Abstract
Introduction
Breast tumors often display an astonishing degree of spatial and temporal heterogeneity, which are associated with cancer progression, drug resistance, and relapse. Triple-negative breast cancer (TNBC) is a particularly aggressive and heterogeneous subtype for which targeted therapies are scarce. Consequently, patients with TNBC have a poorer overall prognosis compared to other breast cancer patients. Within heterogeneous tumors, individual clonal subpopulations may exhibit differences in their rates of growth and degrees of invasiveness. We hypothesized that such phenotypic heterogeneity at the single-cell level may accelerate tumor progression by enhancing the overall growth and invasion of the entire tumor.
Methods
To test this hypothesis, we isolated and characterized clonal subpopulations with distinct morphologies and biomarker expression from the inherently heterogeneous 4T1 mouse mammary carcinoma cell line. We then leveraged a 3D microfluidic tumor model to reverse-engineer intratumoral heterogeneity and thus investigate how interactions between phenotypically distinct subpopulations affect tumor growth and invasion.
Results
We found that the growth and invasion of multiclonal tumors were largely dictated by the presence of cells with epithelial and mesenchymal traits, respectively. The latter accelerated overall tumor invasion, even when these cells comprised less than 1% of the initial population. Consistently, tumor progression was delayed by selectively targeting the mesenchymal subpopulation.
Discussion
This work reveals that highly invasive cells can dominate tumor phenotype and that specifically targeting these cells can slow the progression of heterogeneous tumors, which may help inform therapeutic approaches.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12195-023-00792-w.
Keywords: Epithelial–mesenchymal transition, Interstitial fluid flow, Morphodynamics
Introduction
Human cancers often exhibit inter- and intratumoral heterogeneity that can vary over space and time. Subpopulations of cells within individual tumors can be phenotypically diverse [1], and are likely to alter tumor progression as a function of their phenotypes and abundance within the tumor [2–5]. For instance, clones from single MDA-MB-231 human triple-negative breast cancer (TNBC) cells were shown to display heterogeneity in morphology that corresponded to distinct tumorigenicities and metastatic potentials [6]. Additionally, interactions between subpopulations may impact the effectiveness of therapies [3], so intratumoral heterogeneity will likely need to be considered when selecting a therapeutic approach [7]. Despite diverse mutations across patients, drug-resistant subpopulations within individual tumors have been found to converge at the transcriptomic level, which includes signatures of the epithelial–mesenchymal transition (EMT) [1]. Whereas EMT is a highly plastic process critical during normal development and wound healing, activation of EMT in cancer is thought to facilitate resistance, invasion, and metastasis [8–10]. Thus, subpopulations of cells with EMT signatures represent a critical element of clinically significant phenotypic diversity within a tumor.
Several studies have suggested the importance and complexity of phenotypically distinct subpopulations within a tumor. Pioneering work revealed that “invasion-competent” MDA-MB-231 breast cancer cells could initiate the invasion of ordinarily “invasion-incompetent” nonmalignant MCF10A mammary epithelial cells via leader-follower interactions [11]. This study left unresolved whether observations from 1:1 mixtures of unrelated cell lines were reflective of the phenotypic heterogeneity within a tumor. Indeed, when the same group isolated non-migratory and migratory MDA-MB-231 cells from each other and injected them separately into mice, they uncovered counterintuitive relationships between migration and metastatic capacity: less migratory cells had a higher chance of metastasizing than more migratory cells [12–14]. Complicating the picture even further, a different group isolated single-cell-derived clonal populations from the MDA-MB-231 cell line and used these populations to reveal that interactions between clones could enhance tumorigenicity and aggressiveness [15]. This work suggested that physical interactions and soluble signaling between tumor subpopulations may confer heterogeneous tumors with an enhanced capacity for growth, invasion, and resistance to microenvironmental stressors. These studies highlight critical gaps in our understanding of the mechanisms by which heterogeneous cancer cell subpopulations contribute to tumor growth, invasion, and metastasis.
The 4T1 murine TNBC cell line has been frequently used to study the dynamics of collective invasion and the metastatic cascade, as these cells faithfully reproduce the metastatic profile observed in human breast cancer [16–18]. The 4T1 cell line has also been used as a model of breast cancer heterogeneity: clonal analysis and molecular barcoding demonstrated that this population of cells displays striking phenotypic diversity [19–22]. Studies using 4T1 cells and others have begun to shed light on how intratumoral heterogeneity may contribute to drug resistance and metastasis [10, 19, 20, 23–25] and are consistent with the hypothesis that the diversity of clonal subpopulations may play a critical role in malignant progression.
Thus, elucidating the changes in tumor phenotype that result from intratumoral heterogeneity may help to (1) improve the stratification and characterization of patient disease, (2) provide critical diagnostic and prognostic information, and (3) promote the identification of novel therapies geared at targeting the subpopulations of cells that drive overall tumor behavior. As such, studies are needed to define whether interactions between clonal subpopulations synergistically promote tumor growth, invasion, or metastasis. Here, we establish an engineered model of breast tumor heterogeneity by exploiting the inherently heterogeneous nature of the 4T1 mammary carcinoma cell line. Using this system, we find that clonal subpopulations of cancer cells from along the EMT spectrum exhibit different rates of growth and invasion, which play an important role in directing the behavior of multiclonal tumors. The ability to predict the behavior of such multiclonal tumors may improve the selection of potential targets for personalized medicine.
Materials and Methods
Cell Culture
4T1 murine mammary carcinoma cells (ATCC) were cultured in RPMI-1640 medium (Hyclone) supplemented with 10% fetal bovine serum (heat-inactivated, Atlanta Biologicals) and 1% gentamicin (Gibco). Cells were maintained in a humidified incubator under standard conditions (37°C and 5% CO2).
Generation of Stably Fluorescent 4T1 Clonal Subpopulations
4T1 cells were transfected with high-quality plasmid DNA to enable the expression of nuclear fluorescent proteins. The pNLS-iRFP670 plasmid was a gift from Vladislav Verkhusha [26] (Addgene plasmid #45446). Low-passage (P5) 4T1 cells were transfected with the purified pNLS-iRFP670 plasmid DNA using FuGENE transfection reagents. Culture medium containing G418 antibiotic (800 μg/mL, Sigma) was added after 24 h to select for pNLS-iRFP670-expressing cells. Cells were cultured in G418-containing medium for two weeks, after which time the G418 concentration was reduced to 250 μg/mL for maintenance culture. Stably transfected pNLS-iRFP670-expressing cells were subjected to fluorescence-activated cell sorting (BD FACSAria Fusion Cell Sorter) into 96-well plates containing 50% conditioned medium and 50% fresh growth medium to obtain clonal populations. Clones with homogeneous pNLS-iRFP670 fluorescence and representative morphologies (cobblestone epithelial “E1”, epithelial with peripheral protrusions “E2”, amoeboid “A”, and mesenchymal “M”) were propagated and maintained in culture for subsequent experiments. Limiting-dilution cloning was conducted for each of these morphologically distinct 4T1 clonal subpopulations to validate that individual cells from the respective clones could give rise to clonal progenies with consistent morphological features. To confirm our findings of clonal phenotypes, we repeated the limiting-dilution cloning using the first set of clones to obtain a second set of clonal cell subpopulations, which maintained the expected morphologies. This transfection, selection, and single-cell-cloning process was repeated to generate clones that expressed different fluorescent proteins in the nucleus. Briefly, a clonal line of 4T1 cells with mesenchymal morphology that stably expressed NLS-YFP was selected by culturing the cells in medium containing 400-μg/mL zeocin (Invitrogen). Chronic exposure to zeocin can cause single-strand DNA breaks even in cells expressing the zeocin-resistance gene (Sh ble) [27]; as such, 4T1 cells stably expressing NLS-YFP were routinely cultured in medium without zeocin. We exploited the sensitivity of the NLS-YFP clone to 250-μg/mL G418 to selectively target this population within multiclonal tumors.
Low-passage parental 4T1 cells (passage number less than ten) were also subjected to limiting-dilution cloning into 96-well plates containing 50% conditioned medium and 50% fresh growth medium to characterize the morphological heterogeneity of the parental population. The fraction of clones exhibiting the E1, E2, A, and M morphologies were quantified as a function of the total number of clones for each independent cloning experiment (N = 3).
Immunofluorescence Analysis
For 2D immunostaining experiments, parental 4T1 cells and clonal cell lines were cultured on #1.5-thickness glass coverslips coated with 200 μg/mL (5 μg/cm2) of fibronectin in phosphate-buffered saline (PBS) for 72 h and subsequently fixed for 15 min with 4% paraformaldehyde in PBS at 4°C. Samples were washed with PBS, permeabilized with 0.1% Triton X-100 in PBS, blocked for 1 h at room temperature with 10% goat serum in PBS containing 0.1% Triton X-100, and then incubated in blocking buffer with primary antibodies overnight at 4°C. We used the following antibodies: rabbit anti-E-cadherin (1:200, Cell Signaling Technology #3195) and mouse anti-vimentin (1:200, Millipore Sigma #V2258, clone LN-6). Next, samples were washed with PBS and incubated with Alexa Fluor 488 goat anti-rabbit and Alexa Fluor 594 goat anti-mouse secondary antibodies (Invitrogen), respectively, in blocking buffer for 1 h at room temperature. Samples were then washed with PBS, incubated with Hoechst 33342 (1:2000, Invitrogen) for 20 min at room temperature to label nuclei, washed with PBS, and then mounted on glass slides with Fluoromount-G (SouthernBiotech) for subsequent fluorescence imaging. Samples were visualized using a Nikon Ti-U microscope equipped with a digital CCD camera (Hamamatsu ORCA-03G) and images were acquired using a CFI S Plan Fluor ELWD 20xC/0.45 NA air objective.
