Abstract
Numerous efforts and research have been conducted worldwide to combat the coronavirus disease 2019 (COVID-19) pandemic. In this regard, some researchers have focused on deep and machine-learning approaches to discover more about this disease. There have been many articles on using ensemble learning methods for COVID-19 detection. Still, there seems to be no scientometric analysis or a brief review of these researches. Hence, a combined method of scientometric analysis and brief review was used to study the published articles that employed an ensemble learning approach to detect COVID-19. This research used both methods to overcome their limitations, leading to enhanced and reliable outcomes. The related articles were retrieved from the Scopus database. Then a two-step procedure was employed. A concise review of the collected articles was conducted. Then they underwent scientometric and bibliometric analyses. The findings revealed that convolutional neural network (CNN) is the mostly employed algorithm, while support vector machine (SVM), random forest, Resnet, DenseNet, and visual geometry group (VGG) were also frequently used. Additionally, China has had a significant presence in the numerous top-ranking categories of this field of research. Both study phases yielded valuable results and rankings.
Keywords: Ensemble learning, Ensemble method, Deep learning, Convolutional neural network, COVID-19
1. Introduction
Coronavirus disease 2019 (COVID-19) has been a great challenge for the modern human society. Using smart methods to fight this disease is remarkable. Machine learning approaches have been widely explored to aid in COVID-19 detection, leveraging patterns and features in various types of data, including medical imaging, and clinical and molecular data. So far, there has been a lot of work to detect COVID-19 using machine learning methods, and some review articles or surveys have been published focusing on using machine learning for detecting COVID-19 [[1], [2], [3], [4], [5], [6], [7]]. Also, there are some scientometric studies in this field [[8], [9], [10], [11], [12]].
Detecting COVID-19 has several challenges for the current machine learning approaches, which include limited, imbalanced, noisy, and incomplete data, complex and evolving patterns, overfitting and generalization. In addition, the current machine learning approaches have insufficiencies such as low accuracy, high false-positive rates, and high inter-reader variability [13,14]. Ensemble learning is a powerful approach that combines multiple machine learning models to make predictions or classifications. It has gained significant popularity and has been successfully applied to various domains, including medical diagnosis such as COVID-19 detection. Ensemble learning methods seem to be more effective than the other machine learning methods in detecting COVID-19 [9].
Ensemble methods can leverage the strengths of individual models while mitigating their weaknesses by combining multiple models, each trained on different subsets of data or using different algorithms. This leads to improved accuracy, robustness, and generalization performance [13,15]. Ensemble methods also provide mechanisms for uncertainty estimation, which is crucial in their medical applications such as detecting COVID-19, where the consequences of false positives or false negatives can be severe. These advantages make ensemble methods a valuable tool in the fight against the pandemic, enabling more reliable and effective diagnosis of COVID-19 cases.
Until now, many articles have investigated the application of ensemble learning for detecting COVID-19; however, it seems that none of them have undergone a scientometric analysis and have not been included in a brief review. Hence, we did this research with a combined method, i.e., scientometric analysis and brief review, thereby overcoming the constraints of each method, resulting in improved and more reliable outcomes.
This paper has two sections based on the proposed combined method. The first section presents a brief review of the related published works, summarizing their research methods, datasets, and best performances. The second section uses scientometric methods to provide further insights into the authors and research networks, keyword co-occurrence rates, plus top rated publications, authors, countries, and funding organizations. By examining both the summary of previous works and scientometric analyses, we can gain a deeper understanding of this field.
The research questions were:
-
⬥
What useful information can be extracted from reviewing and analyzing the articles that used the ensemble learning method to learn more about COVID-19?
-
⬥
Are certain algorithms more widely used in ensemble learning for detecting COVID-19 than others? Which algorithms are more common?
-
⬥
Who are the most influential authors, and what are the leading countries, universities, and major funding entities in this field?
-
⬥
Do the results obtained from the scientometric method for determining the most widely used algorithms correspond with those obtained from the brief review?
This research can provide these contributions:
-
⬥
It can provide valuable information and reveal interesting patterns among the studies carried out in this research area.
-
⬥
Usage of co-word analysis aids researchers in quickly grasping the most common methods for COVID-19 analysis using ensemble learning, corresponding datasets, and the highest achieved efficiency.
-
⬥
The scientometric analysis uncovers the leading influential authors and countries, key sponsors, and highlights the top effective publishers and journals that have publications in this area of research.
-
⬥
Usage of co-word analysis on abstracts and review section results, the hybrid approach can provide significant insight into the techniques and algorithms employed in this field.
The rest of this text is structured as follows: a) section 2 explains the research method, b) section 3 presents a brief overview of the related works, c) section 4 gives a scientometric-bibliometric analysis of the extracted metadata, d) section 5 presents a discussion, e) section 6 includes conclusion and f) section 7 has suggestions for future research.
2. Research method
This study aimed to provide insight into the current published research that employed ensemble learning approaches for detecting COVID-19. This research had two phases wherein the related works were briefly reviewed and compared, followed by a scientometric and bibliometric analysis.
It is widely recognized that Scopus is one of the most comprehensive databases available [16]. Compared to other databases, Scopus seems to provide the widest coverage of documents [17,18]. This database contains widest the coverage of documents, peer-reviewed journals and most recent publications [[19], [20], [21], [22]].
While Scopus is a valuable source for conducting literature reviews and obtaining research insights, it is important to consider its limitations and potential biases, particularly when it comes to the coverage and quality of articles in specific domains such as COVID-19 research. Some of the limitations and potential biases of the Scopus database include incomplete coverage, time lag, language bias, quality and peer review variations, publication bias, and access restriction [23,24]. While all databases have their limitations, Scopus boasts the most extensive coverage among them. As such, it has been selected as the preferred database for this research.
To conduct our scientometric and review study, we ran the following queries on keywords and abstracts in the Scopus database: (“ensemble learning” + COVID-19) OR (“ensemble deep learning” + COVID-19) OR (“ensemble model” + COVID-19).
As mentioned, in addition to using it on the keywords, this query was also applied to the abstracts to obtain a greater degree of coverage. Since the validity of the results of papers published in indexed journals is generally higher than those published in conference papers, the above query was applied only to journal papers to have a greater level of confidence. A total of 142 articles matching these characteristics were retrieved on August 1, 2022.
3. Comparing related works
Based on an initial scientometric review in the Scopus database, the peer-reviewed journals with the greatest number of articles published in this research field were identified. Then, journals that had published more than five articles were selected amongst them. Next, the articles that used an ensemble learning method to detect COVID-19 were selected. The best performance, base method, and dataset were extracted from each article. Since presenting all the information in one table would make it hard to read, they were presented in several tables.
Throughout the tables, there was a brief description of the type of ensemble learning method used. In the best performance column, the highest value obtained from a performance criterion was presented. In most of the articles, accuracy was the criterion of performance, but in a few articles, it was also the area under the curve (AOC) or error rate. The dataset field contained a summary of the datasets used in each article. Table 1 shows the related works published by Springer Science and Business Media and Springer. Among the various applied ensemble methods, ResNet, DenseNet, MobileNet, visual geometry group (VGG), and convolutional neural network (CNN) stood out.
Table 1.
Related published research in Springer.
