Abstract
Live cell imaging is a standard technique in experimental biology that enables the observation of isolated cells and tissue slices in real time; and the testing of cellular responses to changes in buffer composition. However, most live cell imaging devices require the use of dedicated microscopes and/or specialized stage adaptors, and come at a reasonably high cost. We employed 3D printing technology to create a low-cost imaging chamber with side ports to exchange fluids, to be used on upright microscopes. The chamber increased the functionality of a standard upright epifluorescent microscope to allow dynamic, real-time calcium imaging of cultured hypothalamic astrocytes from mice, and to test the effects of ATP stimulation upon calcium signaling. It was also used on slices obtained from mouse brain using a brain matrix slicer. The advantages of this chamber include a very simple design that can be used with upright epifluorescence microscopes, does not require any special stage adaptor, and includes ports to permit fluid exchange during imaging. This chamber is ideal for educational settings with undergraduate laboratories that do not have access to dedicated inverted fluorescent microscopes for tissue culture experiments.
Keywords: imaging chamber, astrocyte, tanycyte, cell culture, live cell imaging, calcium imaging
Introduction
Live cell imaging offers unique insight into the physiology of cells, and can be of great educational value when paired with experimental manipulations. The first cell imaging chambers developed in the 1950s simply involved placing a spacer sandwiched between two coverslips (Dailey et al. 2011). This spacer could range from a rubber O-ring or plastic gasket to petroleum gel or modeling clay dam forming a well containing a bath solution.
Limitations of the early live imaging chambers involved short cell survival times and inability to test the effects of experimental substances in cell cultures (Dailey et al. 2011). Scientists explored methods to improve chamber designs. Developing perfusion apparatus for live imaging chambers provided the ability to test substances on living cultures, plus greatly increased cell viability (Dailey et al. 2011). Additionally, oxygenation of the reservoir bath buffers and temperature stabilization of the buffers with a temperature regulator, further improved viability of cells to permit longer imaging sessions (Cole 2014).
The problem that arose with each new development: constant perfusion; constant oxygenation; temperature control; and substance testing, was that specialized equipment to adjust and monitor these parameters was needed. Today, the live cell imaging chambers have become so sophisticated, that they require specialized microscope stages to accommodate them, in addition to the space needed for accessory equipment. With the increased technical abilities, the price for an imaging chamber, its stage adaptor, and monitoring equipment is beyond the budget for many investigators desiring to perform live cell imaging, and beyond the budget for teaching labs at many institutions. Given high set-up costs, the prospect of obtaining pilot data to support a principle employing live-cell imaging experiments and then to apply for funding to acquire such equipment can be challenging. Many laboratories have microscopes used for multiple investigators and procedures, making frequent changes of the stage impractical. Consequently, developing low cost, versatile, and effective live cell imaging chambers will provide greater functionality to current laboratory microscopes, as well as increased educational opportunities for undergraduate and graduate courses in cell biology and neurobiology.
In recent years, 3D printing has emerged as a means by which prototypes of devices are inexpensively produced. In 2015, Wardyn et al reported the development of a 3D chamber used successfully for growth and electrical stimulation of primary neuronal cultures (Wardyn et al. 2015). In the same year, Tyson, Hilton and Andrea developed a brain slice matrix for slicing brain tissue coated with agarose and an imaging chamber designed for both clearing, and antibody staining of the thick brain slices acquired from their matrix (Tyson et al. 2015).
To address the problem of attaining quality live cell imaging of astrocyte cultures, we designed a very simple imaging chamber that was 3D printed. This chamber, having the same x-y dimensions as a standard microscope slide, allows us to calcium image live hypothalamic astrocytes using an upright epifluorescence microscope without an additional stage adaptor. The cell imaging chamber is composed of non-fluorescent acrylonitrile butadiene styrene (ABS). The chamber is designed such that it has an inlet port and an outlet port allowing the ability to exchange fluids in the bath chamber. This inexpensive cell imaging chamber allowed live cell imaging for at least 2 hours and has been implemented in an undergraduate research setting.
