Abstract
DNA topoisomerase I (topo I) from Drosophila melanogaster contains a nonconserved, hydrophilic N-terminal domain of about 430 residues upstream of the conserved core domains. Deletion of this N terminus did not affect the catalytic activity of topo I, while further removal of sequences into the conserved regions inactivated its enzymatic activity. We have investigated the cellular function of the Drosophila topo I N-terminal domain with top1-lacZ transgenes. There was at least one putative nuclear localization signal within the first 315 residues of the N-terminal domain that allows efficient import of the large chimeric proteins into Drosophila nuclei. The top1-lacZ fusion proteins colocalized with RNA polymerase II (pol II) at developmental puffs on the polytene chromosomes. Either topo I or the top1-lacZ fusion protein was colocalized with RNA pol II in some but not all of the nonpuff, interband loci. However, the fusion proteins as well as RNA pol II were recruited to heat shock puffs during heat treatment, and they returned to the developmental puffs after recovery from heat shock. By immunoprecipitation, we showed that two of the largest subunits of RNA pol II coprecipitated with the N-terminal 315-residue fusion protein by using antibodies against β-galactosidase. These data suggest that the topo I fusion protein can be localized to the transcriptional complex on chromatin and that the N-terminal 315 residues were sufficient to respond to cellular processes, especially during the reprogramming of gene expression.
Type I DNA topoisomerase (topo I) catalyzes a cycle of transesterification reactions, during which it generates transient single-strand DNA breaks; these reversible reactions result in the topological transformation of either single-strand or double-strand DNA molecules (8, 20, 55). Biochemical and genetic studies have demonstrated that topo I plays important roles in DNA replication, transcription, and recombination and in chromosome condensation and the maintenance of genome stability. The top1 gene is not essential for viability in yeast, and genetic analysis suggests that some of the biological functions of topo I can be performed by topo II (59). However, recent genetic screens of Saccharomyces cerevisiae have identified four complementation groups of mutants with mutations that, in combination with a top1 mutation, result in a lethal phenotype. Some of these synthetic lethal mutants have mutations in genes other than top2 (44). Therefore, it is possible that not all the biological functions of topo I can be substituted by topo II. Furthermore, because top1 is an essential gene in Drosophila melanogaster (30) and in mice (37), the functions of topo I may not be efficiently substituted by topo II in multicellular organisms.
One of the functions of topo I is to provide a swivel to facilitate the unwinding of the DNA duplex during the process of transcription or replication (56). Diametric DNA supercoiling can transiently accumulate in the front and in the wake of a transcriptional fork as proposed in the twin-domain DNA supercoiling model (33). Eukaryotic topo I can relieve both the positive and negative supercoils generated during the fork movement. By immunological studies, and the use of camptothecin to trap and map topo I cleavages, it has been shown that topo I is concentrated at transcriptionally active loci (15, 16, 49, 61). In addition, topo I can serve as a coactivator of specific gene expression and as a repressor of basal transcription in vitro through its association with the transcription complex TFIID (29, 36). The participation of topo I in transcriptional initiation has been shown by its effect on the assembly of a complex of TFIID and TFIIA on the promoter (45). These data reveal an essential role for topo I in transcription.
Based on the amino acid sequence alignment, eukaryotic topo I enzymes have conserved domains with about 500 amino acid residues in the central and C-terminal portions, while their N termini have little homology and have variable lengths, ranging from 140 residues in S. cerevisiae to 430 residues in D. melanogaster (20, 26). Despite the fact that the N-terminal sequences are not conserved, they are characterized by an abundance of acidic and basic residues, and by their sensitivity to proteolysis (32, 50). Studies with human (10, 50) and yeast (5) enzymes have shown that their N-terminal domains are dispensable for catalytic activity. Although, these N termini are not required for the strand passage activity of topo I, they may have important roles in the in vivo functions of topo I; this remains to be elucidated.
The hydrophilic N-terminal domain of Drosophila topo I is unique, because it is much longer than that of other species, and it contains clusters of serine and histidine residues that are shared by other Drosophila gene products (6, 27). In this paper, we present data relating to the functions of the N terminus of Drosophila topo I. The N terminus was inconsequential for its catalytic activity, as is the case in other species. Furthermore, our studies of N-terminal fusion constructs with a reporter protein, β-galactosidase (β-gal) have demonstrated that the N-terminal domain directs the chimeric products to transcriptionally active loci in the polytene chromosomes and that the fusion proteins are present in a complex that also contains RNA polymerase II (pol II).
MATERIALS AND METHODS
Plasmid constructions.
N-terminal deletion constructs are denoted ND, followed by the number of residues deleted from the N terminus. ND472 and ND582 were made by deleting DNA sequences in pETI.6 (27) from the NheI site near the T7 promoter to the unique SacI and StuI sites, respectively. ND423 was engineered by synthesizing a truncated topo I cDNA fragment with PCR. The primers used in this PCR were 5′GCCAAGGATCCGGAGGTTTGGCGATGG3′ (nucleotides 1528 to 1543 in top1 cDNA) and 5′GAACAGGCCTGGCGGCTC3′ (nucleotides 1994 to 2011). The italicized sequence in the first oligonucleotide is not homologous to topo I cDNA, and the underlined sequences in the oligonucleotides correspond to BamHI and StuI sites, respectively. After digestion with restriction enzymes BamHI and StuI, the PCR fragment was inserted into the BamHI-StuI sites of expression vector pETI.6 to generate ND423. All the constructs were first introduced to the bacterial strain AG1 (Stratagene, La Jolla, Calif.) and sequenced to confirm the junction DNA sequences before they were moved into the expression strain BL21 (DE3, pLysS) (Novagen, Inc., Madison, Wis.).
NF fusion constructs were made by a two-step construction scheme to generate the Drosophila topo I N-terminal fusion constructs. They are designated NF, followed by a number representing the length of the topo I N-terminal sequence. The first step of cloning involved the vector, C4-βgal/BS (a generous gift from P. Schedl, Princeton University), that was generated by inserting the lacZ gene and simian virus 40 poly(A) tail sequences from pC4βgal (52) into polylinker sites of Bluescript plasmid (Stratagene). The ends generated from XmaI digestion of C4-βgal/BS were flushed by treatments of either T4 DNA polymerase to fill in ends or mung bean nuclease to remove protruding ends before the second digestion with XhoI. Two inserts containing topo I N-terminal domains, a 1.87-kb XhoI-StuI fragment and a 3.0-kb XhoI-NdeI fragment, were purified by digestions of the top1 cDNA clone, ctop1.2 (26, 27). The NdeI site of the XhoI-NdeI fragment had been treated with T4 DNA polymerase before the XhoI digestion. The XhoI-StuI fragment was inserted into mung bean nuclease-treated C4-βgal/BS to generate NF582, while the XhoI-NdeI fragment was ligated to the T4 polymerase-treated vector DNA to create NF964.
Two shorter constructs, NF315 and NF438, were constructed from a PCR product, which was amplified by the universal primer and oligonucleotide I-56 (5′GCCAAGGATCCACGCCATCGGCACGCTT3′) from ctop1.2. The 3′ end sequence of oligonucleotide I-56 corresponds to nucleotides 1572 to 1556 in top1 cDNA. The italicized sequence in the 5′ end is the nonhomologous part, and the underlined portion is a BamHI site. The 1.45-kb PCR product was digested with two sets of enzymes: XhoI plus BamHI and XhoI plus BclI. The resulting 1.4-kb XhoI-BamHI fragment and 1.2-kb XhoI-BclI fragment were cloned into XhoI and BamHI sites of C4-βgal/BS to generate NF438 and NF315, respectively.
These four constructs were verified by sequencing analysis at the cloning junctions to ensure maintenance of the correct reading frame. Furthermore, regions of top1 segments in three fusion constructs, NF582, NF438, and NF315, were sequenced throughout to confirm that no mutations were introduced during the cloning process. All constructs were then subcloned into NotI and XhoI sites of pCaSpeRhsp83, a derivative of pCaSpeR (40). Consequently, the resulting fusion genes are under the control of the hsp83 promoter (25).
Expression of topo I and its truncation mutants in BL21 (DE3, pLysS).
