Abstract
Reb1p is a DNA binding protein of Saccharomyces cerevisiae that has been implicated in the activation of transcription by polymerase (Pol) II, in the termination of transcription by Pol I, and in the organization of nucleosomes. Studies of the transcriptional control of the REB1 gene have led us to identify three Reb1p binding sites in the 5′ region of the its gene, termed A, B, and C, at positions −110, −80, and +30 with respect to transcription initiation. In vitro, Reb1p binds to the three sites with the relative affinity of A ≥ C > B. Kinetic parameters suggest that when both A and C sites are present on the same DNA molecule, the C site may recruit Reb1p for the A site. In vivo the A and B sites each contribute to the transcription activity of REB1 in roughly additive fashion. Mutation of both A and B sites abolishes transcription. On the other hand, the C site is a negative element, reducing transcription by 40%. In cells overexpressing Reb1p, the C site reduces transcription by more than 80%. This effect can be transposed to another transcription unit, demonstrating that the effect of Reb1p binding at the C site does not depend on interaction with upstream Reb1p molecules. Relocation of the C site to a position 105 bp downstream of the transcription initiation site abolishes its effect, suggesting that it does not act as a conventional attenuator of transcription. We conclude that binding of Reb1p at the C site hinders formation of the initiation complex. This arrangement of Reb1p binding sites provides a positive and negative mechanism to autoregulate the expression of REB1. Such an arrangement could serve to dampen the inevitable fluctuation in Rep1p levels caused by the intermittent presence of its mRNA within an individual cell.
Reb1p of Saccharomyces cerevisiae is an essential protein of 810 amino acids (28), with an unusual DNA binding region consisting of two Myb-like domains separated by nearly 150 amino acids (42). It binds to the 5′ regions of numerous genes (8, 19, 26, 38, 46, 49, 54, 61). While the core consensus sequence for Reb1p binding has been identified as CGGGTAA (9, 37, 41, 61), substantial flexibility is permissible, and the adjacent nucleotides influence the binding affinity. (Reb1p has also been studied under the names Grf2p [9] and QBP [4].)
Reb1p plays a role in transcription by RNA polymerase II (Pol II). For instance, it is the major transcription factor for the ACT1 gene (38). Reb1p cooperates with other transcription factors, i.e., Rap1p and Abf1p, in regulating transcription of several glycolytic genes (46, 54). A Reb1p binding site is necessary for the threefold induction of the ILV1 gene in cells deprived of isoleucine and valine (49). In many other cases, Reb1p modestly influences the expression of genes by as-yet-undetermined mechanisms. In certain cases at least, Reb1p appears to play a role in chromatin organization. With a footprint of only 20 to 25 bp, the binding of Reb1p to sequences in the GAL1-10 upstream activation sequence (UAS) creates a 200-bp nucleosome-free region (17), although this is not the case for the UAS of the HSC82 gene (13).
Reb1p also plays one or more roles in the transcription of rRNA by RNA Pol I. It binds not only to the transcriptional enhancer, immediately downstream of the 3′ end of the 35S rRNA transcript, but also just upstream of the promoter, about 200 nucleotides 5′ of the start site of transcription (40, 41). In a construct placed within the ribosomal DNA (rDNA) locus, deletion of the two Reb1p binding sites causes a substantial reduction in transcription (31). However, deletion of the Reb1p binding sites has little effect on Pol I transcription of test genes on a variety of plasmid constructs (6, 32). These results imply that Reb1p might be involved in the three-dimensional arrangement of the rDNA within the nucleolus (31, 62). The binding of Reb1p has also been implicated in specifying the termination of the rRNA transcript by causing the polymerase molecule to pause. Release of the completed transcript soon follows (33–35). The mammalian counterpart to Reb1p is TTF1, identified by the Grummt laboratory as binding to the “Sal boxes” downstream of the human and mouse rRNA genes (3, 14, 15, 22). The binding of TTF1 not only is responsible for the termination of Pol I transcription (3, 14, 15) but also plays a role in blocking the DNA replication fork initiated from the intergenic region of rRNA genes (18). As in S. cerevisiae, the mouse rRNA genes have a TTF1 binding site just upstream of the Pol I promoter (21). The binding of TTF1 at that site can overcome chromatin-mediated repression of rRNA transcription (36), suggesting that TTF1 also shares with Reb1p the ability to exclude nucleosomes.
In this report on the regulation of transcription of the REB1 gene, we show that Reb1p binds to three sequences of the 5′ region of its own gene. Two of the sites are upstream of the transcription initiation site. Each contributes to effecting transcription. Deletion of both abolishes transcription. The third is 30 bp downstream of the site of transcription initiation. Binding of Reb1p to the downstream site reduces transcription, probably by preventing the formation of a functional initiation complex. The concentration of REB1 mRNA is, on average, ≅1 molecule per cell (60, 63). The statistical fluctuation of the number of molecules in an individual cell could lead to substantial fluctuation in the concentration of Reb1p. We suggest that this arrangement of sites at the REB1 gene has evolved to minimize such fluctuations.
(This research is from a thesis submitted to the Sue Golding Division of the Albert Einstein College of Medicine in partial fulfillment of the requirements for the Ph.D. degree by Kevin L.-C. Wang.)
MATERIALS AND METHODS
Strains, media, and general methods.
All strains of S. cerevisiae used in this report are derived from W303a (MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 ura3-1 can1-100) (59) and are listed in Table 1. Yeast cells were grown at 30°C in either YPD (1% yeast extract, 2% peptone, and 2% dextrose) or synthetic complete minimal medium with the necessary supplements. Strain DH5α was used for all DNA manipulations in Escherichia coli. Recombinant DNA methods and yeast manipulations are based on established protocols (50, 52).
TABLE 1.
Yeast strains used in this study
| Yeast strain | Reb1p sitea | Genotypeb |
|---|---|---|
| W303a | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100c | |
| WY119 | W303a (pLC31) | |
| WY123 | Vector | W303a (pLC31), URA3::YIp357R |
| WY83 | ABC | W303a (pLC31), URA3::pLC6 |
| WY84 | BC | W303a (pLC31), URA3::pLC7 |
| WY85 | AC | W303a (pLC31), URA3::pLC8 |
| WY90 | AB | W303a (pLC31), URA3::pLC104 |
| WY102 | A | W303a (pLC31), URA3::pLC131 |
| WY103 | B | W303a (pLC31), URA3::pLC130 |
| WY86 | C | W303a (pLC31), URA3::pLC9 |
| WY105 | ABC+75 | W303a (pLC31), URA3::pLC128 |
| WY101 | Null | W303a (pLC31), URA3::pLC107 |
| WY183 | W303a (pLC30) | |
| WY189 | W303a (pLC30, pLC151) | |
| WY190 | W303a (pLC30, pLC151C) |
Plasmids.
Plasmids used in this study are listed in Table 2. Plasmid YIp357R (24) was used as the backbone for the integrating lacZ reporter gene constructs. A 1.2-kb DNA fragment of REB1, including 615 bp of the coding sequence, was subcloned to the BamHI and EcoRI sites of YIp357R, yielding a translational fusion of the REB1′-lacZ reporter gene in plasmid pLC6. This is the parental plasmid for all the other mutant constructs of the Reb1p binding site (Fig. 1A).
TABLE 2.
