Abstract
Cell memory refers to the capacity of cells to maintain their gene expression program once the initiating environmental signal has ceased. This exceptional feature is key during the formation of mammalian organisms, and it is believed to be in part mediated by epigenetic factors that can endorse cells with the landmarks required to maintain transcriptional programs upon cell duplication. Here, we review current literature analyzing the molecular basis of epigenetic memory in mammals, with a focus on the mechanisms by which transcriptionally repressive chromatin modifications such as methylation of DNA and histone H3 are propagated through mitotic cell divisions. The emerging picture suggests that cellular memory is supported by an epigenetic cycle in which reversible activities carried out by epigenetic regulators in coordination with cell cycle transition create a multiphasic system that can accommodate both maintenance of cell identity and cell differentiation in proliferating stem cell populations.
Cell memory is supported by the activity of epigenetic regulators in coordination with cell cycle transition.
INTRODUCTION
The zygote is a diploid cell that is formed upon the fusion of one haploid oocyte and one haploid sperm cell, and that harbors the potential to produce a new complete complex adult organism through a vast number of cell proliferation and differentiation rounds. The adult human body is composed by trillions of zygote-derived cells that are genetically equivalent but functionally distinct, and that cooperate to allow the formation of tissues and organs. Tissue specialization relies on the ability of cells to express a coordinated set of genes that endorses them with specific functions. Many cell type–specific gene expression patterns are established during embryo development but are maintained for a lifetime, although the original differentiation signal instructing the change in gene expression is no longer present during the adult life (1, 2). This type of cellular memory is in part enabled by epigenetic mechanisms that involve processes that ensure inheritance of phenotypic variation beyond (“epi-“) changes in the DNA sequence (“-genetics”) (3, 4). Solid proof of the existence of epigenetics is provided by studies in which two identical DNA sequences are differentially regulated in the nucleus. For example, in the case of gene imprinting and inactivation of the X chromosome, cis-acting epigenetic mechanisms facilitate the monoallelic expression of genes in diploid mammalian cells (5, 6). Deciphering the nature of the epigenetic mechanisms that mediate the ability of cells to have transcriptional memory is key to understanding how complex organisms such as humans are formed.
Epigenetic signals can be conceptually classified into two major groups (7). A given transcriptional response might be self-sustained in the absence of the originating stimulus through self-propagating (i) trans-acting mechanisms or by (ii) cis-acting molecular signals associated with the DNA sequence that they regulate. An example of a trans-acting mechanism would be the positive feedback loops of regulatory molecules that remain soluble in the cytoplasm during mitosis but can rapidly engage in transcriptional regulation upon cell division. While this type of mechanism will contribute to support transcriptional memory across cell generations, it cannot explain many aspects of gene regulation (i.e., monoallelic expression) and it is currently accepted that trans-acting mechanisms must be complemented by cis-acting epigenetic signals that are inherited through constant physical contact with the DNA sequence on which they act (4, 7). The most widely studied cis-acting mechanisms studied so far are covalent modification of DNA, histone variants, and posttranslational modifications (PTMs) of nucleosomes (8). In addition, information might also be encoded in other cis-acting signals such as the position of nucleosomes relative to DNA sequences, stable association of non-histone proteins, and higher-order chromatin structure (8).
In this review, we provide insights into the molecular mechanisms by which epigenetic factors might facilitate mitotically heritable transcriptional states in mammals. In fitting with strictest definitions of epigenetics—an epigenetic trait is a stably heritable phenotype resulting from changes in a chromosome without alterations in the DNA sequence (9)—we focus on cis-acting mechanisms. Among these, we have centered our discussion on methylation of DNA, lysine 27 of histone H3 (H3K27), and lysine 9 of histone H3 (H3K9) because they are three epigenetic signals widely studied so far (8). To complement our view, we refer the reader to recent reviews about epigenetics from a general angle that include nonmammalian systems (8, 10), with a focus on epigenetic changes in response to out-of-the-body environmental stimuli in adults (11), or with an emphasis toward transgenerational epigenetic inheritance (12).
METHYLATION OF DNA, H3K27, AND H3K9 ARE CANDIDATES TO SUPPORT TRANSCRIPTIONAL MEMORY
The term “epigenetics” was coined by C. H. Waddington in 1942, and it provided an explanation as to how the phenotypes of evolving organisms can respond to the environment without changing the genotype (13, 14). Nowadays, it is often accepted that epigenetic mechanisms allow cells to respond to the environment by inducing heritable chemical changes on chromatin that modulate gene expression without altering the sequence encoded on DNA (8). However, we are just beginning to understand what the molecular foundation of epigenetic memory is. Gene expression requires interaction between the transcriptional machinery and target DNA, which is wrapped around nucleosomes that can obstruct the accessibility of transcriptional regulators to their target DNA sequence (15). Therefore, it is perhaps not surprising that the best current candidates to uphold transcriptional memory are chromatin factors that chemically modify DNA without altering its sequence (DNA methylation), or that posttranslationally amend histone proteins that compose the nucleosome (PTMs of histones).
DNA methylation is a major regulator of epigenetic memory in mammals (8). Cytosines present in the DNA sequence in the context of a cytosine-guanine (CpG) dinucleotide can be methylated to produce 5-methylcitosine (5mC) (16). CpG sites are widespread throughout the genome but are accumulated at the so-called CpG islands, which are usually part of regulatory regions such as gene promoters or enhancers (16). Methylation of CpG sites is associated to transcriptional repression of transposable elements and cell type–specific genes (17). The implication of 5mC in cellular memory is exemplified by its role in maintaining transcriptional repression of parentally imprinted loci and the inactive X chromosome, which are expressed in a monoallelic way and therefore are hallmarks of epigenetic control in eukaryotes (5, 6, 18, 19). Although it is evident that DNA methylation supports transcriptional memory of many genomic regions, it is not undisputed whether this holds generally true for all loci across the genome and for all different cell types that compose the embryo and the adult body. Recent studies support that in some cell types such as embryonic stem cells (ESCs), CpG methylation displays very high turnover (20, 21) and that some genomic regions display lower maintenance efficiency as compared to the rest of the genome (22, 23). Mutant mouse ESCs (mESCs) with extremely low levels of DNA methylation can maintain their transcriptional program and self-renew in an undifferentiated state (24). Thus, in some contexts, DNA methylation is more dynamic than expected and might not be critical to maintain transcriptional memory [as discussed in (25, 26)].
Inheritance of nucleosomes containing posttranslationally modified histones is another major mechanism proposed to encode epigenetic memory in cis (8). Different histone PTMs—including acetylation, methylation, ubiquitylation, phosphorylation, and sumoylation—can be added at specific histone residues (27). While many histone PTMs have been shown to regulate gene activity in different cell systems (28), it is not clear which of them are simply regulators of gene expression and which truly constitute a heritable epigenetic mark that carry over functional information across mitotic cell generations. It has been proposed that, among all histone PTMs characterized so far, only two candidates might constitute bona fide epigenetic signals of the mammalian genome (29). These are methylation of H3K27 and H3K9 because they are the only histone PTM systems in which the machinery that deposit the histone marks can “read-and-write” the mark that they catalyze, and they have the capacity to create a positive feedback loop that might become independent of the information encoded on their target DNA sequence [as discussed in (10, 29)].
Methylation of H3K27 is deposited by Polycomb proteins, which are evolutionary conserved regulators of the genome that repress transcription of large sets of developmentally regulated genes (30, 31). Pioneering studies showed that Polycomb proteins maintain the transcriptional repression of Hox homeotic genes during fly development (32–35), whose pattern of expression becomes mitotically heritable upon transient exposure to segmentation transcription factors (TFs) (1). It was thereafter demonstrated that H3K27me3 is part of the mechanism by which Polycomb proteins can endorse transcriptional memory in cis: H3K27me3 can maintain the repressed state of target loci across cell generations in flies (36, 37), and H3K27me3 facilitates monoallelic expression of the X-chromosome during development in Caenorhabditis elegans (38). Notwithstanding, whether H3K27me3 instructs transcriptional memory is still under debate. For example, Polycomb-mediated inheritance of transcriptional states is partly dependent on the DNA in flies (36, 37). In addition, although H3K27me3 is required to maintain transcriptional memory in cis in mammalian somatic cells, it is not essential in pluripotent cells (39, 40).
H3K9me3 is also an evolutionary conserved histone modification associated with heterochromatin and transcriptional repression (41). Methylation of H3K9 is associated not only with gene repression but also with hindering RNA synthesis coming from DNA repetitive sequences such as transposable elements, centromeres, and telomeres (41). In fission yeast, H3K9me3 heterochromatin can be inherited in cis to facilitate transmission of gene repression for many cell generations (42–47). However, like in the case of H3K27me3, it is not clear to what extent the inheritance of H3K9 methylation is completely independent of DNA and RNA sequences (48, 49).
There is currently an open debate as to whether and how chromatin modifications provide cellular memory [i.e., (29, 50)]. However, substantial evidence supports that methylation of DNA, H3K27, and H3K9 provide transcriptional memory in mammals and that their mode of action includes, but is not restricted to, a read-and-write mechanism that facilitates their self-propagation through the mitotic cycle. In the following sections, we provide an overview of the key cellular and molecular aspects that support that methylation of DNA, H3K27, and H3K9 are cis-acting mechanisms that facilitate transmission of transcriptional memory through the mitotic cell cycle.
METHYLATION OF DNA, H3K27, AND H3K9 ARE REQUIRED TO MAINTAIN GENE EXPRESSION PROGRAMS AND CELL IDENTITY
Genetic removal of enzymes involved in methylation of DNA (51, 52), H3K27 (53–55), or H3K9 (56–59) leads to severe abnormalities during embryonic development. For example, deletion of the maintenance DNA methyltransferase DNMT1 causes global demethylation associated to gene derepression and cell death in different cell types (51, 60–64) and embryonic lethality during midgestation in mice (51). Similarly, deletion of Polycomb repressive complex 2 (PRC2) core subunits promotes a global loss of H3K27me3 coupled to misexpression of lineage-specific genes (65–68) and developmental arrest during embryo peri-implantation (53–55). Accordingly, mutation of lysine 27 residue of histone H3 largely phenocopies PRC2 loss of function in mESCs (69). Likewise, deletion of enzymes involved in methylation of H3K9 also promote developmental arrest during early development (56–59). For example, deletion of SetDB1 in mESCs or morula stage leads to up-regulation of trophoblast-specific genes and differentiation toward trophectoderm (70–73), leading to peri-implantation lethality during mouse development (57). Thus, in fitting with their role as regulators of cell memory, DNA methylation, H3K27me3, and H3K9me3 contribute to maintain cell type–specific gene expression programs and are essential for correct embryo development.
METHYLATION OF DNA, H3K27, AND H3K9 CAN BE ERASED FROM THE GENOME BY DEMETHYLASES
Chromatin modifications were originally perceived as stable marks on the genome that, once established, could only be removed by histone eviction or by dilution upon genome duplication in proliferating cells. However, later studies identified enzymes that can remove PTMs from DNA and histones (74–76). In the case of DNA methylation, the steady state of CpG methylation depends on the balance of the DNMTs and demethylases enzymes of the family of Ten-eleven translocation (TET) (77). DNMTs transfer a methyl group to the fifth carbon of cytosines in CpG dinucleotides and produce 5mC. DNMT3A, DNMT3B, and DNMT3C (rodent-specific) methylate previously unmodified CpG sites de novo, while DNMT1 methylates hemi-methylated DNA to maintain CpG methylation of both strands of DNA upon DNA replication (26, 77). The methyl group on carbon five of cytosines can be actively removed following a two-step mechanism. First, TET1, TET2, or TET3 enzymes oxidize 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) (75, 77). Oxidized cytosines are then removed and replaced by unmodified ones through thymine DNA glycosylase (TDG)–dependent base excision repair pathways (75).
Likewise, lysine methylation on histone tails is also the result of the balance between methylation and demethylation carried out by highly regulated and specialized enzymes. H3K27 methylation is catalyzed by PRC2. PRC2 is typically composed of core subunits EED, SUZ12, RBBP4/7, and EZH1/2, of which the last one harbors the histone methyltransferase catalytic function (31). PRC2 function is regulated by nonstoichiometric accessory subunits that form functionally specialized complexes PRC2.1 (PCL1-3, LCOR, and EPOP) and PRC2.2 (JARID2 and AEBP2) (31). Deposition of H3K27me3 by PRC2 can be counteracted by histone demethylases KDM6A/UTX and KDM6B/JMJD3, which belong to the family of Jumonji C (JmjC) domain–containing proteins that carry out demethylation through an oxidative reaction that requires iron Fe(II) and α-ketoglutarate as cofactors (78, 79). In the case of H3K9 methylation, the enzymes responsible for its catalysis are SUV39H1, SUV39H2, SETDB1, SETDB2, G9A, and GLP, which are differentially expressed during development and display different target specificity and methylation capabilities (41). Like in the case of H3K27me3, H3K9me3 can also be erased by the methylase activity of Jumonji proteins that belong to the groups of JMJD1/KDM3 and JMJD2/KDM4 enzymes (80, 81).
Thus, DNA, H3K27, and H3K9 methylation can be actively erased from chromatin through specific demethylases, which are differentially expressed during development and target cell type–specific genomic regions (77, 80, 81). Likewise, methyltransferase complexes target genomic regions in a cell type–specific fashion (77, 82, 83). In addition, nucleosome turnover regulates histone methylation patterns, probably through mechanisms that involve maintenance of low-level histone acetylation (84–86). Therefore, turnover rates of methyl groups on DNA and histones are the result of the balance between nucleosome turnover, methylation, and demethylation processes that can be locus and cell type specific (77, 87, 88). The maintenance of faithful DNA methylation patterns is not homogeneous across the genome because regions with lower DNA methylation levels display reduced maintenance efficiency (23). These low maintenance regions are associated to cancer and aging, suggesting that differential turnover rates of epigenetic information laid on some regions of the genome might lead to loss of cell identity and disease etiology (23).
THE CURRENT MODEL OF EPIGENETIC MEMORY: READ-WRITE DURING S PHASE AND PERSISTENCE THROUGH MITOSIS
To understand the molecular basis of cellular memory, it is essential to decipher how epigenetic signals are passed from one mitotic generation to the next. However, we are just beginning to comprehend how epigenetic regulators function in coordination with cell cycle transition. The eukaryotic cell cycle is divided into four phases: gap 1 (G1), DNA synthesis (S), gap 2 (G2), and mitosis (M) (89). Current studies support a model in which cis-acting epigenetic signals present in G1 must be duplicated and incorporated in the new chromosome during S phase. After progressing through G2, epigenetic marks need to be maintained on chromosomes and distributed between daughter cells during M phase. Upon mitosis exit, epigenetic determinants must facilitate efficient restoration of the cell type–specific gene expression program. Despite that this mode of action is often assumed for chromatin factors, whether epigenetic regulators truly fit this model as well as what is the underlying molecular mechanisms are just beginning to be addressed.
