Abstract
A subset of xeroderma pigmentosum (XP) group E cells lack a factor that binds to DNA damaged by UV radiation. This factor can be purified to homogeneity as p125, a 125-kDa polypeptide. However, when cDNA encoding p125 is translated in vitro, only a small fraction binds to UV-damaged DNA, suggesting that a second factor is required for the activation of p125. We discovered that most hamster cell lines expressed inactive p125, which was activated in somatic cell hybrids containing human chromosome region 11p11.2-11cen. This region excluded p125 but included p48, which encodes a 48-kDa polypeptide known to copurify with p125 under some conditions. Expression of human p48 activated p125 binding in hamster cells and increased p125 binding in human cells. No such effects were observed from expression of p48 containing single amino acid substitutions from XP group E cells that lacked binding activity, demonstrating that the p48 gene is defective in those cells. Activation of p125 occurred by a “hit-and-run” mechanism, since the presence of p48 was not required for subsequent binding. Nevertheless, p48 was capable of forming a complex with p125 either bound to UV-damaged DNA or in free solution. It is notable that hamster cells fail to efficiently repair cyclobutane pyrimidine dimers in nontranscribed DNA and fail to express p48, which contains a WD motif with homology to proteins that reorganize chromatin. We propose that p48 plays a role in repairing lesions that would otherwise remain inaccessible in nontranscribed chromatin.
Xeroderma pigmentosum (XP) is an autosomal recessive disease characterized by abnormal sensitivity to UV radiation, predisposition to skin cancer, and defective nucleotide excision repair (8, 10). Cell fusion experiments demonstrate that XP consists of seven complementation groups (A through G). Genes have been identified for each of the groups with the exception of group E.
Human and monkey cells contain a binding activity which has high specificity for DNA damaged by UV but which also recognizes DNA damaged by cisplatin, nitrogen mustard, denaturation, and depurination (6, 7, 19, 33). This UV-damaged-DNA binding (UV-DDB) activity appears to play a role in nucleotide excision repair: it is absent in a subset of XP group E cells, which like all XP cells are deficient in DNA repair (7, 22, 27), and it is expressed at higher levels in human tumor cell lines, which were selected for resistance to cisplatin and which display cross-resistance to UV radiation and enhanced DNA repair (6).
Purification of UV-DDB activity to apparent homogeneity yields a single polypeptide of 125 kDa (p125) (2, 21, 25). When partially purified preparations of p125 were microinjected into XPE− cells (XPE cells without binding activity), nucleotide excision repair was restored to normal levels (26). Tryptic peptides derived from purified p125 were sequenced, and the amino acid sequences were used to isolate monkey (41) and human (12, 20) cDNAs, each containing an open reading frame of the appropriate size. The identity of the p125 cDNA was confirmed by translation in rabbit reticulocyte extracts and demonstration that a small fraction bound specifically to UV-damaged DNA (20). However, the great majority of the in vitro-translated p125 lacked binding activity, suggesting that full activation of p125 might require a second factor that was present in limiting amounts in the rabbit reticulocyte extracts.
Here we report that p125 binding activity is absent in most hamster cell lines but is activated in somatic cell hybrids containing human chromosome 11. Analysis of chromosome 11 fragments excluded the region containing the p125 gene (DDB1, Online Mendelian Inheritance in Man [OMIM] entry 600045) but included the p48 gene (DDB2, OMIM entry 600811), which encodes a 48-kDa polypeptide that copurifies with p125 under some conditions (25). Transfection of the hamster cells with wild-type human p48 cDNA conferred binding activity, demonstrating that p48 is required for activation of p125. On the other hand, transfection with p48 cDNA derived from XPE− cells failed to confer binding activity, demonstrating that the p48 gene is inactivated by mutation in XPE− cells.
MATERIALS AND METHODS
Cell cultures.
Chinese hamster-human hybrid cells (13, 14, 28, 31) were grown either in RPMI medium with 20% fetal bovine serum (FBS) and 1% hypoxanthine-aminopterin-thymidine (HAT) solution (XII-2D-1f HAT and XII-4A-1d-E HAT cells), in RPMI medium with 20% FBS (XVII-10A-12a, XXI-22A-g-1a, XXI-23A-2c, XXI-54B-1i, and XV-18A-7a-N4 cells), or in minimal essential medium with 15% FBS (DonTK− and V79/380-6 cell lines). All other cell lines were grown in Dulbecco’s modified Eagle’s medium with 10% FBS. All media were supplemented with 1% glutamine and 1% antibiotic (penicillin and streptomycin).
Azacytidine treatment.
Azacytidine treatment was carried out as described previously (35). Briefly, 2 × 105 cells were seeded onto a 100-mm-diameter dish and grown for 15 h in normal medium, for 15 h in medium containing 3 μg of freshly dissolved 5-azacytidine (Sigma, St. Louis, Mo.) per ml, for 24 h in normal medium, and then for 15 h in 5-azacytidine-containing medium again. Whole-cell extract was prepared 48 h after the second azacytidine treatment.
Preparation of whole-cell extract.
Confluent cells in a 100-mm-diameter dish were harvested in 1 ml of ice-cold phosphate-buffered saline and pelleted by centrifugation for 1 min at 12,000 × g. The pellet was resuspended in lysis buffer (700 mM NaCl, 1 mM EGTA, 1 mM EDTA, 10 mM β-glycerophosphate, 2 mM MgCl2, 10 mM KCl, 1 mM sodium vanadate, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, 0.1% Nonidet P-40, and pepstatin, leupeptin, and aprotinin [each at 10 μg/ml]) and incubated at 4°C for 30 min with gentle shaking. The supernatant was saved after centrifugation at 12,000 × g for 30 min at 4°C. Protein concentrations were measured by a modification of the Bradford method (4).
Electrophoretic mobility shift assay for binding activity.
Whole-cell extracts were assayed for UV-DDB activity by an electrophoretic mobility shift assay as described previously (21). Briefly, the 32P-labeled 148-bp DNA probe (f148) was prepared and either left intact or damaged with a UV dose of 300 or 5,000 J/m2. Extracts were incubated with 0.2 ng of DNA probe and a mixture of unlabeled salmon sperm DNA (500 ng) and poly(dI-dC) (1,000 ng) to mask the effects of nonspecific DNA binding proteins. The mixture was then resolved by nondenaturing gel electrophoresis, and the gel was dried and exposed to X-ray film.
Anti-FLAG (M2; IBI-Kodak, New Haven, Conn.) or antihemagglutinin (HA) (12CA5; Boehringer Mannheim, Indianapolis, Ind.) antibodies were used in a mobility supershift assay as described previously (36). Briefly, cell extracts were preincubated at room temperature for 20 min with serial dilutions of antibodies (10−1 to 10−3), incubated at room temperature for 30 min with DNA probe, and then resolved by nondenaturing gel electrophoresis.