Plasmids used in this study
| Plasmid | Descriptiona | Source |
|---|---|---|
| pET-His | E. coli expression vector with six-His tag | 10 |
| pBM272 | CEN, URA3, GAL1-GAL10 promoter | 27 |
| YIp357R | Integration vector, URA3, lacZ as reporter gene | 24 |
| pLC25 | pET-6xHis-REB1 | |
| pLC6 | YIp357R-(ABC) REB1′-lacZ | |
| pLC7 | YIp357R-(BC) REB1′-lacZ | |
| pLC8 | YIp357R-(AC) REB1′-lacZ | |
| pLC104 | YIp357R-(AB) REB1′-lacZ | |
| pLC131 | YIp357R-(A) REB1′-lacZ | |
| pLC130 | YIp357R-(B) REB1′-lacZ | |
| pLC9 | YIp357R-(C) REB1′-lacZ | |
| pLC128 | YIp357R-(ABC+75)-REB1′-lacZ | |
| pLC107 | YIp357R-(null)-lacZ | |
| pLC30 | pBM272-REB1 (URA3) | |
| pLC31 | pBM272-REB1 (URA3 is replaced by HIS3) | |
| pLC151 | CEN TRP1 RPL32′-GFP | |
| pLC151C | CEN TRP1 RPL32′-CGFP |
Reb1p sites are indicated for YIp357R plasmids.
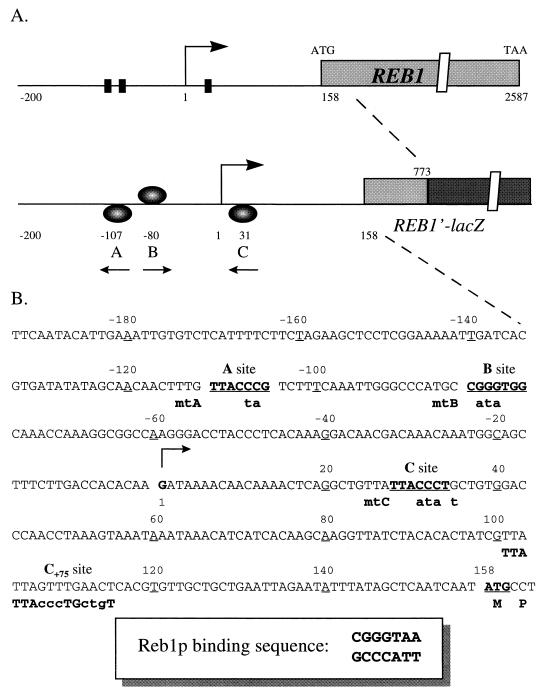
FIG. 1.
Elements controlling REB1 transcription. (A) Three potential Reb1p binding sites upstream of the REB1 ORF. The sequence from −200 to +158 is drawn to scale. The transcription initiation site (28) is designated as 1. A 1.2-kb DNA fragment of REB1, including 615 bp of the coding sequence, was fused to E. coli lacZ as a reporter gene. The orientation and relative distances to the transcription initiation site of the Reb1p binding sites A, B, and C are diagrammed in the REB1′-lacZ construct. (B) Sequence of the region upstream of the REB1 ORF. The A, B, and C sites are indicated. A consensus sequence of Reb1p binding sites is indicated in the lower box (37, 41). Mutations generated at individual sites are designated mtA, mtB, and mtC. The mutated nucleotides of each site are indicated with lowercase letters. Each mutated site was created to embed a restriction enzyme site for convenient verification: SpeI for mtA, NdeI for mtB, and SspI for mtC. For the C+75 construct, the original C site was mutated and a sequence identical over 15 bp with the original C site was constructed as shown at a position 105 bp downstream of the transcription initiation site.
To overexpress Reb1p in yeast cells, we constructed pLC30, which carries a transcriptional fusion of the GAL1 promoter to the coding sequence of REB1, on a CEN plasmid (a YCp50-based vector with a URA3 marker) (27). pLC31 is identical to pLC30 except the URA3 marker is replaced with HIS3. The TATA element and transcription initiation site of both plasmids are derived from the GAL1 gene. No Reb1p binding sites of REB1 are present in these plasmids.
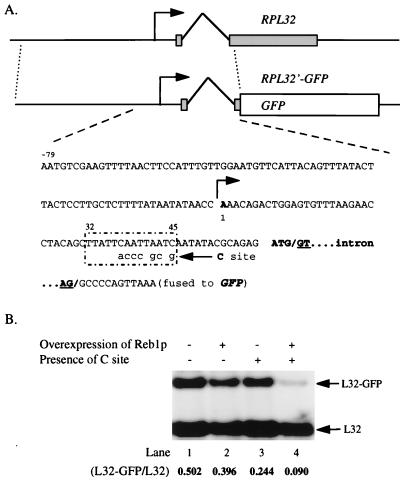
To construct pLC151 (pRS314-RPL32′-GFP) (57), 860 bp of RPL32 DNA was fused to the 5′ terminus of GFP as a reporter gene at an in-frame KpnI site (see Fig. 6A). To create a wild-type C site for Reb1p binding (Fig. 1B), site-directed mutagenesis by PCR was used to introduce the substitutions at the +32-to-+45 region of the RPL32 transcription unit (see Fig. 6A), yielding pLC151C.
FIG. 6.
Reb1p bound at the C site can block transcription of a heterologous promoter. (A) Diagram and partial sequence of the reporter genes. An 860-bp fragment of RPL32 of S. cerevisiae fused to GFP was used as the backbone for the reporter gene, pLC151 (pRS314-RPL32′-GFP). The transcription initiation site is indicated as 1 (11). The nucleotides designating the 5′ (GT) and 3′ (AG) sites of the RPL32 intron are underlined. By substitution of only seven nucleotides it was possible to generate a sequence identical to the original C site over 14 bp at positions +32 to +45 of the RPL32 transcription unit, as shown in the box. The resulting reporter gene was designated pLC151C. (B) Decreased transcription of RPL32′-GFP by Reb1p binding to the C site. Total RNA prepared from WY189 (with pLC151, lanes 1 and 2) and WY190 (with pLC151C, lanes 3 and 4) were subjected to Northern blot analysis and probed with an end-labeled oligonucleotide (JW969 [+31 to +10]; panel A) that will detect transcripts of both RPL32 and RPL32′-GFP (arrows). Both strains contained pLC30 (pGAL1-REB1, see Tables 1 and 2), in which overexpression of Reb1p can be induced by using galactose as the carbon source (lanes 2 and 4). The relative amounts of the two mRNAs, as determined by PhosphorImager analysis, are indicated. Three independent clones from each strain were used for this set of experiments. One representative example of each strain is shown.
Site-directed mutagenesis by PCR.
Mutations were introduced at the sites indicated in Fig. 1B by the megaprimer method of PCR mutagenesis (1). In order to screen the mutated Reb1p binding sites easily, new restriction enzyme sites were created as follows: SpeI for the mutation at the A site, NdeI for the B site, and SspI for the C site. The resulting sequences, with mutated Reb1p binding sites, were used to replace the corresponding region of pLC6 (YIp357R-REB1′-lacZ) by an XbaI fragment (−162 to +240), which includes about 80 bp downstream of ATG. All the mutations were confirmed by sequencing. All the PCR-based mutagenesis procedures in this study were designed to substitute nucleotides without altering the relative spacing of the Reb1p binding sites.
Construction of integration reporter genes and strains.
For the reporter gene constructs with REB1′-lacZ in the YIp357R vector (listed in Table 1), the unique StuI site within the URA3 gene was used to linearize the REB1′-lacZ constructs, which were then transformed into WY119 for targeted integration to the URA3 locus (Table 1) (51). Ura+ cells were selected for Southern blot analysis to screen for single-copy integration of reporter genes. Two to three independent clones with a single integrated reporter gene were used for the β-galactosidase activity assays (see Table 3). Triplicate data from every clone used for the β-galactosidase assays were averaged to determine the final activity.