RESTORATION OF EPIGENETIC PATTERNS UPON DNA REPLICATION
To maintain epigenetic information in sister chromatids after genome duplication, the DNA molecule and associated epigenetic information encoded in cis must be duplicated to produce a new identical chromosome during DNA replication in S phase. DNA replication starts stochastically from hundreds of positions in the genome called DNA replication origins (90), from which a new unmethylated complementary DNA is synthesized by the replisome, which requires unraveling of one or two nucleosomes ahead of the replication fork (91). Thus, upon DNA replication, DNA methylation has been diluted into newly synthesized DNA strands, and the nucleosome landscape has been disrupted genome-wide. How is epigenetic information encoded on DNA methylation and histone PTMs copied on the new replicated chromosomes?
Maintenance of DNA methylation upon S phase
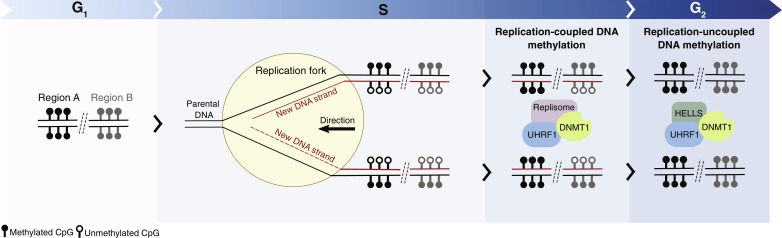
As the replication fork progresses and DNA is replicated, the resulting DNA molecule is hemi-methylated: made of a parental strand in the original methylation state and a newly synthesized unmethylated strand (Fig. 1). Preservation of the methylation state in both DNA molecules relies on the activity of DNMT1, which displays high affinity for hemi-methylated DNA and can restore CpG methylation on the unmethylated strand (26). The function of DNMT1 depends on UHRF1, which also binds hemi-mCpGs through its SET and RING-associated (SRA) domain (92, 93), adding an extra layer of specificity to the activity of DNMT1 on hemi-methylated CpGs. Restoration of DNA methylation patterns is achieved in two phases. A rapid replication-coupled phase within minutes of fork passage and a gradual replication-uncoupled phase that extends outside of the S phase (Fig. 1) (23). During the replication-coupled phase, DNMT1 is recruited to the replication forks by interaction with the DNA replication machinery. These include protein-protein interactions between DNMT1 and PCNA, and between UHRF1 and LIG1 (94, 95). Although this mechanism promotes quick recovery of the methylation pattern for many regions of the genome, other CpG sites can remain hemi-methylated for hours after genome duplication (96–98). The delay between DNA replication and the restoration of DNA methylation is an important challenge for the transmission of epigenetic information because nucleosomes are associated within minutes to nascent DNA (99) and block the activity of DNA methylases (100, 101). How is DNA methylation restored at hemi-methylated CpG sites that were not fully methylated by the replication-coupled machinery? In addition to contain domains that facilitate the binding of UHFR1 to nucleosome-free hemi-methylated CpG sites, UHFR1 harbors TTD (tandem Tudor domain) and PHD (plant homeodomain) domains that recognize histone H3 tail with a preference toward H3K9me2/3, facilitating recruitment of DNMT1 to chromatin independently of the machinery present in the replication forks (102, 103). Thereafter, the activity of the SNF2-familiy chromatin remodeler HELLS/LSH seems to be part of the mechanism facilitating access of DNA methylases to CpG sites and recovery of full methylation patterns (23, 104). The replication-uncoupled mechanism is probably essential to avoid the loss of epigenetic information across cell generations since the replication-coupled mechanism has a too narrow window of opportunity to restore DNA methylation before nucleosomes are reassembled upon DNA synthesis. This mechanism is particularly relevant in isolated CpG sites (with no neighboring CpG sites around 70 base pairs) because these are the DNA methylated sites that display the slowest maintenance kinetics in the genome and have a higher tendency to lose their methylated state across mitotic cell divisions (23, 105).
Fig. 1. Model of the maintenance of DNA methylation upon genome duplication.
The methylated (filled lollipops) or unmethylated (empty lollipops) status of CpG sites are depicted during G1, S, and G2 phases. Proteins involved in restoration of methylation patterns are represented as purple (replisome), green (HELLS), blue (UHRF1), and green (DNMT1) circles.
Maintenance of histone PTMs upon S phase
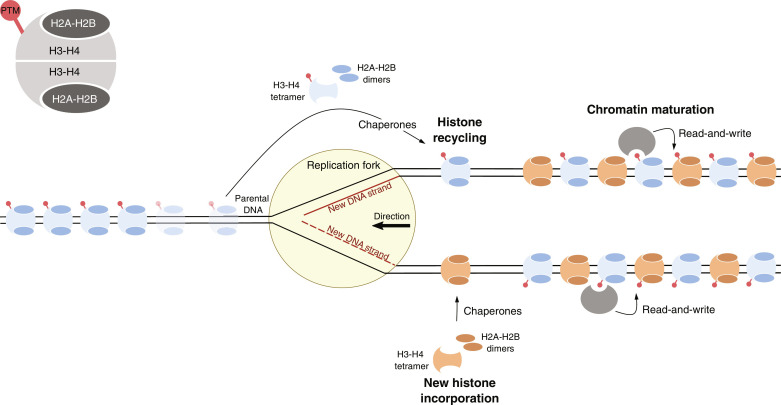
Nucleosomes are dissociated from DNA ahead of the replication fork (91, 106), and therefore, to maintain their encoded information, they would need to be reincorporated at the same genomic position after DNA synthesis. Initial studies showed that after fork passage, histones composing detached nucleosomes are refolded on chromatin with an equal ratio of old to newly synthesized histones H3 and H4 on both synthesized DNA molecules (91, 106), suggesting that histone-encoded epigenetic information is passed to the daughter DNA molecules. It was later demonstrated that disassembly of the nucleosome during DNA replication leads to the release of stable H3-H4 tetramers that do not mix with newly synthesized histones, as opposed to more dynamic H2A-H2B dimers that can mix with new histones and partner with old and new H3-H4 tetramers (91, 106). This highlighted H3-H4 tetramers as good candidates to transmit transcriptional memory across DNA replication. In fitting with this idea, histone chaperones integrated within the replisome ensure that parental histone H3-H4 tetramers are symmetrically distributed on replicated chromosomes (Fig. 2) (107). In particular, MCM2 in association with DNA polymerase α and CTF4 acts as a histone chaperone promoting the recycling of old H3-H4 tetramers into the lagging strand (108, 109), whereas the DNA polymerase ε subunits POLE3 and POLE4 mediate the transfer of parental H3-H4 tetramers onto the leading strand (110, 111). Perturbation of these recycling mechanisms hinders symmetric inheritance of H3K9me3 and H3K27me3, causes derepression of developmental genes, and compromises stem cell potential (112, 113). In addition, a recent report shows that modified H2A-H2B dimers are also segregated symmetrically to daughter strands via POLA1 on the lagging strand (114), further supporting that maintenance of chromatin state through DNA replication is mediated by symmetrical inheritance of modified histones that formed the parental nucleosomes.
Fig. 2. Model of the maintenance of histone PTMs after the passage of the replication fork.
Gray nucleosome displaying typical configuration of histone proteins is depicted on the top left corner. PTMs are represented as a red lollipop. Parental (blue) and newly synthesized (orange) nucleosomes are depicted. Epigenetic machinery with read-and-write capacity during chromatin maturation is represented in gray.
To preserve transcriptional memory, the modified parental histones that are inherited must remain bound to the same genomic position after passage of the replication fork (Fig. 2). This hypothesis has been addressed using new techniques that allow the labeling of nucleosomes ahead of the replication fork and tracking their fate onto the replicated DNA molecule. Parental histones with different types of PTMs, including H3K9me3 and H3K27me3, can be incorporated on newly replicated DNA (88, 115) and reproduce the same genome-wide patterns observed before DNA replication–coupled dissociation (88, 116, 117). Recycling of parental histones to the same genomic position is dissimilar across the genome, and it has been proposed to be more effective at transcriptional repressed genes enriched for H3K27me3 as compared to active ones displaying H3K4me3 (117). In fitting with this idea, the histone chaperone NPM1 interacts with PRC2 and MCM2, and it is required for inheritance of parental nucleosomes at PRC2-repressed chromatin domains during DNA replication (118). The observation that recycling of parental histones is more accused at repressed chromatin as compared to transcriptionally active regions supports that histone PTMs associated with gene repression rather than with transcriptional activity transmit transcriptional memory across genome duplication.
Because the density of nucleosomes on DNA must be maintained upon DNA duplication, recycling of parental nucleosomal histones can only partially restore the initial chromatin landscape. In agreement, global levels of histone PTMs on chromatin, including H3K9me3 and H3K27me3, are diluted twofold after DNA duplication (115). New synthesized histones incorporated onto the genome, generally hypomethylated and hyperacetylated (119, 120), must be “matured” to restore the levels of histone PTMs displayed in G1 phase (Fig. 2). This process is heterogeneous and displays modification and locus-specific kinetics (88, 115). As compared to other PTMs, H3K27me3 displays a globally slower recovery kinetics after replication (88, 114, 115). However, restoration of H3K27me3 levels is faster at sites with high PRC2 occupancy and might be stimulated by the presence of post-replicative H2AK119ub (88, 114). Faster restoration kinetics at these sites is probably mediated by enhanced recruitment of EZH2 through H3K27me3 read-write mechanisms present in PRC2 (121, 122) and JARID2-mediated recruitment to H2AK119ub (123). In fitting, binding of EZH2 to target regions is augmented during S and G2 phases as compared to G1 phase (124). Together, these studies support that H3K27me3 genome-wide distribution is restored on sister chromatids through effective recycling of H3K27me3-modified parental histones that facilitate the action of positive feedback loops that enhance recruitment of PRC2 and restoration of H3K27me3 levels (Fig. 2). Because the enzymatic activities of complexes involved in H3K27me3 methylation and demethylation are regulated during development and disease (80, 82), it is probable that the kinetics of H3K27me3 restoration after DNA replication will vary among different cell types and in some pathological contexts such as cancer.
MAINTENANCE OF EPIGENETIC INFORMATION ON MITOTIC CHROMOSOMES
If DNA methylation, H3K9me3, and H3K27me3 were fully restored at their genomic sites upon genome duplication, to be truly epigenetic mechanisms in cis, they would need to be maintained on mitotic chromosomes, segregated into daughter cells, and instruct the reestablishment of the transcriptional program in the subsequent G1 phase of the cell cycle. This is not a trivial challenge because during mitosis chromosomes are subjected to drastic structural and functional changes. Chromosomes are condensed 10,000-fold, and the nuclear envelope is disassembled (125). This is coupled to disruption of higher-order chromatin organization (126, 127), dissociation of many TFs (128), inactivating phosphorylation of RNA polymerase II (RNAPII) (129), and a general decrease of gene transcription (130). Mitotic chromosomes display widespread alteration on chromatin modifications, including reduced acetylation of histone tails that facilitates chromatin compaction, and accumulation of mitosis-regulatory chromatin marks [i.e., phosphorylation of H3 on serine-10 and serine-28 (131)]. Nevertheless, it is often assumed that once epigenetic marks are duplicated during S phase, their genome-wide distribution present in G2 phase remains unaltered on mitotic chromosomes, and it is inherited by cells in G1 phase without interfering or being affected by regulatory processes required for mitosis completion. Although this constitutes a feasible model to explain epigenetic memory, reality is probably more complex than this, and alternative mechanisms might be operating or coexist.
The genome-wide patterns of methylated DNA, H3K9me3, and H3K27me3 are generally presumed to remain unaltered through mitosis, but detailed molecular and functional characterization of these epigenetic systems during cell division is lacking. Initial cell fractionation and imaging analyses showed that condensed chromosomes harbor DNA methylation (132) and some histone PTMs typically associated with the regulation of gene expression during interphase [including H3K27me3 (133) and H3K9me3 (134, 135)] (Table 1) (131). This led to the hypothesis that a substantial part of the epigenetic landscape present during interphase might be maintained on mitotic chromosomes providing bookmarks that aid to the restoration of gene expression upon mitosis exit (131). Later studies using proteomics have reinforced this idea because they have shown that while histone acetylation marks are decreased during mitosis, methylation marks (including H3K9me3 and H3K27me3) (136) are globally maintained (Table 1). Complementarily, proteomic analyses indicate that most of the chromatin machinery required to sustain DNA methylation, H3K27me3, and H3K9me3 remains bound to chromosomes during mitosis (i.e., DNMT1/3a/3b, several PRC2 subunits, G9A, and SUV39H1/H2) (137, 138). Hence, a substantial part of the molecular machinery required to support epigenetic memory remains bound to chromosomes upon chromosome condensation in metaphase. However, binding of epigenetic factors to mitotic chromosomes does not necessarily imply that their genome-wide localization or functional state is maintained during mitosis, and studies analyzing the genomic localization of the epigenetic machinery are unavailable (Table 1). Genome-wide DNA methylation patterns seem be only partially restored on mitotic chromosomes (23, 97, 139), and this has been proposed to underlie cancer and aging-associated diseases (23). Chromatin immunoprecipitation followed by sequencing (ChIP-seq) analyses of a set of histone PTMs in HeLa cells have shown that the genomic distribution of H3K27me3 and other histone methylation marks (i.e., H3K36me3 and H3K4me3) are similar in asynchronous and metaphase-arrested cells (136), supporting that the Polycomb system might transmit epigenetic information through stable methylation of H3K27 through mitosis. However, characterization of the activity of PRCs during interphase transition supports that polycomb domains build up in every cell cycle in mESCs (124, 140, 141), suggesting that the genomic distribution of H3K27me3 is perturbed during mitosis. These studies have been carried out in different systems, and therefore, they raise the possibility that the mechanisms that endorse epigenetic memory are not identical in different cell types (human cancer HeLa-S3 cells versus mouse pluripotent ESCs). In consonance with this idea, changes in the genomic distribution of histone PTMs upon mitosis entrance differ depending on the analyzed cell type (Table 1). For example, the distribution of H3K27ac on mitotic chromosomes can (i) uniformly increase in breast cancer MCF-7 cells (142), (ii) decrease in U2OS, RPE1, and HeLa-S3 cells (136, 143), or (iii) be reorganized in progenitor G1E erythroblasts (144) and in pluripotent stem cells (145), where its maintenance at specific sites facilitates reactivation of stem cell–associated genes (146).