Materials and Methods
The live cell imaging chamber was designed in consultation with Idea Zoo Inc. in Lafayette, Louisiana. A 3D printer, Zortrax M200, was used to manufacture the live cell imaging chamber with non-fluorescing ABS (Figure 1, computer aided drafting and 3D printing files in multiple formats available as Supplemental Figures 1–4).
Figure 1. 3D printed Live Cell Imaging Chamber.
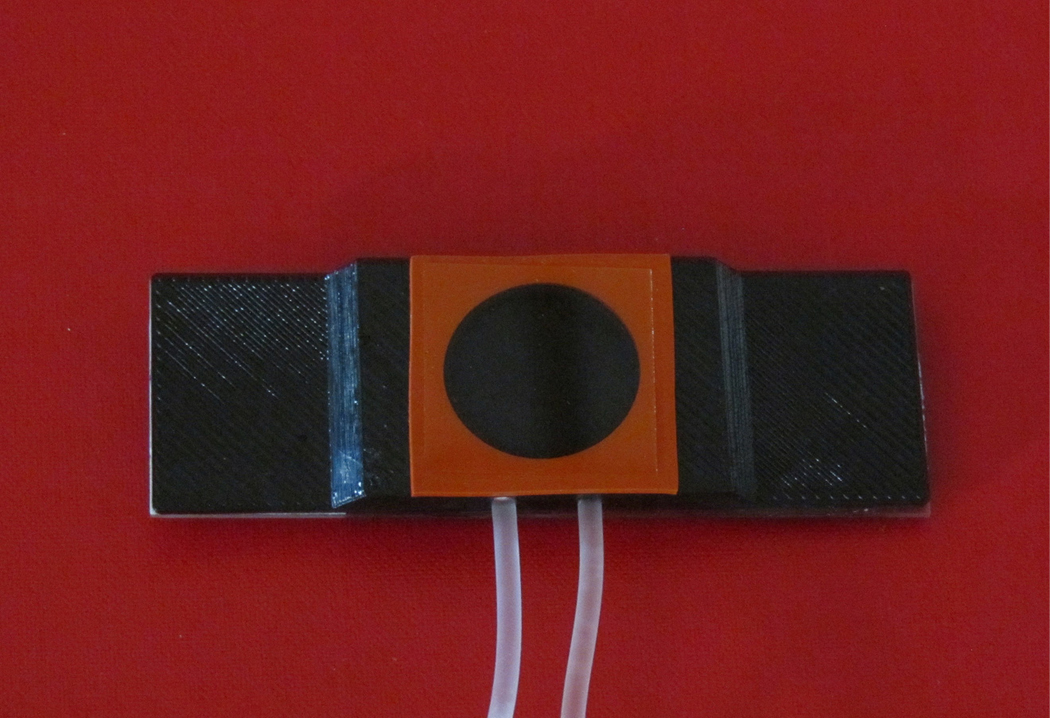
Base Dimensions are 75 mm x 25 mm, allowing it to fit on any upright epifluorescence microscope. It was 3D printed using black plastic without any auto-fluorescence. Cells are grown on the coverslip, stained with a fluorescent dye, and then inverted on top of a well filled with a physiological buffer solution.
The imaging chamber base was the same size as a standard glass microscope slide, 75 mm x 25 mm x 1 mm, allowing it to fit easily on any microscope stage. The total height of the chamber was 4 mm. It was glued on top of a regular glass slide for greater stability. This addition to the chamber only increased the height of the imaging chamber by 1 mm and was easily accommodated by lowering the stage.
The well of the imaging chamber measured 20 mm in diameter and 3 mm in depth. When the silicon isolator (Grace Bio-Labs, Bend, Oregon) was placed on top of the chamber platform, it formed a watertight seal for the 22 mm x 22 mm coverslip. The entire height of the apparatus was slightly greater than 6 mm. The volume of the well was approximately 1 ml.
