Abstract
Nonmethylated CpG islands are generally located at the 5′ ends of genes, but a CpG island in the mouse major histocompatibility complex class II I-Aβ gene is remote from the promoter and covers exon 2. We have found that this CpG island includes a novel intronic promoter that is active in embryonic and germ cells. The resulting transcript potentially encodes a severely truncated protein which would lack the signal peptide and external β1 domains. The functional significance of the internal CpG island may be to facilitate gene conversion, thereby sustaining the high level of polymorphism seen at exon 2. Deletions of the I-Aβ CpG island promoter reduce transcription and frequently lead to methylation of the CpG island in a transgenic mouse assay. These and other results support the idea that all CpG islands arise at promoters that are active in early embryonic cells.
CpG islands are discrete clusters of nonmethylated CpG dinucleotides that altogether make up about 1 to 2% of the mammalian genome (4, 7). About 56% of sequenced human genes have CpG islands near their 5′ ends, including all those that are ubiquitously expressed (housekeeping genes) plus many genes with a tissue-restricted pattern of expression (4, 24, 25, 43). Promoters are normally located at the upstream edge of the CpG island, with the result that one or more of the 5′ exons of the gene generally fall within the island region. In spite of the high density of the methylatable sequence CpG, most CpG islands are nonmethylated in all tissues, including those in which the gene is silent. A small proportion of islands become methylated during development, however, notably those associated with genes of the inactive X chromosome and some parentally imprinted genes (22, 55, 63).
The existence of CpG islands depends on their ability to remain methylation free in the germ line. In the event of methylation, CpGs would be lost, as 5-methylcytosine is a hot spot for mutation and the island region would eventually become indistinguishable from bulk DNA (34). There is evidence for CpG island loss of this kind at the α1-globin pseudogene in humans (8). What is the mechanism which normally protects this subset of CpGs from methylation? The simple idea that CpG islands are intrinsically unmethylatable is opposed by the finding that some become methylated during development and many acquire methylation in permanent cell lines or tumors (3, 31, 33, 41). Similarly, there is little evidence for restricted access of the DNA methyltransferase to CpG island chromatin, as these regions are in fact hyperaccessible to exogenous nucleases (2). Progress in understanding the origin of CpG islands has been made through studies of the mouse and hamster aprt genes. Macleod et al. (47) located the boundaries of the methylation-free island, the positions of nucleosomes, and the sites of protein factor binding. Three GC boxes that interacted with the transcription factor Sp1 in vitro (20) were found to be occupied in vivo, and site-directed mutagenesis showed that these sites were essential to protect the CpG island from methylation. The Sp1 binding sites that were mutated in this study had been previously shown to drive transcription in vitro (20). Mutation of GC boxes within hamster aprt transgenes also resulted in methylation of this CpG island (11).
Although the results for hamster and mouse aprt genes were similar, two somewhat different models were put forward to account for them. Brandeis et al. (11) proposed that Sp1 binding may exert a localized demethylating effect that is independent of its effect on transcription, by directly protecting CpGs from the DNA methyltransferase or by interacting with other factors to actively demodify methylated CpG. Demethylation of the entire island region would in this case depend on occupation of GC boxes over the length of the CpG island. On the other hand, Macleod et al. (47) suggested that the methylation-free status of the aprt CpG island depended on the presence of a functional promoter at the 5′ edge of the island. This proposal is compatible with the preferential location of CpG islands at promoters, although it does not account for their origin in detail.
If all CpG islands are dependent on promoter function, then they should be associated with demonstrable promoter activity in vivo. This prediction is easily met by CpG islands at housekeeping genes, which are transcribed in all tissues. It is less obvious, however, that CpG islands at highly tissue restricted genes or those that are remote from a known promoter should maintain their methylation-free status via a mechanism that depends on transcription. To test the “CpG island promoter” hypothesis, we sought a model gene with highly tissue restricted expression that possessed a CpG island. It has been noted previously that many of the human and mouse major histocompatibility locus (MHC) genes contain CpG-rich regions (64). Class II genes encode α and β peptide chains which form heterodimers on the cell surface, and their expression is primarily restricted to cells of the immune system, such as B cells and macrophages, although low levels of antigen have been found in several other cell types (36, 61). An analysis of human and mouse genomic DNA sequences for class II genes shows that only the β-chain genes have CpG-rich regions and in these cases the CpG island does not include the gene promoter but is located well downstream of it, spanning the second exon. It appeared unlikely at first sight that the presence of the CpG island could be related to transcription. We therefore chose to examine the mouse MHC class II I-Aβ gene in detail, as it presents an interesting example of a gene, expressed in only a subset of tissues, which contains a CpG island in an apparently misplaced location relative to the gene’s promoter.
MATERIALS AND METHODS
Nucleic acid preparation.
DNA was isolated from various mouse tissues by homogenization in 50 mM Tris-HCl (pH 8)–100 mM EDTA–100 mM NaCl. Proteinase K was added to 100 μg/ml, and sodium dodecyl sulfate (SDS) was added to 1%. Samples were incubated for 16 h and then extracted with 1:1 phenol-chloroform and precipitated with isopropanol. RNA was extracted by the method of Chomczynski and Sacchi (13). RNA from 25 3.5-day (3.5d) blastocysts was prepared by a modification of the above method (37).
Analysis of gene expression. (i) Northern blot hybridization.
Generally 10 μg of total RNA/track was run on 1.2% agarose-formaldehyde gels, blotted onto Hybond N+ nylon membranes (Amersham), and UV cross-linked. Random-primed DNA probes were made (21) by using a kit (Boehringer). Hybridization of filters was at 68°C in 0.5 M NaPi (pH 7.2)–7% SDS–1 mM EDTA (14). Filters were washed at 68°C in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS for 30 min twice and then three times for 30 min in 0.1× SSC–0.1% SDS at 68°C. Hybridized filters were exposed to Kodak XAR-5 autoradiographic film. Blots were rehybridized with an S26 ribosomal protein probe (65).
(ii) RNase protection.
A SmaI subclone from intron 1 of the I-Aβd gene (positions 1385 to 1562 [see Fig. 3c]) was cloned into BlueScribe vector (Stratagene). The vector was linearized with HindIII and used to make a 32P-labeled RNA probe by using T7 RNA polymerase. The in vitro reaction mix contained 1 μg of DNA, 40 mM Tris-HCl (pH 7.9), 6 mM MgCl2, 2 mM spermidine, 0.5 mM GTP, UTP, and ATP, 100 μM CTP, 10 mM dithiothreitol, 40 U of RNasin (Promega), 50 μCi of [32P]CTP (3,000 Ci/mmol; Amersham), and 20 U of T7 RNA polymerase (Promega) in 20 μl. Labeled RNA was purified on an acrylamide-urea gel and eluted from the gel slice in 0.5 M ammonium acetate–1 mM EDTA at 37°C for 3 h; 100,000 cpm of labeled RNA was used in each RNase protection reaction. Generally 40 μg of total RNA from mouse tissues or tRNA was used. RNA samples were ethanol precipitated with the probe, and the dried pellet was resuspended in 20 μl of 40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 7)–200 mM sodium acetate–80% formamide and denatured at 90°C for 3 min before incubation at 50°C for 16 h. Reaction mixes were placed on ice, and 300 μl of RNase mix (10 mM Tris-HCl [pH 7.5], 5 mM EDTA, 300 mM NaCl, 12 μg of RNase A, 300 U of RNase T1 [Gibco/BRL]) was added. Samples were incubated at 37°C for 15 to 60 min before addition of 10 μl of 20% SDS and 50 μg of proteinase K and continuation of incubation at 37°C for 15 min before phenol-chloroform extraction and ethanol precipitation with 10 μg of carrier tRNA. Dried samples were resuspended in 80% formamide-dye mix and run on 10% acrylamide–7 M urea gels, which were dried and exposed to Kodak XAR-5 autoradiograph film.
