Abstract
In glial C6 cells constitutively expressing wild-type p53, synthesis of the calcium-binding protein S100B is associated with cell density-dependent inhibition of growth and apoptosis in response to UV irradiation. A functional interaction between S100B and p53 was first demonstrated in p53-negative mouse embryo fibroblasts (MEF cells) by sequential transfection with the S100B and the temperature-sensitive p53Val135 genes. We show that in MEF cells expressing a low level of p53Val135, S100B cooperates with p53Val135 in triggering calcium-dependent cell growth arrest and cell death in response to UV irradiation at the nonpermissive temperature (37.5°C). Calcium-dependent growth arrest of MEF cells expressing S100B correlates with specific nuclear accumulation of the wild-type p53Val135 conformational species. S100B modulation of wild-type p53Val135 nuclear translocation and functions was confirmed with the rat embryo fibroblast (REF) cell line clone 6, which is transformed by oncogenic Ha-ras and overexpression of p53Val135. Ectopic expression of S100B in clone 6 cells restores contact inhibition of growth at 37.5°C, which also correlates with nuclear accumulation of the wild-type p53Val135 conformational species. Moreover, a calcium ionophore mediates a reversible G1 arrest in S100B-expressing REF (S100B-REF) cells at 37.5°C that is phenotypically indistinguishable from p53-mediated G1 arrest at the permissive temperature (32°C). S100B-REF cells proceeding from G1 underwent apoptosis in response to UV irradiation. Our data support a model in which calcium signaling and S100B cooperate with the p53 pathways of cell growth inhibition and apoptosis.
Calcium as a ubiquitous second messenger regulates many cellular functions, including cell growth, differentiation, and apoptosis (15, 35). The S100 family of EF-hand calcium-binding proteins is thought to play a role in mediating calcium signals in cell growth, differentiation, and motility (reviewed in reference 42). To date, 17 different proteins have been assigned to the S100 protein family. They show different degrees of homology, ranging from 25 to 65% identity at the amino acid level. Most of the S100 proteins have been isolated in screens for mRNAs or proteins whose expression is regulated by the state of cellular growth, transformation, or differentiation, suggesting a direct implication of the S100 proteins in cell cycle regulation. The S100B protein is a Ca2+- and Zn2+-binding protein (6) which is expressed at high levels in the vertebrate nervous system, where it is found in the cytoplasm of glial cells (21). In whole rat brain, the S100B level is low at birth and begins to increase abruptly after 12 to 15 days, when rapid differentiation occurs (25). The gene for human S100B maps to the Down’s syndrome (DS) region of chromosome 21 (1). Overexpression of S100B in the brains of patients with DS and Alzheimer’s disease (20, 33, 46), and in the brains of patients with AIDS (47), has led to the hypothesis that S100B plays a contributory, perhaps causal, role in common neuropathologies associated with these diseases. Although the majority of S100B in the brain is cytoplasmic, some data suggest that S100B may be secreted in an oxidized form and that extracellular oxidized S100B has neurotrophic and mitogenic activity (27, 44). In the sympathetic PC12 cell line, high concentrations of extracellular S100B protein are able to inhibit proliferation followed by apoptosis (17). In cultured glioma C6 cells, cytoplasmic accumulation of S100B correlates with contact-dependent inhibition of growth, cell differentiation (29, 30), and increased sensitivity of the cells to UV-induced apoptosis (this study). On the other hand, in human melanoma cells, overproduction of S100B protein in G1 phase is linked with progression through the cell cycle (32). These apparent contradictions suggest that alternative functions for intracellular S100B in negative and positive cell growth regulation might depend on other, as yet unidentified cellular cofactors. We have previously identified the tumor suppressor p53 protein as a putative cellular target for the S100B protein (9). In vitro, S100B interacts in a calcium-dependent manner with p53 to protect p53 from thermal denaturation and aggregation (9). The possible involvement of S100B in cell density-dependent inhibition of growth of glial C6 cells (this study), together with the fact that the major phenotype of cultured astrocytes derived from p53-deficient mice is altered growth inhibition at high density (49), has led us to envision a synergism between S100B and the p53 pathways of cell growth inhibition and apoptosis. To test this hypothesis, we have analyzed the effect of ectopic expression of S100B on the growth properties of two fibroblast cell lines with different genetic backgrounds but expressing the temperature-sensitive (ts) p53Val135 mutant.
p53 has been implicated in cell differentiation (2, 41), cell contact inhibition of growth (49), and protection of the cell from the acquisition of genomic abnormalities (50). The mechanisms by which p53 carries out these functions seem to be related to its ability to induce cell cycle arrest and/or apoptosis (11, 36, 39, 50). In normal or untransformed cells, the half-life of wild-type p53 protein is very short (on the order of 5 to 20 min), making it very difficult to study this protein in these cells. Moreover, accumulation of wild-type p53 correlates with cell growth arrest or apoptosis, preventing establishment of stable cell lines expressing high levels of the protein. The murine ts mutant p53Val135 protein has been developed to overcome these problems and is widely used as an experimental tool in analyzing the regulation and mode of action of p53 in cell proliferation, differentiation, and apoptosis (2, 5, 11, 18, 19, 28, 34, 36, 41, 48, 50). At the nonpermissive temperature (37.5°C), the mutant p53Val135 conformational species predominates over wild-type p53Val135. At the permissive temperature (32°C), the p53Val135 protein primarily folds into a wild-type conformation and is translocated into the cell nucleus, where it can function as a growth suppressor (18, 28, 36) or induce apoptosis (11, 19, 50). Conformational flexibility that characterizes the ts p53Val135 mutant is not specific to this mutation and is representative of the conformational flexibility that also characterizes wild-type p53 (37). Conformational shift between the wild-type and mutant conformations (recognized by monoclonal antibodies PAb246 and PAb240, respectively) is a mechanism regulating wild-type p53 during embryonic differentiation (41).
We show here that in mouse embryo fibroblasts (MEF cells) expressing a low level of the p53Val135 and in rat embryo fibroblast (REF) cell line clone 6, transformed with oncogenic Ha-ras and overexpressing the p53Val135 (36), S100B cooperates with calcium to rescue a p53-dependent G1 checkpoint control at 37.5°C. This study provides the first direct evidence for a role of S100B in calcium signaling linked to activation of a p53-dependent pathway of cell growth inhibition and apoptosis. The apoptosis mediated by calcium and S100B may be relevant in neurodegenerative diseases in which S100B is overexpressed.
MATERIALS AND METHODS
Cell cultures and transfection experiments.
The clonal rat glial C6 cells were grown in DMEM (Dulbecco modified Eagle medium)-Glutamax (Gibco) supplemented with 10% fetal calf serum (FCS; Seromed) at 37.5°C. Clone 6 cells and transfected derivatives were grown in RPMI-Glutamax (Gibco) supplemented with 5% FCS (Seromed) at 37.5°C. p53−/− MEF cells and transfected derivatives were grown in DMEM-Glutamax supplemented with 10% FCS (HyClone) at 37.5°C.
Human S100B cDNA (38) was subcloned into the pcDNA I Neo expression system (Invitrogen). Clone 6 cells were transfected by lipofection using DOTAP (Boehringer) according to the manufacturer’s instructions. After 48 h, cells were selected with neomycin (G418; 500 μg/ml). p53−/− MEF cells were a gift from P. Andreassen (Institut de Biologie Structural, CEN-G, Grenoble, France). MEF cells were doubly transfected with pcDNA-S100B and a vector carrying hygromycin B resistance (ratio, 10:1). Stably transfected clones were selected in hygromycin B (200 μg/ml).
Retrovirus-mediated gene transfer.
MEF cells and MEF cells expressing S100B (S100B-MEF cells; clone J-β) were plated at 5 × 105 cells per 10-cm-diameter dish and incubated overnight. For infections, cells were incubated with culture medium containing the recombinant retrovirus pLXSNp53val135 (19). To generate clones, infected cells were subjected to limiting dilutions.