Bacterial cultures started from a 50-fold dilution of overnight cultures were allowed to grow at 37°C for 2 h before induction by adding isopropyl-β-d-thiogalactoside (IPTG) to 1 mM. The induction proceeded for 5 h at 30°C, and the cultures were pelleted by centrifugation and stored at −70°C. The frozen bacterial pellet was thawed in lysis buffer (20 mM NaPO4 [pH 7.0], 0.5 M NaCl, 2.5 mM EDTA, 5% glycerol, 1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin and pepstatin per ml). The bacterial lysate was sonicated to ensure complete lysis and to reduce its viscosity. The cleared lysate was prepared by centrifugation at 12,000 × g for 20 min. The expression of Drosophila topo I was monitored by assaying supercoil relaxation activity and by immunoblotting with an antibody against Drosophila topo I in accordance with the procedures described earlier (26).
Drosophila germ line transformation.
P-element-mediated transformation of Drosophila was carried out as described by Rubin and Spradling (43, 48), with the modification of using a strain carrying a stable genomic source of transposase (42). Second-chromosome-linked homozygous transformants were used in the immunofluorescence microscopy because the cytological mapping of chromosome puffs is more straightforward in these strains. The lacZ control fly, 28-1 (generously provided by P. Schedl, Princeton University), which expresses β-gal protein under the control of the hsp83 promoter, was generated with transformation vector pC4-hsp83AUGβgal, which was derived from pC4-AUG-βgal plasmid (52).
Salivary gland staining.
Staining of salivary glands was based on a method previously described (17). Briefly, third-instar larvae were dissected in buffered 1% glutaraldehyde. The color development of the tissue was carried out in 0.2% X-Gal (5-bromo-4-chloro-3-indolyl-β-d galactopyranoside) (Boehringer Mannheim Biochemicals, Indianapolis, Ind.) for 60 to 120 min. The stained salivary glands were stored in 100% glycerol and imaged on a Leitz DMIL inverted microscope equipped with a Hamamatsu C5810 camera.
Antibodies.
Rabbit antibodies against Drosophila topo I were affinity purified on a Reacti-Gel agarose column (Pierce Chemical Co., Rockford, Ill.) with covalently cross-linked recombinant proteins, either full-length topo I or truncated ND423 protein. The affinity-purified ND423 antibody was used to label the endogenous topo I, since it could not react with the topo I N-terminal fusion proteins. Goat affinity-purified immunoglobulin G’s (IgGs) specific for the hyperphosphorylated carboxy-terminal domain in the largest subunit of RNA pol II (anti-IIo), exon 2 of the largest subunit (anti-E2), and the second-largest subunit of RNA pol II (anti-IIc) were generous gifts from A. Greenleaf (Duke University). Rabbit anti-β-gal IgG was purchased from Cappel Research Products (Organon Teknika Corporation, Durham, N.C.). Similar immunofluorescence data were also obtained with mouse anti-β-gal monoclonal antibodies (Boehringer Mannheim Biochemicals). Species-specific secondary antibodies included affinity-purified donkey IgGs (multiple-labeling grade) labeled with either fluorescein isothiocyanate or indocarbocyanine (Cy3) (Jackson ImmunoResearch Laboratories, Inc., West Grove, Pa.).
Polytene chromosome squashes and immunofluorescence microscopy.
Preparations of polytene chromosome squashes and immunofluorescence staining were as described by Weeks et al. (57, 58). Briefly, salivary glands from late-third-instar larvae were dissected in phosphate-buffered saline–0.5% Triton X-100 (PBS-T) and fixed in 3.7% formaldehyde–PBS-T. Polytene chromosomes were then squashed in 45% acetic acid–1% formaldehyde. For heat shock treatment, third-instar larvae were placed inside microcentrifuge tubes with small ventilation holes on the lids and submerged in a 37°C water bath for the desired length of time.
The dilutions of primary antibodies used in immunofluorescence staining were as follows: rabbit anti-topo I, 1:200; rabbit anti-ND423, 1:100; rabbit anti-β-gal, 1:100 to 1:200; mouse monoclonal anti-β-gal, 25 μg/ml; and goat anti-IIo, 1:25. The secondary antibodies were adjusted to a 1-μg/ml final concentration. Normal donkey serum (4%) was used as blocking reagent before primary- and secondary-antibody incubations. Chromosome squashes were stained with 20 ng of DAPI (4′,6-diamidino-2-phenylindole dihydrochloride) per ml for 60 s before being mounted. Fluorescence images were viewed under a Zeiss Axiophot microscope through a Zeiss ×40 Plan-NEOFLUAR objective (oil immersion; NA 1.4), and captured with a Photometrics STAR I cooled charge-coupled device imaging system. Image manipulations were carried out with Adobe Photoshop version 3.0.4 software.
Immunoprecipitation of β-gal fusion proteins.
Overnight embryos were homogenized in a lysis buffer (50 mM Tris [pH 8.0], 1% Nonidet P-40, 500 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol), supplemented with 1 mM phenylmethylsulfonyl fluoride and 1 μg of pepstatin, leupeptin, and aprotinin per ml. Lysates were spun in a microcentrifuge for 30 min at 4°C, and the supernatant was used for the immunoprecipitation reaction. These extracts were precleared with protein A agarose beads (Sigma, St. Louis, Mo.; catalog no., P9424) at 4°C for 20 min. Specific antibodies and protein A beads were added to the precleared extracts, and the incubation continued for 1 h at 4°C; then the beads were washed twice with lysis buffer and twice with radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Nonidet P-40, 0.5% Na deoxycholate, 0.1% sodium dodecyl sulfate [SDS]). The washed products were then analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting.
RESULTS
Amino-terminal truncations of Drosophila DNA topo I.
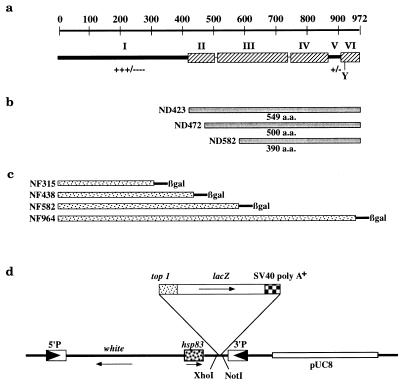
To test the possible biochemical functions of the nonconserved N-terminal domain of the Drosophila enzyme, we constructed a series of N-terminal truncation mutants: ND423, ND472, and ND582 (see Materials and Methods). ND423 lacks the first 423 residues of topo I and contains 549 residues corresponding to the highly homologous domains. ND472 and ND582 lack additional N-terminal sequences and portions of the conserved sequences (Fig. 1a and b). These constructs and a full-length topo I cDNA were cloned into inducible bacterial expression vector pET3b under the control of a phage T7 promoter (51).
FIG. 1.
Drosophila topo I N-terminal deletion and top1-lacZ fusion constructs. (a) Schematic diagram of the structural features of the enzyme. The top of the panel indicates the number of amino acid residues. In the lower part of the panel, the hatched boxes (II, III, IV, and VI) represent regions of amino acid sequence homology among topo I proteins from humans, Drosophila melanogaster, Arabidopsis thaliana, and budding and fission yeasts (26). The active-site tyrosine is shown as Y, and +/− and +++/−−− refer to charged and hydrophilic residues. (b) ND423, ND472, and ND582 are three proteins with N-terminal deletions; they have codons for amino acid residues from 1 to 423, 472, and 582, respectively, removed from the full-length top1 cDNA. (c) Fusion constructs of N-terminal segments of topo I and β-gal. NF stands for N-terminal fusion, and the suffix numbers are the numbers of amino acid residues of topo I remaining in the fusion constructs. (d) Linear representation of pCaSpeRhsp83 plasmid with the top1-lacZ insert (not drawn to scale). The solid arrowheads indicate the 5′ and 3′ terminal repeats of the P element, and the thin arrows show the direction of transcription.