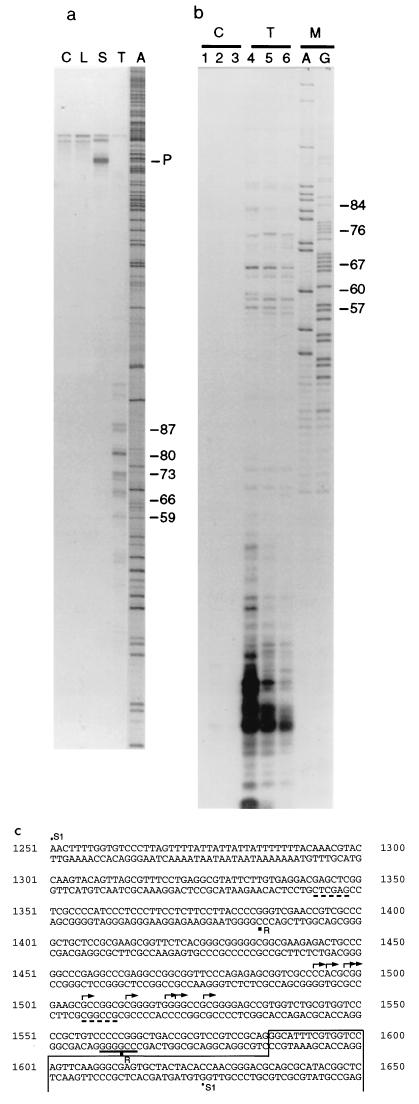
FIG. 3.
Mapping of transcriptional start sites originating from the I-Aβ CpG island. (a) Autoradiograph showing the results of S1 mapping using a 379-bp end-labeled DNA probe (see panel c) annealed with 50 μg of total RNA in each reaction. Samples were control tRNA (C), liver RNA (L), spleen RNA (S), testis RNA (T), and marker (A). The arrowed band (P) indicates the position of the full-sized probe which is fully protected in the spleen sample. The protected bands in the testis RNA track are numbered according to distance (in bases) from the intron 1/exon 2 boundary (see below). Faint bands at the top are artifacts arising in the probe preparation. Samples were run on a 6% acrylamide–7 M urea gel. (b) Autoradiograph showing the results of RNase protection using a labeled antisense RNA probe prepared from the SmaI fragment from intron 1. The probe was hybridized to 50 μg of tRNA (C) or total testis RNA (T). Tracks A and G are random DNA sequence reactions used as a marker (M). Reactions were treated with RNase for 15 min (tracks 1 and 4), 30 min (tracks 2 and 5), or 60 min (tracks 3 and 6). As in panel a, numbers at the right indicate distance of protected fragments from the intron 1/exon 2 boundary. The difference in sizes of protected fragments obtained by RNase protection and S1 nuclease protection reflects the different mobilities of RNA and DNA molecules in denaturing gels. Samples were electrophoresed on 10% acrylamide–7 M urea gels. (c) Part of the mouse MHC class II I-Aβ gene sequence (48). Arrows show the positions of transcriptional start sites in testis as determined by S1 protection. The boxed region indicates exon 2. The positions of the restriction endonuclease sites SacI and NaeI, which were used to construct the pAβΔSN transgene, are underlined with a dashed line (deletion from 1348 to 1511). The SmaI site, used to construct pAβΔPS, is underlined (all sequences upstream of position 1565 are deleted in this construct). The probe used for S1 analysis (S1) extends from 1251 to 1625; the antisense RNase protection probe (R) extends between 1562 and 1383.
(iii) S1 protection.
An end-labeled probe was made by kinase labeling 100 ng of exon 2 primer MHX8 (5′-GGT GTA GTA GCA CTC GCC CTT GAA CTG G-3′, positions 1629 to 1602 [see Fig. 3c]) with [γ-32P]ATP. This was used with upstream primer MHC2 (5′-CCC AAC TTT TGG TGT CCC TTA G-3′) in a PCR for 20 cycles (see below for conditions) using plasmid pAβpolyA as a template. The 379-bp labeled fragment was purified on an acrylamide-urea gel, and 100,000 cpm of the probe was precipitated with 40 μg of tRNA or total RNA isolated from spleen, liver, or testis. The dried pellet was resuspended in 30 μl of 40 mM PIPES–1 mM EDTA–0.4 M NaCl–80% formamide and overlaid with 30 μl of mineral oil (6). Samples were denatured at 90°C for 3 min before incubation overnight at 50°C. S1 nuclease digestion of the hybrid molecules was achieved by dilution of the reaction mixtures with 300 μl of 0.28 M NaCl–50 mM sodium acetate (pH 4.5)–4.5 mM zinc sulfate–20 μg of sonicated salmon sperm DNA per ml–100 U of S1 nuclease and incubation at 45°C for 90 min. The amount of S1 nuclease was determined empirically. Hybrids were ethanol precipitated with 20 μg of carrier tRNA and electrophoresed on a 6% acrylamide–7 M urea gel, which was dried and exposed to Kodak XAR-5 autoradiographic film.
(iv) RT-PCR.
One microgram of total RNA from mouse tissues, or approximately 100 ng of total RNA from mouse 3.5d blastocysts, was annealed to 0.5 μg of oligo(dT) (18-mer) primer in a volume of 12 μl with diethylpyrocarbonate-treated water. The sample was heated at 70°C for 10 min and chilled on ice. The reaction mix, in a volume of 20 μl, contained 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 0.5 mM deoxynucleoside triphosphates (dNTPs), 40 U of RNasin (Promega), and 200 U of Moloney murine leukemia virus reverse transcriptase (Gibco/BRL) and was incubated at 42°C for 2 h. Reactions were stopped by heating at 70°C for 15 min, and 2 μl of the cDNA sample was used in reverse transcription-PCR (RT-PCR) using gene-specific primer pairs in a 50-μl reaction mix containing 20 mM ammonium sulfate, 75 mM Tris-HCl (pH 9), 0.001% (wt/vol) Tween, 1.5 mM MgCl2, 0.2 mM dNTP, 50 pM each primer, and 1 U of RedHot DNA polymerase (Advanced Biotechnologies). In some cases (aprt), 10% dimethyl sulfoxide was also added. Primers were chosen from different exons to avoid misinterpretation of results due to possible contaminating genomic DNA. The reaction mixtures were denatured at 94°C for 2 min before cycling 25 times (92°C for 30 s, 55°C for 30 s, and 2 min at 72°C), with an additional 10-min extension at 72°C; 20 μl of each reaction product was run on 1.5% agarose gels. Ethidium bromide-stained PCR bands were visualized under UV, and in all cases a PCR product of the expected size was confirmed by Southern blotting and hybridization to a 32P-labeled internal oligomer. Hybridization buffer was as above, with incubation at 55°C overnight. Several washes were performed with 2× SSC at room temperature.
Primers were chosen from different exons so that the correctly spliced RT-PCR products could be distinguished from unprocessed transcripts or contaminating DNA.
Primers used for the MHC I-Aβ gene (48) were exon 1 forward primer MRT1 (5′-CAC AGC AGG TGT GAG TCC TG-3′), exon 3 reverse primer MRT4 (5′-GGG AGA TGG CGA CAT TGG GC-3′), intron 1 forward primer MRT3 (5′-GGG CTG ACC GCGTCC GTC CG-3′), and exon 2 internal primer MRT-2 (5′-CGC TCC AGG ATC TCC GGC TG-3′). Primers used for the aprt gene (19) were exon 1 forward primer ART-1 (5′-CGG AAC CTG AGT TGA AAC TG-3′), exon 5 reverse primer ART-2 (5′-GGT CCT AGC CTC TCC CTG CCC-3′), and exon 5 internal primer ART3 (5′-GTC AGC TCC ACC AGG CTC AC-3′). Primers for the mouse 68-kDa neurofilament gene (5′-CGC CGA AGA GTG GTT CAA GAG-3′ and 5′-GTA GGA GCT GCT CTG CAA GCC-3′ and internal primer 5′-GCT CTG AGA GTA GCC GCT GG-3′) (45) gave a 472-bp product. Primers for the mouse SCL (stem cell leukemia) gene (5′-GCC GGT CTG CCT ACA CCG GC-3′ and 5′-CCC CGA AGC TGG GTT TCC CGG-3′ and internal primer 5′-GGA CTC TTG GTG GAC AGG ACC-3′) (5) gave a 245-bp product. Primers for the mouse Twist gene (5′-CCA GGT ACA TCG ACT TCC TG-3′ and 5′-CTG TCC ACG GGC CTG TCT CGC-3′ and internal primer 5′-CTT CTC CGT CTG GAG GAT GG-3′) (67) gave a 372-bp product. Primers for the mouse opsin gene (5′-CTA CAT CCT GCT CAA CTT GGC-3′ and 5′-TGA CAA AGG TAA CGT TGT TGA C-3′ and internal primer 5′-TCG GGG AGA ATC ACG CTA TC-3′) (1) gave 396- and ∼200-bp products due to differential splicing (1). Primers for the mouse β-globin gene (5′-GAC CTA TCC TCT GCC TCT GCT A-3′ and 5′-AGC ACA ATC ACG ATC ATA TTG C-3′ and internal primer 5′-TGC AGC TTG TCA CAG TGG AG-3′) (40) gave a 203-bp product. Primers for the mouse casein gene (5′-TGA ATC TCA TGG GAC AGC TG-3′ and 5′-GGA TTC CAG TTC AGG AGA AAT G-3′ and internal primer 5′-TCA CTC CAG CAT CCA GTC AC-3′) (70) gave a 226-bp product. Primers for the mouse skeletal muscle α-actin gene (5′-GCG AGG TAT CCT GAC CCT GA-3′ and 5′-CTG GAC CTG GCC GGT CGC GAC-3′ and internal primer 5′-GCT ATG TGG CCC TGG ACT TC-3′) (29) gave a 744-bp product.
