Abstract
The RFA1 gene encodes the large subunit of the yeast trimeric single-stranded DNA binding protein replication protein A (RPA), which is known to play a critical role in DNA replication. A Saccharomyces cerevisiae strain carrying the rfa1-44 allele displays a number of impaired recombination and repair phenotypes, all of which are suppressible by overexpression of RAD52. We demonstrate that a rad52 mutation is epistatic to the rfa1-44 mutation, placing RFA1 and RAD52 in the same genetic pathway. Furthermore, two-hybrid analysis indicates the existence of interactions between Rad52 and all three subunits of RPA. The nature of this Rad52-RPA interaction was further explored by using two different mutant alleles of rad52. Both mutations lie in the amino terminus of Rad52, a region previously defined as being responsible for its DNA binding ability (U. H. Mortenson, C. Beudixen, I. Sunjeuaric, and R. Rothstein, Proc. Natl. Acad. Sci. USA 93:10729–10734, 1996). The yeast two-hybrid system was used to monitor the protein-protein interactions of the mutant Rad52 proteins. Both of the mutant proteins are capable of self-interaction but are unable to interact with Rad51. The mutant proteins also lack the ability to interact with the large subunit of RPA, Rfa1. Interestingly, they retain their ability to interact with the medium-sized subunit, Rfa2. Given the location of the mutations in the DNA binding domain of Rad52, a model incorporating the role of DNA in the protein-protein interactions involved in the repair of DNA double-strand breaks is presented.
The yeast single-stranded DNA binding protein replication protein A (RPA) is a multisubunit complex containing three polypeptides of 70, 30, and 14 kDa. This heterotrimeric structure is conserved across all eukaryotic species where the protein is found. In Saccharomyces cerevisiae, deletion of any one of the three subunits is lethal (3), a reflection of the critical role the protein complex plays in DNA replication. RPA’s role in yeast is not limited to replication, however; it participates in repair and recombination as well. RPA’s involvement in yeast recombination is revealed in biochemical studies of Rad51, which show that RPA is required for the Rad51-catalyzed formation of both joint molecules and fully exchanged products from single-stranded circular DNA and linear double-stranded DNA with an overhanging complementary end (31, 38, 39).
Genetic analysis of yeast has underscored the importance of RPA in recombination, as mutations in the gene RFA1, encoding the large (70-kDa) subunit, affect recombination ability (8, 24, 37). One of these mutations, the rfa1-44 allele, results in a 390-fold reduction in a recombination assay that is based on the recombinational repair of HO-endonuclease-induced double-stranded breaks (DSBs) (8). In fact, the rfa1-44 mutant strain is approximately as deficient in its ability to repair a DSB—whether induced by the action of HO endonuclease or by exposure to X rays—as the rad55 and rad57 mutants tested (8, 13, 14). A further indication of the reduced recombinational activity of the rfa1-44 mutant is the 25-fold reduction in the sporulation efficiency and spore viability of the strain compared to those of the wild type (8). The rfa1-44 mutant shows moderate UV radiation sensitivity as well (8).
Interestingly, this mutation does not appear to have an effect on DNA replication, as the [3H]uracil incorporation and growth rates of the rfa1-44 mutant and wild-type strains are not substantially different (data not shown). Accordingly, we have previously suggested that the rfa1-44 mutation is a separation-of-function allele (8).
Earlier evidence stemming from the experiments on the recombination and repair defects of the rfa1-44 mutant indicated the possibility of an interaction between RFA1 and RAD52. First, the rfa1-44 mutant’s recombination and repair defects are suppressible by dose-dependent overexpression of RAD52 (8), indicative of a genetic interaction between RAD52 and RFA1. In addition, an allele-specific genetic interaction between rfa1-44 and one of the rad52 mutants in our collection was observed. Briefly, a diploid strain carrying both the rfa1-44 and rad52-34 mutations (as well as the corresponding wild-type copies of the genes) is only partially complemented with respect to X-ray survival (8). Diploids carrying both the rfa1-44 mutation and any of the other rad52 mutations in our collection show the expected full complementation (7, 8). Given these genetic interactions, we surmised that the RPA and Rad52 proteins might interact physically as well. Such a direct interaction between the mammalian forms of Rad52 and RPA has been demonstrated (32).
