Abstract
The polyomavirus enhancer binding protein 2 (PEBP2)/core binding factor (CBF) is a transcription factor composed of two subunits, α and β. The gene encoding the β subunit is disrupted by inv(16), resulting in the formation of a chimeric protein, β-SMMHC, which is associated with acute myelogenous leukemia. To understand the effect of β-SMMHC on PEBP2-mediated transactivation, we used a luciferase assay system in which contribution of both the α and β subunits was absolutely required to activate transcription. Using this system, we found that the minimal region of the β subunit required for transactivation resides between amino acid 1 and 135, which is known to dimerize with the α subunit. In contrast, β-SMMHC, despite having this minimal region for dimerization and transactivation, failed to support transcription with the α subunit. Furthermore β-SMMHC blocked the synergistic transcription achieved by PEBP2 and CCAAT/enhancer binding protein α. By using a construct in which the PEBP2 α subunit was fused to the glucocorticoid receptor ligand binding domain, we demonstrated that coexpressed β-SMMHC tightly sequestered the α subunit in the cytoplasm and blocked dexamethasone-dependent nuclear translocation of the α subunit. Thus, the result suggess that β-SMMHC inhibits PEBP2-mediated transcription via cytoplasmic sequestration of the α subunit. Lastly proliferation of ME-1 cells that harbor inv(16) was blocked by an antisense oligonucleotide complementary to the junction of the chimeric mRNA, suggesting that β-SMMHC contributes to leukemogenesis by blocking the differentiation of myeloid cells.
A member of the transcription factor family of proteins called polyomavirus enhancer binding protein 2 (PEBP2) or core binding factor (CBF) is composed of dimers of α and β subunits. The α subunit possesses regions responsible for direct DNA binding and transactivation, while the β subunit facilitates DNA binding by the α subunit (12, 32). Gene cloning studies revealed three α subunit encoding genes, PEBP2αA/ CBFA1/AML3, PEBP2αB/CBFA2/AML1, and PEBP2αC/ CBFA3/AML2, in mammals, and two genes, runt and lozenge, in Drosophila. The β subunit is encoded by a single gene, PEBP2β/CBFB, in mammals, and by two genes, brother and big brother, in Drosophila.
Both the α and β subunit-encoding genes are independently rearranged in the chromosomal abnormalities associated with leukemia. Chromosomal translocations involving AML1 include t(8;21) in the French-American-British (FAB) M2 subtype of acute myelogenous leukemia (AML) and t(12;21) in childhood acute lymphoblastic leukemia (ALL). The t(8;21) and t(12;21) translocations produce the chimeric proteins AML1/ETO(MTG8) and TEL-AML1, respectively (5, 9, 23). The β subunit is rearranged in inv(16) of the FAB M4Eo subtype of AML, producing a chimeric protein, CBF/PEBP2β-SMMHC (18) (referred to here as β-SMMHC). The incidences of these fusion proteins involving PEBP2 subunits in leukemias are as follows: AML1/ETO, 12% of AML; β-SMMHC, 12% of AML; and TEL-AML1, 20% of ALL. These values represent a significant contribution to human leukemia in general (19).
Recently, both the α and β subunits were shown to be essential for hematopoiesis in mice. Targeted disruption of either the PEBP2αB/CBFA2/AML1 or PEBP2β/CBFB gene resulted in nearly identical phenotypes of embryonic lethality with accompanying hemorrhage of the central nervous system and defects in fetal liver definitive hematopoiesis (25, 28, 30, 37, 38). These studies thus proved that both subunits of PEBP2 are indispensable for its in vivo function. Interestingly, heterozygous mice having targeted insertion (knockin) of genes complementary to AML1/ETO and CBF/PEBP2β-MYH11 (coding for the β-SMMHC protein) displayed phenotypes similar to those of the corresponding targeted disruptions (4, 41). This finding indicated that both chimeric proteins acted as dominant negative effectors.
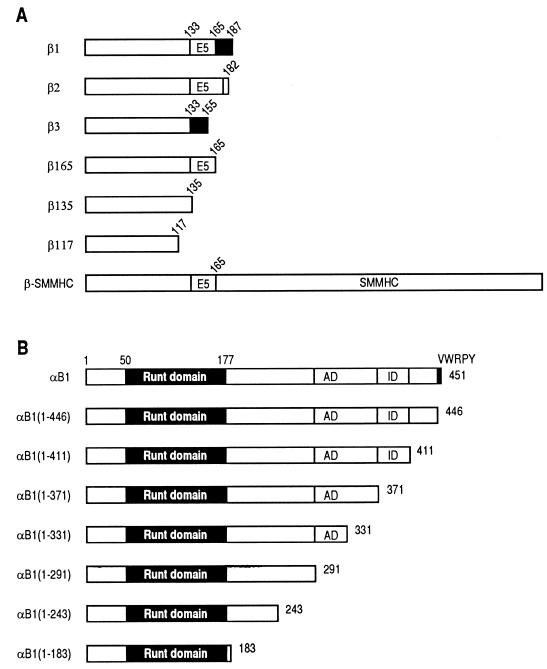
The β subunit has been shown by electrophoretic mobility shift assays (EMSA) to act as a cofactor for DNA binding. This subunit is derived from the single PEBP2β/CBFB gene by alternative splicing to give at least three isoforms, termed β1, β2, and β3 (26, 36). None of the isoforms exhibited DNA binding by themselves. β1 and β2 intensified DNA binding of the α subunit and gave a characteristic band supershift. In contrast, β3 only mildly intensified DNA binding of the α subunit, and the band supershift was not readily apparent. Therefore, it was tentatively concluded that β1 and β2 were functional but β3 was not (13, 26, 38). The only structural difference between β1 and β3 is the absence of the exon 5-encoded region from β3 (26) (Fig. 1A). Accordingly, two of the three independent β-gene knockout studies adapted a strategy to target exon 5 of the gene, leaving only the β3 isoform to be expressed (30, 38). These mice exhibited phenotypes identical to those observed when exon 1 was targeted to achieve the authentic null condition (25), a result that may simply imply that β3 is not functional in vivo either. However, well-controlled analysis of transcriptional potential of β1, β2, and β3 will be required before any firm conclusions can be drawn.
FIG. 1.
Schematic illustration of the structures of full-length and deletion derivatives of the β (A) and α (B) subunits of PEBP2. Numbers denote the positions of amino acids. (A) β1, β2, and β3 are naturally occurring isoforms of the β subunit. The structure of β-SMMHC is also shown. E5 denotes the region encoded by exon 5. Two different splice donor sites are present in exon 5. The black regions in β1 and β3 are encoded by exon 6 but translated in a frame different from that of β2. (B) The Runt domain spans aa 50 to 177, and VWRPY is a stretch of the last 5 aa of αB1. Transactivation domain (AD) and inhibitory domain (ID) are also shown.