| # | Title | Best Performance | Ensemble Method | Dataset |
|---|---|---|---|---|
| 1 | Analysis of origin, risk factors influencing COVID-19a cases in India and its prediction using ensemble learning [25] | The highest accuracy: 84.37% | Naive Bayes, Decision tree, SVMb, KNNc, Neural Network | Kaggle, including 5000 samples from the first and another 5000 samples from the second peaks of COVID-19 from India |
| 2 | Automatic COVID-19 detection from X-ray images using ensemble learning with convolutional neural network [26] | The highest accuracy: 91.62% | CNNd - DenseNet201 Resnet50V2 Inceptionv3 | Two open sources, 1004 chest X-ray images of patients in Europe |
| 3 | COVID_SCREENET: COVID-19 Screening in Chest Radiography Images Using Deep Transfer Stacking [27] | The highest accuracy: 100% | model by transfer stacking approach | 7725 chest X-ray images from three hospitals in India. Sources: Kaggle, Mendeley, and sirm.com |
| 4 | Design ensemble deep learning model for pneumonia disease classification [28] | The highest accuracy: 95.05% F-measure: 94.84% |
InceptionResNet_V2 ResNet50 MobileNet_V2 |
Two datasets from the University of San Diego, California: a CTe scan dataset with 5856 images and a COVID-19 chest X-ray dataset with 231 images |
| 5 | Novel deep transfer learning model for COVID-19 patient detection using X-ray chest images [29] | The highest accuracy for multiclass: 99.21% for two-class: 98.95% | EfficientNet GoogLeNet XceptionNet | 2869 chest X-ray images from Kaggle, Mporas, and Naronglerdrit |
| 6 | COVIDScreen: explainable deep learning framework for differential diagnosis of COVID-19 using chest X-rays [30] | The highest accuracy: 98.67% F1: 100%, 98%, and 98% for COVID-19, normal, and pneumonia, respectively. |
VGGf-19, VGG-16 ResNet-50 DenseNet161 DenseNet-169 |
Normal and pneumonia samples extracted from the open-source NIHg chest X-ray dataset used in the RSNAh pneumonia detection challenge on Kaggle |
| 7 | Stacking Deep Learning for Early COVID-19 Vision Diagnosis [31] |
The highest accuracy: 98.6% | MobileNet, InceptionResNetV2 ResNet50 |
500 chest X-ray images (unknown source) |
| 8 | Real-time internet of medical things framework for early detection of Covid-19 [32] | The highest accuracy: 95.3% | Random Forest (RF), Gradient Boosted Tree (GBT) | 278,848 records in two categories of COVID-19 patients and healthy individuals in Israel |
| 9 | Inverted bell-curve-based ensemble of deep learning models for detection of COVID-19 from chest X-rays [33] | The highest accuracy: 99.54% | DenseNet-161, ResNet-8, VGG-16 | Two public datasets: 1) the COVID-19 radiography dataset and 2) the chest X-ray images from IEEE |
| 10 | Decision and feature level fusion of deep features extracted from public COVID-19 datasets [34] | The highest accuracy: 90.84% | MobileNetV2, VGG16, ResNet50 and ResNet101, NasNet, InceptionV3, Xception | A database called DB1 containing 125 chest X-ray images of COVID-19 cases and 1000 images of non-COVID-19 cases. 353 new COVID-19 scans were added to the DB1 database (named as DB2 database). DB3 database contained 113 COVID-19 scans in addition to DB2 database |
| 11 | Adaptive UNet-based Lung Segmentation and Ensemble Learning with CNN-based Deep Features for Automated COVID-19 Diagnosis [35] | The highest accuracy: 97.09% | SVM, Naïve Bayes Autoencoder |
Chest X-ray images dataset including 481 and 183 records from the IEEE8023 and GitHub repositories |
| 12 | A multichannel EfficientNet deep learning-based stacking ensemble approach for lung disease detection using chest X-ray images [36] | The highest accuracy: 98% | Random forest, SVM, logistic regression | Mendeley-data-V3 dataset, including 4676 records of healthy individuals and 2004 records of people infected with COVID-19 |
| 13 |
A deep learning algorithm using CT images to screen for Coronavirus disease (COVID-19) [37] |
The highest accuracy: 89.5% (internal validation) 79.3% (external validation) |
Using transfer learning (unknown models) | 1065 CT scan images collected from hospitals in China |
| 14 | Automated detection of COVID-19 using ensemble of transfer learning with deep convolutional neural network based on CT scans [38] | The highest accuracy: 85.2% | EfficientNetB0 EfficientNetB3 EfficientNetB5 nception_resnet_v2 Xception | 349 CT scan images of COVID-19 positive cases and 397 COVID-19 negative cases from Tongji Hospital, Wuhan, China |
| 15 | Densely connected convolutional networks-based COVID-19 screening model [39] | The highest accuracy: 98.83% | Densely connected convolutional networks, ResNet152V2, VGG16 | Chest X-ray images including 2373 COVID-19 cases in Wuhan, 2890 cases of pneumonia, 3193 cases of pneumonia from North America, and 3038 cases of healthy individuals |
| 16 | Deep-LSTM ensemble framework to forecast Covid-19: an insight to the global pandemic [40] | The highest accuracy: 97.59% | Convolutional LSTMi, bi-directional LSTM | Unclear dataset from India |
| 17 | Internet of Medical Things-Based COVID-19 Detection in CT Images Fused with Fuzzy Ensemble and Transfer Learning Models [41] | The highest accuracy: 99.15% | MobileNetV2, Sugeno fuzzy integral | CT scan images of 650 cases of pneumonia, COVID-19, and healthy individuals from the National Center for Biological Information, China |
| 18 | Automatic detection of COVID-19 from chest CT scan and chest X-Rays images using deep learning, transfer learning and stacking [42] | The highest accuracy: 99.75% | DenseNet169, VGG19 | Five datasets: a) 746 COVID-19 CT scan images from Github b) 579 chest X-ray images from Github c) 12058 CT scan images from Github d) 2541 chest X-ray images from Kaggle e) 2482 CT scan images from Kaggle |
| 19 | Classifying chest CT images as COVID-19 positive/negative using a convolutional neural network ensemble model and uniform experimental design method [43] | Highest accuracy: 96.7% | A CNN ensemble model including VGG-19, ResNet-101 and DenseNet201 models, and Inception-v3 and Inception-ResNet-v2 | 612 CT scan images, 309 COVID-19 and 303 healthy cases |
Coronavirus Disease 2019.
Support Vector Machine.
K-Nearest Neighbors.
Convolutional Neural Network.
Computed Tomography.
Visual Geometry Group.
National Institute of Health.
Radiological Society of North America.
Long Short-Term Memory.
Ensemble learning approaches have advantages over individual machine learning algorithms in detecting COVID-19 [44]. They can improve accuracy, handle uncertainty, enhance robustness, and capture diverse patterns. Individual algorithms, on the other hand, maybe computationally efficient, easier to interpret, and provide a clearer understanding of the underlying patterns in certain cases. However, they may lack the ability to capture complex relationships or generalize well to new data, especially in the presence of limited or noisy data.
Table 2 shows the related works published by Elsevier. ResNet, DenseNet, VGG, CNN, and support vector machine (SVM) have been at the forefront of the various machine learning methods used.
Table 2.
Related published research in Elsevier.
| # | Title | Best Performance | Ensemble Method | Dataset |
|---|---|---|---|---|
| 1 | CoVNet-19: A Deep Learning model for the detection and analysis of COVID-19 patients [45] |
Highest accuracy: For three-classification 98.2% Binary classification 99.71% |
Two-stage stack ensemble model, which uses VGG19 and DenseNet121 in the first stage, and classification by SVM in the second stage. | Five datasets: a) COVID-19 radiography database from Kaggle b) Chest X-ray images (pneumonia) from Kaggle c) From the GitHub repository: COVID-chestxray-dataset d) From GitHub, the COVID-19 chest X-ray dataset e) COVID-19 X-ray dataset from Kaggle |
| 2 | Hybrid ensemble model for differential diagnosis between COVID-19 and common viral pneumonia by chest X-ray radiograph [46] | Highest accuracy: 98.64% |
Three-stage dual ensemble model: AlexNet for feature extraction, Relief algorithm for feature selection, and SVM for final classification |
1743 CXRa images. The normal CXRs and viral pneumonia CXRs were obtained from the NIH Chest X-ray database, and the Covid-19 CXRs were collected from GitHub |
| 3 | The ensemble deep learning model for novel COVID-19 on CT images [47] | Highest accuracy: 99.5% |
The model is called EDL-Covid consists of three models AlexNet, GoogleNet, ResNet |
2933 chest X-ray images from public databases |
| 4 | Ensemble learning model for diagnosing COVID-19 from routine blood tests [48] | Highest accuracy: 99.8% |
two-stage stack model: extra trees, RF, and LR were used in the first stage, and the XGBoost class was used in the second stage | 56,444 records (blood tests) obtained from the Albert Einstein Hospital in Brazil, including 559 positive results for COVID-19 |
| 5 | TSRNet: Diagnosis of COVID-19 based on self-supervised learning and hybrid ensemble model [49] | Highest accuracy: 99.8% |
A hybrid ensemble model (TSRNet), new pre-training method based on transfer learning with self-supervised learning, a new CNN based on attention mechanism and deep residual network (RANet) for feature extraction | Four datasets: a) ImageNet b) COVID-19 dataset (including 1252 COVID-19 and 1229 lung CT scan images from healthy individuals) c) Use of lung nodule analysis (LUNA) as a source of unlabeled CT scan images, including 1000 images d) Transfer learning to evaluate the effect of the difference between the source and target domains |
| 6 | Complex features extraction with deep learning model for the detection of COVID19 from CT scan images using ensemble based machine learning approach [50] | Highest accuracy: 99.37% |
CLAHEb in pre-processing step to increase the quality of the images, a new CNN: Gaussian Naïve Bayes (GNB), SVM, DTc, LRd and RFe |
Collection of CT scan data for SARS-CoV-2f from Kaggle, original dataset is CT scan from São Paulo Hospitals |
| 7 | A multi model ensemble based deep convolution neural network structure for detection of COVID19 [51] | Highest accuracy: 88.98% |
deep CNN, namely VGGNet, GoogleNet, DenseNet, and NASNet | Kaggle's chest X-ray images from Indore Hospital |
| 8 | An efficient hardware architecture based on an ensemble of deep learning models for COVID -19 prediction [52] | Highest accuracy: 98% |
Five deep learning models namely ResNet, Fitness, IRCNNg and Primary Recurrent Convolutional Neural Network | A database of chest X-ray images for COVID-19 positive cases along with images of normal and viral pneumonia |
Chest X-ray.
Contrast Limited Histogram Equalization.
Decision Tree.
Logistic Regression.
Random Forest.
Severe Acute Respiratory Syndrome Coronavirus 2.
Inception-Recurrent Convolutional Neural Network.
Table 3 shows the related works published by Nature. It seems that CNN remains at the forefront of the various ensemble learning approaches.
Table 3.