TABLE 3.
β-Galactosidase activity assay of REB1′-lacZ reporter genesa
| Strain | REB1 siteb | Result in:
|
|||
|---|---|---|---|---|---|
| DEX medium
|
GAL medium
|
||||
| Activity | % of wild typec | Activity | % of wild typec | ||
| WY83 | ABC | 2.52 ± 0.08 | 100 | 0.67 ± 0.02 | 100 |
| WY85 | AC | 1.96 ± 0.09 | 78 | 0.54 ± 0.02 | 81 |
| WY84 | BC | 1.07 ± 0.02 | 42 | 0.36 ± 0.01 | 54 |
| WY86 | C | 0.09 ± 0.00 | 4 | 0.05 ± 0.00 | 8 |
| WY90 | AB | 4.38 ± 0.08 | 174 | 3.43 ± 0.05 | 512 |
| WY105 | ABC+75 | 4.11 ± 0.09 | 163 | 2.70 ± 0.08 | 403 |
| WY102 | A | 2.12 ± 0.00 | 84 | 2.00 ± 0.04 | 286 |
| WY103 | B | 1.04 ± 0.03 | 41 | 2.18 ± 0.03 | 325 |
| WY101 | Null | 0.06 ± 0.00 | 2 | 0.05 ± 0.01 | 8 |
| WY123 | Vector | 0.04 ± 0.00 | 1 | 0.04 ± 0.00 | 6 |
Strains with a single REB1′-lacZ reporter gene integrated into the URA3 locus were used for the β-galactosidase activity assay as described in Materials and Methods. At least two independent clones with an integrated reporter gene of each construct were assayed. Triplicate data from each clone were used to average the activity; the standard error for each construct was <5%. The entire set of experiments was reproduced at least twice in Ura−, His− dropout medium containing either 2% dextrose (DEX) or 2% galactose (GAL) as the sole carbon source. Reb1p binding sites are indicated to represent the differences between the otherwise isogenic strains. Null is a construct with all three sites mutated; vector is a construct with no promoter in front of the lacZ sequences.
Reb1p binding sites in the REB1′-lacZ reporter genes.
The activity of strains with wild-type Reb1p binding sites is used as 100% in the DEX and GAL media.
Southern and Northern blot analysis.
Both methods followed protocols described previously (50, 52). Genomic DNA was isolated from mid-log-phase cells, digested with HindIII, separated by electrophoresis on a 1% agarose gel, and transferred to a nitrocellulose membrane. The blots were probed with a [32P]dCTP-labeled fragment of the 1.1-kb URA3 gene. The results of Southern blot analysis were used to screen and verify the single-copy integration of the REB1′-lacZ reporter gene at the URA3 locus.
Total RNA was isolated from cells growing in either dextrose- or galactose-containing medium at an optical density at 600 nm (OD600) of 1 to 2 as described previously (53). About 20 μg of RNA was fractionated on a 1.5% agarose gel with 6% formaldehyde and transferred to a nylon membrane. The blot was probed with an oligonucleotide, JW969, complementary to nucleotides +31 to +10 of the RPL32 transcript (see Fig. 6A) and end labeled with [γ-32P]ATP and was then quantitated by PhosphorImager analysis.
Protein purification.
To construct plasmids for overexpressing Reb1p in the bacterial strain BL21(DE3), the 2.4-kb coding sequence of REB1 was generated by PCR by using Pfu DNA polymerase (Stratagene) with NdeI and BamHI sites for subcloning to pET11a (58). His6-Reb1p was generated in a similar way by subcloning the coding sequence of REB1 to pET-His (10) at the XhoI and BamHI sites, leading to a fusion protein with six histidine residues fused to the N terminus of Reb1p. Partially purified Reb1p was prepared by 75% ammonium sulfate precipitation as previously described (42). His6-Reb1p was further purified by using nickel-nitriloacetic acid resin according to the procedures of the manufacturer (Qiagen). The purity of the fusion protein was estimated to be 90% by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by staining with Coomassie blue.
Electrophoretic mobility shift assay (EMSA).
For the binding assay, the following components were added sequentially: 5 μl of 5× DNA binding buffer (100 mM Tris-HCl, pH 7.5; 250 mM KCl; 5 mM EDTA; gelatin, 500 μg/ml; 40% glycerol), 2,000 to 3,000 cpm of DNA probe, 1.0 μg of nonspecific double-stranded competitor [poly(dI-dC) · poly(dI-dC); Pharmacia], H2O (to make up the final 25-μl reaction volume), and the desired amount of Reb1p. The binding assay mixture was incubated on ice for 20 min before loading onto a TBE (90 mM Tris-borate, 1 mM EDTA; pH 8.0) native gel containing 5% polyacrylamide. Gel electrophoresis was run at room temperature at 110 V for 1.5 to 3 h, depending on the size of probes. The gel was dried and subjected to autoradiography and PhosphorImager analysis.
To determine the off-rate of Reb1p binding (see Fig. 5), minor modifications were made as follows. A master tube with 50 μl of reaction mixture was incubated on ice for 20 min. Aliquots (8 μl) from the master tube were distributed to six tubes with 4 μl of H2O (control, time zero) or specific competitor and incubated at room temperature for the times indicated (see Fig. 5). The specific competitor was a double-stranded DNA produced by annealing two oligonucleotides, JW106 (5′-GATCTACTGGG TTACCCGG GGCACCTG) and JW107 (5′-GATCCAGGTGCC CCGGGTAA CCCAGTA), that contain the Reb1p binding site (underlined) from the promoter of rDNA (42).
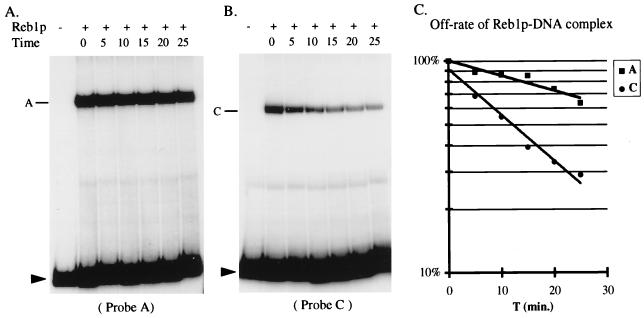
FIG. 5.
The off-rate of Reb1p bound at the A and the C sites. Purified His6-Reb1p was added to the master tubes containing either probe A or probe C (see Fig. 2A). After 20 min of incubation, specific competitor was added, and samples were then loaded onto a running gel at the indicated times (in minutes) (see Materials and Methods for details). Results for the A and C sites are shown in panels A and B, respectively. PhosphorImager quantitation is shown in panel C, where the lines represent the best fit to a single decay rate. The estimated half-lives for the A and C complexes are 43 and 12 min, respectively.
β-Galactosidase activity assay.
Yeast cells were inoculated from fresh overnight cultures into 10 ml of medium with either 2% dextrose or 2% galactose. For dextrose medium, cells were collected when the OD600 reached between 1 and 2. Overexpression of Reb1p in the cells slows growth, with the doubling time estimated to be about 6 h. Cells were collected from galactose medium at 12 h after the inoculation (final OD600 between 1 and 2). The cells were washed once with Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 10 mM MgSO4, 50 mM β-mercaptoethanol; adjusted to pH 7.0) and resuspended in 5 ml of Z buffer. Triplicate (1 ml each) samples from each strain were assayed. β-Galactosidase activity was calculated as follows: OD420/(OD600 of assayed culture × volume assayed × time) (50).