A number of proteins involved in yeast DSB repair appear to function as a complex (4, 14, 17, 18, 25, 27, 36) that has been dubbed a “recombinosome” (8). Relying on earlier successes in which interactions between Rad52 and Rad51 were detected by using the yeast two-hybrid system (6), we used the same approach to search for interactions between Rad52 and the individual subunits of RPA. Additionally, we used the same assay to examine interactions between two Rad52 mutant proteins, Rad52-34 and Rad52-38 (8), each of whose mutations reside in the region believed to be responsible for Rad52’s DNA binding activity (28). Our results indicate that these mutations affect the ability of Rad52 to interact with Rad51 and Rfa1 but do not affect the interaction with Rfa2. We suggest a model in which DNA binding plays a role in facilitating the protein interactions that define the putative recombinosome.
MATERIALS AND METHODS
Yeast strains and media.
Yeast strains used in this study are listed in Table 1. Strains YAF5 and YAF22 (8) were the respective sources of the rad52-34 and rad52-38 mutations. Cells were grown according to standard techniques (35), and YPD plates were routinely supplemented with 40 μg of adenine/ml.
TABLE 1.
Yeast strains used in this study
| Straina | Origin or source | Genotype |
|---|---|---|
| YME2b | 5 | MATa ade2-Δ1 his3-Δ187 leu2-Δ1 ura3-52 |
| YAF44 | 8 | rfa1-44; isogenic to YME2 |
| YAF5 | 8 | rad52-34; isogenic to YME2 |
| YAF22 | 8 | rad52-38; isogenic to YME2 |
| YAF100 | This study | rfa1-44 rad52-38; isogenic to YME2 |
| Y190 | 11 | MATa ade2-101 his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-901 tyr1-501 canR gal4Δ gal80Δ cyhS URA3::GAL-lacZ LYS2::GAL-HIS3 |
All strains are heterothallic.
YME2 is a derivative of strain S288C.
Plasmids.
Plasmids used for two-hybrid analysis in this study are listed in Table 2. Plasmid pCAD-1 (33) was used to create the carboxy-terminal fusions of Rfa1 and Rad52 to the amino-terminal end of the Gal4 activation domain. Plasmids pSLH128 and pSLH129 are rad52-34 and rad52-38 derivatives, respectively, of pSLH127 (14), which contains a Rad52-Gal4 DNA binding domain fusion. Plasmids pSLH217, pSLH218, and pSLH219 contain the corresponding Gal4 activation domain fusions. Other two-hybrid plasmids are described in reference 14. pAF50 is a YCplac33 (9)-derived plasmid carrying an EcoRI-SalI segment containing the RAD52 coding and promoter sequences (8). More detailed descriptions of plasmid constructs are available upon request.
TABLE 2.
Two-hybrid plasmids used in this study
| Plasmid | Source | Description |
|---|---|---|
| pAS1-CYH2 | 11 | Plasmid used for constructing fusions of proteins via their N termini to the DNA binding domain (DBD) of the Gal4 protein |
| pBG4D-1 | R. Brazas | Plasmid used for constructing fusions of proteins via their C termini to the DBD of the Gal4 protein |
| pGAD-GH | G. Hannon | Plasmid used for constructing fusions of proteins via their N termini to the transcriptional activation domain (TAD) of the Gal4 protein |
| pCAD-1 | 33 | Plasmid used for constructing fusions of proteins via their C termini to the TAD of the Gal4 protein |
| pSLH99 | 14 | GAL4-RAD51 TAD fusion expression vector made with pGAD-GH |
| pSLH127 | 14 | RAD52-GAL4 DBD fusion expression vector made with pBG4D |
| pSLH128 | This study | rad52-34–GAL4 DBD fusion expression vector made with pBG4D |
| pSLH129 | This study | rad52-38–GAL4 DBD fusion expression vector made with pBG4D |
| pSLH217 | This study | RAD52-GAL4 TAD fusion expression vector made with pCAD-1 |
| pSLH218 | This study | rad52-34–GAL4 TAD fusion expression vector made with pCAD-1 |
| pSLH219 | This study | rad52-38–GAL4 TAD fusion expression vector made with pCAD-1 |
| pSLH106 | 14 | GAL4-RAD55 TAD fusion expression vector made with pGAD-GH |
| pSLH108 | 14 | GAL4-RAD57 TAD fusion expression vector made with pGAD-GH |
| pSLH223 | This study | RFA1-GAL4 DBD fusion expression vector made with pBG4D-1 |
| pRB3 | This study | RFA2-GAL4 DBD fusion expression vector made with pBG4D-1 |
| pRB5 | This study | RFA3-GAL4 DBD fusion expression vector made with pBG4D-1 |
Cloning of the rad52-34 and rad52-38 alleles.
