Abstract
The editing of apolipoprotein B (apo-B) mRNA involves the site-specific deamination of cytidine to uracil. The specificity of editing is conferred by an 11-nucleotide mooring sequence located downstream from the editing site. Apobec-1, the catalytic subunit of the editing enzyme, requires additional proteins to edit apo-B mRNA in vitro, but the function of these additional factors, known as complementing activity, is not known. Using RNA affinity chromatography, we show that the complementing activity binds to a 280-nucleotide apo-B RNA in the absence of apobec-1. The activity did not bind to the antisense strand or to an RNA with three mutations in the mooring sequence. The eluate from the wild-type RNA column contained a 65-kDa protein that UV cross-linked to apo-B mRNA but not to the triple-mutant RNA. This protein was not detected in the eluates from the mutant or the antisense RNA columns. Introduction of the mooring sequence into luciferase RNA induced cross-linking of the 65-kDa protein. A 65-kDa protein that interacted with apobec-1 was also detected by far-Western analysis in the eluate from the wild-type RNA column but not from the mutant RNA column. For purification, proteins were precleared on the mutant RNA column prior to chromatography on the wild-type RNA column. Silver staining of the affinity-purified fraction detected a single prominent protein of 65 kDa. Our results suggest that the complementing activity may function as the RNA-binding subunit of the holoenzyme.
The posttranscriptional editing of apolipoprotein B (apo-B) mRNA in humans results in the synthesis of two forms of the protein, apo-B100 and apo-B48 (reviewed in reference 6). These two proteins perform distinct functions in lipoprotein structure and metabolism. Apo-B100 (512 kDa) is the full-length protein that is secreted from the liver as a component of low-density and very-low-density lipoproteins (6). apo-B48 (242 kDa) is translated from an edited RNA in which the cytidine at nucleotide (nt) 6666 is deaminated to uracil (18). This modification changes the codon at position 2153 from CAA encoding glutamine to UAA, a premature translational stop codon (7, 27). The truncated apo-B48 form is secreted from enterocytes in the small intestine and is involved in the absorption of dietary lipid and the assembly of chylomicrons (6).
apo-B mRNA contains well-defined sequence elements that are recognized by the proteins involved in editing (3, 4). A short cassette of 26 nt flanking the edited site (nt 6662 to 6687) is sufficient for specific editing in transfected rat hepatoma cells (8). This cassette contains an 11-nt mooring sequence (nt 6671 to 6681) which is critical for editing, since mutations in this sequence drastically reduced or abolished editing (29). A minimal sequence containing the mooring sequence and a 4-nt spacer element supported low-level editing of an upstream C when inserted elsewhere in apo-B mRNA (5) or into a heterologous mRNA (2, 10). The AU-rich sequences flanking this minimal cassette comprise a poorly understood “bulk” RNA context that improves the efficiency of editing (5).
The editing enzyme is a multiprotein complex which does not require divalent cations, nucleoside triphosphates, or RNA as cofactors (15, 17). The holoenzyme has been partially purified from baboon (9), rat (15), and rabbit (13) enterocytes, but the molecular composition and size of this activity remain controversial. Glycerol gradient centrifugation of rat intestinal extracts showed that editing occurs in a 27S particle called the editosome (30), while gel filtration chromatography of baboon and rat enterocyte extracts indicated a minimal size of 120–125 kDa for the holoenzyme (9, 26). One subunit of the enzyme complex, a 27-kDa protein called apobec-1 (apo-B RNA editing enzyme catalytic polypeptide 1), was identified by expression cloning (31). Apobec-1 has limited homology to the active sites of other known cytidine deaminases and dCMP deaminases and has been reported to have cytidine deaminase activity (22, 25). However, apobec-1 alone cannot edit apo-B mRNA in vitro and requires additional factors referred to as complementing activity or auxiliary factors (14, 31). Since apobec-1 has only a weak nonspecific RNA binding activity (1, 24), it has been proposed that the complementing activity may represent the RNA-binding subunit of the editing enzyme.
The protein(s) which complement recombinant apobec-1 were initially detected in cells that synthesize apo-B but lack editing activity (14, 31). Later studies showed the widespread but not ubiquitous expression of complementing activity in baboon (12) and rabbit (33) tissues. This activity has not been purified but UV-cross-linking experiments detected several candidate proteins in extracts from tissues that contain the holoenzyme. Proteins of 60 and 43 kDa from rat enterocyte S100 extracts cross-linked specifically to apo-B mRNA (26). Editosomes that were assembled on apo-B RNA from rat hepatoma cell extracts showed the presence of multiple proteins ranging in size from 20 to 260 kDa. Of these, only the 66- and 44-kDa proteins were shown to specifically cross-link to apo-B mRNA (16, 34). None of these proteins were shown to possess complementing activity. Two proteins that regulate the editing activity of the holoenzyme have been identified: a 240-kDa protein identified by monoclonal antibodies raised against rat liver editosomes (28) and a 49-kDa enhancement factor that was partially purified from chick enterocytes (32). An apobec-1 binding protein, ABBP-1, was recently identified in a yeast two-hybrid screen (21). ABBP-1 is an alternatively spliced form of hnRNP A/B that also interacts with apo-B RNA, but its role in editing is currently speculative.
We previously identified a complementing activity in partially purified baboon kidney extracts that functionally complemented apobec-1 (23). This activity had a native molecular mass of 65 ± 10 kDa and physically interacted with apobec-1 in an in vitro binding assay. However, the ability of the complementing activity to bind to apo-B mRNA has not been established. In the present study, we demonstrate that the complementing activity binds to apo-B mRNA in the absence of apobec-1 and that binding requires an intact mooring sequence. Our results suggest that this protein may represent the RNA-binding subunit of the holoenzyme.
