Abstract
In this study, we introduce a novel approach for the analysis and interpretation of 3D shapes, particularly applied in the context of neuroscientific research. Our method captures 2D perspectives from various vantage points of a 3D object. These perspectives are subsequently analyzed using 2D Convolutional Neural Networks (CNNs), uniquely modified with custom pooling mechanisms.
We sought to assess the efficacy of our approach through a binary classification task involving subjects at high risk for Autism Spectrum Disorder (ASD). The task entailed differentiating between high-risk positive and high-risk negative ASD cases. To do this, we employed brain attributes like cortical thickness, surface area, and extra-axial cerebral spinal measurements. We then mapped these measurements onto the surface of a sphere and subsequently analyzed them via our bespoke method.
One distinguishing feature of our method is the pooling of data from diverse views using our icosahedron convolution operator. This operator facilitates the efficient sharing of information between neighboring views. A significant contribution of our method is the generation of gradient-based explainability maps, which can be visualized on the brain surface. The insights derived from these explainability images align with prior research findings, particularly those detailing the brain regions typically impacted by ASD. Our innovative approach thereby substantiates the known understanding of this disorder while potentially unveiling novel areas of study.
Keywords: Shape Analysis, ASD, brain
1. Introduction
Autism Spectrum Disorder (ASD) is a condition related to the development of the brain that causes differences in neurological functioning. Subjects with ASD have deficits in social communication skills and behavior, the presence of repetitive behavior, restricted interests, hyper- or hypo-sensitivity to sensory stimuli, and an insistence on sameness or strict adherence to routine [23, 26].
Diagnosis of ASD is difficult as there is no medical/lab test to reliably assess the condition. Various studies have found the average age of diagnosis to be 3 years, although the vast majority of children manifest developmental problems between 12 and 24 months, with some showing abnormalities before 12 months[3]. Early diagnosis of ASD before 2.5 years of age is associated with considerable benefits for children who may “outgrow” the condition through therapy[12].
Detecting ASD bio-markers from neuroimaging data is a challenging task owing to the considerable variability in cortical shape and functional organization across individuals, which hinders the ability to make accurate comparisons of brains[16, 13]. There is a clear need for precision analysis tools that are robust to these factors and the discovery of distinct features to characterize ASD.
The main contribution of this paper can be summarized into two key aspects. Firstly, it introduces a new deep-learning framework for general shape analysis that utilizes a multi-view approach. Secondly, it incorporates an explainability component that identifies crucial brain regions at the vertex level and visualizes them on the cortical surface highlighting relevant brain regions for the classification task.
The proposed method was evaluated using Precision, Recall, F1-score, and Accuracy metrics through five 5-fold cross-validation experiments on a cohort of High-Risk Positive (HR+) ASD versus High-Risk Negative (HR−) ASD patients. We implement different pooling layers for our model and compare them against Spherical-U-Net[37] and Spectformer [2], methods designed for brain shape analysis and spectral analysis respectively. We also test our approach against a Random Forest classifier that uses learned shape features and demographic information combined.
2. Related work
2.1. ASD Classification
Several studies have addressed the question of ASD classification using Machine Learning (ML) models. The majority of studies used the ABIDE I/II [15] data set which includes resting state functional (rsfMRI), structural T1/T2 (sMRI) Magnetic Resonance Images, and Diffusion (dMRI) Magnetic Resonance Images. This data set contains data from individuals with autism spectrum disorder (ASD) and typically developing individuals. We point the reader to [21, 10] for a comprehensive review of the literature on ASD classification. It has been demonstrated that different machine learning models can effectively distinguish between individuals with typical development and those with ASD. However, the data used in our study differs, as it includes HR subjects who have not yet developed the condition. This adds a layer of complexity to the analysis, as it requires the identification of biomarkers that can reliably predict ASD in the future.
2.2. 3D shape analysis
Among the different approaches for shape analysis, learning-based methods are currently the most sophisticated ones. There are mainly 4 types of learning-based methods: multi-view, volumetric, parametric, and multi-layer-perceptrons (MLP).
Multi-view approaches adapt state-of-the-art 2D CNNs to work on 3D shapes as the arbitrary structures of 3D models, which are usually represented by point clouds or triangular meshes, are incompatible with convolutional operators that require regular grid-like structures. By rendering 3D objects from different viewpoints, features are extracted using 2D CNNs[33, 20]. On the other hand, volumetric approaches use 3D voxel grids to represent the shape and apply 3D convolutions to learn shape features[35, 29]. Parametric methods require shapes with spherical topology and the convolution is applied directly to the spherical representation of the shape[17, 37]. Finally, other approaches consume the point clouds directly and implement multi-layer-perceptrons and/or transformer architectures[25, 34].