Genomic DNA from strains YAF5 (rad52-34) and YAF22 (rad52-38) was digested with EcoRI and SalI restriction endonucleases, which cut in the sequences flanking the RAD52 coding and regulatory regions. The DNA from these reactions was subsequently digested with XbaI, XhoI, EcoRV, HindIII, StuI, and PvuI endonucleases, none of which cuts within the RAD52 EcoRI-SalI fragment. The digested DNA was electrophoresed on a 0.8% agarose gel; bands ranging from 3.0 to 3.4 kbp of DNA, the approximate size of the RAD52 EcoRI-SalI fragment, were excised, and the DNA was isolated from these gel fragments. This DNA was then ligated to EcoRI-SalI-restricted YCplac111 (9) and transformed into Escherichia coli DH5α. Colonies containing a plasmid bearing the ∼3.2-kbp RAD52 fragment were identified by using a colony lift assay (34); hybridization with a 32P-labeled RAD52 probe (EcoRI-SalI fragment) was employed to detect plasmids containing RAD52 coding sequences. Plasmid DNA from positively hybridizing colonies was prepared and subjected to restriction analysis to validate the presence of the RAD52 coding sequence. Plasmids yielding appropriate restriction patterns were used in domain swap experiments (see below) and for creating the mutant rad52 two-hybrid fusions.
Determination of the location of the rad52-34 and rad52-38 mutations.
DNA from the plasmids described above, which carry the EcoRI-SalI fragment corresponding to RAD52 DNA, were digested with AgeI and BamHI, or BamHI and MluI, endonucleases, effectively dividing the mutant rad52 coding sequence into two fragments. These fragments were used to replace the same fragments on pAF50, resulting in a swap between DNA from the mutant alleles and the wild-type allele carried on pAF50. These plasmids were used to transform rad52 mutant strains to determine whether complementation of the mutant’s X-ray sensitivity occurred. Failure of a plasmid to complement the rad52 mutant was taken to indicate that the mutation resided in the “swapped” region. Regions believed to contain a mutation were transferred to pBluescript (Stratagene) for DNA sequencing (done at the Protein and Nucleic Acid Facility, Center for Molecular and Genetic Medicine, Stanford University, Stanford, Calif.).
Two-hybrid analysis.
Two-hybrid analysis was performed as previously described (14).
RESULTS
RFA1 is a member of the RAD52 epistasis group.
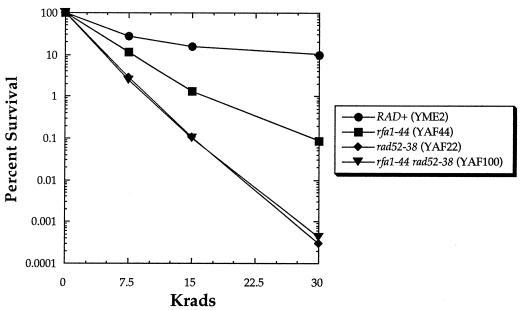
To determine if RFA1 is a new member of the RAD52 epistasis group, classical genetic epistasis analysis was performed by monitoring X-ray survival in strains with the rfa1-44 mutation, the rad52-38 mutation, or both. Figure 1 shows the rfa1-44 mutant’s moderate sensitivity to X rays and the rad52-38 mutant’s substantially greater sensitivity. The double mutant is no more sensitive to X rays than the rad52 single mutant, indicating that the rad52 and rfa1 mutations affect the same pathway.
FIG. 1.
Results of X-ray survival studies of strains bearing either the rfa1-44 mutation, the rad52-38 mutation, or both.
Rad52 and Rfa1 proteins interact in vivo.
Given the genetic evidence indicating an interaction between RFA1 and RAD52 (8), it seemed likely that the Rad52 and Rfa1 proteins could be interacting physically as well. Nevertheless, initial two-hybrid analysis failed to detect such an interaction (12, 17). Because Rfa1 might interact with Rad52 via its relatively nonconserved amino terminus, we suspected that fusion of the Gal4 DNA binding domain to the amino-terminal end of Rfa1 might be blocking its ability to interact with Rad52 and possibly other proteins.