MATERIALS AND METHODS
Plasmids.
All plasmids used in this study have been described previously (10). Plasmid pB2 contains 280 bp of baboon apo-B100 cDNA (nt 6504 to 6784), and plasmid 124 is a triple mooring sequence mutant of pB2 (10). Plasmid Luc 1 contains 480 nt of wild-type luciferase cDNA subcloned into pSK+ (Stratagene), and plasmids Luc 3 and Luc 4 are editing cassette translocation derivatives of Luc 1. For bacterial expression, the rat apobec-1 cDNA was cloned in pQE32 (Qiagen) as a His6-tagged protein (23). The apobec-1 cDNA was also cloned in pGEM3zf+ (Promega) for in vitro transcription and translation.
Synthesis of RNAs and radiolabeled probes.
Linearized plasmid DNAs were transcribed with T7 or SP6 RNA polymerase to generate sense or antisense transcripts, respectively. Large-scale RNA synthesis was done with a Ribomax RNA transcription kit (Promega). Radiolabeled probes were synthesized in the presence of [α-32P]UTP and purified by Sephadex G-50 spin-column chromatography. apo-B RNA probes (280 nt) contained sequences from nt 6531 to 6782 (9).
Production of recombinant apobec-1.
Recombinant His6-tagged apobec-1 was purified from IPTG (isopropyl-β-d-galactopyranoside)-induced bacterial cultures by using Ni-nitrilotriacetic acid chromatography as described earlier (23). For far-Western analysis, synthetic apobec-1 RNA was translated in vitro in the presence of [35S]cysteine by using a rabbit reticulocyte translation kit (Promega) according to manufacturer’s instructions. Proteins were separated from unincorporated [35S]cysteine by Sephadex G-50 spin-column chromatography.
Purification of complementing activity.
Complementing activity was partially purified from baboon kidney whole-cell extracts by precipitation with 15 to 30% ammonium sulfate followed by gel filtration on Sephacryl S300 column as described previously (23). Active fractions were dialyzed against buffer D (20 mM HEPES [pH 7.9], 2.5 mM MgCl2, 100 mM KCl, 20% glycerol, 0.5 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride) prior to RNA affinity chromatography.
RNA affinity chromatography.
Biotinylated RNA transcripts were immobilized on Streptavidin beads (Dynal, Inc.) as described by the manufacturer. Briefly, biotinylated RNA (5 μg) was incubated with 1 mg of Streptavidin-coated beads in buffer B (5 mM Tris-Cl, pH 7.5; 0.5 mM EDTA, 1 M NaCl) for 1 h at room temperature. The unbound fraction was removed, and the beads were washed 10 times in buffer B. The RNA affinity beads were equilibrated in buffer E (buffer D containing 0.5% Nonidet P-40, heparin [0.2 mg/ml], tRNA [0.02 mg/ml], and salmon sperm DNA [0.2 mg/ml]). All subsequent procedures were performed at 4°C. Protein (20 mg) from the gel filtration fraction was incubated with the affinity resin for 2 h. Unbound material was removed, and the resin was washed with 100 volumes of buffer E. Bound proteins were step eluted with buffer D containing 0.2, 0.5, or 1 M NaCl as described in the figure legends.
Assays.
In vitro editing assays were performed as previously described (11, 23). Complementing activity (10 μg of the gel filtration fraction or ∼1 ng of the 0.5 M NaCl eluates from the RNA affinity column) was assayed in reaction mixtures containing 1 ng of synthetic apo-B RNA and 1 μg of recombinant His6-tagged apobec-1. The apo-B48 and apo-B100 primer extension products were quantified by using the NIH ImageQuant software.
For UV cross-linking, proteins eluted from RNA affinity columns were incubated with 1 ng of 32P-labeled RNA for 1 h at 30°C in a final volume of 30 μl containing buffer E. Reaction mixtures were UV irradiated at 254 nm for 10 min (Stratalinker 1800) in a 96-well tissue culture plate (Costar). The reaction mixtures were then treated with RNase A (2 mg/ml) for 30 min at 37°C. Samples were analyzed on 7.5, 8, or 10% gels by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as indicated in the legends and then subjected to autoradiography. Based on a calibration curve of the log molecular mass of known protein standards, the cross-linked protein, p65, migrated with a molecular mass ranging from 64.7 to 66 kDa in different experiments. Figures were generated with ScanWizard Microscan 1.05 (Microtek) and Adobe Photoshop 3.05.
For far-Western analysis, purified proteins were resolved by SDS–8% PAGE and transferred to polyvinylidene difluoride membranes. The proteins immobilized on the filters were denatured in 6 M guanidine hydrochloride in buffer E for 1 h at room temperature. Proteins were renatured by diluting the denaturation buffer 1:1 with buffer E for 12 cycles of 10 min each (19). Filters were blocked overnight and incubated with 35S-labeled apobec-1 (5 × 105 cpm/ml) in buffer E. Membranes were washed three times in buffer E for 60 min, dried, and autoradiographed.
RESULTS
Binding of complementing activity to apo-B mRNA requires an intact mooring sequence.
