Summary
Moderate activation of IFN-I contributes to the body’s immune response, but its abnormal expression, stimulated by oxidative stress or other factors causes pathological damage. Heme oxygenase-1 (HO-1), induced by stress stimuli in the body, exerts a central role in cellular protection. Here we showed that HO-1 could promote IFN-1 under Spring Viremia of Carp virus (SVCV) infection and concomitantly attenuate the replication of SVCV. Further characterization of truncated mutants of HO-1 confirmed that intact HO-1 was essential for its antiviral function via IFN-I. Importantly, HO-1 inhibited the IFN-I signal by degrading the IRF3/7 through the autophagy pathway when it was triggered by H2O2 treatment. The iron ion-binding site (His28) was critical for HO-1 to degrade IRF3/7. HO-1 degradation of IRF3/7 is conserved in fish and mammals. Collectively, HO-1 regulates IFN-I positively under viral infection and negatively under oxidative stress, elucidating a mechanism by which HO-1 regulates IFN-I signaling in bi-directions.
Subject areas: Biological sciences, Biochemistry, Molecular biology
Graphical abstract

Highlights
-
•
Intact HO-1 exhibits the positive regulation of the IFN-1 signal under SVCV infection
-
•
HO-1 exhibits the negative regulation of the IFN-1 signal under oxidative stress
-
•
HO-1 inhibits IFN-1 by degrading IRF3/7 through the autophagy pathway
-
•
The iron ion-binding site is critical for HO-1 to degrade IRF3/7
Biological sciences; Biochemistry; Molecular biology
Introduction
The type-Ⅰ interferon (IFN-Ⅰ) is a crucial regulator of the host innate immune system that can be stimulated by microorganisms or external factors. Signaling cascades occur in different pathways, ultimately promoting the expression of interferon regulatory factors 3/7 (IRF3/7), which in turn translocate into the nucleus and activate the expression of IFN-Ⅰ.1,2,3 IFN-Ⅰ stimulates the transcription of immunogens by paracrine and autocrine means, typically by binding to the heterodimeric complex receptor IFNAR, which activates a class of genes called interferon-stimulated genes (ISGs) via signal transducer and activator of transcription (STAT)-dependent or STAT-independent pathways.4,5 IFN-Ⅰ exerts immune modulation such as antiviral, antitumor, antibacterial, and antioxidant effects.6,7 In addition to the classical pathway described above, IFN-Ⅰ induces a series of deleterious effects that lead to immune imbalance by changing the function of vital effector cells such as B cells, T cells, and plasmacytoid dendritic cells (pDCs).8,9,10
Moderate activation of IFN-Ⅰ contributes to the body’s immune response, but overexpressed IFNs are associated with the development of autoimmune diseases.11 Studies have shown that both pathogenic infections and auto-associated genetic variants affect the cyclic guanosine monophosphate adenosine monophosphate synthase-stimulator of interferon genes (cGAS-STING) pathway, which drives deleterious IFN-Ⅰ activation in multiple classes of autoimmune diseases, including systemic lupus erythematosus (SLE), systemic sclerosis (SSC), rheumatoid arthritis (RA), and sjögren’s syndrome (SS).12,13,14
Oxidative stress has also been found to cause excessive activation of IFN-Ⅰ. Recent studies have shown that reactive oxygen species (ROS)-induced activation of the cGAS/STING signal and the IFN-Ⅰ response, considering raised IFN-Ⅰ response with declined development, is indicative of aging brains.15 UV light can also induce the release of reactive oxygen species, which leads to DNA strand breaks and the formation of pyrimidine dimers in DNA, increasing the production of IFN-Ⅰ.16 Oxidative stress is linked with apoptosis or autophagy and even causes various diseases such as cancer, cardiovascular disease, and atherosclerosis.17,18 Vildagliptin and metformin work together to lessen the oxidative stress that type 2 diabetes mellitus generates.19 Studies have shown that oxidative stress is induced in the presence of dysregulated internal organ microflora and an impaired internal organ barrier, which leads to accrued levels of IFN-Ⅰ and promotes the possibility of SLE disease induction.20 Therefore, the exploration of the regulation of IFN-Ⅰ responses under oxidative stress, especially the negative regulation, may provide a candidate therapeutic target for maintaining IFN-Ⅰ homeostasis.
Heme oxygenase 1 (HO-1) is the inducible enzyme responsible for heme catabolism.21,22 In addition to hemoglobin, there are various factors that affect HO-1, including hypoxia, oxidative stress, and heat shock.23,24,25 HO-1 is also triggered by pathogens to exert anti-microorganism functions.26 It has been demonstrated that HO-1 regulates the IFN-Ⅰ signaling pathway and thus restrains the replication of many viruses, including Zika virus (ZIKV), Influenza A virus (IAV), Ebola virus (EBOV), and Dengue virus (DENV).27 Studies have shown that HO-1 increased the viability of rat alveolar macrophages, significantly inhibited ROS and malondialdehyde (MDA) content, and could weaken oxidative stress and apoptosis.28,29 In teleost fish, our preliminary studies demonstrated that CO, a byproduct of HO-1, can activate the cyclic GMP/protein kinase G (cGMP/PKG) signaling pathway, which in turn prevents Spring Viremia of Carp virus (SVCV) replication.30 In consideration of the central role of HO-1 in redox homeostasis, whether HO-1 is involved in the regulation of IFN-Ⅰ remains unknown.
In this study, we aimed to investigate the regulative role of HO-1 on IFN-Ⅰ under viral infection and oxidative stress. We found that HO-1 displayed the opposite regulation of IFN-Ⅰ signaling under viral infection or oxidative stress through IRF3/7, providing insights into HO-1 function and IFN regulation as well.
Results
Heme oxygenase-1 bi-directionally regulates interferon-1 during poly(I:C) transfection, Spring Viremia of Carp virus infection, and H2O2 treatment
It was previously recognized that Cobaltic protoporphyrin IX chloride (CoPP) was a strong HO-1 inducer through the Nrf2/ARE pathway. CoPP was used as an endogenous HO-1 activator in this study.27 In order to confirm if Ho-1 can regulate INF-1 signaling to participate in the antiviral response. The effect of HO-1 on the promoter activity of IFN-1 and ISRE was first examined using a dual luciferase reporter assay. The results showed that 40 μM CoPP treatment significantly enhanced the promoter activities of IFN-1 and ISRE in the presence of poly(I:C) and SVCV stimulation (Figures 1A and 1B). The effects of HO-1 on IFN-1 expression under different doses of poly(I:C) stimulation and different processes infected with SVCV were further explored. The transcriptional levels of major Ifn-1 signaling downstream surrogates (including Mx1, Viperin and Isg15) and IFN-1 protein levels increased with increasing doses of poly(I:C) (Figures 1C, 1D, and S1A). Consistently, the transcriptional levels of major Ifn-1 signaling downstream surrogates (including Mx1, Viperin, and Isg15) increased with the duration of SVCV infection (Figures 1E and S1B). The expression levels of SVCV-G increased in the SVCV-infected group. Interestingly, the CoPP-treated group was observed significantly reduced the expression levels of SVCV-G (Figure 1F) and SVCV titer (Figure 1G). The above results indicate that HO-1 positively regulates IFN-1 signaling in different viral infection processes.
Figure 1.
HO-1 promotes IFN-1 during poly(I:C) treatment and SVCV infection
(A and B) FHM cells seeded in 24-well plates overnight were and transfected with 500 ng IFN-1pro-Luc or ISREpro-Luc, and 50 ng pRL-TK was used as an internal control, subsequently, incubated with CoPP (40μM) or DMSO. At 24 h posttransfection, cells were treated with poly(I:C) (1 μg/mL), SVCV (0.1 MOI) and left untreated (control). The luciferase assay was performed 24 h after stimulation.
(C) FHM cells seeded in 6-well plates overnight were incubated with CoPP (40 μM) or DMSO. After 24 h, cells were treated with poly(I:C) (2 μg/mL). After 24 h, total RNA was prepared and analyzed by RT-qPCR for the expressions of Ho-1, Ifn-1, Mx1, Viperin, Isg15.
(D) FHM cells seeded in 6-well plates overnight were incubated with CoPP (40 μM) or DMSO. After 24 h, cells were treated with poly(I:C) (1 μg/mL, 2 μg/mL, 5 μg/mL). After 24 h, the cells were harvested for whole-cell extract preparation and Western blotting to detect IFN-1 protein.
(E) FHM cells seeded in 6-well plates overnight were incubated with CoPP (40μM) or DMSO. After 24 h, cells were infected with SVCV (MOI, 0.1). After 12 h, total RNA was prepared and analyzed by RT-qPCR for the expressions of Ho-1, Ifn-1, Mx1, Viperin, and Isg15.
(F and G) FHM cells seeded in 6-well plates overnight were incubated with CoPP (40μM) or DMSO. After 24 h, cells were infected with SVCV (MOI, 0.1). At the indicated time points (3 h, 6 h, 12 h, 24 h), the cells were harvested for whole-cell extract preparation and Western blotting to detect IFN-1 and SVCV-G protein (F). Plaque assay was performed to measure virus titer (G). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.001 in the comparison between the indicated groups.
In contrast, HO-1 induced by CoPP significantly inhibited IFN-1 and ISRE promoter activities after H2O2 treatment (Figure 2A). Ho-1 mRNA and IFN-1 protein levels increased with the extension of H2O2 simulation time (Figures 2B and 2C). Furthermore, in the H2O2 treatment group, the HO-1 mRNA levels significantly increased during CoPP treatment (Figure 2D). At the same time, IFN-1 protein levels decreased with increasing concentrations of CoPP administration (Figure 2E). Furthermore, low concentrations of H2O2 treatment promoted IFN-1 transcription, while high concentrations of H2O2 treatment significantly inhibited IFN-1 transcription in FHM cells (Figure 2F). Totally, these results suggest that HO-1 bi-directionally regulates IFN-1.
