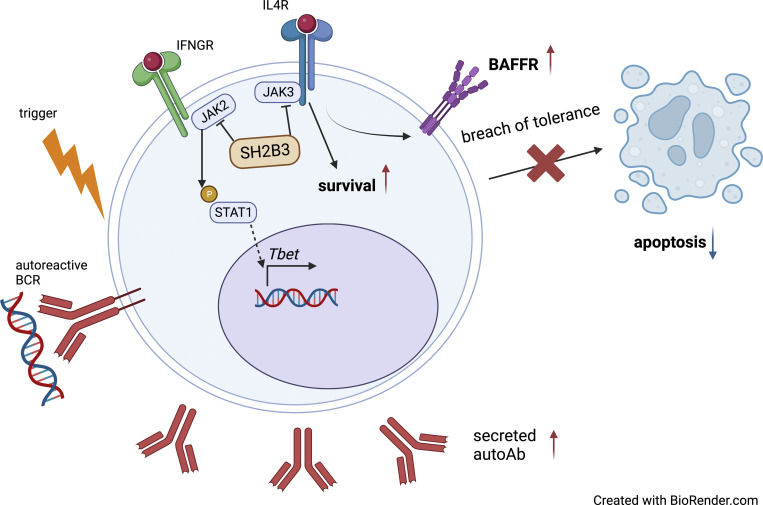
Zhang et al. reveal a role for hypomorphic SH2B3 in lupus risk. The study shows that rare, damaging variants in lupus patients enable breach of B cell immune tolerance checkpoints and suggests involvement for dysregulated IL-4R signaling and BAFF-R expression.
Abstract
Systemic lupus erythematosus (SLE) is a heterogeneous autoimmune disease with a clear genetic component. While most SLE patients carry rare gene variants in lupus risk genes, little is known about their contribution to disease pathogenesis. Amongst them, SH2B3—a negative regulator of cytokine and growth factor receptor signaling—harbors rare coding variants in over 5% of SLE patients. Here, we show that unlike the variant found exclusively in healthy controls, SH2B3 rare variants found in lupus patients are predominantly hypomorphic alleles, failing to suppress IFNGR signaling via JAK2-STAT1. The generation of two mouse lines carrying patients’ variants revealed that SH2B3 is important in limiting the number of immature and transitional B cells. Furthermore, hypomorphic SH2B3 was shown to impair the negative selection of immature/transitional self-reactive B cells and accelerate autoimmunity in sensitized mice, at least in part due to increased IL-4R signaling and BAFF-R expression. This work identifies a previously unappreciated role for SH2B3 in human B cell tolerance and lupus risk.
Graphical Abstract
Introduction
Systemic lupus erythematosus (SLE) is the prototypic systemic autoimmune disease with diverse clinical manifestations driven by a combination of genetic and environmental factors. Despite its heterogeneous nature, SLE patients share some common features and pathogenic mechanisms. Lupus is characterized by the presence of autoantibodies, especially antinuclear antibodies (ANAs) (Tsokos et al., 2016), and the deposition of immune complexes (IC) that leads to organ damage (Koffler et al., 1971). Autoantibodies are secreted by autoreactive B cells that evade central and peripheral checkpoints required for establishing self-tolerance (Yurasov et al., 2005). This can be a result of multiple factors including aberrant Toll-like receptor (TLR) signaling (Fillatreau et al., 2021), dysregulated cytokines, and cytokine receptor signaling (Batten et al., 2000; Granato et al., 2014; Thien et al., 2004), as well as impaired apoptosis and apoptotic cell clearance (Liu et al., 2006; Sisirak et al., 2016).
A role for genetic factors in the pathogenesis of SLE is supported by results from twin concordance studies (Block et al., 1975; Deapen et al., 1992; Ulff-Møller et al., 2018), and >100 susceptibility loci have been identified by genome-wide association studies (GWAS) (Kwon et al., 2019). More recently, whole genome/exome sequencing (WGS/WES) has enabled the identification of highly penetrant and damaging rare genetic variants that cause monogenic forms of SLE (Ellyard et al., 2014; Lo, 2016; Omarjee et al., 2019). We previously investigated the presence of rare coding variants in lupus-associated genes—many of them discovered through GWAS—in SLE patients and established that the majority of patients carry one or more such rare variants (Jiang et al., 2019). Functional studies of rare coding variants in BLK, BANK1, P2RY8, and TLR7 have provided mechanistic insights into disease pathogenesis (Brown et al., 2022; He et al., 2021; Jiang et al., 2019). Our study also revealed that 5.26% of SLE-patients carried rare variants in SH2B3 (Jiang et al., 2019). Here, we describe the impact of these variants on protein function and predisposition to autoimmunity.
SH2B3 encodes the SH2B adaptor protein 3 (SH2B3, also known as LNK), a negative regulator of numerous cytokine and growth factor receptors transduced by Janus kinases JAK2 (Bersenev et al., 2008) and JAK3 (Cheng et al., 2016) as well as receptor tyrosine kinases c-KIT (Simon et al., 2008) and FLT3 (Lin et al., 2012). Variants in SH2B3 have been associated with myeloproliferative neoplasms (Coltro et al., 2019), idiopathic erythrocytosis (McMullin et al., 2011), and autoimmune diseases including SLE (Alcina et al., 2010; Bentham et al., 2015; Morris et al., 2016; Wang et al., 2021), rheumatoid arthritis (RA) (Okada et al., 2014), type 1 diabetes (Steck et al., 2017), and multiple sclerosis (Alcina et al., 2010). A 2013 study on two siblings with homozygous SH2B3 variant D231Profs*38 reported the development of precursor B cell acute lymphoblastic leukemia (ALL) with Hashimoto thyroiditis and suspected autoimmune hepatitis (Perez-Garcia et al., 2013), while a recent case report described a novel clinical syndrome involving myeloproliferative and multiorgan autoimmunity in unrelated patients carrying homozygous SH2B3 variants R148Profs*40 and V402M (Blombery et al., 2022). Sh2b3−/− mice display dysregulation of numerous hematopoietic cell types including hematopoietic stem cells (Bersenev et al., 2008), B and T lymphocytes (Katayama et al., 2014; Mori et al., 2014; Takaki et al., 2000), platelets (Takizawa et al., 2010), dendritic cells (DCs) (Mori et al., 2014), and neutrophils (Laroumanie et al., 2018). Although SH2B3 variants have been associated with autoimmune diseases, the cellular mechanisms by which they contribute to autoimmune pathogenesis are yet to be elucidated.
Using in vitro assays and mouse models engineered with CRISPR/Cas9 to harbor patient Sh2b3 variants, we present data showing that rare SH2B3 variants act as hypomorphic alleles that impair B cell tolerance mechanisms predisposing to autoimmunity.
Results
Loss-of-function SH2B3 variants in SLE patients
Previous bioinformatic analysis of WGS/WES in our cohort of 132 SLE patients and 97 healthy controls (HCs) revealed that 5.26% and 2.06% of patients and HCs, respectively, carried rare (minor allele frequency, MAF < 0.005) SH2B3 coding variants. Clinical information for the patients is shown in Table S1. Additional analyses for rare variants in 22 genes that can cause human SLE when mutated (Table S2) revealed that one patient (D.II.1) was compound heterozygous for ACP5 (a pathogenic variant p.G109R [Lausch et al., 2011] and a novel variant p.C238R). This patient also had a variant of unknown significance in IFIH1 p.A542E. Another patient (A.II.1) was heterozygous for a variant of unknown significance in ADAR, p.I939V. No additional mutations in known SLE-causing genes were identified in the other patients.
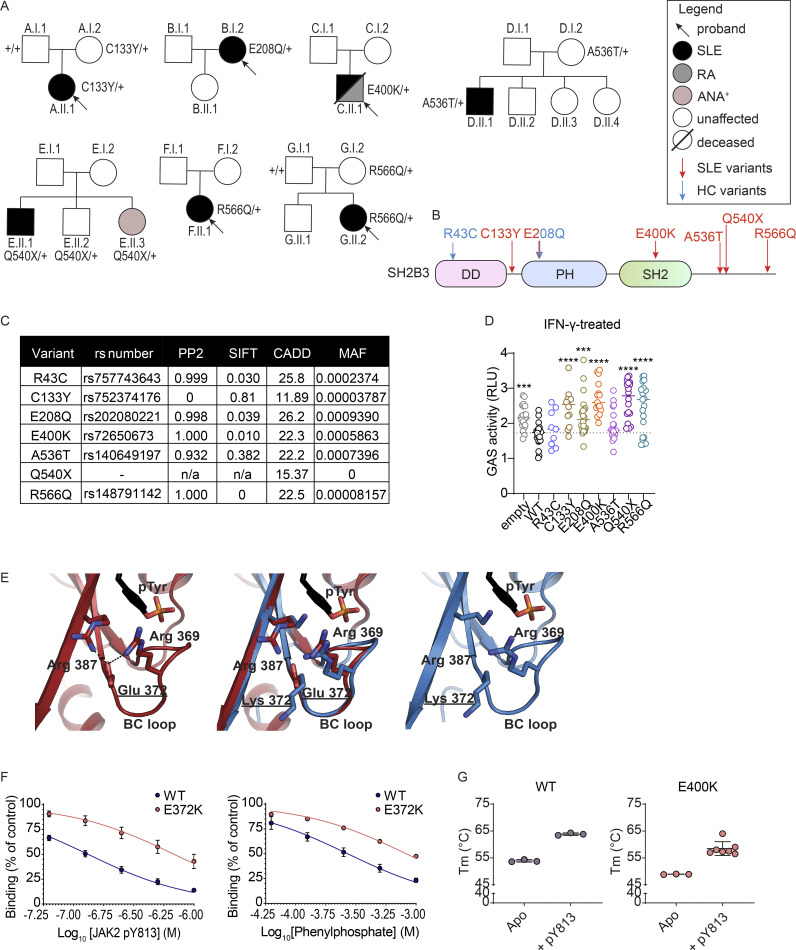
Of the seven rare heterozygous SH2B3 variants identified, five (C133Y, E400K, A536T, Q540X, R566Q) were exclusive to SLE kindreds and one (E208Q) was found in both SLE and HC. The seventh variant (R43C) was identified only in a HC (Fig. 1 A). Whereas the SLE-associated variants occurred in the Pleckstrin homology (PH) domain (E208Q), SH2 domain (E400K), and near the C terminus of the protein (Fig. 1 B), the R43C variant identified in the HC localized to the dimerization domain.
Figure 1.
Rare SH2B3 variants were identified in SLE patients and HCs. (A) Pedigrees of families with rare single nucleotide variants (SNV) in SH2B3 associated with autoimmunity. Black arrows indicate the proband of each cohort. Individuals colored black are affected by SLE, and individuals colored gray are affected by RA. The amino acid changes in SH2B3 protein are shown next to sequenced individuals, with “+” sign indicating WT. (B) Location of patient-specific and HC SNVs in the schematic diagram of SH2B3 amino acid sequence. DD: dimerization domain, SH2: Src homology 2 domain, PH: Pleckstrin homology domain. (C) Summary of variant information, in silico deleteriousness prediction, and MAF of each SNV from HC and SLE patients. PP2: PolyPhen-2. (D) Relative activity of STAT1-GAS in HEK293 cells overexpressing empty vector, WT, or mutant human SH2B3 in the presence of IFN-γ (50 ng/ml) stimulation. Relative activity was calculated by normalizing all STAT1-GAS-Firefly/CMV-Renilla ratios to the average ratios in the unstimulated cells transfected with empty vectors. Results were pooled from seven independent experiments. Sample numbers for each condition are listed as follows: empty (n = 21), WT (n = 21), R43C (n = 9), C133Y (n = 12), E208Q (n = 21), E400K (n = 15), A536T (n = 18), Q540X (n = 18), and R566Q (n = 21). Means are shown as bars, and all conditions were compared with cells transfected with WT SH2B3. lmer with emmeans using the experiment as the blocking factor was used for the statistical analyses. ***: P < 0.001, ****: P < 0.0001. (E) Hydrogen bonding network between Arg369-Glu372-Arg387 (left) that helps position the BC loop relative to the central β-sheet and disrupted hydrogen bond formation (right) when Glu372 is replaced with Lys. (F) Percentage of binding of murine WT or E372K SH2B3 to IL6ST pY757 peptide chip after preincubation with varying concentrations of mutant JAK pY813 peptides (left; WT n = 8, E372K: n = 6) or phenylphosphate (right; WT: n = 4, E372K: n = 2). Data are shown as the mean ± SD of technical replicates from at least three independent experiments. (G) Melting temperatures of WT (left) and E400K (right) SH2B3 protein at apo state (unbound) or bound to pY813 JAK2. Data are displayed as the mean ± SD of technical replicates from at least three independent experiments. WT Apo, WT + pY813, and E400K Apo: n = 3; E400K pY813: n = 7.
We asked whether the rare variants observed in SLE patients conferred functional defects. All variants, except the novel nonsense mutation Q540X, were predicted to be damaging by at least one in silico prediction algorithm (Fig. 1 C). To assess protein function, we examined IFNGR signaling that is in part transduced by JAK2, a target of SH2B3 negative regulation (Igarashi et al., 1994). While overexpression of WT SH2B3 was able to partially suppress STAT1-mediated γ-activated sequence (GAS) activity in HEK293 cells stimulated with IFN-γ, this regulation was lost in cells overexpressing all rare SLE patient variants, except A536T (Fig. 1 D and Fig. S1 A). The A536T variant was present in a proband that also carried biallelic variants in ACP5, which is a likely cause of the patient’s disease (Kara et al., 2020). GAS activity was also suppressed in cells expressing the SH2B3 R43C variant present in the HC. We further examined the suppressive ability of all additional common variants in the MANE (canonical) SH2B3 transcript found in the gnoMAD database with an MAF > 0.001. Of these eight common variants, only T165S showed a decrease in function (Fig. S1 B).
Figure S1.
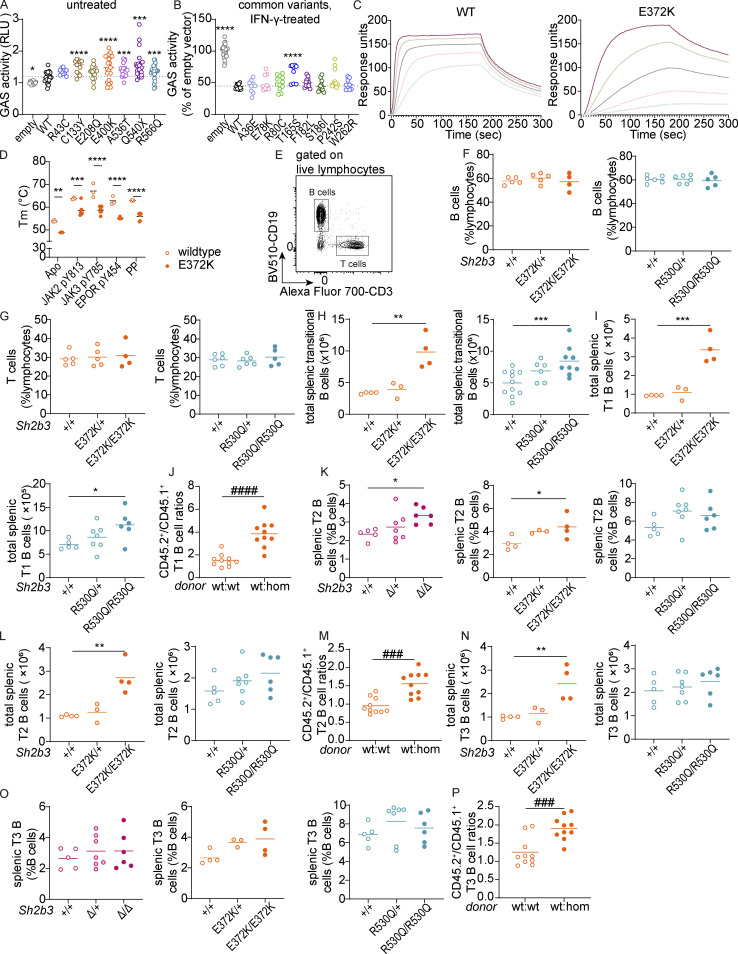
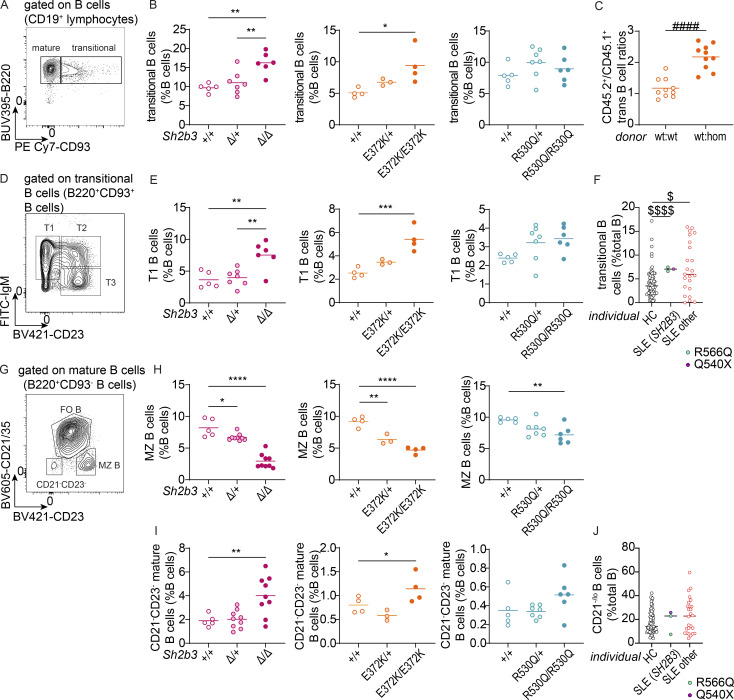
Human and murine SH2B3 protein functionality and gross phenotypes of Sh2b3Δ, Sh2b3E372K, and Sh2b3R530Q mice and transitional B cell phenotypes in Sh2b3E372K and Sh2b3R530Q mice, and chimeras of Sh2b3E372K mice. (A) Relative GAS activity of unstimulated HEK293 cells overexpressing WT or SH2B3 variants identified in SLE and HC cohorts. Sample numbers for each condition are listed as follows: empty (n = 21), WT (n = 21), R43C (n = 9), C133Y (n = 12), E208Q (n = 21), E400K (n = 15), A536T (n = 18), Q540X (n = 18), and R566Q (n = 21). Means are shown as bars, and all conditions were compared to cells transfected with WT SH2B3. lmer with emmeans using the experiment as a blocking factor was used for the statistical analyses. *: P < 0.05, ***: P < 0.001, ****: P < 0.0001. (B) Relative GAS activity of HEK293T cells overexpressing WT or SH2B3 variants listed in gnomAD database at MAF > 0.001 in the presence of IFN-γ (50 ng/ml) stimulation. Results are pooled from four independent experiments and shown as the percentage of GAS activity compared to the EV control. Each dot is a biological replicate and the mean of three technical replicates. Sample numbers for each condition are listed as follows: empty (n = 30), WT (n = 11), W262R (n = 9), T165S (n = 9), F182L (n = 12), S186I (n = 12), P242S (n = 12), R80C (n = 12), A36E (n = 9), E78K (n = 9). lmer with emmeans using the experiment as the blocking factor was used for the statistical analyses. ****: P < 0.0001. (C) Representative direct binding curves for the WT and E372K SH2B3 SH2 domains to a JAK2 pY813 peptide. Results are representative of two independent experiments. (D) Melting temperatures for WT (unfilled) and E372K (filled) SH2B3 SH2 domains in their apo form and after the addition of the phosphomimetic phenyl phosphate (PP), JAK2 pY813, JAK3 pY785, and EPOR pY454 peptides. Results are pooled from at least three independent experiments. All WT: n = 3; E372K: apo n = 3, JAK2 n = 7, JAK3 n = 6, EPOR n = 4, PP n = 6. Means are shown as bars. Two-way ANOVA with Šídák’s adjustment was used for statistical analyses. *: P < 0.05, **: P < 0.01, ***: P < 0.001, ****: P < 0.0001. (E) Representative flow cytometric plot showing the gating strategy of B cells (CD19+CD3−) and T cells (CD19−CD3+). (F and G) Dot plots showing frequencies of B (F) and T cells (G) as percentages of total splenic lymphocytes in Sh2b3E372K (left panel) and Sh2b3R530Q (right panel) mice. (H and I) Total numbers of splenic transitional B cells (H) and T1 B cells (I) in Sh2b3E372K (left) and Sh2b3R530Q (right) mice. (J) CD45.2+/CD45.1+ T1 B cell ratios among splenic B cells in 50:50 BM chimeras of CD45.1-Sh2b3+/+ and CD45.2-Sh2b3+/+/Sh2b3E372K/E372K mice. (K) Frequencies of T2 B cells as percentages of splenic B cells in Sh2b3Δ (left), Sh2b3E372K (middle), and Sh2b3R530Q (right) mice. (L) Total numbers of splenic T2 B cells in Sh2b3E372K (left) and Sh2b3R530Q (right) mice. (M) CD45.2+/CD45.1+ T2 B cell ratios among splenic B cells in 50:50 BM chimeras of CD45.1-Sh2b3+/+ and CD45.2-Sh2b3+/+/Sh2b3E372K/E372K mice. (N) Total numbers of splenic T3 B cells in Sh2b3E372K (left), and Sh2b3R530Q (right) mice. (O) Frequencies of of splenic T3 B cells in Sh2b3Δ (left), Sh2b3E372K (middle), and Sh2b3R530Q (right) mice. (P) Frequencies of (P) CD45.2+/CD45.1+ T3 B cell ratios among splenic B cells in 50:50 BM chimeras of CD45.1-Sh2b3+/+ and CD45.2-Sh2b3+/+/Sh2b3E372K/E372K mice. Results in F–P are representative of two to three independent experiments. Each dot represents one mouse. Sample numbers for each group in F–I, K, L, N, and O are listed as follows: Sh2b3Δ panel: +/+ (n = 5), Δ/+ (n = 7), Δ/Δ (n = 6); Sh2b3E372K panel: +/+ (n = 5 in F and G, n = 4 in others), E372K/+ (n = 5 in F and G, n = 3 in others), E372K/E372K (n = 4); Sh2b3R530Q panels: +/+ (n = 6 in F and G, n = 5 in others), R530Q/+ (n = 6 in F–H, n = 7 in others), and R530Q/R530Q (n = 5–9). n = 10 in J and P. Means are indicated as bars. Student’s t tests were used for the statistical analysis in J, M, and P. One-way ANOVA was used for the statistical analyses in F–I, K, L, N, and O. Significance levels in lmer ANOVAs are indicated with asterisks, while those in multiple Student’s t tests are indicated with hashes. *: P < 0.05, **: P < 0.01, ***/###: P < 0.001, ****/####: P < 0.0001.
