Abstract
Solid tumors are characterized by hypoxic areas, which are prone for macrophage infiltration. Once infiltrated, macrophages polarize to tumor associated macrophages (TAM) to support tumor progression. Therefore, the crosstalk between TAMs and tumor cells is of current interest for the development of novel therapeutic strategies. These may comprise induction of an iron- and lipid peroxidation-dependent form of cell death, known as ferroptosis. To study the macrophage - tumor cell crosstalk we polarized primary human macrophages towards a TAM-like phenotype, co-cultured them with HT1080 fibrosarcoma cells, and analyzed the tumor cell response to ferroptosis induction. In TAMs the expression of ceruloplasmin mRNA increased, which was driven by hypoxia inducible factor 2 and signal transducer and activator of transcription 1. Subsequently, ceruloplasmin mRNA was transferred from TAMs to HT1080 cells via extracellular vesicles. In tumor cells, mRNA was translated into protein to protect HT1080 cells from RSL3-induced ferroptosis. Mechanistically this was based on reduced iron abundance and lipid peroxidation. Interestingly, in naïve macrophages also hypoxia induced ceruloplasmin under hypoxia and a co-culture of HT1080 cells with hypoxic macrophages recapitulated the protective effect observed in TAM co-cultures. In conclusion, TAMs provoke tumor cells to release iron and thereby protect them from lipid peroxidation/ferroptosis.
Keywords: Extracellular vesicles, Iron, HIF-2, STAT1, Hypoxia
Graphical abstract
Highlights
-
•
TAM polarization induces CP RNA.
-
•
CP RNA expression is regulated by HIF-2 and STAT1.
-
•
CP RNA is transferred from TAMs to HT1080 cells.
-
•
CP RNA is translated by HT1080 cells and protects from ferroptosis.
-
•
Co-cultured HT1080 cells decrease iron and lipid peroxidation.
1. Introduction
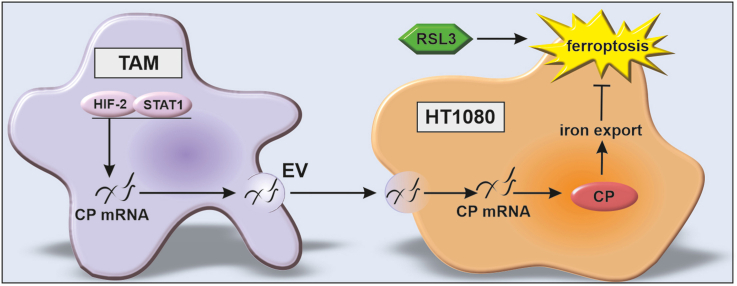
Tumors are characterized by a unique microenvironment including hypoxic regions, which attract various immune cells [1]. Among others, monocytes invade tumors where they differentiate to tumor associated macrophages (TAMs), which are known to promote tumor growth and survival [2]. While the interaction of TAMs with other cells in the tumor is extensively documented, the role of TAMs in affecting tumor cell ferroptosis was rarely studied so far. Ferroptosis is a form of cell death, characterized by increased free iron and lipid peroxidation [3,4]. Lipid peroxidation occurs when ferrous iron (Fe(II)) and H2O2 undergo Fenton chemistry, resulting in hydroxyl radicals, which react with phospholipid hydroperoxides to form phospholipid hydroperoxide radicals. These radicals cause membrane damage and consequently cell death. Since this reaction depends on iron- and lipid metabolism as well as counteracting antioxidant systems, cellular sensitivity is subjected to several regulatory rheostats. Ferritins store cellular iron as Fe(III), thereby limiting the labile iron pool and protecting macrophages from ferroptosis [5]. Depending on their state of polarization macrophages are master regulators of iron homeostasis. Inflammatory macrophages sequester iron, while alternatively polarized macrophages provoke iron release [6]. Various proteins tightly control iron storage (ferritins), uptake (transferrin), and release (ferroportin). Another protein associated with iron release and transport is ceruloplasmin (CP) [7]. CP can be linked to the membrane by a glycosylphosphatidylinositol anchor or can be secreted into the extracellular space, where it functions as a ferroxidase. The oxidation of Fe(II) to Fe(III) constitutes an imported step for transferrin loading and finally iron transport [8]. It appears that CP protects cells from ferroptosis by enhancing iron export [9]. Moreover, iron metabolism is affected by the cellular microenvironment including hypoxia [10]. The response to hypoxia is dominated by signaling of hypoxia inducible factors (HIF), adapting metabolism, i.e. increasing glycolysis, decreasing mitochondrial respiration or affecting iron homeostasis by decreasing iron import (transferrin) and increasing its export (CP and ferroportin) [11]. Iron also acts as cofactor for prolyl hydroxylases (PHD), which degrade HIF. Therefore, iron depletion functionally inactivates PHDs and stabilizes HIF [12]. Hypoxia causes multiple cellular adaptations and hypoxia, respectively HIFs, are shown to either inhibit or facilitate ferroptosis, dependent on the cell type and distinct microenvironments [13]. Most studies addressing ferroptosis, iron metabolism, or hypoxia focus on a single cell, underestimating cellular crosstalk, e.g. between tumor cells and macrophages. Irrespective of the exact model system, it appears of utmost importance to understand the role of macrophages on tumor cell ferroptosis to develop tailored treatment strategies. In the present study we demonstrate such intercellular communication where TAMs express high levels of CP mRNA, which is packed into extracellular vesicles (EVs) and transferred to HT1080 fibrosarcoma cells, where the transcribed protein facilitates iron export and in turn protects HT1080 cells from lipid peroxidation and ferroptosis.
2. Material and methods
2.1. Isolation of primary human macrophages
Primary human macrophages were isolated from Buffy coats using Leucosep tubes (Greiner bio-one, Frickenhausen, Germany) and Biocoll Separating Solution (Biochrom, Berlin, Germany). Cells were washed three times with PBS and were allowed to adhere to 6-well plates, 6 cm dishes, or 48-well plates (Cell+, Sarstedt, Nümbrecht, Germany) for 1 h at 37 °C. Non-adherent cells were removed and remaining monocytes were incubated for at least 7 days with RPMI 1640 medium containing 3% human serum and penicillin/streptomycin. Macrophages were used at a density of approximately 80%. Polarization to tumor associated macrophages was performed by incubating naïve macrophages for 5 days with UV-irradiated MCF-7 cells.
2.2. Cell culture HT1080
HT1080 fibrosarcoma cells were purchased from ATCC and cultured in DMEM medium containing 10% fetal calf serum and penicillin/streptomycin. Cells were seeded 24 h before the experiment to ensure attachment. Co-cultures were performed by seeding and differentiating macrophages on a 6 well plate. HT1080 cells were seeded into transwells (0.4 μm, Sarstedt) and were incubated for 24 h. Afterwards transwells with HT1080 cells were placed on top of macrophages and treated as outlined in the experiments.
2.3. siRNA transfection
Primary human macrophages were incubated in RPMI without human serum 16 h prior transfection. Cells were transfected with 50 nM ON-TARGET plus siRNA against HIF-1α (L-004018-00-0010), HIF-2α (L-004814-00-0020), CP (L-009303-00-0010), and STAT1 (L-003543-00-0005) purchased from Horizon Discovery (Cambridge, UK) using HiPerFect transfection reagent (Qiagen, Hilden, Germany).
2.4. CRISPRa
HT1080 cells were transduced with the CRISPRa lentiviral dCas9-VPR hCMV (Horizon, CAS11914) and after blasticidin selection with 4 different guide RNAs for ceruloplasmin (Horizon, CRISPRmod CRISPRa Human CP Lentiviral sgRNA, GSGH11890-EG1356). Control cells were transduced with a non-targeting guide RNA (Horizon, CRISPRmod CRISPRa lentiviral sgRNA non-targeting controls, GSGC11913). Following transduction, cells were selected with puromycin.
2.5. Treatments
Cells were treated with RSL3 and liproxstatin-1 (both from Cayman Chemicals, Ann Arbor, USA) for indicated times and concentrations. Hypoxic incubations were performed in a SciTive Workstation (Baker Ruskinn, Leeds, UK) at 1% O2 and 5% CO2 for times indicated.
2.6. Western analysis
Cells were lysed in a buffer containing 4% SDS, 150 mM NaCl, and 100 mM Tris/HCl, pH 7.4, and sonicated. For Western analysis of supernatants, cells were incubated in serum free medium for 24 h and proteins were precipitated from the supernatants. Protein content was determined by a protein assay kit (Bio-Rad, Munich, Germany) and 60 μg protein were loaded on a SDS gel. Gels were blotted using a Trans Blot Turbo blotting system (Bio-Rad). Before blocking, membranes were stained using the Revert™ 700 Total Protein Stain kit (Licor, Lincoln, USA) according to manufacturer's advice. Afterwards, membranes were blocked in 5% milk in TBS-T for CP (D7Q5W) (98971S, Cell Signaling, Frankfurt, Germany) and 5% BSA for phospho-Stat1 (Ser727) (D3B7) (8826S, Cell Signaling) and Stat1 (9172, Cell Signaling). Fluorescence signal was detected on an Odyssey scanner (Licor) and quantified with Image Studio Digits 5.0 (Licor). For each lane the lane normalization factor (LNF) was calculated (intensity of a complete lane divided by the intensity of the lane with the maximal intensity) and used for normalization of the signal. Complete pictures of total protein stains are shown in Supplementary Fig. 4.
