Abstract
Recent studies have shown that the histone-modifying enzymes histone acetyltransferase (HAT) and histone deacetylase (HDAC) are involved in transcriptional activation and repression, respectively. However, little is known about the endogenous genes that are regulated by these enzymes or how specificity is achieved. In the present report, we demonstrate that HAT and HDAC activities modulate transcription of the P-glycoprotein-encoding gene, MDR1. Incubation of human colon carcinoma SW620 cells in 100-ng/ml trichostatin A (TSA), a specific HDAC inhibitor, increased the steady-state level of MDR1 mRNA 20-fold. Furthermore, TSA treatment of cells transfected with a wild-type MDR1 promoter/luciferase construct resulted in a 10- to 15-fold induction of promoter activity. Deletion and point mutation analysis determined that an inverted CCAAT box was essential for this activation. Consistent with this observation, overexpression of p300/CREB binding protein-associated factor (P/CAF), a transcriptional coactivator with intrinsic HAT activity, activated the wild-type MDR1 promoter but not a promoter containing a mutation in the CCAAT box; deletion of the P/CAF HAT domain abolished activation. Gel shift and supershift analyses identified NF-Y as the CCAAT-box binding protein in these cells, and cotransfection of a dominant negative NF-Y expression vector decreased the activation of the MDR1 promoter by TSA. Moreover, NF-YA and P/CAF were shown to interact in vitro. This is the first report of a natural promoter that is modulated by HAT and HDAC activities in which the transcription factor mediating this regulation has been identified.
Transcriptional control is mediated by a hierarchy of regulatory components. Although the interplay between DNA elements and transcription factors occurs within the presence of a complex chromosomal architecture, the contribution of chromatin to transcriptional regulation is not fully understood. However, a heightened interest in this area has been spurred by the recent cloning of the histone-modifying enzymes histone acetyltransferases (HATs) and histone deacetylases (HDACs) (5, 29, 39, 45), enzymes with opposing effects on chromatin organization. HATs specifically catalyze the acetylation of the ɛ-amino group of lysine residues at the N-terminal domain of histones, weakening histone-DNA interactions and leading to a destabilization of nucleosome structure (open chromatin), while HDACs remove the acetyl group, leading to a more closed chromatin configuration. It has been proposed that this restructuring of chromatin regulates accessibility of transcription factors to their DNA targets, whereby open chromatin allows for factor binding and closed chromatin does not (43, 44). While this is likely to be an oversimplified model for the role that these enzymes play in transcriptional regulation, there is a plenitude of evidence that supports a role for HATs and HDACs in gene expression and indicates a general correlation between the level of acetylation and the transcriptional activity of a chromosomal domain (19), with hyperacetylated histones accumulated in regions of active chromatin (14) and hypoacetylated histones concentrated in transcriptionally silenced domains (4).
Recent studies have suggested that histone acetylation and deacetylation are involved in the process of chromatin assembly (20, 39). Moreover, HATs and HDACs have been found to be components of some of the general transcriptional coactivator and corepressor complexes, respectively. For example, the yeast ADA complex, which is required for the function of some acidic activators such as VP16, contains GCN5, a subunit with intrinsic HAT activity that is indispensable for transcriptional activation (7, 42). In mammalian cells, one of the general coactivator complexes contains the CREB binding protein (CBP) (or its homolog p300) and P/CAF (p300/CBP-associated factor), both of which have intrinsic HAT activity (33, 45). This complex interacts with NcoA (nuclear coactivator) to mediate nuclear receptor functions (40). p300 and CBP can also interact with a variety of other transcription factors, including AP-1, YY-1, and SP-1, and it has been proposed that their recruitment to a subset of promoters by these factors confers some specificity to their activity. Conversely, HDAC activity is an inherent component of a general transcriptional corepressor complex which interacts with NcoR (nuclear corepressor) and SMRT (silencing mediator of receptor transcription) to mediate nuclear receptor repression (1, 13, 15, 31), as well as with the Mad-Max complex to confer transcriptional repression (21, 48). In these cases, repression can be relieved by exposure to HDAC-specific inhibitors such as trichostatin A (TSA) and tripoxin, indicating an essential role for HDAC in this process. Taken together, these observations provide a strong link between the activities of the histone-modifying enzymes and gene activation and repression. It is now apparent that transcriptional regulation by a sequence-specific DNA binding factor can be mediated by the recruitment of a histone acetylase or deacetylase to the promoter. While it has been proposed that this occurs via a chromatin-specific mechanism involving modification of core histones, further studies are required to substantiate this model.
An overwhelming majority of the studies that have led to the present models for HAT and HDAC function have been carried out using synthetic promoter/reporter constructs for which there is no endogenous counterpart. Therefore, while these studies have been quite valuable models for providing the framework within which to study the biochemistry of HAT and HDAC, results obtained have been limited in their application to cellular events. It is therefore important to identify systems in which both the endogenous gene and natural promoter constructs are similarly regulated by histone-modifying enzymes in order to provide models in which to study the role of these enzymes in a physiologically relevant setting.
P-glycoprotein is a highly conserved 180-kDa membrane protein that was first identified by virtue of its overexpression in cell lines which exhibited a multidrug resistance (MDR) phenotype; it was later shown that P-glycoprotein is involved in the transport of small molecules and mediates the efflux of drugs from MDR cells (11). The gene that encodes human P-glycoprotein, MDR1, is subject to control by a variety of internal and external stimuli, including differentiation signals, heat shock, cytokines, hormones, and a variety of toxic insults (18). A previous analysis of MDR1 activation by differentiation agents showed that gene expression could be activated by sodium butyrate (NaB), a pleiotropic reagent whose multiple functions include the noncompetitive inhibition of HDAC (27, 30). In the present report, we have expanded upon this early observation and have directly evaluated the effect of the highly specific HDAC inhibitor, TSA, on MDR1 activity. We show that the endogenous MDR1 promoter and MDR1 promoter/reporter constructs are activated to similar extents and within similar time frames by TSA and NaB. Activation can also be achieved through the overexpression of P/CAF and requires the intrinsic HAT activity of this factor. Moreover, we show that activation by HDAC inhibitors or P/CAF is dependent on both an intact inverted CCAAT box and the transcription factor NF-Y. Lastly, we demonstrate the interaction of NF-YA with P/CAF in vitro and discuss models by which activation may be mediated. Thus, the MDR1 promoter represents one of the few known natural model systems in which to study the effects of HAT and HDAC in vivo.
MATERIALS AND METHODS
Cell lines, plasmids, and transfections.