The inv(16) chromosome rearrangement produces fusion proteins collectively called β-SMMHC (18). The protein is composed of the amino-terminal portion of the PEBP2 β subunit and the carboxy-terminal portion of the smooth muscle myosin heavy chain (SMMHC). Major chromosomal breakpoints occur in intron 5 of the PEBP2β gene, which result in the conservation of the first 165 amino acids (aa) of the β subunit in the β-SMMHC protein. The function of β-SMMHC has been studied by EMSA using nuclear extracts from β-MYH11 gene-transfected cells (2). The study showed a reduction in DNA binding of the normal α-β dimers and simultaneous appearance of α–β-SMMHC complexes. However, the function of β-SMMHC with respect to transactivation activity of PEBP2 was not addressed.
Analysis of the function of the β subunit as well as that of β-SMMHC in transactivation requires experimental systems that meet two criteria: little or no transactivation by either subunit alone and strong transactivation when both are present together. It is frequently observed that the α subunit alone apparently induces PEBP2 site-dependent transactivation (1, 6, 17, 22, 27, 33, 42). The nature of β-subunit-independent transactivation is unknown, especially in terms of the extent to which this activity depends on the endogenous β subunit. This latter point needs to be emphasized, as it is possible that other transcription factors can also facilitate DNA binding of the α subunit. Also, an earlier study on the effect of the β subunit was undermined by the relatively high activity obtained with transfection of the α subunit alone (42). Recently, we have found that luciferase assays performed in Jurkat T cells by using a reporter containing a regulatory element from the macrophage colony-stimulating factor (M-CSF) receptor promoter meet the above criteria (14). In the present study, we have analyzed the transcriptional properties of the β subunit as well as those of β-SMMHC. We show that β-SMMHC fails to support PEBP2-dependent transactivation despite the fact that it contains the region essential for β-subunit function. Subsequently we show that β-SMMHC sequesters the α subunit in the cytoplasm, thereby precluding it from acting in the nucleus.
MATERIALS AND METHODS
Plasmid construction.
Mammalian expression vector pEF-BOS (24) was used to make a series of expression plasmids: pEF-αB1, pEF-αB1(1–243), and pEF-αB1(1–183) (1); pEF-αB1(1–446), pEF-β1, pEF-β2, pEF-β3, and pEF-β-SMMHC (previously referred to as pEF-b/MYH11) (20); pEF-AML1(453) (previously referred to as AML1b) (45); and pEF-αB1(1–411), pEF-αB1(1–371), pEF-αB1(1–331), and pEF-αB1(1–291) (14). For the β-deletion constructs [pEF-β165 (human), pEF-β135, and pEF-β117], corresponding regions of the β subunit were PCR amplified and ligated into the XbaI site of pEF-BOS. The authenticity of the PCR-amplified sequences was confirmed by sequencing. pMSV-C/EBPα (3) and the luciferase reporter pM-CSF-R-luc (43) have already been described. pME-αB1-GRLBD was constructed as follows. The glucocorticoid receptor ligand binding domain (GRLBD) sequence was cut out from pRShGRNX (7) by XhoI/BamHI digestion and ligated to the carboxy-terminal sequence of αB1 by using the internal BssHII site and a synthetic linker to adjust the reading frame. The entire αB1-GRLBD sequence was then cloned into pME18S, an SRα promoter-driven expression plasmid (34).
Transfection and luciferase assays.
Jurkat human T cells were maintained in RPMI 1640 supplemented with 10% fetal calf serum (FCS) and antibiotics. The cells were transfected with a reporter plus expression plasmids via electroporation, and relative luciferase activities were assayed as described previously (14). The compositions of the transfected plasmids are described in the figure legends. The total amount of transfected DNA was always kept at 10 μg, using the empty vector pEF-BOS.
Indirect immunofluorescence staining.
REF52 rat fibroblasts were cultured in Dulbecco modified Eagle medium supplemented with 10% FCS and antibiotics. Cells were trypsinized and electroporated with 15 μg of each expression plasmid. The transfected cells were seeded onto chamber slides (Nalge Nunc, Naperville, Ill.). After 48 h, cells were either untreated or treated with 1 μM dexamethasone (DEX) for 1 h, fixed, and stained with a rabbit antiserum raised against the Escherichia coli-expressed αB1 and/or a hamster antiserum raised against the E. coli-expressed β2 as described previously (20, 45). Jurkat T cells were electroporated with 15 μg of each expression plasmid and after 48 h either untreated or treated with 1 μM DEX for 1 h. The cells were then cytospun, fixed, and stained.
Proliferation assay of ME-1 cells.
ME-1 cells (40) were grown in RPMI 1640 supplemented with 10% FCS and antibiotics. Proliferation assays were performed with synthesized sense or antisense oligonucleotides as previously described (29). The sequence of the sense oligonucleotide corresponding to the junctional region of β-MYH11 (18) is GAAATGGAGGTCCATGAG. The sequence of the corresponding antisense oligonucleotide is CTCATGGACCTCCATTTC.
RESULTS
β-SMMHC is required for maintenance of the proliferative state of ME-1 leukemic cells with inv(16).
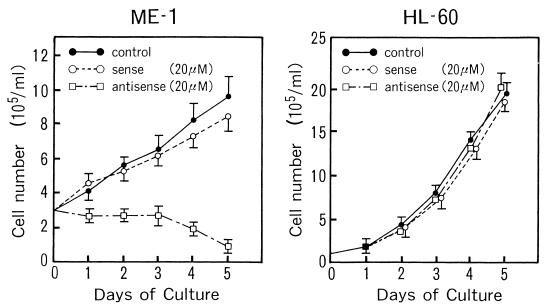
The leukemic cell line ME-1 (40) was established from a leukemic patient with inv(16), and it was from this cell line that β-MYH11 (encoding β-SMMHC) was originally cloned (18). We examined whether the expression of β-SMMHC is essential for the maintenance of the proliferative state of this cell line. We incubated ME-1 cells with an antisense oligonucleotide complementary to the junctional sequence of the fusion transcript. As shown in Fig. 2, the antisense oligonucleotide, but not the corresponding sense oligonucleotide, blocked the proliferation of ME-1 cells. In contrast, neither the antisense nor the sense oligonucleotide blocked proliferation of HL-60 leukemic cells, which do not express β-MYH11. After the antisense oligonucleotide treatment, ME-1 cells showed an increase in the population of differentiated CD13-positive cells (data not shown). The results suggest that the fusion product, β-SMMHC, may act dominantly over the normal β subunit and prevent myeloid cells from entering into terminal differentiation, thereby supporting their continued proliferation.
FIG. 2.
Expression of β-SMMHC is necessary for proliferation of ME-1 leukemic cells with inv(16). ME-1 cells (left) or HL-60 cells (right) were grown in the absence or presence of the sense or antisense oligonucleotides complementary to the junctional region of the β-MYH11 transcript. The results represent three independent experiments.
The naturally occurring three isoforms of the β subunit function as transactivational cofactors of PEBP2.