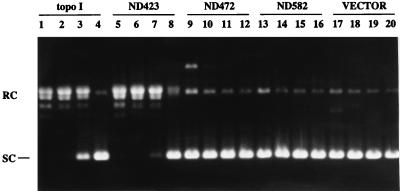
The expression levels of these topo I constructs after IPTG induction were comparable based on immunoblot analysis of the bacterial lysates. Furthermore, the sizes of the expressed peptides were consistent with the calculated molecular masses (135, 61, 55, and 44 kDa for full-length topo I, ND423, ND472, and ND582, respectively; data not shown). The catalytic activities of these truncated topo I peptides were evaluated by supercoil relaxation analyses. The assays were carried out in the presence of 1 mM EDTA and in the absence of any divalent cations to inactivate the endogenous bacterial topo I (9, 56). Bacterial lysates containing the full-length protein were active up to a 1:100 dilution (Fig. 2, lanes 1 to 4). ND423-containing lysates were also active up to a 1:100 dilution (Fig. 2, lanes 5 to 8); however, lysates from cells with ND472, ND582, and the vector control were inactive even when assayed undiluted (Fig. 2, lanes 9, 13, and 17). We confirmed the activities of the full-length and ND423 proteins with partially purified extracts fractionated on cation exchange column Bio-Rex 70 (data not shown). These results indicate that the N-terminal 423 residues of topo I are dispensable for strand passage activity and that further removal of sequences into the conserved domains, as in ND472 and ND582, abolishes catalytic function entirely.
FIG. 2.
Supercoil relaxation activities of bacterial extracts expressing topo I and truncation mutants. Supercoiled plasmid DNA substrate was incubated with extracts prepared from bacteria expressing the full-length protein (lanes 1 to 4), ND423 (lanes 5 to 8), ND472 (lanes 9 to 12), ND582 (lanes 13 to 16), and vector pET3b (lanes 17 to 20). The DNA products were analyzed by agarose gel electrophoresis, and the positions of supercoiled (SC) as well as relaxed-circular (RC) DNA are indicated on the left side. Four successive 10-fold dilutions were carried out in each set of the activity assays. Lanes 1, 5, 9, 13, and 17 show the assays of undiluted extracts, while lanes 4, 8, 12, 16, and 20 show the assays with 1,000-fold-diluted extracts. Expression plasmid DNAs, present in higher concentrations in the reaction mixtures, were visible in lanes 9 and 17.
Construction and expression of top1-lacZ fusion proteins.
Although the hydrophilic N terminus is not required for in vitro topo I activity, it may contain important properties that are essential in vivo. We explored this possibility by constructing vectors expressing fusion peptides of topo I with β-gal in Drosophila cells. Four fusion constructs, NF315, NF438, NF582, and NF964, in which increasing amounts of amino-terminal sequence were fused to β-gal by in-frame fusion, were generated (Fig. 1c and d; see details in Materials and Methods). These chimeric constructs were under the control of the hsp83 promoter and were cloned into a P-element transposon vector derived from pCaSpeR (Fig. 1d). Stable transgenic lines were obtained by microinjecting vector DNAs into fly embryos and following established germ line transformation procedures (43, 48).
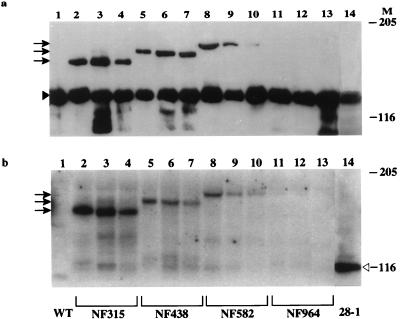
Multiple lines of transformants were obtained from each fusion construct. The expression of these fusion products in the independent transformants was examined by Western analysis using antibodies against either topo I (Fig. 3a) or β-gal (Fig. 3b). All the samples contained endogenous topo I (135 kDa), and additional species of 158, 174, and 196 kDa were also detected in extracts from strains NF315, NF438, and NF582, respectively (Fig. 3a, lanes 2 to 10). The sizes of these fusion products were consistent with the predicted molecular masses. These fusion peptides also reacted with anti-β-gal antibody (Fig. 3b, lanes 2 to 10). Three independent transformants were analyzed for each fusion construct, and all showed fusion products of the same size for each construct.
FIG. 3.
Expression of top1-lacZ fusion proteins in transgenic flies. Protein extracts from adult flies were prepared as described previously (30), and about 50 μg of total protein from each transgenic sample was loaded on an SDS–5% PAGE gel. Western blots were probed with either affinity purified rabbit anti-topo I antibody (a) or rabbit anti-β-gal antibody (b). Lane 1 contains a protein extract from wild-type flies, and lane 14 contains a protein extract from 28-1 flies (a transgenic strain transformed with a control construct of β-gal). Lanes 2 to 13 contain adult fly extracts from the following independent transformants: NF315 (lanes 2, 3, and 4), NF438 (lanes 5, 6, and 7), NF582 (lanes 8, 9, and 10), and NF964 (lanes 11, 12, and 13). Arrows indicate the migration positions of different fusion products. The solid and open arrowheads label the positions of endogenous topo I and β-gal, respectively. Molecular masses for size markers in kilodaltons are indicated under M.
None of the NF964 transformants gave the expected 250-kDa fusion products according to the immunoblot analyses (Fig. 3a and b, lanes 11 to 13). However, the existence of the transgene in the NF964 flies was evident not only from the dominant eye color change caused by the white gene in the P-element vector but also by the presence of a PCR fragment generated with a pair of oligomers in the top1 and lacZ region, which amplified a DNA fragment of the expected size (data not shown). It is not known why the expression of the fusion gene was not observed, but it is possible that the folding and stability of this large fusion polypeptide were anomalous and that no functional, intact protein was produced in these fly strains.
The levels of expression of the fusion proteins in NF315, NF438, and NF582 appeared to be lower than that of endogenous topo I based on immunoblot analyses. It is likely that the efficiency of electrophoretic transfer was less for the larger polypeptides. Although the data shown in Fig. 3 were from flies without heat treatment, heat shock treatment of the transgenic flies had only a modest effect on the induction of fusion proteins (data not shown). This result is consistent with the observation that the hsp83 gene is constitutively expressed in all developmental stages, and is not restricted to heat shock induction (35, 62).
Nuclear localization of top1-lacZ fusion proteins.
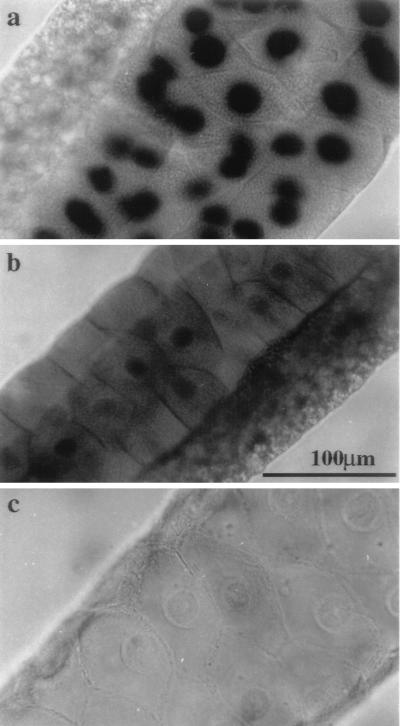
Since topo I is a nuclear enzyme, it is possible that one of the functions residing in the nonconserved N-terminal domain is nuclear localization. We examined the subcellular distribution of the fusion proteins by a histochemical β-gal staining assay, which converted X-Gal into blue precipitates. While salivary glands from the wild-type control showed no endogenous β-gal activity (Fig. 4c), the transgenic fly (28-1), which carried β-gal driven by an hsp83 promoter, showed an even distribution of the stains throughout the salivary gland (Fig. 4b). In this nonfusion β-gal control, nuclear staining varied from a slight accumulation to exclusion from the nucleus. Therefore, β-gal does not have a specific nuclear localization function, a finding consistent with the earlier observations that β-gal expressed in yeast cells was not imported into nuclei (39, 47). In a marked contrast, specific and intense staining was restricted to the nuclei in the salivary gland and surrounding tissue in the NF315 transgenic fly (Fig. 4a). Similar staining patterns were also observed from NF438 and NF582 (data not shown). Consistent with the immunoblot experiments, no staining was observed in the NF964 fly, suggesting that there is very little or no expression of the functional fusion peptide from NF964. The clear differentials between the cytoplasmic and nuclear staining in NF315 suggest that the N-terminal 315 residues in topo I contain a signal for nuclear localization to allow efficient import of large β-gal fusion proteins into nuclei.
FIG. 4.