The human colon carcinoma cell line SW620 (ATCC CCL227) was maintained on RPMI 1640 medium supplemented with 10% fetal calf serum and 2.0 mM glutamine. All MDR1 promoter/luciferase deletion constructs and CCAAT-box mutants were derived from pMDR1(−1202), which was created by inserting an XmaI-NheI MDR1 promoter sequence (−1202 to +118) into the luciferase vector, pGL2B (Promega, Madison, Wis.), between the SmaI and NheI sites. The CCAAT-box mutant constructs pMDR1(MUTC1) and pMDR1(MUTC2) were generated by site-directed mutagenesis, using conditions described previously (16, 17). Briefly, single-stranded plasmid pMDR1(−1202) was prepared as recommended by Promega (35). Oligonucleotides used for mutagenesis were MUT C1 (5′-CTG TTC CTG CCC AGC gAg TCA GCC TCA CCA CAG-3′), MUT C2 (5′-CTG TTC CTG CCC AGC acA TCA GCC TCA CCA CAG-3′) (mutated sequences indicated in lowercase), and a selection oligonucleotide which converted the unique BamHI site in pMDR1(−1202) to a PvuII site. Oligonucleotides and single-stranded pMDR1(−1202) were annealed, followed by second-strand DNA synthesis using T4 DNA polymerase. The resulting mix was used to transform BMH71-17 MutS cells (United States Biochemical Corp., Cleveland, Ohio). Miniprep plasmid was digested with BamHI to eliminate wild-type plasmid and transformed into Escherichia coli JM109. Both deletion and site-directed mutants were confirmed by DNA sequencing using a CircumVent Thermal Cycle DNA sequencing kit (New England Biolabs, Inc., Beverly, Mass.).
SW620 cells were transfected by the calcium phosphate-DNA precipitation method as described previously (30). Briefly, for transient transfection, cells were seeded in six-well plates at a density of 1 × 105 to 2 × 105 cells/well. The following day, cells were transfected with the indicated pMDR1 construct (0.6 to 1.0 μg/well) or cotransfected with 0.6 to 1.0 μg of a pMDR1 construct and 0.8 to 1.0 μg of P/CAF expression vector (45) or the indicated amount of NF-YA29 expression vector (26). The total amount of DNA per well was adjusted to 2.0 μg by the addition of sonicated salmon sperm DNA. Cells were incubated for 24 h prior to treatment with TSA or NaB. Cells were treated for 24 h before harvesting. For stable transfection, cells were seeded in 150-mm-diameter plates at a density of 1 × 106 to 3 × 106 cells per plate. After 24 h, cells were transfected with pMDR1(−1202) (20 μg/plate) and p308 (ATCC 37613), which encodes the gene conferring resistance to G418 (1 μg/plate). After 2 days, cells were split at a ratio of 1:10, and stable transfectants were selected in G418 (800 μg/ml; Life Technologies, Inc., Gaithersburg, Md.) for 3 weeks. Resultant colonies were pooled for further analysis. Luciferase assays were performed as recommended by the vendor (Promega) and normalized relative to protein concentration as determined by the bicinchoninic acid protein assay (Pierce, Rockford, Ill.). The promoter activity was then expressed as luminescence units, which is the ratio of luminescence counts of 10 μl of cell lysate and the absorbance at 595 nm for the same amount of cell lysate stained with 250 μl of bicinchoninic acid protein assay reagent.
RNase protection assay.
pWEB, the plasmid used for synthesis of the riboprobe specific for MDR1, was constructed by cloning an EcoRI-XmnI fragment of the MDR1 cDNA (+1176 to +1459) into EcoRI/HincII-digested pBluescript II KS(+) (Stratagene, La Jolla, Calif.). pSGX was generated by inserting an EcoRI/XbaI fragment of human GAPDH cDNA (+1 to +777) into pBluescript II SK(−) (Stratagene). RNA was extracted from either untreated or treated (with TSA or NaB) SW620 cells as described previously (8) and subjected to RNase protection analysis (16, 17). Briefly, radiolabeled riboprobes were synthesized from EcoRI-digested pWEB or EcoNI-digested pSGX with T3 RNA polymerase as recommended by the vendor (Promega). Cellular RNA (20 to 40 μg for MDR1 mRNA detection or 0.6 μg for GAPDH mRNA detection) and 1 × 105 to 5 × 105 cpm of riboprobe were mixed in a total volume of 30 μl of hybridization buffer containing 40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 6.4), 1 mM EDTA (pH 8.0), 0.4 M NaCl, and 80% formamide. The nucleic acid mixture was denatured at 85°C for 5 to 10 min, followed by annealing at 50°C for 12 to 15 h. Then 270 μl of RNase digestion mix containing 50 mM sodium acetate (pH 4.4), 100 mM NaCl, 10 mM EDTA (pH 8.0), and 30 U of RNase T2 (Life Technologies, Inc.) per ml was added to the mixture, which was incubated at 37°C for 1 h; 300 μl of solution D (4 M guanidinium thiocyanate, 25 mM sodium citrate [pH 7.0], 0.5% Sarkosyl, 0.1 M 2-mercaptoethanol), 20 μg of tRNA, and 600 μl of isopropanol were added to the mixture, and the RNA was precipitated at −20°C for 1 h. Samples were denatured at 85 to 90°C for 3 min and resolved on a 4% denaturing polyacrylamide gel. Bands were visualized by autoradiography and quantitated by Betascope (Betagen Corp., Waltham, Mass.) radioimaging.
Gel mobility shift assays.
Nuclear extracts were prepared from SW620 cells essentially as described previously (22). Approximately 10 μg of nuclear extract was preincubated at room temperature for 15 min, with or without various unlabeled competitor DNAs, in 100 mM KCl–10 mM HEPES (pH 7.9)–2.5 mM MgCl2–0.5 mM dithiothreitol–25 to 50 μg of poly(dI-dC) per ml in a total volume of 20 μl; 20,000 cpm (∼0.5 ng) of 5′-end-labeled probe was then added and the mixture was incubated at room temperature for an additional 20 min. Complexes were resolved on a 4% nondenaturing polyacrylamide gel in 0.5× Tris-borate-EDTA. For gel mobility supershift assays, nuclear extracts were preincubated with mouse monoclonal anti-NF-YA antibody (25) or one of two different rabbit polyclonal anti-NF-YB antibodies (25, 32) at 4°C for 3 h under the conditions described above, prior to the addition of the labeled oligonucleotide probe. The sequences of the upper strands of the double-stranded oligonucleotides used as competitors are as follows: WT (wild type), 5′-GGTGAGGCTGATTGGCTGGGCAGGA-3′; MDR MUT C1, 5′-GGTGAGGCTGAcTcGCTGGGCAGGA-3′; MDR MUT C2, 5′-GGTGAGGCTGATgtGCTGGGCAGGA-3′; and NS (nonspecific), 5′-CATGCACATTTGTTTAACATTTGTCTTGCACAATTG-3′.