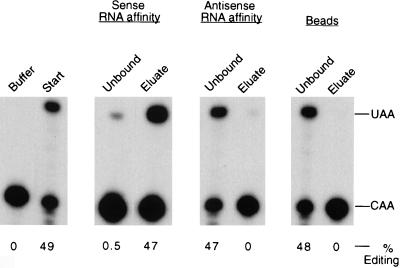
To determine whether the complementing activity binds to apo-B mRNA, we performed RNA affinity chromatography with the sense and antisense strands of a 280-nt baboon apo-B RNA spanning the editing site. Affinity resins were generated by immobilizing in vitro-transcribed biotinylated RNAs on Streptavidin-coated beads. Partially purified complementing activity was incubated with resin alone, the apo-B RNA sense affinity resin, or the antisense RNA resin for 2 h, and the unbound proteins were removed. The beads were washed extensively, and the bound proteins were eluted with 0.5 M NaCl. Aliquots of the starting material (10 μg), the unbound fractions (10 μg), and the eluates (∼1 ng) were analyzed for complementing activity (Fig. 1). More than 95% of the complementing activity in the starting material bound to the wild-type apo-B RNA affinity resin, and the bound activity was eluted with 0.5 M salt. There was no significant binding of the complementing activity to the antisense RNA affinity resin or to the beads alone.
FIG. 1.
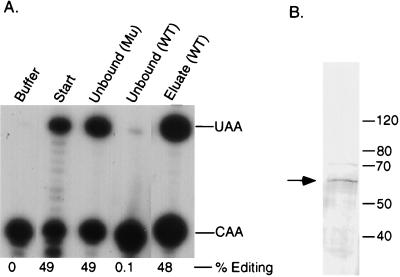
Complementing activity interacts with apo-B RNA in vitro. Partially purified complementing activity (10 mg) was incubated with the sense or antisense apo-B RNA affinity resins or with the beads alone as described in Materials and Methods. After an extensive washing, proteins were eluted with 0.5 M NaCl. The unbound fractions (unbound protein plus the first wash, 10 μg) and eluates (∼1 ng) were assayed for complementing activity in the presence of purified His6-tagged apobec-1. The buffer control and the starting material (10 μg) are shown in the first panel. The positions of the primer extension products from the edited (UAA) and unedited (CAA) RNAs are marked on the right. The data were quantified by using NIH ImageQuant software, and the results are expressed as the percent editing, i.e., UAA/(UAA + CAA).
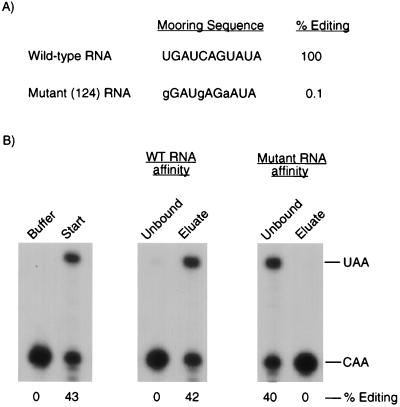
Previous studies have shown that the mooring sequence (UGAUCAGUAUA) is required for the editing of apo-B mRNA (29). To test whether the mooring sequence is necessary for the binding of complementing activity to apo-B RNA, we used a mutant RNA that contains three mutations (gGAUgAGaAUA) and that was not detectably edited by the native editing enzyme (10). Similar results were obtained when editing assays were performed with His6-tagged apobec-1 and partially purified complementing protein (Fig. 2A). When the triple-mutant RNA was used as the ligand in RNA affinity chromatography, there was no significant binding of the complementing activity to the affinity resin (Fig. 2B). Similar experiments were done with a double mutant (UGgUCAGUuUA) RNA that was edited to 13% of the wild-type RNA. Only 20% of the complementing activity bound to the double-mutant RNA affinity column (data not shown).
FIG. 2.
Binding of the complementing activity to apo-B RNA requires the mooring sequence. (A) The percent editing of the wild-type and triple-mutant apo-B RNAs was determined in editing reaction mixtures containing partially purified complementing activity and His6-tagged apobec-1. (B) In vitro binding experiments were performed with partially purified complementing activity and the wild-type (WT) or mutant apo-B RNA affinity columns. The unbound and eluted proteins were assayed for complementing activity as described in the legend to Fig. 1.
The complementing activity copurifies with a 65-kDa protein that specifically cross-links to apo-B RNA.
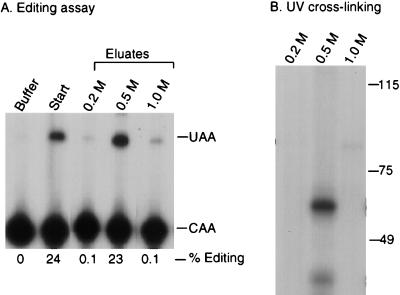
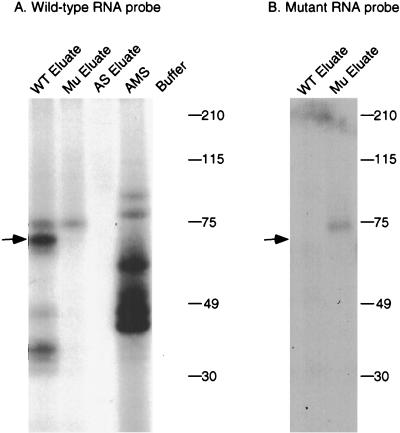
To further characterize the complementing activity, UV cross-linking studies were performed. Proteins bound to the wild-type RNA affinity resin were step eluted with 0.2, 0.5, and 1 M NaCl. As shown in Fig. 3A, most of the complementing activity eluted in the 0.5 M fraction. The eluted proteins were incubated with 32P-labeled apo-B RNA, followed by cross-linking with short-wave UV. After treatment with RNase A, the samples were analyzed by SDS-PAGE and autoradiography. A prominent protein of 65 kDa that cross-linked to apo-B mRNA was detected in the 0.5 M-salt-eluted fraction but not in the 0.2 or 1.0 M fractions (Fig. 3B). This protein was not detected in the 0.5 M eluates from the triple mutant or the antisense RNA affinity columns (Fig. 4A). The original ammonium sulfate fraction did not contain detectable amounts of this protein (Fig. 4A). This may be due to low abundance since the specific activity of this fraction was ∼10,000-fold less than the 0.5 M eluate. The 65-kDa protein did not cross-link to the 32P-labeled triple-mutant apo-B RNA (Fig. 4B). Several smaller proteins ranging from 35 to 45 kDa copurified with the 65-kDa protein and also cross-linked to apo-B mRNA (Fig. 3B and 4). This group of proteins bound to apo-B mRNA in a sequence-specific manner since they were not detected in the eluate from the mutant RNA column (Fig. 4). However, these proteins varied in proportion between different preparations. We also occasionally detected a cross-linking protein of 75 kDa in the eluates from the wild-type and triple-mutant affinity resins, but this finding was not reproducible (see Fig. 5 and 6).
