Abstract
Upon insertion, transposable elements can disrupt or alter gene function in various ways. Transposons moving through a cut-and-paste mechanism are in addition often mutagenic when excising because repair of the empty site seldom restores the original sequence. The characterization of numerous excision events in many eukaryotes indicates that transposon excision from a given site can generate a high degree of DNA sequence and phenotypic variation. Whether such variation is generated randomly remains largely to be determined. To this end, we have exploited a well-characterized system of genetic instability in the fungus Ascobolus immersus to perform an extensive study of excision events. We show that this system, which produces many phenotypically and genetically distinct derivatives, results from the excision of a novel Ds-like transposon, Ascot-1, from the spore color gene b2. A unique set of 48 molecularly distinct excision products were readily identified from a representative sample of excision derivatives. Products varied in their frequency of occurrence over 4 orders of magnitude, yet most showed small palindromic nucleotide additions. Based on these and other observations, compelling evidence was obtained for intermediate hairpin formation during the excision reaction and for strong biases in the subsequent processing steps at the empty site. Factors likely to be involved in these biases suggest new parallels between the excision reaction performed by transposons of the hAT family and V(D)J recombination. An evaluation of the contribution of small palindromic nucleotide additions produced by transposon excision to the spectrum of spontaneous mutations is also presented.
Transposons are ubiquitous components of both prokaryotic and eukaryotic genomes, contributing to structural organization, gene activity, and evolution. Two classes of transposons have been distinguished according to their modes of transposition (12). Class I transposons, or retroelements, move by reverse transcription of their RNA, whereas class II transposons are mobilized by either excision/reinsertion or cointegration. Although class I and class II transposons generate variability primarily by their insertion into host DNA, class II elements that move by a cut-and-paste mechanism often produce additional variability at a given site as a result of the excision footprint left at that site. Understanding how these footprints are formed is therefore essential for the proper evaluation of the full mutational impact of the class II transposons.
The excision reaction can be separated into two steps, double-strand break formation at both ends of the transposon and repair of the gapped molecule. In vivo and in vitro studies performed with bacterial and animal DNA transposons have shown that all transposases examined cleave precisely at the 3′OH ends of the element, which serve for the subsequent strand transfer reaction. On the other hand, the position of the 5′ cleavage required for the excision of the element need not occur precisely at the transposon ends. It varies between transposon types and may not always be fixed for a given transposon species. Thus, 5′ cleavage occurs directly opposite 3′ cleavage for the bacterial transposon Tn10 (6), but is staggered inward by 2 nucleotides for the Caenorhabditis elegans transposons Tc3 and Tc1 (28, 29) and by 17 (and sometimes more) nucleotides for the Drosophila P element (4), and is staggered outward by 3 nucleotides for the bacterial transposon Tn7 (2).
The presence of transposon-derived sequences at the borders of the gap produced by excision of the Tc1, Tc3, and P elements readily accounts for the many excision products they generate that retain the two copies of the target site duplication as well as terminal sequences of the element. Indeed, these products are those expected if repair of the empty site occurs by a simple end-joining reaction (4, 28, 29). Variability among products would result from variations in the position of 5′ cleavage, from exonucleolytic degradation of the 5′ or 3′ extensions of the gapped DNA, and from annealing of small complementary sequence motifs between the 5′ or 3′ extensions prior to filling in and ligation.
In contrast, class II transposons of the hAT family, represented by the Drosophila element hobo and the plant transposons Ac/Ds and Tam3 (7), generate excision products that in many cases cannot be accounted for by a simple end-joining reaction. These products have lost all traces of the element and contain instead an inverted duplication of the transposon-proximal part of one or the other of the two copies of the target site duplication (for examples, see references 1, 9, and 14). In the absence of data concerning the intermediates formed during the excision reaction of transposons of the hAT family, two radically distinct models have been proposed to explain inverted duplication formation. In one model, outward-staggered cuts along the length of the target site duplication are made at each end of the transposon, and inverted duplications are formed during gap repair as a result of the DNA polymerase switching template from the flank DNA to the target sequence still attached to the 5′ ends of the transposon (21, 24). According to this model, variability in the excision footprints generated from a given site would primarily result from a combination of exonuclease activity on the flank DNA before gap repair and of variable DNA synthesis past the template switching point. In the other model, inverted duplications are caused by the formation of hairpins at the flanks and by their subsequent resolution elsewhere in the sequence, before gap repair (9, 21). In the more recent version of this model, based on a larger set of excision footprints, variability would primarily result from resolution of the hairpins at different points within the structure and from a possible exonucleolytic activity on the resolved hairpins (9).
Strong support for the hairpin model of transposition excision comes from the recently deciphered mechanism of V(D)J coding joint formation, responsible for the production of functional immunoglobulin and T-cell receptor genes during vertebrate lymphoid development (for a review, see reference 20). Although hairpin formation in the transposon excision model is assumed to occur by free-end ligation (9), one could equally envisage that it occurs by direct transesterification, as in V(D)J recombination (27). Following hairpin formation, processing relies on similar enzymatic activities in both cases, and it is the interplay of these activities which creates diversity in the hairpin model of transposition, just as it does in V(D)J recombination.
Previously, V(D)J recombination was thought to create junctional diversity in a random fashion, but recent evidence contradicts this. In particular, the nucleotide sequence of the hairpinned coding ends has been shown to play a determinant role in their own processing and thus in the variety of junctions that can be generated at any given V(D)J coding joint (11, 13, 17, 18). An influence of adjacent sequences on the footprints generated on transposon excision has also been documented for the maize transposon Ds, based on an analysis of footprints produced from six different excision sites (26). No attempt, however, has yet been made to evaluate precisely the diversity of excision footprints that can be generated from a given site and the origin of potential biases for any hAT transposon.
We set out to obtain such an evaluation by performing an extensive analysis of the excision products generated by Ascot-1, a newly discovered Ds-like transposon in the fungus Ascobolus immersus. Several key factors made this analysis unique a priori. First, since Ascot-1 was isolated from an unstable mutant allele (b2-G0) of a spore color gene, b2, its excision could be monitored in single cells (16, 19). Second, as Ascot-1 excision from b2-G0 is restricted to a very narrow window during the A. immersus life cycle, just before meiosis (19), excision could be detected just one or two cell divisions after it occurred, limiting the risks of clonal origin. Third, given the ease with which hundreds of thousands of asci can be scored for spore color in A. immersus, hundreds of revertants of independent origin could be identified rapidly.
We characterized from the same b2 site a total of 48 molecularly distinct excision footprints whose frequencies of occurrence ranged over at least 4 orders of magnitude. Small palindromic nucleotide additions indicative of the formation of a hairpin intermediate during the excision reaction at one or the other of the two flanking ends were found in 43 of the 48 footprints. Processing of the two flanking ends differed, and biases including the addition of a nontemplated nucleotide(s) were observed at each processing step. Taken together, these observations provide compelling evidence for strong nonrandomness of the excision reaction and suggest new parallels between the excision reaction performed by transposons of the hAT family and V(D)J recombination. In addition, a disproportionate association between transposon excision and reinsertion was found for one of several predominant products tested. This result indicates for the first time that the precise fate of an element following excision may be affected by the nature of rearrangements taking place at the excision site.