Spheroid arrays were fixed in 4% paraformaldehyde in PBS for 40 min at 4°C. Samples were then washed with PBS, blocked and permeabilized using 10% goat serum in PBS containing 0.1% Triton X-100, and stained with Alexa Fluor-phalloidin 568 (1:200, Invitrogen) to label the actin cytoskeleton. Samples were counterstained with Hoechst 33342 (1:2000) for several hours at room temperature to label nuclei. Samples were washed several times in PBS for 30 min each and then visualized on a Nikon Ti-U microscope equipped with a spinning disk (X-light V2, Crest Optics) and EM-CCD camera (Hamamatsu ImageEM) using a CFI S Plan Fluor ELWD 20xC/0.45 NA air objective.
Microfabricated Spheroids and 3D Microfluidic Tumor Models
3D microfabricated arrays of spheroids with defined size and shape were engineered in type I collagen gel as described previously [28]. Briefly, a 4-mg/mL solution of neutralized bovine type I collagen (Koken KOU-IAC-50) was gelled at 37°C against a poly(dimethylsiloxane) stamp with defined micrometer-scale features (100-μm-diameter circular posts with a height of 180 μm). Following trypsinization, parental 4T1 cells or clonal cell subpopulations were suspended in HBSS at ~ 5 × 106 cells/mL. A 50-μL droplet of the concentrated cell suspension was added on top of the molded collagen array and cells were allowed to settle into the cylindrical microwells. Cells that landed in the cylindrical microwells eventually formed aggregates referred to as “spheroids”. A droplet of neutralized collagen was then added on top of the cell-filled collagen cavities to fully encapsulate the aggregates. Samples were incubated for 30 h to permit the formation of cell–cell and cell–matrix adhesions, and then evaluated by time-lapse microscopy to visualize cell migration and invasion. Similarly, solid tumor aggregates were engineered by molding a 4-mg/mL solution of type I collagen around a 120-µm-diameter acupuncture needle centered within a microfluidic channel, as described previously [29, 30]. Separately, 3D multicellular aggregates were generated by following the hanging-drop method [31, 32]. Briefly, cells were suspended in medium at a concentration of 5 × 104 cells/mL and 10-μL droplets of the suspension were placed on the lid of a Petri dish. Intercellular cohesiveness was evaluated after 24 h by imaging the multicellular aggregates on a fluorescence stereomicroscope (Leica, M205FA) equipped with a monochrome camera (Leica, DFC7000 GT).
Real-Time Microscopy
Arrays of spheroids comprised of 4T1-derived iRFP670-expressing clonal cell lines were subjected to confocal fluorescence time-lapse microscopy using a Nikon Ti-U microscope equipped with a humidified environmental chamber (37°C and 5% CO2, Pathology Devices), spinning disk (X-light V2, Crest Optics), EM-CCD camera (Hamamatsu ImageEM), and motorized stage (Ludl Electronic Products). 3D confocal stacks of individual spheroids (z-range of 120 µm, z-step of 5 µm; 5 spheroids per condition) were acquired every 30 min over a 30-h period with a CFI S Plan Fluor ELWD 20xC / 0.45 NA air objective. At the end of time-lapse imaging experiments, TIFF stacks were imported into Imaris (Bitplane) and the 3D motion of individual nuclei were tracked using a built-in Autoregressive Motion tracking algorithm. The resulting tracking data, including nuclear positions, track speed, track speed variation, and track straightness, were exported for subsequent analysis using custom-written MATLAB (Mathworks) code. For microfluidic tumor samples, phase-contrast images were routinely captured to monitor tumor growth, invasion, and cancer cell escape over two weeks. Escape was defined as the movement of cells into the empty cavity in the collagen gel.
Image Analysis of Tumor Cell Invasion
Custom code was written in MATLAB to analyze the positions of nuclei over time to evaluate the dynamics of cell invasion within microfabricated arrays of spheroids. The positions of nuclei detected from time-lapse experiments and immunostained samples were imported from Imaris. The relative positions of nuclei were used to distinguish between two regional subpopulations of cells: those in the spheroid core and those in invasions, based on a circular ROI defined by user input of the spheroid shape. Core cells were generally detected within a radius of ~ 50 μm from the center of the spheroid, while invading cells were > 50 μm from the center. The nuclear positions were re-centered to 0, 0 (x,y coordinates) relative to the center of the circular ROI. The movement and positions of individual cells in the whole population, spheroid core, and invasions were then analyzed. The invasion distance was calculated as the distance of the invading cell to the spheroid core minus the spheroid radius.
For engineered tumors contained within microfluidic devices, custom MATLAB code was written to segment phase-contrast images to obtain morphological metrics to describe tumor growth and invasion (see Supplementary Methods). Briefly, segmented images of entire tumors were generated by detecting changes in contrast observed at the periphery of tumors. The regions comprising the tumor core and invasions were obtained to analyze tumor growth and invasion, respectively. The areas of the invasions positive for the fluorescent reporters iRFP and YFP of the E2 and YFP M cells, respectively, were analyzed in MATLAB by applying a mask of the invasions over the fluorescent images and determining the area of image with fluorescent signal above background (see Supplementary Methods).
Chick Chorioallantoic Membrane (CAM) Assay
Fertilized Broiler chicken eggs (Gallus gallus variant domesticus) were obtained from Moyer’s Chicks (Quakertown, PA). All experiments with fertilized eggs complied with ethical regulations for the care and use of animals and were approved by the Institutional Animal Care and Use Committee (IACUC) of Princeton University. Eggs were incubated on their sides at 37.5°C and 55% humidity in a standard egg incubator (GQF Manufacturing) and automatically turned at regular intervals. On day 3 of incubation, eggs were windowed to permit access to the late-stage embryo, as described [33]. Briefly, 3 mL of albumen was removed from the narrow end of each fertilized egg, and a window was generated on the top side of the egg. Windows were subsequently sealed with transparent tape and the eggs were re-incubated without turning. On day 9 of incubation, the tape was cut to expose the CAM. 4T1 cells were then xenografted onto a gently traumatized CAM. Specifically, the CAM was abraded using a 30-gauge needle, causing a small bleed, and a 25-μL droplet containing 2 × 106 cells in serum-free RPMI-1640 medium was deposited onto the injured region of the CAM. Eggs were re-sealed, and cells were allowed to settle onto the CAM for 15 min before the eggs were returned to the incubator. On day 18, eggs were chilled on ice for 20 min to minimize embryo movements. Tumors that were visible on the CAM were imaged at 2x magnification using a camera attached to a stereomicroscope. Tumor growth was quantified by measuring the area of the primary tumor from images of the chick CAM.
Quantitative RT-PCR Analysis
Cells were cultured in 6-well tissue-culture plates and lysed after 72 h by the addition of TRIzol Reagent (Invitrogen). RNA was isolated from TRIzol samples following the manufacturer’s instructions. Genomic DNA impurities were removed from RNA samples through the use of Turbo DNase Treatment and Removal Reagents (Ambion). The quality and yield of the resulting RNA was then determined by measuring sample absorbance at 260 nm and 280 nm on a NanoVue Plus spectrophotometer (GE Healthcare). Next, cDNA was synthesized from 1 μg of RNA for each sample using a Verso cDNA Synthesis Kit (Thermo Scientific). Quantitative PCR (qPCR) was performed using iTaq Universal SYBR Green Supermix (Bio-Rad) on a StepOnePlus Real-time PCR System (Applied Biosystems). Amplification of a single product was verified by melt-curve analysis. Primers were determined to be target-specific by BLAST (Supplementary Table S1). Expression of each target gene was normalized to that of 18S ribosomal RNA for each sample.
Bulk RNA-Sequencing Analysis
Cells were cultured in 6-well tissue-culture plates for 72 h, at which time they had reached ~ 80% confluency. RNA was extracted and isolated using an RNeasy Mini Kit (Qiagen) following the manufacturer’s instructions. The extracted RNA was processed by the Princeton University Genomics Core Facility. RNA quality was assessed using an Agilent Bioanalyzer. RNA-Seq directional library preparation for all samples was performed on an Apollo 324 Robot. The samples were run in a single lane on a NovaSeq SP 100 nucleotide Flowcell v1.5 with a read depth of ~ 650–800 million reads. Analysis producing the PCA plot was performed in Galaxy using DESeq2. Z-scores were computed using the normalized counts from DESeq2 and data shown were not scaled.
