Abstract
The application of chondroitinase requires consideration of the complex microenvironment of the target. Our previous research reported a marine-derived sodium dodecyl sulfate (SDS)-resistant chondroitinase VhChlABC. This study further investigated the mechanism of VhChlABC resistance to SDS. Focusing on the hydrophobic cluster on its strong hydrophilic surface, it was found that the reduction of hydrophobicity of surface residues Ala181, Met182, Met183, Ala184, Val185, and Ile305 significantly reduced the SDS resistance and stability. Molecular dynamics (MD) simulation and molecular docking analysis showed that I305G had more conformational flexibility around residue 305 than wild type (WT), which was more conducive to SDS insertion and binding. The affinity of A181G, M182A, M183A, V185A and I305G to SDS was significantly higher than that of WT. In conclusion, the surface hydrophobic microenvironment composed of six residues was the structural basis for SDS resistance. This feature could prevent the binding of SDS and the destruction of hydrophobic packaging by increasing the rigid conformation of protein and reducing the binding force of SDS-protein. The study provides a new idea for the rational design of SDS-resistant proteins and may further promote chondroitinase research in the targeted therapy of lung diseases under the pressure of pulmonary surfactant.
Supplementary Information
The online version contains supplementary material available at 10.1007/s42995-023-00201-1.
Keywords: Hydrophobic cluster, Protein surface, SDS-resistance, Chondroitinase
Introduction
Chondroitin sulfate (CS) is an important member of sulfated glycosaminoglycans (GAGs), and its polysaccharide chain is formed by repeated disaccharide units composed of glucuronic acid (GlcA) and N-acetyl-galactosamine (GalNAc) connected by β-1,4 glycoside bonds (Abdallah et al. 2020; Mishra and Ganguli 2021). CS is widely present in the extracellular matrix of vertebrates in the form of CS proteoglycans (CSPGs), especially in cartilage, ligaments, cornea, skin and other tissues (Morla 2019). CS plays an important role in a variety of biological processes, including cartilage repair and regeneration, as an antioxidant and anti-inflammatory molecule, as well as in the regulation of various pathological conditions such as osteoarthritis and nervous system diseases and physiological processes such as aging (Agiba et al. 2018; Collin et al. 2017; Djerbal et al. 2017; Ishimaru et al. 2014; Mizuguchi et al. 2003; Mou et al. 2018; Nandini and Sugahara 2006; Sato et al. 2008; Sugahara and Mikami 2007; Volpi 2011). Stromal and cell surface CSPGs play important roles in tumor growth, vascularization and metastasis (Khan et al. 2020). The pathogenesis of many tumors, including melanoma, triple-negative breast cancer, glioblastoma, ovarian carcinoma, neuroblastoma, osteosarcoma, and chondrosarcoma, have been confirmed to be closely related to the overexpression of CSPGs (Ilieva et al. 2017).
Chondroitinase can degrade CS and its biological activity is due to its ability to act on CSPGs (Kasinathan et al. 2016). CSPGs deposition in a variety of pathological conditions, including spinal cord injury, citreous adhesion, and cancer, has attracted more and more attention for CS-targeted therapy. Studies have shown that chondroitinase can remove the glial scar formed in the spinal primary injury site and restore the regeneration function of axons (Bradbury et al. 2002; Moon et al. 2001). Enzyme-assisted vitrectomy can greatly reduce the chance of retinal damage during eye surgery (Gandorfer 2008; Staubach et al. 2004). In the treatment of oncolytic virus mediated astrocytoma, chondroitinase increases the spread and infection of virus in cancer cells by removing tumor CSPGs (Dmitrieva et al. 2011). Currently, the application of chondroitinase in tumor therapy is still restricted by many factors, including the effectiveness and stability of the enzyme in specific microenvironments in vivo. Drugs for lung tumors are bound to face pressure from lung surfactants, the detergent-like materials that could produce a sodium dodecyl sulfate (SDS)-like stimulus (Dan et al. 2009; Manganelli et al. 1999). Perhaps, the lack of surfactant-resistant enzymes is one of the essential reasons for the lack of research on targeted lung tumor therapy combined with chondroitinase.
Although considerable progress had been made in GAG lyases with novel structures and functions, there were few reports on SDS-resistant GAG-degrading enzyme, and the understanding of its mechanisms was almost nonexistent. The chondroitinase B of PL6 family from marine bacterium Microbulbifer sp. ALW1 had been found to be surfactant-stable, but the mechanism remained unknown (Mou et al. 2022). In addition, almost all members of the PL6, PL8, PL16, PL30 and PL33 families in the CAZY database were sensitive to SDS. The activities of enCSase from Photobacterium sp. QA16, Vpa_0049 from Vibrio sp. QY108, and ChSase ABC from Acinetobacter sp. C26 were almost completely inhibited by SDS with a concentrations of less than 5 mmol/L (Zhang et al. 2020a, b; Zhu et al. 2017). Lipase is an important model for studying detergent tolerance. A systematic mutagenesis study for Bacillus subtilis lipase A had revealed that surface remodeling was an effective strategy to optimize the stability of the enzyme in detergents, including SDS, CTAB, Tween 80, and sulfobetaine; however, the rationale behind the increased tolerance obtained with amino acid substitutions and its applicability to other enzyme modifications were unclear (Fulton et al. 2015).