Along the side of the chamber there were two ports leading into the well, 1 mm outer diameter tubing was inserted into the port holes. This allowed for the exchange of fluids in the chamber without greatly disturbing the cells at the coverslip and media interface. This exchange of media was accomplished by using syringes; one gently pushing new solutions into the chamber while the other syringe was simultaneously slowly removing fluid from the chamber.
Experiments were performed using hypothalamic astrocytes harvested from p2-p4 mouse pups as previously described (Smith et al. 2014). Astrocytes were grown in small (T25) flasks for 7–14 days using DMEM- F12 media (Thermo Fisher #10565042, Waltham, MA) enhanced with Fetal Bovine Serum (Life Technologies, Thermo Fisher #16140071, or Atlanta Biologicals # S11550H) and Antibiotic-Antimycotic (Life Technologies, Thermo Fisher #15240062) and incubated at 37⁰ C with 5% CO2. At 80–90% confluence, they were trypsinized (0.25% Trypsin, Life Technologies, Thermo Fisher) to remove them from the flasks and plated onto square 22 mm, Poly Lysine D (Sigma Aldrich P0899, St. Louis, MO) coated glass coverslips (Assist-Brand Deutsche Spiegelglas #633075, Carolina Biological, Burtlington, NC) at a density of 50,000 cells per coverslip. The cells were grown in the incubator for a week before imaging with Fluo 3-AM (Biotum, Inc # 50016, Fremont,CA) (Smith et al. 2014).
To image hypothalamic astrocytes, they were stained with a calcium indicator fluorescent dye solution consisting of 4 μM Fluo 3 AM in a 3 μM Pluronic 127 (Biotum #59000)- 2 mM DMSO (Sigma Aldrich # D2650)-HBSS buffer and 1 μM SR101 for 20 minutes at 37⁰ C. The Fluo 3 AM solution was removed with two rinses of 1% FBS in HBSS and 4 rinses of HEPES buffer. The coverslip was inverted on top of the chamber containing HEPES buffer, so the astrocytes were in constant contact with the HEPES on one side and in direct contact with the coverslip on the other side. Buffer exchange with 25μM ATP in HEPES was accomplished through side ports in the chamber. The imaging chamber was returned to the incubator for 10 minutes to allow de-esterification of the Fluo 3 AM by cytoplasmic esterases.
Imaging was performed on an upright epifluorescence microscope, the Zeiss axioimager microscope, with AxioCam MRm camera (the apotome 2 slider was removed for imaging). Zen software (Zeiss) was used to capture a series of images to generate time lapse data. The SOLA light source was controlled with a graphic user interface (GUI). To minimize photobleaching, the light intensity was set below 5%.
Another useful modification of the imaging chamber was to insert a thin glass disc into the well, to serve as a spacer when imaging tissue slices. The spacer brought the tissue closer to the underside of the coverslip in order to minimize imaging artifacts caused by having a large water gap between the cells and the coverslip. To construct the spacer, a circular disc the size of the well was cut from a glass slide with a diamond pen. By placing the glass disc into the well, we imaged calcium transients from Fluo 3 AM loaded hypothalamic brain slices. The brain slices were taken from a p1 mouse pup. Using a 1 mm slices from Zivic Brain Slice Matrix (Neonatal Mouse Brain Slicer Matrix BSMNS001–1, Pittsburg, PA), the slices were placed in ice cold oxygenated Artificial Cerebral Spinal Fluid: 124 mM NaCl, 5 mM KCl, 2 mM MgSO4, 10 mM Glucose, 26 mM NaH2CO3 and 2mM CaCl2. (Wong & Watkins 1982) We followed the same staining procedure for Fluo 3AM as was described above except for extending the dye incubation period from 20 minutes to 30 minutes to permit diffusion of the dye into cells of the thicker tissue slice.