Statistical Analysis
Statistical analyses were performed using Prism (GraphPad) or MATLAB. Expression of EMT biomarkers were compared using repeated measures one-way analysis of variance (ANOVA) with Tukey’s multiple comparison test in Prism. The sizes of tumors on the upper CAM were compared using one-way ANOVA with Tukey’s multiple comparison test in Prism. Individual cell movement in the spheroid microarray was analyzed using the Kolmogorov–Smirnov test in MATLAB. Morphologies of tumors within the microfluidic system were compared in Prism, using Bartlett’s test to determine if the conditions had equal variance. If Bartlett’s test was not significant (p > 0.05), then one-way ANOVA was performed with Tukey’s multiple comparison test. If Bartlett’s test was significant (p < 0.05), then the Brown–Forsythe ANOVA test and Dunnett’s T3 multiple comparisons test were performed. Rates of invasion and escape were compared using the log-rank (Mantel–Cox) test in Prism. The mechanisms of escape by 0-10% M tumors were compared using ordinary one-way ANOVA with Tukey’s multiple comparisons in Prism. The days of escape for different escape mechanisms were compared using Welch's t-test based on normal distribution with unequal variances. Adjustments were made for multiple comparisons and significance was considered p < 0.05. The E2-positive and M-positive areas in 50/50 tumors were compared using a t-test. The E2-positive and M-positive areas in E2 tumors containing relatively rare (1–10%) populations of M cells were compared using two-way ANOVA with Tukey’s multiple comparison test.
Results
Isolation of Clonal Subpopulations from a Heterogeneous TNBC Cell Line
Breast tumors are heterogeneous across and within individual patients, which poses a significant challenge for successful treatment of the disease. Intratumoral heterogeneity may evolve as the disease progresses and in response to microenvironmental stressors. Of note, morphological diversity is prevalent in samples from breast cancer patients and may serve as an important readout of phenotypic heterogeneity [34]. However, it remains unclear whether and how individual clonal subpopulations contribute to malignant progression in heterogeneous tumors. Here, we established a reverse-engineering approach to generate heterogeneous tumors from clonally derived subpopulations of 4T1 murine TNBC cells and track growth and invasion as a function of the initial composition of the tumor.
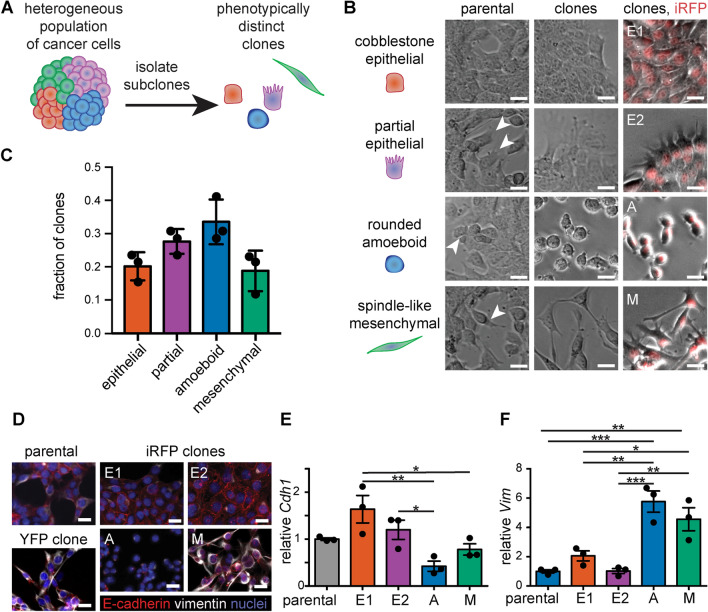
First, we conducted routine culture of parental 4T1 cells and identified four major morphologies in the population: (1) cobblestone epithelial, (2) partial epithelial with extensive protrusions, (3) rounded amoeboid, and (4) spindle-shaped mesenchymal (Fig. 1A, B). 4T1 cells plated at low density were observed to form colonies with these respective morphologies (Fig. S1), suggesting that they were generated through the clonal proliferation of individual cells within a multiclonal parental population. To confirm this observation, we generated clonal subpopulations of the parental 4T1 cell line through several rounds of limiting-dilution culture. The resulting clonal morphologies were binned into the major cell shapes present and included epithelial, partial epithelial, amoeboid, and mesenchymal (Fig. 1B, C). The epithelial and partial epithelial morphologies were similar in abundance, representing ~ 20% and ~ 25% of the clones, respectively. The amoeboid morphology was the most abundant, representing ~ 35% of the clones. The mesenchymal morphology was the least abundant, representing ~ 15% of the clones. Cells within each clonal population had similar morphologies that were maintained over repeated cloning, and there was little overlap in morphologies between different subtypes. The parental 4T1 cell line therefore appears to be a mixture of subpopulations with diverse morphologies, from epithelial to mesenchymal.
Fig. 1.
The 4T1 TNBC cell line is comprised of subpopulations with distinct morphologies and expression of EMT biomarkers. A Schematic of the process used to isolate morphologically distinct clonal subpopulations from the heterogeneous 4T1 murine mammary carcinoma cell line. B Phase-contrast images of the parental 4T1 population (left), single-cell-derived clones (middle), and stable iRFP-expressing clones (right; iRFP in red). C Graph showing the relative abundance of clones with cobblestone epithelial, partial epithelial, rounded amoeboid, and spindle-like mesenchymal morphologies; shown are mean ± SD for n = 3 experiments. D Immunofluorescence analysis for the EMT biomarkers E-cadherin (red) and vimentin (white) and E, F qRT-PCR analysis for E-cadherin (Cdh1) and vimentin (Vim) in the parental 4T1 cells and clones; shown are mean ± SEM for n = 3 experiments. Samples in (D) are counterstained with Hoechst (blue). Scale bars 25 µm. *p < 0.05; **p < 0.01; ***p < 0.001
Characterization of EMT Biomarker Levels in 4T1 Clones
To more closely examine the differences between these morphologically distinct subpopulations, we generated 4T1 clonal cell lines that stably express a near-infrared fluorescent protein in the nucleus (pNLS-iRFP670) [26]. We isolated and characterized iRFP-expressing clones with each of the four morphologies observed within the parental 4T1 cell line (Fig. 1B). Specifically, we isolated two epithelial subpopulations “E1” with characteristic cobblestone morphology and “E2” with extensive filopodial protrusions, an amoeboid-shaped subpopulation “A” with compact, rounded morphology, and a mesenchymal-shaped subpopulation “M” with spindle-like morphology (Fig. 1B). We conducted all experiments within the first twenty passages after transfection and observed that the subpopulations remained phenotypically stable. Additionally, when cells from these subpopulations were seeded as single cells again via limiting-dilution cloning, they maintained the same phenotype as the original subpopulations (Fig. S2). RNA-sequencing analysis confirmed that samples of each clone were similar to each other and distinct from the other clonal populations (Fig. S3A). We hypothesized that variation in EMT status may account for the distinct morphologies of the 4T1 clones. Immunostaining for the classical EMT biomarkers E-cadherin and vimentin demonstrated that the parental 4T1 cell line and iRFP-expressing clones have different EMT signatures (Fig. 1D). The E1 clone displayed a high level of junctional E-cadherin and low level of vimentin, consistent with an epithelial phenotype. The E2 clone spread more than the compact E1 clone and displayed strong junctional E-cadherin and intermediate levels of vimentin, consistent with a partial epithelial phenotype. The A and M clones lacked junctional E-cadherin and presented with a scattered phenotype. The A clone spread the least, with nearly undetectable levels of E-cadherin and noticeable vimentin. The M clone exhibited cytoplasmic puncta of E-cadherin and high levels of vimentin throughout the cytoskeleton, reminiscent of a partial mesenchymal phenotype [35, 36]. In contrast, the parental 4T1 population displayed mixed phenotypes along the EMT spectrum, with regions containing cells of cobblestone morphology that expressed high levels of junctional E-cadherin and low levels of vimentin, surrounded by regions containing spindle-shaped cells that expressed lower levels of E-cadherin and higher levels of vimentin (Fig. 1D).
The trends that we observed in the protein levels of E-cadherin and vimentin were confirmed at the transcript level using qRT-PCR analysis (Fig. 1E, F). E-cadherin transcript levels were similar for 4T1 clones and parental cells but varied subtly across individual clones: the E1 and A clones displayed the highest and lowest transcript levels of Cdh1, respectively (Fig. 1E). Vimentin transcript levels were similar for parental cells, the E1 clone, and the E2 clone. In contrast, vimentin was expressed at high levels in the A and M clones, which displayed ~ 5-fold higher levels of Vim transcript compared to parental cells (Fig. 1F). Thus, 4T1 clones can be binned into two major groups based on their relative expression of EMT biomarkers: E-cadherin-high/vimentin-low (E1 and E2) and E-cadherin-low/vimentin-high (A and M). These data were confirmed by RNA-sequencing analysis, which revealed that the M clone expressed low levels of epithelial markers like Cdh1 and high levels of mesenchymal markers like Vim (Fig. S3B). Our findings thus suggest that the parental 4T1 cell line can be described as a heterogeneous mixture of subpopulations with phenotypes that vary along the EMT spectrum.
