Abstract
Gene targeting of transcription factor PU.1 results in an early block to fetal hematopoiesis, with no detectable lymphoid or myeloid cells produced in mouse embryos. Furthermore, PU.1−/− embryonic stem (ES) cells fail to differentiate into Mac-1+ and F4/80+ macrophages in vitro. We have previously shown that a PU.1 transgene under the control of its own promoter restores the ability of PU.1−/− ES cells to differentiate into macrophages. In this study, we take advantage of our PU.1−/− ES cell rescue system to genetically test which previously identified PU.1 functional domains are necessary for the development of mature macrophages. PU.1 functional domains include multiple N-terminal acidic and glutamine-rich transactivation domains, a PEST domain, several serine phosphorylation sites, and a C-terminal Ets DNA binding domain, all delineated and characterized by using standard biochemical and transactivational assays. By using the production of mature macrophages as a functional readout in our assay system, we have established that the glutamine-rich transactivation domain, a portion of the PEST domain, and the DNA binding domain are required for myelopoiesis. Deletion of three acidic domains, which exhibit potent transactivation potential in vitro, had no effect on the ability of PU.1 to promote macrophage development. Furthermore, mutagenesis of four independent sites of serine phosphorylation also had no effect on myelopoiesis. Collectively, our results indicate that PU.1 interacts with important regulatory proteins during macrophage development via the glutamine-rich and PEST domains. The PU.1−/− ES cell rescue system represents a powerful, in vitro strategy to functionally map domains of PU.1 essential for normal hematopoiesis and the generation of mature macrophages.
Hematopoiesis is a dynamic developmental process that ensures a sufficient supply of terminally differentiated blood cell types essential for survival. Mature blood cells include erythrocytes, megakaryocytes, monocytes, granulocytes and lymphocytes, derived from a population of pluripotent hematopoietic stem cells. Regulation of this process is controlled by an array of extracellular and intracellular signals acting in the context of a multicellular microenvironment. Transcription factors are crucial intracellular signals regulating hematopoietic development by initiating and modulating gene expression required for growth and differentiation. The Ets transcription factor PU.1 (Spi-1) is specifically expressed in hematopoietic organs, with the highest levels detected in lymphoid and myeloid cells (11, 15). We have previously shown that mutagenesis of PU.1 results in a cell-intrinsic block to the development of B cells, T cells, macrophages, and neutrophils in the yolk sac and fetal liver without affecting the generation of erythrocytes or megakaryocytes (29, 43, 44). The primary target appears to be a multipotential lymphoid-myeloid progenitor population, since such cells are significantly reduced in PU.1 mutant fetal livers and do not differentiate into B cells or macrophages in vitro (43). These results suggest that PU.1-dependent genes are involved either in the production of lymphoid-myeloid progenitors from hematopoietic stem cells or in the proliferative and differentiative potential of such progenitors.
The embryonic lethal nature of the PU.1 mutation has made further analysis of PU.1 function during hematopoietic development difficult to study. The ability of embryonic stem (ES) cells to differentiate in culture offers an attractive in vitro system to further understand the function of PU.1 (reviewed in reference 46). In the absence of leukemia-inhibitory factor and stromal cell contact, ES cells form embryoid bodies and differentiate into hematopoietic cells including erythrocytes, granulocytes, macrophages, mast cells, and megakaryocytes (3, 6, 17, 41, 47). The differentiation program of ES cells is thought to resemble that which occurs during yolk sac and fetal liver hematopoiesis.
The ability of PU.1 to promote expression of lymphoid and myeloid genes has prompted several groups to define the functional domains of PU.1 by using both nonhematopoietic and B-cell lines. Multiple transactivation domains in the N terminus (amino acids 7 to 100) of the protein have been identified (12, 20, 23, 45). Three domains are rich in acidic amino acids (amino acids 7 to 32, 33 to 55, and 56 to 74), while a fourth is glutamine rich (amino acids 75 to 100). One of the acidic activation domains contains two serine phosphorylation sites (amino acids 41 and 45) determined to be necessary for macrophage colony-stimulating factor (M-CSF)-dependent proliferation of bone marrow macrophages (4). Downstream of the transactivation region is the PEST domain of PU.1 (amino acids 118 to 160), which is rich in proline (P), glutamic acid (E), serine (S), and threonine (T) residues. PEST domains are thought to be involved in controlling protein stability (38) and can target proteins for proteolytic degradation (36). The PEST region is not required for transactivation as measured by transient transfection assays in tumor-derived cell lines (9, 12, 20, 23). However, a phosphorylation site at serine 148 in the PEST domain mediates the interaction between PU.1 and the PU.1-interacting partner (Pip) in vitro (32, 33). Pip, a member of the interferon stimulatory family of transcription factors, is expressed predominantly in lymphoid cells (8, 25, 49). The PU.1-Pip interaction is required for optimal transactivation of 3′ enhancer regulatory elements located in the kappa and lambda immunoglobulin light-chain genes in nonhematopoietic cell lines (8, 33). Reduced serum immunoglobulin levels and impaired humoral responses in Pip-deficient mice confirm that both PU.1 and Pip are critical for B cell function in vivo (26). DNA binding activity resides in the 85-amino-acid Ets domain located in the carboxy terminus of PU.1 (16, 19). Recently, a second transcription factor, NF-IL6β (C/EBPδ [CAAT enhancer binding protein]), has been shown to interact directly with PU.1 and synergistically activate transcription (28). The interaction site for NF-IL6β maps to the C-terminal 28 amino acids of PU.1.
We and others have demonstrated that PU.1−/− ES cells fail to differentiate into macrophages in vitro (13, 29). The failure of PU.1−/− ES cells to differentiate into macrophages is complemented by a PU.1 transgene under the control of its own promoter (29), proving that PU.1 promotes macrophage differentiation from totipotent ES cells. We now exploit this in vitro differentiation system to examine a series of mutant PU.1 transgenes and assess which previously identified functional domains of PU.1 are necessary for myeloid differentiation. Our study represents the first time that a protein has been functionally dissected in the context of normal differentiating ES cells. The PU.1−/− ES cell in vitro differentiation experimental system is unique for the following two reasons: (i) we have not used a virally transformed cell line derived from ES cells, thus eliminating the unknown effects of the transforming agent on the differentiative program under study (46); and (ii) expression of the mutant PU.1 transgenes was placed under the control of the endogenous PU.1 promoter without the addition of heterologous regulatory regions (2). The goal is that such transgenes will be expressed similarly to the endogenous PU.1 gene. The importance of proper PU.1 expression is highlighted by studies that have demonstrated that expression of PU.1 outside the hematopoietic system or abnormally high levels of ectopically expressed PU.1 within the hematopoietic system can result in apoptosis (42, 48).
Our results show that a subregion of the PEST domain is required for the differentiation of Mac-1+ and F4/80+ macrophages from ES cells. This does not involve the previously identified serine 148 residue which is important for controlling gene expression in the B-cell lineage. In addition, the glutamine-rich transactivation region was essential for myelopoiesis. The DNA binding domain alone was incapable of promoting macrophage development. Surprisingly, none of the acidic transactivation regions had a role in myeloid development. Our inability to demonstrate the necessity of the acidic transactivation domains suggests a possible specialized role for such domains in controlling genes not involved in myeloid development. Identifying both the PEST and glutamine-rich regions as being functionally important allows us to present a working model of how PU.1 is able to regulate genes that are critical for myeloid development via protein-protein interactions with these domains.
MATERIALS AND METHODS
In vitro transcription and translation.
All mutants in the context of plasmid pBluescript KS+ were transcribed with T3 polymerase and translated with a rabbit reticulocyte lysate (Promega) in the presence of [35S]methionine according to the manufacturer’s specifications. Translation products were subjected to electrophoresis on sodium dodecyl sulfate (SDS)-polyacrylamide gels (PAGE) and visualized by autoradiography.
Metabolic labeling and immunoprecipitation experiments.
A modification of the method of Luscher and Eisenman (23a) was used. NIH 3T3 cells were transfected by the calcium phosphate coprecipitation method (11b) with plasmids expressing PU.1 or derivatives. After 24 h, cells were incubated for 2 h with 0.2 mCi of 35S-labeling mix (NEN Expre[35S][35S]) per ml. Cells were harvested in 20 mM Tris-HCl (pH 7.5)–50 mM NaCl–0.5% SDS–0.5% deoxycholate–1 mM dithiothreitol–leupeptin (10 μg/ml)–Nα-p-tosyl-l-arginine methyl ester (TAME) (10 μg/ml)–pepstatin (1 μg/ml)–1 mM phenylmethylsulfonyl fluoride, sonicated, and centrifuged. Samples were immunoprecipitated with anti-PU.1 antibodies (Santa Cruz Biotechnology) and resolved by SDS-PAGE.
