Highlights
-
•
The first PTC patient classifier defined by tumor macrophage differentiation.
-
•
Integrated sequencing data identifies key gene sets.
-
•
Macrophage states in PTC link to immune/metabolic shifts.
-
•
Poor prognosis PTC patient group shows M2 macrophage traits.
Keywords: Multi-omics, Single-cell RNA sequencing, Papillary thyroid carcinoma, Tumor immune microenvironment, Macrophage differentiation
Abstract
Background
The reclassification of Papillary Thyroid Carcinoma (PTC) is an area of research that warrants attention. The connection between thyroid cancer, inflammation, and immune responses necessitates considering the mechanisms of differential prognosis of thyroid tumors from an immunological perspective. Given the high adaptability of macrophages to environmental stimuli, focusing on the differentiation characteristics of macrophages might offer a novel approach to address the issues related to PTC subtyping.
Methods
Single-cell RNA sequencing data of medullary cells infiltrated by papillary thyroid carcinoma obtained from public databases was subjected to dimensionality reduction clustering analysis. The RunUMAP and FindAllMarkers functions were utilized to identify the gene expression matrix of different clusters. Cell differentiation trajectory analysis was conducted using the Monocle R package. A complex regulatory network for the classification of Immune status and Macrophage differentiation-associated Papillary Thyroid Cancer Classification (IMPTCC) was constructed through quantitative multi-omics analysis. Immunohistochemistry (IHC) staining was utilized for pathological histology validation.
Results
Through the integration of single-cell RNA and bulk sequencing data combined with multi-omics analysis, we identified crucial transcription factors, immune cells/immune functions, and signaling pathways. Based on this, regulatory networks for three IMPTCC clusters were established.
Conclusion
Based on the co-expression network analysis results, we identified three subtypes of IMPTCC: Immune-Suppressive Macrophage differentiation-associated Papillary Thyroid Carcinoma Classification (ISMPTCC), Immune-Neutral Macrophage differentiation-associated Papillary Thyroid Carcinoma Classification (INMPTCC), and Immune-Activated Macrophage differentiation-associated Papillary Thyroid Carcinoma Classification (IAMPTCC). Each subtype exhibits distinct metabolic, immune, and regulatory characteristics corresponding to different states of macrophage differentiation.
Introduction
Recent worldwide epidemiological data indicate a notable rise in the prevalence of thyroid cancer over the past few decades [1]. Thyroid cancer (TC) is the most widespread endocrine tumor and ranks as the eighth most common cancer globally [2,3]. Although thyroid cancer has long been recognized as a less malignant malignancy with a better prognosis, its high incidence still leads to a significant number of patients experiencing poorer outcomes. Different TC types originating from the same differentiated cells show varying prognoses. Except for medullary thyroid carcinoma (MTC), which originates from parafollicular cells, most TCs develop from follicular epithelial cells. The majority of cases of thyroid carcinoma consist of papillary thyroid cancer (PTC) and follicular thyroid cancer (FTC). Despite being less common, anaplastic thyroid cancer (ATC) plays a significant role in the mortality associated with thyroid cancer [4,5]. However, despite the generally favorable prognosis associated with PTC, a subset of patients still experience adverse outcomes, including recurrence and metastasis [6].
PTC, FTC, and ATC exhibit distinct molecular mechanisms despite their shared origin. These differences are also believed to potentially demonstrate a progressive nature [7]. The variations in prognosis observed within the same pathological type of TC can be attributed, on the one hand, to clinical characteristics and, on the other hand, to molecular mechanisms that influence tumor prognosis. However, taking PTC as an example, there is still no consensus on whether molecular pathogenic mechanisms contribute to prognostic differences, as evidenced by the ongoing debate regarding the impact of BRAF gene mutations on PTC prognosis [8]. Therefore, we speculate that progressive molecular mechanisms may also be present within the same pathological types of tumors.
At this juncture, considering the prognosis differences of tumors from an immunological perspective offers a new approach to unraveling this complexity. Ever since Rudolf Virchow proposed the connection between inflammation, immune response, and cancer in 1863, understanding how the immune system intervenes in tumor progression has been a crucial direction in exploring the pathogenic mechanisms of cancer [9]. This direction was further explored when Hanahan and Weinberg defined the characteristics of tumors [10]. The notion of the tumor microenvironment was presented, emphasizing its crucial function in the onset and progression of tumors. The normal tissue microenvironment can suppress malignant tumors, while certain pathological tissue features are crucial for tumor progression [11,12]. It is of paramount importance to simultaneously acquire a comprehensive understanding of the tumor microenvironment and delineate the immunological aspects of the tumorigenic process to assist in halting tumor progression via immune surveillance. Given the numerous connections between thyroid cancer, inflammation, and immune response [13], it becomes increasingly crucial to focus on changes in the immune environment within the realm of TC. Tumor-infiltrating myeloid cells (TIMs) [14] are crucial regulators in tumor progression, operating within the intricate cellular networks of the tumor microenvironment. They have significant involvement in modulating tumor inflammation and angiogenesis, as highlighted by studies [15,16]. Macrophages, as important constituents of TIMs, are involved in various aspects of tumor immunity. Furthermore, macrophages have become a central focus in tumor immunology research due to their remarkable adaptability to environmental cues and their complex roles influenced by both temporal factors and the surrounding microenvironment [17,18].
The objective of this research was to uncover the complete picture of tumor pathogenicity by utilizing single-cell RNA sequencing (scRNA-Seq) technology. Our focus was on examining the immune microenvironment to elucidate the prognostic differences in PTC. Based on the differentiation of tumor-infiltrating macrophages and their immunological status within the tumor microenvironment, we established a clinical classification of PTC, termed Immune status and macrophage differentiation-associated papillary thyroid cancer classification (IMPTCC). Based on the immune progression status, we classified IMPTCC into three subtypes: Immune-suppressive macrophage differentiation-associated papillary thyroid carcinoma classification (ISMPTCC), Immune-neutral macrophage differentiation-associated papillary thyroid carcinoma classification (INMPTCC), and Immune-activated macrophage differentiation-associated papillary thyroid carcinoma classification (IAMPTCC). Each subtype exhibits distinct pathogenic mechanisms based on differences in the immune environment and molecular regulatory networks. Moreover, these subtypes provide novel insights into the potential pathogenic mechanisms of thyroid cancer at the levels of the immune microenvironment and metabolism.
Methods
Data acquisition
The scRNA-seq information of thyroid macrophages was acquired from the GSE154763 database, comprising gene expressions of 5312 cells from 10 patients who were diagnosed with papillary thyroid carcinoma. The Cancer Genome Atlas (TCGA) database (https://cancerdata.nci.nih.gov/) provided data for 512 tumor tissues and 5 metastases, along with genomic profiles, RNA sequencing profiles, Reverse Phase Protein Array (RPPA) profiles, Transposase-Accessible Chromatin sequencing (ATAC-seq) profiles, population statistics, and clinical information including overall survival rates. From 2016 to 2018, the Tenth People's Hospital affiliated with Tongji University recruited a total of 72 patients who were diagnosed with PTC for histopathological validation. Furthermore, all primary data were obtained from openly accessible repositories, and no further ethical clearance was necessary. Table S1 lists all the datasets, code packages, and software information used in this study. Supplementary materials containing clinical data confirmed by pathological histology and the original code of this study have been uploaded.
Batch correction and data quality control
Our scRNA-seq data were obtained from batch-corrected PTC sample datasets publicly released by Professor Zemin Zhang's research [19]. These datasets were processed using Scanorama [20], an algorithm designed to identify and merge shared cell types across multiple datasets, thus eliminating batch effects within the datasets. The sequencing data from the GEO database, consisting of scRNA-seq information, were imported into the R software package. After applying the CreateSeuratObject function, the data were converted into a Seurat object, followed by quality control analysis using the Seurat R package. For further analysis, only cells that expressed more than 100,000 transcripts and had less than 10 % mitochondrial genes were considered. Further analysis also included genes that were expressed in a minimum of 3 cells. After applying the NormalizeData function, the data were normalized and the resulting values saved in the pbmc dataset. The function FindVariableFeatures was utilized for the identification of genes that exhibit high variability. The vst technique was utilized to identify the top 2000 genes with significant variation among all the genes, which were then employed for subsequent analysis to reduce dimensionality. Subsequently, the ScaleData function was used to linearly transform the data, normalizing the expression values of each gene to a mean of 0 and a variance of 1, thereby eliminating scale differences between different genes.
Data dimension reduction
The data from individual cells were subjected to principal component analysis (PCA) using the RunPCA function. To find an optimal number of principal components for further analysis, the ElbowPlot function was used and detected a notable change in direction at the 10th PC. Subsequently, the Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) nonlinear dimensionality reduction technique was employed using the RunUMAP function to visually represent the complex structure of the data and the outcomes of clustering. UMAP was performed using the initial 20 principal components as input. The clustering process used the FindNeighbors function to perform clustering, while the FindClusters function was employed to group cells at a clustering resolution of 0.5. The differential gene expression analysis involved the utilization of the FindAllMarkers function to identify markers specific to each cell type. To select genes showing differential expression, certain criteria were considered: a positive correlation, expression in a minimum of 25 % of cells, and a log2 fold change threshold of at least 0.25.
Cell cycle analysis
Cell cycle scoring of the scRNA-seq data was conducted using the Seurat package, employing well-established gene sets associated with the G2/M and S phases. The CellCycleScoring function within Seurat was utilized to convert gene expression values into G2M and S phase scores. Additionally, the VlnPlot function was used to generate violin plots of the G2M and S phase scores, facilitating visualization of score distribution across distinct cell populations.
Cell subpopulation annotation
The annotation of cells within each UMAP cluster was achieved using established cell surface markers, derived from the differential expression genes in each cluster and the CellMarker database [21]. The identification of marker genes for each cell type was conducted using the FindAllMarkers function of the Seurat package. The visualization of the expression trends of these markers across various cell subpopulations was achieved through the generation of heatmaps. The normalized data were represented through feature plots or violin plots. To evaluate the spatiotemporal distribution of distinct subpopulations within each cell type, individual cell types were isolated and organized as Seurat objects for subgroup analysis. The RunUMAP function, in conjunction with the FindAllMarkers function and Wilcoxon rank-sum test, was employed to detect differentially expressed genes (DEGs) among subgroups of the 13 cell types. Ultimately, the variations in spatial distribution and expression traits of the 13 cell subpopulations were depicted through cell feature plots, spatial feature plots, and heatmaps.
Macrophage subtyping and differentiation trajectory analysis
To explore the distribution of macrophage subtypes at different differentiation stages in thyroid cancer, we first employed the RunUMAP and FindAllMarkers functions from the Seurat package, along with the Wilcoxon rank-sum test, to construct an independent Seurat object for all macrophages across the samples. Macrophage subtyping was achieved using the Seurat package's FindClusters function, following PCA dimensionality reduction. We employed the first 20 principal components to discern co-expression patterns among cells. Subsequently, cells were clustered at a resolution of 0.1, leading to the identification of distinct macrophage subtypes. These subtypes were visualized and differentiated using UMAP plots, reflecting their unique expression profiles. The identification of differentially expressed genes (DEGs) within macrophage subtypes 1–5 was performed using the FindAllMarkers function. This function identifies genes significantly upregulated within each subgroup compared to all other cells, based on predefined thresholds of minimum percentage expression (min. pct = 0.25) and log fold change (logfc. threshold = 0.25). This analysis entails comparing each macrophage subgroup against the entirety of other cells in the dataset. We then applied additional filtering criteria, selecting genes with an average log2 fold change (avg_log2FC) greater than 0.5 to pinpoint the most significant DEGs. For each macrophage subgroup, we extracted the top five genes exhibiting the highest avg_log2FC changes as characteristic DEGs, providing a detailed insight into the unique expression profiles of each subgroup. Subsequently, we performed a subpopulation analysis of the data. Next, we utilized the Monocle package in R (version: 2.18.0) to analyze the differentiation trajectory of macrophages [22]. We employed the newCellDataSet function to categorize the UMI expression matrix, sample metadata, and gene expression information of all macrophages based on their differentiation trajectory. Unsupervised ordering was applied to all cells. The estimateSizeFactors function was used for data normalization. The detectGenes function was employed to filter out genes with low expression levels. Cell numbers at each time point were calculated to determine the developmental or differentiation starting point. After performing gene filtering and identifying differentially expressed genes, a total of 1000 genes were obtained as the foundation for cell ordering. Subsequently, the DDRTree method was used to reduce the dimensionality of cell clusters. The main chart was integrated into the high-dimensional scRNA-seq dataset, revealing the correlation between the gene expression space with many dimensions and a space with fewer dimensions. The differentiation trajectory between cells was built by projecting the expression data onto the low-dimensional space through the construction of a Minimum Spanning Tree (MST) using DDRTree. MST was recursively calculated to assign pseudotime to cells for ordering. The plot_cell_trajectory method was employed to display the developmental path of the cells, illustrating their various stages of differentiation.
Furthermore, we applied the Branch expression analysis modeling (BEAM) approach to infer the cell differentiation trajectory, comparing the differences between branching points and terminal branches to hypothesize cell fate in differentiation. Finally, we computed the differentially expressed genes (DEGs) among macrophage subtypes in different differentiation stages (States) and defined them as macrophage differentiation-related genes (MDGs). We selected the MDGs based on a q-value < 0.05 derived from the DDRTree algorithm and used them to sort the differentiation trajectory.
