Abstract
Rat PEX12 cDNA was isolated by functional complementation of peroxisome deficiency of a mutant CHO cell line, ZP109 (K. Okumoto, A. Bogaki, K. Tateishi, T. Tsukamoto, T. Osumi, N. Shimozawa, Y. Suzuki, T. Orii, and Y. Fujiki, Exp. Cell Res. 233:11–20, 1997), using a transient transfection assay and an ectopic, readily visible marker, green fluorescent protein. This cDNA encodes a 359-amino-acid membrane protein of peroxisomes with two transmembrane segments and a cysteine-rich zinc finger, the RING motif. A stable transformant of ZP109 with the PEX12 was morphologically and biochemically restored for peroxisome biogenesis. Pex12p was shown by expression of bona fide as well as epitope-tagged Pex12p to expose both N- and C-terminal regions to the cytosol. Fibroblasts derived from patients with the peroxisome deficiency Zellweger syndrome of complementation group III (CG-III) were also complemented for peroxisome biogenesis with PEX12. Two unrelated patients of this group manifesting peroxisome deficiency disorders possessed homozygous, inactivating PEX12 mutations: in one, Arg180Thr by one point mutation, and in the other, deletion of two nucleotides in codons for 291Asn and 292Ser, creating an apparently unchanged codon for Asn and a codon 292 for termination. These results indicate that the gene encoding peroxisome assembly factor Pex12p is a pathogenic gene of CG-III peroxisome deficiency. Moreover, truncation and site mutation studies, including patient PEX12 analysis, demonstrated that the cytoplasmically oriented N- and C-terminal parts of Pex12p are essential for biological function.
Peroxisomes are present in a wide variety of eukaryotic cells, from yeast to human. Peroxisomes are formed by division of preexisting peroxisomes after posttranslational import of newly synthesized proteins (21). Peroxisomal proteins, including membrane proteins, are encoded by nuclear genes, translated on free polyribosomes in the cytosol, mostly at their final sizes, and posttranslationally translocated to preexisting peroxisomes (21). cis-acting peroxisomal targeting signals (PTSs) have been identified: the C-terminal SKL motif (PTS1) and N-terminal cleavable presequence (PTS2) of several proteins, such as 3-ketoacyl coenzyme A (3-ketoacyl-CoA) thiolase (10, 46). Genetic analyses of peroxisome-deficient mutants of yeast and mammalian cells have led to identification of a number of protein factors essential for peroxisome biogenesis (6, 10, 20, 46).
The primary cause for the peroxisome deficiency in fatal genetic diseases such as Zellweger syndrome was thought to be the failure of peroxisome biogenesis (11, 22, 43). Genetic heterogeneities are seen in subjects with these peroxisome deficiency disorders. Cell fusion studies of fibroblasts from these patients identified at least 10 different complementation groups (CGs) (27, 37, 43, 64). Furthermore, more than 12 genes are likely to be involved in mammalian peroxisome biogenesis, as deduced from the studies of CG analysis of patient fibroblasts and CHO cell mutants (32, 43, 52, 57, 65), including two newly identified CGs (50).
We previously isolated and characterized three distinct CHO cell mutants, Z65, Z24, and ZP92 (43, 57). A 35-kDa peroxisome integral membrane protein, Pex2p (formerly peroxisome assembly factor 1 [PAF-1]), was identified in Z65 (54) by stable transfection of a rat cDNA expression library in the pcD2 vector. A nonsense mutation of the PEX2 gene causes Zellweger syndrome of CG-F (the same group as CG-X in the United States and CG-5 in Europe) (44). Rat PAF-2 cDNA (termed PEX6), encoding a member of the ATPase family, was isolated by functional complementation of peroxisome deficiency of a CHO mutant, ZP92, using a transient transfection assay (55). PEX6 was shown to be responsible for Zellweger syndrome of CG-C (the same as CG-IV in the United States) (13, 63). Pex5p (PTS1 receptor) (7, 9, 61) was found to be defective in CHO mutants such as ZP102 (52), ZP105, and ZP139 of CG-II (35). Dysfunction and mutations of PEX5 were found in CG-II patients (7, 61). Very recently, we cloned human PEX1 by genetic complementation assay using a CHO cell mutant, ZP107, and demonstrated that PEX1 is responsible for peroxisome deficiency disorders of CG-I (the same as CG-E in Japan) (48). Thus, peroxisome assembly-defective CHO cell mutants are indeed useful for studies of peroxisome biogenesis and for elucidating primary defects of human peroxisome biogenesis disorders. We recently isolated peroxisome biogenesis-defective CHO mutants ZP104 and ZP109, which belonged to CG-III of the human peroxisome deficiency condition Zellweger syndrome (32).
We herein identified rat PEX12, which restored peroxisome biogenesis in CHO cell mutant ZP109, by a genetic complementation cloning strategy (13, 44, 54, 55) using green fluorescent protein (GFP) (3). Peroxisomes were also complemented in CG-III human peroxisome-deficient fibroblasts by transfection of human and rat PEX12. In two peroxisome deficiency patients, we found homozygous mutations that inactivate PEX12, demonstrating that PEX12 is the causal gene for CG-III peroxisome deficiency. A RING finger was required for the function of Pex12p.
MATERIALS AND METHODS
Plasmids and cDNA library construction.
A mammalian expression vector, pUcD2SRαMCSHyg, was constructed by replacing the BamHI fragment containing the neo gene of pUcD2SRαMCS (55) with the SalI (synthetic site with PCR technique)-BglII fragment containing the hyg gene of the pSV2hyg vector. Plasmid pUcD2Hyg · RnPEX12 was generated by inserting an XhoI-ApaI fragment (−163 to 1615) of the original rat (Rattus norvegicus) PEX12 (RnPEX12) clone, pBK-CMV · 46, into the pUcD2SRαMCSHyg vector. pUcD2Hyg · RnPEX12 was used to establish PEX12 stable transformants of ZP109 cell by selection in the presence of hygromycin B (200 μg/ml; Sigma, St. Louis, Mo.).
RNA was prepared from the liver of a male F344 rat by the phenol extraction method (12). Poly(A)+ RNA was purified with Oligotex dT-30 latex (Takara, Tokyo, Japan), which was used for cDNA synthesis. cDNA was synthesized by using a cDNA synthesis kit (Stratagene, La Jolla, Calif.) with Superscript II reverse transcriptase (Gibco BRL, Gaithersburg, Md.) and EcoRI adapter (Takara) and was size fractionated on a cDNA size fractionation column (Gibco BRL). cDNA of a larger size was ligated with the ZAP Express predigested vector (EcoRI-XhoI digested, calf intestine alkaline phosphatase treated) (Stratagene) to unidirectionally insert cDNA under control of the cytomegalovirus promoter. Cloned cDNA in the ZAP Express phage vector was packaged with Gigapack III Gold packaging extract (Stratagene) and plated on an XL1-Blue MRF′ host strain. The mean length of cDNA inserts of this library was ∼2.0 kb. The library was divided into small pools, each containing about 2,000 clones. In vivo excision of pBK-CMV phagemid vector from the amplified phage pools was done separately, as recommended by the manufacturer. Combined cDNA pools consisted of a mixture of equal amounts of three small pools, containing approximately 6,000 independent clones.
GFP transformant of CHO cells.
Plasmid containing cDNA encoding a GFP variant, GFP-TT, with the two amino acid substitutions Ser65Thr and Ile167Thr was a generous gift from Y. Jin and H. R. Horvitz. Plasmids for GFP variants pGFP-AKL, encoding GFP with C-terminal PTS1 (-Ala-Lys-Leu) and pGFP-Stop for GFP with no signal, were from J. M. Goodman. Plasmids expressing Ser65Thr-GFP and Ile167Thr-GFP and that with PTS1, GFP(TT)-AKL, were constructed in the mammalian expression vector pUcD2SRαMCSHyg [pUcD2HygGFP(TT)-AKL] as follows. A BalI-NspV fragment, including two amino acid substitutions of GFP-TT, was inserted in place of that of pGFP-AKL. An NheI (blunted)-BamHI fragment of pGFP(TT)-AKL was ligated in pUcD2SRαMCSHyg vector doubly digested with NotI (blunted) and BamHI. Wild-type CHO-K1 and a peroxisome-deficient CHO mutant, ZP109, were transfected with pUcD2HygGFP(TT)-AKL. Respective stable transformants were selected in the presence of hygromycin B (200 μg/ml), and highly GFP-expressing cells were cloned by limiting dilution.
Screening of a cDNA library.
For transfection of the cDNA library, GFP-transformed ZP109 (ZP109G1) cells were cultured in Ham’s F12 medium supplemented with 10% fetal calf serum (FCS). Cells that had been plated on a coverslip 1 day before transfection at 105 cells per well of six-well plate were washed twice with serum-free F12 medium. One microgram of plasmid DNA and 12 μg of Lipofectamine (Gibco BRL) were separately diluted with 100 μl of serum-free medium (OPTI-MEM; Gibco BRL). Both solutions were mixed and incubated for 30 min at room temperature before being poured onto the cells. After incubation for 4 h at 37°C, the DNA-liposome mixture was aspirated off and fresh F12 medium with FCS was added. The cells were cultured for 2 days and further incubated overnight in 2 ml of serum-free F12 medium. GFP in unfixed cells was visualized under a Zeiss Axioskop FL microscope (Carl Zeiss, Oberkochen, Germany) as described below. Among the cDNA pools examined, a positive one (B4) which restored peroxisomes in ZP109G1 was further divided into subpools and screened. A cDNA fragment (EcoRI-XhoI fragment) of an isolated clone, pBK-CMV · 46, was subcloned into the site between EcoRI and XhoI of pBluescript II SK(−) (Stratagene). The nucleotide sequences of both strands were determined by the dideoxy-chain termination method using a Dye-Terminator DNA sequence kit (Applied Biosystems, Foster City, Calif.). Alignment was done with the GENETYX-Mac program (SDC, Tokyo, Japan).
Stable transfection of PEX12.
CHO cell mutants Z24, Z65, ZP92, ZP105, ZP104, and ZP109 were stably transfected with the rat PEX12 expression plasmid pUcD2Hyg · RnPEX12 as described above. The hygromycin B-resistant colonies formed on the coverslips were examined for peroxisomes by staining with anti-rat catalase antibody 6 days after transfection. Peroxisome-restored colonies were counted. To isolate a stable clone of RnPEX12 transformants of mutant ZP109, pUcD2Hyg · RnPEX12 was transfected into ZP109 cells as described above. Three of seven transformants isolated were peroxisome positive, on the basis of immunostaining, and one of the three, named 109P3, was further cloned by the limiting-dilution method.
Morphological analysis.