Experiments were performed in compliance with University of Louisiana at Lafayette IACUC and utilizing the recommendations of The Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Animals were euthanized under the University of Louisiana at Lafayette IACUC committee APS numbers 2015–8717-048 and 2016–8717-031.
Results
We successfully used this chamber to image calcium waves in hypothalamic astrocyte cultures (Figure 2). To test the longevity and viability of the hypothalamic astrocyte cultures, the astrocytes were stained with the calcium indicator dye, Fluo 3 AM. At the start of the imaging session, spontaneous calcium waves were recorded and found to occur at 1–2/minute (Supplementary Movie 1). ATP has been reported to stimulate hypothalamic tanycytes in brain slices (Frayling et al. 2011). We tested 25μM ATP in our imaging system. In order to change fluids in the chamber’s well, the two side ports were designated as “in” or “out”. A syringe attached to tubing inserted into the designated “in” port was used to introduce the new solution into the chamber, whereas the syringe attached to tubing inserted into the designated “out” port was used as a gentle suction to remove excess solution. No adhesives were needed to secure the tubing. The well held 1 ml of fluid and was likely to be completely exchanged with 3–5 ml of new solution. The most common pitfalls associated with injecting fluids through the imaging chamber involved introducing air bubbles into the chamber that inadvertently displaced cells off the coverslip or involved exchanging the solutions too rapidly/forcefully, thereby producing turbulence that dislodged some of the cells from the coverslip. A gentle, even transfer of fluid through the imaging chamber that would not disrupt the cells, was readily achieved with practice.
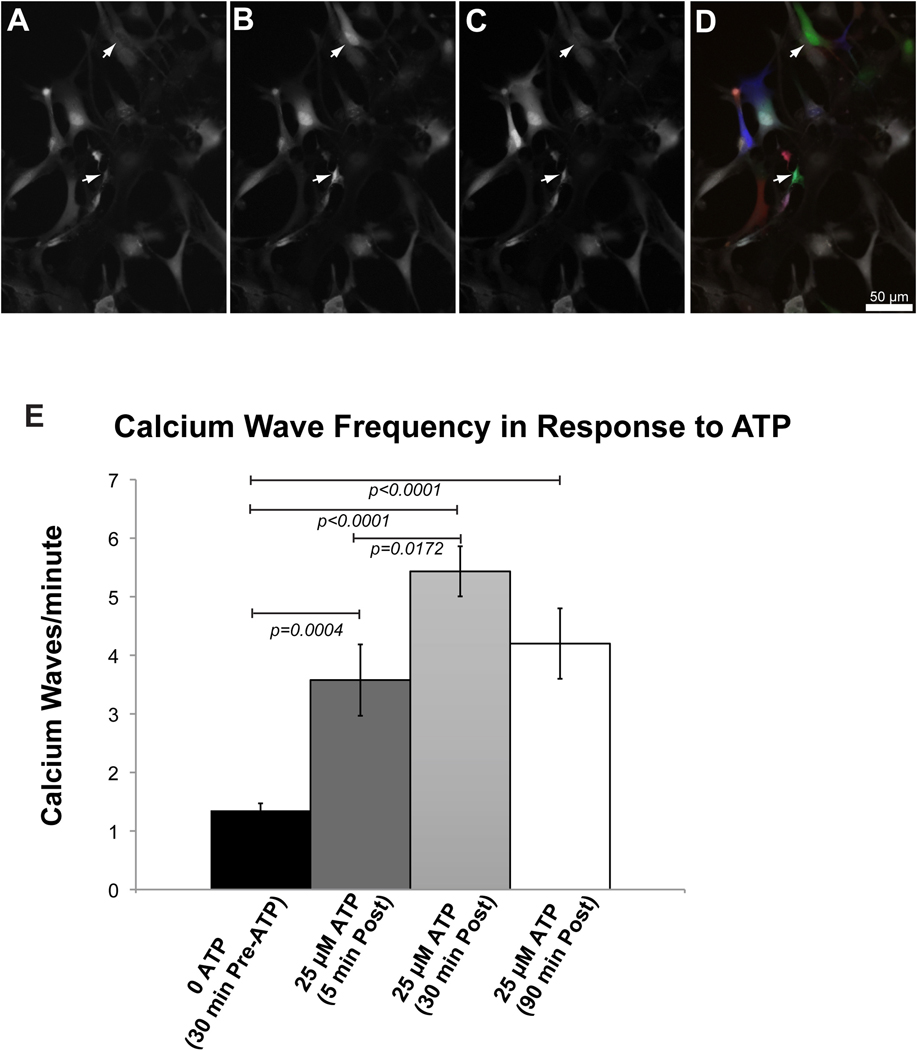
Figure 2. 2-hour imaging of hypothalamic astrocyte cultures using the 3D printed imaging chamber.