Table 1. Mitotic bookmarking by epigenetic factors.
Table summarizing bibliography supporting the presence of epigenetic regulators and histone PTMs on mitotic chromosomes.
| System | Protein | Global decoration of mitotic chromosomes* | Genome-wide localization during mitosis † |
| DNA methylation | CpG methylation | mESCs (132, 139, 217), mouse embryos (218), hESCs (219) | Reduced levels [hESCs (97), mESCs (139), HCT116 (97), HeLa-S3 (23)] |
| DNMT1 | HeLa (220), C2C12 (220), mESCs (138), T98G (137) | – | |
| DNMT3A | mESCs (138) | – | |
| DNMT3B | mESCs (138) | – | |
| MECP2 | C2C12 (221) | – | |
| UHRF1 | MEFs (222) | – | |
| H3K27me3 | H3K27me3 | ESCs (145, 157, 180), MEFs (157), HeLa-S3 (136) | Similar [hESCs (180), HeLa-S3 (136)] |
| EZH2 | HeLa (133), NIH3T3 (133), mESCs (138, 155), T98G (137) | – | |
| SUZ12 | HeLa (133), NIH3T3 (133), mESCs (138, 155), T98G (137) | – | |
| EED | mESCs (138, 155), T98G (137) | – | |
| JARID2 | mESCs (138) | – | |
| MTF2 | mESCs (138) | – | |
| H3K9me3 | H3K9me3 | mESCs (145, 157, 223), MEFs (157), HeLa-S3 (136), HeLa (224), A549 (224) | – |
| SUV39H1 | mESCs (138, 157, 223), MEFs (157), T98G (137), HeLa (225) | – | |
| SUV39H2 | mESCs (138, 157, 223), MEFs (157) | – | |
| GLP/EHMT1 | T98G (137), mESCs (157), MEFs (157) | – | |
| G9A/EHMT2 | T98G (137), mESCs (157), MEFs (157) | – | |
| HP1 | mESCs (157), MEFs (157), HeLa (226) | – | |
| System | Other PTMs | Global decoration of mitotic chromosomes* | Genome-wide localization during mitosis † |
| Histones | H3K36me3 | HeLa-S3 (136) | Similar [HeLa-S3 (136)] |
| H3K4me1 | ESCs (145), HeLa-S3 (136), HeLa (224), A549 (224), U2OS (143), RPE1 (143) | Similar [U2OS (143), RPE1 (143)], reduced levels [HeLa-S3 (136)] | |
| H3K4me2 | ESCs (145), HeLa (224), A549 (224) | Increased levels [MCF-7 (142)] | |
| H3K4me3 | HeLa-S3 (136), HeLa (224), A549 (224), U2OS (143), RPE1 (143) | Similar [HeLa-S3 (136), U2OS (143), RPE1 (143)], reorganized [hESCs (180)] | |
| H3K9ac | ESCs (145), HeLa-S3 (136) | Similar [U2OS (143)], reduced levels [HeLa-S3 (136)] | |
| H3K27ac | ESCs (145), HeLa-S3 (136), U2OS (143), RPE1 (143), G1E-ER4 (161) | Increased levels [MCF-7 (142)], reduced levels [HeLa-S3 (136), G1E-ER4 (161), U2OS (143), RPE1 (143)], reorganized [ESCs (145), G1E-ER4 (144), miPSCs (160)] |
*Global decoration of mitotic chromosomes has been determined by any of the following approaches: live or fixed imaging, mass spectrometry, or biochemical fractionation.
†Genome-wide maps during mitosis have been obtained by ChIP-seq or Cut&Run.
While the importance of methylation of DNA, H3K27, and H3K9 on mitotic bookmarking remains an open question, emerging evidence indicates that some TFs remain bound to mitotic chromosomes, and that this can influence efficient transmission of transcriptional memory (147). During mitosis, higher-order chromatin structure [A/B compartments, topologically associating domains (TADs), and DNA loops] is altered during chromosome condensation (127), but local accessibility to DNA sequences is generally maintained (148, 149). In agreement, some TFs—for example, GATA1 (150) in erythroid cells, FOXA1 (151) in liver cells, or ESRRβ (152) and OCT4 (145) in mESCs—remain bound to a subset of their genomic regions on mitotic chromosomes. The binding of GATA1 and ESRRβ to condensed chromosomes is associated to efficient activation of target genes upon mitosis exit (150, 152), and mitotic bookmarking by the pluripotent factor OCT4 facilitates maintenance of cell identity in mESCs (145). This supports that stable binding of TFs to target genomic regions during cell cycle transition can endorse transcriptional memory to proliferating cells. However, the distribution of the pluripotency TFs OCT4 and SOX2 on mitotic chromosomes can vary depending on the experimental conditions used, which probably reflects the difficulty of capturing dynamic interactions between TFs and mitotic chromatin (145, 148, 153–156). This new role of TFs as mitotic bookmarks might prompt the conclusion that TFs endorse epigenetic memory to the genome, however their mode of action should not be considered an epigenetic mechanism because (i) it is highly dependent on their sequence-specific binding, and (ii) they constitute at the same time the initial trigger that changes gene expression and the mechanism that facilitates its maintenance. Notwithstanding, the role of TFs and epigenetic regulators in mitotic bookmarking might be intermingled because deletion of PRC2, DNMTs, or SUV39H1/H2 in mESCs affects chromosome compaction during mitosis and indirectly alters the binding of bookmarking TFs to mitotic chromatin (138, 157). In consonance, a recent report indicates that core subunits of the chromatin remodeler SWI/SNF bookmark gene promoters on mitotic chromosomes and facilitate TF binding and gene activation of neural differentiation inhibitors upon mitosis exit in mESCs (155).
REACTIVATION OF GENE TRANSCRIPTION DURING MITOTIC EXIT
Following chromosome segregation, during telophase and early G1, cells need to restore their chromatin setup and gene expression level to facilitate housekeeping and cell type–specific cellular functions. The molecular events driving the restoration of gene expression programs are poorly understood (158). Pioneering studies measuring nascent RNA in nocodazole-induced mitotic arrest cells showed that there is a global transient hyperactivation of the genome at 80 to 120 min after prometaphase-arrested cells are released to progress through mitosis (143, 144, 159, 160). Transcriptional reactivation does not occur homogeneously across the genome (143, 144, 159, 160). Enhancer recommissioning [measured as enhancers producing enhancer RNAs (eRNAs)] occurs at the same time of their putative targets, but enhancer-promoter interactions do not spike (143, 144, 159), suggesting that the initial transcriptional burst is not mediated by enhancer-promoter interactions. Although there is a global loss of deacetylation of H3K27 during mitosis, the genomic sites that retain higher levels of this mark are more prone to display a fast transcriptional reactivation (144, 160, 161), supporting the general principle that retention of particular histone PTMs on key genomic regions during mitosis is important for efficient recruitment of transcriptional machinery and restoration of gene expression programs. However, the possibility that the postmitotic burst in gene reactivation is related to a general mechanism based on differential decondensation kinetics of different parts of the genome cannot be discarded. In agreement with this idea, in mESCs where chromatin is particularly dynamic and plastic (162), hyperactivation of the genome occurs faster (163) than in other cell types (143, 144, 159), and it does not show a delay between the activation of housekeeping and stem cell maintenance genes (160, 163). Moreover, a recent report using a drug-free approach in mESCs supports the possibility that the transient hyperactivation of the genome during mitosis exit might be an unphysiological consequence of nocodazole treatment (156). Thus, future studies will need to clarify the molecular underpins of post-mitotic gene reactivation as well as document the relevance of epigenetic marks such as DNA methylation, H3K9me3, and H3K27me3 in the restoration of gene expression programs.
TRANSCRIPTIONAL ACTIVATION OF LINEAGE SPECIFIER GENES IS REGULATED DURING CELL CYCLE TRANSITION IN STEM CELLS
Epigenetic memory is particularly relevant in the context of proliferating stem cells because they need to maintain transcriptional memory but accommodate the possibility of circumventing the maintenance mechanism to allow changes in cell identity. Because epigenetic mechanisms in cis facilitate maintenance of gene expression programs and cell identity across cell duplications, perturbation of the chromatin landscape during DNA replication and chromosome condensation are usually proposed as potential windows of opportunities to initiate the transcriptional changes that lead to lineage transition (89, 147, 164, 165). In agreement, the response of cells to differentiation signals is dependent on the phase of the cell cycle in which the cells reside in several tissues and organisms including yeast (166), amoebas (167), worms (168), flies (169) and mammals (170–172). The phase of the cell cycle that leads to more efficient cell differentiation varies in these biological systems, suggesting that there is not a unique universal mechanism of cell cycle–dependent regulation of cell differentiation.
In the case of mammalian ESC populations, cells in G1 are more prone to induce expression of developmental genes and effectively differentiate than cells in S and G2 phases (173–176). The higher tendency of cells in G1 to exit pluripotency depends on the combined action of several mechanisms (177), of which current studies suggest that the TF SMAD2/3 (175), TDG (178), and the chromatin proteins trithorax (179) and polycomb (124, 140) are key regulators. The repressive activity of PRC1 and PRC2 on lineage specifier genes is partially alleviated during the G1 phase (124, 140, 160). Concordantly, cells in G1 phase accumulate higher levels of the transcriptional activating mark H3K4me3 and its methyltransferase enzyme KMT2B at the promoter of lineage specifier genes (179, 180). This is associated to a higher level of transcriptional activation of PRC-repressed lineage specifier genes during the G1 phase (124, 160, 181) and increased cell differentiation propensity (140, 173–176). The preference of pluripotent cells to enter lineage transition during G1 might also be favored by increased UTX-mediated H3K27me3 demethylation activity during S phase, facilitating the engagement of the newly G1-synthesized lineage specifier TFs on their target genes (182). Notwithstanding, stimulation of mESCs in G2-M with retinoic acid induces the activation of genes involved in extraembryonic development instead of genes enabling the formation of the embryo (183), indicating that mESCs in G2 are still responsive to differentiation signals to some extent, and further supporting that the phase of the cell cycle determines cell fate in pluripotent cells. Thus, together, current studies support that changes in chromatin occurring during mitosis lead to a transient destabilization of the epigenetic mechanisms that ensure maintenance of cell type–specific transcriptional programs in the subsequent G1 phase of the cell cycle in pluripotent cells and that this creates a window of opportunity for efficient activation of the otherwise repressed master regulators of cell differentiation.
THE EPIGENETIC MACHINERY CAN BE DIRECTLY REGULATED BY CELL CYCLE KINASES
Comprehension of the molecular basis of epigenetic memory will require deciphering how epigenetic factors self-perpetuate in coordination with master regulators of cell cycle transition. The eukaryotic cell cycle is governed by the activity of serine-threonine-protein kinases that orchestrated phosphorylation of hundreds of substrate proteins and regulate transition through S and mitosis phases (184). Therefore, a plausible scenario is that cell cycle kinases directly phosphorylate and modulate the function of chromatin factors to coordinate transmission of epigenetic memory. In agreement with this idea, the molecular machinery involved in methylation of DNA, H3K27, and H3K9 is phosphorylated in numerous residues, and many of these phosphosites are bioinformatically predicted [recognition motif identified using kinase prediction tool (185) at www.phosphosite.com] to be substrates of kinases that regulate cell cycle transition and mitosis progression [CDK1/2/4/6 (186), Aurora kinases A/B/C (187), and PLK1/2/3 (188)] (Table 2). Experimental examination of the functional relevance of some of these residues confirms that they are substrates of different cell cycle kinases and that their phosphorylation affects their epigenetic function: DNA methylation (189–191), H3K27me3 (192–197), and H3K9me3 (198, 199) (Table 2).
Table 2. Regulation of epigenetic factors by cell cycle kinases.
Table summarizing phosphoresidues on epigenetic regulators, putative regulating cell cycle kinases, and current functional validation studies. Phosphoresidues and putative cell cycle kinases were identified using the curated database www.phosphosite.com. Only phosphosites that have been described in at least five mass spectrometry–based discovery studies and cell cycle serine-threonine kinases that were highly ranked (site percentile within top 6%) are shown.