GFP-AKL in cells grown a coverglass was observed without fixation under a Zeiss Axioskop FL microscope using a no. 17 filter. Peroxisomes in CHO cells and human fibroblasts were visualized by indirect immunofluorescence light microscopy as described elsewhere (43). We used rabbit antibodies to rat liver catalase (57), human catalase (43), PTS1 peptide comprising the C-terminal 10 amino acid residues of rat acyl-CoA oxidase (35), 3-ketoacyl-CoA thiolase (57), and 70-kDa peroxisomal integral membrane protein (PMP70) (57). Anti-Pex12p antibody was raised in rabbits by immunizing them with a synthetic peptide comprising the C-terminal 21-amino-acid sequence (see Fig. 2A) that had been linked to keyhole limpet hemocyanin (30, 57). Goat anti-rat catalase antibody was raised by conventional subcutaneous injection. Antigen-antibody complex was detected by fluorescein isothiocyanate-labeled sheep anti-rabbit immunoglobulin G (IgG) antibody (Cappel, Durham, N.C.) or donkey anti-goat IgG antibody conjugated to rhodamine (Chemicon, Pittsburgh, Pa.), under a Zeiss Axioskop FL microscope.
FIG. 2.
Deduced amino acid sequence of rat (R. norvegicus [Rn]) PEX12. (A) Alignment with human (Homo sapiens [Hs]) and P. pastoris (Pp) Pex12p. Dashes represent spaces. The putative membrane-spanning segment is overlined; the dashed line indicates the sequence used for chemical synthesis of the Pex12p peptide. Identical amino acids between two or more species are shaded. Conserved cysteine residues of the RING finger are designated by asterisks. Open and solid arrowheads indicate the positions of mutations in CG-III patients PBD3-01 and PBD3-03, respectively. (B) Conserved RING finger in the C-terminal regions of the peroxins required for peroxisome assembly. RING fingers were found in Pex2p of mammals (44, 54, 56), P. pastoris (59), Yarrowia lipolytica (Yl) (8), and Podospora anserina (Pa) (1), Pex10p of P. pastoris (19) and Hansenula polymorpha (Hp) (49), and Pex12p of P. pastoris (18). Italic numbers designate positions in respective deduced amino acid sequence; Xn, numbers of amino acid residues. Consensus cysteine and histidine residues in the RING are shaded and numbered (see text).
Identification of patient mutations.
Total RNA was obtained from cultured patient fibroblasts by the acid guanidinium-phenol-chloroform method (5). Reverse transcription-PCR using total RNA was done with a pair of human PEX12-specific PCR primers, HsPEX12.f and HsPEX12.r (Table 1), covering the full length of the human PEX12 open reading frame (33). Nucleotide sequences of the PCR products cloned into pBluescript were determined as described above.
TABLE 1.
Synthetic oligonucleotide primers used in this study
| Codea | Sequence (5′ to 3′) | Underlined |
|---|---|---|
| HsPEX12.f | GCGCGGATCCCTATGGCTGAGCACGGG | Initiation codon |
| HsPEX12.r | GCGCGGATCCTCAGTTCTCAGGGGAG | Termination codon |
| Myc.r | CGGTACCGGGCCCTCACAAGTCTTCTTCAGAAATAAGC-TTTTGTTCGTTCTCTGGAGAGTAGAG | Myc epitope |
| Flag.f | GCTCTAGAGCGGCCGCCACCATGGATTACAAGGACGAC-GACGATAAGCTTGGGATCCAGGCTGAGCATGG-GGCTC | Flag epitope |
| Flag.r | GTGTTCAGAAAATGAGGC | |
| 46PstI.f | GTGTATTCTTCCTGCAG | |
| C304S.f | GTGTCTCCACTGTGTCG | Codon for 304Cys (C1) mutated to Ser |
| C304S.r | GTGGAGACACTGTTTTC | Codon for 304Cys (C1) mutated to Ser |
| C307S.f | CCACTGTCTCGTAAAGC | Codon for 307Cys (C2) mutated to Ser |
| C307S.r | GCTTTACGAGACAGTGG | Codon for 307Cys (C2) mutated to Ser |
| C325S.f | TCTCTTACCGCTGTGTG | Codon for 325Cys (C5) mutated to Ser |
| C325S.r | GTAAGAGAATACATAGC | Codon for 325Cys (C5) mutated to Ser |
| C328S.f | TTACCGCTCTGTGTTTA | Codon for 328Cys (C6) mutated to Ser |
| C328S.r | TAAACACAGAGCGGTAA | Codon for 328Cys (C6) mutated to Ser |
| H336S.f | GAGAAGTTCTCAAGCCT | Codon for 336His mutated to Ser |
| H336S.r | AGGCTTGAGAACTTCTC | Codon for 336His mutated to Ser |
| C339S.f | GCCTCTCCCATTACAGG | Codon for 339Cys (C7) mutated to Ser |
| C339S.r | GGAGAGGCTTGGTGACT | Codon for 339Cys (C7) mutated to Ser |
| P3/Myc/C.r | GTACCGGGCCCTCACAA |
f and r, forward and reverse primers, respectively.
Patient fibroblasts and transfections.
Fibroblasts from a patient (PBD3-01) with Zellweger syndrome and a patient (PBD3-03) with peroxisome deficiency disorder were classified as CG-III by complementation cell fusion analysis (44a) as described previously (64). Primary fibroblasts from patients were transformed as described previously (31). Fibroblasts were cultured in Dulbecco’s modified Eagle medium (Gibco BRL) supplemented with amino acids and 10% FCS. Patient fibroblasts were transfected with pUcD2Hyg · RnPEX12 by using Lipofectamine and stained with rabbit antiserum against human catalase. Patient-derived PEX12 cDNA was cloned into pCMVSPORT I vector (Gibco BRL) by replacing the BglII-BamHI fragment of PEX12 from a control (33) with that of patient PEX12 in pBluescript. Transfections of the patient PEX12 were done on fibroblasts (106 cells) with 20 μg of plasmid DNA, using a Gene Pulser II electroporator (Bio-Rad, Hercules, Calif.) at settings of 320 V and 500 μF.
Expression of epitope-tagged Pex12p.
A Myc epitope tag was added to the C terminus of rat Pex12p by using a PCR-based technique and forward and reverse primers 46PstI.f and Myc.r, containing the Myc epitope and a stop codon, respectively. Myc-tagged PEX12 was cloned into pBluescript II SK(−) (pBS · 46myc) by replacing the PstI-ApaI fragment of pBS · 46 with the amplified fragment after PCR, using pBS · 46 as the template, following digestion with PstI and ApaI. Plasmid pUcD2Hyg · RnPEX12-myc, constructed by inserting the XhoI-ApaI fragment of pBS · 46myc into pUcD2SRαMCSHyg vector, was transfected into wild-type CHO-K1 and mutant cells. Flag tagging at the N terminus of RnPEX12-encoded protein was likewise done by PCR using forward primer Flag.f, containing a flag epitope, and reverse primer Flag.r. Flag-tagged RnPEX12 in pUcD2Hyg · SRα vector (pUcD2Hyg · flag-RnPEX12) was constructed by replacing the NotI-SpeI fragment of pUcD2Hyg · RnPEX12 with a NotI-SpeI fragment of the PCR product. Pex12p-Myc and flag-Pex12p were detected by using mouse monoclonal antibodies to human c-Myc (9E10) (Santa Cruz Biotechnology, Santa Cruz, Calif.) and flag (M2) (Scientific Imaging Systems, New Haven, Conn.), and Texas red-labeled sheep anti-mouse IgG secondary antibody (Amersham, Buckinghamshire, England), in cells that had been permeabilized with either 25 μg of digitonin per ml or 1% Triton X-100.
Truncation and site mutation studies.
An N-terminal truncation mutant of rat Pex12p, termed ΔN1, was constructed by self-ligation of the BamHI-XbaI fragment of pUcD2Hyg · flag-RnPEX12, adjusting a reading frame by using a 12-mer BamHI linker. ΔN1 designates rat flag-Pex12p with a deletion from the N terminus to amino acid residue 76. Site mutations in the RING finger motif, including Cys at residue 304 to Ser (C304S; termed mut1), C325S (mut5), and C339S (mut7), were generated by two steps of PCR. First, PCR was done with pBS · 46myc as a template using six sets of primers, 46PstI.f and C304S.r, 46PstI.f and C325S.r, 46PstI.f and C339S.r, P3/Myc/C.r and C304S.f, P3/Myc/C.r and C325S.f, and P3/Myc/C.r and C339S.f. The second PCR for each site mutation was done with primers 46PstI.F and P3/Myc/C.r, using as a template a pair of the first PCR products containing the same one-site mutation. After digestion with PstI and ApaI, the amplified fragments were subcloned into pBS by replacing with PstI-ApaI fragment of pBS · 46myc. Resulting mutated forms of RnPEX12-myc cDNA were each cloned into the XhoI-ApaI site of pUcD2Hyg · SRα vector. Two-site mutations were likewise introduced by using a set of templates each containing a single mutation at a different site: C307S.f, C307S.r, C328S.f, C328S.r, H336S.f, and H336S.r. N-terminal flag tagging to C304S-Pex12p (mut1) and C339S-Pex12p (mut7) was done by replacing the SpeI-KpnI fragment of pUcD2Hyg · flag-RnPEX12 with the SpeI-KpnI fragment of pUcD2Hyg · RnPEX12-myc mutated to C304S or C339S. All constructions were assessed by nucleotide sequencing.
Other methods.
Continuous cell labeling was done by culturing cells in medium containing 20 μCi of [35S]methionine and [35S]cysteine (Amersham) per ml for 24 h. Immunoprecipitation of proteins and catalase latency assays using digitonin were done as described previously (57). For Northern blotting, liver RNA was isolated from a rat treated for 7 days with clofibrate and from an untreated rat. The blot was hybridized with the SpeI-PstI fragment (residues at 232 to 772) of rat PEX12 labeled with [α-32P]dCTP (Amersham). In vitro transcription and translation were done as described previously (25). Western blot analysis on a polyvinylidene difluoride membrane (Bio-Rad) was done with a second antibody, donkey anti-rabbit IgG antibody conjugated to horseradish peroxidase, with ECL Western blotting detection reagent (Amersham). Resistance to P12/UV [12-(1′-pyrene)dodecanoic acid/long-wavelength UV light] and P9OH [9-(1′-pyrene)nonanol]/UV treatments was determined under conditions of 2 μM/1.5 min and 6 μM/2 min (43), respectively. Peroxisomal dihydroxyacetone phosphate acyltransferase (DHAP-ATase) activity was measured as described previously (55), using [14C]labeled DHAP as a substrate.