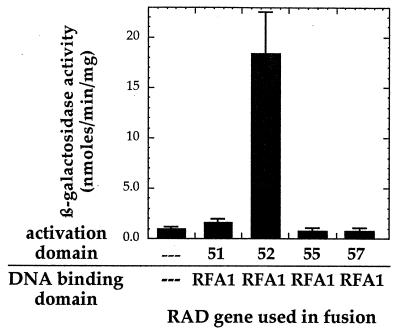
Accordingly, a new Rfa1 fusion construct was prepared in which the Gal4 DNA binding domain was fused to the carboxy terminus of Rfa1, freeing the amino terminus for possible protein-protein interactions. The two-hybrid assay with this new construct indicates that Rfa1 interacts with Rad52 but not with Rad51, Rad55, or Rad57 (Fig. 2), other members of the RAD52 epistasis group known to interact with each other (14, 18). Others have also reported findings regarding the influence of the nature of the fusion constructs on the ability of proteins to interact (33). While freeing the amino terminus may account for the detection of the Rfa1-Rad52 interaction, we cannot discount the possibility that the new construct is active in the two-hybrid assay simply because it is expressed in greater amounts, is more stable, or is folded differently than the amino-terminal fusion.
FIG. 2.
Results of two-hybrid study of Rfa1 and either Rad52, Rad51, Rad55, or Rad57. The error bars represent the standard deviations for at least three repetitions.
Rad52 also interacts with the medium and small subunits of RPA.
Because Rfa1 is part of a heterotrimeric protein complex, the two-hybrid assay allowed us to determine if Rad52 interacts with the other two subunits of RPA: Rfa2 and Rfa3. Appropriate Gal4 fusions to Rfa2 and Rfa3 were constructed and assayed in the two-hybrid system for their ability to interact with one another, as well as with Rfa1. As expected, the members of this complex appear to interact with each other in this assay (data not shown).
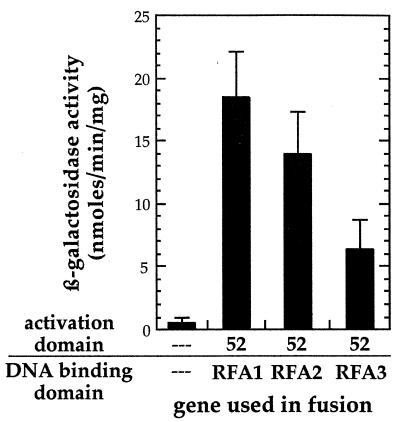
Pairing the Rfa2 and Rfa3 fusions with the Gal4 DNA binding domain in combination with Rad52 fused to the Gal4 transactivation domain indicates that Rad52 interacts not only with the large subunit of RPA, Rfa1, but also with the medium and small subunits, Rfa2 and Rfa3, respectively (Fig. 3).
FIG. 3.
Results of two-hybrid analysis to detect Rad52 interactions with Rfa1, Rfa2, and Rfa3. The error bars represent the standard deviations for at least three repetitions.
In principle, the two-hybrid assay does not allow the conclusion that Rad52 and RPA interact directly; they might appear to do so because of a shared interaction with a common substrate, such as DNA. Both Rad52 (28) and RPA bind single-stranded DNA, the latter through Rfa1 (19, 20, 40). However, the two-hybrid analysis with the rad52-34 and rad52-38 mutants (see below), each of which has a mutation in the well-conserved DNA binding domain of Rad52 (28), appears to rule out this possibility.
Isolation of the rad52-34 and rad52-38 alleles.
The rad52-34 allele, which exhibits the previously described genetic interaction with the rfa1-44 allele (8), was cloned to determine whether this mutant’s ability to interact with other members of the RAD52 epistasis group was affected. A second rad52 mutant, rad52-38, chosen at random from our collection, was examined in parallel. DNA fragments containing the mutant alleles were retrieved from the genome of yeast bearing the mutation, and domain swaps of the mutant alleles were performed in order to locate the region of the coding sequence in which these mutations were located (see Materials and Methods). DNA sequencing of these regions revealed that the mutations result in single base changes leading to single amino acid substitutions. The rad52-34 mutation creates a glycine-to-glutamate change at residue 121; a change from glycine to aspartate at residue 142 constitutes the rad52-38 mutation.
Interactions between the mutant and wild-type forms of Rad52.