Of the SLE patient variants, Q540X, R566Q, and E400K exerted the strongest functional defects. We were particularly interested in the variant E400K as it is a key residue within the SH2 binding pocket that binds phosphotyrosine (pTyr) residues of target proteins (Morris et al., 2021). E400 forms part of a hydrogen bonding network that appears to be required for the stability of the BC loop (connects βB and βC sheets) that forms the base of the pTyr binding pocket. To confirm the impaired function observed in the luciferase assays, we performed a structural analysis of the murine SH2B3 SH2 domain containing the orthologous E372K mutation (Table S3). The murine and human SH2B3 SH2 domains share 92.9% identity with high conservation across peptide-binding residues. Cocrystallization with the JAK2 pY813 motif showed that whilst the motif could still bind, a substitution of the glutamate with a lysine resulted in the loss of the hydrogen bonding network with Arg 369 and Arg 387, potentially destabilizing the BC loop (Fig. 1 E; WT: PDB ID 7R8W, E372K: PDB ID: 8CZ9). Supporting this hypothesis, a decrease in the affinity of the SH2B3/pY813 interaction was observed using a competition assay (E372K IC50 = 685 nM compared with WT = 127 nM), as was the affinity for the pTyr mimetic, phenyl phosphate (Fig. 1 F). The reduction in affinity for JAK2 pY813 was due to a slower on-rate rather than a compromised off-rate (Fig. S1 C).
In addition, the E372K SH2 domain showed decreased thermal stability compared with WT SH2 domain both alone (apo form) or when bound to JAK2 pY813 (Fig. 1 G) or other peptides (Fig. S1 D). These findings suggest that E400K SH2B3 impairs protein function as a result of both reduced affinity to pTyr targets and reduced stability due to the loss of the hydrogen bonding network that requires Glu372. Together, these results indicate that the majority of SH2B3 variants found in SLE patients are hypomorphic alleles.
SLE patient SH2B3 variants behave as hypomorphs in mice
To understand whether these variants conferred phenotypes in vivo, we used CRISPR/Cas9 gene editing technology to generate two mouse models, one carrying the E400K ortholog E372K allele (Sh2b3E372K), which was shown to be important in pTyr binding of the SH2 domain, and another carrying the R566Q ortholog R530Q allele (Sh2b3R530Q), which showed profound loss of activity (Fig. 1 D) and was identified in two unrelated patients (Fig. 1 A). A third strain containing a two-base-pair deletion (c.1107_1108del, coding RefSeq NM_001306127 or p.G371RfsTer56) causing a frameshift and premature stop codon expected to result in a null protein was also generated for comparison.
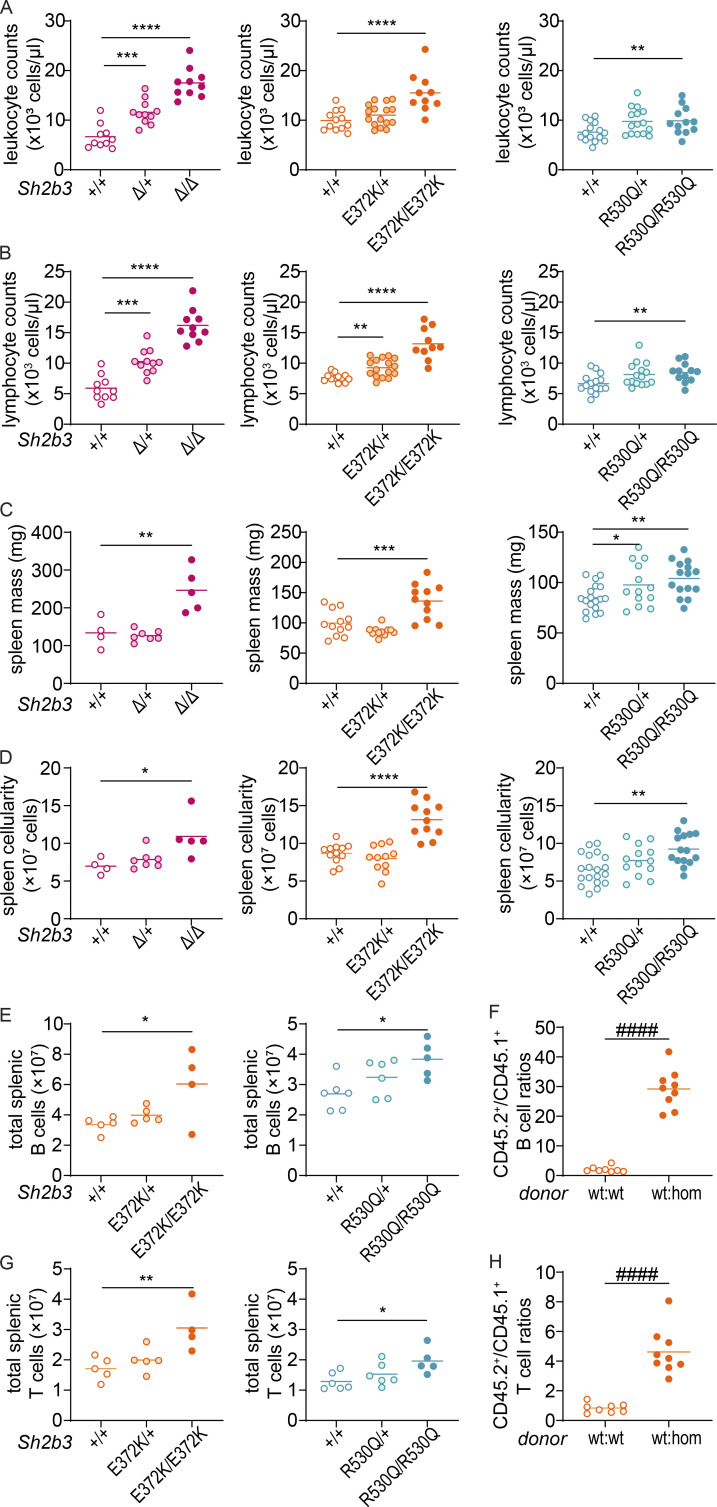
Previous studies have indicated that Sh2b3-deficient (Sh2b3−/−) mice exhibit defects in hematopoiesis, especially lymphopoiesis (Velazquez et al., 2002). Consistent with those reports, we observed a large increase in peripheral blood leukocyte and lymphocyte counts in our Sh2b3-deficient mice (Sh2b3Δ), with a similar but weaker increase in Sh2b3E372K and Sh2b3R530Q strains, which was statistically significant only in homozygous mice (Fig. 2, A and B). Homozygous mice from all three strains also presented with mild splenomegaly (Fig. 2 C) and increased spleen cellularity (Fig. 2 D). Next, we examined splenic B and T cells and observed that although frequencies were unchanged (Fig. S1, E–G), the total number of both B and T cells increased (Fig. 2, E and G) consistent with the overall increase in lymphocyte counts in the spleen and blood. Furthermore, mixed bone marrow (BM) chimeras indicated that Sh2b3E372K/E372K B and T cells displayed a competitive advantage over Sh2b3+/+ cells in repopulating sublethally irradiated Rag1−/− mice (Fig. 2, F and H).
Figure 2.
Gross phenotypes of Sh2b3Δ, Sh2b3E372K, and Sh2b3R530Q mice. (A and B) Peripheral blood leukocyte (A) and lymphocyte (B) counts determined by ADVIA hematological analyzer. Each dot represents one mouse. Sh2b3Δ panels: +/+ (n = 10), Δ/+ (n = 11), Δ/Δ (n = 10); Sh2b3E372K panels: +/+ (n = 12), E372K/+ (n = 16), E372K/E372K (n = 10); Sh2b3R530Q panels: +/+ (n = 15), R530Q/+ (n = 15), R530Q/R530Q (n = 12). (C and D) Spleen mass (C) and cellularity (D). Each dot represents one mouse. Sh2b3Δ panels: +/+ (n = 4), Δ/+ (n = 7), Δ/Δ (n = 5); Sh2b3E372K panels: +/+ (n = 12), E372K/+ (n = 11), E372K/E372K (n = 11); Sh2b3R530Q panels: +/+ (n = 19), R530Q/+ (n = 13), R530Q/R530Q (n = 15). Results in A–D were pooled from two to four independent experiments. Means are shown as bars. The statistical comparison was performed using lmer model ANOVA with multiple comparisons and Tukey’s correction. *: P < 0.05, **: P < 0.01, ***: P < 0.001, ****: P < 0.0001. (E and G) Total splenic B cells (E) and T cells (G) in Sh2b3E372K and Sh2b3R530Q mice. Each dot represents one mouse. Sh2b3E372K panels: +/+ (n = 5), E372K/+ (n = 5), E372K/E372K (n = 4); Sh2b3R530Q panels: +/+ (n = 6), R530Q/+ (n = 6), R530Q/R530Q (n = 5). Results are representative of two independent experiments. Means are shown as bars. Statistical comparison was performed using one-way ANOVA with multiple comparison and Dunnett’s correction. *: P < 0.05, **: P < 0.01. (F and H) Ratios of CD45.2+/CD45.1+ B cell percentages (F) and T cells (H) in the spleens of 50:50 BM chimeras from CD45.1-Sh2b3+/+ and CD45.2-Sh2b3+/+ (n = 8) or Sh2b3E372K/E372K (n = 9) mice. Each dot represents one mouse. Results are representative of two independent experiments. Statistical comparison performed using Student’s t test; ####: P < 0.0001.
Sh2b3E372K and Sh2b3R530Q mice show dysregulated B cell development
Previous studies on Sh2b3−/− mice have reported accumulation of transitional B cells in the spleen, as well as pro- and pre-B cells in the BM due to unrestrained c-KIT and IL-7R signaling (Cheng et al., 2016; Takaki et al., 2000). As expected, we observed increased frequencies of splenic transitional B cells in Sh2b3Δ/+ and Sh2b3Δ/Δ mice (Fig. 3, A and B). We also observed increased frequency and total number of transitional B cells in both Sh2b3E372K/E372K and Sh2b3R530Q/R530Q mice (Fig. 3, A and B; and Fig. S1 H). This increase was due to B cell–intrinsic SH2B3 signaling (Fig. 3 C), and it occurred primarily within the transitional 1 (T1) B cell compartment (Fig. 3, D and E; and Fig. S1, I and J) with a milder increase in transitional 2 (T2) and transitional 3 (T3) B cells (Fig. S1, K–P).
Figure 3.
B cell phenotypes in the spleens of Sh2b3Δ, Sh2b3E372K, and Sh2b3R530Q mice, and peripheral blood of SLE patients carrying SH2B3 variants R566Q and Q540X. (A) Representative flow cytometric plot showing the gating of mature (B220+CD93−) and transitional (B220+CD93+) B cells. (B) Dot plots showing the percentages of splenic transitional B cells in Sh2b3Δ (left), Sh2b3E372K (middle), and Sh2b3R530Q (right) mice. (C) Ratios of CD45.2+/CD45.1+ transitional B cell percentages in the spleens of 50:50 BM chimeras from CD45.1-Sh2b3+/+ and CD45.2-Sh2b3+/+ (n = 10) or Sh2b3E372K/E372K (n = 10) mice. Student’s t test was used for statistical analysis. ####: P < 0.0001. Results are representative of two independent experiments. (D) Representative flow cytometric plot showing the gating of T1 (IgM+CD23−), T2 (IgM+CD23+) and T3 (IgM−CD23+) B cells. (E) Dot plots showing the percentages of splenic T1 B cells in Sh2b3Δ (left), Sh2b3E372K (middle), and Sh2b3R530Q (right) mice. (F) Percentages of human transitional B cells in the peripheral blood of HCs (n = 77), SLE patients carrying SH2B3 variants (R566Q in turquoise, n = 2, and Q540X in purple, n = 1) and other SLE patients (n = 23). (G) Representative flow cytometric plot showing the gating of CD21−CD23− mature B cells and MZ B cells (CD21/35−CD23+). (H) Dot plots showing the frequencies of MZ B cells as percentages of total splenic B cells in Sh2b3Δ (left), Sh2b3E372K (middle), and Sh2b3R530Q (right) mice. (I) Frequencies of CD21−CD23− mature B cells as percentages of total splenic B cells in Sh2b3Δ (left), Sh2b3E372K (middle), and Sh2b3R530Q (right) mice. (J) Percentages of human CD21lo/− B cells in the peripheral blood of HCs (n = 77), SLE patients carrying SH2B3 variants (R566Q in turquoise, n = 2, and Q540X in purple, n = 1), and other SLE patients (n = 23). For panels B, E, H, and I, sample numbers are listed as follows: Sh2b3Δ panels: +/+ (n = 5), Δ/+ (n = 7 in B and E, n = 9 in H and I), Δ/Δ (n = 6 in B and E, n = 9 in H and I); Sh2b3E372K panels: +/+ (n = 4), E372K/+ (n = 3), E372K/E372K (n = 4); Sh2b3R530Q panels: +/+ (n = 5), R530Q/+ (n = 7), R530Q/R530Q (n = 6). Each dot represents one mouse, and means are shown as bars. Results are representative of three independent experiments. One-way ANOVA with multiple comparison and Dunnett’s correction was used for statistical analysis. Significance levels are indicated with asterisks. For panels F and J: Brown–Forsythe and Welch ANOVA tests were used for statistical analysis. Dollar signs are used to indicate significance levels. Significance level criteria are described as follows: */$: P < 0.05, **: P < 0.01, ***: P < 0.001, ****/####/$$$$: P < 0.0001.