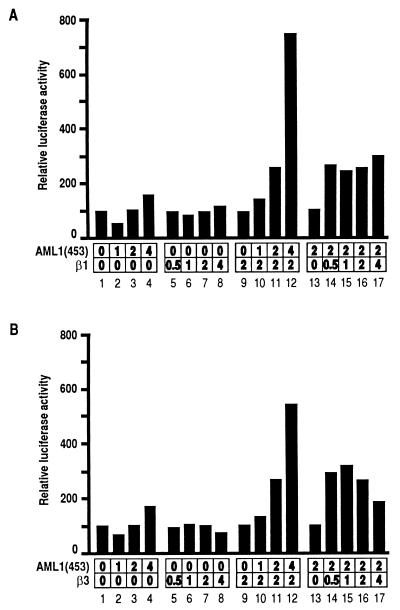
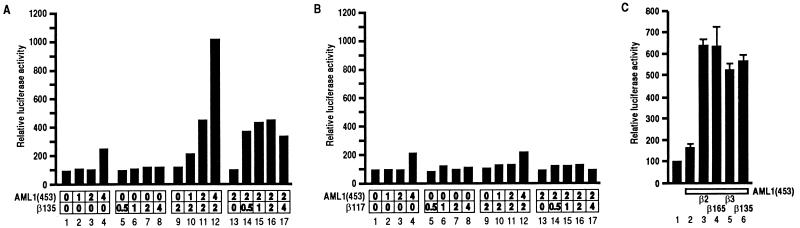
The result described above motivated us to investigate the function of β-SMMHC at the molecular level. We first tried to address the function of β-SMMHC and its normal counterpart, the β subunit, in terms of PEBP2-mediated transactivation. Using the M-CSF receptor promoter linked to the luciferase gene, we previously established a reporter assay system in Jurkat T cells, in which the function of the β subunit can be analyzed. This system requires both the α and the β subunits for significant transactivation, as we have shown by using β2 as the β subunit (14). Here we performed similar function assays for the β1 and β3 isoforms. Neither β1 nor β3 alone activated the promoter to any extent (Fig. 3A and B, lanes 5 to 8). Increasing amounts of AML1(453) [herein, we refer to full-length human and mouse proteins encoded by the PEBP2αB/CBFA2/AML1 gene as AML1(453) and αB1, respectively] only marginally transactivated the reporter activity when expressed alone (Fig. 3A and B, lanes 1 to 4). However, in the presence of β1 or β3, AML1(453) transactivated the promoter in a dose-dependent manner (Fig. 3A and B, lanes 9 to 12). Conversely, in the presence of a fixed amount of AML1(453), a wide range of β1 or β3 concentrations enhanced AML1(453)-dependent transactivation, suggesting that the effect of β1 or β3 was saturating above a certain concentration (Fig. 3A and B, lanes 13 to 17). Therefore, the naturally occurring isoforms of the β subunit, β1, β2, and β3, are all efficient for cooperative transactivation with the α subunit in vivo. Cooperation between the α subunit and the different β subunits was not confined to AML1(453) alone but was a general property of other members of the α subunit family, since similar results were also obtained using PEBP2αA or PEBP2αC as the α subunit (data not shown).
FIG. 3.
Cooperative activation of the M-CSF receptor promoter by the α and β subunits of PEBP2. Jurkat T cells were transfected with 4 μg of pM-CSF-R-luc and indicated amounts (micrograms) of pEF-AML1(453) and pEF-β1 (A) or pEF-β3 (B). Relative luciferase activities obtained with pM-CSF-R-luc and an empty plasmid were set at 100. For presentation, lanes 9, 13, and 16 are duplicated from lanes 7, 3, and 11, respectively.
β-SMMHC does not support cooperative transactivation with full-length αB1 or its deletion derivatives.
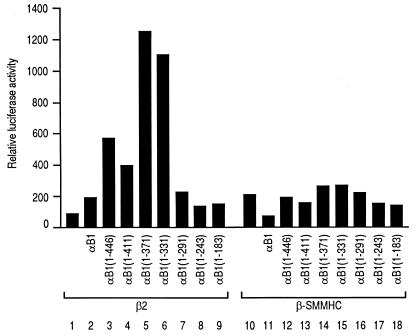
When β-SMMHC was used instead of the naturally occurring β subunit isoforms, β-SMMHC did not cooperate with the α subunit (Fig. 4). To compare the effects of β2 and β-SMMHC, we examined a series of deletion constructs of αB1 (Fig. 1B) in the presence of β2 or β-SMMHC. With β2, peak activities were obtained with αB1(1–371) and αB1(1–331) (Fig. 4, lanes 5 and 6), confirming that the transactivation domain lies between aa 291 and 371 (14). In contrast, β-SMMHC did not lead to strong transactivation with any of the αB1 deletion constructs, demonstrating a clear difference from β2. All of the data suggest that β-SMMHC interferes with the α subunit function.
FIG. 4.
Effects of β2 and β-SMMHC on transactivation with full-length αB1 and its deletion derivatives. Jurkat cells were transfected with 4 μg of pM-CSF-R-luc, 1.5 μg of pEF-β2 (lanes 1 to 9) or pEF-β-SMMHC (lanes 10 to 18), and 4.5 μg of the pEF-BOS-based expression plasmid for αB1 or its deletion derivative as indicated. For lanes 1 and 10, 4.5 μg of an empty vector, pEF-BOS, was used. Relative luciferase activity obtained with cells transfected with 4 μg pM-CSF-R-luc and 6 μg of pEF-BOS was set at 100.
Minimal region of the β subunit required for α-β cooperative transactivation.
β-SMMHC consists of the amino-terminal 165 aa of the β subunit and the coiled-coil tail structure of SMMHC (18, 21) (Fig. 1A). To examine whether the inability of β-SMMHC to cooperate with the α subunit is due to the lack of the carboxy-terminal sequences present in the normal isoforms of the β subunit, we constructed a series of carboxy-terminal deletion constructs of the β subunit as shown in Fig. 1A in expression plasmids and tested their activities. As shown in Fig. 5A and B, β135 enhanced AML1(453)-dependent transactivation, in contrast to β117, which did not exhibit any significant effect. Figure 5C shows a direct comparison of the transactivation abilities of β2, β165, β3, and β135. The results show that these constructs are equally functional, indicating that the amino-terminal 135-aa region in the β subunit is the minimal region required for α-β cooperation. Therefore, the inability of β-SMMHC to cooperate with the α subunit cannot be due to the absence of the cooperation domain with the α subunit. Rather, the results seem to suggest that the presence of the SMMHC region prevents the β-subunit region from properly cooperating with the α subunit.
FIG. 5.
Comparison between the natural isoforms and deletion derivatives of the β subunit with respect to their capacity to cooperate with the α subunit in transactivation. (A and B) Jurkat cells were transfected with 4 μg of pM-CSF-R-luc and indicated amounts (micrograms) of pEF-AML1(453) and pEF-β135 (A) or pEF-β117 (B). As in Fig. 3, lanes 9, 13, and 16 are duplicated from lanes 7, 3, and 11, respectively, for presentation. (C) Jurkat cells were transfected with 4 μg of pM-CSF-R-luc, 4 μg of pEF-AML1(453), and 2 μg of pEF-β2 (lane 3), pEF-β165 (lane 4), pEF-β3 (lane 5), or pEF-β135 (lane 6) as indicated. Relative luciferase activities obtained with pM-CSF-R-luc and an empty plasmid (pEF-BOS) were set at 100, and the results represent three independent experiments.
