supplemental text, Fig. S1 and S2, and Tables S1 to S4.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64:134–140. doi: 10.1093/cid/ciw691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Suphavilai C, Ki Ko KK, Lim KM, Tan MG, Boonsimma P, Keat Chu JJ, Goh SS, Rajandran P, Lee LC, Tan KY, et al. 2023. Discovery of the sixth Candida auris clade in Singapore. medRxiv. doi: 10.1101/2023.08.01.23293435 [DOI] [PubMed] [Google Scholar]
- 3. Escandón P, Chow NA, Caceres DH, Gade L, Berkow EL, Armstrong P, Rivera S, Misas E, Duarte C, Moulton-Meissner H, Welsh RM, Parra C, Pescador LA, Villalobos N, Salcedo S, Berrio I, Varón C, Espinosa-Bode A, Lockhart SR, Jackson BR, Litvintseva AP, Beltran M, Chiller TM. 2019. Molecular epidemiology of Candida auris in Colombia reveals a highly related, countrywide colonization with regional patterns in amphotericin B resistance. Clin Infect Dis 68:15–21. doi: 10.1093/cid/ciy411 [DOI] [PubMed] [Google Scholar]
- 4. Escandón P, Cáceres DH, Lizarazo D, Lockhart SR, Lyman M, Duarte C. 2022. Laboratory-based surveillance of Candida auris in Colombia, 2016-2020. Mycoses 65:222–225. doi: 10.1111/myc.13390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Escandón P, Editor . 2016. Alerta Por emergencia global de infecciones invasivas causadas por la levadura multirresistente, Candida auris, I.N.d.S. In Grupo de Microbiologia [Google Scholar]
- 6. Chow NA, Muñoz JF, Gade L, Berkow EL, Li X, Welsh RM, Forsberg K, Lockhart SR, Adam R, Alanio A, Alastruey-Izquierdo A, Althawadi S, Araúz AB, Ben-Ami R, Bharat A, Calvo B, Desnos-Ollivier M, Escandón P, Gardam D, Gunturu R, Heath CH, Kurzai O, Martin R, Litvintseva AP, Cuomo CA. 2020. Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. mBio 11:e03364-19. doi: 10.1128/mBio.03364-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li J, Coste AT, Liechti M, Bachmann D, Sanglard D, Lamoth F. 2021. Novel ERG11 and TAC1B mutations associated with azole resistance in Candida auris. Antimicrob Agents Chemother 65. doi: 10.1128/AAC.02663-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental text, Fig. S1 and S2, and Tables S1 to S4.
Data Availability Statement
The data underlying this study are available in the GenBank Nucleotide Database at https://www.ncbi.nlm.nih.gov/bioproject/ , and can be accessed with the accession number PRJNA1003896.