Abstract
Germline pathogenic variants in the BRCA2 gene are strongly correlated with an elevated risk of developing breast cancer. Two specific BRCA2 variants, c.8167G>C (p.Asp2723His) and c.1583del (p.Asn528fs), have been identified from individuals with a family history of breast cancer. Here we generated two iPSC lines from breast cancer patients who are heterozygous carriers of these two variants. These iPSCs exhibit pluripotency and demonstrate the capability to differentiate into three germ layers. These iPSC lines represent a valuable resource for personalized pre-clinical research, offering new opportunities to explore the underlying mechanisms of breast cancer and develop targeted therapeutic approaches.
Resource utility
The pathogenic variant BRCA2 c.8167 (p.Asp2723His) (ClinVar ID:52515) and c.1583del (p.Asn528fs) (ClinVar ID:1190132) have been detected in individuals with breast cancer. Human iPSC lines harboring these mutations are valuable resources for tissue-specific cell types that can be used for investigating disease mechanisms and drug screening.
Resource Details
BRCA2 is a critical tumor suppressor that maintains genome integrity by regulating DNA repair and stabilizing the replication forks under stress (Gudmundsdottir and Ashworth, 2006). Pathogenic germline BRCA2 variants confer a high risk of breast and ovarian cancer (Loibl et al., 2021). Indeed, individuals carrying these variants have a lifetime risk of 55% for breast cancer and 16.5% for ovarian cancer (Mavaddat et al., 2013). Consequently, genetic testing for BRCA2 variants has become integral to clinical practice. However, the specific cancer risk associated with different locations of mutations and their functional effects are not yet fully understood (Guidugli et al., 2014). This knowledge gap often causes anxiety and uncertainty among individuals with BRCA2 variants. Therefore, there is an urgent need for experimental evidence to comprehensively investigate the contribution of BRCA2 variants to the initiation and progression of breast and ovarian cancers. In this study, we have successfully generated two human iPSC lines from individuals carrying heterozygous pathogenic variants in the BRCA2 gene. These iPSC lines represent a valuable resource for future investigations on the effects of BRCA2 mutations on the development and progression of breast cancer, and they can also be used for drug screening in precision medicine applications.
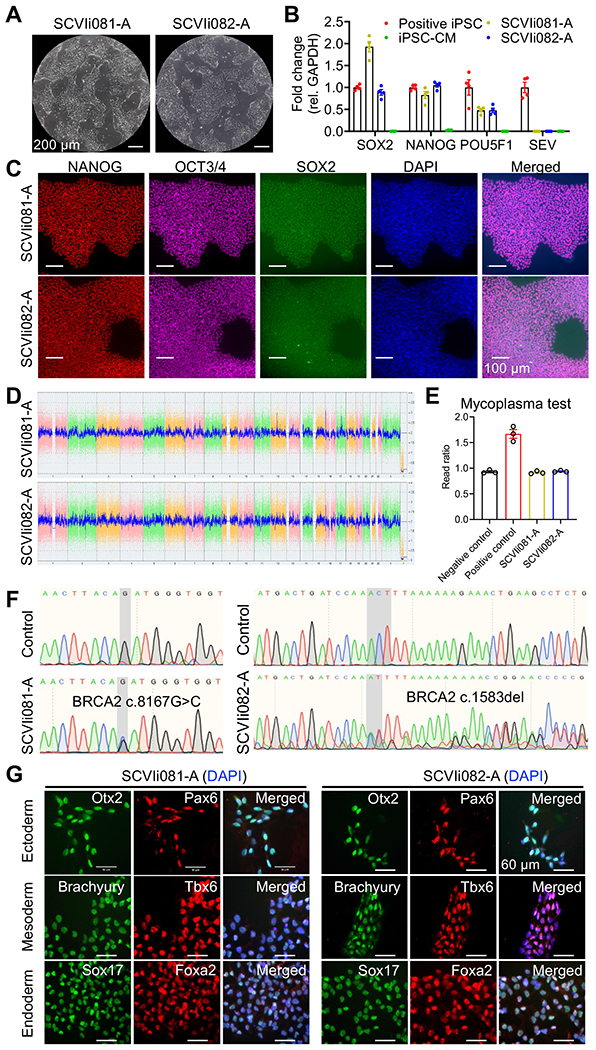
We recruited two breast cancer patients, a 54-year-old white female who developed breast cancer with stage IIIB T4N1M0 of the left breast and a 43-year-old East Asian female who developed breast cancer with stage IIB cT2N1M0 of the right breast, carrying c.8167G>C and c.1583del respectively. Using the peripheral blood mononuclear cells (PBMCs) obtained from these patients (Jahng et al., 2021), we successfully generated two human iPSC lines named SCVIi081-A and SCVIi082-A. These iPSC lines exhibited the typical morphology of iPSCs (Fig. 1A) and robust expression of pluripotency markers such as SOX2, NANOG, and POU5F1 (Fig. 1B), which was further confirmed by immunofluorescence (Fig. 1C). Importantly, at passage 17, the expression of the non-integrating Sendai virus used for reprogramming was no longer detectable