2.6.1. Real time PCR
RNA was isolated using peqGold (Peqlab, Erlangen, Germany) and measured using a Nanodrop ND-1000 spectrophotometer (Peqlab). Reverse transcription was performed with the Maxima First Strand cDNA Synthesis Kit for RT-PCR (Thermo Fisher Scientific, Waltham, USA). RNA expression of IL-1β, IL-8, IL-10, ALOX15, CP, FPN, FTH, FTL, TfR, STEAP3, PHD2, and Glut1 was analyzed using PowerUp SYBR Green Master Mix (Applied Biosystems, Thermo Fisher Scientific) on a QuantStudio 3 PCR Detection System (Applied Biosystems, Thermo Fisher Scientific) and normalized to TBP or 18S. Primers are listed in Table 1.
Table 1.
List of primers.
| TARGET | FORWARD (5′-3′) | REVERSE (5′-3′) |
|---|---|---|
| ALOX15A | TGGAAGGACGGGTTAATTCTGA | GCGAAACCTCAAAGTCAACTCT |
| CP | CTTTCCTGCTACCCTGTTTGATGC | CTTGCAAACCGGCTTTCAGA |
| FTH | TGTGGCGGAGCTGCTGGGTAA | CGAGAGGTGGATACGGCTGCT |
| FTL | AGCCTTCTTTGTGCGGTCGGGTAA | ACGCCTTCCAGAGCCACATCAT |
| FPN | TGAGCCTCCCAAACCGCTTCCATA | GGGCAAAAAGACTACAACGACGACTT |
| Glut1 | CACACACCCCTGACACACC | ACTCCCACTGCGACTCTGAC |
| IL-1Β | AGAAGTACCTGAGCTCGCCA | CAGGTCCTGGAAGGAGCACT |
| IL-8 | CAGGAATTGAATGGGTTTGC | AGCAGACTAGGGTTGCCAGA |
| IL-10 | AAGCCTGACCACGCTTTCTA | TAGCAGTTAGGAAGCCCCAA |
| PHD2 | GAAAGCCATGGTTGCTTGTT | TTGGGTTCAATGTCAGCAAA |
| STEAP3 | CTGCTGTTGCCACCAAAATGC | TTGGCAAGGCTACTATCGCT |
| TBP | GCATCACTGTTTCTTGGCGT | CGCTGGAACTCGTCTCACTA |
| TfR | GAGCGTCGGGATATCGGGT | CAGGATGAAGGGAGGACACG |
| 18s | GTAACCCGTTGAACCCCATT | CCATCCAATCGGTAGTAGCG |
2.7. Iron determination
Iron amount of cell lysates was assessed using the iron assay kit (ab83366, abcam, Berlin, Germany). Samples were prepared according to manufacturer's instructions and measured on a Tecan spark plate reader (Tecan, Männedorf, Switzerland).
2.8. Ceruloplasmin ELISA
Ceruloplasmin ELISA (G-HUFI00479.96, Assay Genie, Dublin, Ireland) was performed from supernatants of control macrophages and TAMs according to manufacturer's instructions.
2.9. Vitality assay
To analyze viability, cells were stained with CellTiter Blue (Promega, Walldorf, Germany) and incubated for 1 h under cell culture conditions. Afterwards, fluorescence was measured on a Tecan spark plate reader (Tecan).
2.10. Isolation of extracellular vesicles
Extracellular vesicles were isolated as reported in Ref. [14]. Briefly, cells were cultured for 48 h in serum free medium. Cell debris was removed by centrifugation and supernatants were concentrated using a 30 kDa molecular cut-off (Centriprep 30 k filters). Afterwards vesicles were isolated by size exclusion chromatography using Sepharose CL-2B columns. RNA was isolated using the total exosome RNA and protein isolation kit (Invitrogen, Thermo Fisher Scientific).
2.11. Imaging flow cytometry
Lipid peroxidation was analyzed by flow cytometric imaging (Amnis ImageStreamX Mk II, Cytek, Amsterdam, Netherlands). 2 × 106 cells were seeded and stained with 5 μM BODIPY C11 (Invitrogen, Thermo Fisher Scientific), washed and resuspended in PBS containing 2% FCS. Cell Staining of macrophages was performed using CellTracker Green CMFDA dye (Invitrogen, Thermo Fisher Scientific). Macrophages were stained with 0.5 μM CellTracker in PBS. Cells were then washed three times and co-cultured with HT1080 cells. From each sample 100.000 cells, which were identified as single cells and in focus, were measured and used for analyses. Pictures were obtained with 60x magnification. Data were analyzed with IDEAS® Image Analysis Software.
2.12. Statistics
All experiments were performed at least three times. Each biological replicate contains a control and was normalized accordingly. Statistics were performed with GraphPad Prism 10.0.2. Data are expressed as mean values ± SEM. Statistically significant differences were calculated after analysis of variance (ANOVA) and Bonferroni's test or Students t-test; p < 0.05 was considered significant.
3. Results and discussion
3.1. Ceruloplasmin expression in TAMs
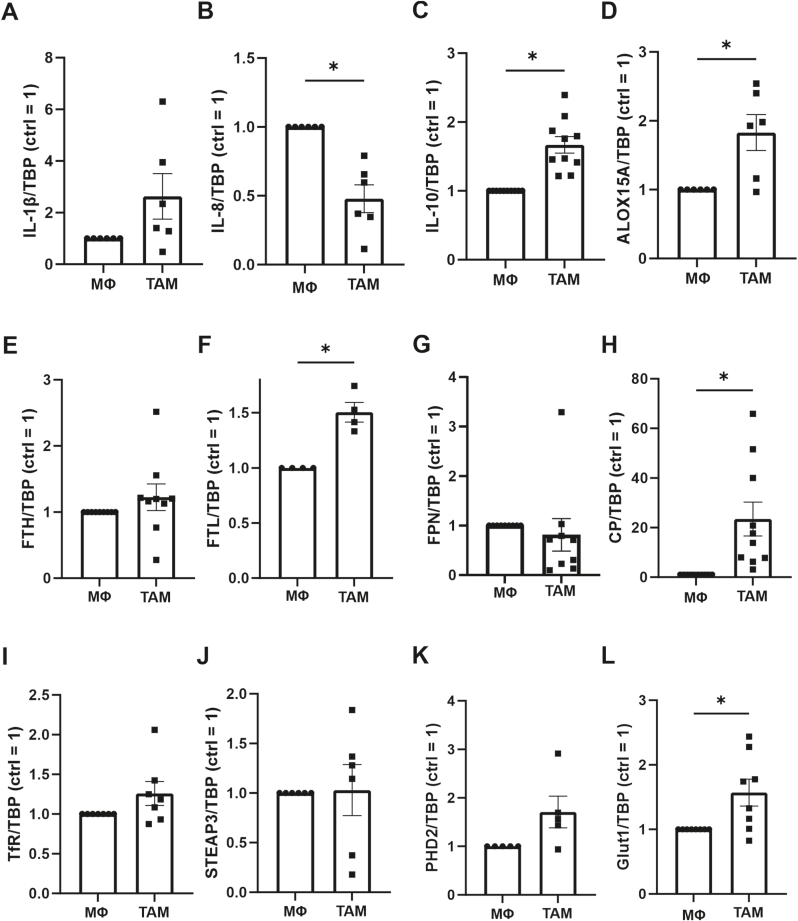
To question the role of tumor associated macrophages (TAM) for tumor cell ferroptosis, we polarized primary human macrophages with MCF-7 cells for 5 days. The resulting TAM-phenotype was characterized by analyzing mRNA expression of interleukin (IL)-1β, IL-8, IL-10, and 15-lipoxygenase (ALOX15A) (Fig. 1A–D). While IL-1β was not significantly altered, IL-8 decreased, while IL-10 as well as ALOX15A significantly increased compared to unstimulated macrophages. This expression pattern is, in accordance with literature, indicative of an alternatively activated, anti-inflammatory, TAM phenotype [15,16]. Since iron metabolism and ferroptosis are closely intertwined and macrophages actively participate in regulating iron homeostasis, we explored mRNAs of genes affecting iron storage or release in TAMs compared to naïve macrophages (Fig. 1E–J). While the mRNA of the iron storage protein ferritin heavy chain (FTH) remained unchanged, ferritin light chain (FTL) mRNA significantly increased. No changes were detected for the iron exporter ferroportin (FPN), the iron import receptor transferrin (TfR), or STEAP3 metalloreductase (STEAP3). However, we noticed a particularly strong induction of ceruloplasmin (CP) mRNA. A slight positive modulation of prolyl hydroxylase (PHD) 2 and a significant increase of solute carrier family 2A1 (glucose transporter 1, Glut1) mRNA pointed towards activation of HIF-mediated transcription. CP is a ferroxidase, which either is released from cells or membrane anchored [17]. The protein promotes iron export and transport and apparently conveys anti-inflammatory properties [18]. Specifically, macrophage-derived CP increased the survival of mice suffering from inflammatory bowel disease [19]. This disease is also antagonized when interfering with lipid peroxidation by liproxstatin, ferrostatin, or vitamin E, suggesting underlying ferroptotic mechanisms [20]. Therefore, we hypothesized that increased CP production by TAMs has an anti-ferroptotic potential.