Nucleotide sequence accession numbers.
The GenBank database accession numbers for rat and human PEX12 genes are AB002111 and AB004546, respectively.
RESULTS
Cloning of a rat PEX12 cDNA.
We used a transient expression assay as a cDNA cloning strategy (55). A rat liver cDNA library divided into small pools was transfected into ZP109G1, a stable transformant of ZP109 transfected with a plasmid expressing GFP fused to PTS1 tripeptide AKL (GFP-AKL). GFP-AKL was properly localized in peroxisomes in the wild-type CHO-K1 cells, as assessed by colocalization with a peroxisomal enzyme, catalase, whereas it was diffused in the cytosol and nucleus in ZP109G1 (Fig. 1a to d). This result indicated that GFP tagged with PTS1 is a useful ectopic marker for peroxisomal protein import with no need for cell fixation. Peroxisome-restoring positive cDNA clones were searched for by directly observing peroxisomal localization of GFP-AKL in unfixed cells, without use of fluorescence-emitting chemicals such as fluorescein isothiocyanate. Twenty-four combined pools (see Materials and Methods) which contained about 1.5 × 105 independent clones were screened, and one combined pool (B4) yielded several peroxisome-restored cells of ZP109G1 in a single dish (Fig. 1e, arrows). After a fourth round of screening, we isolated one positive clone named pBK-CMV · 46, which restored peroxisome biogenesis as assessed by fluorescence microscopy by virtue of GFP-AKL (data not shown). The cDNA portion of pBK-CMV · 46 was sequenced on both strands, indicating that the cDNA was 2,365 bp in length with an open reading frame encoding a protein consisting of 359 amino acids (Fig. 2A). The calculated molecular mass was 40,380 Da. A homology search suggested that this cDNA was likely to be related to Pichia pastoris PEX12 (18), with 27% amino acid identity (Fig. 2A); we therefore termed the protein encoded by this cDNA Pex12p. Rat PEX12 apparently complemented peroxisomal import of GFP-PTS1 in ZP109G1 (Fig. 1f). The rat PEX12 and very recently isolated human PEX12 (4, 33) showed high homology, 87 and 89% at the nucleotide and deduced amino acid sequence levels, respectively (Fig. 2A). Both sequences were more conserved in the C-terminal region and included two putative membrane-spanning segments and a cysteine-rich region, residues 304 through 342, with a good alignment of five cysteine residues to the consensus RING finger motif (42), C3HC4, although three other conserved residues, including histidine, were replaced (Fig. 2B). RING finger motifs were found in a number of proteins apparently involved in DNA binding, DNA recombination, and other cellular events (42), including peroxisome biogenesis factors such as mammalian Pex2p (44, 51, 54, 56).
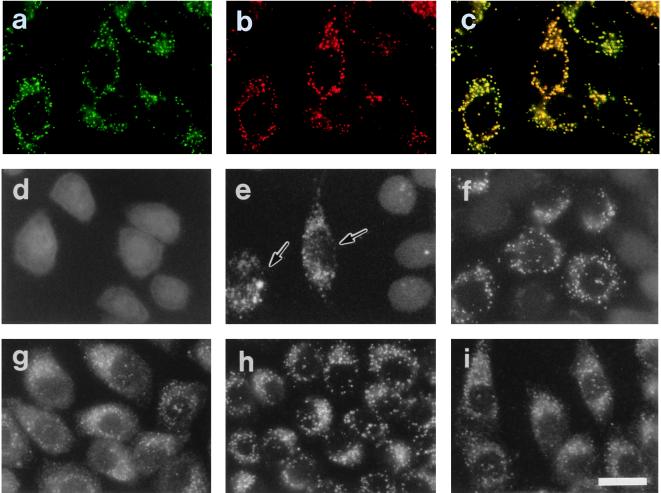
FIG. 1.
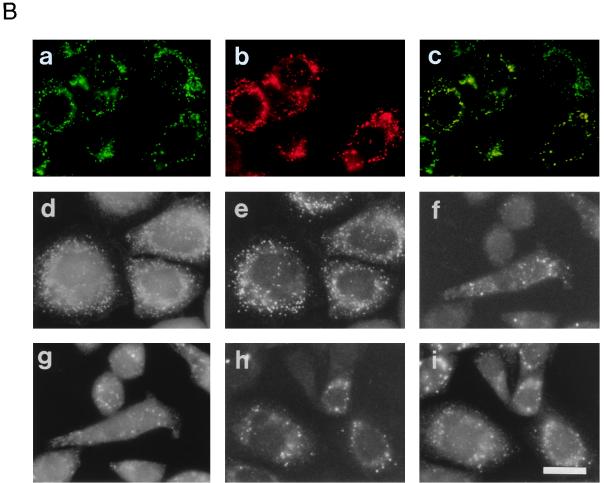
Restoration of peroxisomes in CHO mutant cells. (a and d to f) Fluorescence microscopy of peroxisomes. Intracellular location of GFP in CHO cells stably expressing GFP tagged with tripeptide PTS1 (AKL) was monitored on unfixed cells, except for panel a, under fluorescence microscopy. (b and g to i) Immunofluorescent staining of peroxisomes. (a) Wild-type CHOG1 cells; (b) CHOG1 stained with goat anti-rat catalase antibody; (c) merged view of panels a and b; (d) peroxisome-deficient mutant ZP109G1 cells; (e) peroxisome-restored ZP109G1, after lipofection with a combined pool (B4) of rat cDNA library. Arrows indicate the complemented cells. Cytosolic appearance of GFP-AKL was apparent in the other cells. (f) ZP109G1 transfected with plasmid pUcD2Hyg · RnPEX12. (g to i) 109P3 cells (stable transformants of ZP109 cells obtained with pUcD2Hyg · RnPEX12) were stained with antisera to catalase, PTS1 peptide, and PMP70, respectively. Magnification, ×570; bar, 20 μm.
PEX12 restored peroxisome biogenesis in ZP109.
Several phenotypic abnormalities due to peroxisome deficiency were found in the mutant cell line ZP109 (32). ZP109 was found to lack peroxisomes on immunofluorescence staining, using antibodies to catalase and 3-ketoacyl-CoA thiolase (PTS2 protein). Peroxisomal ghosts were also noted with antibody to PMP70, as in other CHO mutants (43) and patient fibroblasts (41, 60, 62). To determine whether rat PEX12 could correct these phenotypes, a stable PEX12 transformant of ZP109, named 109P3, was isolated by transfecting with pUcD2Hyg · RnPEX12 and then characterized. Catalase was noted in numerous vesicular structures, presumably peroxisomes, when stained with anticatalase antibody (Fig. 1g). PTS1 proteins were likewise localized in peroxisomes (Fig. 1h). Import of a peroxisomal PTS2 protein, thiolase, was also restored (data not shown). PMP70-positive particles were as numerous as those detected by immunostaining of soluble proteins (Fig. 1i). These results strongly suggested that 109P3 cells had morphologically normal peroxisomes, as seen in the wild-type CHO-K1 cells.
In peroxisome-deficient cells, peroxisomal proteins are mislocalized to the cytosol, rapidly degraded, or not converted to mature forms, despite normal synthesis (43, 57). In the digitonin titration assay, catalase was present in peroxisomes in the wild-type cells, and nearly 60% of catalase activity was latent at the digitonin concentration of 100 μg/ml (Fig. 3A). In ZP109 cells, catalase was apparently present in the cytosol, as was the case for a cytosolic marker enzyme, lactate dehydrogenase, in CHO-K1; hence catalase was not latent, a finding consistent with our earlier observation (32). In 109P3 cells, catalase showed almost the same latency as in the wild-type CHO-K1 cells.
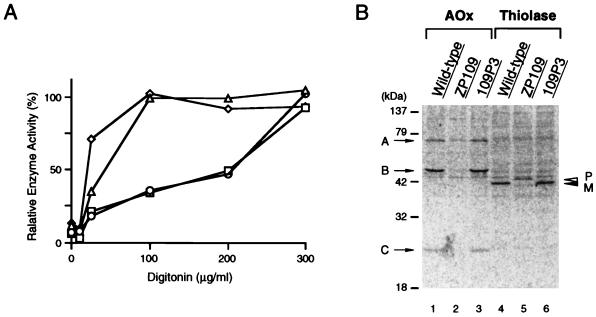
FIG. 3.
Complementation of biogenesis of peroxisomal enzymes. (A) Latency of catalase activity in CHO-K1, ZP109, and 109P3 cells. Circles, CHO-K1; triangles, ZP109; squares, 109P3, stable rat PEX12 transformant of ZP109; diamond, lactate dehydrogenase in CHO-K1. Relative free enzyme activity is expressed as a percentage of the total activity measured in the presence of 1% Triton X-100 (57). The results represent means of duplicate assays. (B) Biogenesis of peroxisomal proteins. Cells were labeled for 24 h with [35S]methionine and [35S]cysteine. Cell types are indicated at the top. Immunoprecipitation was done with rabbit anti-rat acyl-CoA oxidase (AOx) and 3-ketoacyl-CoA thiolase (Thiolase) antibodies. Immunoprecipitates were analyzed by SDS-PAGE (12% polyacrylamide gel). Radioactive polypeptide bands were detected using a FujiX BAS1000 Bio-Imaging Analyzer (Fuji Photo Film, Tokyo, Japan). Exposure time was 14 h. Arrows show the positions of AOx components A, B, and C; open and solid arrowheads indicate a larger precursor (P) and mature protein (M) of 3-ketoacyl-CoA thiolase, respectively.
Acyl-CoA oxidase, the first enzyme of peroxisomal fatty acid β-oxidation system, is synthesized as a 75-kDa polypeptide (component A) and is proteolytically converted into 53- and 22-kDa polypeptides components (B and C, respectively) in peroxisomes (24, 25). All three polypeptide components were evident in CHO-K1 cells (Fig. 3B, lane 1), but only component A was seen in ZP109, and in a much smaller amount, probably due to rapid degradation (57) (lane 2). The three components of acyl-CoA oxidase were found in 109P3, as in CHO-K1 cells (Fig. 3B, lane 3). Peroxisomal 3-ketoacyl-CoA thiolase, the third enzyme of the peroxisomal β-oxidation system, is synthesized as a larger precursor with an amino-terminal presequence, PTS2 (34, 47), and is converted to a mature form in peroxisomes (16, 23, 53). In wild-type CHO-K1 cells, only the mature thiolase was detected on continuous cell labeling followed by immunoprecipitation and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 3B, lane 4), thereby reflecting rapid processing of the precursor form. In ZP109 cells, only the larger precursor was found (lane 5), implying the absence of processing activity. 109P3 cells showed only the mature form of thiolase (Fig. 3B, lane 6).