FIG. 3.
UV-crosslinking of a 65-kDa protein correlates with complementing activity. (A) RNA affinity chromatography was performed as described in Materials and Methods. Editing assays were performed on the 0.2, 0.5, or 1 M-salt-eluted fractions from the wild-type RNA affinity resin. Editing assays were performed as described in the legend to Fig. 1. (B) UV cross-linking was performed with the salt-eluted fractions. Proteins were incubated with 32P-labeled wild-type RNA and exposed to short-wave UV light for 10 min. After treatment with RNase A, samples were analyzed by SDS–7.5% PAGE and autoradiography. Molecular size standards (in kilodaltons) are indicated on the right of each panel. The figure was generated by using ScanWizard 1.05 (Microtek) and Adobe Photoshop 3.05.
FIG. 4.
UV cross-linking of a 65-kDa protein in the eluates from RNA affinity resins. UV cross-linking studies were performed as described in the legend to Fig. 3 with the 15 to 30% ammonium sulfate fraction (AMS) or with proteins eluted from the wild-type (WT eluate), triple-mutant (Mu eluate), or antisense (AS eluate) RNA affinity columns. 32P-labeled wild-type apo-B RNA (A) or mutant apo-B RNA (B) was used as the probe. Samples were analyzed by SDS–10% PAGE and autoradiography.
FIG. 5.
Competition for cross-linking of the 65-kDa protein. The 32P-labeled wild-type apo-B RNA was incubated with proteins eluted from the wild-type RNA affinity column in the absence of competitor (none) or in the presence of a 2.5-, 5-, 7.5-, 10-, or 50-fold molar excess of unlabeled wild-type apo-B RNA; a 500- or 1,000-fold molar excess of the triple-mutant RNA; or a 500-fold molar excess of the antisense apo-B RNA. UV cross-linking and SDS–8% PAGE were performed as described in the legend to Fig. 3. The position of the 65-kDa protein is indicated by an arrow. Molecular size markers (in kilodaltons) are indicated on the right.
FIG. 6.
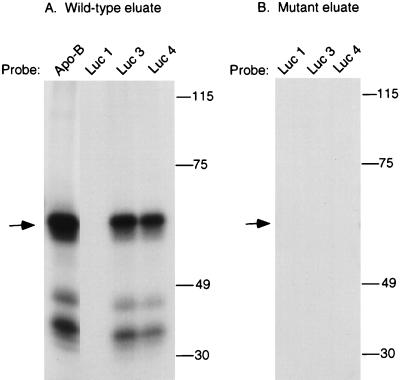
UV cross-linking to luciferase translocation mutants. UV cross-linking experiments were performed with proteins eluted from the wild-type (A) or mutant (B) apo-B RNA affinity columns. Proteins were incubated with 32P-labeled apo-B RNA, wild-type luciferase RNA (Luc 1), or the luciferase translocation mutants Luc 3 and Luc 4, which contain 15 and 25 nt of the apo-B editing cassette, respectively. UV cross-linking was performed as described in Materials and Methods, and the samples were analyzed by SDS–7.5% PAGE. The position of the molecular size standards (in kilodaltons) are indicated on the right, and the position of the 65-kDa protein is indicated by an arrow.
To assess specificity, competition experiments were performed with unlabeled synthetic transcripts (Fig. 5). Cross-linking of the 65-kDa protein and the 35- to 45-kDa proteins was significantly reduced by a 10-fold molar excess and abolished by a 50-fold molar excess of wild-type apo-B RNA. In contrast, a 500- to 1,000-fold molar excess of the triple-mutant apo-B RNA was required to effectively compete for cross-linking. No competition was effected by a 500-fold molar excess of the antisense RNA.
Cross-linking of the 65-kDa protein is dependent on the mooring sequence.