Fig. 1.
Characterization of tumor associated macrophages Primary human macrophages (MΦ) were incubated for 5 days with apoptotic MCF-7 cells to obtain tumor associated macrophages (TAM). RNA of (A) interleukin (IL)-1β, (B) IL-8, (C) IL-10, (D) 15-lipoxygenase (ALOX15), (E) ferritin heavy chain (FTH), (F) ferritin light chain (FTL), (G) ferroportin (FPN), (H) ceruloplasmin (CP), (I) transferrin receptor (TfR), (J) STEAP3 metalloreductase (STEAP3), (K) prolyl hydroxylase 2 (PHD2), and (L) glucose transporter 1 (Glut1) were analyzed. Data were normalized to TATA box binding protein (TBP) and are expressed as mean values ± SEM, *p ≤ 0.05.
3.2. TAMs reduce ferroptosis of HT1080 cells
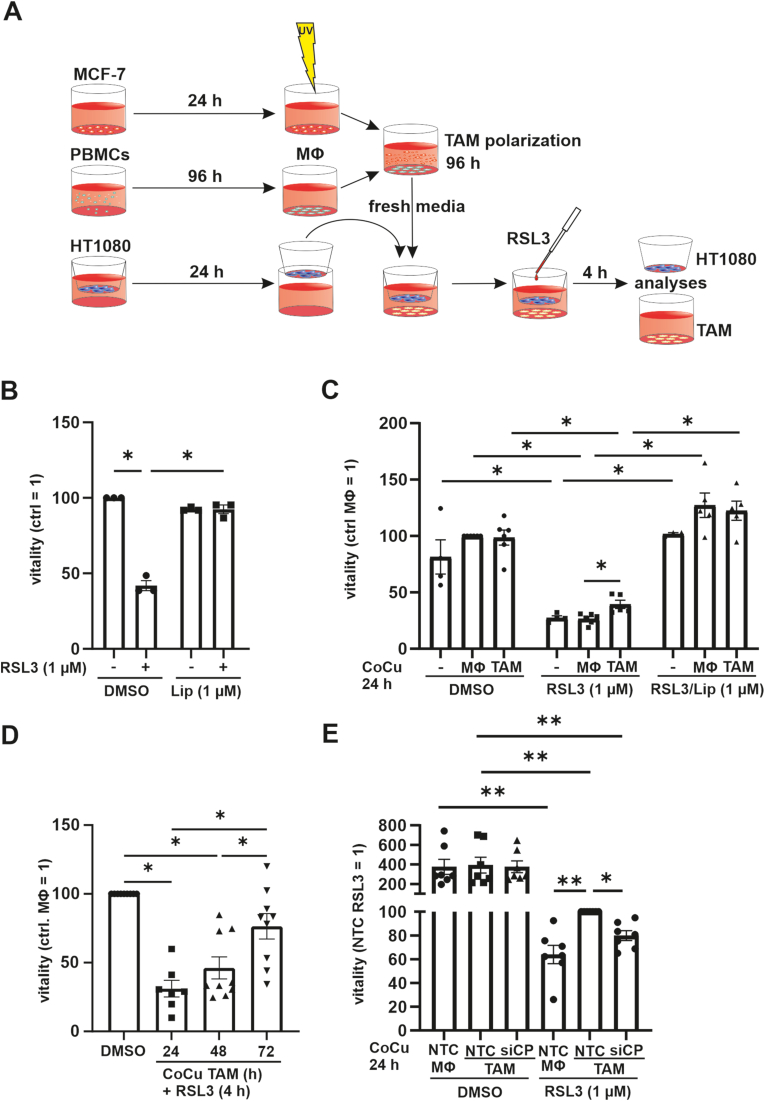
To analyze the impact of TAMs on tumor cell ferroptosis, we polarized macrophages to TAMs and subsequently co-cultured them with HT1080 fibrosarcoma cells using a transwell system. As controls, we either used naïve macrophages in the transwell set up or HT1080 cells alone (Fig. 2 A). In initial experiments incubation of HT1080 cells for 4 h with 1 μM RSL3 substantially induced ferroptosis. This was completely antagonized by 1 μM liproxstatin-1 which interferes with lipid peroxidation and protects cells from ferroptosis (Fig. 2 B). In the following experiments, naïve macrophages or TAMs were co-cultured with HT1080 cells for 24 h in a transwell setting, followed by treatment with RSL3 for 4 h (Fig. 2C). Viability of HT1080 co-cultured with naïve macrophages was reduced to the same extent as in HT1080, while the co-culture of HT1080 cells with TAMs significantly increased the viability of the tumor cells. Liproxstatin-1 protected HT1080 cells, indicating that cell death resulted from ferroptosis. Interestingly, the protective effect of TAM co-culture increased with prolonged co-culture times from 24 to 72 h (Fig. 2 D, Fig. S1 A). To decipher whether increased CP expression in TAMs contributes to enhanced viability of the co-cultured tumor cells, we reduced CP (siCP) expression in TAMs using siRNA technology. Compared to a non-targeting control (NTC) CP mRNA as well as protein were reduced in siCP treated TAMs (Suppl. Fig. 1 B and C). Having validated the knockdown, NTC and siCP treated TAMs were co-cultured with HT1080 cells in the transwell set-up (Fig. 2 E). Viability measurements showed no differences comparing HT1080 cells co-cultured with NTC- or siCP-treated TAMs. Thus, the knockdown of CP in TAMs itself did not effect on viability of naïve HT1080 cells. Compared to DMSO controls, RSL3-treated samples showed reduced viability, indicating effective induction of ferroptosis. Furthermore, HT1080 cells, co-cultured with NTC-treated macrophages were less viable after the addition of RSL3 compared to HT1080 cells co-cultured with TAM, which underscores the initial effect that TAMs protect HT1080 cells from ferroptosis (Fig. 2C). When HT1080 cells were co-cultured with siCP treated TAMs, tumor cell viability was lower compared to the same set up using NTC treated TAMs. Apparently, TAM-derived CP communicates tumor cell protection against ferroptosis.
Fig. 2.
Tumor associated macrophages protect HT1080 cells from ferroptosis
(A) A co-culture system with tumor associated macrophages (TAM) was established. PBMCs were isolated and differentiated for 96 h to macrophages. Afterwards they were incubated with UV irradiated MCF-7 cells for additional 96 h and subsequentially co-cultured with HT1080 cells in a transwell set up and then treated with 1 μM RSL3. (B) HT1080 cells were treated with 1 μM RSL3 and 1 μM liproxstatin-1 (Lip) for 4 h and vitality was measured. (C) HT1080 cells alone or in co-culture with TAMs or naïve macrophages (MΦ) were incubated for 24 h, treated with 1 μM RSL3 and 1 μM Lip for 4 h, and vitality was analyzed. Data were normalized to MΦ control. (D) HT1080 cells were co-cultured with TAMs, incubated for indicated time points, and then treated with 1 μM RSL3 for 4 h. Vitality was analyzed and values were normalized to the corresponding DMSO control. (E) TAMs were generated and transfected with siRNA against ceruloplasmin (siCP) or a non-targeting control (NTC). NTC transfected MΦ served as controls. After 48 h, transfected TAMs and MΦ were co-cultured with HT1080 cells for 24 h, which then were treated with DMSO or RSL3 for 4 h. Vitality was measured and data were normalized to NTC TAMs. All data are expressed as mean values ± SEM, *p ≤ 0.05.
3.3. CP protein expression and release from TAMs
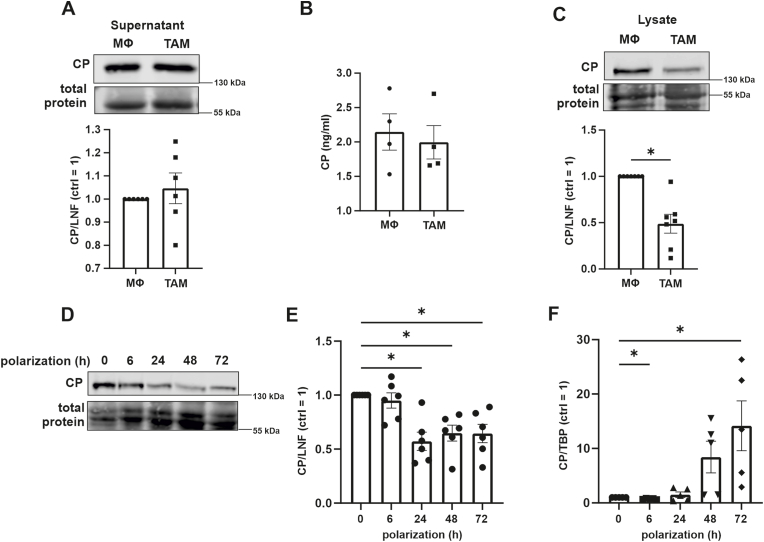
CP is either released from cells or bound to membranes via a glycosylphosphatidylinositol anchor [21]. Since TAMs protect HT1080 cells from ferroptosis without requiring cell-cell contact, we assumed that TAMs secret CP. However, neither Western analysis nor ELISA favored enhanced secretion as the supernatant of naive macrophages and TAMs showed similar levels (Fig. 3 A and B). Albeit, Western analysis of CP in cell lysates indicated a significantly reduced intracellular protein abundance (Fig. 3C). Kinetics of CP expression during TAM polarization revealed a decrease from 24 h onwards (Fig. 3D and E), while CP mRNA started to increase after 24–48 h (Fig. 3 F). Reduced protein expression of CP corroborates previous studies, which identified CP as bona fide target of the IFN-γ-activated inhibitor of translation (GAIT) complex [22]. This complex is known to bind to distinct GAIT elements within the 3′UTR of target mRNAs and blocks translation [23]. Possibly, CP is stable in the supernatant and does not reflect the intra-cellular decrease at the examined time point. Importantly, CP mRNA increased during TAM polarization and we hypothesized that CP mRNA protected tumor cells from ferroptosis.