Antibodies.
Affinity-purified polyclonal rabbit anti-p21, anti-mdm2, anti-GADD45, and anti-p27 were from Santa Cruz Biotechnology. Anti-Rb monoclonal antibody (clone PMG3-245) was from Pharmingen. Anti-β-tubulin was a gift from L. Paturle and D. Job. Anti-p53 monoclonal antibodies PAb240, PAb246, and PAb421 were purified from mouse ascites fluid. Affinity-purified polyclonal rabbit anti-S100B antibodies were purified on an S100B-Sepharose column.
Protein concentration and Western blot analysis.
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer, and protein concentration was estimated by the bicinchoninic acid method (Pierce), with bovine serum albumin (BSA) as a standard. Western blot analysis utilized cell extracts in RIPA buffer mixed with an equal volume of 1% sodium dodecyl sulfate (SDS) containing 20% glycerol, 50 mM dithiothreitol, and a trace of bromophenol blue. For S100B analysis, an SDS-Tris-Tricine–11% polyacrylamide gel was used (48). For p21, p27, p53, and tubulin, proteins were separated on SDS–13% polyacrylamide gels. For Rb and mdm2, proteins were separated on SDS–7.5% polyacrylamide gels. S100B was detected with affinity-purified polyclonal rabbit anti-S100B antibodies. p53 was detected with a mixture of monoclonal antibodies PAb421 and PAb240. p21, p27, Rb, and mdm2 were detected with commercial antibodies.
Immunoprecipitation analysis.
For S100B immunoprecipitation analysis, cells were labeled with [35S]Met-Cys mix (100 μCi/ml) for 6 h prior to lysis. Cell lysates were incubated with affinity-purified rabbit polyclonal anti-S100B antibodies (10 μg) and protein A-agarose. Immunoprecipitates were washed with lysis buffer, and the immunoprecipitated proteins were separated on an SDS-Tris-Tricine–11% polyacrylamide gel (43).
For immunoprecipitation of nuclear 35S-labeled p53Val135, cells were labeled with [35S]Met-Cys mix (100 μCi/ml). Nuclear extracts were prepared in buffer containing nonionic detergent as described in reference 14.
Soft agar assays.
For plating in soft agar, 104 cells were resuspended in 2 ml of 0.35% (wt/vol) agar solution containing DMEM plus 20% FCS and overlaid on a 0.5% (wt/vol) agar solution in a 35-mm-diameter plate. Two days after incubation, 2 ml of DMEM supplemented with 20% FCS was added. The experiments were done twice in duplicate.
Flow cytometry.
Cell cycle analysis by flow cytometry was performed on a FACStar+ (Becton Dickinson). For cell cycle parameter analysis, cells were collected in phosphate-buffered saline (PBS) and vortexed with 0.2% Triton X-100, and nuclei were fixed with 4% formaldehyde. DNA was stained with Hoechst 33258 (Hoechst; 2 μg/ml) just prior to flow cytometry analysis.
Cell cycle analysis.
Cells were labeled for 30 min with 10 μM bromodeoxyuridine (BrdU). Cells were detached with trypsin, fixed in 70% ethanol, and treated for 15 min with 2 N HCl. After centrifugation, cells were resuspended in sodium tetraborate (0.1 M, pH 8.5), centrifuged again, and washed once with PBS–0.5% Tween 20–0.5% BSA–0.1% glucose. Cells were then incubated with fluorescein isothiocyanate-conjugated anti-BrdU in PBS–0.5% Tween 20–0.5% BSA–0.1% glucose. Nuclei were stained with Hoechst (2 μg/ml), and samples were analyzed by two-dimensional flow cytometry.
Immunofluorescence cell staining.
Cells were grown on Permanox slides (Nunc, Inc.), fixed with 4% paraformaldehyde for 30 min, and permeabilized for 3 min with 0.2% Triton. After being washed with PBS, cells were incubated for 2 h in PBS containing 5% goat serum with purified wild-type specific monoclonal antibody PAb246. The cells were then washed five times with PBS and incubated for 1 h with fluorescein isothiocyanate- or cyanine 3-conjugated secondary antibodies. Coverslips were incubated in a solution of Hoechst (2 μg/ml) for 2 min for DNA staining, mounted in aquamount, and observed on a Zeiss fluorescence microscope (magnification of ×40).
RESULTS
Endogenous S100B expression in glial C6 cells correlates with cell contact inhibition of growth and UV-dependent apoptosis.
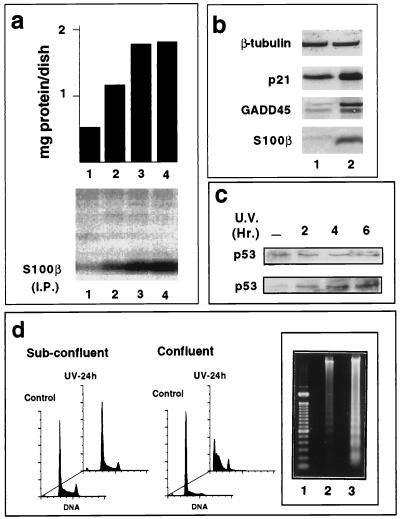
Clonal glial C6 cells were grown to confluence, and expression of S100B was analyzed by immunoprecipitation of [35S]methionine-labeled S100B (Fig. 1a) or Western blot analysis (Fig. 1b). A strong induction of S100B accompanied cell contact-dependent inhibition of growth. S100B synthesis at confluence correlates with accumulation of the cells in the G1 phase of the cell cycle (Fig. 1d, front panels) and with the induction of p21WAF1 and GADD45 proteins (Fig. 1b), two putative mediators of p53-dependent growth arrest (3, 16). We have determined by immunoprecipitation that in glial C6 cells the p53 protein exists in a wild-type conformation (not shown). Only the pan-specific PAb421, not mutant-specific PAb240, immunoprecipitated a protein that migrated at the position of p53 in nondenaturing conditions. This result corroborates a previous study showing that C6 cells express wild-type p53 mRNA (4). S100B synthesis at confluence also correlates with increased sensitivity of the cells to UV-induced apoptosis (Fig. 1d). Internucleosomal DNA cleavage in apoptotic cells revealed by fluorescence-activated cell sorting (FACS) analysis was confirmed by agarose gel electrophoresis (Fig. 1d, inset). The p53 level was not significantly modified as glial C6 cells progressed from low cell density to confluence (Fig. 1c), but UV irradiation of confluent cells produced a significant increase in the p53 level (Fig. 1c), consistent with a role of p53 in glial cell apoptosis.
FIG. 1.
S100B synthesis in glial C6 cells is correlated with cell contact inhibition of growth and UV-induced apoptosis. (a) S100B synthesis in glial C6 cells is correlated with cell contact inhibition of growth. Glial C6 cells were seeded at 5 × 105 cells/ml in 35-mm-diameter dishes. After 24 h (lane 1), 48 h (lane 2), 72 h (lane 3), and 82 h (lane 4), cells were metabolically labeled with [35S]Met-Cys mix for 6 h prior to harvesting. The upper panel shows the quantity of total protein per dish; the lower panel shows S100B protein immunoprecipitated (I.P.) from 1 mg of protein. (b) Western blot analysis for β-tubulin, p21WAF1, GADD45, and S100B in 50 μg of glial C6 cell extracts from exponentially growing cultures (lane 1) or from those at the end of logarithmic growth (lane 2). (c) Time course of p53 induction in subconfluent (upper panel) or confluent (lower panel) glial C6 cells after UV irradiation (20 J/m2). (d) Cell density-dependent inhibition of growth of glial C6 cells is correlated with sensitivity to UV-induced apoptosis. Exponentially growing cultures (Sub-confluent) or those at the end of logarithmic growth (Confluent) were left untreated (Control) or UV irradiated (30 J/m2) (UV-24 h). Cell DNA was analyzed after 24 h by flow cytometry analysis or on an agarose gel. (Inset) Lane 1, DNA markers; lane 2, exponentially growing cells; lane 3, cells at the end of logarithmic growth.