CAT assays.
Transient expression assays were performed by the calcium phosphate coprecipitation method of Graham and van der Eb (11b). Cell lysates were prepared and chloramphenicol acetyltransferase (CAT) assays were performed as described by Gorman et al. (11a).
Generation of the PURI cassette.
With the exception of the serine 148 mutant, PU.1 mutant cDNAs were subcloned as EcoRI fragments into the PURI expression vector (see Fig. 2A). A previously described PU.1 transgene called Putrans (29) containing exon 1 of PU.1, 6 kb of 5′ flanking sequences, and the human growth hormone (hGH) polyadenylation region was modified as follows. The EcoRI site located at the 5′ boundary of the 5′ flanking region was eliminated by a Klenow fill-in reaction, leaving a unique EcoRI site at the 3′ end of the PU.1 cDNA. A novel EcoRI site was engineered into the proximal 5′ flanking region (+95 to +100) (27) by site-directed mutagenesis using the Sculpture in vitro mutagenesis system (Amersham International, Buckinghamshire, England) according to the manufacturer’s instructions. Briefly, mutagenesis reactions were initiated by using a single-stranded DNA template isolated from a PU.1 M13mp19 recombinant along with a mutant primer containing a novel EcoRI site (5′-GGG TCC GCCTgaAtTCCC ACC GAA GCA CC-3′) (the EcoRI recognition site is underlined; lowercase letters indicate mutations). This PU.1 M13 clone was prepared by subcloning a 3.0-kb XbaI fragment containing the PU.1 cDNA, hGH polyadenylation region, and proximal 5′ flanking region. The resulting mutagenized XbaI fragment was inserted into a partially XbaI digested Putrans to generate the PURI cassette, which was characterized by restriction enzyme and DNA sequence analyses.
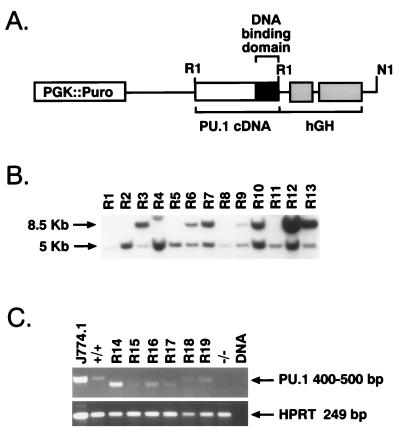
FIG. 2.
Rescue of macrophage development in PU.1−/− ES cells. (A) Design of the PURI expression vector. This construct includes 6.0 kb of the PU.1 5′ region, 135 nucleotides of PU.1 5′ untranslated sequences, exons 4 and 5 of the hGH gene (including the polyadenylation signal), and EcoRI sites flanking a wild-type PU.1 cDNA to allow easy replacement of this region with mutant cDNAs. (B) Southern blot analysis of ES cells transformed with the PU.1 complementation vector containing the serine 148 mutation. Of the 13 independent rescued PU.1−/− clones shown (R1 to R13), R3, R6, R7, R9, R10, R12, and R13 harbor intact PU.1 transgenes based on the presence of the indicated 8.5-kb EcoRI restriction fragment. The 5-kb restriction fragment represents the mutant PU.1 allele generated through gene targeting (44). The serine 148 mutant was subcloned into the original PU.1 promoter construct (29) which lacks the upstream EcoRI sites depicted in panel A. Therefore, EcoRI-restricted DNA from S148A transformants exhibit an 8.5-kb fragment. DNAs from other ES clones contained R1 fragments of various sizes depending on the amount of coding sequences deleted from each PU.1 cDNA (see Materials and Methods). (C) RT-PCR of rescued clones transfected with Δ75-100 (R14, R15, R16, and R17) and Δ2-30 (R18 and R19) showing expression of PU.1 mRNA. RNA isolated from the mature macrophage cell line J774.1 was used as a positive control; ES DNA serves as the negative control for RT-PCR. As expected, RNA harvested from PU.1+/+ ES cells contained PU.1 mRNA, while PU.1−/− cells did not exhibit PU.1 transcripts. RT products were normalized by HPRT expression.
PU.1 mutant transgenes.
PU.1 cDNA deletions were constructed via PCR amplification using Pfu DNA polymerase (Stratagene) or AmpliTaq DNA polymerase (Perkin-Elmer/Cetus) and the wild-type PU.1 cDNA fragment as a template (19). The N-terminal transactivation deletion Δ2-30 or deletion Δ2-167, containing the DNA binding domain alone, was generated with the appropriate primer pairs listed in Table 1. PU.1 internal deletions removing transactivation domains (Δ3-21, Δ33-74, and Δ75-100) or portions of the PEST domain (Δ141, Δ118-167, Δ118-132, and Δ141-167) were produced by using a pair of internal primers listed in Table 1 along with a set of external primers. Transactivation mutant Δ33-100 was constructed as previously described (32). Transactivation mutant Δ33-100, NF-IL6β/C/EBPδ binding mutant (Δ245-272), and serine phosphorylation mutants (S148A and S41/45) were created as previously discussed (32, 33). The integrity of each PU.1 mutant construct was confirmed by DNA sequence analysis, and all deletion mutations were subcloned as full-length clones into the R1 site of the PURI cassette.
TABLE 1.
Oligonucleotide primers used for generating PU.1 mutants
| Category | Sequence |
|---|---|
| Transactivation mutants | |
| Δ3-21 | |
| Forward | 5′-CCC GAG CTC AGC TGG ATG TTA GTT ACT TAC GAT TCAG AGC TA-3′ |
| Reverse | 5′-CCC GAA TTC GGG AGA ATA GCT GTC AAT TTT AC-3′ |
| Δ2-30 | |
| Forward | 5′-GCG TCT AGA GGA TGC CAA TGC ATG ACT ACT ACT CC-3′ |
| Reverse | 5′-GCG TCT AGA CCA GCT GAG CTC CAG GTT GTT-3′ |
| Δ33-74 | |
| Forward | 5′-CGG GAT CCC AGA GCG TGC AGC CCC C-3′ |
| Reverse | 5′-CGG GAT CCC ATT GGA GGT TGG TAT AGC-3′ |
| Δ75-100 | |
| Forward | 5′-CGG GAT CCA TGG TGC CAC CCC ACA CCG GC-3′ |
| Reverse | 5′-CGG GAT CCC AGC TCT GTG AAG TGG-3′ |
| Transactivation domain | 5′-GCA GGA ATT CAT TTA GGT G-3′ |
| External primers | 5′-GGG GGA TCC GCC TGA ATT CCC ACC GAA GCA 3-′ |
| PEST domain mutants | |
| Δ118-167 | |
| Forward | 5′-GCG AGA TCT AAG AAA AAG ATT CGC CTG TAC CAG-3′ |
| Reverse | 5′-GCG AGA TCT GGG CAT GTA GGA AAC CTG GTG-3′ |
| Δ118-132 | |
| Forward | 5′-GCG AGA TCT GAT GAG GAG GGT GAG AGG-3′ |
| Reverse | 5′-GCG AGA TCT GGG CAT GTA GGA AAC CTG GTG-3′ |
| Δ129-141 | |
| Forward | 5′-GCG AAG CTT GCT AGC CCT CCC CTG GAG GTG TGT GATG-3′ |
| Reverse | 5′-GCG AAG CTT GCT AGC TGG GGA CAA GGTTTG ATA AGG-3′ |
| Δ141-167 | |
| Forward | 5′-GCG ATA TCT AAG AAA AAG ATT CGC CTG TAC CAG-3′ |
| Reverse | 5′-GCG AGA TCT CTC ACC CTC CTC CTC ATC TGA-3′ |
| PEST domain | 5′-AATTAA CCCTCA CTA AAG GG-3′ |
| External primers | 5′-GCG TCT AGA CGG TCT CTG GGG GCG ATC AGT GGG G-3′ |
| DNA binding domain mutant | |
| Δ5-167 | |
| Forward | 5′-CCC GAGCTC AGC TGG ATG AGC AAG AAA AAG AAG ATT CGC CTG TAC-3′ |
| Reverse | 5′-CCC CTG CAG AAG GAA GCA CCG TGG CCA CTA AG-3′ |
A puromycin resistance (Purr) gene regulated by the PGK promoter was subcloned into a unique XhoI site in all PU.1 transgenes. Following linearization with NotI, mutant transgenes were electroporated into PU.1−/− ES cells (29). ES cell stable transformants underwent puromycin selection (2 μg/ml) for 7 to 10 days.