Coexpression of β-SMMHC inhibits normal α-β cooperative transactivation.
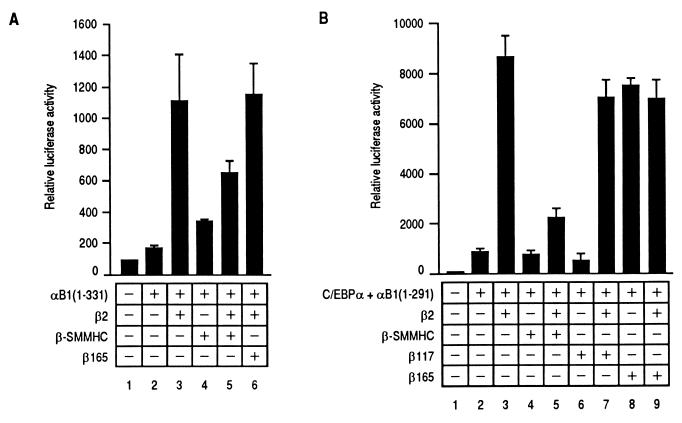
There exist at least two possible mechanisms to explain the observed negative effect of the SMMHC region. One is that β-SMMHC may not be able to interact with the α subunit because of steric inhibition by the SMMHC region. Another possibility is that complexes of the α subunit and β-SMMHC, if formed, may not be able to transactivate, which seems more likely based on already published observations (2, 4). In the former case, β-SMMHC should not interfere with the normal α-β interaction, predicting that coexpression of the β-SMMHC should not affect α-β cooperative transactivation. In contrast, in the latter case, β-SMMHC would compete with the normal β subunit for heterodimerization with the α subunit, thus inhibiting transactivation. To test these possibilities, we examined the effect of β-SMMHC on cooperative transactivation by αB1(1–331) and β2. αB1(1–331) was used as the α subunit because it exhibited higher activity than full-length αB1 in the presence of β2 (Fig. 4), but similar results were obtained with full-length αB1. As shown in Fig. 6A, β-SMMHC inhibited transactivation by αB1(1–331) and β2, suggesting that β-SMMHC competed with β2 for interaction with αB1(1–331), a result that favors the latter possibility. In contrast, β165, which contains the exact β-subunit region of β-SMMHC and is capable of dimerizing with the α subunit, did not inhibit transactivation (lane 6). This is because the α subunit can cooperate with the functional β subunit regardless of whether it is β2 or β165 (Fig. 5C). Furthermore, we examined the effect of β-SMMHC on another mode of transactivation. The M-CSF receptor promoter contains a binding site for CCAAT/enhancer binding protein α (C/EBPα) in close vicinity to the PEBP2 binding site, and C/EBPα and PEBP2 synergistically activate the promoter (44). We have found that the transactivation domain of αB1 (aa 291 to 371) is dispensable for synergistic transactivation and that the region of αB1 from aa 1 to 291 suffices for synergy with C/EBPα (15). αB1(1–291) plus β2, which showed only moderate transactivation (Fig. 4, lane 7), exhibited a remarkable increase in transactivation activity in the presence of C/EBPα (Fig. 6B, lane 3). In this transactivation mediated by synergistic cooperation between αB1(1–291) and C/EBPα, the presence of the β subunit is absolutely required. β-SMMHC not only failed to substitute for the β subunit for the function (lane 4) but also inhibited the effect of β2 when coexpressed (lane 5). In contrast, β165 exhibited cooperativity similar to that of β2 and did not inhibit the effect of β2 (lanes 8 and 9). Again, this is probably because of cooperation of the α subunit with the functional β subunit regardless of whether it is β2 or β165. On the other hand, β117 neither exhibited the same cooperativity as β2 nor inhibited the effect of β2 (lanes 6 and 7), consistent with the fact that β117 does not dimerize with the α subunit. This makes a clear distinction with β-SMMHC, which dimerizes with the α subunit and inhibits transcription.
FIG. 6.
Comparison of effects of β-SMMHC and β165 on two modes of transactivation achieved by αB1 deletion derivatives and β2. (A) Jurkat cells were transfected with 3 μg of pM-CSF-R-luc and different combinations of expression plasmids as indicated [pEF-αB1(1–331), 4.2 μg; pEF-β2, 1.4 μg; pEF-β-SMMHC, 1.4 μg; and pEF-β165, 1.4 μg]. The results represent two independent experiments, in each of which the relative luciferase activity obtained with 3 μg of pM-CSF-R-luc and 7 μg of pEF-BOS was set at 100. (B) Jurkat cells were transfected with 3 μg of pM-CSF-R-luc and different combinations of expression plasmids as indicated [pMSV-C/EBPα, 1 μg; pEF-αB1(1–291), 3.6 μg; pEF-β2, 1.2 μg; pEF-β-SMMHC, 1.2 μg; pEF-β117, 1.2 μg; and pEF-β165, 1.2 μg]. The results represent two independent experiments.
β-SMMHC sequesters the α subunit in the cytoplasm.