| System | Protein | Phosphorylated residues | Putative cell cycle kinase | Functionally validated | Refs. |
| DNA methylation | DNMT1 | S127, S133, T137, S141, S143, S152, S154, S189, S192, S209, S288, S312, S394, S398, Y399, S714, S732, S954, Y969, S1105, Y1405 | CDK1/2 (S127), CDK1/2/4/6 (T137, S714, S954, S1105), CDK1 (S152, S154), AurA/B/C (S732), AurB/C (S141), AurC (S143, S288), PLK1/2/3 (S394), CK2A1/2 (S143, S394) | CDK1/2 (S154), CDK4 (S127, S954) | (189, 190) |
| DNMT3A | S105, S243, S255, T261, S377, S390 | CDK1 (S105), CK2A1/2 (S390) | CK2A1 (S390, S393) | (191) | |
| DNMT3B | S51, S52, S53, S98, S100, S136, S202, T383 | Aur A/B/C (S51, S52, T383) | – | ||
| TET1 | T30, T31, S871 | PLK4 (S871) | – | ||
| TET2 | S38, S75, S99, S696, S1107, T1114, T1122 | CDK2 (S1107), CDK2/6 (S1114, T1122) | – | ||
| TET3 | T441, S504, S801, T816 | CDK1/2/6 (S801, S816) | – | ||
| H3K27me1/2/3 | EZH2 | T339, T345, S362, S363, S366, T367, T369, S380, S412, T416, T487 | CDK1/2/4/6 (T345), CDK2/4/6 (T416), CDK4 (T367), CDK1 (T487), AurB (S366), CK2A1/2 (S380) | CDK1 (T345, T487), CDK2 (T345, T416) | (192–196) |
| EZH1 | – | – | – | ||
| SUZ12 | S541, S546, S583, S695, S726 | PLK2 (S546, S726, S583), CK2A1/2 (S546, S583), PLK3(S583) | PLK1 (S539, S541, S546), CK2A1/2 (S583) | (197, 227) | |
| EED | S2, S34, T55 | PLK3 (S34), CK2A1/2 (S2, S34) | – | ||
| RBBP4 | S110, T144, S146, S355 | PLK2 (S355) | – | ||
| RBBP7 | S3, S95, T143, S145, S354 | PLK1/3 (S95), PLK2/3 (S354) | – | ||
| AEBP2 | S24, S139, S141, S167, S206, S210, S211, T242, S390 | CDK1 (S24), AurA/B/C (S167, S206, S390), PLK1/3 (S210), PLK2 (S211), CK2A1/2 (S211) | – | ||
| JARID2 | S331, S455 | CDK1/2/4/6 (S331, S455) | – | ||
| EPOP | – | – | – | ||
| LCOR | S37, S42, S249 | – | – | ||
| PHF1 | S20, S515, S522 | CDK1 (S20), CDK1/4/6 (S515) | – | ||
| MTF2 | T24, S488 | – | – | ||
| PHF19 | Y45 | – | – | ||
| KDM6A | S769, S818, S829 | – | – | ||
| KDM6B | S224, T637 | CDK1 (S224) | – | ||
| H3K9me1/2/3 | SUV39H1 | S391 | CDK1/2/4/6 (S391) | CDK1/2 (S391) | (198) |
| SUV39H2 | S381, S384, S388 | CDK1/2/6 (S388) | – | ||
| SETDB1 | S1066 | CDK1/2/4/6 (S1066) | – | ||
| SETDB2 | – | – | – | ||
| GLP/EHMT1 | S435 | – | – | ||
| G9A/EHMT2 | S40, T44, S118, S119, S121, S133, S140, S153, S173, S232, S237, S246, S350, S413, T555, S569 | CDK1/2/4/6 (S119), AurA/B/C (S246), AurB/C (S133), PLK2 (S413) | CK2A1 (S211) | (199) | |
| JMJD1A | S463, S264, S265, S325, S373, S445 | AurA/B/C (S264), AurB/C (S265) | – | ||
| JMJD2 | T155, S502, S523 | CK2A1/2 (S523) | – | ||
| H3 | H3.1 | S10, T11, S28, Y41, T45, S57, T58, T80, S86, Y99 | AurB/C (S10, T11, S28) | AurA/B/C (S10), AurB (S28) | (228, 229) |
Pioneering studies analyzing the regulation of PRC2 showed that phosphorylation of T345 and T416 by CDK1/2 favors recruitment of PRC2 and maintenance of H3K27me3 levels at target loci (192, 193, 200, 201). In contrast, CDK1-mediated phosphorylation of T487 disrupts interaction of the PRC2 catalytic subunit EZH2 with core proteins SUZ12 or EED, inhibiting PRC2 methyltransferase activity (194, 195). These studies support that binding of EZH2 to target genes might build up during cell cycle transition as CDK2-CyclinA and CDK1-CyclinB are activated during S and G2 phases, respectively. At later stages of the cell cycle when CDK2-CyclinA is no longer active, CDK1-CyclinB phosphorylates T487 and might promote disassembly of PRC2 complex during metaphase. In fitting with these observations, Polycomb repressive activity on lineage specifier genes is enhanced in G2 phase as compared to cells in G1 phase in mESCs (124, 140). In the case of the DNA methylation and H3K9me3 systems, evidence linking cell cycle machinery and methylation of DNA or H3K9 is scarcer. It has been shown that the activity of DNMT1 (residue S154) (189) and DNMT3A (residues S386 and S389) (191) is regulated by phosphorylation through CDK1/2/5 and CK2, respectively. Likewise, SUV39H1 (residue S391) (198) and G9a (residue S211) (199) are phosphorylated by CDK2 or CK2, respectively, to modulate their recruitment to chromatin. Although these studies start to reveal a direct regulation of epigenetic factors by cell cycle kinases, they are probably just the tip of the iceberg, and future analyses will probably disclose that a solid cross-talk between cell cycle kinases and chromatin factors is an essential property of bona fide epigenetic regulators.
CONCLUDING REMARKS
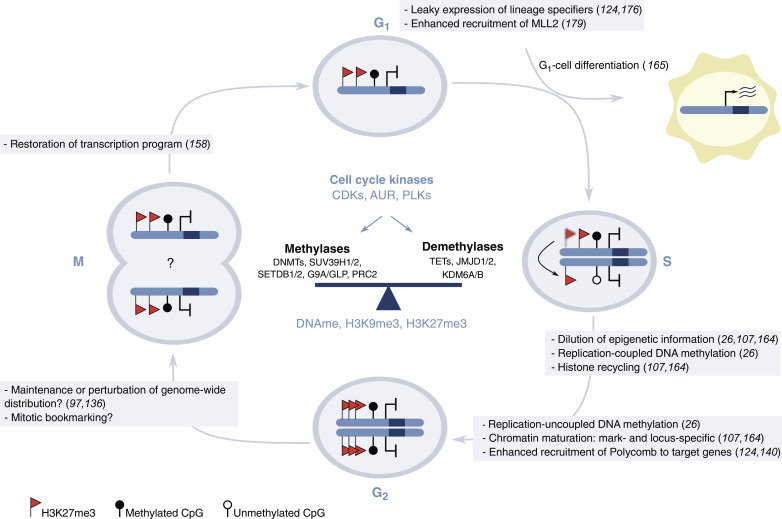
Current studies indicate that transmission of epigenetic memory is more dynamic than originally anticipated. Methyl groups on DNA cytosines, H3K27, and H3K9 are deposited by functionally redundant systems (30, 41, 77), can be actively erased from the genome by specialized enzymes (74, 75), are restored upon genome duplication with mark and locus-specific kinetics (26, 107, 164), display cell cycle phase–specific genome-wide patterns of epigenetic regulators (124, 140), and are directly phosphorylated and regulated by cell cycle kinases (189–199). Despite some of these findings require validation in a wider set of biological systems, current evidence supports that maintenance of the transcriptional program across cell duplication might not rely on a static presence of an unaltered epigenome during cell cycle transition. Instead, epigenetic memory seems to be encoded by an epigenetic cycle in which dynamic reversible activities carried out by epigenetic regulators are balanced and coordinated with cell cycle machinery (Fig. 3). Proliferating stem cells need to self-perpetuate their gene expression program and yet be able to change it during cell differentiation in response to developmental cues. The dynamic nature of the epigenetic cycle predicts a multiphasic system that is particularly well-suited to explain the biology of stem cells because it can easily accommodate both maintenance of cell identity and circumvention of self-renewal during windows of opportunity where epigenetic signals are temporarily weakened. For example, a partial alteration of the epigenome during mitosis might underlie the increased differentiation tendency of pluripotent cells in the G1 phase of the cell cycle (173–176). Although in this review we highlight evidence that allowed us to propose a mechanistic model as to how methylation of DNA and histone H3 support transcriptional memory, other studies support that these chromatin modifications might not be essential to maintain transcriptional memory in some experimental systems (10, 24, 39, 40). This suggest that in addition to chromatin covalent modifications, different components (i.e., other chromatin modifications, chromatin-associated RNA, or proteins) of the epigenetic systems contribute to maintain transcriptional memory and that compensatory mechanisms linking different epigenetic systems probably exist.
Fig. 3. Model of transmission of epigenetic memory in pluripotent cells: The epigenetic cycle.
Undifferentiated cell nuclei during cell cycle transition (G1, S, G2, and M phases) are represented as light blue circles, while differentiated cells are depicted in yellow. Blue bars inside the nuclei represent a chromatid in which a gene encoding for a lineage-specifier protein is highlighted in dark blue. Promoters are indicated by black lines. Methylated and unmethylated CpGs are represented as filled or empty lollipops, respectively. Red flags indicate H3K27me3.
FUTURE DIRECTIONS
The development and combination of high-throughput sequencing and genome editing techniques during the past two decades has facilitated the identification of many chromatin modifications and associated writer, eraser, and reader proteins that regulate gene expression in mammals (8, 202). However, which of these chromatin modifications are simply regulators of gene expression and which are bona fide epigenetic regulators of the genome that preserve transcriptional memory across mitotic cell divisions is currently an open debate (29, 50). This gap in the knowledge is probably a consequence of the fact that most studies analyzing the epigenome use asynchronously growing populations of cells, and this approach can only provide an averaged static view of a dynamic regulatory activity that takes place during cell cycle transition. Experimental setups that facilitate interrogation of the function of chromatin factors during cell cycle transition have been recently developed (Table 3). These techniques will be essential to design experimental settings that allow us to carefully decipher how epigenetic regulators maintain cellular memory during mitotic cell duplication. For example, in addition to traditional drug-based protocols to enrich for cells in particular phases of the cell cycle [i.e. thymidine block (203) or nocodazole (204)], drug-free methods based on centrifugation (205) or fluorescence-based flow cytometry cell sorting (206) can be used to isolate cells in different phases of the cell cycle. These systems allow isolation of up to millions of cells, and thus, cell cycle–separated cells can be analyzed using standard genome-wide methods. Moreover, different setups have been developed to specifically study the dynamics of DNA methylation and chromatin factors upon genome duplication using cell populations [i.e., Hammer-seq (207), ChOR-seq (208), or CRISPR-biotinylation (117)]. Emerging live super-resolution imaging and multi-omics single-cell techniques provide complementary approaches to study the dynamics of chromatin during cell cycle transition (209, 210). These platforms can be used in the context of cell cycle phase–specific perturbation experiments because of the development of degron systems that permit rapid and reversible degradation of target proteins in a cell cycle phase–specific way [i.e., AID (211), dTAG (212), or cyclin B (150)]. In addition, reversible perturbations in cell cycle–sorted cells can also be studied using chemical inhibitors against epigenetic regulators [i.e., EZH2 (213), SUV39 (214), G9A/GLP (215), or DNMT1 (216)]. Because drastic changes in the amount of DNA and chromatin factor occur during cell cycle transition, the field would generally benefit from the establishment of standardized normalization methods that facilitate comparison of findings observed across different studies. Overall, we envision that future studies combining these tools and addressing the function of epigenetic regulators at specific phases of the cell cycle will reveal key insights as to how mammalian cells perpetuate their gene expression program and facilitate maintenance of cellular memory in mammals.
Table 3. Techniques to study epigenetic regulation during cell cycle transition.
Table summarizing current methods to isolate cells in different phases of the cell cycle, study chromatin dynamics after DNA replication, or conditionally perturb epigenetic regulators.
| Method | Application | Ref. | |
| Cell cycle phase isolation | Chemical agents | ||
| Mimosine | Arrest cells in late G1 | (230) | |
| Thymidine | Arrest cells in S phase | (203) | |
| Aphidicolin | Arrest cells in early S phase | (231) | |
| Hydroxyurea | Arrest cells in early S phase | (232) | |
| RO-3306 | Arrest cells in G2-M phase | (233) | |
| Nocodazole | Arrest cells in prometaphase. Usually combined with mitotic shake-off | (204) | |
| Colcemid | Arrest cells in metaphase | (234) | |
| Drug-free | |||
| Serum starvation | Arrest cells at G0/G1 phase | (235) | |
| Centrifugal elutriation | Centrifugation-based separation of asynchronous populations in G1, S, and G2 fractions | (205) | |
| FUCCI | Fluorescence-based separation of asynchronous populations in G1, S, and G2 fractions | (206) | |
| Vybrant DyeCycle | Fluorescence-based separation of asynchronous populations in G1, S, and G2 fractions | (236) | |
| S-phase chromatin dynamics | DNA methylation | ||
| Repli-BS | Measures DNA methylation on newly synthesized daughter strands | (97) | |
| nasBS-seq | Measures DNA methylation in a strand-specific fashion on newly synthesized DNA | (237) | |
| Hammer-seq | Measures the methylation status of both strands on newly synthesized DNA within the same molecule | (23) | |
| iDEMS | Measures DNA modifications on metabolically labeled DNA by mass spectrometry | (139) | |
| Chromatin factors | |||
| NCC + tripleSILAC | Measures the composition of nascent chromatin on newly replicated DNA by mass spectrometry | (238) | |
| ChOR-seq | Measures chromatin factor binding on nascent chromatin after DNA replication | (208) | |
| SCAR-seq | Measures chromatin factor binding on nascent chromatin of sister chromatids after DNA replication | (208) | |
| CRISPR-biotinylation | Tracks the parental nucleosome localization after DNA replication in a locus-specific manner | (117) | |
| CUT&FLOW | Measures chromatin factor binding by CUT & Tag in nuclei sorted in cell cycle fractions by flow cytometry | (141) | |
| Conditional perturbation | Degrons | ||
| Auxin-inducible | Allows rapid degradation of the protein of interest upon addition of auxin-family plant hormones | (211) | |
| dTAGs | Allows rapid degradation of the protein of interest upon addition of the dTAG | (212) | |
| Cyclin B | Temporal loss of function of the protein of interest during M-to-G1 transition | (150) | |
| Light-inducible | Controls the expression of the protein of interest in a tight spatial and temporal manner | (239) | |
| Inhibitors | |||
| EZH2 inhibitors | Inhibits EZH2 methyltransferase activity | (213) | |
| RB-3 | Inhibits recruitment of RING1B to chromatin | (240) | |
| Chaetocin | Inhibits SUV39H1 and G9a methyltransferase activity | (214) | |
| UNC0638 | Inhibits GLP and G9a methyltransferase activity | (215) | |
| Hypomethylating agents | Nucleoside analogs (i.e. decitabine or azacytidine) that irreversibly inhibit DNMT1, 3A and 3B | (241, 242) | |
| GSK-3482364 | Selective, reversible, and non-covalent inhibitor of DNMT1 | (243) | |
| GSK-3685032 | Selective, reversible, and non-covalent inhibitor of DNMT1 | (216) | |
Acknowledgments
Funding: The Landeira laboratory is supported by grant (EUR2021-122005) funded by MCIN/AEI /10.13039/501100011033, the Spanish Ministry of Science and Innovation (PID2022-137060NB-I00 and PID2019-108108-100), the Instituto de Salud Carlos III (IHRC22/00007), the Andalusian regional government (PC-0246-2017, PIER-0211-2019, and PY20_00681), and the University of Granada (A-BIO-6-UGR20) grants.
Author contributions: D.L. conceived the manuscript. M.E.-M., M.A.-F., and D.L. wrote the original manuscript and edited subsequent versions during the review process. M.E.-M. and M.A.-F. produced figures and tables. D.L. obtained funding and supervised writing of the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper.