in SCVIi081-A and SCVIi082-A (Fig. 1B). Karyotyping analysis confirmed that both iPSC lines had normal chromosome profiles in both cell lines (Fig. 1D). To ensure the quality of the iPSC lines, we tested them for mycoplasma contamination, and both lines were found to be mycoplasma-negative (Fig. 1E). The presence of the BRCA2 mutations was confirmed by Sanger sequencing. SCVIi081-A exhibited double peaks (G and C) at position c.8167 (Fig. 1F left panel), indicating a heterozygous mutation from G to C, resulting in a missense mutation at the protein level. In SCVIi082-A, the sequencing showed double peaks (A and C) at position c.1583, followed by continuous doublets (Fig. 1F right panel), indicating a heterozygous deletion of A and resulting in a frameshift mutation. Short tandem repeat analysis confirmed that SCVIi081-A and SCVIi082-A had identical DNA profiles to their donor PBMCs (Submitted in the archive with the journal). Furthermore, we assessed the ability of these iPSC lines to differentiate into the three germ layers. The results demonstrated that both SCVIi081-A and SCVIi082-A retained the capacity to differentiate into ectoderm, endoderm, and mesoderm lineages (Figure 1G).
Materials and Methods
1. Generation of human induced pluripotent stem cells
The donors’ blood samples were collected upon signed informed consent at Stanford University. PBMCs were isolated and purified from the blood samples by PercollR gradient separation and then maintained in the StemPro™-34 SFM medium (100 ng/mL SCF, 100 ng/ mL FLT3, 20 ng/mL IL-3, 20 ng/mL IL-6, and 20 ng/mL EPO). For reprogramming, PBMCs were plated at a density of 3 X 104 cells per cm2 and transduced using the CytoTune™-iPS 2.0 Sendai Reprogramming Kit (ThermoFisher Scientific). At day 5 post-transduction, cells were resuspended and plated on Matrigel-coated plates with StemPro™-34 medium. At day 7 post-transduction, the media was changed to 1:1 mix of StemMACS™ iPS-Brew medium (Miltenyi Biotec) and StemPro™-34 medium until colonies appeared. On day 8, the culture medium was fully switched to the Brew medium. At around day 10–15 post-transduction, iPSC colonies appeared and were picked under the microscope for expansion.
2. RT-qPCR
RNA was extracted by using the miRNeasy Micro Kit (Qiagen). The cDNA was generated from 1 μg RNA with the iScript™ Reverse Transcription Supermix (BIO-RAD). The expression levels of target genes were examined using commercially available probes (Table 2) and the TagMan™ Universal PCR Master Mix ( ThermoFisher Scientific).
Table 2:
Reagents details
| Antibodies used for immunocytochemistry/flow-cytometry | ||||
|---|---|---|---|---|
| Antibody | Dilution | Company Cat # | RRID | |
| Pluripotency Markers | Rabbit Anti-Nanog | 1:100 | Proteintech Cat# 14295-1-AP |
RRID: AB_1607719 |
| Mouse IgG2b κOct3/4 antibody | 1:100 | Santa Cruz Biotechnology Cat# sc-5279 |
RRID: AB_628051 | |
| Goat IgG anti Sox2 | 1:100 | R and D Systems Cat# AF2018 |
RRID: AB_355110 | |
| Ectoderm Markers | Goat Anti-Otx2 | 1:200 | R and D Systems Cat# AF1979 |
RRID: AB_2157172 |
| Rabbit Anti-Pax6 | 1:200 | Thermo Fisher Scientific Cat# 42-6600 |
RRID: AB_2533534 | |
| Endoderm Markers | Goat Anti-Sox17 | 1:200 | R and D Systems Cat# AF1924 |
RRID: AB_355060 |
| Rabbit Anti-Foxa2 | 1:250 | Thermo Fisher Scientific Cat# 701698 |