In a previous study, we identified an SDS-resistant chondroitinase VhChlABC from the marine bacterium Vibrio hyugaensis LWW-1, which retained more than 50% activity at SDS concentrations up to 10% (w/v) (Su et al. 2021). The enclosed study further explored the mechanism of VhChlABC tolerance to SDS, and revealed that the hydrophobic microenvironment on the surface of VhChlABC was the key structural basis. This is the first report on the mechanism of GAG-degrading enzyme resistance to SDS. It provides a new strategy for rational design of SDS-resistant proteins, which is beneficial to obtain the phenotype more conveniently and efficiently. Additionally, the study may advance the research process of chondroitinase in the treatment of lung diseases, which might be constrained by the abundance of surfactants in the lung microenvironment.
Materials and methods
Hydrophilic and hydrophobic analysis of VhChlABC
The online tool ProtScale (https://web.expasy.org/protscale/) was used to map the hydrophilicity and hydrophobicity of the protein. The amino acid scale was selected with the default option “Hphob./Kyte & Doolittle”, the sliding window size was set to 9, and the linear weighted model was selected. The positive values are positively correlated with the hydrophobicity of amino acids. The negative values are negatively correlated with the hydrophilicity of amino acids. Additionally, the hydrophilic and hydrophobic surfaces of the VhChlABC were visualized by PyMOL.
Site-directed mutagenesis of VhChlABC
This study focused on the hydrophobic cluster consisting of six amino acids on the surface of VhChlABC. The site-directed mutation strategy was performed to investigate the effect of the surface hydrophobic cluster on SDS resistance. The mutant expression vector was constructed by the Gibson assembly method on the basis of plasmid pET-28a (+)-VhChlABC (WT) (Su et al. 2021), using the ClonExpress MultiS One Step Cloning Kit (Vazyme, Nanjing, China). The mutant gene was ligated into BglII and XhoI sites of the pET-28a (+) expression vector and verified by DNA sequencing. All primers (Supplementary Table S1) used in this work were ordered from Tsingke Biotech Co., Ltd. (Qingdao, China).
Expression, purification and activity assay of VhChlABC and its mutants
The expression and purification of VhChlABC and its mutants and the A232 enzyme activity assay method were performed as previously described (Su et al. 2021).
Biochemical characterization of VhChlABC and its mutants
Biochemical characterization was determined using CS-A as the substrate. The A232 method was performed for this test. The detection of optimum temperature, thermostability, optimum pH, and pH stability of VhChlABC and its mutants were determined as previously described (Su et al. 2021).
Effects of SDS on VhChlABC and its mutants were examined by measuring the activity in 20 mmol/L phosphate buffer (pH 7.0) in the presence of different concentrations of SDS. To examine the effect of SDS on the stability of the enzymes, the residual activity of VhChlABC/mutants was measured after incubation with 0.1% (w/v) SDS at 0 °C for 1 h.
Homology modeling, MD simulation and molecular docking analysis
SWISS-MODEL server (https://swissmodel.expasy.org/interactive) was used to construct a structural model of VhChlABC using chondroitin sulfate lyase abc (PDB ID: 2q1f.1.A) from Bacteroides thetaiotaomicron wal2926 as the template (Waterhouse et al. 2018). The quality check of the homology protein models was carried out using Verify 3D. PyMOL was used to create the structure of A181G, M182A, M183A, A184G, V185A and I305G mutated from the structure of VhChlABC. The structure of SDS was obtained from PubChem (CID: 3423265), and AutoDock (Trott and Olson 2010) was used to dock this ligand into VhChlABC and its mutants (Fischer and Smieško 2021). Molecular dynamics simulations were performed using the GROMACS 2020.6 package, the protein system used Amberff99SB-ildn force field, and the ligands used the general amber force field (GAFF) (Abraham et al. 2015). The TIP3P water model with an edge of ~ 10 Å was used to solvate the complex system in a cubic box. Sodium ions were added to neutralize the charge. The long-range electrostatic interactions were treated using the Particle Mesh Ewald (PME) method with a 1.0 nm cutoff. Minimization was performed by 5000 steps of steepest-descent minimization procedure. Subsequently, the NVT (constant number of atoms, volume, and temperature) and NPT (constant number of atoms, pressure, and temperature) ensemble were used to carry out a 100 ps restricted simulation. Finally, follow-up analysis was performed using the trajectory between 40 and 50 ns. The maximum cluster of structures in the trajectory between 40 and 50 ns of simulations was used for structure analysis. VMD and PyMOL was used for structural visualization and structural analysis.
Molecular mechanics Poisson–Boltzmann surface area (MM-PBSA) method
The GMX MMPBSA was used to calculate the binding affinity of SDS to WT and mutated proteins (Chen et al. 2022; Valdés-Tresanco et al. 2021). For each system, the binding free energy (ΔGbind) was extracted from the last 10 ns of the MD trajectory. In the MM-PBSA scheme, the ΔGbind was computed according to the reported method (Chen et al. 2015).