Northern blot analysis.
Total RNA was prepared and analyzed by Northern blotting as described previously (37). Probe for p125 mRNA was prepared by digesting p125 cDNA with the restriction enzymes EcoRI and EagI and then eluting a 527-bp DNA fragment from a 1% agarose gel. RNA was denatured, resolved on a 0.9% formaldehyde agarose gel, transferred to a Hybond N nylon membrane (Amersham, Arlington Heights, Ill.), and cross-linked with UV (Stratalinker UV source; Stratagene, La Jolla, Calif.). The membrane was hybridized to 32P-labeled probe at 65°C for 24 h in 1× nylon wash buffer (14% sodium dodecyl sulfate [SDS], 130 mM Na2HPO4, 14 mM EDTA, and 0.2% Triton X-100) prewarmed to 65°C. The membrane was washed twice in 0.5× nylon wash buffer at 65°C for 10 min and then exposed to X-ray film.
Isolation of p48 and p125 cDNAs.
Human p48 cDNA was isolated from a Jurkat T-cell leukemia cDNA library by two-step PCR amplification. The oligonucleotide primers were synthesized from the reported p48 nucleotide sequence as follows: 5′ outer primer, 5′-CCCGCCTTGTTTCTCCCCAG; 3′ outer primer, 5′-CTTGCAGGACTTGATCCCATGTG; 5′ inner primer, 5′-ACGGAGAGTACTATGGCTCCCAAGAAA; 3′ inner primer, 5′-TGGGCTCCAAGGCCTTGTCTGGC; 5′ innermost primer, 5′-TGTCCAGCAGGGGCTCCAGCA; and 3′ innermost primer, 5′-GCCTTGATGGGTGTGAGGTGC. In the first amplification step the 5′ outer primer and the 3′ outer primer were used, and in the second step 1/10 of the first PCR product and the 5′ inner primer and 3′ inner primer were used. The amplified p48 cDNA was cloned into the pBJ5 mammalian expression vector (40) either by itself or with the FLAG epitope sequence (45) fused to the region encoding the N terminus. The cDNA expression vector for p48 without FLAG contains two base changes leading to two amino acid changes near the N terminus, L53P and F110L, due to errors occurring during PCR amplification. The FLAG-p48 cDNA does not contain any base changes compared to the reported p48 cDNA sequence (12). The cDNAs containing the nucleotide changes found in two types of XPE cells (resulting in R273H in XP2415 cells and K244E in XP82TO cells) (30) were isolated by reverse transcription-PCR (RT-PCR), as follows. Total RNA was prepared from XP2415 and XP82TO cells as described previously (37), and 1 μg of the total RNA was reverse transcribed from random hexamer primers and amplified by PCR in two steps. The first step used the 5′ outer primer and 3′ outer primer, and the second step used 1/10 of the first PCR product and the 5′ innermost primer and 3′ innermost primer. The amplified PCR product was digested with EcoRV and HincII and then used to replace the corresponding EcoRV-to-HincII fragment in the p48 expression vectors. The nucleotide changes reported for XP2415 and XP82TO cells were confirmed by DNA sequencing.
Human p125 cDNA was isolated from a Jurkat T-cell leukemia cDNA library as described previously (20). The cDNA was cloned into pBJ5 either alone or with the FLAG epitope sequence fused to the region encoding the N terminus or the HA epitope sequence fused to the region encoding the C terminus.
Transfection.
Cells (2 × 106) were seeded onto a 100-mm-diameter dish and transfected as a monolayer after overnight growth by coprecipitation of DNA with calcium phosphate transfection as described previously (9). The cells were transiently transfected with 24 μg of pBJ5-derived expression vector and 1 μg of pRSV-luciferase (11) to normalize the transfection efficiencies. Whole-cell extracts were prepared 24 or 48 h after transfection to measure the binding activity, luciferase activity, and the expression of FLAG-p48 by immunoblotting.
Immunoprecipitation.
Whole-cell extracts (210 μg) were made from 293T cells transfected with expression vectors for p125 and p48 and then incubated at 4°C for 3 h with 5 μg of anti-FLAG or anti-HA antibodies. Proteins bound to the antibodies were precipitated by incubation with 10 μl of protein G-Sepharose (Pharmacia, Piscataway, N.J.) at 4°C for 1 h with shaking. After the protein G-Sepharose was washed, the bound proteins were eluted by being boiled for 5 min in 30 μl of SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer.
Immunoblot analysis.
Whole-cell extracts (10 to 50 μg) and immunoprecipitated proteins were resolved by SDS-PAGE, transferred to a nitrocellulose membrane (Schleicher & Schuell, Keene, N.H.), and probed with mouse anti-FLAG immunoglobulin G (IgG) (1:250 dilution) or mouse anti-HA IgG (1:250 dilution) antibodies and then with horseradish peroxidase-conjugated goat anti-mouse IgG antibodies (1:1,000 dilution; Vector Laboratories, Burlingame, Calif.). Antibody binding was detected by enhanced chemiluminescence (ECL reagents; Amersham, Little Chalfont, Buckinghamshire, England).
RESULTS
p125 is expressed but inactive in hamster cell lines.
In the course of screening different animal cells for UV-DDB activity, we found that monkey and rabbit cells had levels similar to those of wild-type human cells. Surprisingly, mouse cells had much lower levels, and of 10 hamster cell lines tested, 9 showed no detectable activity and 1 (DonTK−) showed only a low level of activity (see Fig. 1A and 4A; other data not shown).
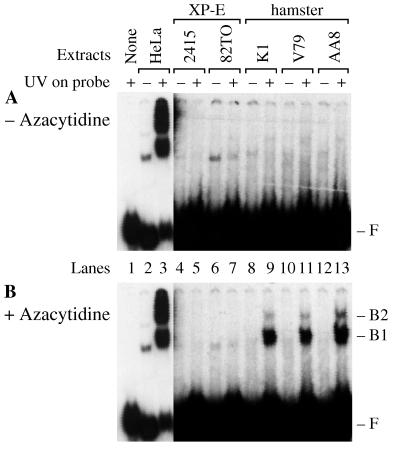
FIG. 1.
Azacytidine activates p125 in hamster cells. Human cell lines (HeLa and XPE− cell lines, 2415 and 82TO) and wild-type hamster cell lines (K1, V79, and AA8) were grown in medium with azacytidine omitted (A) or added (B). Whole-cell extracts (2 μg) were then assayed for damage-specific binding activity with DNA probes either damaged with 5,000 J/m2 UV (+) or left undamaged (−). Panels A and B each show one gel exposed to X-ray film at −80°C for 12 h (lanes 1 to 3) or 72 h (lanes 4 to 13). F marks the migration of free DNA probe, and B1 and B2 mark the mobilities of protein-DNA complexes specific for UV damage. The protein-DNA complexes in lanes 2 and 6 are nonspecific, since they are observed for the undamaged DNA probes.