The first and second steps of synthesis of ether phospholipids proceed in peroxisomes (58). The activity of the first enzyme, DHAP-ATase, was severely diminished in ZP109 mutant cells but was fully restored in 109P3 cells (Table 2). The reason for the nearly twofold-greater activity in 109P3 than in wild-type cells is unknown. P12/UV selection specifically kills peroxisome-deficient mutants, as there is no synthesis of plasmalogen (43, 66), whereas P9OH/UV selection kills wild-type cells which incorporate this fatty alcohol analog into plasmalogen molecules (26, 43). 109P3 cells demonstrated P12/UV-resistant and P9OH/UV-sensitive phenotypes, as was the case for CHO-K1 cells (Table 2).
TABLE 2.
Properties of wild-type CHO-K1, ZP109, and RnPEX12-transfected ZP109 (109P3) cellsa
| Cell line | Peroxisome | Catalase latency (%) | DHAP-ATase (%) | P12/UV (%) | P9OH/UV (%) |
|---|---|---|---|---|---|
| CHO-K1 | + | 64 | 100 | 83 | <0.01 |
| ZP109 | − | 1.4 | 13 | <0.01 | 92 |
| 109P3 | + | 66 | 220 | 73 | <0.01 |
Catalase latency represents peroxisomal catalase, calculated from Fig. 3A as described previously (57). Peroxisomal DHAP-ATase was expressed as a percentage relative to that of wild-type CHO-K1 cells. For determination of P12/UV or P9OH/UV resistance, 200 or 105 cells were inoculated into 60-mm-diameter dishes and selected (43). The numbers of colonies were counted in triplicate experiments and expressed as percentages of that of the unselected control.
Taken together, these results indicate that PEX12 can fully complement the ZP109 mutation.
PEX12 specifically complements fibroblasts from patients with CG-III Zellweger syndrome.
Five CGs of peroxisome-deficient CHO cell mutants, Z24, Z65, ZP92, ZP105, ZP104, and ZP109, were stably transfected with rat PEX12 cDNA. Peroxisome assembly was restored only in one group of CHO mutants, ZP104 and ZP109, confirming that Pex12p is a peroxisome biogenesis factor for this group (Fig. 1e to i; Table 3). ZP104 and ZP109 were found by cell fusion analysis to belong to the same CG as the human CG-III Zellweger syndrome (32). Next, rat PEX12 cDNA was introduced into fibroblasts of 10 human CGs of peroxisomal disease, i.e., CGs A, B, C, D, E, F, and G of Gifu University, Gifu, Japan, and CGs II, III, and VI of the Kennedy-Krieger Institute, Baltimore, Md. As expected, the fibroblasts derived from two CG-III patients (PBD3-01 and PBD3-03) were morphologically restored for peroxisome assembly, as assessed by immunostaining with anti-human catalase antibody (PBD3-01 [Fig. 4A, a and b), but none of the fibroblasts from the other nine CGs was complemented (Table 3). Moreover, fibroblasts from PBD3-01 and PBD3-03 were likewise complemented for peroxisomes by transfection of human PEX12 (33) (PBD3-01 [Fig. 4A, c). These results, together with those described in above, strongly suggest that PEX12 is the causal gene of peroxisome deficiency of CHO mutants ZP104 and ZP109 and of human CG-III disorders.
TABLE 3.
Complementation of CHO mutants and patient fibroblasts by RnPEX12a
| CHO mutant | No. of peroxisome-positive clones | Patient fibroblasts from complementation group(s) | Peroxisome positiveb |
|---|---|---|---|
| A, VIII | − | ||
| B, VII | − | ||
| ZP92 | 0 | C, IV | − |
| D, IX | − | ||
| Z24 | 0 | E, I | − |
| Z65 | 0 | F, X | − |
| ZP105 | 0 | II | − |
| ZP104 | 24 | III | + |
| ZP109 | 20 | III | + |
| VI | − | ||
| G | − |
Peroxisome-deficient CHO mutants of five CGs were transfected with pUcD2Hyg · RnPEX12, and stable transformants were selected with hygromycin B for 5 days. Transfectants were stained with anti-rat catalase antibody. Peroxisome-positive colonies among 30 hygromycin-resistant colonies were counted on day 6 posttransfection. Patient fibroblasts of 10 CGs of peroxisomal diseases, i.e., CGs A, B, C, D, E, F, and G of Gifu University, Gifu, Japan, and CGs II, III (patients PBD3-01 and PBD3-03), and VI of the Kennedy-Krieger Institute, Baltimore, Md., were transfected with pUcD2Hyg · RnPEX12 and examined for peroxisome assembly by immunostaining with anti-human catalase antibody on day 4 posttransfection.
+, complemented; −, not complemented.
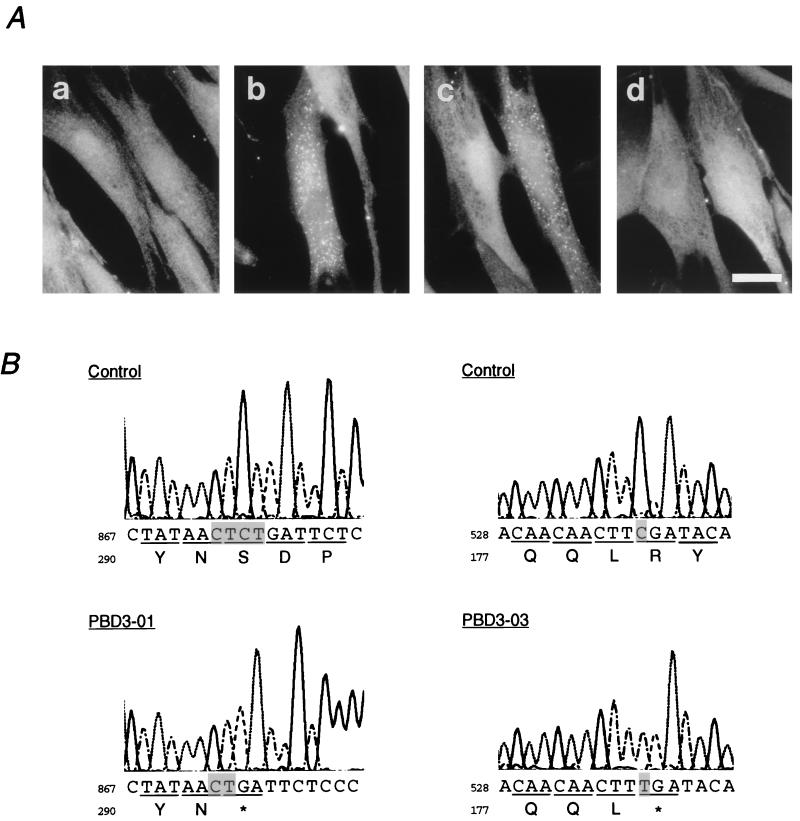
FIG. 4.
Mutation analysis of PEX12 from CG-III patients. (A) Complementation of peroxisome assembly in CG-III fibroblasts. Transfection to fibroblasts from patient PBD3-01 of complementation CG-III (a) was done with rat and human PEX12 (b and c) or a PEX12 isolated from the PBD3-01 fibroblasts (d). Immunofluorescence microscopic analysis was done as for Fig. 1, using anti-human catalase antibody. Numerous peroxisomes were present in the cytoplasm of the transfected cells in panels b and c but not in panel d. Magnification, ×630; bar, 30 μm. (B) Nucleotide sequence analysis of PEX12 isolated from CG-III patients and a control. Partial sequence and deduced amino acid sequence are shown. A two-base deletion in the nucleotide sequence at residues 873 to 876 created a codon 292 (open arrowhead in Fig. 2A) for termination in patient PBD3-01 (left); a point mutation at nucleotide residue 538 changes a codon 180 for Arg (solid arrowhead in Fig. 2A) to a termination codon in a patient (PBD3-03) with Zellweger syndrome (right).
CG-III patient analysis.
To determine the dysfunction of PEX12 in these two patients, we isolated PEX12 cDNA from their fibroblasts by means of reverse transcription-PCR and subsequent cloning. The nucleotide sequences of six independent cDNA clones from these patients were analyzed. In patient PBD3-01, two bases, CT or TC, were deleted in the nucleotide sequence AACTCT at residues 871 to 876, codons for 291Asn and 292Ser, thereby creating an apparently unchanged codon for Asn and a codon 292 for termination (Fig. 4B, left). In patient PBD3-03, nucleotide C at position 538 in a codon 180 for Arg was mutated to T, resulting in creation of termination codon TGA (Fig. 4B, right). All of the six cDNA clones examined from each patient showed the same site mutation, suggesting that the patients were homozygous for the mutation. When PEX12 from the PBD3-01 fibroblasts was transfected back into fibroblasts of this patient, none of the cells was complemented in peroxisomes, indicating dysfunction for patient PEX12, presumably due to a premature termination (Fig. 4A, d). Dysfunction of PEX12 from patient PBD3-03 was likewise assessed by back-transfection to fibroblasts, where all cells were peroxisome negative (data not shown). Moreover, the PBD3-01- and PBD3-03-derived PEX12 could not complement ZP109 (data not shown). These data demonstrate that mutations in the PEX12 gene are responsible for the peroxisome deficiency in CG-III disorders.
Expression of PEX12.
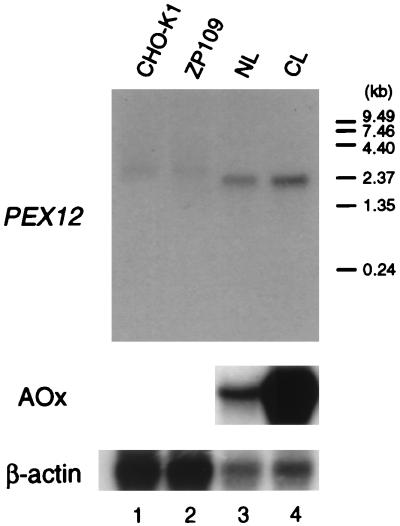
On Northern blotting, PEX12 mRNA was detected as a single band of about 2.3 kb in CHO-K1 and ZP109 cells but at much lower levels and with an apparently larger size compared with those in rat liver [Fig. 5; note that poly(A)+ RNA was loaded on lanes 1 and 2]. The amount and size of PEX12 mRNA were indistinguishable between mutant and wild-type cells, suggesting that the alteration in the PEX12 gene in ZP109 was a point mutation rather than at the transcriptional level. Peroxisomes and peroxisomal proteins are induced in rat liver by administration of hypolipidemic compounds such as clofibrate (15, 39). PEX12 mRNA was not induced in rats by clofibrate (Fig. 5, lanes 3 and 4, upper panel), under which acyl-CoA oxidase mRNA was highly (about 50-fold) induced (middle panel), as estimated by dot blot analysis (data not shown) and consistent with previous observations (39, 56).