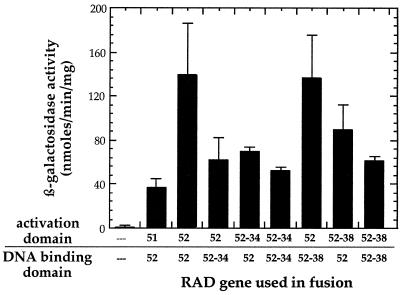
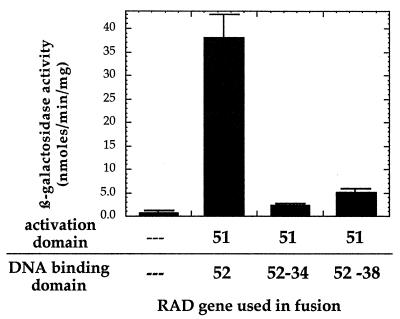
Rad52 is reported to be involved in homotypic interactions (2, 4). We used the yeast two-hybrid system to determine if the Rad52-34 and Rad52-38 proteins are able to interact with themselves and with wild-type Rad52. Rad52 self-interaction is clearly evident from the high levels of β-galactosidase produced in strains carrying fusions of Rad52 to the Gal4 DNA binding and activation domains (Fig. 4). Furthermore, each of the mutants appears to interact with wild-type Rad52 as well as with itself. Experiments to determine whether there is an interaction between the two mutant forms of Rad52 were inconclusive, with the interaction being dependent on the fusion of the mutant proteins to either the Gal4 activation domain or the DNA binding domain (data not shown). This may be the result of differential effects of the two Gal4 domains on each of the mutant Rad52 proteins.
FIG. 4.
Results of two-hybrid analysis to detect possible homotypic interactions of Rad52-34, Rad52-38, and wild-type Rad52. The error bars represent the standard deviations for at least three repetitions.
Interactions between the mutant forms of Rad52 and Rad51, Rfa1, Rfa2, and Rfa3.
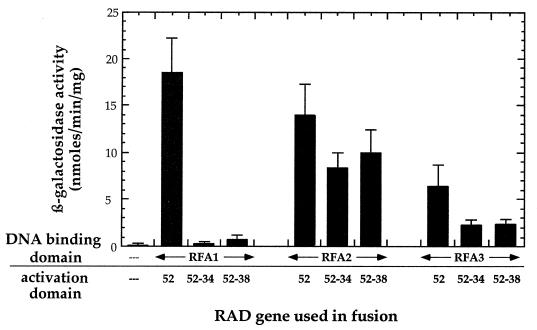
We and others have used two-hybrid analysis to show that Rad52 interacts with Rad51, and Fig. 2 and 3 indicate that Rad52 interacts with Rfa1, Rfa2, and Rfa3. We next sought to determine if the mutant forms of Rad52, Rad52-34 and Rad52-38, are able to interact with Rad51 and with the RPA subunits. In contrast to their self-interaction, the mutant Rad52 proteins are unable to interact with Rad51 (Fig. 5). Interestingly, the mutant forms of Rad52 also fail to interact with the large subunit of RPA, Rfa1, but remain active in their interaction with Rfa2 (Fig. 6), which has no DNA binding activity (16). This implies that Rad52 can interact with RPA in the absence of simultaneous binding to DNA. These results also suggest that the interaction of Rad52 with Rfa1 is not mediated by endogenous Rfa2. Such a “bridging” event has been noted before, when the bridge protein is overexpressed (23). In the present case, however, if the endogenous Rfa2 served as a bridge, then the competence of the Rad52-Rfa2 interaction should be sufficient to allow the Rad52-Rfa1 interaction to be detected in our two-hybrid assays. Since the rad52-34 and rad52-38 mutations appear to eliminate the mutant proteins’ interaction with Rfa1 but still allow for interaction with Rfa2 (Fig. 6), we regard the bridging explanation as unlikely. In addition, the fact that the Rad52 mutant proteins retain their ability to interact with Rfa2, wild-type Rad52, and themselves makes it unlikely that the mutant proteins are grossly misfolded and therefore unable to participate in protein-protein interactions.
FIG. 5.
Results of two-hybrid analysis for possible interactions between Rad52-34 and Rad52-38 with Rad51. The error bars represent the standard deviations for at least three repetitions.
FIG. 6.
Results of test for two-hybrid interactions of Rad52-34 and Rad52-38 with Rfa1, Rfa2, and Rfa3. The error bars represent the standard deviations for at least three repetitions.
Given that the interactions between Rfa3 and the Rad52 mutant proteins are barely at the threshold for positive identification of an interaction, we cannot determine with any certainty if the reduced interactions seen between the mutant forms of Rad52 and Rfa3 are significant. Thus, the question of whether the mutant forms of Rad52 interact with Rfa3 remains ambiguous.
Because of the interesting genetic interactions between the rfa1-44 and rad52-34 mutants, we were particularly interested in studying the corresponding protein fusions in the two-hybrid system. Accordingly, an rfa1-44–GAL4 DNA binding domain fusion expression vector was constructed. For reasons that are not clear, the resulting fusion was toxic in the yeast strain used for our two-hybrid experiments, as indicated by the fact that yeast carrying the fusion expression vector grew extremely poorly (data not shown). For this reason, we were unable to ascertain the existence of interactions by using the Rfa1-44 mutant protein in the two-hybrid system.