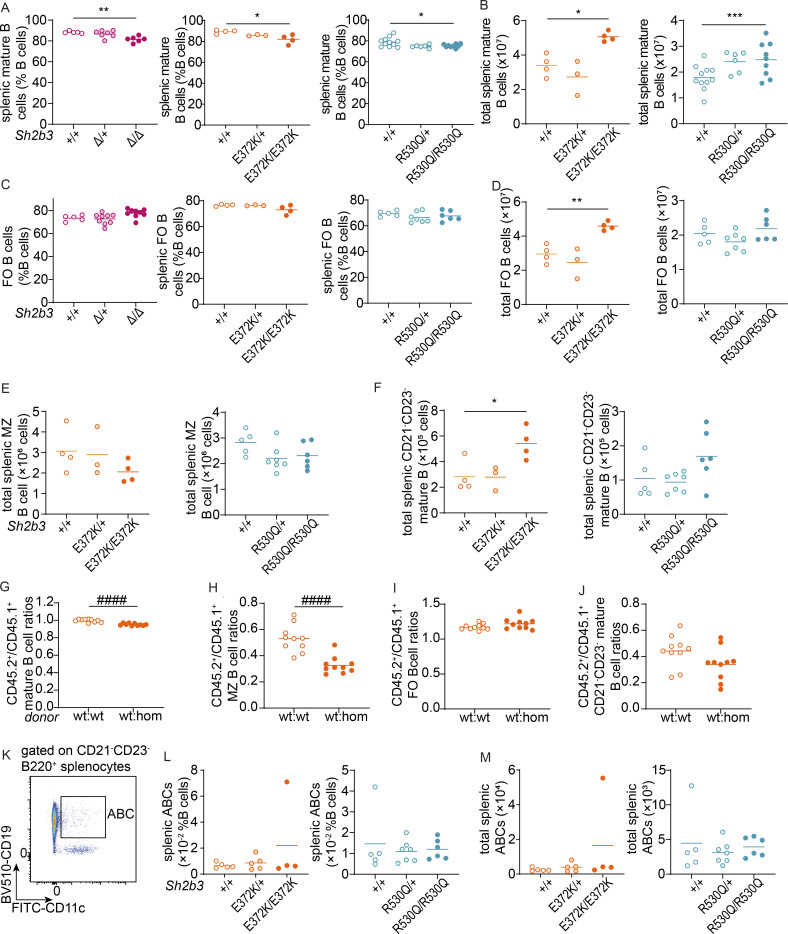
Whilst the frequency of mature B cells was decreased in Sh2b3E372K/E372K and Sh2b3R530Q/R530Q mice, total numbers remained elevated as a result of the overall increase in splenic B cells (Fig. S2, A–D). Marginal zone (MZ) B cells in both Sh2b3E372K/E372K, Sh2b3R530Q/R530Q, and Sh2b3Δ/Δ mice were also decreased, with heterozygous Sh2b3E372K/+ and Sh2b3R530Q/+ displaying an intermediate phenotype (Fig. 3, G and H). This change was not due to a reduction of MZ B cell numbers (Fig. S2 E), but an increase in the other mature B cell subsets (Fig. S2, D and F). Analysis of 50:50 mixed BM chimeras revealed that the decrease in MZ B cells was cell-intrinsic (Fig. S2 H). This phenotype has not previously been reported in Sh2b3−/− mice, but the loss of MZ B cells has; although a decrease in MZ B cells has been observed in lupus-prone B6.Yaa mice, this was cell-extrinsic as a result of enhanced activation by antigen-bearing DCs (Santiago-Raber et al., 2010). We also examined CD21/35−CD23− mature B cells, a B cell subset enriched in atypical memory B cells (ABCs) that were found to be elevated in scenarios of chronic antigen stimulation such as patients with autoimmunity (Rubtsov et al., 2011; Wehr et al., 2004). There was a slight but not statistically significant cell-extrinsic increase of CD21/35−CD23− mature B cells in both Sh2b3E372K/E372K, Sh2b3R530Q/R530Q, and Sh2b3Δ/Δ mice (Fig. 3 I and Fig. S2 J). No difference in ABCs was observed (Fig. S2, K–M).
Figure S2.
Mature B cell phenotypes in Sh2b3Δ, Sh2b3E372K, and Sh2b3R530Q mice, and chimeras of Sh2b3E372K mice. (A) Frequencies of mature B cells as percentages of splenic B cells. (B) Total numbers of splenic mature B cells. (C) Frequencies of FO B cells as percentages of splenic B cells. (D) Total splenic FO B cells. (E and F) Total splenic MZ B cells (E) and CD21−CD23− mature B cells (F). Each dot represents one mouse. Sample numbers for each group in A–F are listed as follows: Sh2b3Δ panels: +/+ (n = 5), Δ/+ (n = 7), Δ/Δ (n = 6); Sh2b3E372K panels: +/+ (n = 4), E372K/+ (n = 3), E372K/E372K (n = 4); Sh2b3R530Q panels: +/+ (n = 5–11), R530Q/+ (n = 6–7), R530Q/R530Q (n = 6–9). Means are shown as bars. One-way ANOVA was used for statistical analysis. (G–J) CD45.2+/CD45.1+ ratios of frequencies of splenic mature B cells (G), MZ B cells (H), FO B cells (I), and CD21/35−CD23− mature B cells (J) in 50:50 BM chimeras of CD45.1-Sh2b3+/+ and CD45.2-Sh2b3+/+ (n = 10) or Sh2b3E372K/E372K (n = 10) mice. Statistical analysis was performed using Student’s t test. Results are representative of two independent experiments. (K) Representative flow cytometric plot of splenic ABCs (B220+CD21/35−CD23−CD11c+CD19+). (L and M) Frequencies (L) and total numbers (M) of splenic ABCs. Each dot represents one mouse. Sample numbers for each group in L and M are listed as follows: Sh2b3E372K panels: +/+ (n = 4), E372K/+ (n = 5), E372K/E372K (n = 4); Sh2b3R530Q panels: +/+ (n = 5–11), R530Q/+ (n = 7), R530Q/R530Q (n = 6). Results are representative of two to three independent experiments. Means are shown as bars. One-way ANOVA was used for statistical analysis of immunophenotyping data (A–F, L, and M), while Student’s t test was used for analyzing data from BM chimera experiments (G–J). Significance levels of one-way ANOVAs are indicated with asterisks, while those of Student’s t tests are indicated with hashes. Significance level criteria are indicated as follows: *: P < 0.05, **: P < 0.01, ***: P < 0.001, ####: P < 0.0001.
Phenotyping of the three human SLE patients with rare variants in SH2B3 (one carrying Q540X and two carrying R566Q) (Fig. 1 A) using our established protocols (Ellyard et al., 2019) revealed a higher frequency of transitional B cells compared with HCs, which was also observed in other SLE patients (Fig. 3 F) and reported previously (Sims et al., 2005). CD21−/lo B cells, a population also reported to be elevated in SLE patients (Wehr et al., 2004), showed a trend of increase in two SLE patients carrying rare SH2B3 variants and other SLE patients (Fig. 3 J).
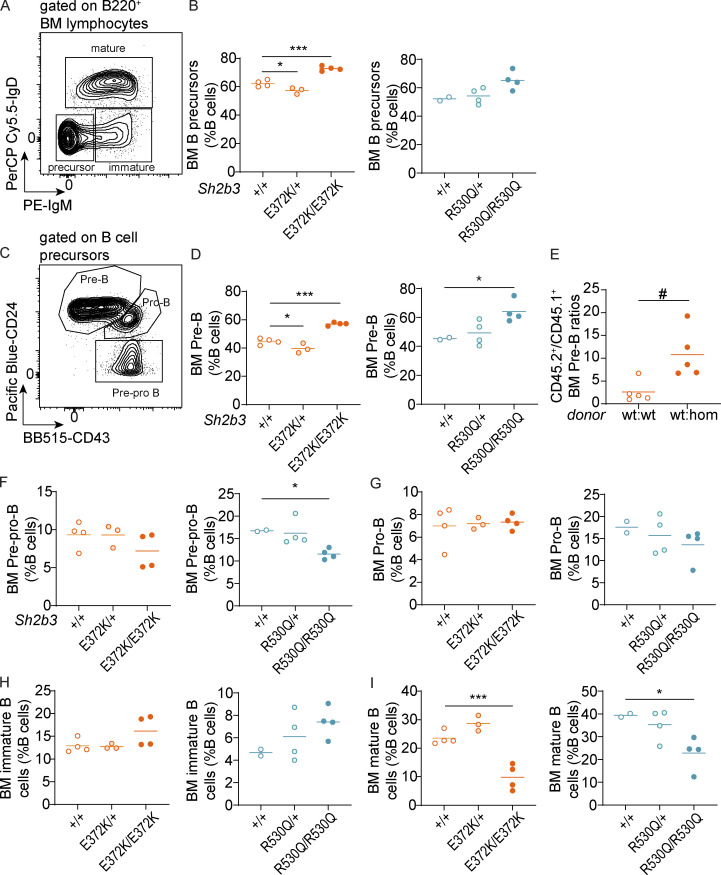
Having established a dysregulation in peripheral B cells, we investigated B cell development in the BM. Consistent with reports from Sh2b3−/− mice (Takaki et al., 2000), we observed a significant increase of B cell precursors as a percentage of BM B cells in Sh2b3E372K/E372K mice and a similar trend in Sh2b3R530Q/R530Q mice (P = 0.0536) (Fig. S3, A and B). Among B cell precursors, there was a cell-intrinsic increase of pre-B cell percentages in both strains (Fig. S3, C–E), but no consistent differences in pre-pro B or pro-B cells (Fig. S3, F and G). We also identified a trend of increased immature B cells (Fig. S3 H) and a significant decrease in mature B cells (Fig. S3 I). Our results are consistent with the published data on Sh2b3−/− mice and suggest that impaired negative regulation of growth factor/cytokine receptor signaling in BM B cells causes increased B cell generation and therefore accumulation in the BM.
Figure S3.
BM B cell phenotypes in Sh2b3E372K and Sh2b3R530Q mice. (A–I) BM B cell phenotypes. (A) Representative flow cytometric plot showing the gating of B cell precursors (IgM−IgD−), immature (IgM+IgD−), and mature (IgD+) B cells. Cells were pregated on B220+ lymphocytes. (B) Frequencies of B cell precursors as percentages of BM B cells. (C) Representative flow cytometric plot showing the gating of pre-pro (CD24−CD43+), pro- (CD24hiCD43+), and pre-B cells (CD24+CD43−/lo). Frequencies of pre-B cells as percentages of BM B cells in (D) Sh2b3E372K (left) and Sh2b3R530Q (right) mice and (E) 50:50 BM chimeras of CD45.1-Sh2b3+/+ and CD45.2-Sh2b3+/+/Sh2b3E372K/E372K mice (n = 5). Frequencies of (F) pre-pro and (G) pro-, (H) immature, and (I) mature cells as percentages of BM B cells. Lines in all dot plots show means, and results are representative of two independent experiments. Each dot represents one mouse, and sample numbers for each group in B, D, and F–M are listed as follows: Sh2b3E372K panels: +/+ (n = 4), E372K/+ (n = 3), E372K/E372K (n = 4); Sh2b3R530Q panels: +/+ (n = 2), R530Q/+ (n = 4), R530Q/R530Q (n = 4). One-way ANOVA was used for statistical analysis of immunophenotyping data (B, D, and F–M), and Student’s t test was used for analyzing data from BM chimera experiments (E). Significance levels of one-way ANOVAs are indicated with asterisks, while those of Student’s t tests are indicated with hashes. Significance level criteria are indicated as follows: */#: P < 0.05, ***: P < 0.001.
Increased T cell activation in Sh2b3E372K and Sh2b3R530Q mice
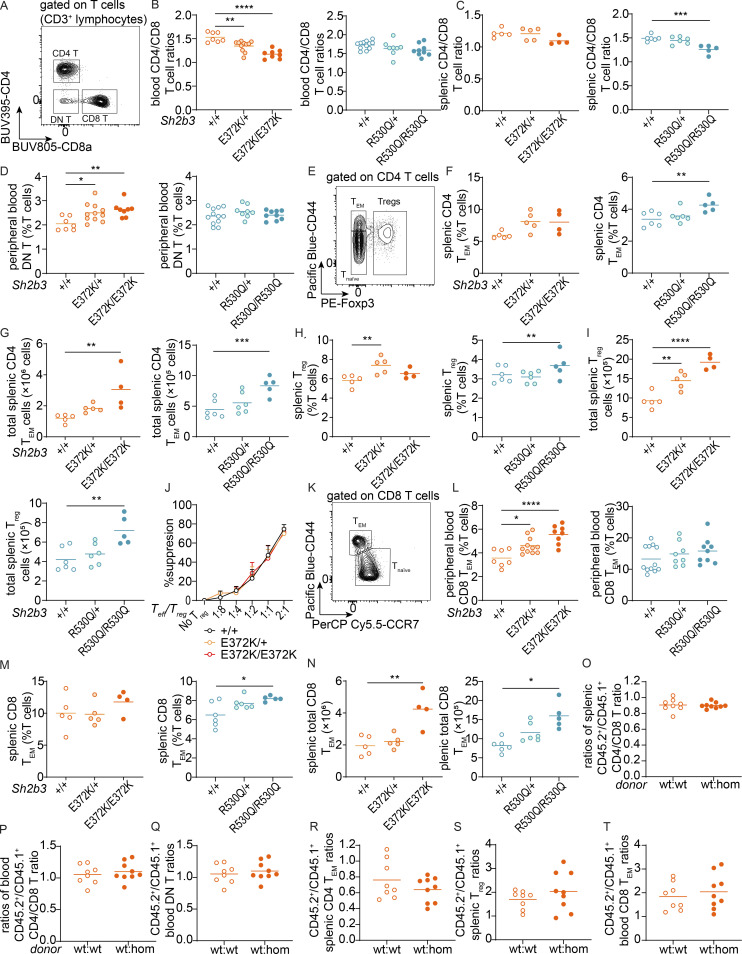
Studies in human cell lines have suggested that SH2B3 may play important roles in T cell activation by negatively regulating components of the TCR signaling pathway through interactions with PLCγ, GRB2, PI3K, and FLNA (He et al., 2000; Huang et al., 1995; Takaki et al., 1997), and indirectly repress T cell activation through cytokine receptor signaling including IL-7R and IL-15R in DCs (Katayama et al., 2014; Lawson et al., 2015). We observed decreased CD4/CD8 T cell ratios in the peripheral blood of Sh2b3E372K/+, Sh2b3E372K/E372K (Fig. S4, A and B), and spleens of Sh2b3R530Q/R530Q mice (Fig. S4 C), but not in the peripheral blood of Sh2b3R530Q mice (Fig. S4 B) and only a slight trend in the spleens of Sh2b3E372K/E372K mice (Fig. S4 C). Such phenotype has been reported among SLE patients (Maeda et al., 1999). Sh2b3E372K/+ and Sh2b3E372K/E372K, but not Sh2b3R530Q mice, also presented with increased percentages of double negative (DN: CD4−CD8−) T cells in peripheral blood (Fig. S4 D). This subset has also been found to be expanded in some SLE patients (Crispín et al., 2008), although it was not present in the SLE patients with SH2B3 rare variants (data not shown). Within CD4 T cells, we observed increased percentages and total numbers of effector memory CD4 T (CD4 TEM) cells and regulatory T (Treg) cells in the spleens of Sh2b3E372K/+ and Sh2b3R530Q/R530Q mice (Fig. S4, E–I). Treg suppressive function was normal (Fig. S4 J). Among CD8 T cells, we observed increased percentages of CD8 TEM cells in the peripheral blood of Sh2b3E372K/+ and Sh2b3E372K/E372K mice (Fig. S4, K and L) and the spleens of Sh2b3R530Q/R530Q mice (Fig. S4, M and N). Analysis of 50:50 mixed BM chimeras revealed that these T cell phenotypes were largely cell-extrinsic (Fig. S4, O–T). Together, our findings reveal a trend toward increased activated and/or effector T cells in mice carrying hypomorphic Sh2b3 variants.
Figure S4.
Peripheral blood and splenic T cell phenotypes in Sh2b3E372K and Sh2b3R530Q mice. (A) Representative flow cytometric plot showing the gating of double-negative (DN; CD4−CD8−), CD4+ (CD4+CD8−), and CD8+ (CD4−CD8+) T cells. (B and C) Dot plots showing the ratios of (B) blood and (C) splenic CD4/CD8 T cells in Sh2b3E372K mice and splenic CD4/CD8 T cells in Sh2b3R530Q mice. (D) Frequencies of DN T cells in the peripheral blood of Sh2b3E372K mice. (E) Representative flow cytometric plot showing the gating of CD4 naïve T (Tnaïve: CD44lo/−Foxp3−), TEM (CD44hiFoxp3−), and Treg (Foxp3+) cells. (F and G) Frequencies (F) and total numbers (G) of CD4 TEM cells in the spleens of Sh2b3E372K and Sh2b3R530Q mice. (H and I) Frequencies (H) and total numbers (I) of Treg cells in the spleen of Sh2b3E372K (left) and Sh2b3R530Q (right) mice. (J) Percentage suppression of effector T cells (Teff) by Treg cells in culture at various Teff/Treg ratios by using Treg cells sorted from the spleens of Sh2b3+/+ (n = 3), Sh2b3E372K/+ (n = 3), and Sh2b3E372K/E372K (n = 4) mice. (K) Representative flow cytometric plot showing the gating of CD8 Tnaïve (CD44lo/−CCR7lo) and TEM (CD44hiCCR7−) cells. (L and M) Frequencies of (L) peripheral blood and (M) splenic CD8 TEM cells in Sh2b3E372K and Sh2b3R530Q mice. (N) Total numbers of splenic CD8 TEM in Sh2b3E372K, and Sh2b3R530Q mice. (O–T) Ratios of CD45.2+/CD45.1− (O) splenic and (P) peripheral blood CD4/CD8 T ratios, (Q) peripheral blood DN T cells, (R) splenic TEM cells, (S) Treg cells, and (T) peripheral blood CD8 TEM cells in 50:50 BM chimeras of CD45.1-Sh2b3+/+ and CD45.2-Sh2b3+/+/Sh2b3E372K/E372K mice (n = 8). Results in B–D, F–J, L–N, and S are representative of two independent experiments. Results in O–R and T are from a single experiment. Each dot represents one mouse, and sample numbers for each group in B–D, F–J, and L–N are listed as follows: Sh2b3E372K panels: +/+ (spleen: n = 5, blood: n = 7), E372K/+ (spleen: n = 5, blood: n = 11), E372K/E372K (spleen: n = 4, blood: n = 9); Sh2b3R530Q panels: +/+ (spleen: n = 6, blood: n = 12), R530Q/+ (spleen: n = 6, blood: n = 8), R530Q/R530Q (spleen: n = 5, blood: n = 9). One-way ANOVA was used for statistical analysis of immunophenotyping data (B–D, F–I, and L-N), while Student’s t test was used for analyzing data from BM chimera experiments (O–T). Significance levels of one-way ANOVAs are indicated with asterisks as follows: *: P < 0.05, **: P < 0.01, ***: P < 0.001, ****: P < 0.0001.