Related published research in Nature.
| # | Title | Best Performance | Ensemble Method | Dataset |
|---|---|---|---|---|
| 1 | Spatio-temporal prediction of the COVID-19 pandemic in US counties: modeling with a deep LSTM neural network [53] | Root mean square error (RMSE) improvement | LSTM | COVID-19 cases, deaths, and foot traffic at the county level in 33 weeks obtained from the Center for Systems Science and Engineering at Johns Hopkins University and SafeGraph's Places Schema dataset |
| 2 | Fuzzy rank-based fusion of CNN models using Gompertz function for screening COVID-19 CT-scans [54] | Highest accuracy: 99.2% | Three transfer learning-based CNN models were used, namely VGG-11, Wide ResNet-50-2, and Inception v3 | SARS-COV-2 dataset, Harvard Dataverse chest CT scan dataset |
| 3 | Machine learning based early warning system enables accurate mortality risk prediction for COVID-19 [55] | Area under curve of 96.21% | Four techniques: Logistic Regression, SVM, Gradient Boosted Decision Tree, and Neural Network | 2520 COVID-19 patients from two hospitals of Tongji Medical College, Huazhong University of Science and Technology, China |
| 4 | EpistoNet: an ensemble of Epistocracy-optimized mixture of experts for detecting COVID-19 on chest X-ray images [56] | Highest accuracy: 95% |
A decision tree-based ensemble model consisting of two distinct expert combinations called EpistoNet, from the Epistocrac algorithm. Several deep CNNs in each cluster |
2500 X-ray images including 1250 COVID-19 cases and 1250 non-COVID-19 cases |
| 5 | An ensemble learning approach to digital corona virus preliminary screening from cough sounds [57] | curve = 0.77, precision = 0.80, recall = 0.71, F1 measure = 0.75, Kappa = 0.53 |
Cough sound samples by splitting/separating the cough sound. CNN for modelling. Shallow machine learning, CNN, and pre-trained CNN models | Crowdsourced respiratory sounds collected to detect COVID-19. The authors used breathing and cough to distinguish COVID-19 sounds from asthma patients or healthy individuals |
Table 4 shows the related works published by IEEE. Among the employed ensemble learning approaches, ResNet, DenseNet, VGG, CNN, and SVM were more common.
Table 4.
Related published research in IEEE.
| # | Title | Best Performance | Ensemble Method | Dataset |
|---|---|---|---|---|
| 1 | EDL-COVID: Ensemble deep learning for COVID-19 case detection from chest X-ray images [58] | Highest accuracy: 96.4% | Deep CNN | The latest COVIDx dataset by Wang et al. containing 15,477 CXR images from 13,870 cases, including 6053 pneumonia cases, 8851 normal cases, and 573 COVID-19 cases. The COVIDx datasets were extracted from GitHub and Kaggle |
| 2 | Ensemble learning‐based COVID‐19 detection by feature boosting in chest X‐ray images [59] | Highest accuracy: 99.8% |
VGG-16 (base) + logistic regression (meta) | 5863 CXR images of normal and pneumonia cases obtained from Kaggle |
| 3 | Iteratively pruned deep learning ensembles for COVID-19 detection in chest X-rays [60] | Highest accuracy: 97% |
A custom CNN and a set of pre-trained ImageNet patient-level models were trained and evaluated. Knowledge learned was transferred and adjusted to improve performance. The following models were used: VGG-16 VGG-19, Inception-V3 |
Four datasets: a) The data collected from Guangzhou Women and Children's Medical Center in Guangzhou, China, the anteriorposterior CXRs of children from one to five years old, showing normal lungs, bacterial pneumonia, and non-COVID-19 viral pneumonia b) normal CXRs and abnormal images with non-pneumonia and pneumonia-like opacities from the National Institute of Health CXR-14 dataset c) Twitter COVID-19 CXR dataset d) MONTREAL COVID-19 CXR dataset from GitHub |
| 4 | Iteratively pruned deep learning ensembles for COVID-19 detection in chest X-rays [61] | Highest accuracy: 98.33% for binary 92.36% for multi-classes |
Several classifications such as decision tree, KNN, SVM, autoencoder, Boltzmann machine, CNN, Inceptionv3, DenseNet121, Xception, Inception, ResNetv2 | Chest X-ray image. COVID-19 and non-COVID-19 datasets obtained from different sources plus the pneumonia dataset from Kaggle. This dataset consists of 10,000 CXR images, of which 2022 were for pneumonia, 2161 for COVID-19, and 5863 were for non-COVID-19 |
| 5 | Choquet integral and coalition game-based ensemble of deep learning models for covid-19 screening from chest x-ray images [62] | highest accuracy: 97% |
A hybrid model based on lambda fuzzy from DCNN, VGG16, Xception, InceptionV3 architectures, using Choquet Integral | A new dataset of CXR images by combining three publicly available datasets from Kaggle and GitHub |
| 6 | Covid‐19 detection from radiographs by feature‐reinforced ensemble learning [63] | Highest accuracy: 98.47% |
Combination of SVM, linear discriminant analysis, KNN new bayes, decision tree, ResCNN, local binary patterns, histogram of oriented gradients, majority voting | 5228 chest X-ray images extracted from Kaggle, a subsidiary of Google LLC. The images were categorized into three categories: natural, pneumonia, and COVID-19 |
| 7 | COVID-19 detection using integration of deep learning classifiers and contrast-enhanced canny edge detected X-ray images [64] | Highest accuracy: 97.9% |
VGG16, InceptionV3 | 588 cases of positive COVID-19 and positive pneumonia obtained from GitHub, Radiopedia, and SIRM |
Table 5 shows the related works published by open-access publications such as MDPI, NLM, JAMIR, and Hindawi. Among the various ensemble methods, ResNet, DenseNet, VGG, CNN, and SVM have been at the forefront.
Table 5.
Related published research by open access publishers.
| # | Title | Best Performance | Ensemble Method | Dataset |
|---|---|---|---|---|
| 1 | Development of Machine-Learning Model to Predict COVID-19 Mortality: Application of Ensemble Model and Regarding Feature Impacts [65] | The highest accuracy: 85% |
Combination of deep and machine learning: MLP with SVM, XGBoosta and Random Forest | 203 patients with severe and moderate levels of COVID-19 in South Korea |
| 2 | An Improved Machine-Learning Approach for COVID-19 Prediction Using Harris Hawks Optimization and Feature Analysis Using SHAP [66] | The highest accuracy: 92.38% |
Applying the Harris Hawkes optimization algorithm to XGBoost, light gradient boosting, classification boosting, RF and SVM classifiers | Data collected by the Nexoid research team in London consisting information collected from 1,023,426 individuals, 98.80% of whom had negative COVID-19 results and 1.20% had positive results |
| 3 | A Novel βSA Ensemble Model for Forecasting the Number of Confirmed COVID-19 Cases in the US [67] | Not clear (reducing error rate) | Combination of α-Sutte indicator and ARIMAb with error-based dynamic weighting method | Collected by Centers for Disease Control and Prevention in the United States in July 2021 |
| 4 | An Ensemble Learning Model for COVID-19 Detection from Blood Test Samples [68] | Highest accuracy: 99.4% |
CNN as the first stage classifier was combined with 15 supervised machine learning algorithms: SVM, Bayes, DT, RF, MLPc, Addabost, LR, LDAd/QDAe, and stochastic gradient descent |
Data from 279 blood tests taken at the San Raffaele Hospital in Milan, Italy |
| 5 | Forecasting the Potential Number of Influenza-like Illness Cases by Fusing Internet Public Opinion [69] | Reducing error rate from 6.48% to 2.68% | Combination of XGBoost, RF and SVRf | The data collected at the Yuan Environmental Protection Center and the Statistics Office of the Ministry of the Interior of Taiwan |
| 6 | Deep Ensemble Learning-Based Models for Diagnosis of COVID-19 from Chest CT Images [70] | Best accuracy: 99.2% |
Transfer learning by pre-trained CNN, stacking and Weighted Aged Ensemble to combine the performance of three base classifier VGG19, ResNet50 and DenseNet201 | 2482 CT scan images obtained from 120 patients in So Paulo, Brazil. 1252 COVID-19 positive cases and 1230 COVID-19 negative cases |
| 7 | Deep Ensemble Model for COVID-19 Diagnosis and Classification Using Chest CT Images [71] | Highest accuracy: 98.59% |
Preprocessing based on Gaussian filter to remove noise. Using a shark optimization algorithm and improved bat algorithm with multi-class SVM | A collection of CT chest images uploaded to GitHub, including 349 images from 216 patients in several hospitals in China |
| 8 | Automated diagnosis of childhood pneumonia in chest radiographs using modified densely residual bottleneck-layer features [72] | Highest accuracy: 99.6% |
Adaboost | 5232 CXR images for children aged one to five years old from Kaggle, of which 3883 were infected and 1349 were normal |
| 9 | Multi-channel transfer learning of chest x-ray images for screening of COVID-19 [73] | Highest accuracy: 94% Recall: 100% |
Three ResNet-based models for one-to-all classification | Chest X-ray images including 1579 normal, 4245 pneumonia and 184 COVID-19 cases |
| 10 | Ensemble learning for poor prognosis predictions: A case study on SARS-CoV-2 [74] | Highest accuracy: 95% |
Seven prediction models defined in China named Dong, Shi, Gong, Lu, Yan, Xie, and Levy |
5394 cases in two hospitals in China, London King's College Hospital and University Hospitals Birmingham |
| 11 | Using Automated Machine Learning to Predict the Mortality of Patients With COVID-19: Prediction Model Development Study [75] | Highest accuracy for stacking model: 79.1% |
Using a real-time method and building 20 different machine learning models through auto ML. Model interpretation through Shapley's additive explanation and dependency graphs for extracting 10 influential variables - using a binary classifier | 4313 cases in Albert Einstein College of Pharmaceutical Sciences in New York |
| 12 | COVID-19 epidemic: analysis and prediction [76] | Highest accuracy: 99.94% |
Using linear regression, polynomial regression and SVM | 14,654 Indian patients' data from Johns Hopkins University Center for Science and Engineering |
| 13 | Deep ensemble model for classification of novel coronavirus in chest X-ray images [77] | Deep CNN model namely MobileNet, ResNet50 and InceptionV3. | Four classes of chest X-rays images (natural, bacterial, viral, and COVID-19), each containing 1050 images obtained from Kaggle | |
| 14 | Ensemble deep learning and internet of things-based automated COVID-19 diagnosis framework [78] | Highest accuracy: 99.12% |
CNN, transfer learning and three types of deep learning: ResNet152V2, DenseNet201 and InceptionResNetV2 (IRNV2). | CT scan datasets for CNN, deep learning, and transfer learning from a variety of sources |
| 15 | Artificial intelligence and medical internet of things framework for diagnosis of coronavirus suspected cases [79] | Highest accuracy: 99.2% |
ResNet152V2, InceptionResNetV2, VGG16, DenseNet201 | 1663 COVID-19 positive patients, 401 cases of pneumonia (viral and bacterial), 394 tuberculosis patients, and 2039 images of healthy people |
| 16 | On the detection of covid-19 from chest x-ray images using cnn-based transfer learning [80] | Highest accuracy for ResNet50V2 | Using five models: VGG16, ResNet50V2, Xception, MobileNet, DenseNet121 |
X-ray images of 678 patients with or without COVID-19 obtained from GitHub |
| 17 | COVID-19 cases prediction in Saudi Arabia using tree-based ensemble Models [81] |
Highest accuracy: For XGBoost MAE: 4.41, RMSE: 7.11 MAPE: 0.95% |
Four ensemble methods based on tree: Gradient Tree Boosting Random Forest Extreme Gradient Boosting Voting Regressor |
The data by Our World in Data (OWID) in Saudi Arabia. This dataset contains 59 parameters, updated daily, including confirmed cases, deaths, and testing |
| 18 | FLANNEL: Focal Loss Based Neural Network Ensemble for COVID-19 Detection [82] |
Highest accuracy: Macro-F1 91.7% |
FLANNEL model consists of two stages; in the first stage, Inception v3, VGG 19-bn, ResNetXt101, Resnet152, and Densenet161. in the second stage, Neural Weight Module | The COVID and Kaggle chest X-ray images (pneumonia) dataset containing 5508 images from 2874 patients in four categories |
Extreme Gradient Boosting.
