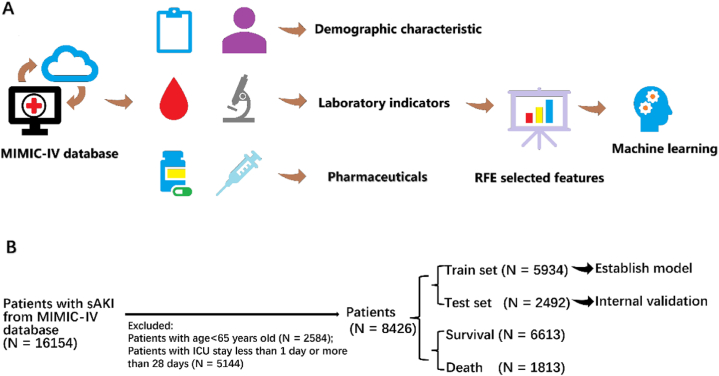
Abstract
Background
Sepsis-associated acute kidney injury (SA-AKI) is a severe complication associated with poorer prognosis and increased mortality, particularly in elderly patients. Currently, there is a lack of accurate mortality risk prediction models for these patients in clinic.
Objectives
This study aimed to develop and validate machine learning models for predicting in-hospital mortality risk in elderly patients with SA-AKI.
Methods
Machine learning models were developed and validated using the public, high-quality Medical Information Mart for Intensive Care (MIMIC)-IV critically ill database. The recursive feature elimination (RFE) algorithm was employed for key feature selection. Eleven predictive models were compared, with the best one selected for further validation. Shapley Additive Explanations (SHAP) values were used for visualization and interpretation, making the machine learning models clinically interpretable.
Results
There were 16,154 patients with SA-AKI in the MIMIC-IV database, and 8426 SA-AKI patients were included in this study (median age: 77.0 years; female: 45%). 7728 patients excluded based on these criteria. They were randomly divided into a training cohort (5,934, 70%) and a validation cohort (2,492, 30%). Nine key features were selected by the RFE algorithm. The CatBoost model achieved the best performance, with an AUC of 0.844 in the training cohort and 0.804 in the validation cohort. SHAP values revealed that AKI stage, PaO2, and lactate were the top three most important features contributing to the CatBoost model.
Conclusion
We developed a model capable of predicting the risk of in-hospital mortality in elderly patients with SA-AKI.
Keywords: Machine learning, Model interpretation, Sepsis, Acute kidney injury, Elderly
1. Introduction
Sepsis is a life-threatening organ dysfunction caused by a dysregulated host response to infection, and it remains the primary cause of death in critically ill patients [1]. Although advances in antibiotics and intensive supportive measures have contributed to a decrease in sepsis mortality rates, the long-term prognosis remains unfavorable. Approximately 48.9 million cases of sepsis and 11 million sepsis-related deaths are reported annually, accounting for 19.7% of all deaths [2].
Acute kidney injury (formerly known as acute renal failure) is a syndrome characterized by the rapid loss of the kidney's excretory function, which is defined as any of the following (Not Graded):(1)Increase in SCr by ≥ 0.3 mg/dl (≥26.5 lmol/l) within 48 h; or(2) Increase in SCr to ≥1.5 times baseline, which is known or presumed to have occurred within the prior 7 days; or(3) Urine volume <0.5 ml/kg/h for 6 h [3]. Acute kidney injury (AKI) has long been recognized as a common and complex clinical complication of sepsis, affecting up to 60% of patients with sepsis [4,5]. Sepsis-associated AKI (SA-AKI) is closely associated with a worse prognosis, extended intensive care unit (ICU) stays, diminished quality of life, and a higher risk of mortality compared to non-AKI patients [[6], [7], [8]].
Between 2000 and 2019, the average global life span increased by over six years, with life expectancy rising from 66.8 to 73.4 years [9]. As the world's population ages, approximately 700 million people are now aged 65 years and older, comprising 9% of the total population. This population segment is projected to increase to 17% by 2050. Age is considered an independent risk factor for mortality in both sepsis and AKI [8,9]. Studies have demonstrated that sepsis mortality increases from 10% in children to 26% in patients aged 60–64, and 38% in those over 85 [10]. Schmitt et al. found that patients aged over 65 had a significantly worse prognosis of AKI compared to younger patients [11].
Considering the critical condition of elderly patients with SA-AKI, early identification of their outcomes is essential. Mortality prediction is a common clinical task for quantifying the severity of a patient's physiological condition, and it can help clinicians improving clinical decision-making [12], particularly in the ICU. Presently, severity scoring systems, such as the Simplified Acute Physiology Score II (SAPS-II) and Sequential Organ Failure Assessment (SOFA) score, are widely used for mortality prediction. However, their performance is often unsatisfactory in various clinical situations due to the non-specificity and linearity characteristics of the employed models [13,14].
Recently, with the rapid advancement of artificial intelligence (AI), machine learning (ML) models have achieved state-of-the-art predictive performance [15,16]. Through appropriate "learning" and "interpreting," an explainable ML model can efficiently process and visualize clinical information, allowing clinicians to provide better healthcare to patients. The effectiveness of interpretable ML models in addressing specific ICU-related tasks has been demonstrated and gained satisfactory performance. Therefore, this study aims to develop an explainable ML model to predict mortality risk for elderly patients with SA-AKI in the ICU. Using the Medical Information Mart for Intensive Care IV (MIMIC-IV) population, we employed recursive feature elimination (RFE) to identify key features. Eleven machine learning models were performed, with the best selected for further validation. The SHapley Additive exPlanations (SHAP) values were then used to visualize the relationships between these features and the outcomes.
2. Materials and methods
Data Source: We conducted this retrospective study utilizing the MIMIC-IV database, which comprises comprehensive and high-quality data of patients admitted to ICUs at the Beth Israel Deaconess Medical Center between 2008 and 2019. The institutional review boards of Beth Israel Deaconess Medical Center and the Massachusetts Institute of Technology approved the establishment of this database. All patients in this study were anonymized, eliminating the need for informed consent and an ethical approval statement.
Study Population: In MIMIC-IV, all patients aged over 65 years who met the definition of SA-AKI between 2008 and 2019 were included. According to the consensus report of the 28th Acute Disease Quality Initiative workgroup, SA-AKI can be diagnosed when the diagnostic criteria for both sepsis and AKI were met [17]. Sepsis was defined as a suspected infection combined with an acute increase in Sequential Organ Failure Assessment (SOFA) score ≥2, according to the Sepsis-3 criteria [18]. AKI was determined based on the 2012 Kidney Disease: Improving Global Outcomes (KDIGO) guidelines, including an increase in creatinine or a decrease in urine output. The exclusion criteria were: (1) Patients with ICU stays shorter than 24 h or longer than 28 days; (2) Patients with a prior history of AKI; (3) Patients with over 5% missing data, which were excluded from the analysis.
Endpoints: The primary endpoint was death during hospitalization.
Predictor Selection: Clinical and laboratory variables were collected within 24 h of admission. For the prediction of mortality in SA-AKI, 66 variables were gathered, including patient characteristics (age, gender, etc.), clinical signs (temperature, blood pressure, respiratory rate, heart rate, comorbidity), medication history (norepinephrine dose), laboratory data (blood gas, routine blood analysis, liver function, renal function, and coagulation profile), and scale scores (Simplified Acute Physiology Score II (SAPS-II), SOFA, Glasgow Coma Scale (GCS), Charlson Comorbidity Index (CCI)). The recursive feature elimination (RFE) algorithm was employed to select key features. Data extraction was conducted by a full-time researcher and a medical student. The K-nearest neighbors’ algorithm was utilized to fill in missing values.