Figure 2.
HO-1 negatively regulates IFN-1 during H2O2 treatment
(A) FHM cells seeded in 24-well plates overnight were and transfected with 500 ng IFN-1pro-Luc or ISREpro-Luc, and 50 ng pRL-TK was used as an internal control, subsequently, incubated with CoPP (40 μM) or DMSO. At 24 h posttransfection, cells were incubated with H2O2 (500 μM) and left untreated (control). The luciferase assay was performed 24 h after stimulation.
(B and C) FHM cells seeded in 6-well plates overnight were incubated with H2O2 (500 μM), total RNA was prepared and analyzed by RT-qPCR for the expressions of Ho-1 (B) and immunoblot analysis for IFN-1 (C) for the indicated times.
(D and E) FHM cells seeded in 6-well plates overnight were FHM cells were firstly incubated with CoPP (20 μM, 40 μM, 80 μM) or DMSO, respectively. After 24 h, were incubated with H2O2 (500 μM). Detection of mRNA levels of Ho-1 (D) and protein expression of IFN-1 (E). (F) Detection of Ifn-1 mRNA expression from FHM cells incubated with different concentrations of H2O2 for 24 h. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.001 in the comparison between the indicated groups.
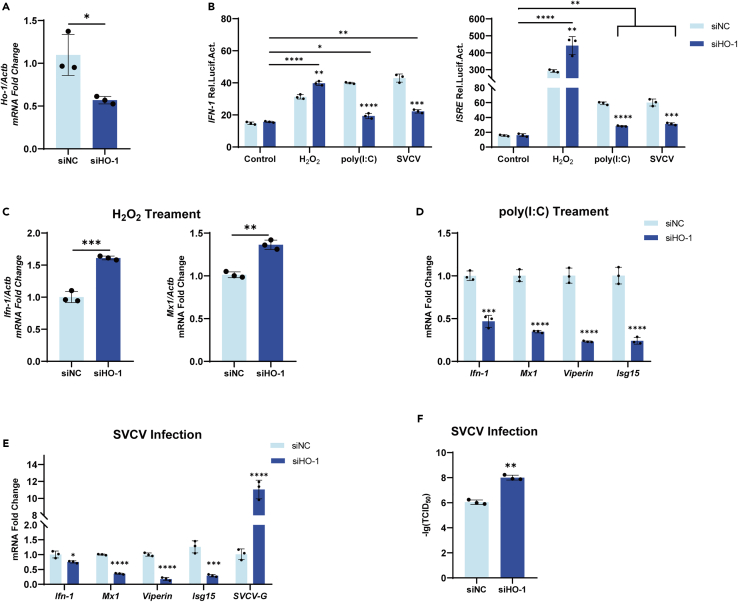
Knocking down of heme oxygenase-1 reverses the regulation of interferon-1 during poly(I:C) transfection, Spring Viremia of Carp virus infection, and H2O2 treatment
To confirm HO-1’s involvement in the regulation of IFN-1, FHM cells were transfected with HO-1-specific siRNA to knockdown HO-1 and then stimulated with poly(I:C) transfection, SVCV infection, or H2O2 treatment, respectively. The results showed that the transcript levels of HO-1 were significantly decreased by specific siRNA transfection when compared with those in the group transfected with control siRNA (Figure 3A). Knocking down HO-1 reversed the tendency of the regulated IFN-1 and ISRE promoters to be in their original states under poly(I:C) transfection, SVCV infection, or H2O2 treatment, respectively (Figure 3B). Consistent with these results, a significant increase in Ifn-1 and Mx1 mRNA levels was observed under H2O2 treatment (Figure 3C). After stimulation with poly(I:C), the expression levels of Ifn-1, Mx1, Viperin, and Isg15 were significantly lower than mock (Figure 3D). Similar results were obtained when SVCV was used as a stimulus (Figure 3E), meanwhile, expression levels of SVCV-G were significantly increased after the knockdown of HO-1. We further compared the titers of SVCV in the groups with or without HO-1 knocking down, and the results showed that the knocking down of HO-1 significantly promoted the proliferation of SVCV (Figure 3F). All these in vitro results indicated that HO-1 was a critical regulator of IFN-1.
Figure 3.
Knocking down of HO-1 on the regulation of IFN-1 during poly(I:C) transfection, SVCV infection, and H2O2 treatment
(A) FHM cells were seeded in 6-well plates and transfected the next day with siHO-1 and siNC, total RNA was prepared and analyzed by RT-qPCR for the expressions of Ho-1 after 24h.
(B) FHM cells were seeded in 24-well plates and transfected the next day with 250 ng IFN-1pro-Luc, and ISREpro-Luc, respectively, and 25 ng pRL-TK plus 250 ng siHo-1 or Control. At 24 h posttransfection, cells were treated with poly(I:C) (1 μg/ml), SVCV (MOI, 0.1), H2O2 (500μM), respectively. The luciferase activities were monitored at 24 h after stimulation.
(C) FHM cells were seeded in 6-well plates and transfected the next day with siHO-1 and siNC, after 24 h, FHM cells were incubated with H2O2 (500 μM). Total RNA was prepared and analyzed by RT-qPCR for the expressions of ifn-1 and mx1 after 24h.
(D) FHM cells were seeded in 6-well plates and transfected the next day with siHO-1 and siNC, after 24 h, FHM cells were transfected with poly(I:C) (1 μg/ml), SVCV (0.1 MOI) were firstly transfected with siHO-1 and siNC, respectively. Total RNA was prepared and analyzed by RT-qPCR for the expressions of Ifn-1, Mx1, Viperin, and Isg15.
(E–F) FHM cells were seeded in 6-well plates and transfected the next day with siHO-1 and siNC, after 24 h, FHM cells were infected with SVCV (0.1 MOI) were firstly transfected with siHO-1 and siNC, respectively. Total RNA was prepared and analyzed by RT-qPCR for the expressions of Ifn-1, Mx1, Viperin, Isg15 and SVCV-G after 24h (E). Plaque assay was performed to measure virus titer (F). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.001 in the comparison between the indicated groups.
Intact heme oxygenase-1 promotes interferon-1 by binding with interferon regulatory factor 3
To search for the regulatory region where HO-1 mediates IFN-1, PpHO-1 truncates were constructed, the red color of the N-terminus (1-193aa) is the enzyme activity region, and the blue color of the C-terminus (194-286aa) region has no active binding site (Figure 4A). To explore the function domain of PpHO-1 for IFN-1 regulation, plasmids expressing truncated PpHO-1, including HO-1-N (1-193aa) and HO-1-C (194-286aa) were constructed, respectively. PpHO-1, PpHO-1-N, PpHO-1-C, and an empty vector were each transfected into FHM cells for 24 h before the cells were infected with 0.1 MOI SVCV. The results showed that the overexpression of PpHO-1 significantly promoted the mRNA levels of Irf3, Irf7, Ifn-1, and Mx1, consequently, the mRNA levels of SVCV-G were inhibited (Figure 4B). However, PpHO-1-N and PpHO-1-C showed no effect on Irf3, Irf7, Ifn-1, and Mx1. And no antiviral activity was observed. These data demonstrated that the antiviral function of PpHO-1 depends on its structural integrity.
Figure 4.
HO-1 inhibits SVCV-induced antiviral gene expression and interacts with IRF3
(A) Schematic diagram of HO-1 mutant constructs.
(B) FHM cells were seeded in 6-well plates and transfected the next day with 1μg PpHO-1-N, 1μg PpHO-1-C, and 1μg PpHO-1, or 1μg empty vector plasmid, then infected with SVCV (MOI, 0.1). Cell samples were collected at 24 h. The mRNA expression of Irf3, Irf7, Ifn-1, Mx1 and SVCV-G was detected by RT-qPCR.
(C and D) Empty vector and 5 μg HO-1-HA plasmid were co-transfected with 5 μg IRF3-Flag into FHM cells for 24 h. After 24 h overexpression, cell lysates were immuno-precipitated (IP) with anti-Flag-agarose or anti-HA-agarose conjugate and immunoblotted with anti-flag, anti-HA.
(E and F) Empty vector and 5 μg HO-1-HA plasmid were co-transfected with 5 μg IRF7 into FHM cells for 24 h. After 24 h overexpression, cell lysates were immuno-precipitated (IP) with anti-Flag-agarose or anti-HA-agarose conjugate and immunoblotted with anti-flag, anti-HA.
(G) Empty vector and 5 μg IRF3-His plasmid were co-transfected with 5 μg HO-1-HA and 5 μg IRF7-Flag into FHM cells for 24 h. After 24 h overexpression, cell lysates were immuno-precipitated (IP) with anti-His-agarose and immunoblotted with anti-flag, anti-HA. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.001 in the comparison between the indicated groups.
IRF3 is phosphorylated and nuclear translocated as a result of HO-1’s interaction with it, which in turn mediates the type I IFN response and exerts antiviral effects. Whether PpHO-1 regulates IRF3/7 in fish was further determined. Firstly, we confirmed that PpHO-1 precipitated with IRF3 by Co-IP and reverse Co-IP assays (Figures 4C and 4D). However, PpHO-1 did not precipitate with IRF7 (Figures 4E and 4F). This situation was reversed in the presence of IRF3 (Figure 4G). CO-IP experiments on PpHO-1-N and PpHO-1-C with IRF3/7 were performed, respectively.