AutoRegressive Integrated Moving Average.
Multilayer Perceptron.
Linear Discriminant Analysis.
Quadratic Discriminant Analysis.
Support Vector Regression.
4. Scientometric and bibliometric analysis
Bibliometric search retrieves data for required documents that have an academic structure [83,84]. VOSviewer [85] and BibExcel [86] softwares are used for doing the scientometric-bibliometric analysis. These two softwares have been widely used in recent scientometric-bibliometric studies [[87], [88], [89], [90], [91]].
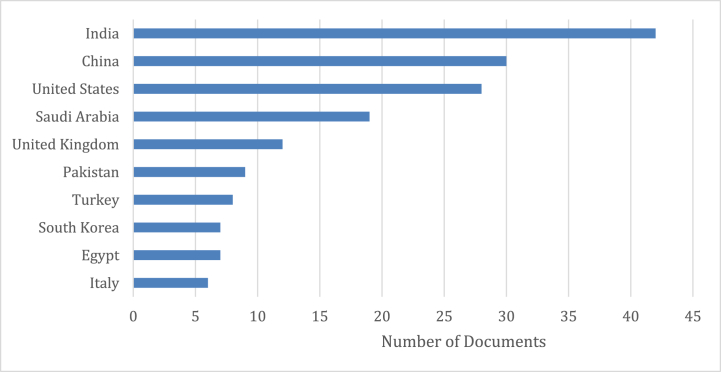
As mentioned earlier, a total of 142 journal papers (records) were extracted from the Scopus database based on our criteria including ensemble learning and COVID-19 keywords. This section attempts to capture an overall picture of ensemble learning and COVID-19. The top 10 most influential countries have been India, China, and the United States which have published the greatest number of journal papers (Fig. 1).
Fig. 1.
Top influential countries in this field of research.
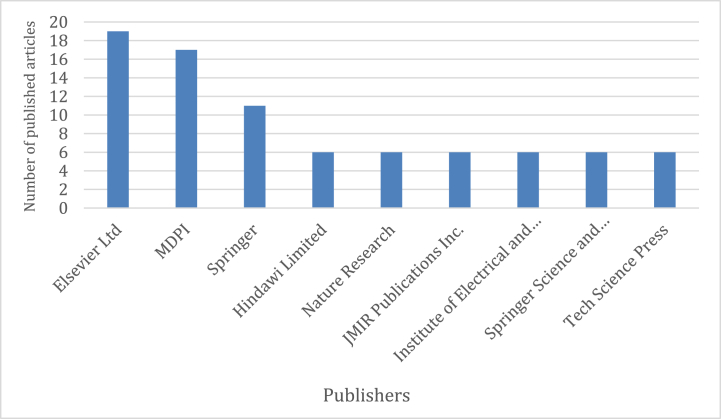
Fig. 2 shows the top 10 most influential publishers that have published more than five articles in this research field. Elsevier, MDPI, and Springer have published the greatest number of papers. Although the number of articles published by Elsevier is relatively higher than MDPI, MDPI has published more open-access articles than Elsevier in this field of research.
Fig. 2.
Number of published articles by each of the studied publishers.
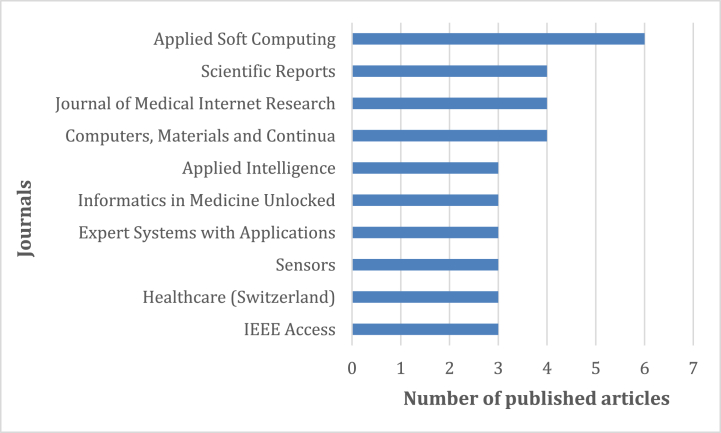
Fig. 3 shows the top 10 influential journals based on the number of published papers. Applied Soft Computing, Scientific Report, Journal of Medical Internet Research and Computers, Materials and Continua are the top journals in this field of research.
Fig. 3.
Number of documents per journal.
Table 6 shows the 10 top-cited papers. The most cited paper was [92] authored by Ribeiro et al. with 230 citations. The second place belonged to Ref. [60] by Rajaraman with 129 citations. The next most cited paper was [55] by Gao with 96 citations. The average number of citations from January 2020 to August 1, 2022 was 10.78.
Table 6.
Ten top-cited papers.
| Title | Citations | Reference No. |
|---|---|---|
| Short-term forecasting COVID-19 cumulative confirmed cases: perspectives for Brazil | 230 | [92] |
| Iteratively pruned deep learning ensembles for COVID-19 detection in chest X-rays | 129 | [60] |
| Machine learning based early warning system enables accurate mortality risk prediction for COVID-19 | 96 | [93] |
| The ensemble deep learning model for novel COVID-19 on CT images | 83 | [47] |
| Assessment of lockdown effect in some states and overall India: A predictive mathematical study on COVID-19 outbreak | 77 | [94] |
| Densely connected convolutional networks-based COVID-19 screening model | 52 | [39] |
| Lies Kill, Facts Save: Detecting COVID-19 Misinformation in Twitter | 47 | [95] |
| Trends in US pediatric hospital admissions in 2020 compared with the decade before the COVID-19 pandemic | 47 | [96] |
| Ensemble learning model for diagnosing COVID-19 from routine blood tests | 37 | [97] |
| Full-coverage mapping and spatiotemporal variations of ground-level ozone (O3) pollution from 2013 to 2020 across China | 37 | [98] |
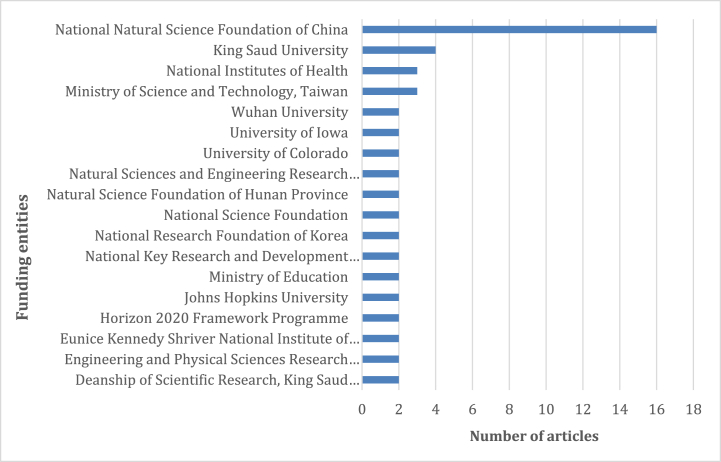
Fig. 4 shows the number of papers funded by each funding entity. The National Natural Science Foundation of China dedicated the largest sum of research funds during our study period. Meanwhile, the investment of Saudi Arabia was also impressive. Fig. 1, Fig. 4 demonstrate that countries with more investments on researching in this area have published more scientific articles, including China and Saudi Arabia.