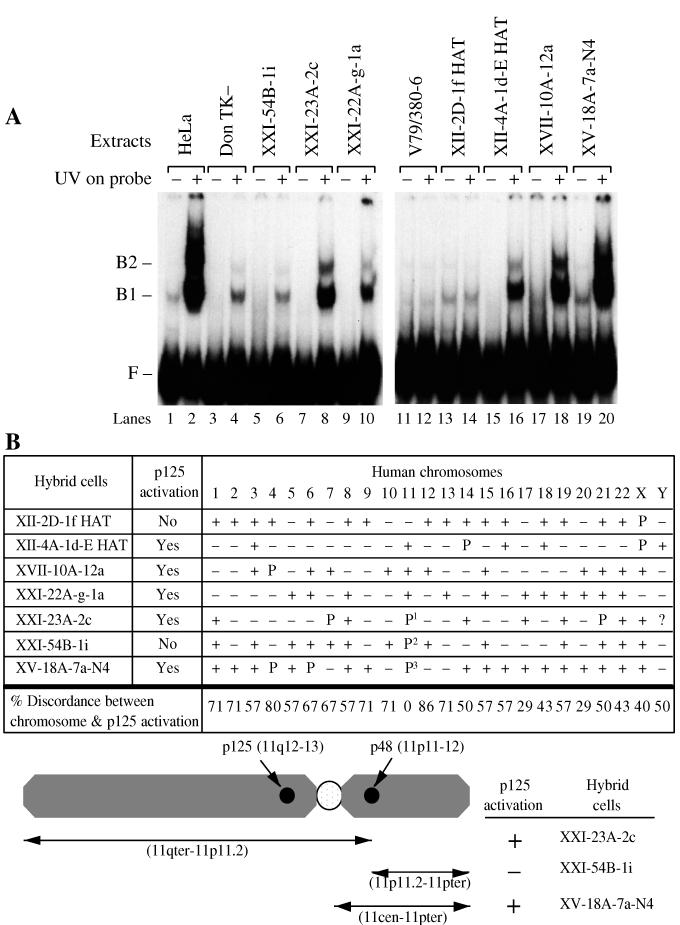
FIG. 4.
Human chromosome region 11p11.2-11cen activates p125 in hamster cells. (A) Binding activity. Extracts (2 μg) were assayed for p125 binding activity in three hybrid cell lines (lanes 5 to 10) derived from the DonTK− hamster line, which has a small amount of binding activity, and four hybrid cell lines (lanes 13 to 20) derived from the V79/380-6 hamster cell line, which does not have any detectable binding activity. (B) Human chromosomes contained in the hybrid cell lines. +, chromosome is present in at least 10% of the cells; P, part of the chromosome is present; P1, 11p11.2-11qter is present; P2, the reciprocal chromosome containing 11pter-11p11.2, is present; P3, 11pter-11cen is present. Discordance analysis was performed for the presence of each chromosome and activation of p125. The analysis included complete human chromosomes but not parts of chromosomes. Discordance was defined as either the absence of p125 activation in the presence of the chromosome or the presence of p125 activation in the absence of the chromosome. The percent discordance was calculated by dividing the number of discordant hybrids by the number of informative hybrids and multiplying by 100. F, free DNA probe migration; B1 and B2, UV damage-specific protein-DNA complex mobilities.
UV-DDB activity in human and monkey cells copurifies with a single 125-kDa polypeptide (p125), encoded by a cDNA containing an open reading frame of the expected size (12, 20, 41). Expression of p125 cDNA in XP group E cells was problematic, since the only available cells are primary fibroblasts or lymphoblastoid cells, which we have been unable to transfect efficiently by either calcium phosphate coprecipitation or electroporation. To circumvent this problem, we attempted to exploit our discovery that most hamster cell lines lack binding activity. Nevertheless, transfection of several hamster cell lines with any of several different expression vectors for p125 cDNA failed to confer binding activity (see Fig. 7A; other data not shown).
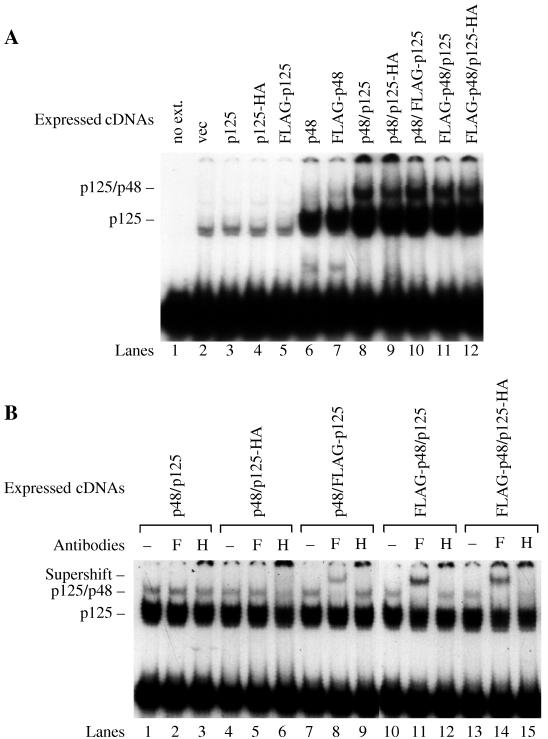
FIG. 7.
p125-p48 complex binds to UV-damaged DNA. (A) Coexpression of p125 with p48 increases the levels of both complexes. Expression vectors for p125 (p125, p125-HA, or FLAG-p125) and p48 (p48 or FLAG-p48) were transfected into 293T cell lines. Whole-cell extracts from the transfectants (2 μg) were assayed for damaged-DNA-specific binding activity with DNA probes damaged at a low dose (300 J/m2) of UV. (B) Whole-cell extracts (2 μg) were preincubated with anti-FLAG (F) or anti-HA (H) antibodies (0.02 μg/ml) at room temperature for 20 min before initiation of the binding reaction by addition of DNA probe damaged at the same dose of UV.
To determine whether hamster cells are capable of activating a latent binding activity in p125, three cell lines were treated with azacytidine, a demethylating agent that can induce the expression of alleles otherwise silenced by methylation. Azacytidine induced significant levels of binding activity, which was detected as protein-DNA complexes at positions B1 and B2 in an electrophoretic mobility shift assay (Fig. 1). These complexes were specific for UV-damaged DNA, migrated with precisely the same mobility as the binding activity in human cells (Fig. 1), and showed the same strong preference for UV-damaged DNA over undamaged DNA (Fig. 2).