In vitro transcription-translation and pull-down assay.
pNF-YA (Genome Systems, Inc., St. Louis, Mo.), the plasmid used in in vitro transcription-translation assays, contains the human NF-YA cDNA cloned downstream of the T7 promoter in the pT7T3D-Pac vector (Pharmacia Biotech, Piscataway, N.J.). pKS-flag-P/CAF was constructed by inserting the EcoRI-HindIII fragment of pCX-flag-P/CAF (45) into pBluescript II KS(+) (Stratagene). NF-YA and flag-tagged P/CAF were separately translated in the presence of [35S]methionine in a 50-μl volume reaction, using a TNT T7 Quick coupled transcription-translation system (Promega) under conditions recommended by the vendor. Then 50 μl of NF-YA and 25 μl of flag-P/CAF transcription-translation mixes (or 1 μg of flag-tagged bacterial alkaline phosphatase [flag-BAP] protein as a control) were pooled and incubated at 4°C for 1 h in 1.0-ml pull-down buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and protease inhibitor cocktail [Complete; Boehringer Mannheim, Indianapolis, Ind.]); 20 μl (bed volume) of M2 agarose beads (Eastman Kodak, New Haven, Conn.) was added to the mix, and incubation continued overnight at 4°C. The beads were recovered by centrifugation and washed twice with the same pull-down buffer with 2% bovine serum albumin followed by two washes with pull-down buffer without bovine serum albumin. The pull-down was analyzed by sodium dodecyl sulfate (SDS)–10% polyacrylamide gel electrophoresis followed by radioautography. The integrity of the in vitro-translated NF-YA and that of flag-P/CAF were confirmed by Western blot analysis using anti-NF-YA antibody (25) and anti-P/CAF antibody (45).
RESULTS
MDR1 gene expression is specifically induced upon exposure to TSA.
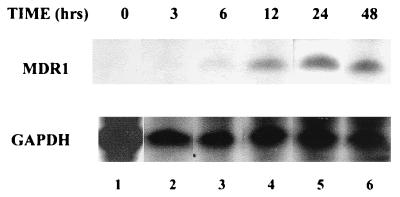
Previous studies had indicated that transcription of MDR1 is induced by NaB, an agent with pleiotropic cellular effects including the inhibition of HDAC (30). To determine whether histone acetylation/deacetylation was the mechanism underlying MDR1 induction, studies were carried out with TSA, a well-characterized, potent, and specific mammalian HDAC inhibitor (46). SW620 colon carcinoma cells were grown to 40 to 50% confluence and treated with TSA (100 ng/ml [∼0.3 μM]) for various time periods. Total RNA was isolated and analyzed by RNase protection using an MDR1-specific probe; levels of GAPDH mRNA were also determined and used for normalization. As shown in Fig. 1, increased levels of MDR1 mRNA could be detected as early as 6 h (lane 3, upper panel), reaching a maximum increase of ∼20-fold by 24 h (lane 5, upper panel).
FIG. 1.
Time course of MDR1 mRNA induction by TSA in SW620 cells. SW620 cells were treated with TSA (100 ng/ml) for 0, 3, 6, 12, 24, and 48 h as indicated. Total RNA prepared from these cells was analyzed by RNase protection using antisense RNA probes specific for MDR1 (upper panel; 20 μg of total RNA/lane) or GAPDH (lower panel; 0.6 μg of total RNA/lane). The levels of MDR1 and GAPDH were quantified with a radioimager (Betascope; Betagen).
An MDR1 promoter/reporter construct is activated by TSA.
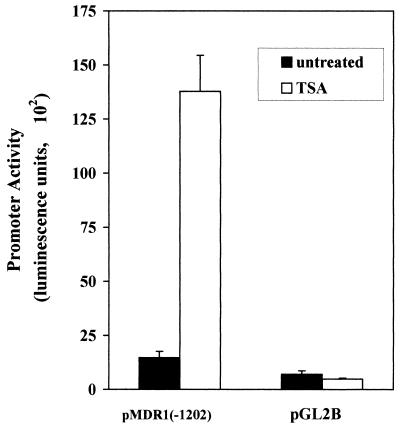
Since the primary effect of inhibiting histone deacetylation has been reported to be modulation of the transcription of a subset of genes (see references 43 and 44 for reviews), we next determined whether TSA modulated activity of the MDR1 promoter. An MDR1/luciferase reporter construct [pMDR1(−1202)], containing promoter sequences from −1202 to +118, was stably transfected into human colon carcinoma SW620 cells as described in Materials and Methods. As a control, we also selected transfectants which contained pGL2B, a promoterless luciferase vector. Both sets of transfectants were exposed to TSA (100 ng/ml) for 24 h, after which luciferase activity was determined. As shown in Fig. 2, luciferase expression driven by the MDR1 promoter increased 10- to 14-fold following TSA treatment, while virtually no change in luciferase activity was observed in transfectants containing the control pGL2B vector. Promoter activation could be detected with TSA concentrations as low as 10 ng/ml (data not shown). These results indicate that the effect of TSA (and, by inference, inhibition of HDAC) was realized at the level of MDR1 transcription.
FIG. 2.
Activation of stably integrated MDR1 promoter by TSA. SW620 cells were stably transfected with either an MDR1 promoter/luciferase reporter construct [pMDR1(−1202)] or with vector alone (pGL2B). The cells were not treated or treated with TSA (100 ng/ml) for 24 h. Luciferase activity of lysed cells was measured and normalized against protein concentration. The data represent three independent experiments performed in triplicate.
An inverted CCAAT-box element is necessary for promoter activation by TSA.