Analysis of ES cell clones.
Southern blot analysis of EcoRI-digested genomic DNA isolated from Purr clones was performed with a PU.1 cDNA probe (19). ES cell clones were differentiated in vitro for 11 days in 0.9% methylcellulose medium containing 10% fetal bovine serum (HyClone), human interleukin-1α (IL-1α; 7.5 × 102 U/ml), murine stem cell factor (SCF; 10 ng/ml), murine granulocyte-macrophage colony-stimulating factor (GM-CSF; 0.5 ng/ml), murine IL-3 (5 ng/ml), human erythropoietin (EPO; 2 U/ml; Amgen, Thousand Oaks, Calif.), and 4.5 × 10−4 M α-monothiaglycerol (29). The IL-1α, SCF, GM-CSF, and IL-3 were a gift of Genetics Institute (Boston, Mass.). At day 11, embryoid bodies (EBs) were picked and erythrocytes, macrophages, neutrophils, and early megakaryocytes were identified by cytospin preparations stained with May-Grunwald-Giemsa (MGG) stain.
CD11b and F4/80 expression was analyzed after culturing cell suspensions prepared from EBs in eight-chamber slides (Nunc, Naperville, Ill.) for 24 to 48 h at 37°C as described previously (29). Briefly, fixed adherent cells were stained with rat monoclonal anti-mouse CD11b (PharMingen; San Diego, Calif.) or F4/80 (Caltag, San Francisco, Calif.) antibodies, using either a Vectastain ABC-alkaline phosphatase or horseradish peroxidase kit (Vector Laboratories, Inc., Burlington, Calif.).
Total cellular RNA was isolated from day 11 EBs by using RNAzol (Tel-Test, Woodlands, Tex.) or TRIzol (Gibco BRL, Grand Island, N.Y.) according to the manufacturer’s specifications. cDNA was synthesized from total RNA by using a First Strand cDNA synthesis kit (Pharmacia Biotech). Reverse transcription (RT)-PCR analysis to detect PU.1, HPRT, and M-CSF receptor (M-CSFR) RNA levels was performed as described by Olson et al. (29) except that a 5′ primer (5′-GGG GGA TCC GCC TGA ATT CCC ACC GAA GCA GG-3′) was used for detecting PU.1 mRNA in the Δ33-74 RNA samples.
RESULTS
Strategy to identify PU.1 functional domains.
We had previously observed that PU.1−/− ES cells are incapable of differentiating into macrophages in vitro. However, macrophage production is restored in the PU.1−/− ES cells upon introduction of a PU.1 cDNA expression construct controlled by approximately 6 kb of 5′ flanking region from the PU.1 gene (29). Interestingly, the 6-kb native promoter was the only promoter that rescued myeloid development. The 6-kb promoter restored expression of PU.1 mRNA to wild-type levels in an appropriate temporal fashion during in vitro differentiation. The wild-type PURI expression cassette (see Fig. 2) rescues myeloid development as well as a 100-kb P1 clone containing the entire PU.1 locus, suggesting that cDNA expression is regulated similarly to the native locus (data not shown). In contrast, approximately 1 kb of promoter was insufficient for high levels of PU.1 expression (data not shown). Furthermore, multiple heterologous promoters expressing the PU.1 cDNA failed to promote myeloid development and were actually deleterious to the PU.1−/− ES cells. The generation of macrophages from totipotent, undifferentiated ES cells is monitored by the expression of gene products associated with terminal myeloid differentiation, including CD11b, F4/80, and M-CSFR. To further our understanding of how PU.1 may be involved in committing cells to the myeloid lineage, we sought to identify which previously defined functional domains of PU.1 are required to promote the differentiation of ES cells along the myeloid pathway.
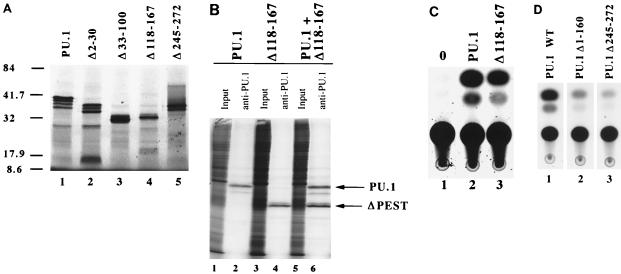
As described above, PU.1 contains three functionally characterized domains. These include the transcriptional activation domain (residues 7 to 100), the PEST domain (residues 118 to 160), and the Ets DNA binding domain (residues 168 to 255) (9, 12, 20, 22, 32, 45). PU.1 mutant cDNA derivatives containing various deletions within each of these functional domains were generated. The integrity of each PU.1 mutant construct was confirmed by the ability to produce the appropriate-size protein upon in vitro transcription, translation, and SDS-PAGE. Selected mutants are shown in Fig. 1A. To confirm that each deletion mutant retains the ability to bind DNA, electrophoretic mobility shift assays (EMSAs) were performed with a PU.1 binding site probe (data not shown). Furthermore, mutants were tested for their stability when expressed in transfected cells. Figure 1B shows immunoprecipitation of metabolically labeled PU.1 proteins produced by transient transfection of NIH 3T3 cells. Importantly, the Δ118-167 deletion mutant, which is lacking the entire PEST domain, shows protein stability characteristics similar to those of wild-type PU.1 protein (Fig. 1B, lane 6). Where appropriate, mutants were also tested for the ability to activate expression of a PU.1-dependent CAT reporter construct. Figure 1C illustrates the ability of Δ118-167 to activate transcription in this assay. In all cases, the mutants used in this study expressed stable proteins and displayed functional properties that were consistent with the mutation.
FIG. 1.
Stable expression of and transactivation by PU.1 mutant proteins. (A) SDS-PAGE of in vitro-transcribed and -translated PU.1 mutant proteins. All constructs produced proteins of the predicted molecular sizes (indicated in kilodaltons) based on the engineered mutation. (B) Immunoprecipitation of metabolically labeled PU.1 and Δ118-167 proteins. Lanes 1, 3, and 5, total labeled protein prior to immunoprecipitation; lanes 2, 4, and 6, anti-PU.1 antibody-precipitated proteins of the predicted molecular weights for the engineered mutation. All PU.1 variants were equivalently expressed and stable in NIH 3T3 cells. (C) CAT transactivation assays of PU.1 and Δ118-167, using a multimerized PU.1 binding site from the immunoglobulin kappa 3′ enhancer region (34). Lane 1, cells containing the reporter construct and no PU.1 protein; lane 2, activation by the wild-type PU.1 protein; lane 3, activation by Δ118-167 mutant protein. (D) CAT transactivation of the same kappa 3′ enhancer element, using wild-type (WT) and mutant PU.1 proteins with Pip, c-Fos, and c-Jun as described elsewhere (35).
The PURI expression vector (Fig. 2A) was designed to facilitate mutant cDNA transgene construction (see Materials and Methods). PU.1−/− ES cells were initially generated by sequential gene targeting of each allele of the murine PU.1 locus (29). Since PU.1−/− ES cells are G418r/Hygr, a cDNA encoding Purr was incorporated into PURI to select for ES transfectants (Fig. 2A). Expression plasmids encoding PU.1 deletion mutants were transfected into PU.1−/− ES cells, and stable integrants were selected for Purr. For each mutation, 24 clones were isolated, expanded, and analyzed for integrity and copy number of the inserted transgenes. The Southern blot in Fig. 2B shows a characteristic hybridization pattern for Purr PU.1−/− ES clones stably transfected with the PU.1 expression vector. RNA was isolated from ES cell transfectants differentiated in vitro for 11 days in methylcellulose cultures containing the cytokines EPO, SCF, GM-CSF, IL-1α, and IL-3. RT-PCR was used to detect the relative level of PURI transgene expression (Fig. 2C). As expected, the size of the PCR products varied depending on the extent of the deleted region. Due to the small proportion (20 to 30% on average) of macrophages in our day 11 methylcellulose cultures and the great difficulty of detecting PU.1 protein levels in normal cells using either in situ immunohistochemistry or Western blotting with the available anti-PU.1 immunological reagents, we relied on RT-PCR to demonstrate that ES cell transfectants containing mutant PU.1 transgenes were being expressed at wild-type levels. For each mutant PU.1 transgene, four ES clones expressing wild-type levels were selected for further study (15, 43).