DISCUSSION
RPA has a well-documented role in DNA replication. Conceivably, its participation in recombination and repair is the result of its role in DNA synthesis during the replacement of missing genetic information at the site of a DSB or in the course of filling the gap at the site of an excised lesion. However, previous analysis indicated that the rfa1-44 mutation does not result in a dramatic increase in the cell’s doubling time, and the rate of [3H]uracil incorporation, a more direct measure of DNA synthesis, is not substantially reduced in a rfa1-44 mutant strain compared to that in wild-type yeast (data not shown). These findings suggest that the rfa1-44 mutant’s reduced capacity for recombinational repair of DSBs is not due primarily to an impairment of DNA synthesis. Thus, the rfa1-44 allele appears to be a true separation-of-function allele which affects RPA’s role in recombination and repair without dramatically affecting its role in DNA replication.
What might Rfa1’s role in recombination be? Our present experiments indicate that RAD52 is epistatic to RFA1 for recombinational repair of DSBs (Fig. 1). Furthermore, there is growing evidence that the proteins encoded by members of the RAD52 epistasis group function as a multiprotein complex (4, 14, 17, 18, 25, 27, 36), which may be considered a recombinosome (8). It seems plausible that the rfa1-44 mutant’s inability to properly interact with one or more members of the recombinosome, but particularly with a critical participant, Rad52, could explain the defect. The region of RFA1 in which the rfa1-44 mutation occurs is not highly conserved among species, perhaps because this protein domain is involved in species-specific protein-protein interactions integral to recombination or repair pathways rather than with the more highly conserved system for DNA replication. For that reason we examined some of these protein-protein interactions in greater detail.
Our two-hybrid analysis indicates that Rad52 does in fact interact with Rfa1 (Fig. 2). Rad52 also interacts with Rfa2 and Rfa3 (Fig. 3), raising the possibility that Rad52 makes multiple contacts with the RPA heterotrimer. In contrast, no interactions were detected between Rfa1 and the other members of the Rad52 epistasis group tested, Rad51, Rad55, and Rad57 (Fig. 2).
One possibility for the role of the RPA-Rad52 interaction is that RPA helps to recruit a DNA polymerase to the site of a DSB via simultaneous interactions with Rad52 and a repair polymerase. A second possibility, given RPA’s role in Rad51-mediated strand exchange (31, 38, 39), is that a complex of Rad51-Rad52 recruits RPA to the site of a DSB or other recombinogenic lesion in preparation for RPA’s role in the strand exchange event. Finally, it could be that RPA, alone or in concert with Rad52, binds the single-stranded overhangs at the site of a DSB, perhaps protecting the exposed single-stranded ends from cellular nucleases. Localized to such a site, RPA, through its interaction with Rad52, might nucleate the formation of a recombination complex, or recombinosome, that would carry out DSB repair. Such a role for RPA might be considered analogous to the role of human RPA in nucleotide excision repair, where RPA interacts with the repair proteins XPA and XPG (15, 21, 22, 26, 30) and this interaction is necessary both for XPA to bind to damaged DNA and for the action of the XPG nuclease. In this same system, RPA also plays a role in the subsequent gap-filling reaction (1, 10, 29).
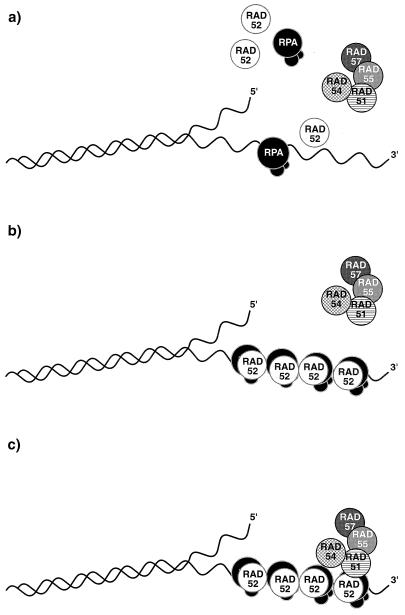
The inability of the Rad52-34 and Rad52-38 mutants, whose sequence changes lie in the presumptive DNA binding domain of Rad52 (28), to interact specifically with Rfa1 and Rad51 raises the possibility that these interactions require the integrity of the Rad52 DNA binding domain while the interaction with Rfa2 does not. Thus, association between Rad52 and DNA might conceivably be necessary for the proper interaction of Rad52 with the Rfa1 subunit of RPA. Similarly, the interaction of Rad52 with Rad51, itself in a complex with its homologs, Rad55 and Rad57, and the Rad54 helicase, might also be dependent on the DNA binding ability of Rad52. Such a model, in which Rad52, in concert with RPA, nucleates the assembly of the recombinosome, is depicted in Fig. 7.