Consistency cluster analysis
To develop the immune status in Macrophage-infiltrated Thyroid Cancer Classification (IMPTCC), additional screening of MDGs is necessary. This screening procedure involved the integration of bulk-RNA sequencing data sourced from the TCGA database. Through the correlation of gene expression data with prognostic information derived from clinical samples, we utilized the survminer and limma packages to calculate Cox proportional hazards models for each gene. Genes with KM p-value < 0.05 and Cox regression p-value < 0.05 were selected for inclusion in the IMPTCC. Integration of significantly expressed genes from Bulk-RNA sequencing data analysis and scRNA-seq data pseudotime analysis retained genes that satisfied the following four criteria: [1] MDGs with q-values < 0.05 obtained through DDRTree algorithm and genes used for differentiation trajectory order; [2] MDGs exhibiting significant (non-parametric test p-value < 0.05) correlation between gene expression and pseudotime of PTC samples; [3] Genes showing significant (non-parametric test p-value < 0.05) results in Kaplan-Meier survival analysis; [23] Genes demonstrating significant (non-parametric test p-value < 0.05) results in univariate Cox survival analysis. Consequently, the integration of cellular differentiation features and clinical prognostic features yielded Macrophage differentiation and prognosis-related genes (MD&PRGs).
Subsequently, we employed the packages igraph, psych, reshape2, and RColorBrewer to construct and visualize gene co-expression networks for 19 MD&PRGs. Initially, the correlation between gene expressions was computed, and significant gene pairs were selected based on a p-value threshold; the correlation coefficients and p-values stored in a matrix. Next, the igraph package was utilized to generate the co-expression network graph. Nodes were assigned colors based on the grouping of genes according to their cellular differentiation states and risk factors, while node sizes were allocated based on the results of Cox proportional hazards regression analysis. The relationships between genes were represented using line attributes, with line width indicating the magnitude of p-values, and line color indicating the direction of correlation (pink for positive correlation and blue for negative correlation).
By using the ConsensusClusterPlus package in the R programming language, the consensus clustering analysis (CCA) algorithm was applied to cluster the observed samples based on the expression profiles of key genes and to integrate their intrinsic characteristics. Observational samples with similar features, as determined by their close Euclidean distances, were defined as belonging to the same cluster subtype. To enhance clustering stability and representative grouping, the optimal clustering model was selected by constraining the cumulative distribution function (CDF) growth rate, the proportions of samples within each group, and both intra-group and inter-group correlations [24].
Principal component analysis and clinical correlation analysis
To convert the clustering results into quantitative variables, we performed PCA analysis on the sample data using the prcomp and predict functions. The gene expression data of the samples were dimensionally reduced, and the scores of each sample on the first two principal components were computed, yielding PCA scores for each sample. Based on these scores, the samples were divided into high and low groups. The surv_cutpoint, survdiff, and survfit functions were applied to integrate the survival data of the samples for survival analysis, and the ggsurvplot and ggplot functions were used for visualization. The results were visualized using box plots, bar graphs, violin plots, and heatmaps (P<0.05 indicating statistical significance). To obtain more reliable results, we chose progression-free survival (PFS) as the clinical survival outcome endpoint for analysis. Furthermore, with the aid of PCA scores, we conducted multi-omics analysis at the genomic, transcriptomic, proteomic, and epigenomic levels for the 19 MD&PRGs.
Survival analysis and prediction model construction and diagnosis
Differential expression analysis was performed using primary tumor and metastasis RNA-seq data from the TCGA database. Survival-associated genes were identified through univariate Cox regression analysis, and the top 20 prognostic genes were determined using Lasso regression based on the survival-associated genes. A multifactorial prognostic model was established by identifying the differentiation cell fate genomic signature in THCA. Subsequently, we employed random number table allocation to divide all samples into a training set and a validation set in a 6:4 ratio, and the prior model was trained based on the training set. Finally, the hypothesis was validated using the cross-validation set to determine the optimal Receiver Operating Characteristic (ROC) area. Samples were scored based on the median risk score, and risk curves, scatter plots, and expression heatmaps were generated. The area under the ROC curve was used to evaluate the accuracy of the model. Univariate and multivariate Cox regression analyses were applied to evaluate whether the risk score, age, gender, and TNM staging were independent prognostic factors.
Genomics correlation analysis
To calculate the percentage of copy number gains and losses for important genes in the samples, we utilized Perl scripts and R code for copy number variation (CNV) analysis. Circular chromosome plots displaying the chromosomal location information of the key genes were generated using the RCircos package. To analyze the correlation between tumor mutation burden (TMB) and PCA scores, as well as microsatellite instability (MSI), and to evaluate the prognostic differences of the samples by integrating TMB and PCA scores, the ggpubr and reshape2 packages were utilized. The oncoplot package was utilized to visualize and analyze gene mutation data.
Gene-set enrichment analysis
In this study, we used gene set variation analysis (GSVA) [25], gene set enrichment analysis (GSEA) [26], and Overall Represent analysis (ORA) [27] methods to better explore gene expression differences and their underlying functional implications.
The gene sets utilized in this research were acquired from the Molecular Signatures Database (MSigDB) (version 7.1) [28]. To assess the variation in the activity of each sample within gene sets, we initially utilized GSVA to produce enrichment scores for hallmark gene sets for each sample using the ordered gene expression profiles. GSVA assesses the variation of biological processes without predefined phenotype information, thus utilizing all available gene expression information. The GSVA package was utilized to conduct GSVA analysis on the gene expression matrix, whereas the limma package was employed for differential analysis, filtering significantly differentially expressed pathways using a logFC cutoff of 0.0001 and adjPvalue cutoff of 0.05. The ggplot2 package was used to create stacked bar plots and volcano plots, while the pheatmap package was employed to visualize the GSVA analysis results using heatmap plots.
Furthermore, we utilized the ORA technique to investigate genes exhibiting notable variations in expression. To assess the degree of enrichment in a particular set of genes, the comparison was made based on the proportion of genes that showed significant differential expression within the said gene set. By utilizing the ORA analysis, we conducted a comparison between the 19 essential genes and the gene sets in the reference. We evaluated the enrichment of the important genes in nine sets of genes by conducting hypergeometric distribution testing. The gene sets were classified into nine functional categories according to various functional characteristics, which include groups of gene sets labeled as C1-C8 and the HALLMARK gene set group.
To further elucidate underlying biological phenomena and mitigate biases arising from threshold selection, we employed the GSEA method to identify gene sets with similar expression patterns and examine their enrichment between the high-risk and low-risk groups. Specifically, we conducted GSEA analyses on the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) gene set databases. Significantly enriched gene sets with p-values less than 0.05 were identified for each database. GSEA plots depicting the top five significantly enriched gene sets were generated separately for the high-risk and low-risk groups [29,30].
Immune infiltration and immune checkpoint analysis
We analyzed the correlation between the expression of the PD-L1 gene and PCA scores by utilizing data acquired from The Cancer Immunome Atlas (TCIA) [31]. Furthermore, we investigated the variances in the quantities of immune cells infiltrating tumors among PCA score groups categorized as high and low. This analysis involved assessing the levels of expression for two immune checkpoint proteins, namely CTLA-4 and PD-L1. The limma and ggpubr packages were utilized for the analysis of immune checkpoint data processing and visualization.
We employed single-sample gene set enrichment analysis (ssGSEA) [32] to assess the immune infiltration status and accurately evaluate the differences in immune expression among different subtypes and between primary and metastatic samples. By utilizing known immune-related gene sets, we calculated immune infiltration scores for each sample to evaluate the degree of immune infiltration. The immune infiltration status was visualized using the corrplot and ggpubr packages, allowing for intuitive observation and comparison of the differences in immune infiltration levels among different subtypes.
To investigate the composition of infiltrating immune cells in complex tissues, we employed a technique known as Cell-type Identification By Estimating Relative Subsets Of RNA Transcripts (CIBERSORT) [33]. This method relies on the characteristics of gene expression profiles to estimate the relative subsets of RNA transcripts. To determine the correlation between MDGs and immune cell infiltration in PTC tissues, we uploaded the microarray expression data and RNA-seq gene expression matrix data to the Cell-type Identification By Estimating Relative Subsets Of RNA Transcripts (CIBERSORT) database to assess the immune infiltration status of the samples.
Proteomic analysis
Protein expression values were calculated using gene expression profile data and Reverse Phase Protein Array (RPPA) data. Furthermore, the correlation between the gene expression profile and protein expression values was computed.
Regulatory network construction
To further explore the potential regulatory mechanisms of various subtypes of IMPTCC, a standardized approach was used to analyze quantitative results obtained from MD&PRGs, transcription factors, immune cells/immune function, RPPA protein arrays, and signaling pathways. We performed co-expression analysis by utilizing Pearson/Spearman correlation analysis. The analysis was based on data normality and rank, with a threshold set at R > 0.300 and P 〈 0.001. With minimum screening criteria of R 〉 0.300 and P < 0.001, we constructed an intricate regulatory network focused on the MD&PRGs of every IMPTCC subtype. The regulatory network was visualized using the igraph package in the R programming language (http://igraph.sourceforge.net) [34].
Targeted drug prediction analysis for each subtype
The Connectivity Map (Cmap) is an algorithm that explores the statistical matching relationship between small molecule active drugs and target genes based on whole-genome transcriptional expression data from human cells treated with small molecule compounds. The Cmap algorithm incorporates over 7000 cell culture systems and 1309 small molecule active drugs, all of which are FDA-approved drugs, and establishes a drug-function gene-disease network. This network provides a statistical basis for identifying small molecule inhibitors targeting genes associated with therapeutically relevant diseases (https://portals.broadinstitute.org/cmap/) [35]. In this study, the Cmap algorithm was employed to explore small molecule drugs targeting the regulatory networks of different IMPTCC subtypes. Subsequently, a dot plot heatmap analysis was used to identify the top 10 drugs with the lowest p-values as targeted drugs for each IMPTCC subtype. Furthermore, by integrating the gene expression profiles with the subtype classification, the most effective active drugs were selected. Additionally, we linked pharmacological data with genomic information using information from eight common thyroid cancer cell lines and visualized the data as heatmaps and box plots. Moreover, GDSC genomic data were utilized for predictive modeling of pharmacological sensitivity [36].
Chromatin accessibility assessment
ATAC-seq [37] is a remarkable, versatile, uncomplicated, and potent approach for the comprehensive characterization of chromatin regions across the genome, in contrast to traditional methodologies such as functional assays or sequence conservation analyses. To establish the accessibility of crucial genes on chromatin, we procured ATAC-seq data pertaining to these genes.
Histopathological validation
Approval for histopathological validation using clinical samples was obtained from the Clinical Research Institution Review Committee of the Tenth People's Hospital, which is affiliated with Tongji University in Shanghai. Before participation, all patients provided informed written consent. Samples for additional validation were gathered from the Tenth People's Hospital, affiliated with Tongji University, from 2016 to 2018.
Samples of initial tumors collected from a group of 72 patients diagnosed with papillary thyroid carcinoma underwent fixation and were then embedded in formalin and paraffin. Subsequently, these specimens underwent standard procedures of sectioning, dehydration, antigen retrieval through rehydration, and blocking, and were sliced into sections measuring 4 µm in thickness. The sections underwent incubation with an APOE antibody (1:3000 proteintech) at 4 °C overnight. Afterward, polymer HRP was used to label all sections for 40 min and then counterstained with hematoxylin for 5 min at room temperature. The evaluation of all slides was conducted by two pathologists, whereby positive cytoplasmic staining in the cancer cells was defined. It is worth noting that both cytoplasmic and membranous APOE were stained utilizing the same antibody. To create a negative control, the primary antibody was replaced with a buffer solution. We utilized non-parametric tests and Spearman's correlation tests to assess the association between the APOE staining intensity scores and the clinical features of the individuals.
Statistical analysis
The R programming language was utilized for processing and analyzing all the data. The normality of quantitative variables was assessed using the Shapiro-Wilk test, while Levene's test was employed to examine the homogeneity of variances. Comparisons between the two groups were conducted using either the Mann-Whitney U test (also known as the Wilcoxon rank sum test) or the t-test, depending on the distribution of the data. The chi-squared test or Fisher's exact test was used for categorical variables, depending on the anticipated frequencies. The level of statistical significance was determined to be p<0.05.
Result
ScRNA-seq cell subtype annotation
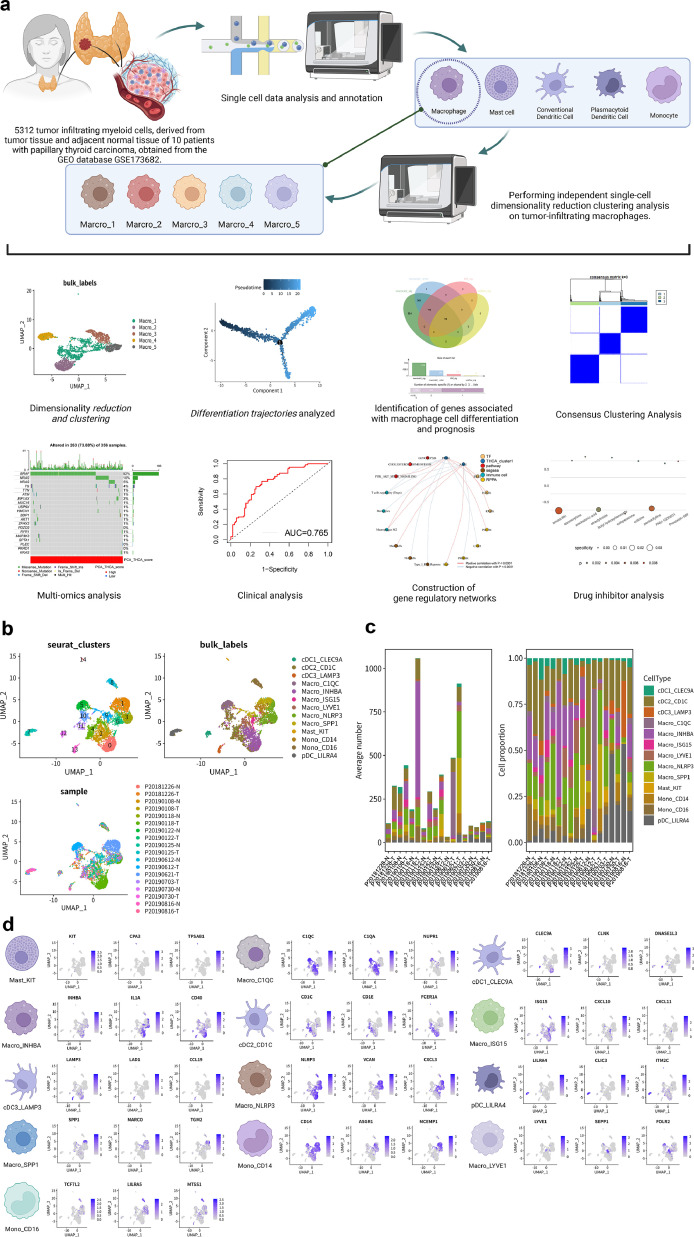
Fig. 1a and Figure S1 display the workflow and schematic of the article. To explore the tumor microenvironment and immune landscape of thyroid cancer, we analyzed 5312 TIMs from 10 thyroid cancer patients obtained from the GEO database. Dimensionality reduction and clustering of scRNA-seq data using the Uniform Manifold Approximation and Projection method identified 15 unsupervised clusters, comprising 13 unique cell populations, and 5 cell types (monocytes, adipocytes, macrophages, cDC, and pDC) (Fig. 1b). The average number and percentage distribution of the 13 cell subgroups in 10 cancer and adjacent samples are displayed in a bar chart (Fig. 1c). It can be observed that, compared to normal tissues, tumor tissues exhibit significant macrophage infiltration. Similarly, the bar chart in Figure S2a also demonstrates a comparable distribution, and the Cleveland dot plot analysis of three marker genes further corroborates this observation. The macrophage-associated marker, CD68[38], exhibits high expression within the macrophage-related subgroups, while the mast cell-associated marker, KIT[39], shows elevated expression within the mast cell subgroups. As observed from the bar graph, tumor tissues distinctly possess a higher expression proportion within the macrophage subgroups. The classical macrophage marker gene CD68 is significantly overexpressed in the macrophage subgroup, while ITGAX is expressed across all subtypes. The expression patterns of marker genes for each cell subtype are visualized in Fig. 1d. Considering the significant contribution of macrophages that infiltrate tumors to the advancement of tumors, we isolated 3326 macrophages individually and conducted separate analyses to reduce their dimensions and cluster them. This process sets the stage for additional investigation into the characteristics of macrophages that infiltrate tumors.