During in vitro differentiation, ES cells form EBs, which are three-dimensional structures that include a variety of hematopoietic cells such as primitive erythrocytes, definitive erythrocytes, macrophages, neutrophils, and megakaryocytes (6, 17). After 11 days in culture, PU.1+/+ and PU.1−/− EBs and EBs produced from various rescued ES clones were scored for the presence of macrophages. Successful rescue of macrophage differentiation was defined by cell morphology following MGG staining and immunohistochemical detection of Mac-1 and F4/80 surface expression (Fig. 3, 4, and 5). Mac-1 (CD11b), an integrin cell surface protein, is expressed by both mature macrophages and granulocytes, and its expression increases during myeloid differentiation (39). F4/80, a member of the seven-transmembrane region family of cell surface molecules, is expressed specifically on macrophages (1). Further evidence of successful macrophage development was the expression of M-CSFR RNA as measured by RT-PCR analysis. M-CSFR is a myeloid lineage-specific marker and may be involved in committing granulomonocytic progenitors into monocytes (30). Wild-type EBs produced abundant macrophages, whereas PU.1−/− EBs did not (Fig. 3A and C; Fig. 4A to D). PU.1−/− ES cells that carried the wild-type PU.1 cDNA transgene rescued macrophage development (Fig. 3E, 4E, and 4F). Unlike PU.1−/− EBs, the rescued EBs also expressed the M-CSFR based on RT-PCR analysis (data not shown). Importantly, each independent PU.1−/− ES clone harboring a PU.1 cDNA transgene analyzed differentiated into macrophages in vitro.
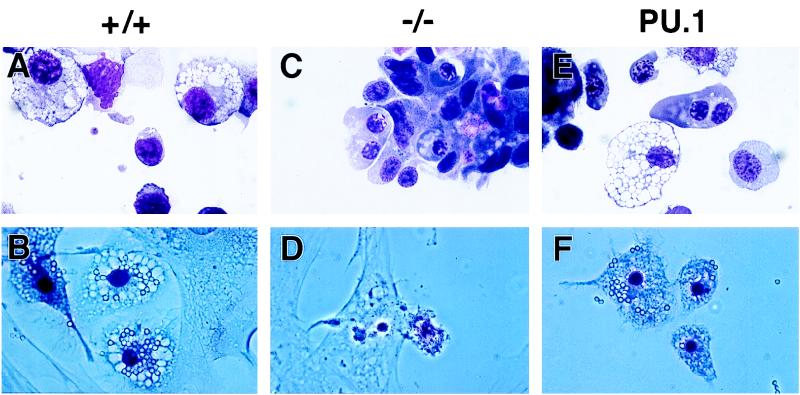
FIG. 3.
Representative cells from day 11 EB cultures. MGG-stained hematopoietic cells in cultures derived from PU.1+/+ ES cells (+/+), PU.1−/− ES cells (−/−), and a PU.1−/− ES cell clone transformed with the wild-type PU.1 transgene (PU.1) are shown. Macrophages capable of phagocytizing latex beads (after 4 h at 37°C) are clearly apparent in the PU.1+/+ and rescued cultures and absent from the PU.1−/− cultures. Light-field photomicrographs are depicted in panels A, C, and E, while phase-contrast photomicrographs are shown in panels B, D, and F. Magnification, ×69.
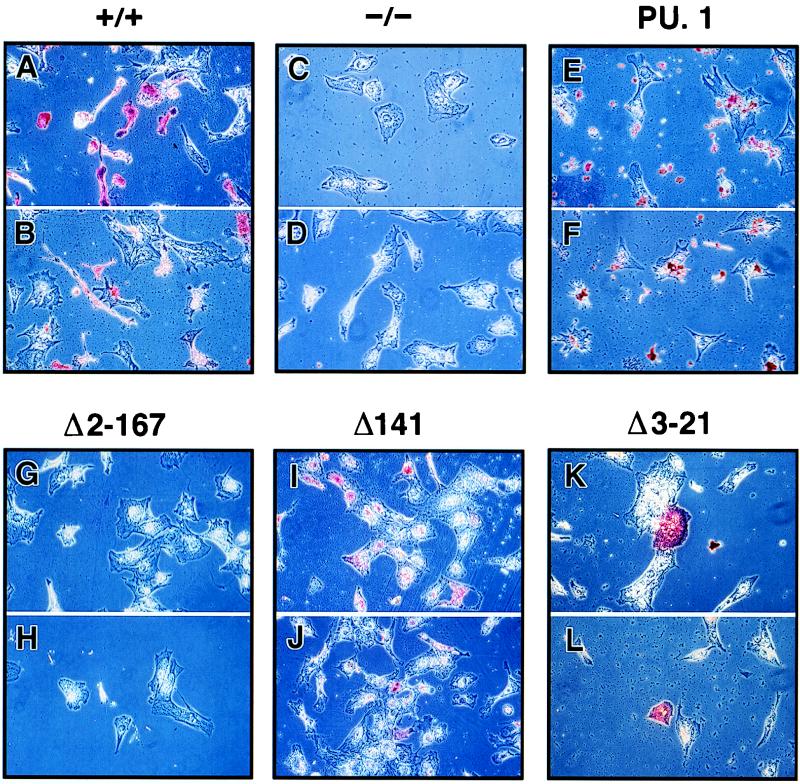
FIG. 4.
Identification of macrophages by immunocytochemical staining. Phase-contrast photomicrographs of staining by antibodies for CD11b (A, C, E, G, I, and K) and F4/80 (B, D, F, H, J, and L) are shown for differentiated PU.1+/+ ES cells, PU.1−/− cells, and transformants containing wild-type (PU.1), Δ2-167, Δ141, and Δ3-21 cDNAs. Magnifications: A to J and L, ×36; K, ×90.
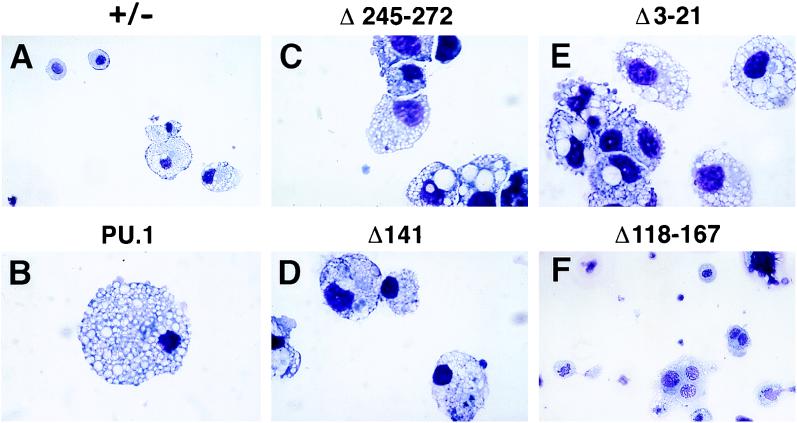
FIG. 5.
Representative macrophages obtained from day 11 EB cultures derived from ES clones transfected with wild-type and mutant cDNAs. MGG cytological stains for macrophages detected in PU.1+/− cultures and cultures transfected with wild-type (PU.1), Δ245-272, Δ141, Δ3-21, and Δ2-167 transgenes are shown. Magnifications: A and F, ×27; B to E, ×68.
As a final test of our rescue system, macrophages produced by in vitro differentiation were tested for the ability to phagocytose latex beads (Fig. 3). PU.1+/+ ES cells differentiated into mature macrophages, as demonstrated by MGG staining (Fig. 3A), were also capable of phagocytosing latex beads (Fig. 3B). In contrast, PU.1−/− ES cells do not produce functional macrophages upon differentiation (Fig. 3C and D). Introduction of the wild-type PU.1 transgene, however, rescues the ability of PU.1−/− ES cells to produce cells with the morphology of mature macrophages capable of phagocytosis (Fig. 3E and F). Therefore, our rescue system is capable of assaying the ability of mutant PU.1 transgenes to support the development of mature macrophages from totipotent ES cells.
The Ets domain alone is insufficient for macrophage differentiation.
PU.1 binds to DNA via its carboxy-terminal Ets domain. Since this domain is crucial for DNA binding, one would expect the Ets domain to be required for macrophage differentiation. It was recently reported that the DNA binding domain of the GATA-1 transcription factor alone is sufficient to alleviate the block in terminal differentiation of the GATA-1− G1E erythroblastic cell line (46). To determine if a similar situation holds true for PU.1, we tested a PU.1 cDNA mutant which encodes the DNA binding domain but not the transactivation and PEST domains (Δ2-167).
As shown in Fig. 4G, 4H, and 6, Δ2-167 failed to restore terminal macrophage development to PU.1−/− cells. None of the Δ2-167 ES clones were able to produce CD11b+ or F4/80+ cells or cells with macrophage morphology, based on MGG staining. Therefore, the DNA binding domain of PU.1 alone is not sufficient to generate myeloid differentiation. This finding suggests that additional domains of PU.1 are required to work in conjunction with the Ets DNA binding domain to promote macrophage differentiation.