Because S100B interacts with p53 in vitro, we have envisioned a possible synergism between S100B and the p53 pathways of cell growth inhibition and apoptosis in glial cells.
Characterization of p53-negative MEF cells expressing S100B and p53Val135.
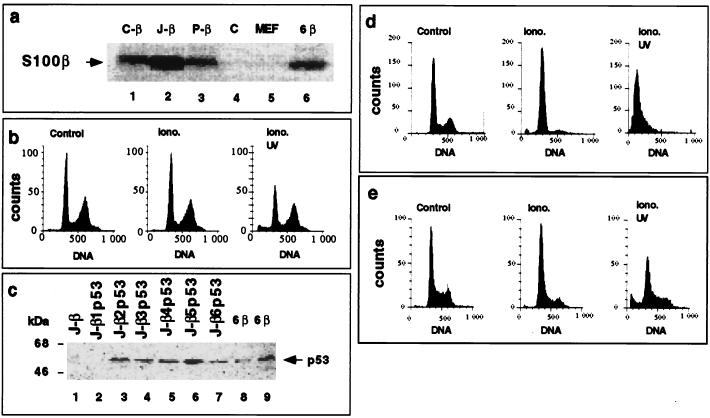
To investigate a possible functional interaction between S100B and the p53-dependent pathway of cell growth arrest and cell death, we introduced S100B and the ts p53Val135 into p53-negative MEF cells lacking both components. We first doubly transfected p53−/− MEF cells with a pcDNA-Neo expression vector bearing the gene for S100B and with a plasmid sequence containing the hygromycin resistance gene. Three hygromycin-resistant clones expressing S100B (C-β, J-β, and P-β), and one hygromycin-resistant clone not expressing S100B (C) were selected (Fig. 2a, lanes 1 to 4). Selected S100B-MEF clones express S100B at levels similar to those for S100B-REF cells (lane 6) used later in this study.
FIG. 2.
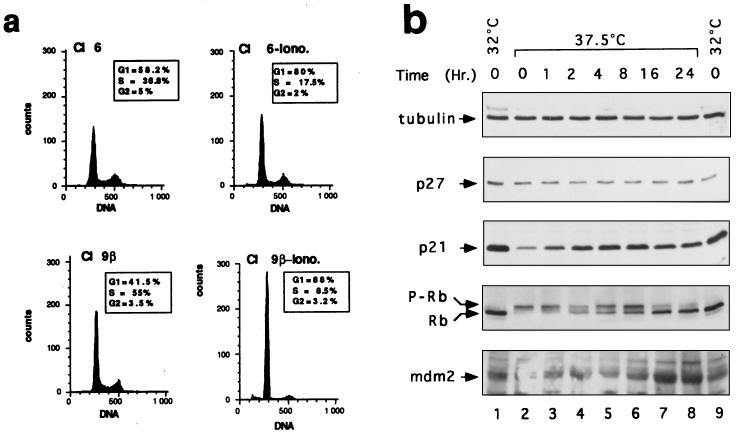
Ectopic expression of S100B and p53Val135 in p53-negative MEF cells activates a calcium-dependent G1 checkpoint at the nonpermissive temperature. (a) Comparison of S100B synthesis in hygromycin-resistant MEF cell clones C-β (lane 1), J-β (lane 2), and P-β (lane 3), hygromycin-resistant MEF cell clone C (lane 4), parental MEF cells (lane 5), and REF clone 6β cells (lane 6). Cells were metabolically labeled with [35S]Met-Cys mix and lysed in RIPA buffer. Extracts, corresponding to 1 mg of protein, were incubated with affinity-purified rabbit polyclonal anti-S100B. (b) Flow cytometry analysis of the DNA content of S100B-MEF J-β cells grown at 37.5°C not stimulated (Control), stimulated with ionomycin (Iono.) for 24 h, or stimulated for 24 h with ionomycin and UV irradiated (Iono. UV). (c) Western blot analysis of the p53Val135 content in total cell extracts of S100B-MEF J-β cells infected with pLXSNp53val135 recombinant retrovirus. S100B-MEF J-β cells (lane 1) and six derived clones (J-β1p53 to J-β6p53) were selected by limiting dilution (lanes 2 to 7). Fifty micrograms of protein was loaded in each lane. In lanes 8 and 9, 0.05 and 0.1 μg of total protein from clone 6β cell extract were loaded for comparison. (d and e) Flow cytometry analysis of the DNA content of clone J-β2p53 cells (d) and clone Cp53 cells (e) grown at 37.5°C not stimulated (Control), stimulated with ionomycin (Iono.) for 36 h, or stimulated with ionomycin for 24 h and UV irradiated (Iono. UV). Clone Cp53 was generated by transfection of clone C (panel a, lane 4) with p53Val135 recombinant retrovirus and limiting dilution. In panels b, d, and e, ionomycin was used at 1 μM (final concentration). The irradiation dose was 10 J/m2, and cells were analyzed 24 h postirradiation.
Parental MEF cells underwent G1 arrest at confluence (not shown). Hence, the effect of S100B expression on cell contact inhibition of growth could not be evaluated.
S100B is a calcium-binding protein whose functions are activated upon binding calcium (6 to 9). Because cell contact induces an elevation in cytosolic calcium concentration that can be mimicked by the calcium ionophore ionomycin (13, 15), we evaluated the effect of ionomycin on the pattern of the cell cycle distribution of exponentially growing clone J-β cells. As shown in Fig. 2b, ionomycin produced no significant effect on the cell cycle parameters, suggesting that S100B expression alone is not sufficient for triggering MEF cell growth arrest. Moreover, S100B-MEF cells showed no apoptosis upon UV irradiation.
Hygromycin-resistant MEF cells not expressing S100B (clone C [Fig. 2a, lane 4]) or expressing S100B (clone J-β [Fig. 2a, lane 2]) were then transfected with recombinant pLXSNp53Val135 retroviruses (19). Stably transfected clones that express p53Val135 at equivalent levels were selected by limiting dilution. One clone (Cp53 [Fig. 3]) derived from MEF cells and five clones (J-β2p53 to J-β6p53) derived from S100B-MEF cells were selected (Fig. 2c and 3). We estimated the amount of p53Val135 in S100B-MEF cells to be 500- to 1,000-fold less than in REF clone 6 cells overexpressing the p53Val135 used later in this study (Fig. 2c; compare lanes 3 to 7 to lanes 8 and 9). The low level of p53Val135 expression in S100B-MEF cells was nevertheless sufficient to rescue a calcium-dependent G1 checkpoint control in all the selected S100B-MEF clones at the nonpermissive temperature (37.5°C). As shown in Fig. 2d, ionomycin stimulation of clone J-β2p53 cells produced an accumulation of the cells in the G1 phase. Moreover, S100B-MEF cells expressing p53Val135 rapidly died upon a low dose of UV irradiation (Fig. 2d). In contrast, ionomycin stimulation of clone Cp53 cells was not able to promote G1 arrest and to sensitize cells to UV-mediated apoptosis (Fig. 2e).
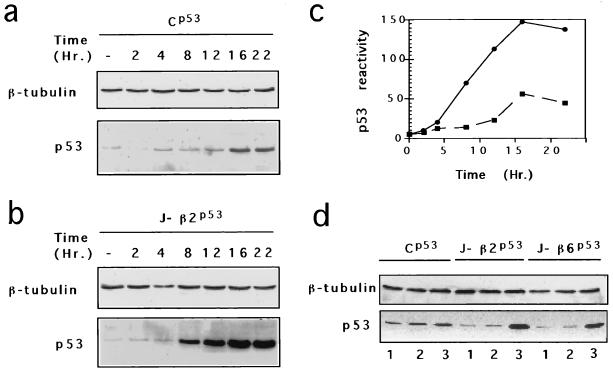
FIG. 3.