Fig. 1.
scRNA-seq analysis of tumor-infiltrating myeloid cells
(a) The experimental design flow. The figure was created using BioRender.com.
(b) The UMAP scatter plot illustrates the results of dimensionality reduction clustering, annotations of cellular subtypes, and the distribution of samples. Notably, 5312 cells from 10 papillary thyroid cancer patients were clustered into 15 unsupervised clusters, which were subsequently annotated as 13 cell populations (Macro_NLRP3, Macro_INHBA, Macro_ISG15, Macro_LYVE1, Macro_C1QC, Macro_SPP1, cDC1_CLEC9A, cDC2_CD1C, cDC3_LAMP3, Mono_CD14, Mono_CD16, Mast_KIT, pDC_LILRA4). The distribution of tumor and normal tissue samples from the 10 patients exhibited differential patterns across these cellular subgroups.
(c) The bar graph depicting the distribution of 13 cell populations across 10 sample sources.
(d) The feature map illustrating the expression patterns of signature genes within 13 cell populations.
Differential expression analysis and cell cycle analysis for each cell
To provide a deeper understanding of the immune composition in the 5312 individual cell samples, we conducted an analysis of differentially expressed genes (DEGs) on the top 2000 genes that exhibit high variability across all samples. A heatmap was created to exhibit the levels of expression of the top 5 differentially expressed genes (DEGs) for every subtype in every cell type (Figure S2b). To visualize the communication among the 13 subtypes of cells, cell communication networks and circular plots were employed (Figure S2c-d). Analysis of the cell cycle (Figure S2e-f) revealed that macrophages were primarily active during the G1 and G2M stages.
Macrophage subtyping analysis
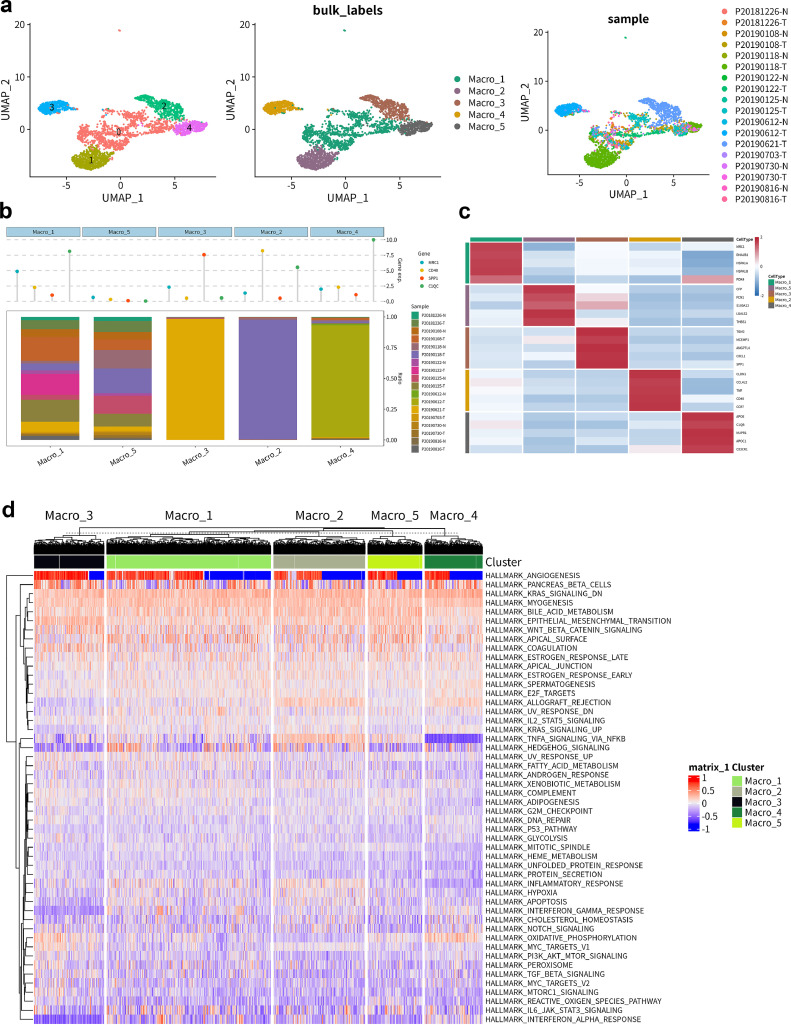
The independent dimensionality reduction and clustering analysis of macrophages was performed by constructing separate Seurat objects for all samples. This analysis identified five subtypes of macrophages (Macro_1–5) (Fig. 2a). The distribution of 10 tumors and adjacent non-tumor samples within the five macrophage clusters was visualized using a bar plot (Fig. 2b). Among them, macrophages 2–4 predominantly originated from tumor tissue, while macrophages 1 and 5 exhibited a more complex origin, including both tumor tissue and adjacent normal tissue. The Cleveland dot plot depicted the expression levels of four classical marker genes associated with immune regulation and immune response (MRC1, CD40, SPP1, C1QC), revealing distinct immunological expression profiles among the five macrophage subtypes. Fig. 2c presents the top 5 differentially expressed genes (DEGs) within macrophages 1–5. Based on the gene expression characteristics in Fig. 2c, we functionally categorized five macrophage subgroups. Macro_1 expresses M2-related marker MRC1 [40] and stress-response genes HSPA1A and HSPA1B, indicating potential dual roles in pro- and anti-inflammatory processes [41]. Macro_2 is characterized by the expression of TNF and CD40, associated with pro-inflammatory responses [42]. Macro_3 expresses SPP1 and CXCL1, underscoring its role in tissue repair and pro-inflammatory processes [43,44]. Macro_4 exhibits lipid metabolism genes APOE and APOC1, involved in lipid transport dysregulation and inflammation [45]. Finally, Macro_5 expresses CFP and FCN1, consistent with its function in complement activation and inflammation [46,47]. These subgroup characteristics emphasize the versatility of macrophages in the PTC microenvironment and their potential roles in disease progression. Finally, to further elucidate the malignancy-associated expression features of the five macrophage subtypes, Gene Set Variation Analysis (GSVA) was applied to generate hallmark gene set expression scores based on ranked gene changes in the sample expression profiles. In the results of gene set enrichment analysis shown in Fig. 2d, Macro_2 specifically exhibits a high expression of the classical inflammation-related TNFα pathway, highlighting its key role in pro-inflammatory responses [42]. Macro_4 shows elevated expression of the oxidative phosphorylation pathway, emphasizing its specialized function in energy metabolism. The IL6-JAK-STAT3 signaling pathway, associated with immune response and inflammation regulation [48], aligns with the functions of Macro_1. The commonly high expression of downregulated KRAS signaling and WNT/β-catenin pathways across all subtypes indicates a shared mechanism in regulating the PTC tumor microenvironment, consistent with our following co-expression network analysis results. These differences will be comprehensively explained in subsequent subtype analyses.
Fig. 2.
scRNA-seq analysis of macrophage subpopulations.
(a) The UMAP scatter plot revealing the dimensionality-reduced clustering of isolated macrophages, with annotations for cell subtypes and sample distributions. A total of 3326 cells were clustered into 5 cell subgroups (Macro_1, Macro_2, Macro_3, Macro_4, Macro_5). Tumoral and adjacent non-tumoral samples from the 10 sources exhibit differential distribution across these cell subgroups.
(b) The bar graph presenting the distribution of 10 tumor and adjacent non-tumor samples across the 5 macrophage clusters. Cleveland dot plot delineates the expression levels of four canonical markers (MRC1, CD40, SPP1, C1QC) associated with immunoregulation and immune response.
(c) The heat map demonstrating the top 5 marker DEG expression levels for each cell type.
(d) The heatmap displays the GSVA gene set enrichment analysis results for the 5 macrophage subpopulations.
Macrophage differentiation trajectory analysis
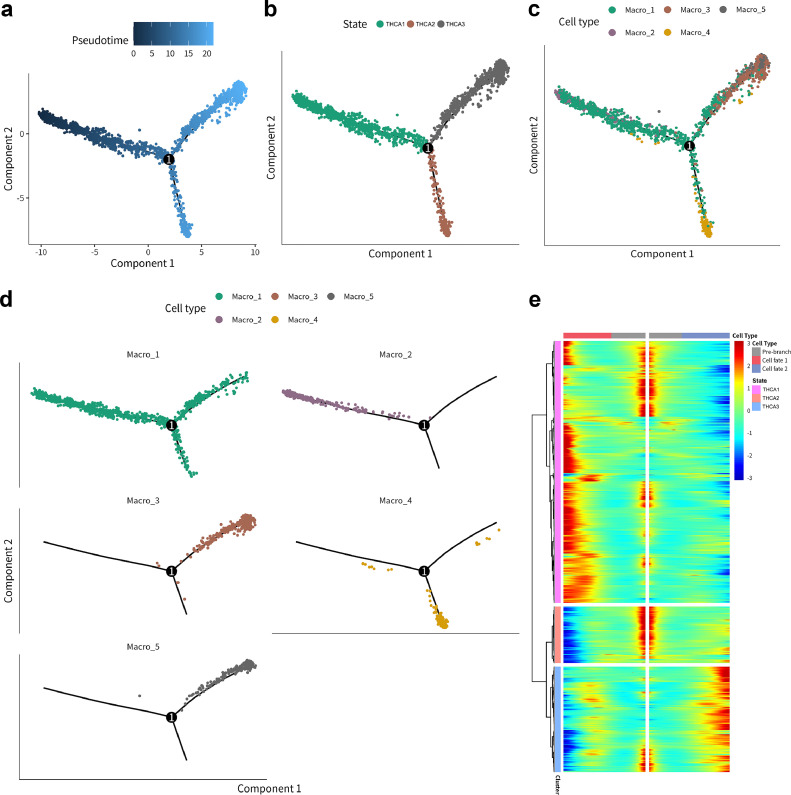
To delineate the temporal changes in the immune microenvironment across the five macrophage subtypes, the Monocle package in the R language was employed to infer the differentiation order and formulate cell fate hypotheses. Pseudotime analysis, visualized through cell developmental trajectories, was used to depict the differentiation states of the macrophage subtypes. Figs. 3a-d depict the distribution and differentiation trajectory of macrophage subtypes along three distinct cell fates. Each macrophage subgroup corresponds to a different state of cell differentiation. Macro_1 is involved throughout the differentiation process, while Macro_2 primarily exists in THCA 1, Macro_4 in THCA 2, and Macro_3 and Macro_5 in THCA 3. The time distribution suggests that THCA 1–3 represent early, mid, and late stages of pseudotime differentiation. With a polar division based on the key differentiation node 1 in Fig. 3a, two cell fates (cell fate 1, cell fate 2) are identified, as shown in the heatmap of Fig. 3e. THCA 1 and THCA 3 exhibit different cell fates, while THCA 2 appears to be in a transitional state. This comprehensive analysis led to a hypothesis about the sequence of macrophage subtype differentiation: THCA1 (early differentiation) corresponds to Macrophages 1 and 2; THCA2 (early/mid-differentiation) to Macrophage 4; and THCA3 (late differentiation) to Macrophages 1, 3, and 5.
Fig. 3.
Pseudo-time analysis traces the differentiation trajectory of macrophage subpopulations
(a) Pseudo-time analysis reveals the order in which cell fate differentiation takes place.
(b) Pseudo-time analysis illustrates the distribution of three cell fates.
(c-d) Pseudo-time analysis demonstrates the distribution of 5 macrophage subpopulations.
(e) The heatmap illustrates two distinct cell differentiation fate directions in the overall presentation of three cellular differentiation states (THCA1–3).