Clonal Subpopulations Exhibit Diverse Behaviors in 3D Microenvironments
Given the relationship between cell form and function [37], we hypothesized that the morphological heterogeneity across the 4T1 clones may translate into distinct behaviors associated with tumor progression. To test this hypothesis, we conducted a range of assays to interrogate tumorigenicity, cohesiveness, growth, and invasiveness. First, we leveraged the chick chorioallantoic membrane (CAM) assay to determine the tumorigenicity of the parental 4T1 cells and clones (Fig. S4). We confirmed that all four 4T1 clones (E1, E2, A, and M) could form tumors on the chick CAM (Fig. 2A), which were similar in size to tumors generated by the parental cell line (Fig. 2B). Then, we used the hanging-drop assay to characterize the cohesiveness of the parental 4T1 cells and clones [31, 32]. We found that the parental line and the E1, E2, and M clones all formed compact aggregates that resembled multicellular spheroids within 24 h (Fig. 2C). In contrast, the A clone failed to form a compact spheroid and instead remained as a loose suspension of cells. The capacity of cells to aggregate, compact, and form spheroids is facilitated by cell-surface glycoproteins and integral membrane proteins, which enable the formation of intercellular junctions [38]. As such, these results suggest that the parental, E1, E2, and M clones form significantly stronger cell–cell adhesions than the A clone, consistent with the ameboid morphology of the latter [39].
Fig. 2.
Epithelial and mesenchymal clones exhibit distinct behaviors in 3D culture. A Bright field images of tumors comprised of parental 4T1 cells and clones that were generated on the upper CAM; scale bars 2 mm. B Graph showing area of tumors on the upper CAM; shown are mean ± SEM for at least n = 3 experiments. C Schematic (left) and phase-contrast images (right) of hanging-drop assay of parental 4T1 cells and clones after 24 h; scale bars 200 µm. D Fluorescence images of representative micropatterned spheroids within 3D microarrays, labeled with phalloidin and Hoechst after 72 h of culture. Shown are maximum-intensity projections; arrowheads indicate prominent invasions; scale bars 50 µm. E Representative image of nuclear tracking depicting the regions of micropatterned spheroids containing core cells and invading cells, based on an initial fit at time 0; scale bar 25 µm. F Frequency plot of nuclear positions across time for clonal spheroids show the noninvasive (E1/E2) and invasive (A/M) phenotypes; blue–red color bar indicates log-scaled bin counts, where dark blue = 0 counts; scale bars 25 µm. G Graph showing distance of nuclei from the spheroid center for all cells and timepoints. H Graph showing maximum distance of nuclei of invaded cells from the spheroid periphery. Shown are mean ± SD for n = 3 independent experiments. I Violin plot showing mean speed for cells located in the core or invasions (inv) of A or M spheroids. Analyses in F–I depict data pooled for 15 spheroids at 60 h taken from n = 3 independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
The hanging-drop assay is often used to generate clusters of tumor cells that are subsequently embedded within 3D extracellular matrix. Using this method, we found that the loose suspension of A cells readily dispersed; as such, the hanging-drop approach could not be used to compare the 3D behaviors of the clonal subpopulations derived here. To circumvent this issue, we used a microfabrication-based approach to engineer 3D aggregates of tumor cells [28], which permits the generation of a microarray of spheroids with controlled shape and size irrespective of the underlying cohesiveness of the constituent cells (Fig. S5A). This approach revealed that spheroids generated from suspensions of parental 4T1 cells, A cells, or M cells readily extended protrusions into the surrounding matrix within 30 h and invaded substantially by 60 h, whereas those generated from E1 or E2 cells were neither protrusive nor invasive over this timeframe (Fig. S5B). Fluorescent labeling of actin and nuclei confirmed the presence of many actin-rich protrusions and collective invasions emanating from microfabricated spheroids comprised of parental 4T1 cells and the A and M clones (Fig. 2D).
To further characterize the differences in invasive behaviors across spheroids comprised of the clonal cell lines, we carried out time-lapse imaging analysis to track the positions of fluorescent nuclei (Fig. 2E; Fig. S6). We found that clonal cells exhibited distinct motility signatures (Fig. S6): E1 and E2 cells moved slower on average (Fig. S6A) and with more variance in speed (Fig. S6C) than A and M cells. Frequency maps of nuclei from fifteen spheroids from each condition revealed that E1 and E2 cells rarely invaded into the surrounding matrix (Fig. 2F). In contrast, A and M spheroids exhibited long collective invasions, which tended to be broader in M spheroids compared to the more single-file strands observed in A spheroids.
We then carried out quantitative image analysis to track the motion of individual cells within the spheroid core or invading strands (Fig. 2E). This analysis confirmed that, in contrast to E1 and E2 spheroids, A and M spheroids displayed robust invasive behaviors (Fig. 2G), with cells invading up to ~ 100 μm from the periphery of the spheroids after 60 h of culture (Fig. 2H). Furthermore, while the average speed of cells in A and M spheroids was higher than that of cells in E1 and E2 spheroids (Fig. S6A), the speed of cells within invasions was dramatically lower than that of cells within the core of the same spheroid (Fig. 2I). These data indicate that cells in the core of A and M spheroids moved the fastest prior to invasion, and then slowed significantly after they invaded into the dense 3D matrix. Together, these results demonstrate that the diversity in morphology and expression of EMT biomarkers observed across 4T1 clones correlate with distinct invasive behaviors in 3D microenvironments.
Microfluidic Tumor Model Reveals Different Rates of Growth and Invasion
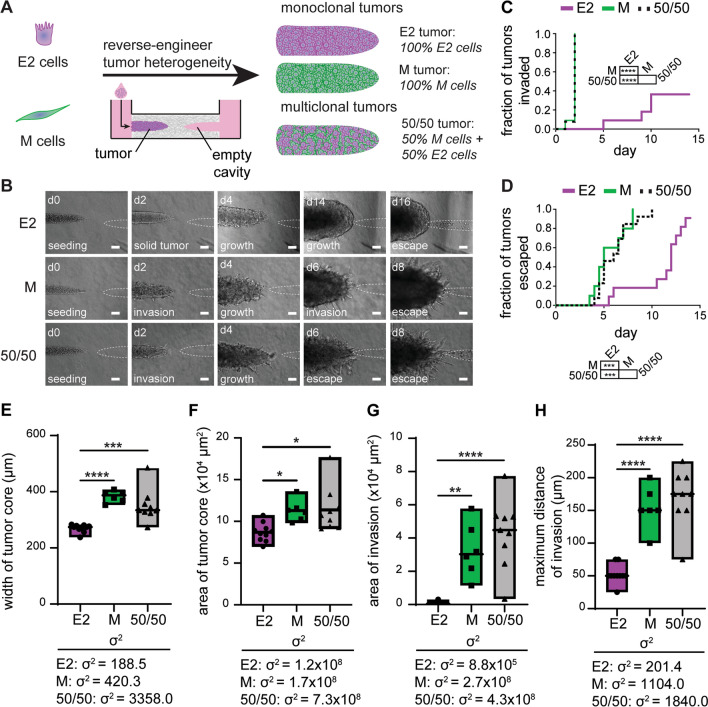
Given the apparent differences in cell morphology, cohesiveness, and invasive behavior across the 4T1 clones, we aimed to define how such intratumoral heterogeneity may contribute to the rates of tumor progression. We took advantage of a microfluidic engineered tumor model that supports extended 3D culture and permits observation of cancer cell behaviors for up to two weeks [30, 40]. We began by engineering tumors that consisted of a solid aggregate of cancer cells within a collagen gel (Fig. 3A, i–ii). Next, we observed cells as they invaded from the solid aggregate (Fig. 3A, iii) and later escaped into an adjacent empty cavity (Fig. 3A, iv), which mimics the earliest stages of the metastatic cascade [29, 40, 41]. We generated tumors from suspensions of the parental 4T1 cells and each clonal line. We found that 80% of parental tumors invaded by day 3.5 (Fig. 3B, C), and all of the parental tumors reached the empty cavity (escaped) by day 10.5 (Fig. 3D). A and M tumors exhibited similar rates of invasion and escape as the parental tumors (Fig. 3B–D). Surprisingly, E1 tumors often formed invasions at similar times as parental, A, and M tumors and had similar rates of escape. However, E1 tumors appeared to exhibit fewer invasions than the A and M tumors, which was consistent with the epithelial appearance in 2D and lack of invasion from E1 spheroids. In contrast, E2 tumors maintained a smooth surface without invasions for much of the assay (Fig. 3B). Consistently, E2 tumors had a significantly slower rate of invasion than the A and M tumors and a significantly slower rate of escape than the parental tumors. As such, we found that E2 tumors were the least aggressive and generally maintained a smooth perimeter without invasions.
Fig. 3.