Representative images of calcium imaging in hypothalamic astrocytes at the beginning (A), peak (B), and end (C), of a calcium wave in two cells (arrows). The images in panels in A-C, were falsely colored red (A), green (B) and blue (C) and overlaid to make panel D. This panel demonstrates the dynamic nature of signaling, with colored cells representing actively signaling cells, and white cells representing cells between calcium waves. Initial spontaneous background calcium waves in astrocytes represented at 0 ATP (17 cells, 3 biological replicates). After 30 minutes imaging, 25 μM ATP stimulation was preformed via the exchange of fluid in the chamber (19 cells, 3 biological replicates). Imaging was also performed 30 minutes (15 cells, 3 biological replicates) and 90 minutes (10 cells, 2 biological replicates) after this one-time application of 25 μM ATP.
The viability of the astrocytes continued for 2 hours as evidenced by the presence of intracellular calcium waves during the ATP stimulated condition (Figure 2). Thirty minutes after the start of the imaging experiment, the 25 μM ATP was added. Within a few minutes after loading the chamber with the 25 μM ATP solution, the frequency of the waves increased to 3–4/minute (p=0.004, see Supplementary Movie 2). After 30 min of ATP stimulation, the transients significantly increased to 5–6/minute compared to the non-stimulated condition (p<0.0001), and compared to the first few minutes of ATP stimulation (p=0.0172, Supplementary Movie 3). However, the first and second ATP stimulated time points were not significantly different from each other after applying a Bonferroni correction for multiple comparisons. Finally, after 2 hours of imaging and 90 minutes of ATP stimulation, we found that the cells were still viable, and that there were significantly more calcium transients compared to non ATP stimulated cells (p<0.0001), but not significantly different from either of the previous two ATP stimulated time points (Supplemental Movie 4 Supplemental Movie 4).
Our observation of increased frequencies of calcium signaling transients in hypothalamic cell cultures in response to 25μM ATP was consistent with reported hypothalamic tanycyte activity in culture and slice preparations from other investigators (Frayling et al. 2011; Orellana et al. 2012). The third movie showed a continuation of calcium wave activity in the same hypothalamic astrocyte culture after 2 hours of imaging. During this 2 h incubation time, there was no further exchange of media. Thus, the 25μM ATP induced increase in the frequency of calcium transients persisted for at least 1.5 hours until the imaging session concluded at 2 hours total imaging time. The chamber, with the addition of the glass disk in the base of the well, may also be used to image brain slices, which would allow slicing and imaging of calcium waves within the time frame of a 3 hour laboratory period (Supplemental Movie 5).