Nuclear localization of the top1-lacZ fusion protein in salivary glands detected by X-Gal staining for β-gal activity. Salivary glands from third-instar larvae of NF315 (a), 28-1 (b), and wild-type (c) flies were prepared as described in Materials and Methods. The β-gal activity is readily detected in the nuclei of NF315 transgenic animals and is shown as the dark staining. Note that the smaller nuclei of adjacent tissue also exhibit the activity (a). Scale bar (b), 100 μm.
Localization of top1-lacZ fusion proteins on the developmental puffs in polytene chromosomes.
Both biochemical and genetic studies have suggested that there are important functions for topo I in transcription (20, 55). For example, topo I is associated with the chromatin domains, where there is active transcription (38, 49, 61). Furthermore, topo I has been shown to colocalize with RNA pol II in developmental puffs of the polytene chromosomes by immunofluorescence staining (15). Since the top1-lacZ fusion proteins were efficiently transported into nuclei, we proceeded to examine if these proteins can be targeted to regions of transcription just like the full-length topo I.
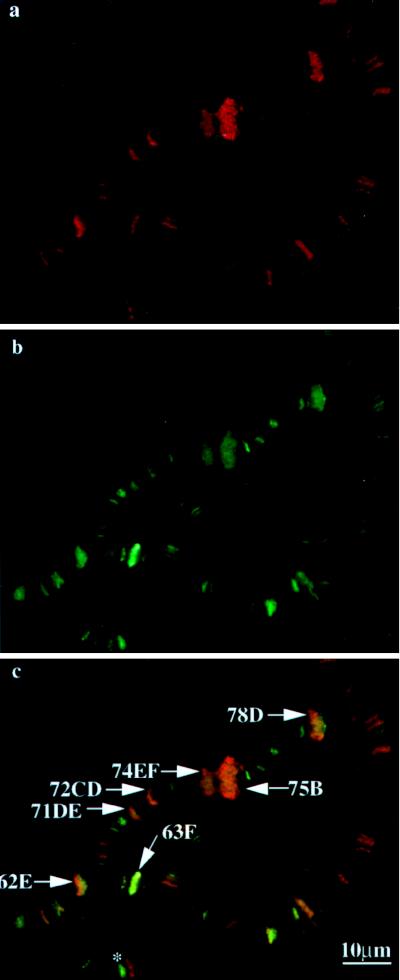
Polytene chromosome squashes prepared as described previously (58) were reacted with primary antibodies against either β-gal (anti-β-gal) or the largest subunit of RNA pol II (anti-IIo) and then with the appropriate secondary antibodies with different fluorescence labels to reveal the localization of antigens in the same polytene chromosome preparation. Control experiments using either preimmune serum or normal serum before the secondary antibody or secondary antibody alone did not reveal any specific signals (data not shown). However, specific fluorescence signals were observed with either anti-β-gal or anti-IIo antibodies. A typical experiment with the chromosome squashes obtained from the NF438 transgenic fly is shown in Fig. 5. The localization of the NF438 fusion protein was revealed by the green fluorescence resulting from fluorescein isothiocyanate-conjugated anti-rabbit IgGs reacting with the anti-β-gal antibodies (Fig. 5b). The transcriptionally active chromosome puffs are readily identified by the morphology of the chromosome banding patterns, as well as by the RNA pol IIo immunofluorescence staining, shown here as the red fluorescence from the Cy3-conjugated secondary antibodies (Fig. 5a). Earlier work has established that this affinity-purified anti-IIo antibody selectively reacts with the phosphorylated form of the 215-kDa subunit of RNA pol II, which is highly enriched in the chromosome puffs (58). The colocalization of the NF438 and RNA pol IIo was evident in the merged images as the orange-yellow staining (Fig. 5c).
FIG. 5.
top1-lacZ fusion products colocalize with RNA pol IIo on developmental puffs. Preparations of chromosome squashes from transgenic flies carrying NF438 were reacted with goat anti-IIo and rabbit anti-β-gal, followed by species-specific, fluorescence-tagged secondary antibody. Red fluorescence for RNA pol IIo (a) and green fluorescence for β-gal (b) are shown. Panel c is a superimposed fluorescence image of panels a and b, showing blended colors of green and red at the indicated developmental puffs. The asterisk in panel c marks cytological position 66B, which is enriched in topo I fusion protein but not RNA pol IIo.
The major developmental puffs, 74EF and 75B, as well as the minor puffs, 62E, 63F, 71DE, 72CD, and 78D, were clearly enriched for both topo I fusion proteins and RNA pol IIo (Fig. 5c) (see reference 3 for a summary of the developmental puffs). Not shown in this image were the similar colocalization patterns of other puffs, such as 85F and 88EF, as well as those of 2B, 5B, and 68C from third-instar larvae at slightly different developmental stages. While there was a clear colocalization of topo I fusion proteins and RNA pol IIo in all the developmental puffs, this colocalization was apparent in some but not all of the chromosome loci (red or green staining versus yellow-orange staining in Fig. 5c). It should be pointed out that the differences in the degrees of staining among the interband loci are less than that between the puff and nonpuff sites and are sensitive to the experimental conditions and the stages of larvae. The biochemical basis for the differential distributions of topo I fusion proteins and RNA pol IIo in some of these nonpuff chromosome loci is unknown. It is possible that these loci may represent regions of active transcription by a polymerase other than RNA pol IIo, such as RNA pol IIa (the hypophosphorylated form of the largest subunit of RNA pol II) and RNA pol III. For example, cytogenetic position 66B (Fig. 5c) is known to contain a cluster of tRNA genes (14, 18), and this region is enriched in the top1-lacZ fusion protein but not in RNA pol IIo. It remains to be demonstrated if this locus is truly enriched in RNA pol III.
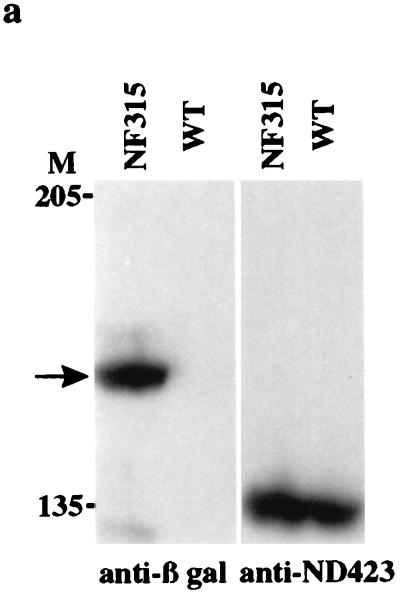
Although the top1-lacZ fusion protein was localized to the major transcriptionally active loci, further investigation is needed to demonstrate that topo I N-terminal fusion proteins are colocalized with the endogenous topo I. It has been shown that topo I is targeted to the transcriptionally active loci, including developmental puffs and as well as those induced by heat shock treatment (15). We proceeded to investigate whether the endogenous topo I behaves in a manner similar to that of the top1-lacZ fusion protein under our experimental conditions. To be able to distinguish endogenous topo I from the fusion products, we prepared anti-ND423 antibodies by affinity purification (see Materials and Methods). The affinity-purified antibody against ND423 should be specific for the C-terminal half of topo I; thus it should be reactive only toward endogenous topo I. As shown by an immunoblot (Fig. 6a), anti-ND423 antibody recognized the 135-kDa endogenous topo I in both wild-type and NF315 transgenic flies but did not detect the 158-kDa N-terminal topo I fusion protein that was recognized by the anti-β-gal antibody.
FIG. 6.
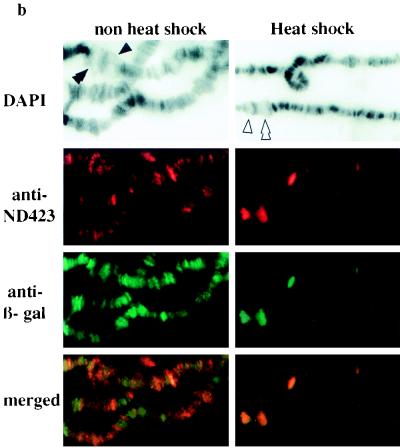
top1-lacZ fusion products are recruited to chromosome puffs as the endogenous topo I. (a) Extracts from wild-type (WT) and NF315 adult flies were separated on an SDS–7% PAGE gel. Western blots were probed with either mouse anti-β-gal or affinity-purified rabbit anti-ND423 antibodies. The arrow indicates the top1-lacZ fusion protein. The size markers are marked on the left of the Western blot. (b) Immunofluorescence images of both untreated and heat-shocked polytene chromosomes double stained with affinity-purified rabbit anti-ND423 and mouse anti-β-gal. DAPI-stained fluorescence images were used to identify chromosome bands and puffs. The symbols ▴ and ▴▴ identify developmental puffs 74EF and 75B, respectively; the symbols ▵ and ▵▵ indicate heat shock puffs 87A and 87C, respectively.