Sensitized Sh2b3E372K mice manifest accelerated autoimmunity
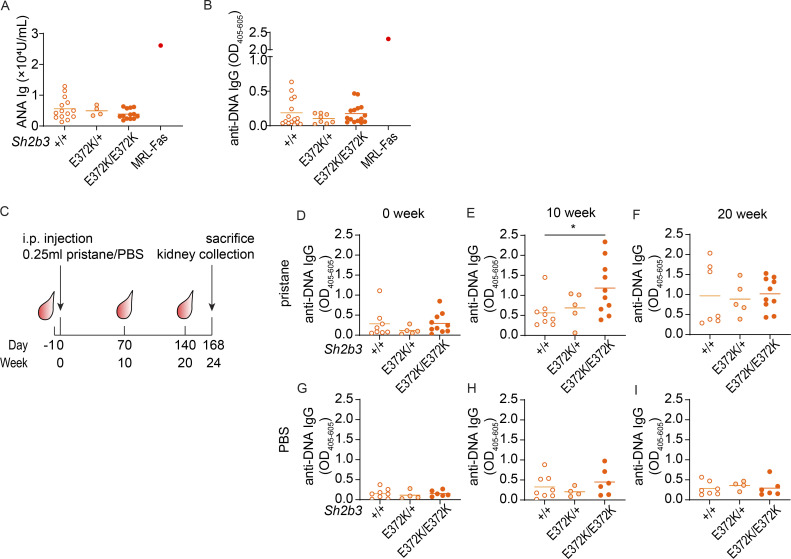
Having demonstrated that SH2B3 variants identified in SLE patients are hypomorphic alleles resulting in immune dysregulation, we next asked whether they confer susceptibility to autoimmunity. A hallmark of SLE is the production of ANAs, particularly against double-stranded DNA. We analyzed levels of ANAs and anti-DNA antibodies in the sera of Sh2b3E372K mice but found no substantial differences compared with WT littermate controls (Fig. 4, A and B). Hence, our results indicate that Sh2b3E372K mice do not develop autoimmunity spontaneously.
Figure 4.
Autoimmune triggers induced exacerbated autoimmunity in Sh2b3E372K mice. (A and B) Serum ANAs (A; +/+: n = 14, E372K/+: n = 4, E372K/E372K: n = 13) and anti-DNA IgG (B; +/+: n = 15, E372K/+: n = 8, E372K/E372K: n = 15) determined by ELISA. One-way ANOVA was used for statistical analyses. Results are pooled from two to four separate mouse cohorts and representative to two independent experiments. (C) Experimental plan of pristane-induced lupus-like autoimmune model in Sh2b3E372K mice. 14-wk-old female Sh2b3E372K mice were bled 1 day before the IP injection of 0.25-ml pristane or PBS (control) and later 2-, 10-, and 20-wk after injection and sacrificed 24-wk after injection for spleen and kidney analysis. (D–I) Levels of serum anti-DNA IgG in pristane-treated mice at 0 (D), 10 (E), and 20 (F) weeks after treatment, and PBS-treated mice at 0 (G), 10 (H), and 20 (I) weeks after treatment. Means are shown as bars in all dot plots. Data were pooled from two independent experiments, +/+: n = 8, E372K: n = 5, E372K/E372K: n = 11. Statistical analyses performed using lmer ANOVA; *: P < 0.05.
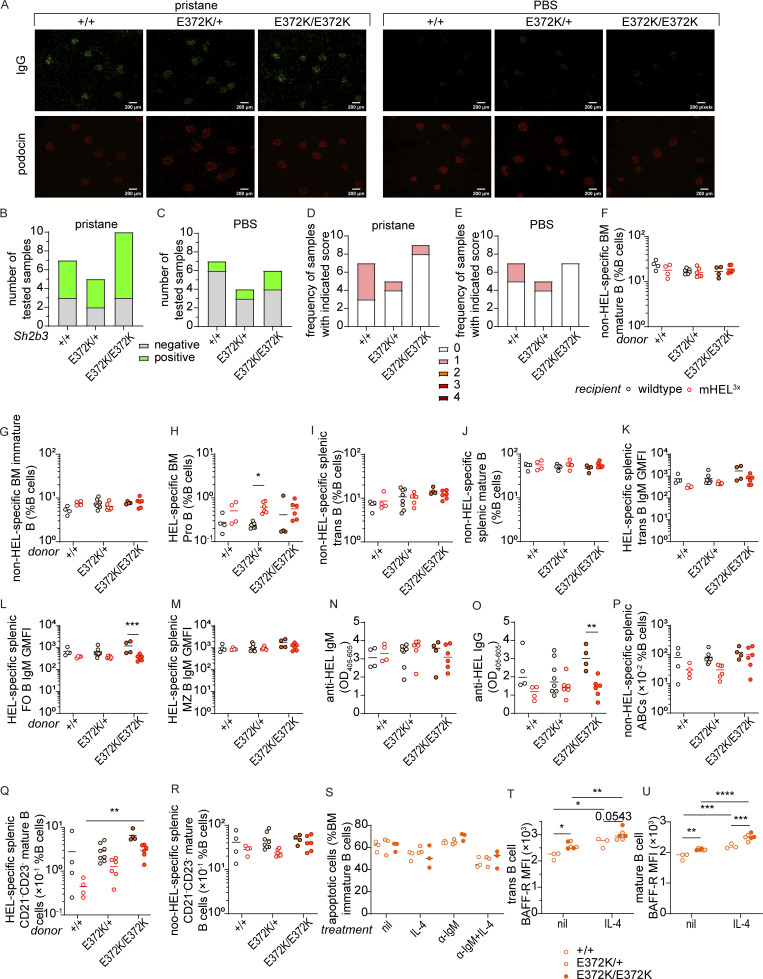
We then considered whether autoimmunity may be accelerated or more severe in a sensitized background. Pristane, a hydrocarbon compound derived from shark liver oil and mineral oil (Ackman, 1971), also known as 2,6,10,14-tetramethylpentadecane, is a well-established inducer of lupus-like disease when administrated intraperitoneally (IP) in mice (Reeves et al., 2009). We injected 12-wk-old Sh2b3E372K mice IP with either pristane or PBS control and monitored the development of anti-DNA autoantibodies in the serum (Fig. 4 C). Sh2b3E372K/E372K mice developed significantly higher titers of anti-DNA IgG 10 wk after pristane treatment (Fig. 4, D and E). However, this difference was lost by 20 wk when pristane-treated Sh2b3+/+, Sh2b3E372K/+, and Sh2b3E372K/E372K mice all displayed similar titers of anti-DNA autoantibodies (Fig. 4 F). Pristane treatment also caused IgG IC deposition in the glomeruli of mice at 24 wk (Fig. S5, A and B), with some mice also displaying minimal glomerular leukocyte infiltration (score 1 out of 4) (Fig. S5 D). We did not observe an increase in proteinuria in pristane-treated mice compared with the PBS treatment group (data not shown). Anti-DNA IgG titers did not change significantly over time in PBS-treated control mice regardless of genotype (Fig. 4, G–I) with few mice displaying glomerular IgG IC deposition (Fig. S5, A and C) or minimal glomerular infiltration (Fig. S5 E).
Figure S5.
Spontaneous autoimmunity and B cell tolerance in Sh2b3E372K mice. (A–C) IgG immune-complex deposits in mice treated with PBS or pristane. (A) Representative IF images showing IgG (green) and podocin (red) staining in the kidney sections of pristane/PBS-treated Sh2b3E372K mice 20 wk following treatment with pristane and PBS. (B and C) Numbers of kidney sections that are positive and negative for IgG ICs in the glomeruli of Sh2b3E372K mice treated with (B) pristane (+/+: n = 7, E372K/+: n = 5, E372K/E372K: n = 10) and (C) PBS (+/+: n = 7, E372K/+: n = 4, E372K/E372K: n = 6). (D and E) Numbers of kidney sections with indicated glomerular score 20 wk following treatment with (D) pristane (+/+: n = 7, E372K/+: n = 5, E372K/E372K: n = 9) and (E) PBS (+/+: n = 7, E372K/+: n = 5, E372K/E372K: n = 7). (F–R) Cellular and serological phenotypes of SWHEL-mHEL3× chimeric mice. Mouse numbers for each group are listed as follows: +/+ WT (n = 4), +/+ mHEL3× (n = 4), E372K/+ WT (n = 8), E273K/+ mHEL3× (n = 6), E372K/E372K WT (n = 4), and E372K/E372K mHEL3× (n = 6). Frequencies of non-HEL-specific BM (F) mature and (G) immature B cells, (H) HEL-specific pro-B cells, non-HEL-specific splenic (I) transitional and (J) mature B cells, relative surface IgM expression on HEL-specific splenic (K) transitional, (L) FO, and (M) MZ B cells in WT (black circles) or mHEL3× (red circles) recipients receiving BM from SWHEL-Sh2b3+/+ (unfilled), SWHEL-Sh2b3E372K/+ (yellow filling), or SWHEL-Sh2b3E372K/E372K (orange filling) donors. Levels of anti-HEL (N) IgM and (O) IgG in WT and mHEL3× mice receiving BM from SWHEL-Sh2b3+/+, -Sh2b3E372K/+ and, -Sh2b3E372K/E372K donors measured as OD405–605 by ELISA. Frequencies of (P) non-HEL-specific ABCs, (Q) HEL-specific and (R) non-HEL-specific CD21−CD23− mature B cells as percentages of splenic lymphocytes. (S) Percentages of apoptotic BM immature B cells in untreated, 20 ng/ml IL-4 only, 5 μg/ml anti-IgM (α-IgM) only, and 5 μg/ml α-IgM + 20 ng/ml IL-4 conditions (n = 3). (T and U) BAFF-R surface expression on (T) transitional and (U) mature B cells measured by flow cytometry as median fluorescence intensity (MFI) (n = 3). Data in A–E is pooled from two independent experiments. Results in F–U are representative of two to three independent experiments. Fisher’s exact test was used for the statistical analyses in B–E. Means in F–U are shown as bars, and two-way ANOVA was used for statistical analysis in F–U. Significance levels for two-way ANOVAs are indicated with asterisks. *: P < 0.05, **: P < 0.01, ***: P < 0.001, ****: P < 0.0001.
Decreased SH2B3 function compromises B cell tolerance at multiple immune checkpoints
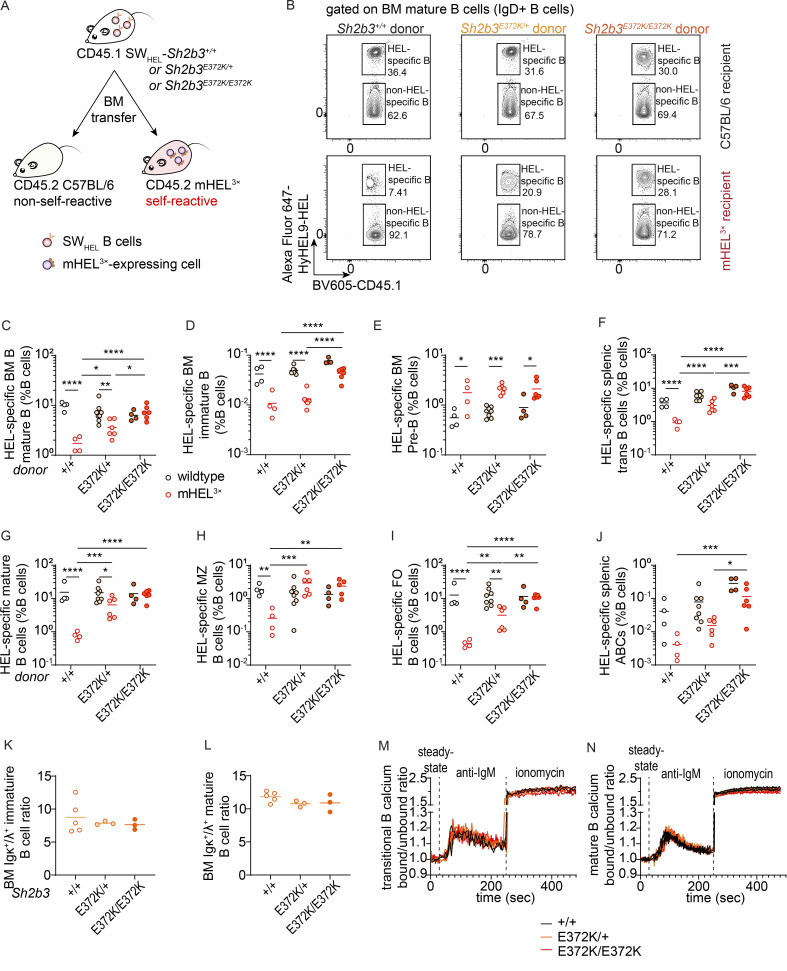
The fact that sensitizing Sh2b3E372K/E372K mice led to accelerated development of autoantibodies together with an increase in immature/transitional B cells that are known to harbor autoreactivity (Wardemann et al., 2003) suggests that hypomorphic Sh2b3 variants promote the escape of autoreactive B cells. To examine this, we took advantage of the SWHEL-mHEL3× mixed chimera model, in which BM with B cells expressing transgenic immunoglobulin heavy and light chains specific for hen egg lysozyme (HEL) from SWHEL mice is engrafted into irradiated mHEL3× mice that express HEL as a membrane-bound self-antigen (Burnett et al., 2018). We crossed the Sh2b3E372K mice to SWHEL mice (hereinafter referred to as SWHEL-Sh2b3E372K mice) and transplanted BM cells from these crosses into irradiated WT (non-self-reactive) or mHEL3× mice (self-reactive) recipients (Fig. 5 A). Consistent with previous reports (Lau et al., 2019), we observed an 80–90% decrease in the frequency of HEL-specific mature Sh2b3+/+ (WT) B cells in the BM of mHEL3× recipients compared with WT recipients, evidence that autoreactive HEL-binding B cells are purged from the recirculating B cell repertoire (Fig. 5, B and C). In contrast, no decrease in self-reactive B cells was observed in recipients of HEL-binding Sh2b3E372K/E372K BM, suggesting that reduced SH2B3 function prevents the removal of autoreactive B cells. Interestingly, there appeared to be a partial negative selection of HEL-binding Sh2b3E372K/+ B cells, indicating an intermediate phenotype. No difference was seen in non-HEL binding mature B cells, indicating that the purging of WT B cells observed in HEL3× recipients is antigen specific (Fig. S5 F).
Figure 5.
B cell tolerance breach in Sh2b3E372K mice. (A) Design of SWHEL-mHEL3× BM chimera experiment for assessing B cell tolerance in Sh2b3+/+, Sh2b3E372K/+, and Sh2b3E372K/E372K mice. (B) Representative flow cytometric plots showing the gating of HEL-specific (self-reactive) and non-HEL-specific (non-self-reactive) BM mature B cells in WT or mHEL3× mice receiving BM from SWHEL-Sh2b3+/+, SWHEL-Sh2b3E372K/+, or SWHEL-Sh2b3E372K/E372K donors. (C–J) Dot plot demonstrating the frequencies of HEL-specific (C) BM mature B cells (D) BM immature B cells, (E) BM pre-B cells, (F) splenic transitional B cells, (G) splenic mature B cells, and (H) splenic MZ B cells, (I) splenic FO B cells, and (J) splenic ABCs as percentages of total lymphocytes in WT or mHEL3× recipient mice receiving BM from SWHEL-Sh2b3+/+, SWHEL-Sh2b3E372K/+, or SWHEL-Sh2b3E372K/E372K mice. +/+ WT (n = 4), +/+ mHEL3× (n = 4), E372K/+ WT (n = 8), E372K/+ mHEL3× (n = 6), E372K/E372K WT (n = 4), E372K/E372K mHEL3× (n = 6). Means are shown as bars. Results are representative of two independent experiments. Two-way ANOVA was performed on log10-transformed data to meet the variance homoscedasticity; *: P < 0.05, **: P < 0.01, ***: P < 0.001, ****: P < 0.0001. (K–L) Ratios of BM Igκ+/Igλ+ (K) immature and (L) mature B cells in Sh2b3E372K mice (+/+: n = 5, E372K/+: n = 3, E372K/E372K: n = 3). Means are shown as bars in C–L. One-way ANOVA was used for statistical analysis. Results are representative of two independent experiments. (M and N) Calcium influx into the cytoplasm of peripheral blood (M) transitional and (N) mature B cells of Sh2b3E372K mice (+/+: n = 2, E372K/+: n = 3, E372K/E372K: n = 3) at steady state, following treatment with anti-IgM antibody and treatment with ionomycin measured by the ratios of bound/unbound indo-1 AM ester to Ca2+. Results are representative of two independent experiments.
To determine at which immune checkpoint tolerance is initially breached, we first looked at B cells developing in the BM. Autoreactive B cell clones are typically removed through negative selection at the pre-B (the first stage in B cell development at which pre-BCR are expressed on most B cells) to the immature development stage. Similar to mature B cells, there was a failure to delete immature Sh2b3E372K/E372K HEL-specific B cells in HEL3× recipients, although this was not observed for B cells heterozygous for the Sh2b3E372K allele (Fig. 5 D). In contrast, HEL-specific pre-B cells were not deleted in mHEL3× recipients, consistent with these cells not having undergone negative selection (Fig. 5 E), and, in fact, we observed an increase in the mHEL3× autoreactive environment regardless of Sh2b3 genotype.
We next examined immune tolerance checkpoints in the spleen, where autoreactive transitional B cells are negatively selected and inhibited from entering the mature follicular (FO) B cell pool. We observed a failure to remove HEL-specific transitional Sh2b3E372K/E372K B cells in mHEL3× recipients (Fig. 5 F), with heterozygous Sh2b3E372K/+ B cells leading to an intermediate phenotype. This same enrichment in self-reactive B cells was also observed amongst HEL-specific MZ and FO B cells (Fig. 5, G–I) in mHEL3× mice receiving SWHEL-Sh2b3E372K/E372K or Sh2b3E372K/+ BM, whereas these compartments were significantly devoid of self-reactive B cells in mHEL3× mice receiving SWHEL-Sh2b3+/+ BM. This indicates a specific defect in the tolerance checkpoint between splenic transitional and mature B cells. Downregulation of IgM surface expression is an indicator of recent BCR engagement and thus often used as a measure of B cell anergy (Goodnow et al., 1989), although it can also indicate activation. Consistently, IgM expression in the chimeras was decreased on HEL-specific Sh2b3+/+ transitional and FO B cells in mHEL3× recipient mice compared with WT recipients (Fig. S5, K and L). Similarly, IgM was significantly downregulated in HEL-specific Sh2b3E372K/E372K FO B cells in the mHEL3× recipient environment and IgM downregulation on transitional Sh2b3E372K/E372K B cells was less affected. Interestingly, as previously noted (Lau et al., 2019), an autoreactive environment does not appear to modulate IgM expression in MZ B cells regardless of genotype (Fig. S5 M).