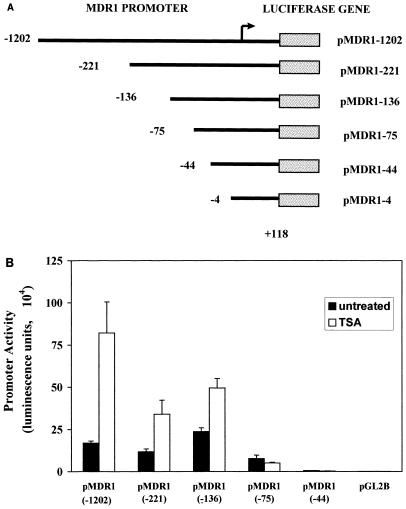
Although some reports have indicated that intact chromatin mediates the action of HAT and HDAC on transcription, and therefore only transcription of promoters which are integrated into chromosomes could be altered by HATs and HDACs (2, 41), several other studies have shown that although HATs and HDACs work most effectively on a nucleosomal template, the activity of transiently transfected promoters could also be efficiently modulated by HATs and HDACs (1, 13, 15, 31, 40). We therefore tested the TSA inducibility of a transiently transfected MDR1 promoter construct. As shown in Fig. 3B, the transiently transfected construct was also activated upon addition of TSA, albeit to a lesser degree than the stable transfectant. Therefore, to further define the promoter region required for TSA activation, a series of 5′ promoter deletion constructs was generated (Fig. 3A) and transiently transfected into SW620 cells. Following transfection, cells were exposed to TSA for 24 h and then analyzed for luciferase activity (Fig. 3B). Deletion of sequences to −136 increased basal expression ∼2-fold, suggesting the presence of a repressor binding site within this region. TSA inducibility was also decreased by this deletion. However, deletion of sequences from −136 to −75 reduced basal activity to ∼50% and, most significantly, abolished activation by TSA. Basal activity was drastically affected when sequences from −75 to −44 were removed, and only background activity was supported by a −4/+118 construct (data not shown), which is comparable to the promoterless construct pGL2B, consistent with results reported previously (27, 28, 30, 34, 38).
FIG. 3.
Activation of MDR1 promoter deletion constructs by TSA in SW620 cells. (A) MDR1 promoter deletion constructs. MDR1 5′ deletion promoters (5′ ends and 3′ ends as indicated) were inserted into the pGL2B vector directly upstream of the luciferase gene (stippled box). The arrow indicates the major MDR1 transcription initiation site. (B) Activation of MDR1 promoter deletion constructs determined by transient transfection assays. SW620 cells were transiently transfected with 1.0 μg of the indicated deletion construct and not treated or treated with TSA (100 ng/ml) for 24 h. Luciferase activity was determined and normalized to protein concentration. The data represent three independent experiments performed in triplicate.
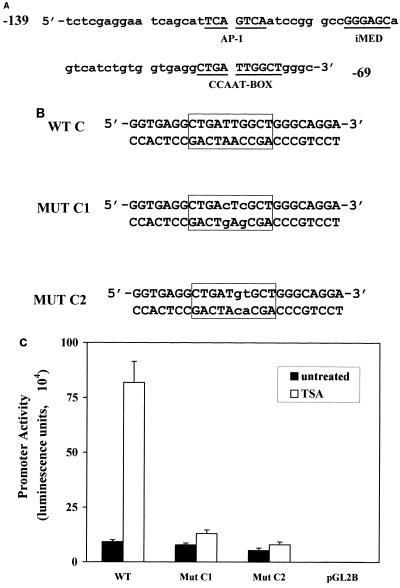
Examination of the sequence between −136 and −75 revealed the presence of three potential transcription factor binding sites (Fig. 4A): a putative AP1 site (−121 to −115), an inverted MED1 element (−105 to −100 [16]), and an inverted CCAAT-box element (−82 to −73) which had previously been reported to be involved in basal transcription of the MDR1 gene (27, 28, 30, 34, 38) as well as in induction by UV irradiation (34), chemotherapeutics (34), differentiation agents (27, 30), and heat shock (28). To determine which, if any, of these elements were required for TSA induction, the activities of promoter/luciferase constructs in which each element was independently mutated within the context of the −1202 promoter were determined following transient transfection into SW620 cells. Mutation of either the AP1-like or inverted MED1 element had no effect on TSA inducibility (data not shown). However, two different mutations in the CCAAT-box element (Mut C1 and Mut C2 [Fig. 4B]) abolished promoter response to TSA (Fig. 4C), indicating an absolute requirement for this element in the induction response.
FIG. 4.
The CCAAT-box element is required for TSA activation. (A) MDR1 promoter sequence from −139 to −69 relative to the major transcription site. Underlined sequences are putative transcription factor binding sites as indicated. (B) Double-stranded oligonucleotides used for site-directed mutagenesis and gel mobility shift assays. WT is the wild-type probe corresponding to the MDR1 promoter sequence from −89 to −64. MUT C1 and MUT C2 contain mutations in the inverted CCAAT box (outlined) as indicated by lowercase letters. (C) Mutation of the CCAAT box decreases MDR1 promoter activation by TSA. The MDR1 promoter wild-type construct pMDR(−1202) (WT) or two CCAAT-box mutant constructs (Mut C1 and Mut C2) were transfected into SW620 cells with or without TSA (100 ng/ml) treatment. Results represent three independent experiments performed in triplicate.
Activation of the MDR1 promoter by P/CAF, a transcriptional coactivator with intrinsic HAT activity, is dependent on an intact CCAAT box and the P/CAF HAT domain.
Since the only known function of TSA is the inhibition of HDAC activity (46, 47), our results thus far strongly suggested that activation of the MDR1 promoter by TSA involved a disruption of the normal ratio of HAT to HDAC activity; i.e., inhibition of HDACs led to hyperacetylation, which resulted in activation. To strengthen this observation, a second method was used to alter the HAT/HDAC ratio: rather than decrease HDAC activity, HAT activity was increased. To accomplish this, a construct expressing P/CAF, a p300/CBP-associated factor with intrinsic HAT activity, was cotransfected with either a wild-type [pMDR1(−1202)] or CCAAT-box-mutated [pMDR1(mutC1) or pMDR1(mutC2)] construct into SW620 cells. While the overexpression of P/CAF activated the wild-type promoter ∼6-fold, it had no effect on the CCAAT-box mutants (Fig. 5A).
FIG. 5.
P/CAF requires an intact CCAAT box and the HAT domain for MDR1 promoter activation. (A) A 1.0-μg aliquot of wild-type (WT) or CCAAT mutant (MutC1 or MutC2) MDR1 reporter construct was transfected into SW620 cells with or without 0.8 μg of P/CAF expression vector. Luciferase activity was determined and normalized to protein concentration. Results represent the average of two independent experiments performed in triplicate. (B) A 0.8-μg aliquot of wild-type MDR1 reporter construct was cotransfected with 1 μg of either empty cloning vector (pCI), wild-type pCAF (pCI-P/CAF), or a mutant P/CAF expression vector lacking the HAT domain (pCI-P/CAF-Δ579-608). Luciferase activity was determined and normalized to protein concentration. Experiments were performed in triplicate.