FIG. 5.
Northern blot analysis of PEX12 mRNA. RNA was separated, transferred to a Zeta-Probe GT membrane (Bio-Rad), and hybridized with 32P-labeled cDNA probes for rat PEX12 (top) and rat acyl-CoA oxidase (AOx; middle), respectively. Human β-actin (bottom) cDNA was used as a control probe to check the amount of RNA loaded. Washing was done twice with 0.15 M NaCl–10 mM sodium phosphate (pH 7.4)–1 mM EDTA–0.5% SDS at 60°C. Lanes: 1 and 2, poly(A)+ RNA (5 μg) from wild-type CHO-K1 and ZP109 cells, respectively; 3 and 4, total RNA (20 μg) from the livers of a normal (NL) and a clofibrate-treated (CL) rat, respectively. Exposure times were 90 h (top), 28 h (middle), and 24 h (bottom).
Pex12p is localized to peroxisomes.
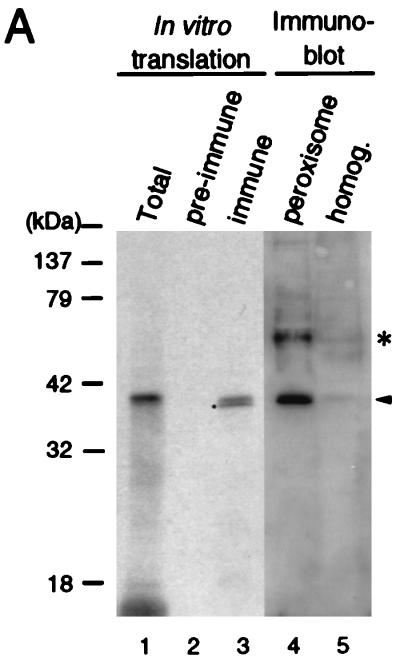
The C-terminal peptide of Pex12p (residues 339 to 359) was used to raise rabbit antibodies. This antibody reacted with only a single protein of the predicted size in immunoblots of purified peroxisomes and homogenates of rat liver, suggesting localization of Pex12p in peroxisomes (Fig. 6A, lanes 4 and 5). The size of Pex12p was exactly the same as that of Pex12p synthesized in vitro by coupled transcription-translation, which was specifically immunoprecipitated by anti-Pex12p antibody (lanes 1 to 3), thereby indicating that a cloned PEX12 cDNA encodes bona fide Pex12p. This result implies the synthesis of Pex12p at its final size, consistent with a general rule for peroxisomal proteins (21).
FIG. 6.
Intracellular localization of Pex12p. (A) Size comparison of rat liver Pex12p and in vitro transcription-translation product of Pex12p cDNA. The [35S]methionine-labeled in vitro transcription-translation product of rat PEX12 and rat liver fractions were subjected to SDS-PAGE and transferred to a polyvinylidene difluoride membrane. The autoradiograph (lanes 1 to 3) was exposed for 3 days; immunodetection (lanes 4 and 5) was done with rabbit anti-Pex12p peptide antibody. Lanes: 1, in vitro transcription-translation product (1 μl) of PEX12 cDNA; 2 and 3, immunoprecipitation of 35S-Pex12p (1 μl) with preimmune and anti-Pex12p immune sera, respectively; 4, peroxisomes (20 μg); 5, liver homogenates (50 μg). The dot indicates an apparently cleaved product of 35S-Pex12p during the immunoprecipitation; an unidentified band is shown by an asterisk. (B) Morphological analysis. Myc-tagged rat Pex12p was expressed in CHO-K1, Z65, and ZP92 cells. ZP109 was transfected with rat PEX12. (a and b) CHO-K1 cells stained with anti-Myc antibody and anti-PTS1 peptide antibody, respectively; (c) merged view of panels a and b; (d and e) ZP109 cells stained with antisera against Pex12p and catalase, respectively; (f and g) Z65 cells stained with antibodies to Myc and PMP70, respectively; (h and i) ZP92 cells stained with antibodies to Myc and PMP70, respectively. Magnification, ×500; bar, 20 μm.
The intracellular localization of Pex12p was analyzed by immunofluorescence microscopy, using rat Pex12p tagged with Myc at its C terminus. The Pex12p-Myc fully restored peroxisome assembly in ZP109 as efficiently as PEX12, indicating that the C-terminal tagging did not interfere with PEX12 function (data not shown). In wild-type CHO-K1 cells transfected with PEX12-myc, Pex12p was detected in a punctate staining pattern, using anti-Myc antibody (Fig. 6B, a). The pattern was superimposable on that obtained by using anti-PTS1 antibody, suggesting that Pex12p-Myc was targeted to peroxisomes (Fig. 6B, b and c). Similar results were obtained when rat Pex12p tagged with flag at the N terminus was expressed in CHO-K1 (see below). Endogenous Pex12p was not detectable in CHO-K1, due to a lower level of Pex12p expression, as found by Northern analysis (Fig. 5). The possibility that Chinese hamster Pex12p may not be recognized by anti-rat Pex12p peptide antibody would need to be excluded (data not shown). When rat PEX12 was expressed in ZP109, peroxisomes were restored and Pex12p was also found in the same vesicular structures as those detected with anticatalase antibody (Fig. 6B, d and e). Moreover, essentially the same results were obtained when PEX12-myc was expressed in ZP109 (see below). These results were interpreted to mean that flag- and myc-tagged Pex12p and Pex12p were translocated to peroxisomes. It is noteworthy that Pex12p-myc was colocalized with PMP70 in peroxisome-like ghosts in other CGs of CHO cell mutants, Pex2p-defective Z65 (54) and Pex6p-defective ZP92 (55) (Fig. 6B, f to i). Translocation of peroxisomal membrane proteins, including Pex12p, does not seem to be impaired in these mutant cells. A subcellular fractionation study using rat liver indicated that Pex12p is a peroxisomal integral membrane protein (33), consistent with the presence of two putative transmembrane segments deduced by hydropathy analysis (Fig. 2A).
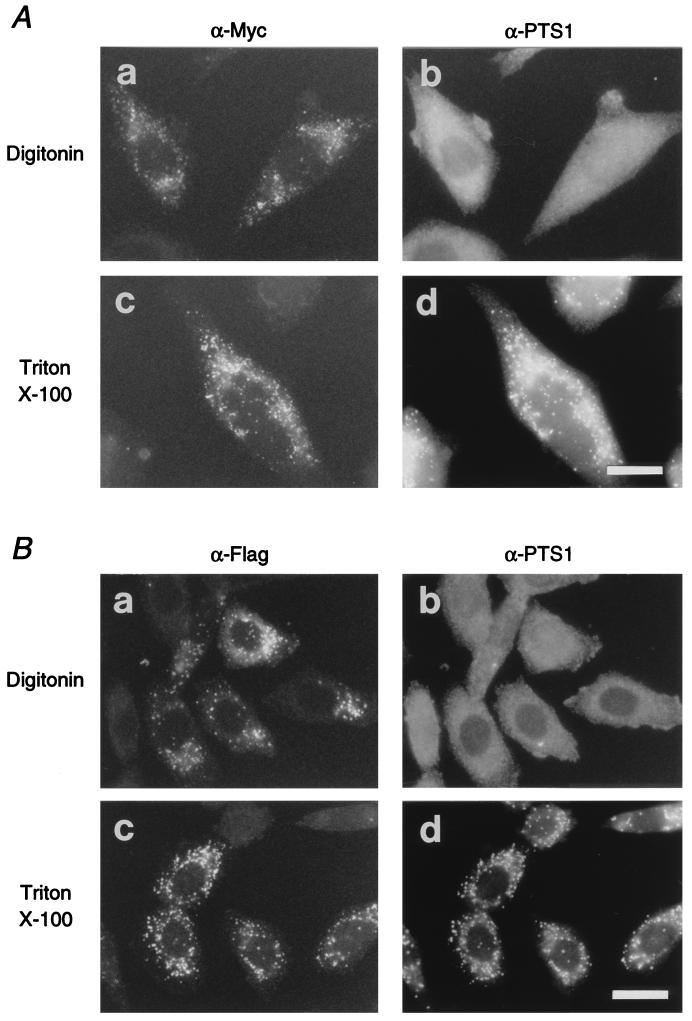
Pex12p is exposed to cytosol.
Membrane topology of Pex12p was determined by a differential permeabilization procedure whereby Pex12p was detected with antibodies to epitope tags Myc and flag. The wild-type CHO-K1 cells expressing rat Pex12p-Myc were permeabilized with 25 μg of digitonin per ml, under which plasma membranes are selectively permeabilized and intraperoxisomal proteins are inaccessible to exogenous antibodies (28). Pex12p-Myc was observed in a punctate staining pattern, whereas there was almost no staining of cells with anti-PTS1 antibody (Fig. 7A, a and b). When the cells were treated with Triton X-100, which solubilizes all cellular membranes, both PTS1 proteins and Pex12p-Myc were detected in particulates, presumably peroxisomes, similar in number (Fig. 7A, c and d). Similar results were obtained in studies of rat flag-Pex12p-expressing CHO-K1 cells, using antibodies to flag tag and PTS1 peptide (Fig. 7B). Taken together, these results strongly suggest that the both N- and C-terminal parts of Pex12p are exposed to the cytosol, presumably anchored by two membrane-spanning segments (Fig. 2A). Studies of flag-Pex12p- or Pex12p-Myc-transfected ZP109 cells, either with a set of antibodies to the flag and PTS1 or with a set of anti-Myc and -PTS1 antibodies, gave essentially the same results as found for CHO-K1 cells (not shown). This finding confirms the peroxisomal localization and topology of Pex12p.
FIG. 7.
Topology of Pex12p. (A, a and b), CHO-K1 cells transfected with rat PEX12-myc were treated with 25 μg of digitonin per ml, under which the plasma membrane was permeabilized (28). (c and d) PEX12-myc-transfected CHO-K1 cells were treated with 1% Triton X-100. Cells were stained with anti-Myc antibody (a and c) and anti-PTS1 peptide antibody (b and d). Note that Pex12p-Myc was detected after both types of treatments. Magnification, ×630; bar, 20 μm. (B) CHO-K1 cells transfected with N-terminally flag-tagged rat PEX12 were treated with 25 μg of digitonin per ml (a and b) or with 1% Triton X-100 (c and d). Cells were stained with rabbit anti-flag antibody (a and c) and anti-PTS1 antibody (b and d). Magnification, ×630; bar, 20 μm.