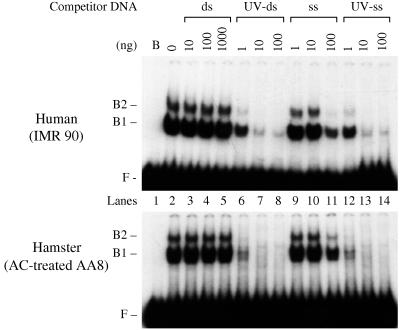
FIG. 2.
Human and hamster cells each contain a factor with the same high affinity for UV-damaged DNA. Whole-cell extracts from IMR90 cells (0.5 μg) and azacytidine (AC)-treated AA8 cells (2 μg) were incubated with DNA probe damaged with UV at 5,000 J/m2, unlabeled salmon sperm, and poly(dI-dC) competitor DNA, plus additional unlabeled competitor DNA in the form of pUC18 linearized by digestion with HindIII. The pUC18 DNA was added as intact double-stranded DNA (ds), UV-irradiated double-stranded DNA (UV-ds), single-stranded DNA (ss), or UV-irradiated single-stranded DNA (UV-ss). Single-stranded DNA was prepared by heating at 95°C for 5 min followed by rapid cooling. The lanes marked B and 0 correspond to control reactions with extract omitted and with extract present but no pUC18 competitor DNA, respectively. F, free DNA probe migration; B1 and B2, UV damage-specific protein-DNA complex mobilities.
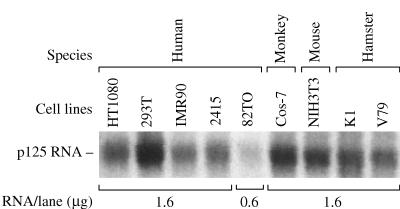
High-stringency Northern blotting with a human p125 cDNA probe demonstrated that human and hamster cells express mRNAs of identical size (Fig. 3). Thus, hamster cells express a gene highly homologous to the human p125 gene, and azacytidine treatment activated a factor with the same biochemical characteristics as human p125 protein. We conclude that this factor was the hamster homolog of human p125. Finally, hamster p125 mRNA levels did not change with azacytidine treatment (data not shown), indicating that activation of p125 was mediated by induction of a gene other than p125.
FIG. 3.
p125 RNA message is present in hamster and XPE− cells. Northern blotting was carried out with total RNA from human, monkey, mouse, and hamster cell lines. Each lane contained 1.6 μg of total RNA except lane 82TO, which contained 0.6 μg of total RNA.
Human chromosome 11p11.2-11cen activates p125 in hamster cells.
To identify this activating gene, binding activity was measured in several hamster-human hybrid cell lines (13, 14, 28, 31) (Fig. 4A). Binding activity was expressed only in hybrids containing human chromosome 11, while all other chromosomes were excluded based on discordance of p125 activation in hybrids (Fig. 4B).
The activating gene was mapped more precisely in three hybrids containing fragments of chromosome 11, constraining the gene to the region 11p11.2-11cen. This region excluded the p125 gene (11q12-q13), but included the p48 gene (11p12-p11) (12), which encodes a polypeptide originally identified by copurification with p125 (25).
It remains possible that p125 is activated by either of two factors, one factor encoded in the region 11p11.2-11cen and a second factor encoded outside this region. However, this was ruled out for every chromosome except chromosomes 17, 20, Y, and 11q, since those regions were absent in the two hybrids that lacked binding activity.
p125 is activated by transfection of p48 cDNA.
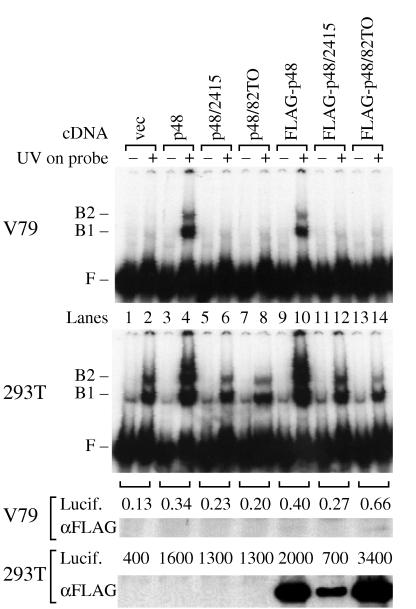
The levels of p48 mRNA in the hamster cells were too low to be detected by Northern blotting either before or after azacytidine treatment (data not shown). Therefore, to directly test the possibility that expression of p48 activates p125, human p48 cDNA and FLAG-p48 cDNA were constructed by PCR amplification from a human Jurkat cell cDNA library and transiently transfected into hamster (V79) and human (293T) cells (Fig. 5). Both cDNA constructs conferred binding activity on V79 cells, demonstrating that p48 is required for p125 activation in hamster cells. Furthermore, both cDNA constructs increased binding activity in 293T cells, demonstrating that p48 is limiting for binding activity in human cells (Fig. 5; compare lane 2 to lanes 4 and 10). Note that the greatly increased binding activity in the 293T transfectants was accompanied by the appearance of additional bands above position B2. These correspond, at least in part, to multiple binding events of p125 to different lesions on the same molecule of probe DNA, a phenomenon previously characterized in experiments with purified preparations of p125 (21).
FIG. 5.
p125 is activated by wild-type p48, but not by p48 from XPE− cells. Wild-type p48 cDNAs (p48 and FLAG-p48) and p48 cDNAs encoding amino acid changes found in XPE− cells (2415 and 82TO) were cloned into an expression vector and transfected into V79 (hamster) and 293T (human) cells. The expression vector in lanes vec contained no cDNA. After transfection, cell extracts (2 μg) were assayed for UV-specific binding activity with DNA probe damaged with UV at 5,000 J/m2 (two upper panels). A protein-DNA complex in 293T cells migrates to slightly below B1 but is present whether or not the probe DNA is damaged with UV. Transfection efficiency was measured by analyzing the activity from a cotransfected luciferase reporter gene (102/μg). Levels of p48 protein were measured by immunoblotting with anti-FLAG antibody (2 μg/ml) after SDS-PAGE of V79 (50 μg) and 293T (10 μg) extracts. F, free DNA probe migration; B1 and B2, UV damage-specific protein-DNA complex mobilities.
p48 is inactivated by mutations in XPE− cells.
The p48 cDNAs from two XPE− cell lines each encode a single amino acid substitution (R273H in XP2415 cells and K244E in XP82TO cells) (30). To rule out the possibility that these amino acid substitutions are merely innocent polymorphisms, cDNA expression vectors were made by RT-PCR amplification of mRNA harvested from XP2415 and XP82TO cells. The sequencing of these cDNAs confirmed the nucleotide differences previously reported (30).