The rabbit anti-ND423 and mouse anti-β-gal antibodies were used in the immunofluorescence experiments to monitor the localization of endogenous topo I and the top1-lacZ fusion protein on the polytene chromosomes of untreated and heat-treated third-instar larvae. The endogenous topo I colocalized with the NF315 fusion protein at the major developmental puffs (left panels of Fig. 6b). Similar to what was observed between top1-lacZ fusion protein and RNA pol IIo, the colocalization between endogenous topo I and topo I fusion protein was apparent in some but not all of the nonpuff loci. The longer fusion constructs, including NF438 and NF582, did not have significantly different staining patterns for these interband, nonpuff loci (data not shown). However, strict colocalization of these two antigens was found at chromosomal puffs induced by heat shock at 37°C for 20 min (right panels of Fig. 6b). These experiments demonstrate that both the top1-lacZ fusion protein and endogenous topo I have similar patterns of association with chromosomal puffs in these transgenic flies. Moreover, they provide an additional control that the localization of the N-terminal topo I protein at the transcriptionally active loci is unlikely due to a nonspecific interaction.
Time course of recruitment of top1-lacZ fusion proteins upon heat shock treatment.
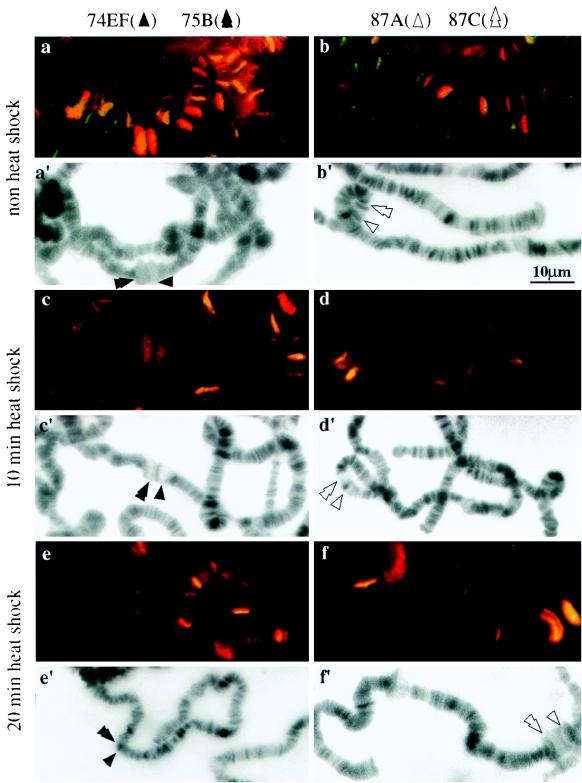
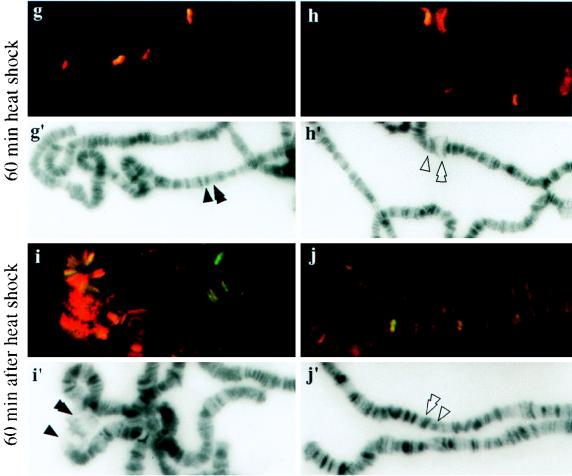
The above experiments demonstrate that the N terminus of topo I is capable of recruiting top1-lacZ fusion proteins to transcriptionally active loci induced by heat shock. An environmental stress like heat shock can repress the developmental puffs and induce new chromosomal puffs (3), thus providing an excellent system to monitor the movement of topo I fusion proteins during the reprogramming of gene expression. We examined the rate at which this recruitment takes place and whether the N-terminal domain could mobilize the fusion proteins from heat shock loci during the recovery from heat treatment. Chromosome squashes prepared from transgenic strain NF315 that had undergone heat shock treatment for various times were used in these studies. Only the merged images are shown here. Yellow-orange staining shows the colocalization of RNA pol IIo and top1-lacZ fusion protein (Fig. 7). Fluorescence images of DAPI-stained squashes were also recorded to facilitate the identification of chromosome bands and puffs (Fig. 7a′ to j′). Before heat shock, strong ecdysone-induced puffs 74EF and 75B were clearly marked with both antigens (Fig. 7a), while the heat shock loci, 87A and 87C, were not labeled with either antigen (Fig. 7b). The staining of 74EF and 75B was significantly reduced after 10 min of heat shock (Fig. 7c), and staining disappeared completely in the samples with 20 (Fig. 7e) and 60 (Fig. 7g) min of heat treatment. There was a parallel accumulation of both proteins at heat shock loci after 10 (Fig. 7d) and 20 (Fig. 7f) min of heat shock treatment. After a prolonged heat treatment, heat shock puffs regressed slightly, and the presence of both proteins persisted, albeit in lesser amounts (60-min point in Fig. 7h). The dynamic movement of these proteins at chromosome puffs was also apparent during the cessation of heat treatment. After 60 min of recovery from heat treatment, the program for normal developmental expression resumed, with both proteins reentering the 74EF and 75B puffs (Fig. 7i) along with their coordinated departure from the heat shock loci (Fig. 7j). We only highlighted 74EF and 75B for developmental puffs and 87A and 87C for heat shock puffs in these experiments, but similar dynamic movement of these proteins was also observed on other major developmental and heat shock puffs. These results show that the N-terminal 315 residues can mobilize the fusion proteins to new loci during the reprogramming of gene expression as efficiently as the holoenzyme of topo I.
FIG. 7.
Localization of top1-lacZ fusion products to the developmental and heat shock-induced puffs during heat treatment. Polytene chromosome squashes were prepared from NF315 flies with no heat shock (a and b); heat treatment for 10 (c and d), 20 (e and f), or 60 (g and h) min; or recovery at 25°C for 60 min after heat shock for 60 min (i and j). They were then immunostained with anti-β-gal and anti-IIo antibodies. Merged images are shown here for comparing the colocalizations of topo I fusion protein and RNA pol IIo. Panels a′ to j′ are identical to panels a to j except that DAPI fluorescence micrographs are presented for identifying chromosome puffs.
Coimmunoprecipitation of RNA pol II with top1-lacZ fusion protein.
Although we have demonstrated the colocalization of top1-lacZ fusion protein with RNA pol II by immunofluorescence microscopy, the coincidence of these two proteins at the level of immunofluorescence experiments may not necessarily imply that the N-terminal topo I fusion protein participates in a transcriptional complex. Therefore, we exploited an immunoprecipitation assay to determine whether the N-terminal topo I fusion protein is a part of a macromolecular transcriptional complex.