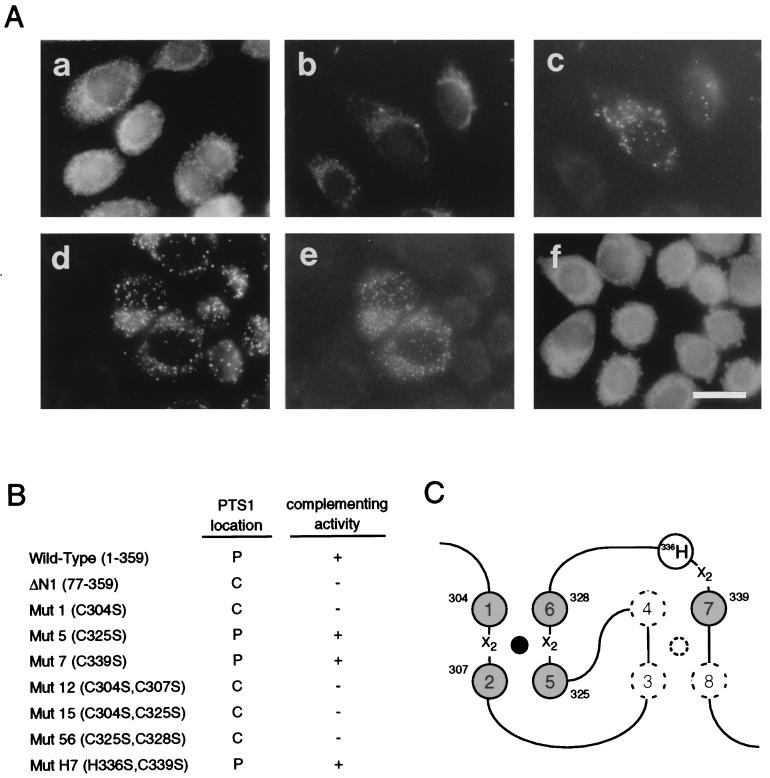
N-terminal deletion and RING finger mutagenesis.
A Pex12p mutant, ΔN1, truncated from the N terminus to residue 76 was defective in restoring peroxisomes in ZP109, suggesting that the N-terminal part was also important for the function of Pex12p (Fig. 8A, a). To determine which residue(s) of five cysteines in a putative RING finger motif is essential for the biological activity of Pex12p, ZP109 cells were transfected with RING mutant PEX12, in which the conserved cysteines had been mutated to serine. ZP109 was restored in PTS1 protein import by mutant PEX12 with a mutation C339S (mut7), whereas peroxisomes were not complemented by mutant PEX12 with C304S (mut1) or two-site mutation C325S/C328S (mut56) (Fig. 8A, b, d, and f). Mutant forms C304S-Pex12p (mut1) and C339S-Pex12p (mut7) were expressed when transfected into ZP109, as assessed by cell staining using anti-flag antibody (Fig. 8A, c and e; note that only the cells highly expressing epitope-tagged Pex12p were discernible). C304S-Pex12p was colocalized with PMP70 in ZP109, indicating that Pex12p is targeted to peroxisomal ghosts (data not shown). Colocalization of C304S-Pex12p and C339S-Pex12p each with PMP70 in peroxisomes was likewise noted upon expression in CHO-K1 cells (data not shown). RING mutations at C304S/C307S (mut12) or C304S/C325S (mut15) abolished complementing activity (Fig. 8B). On the other hand, Pex12p mutants with C325S (mut5), C339S (mut7), or C339S plus nonconserved H335S (mutH7) were functional in restoring peroxisomes. Collectively, RING residues C1 (at 304C), C2 (307C), C5 (325C), and C6 (328C) appear to be required for the biological activity, apparently by forming a zinc finger, although mut5 has no effect on the activity (Fig. 8B and C). We cannot exclude the possibility that the expression level of each RING mutant varies, even though all variants were constructed in the same expression vector.
FIG. 8.
N-terminal deletion and RING finger mutagenesis of Pex12p. (A) ΔN1, an N-terminally flag-tagged Pex12p variant lacking amino acid residues from the N terminus to position 76. Conserved cysteine residues of the RING finger were mutated to serine: C304S (mut1), C339S (mut7), and C325S/C328S (mut56). mut1 and mut7 were epitope tagged with flag and Myc at the N and C termini, respectively; mut56 was C-terminally tagged with Myc. ZP109 cells were transfected by PEX12 mutant ΔN1 (a), mut1 (b and c), mut7 (d and e), or mut56 (f). Cells were stained with antibodies to PTS1 (a, b, d, and f) and flag (c and e). Magnification, ×630; bar, 20 μm. (B) Complementing activity of Pex12p mutants. ZP109 was complemented (+) or not complemented (−), as assessed by PTS1 protein import. (C) Schematic view of the RING finger. One zinc finger is formed in Pex12p by four cysteine residues, C1 (at 304C), C2 (307C), C5 (325C), and C6 (328C), corresponding to those in authentic RING, C3HC4 (Fig. 2B). Solid circle, a divalent zinc ion; X2, two amino acids.
DISCUSSION
In the present work, we isolated a rat Pex12p cDNA by functional complementation of peroxisome-deficient CHO cell mutant ZP109. ZP109 cells were restored for peroxisome assembly by both morphological and biochemical criteria. Complementation of peroxisome deficiency by rat PEX12 transfection only in CG-III fibroblasts strongly suggested that PEX12 is the causal gene of this group, compatible with our recent findings that human PEX12 expression restored peroxisome assembly in ZP109 (33). We delineated the mutation sites in the PEX12 gene in CG-III patients: a one-base transition mutation and a two-base deletion. The findings by back-transfection of these mutated forms of PEX12 into patient fibroblasts and CHO mutant ZP109 confirmed that PEX12 is the causal gene in peroxisome biogenesis disorders of CG-III. Recently, Chang et al. (4) isolated human PEX12 by a homology search using yeast PEX12 on an expressed sequence tag database and showed its ability to complement peroxisome deficiency of complementation CG-III disorder. PEX12 is the fifth gene thus far identified as being responsible for peroxisome deficiency diseases.
Rat and the very recently identified human (4, 33) Pex12 proteins contain RING fingers with only five conserved cysteines of the motif C3HC4 (42), very similar to that found in Pichia pastoris Pex12p (18) (Fig. 2). Four cysteine residues, at positions 304, 307, 325, and 328, of the five are likely to bind a zinc ion and form a finger, as noted for P. pastoris Pex12p (18), based on the site mutation study in the present work and the three-dimensional structure of RING, although the full motif, C3HC4, can bind two zinc molecules (42). Whether one zinc finger and two of RING have distinct functions in peroxisome biogenesis such as those in interactions with other proteins may be investigated by using presently available peroxisome-defective cell mutants and their complementing RING finger proteins, Pex2p (1, 8, 44, 54, 56, 59), Pex10p (19, 49), Pex12p (4, 18, 33), and Saccharomyces cerevisiae PAS4 and PAS5 (17). Moreover, it is noteworthy that Pex12p was terminated at alignment position 292 in patient PBD3-01, including two putative transmembrane segments but excluding a RING finger. This finding as well as those obtained in the site mutation study imply that a RING-containing C-terminal region exposed to the cytosol, comprising the residues from alignment position 292 to the C terminus, plays an indispensable role in Pex12p function. The functional significance of the cytoplasmically exposed N-terminal part of Pex12p is also inferred from a truncated mutant, ΔN1, lacking amino acid residues 1 to 76. Delineation of the minimal sequence required for Pex12p function can be readily done by deletion analysis. It would be also intriguing to investigate structural and functional aspects of Pex12p in relation to the clinical phenotype.
We have shown previously (54, 55) and in this work that a cloning strategy using a mammalian expression vector is highly efficient in delineating peroxisome biogenesis factors in mammals. We introduced GFP to further accelerate the screening process for the cDNA clone that complements mutant cells in peroxisome assembly. We detected such cells without treatment with fixatives such as formaldehyde. Expression of GFP is also applicable to dual staining with immunoflorescence methods, as shown in Fig. 1. We have recently isolated two newly identified peroxisome-deficient CHO mutants distinct from any of 10 human CGs (50). With possession of these mutants in addition to our earlier mutant Z24 and a modified, efficient functional complementation assay described in this report, cDNA cloning of other mammalian peroxisome assembly factors can be readily done.
Generally, isolation of mutants and gene cloning by genetic complementation are achieved much more readily with yeast than with mammalian cells. Human PEX cDNAs such as PEX5 (7), PEX6 (63), and PEX1 (36, 40), restoring peroxisome biogenesis in CG-II, CG-IV, and CG-I disorders, respectively, were recently identified by finding in a DNA database a human expressed sequence tag sequence related to yeast PEX5, PEX6, and PEX1. However, it seems generally difficult to clone human counterparts of yeast genes, particularly those expressed at lower levels, by such a homology-based method. Our strategy involves use of a more direct, functional complementation assay and mammalian somatic cell mutants (references 13, 44, 48, 54, and 55 and this study).
A peroxisomal protein import assay using permeabilized human fibroblasts demonstrated that the molecular defect of CG-III fibroblasts was associated with the organelle while cytoplasmic factors seemed to be functionally normal (60). These observations agree well with the findings that Pex12p is located in the peroxisomal membrane (references 4 and 33 and this study) as an integral membrane protein (33). While peroxisomal transport of membrane proteins, including PMP70, appears to be normal in ZP104 and ZP109, import of soluble proteins such as PTS1 and PTS2 proteins is defective, consistent with the finding in fibroblasts from CG-III patients (45). Pex12p may function in peroxisome assembly processes as a component of the matrix protein import machinery. We would also want to determine if Pex12p interacts with other peroxisome biogenesis factors, such as Pex1p (36, 40, 48), Pex2p (44, 54, 56), Pex5p (PTS1 receptor) (7, 9, 61), Pex6p (13, 55, 63), Pex7p (PTS2 receptor) (2, 29, 38), and Pex13p (PTS1 receptor docking protein) (14).
ACKNOWLEDGMENTS
We thank T. Hashimoto for anti-human catalase antiserum and T. Harano, K. Mizuno, and M. Ohara for comments.
This work was supported in part by a CREST grant (to Y.F.) from the Science and Technology Corporation of Japan; Grants-in-Aid for Scientific Research (07408016, 08249232, and 08557011 to Y.F.) from the Ministry of Education, Science, Sports and Culture; and grants (to Y.F.) from the Mitsubishi Foundation, the Uehara Foundation, and the Nagase Science and Technology Foundation.