Machine Learning Model Comparison: We established and validated eleven ML models, including Logistic Regression (LR), Support Vector Machine (SVM), Gradient Boosting Machine (GBM), Adaptive Boosting (AdaBoost), Extreme Gradient Boosting (XGBoost), Category Boosting (CatBoost), Naïve Bayesian (NB), Neural Network (NN), Multilayer Perceptron (MLP), K-nearest neighbors (KNN), and Random Forest (RF) Classifier. A total of 70% of the study population was randomly selected as the training set, with the remaining 30% used for internal validation. We evaluated the performance of models using areas under curves (AUCs) and decision curve analysis (DCA).
Model Explainability: We employed the SHAP values for interpretability of the best-performing ML model. The SHAP values can help quantify the contribution of each feature and explain how the features influence the output of the optimal prediction model. To determine the importance of each feature at the overall level, the SHAP values of all features for all samples were plotted and sorted in descending order according to the sum of the SHAP values. The color represents the feature's importance (yellow for high and purple for low), with each point representing a sample.
Statistical Analysis: Baseline characteristics were compared between two groups in MIMIC-IV. Values are presented as means [standard deviations] (if normally distributed) or medians [interquartile ranges] (if non-normally distributed) for continuous variables and total numbers [percentages] for categorical variables. Comparisons were made using the Student's t-test or rank-sum test for continuous variables and the Chi-square test or Fisher's exact test for categorical variables, as appropriate. A two-tailed P value less than 0.05 was considered statistically significant. All statistical analyses were performed with R 4.1.3 and Rstudio 1.1.463. The R package "caret" was used for data pre-processing, parameter tuning, and model training. The R package "shapviz" was employed for evaluating SHAP values and feature importance. The R package "DMwR2″ was utilized to fill in the missing values of features. RFE was conducted using the package "caret". Calibration curves were generated with the R package "rms". ROC curve analysis and AUC calculations were performed using the pROC and ggplot2 packages to compare the efficacy of each model.
3. Results
3.1. Baseline characteristics
After screening based on the inclusion and exclusion criteria, a total of 8426 elderly patients with SA-AKI were included in this study. The study flowchart is presented in Fig. 1. As shown in Table 1, 1813 patients died during ICU admission, while 6613 patients survived. Among the 8426 patients, 4634 (55%) were female, and the median age was 77 (71, 84) years. Compared to the survivors group, non-survivors were older (77 [70, 83] vs. 79 [72, 85], P < 0.001), had higher CCI scores (7 [5,9] vs. 8 [6,19], P < 0.001), and were more likely to have other comorbidities such as chronic heart failure (41% vs. 46%, P < 0.001), myocardial infarction (13% vs. 19%, P < 0.001), and chronic kidney disease (34% vs. 40%, P < 0.001). Furthermore, non-survivors exhibited poorer kidney function, including higher serum creatinine concentration (1.2 [0.9, 2] vs. 1.8 [1.2, 2.9], P < 0.001), lower urine output on day 1 (1135 [715, 1688] vs. 665 [290, 1135], P < 0.001), a higher percentage of patients in AKI stage 3 (28% vs. 65%, P < 0.001) and those requiring renal replacement therapy (6% vs. 11%, P < 0.001). Moreover, non-survivors had higher organ injury scores, such as SAPS II (43 [36, 51] vs. 54 [46, 65], P < 0.001) and SOFA (5 [4,7] vs. 8 [5,10], P < 0.001) scores.
Fig. 1.
Flowchart of this study.
Table 1.
Baseline characteristics of the study cohort.
| Characteristics | Total (n = 8426) | Survivors (n = 6613) | Non-survivors (n = 1813) | P value |
|---|---|---|---|---|
| Demographic | ||||
| Age, year | 77 (71, 84) | 77 (70, 83) | 79 (72, 85) | <0.001 |
| Gender, n(%) | 0.173 | |||
| - Male | 3792 (45) | 2950 (45) | 842 (46) | |
| - Female | 4634 (55) | 3663 (55) | 971 (54) | |
| Weight, kg | 81 (68, 96) | 82 (69, 97) | 76 (64, 90) | <0.001 |
| Height, cm | 168 (163, 175) | 168 (163, 175) | 168 (160, 173) | <0.001 |
| BMI | 29 (26, 33) | 29 (26, 33) | 28 (25, 31) | <0.001 |
| Comorbidities | ||||
| Hypertension, n (%) | 6613 (78) | 5257 (79) | 1356 (75) | <0.001 |
| Diabetes, n (%) | 1766 (21) | 1370 (21) | 396 (22) | 0.312 |
| COPD, n (%) | 1468 (17) | 1142 (17) | 326 (18) | 0.501 |
| Chronic heart failure, n (%) | 3557 (42) | 2715 (41) | 842 (46) | <0.001 |
| Myocardial infarction, n (%) | 1188 (14) | 845 (13) | 343 (19) | <0.001 |
| Chronic kidney disease, n (%) | 2944 (35) | 2219 (34) | 725 (40) | <0.001 |
| Vital signs on day 1 | ||||
| Heart rate, bpm | 101 (88, 117) | 99 (88, 114) | 110 (94, 126) | <0.001 |
| Systolic blood pressure, mmHg | 144 (132, 159) | 145 (133, 160) | 141 (128, 157) | <0.001 |
| Diastolic blood pressure, mmHg | 81 (70, 95) | 80 (70, 95) | 84 (71, 97) | <0.001 |