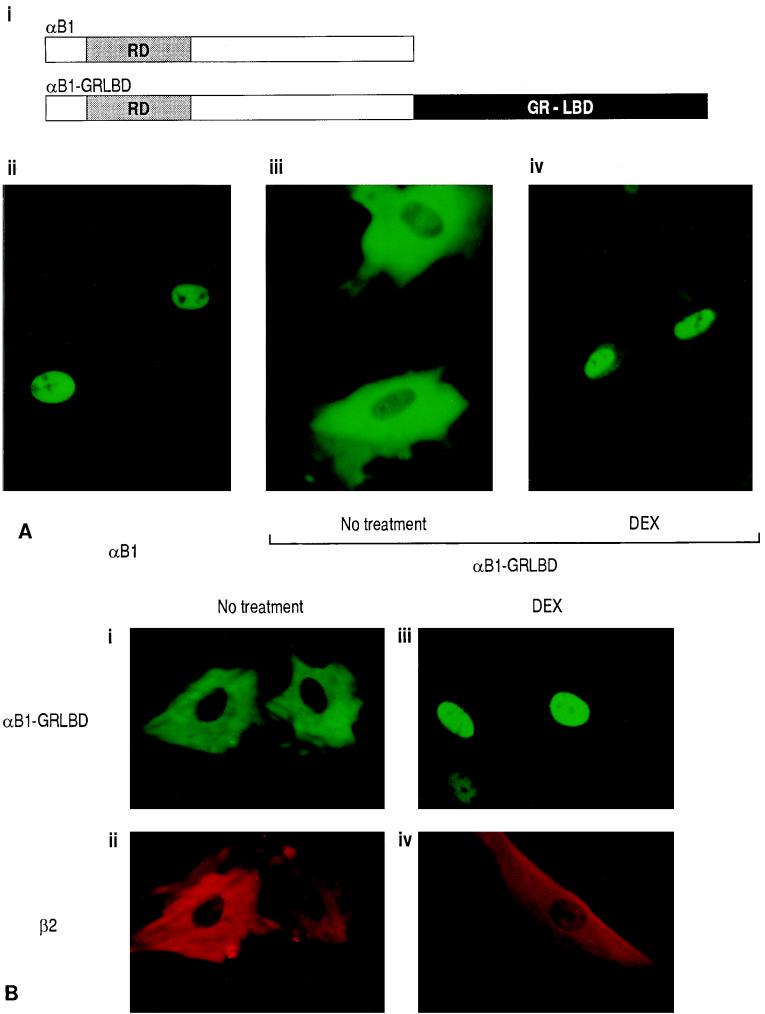
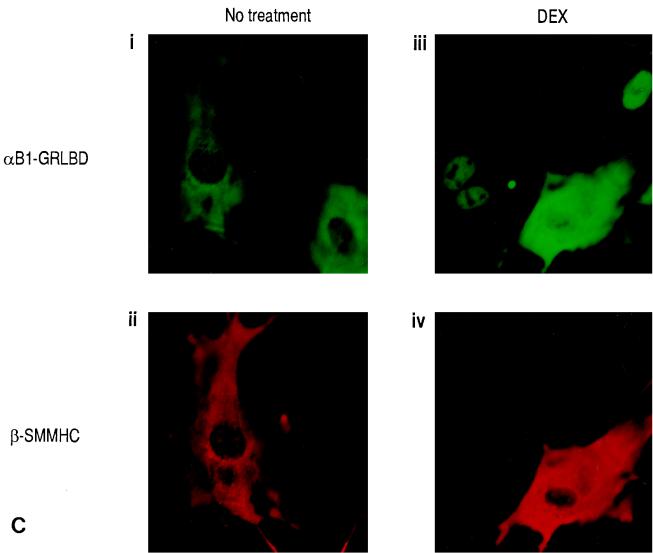
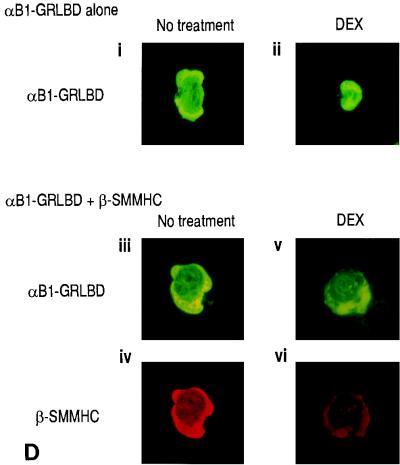
The next question was why dimers composed of the α subunit and β-SMMHC cannot transactivate. We previously observed that the α subunit and β-SMMHC localized both in the cytoplasm and in the nucleus when expressed together (20). When expressed separately, the α subunit was exclusively nuclear whereas β-SMMHC was cytoplasmic. Thus, it seems that coexpression of the β-SMMHC affected subcellular localization of the α subunit, but no direct experimental evidence for this hypothesis was provided (20). To examine this hypothesis more clearly, we established a system in which translocation of one factor can be controlled by an addition of a ligand. We constructed an expression plasmid coding for the fusion protein αB1-GRLBD. The intracellular localization of αB1-GRLBD can be controlled by DEX treatment (Fig. 7A). While parental αB1 by itself is localized exclusively to the nucleus (Fig. 7A, ii), αB1-GRLBD was localized to the cytoplasm in the absence of DEX treatment, because it was tethered there by GRLBD (Fig. 7A, iii). With DEX treatment, αB1-GRLBD translocated into the nucleus (Fig. 7A, iv). When αB1-GRLBD and β2 were coexpressed, both were localized to the cytoplasm in the absence of DEX treatment (Fig. 7B, left panels). With DEX treatment, αB1-GRLBD was mostly translocated into the nucleus along with a small fraction of β2 (Fig. 7B, right panels). It is of note that most of the β subunit remained in the cytoplasm. When αB1-GRLBD and β-SMMHC were coexpressed, both were localized to the cytoplasm without DEX treatment, as expected (Fig. 7C, left panels). Interestingly, with DEX treatment, the majority of αB1-GRLBD still remained in the cytoplasm together with β-SMMHC (Fig. 7C, right panels; see the cell in which both were expressed). In this experiment, DEX treatment was sufficient to induce ligand-dependent nuclear translocation, because in other cells where only αB1-GRLBD was expressed, αB1-GRLBD completely translocated it into the nucleus upon DEX treatment. These results show that nuclear translocation of αB1-GRLBD upon DEX treatment was blocked by β-SMMHC but not by wild-type β2. The same was found for Jurkat T cells, where DEX-dependent nuclear translocation of αB1-GRLBD was blocked by β-SMMHC (Fig. 7D).
FIG. 7.
DEX-dependent nuclear translocation of αB1 is blocked by β-SMMHC. (A) Schematic illustration of the αB1-GRLBD construct and effect of DEX on the subcellular localization of αB1-GRLBD. REF52 cells were transfected with pEF-αB1 (ii) or pME-αB1-GRLBD (iii and iv) and after 48 h either untreated (ii and iii) or treated with 1 μM DEX for 1 h (iv). Cells were stained with a rabbit anti-αB1 antiserum and a fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit antiserum. (B) REF52 cells were transfected with expression plasmids for αB1-GRLBD (pME-αB1-GRLBD) and β2 (pEF-β2) and either untreated (left panels) or DEX treated (right panels). Cells were double stained with a rabbit anti-αB1 antiserum followed by an FITC-conjugated goat anti-rabbit antiserum and with a hamster anti-β2 antiserum followed by a rhodamine-conjugated goat anti-hamster antiserum. Signals from FITC were detected by using a 495-nm-wavelength filter, and those from rhodamine were detected by using a 555-nm-wavelength filter; upper panels represent FITC signals (αB1-GRLBD), and lower panels represent rhodamine signals (β2). (C) REF52 cells were transfected with expression plasmids for αB1-GRLBD (pME-αB1-GRLBD) and β-SMMHC (pEF-β-SMMHC) and either untreated (left panels) or DEX treated (right panels). For double-stained cells, upper panels show FITC signals (αB1-GRLBD) and lower panels show rhodamine signals (β-SMMHC). Note that some cells expressed αB1-GRLBD but not β-SMMHC. (D) Jurkat T cells were transfected with pME-αB1-GRLBD alone (i and ii) or pME-αB1-GRLBD plus pEF-β-SMMHC (iii to vi) and either untreated (left panels) or DEX-treated (right panels). Panels iii and iv and panels v and vi show double staining of the same cells. i, ii, iii, and v, FITC signals (αB1-GRLBD); iv and vi, rhodamine signals (β-SMMHC).