REFERENCES AND NOTES
- 1.Steffen P. A., Ringrose L., What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat. Rev. Mol. Cell Biol. 15, 340–356 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Atlasi Y., Stunnenberg H. G., The interplay of epigenetic marks during stem cell differentiation and development. Nat. Rev. Genet. 18, 643–658 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Elsherbiny A., Dobreva G., Epigenetic memory of cell fate commitment. Curr. Opin. Cell Biol. 69, 80–87 (2021). [DOI] [PubMed] [Google Scholar]
- 4.D'Urso A., Brickner J. H., Mechanisms of epigenetic memory. Trends Genet. 30, 230–236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferguson-Smith A. C., Genomic imprinting: The emergence of an epigenetic paradigm. Nat. Rev. Genet. 12, 565–575 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Loda A., Collombet S., Heard E., Gene regulation in time and space during X-chromosome inactivation. Nat. Rev. Mol. Cell Biol. 23, 231–249 (2022). [DOI] [PubMed] [Google Scholar]
- 7.Bonasio R., Tu S., Reinberg D., Molecular signals of epigenetic states. Science 330, 612–616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allis C. D., Jenuwein T., The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Berger S. L., Kouzarides T., Shiekhattar R., Shilatifard A., An operational definition of epigenetics. Genes Dev. 23, 781–783 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grewal S. I. S., The molecular basis of heterochromatin assembly and epigenetic inheritance. Mol. Cell 83, 1767–1785 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavalli G., Heard E., Advances in epigenetics link genetics to the environment and disease. Nature 571, 489–499 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Fitz-James M. H., Cavalli G., Molecular mechanisms of transgenerational epigenetic inheritance. Nat. Rev. Genet. 23, 325–341 (2022). [DOI] [PubMed] [Google Scholar]
- 13.Waddington C. H., The epigenotype. Endeavour 1, 18–20 (1942). [Google Scholar]
- 14.Waddington C. H., Canalization of development and the inheritance of acquired characters. Nature 150, 563–565 (1942). [Google Scholar]
- 15.Cramer P., Organization and regulation of gene transcription. Nature 573, 45–54 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Jones P. A., Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Greenberg M. V. C., Bourc'his D., The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 20, 590–607 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Li E., Beard C., Jaenisch R., Role for DNA methylation in genomic imprinting. Nature 366, 362–365 (1993). [DOI] [PubMed] [Google Scholar]
- 19.Mohandas T., Sparkes R. S., Shapiro L. J., Reactivation of an inactive human X chromosome: Evidence for X inactivation by DNA methylation. Science 211, 393–396 (1981). [DOI] [PubMed] [Google Scholar]
- 20.Zhao L., Sun M. A., Li Z., Bai X., Yu M., Wang M., Liang L., Shao X., Arnovitz S., Wang Q., He C., Lu X., Chen J., Xie H., The dynamics of DNA methylation fidelity during mouse embryonic stem cell self-renewal and differentiation. Genome Res. 24, 1296–1307 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shipony Z., Mukamel Z., Cohen N. M., Landan G., Chomsky E., Zeliger S. R., Fried Y. C., Ainbinder E., Friedman N., Tanay A., Dynamic and static maintenance of epigenetic memory in pluripotent and somatic cells. Nature 513, 115–119 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Wang Q., Yu G., Ming X., Xia W., Xu X., Zhang Y., Zhang W., Li Y., Huang C., Xie H., Zhu B., Xie W., Imprecise DNMT1 activity coupled with neighbor-guided correction enables robust yet flexible epigenetic inheritance. Nat. Genet. 52, 828–839 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Ming X., Zhang Z., Zou Z., Lv C., Dong Q., He Q., Yi Y., Li Y., Wang H., Zhu B., Kinetics and mechanisms of mitotic inheritance of DNA methylation and their roles in aging-associated methylome deterioration. Cell Res. 30, 980–996 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsumura A., Hayakawa T., Kumaki Y., Takebayashi S., Sakaue M., Matsuoka C., Shimotohno K., Ishikawa F., Li E., Ueda H. R., Nakayama J., Okano M., Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 11, 805–814 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Wei A., Wu H., Mammalian DNA methylome dynamics: Mechanisms, functions and new frontiers. Development 149, dev182683 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ming X., Zhu B., Li Y., Mitotic inheritance of DNA methylation: More than just copy and paste. J. Genet. Genomics 48, 1–13 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Kouzarides T., Chromatin modifications and their function. Cell 128, 693–705 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Bannister A. J., Kouzarides T., Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reinberg D., Vales L. D., Chromatin domains rich in inheritance. Science 361, 33–34 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Blackledge N. P., Klose R. J., The molecular principles of gene regulation by polycomb repressive complexes. Nat. Rev. Mol. Cell Biol. 22, 815–833 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuettengruber B., Bourbon H. M., Di Croce L., Cavalli G., Genome regulation by polycomb and trithorax: 70 years and counting. Cell 171, 34–57 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Struhl G., Akam M., Altered distributions of ultrabithorax transcripts in extra sex combs mutant embryos of Drosophila. EMBO J. 4, 3259–3264 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones R. S., Gelbart W. M., Genetic analysis of the enhancer of zeste locus and its role in gene regulation in Drosophila melanogaster. Genetics 126, 185–199 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simon J., Chiang A., Bender W., Ten different Polycomb group genes are required for spatial control of the abdA and AbdB homeotic products. Development 114, 493–505 (1992). [DOI] [PubMed] [Google Scholar]
- 35.Beuchle D., Struhl G., Müller J., Polycomb group proteins and heritable silencing ofDrosophilaHox genes. Development 128, 993–1004 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Laprell F., Finkl K., Müller J., Propagation of polycomb-repressed chromatin requires sequence-specific recruitment to DNA. Science 356, 85–88 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Coleman R. T., Struhl G., Causal role for inheritance of H3K27me3 in maintaining the OFF state of a Drosophila HOX gene. Science 356, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaydos L. J., Wang W., Strome S., Gene repression. H3K27me and PRC2 transmit a memory of repression across generations and during development. Science 345, 1515–1518 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Højfeldt J. W., Laugesen A., Willumsen B. M., Damhofer H., Hedehus L., Tvardovskiy A., Mohammad F., Jensen O. N., Helin K., Accurate H3K27 methylation can be established de novo by SUZ12-directed PRC2. Nat. Struct. Mol. Biol. 25, 225–232 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holoch D., Wassef M., Lövkvist C., Zielinski D., Aflaki S., Lombard B., Héry T., Loew D., Howard M., Margueron R., A cis-acting mechanism mediates transcriptional memory at Polycomb target genes in mammals. Nat. Genet. 53, 1686–1697 (2021). [DOI] [PubMed] [Google Scholar]
- 41.Padeken J., Methot S. P., Gasser S. M., Establishment of H3K9-methylated heterochromatin and its functions in tissue differentiation and maintenance. Nat. Rev. Mol. Cell Biol. 23, 623–640 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grewal S. I., Klar A. J., Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis. Cell 86, 95–101 (1996). [DOI] [PubMed] [Google Scholar]
- 43.Hall I. M., Shankaranarayana G. D., Noma K., Ayoub N., Cohen A., Grewal S. I., Establishment and maintenance of a heterochromatin domain. Science 297, 2232–2237 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Audergon P. N., Catania S., Kagansky A., Tong P., Shukla M., Pidoux A. L., Allshire R. C., Epigenetics. Restricted epigenetic inheritance of H3K9 methylation. Science 348, 132–135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ragunathan K., Jih G., Moazed D., Epigenetics. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science 348, 1258699 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cutter DiPiazza A. R., Taneja N., Dhakshnamoorthy J., Wheeler D., Holla S., Grewal S. I. S., Spreading and epigenetic inheritance of heterochromatin require a critical density of histone H3 lysine 9 tri-methylation. Proc. Natl. Acad. Sci. U.S.A. 118, e2100699118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hathaway N. A., Bell O., Hodges C., Miller E. L., Neel D. S., Crabtree G. R., Dynamics and memory of heterochromatin in living cells. Cell 149, 1447–1460 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X., Paulo J. A., Li X., Zhou H., Yu J., Gygi S. P., Moazed D., A composite DNA element that functions as a maintainer required for epigenetic inheritance of heterochromatin. Mol. Cell 81, 3979–3991.e4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu R., Wang X., Moazed D., Epigenetic inheritance mediated by coupling of RNAi and histone H3K9 methylation. Nature 558, 615–619 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henikoff S., Greally J. M., Epigenetics, cellular memory and gene regulation. Curr. Biol. 26, R644–R648 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Li E., Bestor T. H., Jaenisch R., Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926 (1992). [DOI] [PubMed] [Google Scholar]
- 52.Okano M., Bell D. W., Haber D. A., Li E., DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999). [DOI] [PubMed] [Google Scholar]
- 53.O'Carroll D., Erhardt S., Pagani M., Barton S. C., Surani M. A., Jenuwein T., The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 21, 4330–4336 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Faust C., Lawson K. A., Schork N. J., Thiel B., Magnuson T., The polycomb-group gene eed is required for normal morphogenetic movements during gastrulation in the mouse embryo. Development 125, 4495–4506 (1998). [DOI] [PubMed] [Google Scholar]
- 55.Pasini D., Bracken A. P., Jensen M. R., Lazzerini Denchi E., Helin K., Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 23, 4061–4071 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peters A. H., O'Carroll D., Scherthan H., Mechtler K., Sauer S., Schöfer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A., Opravil S., Doyle M., Sibilia M., Jenuwein T., Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337 (2001). [DOI] [PubMed] [Google Scholar]
- 57.Dodge J. E., Kang Y. K., Beppu H., Lei H., Li E., Histone H3-K9 methyltransferase ESET is essential for early development. Mol. Cell. Biol. 24, 2478–2486 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tachibana M., Sugimoto K., Nozaki M., Ueda J., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H., Shinkai Y., G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 16, 1779–1791 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tachibana M., Nozaki M., Takeda N., Shinkai Y., Functional dynamics of H3K9 methylation during meiotic prophase progression. EMBO J. 26, 3346–3359 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liao J., Karnik R., Gu H., Ziller M. J., Clement K., Tsankov A. M., Akopian V., Gifford C. A., Donaghey J., Galonska C., Pop R., Reyon D., Tsai S. Q., Mallard W., Joung J. K., Rinn J. L., Gnirke A., Meissner A., Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat. Genet. 47, 469–478 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fan G., Beard C., Chen R. Z., Csankovszki G., Sun Y., Siniaia M., Biniszkiewicz D., Bates B., Lee P. P., Kuhn R., Trumpp A., Poon C., Wilson C. B., Jaenisch R., DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J. Neurosci. 21, 788–797 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jackson-Grusby L., Beard C., Possemato R., Tudor M., Fambrough D., Csankovszki G., Dausman J., Lee P., Wilson C., Lander E., Jaenisch R., Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 27, 31–39 (2001). [DOI] [PubMed] [Google Scholar]
- 63.Sen G. L., Reuter J. A., Webster D. E., Zhu L., Khavari P. A., DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature 463, 563–567 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trowbridge J. J., Snow J. W., Kim J., Orkin S. H., DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell 5, 442–449 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boyer L. A., Plath K., Zeitlinger J., Brambrink T., Medeiros L. A., Lee T. I., Levine S. S., Wernig M., Tajonar A., Ray M. K., Bell G. W., Otte A. P., Vidal M., Gifford D. K., Young R. A., Jaenisch R., Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Shen X., Liu Y., Hsu Y. J., Fujiwara Y., Kim J., Mao X., Yuan G. C., Orkin S. H., EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 32, 491–502 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pasini D., Bracken A. P., Hansen J. B., Capillo M., Helin K., The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 27, 3769–3779 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ezhkova E., Pasolli H. A., Parker J. S., Stokes N., Su I. H., Hannon G., Tarakhovsky A., Fuchs E., Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 136, 1122–1135 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sankar A., Mohammad F., Sundaramurthy A. K., Wang H., Lerdrup M., Tatar T., Helin K., Histone editing elucidates the functional roles of H3K27 methylation and acetylation in mammals. Nat. Genet. 54, 754–760 (2022). [DOI] [PubMed] [Google Scholar]
- 70.Bilodeau S., Kagey M. H., Frampton G. M., Rahl P. B., Young R. A., SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 23, 2484–2489 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lohmann F., Loureiro J., Su H., Fang Q., Lei H., Lewis T., Yang Y., Labow M., Li E., Chen T., Kadam S., KMT1E mediated H3K9 methylation is required for the maintenance of embryonic stem cells by repressing trophectoderm differentiation. Stem Cells 28, 201–212 (2010). [DOI] [PubMed] [Google Scholar]