MATERIALS AND METHODS
Culture media, crossing conditions, and strains.
Standard media and procedures were used (19). The b2-G0, b2-G1, and b2-G234 alleles have been described previously (19).
DNA isolation and manipulation.
The procedures were as described previously (10). The Ascot-1 probe was obtained by PCR (for conditions, see below), using the single primer pITR corresponding to the sequence of the left terminal repeat (Fig. 1A): 5′CAGTGTTCTCAACAGTCAGTCCGGC3′.
FIG. 1.
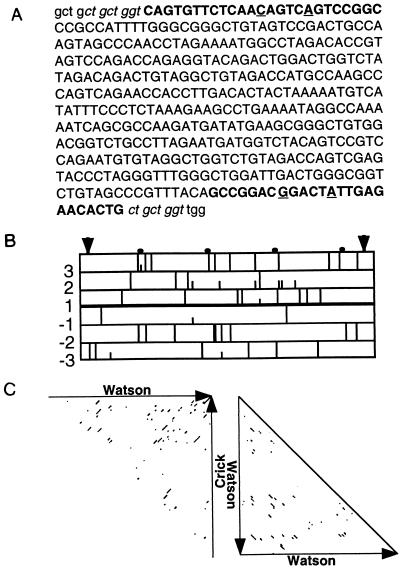
(A) Sequence of the 417-bp insertion present in b2-G0. The near-perfect 25-bp terminal inverted repeats are in boldface (the two mismatches are underlined); the 8-bp target site duplication of the b2 sequence is in italics. The insertion is located between codons 849 (ggt) and 850 (tgg) of b2. (B) Ascot-1 possesses no significant open reading frame (small vertical bars, ATG; large vertical bars, stop codons) and introduces eight stop codons in the b2 reading frame (frame 1). Vertical arrows delineate the 417-bp insertion. (C) Dot plot analysis (window 12, stringency 9) of Ascot-1 reveals the presence of numerous small inverted (left) and direct (right) repeats.
PCR analysis and sequencing of b2-G0 and of individually isolated derivatives.
Amplification of genomic DNA (50 to 100 ng) was carried out by using one step of 94°C (4 min), followed by 35 cycles of 94°C (1 min)-63°C (30 s)-72°C (4 min) and one last step of 72°C (7 min). Amplification products were polyethylene glycol purified (22) and then run on a gel to estimate their sizes and quantities. Direct sequencing of the purified products was performed at least once on both strands on an ABI 372A machine, using ABI Dye Terminator Cycle Sequencing kits (Perkin-Elmer).
The following primers were used to locate and sequence Ascot-1 within the 2.4-kb NdeII fragment of b2-G0. They are named according to the position of their 5′ nucleotides (upper primers) or their 3′ nucleotides (lower primers) on the sequenced fragment containing the b2 gene (9b). The 8-bp Ascot-1 target site within b2 is located between positions 5323 and 5330. The upper primers were p4714 (5′ACCGTCGTCCAAGTCCTCA3′) and p5189 (5′GGGAGGTGGCGATTATGAT3′). The lower primers were pR5472 (5′ATAGGAACCCGCCTTATCG3′), pR5932 (5′CAGCGGCGGCATAGTTGTT3′), pR6404 (5′GCATCTTCGTCGGAGGAGT3′), and pR6737 (5′CCAAGACCTCGTCCCAAGC3′).
The various excision products were amplified with primers p4714 and pR6737, except for those corresponding to products 35 to 42 (Fig. 2B), which were amplified with a different upper primer: p4323 (5′CTGAGGAGGACGACTACCA3′; for products 35 to 41) or p2805 (5′GCCTCCGTCCTTTTCGTCA3′; for product 42).
FIG. 2.
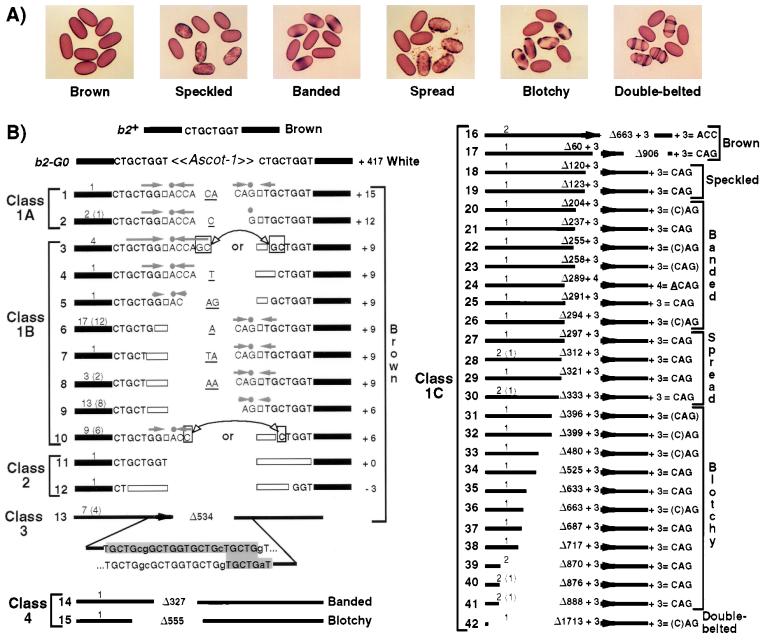
Spore color phenotype and DNA sequence of the excision footprint of 96 b2-G0 colored derivatives. (A) Phenotypes were assessed through crosses with the wild type (brown). (B) The 96 colored derivatives correspond to 42 distinct excision products, numbered 1 to 42. The number of derivatives corresponding to a given product is indicated above that product, on the left. When molecularly identical derivatives were not all isolated from independent crosses, the numbers of those that were (and that are therefore unambiguously nonclonal) are indicated in brackets. The b2 8-bp target site and its duplication upon Ascot-1 integration are shown at the top of the left panel. For the products of classes 1A, 1B, and 2, the remnant of the target site duplication generated upon Ascot-1 integration is indicated and missing nucleotides are shown as white boxes. A grey arrow and a dot designate an inverted duplication of part of the target site and a transversion of its transposon-proximal nucleotide, respectively (the nucleotides of products 3 and 10 that could belong either to the left inverted duplication or to the right copy of the target site duplication are boxed). Additional, nontemplated nucleotides are underlined. Size differences with the wild-type b2 sequence are indicated on the right. For the products of the other classes, deletion sizes are indicated centrally and deletions are drawn to scale, except for products 16, 17, and 42. The structure of the deletion ends of the class 3 product is shown in grey. The nearly intact copy of the target site duplication which is retained in products of class 1C and class 3 is indicated by a black arrow. The inverted duplication and the transition associated with this copy in class 1C products are shown on the right: CAG for the remnant right copy TGCTGGT and ACC for the remnant left copy CTGCTGG. Nucleotides are shown in brackets when they could alternatively correspond to the first b2 nucleotide past the 5′ deletion endpoint. The exceptional double-deletion product 17 corresponds to the b2-G234 derivative. The upstream deletion endpoints of products 14 and 15 are 295 and 448 bp away from the excision site.