Statistical analysis
Statistical analysis was performed using the two-tailed unpaired Student’s t test. * indicates p ≤ 0.05; ** indicates p ≤ 0.01.
Results and discussion
Hydrophilic and hydrophobic characteristics of VhChlABC
The hydrophilic/hydrophobic map of VhChlABC showed that the number of amino acids with a hydrophobic index less than zero was significantly more than those with a hydrophobic index greater than zero, indicating that the overall hydrophilicity of VhChlABC was strong (Fig. 1A). Moreover, the large number of strong hydrophilic amino acids on the protein surface formed a strong hydrophilic surface layer of VhChlABC (Fig. 1B). The visualization of the hydrophilic and hydrophobic surface by PyMOL (PyMOL Molecular Graphics System, ver. 2.0, Schrödinger, LLC.) also showed the typical characteristics of the strong hydrophilic surface of VhChlABC (Fig. 1C). As reported, SDS consists of a hydrophilic polar head and a hydrophobic non-polar tail, in which the hydrophobic part can be inserted into the hydrophobic interior of the protein to destroy the spatial structure of the protein and cause it to be denatured (Khan et al. 2019; Zaidi et al. 2014). Although there is no absolute correlation between hydrophobicity and stability, the regular arrangement of non-polar amino acids in the protein sphere is one of the reasons for stability. Improving hydrophobic core packing is one of the practical ways to stabilize proteins (Tanaka et al. 2005; Tych et al. 2016). Accordingly, we hypothesized that the strong hydrophilic surface of VhChlABC may better protect the internal hydrophobic center, thus playing an important role in the stability of its overall structure, including a wide pH tolerance range, temperature stability, and strong SDS resistance (Su et al. 2021).
Fig. 1.
Hydrophilicity/hydrophobicity analysis of VhChlABC. A Hydrophilic/hydrophobic map. The vertical axis represents the hydrophobic fraction, and the horizontal axis represents the amino acid residues at the corresponding positions. The online tool ProtScale was used to map the hydrophilicity and hydrophobicity of VhChlABC as described in the “Materials and Methods”. B 3D-distribution map of strongly hydrophilic/hydrophobic amino acids. Amino acids with hydrophobicity index higher than 1 and less than − 1.5 were defined as strongly hydrophobic (red) and strongly hydrophilic (green), respectively. C The hydrophilic/hydrophobic surface, the surface hydrophobic cluster and the critical catalytic residues. PyMOL was used for visualization of hydrophilic/hydrophobic surface of VhChlABC. The darker the red, the more hydrophobic the amino acid. The six amino acids that form the hydrophobic cluster were represented as spheres. The critical catalytic residues were shown as mesh
As described above, SDS can bind to proteins by predominantly hydrophobic interactions, causing unfolding of the tertiary structure (Zaidi et al. 2014). For proteins with a strong hydrophilic protective layer, a hydrophobic cluster on the protein surface may be a breakthrough for SDS to “destroy” the protein. It was worth noting that there was a strong hydrophobic cluster on the surface of VhChlABC, which was mainly composed of six hydrophobic amino acids: Ala181, Met182, Met183, Ala184, Val185, and Ile305 (Fig. 1C). Obviously, SDS did not use this hydrophobic cluster as a “break point”, suggesting that this cluster may have special structural features that prevent SDS binding.
Acquisition of site-directed mutants of VhChlABC
In order to further explore the relationship between SDS resistance and the hydrophobic cluster on the surface of VhChlABC, the six amino acids forming the hydrophobic cluster were respectively mutated to reduce the hydrophobicity of this part. The 16 mutants designed were shown in Table 1.
Table 1.
Mutation sites and mutants of VhChlABC
| Site | Mutant | Hydrophobicity fraction | |
|---|---|---|---|
| The former | The latter | ||
| Ala181 | A181G | 1.8 | − 0.4 |
| A181S | 1.8 | − 0.8 | |
| Met182 | M182A | 1.9 | 1.8 |
| M182G | 1.9 | − 0.4 | |
| M182S | 1.9 | − 0.8 | |
| Met183 | M183A | 1.9 | 1.8 |
| M183G | 1.9 | − 0.4 | |
| M183S | 1.9 | − 0.8 | |
| Ala184 | A184G | 1.8 | − 0.4 |
| A184S | 1.8 | − 0.8 | |
| Val185 | V185A | 4.2 | 1.8 |
| V185G | 4.2 | − 0.4 | |
| V185S | 4.2 | − 0.8 | |
| Ile305 | I305A | 4.5 | 1.8 |
| I305G | 4.5 | − 0.4 | |
| I305S | 4.5 | − 0.8 | |
The expression vectors of all mutants were listed in Table S2
A alanine (Ala), G glycine (Gly), S serine (Ser), M methionine (Met), V valine (Val), I isoleucine (Ile)
Under the same conditions as VhChlABC (Su et al. 2021), the mutant proteins were expressed in soluble form in the pET-28a (+)/E.coli BL21(DE3) system and purified by nickel affinity chromatography. Mutant I305A was not expressed after IPTG induction (there was no obvious band of corresponding size in the crude enzyme solution; data not shown). The reason for the lack of soluble expression was unclear, so this mutant was not studied further in this research. As shown in Supplementary Fig. S1, the target proteins with a molecular weight of about 110 kDa were identified.