Fig. 4.
Top funding entities in this field of research.
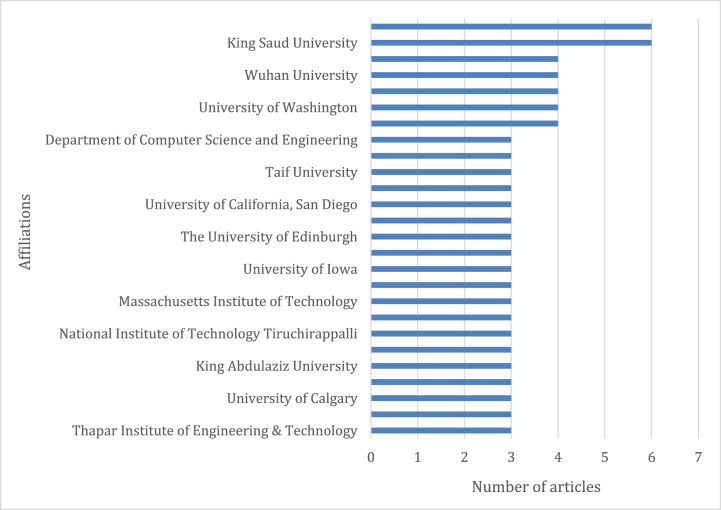
An investigation of affiliations reveal that the Ministry of Education of China and King Saud University in Saudi Arabia have had the most contributions (Fig. 5). There was a significant relationship between the top 10 counties, funding entities, and affiliations.
Fig. 5.
Number of documents based on affiliation of the published research.
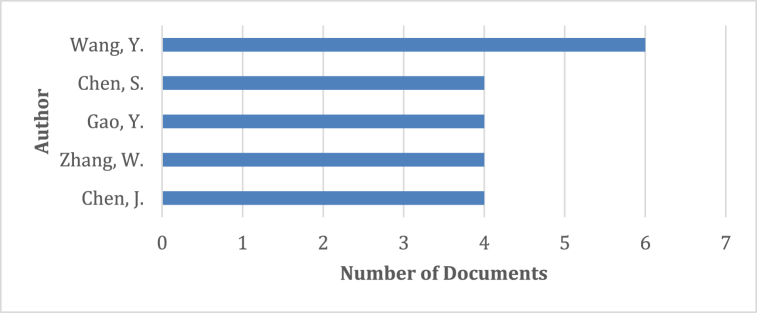
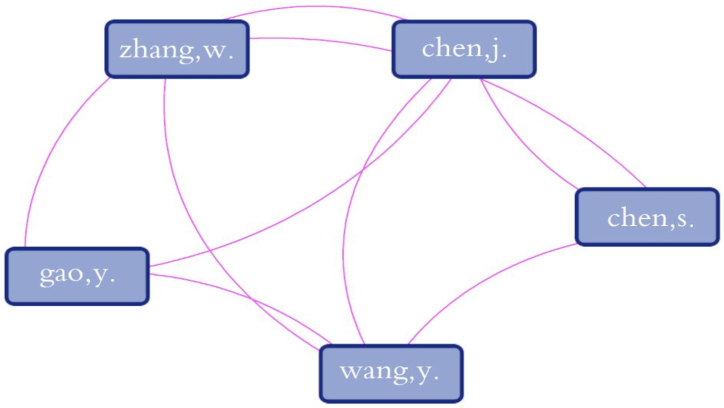
The most effective authors are shown in Fig. 6. These authors have published more than three papers in this research area. In addition, it is necessary to examine the co-authorship map (Fig. 7). They have formed a network of collaboration among themselves, as shown in the diagram below.
Fig. 6.
Top five influential authors in this research field.
Fig. 7.
Co-authorship map for the top five influential authors.
Fig. 7 shows the co-authorship map between the top authors. This figure shows that there are strong cooperation networks between the top influential authors.
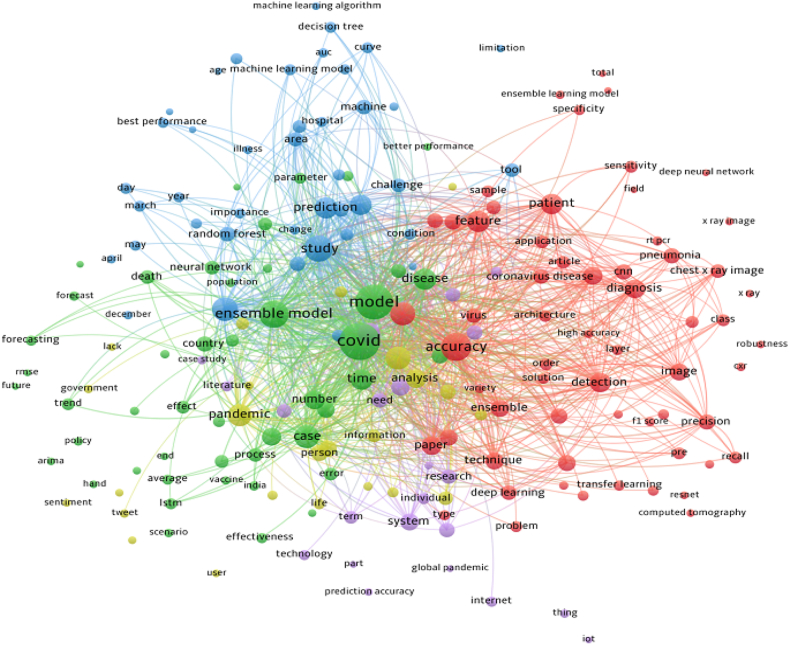
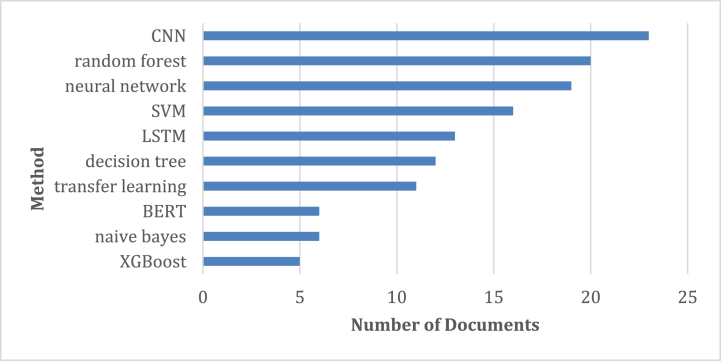
Fig. 8 presents a word occurrence map that was extracted from the abstracts. Besides general keywords such as COVID-19, ensemble learning, and models, some of the specialized keywords were particularly noteworthy. In the red cluster, for example, CNN, transfer learning, and Resnet appear to be important. LSTM is prominent in the green cluster while the decision tree and random forest are prominent in the blue cluster. Fig. 9 shows the top algorithms used in the article. This figure is drawn based on the number of frequent words in the abstract of the articles. CNN, random forest, neural network, SVM, and LSTM were among the top algorithms/techniques.
Fig. 8.
Word occurrence in abstracts.
Fig. 9.
Top 10 used methods/algorithms.
5. Discussion
This research was conducted in two phases. The first stage involved a general review and extraction of data from each article in tables, including the general research method, best performance, and dataset.
Then in the brief review section, an attempt was made to provide an overview of the related works. It is beneficial since it allows researchers to understand the types of research already done in the form of a table more readily and easily. A major objective of this research was to obtain the most common algorithms n ensemble learning methods. CNN was the most frequently used algorithm in all the studied publications. SVM was also applied often. Interestingly, other methods such as Resnet, Densenet, and VGG were also widely used (Table 7).
Table 7.
Most frequent algorithms/techniques extracted from the literature review.
| Most frequent techniques | Publisher |
|---|---|
| ResNet, DenseNet, MobileNet, VGG and CNN | Springer |
| ResNet, DenseNet, VGG, CNN and SVM | Elsevier |
| CNN | Nature |
| ResNet, DenseNet, VGG, CNN and SVM | IEEE |
| ResNet, DenseNet, VGG, CNN and SVM | Open Access Publications |
CNN has been frequently used in related works for the following reasons:
-
●
Feature extraction: CNNs are designed to automatically learn hierarchical representations of data. They excel at extracting relevant features from raw input data, especially in the case of images. In detecting COVID-19 infection, CNNs can effectively identify distinctive patterns or features from chest X-ray or computed tomography scan images.
-
●
Spatial hierarchies: Spatial hierarchies can be captured in data with CNNs. In images, they can learn to recognize low-level features such as edges, textures, and shapes, and gradually build up to higher-level representations. It is beneficial to extract hierarchical features for detecting COVID-19, since certain visual abnormalities associated with the disease may be present at different spatial scales.
-
●
Deep learning capabilities: As a type of deep learning model, CNNs are capable of learning complex relationships and representations from large amounts of data. By automating feature learning, they eliminate the need for manual feature engineering, which is time-consuming and may not capture all the information needed. Due to the availability of large datasets of COVID-19 cases, CNNs can effectively take advantage of this depth to learn discriminative patterns.