Our method falls in the multi-view category. We render the object and capture 2D images from different viewpoints following an icosahedron subdivision. The multiple captures ensure coverage of the whole object. One of the primary benefits of multi-view approaches is their ability to operate on surfaces with any topology, including those with missing data or holes. Our method is tested on spheres derived from brain cortical gray/white matter surfaces.
2.3. Explainable Artificial Intelligence
ML systems are becoming increasingly ubiquitous and they outperform humans on a variety of specific tasks. There is increasing concern related to the deployment of such complex applications that have a direct impact on human lives. Such systems must be able to explain the basis for their decision to any impacted individual in terms understandable to a layperson, this is especially the case in the field of medical imaging. Explainability methods fall into 3 categories: visualization, model distillation, and intrinsic[27]. To the best of our knowledge, we found 2 methods for cortical surface analysis and explainability. First, a perturbation-based method for geometric deep learning of retinotopy through systematic manipulations of the input data and measurement of changes in the model’s output [28]. Second, NeuroExplainer [36] a method that uses spherical surfaces of the brain hemispheres with cortical attributes (thickness, mean curvature, and convexity), and a spherical convolution block in an encoder/decoder architecture that propagates the vertex-wise attributes and captures fine-grained explanations for a classification task.
Our explainability model is agnostic to the input data and does not require systematic perturbations to produce explanations. Moreover, it does not require subsampling the input data through encoder/decoder architectures and does not require shapes with a spherical topology or specialized operators such as spherical convolutions. In our experiments, we use spheres of +160000 vertex at full resolution.
3. Materials
Infants at high and low familial risk for ASD were enrolled at four clinical sites (University of North Carolina, University of Washington, Washington University, and Children’s Hospital of Philadelphia) [31, 14]. HR infants had an older sibling with a clinical diagnosis of ASD, corroborated by the Autism Diagnostic Interview–Revised [19]. LR infants had a typically developing older sibling and no first- or second-degree relatives with intellectual/psychiatric disorders [11]. Infants were assessed at 6, 12, and 24 months with magnetic resonance imaging (MRI) scans and a behavioral battery that included measures of cognitive development [22] and adaptive functioning[32]. DSM-IV-TR criteria [1] and the Autism Diagnostic Observation Schedule–Generic [18] were administered to all participants at 24 months. The Autism Diagnostic Interview–Revised was administered at 24 months to all parents of high-risk infants and to all low-risk infants with clinical concerns. At 24 months, infants were classified as having ASD based on expert clinical judgment and all available clinical information.
In our experiments, we use a subset of HR infants only and compare a group of 760 HR+ v.s. 202 HR−. We include demographic data in our analysis by combining image features and demographics through a separate branch that concatenates with the output of the features computed by the NN.
The demographics include gender, visit age for MRI, volume measurements for subcortical structures (amygdala, hippocampus, lateral ventricles), intracranial volume (ICV), and cerebrum and cerebellum volume.
4. Method description
4.1. Rendering the 2D views
The Pytorch3D6 framework allows rendering and training in an end-to-end fashion. The rendering engine provides a map that relates pixels (pix2face) in the images to faces in the mesh and allows rapid extraction of point data as well as setting information back into the mesh after inference. We use pix2face to extract values for the 3 brain features namely: extra-axial cerebral spinal fluid (EA-CSF), surface area (SA), and cortical thickness. The EACSF features are precomputed via a probabilistic brain tissue segmentation, cortical surface reconstruction, and streamline-based local EA-CSF quantification[8]. SA and CT are precomputed via CIVET [6]. The pix2face map allows us to extract the vertex information and map them into 2D images set to 224px resolution. These images are then fed to the NN for feature extraction.
4.2. Architecture
We developed a novel NN architecture called BrainIcoNet and perform extensive experiments with a variety of feature extraction layers and IcoConv operators.
Figure 1 shows the general architecture of our approach which consists of a feature extraction step followed by our IcoConv operator. Figure 2 shows the different IcoConv operators.
Fig. 1.