Ionomycin stimulates p53Val135 accumulation in S100B-MEF cells. (a to c) Time course of p53Val135 protein induction in clone Cp53 (a) and clone J-β2p53 (b) cells after ionomycin stimulation. (c) p53Val135 immunoreactivity quantified for clone Cp53 (▪) and clone J-β2p53 (•). (d) Comparison of p53Val135 contents in clone Cp53, clones J-β2p53, and clone J-β6p53 grown at 37.5°C (lanes 1) or 32°C (lanes 2) or stimulated with ionomycin for 18 h at 37.5°C (lanes 3).
Calcium promotes nuclear accumulation of wild-type p53Val135 in S100B-MEF cells at the nonpermissive temperature.
To assess the mechanism by which calcium and S100B rescue p53Val135-dependent G1 arrest in MEF cells at 37.5°C, we first compared the effects of ionomycin stimulation on p53Val135 accumulation in control (clone Cp53) and S100B-MEF cells by Western blot analysis (Fig. 3). Ionomycin stimulation of Cp53 cells produced only limited p53 accumulation (Fig. 3a and d). In contrast, ionomycin-mediated G1 arrest of S100B-MEF cells correlates with a strong induction of the p53Val135 protein (Fig. 3b to d). This accumulation was dependent on calcium and S100B, as shifting cells to 32°C was not sufficient to promote p53Val135 accumulation in MEF or S100B-MEF cells (Fig. 3d).
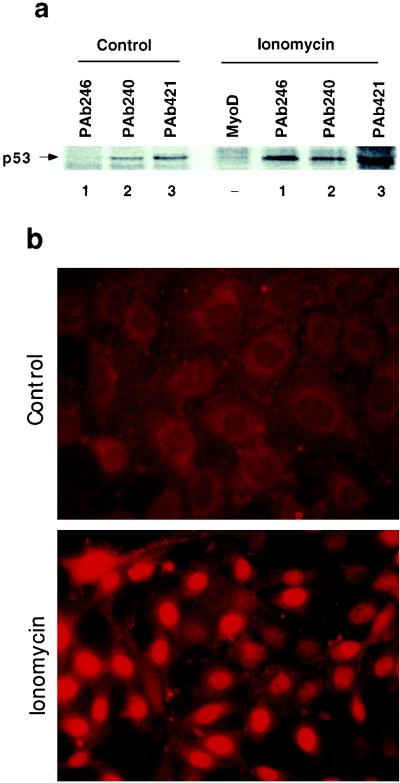
We next analyzed the conformational status and the subcellular localization of p53Val135 in S100B-MEF J-β2p53 cells before and after ionomycin stimulation (Fig. 4a). Cells were labeled with [35S]methionine, and 35S-labeled p53Val135 was immunoprecipitated from nuclear extracts with either wild-type-specific monoclonal antibody PAb246 (lanes 1), mutant-specific monoclonal antibody PAb240 (lanes 2), or the pan-specific PAb421 (lanes 3). In asynchronously growing cells, most of the nuclear p53Val135 is in a mutant conformation recognized by PAb240. Ionomycin stimulation produced a drastic increase in the amount of nuclear wild-type p53Val135 species recognized by the PAb246, resulting in a high wild-type/mutant ratio. These results were confirmed by indirect immunofluorescence analysis (Fig. 4b). In asynchronously growing clone J-β2p53 cells, wild-type p53Val135 immunoreactivity (PAb246) was weak and mostly cytoplasmic. Upon ionomycin stimulation, nuclear wild-type p53Val135 immunoreactivity drastically increased (Fig. 4b). In contrast, ionomycin stimulation produced only a limited increase in nuclear PAb240 immunoreactivity (not shown).
FIG. 4.
Ionomycin promotes nuclear accumulation of the wild-type p53Val135 species in clone J-β2p53 cells. (a) Clone J-β2p53 cells grown at 37.5°C were not stimulated (Control) or stimulated for 18 h with ionomycin (Ionomycin) prior to labeling with [35S]Met-Cys mix (100 μCi/ml) for 3 h. Nuclear extracts were prepared and 35S-labeled p53Val135 was immunoprecipitated with the wild-type-specific PAb246 (lanes 1), the mutant-specific PAb240 (lanes 2), or the pan-specific PAb421 (lanes 3). Anti-MyoD immunoglobulin G was used as a control. (b) Immunofluorescence analysis of PAb246 immunoreactivities in subconfluent clone J-β2p53 cells grown at 37.5°C not stimulated (Control) or stimulated with 1 μM ionomycin (Ionomycin) for 20 h.
Characterization of transfected clone 6 cells expressing S100B (S100B-REF).
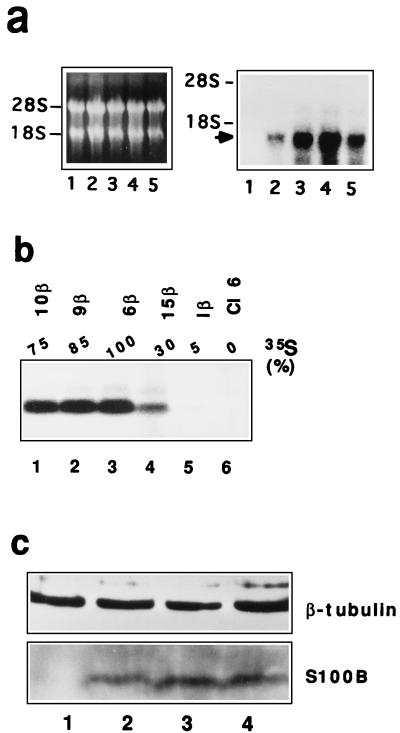
To confirm a role for S100B as a modulator of wild-type p53Val135 functions, we next analyzed the effect of ectopic S100B expression on the growth properties of a transformed REF cell line, clone 6 (36), that lacks endogenous S100B protein but overexpresses the ts p53Val135 mutant. Clone 6 cells were transfected with the pcDNA-Neo expression vector containing the S100B cDNA, followed by selection with G418. Four transfected clones resistant to G418, clones 6β, 9β, 10β, and 15β, that express S100B mRNA and protein were selected (Fig. 5a and b). The synthesis of S100B protein in clones positive for S100B mRNA was assessed by metabolic labeling of exponentially growing cells with [35S]Met-Cys and immunoprecipitation of labeled S100B (Fig. 5b). Quantification of the 35S incorporated into the S100B protein band with a phosphorimager indicated that maximal S100B production was in clone 6β (Fig. 5b). Clone 1β, used as a control, is a resistant clone that does not express a significant amount of S100B. Note that clone 6β cells express S100B at a level similar to those for S100B-MEF cells (Fig. 2a) and confluent glial C6 cells (Fig. 5c).
FIG. 5.
Expression of S100B in clone 6 cells. (a) Northern blot analysis for S100B mRNA in clone 6 cells (lane 1) and transfected clone 6β (lane 4), 9β (lane 3), 10β (lane 5), and 15β (lane 2) cells. In the left panel is ethidium bromide staining of electrophoresed RNAs; the right panel shows the autoradiograph of the hybridized membrane. Positions of the 18S and 28S RNAs are shown. The arrow points to S100B mRNA. (b) Immunoprecipitation analysis of 35S-labeled S100B in clone 6 cells (lane 6) and transfected clone 1β (lane 5), 15β (lane 4), 6β (lane 3), 9β (lane 2), and 10β (lane 1) cells. Cells were lysed in RIPA buffer. Extracts, corresponding to 1 mg of protein, were incubated with affinity-purified rabbit polyclonal anti-S100B. The relative percentage of 35S incorporated into the S100B protein as quantified with a phosphorimager is shown above each lane. (c) Western blot analysis of β-tubulin and S100B protein in total protein extracts (50 μg) from exponentially growing (lanes 1 and 3) and confluent (lanes 2 and 4) glial C6 cells (lanes 1 and 2) and clone 6β cells (lanes 3 and 4).