MD&PRGs identification
During the analysis of pseudotime, we incorporated differentially expressed genes across various differentiation stages of macrophage subtypes, as well as relevant genes utilized for trajectory ordering through the DDRTree algorithm. This approach allowed us to identify crucial genes that are associated with the process of macrophage differentiation. Additionally, we obtained bulk RNA sequencing data from the TCGA database, matching gene expression data with clinical sample prognosis information, to identify prognostically relevant key genes. By intersecting these two gene sets, we effectively screened genes based on four criteria: [1] Macrophage Differentiation Genes (MDGs) with q-values < 0.05 obtained through the DDRTree algorithm and genes used for differentiation trajectory ordering from scRNA-seq samples; [2] MDGs showing significant correlation (non-parametric test p-value < 0.05) with the pseudotime of Papillary Thyroid Cancer (PTC) samples in scRNA-seq; [3] Genes exhibiting significant results in Kaplan-Meier survival analysis (non-parametric test p-value < 0.05) from TCGA thyroid cancer samples; [23] Genes demonstrating significant results in univariate Cox survival analysis (non-parametric test p-value < 0.05) from TCGA thyroid cancer samples. This process led to the identification of a key gene set, termed Macrophage Differentiation and Prognosis-Related Genes (MD&PRGs), including 19 critical genes (Figure S3a). The prognostic information of these 19 genes is displayed in Figure S3b-c.
Construct imptcc using consistent cluster analysis
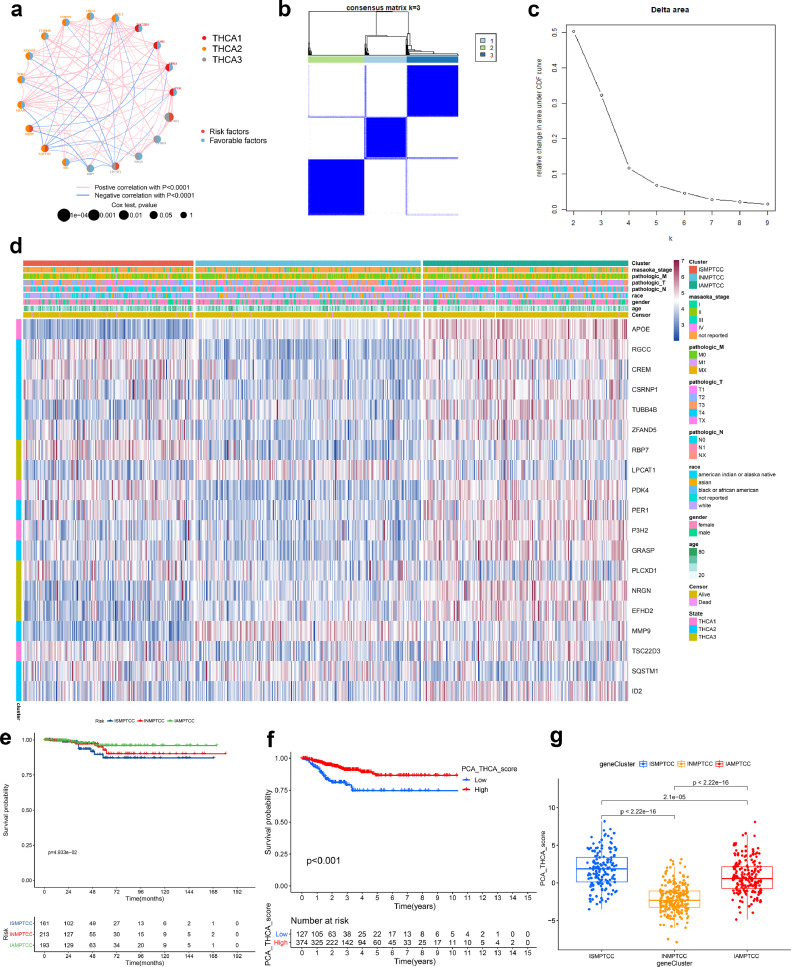
By conducting a correlation analysis on MD&PRGs, we performed a co-expression network analysis to elucidate the expression patterns of these genes across three cellular differentiation states (THCA1–3) (Fig. 4a). The ConsensusClusterPlus package in R was used, applying the consensus clustering algorithm, to construct the IMPTCC based on the expression profiles of the MD&PRGs. Subsequently, an analysis of feature proportions within the MD&PRGs gene set was conducted, with features subsampled for all specimens. The results suggest that k = 3 is the most optimal value (Figs. 4b-c). When there are three subtypes in IMPTCC, their respective expression statuses of the 19 MD&PRGs in each sample are visualized via a heatmap (Fig. 4d).
Fig. 4.
Identification of MD&PRGs and IMPTCC construction
(a) Co-expression network analysis elucidating the co-expression relationships of 19 MD&PRGs across three cellular differentiation states.
(b-c) Bivariate clustering results from consistency matrices suggest optimal clustering into three groups at k = 3, evident from pronounced inter-group differences, minimal intra-group heterogeneity, and an expansive area under the CDF curve. Considering clustering distinction and maximal curve area principles, k = 3 is deemed optimal.
(d) The heatmap displays the differential expression of 19 MD&PRGs across three IMPTCC clusters and three cellular differentiation states, correlating with their respective clinical characteristics.
(e) Kaplan-Meier survival curves contrasting survival outcomes of the three clusters.
(f) Kaplan-Meier survival curves comparing survival outcomes between high and low PCA groups.
(g) Box plot delineating PCA score variations among the three clusters.
Integrating Macro_1–5, derived from dimensionality reduction clustering, with the cell fate classifications THCA 1–3 from further analysis, we developed the IMPTCC. This classification, grounded in macrophage differentiation characteristics within the tumor microenvironment and correlated with clinical prognosis, reveals significant clinical and environmental implications. Fig. 4d demonstrates potential correlations between the three IMPTCC subtypes (Cluster 1–3) and the THCA1–3 cell fates, with Subtype 3 aligning with THCA1–2, Cluster 1 with THCA3, and Cluster 2 appearing in a quiescent phase of differentiation. This correlation will be further elucidated in upcoming co-expression network analyses. Further, Kaplan-Meier survival analysis confirmed that patients in Cluster 1 had the poorest prognosis, whereas those in Cluster 3 exhibited the best prognosis (Fig. 4e).
Considering that the categorical results of IMPTCC obtained from consensus clustering might introduce bias in further analyses, we employed the Principal Component Analysis (PCA) algorithm to compute the PCA scores of the samples. All samples were categorized into high/low PCA score groups, and as depicted in Fig. 4f, there was a notable correlation between the PCA scores and prognosis. Furthermore, Fig. 4g and Figure S4a showed the PCA scores of the three IMPTCC subtypes. Figure S4b visually displays the prognostic outcomes of the three subtypes through a Sankey diagram.
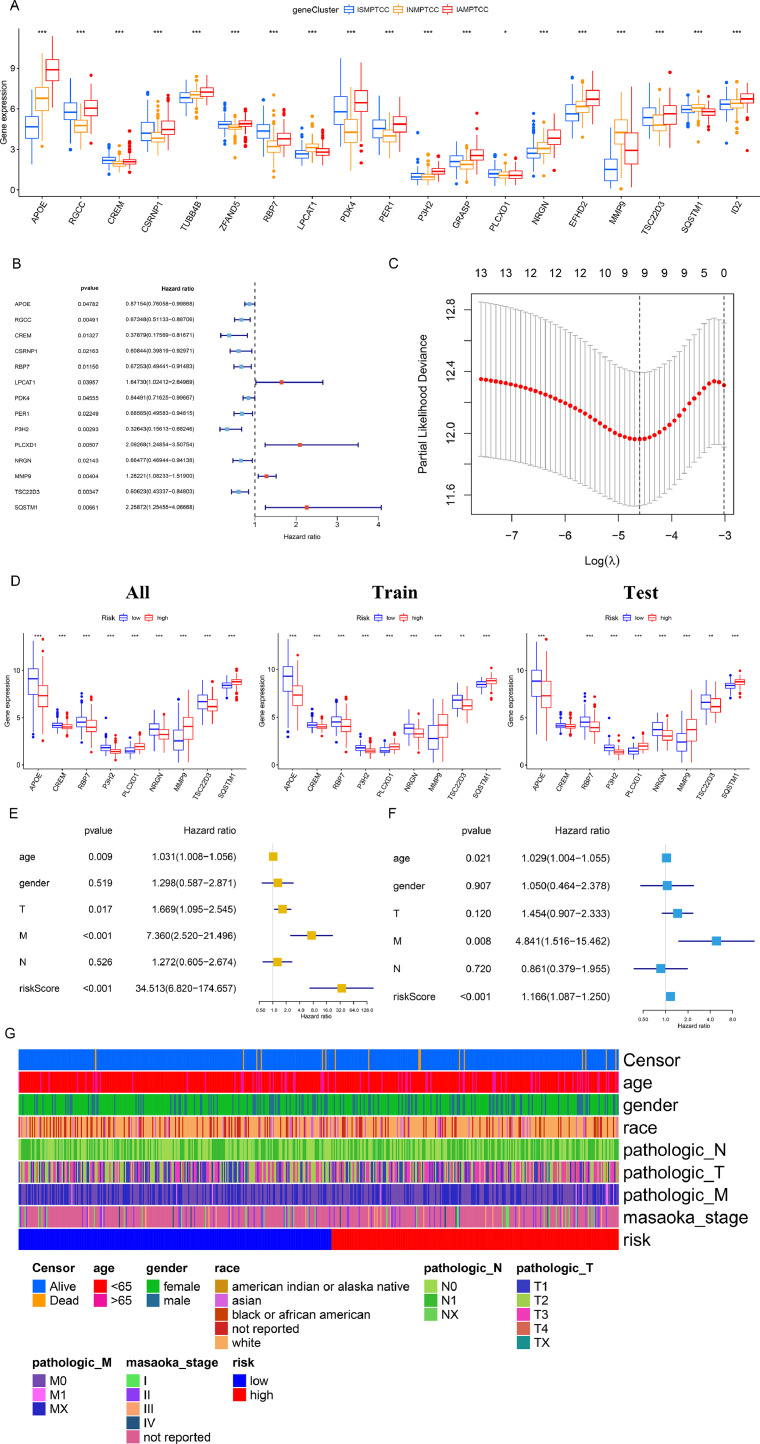
Construction, validation, and diagnosis of the predictive model
Fig. 5a presents a box plot illustrating the differential expression of MD&PRGs among the three clinical subtypes. To confirm the accuracy of MD&PRGs in predicting the clinical results of thyroid cancer, we combined the prognostic data of the expression pattern of 19 MD&PRGs from 503 TCGA database samples, excluding those without available metastasis information. This was subjected to univariate Cox regression, and 14 genes with statistical significance were incorporated into the subsequent analysis (Fig. 5b). Using Lasso regression, we identified that the ideal number of variables was nine (Fig. 5c, S4c). Following the analysis of multiple variables, a regression model was performed to create the predictive framework, affirming its significance as a distinct prognostic determinant (Figs. 5d-e). By employing a random number technique, the entire set of samples was partitioned into training and validation sets with a proportion of 6 to 4. The nine key genes exhibited differential expression patterns across the full sample set, training set, and validation set (Fig. 5f). To test the hypothesis, cross-validation was used, resulting in the optimal area under the ROC curve. Afterward, samples were classified into high-risk and low-risk groups based on the median risk score. The KM survival curve (Figure S4d), expression heatmap (Figure S4e), scatter plot (Figure S4f), and risk curve (Figure S4g) indicate that the risk model effectively differentiates the high-risk and low-risk groups. Figure S4h illustrates the quantitative evaluation of the model's accuracy through the area beneath the ROC curve. The heatmap elucidates the baseline distribution of samples within the risk model (Fig. 5g).
Fig. 5.
Construction, Validation, and Diagnosis of the Predictive Model
(a) Box plot illustrating the expression level disparities of 19 MD&PRGs across three subtypes. (Statistical significance denoted as: *** for p < 0.001, ** for p < 0.01, and * for p<0.05).
(b) Forest plot detailing the univariate Cox regression analysis of the 19 MD&PRGs, with 14 genes displaying statistical significance earmarked for subsequent selection.
(c) Lasso path diagram demonstrating that post-lasso regression, the number of key genes incorporated into the predictive model was finalized at nine.
(d) Box plot showcasing the differential expression of the nine pivotal genes across all sample groups, training sets, and validation sets, stratified by risk.
(e) Univariate Cox regression analysis validates the risk score, computed from the predictive model, as a potential independent prognostic factor for PTC.
(f) Multivariate Cox regression analysis confirms the risk score, derived from the predictive model, as an independent prognostic determinant for PTC.
(g) Heatmap elucidates the baseline distribution of samples within the risk model.
Multi-omics analysis
Using PCA scores, we performed a co-expression analysis of MD&PRGs at the multi-omics level. The analysis of mutation probability visualized the patterns of gene mutation in the groups with high and low PCA scores, suggesting notable disparities in gene expression patterns between the two groups. Significantly, the group with the highest score displayed an increased likelihood of mutation in the RAS gene, aligning with the unfavorable prognosis observed in the high PCA group (Figure S5a-b). Based on the co-expression analysis presented in Figures S5c-d, it is inferred that the group with a low PCA score exhibits a higher TMB score. This finding suggests that this particular group may display increased sensitivity to immunotherapeutic agents like immune checkpoint inhibitors (for example, PD-1/PD-L1 inhibitors). Figures S5e and S5f respectively display the copy number variations of key genes and their chromosomal location information.
From the results in Figure S6, the application of ORA and GSEA for enrichment analysis of various gene sets indicates that key genes consistently show enrichment for inflammation and immune-related gene sets across multiple databases.