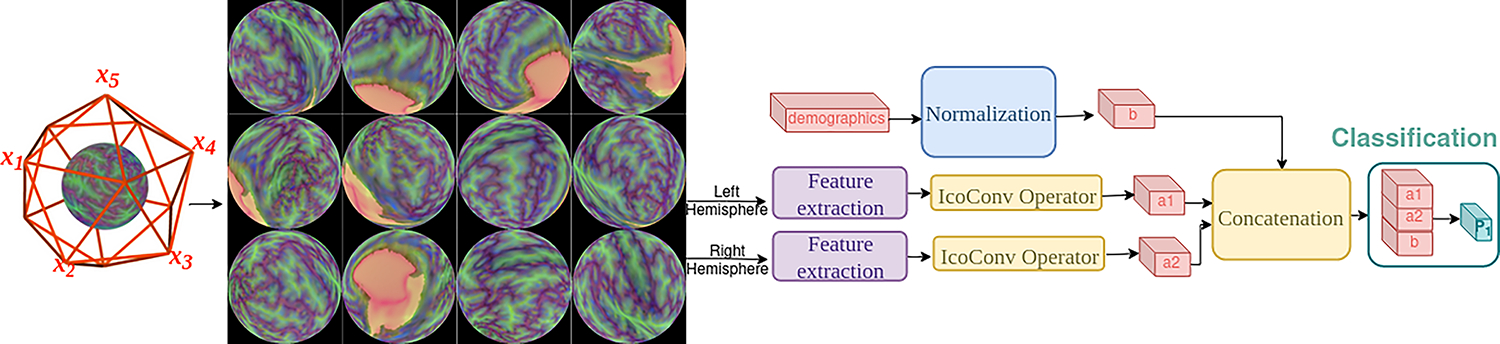
Architecture for the ASD classification task. To initiate our analysis, we begin by capturing views of the unique characteristics of each cerebral hemisphere - the left and the right - as they are projected onto the spherical surface. The vantage point follow an icosahedron subdivision. We use a feature extraction network (resnet18, SpectFormer) on each individual view. We experiment with different IcoConv (IcoConv for icosahedron and convolution) operators that pool the information from all views. Finally, we concatenate the left/right outputs and normalized demographics. We perform a final linear layer for the classification.
Fig. 2.
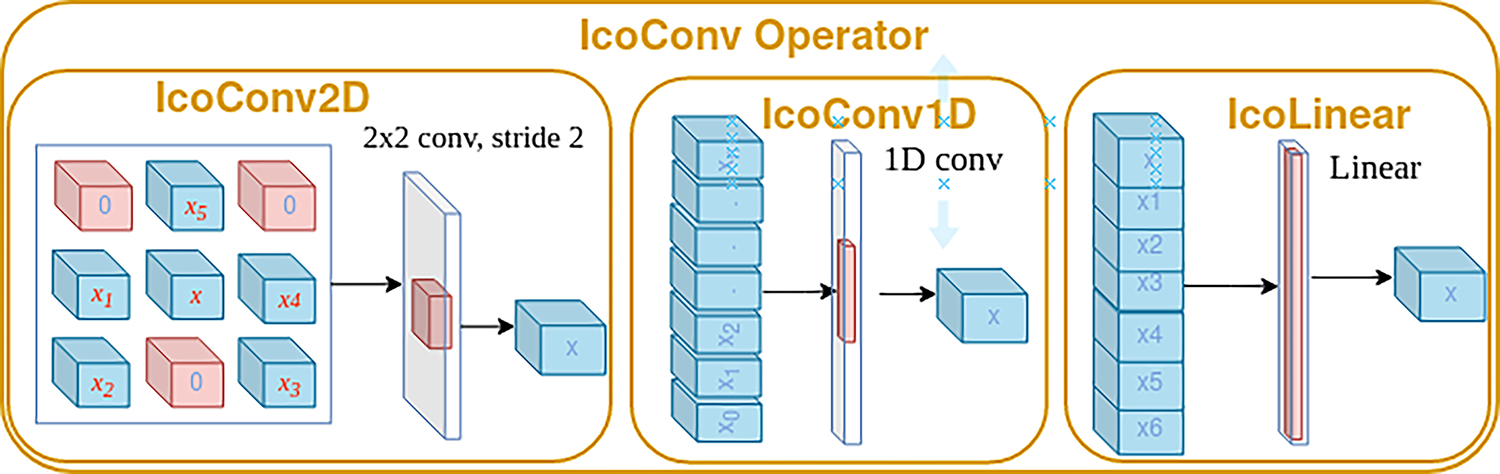
Different IcoConv operators. IcoConv2D arranges the features extracted from adjacent views in 3×3 grid and performs an additional 2D Convolution. IcoConv1D aranges the features and performs a 1D Convolution followed by Average/Max pooloing. IcoLinear stacks the features and performs a Linear layer.
In our experimental setup, we change the number of views, evaluating both 12 and 42 perspectives, and alter the radius of the icosahedron to adjust the proximity to the 3D object. A smaller radius sets the view closer to the 3D object thereby restricting the breadth of the captured view but acquiring finer detailed information.
The captured views are then fed to the feature extraction layer. We use two distinct branches, each dedicated to a specific hemisphere. The assumption is that the left and right hemispheres exhibit unique features that should be treated separately.