Sequencing of products of apparent wild-type size was performed on both strands with primers p5189 and pR5472, respectively. Shorter products were sequenced with one or two of the other primers, chosen on an ad hoc basis (the upper strand of product 42 was sequenced with yet another primer, p3530 [5′CCTCCCCTCCGAATACGAA3′]).
PCR isolation and sequencing of mutant derivatives obtained after en masse harvesting of progeny of b2-G0 × b2-G0 crosses.
To avoid amplification of the vastly more abundant b2-G0 allele or across the stem-loop structure that could be potentially formed by the Ascot-1 sequence, DNA isolated both from the parental strains and following en masse germination of nonrevertant progeny spores was first digested to completion with the enzyme ScfI, whose recognition site CTPuPyAG is present seven times within Ascot-1 but absent from the entire b2 sequence (data not shown). Digests (∼25 ng, ∼5 × 105 genomes) were then subjected to PCR with primers p4717 and pR5932, each approximately 600 bp away from the Ascot-1 integration site.
Nucleotide sequence accession number.
The 409-bp Ascot-1 sequence has been submitted to GenBank under accession no. AF054897.
RESULTS
The unstable white-spore b2-G0 mutation results from the insertion of a 409-bp nonautonomous transposon into the b2 coding region.
The approximately 400-bp insertion present in the 2.4-kb MboI fragment of b2-G0 (10) was further localized by PCR using various pairs of primers derived from the corresponding wild-type fragment (results not shown). Direct sequencing of chosen amplification products indicated that the insertion is 417 bp long and located within the b2 coding sequence (Fig. 1A). Several in-frame stop codons are created as a result, suggesting that the b2-G0 white-spore mutant phenotype is caused by the synthesis of a truncated b2 gene product (Fig. 1B). Further examination of the insertion shows that it is composed of a noncoding 409-bp sequence with near-perfect 25-bp inverted terminal repeats and several internal direct or inverted repeats (Fig. 1C), flanked by an 8-bp duplication of the b2 sequence. Considered together, these and other observations (see below) designate the 409-bp sequence as a nonautonomous class II transposon (12), which we have called Ascot-1 (Ascobolus transposon-1).
The b2-G0 instability generates at least six phenotypically distinct colored derivatives.
The system of spore color instability associated with the b2-G0 mutation was reconstituted by setting up multiple crosses between two b2-G0 strains selected for high fertility. Among the approximately 7 × 105 asci obtained, 1,119 contained at least one colored spore. Spore color segregated 4:4 in 94% of the 1,119 asci, indicating premeiotic reversion of the b2-G0 mutation, and mostly 6:2 or 2:6 otherwise, as expected if gene conversion occurred during meiosis following premeiotic reversion in one parent. Taken together, these results give an apparent reversion frequency of 0.8 × 10−3, a value similar to that reported originally (0.4 × 10−3 to 0.5 × 10−3 [19]). Whereas the colored spores of 1,047 of the revertant asci were phenotypically indistinguishable from wild-type brown spores, those of the other 72 revertant asci showed only partial pigmentation. To determine spore color unambiguously, we germinated one partially colored spore from each of the 72 relevant asci and one wild-type-like spore from a sample of 93 of the 1,047 other revertant asci. Crosses were performed between the resulting strains and a wild-type tester strain of the opposite mating type. Of the 72 strains derived from partially colored spores, 68 bred true and 4 yielded only phenotypically wild-type spores. As for the 93 strains derived from fully colored spores, all but one bred true, the exceptional one yielding only partially colored spores. Thus, spore color was correctly determined in over 90% of cases directly from the original crosses and was always stable. Based on this result, it can be estimated that 7% (79/1,119) of all colored derivatives exhibit partial coloration. Significantly, more precise analysis of spore color revealed an extended range of partial color phenotypes: besides known color variants described previously as banded, spread, blotchy, and double-belted, a new one, termed speckled, was found (Fig. 2A), as well as a whole spectrum of intermediate stable phenotypes ranging from spread to blotchy on one side and from blotchy to double-belted on the other side (not shown).
The b2-G0 instability results from the excision of Ascot-1 and generates dozens of molecularly distinct colored derivatives.
To determine whether fully colored derivatives of b2-G0 were genetically true wild type, 16 were picked at random and crossed with a b2-G1 mutant strain. When crossed with the wild type, b2-G1 gives approximately 10% of asci with a non-4- brown:4-white (4B:4W) segregation of spore color, and of these, over 80% show postmeiotic segregation (5B:3W and 3B:5W) (19). In the 16 crosses performed, the frequency of postmeiotic segregation was always much lower than 80% of the non-4B:4W segregation (data not shown), indicating that none of the 16 fully colored derivatives that were analyzed genetically carried a true wild-type b2 allele. Likewise, genetic analysis of 30 derivatives covering the whole spectrum of partial color produced several examples of distinct patterns of gene conversion for a given phenotypic class in crosses with b2-G1 (data not shown). These results are in line with those obtained previously on another series of colored b2-G0 derivatives (19).
To investigate the nature of the molecular diversity revealed by the combined phenotypic and genetic analysis, total DNA was extracted from the mycelia of the 46 colored derivatives that were analyzed genetically (16 brown and 30 partially colored) as well as from the mycelia of 48 brown and 2 partially colored derivatives that were obtained in other b2-G0 × b2-G0 crosses (including the previously described b2-G234 brown derivative [19]). DNAs were amplified with primers flanking the Ascot-1 insertion site in b2-G0 and then directly sequenced. No Ascot-1 sequence was present in any of the amplified products, indicating that all of the 96 colored derivatives result from Ascot-1 excision (Fig. 2B). Together with the observation that Ascot-1 was found reinserted elsewhere in the genome in a fraction of the colored derivatives (see Fig. 4 and below), this provides definitive evidence that Ascot-1 is mobile and that it moves through a cut-and-paste mechanism.
FIG. 4.
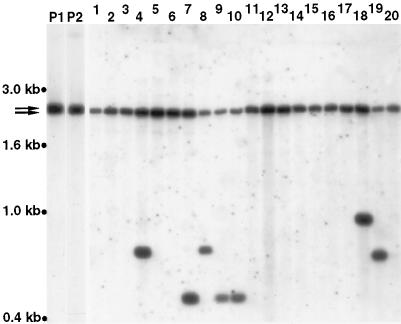
Example of the detection of Ascot-1 transposition in b2-G0 derivatives. Southern blots of genomic DNA digested with NdeII were probed with the 409-bp Ascot-1 sequence. Lanes P1 and P2, b2-G0 parental strains, containing two copies of Ascot-1 (arrows). The b2-interrupting copy is present on the larger of the two nearly comigrating NdeII fragments. Lanes 1 to 20, colored derivatives obtained from several independent crosses between P1 and P2. In all 20 derivatives, the larger NdeII fragment is missing, indicating full excision of Ascot-1 from b2. In seven derivatives, a new hybridizing NdeII fragment indicative of Ascot-1 reinsertion elsewhere in the genome can be seen. Derivatives in lanes 4, 7, and 8 are independent and correspond to product 9; those in lanes 9 and 10 were isolated from the same fruiting body and correspond to product 16. Derivatives in lanes 18 and 19 correspond to products 34 and 19, respectively.