To determine whether the mooring sequence was sufficient for cross-linking of the 65-kDa protein, apo-B editing cassettes were inserted into a heterologous RNA. Luc 1 is a 480-nt luciferase RNA with an AU content similar to that of apo-B mRNA, which is 78% AU-rich in the region of the editing site (10). Luc 3 contains the mooring sequence inserted 5 nt downstream of a cytidine, whereas Luc 4 contains both the mooring sequence and 10 nt of upstream apo-B sequence. We previously showed that insertion of these cassettes induced editing of an upstream cytidine by the enterocyte holoenzyme (10). Similar results were obtained with the reconstituted enzyme, which edited both Luc 3 and Luc 4 RNAs to 20% relative to wild-type apo-B RNA (data not shown). As shown in Fig. 6A, the 65-kDa protein and the 35- to 45-kDa proteins that eluted from the wild-type RNA affinity column cross-linked to Luc 3 and Luc 4 RNAs but not to Luc 1 RNA. No cross-linking proteins were detected in the eluate from the mutant apo-B RNA affinity column (Fig. 6B).
Identification of a 65-kDa protein that interacts with apobec-1 and copurifies with the complementing activity.
For activity-band correlation, the affinity-purified proteins were resolved on SDS-PAGE and analyzed by silver staining. Although several proteins were common to the eluates from the wild-type and mutant RNA affinity columns, a protein of 65 kDa was present only in the wild-type eluate (Fig. 7A). UV cross-linking reactions that were run in parallel lanes of the same gel showed comigration of the 65-kDa protein with the cross-linked band (Fig. 7A). Although the 35- to 45-kDa proteins were detected by cross-linking, these proteins were not detected by silver staining. In Fig. 7B, we performed a far-Western analysis using radiolabeled apobec-1 as a probe. Proteins were resolved by SDS-PAGE and transferred to PVDF membranes. After denaturation and renaturation, the membranes were incubated with in vitro translated 35S-labeled apobec-1. Apobec-1 bound only to a 65-kDa protein that was present in the eluate from the wild-type apo-B RNA affinity column but not in the eluate from the mutant column (Fig. 7B). The labeled apobec-1 did not bind any proteins in the less-purified gel filtration fraction (data not shown). Interestingly, apobec-1 also did not bind to the 35- to 45-kDa proteins which cross-linked to apo-B mRNA.
FIG. 7.
Correlation of complementing activity with a 65-kDa protein. (A) Proteins eluted from the wild-type (WT) or mutant RNA affinity columns were analyzed either directly or after UV cross-linking to labeled apo-B RNA. Untreated and UV cross-linked samples were electrophoresed in parallel on the same SDS–8% PAGE gel. One-half of the gel was silver stained, while the other half was developed by autoradiography. The position of the 65-kDa protein is indicated by an arrow. (B) Results of far-Western analysis with [35S]cysteine-labeled apobec-1 as the probe. The affinity-purified proteins from the wild-type or mutant RNA columns were resolved by SDS–8% PAGE and transferred to polyvinylidene difluoride membranes as described in Materials and Methods. Proteins were subjected to multiple cycles of denaturation and renaturation and probed with [35S]cysteine-labeled apobec-1.
Further purification of the complementing activity by RNA affinity chromatography.
In addition to the 65-kDa protein, the eluate from the wild-type RNA column contained several other prominent proteins of 123, 118, 110, and 75 kDa which also bound to the mutant RNA column (Fig. 7A). To further purify the complementing activity, whole-cell extracts from 20 g of baboon kidney were fractionated by ammonium sulfate precipitation (15 to 30%) and gel filtration chromatography on Sephacryl S300. Active fractions were precleared by incubation with the mutant apo-B RNA affinity beads, which did not deplete complementing activity (Fig. 8A). The unbound proteins were loaded onto the wild-type RNA affinity resin, and the bound proteins were eluted with 0.5 M salt. Due to the low yield, the amount of protein in this fraction was roughly estimated by silver staining in comparison with known standards. As shown in Table 1, this scheme resulted in an approximately 140,000-fold purification with respect to the starting material, with an overall recovery of 3.6%. The purified complementing activity contained a single prominent protein of 65 kDa, which was detected by silver staining (Fig. 8B). The other proteins were eliminated or greatly reduced by the preclearing step, which suggests that they are not required for complementing activity in stoichiometric amounts.
FIG. 8.
Purification of complementing activity by RNA affinity chromatography. Partially purified complementing activity was incubated with the triple-mutant (Mu) RNA affinity resin. After this preclearing step, the unbound fraction was loaded onto the wild-type (WT) RNA affinity resin. Bound proteins were eluted with 0.5 M-salt-containing buffer. (A) The unbound fractions and the 0.5 M-salt-eluted fraction were assayed for complementing activity. (B) An aliquot of the affinity-purified fraction was also resolved by SDS–8% PAGE and analyzed by silver staining.
TABLE 1.
Purification of complementing activity from baboon kidney extracta
| Step | Total protein (mg) | Total activityb (U) | Sp act (U/mg) | Purification (fold) | Recovery (%) |
|---|---|---|---|---|---|
| 1. WCE | 952 | 15,001 | 15.7 | 1 | 100 |
| 2. 15 to 30% AMS | 115 | 12,340 | 107 | 6.8 | 82 |
| 3. Sephacryl S300 | 54 | 9,600 | 178 | 11.3 | 64 |
| 4. RNA affinity | 0.00025c | 550 | 2,200,000 | 140,127 | 3.6 |
Complementing activity was purified from whole-cell extracts (WCE) obtained from 20 g of baboon kidney by precipitation with 15 to 30% ammonium sulfate (AMS) and gel filtration on Sephacryl S300. For RNA affinity chromatography, proteins were precleared with a mutant apo-B RNA column. The unbound fraction was incubated with the wild-type RNA affinity column, and the bound proteins were eluted with 0.5 M NaCl.
One unit is defined as the amount of complementing activity that edits 1 fmol of apo-B RNA/h in the presence of recombinant apobec-1.