Each branch uses resnet18 or a SpectFormer block for feature extraction. The features are then arranged and passed to the IcoConv block. We experiment with 2D/1D Convolutions and a Linear layer. The IcoConv2D operator was designed to allow sharing of information across adjacent views only. As demonstrated by the results, the explainability maps are localized and corroborate previous findings about brain regions affected by ASD.
Finally, we use a linear layer for the binary classification task.
Additional experiments were conducted including demographic information which is normalized and concatenated to the left/right brain hemispheres. We train a random forest classifier and perform a feature importance analysis.
4.3. Training the models
We perform a 5 fold cross-validation training for every model. We use a series of augmentation techniques including random rotations of the input sphere, a dropout layer with p = 20% just before our linear layer for classification, and Gaussian noise applied on each image as well as the coordinates of the sphere points, i.e., a small perturbation which is then normalized back on the spherical surface.
Training is done on an NVIDIA RTX6000 GPU with a batch size of 10, learning rate 1e−4, the AdamW optimizer, and use the early stopping criteria to stop training automatically (patience 100) and keep the best performing model. To account for the highly imbalanced nature of our dataset classes during training, we utilize a sampling approach and ensure that each batch is balanced during training.
4.4. Explainability maps
To find out what are the relevant areas for the classification task we use GradCam [30]. This technique utilizes the gradients of the classification score with respect to the final feature map, thus, identifying which regions of the image contribute to the final classification score. We project each explainability map back to the 3D-object/sphere and apply a median filter with neighboring vertices to remove noise. The projection of these maps onto the spherical surface enables the visualization of explainability maps directly on the inflated cortical surfaces. This approach simplifies the task of identifying the regions impacted by the condition under study. By utilizing this 3D spatial representation, we are effectively able to correlate the intricate details from our explainability maps with specific locations on the brain’s surface, providing a clear illustration of affected areas.
5. Results
The results in this section are computed using the test set from each fold and are reported for the whole population.
Table 1 shows the mean and standard deviation for precision, recall, f1-score, and accuracy for the 5 folds.
Table 1.
Classification report for the 5 folds. We report the mean and standard deviation for each metric. 42V = 42 views (icoshedron subdivision level 2). MAR = Macro Average Recall.
| Approach | Class | Precision | Recall | FI Score | MAR | Accuracy |
|---|---|---|---|---|---|---|
|
| ||||||
| S-Unet | No ASD | 0.83 ± 0.02 | 0.88 ± 0.05 | 0.85 ± 0.02 | ||
| ASD | 0.37 ± 0.13 | 0.29 ±0.07 | 0.33±0.08 | 0.585 ±0.04 | 0.76 ±0.03 | |
|
| ||||||
| IcoCo2D | No ASD | 0.83 ± 0.01 | 0.88 ±0.05 | 0.86±0.03 | ||
| ASD | 0.38±0.19 | 0.28 ±0.07 | 0.32±0.11 | 0.58 ±0.05 | 0.76±0.04 | |
|
| ||||||
| IcoColD | No ASD | 0.84 ± 0.02 | 0.87± 0.05 | 0.85±0.03 | ||
| ASD | 0.374= 0.15 | 0.32± 0.11 | 0.34± 0.11 | 0.595 ±0.05 | 0.76± 0.05 | |
|
| ||||||
| IcoCoLinear | No ASD | 0.84 ± 0.03 | 0.84± 0.04 | 0.84± 0.01 | ||
| ASD | 0.374= 0.12 | 0.37±0.17 | 0.37± 0.13 | 0.605± 0.07 | 0.75± 0.02 | |
|
| ||||||
| IcoCoLinearinf | No ASD | 0.85 ±0.01 | 0.85 ± 0.04 | 0.85± 0.02 | ||
| ASD | 0.39± 0.11 | 0.39± 0.07 | 0.39± 0.09 | 0.62±0.04 | 0.75± 0.03 | |
|
| ||||||
| Spcct 42V | No ASD | 0.84 ±0.03 | 0.86±0.05 | 0.85± 0.02 | ||
| ASD | 0.38± 0.12 | 0.35±0.07 | 0.36±0.05 | 0.605±0.02 | 0.76±0.03 | |
|
| ||||||
| SpcctICo 42V | No ASD | 0.85± 0.02 | 0.83±0.05 | 0.84±0.02 | ||
| ASD | 0.37±0.1 | 0.41± 0.05 | 0.39±0.05 | 0.62 ±0.02 | 0.74± 0.03 | |
|
| ||||||
| IcoCo2D 42V | No ASD | 0.85± 0.03 | 0.86± 0.05 | 0.85±0.02 | ||
| ASD | 0.42± 0.08 | 0.41± 0.1 | 0.42± 0.06 | 0.635±0.04 | 0.77±0.02 | |
We perform extensive experiments with S-Unet, each IcoConv operator, different feature extraction layers namely resnet18 and SpectFormer, and increasing the number of views and reducing the radius to capture finer details.