DISCUSSION
A system for in vivo functional analysis of the β subunit has revealed that β1, β2, and β3 isoforms are all transcriptionally functional.
Previously the function of the β subunit was studied mostly by in vitro EMSA (13, 26, 38). In the present study, the M-CSF receptor promoter-luciferase reporter was used to assay β-subunit function in Jurkat T cells, and the results showed that the three naturally occurring isoforms of the β subunit, β1, β2, and β3, are all effective in supporting α-β cooperative transactivation. Previously we and others observed prominent band supershifts of α-subunit–DNA complexes with β1 and β2 but not with β3 in EMSA (13, 26, 38). However, β3 did have an effect of intensifying DNA binding of the α subunit. Thus, α-β dimers may be stable enough for enhanced transactivation in vivo but not stable enough to produce a supershift in EMSA.
On the basis of the previous EMSA study (13), we concluded that the amino-terminal 135-aa region of the β subunit (β135) is sufficient for heterodimerization with the α subunit. By transactivation assays performed in the present study, we found that the same 135-aa region was sufficient for cooperation with the α subunit to transactivate. In these assays, it seemed that variable regions of the β subunit more carboxy terminal to the first 135-aa region did not contribute to the transactivation function. Further deletion of 18 aa (β117) from β135 completely abolished β-subunit function in both assays. Shurtleff et al. reported that the amino-terminal 133-aa region of the β subunit exhibited weaker DNA binding activity than β165 when mixed with the α subunit (31). Also, deletion analysis of Drosophila Brother revealed that a construct corresponding to the amino-terminal 132 aa of the mouse β subunit exhibited weaker DNA binding activity in the presence of Runt than the corresponding amino-terminal 137 aa of the β subunit (8). It will be necessary to reevaluate these findings by transactivation assays.
Properties of β-SMMHC.
We have found that β-SMMHC does not cooperate with the α subunit for transactivation despite the presence of the minimal region for β-subunit function. Rather, β-SMMHC seems to inhibit normal α-β cooperative transactivation. The results suggest that β-SMMHC can efficiently dimerize with the α subunit, but that SMMHC region actively participates in undermining the proper functioning of these complexes.
How then does the carboxyl-terminal SMMHC region interfere with the potential harbored in the β-subunit region (identical to β165)? One possible mechanism is the sequestration of the otherwise nucleus-located α subunit in the cytoplasm. We showed that β-SMMHC blocked nuclear translocation of αB1-GRLBD upon DEX treatment, whereas β2 did not. With this GRLBD fusion system, the majority of the α–β-SMMHC complexes were retained in the cytoplasm. As such, the α–β-SMMHC complexes can no longer participate in DNA binding and transactivation. This constitutes a form of dominant negative inhibition. It is important to note that colocalization of β-SMMHC and the α subunit was observed both in this and the previous (20) study. However, in conventional cotransfection assays using β-SMMHC and wild-type αB1, complexes of both proteins were found to be localized in the nucleus as well as in the cytoplasm (20). We explain this as follows. When expressed separately, the α subunit is localized in the nucleus because of the presence of nuclear localization signals (NLSs) (14, 20), while β-SMMHC is localized in the cytoplasm (20), probably because it lacks an NLS. By analogy to normal myosin structure, two molecules of β-SMMHC would be packed into a monomer by forming an α-helical coiled-coil rod. When both the α subunit and β-SMMHC are coexpressed at a low level, α–β-SMMHC complexes with a monomeric SMMHC tail structure would form and enter into the nucleus, utilizing the NLS of the α subunit, and would bind to DNA (2). However, with increasing levels of expression over a “critical monomer concentration” (16), the SMMHC portion would begin to form filaments via the carboxy-terminal nonhelical tailpiece (11). These α–β-SMMHC complexes polymerized through SMMHC tails would be precluded from entering into the nucleus. With the GRLBD fusion system, in contrast, most of the complexes would form polymers being retained in the cytoplasm via the GRLBD portion before DEX treatment. Thus, polymerized α–β-SMMHC complexes would not be able to enter the nucleus upon DEX treatment.
There is a precedent for a leukemogenic fusion protein that causes abnormal localization of transcription factors. Acute promyelocytic leukemia retinoic acid receptor alpha (PML-RARα) is localized to discrete subnuclear compartments called nuclear bodies, and it subsequently causes disruption of nuclear bodies (39). As a result of this aberrant localization, retinoid X receptors, which are the heterodimeric partners of RARα, are sequestered to these compartments, causing attenuated responses to retinoids and vitamin D3 (10, 35, 39). The situation is quite analogous to the sequestration of the α subunit by β-SMMHC to the cytoplasm described in the present study. In the case of PML-RARα, retinoic acid can restore the structure of the nuclear bodies and differentiation responses to vitamin D3 (35, 39). By analogy, it would be intriguing to find the molecular mechanisms of cytoplasmic retention by β-SMMHC with a view of developing specific therapeutic agents that induce the release of the α subunit.
Effect of β-SMMHC on proliferation.
The present study suggests that β-SMMHC plays a key role in the maintenance of the proliferative state of ME-1 cells which have inv(16). In contrast, Cao et al. recently reported that β-SMMHC blocks proliferation of 32D myeloid cells at the G1/S transition (2). Further studies will be needed to address this apparent discrepancy between the two different systems. Nonetheless, it is interesting that proliferation of Kasumi-1 cells, in which AML1/ETO is produced as a result of the t(8;21) translocation, was specifically blocked by an antisense oligonucleotide complementary to the junction region of the AML1/ETO mRNA (29). Thus, β-SMMHC and AML1/ETO are analogous in some respects: both are dominant negative inhibitors in developing embryos (4, 41), and both are essential for the proliferative properties of leukemic cells that have the corresponding chromosomal abnormalities.
Alternative interpretation of the β knockout and the β-SMMHC knockin studies.
Previously, the β3 isoform was thought to be inactive as discussed above. Accordingly, the β knockout mice were produced via targeting of exon 5, leaving the β3 isoform to be expressed normally or increased in a compensatory fashion (30, 38). These mice exhibited nearly the same phenotypes as the mice that were targeted in exon 1, which represents the authentic knockout (25). Together, these observations suggest that β3 is inactive in vivo. In fact, however, alternative splicing with the skipping of exon 5 in the former group of mice did not occur so as to quantitatively compensate for the loss of β1 and β2 (reference 38; our unpublished observation [15] on mice generated by Sasaki et al. [30]). Thus the observed phenotypes may be attributed to a dosage effect but not to the disabled function of β3 per se.
The present study revealed that β3 is effective in transactivation assays. Therefore, if impaired transcriptional activity is responsible for the knockout phenotypes, the impaired development of the exon 5-targeted mice is more likely to be due to the lack of a compensatory increase in β3 levels. Low levels of β3, such as those naturally present, may be insufficient, and we speculate that there might exist a threshold level of the β subunit that allows fetal liver definitive hematopoiesis to emerge. Such a threshold level would be less than 50% of the normal level, because heterozygous mice targeted in exon 1 were phenotypically normal. The results of the present study suggest that one of the mechanisms underlying the phenotypes of the β-SMMHC knockin mice may be a decrease in the available amount of the α subunit due to its aberrant sequestration. The amount of the α subunit has been reported to be limiting in fetal hematopoietic tissues, because heterozygous knockout mice for the α subunit exhibited a minor but still significant abnormality in in vitro colony assays (37).