Effects of SDS on activity and stability of VhChlABC and mutants
The specific activity of each mutant under optimal reaction conditions was first determined as previously reported (Su et al. 2021). Although the surface hydrophobic cluster was away from the catalytic center (Fig. 1C), it still had a certain effect on the activity. As shown in Fig. 2, mutants A181S, M182G, M182S, M183G, M183S, A184S, V185G, V185S and I305S showed a significant decrease in activity. We further investigated the effects of SDS on the activity and stability of mutants A181G, M182A, M183A, A184G, V185A and I305G, whose specific activity was not significantly reduced compared with the wild-type (WT) enzyme. The effect of different concentrations of SDS on enzyme activity was elucidated. For all conditions, WT showed higher activity than that of the mutants (Fig. 3A). Apparently, I305G was completely inactivated in the presence of 0.1% (w/v) SDS (Fig. 3A). For the SDS stability study, all enzymes were incubated in the presence of 0.1% (w/v) SDS for 1 h, and the remaining activity was measured. The result indicated that the SDS stability of all the mutants was significantly lower than that of WT (Fig. 3B). The hydrophobic pocket formed by these hydrophobic amino acids was important for the SDS stability of VhChlABC.
Fig. 2.
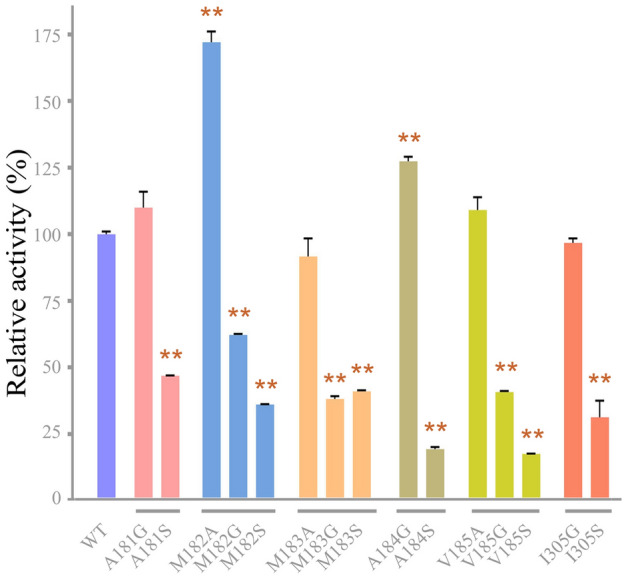
Relative specific activity of WT and mutants. Error bars indicated standard deviation (n = 3). **Represents an extremely significant difference, p < 0.01
Fig. 3.
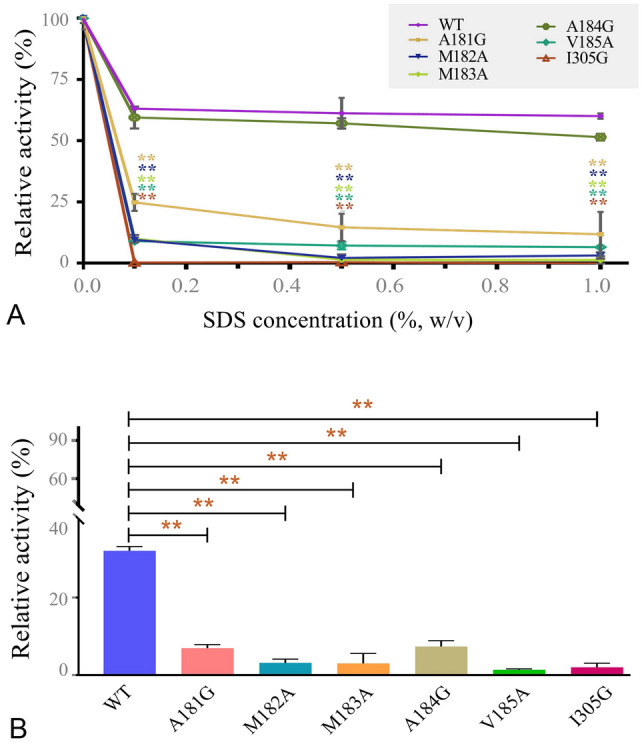
Effects of SDS on activity and stability of WT and its mutants. A Relative activity of enzymes in different concentrations of SDS. B Stability of enzymes pre-incubated with 0.1% (w/v) SDS for 1 h. Error bars indicated standard deviation (n = 3). **Represents an extremely significant difference, p < 0.01. Enzyme activity without SDS pre-incubation was defined as 100%
Molecular dynamics (MD) simulation and molecular docking
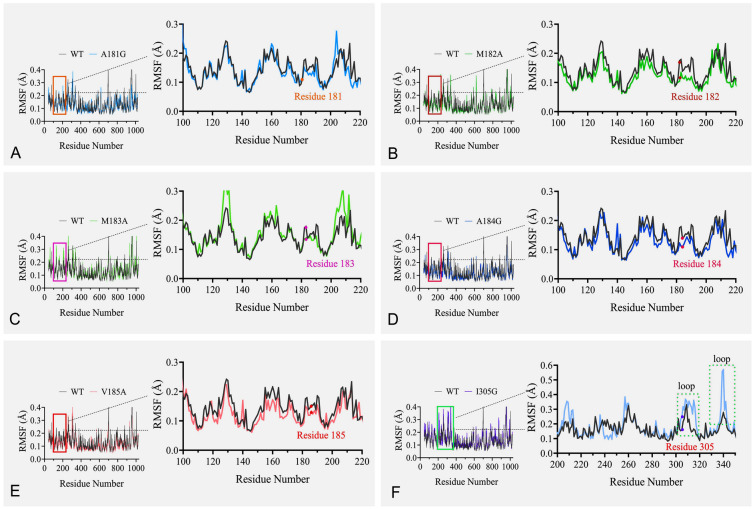
To shed light on the mechanisms responsible for differences in SDS resistance, MD simulations were carried out on WT and mutants A181G, M182A, M183A, A184G, V185A and I305G. Supplementary Fig. S2 showed all root-mean-square deviation (RMSD) reached a stationary shape after 40 ns. The analysis of the root-mean-square fluctuation (RMSF, as shown in Fig. 4) showed that the fluctuation trend of mutants was roughly the same as that of WT. There was no obvious fluctuation near the corresponding mutation sites in mutants A181G, M182A, M183A, A184G and V185A (Fig. 4A–E). The conversion of isoleucine to glycine did not cause significant fluctuations in the RMSF value of the target site. However, there was an obvious fluctuation on the loop formed by Gly304-Asp316 and Ser338-Glu342, which might lead to the swing of Gly305 (Fig. 4F and Supplementary Fig. S3). This suggested that I305G became more flexible around this site, which may be more conducive to SDS insertion and binding.
Fig. 4.