Microfluidic tumor model reveals the rates of invasion and escape for epithelial and mesenchymal clones. A Schematic depicting the process used to generate solid microtumors within type I collagen cultured under interstitial fluid flow. B Phase-contrast images of parental 4T1 and clonal tumors at different days after seeding; inset highlights cells that have invaded into the surrounding collagen (blue arrowheads) and cells that have entered an escape cavity (orange asterisks); scale bars 100 µm. Kaplan–Meier plots of C invasion and D escape of parental 4T1 and clonal tumors. Shown are the results of 9–10 tumors for each condition, pooled over n = 3 independent experiments. **p < 0.01; ***p < 0.001; ****p < 0.0001
Reverse Engineering Intratumoral Heterogeneity Reveals That Mesenchymal Populations Hasten Rates of Invasion and Escape
Our data suggest that the parental 4T1 cell line is comprised of distinct subpopulations that exhibit a range of invasiveness when isolated from each other. We therefore hypothesized that heterogeneous tumors comprised of both noninvasive and invasive clonal cells from different positions along the EMT spectrum may exhibit different rates of invasion and escape compared to tumors of monoclonal composition. Given that the E2 clone exhibited a relatively noninvasive epithelial phenotype, while the M clone exhibited a highly invasive mesenchymal phenotype, we tested our hypothesis by engineering heterogeneous tumors containing mixtures of these two clones (Fig. 4A). To distinguish between the two populations, we isolated a stable clone of M cells that expresses YFP (YFP-M) and generated monoclonal and multiclonal tumors using the iRFP-E2 and YFP-M clonal lines. We found that multiclonal tumors comprised of 50% iRFP-E2 and 50% YFP-M cells (50/50) largely resembled the invasive phenotype of M monoclonal tumors (Fig. 4B), which both invaded and escaped more rapidly than E2 monoclonal tumors (Fig. 4C, D). Strikingly, the invasion and escape kinetics of 50/50 multiclonal and M monoclonal tumors were indistinguishable from each other. To more closely examine how clonal composition may contribute to the degree of tumor invasiveness, we generated an image-analysis scheme to extract morphological features of tumors from phase-contrast images (Fig. S7). Specifically, we segmented the tumor core to analyze its area, length, and width as metrics of tumor growth. We also segmented the tumor periphery to analyze the area occupied by invasions and the maximum distances of the invasions (Fig. S7). This analysis revealed that 50/50 multiclonal and M monoclonal tumors grew (Fig. 4E, F) and invaded (Fig. 4G, H) more than E2 monoclonal tumors. Nonetheless, the 50/50 multiclonal tumors could still be distinguished from the M monoclonal tumors, as the former had a higher variance between replicates in all metrics of growth and invasion (Fig. 4E–H). Additionally, when we examined the relative density of the fluorescent reporters of the two cell populations, we found that ~ 90% of the cells in the invasions expressed YFP (Fig. S8), despite the fact that the M cells only comprised 50% of the tumor upon initial seeding. Importantly, this analysis also revealed that E2 cells were present in the invasions emanating from 50/50 tumors, even though these cells were slow to form invasions in monoclonal tumors.
Fig. 4.
Mesenchymal cells dominate growth and invasion kinetics of 50/50 multiclonal tumors. A Schematic depicting the process used to generate multiclonal tumors by mixing E2 and M cells. B Phase-contrast images of E2 monoclonal, M monoclonal, and 50/50 multiclonal tumors at different days after seeding; scale bars 100 µm. Kaplan–Meier plots of C invasion and D escape of E2 monoclonal, M monoclonal, and 50/50 multiclonal tumors. Shown are results from 14 to 18 tumors for each condition, pooled over n = 3 independent experiments. Graphs of the E width of tumor cores, F area of tumor cores, G area of invasions, and H maximum distance of invasions in E2 monoclonal, M monoclonal, and 50/50 multiclonal tumors on day 6 of culture. Shown are results from 6 to 10 tumors for each condition, pooled over n = 3 independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
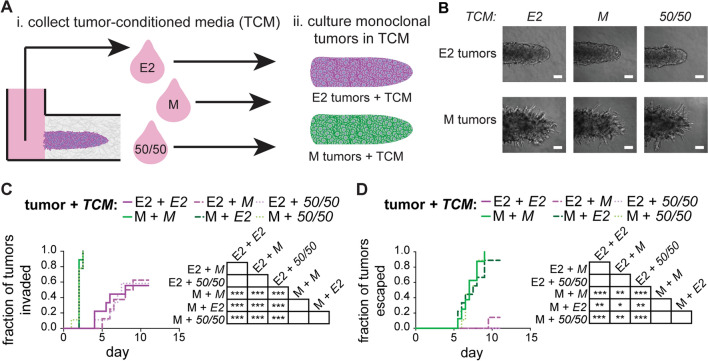
These data suggested that the M cells might promote the proliferation and invasion of the E2 cells. To determine whether these effects were downstream of soluble signals (secreted factors), we collected conditioned media from donor E2 and M monoclonal tumors as well as 50/50 multiclonal tumors (Fig. 5A, i). Subsequently, we generated a separate set of recipient E2 and M monoclonal tumors and cultured these in the presence of the different conditioned media (Fig. 5A, ii). We found that the morphologies of monoclonal tumors were unaffected by culture in E2, M, or 50/50 tumor-conditioned media: E2 tumors were noninvasive in their appearance, while M tumors invaded extensively, regardless of the media conditions (Fig. 5B). Similarly, we found that the rates of invasion (Fig. 5C) and escape (Fig. 5D) of the E2 and M tumors were independent of the conditioned media applied, suggesting that physical—not paracrine—interactions dictated the behavior of 50/50 multiclonal tumors.
Fig. 5.
Conditioned media from E2 or M tumors do not influence the rates of invasion or escape. A Schematic depicting the process used to (i) collect tumor-conditioned media (TCM) from E2 monoclonal, M monoclonal, or 50/50 multiclonal tumors and (ii) apply it to recipient tumors. B Phase-contrast images of E2 or M monoclonal tumors treated with E2, M, or 50/50 TCM; scale bars 100 µm. Kaplan–Meier plots of C invasion and D escape of E2 or M monoclonal tumors treated with E2, M, or 50/50 TCM; shown are results from 8 to 9 tumors for each condition, pooled over n = 3 independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001
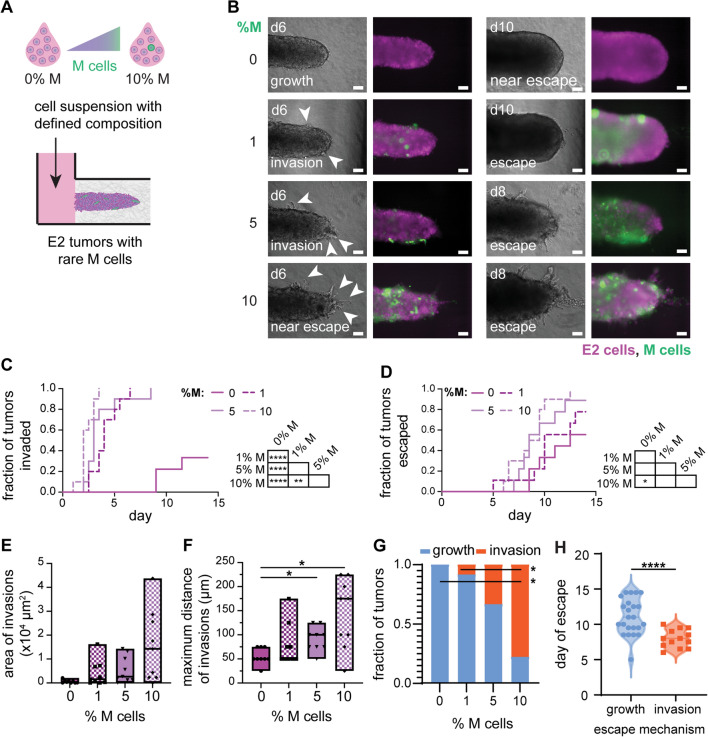
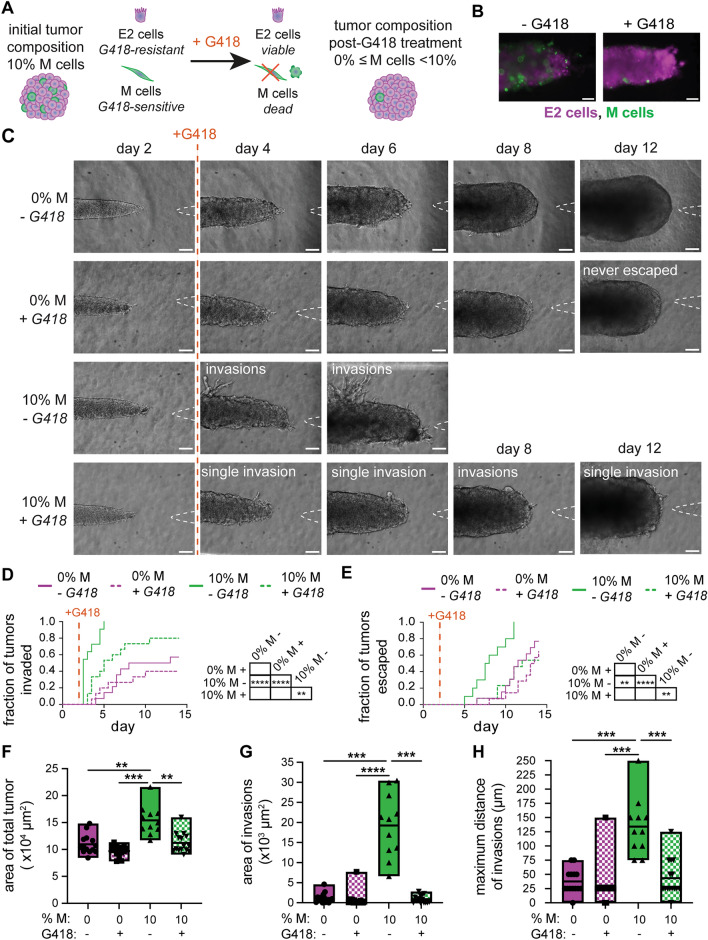
Based on the results of the conditioned-media experiments, we hypothesized that the emergence of an aggressive clonal subpopulation might be sufficient to promote the growth and invasion of an otherwise noninvasive tumor. To test this hypothesis, we engineered multiclonal tumors containing minor fractions of YFP-M cells amongst a majority of iRFP-E2 cells. Specifically, we generated tumors with 1, 5, or 10% M cells to determine whether a relatively rare population of invasive cells could augment invasion and escape (Fig. 6A). We found that multiclonal tumors comprised of 1–10% M cells displayed dramatically accelerated invasion compared to E2 monoclonal tumors (0% M cells) (Fig. 6B, C). Although all multiclonal tumors exhibited accelerated rates of invasion, only tumors with 10% M cells displayed accelerated rates of escape. Notably, 10% M tumors escaped the most rapidly, with half escaping by day 8, in stark contrast to 0% M tumors for which half escaped by day 14 (Fig. 6D). Thus, although multiclonal tumors invaded at accelerated rates, these data suggest that there may be a critical minimum threshold of M cells required to increase the rate of tumor escape.