Heme oxygenase-1 negatively regulates interferon-1 by inducing interferon regulatory factors 3/7 degradation
To investigate whether PpHO-1 inhibits IFN-1 by affecting IRF3 and IRF7 under oxidative stress, FHM cells were incubated with CoPP (40μM) for 24 h and then stimulated with H2O2 cells for 24 h. The mRNA levels of Irf3, Irf7, Ifn-1, and Mx1 were tested 24 h later. The results showed (Figure 5A) that H2O2 treatment of cells significantly upregulated the mRNA levels of Ifn-1 and Mx1 and had no effect on the transcriptional levels of IRF3 and IRF7, which indicated that HO-1 showed no effect on the mRNA levels of irf3 and irf7. To investigate the effect of HO-1 on the protein levels of IRF3 and IRF7, recombinant plasmids for IRF3-Flag and IRF7-Flag, respectively, were transfected into FHM cells and then treated with different concentrations of CoPP. IRF3 (Figures 5B and 5C) and IRF7 (Figures 5D and 5E) protein levels were considerably decreased as CoPP concentrations rose. Furthermore, the protein levels of IRF3 (Figures 5F–5G) and IRF7 (Figures 5H and 5I) were reduced with the extension of the incubating time. To determine whether CoPP suppresses IRF3 and IRF7 protein levels through PpHO-1, knockdown of PpHO-1 was performed with siRNA transfection. In the control cells, by transfecting control siRNA, CoPP treatment significantly inhibited the protein expression of IRF3 (Figures 5J and 5K) and IRF7 (Figures 5L and 5M), respectively. By comparison, knocking down PpHO-1 dampened the enhancement of IRF3 and IRF7. These results suggest that HO-1 promotes the degradation of IRF3 and IRF7 proteins.
Figure 5.
HO-1 degrades IRF3/7 during H2O2 treatment
(A) FHM cells incubated with DMSO and CoPP (40 μM) for 24 h, and then incubated with H2O2 (500 μM) for another 24 h, Ho-1, Irf3, Irf7, Ifn-1, and Mx1 mRNA expression as measured by RT-qPCR.
(B–E) FHM cells were seeded in 6-well plates overnight, empty vector and 1 μg IRF3-Flag or 1 μg IRF7-Flag plasmid were transfected into FHM cells, respectively, then FHM cells were incubated with CoPP (0 μM, 10 μM, 20 μM, 40 μM, 80 μM). After 24 h overexpression, the mRNA level of Ho-1 (B) and IRF3 protein expression (C) transfected with IRF3-Flag group were detected, the mRNA level of Ho-1 (D) and IRF7 protein expression (E) transfected with IRF7-Flag group were detected.
(F–I) FHM cells were seeded in 6-well plates overnight, empty vector and 1 μg IRF3-Flag or 1 μg IRF7-Flag plasmid were transfected into FHM cells, respectively, then FHM cells were incubated with CoPP (40 μM). Cell samples were collected at the indicated time points (0 h, 3 h, 6 h, 12 h, 24 h, 48 h). The mRNA level of Ho-1 (F) and IRF3 protein expression (G) transfected with IRF3-Flag group were detected, the mRNA level of Ho-1 (H) and IRF7 protein expression (I) transfected with IRF7-Flag group were detected.
(J–M) FHM cells were seeded in 6-well plates and transfected the next day with 1 μg IRF3-Flag or 1 μg IRF7-Flag plus siHO-1 or Control, subsequently, incubated with CoPP (40 μM) or DMSO, after 48 h, total RNA of Ho-1 was prepared and analyzed by RT-qPCR (J, L), and IB was conducted to detect the protein level of IRF3 (K) and IRF7 (M). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.001 in the comparison between the indicated groups.
Heme oxygenase-1 induces the degradation of interferon regulatory factors 3/7 through the autophagy pathway
The current studies on protein degradation have focused on three regulatory pathways: the ubiquitin-proteasome pathway (UPP), the lysosomal pathway, and the autophagosome pathway. To elucidate the pathway for HO-1 to degrade IRF3 and IRF7, the FHM cells were treated with the indicated inhibitors. The autophagosome inhibitor 3-MA completely prevented the HO-1-induced degradation of IRF3, but not MG132 or NH4Cl, indicating that HO-1 was responsible for the degradation of IRF3 through the autophagosome pathway (Figures 6A, 6C, and 6E). The lysosomal inhibitor NH4Cl and the autophagosome inhibitor 3-MA completely prevented the HO-1-induced degradation of IRF7 but not MG132, demonstrating that HO-1 was responsible for both the autophagosome pathway and the lysosomal pathway in the degradation of IRF7 (Figures 6B, 6D, and 6F). Additionally, IRF3/7 levels were gradually restored with increasing 3-MA concentrations (Figures 6G and 6H). To explore whether autophagy was induced by HO-1, the levels of MAP1LC3 (microtubule-associated protein 1 light chain 3), a marker protein of autophagy, were tested through Western blotting. Consistent with previous results, HO-1 inhibited the expression of IRF3/7, and HO-1 significantly upregulated LC3-II/LC3-I (Figures 6I and 6J), indicating HO-1’s role in mediating IRF3/7 degradation via an autophagosome-dependent mechanism.
Figure 6.
HO-1 degrades IRF3/7 through the autophagy pathway
(A and B) FHM cells were seeded in 6-well plates and transfected the next day with 1 μg IRF3-Flag or 1 μg IRF7-Flag plus 1 μg HO-1-HA or empty vector. The MG132 (20 μM) or PBS was incubated as indicated at 24 h posttransfection and treated for another 8 h. The IRF3-Flag (A) or IRF7-Flag (B) protein patterns were assessed by immunoblotting.
(C and D) FHM cells were seeded in 6-well plates and transfected the next day with 1 μg IRF3-Flag or 1 μg IRF7-Flag plus 1 μg HO-1-HA or empty vector. The NH4CL (20 mM) or PBS was incubated as indicated at 24 h posttransfection and treated for another 8 h. The IRF3-Flag (C) or IRF7-Flag (D) protein patterns were assessed by immunoblotting.
(E and F) FHM cells were seeded in 6-well plates and transfected the next day with 1 μg IRF3-Flag or 1 μg IRF7-Flag plus 1 μg HO-1-HA or empty vector. The 3-MA (5 μM) or PBS was incubated as indicated at 24 h posttransfection and treated for another 8 h. The IRF3-Flag (E) or IRF7-Flag (F) protein patterns were assessed by immunoblotting.
(G and H) FHM cells were seeded in 6-well plates and transfected the next day with 1 μg IRF3-Flag or 1 μg IRF7-Flag plus 1 μg HO-1-HA or empty vector. The 3-MA (0 μM, 5 μM, 10 μM, 20 μM) or PBS was incubated as indicated at 24 h posttransfection and treated for another 8 h. The IRF3-Flag (G) or IRF7-Flag (H) protein patterns were assessed by immunoblotting.
(I and J) FHM cells were seeded in 6-well plates and transfected the next day with 1 μg IRF3-Flag or 1 μg IRF7-Flag plus 1 μg HO-1-HA or empty vector for 48 h. Then, the cells were harvested for immunoblotting with anti-Flag, anti-HA, anti-LC3 and anti–β-actin Abs. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.001 in the comparison between the indicated groups.
His28 of heme oxygenase-1 is the key to inducing the degradation of interferon regulatory factors 3/7
To explore the functional domain of PpHO-1 for IRF3/7 degradation, GFP-tagged full-length HO-1 and a series of truncations were prepared as indicated (Figure 7A). Firstly, all truncated and full-length PpHO-1 were co-transfected with IRF3-Flag recombinant plasmids in FHM cells. As shown in Figure 7B, GFP-HO-11−95aa, GFP-HO-11−193aa, and GFP-HO-1 significantly reduced the protein level of IRF3, suggesting that the ferric ion-containing binding site is associated with the degradation of IRF3. The expression of PpHO-1 and its mutants on IRF7 was probed in the same way as above. Interestingly, HO-1194−286aa lacking the ferric ion-containing binding site can still degrade IRF7 (Figure 7C), but the degradation efficiency is lower than that of PpHO-1 truncation containing His28, indicating that PpHO-1 may implement the degradation of IRF7 in a manner different from that of IRF3. His28 of PpHO-1 is an important functional site for the degradation of heme to release iron ions.31 In order to further investigate the key site for the degradation of IRF3/7, the mutant HO-1-H28A was constructed and then co-transfected with IRF3 (Figures 7D and 7E). The results showed that HO-1-H28A enhanced the expression of Irf3, Ifn-1, and Mx1, the same results were presented with IRF7 (Figures 7F and 7G), reversing the negative regulation of showing an opposite regulation on IFN-I signaling for wild-type PpHO-1. These findings reveal that His28 is a key site for PpHO-1 to negatively regulate IFN-I signaling under oxidative stress.
Figure 7.
HO-1 and its N terminal degrade IRF3/7
(A) Schematic representation of HO-1 and its mutants.
(B and C) The 1 μg plasmids of full-length HO-1 or its constructs were transfected together with 1 μg IRF3-Flag or 1 μg IRF7-Flag into FHM cells for 48 h, cells were harvested for the Western blot analysis of the whole-cell extracts with the indicated Abs.
(D–G) 1 μg IRF3-Flag or 1 μg IRF7-Flag was transfected together with 1 μg HO-1 or 1 μg HO-1-H28A in FHM cells. At 48 h post-transfection, the IRF3 protein expression (D) and the mRNA level Ifn-1 and Mx1 (E) were detected, the IRF7 protein expression (F) and the mRNA level ifn-1 and mx1 (G) were detected. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.001 in the comparison between the indicated groups.
The pervasiveness of heme oxygenase-1 on interferon-I regulation
To explore whether the function of PpHO-1 in IFN-I regulation is conserved across different species, the amino acid sequences of representative PpHO-1 from 10 species were aligned. The phylogenetic tree analysis showed that the P. promelas HO-1 clustered with grass carp, zebrafish, and other Osteichthyes, which were far distant from human or mouse HO-1. The P. promelas HO-1 shared a 33%–86% amino acid identity with the indicated HO-1in other species. (Figure 8A). However, the functional domains of PpHO-1 among different species were conserved, implying that the function of PpHO-1 is conserved among different species (Figure 8B).
Figure 8.