RRID: AB_2576439 | |
| Mesoderm Markers | Goat Anti-Brachyury | 1:200 | R and D Systems Cat# AF2085 |
RRID: AB_2200235 |
| Rabbit Anti-Tbx6 | 1:200 | Thermo Fisher Scientific Cat# PA5-35102 |
RRID: AB_2552412 | |
|
Secondary antibodies |
Alexa Fluor 647 Goat Anti-Mouse IgG2b | 1:250 | Thermo Fisher Scientific Cat# A-21242 |
RRID: AB_2535811 |
| Alexa Fluor 555 Goat Anti-Rabbit IgG (H+L) | 1:500 | Thermo Fisher Scientific Cat# A-21428 |
RRID: AB_141784 | |
| Alexa Fluor 488 Donkey Anti-Goat IgG | 1:1000 | Thermo Fisher Scientific Cat# A-11055 |
RRID: AB_2534102 | |
| Primers | ||||
| Target | Size of band | Forward/Reverse primer (5′-3′) | ||
| Sendai virus plasmid (qPCR) | Sendai virus genome | 181bp | Mr042698800_mr (Thermo Fisher Scientific) | |
| Pluripotency Markers (qPCR) | NANOG | 109bp | Hs02387400_g1 (Thermo Fisher Scientific) | |
| House-Keeping Genes (qPCR) | POU5F1 | 77bp | Hs00999632_g1 (Thermo Fisher Scientific) | |
| Pluripotency Markers (qPCR) | SOX2 | 86bp | Hs04234836_s1 (Thermo Fisher Scientific) | |
| House-Keeping Genes (qPCR) | GAPDH | 157bp | Hs02786624_g1 (Thermo Fisher Scientific) | |
| Genotyping | BRCA2 c.8167G>C | 350bp | Forward: 5’-TCACTTTTAGATATGATACG-3’ Reverse: 5’-TTCTGGGGCTTCAAGAGGTG-3’ |
|
| Genotyping | BRCA2 c.1583del | 350bp | Forward: 5’-AGGAAACAGTGGTAAATAAG-3’ Reverse: 5’-TGCATTCTTCAAAGCTACAG-3’ |
|
3. Immunofluorescence staining
At room temperature, cells were fixed with 4% paraformaldehyde for 20 minutes. After twice washes with DPBS, the fixed cells were incubated with DPBS containing 0.1% TritonX100 for 10 minutes, followed by incubation with the blocking solution (DPBS with 1% goat serum) for 1 hour. After three times washes, the cells were incubated with the primary antibodies (Table 2) overnight at 4 °C. On day 2, the cells were washed three times with DPBS and incubated with the second antibodies (Table 2) for 1 hour. After three times washes with DPBS, nuclei were counterstained with NucBlue Probes (ThermoFisher Scientific) for imaging.
4. Karyotyping
Around 2 million cells were collected at passage 17 and then subjected to the KaryoStat™ assay (ThermoFisher Scientific) analysis.
5. Targeted sequencing
The genomic DNA from each cell line was extracted using the QuickExtract™ DNA Extraction Solution. The PCR assay was performed with the PrimeSTAR GXL DNA Polymerase (Clontech) and the primers in Table 2. The PCR products were purified and sequenced by the Stanford Protein and Nuclear Acid (PAN) facility.
6. Mycoplasma detection
The culture medium from full confluent iPSCs was collected for the mycoplasma test with the MycoAlert™ Detection Kit (Lonza).
7. Trilineage differentiation
The StemXVivo Ectoderm kit (R & D systems) and the StemDiff™ Definitive Endoderm differentiation kit (STEMCELL™ Technologies) were used for ectoderm and endoderm differentiation. Mesoderm differentiation was induced by RPMI media (B27 Minus Insulin supplement with 6 μM CHIR (Selleck Chemicals)) for 48 hours.
8. STR analysis
Genomic DNA was extracted with the DNeasy Blood & Tissue Kit (Qiagen). One ng genomic DNA was used for the PCR assay with the CLA IdentiFiler™ Direct PCR Amplification Kit (ThermoFisher Scientific). Capillary electrophoresis was performed on ABI3130xl by the Stanford PAN facility for the fragmental analysis.