Fig. 3.
Ceruloplasmin expression by tumor associated macrophagesTumor associated macrophages (TAM) were generated and (A) supernatants were analyzed for ceruloplasmin (CP) by Western analyses and compared to naïve macrophages (MΦ). Data were normalized to lane normalization factor (LNF). (B) Supernatants of MΦ and TAMs were analyzed for CP by ELISA. (C) Cell lysates were analyzed for ceruloplasmin (CP) by Western analyses and compared to naïve macrophages (MΦ). Data were normalized to lane normalization factor (LNF). (D) Primary human macrophages were incubated with apoptotic MCF-7 cells for times indicated and CP protein was measured by Western analyzes. (E) Quantification of (D). (F) Primary human macrophages were incubated with apoptotic MCF-7 cells for times indicated and CP mRNA was measured and normalized to TATA box binding protein (TBP). All data are expressed as mean values ± SEM, *p ≤ 0.05.
3.4. CP mRNA regulation in TAMs
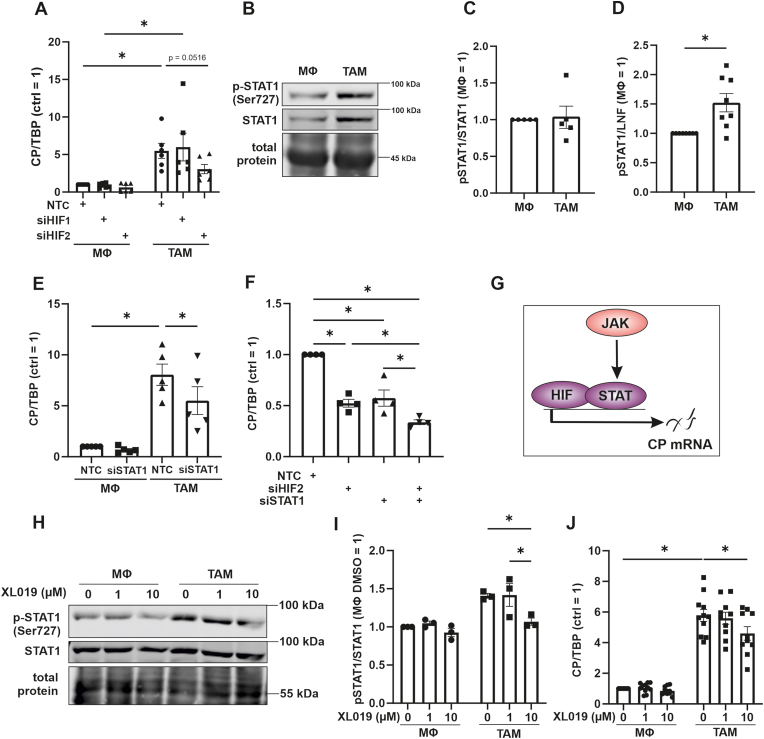
Next, we investigated molecular mechanisms to explain increased CP mRNA expression in TAMs. The presence of hypoxia response elements in the CP promotor is well established [24]. This together with the induction of the putative HIF-target Glut1 in TAMs (Fig. 1L), we questioned the potential role of HIF in this process. HIF is known to be stabilized in TAMs [25,26]. Hereby a knockout of HIF-1α shifted TAM polarization towards a M2 phenotype and attenuated their pro-angiogenetic functions. To explore a role of HIF, we transfected naïve macrophages and TAMs with siRNA against HIF-1α (siHIF1) or HIF-2α (siHIF2) (Suppl. Fig. 1 D and E). While the knockdown of HIF-1α left CP expression unaltered, the knockdown of HIF-2α decreased the CP mRNA amount in TAMs (Fig. 4 A). Interestingly, HIF-2 appears to have a pivotal function in human macrophages and was correlated to angiogenesis in primary invasive breast carcinomas [27]. Studies in primary human macrophages showed that HIF-2 acts by binding to enhancer regions within target genes and thereby facilitates their expression [28,29]. According to the literature, HIF-1α appears to be the major regulator of CP in vitro and in vivo [30]. In contrast, our study suggests that HIF-2 mediates CP expression in macrophages, which points to a cell type-dependent role of HIF-isoforms in inducing CP expression. Nevertheless, HIF-1 was shown to enhance CP transcription and to reduce sensitivity of HCC cells towards ferroptosis and radiotherapy [31]. This study supports the protective role of CP and highlights that the isoform of HIF, which promotes CP expression, can be cell type specific. TAMs are also known to activate Janus kinase (JAK)/signal transducer and activator of transcription 1 (STAT1) [32]. Western analyses indeed revealed that phosphorylated as well as total STAT1 increased in TAMs compared to naïve macrophages (Fig. 4B–D). Despite the fact that the pSTAT1/STAT1 ratio was unaltered, more phosphorylated and thus, active STAT1 characterized TAMs in our system. Knocking down STAT1, significantly decreased CP mRNA, suggesting a regulatory function of not only HIF-2 but also STAT1 (Fig. 4 E and G, Suppl Fig. 1F and G). To elucidate additive effects of HIF-2 and STAT1, we approached a knockdown of both transcription factors (Fig. 4 F). While a single knockdown of either HIF-2α or STAT1 decreased CP mRNA to roughly 50%, the double knockdown lowered CP mRNA abundance further, i.e. to roughly 30%. As the JAK-signaling cascade likely activates STAT1, we inhibited JAK with XL019, which reduced STAT1 phosphorylation, thereby validating the potency of the inhibitor (Fig. 4H and I). Measuring CP mRNA expression in TAMs with JAK being inhibited lowered CP abundance compared to the control, while no effect of the JAK inhibitor was apparent in naïve macrophages. Besides regulation of CP by HIF-2, HIF-2 was also shown to be regulated by CP in lung cancer. Enhanced CP expression increased Fe(III), reduced PHD activity, and finally stabilized HIF-2α [33]. As PHDs are not specific for only one HIF isoform, this mechanism was also observed for HIF-1α in epithelial cells [34]. Our results point to HIF-2 and STAT1 as regulators of CP transcription in TAMs.
Fig. 4.
Ceruloplasmin is regulated by HIF-2 and STAT1 in tumor associated macrophages
(A) Control macrophages (MΦ) and tumor associated macrophages (TAM) were transfected with siRNA against hypoxia inducible factor (HIF)-1α (siHIF1), HIF-2α (siHIF2), or a non-targeting control (NTC). Ceruloplasmin (CP) mRNA was measured and normalized to TATA box binding protein (TBP). (B) Signal transducer and activator of transcription 1 (STAT1) and phosphorylated STAT1 (p-STAT1, Ser727) was analyzed in MΦ and TAMs by Western analyzes. (C) Ratio of pSTAT1 to STAT1. (D) p-STAT1 was quantified and normalized to the lane normalization factor (LNF). (E) MΦ and TAMs were transfected with siRNA against STAT1 (siSTAT1) or NTC. CP mRNA was measured and normalized to TBP. MΦ and TAMs were transfected with siRNA against HIF-2α (siHIF2), STAT1 (siSTAT1) or NTC. CP mRNA was measured and normalized to TBP. (G) CP is regulated by HIF-2 and STAT1. (H) MΦ and TAMs were treated with indicated concentrations of XL019 and incubated for 24 h. Afterwards, STAT1 and p-STAT1 protein was analyzed by Western analysis. (I) Quantification of (H). p-STAT was normalized to total STAT1. (J) MΦ and TAMs were treated with indicated concentrations of XL019, incubated for 24 h and CP mRNA was measured. Data were normalized to TBP. All data are expressed as mean values ± SEM, *p ≤ 0.05.