S100B expression rescues cell contact inhibition of growth in REF clone 6 cells.
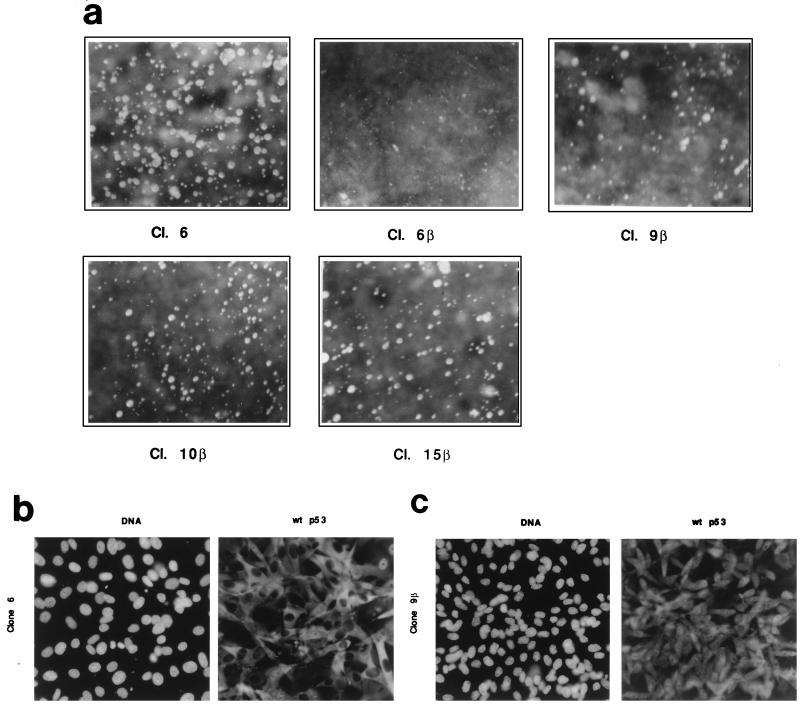
It has been shown previously that both wild-type and mutant conformational forms of p53Val135 are present at the nonpermissive temperature (37.5°C) in clone 6 cells and other clones of REF cells overexpressing p53Val135 (14, 18, 34). Wild-type p53Val135 is inactivated through cytoplasmic sequestration (18, 28, 34), and the mutant form cooperates with oncogenic ras in cell transformation, resulting in the loss of cell contact inhibition of growth (34, 36). In glial C6 cells, S100B synthesis is linked with cell contact inhibition of growth (Fig. 1). Thus, we first evaluated the effects of S100B expression on the ability of clone 6 cells to grow in soft agar (Fig. 6a), a property that characterizes cells which escape cell contact-dependent inhibition of growth. Two independent experiments carried out in duplicate showed that S100B inhibits the anchorage-independent growth that characterizes untransfected cells. The extent of inhibition varied: 100% with clone 6β, 75% ± 5% with clone 9β, 60% ± 5% with clone 10β, and 50% ± 5% with clone 15β. A net correlation exists between the amount of S100B expression and the inhibition of colony formation in soft agar. Consistent with the soft agar assay, at confluence S100B-REF cells accumulate in G1 and only confluent S100B-REF cells expressed the p21 cyclin-dependent kinase (CDK) inhibitor protein, a molecular marker of cell growth inhibition (data not shown). Moreover, cell contact inhibition of growth characterizing S100B-REF cells correlates with nuclear accumulation of the wild-type p53Val135 (Fig. 6b). As previously shown (18, 28), wild-type p53 immunoreactivity accumulates within the cytoplasm of most clone 6 cells at confluence. Confluent S100B-REF clone 9β cells showed both cytoplasmic and nuclear staining (Fig. 6b), as did S100B-REF clone 6β cells (not shown). Hence, S100B cooperates with a cell contact-signaling pathway to rescue wild-type p53Val135 nuclear translocation.
FIG. 6.
S100B expression promotes cell contact inhibition of growth of clone 6 cells. (a) S100B expression inhibits anchorage-independent growth of clone 6 cells. Clone (Cl.) 6, clone 6β, clone 9β, clone 10β and clone 15β cells were tested for anchorage-independent growth in soft agar. Colonies were photographed 15 days after plating. The results are representative of two different experiments carried out in duplicate. (b and c) p53Val135 is located in the nuclei of confluent clone 9β cells. Clone 6 (b) and clone 9β (c) cells were grown to confluence, and p53Val135 was immunostained with purified wild-type (wt)-specific monoclonal antibody PAb246 (right panels). Left panels show DNA staining with Hoechst. Magnification, ×36.
S100B expression rescues a calcium-dependent G1 checkpoint control in REF clone 6 cells.
The calcium ionophore ionomycin substitutes for the cell contact-signaling pathway to activate S100B-dependent wild-type p53Val135 nuclear translocation and G1 arrest in MEF cells grown at 37.5°C (Fig. 2 to 4). Similarly, ionomycin stimulation of S100B-REF cells produced an increase in cells positioned in the G1 phase of the cell cycle after 24 h, with few cells remaining in S and G2/M phases (Fig. 7a). Analysis of the cell cycle parameters of clone 9β by pulsing the cells for 30 min with BrdU demonstrated that ionomycin stimulation induced a total arrest in G1 (the G1/S-phase ratio increased by a factor of 10 (Fig. 7a, insets). Ionomycin only produced a general decrease in the rate of the cell cycle in parental REF cells (the ratio of G1 to S-phase populations increased by a factor of 3). We determined that the ionomycin-dependent growth arrest of S100B-REF is fully reversible (not shown). Ionomycin-mediated G1 arrest of S100B-REF cells at 37.5°C is phenotypically indistinguishable from p53Val135-mediated G1 arrest at the permissive temperature (32°C). Both arrests are characterized by induction of the p21 CDK inhibitor protein and mdm2, two proteins that are normally induced by wild-type p53Val135 in REF cells (5–16), and with dephosphorylation of Rb protein (Fig. 7b). Note that the induction of mdm2 by ionomycin is significantly greater than that produced upon temperature shift (Fig. 6c; compare lanes 7 and 8 with lanes 1 and 9). In contrast to p21, the p27 protein, another CDK inhibitor that is not regulated by p53, is not induced upon ionomycin stimulation or temperature shift to 32°C.
FIG. 7.
S100B cooperates with ionomycin to rescue a G1-phase growth arrest in REF clone 6 cells at the nonpermissive temperature (37.5°C). (a) Flow cytometry analysis of the DNA content of clone 6 and clone 9β cells stimulated with 1 μM ionomycin (−Iono) for 24 h as indicated. Insets show the cell cycle parameters of clone 6 and clone 9β cells unstimulated or stimulated for 20 h with ionomycin as determined by pulsing cells with BrdU. (b) Effects of temperature shift and ionomycin on cell cycle regulatory proteins in clone 6β cells. Immunoblots show cellular lysates corresponding to clone 6β cells growth arrested by shifting the temperature to 32°C for 24 h (lanes 1 and 9) or grown at 37.5°C (lane 2) and stimulated with ionomycin for 1 h (lane 3), 2 h (lane 4), 4 h (lane 5), 8 h (lane 6), 16 h (lane 7), and 24 h (lane 8) prior lysis in SDS sample buffer. Cl, clone.