| Mean arterial pressure, mmHg | 98 (88, 112) | 98 (88, 111) | 98 (88, 113) | 0.664 |
| Respiratory rate | 28 (24, 32) | 27 (24, 31) | 30 (26, 34) | <0.001 |
| Body temperature, °C | 37.3 (36.9, 37.7) | 37.3 (36.9, 37.7) | 37.2 (36.9, 37.8) | 0.137 |
| SpO2, % | 97 (96, 99) | 97 (96, 99) | 97 (96, 99) | 0.127 |
| Laboratory results on day 1 | ||||
| White blood cell, × 109/L | 13.6 (9.8, 18.5) | 13.3 (9.7, 18) | 15 (10.5, 20.5) | <0.001 |
| Hemoglobin, g/dL | 10.6 (9.4, 11.8) | 10.6 (9.5, 11.8) | 10.4 (9.1, 11.8) | <0.001 |
| Hematocrit, % | 32.4 (29.2, 36.2) | 32.4 (29.4, 36) | 32.32 (28.7, 37) | 0.98 |
| Platelets, × 109/L | 190 (140, 257) | 189 (142, 253) | 197 (132, 274) | 0.31 |
| International normalized ratio | 1.4 (1.2, 1.8) | 1.4 (1.2, 1.7) | 1.6 (1.3, 2.2) | <0.001 |
| Prothrombin time, s | 15.6 (13.7, 19.2) | 15.4 (13.6, 18.2) | 17.3 (14.2, 24) | <0.001 |
| Partial thromboplastin time, s | 34.8 (29.8, 45.4) | 34.1 (29.6, 42.9) | 39.0 (31.2, 57.7) | <0.001 |
| Blood urea nitrogen, mg/dL | 28 (19, 46) | 26 (18, 41) | 39 (26, 59) | <0.001 |
| Serum creatinine, mg/dL | 1.3 (0.9, 2.3) | 1.2 (0.9, 2) | 1.8 (1.2, 2.9) | <0.001 |
| Lactate, mmol/L | 1.5 (1.2, 2.0) | 1.5 (1.2, 1.9) | 1.7 (1.3, 2.6) | <0.001 |
| pH | 7.42 (7.38, 7.45) | 7.42 (7.39, 7.45) | 7.4 (7.35, 7.44) | <0.001 |
| PaO2, mmHg | 202 (124, 310) | 221 (133, 327) | 160 (106, 223) | <0.001 |
| PCO2, mmHg | 45 (41, 50) | 45 (41, 50) | 45 (39, 51) | <0.001 |
| FiO2, mmHg | 70 (53, 100) | 71 (55, 100) | 66 (50, 100) | <0.001 |
| Base excess | 0.3 (−1.0, 2.1) | 0.9 (−0.6, 2.6) | −0.9 (−4.0, 1.1) | <0.001 |
| Anion gap | 16 (13, 19) | 15 (13, 18) | 18 (15, 22) | <0.001 |
| Bicarbonate, mmol/L | 24 (21, 26) | 24 (22, 26) | 22 (19, 26) | <0.001 |
| Serum calcium, mmol/L | 8.3 (7.7, 8.8) | 8.3 (7.7, 8.8) | 8.4 (7.9, 8.9) | <0.001 |
| Serum chloride, mmol/L | 106 (102, 110) | 107 (102, 110) | 105 (100, 110) | <0.001 |
| Serum potassium, mmol/L | 4.5 (4.1, 5) | 4.5 (4.1, 4.9) | 4.6 (4.1, 5.2) | <0.001 |
| Blood glucose, mg/dL | 142 (114, 190) | 139 (113, 181) | 162 (123, 221) | <0.001 |
| AKI stage, n(%) | <0.001 | |||
| - Stage 1 | 1459 (17) | 1303 (20) | 156 (8) | |
| - Stage 2 | 3925 (47) | 3460 (52) | 465 (26) | |
| - Stage 3 | 3042 (36) | 1850 (28) | 1192 (65) | |
| Urine output on day 1, mL | 1040 (598, 1595) | 1135 (715, 1688) | 665 (290, 1135) | <0.001 |
| Treatments | ||||
| Renal replacement therapy, n (%) | 593 (7) | 390 (6) | 203 (11) | <0.001 |
| Invasive mechanical ventilation, n (%) | 4820 (57) | 3710 (56) | 1110 (61) | <0.001 |
| Rate of norepinephrine | 0 (0, 0.08) | 0 (0, 0.03) | 0.05 (0, 0.3) | <0.001 |
| Severity of illness scores | ||||
| SAPS Ⅱ | 45 (37, 54) | 43 (36, 51) | 54 (46, 65) | <0.001 |
| SOFA | 6 (4, 8) | 5 (4, 7) | 8 (5, 11) | <0.001 |
| GCS | 15 (13, 15) | 15 (13, 15) | 15 (12, 15) | 0.025 |
| CCI | 7 (5, 9) | 7 (5, 9) | 8 (6, 10) | <0.001 |
Data are presented as a number with the percentage in parentheses, or as the median with the interquartile range (IQR) in parentheses.
SAPS II Simplified Acute Physiology Score II, SOFA Sequential Organ Failure Assessment, GCS Glasgow coma scale, CCI Charlson Comorbidity Index.
3.2. Selection of key features
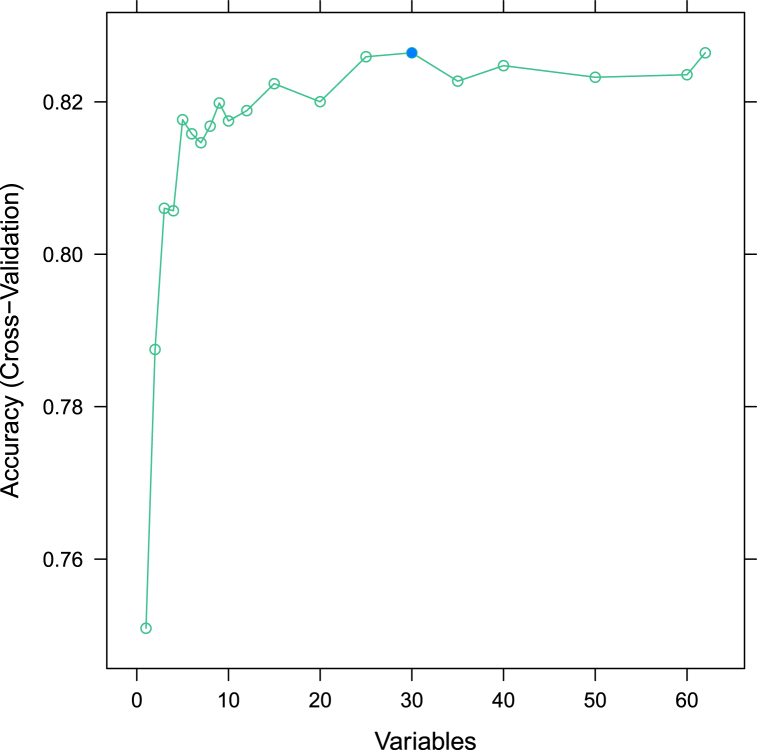
On the first day after ICU admission, 66 variables were collected and analyzed using the RFE algorithm. Nine key features with an accuracy of 82% were selected, including AKI stage, partial pressure of oxygen (PaO2), lactate, urine output, rate of norepinephrine, blood urea nitrogen (BUN) level, invasive mechanical ventilation (IMV), base excess, and anion gap (AP) (Fig. 2). These features were identified as risk factors for mortality in patients with SA-AKI during ICU admission and were used for subsequent modeling.
Fig. 2.
Feature elimination of the 66 variables using the RFE algorithm. RFE: recursive feature elimination.
3.3. Development and comparison of machine learning models
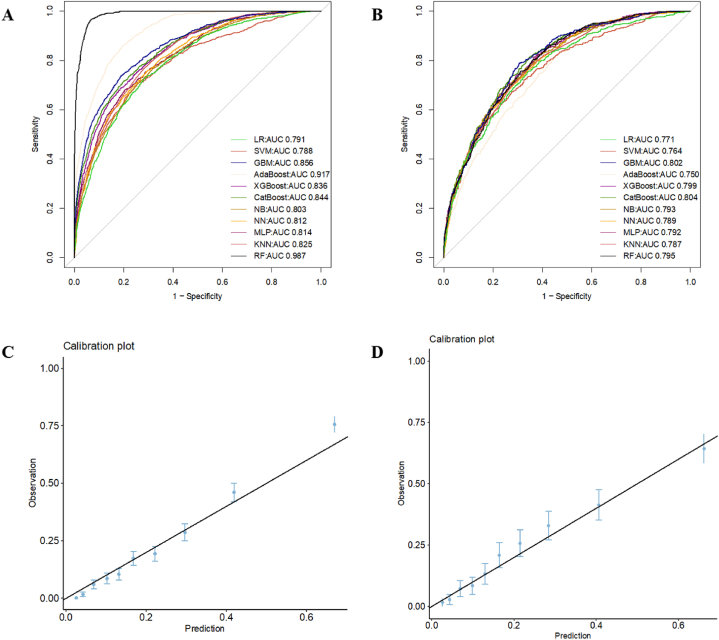
The total 8426 patients were randomly divided into two subgroups: 5934 patients in a training cohort (training set) and 2492 in a validation cohort (test set). No significant differences in baseline features were observed between the two subgroups (Supplementary Table 2 and Supplementary Table 3 for variable data). Eleven machine learning models were employed in this study. The relevant model data obtained for the training cohort and validation cohort: The cutoff value, Youden index, F1, accuracy, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) are shown in the following tables (Table 2 for the training cohort and Table 3 for the validation cohort). The CatBoost algorithm had the best discrimination ability with AUC = 0.844 in the training cohort and AUC = 0.804 in the validation cohort (Fig. 3A and B). The calibration plot indicated that the CatBoost algorithm was relatively stable (Fig. 3C and D). The data of other ML models in the training and validation cohorts are shown in Table 2, Table 3, respectively.