The task of classifying HR+ v.s. HR− subjects presents a challenge. This is largely due to the fact that at this early stage, the brains of the subjects often do not exhibit explicit or easily distinguishable characteristics associated with the condition. Consequently, subtle nuances and variations may be critical in this classification task, underscoring the need for advanced and sensitive analytical methods.
We underscore that this dataset is highly imbalanced and achieving a high recall ensures that the model does not merely predict the majority class and miss the minority class instances.
The best performing model is the IcoConv2D with 42 views and the explainability maps are generated with it.
The explainability maps are shown in Figure 3. Interestingly, the model favors features from the right hemisphere over the left ones. Furthermore, our findings support previous research[14] that highlights the significance of similar brain regions sensitive to this classification task.
Fig. 3.
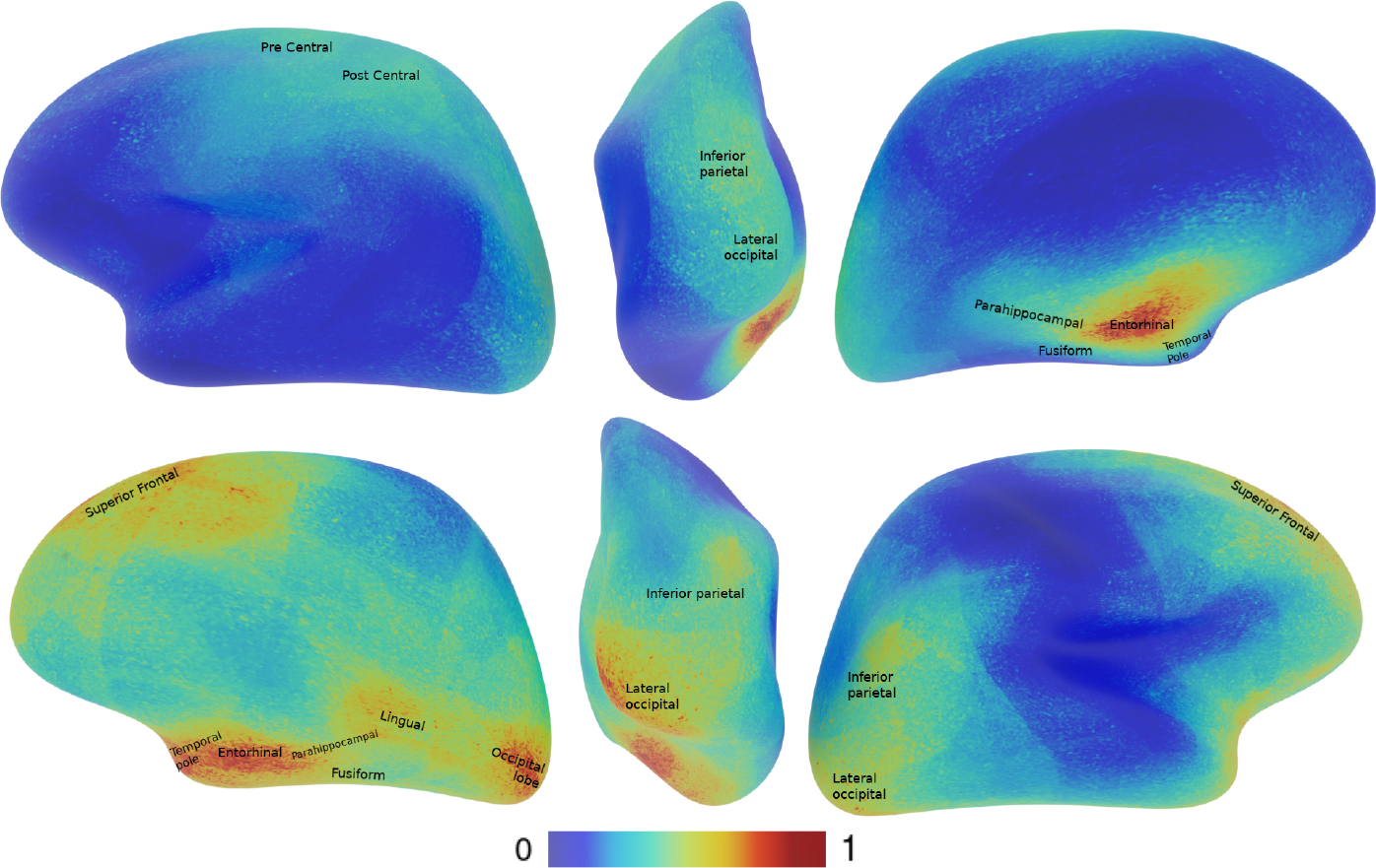
Left, posterior, right views for the left hemisphere above and right hemisphere below. The gradcam maps are generated using only the correctly classified HR+ subjects and using HR+ as the target class. It indicates that features from the right hemisphere are preferred over the left ones. The name of the area are based on this labeling map [9]
We use the Desikan parcellation [9] to identify the affected brain regions that appear in our explainability maps. Similar activation maps appear on both hemispheres centered on the entorhinal spreading to the parahippocampal, temporal pole, and fusiform. The right hemisphere present higher activity in the lingual and occipital lobe, with some activation in the right and left for the inferior parietal and superior frontal regions. These specific areas have been reported in previous studies, the entorhinal cortex, lingual and fusiform have been reported respectively by [5, 7, 24] to be areas impacted by ASD.