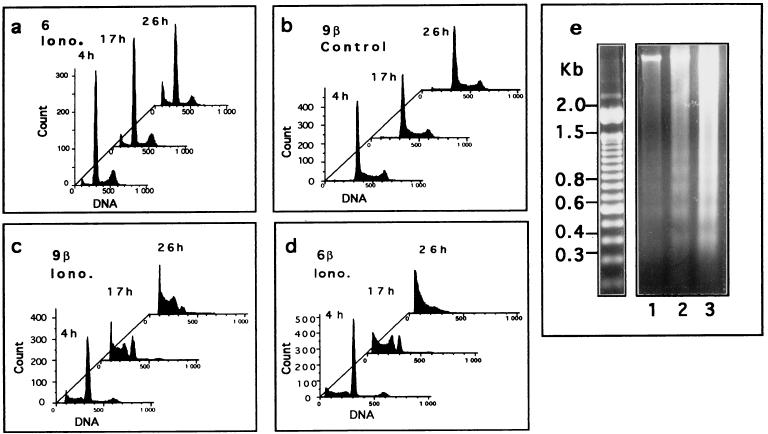
Calcium and S100B also modulate the sensitivity of REF cells to a low dose of UV irradiation (10 J/m2) (Fig. 8). Parental REF cells showed only moderate apoptosis upon UV irradiation, and apoptosis was not significantly increased if cells were pretreated with ionomycin (Fig. 8a). Exponentially growing S100B-REF clone 6β or 9β cells were also insensitive to UV irradiation (Fig. 8b) but showed full apoptotic response if they were first G1 arrested by stimulation with ionomycin for 20 h prior to UV irradiation (Fig. 8c to e). Internucleosomal DNA cleavage characterizing apoptotic cells was revealed by FACS analysis (Fig. 8a to d) and confirmed by agarose gel electrophoresis (Fig. 8e). Apoptosis was rapid and maximal 24 h postirradiation (Fig. 8c to e).
FIG. 8.
S100B cooperates with ionomycin to promote UV-dependent apoptosis of clone 6 cells at the nonpermissive temperature. (a) Clone 6 cells stimulated for 24 h with 1 μM ionomycin (Iono.) and UV irradiated. (b) Unstimulated clone 9β cells UV irradiated. (c and d) Clone 9β and 6β cells, respectively, stimulated for 24 h with 1 μM ionomycin and UV irradiated. Cells were collected after 4, 17, and 26 h and analyzed for DNA content by FACS analysis. (e) Agarose gel analysis of DNA fragmentation in apoptotic clone 9β cells 4 h (lane 1), 17 h (lane 2), and 26 h (lane 3) after UV irradiation. In all experiments, after irradiation (10 J/m2), cells were grown in medium without ionomycin.
DISCUSSION
S100B and calcium regulate p53-mediated cell growth inhibition.
A role for wild-type p53 in cell contact inhibition of growth of glial cells has been previously demonstrated (49). In the first part of this study, we showed that in glial C6 cells expressing a constitutive wild-type p53, a tight correlation exists between S100B synthesis, cell contact inhibition of growth, and apoptosis in response to UV irradiation. The peak of S100B synthesis in the G1 phase of the cell cycle is synchronized with proper timing for p53 nuclear translocation and activation. Modulation of p53 subcellular trafficking during a critical period in G1 is a potential mechanism by which cells constitutively expressing p53 regulate the growth suppressor and apoptotic functions of the p53 protein (40). Hence, we have hypothesized that in glial cells, S100B could synergize with the p53-dependent growth arrest and apoptosis pathways by favoring p53 nuclear translocation in the G1 phase of the cell cycle.
To test this hypothesis, we analyzed the effect of ectopic S100B expression on the growth properties of two fibroblast cell lines with different genetic backgrounds but expressing the ts p53Val135 mutant. We have shown that in p53−/− MEF cells expressing a low level of p53Val135, S100B cooperates with calcium in stimulating nuclear translocation and stabilization of the wild-type p53Val135 conformational species at the nonpermissive temperature. Nuclear accumulation of wild-type p53Val135 correlates with G1 cell growth arrest and cell death in response to UV irradiation.
In the REF cell line clone 6, which is transformed by oncogenic Ha-ras and the murine ts p53Val135 mutant, ectopic expression of S100B at levels comparable to that found in confluent glial cells restores cell contact inhibition of growth (Fig. 6a). Contact inhibition of growth correlates with nuclear accumulation of the wild-type p53Val135 conformational species (Fig. 6b). With this cell line, we have confirmed that the calcium ionophore substitutes for the cell contact-signaling pathway to cooperate with S100B in activation of p53-dependent cell growth arrest and apoptosis (Fig. 7 and 8).
Several hypotheses can be envisioned to explain the rescue of wild-type p53 nuclear translocation and functions by S100B in MEF and REF cells. The loss of growth suppressor function of the wild-type p53Val135 species at 37.5°C is partially due to its cytoplasmic sequestration through stable oligomer formation with the mutant species and/or interaction with cytoplasmic anchor proteins (14, 18, 28, 34). The effect is also due to a high mutant/wild-type conformation ratio, which probably acts in a negative dominant fashion even inside the nucleus and abolishes the biochemical functions of the wild-type p53 subpopulation. In vitro, S100B interacts in a calcium-dependent manner with p53 to protect p53 from thermal denaturation and aggregation (9). Hence, S100B could interfere with cytoplasmic hetero-oligomerization and inactivation of the p53Val135. By stabilizing the nascent p53Val135 protein in a wild-type conformation, S100B could subsequently favor nuclear accumulation of wild-type p53Val135. This hypothesis is consistent with the fact that in S100B-MEF cells, ionomycin stimulation produced a drastic increase in nuclear wild-type p53Val135 immunoreactivity (Fig. 4). Conformational modulation of p53 between wild-type and mutant-like conformations has recently emerged as a possible mechanism for regulation of p53 functions (41). It would be now worthwhile to test if calcium signaling and S100 proteins are more widely implicated in a conformational switch that regulates p53 activities. Alternatively, S100B could also indirectly regulate the interaction of wild-type p53 with cytoplasmic anchor proteins to favor its nuclear translocation. Once transported into the nucleus, the wild-type p53 is then stabilized. We are currently pursuing these directions of inquiry.
S100B cooperates with calcium to specifically rescue p53-dependent apoptosis.
In glial C6 cells, S100B synthesis is associated with apoptosis in response to UV irradiation (Fig. 1). In MEF and REF cells, S100B expression was also associated with the rescue of p53 apoptotic function. S100B-MEF and S100B-REF cells proceeding from G1 underwent apoptosis in response to UV irradiation at 37.5°C (Fig. 2c and 8). When shifted to 32°C, S100B-REF cells arrest primarily at the G1/S and G2/M transitions (36). However, in contrast to ionomycin-mediated growth inhibition at 37.5°C, permanently G1-arrested cells at 32°C show only limited apoptosis response to UV irradiation (data not shown). Thus, two p53-dependent growth arrest pathways in REF cells exist. One, at 37.5°C, is tightly regulated by S100B protein and calcium and is linked with cell apoptosis. The other, at 32°C, protects cells from apoptosis. A protective effect of p53Val135 at 32°C toward the cytotoxic effects of anticancer agents such as doxorubicin also has been reported (48). Growth arrest and apoptosis are now considered as two genetically separable functions of p53 that may act independently of each other (12, 22, 39). In some systems, p53-dependent cell cycle arrest inhibits apoptosis (12, 39). A central question that must be resolved in order to optimize anticancer therapies is how a cell decides whether to undergo a viable growth arrest and/or apoptosis when p53 is activated. Future experiments to understand the molecular mechanism by which calcium and S100B specifically restore p53 apoptotic function in MEF and REF cells at 37.5°C could shed new light on the mechanisms underlying activation of the p53-dependent apoptosis pathway and the role of calcium in regulating apoptosis in general (35).
Conclusions.