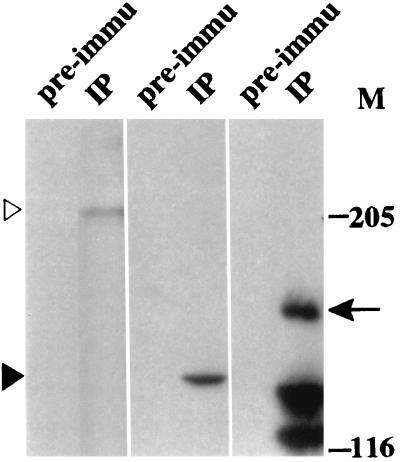
Embryo extracts from NF315 were processed for immunoprecipitation by using either rabbit anti-β-gal antibodies or a preimmune serum. The immunoprecipitated complex was washed and analyzed by Western blotting; the blots were probed with antibodies against the largest (IIa) and the second-largest (IIc) subunits of RNA pol II and β-gal (left, middle, and right panels, respectively, of Fig. 8). These panels show that anti-β-gal antibodies immunoprecipitated the 158-kDa top1-lacZ fusion protein and that both the 215- and 140-kDa subunits of RNA pol II were present in this immunocomplex. The preimmune serum did not precipitate either the top1-lacZ fusion or the RNA pol II subunits (Fig. 8). Furthermore, when embryo extracts from wild-type flies were used in the immunoprecipitation, no RNA pol II signal was detected (data not shown). These results demonstrate the presence of the topo I fusion protein in an immunocomplex containing RNA pol II, and imply a physical interaction between the N-terminal domain of topo I and the macromolecular complex involved in transcription.
FIG. 8.
Coimmunoprecipitation of RNA pol II with the top1-lacZ fusion product. Samples from immunoprecipitation with either rabbit preimmune serum (pre-immu) or rabbit anti-β-gal (IP) were separated on an SDS–7% PAGE gel. Western blots were probed with either affinity-purified goat anti-E2 (exon 2 of RNA pol IIa; left panel), anti-IIc (middle panel), or mouse anti-β-gal (right panel). The open arrowhead indicates RNA pol IIa, while the solid arrowhead designates IIc, the second-largest subunit of RNA pol II. The arrow indicates the top1-lacZ fusion product. Degradation products from the top1-lacZ protein were also detected in an anti-β-gal blot.
DISCUSSION
The function of the hydrophilic N termini in eukaryotic topo I presents an interesting question. The nonconserved N terminus accounts for nearly half of the molecule in Drosophila topo I. As indicated by earlier results with yeast and human topo I (5, 10, 50), topo I from the fruit fly does not require its N terminus for its catalytic activity. It is conceivable that the N terminus may play a regulatory role(s) for topo I activity in vivo. One of the phosphorylation sites in topo I has been mapped to a serine residue in the N terminus (12), and the phosphorylation of serine and threonine residues can stimulate supercoil relaxation and DNA cleavage activities (13, 28, 41, 54). It is also possible that the N terminus has additional roles that are important for the intracellular function of topo I.
Since topo I is a nuclear protein, its N terminus may function in nuclear import. In a heterologous yeast expression system, full-length human topo I can be imported into yeast nuclei, while removal of a 70-residue segment in the N terminus of human topo I results in its distribution in the cytoplasm (1). Using a chromogenic assay of β-gal to detect the localization of top1-lacZ fusion constructs, we have shown that the N-terminal 315 residues are able to direct the chimeric protein into Drosophila nuclei. It has been documented that the efficiency of nuclear import is very sensitive to the context of the nuclear localization sequence that is linked to this bacterial enzyme (39). Therefore, the N-terminal 315 residues of Drosophila topo I must contain potent nuclear localization sequences which are competent in importing these large β-gal fusion proteins into nuclei. While the nuclear localization signal (NLS) in the N terminus of topo I remains to be defined, sequence analysis revealed a possible candidate NLS from residues 135 to 150 (KRSSKDKERRDKDKDR). This putative NLS contains two clusters of basic amino acids spaced nine residues apart, in agreement with previously described bipartite NLSs (11, 22). We have made a construct that expresses, in yeast, a green fluorescent protein with these 16 amino acid residues placed at its C terminus and have found that it was not imported into yeast nuclei (data not shown). This result does not necessarily address the issue of whether this sequence can serve as an NLS in Drosophila cells. It is possible that other basic residues in this hydrophilic tail may serve such a function. Alternatively, the NLS may require adjacent residues in addition to the 16-mer sequence tested here.
We have also demonstrated that the N-terminal 315 residues can target top1-lacZ fusion proteins to transcriptionally active loci, where the functions of topo I are required. Mapping the topo I binding site by DNA cleavage reactions has shown that the distribution of topo I is enriched in the regions of chromatin with active transcription (38, 49, 61). Immunofluorescence experiments have also demonstrated that topo I is concentrated at the chromosome puffs in Drosophila salivary glands (15). Our data indicated that the N-terminal 315 residues were sufficient to direct a large bacterial protein to the puffs where the hyperphosphorylated form of RNA pol II was also present. While RNA pol IIo is always concentrated in the chromosome puffs, its distribution in the interbands is not uniform and does not coincide with the distribution of the hypophosphorylated form of RNA pol II (58). The interband distributions of both topo I and the top1-lacZ fusion protein (Fig. 5) also were not always coincident with that of RNA pol IIo. Earlier work also showed that the distribution of topo I holoenzyme in the chromatin interband is not completely coincident with that of RNA pol II (15). The biochemical bases for these different patterns of interband distribution are unclear. Our data did not address the localizations of RNA pol I, pol IIa, or pol III at the transcriptionally active loci. However, the fusion protein is also found in the nucleolus organizers in the polytene chromosome squashes (data not shown), regions where ribosomal DNA (rDNA) undergoes active transcription. It is noteworthy that many lines of evidence have linked topo I to rDNA transcription (15, 23, 38, 61). Therefore, the N-terminal topo I sequence may be capable of targeting to rDNA transcription loci.
As a reaction to an environmental stress like heat shock, chromatin undergoes rapid and dynamic remodeling to turn off most of the gene expression and to turn on the genes specifically associated with heat shock responses (reviewed in reference 31). The induction of heat shock loci, which results in the synthesis of heat shock proteins and RNA, is accomplished by the recruitment of RNA pol II, transcription factors, and other chromosomal proteins including topo I to heat shock puffs (15, 19, 24, 46, 53, 58). Our immunofluorescence data demonstrated that the topo I N-terminal sequence enables rapid mobilization of topo I to the heat shock puffs. This dynamic mobilization of top1-lacZ fusion proteins was also observed when heat shock puffs regressed during a prolonged heat treatment and when the developmental puffs were reestablished after the cessation of heat shock. Therefore, the ability of topo I to respond to cellular processes, especially during the reprogramming of gene expression, may reside completely in the N-terminal 315 residues.
The biochemical basis for the recruitment of topo I to transcriptionally active loci is not yet established. There are a number of possible mechanisms underlying the association of topo I with a transcriptional complex. First, the binding might be mediated through RNA transcripts. In this case, one would expect a lag between the localization of the topo I fusion protein and RNA pol II. However, the topo I fusion protein appears to colocalize with RNA pol II in the newly established heat shock loci even at the earliest detectable point of the heat treatment (Fig. 7d). While the developmental puffs have not yet regressed, both topo I fusion protein and RNA pol II have already begun to depart from these puffs (compare Fig. 7c′ and c). These experiments suggest that there is no significant lapse in time between the movements of the topo I fusion protein and RNA pol II from the chromosome puffs. Furthermore, by using a procedure of RNase treatment that can remove nuclear ribonucleoprotein complexes from polytene chromosome squashes (2), it was found that immunostaining signals of both topo I fusion protein and RNA pol II were not diminished in comparison with those from the control experiment without RNase treatment (data not shown).
A second possible mechanism for topo I targeting to the transcriptional complex is that topo I may recognize special DNA structures associated with a chromatin region undergoing active transcription. Such structures could be, for example, the positive and negative supercoils present in the front and in the wake, respectively, of a transcription fork (33). Eukaryotic topo I is known to be able to recognize and bind to both positively and negatively supercoiled DNA (7, 34, 38, 60). However, this explanation can be ruled out, since preferential binding of topo I to supercoiled DNA is mediated through the conserved catalytic core domains (34) and the N-terminal 315 residues lack the conserved core domains.