- 72.Yeap L. S., Hayashi K., Surani M. A., ERG-associated protein with SET domain (ESET)-Oct4 interaction regulates pluripotency and represses the trophectoderm lineage. Epigenetics Chromatin 2, 12 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yuan P., Han J., Guo G., Orlov Y. L., Huss M., Loh Y. H., Yaw L. P., Robson P., Lim B., Ng H. H., Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev. 23, 2507–2520 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Klose R. J., Kallin E. M., Zhang Y., JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7, 715–727 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Wu X., Zhang Y., TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat. Rev. Genet. 18, 517–534 (2017). [DOI] [PubMed] [Google Scholar]
- 76.Tsukada Y., Fang J., Erdjument-Bromage H., Warren M. E., Borchers C. H., Tempst P., Zhang Y., Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816 (2006). [DOI] [PubMed] [Google Scholar]
- 77.Parry A., Rulands S., Reik W., Active turnover of DNA methylation during cell fate decisions. Nat. Rev. Genet. 22, 59–66 (2021). [DOI] [PubMed] [Google Scholar]
- 78.Hong S., Cho Y. W., Yu L. R., Yu H., Veenstra T. D., Ge K., Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc. Natl. Acad. Sci. U.S.A. 104, 18439–18444 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lan F., Bayliss P. E., Rinn J. L., Whetstine J. R., Wang J. K., Chen S., Iwase S., Alpatov R., Issaeva I., Canaani E., Roberts T. M., Chang H. Y., Shi Y., A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449, 689–694 (2007). [DOI] [PubMed] [Google Scholar]
- 80.Cloos P. A., Christensen J., Agger K., Helin K., Erasing the methyl mark: Histone demethylases at the center of cellular differentiation and disease. Genes Dev. 22, 1115–1140 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nottke A., Colaiácovo M. P., Shi Y., Developmental roles of the histone lysine demethylases. Development 136, 879–889 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Piunti A., Shilatifard A., The roles of polycomb repressive complexes in mammalian development and cancer. Nat. Rev. Mol. Cell Biol. 22, 326–345 (2021). [DOI] [PubMed] [Google Scholar]
- 83.Ninova M., Fejes Toth K., Aravin A. A., The control of gene expression and cell identity by H3K9 trimethylation. Development 146, dev181180 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chory E. J., Calarco J. P., Hathaway N. A., Bell O., Neel D. S., Crabtree G. R., Nucleosome turnover regulates histone methylation patterns over the genome. Mol. Cell 73, 61–72.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Deal R. B., Henikoff J. G., Henikoff S., Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science 328, 1161–1164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Aygün O., Mehta S., Grewal S. I., HDAC-mediated suppression of histone turnover promotes epigenetic stability of heterochromatin. Nat. Struct. Mol. Biol. 20, 547–554 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dobrinić P., Szczurek A. T., Klose R. J., PRC1 drives polycomb-mediated gene repression by controlling transcription initiation and burst frequency. Nat. Struct. Mol. Biol. 28, 811–824 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reverón-Gómez N., González-Aguilera C., Stewart-Morgan K. R., Petryk N., Flury V., Graziano S., Johansen J. V., Jakobsen J. S., Alabert C., Groth A., Accurate recycling of parental histones reproduces the histone modification landscape during DNA replication. Mol. Cell 72, 239–249.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu L., Michowski W., Kolodziejczyk A., Sicinski P., The cell cycle in stem cell proliferation, pluripotency and differentiation. Nat. Cell Biol. 21, 1060–1067 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hu Y., Stillman B., Origins of DNA replication in eukaryotes. Mol. Cell 83, 352–372 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Šviković S., Sale J. E., The effects of replication stress on S phase histone management and epigenetic memory. J. Mol. Biol. 429, 2011–2029 (2017). [DOI] [PubMed] [Google Scholar]
- 92.Bostick M., Kim J. K., Estève P. O., Clark A., Pradhan S., Jacobsen S. E., UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760–1764 (2007). [DOI] [PubMed] [Google Scholar]
- 93.Sharif J., Koseki H., Recruitment of Dnmt1. Prog. Mol. Biol. Transl. Sci. 101, 289–310 (2011). [DOI] [PubMed] [Google Scholar]
- 94.Chuang L. S., Ian H. I., Koh T. W., Ng H. H., Xu G., Li B. F., Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science 277, 1996–2000 (1997). [DOI] [PubMed] [Google Scholar]
- 95.Ferry L., Fournier A., Tsusaka T., Adelmant G., Shimazu T., Matano S., Kirsh O., Amouroux R., Dohmae N., Suzuki T., Filion G. J., Deng W., de Dieuleveult M., Fritsch L., Kudithipudi S., Jeltsch A., Leonhardt H., Hajkova P., Marto J. A., Arita K., Shinkai Y., Defossez P. A., Methylation of DNA ligase 1 by G9a/GLP recruits UHRF1 to replicating DNA and regulates DNA methylation. Mol. Cell 67, 550–565.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 96.Liang G., Chan M. F., Tomigahara Y., Tsai Y. C., Gonzales F. A., Li E., Laird P. W., Jones P. A., Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol. Cell. Biol. 22, 480–491 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Charlton J., Downing T. L., Smith Z. D., Gu H., Clement K., Pop R., Akopian V., Klages S., Santos D. P., Tsankov A. M., Timmermann B., Ziller M. J., Kiskinis E., Gnirke A., Meissner A., Global delay in nascent strand DNA methylation. Nat. Struct. Mol. Biol. 25, 327–332 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Woodcock D. M., Simmons D. L., Crowther P. J., Cooper I. A., Trainor K. J., Morley A. A., Delayed DNA methylation is an integral feature of DNA replication in mammalian cells. Exp. Cell Res. 166, 103–112 (1986). [DOI] [PubMed] [Google Scholar]
- 99.Ramachandran S., Henikoff S., Transcriptional regulators compete with nucleosomes post-replication. Cell 165, 580–592 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Okuwaki M., Verreault A., Maintenance DNA methylation of nucleosome core particles. J. Biol. Chem. 279, 2904–2912 (2004). [DOI] [PubMed] [Google Scholar]
- 101.Felle M., Hoffmeister H., Rothammer J., Fuchs A., Exler J. H., Längst G., Nucleosomes protect DNA from DNA methylation in vivo and in vitro. Nucleic Acids Res. 39, 6956–6969 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gelato K. A., Tauber M., Ong M. S., Winter S., Hiragami-Hamada K., Sindlinger J., Lemak A., Bultsma Y., Houliston S., Schwarzer D., Divecha N., Arrowsmith C. H., Fischle W., Accessibility of different histone H3-binding domains of UHRF1 is allosterically regulated by phosphatidylinositol 5-phosphate. Mol. Cell 54, 905–919 (2014). [DOI] [PubMed] [Google Scholar]
- 103.Fang J., Cheng J., Wang J., Zhang Q., Liu M., Gong R., Wang P., Zhang X., Feng Y., Lan W., Gong Z., Tang C., Wong J., Yang H., Cao C., Xu Y., Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition. Nat. Commun. 7, 11197 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lyons D. B., Zilberman D., DDM1 and Lsh remodelers allow methylation of DNA wrapped in nucleosomes. eLife 6, e30674 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhou W., Dinh H. Q., Ramjan Z., Weisenberger D. J., Nicolet C. M., Shen H., Laird P. W., Berman B. P., DNA methylation loss in late-replicating domains is linked to mitotic cell division. Nat. Genet. 50, 591–602 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Annunziato A. T., Assembling chromatin: The long and winding road. Biochim. Biophys. Acta 1819, 196–210 (2013). [DOI] [PubMed] [Google Scholar]
- 107.Escobar T. M., Loyola A., Reinberg D., Parental nucleosome segregation and the inheritance of cellular identity. Nat. Rev. Genet. 22, 379–392 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Petryk N., Dalby M., Wenger A., Stromme C. B., Strandsby A., Andersson R., Groth A., MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science 361, 1389–1392 (2018). [DOI] [PubMed] [Google Scholar]
- 109.Gan H., Serra-Cardona A., Hua X., Zhou H., Labib K., Yu C., Zhang Z., The Mcm2-Ctf4-Polα axis facilitates parental histone H3-H4 transfer to lagging strands. Mol. Cell 72, 140–151.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yu C., Gan H., Serra-Cardona A., Zhang L., Gan S., Sharma S., Johansson E., Chabes A., Xu R. M., Zhang Z., A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science 361, 1386–1389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bellelli R., Belan O., Pye V. E., Clement C., Maslen S. L., Skehel J. M., Cherepanov P., Almouzni G., Boulton S. J., POLE3-POLE4 is a histone H3-H4 chaperone that maintains chromatin integrity during DNA replication. Mol. Cell 72, 112–126.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wenger A., Biran A., Alcaraz N., Redó-Riveiro A., Sell A. C., Krautz R., Flury V., Reverón-Gómez N., Solis-Mezarino V., Völker-Albert M., Imhof A., Andersson R., Brickman J. M., Groth A., Symmetric inheritance of parental histones governs epigenome maintenance and embryonic stem cell identity. Nat. Genet. 55, 1567–1578 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wen Q., Zhou J., Tian C., Li X., Song G., Gao Y., Sun Y., Ma C., Yao S., Liang X., Kang X., Wang N., Yao Y., Wang H., Liang X., Tang J., Offer S. M., Lei X., Yu C., Liu X., Liu Z., Wang Z., Gan H., Symmetric inheritance of parental histones contributes to safeguarding the fate of mouse embryonic stem cells during differentiation. Nat. Genet. 55, 1555–1566 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Flury V., Reverón-Gómez N., Alcaraz N., Stewart-Morgan K. R., Wenger A., Klose R. J., Groth A., Recycling of modified H2A-H2B provides short-term memory of chromatin states. Cell 186, 1050–1065.e19 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Alabert C., Barth T. K., Reveron-Gomez N., Sidoli S., Schmidt A., Jensen O. N., Imhof A., Groth A., Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev. 29, 585–590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schlissel G., Rine J., The nucleosome core particle remembers its position through DNA replication and RNA transcription. Proc. Natl. Acad. Sci. U.S.A. 116, 20605–20611 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Escobar T. M., Oksuz O., Saldaña-Meyer R., Descostes N., Bonasio R., Reinberg D., Active and repressed chromatin domains exhibit distinct nucleosome segregation during DNA replication. Cell 179, 953–963.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Escobar T. M., Yu J. R., Liu S., Lucero K., Vasilyev N., Nudler E., Reinberg D., Inheritance of repressed chromatin domains during S phase requires the histone chaperone NPM1. Sci. Adv. 8, eabm3945 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Loyola A., Bonaldi T., Roche D., Imhof A., Almouzni G., PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol. Cell 24, 309–316 (2006). [DOI] [PubMed] [Google Scholar]
- 120.Sobel R. E., Cook R. G., Perry C. A., Annunziato A. T., Allis C. D., Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl. Acad. Sci. U.S.A. 92, 1237–1241 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hansen K. H., Bracken A. P., Pasini D., Dietrich N., Gehani S. S., Monrad A., Rappsilber J., Lerdrup M., Helin K., A model for transmission of the H3K27me3 epigenetic mark. Nat. Cell Biol. 10, 1291–1300 (2008). [DOI] [PubMed] [Google Scholar]
- 122.Margueron R., Justin N., Ohno K., Sharpe M. L., Son J., Drury W. J. III, Voigt P., Martin S. R., Taylor W. R., De Marco V., Pirrotta V., Reinberg D., Gamblin S. J., Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–767 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cooper S., Grijzenhout A., Underwood E., Ancelin K., Zhang T., Nesterova T. B., Anil-Kirmizitas B., Bassett A., Kooistra S. M., Agger K., Helin K., Heard E., Brockdorff N., Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between polycomb complexes PRC1 and PRC2. Nat. Commun. 7, 13661 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Asenjo H. G., Gallardo A., López-Onieva L., Tejada I., Martorell-Marugán J., Carmona-Sáez P., Landeira D., Polycomb regulation is coupled to cell cycle transition in pluripotent stem cells. Sci. Adv. 6, eaay4768 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Güttinger S., Laurell E., Kutay U., Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat. Rev. Mol. Cell Biol. 10, 178–191 (2009). [DOI] [PubMed] [Google Scholar]
- 126.Gibcus J. H., Samejima K., Goloborodko A., Samejima I., Naumova N., Nuebler J., Kanemaki M. T., Xie L., Paulson J. R., Earnshaw W. C., Mirny L. A., Dekker J., A pathway for mitotic chromosome formation. Science 359, eaao6135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Naumova N., Imakaev M., Fudenberg G., Zhan Y., Lajoie B. R., Mirny L. A., Dekker J., Organization of the mitotic chromosome. Science 342, 948–953 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Martínez-Balbás M. A., Dey A., Rabindran S. K., Ozato K., Wu C., Displacement of sequence-specific transcription factors from mitotic chromatin. Cell 83, 29–38 (1995). [DOI] [PubMed] [Google Scholar]
- 129.Gottesfeld J. M., Forbes D. J., Mitotic repression of the transcriptional machinery. Trends Biochem. Sci. 22, 197–202 (1997). [DOI] [PubMed] [Google Scholar]
- 130.Prescott D. M., Bender M. A., Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp. Cell Res. 26, 260–268 (1962). [DOI] [PubMed] [Google Scholar]
- 131.Wang F., Higgins J. M., Histone modifications and mitosis: Countermarks, landmarks, and bookmarks. Trends Cell Biol. 23, 175–184 (2013). [DOI] [PubMed] [Google Scholar]
- 132.Ito T., Kubiura-Ichimaru M., Murakami Y., Bogutz A. B., Lefebvre L., Suetake I., Tajima S., Tada M., DNMT1 regulates the timing of DNA methylation by DNMT3 in an enzymatic activity-dependent manner in mouse embryonic stem cells. PLOS ONE 17, e0262277 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Aoto T., Saitoh N., Sakamoto Y., Watanabe S., Nakao M., Polycomb group protein-associated chromatin is reproduced in post-mitotic G1 phase and is required for S phase progression. J. Biol. Chem. 283, 18905–18915 (2008). [DOI] [PubMed] [Google Scholar]
- 134.McManus K. J., Biron V. L., Heit R., Underhill D. A., Hendzel M. J., Dynamic changes in histone H3 lysine 9 methylations. J. Biol. Chem. 281, 8888–8897 (2006). [DOI] [PubMed] [Google Scholar]
- 135.Peters A. H., Mermoud J. E., O'Carroll D., Pagani M., Schweizer D., Brockdorff N., Jenuwein T., Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat. Genet. 30, 77–80 (2002). [DOI] [PubMed] [Google Scholar]