In conclusion, this study provides the first direct evidence for a function of the intracellular calcium-binding protein S100B in negative cell growth regulation. Hence, S100B could be classified as a growth suppressor, as has been previously suggested for S100A2 (also called S100L and CaN19) (31). It is noteworthy than more that 50% identity exists between S100A2 and S100B at the amino acid level and that most of the other amino acids are homologous, strongly suggesting similar structures and functions. The finding that the intracellular level of S100B could have a direct effect on the sensitivity of cells to UV-mediated apoptosis may provide a link between overexpression of S100B and neurodegenerative diseases such as DS, Alzheimer’s disease, and AIDS. Apoptosis plays a critical role during central nervous system development, but in the adult brain glial cell apoptosis is associated only with neurodegenerative diseases such as AIDS (23). Altered calcium homeostasis and oxidative stress both contribute to these neurodegenerative disorders (24, 45). Oxidative stresses represent internal sources of DNA damage similar to those induced by UV irradiation (10). We suggest that oxidative stress-mediated DNA damage and altered calcium homeostasis may activate the S100B apoptosis pathway contributing to the abnormal apoptotic death of glial cells in patients with AIDS (23) and probably in other neurodegenerative diseases where S100B is overexpressed. It is noteworthy that in the brains of patients with Alzheimer’s disease, p53 is also overproduced in glial cells and may be directly involved in glial cell apoptosis (26).
ACKNOWLEDGMENTS
We thank A. M. Chinn and R. Griffin for critical reading of the manuscript, R. Kuwano (Niigata University, Niigata, Japan) for providing human S100B cDNA, E. Gottlieb and M. Oren for providing recombinant p53Val135 retrovirus and clone 6 cells, Paul Andreassen for providing p53−/− MEF, H. Y. Peng for help in constructing the S100B plasmid, D. Grunwald for FACS analysis, and B. Justine and B. Clemence for stimulating discussions.
This work was supported by grants from Association pour la Recherche sur le Cancer and Ligue National Contre le Cancer to J.B.
REFERENCES
- 1.Allore R, O’Hanlon D, Price R, Neilson K, Willard H F, Cox D R, Marks A, Dunn R J. Gene encoding the β-subunit of S100 protein is on chromosome 21: implication for Down syndrome. Science. 1988;239:1311–1313. doi: 10.1126/science.2964086. [DOI] [PubMed] [Google Scholar]
- 2.Aloni-Grinstein R, Schwartz D, Rotter V. Accumulation of wild-type p53 protein upon γ-irradiation induces a G2 arrest-dependent immunoglobulin κ light chain gene expression. EMBO J. 1995;14:1392–1401. doi: 10.1002/j.1460-2075.1995.tb07125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Artuso M, Esteve A, Brésil H, Vuillaume M, Hall J. The role of the Ataxia telangiectasia gene in the p53, WAF1/CIP1 (p21) and GADD45-mediated response to DNA damage produced by ionising radiation. Oncogene. 1995;11:1427–1435. [PubMed] [Google Scholar]
- 4.Asai A, Miyagi Y, Sugiyama A, Gamanuma M, Hong S I, Takamoto S, Nomura K, Matsutani M, Takakura K, Kuchino Y. Negative effects of wild-type p53 and s-Myc on cellular growth and tumorigenicity of glioma cells. J Neuro-Oncol. 1994;19:259–268. doi: 10.1007/BF01053280. [DOI] [PubMed] [Google Scholar]
- 5.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baudier J, Glasser N, Gérard D. Ions binding to S100 protein. Calcium and zinc-binding properties of bovine brain S100αα, S100a (αβ), and S100B (ββ) protein: Zn2+ regulates Ca2+ binding on S100B protein. J Biol Chem. 1986;261:8192–8203. [PubMed] [Google Scholar]
- 7.Baudier J, Mochly-Rosen D, Newton A, Lee S H, Koshland D E, Cole R D. Comparison of S100b protein with calmodulin: interactions with melitin and microtubule-associated τ proteins and inhibition of phosphorylation of τ proteins by protein kinase C. Biochemistry. 1987;26:2886–2893. doi: 10.1021/bi00384a033. [DOI] [PubMed] [Google Scholar]
- 8.Baudier J, Bergeret E, Bertacchi N, Weintraub H, Gagnon J, Garin J. Interactions of myogenic bHLH transcription factors with calcium-binding calmodulin and S100a (αα) proteins. Biochemistry. 1995;34:7834–7846. doi: 10.1021/bi00024a007. [DOI] [PubMed] [Google Scholar]
- 9.Baudier J, Delphin C, Grunwald D, Khoshbin S, Lawrence J J. Characterization of the tumor suppressor protein p53 as a protein kinase C substrate and a S100B-binding protein. Proc Natl Acad Sci USA. 1992;89:11627–11631. doi: 10.1073/pnas.89.23.11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonfoco E, Krainc D, Ankarcrona M, Nicotera P, Lipton S A. Apoptosis and necrosis: two distinct events induced, respectively, by mild and intense insults with N-methyl-d-aspartate or nitric oxyde/superoxide in cortical cell cultures. Proc Natl Acad Sci USA. 1995;92:7162–7166. doi: 10.1073/pnas.92.16.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caelles C, Heimberg A, Karim M. p53 dependent apoptosis in the absence of transcriptional activation of p53-target genes. Nature. 1994;370:220–224. doi: 10.1038/370220a0. [DOI] [PubMed] [Google Scholar]
- 12.Chen X, Ko L J, Jayaraman L, Prive C. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 1996;10:2438–2451. doi: 10.1101/gad.10.19.2438. [DOI] [PubMed] [Google Scholar]
- 13.Coppolino M, Woodside M J, Demaurex N, Grinstein S, St-Arnaud R, Dedhar S. Calreticulin is essential for integrin-mediated calcium signalling and cell adhesion. Nature. 1997;286:843–847. doi: 10.1038/386843a0. [DOI] [PubMed] [Google Scholar]
- 14.Delphin C, Baudier J. The protein kinase C activator, phorbol ester, cooperates with the wild-type p53 species of ras-transformed embryo fibroblasts growth arrest. J Biol Chem. 1994;269:29579–29587. [PubMed] [Google Scholar]
- 15.Dolmetsch R E, Lewis R S, Goodnow C C, Healy J I. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature. 1997;286:855–858. doi: 10.1038/386855a0. [DOI] [PubMed] [Google Scholar]
- 16.El-Deiry W S, Tokino T, Velculenscu V E, Levy D B, Parson R, Trent J M, Lin D, Mercer E W, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumour suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 17.Fano G, Mariggio M A, Angelella P, Nicoletti I, Antonica A, Fulle S, Calissano P. The S100 protein causes an increase of intracellular calcium and death of PC12 cells. Neuroscience. 1993;53:919–925. doi: 10.1016/0306-4522(93)90477-w. [DOI] [PubMed] [Google Scholar]
- 18.Gannon J V, Lane D P. Protein synthesis required to anchor a mutant p53 protein which is temperature-sensitive for nuclear transport. Nature. 1991;349:802–805. doi: 10.1038/349802a0. [DOI] [PubMed] [Google Scholar]
- 19.Gottlieb E, Haffner R, Von Rüde T, Wagner E F, Oren M. Down-regulation of wild-type p53 activity interferes with apoptosis of Il-3-dependent hematopoietic cells following Il-3 withdrawal. EMBO J. 1994;13:1368–1374. doi: 10.1002/j.1460-2075.1994.tb06390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffin W S T, Stanley L C, Ling C, White L, MacLeod V, Perrot L J, White C L, Araoz C. Brain interleukin 1 and S100 immunoreactivity are elevated in Down syndrome and Alzheimer disease. Proc Natl Acad Sci USA. 1989;86:7611–7615. doi: 10.1073/pnas.86.19.7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haglid K, Hamberger A, Hansson H, Hyden H, Persson L, Ronnbach L. Cellular and subcellular distribution of the S100 protein in rabbit and rat central nervous system. J Neurosci Res. 1976;2:175–192. doi: 10.1002/jnr.490020302. [DOI] [PubMed] [Google Scholar]