As shown in Fig. 2B, the 96 colored derivatives corresponded to 42 molecularly distinct excision products. As expected, all restored an uninterrupted reading frame, yet large differences existed between products. Phenotypic and molecular differences correlated in some instances. Thus, all of the 27 different excision products (14, 15, and 18 to 42) that corresponded to the 32 partially colored derivatives exhibited deletions. These extend 120 bp or more 5′ from the excision site, and color defects increase with deletion size. Conversely, 3 of the 15 different excision products defined by the 64 brown derivatives exhibited large deletions of the b2 coding sequence 3′ to the target site (products 13, 16, and 17), another 10 exhibited small sequence additions (6 to 15 bp, products 1 to 10), 1 exhibited a 3-bp deletion (product 12), and 1 restored the wild-type b2 sequence (product 11).
Revertant excision products fall into four classes.
The 42 excision products obtained can be grouped into several classes and subclasses (Fig. 2B). Class 1 products (37 in total) shared three features: (i) they retained nearly intact one or the other of the two copies of the target site duplication, (ii) the nearly intact copy always contained as the only modification a transversion of the transposon-proximal nucleotide, and (iii) it was always associated with an inverted partial duplication centered on the transversion site. Class 1 products can be further subdivided into three subclasses, according to the fate of the second copy of the original target site duplication. Class 1A products (2 of 37) retained a second copy showing (like the first one) a transversion of the transposon-proximal nucleotide, plus an inverted duplication of two nucleotides in one case, but no other modification. In contrast, class 1B products (8 of 37) contained a second copy which lacks two to four nucleotides from the transposon-proximal end. Significantly, this partly deleted copy was not associated with any inverted duplication. Finally, class 1C products (27 of 37) completely lacked the second copy of the target site duplication. In fact, class 1C products showed deletions that extend to various lengths past the second copy into the b2 flanking sequence, and one (product 17, corresponding to derivative b2-G234) showed, in addition, a second deletion located 180 bp 3′ to the first one. Among the class 1 products, 8 of 37 were also distinguished by the presence of one or two nontemplated nucleotides (underlined in Fig. 2B), which abut the inverted duplication associated with the near intact copy(ies) of the target site duplication. Incidentally, this addition of nontemplated nucleotides was seen in the 2 products of class 1A and 5 of 8 (products 4 to 8) of class 1B, but only 1 of 27 (product 24) of class 1C.
In contrast to the class 1 products, the other five excision products obtained from our sample of colored derivatives all corresponded to simple deletions. Class 2 products had deletion endpoints that lie within the target site duplication. This led in one case (product 11) to the restoration of the wild-type b2 sequence and in the other case (product 12) to a 3-bp deletion. As for the deletion present in the single class 3 product 13, its endpoints are within two near identical b2 sequences of substantial length (25 bp). Significantly, the 3′ end of the upstream copy, which is separated by 534 bp from the downstream copy in the wild-type b2 allele, corresponds precisely to the target site and is where the deletion endpoint lies within the 25-bp sequence (Fig. 2B). Lastly, two products (14 and 15) composed class 4: they exhibited a bidirectional deletion that extends beyond the target site duplication and ends within sequences of little (2 bp) or no homology.
Ascot-1 excision leads to hundreds of minor revertant products and to five predominant ones.
Although the 96 colored derivatives that were analyzed molecularly originated from 48 independent b2-G0 × b2-G0 crosses (data not shown), 22 cases of a derivative molecularly identical to one or more derivatives of the same cross were found (Fig. 2B). To explore the possibility that Ascot-1 excision occurs earlier than during the last mitotic cycle before meiosis and that clonally related derivatives could arise as a result, several individual fruiting bodies with one 4B:4W ascus already protruding were isolated from a crossing plate. Among these, one gave ultimately another 4B:4W ascus and several 0B:8W asci. Sequencing of the two colored derivatives isolated from the same fruiting body indicated they were identical to each other and hence clonally related (not shown).
Given this last result, we have only considered in the following analysis the 74 derivatives that were unambiguously nonclonal (28 partially colored and 46 phenotypically wild type). Hence, we have assumed a true frequency of reversion in b2-G0 × b2-G0 crosses of 0.5 × 10−3 instead of the apparent one of 0.8 × 10−3.
The 28 partially colored derivatives correspond to 27 distinct deletion products, 25 of class 1C, and 2 of class 4. Given that these products exhibited deletions ranging from 120 to 1,713 bp and that they were represented only once, with one exception (product 39), it is therefore likely that at least 531 [(1,713 − 120)/3] and probably many more distinct products giving partially colored spores could be generated upon Ascot-1 excision. Since partially colored derivatives represent an estimated 7% of all the colored derivatives generated upon Ascot-1 excision (see above), each of the predicted 531 or more products giving partially colored spores would arise at a frequency of about 0.7 × 10−7 or less (0.5 × 10−3 × 7% × <1/531) in b2-G0 × b2-G0 crosses when of class 1C and at least 10-fold less frequently when of class 4.
In contrast to the situation observed for the 28 partially colored derivatives, the 46 phenotypically wild-type derivatives correspond to 15 distinct products only. These products are not all equally abundant, since 5 were represented by four derivatives or more, while the other 10 were represented by one or two derivatives only. The five predominant products are the single class 3 product (product 13) and four of the class 1B products (products 3, 6, 9, and 10). Given that these products were represented by between 4 (9%) and 12 (26%) of the 46 fully colored derivatives, their frequency of occurrence in a cross can be estimated to be approximately 0.4 × 10−4 to 1.2 × 10−4 (0.5 × 10−3 × 93% × 9 to 26%). As for the 10 minor products, 2 belong to class 1A, 4 belong to class 1B, 2 belong to class 1C, and 2 belong to class 2. This suggests that Ascot-1 excision can generate dozens or more of phenotypically wild-type but molecularly diverse products in addition to the five predominant ones. Their frequency of occurrence in b2-G0 × b2-G0 crosses is likely to vary greatly between classes, from 10−8 or less for class 4 products (yet to be documented) to perhaps as much as 10−4 or so for the class 1 products 8 and 14.
Ascot-1 excision also leads preferentially to a limited number of stable mutant products.
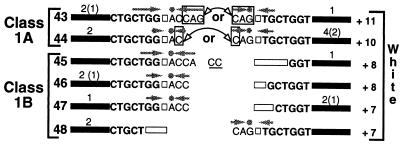
Previous studies have shown that b2-G0 also gives rise to white derivatives at high frequency (approximately 5 × 10−3 in b2-G0 × b2+ crosses), which can be distinguished from the white-spore b2-G0 mutation on the basis of genetic criteria (19). To investigate the molecular nature of these derivatives, eight were isolated at random from two independent b2-G0 × b2+ crosses and sequenced. Sequencing was also performed on one of the originally described white-spore derivatives, b2-G1 (19). As shown in Fig. 3, the nine derivatives corresponded to five distinct excision products, which added 7, 8, 10, or 11 nucleotides to the wild-type b2 sequence. Although these five products disrupted the b2 reading frame, as expected from their mutant phenotype, they exhibited the same molecular features as the phenotypically wild-type products of classes 1A and 1B. They were thus designated either class 1A or class 1B products (Fig. 3).