Because of the low yield, the total protein in this fraction was estimated by silver staining in comparison with known standards.
DISCUSSION
Although it is well established that the sequence-specific recognition of apo-B mRNA by the editing enzyme requires the mooring sequence, the mechanism by which the holoenzyme recognizes its target RNA is not well understood. Apobec-1, the catalytic subunit, was reported to have a novel RNA-binding domain which overlaps with residues in the active site (24). Because this RNA-binding activity was of low specificity with a preference for AU-rich sequences, it was suggested that the recognition of apo-B mRNA requires the complementing activity. There was, however, no evidence for the validity of this model. The present study is the first direct demonstration that the complementing activity has an RNA-binding activity and that this activity binds to apo-B mRNA in a mooring-sequence-specific manner. Using RNA affinity chromatography, we show that the complementing activity recognizes the sequences critical for the editing of apo-B mRNA in the absence of apobec-1. We had previously provided evidence for the physical association of complementing activity with recombinant apobec-1 in vitro in the absence of apo-B mRNA (23). Taken together, these results support the hypothesis that the complementing activity may represent the RNA-binding subunit of the editing enzyme which interacts with apobec-1 and docks it to edit apo-B mRNA. However, our data do not eliminate the possibility that apobec-1 or additional proteins may increase the affinity or specificity of the complementing activity for apo-B mRNA.
Another significant finding is that RNA affinity chromatography was highly effective in purification of the complementing activity. Based on SDS-PAGE and silver staining, the most purified fraction contained a prominent protein of 65 kDa. This result is in good agreement with our previous study which showed that the native molecular mass of the complementing activity is 65 ± 10 kDa by gel filtration chromatography. Several other lines of evidence suggest that the complementing activity correlates with the presence of a 65-kDa protein. Both the complementing activity and a 65-kDa protein bound to the wild-type apo-B RNA affinity column but not to a triple-mutant RNA column. Based on silver staining, this 65-kDa protein was the only detectable difference between eluates from the wild-type and mutant columns. Secondly, a 65-kDa protein that specifically UV cross-linked to apo-B mRNA was detected in the eluate from the wild-type RNA affinity column but not in that from the mutant RNA column. Thirdly, the affinity-purified fraction contained a 65-kDa protein that interacted with apobec-1 in a far-Western analysis. These results strongly suggest that the complementing activity is a 65-kDa protein that has the ability to bind to apo-B mRNA and interact with apobec-1. However, cloning and expression studies will be required in the future to establish that these functions are encoded by a single polypeptide.
Our competition studies show that the mooring sequence is essential for UV cross-linking of a 65-kDa protein compared to the flanking sequences which are identical in both the wild-type and the mutant RNAs. More importantly, introduction of the mooring sequence as a 15- or 25-nt cassette into the AU-rich background of luciferase RNA induced cross-linking of this protein. This construct was comparable to apo-B mRNA since the sequences flanking the editing site are rich in A and U residues. These results suggest that cross-linking of the 65-kDa protein is not solely due to AU content and requires mainly the presence of the mooring sequence. However, whether an AU-rich background contributes to sequence-specific recognition is an open question. There is no evidence for the existence of any secondary structure in the 74% AU-rich region of apo-B mRNA encompassing the editing site. It is possible that this lack of secondary structure may be critical in directing the editing holoenzyme to the editing site in vivo.
Our finding that a 65-kDa protein specifically cross-links to apo-B mRNA is consistent with previous studies. UV cross-linking of proteins of 60 to 66 kDa (p60) in rat tissue extracts has been reported (16, 26). We currently do not know whether the 65-kDa protein in our affinity-purified fraction is the same as p60. First, the ability of p60 to complement apobec-1 could not be tested since these studies were done with partially purified holoenzyme or assembled editosomes, both of which contain apobec-1. Secondly, the binding site of p60 has not been well defined. Mutagenesis of nt 6671 to 6674, the first 4 nt of the mooring sequence, abolished or reduced editing. However, single point mutations in this region resulted in only a modest (<2-fold) reduction in p60 binding as determined by competition experiments (26). Finally, the studies on p60 were done with partially purified holoenzyme, and the binding site of p60 alone has not been established.
We also detected a group of proteins ranging in size from 35 to 45 kDa that UV cross-linked to apo-B mRNA and exhibited binding characteristics similar to that of the 65-kDa protein. The 35- to 45-kDa proteins did not cross-link consistently between experiments, were not detectable by staining with silver (Fig. 7A) or Coomassie blue, and were not generated during the cross-linking reaction (23a). These proteins may represent breakdown products of the 65-kDa protein. This hypothesis is consistent with an earlier study of the rat enterocyte holoenzyme which suggested that p40 may represent the proteolytic product of p60 (26). This may also explain why the 35- to 45-kDa proteins did not interact with apobec-1 in a far-Western analysis. Alternatively, these proteins may be associated with the 65-kDa protein in substoichiometric amounts.
The editing complex in baboon (9) and rat (26) enterocytes was shown to possess a minimal molecular mass of 125 ± 5 kDa. Apobec-1 is a 27-kDa protein which can dimerize in vitro (20). The combined sizes of an apobec-1 dimer and the 65-kDa complementing protein add up to the expected size of the holoenzyme. However, the molecular composition and size of the holoenzyme remain controversial. Smith and colleagues have shown that editing may involve a macromolecular complex or editosome (30). In support of this hypothesis, an increasing number of proteins have been shown to interact with apo-B mRNA or apobec-1 (34, 35). In addition to the complementing activity, these include p60 and p40, which cross-link to apo-B mRNA; ABBP-1, which interacts with apobec-1 (21); and a 240-kDa protein that is associated with the editosome and may regulate the efficiency of editing (28). However, the role of these proteins remains ambiguous. Although our results indicate that the mooring sequence is specifically recognized by the complementing activity and that a minimal holoenzyme is sufficient for editing in vitro, they do not rule out the role of additional proteins in the sequence-specific recognition of apo-B mRNA. The identification of the editing site in vivo is likely to be more complex since editing of the ∼14,500-nt apo-B mRNA occurs in the middle of exon 26, which is 7.5 kb long.