Given this escape of autoreactive Sh2b3E372K/E372K B cells into the mature recirculating pool, we next examined whether they were also activated in the periphery to become an autoantibody-secreting cell, an additional checkpoint in B cell tolerance. Initially, we examined the generation of HEL-specific antibodies. We did not observe an increase in serum HEL-specific IgM antibodies in the autoreactive mHEL3× background (Fig. S5 N), whilst Sh2b3E372K/E372K B cells produced less HEL-specific IgG antibodies in mHEL 3× mice compared with WT recipients (Fig. S5 O). Interestingly, we did observe a propensity for Sh2b3E372K/E372K B cells to differentiate ABCs that can be activated to produce pathogenic autoantibodies (Rubtsov et al., 2011; Wang et al., 2018). Whilst, Sh2b3+/+ and Sh2b3E372K/+ B cells appeared to be prevented from differentiating into ABCs in the mHEL3× recipient environment, Sh2b3E372K/E372K B cells gave rise to a higher frequency of HEL-specific ABCs, suggesting they may not be as effectively removed (Fig. 5 J). A similar trend was observed for the parent population CD21−CD23− mature B cells (Fig. S5 Q). However, the amount of HEL-specific ABC or CD21−CD23− mature B cells was generally low compared with non-HEL-specific populations (Fig. S5, P and R).
To determine whether changes in receptor editing in SH2B3 hypomorphic B cells may contribute to the breach of central tolerance, we stained for immunoglobulin κ and λ light chains in BM B cells from Sh2b3+/+, Sh2b3E372K/+, and Sh2b3E372K/E372K mice but did not observe any difference in the Igκ/Igλ ratios between genotypes for either immature or mature B cells (Fig. 5, K and L). Changes in the BCR signaling strength may also influence B cell negative selection (Lam et al., 1997; Nemazee and Bürki, 1989). We performed a calcium flux assay to determine if the E372K allele could regulate BCR signal strength but observed no difference in the level of calcium flux for Sh2b3E372K/+ or Sh2b3E372K/E372K transitional or mature B cells compared with Sh2b3+/+ controls (Fig. 5, M and N).
Sh2b3E372K B cells are hyperresponsive to IL-4–induced rescue
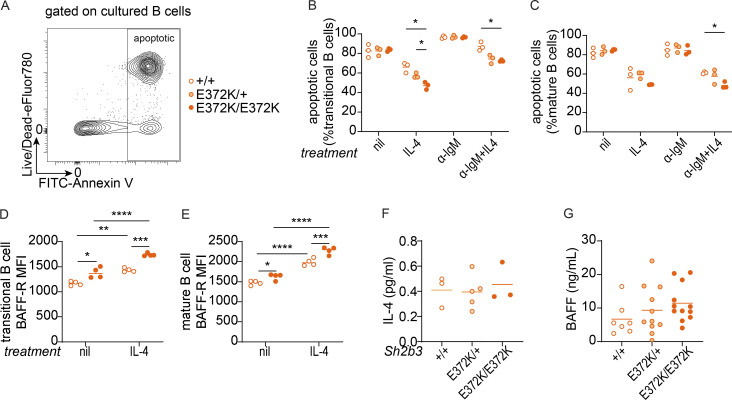
IL-4 is known to exert anti-apoptotic effects on B cells during BCR-mediated negative selection (Granato et al., 2014). Since IL-4R signaling is in part transduced by JAK3 (Malabarba et al., 1995), which is regulated by SH2B3, we examined whether hypomorphic SH2B3 altered B cell apoptosis following BCR ligation. Treating transitional or mature B cells with IL-4 increased the survival of B cells homozygous or heterozygous for the E372K allele to a greater extent than WT cells (Fig. 6, A–C). In contrast, IL-4 did not increase the survival of immature B cells isolated from BM (Fig. S5 S). Thus, augmented IL-4R signaling may contribute to defective negative selection in the spleen and thus the breach in B cell tolerance seen in Sh2b3E372K mice.
Figure 6.
IL-4R and BAFF-R signaling in the survival in IgM-crosslinked splenic B cells of Sh2b3+/+, Sh2b3E372K/+, and Sh2b3E372K/E372K mice. (A) Representative flow cytometry plot showing the gating strategy of apoptotic (Annexin+7-AAD−/+) cultured, sorted splenic B cells. (B and C) Frequencies of apoptotic transitional (B) and mature B cells (C) in untreated, 20 ng/ml IL-4 only, 5 μg/ml anti-IgM (α-IgM) only, and 5 μg/ml α-IgM + 20 ng/ml IL-4 conditions (n = 3 in each group). Two-way ANOVA with Šídák’s adjustment was used for statistical analyses; *: P < 0.05. (D and E) BAFF-R expression was measured by GMFI in transitional (D) and mature (E) B cells in untreated or 20 ng/ml IL-4 conditions (n = 4 in each group). Two-way ANOVA with Šídák’s adjustment was used for statistical analyses. *: P < 0.05, **: P < 0.01, ***: P < 0.001, ****: P < 0.0001. Results are representative of three independent experiments. (F) Plasma IL-4 measured by Meso Scale Discovery assay (+/+: n = 3, E372K/+: n = 5, E372K/E372K: n = 3). One-way ANOVA was used for statistical analyses. Results are from a single experiment. (G) Serum BAFF measured by bead-based immunoassay (+/+: n = 7, E372/+: n = 11, E372K/E372K: n = 12). Results are pooled from two independent experiments. One-way ANOVA was used for statistical analyses.
Another factor important for transitional B cell survival is BAFF-R signaling (Batten et al., 2000; Schiemann et al., 2001). We examined BAFF-R expression on splenic B cells following in vitro culture of transitional and mature B cells with and without IL-4 and were surprised to observe an increase in BAFFR expression (Fig. 6, D and E; and Fig. S5, T and U). This effect appeared to be greater in Sh2b3E372K/E372K and Sh2b3E372K/+ cells, providing a possible mechanistic link between IL-4 signaling and accumulation of immature/transitional B cells in Sh2b3E372K/E372K and Sh2b3E372K/+ mice. Serum concentrations of IL-4 and BAFF in Sh2b3E372K/E372K and Sh2b3E372K/+ were similar to Sh2b3+/+ (Fig. 6, F and G), suggesting that cellular phenotypes and the breach in tolerance are driven by receptor hyperresponsiveness or expression rather than an increase in available cytokines or growth factors.
Discussion
In conjunction with previous work (Jiang et al., 2019), this study demonstrates that rare functional variants occurring in GWAS-associated SLE genes may contribute to disease pathogenesis. Often, the strongest evidence for an association between a particular gene and a disease comes from GWAS—with many monogenic, disease-causing mutations occurring in genes that have also been identified by well-powered common variant associations. SH2B3 has already been robustly linked to SLE by GWAS with the lead common variant (rs10774625) tagging a haplotype block that encompasses the whole of SH2B3, being a strong expression quantitative trait loci (Alcina et al., 2010; Bentham et al., 2015; Morris et al., 2016; Wang et al., 2021), and being in linkage with a relatively common missense SH2B3 variant, rs3184504 (r2 = 0.92), encoding SH2B3 W262R. Here, we add weight to the link between SH2B3 and SLE pathogenesis by showing that SLE patient–specific novel and ultrarare variants in SH2B3 encode proteins with impaired function leading to dysregulated B cell development and tolerance that predisposes carriers to autoimmunity. The presence of the hypomorphic allele alone is insufficient to cause disease, consistent with occurrence in some unaffected family members. Instead, clinical disease is likely to require additional environmental or genetic triggers.
Five out of the six rare patient-specific variants impaired the ability of SH2B3 to suppress IFNGR signaling. Interestingly, the patient (D.II.1) who carried the A536T variant that retained normal suppressive function was also compound heterozygous for ACP5 (Table S2), which is a known cause of spondyloenchondrodysplasia with immune dysregulation that shares features with SLE. In contrast, the ADAR variant found in patient A.II.1 is unlikely to be sufficient to cause SLE in the heterozygous state, and there may be an additional role for SH2B3. Unlike the majority of SLE variants, both the R43C variant found exclusively in the HCs and the majority of more common SH2B3 variants (MAF > 0.001) did not impair the negative regulatory function of SH2B3. The exception to this was T165S. According to the gnomAD database, the global MAF for the allele that encodes this p.T165S variant is low frequency (MAF = 0.019), although the frequency is considerably higher (MAF = 0.356) in the African/African American genetic ancestry group. Interestingly, individuals of African ancestry in America have an increase in the incidence of SLE compared with those of Caucasian ancestry (Izmirly et al., 2021). Within our SLE patient cohort (Jiang et al., 2019), we identified one proband who was heterozygous for the T165S variant. The computed ethnic ancestry of this individual was African. We did not identify this variant in genomes from our 97 HCs.
Through structural studies, we showed that the substitution of key residues responsible for forming the hydrogen-bonding network of the BC loop in the SH2 domain can result in reduced binding to target pTyr and domain instability. Indeed, our previous work on the SH2 domain of SH2B3 revealed similar examples of functional impairment, with V402M affecting more severely the binding to pTyr and inhibition of IFNGR signaling (Morris et al., 2021). Nevertheless, we were only able to cocrystallize the murine SH2 domain of SH2B3 with pY813 JAK2 as the expression of human SH2B3 was unsuccessful. In addition, we showed that patient-specific variants outside known functional domains (C133Y and R566Q) also impaired SH2B3 protein function. How these variants affect protein function will require future study on unannotated regions of the SH2B3 protein.
Using the SWHEL-mHEL3× mouse model, we demonstrated an incomplete negative selection of self-reactive Sh2b3E372K B cells at multiple immune checkpoints. This tolerance breach is also likely to be applicable in a non-transgenic setting since 55–75% of immature B cells have been shown to harbor autoreactivity in healthy individuals (Wardemann et al., 2003). In most individuals these autoreactive B cells are removed through tolerance checkpoint mechanisms, but this has been shown to be impaired in individuals with SLE (Yurasov et al., 2005). While evasion of normal tolerance mechanisms was not sufficient to cause spontaneous autoimmunity in Sh2b3E372K/E372K mutant mice, sensitization with pristane treatment led to accelerated autoantibody production, suggesting that autoimmunity may occur in the presence of additional triggers.
Our data suggest that elevated IL-4R signaling and BAFF-R expression may be in part responsible for the impaired negative selection in the periphery. Engagement of BCR on developing B cells can either induce negative selection or facilitate B cell maturation depending on the strength of BCR signal, as well as positive signals from other receptors including cytokine receptors, such as IL-4R and BAFF-R (Granato et al., 2014). Signaling through IL-4R provides pro-survival signals to BM immature and splenic transitional B cells by impeding the upregulation of Bim following BCR engagement (Granato et al., 2014). Consistently, we observed that IL-4 treatment of splenic transitional and mature WT B cells could rescue them from apoptosis following IgM-crosslinking, while the hyperresponsiveness of Sh2b3E372K/E372K B cells to IL-4 signaling appears to provide autoreactive B cells with higher survival potential.
The binding of BAFF to BAFF-R supports the differentiation of T1 into T2 B cells and T2 B cell survival, as well as their further differentiation into mature naïve B cells and subsequent maintenance (Sasaki et al., 2004). Anergic autoreactive T1 B cells are more dependent on BAFF signaling due to elevated Bim expression and thus compete less well for selection into the recirculating follicular pool compared with non-autoreactive T1 B cells (Lesley et al., 2004). Therefore, enhanced IL-4R signaling and subsequent restrained Bim expression in autoreactive Sh2b3E372K/E372K B cells may mean they are not as dependent on BAFF as their WT counterparts. Unexpectedly, we also observed the elevation of surface BAFF-R expression on transitional B cells in Sh2b3E372K/E372K mice in cell culture, which may further increase their competitive survival advantage in vivo.
Interestingly, we have found that IL-4 treatment can upregulate BAFF-R expression in both splenic transitional and mature B cells. In contrast, IL-4 treatment has been reported to have no effect on BAFF-R expression on CD23+ immature B cells isolated from the BM (Granato et al., 2014), which may suggest that splenic transitional B cells respond differently to those in the BM. Regardless, our observations suggest that heightened IL-4R signaling in Sh2b3E372K/+ and Sh2b3E372K/E372K mice may lead to increased BAFF-R expression and contribute to the breach of immune tolerance checkpoints. Notably, BAFF was not higher in the serum of Sh2b3E372K/+ or Sh2b3E372K/E372K mice, suggesting that any survival advantage is not a result of increased bioavailability of BAFF.
Increases in BAFF-R and IL-4R are unlikely to be the sole mechanisms that increase autoimmune susceptibility in carriers of SH2B3 hypomorphic variants; we showed that IL-4 did not promote the survival of immature B cells isolated from BM. Previous studies have suggested that the overproduction of immature B cells in Sh2b3−/− mice is a consequence of dysregulated IL-7R and c-KIT signaling (Cheng et al., 2016; Takaki et al., 2000). Unrestrained IL-7R signaling promotes B cell precursor proliferation leading to increased B cell production in humans (Parrish et al., 2009), and gain-of-function IL-7R mutations increase peripheral B cell numbers and contribute to precursor B cell ALL (Almeida et al., 2021; Shochat et al., 2011). Other cytokine responses that may be enhanced in the absence of SH2B3 and also impair central and/or peripheral tolerance include IL-2 and IL-15, both of which promote the proliferation of activated B cells via IL-2R signaling (Armitage et al., 1995; Mingari et al., 1984). Additional candidates are IL-3, which support the proliferation of murine (Palacios et al., 1984) and human B cell precursors (Crooks et al., 2000), as well as the survival and maturation of plasmacytoid DCs; IL-6, which drives pristane-induced anti-DNA IgG and facilitates spontaneous germinal centers (GCs) in autoimmune models (Richards et al., 1998); and IFN-γ, which induces T-bet expression in B cells and drives spontaneous GC formation in autoimmune models (Domeier et al., 2016; Jackson et al., 2016) and ABC formation (Naradikian et al., 2016; Peng et al., 2002).
Contrary to the leukopenia and lymphopenia observed in some SLE patients (Rivero et al., 1978), our SH2B3 mutant mouse strains exhibited increased lymphocytes. This is perhaps unsurprising since lymphopenia is usually related to disease activity in SLE (Lu et al., 2021; Sapartini et al., 2018), and mice with hypomorphic Sh2b3 mutations do not spontaneously develop an autoimmune phenotype. Lymphocytosis is also observed in individuals with compromised lymphocyte apoptosis such as in autoimmune lymphoproliferative syndrome patients and in the corresponding Faslpr lupus-like mouse model (Adachi et al., 1996; Völkl et al., 2016). Our studies showed that hypomorphic Sh2b3 can protect B cells from apoptosis in vitro, possibly preventing lymphopenia. Clinical investigations (Table S1) indicated that only two of the seven SLE patients with SH2B3 mutations in this study displayed lymphopenia.
Taken together, our study uncovers a role for SH2B3 in mouse and human B cell tolerance due to negative regulation of cytokine receptor signaling. It also provides a rationale for the use of JAK inhibitors (Jakinibs) already approved for several autoimmune conditions and currently in trials for SLE (Tanaka et al., 2022). The convergence between IL-4R signaling and BAFF-R expression in promoting B cell survival suggests that Jakinib therapy may not only restrain aberrant JAK-STAT signaling but potentially dampen BAFF-R expression, limiting aberrant selection and/or survival of autoreactive B cells. Overall, this study highlights the importance of studying rare variants in SLE patients to understand mechanisms that drive or contribute to the loss of tolerance and disease pathogenesis, which may be targeted to prevent autoimmune development.
Materials and methods
Site-directed mutagenesis (SDM)
Primer pairs for SDM were designed using Agilent’s primer design program (https://www.agilent.com/store/primerDesignProgram.jsp) following QuikChange XL II kit recommendations and ordered as 25 nmole DNA oligos (Table S4) from Integrated DNA Technologies. The oligos were resuspended at 100 μM in UltraPure DNase/RNase-free distilled water and then reconstituted to a working concentration of 125 ng/μl. DNA templates were reconstituted at 50 ng/μl. The reagents were mixed as per the QuikChange II XL Site-Directed Mutagenesis Kit instructions and loaded into a Mastercycler pro S (Eppendorf) with the settings recommended by the manual. After PCR, 1 μl of DpnI restriction endonuclease (cat # R0176L; New England Biolabs) was added to each reaction tube to digest DNA templates. The resulting products were transformed into DH5α competent Escherichia coli cells for amplification, Sanger sequencing, and selection.
Sanger sequencing
To prepare primers for Sanger sequencing, DNA oligos (Table S5; Integrated DNA Technologies) were resuspended at 100 μM and then further diluted into 1.6 pmol/μl (μM). Plasmid DNA templates were diluted to 250 ng/μl. The following reagents were mixed in each well of a 96-well PCR plate (cat # AB-0600; Thermo Fisher Scientific): 3.5 μl of 5× sequencing buffer, 1 μl BigDye Terminator 3.1 (cat # 4337455; Applied Biosystems), 1.6 pmol/μl sequencing primer, 5% (vol/vol) DMSO, if the sequence was GC-rich, and UltraPure DNase/RNase-free distilled water (cat # 10977015; Invitrogen) to a final volume of 20 μl. Thermocycling was performed with the following cycling conditions: 1 cycle of initial denaturation at 94°C for 5 min, 30 cycles of denaturation (96°C, 10 sec), annealing (50°C, 5 s), and extension (60°C, 4 min), and hold at 4°C when all steps were complete. The resulting amplicons were purified using ethanol and EDTA before undergoing capillary sequencing at the Biomolecular Resource Facility, John Curtin School of Medical Research (JCSMR). Results were analyzed using Sequencher (Gene Codes).
Cell culture and luciferase assay
HEK293 cells (CRL-1573; ATCC) and HEK293T cells (CRL-3216; ATCC) were cultured in complete DMEM (cDMEM) in Nunc EasYFlask T75 cell-culture flasks (cat # 156499; Thermo Fisher Scientific) and housed in CO2 incubator (5% CO2) at 37°C, with passaging every 3 days by a 1 in 10 split. All cell lines tested negative for mycoplasma contamination using PlasmoTest Mycoplasma Detection Kit (cat # rep-pt1; InvivoGen).
To set up transfection for luciferase assays, cells seeded in 24-well plates were transfected with STAT1BS-Firefly (145 ng), pRL-CMV-Renilla (5 ng), and pReceiver-M12-SH2B3 (100 ng for HEK293 cells and 1 ng for HEK293T cells) plasmids using Lipofectamine 2000 (cat # 11668019; Invitrogen) resuspended in Opti-MEM (cat # 31985070; Gibco) following the manufacturer’s instructions. The STAT1BS-GAS Firefly luciferase reporter is a gift from V. Athanasopoulos. Transfected cells were incubated for 24 h before treatment with 50 ng/ml recombinant human IFN-γ (cat # 300-02; Peprotech) or cDMEM-only control for 24 h. Cell lysates were analyzed with Luc-Pair Duo-Luciferase assay kit (cat # LF003; GeneCopoeia) using a VICTOR Nivo multimode microplate reader (cat # HH35000500; PerkinElmer).