In conclusion, β-SMMHC inhibits PEBP2-mediated transactivation via cytoplasmic sequestration of the α subunit, which is limiting in cells. Whereas the β subunit is relatively abundant, a level as low as that of the natural amount of β3 may not be sufficient to support the function(s) of PEBP2 in vivo. An alteration in the available amounts of PEBP2 subunits at sites of transcription could have a significant effect on hematopoiesis and eventually lead to leukemia.
ACKNOWLEDGMENTS
We thank K. Umesono for pRShGRNX, D.-E. Zhang for pM-CSF-R-luc, A. D. Friedman for pMSV-C/EBPα, and K. Yanagisawa for ME-1 cells. We also thank T. Komori and K. Sasaki for making PEBP2β knockout mice available to us. We thank M. Osato for helpful discussion.
The work was supported in part by a grant (FY1995, B-333) from the New Energy and Industrial Technology Development Organization and by Grant-in-Aid 0925322 for Priority Area on Cancer Research from the Minister of Education, Science and Culture, Japan.
REFERENCES
- 1.Bae S C, Ogawa E, Maruyama M, Oka H, Satake M, Shigesada K, Jenkins N A, Gilbert D J, Copeland N G, Ito Y. PEBP2αB/mouse AML1 consists of multiple isoforms that possess differential transactivation potentials. Mol Cell Biol. 1994;14:3242–3252. doi: 10.1128/mcb.14.5.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao W, Britos-Bray M, Claxton D F, Kelley C A, Speck N A, Liu P P, Friedman A D. CBFb-SMMHC, expressed in M4Eo AML, reduced CBF DNA-binding and inhibited the G1 to S cell cycle transition at the restriction point in myeloid and lymphoid cells. Oncogene. 1997;15:1315–1327. doi: 10.1038/sj.onc.1201305. [DOI] [PubMed] [Google Scholar]
- 3.Cao Z, Umek R M, MaKnight S L. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 4.Castilla L H, Wijmenga C, Wang Q, Stacy T, Speck N A, Eckhaus M, Marin-Padilla M, Collins F S, Wynshaw-Boris A, Liu P P. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell. 1996;87:687–696. doi: 10.1016/s0092-8674(00)81388-4. [DOI] [PubMed] [Google Scholar]
- 5.Erickson P, Gao J, Chang K S, Look T, Whisenant E, Raimondi S, Lasher R, Trujillo J, Rowley J, Drabkin H. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992;80:1825–1831. [PubMed] [Google Scholar]
- 6.Frank R, Zhang J, Uchida H, Meyers S, Hiebert S W, Nimer S D. The AML1/ETO fusion protein blocks transactivation of the GM-CSF promoter by AML1B. Oncogene. 1995;11:2667–2674. [PubMed] [Google Scholar]
- 7.Giguere V, Ong E S, Segui P, Evans R M. Identification of a receptor for the morphogen retinoic acid. Nature. 1987;330:624–629. doi: 10.1038/330624a0. [DOI] [PubMed] [Google Scholar]
- 8.Golling G, Li J-H, Pepling M, Stebbins M, Gergen J P. Drosophila homologs of the proto-oncogene product PEBP2/CBFβ regulate the DNA-binding properties of Runt. Mol Cell Biol. 1996;16:932–942. doi: 10.1128/mcb.16.3.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golub T R, Barker G F, Bohlander S K, Hiebert S W, Ward D C, Bray-Ward P, Morgan E, Raimondi S C, Rowley J D, Gilliland D G. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 1995;92:4917–4921. doi: 10.1073/pnas.92.11.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grignani F, Ferrucci P F, Testa U, Talamo G, Fagioli M, Alcalay M, Mencarelli A, Peschle C, Nicoletti I, Pelicci P G. The acute promyelocytic leukemia-specific PML-RARα fusion protein inhibits differentiation and promotes survival of myeloid precursor cells. Cell. 1993;74:423–431. doi: 10.1016/0092-8674(93)80044-f. [DOI] [PubMed] [Google Scholar]
- 11.Hodge T P, Cross R, Kendrick-Jones J. Role of the COOH-terminal nonhelical trailpiece in the assembly of a vertebrate nonmuscle myosin rod. J Cell Biol. 1992;118:1085–1095. doi: 10.1083/jcb.118.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito Y, Bae S-C. The Runt domain transcription factor, PEBP2/CBF, and its involvement in human leukemia. In: Yaniv M, Ghysdael J, editors. Oncogenes as transcriptional regulators. Vol. 2. Basel, Switzerland: Birkhauser Verlag; 1997. pp. 107–132. [Google Scholar]
- 13.Kagoshima H, Akamatsu Y, Ito Y, Shigesada K. Functional dissection of the alpha and beta subunits of transcription factor PEBP2 and the redox susceptibility of its DNA binding activity. J Biol Chem. 1996;271:33074–33082. doi: 10.1074/jbc.271.51.33074. [DOI] [PubMed] [Google Scholar]
- 14.Kanno T, Kanno Y, Chen L-F, Ogawa E, Kim W-Y, Ito Y. Intrinsic transcriptional activation-inhibition domains of the polyomavirus enhancer binding protein 2/core binding factor α subunit revealed in the presence of the β subunit. Mol Cell Biol. 1998;18:2444–2454. doi: 10.1128/mcb.18.5.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanno, Y., T. Kanno, and Y. Ito. Unpublished data.