DNA methylation analysis.
Samples of DNA (10 μg) from various mouse tissues were digested with an excess of restriction enzyme overnight, using buffers and enzymes supplied by the manufacturer (New England Biolabs). A sample of each reaction mix (1/10) was added to 0.1 μg of plasmid DNA as a control for completeness of digestion. Samples were electrophoresed on 1.5% agarose gels by using standard methods and Southern blotted to Hybond N+ nylon membranes (Amersham). Probes and hybridization conditions were as for Northern blotting. Digests were repeated several times to confirm results.
Production of transgenic mice.
Transgenic mice were produced by microinjection of linearized transgene fragments at 1 μg/ml in 10 mM Tris-HCl (pH 7.5)–0.1 mM EDTA, using standard procedures (26). Donor mice were F2 animals from a C57BL/6 × CBA/Ca cross.
RESULTS
The mouse I-Aβ gene CpG island is nonmethylated in expressing and nonexpressing tissues.
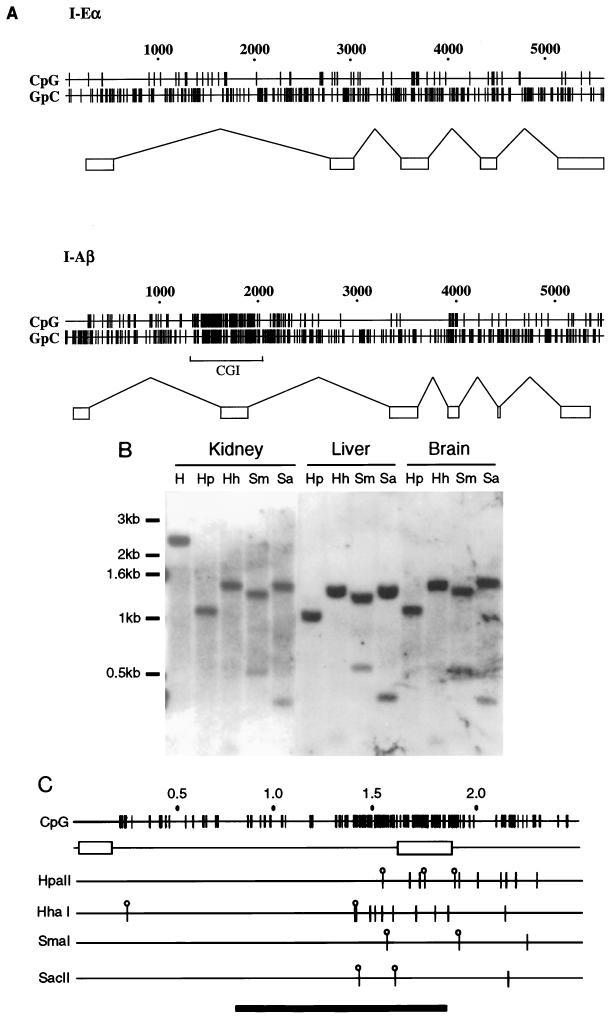
Figure 1A shows a plot of CpG and GpC distribution through the mouse I-Eα gene and I-Aβ gene DNA sequences (30, 48). A 600-bp CpG cluster can be seen in the I-Aβ gene, which incorporates exon 2. It is unlikely that the CpG-rich character of exon 2 is maintained due to a selective requirement for CpG-containing codons, as alternative CpG-poor codons could specify the same amino acid sequence. For example, 10 of the 13 codons specifying arginine have CG rather than AG as the first two nucleotides of the codon. In the remainder of the protein, only 4 of 11 arginines use the CG-containing codon. There is no clustering of CpG dinucleotides in the I-Eα gene sequence. The position of the CpG island is unusual, as these regions are normally located at the 5′ end of the gene. To determine whether the I-Aβ CpG island is nonmethylated, DNA samples from various adult tissues were tested by using methylation-sensitive restriction enzymes (Fig. 1B). Sites within the island were found to be nonmethylated in all tissues examined. Figure 1B shows some of this analysis using DNA extracted from an expressing tissue (kidney) and from nonexpressing tissues (liver and brain). All of these samples are cleaved with the methylation-sensitive enzymes, as were DNA samples from spleen and testis DNA (data not shown). The I-Aβ CpG island is therefore nonmethylated in all tissues tested, regardless of expression.
FIG. 1.
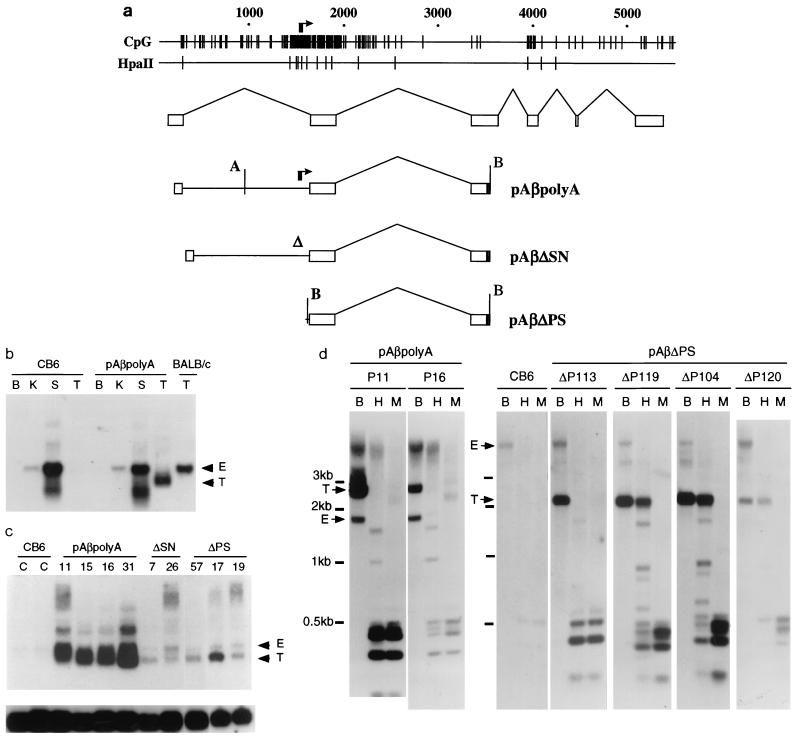
The I-Aβ gene contains a nonmethylated CpG island over the second exon. (A) Map plots of the mouse MHC class II I-Eα and I-Aβ gene sequences (30, 48), represented by horizontal lines numbered above in base pairs. The short vertical lines indicate the positions of CpG or GpC dinucleotides in the DNA sequence. The rectangular boxes represent gene exon arrangements, and the bracket under the I-Aβ map plot shows the position of the CpG island. (B) Southern blot of DNA isolated from various adult BALB/c mouse tissues hybridized to a BsaI probe from the I-Aβ gene (see below). Digests were carried out with 10 μg of DNA/track with HindIII (H) or HindIII in addition to HpaII (Hp), HhaI (Hh), SmaI (Sm), or SacII (Sa). The HindIII-alone digest is only shown in the kidney sample (first track). DNA marker sizes are noted at the left. (C) Map plot of the I-Aβ gene HindIII fragment showing the position of the CpG cluster over the second exon and the relative positions of the methylation-sensitive restriction enzyme sites used in the methylation analysis. Although it is likely that all of the HpaII and HhaI sites are cleaved within the CpG island, only sites which are resolved by electrophoresis are indicated by lollipops. A bar at the bottom shows the position of the BsaI probe. Exons are indicated by rectangles.
The MHC class II I-Aβ gene uses an alternative CpG island promoter in testis.