Expression and purification of LNK and SH2B SH2 domains
DNA encoding the mouse E372K and WT SH2B3 SH2 domain (residues 324–446) and an N-terminal NusA fusion separated by a TEV cleavage in a pET-50b (+) was transformed into tuner (DE3) (cat # 70623; Novagen), BL21(DE3) E. coli cells and expression was induced by addition of 1 mM IPTG at 18°C overnight. Cells were collected by centrifugation and frozen at −30°C. Cells from 1 liter culture were resuspended in 40 ml lysis buffer (20 mM Tris [pH 8.0], 10 mM imidazole [pH 8.0], 300 mM NaCl, 2 mM TCEP (tris(2-carboxyethyl)phosphine), 5 mM phenyl phosphate, 1 U DNase, 1 mM PMSF, and 20 mg lysozyme) containing EDTA-free protease inhibitor cocktail (cat # 11697498001; Sigma-Aldrich) and lysed by sonication. Lysate was clarified by spinning cells for 10 min at 20,000 × g before loading supernatant onto cOmplete His-Tag purification resin (cat # 5893682001; Merck). Bound proteins were washed with 20 mM Tris (pH 8.0), 10 mM imidazole (pH 8.0) and 300 mM NaCl, and 5 mM phenyl phosphate followed by 20 mM Tris (pH 8.0), 30 mM imidazole (pH 8.0) and 300 mM NaCl, and 2 mM TCEP 5 mM phenyl phosphate. Protein was eluted in 20 mM Tris (pH 8.0), 250 mM imidazole (pH 8.0) and 300 mM NaCl, 5 mM phenyl phosphate, and 2 mM TCEP. Eluate was then cleaved with TEV (tobacco etch virus) protease overnight at 4°C and subsequently purified further by size exclusion chromatography (Superdex 200 26/600 from cat # GE28-9893-36; GE Healthcare) in TBS, 2 mM TCEP, and 5 mM phenyl phosphate.
Thermostability assays
WT and E374K SH2B3 SH2 domains were desalted into 100 mM NaCl, 20 mM Tris (pH 8.0), and 2 mM TCEP buffer and diluted to 100 μM. Where peptides were used, a fivefold molar excess of peptide was used, and in phenyl phosphate conditions, concentration was 8 mM. Each sample was transferred into a capillary and measured from 35°C to 95°C using a Tycho N6T (Nanotemper). Data were analyzed in Prism (GraphPad Software).
Surface plasmon resonance (SPR) assays
Direct binding experiments were performed on a Biacore 4000 (GE Healthcare) in 10 mM HEPES (pH 7.4), 150 mM NaCl, 3.4 mM EDTA, and 0.005% Tween 20 using a streptavidin-coated chip and were regenerated in 50 mM NaOH and 1 M NaCl. SH2B3 SH2 domains were flowed over a streptavidin-coated chip for 420 s at 30 μl/min with immobilized biotinylated JAK2 pY813. A reference flow cell was included by passing buffer without protein over a single lane, and the sensorgrams from the reference cell were subtracted from the experimental flow cell analyses. Data were subsequently plotted in Prism.
SPR competition assays were performed on a Biacore 4000 (GE Healthcare) in 10 mM HEPES (pH 7.4), 150 mM NaCl, 3.4 mM EDTA, and 0.005% Tween 20 using a streptavidin-coated chip and were regenerated in 50 mM NaOH and 1 M NaCl. A biotinylated peptide representing the IL6ST pY757 sequence was immobilized to the chip by passing over 1 μg/ml of peptide dissolved in 10 mM HEPES pH 7.4, 150 mM NaCl, 3.4 mM EDA, 0.005% Tween 20. 0.1–0.5 μM SH2B3 SH2 domain was preincubated with phosphopeptides or phenylphosphate before being flowed over the chip for 240–720 s at 30 μl/min. Data were analyzed using Prism, and the response of SH2B3 in the presence of the peptide was normalized to an SH2B3-only control and then fitted as an IC50 curve via nonlinear regression.
Crystallography
The E372K SH2B3 SH2 domain was buffer-exchanged into a low-salt buffer (20 mM Tris [pH 8.0], 2 mM TCEP, and 100 mM NaCl), and crystal trays were set up with 5 mg/ml of protein and a twofold molar excess of peptide using vapor diffusion hanging drop experiments set up in-house. The E372K SH2B3 SH2 domain crystallized in 18% PEG 8000, 0.05 M MgAc, 0.1 M Tris, pH 8.5 and was cryoprotected in paratone and immediately snap-frozen in liquid nitrogen. Data were collected at the MX2 beamline at the Australian Synchrotron. Data reduction, scaling, and integration were performed using XDS. The crystal structure of the E372K SH2B3 SH2 domain was solved by molecular replacement (search model PDB ID: 7R8W) using Phaser as implemented in PHENIX (Liebschner et al., 2019). All structures were refined using PHENIX and model building was performed in COOT (Emsley and Cowtan, 2004; Emsley et al., 2010). Structures were visualized using PyMOL (Schrodinger, 2015) (Schrödinger, Inc.).
Human patients and HCs
Written informed consent was obtained as part of the Australian Point Mutation in Systemic Lupus Erythematosus study, the Centre for Personalised Immunology program, and the Healthy Blood Donors register (The Canberra Hospital, Garran, Australia). This study was approved by Australian National University (ANU) and ACT Health Human Ethics Committees and conducted in accordance with the guidelines of the National Statement on Ethical Conduct in Human Research (2007).
Whole blood of patients, relatives, and healthy donors was collected by venipuncture at the antecubital area into 9 ml Vacutainer ACD-A collection tubes (cat # 455055; BD) and processed within 24 h of collection. Peripheral blood mononuclear cells (PBMCs) were isolated by layering whole blood over Lymphoprep density gradient media (cat # 07851; Stemcell Technologies), resuspended in freezing media consisting of 90% fetal bovine serum (FBS) and 10% dimethyl sulfonate (DMSO), and then stored at −80°C cryogenic freezer prior to analysis.
Mouse models
All mice were bred and housed in specific pathogen–free environments at the Australian Phenomics Facility (APF), ANU, Acton, Australia. All mouse-related procedures were approved by the Animal Experimentation Ethics Committees of the ANU.
Mouse models carrying orthologous mutations of SLE patients were generated in C57BL/6 mice (Charles River Laboratories) via CRISPR-Cas9 genome editing technology according to Jiang et al. (2019) and Gurumurthy et al. (2019). Three mouse models were generated: Sh2b3E372K, Sh2b3Δ, and Sh2b3R530Q (Table S6). Briefly, single-stranded oligonucleotides (ssODN) and single-guide RNA (sgRNA) were purchased from IDT and delivered to the C57BL/6Ncrl fertilized mouse zygotes as a ribonucleoprotein complex with the following concentrations: Cas9 protein (50 ng/µl), sgRNA (2.5 ng/µl), and ssODN (50 ng/µl). The edited mouse zygotes were surgically transferred into the uterine horn of the pseudopregnant CFW/crl mice. Founder mice were genotyped by Sanger sequencing using the primer pairs listed in the Table S6. B6.129-Rag1tm1Mom (Rag1−/−) mice were used as BM recipients in our BM chimera experiments. C57BL/6-Ptprca mice were used as WT congenic (CD45.1) BM donors in the BM chimera experiments.
The mice were cohoused by litters and sexes with mixed genotypes.
50:50 mixed BM chimera
BM cells were collected from CD45.1+ WT, CD45.2+-Sh2b3+/+, and CD45.2+- Sh2b3E372K/E372K mice. Cells from CD45.1+ WT and CD45.2+-Sh2b3+/+ or CD45.2+- Sh2b3E372K/E372K mice were mixed at 1:1 ratio, respectively, and 2 × 106 BM cell mix injected into the tail vein of each corresponding Rag1tm1Mom (hereinafter referred to as Rag−/−) mouse that had received 500 cGy radiation from the RS2000 small animal irradiator (Rad Source Technologies). Injected mice were allowed 10 wk for BM reconstitution before analysis.
SWHEL-mHEL3× BM chimera
Sh2b3E372K/+ (CD45.2) mice were crossed to SWHEL mice (CD45.1) mice to generate Sh2b3+/+-SWHELHc/−,Lc/− (CD45.1/2) and Sh2b3E372K/E372K-SWHELHc/−,Lc/− (CD45.1/2) BM donor mice. Recipient membrane-bound HEL3× (mHEL3×) mice (on CD45.1 congenic background) were a kind gift from Professor Robert Brink (Garvan Institute, Sydney, Australia). The mHEL3× expressed in the mutant mice served as a ubiquitously expressed self-antigen.
BM was collected from donor mice and cryopreserved in a freezing medium (90% FBS with 10% DMSO) at −80°C until needed. BM ablation of recipient mice was performed by double irradiation (450 cGy each time) 4 h apart in an RS5000 small animal irradiator. Irradiated mice were injected with 2 × 106 thawed donor BM cells and allowed for reconstitution 8 wk before analysis.
Pristane-induced murine SLE model
12-wk-old Sh2b3E372K mice were injected with either 0.25 ml pristane or PBS IP (randomly assigned). Retro-orbital blood was collected from injected mice before pristane injection and periods were set thereafter to measure serum immunoglobulins and ANAs by ELISA and assess cellular phenotypes. 6 mo after pristane injection, mice were euthanized. Kidneys were either fixed on 10% formalin and stained for H&E to look at renal pathology or cryopreserved in optimal cutting temperature compound (OCT) for immunofluorescence (IF) analysis.
Hematology analysis
200 μl mouse blood was collected via retro-orbital bleed into a Monovette 200 μl EDTA tube (cat # 20.1288; Sarstedt). Following collection, the blood samples were deidentified and assigned sample numbers during preparation and acquisition to achieve blinding. 100 μl of the anticoagulated blood was transferred into Axygen 1.1 ml polypropylene cluster tubes (cat # MTS-11-C; Corning) containing 100 μl FACS wash. Samples were analyzed on an ADVIA 2120 hematology system (Siemens Healthineers).
IL-4–supplemented IgM crosslinking of B cells
Spleens were collected from Sh2b3+/+, Sh2b3E372K/+, and Sh2b3E372K/E372K mice and meshed to generate single-cell suspensions, which were stained with antibody cocktail containing BV480-CD93 (cat # 136507; BioLegend), PE-CD19 (cat # 115508; BioLegend), Alexa Fluor 647-B220 (cat # 103226; BioLegend), and Alexa Fluor 700-CD3 (cat # 100216; BioLegend) antibodies and then with 7-AAD (cat # A1310; Invitrogen). The cells were sorted into live (7-AAD−) transitional (CD19+CD3−B220+CD93+) and mature B cells (CD19+CD3−B220+CD93−) on BD FACSAria Fusion and FACSAria II (BD) before being cultured in complete RPMI-1640 media without/with 5 μg/ml goat anti-mouse IgM (cat # 115-006-075; Jackson ImmunoResearch) and/or recombinant murine 25 ng/ml IL-4 (cat # 214-14; Peprotech) for 16 h at 37°C/5% CO2 before staining with fixable viability dye eFluor 780 (cat # 65-0865-14; Invitrogen) and FITC-Annexin V (cat # 556419; BD) for apoptosis analysis on a LSRFortessa X-20 flow cytometer (BD). To determine BAFF-R experssion, sorting and cell culture were performed as described above for the apoptosis assay. Following a 16-h incubation at 37°C/5% CO2, the cultured B cells were stained with Fc block followed by fixable viability dye eFluor 780 and FITC BAFF-R monoclonal antibody (cat #11-5943-81; Invitrogen) and analyzed on an LSRFortessa X-20 flow cytometer (BD).
Flow cytometry
To prepare mouse spleens for flow cytometry analysis, collected spleens were isolated as single-cell suspensions. For BM cell isolation, claws were removed by incising the ankles of each leg, while muscles, ligaments, and tendons were removed from the tibiae and femora with a scalpel. Joints on one end of the cleaned tibiae and femora were cleaved. BM was collected with a 25G needle attached to a 1-ml syringe. For preparing murine blood for flow cytometry antibody staining, plasma-separated mouse blood with anticoagulant (EDTA) was washed with FACS wash. To lyse red blood cells in these samples, RBC lysis buffer (168 mM NH4Cl, 10 mM KHCO3, 100 μM Na2EDTA, pH7.3) was added and incubated for 3–5 min before washing. The resulting cell suspension was enumerated and 3–5 × 106 cells were plated in 96-well round bottom plates for staining. The samples were deidentified during collection and assigned numbers during preparation and acquisition to achieve blinding. Prior to antibody staining, cells were treated with TruStain FcX rat anti-mouse CD16/32 antibodies (cat # 101320; BioLegend) or human TruStain FcX CD16/32/64 antibodies (cat # 422302; BioLegend) to block Fc receptors. The cells were then stained with an antibody cocktail (Table S7) along with LIVE/DEAD Fixable Aqua Dead Cell Stain (cat # L34957; Invitrogen) or eBioscience Fixable eFluor780 viability dye (cat # 65-0865-14; Invitrogen). To fix and permeabilize samples, eBioscience Foxp3/transcription factor staining buffer set (cat # 00-5523-00; Invitrogen) was used per the manufacturer’s instructions. Staining of intracellular markers was performed following permeabilization. Single-color controls and samples were acquired on a LSRFortessa X-20 flow cytometer (BD), while results were analyzed using FlowJo software (BD).
Flow cytometric analysis of human PBMCs was performed following methods described in the study by Ellyard et al. (2019).
Calcium flux
Mouse peripheral blood was processed to remove RBC via lysis prior to Fc blocking and then stained with antibodies against surface markers including biotin-CD93 (cat # 13-5892-85; Invitrogen), BV605-CD19 (cat # 115539; BD), APC Cy7-CD3 (cat # 100222; BioLegend), Alexa Fluor 647-B220 (cat # 103226; BioLegend), and PE Cy7-streptavidin (cat # 405206; BioLegend). The cells were washed with complete RPMI-1640 prewarmed to 37°C and stained with Indo-1 AM (cat # I1223; Invitrogen) diluted to 2 μM in warm RPMI-1640 and incubated at 37°C for 30 min before being washed and stained with 7-AAD (cat # A1310; Invitrogen).
For acquisition, data scales of the detectors for the BUV395 (379/28) and BUV496 (515/30 450LP) filters were set to linear. Samples were kept at 37°C prior to and during acquisition. The baseline was recorded for 30 s, followed by the addition of anti-mouse IgM (cat # 115-006-075; Jackson ImmunoResearch) to a final concentration of 10 μg/ml, immediately mixed before resuming acquisition for further 3.5 min to measure the calcium flux following BCR ligation. At 4 min after the acquisition, ionomycin (cat # I24222; Invitrogen) was added to the sample tube to a final concentration of 1 μg/ml and immediately mixed before resuming acquisition for another 4 min to measure maximal Ca2+ flux output.
Bead-based immunoassay
Serum BAFF quantitation was performed on plasma samples from 12- to 14 wk-old Sh2b3E372K mice using the LEGENDplex Mouse B cell Panel – S/P (1-plex) w/VbP kit (cat # 740983; BioLegend) following the manufacturer’s instructions. Data were analyzed using the LEGENDplex Data Analysis Software Suite (BioLegend, https://www.biolegend.com/en-us/immunoassays/legendplex/support/software).
Meso Scale Discovery
Plasma samples from Sh2b3E372K mice were plated for the mouse U-plex Biomarker Group 1 Multiplex Assay Box 2 10-Assay Plate (cat #K15322K; Meso Scale Discovery) and the assay was performed according to the manufacturer’s instructions. The plate was read on a Meso Sector S 600 (Meso Scale Discovery).
Treg suppression assay
Tregs (CD4+CD25+), naïve T cells (CD4+CD62L+CD44lo), and antigen-presenting cells (APC; CD3−) were purified by flow cytometry. Naïve T cells were labeled with Cell Trace Violet (cat # C34557; Thermo Fischer Scientific) according to the manufacturer’s instructions. Naïve T cells (2 × 104) were cocultured with Tregs at the rations shown in the presence of APCs (4 × 104 cells) and anti-CD3 (2 μg/ml) for 72 h at 37°C/5% CO2. T cell proliferation was measured by flow cytometry in the presence of 7AAD to allow for the discrimination of live cells.
ELISA
Flat-bottom 96-well plates were pretreated with 0.002% (wt/vol) poly-L-lysine solution for 5 h at room temperature before coating with 50 μl of 50 ng/μl calf thymus DNA (cat # D8661; Sigma-Aldrich) or 5 ng/μl HEL and resuspended in ELISA coating buffer (50 mM carbonate buffer, pH 9.6) overnight at 4°C in a humidified chamber. Coated plates were blocked with 150 μl of blocking buffer (1× PBS, 1% wt/vol BSA, and 0.5 vol/vol Tween20). Plasma samples diluted at 1:40 and 1:80 or positive controls (sera from MRL-Fas mice) diluted at 1:80, 1:160, 1:320, 1:640, and 1:1,280 were added to the wells and incubated overnight at 4°C. The plates were washed and incubated with either AP-conjugated goat anti-mouse IgG or IgM antibody (cat # 1030-04, 1020-04; SouthernBiotech) (Table S8), washed again, and incubated with phosphatase substrate (cat # S0942; Sigma-Aldrich) dissolved in developing buffer (0.1 M glycine, 0.1 mM ZnCl2, 1.0 M MgCl2·6H2O). Absorbances at 405 and 605 nm (reference wavelength) were measured using an Infinite 200 Pro plate reader (Tecan).
The ANA ELISA was performed on sera collected from 17-wk-old female Sh2b3E372K mice using a mouse antinuclear antigen (ANA/ENA) total immunoglobulins (IgA+IgG+IgM) ELISA kit (cat # 5210; Alpha Diagnostic) following the manufacturer’s instruction.
IF
OCT-embedded, cryopreserved mouse kidneys were cut into 7-μm sections, fixed with acetone, blocked with blocking buffer (1× PBS, 3% BSA, 0.5% Triton X-100), and stained with goat anti-mouse podocin antibody (cat # sc-22296; Santa Cruz Biotechnology) (Table S8) and then with Alexa Fluor 594–conjugated donkey anti-goat IgG (H+L) and Alexa Fluor 488–conjugated donkey anti-mouse IgG (H+L) antibodies (cat # A-21202; Invitrogen). The stained sections were preserved in Vectashield antifade mounting medium with DAPI (cat # H-1200-10; Vector Laboratories) and imaged using an Olympus DP70 camera system attached to an IX70 fluorescence microscope (Olympus Life Science). A 200× magnification was used with a 1/200-s exposure for the green channel (IgG) and a 1/50-s exposure for the red channel (podocin). IgG IC deposition was scored blindly by the investigator following imaging.