3.5. TAM-derived CP mRNA is transferred to HT1080 cells and translated into protein
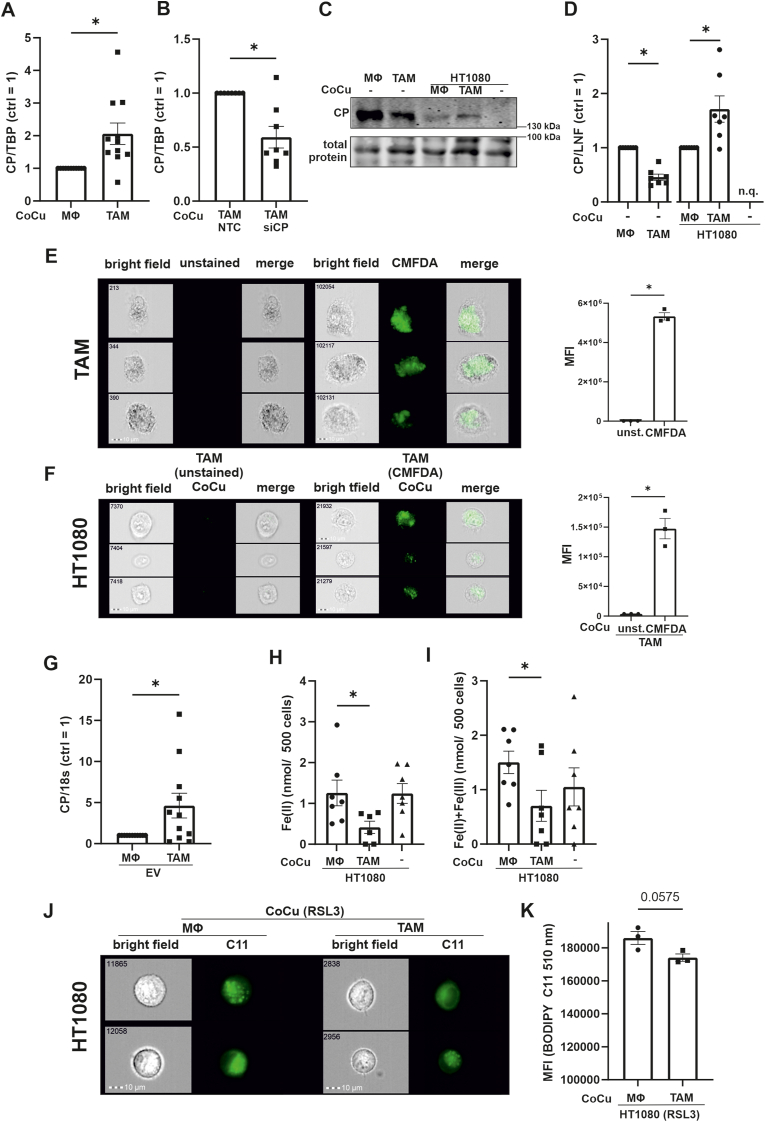
Next, we asked how TAM-derived CP might antagonize ferroptosis in HT1080 cells. To exclude endogenous expression of CP mRNA in HT1080 cells, we measured CP mRNA abundance and detected CT values of around 35, indicating that CP is expressed in tumor cells at very low level, being virtually absent (Suppl. Fig. 2). We then used the transwell set-up to co-culture HT1080 cells with naïve macrophages or TAMs for 24 h and analyzed CP mRNA (Fig. 5 A). HT1080 tumor cells co-cultured with TAMs contained significantly more CP mRNA compared to those in the transwell set-up with naïve macrophages. To support the idea of mRNA, transfer from TAMs to tumor cells, we used siRNA to reduce CP mRNA in TAMs and co-cultured these cells with HT1080 (Fig. 5 B). A knockdown of CP in TAMs significantly lowered CP mRNA abundance in tumor cells compared to NTC controls. Following that, we analyzed if CP mRNA was translated into protein (Fig. 5C and D), again using naïve macrophages and TAMs in the transwell set-up with HT1080 cells. As seen before (Fig. 4 A), TAMs express less CP protein compared to naïve macrophages. Protein expression in HT1080 cells was significantly higher when co-cultured with TAMs compared to control macrophages, while HT1080 cells alone did not express CP protein at all. We suggest that CP mRNA is transferred from TAMs to HT1080 cells, which then translate CP mRNA into protein.
Fig. 5.
Ceruloplasmin is transferred from TAMs to HT1080 cells
(A) HT1080 cells were incubated with naïve macrophages (MΦ) and tumor associated macrophages (TAM) in a transwell system and mRNA was isolated from HT1080 cells. Ceruloplasmin (CP) mRNA was measured and normalized to TATA box binding protein (TBP). (B) TAMs were transfected with siRNA against CP (siCP) or a non-targeting control (NTC) and co-cultured with HT1080 cells. mRNA was isolated from HT1080 cells and analyzed for CP, which was normalized to TBP. (C) MΦ and TAMs as well as HT1080 cells, either alone or co-cultured with MΦ respectively TAMs, were analyzed for CP protein expression by Western analyzes. (D) Blots from (C) were quantified and data were normalized to lane normalization factor (LNF). CP expression in HT1080 cells alone could not be quantified (n.q.). (E) TAMs were generated and stained with Cell Tracker CMFDA dye. Cells were measured by FACS-imaging. Mean fluorescence intensity (MFI) was compared between unstained and stained TAMs. Each dot in the graph represents the mean of 100.000 single cells. Pictures of three representative cells are shown. Numbers included in the picture indicate the event count of a flow cytometer. (F) HT1080 cells were co-cultured with unstained and Cell Tracker CMFDA stained TAMs for 24 h and measured by FACS imaging. Mean fluorescence intensity (MFI) was compared between HT1080 cells co-cultured with unstained and stained TAMs. The graph was composed as described for (E). (G) Extracellular vesicles (EV) were isolated from MΦ and TAM supernatants and analyzed for CP mRNA content. CP mRNA was normalized to 18s RNA (18s). (H and I) Iron was measured in HT1080 cells, which were co-cultured with MΦ or TAMs and in HT1080 cells alone. (H) shows Fe(II) and (I) total iron. (J) HT1080 cells were co-cultured with MΦ and TAMs and treated with RSL3 for 4 h. Afterwards, HT1080 cells were stained with BODIPY C11 and analyzed by FACS imaging. Pictures of two representative cells are shown. The numbers indicate the event count of a flow cytometer. (K) MFI was compared between RSL3 treated HT1080 cells after co-culture with MΦ and TAMs. The graph was composed as described for (E). All data are expressed as mean values ± SEM, *p ≤ 0.05.
3.6. TAMs deliver CP mRNA to HT1080 cells via extracellular vesicles
To search for potential CP mRNA transport mechanisms from TAMs to tumor cells, we stained TAMs with CellTracker Green CMFDA, as validated by flow cytometric imaging (Fig. 5 E). The dye stains extra- and intracellular membranes. TAMs showed a dominant intracellular staining, likely including the ER, which is important for vesicle production. After washing TAMs and replacing media, we seeded HT1080 cells into transwells and co-cultured them with CMFDA-stained TAMs for 24 h (Fig. 5 F). HT1080 cells remained unstained when co-cultured with unlabeled TAMs, but became CMFDA positive upon their co-culture with labelled TAMs. Apparently, the dye was transferred from TAMs to HT1080 cells, which opens up the possibility for the transfer of proteins and/or mRNA between these cells. With these observations in mind, we hypothesize that TAMs release CP mRNA, which is taken up by HT1080 cells. We analyzed CP mRNA abundance in extracellular vesicles (EV), following EV isolation by size exclusion chromatography as described by Baer et al. [14]. CP mRNA amount in EV from TAMs was significantly higher compared to EV from control macrophages (Fig. 5 G). While the transport of the membrane bound form of CP in vesicles was reported [35], the transfer of CP mRNA from TAMs to tumor cells including its translation has so far not been described. We propose the transfer of EVs packed with CP mRNA generated by TAMs to HT1080 cells, which subsequently translate mRNA to CP protein.
3.7. TAMs decrease iron and lipid peroxidation of HT1080 cells
Since CP facilitates iron export, we measured iron in HT1080 cells alone or in the transwell set-up with macrophages/TAMs (Fig. 5H and I) [36]. HT1080 cells alone or in co-culture with macrophages showed the same iron content, while TAMs significantly reduced total iron as well as reactive Fe2+ in co-cultured tumor cells. We anticipate that reduced intracellular iron in HT1080 results from enhanced export due to higher CP protein expression. CP facilitates iron export, which may explain viability data shown in Fig. 2 E, considering that Fe2+ in the labile iron pool executes Fenton reaction and promotes ferroptosis [37]. Marques and co-workers showed that various cell types express glycosylphosphatidylinositol-anchored CP in lipid rafts within the membrane in close proximity to ferroportin [38], supporting a role of CP in iron export. Moreover, CP was also shown to facilitate iron transport by transferrin [39]. Protective capabilities of CP were proposed by a study showing that CP supplementation prevented ferroptosis upon ischemic stroke by elevating iron export and thus, reducing the labile iron pool in mice brain [37]. Systemically, mutations in the CP gene increased serum and brain lipid peroxidation [40,41]. These data suggest an antioxidative function of CP by preventing iron mediated oxidative stress [42]. To substantiate this idea, we analyzed lipid peroxidation, another hallmark of ferroptosis (Fig. 5 J and K). HT1080 cells, co-cultured in the transwell set-up with TAMs and treated with RSL3, showed less lipid peroxidation compared to their co-culture with naïve macrophages. RSL3 induced lipid peroxidation was quantified with BODIPY C11 via imaging flow cytometry and supported our assumption that CP delivered by TAMs protects HT1080 cells from ferroptosis.