Calculation RMSF (Å) for VhChlABC (WT) and its mutants with a 40–50 ns trajectory. WT is shown in gray; A181G (A), M182A (B), M183A (C), A184G (D), V185A (E) and I305G (F) are shown in light blue, green, cyan, dark blue, orange and pink, respectively. The corresponding mutant sites are highlighted with red dots in the local RMSF map, and the two loops formed by Gly304-Asp316 and Ser338-Glu342 are marked by dashed box (F)
Hydrophobic regions usually refer to ligand-binding pockets in proteins, which are of great significance for proteins to perform their biological functions. For example, mammalian sugar-recognizing F-box proteins commonly bind the N-glycans through a unique small hydrophobic pocket in their loops, initiating the degradation process of glycoproteins (Yoshida et al. 2019). Capsid proteins (CP) of many viruses have conserved hydrophobic pockets that play a crucial role in capsid assembly and virus germination (Aggarwal et al. 2017; Kumar et al. 2021). SDS could bind to proteins by predominantly hydrophobic interactions, and it was speculated that the resistance of VhChlABC to SDS might be due to the inhibition of their binding. In order to evaluate the interaction between enzyme and SDS, AutoDock was used to dock the SDS into WT and its mutants. Take the mutant I305G, for example, the trajectory between 40 and 50 ns of simulations were selected for further analyses. The maximum cluster of structures in the simulated systems was used for molecular docking. SDS was predicted to bind in the hydrophobic pocket as illustrated in Fig. 5. More notably, the reduced hydrophobicity of residue 305 (I → G) affected the binding conformation between protein and SDS. Compared with WT, the hydrophobic hydrocarbon chain of SDS docked in I305G was inserted deeper into the hydrophobic pocket (Fig. 5). Furthermore, the RMSD analysis of the last 10 ns trajectory of SDS-protein complexes showed that the RMSD of SDS-mutants was lower than that of SDS-WT, and the fluctuation was also significantly reduced, especially for the SDS-I305G complex (Supplementary Fig. S4). This result suggested that SDS could bind to the surface of mutant I305G more stably, which might lead to its SDS sensitivity.
Fig. 5.
Binding conformation of SDS on the hydrophobic surface of enzyme molecules. A WT, B I305G
Additionally, the binding free energy (ΔGbind) of SDS to enzyme was calculated based on MD simulations of SDS-enzyme complex using the gmx_MMPBSA (Valdés-Tresanco et al. 2021). As the result, except for A184G, the affinity of the mutants with SDS was significantly higher than that of WT, and I305G showed the highest affinity (Fig. 6), which might be the key factor leading to the inhibition of their activities by SDS. Additionally, the average ΔGbind of mutant A184G to SDS was not significantly different from that of WT, and its activity in the presence of SDS was close to that of WT (Fig. 3A). However, the SDS stability of mutant A184G was significantly lower than that of WT (Fig. 3B), indicating that residue Ala184 played an important role in maintaining the stability of the enzyme in SDS.
Fig. 6.