-
●
Ensemble diversity: Ensemble learning benefits from combining diverse models. CNNs can contribute to ensemble diversity by being trained with different architectures, hyperparameters, or subsets of data. This diversity helps mitigate overfitting and improves the generalization ability of the ensemble, making it more robust in COVID-19 detection tasks.
-
●
State-of-the-art performance: CNNs have demonstrated excellent performance in various image classification tasks, surpassing human-level performance in some cases. Their ability to learn complex representations and generalize well from large datasets has made them a popular choice for COVID-19 detection, where an accurate and reliable diagnosis is crucial [99,100].
The scientometric analysis section also shows similar results by extracting keywords from the abstracts (Fig. 9). Its findings have many similarities with those of the brief review. Still, there are frequently used algorithms that were revealed by the brief review method, but we were unable to uncover them using the scientometric method. Our findings indicate that scientometric methods and the evaluation of abstracts and keywords cannot provide a complete picture of the content of articles. Yet, they can provide useful information as a complementary method.
A topic that emerged in our study was the frequent appearance of certain countries in the top rankings. Although India has the highest number of publications, China appears in most of the top lists. In terms of the number of published articles, China ranks second. China is the top sponsor for research in this area. Furthermore, some of the top affiliations also belong to China. In evaluating the top influential authors, we found that most are Chinese, but they also have strong co-authorship and cooperation networks. The review of the used datasets reveal that an extensive number of them were prepared in China. It is also necessary to highlight that Saudi Arabia has had the most investments for research in this field after China and the highest level of affiliation was with Saudi universities. The United States was also listed among the top countries after China and Saudi Arabia. Considering the number of published articles, the United States ranks third. American universities are present in the top 10 list of affiliations.
6. Conclusion
We did a brief review and a scientometric-bibliometric analysis of the published research on detecting COVID-19 with ensemble learning approaches. The data were extracted from the articles published in journals that are indexed in the Scopus database using relevant queries. According to the preliminary bibliographic analysis, China and Saudia Arabia had allocated the greatest number of research funds and had the most affiliations. India, China, and Saudi Arabia have had the greatest number of published articles in this field of research. It was found that four authors had significant co-authorship relationships. Among the most commonly used algorithms in ensemble learning approaches, CNN was the most frequently used in the studied published articles. In addition, SVM and random forest method were among the most commonly used methods. A review evaluation also revealed that Resnet, DenseNet, and VGG methods were frequently employed.
7. Challenges and future works
Ensemble learning can be challenging to implement in real-world settings due to the following reasons [14]:
-
⬥
Ensemble models are computationally expensive and require more resources than single models.
-
⬥
Ensemble models are more complex and difficult to interpret than single models.
-
⬥
Ensemble models require more data than single models.
To address these challenges, researchers have proposed several solutions such as using pre-training specific to chest X-ray images in transferring and fine-tuning the learned knowledge toward improving COVID-19 detection performance; using ensembles of the fine-tuned models to further improve performance over individual constituent models; performing feature selection to reduce the dimensionality of the data; and using transfer learning [101].
Further research is needed in several areas based on our findings, including:
-
⬥
Standardization and benchmarking: There is a need for standardized datasets, evaluation metrics, and benchmarking protocols to facilitate fair comparisons and reproducibility of ensemble learning models for COVID-19 detection.
-
⬥
Interpretability and transparency: Research should focus on improving the interpretability and transparency of ensemble models to gain insights into their decision-making processes, feature importance, and potential biases.
-
⬥
Real-world deployment: Investigating the feasibility of deploying ensemble learning models in real-world healthcare settings, considering computational requirements, integration with existing systems, and validation in diverse clinical environments.
-
⬥
Addressing data limitations: Addressing data limitations, including imbalanced datasets, lack of diversity in demographic representation, and potential biases, to enhance the robustness and generalizability of ensemble models.
Funding
The author did not receive any financial support.
Data availability
Data will be available upon request.
CRediT authorship contribution statement
Mohammad Javad Shayegan: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Al-Emran M., Al-Kabi M.N., Marques G. A survey of using machine learning algorithms during the COVID-19 pandemic. Emerging technologies during the era of COVID-19 pandemic. 2021:1–8. [Google Scholar]
- 2.Alballa N., Al-Turaiki I. Machine learning approaches in COVID-19 diagnosis, mortality, and severity risk prediction: a review. Inform. Med. Unlocked. 2021;24 doi: 10.1016/j.imu.2021.100564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Rashidy N., et al. Comprehensive survey of using machine learning in the COVID-19 pandemic. Diagnostics. 2021;11(7):1155. doi: 10.3390/diagnostics11071155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lalmuanawma S., Hussain J., Chhakchhuak L. Applications of machine learning and artificial intelligence for Covid-19 (SARS-CoV-2) pandemic: a review. Chaos, Solit. Fractals. 2020;139 doi: 10.1016/j.chaos.2020.110059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li W.T., et al. Using machine learning of clinical data to diagnose COVID-19: a systematic review and meta-analysis. BMC Med. Inf. Decis. Making. 2020;20(1):1–13. doi: 10.1186/s12911-020-01266-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahimi I., Chen F., Gandomi A.H. A review on COVID-19 forecasting models. Neural Comput. Appl. 2021:1–11. doi: 10.1007/s00521-020-05626-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tiwari S., Chanak P., Singh S.K. IEEE Transactions on Artificial Intelligence; 2022. A Review of the Machine Learning Algorithms for COVID-19 Case Analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atlasi R., Noroozi Chakoli A., Ramezani A., Tabatabaei-Malazy O., Larijani B. Scientometric analyzing the output of researchers and organizations on COVID-19 for better conducting the scientific efforts: with a glance to endocrinology. J. Diabetes Metab. Disord. 2021;20(1):107–118. doi: 10.1007/s40200-020-00718-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beranová L., Joachimiak M.P., Kliegr T., Rabby G., Sklenák V. Why was this cited? Explainable machine learning applied to COVID-19 research literature. Scientometrics. 2022:1–37. doi: 10.1007/s11192-022-04314-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colavizza G., Costas R., Traag V.A., van Eck N.J., van Leeuwen T., Waltman L. A scientometric overview of CORD-19. PLoS One. 2021;16(1) doi: 10.1371/journal.pone.0244839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunningham E., Smyth B., Greene D. Collaboration in the time of COVID: a scientometric analysis of multidisciplinary SARS-CoV-2 research. Humanities and Social Sciences Communications. 2021;8(1):1–8. doi: 10.1057/s41599-021-00957-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodríguez-Rodríguez I., Rodríguez J.-V., Shirvanizadeh N., Ortiz A., Pardo-Quiles D.-J. Applications of artificial intelligence, machine learning, big data and the internet of things to the covid-19 pandemic: a scientometric review using text mining. Int. J. Environ. Res. Publ. Health. 2021;18(16):8578. doi: 10.3390/ijerph18168578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho T.K.K., Gwak J. Feature-level ensemble approach for COVID-19 detection using chest X-ray images. PLoS One. 2022;17(7) doi: 10.1371/journal.pone.0268430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajaraman S., Sornapudi S., Alderson P.O., Folio L.R., Antani S.K. Analyzing inter-reader variability affecting deep ensemble learning for COVID-19 detection in chest radiographs. PLoS One. 2020;15(11) doi: 10.1371/journal.pone.0242301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O. O. Abayomi-Alli, R. Damaševičius, R. Maskeliūnas, and S. Misra, "An ensemble learning model for COVID-19 detection from blood test Samples," Sensors, vol. 22, no. 6, doi: 10.3390/s22062224.. [DOI] [PMC free article] [PubMed]
- 16.Bakkalbasi N., Bauer K., Glover J., Wang L. Three options for citation tracking: google scholar, Scopus and web of science. Biomed. Digit Libr. 2006;3(1):1–8. doi: 10.1186/1742-5581-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Al-Sharafi M.A., Al-Qaysi N., Iahad N.A., Al-Emran M. Evaluating the sustainable use of mobile payment contactless technologies within and beyond the COVID-19 pandemic using a hybrid SEM-ANN approach. Int. J. Bank Market. 2021;40(5):1071–1095. [Google Scholar]
- 18.Mongeon P., Paul-Hus A. The journal coverage of Web of Science and Scopus: a comparative analysis. Scientometrics. 2016;106(1):213–228. [Google Scholar]