3.8. Overexpression of CP in HT1080 cells
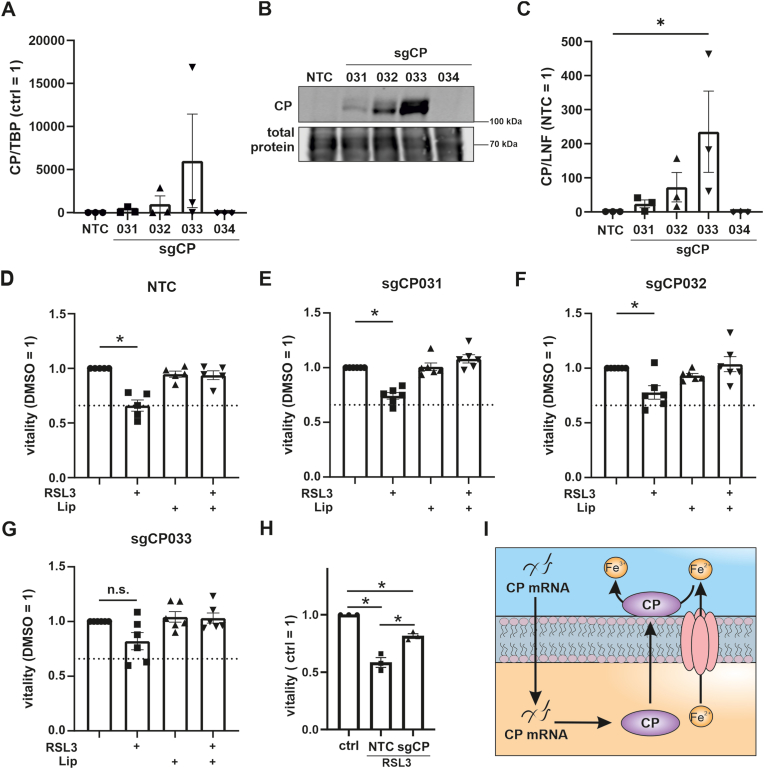
To reassure the anti-ferroptotic function of CP we expressed CP in HT1080 cells using CRISPRa technology. First cells were transduced to stably express dCas9 VPR, an inactive Cas9, which was fused to the transcriptional activator VP64, transcription factor p65, and the replication and transcription activator (Rta). Cells were then transduced with vectors containing 4 guide RNAs, which facilitate translocation of the dCas9 VPR to the promotor of CP (sgCP), to enhance its endogenous transcription. First, we validated expression of CP at mRNA and protein level. CP mRNA was elevated in sgCP031, 032 and 033 transduced cells, while sgCP034 showed no induction (Fig. 6 A). This pattern was reflected at protein level (Fig. 6 B and C). While sgCP031 and 032 evoked low CP expression, sgCP033 significantly enhanced it. As CP was not expressed with sg034 the clone was excluded from further analyses. We went on to analyze viability of NTC, sgCP031, 032, and 033 transfected cells (Fig. 6D–G). Viability of NTC cells decreased significantly when treated with RSL3. Although viability of sgCP031 and 032 also decreased, viability was higher compared to NTC and correlated with CP protein expression. Viability of sgCP033 cells, showing the highest CP expression, was not reduced by RSL3, underscoring a protective effect of CP expression. To explore a combinatory effect of all three sgRNAs we analyzed mean data of all experiments (every dot represents one of the three sgRNAs) and compared RSL3-treated NTC vs. sgRNA cells (Fig. 6H). Analyzing data in this way indicated that sgCP-treated cells were significantly less sensitive towards ferroptosis, underscoring the protective impact of CP against ferroptosis (Fig. 6 I). These results corroborate literature data that ferroptosis antagonizes function of CP when being overexpressed in hepatocellular carcinoma cell lines [9]. A crucial role of CP also became apparent when its overexpression reduced migration, invasion, and survival of nasopharyngeal carcinoma cells [43].
Fig. 6.
Overexpression of ceruloplasmin in HT1080 cellsHT1080 cells were stably transduced with a vector expressing dCas9 VPR. After selection dCas9 VPR expressing cells they were transduced with control guide RNA (NTC) and 4 guide RNAs (sg) targeting the ceruloplasmin (CP) promotor region (sgCP 031, 032, 033, 034). (A) CP mRNA was analyzed and normalized to TATA box binding protein (TBP). (B) CP protein was measured by Western analyzes and (C) quantified. Data were normalized to the lane normalization factor. (D–G) NTC, sgCP031, 032, 033, and 034 transduced HT1080 cells were treated with RSL3 and liproxstatin-1 (Lip) and vitality was measured. (H) CP overexpressing and NTC HT1080 cells were stimulated with RSL3. Each dot represents the mean of n = 5 for one single guide RNA. Data were normalized to RSL3 treated NTC. (I) Proposed mechanism for CP-mediated protection against ferroptosis.
3.9. Hypoxic macrophages protect tumor cells from ferroptosis
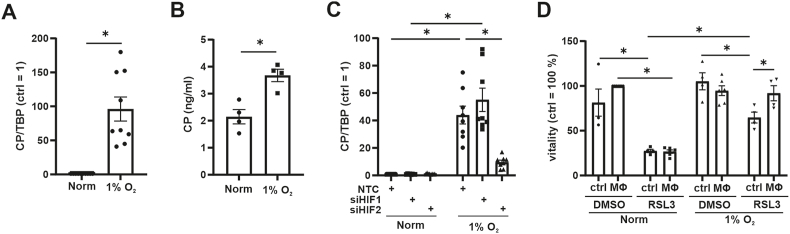
Our data suggest that HIF-2 regulates CP in TAMs. As macrophages invade hypoxic regions of tumors, we asked whether the lack of oxygen in macrophages causes CP expression in sufficient amounts to protect tumor cells from ferroptosis. The relevance of CP in the cancer context became obvious in invasive breast carcinoma, which is correlated with infiltration of immune cells, among others also macrophages [44]. Interestingly, CP expression is discussed as a prognostic marker [45,46], with low CP abundance correlating with a favorable patient prognosis. To explore the link between hypoxia and CP expression we incubated naïve macrophages for 24 h under hypoxia (1% O2) followed by CP mRNA analysis (Fig. 7 A). Hypoxia significantly increased CP mRNA abundance in naïve macrophages. In contrast, protein expression was not significantly altered by hypoxia (Suppl. Fig. 3 A and B) but protein secretion increased (Fig. 7 B). To test whether HIF-1 or HIF-2 mediates CP expression under hypoxia, we used siRNA for both and noticed a significant decrease in CP expression only in HIF-2α knockdown cells (Fig. 7C). Having established that hypoxia provokes a CP mRNA increased in naïve macrophages, we postulated that these cells should protect HT1080 cells from ferroptosis. Using the established transwell co-culture system under hypoxia, RSL3 reduced viability of HT1080 (Fig. 7 D). As reported previously, hypoxia protected HT1080 cells from ferroptosis [5]. Co-culture of HT1080 cells with hypoxic macrophages antagonized ferroptosis as observed for co-cultures with TAMs. Conclusively, a physiological stimulus such as hypoxia, which modulates inter-cellular communication, suffices to induce CP, which in turn protects tumor cells from apoptosis. Indeed, hypoxia was shown to protect from ferroptosis in various cell types. Hypoxia via HIF-1 increased the expression of SLC7A11 in glioblastoma cells, which suppressed the induction of ferroptosis by sulfasalazine [47]. In hepatocellular carcinoma cells HIF-1 suppressed methyltransferase 14, which blocked SLC7A11 expression [48]. These data were supported by a study in HSC-T6 cells, which were protected from ferroptosis by a HIF-1/SLC7A11 axis upon sorafinib-treatment [49]. These studies show that hypoxia appears to protect cells by intra-cellular signaling cascades involving SLC7A11. Our data extend these findings by showing that the secretome of hypoxic macrophages protect tumor cells from ferroptosis. These data indicate that macrophages, which reside in hypoxic areas of a tumor, may protect surrounding tumor cells from ferroptosis.
Fig. 7.
Macrophages protect HT1080 cell from ferroptosis under hypoxia
(A-D) Macrophages (MΦ) were incubated for 24 h under hypoxia (1% O2). (A) Ceruloplasmin (CP) mRNA was measured and normalized to TATA box binding protein (TBP). (B) Supernatants of MΦ were analyzed for CP by ELISA. (C) MΦ were transfected with siRNA against hypoxia inducible factor (HIF)-1α (siHIF1), HIF-2α (siHIF2), or a non-targeting control (NTC). Afterwards cells were incubated for 24 h under hypoxia. CP mRNA was measured and normalized to TBP. (D) MΦ were co-cultured with HT1080 cells for 24 h under hypoxia and then treated with RSL3 for 4 h. As a control HT1080 cells alone were incubated under hypoxia. Vitality was measured and data were normalized to DMSO control. All data are expressed as mean values ± SEM, *p ≤ 0.05.