Finally, we test the feature importance with a random forest classifier trained using scikit-learn version 1.1.1. Figure 4 shows that gender is the most important demographic and amygdala is the most important if the gender is removed from the analysis.
Fig. 4.
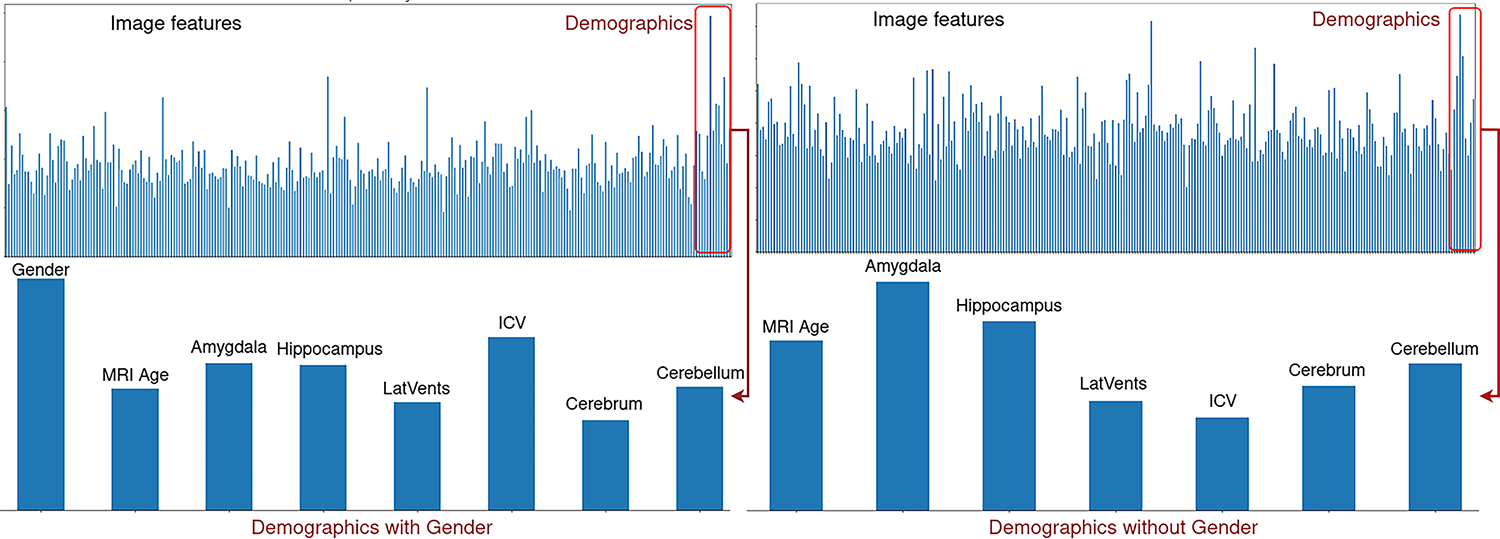
The top left figure shows a plot of importance for features concatenated with normalized demographic values. The bottom left is only demographics to highlight that gender is the most important feature for the Random Forest classifier. The right plot shows an experiment with the gender removed from the analysis and shows that the amygdala is the most important feature in the demographics for the classification task.
6. Conclusion
In conclusion, we have created a framework for shape analysis and explainability that is agnostic to the neural network model and the shape topology of the input meshes.
Our first contribution is a novel approach for shape analysis that does not require shapes with specific spherical topology or any form of subsampling of the mesh. Our shape analysis framework offers a significant advantage as it can handle meshes that are not in spherical topology or have holes, which is a requirement for S-Unet. We demonstrate this crucial feature by performing an experiment using subject specific inflated cortical surfaces.