- 19.Arpaci I., Al-Emran M., Al-Sharafi M.A. The impact of knowledge management practices on the acceptance of Massive Open Online Courses (MOOCs) by engineering students: a cross-cultural comparison. Telematics Inf. 2020;54 [Google Scholar]
- 20.Chadegani A.A., et al. 2013. A Comparison between Two Main Academic Literature Collections: Web of Science and Scopus Databases. arXiv preprint arXiv:1305.0377. [Google Scholar]
- 21.Martín-Martín A., Orduna-Malea E., Thelwall M., López-Cózar E.D. Google Scholar, Web of Science, and Scopus: a systematic comparison of citations in 252 subject categories. Journal of informetrics. 2018;12(4):1160–1177. [Google Scholar]
- 22.Mok K.Y., Shen G.Q., Yang J. Stakeholder management studies in mega construction projects: a review and future directions. Int. J. Proj. Manag. 2015;33(2):446–457. [Google Scholar]
- 23.Gusenbauer M. Search where you will find most: comparing the disciplinary coverage of 56 bibliographic databases. Scientometrics. 2022/05/01 2022;127(5):2683–2745. doi: 10.1007/s11192-022-04289-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh V.K., Singh P., Karmakar M., Leta J., Mayr P. The journal coverage of Web of Science, Scopus and Dimensions: a comparative analysis. Scientometrics. 2021/06/01 2021;126(6):5113–5142. doi: 10.1007/s11192-021-03948-5. [DOI] [Google Scholar]
- 25.Rajesh N., Christodoss P.R. Analysis of origin, risk factors influencing COVID-19 cases in India and its prediction using ensemble learning. International Journal of System Assurance Engineering and Management. 2021:1–8. [Google Scholar]
- 26.Das A.K., Ghosh S., Thunder S., Dutta R., Agarwal S., Chakrabarti A. Automatic COVID-19 detection from X-ray images using ensemble learning with convolutional neural network. Pattern Anal. Appl. 2021;24(3):1111–1124. [Google Scholar]
- 27.Elakkiya R., Vijayakumar P., Karuppiah M. COVID_SCREENET: COVID-19 screening in chest radiography images using deep transfer stacking. Inf. Syst. Front. 2021;23(6):1369–1383. doi: 10.1007/s10796-021-10123-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El Asnaoui K. Design ensemble deep learning model for pneumonia disease classification. International Journal of Multimedia Information Retrieval. 2021;10(1):55–68. doi: 10.1007/s13735-021-00204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar N., Gupta M., Gupta D., Tiwari S. Novel deep transfer learning model for COVID-19 patient detection using X-ray chest images. J. Ambient Intell. Hum. Comput. 2021:1–10. doi: 10.1007/s12652-021-03306-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh R.K., Pandey R., Babu R.N. COVIDScreen: explainable deep learning framework for differential diagnosis of COVID-19 using chest X-rays. Neural Comput. Appl. 2021;33(14):8871–8892. doi: 10.1007/s00521-020-05636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammam A.A., Elmousalami H.H., Hassanien A.E. Big Data Analytics and Artificial Intelligence against COVID-19: Innovation Vision and Approach. Springer; 2020. Stacking deep learning for early COVID-19 vision diagnosis; pp. 297–307. [Google Scholar]
- 32.Yildirim E., Cicioğlu M., Çalhan A. Real-time internet of medical things framework for early detection of Covid-19. Neural Comput. Appl. 2022:1–14. doi: 10.1007/s00521-022-07582-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paul A., Basu A., Mahmud M., Kaiser M.S., Sarkar R. Inverted bell-curve-based ensemble of deep learning models for detection of COVID-19 from chest X-rays. Neural Comput. Appl. 2022:1–15. doi: 10.1007/s00521-021-06737-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ilhan H.O., Serbes G., Aydin N. Decision and feature level fusion of deep features extracted from public COVID-19 data-sets. Appl. Intell. 2022;52(8):8551–8571. doi: 10.1007/s10489-021-02945-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das A. Adaptive UNet-based lung segmentation and ensemble learning with CNN-based deep features for automated COVID-19 diagnosis. Multimed. Tool. Appl. 2022;81(4):5407–5441. doi: 10.1007/s11042-021-11787-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ravi V., Acharya V., Alazab M. A multichannel EfficientNet deep learning-based stacking ensemble approach for lung disease detection using chest X-ray images. Cluster Comput. 2022:1–23. doi: 10.1007/s10586-022-03664-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S., et al. A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19) Eur. Radiol. 2021;31(8):6096–6104. doi: 10.1007/s00330-021-07715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shalbaf A., Vafaeezadeh M. Automated detection of COVID-19 using ensemble of transfer learning with deep convolutional neural network based on CT scans. Int. J. Comput. Assist. Radiol. Surg. 2021;16(1):115–123. doi: 10.1007/s11548-020-02286-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh D., Kumar V., Kaur M. Densely connected convolutional networks-based COVID-19 screening model. Appl. Intell. 2021;51(5):3044–3051. doi: 10.1007/s10489-020-02149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shastri S., Singh K., Kumar S., Kour P., Mansotra V. Deep-LSTM ensemble framework to forecast Covid-19: an insight to the global pandemic. Int. J. Inf. Technol. 2021;13(4):1291–1301. doi: 10.1007/s41870-020-00571-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahanty C., Kumar R., Patro S. Internet of medical things-based COVID-19 detection in CT images fused with fuzzy ensemble and transfer learning models. New Generat. Comput. 2022:1–17. doi: 10.1007/s00354-022-00176-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jangam E., Barreto A.A.D., Annavarapu C.S.R. Automatic detection of COVID-19 from chest CT scan and chest X-Rays images using deep learning, transfer learning and stacking. Appl. Intell. 2022;52(2):2243–2259. doi: 10.1007/s10489-021-02393-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y.-M., Chen Y.J., Ho W.-H., Tsai J.-T. Classifying chest CT images as COVID-19 positive/negative using a convolutional neural network ensemble model and uniform experimental design method. BMC Bioinf. 2021;22(5):1–19. doi: 10.1186/s12859-021-04083-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar M., Atalla S., Almuraqab N., Moonesar I.A. Detection of COVID-19 using deep learning techniques and cost effectiveness evaluation: a survey. Frontiers in Artificial Intelligence. May-27 2022;5:2022. doi: 10.3389/frai.2022.912022. (in English) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Priyansh Kedia A., Katarya Rahul. CoVNet-19: a Deep Learning model for the detection and analysis of COVID-19 patients. Applied Soft Computing Journal. 2021;104 doi: 10.1016/j.asoc.2021.107184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiqiu Jin S.D., Dong Changzi, Ye Xiaodan. Hybrid ensemble model for differential diagnosis between COVID-19 and common viral pneumonia by chest X-ray radiograph. Comput. Biol. Med. 2021;131 doi: 10.1016/j.compbiomed.2021.104252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.L. H. Zhou Tao , Yang Zaoli , Qiu Shi , Huo Bingqiang , Dong Yali, "The ensemble deep learning model for novel COVID-19 on CT images," Applied Soft Computing Journal, doi: 10.1016/j.asoc.2020.106885.. [DOI] [PMC free article] [PubMed]
- 48.Maryam AlJame I.A., Imtiaz Ayyub, Mohammed Ameer. Ensemble learning model for diagnosing COVID-19 from routine blood tests. Inform. Med. Unlocked. 2020;21 doi: 10.1016/j.imu.2020.100449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun J., Pi P., Tang C., Wang S.-H., Zhang Y.-D. TSRNet: diagnosis of COVID-19 based on self-supervised learning and hybrid ensemble model. Comput. Biol. Med. 2022;146 doi: 10.1016/j.compbiomed.2022.105531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Islam M.R., Nahiduzzaman M. Complex features extraction with deep learning model for the detection of COVID19 from CT scan images using ensemble based machine learning approach. Expert Syst. Appl. 2022;195 doi: 10.1016/j.eswa.2022.116554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deb S.D., Jha R.K., Jha K., Tripathi P.S. A multi model ensemble based deep convolution neural network structure for detection of COVID19. Biomed. Signal Process Control. 2022;71 doi: 10.1016/j.bspc.2021.103126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thaseen S.R., V I.S.M., M D., M A., R M., Mahendran A., Alnumay W., Chatterjee P. An efficient hardware architecture based on an ensemble of deep learning models for COVID -19 prediction. Sustain. Cities Soc. 2022;80 doi: 10.1016/j.scs.2022.103713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nikparvar B., Rahman M., Hatami F., Thill J.-C. Spatio-temporal prediction of the COVID-19 pandemic in US counties: modeling with a deep LSTM neural network. Sci. Rep. 2021;11(1):1–12. doi: 10.1038/s41598-021-01119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kundu R., Basak H., Singh P.K., Ahmadian A., Ferrara M., Sarkar R. Fuzzy rank-based fusion of CNN models using Gompertz function for screening COVID-19 CT-scans. Sci. Rep. 2021;11(1):1–12. doi: 10.1038/s41598-021-93658-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao Y., et al. "Machine learning based early warning system enables accurate mortality risk prediction for COVID-19. Nat. Commun. 2020;11:5033. doi: 10.1038/s41467-020-18684-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mousavi Mojab S.Z., Shams S., Fotouhi F., et al. EpistoNet: an ensemble of Epistocracy-optimized mixture of experts for detecting COVID-19 on chest X-ray images. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-00524-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mohammed E.A., Keyhani M., Sanati-Nezhad A., et al. An ensemble learning approach to digital corona virus preliminary screening from cough sounds. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-95042-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang S., et al. EDL-COVID: ensemble deep learning for COVID-19 case detection from chest X-ray images. IEEE Trans. Ind. Inf. 2021;17(9):6539–6549. doi: 10.1109/TII.2021.3057683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Upadhyay K., Agrawal M., Deepak D. Ensemble learning‐based COVID‐19 detection by feature boosting in chest X‐ray images. IET Image Process. 2020;14(16):4059–4066. [Google Scholar]