4. Conclusion
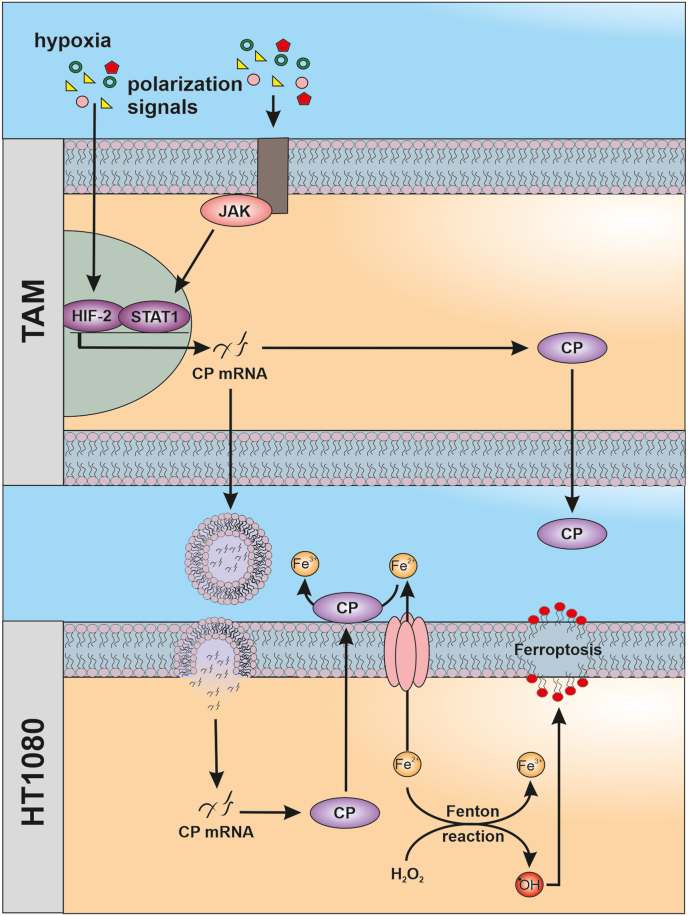
Our study aimed at understanding the impact of macrophages on tumor cell ferroptosis (Fig. 8). Therefore, we established a transwell co-culture set-up using primary human macrophages and HT1080. Macrophages, polarized towards a TAM phenotype, efficiently protected HT1080 from RSL3 induced ferroptosis compared to naïve macrophages. Mechanistically, TAM-derived CP mRNA was transferred to HT1080 cells and there translated into protein. As a result, iron content and lipid peroxidation after RSL3 treatment of HT1080 cells were reduced. In hypoxic macrophages, the TAM phenotype with regard to CP formation and function was phenocopied. In conclusion, macrophages polarized to tumor-associated macrophages in the tumor microenvironment and/or macrophages in a hypoxic environment counteract the ability of tumor cells to undergo ferroptosis. This study highlights the role of CP in the cross-talk of macrophages with tumor cells and adds to the multitude of pathways determining ferroptosis.
Fig. 8.
Scheme of the proposed mechanismCeruloplasmin (CP) mRNA is expressed in tumor associated macrophages (TAM) dependent on hypoxia inducible factor 2 (HIF-2) and Janus kinase (JAK)/signal transducer and activator of transcription 1 (STAT1). CP mRNA is packed into extracellular vesicles and transferred to HT1080 cells were mRNA is translated into protein to facilitates iron export. Thereby, ferroptosis sensitivity of HT1080 cells was decreased likely by lowering iron-mediated production of hydroxyl radicals (•OH) and lipid peroxidation.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation), project 461431951 (SPP 2306) and SFB 1039/TP 04.
CRediT authorship contribution statement
Anna Schwantes: Investigation, Data curation. Anja Wickert: Investigation. Sabrina Becker: Investigation, Formal analysis, Data curation. Patrick C. Baer: Methodology, Investigation. Andreas Weigert: Writing – review & editing, Conceptualization. Bernhard Brüne: Writing – review & editing, Funding acquisition, Conceptualization. Dominik C. Fuhrmann: Writing – original draft, Project administration, Funding acquisition, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
We thank Tanja Keppler for excellent technical assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2024.103093.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Sormendi S., Wielockx B. Hypoxia pathway proteins as central mediators of metabolism in the tumor cells and their microenvironment. Front. Immunol. 2018;9:40. doi: 10.18632/oncotarget.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Y., Zhao L., Li X.-F. Hypoxia and the tumor microenvironment. Technol. Cancer Res. Treat. 2021;20 doi: 10.1177/15330338211036304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stockwell B.R. Ferroptosis turns 10: emerging mechanisms, physiological functions, and therapeutic applications. Cell. 2022;185:2401–2421. doi: 10.1016/j.cell.2022.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuhrmann D.C., Brüne B. A graphical journey through iron metabolism, microRNAs, and hypoxia in ferroptosis. Redox Biol. 2022;54 doi: 10.1016/j.redox.2022.102365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuhrmann D.C., Mondorf A., Beifuß J., Jung M., Brüne B. Hypoxia inhibits ferritinophagy, increases mitochondrial ferritin, and protects from ferroptosis. Redox Biol. 2020;36 doi: 10.1016/j.redox.2020.101670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung M., Mertens C., Brüne B. Macrophage iron homeostasis and polarization in the context of cancer. Immunobiology. 2015;220:295–304. doi: 10.1016/j.imbio.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 7.de Silva D.M., Aust S.D. Ferritin and ceruloplasmin in oxidative damage: review and recent findings. Can. J. Physiol. Pharmacol. 1993;71:715–720. doi: 10.1139/y93-107. [DOI] [PubMed] [Google Scholar]
- 8.Frazer D.M., Anderson G.J. The regulation of iron transport. Biofactors. 2014;40:206–214. doi: 10.1002/biof.1148. [DOI] [PubMed] [Google Scholar]
- 9.Shang Y., Luo M., Yao F., Wang S., Yuan Z., Yang Y. Ceruloplasmin suppresses ferroptosis by regulating iron homeostasis in hepatocellular carcinoma cells, Cell. Signal. 2020;72 doi: 10.1016/j.cellsig.2020.109633. [DOI] [PubMed] [Google Scholar]
- 10.Haase V.H. Hypoxic regulation of erythropoiesis and iron metabolism. Am. J. Physiol. Ren. Physiol. 2010;299:F1–F13. doi: 10.1152/ajprenal.00174.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peyssonnaux C., Nizet V., Johnson R.S. Role of the hypoxia inducible factors HIF in iron metabolism. Cell Cycle. 2008;7:28–32. doi: 10.4161/cc.7.1.5145. [DOI] [PubMed] [Google Scholar]
- 12.Baek J.H., Reiter C.E.N., Manalo D.J., Buehler P.W., Hider R.C., Alayash A.I. Induction of hypoxia inducible factor (HIF-1α) in rat kidneys by iron chelation with the hydroxypyridinone, CP94. Biochim. Biophys. Acta. 2011;1809:262–268. doi: 10.1016/j.bbagrm.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 13.Zheng X., Liang Y., Zhang C. Ferroptosis regulated by hypoxia in cells. Cells. 2023;12 doi: 10.3390/cells12071050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geßner A., Koch B., Klann K., Fuhrmann D.C., Farmand S., Schubert R., Münch C., Geiger H., Baer P.C. Characterization of extracellular vesicles from preconditioned human adipose-derived stromal/stem cells. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22062873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han S., Wang W., Wang S., Yang T., Zhang G., Wang Di, Ju R., Lu Y., Wang H., Wang L. Tumor microenvironment remodeling and tumor therapy based on M2-like tumor associated macrophage-targeting nano-complexes. Theranostics. 2021;11:2892–2916. doi: 10.7150/thno.50928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weigert A., Strack E., Snodgrass R.G., Brüne B. mPGES-1 and ALOX5/-15 in tumor-associated macrophages. Cancer Metastasis Rev. 2018;37:317–334. doi: 10.1007/s10555-018-9731-3. [DOI] [PubMed] [Google Scholar]
- 17.Banha J., Marques L., Oliveira R., Martins M.d.F., Paixão E., Pereira D., Malhó R., Penque D., Costa L. Ceruloplasmin expression by human peripheral blood lymphocytes: a new link between immunity and iron metabolism. Free Radic. Biol. Med. 2008;44:483–492. doi: 10.1016/j.freeradbiomed.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 18.Musci G., Polticelli F., Di Bonaccorsi Patti M.C. Ceruloplasmin-ferroportin system of iron traffic in vertebrates, World. J. Biol. Chem. 2014;5:204–215. doi: 10.4331/wjbc.v5.i2.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bakhautdin B., Febbraio M., Goksoy E., de La Motte C.A., Gulen M.F., Childers E.P., Hazen S.L., Li X., Fox P.L. Protective role of macrophage-derived ceruloplasmin in inflammatory bowel disease. Gut. 2013;62:209–219. doi: 10.1136/gutjnl-2011-300694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ocansey D.K.W., Yuan J., Wei Z., Mao F., Zhang Z. Role of ferroptosis in the pathogenesis and as a therapeutic target of inflammatory bowel disease. Int. J. Mol. Med. 2023;51 doi: 10.3892/ijmm.2023.5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hellman N.E., Gitlin J.D. Ceruloplasmin metabolism and function. Annu. Rev. Nutr. 2002;22:439–458. doi: 10.1146/annurev.nutr.22.012502.114457. [DOI] [PubMed] [Google Scholar]
- 22.Sampath P., Mazumder B., Seshadri V., Fox P.L. Transcript-selective translational silencing by gamma interferon is directed by a novel structural element in the ceruloplasmin mRNA 3' untranslated region. Mol. Cell Biol. 2003;23:1509–1519. doi: 10.1128/MCB.23.5.1509-1519.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arif A., Yao P., Terenzi F., Jia J., Ray P.S., Fox P.L. The GAIT translational control system. Wiley Interdiscip Rev RNA. 2018;9 doi: 10.1002/wrna.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukhopadhyay C.K., Mazumder B., Fox P.L. Role of hypoxia-inducible factor-1 in transcriptional activation of ceruloplasmin by iron deficiency. J. Biol. Chem. 2000;275:21048–21054. doi: 10.1074/jbc.M000636200. [DOI] [PubMed] [Google Scholar]