We employed the CIBERSORT algorithm and the ssGSEA algorithm to quantify 23 types of immune cells and immune functions in THCA and compared the correlation of these immune cells/features with MD&PRGs. The results reveal that most immune cells/functions exhibit significant differences across different IMPTCC subtypes (Figure S7a). In the analysis related to immune infiltration, subtype 2 demonstrated relatively active immune cell expression levels, consistent with our definition of subtype 2, suggesting that it might have relatively high immune cell expression levels. However, the intermediate prognosis outcome of subtype 2 suggests that this active immune state does not directly lead to tumor progression. In the combined PCA analysis, consistent with the TMB analysis results, the PD-L1 expression level in the low PCA score group was significantly elevated (Figure S7b). Concurrently, the combined analysis of PD-L1 and CTLA-4 indicates that the low PCA group has a better immune therapy response (Figures S7c-f). Consistently, PCA scores are negatively correlated with immune cell expression levels (Figure S7g).
Construction of the co-expression network
Upon determining the immune infiltration related to key genes and the enrichment of gene sets, we utilized the transcription factor data from the Cistrome database and the RPPA protein chip data obtained from the TCGA database to identify the related transcription factors upstream of the key genes and the downstream RPPA protein expression. In Figure S8, we visualized the key differential genes and the key components of the multi-omics analysis, including transcription factors, immune cells, and hallmark gene sets, using volcano plots and heatmaps. As a result, we discerned the relationships among the key components required to construct the regulatory network, including DETFs, MD&PRGs, immune cells/immune functions, and signaling pathways.
Construction of the regulatory network for each IMPTCC subtype
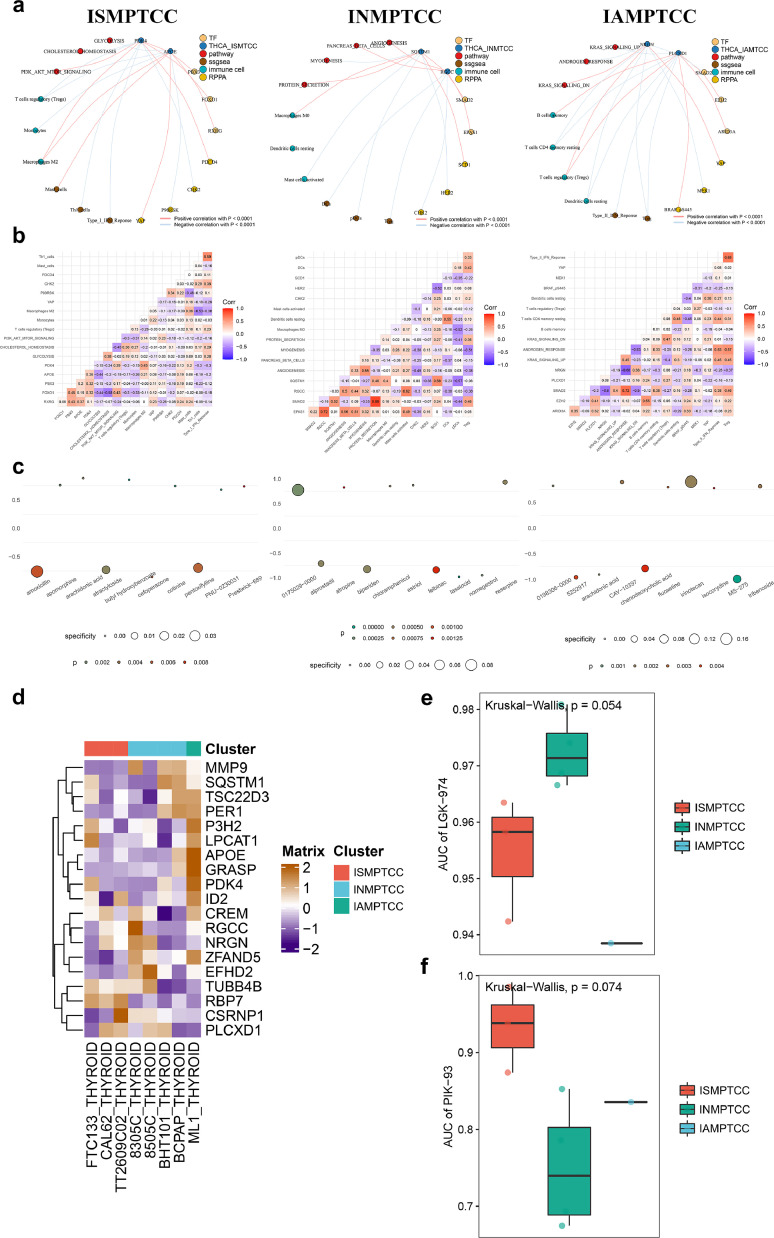
To further investigate the potential key regulatory mechanisms of each IMPTCC subtype, we conducted a co-expression analysis based on the standardized DETFs, MD&PRGs, immune cells/immune functions, and signaling pathways quantitative results for each IMPTCC subtype. Using a correlation coefficient R>0.300 and P<0.001 as the minimum screening criteria, we constructed three complex regulatory networks centered on MD&PRGs, which encompassed upstream TFs, downstream signaling pathways, and potential regulatory immune cells/functions (Fig. 6a). Based on gene expression patterns, inhibitor analysis, and integrated regulatory network analysis, we named the three types from cluster1–3 as Immune-Suppressive Macrophage-Infiltrated Thyroid Carcinoma Classification (ISMPTCC), Immune-Neutral Macrophage-Infiltrated Thyroid Carcinoma Classification (INMPTCC), and Immune-Activated Macrophage-Infiltrated Thyroid Carcinoma Classification (IAMPTCC). We also used a co-expression heatmap to display the specific correlation coefficients in each regulatory network (Fig. 6b). The detailed regulatory relationships within the co-expression network will be further elaborated in the discussion section. At this stage, we have understood the expression characteristics of the three IMPTCC subtypes. This clarifies the unique correspondence among macrophage subtypes (Macro_1–5), cell fate differentiation states (THCA1–3), and IMPTCC subtypes: IAMPTCC (related to M1 macrophage differentiation direction) correlates with THCA1 and corresponds to macrophages 1, 2; ISMPTCC (related to M2 macrophage differentiation direction) with THCA3, corresponding to macrophages 1, 3, 5. INMPTCC, without a clear macrophage subgroup correspondence, suggests an intermediary differentiation state. Integrating these relationships, we find that macrophages infiltrating PTC exhibit characteristics of transitioning from M1 polarized macrophages (anti-cancer, pro-inflammatory) to M2 polarized macrophages (anti-inflammatory, pro-cancer). This aligns with our definitions of macrophage subtypes, cell fate differentiation subtypes, and IMPTCC subtypes.
Fig. 6.
Construction of multi-omics regulatory network and target drug prediction for three subtypes in IMPTCC typing
(a) Multi-omics analysis results, amalgamating both scRNA-seq and bulk-sequencing data, utilizing a minimum selection criterion of correlation coefficient R>0.400 and P<0.001, structured around the MD&PRGs to form intricate regulatory networks—consisting of upstream TFs, downstream signaling pathways, and potential immune cell/function modulators—for the ISMPTCC, INMPTCC, and IAMPTCC subtypes.
(b) Co-expression heatmap manifesting specific correlation coefficients within each regulatory network.
(c) Dot map spotlighting target small molecule drugs identified for each IMPTCC subtype's regulatory network via the Cmap algorithm, with the ten drugs boasting the lowest P-values proposed as prospective inhibitors for each IMPTCC subtype.
(d-f) Utilizing data from the CCLE tumor cell line database for drug susceptibility analysis on genes affiliated with cellular destiny, it was discerned that the inhibitor PIK-93 exhibits specificity within ISMPTCC, while LGK-974 is specific to INMPTCC.
Identification of inhibitors through C-map and CCLE algorithms
To further integrate our proposed clinical staging with clinical treatment decisions, we initially employed the Cmap algorithm to identify small molecule drugs targeting MD&PRGs and TFs within the regulatory network of each IMPTCC subtype. We then selected the top 10 drugs with the smallest P-values and visualized them using a dot heatmap as target drugs for each IMPTCC subtype (Fig. 6c). Subsequently, with the assistance of the GDSC database, an analysis of drug sensitivity related to cell fate genes identified PIK-93 as a specific inhibitor for ISMPTCC (Figs. 6d-f). By integrating the results from both Cmap and GDSC databases, PI3K pathway inhibitor PIK93 [49], WNT pathway inhibitor LGK-974 [50,51], and M2 polarization inhibitor MS-275 [52] were found to be specifically effective against ISMPTCC, INMPTCC, and IAMPTCC, respectively.
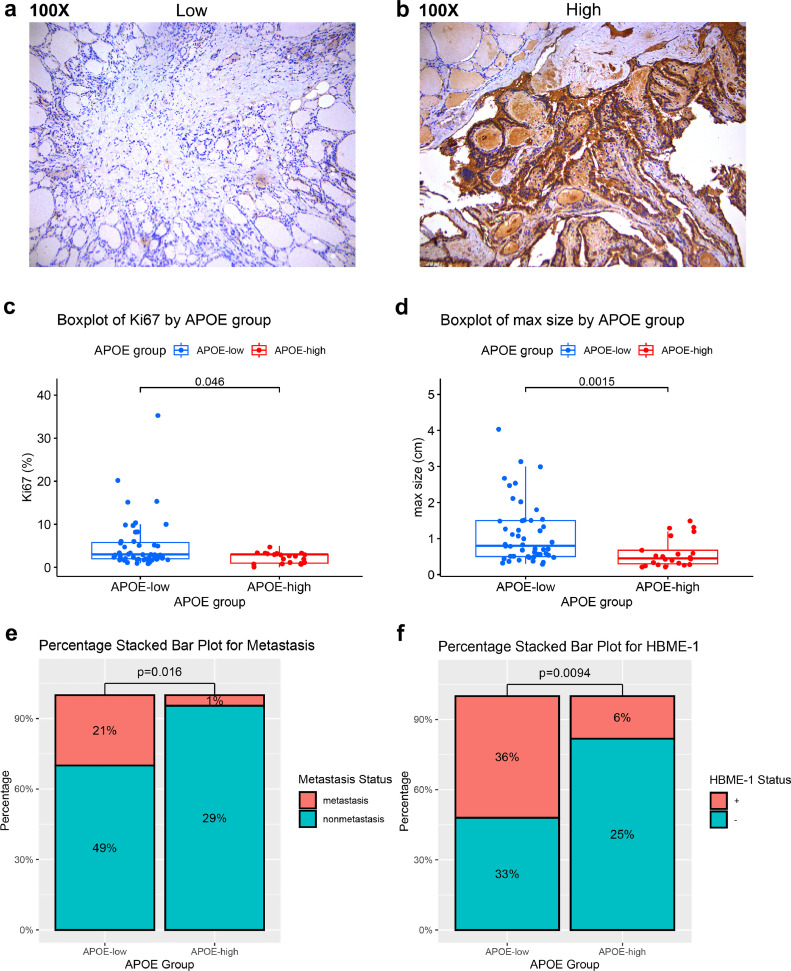
Histopathological validation
To enhance the clinical applicability of IMPTCC subtyping, the present study adopted the differential expression of the APOE gene as a marker to delineate the three subtypes of IMPTCC. A total of 72 patients with a confirmed diagnosis of PTC were included, and based on the staining results of tumor pathological sections, they were categorized into groups with high and low staining intensities for APOE (Fig. 7a-b). Detailed clinical and immunohistochemical data were carefully recorded for all samples. To discern disparities in clinical information and prognosis between the two groups, we conducted an association analysis concerning quantitative variables such as age, maximal dimension, and Ki67, along with categorical variables like metastasis, BRAF, and TERT, contingent on the high and low APOE expression categorizations. Given the inherent characteristics of the data distribution, the Mann-Whitney U test was deployed for quantitative variable comparisons. Categorical variables were evaluated using chi-square tests or Fisher's exact tests. Table 1 showcases the baseline clinical and statistical analysis outcomes of the 72 PTC samples. We focused on the intensity of Ki67 expression, an immunohistochemical index associated with the prognosis of PTC [53], and the average intensity of Ki67 expression was higher in the APOE low expression group than in the APOE high expression group (Fig. 7c). Regarding clinical features, the percentage of lymph node metastasis and the mean tumor diameter were significantly higher in the APOE low expression group than in the APOE high expression group (Fig. 7d-e). Additionally, there was a significant difference in the positivity rate of HBME-1, an immunohistochemical marker used for the diagnosis of PTC [54], between the APOE high/low groups (Fig. 7f). These observations resonate with our antecedent subtype analysis, suggesting an association between diminished APOE expression and a less favorable clinical prognosis.
Fig. 7.
Histopathological Validation
(a) Representative H&E images for the APOE-low staining group.
(b) Representative H&E images for APOE-high staining group.
(c) Stacked bar chart showing the differences in metastasis rates between the APOE high and low staining groups.
(d) Stacked bar chart showing the differences in HBME-1 positivity rates between the APOE high and low staining groups.
(e) Boxplot showing the differences in maximum tumor diameter between the APOE high and low staining groups.
Table 1.
Clinical baseline information and differential analysis results of PTC patients with high/low APOE expression.