Statistical analyses
One-way ANOVA using Dunnett’s test or two-way ANOVA with Šídák correction was used for comparing differences among three or more groups of data from single experiments. Brown–Forsythe and Welch ANOVA tests were used for analyzing human immunophenotyping data. Student’s t test was used for comparing the differences between CD45.2+/CD45.1+ cell percentage ratios in two types of mixed BM chimeras (wt:wt and hom:wt, of the Sh2b3E372K strain). Linear mixed-effects model (lmer) with estimated marginal means (emmeans) using the experiment as a blocking factor was used for analyzing luciferase data. lmer ANOVA using the experiment as a blocking factor was used for analyzing ADVIA and gross phenotype data pooled from multiple experiments. Fisher’s exact tests were used for analyzing categorical data in the IF experiment and glomerular scores. Variances of data from the SWHEL-mHEL3× were not homoscedastic, and therefore the data were log10-transformed before two-way ANOVAs to fulfill the assumption of homoscedasticity. All Student’s t tests, one-way ANOVAs, two-way ANOVAs, Brown–Forsythe, and Welch ANOVAs were performed in Prism (GraphPad Software). Two-way ANOVAs (for SWHEL-mHEL3× data), lmer emmeans, lmer ANOVAs, and Fisher’s exact tests were performed using R (R Foundation for Statistical Computing).
For testing the assumptions for each statistical test used, we followed the following scheme: in each one-way ANOVA, Bartlett’s test of homogeneity of variance was used for testing the assumption of equal variances, a residual plot was generated to examine the fit for a linear model, and a Q-Q plot was generated to assess the normality of data distribution; for two-way ANOVAs, residual plots and Q-Q plots were used for assessing the model assumptions. Since Brown–Forsythe and Welch ANOVA tests do not assume equal variance but assume a normal distribution, Q-Q plots were generated to assess the normality of data distribution; lmer-based tests were utilized for analyzing pooled data from independent experiments to estimate and remove random effects due to the variability introduced during independent experiment setups, and residual plots were generated to assess the assumptions of constant variance and linearity of effects.
Online supplemental material
Fig. S1 shows additional SH2B3 protein functionality and transitional B cell phenotype information. Fig. S2 shows additional data on mature B cell phenotypes in Sh2b3Δ, Sh2b3E372K, and Sh2b3R530Q mice. Fig. S3 shows BM B cell phenotypes in Sh2b3E372K and Sh2b3R530Q mice. Fig. S4 shows peripheral blood and splenic T cell phenotypes in Sh2b3E372K and Sh2b3R530Q mice. Fig. S5 shows IgG complexes in pristane-treated mice, cellular and serological phenotypes of SWHEL-mHEL3× chimeric mice, and analysis of apoptosis in BM immature B cells and additional data of BAFFR expression on transitional and mature B cells. Table S1 shows the diagnosis and clinical manifestations of probands carrying SH2B3 variants. Table S2 shows rare variants in genes known to cause human SLE in the probands identified through WES. Table S3 shows data collection and refinement statistics for the SH2 domains of murine SH2B3 protein with phosphopeptides bound. Table S4 lists the primers used for introducing patient-specific variants and other published variants into mammalian expression vectors of human SH2B3. Table S5 shows sequencing primers designed for validating open reading frame (ORF) sequences in the mammalian expression vectors of human SH2B3 via Sanger sequencing. Table S6 lists the guide RNAs (gRNAs), ssODNs for CRISPR/Cas9 gene editing of mouse models, and oligos for validating the editing results by Sanger sequencing. Table S7 shows antibodies, fluorochrome-conjugated streptavidin, and viability dyes used in flow cytometry. Table S8 shows primary and secondary antibodies used for ELISA and IF.
Supplementary Material
shows the diagnosis and clinical manifestations of probands carrying SH2B3 variants.
shows rare variants in genes known to cause human SLE in the probands identified through WES.
shows data collection and refinement statistics for the SH2 domains of murine SH2B3 protein with phosphopeptides bound.
lists the primers used for introducing patient-specific variants and other published variants into mammalian expression vectors of human SH2B3.
shows sequencing primers designed for validating ORF sequences in the mammalian expression vectors of human SH2B3 via Sanger sequencing.
lists the guide RNAs (gRNAs), ssODNs for CRISPR/Cas9 gene editing of mouse models, and oligos for validating the editing results by Sanger sequencing.
shows antibodies, fluorochrome-conjugated streptavidin, and viability dyes used in flow cytometry.
shows primary and secondary antibodies used for ELISA and IF.
Acknowledgments
We thank the personnel of the Australian Cancer Research Foundation Biomolecular Resource Facility (JCSMR) for Sanger sequencing; Jing Gao, Nikki Ross, Jenna Lowe, Nay Chi Khin, and Lora Starrs for assistance with the generation of CRISPR mouse models; Robert Tunningley for assisting with an ADVIA experiment; staff of the APF for the care of experimental animals; Dr. Harpreet Vohra, Michael Devoy, and Cathy Gillespie from the Microscopy and Flow Cytometry Facility, JCSMR for assistance with flow cytometry and imaging; Koula Diamand for technical assistance running the Meso Scale Discovery platform; Jessica Lovell of the Bruestle Group, JCSMR for assistance with cryosectioning; Dr Yafei Zhang of the APF NGS Team for WES of CRISPR mice; and Pablo Fernández de Cañete Nieto for conjugation of HyHEL9-Alexa Fluor 647. Dr. Teresa Neeman of the Biological Data Science Institute of the ANU provided consultation on the statistical analyses in this study.
This research/project was undertaken with the assistance of resources and services from the National Computational Infrastructure (NCI) and National Collaborative Research Infrastructure Strategy (NCRIS) via Phenomics Australia (to J.I. Ellyard). This project was funded by the Australian National Health and Medical Research Council Fellowship and program grants to C.G. Vinuesa as well as funding from the Francis Crick Institute; and funding from the Pryor Bequest to ANU from Jenny and Bruce Pryor. This research project was undertaken with the assistance of computational resources and services from the NCI an NCRIS-enabled capability supported by the Australian Government, and the Phenomics Translational Initiative funded by the Medical Research Future Fund.
Author contributions: Supervision: J.I. Ellyard and C.G. Vinuesa; Conceptualization: J.I. Ellyard, C.G. Vinuesa, J.J. Babon, Y. Zhang, R. Morris, and G.J. Brown; Investigation: Y. Zhang, R. Morris, G.J. Brown, A.M.D. Lorenzo, X. Meng, N.J. Kershaw, P. Kiridena, S. Gross, J.Y. Cappello, Q. Shen, H. Wang, C. Turnbull, T. Lea-Henry, M. Stanley, Z.Yu, F.D. Ballard, J.C. Lee, and D.A. Fulcher; Resources: A. Enders, S.L. Masters, V. Athanasopoulos, J.C. Lee, A-M. Hatch, P. Trnka, D. Mallon, J.T. Fletcher, G.D. Walters, M. Sestan, M. Jelušić, and M.C. Cook; Writing: J.I. Ellyard, C.G. Vinuesa, Y. Zhang, and R. Morris; Funding acquisition: C.G. Vinuesa.
Data availability
The data underlying figures are available in the published article and its online supplemental material. Data used in Fig. 1 E have been deposited in the PBD archive (PDB ID 7R8W, E372K: PDB ID: 8CZ9).
References
- Ackman, R.G. 1971. Pristane and other hydrocarbons in some freshwater and marine fish oils. Lipids. 6:520–522. 10.1007/BF02531239 [DOI] [PubMed] [Google Scholar]
- Adachi, M., Suematsu S., Suda T., Watanabe D., Fukuyama H., Ogasawara J., Tanaka T., Yoshida N., and Nagata S.. 1996. Enhanced and accelerated lymphoproliferation in Fas-null mice. Proc. Natl. Acad. Sci. USA. 93:2131–2136. 10.1073/pnas.93.5.2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcina, A., Vandenbroeck K., Otaegui D., Saiz A., Gonzalez J.R., Fernandez O., Cavanillas M.L., Cénit M.C., Arroyo R., Alloza I., et al. 2010. The autoimmune disease-associated KIF5A, CD226, and SH2B3 gene variants confer susceptibility for multiple sclerosis. Genes Immun. 11:439–445. 10.1038/gene.2010.30 [DOI] [PubMed] [Google Scholar]
- Almeida, A.R.M., Neto J.L., Cachucho A., Euzébio M., Meng X., Kim R., Fernandes M.B., Raposo B., Oliveira M.L., Ribeiro D., et al. 2021. Interleukin-7 receptor α mutational activation can initiate precursor B-cell acute lymphoblastic leukemia. Nat. Commun. 12:7268. 10.1038/s41467-021-27197-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage, R.J., Macduff B.M., Eisenman J., Paxton R., and Grabstein K.H.. 1995. IL-15 has stimulatory activity for the induction of B cell proliferation and differentiation. J. Immunol. 154:483–490. 10.4049/jimmunol.154.2.483 [DOI] [PubMed] [Google Scholar]
- Batten, M., Groom J., Cachero T.G., Qian F., Schneider P., Tschopp J., Browning J.L., and Mackay F.. 2000. BAFF mediates survival of peripheral immature B lymphocytes. J. Exp. Med. 192:1453–1466. 10.1084/jem.192.10.1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentham, J., Morris D.L., Graham D.S.C., Pinder C.L., Tombleson P., Behrens T.W., Martín J., Fairfax B.P., Knight J.C., Chen L., et al. 2015. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat. Genet. 47:1457–1464. 10.1038/ng.3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersenev, A., Wu C., Balcerek J., and Tong W.. 2008. Lnk controls mouse hematopoietic stem cell self-renewal and quiescence through direct interactions with JAK2. J. Clin. Invest. 118:2832–2844. 10.1172/JCI35808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block, S.R., Winfield J.B., Lockshin M.D., D’Angelo W.A., and Christian C.L.. 1975. Studies of twins with systemic lupus erythematosus. A review of the literature and presentation of 12 additional sets. Am. J. Med. 59:533–552. 10.1016/0002-9343(75)90261-2 [DOI] [PubMed] [Google Scholar]
- Blombery, P., Pazhakh V., Albuquerque A., Maimaris J., Tu L., Briones B., Evans F., Thompson E., Carpenter B., Curtin J., et al. 2022. Biallelic deleterious germline SH2B3 variants cause a novel clinical syndrome of myeloproliferation and multi-organ autoimmunity. Blood. 140:992–993. 10.1182/blood-2022-163097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, G.J., Cañete P.F., Wang H., Medhavy A., Bones J., Roco J.A., He Y., Qin Y., Cappello J., Ellyard J.I., et al. 2022. TLR7 gain-of-function genetic variation causes human lupus. Nature. 605:349–356. 10.1038/s41586-022-04642-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett, D.L., Langley D.B., Schofield P., Hermes J.R., Chan T.D., Jackson J., Bourne K., Reed J.H., Patterson K., Porebski B.T., et al. 2018. Germinal center antibody mutation trajectories are determined by rapid self/foreign discrimination. Science. 360:223–226. 10.1126/science.aao3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y., Chikwava K., Wu C., Zhang H., Bhagat A., Pei D., Choi J.K., and Tong W.. 2016. LNK/SH2B3 regulates IL-7 receptor signaling in normal and malignant B-progenitors. J. Clin. Invest. 126:1267–1281. 10.1172/JCI81468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coltro, G., Lasho T.L., Finke C.M., Gangat N., Pardanani A., Tefferi A., Jevremovic D., Altman J.K., and Patnaik M.M.. 2019. Germline SH2B3 pathogenic variant associated with myelodysplastic syndrome/myeloproliferative neoplasm with ring sideroblasts and thrombocytosis. Am. J. Hematol. 94:E231–E234. 10.1002/ajh.25552 [DOI] [PubMed] [Google Scholar]
- Crispín, J.C., Oukka M., Bayliss G., Cohen R.A., Van Beek C.A., Stillman I.E., Kyttaris V.C., Juang Y.-T., and Tsokos G.C.. 2008. Expanded double negative T cells in patients with systemic lupus erythematosus produce IL-17 and infiltrate the kidneys. J. Immunol. 181:8761–8766. 10.4049/jimmunol.181.12.8761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks, G.M., Hao Q.-L., Petersen D., Barsky L.W., and Bockstoce D.. 2000. IL-3 increases production of B lymphoid progenitors from human CD34+CD38− cells. J. Immunol. 165:2382–2389. 10.4049/jimmunol.165.5.2382 [DOI] [PubMed] [Google Scholar]
- Deapen, D., Escalante A., Weinrib L., Horwitz D., Bachman B., Roy-Burman P., Walker A., and Mack T.M.. 1992. A revised estimate of twin concordance in systemic lupus erythematosus. Arthritis Rheum. 35:311–318. 10.1002/art.1780350310 [DOI] [PubMed] [Google Scholar]
- Domeier, P.P., Chodisetti S.B., Soni C., Schell S.L., Elias M.J., Wong E.B., Cooper T.K., Kitamura D., and Rahman Z.S.M.. 2016. IFN-γ receptor and STAT1 signaling in B cells are central to spontaneous germinal center formation and autoimmunity. J. Exp. Med. 213:715–732. 10.1084/jem.20151722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellyard, J.I., Jerjen R., Martin J.L., Lee A.Y.S., Field M.A., Jiang S.H., Cappello J., Naumann S.K., Andrews T.D., Scott H.S., et al. 2014. Identification of a pathogenic variant in TREX1 in early-onset cerebral systemic lupus erythematosus by Whole-exome sequencing. Arthritis Rheumatol. 66:3382–3386. 10.1002/art.38824 [DOI] [PubMed] [Google Scholar]
- Ellyard, J.I., Tunningley R., Lorenzo A.M., Jiang S.H., Cook A., Chand R., Talaulikar D., Hatch A.-M., Wilson A., Vinuesa C.G., et al. 2019. Non-parametric heat map representation of flow cytometry data: Identifying cellular changes associated with genetic immunodeficiency disorders. Front. Immunol. 10:2134. 10.3389/fimmu.2019.02134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley, P., and Cowtan K.. 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Emsley, P., Lohkamp B., Scott W.G., and Cowtan K.. 2010. Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillatreau, S., Manfroi B., and Dörner T.. 2021. Toll-like receptor signalling in B cells during systemic lupus erythematosus. Nat. Rev. Rheumatol. 17:98–108. 10.1038/s41584-020-00544-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodnow, C.C., Crosbie J., Jorgensen H., Brink R.A., and Basten A.. 1989. Induction of self-tolerance in mature peripheral B lymphocytes. Nature. 342:385–391. 10.1038/342385a0 [DOI] [PubMed] [Google Scholar]
- Granato, A., Hayashi E.A., Baptista B.J.A., Bellio M., and Nobrega A.. 2014. IL-4 regulates Bim expression and promotes B cell maturation in synergy with BAFF conferring resistance to cell death at negative selection checkpoints. J. Immunol. 192:5761–5775. 10.4049/jimmunol.1300749 [DOI] [PubMed] [Google Scholar]
- Gurumurthy, C.B., O’Brien A.R., Quadros R.M., Adams J. Jr., Alcaide P., Ayabe S., Ballard J., Batra S.K., Beauchamp M.-C., Becker K.A., et al. 2019. Reproducibility of CRISPR-cas9 methods for generation of conditional mouse alleles: A multi-center evaluation. Genome Biol. 20:171. 10.1186/s13059-019-1776-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X., Li Y., Schembri-King J., Jakes S., and Hayashi J.. 2000. Identification of actin binding protein, ABP-280, as a binding partner of human Lnk adaptor protein. Mol. Immunol. 37:603–612. 10.1016/S0161-5890(00)00070-5 [DOI] [PubMed] [Google Scholar]
- He, Y., Gallman A.E., Xie C., Shen Q., Ma J., Wolfreys F.D., Sandy M., Arsov T., Wu X., Qin Y., et al. 2021. P2RY8 variants in lupus patients uncover a role for the receptor in immunological tolerance. J. Exp. Med. 219:e20211004. 10.1084/jem.20211004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X., Li Y., Tanaka K., Moore K.G., and Hayashi J.I.. 1995. Cloning and characterization of Lnk, a signal transduction protein that links T-cell receptor activation signal to phospholipase C gamma 1, Grb2, and phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. USA. 92:11618–11622. 10.1073/pnas.92.25.11618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi, K., Garotta G., Ozmen L., Ziemiecki A., Wilks A.F., Harpur A.G., Larner A.C., and Finbloom D.S.. 1994. Interferon-gamma induces tyrosine phosphorylation of interferon-gamma receptor and regulated association of protein tyrosine kinases, Jak1 and Jak2, with its receptor. J. Biol. Chem. 269:14333–14336. 10.1016/S0021-9258(17)36621-8 [DOI] [PubMed] [Google Scholar]
- Izmirly, P.M., Parton H., Wang L., McCune W.J., Lim S.S., Drenkard C., Ferucci E.D., Dall’Era M., Gordon C., Helmick C.G., and Somers E.C.. 2021. Prevalence of systemic lupus erythematosus in the United States: Estimates from a meta-analysis of the centers for disease control and prevention national lupus registries. Arthritis Rheumatol. 73:991–996. 10.1002/art.41632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, S.W., Jacobs H.M., Arkatkar T., Dam E.M., Scharping N.E., Kolhatkar N.S., Hou B., Buckner J.H., and Rawlings D.J.. 2016. B cell IFN-γ receptor signaling promotes autoimmune germinal centers via cell-intrinsic induction of BCL-6. J. Exp. Med. 213:733–750. 10.1084/jem.20151724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, S.H., Athanasopoulos V., Ellyard J.I., Chuah A., Cappello J., Cook A., Prabhu S.B., Cardenas J., Gu J., Stanley M., et al. 2019. Functional rare and low frequency variants in BLK and BANK1 contribute to human lupus. Nat. Commun. 10:2201. 10.1038/s41467-019-10242-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kara, B., Ekinci Z., Sahin S., Gungor M., Gunes A.S., Ozturk K., Adrovic A., Cefle A., Inanç M., Gul A., and Kasapcopur O.. 2020. Monogenic lupus due to spondyloenchondrodysplasia with spastic paraparesis and intracranial calcification: Case-based review. Rheumatol. Int. 40:1903–1910. 10.1007/s00296-020-04653-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama, H., Mori T., Seki Y., Anraku M., Iseki M., Ikutani M., Iwasaki Y., Yoshida N., Takatsu K., and Takaki S.. 2014. Lnk prevents inflammatory CD8⁺ T-cell proliferation and contributes to intestinal homeostasis. Eur. J. Immunol. 44:1622–1632. 10.1002/eji.201343883 [DOI] [PubMed] [Google Scholar]