Fig. 6.
Relatively rare populations of M cells accelerate invasion of E2 tumors. A Schematic depicting the process used to generate multiclonal tumors containing relatively rare M cells; initial tumor composition represented by the percentage of M cells. B Phase-contrast (left) and fluorescence (right) images of multiclonal tumors at different days after seeding (green, YFP-M cells; magenta, E2 cells). Invasions are indicated in day 6 images (white arrowheads). Scale bars 100 µm. Kaplan–Meier plots of C invasion and D escape for monoclonal (0% M) and multiclonal (1–10% M) tumors. Shown are results from 10 to 12 tumors for each condition, pooled over n = 3 independent experiments. Graphs of E area of invasions, F maximum distance of invasions, and G fraction of tumors that have escaped via growth or invasion as a function of %M cells; analysis of morphology on day 6. H Violin plots showing day of escape as a function of the mechanism of escape for pooled tumors ranging from 0 to 10% M cells. Morphometric analyses show results from 9 tumors for each condition, pooled over n = 3 independent experiments. *p < 0.05; **p < 0.01; ****p < 0.0001
To further delineate how a rare population of M cells could promote the invasiveness of E2 tumors, we analyzed the morphologies of tumors over time. This analysis revealed that the area (Fig. S9A), length (Fig. S9B), and width (Fig. S9C) of the tumor core were comparable for tumors containing 0-10% M cells. Rare populations of M cells thus appear to have little to no effect on the rate of tumor growth, which appears to be governed by the dominant E2 population. Although the area occupied by invasions (Fig. 6E) was not significantly different between the tumors, the maximum distance of invasions (Fig. 6F) was significantly higher in 10% M tumors than in 0% and 1% M tumors. These data suggest that relatively rare subpopulations of M cells can enhance the elongation of invasions from otherwise noninvasive tumors. Additionally, analysis of the relative densities of the fluorescent reporters of the revealed that ~ 60% of the cells in the invasions expressed YFP (Fig. S10), despite the fact that the M cells were 1-10% of the population upon initial seeding. The remaining ~ 40% of the cells in the invasions expressed iRFP, indicating that the presence of the relatively rare M cells had promoted invasion of the E2 cells.
Consistently, we found that tumors containing 10% M cells exhibited a switch in the mechanism by which they escaped into the empty cavity (Fig. 6G). Specifically, tumors with 10% M cells escaped by invasion, whereas those with 0–5% M cells escaped primarily by overgrowing into the cavity. Across all conditions examined (0–10% M cells), tumors for which cancer cells escaped by invasion did so at earlier times (~day 8) than those that escaped by overgrowing the cavity (~day 11) (Fig. 6H). Therefore, tumors with as low as 1% or 10% mesenchymal cells increase their rates of invasion or escape, respectively, and these changes are accompanied by changes in tumor growth and in the mechanism of escape.
Selectively Killing the Invasive Clonal Subpopulation Slows Tumor Progression
Given that the presence of a minor subpopulation of invasive cells within a tumor can accelerate its invasion, we hypothesized that selectively targeting these cells may slow tumor progression. To test this hypothesis, we again leveraged the iRFP-E2 and YFP-M clonal lines, which stably express different resistance genes. Specifically, iRFP-E2 cells express the dominant-acting resistance gene for geneticin (G418), whereas YFP-M cells express the dominant-acting resistance gene for zeocin. As such, we applied G418 to multiclonal tumors comprised of E2 and M cells to selectively kill the M-cell subpopulation (Fig. 7A), which could be visualized by the reduction in YFP fluorescence observed after treatment (Fig. 7B). As a control, we also generated E2 monoclonal tumors (0% M cells), which should be impervious to treatment due to the inherent G418 resistance of the iRFP-E2 cells. To selectively target the M cells prior to invasion, we added G418 on day 2 of culture. As expected, 10% M tumors invaded extensively in the absence of G418 (Fig. 7C, D). Further, E2 (0% M) tumors exhibited few invasions with or without G418 (Fig. 7C, D). Targeting the M-cell subpopulation with G418 significantly delayed tumor invasion relative to untreated controls (Fig. 7D): 10% M tumors treated with G418 formed few invasions (Fig. 7C) at a slower rate (Fig. 7D), similar to 0% M tumors. In addition, treatment with G418 delayed the escape of 10% M tumors compared to untreated controls, in a manner similar to 0% M tumors (Fig. 7E). Treatment with G418 also significantly decreased expansion of the 10% M tumors, which had a smaller total tumor area than the untreated 10% M tumors on day 6 of culture (Fig. 7F). Quantitative image analysis yielded data consistent with these conclusions. Treatment with G418 resulted in a significantly smaller area (Fig. 7G) and maximum distance of invasions (Fig. 7H) in 10% M tumors. However, the area, length, and width of the core of 10% M tumors were unaffected by treatment with G418 (Fig. S11), consistent with our conclusions that tumor growth is dominated by the E2 subpopulation (Fig. S9). Overall, we found that depleting the aggressive M-cell subpopulation at early timepoints reduced invasion of the tumors and slowed the rate of escape.
Fig. 7.
Targeting the relatively rare, invasive population delays invasion and escape. A Schematic depicting the process used to apply G418 to selectively kill M cells within multiclonal tumors. B Fluorescence images of multiclonal tumors with or without G418 (green, YFP-M cells; magenta, E2 cells). C Phase-contrast images of 0% or 10% M tumors treated with or without G418 on day 2 show decreased invasions over time. Kaplan–Meier plots of D invasion and E escape for 0% or 10% M tumors treated with or without G418 on day 2. Graphs of F area of the total tumor, G area of invasions, and H maximum distance of invasions of tumors treated with or without G418. Shown are data from 11 to 15 tumors for each condition, pooled over n = 3 independent experiments. **p < 0.01; ***p < 0.001; ****p < 0.0001
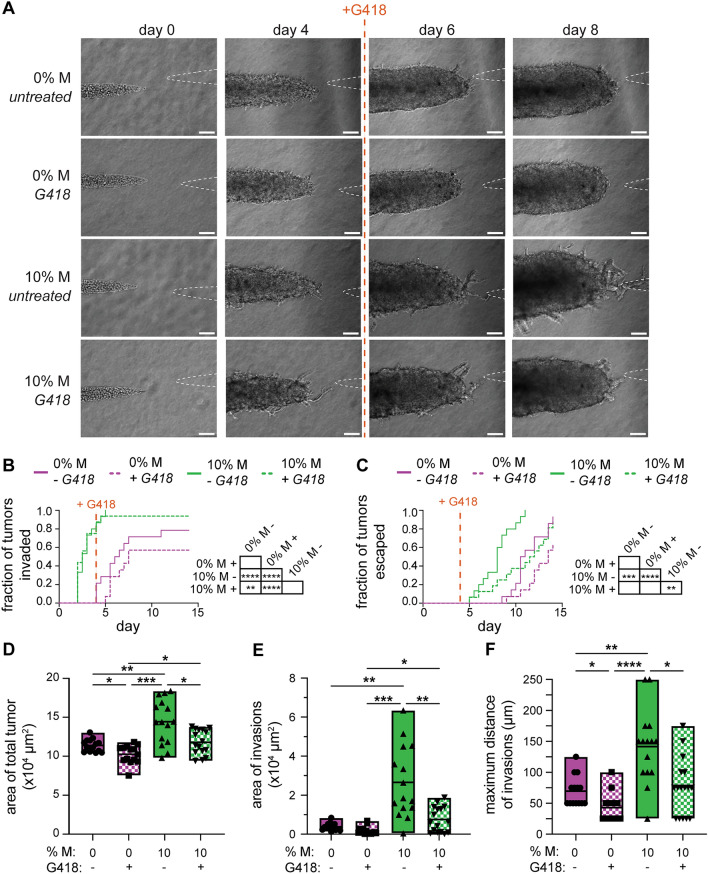
Based on the above results, we hypothesized that selectively targeting the M cells at timepoints after invasion might also cause a delay in tumor progression. To test this hypothesis, we treated tumors with G418 on day 4 of culture, after the 10% M tumors had extended their first invasions but had not yet escaped (Fig. 8A–C). As when the tumors were treated with G418 at an earlier timepoint, treatment on day 4 of culture did not affect the area, length, or width of the tumor core (Fig. S12). However, we found that treatment with G418 decreased the area of the total tumor (Fig. 8D), area of invasions (Fig. 8E), and maximum distance of invasions (Fig. 8F) on day 6 of culture. Moreover, we found that treatment with G418 delayed the escape of 10% M tumors such that they had similar escape kinetics as 0% M tumors (Fig. 8C). These data suggest that selectively depleting mesenchymal cells reduces the formation of invasions as well as the elongation of existing invasions. Notably, depleting the M-cell subpopulation using G418 was sufficient to delay cancer cell escape, even when this treatment was applied after tumors had already invaded. Thus, selectively reducing a target-cell subpopulation might be beneficial for patients presenting with tumors that contain a mixture of cells with varying degrees of invasiveness.