FIG. 3.
DNA sequences of the excision footprints of 17 white derivatives of b2-G0. A total of six molecularly distinct excision products, numbered 43 to 48, were obtained from the sample. Representation is as in Fig. 2B, except that the numbers shown above the products refer to derivatives isolated by means of genetic criteria (left) or by PCR (right). Nucleotides that could be part of either of the two inverted duplications of the two class 1A products are boxed. Note that the predominant product 44 could be drawn alternatively with only one inverted duplication, CAG, associated with a nontemplated addition of an A. This would make this product resemble more closely class 1B product 6, which is predominant among products that restore spore color partially or fully (Fig. 2B). The mutant derivative b2-G1 corresponds to product 48.
White derivatives of b2-G0 were also analyzed by PCR performed on DNA isolated en masse from the progeny of b2-G0 × b2-G0 crosses (see Materials and Methods). No excision products were detected in DNA from two parental b2-G0 strains, confirming that Ascot-1 excision does not occur at detectable levels during vegetative growth. In contrast, PCR amplification of DNA from germinated spores of approximately 7,000 and 22,000 0B:8W asci issued from two independent crosses always yielded single bands of wild-type size on gels (not shown). Given that primers were located approximately 600 bp from the Ascot-1 insertion site, this result implies that the noncolored derivatives of b2-G0 correspond predominantly to wild-type size excision products. Direct sequencing of the wild-type size bands revealed a mixture of distinct products (not shown). To document this further, the amplified DNA was cloned and eight inserts were sequenced. Four different excision products were identified, three of which were previously identified in b2-G0 × b2+ crosses by genetic criteria (products 43, 44, and 47 [Fig. 3]). The fourth product, 45, added eight nucleotides to the wild-type b2 sequence and was assigned to class 1B (Fig. 3).
In conclusion, our analysis of white derivatives of b2-G0 isolated either genetically or directly by PCR yielded six distinct excision products, all disrupting the b2 reading frame and belonging to class 1A or 1B. At least four of these products must occur frequently since they were recovered more than once from independent crosses in the small sample analyzed (Fig. 3). Conversely, as no products of classes 1C, 2, and 4 were recovered in our analysis of white derivatives, it is likely that these classes are also rare among products giving white spores.
Ascot-1 excision is accompanied by its reinsertion elsewhere in the genome.
To obtain conclusive evidence that Ascot-1 is a bona fide class II transposon, the fate of the element present in b2-G0 was investigated on a sample of 61 colored derivatives, including one clonal pair, and corresponding to 34 distinct products in total. DNAs were digested with NdeII, a restriction enzyme that does not cut within the 409-bp Ascot-1 element, and subjected to Southern blot analysis using the entire 409-bp element as a probe. This analysis was facilitated by the fact that all of the parental b2-G0 strains contained only two copies of Ascot-1, one residing in b2 and one residing elsewhere in the genome (Fig. 4 shows examples). A total of 14 derivatives showed a new hybridizing NdeII fragment and thus proof of Ascot-1 mobility (see Fig. 4 for examples and Table 1 for a summary). Furthermore, the two clonal derivatives were among these, and both exhibited the same new hybridizing band, as expected.
TABLE 1.
Analysis of Ascot-1 reinsertion in colored b2-G0 derivatives
| Product no. | No. of derivatives tested | No. of derivatives showing Ascot-1 reinsertion |
|---|---|---|
| 1 | 1 | 0 |
| 3 | 3 | 0 |
| 6 | 5 | 1 |
| 9 | 8 (7a) | 6b |
| 10 | 4 | 0 |
| 13 | 5 (4a) | 1 |
| 14, 15 | 2 | 0 |
| 16 | 3 (2a)c | 2d |
| 18–42 | 30 (26a) | 4 (products 19, 34, 37, and 42) |
Number of derivatives isolated from independent crosses (and thus unambiguously nonclonal).
Two of the six reinsertions involve the two derivatives isolated from the same cross. These two reinsertions are nevertheless nonidentical, which indicates that the two derivatives are nonclonal.
The two derivatives isolated from the same cross were in fact produced by the same fruiting body and were clonal.
The two reinsertion events are identical and involve the two clonal derivatives.
The 13 reinsertions involved 8 of the 34 products. These eight products belonged to classes 1B, 1C, and 3 (Table 1 and Fig. 2B). Strong distortions were observed, however, since 6 of 13 reinsertions involved a single class 1B product (product 9 [Table 1]).
DISCUSSION
We have identified the first class II transposon of the fungus A. immersus and have performed an extended analysis of the spectrum of footprints produced by its excision from a given site within the spore color gene b2. Whereas an unbiased PCR approach would preferentially identify those excision products that occur most frequently and/or would be constrained by the choice of primers, the use of spore color as a selection criterion and the wide range of rare partial revertant phenotypes produced upon Ascot-1 excision ensured that excision footprints were identified essentially independently of their frequency of occurrence or of their nature (large deletions versus small alterations). Thus, from a selected sample of around 100 b2-G0 derivatives, a total of 48 molecularly distinct excision products were obtained, varying in their individual frequency of occurrence over at least 4 orders of magnitude and in their nature from the addition of a few base pairs with respect to the wild-type sequence to the deletion of over 1.7 kb. This data set provides the most comprehensive picture so far of the type and range of excision products that can be generated by transposon excision from a given site.
Despite varying greatly in frequency of occurrence, the vast majority of the excision products identified (43 of 48) belong to a single class (class 1). All class 1 products present at least one nearly intact copy of the target site duplication (with a transversion of the transposon-proximal nucleotide as sole modification), and they all display an inverted partial duplication of this copy, centered on the transversion site (Fig. 2B and 3). These features are entirely consistent with the intermediate hairpin formation model proposed to account for many of the excision footprints left by the hAT family of transposons (9). Moreover, since these features are shared by all but one of the most frequently generated Ascot-1 excision products, it is likely that Ascot-1 excision occurs predominantly by this pathway. Thus, Ascot-1 resembles most closely nonautonomous transposons of the hAT family, such as the Ds element of maize.
In the hairpin pathway, cleavage would occur precisely at the 3′ ends of Ascot-1, as in any other pathway (see the introduction), and would be staggered outward from the transposon 5′ ends by one nucleotide so that an appropriate hairpin is formed at each flank. Depending on how cleavage of the element is effected, hairpin formation would result either from ligation of two free ends at each flank or else, as in V(D)J recombination, from direct transesterification (Fig. 5A). Resolution and further processing of the two hairpin structures would follow as in V(D)J recombination and would involve the recruitment of different end-joining activities that may or may not be specific (23).