RESULTS
Three Reb1p binding sites are located upstream of the REB1 open reading frame (ORF).
Examination of the upstream non-coding sequence of REB1 reveals three sites that can potentially bind Reb1p (Fig. 1): sites A and B, 110 and 80 bp upstream of the transcription initiation site (28), respectively, and site C, 30 bp downstream of it. To verify the authenticity of these sites, we generated several DNA fragments in which certain of the sites had been mutated (Fig. 2A). The mutations altered only the sequences of the sites without changing the distance between them (Fig. 1B). The binding of purified recombinant Reb1p to fragments containing the three individual sites is shown in the three left panels of Fig. 2B. It is clear that Reb1p binds to each of the putative sites, with relative affinities as follows: A ≥ C ≫ B. The dissociation constants (Kd) of the Reb1p binding sites were estimated to be about 25 and 70 nM for the A and C sites, respectively (7). The Kd value for the B site is roughly 10-fold higher.
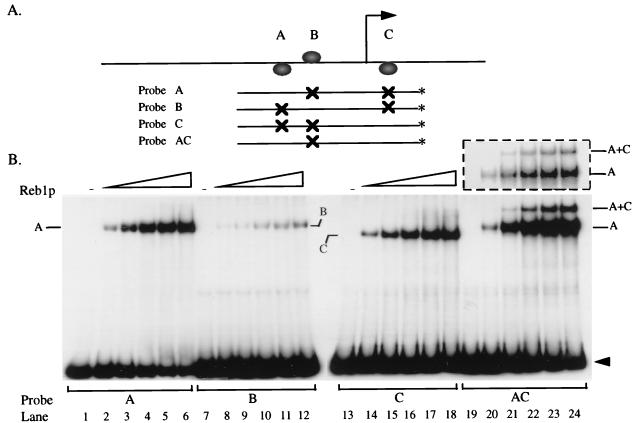
FIG. 2.
A, B, and C are authentic Reb1p binding sites. (A) The indicated probes were generated by PCR with an oligonucleotide end labeled (∗) with [γ-32P]ATP as one of the primers (JW202 [−203 to −185] and JW243 [+68 to +87]). Thus, the specific activities of the probes should be identical. Plasmids containing mutations in the Reb1p binding sites (Fig. 1B) were used as templates in the PCR. (B) Each of the four probes was incubated with increasing amounts of purified His6-Reb1p (i.e., 0, 4.5, 9, 13.5, 18, and 22.5 nM as indicated) and subjected to EMSA. Positions of single-bound (A, B, and C) and double-bound (AC) complexes are indicated. Free probes are denoted by arrowheads. The dashed box exhibits a shorter exposure of lanes 19 to 24.
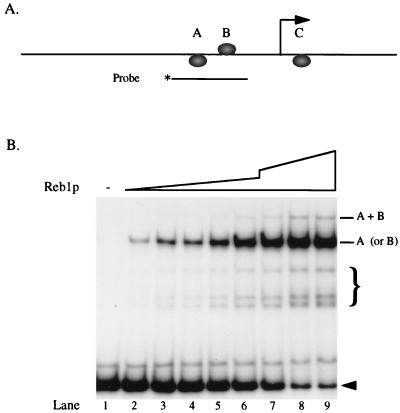
Reb1p protects about 20 to 25 bp when it binds to DNA (41, 42, 61), and the distance between the A and B sites is only 25 bp. Does this proximity lead either to cooperativity or to interference in binding? The experiment shown in Fig. 3 shows that neither of these occurs. Both the A site (single shift for the most part) and the B site (double shift) are filled at about the same rate as when they are alone (Fig. 2B). Clearly, there is no cooperativity and little if any interference, although small interference effects would be difficult to detect.
FIG. 3.
Binding of Reb1p to the A and B sites. (A) The probe was generated by PCR with an end-labeled (∗) oligonucleotide (JW821 [−162 to −143]; Fig. 1B) as one of the primers. (B) A fixed amount of probe was incubated with partially purified Reb1p (0, 0.6, 1.2, 1.8, 2.4, 3.0, 4.5, 6.0, and 7.5 μg in lanes 1 to 9, respectively) and subjected to EMSA. Positions of bound probes (single and double) are indicated. Free probe is denoted by an arrowhead. The faster-migrating complexes indicated by the brace are due to degradation of the partially purified Reb1p (42).
Reb1p binds preferentially to the A site when both the A and C sites are on the same molecule.
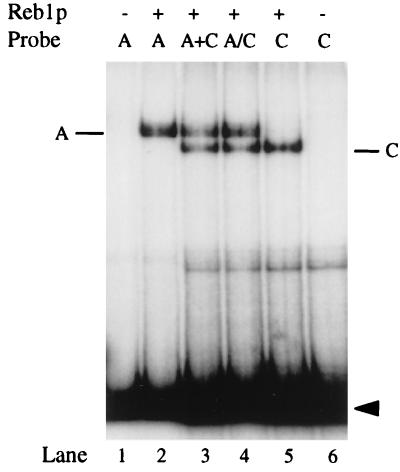
Careful examination of Fig. 2 reveals that although all probes are the same size, the Reb1p-C complex migrates somewhat faster than either the A or the B complex. This is presumably due to bending of DNA at a site relatively near the end of the molecule (64). In any case, the difference in migration provides a means to distinguish binding at the A site from that at the C site. At low concentrations of Reb1p, a probe carrying both A and C sites shows only complexes corresponding to binding at the A site. At higher concentrations there are complexes due to binding at both sites (Fig. 2B, lanes 19 to 24). In no case is there a band corresponding to binding at the C site alone, as is clear in the brief exposure (above lanes 19 to 24). This is a surprising result since the two sites differ in Kd by only a factor of three. To determine whether these observations depend on the A and C sites being on the same molecule, we incubated Reb1p with two separate fragments, each containing one of the sites. As shown in Fig. 4, lanes 3 and 4, Reb1p binds to both the A and the C sites when they are on different molecules. Thus, the apparent loss of C site binding occurs only when the two sites are on the same molecule of DNA. In contrast, when the B and C sites are on the same molecule, the C site is filled at a normal rate (data not shown).
FIG. 4.
Binding of Reb1p to the A and C sites on separate probes. The end-labeled probes A and C (see Fig. 2A) were incubated with purified His6-Reb1p as indicated (lanes 2 and 5). For lane 3 (A+C), individual A and C probes were mixed and then incubated with His6-Reb1p. For lane 4 (A/C), the A and C probes were incubated separately with Reb1p and were then mixed just before electrophoresis. All samples were subjected to EMSA. Positions of bound complexes are indicated. Free probes are indicated by the arrowhead.