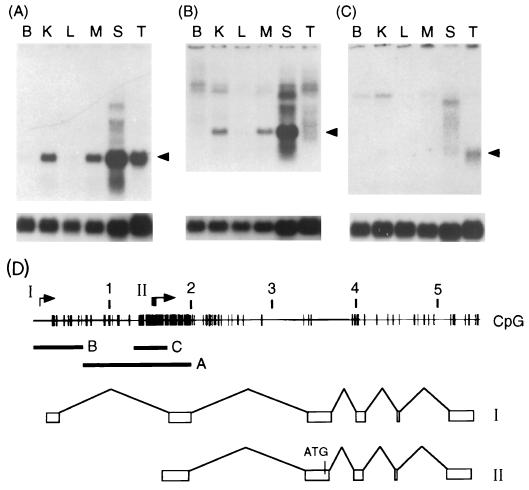
To test the CpG island promoter model, we analyzed RNA samples from various adult tissues by Northern blot hybridization. A DNA probe which includes the second exon of the I-Aβ gene (Fig. 2D) detected a transcript of the expected size (1.4 kb) in total RNA samples from spleen, kidney, maxillary salivary gland, and testis. No expression was seen in brain or liver (Fig. 2A). Low levels of I-Aβ protein have been detected in rat renal tubule cells (27), but to our knowledge, expression in salivary gland has not been reported. An exon 1 probe (Fig. 2D) detected expression in the same tissues but, surprisingly, not in testis RNA (Fig. 2B). This result suggested that an alternative promoter could be used in testis. As transcription initiates at the 5′ edge of the CpG island in many genes, it was possible that transcription may also initiate at this position in the I-Aβ island, within the first intron. An intronic probe (Fig. 2D) indeed detected a 1.4-kb transcript in testis but not in the other tissues (Fig. 2C).
FIG. 2.
An alternative CpG island-specific promoter is used in BALB/c testis. (A to C) Autoradiographs showing Northern blot analysis using for each track 10 μg of total RNA isolated from BALB/c mouse brain (B), kidney (K), liver (L), maxillary salivary gland (M), spleen (S), or testis (T) and probed with probe A, B, or C (see panel D). The arrow indicates the position of the 1.4-kb transcript, and the lower panels show control hybridization with an S26 ribosomal protein probe. (D) CpG map plot of the MHC class II I-Aβ gene showing positions of probes A to C. The scale is shown in kilobase pairs. Probe A is a BsaI fragment used to detect transcripts containing exon 2; probe B is a HindIII/NdeI probe used to detect exon 1; probe C is a SmaI fragment used to detect transcripts which include intron 1. The transcriptional start sites for the normal (I) and CpG island (II) promoters are indicated. The spliced exon arrangement for each transcript is shown below. The position of an in-frame ATG is indicated in transcript II.
The transcriptional initiation sites were exactly mapped within the first intron by S1 protection (Fig. 3a) and confirmed by RNase protection (Fig. 3b). This analysis showed multiple transcriptional start sites within the first intron in testis which were located 60 to 90 bp upstream of the I-Aβ exon II boundary (Fig. 3c). In agreement with Northern blot analysis, S1 protection showed that the CpG island promoter is not detectably active in spleen cells (Fig. 3a) where the normal I-Aβ promoter is used. A larger hybridizing transcript in the spleen sample, which fully protected the S1 probe, may be due to unprocessed mRNA transcribed from the upstream promoter (Fig. 3a). Although different promoters are used, the testis and spleen transcripts are approximately the same size, as predicted by Northern blot hybridization (Fig. 2A). The testis transcripts do not contain exon 1 and are believed to be correctly processed based on transcript size and RT-PCR analysis (see below). Transcripts were located in late spermatogenic germ cells by in situ analysis (data not shown). If the transcript produced in the germ cells were to be translated, the truncated peptide would not contain the putative signal peptide sequence or the external β1 domain (48), as the next in-frame ATG is in the third exon (Fig. 2).
The I-Aβ CpG island promoter is also active in embryonic cells.
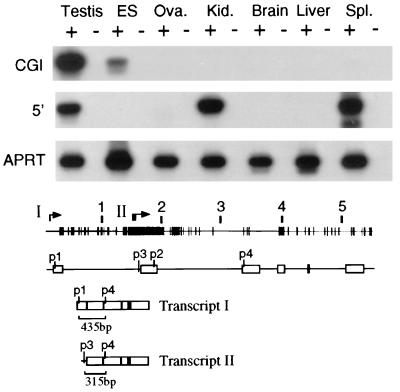
We were interested to know if the I-Aβ CpG island promoter is active at low levels in other tissues. We detected low levels of expression with an exon 2 probe in cultured embryonic stem (ES) cells by Northern blot hybridization (data not shown), but lack of sensitivity did not allow us to determine if this hybridization was due to activity of the normal or CpG island promoter. We therefore chose RT-PCR as a more sensitive assay method. Two sets of DNA primers were used; primers p1 and p4 (exon 1 and exon 3) detect spliced transcripts originating from the upstream promoter, and primers p3 and p4 (intron 1 and exon 3) detect spliced transcripts initiating from the CpG island promoter (Fig. 4). First-strand cDNA was made from a variety of RNA samples primed with oligo(dT). Amplified DNA molecules of the correct size were detected by ethidium bromide staining following gel electrophoresis and confirmed by Southern blot hybridization to an end-labeled internal DNA oligomer. This analysis was repeated several times and revealed that, in addition to being active in testis, the I-Aβ CpG island promoter was active in ES cells but was not detectably active, after 25 cycles of PCR, in cDNA samples from ovary, kidney, brain, liver, or spleen (Fig. 4). Activity of the normal I-Aβ promoter was not detected in ES cells but was strongly detected, as expected, in spleen and kidney. A weaker PCR band was also detected in testis even though Northern hybridization showed only trace amounts (Fig. 2B). In all experiments, the correct fragments were detected only in samples which contained reverse transcriptase in the cDNA synthesis reaction. The same cDNA samples were also used to amplify transcripts from the mouse aprt gene as a control (Fig. 4).
FIG. 4.
The CpG island promoter of I-Aβ is active in testis and ES cells. RT-PCR analysis using cDNA samples prepared from testis, ES cells, ovary (Ova.), kidney (Kid.), brain, liver, and spleen (Spl.) RNA derived from BALB/c mice; + and − indicate with or without reverse transcriptase in the preparation of cDNA. The results are presented as autoradiographs hybridized to an internal labeled oligomer. PCR bands amplified from the CpG island (CGI) promoter (top row) were obtained by using primers p3 and p4; those from the normal 5′ promoter (middle row) were obtained with primers p1 and p4. A labeled primer (p2) was used for Southern blot hybridization. The bottom row shows control RT-PCR using the same cDNA samples, with primers from the mouse aprt gene. The lower portion shows a CpG map plot of the I-Aβ gene. The exon arrangement of the gene is shown under the CpG plot, and the positions of primers p1 to p4 used for the RT-PCR analysis are numbered. Arrows above the CpG plot refer to the transcriptional start sites for the normal promoter (I) and the CpG island promoter (II). The spliced exon arrangements are shown underneath, and the sizes of the RT-PCR bands with each of the primer sets are noted. Transcript I is derived from the normal promoter, and transcript II is derived from the CpG island promoter.
Are most CpG island promoters active in ES cells?
We wished to examine whether other CpG island promoters of tissue specific genes were also expressed in ES cells. Primers were designed from a variety of CpG island-associated mouse genes that are expressed tissue specifically in the adult. RT-PCR analysis was carried out on ES cell cDNA by using primers from the 68-kDa neurofilament gene (45), the SCL gene, which is expressed during hematopoiesis (5), the Twist gene, which is expressed during embryogenesis and in adult skin (67), and the MyoD1 gene, which is expressed during myogenesis (17, 72). In addition we tested genes which did not contain CpG islands: those encoding β-globin (40), opsin (1), skeletal muscle-specific α-actin (29), and casein (70). The results are shown in Table 1. Transcripts were detected in ES cells from three of the four genes which had CpG islands; SCL, 68-kDa neorofilament, and Twist. We were unable to detect transcripts of the myogenesis gene MyoD1 in ES cells although it possesses a CpG island. Conversely, transcripts from the β-globin, opsin, skeletal muscle-specific α-actin, and casein genes, which are highly tissue specific but lack CpG islands, were absent under the same conditions.
TABLE 1.