- 25.Werno C., Menrad H., Weigert A., Dehne N., Goerdt S., Schledzewski K., Kzhyshkowska J., Brune B. Knockout of HIF-1alpha in tumor-associated macrophages enhances M2 polarization and attenuates their pro-angiogenic responses. Carcinogenesis. 2010;31:1863–1872. doi: 10.1093/carcin/bgq088. [DOI] [PubMed] [Google Scholar]
- 26.Susen R.M., Bauer R., Olesch C., Fuhrmann D.C., Fink A.F., Dehne N., Jain A., Ebersberger I., Schmid T., Brüne B. Macrophage HIF-2α regulates tumor-suppressive Spint1 in the tumor microenvironment. Mol. Carcinog. 2019;58:2127–2138. doi: 10.1002/mc.23103. [DOI] [PubMed] [Google Scholar]
- 27.Leek R.D., Talks K.L., Pezzella F., Turley H., Campo L., Brown N.S., Bicknell R., Taylor M., Gatter K.C., Harris A.L. Relation of hypoxia-inducible factor-2 alpha (HIF-2 alpha) expression in tumor-infiltrative macrophages to tumor angiogenesis and the oxidative thymidine phosphorylase pathway in Human breast cancer. Cancer Res. 2002;62:1326–1329. [PubMed] [Google Scholar]
- 28.Tausendschon M., Rehli M., Dehne N., Schmidl C., Doring C., Hansmann M.L., Brune B. Genome-wide identification of hypoxia-inducible factor-1 and -2 binding sites in hypoxic human macrophages alternatively activated by IL-10. Biochim. Biophys. Acta. 2015;1849:10–22. doi: 10.1016/j.bbagrm.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 29.Fuhrmann D.C., Tausendschon M., Wittig I., Steger M., Ding M.G., Schmid T., Dehne N., Brune B. Inactivation of tristetraprolin in chronic hypoxia provokes the expression of cathepsin B. Mol. Cell Biol. 2015;35:619–630. doi: 10.1128/mcb.01034-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin F., Linden T., Katschinski D.M., Oehme F., Flamme I., Mukhopadhyay C.K., Eckhardt K., Tröger J., Barth S., Camenisch G., Wenger R.H. Copper-dependent activation of hypoxia-inducible factor (HIF)-1: implications for ceruloplasmin regulation. Blood. 2005;105:4613–4619. doi: 10.1182/blood-2004-10-3980. [DOI] [PubMed] [Google Scholar]
- 31.Yang M., Wu X., Hu J., Wang Y., Wang Y., Zhang L., Huang W., Wang X., Li N., Liao L., Chen M., Xiao N., Dai Y., Liang H., Huang W., Yuan L., Pan H., Li L., Chen L., Liu L., Liang L., Guan J. COMMD10 inhibits HIF1α/CP loop to enhance ferroptosis and radiosensitivity by disrupting Cu-Fe balance in hepatocellular carcinoma. J. Hepatol. 2022;76:1138–1150. doi: 10.1016/j.jhep.2022.01.009. [DOI] [PubMed] [Google Scholar]
- 32.Kusmartsev S., Gabrilovich D.I. STAT1 signaling regulates tumor-associated macrophage-mediated T cell deletion. J. Immunol. 2005;174:4880–4891. doi: 10.4049/jimmunol.174.8.4880. [DOI] [PubMed] [Google Scholar]
- 33.Tsai Y.-M., Wu K.-L., Chang Y.-Y., Chang W.-A., Huang Y.-C., Jian S.-F., Tsai P.-H., Lin Y.-S., Chong I.-W., Hung J.-Y., Hsu Y.-L. Loss of miR-145-5p causes ceruloplasmin interference with PHD-iron Axis and HIF-2α stabilization in lung adenocarcinoma-mediated angiogenesis. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21145081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harned J., Ferrell J., Nagar S., Goralska M., Fleisher L.N., McGahan M.C. Ceruloplasmin alters intracellular iron regulated proteins and pathways: ferritin, transferrin receptor, glutamate and hypoxia-inducible factor-1α. Exp. Eye Res. 2012;97:90–97. doi: 10.1016/j.exer.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gangoda L., Liem M., Ang C.-S., Keerthikumar S., Adda C.G., Parker B.S., Mathivanan S. Proteomic profiling of exosomes secreted by breast cancer cells with varying metastatic potential. Proteomics. 2017;17 doi: 10.1002/pmic.201600370. [DOI] [PubMed] [Google Scholar]
- 36.Olivieri S., Conti A., Iannaccone S., Cannistraci C.V., Campanella A., Barbariga M., Codazzi F., Pelizzoni I., Magnani G., Pesca M., Franciotta D., Cappa S.F., Alessio M. Ceruloplasmin oxidation, a feature of Parkinson's disease CSF, inhibits ferroxidase activity and promotes cellular iron retention. J. Neurosci. 2011;31:18568–18577. doi: 10.1523/JNEUROSCI.3768-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tuo Q.-Z., Lei P., Jackman K.A., Li X.-L., Xiong H., Liuyang Z.-Y., Roisman L., Zhang S.-T., Ayton S., Wang Q., Crouch P.J., Ganio K., Wang X.-C., Pei L., Adlard P.A., Lu Y.-M., Cappai R., Wang J.-Z., Liu R., Bush A.I. Tau-mediated iron export prevents ferroptotic damage after ischemic stroke. Mol. Psychiatr. 2017;22:1520–1530. doi: 10.1038/mp.2017.171. [DOI] [PubMed] [Google Scholar]
- 38.Marques L., Auriac A., Willemetz A., Banha J., Silva B., Canonne-Hergaux F., Costa L. Immune cells and hepatocytes express glycosylphosphatidylinositol-anchored ceruloplasmin at their cell surface. Blood Cells Mol. Dis. 2012;48:110–120. doi: 10.1016/j.bcmd.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 39.Eid C., Hémadi M., Ha-Duong N.-T., El Hage Chahine J.-M. Iron uptake and transfer from ceruloplasmin to transferrin. Biochim. Biophys. Acta. 2014;1840:1771–1781. doi: 10.1016/j.bbagen.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 40.Miyajima H., Takahashi Y., Serizawa M., Kaneko E., Gitlin J.D. Increased plasma lipid peroxidation in patients with aceruloplasminemia. Free Radic. Biol. Med. 1996;20:757–760. doi: 10.1016/0891-5849(95)02178-7. [DOI] [PubMed] [Google Scholar]
- 41.Yoshida K., Kaneko K., Miyajima H., Tokuda T., Nakamura A., Kato M., Ikeda S. Increased lipid peroxidation in the brains of aceruloplasminemia patients. J. Neurol. Sci. 2000;175:91–95. doi: 10.1016/s0022-510x(00)00295-1. [DOI] [PubMed] [Google Scholar]
- 42.Di Bonaccorsi Patti M.C. The ferroportin-ceruloplasmin system and the mammalian iron homeostasis machine: regulatory pathways and the role of lactoferrin. Biometals. 2018:1–16. doi: 10.1007/s10534-018-0087-5. [DOI] [PubMed] [Google Scholar]
- 43.Yang H., Bao Y., Jin F., Jiang C., Wei Z., Liu Z., Xu Y. Ceruloplasmin inhibits the proliferation, migration and invasion of nasopharyngeal carcinoma cells and is negatively regulated by miR-543. Nucleos Nucleot. Nucleic Acids. 2022;41:474–488. doi: 10.1080/15257770.2022.2052314. [DOI] [PubMed] [Google Scholar]
- 44.Chen F., Han B., Meng Y., Han Y., Liu B., Zhang B., Chang Y., Cao P., Fan Y., Tan K. Ceruloplasmin correlates with immune infiltration and serves as a prognostic biomarker in breast cancer. Aging (Albany NY) 2021;13:20438–20467. doi: 10.18632/aging.203427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han I.W., Jang J.-Y., Kwon W., Park T., Kim Y., Lee K.B., Kim S.-W. Ceruloplasmin as a prognostic marker in patients with bile duct cancer. Oncotarget. 2017;8:29028–29037. doi: 10.18632/oncotarget.15995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manjula S., Aroor A.R., Raja A., Rao S.N., Rao A. Elevation of serum ceruloplasmin levels in brain tumours. Acta Neurol. Scand. 1992;86:156–158. doi: 10.1111/j.1600-0404.1992.tb05058.x. [DOI] [PubMed] [Google Scholar]
- 47.Sun S., Guo C., Gao T., Ma D., Su X., Pang Q., Zhang R. Hypoxia enhances glioma resistance to sulfasalazine-induced ferroptosis by upregulating SLC7A11 via PI3K/AKT/HIF-1α Axis. Oxid. Med. Cell. Longev. 2022;2022 doi: 10.1155/2022/7862430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan Z., Yang G., Zhang W., Liu Q., Liu G., Liu P., Xu L., Wang J., Yan Z., Han H., Liu R., Shu M. Hypoxia blocks ferroptosis of hepatocellular carcinoma via suppression of METTL14 triggered YTHDF2-dependent silencing of SLC7A11. J. Cell Mol. Med. 2021;25:10197–10212. doi: 10.1111/jcmm.16957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan S., Wei C., Liu G., Zhang L., Li J., Li L., Cai S., Fang L. Sorafenib attenuates liver fibrosis by triggering hepatic stellate cell ferroptosis via HIF-1α/SLC7A11 pathway. Cell Prolif. 2022;55 doi: 10.1111/cpr.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.