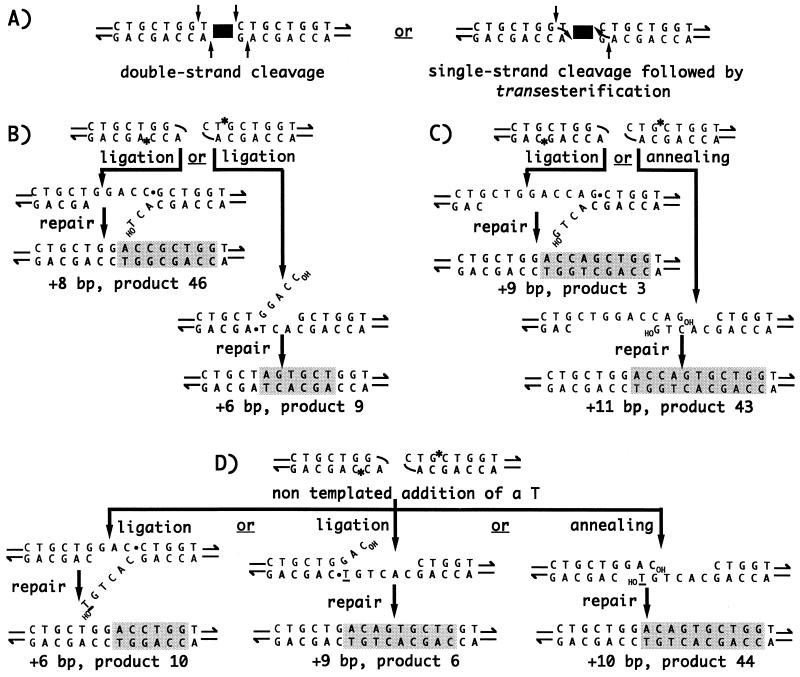
FIG. 5.
Models for hairpin formation and footprints generated by Ascot-1 excision. (A) Hairpins may be formed either by staggered double-strand cleavage (left; vertical arrows) and ligation of the ends released at each flank or by single-strand cleavage followed by direct transesterification (right; vertical and curved arrows, respectively). In the latter case, nucleophilic attack by the exposed 3′OH flank ends does not occur on the phosphodiester bond directly opposite, as in V(D)J recombination, but on the bond immediately 5′ to it. (B to D) The resulting left and right hairpins are indicated at the top of each section. Asterisks indicate the position of the nick used for hairpin resolution. All nicks have the same polarity and produce 3′ overhangs, which may favor the nontemplated addition of nucleotides by a terminal transferase-like activity (8). Ligation is drawn arbitrarily as occurring on only one strand, followed by repair. Alternatively, ligation could involve both strands and would be followed by repair of the mismatched DNA duplex. (B) Products 9 and 46 are reciprocal ligation/repair products. Products 47 and 48 (Fig. 3) are also reciprocal ligation/repair products, generated when the resolving nicks occur as in panel B for the left hairpin and as in panel C for the right hairpin. (C) Depending on whether the resolved hairpins are directly ligated or annealed first, product 3 or 43, respectively, is generated. (D) Three possible outcomes from the same pair of resolved hairpins, following the nontemplated addition of a T (underlined) at the 3′ end of the right flank. Alternative ligation/repair gives products 6 and 10, while annealing gives product 44.
A common origin for small palindromic additions and small deletions?
The hairpin model is attractively economical in explaining all of the predominant class 1 products of Ascot-1 excision (Fig. 5). Thus, products 9 and 46 (Fig. 5B) and products 47 and 48 (see the legend to Fig. 5B) can most simply be viewed as corresponding to the two alternative outcomes possible after direct ligation of the nicked hairpins and repair. On the other hand, products 3 and 43 (Fig. 5C) could arise as a result of two simple alternative reactions, direct ligation and annealing, respectively (the alternative ligation/repair outcome, a 5-bp product, was not observed either because of the limited number of stable mutant derivatives analyzed and/or because of bias in the ligation/repair reaction). As for the three products shown in Fig. 5D, they could result from the nontemplated addition of a T at the 3′ end of the resolved right hairpin: subsequent ligation/repair would give alternatively products 6 and 10, and annealing would give product 44.
As developed in Fig. 5, the hairpin model suggests that the small deletions which distinguish class 1B from class 1A products originate by single-strand nicking of the corresponding hairpins rather than by double-strand nicking. Thus, the small deletions shown by the class 1B products may have the same origin as the small palindromic additions exhibited by all class 1 products. Moreover, by indicating that nicking of the two hairpins must occur on opposite strands, this model suggests the existence of steric constraints also at this stage.
The formation of the less frequently observed class 1 products displaying small size alterations are also readily explained by the hairpin model. Products 7 and 8 could both be related to products 6 and 9, if two nontemplated nucleotides are added to the nicked right hairpin shown in Fig. 5D before ligation to the nicked left hairpin shown in Fig. 5B. Similarly, products 1, 2, 4, 5, and 45 might all result from the nontemplated addition of nucleotide(s), prior to either ligation (products 4, 5, and 45) or filling in of the recessed 5′ ends and ligation (products 1 and 2) (not shown). Alternative schemes, such as the formation, in rare cases, of a noncanonical hairpin at one end, can clearly be envisaged. Product 5, for example, could result from the nicking of a right hairpin formed following an outward 5′ cleavage staggered by two nucleotides instead of one at the downstream transposon end (not shown). As for the two products of the rare class 2, little is revealed by their structure regarding the nature of the molecular events that might have led to their synthesis.
Unidirectional, large deletions are probably caused by an exonuclease activity.
Among the less frequent class 1 products, class 1C products are distinguished by the unique feature of exhibiting large deletions on one side of the excision site (the origin of the second deletion shown by product 17 is not known and will not be discussed, as it may be unrelated to the excision event; product 17 corresponds to derivative b2-G234, crossed many times since first isolated). Our data set provides interesting information as to the likely cause of these deletions. Comparison of products 17 to 42 indicates that their upstream deletion endpoints are distributed uniformly along the sequence and thus strongly suggests the action of an exonuclease from the resolved left hairpin prior to ligation and repair. Remarkably, products 17 to 42 all exhibit a sequence motif (CAGTGCTGGT) at the downstream end of the deletion that potentially results from the right hairpin being resolved at the same position in every case. Moreover, with the exception of product 24, none of these products exhibits a nontemplated addition of a nucleotide at the 3′ end of the putative resolved right hairpin. These observations strongly suggest that the vulnerability of the resolved left hairpin to exonucleolytic degradation is not determined randomly. The factor(s) that may govern this vulnerability could be the sequence or structure of the resolved left hairpin itself and/or the type and order of processing events at the right hairpin. Whether the same or other factors govern the vulnerability of the right hairpin and cause large downstream deletions (as in product 16 [Fig. 2B]) can be determined only by systematically searching for these deletions. The frequent occurrence of the only other large downstream deletion identified in our sample (product 13 [Fig. 2B]) suggests, however, that this search might be in vain. Indeed, the deletion exhibited by product 13 is unique in pointing to an end-joining reaction directed by significant sequence homology. It is therefore possible that whenever exonucleolytic degradation involves at least 534 nucleotides, annealing between the left resolved hairpin and the nearly identical 23-nucleotide-long motif thus exposed will occur preferentially to end ligation.
Processing of the two flanks at the excision site exhibits strong biases.