REFERENCES
- 1.Berteaux-Lecellier V, Picard M, Thompson-Coffe C, Zickler D, Panvier-Adoutte A, Simonet J-M. A nonmammalian homolog of the PAF1 gene (Zellweger syndrome) discovered as a gene involved in caryogamy in the fungus Podospora anserina. Cell. 1995;81:1043–1051. doi: 10.1016/s0092-8674(05)80009-1. [DOI] [PubMed] [Google Scholar]
- 2.Braverman N, Steel G, Obie C, Moser A, Moser H, Gould S J, Valle D. Human PEX7 encodes the peroxisomal PTS2 receptor and is responsible for rhizomelic chondrodysplasia punctata. Nat Genet. 1997;15:369–376. doi: 10.1038/ng0497-369. [DOI] [PubMed] [Google Scholar]
- 3.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science (Washington, DC) 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 4.Chang C-C, Lee W-H, Moser H, Valle D, Gould S J. Isolation of the human PEX12 gene, mutated in group 3 of the peroxisome biogenesis disorders. Nat Genet. 1997;15:385–388. doi: 10.1038/ng0497-385. [DOI] [PubMed] [Google Scholar]
- 5.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-PhOH-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 6.Distel B, Erdmann R, Gould S J, Blobel G, Crane D I, Cregg J M, Dodt G, Fujiki Y, Goodman J M, Just W W, Kiel J A K W, Kunau W-H, Lazarow P B, Mannaerts G P, Moser H, Osumi T, Rachubinski R A, Roscher A, Subramani S, Tabak H F, Tsukamoto T, Valle D, van der Klei I, van Veldhoven P P, Veenhuis M. A unified nomenclature for peroxisome biogenesis factors. J Cell Biol. 1996;135:1–3. doi: 10.1083/jcb.135.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dodt G, Braverman N, Wong C S, Moser A, Moser H W, Watkins P, Valle D, Gould S J. Mutations in the PTS1 receptor gene, PXR1, define complementation group 2 of the peroxisome biogenesis disorders. Nat Genet. 1995;9:115–125. doi: 10.1038/ng0295-115. [DOI] [PubMed] [Google Scholar]
- 8.Eitzen G A, Titorenko V I, Smith J J, Veenhuis M, Szilard R K, Rachubinski R A. The Yarrowia lipolytica gene PAY5 encodes a peroxisomal integral membrane protein homologous to the mammalian peroxisome assembly factor PAF-1. J Biol Chem. 1996;271:20300–20306. doi: 10.1074/jbc.271.34.20300. [DOI] [PubMed] [Google Scholar]
- 9.Fransen M, Brees C, Baumgart E, Vanhooren J C, Baes M, Mannaerts G P, Veldhoven P P V. Identification and characterization of the putative human peroxisomal C-terminal targeting signal import receptor. J Biol Chem. 1995;270:7731–7736. doi: 10.1074/jbc.270.13.7731. [DOI] [PubMed] [Google Scholar]
- 10.Fujiki Y. Approaches to studies on peroxisome biogenesis and human peroxisome-deficient disorders. Ann N Y Acad Sci. 1996;804:491–501. doi: 10.1111/j.1749-6632.1996.tb18639.x. [DOI] [PubMed] [Google Scholar]
- 11.Fujiki Y. Molecular defects in genetic diseases of peroxisomes. Biochim Biophys Acta. 1997;1361:235–250. doi: 10.1016/s0925-4439(97)00051-3. [DOI] [PubMed] [Google Scholar]
- 12.Fujiki Y, Rachubunski R A, Lazarow P B. Synthesis of a major integral membrane polypeptide of rat liver peroxisomes on free polysomes. Proc Natl Acad Sci USA. 1984;81:7127–7131. doi: 10.1073/pnas.81.22.7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukuda S, Shimozawa N, Suzuki Y, Tomatsu S, Tsukamoto T, Hashiguchi N, Osumi T, Masuno M, Imaizumi K, Kuroki Y, Fujiki Y, Orii T, Kondo N. Human peroxisome assembly factor-2 (human PAF-2): a gene responsible for group C peroxisome biogenesis disorder in humans. Am J Hum Genet. 1996;59:1210–1220. [PMC free article] [PubMed] [Google Scholar]
- 14.Gould S J, Kalish J E, Morrell J C, Bjorkman J, Urquhart A J, Crane D I. Pex13p is an SH3 protein of the peroxisome membrane and a docking factor for the predominantly cytoplasmic PTS1 receptor. J Cell Biol. 1996;135:85–95. doi: 10.1083/jcb.135.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hess R, Staubli W, Reiss W. Nature of the hepatomegalic effect produced by ethyl-chlorophenoxyisobutyrate in the rat. Nature. 1965;208:856–858. doi: 10.1038/208856a0. [DOI] [PubMed] [Google Scholar]
- 16.Hijikata M, Ishii N, Kagamiyama H, Osumi T, Hashimoto T. Structural analysis of cDNA for rat peroxisomal 3-ketoacyl-CoA thiolase. J Biol Chem. 1987;262:8151–8158. [PubMed] [Google Scholar]
- 17.Hoehfeld J, Mertens D, Wiebel F F, Kunau W-H. Defining components required for peroxisome assembly in Saccharomyces cerevisiae. In: Neupert W, Lill R, editors. Membrane biogenesis and protein targeting. Amsterdam, The Netherlands: Elsevier Science Publishers B.V.; 1992. pp. 185–207. [Google Scholar]
- 18.Kalish J E, Keller G A, Morrell J C, Mihalik S J, Smith B, Cregg J M, Gould S J. Characterization of a novel component of the peroxisomal protein import apparatus using fluorescent peroxisomal proteins. EMBO J. 1996;15:3275–3285. [PMC free article] [PubMed] [Google Scholar]
- 19.Kalish J E, Theda C, Morrell J C, Berg J M, Gould S J. Formation of the peroxisome lumen is abolished by loss of Pichia pastoris Pas7p, a zinc-binding integral membrane protein of the peroxisome. Mol Cell Biol. 1995;15:6406–6419. doi: 10.1128/mcb.15.11.6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunau W-H, Beyer A, Franken T, Goette K, Marzioch M, Saidowsky J, Skaletz-Rorowski A, Wiebel F F. Two complementary approaches to study peroxisome biogenesis in Saccharomyces cerevisiae: forward and reversed genetics. Biochimie. 1993;75:209–224. doi: 10.1016/0300-9084(93)90079-8. [DOI] [PubMed] [Google Scholar]
- 21.Lazarow P B, Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- 22.Lazarow P B, Moser H W. Disorders of peroxisome biogenesis. In: Scriver C R, Beaudet A I, Sly W S, Valle D, editors. The metabolic basis of inherited disease. 7th ed. New York, N.Y: McGraw-Hill; 1995. pp. 2287–2324. [Google Scholar]
- 23.Miura S, Miyazawa S, Osumi T, Hashimoto T, Fujiki Y. Post-translational import of 3-ketoacyl-CoA thiolase into rat liver peroxisomes in vitro. J Biochem. 1994;115:1064–1068. doi: 10.1093/oxfordjournals.jbchem.a124458. [DOI] [PubMed] [Google Scholar]
- 24.Miyazawa S, Hayashi H, Hijikata M, Ishii N, Furuta S, Kagamiyama H, Osumi T, Hashimoto T. Complete nucleotide sequence of cDNA and predicted amino acid sequence of rat acyl-CoA oxidase. J Biol Chem. 1987;262:8131–8137. [PubMed] [Google Scholar]
- 25.Miyazawa S, Osumi T, Hashimoto T, Ohno K, Miura S, Fujiki Y. Peroxisome targeting signal of rat liver acyl-coenzyme A oxidase resides at the carboxy terminus. Mol Cell Biol. 1989;9:83–91. doi: 10.1128/mcb.9.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morand O H, Allen L-A H, Zoeller R A, Raetz C R H. A rapid selection for animal cell mutants with defective peroxisomes. Biochim Biophys Acta. 1990;1034:132–141. doi: 10.1016/0304-4165(90)90066-6. [DOI] [PubMed] [Google Scholar]
- 27.Moser A B, Rasmussen M, Naidu S, Watkins P A, McGuiness M, Hajra A K, Chen G, Raymond G, Liu A, Gordon D, Garnaas K, Walton D S, Skjeldal O H, Guggenheim M A, Jackson L G, Elias E R, Moser H W. Phenotype of patients with peroxisomal disorders subdivided into sixteen complementation groups. J Pediatr. 1995;127:13–22. doi: 10.1016/s0022-3476(95)70250-4. [DOI] [PubMed] [Google Scholar]
- 28.Motley A, Hettema E, Distel B, Tabak H. Differential protein import deficiencies in human peroxisome assembly disorders. J Cell Biol. 1994;125:755–767. doi: 10.1083/jcb.125.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Motley A M, Hettema E H, Hogenhout E M, Brites P, ten Asbroek A L M A, Wijburg F A, Baas F, Heijmans H S, Tabak H F, Wanders R J A, Distel B. Rhizomelic chondrodysplasia punctata is a peroxisomal protein targeting disease caused by a non-functional PTS2 receptor. Nat Genet. 1997;15:377–380. doi: 10.1038/ng0497-377. [DOI] [PubMed] [Google Scholar]
- 30.Niman H L, Houghten R A, Walker L E, Reisfeld R A, Wilson I A, Hogle J M, Lerner R A. Generation of protein-reactive antibodies by short peptides is an event of high frequency: implications for the structural basis of immune recognition. Proc Natl Acad Sci USA. 1983;80:4949–4953. doi: 10.1073/pnas.80.16.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okamoto H, Suzuki Y, Shimozawa N, Yajima S, Masuno M, Orii T. Transformation and characterization of mutant human fibroblasts defective in peroxisome assembly. Exp Cell Res. 1992;201:307–312. doi: 10.1016/0014-4827(92)90278-g. [DOI] [PubMed] [Google Scholar]
- 32.Okumoto K, Bogaki A, Tateishi K, Tsukamoto T, Osumi T, Shimozawa N, Suzuki Y, Orii T, Fujiki Y. Isolation and characterization of peroxisome-deficient Chinese hamster ovary cell mutants representing human complementation group III. Exp Cell Res. 1997;233:11–20. doi: 10.1006/excr.1997.3552. [DOI] [PubMed] [Google Scholar]
- 33.Okumoto K, Fujiki Y. PEX12 encodes an integral membrane protein of peroxisomes. Nat Genet. 1997;17:265–266. doi: 10.1038/ng1197-265. [DOI] [PubMed] [Google Scholar]
- 34.Osumi T, Tsukamoto T, Hata S, Yokota S, Miura S, Fujiki Y, Hijikata M, Miyazawa S, Hashimoto T. Amino-terminal presequence of the precursor of peroxisomal 3-ketoacyl-CoA thiolase is a cleavable signal peptide for peroxisomal targeting. Biochem Biophys Res Commun. 1991;181:947–954. doi: 10.1016/0006-291x(91)92028-i. [DOI] [PubMed] [Google Scholar]