The localization of topo I to the transcriptionally active loci could be mediated through protein-protein interactions between the N terminus of topo I and the transcriptional complexes. The hydrophilic patches in the topo I N terminus may provide a surface for such protein-protein interactions. Recent biochemical data demonstrated an interaction of the human topo I N terminus with either an abundant nucleolar protein, nucleolin, or Simian virus 40 T antigen (4, 21). By immunoprecipitation assays, we have demonstrated that the two largest subunits of RNA pol II coprecipitated with the top1-lacZ fusion protein. This result does not address the question of whether topo I is directly associated with RNA pol II. However, topo I has been shown to collaborate with transcription factor TFIID and to function as a coactivator of specific activation and as a repressor in in vitro transcription reactions (29, 36). This coactivation by topo I is due to the participation of topo I in the formation of an active TFIID-TFIIA initiation complex (45). Interestingly, a topo I mutant with the active-site tyrosine replaced with phenylalanine still functions as a coactivator, suggesting that coactivation does not require the catalytic activity of topo I (36, 45). Together, these data suggest that the colocalization of topo I with RNA pol II at chromosome puffs could be mediated through the protein-protein interaction within the transcriptional initiation complex and that this interaction surface may only involve the N-terminal 315 residues. Although we have not yet identified the minimal interaction sequence, further investigation of the physical contact between the N terminus of topo I and the transcriptional complex will provide insight into the molecular mechanism of the role of topo I in gene expression.
In summary, we have generated a series of Drosophila transgenes of the N-terminal topo I sequences linked to β-gal and have used them to demonstrate that the N-terminal 315 residues of topo I can target the fusion proteins to transcriptionally active loci on chromosomes. These topo I fusion transgenes may provide useful biochemical and genetic tools to further probe the biological functions of topo I.
ACKNOWLEDGMENTS
We are grateful to Paul Schedl for helpful discussion and for providing plasmid vectors as well as a fly strain carrying the β-gal transgene. We thank Arno Greenleaf for generously providing us the affinity-purified antibodies against RNA pol II. We appreciate Alice Chen and Steve Chang for their excellent technical assistance, and acknowledge the Department of Cell Biology at Duke University for the use of its fluorescence microscope facility.
This work is supported by a grant GM29006 from NIH.
REFERENCES
- 1.Alsner J, Svejstrup J Q, Kjeldsen E, Sorensen B S, Westergaard O. Identification of an N-terminal domain of eukaryotic DNA. J Biol Chem. 1992;267:12408–12411. [PubMed] [Google Scholar]
- 2.Amero S A, Matunis M J, Matunis E L, Hockensmith J W, Raychaudhuri G, Beyer A L. A unique ribonucleoprotein complex assembles preferentially on ecdysone-responsive sites in Drosophila melanogaster. Mol Cell Biol. 1993;13:5323–5330. doi: 10.1128/mcb.13.9.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashburner M, Bonner J J. The induction of gene activity in Drosophila by heat shock. Cell. 1979;17:241–254. doi: 10.1016/0092-8674(79)90150-8. [DOI] [PubMed] [Google Scholar]
- 4.Bharti A K, Olson M O J, Kufe D M, Rubin E H. Identification of a nucleolin binding site in human topoisomerase I. J Biol Chem. 1996;271:1993–1997. doi: 10.1074/jbc.271.4.1993. [DOI] [PubMed] [Google Scholar]
- 5.Bjornsti M A, Wang J C. Expression of yeast DNA topoisomerase I can complement a conditional-lethal DNA topoisomerase I mutation in Escherichia coli. Proc Natl Acad Sci USA. 1987;84:8971–8975. doi: 10.1073/pnas.84.24.8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown S D, Zhang C, Chen A, Hsieh T-S. Structure of the Drosophila DNA topoisomerase I gene and expression of messages with different lengths in 3′ untranslated region. Gene. 1998;211:195–203. doi: 10.1016/s0378-1119(98)00119-x. [DOI] [PubMed] [Google Scholar]
- 7.Caserta M, Amadei A, Camilloni G, Di Mauro E. Regulation of the function of eukaryotic DNA topoisomerase I: analysis of the binding step and of the catalytic constants of topoisomerization as a function of DNA topology. Biochemistry. 1990;29:8152–8157. doi: 10.1021/bi00487a024. [DOI] [PubMed] [Google Scholar]
- 8.Champoux J J. Mechanistic aspects of type-I topoisomerases. In: Cozzarelli N R, Wang J C, editors. DNA topology and its biological effects. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. pp. 217–242. [Google Scholar]
- 9.Champoux J J, Dulbecco R. An activity from mammalian cells that untwists superhelical DNA—a possible swivel for DNA replication. Proc Natl Acad Sci USA. 1972;69:143–146. doi: 10.1073/pnas.69.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Arpa P, Machlin P S, de Ratrie H, Rothfield N F, Cleveland D W, Earnshaw W C. cDNA cloning of human DNA topoisomerase I: catalytic activity of a 67.7-kDa carboxyl-terminal fragment. Proc Natl Acad Sci USA. 1988;85:2543–2547. doi: 10.1073/pnas.85.8.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 12.Durban E, Goodenough M, Mills J, Busch H. Topoisomerase I phosphorylation in vitro and in rapidly growing Novikoff hepatoma. EMBO J. 1985;4:2921–2926. doi: 10.1002/j.1460-2075.1985.tb04024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durban E, Mills J S, Roll D, Busch H. Phosphorylation of purified Novikoff hepatoma topoisomerase I. Biochem Biophys Res Commun. 1983;111:897–905. doi: 10.1016/0006-291x(83)91384-0. [DOI] [PubMed] [Google Scholar]
- 14.Elder R T, Uhlenbeck O C, Szabo P. 4S RNA gene organization in Drosophila melanogaster. In: Söll D, Abelson J N, Schimmel P R, editors. Transfer RNA: biological aspects. 9B. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. pp. 317–323. [Google Scholar]
- 15.Fleischmann G, Pflugfelder G, Steiner E K, Javaherian K, Howard G C, Wang J C, Elgin S C R. Drosophila DNA topoisomerase I is associated with transcriptionally active regions of the genome. Proc Natl Acad Sci USA. 1984;81:6958–6962. doi: 10.1073/pnas.81.22.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilmour D S, Pflugfelder G, Wang J C, Lis J T. Topoisomerase I interacts with transcribed regions in Drosophila cells. Cell. 1986;44:401–407. doi: 10.1016/0092-8674(86)90461-7. [DOI] [PubMed] [Google Scholar]
- 17.Glaser R L, Wolfner M F, Lis J T. Spatial and temporal pattern of hsp26 expression during normal development. EMBO J. 1986;5:747–754. doi: 10.1002/j.1460-2075.1986.tb04277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glew L, Lo R, Nichols M, Söll D, Bell J. The nucleotide sequence, localization and transcriptional properties of a tRNALeu gene from Drosophila melanogaster. Gene. 1986;44:307–314. doi: 10.1016/0378-1119(86)90195-2. [DOI] [PubMed] [Google Scholar]
- 19.Greenleaf A L, Plagens U, Jamrich M, Bautz E K. RNA polymerase B (or II) in heat induced puffs of Drosophila polytene chromosomes. Chromosoma. 1978;65:127–136. doi: 10.1007/BF00329465. [DOI] [PubMed] [Google Scholar]
- 20.Gupta M, Fujimori A, Pommier Y. Eukaryotic DNA topoisomerases I. Biochim Biophys Acta. 1995;1262:1–14. doi: 10.1016/0167-4781(95)00029-g. [DOI] [PubMed] [Google Scholar]
- 21.Haluska P, Jr, Saleem A, Edwards T K, Rubin E H. Interaction between the N-terminus of human topoisomerase I and SV40 large T antigen. Nucleic Acids Res. 1998;26:1841–1847. doi: 10.1093/nar/26.7.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hicks G R, Raikhel N V. Protein import into the nucleus: an integrated view. Annu Rev Cell Dev Biol. 1995;11:155–188. doi: 10.1146/annurev.cb.11.110195.001103. [DOI] [PubMed] [Google Scholar]
- 23.Higashinakagawa T, Wahn H, Reeder R H. Isolation of ribosomal gene chromatin. Dev Biol. 1977;55:375–386. doi: 10.1016/0012-1606(77)90180-4. [DOI] [PubMed] [Google Scholar]