Resource Table:
| Unique stem cell lines identifier | 1. SCVIi081-A 2. SCVIi082-A |
| Alternative name(s) of stem cell lines | 1. SCVI2535 (SCVIi081-A) 2. SCVI2838 (SCVIi082-A) |
| Institution | Stanford Cardiovascular Institute, Stanford, CA, US |
| Contact information of distributor | Joseph C. Wu, joewu@stanford.edu |
| Type of cell lines | iPSC |
| Origin | Human |
| Additional origin info required for human ESC or iPSC |
Age: 54 (SCVIi081-A) and 43 (SCVIi082-A)
Sex: Female Ethnicity: White (SCVIi081-A) and East Asian (SCVIi082-A) |
| Cell Source | PBMCs |
| Clonality | Clonal |
| Method of reprogramming | Nonintegrating Sendai virus expression of human OCT4, SOX2, KLF4, and c-MYC |
| Genetic Modification | YES |
| Type of Genetic Modification | Spontaneous/naturally occurred mutation |
| Evidence of the reprogramming transgene loss | RT-qPCR |
| Associated disease | Breast cancer |
| Gene/locus |
BRCA2: 13q13.1
SCVIi081-A: Chr13:32363369 (GRCh38) SCVIi082-A: Chr13:32333060 (GRCh38) |
| Date archived/stock date | 05/07/2023 |
| Cell line repository/bank |
https://hpscreg.eu/cell-line/SCVIi081-A
https://hpscreg.eu/cell-line/SCVIi082-A |
| Ethical approval | The generation of these iPSC lines was approved by the Administrative Panel on Human Subjects Research under Institutional Review Board (IRB) #29904 “Derivation of Human Induced Pluripotent Stem Cells (Biorepository)” |
Table 1:
Characterization and validation
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography Bright field | Visual record of the line: normal | Figure 1 panel A |
| Phenotype | Qualitative analysis: Immunofluorescence staining |
Positive expression of pluripotency markers: Oct3/4, Nanog, Sox2. | Figure 1 panel C |
| Quantitative analysis: RT-qPCR |
High expression levels of pluripotency markers (Nanog, Sox2, and Pou5f1) in iPSCs but absent in differentiated iPSC-CMs | Figure 1 panel B | |
| Genotype | Karyotype (G-banding) and resolution |
Karyostat™ Assay, resolution 1-2Mb:
Normal karyotype: 46, XX for both iPSC lines |
Figure 1 panel D |
| Identity | Microsatellite PCR (mPCR) OR STR analysis |
N/A | N/A |
| 16 loci tested, 100% identical | Submitted in archive with journal | ||
| Mutation analysis (IF APPLICABLE) | Sequencing |
Heterozygous for both lines
SCVIi081-A: c.8167G>C SCVIi082-A: c.1583del |
Figure 1 panel F |
| Southern Blot OR WGS | N/A | N/A | |
| Microbiology and virology | Mycoplasma | Mycoplasma testing by luminescence: Negative | Figure 1 panel E |
| Differentiation potential | Directed differentiation | Positive IF staining of three germ layer markers | Figure 1 panel G |
| List of recommended germ layer markers | Expression of these markers has to be demonstrated at mRNA (RT PCR) or protein (IF) levels, at least 2 markers need to be shown per germ layer |
Positive expression of germ layer markers: Ectoderm: PAX6, OTX2; Endoderm: SOX17, FOXA2; Mesoderm: BRACHYURY, TBX6 |
Figure 1 panel G |
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | N/A | N/A |
| Genotype additional info (OPTIONAL) | Blood group genotyping | N/A | N/A |
| HLA tissue typing | N/A | N/A |
Acknowledgments
This work was supported by National Institutes of Health 75N92020D00019, R01 HL130020, R01 HL141371, R01 HL150693, R01 HL163680 (JCW), and the Tobacco-Related Disease Research Program(TRDRP) T32FT4853 (M. Zhang).
Footnotes
Declaration of competing interest
J.C.W. is a co-founder of Greenstone Biosciences. The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Gudmundsdottir K, Ashworth A, 2006. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene 25, 5864–5874. 10.1038/sj.onc.1209874 [DOI] [PubMed] [Google Scholar]
- Guidugli L, Carreira A, Caputo SM, Ehlen A, Galli A, Monteiro ANA, Neuhausen SL, Hansen TVO, Couch FJ, Vreeswijk MPG, ENIGMA consortium, 2014. Functional assays for analysis of variants of uncertain significance in BRCA2. Hum Mutat 35, 151–164. 10.1002/humu.22478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahng JWS, Black KE, Liu L, Bae HR, Perez M, Ashley EA, Sallam K, Wu JC, 2021. Generation of three induced pluripotent stem cell lines, SCVIi003-A, SCVIi004-A, SCVIi005-A, from patients with ARVD/C caused by heterozygous mutations in the PKP2 gene. Stem Cell Res 53, 102284. 10.1016/j.scr.2021.102284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loibl S, Poortmans P, Morrow M, Denkert C, Curigliano G, 2021. Breast cancer. The Lancet 397, 1750–1769. 10.1016/S0140-6736(20)32381-3 [DOI] [PubMed] [Google Scholar]
- Mavaddat N, Peock S, Frost D, Ellis S, Platte R, Fineberg E, Evans DG, Izatt L, Eeles RA, Adlard J, Davidson R, Eccles D, Cole T, Cook J, Brewer C, Tischkowitz M, Douglas F, Hodgson S, Walker L, Porteous ME, Morrison PJ, Side LE, Kennedy MJ, Houghton C, Donaldson A, Rogers MT, Dorkins H, Miedzybrodzka Z, Gregory H, Eason J, Barwell J, McCann E, Murray A, Antoniou AC, Easton DF, EMBRACE, 2013. Cancer risks for BRCA1 and BRCA2 mutation carriers: results from prospective analysis of EMBRACE. J Natl Cancer Inst 105, 812–822. 10.1093/jnci/djt095 [DOI] [PubMed] [Google Scholar]