- 35.Otera H, Tateishi K, Okumoto K, Ikoma Y, Matsuda E, Nishimura M, Tsukamoto T, Osumi T, Ohashi K, Higuchi O, Fujiki Y. Peroxisome targeting signal type 1 (PTS1) receptor is involved in import of both PTS1 and PTS2: studies with PEX5-defective CHO cell mutants. Mol Cell Biol. 1998;18:388–399. doi: 10.1128/mcb.18.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Portsteffen H, Beyer A, Becker E, Epplen C, Pawlak A, Kunau W-H, Dodt G. Human PEX1 is mutated in complementation group 1 of the peroxisome biogenesis disorders. Nat Genet. 1997;17:449–452. doi: 10.1038/ng1297-449. [DOI] [PubMed] [Google Scholar]
- 37.Poulos A, Christodoulou J, Chow C W, Goldblatt J, Paton B C, Orii T, Suzuki Y, Shimozawa N. Peroxisomal assembly defects: clinical, pathologic, and biochemical findings in two patients in a newly identified complementation group. J Pediatr. 1995;127:596–599. doi: 10.1016/s0022-3476(95)70121-4. [DOI] [PubMed] [Google Scholar]
- 38.Purdue P E, Zhang J W, Skoneczny M, Lazarow P B. Rhizomelic chondrodysplasia punctata is caused by deficiency of human PEX7, a homologue of the yeast PTS2 receptor. Nat Genet. 1997;15:381–384. doi: 10.1038/ng0497-381. [DOI] [PubMed] [Google Scholar]
- 39.Rachubinski R A, Fujiki Y, Mortensen R M, Lazarow P B. Acyl-CoA oxidase and hydratase-dehydrogenase, two enzymes of the peroxisomal β-oxidation system, are synthesized on free polysomes of clofibrate-treated rat liver. J Cell Biol. 1984;99:2241–2246. doi: 10.1083/jcb.99.6.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reuber B E, Germain-Lee E, Collins C S, Morrell J C, Ameritunga R, Moser H W, Valle D, Gould S J. Mutations in PEX1 are the most common cause of peroxisome biogenesis disorders. Nat Genet. 1997;17:445–448. doi: 10.1038/ng1297-445. [DOI] [PubMed] [Google Scholar]
- 41.Santos M J, Hoefler S, Moser A B, Moser H W, Lazarow P B. Peroxisome assembly mutations in humans: structural heterogeneity in Zellweger syndrome. J Cell Physiol. 1992;151:103–112. doi: 10.1002/jcp.1041510115. [DOI] [PubMed] [Google Scholar]
- 42.Saurin A J, Borden K L B, Boddy M N, Freemont P S. Does this have a familiar RING? Trends Biochem Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- 43.Shimozawa N, Tsukamoto T, Suzuki Y, Orii T, Fujiki Y. Animal cell mutants represent two complementation groups of peroxisome-defective Zellweger syndrome. J Clin Invest. 1992;90:1864–1870. doi: 10.1172/JCI116063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimozawa N, Tsukamoto T, Suzuki Y, Orii T, Shirayoshi Y, Mori T, Fujiki Y. A human gene responsible for Zellweger syndrome that affects peroxisome assembly. Science (Washington, DC) 1992;255:1132–1134. doi: 10.1126/science.1546315. [DOI] [PubMed] [Google Scholar]
- 44a.Shimozawa, N., et al. Unpublished data.
- 45.Slawecki M L, Dodt G, Steinberg S, Moser A B, Moser H W, Gould S J. Identification of three distinct peroxisomal protein import defects in patients with peroxisome biogenesis disorders. J Cell Sci. 1995;108:1817–1829. doi: 10.1242/jcs.108.5.1817. [DOI] [PubMed] [Google Scholar]
- 46.Subramani S. Protein import into peroxisomes and biogenesis of the organelle. Annu Rev Cell Biol. 1993;9:445–478. doi: 10.1146/annurev.cb.09.110193.002305. [DOI] [PubMed] [Google Scholar]
- 47.Swinkels B W, Gould S J, Bodnar A G, Rachubinski R A, Subramani S. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 1991;10:3255–3262. doi: 10.1002/j.1460-2075.1991.tb04889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamura S, Okumoto K, Toyama R, Shimozawa N, Tsukamoto T, Suzuki Y, Osumi T, Kondo N, Fujiki Y. Human PEX1 cloned by functional complementation on a CHO cell mutant is responsible for peroxisome-deficient Zellweger syndrome of complementation group I. Proc Natl Acad Sci USA. 1998;95:4350–4355. doi: 10.1073/pnas.95.8.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan X, Waterham H R, Veenhuis M, Cregg J M. The Hansenula polymorpha PER8 gene encodes a novel peroxisomal integral membrane protein involved in proliferation. J Cell Biol. 1995;128:307–319. doi: 10.1083/jcb.128.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tateishi K, Okumoto K, Shimozawa N, Tsukamoto T, Osumi T, Suzuki Y, Kondo N, Okano I, Fujiki Y. Newly identified Chinese hamster ovary cell mutants defective in peroxisome biogenesis represent two novel complementation groups in mammals. Eur J Cell Biol. 1997;73:352–359. [PubMed] [Google Scholar]
- 51.Thieringer R, Raetz C R H. Peroxisome-deficient Chinese hamster ovary cells with point mutations in peroxisome assembly factor-1. J Biol Chem. 1993;268:12631–12636. [PubMed] [Google Scholar]
- 52.Tsukamoto T, Bogaki A, Okumoto K, Tateishi K, Fujiki Y, Shimozawa N, Suzuki Y, Kondo N, Osumi T. Isolation of a new peroxisome deficient CHO cell mutant defective in peroxisome targeting signal-1 receptor. Biochem Biophys Res Commun. 1997;230:402–406. doi: 10.1006/bbrc.1996.5971. [DOI] [PubMed] [Google Scholar]
- 53.Tsukamoto T, Hata S, Yokota S, Miura S, Fujiki Y, Hijikata M, Miyazawa S, Hashimoto T, Osumi T. Characterization of the signal peptide at the amino terminus of the rat peroxisomal 3-ketoacyl-CoA thiolase precursor. J Biol Chem. 1994;269:6001–6010. [PubMed] [Google Scholar]
- 54.Tsukamoto T, Miura S, Fujiki Y. Restoration by a 35K membrane protein of peroxisome assembly in a peroxisome-deficient mammalian cell mutant. Nature. 1991;350:77–81. doi: 10.1038/350077a0. [DOI] [PubMed] [Google Scholar]
- 55.Tsukamoto T, Miura S, Nakai T, Yokota S, Shimozawa N, Suzuki Y, Orii T, Fujiki Y, Sakai F, Bogaki A, Yasumo H, Osumi T. Peroxisome assembly factor-2, a putative ATPase cloned by functional complementation on a peroxisome-deficient mammalian cell mutant. Nat Genet. 1995;11:395–401. doi: 10.1038/ng1295-395. [DOI] [PubMed] [Google Scholar]
- 56.Tsukamoto T, Shimozawa N, Fujiki Y. Peroxisome assembly factor 1: nonsense mutation in a peroxisome-deficient Chinese hamster ovary cell mutant and deletion analysis. Mol Cell Biol. 1994;14:5458–5465. doi: 10.1128/mcb.14.8.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsukamoto T, Yokota S, Fujiki Y. Isolation and characterization of Chinese hamster ovary cell mutants defective in assembly of peroxisomes. J Cell Biol. 1990;110:651–660. doi: 10.1083/jcb.110.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van den Bosch H, Schutgens R B H, Wanders R J A, Tager J M. Biochemistry of peroxisomes. Annu Rev Biochem. 1992;61:157–197. doi: 10.1146/annurev.bi.61.070192.001105. [DOI] [PubMed] [Google Scholar]
- 59.Waterham H R, de Vries Y, Russel K A, Xie W, Vennhuis M, Cregg J M. The Pichia pastoris PER6 gene product is a peroxisomal integral membrane protein essential for peroxisome biogenesis and has sequence similarity to the Zellweger syndrome protein PAF-1. Mol Cell Biol. 1996;16:2527–2536. doi: 10.1128/mcb.16.5.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wendland M, Subramani S. Presence of cytoplasmic factors functional in peroxisomal protein import implicates organelle-associated defects in several human peroxisomal disorders. J Clin Invest. 1993;92:2462–2468. doi: 10.1172/JCI116854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wiemer E A, Nuttley W M, Bertolaet B L, Li X, Francke U, Wheelock M J, Anne U K, Johnson K R, Subramani S. Human peroxisomal targeting signal-1 receptor restores peroxisomal protein import in cells from patients with fatal peroxisomal disorders. J Cell Biol. 1995;130:51–65. doi: 10.1083/jcb.130.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wiemer E A C, Brul S, Just W W, van Driel R, Brouwer-Kelder E, van den Berg M, Weijers P J, Schutgens R B H, van den Bosch H, Schram A, Wanders R J A, Tager J M. Presence of peroxisomal membrane proteins in liver and fibroblasts from patients with the Zellweger syndrome and related disorders: evidence for the existence of peroxisomal ghosts. Eur J Cell Biol. 1989;50:407–417. [PubMed] [Google Scholar]
- 63.Yahraus T, Braverman N, Dodt G, Kalish J E, Morrell J C, Moser H W, Valle D, Gould S J. The peroxisome biogenesis disorder group 4 gene, PXAAA1, encodes a cytoplasmic ATPase required for stability of the PTS1 receptor. EMBO J. 1996;15:2914–2923. [PMC free article] [PubMed] [Google Scholar]
- 64.Yajima S, Suzuki Y, Shimozawa N, Yamaguchi S, Orii T, Fujiki Y, Osumi T, Hashimoto T, Moser H W. Complementation study of peroxisome-deficient disorders by immunofluorescence staining and characterization of fused cells. Hum Genet. 1992;88:491–499. doi: 10.1007/BF00219334. [DOI] [PubMed] [Google Scholar]
- 65.Zoeller R A, Allen L-A H, Santos M J, Lazarow P B, Hashimoto T, Tartakoff A M, Raetz C R H. Chinese hamster ovary cell mutants defective in peroxisome biogenesis. Comparison to Zellweger syndrome. J Biol Chem. 1989;264:21872–21878. [PubMed] [Google Scholar]
- 66.Zoeller R A, Morand O H, Raetz C R H. A possible role for plasmalogens in protecting animal cells against photosensitized killing. J Biol Chem. 1988;263:11590–11596. [PubMed] [Google Scholar]