The pervasiveness of HO-1 on IFN regulation
(A) Phylogenetic analysis based on the amino acid sequences. The HO-1 sequences from different species (Homo sapiens: GenBank: NP_002124.1; Xenopus laevis: GenBank: NP_001089909.1; Mus musculus: GenBank: NP_034572.1; Triplophysa tibetana: GenBank: KAA0701349.1; Gallus: GenBank: NP_990675.2; Tetraodon nigroviridis: GenBank: CAF95107.1; Danio rerio: GenBank: AOT80451.1; Megalobrama amblycephala: GenBank: ANP95035.1; Ctenopharyngodon Idella: GenBank: ART92582.1; Carassius auratus: GenBank: XP_026116539.1; Pimephales promelas: GenBank: XP_039512543.1) were downloaded from NCBI, and the software MEGA7.0 was performed to analysis.
(B) Amino acid sequence alignment of HO-1. ★: Heme binding sites. ▲: Fe2+ binding sites. ▭: Proximal helices.  : Distal helices.
: Distal helices.
(C) CIK cells were seeded in 6-well plates and transfected the next day with 1 μg CiIRF3-Flag or 1 μg CiIRF7-Flag, subsequently, incubated with CoPP (40 μM) or DMSO, after 24 h, cells were harvested for the Western blot analysis of the whole-cell extracts with the indicated Abs.
(D) ZFL cells were seeded in 6-well plates and transfected the next day with 1 μg DrIRF3-His or 1 μg DrIRF7-His, subsequently, incubated with CoPP (40 μM) or DMSO, after 24 h, cells were harvested for the Western blot analysis of the whole-cell extracts with the indicated Abs.
(E) 293T cells were seeded in 6-well plates and transfected the next day incubated with CoPP (40 μM) or DMSO, after 24 h, 293T cells were treated with H2O2 (500 μM) for 24 h, and the endogenous IRF3, IRF7 and IFN-1 protein levels were detected by Western blot assay.
To reveal whether the PpHO-1 degradation of IRF3 and IRF7 is conserved in different species, recombinant plasmids expressing grass carp IRF3 (p3xFlag-CMV-10-CiIRF3) and IRF7 (p3xFlag-CMV-10-CiIRF7) and zebrafish IRF3 (pcDNA4.0-DrIRF3) and IRF7 (pcDNA4.0-DrIRF7) were constructed, respectively. To explore whether PpHO-1 from grass carp degraded IRF3 and IRF7, CIK cells were pretreated with CoPP (40μM) for 24 h and then transfected with p3xFlag-CMV-10-CiIRF3 and p3xFlag-CMV-10-CiIRF7 for another 24 h. These cells were collected for protein immunoblotting analysis. The results showed that the overexpression of grass carp HO-1 inhibited CIK-IRF3-Flagand CIK-IRF7-Flag protein levels in CIK cells (Figure 8C). Also, endogenous overexpression of HO-1 in ZFL cells inhibited Dr-IRF3-His and Dr-IRF7-His protein levels (Figure 8D). 293T cells were also used to detect the function of HO-1 in the degradation of endogenous IRF3 and IRF7. The results showed (Figure 8E) that IRF3 and IRF7 were significantly down-regulated in COPP-treated cells when compared with vertical controls. Consequently, the IFN-α levels in these COPP-treated cells decreased significantly when compared with control cells. These findings suggest that HO-1 negatively regulates IRF3 and IRF7 universally in fish and mammals under oxidative stress.
Discussion
IFN-I plays an important role in inflammatory responses, cancer immunity, cell proliferation, and apoptosis.32,33 Abnormalities in IFN-I signaling disturb the host immune system and induce the development of autoimmune diseases.12,13,14 Homeostatic regulation of IFN-I signaling is tightly linked with host fitness. Our study first demonstrated the bi-directional regulation of IFN-I signaling by HO-1 under different stimuli and identified a negative regulation of IFN-I signaling by the degradation of IRF3/7 via HO-1, elucidating a homeostatic regulation of IFN-I signaling under oxidative stress.
The antiviral activity of HO-1 has been widely reported in mammals, which depends on its enzymatic reaction by-products, including biliverdin (BV), CO, and Fe2+, or on the direct regulation of the IFN-I response. It was shown that HO-1 inhibited porcine reproductive and respiratory syndrome virus (PRRSV) through the metabolite BV and the secondary metabolite bilirubin (BR) via a nitric oxide (NO)-dependent cGMP/PKG signaling pathway.34 The effect of HO-1 on hepatitis C virus (HCV) replication may be partially attributed to the iron-dependent inactivation of the HCV RNA polymerase NS5B and BV-mediated IFN-I-induced antiviral responses.35 On the other hand, HO-1 inhibited influenza virus replication by interacting with IRF3 to activate the IFN-I response.36 Our study showed that HO-1 significantly promoted the activation of IFN-I signaling and inhibited the replication of SVCV, either by SVCV challenge or poly(I:C) stimulation, providing a potent target for antiviral agent screening.
IFN-I is regulated by two main families of transcription factors: nuclear factor kappa-light chain-activated B-cell enhancers (NF-κB) and interferon regulatory factors (IRFs), of which IRF3/7 is the most potent player in the regulation of IFN-I in the innate immune system. After being subjected to cellular signaling, IRF3/7 is phosphorylated and translocated to the nucleus to activate IFN-I transcription. Our findings indicated that only full-length PpHO-1 promoted the transcription of Irf3/7, Ifn-1, Mx1, Viperin, and Isg15, thus inhibited SVCV replication. We further confirmed that only full-length PpHO-1 interacted directly with IRF3. However, IRF7 could be pulled down by PpHO-1 in the presence of IRF3, probably because IRF3 and IRF7 could form a heterodimer. These findings indicated that the conformation of PpHO-1 is important to bind with IRF3 and activate the IFN-1 signal, which is consistent with homo sapiens HO-1.37 The fold of HO-1 is a single compact domain consisting of mostly α helices and two helices.38 The two helices in HO-1, known as the proximal and distal helices, contribute most of the contacts to the heme group.31 As for the incompetency of IRF3 binding for the NTD or CTD of HO-1, we suspected that the two proximal and distal helices might be important for IRF3 binding, which might explain why the full-length but not the truncated HO-1 could bind with IRF3.
Oxidative stress and viral infection are closely related. The SARS-CoV-2 virus NSP14 targets SIRT1 to suppress Nrf2-HO-1 antioxidant signaling, and allopurinol can effectively alleviate its induced oxidative stress.39,40 DENV also promotes Nrf2 degradation to increase oxidative stress and encourage virus reproduction.41 On the other hand, pathologically induced oxidative stress can activate IFN-I signaling through various pathways such as STING, forkhead box protein O3 (FOXO3), or SUMO/specific protease 3 (SENP3).15,16,42 HO-1, as an antioxidant protein, contributes to the repair of ROS-induced oxidative damage and attenuates the oxidative stress response. Our study showed that H2O2-induced oxidative stress significantly upregulated IFN-1 signaling in FHM cells. We found that CoPP significantly inhibited oxidative stress-induced IFN-1 signaling in H2O2-treated FHM cells, while the IFN-1 signaling rebounded after knocking down PpHO-1 by siRNA transfection (Figures 2 and 3). For different intensities of oxidative stress, the host responds differently in a dose-dependent manner. At low levels of oxidative stress, genes encoding antioxidant enzymes are upregulated through the Nrf2/kelch-like ECH-associated protein 1 (Keap1/Nrf2) system, but when oxidative stress levels are aggravated, inflammatory factors as well as heat shock proteins are induced, and even programmed cell death is initiated through signaling pathways such as NF-κB and activating protein-1 (AP1).43 Interestingly, under a resting state or low concentration of H2O2 stimulation, PpHO-1 promotes IFN-1 signaling. While at high concentrations of H2O2 stimulation, PpHO-1 shifted to negatively regulate IFN-1 signaling (Figure 2F). H2O2 is not only a toxic substance but also serves as a secondary messenger for its relative stability. We suspected that intracellular levels of H2O2 may play a crucial role in the shift of PpHO-1 in IFN-1 regulation. It has been shown that IFN-I signaling plays an important role in oxidative stress-mediated tissue damage, and oxidative stress-induced oxidized mitochondrial DNA activates IFN-I signaling through the STING pathway.44 We hypothesize that IFN-1 signaling at high concentrations of H2O2 is passively activated by oxidative stress-induced DNA damage and that HO-1 drives the negative regulation of IFN-1 signaling to maintain intracellular environmental homeostasis.
IRF3/7 can be degraded through multiple pathways. The various E3 ubiquitin ligases, including RTA-associated ubiquitin ligase (RAUL), casitas b-lineage lymphoma (c-Cbl), peptidyl-prolyl cis-trans isomerase nima-interacting 1 (Pin1), and tripartite motif-containing protein 21 (TRIM21), can promote ubiquitination and subsequent proteasomal degradation.45,46,47,48 The phospholipase A/acyltransferase 1 (PLAAT1) and NBR1 autophagy cargo receptor (NBR1) degrade IRF3/7 through the autophagosomal pathway.49,50 The mucin-domain containing molecule-3 (Tim-3) and zebrafish’s transcript isoform of the TANK binding kinase 1 (TBK1) heterodimer can degrade IRF3/7 through the lysosomal pathway.51,52 PpHO-1 was found to promote the degradation of IRF3/IRF7 through the autophagic pathway, providing evidence that autophagy is involved in the regulation of IFN-1 homeostasis. ROS can induce autophagy through various pathways, such as Beclin-1 and mitogen activated kinase-like proteins (MAPKs).53,54 As reported, the cargo receptor sequestosome 1 (SQSTM1/p62) play a pivotal role in delivering substrates for autophagic degradation.55 For instance, the deubiquitinase OTU deubiquitinase 7B (OTUD7B) interacts with IRF3 and activates p62 oligomerization to promote the selective autophagic degradation of IRF3.56 The release and activation of Nrf2, an important regulator of HO-1, is also achieved by ubiquitin-binding protein p62-dependent autophagy of Keap1.57 Whether HO-1 depends on p62 to promote the selective autophagic degradation of IRF3/7 need to be further explored.