Table 2.
Performance comparison of the machine learning models in the training cohort.
| model | cutoff value | Youden index | F1 | accuracy | sen | spe | PPV | NPV |
|---|---|---|---|---|---|---|---|---|
| AdaBoost | 0.45 | 0.66 | 0.646 | 0.87 (0.86, 0.88) | 0.56 (0.53, 0.58) | 0.95 (0.95, 0.96) | 0.77 (0.74, 0.79) | 0.89 (0.88, 0.90) |
| GBM | 0.24 | 0.55 | 0.526 | 0.84 (0.84, 0.85) | 0.40 (0.37, 0.43) | 0.97 (0.96, 0.97) | 0.76 (0.72, 0.79) | 0.86 (0.85, 0.87) |
| KNN | 0.20 | 0.49 | 0.382 | 0.82 (0.81, 0.83) | 0.26 (0.24, 0.28) | 0.97 (0.97, 0.98) | 0.72 (0.68, 0.77) | 0.83 (0.82, 0.84) |
| LR | 0.21 | 0.44 | 0.355 | 0.81 (0.80, 0.82) | 0.25 (0.22, 0.27) | 0.96 (0.95, 0.96) | 0.62 (0.58, 0.67) | 0.82 (0.81, 0.83) |
| MLP | 0.25 | 0.48 | 0.472 | 0.82 (0.81, 0.83) | 0.38 (0.35, 0.40) | 0.94 (0.93, 0.95) | 0.63 (0.60, 0.67) | 0.85 (0.84, 0.86) |
| NB | 0.02 | 0.45 | 0.388 | 0.82 (0.81, 0.83) | 0.27 (0.25, 0.30) | 0.96 (0.96, 0.97) | 0.67 (0.63, 0.71) | 0.83 (0.82, 0.84) |
| NN | 0.21 | 0.46 | 0.440 | 0.82 (0.81, 0.83) | 0.34 (0.31, 0.36) | 0.95 (0.94, 0.95) | 0.64 (0.60, 0.67) | 0.84 (0.83, 0.85) |
| RF | 0.31 | 0.90 | 0.764 | 0.92 (0.91, 0.92) | 0.63 (0.61, 0.66) | 0.99 (0.99, 1.00) | 0.97 (0.95, 0.98) | 0.91 (0.90, 0.92) |
| SVM | 0.17 | 0.45 | 0.368 | 0.82 (0.81, 0.82) | 0.25 (0.23, 0.28) | 0.97 (0.96, 0.97) | 0.68 (0.64, 0.73) | 0.83 (0.82, 0.84) |
| XGBoost | 0.27 | 0.50 | 0.462 | 0.83 (0.82, 0.84) | 0.35 (0.32, 0.37) | 0.96 (0.95, 0.97) | 0.70 (0.66, 0.74) | 0.84 (0.83, 0.85) |
| CatBoost | 0.25 | 0.52 | 0.489 | 0.84 (0.83, 0.85) | 0.36 (0.34, 0.39) | 0.97 (0.96, 0.97) | 0.74 (0.71, 0.78) | 0.85 (0.84, 0.86) |
Table 3.
Performance comparison of the machine learning models in the test cohort.
| model | cutoff value | Youden index | F1 | accuracy | sen | spe | PPV | NPV |
|---|---|---|---|---|---|---|---|---|
| AdaBoost | 0.391 | 0.37 | 0.405 | 0.78 (0.76, 0.80) | 0.34 (0.30, 0.38) | 0.90 (0.89, 0.92) | 0.50 (0.44, 0.55) | 0.83 (0.81, 0.85) |
| GBM | 0.168 | 0.48 | 0.401 | 0.80 (0.79, 0.82) | 0.30 (0.26, 0.34) | 0.94 (0.93, 0.95) | 0.60 (0.54, 0.66) | 0.83 (0.81, 0.84) |
| KNN | 0.160 | 0.42 | 0.321 | 0.80 (0.79, 0.82) | 0.21 (0.18, 0.25) | 0.97 (0.96, 0.98) | 0.67 (0.59, 0.74) | 0.82 (0.80, 0.83) |
| LR | 0.176 | 0.41 | 0.352 | 0.80 (0.79, 0.82) | 0.25 (0.21, 0.28) | 0.96 (0.95, 0.97) | 0.62 (0.55, 0.68) | 0.82 (0.80, 0.84) |
| MLP | 0.182 | 0.45 | 0.429 | 0.80 (0.79, 0.82) | 0.34 (0.30, 0.38) | 0.93 (0.92, 0.94) | 0.59 (0.53, 0.64) | 0.83 (0.82, 0.85) |
| NB | 0.023 | 0.44 | 0.379 | 0.81 (0.79, 0.82) | 0.27 (0.23, 0.31) | 0.96 (0.95, 0.97) | 0.63 (0.57, 0.70) | 0.82 (0.81, 0.84) |
| NN | 0.158 | 0.42 | 0.403 | 0.80 (0.79, 0.82) | 0.31 (0.27, 0.35) | 0.94 (0.93, 0.95) | 0.59 (0.53, 0.64) | 0.83 (0.81, 0.84) |
| RF | 0.210 | 0.44 | 0.395 | 0.81 (0.79, 0.82) | 0.29 (0.25, 0.33) | 0.95 (0.94, 0.96) | 0.62 (0.55, 0.68) | 0.83 (0.81, 0.84) |
| SVM | 0.165 | 0.40 | 0.344 | 0.80 (0.79, 0.82) | 0.24 (0.20, 0.27) | 0.96 (0.95, 0.97) | 0.64 (0.57, 0.71) | 0.82 (0.80, 0.83) |
| XGBoost | 0.191 | 0.44 | 0.394 | 0.81 (0.79, 0.82) | 0.29 (0.25, 0.33) | 0.95 (0.94, 0.96) | 0.63 (0.57, 0.69) | 0.83 (0.81, 0.84) |
| CatBoost | 0.163 | 0.47 | 0.402 | 0.81 (0.79, 0.83) | 0.29 (0.25, 0.33) | 0.95 (0.94, 0.96) | 0.64 (0.58, 0.70) | 0.83 (0.81, 0.84) |
Fig. 3.
ROC curves and model evaluation of eleven ML models. (A) ROC curves of ML models in training cohort; (B) ROC curves of ML models in validation cohort; (C) Calibration plot of ML models in training cohort; (D) Calibration plot of ML models in validation cohort. ROC: receiver operating characteristic, ML: machine learning.
In the validation set (Table 3), The cutoff value and Youden index of CatBoost model were 0.163 and 0.47, which were better at discriminating real patients from nonpatients compared to other models, and the F1 value of 0.402 suggested that the model had better positive prediction value and sensitivity, and the actual values of positive prediction value and sensitivity, 0.64 and 0.29, were higher relative to the other models as well illustrates this point. In addition, the higher accuracy, specificity, and negative prediction rate of this model suggested that CatBoost model had better predictive performance compared to the other 10 models.