Fig. 8.
Targeting the relatively rare, invasive population after invasion delays escape. A Phase-contrast images of 0% or 10% M tumors treated with or without G418 on day 4. Kaplan–Meier plots of B invasion and C escape for 0% or 10% M tumors treated with or without G418 on day 4. Graphs of D area of the total tumor, E area of invasions, and F maximum distance of invasions of tumors treated with or without G418. Shown are data from 14 to 16 tumors for each condition, pooled over n = 3 independent experiments. Scale bars 100 µm. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
Discussion
We engineered a model of intratumoral heterogeneity that (1) enables control over initial tumor composition and (2) supports the longitudinal tracking of samples over time. We took advantage of the inherently heterogeneous 4T1 TNBC cell line [19, 20] to derive clonal subpopulations with distinct morphologies. We found that the parental 4T1 population can be described as a mixture of clonal subpopulations with phenotypes that vary along the epithelial–mesenchymal spectrum. To model tumor heterogeneity, we incorporated these clonal subpopulations into a microfluidic platform to engineer solid tumors within 3D matrix and tracked tumor growth, invasion, and cancer cell escape over two weeks. This analysis revealed that monoclonal tumors comprised of mesenchymal (M) cells were highly invasive and escaped quickly, whereas those comprised of epithelial (E2) cells exhibited negligible invasion and escaped gradually via tumor growth. When both mesenchymal and epithelial cells were present in either a parental 4T1 tumor or an engineered multiclonal tumor, the tumor exhibited similar rates of invasion and escape as monoclonal mesenchymal tumors did. These observations suggest that cells with mesenchymal traits dominate the invasive phenotype of heterogeneous tumors.
The presence of both epithelial and mesenchymal cells in tumors is common. One study profiled the tumors of five TNBC patients and found epithelial-like and mesenchymal-like cells present in every tumor [42]. When the cells from all these tumors were combined, the numbers of epithelial-like and mesenchymal-like cells were roughly equal [42]. To better understand the effects of these mixed populations of cells, we engineered multiclonal tumors comprised of varying ratios of noninvasive (E2) and invasive (M) cells. These multiclonal tumors invaded and escaped at rates similar to M monoclonal tumors, but the tumor cores grew at rates similar to E2 monoclonal tumors. Further, we found that tumor-conditioned media did not affect the invasion or escape of monoclonal tumors, suggesting that physical interactions between clonal subpopulations were critical in driving tumor invasion. Importantly, we found that the presence of a relatively rare population of M cells was sufficient to accelerate invasion and dictate the mechanism of escape, thus enhancing tumor progression. We also found that the presence of M cells was sufficient to promote the invasion of E2 cells, which were largely noninvasive on their own. It is possible that the more invasive M cells may be leading the less invasive E2 cells through the surrounding extracellular matrix, as has been reported by others [43, 44]. Indeed, RNA-sequencing analysis revealed that M cells express low levels of epithelial cell–cell adhesion proteins, so the observed effects may be due to adhesion between the two clonal populations. The increased invasion could be disrupted by selectively eliminating the relatively rare M cells. Our results thus suggest that the selective reduction of a target-cell subpopulation could have therapeutic benefit for patients presenting with heterogeneous tumors.
Some of our experimental findings were recently predicted by in silico models of tumor heterogeneity. One model showed that small populations of cancer cells capable of producing large motile forces could initiate collective invasion and escape of cancer cells from tumors [45]. Moreover, computational models have shown that whereas tumor subpopulations can display different sensitivities to drug treatment, therapies geared at targeting the predominant subpopulation may be ineffective in heterogeneous tumors [46, 47]. Attempts to ubiquitously kill all cells within heterogeneous tumors with a single therapy are often ineffective, as subpopulations of cancer cells can acquire resistance to treatment [3, 7, 48–53]. Furthermore, therapeutically altering the tumor microenvironment can drive tumor evolution and enhance intratumoral heterogeneity [54]. Treatment of a heterogeneous tumor can then drive the selection of resistant subpopulations [3, 7, 48–51]. These computational studies are congruent with our experimental findings and underscore the importance of accurately detecting tumor heterogeneity in patient biopsies.
Overall, we have established a model to reverse-engineer TNBC heterogeneity by generating tumors from clonally-derived subpopulations of cancer cells with disparate phenotypes. This system enables the fine tuning of tumor composition to elucidate how intratumoral heterogeneity contributes to cancer progression. Our findings complement past studies of tumor heterogeneity and reveal new insights into reciprocal interactions between clonal subpopulations that may affect tumor phenotype. Future work will apply these tools to characterize patient-derived tumor tissue and generate a personalized atlas of cancer progression, which could strengthen existing clinical measures to aid in stratifying breast cancer and selecting personalized therapies. Personalized approaches to treatment may enhance the effectiveness of treatments for heterogeneous tumors [7]. Alternatively, the sensitivities of more benign or malignant populations could be exploited to prevent tumor progression through timing of treatments targeting specific populations [3].
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported in part by grants from the National Institutes of Health (CA187692, CA214292), the New Jersey Health Foundation, and a Faculty Scholars Award from the Howard Hughes Medical Institute. SEL was supported in part by the National Center for Advancing Translational Sciences of the National Institutes of Health (TL1TR003019, UL1TR003017). MCB was supported in part by the Ludwig Princeton Branch and by a New Jersey Commission on Cancer Research Predoctoral Fellowship (COCR22PRF009).
Abbreviations
- CAM
Chorioallantoic membrane
- EMT
Epithelial–mesenchymal transition
- TNBC
Triple-negative breast cancer
Data availability
RNA-seq data have been deposited at GEO under accession number GSE252177 and are available as of the date of publication.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Susan E. Leggett and Molly C. Brennan have contributed equally.
References
- 1.Kim C, Gao R, Sei E, Brandt R, Hartman J, Hatschek T, et al. Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell. 2018;173(4):879–893. doi: 10.1016/j.cell.2018.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torab P, Yan Y, Ahmed M, Yamashita H, Warrick JI, Raman JD, et al. Intratumoral heterogeneity promotes collective cancer invasion through NOTCH1 variation. Cells. 2021 doi: 10.3390/cells10113084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochim. Biophys. Acta. 2010;1805(1):105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inda MM, Bonavia R, Mukasa A, Narita Y, Sah D, Vandenberg S, et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010 doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Summerbell ER, Mouw JK, Bell JSK, Knippler CM, Pedro B, Arnst JL, et al. Epigenetically heterogeneous tumor cells direct collective invasion through filopodia-driven fibronectin micropatterning. Sci. Adv. 2020 doi: 10.1126/sciadv.aaz6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu PH, Gilkes DM, Phillip JM, Narkar A, Cheng TW, Marchand J, et al. Single-cell morphology encodes metastatic potential. Sci. Adv. 2020 doi: 10.1126/sciadv.aaw6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fedele C, Tothill RW, McArthur GA. Navigating the challenge of tumor heterogeneity in cancer therapy. Cancer Discov. 2014;4(2):146–148. doi: 10.1158/2159-8290.CD-13-1042. [DOI] [PubMed] [Google Scholar]
- 8.Kalluri R, Weinberg R. The basics of epithelial–mesenchymal transition. J. Clin. Investig. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J, Antin P, Berx G, Blanpain C, Brabletz T, Bronner M, et al. Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020;21(6):341–352. doi: 10.1038/s41580-020-0237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X, Li J, Cadilha BL, Markota A, Voigt C, Huang Z, et al. Epithelial-type systemic breast carcinoma cells with a restricted mesenchymal transition are a major source of metastasis. Sci. Adv. 2019 doi: 10.1126/sciadv.aav4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carey SP, Starchenko A, McGregor AL, Reinhart-King CA. Leading malignant cells initiate collective epithelial cell invasion in a three-dimensional heterotypic tumor spheroid model. Clin. Exp. Metastasis. 2013 doi: 10.1007/s10585-013-9565-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hapach LA, Carey SP, Schwager SC, Taufalele PV, Wang WW, Mosier JA, et al. Phenotypic heterogeneity and metastasis of breast cancer cells. Cancer Res. 2021 doi: 10.1158/0008-5472.CAN-20-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young KM, Reinhart-King CA. Phenotypic heterogeneity and cooperation in the metastatic cascade. Oncoscience. 2022;9:55–56. doi: 10.18632/oncoscience.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hapach LA, Wang W, Schwager SC, Pokhriyal D, Fabiano ED, Reinhart-King CA. Phenotypically sorted highly and weakly migratory triple negative breast cancer cells exhibit migratory and metastatic commensalism. Breast Cancer Res. 2023;25(1):102. doi: 10.1186/s13058-023-01696-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martín-Pardillos, A., Á. Valls Chiva, G. Bande Vargas, P. Hurtado Blanco, R. Piñeiro Cid, P. J. Guijarro, et al. The role of clonal communication and heterogeneity in breast cancer. BMC Cancer 19(1):1–26, 2019. 10.1186/s12885-019-5883-y. [DOI] [PMC free article] [PubMed]