An interesting issue is the functional domain(s) of HO-1 for the bi-directional regulation of IFN under different conditions. We verified that only full-length PpHO-1 bound to IRF3 and promoted IFN-1. However, the N terminal (1–95 aa) of PpHO-1 containing the iron ion-binding site was critical for PpHO-1 to degrade IRF3 (Figure 7B). The Ala substitution at His28 completely blocked the degradation of HO-1 on IRF3, indicating that His28 is the key site for PpHO-1 to degrade IRF3 (Figure 7D). The iron ion-binding site His28 interacts with iron to release ferrous ions from heme and store them in ferritin to maintain intracellular iron homeostasis.31 Iron ions can activate cGAS-STING to promote IFN-1 expression.58 In addition, the absence of glutathione peroxidase 4 (GPX4), the core regulatory molecule of ferroptosis, promotes STING carboxylation and inhibits downstream IFN-1 signaling, suggesting a cross-linking of iron ions with IFN-1 signaling.59 In human lupus nephritis, HO-1 and ferritin are colocalized within the glomeruli and expressed with similar patterns, and knockdown of either impairs host anti-oxidative stress, implying ferritin mediates the protective effect of PpHO-1 in response to oxidative stress.60 Whether HO-1-mediated autophagic degradation of IRF3 is related to intracellular iron ion concentration or ferritin remains to be explored. Considering that only full-length PpHO-1 could bind with IRF3, we speculated that the degradation activity of PpHO-1 was independent of its binding with IRF3. On the other hand, HO-1-truncated bodies containing only the C-terminal can also induce the degradation of IRF7, while H28A-HO-1 cannot (Figures 7D and 7F). Previous studies have shown that the intact conformation is crucial for HO-1 to exert the positive regulation of IFN-1. Therefore, we speculate that H28A-HO-1 inhibits the ability of HO-1-C to induce the degradation of IRF7 due to its ability to retain the positive regulation of IFN-1. The above speculation and whether other unknown proteins contribute to the degradation of IRF7 along with HO-1 need further exploration. The ferric ion binding sites of HO-1 are highly conserved among different species. Accordingly, the degradation of IRF3/7 by HO-1 under oxidative stress was equally conserved across species, which expands the function of HO-1 in the regulation of IFN-I homeostasis (Figure 8).
In summary, our findings indicate that HO-1 maintains host homeostasis by directly interacting with IRF3 to promote IFN-I against viral pathogens and inducing the autophagic degradation of IRF3/7 to inhibit IFN-I under oxidative stress. Our work lays the groundwork for exploring targets for the treatment of autoimmune illnesses and for the screening of antiviral medications that target HO-1.
Limitations of the study
One limitation of our study is that we do not elucidate how HO-1 senses the different states of cells. Our preliminary results indicated that the regulation tendency of HO-1 on IFN-I depended on the concentration of H2O2 treatment (Figure 2F), suggesting that the intensity of oxidative stress might be important for the function of HO-1. Further experiments are needed to answer this interesting but intricate question. Another limitation of our study is we failed to uncover the molecular mechanism underlying the degradation of IRF3/7 depending on the HO-1’s iron-binding sites (His28), although we revealed the key domain of HO-1 for IRF3/7 degradation under oxidative stress. HO-1 degrades heme and releases iron ions to participate in cellular iron metabolism. We speculated whether the mechanism of HO-1 degrading IRF3/7 was related to cellular iron metabolism or ferroptosis. However, the following projects are needed to investigate the cross-linking effect of HO-1 between the IFN-I pathway and the iron metabolism pathway.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti HA-Tag mAb | ABclone | Cat#AE008; RRID:AB_2770404 |
| Mouse anti DDDDK-Tag mAb | ABclone | Cat#AE005; RRID:AB_2770401 |
| Mouse anti His-Tag mAb | ABclone | Cat#AE003; RRID:AB_2728734 |
| Mouse anti GFP-Tag mAb | ABclone | Cat#AE012; RRID:AB_2770402 |
| Heme Oxygenase 1 Rabbit pAb | ABclone | Cat#A1346; RRID:AB_2760320 |
| IRF3 Rabbit mAb | ABclone | Cat#A19717; RRID:AB_2862750 |
| IRF7 Rabbit pAb | ABclone | Cat#A0159; RRID:AB_2756990 |
| IFN-α1 Rabbit mAb | ABclone | Cat#A0285; RRID:AB_2757097 |
| β-Actin Rabbit mAb | ABclone | Cat#AC026; RRID:AB_2768234 |
| LC3B Rabbit mAb | ABclone | Cat# A19665; RRID:AB_2862723 |
| HRP Goat Anti-Mouse IgG (H+L) | ABclone | Cat#AS003; RRID:AB_2769851 |
| HRP Goat Anti-Rat IgG (H+L) | ABclone | Cat#AS028; RRID:AB_2769855 |
| SVCV-G | Huazhong Agricultural University | N/A |
| IFN-1(Pimephales promelas) | Huazhong Agricultural University | N/A |
| HO-1-GC | Zhongkai University of Agriculture and Engineering | N/A |
| Bacterial and virus strains | ||
| Spring viremia of carp virus | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Medium 199 | Gibco | Cat#11150067 |
| Dulbecco’s Modified Eagle Medium | Gibco | Cat#10566016 |
| Fetal bovine serum (FBS) | Gibco | Cat#10091-148 |
| 3-Methyladenine | MedChemExpress | Cat#HY-19312 |
| MG-132 | MedChemExpress | Cat#HY-13259 |
| Polyinosinic-polycytidylic acid | MedChemExpress | Cat#HY-107202 |
| NH4Cl | Sigma-Aldrich | Cat#213330 |
| Cobaltic protoporphyrin IX chloride (CoPP) | Sigma-Aldrich | Cat#102601-60-5 |
| DMSO | Sigma-Aldrich | Cat#67-68-5 |
| Critical commercial assays | ||
| RNA Extraction Kit | Takara | Cat# A2270 |
| Reverse transcription reagent Kit | Takara | Cat#R22301 |
| Quantitative PCR (RT-qPCR) Kit | Vazyme | Cat#Q71102 |
| Renilla-Firefly Luciferase Dual Assay Kit | MedChemExpress | Cat#HY-K1013 |
| DAPI | Beyotime | Cat#P0131 |
| 4% formaldehyde | Biosharp | Cat#BL539A |
| Experimental models: Cell lines | ||
| Fathead minnow muscle cell line (FHM) | ATCC | CCL-42 |
| Human embryonic kidney 293T cell line (HEK293T) | ATCC | CRL-11268 |
| Zebrafish liver cell line (ZFL) | ATCC | CRL-2643 |
| Ctenopharyngodon idellus kidney cell line (CIK) | Huazhong Agricultural University | N/A |
| Oligonucleotides | ||
| HO-1-F Reverse primer (5′-3′) ATGGAGACGATTGAGAAAAC |
This paper | N/A |
| HO-1-R Reverse primer (5′-3′) TTAAGCGTAGTCTGGGACGT |
This paper | N/A |
| HO-1-HA-F Reverse primer (5′-3′) CCGGAATTCCCGCCACCATGGAGACGATTGAGAAAAC |
This paper | N/A |
| HO-1-HA-R Reverse primer (5′-3′) CCGCTCGAGTTAAGCGTAGTCTGGGA CGTCGTATGGTAAAAGGCGTAGACCATCCCGA |
This paper | N/A |
| HO-1-GFP-F Reverse primer (5′-3′) CCGGAATTCGCCACCATGGAGACGATTGAGAAAAC |
This paper | N/A |
| HO-1-GFP-R Reverse primer (5′-3′) CGGGGTACCTCAAAGGCGTAGACCATCCCGAC |
This paper | N/A |
| HO-11-95aa-GFP-F Reverse primer (5′-3′) CCGGAATTCGCCACCATGGAGACGATTGAGAAAAC |
This paper | N/A |
| HO-11-95aa-GFP-R Reverse primer (5′-3′) CGGGGTACCTCGTCTTTCCCGACGGACTCCAG |
This paper | N/A |
| HO-1194-286aa-GFP-F Reverse primer (5′-3′) CCGGAATTCGCCACCATGACGGAGGAGCAGAGGAG |
This paper | N/A |
| HO-1194-286aa-GFP-R Reverse primer (5′-3′) CGGGGTACCTCAAAGGCGTAGACCATCCCGAC |
This paper | N/A |
| HO-11-193aa-GFP-F Reverse primer (5′-3′) CCGGAATTCGCCACCATGGAGACGATTGAGAAAAC |
This paper | N/A |
| HO-11-193aa -GFP-R Reverse primer (5′-3′) CGGGGTACCTCCAGCTCGATGCTGTT CATGCG |
This paper | N/A |
| HO-196-286aa-GFP-F Reverse primer (5′-3′) CCGGAATTCGCCACCATGGAGACGATTGAGAAAAC |
This paper | N/A |
| HO-196-286aa-GFP-R Reverse primer (5′-3′) CGGGGTACCTCCAGCTCGATGCTGTT CATGCG |
This paper | N/A |
| IRF3-RFP-F Reverse primer (5′-3′) CCGCTCGAGCCGCCACCATGACCCAA CCAAAGCCGCTCTT |
This paper | N/A |
| IRF3-RFP-R Reverse primer (5′-3′) CCGGGTACCACACAACTCCATCATTTGCTCCAG |
This paper | N/A |
| IRF3-Flag-F Reverse primer (5′-3′) CCGAAGCTTGCCACCATGACCCAACCAAAGCCGCT |
This paper | N/A |
| IRF3-Flag-R Reverse primer (5′-3′) CCGAAGCTTGCCACCATGCAGAGCACGATGGGCAAAC |
This paper | N/A |
| IRF7-Flag-F Reverse primer (5′-3′) CCGAAGCTTGCCACCATGCAGAGCACGATGGGCAAAC |
This paper | N/A |
| IRF7-Flag-R Reverse primer (5′-3′) CGGGGTACCGAGTCCATTGAAGGCAGACCCAAAG |
This paper | N/A |
| IRF7-RFP-F Reverse primer (5′-3′) CCGGAATTCCGCCACCATGCAGAGCACGATGGGCAA |
This paper | N/A |
| IRF7-RFP-R Reverse primer (5′-3′) CGGGGTACCTTAGTCCATTGAAGGCAGAC |
This paper | N/A |
| DrIRF3-His-F Reverse primer (5′-3′) CCGGAATTCGCCACCATGACTCAAGCAAAACCGCTG |
This paper | N/A |
| DrIRF3-His-R Reverse primer (5′-3′) CGGGGTACCGAGCAGAGCTCCATCATTTGC |
This paper | N/A |
| DrIRF7-His-F Reverse primer (5′-3′) CCGGAATTCGCCACCATGCAGAGCACA AATGCC |
This paper | N/A |
| DrIRF7-His-R Reverse primer (5′-3′) CGGGGTACCGACTGGGTCTGCCTTCAGTGGAA |
This paper | N/A |
| CiIRF3-Flag-F Reverse primer (5′-3′) CCGAATTCGCCACCATGACCCATCCAAAACCG |
This paper | N/A |
| CiIRF3-Flag-R Reverse primer (5′-3′) CCGGTACCTCCTTGGTGTCACACAAC |
This paper | N/A |
| CiIRF7-Flag-F Reverse primer (5′-3′) CCGAATTCGCCACCATGGCAGCGATGCAGAGCAC |
This paper | N/A |
| CiIRF7-Flag-R Reverse primer (5′-3′) GGGGTACCTTAGTCCATTGAAGGCAGAC |
This paper | N/A |
| Primers for Ho-1, see Table S1 | This paper | N/A |
| Primers for Irf3, see Table S1 | This paper | N/A |
| Primers for Irf7, see Table S1 | This paper | N/A |
| Primers for Ifn-1, see Table S1 | This paper | N/A |
| Primers for Mx1, see Table S1 | This paper | N/A |
| Primers for Viperin, see Table S1 | This paper | N/A |
| Primers for Isg15, see Table S1 | This paper | N/A |
| Primers for SVCV-G, see Table S1 | This paper | N/A |
| Primers for β-actin, see Table S1 | This paper | N/A |
| Primers for siRNA Negative control, see Table S1 | This paper | N/A |
| Primers for Ho-1-siRNA, see Table S1 | This paper | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents, please contact Junfa Yuan and will be fulfilled (jfyuan@mail.hzau.edu.cn).