3.4. Model Explainability
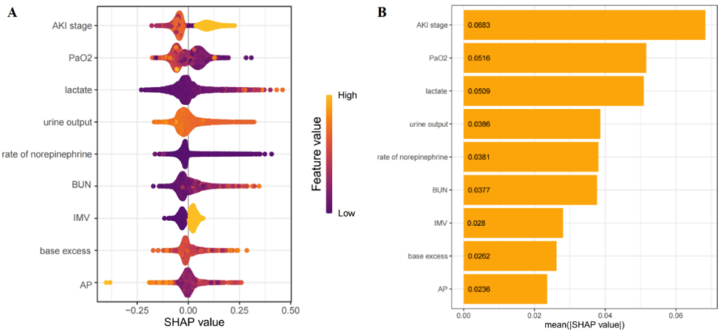
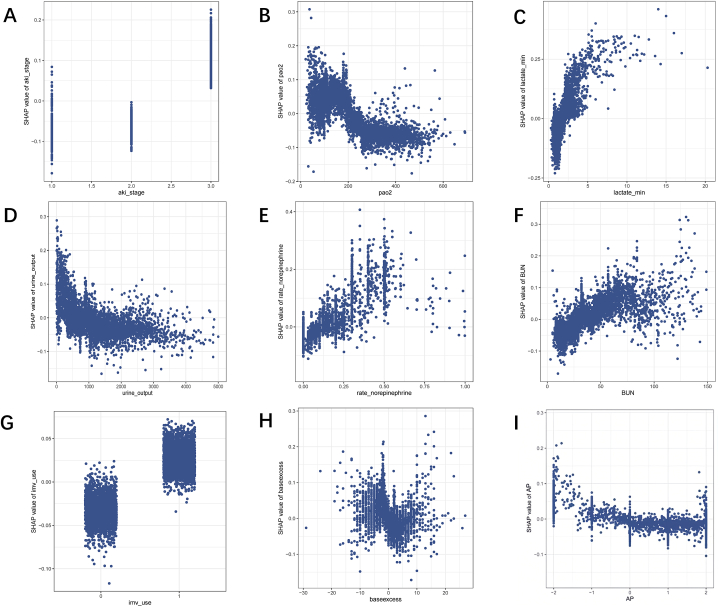
To enhance the visualization of the model's output, we employed SHAP to identify features with the greatest correlation to hospital mortality. The SHAP summary plot is shown in Fig. 4A, with the color scale indicating whether the value corresponds to a high or low value of the feature. The results of the analysis for the nine features with the highest impact on mortality are displayed in descending order (Fig. 4B). To better understand the correlation between these features and mortality risk, Fig. 5A–I illustrates the relationship between these features and the prediction of patient outcomes. The SHAP values for these features exceed zero, representing an increased risk of mortality. Elevated AKI stage, lactate, rate of norepinephrine, BUN, and IMV showed a positive correlation with mortality due to SA-AKI, while increased PaO2, urine output, and AP demonstrated a negative correlation. Additionally, base excess values greater than 5 or less than −1 tended to increase the risk of mortality.
Fig. 4.
(A) SHAP summary plot of the CatBoost model. A higher SHAP value indicates a higher mortality risk. For each feature, each dot is created according its positive (yellow color) or negative (purple color) contribution to the predictive model of mortality for each patient, creating the dots line for each feature of all patients. (B) The rank of these features in a decreasing order.
Fig. 5.
SHAP dependence plot of the CatBoost model. (A) AKI stage; (B) PaO2; (C) lactate; (D) urine output; (E) rate of norepinephrine; (F) BUN level; (G) IMV; (H) base excess; (I) AP. BUN: blood urea nitrogen; IMV: invasive mechanical ventilation; AP: anion gap.
3.5. Application of model predictive performance
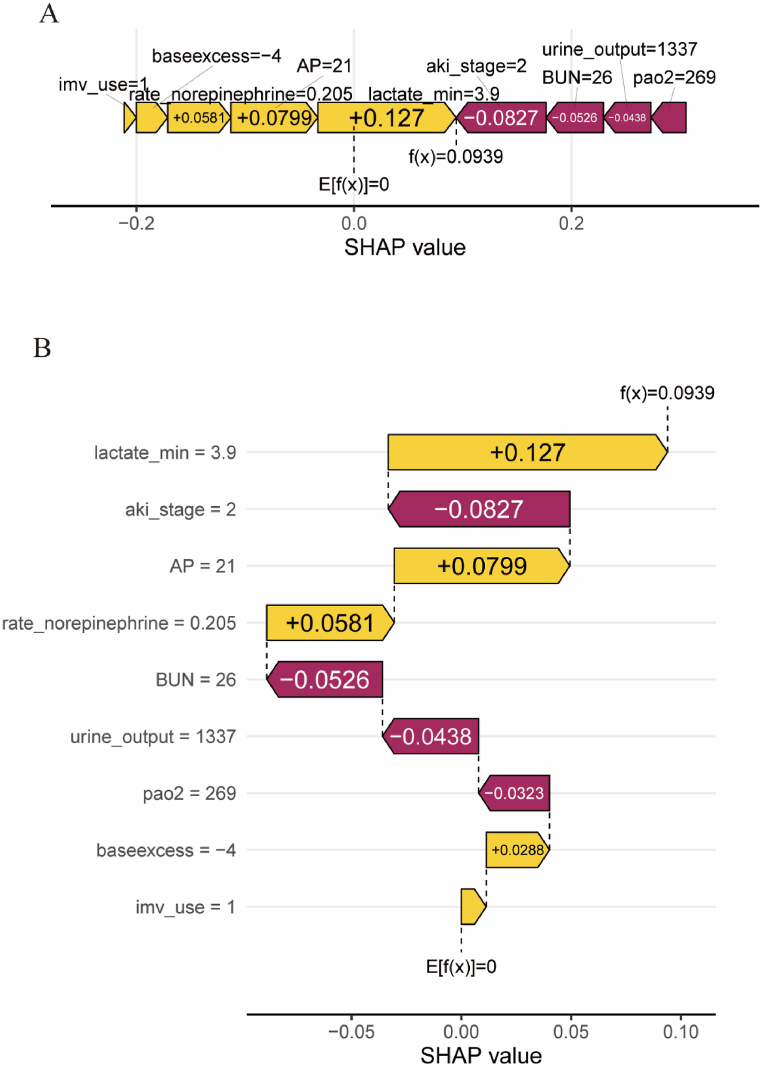
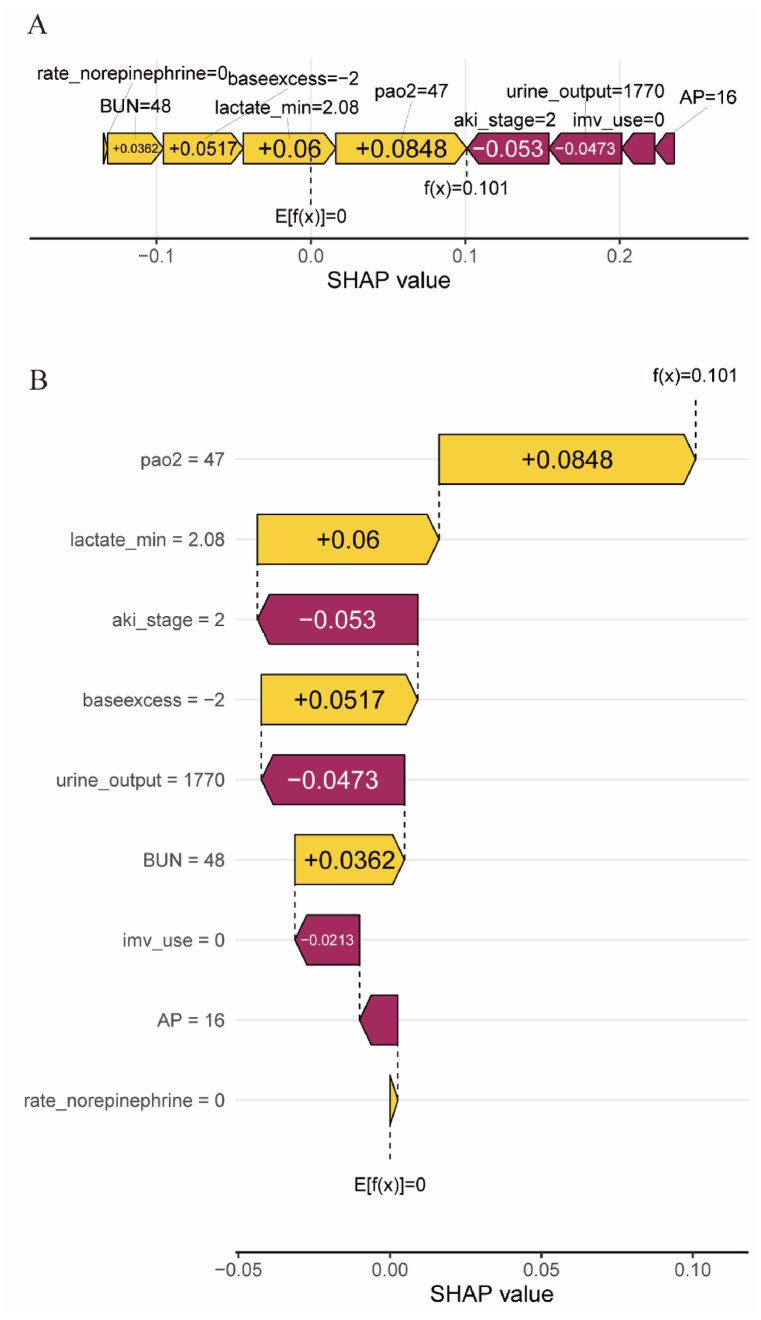
To further clarify the clinical application of the CatBoost model, we randomly selected a patient from the validation cohort. The force plot and waterfall plot displayed the contributions of features for this patient, with the yellow and purple bars representing positive and negative contributions to in-hospital mortality, respectively (Fig. 6, Fig. 7B). For patient NO.one (Fig. 6), the CatBoost model predicted a mortality rate of 28%. The results showed that a lactate level of 3.9 mmol/L, AKI of stage 2, and AP = 21 were the top three factors contributing to this prediction. Fig. 6A and B illustrates the specific impact of these factors on the predicted outcome, which actually resulted in an eventual in-hospital death for this patient. For patient NO. two (Fig. 7), the CatBoost model predicted a mortality rate of 1.5%. The results showed that an arterial partial pressure of oxygen (PaO2) of 47 mmHg, a lactate level of 2.08 mmol/L, and AKI stage 2 were the top three factors contributing to this prediction. Fig. 7A and B illustrates the specific impact of these factors on the prediction that the patient actually survived the hospitalization.
Fig. 6.
The forceplot (A) and waterfall plot (B) for explaining the contribution of features on a certain patient (the patient one). the yellow bar and purple bar represented a positive and negative contribution to in-hospital mortality, respectively.
Fig. 7.
The forceplot (A) and waterfall plot (B) for explaining the contribution of features on a certain patient (the patient two). the yellow bar and purple bar represented a positive and negative contribution to in-hospital mortality, respectively.
4. Discussion
In this study, we developed and validated eleven ML models to predict the prognosis of elderly patients with SA-AKI admitted to the ICU. The data on elderly patients with SA-AKI were obtained from the MIMIC-IV database, which was used in dozens of articles on hospitalized mortality analysis. In view of this, we believe that it is desirable and plausible that the MIMIC-IV database can be used to construct a model for predicting the hospitalized mortality in our research [[20], [21], [22]]. The validity of the RFE methodology has been demonstrated in a variety of medical studies [[23], [24], [25], [26]], including the CatBoost model [[27], [28], [29]]. The CatBoost model demonstrated the best performance. The RFE algorithm selected nine key features from 66 variables, including AKI stage, PaO2, lactate, urine output, rate of norepinephrine, BUN, IMV, base excess, and AP. We examined how these features influenced the CatBoost model. The SHAP value was implemented to visualize individualized predictions of mortality for patients with SA-AKI. Furthermore, individual explanations constructed by SHAP force analysis can help clinicians understanding how the model makes specific recommendations for high-risk decisions. Overall, our study demonstrated a reliable predictive ML model of mortality risk for elderly patients with SA-AKI.