To explain these observations, we suggest that Reb1p can bind either the A site or the C site but that during the electrophoretic assay the bound protein migrates from the C to the A site, presumably by dissociation and reassociation. Note that the two sites are only 140 bp, i.e., <500 Å, apart. Stable occupancy of the C site apparently occurs only once the A site is fully occupied. To substantiate the hypothesis that Reb1p can migrate from the C site to the A site, we measured the off-rate for binding at the two sites by challenge with excess competitor at different times before the EMSA was conducted (Fig. 5A and B). It is apparent that Reb1p dissociates far faster from the C site than from the A site. The half-life for dissociation is estimated to be 43 min for the A site and 12 min for the C site (Fig. 5C). These data are consistent with the notion that a Reb1p molecule can migrate from the C site to the A site when they are on a single fragment of DNA. Careful examination of the bands in the shorter exposure in Fig. 2 (lanes 19 to 24) reveals a slight forward spreading from the A site when the C site is present that is not observed when the A site is alone. This would be expected for molecules on which Reb1p moved from the C site to the A site during the electrophoresis. By definition, Kd equals the “off-rate” divided by the “on-rate.” Since the Kd for the C site is about one-third that for the A site but the off-rate is four times that for the A site, we conclude that the on-rate at the C site is greater than or equal to that for the A site. Thus, the presence of the C site could enhance the rate of binding of Reb1p to the A site by acting as a recruiter. While this would not be apparent in the pseudo-equilibrium conditions of an EMSA, the C site may play such a role in the natural context of the REB1 promoter.
Transcriptional activity of Reb1p binding sites.
To study the role of Reb1p in the transcription of its own gene, we constructed a chimeric reporter gene in which 1.2 kb of REB1, including 615 bp of the coding region, was fused upstream of the β-galactosidase ORF (see Fig. 1A). Several constructs that included the mutations of the Reb1p binding sites used for the binding studies (see Fig. 1B) were integrated as a single copy at the URA3 locus as described in Materials and Methods. The wild-type REB1 sequence (ABC) supports the expression of the lacZ reporter gene (Table 3). Mutation of either the B site (AC) or the A site (BC) reduces expression partially. Mutation of both the A and the B sites (C) abolishes expression almost completely. Thus, these two sites are essential for REB1 transcription. The presence of either the A site or the B site leads to significant expression; they are approximately additive (Table 3, compare AC + BC with ABC). It is interesting that the B site alone, with less than one-tenth the affinity for Reb1p binding in vitro, has nearly half the transcription activity of the A site, suggesting that the intrinsic activity of a Reb1p molecule is greater when bound at that position. These results are reminiscent of the situation of the PDR3 gene, in which two Pdr3p binding sites not far upstream from the transcription initiation site are essential for transcription and provide additive effects (12).
Strikingly, mutation of the C site leads to a nearly 75% increase in transcription activity (Table 3, compare AB with ABC), suggesting that this element can repress REB1 expression. Thus, Reb1p plays both a positive and a negative role in the transcription of its own gene. If the arrangement of these sites represents a balance to ensure a relatively constant amount of Reb1p, one would predict that an artificial elevation of the concentration of Reb1p would lead to reduced transcription from the REB1 promoter. To test this prediction, we introduced a CEN plasmid carrying the coding region of REB1 under control of the inducible GAL1 promoter. Western analysis shows that in such cells, when growing on 2% galactose as the sole carbon source, the level of Reb1p is elevated about 10-fold (data not shown). In such cells the expression of the REB1′-lacZ reporter is reduced by nearly 75% (Table 3, compare ABC in dextrose [DEX] with galactose [GAL]). Ablation of the C site leads to a fivefold increase of transcription (compare AB versus ABC in GAL), demonstrating that the C site can serve as a very effective repressing element when the concentration of Reb1p rises. Although the cultures are not strictly comparable, with different growth rates due to different carbon sources and to the inhibitory effect of excess Reb1p, this result suggests that under normal growth conditions the REB1 promoter is nearly saturated with Reb1p (compare AB in DEX and GAL media). However, under normal growth conditions there appears to be only partial occupancy of the C site since the increased level of Reb1p in galactose medium leads to substantially greater repression of transcription unless the C site is mutated. This is true also of transcription due to the A and B sites separately (compare AC with A and BC with B in GAL). Surprisingly, however, the presence of the C site seems to have little effect on transcription arising from the A or the B site alone (compare AC with A and BC with B in DEX medium). Whether this result represents a complex interaction between the sites, an effect of the rare transcription of this gene, or some more subtle effect is not clear.
Reb1p blocks transcription at the initiation step.
We sought to determine whether Reb1p binding at the C site prevents initiation of transcription or acts as an attenuator of ongoing transcription. To do so, we moved the C site. Starting with a reporter gene that had a mutant C site (AB in Table 3), we generated a 15-bp sequence identical to the C site 105 bp downstream of the transcription initiation site (C+75 in Fig. 1B). EMSA was used to verify the binding of Reb1p to the C+75 site (data not shown). If the C site is an attenuator (30), it should block the elongation of transcription even at a downstream site. As shown in Table 3, moving the C site downstream had nearly the same effect on transcription as did mutation of the C site (compare ABC, AB, and ABC+75 in both the DEX and the GAL columns). This result suggests that the repressive effect of Reb1p occupying the C site depends on its proximity to the transcription initiation site. Thus, we conclude that Reb1p binding to the C site blocks transcription at a step prior to elongation, perhaps by preventing the proper alignment of a competent transcription initiation complex.
A Reb1p binding site can block transcription dependent on a heterologous UAS.
Does the inhibitory effect of an occupied C site depend on the presence of the A and/or B site upstream? In short, is it specific for the REB1 gene? To answer this question, we introduced a C site 35 bp downstream of the transcription initiation site of the RPL32 gene, which encodes ribosomal protein L32 (11). RPL32 has no apparent Reb1p binding sites. In this reporter construct, the ORF of GFP was fused to the second exon of RPL32 (Fig. 6A).
The level of the RPL32′-GFP mRNA was compared with that of RPL32 itself, which has an identical promoter (Fig. 6B). It is clear that the presence of the C site reduces the RPL32′-GFP mRNA by about 50% in normal cells (compare lanes 3 and 1). In cells overproducing Reb1p, the presence of the C site reduces the RPL32′-GFP mRNA by more than 75% (compare lanes 4 and 2). These values are very close to the values of 43 and 80%, respectively, observed for repression of the REB1 gene itself (Table 3). Thus, a filled C site does not depend on the presence of upstream Reb1p for its effect. Presumably, it inhibits transcription by direct interference with the transcription apparatus. Furthermore, estimates of protein levels and of mRNA copy number (60) indicate that each ribosomal protein gene is at least 50 times more active than REB1, suggesting that the RPL32 promoter is probably at least 50-fold stronger than the REB1 promoter. Therefore, we conclude that the effect of Reb1p bound to the C site is independent of the strength of the promoter.
DISCUSSION
The presence within a small region of DNA of three sites that bind Reb1p, all with different properties, provides an opportunity to consider the relationship among adjacent sites from a new perspective. On the one hand it is interesting that two sites within 30 bp of each other can bind with neither interference nor cooperativity. The 30 bp of extended DNA extends over about 100 Å, although we expect it to be far more tightly coiled in vivo. With 810 amino acids, Reb1p would fill a sphere with a diameter of ca. 60 Å, although it may, of course, be elongated. In vitro, however, we find that the presence of a filled A site has little effect on the binding of Reb1p to the considerably weaker B site (Fig. 3).
The second point relates to the relationship between the A and the C sites. Reb1p has similar affinities for these two sites when they are on different molecules, yet when they are on the same molecule, the C site is not filled in the presence of the A site under the nonequilibrium conditions of an EMSA (Fig. 2B). We interpret this result as implying that the C site can donate its Reb1p ligand to the A site. That is, Reb1p binds to the C site but over time, because the off-rate is greater at the C site, the Reb1p molecule migrates to the A site. This migration could represent either a true release followed by rebinding or a partial release followed by a sliding from the C to the A site. While this migration has been observed only in vitro, it seems likely that the environment in vivo, with its high concentration of proteins and nucleic acids, would partially mimic the “cage” effect of a polyacrylamide gel lattice. We consider below the biological implications of this arrangement.