Materials availability
This study did not generate new unique materials.
Data and code availability
-
•
Data: All data reported in this paper will be shared by the lead contact upon request.
-
•
Code: This paper does not report original code.
-
•
Other items: Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Cell lines and viruses
The fathead minnow muscle cell line (FHM, ATCC: CCL-42), the human embryonic kidney 293T cell line (HEK293T, ATCC: CRL-11268), and the zebrafish liver cell line (ZFL, ATCC: CRL-2643) were provided by the American Type Culture Collection (ATCC). The Ctenopharyngodon idellus kidney cell line (CIK) was kindly provided by Prof. Jianguo Su (Huazhong Agricultural University, China). The spring viremia of carp virus (SVCV) was stored in our lab.
Reagents and Abs
The MG132, 3-Methyladenine (3-MA), and ammonium chloride (NH4Cl) were from MCE (New Jersey, USA). The poly(I:C), COPP, and 3% H2O2 were purchased from Merck (Darmstadt, Germany).
Mouse anti-HA (1:5000), anti-Flag (1:5000), anti-His (1:3000), anti-GFP (1:5000), and rabbit anti-β-actin (1:10000), anti-HO-1 (1:1000), anti-IRF3 (1:1000), anti-IRF7 (1:1000), anti-IFN-α (1:500), and anti-LC3 (1:2000), were ordered from ABclone (Boston, USA). Secondary antibodies against mouse (1:5000) and rabbit (1:5000) were provided by Abcam (Cambridge, UK). Rabbit polyclonal anti-Pimephales promelas IFN antiserum was prepared by immunization of animals with prokaryotic-expressed proteins previously made in our laboratory. The rabbit anti-SVCV-G and mouse anti-Grass Carp HO-1 antiserum were kindly gifted by Prof. Xueqin Liu (Huazhong Agricultural University) and Prof. Zhendong Qin (Zhongkai University of Agriculture and Engineering), respectively.
Method details
Cell culture and viruses
The medium M199 containing 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 U/ml streptomycin was used for the culture of FHM, CIK, and ZFL cells, respectively. 293T cells were cultured at 37°C with 5% CO2 in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS, 100 U/ml penicillin, and 100 U/ml streptomycin.
The spring viremia of carp virus (SVCV) was propagated in FHM cells at 28°C and harvested after >80% cytotoxicity was observed.
Plasmid construction, in vitro RNAi, and transfection
The open reading frame (ORF) of HO-1 (PpHO-1, GenBank: XM039656609.1), IRF3 (PpIRF3, GenBank: MN781134.1), and IRF7 (PpIRF7, GenBank: MN781135.1) from Pimephales promelas were cloned into the pcDNA4.0, pEGFP-N1, p3flag-CMV-10, or pDsRed1-N1 vector, respectively. The ORFs IRF3 and IRF7 from Danio rerio (DrIRF3, GenBank: XM017358468.2; DrIRF7, GenBank: NM200677.2) and Ctenopharyngodon idellus (CiIRF3, GenBank: KC898261.1; CiIRF7, GenBank: KY613780.1) were also cloned into the pcDNA4.0 and p3flag-CMV-10 vector, respectively. IFN-1 pro-Luc and ISRE pro-Luc promoter vectors were previously constructed in our laboratory. key resources table lists the primers used for plasmid synthesis as well as the restriction enzyme cutting sites.
To knockdown HO-1 in FHM cells, the HO-1 siRNA sequences were designed by GenePharma according to our lab (Table S1).
FHM, CIK, ZFL, or 293T cells were cultured in plates for 16 h, and plasmids or siRNAs were transfected into cells using FishTrance or PEI transfection reagents according to manufacturer instructions. Gene expression or function investigation was performed 24 h after challenge or stimulation (unless stated otherwise).
Luciferase activity assay
FHM cells were seeded in 24-well plates at a density of 5 × 105 cells/ml for 24 h. Then co-transfection was performed with 500-ng expression plasmid, 500-ng target promoter-luciferase plasmid, and 50-ng pRL-TK for 24 h, stimulated with incubated with 0.1 MOI SVCV, 500 μM H2O2, or 1 μg/mL poly(I:C), respectively. After 24 h, the cells were rinsed with PBS and lysed before being measured for luciferase activity. The luciferase reading of each sample was first normalized against the pRL-TK level, and the relative light unit intensity was presented as the ratio of firefly luciferase to renilla luciferase. All experiments were performed in triplicate.
RT-qPCR
RNA Extraction Kit (Takara, China) was used to extract total RNA, and cDNA was synthesized using random hexamer primers with a reverse transcription kit (Takara, Dalian, China). RT-qPCR was performed with High ROX Premixed (Vazyme, China) on the CFX Opus Real-Time PCR Systems (Bio-Rad, California, USA). PCR conditions were as follows: 95°C for 5 min and then 40 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 20 s mRNA expression levels were normalized to the expression of β-action in cells and the RT-qPCR primers that were employed in Table S1. Statistical analysis was performed using method 2−ΔΔCt.
Immunoblotting (IB) and coimmunoprecipitation (co-IP) analysis
Protein extracts were transferred onto Immobilon transfer membranes after being separated on 10–15% SDS-PAGE gels for IB analysis. The membranes were incubated with 5% TBS-T containing skim milk powder (Biofroxx, Beijing, China) at 37°C for 2 h. The membranes were washed with TBS-T and then incubated with primary and secondary Ab antibodies at 37° for 2 h and 45 min, respectively, during which the membranes were washed with TBS-T. Following three TBST buffer washes, depending on the size of the membrane, BeyoECL Plus working solution was added dropwise to the membrane at a ratio of 1 mL per 10 cm2 of membrane. After incubating for 1–2 min, the Amersham Imager 600 system (GE Healthcare, New York, USA) was used to scan and image the transfer membrane.
For co-IP analysis, FHM cells in 15-cm2 Petri dish were co-transfected with the indicated plasmids for 48 h. The cells were lysed for 30 min on ice in IP lysis buffer (Beyotime Biotechnology, Beijing, China), and the cellular debris was removed by centrifugation at 12,000 g for 30 min at 4°C. The supernatant was transferred to a fresh tube and incubated with 1 μg of Ab with gentle shaking overnight at 4°C. Protein A+G Sepharose beads were added to the mixture and incubated for 6 h at 4°C. After centrifugation at 3,000 g for 5 min, the beads were then collected, washed five times with lysis buffer, suspended in 20-μl 2 × SDS loading buffer, and denatured at 95°C for 10 min, followed by immunoblotting.
Virus infection and virus titration
For viral infection, FHM cells were plated for 24 h in advance and then infected with SVCV, at a multi-plicity of infection (MOI) of 0.1. For virus titration, FHM cells (1 × 106 cells/well) were transfected with indicated plasmids or siRNA and then infected with SVCV at a MOI of 0.1 for 24 h.