FIG. 7.
A model in which the interaction of Rad52 with DNA occurs via interaction with all three subunits of RPA, which is bound to single-stranded DNA. RPA alone binds to single strands created at a DSB followed by association with Rad52 (a) or a complex of RPA and Rad52 binds at such single-stranded ends (b); a subassembly of Rad51 associated with Rad54 and Rad55 and, through Rad55, with Rad57 is presumed to bind to the Rad52-RPA complex (c).
In this model, we suppose that RPA alone binds to single strands created at a DSB, followed by association with Rad52, or that a complex of RPA and Rad52 binds at such single-stranded ends. We surmise that Rad51, along with the Rad54 and the complex of Rad55 and Rad57, then associates through the interacting domains of Rad51 and Rad52. It is also conceivable that homotypic Rad52 interactions bring the broken ends in apposition and that the Rad51 positioned at or near the end promotes strand invasion to initiate the repair process. Further information about the assembly of the putative recombinosome awaits further genetic and biochemical analysis.
ACKNOWLEDGMENTS
We thank Steve Brill and Bruce Stillman for kindly providing the RFA2 and RFA3 plasmids used to create the two-hybrid vectors and Robert Brazas and Greg Hannon for other plasmids.
REFERENCES
- 1.Aboussekhra A, Biggerstaff M, Shivji M K, Vilpo J A, Moncollin V, Podust V N, Protic M, Hubscher U, Egly J M, Wood R D. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 2.Boundy M K, Livingston D M. A Saccharomyces cerevisiae RAD52 allele expressing a C-terminal truncation protein: activities and intragenic complementation of missense mutations. Genetics. 1993;133:39–49. doi: 10.1093/genetics/133.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brill S J, Stillman B. Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 1991;5:1589–1600. doi: 10.1101/gad.5.9.1589. [DOI] [PubMed] [Google Scholar]
- 4.Donovan J W, Milne G T, Weaver D T. Homotypic and heterotypic protein associations control Rad51 function in double-strand break repair. Genes Dev. 1994;8:2552–2562. doi: 10.1101/gad.8.21.2552. [DOI] [PubMed] [Google Scholar]
- 5.Elias-Arnanz M, Firmenich A A, Berg P. Saccharomyces cerevisiae mutants defective in plasmid-chromosome recombination. Mol Gen Genet. 1996;252:530–538. doi: 10.1007/BF02172399. [DOI] [PubMed] [Google Scholar]
- 6.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 7.Firmenich, A. A. Unpublished data.
- 8.Firmenich A A, Elias-Arnanz M, Berg P. A novel allele of Saccharomyces cerevisiae RFA1 that is deficient in recombination and repair and suppressible by RAD52. Mol Cell Biol. 1995;15:1620–1631. doi: 10.1128/mcb.15.3.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 10.Guzder S N, Habraken Y, Sung P, Prakash L, Prakash S. Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A, and transcription factor TFIIH. J Biol Chem. 1995;270:12973–12976. doi: 10.1074/jbc.270.22.12973. [DOI] [PubMed] [Google Scholar]
- 11.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 12.Hays, S. L. Unpublished data.
- 13.Hays, S. L., and A. A. Firmenich. Unpublished data.