To determine whether activation of the MDR1 promoter was dependent on the acetyltransferase activity of P/CAF, a mutant P/CAF protein lacking HAT activity as a result of a partial deletion of the HAT domain (pCI-flag-P/CAFΔ579-608 [36]) was cotransfected with the MDR1 reporter construct (Fig. 5B) into SW620 cells. The mutant P/CAF had no effect on MDR1 promoter activity, consistent with the prediction that activation was mediated by acetylation. Western blot analysis indicated that the wild-type and mutant P/CAF proteins were expressed to the same approximate levels following transfection into SW620 cells (data not shown). Taken together, these data strongly support both an involvement of HAT in MDR1 activation and a requirement for the CCAAT box in response to changes in the relative activity of the histone-modifying enzymes.
NF-Y mediates the MDR1 promoter response to TSA.
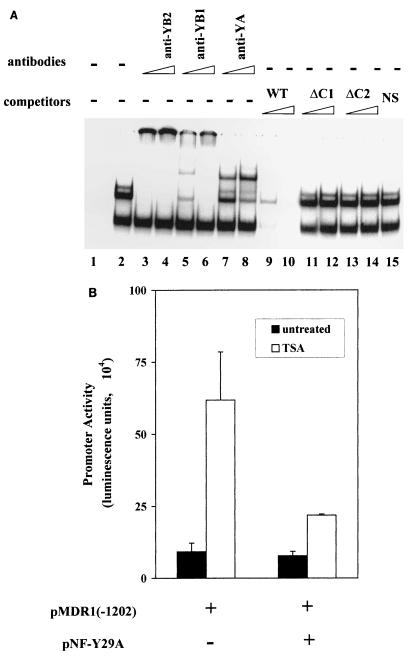
Several transcription factors have been reported to interact with the CCAAT box in various promoters and cell lines (9, 38). To begin to investigate which, if any, of these factors was involved in the TSA response in SW620 cells, gel mobility shift assays were performed with SW620 nuclear extracts prepared from TSA-treated and untreated cells. With the wild-type oligonucleotide depicted in Fig. 4B as a probe, one minor (C1) and two major (C2 and C3) protein-DNA complexes were observed (Fig. 6A, lane 2); formation of all three complexes was competed for by addition of the wild-type oligonucleotide (lanes 9 and 10) but not by the addition of oligonucleotides containing the Mut C1 or Mut C2 mutation (lanes 11 to 14) or nonspecific sequence (lane 15). Whether the cells had been treated with TSA prior to preparation of nuclear extract had no apparent effect on complex formation (data not shown).
FIG. 6.
Binding of NF-Y to the CCAAT box mediates TSA induction. (A) Gel mobility shift assay indicates specific complex formation at the CCAAT box. 32P-labeled wild-type oligonucleotide (WT; 0.5 ng [20,000 cpm]; lane 1) was incubated with nuclear extracts prepared from untreated SW620 cells (lane 2) with 25 to 50 ng of the indicated competitors (lanes 9 to 15). For supershift analysis, nuclear extracts were preincubated with the indicated antibodies before the addition of the probe: polyclonal anti-NF-YB2 (lanes 3 [50 ng] and 4 [100 ng]), polyclonal anti-NF-YB1 (lanes 5 [50 ng] and 6 [100 ng]), or monoclonal anti-NF-YA (lanes 7 [100 ng] and 8 [200 ng]) antibody. (B) Overexpression of a dominant negative NF-Y construct (NF-YA29) decreased promoter induction by TSA. pMDR1(−1202) (1.0 μg) was cotransfected with or without 0.6 μg of NF-YA29 expression vector as indicated. Transfectants were then not treated or treated with TSA (100 ng/ml) for 24 h. Results shown represent three independent experiments performed in triplicate.
The identity of the transcription factor interacting with the MDR1 CCAAT box was determined by supershift analysis. Since it had previously been reported that NF-Y interacted with the MDR1 CCAAT-box element in HCT 116 and HepG2 nuclear extracts (38), we began by determining whether NF-Y was also the interacting species in SW620 cells. Mammalian NF-Y is comprised of three subunits, NF-YA, NF-YB, and NF-YC (24, 37), which are highly conserved throughout evolution. Antibodies against either NF-YA or NF-YB were therefore used to investigate the presence of NF-Y in the gel shift complexes (Fig. 6A). Preincubation with either of the two polyclonal anti-NF-YB antibodies (lanes 3 to 6) or a mouse anti-NF-YA monoclonal antibody (lanes 7 and 8) resulted in supershift of the C1 and C2 complexes (lanes 7 and 8). Higher concentrations of anti-NF-YB antibodies (lanes 3, 4, and 6) aggregated the NF-Y complex and prevented the probe from entering the gel, consistent with the initial observations made with these antibodies (25). Essentially the same results were observed whether the nuclear extract was prepared from treated or untreated cells. Moreover, neither preimmune sera nor antibodies against two other CCAAT-box binding proteins, C/EBP and YB-1, altered complex formation (data not shown).
To investigate the role of NF-Y in promoter response to TSA in vivo, a dominant negative NF-YA expression vector, NF-YA29 (26), was cotransfected with the pMDR1(−1202) reporter into SW620 cells. NF-YA29 is a dominant negative form of NF-YA in which three amino acids in the DNA binding domain are mutated. NF-YA29 forms a complex with NF-YB (and NF-YC), but the complex fails to bind to the CCAAT box and is therefore functionally inactive (26). As shown in Fig. 6B, introduction of NF-YA29 effectively decreased promoter activation by TSA. Taken together, these data support a role for NF-Y in induction of MDR1 transcription by inhibition of HDAC.
NF-YA and P/CAF interact in vitro.
Overexpression of P/CAF activated the MDR1 promoter through the CCAAT box, and NF-Y interacted with the CCAAT box, implicating a linkage between P/CAF and NF-Y. To address the question whether NF-Y and P/CAF interact with each other directly, pull-down experiments were performed. Flag-tagged P/CAF and NF-YA were separately translated in vitro and then coincubated as described in Materials and Methods. As shown in Fig. 7 (lane 5), both flag-P/CAF and NF-YA were pulled down by M2-agarose beads (agarose beads cross-linked with an antiflag antibody). In contrast, when the same experiment was performed with flag-BAP in place of flag-P/CAF (lane 4) or without any flag-tagged protein (lane 3), NF-Y was not present in the pull-down, indicating that the P/CAF protein was required for the interaction to occur. The gel shown in Fig. 7 was stained with Coomassie blue, which demonstrated that all lanes were equally loaded and that the unlabeled flag-BAP was present in the pull-down in lane 4 (data not shown). Similar experiments were performed with NF-YB and NF-YC subunits; however, there was no interaction between these peptides and flag-P/CAF (data not shown).
FIG. 7.