RT-PCR data showing whether a specific gene is expressed in a particular mouse tissue or in ES cells
| Type of gene | Expressiona
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Testis | ES cells | Kidney | Brain | Spleen | Muscle | Skin | Liver | Eye | Mammary gland | |
| With CpG islands | ||||||||||
| aprt | + | + | + | + | + | + | ||||
| 68-kDa neurofilament | + | + | − | + | ||||||
| SCL | − | + | − | − | + | |||||
| Twist | − | + | − | − | + | |||||
| MyoD1 | − | − | − | − | + | |||||
| Without CpG islands | ||||||||||
| Opsin | − | − | − | − | + | |||||
| Casein | − | − | − | − | + | |||||
| Skeletal α-actin | − | − | − | − | + | |||||
| β-Globin | + | − | + | + | + | |||||
+, expression is detected by RT-PCR (observed by ethidium bromide staining and confirmed by using a labeled internal oligomer hybridized to Southern blots). −, no PCR band is detectable. Samples which were not tested are assigned no symbol. In addition, each sample of RNA was incubated without reverse transcriptase as a control; these samples did not show any PCR product in all cases.
Promoter inactivation can lead to methylation of the I-Aβ CpG island.
Previous analyses have shown that embryonic cells (ES cells and F9 cells) and transgenic mice have the ability to recognize a CpG island and keep it free of methylation (11, 23, 39, 47). We wanted to test if this was also the case for the mouse I-Aβ CpG island and, in addition, if lack of methylation was related to expression. A truncated I-Aβ gene construct was made by using a 3,364-bp fragment from the I-Aβd gene (48) which extends from a PstI site within exon 1 to an AccI site in exon 3. The AccI end was ligated to a fragment from simian virus 40 containing a polyadenylation initiation sequence. The resulting construct (pAβpolyA) contained only the CpG island promoter, as the normal promoter was deleted (Fig. 5a), and transcripts originating from this promoter should be, on average, 720 bp. Thus, the transgene-specific transcripts could be distinguished in size from the endogenous 1.4-kb transcript. We produced six transgenic mouse lines with the pAβpolyA construct, five of which were subjected to expression analysis. Like the endogenous CpG island promoter, the transgene transcript was highly expressed in testis (Fig. 5b and c) but was not present in spleen, kidney, or brain (Fig. 5b). The transgene was also transcribed at lower levels in the ovaries of founder animals (data not shown).
FIG. 5.
Reduced promoter activity can lead to increased de novo methylation of the I-Aβ gene CpG island. (a) Transgene constructs based on the mouse MHC class II I-Aβ gene. The distribution of CpG dinucleotides in the I-Aβ gene, indicated by short vertical lines, is shown at the top. The clustered arrows indicate positions of the transcriptional start sites in testis. The positions of HpaII sites relative to the gene sequence are also shown. Underneath the map plot is the normal exon arrangement of the endogenous gene, and below this is shown the arrangement contained in each of the transgenes. The normal gene promoter is deleted in each transgene. Positions of the BamHI (B) and AflII (A) sites are indicated. (b) Analysis of expression in transgenic mice. Total RNA extracted from brain (B), kidney (K), spleen (S), and testis (T) hybridized to probe A from exon 2 (Fig. 2). Samples were derived from nontransgenic (C57BL6 × CBA/Ca)F2 animals (CB6), BALB/c mice, or a transgenic founder (P19) containing the pAβpolyA transgene. Note that BALB/c testis expression of the endogenous I-Aβ gene is much higher than that of the nontransgenic (CB6) littermates (undetectable at this exposure). Positions of the endogenous (E) and transgenic (T) transcripts are indicated. (c) Analysis of expression from the testes of nontransgenic CB6 mice (C) or transgenic lines (numbered). In all cases, 10 μg of RNA was loaded except for the last two tracks (17 and 19), where 7 μg was loaded. The transgenes (Fig. 5a) are pAβpolyA, pAβΔSN, and pAβΔPS. Positions of the endogenous (E) and transgenic (T) transcripts are indicated. The lower panel shows the same filter hybridized to the S26 ribosomal gene control probe. (d) Analysis of DNA methylation in animals carrying the undeleted control transgene pAβpolyA (left; P11 and P16) or from the deleted construct pAβΔPS (right). DNA from a nontransgenic littermate is also included in the right-hand panel (CB6). DNA (10 μg) was cleaved in the pAβpolyA samples with AflII and BamHI or in the pAβΔPS samples with BamHI alone (tracks B) or in addition to the methylation-sensitive HpaII (tracks H) or its methylation-insensitive isoschizomer MspI (tracks M). Southern blots were probed with the BsaI fragment (Fig. 2D). In both panels, positions of the transgene-specific band (T) and the endogenous band (E) are indicated. Positions of molecular weight markers are shown at the left. Note that these are composite figures obtained from different gels and unequal exposures, and lower-molecular-weight bands do not always align exactly.
It is interesting that the endogenous I-Aβ gene in nontransgenic littermates was expressed at much lower levels in the testis compared with BALB/c mice (Fig. 5b). These mice are derived from an F2 cross between C57BL/6 and CBA/Ca parents and provided the fertilized eggs which are the recipients of the transgene DNA. Lower levels of endogenous gene expression in testis of nontransgenic littermates may be due to sequence differences in the intronic CpG island promoter region. The transgenes are of the Aβd haplotype, cloned from BALB/c mice (48), and the C57BL/6 and CBA/Ca mice have the I-Aβb and I-Aβk haplotypes, respectively (38). A comparison between the available genomic sequences, the I-Aβd (48) and I-Aβb (42) haplotypes, revealed several polymorphisms in this region, some of which may contribute to differences in expression.
Samples of DNA from the tails of pAβpolyA founder animals or from adult kidney, testis, or spleen were tested for transgene methylation by using methylation-sensitive restriction enzymes HpaII, SacII, and HhaI, which have multiple sites within the CpG island (Fig. 1B). The transgenic DNA from all six transgenic lines was cleaved with these enzymes and is therefore nonmethylated at the tested sites. Examples of DNA digested with methylation-sensitive HpaII and its methylation-insensitive isoschizomer MspI are shown in Fig. 5d. Thus, the integrated pAβpolyA transgenes have nonmethylated CpG islands as does the endogenous gene.
To determine whether transcriptional activity is required to prevent methylation, we attempted to inactivate the CpG island promoter by making promoter-proximal deletions of pAβpolyA. The pAβΔSN transgene contains a 113-bp SacI/NaeI deletion which removes some of the transcriptional start sites and the region immediately upstream (Fig. 3c). In the pAβΔPS construct, all sequences upstream of a SmaI site (23 bp upstream of the second exon) were removed from pAβpolyA (Fig. 3c), thereby deleting all of the testis-specific initiation sites and potential transcription factor binding sites upstream. Transgenic lines were produced from these constructs, and transgene expression in adult testes was examined by Northern blot hybridization (Fig. 5). Transgene transcription was detected in the testis samples from all three founder pAβΔSN transgenic lines and three tested founder pAβΔPS lines. However, the level of expression is much lower than in the pAβpolyA transgenic mice (Fig. 5c). Methylation of the DNA from transgenic animals was analyzed by using methylation-sensitive enzymes. Transgenic DNAs from all six founder animals produced with the pAβΔSN transgene were found to be nonmethylated (data not shown). However, 4 of 10 founder animals produced with the pAβΔPS construct showed extensive methylation of the transgene, based on their resistance to cleavage by HpaII (Fig. 5d). The results indicate that deletions of the CpG island promoter and upstream regions are associated with an increased frequency of de novo methylation of the CpG island.
DISCUSSION
A promoter within a mouse MHC class II gene.
Our motivation for studying the MHC class II I-Aβ gene was to test the proposition that all CpG islands mark promoters and depend for their existence on transcription. This well-studied gene was thought to represent a serious challenge to the hypothesis for two reasons. First, the I-Aβ CpG island, like most other islands, is nonmethylated in all tested tissues, including those that do not express the I-Aβ gene. The correlation between transcription and lack of methylation was therefore absent. Second, the CpG island is in an unusual place, far from the I-Aβ gene promoter, which is itself not associated with a CpG island. We have demonstrated that this CpG island does in fact harbor an alternative promoter which lies within intron 1 and is active in embryonic and germ cells. The promoter is located at the 5′ edge of the CpG island, which extends downstream, encompassing exon 2. Our finding of this novel promoter strengthens the argument that CpG islands invariably contain promoters. Moreover, the position of transcription initiation, close to the periphery of the CpG island, is typical.