FIG. 6.
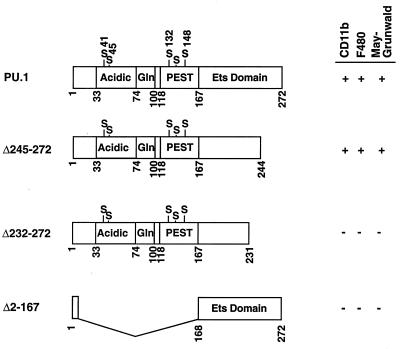
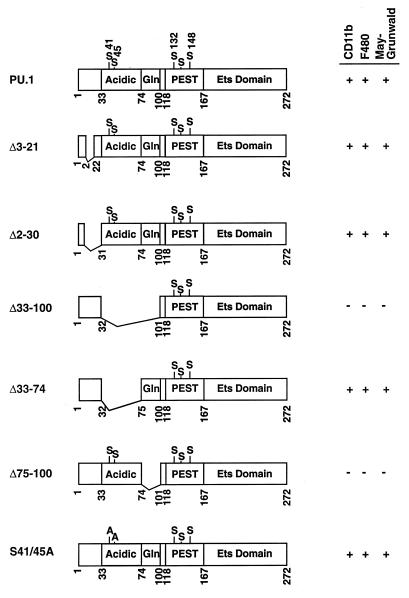
Diagram of wild-type and deletion mutant PU.1 cDNAs tested in the ES cell system. Numbers indicate the positions of amino acid residues in PU.1; the acidic, glutamine-rich, PEST, and Ets-type DNA-binding regions are represented as boxes. Serine residues thought to be phosphorylated are also indicated. PURI constructs capable of promoting myeloid development were scored for the presence of CD11b+ or F4/80+ cells or cells with the morphology of macrophages upon MGG staining.
An additional PU.1 mutation (Δ245-272) that deletes the NF-IL6β/C/EBPδ binding site (28) and carboxyl 10 amino acids of the Ets DNA binding domain (extending from residues 168 to 255) was examined. The Δ245-272 PU.1 protein also fails to bind DNA as assayed by EMSA (32). Interaction of PU.1 and C/EBP family members is thought to play a critical role in the expression of multiple myeloid genes (reviewed in reference 10). Surprisingly, the Δ245-272 mutant was capable of supporting macrophage differentiation by all criteria examined, including the production of CD11b+, F4/80+, and MGG+ cells (Fig. 5C and 6). These results suggest that Δ245-272 is still capable of interacting with other proteins in vivo to promote myeloid gene expression. It is possible that this mutant is capable of independently binding DNA in vivo under conditions that are not mimicked during in vitro EMSA. Alternatively, PU.1 is recruited to myeloid promoters in vivo by protein-protein interactions involving factors other than C/EBP family members. Previously, we showed that the immunoglobulin kappa enhancer can be activated in NIH 3T3 cells by cotransfection with PU.1, Pip, c-Fos, and c-Jun (35). Pip, c-Fos, and c-Jun support a low level of enhancer activity which is greatly stimulated by the addition of PU.1. This stimulation requires both the DNA binding and transactivation domains of PU.1. We tested the ability of our Δ245-272 deletion to transactivate a 3′ enhancer-dependent reporter plasmid in NIH 3T3 cells (Fig. 1D). A wild-type PU.1 transfection served as our positive control, and a construct expressing just the Ets domain (Δ1-160) was our negative control for PU.1 transactivation. The Δ245-272 mutant was unable to activate transcription of the immunoglobulin kappa 3′ enhancer. However, the rescue of myeloid differentiation is unaffected by the Δ245-272 truncation. It seems possible that these results reflect a difference between the regulation of lymphoid and myeloid genes by PU.1. ES cells expressing Δ245-272 activate M-CSFR, CD11b, and F4/80 gene expression. In contrast to Δ245-272, a Δ232-272 mutant protein failed to produce macrophages when introduced into PU.1−/− ES cells (Fig. 6). Therefore, more than 64 amino acids of the 85-amino-acid Ets region are essential for myelopoiesis.
Role of the PEST domain of PU.1 in macrophage differentiation.
The central portion of PU.1 (amino acids 118 to 167) contains a high content of proline, glutamic acid, serine, and threonine. As described earlier, the PU.1 PEST domain is known to function in B cells to recruit a second binding protein, Pip, to DNA. This recruitment requires phosphorylation of serine 148 within the PU.1 PEST domain (33). Macrophages, however, do not express Pip (7, 25, 49). Therefore, the PU.1 PEST domain cannot function via Pip recruitment in the macrophage lineage. Thus, it is possible that the entire PEST domain is dispensable for macrophage differentiation. This would be consistent with the observation that transactivation of PU.1-dependent reporter constructs does not require the PEST domain (9, 12, 20, 23). Alternatively, the PEST domain could be required for myeloid-specific protein-protein interactions.
To test these alternatives, a mutant expression construct lacking the entire PU.1 PEST domain (Δ118-167) was assayed in PU.1−/− ES cells. Interestingly, Δ118-167 failed to rescue macrophage differentiation (Fig. 7). Although PEST sequences have been implicated in targeting proteins for degradation (38), deletion of amino acids 118 to 167 did not increase PU.1 stability (Fig. 1B and C). These data demonstrate a necessary role for the PU.1 PEST domain during macrophage development and suggest that one or more myeloid proteins may interact with PU.1 in this region.
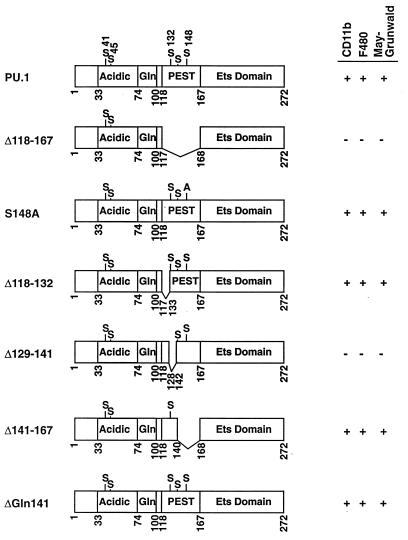
FIG. 7.
Diagram of wild-type and PEST region PU.1 mutant cDNAs examined in the ES cell rescue assay. As shown in Fig. 6, constructs that promoted macrophage differentiation were scored for the presence of CD11b+, F4/80+, and MGG+ cells.
Since mutation of serine 148 to alanine is known to inhibit the PU.1-Pip interaction in B cells (33), we tested the ability of the S148A mutant to rescue macrophage differentiation. In contrast to the results with the Δ118-167 mutant construct, mutation of serine 148 to alanine had no effect on macrophage differentiation (Fig. 7, S148A). To better define the PEST sequences necessary for macrophage differentiation, three deletions (Δ118-132, Δ129-141, and Δ141-167) that collectively span the PEST domain were assayed. Deletions of amino acids 118 to 132 and 141 to 167 had no effect on macrophage differentiation, whereas deletion of residues 129 to 141 yielded a mutant PU.1 protein incapable of supporting differentiation, as evidenced by absence of CD11b+ and F4/80+ cells and no expression of M-CSFR RNA (Fig. 7 and 8). The only known functional motifs of this PU.1 sequence are serine phosphorylation sites at residues 132 and 133 (33). Since serine 132 was deleted in the Δ118-132 mutant that promoted myelopoiesis, it cannot be a required residue. Furthermore, the overlapping nature of our PEST deletions narrows the required residues to an eight-amino-acid region from 133 to 140 that is highly acidic. Of the eight residues, four are glutamic acid, one is aspartic acid, and one is glutamine. It will be of interest in the future to determine whether phosphorylation of serine 133 or the acidic nature of these residues is necessary for macrophage differentiation.
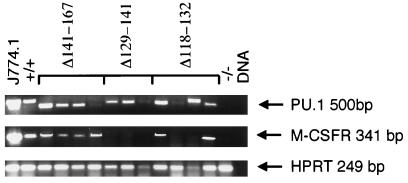
FIG. 8.
Gene expression in day 11 EB cultures derived from ES transformants containing the Δ141-167, Δ129-141, and Δ118-132 cDNA transgenes. Four independent clones were analyzed for the Δ118-132 and Δ141-167 constructs, while three independent clones were analyzed for the Δ129-141 transgene. EB assays were harvested for total cellular RNA, and RT-PCR analyses for PU.1, M-CSFR, and HPRT transcripts were performed. While a 500-bp RT-PCR product was detected in J774.1 and PU.1+/+ ES RNAs, PCR products in clones transfected with the Δ141-167 construct were smaller due to the deletion introduced into the PU.1 cDNA.