When the altered cDNAs were tested by transfection of hamster and human cells, either amino acid substitution inactivated p48 (Fig. 5, lanes 6, 8, 12, and 14). Successful transfection of the cDNA was verified by expression of a cotransfected luciferase gene. The extremely high level of luciferase expression in 293T cells was due to the high transfection efficiency of these cells and the expression of simian virus 40 T antigen, which drove the replication of the episomal pBJ5 vector to a high copy number. Successful expression of FLAG-p48 in 293T cells was confirmed by immunoblotting with anti-FLAG antibodies (Fig. 5), demonstrating that the p48 genes from the two XPE− cell lines were expressed but nonfunctional.
Failure to detect FLAG-p48 in the transiently transfected V79 cells was consistent with the 2,500-fold- to 6,500-fold-lower level of expression of the cotransfected luciferase reporter gene in those cells. V79 cells can indeed support expression of FLAG-p48 protein, since the protein was detected by immunoblotting in two independent V79 clonal cell lines stably transformed with the FLAG-p48 expression vector (data not shown). Thus, even the low levels of FLAG-p48 in transiently transfected V79 cells were sufficient to activate detectable levels of binding activity.
p48 forms a complex with p125 on damaged DNA.
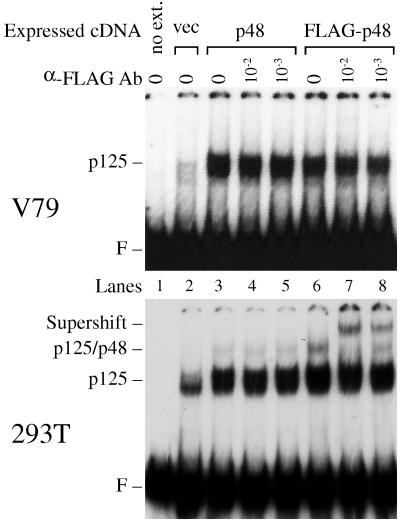
Protein–DNA complexes dependent on p48 and p125 were examined further by using transfection experiments and anti-FLAG antibodies. Extracts (2 μg) from the 293T cells produced at least three shifted bands, and extracts (2 μg) from V79 cells produced two shifted bands (Fig. 5), corresponding to multiple binding events to each molecule of DNA (21). To simplify the analysis, f148 DNA was UV irradiated with 300 J/m2, a dose that produced one or no lesions on each DNA molecule (21). In V79 extracts, only one protein-DNA complex appeared, and this complex was greatly increased after transient transfections with p48 or FLAG-p48 expression vectors (Fig. 6). On the other hand, when 293T cells were transfected with p48 or FLAG-p48 expression vectors, a second protein-DNA complex was observed (Fig. 6), consistent with the formation of a larger complex. Furthermore, addition of anti-FLAG antibody specifically supershifted the larger complex to a new position higher in the gel (Fig. 6), demonstrating that this complex contained p48. By contrast, the major protein-DNA complex, which migrated at a position lower in the gel, was unaffected.
FIG. 6.
p48 can be contained in a complex with damaged DNA. V79 cells or 293T cells were transfected with vectors expressing p48 or FLAG-p48. Binding activity was measured with DNA probe damaged at a low dose (300 J/m2) of UV. Extracts (2 μg) from the V79 cells or 293T cells were preincubated with different concentrations of anti-FLAG antibodies (Ab) (10−2 and 10−3 dilutions of stock solution [2 μg/ml]) before initiation of the binding reaction by addition of UV-damaged DNA probe. The upper (V79) and lower (293T) gels were exposed to X-ray film at −80°C for 100 and 16 h, respectively. For the longer exposure, a protein-DNA complex appears in lane 2 of V79 extracts, but this complex was nonspecific, since it was also observed for an undamaged DNA probe (Fig. 1 and 4). F, free DNA probe migration.
To further demonstrate the existence of two distinct complexes, 293T cells were transfected with expression vectors for p125 (p125, p125-HA, and FLAG-p125) and/or p48 (p48, and FLAG-p48). Binding activity did not increase after transfection with p125 vector alone (Fig. 7A, lanes 3 to 5) but did increase after transfection with p48 vector alone (Fig. 7A, lanes 6 and 7). When p48 and p125 vectors were transfected together, both complexes were further increased (Fig. 7A, lanes 8 to 12). Thus, both complexes are dependent on p125 expression, provided that p48 is coexpressed so as not to be rate limiting for binding activity.
When either p48 or FLAG-p48 vector was cotransfected with p125-HA vector, preincubation of protein extracts with anti-HA antibodies inhibited the formation of the upper protein-DNA complex (Fig. 7B, lanes 6 and 15). When p48 and FLAG-p125, FLAG-p48 and p125, or FLAG-p48 and p125-HA were expressed, the addition of anti-FLAG antibodies during the binding assay specifically supershifted most of the upper complex (Fig. 7B, lanes 8, 11, and 14). Thus, the upper complex contained both p125 and p48.
The lower complex must contain p125, due to several lines of evidence. More of the complex is expressed after p125 and p48 cotransfection than after p48 transfection (Fig. 7A). Purification based on activity in the lower complex yields the p125 polypeptide (2, 21). Finally, antibodies against p125 will react with the complex (41). On the other hand, the lower complex is very likely devoid of p48: purified p125 devoid of p48 forms a protein-DNA complex that migrates to exactly the same position in the gel (21), and anti-FLAG antibodies directed against FLAG-p48 fail to supershift the lower complex while successfully supershifting the upper complex.
It is noteworthy that the lower complex failed to react with anti-FLAG and anti-HA antibodies after transfection with FLAG-p125 or p125-HA expression vectors (Fig. 7B, lanes 6, 8, and 15). This may have occurred because the lower complex consisted mostly of p125 endogenously expressed by the 293T cells (Fig. 7A, compare lanes 6 and 7 to lanes 8 to 12), making it difficult to detect an antibody interaction with the fraction of the lower complex containing the tagged p125 molecules. Alternatively, the p125-DNA complex in the absence of p48 might fold into a conformation in which the N and C termini of p125 are inaccessible to the antibodies, while the p125–p48-DNA complex folds into a different conformation in which the N and C termini are accessible (Fig. 7B, lanes 6, 8, and 15).
p48 forms a complex with p125 in solution.
To obtain further evidence for an interaction between p48 and p125 in vivo, 293T cells were cotransfected with expression vectors for FLAG-p48 and p125-HA (Fig. 8). Extracts from the transfected cells were then incubated with anti-FLAG or anti-HA antibodies to immunoprecipitate FLAG-p48 or p125-HA and interacting proteins. When the immunoprecipitated proteins were resolved by SDS-PAGE and blotted with anti-FLAG and anti-HA antibodies, both FLAG-p48 and p125-HA were detected when either of the antibodies was used for immunoprecipitation (Fig. 8). Thus, even in crude extracts and in the absence of DNA substrate, p48 will interact with p125.