Average binding free energy (ΔGbind) analysis of SDS to WT and its mutants. Error bars indicated standard deviation (n = 3). **Represents an extremely significant difference, p < 0.01
Effects of temperature and pH on VhChlABC and mutants
Other biochemical characterizations of mutants, including optimum temperature, thermostability, optimum pH and pH stability, were also examined in this study. As the result, mutants A181G, M182A, M183A, A184G, V185A and I305G all exhibited the highest activity towards CS-A at 40 °C (Supplementary Fig. S5), and their initial activity maintained more than 90% and more than 60% post pre-incubation at 0–30 °C and at 40 °C for 1 h (Supplementary Fig. S5), respectively, which were consistent with the VhChlABC. The decrease in SDS tolerance of the mutants was not accompanied by a decrease in temperature stability, highlighting the close association between SDS tolerance and hydrophobicity of the residues at these sites. The optimal pH system of these six mutants was the same as that of WT, showing the highest activity in 50 mmol/L Na2HPO4-NaH2PO4 buffer at pH 7.0 (Supplementary Fig. S6). However, the stability of the mutants rapidly decreased in glycine–NaOH buffer at a pH higher than 9.0 (Supplementary Fig. S6), suggesting that the decrease in hydrophobicity of the corresponding mutant residue also played an important role in the stability of the enzyme in strongly alkaline solutions. Since the SDS-related tests in this study were carried out in Na2HPO4-NaH2PO4 buffer with pH 7.0, the relationship between pH stability and SDS tolerance was not explored in depth.
Conclusion
In this study, the mechanism of VhChlABC resistance to SDS was investigated by combining bioinformatics analysis, site-directed mutagenesis, MD simulation and molecular docking methods. Altogether, it was indicated that the hydrophobic microenvironment composed of residues Ala181, Met182, Met183, Ala184, Val185, and Ile305 on the surface of VhChlABC could prevent SDS from binding to this region and destroying its hydrophobic packaging by increasing the rigid conformation of the hydrophobic pocket on the protein surface and decreasing the binding force between SDS and protein, which was the key structural basis for determining the resistance of protein to SDS.
The study on the SDS-resistant mechanism of VhChlABC provides a new strategy for the rational design of SDS-resistant proteins, which is beneficial to obtain the phenotype more conveniently and efficiently. Moreover, this study may further advance the research process of chondroitinase in the targeted therapy of lung diseases, which might be constrained by the abundance of surfactants in the lung microenvironment.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by Qingdao Marine Science and Technology Center (2022QNLM030003-1), Shandong Province Technology Innovation Guidance Program (2018YFC0311105) and Shandong Provincial Natural Science Foundation (ZR2019ZD18).
Author contributions
JS methodology, software, validation, formal analysis, investigation, data curation, visualization, writing original draft. HW software, conceptualization, methodology, validation, formal analysis, investigation, data curation. CY software, investigation, data curation. FZ investigation, data curation. FH conceptualization, supervision, project administration, funding acquisition, writing-review and editing. WY supervision, project administration, writing-review & editing. The final manuscript was approved by all the authors.
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Conflict of interest
All authors declare no conflicts of interests. Author Wengong Yu is one of the Editorial Board Members, but he was not involved in the journal’s review of, or decision related to, this manuscript.
Human and animal rights
This article does not contain any studies with human participants and animal experiments.
Footnotes
Juanjuan Su and Hao Wu contributed equally to this work.
Contributor Information
Feng Han, Email: fhan@ouc.edu.cn.
Wengong Yu, Email: yuwg66@ouc.edu.cn.
References
- Abdallah MM, Fernández N, Matias AA, Bronze MDR. Hyaluronic acid and chondroitin sulfate from marine and terrestrial sources: extraction and purification methods. Carbohydr Polym. 2020;243:116441. doi: 10.1016/j.carbpol.2020.116441. [DOI] [PubMed] [Google Scholar]
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- Aggarwal M, Kaur R, Saha A, Mudgal R, Yadav R, Dash PK, Parida M, Kumar P, Tomar S. Evaluation of antiviral activity of piperazine against Chikungunya virus targeting hydrophobic pocket of alphavirus capsid protein. Antivir Res. 2017;146:102–111. doi: 10.1016/j.antiviral.2017.08.015. [DOI] [PubMed] [Google Scholar]