- 16.Kendrick-Jones J, Smith R C, Craig R, Citi S. Polymerization of vertebrate non-muscle and smooth muscle myosins. J Mol Biol. 1987;198:241–252. doi: 10.1016/0022-2836(87)90310-x. [DOI] [PubMed] [Google Scholar]
- 17.Kurokawa M, Tanaka T, Tanaka K, Ogawa S, Mitani K, Yazaki Y, Hirai H. Overexpression of the AML1 proto-oncoprotein in NIH3T3 cells leads to neoplastic transformation depending on the DNA-binding and transactivational potencies. Oncogene. 1996;12:883–892. [PubMed] [Google Scholar]
- 18.Liu P, Tarle S A, Hajra A, Claxton D F, Marlton P, Freedman M, Siciliano M J, Collins F S. Fusion between transcription factor CBFβ/PEBP2β and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261:1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- 19.Look A T. Oncogenic transcription factors in the human acute leukemias. Science. 1997;278:1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 20.Lu J, Maruyama M, Satake M, Bae S C, Ogawa E, Kagoshima H, Shigesada K, Ito Y. Subcellular localization of the α and β subunits of the acute myeloid leukemia-linked transcription factor PEBP2/CBF. Mol Cell Biol. 1995;15:1651–1661. doi: 10.1128/mcb.15.3.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuoka R, Yoshida M O, Furutani Y, Imamura S, Kanda N, Yanagisawa M, Masaki T, Takao A. Human smooth muscle myosin heavy chain gene mapped to chromosomal region 16q12. Am J Med Genet. 1993;46:61–67. doi: 10.1002/ajmg.1320460110. [DOI] [PubMed] [Google Scholar]
- 22.Meyers S, Lenny N, Hiebert S W. The t(8;21) fusion protein interferes with AML-1B-dependent transcriptional activation. Mol Cell Biol. 1995;15:1974–1982. doi: 10.1128/mcb.15.4.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyoshi H, Kozu T, Shimizu K, Enomoto K, Maseki N, Kaneko Y, Kamada N, Ohki M. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J. 1993;12:2715–2721. doi: 10.1002/j.1460-2075.1993.tb05933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niki M, Okada H, Takano H, Kuno J, Tani K, Hibino H, Asano S, Ito Y, Satake M, Noda T. Hematopoiesis in the fetal liver is impaired by targeted mutagenesis of a gene encoding a non-DNA binding subunit of the transcriptional factor, polyomavirus enhancer binding protein 2/core binding factor. Proc Natl Acad Sci USA. 1997;94:5697–5702. doi: 10.1073/pnas.94.11.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogawa E, Inuzuka M, Maruyama M, Satake M, Naito-Fujimoto M, Ito Y, Shigesada K. Molecular cloning and characterization of PEBP2β, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2α. Virology. 1993;194:314–331. doi: 10.1006/viro.1993.1262. [DOI] [PubMed] [Google Scholar]
- 27.Ogawa E, Maruyama M, Kagoshima H, Inuzuka M, Lu J, Satake M, Shigesada K, Ito Y. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci USA. 1993;90:6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okuda T, van Deursen J, Hiebert S W, Grosveld G, Downing J R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 29.Sakakura C, Yamaguchi-Iwai Y, Satake M, Bae S C, Takahashi A, Ogawa E, Hagiwaga A, Takahashi T, Murakami A, Makino K, Nakagawa T, Kamada N, Ito Y. Growth inhibition and induction of differentiation of t(8;21) acute myeloid leukemia cells by the DNA binding domain of PEBP2 and the AML1/MTG8(ETO) specific antisense oligonucleotide. Proc Natl Acad Sci USA. 1994;91:11723–11727. doi: 10.1073/pnas.91.24.11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasaki K, Yagi H, Bronson R T, Tominaga K, Matsunashi T, Deguchi K, Tani Y, Kishimoto T, Komori T. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc Natl Acad Sci USA. 1996;93:12359–12363. doi: 10.1073/pnas.93.22.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shurtleff S A, Meyers S, Hiebert S W, Raimondi S C, Head D R, Willman C L, Wolman S, Slovak M L, Caroll A J, Behm F, Hulshof M G, Motroni T A, Okuda T, Liu P, Collins F S, Downing J R. Heterogeneity in CBFβ/MYH11 fusion messages encoded by the inv(16)(p13q22) and the t(16;16)(p13;q22) in acute myelogenous leukemia. Blood. 1995;85:3695–3703. [PubMed] [Google Scholar]
- 32.Speck N A, Stacy T. A new transcription factor family associated with human leukemias. Crit Rev Eukaryotic Gene Expr. 1995;5:337–364. doi: 10.1615/critreveukargeneexpr.v5.i3-4.60. [DOI] [PubMed] [Google Scholar]
- 33.Takahashi A, Satake M, Yamagichi-Iwai Y, Bae S C, Lu J, Maruyama M, Zhang Y W, Oka H, Arai N, Arai K, Ito Y. Positive and negative regulation of granulocyte-macrophage colony-stimulating factor promoter activity by AML1-related transcription factor, PEBP2. Blood. 1995;86:607–616. [PubMed] [Google Scholar]
- 34.Takebe Y, Seiki M, Fujisawa J, Hoy P, Yokota K, Arai K, Yoshida M, Arai N. SRα promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Testa U, Grignani F, Barberi T, Fagioli M, Masciulli R, Ferruci P F, Seripa D, Camagna A, Alcalay M, Pelicci P G, Peschle C. PML/RARα+ U937 mutant and NB4 cell lines: retinoic acid restores the monocytic differentiation response to vitamin D3. Cancer Res. 1994;54:4508–4515. [PubMed] [Google Scholar]
- 36.Wang S, Wang Q, Crute B E, Melnikova I N, Keller S R, Speck N A. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol Cell Biol. 1993;13:3324–3339. doi: 10.1128/mcb.13.6.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe A H, Speck N A. Disruption of the cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci USA. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Q, Stacy T, Miller J D, Lewis A F, Gu T-L, Huang X, Bushweller J H, Bories J-C, Alt F W, Ryan G, Liu P P, Wynshaw-Boris A, Binder M, Marin-Padilla M, Sharpe A H, Speck N A. The CBFβ subunit is essential for CBFα2 (AML1) function in vivo. Cell. 1996;87:697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- 39.Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RARα in acute promyelocytic leukemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 40.Yanagisawa K, Horiuchi T, Fujita S. Establishment and characterization of a new human leukemia cell line derived from M4Eo. Blood. 1991;78:451–457. [PubMed] [Google Scholar]
- 41.Yergeau D A, Hetherington C J, Wang Q, Zhang P, Sharpe A H, Binder M, Marin-Padilla M, Tenen D G, Speck N A, Zhang D E. Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1-ETO fusion gene. Nat Genet. 1997;15:303–306. doi: 10.1038/ng0397-303. [DOI] [PubMed] [Google Scholar]
- 42.Zaiman A L, Lewis A F, Crute B E, Speck N A, Lenz J. Transcriptional activity of core binding factor α (AML1) and β subunits on murine leukemia virus enhancer cores. J Virol. 1995;69:2898–2906. doi: 10.1128/jvi.69.5.2898-2906.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang D E, Hetherington C J, Chen H M, Tenen D G. The macrophage transcription factor PU.1 directs tissue-specific expression of the macrophage colony-stimulating factor receptor. Mol Cell Biol. 1994;14:373–381. doi: 10.1128/mcb.14.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang D E, Hetherington C J, Meyers S, Rhoades K L, Larson C J, Chen H M, Hiebert S W, Tenen D G. CCAAT enhancer-binding protein (C/EBP) and AML1 (CBF alpha2) synergistically activate the macrophage colony-stimulating factor receptor promoter. Mol Cell Biol. 1996;16:1231–1240. doi: 10.1128/mcb.16.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y W, Bae S C, Huang G, Fu Y X, Lu J, Ahn M Y, Kanno Y, Kanno T, Ito Y. A novel transcript encoding an N-terminally truncated AML1/PEBP2αB protein interferes with transactivation and blocks granulocytic differentiation of 32Dcl3 myeloid cells. Mol Cell Biol. 1997;17:4133–4145. doi: 10.1128/mcb.17.7.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]