ACKNOWLEDGMENTS
This work was supported by NIH grant HL-45478 and an Established Investigator Award from the American Heart Association (D.M.D.). Tissues were obtained from the Regional Primate Research Center at the University of Washington, which is supported by grant RR00166.
We thank Paul Copeland and Shigenori Murata for advice and reading of the manuscript; Bella Gorbatcheva for technical assistance; and Karen Rice, Southwest Foundation for Biomedical Research, for providing tissues.
REFERENCES
- 1.Anant S, MacGinnitie A J, Davidson N O. Apobec-1, the catalytic subunit of the mammalian apolipoprotein B mRNA editing enzyme, is a novel RNA-binding protein. J Biol Chem. 1995;270:14762–14767. [PubMed] [Google Scholar]
- 2.Backus J W, Schock D, Smith H C. Only cytidines 5′ of the apolipoprotein B mRNA mooring sequence are edited. Biochim Biophys Acta. 1994;1219:1–14. doi: 10.1016/0167-4781(94)90240-2. [DOI] [PubMed] [Google Scholar]
- 3.Backus J W, Smith H C. Apolipoprotein B mRNA sequences 3′ of the editing site are necessary and sufficient for editing and editosome assembly. Nucleic Acids Res. 1991;19:6781–6786. doi: 10.1093/nar/19.24.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backus J W, Smith H C. Three distinct RNA sequence elements are required for efficient apolipoprotein B (apoB) RNA editing in vitro. Nucleic Acids Res. 1992;20:6007–6014. doi: 10.1093/nar/20.22.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Backus J W, Smith H C. Specific 3′ sequences flanking a minimal apolipoprotein B (apoB) mRNA editing ’cassette’ are critical for efficient editing in vitro. Biochim Biophys Acta. 1994;1217:65–73. [PubMed] [Google Scholar]
- 6.Chan L. Apolipoprotein B, the major protein component of triglyceride-rich and low density lipoproteins. J Biol Chem. 1992;267:25621–25624. [PubMed] [Google Scholar]
- 7.Chen S H, Habib G, Yang C Y, Gu Z W, Lee B R, Weng S A, Silberman S R, Cai S J, Deslypere J P, Rosseneu M, et al. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon. Science. 1987;238:363–366. doi: 10.1126/science.3659919. [DOI] [PubMed] [Google Scholar]
- 8.Davies M S, Wallis S C, Driscoll D M, Wynne J K, Williams G W, Powell L M, Scott J. Sequence requirements for apolipoprotein B RNA editing in transfected rat hepatoma cells. J Biol Chem. 1989;264:13395–13398. [PubMed] [Google Scholar]
- 9.Driscoll D M, Casanova E. Characterization of the apolipoprotein B mRNA editing activity in enterocyte extracts. J Biol Chem. 1990;265:21401–21403. [PubMed] [Google Scholar]
- 10.Driscoll D M, Lakhe-Reddy S, Oleksa L M, Martinez D. Induction of RNA editing at heterologous sites by sequences in apolipoprotein B mRNA. Mol Cell Biol. 1993;13:7288–7294. doi: 10.1128/mcb.13.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Driscoll D M, Wynne J K, Wallis S C, Scott J. An in vitro system for the editing of apolipoprotein B mRNA. Cell. 1989;58:519–525. doi: 10.1016/0092-8674(89)90432-7. [DOI] [PubMed] [Google Scholar]
- 12.Driscoll D M, Zhang Q. Expression and characterization of p27, the catalytic subunit of the apolipoprotein B mRNA editing enzyme. J Biol Chem. 1994;269:19843–19847. [PubMed] [Google Scholar]
- 13.Garcia Z C, Poksay K S, Bostrom K, Johnson D F, Balestra M E, Shechter I, Innerarity T L. Characterization of apolipoprotein B mRNA editing from rabbit intestine. Arterioscler Thromb. 1992;12:172–179. doi: 10.1161/01.atv.12.2.172. [DOI] [PubMed] [Google Scholar]
- 14.Giannoni F, Bonen D K, Funahashi T, Hadjiagapiou C, Burant C F, Davidson N O. Complementation of apolipoprotein B mRNA editing by human liver accompanied by secretion of apolipoprotein B48. J Biol Chem. 1994;269:5932–5936. [PubMed] [Google Scholar]
- 15.Greeve J, Navaratnam N, Scott J. Characterization of the apolipoprotein B mRNA editing enzyme: no similarity to the proposed mechanism of RNA editing in kinetoplastid protozoa. Nucleic Acids Res. 1991;19:3569–3576. doi: 10.1093/nar/19.13.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris S G, Sabio I, Mayer E, Steinberg M F, Backus J W, Sparks J D, Sparks C E, Smith H C. Extract-specific heterogeneity in high-order complexes containing apolipoprotein B mRNA editing activity and RNA-binding proteins. J Biol Chem. 1993;268:7382–7392. [PubMed] [Google Scholar]
- 17.Hodges P, Scott J. Apolipoprotein B mRNA editing: a new tier for the control of gene expression. Trends Biochem Sci. 1992;17:77–81. doi: 10.1016/0968-0004(92)90506-5. [DOI] [PubMed] [Google Scholar]