A single amino acid deletion at position 141 within the PEST domain was also tested (Fig. 7, Δ141). This mutation is known to recruit Pip to DNA more efficiently than wild-type PU.1 (31). Interestingly, based on the production of CD11b+, F4/80+, and MGG+ cells, this mutation was more efficient at inducing macrophage differentiation than the wild-type PU.1 protein (Fig. 4I, 4J, and 5D). The reason for this increased capacity for differentiation is presently unclear.
Only the glutamine-rich region of the PU.1 transactivation domain is required for normal macrophage differentiation.
The work of Klemsz and Maki (20) showed that the PU.1 transactivation domain encompasses residues 7 to 100. Residues 7 to 30 are moderately acidic, while 33 to 54 and 55 to 74 are highly acidic and exhibit potent transactivation potential in HeLa cells (20). The glutamine-rich region (amino acids 75 to 100) has transactivation activity in both HeLa cells and the lymphoid S194 cell line (20, 45). However, deletion of the glutamine-rich domain results in a modest effect on transactivation by PU.1, while the highly acidic residues of amino acids 33 to 74 are strictly required (20).
We tested whether the N terminus of PU.1 is necessary for myeloid maturation. Truncation of the first 20 amino acid residues results in a decrease in the ability of PU.1 to transactivate reporter genes in HeLa cells (20). Surprisingly, PU.1 proteins lacking the first 20 (Δ3-21) or 30 (Δ2-30) amino acids rescued macrophage development to the same extent as the wild-type protein (Fig. 4K, 4L, 5E, and 9). Both mutants promoted myeloid differentiation by all criteria examined, including the production of CD11b+ and F4/80+ cells and the characteristic macrophage morphology following MGG staining.
FIG. 9.
Diagram of mutant PU.1 cDNAs containing mutations in the transactivation domain (amino acids 1 to 102).
Deletion of both the acidic and glutamine rich regions (Δ33-100) abolished rescue (Fig. 9). In contrast to the in vitro transactivation data, we found that deletion of the two acidic regions from residues 33 to 74 had no effect on macrophage differentiation (Fig. 9, Δ33-74). Instead, deletion of the glutamine-rich domain abolished the ability of these mutant ES cells to differentiate into macrophages (Fig. 9, Δ75-100). The Δ75-100 PU.1 protein was capable of recruiting Pip to DNA and stimulating expression of a reporter construct in transient transfection assays (data not shown). The data presented above indicate that a large portion of the PU.1 protein defined as necessary for transcriptional activation by transient expression assays is dispensable for macrophage differentiation. Only the glutamine-rich sequence from 75 to 100 in the PU.1 transactivation domain is required to support macrophage development.
Two in vivo-phosphorylated serine residues (41 and 45) within the PU.1 activation domain have been implicated in the M-CSF-dependent proliferation of murine bone marrow macrophages (4). These residues are within the dispensable region identified above. We tested whether mutation of these serines to alanines would affect macrophage rescue. As expected, this mutant supported macrophage differentiation, indicating that serine residues 41 and 45 are not critical (Fig. 9).
In summary, our results indicate that at least three segments of PU.1 are necessary for macrophage differentiation. Functional PU.1 regions include amino acids 75 to 100, 133 to 140, and 232 to 245, which lie within the activation, PEST, and DNA binding domains, respectively. Surprisingly, each of these functionally important segments is much smaller than the functional domains identified by either transient transfection or biochemical assays. Large segments of each PU.1 domain can be deleted with no adverse consequences for differentiation of the macrophage lineage in vitro. The significance of these observations is discussed below.
DISCUSSION
ES cell in vitro differentiation offers a useful experimental model to address the role of transcription factors during fetal hematopoiesis. This strategy is of particular use for knockout mice that contain a prenatal lethal mutation as is the case for the Ets family transcription factor PU.1 (44). Our previous studies have shown that deleting the entire DNA recognition domain (amino acids 200 to 272) of the PU.1 gene by homologous recombination blocks normal macrophage development in the fetal liver and yolk sac in vivo and from in vitro-differentiated ES cells (29, 44). Expression of a wild-type PU.1 cDNA transgene rescues the ability of totipotent PU.1−/− ES cells to differentiate into Mac-1+ and F4/80+ macrophages. Here we have reengineered our PU.1 cDNA expression vector to express a panel of mutant transgenes under the control of the endogenous PU.1 promoter. This body of work represents the first study that has systematically characterized the functional domains of a transcription factor required for normal myeloid development from ES cells. By monitoring the generation of mature macrophages in vitro, we have been able to identify multiple domains of PU.1 that are required to complete the myeloid differentiation program.
The DNA binding domain alone does not rescue myeloid development.
A limited set of N-terminal and C-terminal PU.1 deletions were analyzed by gel electrophoresis DNA binding assays to localize amino acids required for DNA binding to residues 170 to 269 (19, 32). Recent X-ray crystallographic analyses of the PU.1 DNA binding domain spanning amino acids 171 to 258 have determined that PU.1 binds its recognition binding site(s) via a winged helix-turn-helix structure (21). Studies of the μ70 enhancer from the immunoglobulin heavy-chain gene showed that the DNA binding domain of PU.1 is sufficient to promote enhancer activity in vitro (9). Likewise, the DNA binding and PEST domains of PU.1 are sufficient to transactivate the kappa light-chain immunoglobulin 3′ core region (32). Both of these studies utilized transient transfection assays for their readout of PU.1 activity. Using an immortalized GATA-1− proerythroblastic cell line, Weiss et al. demonstrated that the zinc finger DNA binding domain alone of transcription factor GATA-1 promotes erythroid maturation in vitro (46). In contrast, PU.1 mutant Δ2-167 failed to rescue macrophage differentiation in mutant ES cells, implying that PU.1 functions in a fundamentally different manner from GATA-1. Furthermore, our results do not correlate with the study of PU.1 function at the B-cell μ70 enhancer. This discrepancy may reflect an intrinsic difference between the mechanisms of how PU.1 modulates gene expression in the lymphoid lineage versus the myeloid lineage. The majority of PU.1 binding sites for the myeloid lineage are TATA-less promoter regions containing Sp1 binding sites. In the lymphoid lineage, the majority of the binding sites are in enhancer regions that assemble multiprotein higher-order complexes (reviewed in reference 10). The PU.1 DNA binding domain is required for myeloid development, as demonstrated by the lack of rescue by the Δ232-272 mutant. Therefore, the DNA binding domain is necessary but not sufficient for myeloid development.
The carboxy terminus of PU.1 is known to interact with several proteins including the transcription factor NF-IL6β/C/EBPδ. C/EBPδ is a member of the leucine zipper family of transcription factors and is induced during inflammation (18). C/EBP binding sites are found adjacent to PU.1 binding sites in many myeloid promoters, and PU.1 and C/EBP family members function together to promote gene expression (10). Deletion of amino acids 245 to 272 prevents this interaction in vitro and blocks DNA binding in EMSA studies (35), consistent with loss of the wing structure formed between β strands 3 and 4 which interacts with the mirror groove of DNA. Surprisingly, when Δ245-272 was expressed in PU.1−/− ES cells, macrophage development was restored. The discrepancies between the in vivo rescue and the EMSA results are unclear but may point to a PU.1-protein interaction (other than C/EBPδ) in vivo that stabilizes the binding of the Δ245-272 PU.1 mutant to target binding sites. The DNA recognition motif as determined from X-ray crystallographic analysis of one PU.1 Ets domain-DNA complex in vitro may not reflect the requirements of the full-length PU.1 protein binding target sites in vivo. However, transient transfection assays with Δ245-272 failed to demonstrate DNA binding and transactivation in NIH 3T3 cells (Fig. 1D). Collectively, these results suggest that PU.1-promoter sequence interactions occur that are specific to myeloid cells.
A subregion of the PEST domain is required for myelopoiesis.