SA-AKI is a common sepsis complication and is closely associated with a higher risk of mortality [7,8]. Studies have shown that elderly patients are more likely to develop SA-AKI and experience significantly worse prognoses [6,7]. Thus, predicting the mortality risk of elderly patients with SA-AKI could provide valuable guidance for clinical healthcare. However, a reliable risk assessment tool for this patient population is lacking in clinical practice. Existing ICU scoring systems, such as SAPS-II and SOFA score, are widely used for outcome prediction, but their poor specificity and linearity characteristics limit their predictive power [16].
With the advancement of AI, ML models have become increasingly essential tools in critical care settings [30]. The advantage of ML lies in its ability to learn from vast quantities of complex, linear, and nonlinear data, outputting valuable results that conventional regression analyses might overlook. ML models have demonstrated strong performance in areas such as diagnosis, mortality prediction, and readmission rates [31]. In our study, we found that the CatBoost model outperformed other ML models. CatBoost, a member of the gradient boosting algorithm family, has been shown to surpass other ML models in various tasks in previous studies [32]. The primary advantage of CatBoost is its ability to handle categorical features and missing values automatically and effectively during training, rather than preprocessing. Consequently, categorical features no longer require encoding, and missing values do not need imputation. Another benefit of the algorithm is its use of a novel schema to calculate leaf values when selecting the tree structure, reducing overfitting—a major issue that constrains the generalization ability of machine learning models [33].
ML models are often regarded as black boxes, so that we employed the SHAP value to elucidate the model's inner workings. In our study, the SHAP summary analysis revealed that AKI stage, PaO2, and lactate were the top three most important features contributing to the CatBoost model. AKI stage is the strongest predictive variable in this model, particularly AKI stage 3, which is associated with a high mortality risk. Our study is supported by data from previous research, indicating that early recognition of AKI can improve the prognosis of SA-AKI [34,35]. PaO2 is another crucial predictor for SA-AKI, and several studies have established the link between PaO2 and AKI outcomes [36]. As such, it is important to emphasize respiratory functions in elderly patients with SA-AKI. High lactate levels are associated with tissue hypoperfusion or organ dysfunction from sepsis, our study found that lactate was closely related to mortality in patients with SA-AKI.
There are some limitations to our study that should be acknowledged. First, our research was a retrospective analysis, which limited our ability to determine causal relationships between features and outcomes. Therefore, further prospective randomized clinical trials are needed to validate our model's effectiveness. Second, our predictive models lack proper external validation, which may impact the credibility of the CatBoost model. Third, our study extracted clinical data within the first 24 h after ICU admission, potentially overlooking the dynamic changes of these features over time.
In conclusion, we have demonstrated that the CatBoost had a good prognostic prediction ability in elderly patients with SA-AKI during ICU admission. It may facilitate earlier intervention, optimize treatment, and improve the prognosis for elderly patients with SA-AKI.
Ethics requirements
The Xiangya Hospital of Central South University Ethics Committee reviewed and approved this study on 27 April 2022 (protocol number 202204101).
Data availability statement
Further inquiries can be directed to the corresponding authors. The name of the repository: MIMIC-IV database. The accession number: 42039813.