Although our data provide ample evidence that the excision reaction can generate extensive diversity at the excision site, they also show that it does so in a highly nonrandom manner. For example, the putative right hairpin appears to be preferentially nicked as shown in Fig. 5C and D, since the right palindrome most frequently observed, either directly or indirectly as a result of alternative ligation/repair, is CAGTG (Fig. 2B, 3, and 5). Similarly, nicking of the putative left hairpin would exhibit some preference, although less marked than for the right hairpin. Thus, the most frequently observed left palindrome is GGACC (corresponding to a nicking of the left hairpin as in Fig. 5B), but other palindromes are also frequent (Fig. 2B, 3, and 5). Nonrandom positioning of the single-strand nick required for hairpin resolution has been recently demonstrated for V(D)J recombination, based on extensive in vitro studies performed with various V(D)J substrates (11, 13, 17, 18). Furthermore, these studies have led to the conclusion that hairpin sequences play a determinant role in dictating where resolution takes place. Although the two hairpins formed at the excision site are related in sequence (in their terminal part) by virtue of the target site duplication that was created during transposon insertion, they differ because of their opposite polarities (Fig. 5). Thus, it is likely that the nonrandom, distinct patterns of resolution observed for the putative left and right hairpins are determined in large part by their sequences. In turn, this implies that the predominant excision footprints produced at different locations by the same hAT transposon may differ with respect to the length of the palindrome(s) they may contain. Data compatible with this prediction have recently been obtained for Ds transposon in maize (26).
As alluded to above, nonrandomness in the processing of the two putative hairpins formed during Ascot-1 excision is also overtly apparent at a later step. Thus, nontemplated addition of nucleotides is frequently observed upstream of the most frequent right palindrome CAGTGC, but only when this palindrome is not associated with a large deletion of the left flank (18 of 19 class 1A or 1B products but only 1 of 25 class 1C products [Fig. 2B and 3]). Moreover, this nontemplated addition is highly biased, since the first nucleotide added in every single case is a T (shown on the opposite strand as an A in Fig. 2B and 3). Nontemplated addition may also occur preferentially at the end of the left palindrome TGGACCA, but data are too limited for this observation to be significant. Noticeably, however, addition of the first residue is not restricted to the nucleotide T this time. Whether these nontemplated additions are effected by a terminal transferase is an open question, as this enzyme has only been described to date in mammals, exclusively in cells actively engaged in V(D)J recombination (8). A putative Ascobolus terminal transferase activity would nevertheless differ from that of mammals, since the latter shows a marked preference for the insertion of G, not T, residues (3). Alternatively, nonpalindromic nucleotides may be added at the excision site by other mechanisms, such as those proposed on an ad hoc basis to explain the presence of filler DNA at the junctions formed by end joining in nonlymphoid mammalian cells (23).
Transposon fate is linked to the processing of the excision site.
Transposon reinsertion was detected in approximately a quarter (13 of 54) of independent derivatives tested (Table 1). Given that reinsertion was looked for in only one of the four meiotic products of each revertant ascus and that Ascot-1 transposition takes place preferentially during the last mitotic cycle before meiosis, this proportion is compatible with transposition occurring from unreplicated sites into replicated sites that are genetically unlinked. Statistically significant departure from this possible scenario, however, was observed for the predominant excision product 9, for which reinsertion was detected in six of seven independent cases, compared with reinsertion in one of five independent cases for the predominant product 6 and in none of four cases for each of the predominant products 10 and 13. This disproportionately high level of association between excision and reinsertion observed for product 9 indicates that reinsertion must occur frequently to neighboring locations whenever excision gives rise to this product. This result would therefore imply that whether reinsertion of an element occurs to linked or unlinked sites depends on the exact nature of the processing events at the excision site.
Evolutionary considerations.
Like other small-size DNA alterations and point mutations, small palindromic additions create variability. Intriguingly, they appear to be produced exclusively by V(D)J recombination or excision of transposons of the hAT family, as they are not otherwise within the range of DNA alterations produced by repair of chromosomal double-strand breaks (15, 25, 30), nor are they produced by replication errors (5). Since V(D)J recombination is a somatic process, however, the mutations that it produces do not serve as raw material for evolution. In marked contrast, transposon excision can occur in the germ line, as in the case of Ascot-1. Thus, it is legitimate to ask to what extent excision footprints containing small palindromic additions contribute to mutation pressure. Based on a series of crosses related to those performed in the present study, 5% of all spontaneous spore color mutations in Ascobolus were found to be due to the insertion of Ascot-1 (9a). Given that the frequency of spore color mutants in these crosses was approximately 2.5 × 10−4 per meiosis and that 18 genes in addition to b2 control spore color, this gives an absolute frequency of forward mutations caused by Ascot-1 of 6.6 × 10−7 per gene per meiosis. With a frequency of excision events restoring the reading frame >0.5 × 10−3 (see Results) and a proportion of excision footprints with small palindromic insertions of ≥50% (Fig. 2B and 3), alterations of this type will arise at a combined frequency of >1.6 × 10−10 per gene per meiosis. As these excision footprints can be expected to add several new codons to a gene sequence (Fig. 2B), it is interesting to compare their frequency to that of two intersuppressing frameshift mutations, since these may also change the coding sequence locally (note, however, that intersuppressing frameshift mutations rarely lead to codon addition and allow a much narrower range of codon changes overall). Assuming that all mutations except those caused by Ascot-1 are frameshift mutations, their frequency is 1.2 × 10−5 per gene per meiosis. The frequency of obtaining a second, suppressing frameshift mutation in a given gene can be estimated to be 1.2 × 10−7 [(1.2/2) × 10−5 × (60/3,000)], assuming that all frameshift mutations are divided randomly between +1 bp and −1 bp, that the gene size is 3 kb, and that intergenic suppression can take place between sites located no more than 60 nucleotides apart. This gives, therefore, an estimated combined frequency of at most 1.4 × 10−12 per gene per meiosis for two intersuppressing frameshifts, compared to an estimated frequency of 1.6 × 10−10 for palindrome-containing footprints that restore the reading frame. Although based on many approximations, these results strongly argue in favor of the importance of small palindromic additions produced by excision of transposons of the hAT family as generators of genotypic and phenotypic variability.
ACKNOWLEDGMENTS
We thank Anne Dumay, Fatima Graia, and Marc-André Sélosse for their contributions to the work, Denise Zickler for help with photography, and Christian Cibert, Shelly Esposito, Vincenzo Rocco, and François Taddei for valuable suggestions during preparation of the manuscript. Special thanks are due to Michael Ronemus for critical reading of the manuscript.
This work was supported by grants from the Association pour la Recherche sur le Cancer (contract 6200) and the Groupement de Recherches et d’Etudes sur les Génomes (contract 44/95).
ADDENDUM
Since this paper was submitted for review, evidence for the formation of 3′ overhangs as a result of hairpin resolution has been obtained for V(D)J recombination (25a).