Transcripts from this island promoter are abundant in BALB/c testis but are also found in ES cells. In addition, we have detected expression from the CpG island promoter in 3.5d blastocysts by RT-PCR (data not shown). The normal promoter is not active in ES cells, but low levels of expression can be detected in testis by RT-PCR. Transcripts have previously been detected by using the I-Aβ cDNA as a probe on RNA isolated from mouse epididymal sperm cells (52). In these experiments, the cDNA probe does not discriminate between transcripts originating from the normal promoter and the CpG island promoter. We have shown that the great majority of these transcripts are derived from the island promoter. It is still controversial as to whether the I-Aβ protein is located on mouse sperm heads (44). Transcripts derived from the CpG island promoter do not encode exon 1, and as the next initiation codon is in exon 3, a putative peptide would lack the signal sequence and external β1 domain (48). However, we cannot rule out the possibility that such a truncated protein, containing only the dimerization and transmembrane domains, has a function. Certainly any function of a putative peptide would not be essential, as mice lacking MHC class II molecules have a depleted T-cell repertoire and can show reduced growth but are otherwise viable. However, these mice do not breed well (16).
Downstream CpG islands have been observed in a variety of genes, notably those that are tissue specifically expressed (24, 25, 43). In the case of the pro-opiomelanocortin (POMC) gene and the apolipoprotein-AI and -E genes, the islands are located over the extreme 3′ exons of the genes concerned. Short POMC transcripts that initiate at the edge of the downstream POMC CpG island have been observed in various mammalian species (see reference 25 for references). Notably, this CpG island promoter is active in testis, ovary, and embryonic cells of the mouse as well as a subset of other tissues (25). The resulting transcript could in theory give rise to some of the neuropeptides that are normally processed from the translated product of this gene, but the translation product appears to be unprocessed in testis (15). Thus, as proposed for the I-Aβ CpG island transcript, it is possible that the POMC island-derived transcript does not lead to a functional protein product. It has recently been found that the intronic CpG island (region 2) of the Igf2r gene is also associated with promoter activity (27a, 68). These examples further strengthen the connection between promoter activity, particularly in germ cells and embryonic cells, and CpG island formation.
We attempted to test the significance of the I-Aβ CpG island promoter by mutagenesis. Neither minor nor major deletions in the presumed promoter region abolished transgene expression, but both led to significantly reduced levels of transcription in testes of founders. All 6 founders with the small deletion (pAβΔSN) produced the nonmethylated CpG island, but 4 of 10 founders lacking the entire upstream region (pAβΔPS) showed extensive island methylation. Loss of immunity to methylation with the large deletion, but not the small deletion, may suggest the presence of a cis-acting sequence that persists in ΔSN but is absent in ΔPS. As the effects of the two deletions on transcription appears to be approximately similar in testes, the putative demethylating sequence may not act via an effect on promoter activity. In our view this interpretation is premature, as the number of founders is too low to be sure that the probability of CpG island methylation differs significantly between the ΔSN and ΔPS constructs. In addition, transcription of the two transgene constructs would be more usefully assayed in early embryos rather than testes. We have unfortunately been unable to detect transcription of the mutated or unmutated transgenes in blastulae of transgenic mice by RT-PCR. It is not clear whether the transgenes are inactive at this stage or are transcribed at very low levels. In summary, the mutagenesis experiments with the I-Aβ CpG island are less clear-cut than those with the aprt island (47), as point mutations in the latter promoter reliably led to methylation of the CpG island. In view of these uncertainties, the evidence for a causal relationship between transcription of the intronic promoter and lack of methylation at the I-Aβ CpG island must be considered preliminary.
An embryonic origin for CpG islands.
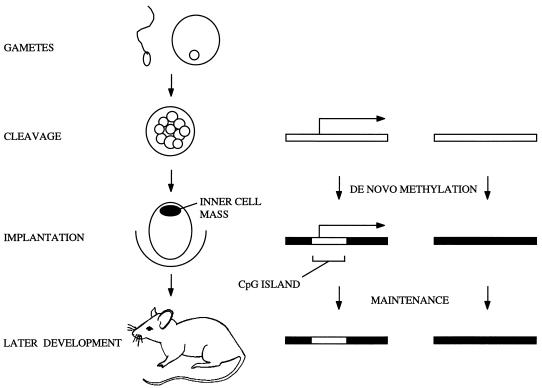
If CpG islands arise as a consequence of promoter activity, why do they remain methylation free in tissues where the CpG island promoter is inactive? For example, the I-Aβ island is nonmethylated in liver, yet no CpG island-derived transcript is observed even by RT-PCR. Also, the POMC CpG island transcript is apparently absent in several tissues, and the human α-globin gene, which falls entirely within a constitutively nonmethylated CpG island (8), is silent in nonerythroid cells (69). The finding that CpG island promoters are active in germ cells and embryos offers a plausible explanation for the discrepancy, as all somatic cell types are descendants of these lineages. Global de novo methylation changes take place during gametogenesis and mammalian embryonic development (35, 51, 58), and transcription may protect CpG island sequences from methylation at these developmental stages. Methylation analysis of founder transgenic animals shows that protection from de novo methylation must occur in the early embryonic stages, as the transgenes have not yet passed through the germ line (11, 39, 47). Thus, the nonmethylated state of the CpG island may be established in the early embryo and copied to the somatic lineages by maintenance methylation. According to this scenario, the decision to create a methylation-free island would be taken only in totipotent cells.
We suggest that promoters that are active at the appropriate totipotent stages invariably give rise to nonmethylated CpG islands (Fig. 6). Consistent with this view is evidence for embryonic transcription of a variety of CpG island-associated genes that are expressed tissue specifically in the adult (see also reference 24). MyoD1 is an exception, but it may be relevant that in the Xenopus system, ubiquitous transient expression of MyoD1 has been observed at the midblastula transition (57). Transcriptional silence at this stage would, on the other hand, invite de novo methylation. A subset of CpG islands that become de novo methylated during mouse development, including those at X-linked genes as well as transgenes, retroviral proviruses, and some imprinted genes such as Xist, acquire their methylation during the early stages of mouse embryogenesis (12, 32, 53, 56). In the cases of retroviral proviruses, inactive X genes, and the inactive copy of the Xist gene, silencing appears to precede de novo methylation (32, 46). Some other imprinted genes with CpG islands remain partially methylated at the blastocyst stage, and methylation becomes more extensive after implantation (10, 62). Thus de novo methylation in the early embryo has the character of a default condition (59), affecting all sequences that are not specifically protected.
FIG. 6.
Transcription-related origin for CpG islands. Transcription of CpG island promoters in the early cleavage stages would protect these regions from the global de novo methylation which occurs at implantation. This nonmethylated profile, once established, would then be maintained in the somatic tissues of the adult mouse. The open rectangles represent nonmethylated DNA; the filled areas represent methylated regions. The horizontal arrows indicate active transcription.
The mechanism that might connect embryonic transcription with absence of methylation at the CpG island is obscure. It has been suggested that active demethylation, perhaps via an RNA intermediate, is responsible (64). It is also possible that the demethylating mechanism is passive, due to exclusion of the methyltransferase from a transcriptionally active CpG island (50). Selker (59) proposed that bound transcription factors (and other sequence-specific proteins) that alter chromatin structure or the timing or mechanistic requirements of DNA replication could determine the methylation status of DNA. Evidence that CpG islands depend on transcription renders less likely the idea that transcription factors such as Sp1 induce local demethylation by a mechanism that is independent of transcription but does not rule it out. It is already clear that Sp1 is not necessary for the demethylation process, as CpG islands are methylation free in mouse embryos that have no Sp1 gene (49). The CpG island promoter model is compatible with this result, as it requires transcription per se for CpG island formation rather than one particular transcription factor. Recently, data indicating that CpG island promoters serve as origins of replication have been presented (18). This finding raises the intriguing possibility that CpG islands are footprints of replication initiation at embryonic promoters.
Functional significance of the MHC class II CpG islands.
A possible function of CpG islands is to ensure that the promoters of the associated genes are free of high-density methylation which will silence transcription. Preservation of the downstream CpG island of the MHC class II I-A β-chain gene has a less obvious rationale, however, as the major promoter of the gene is remote from the CpG island. A plausible explanation for its existence is that the presence of the CpG island is involved in maintenance of polymorphism of amino acids encoded by exon 2. Colocalization of the CpG island with the polymorphic exon was first noted by Tykocinski and Max (64). Later data emphasized the correlation, as the homologous human MHC genes which encode the HLA-DQ, -DP, and -DR β chains all contain CpG islands over the second exon, based on CpG frequency. The polymorphic regions of the class I chains also lie within CpG islands, and it has been suggested that these GC-rich regions within class I and II genes may be important in promoting polymorphism (64). Base substitutions (within the island) of mouse class I genes may be due to gene conversion (54). Also, polymorphisms in class II β chain genes, in the second exon of the mouse I-Aβ chain gene (60), and between alleles of the HLA-DPB1 gene in the human germ line (71) have been attributed to interallelic gene conversion. We suggest that germ line transcription facilitates genetic exchange between related sequences, thereby sustaining polymorphism of exon 2. Specifically, exposure of the DNA for strand invasion may be aided by the relatively decondensed nature of transcribed CpG island chromatin (63). According to this view, the functional significance of this CpG island would be confined to totipotent cells.