- 24.Holt T K. Local protein accumulation during gene activation. I. Quantitative measurements on dye binding capacity at subsequent stages of puff formation in Drosophila hydei. Chromosoma. 1970;32:64–78. doi: 10.1007/BF00334011. [DOI] [PubMed] [Google Scholar]
- 25.Horabin J I, Schedl P. Regulated splicing of the Drosophila sex-lethal male exon involves a blockage mechanism. Mol Cell Biol. 1993;13:1408–1414. doi: 10.1128/mcb.13.3.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsieh T, Lee M P, Brown S D. Structure of eukaryotic type I DNA topoisomerase. Adv Pharmcol. 1994;29:191–200. doi: 10.1016/s1054-3589(08)60546-3. [DOI] [PubMed] [Google Scholar]
- 27.Hsieh T-S, Brown S D, Huang P, Fostel J. Isolation and characterization of a gene encoding DNA topoisomerase I in Drosophila melanogaster. Nucleic Acids Res. 1992;20:6177–6182. doi: 10.1093/nar/20.23.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaiserman H B, Ingebritsen T S, Benbow R M. Regulation of Xenopus laevis DNA topoisomerase I activity by phosphorylation in vitro. Biochemistry. 1988;27:3216–3222. doi: 10.1021/bi00409a014. [DOI] [PubMed] [Google Scholar]
- 29.Kretzschmar M, Meisterernst M, Roeder R G. Identification of human DNA topoisomerase I as a cofactor for activator-dependent transcription by RNA polymerase II. Proc Natl Acad Sci USA. 1993;90:11508–11512. doi: 10.1073/pnas.90.24.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee M P, Brown S D, Chen A, Hsieh T-S. DNA topoisomerase I is essential in Drosophila melanogaster. Proc Natl Acad Sci USA. 1993;90:6656–6660. doi: 10.1073/pnas.90.14.6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lis J T, Wu C. Protein traffic on the heat shock promoter: parking, stalling, and trucking along. Cell. 1993;74:1–4. doi: 10.1016/0092-8674(93)90286-y. [DOI] [PubMed] [Google Scholar]
- 32.Liu L F, Miller K G. Eukaryotic DNA topoisomerases: two forms of type I DNA topoisomerases from HeLa cell nuclei. Proc Natl Acad Sci USA. 1981;78:3487–3491. doi: 10.1073/pnas.78.6.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu L F, Wang J C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madden K R, Stewart L, Champoux J J. Preferential binding of human topoisomerase I to superhelical DNA. EMBO J. 1995;14:5399–5409. doi: 10.1002/j.1460-2075.1995.tb00224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mason P J, Hall L M C, Gausz J. The expression of heat shock genes during normal development in Drosophila melanogaster (heat shock/abundant transcripts/developmental regulation) Mol Gen Genet. 1984;194:73–78. [Google Scholar]
- 36.Merino A, Madden K R, Lane W S, Champoux J J, Reinberg D. DNA topoisomerase I is involved in both repression and activation of transcription. Nature. 1993;365:227–232. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 37.Morham S G, Kluckman K D, Voulomanos N, Smithies O. Targeted disruption of the mouse topoisomerase I gene by camptothecin selection. Mol Cell Biol. 1996;16:6804–6809. doi: 10.1128/mcb.16.12.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muller M T, Pfund W P, Mehta V B, Trask D K. Eukaryotic type I topoisomerase is enriched in the nucleolus and catalytically active on ribosomal DNA. EMBO J. 1985;4:1237–1243. doi: 10.1002/j.1460-2075.1985.tb03766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson M, Silver P. Context affects nuclear protein localization in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:384–389. doi: 10.1128/mcb.9.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pirrotta V. Vectors for P-mediated transformation in Drosophila. In: Rodriguez R L, Denhardt D T, editors. Vectros, a survey of molecular cloning vectors and their uses. Boston, Mass: Butterworths; 1988. pp. 437–456. [DOI] [PubMed] [Google Scholar]
- 41.Pommier Y, Kerrigan D, Hartman K D, Glazer R I. Phosphorylation of mammalian DNA topoisomerase I and activation by protein kinase C. J Biol Chem. 1990;265:9418–9422. [PubMed] [Google Scholar]
- 42.Robertson H M, Preston C R, Phillis R W, Johnson-Schlitz D M, Benz W K, Engels W R. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rubin G M, Spradling A C. Genetic transformation of Drosophila with transposable element. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 44.Sadoff B U, Heath-Pagliuso S, Castano I B, Zhu Y, Kieff F S, Christman M F. Isolation of mutants of Saccharomyces cerevisiae requiring DNA topoisomerase I. Genetics. 1995;141:465–479. doi: 10.1093/genetics/141.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shykind B M, Kim J, Stewart L, Champoux J J, Sharp P A. Topoisomerase I enhances TFIID-TFIIA complex assembly during activation of transcription. Genes Dev. 1997;11:397–407. doi: 10.1101/gad.11.3.397. [DOI] [PubMed] [Google Scholar]
- 46.Silver L M, Elgin S C. Immunofluorescent analysis of chromatin structure in relation to gene activity: a speculative essay. Curr Top Dev Biol. 1979;13:71–88. doi: 10.1016/s0070-2153(08)60690-0. [DOI] [PubMed] [Google Scholar]
- 47.Silver P A, Keegan L, Ptashne M. Amino terminus of the yeast GAL4 gene product is sufficient for nuclear localization. Proc Natl Acad Sci USA. 1984;81:5951–5955. doi: 10.1073/pnas.81.19.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spradling A C, Rubin G M. Transposition of cloned P elements into Drosophila germ line. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 49.Stewart A F, Herrera R E, Nordheim A. Rapid induction of c-fos transcription reveals quantitative linkage of RNA polymerase II and DNA topoisomerase I enzyme activities. Cell. 1990;60:141–149. doi: 10.1016/0092-8674(90)90724-s. [DOI] [PubMed] [Google Scholar]
- 50.Stewart L, Ireton G C, Champoux J J. The domain organization of human topoisomerase I. J Biol Chem. 1996;271:7602–7608. doi: 10.1074/jbc.271.13.7602. [DOI] [PubMed] [Google Scholar]
- 51.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 52.Thummel C S, Boulet A M, Lipshitz H D. Vectors for Drosophila P-element-mediated transformation and tissue culture transfection. Gene. 1988;74:445–456. doi: 10.1016/0378-1119(88)90177-1. [DOI] [PubMed] [Google Scholar]
- 53.Tsukiyama T, Becker P B, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter binding of GAGA transcription factor. Nature. 1994;367:525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- 54.Turman M A, Douvas A. A casein kinase type II (CKII)-like nuclear protein kinase associates with, phosphorylates, and activates topoisomerase I. Biochem Med Metab Biol. 1993;50:210–225. doi: 10.1006/bmmb.1993.1063. [DOI] [PubMed] [Google Scholar]
- 55.Wang J C. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 56.Wang J C. Interaction between DNA and Escherichia coli protein ω. J Mol Biol. 1971;55:523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]
- 57.Weeks J R, Coulter D E, Greenleaf A L. Immunological studies of RNA polymerase II using antibodies to subunits of Drosophila and wheat germ enzyme. J Biol Chem. 1982;257:5884–5892. [PubMed] [Google Scholar]
- 58.Weeks J R, Hardin S E, Shen J, Lee J M, Greenleaf A L. Locus-specific variation in phosphorylation state of RNA polymerase II in vivo: correlations with gene activity and transcript processing. Genes Dev. 1993;7:2329–2344. doi: 10.1101/gad.7.12a.2329. [DOI] [PubMed] [Google Scholar]
- 59.Yanagida M, Sternglanz R. Genetics of DNA topoisomerases. In: Cozzarelli N R, Wang J C, editors. DNA topology and its biological effects. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. pp. 299–320. [Google Scholar]
- 60.Zechiedrich E L, Osheroff N. Eukaryotic topoisomerases recognize nucleic acid topology by preferentially interacting with DNA crossovers. EMBO J. 1990;9:4555–4562. doi: 10.1002/j.1460-2075.1990.tb07908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang H, Wang J C, Liu L F. Involvement of DNA topoisomerase I in transcription of human ribosomal RNA genes. Proc Natl Acad Sci USA. 1988;85:1060–1064. doi: 10.1073/pnas.85.4.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zimmerman J L, Petri W, Meselson M. Accumulation of a specific subset of D. melanogaster heat shock mRNAs in normal development without heat shock. Cell. 1983;32:1161–1170. doi: 10.1016/0092-8674(83)90299-4. [DOI] [PubMed] [Google Scholar]