FIG. 8.
p125 interacts with p48 in free solution. Whole-cell extracts from 293T cells transfected with p125 (p125 or p125-HA) and FLAG-p48 were immunoprecipitated (IP) with anti-FLAG or anti-HA antibodies (Abs). The immunoprecipitated proteins (lanes 1 to 4) and 10 μg of whole-cell extract (lanes 5 to 7) were resolved by SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was stained with Ponceau S to visualize the molecular weight markers and then cut into the two pieces. The top half, which contained proteins larger than 65 kDa, was immunoblotted (IB) with anti-HA antibodies, and the bottom half was immunoblotted with anti-FLAG antibodies.
DISCUSSION
p48 is required for the activation of p125.
Purification of UV-DDB activity to homogeneity yields a single 125-kDa polypeptide (p125) (2, 21, 25). However, two lines of evidence indicate that p125 must be activated by a second factor before it can bind to UV-damaged DNA. First, only a small fraction of p125 expressed in vitro has binding activity (20). Second, we discovered that although Chinese hamster cell lines express p125 mRNA, the p125 protein is completely or largely inactive.
Significantly, the hamster cells were fully competent to express active p125, since treatment with azacytidine induced binding activity. Therefore, a panel of hamster-human somatic cell hybrids was tested to identify the human chromosomal fragment that would activate p125. The somatic cell hybrids identified an activating region at 11p11.2-11cen. Surprisingly, this region included the p48 gene, which encodes a polypeptide that copurifies with p125 under some conditions but is not required for binding activity in vitro, since fractions containing p125 but not p48 remain active for binding (2, 21, 25).
We proved that p48 is required for the activation of p125 in vivo, since the transfection of hamster cells with a p48 cDNA expression vector induced damage-specific binding activity. Furthermore, the transfection of human cells with p48 cDNA led to a marked increase in binding activity, demonstrating that p48 is limiting for binding activity in human cells.
How does p48 activate p125? Significantly, p48 is not contained in the complexes of activated p125 with UV-damaged DNA in transfected hamster cells, and it is contained only in a minor component of the binding activity in transfected 293T cells (Fig. 6 and 7). It appears that p48 associates with p125, mediates p125 activation, and then dissociates from the activated p125 by a “hit-and-run” mechanism. Thus, p48 may mediate posttranslational modification of p125, either by acting directly on p125 or by acting indirectly by presenting p125 for modification by a third protein.
Although most of the binding activity in extracts does not contain p48 and is due to a “hit-and-run” activation of p125, we have demonstrated that a small fraction of the binding activity is due to a p48-p125 complex bound to UV-damaged DNA. This complex is almost certainly due to a cooperative interaction between p48 and p125 rather than to separate binding events, since we have demonstrated that p48 and p125 will interact with each other in an unbound state, and since the addition of purified p48 will alter the characteristics of the DNA-binding footprint of purified p125 without changing its size (38). Nevertheless, the physiological significance of the p48-p125-DNA complex in vivo remains to be determined.
Roles of p48 and p125 in nucleotide excision repair.
What are the roles of p48 and p125 in DNA repair? In vitro, the p125 protein binds to UV-damaged dinucleotides with a 500,000-to-1 preference over undamaged dinucleotides (21) but confers only a twofold increase in the repair of naked DNA reconstituted from purified proteins (1). In vivo, hamster cells differ from human cells not only by failing to express p48 but also by failing to efficiently repair UV-induced cyclobutane pyrimidine dimers in nontranscribed DNA (3, 16). To explain these observations, we hypothesize that p48, perhaps in association with p125, plays a role in disassembly of nontranscribed chromatin at the sites of cyclobutane pyrimidine dimers.
Results of a database search are at least consistent with this hypothesis: p48 contains a WD motif with 24 to 49% identity to the WD motifs in a subfamily of WD repeat proteins involved in the reorganization of chromatin (Fig. 9). Included in this subfamily are subunits of chromatin assembly factors (CAFs) for humans (CAF-1 p48) (44), Drosophila melanogaster (CAF-1 p55) (43), and Saccharomyces cerevisiae (Msi1p) (23), as well as yeast Hat2p, the histone recognition subunit of a histone H4 acetyltransferase (32). Notably, human CAF-1 is required for reassembly of chromatin coupled to nucleotide excision repair (15), and disruption of yeast Msi1p confers hypersensitivity to UV radiation (23).
FIG. 9.
p48 contains a WD motif homologous to the WD motif in a subfamily of proteins involved in chromatin reorganization. Homologous genes were identified by BLAST search and aligned by the BOXSHADE program. Identical amino acids are highlighted in black. Functionally conserved amino acids are highlighted in gray. Conserved amino acids are classified as follows: V, I, L, and M; D, E, Q, and N; F, Y, and W; G, S, T, P, and A; and K, R, and H. The positions of the mutated amino acids of p48 in XPE− cells are marked as # (K-to-E change in 82TO) and ∗ (R-to-H change in 2415). The proteins are as follows: CAF-1 p48, chromatin assembly factor p48 (EMBL X74262) (44); RbA p46, retinoblastoma-associated protein (EMBL X72841) (34); CAF-1 p55 (GenBank U62388) (43); K07A1.12 and K07A1.11, identified by Caenorhabditis elegans genome sequencing (EMBL Z81097); XPE p48 (GenBank U18300) (12); CSA, Cockayne syndrome A protein (GenBank U28413) (18); YDL156w, identified by S. cerevisiae genome sequencing (EMBL Z74204); Msi1p, identified as a negative regulator of the Ras-cyclic AMP pathway (GenBank M27300) (39); and Hat2p, histone acetyltransferase (GenBank U18795) (32). The position of the first amino acid (a.a.) of each WD motif is shown to the left of the amino acid sequence, and the percent identity to p48, starting at the conserved His241, is shown to the right.
The hypothesized role for p48 and/or p125 in chromatin disassembly does not rule out other roles for p125 in DNA repair, since purified p125 enhances twofold the repair of naked UV-irradiated DNA in a reconstituted nucleotide excision repair system (1). Although human and hamster cells repair cyclobutane pyrimidine dimers on the transcribed DNA strand with equal efficiency, hamster cells are significantly less efficient in repairing the nontranscribed strand (29). This difference between human and hamster cells may be explained if p125 plays a role in recognizing UV damage on the nontranscribed strand even in the absence of chromatin while RNA polymerase II plays that role for the transcribed strand.