- Agiba AM, Nasr M, Abdel-Hamid S, Eldin AB, Geneidi AS. Enhancing the intestinal permeation of the chondroprotective nutraceuticals glucosamine sulphate and chondroitin sulphate using conventional and modified liposomes. Curr Drug Deliv. 2018;15:907–916. doi: 10.2174/1567201815666180123100148. [DOI] [PubMed] [Google Scholar]
- Bradbury EJ, Moon LD, Popat RJ, King VR, Bennett GS, Patel PN, Fawcett JW, McMahon SB. Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature. 2002;416:636–640. doi: 10.1038/416636a. [DOI] [PubMed] [Google Scholar]
- Chen J, Wang X, Zhu T, Zhang Q, Zhang JZ. A comparative insight into amprenavir resistance of mutations V32I, G48V, I50V, I54V, and I84V in HIV-1 protease based on thermodynamic integration and MM-PBSA methods. J Chem Inf Model. 2015;55:1903–1913. doi: 10.1021/acs.jcim.5b00173. [DOI] [PubMed] [Google Scholar]
- Chen J, Zeng Q, Wang W, Sun H, Hu G. Decoding the identification mechanism of an SAM-III riboswitch on ligands through multiple independent gaussian-accelerated molecular dynamics simulations. J Chem Inf Model. 2022;62:6118–6132. doi: 10.1021/acs.jcim.2c00961. [DOI] [PubMed] [Google Scholar]
- Collin EC, Carroll O, Kilcoyne M, Peroglio M, See E, Hendig D, Alini M, Grad S, Pandit A. Ageing affects chondroitin sulfates and their synthetic enzymes in the intervertebral disc. Signal Transduct Target Ther. 2017;2:17049. doi: 10.1038/sigtrans.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan L, Jianping X, Ruzhen G, Honghai W. Cloning and characterization of Rv0621 gene related to surfactant stress tolerance in Mycobacterium tuberculosis. Mol Biol Rep. 2009;36:1811–1817. doi: 10.1007/s11033-008-9384-3. [DOI] [PubMed] [Google Scholar]
- Djerbal L, Lortat-Jacob H, Kwok J. Chondroitin sulfates and their binding molecules in the central nervous system. Glycoconj J. 2017;34:363–376. doi: 10.1007/s10719-017-9761-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitrieva N, Yu L, Viapiano M, Cripe TP, Chiocca EA, Glorioso JC, Kaur B. Chondroitinase ABC I-mediated enhancement of oncolytic virus spread and antitumor efficacy. Clin Cancer Res. 2011;17:1362–1372. doi: 10.1158/1078-0432.CCR-10-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Smieško M. Decision making in structure-based drug discovery: visual inspection of docking results. J Med Chem. 2021;64:2489–2500. doi: 10.1021/acs.jmedchem.0c02227. [DOI] [PubMed] [Google Scholar]
- Fulton A, Frauenkron-Machedjou VJ, Skoczinski P, Wilhelm S, Zhu L, Schwaneberg U, Jaeger KE. Exploring the protein stability landscape: Bacillus subtilis lipase A as a model for detergent tolerance. ChemBioChem. 2015;16:930–936. doi: 10.1002/cbic.201402664. [DOI] [PubMed] [Google Scholar]
- Gandorfer A. Enzymatic vitreous disruption. Eye (lond) 2008;22:1273–1277. doi: 10.1038/eye.2008.29. [DOI] [PubMed] [Google Scholar]
- Ilieva KM, Cheung A, Mele S, Chiaruttini G, Crescioli S, Griffin M, Nakamura M, Spicer JF, Tsoka S, Lacy KE, Tutt ANJ, Karagiannis SN. Chondroitin sulfate proteoglycan 4 and its potential as an antibody immunotherapy target across different tumor types. Front Immunol. 2017;8:1911. doi: 10.3389/fimmu.2017.01911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru D, Sugiura N, Akiyama H, Watanabe H, Matsumoto K. Alterations in the chondroitin sulfate chain in human osteoarthritic cartilage of the knee. Osteoarthritis Cartil. 2014;22:250–258. doi: 10.1016/j.joca.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Kasinathan N, Volety SM, Josyula VR. Chondroitinase: a promising therapeutic enzyme. Crit Rev Microbiol. 2016;42:474–484. doi: 10.3109/1040841X.2014.959893. [DOI] [PubMed] [Google Scholar]
- Khan S, Khan P, Hassan MI, Ahmad F, Islam A. Protein stability: determination of structure and stability of the transmembrane protein Mce4A from M. tuberculosis in membrane-like environment. Int J Biol Macromol. 2019;126:488–495. doi: 10.1016/j.ijbiomac.2018.12.183. [DOI] [PubMed] [Google Scholar]
- Khan AR, Yang X, Du X, Yang H, Liu Y, Khan AQ, Zhai G. Chondroitin sulfate derived theranostic and therapeutic nanocarriers for tumor-targeted drug delivery. Carbohydr Polym. 2020;233:115837. doi: 10.1016/j.carbpol.2020.115837. [DOI] [PubMed] [Google Scholar]
- Kumar R, Nehul S, Singh A, Tomar S. Identification and evaluation of antiviral potential of thymoquinone, a natural compound targeting Chikungunya virus capsid protein. Virology. 2021;561:36–46. doi: 10.1016/j.virol.2021.05.013. [DOI] [PubMed] [Google Scholar]
- Manganelli R, Dubnau E, Tyagi S, Kramer FR, Smith I. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol Microbiol. 1999;31:715–724. doi: 10.1046/j.1365-2958.1999.01212.x. [DOI] [PubMed] [Google Scholar]
- Mishra S, Ganguli M. Functions of, and replenishment strategies for, chondroitin sulfate in the human body. Drug Discov Today. 2021;26:1185–1199. doi: 10.1016/j.drudis.2021.01.029. [DOI] [PubMed] [Google Scholar]