- 14.Hays S L, Firmenich A A, Berg P. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc Natl Acad Sci USA. 1995;92:6925–6929. doi: 10.1073/pnas.92.15.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Z, Henricksen L A, Wold M S, Ingles C J. RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature. 1995;374:566–569. doi: 10.1038/374566a0. [DOI] [PubMed] [Google Scholar]
- 16.Henricksen L A, Umbricht C B, Wold M S. Recombinant replication protein A: expression, complex formation, and functional characterization. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 17.Jiang H, Xie Y, Houston P, Stemke-Hale K, Mortensen U H, Rothstein R, Kodadek T. Direct association between the yeast Rad51 and Rad54 recombination proteins. J Biol Chem. 1996;271:33181–33186. doi: 10.1074/jbc.271.52.33181. [DOI] [PubMed] [Google Scholar]
- 18.Johnson R D, Symington L S. Functional differences and interactions among the putative RecA homologs Rad51, Rad55, and Rad57. Mol Cell Biol. 1995;15:4843–4850. doi: 10.1128/mcb.15.9.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenny M K, Schlegel U, Furneaux H, Hurwitz J. The role of human single-stranded DNA binding protein and its individual subunits in simian virus 40 DNA replication. J Biol Chem. 1990;265:7693–7700. [PubMed] [Google Scholar]
- 20.Kim C, Snyder R O, Wold M S. Binding properties of replication protein A from human and yeast cells. Mol Cell Biol. 1992;12:3050–3059. doi: 10.1128/mcb.12.7.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee S H, Kim D K, Drissi R. Human xeroderma pigmentosum group A protein interacts with human replication protein A and inhibits DNA replication. J Biol Chem. 1995;270:21800–21805. doi: 10.1074/jbc.270.37.21800. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Lu X, Peterson C A, Legerski R J. An interaction between the DNA repair factor XPA and replication protein A appears essential for nucleotide excision repair. Mol Cell Biol. 1995;15:5396–5402. doi: 10.1128/mcb.15.10.5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Y L, Chen C, Keshav K F, Winchester E, Dutta A. Dissection of functional domains of the human DNA replication protein complex replication protein A. J Biol Chem. 1996;271:17190–17198. doi: 10.1074/jbc.271.29.17190. [DOI] [PubMed] [Google Scholar]
- 24.Longhese M P, Plevani P, Lucchini G. Replication factor A is required in vivo for DNA replication, repair, and recombination. Mol Cell Biol. 1994;14:7884–7890. doi: 10.1128/mcb.14.12.7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lovett S T, Mortimer R K. Characterization of null mutants of the RAD55 gene of Saccharomyces cerevisiae: effects of temperature, osmotic strength and mating type. Genetics. 1987;116:547–553. doi: 10.1093/genetics/116.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuda T, Saijo M, Kuraoka I, Kobayashi T, Nakatsu Y, Nagai A, Enjoji T, Matsutani C, Sugasawa K, Hanaoka F, et al. DNA repair protein XPA binds replication protein A (RPA) J Biol Chem. 1995;270:4152–4157. doi: 10.1074/jbc.270.8.4152. [DOI] [PubMed] [Google Scholar]
- 27.Milne G T, Weaver D T. Dominant negative alleles of RAD52 reveal a DNA repair/recombination complex including Rad51 and Rad52. Genes Dev. 1993;7:1755–1765. doi: 10.1101/gad.7.9.1755. [DOI] [PubMed] [Google Scholar]
- 28.Mortensen U H, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc Natl Acad Sci USA. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mu D, Park C H, Matsunaga T, Hsu D S, Reardon J T, Sancar A. Reconstitution of human DNA repair excision nuclease in a highly defined system. J Biol Chem. 1995;270:2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 30.Nagai A, Saijo M, Kuraoka I, Matsuda T, Kodo N, Nakatsu Y, Mimaki T, Mino M, Biggerstaff M, Wood R D, et al. Enhancement of damage-specific DNA binding of XPA by interaction with the ERCC1 DNA repair protein. Biochem Biophys Res Commun. 1995;211:960–966. doi: 10.1006/bbrc.1995.1905. [DOI] [PubMed] [Google Scholar]
- 31.Namsaraev E, Berg P. Characterization of strand exchange activity of yeast Rad51 protein. Mol Cell Biol. 1997;17:5359–5368. doi: 10.1128/mcb.17.9.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park M S, Ludwig D L, Stigger E, Lee S H. Physical interaction between human RAD52 and RPA is required for homologous recombination in mammalian cells. J Biol Chem. 1996;271:18996–19000. doi: 10.1074/jbc.271.31.18996. [DOI] [PubMed] [Google Scholar]
- 33.Printen J A, Sprague G F., Jr Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics. 1994;138:609–619. doi: 10.1093/genetics/138.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 36.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 37.Smith J, Rothstein R. A mutation in the gene encoding the Saccharomyces cerevisiae single-stranded DNA binding protein Rfa1 stimulates a RAD52-independent pathway for direct-repeat recombination. Mol Cell Biol. 1995;15:1632–1641. doi: 10.1128/mcb.15.3.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 39.Sung P, Robberson D L. DNA strand exchange mediated by a Rad51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 40.Wold M S, Weinberg D H, Virshup D M, Li J J, Kelly T J. Identification of cellular proteins required for simian virus 40 DNA replication. J Biol Chem. 1989;264:2801–2809. [PubMed] [Google Scholar]