In addition to cyclobutane pyrimidine dimers, p125 also recognizes 6-4 photoproducts (42). Furthermore, extracts from XPE+ and XPE− cells were severely defective in excising 6-4 photoproducts from naked DNA (24). Surprisingly, both XPE+ and XPE− extracts were rescued by purified human RPA (the single-stranded DNA binding protein, replication protein A [RPA]) but not by purified p125, even though no mutations were found in the RPA gene (24). A possible explanation for this puzzling result is that 6-4 photoproducts are targeted for excision repair by two different mechanisms, one involving RPA and one involving p125. RPA will bind directly to 6-4 photoproducts (5) and can interact cooperatively with XPA protein in binding to damaged DNA in vitro (17). Perhaps these properties are sufficient to repair 6-4 photoproducts when RPA is added to biochemical excess in a cell-free system. On the other hand, p125 may require further activation beyond its DNA binding activity in order to mediate the repair of 6-4 photoproducts, and this activation did not occur in the XPE extracts.
p48 mutations in XPE− cells.
The p125 cDNA sequences are identical in HeLa cells, three XPE+ cell lines, and two XPE− cell lines (30). On the other hand, the p48 cDNA sequences in the two XPE− cell lines encode single amino acid substitutions (R273H in XP2415 cells and K244E in XP82TO cells) (30), both of which are located in the WD motif. These substitutions may not have had functional consequences, since the analysis included only four cell lines with intact binding activity (HeLa cells and the three XPE+ cell lines). Furthermore, R273H replaces the highly conserved arginine in the WD motif with histidine, another basic amino acid (Fig. 9). K244E replaces a lysine shared by another member of the WD motif subfamily with a glutamate conserved in six other members. In the absence of functional studies, these subtle substitutions might very well have proven to be innocent polymorphisms. This study demonstrates that both amino acid substitutions in p48 abolish the activation of p125, proving that they are indeed inactivating mutations rather than polymorphisms. Thus, XPE− cells have mutations in the p48 gene. On the other hand, XPE+ cells do not appear to have mutations in either p48 or p125 and therefore have mutations at another locus that remains to be identified.
Because the p48 mutations are subtle, they may preserve residual function in p48, perhaps explaining why XPE patients have mild disease. Null mutations might produce a far more severe phenotype, and p48 and/or p125 may prove to be more important for nucleotide excision repair than previously believed.
ACKNOWLEDGMENTS
This work was supported by grant DAMD 17-94-J-4350 from the U.S. Army Medical Research and Material Command and funds from Graham and Jane Nissen (to G.C.) and the Howard Hughes Medical Institute (to U.F.).
We thank Vaughn Smider, Virginia Goss, Ola Hammarsten, and Jean Tang for helpful discussions and Yun Kee for helping with sequence analysis.
REFERENCES
- 1.Aboussekhra A, Biggerstaff M, Shivji M, Vilpo J, Moncollin V, Podust V, Protic M, Hubscher U, Egly J, Wood R. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 2.Abramic M, Levine A, Protic M. Purification of an ultraviolet-inducible, damage-specific DNA-binding protein from primate cells. J Biol Chem. 1991;266:22493–22500. [PubMed] [Google Scholar]
- 3.Bohr V, Smith C, Okumoto D, Hanawalt P. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Burns J L, Guzder S N, Sung P, Prakash S, Prakash L. An affinity of human replication protein A for ultraviolet-damaged DNA. J Biol Chem. 1996;271:11607–11610. doi: 10.1074/jbc.271.20.11607. [DOI] [PubMed] [Google Scholar]
- 6.Chu G, Chang E. Cisplatin-resistant cells express increased levels of a factor that recognizes damaged DNA. Proc Natl Acad Sci USA. 1990;87:3324–3327. doi: 10.1073/pnas.87.9.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu G, Chang E. Xeroderma pigmentosum group E cells lack a nuclear factor that binds to damaged DNA. Science. 1988;242:564–567. doi: 10.1126/science.3175673. [DOI] [PubMed] [Google Scholar]
- 8.Chu G, Mayne L. Xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy: do the genes explain the disease? Trends Genet. 1996;12:187–192. doi: 10.1016/0168-9525(96)10021-4. [DOI] [PubMed] [Google Scholar]
- 9.Chu G, Sharp P A. SV40 DNA transfection of cells in suspension: analysis of the efficiency of transcription and translation of T antigen. Gene. 1981;13:197–202. doi: 10.1016/0378-1119(81)90008-1. [DOI] [PubMed] [Google Scholar]
- 10.Cleaver J E. Defective repair replication of DNA in xeroderma pigmentosum. Nature. 1968;218:652–656. doi: 10.1038/218652a0. [DOI] [PubMed] [Google Scholar]
- 11.de Wet J R, Wood K V, DeLuca M, Helinski D R, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dualan R, Brody T, Keeney S, Nichols A, Admon A, Linn S. Chromosomal localization and cDNA cloning of the genes (DDB1 and DDB2) for the p127 and p48 subunits of a human damage-specific DNA binding protein. Genomics. 1995;29:62–69. doi: 10.1006/geno.1995.1215. [DOI] [PubMed] [Google Scholar]
- 13.Francke U, Busby N, Shaw D, Hansen S, Brown M G. Intrachromosomal gene mapping in man: assignment of nucleoside phosphorylase to region 14cen-14q21 by interspecific hybridization of cells with a t(X;14) (p22;q21) translocation. Somatic Cell Genet. 1976;2:27–40. doi: 10.1007/BF01539240. [DOI] [PubMed] [Google Scholar]
- 14.Francke U, Francke B. Requirement of the human chromosome 11 long arm for replication of herpes simplex virus type 1 in nonpermissive Chinese hamster × human diploid fibroblast hybrids. Somatic Cell Genet. 1981;7:171–191. doi: 10.1007/BF01567656. [DOI] [PubMed] [Google Scholar]
- 15.Gaillard P-H L, Martini E M-D, Kaufman P D, Stillman B, Moustacchi E, Almouzni G. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor 1. Cell. 1996;86:887–896. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- 16.Hanawalt P. Transcription-coupled repair and human disease. Science. 1994;266:1957–1958. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- 17.He Z, Henricksen L, Wold M, Ingles C. RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature. 1995;374:566–569. doi: 10.1038/374566a0. [DOI] [PubMed] [Google Scholar]