ACKNOWLEDGMENTS
We thank Deborah Fowlis for tutoring in the production of transgenic animals, Susan Carson for the MHC clones, Austin Smith for the BALB/c ES cells, and Aileen Greig, Joan Davidson, and the staff of the Ann Walker Building for technical support. We also thank our photographic department for help in producing the figures and Brian Hendrich for helpful comments on the manuscript.
This work was supported by The Welcome Trust and The Howard Hughes Medical Institute.
REFERENCES
- 1.Al-Ubaldi M R, Pittler S J, Champagne M S, Triantafyllos J T, McGinnis J F, Baehr W. Mouse opsin. Gene structure and molecular basis of multiple transcripts. J Biol Chem. 1990;265:20563–20569. [PubMed] [Google Scholar]
- 2.Antequera F, Macleod D, Bird A P. Specific protection of methylated CpGs in mammalian nuclei. Cell. 1989;58:509–517. doi: 10.1016/0092-8674(89)90431-5. [DOI] [PubMed] [Google Scholar]
- 3.Antequera F, Boyes J, Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990;62:503–514. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- 4.Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci USA. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Begley C G, Robb L, Rockman S, Visvader J, Bockamp E O, Chan Y S, Green A R. Structure of the gene encoding the murine SCL protein. Gene. 1994;138:93–99. doi: 10.1016/0378-1119(94)90787-0. [DOI] [PubMed] [Google Scholar]
- 6.Berk A J, Sharp P A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1997;12:721. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- 7.Bird A P. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 8.Bird A P, Taggart M H, Nicholls R D, Higgs D R. Non-methylated CpG-rich islands at the human alpha-globin locus: implications for evolution of the alpha-globin pseudogene. EMBO J. 1987;6:999–1004. doi: 10.1002/j.1460-2075.1987.tb04851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyes J, Bird A. Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J. 1992;11:327–333. doi: 10.1002/j.1460-2075.1992.tb05055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brandeis M, Kafri T, Ariel M, Chaillet J R, McCarrey J, Razin A, Cedar H. The ontogeny of allele-specific methylation associated with imprinted genes in the mouse. EMBO J. 1993;12:3669–3677. doi: 10.1002/j.1460-2075.1993.tb06041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brandeis M, Frank D, Keshet I, Siegfreid Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 12.Chaillet J R, Vogt T F, Beler D R, Leder P. Parental-specific methylation of an imprinted transgene is established during gametogenesis and progressively changes during embryogenesis. Cell. 1991;66:77–83. doi: 10.1016/0092-8674(91)90140-t. [DOI] [PubMed] [Google Scholar]
- 13.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 14.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark A J L, Lavender P M, Coates P, Johnson M R, Rees L H. In vitro and in vivo analysis of the processing and fate of the peptide products of the short pro-opriomelanocortin mRNA. Mol Endocrinol. 1990;4:1737–1743. doi: 10.1210/mend-4-11-1737. [DOI] [PubMed] [Google Scholar]
- 16.Cosgrove D, Gray D, Dierich A, Kaufman J, Lameur M, Benoist C, Mathis D. Mice lacking MHC class II molecules. Cell. 1991;66:1051–1066. doi: 10.1016/0092-8674(91)90448-8. [DOI] [PubMed] [Google Scholar]
- 17.Davis R L, Weintraub H, Lassar A B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;31:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 18.Delgado S, Gomez M, Bird A, Antequera F. Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J. 1998;17:2426–2435. doi: 10.1093/emboj/17.8.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dush M K, Sikela J M, Khan S A, Tischfield J A, Stambrook P J. Nucleotide sequence and organisation of the mouse adenine phosphoribosyltransferase gene: presence of a coding region common to animal and bacterial phosphoribosyltransferases that has a variable intron/exon arrangement. Proc Natl Acad Sci USA. 1985;82:2731–2735. doi: 10.1073/pnas.82.9.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dush M K, Briggs M R, Royce M E, Schaff D A, Khan S A, Tischfield J A, Stambrook P J. Identification of DNA sequences required for mouse APRT gene expression. Nucleic Acids Res. 1988;16:8509–8525. doi: 10.1093/nar/16.17.8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1984;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 22.Ferguson-Smith A C, Sasaki H, Cattanach B M, Surani M A. Parental-origin-specific epigenetic modification of the mouse H19 gene. Nature. 1993;362:751–755. doi: 10.1038/362751a0. [DOI] [PubMed] [Google Scholar]
- 23.Frank D, Keshet I, Shani M, Levine A, Razin A, Cedar H. Demethylation of CpG islands in embryonic cells. Nature. 1991;351:239–241. doi: 10.1038/351239a0. [DOI] [PubMed] [Google Scholar]
- 24.Gardiner-Gardner M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 25.Gardiner-Gardner M, Frommer M. Transcripts and CpG islands associated with the pro-opiomelanocortin gene and other neurally expressed genes. J Mol Endocrinol. 1994;12:365–382. doi: 10.1677/jme.0.0120365. [DOI] [PubMed] [Google Scholar]
- 26.Hansen R S, Gartler S M. 5-Azacytidine-induced reactivation of the human X chromosome-linked PGK1 gene is associated with a large region of demethylation in the 5′ CpG island. Proc Natl Acad Sci USA. 1990;87:4174–4178. doi: 10.1073/pnas.87.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hart D N J, Fabre J W. Endogenously produced Ia antigen within the cells of convoluted tubules of rat kidney. J Immunol. 1981;126:2109–2113. [PubMed] [Google Scholar]
- 27a.Hendrich, B. Unpublished data.