Our second contribution is the visualization of explainability maps on complex shapes such as cortical surfaces. We tested our approach on a challenging classification task using subjects with high risk of developing autism and comparing HR+ v.s. HR−.
Our study utilized distinct neural networks for each hemisphere of the brain. Our results reveal that certain characteristics in the right hemisphere of the brain play a significant role in the classification of ASD. Our approach has identified brain regions and corroborate previous findings [5, 7, 24] of ASD-affected brain regions.
Finally, our approach allows including demographic information and highlights the amygdala volume as an important predictor for ASD. This finding also been corroborated in a previous study [4].
In future work, we will extend our analysis to other neuro-psychiatric disorders such as schizophrenia, attention deficit hyper-activity disorder, and bipolar.
Supplementary Material
7. Acknowledgment
This work was supported by grants R01EB021391, P50HD103573, and the Foundation of Hope.
Footnotes
References
- 1.Association AP, et al. : Diagnostic and statistical manual of mental disorders, text revision (dsm-iv-tr®) (2010) [Google Scholar]
- 2.Badri Narayana Patro VPN, Agneeswaran VS: Spectformer: Frequency and attention is what you need in a vision transformer (2023) [Google Scholar]
- 3.Barbaro J, Dissanayake C: Autism spectrum disorders in infancy and toddlerhood: a review of the evidence on early signs, early identification tools, and early diagnosis. Journal of Developmental & Behavioral Pediatrics 30(5), 447–459 (2009) [DOI] [PubMed] [Google Scholar]
- 4.Bellani M, Calderoni S, Muratori F, Brambilla P: Brain anatomy of autism spectrum disorders ii. focus on amygdala. Epidemiology and psychiatric sciences 22(4), 309–312 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blatt GJ: The neuropathology of autism p. 6 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boucher M, Whitesides S, Evans A: Depth potential function for folding pattern representation, registration and analysis. Medical image analysis 13(2), 203–214 (2009) [DOI] [PubMed] [Google Scholar]
- 7.Chandran VA, Pliatsikas C, Neufeld J, O’Connell G, Haffey A, DeLuca V, Chakrabarti B: Brain structural correlates of autistic traits across the diagnostic divide: A grey matter and white matter microstructure study. NeuroImage: Clinical 32, 102897 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deddah T, Styner M, Prieto J: Local extraction of extra-axial csf from structural mri. In: Medical Imaging 2022: Biomedical Applications in Molecular, Structural, and Functional Imaging. vol. 12036, pp. 29–34. SPIE; (2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, et al. : An automated labeling system for subdividing the human cerebral cortex on mri scans into gyral based regions of interest. Neuroimage 31(3), 968–980 (2006) [DOI] [PubMed] [Google Scholar]
- 10.Eslami T, Almuqhim F, Raiker JS, Saeed F: Machine learning methods for diagnosing autism spectrum disorder and attention-deficit/hyperactivity disorder using functional and structural mri: a survey. Frontiers in neuroinformatics 14, 575999 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Estes A, Zwaigenbaum L, Gu H, St John T, Paterson S, Elison JT, Hazlett H, Botteron K, Dager SR, Schultz RT, et al. : Behavioral, cognitive, and adaptive development in infants with autism spectrum disorder in the first 2 years of life. Journal of neurodevelopmental disorders 7(1), 1–10 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabbay-Dizdar N, Ilan M, Meiri G, Faroy M, Michaelovski A, Flusser H, Menashe I, Koller J, Zachor DA, Dinstein I: Early diagnosis of autism in the community is associated with marked improvement in social symptoms within 1–2 years. Autism 26(6), 1353–1363 (2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, Ugurbil K, Andersson J, Beckmann CF, Jenkinson M, et al. : A multi-modal parcellation of human cerebral cortex. Nature 536(7615), 171–178 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hazlett HC, Gu H, Munsell BC, Kim SH, Styner M, Wolff JJ, Elison JT, Swanson MR, Zhu H, Botteron KN, et al. : Early brain development in infants at high risk for autism spectrum disorder. Nature 542(7641), 348–351 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinsfeld AS, Franco AR, Craddock RC, Buchweitz A, Meneguzzi F: Identification of autism spectrum disorder using deep learning and the abide dataset. NeuroImage: Clinical 17, 16–23 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong Y, Gao J, Xu Y, Pan Y, Wang J, Liu J: Classification of autism spectrum disorder by combining brain connectivity and deep neural network classifier. Neurocomputing 324, 63–68 (2019) [Google Scholar]