As described above, the binding of Reb1p has been experimentally implicated in a variety of functions involving both RNA Pol I and II. The results presented here demonstrate two additional functions of Reb1p, first as an essential transcription factor for its own gene (occupying sites A and B) and also as an obstruction to transcription initiation (occupying site C). Considering first the contribution of the individual sites to the transcription of REB1, we will then attempt to integrate the role of all three sites. The A site has the highest affinity and a very low off-rate. Loss of the A site leads to loss of about 60% of transcription of the gene. Does Reb1p bound at the A site activate transcription by providing an activation domain? Indeed, the N-terminal region of Reb1p can serve as an activator (unpublished data) and contains a glutamine-rich region between residues 60 and 200. Alternatively, Reb1p binding at the A site could recruit another transcription factor that itself activates transcription. Finally, Reb1p binding at the A site could alter the chromatin structure of the region sufficiently to permit the binding of TATA binding protein and the associated factors necessary to initiate transcription, no true activator being necessary for such an infrequent transcription event (see below).
The B site remains puzzling. Although Reb1p binds to it very weakly in vitro, it provides a major element of transcription activation in vivo. At normal levels of Reb1p it has half the activity of the A site; at elevated levels of Reb1p it has activity equal to that of the A site. One possible explanation is that binding of Reb1p to the B site leads to an intrinsically higher activation of transcription. Alternatively, another protein may enhance binding of Reb1p at the B site in vivo, as the SWI-SNF complex appears to do for a weak Gal4p binding site (5).
Mutation of both the A and the B sites leads to almost complete inhibition of transcription of REB1. Thus, Reb1p is an essential factor for the transcription of its own gene. Yet these two sites are not sufficient to replace the UAS of the CYC1 gene (data not shown). Indeed, it should be noted that the location of the A and B sites is much closer to the site of initiation than most transcription activators, which usually bind 150 to 500 bp upstream. In the case of the REB1 gene, perhaps the presence of at least one Reb1p molecule prevents the chromatin structure from becoming so condensed that no activation can occur.
Although one could raise the hypothesis that Reb1p bound to the A or the B site is somehow necessary to overcome the repression of transcription due to the occupancy of the C site, the data in Table 3 show that such is not the case. The transcription from a construct missing all three sites is no greater than that from a construct missing just the A and B sites.
As a negative element just downstream of transcription initiation, the C site plays an unusual role in the transcription of REB1. While transcription can be inhibited by the binding of a factor to a site artificially placed downstream of the transcription initiation site (25, 39, 47, 55), REB1 is unusual in having such an arrangement in its natural environment. The binding of Reb1p to an element of the rRNA gene plays a role in the termination of Pol I transcription by leading to a transcriptional pause (33). Yet, that seems not to be the mechanism at work here because moving the C site downstream, away from the initiation site, abolishes its effect. Such a result suggests that occupancy of the C site prevents the polymerase complex from aligning at the proper site to initiate transcription. This conclusion is consistent with a recent report based on intermolecular cross-linking that the Pol II preinitiation complex extends to at least position +20 (29). The effect of a bound Reb1p molecule seems to be general, for transcription is also reduced when the Reb1p binding site is inserted at position +30 in a different gene (Fig. 6). This result demonstrates two points. Occupancy of the C site represses transcription independent of the presence of other Reb1p molecules acting to activate transcription. Furthermore, occupancy of the C site can inhibit transcription from a very active gene to the same degree as from a modestly active gene.
In considering the biological role of the positive and negative regulation of REB1 transcription by Reb1p, it is important to bear in mind the dynamic state of both REB1 and its product within the cell. We have estimated that a cell has 2,000 to 3,000 molecules of Reb1p (41), which is abundant for a transcription factor but only 1 to 2% of the level of ribosomal proteins, for example. Recent measurements of the abundance of mRNAs in S. cerevisiae (60) found only a single REB1 mRNA among 60,000 mRNAs tabulated (our analysis of the complete data set, kindly provided by V. E. Velculescu). This result suggests that on average there is less than one mRNA per cell. An alternative approach led to a value of about one REB1 mRNA per cell (37a, 63). Although one might initially think that these mRNA values seem somewhat low, they are consistent with the relative values obtained for ribosomal protein mRNAs. These values have two important implications.
We have measured the half-life of the REB1 mRNA by using the rpb1-1 mutant allele of RNA Pol II (44) and obtained a value of 15 to 20 min (data not shown), a reasonable value for an mRNA of S. cerevisiae (23). If there is only one mRNA per cell, then transcription of the REB1 gene occurs, on average, only once every 30 to 40 min. While we do not know the rate at which association and dissociation of transcription factors and DNA occur in the complex milieu of the nucleus, it is clear that this is within the time frame of the dissociation we observed in vitro. Thus, consideration of the influence of the association and dissociation of the Reb1p molecules at the REB1 gene should incorporate the fact that the gene is idle most of the time. The apparently anomalous activity of Reb1p bound to the B site could be a manifestation of such a nonequilibrium situation.
A second implication of the very small number of REB1 mRNAs per cell is that there will necessarily be substantial fluctuations in the number of mRNAs in an individual cell, leading to fluctuations, albeit damped, in the level of Reb1p itself.
Numerous instances of transcription factors controlling their own synthesis have been reported. The proto-oncogenes myb and jun positively regulate their own expression (2, 43); how that expression is extinguished is unclear. In contrast, fos and myc negatively regulate their own transcription, and loss of this control through mutation or genomic rearrangement leads to oncogenesis (16, 45). An interesting example in S. cerevisiae is Rap1p, which can partially repress the transcription of its own gene (20). The amplification implicit in positive autoregulation is used in Drosophila spp. to establish programmed cell types during development (reviewed in reference 56). Similarly, it is used to mount a vigorous response to stress, e.g., in the case of the AMT1 gene of Candida glabrata in responding to heavy metals (65) and perhaps in the case of the drug resistance gene PDR3 of S. cerevisiae (12).
On the other hand, examples of a transcription factor regulating its own gene in both a positive and negative way are rare. One that does is the cI gene, encoding the λ repressor, which also uses three sites to maintain a constant, tightly controlled level of repressor. Cooperative binding of repressor to the two distal sites stimulates transcription of the cI gene. As the level of repressor rises, it binds to the third site, very close to transcription initiation, where it blocks the entry of polymerase (reviewed in reference 48). With the exception of the cooperative binding at the upstream sites, REB1 shares these elements.
Let us now consider the REB1 gene as a biological entity. Reb1p can bind to at least several hundred sites within the genome, participating in a number of important transcriptional events. We suggest that the regulatory elements of REB1 have evolved so as to maintain a stable production of Reb1p, since either overproduction or underproduction of Reb1p is deleterious for growth (unpublished data). At first glance it would seem that having the gene product as an essential component for its transcription would lead to an unstable situation, one that is autocatalytic in either direction. However, the presence of the C site changes that scenario entirely, as we see from Table 3, where overproduction of Reb1p leads to severe repression of REB1 transcription. Under normal growing conditions, it appears that the A site is more or less fully occupied, since in strain WY102 there is little difference in lacZ activity whether or not Reb1p is abundant. On the other hand, it appears that the C site is occupied far less, since the repression due to the C site is much greater when Reb1p is overproduced. We suggest that the arrangement of sites has evolved to buffer the fluctuation of Reb1p concentration that must occur due to the statistical nature of transcription, given the small average number of REB1 mRNA molecules per cell. As the level of Reb1p falls, the C site can serve as a recruiter for Reb1p, transferring it to the A site and even perhaps to the B site, where it can activate transcription. As the level of Reb1p rises, the C site becomes more fully occupied because the occupied A site is no longer available as a sink, and transcription is repressed. The more rapid on- and off-rates of the C site will render it more responsive to changing levels of Reb1p.