NF-YA and P/CAF interact in vitro. In vitro-translated and 35S-labeled NF-YA was incubated with either in vitro-translated and 35S-labeled flag-tagged P/CAF (lane 5) or 1 μg of unlabeled flag-BAP (lane 4), followed by a pull-down on M2-agarose beads containing cross-linked flag antibody (see Materials and Methods for experimental details). A pull-down using M2 beads in the absence of any flag-tagged protein is shown in lane 3; 2.0% of input NF-YA and 6.0% of P/CAF input are shown in lanes 1 and 2, respectively. Pull-down proteins were resolved on an SDS–10% polyacrylamide gel and subjected to autoradiography.
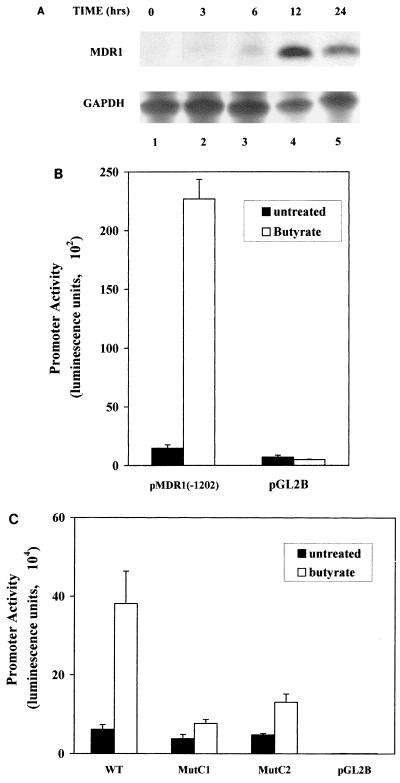
TSA and NaB have similar effects on MDR1 transcription.
Previous studies had indicated that NaB could activate MDR1 gene expression at the level of transcription (30). However, the mechanism by which this was accomplished was unclear, since at the concentrations used NaB has a variety of cellular effects, including alteration of cell proliferation, differentiation, and apoptotic pathways, as well as modulation of receptor abundance and activity of key enzymes (27, 30, 46). One purported effect of NaB at millimolar concentrations is the noncompetitive inhibition of HDAC, leading us to investigate the possibility that activation of MDR1 transcription by NaB occurred through a mechanism similar to that of TSA. First, the time course of induction of MDR1 RNA levels by NaB was investigated. As shown in Fig. 8A, increased steady-state levels of MDR1 could be observed as early as 6 h, with maximum levels (∼20-fold) achieved by 12 to 24 h. Therefore, both the degree and kinetics of induction are similar to those of TSA. We also investigated the effect of NaB on activity of the stably integrated pMDR1(−1202); exposure to 2.0 mM NaB for 24 h resulted in a 16-fold increase in promoter activity (Fig. 8B), similar to what was observed in the presence of TSA (13-fold induction). Promoter activation by NaB was also dependent on an intact CCAAT box, as demonstrated by transient transfection analysis of the wild-type and CCAAT-box mutant promoter constructs in the presence or absence of NaB (Fig. 8C). However, it should be noted that unlike what was observed for TSA-mediated induction, the CCAAT-box mutations did not entirely eliminate activation by NaB, suggesting that NaB may have additional effects on promoter activity. Nevertheless, taken together, these data indicate that NaB activates MDR1 transcription primarily through an HDAC-dependent mechanism.
FIG. 8.
NaB exerts its effect through the CCAAT box. (A) Time course of NaB induction. SW620 cells were treated with NaB (100 ng/ml) for 0, 3, 6, 12, or 24 h as indicated. Total RNA prepared from these cells was analyzed by RNase protection using antisense RNA probes specific for MDR1 (upper panel; 20 μg of total RNA/lane) or GAPDH (lower panel; 0.6 μg of total RNA/lane). The levels of MDR1 and GAPDH were quantified with a radioimager (Betascope). (B) A stably integrated MDR1 promoter is activated by NaB. SW620 cells were stably transfected either with an MDR1 promoter/luciferase reporter construct [pMDR1(−1202)] or with vector alone (pGL2B). The cells were not treated or treated with 2.0 mM NaB for 24 h. Luciferase activity of lysed cells was measured and normalized against protein concentration. The data represent three independent experiments performed in triplicate. (C) Mutation of the CCAAT box reduces promoter activation by NaB. SW620 transiently transfected with wild-type (WT) or CCAAT-box-mutated promoter constructs (MutC1 or MutC2) were not treated or treated with 2.0 mM NaB. Results presented represent three independent experiments performed in triplicate.
DISCUSSION
One recent area of interest in the study of transcriptional regulation is the role played by the histone-modifying enzymes HATs and HDACs. While there is considerable literature supporting the hypothesis that HATs and HDACs have opposing effects on the stabilization and destabilization of chromatin structure, the intricacies of this interplay are not yet understood. Perhaps the greatest conundrum lies in the question of specificity; i.e., how is activation localized to a particular promoter or subset of promoters? At least part of the answer lies in the observation that many transcription factors are able to specifically interact with HATs and HDACs. Moreover, a recent study indicates that histones may not be the only substrates of HATs (12), adding further complexity to the role(s) that these enzymes play in transcriptional regulation.
The large majority of studies of HATs and HDACs in the regulation of transcription have used artificial promoters for analyses. While these studies clearly provide considerable and valuable information, questions regarding promoter specificity may best be addressed by using natural promoters, where the influence of adjacent regulatory elements and factors on HAT and HDAC function can be evaluated. In the present study, we have examined the effects of HAT and HDAC activities on the human MDR1 promoter. Both the endogenous MDR1 gene and transfected MDR1/reporter constructs were activated by HDAC inhibitors; the transfected constructs were also activated by the introduction of the HAT-containing transcription coactivator, P/CAF. In both cases, this activation was found to be dependent on an intact CCAAT box (−82 to −73) and the transcription factor NF-Y. Thus, within the context of the 1.2-kb MDR1 promoter sequence, activation by HAT and HDAC activities is mediated by a single transcription factor binding site. In vitro interaction between NF-YA and P/CAF and preliminary data showing that HAT activity associates with immunoprecipitated NF-Y (19a) suggest that a direct interaction between NF-Y and P/CAF mediates transcriptional activation.
Recent reports in the literature and our data suggest models whereby HATs and HDACs may work to maintain the activity of the MDR1 promoter. In the first and most straightforward model, NF-Y recruits HAT activity to the CCAAT box by virtue of its interaction with P/CAF. The binding of NF-Y would thereby result in histone acetylation, presumably leading to a local disruption of nucleosome structure resulting in transcriptional activation. Under normal growth conditions, the function of HAT would be partially antagonized by an HDAC; upon TSA treatment, HDAC would be specifically inhibited and this antagonism would be relieved. In support of a role for NF-Y in the remodeling of chromatin structure, it is interesting that in the major histocompatibility complex II-associated variant chain promoter, NF-Y was shown to be required for in vivo assembly of a nucleoprotein complex which is crucial for activation by interferon (23); in this case, mutation of the CCAAT box not only abolished interaction with NF-Y but also affected binding of factors to sequences 150 bp upstream.