CRediT authorship contribution statement
Jie Tang: Writing – review & editing, Writing – original draft. Jian Huang: Data curation. Xin He: Investigation. Sijue Zou: Investigation. Li Gong: Investigation. Qiongjing Yuan: Supervision. Zhangzhe Peng: Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This project was supported by the Major Program of the National Natural Science Foundation of China (82090024), the General Programs of the National Natural Science Foundation of China (82173877 and 82073918), the Key Research and Development Program of Hunan Province (2021SK2015), and the Outstanding Youth Foundation of the Natural Science Foundation of Hunan Province (2022JJ10100).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e26570.
Contributor Information
Qiongjing Yuan, Email: yuanqiongjing@csu.edu.cn.
Zhangzhe Peng, Email: pengzhangzhe@csu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Singer M., Deutschman C.S., Seymour C.W., Shankar-Hari M., Annane D., Bauer M., Bellomo R., Bernard G.R., Chiche J.D., Coopersmith C.M., Hotchkiss R.S., Levy M.M., Marshall J.C., Martin G.S., Opal S.M., Rubenfeld G.D., van der Poll T., Vincent J.L., Angus D.C. The third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315(8):801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudd K.E., Johnson S.C., Agesa K.M., Shackelford K.A., Tsoi D., Kievlan D.R., Colombara D.V., Ikuta K.S., Kissoon N., Finfer S., Fleischmann-Struzek C., Machado F.R., Reinhart K.K., Rowan K., Seymour C.W., Watson R.S., West T.E., Marinho F., Hay S.I., Lozano R., Lopez A.D., Angus D.C., Murray C.J.L., Naghavi M. Global, regional, and national sepsis incidence and mortality, 1990-2017: analysis for the Global Burden of Disease Study. Lancet. 2020;395(10219):200–211. doi: 10.1016/S0140-6736(19)32989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellomo R., Kellum J.A., Ronco C. Acute kidney injury. Lancet. 2012;380(9843):756–766. doi: 10.1016/S0140-6736(11)61454-2. [DOI] [PubMed] [Google Scholar]
- 4.Uchino S., Kellum J.A., Bellomo R., Doig G.S., Morimatsu H., Morgera S., Schetz M., Tan I., Bouman C., Macedo E., Gibney N., Tolwani A., Ronco C. Beginning and Ending Supportive Therapy for the Kidney (BEST Kidney) Investigators. Acute renal failure in critically ill patients: a multinational, multicenter study. JAMA. 2005;294(7):813–818. doi: 10.1001/jama.294.7.813. [DOI] [PubMed] [Google Scholar]
- 5.Bagshaw S.M., Lapinsky S., Dial S., Arabi Y., Dodek P., Wood G., Ellis P., Guzman J., Marshall J., Parrillo J.E., Skrobik Y., Kumar A. Cooperative Antimicrobial Therapy of Septic Shock (CATSS) Database Research Group. Acute kidney injury in septic shock: clinical outcomes and impact of duration of hypotension prior to initiation of antimicrobial therapy. Intensive Care Med. 2009;35(5):871–881. doi: 10.1007/s00134-008-1367-2. [DOI] [PubMed] [Google Scholar]
- 6.Zarbock A., Nadim M.K., Pickkers P., Gomez H., Bell S., Joannidis M., Kashani K., Koyner J.L., Pannu N., Meersch M., Reis T., Rimmelé T., Bagshaw S.M., Bellomo R., Cantaluppi V., Deep A., De Rosa S., Perez-Fernandez X., Husain-Syed F., Kane-Gill S.L., Kelly Y., Mehta R.L., Murray P.T., Ostermann M., Prowle J., Ricci Z., See E.J., Schneider A., Soranno D.E., Tolwani A., Villa G., Ronco C., Forni L.G. Sepsis-associated acute kidney injury: consensus report of the 28th Acute Disease Quality Initiative workgroup. Nat. Rev. Nephrol. 2023;19(6):401–417. doi: 10.1038/s41581-023-00683-3. [DOI] [PubMed] [Google Scholar]
- 7.Bagshaw S.M., Uchino S., Bellomo R., Morimatsu H., Morgera S., Schetz M., Tan I., Bouman C., Macedo E., Gibney N., Tolwani A., Oudemans-van Straaten H.M., Ronco C., Kellum J.A. Beginning and Ending Supportive Therapy for the Kidney (BEST Kidney) Investigators. Septic acute kidney injury in critically ill patients: clinical characteristics and outcomes. Clin. J. Am. Soc. Nephrol. 2007;2(3):431–439. doi: 10.2215/CJN.03681106. [DOI] [PubMed] [Google Scholar]
- 8.Bouchard J., Acharya A., Cerda J., Maccariello E.R., Madarasu R.C., Tolwani A.J., Liang X., Fu P., Liu Z.H., Mehta R.L. A prospective international multicenter study of AKI in the intensive care unit. Clin. J. Am. Soc. Nephrol. 2015;10(8):1324–1331. doi: 10.2215/CJN.04360514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chronopoulos A., Cruz D.N., Ronco C. Hospital-acquired acute kidney injury in the elderly. Nat. Rev. Nephrol. 2010;6(3):141–149. doi: 10.1038/nrneph.2009.234. [DOI] [PubMed] [Google Scholar]
- 10.Starr M.E., Saito H. Sepsis in old age: review of human and animal studies. Aging Dis. 2014;5(2):126–136. doi: 10.14336/AD.2014.0500126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitt R., Coca S., Kanbay M., Tinetti M.E., Cantley L.G., Parikh C.R. Recovery of kidney function after acute kidney injury in the elderly: a systematic review and meta-analysis. Am. J. Kidney Dis. 2008;52(2):262–271. doi: 10.1053/j.ajkd.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 12.Schvetz M., Fuchs L., Novack V., Moskovitch R. Outcomes prediction in longitudinal data: study designs evaluation, use case in ICU acquired sepsis. J. Biomed. Inf. 2021;117 doi: 10.1016/j.jbi.2021.103734. [DOI] [PubMed] [Google Scholar]
- 13.da Hora Passos R., Ramos J.G., Mendonça E.J., Miranda E.A., Dutra F.R., Coelho M.F., Pedroza A.C., Correia L.C., Batista P.B., Macedo E., Dutra M.M. A clinical score to predict mortality in septic acute kidney injury patients requiring continuous renal replacement therapy: the HELENICC score. BMC Anesthesiol. 2017;17(1):21. doi: 10.1186/s12871-017-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu H., Li L., Zhang Y., Sha T., Huang Q., Guo X., An S., Chen Z., Zeng Z. A prediction model for assessing prognosis in critically ill patients with sepsis-associated acute kidney injury. Shock. 2021;56(4):564–572. doi: 10.1097/SHK.0000000000001768. [DOI] [PubMed] [Google Scholar]
- 15.Moll M., Qiao D., Regan E.A., Hunninghake G.M., Make B.J., Tal-Singer R., McGeachie M.J., Castaldi P.J., San Jose Estepar R., Washko G.R., Wells J.M., LaFon D., Strand M., Bowler R.P., Han M.K., Vestbo J., Celli B., Calverley P., Crapo J., Silverman E.K., Hobbs B.D., Cho M.H. Machine learning and prediction of all-cause mortality in COPD. Chest. 2020;158(3):952–964. doi: 10.1016/j.chest.2020.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou N., Li M., He L., Xie B., Wang L., Zhang R., Yu Y., Sun X., Pan Z., Wang K. Predicting 30-days mortality for MIMIC-III patients with sepsis-3: a machine learning approach using XGboost. J. Transl. Med. 2020;18(1):462. doi: 10.1186/s12967-020-02620-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zarbock A., Nadim M.K., Pickkers P., Gomez H., Bell S., Joannidis M., Kashani K., Koyner J.L., Pannu N., Meersch M., Reis T., Rimmelé T., Bagshaw S.M., Bellomo R., Cantaluppi V., Deep A., De Rosa S., Perez-Fernandez X., Husain-Syed F., Kane-Gill S.L., Kelly Y., Mehta R.L., Murray P.T., Ostermann M., Prowle J., Ricci Z., See E.J., Schneider A., Soranno D.E., Tolwani A., Villa G., Ronco C., Forni L.G. Sepsis-associated acute kidney injury: consensus report of the 28th Acute Disease Quality Initiative workgroup. Nat. Rev. Nephrol. 2023;19(6):401–417. doi: 10.1038/s41581-023-00683-3. [DOI] [PubMed] [Google Scholar]