ACKNOWLEDGMENTS
We are grateful to John Blanchard for discussion and to Ron Reeder, Josep Vilardell, and Marie Nierras for their cogent comments on the manuscript. V. E. Velculescu and D. J. Lockhart kindly provided raw data from their analyses of the “transcriptome” of S. cerevisiae.
This research was partially supported by NIH grants GM25532 to J.R.W. and CA13330 to the Albert Einstein Cancer Center.
REFERENCES
- 1.Aiyar A, Leis J. Modification of the megaprimer method of PCR mutagenesis: improved amplification of the final product. BioTechniques. 1993;14:366–368. [PubMed] [Google Scholar]
- 2.Angel P, Hattori K, Smeal T, Karin M. The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell. 1988;55:875–885. doi: 10.1016/0092-8674(88)90143-2. [DOI] [PubMed] [Google Scholar]
- 3.Bartsch I, Schoneberg C, Grummt I. Purification and characterization of TTFI, a factor that mediates termination of mouse ribosomal DNA transcription. Mol Cell Biol. 1988;8:3891–3897. doi: 10.1128/mcb.8.9.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandl C J, Struhl K. A nucleosome-positioning sequence is required for GCN4 to activate transcription in the absence of a TATA element. Mol Cell Biol. 1990;10:4256–4265. doi: 10.1128/mcb.10.8.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burns L G, Peterson C L. The yeast SWI-SNF complex facilitates binding of a transcriptional activator to nucleosomal sites in vivo. Mol Cell Biol. 1997;17:4811–4819. doi: 10.1128/mcb.17.8.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butlin M, Quincey R. Activity of promoter mutants of the yeast ribosomal RNA gene with and without the enhancer. Yeast. 1991;7:679–689. doi: 10.1002/yea.320070703. [DOI] [PubMed] [Google Scholar]
- 7.Carey J. Gel retardation at low pH resolves trp repressor-DNA complexes for quantitative study. Proc Natl Acad Sci USA. 1988;85:975–979. doi: 10.1073/pnas.85.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carmen A A, Holland M J. The upstream repression sequence from the yeast enolase gene ENO1 is a complex regulatory element that binds multiple trans-acting factors including REB1. J Biol Chem. 1994;269:9790–9797. [PubMed] [Google Scholar]
- 9.Chasman D I, Lue N F, Buchman A R, LaPointe J W, Lorch Y, Kornberg R D. A yeast protein that influences the chromatin structure of UASG and functions as a powerful auxiliary gene activator. Genes Dev. 1990;4:503–514. doi: 10.1101/gad.4.4.503. [DOI] [PubMed] [Google Scholar]
- 10.Chen B P, Hai T. Expression vectors for affinity purification and radiolabeling of proteins using Escherichia coli as host. Gene. 1994;139:73–75. doi: 10.1016/0378-1119(94)90525-8. [DOI] [PubMed] [Google Scholar]
- 11.Dabeva M D, Warner J R. The yeast ribosomal protein L32 and its gene. J Biol Chem. 1987;262:16055–16059. [PubMed] [Google Scholar]
- 12.Delahodde A, Delaveau T, Jacq C. Positive autoregulation of the yeast transcription factor Pdr3p, which is involved in control of drug resistance. Mol Cell Biol. 1995;15:4043–4051. doi: 10.1128/mcb.15.8.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erkine A M, Adams C C, Diken T, Gross D S. Heat shock factor gains access to the yeast HSC82 promoter independently of other sequence-specific factors and antagonizes nucleosomal repression of basal and induced transcription. Mol Cell Biol. 1996;16:7004–7017. doi: 10.1128/mcb.16.12.7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evers R, Grummt I. Molecular coevolution of mammalian ribosomal gene terminator sequences and the transcription termination factor TTF-I. Proc Natl Acad Sci USA. 1995;92:5827–5831. doi: 10.1073/pnas.92.13.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evers R, Smid A, Rudloff U, Lottspeich F, Grummt I. Different domains of the murine RNA polymerase I-specific termination factor mTTF-I serve distinct functions in transcription termination. EMBO J. 1995;14:1248–1256. doi: 10.1002/j.1460-2075.1995.tb07108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Facchini L M, Chen S, Marhin W W, Lear J N, Penn L Z. The Myc negative autoregulation mechanism requires Myc-Max association and involves the c-myc P2 minimal promoter. Mol Cell Biol. 1997;17:100–114. doi: 10.1128/mcb.17.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fedor M J, Lue N F, Kornberg R D. Statistical positioning of nucleosomes by specific protein-binding to an upstream activating sequence in yeast. J Mol Biol. 1988;204:109–127. doi: 10.1016/0022-2836(88)90603-1. [DOI] [PubMed] [Google Scholar]
- 18.Gerber J K, Gogel E, Berger C, Wallisch M, Muller F, Grummt I, Grummt F. Termination of mammalian rDNA replication: polar arrest of replication fork movement by transcription termination factor TTF-I. Cell. 1997;90:559–567. doi: 10.1016/s0092-8674(00)80515-2. [DOI] [PubMed] [Google Scholar]
- 19.Graham I R, Chambers A. The Reb1p-binding site is required for efficient activation of the yeast RAP1 gene, but multiple binding sites for Rap1p are not essential. Mol Microbiol. 1994;12:931–940. doi: 10.1111/j.1365-2958.1994.tb01081.x. [DOI] [PubMed] [Google Scholar]
- 20.Graham I R, Chambers A. Rap1p is a negative regulator of the RAP1 gene. Curr Genet. 1996;30:93–100. doi: 10.1007/s002940050106. [DOI] [PubMed] [Google Scholar]
- 21.Grummt I, Kuhn A, Bartsch I, Rosenbauer H. A transcription terminator located upstream of the mouse rDNA initiation site affects rRNA synthesis. Cell. 1986;47:901–911. doi: 10.1016/0092-8674(86)90805-6. [DOI] [PubMed] [Google Scholar]
- 22.Grummt I, Maier U, Ohrlein A, Hassouna N, Bachellerie J P. Transcription of mouse rDNA terminates downstream of the 3′ end of 28S RNA and involves interaction of factors with repeated sequences in the 3′ spacer. Cell. 1985;43:801–810. doi: 10.1016/0092-8674(85)90253-3. [DOI] [PubMed] [Google Scholar]
- 23.Herrick D, Parker R, Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill J E, Myers A M, Koerner T J, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 25.Hu M C, Davidson N. The inducible lac operator-repressor system is functional in mammalian cells. Cell. 1987;48:555–566. doi: 10.1016/0092-8674(87)90234-0. [DOI] [PubMed] [Google Scholar]
- 26.Huie M A, Scott E W, Drazinic C M, Lopez M C, Hornstra I K, Yang T P, Baker H V. Characterization of the DNA-binding activity of GCR1: in vivo evidence for two GCR1-binding sites in the upstream activating sequence of TPI of Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2690–2700. doi: 10.1128/mcb.12.6.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnston M, Davis R W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ju Q, Morrow B E, Warner J R. REB1, a yeast DNA-binding protein with many targets, is essential for growth and bears some resemblance to the oncogene myb. Mol Cell Biol. 1990;10:5226–5234. doi: 10.1128/mcb.10.10.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim T, Lagrange T, Wang Y, Griffith J D, Reinberg D, Ebright R H. Trajectory of DNA in the RNA polymerase II transcription preinitiation complex. Proc Natl Acad Sci USA. 1997;94:12268–12273. doi: 10.1073/pnas.94.23.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krumm A, Meulia T, Groudine M. Common mechanisms for the control of eukaryotic transcriptional elongation. Bioessays. 1993;15:659–665. doi: 10.1002/bies.950151005. [DOI] [PubMed] [Google Scholar]