A second model is suggested by the observation that the inhibition of HDAC and the introduction of P/CAF led to activation of the MDR1 promoter in both transiently and stably transfected cells. Moreover, the majority of studies of HAT and HDAC effects on transiently transfected constructs have reported similar observations (1, 13, 15, 31, 40). However, there are several studies documenting the lack of accurate nucleosome structure associated with transiently transfected constructs. A possible explanation for transcriptional modulation of transiently transfected promoters by HAT and HDAC activities is that although the nucleosomes on transiently transfected plasmids are not normal, sufficient histone association occurs such that acetylation can result in a more open structure. However, a second, more intriguing possibility stems from the recent observation of Gu and Roeder (12), who demonstrated that the HAT-containing factor, p300, specifically acetylates the tumor suppressor gene p53 both in vitro and in vivo, thereby altering its DNA binding capacity. This suggests a model whereby other proteins might be the targets for the HATs and HDACs. It is therefore intriguing to speculate that NF-YB and NF-YC, which contain histone-like motifs and have been suggested to interact in a nucleosome-like structure (3, 37), could be targets for acetylation/deacetylation by the histone-modifying enzymes.
A role for the inverted CCAAT box in basal transcription of the MDR1 gene has been demonstrated previously (30, 38). In one study (38), gel shift analysis indicated that the MDR1 CCAAT-box binding protein in HCT116 and HepG2 cells was NF-Y, although the role of NF-Y in vivo was not addressed. Furthermore, the MDR1 promoter can be induced by a variety of stress-inducing agents, including UV irradiation, serum starvation, heat shock, and chemotherapeutics (10, 27, 28, 30, 34). Interestingly, in all four cases, activation requires sequences between −136 and −75 within the promoter. It will be interesting to determine whether these stress stimuli exert their effects through the binding of NF-Y to the inverted CCAAT box and whether they have a direct effect on the activity of HATs and HDACs in this system.
Our results indicate that NaB and TSA have the same effect on transcription of the MDR1 promoter with respect to the level and time course of endogenous mRNA induction, the degree and time course of promoter activation, and dependence on an intact CCAAT box. Although it had been previously shown that NaB activates MDR1 transcription (10, 30), the mechanism whereby this occurred was unknown, due to the diverse effects of NaB on cellular processes. These effects include the induction of differentiation and apoptosis, alteration in levels of membrane receptors, and changes in the activity of many key enzymes. While the concentration used to activate the MDR1 promoter in SW620 colon carcinoma cells seems high (2.0 mM), the endogenous concentration of NaB in the normal human colon can be as high as 20 mM and is believed to play a role in regulating colon epithelial cell maturation. Mature colon epithelial cells have high levels of expression of MDR1 relative to most other cell types, and it is intriguing to speculate that this high intrinsic expression could be modulated by the effect of NaB on HAT and HDAC activities.
In conclusion, we have demonstrated that proteins capable of modifying histones are involved in the modulation of the MDR1 promoter. This is, to our knowledge, the first report on the regulation of a natural promoter by HAT and HDAC activities in which the transcription factor mediating regulation has been identified. Moreover, this is the first time that the ubiquitous transcription factor NF-Y has been implicated in regulation by HAT and HDAC activities. These results may explain a previously reported role for NF-Y in chromatin assembly (23) and shed light on the regulation of MDR1 gene expression in certain tissue types. Therefore, the MDR1 promoter provides a new, physiologically relevant model in which to study transcriptional activation and repression by HATs and HDACs.
ACKNOWLEDGMENTS
We thank Y. Nakatani for the P/CAF wild-type and HAT deletion expression vectors, R. Mantovani and R. A. Currie for antibodies against NF-YA and NF-YB, R. Mantovani for the NF-YA29 dominant negative construct, and Victoria Richon and Steven Swendenman for helpful discussions. We also thank the members of our laboratory, particularly Tan Ince, for critical review of the manuscript, Sarah Thayer for MDR1 promoter constructs, and Yixing Lin and Kirk Pabon for gel shift studies.
This work was supported by National Cancer Institute grants P30-CA-08748 (Memorial Sloan-Kettering Cancer Center) and RO1-CA-57307 (K.W.S.).
ADDENDUM IN PROOF
While this paper was under review, R. A. Currie also described a direct physical interaction between NF-Y and both P/CAF and GCN5 (J. Biol. Chem. 273:1430–1434, 1998).
REFERENCES
- 1.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, Depinho R A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 2.Bartsch J, Truss M, Bode J, Beato M. Moderate increase in histone acetylation activates the mouse mammary tumor virus promoter and remodels its nucleosome structure. Proc Natl Acad Sci USA. 1996;93:10741–10746. doi: 10.1073/pnas.93.20.10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baxevanis A D, Arents G, Moudrianakis E N, Landsman D. A variety of DNA-binding and multimeric proteins contein the histone fold motif. Nucleic Acids Res. 1995;23:2685–2691. doi: 10.1093/nar/23.14.2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 5.Brownell J, Zhou J, Ranalli T, Kobayashi R, Edmondson D, Roth S, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 6.Bucher P. Weight matrix description of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J Mol Biol. 1990;212:563–579. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- 7.Candau R, Zhou J X, Allis C D, Berger S L. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 1997;16:555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single-step RNA isolation from cultured cells or tissue. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1996. pp. 4.2.2–4.2.3. [Google Scholar]
- 9.Dorn A, Bollekens J, Staub A, Benoist C, Mathis D. A multiplicity of CCAAT box-binding proteins. Cell. 1987;50:863–872. doi: 10.1016/0092-8674(87)90513-7. [DOI] [PubMed] [Google Scholar]
- 10.Frommel T O, Coon J S, Tsuruo T, Roninson I B. Variable effects of sodium butyrate on the expression and function of the MDR1 (P-glycoprotein) gene in colon carcinoma cell lines. Int J Cancer. 1993;55:297–302. doi: 10.1002/ijc.2910550221. [DOI] [PubMed] [Google Scholar]
- 11.Germann U A, Pastan I, Gottesman M. P-glycoproteins: mediators of multidrug resistance. Semin Cell Biol. 1993;4:63–76. doi: 10.1006/scel.1993.1008. [DOI] [PubMed] [Google Scholar]
- 12.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 13.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Histone deacetylase activity is required for full transcriptional repression by msin3a. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 14.Hebbes T R, Clayton A L, Thorne A W, Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinzel T, Lavinsky R M, Mullen T-M, Soderstrom M, Laherty C D, Torchia J, Yang W-M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 16.Ince T A, Scotto K W. A conserved downstream element defines a new class of RNA polymerase II promoters. J Biol Chem. 1995;270:30249–30252. doi: 10.1074/jbc.270.51.30249. [DOI] [PubMed] [Google Scholar]
- 17.Ince T A, Scotto K W. Stable transfection of the P-glycoprotein promoter reproduces the endogenous overexpression phenotype: the role of MED-1. Cancer Res. 1996;56:2021–2024. [PubMed] [Google Scholar]
- 18.Ince T A, Scotto K W. Transcriptional regulation of multidrug resistant genes. In: Bertino J R, editor. Encyclopedia of cancer. Vol. 3. San Diego, Calif: Academic Press; 1997. pp. 1751–1764. [Google Scholar]
- 19.Jeppesen P, Turner B M. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- 19a.Jin, S., and K. W. Scotto. Unpublished data.