For virus titration, the supernatant was collected according to the above experiment, sequentially diluted 10-fold (10−1 to 10−8), and incubated with FHM cells in a 96-well plate at 28°C for 7 d to determine the 50% tissue culture–infective dose (TCID50). On day 7, the plates were examined under the microscope for the presence of cytopathic effects (CPE). Recovered viruses were 10-fold diluted and inoculated onto FHM cells in 12-well plates. After 1 h of viral adsorption, cells were washed with serum-free M199 and cultured in M199 containing 5% FBS and 1.5% sodium carboxymethyl cellulose. At 72 h post-infection, infected cells were washed in PBS, fixed in 4% paraformaldehyde, and stained in 1% crystal violet to identify CPE.
Quantification and statistical analysis
All assays were repeated three times using independently collected or prepared samples. The SEM is shown in error bars (n = 3, physiologically separate samples). The gray values of protein expression and relative plaque intensity were quantified by ImageJ. All results were presented as mean ± SD. The mean values from each group were compared by Student’s t test. p value <0.05 was considered a statistically significant difference (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.001).
Acknowledgments
This work was supported by the National Natural Science Foundation of China 31872598.
Author contributions
M.M.W. and Y.H.F. performed the experiments, analyzed the data, and wrote the article. L.J.L. provided technical support and reviewed the article. J.F.Y. performed project administration, writing—review and editing, read and approved the final article. All authors approved the final article.
Declaration of interests
The authors declare no potential competing interests.
Published: February 8, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109185.
Supplemental information
References
- 1.Du L., Liu W., Rosen S.T., Chen Y. Mechanism of SUMOylation-Mediated Regulation of Type I IFN Expression. J. Mol. Biol. 2023;435 doi: 10.1016/j.jmb.2023.167968. [DOI] [PubMed] [Google Scholar]
- 2.Hervas-Stubbs S., Perez-Gracia J.L., Rouzaut A., Sanmamed M.F., Le Bon A., Melero I. Direct effects of type I interferons on cells of the immune system. Clin. Cancer Res. 2011;17:2619–2627. doi: 10.1158/1078-0432.CCR-10-1114. [DOI] [PubMed] [Google Scholar]
- 3.Lazear H.M., Schoggins J.W., Diamond M.S. Shared and Distinct Functions of Type I and Type III Interferons. Immunity. 2019;50:907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu L., Zhou X., Wang W., Wang Y., Yin Y., Laan L.J.W.v.d., Sprengers D., Metselaar H.J., Peppelenbosch M.P., Pan Q. IFN regulatory factor 1 restricts hepatitis E virus replication by activating STAT1 to induce antiviral IFN-stimulated genes. Faseb. J. 2016;30:3352–3367. doi: 10.1096/fj.201600356R. [DOI] [PubMed] [Google Scholar]
- 5.Olson G.S., Murray T.A., Jahn A.N., Mai D., Diercks A.H., Gold E.S., Aderem A. Type I interferon decreases macrophage energy metabolism during mycobacterial infection. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.109195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lam K.C., Araya R.E., Huang A., Chen Q., Di Modica M., Rodrigues R.R., Lopès A., Johnson S.B., Schwarz B., Bohrnsen E., et al. Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell. 2021;184:5338–5356.e21. doi: 10.1016/j.cell.2021.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi M., Lio C.W.J., Campeau A., Steger M., Ay F., Mann M., Gonzalez D.J., Jain M., Sharma S. The tumor suppressor kinase DAPK3 drives tumor-intrinsic immunity through the STING-IFN-β pathway. Nat. Immunol. 2021;22:485–496. doi: 10.1038/s41590-021-00896-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarousse N., Trujillo D.L., Wilcox-Adelman S., Coscoy L. Virally-induced upregulation of heparan sulfate on B cells via the action of type I IFN. J. Immunol. 2011;187:5540–5547. doi: 10.4049/jimmunol.1003495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.González-Navajas J.M., Lee J., David M., Raz E. Immunomodulatory functions of type I interferons. Nat. Rev. Immunol. 2012;12:125–135. doi: 10.1038/nri3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaudhary V., Ah Kioon M.D., Hwang S.M., Mishra B., Lakin K., Kirou K.A., Zhang-Sun J., Wiseman R.L., Spiera R.F., Crow M.K., et al. Chronic activation of pDCs in autoimmunity is linked to dysregulated ER stress and metabolic responses. J. Exp. Med. 2022;219 doi: 10.1084/jem.20221085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez-Ruiz R., Niewold T.B. Type I Interferons in Autoimmunity. J. Invest. Dermatol. 2022;142:793–803. doi: 10.1016/j.jid.2021.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Postal M., Vivaldo J.F., Fernandez-Ruiz R., Paredes J.L., Appenzeller S., Niewold T.B. Type I interferon in the pathogenesis of systemic lupus erythematosus. Curr. Opin. Immunol. 2020;67:87–94. doi: 10.1016/j.coi.2020.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang J., Zhao M., Chang C., Wu H., Lu Q. Type I Interferons in the Pathogenesis and Treatment of Autoimmune Diseases. Clin. Rev. Allergy Immunol. 2020;59:248–272. doi: 10.1007/s12016-020-08798-2. [DOI] [PubMed] [Google Scholar]
- 15.Hwang I., Uchida H., Dai Z., Li F., Sanchez T., Locasale J.W., Cantley L.C., Zheng H., Paik J. Cellular stress signaling activates type-I IFN response through FOXO3-regulated lamin posttranslational modification. Nat. Commun. 2021;12:640. doi: 10.1038/s41467-020-20839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolf S.J., Estadt S.N., Theros J., Moore T., Ellis J., Liu J., Reed T.J., Jacob C.O., Gudjonsson J.E., Kahlenberg J.M. Ultraviolet light induces increased T cell activation in lupus-prone mice via type I IFN-dependent inhibition of T regulatory cells. J. Autoimmun. 2019;103 doi: 10.1016/j.jaut.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang S., Kang L., Dai X., Chen J., Chen Z., Wang M., Jiang H., Wang X., Bu S., Liu X., et al. Manganese induces tumor cell ferroptosis through type-I IFN dependent inhibition of mitochondrial dihydroorotate dehydrogenase. Free Radic. Biol. Med. 2022;193:202–212. doi: 10.1016/j.freeradbiomed.2022.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Tsai C.Y., Hsieh S.C., Lu C.S., Wu T.H., Liao H.T., Wu C.H., Li K.J., Kuo Y.M., Lee H.T., Shen C.Y., Yu C.L. Cross-Talk between Mitochondrial Dysfunction-Provoked Oxidative Stress and Aberrant Noncoding RNA Expression in the Pathogenesis and Pathophysiology of SLE. Int. J. Mol. Sci. 2019;20:5183. doi: 10.3390/ijms20205183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Kuraishy H.M., Sami O.M., Hussain N.R., Al-Gareeb A.I. Metformin and/or vildagliptin mitigate type II diabetes mellitus induced-oxidative stress: The intriguing effect. \"J. Adv. Pharm. Technol. Research\"\" (JAPTR)\". 2020;11:142–147. doi: 10.4103/japtr.JAPTR_18_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H., Wang G., Banerjee N., Liang Y., Du X., Boor P.J., Hoffman K.L., Khan M.F. Aberrant Gut Microbiome Contributes to Intestinal Oxidative Stress, Barrier Dysfunction, Inflammation and Systemic Autoimmune Responses in MRL/lpr Mice. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.651191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soares M.P., Marguti I., Cunha A., Larsen R. Immunoregulatory effects of HO-1: how does it work? Curr. Opin. Pharmacol. 2009;9:482–489. doi: 10.1016/j.coph.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 22.Chiang S.K., Chen S.E., Chang L.C. The Role of HO-1 and Its Crosstalk with Oxidative Stress in Cancer Cell Survival. Cells. 2021;10:2401. doi: 10.3390/cells10092401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y., Yang C., Elsheikh N.A.H., Li C., Yang F., Wang G., Li L. HO-1 reduces heat stress-induced apoptosis in bovine granulosa cells by suppressing oxidative stress. Aging. 2019;11:5535–5547. doi: 10.18632/aging.102136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X., Yu J., Gong L., Zhang Y., Dong S., Shi J., Li C., Li Y., Zhang Y., Li H. Heme oxygenase-1(HO-1) regulates Golgi stress and attenuates endotoxin-induced acute lung injury through hypoxia inducible factor-1α (HIF-1α)/HO-1 signaling pathway. Free Radic. Biol. Med. 2021;165:243–253. doi: 10.1016/j.freeradbiomed.2021.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang W., Wang Y., Zhang P., Sun X., Chen X., Yu J., Shi L., Yin Y., Tao K., Li R. Immune-responsive gene 1 protects against liver injury caused by concanavalin A via the activation Nrf2/HO-1 pathway and inhibition of ROS activation pathways. Free Radic. Biol. Med. 2022;182:108–118. doi: 10.1016/j.freeradbiomed.2022.02.030. [DOI] [PubMed] [Google Scholar]
- 26.Zou L., Lei H., Shen J., Liu X., Zhang X., Wu L., Hao J., Jiang W., Hu Z. HO-1 induced autophagy protects against IL-1 β-mediated apoptosis in human nucleus pulposus cells by inhibiting NF-κB. Aging. 2020;12:2440–2452. doi: 10.18632/aging.102753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Espinoza J.A., González P.A., Kalergis A.M. Modulation of Antiviral Immunity by Heme Oxygenase-1. Am. J. Pathol. 2017;187:487–493. doi: 10.1016/j.ajpath.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 28.Zu G., Zhou T., Che N., Zhang X. Salvianolic Acid A Protects Against Oxidative Stress and Apoptosis Induced by Intestinal Ischemia-Reperfusion Injury Through Activation of Nrf2/HO-1 Pathways. Cell. Physiol. Biochem. 2018;49:2320–2332. doi: 10.1159/000493833. [DOI] [PubMed] [Google Scholar]