Significant statistical differences were observed between patients with high and low APOE expression in terms of metastasis rate, maximum tumor diameter, Ki67 mutation proportion, and HBME-1 positivity rate.
| APOE-low (N = 50) |
APOE-high (N = 22) |
P-value | |
|---|---|---|---|
| Metastasis | |||
| Metastasis | 15 (30.0 %) | 1 (4.5 %) | 0.0161 |
| Nonmetastasis | 35 (70.0 %) | 21 (95.5 %) | |
| Max size (cm) | |||
| Mean (SD) | 1.12 (0.844) | 0.595 (0.408) | 0.00145 |
| Median [Min, Max] | 0.800 [0.300, 4.00] | 0.450 [0.200, 1.50] | |
| Ki67 (%) | |||
| Mean (SD) | 5.00 (5.94) | 2.23 (1.19) | 0.0457 |
| Median [Min, Max] | 3.00 [1.00, 35.0] | 3.00 [0, 5.00] | |
| Age (years) | |||
| Mean (SD) | 48.2 (12.6) | 47.3 (12.5) | 0.651 |
| Median [Min, Max] | 47.5 [22.0, 70.0] | 41.0 [30.0, 69.0] | |
| HBME-1 | |||
| - | 24 (48.0 %) | 18 (81.8 %) | 0.00938 |
| + | 26 (52.0 %) | 4 (18.2 %) | |
| BRAF | |||
| - | 17 (34.0 %) | 8 (36.4 %) | 1 |
| + | 33 (66.0 %) | 14 (63.6 %) | |
| TERT | |||
| - | 50 (100 %) | 22 (100 %) | 1 |
| + | 0 (0 %) | 0 (0 %) | |
| CK19 | |||
| - | 2 (4.0 %) | 1 (4.5 %) | 1 |
| + | 48 (96.0 %) | 21 (95.5 %) | |
| TPO | |||
| - | 44 (88.0 %) | 22 (100 %) | 0.168 |
| + | 6 (12.0 %) | 0 (0 %) | |
| Galectin-3 | |||
| - | 2 (4.0 %) | 0 (0 %) | 1 |
| + | 48 (96.0 %) | 22 (100 %) | |
| CD56 | |||
| - | 46 (92.0 %) | 21 (95.5 %) | 1 |
| + | 4 (8.0 %) | 1 (4.5 %) | |
| 34βE12 | |||
| - | 5 (10.0 %) | 2 (9.1 %) | 1 |
| + | 45 (90.0 %) | 20 (90.9 %) | |
| TG | |||
| - | 2 (4.0 %) | 1 (4.5 %) | 1 |
| + | 48 (96.0 %) | 21 (95.5 %) |
Sequencing to detect transposase accessible chromatin
Moreover, the accessibility of chromatin fragments for important genes (SQSTM1, RGCC, NRGN, PDK4, PLCXD1, APOE) was demonstrated by high-throughput sequencing findings of transposase-accessible chromatin (ATAC-seq) (Figure S9).
Discussion
Tumors exhibit a complicated ecological environment in which heterogeneous malignant cells engage in complex interactions with both immune and non-immune cells, establishing the intricate cell network of the tumor microenvironment [10,55]. In the tumor microenvironment, TIMs [14] are crucial components of the immune ecology of the TME [15]. TIMs consist of several distinct major lineages, including adipocytes, plasmacytoid dendritic cells (pDC), conventional dendritic cells (cDC), monocytes, and macrophages [12]. Macrophages are considered a double-edged sword with dual potential in cancer; their plasticity in response to environmental cues offers rich possibilities for macrophage-related research [[56], [57], [58]]. Owing to their function in mediating anti-tumor activity and inducing adaptive immune responses in immune regulation, macrophages have become one of the primary targets of current checkpoint inhibitor immunotherapy [59,60]. Additionally, studies on colorectal cancer in humans and mice have revealed that different subgroups of myeloid cells, while presenting functional differences, have variable sensitivities to therapeutic strategies targeting immune cells [61]. This result suggests that different differentiation states of macrophages may lead to varied clinical prognostic outcomes and responses to targeted treatments in tumors, which is also one of the significant reasons for our study on macrophage differentiation in the thyroid cancer environment.
During the research on macrophage differentiation, the polarization system of "classically activated" M1 and "alternatively activated" M2 macrophages has been used to describe the in vitro activation status of macrophages [62]. However many studies believe that the binary M1-M2 model is insufficient to describe the activation of macrophages [[63], [64], [65]]. Owing to the plasticity of macrophages and their dependency on the surrounding environment, macrophages in the actual in vivo environment may exhibit more complex differentiation forms [66].
Through the integration of scRNA-seq and bulk RNA sequencing data, we analyzed the differentiation status of macrophages in tumor-related myeloid cells. Based on the differentiation characteristics of macrophages, we further reclassified the immunological environment levels of thyroid cancer. Using multi-omics analysis based on both scRNA-seq and bulk-sequencing data, we categorized the differentiation states of macrophages into three subtypes (ISMPTCC, INMPTCC, and IAMPTCC). These subtypes are classified based on their immune characteristics into immune homeostasis, immune activation, and immune repair states, respectively reflecting the traits of M1-M0-M2 macrophages. However, unlike the classical binary description, this immune progression state, based on co-expression analysis results, does not strictly follow the classical characteristics of macrophage M1/M2 differentiation and shows different molecular mechanisms of action.
ISMPTCC is associated with Type I IFN response, M2 macrophages, and immune cells involved in immune repair. Concurrently, within the co-expression network, genes, transcription factors, and signaling pathways expressed by ISMPTCC manifest cellular metabolic traits. Notably, the expression patterns of glycolytic and lipid metabolic markers align with the anti-inflammatory metabolic characteristics typically seen in classically activated M2 macrophages. Macrophage polarization involves intricate metabolic shifts. Lipid and glucose metabolism are at the forefront of these changes. In M2 polarized macrophages, research indicates certain trends. Compared to M1 polarized macrophages, which have pronounced inflammatory traits, M2 macrophages show decreased glycolytic metabolism levels [67]. They also demonstrate increased lipid metabolism, represented by processes like fatty acid uptake and oxidation [[68], [69], [70]]. These observations match the gene expression traits we identified in our study. Within the co-expression network of Fig. 6, cluster1, key genes PDK4, APOE, transcription factor FOXO1, and the downstream glycolytic pathway, PI3K/AKT/mTOR pathway hint at the potential regulatory mechanisms of this subtype.
The transcription factor FOXO1 is a key member of the Forkhead transcription factors (FOX) family [71]. Depending on the regulatory targets it modulates, FOXO1 exhibits intricate regulatory activities, encompassing processes such as cell apoptosis, cellular metabolism, and immunological processes [72]. Its role in metabolic processes extends to both glucose metabolism and lipid metabolism. In terms of glucose metabolism, FOXO1 has been reported to be activated in diabetic cardiomyocytes and attenuates glycolysis by stimulating PDK4 [73]. PDK4 acts by phosphorylating and inhibiting the activity of another protein kinase, PDH, thereby suppressing glycolysis by limiting pyruvate production [74]. Consistent with this, PDK4 is expressed at high levels in ISMPTCC (Fig. 5a), suggesting that ISMPTCC may exhibit a suppressed state in glycolytic metabolism.
On the other hand, in terms of lipid metabolism regulation, FOXO1 has been reported to modulate lipid synthesis through a principal lipogenesis regulator, PPARγ [75]. Concurrently, in ISMPTCC, RBP7 is distinctly overexpressed (Fig. 5a). RBP7 is a target gene of PPARγ [76]. A dedicated study on this gene revealed that overexpression of RBP7 in adipocytes enhances lipid droplet formation and TG accumulation. This is accompanied by an increased expression of crucial lipogenic proteins including Pparγ, Fabp4, C/ebpα, and AdipoQ [77], suggesting an active state of lipid synthesis in ISMPTCC. Beyond its evident lipid synthesis characteristics, regarding lipid breakdown regulation, APOE is specifically underexpressed in ISMPTCC (Fig. 5a). Apolipoprotein E (apoE) is a multifunctional protein involved in clearing apoB-containing chylomicrons and very low-density lipoprotein (VLDL) remnants from plasma, thereby reducing plasma lipid levels [78]. The diminished expression of APOE indicates reduced lipid transport in ISMPTCC, and lipid retention is one of the crucial metabolic features of tumors [79]. This aligns with the traits of M2 macrophages promoting tumor progression.
The metabolic characteristics of ISMPTCC in terms of carbohydrate and lipid metabolism align with the metabolic features of M2 polarized macrophages, both exhibiting inhibition of glycolysis and enhancement of lipid metabolism. We also observed that the PI3K/AKT/mTOR pathway is an integral part of the ISMPTCC expression network. Intriguingly, both FOXO1 and MTOR are activated by the PI3K/AKT signal [80], and MTOR also plays a role in metabolic regulation [81]. This suggests a consistent role in metabolic regulation by MTOR and FOXO1, both downstream molecules of PI3K/AKT, and hints at a new direction for targeting the pathogenic mechanisms of ISMPTCC. In line with this, based on our drug sensitivity analysis conducted on tumor cell lines, the PI3K inhibitor PIK-93 demonstrated favorable sensitivity [82]. Being an upstream target of both the PI3K/AKT/MTOR and PI3K/AKT/FOXO1 pathways, inhibiting PI3K might play a role in targeting the metabolic regulation of FOXO1 and MTOR, assisting TAMs in adjusting their polarization state.
INMPTCC expresses M0 macrophages and immune cells related to immune activation. Within its co-expression network, the regulatory relationships among molecules suggest that the autophagy mechanism might exert its influence by affecting tumor angiogenesis. Fig. 6, showcasing the INMPTCC co-expression network, highlights key genes SQSTM1, protein SCD1, and transcription factors FOXO1, SMAD2, and EPAS1. The downstream angiogenesis pathway hints at the potential regulatory mechanisms inherent to this subtype.
Autophagy exhibits bidirectional and contradictory roles in both tumor suppression and tumor progression [83]. SMAD2 is a member of the SMAD protein family, mediating the signaling of transforming growth factor-beta (TGF-β) to regulate various cellular processes [84]. Research has identified that the TGF-β signaling pathway can activate the autophagy process in human cancer cells [85], thereby linking SMAD2 to the activation of autophagy. Concurrently, P62/SQSTM1, as a selective autophagy receptor and a crucial molecule in autophagosome formation, extensively participates in the autophagy process as a principal component and an initiator of WNT-regulated autophagy functions [86]. On the other hand, SCD1 has been established as an essential molecule during the early stages of autophagosome formation. The SCD1 inhibitor 28c inhibits the formation of p62/SQSTM1 puncta [87]. The presence of these three molecules in the INMPTCC co-expression network suggests the significance of the autophagy mechanism in the pathogenic mechanism of INMPTCC tumors. Autophagy in macrophages suppresses M1-type polarization of macrophages, alleviating chronic inflammation and organ fibrosis. The autophagy signaling in INMPTCC, at the molecular mechanism level, indicates that INMPTCC may serve as an intermediate state transitioning from M1 polarization to M2 polarization [88].
Further, the presence of the EPAS1 molecule suggests a connection between autophagy and the angiogenic phenotype. Angiogenesis is closely linked with both macrophages and tumors. Tumor-associated macrophages (TAMs) can promote tumor progression by producing angiogenic growth factors [18]. Angiogenesis-associated phenotypes are considered to be prominently enriched pathways in a macrophage-infiltrated immune microenvironment [61]. This perspective is validated in a single-cell study encompassing 15 cancer types, where angiogenesis-promoting tumor-associated macrophages were associated with poor prognoses in multiple tumors [19]. As a member of the hypoxia-inducible factors (HIFs), the family of transcription factors activated by hypoxia [89], EPAS1 can stabilize and activate cancer cells. Here, it induces angiogenesis-associated genes like VEGF [90], promoting angiogenesis and hence tumor progression. Concurrently, EPAS1 has been targeted therapeutically to inhibit vascular and tumor growth [91]. Importantly, autophagy activation involving SQSTM1 has been proven to suppress the angiogenic phenotype by degrading EPAS1, thereby inhibiting renal cell carcinoma. This indicates the adverse effect of autophagy activation on the angiogenic phenotype. In INMPTCC, aside from the high expression of genes associated with angiogenesis inhibition, MMP9 is specifically overexpressed (Fig. 5a). MMP9, a member of the pericellular proteases, plays a crucial role in initiating angiogenesis. It stimulates the recruitment and function of auxiliary cells from blood or bone marrow, enhancing angiogenesis [92]. Concurrently, the secretion of MMP9 is regulated by EPAS1, and overexpression of EPAS1 can boost the secretion of both VEGF and MMP9 [93].
In a comprehensive view, INMPTCC demonstrates intricate characteristics of autophagy and angiogenesis. The suppressive effect on macrophage M1 polarization brought about by autophagy and the negative regulatory influence on the angiogenic phenotype might counterbalance the positive regulatory role of angiogenesis directly mediated by the activated MMP9. Together, these factors might collectively constitute the regulatory milieu of INMPTCC, potentially explaining the intermediate prognostic state of INMPTCC. Concurrently, in our drug analysis, we observed that WNT pathway inhibitors, LGK-974 and WNT-C59, exhibited specificity in this subtype. Given the pivotal role of the WNT pathway in autophagy mechanisms, we hypothesize that the autophagic mechanism in INMPTCC might predominantly manifest tumor-promoting effects. Nonetheless, this conjecture warrants further experimental validation.
IAMPTCC manifests a type II IFN response as well as immune cells associated with other immune homeostatic states. Correspondingly, the molecular regulation evidence in IAMPTCC directly suggests its association with the M1 polarization state of macrophages. IAMPTCC's immune milieu displays characteristics related to immune homeostasis, accompanied by a type II IFN response. Interferons (IFNs) were initially identified as substances that "interfere" with viral replication in vitro, comprising type I interferons (IFN-α/β and related molecules), type II interferon (IFN-γ), and type III interferons (IFN-lambda) [94]. Type II IFN response is considered a classical hallmark of activated M1 macrophage polarization [95,96]. EFHD2, which exhibits specific overexpression in IAMPTCC (Fig. 5a), is believed to be a crucial molecule for the binding of IFN-γ to functional IFN-γR [97]. We also observed the appearance in IAMPTCC's molecular regulatory network of the protein YAP1, known to promote M1 polarization [98], molecules related to the MAPK pathway involved in M1 polarization regulation, namely BRAF, MEK1 [99], and EZH2, which enhances the chemotaxis of inflammatory macrophages in the tumor environment [100].
In terms of pathway expression features, the KRAS pathway emerges in the co-expression network [101], aligning with a breast cancer-related study that found the enrichment of KRAS signaling-associated genes to be associated with an anti-tumor immune microenvironment. Likewise, the androgen receptor pathway, which also appears in the co-expression network, has similar evidence [102]. Evidently, in terms of immune characteristics and gene and pathway expression patterns, IAMPTCC displays differentiation characteristics of M1 polarized macrophages, distinguishing it significantly from subtypes 1 and 2 in molecular expression features.