- 31.Kulkens T, van der Sande C A, Dekker A F, van Heerikhuizen H, Planta R J. A system to study transcription by yeast RNA polymerase I within the chromosomal context: functional analysis of the ribosomal DNA enhancer and the RBP1/REB1 binding sites. EMBO J. 1992;11:4665–4674. doi: 10.1002/j.1460-2075.1992.tb05568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kulkens T, van Heerikhuizen H, Klootwijk J, Oliemans J, Planta R J. A yeast ribosomal DNA-binding protein that binds to the rDNA enhancer and also close to the site of Pol I transcription initiation is not important for enhancer functioning. Curr Genet. 1989;16:351–359. doi: 10.1007/BF00340714. [DOI] [PubMed] [Google Scholar]
- 33.Lang W H, Morrow B E, Ju Q, Warner J R, Reeder R H. A model for transcription termination by RNA polymerase I. Cell. 1994;79:527–534. doi: 10.1016/0092-8674(94)90261-5. [DOI] [PubMed] [Google Scholar]
- 34.Lang W H, Reeder R H. The REB1 site is an essential component of a terminator for RNA polymerase I in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:649–658. doi: 10.1128/mcb.13.1.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lang W H, Reeder R H. Transcription termination of RNA polymerase I due to a T-rich element interacting with Reb1p. Proc Natl Acad Sci USA. 1995;92:9781–9785. doi: 10.1073/pnas.92.21.9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langst G, Blank T A, Becker P B, Grummt I. RNA polymerase I transcription on nucleosomal templates: the transcription termination factor TTF-I induces chromatin remodeling and relieves transcriptional repression. EMBO J. 1997;16:760–768. doi: 10.1093/emboj/16.4.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liaw P C, Brandl C J. Defining the sequence specificity of the Saccharomyces cerevisiae DNA binding protein REB1p by selecting binding sites from random-sequence oligonucleotides. Yeast. 1994;10:771–787. doi: 10.1002/yea.320100608. [DOI] [PubMed] [Google Scholar]
- 37a.Lockhart, D. J. Personal communication.
- 38.McLean M, Hubberstey A V, Bouman D J, Pece N, Mastrangelo P, Wildeman A G. Organization of the Saccharomyces cerevisiae actin gene UAS: functional significance of reiterated REB1 binding sites and AT-rich elements. Mol Microbiol. 1995;18:605–614. doi: 10.1111/j.1365-2958.1995.mmi_18040605.x. [DOI] [PubMed] [Google Scholar]
- 39.Morita T, Shigesada K, Kimizuka F, Aiba H. Regulatory effect of a synthetic CRP recognition sequence placed downstream of a promoter. Nucleic Acids Res. 1988;16:7315–7332. doi: 10.1093/nar/16.15.7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morrow B E, Johnson S P, Warner J R. Proteins that bind to the yeast rDNA enhancer. J Biol Chem. 1989;264:9061–9068. [PubMed] [Google Scholar]
- 41.Morrow B E, Ju Q, Warner J R. Purification and characterization of the yeast rDNA binding protein REB1. J Biol Chem. 1990;265:20778–20783. [PubMed] [Google Scholar]
- 42.Morrow B E, Ju Q, Warner J R. A bipartite DNA-binding domain in yeast Reb1p. Mol Cell Biol. 1993;13:1173–1182. doi: 10.1128/mcb.13.2.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicolaides N C, Gualdi R, Casadevall C, Manzella L, Calabretta B. Positive autoregulation of c-myb expression via Myb binding sites in the 5′ flanking region of the human c-myb gene. Mol Cell Biol. 1991;11:6166–6176. doi: 10.1128/mcb.11.12.6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nonet M, Scafe C, Sexton J, Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987;7:1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ofir R, Dwarki V J, Rashid D, Verma I M. Phosphorylation of the C terminus of Fos protein is required for transcriptional transrepression of the c-fos promoter. Nature. 1990;348:80–82. doi: 10.1038/348080a0. [DOI] [PubMed] [Google Scholar]
- 46.Packham E A, Graham I R, Chambers A. The multifunctional transcription factors Abf1p, Rap1p and Reb1p are required for full transcriptional activation of the chromosomal PGK gene in Saccharomyces cerevisiae. Mol Gen Genet. 1996;250:348–356. doi: 10.1007/BF02174393. [DOI] [PubMed] [Google Scholar]
- 47.Paulmier N, Yaniv M, von Wilcken-Bergmann B, Muller-Hill B. gal4 transcription activator protein of yeast can function as a repressor in Escherichia coli. EMBO J. 1987;6:3539–3542. doi: 10.1002/j.1460-2075.1987.tb02680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ptashne M. A genetic switch: gene control and phage λ. Oxford, England: Cell Press & Blackwell Scientific Publications, Ltd.; 1986. [Google Scholar]
- 49.Remacle J E, Holmberg S. A REB1-binding site is required for GCN4-independent ILV1 basal level transcription and can be functionally replaced by an ABF1-binding site. Mol Cell Biol. 1992;12:5516–5526. doi: 10.1128/mcb.12.12.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 51.Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 53.Schmitt M E, Brown T A, Trumpower B L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scott E W, Baker H V. Concerted action of the transcriptional activators REB1, RAP1, and GCR1 in the high-level expression of the glycolytic gene TPI. Mol Cell Biol. 1993;13:543–550. doi: 10.1128/mcb.13.1.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sellitti M A, Pavco P A, Steege D A. lac repressor blocks in vivo transcription of lac control region DNA. Proc Natl Acad Sci USA. 1987;84:3199–3203. doi: 10.1073/pnas.84.10.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Serfling E. Autoregulation—a common property of eukaryotic transcription factors? Trends Genet. 1989;5:131–133. doi: 10.1016/0168-9525(89)90049-8. [DOI] [PubMed] [Google Scholar]
- 57.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 59.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 60.Velculescu V E, Zhang L, Zhou W, Vogelstein J, Basrai M A, Bassett D E, Jr, Hieter P, Vogelstein B, Kinzler K W. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 61.Wang H, Nicholson P R, Stillman D J. Identification of a Saccharomyces cerevisiae DNA-binding protein involved in transcriptional regulation. Mol Cell Biol. 1990;10:1743–1753. doi: 10.1128/mcb.10.4.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Warner J R. Synthesis of ribosomes in Saccharomyces cerevisiae. Microbiol Rev. 1989;53:256–271. doi: 10.1128/mr.53.2.256-271.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wodicka L, Dong H, Mittmann M, Ho M, Lockhart D J. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 64.Wu H M, Crothers D M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984;308:509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- 65.Zhou P, Thiele D J. Rapid transcriptional autoregulation of a yeast metalloregulatory transcription factor is essential for high-level copper detoxification. Genes Dev. 1993;7:1824–1835. doi: 10.1101/gad.7.9.1824. [DOI] [PubMed] [Google Scholar]