REFERENCES
- 1.Atkinson P, Warren W, O’Brochta D. The hobo transposable element of Drosophila can be cross-mobilized in houseflies and excises like the Ac element of maize. Proc Natl Acad Sci USA. 1993;90:9693–9697. doi: 10.1073/pnas.90.20.9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bainton R, Gamas P, Craig N. Tn7 transposition in vitro proceeds through an excised transposon intermediate generated by staggered breaks in DNA. Cell. 1991;65:805–816. doi: 10.1016/0092-8674(91)90388-f. [DOI] [PubMed] [Google Scholar]
- 3.Basu M, Hegde M, Modak M. Synthesis of compositionally unique DNA by terminal deoxynucleotidyl transferase. Biochem Biophys Res Commun. 1983;111:1105–1112. doi: 10.1016/0006-291x(83)91413-4. [DOI] [PubMed] [Google Scholar]
- 4.Beall E, Rio D. Drosophila P-element transposase is a novel site-specific endonuclease. Genes Dev. 1997;11:2137–2151. doi: 10.1101/gad.11.16.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beckman R, Loeb L. Multi-stage proofreading in DNA replication. Q Rev Biophys. 1993;26:225–331. doi: 10.1017/s0033583500002869. [DOI] [PubMed] [Google Scholar]
- 6.Benjamin H, Kleckner N. Excision of Tn10 from the donor site during transposition occurs by flush double-strand cleavages at the transposon termini. Proc Natl Acad Sci USA. 1992;89:4648–4652. doi: 10.1073/pnas.89.10.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calvi B, Hong T, Findley S, Gelbart W. Evidence for a common evolutionary origin of inverted repeat transposons in Drosophila and plants: hobo, Activator, and Tam3. Cell. 1991;66:465–471. doi: 10.1016/0092-8674(81)90010-6. [DOI] [PubMed] [Google Scholar]
- 8.Chang L, Bollum F. Molecular biology of terminal transferase. Crit Rev Biochem. 1986;21:27–52. doi: 10.3109/10409238609113608. [DOI] [PubMed] [Google Scholar]
- 9.Coen E, Robbins T, Almeida J, Hudson A, Carpenter R. Consequences and mechanisms of transposition in Antirrhinum majus. In: Berg D, Howe M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 413–436. [Google Scholar]
- 9a.Coïc, E., V. Puzos, V. Haedens, and V. Colot. Unpublished data.
- 9b.Colot, V., and O. Lespinet. Unpublished data.
- 10.Colot V, Rossignol J-L. Isolation of the Ascobolus immersus spore color gene b2 and study in single cells of gene silencing by methylation induced premeiotically. Genetics. 1995;141:1299–1314. doi: 10.1093/genetics/141.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ezekiel U, Sun T, Bozek G, Storb U. The composition of coding joints formed in V(D)J recombination is strongly affected by the nucleotide sequence of the coding ends and their relationship to the recombination signal sequences. Mol Cell Biol. 1997;17:4191–4197. doi: 10.1128/mcb.17.7.4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finnegan D. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989;5:103–107. doi: 10.1016/0168-9525(89)90039-5. [DOI] [PubMed] [Google Scholar]
- 13.Gauss G, Lieber M. Mechanistic constraints on diversity in human V(D)J recombination. Mol Cell Biol. 1996;16:258–269. doi: 10.1128/mcb.16.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grappin P, Audeon C, Chupeau M, Grandbastien M. Molecular and functional characterization of Slide, an Ac-like autonomous transposable element from tobacco. Mol Gen Genet. 1996;252:386–397. doi: 10.1007/BF02173003. [DOI] [PubMed] [Google Scholar]
- 15.Haber J E. In vivo biochemistry: physical monitoring of recombination induced by site-specific endonucleases. Bioessays. 1995;17:609–620. doi: 10.1002/bies.950170707. [DOI] [PubMed] [Google Scholar]
- 16.Mekki-Berrada A, Rossignol J-L, Paquette N. Haute fréquence de réversion d’un mutant chez Ascobolus immersus. C R Acad Sci (Paris) 1976;283:971–974. [PubMed] [Google Scholar]
- 17.Nadel B, Feeney A. Influence of coding-end sequence on coding-end processing in V(D)J recombination. J Immunol. 1995;155:4322–4329. [PubMed] [Google Scholar]
- 18.Nadel B, Feeney A. Nucleotide deletion and P addition in V(D)J recombination: a determinant role of the coding-end sequence. Mol Cell Biol. 1997;17:3768–3778. doi: 10.1128/mcb.17.7.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicolas A, Hamza H, Mekki-Berrada A, Kalogeropoulos A, Rossignol J-L. Premeiotic and meiotic instability generates numerous b2 mutation derivatives in Ascobolus. Genetics. 1987;116:33–43. doi: 10.1093/genetics/116.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oettinger M. Cutting apart V(D)J recombination. Curr Opin Genet Dev. 1996;6:141–145. doi: 10.1016/s0959-437x(96)80042-6. [DOI] [PubMed] [Google Scholar]
- 21.Peacock W, Dennis E, Gerlach W, Sachs M, Schwartz D. Insertion and excision of Ds controlling elements in maize. Cold Spring Harbor Symp Quant Biol. 1984;49:347–354. doi: 10.1101/sqb.1984.049.01.041. [DOI] [PubMed] [Google Scholar]
- 22.Rosenthal A, Coutelle O, Craxton M. Large-scale production of DNA sequencing templates by microtitre format PCR. Nucleic Acids Res. 1993;21:173–174. doi: 10.1093/nar/21.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roth D, Chang X, Wilson J. Comparison of filler DNA at immune, nonimmune, and oncogenic rearrangements suggests multiple mechanisms of formation. Mol Cell Biol. 1989;9:3049–3057. doi: 10.1128/mcb.9.7.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saedler H, Nevers P. Transposition in plants: a molecular model. EMBO J. 1985;4:585–590. doi: 10.1002/j.1460-2075.1985.tb03670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sargent R, Brenneman M, Wilson J. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol Cell Biol. 1997;17:267–277. doi: 10.1128/mcb.17.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Schlissel M S. Structure of nonhairpin coding-end DNA breaks in cells undergoing V(D)J recombination. Mol Cell Biol. 1998;18:2029–2037. doi: 10.1128/mcb.18.4.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott L, LaFoe D, Weil C. Adjacent sequences influence DNA repair accompanying transposon excision in maize. Genetics. 1996;142:237–246. doi: 10.1093/genetics/142.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Gent D, Mizuuchi K, Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996;271:1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- 28.van Luenen H, Colloms S, Plasterk R. The mechanism of transposition of Tc3 in C. elegans. Cell. 1994;79:293–301. doi: 10.1016/0092-8674(94)90198-8. [DOI] [PubMed] [Google Scholar]
- 29.Vos J, De Baere I, Plasterk R. Transposase is the only nematode protein required for in vitro transposition of Tc1. Genes Dev. 1996;10:755–761. doi: 10.1101/gad.10.6.755. [DOI] [PubMed] [Google Scholar]
- 30.Wang P, Zhou R H, Zou Y, Jackson-Cook C K, Povirk L F. Highly conservative reciprocal translocations formed by apparent joining of exchanged DNA double-strand break ends. Proc Natl Acad Sci USA. 1997;94:12018–12023. doi: 10.1073/pnas.94.22.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]