- 20.Kaufman P D, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- 21.Laherty C D, Yang W-M, Sun J-M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 22.Lee K A, Green M R. Small-scale preparation of extracts from radiolabeled cells efficient in pre-mRNA splicing. Methods Enzymol. 1990;181:20–30. doi: 10.1016/0076-6879(90)81108-7. [DOI] [PubMed] [Google Scholar]
- 23.Linhoff M W, Wright K L, Ting J P-Y. CCAAT-binding factor NF-Y and RFX are required for in vivo assembly of a nucleoprotein complex that spans 250 base pairs: the invariant chain promoter as a model. Mol Cell Biol. 1997;17:4589–4596. doi: 10.1128/mcb.17.8.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maity S N, Sinha S, Ruteshouser E C, de Crombrugghe B. Three different polypeptides are necessary for DNA binding of mammalian heteromeric CCAAT binding factor. J Biol Chem. 1992;267:16574–16580. [PubMed] [Google Scholar]
- 25.Mantovani R, Pessara U, Tronche F, Li X-Y, Knapp A-M, Pasquali J-L, Benoist C, Mathis D. Monoclonal antibodies to NF-Y define its function in MHC class II and albumin gene transcription. EMBO J. 1992;11:3315–3322. doi: 10.1002/j.1460-2075.1992.tb05410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mantovani R, Li X-Y, Pessara U, van Huisjduijnen R H, Benoist C, Mathis D. Dominant negative analogs of NF-YA. J Biol Chem. 1994;269:20340–20346. [PubMed] [Google Scholar]
- 27.Mickley L A, Bates S E, Richert N D, Currier S, Tanaka S, Foss F, Rosen N, Fojo A T. Modulation of the expression of a multidrug resistance gene (mdr-1/P-glycoprotein) by differentiating agents. J Biol Chem. 1989;264:18031–18040. [PubMed] [Google Scholar]
- 28.Miyazaki M, Kohno K, Uchiumi T, Tanimura H, Matsuo K-I, Nasu M, Kuwano M. Activation of human multidrug resistance-1 gene promoter in response to heat shock stress. Biochem Biophys Res Commun. 1992;187:677–684. doi: 10.1016/0006-291x(92)91248-o. [DOI] [PubMed] [Google Scholar]
- 29.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 30.Morrow C S, Nakagawa M, Goldsmith M E, Madden M J, Cowan K H. Reversible transcriptional activation of mdr1 by sodium butyrate treatment of human colon cancer cells. J Biol Chem. 1994;269:10739–10746. [PubMed] [Google Scholar]
- 31.Nagy L, Kao H-Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S, Evans R E. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 32.Nakshatri H, Bhat-Nakshatri P, Currie R A. Subunit association and DNA binding activity of the heterotrimeric transcription factor NF-Y is regulated by cellular redox. J Biol Chem. 1996;271:28784–28791. doi: 10.1074/jbc.271.46.28784. [DOI] [PubMed] [Google Scholar]
- 33.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 34.Ohga T, Koike K, Ono M, Makino Y, Itagaki Y, Tanimoto M, Kuwano M, Kohno K. Role of the human Y box-binding protein YB1 in cellular sensitivity to the DNA-damaging agents cisplatin, mitomycin C, and ultraviolet light. Cancer Res. 1996;56:4224–4228. [PubMed] [Google Scholar]
- 35.Promega Corporation. WI. Protocols and applications guide. 3rd ed. Madison, Wis: Promega Corp.; 1996. pp. 47–48. [Google Scholar]
- 36.Puri P L, Sartorelli V, Yang X-J, Hamamori Y, Ogryzko V V, Howard B, Kedes L, Wang J Y J, Graessmann A, Nakatani Y, Levrero M. Differential roles of p300 and pCAF acetyltransferases in muscle differentiation. Mol Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 37.Sinha S, Maity S N, Lu J, de Crombrugghe B. Recombinant rat CBF-C, the third subunit of CBF/NFY, allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proc Natl Acad Sci USA. 1995;92:1624–1628. doi: 10.1073/pnas.92.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sundseth R, MacDonald G, Ting J, King A C. DNA elements recognizing NF-Y and Sp-1 regulate the human multidrug-resistance gene promoter. Mol Pharmacol. 1997;51:963–971. doi: 10.1124/mol.51.6.963. [DOI] [PubMed] [Google Scholar]
- 39.Taunton J, Hassig C A, Schreiber S L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 40.Torchia J, Rose D W, Inostroza J, Kamel Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 41.Van Lint C, Emiliani S, Verdin E. Transcriptional activation and chromatin remodeling of the HIV-1 promoter. EMBO J. 1996;15:1112–1120. [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L, Mizzen C, Ying C, Candau R, Barlev N, Brownell J, Allis D, Berger S L. Histone acetyltransferase activity is conserved between yeast and human GCN5 and is required for complementation of growth and transcriptional activation. Mol Cell Biol. 1997;17:519–527. doi: 10.1128/mcb.17.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolffe A P, Pruss D. Targeting chromatin disruption: transcription regulators that acetylate histones. Cell. 1996;84:817–819. doi: 10.1016/s0092-8674(00)81059-4. [DOI] [PubMed] [Google Scholar]
- 44.Wolffe A P. Sinful repression. Nature. 1997;387:16–17. doi: 10.1038/387016a0. [DOI] [PubMed] [Google Scholar]
- 45.Yang X-J, Ogryzko V V, Nishikawa J-I, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 46.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 47.Yoshida M, Horinouchi S, Beppu T. Trichostatin A and Tripoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylase and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]