- 28.Hogan B, Beddington R, Constantini F, Lacy E. Manipulating the mouse embryo. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 29.Hu M C T, Sharp S B, Davidson N. The complete sequence of the mouse skeletal alpha-actin gene reveals several conserved and inverted repeat sequences outside of the coding regions. Mol Cell Biol. 1986;6:15–25. doi: 10.1128/mcb.6.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hyldig-Neilsen J J, Schenning L, Hammerling U, Widmark E, Heldin E, Lind P, Servenius B, Lund T, Flavell R, Lee J S, Trowsdale J, Schreirer H, Zablitzky F, Larhammar D, Peterson P A, Rask L. The complete sequence of the I-Eβd immune response gene. Nucleic Acids Res. 1983;11:5055–5071. doi: 10.1093/nar/11.15.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Issa J-P, Ottaviano Y L, Celo P, Hamilton S R, Davidson N M, Baylin S B. Methylation of the oestrogen receptor CpG island links aging and neoplasia in human colon. Nat Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 32.Jahner D, Jaenisch R. DNA methylation in early mammalian development. In: Razin A, Cedar H, Riggs A D, editors. DNA methylation; biochemistry and biological significance. New York, N.Y: Springer Verlag; 1985. pp. 189–219. [Google Scholar]
- 33.Jones P, Wolkowicz M, Rideout W, Gonzales F, Marziasz C, Coetzee G, Tapscott S. De novo methylation of the MyoD1 CpG island during the establishment of immortal cell lines. Proc Natl Acad Sci USA. 1990;87:6117–6121. doi: 10.1073/pnas.87.16.6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones P A, Rideout W M, Shen J-C, Spruck C H, Tsai Y C. Methylation, mutation and cancer. Bioessays. 1992;14:33–36. doi: 10.1002/bies.950140107. [DOI] [PubMed] [Google Scholar]
- 35.Kafri T, Ariel M, Brandeis M, Shemer R, Urven L, McCarrey J, Cedar H, Razin A. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 1992;6:705–714. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- 36.Kaufman J F, Aufray C, Korman A J, Shackelford D A, Strominger J. The class II genes of the human and murine major histocompatibility complex. Cell. 1984;36:1–13. doi: 10.1016/0092-8674(84)90068-0. [DOI] [PubMed] [Google Scholar]
- 37.Kay G F, Penny G D, Patel D, Ashworth A, Brockdorf N, Rastan S. Expression of Xist during mouse development suggests a role in X chromosome inactivation. Cell. 1993;72:171–182. doi: 10.1016/0092-8674(93)90658-d. [DOI] [PubMed] [Google Scholar]
- 38.Klein J. Congenic and segregating inbred strains. In: Lyon M F, Searle A G, editors. Genetic variants and strains of the laboratory mouse. Oxford, England: Oxford University Press; 1989. pp. 797–842. [Google Scholar]
- 39.Kolsto A-B, Kollias G, Giguere V, Isobe K-I, Prydz H, Grosveld F. The maintenance of methylation-free islands in transgenic mice. Nucleic Acids Res. 1986;14:9667–9678. [PMC free article] [PubMed] [Google Scholar]
- 40.Konkel D A, Tilghman S M, Leder P. The sequence of the chromosomal mouse beta-globin major gene: homologies in capping, splicing and poly(a) sites. Cell. 1978;15:1125–1132. doi: 10.1016/0092-8674(78)90040-5. [DOI] [PubMed] [Google Scholar]
- 41.Laird P W, Jaenisch R. DNA methylation and cancer. Hum Mol Genet. 1994;3:1487–1495. doi: 10.1093/hmg/3.suppl_1.1487. [DOI] [PubMed] [Google Scholar]
- 42.Larhammar D, Hammerling U, Denaro M, Lund T, Flavell R A, Rask L, Peterson A. Structure of the murine immune response I-Aβ locus: sequence of the I-Aβ gene and an adjacent β-chain second domain exon. Cell. 1983;34:179–188. doi: 10.1016/0092-8674(83)90148-4. [DOI] [PubMed] [Google Scholar]
- 43.Larsen F, Solheim G, Lopez R, Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- 44.Lavitrano M, Maione R, Forte E, Francolini M, Sperandio S, Testi R, Spadafora C. The interaction of sperm cells with exogenous DNA: a role of CD4 and major histocompatibility complex class II molecules. Exp Cell Res. 1997;233:56–62. doi: 10.1006/excr.1997.3534. [DOI] [PubMed] [Google Scholar]
- 45.Lewis S A, Cowan N J. Anomalous placement of introns in a member of the intermediate filament multiprotein family: an evolutionary conundrum. Mol Cell Biol. 1986;6:1529–1534. doi: 10.1128/mcb.6.5.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lock L, Takagi N, Martin G. Methylation of the HPRT gene on the inactive X occurs after chromosome inactivation. Cell. 1987;48:39–46. doi: 10.1016/0092-8674(87)90353-9. [DOI] [PubMed] [Google Scholar]
- 47.Macleod D, Charlton J, Mullins J, Bird A. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 48.Malissen M, Hunkapiller T, Hood L. Nucleotide sequence of a light chain gene of the mouse I-A subregion. Aβd. Science. 1983;221:750–754. doi: 10.1126/science.6410508. [DOI] [PubMed] [Google Scholar]
- 49.Marin M, Karis A, Visser P, Grosveld F, Philipsen S. Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell. 1997;89:619–628. doi: 10.1016/s0092-8674(00)80243-3. [DOI] [PubMed] [Google Scholar]
- 50.Matsuo K, Silke J, Georgiev O, Marti P, Giovannini N, Rungger D. An embryonic demethylation mechanism involving binding of transcription factors to replicating DNA. EMBO J. 1998;17:1446–1453. doi: 10.1093/emboj/17.5.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- 52.Mori T, Wu G M, Mori K, Shindo Y, Mori N, Fukuda A, Mori T. Expression of class II major histocompatibility complex antigen on mouse sperm and its roles in fertilization. Am J Reprod Immunol. 1990;24:9–14. doi: 10.1111/j.1600-0897.1990.tb00688.x. [DOI] [PubMed] [Google Scholar]
- 53.Norris D P, Patel D, Kay G F, Penny G D, Brockdorff N, Sheardown S A, Rastan S. Evidence that random and imprinted Xist expression is controlled by preemptive methylation. Cell. 1994;77:41–51. doi: 10.1016/0092-8674(94)90233-x. [DOI] [PubMed] [Google Scholar]
- 54.Pease L R, Schulze D H, Pfaffenbach G M, Nathenson S G. Spontaneous H-2 mutants provide evidence that a single copy mechanism analogous to gene conversion generates polymorphism in the major histocompatability complex. Proc Natl Acad Sci USA. 1983;80:242–246. doi: 10.1073/pnas.80.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riggs A D, Pfeifer G P. X-chromosome inactivation and cell memory. Trends Genet. 1992;8:169–174. doi: 10.1016/0168-9525(92)90219-t. [DOI] [PubMed] [Google Scholar]
- 56.Riggs A D. X-inactivation, differentiation and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- 57.Rupp R A W, Weintraub H. Transcription at the midblastula transition precedes induction-dependent MyoD expression in presumptive mesoderm of X. laevis. Cell. 1991;65:927–937. doi: 10.1016/0092-8674(91)90545-a. [DOI] [PubMed] [Google Scholar]
- 58.Sanford J P, Clark H J, Chapman V M, Rossant J. Differences in DNA methylation during oogenesis and spermatogenesis and their persistence during early embryogenesis in the mouse. Genes Dev. 1987;1:1039–1046. doi: 10.1101/gad.1.10.1039. [DOI] [PubMed] [Google Scholar]
- 59.Selker E U. DNA methylation and chromatin structure: a view from below. Trends Biochem Sci. 1990;15:103–107. doi: 10.1016/0968-0004(90)90193-f. [DOI] [PubMed] [Google Scholar]
- 60.She J X, Boehme S A, Wang T W, Bonhomme F, Wakeland E K. Amplification of major histocompatibility complex class II gene diversity by intraexonic recombination. Proc Natl Acad Sci USA. 1991;88:453–457. doi: 10.1073/pnas.88.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Steinmetz M, Hood L. Genes of the major histocompatibility locus of mouse and man. Science. 1988;222:727–733. doi: 10.1126/science.6356354. [DOI] [PubMed] [Google Scholar]
- 62.Stöger R, Kubicka P, Liu C-G, Kafri T, Razin A, Cedar H, Barlow D P. Maternal specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell. 1993;73:61–71. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- 63.Tazi J, Bird A. Alternative chromatin structure at CpG islands. Cell. 1990;60:909–920. doi: 10.1016/0092-8674(90)90339-g. [DOI] [PubMed] [Google Scholar]
- 64.Tykocinski M L, Max E C. CG clusters in MHC genes and 5′ demethylated genes. Nucleic Acids Res. 1984;12:4385–4396. doi: 10.1093/nar/12.10.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vincent S, Marty L, Fort P. S26 ribosomal protein RNA: an invariant control for gene regulation experiments in eucaryotic cells and tissues. Nucleic Acids Res. 1993;21:1498. doi: 10.1093/nar/21.6.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weiss A, Keshet I, Cedar H. DNA demethylation in vitro: involvement of RNA. Cell. 1996;86:709–718. doi: 10.1016/s0092-8674(00)80146-4. [DOI] [PubMed] [Google Scholar]
- 67.Wolf C, Thisse C, Stoetzel C, Thisse B, Gerlinger P, Perrin-Schmitt F. The M-twist gene of mus is expressed in subsets of mesodermal cells and is closely related to the Xenopus x-twi and drosophila twist genes. Dev Biol. 1991;143:363–373. doi: 10.1016/0012-1606(91)90086-i. [DOI] [PubMed] [Google Scholar]
- 68.Wutz A, Smrzka O W, Schweifer N, Schellander K, Wagner E F, Barlow D P. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature. 1997;389:745–749. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]
- 69.Yagi M, Groudine M. Chromatin structure and developmental expression of the human α-globin cluster. Mol Cell Biol. 1986;6:1108–1116. doi: 10.1128/mcb.6.4.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yoshimura M, Oka T. Isolation and structural analysis of the mouse beta casein gene. Gene. 1989;78:267–275. doi: 10.1016/0378-1119(89)90229-1. [DOI] [PubMed] [Google Scholar]
- 71.Zangenberg G, Huang M-M, Arnheim N, Erlich H. New HLA-DPB1 alleles generated by interallelic gene conversion detected by analysis of sperm. Nat Gen. 1995;10:407–414. doi: 10.1038/ng0895-407. [DOI] [PubMed] [Google Scholar]
- 72.Zingg J-M, Alva G P, Jost J P. Characterisation of a genomic clone covering the structural mouse MyoD1 gene and its promoter region. Nucleic Acids Res. 1991;23:6433–6439. doi: 10.1093/nar/19.23.6433. [DOI] [PMC free article] [PubMed] [Google Scholar]