Discussion
One parameter that commercially available imaging chambers possess is the ability to maintain a constant temperature. As designed, this chamber did not have any external temperature control. We performed imaging sessions at ambient room temperature. Here, we show that murine brain cells remain viable, and suitable for physiological measurements, for at least 2 hours outside the tissue culture incubator. There are many excellent live imaging chambers currently available on the market, yet the cost and required specialized equipment prohibits their widespread implementation. Many laboratories have a microscope set-up dedicated to performing live cell imaging. Since our epifluorescence microscope is also used for stereology and neuronal tracing (Stereo Investigator and Neurolucida, MBF), it cannot be solely dedicated to live cell imaging. We designed a generic multipurpose imaging chamber that can be used on any upright epifluorescence microscope. Using 3D printing and the files in Supplemental Material, investigators can print their own imaging chamber. The cost of this chamber is only a fraction of the specialty marketed live imaging chambers (less than $15.00). This chamber provides a low-cost live cell imaging method, allowing excellent visualization of live cultured cells with viability of the cells lasting at least for several hours. With use of syringes, the ports on the side of the chamber provide an option to exchange the chamber media, adding a greater functionality to this imaging chamber.
Through the addition of a glass disk into the base of the well, we were also able to perform calcium imaging in brain tissue slices obtained with a brain matrix. This will provide experimental conditions that are closer to the in vivo physiology of the entire tissue, and experiments that can be performed in teaching labs without preparation of cell cultures in a previous lab session. The use of a brain matrix to slice tissues is simple, swift, and affordable.
This imaging chamber is well suited for cellular physiology education and could be used for various experimental applications in teaching labs such as imaging cytoskeletal dynamics with stains for actin and tubulin. Developing a lab experience that allows students the opportunity to experience live cell imaging without having to use specialized and cost-prohibited equipment would enrich any cellular physiology and neurobiology-based laboratory course.
Supplementary Material
Acknowledgements
The authors wish to thank Lilian Gathanga, Michael Sides, Michelle Hendrick and Blake Bolgiano for technical assistance. The authors also wish to thank the Microbrightfield staff, including Masha Stern and Geoff Green for helpful advice on adapting our equipment set up for time lapse imaging. The authors appreciate Jacques Dugal from Idea Zoo for providing the Supplemental Data CAD files of the imaging chamber.
Funding
This work was supported by the Brain Behavior Research Foundation Young Investigator Award (KMS), The National Institute of Mental Health (K01MH087845), Ray P. Authement College of Sciences startup funds from the University of Louisiana at Lafayette (KMS), and the Graduate Student Organization at the University of Louisiana at Lafayette (DR).
References
- Cole R, 2014. Live-cell imaging: The cell’s perspective. Cell Adhesion and Migration, 8(5), pp.452–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey ME, Marrs GS & Kurpius D, 2011. Maintaining live cells and tissue slices in the imaging setup. Cold Spring Harbor protocols, (4), pp.1–12. Available at: http://www.ncbi.nlm.nih.gov/pubmed/21460059 [Accessed July 20, 2016]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayling C, Britton R & Dale N, 2011. ATP-mediated glucosensing by hypothalamic tanycytes. The Journal of physiology, 589(Pt 9), pp.2275–86. Available at: http://www.mendeley.com/research/atpmediated-glucosensing-hypothalamic-tanycytes/ [Accessed May 9, 2015]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orellana JA et al. , 2012. Glucose increases intracellular free Ca(2+) in tanycytes via ATP released through connexin 43 hemichannels. Glia, 60(1), pp.53–68. Available at: http://www.ncbi.nlm.nih.gov/pubmed/21987367 [Accessed September 11, 2016]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KM et al. , 2014. Fgfr1 inactivation in the mouse telencephalon results in impaired maturation of interneurons expressing parvalbumin. PLoS ONE, 9(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson AL, Hilton ST & Andreae LC, 2015. Rapid, simple and inexpensive production of custom 3D printed equipment for large-volume fluorescence microscopy. International Journal of Pharmaceutics, 494, pp.651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardyn JD et al. , 2015. Low cost production of 3D-printed devices and electrostimulation chambers for the culture of primary neurons. Journal of Neuroscience Methods, 251, pp.17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong RK & Watkins DJ, 1982. Cellular factors influencing GABA response in hippocampal pyramidal cells. Journal of neurophysiology, 48(4), pp.938–51. Available at: http://www.ncbi.nlm.nih.gov/pubmed/7143035 [Accessed September 9, 2016]. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.