- 136.Javasky E., Shamir I., Gandhi S., Egri S., Sandler O., Rothbart S. B., Kaplan N., Jaffe J. D., Goren A., Simon I., Study of mitotic chromatin supports a model of bookmarking by histone modifications and reveals nucleosome deposition patterns. Genome Res. 28, 1455–1466 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ginno P. A., Burger L., Seebacher J., Iesmantavicius V., Schübeler D., Cell cycle-resolved chromatin proteomics reveals the extent of mitotic preservation of the genomic regulatory landscape. Nat. Commun. 9, 4048 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Djeghloul D., Patel B., Kramer H., Dimond A., Whilding C., Brown K., Kohler A. C., Feytout A., Veland N., Elliott J., Bharat T. A. M., Tarafder A. K., Löwe J., Ng B. L., Guo Y., Guy J., Huseyin M. K., Klose R. J., Merkenschlager M., Fisher A. G., Identifying proteins bound to native mitotic ESC chromosomes reveals chromatin repressors are important for compaction. Nat. Commun. 11, 4118 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Stewart-Morgan K. R., Requena C. E., Flury V., Du Q., Heckhausen Z., Hajkova P., Groth A., Quantifying propagation of DNA methylation and hydroxymethylation with iDEMS. Nat. Cell Biol. 25, 183–193 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Asenjo H. G., Alcazar-Fabra M., Espinosa-Martínez M., Lopez-Onieva L., Gallardo A., Dimitrova E., Feldmann A., Pachano T., Martorell-Marugán J., Carmona-Sáez P., Sanchez-Pozo A., Rada-Iglesias Á., Klose R. J., Landeira D., Changes in PRC1 activity during interphase modulate lineage transition in pluripotent cells. Nat. Commun. 14, 180 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.G. M. B. Veronezi, S. Ramachandran, Nucleation and spreading rejuvenate polycomb domains every cell cycle. bioRxiv 502476 [Preprint] (2022). 10.1101/2022.08.02.502476. [DOI] [PMC free article] [PubMed]
- 142.Liu Y., Chen S., Wang S., Soares F., Fischer M., Meng F., Du Z., Lin C., Meyer C., DeCaprio J. A., Brown M., Liu X. S., He H. H., Transcriptional landscape of the human cell cycle. Proc. Natl. Acad. Sci. U.S.A. 114, 3473–3478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kang H., Shokhirev M. N., Xu Z., Chandran S., Dixon J. R., Hetzer M. W., Dynamic regulation of histone modifications and long-range chromosomal interactions during postmitotic transcriptional reactivation. Genes Dev. 34, 913–930 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hsiung C. C., Bartman C. R., Huang P., Ginart P., Stonestrom A. J., Keller C. A., Face C., Jahn K. S., Evans P., Sankaranarayanan L., Giardine B., Hardison R. C., Raj A., Blobel G. A., A hyperactive transcriptional state marks genome reactivation at the mitosis-G1 transition. Genes Dev. 30, 1423–1439 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liu Y., Pelham-Webb B., Di Giammartino D. C., Li J., Kim D., Kita K., Saiz N., Garg V., Doane A., Giannakakou P., Hadjantonakis A. K., Elemento O., Apostolou E., Widespread mitotic bookmarking by histone marks and transcription factors in pluripotent stem cells. Cell Rep. 19, 1283–1293 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pelham-Webb B., Murphy D., Apostolou E., Dynamic 3D chromatin reorganization during establishment and maintenance of pluripotency. Stem Cell Rep. 15, 1176–1195 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Festuccia N., Gonzalez I., Owens N., Navarro P., Mitotic bookmarking in development and stem cells. Development 144, 3633–3645 (2017). [DOI] [PubMed] [Google Scholar]
- 148.Teves S. S., An L., Hansen A. S., Xie L., Darzacq X., Tjian R., A dynamic mode of mitotic bookmarking by transcription factors. eLife 5, e22280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hsiung C. C., Morrissey C. S., Udugama M., Frank C. L., Keller C. A., Baek S., Giardine B., Crawford G. E., Sung M. H., Hardison R. C., Blobel G. A., Genome accessibility is widely preserved and locally modulated during mitosis. Genome Res. 25, 213–225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kadauke S., Udugama M. I., Pawlicki J. M., Achtman J. C., Jain D. P., Cheng Y., Hardison R. C., Blobel G. A., Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell 150, 725–737 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Caravaca J. M., Donahue G., Becker J. S., He X., Vinson C., Zaret K. S., Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes Dev. 27, 251–260 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Festuccia N., Dubois A., Vandormael-Pournin S., Gallego Tejeda E., Mouren A., Bessonnard S., Mueller F., Proux C., Cohen-Tannoudji M., Navarro P., Mitotic binding of Esrrb marks key regulatory regions of the pluripotency network. Nat. Cell Biol. 18, 1139–1148 (2016). [DOI] [PubMed] [Google Scholar]
- 153.Festuccia N., Owens N., Papadopoulou T., Gonzalez I., Tachtsidi A., Vandoermel-Pournin S., Gallego E., Gutierrez N., Dubois A., Cohen-Tannoudji M., Navarro P., Transcription factor activity and nucleosome organization in mitosis. Genome Res. 29, 250–260 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Deluz C., Friman E. T., Strebinger D., Benke A., Raccaud M., Callegari A., Leleu M., Manley S., Suter D. M., A role for mitotic bookmarking of SOX2 in pluripotency and differentiation. Genes Dev. 30, 2538–2550 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhu Z., Chen X., Guo A., Manzano T., Walsh P. J., Wills K. M., Halliburton R., Radko-Juettner S., Carter R. D., Partridge J. F., Green D. R., Zhang J., Roberts C. W. M., Mitotic bookmarking by SWI/SNF subunits. Nature 618, 180–187 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.S. Placzek, L. Vanzan, D. M. Suter, Orchestration of pluripotent stem cell genome reactivation during mitotic exit. bioRxiv [Preprint] 552605 (2023). 10.1101/2023.01.27.525826. [DOI]
- 157.Djeghloul D., Dimond A., Cheriyamkunnel S., Kramer H., Patel B., Brown K., Montoya A., Whilding C., Wang Y. F., Futschik M. E., Veland N., Montavon T., Jenuwein T., Merkenschlager M., Fisher A. G., Loss of H3K9 trimethylation alters chromosome compaction and transcription factor retention during mitosis. Nat. Struct. Mol. Biol. 30, 489–501 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Palozola K. C., Lerner J., Zaret K. S., A changing paradigm of transcriptional memory propagation through mitosis. Nat. Rev. Mol. Cell Biol. 20, 55–64 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Palozola K. C., Donahue G., Liu H., Grant G. R., Becker J. S., Cote A., Yu H., Raj A., Zaret K. S., Mitotic transcription and waves of gene reactivation during mitotic exit. Science 358, 119–122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pelham-Webb B., Polyzos A., Wojenski L., Kloetgen A., Li J., Di Giammartino D. C., Sakellaropoulos T., Tsirigos A., Core L., Apostolou E., H3K27ac bookmarking promotes rapid post-mitotic activation of the pluripotent stem cell program without impacting 3D chromatin reorganization. Mol. Cell 81, 1732–1748.e8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Behera V., Stonestrom A. J., Hamagami N., Hsiung C. C., Keller C. A., Giardine B., Sidoli S., Yuan Z. F., Bhanu N. V., Werner M. T., Wang H., Garcia B. A., Hardison R. C., Blobel G. A., Interrogating histone acetylation and BRD4 as mitotic bookmarks of transcription. Cell Rep. 27, 400–415.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Meshorer E., Yellajoshula D., George E., Scambler P. J., Brown D. T., Misteli T., Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell 10, 105–116 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chervova A., Festuccia N., Altamirano-Pacheco L., Dubois A., Navarro P., A gene subset requires CTCF bookmarking during the fast post-mitotic reactivation of mouse ES cells. EMBO Rep. 24, e56075 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Stewart-Morgan K. R., Petryk N., Groth A., Chromatin replication and epigenetic cell memory. Nat. Cell Biol. 22, 361–371 (2020). [DOI] [PubMed] [Google Scholar]
- 165.Soufi A., Dalton S., Cycling through developmental decisions: How cell cycle dynamics control pluripotency, differentiation and reprogramming. Development 143, 4301–4311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Haber J. E., Mating-type gene switching INSACCHAROMYCES cerevisiae. Annu. Rev. Genet. 32, 561–599 (1998). [DOI] [PubMed] [Google Scholar]
- 167.Gomer R. H., Firtel R. A., Cell-autonomous determination of cell-type choice in Dictyostelium development by cell-cycle phase. Science 237, 758–762 (1987). [DOI] [PubMed] [Google Scholar]
- 168.Ambros V., Cell cycle-dependent sequencing of cell fate decisions in Caenorhabditis elegans vulva precursor cells. Development 126, 1947–1956 (1999). [DOI] [PubMed] [Google Scholar]
- 169.Hunter G. L., Hadjivasiliou Z., Bonin H., He L., Perrimon N., Charras G., Baum B., Coordinated control of Notch/Delta signalling and cell cycle progression drives lateral inhibition-mediated tissue patterning. Development 143, 2305–2310 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.McConnell S. K., Kaznowski C. E., Cell cycle dependence of laminar determination in developing neocortex. Science 254, 282–285 (1991). [DOI] [PubMed] [Google Scholar]
- 171.Kim Y. H., Larsen H. L., Rue P., Lemaire L. A., Ferrer J., Grapin-Botton A., Cell cycle-dependent differentiation dynamics balances growth and endocrine differentiation in the pancreas. PLOS Biol. 13, e1002111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Mummery C. L., van den Brink C. E., de Laat S. W., Commitment to differentiation induced by retinoic acid in P19 embryonal carcinoma cells is cell cycle dependent. Dev. Biol. 121, 10–19 (1987). [DOI] [PubMed] [Google Scholar]
- 173.Coronado D., Godet M., Bourillot P.-Y., Tapponnier Y., Bernat A., Petit M., Afanassieff M., Markossian S., Malashicheva A., Iacone R., Anastassiadis K., Savatier P., A short G1 phase is an intrinsic determinant of naïve embryonic stem cell pluripotency. Stem Cell Res. 10, 118–131 (2013). [DOI] [PubMed] [Google Scholar]
- 174.Sela Y., Molotski N., Golan S., Itskovitz-Eldor J., Soen Y., Human embryonic stem cells exhibit increased propensity to differentiate during the G1 phase prior to phosphorylation of retinoblastoma protein. Stem Cells 30, 1097–1108 (2012). [DOI] [PubMed] [Google Scholar]
- 175.Pauklin S., Vallier L., The cell-cycle state of stem cells determines cell fate propensity. Cell 155, 135–147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Singh A. M., Chappell J., Trost R., Lin L., Wang T., Tang J., Matlock B. K., Weller K. P., Wu H., Zhao S., Jin P., Dalton S., Cell-cycle control of developmentally regulated transcription factors accounts for heterogeneity in human pluripotent cells. Stem Cell Rep. 1, 532–544 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gonzales K. A., Liang H., Lim Y. S., Chan Y. S., Yeo J. C., Tan C. P., Gao B., Le B., Tan Z. Y., Low K. Y., Liou Y. C., Bard F., Ng H. H., Deterministic restriction on pluripotent state dissolution by cell-cycle pathways. Cell 162, 564–579 (2015). [DOI] [PubMed] [Google Scholar]
- 178.Aranda S., Alcaine-Colet A., Ballaré C., Blanco E., Mocavini I., Sparavier A., Vizán P., Borràs E., Sabidó E., Di Croce L., Thymine DNA glycosylase regulates cell-cycle-driven p53 transcriptional control in pluripotent cells. Mol. Cell 83, 2673–2691.e7 (2023). [DOI] [PubMed] [Google Scholar]
- 179.Singh A. M., Sun Y., Li L., Zhang W., Wu T., Zhao S., Qin Z., Dalton S., Cell-cycle control of bivalent epigenetic domains regulates the exit from pluripotency. Stem Cell Rep. 5, 323–336 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Grandy R. A., Whitfield T. W., Wu H., Fitzgerald M. P., VanOudenhove J. J., Zaidi S. K., Montecino M. A., Lian J. B., van Wijnen A. J., Stein J. L., Stein G. S., Genome-wide studies reveal that H3K4me3 modification in bivalent genes is dynamically regulated during the pluripotent cell cycle and stabilized upon differentiation. Mol. Cell. Biol. 36, 615–627 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Singh A. M., Cell cycle-driven heterogeneity: On the road to demystifying the transitions between “poised” and “restricted” pluripotent cell states. Stem Cells Int. 2015, 219514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Petruk S., Cai J., Sussman R., Sun G., Kovermann S. K., Mariani S. A., Calabretta B., McMahon S. B., Brock H. W., Iacovitti L., Mazo A., Delayed accumulation of H3K27me3 on nascent dna is essential for recruitment of transcription factors at early stages of stem cell differentiation. Mol. Cell 66, 247–257.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Herchcovici Levy S., Feldman Cohen S., Arnon L., Lahav S., Awawdy M., Alajem A., Bavli D., Sun X., Buganim Y., Ram O., Esrrb is a cell-cycle-dependent associated factor balancing pluripotency and XEN differentiation. Stem Cell Rep. 17, 1334–1350 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Fisher D., Krasinska L., Coudreuse D., Novák B., Phosphorylation network dynamics in the control of cell cycle transitions. J. Cell Sci. 125, 4703–4711 (2012). [DOI] [PubMed] [Google Scholar]
- 185.Johnson J. L., Yaron T. M., Huntsman E. M., Kerelsky A., Song J., Regev A., Lin T. Y., Liberatore K., Cizin D. M., Cohen B. M., Vasan N., Ma Y., Krismer K., Robles J. T., van de Kooij B., van Vlimmeren A. E., Andrée-Busch N., Käufer N. F., Dorovkov M. V., Ryazanov A. G., Takagi Y., Kastenhuber E. R., Goncalves M. D., Hopkins B. D., Elemento O., Taatjes D. J., Maucuer A., Yamashita A., Degterev A., Uduman M., Lu J., Landry S. D., Zhang B., Cossentino I., Linding R., Blenis J., Hornbeck P. V., Turk B. E., Yaffe M. B., Cantley L. C., An atlas of substrate specificities for the human serine/threonine kinome. Nature 613, 759–766 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Satyanarayana A., Kaldis P., Mammalian cell-cycle regulation: Several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene 28, 2925–2939 (2009). [DOI] [PubMed] [Google Scholar]
- 187.Willems E., Dedobbeleer M., Digregorio M., Lombard A., Lumapat P. N., Rogister B., The functional diversity of Aurora kinases: A comprehensive review. Cell Div. 13, 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zitouni S., Nabais C., Jana S. C., Guerrero A., Bettencourt-Dias M., Polo-like kinases: Structural variations lead to multiple functions. Nat. Rev. Mol. Cell Biol. 15, 433–452 (2014). [DOI] [PubMed] [Google Scholar]
- 189.Lavoie G., St-Pierre Y., Phosphorylation of human DNMT1: Implication of cyclin-dependent kinases. Biochem. Biophys. Res. Commun. 409, 187–192 (2011). [DOI] [PubMed] [Google Scholar]