- 18.Henning K, Li L, Iyer N, McDaniel L, Reagan M, Legerski R, Stefanini S R M, Lehmann A, Mayne L, Friedberg E. The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell. 1995;82:555–564. doi: 10.1016/0092-8674(95)90028-4. [DOI] [PubMed] [Google Scholar]
- 19.Hirschfeld S, Levine A S, Ozato K, Protíc M. A constitutive damage-specific DNA-binding protein is synthesized at higher levels in UV-irradiated primate cells. Mol Cell Biol. 1990;10:2041–2048. doi: 10.1128/mcb.10.5.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang B, Liao J, Chu G. Isolation of a cDNA encoding a UV-damaged DNA binding factor defective in xeroderma pigmentosum group E cells. Mutat Res. 1996;362:105–117. doi: 10.1016/0921-8777(95)00040-2. [DOI] [PubMed] [Google Scholar]
- 21.Hwang B J, Chu G. Purification and characterization of a protein that binds to damaged DNA. Biochemistry. 1993;32:1657–1666. doi: 10.1021/bi00057a033. [DOI] [PubMed] [Google Scholar]
- 22.Kataoka H, Fujiwara Y. UV damage-specific DNA-binding protein in xeroderma pigmentosum complementation group E. Biochem Biophys Res Commun. 1991;175:1139–1143. doi: 10.1016/0006-291x(91)91684-5. [DOI] [PubMed] [Google Scholar]
- 23.Kaufman P D, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-1. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 24.Kazantsev A, Mu D, Nichols A F, Zhao X, Linn S, Sancar A. Functional complementation of xeroderma pigmentosum complementation group E by replication protein A in an in vitro system. Proc Natl Acad Sci USA. 1996;93:5014–5018. doi: 10.1073/pnas.93.10.5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keeney S, Chang G J, Linn S. Characterization of human DNA damage binding protein implicated in xeroderma pigmentosum E. J Biol Chem. 1993;268:21293–21300. [PubMed] [Google Scholar]
- 26.Keeney S, Eker A, Brody T, Vermuelen W, Bootsma D, Hoeijmakers J, Linn S. Correction of the DNA repair defect in xeroderma pigmentosum group E by injection of a DNA damage-binding protein. Proc Natl Acad Sci USA. 1994;91:4053–4056. doi: 10.1073/pnas.91.9.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keeney S, Wein H, Linn S. Biochemical heterogeneity in xeroderma pigmentosum complementation group E. Mutat Res. 1992;273:49–56. doi: 10.1016/0921-8777(92)90049-9. [DOI] [PubMed] [Google Scholar]
- 28.Martinville B, Giacalone J, Shih C, Weinberg R A, Francke U. Oncogene from human EJ bladder carcinoma is located on the short arm of chromosome 11. Science. 1983;219:498–501. doi: 10.1126/science.6297001. [DOI] [PubMed] [Google Scholar]
- 29.Mellon I, Spivak G, Hanawalt P. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 30.Nichols A, Ong P, Linn S. Mutations specific to the xeroderma pigmentosum group E Ddb− phenotype. J Biol Chem. 1996;271:24317–24320. doi: 10.1074/jbc.271.40.24317. [DOI] [PubMed] [Google Scholar]
- 31.Oliver N, Francke U, Pellegrino M A. Regional assignment of genes for mannose phosphate isomerase, pyruvate kinase-3, and b2-microglobulin expression on human chromosome 15 by hybridization of cells from a t(15;22) (q14;q13.3) translocation carrier. Cytogenet Cell Genet. 1978;22:506–510. doi: 10.1159/000131009. [DOI] [PubMed] [Google Scholar]
- 32.Parthun M R, Widom J, Gottschling D E. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 33.Payne A, Chu G. Xeroderma pigmentosum group E binding factor recognizes a broad spectrum of DNA damage. Mutat Res. 1994;310:89–102. doi: 10.1016/0027-5107(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 34.Qian Y W, Lee E Y H P. Dual retinoblastoma-binding proteins with properties related to a negative regulator of Ras in yeast. J Biol Chem. 1995;270:25507–25513. doi: 10.1074/jbc.270.43.25507. [DOI] [PubMed] [Google Scholar]
- 35.Rathmell W K, Chu G. A DNA end-binding factor involved in double-strand break repair and V(D)J recombination. Mol Cell Biol. 1994;14:4741–4748. doi: 10.1128/mcb.14.7.4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rathmell W K, Chu G. Involvement of the Ku autoantigen in the cellular response to DNA double-strand breaks. Proc Natl Acad Sci USA. 1994;91:7623–7627. doi: 10.1073/pnas.91.16.7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rathmell W K, Kaufmann W K, Hurt J C, Byrd L L, Chu G. DNA-dependent protein kinase is not required for accumulation of p53 or cell cycle arrest after DNA damage. Cancer Res. 1997;57:68–74. [PubMed] [Google Scholar]
- 38.Reardon J T, Nichols A F, Keeney S, Smith C A, Taylor J S, Linn S, Sancar A. Comparative analysis of binding of human damaged DNA-binding protein (XPE) and Escherichia coli damage recognition protein (uvrA) to the major ultraviolet photoproducts: T[c,s]T, T[t,s]T, T[6-4]T, and T[Dewar]T. J Biol Chem. 1993;268:21301–21308. [PubMed] [Google Scholar]
- 39.Ruggieri R, Tanaka K, Nakafuku M, Kaziro Y, Toh-e A, Matsumoto K. MSI1, a negative regulator of the RAS-cAMP pathway in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1989;86:8778–8782. doi: 10.1073/pnas.86.22.8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smider V, Rathmell W K, Lieber M, Chu G. Restoration of X-ray resistance and V(D)J recombination in mutant cells by Ku cDNA. Science. 1994;266:288–291. doi: 10.1126/science.7939667. [DOI] [PubMed] [Google Scholar]
- 41.Takao M, Abramic M, Moos M, Otrin V, Wootton J, McLenigan M, Levine A, Protic M. A 127-kDa component of a UV-damaged DNA-binding complex, which is defective in some xeroderma pigmentosum group E patients, is homologous to a slime mold protein. Nucleic Acids Res. 1993;21:4111–4118. doi: 10.1093/nar/21.17.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Treiber D, Chen Z, Essigmann J. An ultraviolet light-damaged DNA recognition protein absent in xeroderma pigmentosum group E cells binds selectively to pyrimidine (6-4) pyrimidone photoproducts. Nucleic Acids Res. 1992;20:5805–5810. doi: 10.1093/nar/20.21.5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tyler J K, Bulger M, Kamakaka R T, Kobayashi R, Kadonaga J T. The p55 subunit of Drosophila chromatin assembly factor 1 is homologous to a histone deacetylase-associated protein. Mol Cell Biol. 1996;16:6149–6159. doi: 10.1128/mcb.16.11.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 45.Zhang X K, Hoffmann B, Tran B, Magnus P. Retinoid X receptor is an auxiliary protein for thyroid hormone and retinoic acid receptors. Nature. 1992;355:441–446. doi: 10.1038/355441a0. [DOI] [PubMed] [Google Scholar]