- 60.Rajaraman S., Siegelman J., Alderson P.O., Folio L.S., Folio L.R., Antani S.K. Iteratively pruned deep learning ensembles for COVID-19 detection in chest X-rays. IEEE Access. 2020;8:115041–115050. doi: 10.1109/ACCESS.2020.3003810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhardwaj P., Kaur A. A novel and efficient deep learning approach for COVID‐19 detection using X‐ray imaging modality. Int. J. Imag. Syst. Technol. 2021;31(4):1775–1791. doi: 10.1002/ima.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bhowal P., Sen S., Yoon J.H., Geem Z.W., Sarkar R. Choquet integral and coalition game-based ensemble of deep learning models for covid-19 screening from chest x-ray images. IEEE Journal of Biomedical and Health Informatics. 2021;25(12):4328–4339. doi: 10.1109/JBHI.2021.3111415. [DOI] [PubMed] [Google Scholar]
- 63.Elen A. Covid‐19 detection from radiographs by feature‐reinforced ensemble learning. Concurrency Comput. Pract. Ex. 2022:e7179. doi: 10.1002/cpe.7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kieu S.T.H., Bade A., Hijazi M.H.A., Kolivand H. COVID-19 detection using integration of deep learning classifiers and contrast-enhanced canny edge detected X-ray images. It Professional. 2021;23(4):51–56. [Google Scholar]
- 65.Baik S.-M., Lee M., Hong K.-S., Park D.-J. Development of machine-learning model to predict COVID-19 mortality: application of ensemble model and regarding feature impacts. Diagnostics. 2022;12(6):1464. doi: 10.3390/diagnostics12061464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Debjit K., et al. An improved machine-learning approach for COVID-19 prediction using Harris Hawks optimization and feature analysis using SHAP. Diagnostics. 2022;12(5):1023. doi: 10.3390/diagnostics12051023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shih D.-H., Wu T.-W., Shih M.-H., Yang M.-J., Yen D.C. A novel βSA ensemble model for forecasting the number of confirmed COVID-19 cases in the US. Mathematics. 2022;10(5):824. [Google Scholar]
- 68.Abayomi-Alli O.O., Damaševičius R., Maskeliūnas R., Misra S. An ensemble learning model for COVID-19 detection from blood test samples. Sensors. 2022;22(6):2224. doi: 10.3390/s22062224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wei Y.-C., Ou Y.-L., Li J., Wu W.-C. Forecasting the potential number of influenza-like illness cases by fusing internet public opinion. Sustainability. 2022;14(5):2803. [Google Scholar]
- 70.Mouhafid M., Salah M., Yue C., Xia K. Deep ensemble learning-based models for diagnosis of COVID-19 from chest CT images. Healthcare. 2022;10(1):166. doi: 10.3390/healthcare10010166. MDPI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ragab M., et al. Deep ensemble model for COVID-19 diagnosis and classification using chest CT images. Biology. 2021;11(1):43. doi: 10.3390/biology11010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alkassar S., Abdullah M.A., Jebur B.A., Abdul-Majeed G.H., Wei B., Woo W.L. Automated diagnosis of childhood pneumonia in chest radiographs using modified densely residual bottleneck-layer features. Appl. Sci. 2021;11(23) [Google Scholar]
- 73.Misra S., Jeon S., Lee S., Managuli R., Jang I.-S., Kim C. Multi-channel transfer learning of chest X-ray images for screening of COVID-19. Electronics. 2020;9(9):1388. [Google Scholar]
- 74.Wu H., et al. Ensemble learning for poor prognosis predictions: a case study on SARS-CoV-2. J. Am. Med. Inf. Assoc. 2021;28(4):791–800. doi: 10.1093/jamia/ocaa295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ikemura K., et al. Using automated machine learning to predict the mortality of patients with COVID-19: prediction model development study. J. Med. Internet Res. 2021;23(2) doi: 10.2196/23458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Santosini Bhutia B.P., Ray Mitrabinda. COVID-19 epidemic: analysis and prediction. IAES Int. J. Artif. Intell. 2022 doi: 10.11591/ijai.v11.i2.pp736-745. [DOI] [Google Scholar]
- 77.Ahmad F., Farooq A., Ghani M.U. Deep ensemble model for classification of novel coronavirus in chest X-ray images. Comput. Intell. Neurosci. 2021;2021 doi: 10.1155/2021/8890226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kini A.S., et al. vol. 2022. Contrast Media & Molecular Imaging; 2022. (Ensemble Deep Learning and Internet of Things-Based Automated COVID-19 Diagnosis Framework). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Iskanderani A.I., et al. Artificial intelligence and medical internet of things framework for diagnosis of coronavirus suspected cases. Journal of Healthcare Engineering. 2021;2021 doi: 10.1155/2021/3277988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shorfuzzaman M., Masud M. Cmc-Computers Materials & Continua; 2020. On the Detection of Covid-19 from Chest X-Ray Images Using Cnn-Based Transfer Learning; pp. 1359–1381. [Google Scholar]
- 81.Almazroi A.A., Usmani R.S.A. COVID-19 cases prediction in Saudi Arabia using tree-based ensemble models. Intelligent Automation and Soft Computing. 2022:389–400. [Google Scholar]
- 82.Qiao Z., Bae A., Glass L.M., Xiao C., Sun J. FLANNEL (focal loss based neural network ensemble) for COVID-19 detection. J. Am. Med. Inf. Assoc. 2021;28(3):444–452. doi: 10.1093/jamia/ocaa280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baarimah A.O., et al. A bibliometric analysis and review of building information modelling for post-disaster reconstruction. Sustainability. 2021;14(1):393. [Google Scholar]
- 84.Bankar R.S., Lihitkar S.R. Science mapping and visualization tools used for bibliometric and scientometric studies: a comparative study. Journal of Advancements in Library Sciences. 2019;6(1):382–394. [Google Scholar]
- 85.Van Eck N.J., Waltman L. VOSviewer manual. Leiden: Univeristeit Leiden. 2013;1(1):1–53. [Google Scholar]
- 86.Persson O., Danell R., Schneider J.W. How to use Bibexcel for various types of bibliometric analysis. Celebrating scholarly communication studies: A Festschrift for Olle Persson at his 60th Birthday. 2009;5:9–24. [Google Scholar]
- 87.Jemghili R., Taleb A.A., Khalifa M. A bibliometric indicators analysis of additive manufacturing research trends from 2010 to 2020. Rapid Prototyp. J. 2021;27(7):1432–1454. [Google Scholar]
- 88.Kanade S.G., Duffy V.G. International Conference on Digital Human Modeling and Applications in Health, Safety. Ergonomics and Risk Management; Springer: 2022. Use of virtual reality for safety training: a systematic review; pp. 364–375. [Google Scholar]
- 89.Shayegan M.J., Mohammad M.M. 2021 7th International Conference on Web Research (ICWR) IEEE; 2021. Bibliometric of semantic enrichment; pp. 202–205. [Google Scholar]
- 90.Sodhi D., Duffy V. Human-Automation Interaction; 2023. A Systematic Literature Review of Virtual Reality Education and COVID-19 Safety; pp. 627–647. [Google Scholar]
- 91.Xia H., Liu Z., Maria E., Liu X., Lin C. Sustainable Cities and Society; 2022. Study on City Digital Twin Technologies for Sustainable Smart City Design: A Review and Bibliometric Analysis of Geographic Information System and Building Information Modeling Integration. [Google Scholar]
- 92.Ribeiro M.H.D.M., da Silva R.G., Mariani V.C., dos Santos Coelho L. Short-term forecasting COVID-19 cumulative confirmed cases: perspectives for Brazil. Chaos, Solit. Fractals. 2020;135 doi: 10.1016/j.chaos.2020.109853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gao Y., et al. Machine learning based early warning system enables accurate mortality risk prediction for COVID-19. Nat. Commun. 2020;11(1):1–10. doi: 10.1038/s41467-020-18684-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sardar T., Nadim S.S., Rana S., Chattopadhyay J. Assessment of lockdown effect in some states and overall India: a predictive mathematical study on COVID-19 outbreak. Chaos, Solit. Fractals. 2020;139 doi: 10.1016/j.chaos.2020.110078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Al-Rakhami M.S., Al-Amri A.M. Lies kill, facts save: detecting COVID-19 misinformation in twitter. IEEE Access. 2020;8:155961–155970. doi: 10.1109/ACCESS.2020.3019600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pelletier J.H., Rakkar J., Au A.K., Fuhrman D., Clark R.S., Horvat C.M. Trends in US pediatric hospital admissions in 2020 compared with the decade before the COVID-19 pandemic. JAMA Netw. Open. 2021;4(2) doi: 10.1001/jamanetworkopen.2020.37227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.AlJame M., Ahmad I., Imtiaz A., Mohammed A. Ensemble learning model for diagnosing COVID-19 from routine blood tests. Inform. Med. Unlocked. 2020;21 doi: 10.1016/j.imu.2020.100449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wei J., et al. Full-coverage mapping and spatiotemporal variations of ground-level ozone (O3) pollution from 2013 to 2020 across China. Remote Sensing of Environment. 2022;270 [Google Scholar]
- 99.Akl A.A., Hosny K.M., Fouda M.M., Salah A. A hybrid CNN and ensemble model for COVID-19 lung infection detection on chest CT scans. PLoS One. 2023;18(3) doi: 10.1371/journal.pone.0282608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sarki R., Ahmed K., Wang H., Zhang Y., Wang K. Automated detection of COVID-19 through convolutional neural network using chest x-ray images. PLoS One. 2022;17(1) doi: 10.1371/journal.pone.0262052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cheng L., Qiu Y., Schmidt B.J., Wei G.-W. Review of applications and challenges of quantitative systems pharmacology modeling and machine learning for heart failure. J. Pharmacokinet. Pharmacodyn. 2022/02/01 2022;49(1):39–50. doi: 10.1007/s10928-021-09785-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be available upon request.