- 29.Fang Y., Xing C., Wang X., Cao H., Zhang C., Guo X., Zhuang Y., Hu R., Hu G., Yang F. Activation of the ROS/HO-1/NQO1 signaling pathway contributes to the CoPPer-induced oxidative stress and autophagy in duck renal tubular epithelial cells. Sci. Total Environ. 2021;757 doi: 10.1016/j.scitotenv.2020.143753. [DOI] [PubMed] [Google Scholar]
- 30.Li C., Li L., Jin L., Yuan J. Heme Oxygenase-1 inhibits spring viremia of carp virus replication through carbon monoxide mediated cyclic GMP/Protein kinase G signaling pathway. Fish Shellfish Immunol. 2018;79:65–72. doi: 10.1016/j.fsi.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 31.Unno M., Matsui T., Ikeda-Saito M. Structure and catalytic mechanism of heme oxygenase. Nat. Prod. Rep. 2007;24:553–570. doi: 10.1039/b604180a. [DOI] [PubMed] [Google Scholar]
- 32.Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen K., Liu J., Cao X. Regulation of type I interferon signaling in immunity and inflammation: A comprehensive review. J. Autoimmun. 2017;83:1–11. doi: 10.1016/j.jaut.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 34.Zhang A., Duan H., Li N., Zhao L., Pu F., Huang B., Wu C., Nan Y., Du T., Mu Y., et al. Heme oxygenase-1 metabolite biliverdin, not iron, inhibits porcine reproductive and respiratory syndrome virus replication. Free Radic. Biol. Med. 2017;102:149–161. doi: 10.1016/j.freeradbiomed.2016.11.044. [DOI] [PubMed] [Google Scholar]
- 35.Lehmann E., El-Tantawy W.H., Ocker M., Bartenschlager R., Lohmann V., Hashemolhosseini S., Tiegs G., Sass G. The heme oxygenase 1 product biliverdin interferes with hepatitis C virus replication by increasing antiviral interferon response. Hepatology. 2010;51:398–404. doi: 10.1002/hep.23339. [DOI] [PubMed] [Google Scholar]
- 36.Ma L.L., Zhang P., Wang H.Q., Li Y.F., Hu J., Jiang J.D., Li Y.H. heme oxygenase-1 agonist CoPP suppresses influenza virus replication through IRF3-mediated generation of IFN-α/β. Virology. 2019;528:80–88. doi: 10.1016/j.virol.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 37.Feng Y., Guo X., Tian H., He Y., Li Y., Jiang X., Zheng H., Xiao S. Induction of HOXA3 by Porcine Reproductive and Respiratory Syndrome Virus Inhibits Type I Interferon Response through Negative Regulation of HO-1 Transcription. J. Virol. 2022;96 doi: 10.1128/JVI.01863-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lad L., Schuller D.J., Shimizu H., Friedman J., Li H., Ortiz de Montellano P.R., Poulos T.L. Comparison of the heme-free and -bound crystal structures of human heme oxygenase-1. J. Biol. Chem. 2003;278:7834–7843. doi: 10.1074/jbc.M211450200. [DOI] [PubMed] [Google Scholar]
- 39.Zhang S., Wang J., Wang L., Aliyari S., Cheng G. SARS-CoV-2 virus NSP14 Impairs NRF2/HMOX1 activation by targeting Sirtuin 1. Cell. Mol. Immunol. 2022;19:872–882. doi: 10.1038/s41423-022-00887-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Al-Kuraishy H.M., Al-Gareeb A.I., Al-Niemi M.S., Aljowaie R.M., Almutairi S.M., Alexiou A., Batiha G.E.S. The Prospective Effect of Allopurinol on the Oxidative Stress Index and Endothelial Dysfunction in Covid-19. Inflammation. 2022;45:1651–1667. doi: 10.1007/s10753-022-01648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferrari M., Zevini A., Palermo E., Muscolini M., Alexandridi M., Etna M.P., Coccia E.M., Fernandez-Sesma A., Coyne C., Zhang D.D., et al. Dengue Virus Targets Nrf2 for NS2B3-Mediated Degradation Leading to Enhanced Oxidative Stress and Viral Replication. J. Virol. 2020;94:e015511-20. doi: 10.1128/JVI.01551-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Z., Teng X.L., Zhang T., Yu X., Ding R., Yi J., Deng L., Wang Z., Zou Q. SENP3 senses oxidative stress to facilitate STING-dependent dendritic cell antitumor function. Mol. Cell. 2021;81:940–952.e5. doi: 10.1016/j.molcel.2020.12.024. [DOI] [PubMed] [Google Scholar]
- 43.Lushchak V.I. Free radicals, reactive oxygen species, oxidative stress and its classification. Chem. Biol. Interact. 2014;224:164–175. doi: 10.1016/j.cbi.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 44.Lood C., Blanco L.P., Purmalek M.M., Carmona-Rivera C., De Ravin S.S., Smith C.K., Malech H.L., Ledbetter J.A., Elkon K.B., Kaplan M.J. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat. Med. 2016;22:146–153. doi: 10.1038/nm.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu Y., Hayward G.S. The ubiquitin E3 ligase RAUL negatively regulates type i interferon through ubiquitination of the transcription factors IRF7 and IRF3. Immunity. 2010;33:863–877. doi: 10.1016/j.immuni.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao X., Zhu H., Yu J., Li H., Ge J., Chen W. c-Cbl-mediated ubiquitination of IRF3 negatively regulates IFN-β production and cellular antiviral response. Cell. Signal. 2016;28:1683–1693. doi: 10.1016/j.cellsig.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 47.Saitoh T., Tun-Kyi A., Ryo A., Yamamoto M., Finn G., Fujita T., Akira S., Yamamoto N., Lu K.P., Yamaoka S. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat. Immunol. 2006;7:598–605. doi: 10.1038/ni1347. [DOI] [PubMed] [Google Scholar]
- 48.Tan T., Xia L. TRIM21 Aggravates Herpes Simplex Virus Epithelial Keratitis by Attenuating STING-IRF3-Mediated Type I Interferon Signaling. Front. Microbiol. 2020;11:703. doi: 10.3389/fmicb.2020.00703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao X., Huang W., Shi Y., Guo J., Xiao H., Ji N., Feng J., Dang H., Zou J. PLAAT1 inhibits type I interferon response via degradation of IRF3 and IRF7 in Zebrafish. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.979919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cai Y., Zhu Y., Zheng J., Zhang Y., Chen W. NBR1 mediates autophagic degradation of IRF3 to negatively regulate type I interferon production. Biochem. Biophys. Res. Commun. 2022;623:140–147. doi: 10.1016/j.bbrc.2022.07.043. [DOI] [PubMed] [Google Scholar]
- 51.Schwartz J.A., Clayton K.L., Mujib S., Zhang H., Rahman A.K.M.N.U., Liu J., Yue F.Y., Benko E., Kovacs C., Ostrowski M.A. Tim-3 is a Marker of Plasmacytoid Dendritic Cell Dysfunction during HIV Infection and Is Associated with the Recruitment of IRF7 and p85 into Lysosomes and with the Submembrane Displacement of TLR9. J. Immunol. 2017;198:3181–3194. doi: 10.4049/jimmunol.1601298. [DOI] [PubMed] [Google Scholar]
- 52.Zhang J., Wu X.M., Hu Y.W., Chang M.X. A Novel Transcript Isoform of TBK1 Negatively Regulates Type I IFN Production by Promoting Proteasomal Degradation of TBK1 and Lysosomal Degradation of IRF3. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.580864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo Q.Q., Wang S.S., Zhang S.S., Xu H.D., Li X.M., Guan Y., Yi F., Zhou T.T., Jiang B., Bai N., et al. ATM-CHK2-Beclin 1 axis promotes autophagy to maintain ROS homeostasis under oxidative stress. EMBO J. 2020;39 doi: 10.15252/embj.2019103111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang T.L., Yang Y., Guo L., Xia S., Zhang B., Yan M. Sunitinib induced hepatotoxicity in L02 cells via ROS-MAPKs signaling pathway. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.1002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lamark T., Svenning S., Johansen T. Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem. 2017;61:609–624. doi: 10.1042/EBC20170035. [DOI] [PubMed] [Google Scholar]
- 56.Xie W., Tian S., Yang J., Cai S., Jin S., Zhou T., Wu Y., Chen Z., Ji Y., Cui J. OTUD7B deubiquitinates SQSTM1/p62 and promotes IRF3 degradation to regulate antiviral immunity. Autophagy. 2022;18:2288–2302. doi: 10.1080/15548627.2022.2026098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ichimura Y., Waguri S., Sou Y.S., Kageyama S., Hasegawa J., Ishimura R., Saito T., Yang Y., Kouno T., Fukutomi T., et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol. Cell. 2013;51:618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 58.Jia M., Qin D., Zhao C., Chai L., Yu Z., Wang W., Tong L., Lv L., Wang Y., Rehwinkel J., et al. Redox homeostasis maintained by GPX4 facilitates STING activation. Nat. Immunol. 2020;21:727–735. doi: 10.1038/s41590-020-0699-0. [DOI] [PubMed] [Google Scholar]
- 59.Li H., Hu L., Wang L., Wang Y., Shao M., Chen Y., Wu W., Wang L. Iron Activates cGAS-STING Signaling and Promotes Hepatic Inflammation. J. Agric. Food Chem. 2022;70:2211–2220. doi: 10.1021/acs.jafc.1c06681. [DOI] [PubMed] [Google Scholar]
- 60.Cheng H.T., Yen C.J., Chang C.C., Huang K.T., Chen K.H., Zhang R.Y., Lee P.Y., Miaw S.C., Huang J.W., Chiang C.K., et al. Ferritin heavy chain mediates the protective effect of heme oxygenase-1 against oxidative stress. Biochim. Biophys. Acta. 2015;1850:2506–2517. doi: 10.1016/j.bbagen.2015.09.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data: All data reported in this paper will be shared by the lead contact upon request.
-
•
Code: This paper does not report original code.
-
•
Other items: Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.