Amino acids 118 to 167 encode the PEST domain of PU.1. Klemsz and Maki (20) demonstrated that the PEST domain is not required for transactivation during transient transfection assays in HeLa cells. This observation was confirmed in studies using an IL-1β promoter-dependent reporter system or GAL4-PU.1 derivatives tested in nonhematopoietic cells (12, 23). When the entire PEST domain was deleted from PU.1 (Δ118-167), no macrophage development was observed. To determine which residues were important for rescue of differentiation, subdomains of the PEST domain were deleted. Overlapping deletions defined a highly acidic eight-amino-acid region from 133 to 140 required for myelopoiesis. How this region functions is uncertain, but it probably is involved in protein-protein interactions. A novel Δ141 mutation that deletes a glutamine in the PEST region has been determined to increase Pip recruitment by PU.1 in vitro (31). The Δ141 protein was able to increase the number of CD11b+ and F4/80+ macrophages. These observations suggest that deletion of glutamine 141 may increase the affinity of PU.1 for its DNA recognition element and/or its myeloid-specific partner and that the acidic PEST subdomain is the site of interaction. Whether this interaction requires phosphorylation of serine 133 remains to be determined. Currently, we have no data to suggest that phosphorylation of PU.1 plays a role in myeloid development, but phosphorylation of serine 133 and interaction with a myeloid cofactor remain an attractive model for future analyses.
The glutamine-rich domain, unlike the acidic domain, is required for macrophage differentiation.
Removal of both previously described acidic and glutamine rich transactivation domains (12, 20, 23, 45) (Δ33-100) resulted in the complete absence of Mac-1+ and F4/80+ macrophages. In contrast, deletion of an additional putative activation domain encompassing the N-terminal 30 amino acids (Δ2-30) had no effect on myeloid development. Klemsz and Maki (20) showed that removal of the N-terminal 20 amino acids from PU.1 resulted in a 4.5-fold drop in transactivation of a reporter gene following transient transfection into PU.1-deficient HeLa cells. The presence of an N-terminal transactivation domain was confirmed by Kominato et al. (23), using an IL-β promoter-dependent reporter gene in phorbol myristic acid-activated HeLa cells. In their experimental system, they lost approximately half of the transcriptional activity by deleting amino acids 8 through 32 from PU.1. Early studies of the yeast transcription factors GCN4 and GAL4 first identified acidic regions as being responsible for transactivation (14, 24). Similar acidic transactivation domains were later identified in the herpes simplex virus VP16 (40). It has been proposed that acidic activating regions are able to contact one or more of the following proteins: TATA binding protein (TBP), TBP-associated factors (TAFs), TFIIB, and TFIIH (reviewed in reference 37). This leads to the assembly of the transactivational initiation complex consisting of TFIID and RNA polymerase II on DNA. However, by testing additional PU.1 mutants that eliminated either both acidic domains or the glutamine-rich domain, we have determined that only the glutamine-rich domain is important for myeloid development.
In comparison to the acidic activation domain, the glutamine-rich activation region was determined to be weaker or of equal activity in vitro (20, 23). Expression of GAL4-PU.1 deletion mutants in an immortalized skin fibroblast cell line failed to detect transactivation potential in the glutamine-rich domain (12). However, the glutamine-rich region is essential for myeloid differentiation. Our data documenting the importance of the glutamine-rich domain of PU.1 for myeloid development is supported by the work of Shin and Koshland (45), which is the only other existing study where PU.1 transactivation mutant proteins were functionally tested in hematopoietic cells. Their work demonstrated that only a region corresponding to the glutamine-rich domain was able to transactivate multimerized PU.1 binding sites from the J-chain promoter in the terminally differentiated S194B cell line. Glutamine-rich domains functioning as activation domains were first noted for the transcription factor Sp1 (5). It has previously been noted that the PU.1 glutamine-rich domain is similar to activation B domain of Sp1 in which the alternating glutamine and hydrophobic residues may interact with a TAF. TAF proteins in general would serve as a bridge to connect the transactivation domain of PU.1 to the TBP.
Multiple functional domains of PU.1 are required for controlling gene expression in a hematopoietic system.
Our data has demonstrated that the glutamine-rich domain of PU.1 is the only activation domain required for normal myeloid differentiation. The glutamine-rich activation domain works in conjunction with the Ets DNA binding domain and the PEST domain to modulate the activity of PU.1-dependent promoter regions to express genes in the myeloid lineage. Our results obtained from an in vitro differentiation system that mimics normal fetal hematopoiesis have not substantiated the experimental results of others claiming that PU.1 contains an acidic transactivation domain or that the PEST domain is not required for gene expression (12, 20, 23). The short stretch of the PEST domain (amino acids 133 to 140) required implies that a protein-protein interaction critical for myeloid development occurs. This interaction is clearly independent of serine 148, which is important for immunoglobulin 3′ enhancer activity in the B-lymphoid lineage (8, 33). PU.1 and possibly another protein interacting with the PEST domain are able to bind to TBP and TAFs and assemble the transcriptional initiation complex to initiate and modulate the expression of PU.1-dependent myeloid promoters. Since there are no PU.1-specific myeloid enhancer regions identified to date, we do not know if the transactivation domain is expendable as has been demonstrated for B-cell-specific enhancer regions in vitro. We are currently developing strategies to test the functional importance of the glutamine-rich transactivation and PEST domains for proper development and functional integrity of both the B-lymphoid and myeloid lineages in vivo.
ACKNOWLEDGMENTS
R.C.F., M.C.O., E.W.S., and M.C.S. contributed equally to this work.
We thank Brian Keith, Beth McNally, and Michael Parmacek for critical reading of the manuscript, Lisa Gottschalk for graphic illustrations, and Cheryl Small for secretarial assistance.
This research was supported by the Howard Hughes Medical Institute and the National Institutes of Health (grants HL52094 to M.C.S., CA72769 and KL58716 to E.W.S., and GM42415 to M.L.A.).
Footnotes
Corresponding author. Mailing address: Department of Medicine and Department of Molecular Genetics and Cell Biology, The University of Chicago, 5841 South Maryland, N138, MC1028, Chicago, IL 60637. Phone: (773) 702-4721. Fax: (773) 702-0271. E-mail: csimon@medicine.bsd.uchicago.edu.
REFERENCES
- 1.Austyn J, Gordon S. F4/80, a monoclonal antibody directed specifically against the mouse macrophage. Eur J Immunol. 1981;11:805–815. doi: 10.1002/eji.1830111013. [DOI] [PubMed] [Google Scholar]
- 2.Blobel G A, Sieff C A, Orkin S H. Ligand-dependent repression of the erythroid transcription factor GATA-1 by the estrogen receptor. Mol Cell Biol. 1995;15:3147–3153. doi: 10.1128/mcb.15.6.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burkert U, von Ruden R, Wagner E. Early fetal hematopoietic development from in vitro differentiated embryonic stem cells. New Biol. 1991;3:698–708. [PubMed] [Google Scholar]
- 4.Celada A, Borras F E, Soler C, Lloberas J, Klemz M, Vanbeveren C, McKercher S, Maki R A. The transcription factor PU.1 is involved in macrophage proliferation. J Exp Med. 1996;184:61–69. doi: 10.1084/jem.184.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Courey A J, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 6.Doetschman T, Eistetter H, Katz M, Schmidt W, Kemler R. The in vitro development of blastocyst-derived embryonic stem cell lines: formation of visceral yolk sac, blood islands, and myocardium. J Embryol Exp Morphol. 1985;87:27–45. [PubMed] [Google Scholar]
- 7.Eisenbeis C, Singh H, Storb U. PU.1 is a component of a multiprotein complex which binds an essential site in the murine immunoglobulin γ2-4 enhancer. Mol Cell Biol. 1993;13:6452–6461. doi: 10.1128/mcb.13.10.6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisenbeis C F, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- 9.Erman B, Sen R. Context dependent transactivation domains activate the immunoglobulin mu heavy chain gene enhancer. EMBO J. 1996;15:4566–4575. [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher R C, Scott E W. Role of PU.1 in hematopoiesis. Stem Cells. 1998;16:25–27. doi: 10.1002/stem.160025. [DOI] [PubMed] [Google Scholar]