- 16.Tao K, Fang M, Alroy J, Sahagian GG. Imagable 4T1 model for the study of late stage breast cancer. BMC Cancer. 2008;8:228. doi: 10.1186/1471-2407-8-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heppner GH, Miller FR, Shekhar PM. Nontransgenic models of breast cancer. Breast Cancer Res. 2000;2(5):331–334. doi: 10.1186/bcr77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ilina O, Campanello L, Gritsenko PG, Vullings M, Wang C, Bult P, et al. Intravital microscopy of collective invasion plasticity in breast cancer. Dis. Model Mech. 2018 doi: 10.1242/dmm.034330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagenblast E, Soto M, Gutiérrez-Ángel S, Hartl CA, Gable AL, Maceli AR, et al. A model of breast cancer heterogeneity reveals vascular mimicry as a driver of metastasis. Nature. 2015;520(7547):358–362. doi: 10.1038/nature14403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang R, Jin C, Hu X. Evidence of drug-response heterogeneity rapidly generated from a single cancer cell. Oncotarget. 2017;8(25):41113–41124. doi: 10.18632/oncotarget.17064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeo SK, Zhu X, Okamoto T, Hao M, Wang C, Lu P, et al. Single-cell RNA-sequencing reveals distinct patterns of cell state heterogeneity in mouse models of breast cancer. Elife. 2020 doi: 10.7554/eLife.58810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schrors B, Boegel S, Albrecht C, Bukur T, Bukur V, Holtstrater C, et al. Multi-omics characterization of the 4T1 murine mammary gland tumor model. Front. Oncol. 2020;10:1195. doi: 10.3389/fonc.2020.01195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heppner GH, Dexter DL, DeNucci T, Miller FR, Calabresi P. Heterogeneity in drug sensitivity among tumor cell subpopulations of a single mammary tumor. Cancer Res. 1978;38(11 Pt 1):3758–3763. [PubMed] [Google Scholar]
- 24.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52(6):1399–1405. [PubMed] [Google Scholar]
- 25.Bhatia S, Monkman J, Blick T, Duijf PH, Nagaraj SH, Thompson EW. Multi-omics characterization of the spontaneous mesenchymal–epithelial transition in the PMC42 breast cancer cell lines. J. Clin. Med. 2019;8(8):1253. doi: 10.3390/jcm8081253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shcherbakova DM, Verkhusha VV. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat. Methods. 2013;10(8):751–754. doi: 10.1038/nmeth.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliva-Trastoy M, Defais M, Larminat F. Resistance to the antibiotic Zeocin by stable expression of the Sh ble gene does not fully suppress Zeocin-induced DNA cleavage in human cells. Mutagen. 2005;20(2):111–114. doi: 10.1093/mutage/gei016. [DOI] [PubMed] [Google Scholar]
- 28.Piotrowski-Daspit AS, Nelson CM. Engineering three-dimensional epithelial tissues embedded within extracellular matrix. J. Vis. Exp. 2016 doi: 10.3791/54283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piotrowski-Daspit AS, Tien J, Nelson CM. Interstitial fluid pressure regulates collective invasion in engineered human breast tumors via Snail, vimentin, and E-cadherin. Integr. Biol. 2016;8(3):319–331. doi: 10.1039/c5ib00282f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piotrowski-Daspit AS, Simi AK, Pang M, Tien J, Nelson CM. A 3D culture model to study how fluid pressure and flow affect the behavior of aggregates of epithelial cells. In: Martin F, Stein T, Howlin J, editors. Mammary Gland Development: Methods and Protocols (Methods in Molecular Biology Series) New York: Springer; 2017. pp. 245–257. [DOI] [PubMed] [Google Scholar]
- 31.Teng Y. Hanging drop aggregation assay of breast cancer cells. Bio-Protocols. 2015 doi: 10.21769/BioProtoc.1393. [DOI] [Google Scholar]
- 32.Foty R. A simple hanging drop cell culture protocol for generation of 3D spheroids. J. Vis. Exp. 2011 doi: 10.3791/2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spurlin III, J., and P. Lwigale. A technique to increase accessibility to late-stage chick embryos for in ovo manipulations. Dev. Dyn. 2013. 10.1002/dvdy.23907. [DOI] [PMC free article] [PubMed]
- 34.Aleskandarany MA, Vandenberghe ME, Marchiò C, Ellis IO, Sapino A, Rakha EA. Tumour heterogeneity of breast cancer: from morphology to personalised medicine. Pathobiology. 2018;85(1–2):23–34. doi: 10.1159/000477851. [DOI] [PubMed] [Google Scholar]
- 35.Aiello NM, Maddipati R, Norgard RJ, Balli D, Li J, Yuan S, et al. EMT subtype influences epithelial plasticity and mode of cell migration. Dev. Cell. 2018 doi: 10.1016/j.devcel.2018.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mendez MG, Kojima S, Goldman RD. Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB J. 2010 doi: 10.1096/fj.09-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prasad A, Alizadeh E. Cell form and function: interpreting and controlling the shape of adherent cells. Trends Biotechnol. 2019 doi: 10.1016/j.tibtech.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 38.Mueller-Klieser W. Multicellular spheroids. A review on cellular aggregates in cancer research. J. Cancer Res. Clin. Oncol. 1987 doi: 10.1007/BF00391431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graziani V, Rodriguez-Hernandez I, Maiques O, Sanz-Moreno V. The amoeboid state as part of the epithelial-to-mesenchymal transition programme. Trends Cell Biol. 2022;32(3):228–242. doi: 10.1016/j.tcb.2021.10.004. [DOI] [PubMed] [Google Scholar]
- 40.Tien J, Truslow JG, Nelson CM. Modulation of invasive phenotype by interstitial pressure-driven convection in aggregates of human breast cancer cells. PLoS ONE. 2012 doi: 10.1371/journal.pone.0045191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tien J, Ghani U, Dance YW, Seibel AJ, Karakan MC, Ekinci KL, et al. Matrix pore size governs escape of human breast cancer cells from a microtumor to an empty cavity. Iscience. 2020 doi: 10.1016/j.isci.2020.101673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Fang Y, Bao L, Wu F, Wang S, Hao S. Intercellular communication reveals therapeutic potential of epithelial–mesenchymal transition in triple-negative breast cancer. Biomolecules. 2022 doi: 10.3390/biom12101478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Konen J, Summerbell E, Dwivedi B, Galior K, Hou Y, Rusnak L, et al. Image-guided genomics of phenotypically heterogeneous populations reveals vascular signalling during symbiotic collective cancer invasion. Nat. Commun. 2017;8:15078. doi: 10.1038/ncomms15078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saenz-de-Santa-Maria I, Celada L, Chiara MD. The leader position of mesenchymal cells expressing N-cadherin in the collective migration of epithelial cancer. Cells. 2020 doi: 10.3390/cells9030731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hallou A, Jennings J, Kabla AJ. Tumour heterogeneity promotes collective invasion and cancer metastatic dissemination. R. Soc. Open Sci. 2017;4(8):161007. doi: 10.1098/rsos.161007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao B, Pritchard JR, Lauffenburger DA, Hemann MT. Addressing genetic tumor heterogeneity through computationally predictive combination therapy. Cancer Discov. 2014;4(2):166–174. doi: 10.1158/2159-8290.CD-13-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao B, Hemann MT, Lauffenburger DA. Intratumor heterogeneity alters most effective drugs in designed combinations. Proc. Natl Acad. Sci. USA. 2014;111(29):10773–10778. doi: 10.1073/pnas.1323934111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McQuerry JA, Chang JT, Bowtell DDL, Cohen A, Bild AH. Mechanisms and clinical implications of tumor heterogeneity and convergence on recurrent phenotypes. J. Mol. Med. 2017 doi: 10.1007/s00109-017-1587-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kleppe M, Levine RL. Tumor heterogeneity confounds and illuminates: assessing the implications. Nat. Med. 2014 doi: 10.1038/nm.3522. [DOI] [PubMed] [Google Scholar]
- 50.Tammela T, Sage J. Investigating tumor heterogeneity in mouse models. Annu. Rev. Cancer Biol. 2020 doi: 10.1146/annurev-cancerbio-030419-033413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Denison TA, Bae YH. Tumor heterogeneity and its implication for drug delivery. J. Control Release. 2012;164(2):187–191. doi: 10.1016/j.jconrel.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marusyk A, Janiszewska M, Polyak K. Intratumor heterogeneity: The Rosetta Stone of therapy resistance. Cancer Cell. 2020;37(4):471–484. doi: 10.1016/j.ccell.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hayford CE, Tyson DR, Robbins CJ, III, Frick PL, Quaranta V, Harris LA. An in vitro model of tumor heterogeneity resolves genetic, epigenetic, and stochastic sources of cell state variability. PLoS Biol. 2021;19(6):e3000797. doi: 10.1371/journal.pbio.3000797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zellmer VR, Zhang S. Evolving concepts of tumor heterogeneity. Cell Biosci. 2014;4(1):1–8. doi: 10.1186/2045-3701-4-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited at GEO under accession number GSE252177 and are available as of the date of publication.