- Koffler, D., Agnello V., Thoburn R., and Kunkel H.G.. 1971. Systemic lupus erythematosus: Prototype of immune complex nephritis in man. J. Exp. Med. 134:169–179. 10.1084/jem.134.3.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, Y.-C., Chun S., Kim K., and Mak A.. 2019. Update on the genetics of systemic lupus erythematosus: Genome-wide association studies and beyond. Cells. 8:1180. 10.3390/cells8101180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, K.-P., Kühn R., and Rajewsky K.. 1997. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 90:1073–1083. 10.1016/S0092-8674(00)80373-6 [DOI] [PubMed] [Google Scholar]
- Laroumanie, F., Korneva A., Bersi M.R., Alexander M.R., Xiao L., Zhong X., Van Beusecum J.P., Chen Y., Saleh M.A., McMaster W.G., et al. 2018. LNK deficiency promotes acute aortic dissection and rupture. JCI Insight. 3:e122558. 10.1172/jci.insight.122558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, A., Avery D.T., Jackson K., Lenthall H., Volpi S., Brigden H., Russell A.J., Bier J., Reed J.H., Smart J.M., et al. 2019. Activated PI3Kδ breaches multiple B cell tolerance checkpoints and causes autoantibody production. J. Exp. Med. 217:e20191336. 10.1084/jem.20191336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lausch, E., Janecke A., Bros M., Trojandt S., Alanay Y., De Laet C., Hübner C.A., Meinecke P., Nishimura G., Matsuo M., et al. 2011. Genetic deficiency of tartrate-resistant acid phosphatase associated with skeletal dysplasia, cerebral calcifications and autoimmunity. Nat. Genet. 43:132–137. 10.1038/ng.749 [DOI] [PubMed] [Google Scholar]
- Lawson, B.R., Gonzalez-Quintial R., Eleftheriadis T., Farrar M.A., Miller S.D., Sauer K., McGavern D.B., Kono D.H., Baccala R., and Theofilopoulos A.N.. 2015. Interleukin-7 is required for CD4(+) T cell activation and autoimmune neuroinflammation. Clin. Immunol. 161:260–269. 10.1016/j.clim.2015.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesley, R., Xu Y., Kalled S.L., Hess D.M., Schwab S.R., Shu H.-B., and Cyster J.G.. 2004. Reduced competitiveness of autoantigen-engaged B cells due to increased dependence on BAFF. Immunity. 20:441–453. 10.1016/S1074-7613(04)00079-2 [DOI] [PubMed] [Google Scholar]
- Liebschner, D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., Hintze B., Hung L.-W., Jain S., McCoy A.J., et al. 2019. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in phenix. Acta Crystallogr. D Struct. Biol. 75:861–877. 10.1107/S2059798319011471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, D.-C., Yin T., Koren-Michowitz M., Ding L.-W., Gueller S., Gery S., Tabayashi T., Bergholz U., Kazi J.U., Rönnstrand L., et al. 2012. Adaptor protein Lnk binds to and inhibits normal and leukemic FLT3. Blood. 120:3310–3317. 10.1182/blood-2011-10-388611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Karypis G., Hippen K.L., Vegoe A.L., Ruiz P., Gilkeson G.S., and Behrens T.W.. 2006. Genomic view of systemic autoimmunity in MRLlpr mice. Genes Immun. 7:156–168. 10.1038/sj.gene.6364286 [DOI] [PubMed] [Google Scholar]
- Lo, M.S. 2016. Monogenic lupus. Curr. Rheumatol. Rep. 18:71. 10.1007/s11926-016-0621-9 [DOI] [PubMed] [Google Scholar]
- Lu, W., Zhong Y., Zhang Y., Liu Z., and Xue L.. 2021. The clinical characteristics of leukopenia in patients with systemic lupus erythematosus of han ethnicity in China: A cross-sectional study. Rheumatol. Ther. 8:1177–1188. 10.1007/s40744-021-00336-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, N., Sekigawa I., Iida N., Matsumoto M., Hashimoto H., and Hirose S.. 1999. Relationship between CD4+/CD8+ T cell ratio and T cell activation in systemic lupus erythematosus. Scand. J. Rheumatol. 28:166–170. 10.1080/03009749950154248 [DOI] [PubMed] [Google Scholar]
- Malabarba, M.G., Kirken R.A., Rui H., Koettnitz K., Kawamura M., O’Shea J.J., Kalthoff F.S., and Farrar W.L.. 1995. Activation of JAK3, but not JAK1, is critical to interleukin-4 (IL4) stimulated proliferation and requires a membrane-proximal region of IL4 receptor α. J. Biol. Chem. 270:9630–9637. 10.1074/jbc.270.16.9630 [DOI] [PubMed] [Google Scholar]
- McMullin, M.F., Wu C., Percy M.J., and Tong W.. 2011. A nonsynonymous LNK polymorphism associated with idiopathic erythrocytosis. Am. J. Hematol. 86:962–964. 10.1002/ajh.22154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingari, M.C., Gerosa F., Carra G., Accolla R.S., Moretta A., Zubler R.H., Waldmann T.A., and Moretta L.. 1984. Human interleukin-2 promotes proliferation of activated B cells via surface receptors similar to those of activated T cells. Nature. 312:641–643. 10.1038/312641a0 [DOI] [PubMed] [Google Scholar]
- Mori, T., Iwasaki Y., Seki Y., Iseki M., Katayama H., Yamamoto K., Takatsu K., and Takaki S.. 2014. Lnk/Sh2b3 controls the production and function of dendritic cells and regulates the induction of IFN-γ-producing T cells. J. Immunol. 193:1728–1736. 10.4049/jimmunol.1303243 [DOI] [PubMed] [Google Scholar]
- Morris, D.L., Sheng Y., Zhang Y., Wang Y.-F., Zhu Z., Tombleson P., Chen L., Cunninghame Graham D.S., Bentham J., Roberts A.L., et al. 2016. Genome-wide association meta-analysis in Chinese and European individuals identifies ten new loci associated with systemic lupus erythematosus. Nat. Genet. 48:940–946. 10.1038/ng.3603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, R., Zhang Y., Ellyard J.I., Vinuesa C.G., Murphy J.M., Laktyushin A., Kershaw N.J., and Babon J.J.. 2021. Structural and functional analysis of target recognition by the lymphocyte adaptor protein LNK. Nat. Commun. 12:6110. 10.1038/s41467-021-26394-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naradikian, M.S., Myles A., Beiting D.P., Roberts K.J., Dawson L., Herati R.S., Bengsch B., Linderman S.L., Stelekati E., Spolski R., et al. 2016. Cutting edge: IL-4, IL-21, and IFN-γ interact to govern T-bet and CD11c expression in TLR-activated B cells. J. Immunol. 197:1023–1028. 10.4049/jimmunol.1600522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemazee, D.A., and Bürki K.. 1989. Clonal deletion of B lymphocytes in a transgenic mouse bearing anti-MHC class I antibody genes. Nature. 337:562–566. 10.1038/337562a0 [DOI] [PubMed] [Google Scholar]
- Okada, Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S., et al. 2014. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 506:376–381. 10.1038/nature12873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omarjee, O., Picard C., Frachette C., Moreews M., Rieux-Laucat F., Soulas-Sprauel P., Viel S., Lega J.-C., Bader-Meunier B., Walzer T., et al. 2019. Monogenic lupus: Dissecting heterogeneity. Autoimmun. Rev. 18:102361. 10.1016/j.autrev.2019.102361 [DOI] [PubMed] [Google Scholar]
- Palacios, R., Henson G., Steinmetz M., and McKearn J.P.. 1984. Interleukin-3 supports growth of mouse pre-B-cell clones in vitro. Nature. 309:126–131. 10.1038/309126a0 [DOI] [PubMed] [Google Scholar]
- Parrish, Y.K., Baez I., Milford T.-A., Benitez A., Galloway N., Rogerio J.W., Sahakian E., Kagoda M., Huang G., Hao Q.-L., et al. 2009. IL-7 Dependence in human B lymphopoiesis increases during progression of ontogeny from cord blood to bone marrow. J. Immunol. 182:4255–4266. 10.4049/jimmunol.0800489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, S.L., Szabo S.J., and Glimcher L.H.. 2002. T-bet regulates IgG class switching and pathogenic autoantibody production. Proc. Natl. Acad. Sci. USA. 99:5545–5550. 10.1073/pnas.082114899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Garcia, A., Ambesi-Impiombato A., Hadler M., Rigo I., LeDuc C.A., Kelly K., Jalas C., Paietta E., Racevskis J., Rowe J.M., et al. 2013. Genetic loss of SH2B3 in acute lymphoblastic leukemia. Blood. 122:2425–2432. 10.1182/blood-2013-05-500850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves, W.H., Lee P.Y., Weinstein J.S., Satoh M., and Lu L.. 2009. Induction of autoimmunity by pristane and other naturally occurring hydrocarbons. Trends Immunol. 30:455–464. 10.1016/j.it.2009.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, H.B., Satoh M., Shaw M., Libert C., Poli V., and Reeves W.H.. 1998. Interleukin 6 dependence of anti-DNA antibody production: Evidence for two pathways of autoantibody formation in pristane-induced lupus. J. Exp. Med. 188:985–990. 10.1084/jem.188.5.985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero, S.J., Díaz-Jouanen E., and Alarcón-Segovia D.. 1978. Lymphopenia in systemic lupus erythematosus. Clinical, diagnostic, and prognostic significance. Arthritis Rheum. 21:295–305. 10.1002/art.1780210302 [DOI] [PubMed] [Google Scholar]
- Rubtsov, A.V., Rubtsova K., Fischer A., Meehan R.T., Gillis J.Z., Kappler J.W., and Marrack P.. 2011. Toll-like receptor 7 (TLR7)-driven accumulation of a novel CD11c⁺ B-cell population is important for the development of autoimmunity. Blood. 118:1305–1315. 10.1182/blood-2011-01-331462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Raber, M.-L., Amano H., Amano E., Fossati-Jimack L., Swee L.K., Rolink A., and Izui S.. 2010. Evidence that Yaa-induced loss of marginal zone B cells is a result of dendritic cell-mediated enhanced activation. J. Autoimmun. 34:349–355. 10.1016/j.jaut.2010.01.001 [DOI] [PubMed] [Google Scholar]
- Sapartini, G., Ghrahani R., and Setiabudiawan B.. 2018. Association between lymphopenia and clinical manifestations, anti-dsDNA, and disease activity in children with systemic lupus erythematosus. J. Allergy Clin. Immunol. 141:AB119. 10.1016/j.jaci.2017.12.379 [DOI] [Google Scholar]
- Sasaki, Y., Casola S., Kutok J.L., Rajewsky K., and Schmidt-Supprian M.. 2004. TNF family member B cell-activating factor (BAFF) receptor-dependent and -independent roles for BAFF in B cell physiology. J. Immunol. 173:2245–2252. 10.4049/jimmunol.173.4.2245 [DOI] [PubMed] [Google Scholar]
- Schiemann, B., Gommerman J.L., Vora K., Cachero T.G., Shulga-Morskaya S., Dobles M., Frew E., and Scott M.L.. 2001. An essential role for BAFF in the normal development of B cells through a BCMA-independent pathway. Science. 293:2111–2114. 10.1126/science.1061964 [DOI] [PubMed] [Google Scholar]
- Schrodinger, L.L.C. 2015. The PyMOL Molecular Graphics System, Version 1.8. [Google Scholar]
- Shochat, C., Tal N., Bandapalli O.R., Palmi C., Ganmore I., te Kronnie G., Cario G., Cazzaniga G., Kulozik A.E., Stanulla M., et al. 2011. Gain-of-function mutations in interleukin-7 receptor-α (IL7R) in childhood acute lymphoblastic leukemias. J. Exp. Med. 208:901–908. 10.1084/jem.20110580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, C., Dondi E., Chaix A., de Sepulveda P., Kubiseski T.J., Varin-Blank N., and Velazquez L.. 2008. Lnk adaptor protein down-regulates specific Kit-induced signaling pathways in primary mast cells. Blood. 112:4039–4047. 10.1182/blood-2008-05-154849 [DOI] [PubMed] [Google Scholar]
- Sims, G.P., Ettinger R., Shirota Y., Yarboro C.H., Illei G.G., and Lipsky P.E.. 2005. Identification and characterization of circulating human transitional B cells. Blood. 105:4390–4398. 10.1182/blood-2004-11-4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisirak, V., Sally B., D’Agati V., Martinez-Ortiz W., Özçakar Z.B., David J., Rashidfarrokhi A., Yeste A., Panea C., Chida A.S., et al. 2016. Digestion of chromatin in apoptotic cell microparticles prevents autoimmunity. Cell. 166:88–101. 10.1016/j.cell.2016.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steck, A.K., Xu P., Geyer S., Redondo M.J., Antinozzi P., Wentworth J.M., Sosenko J., Onengut-Gumuscu S., Chen W.-M., Rich S.S., et al. 2017. Can non-HLA single nucleotide polymorphisms help stratify risk in TrialNet relatives at risk for type 1 diabetes? J. Clin. Endocrinol. Metab. 102:2873–2880. 10.1210/jc.2016-4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaki, S., Sauer K., Iritani B.M., Chien S., Ebihara Y., Tsuji K., Takatsu K., and Perlmutter R.M.. 2000. Control of B cell production by the adaptor protein lnk. Definition of a conserved family of signal-modulating proteins. Immunity. 13:599–609. 10.1016/S1074-7613(00)00060-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaki, S., Watts J.D., Forbush K.A., Nguyen N.T., Hayashi J., Alberola-Ila J., Aebersold R., and Perlmutter R.M.. 1997. Characterization of Lnk. An adaptor protein expressed in lymphocytes. J. Biol. Chem. 272:14562–14570. 10.1074/jbc.272.23.14562 [DOI] [PubMed] [Google Scholar]
- Takizawa, H., Nishimura S., Takayama N., Oda A., Nishikii H., Morita Y., Kakinuma S., Yamazaki S., Okamura S., Tamura N., et al. 2010. Lnk regulates integrin alphaIIbbeta3 outside-in signaling in mouse platelets, leading to stabilization of thrombus development in vivo. J. Clin. Invest. 120:179–190. 10.1172/JCI39503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, Y., Luo Y., O’Shea J.J., and Nakayamada S.. 2022. Janus kinase-targeting therapies in rheumatology: A mechanisms-based approach. Nat. Rev. Rheumatol. 18:133–145. 10.1038/s41584-021-00726-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thien, M., Phan T.G., Gardam S., Amesbury M., Basten A., Mackay F., and Brink R.. 2004. Excess BAFF rescues self-reactive B cells from peripheral deletion and allows them to enter forbidden follicular and marginal zone niches. Immunity. 20:785–798. 10.1016/j.immuni.2004.05.010 [DOI] [PubMed] [Google Scholar]
- Tsokos, G.C., Lo M.S., Costa Reis P., and Sullivan K.E.. 2016. New insights into the immunopathogenesis of systemic lupus erythematosus. Nat. Rev. Rheumatol. 12:716–730. 10.1038/nrrheum.2016.186 [DOI] [PubMed] [Google Scholar]
- Ulff-Møller, C.J., Svendsen A.J., Viemose L.N., and Jacobsen S.. 2018. Concordance of autoimmune disease in a nationwide Danish systemic lupus erythematosus twin cohort. Semin. Arthritis Rheum. 47:538–544. 10.1016/j.semarthrit.2017.06.007 [DOI] [PubMed] [Google Scholar]
- Velazquez, L., Cheng A.M., Fleming H.E., Furlonger C., Vesely S., Bernstein A., Paige C.J., and Pawson T.. 2002. Cytokine signaling and hematopoietic homeostasis are disrupted in Lnk-deficient mice. J. Exp. Med. 195:1599–1611. 10.1084/jem.20011883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Völkl, S., Rensing-Ehl A., Allgäuer A., Schreiner E., Lorenz M.R., Rohr J., Klemann C., Fuchs I., Schuster V., von Bueren A.O., et al. 2016. Hyperactive mTOR pathway promotes lymphoproliferation and abnormal differentiation in autoimmune lymphoproliferative syndrome. Blood. 128:227–238. 10.1182/blood-2015-11-685024 [DOI] [PubMed] [Google Scholar]
- Wang, S., Wang J., Kumar V., Karnell J.L., Naiman B., Gross P.S., Rahman S., Zerrouki K., Hanna R., Morehouse C., et al. 2018. IL-21 drives expansion and plasma cell differentiation of autoreactive CD11chiT-bet+ B cells in SLE. Nat. Commun. 9:1758. 10.1038/s41467-018-03750-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.-F., Zhang Y., Lin Z., Zhang H., Wang T.-Y., Cao Y., Morris D.L., Sheng Y., Yin X., Zhong S.-L., et al. 2021. Identification of 38 novel loci for systemic lupus erythematosus and genetic heterogeneity between ancestral groups. Nat. Commun. 12:772. 10.1038/s41467-021-21049-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardemann, H., Yurasov S., Schaefer A., Young J.W., Meffre E., and Nussenzweig M.C.. 2003. Predominant autoantibody production by early human B cell precursors. Science. 301:1374–1377. 10.1126/science.1086907 [DOI] [PubMed] [Google Scholar]
- Wehr, C., Eibel H., Masilamani M., Illges H., Schlesier M., Peter H.-H., and Warnatz K.. 2004. A new CD21low B cell population in the peripheral blood of patients with SLE. Clin. Immunol. 113:161–171. 10.1016/j.clim.2004.05.010 [DOI] [PubMed] [Google Scholar]
- Yurasov, S., Wardemann H., Hammersen J., Tsuiji M., Meffre E., Pascual V., and Nussenzweig M.C.. 2005. Defective B cell tolerance checkpoints in systemic lupus erythematosus. J. Exp. Med. 201:703–711. 10.1084/jem.20042251 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the diagnosis and clinical manifestations of probands carrying SH2B3 variants.
shows rare variants in genes known to cause human SLE in the probands identified through WES.
shows data collection and refinement statistics for the SH2 domains of murine SH2B3 protein with phosphopeptides bound.
lists the primers used for introducing patient-specific variants and other published variants into mammalian expression vectors of human SH2B3.
shows sequencing primers designed for validating ORF sequences in the mammalian expression vectors of human SH2B3 via Sanger sequencing.
lists the guide RNAs (gRNAs), ssODNs for CRISPR/Cas9 gene editing of mouse models, and oligos for validating the editing results by Sanger sequencing.
shows antibodies, fluorochrome-conjugated streptavidin, and viability dyes used in flow cytometry.
shows primary and secondary antibodies used for ELISA and IF.
Data Availability Statement
The data underlying figures are available in the published article and its online supplemental material. Data used in Fig. 1 E have been deposited in the PBD archive (PDB ID 7R8W, E372K: PDB ID: 8CZ9).