We acknowledge the limitations of our study. As a project reliant on publicly available data analysis, we utilized thyroid cancer scRNA-seq data from Peking University Cancer Hospital & Institute and bulk RNA sequencing data obtained from the TCGA database. This cross-verification approach may introduce biases stemming from ethnic and regional differences. Additionally, we made limited inferences based on existing research conclusions regarding the regulatory network relationships indicated by our bioinformatics analysis results. Preliminary experimental validations were conducted based on these interpretations. Building upon our preliminary validations, our future research will delve into the transdifferentiation of macrophages from anti-inflammatory to pro-inflammatory states. We plan to conduct comprehensive experiments, encompassing both upstream and downstream validation of the potential molecular regulatory relationships identified in our initial study. This will involve detailed functional assays to understand the mechanistic underpinnings of macrophage behavior in the context of papillary thyroid carcinoma. These studies aim to validate and expand upon our predictive model and candidate molecules, enriching our understanding of their roles in tumor biology.
Conclusion
In summary, through the integration of scRNA-seq data from the PTC environment and bulk RNA-seq data from THCA for multi-omics analysis, our study established a thyroid cancer typing system based on macrophage differentiation characteristics. Guided by intricate upstream and downstream co-expression network regulatory clues, we identified potential thyroid cancer metabolism and regulation features under different macrophage differentiation states. The differentiation variance in macrophage states may indeed offer some discriminative power in the reclassification of PTC. Research into the specific mechanisms underlying the prognostic variations of PTC should consider the tumor immune microenvironment, especially the differential characteristics of tumor-infiltrating macrophages. We hope our analysis can offer new insights and research directions for risk stratification and precision treatment of PTC.
Funding
This study was supported by the General Project of Natural Science Foundation of Henan Province (222300420568); Key Medical Science and Technology Project of Henan Province (SBGJ202101014); Major Scientific Research Projects of Traditional Chinese Medicine in Henan Province (20–21ZYZD14); Cultivation of Young and Middle-aged Health Science and Technology Innovation Leading Talents in Henan Province (YXKC2020015); National Natural Science Foundation of China (Grant No. 82002923) and Shanghai Rising-Star Program (Sailing Special Program) (No. 23YF1458400).
Availability of data and materials
Table S1 displays all the datasets, code packages, and software information employed in this study. The clinical patient information and all the codes generated during the data analysis process in this study are stored in Supplementary material.
Ethics approval and consent to participate
Approval for histopathological validation using clinical samples was obtained from the Clinical Research Institution Review Committee of the Tenth People's Hospital, which is affiliated with Tongji University in Shanghai. Before participation, all patients provided informed written consent. Samples for the additional validation were gathered from the Tenth People's Hospital, which is affiliated with Tongji University, from 2016 to 2018.
Consent for publication
Not applicable.
CRediT authorship contribution statement
Hanlin Sun: Formal analysis, Software, Validation, Writing – original draft. Zhengyan Chang: Funding acquisition, Methodology, Resources, Validation. Hongqiang Li: Resources, Supervision, Writing – review & editing. Yifeng Tang: Resources, Supervision, Writing – review & editing. Yihao Liu: Investigation, Supervision, Writing – review & editing. Lixue Qiao: Investigation, Writing – review & editing. Guicheng Feng: Writing – review & editing. Runzhi Huang: Conceptualization, Data curation, Formal analysis, Methodology, Software. Dongyan Han: Funding acquisition, Supervision. De-tao Yin: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to express our gratitude to the team at TCGA and GEO for allowing us to use their data.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2024.101889.
Contributor Information
Runzhi Huang, Email: runzhihuang2022@163.com.
Dongyan Han, Email: 1500057@tongji.edu.cn.
De-tao Yin, Email: detaoyin@zzu.edu.cn.
Appendix. Supplementary materials
References
- 1.Wiltshire J.J., Drake T.M., Uttley L., Balasubramanian S.P. Systematic review of trends in the incidence rates of thyroid cancer. Thyroid. 2016;26(11):1541–1552. doi: 10.1089/thy.2016.0100. [DOI] [PubMed] [Google Scholar]
- 2.Antonelli A., La Motta C. Novel therapeutic clues in thyroid carcinomas: the role of targeting cancer stem cells. Med. Res. Rev. 2017;37(6):1299–1317. doi: 10.1002/med.21448. [DOI] [PubMed] [Google Scholar]
- 3.Qiu H., Cao S., Xu R. Cancer incidence, mortality, and burden in China: a time-trend analysis and comparison with the United States and United Kingdom based on the global epidemiological data released in 2020. Cancer Commun. (Lond) 2021;41(10):1037–1048. doi: 10.1002/cac2.12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrari S.M., Bocci G., Di Desidero T., Ruffilli I., Elia G., Ragusa F., et al. Vandetanib has antineoplastic activity in anaplastic thyroid cancer, in vitro and in vivo. Oncol. Rep. 2018;39(5):2306–2314. doi: 10.3892/or.2018.6305. [DOI] [PubMed] [Google Scholar]
- 5.Antonelli A., Ferrari S.M., Fallahi P., Berti P., Materazzi G., Marchetti I., et al. Evaluation of the sensitivity to chemotherapeutics or thiazolidinediones of primary anaplastic thyroid cancer cells obtained by fine-needle aspiration. Eur. J. Endocrinol. 2008;159(3):283–291. doi: 10.1530/EJE-08-0190. [DOI] [PubMed] [Google Scholar]
- 6.Davies L., Hoang J.K. Thyroid cancer in the USA: current trends and outstanding questions. Lancet. Diabetes Endocrinol. 2021;9(1):11–12. doi: 10.1016/S2213-8587(20)30372-7. [DOI] [PubMed] [Google Scholar]
- 7.Xing M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat. Rev. Cancer. 2013;13(3):184–199. doi: 10.1038/nrc3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheffel R.S., Dora J.M., Maia A.L. BRAF mutations in thyroid cancer. Curr. Opin. Oncol. 2022;34(1):9–18. doi: 10.1097/CCO.0000000000000797. [DOI] [PubMed] [Google Scholar]
- 9.Balkwill F., Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357(9255):539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Bissell M.J., Hines W.C. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat. Med. 2011;17(3):320–329. doi: 10.1038/nm.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coussens L.M., Zitvogel L., Palucka A.K. Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science. 2013;339(6117):286–291. doi: 10.1126/science.1232227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrari S.M., Fallahi P., Elia G., Ragusa F., Ruffilli I., Paparo S.R., et al. Thyroid autoimmune disorders and cancer. Semin. Cancer Biol. 2020;64:135–146. doi: 10.1016/j.semcancer.2019.05.019. [DOI] [PubMed] [Google Scholar]
- 14.Le Chatelier E., Nielsen T., Qin J., Prifti E., Hildebrand F., Falony G., et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500(7464):541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 15.Engblom C., Pfirschke C., Pittet M.J. The role of myeloid cells in cancer therapies. Nat. Rev. Cancer. 2016;16(7):447–462. doi: 10.1038/nrc.2016.54. [DOI] [PubMed] [Google Scholar]
- 16.Schmid M.C., Varner J.A. Myeloid cells in the tumor microenvironment: modulation of tumor angiogenesis and tumor inflammation. J. Oncol. 2010;2010 doi: 10.1155/2010/201026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ginhoux F., Schultze J.L., Murray P.J., Ochando J., Biswas S.K. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat. Immunol. 2016;17(1):34–40. doi: 10.1038/ni.3324. [DOI] [PubMed] [Google Scholar]
- 18.DeNardo D.G., Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 2019;19(6):369–382. doi: 10.1038/s41577-019-0127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng S., Li Z., Gao R., Xing B., Gao Y., Yang Y., et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell. 2021;184(3):792–809.e23. doi: 10.1016/j.cell.2021.01.010. [DOI] [PubMed] [Google Scholar]
- 20.Hie B., Bryson B., Berger B. Efficient integration of heterogeneous single-cell transcriptomes using Scanorama. Nat. Biotechnol. 2019;37(6):685–691. doi: 10.1038/s41587-019-0113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X., Lan Y., Xu J., Quan F., Zhao E., Deng C., et al. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic. Acids. Res. 2019;47(D1):D721–D7d8. doi: 10.1093/nar/gky900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A., et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14(10):979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tremaroli V., Karlsson F., Werling M., Ståhlman M., Kovatcheva-Datchary P., Olbers T., et al. Roux-en-Y gastric bypass and vertical banded gastroplasty induce long-term changes on the human gut microbiome contributing to fat mass regulation. Cell Metab. 2015;22(2):228–238. doi: 10.1016/j.cmet.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., Mesirov J.P. GenePattern 2.0. Nat. Genet. 2006;38(5):500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 25.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hosack D.A., Dennis G., Jr., Sherman B.T., Lane H.C., Lempicki R.A. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4(10):R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic. Acids. Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Charoentong P., Finotello F., Angelova M., Mayer C., Efremova M., Rieder D., et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 32.Xiao B., Liu L., Li A., Xiang C., Wang P., Li H., et al. Identification and verification of immune-related gene prognostic signature based on ssGSEA for osteosarcoma. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.607622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mora A., Donaldson I.M. iRefR: an R package to manipulate the iRefIndex consolidated protein interaction database. BMC Bioinf. 2011;12:455. doi: 10.1186/1471-2105-12-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamb J., Crawford E.D., Peck D., Modell J.W., Blat I.C., Wrobel M.J., et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 36.Yang W., Soares J., Greninger P., Edelman E.J., Lightfoot H., Forbes S., et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic. Acids. Res. 2013;41(Database issue):D955–D961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10(12):1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y.Y., Jiang H., Pan J., Huang X.R., Wang Y.C., Huang H.F., et al. Macrophage-to-Myofibroblast Transition Contributes to Interstitial Fibrosis in Chronic Renal Allograft Injury. J. Am. Soc. Nephrol. 2017;28(7):2053–2067. doi: 10.1681/ASN.2016050573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilfillan A.M., Rivera J. The tyrosine kinase network regulating mast cell activation. Immunol. Rev. 2009;228(1):149–169. doi: 10.1111/j.1600-065X.2008.00742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niehaus J.K., Taylor-Blake B., Loo L., Simon J.M., Zylka M.J. Spinal macrophages resolve nociceptive hypersensitivity after peripheral injury. Neuron. 2021;109(8):1274. doi: 10.1016/j.neuron.2021.02.018. -82.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu D., Liu Z., Liang M.X., Chen W.Q., Fei Y.J., Yang S.J., et al. Hyperthermia promotes M1 polarization of macrophages via exosome-mediated HSPB8 transfer in triple negative breast cancer. Discov Oncol. 2023;14(1):81. doi: 10.1007/s12672-023-00697-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu P.S., Chen Y.T., Li X., Hsueh P.C., Tzeng S.F., Chen H., et al. CD40 signal rewires fatty acid and glutamine metabolism for stimulating macrophage anti-tumorigenic functions. Nat. Immunol. 2023;24(3):452–462. doi: 10.1038/s41590-023-01430-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yim A., Smith C., Brown A.M. Osteopontin/secreted phosphoprotein-1 harnesses glial-, immune-, and neuronal cell ligand-receptor interactions to sense and regulate acute and chronic neuroinflammation. Immunol. Rev. 2022;311(1):224–233. doi: 10.1111/imr.13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuch K., Wanko B., Ambroz K., Castelo-Rosa A., Moreno-Viedma V., Grün N.G., et al. Osteopontin affects macrophage polarization promoting endocytic but not inflammatory properties. Obesity (Silver Spring) 2016;24(7):1489–1498. doi: 10.1002/oby.21510. [DOI] [PubMed] [Google Scholar]
- 45.Fuior E.V., Gafencu A.V. Apolipoprotein C1: its Pleiotropic Effects in Lipid Metabolism and Beyond. Int. J. Mol. Sci. 2019;20(23) doi: 10.3390/ijms20235939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abe M. [Complement activation and inflammation] Rinsho Byori. 2006;54(7):744–756. [PubMed] [Google Scholar]