- 11.Galson D, Hensold J, Bishop T, Schalling M, D’Andrea A, Jones C, Auron P, Houseman D. Mouse β-globin DNA-binding protein B1 is identical to a proto-oncogene, the transcription factor Spi-1/Pu.1, and is restricted in expression to hematopoietic cells. Mol Cell Biol. 1993;13:2929–2941. doi: 10.1128/mcb.13.5.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11b.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 12.Hagemeier C, Bannister A, Cook A, Kouzarides T. The activation domain of transcription factor PU.1 binds the retinoblastoma (RB) protein and the transcription factor TFIID in vitro: RB shows sequence similarity to TFIID and TFIIB. Proc Natl Acad Sci USA. 1993;90:1580–1584. doi: 10.1073/pnas.90.4.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henkel G W, McKercher S R, Yamamoto H, Anderson K L, Oshima R G, Maki R A. PU.1 but not ets-2 is essential for macrophage development from embryonic stem cells. Blood. 1996;88:2917–2926. [PubMed] [Google Scholar]
- 14.Hope I A, Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986;46:885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- 15.Hromas R, Orazi A, Neiman R, Maki R, Van Beveran C, Moore J, Klemsz M. Hematopoietic lineage- and stage-restricted expression of the ETS oncogene family member PU.1. Blood. 1993;82:2998–3004. [PubMed] [Google Scholar]
- 16.Karim F D, Urness L D, Thummel C S, Klemsz M J, McKercher S R, Celada A, Van Beveren C, Maki R A, Gunther C V, Nye J A, et al. The ETS-domain: a new DNA-binding motif that recognizes a purine-rich core DNA sequence. Genes Dev. 1990;4:1451–1453. doi: 10.1101/gad.4.9.1451. [DOI] [PubMed] [Google Scholar]
- 17.Keller G, Kennedy M, Papayannopoulou T, Wiles M. Hematopoietic commitment during embryonic stem cell differentiation in culture. Mol Cell Biol. 1993;13:473–486. doi: 10.1128/mcb.13.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinoshita S, Akira S, Kishimoto T. A member of the C/EBP family, NF-IL6 beta, forms a heterodimer and transcriptionally synergizes with NF-IL6. Proc Natl Acad Sci USA. 1992;89:1473–1476. doi: 10.1073/pnas.89.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klemsz M, McKercher S, Celada A, Van Beveren C, Maki R. The macrophage and B cell-specific transcription factor PU.1 is related to the Ets oncogene. Cell. 1990;61:113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 20.Klemsz M J, Maki A A. Activation of transcription by PU.1 requires both acidic and glutamine domains. Mol Cell Biol. 1996;16:390–397. doi: 10.1128/mcb.16.1.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kodandapani R, Pio F, Ni C-Z, Piccialli G, Klemz M, McKercher S, Maki R A, Ely K R. A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex. Nature. 1996;380:456–480. doi: 10.1038/380456a0. [DOI] [PubMed] [Google Scholar]
- 22.Kodandapani R, Veerapandian B, Kunicki T J, Ely K R. Crystal structure of the OPG2 Fab. An antireceptor antibody that mimics an RGD cell adhesion site. J Biol Chem. 1995;270:2268–2273. doi: 10.1074/jbc.270.5.2268. [DOI] [PubMed] [Google Scholar]
- 23.Kominato Y, Galson D, Waterman W R, Webb A C, Auron P E. Monocyte expression of the human prointerleukin 1β gene (IL1B) is dependent on promoter sequences which bind the hematopoietic transcription factor Spi-1/PU.1. Mol Cell Biol. 1995;15:58–68. [PMC free article] [PubMed] [Google Scholar]
- 23a.Luscher B, Eisenman R N. c-myc and c-myb protein degradation: effect of metabolic inhibitors and heat shock. Mol Cell Biol. 1998;8:2504–2512. doi: 10.1128/mcb.8.6.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma J, Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987;48:847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- 25.Matsuyama T, Grossman R A, Mittrucker H W, Siderovski D P, Kiefer F, Kawakami T, Richardson C D, Taniguchi T, Yoshinaga S K, Mak T W. Molecular cloning of LSIRF, a lymphoid-specific member of the interferon regulatory factor family that binds the interferon-stimulated response element (ISRE) Nucleic Acids Res. 1995;23:2127–2136. doi: 10.1093/nar/23.12.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mittrucker H W, Matsuyama T, Grossman A, Kundig T M, Potter J, Shahinian A, Wakeham A, Patterson B, Ohashi P S, Mak T W. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997;275:540–543. doi: 10.1126/science.275.5299.540. [DOI] [PubMed] [Google Scholar]
- 27.Moreau-Gachelin F, Ray D, Mattei M, Tambourin P, Tavitian A. The putative oncogene Spi-1: murine chromosomal localization and transcriptional activation in murine acute erythroleukemias. Oncogene. 1989;4:1449–1456. [PubMed] [Google Scholar]
- 28.Nagulapalli S, Pongubala J, Atchison M. Multiple proteins physically interact with PU.1. J Immunol. 1995;155:4330–4338. [PubMed] [Google Scholar]
- 29.Olson M, Scott E W, Hack A, Su G, Singh H, Simon M C. PU.1 is not essential for early myeloid gene expression but is required for terminal myeloid differentiation. Immunity. 1995;3:702–714. doi: 10.1016/1074-7613(95)90060-8. [DOI] [PubMed] [Google Scholar]
- 30.Olweus J, Thompson P A, Lund-Johnson F. Granulocytic and monocytic differentiation of CD34hi cells is associated with distinct changes in the expression of the PU.1-regulated molecules, CD64 and macrophage colony-stimulating factor receptor. Blood. 1996;88:3741–3754. [PubMed] [Google Scholar]
- 31.Perkel, J. M., and M. L. Atchison. 1997. Personal communication.
- 32.Pongubala J, Nagulapalli S, Klemsz M, McKercher S, Maki R, Atchison M. PU.1 recruits a second nuclear factor to a site important for immunoglobulin kappa 3′ enhancer activity. Mol Cell Biol. 1992;12:368–378. doi: 10.1128/mcb.12.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pongubala J, Van Beveren C, Nagulapalli S, Klemsz M, McKercher S, Maki R, Atchison M. Effect of PU.1 phosphorylation on interaction with NF-EM5 and transcriptional activation. Science. 1993;259:1622–1625. doi: 10.1126/science.8456286. [DOI] [PubMed] [Google Scholar]
- 34.Pongubala J M, Atchison M L. Functional characterization of the developmentally controlled immunoglobulin kappa 3′ enhancer: regulation by Id, a repressor of helix-loop-helix transcription factors. Mol Cell Biol. 1991;11:1040–1047. doi: 10.1128/mcb.11.2.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pongubala J M, Atchison M L. PU.1 can participate in an active enhancer complex without its transcriptional activation domain. Proc Natl Acad Sci USA. 1997;94:127–132. doi: 10.1073/pnas.94.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rechsteiner M, Rogers S W. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 37.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 38.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 39.Rosmarin A, Weil S, Rosner G, Griffin J, Arnaout M, Tenen D. Differential expression of CD11b/CD18 (Mol) and myeloperoxidase genes during myeloid differentiation. Blood. 1989;73:131–136. [PubMed] [Google Scholar]
- 40.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL-4 VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 41.Schmitt R, Bruyns E, Snodgrass H. Hematopoietic development of embryonic stem cells in vitro: cytokine and receptor gene expression. Genes Dev. 1991;5:728–740. doi: 10.1101/gad.5.5.728. [DOI] [PubMed] [Google Scholar]
- 42.Schuetze S, Stenberg P, Kabat D. The ETS-related transcription factor PU.1 immoralizes erythroblasts. Mol Cell Biol. 1993;13:5670–5678. doi: 10.1128/mcb.13.9.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott E W, Fisher R, Olson M, Simon M C, Singh H. PU.1 functions in a cell-autonomous manner to control the differentiation of multipotential lymphoid-myeloid progenitors. Immunity. 1997;6:437–447. doi: 10.1016/s1074-7613(00)80287-3. [DOI] [PubMed] [Google Scholar]
- 44.Scott E W, Simon M C, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 45.Shin M, Koshland M. Ets-related protein PU.1 regulates expression of the immunoglobulin J-chain gene through a novel Ets-binding element. Genes Dev. 1993;7:2006–2015. doi: 10.1101/gad.7.10.2006. [DOI] [PubMed] [Google Scholar]
- 46.Weiss M J, Yu C, Orkin S H. Erythroid-cell-specific properties of transcription GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol. 1997;17:1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wiles M, Keller G. Multiple hematopoietic lineages develop from embryonic stem (ES) cells in culture. Development. 1991;111:259–267. doi: 10.1242/dev.111.2.259. [DOI] [PubMed] [Google Scholar]
- 48.Yamada T, Kondoh N, Matsumoto M, Yoshida M, Maekawa A, Oikawa T. Overexpression of PU.1 induces growth and differentiation inhibition and apoptotic cell death in murine erythroleukemia cells. Blood. 1997;89:1383–1393. [PubMed] [Google Scholar]
- 49.Yamagata T, Nishida J, Tanaka S, Sakai R, Mitani K, Yoshida M, Taniguchi T, Yazaki Y, Hirai J. A novel interferon regulatory factor family transcription factor, ICSAT/Pip/LSIRF, that negatively regulates the activity of interferon-regulated genes. Mol Cell Biol. 1996;16:1283–1294. doi: 10.1128/MCB.16.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]