- 18.Singer M., Deutschman C.S., Seymour C.W., Shankar-Hari M., Annane D., Bauer M., Bellomo R., Bernard G.R., Chiche J.D., Coopersmith C.M., Hotchkiss R.S., Levy M.M., Marshall J.C., Martin G.S., Opal S.M., Rubenfeld G.D., van der Poll T., Vincent J.L., Angus D.C. The third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315(8):801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ali T., Khan I., Simpson W., Prescott G., Townend J., Smith W., Macleod A. Incidence and outcomes in acute kidney injury: a comprehensive population-based study. J. Am. Soc. Nephrol. 2007 Apr;18(4):1292–1298. doi: 10.1681/ASN.2006070756. [DOI] [PubMed] [Google Scholar]
- 20.Cai W., Xu J., Wu X., Chen Z., Zeng L., Song X., Zeng Y., Yu F. Association between triglyceride-glucose index and all-cause mortality in critically ill patients with ischemic stroke: analysis of the MIMIC-IV database. Cardiovasc. Diabetol. 2023;22(1):138. doi: 10.1186/s12933-023-01864-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng R., Qian S., Shi Y., Lou C., Xu H., Pan J. Association between triglyceride-glucose index and in-hospital mortality in critically ill patients with sepsis: analysis of the MIMIC-IV database. Cardiovasc. Diabetol. 2023;22(1):307. doi: 10.1186/s12933-023-02041-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Z., Gong H., Kan F., Ji N. Association between the triglyceride glucose (TyG) index and the risk of acute kidney injury in critically ill patients with heart failure: analysis of the MIMIC-IV database. Cardiovasc. Diabetol. 2023;22(1):232. doi: 10.1186/s12933-023-01971-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu C., Song J., Li H., Yu W., Hao Y., Xu K., Xu P. Predicting venous thrombosis in osteoarthritis using a machine learning algorithm: a population-based cohort study. J. Personalized Med. 2022;12(1):114. doi: 10.3390/jpm12010114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinal-Fernandez I., Casal-Dominguez M., Derfoul A., Pak K., Miller F.W., Milisenda J.C., Grau-Junyent J.M., Selva-O'Callaghan A., Carrion-Ribas C., Paik J.J., Albayda J., Christopher-Stine L., Lloyd T.E., Corse A.M., Mammen A.L. Machine learning algorithms reveal unique gene expression profiles in muscle biopsies from patients with different types of myositis. Ann. Rheum. Dis. 2020;79(9):1234–1242. doi: 10.1136/annrheumdis-2019-216599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z., Kaiser L., Holzgreve A., Ruf V.C., Suchorska B., Wenter V., Quach S., Herms J., Bartenstein P., Tonn J.C., Unterrainer M., Albert N.L. Prediction of TERTp-mutation status in IDH-wildtype high-grade gliomas using pre-treatment dynamic [F]FET PET radiomics. Eur. J. Nucl. Med. Mol. Imag. 2021;48(13):4415–4425. doi: 10.1007/s00259-021-05526-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung C.R., Nishihama Y., Nakayama S.F., Tamura K., Isobe T., Michikawa T., Iwai-Shimada M., Kobayashi Y., Sekiyama M., Taniguchi Y., Yamazaki S. Japan Environment and Children's Study Group. Indoor air quality of 5,000 households and its determinants. Part B: volatile organic compounds and inorganic gaseous pollutants in the Japan Environment and Children's study. Environ. Res. 2021;197 doi: 10.1016/j.envres.2021.111135. [DOI] [PubMed] [Google Scholar]
- 27.Bai X., Zhou Z., Su M., Li Y., Yang L., Liu K., Yang H., Zhu H., Chen S., Pan H. Predictive models for small-for-gestational-age births in women exposed to pesticides before pregnancy based on multiple machine learning algorithms. Front. Public Health. 2022;10 doi: 10.3389/fpubh.2022.940182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bayani A., Hosseini A., Asadi F., Hatami B., Kavousi K., Aria M., Zali M.R. Identifying predictors of varices grading in patients with cirrhosis using ensemble learning. Clin. Chem. Lab. Med. 2022;60(12):1938–1945. doi: 10.1515/cclm-2022-0508. [DOI] [PubMed] [Google Scholar]
- 29.Yang H., Bath P.A. The use of data mining methods for the prediction of dementia: evidence from the English longitudinal study of aging. IEEE J Biomed Health Inform. 2020;24(2):345–353. doi: 10.1109/JBHI.2019.2921418. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Q.Y., Wang H., Luo J.C., Luo M.H., Liu L.P., Yu S.J., Liu K., Zhang Y.J., Sun P., Tu G.W., Luo Z. Development and validation of a machine-learning model for prediction of extubation failure in intensive care units. Front. Med. 2021;8 doi: 10.3389/fmed.2021.676343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greener J.G., Kandathil S.M., Moffat L., Jones D.T. A guide to machine learning for biologists. Nat. Rev. Mol. Cell Biol. 2022;23(1):40–55. doi: 10.1038/s41580-021-00407-0. [DOI] [PubMed] [Google Scholar]
- 32.Hancock J.T., Khoshgoftaar T.M. CatBoost for big data: an interdisciplinary review. J Big Data. 2020;7(1):94. doi: 10.1186/s40537-020-00369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Q.Y., Liu L.P., Luo J.C., Luo Y.W., Wang H., Zhang Y.J., Gui R., Tu G.W., Luo Z. A machine-learning approach for dynamic prediction of sepsis-induced coagulopathy in critically ill patients with sepsis. Front. Med. 2021;7 doi: 10.3389/fmed.2020.637434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng X., McMahon G.M., Brunelli S.M., Bates D.W., Waikar S.S. Incidence, outcomes, and comparisons across definitions of AKI in hospitalized individuals. Clin. J. Am. Soc. Nephrol. 2014;9(1):12–20. doi: 10.2215/CJN.02730313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang W., Zhang C., Yu J., Shao J., Zheng R. Development and validation of a nomogram for predicting in-hospital mortality of elderly patients with persistent sepsis-associated acute kidney injury in intensive care units: a retrospective cohort study using the MIMIC-IV database. BMJ Open. 2023;13(3) doi: 10.1136/bmjopen-2022-069824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weng C.H., Chen H.H., Hu C.C., Huang W.H., Hsu C.W., Fu J.F., Lin W.R., Wang I.K., Yen T.H. Predictors of acute kidney injury after paraquat intoxication. Oncotarget. 2017;8(31):51345–51354. doi: 10.18632/oncotarget.17975. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Further inquiries can be directed to the corresponding authors. The name of the repository: MIMIC-IV database. The accession number: 42039813.