- 47.Katayama M., Ota K., Nagi-Miura N., Ohno N., Yabuta N., Nojima H., et al. Ficolin-1 is a promising therapeutic target for autoimmune diseases. Int. Immunol. 2019;31(1):23–32. doi: 10.1093/intimm/dxy056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zegeye M.M., Lindkvist M., Fälker K., Kumawat A.K., Paramel G., Grenegård M., et al. Activation of the JAK/STAT3 and PI3K/AKT pathways are crucial for IL-6 trans-signaling-mediated pro-inflammatory response in human vascular endothelial cells. Cell Commun. Signal. 2018;16(1):55. doi: 10.1186/s12964-018-0268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hanes C.M., D'Amico A.E., Ueyama T., Wong A.C., Zhang X., Hynes W.F., et al. Golgi-Associated Protein Kinase C-ε Is Delivered to Phagocytic Cups: role of Phosphatidylinositol 4-Phosphate. J. Immunol. 2017;199(1):271–277. doi: 10.4049/jimmunol.1700243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang Y., Jiang M., Chen A., Qu W., Han X., Zuo J., et al. Porcupine inhibitor LGK‑974 inhibits Wnt/β‑catenin signaling and modifies tumor‑associated macrophages resulting in inhibition of the malignant behaviors of non‑small cell lung cancer cells. Mol Med Rep. 2021;24(2) doi: 10.3892/mmr.2021.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Y., Ye Y.C., Chen Y., Zhao J.L., Gao C.C., Han H., et al. Crosstalk between hepatic tumor cells and macrophages via Wnt/β-catenin signaling promotes M2-like macrophage polarization and reinforces tumor malignant behaviors. Cell Death. Dis. 2018;9(8):793. doi: 10.1038/s41419-018-0818-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He L., Jhong J.H., Chen Q., Huang K.Y., Strittmatter K., Kreuzer J., et al. Global characterization of macrophage polarization mechanisms and identification of M2-type polarization inhibitors. Cell Rep. 2021;37(5) doi: 10.1016/j.celrep.2021.109955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang J., Gui C., Qiu S., Wang M. The clinicopathological significance of Ki67 in papillary thyroid carcinoma: a suitable indicator? World J Surg Oncol. 2018;16(1):100. doi: 10.1186/s12957-018-1384-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Y.J., Zhao R.M., Zhao Q., Li B.Y., Ma Q.Y., Li X., et al. Diagnostic significance of elevated expression of HBME-1 in papillary thyroid carcinoma. Tumour Biol. 2016;37(7):8715–8720. doi: 10.1007/s13277-015-4169-5. [DOI] [PubMed] [Google Scholar]
- 55.Whitmer R.A., Gunderson E.P., Barrett-Connor E., Quesenberry C.P., Jr., Yaffe K. Obesity in middle age and future risk of dementia: a 27 year longitudinal population based study. BMJ. 2005;330(7504):1360. doi: 10.1136/bmj.38446.466238.E0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mantovani A., Marchesi F., Malesci A., Laghi L., Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 2017;14(7):399–416. doi: 10.1038/nrclinonc.2016.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cassetta L., Pollard J.W. Targeting macrophages: therapeutic approaches in cancer. Nat. Rev. Drug Discov. 2018;17(12):887–904. doi: 10.1038/nrd.2018.169. [DOI] [PubMed] [Google Scholar]
- 58.Locati M., Curtale G., Mantovani A. Diversity, Mechanisms, and Significance of Macrophage Plasticity. Annu Rev Pathol. 2020;15:123–147. doi: 10.1146/annurev-pathmechdis-012418-012718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kryczek I., Zou L., Rodriguez P., Zhu G., Wei S., Mottram P., et al. B7-H4 expression identifies a novel suppressive macrophage population in human ovarian carcinoma. J. Exp. Med. 2006;203(4):871–881. doi: 10.1084/jem.20050930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim I.S., Gao Y., Welte T., Wang H., Liu J., Janghorban M., et al. Immuno-subtyping of breast cancer reveals distinct myeloid cell profiles and immunotherapy resistance mechanisms. Nat. Cell Biol. 2019;21(9):1113–1126. doi: 10.1038/s41556-019-0373-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang L., Li Z., Skrzypczynska K.M., Fang Q., Zhang W., O'Brien S.A., et al. Single-Cell Analyses Inform Mechanisms of Myeloid-Targeted Therapies in Colon Cancer. Cell. 2020;181(2):442. doi: 10.1016/j.cell.2020.03.048. -59.e29. [DOI] [PubMed] [Google Scholar]
- 62.Vogel D.Y., Glim J.E., Stavenuiter A.W., Breur M., Heijnen P., Amor S., et al. Human macrophage polarization in vitro: maturation and activation methods compared. Immunobiology. 2014;219(9):695–703. doi: 10.1016/j.imbio.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 63.Helm O., Held-Feindt J., Grage-Griebenow E., Reiling N., Ungefroren H., Vogel I., et al. Tumor-associated macrophages exhibit pro- and anti-inflammatory properties by which they impact on pancreatic tumorigenesis. Int. J. Cancer. 2014;135(4):843–861. doi: 10.1002/ijc.28736. [DOI] [PubMed] [Google Scholar]
- 64.Kratochvill F., Neale G., Haverkamp J.M., Van de Velde L.A., Smith A.M., Kawauchi D., et al. TNF Counterbalances the Emergence of M2 Tumor Macrophages. Cell Rep. 2015;12(11):1902–1914. doi: 10.1016/j.celrep.2015.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stables M.J., Shah S., Camon E.B., Lovering R.C., Newson J., Bystrom J., et al. Transcriptomic analyses of murine resolution-phase macrophages. Blood. 2011;118(26):e192–e208. doi: 10.1182/blood-2011-04-345330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shalek A.K., Satija R., Shuga J., Trombetta J.J., Gennert D., Lu D., et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510(7505):363–369. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saha S., Shalova I.N., Biswas S.K. Metabolic regulation of macrophage phenotype and function. Immunol. Rev. 2017;280(1):102–111. doi: 10.1111/imr.12603. [DOI] [PubMed] [Google Scholar]
- 68.Martinez F.O., Gordon S., Locati M., Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J. Immunol. 2006;177(10):7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 69.Odegaard J.I., Chawla A. Alternative macrophage activation and metabolism. Annu Rev Pathol. 2011;6:275–297. doi: 10.1146/annurev-pathol-011110-130138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu L., Zhao Q., Yang T., Ding W., Zhao Y. Cellular metabolism and macrophage functional polarization. Int. Rev. Immunol. 2015;34(1):82–100. doi: 10.3109/08830185.2014.969421. [DOI] [PubMed] [Google Scholar]
- 71.Kandula V., Kosuru R., Li H., Yan D., Zhu Q., Lian Q., et al. Forkhead box transcription factor 1: role in the pathogenesis of diabetic cardiomyopathy. Cardiovasc Diabetol. 2016;15:44. doi: 10.1186/s12933-016-0361-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xing Y.Q., Li A., Yang Y., Li X.X., Zhang L.N., Guo H.C. The regulation of FOXO1 and its role in disease progression. Life Sci. 2018;193:124–131. doi: 10.1016/j.lfs.2017.11.030. [DOI] [PubMed] [Google Scholar]
- 73.Yan D., Cai Y., Luo J., Liu J., Li X., Ying F., et al. FOXO1 contributes to diabetic cardiomyopathy via inducing imbalanced oxidative metabolism in type 1 diabetes. J. Cell. Mol. Med. 2020;24(14):7850–7861. doi: 10.1111/jcmm.15418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li Z., Peng Y., Li J., Chen Z., Chen F., Tu J., et al. N(6)-methyladenosine regulates glycolysis of cancer cells through PDK4. Nat. Commun. 2020;11(1):2578. doi: 10.1038/s41467-020-16306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen J., Lu Y., Tian M., Huang Q. Molecular mechanisms of FOXO1 in adipocyte differentiation. J. Mol. Endocrinol. 2019;62(3):R239–Rr53. doi: 10.1530/JME-18-0178. [DOI] [PubMed] [Google Scholar]
- 76.Woll A.W., Quelle F.W., Sigmund C.D. PPARγ and retinol binding protein 7 form a regulatory hub promoting antioxidant properties of the endothelium. Physiol. Genomics. 2017;49(11):653–658. doi: 10.1152/physiolgenomics.00055.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim D.H., Ahn J., Suh Y., Ziouzenkova O., Lee J.W., Lee K. Retinol Binding Protein 7 Promotes Adipogenesis in vitro and Regulates Expression of. Genes Involved in Retinol Metabolism. Front Cell Dev Biol. 2022;10 doi: 10.3389/fcell.2022.876031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Getz G.S., Reardon C.A. Apoprotein E and Reverse Cholesterol Transport. Int. J. Mol. Sci. 2018;19(11) doi: 10.3390/ijms19113479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bian X., Liu R., Meng Y., Xing D., Xu D., Lu Z. Lipid metabolism and cancer. J. Exp. Med. 2021;218(1) doi: 10.1084/jem.20201606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Song M., Bode A.M., Dong Z., Lee M.H. AKT as a Therapeutic Target for Cancer. Cancer Res. 2019;79(6):1019–1031. doi: 10.1158/0008-5472.CAN-18-2738. [DOI] [PubMed] [Google Scholar]
- 81.Feng J., Qiu S., Zhou S., Tan Y., Bai Y., Cao H., et al. mTOR: a Potential New Target in Nonalcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2022;23(16) doi: 10.3390/ijms23169196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Knight Z.A., Gonzalez B., Feldman M.E., Zunder E.R., Goldenberg D.D., Williams O., et al. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell. 2006;125(4):733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eisenberg-Lerner A., Kimchi A. The paradox of autophagy and its implication in cancer etiology and therapy. Apoptosis. 2009;14(4):376–391. doi: 10.1007/s10495-008-0307-5. [DOI] [PubMed] [Google Scholar]
- 84.Abdollah S., Macías-Silva M., Tsukazaki T., Hayashi H., Attisano L., Wrana J.L. TbetaRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2-Smad4 complex formation and signaling. J. Biol. Chem. 1997;272(44):27678–27685. doi: 10.1074/jbc.272.44.27678. [DOI] [PubMed] [Google Scholar]
- 85.Kiyono K., Suzuki H.I., Matsuyama H., Morishita Y., Komuro A., Kano M.R., et al. Autophagy is activated by TGF-beta and potentiates TGF-beta-mediated growth inhibition in human hepatocellular carcinoma cells. Cancer Res. 2009;69(23):8844–8852. doi: 10.1158/0008-5472.CAN-08-4401. [DOI] [PubMed] [Google Scholar]
- 86.Lorzadeh S., Kohan L., Ghavami S., Azarpira N. Autophagy and the Wnt signaling pathway: a focus on Wnt/β-catenin signaling. Biochim. Biophys. Acta. Mol. Cell Res. 2021;1868(3) doi: 10.1016/j.bbamcr.2020.118926. [DOI] [PubMed] [Google Scholar]
- 87.Ascenzi F., De Vitis C., Maugeri-Saccà M., Napoli C., Ciliberto G., Mancini R. SCD1, autophagy and cancer: implications for therapy. J. Exp. Clin. Cancer Res. 2021;40(1):265. doi: 10.1186/s13046-021-02067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wen J.H., Li D.Y., Liang S., Yang C., Tang J.X., Liu H.F. Macrophage autophagy in macrophage polarization, chronic inflammation and organ fibrosis. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.946832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gonzalez F.J., Xie C., Jiang C. The role of hypoxia-inducible factors in metabolic diseases. Nat. Rev. Endocrinol. 2018;15(1):21–32. doi: 10.1038/s41574-018-0096-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu Y., Cox S.R., Morita T., Kourembanas S. Hypoxia regulates vascular endothelial growth factor gene expression in endothelial cells. Identification of a 5′ enhancer. Circ. Res. 1995;77(3):638–643. doi: 10.1161/01.res.77.3.638. [DOI] [PubMed] [Google Scholar]
- 91.Arjaans M., Schröder C.P., Oosting S.F., Dafni U., Kleibeuker J.E., de Vries E.G. VEGF pathway targeting agents, vessel normalization and tumor drug uptake: from bench to bedside. Oncotarget. 2016;7(16):21247–21258. doi: 10.18632/oncotarget.6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van Hinsbergh V.W., Engelse M.A., Quax P.H. Pericellular proteases in angiogenesis and vasculogenesis. Arterioscler. Thromb. Vasc. Biol. 2006;26(4):716–728. doi: 10.1161/01.ATV.0000209518.58252.17. [DOI] [PubMed] [Google Scholar]
- 93.Zhang Q., Lou Y., Zhang J., Fu Q., Wei T., Sun X., et al. Hypoxia-inducible factor-2α promotes tumor progression and has crosstalk with Wnt/β-catenin signaling in pancreatic cancer. Mol. Cancer. 2017;16(1):119. doi: 10.1186/s12943-017-0689-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang S.Y., Boisson-Dupuis S., Chapgier A., Yang K., Bustamante J., Puel A., et al. Inborn errors of interferon (IFN)-mediated immunity in humans: insights into the respective roles of IFN-alpha/beta, IFN-gamma, and IFN-lambda in host defense. Immunol. Rev. 2008;226:29–40. doi: 10.1111/j.1600-065X.2008.00698.x. [DOI] [PubMed] [Google Scholar]
- 95.Wang F., Zhang S., Jeon R., Vuckovic I., Jiang X., Lerman A., et al. Interferon Gamma Induces Reversible Metabolic Reprogramming of M1 Macrophages to Sustain Cell Viability and Pro-Inflammatory Activity. EBioMedicine. 2018;30:303–316. doi: 10.1016/j.ebiom.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pagie S., Gérard N., Charreau B. Notch signaling triggered via the ligand DLL4 impedes M2 macrophage differentiation and promotes their apoptosis. Cell Commun. Signal. 2018;16(1):4. doi: 10.1186/s12964-017-0214-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu X., Xu J., Wu J., Hu Y., Han Y., Gu Y., et al. Phosphorylation-Mediated IFN-γR2 Membrane Translocation Is Required to Activate Macrophage Innate Response. Cell. 2018;175(5):1336. doi: 10.1016/j.cell.2018.09.011. -51.e17. [DOI] [PubMed] [Google Scholar]
- 98.Zhou X., Li W., Wang S., Zhang P., Wang Q., Xiao J., et al. YAP Aggravates Inflammatory Bowel Disease by Regulating M1/M2 Macrophage Polarization and Gut Microbial Homeostasis. Cell Rep. 2019;27(4):1176. doi: 10.1016/j.celrep.2019.03.028. -89.e5. [DOI] [PubMed] [Google Scholar]
- 99.Liu C., Hu F., Jiao G., Guo Y., Zhou P., Zhang Y., et al. Dental pulp stem cell-derived exosomes suppress M1 macrophage polarization through the ROS-MAPK-NFκB P65 signaling pathway after spinal cord injury. J Nanobiotechnology. 2022;20(1):65. doi: 10.1186/s12951-022-01273-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang X., Wang Y., Yuan J., Li N., Pei S., Xu J., et al. Macrophage/microglial Ezh2 facilitates autoimmune inflammation through inhibition of Socs3. J. Exp. Med. 2018;215(5):1365–1382. doi: 10.1084/jem.20171417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tokumaru Y., Oshi M., Katsuta E., Yan L., Satyananda V., Matsuhashi N., et al. KRAS signaling enriched triple negative breast cancer is associated with favorable tumor immune microenvironment and better survival. Am J Cancer Res. 2020;10(3):897–907. [PMC free article] [PubMed] [Google Scholar]
- 102.Salehi R., Asare-Werehene M., Wyse B.A., Abedini A., Pan B., Gutsol A., et al. Granulosa cell-derived miR-379-5p regulates macrophage polarization in polycystic ovarian syndrome. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1104550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Table S1 displays all the datasets, code packages, and software information employed in this study. The clinical patient information and all the codes generated during the data analysis process in this study are stored in Supplementary material.