- Mizuguchi S, Uyama T, Kitagawa H, Nomura KH, Dejima K, Gengyo-Ando K, Mitani S, Sugahara K, Nomura K. Chondroitin proteoglycans are involved in cell division of Caenorhabditis elegans. Nature. 2003;423:443–448. doi: 10.1038/nature01635. [DOI] [PubMed] [Google Scholar]
- Moon LD, Asher RA, Rhodes KE, Fawcett JW. Regeneration of CNS axons back to their target following treatment of adult rat brain with chondroitinase ABC. Nat Neurosci. 2001;4:465–466. doi: 10.1038/87415. [DOI] [PubMed] [Google Scholar]
- Morla S. Glycosaminoglycans and glycosaminoglycan mimetics in cancer and inflammation. Int J Mol Sci. 2019;20:1963. doi: 10.3390/ijms20081963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou J, Li Q, Qi X, Yang J. Structural comparison, antioxidant and anti-inflammatory properties of fucosylated chondroitin sulfate of three edible sea cucumbers. Carbohydr Polym. 2018;185:41–47. doi: 10.1016/j.carbpol.2018.01.017. [DOI] [PubMed] [Google Scholar]
- Mou M, Hu Q, Li H, Long L, Li Z, Du X, Jiang Z. Characterization of a thermostable and surfactant-tolerant chondroitinase B from a marine bacterium Microbulbifer sp. ALW1. Int J Mol Sci. 2022;23:5008. doi: 10.3390/ijms23095008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandini CD, Sugahara K. Role of the sulfation pattern of chondroitin sulfate in its biological activities and in the binding of growth factors. Adv Pharmacol. 2006;53:253–279. doi: 10.1016/S1054-3589(05)53012-6. [DOI] [PubMed] [Google Scholar]
- Sato Y, Nakanishi K, Tokita Y, Kakizawa H, Ida M, Maeda H, Matsui F, Aono S, Saito A, Kuroda Y, Hayakawa M, Kojima S, Oohira A. A highly sulfated chondroitin sulfate preparation, CS-E, prevents excitatory amino acid-induced neuronal cell death. J Neurochem. 2008;104:1565–1576. doi: 10.1111/j.1471-4159.2007.05107.x. [DOI] [PubMed] [Google Scholar]
- Staubach F, Nober V, Janknecht P. Enzyme-assisted vitrectomy in enucleated pig eyes: a comparison of hyaluronidase, chondroitinase, and plasmin. Curr Eye Res. 2004;29:261–268. doi: 10.1080/02713680490516747. [DOI] [PubMed] [Google Scholar]
- Su J, Wang X, Yin C, Li Y, Wu H, Yu W, Han F. Identification and biochemical characterization of a surfactant-tolerant chondroitinase VhChlABC from Vibrio hyugaensis LWW-1. Mar Drugs. 2021;19:399. doi: 10.3390/md19070399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugahara K, Mikami T. Chondroitin/dermatan sulfate in the central nervous system. Curr Opin Struct Biol. 2007;17:536–545. doi: 10.1016/j.sbi.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Igarashi S, Ferri S, Sode K. Increasing stability of water-soluble PQQ glucose dehydrogenase by increasing hydrophobic interaction at dimeric interface. BMC Biochem. 2005;6:1. doi: 10.1186/1471-2091-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tych KM, Batchelor M, Hoffmann T, Wilson MC, Hughes ML, Paci E, Brockwell DJ, Dougan L. Differential effects of hydrophobic core packing residues for thermodynamic and mechanical stability of a hyperthermophilic protein. Langmuir. 2016;32:7392–7402. doi: 10.1021/acs.langmuir.6b01550. [DOI] [PubMed] [Google Scholar]
- Valdés-Tresanco MS, Valdés-Tresanco ME, Valiente PA. gmx_MMPBSA: a new tool to perform end-state free energy calculations with GROMACS. J Chem Theory Comput. 2021;17:6281–6291. doi: 10.1021/acs.jctc.1c00645. [DOI] [PubMed] [Google Scholar]
- Volpi N. Anti-inflammatory activity of chondroitin sulphate: new functions from an old natural macromolecule. Inflammopharmacology. 2011;19:299–306. doi: 10.1007/s10787-011-0098-0. [DOI] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Mizushima T, Tanaka K. Sugar-recognizing ubiquitin ligases: action mechanisms and physiology. Front Physiol. 2019;10:104. doi: 10.3389/fphys.2019.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi N, Nusrat S, Zaidi FK, Khan RH. pH-dependent differential interacting mechanisms of sodium dodecyl sulfate with bovine serum fetuin: a biophysical insight. J Phys Chem B. 2014;118:13025–13036. doi: 10.1021/jp501515g. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Lu D, Wang S, Wei L, Wang W, Li F. Identification and biochemical characterization of a novel chondroitin sulfate/dermantan sulfate lyase from Photobacterium sp. Int J Biol Macromol. 2020;165:2314–2325. doi: 10.1016/j.ijbiomac.2020.10.119. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Su H, Wang X, Tang L, Hu J, Yu W, Han F. Cloning and characterization of a novel chondroitinase ABC categorized into a new subfamily of polysaccharide lyase family 8. Int J Biol Macromol. 2020;164:3762–3770. doi: 10.1016/j.ijbiomac.2020.08.210. [DOI] [PubMed] [Google Scholar]
- Zhu C, Zhang J, Zhang J, Jiang Y, Shen Z, Guan H, Jiang X. Purification and characterization of chondroitinase ABC from Acinetobacter sp. C26. Int J Biol Macromol. 2017;95:80–86. doi: 10.1016/j.ijbiomac.2016.10.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.