- 190.Acevedo M., Vernier M., Mignacca L., Lessard F., Huot G., Moiseeva O., Bourdeau V., Ferbeyre G., A CDK4/6-dependent epigenetic mechanism protects cancer cells from PML-induced senescence. Cancer Res. 76, 3252–3264 (2016). [DOI] [PubMed] [Google Scholar]
- 191.Deplus R., Blanchon L., Rajavelu A., Boukaba A., Defrance M., Luciani J., Rothé F., Dedeurwaerder S., Denis H., Brinkman A. B., Simmer F., Müller F., Bertin B., Berdasco M., Putmans P., Calonne E., Litchfield D. W., de Launoit Y., Jurkowski T. P., Stunnenberg H. G., Bock C., Sotiriou C., Fraga M. F., Esteller M., Jeltsch A., Fuks F., Regulation of DNA methylation patterns by CK2-mediated phosphorylation of Dnmt3a. Cell Rep. 8, 743–753 (2014). [DOI] [PubMed] [Google Scholar]
- 192.Kaneko S., Li G., Son J., Xu C. F., Margueron R., Neubert T. A., Reinberg D., Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 24, 2615–2620 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Chen S., Bohrer L. R., Rai A. N., Pan Y., Gan L., Zhou X., Bagchi A., Simon J. A., Huang H., Cyclin-dependent kinases regulate epigenetic gene silencing through phosphorylation of EZH2. Nat. Cell Biol. 12, 1108–1114 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wei Y., Chen Y. H., Li L. Y., Lang J., Yeh S. P., Shi B., Yang C. C., Yang J. Y., Lin C. Y., Lai C. C., Hung M. C., CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat. Cell Biol. 13, 87–94 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wu S. C., Zhang Y., Cyclin-dependent kinase 1 (CDK1)-mediated phosphorylation of enhancer of zeste 2 (Ezh2) regulates its stability. J. Biol. Chem. 286, 28511–28519 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Minnebo N., Görnemann J., O'Connell N., Van Dessel N., Derua R., Vermunt M. W., Page R., Beullens M., Peti W., Van Eynde A., Bollen M., NIPP1 maintains EZH2 phosphorylation and promoter occupancy at proliferation-related target genes. Nucleic Acids Res. 41, 842–854 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhang H., Diab A., Fan H., Mani S. K., Hullinger R., Merle P., Andrisani O., PLK1 and HOTAIR accelerate proteasomal degradation of SUZ12 and ZNF198 during hepatitis B virus-induced liver carcinogenesis. Cancer Res. 75, 2363–2374 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Park S. H., Yu S. E., Chai Y. G., Jang Y. K., CDK2-dependent phosphorylation of Suv39H1 is involved in control of heterochromatin replication during cell cycle progression. Nucleic Acids Res. 42, 6196–6207 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yang Q., Zhu Q., Lu X., Du Y., Cao L., Shen C., Hou T., Li M., Li Z., Liu C., Wu D., Xu X., Wang L., Wang H., Zhao Y., Yang Y., Zhu W. G., G9a coordinates with the RPA complex to promote DNA damage repair and cell survival. Proc. Natl. Acad. Sci. U.S.A. 114, E6054–E6063 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Nie L., Wei Y., Zhang F., Hsu Y. H., Chan L. C., Xia W., Ke B., Zhu C., Deng R., Tang J., Yao J., Chu Y. Y., Zhao X., Han Y., Hou J., Huo L., Ko H. W., Lin W. C., Yamaguchi H., Hsu J. M., Yang Y., Pan D. N., Hsu J. L., Kleer C. G., Davidson N. E., Hortobagyi G. N., Hung M. C., CDK2-mediated site-specific phosphorylation of EZH2 drives and maintains triple-negative breast cancer. Nat. Commun. 10, 5114 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Caretti G., Palacios D., Sartorelli V., Puri P. L., Phosphoryl-EZH-ion. Cell Stem Cell 8, 262–265 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Musselman C. A., Lalonde M. E., Côté J., Kutateladze T. G., Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 19, 1218–1227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Whitfield M. L., Zheng L. X., Baldwin A., Ohta T., Hurt M. M., Marzluff W. F., Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 20, 4188–4198 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zieve G. W., Turnbull D., Mullins J. M., McIntosh J. R., Production of large numbers of mitotic mammalian cells by use of the reversible microtubule inhibitor nocodazole. Nocodazole accumulated mitotic cells. Exp. Cell Res. 126, 397–405 (1980). [DOI] [PubMed] [Google Scholar]
- 205.Keng P. C., Li C. K., Wheeler K. T., Synchronization of 9L rat brain tumor cells by centrifugal elutriation. Cell Biophys. 2, 191–206 (1980). [DOI] [PubMed] [Google Scholar]
- 206.Sakaue-Sawano A., Kurokawa H., Morimura T., Hanyu A., Hama H., Osawa H., Kashiwagi S., Fukami K., Miyata T., Miyoshi H., Imamura T., Ogawa M., Masai H., Miyawaki A., Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498 (2008). [DOI] [PubMed] [Google Scholar]
- 207.Ming X., Zhu B., Zhang Z., Simultaneously measuring the methylation of parent and daughter strands of replicated DNA at the single-molecule level by Hammer-seq. Nat. Protoc. 16, 2131–2157 (2021). [DOI] [PubMed] [Google Scholar]
- 208.Petryk N., Reverón-Gómez N., González-Aguilera C., Dalby M., Andersson R., Groth A., Genome-wide and sister chromatid-resolved profiling of protein occupancy in replicated chromatin with ChOR-seq and SCAR-seq. Nat. Protoc. 16, 4446–4493 (2021). [DOI] [PubMed] [Google Scholar]
- 209.Kelsey G., Stegle O., Reik W., Single-cell epigenomics: Recording the past and predicting the future. Science 358, 69–75 (2017). [DOI] [PubMed] [Google Scholar]
- 210.Sydor A. M., Czymmek K. J., Puchner E. M., Mennella V., Super-resolution microscopy: From single molecules to supramolecular assemblies. Trends Cell Biol. 25, 730–748 (2015). [DOI] [PubMed] [Google Scholar]
- 211.Nishimura K., Fukagawa T., Takisawa H., Kakimoto T., Kanemaki M., An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 6, 917–922 (2009). [DOI] [PubMed] [Google Scholar]
- 212.Nabet B., Roberts J. M., Buckley D. L., Paulk J., Dastjerdi S., Yang A., Leggett A. L., Erb M. A., Lawlor M. A., Souza A., Scott T. G., Vittori S., Perry J. A., Qi J., Winter G. E., Wong K. K., Gray N. S., Bradner J. E., The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol. 14, 431–441 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kim K. H., Roberts C. W., Targeting EZH2 in cancer. Nat. Med. 22, 128–134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Greiner D., Bonaldi T., Eskeland R., Roemer E., Imhof A., Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nat. Chem. Biol. 1, 143–145 (2005). [DOI] [PubMed] [Google Scholar]
- 215.Vedadi M., Barsyte-Lovejoy D., Liu F., Rival-Gervier S., Allali-Hassani A., Labrie V., Wigle T. J., Dimaggio P. A., Wasney G. A., Siarheyeva A., Dong A., Tempel W., Wang S. C., Chen X., Chau I., Mangano T. J., Huang X. P., Simpson C. D., Pattenden S. G., Norris J. L., Kireev D. B., Tripathy A., Edwards A., Roth B. L., Janzen W. P., Garcia B. A., Petronis A., Ellis J., Brown P. J., Frye S. V., Arrowsmith C. H., Jin J., A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat. Chem. Biol. 7, 566–574 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Pappalardi M. B., Keenan K., Cockerill M., Kellner W. A., Stowell A., Sherk C., Wong K., Pathuri S., Briand J., Steidel M., Chapman P., Groy A., Wiseman A. K., McHugh C. F., Campobasso N., Graves A. P., Fairweather E., Werner T., Raoof A., Butlin R. J., Rueda L., Horton J. R., Fosbenner D. T., Zhang C., Handler J. L., Muliaditan M., Mebrahtu M., Jaworski J. P., McNulty D. E., Burt C., Eberl H. C., Taylor A. N., Ho T., Merrihew S., Foley S. W., Rutkowska A., Li M., Romeril S. P., Goldberg K., Zhang X., Kershaw C. S., Bantscheff M., Jurewicz A. J., Minthorn E., Grandi P., Patel M., Benowitz A. B., Mohammad H. P., Gilmartin A. G., Prinjha R. K., Ogilvie D., Carpenter C., Heerding D., Baylin S. B., Jones P. A., Cheng X., King B. W., Luengo J. I., Jordan A. M., Waddell I., Kruger R. G., McCabe M. T., Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia. Nat. Cancer 2, 1002–1017 (2021). [PMC free article] [PubMed] [Google Scholar]
- 217.Ficz G., Branco M. R., Seisenberger S., Santos F., Krueger F., Hore T. A., Marques C. J., Andrews S., Reik W., Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473, 398–402 (2011). [DOI] [PubMed] [Google Scholar]
- 218.Inoue A., Zhang Y., Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science 334, 194 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Szulwach K. E., Li X., Li Y., Song C. X., Han J. W., Kim S., Namburi S., Hermetz K., Kim J. J., Rudd M. K., Yoon Y. S., Ren B., He C., Jin P., Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLOS Genet. 7, e1002154 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Easwaran H. P., Schermelleh L., Leonhardt H., Cardoso M. C., Replication-independent chromatin loading of Dnmt1 during G2 and M phases. EMBO Rep. 5, 1181–1186 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Brero A., Easwaran H. P., Nowak D., Grunewald I., Cremer T., Leonhardt H., Cardoso M. C., Methyl CpG-binding proteins induce large-scale chromatin reorganization during terminal differentiation. J. Cell Biol. 169, 733–743 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Uemura T., Kubo E., Kanari Y., Ikemura T., Tatsumi K., Muto M., Temporal and spatial localization of novel nuclear protein NP95 in mitotic and meiotic cells. Cell Struct. Funct. 25, 149–159 (2000). [DOI] [PubMed] [Google Scholar]
- 223.Velazquez Camacho O., Galan C., Swist-Rosowska K., Ching R., Gamalinda M., Karabiber F., De La Rosa-Velazquez I., Engist B., Koschorz B., Shukeir N., Onishi-Seebacher M., van de Nobelen S., Jenuwein T., Major satellite repeat RNA stabilize heterochromatin retention of Suv39h enzymes by RNA-nucleosome association and RNA:DNA hybrid formation. eLife 6, e25293 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Park J. A., Kim A. J., Kang Y., Jung Y. J., Kim H. K., Kim K. C., Deacetylation and methylation at histone H3 lysine 9 (H3K9) coordinate chromosome condensation during cell cycle progression. Mol. Cells 31, 343–350 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Chu L., Zhu T., Liu X., Yu R., Bacanamwo M., Dou Z., Chu Y., Zou H., Gibbons G. H., Wang D., Ding X., Yao X., SUV39H1 orchestrates temporal dynamics of centromeric methylation essential for faithful chromosome segregation in mitosis. J. Mol. Cell Biol. 4, 331–340 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Burke L. J., Zhang R., Bartkuhn M., Tiwari V. K., Tavoosidana G., Kurukuti S., Weth C., Leers J., Galjart N., Ohlsson R., Renkawitz R., CTCF binding and higher order chromatin structure of the H19 locus are maintained in mitotic chromatin. EMBO J. 24, 3291–3300 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Gong L., Liu X., Jiao L., Yang X., Lemoff A., Liu X., CK2-mediated phosphorylation of SUZ12 promotes PRC2 function by stabilizing enzyme active site. Nat. Commun. 13, 6781 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Goto H., Yasui Y., Nigg E. A., Inagaki M., Aurora-B phosphorylates Histone H3 at serine28 with regard to the mitotic chromosome condensation. Genes Cells 7, 11–17 (2002). [DOI] [PubMed] [Google Scholar]
- 229.Fischle W., Tseng B. S., Dormann H. L., Ueberheide B. M., Garcia B. A., Shabanowitz J., Hunt D. F., Funabiki H., Allis C. D., Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438, 1116–1122 (2005). [DOI] [PubMed] [Google Scholar]
- 230.Krude T., Mimosine arrests proliferating human cells before onset of DNA replication in a dose-dependent manner. Exp. Cell Res. 247, 148–159 (1999). [DOI] [PubMed] [Google Scholar]
- 231.Levenson V., Hamlin J. L., A general protocol for evaluating the specific effects of DNA replication inhibitors. Nucleic Acids Res. 21, 3997–3404 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Tobey R. A., Crissman H. A., Preparation of large quantities of synchronized mammalian cells in late G1 in the pre-DNA replicative phase of the cell cycle. Exp. Cell Res. 75, 460–464 (1972). [DOI] [PubMed] [Google Scholar]
- 233.Vassilev L. T., Tovar C., Chen S., Knezevic D., Zhao X., Sun H., Heimbrook D. C., Chen L., Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl. Acad. Sci. U.S.A. 103, 10660–10665 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Kleinfeld R. G., Sisken J. E., Morphological and kinetic aspects of mitotic arrest by and recovery from colcemid. J. Cell Biol. 31, 369–379 (1966). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Welsh C. F., Roovers K., Villanueva J., Liu Y., Schwartz M. A., Assoian R. K., Timing of cyclin D1 expression within G1 phase is controlled by Rho. Nat. Cell Biol. 3, 950–957 (2001). [DOI] [PubMed] [Google Scholar]
- 236.Bradford J. A., Whitney P., Huang T., Pinson P., Cheung C.-Y., Yue S., Godfrey W. L., Novel Vybrant® DyeCycle ™ stains provide cell cycle analysis in live cells using flow cytometry with violet, blue, and green excitation. Blood 108, 4234 (2006). [Google Scholar]
- 237.Xu C., Corces V. G., Nascent DNA methylome mapping reveals inheritance of hemimethylation at CTCF/cohesin sites. Science 359, 1166–1170 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Alabert C., Bukowski-Wills J.-C., Lee S.-B., Kustatscher G., Nakamura K., de Lima Alves F., Menard P., Mejlvang J., Rappsilber J., Groth A., Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 16, 281–291 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Renicke C., Schuster D., Usherenko S., Essen L. O., Taxis C., A LOV2 domain-based optogenetic tool to control protein degradation and cellular function. Chem. Biol. 20, 619–626 (2013). [DOI] [PubMed] [Google Scholar]
- 240.Shukla S., Ying W., Gray F., Yao Y., Simes M. L., Zhao Q., Miao H., Cho H. J., González-Alonso P., Winkler A., Lund G., Purohit T., Kim E., Zhang X., Ray J. M., He S., Nikolaidis C., Ndoj J., Wang J., Jaremko Ł., Jaremko M., Ryan R. J. H., Guzman M. L., Grembecka J., Cierpicki T., Small-molecule inhibitors targeting polycomb repressive complex 1 RING domain. Nat. Chem. Biol. 17, 784–793 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Jones P. A., Taylor S. M., Cellular differentiation, cytidine analogs and DNA methylation. Cell 20, 85–93 (1980). [DOI] [PubMed] [Google Scholar]
- 242.Sorm F., Pískala A., Cihák A., Veselý J., 5-Azacytidine, a new, highly effective cancerostatic. Experientia 20, 202–203 (1964). [DOI] [PubMed] [Google Scholar]
- 243.Gilmartin A. G., Groy A., Gore E. R., Atkins C., Long E. R., Montoute M. N., Wu Z., Halsey W., McNulty D. E., Ennulat D., Rueda L., Pappalardi M., Kruger R. G., McCabe M. T., Raoof A., Butlin R., Stowell A., Cockerill M., Waddell I., Ogilvie D., Luengo J., Jordan A., Benowitz A. B., In vitro and in vivo induction of fetal hemoglobin with a reversible and selective DNMT1 inhibitor. Haematologica 106, 1979–1987 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]