- 17.Liu M, Yao F, Choi C, Sinha A, Ramani K: Deep learning 3d shapes using alt-az anisotropic 2-sphere convolution. In: International Conference on Learning Representations (2018) [Google Scholar]
- 18.Lord C, Risi S, Lambrecht L, Cook EH, Leventhal BL, DiLavore PC, Pickles A, Rutter M: The autism diagnostic observation schedule—generic: A standard measure of social and communication deficits associated with the spectrum of autism. Journal of autism and developmental disorders 30(3), 205–223 (2000) [PubMed] [Google Scholar]
- 19.Lord C, Rutter M, Le Couteur A: Autism diagnostic interview-revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. Journal of autism and developmental disorders 24(5), 659–685 (1994) [DOI] [PubMed] [Google Scholar]
- 20.Ma C, Guo Y, Yang J, An W: Learning multi-view representation with lstm for 3-d shape recognition and retrieval. IEEE Transactions on Multimedia 21(5), 1169–1182 (2018) [Google Scholar]
- 21.Moridian P, Ghassemi N, Jafari M, Salloum-Asfar S, Sadeghi D, Khodatars M, Shoeibi A, Khosravi A, Ling SH, Subasi A, et al. : Automatic autism spectrum disorder detection using artificial intelligence methods with mri neuroimaging: A review. arXiv preprint arXiv:2206.11233 (2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mullen EM, et al. : Mullen scales of early learning. AGS Circle Pines, MN (1995) [Google Scholar]
- 23.Nietzel M, Wakefield J: American psychiatric association diagnostic and statistical manual of mental disorders. Contemporary psychology 41, 642–651 (1996) [Google Scholar]
- 24.Pierce K, Redcay E: Fusiform function in children with an asd is a matter of “who” (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi CR, Su H, Mo K, Guibas LJ: Pointnet: Deep learning on point sets for 3d classification and segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp. 652–660 (2017) [Google Scholar]
- 26.Rahman MM, Usman OL, Muniyandi RC, Sahran S, Mohamed S, Razak RA: A review of machine learning methods of feature selection and classification for autism spectrum disorder. Brain sciences 10(12), 949 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ras G, Xie N, van Gerven M, Doran D: Explainable deep learning: A field guide for the uninitiated. Journal of Artificial Intelligence Research 73, 329–397 (2022) [Google Scholar]
- 28.Ribeiro FL, Bollmann S, Cunnington R, Puckett AM: An explainability framework for cortical surface-based deep learning. arXiv preprint arXiv:2203.08312 (2022) [Google Scholar]
- 29.Riegler G, Osman Ulusoy A, Geiger A: Octnet: Learning deep 3d representations at high resolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. pp. 3577–3586 (2017) [Google Scholar]
- 30.Selvaraju RR, Das A, Vedantam R, Cogswell M, Parikh D, Batra D: Gradcam: Why did you say that? arXiv preprint arXiv:1611.07450 (2016) [Google Scholar]
- 31.Shen MD, Kim SH, McKinstry RC, Gu H, Hazlett HC, Nordahl CW, Emerson RW, Shaw D, Elison JT, Swanson MR, et al. : Increased extra-axial cerebrospinal fluid in high-risk infants who later develop autism. Biological psychiatry 82(3), 186–193 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sparrow S, Balla D, Cicchetti D: Vineland scales of adaptive behavior, survey form manual. Circle Pines, MN: American Guidance Service; (1984) [Google Scholar]
- 33.Su H, Maji S, Kalogerakis E, Learned-Miller E: Multi-view convolutional neural networks for 3d shape recognition. In: Proceedings of the IEEE international conference on computer vision. pp. 945–953 (2015) [Google Scholar]
- 34.Wu W, Qi Z, Fuxin L: Pointconv: Deep convolutional networks on 3d point clouds. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. pp. 9621–9630 (2019) [Google Scholar]
- 35.Wu Z, Song S, Khosla A, Yu F, Zhang L, Tang X, Xiao J: 3d shapenets: A deep representation for volumetric shapes. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp. 1912–1920 (2015) [Google Scholar]
- 36.Xue C, Wang F, Zhu Y, Li H, Meng D, Shen D, Lian C: Neuroexplainer: Fine-grained attention decoding to uncover cortical development patterns of preterm infants. arXiv preprint arXiv:2301.00815 (2023) [Google Scholar]
- 37.Zhao F, Xia S, Wu Z, Duan D, Wang L, Lin W, Gilmore JH, Shen D, Li G: Spherical u-net on cortical surfaces: methods and applications. In: Information Processing in Medical Imaging: 26th International Conference, IPMI 2019, Hong Kong, China, June 2–7, 2019, Proceedings 26. pp. 855–866. Springer (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