- 18.Johnson D F, Poksay K S, Innerarity T L. The mechanism for apo-B mRNA editing is deamination. Biochem Biophys Res Commun. 1993;195:1204–1210. doi: 10.1006/bbrc.1993.2172. [DOI] [PubMed] [Google Scholar]
- 19.Kohtz J D, Jamison S F, Will C L, Zuo P, Luhrmann R, Garcia-Blanco M A, Manley J L. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 20.Lau P P, Zhu H J, Baldini A, Charnsangavej C, Chan L. Dimeric structure of a human apolipoprotein B mRNA editing protein and cloning and chromosomal localization of its gene. Proc Natl Acad Sci USA. 1994;91:8522–8526. doi: 10.1073/pnas.91.18.8522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lau P P, Zhu H J, Nakamuta M, Chan L. Cloning of an apobec-1-binding protein that also interacts with apolipoprotein B mRNA and evidence for its involvement in RNA editing. J Biol Chem. 1997;272:1452–1455. doi: 10.1074/jbc.272.3.1452. [DOI] [PubMed] [Google Scholar]
- 22.MacGinnitie A J, Anant S, Davidson N O. Mutagenesis of apobec-1, the catalytic subunit of the mammalian apolipoprotein B mRNA editing enzyme, reveals distinct domains that mediate cytosine nucleoside deaminase, RNA binding, and RNA editing activity. J Biol Chem. 1995;270:14768–14775. [PubMed] [Google Scholar]
- 23.Mehta A, Banerjee S, Driscoll D M. Apobec-1 interacts with a 65-kDa complementing protein to edit apolipoprotein-B mRNA in vitro. J Biol Chem. 1996;271:28294–28299. doi: 10.1074/jbc.271.45.28294. [DOI] [PubMed] [Google Scholar]
- 23a.Mehta, A., and D. M. Driscoll. Unpublished observations.
- 24.Navaratnam N, Bhattacharya S, Fujino T, Patel D, Jarmuz A L, Scott J. Evolutionary origins of apoB mRNA editing: catalysis by a cytidine deaminase that has acquired a novel RNA-binding motif at its active site. Cell. 1995;81:187–195. doi: 10.1016/0092-8674(95)90328-3. [DOI] [PubMed] [Google Scholar]
- 25.Navaratnam N, Morrison J R, Bhattacharya S, Patel D, Funahashi T, Giannoni F, Teng B B, Davidson N O, Scott J. The p27 catalytic subunit of the apolipoprotein B mRNA editing enzyme is a cytidine deaminase. J Biol Chem. 1993;268:20709–20712. [PubMed] [Google Scholar]
- 26.Navaratnam N, Shah R, Patel D, Fay V, Scott J. Apolipoprotein B mRNA editing is associated with UV crosslinking of proteins to the editing site. Proc Natl Acad Sci USA. 1993;90:222–226. doi: 10.1073/pnas.90.1.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Powell L M, Wallis S C, Pease R J, Edwards Y H, Knott T J, Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987;50:831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- 28.Schock D, Kuo S R, Steinburg M F, Bolognino M, Sparks J D, Sparks C E, Smith H C. An auxiliary factor containing a 240-kDa protein complex is involved in apolipoprotein B RNA editing. Proc Natl Acad Sci USA. 1996;93:1097–1102. doi: 10.1073/pnas.93.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shah R R, Knott T J, Legros J E, Navaratnam N, Greeve J C, Scott J. Sequence requirements for the editing of apolipoprotein B mRNA. J Biol Chem. 1991;266:16301–16304. [PubMed] [Google Scholar]
- 30.Smith H C, Kuo S R, Backus J W, Harris S G, Sparks C E, Sparks J D. In vitro apolipoprotein B mRNA editing: identification of a 27S editing complex. Proc Natl Acad Sci USA. 1991;88:1489–1493. doi: 10.1073/pnas.88.4.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teng B, Burant C F, Davidson N O. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993;260:1816–1819. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- 32.Teng B, Davidson N O. Evolution of intestinal apolipoprotein B mRNA editing. Chicken apolipoprotein B mRNA is not edited, but chicken enterocytes contain in vitro editing enhancement factor(s) J Biol Chem. 1992;267:21265–21272. [PubMed] [Google Scholar]
- 33.Yamanaka S, Poksay K S, Balestra M E, Zeng G Q, Innerarity T L. Cloning and mutagenesis of the rabbit ApoB mRNA editing protein. A zinc motif is essential for catalytic activity, and noncatalytic auxiliary factor(s) of the editing complex are widely distributed. J Biol Chem. 1994;269:21725–21734. [PubMed] [Google Scholar]
- 34.Yang Y, Kovalski K, Smith H C. Partial characterization of the auxiliary factors involved in apolipoprotein B mRNA editing through APOBEC-1 affinity chromatography. J Biol Chem. 1997;272:27700–27706. doi: 10.1074/jbc.272.44.27700. [DOI] [PubMed] [Google Scholar]
- 35.Yang Y, Smith H C. In vitro reconstitution of apolipoprotein B RNA editing activity from recombinant APOBEC-1 and McArdle cell extracts. Biochem Biophys Res Commun. 1996;218:797–801. doi: 10.1006/bbrc.1996.0142. [DOI] [PubMed] [Google Scholar]