- 22.Haupt Y, Rowan S, Shaulian E, Vousden K H, Oren M. Induction of apoptosis in HeLa cells by trans-activation-deficient p53. Genes Dev. 1995;9:2170–2183. doi: 10.1101/gad.9.17.2170. [DOI] [PubMed] [Google Scholar]
- 23.He J, De Castro C M, Vandenbark G R, Busciglio J, Gabuzda D. Astrocyte apoptosis induced by HIV-1 transactivation of the c-kit protooncogene. Proc Natl Acad Sci USA. 1997;94:3954–3959. doi: 10.1073/pnas.94.8.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heizmann C W, Braun K. Changes in Ca2+-binding proteins in human neurodegenerative disorders. Trends Neurol Sci. 1992;15:259–264. doi: 10.1016/0166-2236(92)90067-i. [DOI] [PubMed] [Google Scholar]
- 25.Herschman H R, Levine L, De Vellis J. Appearance of a brain-specific antigen (S100-protein) in the developing rat brain. J Neurochem. 1971;18:629–633. doi: 10.1111/j.1471-4159.1971.tb11993.x. [DOI] [PubMed] [Google Scholar]
- 26.Kitamura Y, Shimohama S, Kamoshima W, Matsuoka Y, Nomura Y, Taniguchi T. Changes of p53 in the brains of patients with Alzheimer’s disease. Biochem Biophys Res Commun. 1997;232:418–421. doi: 10.1006/bbrc.1997.6301. [DOI] [PubMed] [Google Scholar]
- 27.Kligman D, Marshak D R. Purification and characterization of a neurite extension factor from bovine brain. Proc Natl Acad Sci USA. 1985;82:7136–7139. doi: 10.1073/pnas.82.20.7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knippschild U, Oren M, Deppert W. Abrogation of wild-type p53 mediated growth-inhibition by nuclear exclusion. Oncogene. 1996;12:1755–1765. [PubMed] [Google Scholar]
- 29.Kolber A R, Permual A S, Goldstein M N, Moore B W. Drug-induced differentiation of a rat glioma in vitro: the expression of S100, a glial specific protein and steroid sulfatase. Brain Res. 1978;143:513–520. doi: 10.1016/0006-8993(78)90361-x. [DOI] [PubMed] [Google Scholar]
- 30.Labourdette G, Marks A. Synthesis of S100 protein in monolayer cultures of rat-glial cells. Eur J Biochem. 1975;58:73–79. doi: 10.1111/j.1432-1033.1975.tb02350.x. [DOI] [PubMed] [Google Scholar]
- 31.Lee S W, Tomasetto C, Swisshelm K, Keyomarsi K, Sager R. Down-regulation of a member of the S100 gene family in mammary carcinoma cells and reexpression by azadeoxycytidine treatment. Proc Natl Acad Sci USA. 1992;89:2504–2508. doi: 10.1073/pnas.89.6.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marks A, Petsche D, O’Hanlon D, Kwong P C, Stead R, Dunn R, Baumal R, Liao S K. S100 protein expression in human melanoma cells: comparison of levels of expression among different cell lines and individual cells in different phases of the cell cycle. Exp Cell Res. 1990;187:59–64. doi: 10.1016/0014-4827(90)90116-r. [DOI] [PubMed] [Google Scholar]
- 33.Marshak D R, Pesce S A, Stanley L C, Griffin W S T. Increased S100β neurotrophic activity in Alzheimer’s disease temporal lobe. Neurobiol Aging. 1991;13:1–7. doi: 10.1016/0197-4580(92)90002-f. [DOI] [PubMed] [Google Scholar]
- 34.Martinez J, Georgoff I, Martinez J, Levine A J. Cellular localization and cell cycle regulation by a temperature-sensitive p53 protein. Genes Dev. 1991;5:151–159. doi: 10.1101/gad.5.2.151. [DOI] [PubMed] [Google Scholar]
- 35.McConkey D J, Orrenius S. Signal transduction pathways to apoptosis. Trends Cell Biol. 1994;4:370–375. doi: 10.1016/0962-8924(94)90087-6. [DOI] [PubMed] [Google Scholar]
- 36.Michalovitz D, Halevy O, Oren M. Conditional inhibition of transformation and of cell proliferation by a temperature-sensitive mutant of p53. Cell. 1990;62:671–680. doi: 10.1016/0092-8674(90)90113-s. [DOI] [PubMed] [Google Scholar]
- 37.Milner J. Flexibility: the key to p53 function? Trends Biochem Sci. 1995;20:49–51. doi: 10.1016/s0968-0004(00)88954-9. [DOI] [PubMed] [Google Scholar]
- 38.Morii K, Tanaka R, Takahashi Y, Kuwano R. Cloning of cDNAs encoding human S-100α and β subunits and their differential expression in human tumor cell lines. J Neurosci Res. 1992;32:27–33. doi: 10.1002/jnr.490320104. [DOI] [PubMed] [Google Scholar]
- 39.Polyak K, Waldman T, He T C, Kinzler K, Vogelstein B. Genetic determinants of p53-induced apoptosis and growth arrest. Genes Dev. 1996;10:1945–1952. doi: 10.1101/gad.10.15.1945. [DOI] [PubMed] [Google Scholar]
- 40.Ryan J J, Prochownik E, Gottlieb C A, Apel I J, Merino R, Nunez G, Clarke M. c-myc and bcl-2 modulate p53 function by altering p53 subcellular trafficking during the cell cycle. Proc Natl Acad Sci USA. 1994;91:5878–5882. doi: 10.1073/pnas.91.13.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabapathy K, Klemm M, Jaenisch R, Wagner E F. Regulation of ES cell differentiation by functional and conformational modulation of p53. EMBO J. 1997;16:6217–6229. doi: 10.1093/emboj/16.20.6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schäfer B W, Heizmann C. The S100 family of EF-hand calcium-binding proteins: functions and pathology. Trends Biochem Sci. 1996;21:134–140. doi: 10.1016/s0968-0004(96)80167-8. [DOI] [PubMed] [Google Scholar]
- 43.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 44.Selinfreund R H, Barger S W, Pledger W J, Van Eldik L. Neurotrophic protein S100b stimulates glial proliferation. Proc Natl Acad Sci USA. 1991;88:3554–3558. doi: 10.1073/pnas.88.9.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sendtner M, Thoenen H. Neurodegenerative disease. Oxidative stress and motoneuron disease. Curr Biol. 1994;4:1036–1039. doi: 10.1016/s0960-9822(00)00237-2. [DOI] [PubMed] [Google Scholar]
- 46.Sheng J G, Mrak R E, Griffin W S T. S100b protein expression in Alzheimer’s disease: potential role in the pathogenesis of neuritic plaques. J Neurosci Res. 1994;39:398–404. doi: 10.1002/jnr.490390406. [DOI] [PubMed] [Google Scholar]
- 47.Stanley L C, Mrak R E, Woody R C, Perrot L J, Zhang S, Marshak D R, Nelson S J, Griffin W S. Glial cytokines as neuropathogenic factors in HIV infection: pathogenic similarities to Alzheimer’s disease. J Neuropathol Exp Neurol. 1994;53:231–238. doi: 10.1097/00005072-199405000-00003. [DOI] [PubMed] [Google Scholar]
- 48.Vikhanskaya F, Erba E, D’Incalci M, Broggini M. Changes in cyclins and cyclin-dependent kinases induced by DNA damaging agents in a human ovarian cancer cell line expressing mutated or wild-type p53. Exp Cell Res. 1996;227:380–385. doi: 10.1006/excr.1996.0288. [DOI] [PubMed] [Google Scholar]
- 49.Yahanda A M, Bruner J M, Donehower L A, Morrison R S. Astrocytes derived from p53-deficient mice provide a multistep in vitro model for development of malignant gliomas. Mol Cell Biol. 1995;15:4249–4259. doi: 10.1128/mcb.15.8.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yonish-Rouach E, Resnitzky D, Lotem J, Sachs L, Kimchi A, Oren M. Wild-type p53 induces apoptosis of myeloid leukaemic cells that is inhibited by interleukin-6. Nature. 1991;352:345–347. doi: 10.1038/352345a0. [DOI] [PubMed] [Google Scholar]